Arabidopsis EDR4 interacts with CLATHRIN HEAVY CHAIN2 and EDR1 and negatively affects plant immunity by regulating the relocation of EDR1 to the penetration site of powdery mildew infection.
Abstract
Obligate biotrophs, such as the powdery mildew pathogens, deliver effectors to the host cell and obtain nutrients from the infection site. The interface between the plant host and the biotrophic pathogen thus represents a major battleground for plant-pathogen interactions. Increasing evidence shows that cellular trafficking plays an important role in plant immunity. Here, we report that Arabidopsis thaliana ENHANCED DISEASE RESISTANCE4 (EDR4) plays a negative role in resistance to powdery mildew and that the enhanced disease resistance in edr4 mutants requires salicylic acid signaling. EDR4 mainly localizes to the plasma membrane and endosomal compartments. Genetic analyses show that EDR4 and EDR1 function in the same genetic pathway. EDR1 and EDR4 accumulate at the penetration site of powdery mildew infection, and EDR4 physically interacts with EDR1, recruiting EDR1 to the fungal penetration site. In addition, EDR4 interacts with CLATHRIN HEAVY CHAIN2 (CHC2), and edr4 mutants show reduced endocytosis rates. Taken together, our data indicate that EDR4 associates with CHC2 and modulates plant immunity by regulating the relocation of EDR1 in Arabidopsis.
INTRODUCTION
Plants protect themselves against various pathogens through multiple layers of defense, including nonhost resistance and host resistance, which involve physical barriers and systems to detect nonself cues, via basal defense, and RESISTANCE (R) gene-mediated defenses (Schulze-Lefert and Panstruga, 2011; Nielsen and Thordal-Christensen, 2013). Detection of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors activates basal defense, which leads to PAMP-triggered immunity (PTI) (Boller and Felix, 2009). Specific recognition of pathogen effectors by R proteins leads to effector-triggered immunity (ETI) (Jones and Dangl, 2006). The defense responses associated with PTI and ETI may share signaling components and often include the activation of PATHOGENESIS-RELATED (PR) genes, mitogen-activated protein kinases (MAPKs), calcium-dependent protein kinases, and the hypersensitive response to restrict pathogen growth and spread (Tena et al., 2011).
Increasing evidence shows that vesicle trafficking plays an important role in the different layers of plant defense. For instance, forward genetic approaches identified Arabidopsis thaliana PENETRATION1 (PEN1), PEN2, and PEN3, which play positive roles in nonhost resistance; loss of function of each PEN results in defects in penetration resistance to the nonadapted powdery mildew pathogen Blumeria graminis f sp hordei (Collins et al., 2003; Lipka et al., 2005; Stein et al., 2006; Underwood and Somerville, 2008). In addition to penetration resistance, vesicle trafficking also plays important roles in PTI and ETI (Robatzek, 2014). For instance, the Arabidopsis PAMP receptor FLAGELLIN SENSING2 (FLS2), a well-characterized pattern recognition receptor, recognizes flagellin, a subunit of the bacterial flagellum (Zipfel et al., 2004), and activates the downstream signaling pathway (Veronese et al., 2006; Chinchilla et al., 2007; Lu et al., 2010; Shi et al., 2013). FLS2 localizes to the plasma membrane, and after activation by flagellin, FLS2 is internalized into the cell (Robatzek et al., 2006). Blocking flagellin-activated FLS2 endocytosis alters defense against bacterial pathogens (Robatzek et al., 2006; Spallek et al., 2013). FLS2 endocytosis requires ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT-I (Spallek et al., 2013). In addition, the ARF-GEF family protein MIN7, a key component of the vesicle-trafficking machinery, is required for PTI and ETI to Pto DC3000 infection in Arabidopsis. Also, the bacterial effector HopM1 targets MIN7 for the suppression of host resistance (Nomura et al., 2006, 2011).
Vesicle trafficking plays important roles, including in disease resistance against filamentous organisms (e.g., fungi and oomycetes, such as powdery mildew) (Yi and Valent, 2013). Powdery mildew pathogens are common and widespread obligate biotrophs that infect large numbers of plants. When a powdery mildew fungal spore infects a plant, the fungus produces a germ tube and appressorium that directly penetrate into epidermal cells and then develops a haustorium (Koh et al., 2005). The haustorium differentiates from the penetration hypha and plays important roles in transporting effectors into the cell and acquiring nutrition from the host (Panstruga and Dodds, 2009; Hückelhoven and Panstruga, 2011). Work in Arabidopsis has identified, in addition to PEN1, PEN2, and PEN3, a number of genes that function in powdery mildew resistance. For instance, ENHANCED DISEASE RESISTANCE1 (EDR1) encodes a Raf-like MAPK kinase kinase that negatively regulates salicylic acid (SA)-inducible defense responses (Frye et al., 2001). The N-terminal regulatory domain of EDR1 physically interacts with MKK4 and MKK5 and negatively affects MKK4/MKK5 and MPK3/6 protein levels. Also, mutations in MPK3, MKK4, or MKK5 can suppress edr1-mediated resistance, indicating that EDR1 fine-tunes plant innate immunity by negatively regulating the MAPK cascade (Zhao et al., 2014). KEEP ON GOING (KEG) encodes a protein containing a RING E3 ligase domain and a kinase domain, and a specific missense mutation in KEG suppresses edr1-associated phenotypes. KEG interacts with EDR1 and recruits it to the trans-Golgi network/early endosome (TGN/EE) vesicles (Wawrzynska et al., 2008; Gu and Innes, 2011). KEG regulates endomembrane protein trafficking and is degraded specifically in cells infected by powdery mildew (Gu and Innes, 2012). Recent work showed that EDR1 also interacts with ARABIDOPSIS TOXICOS EN LEVADURA1 (ATL1), an E3 ubiquitin ligase that positively regulates resistance and cell death. Overexpression of ATL1 causes cell death in Arabidopsis and Nicotiana benthamiana, and the cell death induced by ATL1 can be suppressed by EDR1. In addition, knockdown of ATL1 expression suppressed edr1-mediated powdery mildew resistance, indicating that ATL1 is a potential substrate of EDR1 (Serrano et al., 2014). EDR2 encodes a protein that contains a pleckstrin homology domain and a steroidogenic acute regulatory protein-related lipid-transfer domain (Tang et al., 2005; Vorwerk et al., 2007). EDR2 mainly localizes to the endoplasmic reticulum (ER) and also localizes to the plasma membrane and endosomes (Vorwerk et al., 2007). Genetic analyses showed that mutations in the transcription factor SR1, the 26S proteasome subunit RPN1a, or the receptor-like cytoplasmic kinase BSK1 can suppress edr1- and edr2-mediated resistance (Nie et al., 2012; Yao et al., 2012; Shi et al., 2013). EDR3 encodes DYNAMIN-RELATED-PROTEIN 1E, which is thought to be involved in membrane tubulation, a process essential for vesicle scission (Hong et al., 2003; Tang et al., 2006). Although vesicle trafficking is implicated in powdery mildew resistance mediated by edr1 and edr3, the mechanism by which vesicle trafficking affects resistance remains unclear.
The plasma membrane is a source and a sink for the endocytic and exocytic vesicle-trafficking pathways. Clathrin-mediated endocytosis (CME) is the major route of endocytosis (Dhonukshe et al., 2007). CME initiates at the plasma membrane, where cargo and coat machinery are recruited into clathrin-coated pits (Chen et al., 2011). GTPase dynamins scissor off the clathrin-coated pits after maturation to form clathrin-coated vesicles, which then target to and fuse with the early endosome. In the early endosome, cargo is differentially sorted for recycling back to the plasma membrane or to the vacuole for degradation (McMahon and Boucrot, 2011). CME complex components include clathrin, adaptor protein complexes, and accessory adaptor proteins, which bind directly to cargo or cargo receptors. Various accessory adaptor proteins can transport many different cargoes, making CME very versatile (McMahon and Boucrot, 2011). Clathrin subunits form triskelion, composed of three clathrin heavy chains and three clathrin light chains that self-polymerize (Chen et al., 2011; McMahon and Boucrot, 2011). The Arabidopsis genome has two clathrin heavy chain and three clathrin light chain genes (Chen et al., 2011). Genetic analyses show that those genes are critical for endocytosis and other clathrin-mediated pathways; for instance, clathrin heavy chain2 (chc2) mutants show defects in endocytosis and the internalization of auxin transporters (Kitakura et al., 2011). Recent work showed that SA, which usually associates with biotrophic pathogen defenses, also interferes with clathrin-mediated endocytic trafficking (Du et al., 2013). Although many components of CME remain to be identified, the recent identification and characterization of CME adaptors shed new light on how CME functions in plants (Gadeyne et al., 2014).
To identify components involved in powdery mildew resistance, we screened Arabidopsis mutants for enhanced disease resistance to the powdery mildew pathogen Golovinomyces cichoracearum (Tang et al., 2005). Here, we show that the loss-of-function edr4 mutant displays enhanced disease resistance to powdery mildew. EDR4 interacts with CHC2, one of the heavy chains of clathrin, and the edr4 mutants show reduced endocytosis rates. In addition, EDR4 interacts with EDR1 and recruits EDR1 to the site of fungal penetration, indicating that EDR4 plays an important role in plant immunity. We propose that EDR4 might modulate plant immunity by recruiting and transporting EDR1 to its destination.
RESULTS
The edr4-1 Mutants Display Enhanced Disease Resistance to G. cichoracearum
To study the molecular interactions between Arabidopsis and powdery mildew, we previously screened ethyl methanesulfonate-mutagenized Arabidopsis Columbia-0 (Col-0) plants for enhanced disease resistance to G. cichoracearum (Tang et al., 2005). Among three mutants identified in this screen, edr2 and edr3 have been described (Tang et al., 2005, 2006); here, we describe our characterization of the edr4-1 mutant.
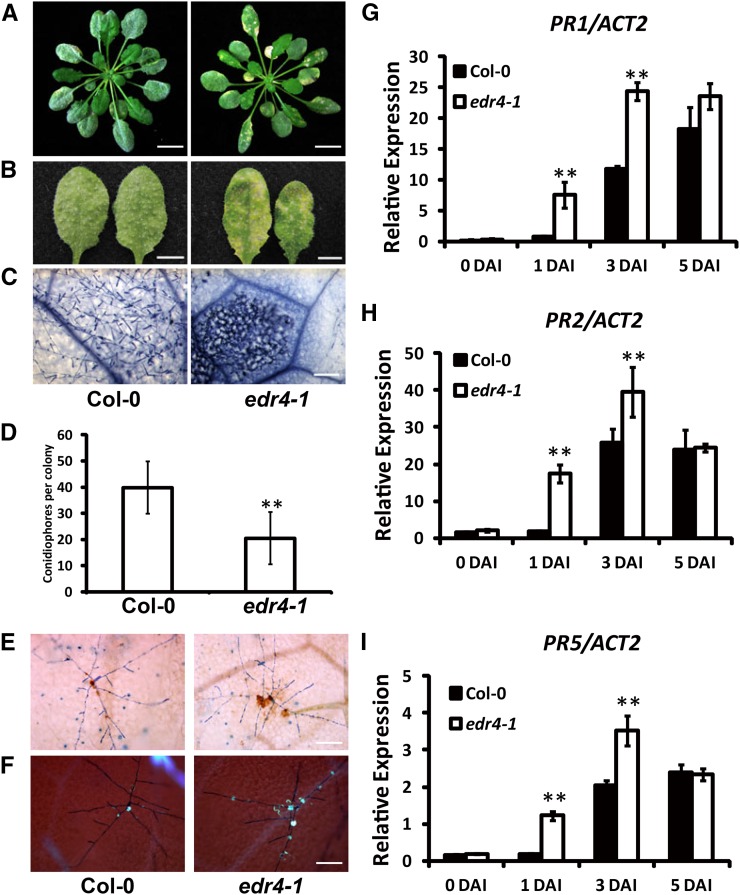
The edr4-1 mutants did not show any growth defects under short-day conditions; however, they displayed mild, hypersensitive response-like lesions under long-day conditions at late growth stages (Supplemental Figure 1). To characterize powdery mildew resistance in edr4-1, we infected 4-week-old plants with G. cichoracearum UCSC1 under short-day conditions. The wild-type plants were susceptible and supported abundant conidiophores on their leaves at 8 d after inoculation (DAI). However, the edr4-1 mutants showed fewer conidiophores and massive necrotic lesions on the leaves at 8 DAI (Figures 1A to 1C). To further monitor fungal reproduction, we counted the conidiophores per colony in wild-type and edr4-1 plants. As shown in Figure 1D, significantly fewer conidiophores were produced on edr4-1 leaves than on wild-type leaves at 6 DAI, indicating that the growth of powdery mildew fungus was inhibited in the edr4-1 mutant.
Figure 1.
The edr4-1 Mutant Exhibited Enhanced Disease Resistance to G. cichoracearum.
(A) Four-week-old wild-type and edr4-1 plants inoculated with G. cichoracearum UCSC1. The photographs were taken at 8 DAI. Bars = 1 cm.
(B) Representative leaves removed from 4-week-old wild-type and edr4-1 plants inoculated with G. cichoracearum. The photographs were taken at 8 DAI. Bars = 0.5 cm.
(C) Trypan blue staining of the leaves shown in (B) to visualize fungal structures and plant cell death. Bar = 100 μm.
(D) Quantitative analysis of conidiophore formation on 4-week-old wild-type and edr4-1 plants at 6 DAI. The bars represent means and sd in one experiment (n = 20). The experiment was repeated three times with similar results. The asterisks indicate a statistically significant difference from the wild type (P < 0.01, Student’s t test).
(E) Trypan blue-DAB staining of 4-week-old wild-type and edr4-1 plants at 2 DAI with G. cichoracearum. Brown staining shows the accumulation of H2O2. Bar = 100 μm.
(F) Aniline blue staining of 4-week-old wild-type and edr4-1 plants at 2 DAI with G. cichoracearum to visualize callose deposition (blue dots). Bar = 100 μm.
(G) to (I) Four-week-old wild-type and edr4-1 plants were inoculated with G. cichoracearum. Relative transcript levels of PR1, PR2, and PR5 were examined at various time points using real-time PCR with ACT2 as an internal control. Bars represent means and sd of values obtained from three experiments. Asterisks indicate significant differences from the wild type (P < 0.01, Student’s t test).
Activation of plant defenses often involves the production of H2O2 and the deposition of callose. To examine whether edr4-1 mutants accumulate more H2O2 than the wild type after infection, we used 3,3′-diaminobenzidine hydrochloride (DAB) to stain infected leaves at 2 DAI with G. cichoracearum. The edr4-1 mutants showed more pronounced H2O2 accumulation in the infection site than did wild-type plants (Figure 1E; Supplemental Figures 2A and 2B). To monitor callose deposition in wild-type and edr4-1 mutant plants, we performed aniline blue staining on the infected leaves at 2 DAI and found that the edr4-1 mutants showed more callose deposition at the infection site than the wild type (Figure 1F; Supplemental Figures 2C and 2D).
To investigate whether the edr4-1 mutation affects the expression of defense-related genes, we examined the transcript levels of PR1, PR2, and PR5 in wild-type and edr4-1 mutant plants at different time points after G. cichoracearum infection using real-time PCR. Both the wild type and edr4-1 showed relatively low accumulation of PR1, PR2, and PR5 transcripts before infection. The transcript levels of PR genes increased after inoculation in both the wild type and edr4-1, but significantly more transcripts of PR genes accumulated in edr4-1 plants than in wild-type plants at 1 and 3 DAI (Figures 1G to 1I). These observations indicate that the edr4-1 mutants induced the defense response more rapidly than the wild-type plants did.
The Enhanced Disease Resistance in edr4-1 Mutants Depends on SA Signaling
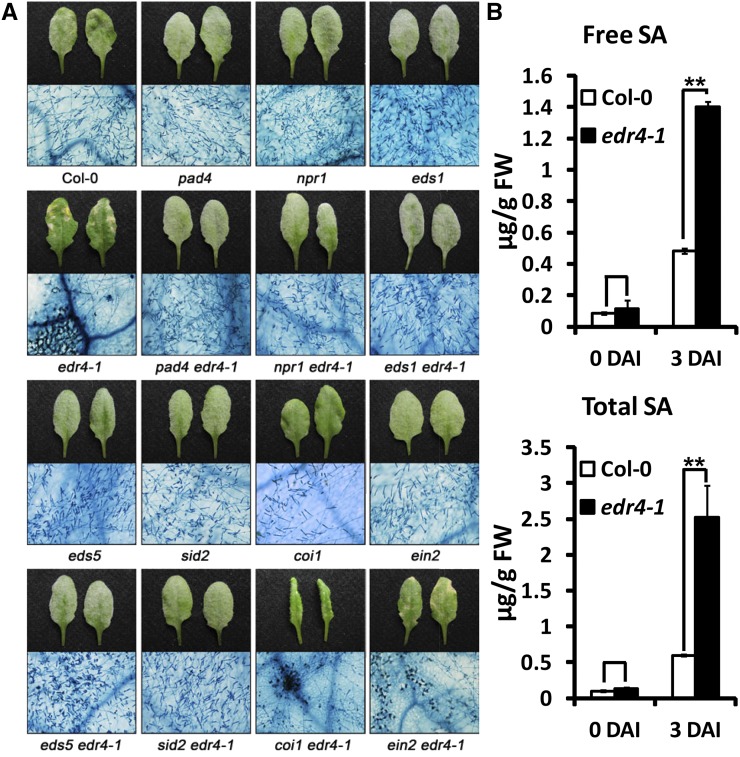
SA, jasmonic acid (JA), and ethylene signaling pathways play important roles in plant defense responses. To examine whether those pathways affect edr4-1-mediated resistance, we constructed double mutants and assessed whether mutations in those pathways alter powdery mildew resistance in edr4-1. The npr1 mutation, which disrupts SA perception, and the pad4, eds1, eds5, and sid2 mutations, which reduce levels of pathogen-induced SA, suppressed edr4-1-mediated enhanced resistance to powdery mildew and mildew-induced cell death (Figure 2A; Supplemental Figure 3). By contrast, mutations in EIN2 or COI1 blocked the ethylene response or the JA response, respectively, and did not affect edr4-1 resistance and mildew-induced cell death (Figure 2A; Supplemental Figure 3). These data indicate that edr4-1-mediated resistance and mildew-induced cell death phenotypes are SA-dependent but ethylene- and JA-independent.
Figure 2.
The Enhanced Disease Resistance Mediated by edr4-1 Requires the SA Signaling Pathway.
(A) Four-week-old plants were inoculated with G. cichoracearum, and representative leaves were removed and photographed (top panels) or stained with trypan blue (bottom panels) at 8 DAI. The edr4-1-mediated resistance is suppressed by pad4, npr1, eds1, eds5, and sid2 but not by coi1 or ein2. Bar = 200 μm.
(B) Free and total SA was extracted from 4-week-old wild-type and edr4-1 leaves at 0 and 3 DAI, then the SA levels were determined by HPLC. The bars represent means and sd (n = 6). The experiment was repeated three times with similar results. The asterisks indicate statistically significant differences from the wild type (P < 0.01, Student’s t test). FW, fresh weight.
To further assess the role of SA in edr4-1-mediated resistance, we measured SA levels in the wild type and edr4-1 before and after infection with G. cichoracearum. In the absence of pathogen, the edr4-1 mutants and wild-type plants had similar levels of free and total SA; by contrast, at 3 DAI, SA accumulated at much higher levels in edr4-1 than in the wild type (Figure 2B). These data indicate that higher SA levels in edr4-1 mutants cause the enhanced disease resistance and mildew-induced cell death.
EDR4 Encodes a Protein of Unknown Function
The edr4-1 mutation was identified by standard map-based cloning (Supplemental Figure 4A). A C-to-T transition at nucleotide 1213 in At5g05190 was found in edr4-1, and this transition causes an early stop (Q405STOP) in the predicted protein sequence (Supplemental Figure 4A). To confirm that At5g05190 is EDR4, a genomic clone of At5g05190 was transformed into edr4-1 plants. Among 25 transgenic plants obtained, 22 displayed the wild-type, susceptible phenotype at 8 DAI when infected with G. cichoracearum, indicating that the At5g05190 genomic clone complemented the edr4-1 phenotype (Supplemental Figures 4B and 4C). As an additional confirmation, we also examined the phenotypes of two T-DNA insertion lines of At5g05190, SALK_048465 (edr4-2) and SALK_009370 (edr4-3) (Supplemental Figure 4A). No full-length transcript of At5g05190 could be detected in edr4-1 or in these two lines, indicating that the expression of EDR4 is disrupted in these three mutant lines (Supplemental Figure 4D). These two mutants showed enhanced disease resistance and mildew-induced cell death upon powdery mildew infection at 8 DAI, which was very similar to the phenotype of edr4-1 (Supplemental Figures 4B and 4C). Taken together, these data indicate that At5g05190 is the gene responsible for the edr4-1 phenotypes; therefore, we designate At5g05190 as EDR4.
EDR4 encodes a protein of 615 amino acids with no assigned function, and no clear EDR4 homolog could be identified in the Arabidopsis genome. A search for motifs in the primary sequence of EDR4, using the Eukaryotic Linear Motif server (http://elm.eu.org/) (Puntervoll et al., 2003), identified three types of structural elements in EDR4: a coiled-coil (CC) domain, four low-complexity regions (LCR), and a Duf3133 domain of unknown function (Supplemental Figure 4E). The CC domain and LCRs often function as protein interaction domains for a wide variety of proteins (Burkhard et al., 2001; Lee et al., 2011).
EDR4 Mainly Localizes at the Plasma Membrane and Endosomal Compartments
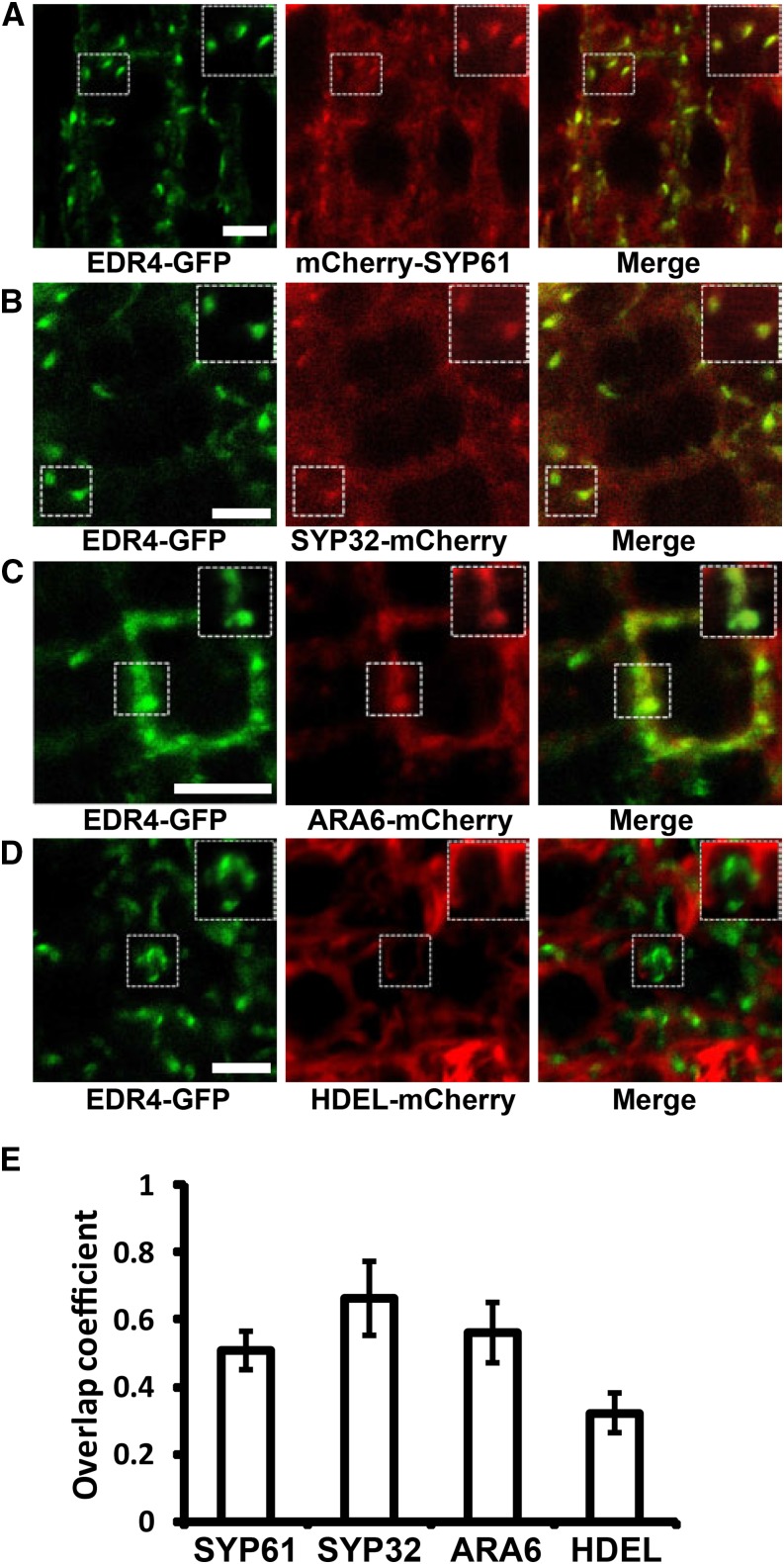
To investigate the subcellular localization of EDR4, we made a fusion of EDR4 to Green Fluorescent Protein (GFP), expressed it from the pEDR4-EDR4-eGFP construct (EDR4-GFP), and transformed it into edr4-1 mutant plants. EDR4-GFP was expressed as a full-length protein and complemented the edr4-1 phenotypes, indicating that EDR4-GFP was functional (Supplemental Figure 5). A representative homozygous edr4-1 EDR4-GFP transgenic line was chosen to examine EDR4 localization. In root tip cells of 5-d-old seedlings, time-lapse analysis revealed that EDR4-GFP displayed dynamic intracellular movement, which partially overlapped with FM4-64 (Supplemental Figure 6A and Supplemental Movie 1). EDR4-GFP appears to localize on the plasma membrane and on dot-like intracellular structures in the cytosol. To determine the subcellular localization of EDR4, we assessed the colocalization of EDR4-GFP with several organelle markers, including mCherry-SYP61 (TGN/EE), ARA6-mCherry (late endosome), SYP32-mCherry (cis-Golgi), and HDEL-mCherry (ER) (Nelson et al., 2007; Geldner et al., 2009; Gu and Innes, 2011, 2012; Sauer et al., 2013), and examined EDR4-GFP in root tips of transgenic plants. Microscopy and quantification analysis showed that a large portion of EDR4-GFP protein overlapped with the TGN/EE marker SYP61, the late endosome marker ARA6, and the cis-Golgi marker SYP32, but a smaller portion of EDR4-GFP overlapped with the ER marker HDEL (Figures 3A to 3E). Taken together, these data indicate that EDR4 mainly localizes at the plasma membrane and in endosomal compartments and displays dynamic movement in cells.
Figure 3.
EDR4 Localizes to the Plasma Membrane, TGN/EE, and cis-Golgi.
(A) to (D) The root tip cells of 5-d-old seedlings that expressed EDR4-GFP and the indicated subcellular markers were imaged using confocal microscopy, and single optical sections are shown. Enlarged images of the boxed areas are shown at top right. The mCherry-SYP61 (A) and ARA6-mCherry (C) constructs were introduced into the EDR4-GFP plants by transformation, while SYP32-mCherry (B) and HDEL-mCherry (D) were introduced into the EDR4-GFP plants by genetic crosses. Bars = 5 μm.
(E) Quantitative analysis of the overlap of EDR4-GFP with the indicated organelle markers. The bars show Mander’s overlap coefficient (Herda et al., 2012), indicating a level of colocalization between EDR4-GFP and the organelle marker. Two independent experiments with 20 to 35 cells for each subcellular marker were evaluated.
EDR4 Accumulates at the Penetration Site of Fungal Infection
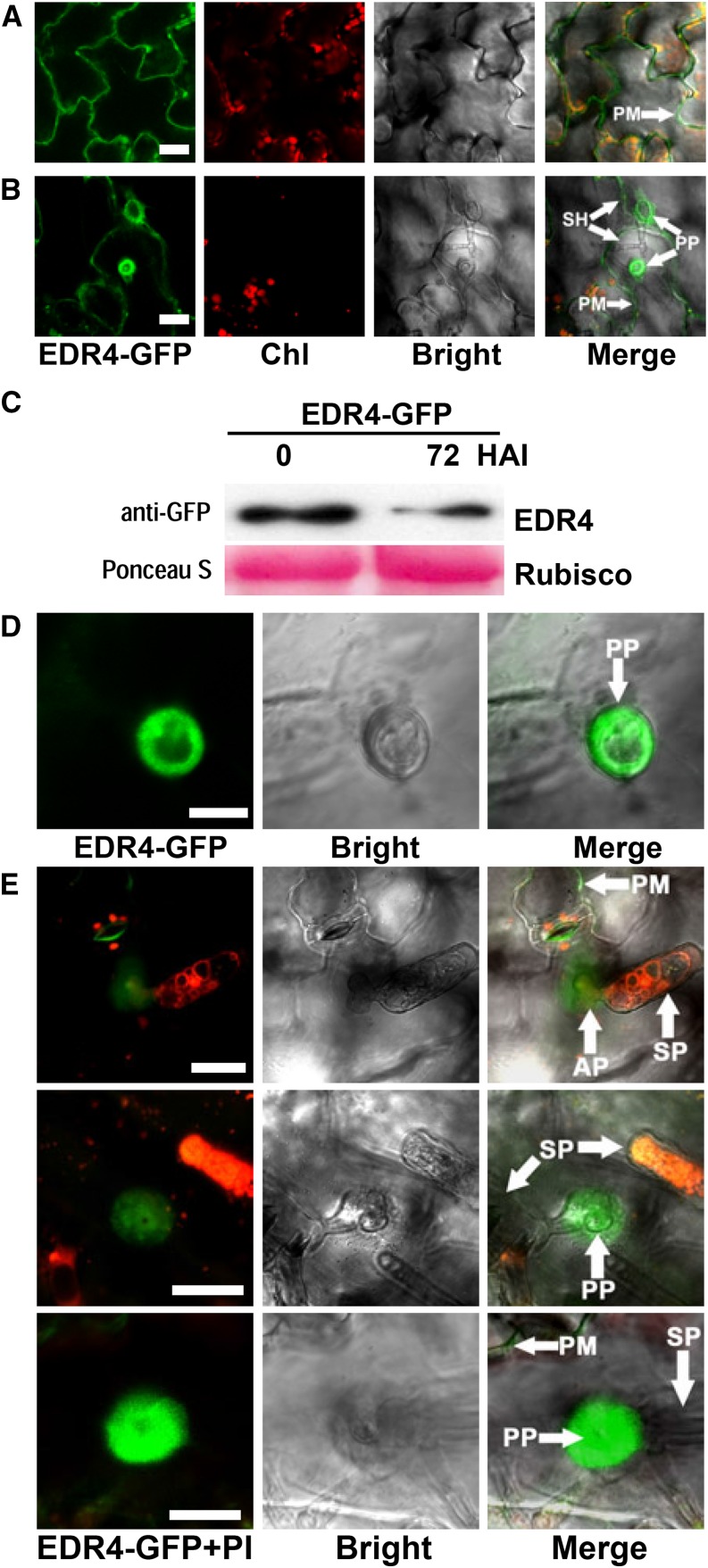
We further examined the subcellular localization of EDR4 in epidermal cells after G. cichoracearum infection using the EDR4-GFP transgenic line. The GFP signal was observed in epidermal cells in uninfected leaves (Figure 4A) and also showed intracellular movement (Supplemental Figure 6B and Supplemental Movie 2). At 48 h after inoculation (HAI) with G. cichoracearum, EDR4-GFP accumulated under the penetration peg at the infection site in epidermal cells (Figure 4B), but overall, the EDR4-GFP protein levels decreased at 72 HAI with G. cichoracearum (Figure 4C). To further confirm the accumulation of EDR4-GFP at the site of infection, we also examined EDR4-GFP localization at 24 HAI, with the cis-Golgi-localized protein N-acetylglucosaminyltransferase I (NAG) fused to GFP (Essl et al., 1999) as a control. Fungal structures were labeled with propidium iodide. We found that EDR4-GFP accumulated at penetration sites at 24 HAI (Figures 4D and 4E), while there was no obvious difference in the subcellular localization of NAG-GFP before and after inoculation (Supplemental Figure 7). To determine whether EDR4 accumulates at the plasma membrane or papillae at the penetration site after infection, we used plasmolysis of infected leaves at 24 HAI. As shown in Supplemental Figure 8A, EDR4-GFP accumulated in the plasma membrane but not in papillae.
Figure 4.
EDR4-GFP Accumulates at the Powdery Mildew Penetration Site in Epidermal Cells.
(A) EDR4-GFP localization in epidermal cells of uninfected leaves. Bar = 25 μm.
(B) EDR4-GFP accumulates under the penetration peg of the secondary hyphal infection site in epidermal cells at 48 HAI with G. cichoracearum. Bar = 25 μm.
(C) Four-week-old transgenic EDR4-GFP plants were infected with G. cichoracearum. Immunoblotting was performed using an anti-GFP antibody. Ponceau S staining of Rubisco is shown as a loading control. The experiment was repeated at least three times with similar results.
(D) and (E) Accumulation of EDR4-GFP around the penetration peg of the appressorium at 24 HAI with G. cichoracearum, shown in different views. Leaves in (E) were stained with propidium iodide (PI) to visualize the fungal structures, which appear red. Bar in (D) = 10 μm; bars in (E) = 20 μm.
AP, appressoria; Chl, chloroplast; PM, plasma membrane; PP, penetration peg; SH, secondary hyphae; SP, spores.
EDR1, EDR4, and KEG Act in the Same Genetic Pathway
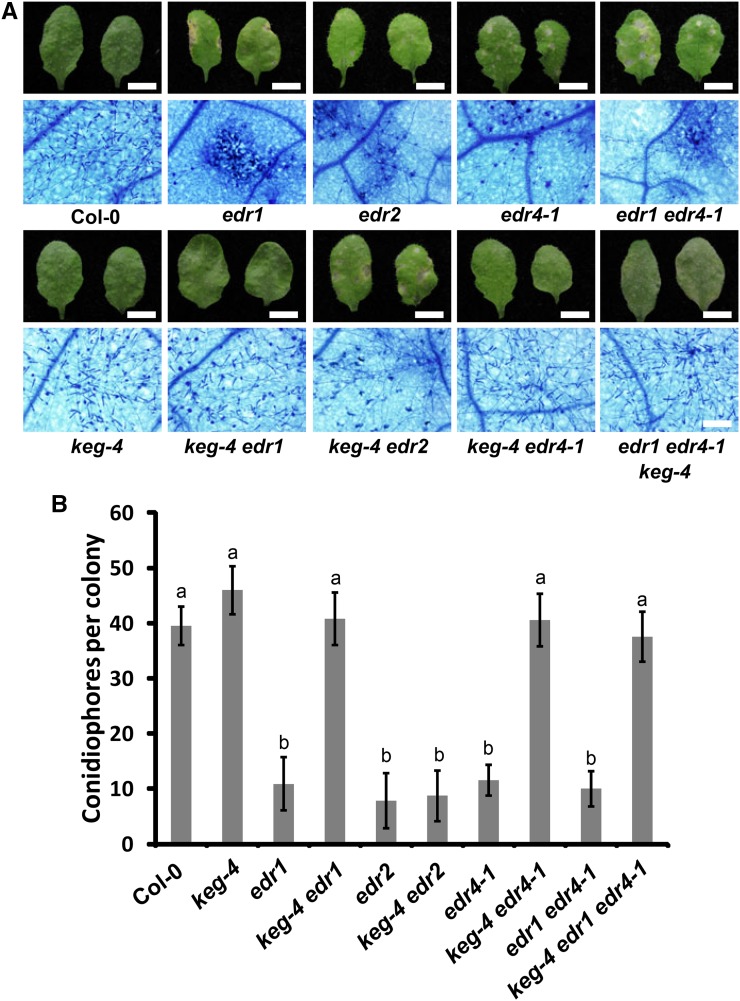
To further characterize the edr4-1 mutant, we crossed edr4-1 with edr1 and edr2, two well-characterized powdery mildew resistance mutants, and examined the phenotypes of the double mutants. The edr2 edr4-1 mutant showed more cell death and fewer fungal spores than the edr2 and edr4-1 single mutants, indicating that the edr2 mutation enhanced the edr4-1 phenotype. However, the edr1, edr4-1, and edr1 edr4-1 mutants showed no significant differences in growth of fungal spores (Supplemental Figure 9). A missense mutation in KEG (keg-4) suppressed the edr1 phenotype (Wawrzynska et al., 2008). To further examine the relationship between EDR1 and EDR4, we crossed edr1, edr4-1, and edr1 edr4-1 with keg-4 and generated double and triple mutants. The keg-4 mutation suppressed the enhanced disease resistance of edr4 and edr1 edr4-1 mutants (Figures 5A and 5B), indicating that edr4-mediated resistance also requires KEG. Previous work showed that keg-4 suppresses edr1-mediated resistance, but not edr2-mediated resistance, indicating that keg-4 specifically suppresses edr1 (Yao et al., 2012). The observations that keg-4 suppressed edr4-1 and edr1 edr4-1, and that edr1, edr4-1, and edr1 edr4-1 displayed similar levels of resistance, suggest that EDR1, EDR4, and KEG act in the same genetic pathway. Consistent with this notion, MPK3 and MPK6 kinase activation increased in edr4-1 mutants upon infection by G. cichoracearum, and mpk3, mkk4, and mkk5 mutations also suppressed powdery mildew resistance in edr4-1 (Supplemental Figure 10), similar to the edr1 mutant (Zhao et al., 2014).
Figure 5.
The edr1 Suppressor keg-4 Suppresses edr4-1-Mediated Powdery Mildew Resistance.
(A) Photograph and trypan blue staining of representative leaves removed from 4-week-old plants at 8 DAI with G. cichoracearum. Bars in top panels = 0.5 cm; bar in bottom panels = 100 μm.
(B) Quantitative analysis of conidiophore formation on infected plants at 6 DAI. The bars represent means and sd (n = 20). The samples are labeled with different letters to indicate statistically significant differences (P < 0.01, one-way ANOVA). The experiments were repeated three times with similar results.
EDR4 Interacts with EDR1
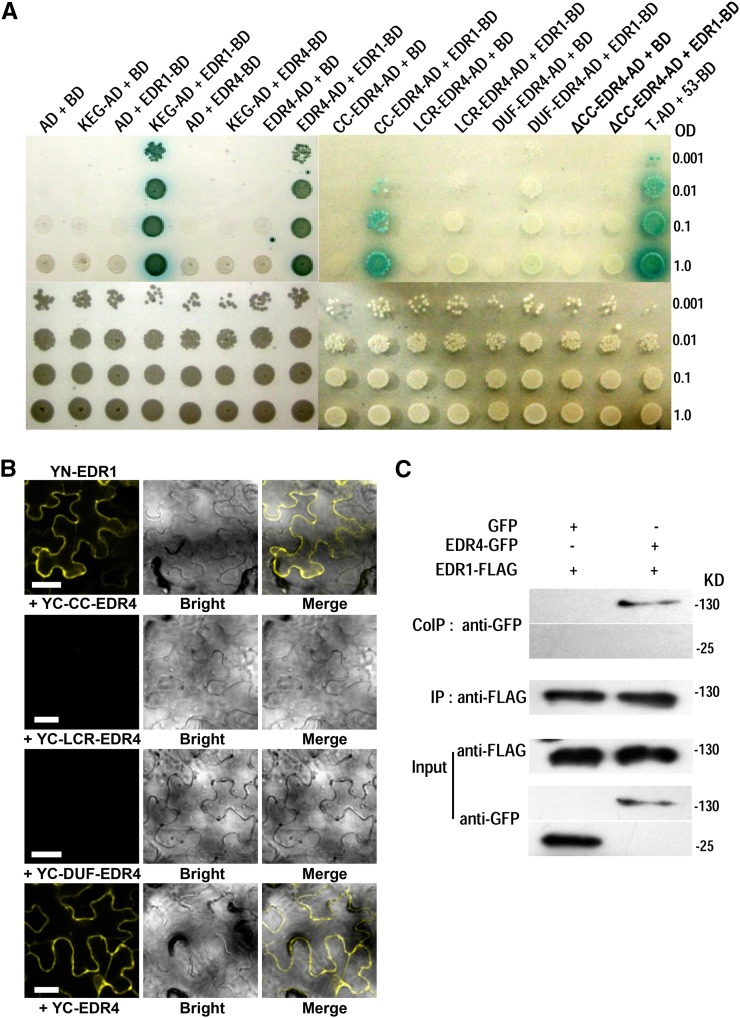
Since edr1 and edr4-1 displayed similar powdery mildew resistance, and EDR1, EDR4, and KEG may act in the same genetic pathway, we hypothesized that EDR4 may associate with EDR1 to modulate plant immunity. To test this hypothesis, we performed yeast two-hybrid assays to examine whether EDR4 interacts with EDR1 and KEG. EDR4 could interact with the N-terminal domain (amino acids 1 to 657) of EDR1 and full-length EDR1 but not with the C-terminal domain of EDR1 (amino acids 658 to 933) or with KEG (Figure 6A; Supplemental Figure 11). To determine which domain of EDR4 is responsible for the interaction with EDR1, we conducted a deletion analysis of EDR4 and examined the interactions of the deleted variants by yeast two-hybrid assays. As shown in Figure 6A, a variant of EDR4 with only the CC domain (amino acids 1 to 149) could interact with EDR1, while the LCR, Duf3133 domain, or CC domain deletion form of EDR4 could not interact with EDR1. These data indicate that the CC domain of EDR4 is necessary and sufficient for interaction with EDR1.
Figure 6.
EDR4 Interacts with EDR1.
(A) EDR4 interacts with EDR1 in yeast two-hybrid assays. Yeast cells containing the indicated plasmids were serially diluted from OD600 = 1 and spotted on control medium and on selective medium supplemented with 25 mM 3-amino-1,2,4-triazole and 40 μg/mL X-α-Gal. AD and BD represent pGADT7 and pGBKT7, respectively. T-AD + 53-BD was used as a positive control. CC-EDR4, amino acids 1 to 149 of EDR4, which includes the CC domain; ΔCC-EDR4, a deletion variant of EDR4 protein, which lacks amino acids 1 to 149; LCR-EDR4, amino acids 150 to 491 of EDR4, which includes the four LCR domains; DUF-EDR4, amino acids 492 to 615 of EDR4, which includes the Duf3133 domain.
(B) EDR4 interacted with EDR1 in BiFC assays in N. benthamiana. EDR1 was fused to the N-terminal fragment of YFP, and EDR4 (full length or deletion form of EDR4) was fused to the C-terminal fragment of YFP. YFP fluorescence indicates an interaction between the two proteins. Bars = 50 μm.
(C) Expression and Co-IP of pEDR1-EDR1-FLAG and pEDR4-EDR4-GFP in Arabidopsis. Total protein was extracted from 4-week-old transgenic plants expressing both EDR1-FLAG and EDR4-GFP. Plants expressing EDR1-FLAG and GFP (left panels) were used as negative controls. The EDR1-FLAG protein was immunoprecipitated with anti-FLAG antibody, and the presence of EDR4-GFP or GFP protein was detected by immunoblot analysis with anti-GFP antibody.
These experiments were repeated three times with similar results.
To further confirm the interactions of EDR1 and EDR4, we performed bimolecular fluorescence complementation (BiFC) by fusing EDR1, EDR4, and EDR4 deletion forms or KEG to the N- or C-terminal fragment of Yellow Fluorescent Protein (YFP) and transiently expressing those constructs in N. benthamiana leaves. We observed reconstituted YFP fluorescence in leaves cotransformed with 35S-driven YN-EDR1 and YC-KEG, as well as YN-EDR1 and YC-EDR4 or the YC-CC-EDR4 domain, but not in leaves cotransformed with YN-KEG and YC-EDR4 or other YC-EDR4 deletions (Figure 6B; Supplemental Figure 12), indicating that the EDR4 CC domain interacts with EDR1 in N. benthamiana, consistent with the results of the yeast two-hybrid assays.
To validate the interaction of EDR4 and EDR1, we also performed coimmunoprecipitation (Co-IP) assays in stable transgenic Arabidopsis plants. We crossed EDR4-GFP plants with plants expressing EDR1-FLAG driven by the native EDR1 promoter and used the F2 plants that express both EDR4-GFP and EDR1-FLAG in Co-IP assays, with plants that expressed both EDR1-FLAG and GFP as a negative control. We extracted total protein and immunoprecipitated EDR1 using anti-FLAG antibody and then detected the presence or absence of EDR4 with anti-GFP antibody. EDR4 was detected only in the precipitates of leaves that express EDR1-FLAG and EDR4-GFP but not in the negative control plants (Figure 6C), indicating that EDR4 interacts with EDR1 in Arabidopsis. Taken together, these observations indicate that EDR4 interacts with EDR1.
Since EDR1 interacts with EDR4, we then examined whether EDR4 colocalizes with EDR1. We first transiently expressed EDR4-GFP and EDR1-mCherry in N. benthamiana leaves. EDR1 appears to colocalize with EDR4 in the plasma membrane and endosomal compartments (Supplemental Figures 13A and 13B). We then examined transgenic plants that coexpressed EDR4-GFP and EDR1-mCherry. Similarly, EDR4-GFP and EDR1-mCherry colocalized in endosomal compartments in root cells (Supplemental Figure 13C).
EDR4 Recruits EDR1 to Powdery Mildew Infection Sites on the Plasma Membrane
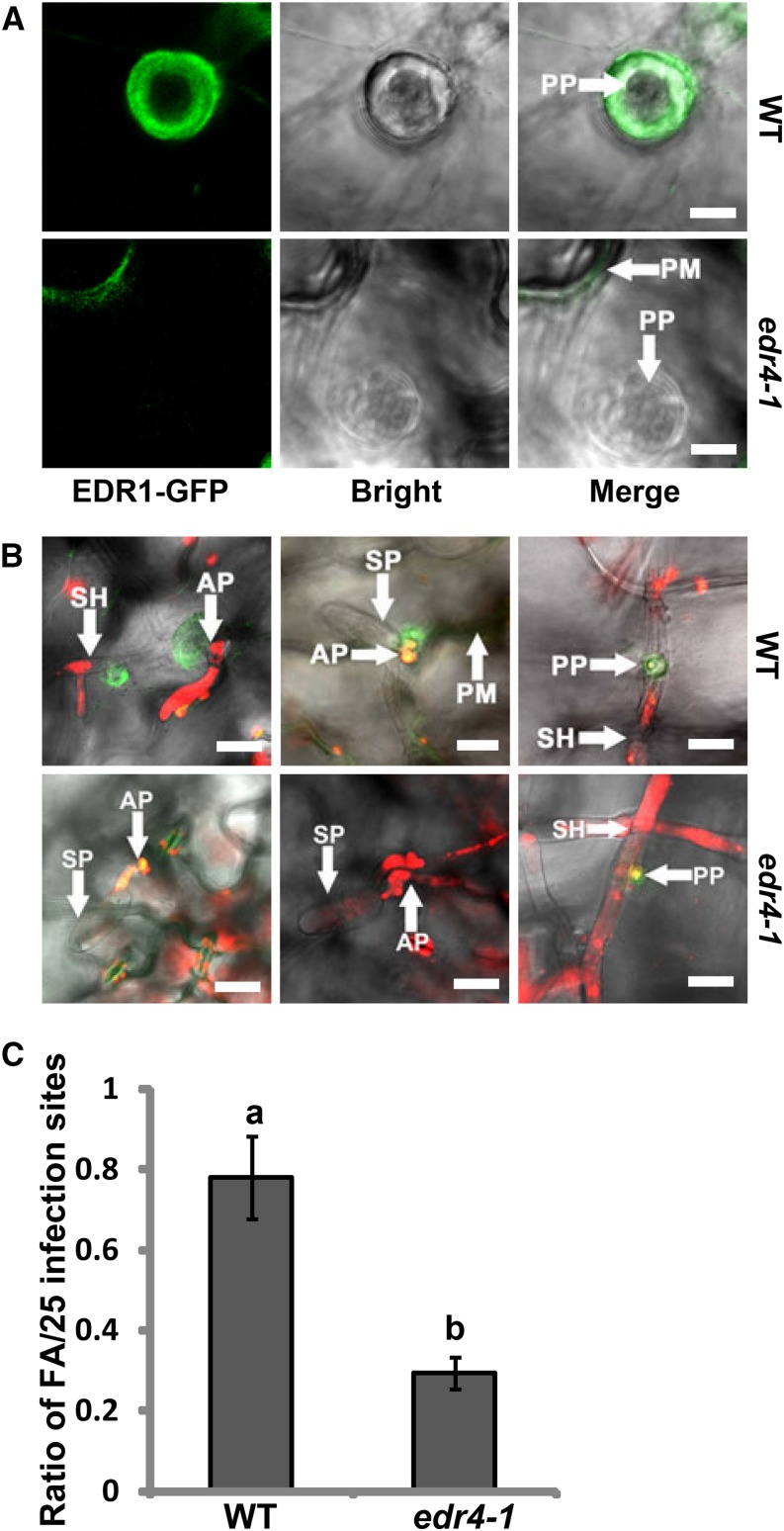
Since EDR1 interacts with EDR4, we next examined whether EDR1 and EDR4 have similar localization patterns. EDR1 also shows dynamic intracellular movement (Supplemental Figure 6C and Supplemental Movie 3). As EDR4 accumulated at powdery mildew infection sites, we then examined whether EDR1 also accumulates at infection sites and whether the edr4-1 mutation affects this localization. We thus crossed the EDR1-GFP transgenic plant with the edr4-1 mutant to generate edr4-1 EDR1-GFP plants. We did not observe an obvious difference in EDR1-GFP localization in wild-type and edr4-1 backgrounds in the absence of powdery mildew pathogen (Supplemental Figure 14A). Upon infection, EDR1-GFP also accumulated at the infection site at 24 HAI in wild type leaves; however, EDR1-GFP accumulated at much lower levels at infection sites in edr4-1 leaves (Figures 7A and 7B). To quantify the level of accumulation of EDR1-GFP, we calculated the frequency of focal accumulations of EDR1-GFP at infection sites in wild-type and edr4-1 plants according to the previously described method (Underwood and Somerville, 2013). As shown in Figure 7C, the frequency of focal accumulations of EDR1-GFP in edr4-1 mutants was much lower than in the wild type, indicating that the edr4-1 mutation compromised the accumulation of EDR1-GFP at the fungal infection site. Similar to EDR4-GFP, plasmolysis analysis showed that EDR1-GFP also accumulated at the plasma membrane, not in papillae, after infection with G. cichoracearum (Supplemental Figure 8B). Taken together, our data suggested that EDR1 moves to the plasma membrane around powdery mildew infection sites and that EDR4 plays an important role in EDR1 relocation.
Figure 7.
edr4-1 Affects EDR1 Focal Accumulation at the Interface between the Powdery Mildew and Arabidopsis.
(A) and (B) Focal accumulation of EDR1-GFP at the pathogen infection site in epidermal cells of 4-week-old wild-type and edr4-1 plants after inoculation with G. cichoracearum, shown in different views. Leaves were stained with propidium iodide in (B). Fungal structures are shown in red. AP, appressoria; PM, plasma membrane; PP, penetration peg; SH, secondary hyphae; SP, spores. Bars = 20 μm.
(C) Quantitative analysis of powdery mildew-induced EDR1-GFP focal accumulations (FA) in wild-type and edr4-1 mutant plants. The frequency was determined by counting the number of focal accumulations present in 25 random successful penetrations by appressoria per leaf, for three leaves per line. Bars represent means and sd of values obtained from three biological replicates per genotype. Statistically significant differences among the samples are labeled with different letters (P < 0.01, one-way ANOVA). The experiments were repeated three times with similar results.
As the CC domain of EDR4 is necessary for interaction with EDR1, we examined whether EDR1 localization was affected in the EDR4 CC domain deletion background. We made an EDR4-CC-domain deletion-FLAG construct and transformed it alone or with the pEDR1-EDR1-GFP construct into edr4-2. The EDR4-CC-domain deletion-FLAG construct did not complement the edr4-2 phenotype, indicating that EDR4 function requires the CC domain (Supplemental Figure 15). We also compared EDR1 localization in wild-type, edr4-2, and edr4-2 EDR4-CC-domain deletion plants after powdery mildew infection. The localization of EDR1-GFP in the edr4-2 and edr4-2 EDR4-CC-domain deletion background was similar, as both show defects in focal accumulation to the site of mildew infection (Supplemental Figure 16), indicating that the EDR4-CC-domain deletion construct could not function as the full-length EDR4 protein. To examine whether edr1 affected EDR4 localization, we examined EDR4-GFP localization in wild-type and edr1 plants after powdery mildew infection at 24 HAI. EDR4-GFP accumulated at the infection site in wild-type and edr1 plants, and no significant difference was observed between the wild type and edr1, indicating that edr1 did not affect the localization of EDR4 (Supplemental Figure 17).
To examine whether powdery mildew triggers continuous accumulation of EDR1 and EDR4, we performed fluorescence recovery after photobleaching (FRAP) (Bhat et al., 2005) of EDR1-GFP and EDR4-GFP leaves at G. cichoracearum penetration sites and distal membrane sites. At the penetration site, the EDR1-GFP signal recovered to less than 40% in 60 min, compared with before bleaching, and EDR4-GFP displayed a similar pattern, recovering to less than 20% in 60 min (Supplemental Figures 18A and 18C). By contrast, at a distal membrane site, the EDR1-GFP and EDR4-GFP signals showed rapid recovery (within 15 min) after bleaching (Supplemental Figures 18B and 18D). These data suggested that EDR1 and EDR4 proteins were not continuously delivered to penetration sites but were delivered one time and retained at these sites. These results are comparable to previous findings that focal accumulations of PEN1 and PEN3 after B. graminis f sp hordei infection result from single delivery events rather than continuous delivery (Bhat et al., 2005; Underwood and Somerville, 2013).
EDR4 Interacts with CHC2, and edr4 Shows Reduced Endocytosis Rates
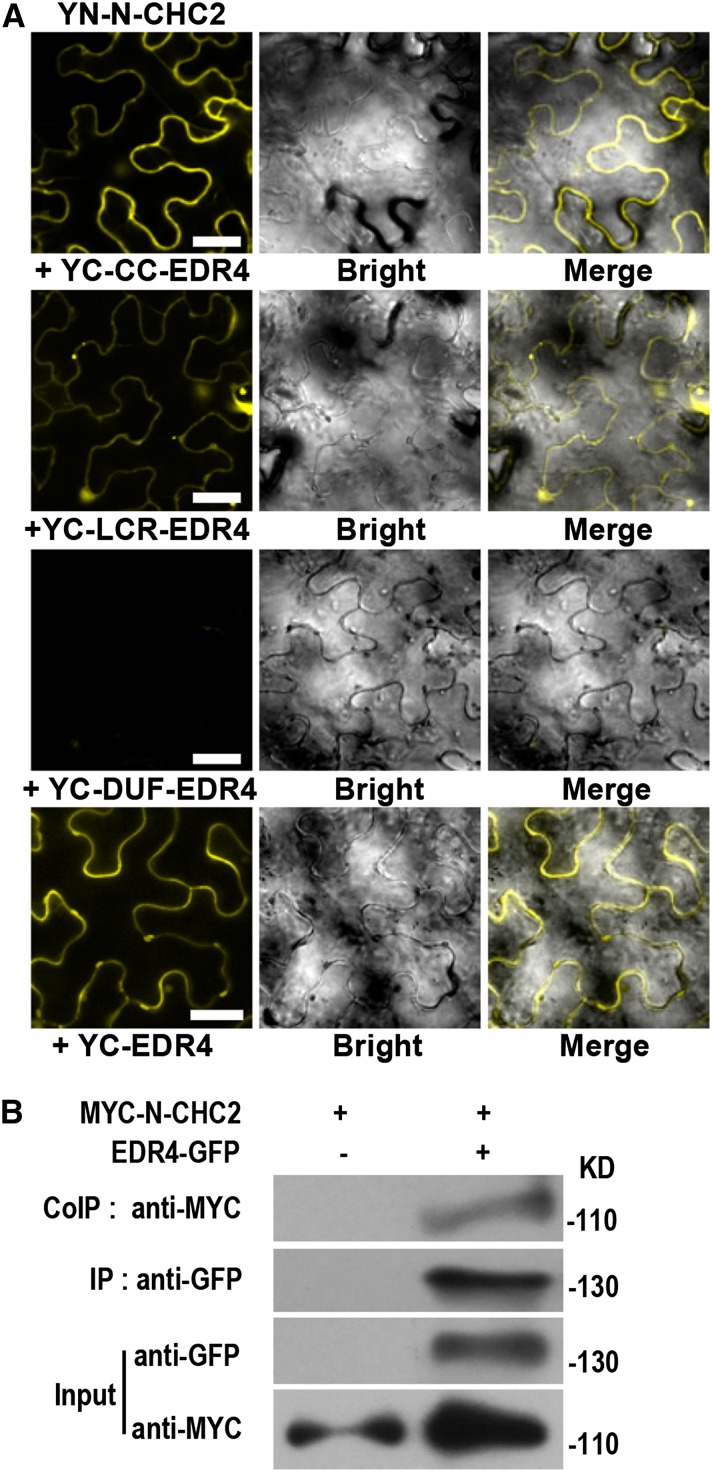
The Arabidopsis Interactome Mapping Consortium (2011) reported that EDR4 could interact with CHC2, a major component of clathrin-coated vesicles in CME and clathrin-mediated pathways in yeast two-hybrid assays. To confirm the interactions between CHC2 and EDR4 in planta, we performed BiFC assays using N. benthamiana. As shown in Figure 8A, we observed YFP signal in leaves coexpressing CHC2 and full-length EDR4, the EDR4 CC domain, or the EDR4 LCR domain but not the EDR4 Duf3133 domain, confirming that EDR4 interacts with CHC2 and that the CC and LCR domains may be responsible for this interaction. The interaction between EDR4 and CHC2 suggested that EDR4 could function in clathrin-mediated pathways. To further confirm the interaction between EDR4 and CHC2, we performed Co-IP assays in stable transgenic Arabidopsis plants. We crossed EDR4-GFP plants with plants expressing MYC-N-CHC2 driven by the 35S promoter and used the F2 plants that expressed both EDR4-GFP and MYC-N-CHC2 in Co-IP assays, with plants that expressed only MYC-N-CHC2 as a negative control. CHC2 was only detected in the precipitates of leaves that express EDR4-GFP and MYC-N-CHC2 but not in the negative control plants (Figure 8B), indicating that EDR4 interacts with CHC2 in Arabidopsis.
Figure 8.
EDR4 Interacts with CHC2 in Vivo.
(A) BiFC assays in N. benthamiana show interactions between YN-N-CHC2 and YC-EDR4. The N-terminal domain (amino acids 1 to 1016) of CHC2 was fused to the N-terminal fragment of YFP, and the fragments, including full-length EDR4, the CC domain, four LCR domains, and the Duf3133 domain of EDR4, were fused to the C-terminal fragment of YFP. YFP fluorescence indicates an interaction between the two proteins. The experiments were repeated three times with similar results. Bars = 30 μm.
(B) Expression and Co-IP of pEDR4-EDR4-GFP and 35S-MYC-N-CHC2 in Arabidopsis. Total protein was extracted from 4-week-old transgenic plants expressing both EDR4-GFP and MYC-N-CHC2. The EDR4-GFP protein was immunoprecipitated with anti-GFP antibody, and the presence of MYC-N-CHC2 protein was detected by immunoblot analysis with anti-MYC antibody. The experiments were repeated three times with similar results.
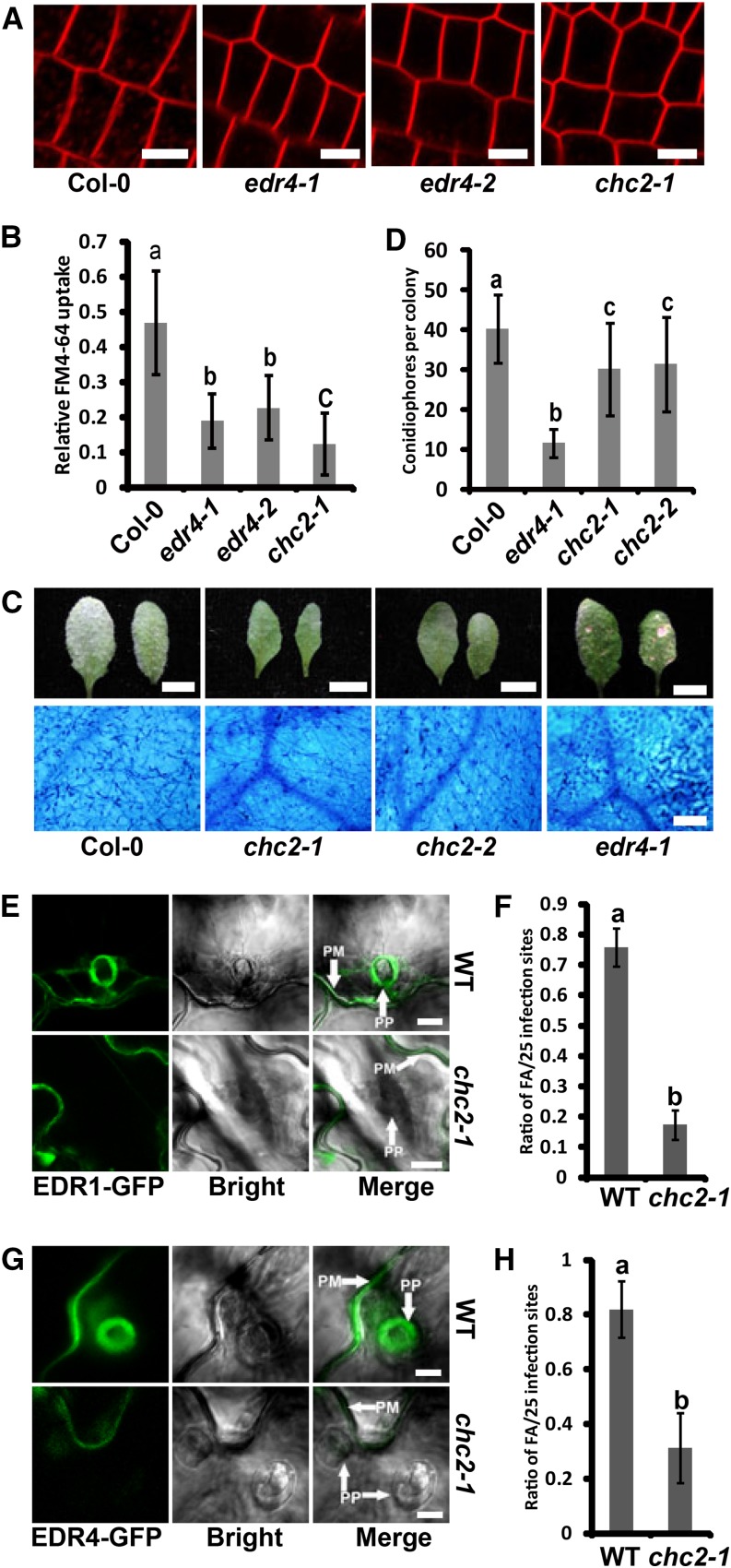
The chc2 mutants display reduced endocytosis rates, as measured by uptake of the endocytic tracer dye FM4-64 (Kitakura et al., 2011). To assess whether EDR4 also functions in endocytosis, we performed FM4-64 uptake tests on wild-type, edr4-1, and edr4-2 plants, using chc2-1 as an endocytosis-deficient control. The intracellular accumulation of FM4-64 showed clear labeling of endosomes in the wild type within 8 min; however, edr4-1, edr4-2, and chc2-1 mutants showed much less labeling at the same time (Figures 9A and 9B), indicating that edr4 mutants show reduced endocytosis rates. By contrast, we did not observe any difference in FM4-64 uptake between the wild type and edr1 mutants (Supplemental Figure 19). To examine whether CHC2 affects powdery mildew resistance, we infected the chc2-1 and chc2-2 mutants with G. cichoracearum. The chc2-1 and chc2-2 mutants supported significantly less fungal growth than the wild type, although the resistance in chc2-1 and chc2-2 mutants was not as strong as in the edr4-1 mutant (Figures 9C and 9D). In addition, the chc2-1 and chc2-2 mutants showed higher H2O2 and callose accumulation and stronger activation of MPKs than the wild type (Supplemental Figure 20). The callose accumulation and activation of MPKs in chc2-1 and chc2-2 were similar to those in edr4-1, but the accumulation of H2O2 was significantly lower than that observed in edr4-1 mutants (Supplemental Figure 20). Taken together, our data indicated that CHC2 contributes to powdery mildew resistance.
Figure 9.
edr4 Mutants Show Defects in FM4-64 Uptake; Focal Accumulation of EDR1 and EDR4 Requires CHC2.
(A) Delay of FM4-64 uptake in the edr4 mutants. Intracellular accumulation of FM4-64 was examined at 8 min after staining with 2 μM FM4-64 in root tip cells of 5-d-old seedlings of the wild type and edr4-1, edr4-2, and chc2-1 mutants. Bars = 10 μm.
(B) Quantification of relative FM4-64 uptake. The bars represent means and sd (n > 50 cells analyzed). Statistically significant differences among the samples are labeled with different letters (P < 0.01, one-way ANOVA). The experiments were repeated three times with similar results.
(C) Photographs and trypan blue staining of representative leaves removed from 4-week-old wild-type, chc2-1, chc2-2, and edr4-1 plants at 8 DAI with G. cichoracearum. Bars in top panels = 0.5 cm; bar in bottom panels = 100 μm.
(D) Quantitative analysis of conidiophore formation on 4-week-old wild-type, chc2-1, chc2-2, and edr4-1 plants inoculated with G. cichoracearum at 6 DAI. The bars represent means and sd. The experiment was repeated three times with similar results (n = 20). Statistically significant differences are labeled with different letters (P < 0.01, one-way ANOVA). The experiments were repeated three times with similar results.
(E) and (G) Focal accumulation of EDR1-GFP (E) or EDR4-GFP (G) at the pathogen infection site in epidermal cells of 4-week-old wild-type and chc2-1 plants after inoculation with G. cichoracearum at 24 HAI. PM, plasma membrane; PP, penetration peg. Bars = 10 μm.
(F) and (H) Quantitative analysis of powdery mildew-induced EDR1-GFP (F) or EDR4-GFP (H) focal accumulations (FA) in wild-type and chc2-1 plants. Bars represent means and sd of values obtained from three biological replicates per genotype. Statistically significant differences among the samples are labeled with different letters (P < 0.01, one-way ANOVA). The experiments were repeated three times with similar results.
As the chc2 mutants displayed an edr4-like phenotype, and CHC2 interacts with EDR4, we then asked whether CHC2 contributes to the relocation of EDR1 and EDR4 upon powdery mildew infection. To address this question, we transformed EDR1-GFP and EDR4-GFP constructs into chc2-1, to generate chc2-1 EDR1-GFP and chc2-1 EDR4-GFP transgenic plants. We then examined the localization of EDR1-GFP and EDR4-GFP in the chc2-1 mutant. The localization of EDR1-GFP and EDR4-GFP was similar in wild-type and chc2-1 plants before powdery mildew infection (Supplemental Figures 14B and 14C). However, both EDR1-GFP and EDR4-GFP accumulated at much lower levels at infection sites in chc2-1 than wild-type leaves at 24 HAI (Figures 9E and 9G). Quantification analysis also confirmed those results (Figures 9F and 9H).
DISCUSSION
The loss-of-function mutants of EDR4 showed enhanced resistance to G. cichoracearum and increased mildew-induced cell death. The edr4-mediated resistance included increased callose deposition, higher levels of H2O2, stronger activation of MPKs, enhanced expression of pathogenesis-related genes, and higher levels of SA, which are very similar to previously identified edr1 mutants (Frye et al., 2001). Genetic analysis showed that edr1- and edr4-mediated resistance both require pathways induced by SA but not by JA or ethylene. In addition, the edr1 edr4-1 double mutant displays a similar phenotype to edr1 and edr4-1 single mutants, and keg-4 suppresses the double and single mutants (Wawrzynska et al., 2008). By contrast, although edr2 also shares a similar phenotype with edr1 and edr4-1, it appears that edr4-1 and edr2 function by different mechanisms, as the edr2 mutation enhances edr4-mediated resistance and mildew-induced cell death phenotypes and keg-4 suppresses edr4-1 but not edr2. In addition, similar to the edr1 mutant, edr4-mediated resistance also requires MPK3, MKK4, and MKK5. Taken together, these observations strongly suggest that EDR1 and EDR4 may function in the same pathway to regulate powdery mildew resistance and cell death, and this pathway requires KEG function.
EDR4 encodes an unknown protein, and its biological function is not clear; however, several lines of evidence suggest that EDR4 may contribute to clathrin-mediated pathways. First, EDR4 interacts with CHC2, the clathrin-coated vesicle component in clathrin-mediated pathways (Arabidopsis Interactome Mapping Consortium, 2011). Second, EDR4-GFP shows intracellular movement. Third, edr4 mutants display defects in FM4-64 uptake, similar to the phenotype of chc2 mutants (Kitakura et al., 2011). In plants, several evolutionarily conserved CME components have already been identified, including clathrin, the actin cytoskeleton, dynamin-related proteins, and accessory adaptor proteins (Chen et al., 2011). One possibility is that EDR4 may function as an accessory adaptor protein, which links cargo to clathrin and transports the associated cargo to its destination, although more evidence is needed to support this hypothesis.
Increasing evidence shows that vesicle trafficking plays important roles in plant defense. For example, PEN1-mediated vesicle trafficking is important for nonhost resistance (Nielsen and Thordal-Christensen, 2013), and MIN7-mediated vesicle trafficking contributes to resistance to bacterial pathogens (Nomura et al., 2011). In addition, internalization of FLS2 by vesicle trafficking is also critical for the activation of FLS2-mediated responses (Robatzek et al., 2006). However, PEN1- and MIN7-mediated vesicle trafficking, as well as internalization of FLS2, all play positive roles in plant immunity. By contrast, loss-of-function mutation of EDR4 leads to enhanced disease resistance, suggesting that EDR4 plays a negative role in plant immunity. Similarly, mutations in CHC2, the heavy chain of clathrin, which associates with EDR4, also show enhanced disease resistance to powdery mildew, suggesting a negative role of CHC2 in powdery mildew resistance. Why does EDR4-mediated vesicle trafficking have negative effects on plant immunity? The simplest explanation is that resistance in edr4 results from the mislocalization of EDR1, a negative regulator of defense responses, as EDR4 may help to transport EDR1 to the infection site. Recently, we showed that EDR1 interacts with MKK4/MKK5 and negatively affects the MAPK cascade to fine-tune plant disease resistance (Zhao et al., 2014). In this context, EDR1 transport to the fungal infection site likely keeps the activation of the MAPK pathway at low levels. By contrast, in edr4 mutants, failure to transport EDR1 to the infection site results in MAPK pathway activation to high levels, which leads to the enhanced resistance and cell death in edr4 mutants, similar to edr1. However, it is worthwhile to note that the chc2 mutant did not show strong resistance to powdery mildew, in contrast with the edr4 mutant, indicating that factors in addition to the reduced recruitment of EDR1 to the sites of penetration cause the enhanced disease resistance phenotype of the edr4 mutants.
Similar to EDR1 and EDR4, PEN1 accumulates at penetration sites upon powdery mildew infection. However, PEN1 accumulates at both papillae and plasma membrane at B. graminis f sp hordei penetration sites (Meyer et al., 2009), while EDR1 and EDR4 accumulate on the plasma membrane but not at papillae. Recent work shows that the accumulation of PEN1 on papillae may have no direct effect on penetration resistance; instead, PEN1 may function on the endosome and trans-Golgi network, and recycling mediated by PEN1 could be an important response to fungal infection (Nielsen and Thordal-Christensen, 2013). In this context, recycling of EDR1 by an EDR4-mediated pathway could play an important role in the negative regulation of powdery mildew resistance. Regarding recycling of EDR1, one important question is why plants need to transport a negative regulator of defense to infection sites to suppress immunity. One possible explanation is that plants have developed sophisticated mechanisms to regulate innate immunity, in which many positive and negative regulators are recruited and deployed at the infection sites, thus allowing the plant to fine-tune its defenses in response to different situations. Deployment of the negative regulators to the infection site could prevent inappropriate activation of plant immunity or limit plant immunity to appropriate levels.
KEG plays a critical role in multiple intracellular trafficking processes, including vacuole biogenesis, targeting of membrane-associated proteins to the vacuole, and secretion of apoplastic proteins (Gu and Innes, 2012). Mutation of KEG specifically suppresses edr1-mediated resistance, but how KEG affects edr1-mediated defenses remains unclear. KEG associates with EDR1 and also moves to the site of infection (Gu and Innes, 2012). Although KEG does not physically associate with EDR4, it will be interesting to see whether the movement of KEG to the G. cichoracearum infection sites requires EDR4. Recently, ATL1 was identified as a potential substrate of EDR1. ATL1 interacts with EDR1 in the TGN/EE and constitutively cycles between the plasma membrane and the TGN/EE (Serrano et al., 2014). It would be interesting to examine whether edr4-mediated powdery mildew resistance requires ATL1 and whether EDR4 contributes to the recycling of ATL1.
The resistance and cell death mediated by edr1 and edr4 require intact SA signaling. SA interferes with endocytosis and affects the cycling of proteins at the plasma membrane (Du et al., 2013). Also, the impact of SA on endocytosis involves clathrin, as clathrin mutants show less sensitivity to SA. Interestingly, the effect of SA on CME does not require NPR1, indicating that SA affects CME by different mechanisms and is not involved in transcriptional regulation by NPR1. This suggests that CME has a complicated role in plant immunity; for example, it remains to be tested whether SA affects an EDR4-mediated pathway and whether the interference of CME by SA represents a novel mechanism for SA regulation of plant immunity.
In this study, we demonstrated that EDR4 plays important roles in plant disease resistance and regulates EDR1 focal accumulation by forming a complex to affect plant defenses. However, several questions remain to be answered. Does EDR4 play a role in clathrin-mediated pathways? What are the other proteins that EDR4 helps to relocate? Does EDR4 contribute to CME? Answering these questions will increase our understanding of the functions of clathrin-mediated pathways in plant immunity.
METHODS
Plant Growth and Materials
Arabidopsis thaliana seeds were surface-sterilized and chilled at 4°C for 3 d, then sown on plates containing half-strength Murashige and Skoog medium with 1% sucrose. For phenotyping, plants were grown in a growth room with a 9-h-light/15-h-dark photoperiod, light intensity of 7000 to 8000 lux, and relative humidity of 50 to 60% at 21 to 24°C. Seedlings were transplanted into soil 7 d after germination. For seed set, the plants were placed in a growth room at 21 to 24°C with a long-day photoperiod (16-h-light/8-h-dark cycle). Nicotiana benthamiana plants used for transient expression were grown under the same short-day conditions as Arabidopsis.
Powdery Mildew Infections
Four-week-old plants were inoculated with Golovinomyces cichoracearum, and the number of conidiophores per colony was counted at 6 DAI, as described previously (Wang et al., 2011). Trypan blue staining was used to monitor the fungal structures and dead plant cells (Frye and Innes, 1998). H2O2 accumulation was detected by staining with DAB (Xiao et al., 2005), and callose deposition was detected by aniline blue staining (Pan et al., 2012). Quantitative analysis of callose and H2O2 accumulation followed the method described previously (Zhang et al., 2007; Yoshimoto et al., 2009). The samples were observed and photographed with an Olympus BX53 microscope.
Construction of Double Mutants
Double and triple mutants were created by standard genetic crosses with previously described mutant alleles, including eds1-2 (Bartsch et al., 2006), eds5-1 (Nawrath et al., 2002), pad4-1 (Jirage et al., 1999), npr1-63 (Alonso et al., 2003), coi1-1 (Xie et al., 1998), ein2-1 (Guzmán and Ecker, 1990), sid2-2 (Wildermuth et al., 2001), edr1 (Frye et al., 2001), edr1-2 (Hiruma et al., 2011), edr2 (Tang et al., 2005), keg-4 (Wawrzynska et al., 2008), chc2-1 and chc2-2 (Kitakura et al., 2011), mpk3-1 and mpk6-3 (Bartels et al., 2009), and mkk4-18 and mkk5-18 (Zhao et al., 2014). All the plants used were in the Col-0 background, except for coi1-1 (in the Columbia-6 background). The genotypes of mutants were confirmed by PCR using the primers listed in Supplemental Table 1.
Map-Based Cloning of EDR4
To map EDR4, edr4-1 was crossed with Landsberg erecta to generate a segregating population. Initially, the edr4-1 mutation was mapped between markers CTR1 and CIW14 on chromosome 5. Fine-mapping narrowed the mutation to a 50-kb interval spanning the 3′ end of BAC clone K2A11 (GenBank accession number AB018111.1) and the 5′ end of BAC K18I23 (GenBank accession number AB010692.1). We subsequently amplified and sequenced all 19 predicted genes in this region.
For complementation analysis, a 5.0-kb genomic DNA of At5g05190 from the wild type was amplified and inserted into the pCAMBIA1300 vector. The constructs were verified by sequencing and introduced into Agrobacterium tumefaciens GV3101, then transformed into edr4-1 plants using the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on half-strength Murashige and Skoog medium containing 50 mg/L hygromycin (Sigma-Aldrich). Transformants were transplanted to soil 7 d after germination, and T1 transgenic plants were used for phenotyping.
The primers used are listed in Supplemental Table 1.
Real-Time PCR Analysis and SA Quantification
Real-time PCR to measure transcript accumulation was performed as described (Nie et al., 2012). SA extraction and measurement were performed as described (Gou et al., 2009).
Confocal Microscopy and FRAP
To produce the pEDR4-EDR4-GFP construct, the genomic sequence or coding sequence (CDS) without the stop codon of EDR4 was amplified (primer sequences are listed in Supplemental Table 1) and cloned into the Gateway entry vector pDONR207 using the BP Clonase kit (Invitrogen). The fragment was then cloned into pMDC107 with the LR Clonase kit (Invitrogen). The constructs were introduced into Agrobacterium strain GV3101 and then transformed into the edr4-1 mutant. Similarly, ΔCC-EDR4 (CC domain deletion) was cloned into pEarleyGate302 vector with EDR4 promoter and C-terminal FLAG fusion. The pEDR1-EDR1-GFP (Zhao et al., 2014) and NAG-GFP (Essl et al., 1999) lines were described previously. The TGN/EE and late endosome markers SYP61 and ARA6 (Gu and Innes, 2011, 2012; Serrano et al., 2014) were introduced into EDR4-GFP plants by transformation, respectively. The ER marker line expressing HDEL-mCherry and the cis-Golgi marker line expressing SYP32-mCherry (Nelson et al., 2007; Geldner et al., 2009) were introduced into EDR4-GFP plants by genetic crosses. Confocal imaging was performed on an inverted Zeiss LSM 710 NLO microscope with ZEN 2009 software. Quantification of colocalization was performed as described previously (Herda et al., 2012). To visualize fungal structures, the infected leaves were stained with 0.01 mg/mL propidium iodide for 15 min (Ramonell et al., 2005). Calculating the focal accumulations of EDR1-GFP upon powdery mildew infection was performed as described previously (Underwood and Somerville, 2013).
FRAP assays were performed on a region of focal accumulation or a distal plasma membrane site as described previously (Bhat et al., 2005). Briefly, Arabidopsis leaves were bleached using the 488-nm argon laser line 50 times at 100% intensity. Images were collected 2 min before bleaching and at different time points after bleaching. Fluorescence intensity at each time point for each bleached site was determined using ImageJ software. Relative intensity values were normalized based on focal accumulation before and after bleaching.
FM4-64 Uptake Assay
The FM4-64 uptake assay was performed as described previously, with minor changes (Kitakura et al., 2011). Five-day-old seedlings were incubated with 2 μM FM4-64 for 8 min. The roots of seedlings were observed by confocal microscopy. The mean pixel intensity of the cytosolic side of cells and the adjacent plasma membrane was measured with ImageJ. Then, the quotients of values for the cytosolic side and the plasma membrane were calculated.
Plasmolysis
The plasmolysis assay was performed as described previously (Underwood and Somerville, 2013). For plasmolysis observation, Arabidopsis leaves infected with G. cichoracearum at 24 HAI were soaked in 0.85 M NaCl for 15 min and then observed by confocal microscopy.
MAPK Assay
Total protein from leaves was extracted as described (Liu et al., 2010). The activated MAPKs were detected using anti-pTEpY primary antibodies (Cell Signaling Technology).
Yeast Two-Hybrid Assays
CDSs of EDR1, EDR1 N-terminal domain, and EDR1 C-terminal domain were cloned into pGBKT7 (Zhao et al., 2014). CDSs of KEG, EDR4, and deletion variants of EDR4 were amplified and ligated into pGADT7. The CDS of EDR4 was also cloned into pGBKT7. The plasmids were cotransformed into yeast strain Y190, and positive clones were selected on SD/-Leu/-Trp medium and then verified by PCR amplification of both cotransformed genes. Verified clones were selected on SD/-Leu/-Trp/-His medium with 25 mM 3-amino-1,2,4-triazole (Sigma-Aldrich) and stained with 40 μg/mL X-Gal (Clontech) to determine β-galactosidase activity.
BiFC Analysis
35S-YN-EDR1 was described previously (Zhao et al., 2014). To generate 35S-YC fusion constructs, CDSs of KEG, EDR4, and the deletion variants of EDR4 were cloned into pSY735 (Bracha-Drori et al., 2004) in frame with YFPC. Then, the fusion sequences were amplified and cloned into the binary vector pMDC32. Similarly, to generate 35S-YN-N-CHC2 and 35S-YN-KEG, CDSs of CHC2 (encoding the N-terminal domain amino acids 1 to 1016 of CHC2) and KEG were first cloned into pSY736 in frame with YFPN and then cloned into pMDC32. The plasmid was subsequently introduced into Agrobacterium strain GV3101. Reconstitution of YFP fluorescence was examined by transient coexpression in 4-week-old N. benthamiana by infiltration with Agrobacterium strain GV3101 carrying different YFPN and YFPC pairs, as described (Zhao et al., 2014). Samples were observed by confocal microscopy 2 DAI.
Protein Extraction and Co-IP Assays
For EDR1 and EDR4 Co-IP assays in Arabidopsis, transgenic edr1 plants expressing pEDR1-EDR1-FLAG (Zhao et al., 2014) were crossed with edr4-1 plants expressing pEDR4-EDR4-GFP or 35S-GFP. The leaf samples were from 4-week-old F2 plants expressing both EDR1-FLAG and EDR4-GFP or F1 plants expressing EDR1-FLAG and GFP. For CHC2 and EDR4 Co-IP assays in Arabidopsis, CDS of CHC2 (encoding the N-terminal domain amino acids 1 to 1016 of CHC2) was cloned into pEarleyGate203 vector with the 35S promoter and N-terminal MYC tag. The derived 35S-MYC-N-CHC2 construct was transformed into Col-0 plants, and the transgenic plants were then crossed with EDR4-GFP plants. The leaves from 4-week-old F2 plants expressing both MYC-N-CHC2 and EDR4-GFP or MYC-N-CHC2 were used for Co-IP assays. The protein was extracted using NB1 buffer plus 0.2% (v/v) IGEPAL CA-630 (Sigma-Aldrich) (Liu et al., 2010), and Co-IP assays were performed as described previously (Shi et al., 2013).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: EDR4 (AT5G05190), SYP32 (AT3G24350), SYP61 (AT1G28490), ARA6 (AT3G54840), PEN1 (AT3G11820), EDR1 (AT1G08720), KEG (AT5G13530), CHC2 (AT3G08530), ACT2 (AT3G18780), PR1 (AT2G14610), PR2 (AT3G57260), and PR5 (AT1G75040).
Supplemental Data
Supplemental Figure 1. The Growth Phenotype of edr4-1 Mutants under Short and Long Day Conditions.
Supplemental Figure 2. Quantification of H2O2 and Callose Accumulation in edr4-1 after G. cichoracearum Infection.
Supplemental Figure 3. Quantitative Analysis of Conidiophore Formation on Inoculated Leaves of Wild Type, edr4-1, and Different Double Mutant Plants with G. cichoracearum at 6 DAI.
Supplemental Figure 4. Identification and Complementation of the edr4-1 Mutation.
Supplemental Figure 5. EDR4-GFP Fusion Protein Complemented the edr4-1 Powdery Mildew Resistance Phenotype.
Supplemental Figure 6. Selected Frames of Supplemental Movies 1, 2, and 3.
Supplemental Figure 7. NAG-GFP Proteins Did Not Accumulate at the Penetration Site upon G. cichoracearum Infection.
Supplemental Figure 8. EDR4 and EDR1 Localize to the Plasma Membrane, Not Papillae.
Supplemental Figure 9. The edr2 Mutation Enhances the edr4-Mediated Powdery Mildew Resistance Phenotype.
Supplemental Figure 10. edr4-Mediated Resistance Requires the MAPK Pathway.
Supplemental Figure 11. EDR4 Interacts with N-Terminal Domain of EDR1 in Yeast Two-Hybrid Assays.
Supplemental Figure 12. EDR4 Interacts with EDR1, Not KEG, in Vivo.
Supplemental Figure 13. EDR4 Colocalizes with EDR1.
Supplemental Figure 14. The edr4-1 and chc2-1 Mutations Did Not Affect EDR1-GFP Localization in Uninfected Leaves.
Supplemental Figure 15. The EDR4 Coiled-Coil Domain Is Required for EDR4 Function.
Supplemental Figure 16. EDR1-GFP Localization in an EDR4-CC-Domain Deletion Background.
Supplemental Figure 17. The edr1-1 Mutation Did Not Affect EDR4-GFP Localization in Uninfected and Infected Leaves.
Supplemental Figure 18. FRAP Analysis of EDR1-GFP and EDR4-GFP Focal Accumulation at Powdery Mildew Penetration Sites.
Supplemental Figure 19. The edr1 Mutant Did Not Show Defects in FM4-64 Uptake.
Supplemental Figure 20. The chc2 Mutants Show Increased Accumulation of H2O2 and Callose Deposition and Stronger MPK Activation.
Supplemental Table 1. Primers Used in This Study.
Supplemental Movie 1. EDR4-GFP Intracellular Trafficking in a Root Cell.
Supplemental Movie 2. EDR4-GFP Intracellular Trafficking in an Epidermal Cell.
Supplemental Movie 3. EDR1-GFP Intracellular Trafficking in an Epidermal Cell.
Supplementary Material
Acknowledgments
We thank Roger Innes for edr4-1 seeds and mCherry-SYP61 and ARA6-mCherry plasmids, Jiri Friml for chc2-1 and chc2-2 seeds, Jinlong Qiu for PEN1-GFP seeds, Lei Ge for HDEL-mCherry and NAG-GFP seeds, and the ABRC and Nottingham Arabidopsis Stock Centre for SYP32-mCherry and T-DNA insertion lines. We thank Hui Liang for assistance with confocal microscopy. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDB11020100), the National Basic Research Program of China (Grant 2015CB910200), and the National Natural Science Foundation of China (Grant 31171160).
AUTHOR CONTRIBUTIONS
D.T. and G.W. initiated the project and designed the experiments. G.W., S.L., Y.Z., and W.W. performed the research. D.T., G.W., and Z.K. analyzed data. D.T. and G.W. wrote the article.
Glossary
- PAMP
pathogen-associated molecular pattern
- PTI
PAMP-triggered immunity
- ETI
effector-triggered immunity
- MAPK
mitogen-activated protein kinase
- SA
salicylic acid
- CME
clathrin-mediated endocytosis
- Col-0
Columbia-0
- DAI
days after inoculation
- DAB
3,3′-diaminobenzidine hydrochloride
- JA
jasmonic acid
- TGN/EE
trans-Golgi network/early endosome
- ER
endoplasmic reticulum
- HAI
hours after inoculation
- BiFC
bimolecular fluorescence complementation
- Co-IP
coimmunoprecipitation
- CDS
coding sequence
- FRAP
fluorescence recovery after photobleaching
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels S., Anderson J.C., González Besteiro M.A., Carreri A., Hirt H., Buchala A., Métraux J.P., Peck S.C., Ulm R. (2009). MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 21: 2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch M., Gobbato E., Bednarek P., Debey S., Schultze J.L., Bautor J., Parker J.E. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat R.A., Miklis M., Schmelzer E., Schulze-Lefert P., Panstruga R. (2005). Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl. Acad. Sci. USA 102: 3135–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Bracha-Drori K., Shichrur K., Katz A., Oliva M., Angelovici R., Yalovsky S., Ohad N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40: 419–427. [DOI] [PubMed] [Google Scholar]
- Burkhard P., Stetefeld J., Strelkov S.V. (2001). Coiled coils: A highly versatile protein folding motif. Trends Cell Biol. 11: 82–88. [DOI] [PubMed] [Google Scholar]
- Chen X., Irani N.G., Friml J. (2011). Clathrin-mediated endocytosis: The gateway into plant cells. Curr. Opin. Plant Biol. 14: 674–682. [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D.G., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Collins N.C., Thordal-Christensen H., Lipka V., Bau S., Kombrink E., Qiu J.L., Hückelhoven R., Stein M., Freialdenhoven A., Somerville S.C., Schulze-Lefert P. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Aniento F., Hwang I., Robinson D.G., Mravec J., Stierhof Y.D., Friml J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17: 520–527. [DOI] [PubMed] [Google Scholar]
- Du Y., Tejos R., Beck M., Himschoot E., Li H., Robatzek S., Vanneste S., Friml J. (2013). Salicylic acid interferes with clathrin-mediated endocytic protein trafficking. Proc. Natl. Acad. Sci. USA 110: 7946–7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essl D., Dirnberger D., Gomord V., Strasser R., Faye L., Glössl J., Steinkellner H. (1999). The N-terminal 77 amino acids from tobacco N-acetylglucosaminyltransferase I are sufficient to retain a reporter protein in the Golgi apparatus of Nicotiana benthamiana cells. FEBS Lett. 453: 169–173. [DOI] [PubMed] [Google Scholar]
- Frye C.A., Innes R.W. (1998). An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10: 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye C.A., Tang D., Innes R.W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. USA 98: 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadeyne A., et al. (2014). The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156: 691–704. [DOI] [PubMed] [Google Scholar]
- Geldner N., Dénervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.D., Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou M., Su N., Zheng J., Huai J., Wu G., Zhao J., He J., Tang D., Yang S., Wang G. (2009). An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J. 60: 757–770. [DOI] [PubMed] [Google Scholar]
- Gu Y., Innes R.W. (2011). The KEEP ON GOING protein of Arabidopsis recruits the ENHANCED DISEASE RESISTANCE1 protein to trans-Golgi network/early endosome vesicles. Plant Physiol. 155: 1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Innes R.W. (2012). The KEEP ON GOING protein of Arabidopsis regulates intracellular protein trafficking and is degraded during fungal infection. Plant Cell 24: 4717–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán P., Ecker J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herda S., Raczkowski F., Mittrücker H.W., Willimsky G., Gerlach K., Kühl A.A., Breiderhoff T., Willnow T.E., Dörken B., Höpken U.E., Rehm A. (2012). The sorting receptor Sortilin exhibits a dual function in exocytic trafficking of interferon-γ and granzyme A in T cells. Immunity 37: 854–866. [DOI] [PubMed] [Google Scholar]
- Hiruma K., Nishiuchi T., Kato T., Bednarek P., Okuno T., Schulze-Lefert P., Takano Y. (2011). Arabidopsis ENHANCED DISEASE RESISTANCE 1 is required for pathogen-induced expression of plant defensins in nonhost resistance, and acts through interference of MYC2-mediated repressor function. Plant J. 67: 980–992. [DOI] [PubMed] [Google Scholar]
- Hong Z., Bednarek S.Y., Blumwald E., Hwang I., Jurgens G., Menzel D., Osteryoung K.W., Raikhel N.V., Shinozaki K., Tsutsumi N., Verma D.P. (2003). A unified nomenclature for Arabidopsis dynamin-related large GTPases based on homology and possible functions. Plant Mol. Biol. 53: 261–265. [DOI] [PubMed] [Google Scholar]
- Hückelhoven R., Panstruga R. (2011). Cell biology of the plant-powdery mildew interaction. Curr. Opin. Plant Biol. 14: 738–746. [DOI] [PubMed] [Google Scholar]
- Jirage D., Tootle T.L., Reuber T.L., Frost L.N., Feys B.J., Parker J.E., Ausubel F.M., Glazebrook J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96: 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kitakura S., Vanneste S., Robert S., Löfke C., Teichmann T., Tanaka H., Friml J. (2011). Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23: 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S., André A., Edwards H., Ehrhardt D., Somerville S. (2005). Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J. 44: 516–529. [DOI] [PubMed] [Google Scholar]
- Lee H.Y., Bowen C.H., Popescu G.V., Kang H.G., Kato N., Ma S., Dinesh-Kumar S., Snyder M., Popescu S.C. (2011). Arabidopsis RTNLB1 and RTNLB2 Reticulon-like proteins regulate intracellular trafficking and activity of the FLS2 immune receptor. Plant Cell 23: 3374–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka V., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183. [DOI] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903. [DOI] [PubMed] [Google Scholar]
- Lu D., Wu S., Gao X., Zhang Y., Shan L., He P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H.T., Boucrot E. (2011). Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12: 517–533. [DOI] [PubMed] [Google Scholar]
- Meyer D., Pajonk S., Micali C., O’Connell R., Schulze-Lefert P. (2009). Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 57: 986–999. [DOI] [PubMed] [Google Scholar]
- Nawrath C., Heck S., Parinthawong N., Métraux J.P. (2002). EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14: 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nie H., Zhao C., Wu G., Wu Y., Chen Y., Tang D. (2012). SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol. 158: 1847–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M.E., Thordal-Christensen H. (2013). Transcytosis shuts the door for an unwanted guest. Trends Plant Sci. 18: 611–616. [DOI] [PubMed] [Google Scholar]
- Nomura K., Debroy S., Lee Y.H., Pumplin N., Jones J., He S.Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223. [DOI] [PubMed] [Google Scholar]
- Nomura K., Mecey C., Lee Y.N., Imboden L.A., Chang J.H., He S.Y. (2011). Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 10774–10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Liu S., Tang D. (2012). HPR1, a component of the THO/TREX complex, plays an important role in disease resistance and senescence in Arabidopsis. Plant J. 69: 831–843. [DOI] [PubMed] [Google Scholar]
- Panstruga R., Dodds P.N. (2009). Terrific protein traffic: The mystery of effector protein delivery by filamentous plant pathogens. Science 324: 748–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntervoll P., et al. (2003). ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 31: 3625–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramonell K., Berrocal-Lobo M., Koh S., Wan J., Edwards H., Stacey G., Somerville S. (2005). Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol. 138: 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S. (2014). Endocytosis: At the crossroads of pattern recognition immune receptors and pathogen effectors. In Applied Plant Cell Biology, Nick P., Opatrny Z., eds (Berlin: Springer: ), pp. 273–297. [Google Scholar]
- Robatzek S., Chinchilla D., Boller T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M., Delgadillo M.O., Zouhar J., Reynolds G.D., Pennington J.G., Jiang L., Liljegren S.J., Stierhof Y.D., De Jaeger G., Otegui M.S., Bednarek S.Y., Rojo E. (2013). MTV1 and MTV4 encode plant-specific ENTH and ARF GAP proteins that mediate clathrin-dependent trafficking of vacuolar cargo from the trans-Golgi network. Plant Cell 25: 2217–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P., Panstruga R. (2011). A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 16: 117–125. [DOI] [PubMed] [Google Scholar]
- Serrano I., Gu Y., Qi D., Dubiella U., Innes R.W. (2014). The Arabidopsis EDR1 protein kinase negatively regulates the ATL1 E3 ubiquitin ligase to suppress cell death. Plant Cell 26: 4532–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Shen Q., Qi Y., Yan H., Nie H., Chen Y., Zhao T., Katagiri F., Tang D. (2013). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25: 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallek T., Beck M., Ben Khaled S., Salomon S., Bourdais G., Schellmann S., Robatzek S. (2013). ESCRT-I mediates FLS2 endosomal sorting and plant immunity. PLoS Genet. 9: e1004035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M., Dittgen J., Sánchez-Rodríguez C., Hou B.-H., Molina A., Schulze-Lefert P., Lipka V., Somerville S. (2006). Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Ade J., Frye C.A., Innes R.W. (2005). Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. Plant J. 44: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Ade J., Frye C.A., Innes R.W. (2006). A mutation in the GTP hydrolysis site of Arabidopsis dynamin-related protein 1E confers enhanced cell death in response to powdery mildew infection. Plant J. 47: 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G., Boudsocq M., Sheen J. (2011). Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 14: 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood W., Somerville S.C. (2008). Focal accumulation of defences at sites of fungal pathogen attack. J. Exp. Bot. 59: 3501–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood W., Somerville S.C. (2013). Perception of conserved pathogen elicitors at the plasma membrane leads to relocalization of the Arabidopsis PEN3 transporter. Proc. Natl. Acad. Sci. USA 110: 12492–12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese P., Nakagami H., Bluhm B., Abuqamar S., Chen X., Salmeron J., Dietrich R.A., Hirt H., Mengiste T. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18: 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorwerk S., Schiff C., Santamaria M., Koh S., Nishimura M., Vogel J., Somerville C., Somerville S. (2007). EDR2 negatively regulates salicylic acid-based defenses and cell death during powdery mildew infections of Arabidopsis thaliana. BMC Plant Biol. 7: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Nishimura M.T., Zhao T., Tang D. (2011). ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 68: 74–87. [DOI] [PubMed] [Google Scholar]
- Wawrzynska A., Christiansen K.M., Lan Y., Rodibaugh N.L., Innes R.W. (2008). Powdery mildew resistance conferred by loss of the ENHANCED DISEASE RESISTANCE1 protein kinase is suppressed by a missense mutation in KEEP ON GOING, a regulator of abscisic acid signaling. Plant Physiol. 148: 1510–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
- Xiao S., Calis O., Patrick E., Zhang G., Charoenwattana P., Muskett P., Parker J.E., Turner J.G. (2005). The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J. 42: 95–110. [DOI] [PubMed] [Google Scholar]
- Xie D.-X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Yao C., Wu Y., Nie H., Tang D. (2012). RPN1a, a 26S proteasome subunit, is required for innate immunity in Arabidopsis. Plant J. 71: 1015–1028. [DOI] [PubMed] [Google Scholar]
- Yi M., Valent B. (2013). Communication between filamentous pathogens and plants at the biotrophic interface. Annu. Rev. Phytopathol. 51: 587–611. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185. [DOI] [PubMed] [Google Scholar]
- Zhao C., Nie H., Shen Q., Zhang S., Lukowitz W., Tang D. (2014). EDR1 physically interacts with MKK4/MKK5 and negatively regulates a MAP kinase cascade to modulate plant innate immunity. PLoS Genet. 10: e1004389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D.G., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.