Evolutionarily conserved Tudor Staphylococcal Nuclease is essential for maintaining the integrity of stress granules and processing bodies and for stress-induced mRNA decapping.
Abstract
Tudor Staphylococcal Nuclease (TSN or Tudor-SN; also known as SND1) is an evolutionarily conserved protein involved in the transcriptional and posttranscriptional regulation of gene expression in animals. Although TSN was found to be indispensable for normal plant development and stress tolerance, the molecular mechanisms underlying these functions remain elusive. Here, we show that Arabidopsis thaliana TSN is essential for the integrity and function of cytoplasmic messenger ribonucleoprotein (mRNP) complexes called stress granules (SGs) and processing bodies (PBs), sites of posttranscriptional gene regulation during stress. TSN associates with SGs following their microtubule-dependent assembly and plays a scaffolding role in both SGs and PBs. The enzymatically active tandem repeat of four SN domains is crucial for targeting TSN to the cytoplasmic mRNA complexes and is sufficient for the cytoprotective function of TSN during stress. Furthermore, our work connects the cytoprotective function of TSN with its positive role in stress-induced mRNA decapping. While stress led to a pronounced increase in the accumulation of uncapped mRNAs in wild-type plants, this increase was abrogated in TSN knockout plants. Taken together, our results establish TSN as a key enzymatic component of the catabolic machinery responsible for the processing of mRNAs in the cytoplasmic mRNP complexes during stress.
INTRODUCTION
Adaptation to environmental conditions depends on the modulation of gene expression. However, changes in gene transcription require longer time than necessary for quick responses to stress challenges. To overcome this limitation, eukaryotes have evolved an alternative mechanism in which, upon perception of a stress stimulus, specific mRNAs can be compartmentalized by RNA binding proteins to cytoplasmic structures known as stress granules (SGs) and processing bodies (PBs) (Anderson and Kedersha, 2008; Uniacke and Zerges, 2008; Thomas et al., 2011; Muench et al., 2012). Both cytoplasmic foci are highly dynamic molecular ensembles acting in the process of continuous exchange of mRNA and proteins with the cytoplasm and between each other (Anderson and Kedersha, 2006). Several works have suggested that both structures can be transiently linked (Kedersha et al., 2005; Wilczynska et al., 2005).
Recent studies in yeast and animal models have led to detailed characterization of the molecular composition of SGs and PBs. The core components of SGs are poly(A)+ mRNA, 40S ribosomal subunits, poly(A) binding protein, and eukaryotic initiation factors (eIFs). These universal factors of SGs coexist with other cell type- and stress stimulus-specific components (Thomas et al., 2011), such as RNA helicases (Yu et al., 2011; Yasuda-Inoue et al., 2013), RNA binding proteins (Anderson and Kedersha, 2008), and regulators of mRNA decay and translation (Tourrière et al., 2003). It was suggested that SGs play a role in translational repression by sequestering, stabilizing, and storing mRNAs and translation factors (Anderson and Kedersha, 2009; Vanderweyde et al., 2013). Furthermore, it has been proposed that SGs can also sequester proapoptotic proteins, thereby protecting cells from death (Eisinger-Mathason et al., 2008; Buchan et al., 2012).
Although PBs are present in unstressed cells, their number increases under stress conditions (Kedersha et al., 2005). Typical components of PBs in yeast and animals are the 5′ cap binding protein 4E, subunits of decapping and exosome complexes, deadenylases, untranslated mRNA, and RNA binding proteins (Franks and Lykke-Andersen, 2008). PBs are thought to function in translational repression and mRNA decay (Kedersha et al., 2005; Xu and Chua, 2011).
The cytoplasmic mRNA decay pathways are conserved in all eukaryotes and can be deadenylation-dependent, deadenylation-independent, and endonucleolytic cleavage-dependent (Nagarajan et al., 2013). In brief, following the removal of the 3′ poly(A) tail by deadenylases, mRNA can be degraded via two distinct routes: either by the exosome complex, which executes 3′→5′ decay, or through the removal of the 5′ cap by decapping enzymes. In the deadenylation-independent pathway, adenylated mRNA with a premature termination codon is degraded through the so-called nonsense-mediated decay. Lastly, endonucleolytic cleavage generates uncapped mRNA that is degraded by the exosome complex.
The process of SG assembly remains poorly understood. It has been reported that this appears to be a stepwise process initiated within minutes of the perception of a stress stimulus and that SGs gradually disassemble during stress recovery (Kedersha et al., 2000; Loschi et al., 2009). In animal systems, SGs are dynamically linked with microtubules (MTs). Accordingly, pharmacological suppression of MT dynamics inhibits the assembly and mobility of SGs (Kwon et al., 2007; Nadezhdina et al., 2010). Interestingly, disruption of MTs promotes the formation of PBs in mammals and yeast (Sweet et al., 2007), suggesting that MTs play distinct roles in the biogenesis of SGs and PBs. In contrast with animals, the movement of PBs in plant cells depends on actin instead of MTs (Hamada et al., 2012; Steffens et al., 2014).
Plants contain genes encoding homologs of bona fide SG and PB constituents and form similar structures (Belostotsky and Sieburth, 2009; Muench et al., 2012). For example, Ubp1b and Ubp1c are SG-nucleating RNA binding proteins, but Ubp1c was found to be a component of the machinery that reprograms posttranscriptional gene expression under hypoxia (Sorenson and Bailey-Serres, 2014). Similarly, the translation initiation factor E (eIF4E) and the component of pre-mRNA splicing machinery Rbp47b localized to Arabidopsis thaliana SGs under heat stress conditions (Lorković et al., 2000; Weber et al., 2008). Furthermore, components of the mRNA decapping complex, Decapping 1 (DCP1), DCP2/Trident, DCP5, and Varicose, as well as both 5′→3′ exoribonuclease 4 (XRN4) and Argonaute 1b, localized to Arabidopsis PBs (Xu et al., 2006; Goeres et al., 2007; Weber et al., 2008; Xu and Chua, 2009, 2011; Pomeranz et al., 2010). Intriguingly, some proteins were found in both SGs and PBs in Arabidopsis (e.g., the CCCH tandem zinc finger proteins TZF1, TZF4, TZF5, and TZF6) (Pomeranz et al., 2010; Bogamuwa and Jang, 2013).
Tudor Staphylococcal Nuclease (TSN) was recently described as a novel component of SGs in animals (Gao et al., 2010, 2014; Weissbach and Scadden, 2012; Zhu et al., 2013) and Arabidopsis (Yan et al., 2014). TSN is conserved in all eukaryotes, with the exception of Saccharomyces cerevisiae (Broadhurst and Wheeler, 2001; Abe et al., 2003; Zhao et al., 2003; Howard-Till and Yao, 2007). In animal systems, TSN functions in several gene expression pathways in both the nucleus and the cytoplasm, including regulation of transcription (Tong et al., 1995; Leverson et al., 1998; Yang et al., 2002; Paukku et al., 2003), stimulation of pre-mRNA splicing (Yang et al., 2007), stabilization of mRNA (Paukku et al., 2008), regulation of RNA silencing (as a component of the RNA-induced silencing complex) (Caudy et al., 2003), and cleavage of hyperedited double-stranded RNA (Scadden, 2005).
The molecular functions of TSN in plants remain poorly understood. Considering that, unlike animal systems, plant TSN is absent from the nucleus (Sundström et al., 2009), its functions could be different. It has been shown that rice (Oryza sativa) TSN is a cytoskeleton-associated RNA binding protein with a role in mRNA transport to the cortical endoplasmic reticulum (Wang et al., 2008) and that Arabidopsis TSN is essential for reproduction and embryogenesis (Sundström et al., 2009). Arabidopsis TSN also promotes stress tolerance through the stabilization of stress-regulated mRNAs encoding secreted proteins (Frei dit Frey et al., 2010) or by modulating mRNA levels of gibberellin 20-oxidase 3, a key enzyme in gibberellin biosynthesis (Yan et al., 2014). Furthermore, Norway spruce (Picea abies) TSN is cleaved and inactivated by metacaspase family proteases during developmental and stress-induced cell death (Sundström et al., 2009), providing further evidence for the cytoprotective role of TSN.
The domain architecture of TSN is conserved in all studied organisms, comprising a tandem repeat of four Staphylococcal Nuclease (SN) domains followed by a Tudor and C-terminal partial SN domain (Abe et al., 2003). While both SN and Tudor domains interact physically with various proteins (Välineva et al., 2006; Yang et al., 2007; Gao et al., 2012), SN domains additionally possess nucleolytic activity and can bind RNA (Caudy et al., 2003; Li et al., 2008). The ability of TSN to cross-link RNA and proteins makes it an ideal scaffolding protein. Here, we report that Arabidopsis TSN is an integral component of both SGs and PBs during the heat stress response. We show that the assembly of plant SGs is a highly dynamic MT-dependent process where TSN plays the roles of both regulatory and scaffolding factors. Finally, we show that TSN functions in mRNA catabolism during stress as a positive regulator of mRNA decapping.
RESULTS
TSN Proteins Localize to SGs and PBs during Heat Stress
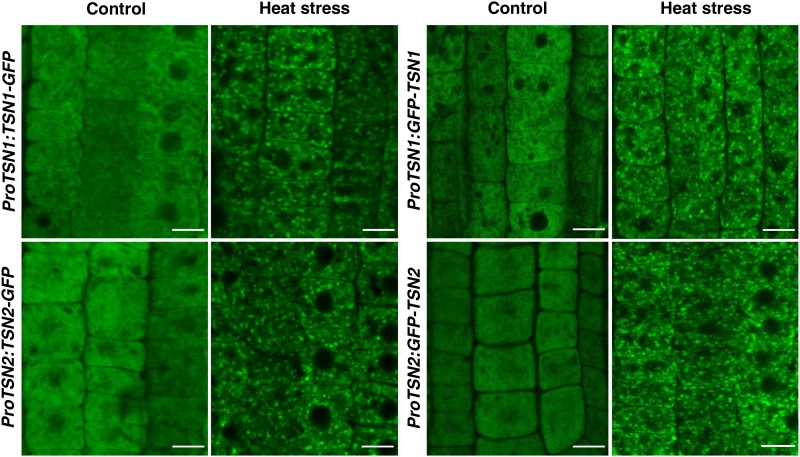
The Arabidopsis genome encodes two functionally redundant TSN homologs, TSN1 (At5g07350) and TSN2 (At5g61780) (Sundström et al., 2009; Frei dit Frey et al., 2010; Liu et al., 2010). Transgenic seedlings expressing N- or C-terminal translational fusions of TSN1 or TSN2 with green fluorescent protein (GFP), under the control of the corresponding native promoters (ProTSN:TSN-GFP and ProTSN:GFP-TSN), were incubated at 39°C for 40 min (heat stress), and the distribution of fluorescent proteins was analyzed in root tip cells. Under control conditions (23°C), TSN1 and TSN2 exhibited diffuse cytoplasmic localization, as reported previously (Frei dit Frey et al., 2010), whereas heat stress treatment induced an association of the GFP signal with prominent cytoplasmic foci (Figure 1).
Figure 1.
Heat Stress Induces Relocalization of TSN1 and TSN2 to the Cytoplasmic Foci.
Localization of TNS is shown in the root cells of 5-d-old Col seedlings expressing ProTSN1:TSN1-GFP, ProTSN2:TSN2-GFP, ProTSN1:GFP-TSN1, and ProTSN2:GFP-TSN2. The seedlings were grown under control conditions (23°C) or incubated for 40 min at 39°C (heat stress). Bars = 10 µm.
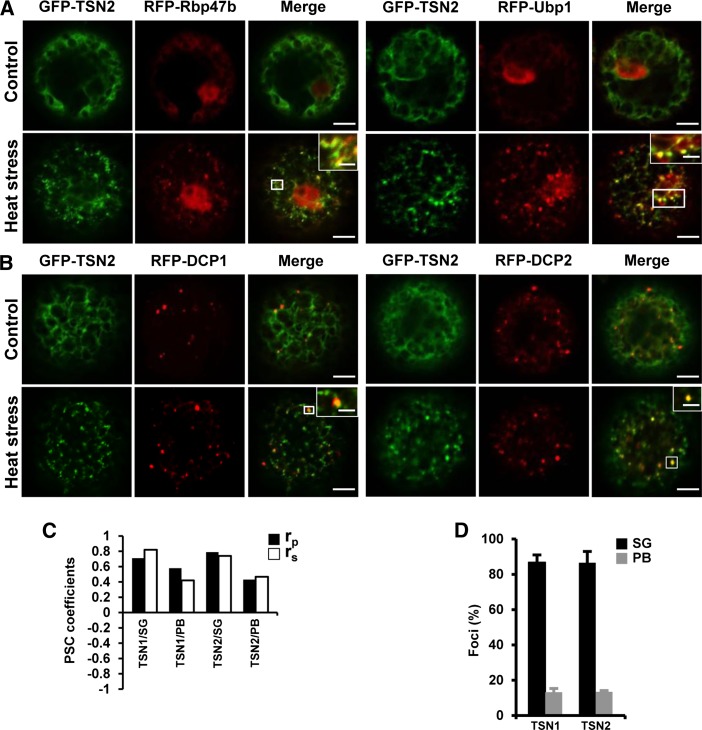
To probe the identity of the TSN foci, protoplasts isolated from Nicotiana benthamiana leaves were cotransformed with GFP-TSN1 or GFP-TSN2 and with marker proteins of Arabidopsis SGs (RFP-Rbp47b and RFP-Ubp1) or PBs (RFP-DCP1 and RFP-DCP2) (Xu et al., 2006; Weber et al., 2008) (Figure 2; Supplemental Figure S1). Under control conditions, Rbp47b localized in the nucleus and the cytoplasm and Ubp1 localized in the nucleus only (Figure 2A; Supplemental Figure S1A), in agreement with data by Lorković et al. (2000), whereas DCP1 and DCP2 localized to PB foci regardless of heat stress exposure (Figure 2B; Supplemental Figure S1B), in agreement with Xu et al. (2006). Similar to root tip cells, both GFP-TSN1 and GFP-TSN2 showed diffused cytoplasmic localization under control conditions and redistribution to foci following heat stress (Figure 2; Supplemental Figure S1). GFP-TSN1 and GFP-TSN2 colocalized with SGs (Figure 2A; Supplemental Figure S1) and PBs (Figure 2B; Supplemental Figure S1B) after heat stress. No colocalization was observed between TSN1 or TSN2 and late endosomes (Rab5 member GTPase, ARA7), Golgi (Ras-related protein, RabD2b), recycling endosomes (Rab GTPase homolog A1e, RabA1e) (Supplemental Figure 2; Geldner et al., 2009), or the membrane-specific dye FM4-64 (Supplemental Figure 2). These analyses demonstrated that TSN is an SG- and PB-associated protein and that it does not colocalize with membranous compartments.
Figure 2.
TSN1 and TSN2 Localize to SGs and PBs.
(A) and (B) Colocalization of GFP-TSN2 (green) with the SG (A) and PB (B) marker proteins (red) RFP-Rbp47b/RFP-Ubp1 and RFP-DCP1/RFP-DCP2, respectively, in protoplasts from N. benthamiana under control conditions (23°C) or after 30 min at 39°C (heat stress). N-terminal GFP and RFP fusion proteins were expressed under the control of the 35S promoter. White boxes indicate the areas that are magnified in the insets (merge). For colocalization of GFP-TSN1 with the SG and PB markers, see Supplemental Figure 1. Bars = 5 μm (2 μm in insets).
(C) Pearson and Spearman coefficients (rp and rs, respectively) of colocalization (PSC; French et al., 2008) of GFP-TSN1 and GFP-TSN2 with the SG marker RFP-Rbp47b and the PB marker RFP-DCP1 in protoplasts after heat stress.
(D) Frequency (%) of colocalization of TSN-containing foci with the SG marker RFP-Rbp47b and the PB marker RFP-DCP1 in protoplasts after heat stress. Data show means ± sd of triplicate experiments, each including 20 protoplasts.
Next, the localization of TSN proteins in SGs and PBs was confirmed using treatment with cycloheximide (CHX), an inhibitor of the translocation step during the elongation phase in protein synthesis, thereby trapping mRNA in polysomes and causing blockage of the SG and PB assembly in mammalian and plant cells (Teixeira et al., 2005; Weber et al., 2008; Wolozin, 2012). Treatment of Arabidopsis protoplasts and 5-d-old seedlings with CHX blocked the heat stress-induced assembly of TSN-positive foci, confirming that the foci were SGs and PBs (Supplemental Figures 3A and 3B). In addition, both TSN2 and Rbp47b foci showed faster disassembly when seedlings were treated with CHX after the removal of heat stress (Supplemental Figure 3B), demonstrating that mRNA present in SGs and PBs is a subject of continuous traffic to polysomes during recovery from stress.
Pixel correlation analysis revealed that both TSN1 and TSN2 colocalized significantly with the SG and PB marker proteins RFP-Rbp47b and RFP-DCP1, respectively (Figure 2C). However, only ∼10% of all TSN-positive foci colocalized with the PB marker protein (Figure 2D), suggesting intrinsic heterogeneity of the PB population following heat stress.
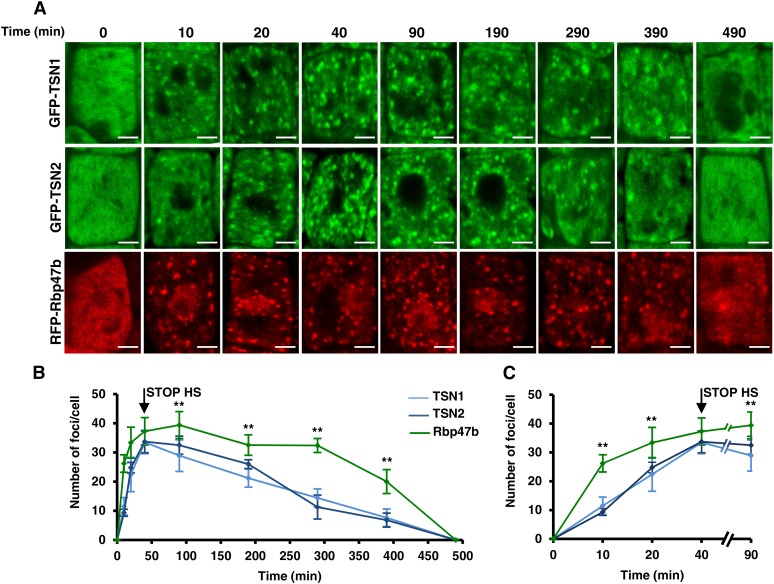
Kinetics of SGs
To examine the kinetics of SG assembly and disassembly, we followed the formation and disappearance of fluorescent foci in 5-d-old seedlings expressing GFP-TSN1, GFP-TSN2, or RFP-Rbp47b during exposure to heat stress for 40 min and a long recovery phase of up to 500 min (Figure 3). This analysis showed that the kinetics of TSN1 and TSN2 foci were indistinguishable (Figure 3). Notably, the overall number of Rbp47b foci was significantly higher than that of TSN foci at all time points except at 40 min (Figures 3B and 3C), indicating that TSN does not associate with a fraction of SG. While disassembly of Rbp47b-positive foci started 50 min after the onset of the recovery phase, disassembly of TSN-positive foci coincided with the onset of the recovery phase (Figure 3B). Yet, both Rbp47b- and TSN-positive foci completed their disassembly by 500 min (Figures 3A and 3C).
Figure 3.
Kinetics of the Assembly and Disassembly of TSN1, TSN2, and Rbp47b Foci in Arabidopsis Root Tip Cells.
(A) Five-day-old Col seedlings expressing ProTSN1:GFP-TSN1, ProTSN2:GFP-TSN2, and Pro35S:RFP-Rbp47b were heat-stressed at 39°C for 40 min, followed by incubation at 23°C for up to 500 min, and images of root tip cells were taken at the indicated time points. Bars = 2 μm.
(B) Kinetics of the assembly and disassembly of TSN1, TSN2, and Rbp47b foci.
(C) Fragment of the graph shown in (B) corresponding to the first 90-min period.
Graphs in (B) and (C) show means ± sd of triplicate experiments, each including 5 to 10 plants per line. STOP HS is the time point when the plants were transferred from 39 to 23°C. **P < 0.05, Student’s t test (Rbp47b versus TSN2).
MTs Control SG Formation
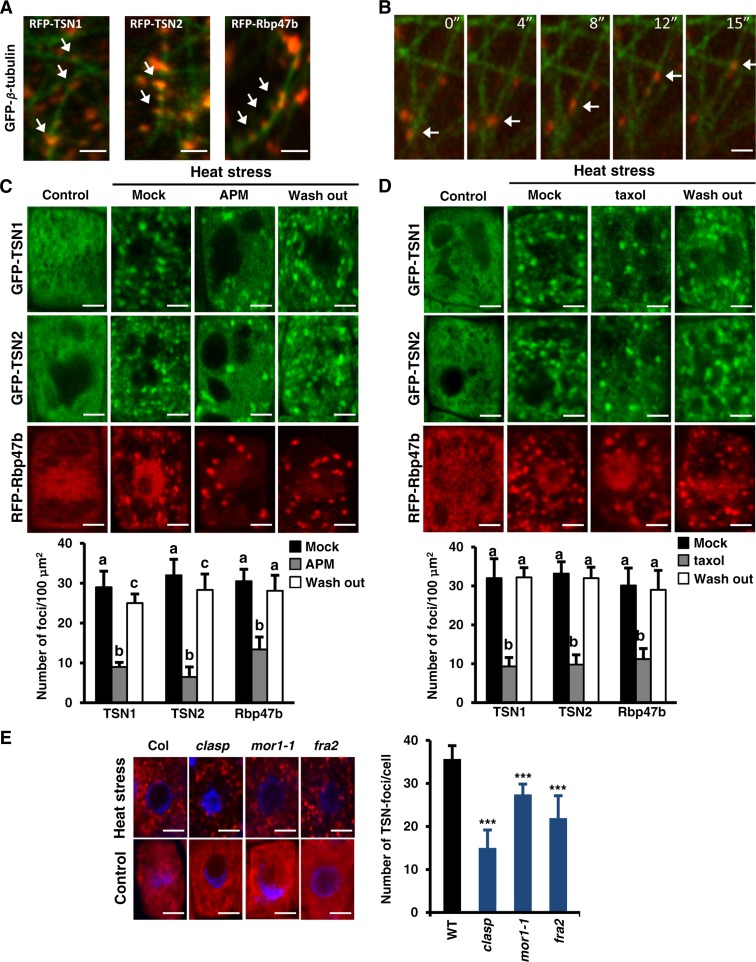
In mammals, MTs are important components of the machinery regulating the assembly, mobility, and dynamics of SGs (Buchan and Parker, 2009; Nadezhdina et al., 2010). Considering that TSN was identified as a cytoskeleton-associated RNA binding protein in rice (Sami-Subbu et al., 2001) and later as a tubulin binding protein in Arabidopsis (Chuong et al., 2004), we examined whether SG assembly and disassembly are MT-dependent. First, we assessed the colocalization of SG and TSN foci with MTs during heat stress in N. benthamiana leaf epidermal cells transiently cotransformed with RFP-TSN1, RFP-TSN2, or RFP-Rbp47b with GFP-β-tubulin. In these assays, Rbp47b and TSN foci colocalized with cortical MTs and moved along MT tracks (Figures 4A and 4B).
Figure 4.
(A) Colocalization of RFP-TSN1 (red), RFP-TSN2 (red), and RFP-Rbp47b (red) with MT labeled by GFP-β-tubulin (green) in N. benthamiana leaf epidermal cells. Two days after infiltration, the leaves were heat-stressed at 39°C for 40 min. N-terminal GFP and RFP fusion proteins were expressed under the control of the 35S promoter. Arrows denote colocalization. Bars = 5 μm.
(B) Selected time frames showing the movement of SGs (red; denoted by arrows) along MTs (green). Bar = 4 μm.
(C) and (D) Treatment with APM (C) or taxol (D) inhibits the formation of TSN and Rbp47b foci in root tip cells. Five-day-old Arabidopsis Col seedlings expressing ProTSN:GFP-TSN and Pro35S:RFP-Rbp47b were treated with APM (10 μM), taxol (20 μM), or DMSO (at the same concentrations; mock) for 5 h at room temperature. APM- and taxol-treated seedlings were washed and incubated for 2 h in liquid MS medium (wash out). Heat stress was induced for 40 min at 39°C. Seedlings grown under 23°C were used as a control. Graphs show mean numbers ± sd of TSN or Rbp47b foci per 100 μm2 of cytoplasm counted in 5 to 10 plants per treatment in three independent experiments. Means with different letters are significantly different at P < 0.05, Student’s t test. Bars = 2 μm.
(E) Formation of TSN foci is inhibited in the Arabidopsis MT mutants clasp, mor1-1, and fra2. Seedlings were heat-stressed at 39°C for 40 min and then immunostained with anti-TSN (red). Control seedlings were grown at 23°C. DNA staining with 4′,6-diamidino-2-phenylindole is shown in blue. The graph shows mean numbers ± sd of TSN foci per cell counted in 5 to 10 plants in each triplicate experiment. ***P < 0.001 versus the wild type, Dunnett’s test. Bars = 2 μm.
Second, we examined the role of MTs in the formation of TSN foci using MT inhibitors. Disruption of the MT network with 10 μM amiprophos-methyl (APM), an herbicide that promotes the depolymerization of MTs, caused a significant reduction in the number of TSN foci (Figure 4C). Similar results were obtained when MTs were stabilized by treatment with 20 μM taxol (Figure 4D). To verify whether MT dynamics are essential only for the recruitment of TSN to cytoplasmic foci or for the formation of SGs, we treated seedlings expressing RFP-Rbp47b with APM or taxol. Both treatments inhibited the formation of Rbp47b foci during heat stress (Figures 4C and 4D), demonstrating that SG assembly requires dynamic MTs. Washing out taxol or APM restored the number of both TSN and Rbp47b foci per 100 μm2 of cytoplasm, confirming that the inhibition of SG formation was the consequence of affected MT dynamics (Figures 4C and 4D, wash out). Treatment of 5-d-old seedlings coexpressing GFP-TSN2 and RFP-Rbp47b confirmed that APM specifically inhibited TSN-positive SGs (Supplemental Figure 4A). In contrast with SGs, the formation of PBs was unaffected by APM, consistent with the fact that PBs are regulated by actin (Supplemental Figure 4B) (Steffens et al., 2014).
The importance of MTs for SG formation was corroborated using several mutants with compromised MT dynamics. An allele of the MT-associated protein CLASP (clasp-1) (Ambrose et al., 2007) exhibits reduced stability and density of interphase MTs and is hypersensitive to MT depolymerization inhibitors. A temperature-sensitive allele of the MT-associated protein MOR1/GEM1 (mor1-1) (Whittington et al., 2001) exhibits a normal MT network that becomes disorganized at mild heat shock (30°C). Since the disorganization of MTs in mor1-1 would coincide with the initiation of SG assembly, we can use this allele to exclude scenarios in which MTs play a role in the formation of SG precursors prior to heat stress. The third mutant, fra2, lacks activity of the MT-severing protein katanin and, consequently, has a more stable MT network (Burk et al., 2001). The assembly of TSN foci was suppressed in all three mutants (Figure 4E), suggesting the importance of a balance between MT polymerization and depolymerization for the assembly of SGs.
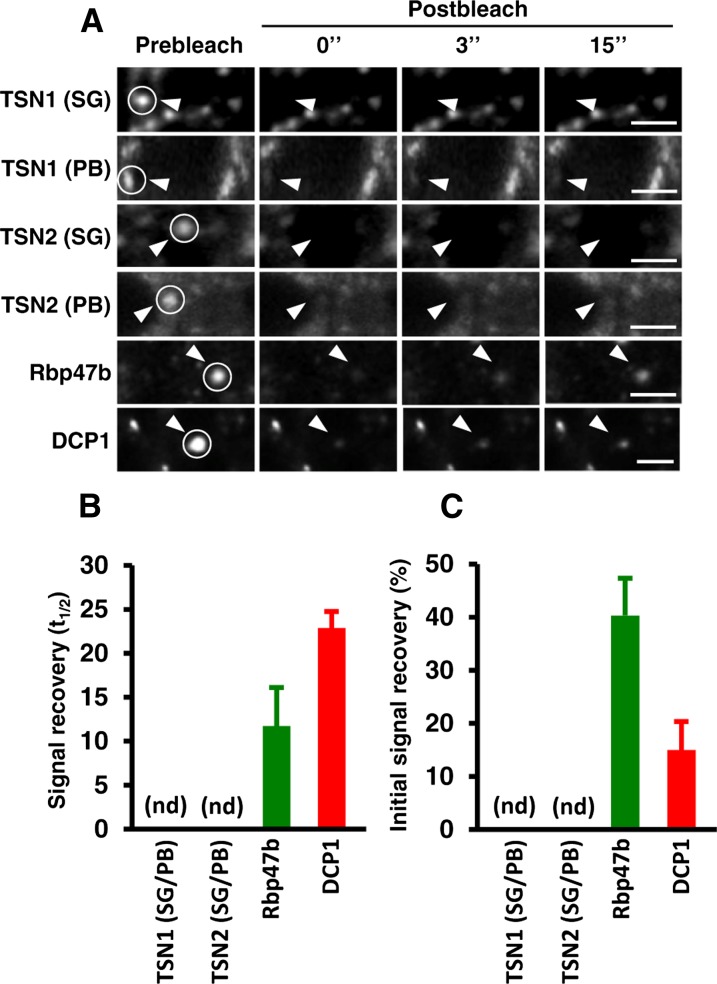
TSN Proteins Are Stably Associated with SGs and PBs
SGs and PBs are dynamic structures exchanging mRNA and proteins with the cytosol (Anderson and Kedersha, 2008; Franks and Lykke-Andersen, 2008; Mollet et al., 2008). We investigated the possibility that TSN associates dynamically with SGs and PBs using fluorescence recovery after photobleaching (FRAP) analysis. Surprisingly, the fluorescent signal of GFP-TSN1 and GFP-TSN2 lacked any apparent recovery after photobleaching (Figures 5A and 5B), pointing out the stable association of TSN proteins with SGs and PBs. In contrast with TSN, interaction of both GFP-Rbp47b and GFP-DCP1 with foci was dynamic (Figures 5A and 5B). Therefore, specific constituents of SGs and PBs can have distinct modes of recruitment and association with these structures. Since the recovery of fluorescence signal of GFP-Rbp47b and GFP-DCP1 was incomplete (Figure 5C), we conclude that a significant proportion of both proteins exchanges with the cytoplasm very slowly or is immobile.
Figure 5.
TSN1 and TSN2 Are Integral Proteins of SGs and PBs.
(A) Selected time frames (0, 3, and 15 s after bleaching) from FRAP analysis of GFP-TSN1, GFP-TSN2, GFP-Rbp47b, and GFP-DCP1 in root tip cells of Col seedlings expressing ProTSN:GFP-TSN, Pro35S:GFP-Rbp47b, and Pro35S:GFP-DCP1 after heat stress (40 min at 39°C). To distinguish between SG- and PB-associated GFP-TSN1 or GFP-TSN2, both proteins were stably coexpressed with RFP-Rbp47b and RFP-DCP1, respectively. Circles and arrowheads indicate the bleached foci. Bars = 3 μm.
(B) Signal recovery rate (t1/2) of GFP fusion proteins. nd, not detected.
(C) Proportion of the initial signal recovered (%).
Data in (B) and (C) represent means ± sd of three independent experiments, each containing at least 10 seedlings.
TSN Deficiency Affects the Integrity and Formation of SGs and PBs
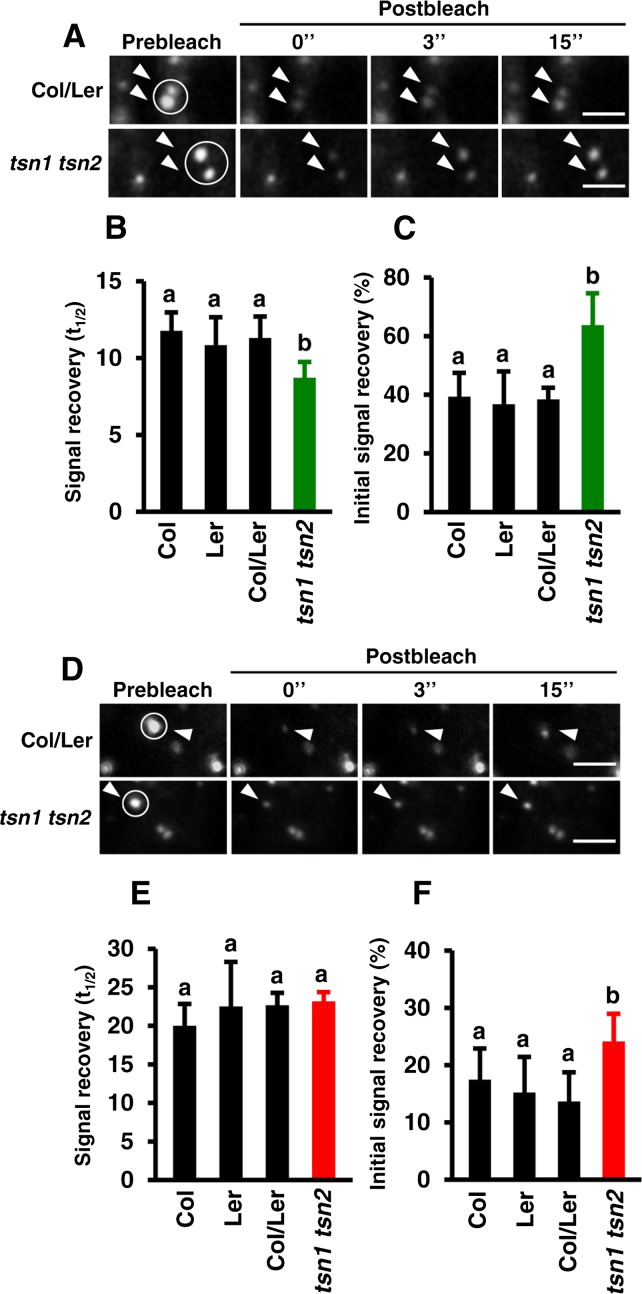
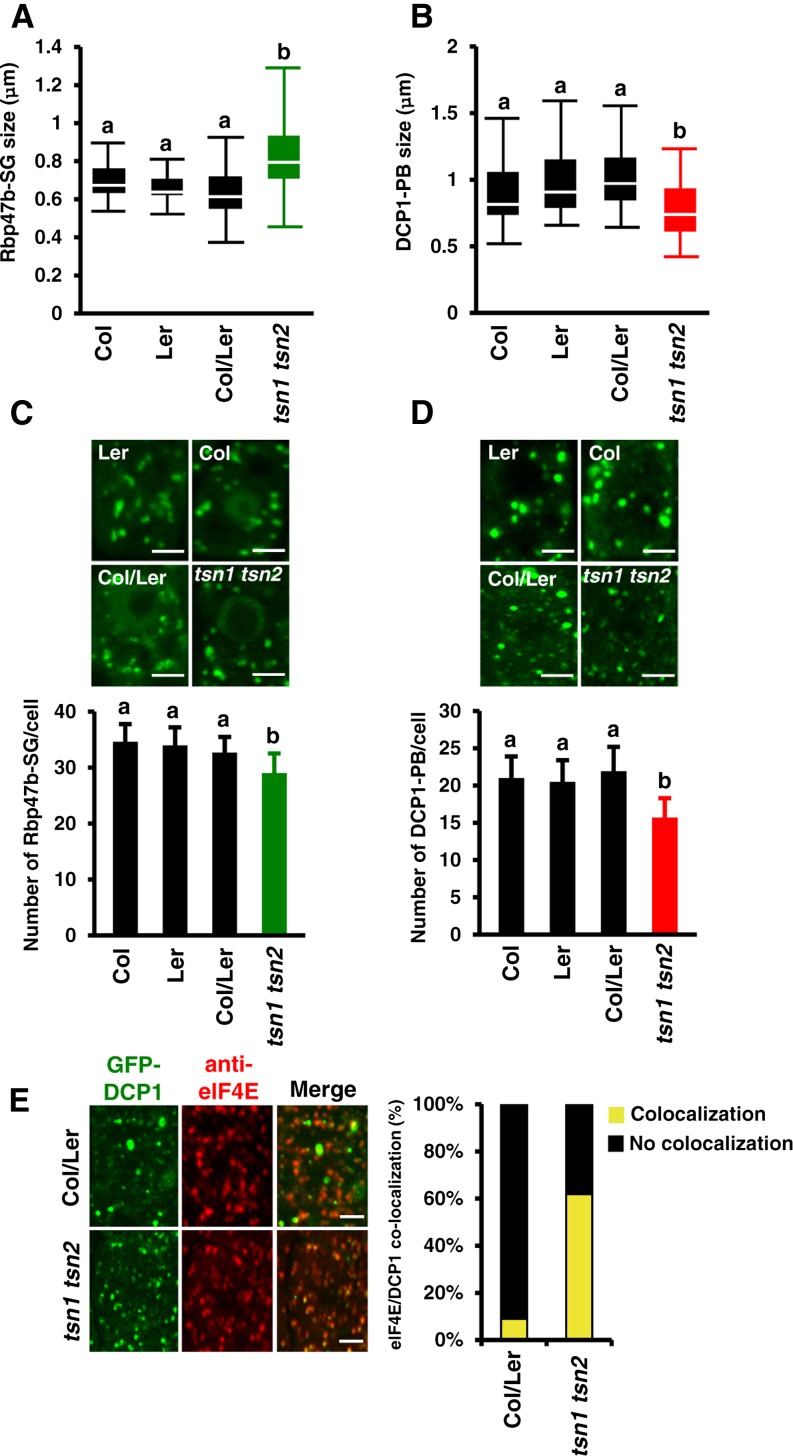
To address the role of TSN in the maintenance of structural integrity of SGs and PBs, we measured shuttling of Rbp47b and DCP1 between foci and the cytoplasm in TSN-deficient plants. Single T-DNA insertion mutants for TSN1 and TSN2 in Landsberg erecta (Ler) and Columbia-0 (Col) backgrounds, respectively, were crossed to generate a tsn1 tsn2 double loss-of-function mutant (Supplemental Figure 5A). The lack of TSN protein in tsn1 tsn2 plants was confirmed by immunoblot analysis using anti-TSN antibody, which recognizes both TSN isotypes (Supplemental Figure 5B; Sundström et al., 2009). Five-day-old seedlings of tsn1 tsn2, Col, Ler, and Col/Ler, all expressing GFP-Rbp47b or GFP-DCP1, were exposed to heat stress for 40 min at 39°C, and the association of both proteins with SGs or PBs was assessed by FRAP. TSN deficiency increased the exchange rate of GFP-Rbp47b between SGs and the cytoplasm (Figures 6A and 6B) and decreased the immobile fraction from 60 to 40% (Figure 6C). The shuffling of GFP-DCP1 between PBs and the cytoplasm was not affected by TSN deficiency (Figures 6D and 6E); however, the immobile fraction of GFP-DCP1 was reduced by almost 2-fold in comparison with the wild type (Figure 6F). The size of SGs and PBs was also affected by TSN deficiency, but in a different way. While SG size increased (Figure 7A), the size of PBs decreased (Figure 7B). Together, these results suggest that TSN can modulate the integrity of SGs and PBs and regulate the retention of their core functional proteins and RNA.
Figure 6.
TSN Mediates the Association of Rbp47b and DCP1 with Cytoplasmic Foci.
(A) Selected time frames (0, 3, and 15 s after bleaching) from FRAP analysis of GFP-Rbp47b in root tip cells of tsn1 tsn2 and Col/Ler seedlings expressing Pro35S:GFP-Rbp47b after heat stress. Bars = 3 μm.
(B) and (C) Signal recovery rate (t1/2; [B]) and proportion of the initial signal recovery (%; [C]) of GFP-Rbp47b in root tip cells of wild-type (Col, Ler, and Col/Ler) and tsn1 tsn2 seedlings expressing Pro35S:GFP-Rbp47b. Green columns correspond to the mutant background (tsn1 tsn2), while black columns correspond to wild-type backgrounds.
(D) Selected time frames (0, 3, and 15 s after bleaching) from FRAP analysis of GFP-DCP1 in root tip cells of tsn1 tsn2 and Col/Ler seedlings expressing Pro35S:GFP-DCP1 after heat stress. Bars = 3 μm.
(E) and (F) Signal recovery rate (t1/2; [E]) and proportion of the initial signal recovery (%; [F]) of GFP-DCP1 in root tip cells of wild-type (Col, Ler, and Col/Ler) and tsn1 tsn2 seedlings expressing Pro35S:GFP-DCP1. Red columns correspond to the mutant background (tsn1 tsn2), while black columns correspond to wild-type backgrounds.
The seedlings were heat-stressed at 39°C for 40 min. Circles and arrowheads in (A) and (D) indicate the bleached foci. Data in (B), (C), (E), and (F) show means ± sd of triplicate experiments, each containing at least 10 seedlings. Means with different letters are significantly different at P < 0.05, Student’s t test.
Figure 7.
TSN Deficiency Affects the Formation and Segregation of SGs and PBs.
(A) and (B) Box plots of the size (diameter) of GFP-Rbp47b (A) and GFP-DCP1 (B) foci in root tip cells of wild-type (Col, Ler, and Col/Ler) and tsn1 tsn2 seedlings expressing Pro35S:GFP-Rbp47b and Pro35S:GFP-DCP1, respectively, after heat stress (40 min at 39°C). Each box plot shows the median (solid line), the 25th and 75th percentiles (boxes), and the 5th and 95th percentiles (error bars).
(C) and (D) Formation of GFP-Rbp47b (C) and GFP-DCP1 (D) foci in root tip cells of wild-type (Col, Ler, and Col/Ler) and tsn1 tsn2 seedlings expressing Pro35S:GFP-Rbp47b and Pro35S:GFP-DCP1, respectively, after heat stress. The graphs show mean numbers ± sd of foci per cell counted in 5 to 10 plants in each triplicate experiment. Green and red columns correspond to the mutant background (tsn1 tsn2), while black columns correspond to wild-type backgrounds. Means with different letters are significantly different at P < 0.05, Student’s t test. Bars = 2 μm.
(E) Colocalization analysis of GFP-DCP1 (green) and eIF4E (red) in root tip cells of Col/Ler and tsn1 tsn2 seedlings expressing Pro35S:GFP-DCP1 after heat stress. The graph displays the frequency of colocalization of DCP1 and eIF4E in wild-type and tsn1 tsn2 plants. Bars = 2 μm.
To investigate whether the altered integrity of SGs and PBs in TSN-deficient plants could have an effect on their formation, we quantified SGs and PBs in the root tip cells of heat-stressed tsn1 tsn2 and wild-type plants. Although SGs and PBs could assemble without TSN, the number of Rbp47b and DCP1 foci was significantly reduced in comparison with the wild type (Figures 7C and 7D). Furthermore, the kinetics of SG assembly in tsn1 tsn2 plants lagged behind that of the wild type, reaching the same density only after 50 min of the recovery phase (Supplemental Figures 6A and 6C). However, the kinetics of SG disassembly was indistinguishable between tsn1 tsn2 and the wild type (Supplemental Figure 6B), indicating that TSN is not required for the disassembly of SGs.
TSN Defines the Identity of Cytoplasmic Foci
Although SGs and PBs are distinct structures, they may share some components (e.g., eIF4E, which is usually found in SGs but can also be detected in some PBs) (Kedersha et al., 2005). In this context, we used eIF4E localization as a marker to investigate the role of TSN in the identity of both structures. Immunostaining of seedlings expressing GFP-DCP1 with anti-eIF4E demonstrated that, similar to animal systems (Kedersha et al., 2005), eIF4E colocalizes with ∼5% of PB. In tsn1 tsn2 plants, this colocalization increased to approximately 60% (Figure 7E). This suggests that TSN could be required for the molecular identity of SGs and PBs.
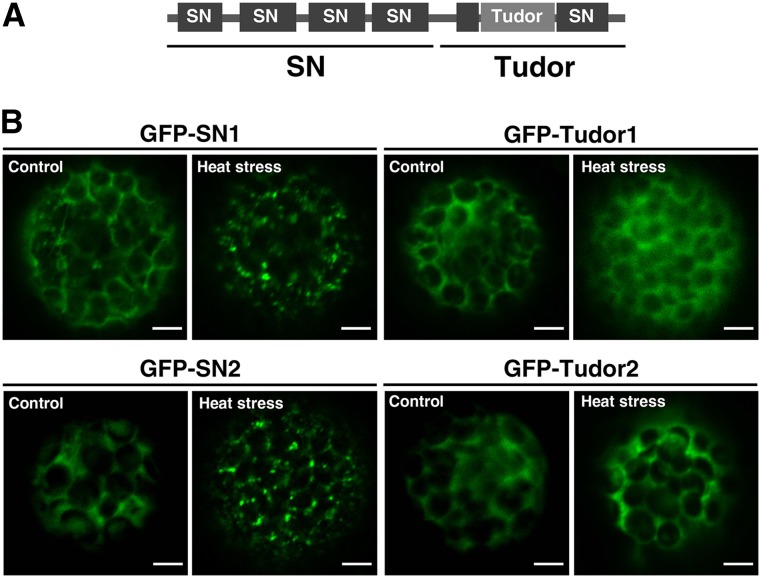
SN Domains Are Required for the Localization of TSN to the Cytoplasmic Foci
To explore how TSN is targeted to SGs and PBs, we compared the localization of GFP fused to a tandem repeat of four N-terminally located SN domains (hereafter designated as SN) and to Tudor and the fifth SN domains (hereafter designated as Tudor) (Figure 8A). Protoplasts from N. benthamiana leaves were transfected with both constructs and after 24 h subjected to heat stress for 30 min. Under control conditions, both SN and Tudor truncated forms were localized in the cytoplasm (Figure 8B), similar to the full-length TSN1 and TSN2. After exposure to heat stress, SN became associated with the foci, whereas the localization of Tudor remained cytoplasmic (Figure 8B). Therefore, TSN is targeted to SGs and PBs by a tandem repeat of four SN domains.
Figure 8.
Tandem Repeat of Four SN Domains Is Required for the Localization of TSN to the Cytoplasmic Foci.
(A) Schematic diagram of TSN protein structure showing SN and Tudor regions.
(B) Localization analysis of GFP-SN1 (amino acids 1 to 723), GFP-Tudor1 (amino acids 724 to 990), GFP-SN2 (amino acids 1 to 720), and GFP-Tudor2 (amino acids 721 to 985) in N. benthamiana leaf protoplasts. Protoplasts were kept under control conditions or heat-stressed at 39°C for 30 min. N-terminal GFP fusion proteins were expressed under the control of the 35S promoter. Bars = 5 μm.
Nucleolytic Activity of TSN Is Required for the Proper Dynamics of Rbp47b in SGs
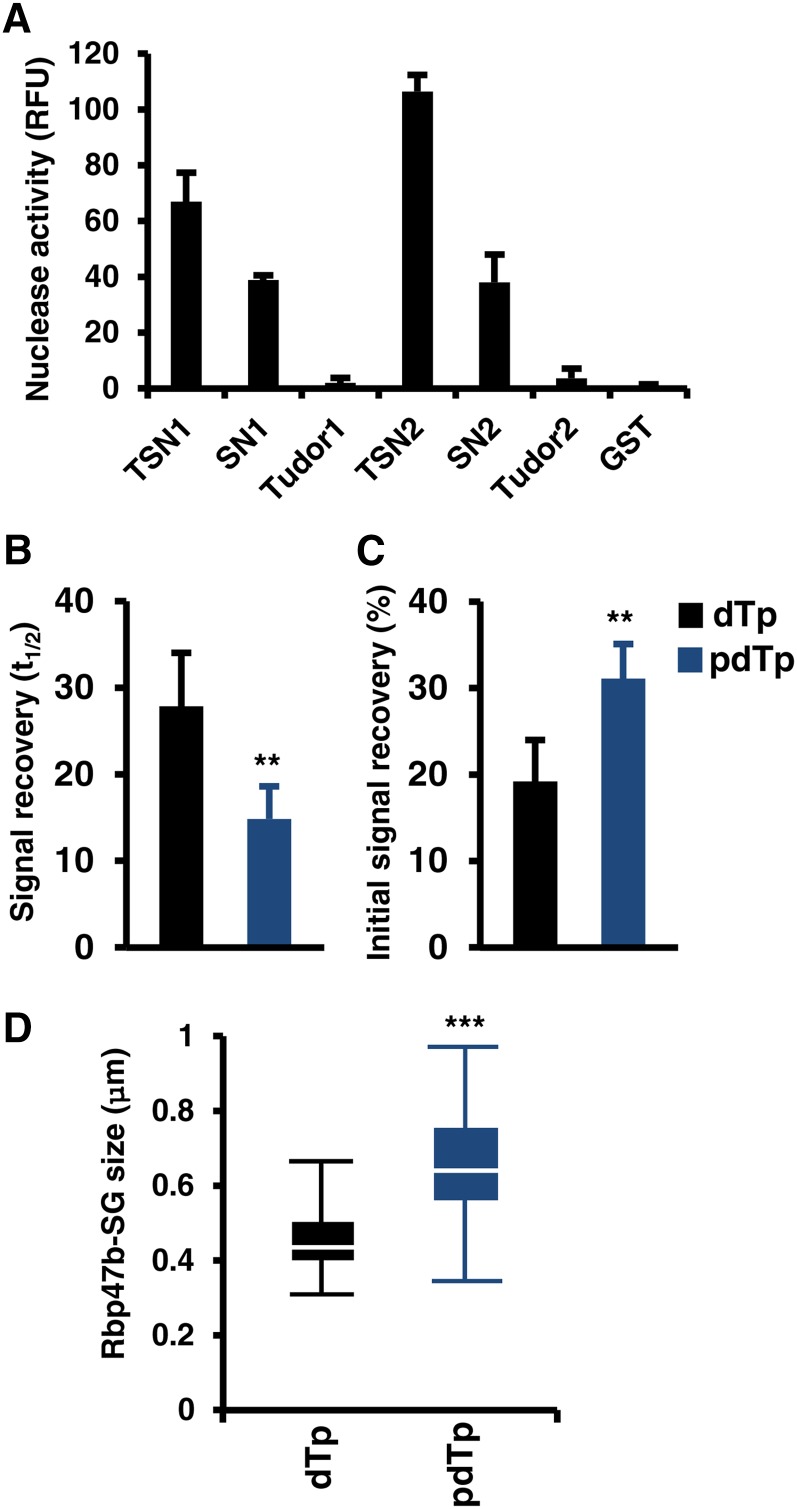
It has been suggested that the nucleolytic activity of TSN in mammals, protozoa, and plants is required for its biological function (Caudy et al., 2003; Hossain et al., 2008; Sundström et al., 2009). Therefore, we investigated how the nucleolytic activity of TSN affects its association with SGs. First, the activity of full-length TSN and its truncations was compared by an in vitro nuclease assay using fluorescent RNA substrate. Full-length proteins and SN1 and SN2 were nucleolytically active, whereas Tudor1 and Tudor2 were inactive (Figure 9A).
Figure 9.
Nucleolytic Activity of TSN Is Required for the Proper Dynamics of Rbp47b in SGs.
(A) In vitro nucleolytic activity of full-length and truncated forms of TSN1 and TSN2. Recombinant GST fusion proteins were purified from E. coli and incubated in nuclease buffer containing fluorescent RNA substrate. RNase activity was determined by a fluorometric assay. GST was used as a negative control. RFU, relative fluorescence units. Data show means ± sd of three independent experiments.
(B) and (C) Signal recovery (t1/2; [B]) and initial signal recovery (%; [C]) of RFP-Rbp47b determined by FRAP analysis in Arabidopsis root tip cells treated with pdTp and dTp. Five-day-old seedlings expressing Pro35S:GFP-Rbp47b were incubated in MS medium supplemented with 50 μM pdTp or dTp (control) for 5 h at 23°C and then heat-stressed for 40 min at 39°C. **P < 0.01, Student’s t test. Data show means ± sd of triplicate experiments, each containing at least 10 seedlings.
(D) Box plot of the size of GFP-Rbp47b foci (SGs) in root tip cells of plants treated with pdTp and dTp and then subjected to heat stress for 40 min at 39°C. Each box plot shows the median (solid line), the 25th and 75th percentiles (boxes), and the 5th and 95th percentiles (error bars). ***P < 0.001, Student’s t test.
Second, the nucleolytic activity of TSN can be suppressed by treatment with the specific inhibitor 3′,5′-deoxythymidine bisphosphate (pdTp) (Caudy et al., 2003; Scadden, 2005; Li et al., 2008; Sundström et al., 2009). Therefore, 5-d-old seedlings expressing RFP-Rbp47b were treated with pdTp (50 μM) for 5 h at 23°C followed by heat stress at 39°C for 40 min. Treatment with an inactive analog of pdTp, 3′-deoxythymidine monophosphate (dTp), was used as a negative control (Sundström et al., 2009). FRAP analyses showed that pdTp treatment resulted in a faster recovery of RFP-Rbp47b signal in SGs and in the reduction of the Rbp47b immobile fraction (Figures 9B and 9C), an effect similar to TSN knockout (Figures 6B and 6C). Moreover, in analogy with TSN knockout, the RFP-Rbp47b foci were larger in the pdTp-treated plants (compare Figures 7A and 9D). Our data demonstrate that the nucleolytic activity of TSN conferred by the tandem repeat of four SN domains is essential for targeting of TSN to SG as well as for the proper dynamics of Rbp47b in SG and their morphology.
Nucleolytic Activity of TSN Is Required for Stress Tolerance
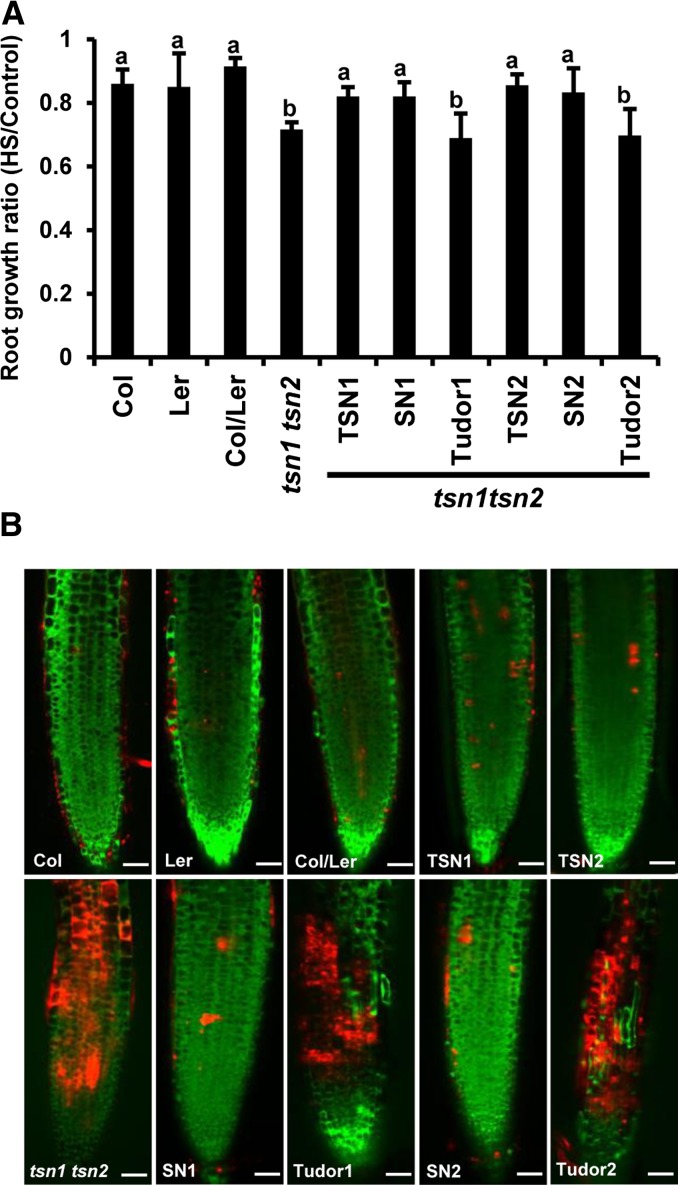
TSN is a cytoprotective protein previously shown to be essential for plant development and salt stress resistance (Sundström et al., 2009; Frei dit Frey et al., 2010; Yan et al., 2014). To establish whether the requirement of TSN for stress tolerance is dependent on its nucleolytic activity, we compared root growth during long-term heat stress of the wild type (Col, Ler, and Col/Ler), tsn1 tsn2, and tsn1 tsn2 expressing full-length or truncated TSN versions (Supplemental Figure 7). The root growth of tsn1 tsn2 plants was significantly suppressed in comparison with the wild type (Figure 10A). This phenotype was rescued by either TSN1 or TSN2, indicating their functional redundancy. The tsn1 tsn2 root phenotype was also rescued by tandem repeats of four SN domains but not by Tudor domains (Figure 10A).
Figure 10.
The Tandem Repeat of Four SN Domains of TSN Is Essential for Stress Tolerance.
Five-day-old seedlings expressing ProTSN:GFP-TSN, Pro35S:GFP-SN, and Pro35S:GFP-Tudor in the tsn1 tsn2 background were compared with the wild type (Col, Ler, and Col/Ler) under long-term (A) and short-term (B) heat stress treatments.
(A) Ratio of root length of seedlings grown for 24 h under heat stress (HS) to root length of control seedlings grown at 23°C. The data show means ± sd of triplicate experiments, each containing 10 seedlings. Means with different letters are significantly different at P < 0.05, Student’s t test.
(B) Root staining of heat-stressed (for 4 h) seedlings with FDA (green staining; indicative of living cells) and SYTOX Orange (red staining; indicative of dead cells). Bars = 65 μm.
These data correlated with cell viability assays. Five-day-old seedlings were exposed to short-term heat stress for 4 h at 39°C, and roots were stained with fluorescein diacetate (FDA), a marker of living cells, and SYTOX Orange, a marker of damaged plasma membrane and dead cells (Truernit and Haseloff, 2008). The roots of tsn1 tsn2 plants exhibited a lower frequency of FDA-negative cells and a higher frequency of SYTOX Orange-positive cells, as compared with the wild type (Figure 10B), showing that TSN deficiency causes ectopic cell death. This cell death phenotype was rescued by full-length TSN and SN domains but not by Tudor domains (Figure 10B). These results demonstrate that a nucleolytically active tandem repeat of four SN domains is essential for TSN-mediated cytoprotection and heat stress tolerance.
TSN Is a Positive Regulator of mRNA Decapping during Stress
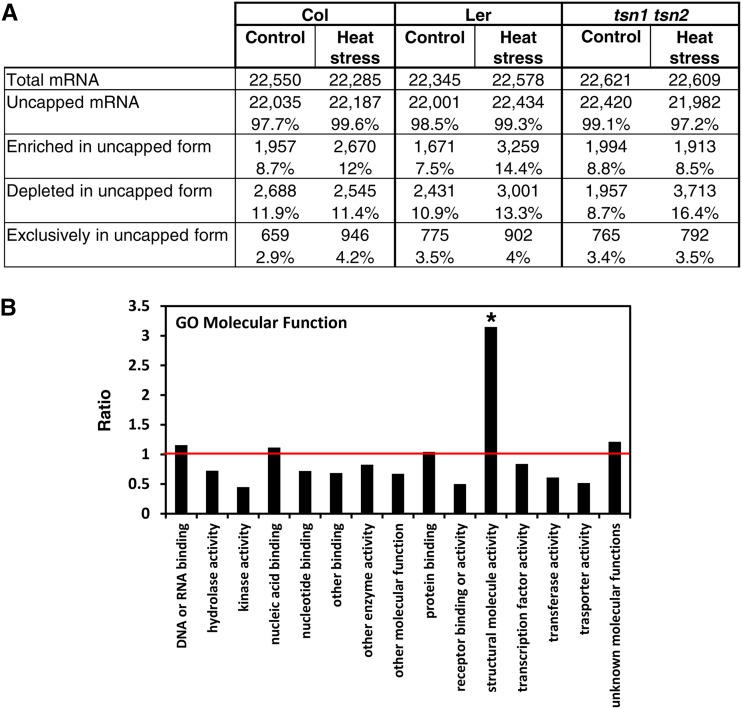
Since mRNA decay represents one of the primary functions of the cytoplasmic messenger ribonucleoprotein (mRNP) complexes and is executed by endonucleases (Tomecki and Dziembowski, 2010; Nagarajan et al., 2013), we reasoned that TSN could be implicated in mRNA decay. To address this possibility, we analyzed the mRNA degradome of wild-type (Col and Ler) and tsn1 tsn2 seedlings grown under heat stress (39°C for 40 min) and control (23°C) conditions. Uncapped mRNA fractions composed of intermediates of the 5′→3′ decay pathway were purified and used for measuring the abundance of individual mRNAs by using cDNA arrays (Jiao et al., 2008; Jiao and Riechmann, 2012). Before microarray hybridization, a control experiment confirmed that there was no contamination from intact 5′ cap mRNAs in our uncapped mRNA preparations (Jiao et al., 2008) (Supplemental Figure 8). The analysis revealed that, regardless of genotype and growth conditions, the vast majority (>97%) of mRNAs exist, at least transiently, in the uncapped form (Figure 11A). Consistent with an earlier report (Jiao et al., 2008), numerous transposable element-related genes and pseudogenes were detected in the uncapped form (Supplemental Figure 9). In our study, we compared the abundance of uncapped individual mRNAs with the levels of the corresponding total (uncapped and capped) mRNAs. This analysis furnished lists of transcripts that were either enriched or depleted in uncapped form in individual genetic backgrounds under specific (control or stress) conditions (Supplemental Data Set 1). Upon heat stress, the fraction of mRNAs enriched in the uncapped form markedly increased in the wild type but remained constant in tsn1 tsn2 plants (Figure 11A, enriched in uncapped form), demonstrating that TSN is required for heat stress-induced mRNA decapping. It is noteworthy that the efficiency of mRNA decapping under control conditions was similar in tsn1 tsn2 and wild-type plants (Figure 11A). Taken together, these data suggest that relocation of TSN to the cytoplasmic foci under stress conditions is required for mRNA decapping.
Figure 11.
TSN Is a Positive Regulator of mRNA Decapping during Heat Stress.
(A) Quantification and relative abundance of uncapped mRNAs in Col, Ler, and tsn1 tsn2 plants grown under control and heat stress conditions. The percentages of mRNAs in each category were calculated relative to the total number of mRNAs.
(B) GO analysis (term “molecular function”) of TSN-dependent mRNA decapping. The chart displays the ratios between the percentage of mRNAs belonging to a particular GO term that show significant depletion in uncapped form in tsn1 tsn2 and the percentage of mRNAs belonging to the same GO term in the total mRNA population under heat stress. The horizontal red axis indicates a ratio value of 1. The overrepresented category is marked with an asterisk (P < 0.05, Fisher’s exact test).
The requirement of TSN for mRNA decapping under heat stress was further validated by comparing the abundance of individual uncapped mRNAs between heat stress and control conditions (Supplemental Figure 10 and Supplemental Data Set 2). We observed an almost 2-fold increase in the frequency and absolute number of transcripts depleted in the uncapped form in tsn1 tsn2 plants upon heat stress (Figures 11A, depleted in uncapped form; Supplemental Figure 10). This might suggest that many mRNAs remain capped in TSN-deficient plants. To characterize the transcripts depleted in the uncapped form in heat-stressed tsn1 tsn2 plants, we grouped them into Gene Ontology (GO) molecular function categories (Figure 11B; Supplemental Data Set 1). When compared with the total mRNA population, the most enriched GO molecular function was that of “structural molecule activity” (Figure 11B). In particular, a substantial enrichment in ribosomal proteins was detected (Supplemental Data Set 1, see “Metabolism, Biosynthesis and Catabolism” category in section C4). These data are consistent with the notion that TSN facilitates the catabolism of specific mRNAs whose expression may compromise stress tolerance.
DISCUSSION
Role of TSN in the Assembly of Stress-Induced Cytoplasmic Foci
Although TSN exhibits evolutionarily conserved primary and secondary structure (Li et al., 2008), little is known about how conserved its functions are. Judging by the variability of intracellular localization, the functions of TSN in different eukaryotic lineages may not be conserved. For example, mammalian TSN homologs localize to both the cytoplasm and nucleus (Caudy et al., 2003; Weissbach and Scadden, 2012), while plant TSN is cytoplasmic (Wang et al., 2008; Sundström et al., 2009; Frei dit Frey et al., 2010). Furthermore, while insect and human TSN localize only to SGs (Gao et al., 2010; Zhu et al., 2013), we show that plant TSN is part of both SGs and PBs.
SGs and PBs belong to the group of mRNP complexes functioning in the posttranscriptional control of protein expression (Anderson and Kedersha, 2009). Temporal interactions between SGs and PBs facilitate the exchange of mRNA and proteins (Parker and Sheth, 2007; Lavut and Raveh, 2012). As a result, the protein composition of SGs and PBs partly overlaps (i.e., both structures contain eIF4E, Fas-activated serine/threonine phosphoprotein [FAST], 5′→3′ XRN1, and tristetrapolin) (Kedersha et al., 2005). However, some proteins are found exclusively in SGs or PBs. SGs assemble in response to stress stimuli and contain components of the translation initiation machinery, whereas PBs are constitutive structures containing components of the mRNA decay machinery. In this work, we observed that Arabidopsis TSN colocalizes with SG and PB markers, Rbp47b/Ubp1 and DCP1/DCP2, respectively, following heat stress. This observation indicates that TSN is a component of both types of cytoplasmic mRNP complexes, but in addition it points to the existence of several classes of stress-induced cytoplasmic complexes of different molecular composition and functions.
The concerted action of several factors that mediate RNA binding (Kedersha et al., 2005; Ohn and Anderson, 2010) promotes the assembly of small mRNP complexes followed by the formation of larger SGs (Kedersha et al., 1999; Mollet et al., 2008). Considering the dynamic nature of SG assembly, the role of individual proteins in this process can be assessed by the timing of their recruitment to the SGs following a stress stimulus (Buchan and Parker, 2009; Buchan et al., 2011). In our experiments, Rbp47b foci became detectable before the appearance of TSN foci, indicating that TSN is not required for the initial steps of SG assembly in Arabidopsis. In agreement with our data, SG assembly in mammals was shown to be dependent on T cell Intracellular Antigen-1 (TIA-1; a homolog of Rbp47b) and TIAR proteins (Gilks et al., 2004; Weber et al., 2008) but not on TSN (Gao et al., 2012).
Analyses of SGs and PBs in nonplant systems revealed that some proteins exhibit rapid dynamic exchange between the cytoplasm and foci (Kedersha and Anderson, 2007). Accordingly, we observed that SGs and PBs in Arabidopsis are also dynamic structures, since substantial fractions of both Rbp47b and DCP1 proteins are present in mobile form and show rapid recovery after FRAP. On the contrary, we detected no exchange of TSN between the cytoplasm and foci. Since the slower exchange of FAST, Fragile X Mental Retardation, and Ribosomal S6 Kinase 2 proteins in mammalian cells was attributed to their putative scaffolding role in the organization of SGs and PBs (Kedersha et al., 2005; Chalupniková et al., 2008; Eisinger-Mathason et al., 2008), TSN could perform a similar function. In agreement with this conclusion, the knockout of both TSN genes resulted in a higher mobility of Rbp47b and DCP1, enlargement of SGs, and condensation of PBs. This demonstrates that, despite the relatively late recruitment of TSN to SGs and PBs in the course of their formation, TSN is required for the maintenance of the structural integrity of both complexes and could also act in their maturation with possible consequences for their functionality.
Requirement of MTs for the Assembly of TSN-Positive SGs
MTs serve as tracks for the intracellular trafficking of membranous and protein structures. In animal models, the assembly of SGs was inhibited by the depolymerization of MTs, while the stabilization of MTs promoted SG formation (Ivanov et al., 2003; Chernov et al., 2009). Interestingly, both destabilization and stabilization of MTs in Arabidopsis inhibited the assembly of TSN and Rbp47b foci. These observations suggest that MT drugs inhibited the process of SG formation rather than the delivery of individual components to the sites of SG assembly. Furthermore, the assembly of TSN foci was inhibited in the mor1-1 and clasp-1 mutants with reduced MT stability and in fra2 with increased MT stability. Thus, contrary to animal models, where the formation of SGs is proportional to the availability of the MT lattice that presumably provides a surface for specific stages of SG assembly, in plants the depolymerization of MTs plays an equally important role. This discrepancy fits well with the model in which discrete stages of the multiphasic pathway of SG assembly require polymerizing and depolymerizing MTs (Anderson and Kedersha, 2008). The early stages of SG assembly take place on the MT surface and, therefore, require the MT lattice, while SG maturation requires their release to the cytoplasm through MT depolymerization. Consistent with the recent report by Steffens et al. (2014), the assembly of PBs was found to be independent of MTs.
Molecular Function of TSN in SGs and PBs
The mechanistic role of plant TSN remains elusive, but considering the high degree of conservation of TSN structure in evolutionarily distant lineages, Arabidopsis TSN could be essential for the similar mRNA metabolic processes in SGs and PBs, as reported in animals. For example, human TSN was shown to catalyze the cleavage of hyperedited double-stranded RNA (Scadden, 2005) and modulate microRNA processing and the expression by ADAR (adenosine deaminase acting on RNA) (Zhao et al., 2003). Interestingly, human TSN colocalizes and interacts with ADAR1 and 3′-untranslated region of angiotensin II receptor, type 1 mRNA in SGs after arsenite-induced oxidative stress (Weissbach and Scadden, 2012; Gao et al., 2014).
Here, we show that the nucleolytic activity of TSN is indispensable for its function in stress tolerance. The N-terminal tandem repeat of four SN domains was sufficient for the targeting of TSN to SGs and for protecting root cells from death during the stress response, while the C-terminal Tudor domain-containing region was dispensable for these processes. The N-terminal SN domains of mammalian and silkworm TSN interact with several SG-associated proteins, suggesting that, in addition to the nucleolytic activity, this part of TSN serves as a protein-protein interface required for the assembly and functions of SGs and PBs (Gao et al., 2010; Zhu et al., 2013).
The main function of PBs is mRNA decay, which requires the activity of the decapping complex (Coller and Parker, 2004; Ling et al., 2011). The eIF4E protein localizes in both PBs and SGs, where it binds to the mRNA 5′-cap, making mRNA resistant to decapping and subsequent degradation (Parker and Sheth, 2007). For example, binding of the yeast eIF4E homolog to the mRNA 5′-cap inhibits DCP1-dependent decapping activity (Vilela et al., 2000). More recently, it was suggested that smaller DCP1-positive PBs in the dcp5-1 knockdown in Arabidopsis might be a consequence of the decreased capacity to retain mRNA (Xu and Chua, 2009). DCP5 of Arabidopsis was required for decapping and the repression of translation in vivo (Xu and Chua, 2009; Maldonado-Bonilla, 2014). Thus, smaller GFP-DCP1 foci and the increased number of eIF4E-positive PBs in tsn1 tsn2 plants suggest that mRNA decapping is impaired.
It is well known that endoribonucleases are actively involved in mRNA decay pathways in eukaryotes (Arraiano et al., 2013; Zhang et al., 2013). Bombyx mori TSN has recently been shown to interact with DCP2, suggesting a role for TSN in mRNA decay (Zhu et al., 2013). Remarkably, we found that heat stress-induced accumulation of uncapped mRNAs is abrogated in tsn1 tsn2 plants. Therefore, we conclude that TSN is a positive regulator of mRNA decapping in PBs under stress. This novel function complements the previously described role of Arabidopsis TSN in stabilizing a subset of mRNAs required for stress tolerance (Frei dit Frey et al., 2010).
Posttranscriptional reprogramming of mRNA catabolism in SGs and PBs tailors the proteome for adaptation to adverse environmental conditions (Anderson and Kedersha, 2008). Here, we report that plant TSN belongs to poorly characterized protein complexes located in SGs and PBs and functions in mRNA catabolism during stress tolerance. In addition, we found that recruitment of TSN to SGs and PBs plays an important scaffolding role in both structures. The next critical step toward understanding the mechanistic role of TSN in plants would be the detailed analysis of its interactome.
METHODS
Plant Material and Growth Conditions
The tsn1 tsn2 mutant of Arabidopsis thaliana was isolated by crossing tsn1 (Ler; line CSHL_ET12646) and tsn2 (Col; line SALK_143497) homozygous T-DNA insertion lines (Sundström et al., 2009), and the double homozygous line was identified after genotyping 90 F2 seedlings by PCR. Col and Ler were crossed to generate the Col/Ler wild-type line. Seedlings were grown on vertical plates containing half-strength Murashige and Skoog (MS) medium supplemented with 1% (w/v) sucrose and 0.7% (w/v) plant agar, at 23°C (control) or 39°C (heat stress), under a 16/8-h light/dark cycle and a light intensity of 150 μE m−2 s−1. For visualization of SGs, Petri dishes with 5-d-old transgenic seedlings expressing GFP or red fluorescent protein (RFP) fusion proteins were incubated for 40 min on a thermoblock at 39°C. For long-term heat stress, 5-d-old seedlings grown on MS plates were transferred to 35°C under a 16/8-h light/dark cycle and a light intensity of 150 μE m−2 s−1 for 24 h. For short-term heat stress, plates with 5-d-old seedlings were incubated for 4 h on a thermoblock at 39°C. The following transgenic fluorescent protein lines in the Col background were used: ProTSN1:GFP-TSN1, ProTSN2:GFP-TSN2, ProTSN1:TSN1-GFP, ProTSN2:TSN2-GFP, and Pro35S:RFP-Rbp47b. The Pro35S:GFP-DCP1 and Pro35S:GFP-Rbp47b lines were in tsn1 tsn2, Ler, Col, and Col/Ler backgrounds. For complementation experiments, the following transgenic fluorescent protein lines in the tsn1 tsn2 background were used: ProTSN1:TSN1-GFP, ProTSN2:TSN2-GFP, Pro35S:SN1-GFP, Pro35S:SN2-GFP, Pro35S:Tudor1-GFP, and Pro35S:Tudor2-GFP. All constructs used in this study are shown in Supplemental Table 1. The following mutant alleles were also used: clasp, fra2, and mor1-1 (Burk et al., 2001; Whittington et al., 2001; Ambrose et al., 2007).
Molecular Biology
All oligonucleotide primers used in this study are shown in Supplemental Table 2. TSN1 (egb1/egb2), TSN2 (egb3/egb4), proTSN1 (egb12/egb13), proTSN2 (egb14/egb15), Rbp47b (egb9/egb10), Ubp1 (egb11/egb12), DCP1 (egb13/egb14), and DCP2 (egb15/egb16) were amplified by PCR from a cDNA derived from 2-week-old leaves of Arabidopsis Col plants. All cDNA sequences were introduced in pDONR/Zeo using Gateway technology (Invitrogen). For the expression of N-terminal GFP and RFP fusions under the control of the 35S promoter, TSN1, TSN2, Rbp47b, Ubp1, DCP1, and DCP2 cDNAs were introduced into the destination vectors pMDC43 and pGWB655 (Curtis and Grossniklaus, 2003; Nakagawa et al., 2007). For proTSN1 and proTSN2 C-terminal GFP fusions, 2-kb promoter regions from both genes were fused to the cDNA of TSN1 or TSN2 using overlay PCR (egb17-20 and egb23-26). N-terminal GFP fusions were assembled by two consecutive rounds of overlay PCR, first between the promoter and GFP and then between promoter-GFP and the corresponding TSN (egb21-22 and egb27-33), and introduced into pDONR/Zeo. GFP was amplified from the vector pGWB6. Fusions were introduced into the destination vectors pGWB1 and pGWB5 for ProTSNs:GFP-TNSs and ProTSNs:TSNs-GFP, respectively (Nakagawa et al., 2007). N-terminally located SN domains (named SN; egb1/egb5 for SN1 and egb3/egb7 for SN2) and Tudor with the last SN domain (named Tudor; egb2/egb6 for Tudor1 and egb4/egb8 for Tudor2) were amplified from TSN full-length cDNA and then introduced into the pDONR/Zeo vector. Finally, cDNAs were introduced into the destination vector pMDC43. All plasmids and derived constructs were verified by sequencing using the M13 forward and reverse primers.
Plant and Protoplast Transformation
Arabidopsis plants were transformed as described previously (Clough and Bent, 1998) using Agrobacterium tumefaciens GV3101. In all experiments, plants from the T2 and T3 generations were used. For transient expression in Nicotiana benthamiana mesophyll cells, Agrobacterium strain GV3101 cells were transformed with the appropriate binary vectors by electroporation as described previously (de la Torre et al., 2013). Agrobacterium-positive clones were grown in Luria-Bertani medium until OD600 = 0.4 and were pelleted after centrifugation at 3000g for 10 min. Cells were resuspended in MM (10 mM MES, pH 5.7, 10 mM MgCl2, supplemented with 0.2 mM acetosyringone) until OD600 = 0.4, incubated at room temperature for 2 h, and infiltrated into N. benthamiana leaves, using a 1-mL hypodermic syringe. Leaves were analyzed after 48 h using a Zeiss 780 confocal microscope with a 40× objective. The excitation/emission wavelengths were 480/508 nm for GFP and 561/610 nm for RFP.
Protoplasts were isolated from leaves of 15- to 20-d-old N. benthamiana, transiently expressing the corresponding fluorescent proteins, as described previously (Wu et al., 2009). The cell walls were digested in enzymatic solution containing 1% (w/v) Cellulose R-10, 0.25% (w/v) Macerozyme R-10, 20 mM MES-HOK, pH 5.7, 400 mM mannitol, 10 mM CaCl2, 20 mM KCl, and 0.1% (w/v) BSA for 60 min. Protoplasts were separated from debris by centrifugation (100g, 3 min, 4°C), washed two times with ice-cold W5 buffer (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES-KOH, pH 5.7), and resuspended in ice-cold W5 buffer at a density of 2.5 × 105 protoplasts/mL. The protoplast suspension was incubated for 15 min on ice before heat stress exposure (30 min at 39°C). Finally, the protoplasts were analyzed using a Zeiss 780 confocal microscope with a 40× water-immersion objective.
Preparation of Antibodies
Full-length TSN2 was introduced into the pDEST15 vector (Invitrogen). The pDEST15-TSN2 construct was transformed into Escherichia coli BL21 (DE3) RIL (Stratagene) cells. Induction of the expression of the recombinant protein was performed as described previously (San-Miguel et al., 2013). Briefly, protein expression was induced at OD600 = 0.5 by adding 1 mM isopropyl β-d-1-thiogalactopyranoside to Luria-Bertani medium supplemented with 100 μg/mL ampicillin and 2 g/L glucose. The cells were harvested by centrifugation at 3000g for 10 min at room temperature and frozen overnight at −80°C. Preparation of GST-tagged recombinant protein from pDEST15-TSN2 was performed according to the manufacturer’s instructions (GE Healthcare). The antiserum was raised in rabbits using recombinant protein as the antigen.
Nuclease Activity Assay
The nuclease activity assay was performed as described previously (Sundström et al., 2009). For the quantification of TSN RNase activity, recombinant full-length TSN and SN and Tudor fragments (2.5 pmol) were incubated with fluorescently labeled single-stranded RNA substrate (1 pmol) from the fluorometric RNase detection assay (Ambion). The reaction volume was adjusted to 45 μL with reaction buffer containing 50 mM HEPES, pH 7.0, 100 mM NaCl, 1 mM DTT, 5 mM CaCl2, and 10% glycerol. The reactions were incubated for 3 h at 28°C. The fluorescence was measured on a VersaFluor fluorometer (Bio-Rad) using excitation/emission wavelengths of 490/520 nm and was directly proportional to the RNase activity.
Immunocytochemistry and Imaging
For immunocytochemistry, 5-d-old Arabidopsis plants were heat-stressed for 40 min on a thermoblock at 39°C. Subsequently, roots were cut and fixed for 60 min at room temperature with 4% (w/v) paraformaldehyde in 50 mM PIPES, pH 6.8, 5 mM EGTA, 2 mM MgCl2, and 0.4% Triton X-100. The fixative was washed away with PBS buffer supplemented with Tween 20 (PBST), and cells were treated for 8 min at room temperature with a solution of 2% (w/v) Driselase (Sigma-Aldrich) in 0.4 M mannitol, 5 mM EGTA, 15 mM MES, pH 5.0, 1 mM phenylmethylsulfonyl fluoride, 10 μg/mL leupeptin, and 10 μg/mL pepstatin A. Thereafter, roots were washed twice for 10 min each in PBST and then in 1% (w/v) BSA in PBST for 30 min before overnight incubation with a primary antibody (rabbit anti-TSN diluted 1:500 or rabbit anti-eIF4E diluted 1:500). The specimens were then washed three times for 90 min in PBST and incubated overnight with goat anti-rabbit rhodamine (Bio-Rad)-conjugated secondary antibody diluted 1:200. After washing in PBST, the specimens were mounted in Vectashield mounting medium (Vector Laboratories).
Staining with FDA and SYTOX Orange (both from Molecular Probes, Invitrogen) was performed on 5-d-old Arabidopsis seedlings. FDA and SYTOX Orange were added to final concentrations of 250 nM and 2 μg/mL, respectively, in water. After 10 min of incubation in the dark, the samples were washed twice with half-strength liquid MS medium (LMS) supplemented with 1% (w/v) sucrose, pH 5.7, and observed immediately. For plasma membrane staining, 5-d-old seedlings were incubated in LMS supplemented with 2 μM FM4-64 (Molecular Probes; made from a 2 mM stock in DMSO) for 15 min. Finally, the roots were washed two times in LMS and observed immediately. The samples were examined using a Zeiss 780 confocal microscope.
Drug Treatments
Taxol (Enzo) was dissolved in DMSO to make a 50 mM stock solution and used at a final concentration of 20 μM. APM (Sigma-Aldrich) was dissolved in DMSO, stored as aliquots of 100 mM stock solutions, and used at a final concentration of 10 μM. CHX (Sigma-Aldrich) was added from a 50 mM aqueous stock to a final concentration of 35 μM. The TSN nuclease activity inhibitor pdTP and its inactive analog dTP were dissolved in water as 100 mM stock solutions and used at a final concentration of 50 μM. Five-day-old seedlings were incubated in LMS containing the corresponding drugs for 5 h and subsequently heat-stressed for 40 min at 39°C. For the CHX assay in protoplasts, the drug was added to the protoplast suspension, incubated for 15 min, and then heat-stressed for 30 min at 39°C. For the SG disassembly assay with CHX, 5-d-old seedlings incubated in LMS were heat-stressed for 40 min at 39°C and subsequently treated with CHX.
Immunoblotting
One hundred milligrams of leaf material was mixed with 300 μL of extraction buffer (100 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 10% glycerol, 0.1% Triton X-100, and 1× Complete protease inhibitor [Roche]) and centrifuged for 15 min at 14,000g. Laemmli sample was added to 100 μL of supernatant and boiled for 5 min. Equal amounts of supernatant were loaded on 9% polyacrylamide gels and blotted on a polyvinylidene difluoride membrane. Anti-TSN and anti-rabbit horseradish peroxidase conjugate (Amersham, GE Healthcare) were used at dilutions of 1:1000 and 1:5000, respectively. The reaction was developed for 1 min using the ECL Prime kit (Amersham, GE Healthcare) and detected in an LAS-3000 luminescent image analyzer (Fujifilm, Fuji Photo Film).
FRAP
The assay was performed as described previously (Moschou et al., 2013). Five-day-old seedlings were grown on sterile CellView cell culture plates with glass bottoms (35 mm; Greiner Bio One) containing 1 mL of half-strength MS, 1% (w/v) sucrose, and 0.3% (w/v) electrophoresis-grade agarose. For heat stress treatment, plates were incubated for 30 min on a thermoblock at 39°C. For inhibitor treatments, plants were placed in Eppendorf tubes containing the corresponding inhibitor dissolved in LMS medium. GFP or RFP fluorescence was detected using a water-corrected 40× objective. During analyses, the FRAP mode of Zeiss 780 ZEN software was set up for the acquisition of one prebleach image, one bleach scan, and 40 postbleach scans. In FRAP of SGs and PBs, the width of the bleached region was 2 μm. The following settings were used for photobleaching: 10 to 20 iterations for GFP; 10 to 60 s per frame; and 75% transmittance with the 458- to 561-nm laser lines of the argon laser. Prebleach and postbleach scans were at the minimum possible laser power (1.4 to 20% transmittance) for 488 or 561 nm and at 0% for all other laser lines, 512 × 512 pixel format, and zoom factor of 5.1. Analyses of fluorescence intensities during FRAP were performed in regions of interest corresponding to the size of the bleached region. One region of interest was measured outside the bleached region to serve as the background. The background values were subtracted from the fluorescence recovery values, and the resulting values were normalized by the first postbleach time point. Initial signal recovery (%) = 100 × (Ifinal,postbleach − Iinitial,postbleach)/(Iprebleach − Iinitial,postbleach), where I is the normalized signal intensity (relative to the background intensity). Values were corrected for the artificial loss of fluorescence using values from the neighboring cells. At least eight cells from different roots were analyzed for each FRAP experiment.
Microarray Analysis
For the microarray studies, Col, Ler, and tsn1 tsn2 plants were grown vertically on MS agar plates under a 16/8-h light/dark cycle and a light intensity of 150 μE m−2 s−1 for 5 d. For heat stress, plates with 5-d-old seedlings were incubated for 40 min on a thermoblock at 39°C. Root tissue was collected and used to purify uncapped and total mRNA. Two independent sets of biological samples were used for the experiments. Uncapped mRNA purification was performed as described previously, using T4 RNA ligase (Ambion) to add an RNA adaptor to the uncapped mRNA (Jiao et al., 2008; Jiao and Riechmann, 2012). Uncapped mRNA was reverse transcribed, and then second-strand cDNA (double-stranded DNA [dsDNA]-uncapped) was synthesized using a specific primer as described previously (Jiao and Riechmann, 2012). The dsDNA samples along with total mRNA samples were sent to OakLabs, where total mRNA samples were reverse transcribed and the second-strand cDNA (dsDNA-total) was synthesized using Agilent’s LIQA kit. Both dsDNA-uncapped and dsDNA-total were used to synthetize copy RNA (cRNA) by in vitro transcription. Subsequently, the cRNA samples were labeled using Cy-3 dye and hybridized on Arabidopsis ArrayXS Thaliana chips (http://www.oak-labs.com/). The samples were normalized using the ranked median quantiles as described previously (Bolstad et al., 2003). Briefly, the mean signal of each target was ranked relative to all other targets. The ranked signal value was replaced with the median value of the sample rank. So the highest value in all samples became the mean of the highest values, the second highest value became the mean of the second highest values, and so on. Before microarray hybridization, the size distribution of cRNA samples from positive and control experiments, with and without T4 RNA ligase, respectively, was evaluated using an Agilent 2100 bioanalyzer (Jiao and Riechmann, 2012). A method described previously was used to define when a probe was to be considered as a positive signal (Ma et al., 2005). Statistical analyses including a Bonferroni adjusted t test (P < 0.05) between cases and identification of false discovery rate (threshold of 5%) were performed in the TIGR MultiExperiment Viewer module of TM4 software (Saeed et al., 2003). To identify differentially expressed genes, P values and log2 fold changes were calculated for groups of biological replicates. The P value cutoff was set at 0.05, and the log2 fold cutoff was set at 2-fold. GO annotations for all gene models were downloaded from TAIR (Rhee et al., 2003). Functional annotation of GO was performed using GOslim terms (Berardini et al., 2004). Identification of transposon-related genes and pseudogenes was performed according to TAIR annotation version 7. The microarray data presented in this article are available in the Gene Expression Omnibus under the accession number GSE63522.
Image and Statistical Analyses
The image analysis was done using ImageJ version 1.41 software (http://rsb.info.nih.gov/ij/index.html). SGs and PBs were scored as positive when they had a minimal size of 0.5 μm. SG and PB counting was performed manually with the Cell Counter plugin of ImageJ (http://rsbweb.nih.gov/ij/plugins/cell-counter.html). The size of foci was measured manually with ImageJ. The recovery rate was calculated by the single exponential fit as described previously (Chang et al., 2005). For colocalization analyses, we calculated the linear Pearson (rp) and nonlinear Spearman's rank (rs) correlation coefficient (PSC) for the pixels representing the fluorescence signals in both channels (French et al., 2008). Levels of colocalization can range from +1 for positive correlation to −1 for negative correlations. Root length was measured by Adobe Illustrator version CS5 on digital photographs. Statistical analysis was performed with JMP version 9 (www.jmp.com).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TSN1 (At5g07350), TSN2 (At5g61780), Rbp47b (At3g19130), DCP1 (At1g08370), DCP2 (At5g13570), Ubp1b (At1g17370), and eIF4E (At4g18040).
Supplemental Data
Supplemental Figure 1. TSN1 Localizes to SG and PB.
Supplemental Figure 2. Colocalization Analysis of GFP-TSN2 with Endosome, Golgi, Late Endosome, and Membrane Markers.
Supplemental Figure 3. Assembly and Disassembly of TSN-Containing Foci Is CHX Dependent.
Supplemental Figure 4. Colocalization Analysis of TSN2 with RFP-Rbp47b and GFP-DCP1 in Heat-Stressed Root Tip Cells after APM Treatment.
Supplemental Figure 5. Characterization of tsn1 tsn2 Double Knockout Arabidopsis Line.
Supplemental Figure 6. Kinetics of Assembly and Disassembly of GFP-Rbp47b Foci in WT (Col/Ler) and tsn1 tsn2 Root Tip Cells.
Supplemental Figure 7. Protein Expression Levels in tsn1 tsn2 Double Knockout Complemented Lines.
Supplemental Figure 8. Bioanalyzer Electropherograms of Amplified cRNA Samples.
Supplemental Figure 9. Distribution Profiles of mRNA Decapping Levels in Heat-Stressed Col Plants.
Supplemental Figure 10. Expression Profile of Uncapped mRNAs in Col, Ler and tsn1 tsn2 Plants under Heat Stress Versus Control Conditions.
Supplemental Table 1. Gene Constructs Used in This Study.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. Table of Contents.
Supplemental Data Set 2. Transcripts Differentially Represented in tsn1 tsn2 Plants under Heat Stress Versus Control Conditions.
Supplementary Material
Acknowledgments
We thank Karen Browning (University of Texas at Austin) for providing eIF4E antibody, Tsuyoshi Nakagawa (Shimane University) for providing Gateway binary vectors that contain the bar gene, which was identified by Meiji Seika Kaisha, Ltd., and both Anders Hafrén and Lars Hennig (Swedish University of Agricultural Sciences) for valuable comments on the manuscript. This work was supported by grants from the Knut and Alice Wallenberg Foundation (to P.V.B.), the Swedish Research Council (to P.N.M. and P.V.B.), the Pehrssons Fund (to P.V.B.), the Swedish Foundation for Strategic Research (to P.V.B.), the Olle Engkvist Foundation (to P.V.B.), and the August T. Larsson Foundation (to A.P.S. and P.V.B.).
AUTHOR CONTRIBUTIONS
E.G.-B., P.N.M., and P.V.B. designed research. E.G.-B. performed research. E.G.-B., P.N.M., A.P.S., and P.V.B. analyzed data. E.G.-B., P.N.M., A.P.S., and P.V.B. wrote the article.
Glossary
- SG
stress granule
- PB
processing body
- MT
microtubule
- CHX
cycloheximide
- APM
amiprophos-methyl
- FRAP
fluorescence recovery after photobleaching
- Ler
Landsberg erecta
- Col
Columbia-0
- pdTp
3′,5′-deoxythymidine bisphosphate
- dTp
3′-deoxythymidine monophosphate
- FDA
fluorescein diacetate
- mRNP
messenger ribonucleoprotein
- GO
Gene Ontology
- MS
Murashige and Skoog
- PBST
PBS buffer supplemented with Tween 20
- dsDNA
double-stranded DNA
- cRNA
copy RNA
- LMS
liquid MS medium
References
- Abe S., Sakai M., Yagi K., Hagino T., Ochi K., Shibata K., Davies E. (2003). A Tudor protein with multiple SNc domains from pea seedlings: Cellular localization, partial characterization, sequence analysis, and phylogenetic relationships. J. Exp. Bot. 54: 971–983. [DOI] [PubMed] [Google Scholar]
- Ambrose J.C., Shoji T., Kotzer A.M., Pighin J.A., Wasteneys G.O. (2007). The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19: 2763–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. (2006). RNA granules. J. Cell Biol. 172: 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. (2008). Stress granules: The Tao of RNA triage. Trends Biochem. Sci. 33: 141–150. [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. (2009). RNA granules: Post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10: 430–436. [DOI] [PubMed] [Google Scholar]
- Arraiano C.M., Mauxion F., Viegas S.C., Matos R.G., Séraphin B. (2013). Intracellular ribonucleases involved in transcript processing and decay: Precision tools for RNA. Biochim. Biophys. Acta 1829: 491–513. [DOI] [PubMed] [Google Scholar]
- Belostotsky D.A., Sieburth L.E. (2009). Kill the messenger: mRNA decay and plant development. Curr. Opin. Plant Biol. 12: 96–102. [DOI] [PubMed] [Google Scholar]
- Berardini T.Z., et al. (2004). Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 135: 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogamuwa S., Jang J.C. (2013). The Arabidopsis tandem CCCH zinc finger proteins AtTZF4, 5 and 6 are involved in light-, abscisic acid- and gibberellic acid-mediated regulation of seed germination. Plant Cell Environ. 36: 1507–1519. [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193. [DOI] [PubMed] [Google Scholar]
- Broadhurst M.K., Wheeler T.T. (2001). The p100 coactivator is present in the nuclei of mammary epithelial cells and its abundance is increased in response to prolactin in culture and in mammary tissue during lactation. J. Endocrinol. 171: 329–337. [DOI] [PubMed] [Google Scholar]
- Buchan J.R., Parker R. (2009). Eukaryotic stress granules: The ins and outs of translation. Mol. Cell 36: 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R., Capaldi A.P., Parker R. (2012). TOR-tured yeast find a new way to stand the heat. Mol. Cell 47: 155–157. [DOI] [PubMed] [Google Scholar]
- Buchan J.R., Yoon J.H., Parker R. (2011). Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J. Cell Sci. 124: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk D.H., Liu B., Zhong R., Morrison W.H., Ye Z.H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13: 807–827. [PMC free article] [PubMed] [Google Scholar]
- Caudy A.A., Ketting R.F., Hammond S.M., Denli A.M., Bathoorn A.M., Tops B.B., Silva J.M., Myers M.M., Hannon G.J., Plasterk R.H. (2003). A micrococcal nuclease homologue in RNAi effector complexes. Nature 425: 411–414. [DOI] [PubMed] [Google Scholar]
- Chalupníková K., Lattmann S., Selak N., Iwamoto F., Fujiki Y., Nagamine Y. (2008). Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J. Biol. Chem. 283: 35186–35198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.Y., Smertenko A.P., Igarashi H., Dixon D.P., Hussey P.J. (2005). Dynamic interaction of NtMAP65-1a with microtubules in vivo. J. Cell Sci. 118: 3195–3201. [DOI] [PubMed] [Google Scholar]
- Chernov K.G., Barbet A., Hamon L., Ovchinnikov L.P., Curmi P.A., Pastré D. (2009). Role of microtubules in stress granule assembly: Microtubule dynamical instability favors the formation of micrometric stress granules in cells. J. Biol. Chem. 284: 36569–36580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong S.D., Good A.G., Taylor G.J., Freeman M.C., Moorhead G.B., Muench D.G. (2004). Large-scale identification of tubulin-binding proteins provides insight on subcellular trafficking, metabolic channeling, and signaling in plant cells. Mol. Cell. Proteomics 3: 970–983. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Coller J., Parker R. (2004). Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73: 861–890. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre F., Gutiérrez-Beltrán E., Pareja-Jaime Y., Chakravarthy S., Martin G.B., del Pozo O. (2013). The tomato calcium sensor Cbl10 and its interacting protein kinase Cipk6 define a signaling pathway in plant immunity. Plant Cell 25: 2748–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger-Mathason T.S., Andrade J., Groehler A.L., Clark D.E., Muratore-Schroeder T.L., Pasic L., Smith J.A., Shabanowitz J., Hunt D.F., Macara I.G., Lannigan D.A. (2008). Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol. Cell 31: 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei dit Frey N., Muller P., Jammes F., Kizis D., Leung J., Perrot-Rechenmann C., Bianchi M.W. (2010). The RNA binding protein Tudor-SN is essential for stress tolerance and stabilizes levels of stress-responsive mRNAs encoding secreted proteins in Arabidopsis. Plant Cell 22: 1575–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T.M., Lykke-Andersen J. (2008). The control of mRNA decapping and P-body formation. Mol. Cell 32: 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French A.P., Mills S., Swarup R., Bennett M.J., Pridmore T.P. (2008). Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat. Protoc. 3: 619–628. [DOI] [PubMed] [Google Scholar]
- Gao X., Ge L., Shao J., Su C., Zhao H., Saarikettu J., Yao X., Yao Z., Silvennoinen O., Yang J. (2010). Tudor-SN interacts with and co-localizes with G3BP in stress granules under stress conditions. FEBS Lett. 584: 3525–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Shi X., Fu X., Ge L., Zhang Y., Su C., Yang X., Silvennoinen O., Yao Z., He J., Wei M., Yang J. (2014). Human Tudor staphylococcal nuclease (Tudor-SN) protein modulates the kinetics of AGTR1-3’UTR granule formation. FEBS Lett. 588: 2154–2161. [DOI] [PubMed] [Google Scholar]
- Gao X., Zhao X., Zhu Y., He J., Shao J., Su C., Zhang Y., Zhang W., Saarikettu J., Silvennoinen O., Yao Z., Yang J. (2012). Tudor staphylococcal nuclease (Tudor-SN) participates in small ribonucleoprotein (snRNP) assembly via interacting with symmetrically dimethylated Sm proteins. J. Biol. Chem. 287: 18130–18141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Denervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.D., Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., Anderson P. (2004). Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15: 5383–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeres D.C., Van Norman J.M., Zhang W., Fauver N.A., Spencer M.L., Sieburth L.E. (2007). Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell 19: 1549–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T., Tominaga M., Fukaya T., Nakamura M., Nakano A., Watanabe Y., Hashimoto T., Baskin T.I. (2012). RNA processing bodies, peroxisomes, Golgi bodies, mitochondria, and endoplasmic reticulum tubule junctions frequently pause at cortical microtubules. Plant Cell Physiol. 53: 699–708. [DOI] [PubMed] [Google Scholar]
- Hossain M.J., Korde R., Singh S., Mohmmed A., Dasaradhi P.V., Chauhan V.S., Malhotra P. (2008). Tudor domain proteins in protozoan parasites and characterization of Plasmodium falciparum tudor staphylococcal nuclease. Int. J. Parasitol. 38: 513–526. [DOI] [PubMed] [Google Scholar]
- Howard-Till R.A., Yao M.C. (2007). Tudor nuclease genes and programmed DNA rearrangements in Tetrahymena thermophila. Eukaryot. Cell 6: 1795–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P.A., Chudinova E.M., Nadezhdina E.S. (2003). Disruption of microtubules inhibits cytoplasmic ribonucleoprotein stress granule formation. Exp. Cell Res. 290: 227–233. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Riechmann J.L. (2012). Genome-wide profiling of uncapped mRNA. Methods Mol. Biol. 876: 207–216. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Riechmann J.L., Meyerowitz E.M. (2008). Transcriptome-wide analysis of uncapped mRNAs in Arabidopsis reveals regulation of mRNA degradation. Plant Cell 20: 2571–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Anderson P. (2007). Mammalian stress granules and processing bodies. Methods Enzymol. 431: 61–81. [DOI] [PubMed] [Google Scholar]
- Kedersha N., Cho M.R., Li W., Yacono P.W., Chen S., Gilks N., Golan D.E., Anderson P. (2000). Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151: 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N.L., Gupta M., Li W., Miller I., Anderson P. (1999). RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147: 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S., Zhang Y., Matthias P. (2007). The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 21: 3381–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavut A., Raveh D. (2012). Sequestration of highly expressed mRNAs in cytoplasmic granules, P-bodies, and stress granules enhances cell viability. PLoS Genet. 8: e1002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverson J.D., Koskinen P.J., Orrico F.C., Rainio E.M., Jalkanen K.J., Dash A.B., Eisenman R.N., Ness S.A. (1998). Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol. Cell 2: 417–425. [DOI] [PubMed] [Google Scholar]
- Li C.L., Yang W.Z., Chen Y.P., Yuan H.S. (2008). Structural and functional insights into human Tudor-SN, a key component linking RNA interference and editing. Nucleic Acids Res. 36: 3579–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S.H., Qamra R., Song H. (2011). Structural and functional insights into eukaryotic mRNA decapping. Wiley Interdiscip. Rev. RNA 2: 193–208. [DOI] [PubMed] [Google Scholar]
- Liu S., Jia J., Gao Y., Zhang B., Han Y. (2010). The AtTudor2, a protein with SN-Tudor domains, is involved in control of seed germination in Arabidopsis. Planta 232: 197–207. [DOI] [PubMed] [Google Scholar]
- Lorković Z.J., Wieczorek Kirk D.A., Klahre U., Hemmings-Mieszczak M., Filipowicz W. (2000). RBP45 and RBP47, two oligouridylate-specific hnRNP-like proteins interacting with poly(A)+ RNA in nuclei of plant cells. RNA 6: 1610–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschi M., Leishman C.C., Berardone N., Boccaccio G.L. (2009). Dynein and kinesin regulate stress-granule and P-body dynamics. J. Cell Sci. 122: 3973–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Sun N., Liu X., Jiao Y., Zhao H., Deng X.W. (2005). Organ-specific expression of Arabidopsis genome during development. Plant Physiol. 138: 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Bonilla L.D. (2014). Composition and function of P bodies in Arabidopsis thaliana. Front. Plant Sci. 5: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet S., Cougot N., Wilczynska A., Dautry F., Kress M., Bertrand E., Weil D. (2008). Translationally repressed mRNA transiently cycles through stress granules during stress. Mol. Biol. Cell 19: 4469–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschou P.N., Smertenko A.P., Minina E.A., Fukada K., Savenkov E.I., Robert S., Hussey P.J., Bozhkov P.V. (2013). The caspase-related protease separase (extra spindle poles) regulates cell polarity and cytokinesis in Arabidopsis. Plant Cell 25: 2171–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muench D.G., Zhang C., Dahodwala M. (2012). Control of cytoplasmic translation in plants. Wiley Interdiscip. Rev. RNA 3: 178–194. [DOI] [PubMed] [Google Scholar]
- Nadezhdina E.S., Lomakin A.J., Shpilman A.A., Chudinova E.M., Ivanov P.A. (2010). Microtubules govern stress granule mobility and dynamics. Biochim. Biophys. Acta 1803: 361–371. [DOI] [PubMed] [Google Scholar]
- Nagarajan V.K., Jones C.I., Newbury S.F., Green P.J. (2013). XRN 5′→3′ exoribonucleases: Structure, mechanisms and functions. Biochim. Biophys. Acta 1829: 590–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Ohn T., Anderson P. (2010). The role of posttranslational modifications in the assembly of stress granules. Wiley Interdiscip. Rev. RNA 1: 486–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Sheth U. (2007). P bodies and the control of mRNA translation and degradation. Mol. Cell 25: 635–646. [DOI] [PubMed] [Google Scholar]
- Paukku K., Kalkkinen N., Silvennoinen O., Kontula K.K., Lehtonen J.Y. (2008). p100 increases AT1R expression through interaction with AT1R 3′-UTR. Nucleic Acids Res. 36: 4474–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paukku K., Yang J., Silvennoinen O. (2003). Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcription 5. Mol. Endocrinol. 17: 1805–1814. [DOI] [PubMed] [Google Scholar]
- Pomeranz M.C., Hah C., Lin P.C., Kang S.G., Finer J.J., Blackshear P.J., Jang J.C. (2010). The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol. 152: 151–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S.Y., et al. (2003). The Arabidopsis Information Resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 31: 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A.I., et al. (2003). TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378. [DOI] [PubMed] [Google Scholar]
- Sami-Subbu R., Choi S.B., Wu Y., Wang C., Okita T.W. (2001). Identification of a cytoskeleton-associated 120 kDa RNA-binding protein in developing rice seeds. Plant Mol. Biol. 46: 79–88. [DOI] [PubMed] [Google Scholar]
- San-Miguel T., Pérez-Bermúdez P., Gavidia I. (2013). Production of soluble eukaryotic recombinant proteins in E. coli is favoured in early log-phase cultures induced at low temperature. Springerplus 2: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden A.D. (2005). The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 12: 489–496. [DOI] [PubMed] [Google Scholar]
- Sorenson R., Bailey-Serres J. (2014). Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 2373–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens A., Jaegle B., Tresch A., Hülskamp M., Jakoby M. (2014). Processing-body movement in Arabidopsis depends on an interaction between myosins and DECAPPING PROTEIN1. Plant Physiol. 164: 1879–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström J.F., et al. (2009). Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat. Cell Biol. 11: 1347–1354. [DOI] [PubMed] [Google Scholar]
- Sweet T.J., Boyer B., Hu W., Baker K.E., Coller J. (2007). Microtubule disruption stimulates P-body formation. RNA 13: 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M.A., Brengues M., Parker R. (2005). Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11: 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M.G., Loschi M., Desbats M.A., Boccaccio G.L. (2011). RNA granules: The good, the bad and the ugly. Cell. Signal. 23: 324–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R., Dziembowski A. (2010). Novel endoribonucleases as central players in various pathways of eukaryotic RNA metabolism. RNA 16: 1692–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X., Drapkin R., Yalamanchili R., Mosialos G., Kieff E. (1995). The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol. Cell. Biol. 15: 4735–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., Tazi J. (2003). The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160: 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Truernit E., Haseloff J. (2008). A simple way to identify non-viable cells within living plant tissue using confocal microscopy. Plant Methods 4: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniacke J., Zerges W. (2008). Stress induces the assembly of RNA granules in the chloroplast of Chlamydomonas reinhardtii. J. Cell Biol. 182: 641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Välineva T., Yang J., Silvennoinen O. (2006). Characterization of RNA helicase A as component of STAT6-dependent enhanceosome. Nucleic Acids Res. 34: 3938–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderweyde T., Youmans K., Liu-Yesucevitz L., Wolozin B. (2013). Role of stress granules and RNA-binding proteins in neurodegeneration: a mini-review. Gerontology 59: 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela C., Velasco C., Ptushkina M., McCarthy J.E. (2000). The eukaryotic mRNA decapping protein Dcp1 interacts physically and functionally with the eIF4F translation initiation complex. EMBO J. 19: 4372–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Washida H., Crofts A.J., Hamada S., Katsube-Tanaka T., Kim D., Choi S.B., Modi M., Singh S., Okita T.W. (2008). The cytoplasmic-localized, cytoskeletal-associated RNA binding protein OsTudor-SN: Evidence for an essential role in storage protein RNA transport and localization. Plant J. 55: 443–454. [DOI] [PubMed] [Google Scholar]
- Weber C., Nover L., Fauth M. (2008). Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 56: 517–530. [DOI] [PubMed] [Google Scholar]
- Weissbach R., Scadden A.D. (2012). Tudor-SN and ADAR1 are components of cytoplasmic stress granules. RNA 18: 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington A.T., Vugrek O., Wei K.J., Hasenbein N.G., Sugimoto K., Rashbrooke M.C., Wasteneys G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411: 610–613. [DOI] [PubMed] [Google Scholar]
- Wilczynska A., Aigueperse C., Kress M., Dautry F., Weil D. (2005). The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 118: 981–992. [DOI] [PubMed] [Google Scholar]
- Wolozin B. (2012). Regulated protein aggregation: Stress granules and neurodegeneration. Mol. Neurodegener. 7: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.H., Shen S.C., Lee L.Y., Lee S.H., Chan M.T., Lin C.S. (2009). Tape-Arabidopsis Sandwich—A simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Chua N.H. (2009). Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell 21: 3270–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Chua N.H. (2011). Processing bodies and plant development. Curr. Opin. Plant Biol. 14: 88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Yang J.Y., Niu Q.W., Chua N.H. (2006). Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18: 3386–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Yan Z., Wang Y., Yan X., Han Y. (2014). Tudor-SN, a component of stress granules, regulates growth under salt stress by modulating GA20ox3 mRNA levels in Arabidopsis. J. Exp. Bot. 65: 5933–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Aittomäki S., Pesu M., Carter K., Saarinen J., Kalkkinen N., Kieff E., Silvennoinen O. (2002). Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J. 21: 4950–4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Välineva T., Hong J., Bu T., Yao Z., Jensen O.N., Frilander M.J., Silvennoinen O. (2007). Transcriptional co-activator protein p100 interacts with snRNP proteins and facilitates the assembly of the spliceosome. Nucleic Acids Res. 35: 4485–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda-Inoue M., Kuroki M., Ariumi Y. (2013). DDX3 RNA helicase is required for HIV-1 Tat function. Biochem. Biophys. Res. Commun. 441: 607–611. [DOI] [PubMed] [Google Scholar]
- Yu S.F., Lujan P., Jackson D.L., Emerman M., Linial M.L. (2011). The DEAD-box RNA helicase DDX6 is required for efficient encapsidation of a retroviral genome. PLoS Pathog. 7: e1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Mao Z., Chong K. (2013). A global profiling of uncapped mRNAs under cold stress reveals specific decay patterns and endonucleolytic cleavages in Brachypodium distachyon. Genome Biol. 14: R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C.T., Shi K.H., Su Y., Liang L.Y., Yan Y., Postlethwait J., Meng A.M. (2003). Two variants of zebrafish p100 are expressed during embryogenesis and regulated by nodal signaling. FEBS Lett. 543: 190–195. [DOI] [PubMed] [Google Scholar]
- Zhu L., Tatsuke T., Mon H., Li Z., Xu J., Lee J.M., Kusakabe T. (2013). Characterization of Tudor-sn-containing granules in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 43: 664–674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.