A trihelix transcription factor downstream of a MAP kinase cascade negatively regulates plant immune gene expression.
Abstract
Proper control of immune-related gene expression is crucial for the host to launch an effective defense response. Perception of microbe-associated molecular patterns (MAMPs) induces rapid and profound transcriptional reprogramming via unclear mechanisms. Here, we show that ASR3 (ARABIDOPSIS SH4-RELATED3) functions as a transcriptional repressor and plays a negative role in regulating pattern-triggered immunity (PTI) in Arabidopsis thaliana. ASR3 belongs to a plant-specific trihelix transcription factor family for which functional studies are lacking. MAMP treatments induce rapid phosphorylation of ASR3 at threonine 189 via MPK4, a mitogen-activated protein kinase that negatively regulates PTI responses downstream of multiple MAMP receptors. ASR3 possesses transcriptional repressor activity via its ERF-associated amphiphilic repression motifs and negatively regulates a large subset of flg22-induced genes. Phosphorylation of ASR3 by MPK4 enhances its DNA binding activity to suppress gene expression. Importantly, the asr3 mutant shows enhanced disease resistance to virulent bacterial pathogen infection, whereas transgenic plants overexpressing the wild-type or phospho-mimetic form of ASR3 exhibit compromised PTI responses. Our studies reveal a function of the trihelix transcription factors in plant innate immunity and provide evidence that ASR3 functions as a transcriptional repressor regulated by MAMP-activated MPK4 to fine-tune plant immune gene expression.
INTRODUCTION
Plants and animals are at constant risk for infections from various microorganisms in their natural habitats. In contrast to animals, plants lack specialized mobile immune cells and the adaptive immune system. In addition to preformed physical barriers, sessile plants largely rely on the innate immune system to launch prompt defense responses in situ to fend off potential infections. Plant innate immunity has been classified as a two-tier defense system (Jones and Dangl, 2006; Dodds and Rathjen, 2010). Perception of pathogen- or microbe-associated molecular patterns (MAMPs) by plasma membrane-resident pattern recognition receptors (PRRs) activates the first line of innate immunity, termed pattern-triggered immunity (PTI), mainly to ward off the attacks from host nonadapted pathogens (Schwessinger and Ronald, 2012; Macho and Zipfel, 2014). Host-adapted pathogens deploy various virulence factors to interfere with PTI and establish successful infections (Dou and Zhou, 2012; Xin and He, 2013). Host plants further evolved the intracellular receptors often encoded by nucleotide binding domain leucine-rich repeat proteins, also named disease resistance proteins, to recognize virulence effectors or sense effector-mediated perturbations of host targets and elicit the second tier of defense responses, termed effector-triggered immunity (Elmore et al., 2011; Gassmann and Bhattacharjee, 2012; Qi and Innes, 2013).
Although the full repertoire of MAMPs perceived by plants remains unknown, several MAMPs, including bacterial flagellin, lipopolysaccharide (LPS), peptidoglycan, elongation factor Tu (EF-Tu), and fungal chitin, have been well characterized to elicit various defense responses in plant cells (Boller and Felix, 2009; Schwessinger and Ronald, 2012). A 22-amino acid peptide corresponding to a region near the N terminus of flagellin, flg22, is perceived by Arabidopsis thaliana PRR FLS2 (FLAGELLIN-SENSING2), a leucine-rich repeat receptor-like kinase (LRR-RLK) that initiates immune signaling by instantaneous heterodimerization with another LRR-RLK, BAK1 (BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1) (Chinchilla et al., 2007; Heese et al., 2007; Sun et al., 2013). BIK1 (BOTRYTIS-INDUCED KINASE1), a receptor-like cytoplasmic kinase, and its homolog PBL1 (PBS1-LIKE1) constitutively associate with FLS2 and BAK1 and are rapidly phosphorylated and released from the receptor complex upon flg22 perception (Lu et al., 2010; Zhang et al., 2010). BIK1 directly phosphorylates plasma membrane-resident NADPH oxidase RESPIRATORY BURST OXIDASE HOMOLOG D for transient production of reactive oxygen species (ROS), an early event triggered by multiple MAMPs (Kadota et al., 2014; Li et al., 2014b). The FLS2 complex is also subjected to layered negative regulations via various mechanisms to fine-tune PTI responses. Two closely related plant U-box E3 ubiquitin ligases PUB12 and PUB13 are recruited to the FLS2 complex via interaction with BAK1 upon flg22 perception and directly ubiquitinate FLS2, which leads to flg22-induced FLS2 degradation (Lu et al., 2011). BAK1-INTERACTING RLK2, an LRR-RLK without detectable kinase activity, constitutively interacts with BAK1 and negatively regulates flg22-induced FLS2-BAK1 heterodimerization (Halter et al., 2014). In addition, PROTEIN PHOSPHATASE 2A regulates the activation of PRR complexes, likely by modulating the phosphorylation status of BAK1 (Segonzac et al., 2014). The calcium-dependent protein kinase CPK28 phosphorylates BIK1 and contributes to BIK1 turnover, thereby negatively regulating PTI signaling (Monaghan et al., 2014).
Rapid activation of mitogen-activated protein kinase (MAPK) cascades has been observed upon perception of various MAMPs (Pitzschke et al., 2009; Rodriguez et al., 2010; Meng and Zhang, 2013). A typical MAPK cascade is composed of three sequentially activated kinases consisting of a MAPK kinase kinase (MAP3K or MEKK), a MAPK kinase (MAP2K or MKK), and a MAPK, which links upstream signals to its downstream targets. Arabidopsis MAPK signaling pathways, with one branch consisting of a MEKK-MKK4/MKK5-MPK3/MPK6 cascade as the positive regulators and another branch of a MEKK1-MKK1/MKK2-MPK4 cascade as the negative regulators, have been implicated in plant immunity (Meng and Zhang, 2013). In general, MAPKs regulate gene expression through phosphorylation of downstream transcription factors. WRKY and ERF transcription factors are two major defense-related transcription factors in plants, and some of them have been shown to be phosphorylated by MAPKs. ERF104 is a substrate of MPK6 activated by flg22, and phosphorylation induces its release from MPK6 to regulate target gene expression (Bethke et al., 2009). WRKY33 is phosphorylated by MPK3 and MPK6 in vivo upon Botrytis cinerea infection, thereby inducing camalexin biosynthetic gene expression to promote phytoalexin biosynthesis (Mao et al., 2011). In addition, ERF6 is phosphorylated by MPK3 and MPK6 and plays an important role in plant defense against fungal pathogen (Meng et al., 2013). It remains unknown whether other types of transcription factors can be regulated by MAPKs in plant immune responses.
To elucidate the signaling networks orchestrating immune gene activation, we developed a genetic screen with an ethyl methanesulfonate-mutagenized population of Arabidopsis pFRK1:LUC transgenic plants expressing a firefly luciferase reporter gene under the control of the FRK1 (FLG22-INDUCED RECEPTOR-LIKE KINASE1) promoter. FRK1 is a specific and early immune-responsive gene activated by multiple MAMPs (Asai et al., 2002; He et al., 2006). A series of mutants with altered FRK1 promoter activity upon flg22 treatment or inoculation with nonpathogenic Pseudomonas syringae pv tomato (Pst) DC3000 type III secretion mutant hrcC were identified and named as Arabidopsis genes governing immune gene expression (aggies) (Li et al., 2014a; Feng et al., 2015). During the map-based cloning of the aggie1 mutant, a collection of homozygous Salk T-DNA insertion mutants for individual genes located in a 110-kb region on Chromosome 2 was analyzed for disease resistance to the virulent bacterium Pst DC3000 infection and flg22-induced FRK1 expression. Interestingly, a knockout line with a T-DNA insertion at At2g33550 exhibited enhanced flg22-induced FRK1 expression and elevated resistance to virulent bacterial pathogens. At2g33550 encodes a plant-specific protein with no significant similarity to any known proteins. The predicted gene product of At2g33550 has two putative nuclear localization signals, a putative trihelix DNA binding motif at its N terminus and a loosely conserved coiled-coil motif at its C terminus. Based on these features, At2g33550 was classified to the SH4 (Shattering 4; a quantitative trait locus controlling grain shattering in rice [Oryza sativa]) clade of the trihelix transcription factor family (Kaplan-Levy et al., 2012). We named this gene ASR3 for ARABIDOPSIS SH4-RELATED3. SH4 was identified as a dominant gene controlling seed shattering in the wild species of rice. The domesticated rice cultivars carry mutations in this gene, thus eliminating seed shattering (Li et al., 2006; Lin et al., 2007). Here, we report that Arabidopsis ASR3 is rapidly phosphorylated upon MAMP treatment downstream of MPK4. Our results provide genetic evidence that a trihelix family transcription factor functions in plant biotic stresses and identify a target of MPK4, which acts as a transcriptional repressor to negatively regulate plant innate immunity and immune gene expression.
RESULTS
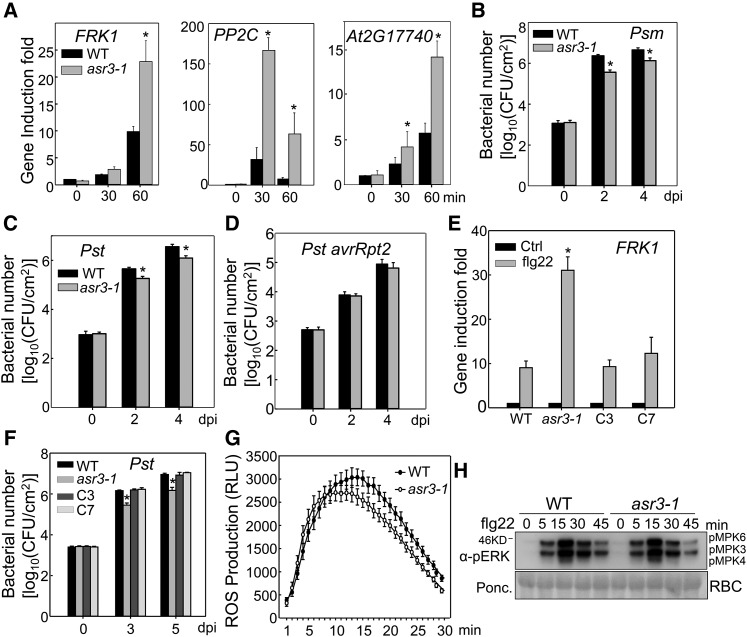
The asr3 Mutant Shows Enhanced Immune Gene Activation and Disease Resistance
We initially isolated two ASR3 T-DNA insertion lines asr3-1 (SALK_112571C) and asr3-2 (SALK_047951C) (Supplemental Figure 1A). Genotyping and RT-PCR analysis confirmed that asr3-1 is a knockout mutant with no detectable full-length transcript. However, asr3-2, with the T-DNA insertion at the stop codon, exhibited the same level of full-length transcript as wild-type plants (Supplemental Figure 1B). Accordingly, the asr3-1 mutant was used for further studies. The asr3-1 mutant displayed an elevated expression of several MAMP marker genes, including FRK1, PP2C (protein phosphatase 2C family protein), and At2G17740 after flg22 treatment compared with wild-type plants (Figure 1A). In addition, the asr3-1 mutant was more resistant to infections by virulent bacteria Pst DC3000 and P. syringae pv maculicola (Psm) ES4326 than were wild-type plants, as indicated by a more than 5-fold smaller bacterial population in the mutant compared with that in wild-type plants 2 and 4 d postinoculation (dpi) (Figures 1B and 1C). The asr3-1 mutant displayed unaltered disease resistance to the avirulent strain Pst DC3000 carrying effector avrRpt2 (Figure 1D). To confirm that the asr3-1 phenotypes were caused by the mutation in the ASR3 gene, we introduced HA epitope-tagged ASR3 under the control of its native promoter (2.1 kb upstream of the translational start site) into the asr3-1 mutant. Two independent complementation lines with similar detectable ASR3 expression (Supplemental Figure 1C) restored the flg22-induced FRK1 induction to the wild-type level (Figure 1E). In addition, the complementation lines restored the susceptibility to Pst infection of the asr3-1 mutant to the wild-type level (Figure 1F). Together, these results suggest that ASR3 negatively regulates immune gene expression and disease resistance to virulent bacterial pathogens. However, flg22-induced ROS burst and MAPK activation did not show a detectable difference in wild-type and asr3-1 plants (Figures 1G and 1H), suggesting that ASR3 functions either downstream or independently of MAPK activation and ROS production in FLS2 signaling.
Figure 1.
The asr3 Mutant Displays Enhanced Disease Resistance and Immune Gene Activation.
(A) The flg22-induced marker gene expression in the wild type and asr3-1 mutant. Ten-day-old seedlings were treated with 100 nM flg22 for 30 and 60 min for qRT-PCR analysis.
(B) and (C) The asr3-1 mutant is more resistant to Psm and Pst infections. Four-week-old wild-type and asr3-1 mutant plants were hand-inoculated with bacterial suspension at a density of 5 × 105 cfu/mL, and bacterial population was quantified at 0, 2, and 4 dpi.
(D) Bacterial growth of avirulent strain Pst avrRpt2.
(E) ASR3 complements asr3-1 mutant for FRK1 gene induction. Ten-day-old seedlings from Col-0 wild type, asr3-1 mutant, and complementation lines C3 and C7 were treated with 100 nM flg22 for 60 min for qRT-PCR analysis.
(F) ASR3 complements asr3-1 mutant in Pst-mediated pathogen infection. Four-week-old plants were spray-inoculated with Pst at 108 cfu/mL, and bacterial counting was performed at 0, 3, and 5 dpi.
(G) flg22-induced ROS burst in the wild type and asr3-1 mutant. Leaf discs from 5-week-old plants were treated with water or 100 nM flg22 over 30 min. The data are shown as means ± se from 24 leaf discs.
(H) flg22-induced MAPK activation in the wild type and asr3-1 mutant. Ten-day-old seedlings were treated with 100 nM flg22 and collected at the indicted time points. MAPK activation was analyzed by immunoblot with α-pERK antibody (top panel), and the protein loading is shown by Ponceau S staining for Rubisco (RBC) (bottom panel).
The data in (A) to (F) are shown as mean ± sd from three independent repeats, and the asterisk indicates a significant difference with a Student’s t test (P < 0.05) when compared with the wild type. The above experiments were repeated three times with similar results.
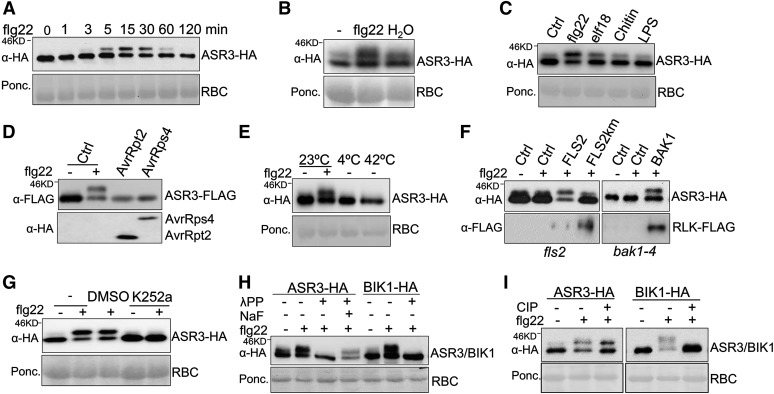
The flg22 Perception Induces ASR3 Phosphorylation
To reveal the underlying mechanism of ASR3 in plant immune signaling, we ectopically expressed ASR3 in Arabidopsis protoplasts and transgenic plants. Interestingly, when expressed in protoplasts, the ASR3 protein displayed a rapid and dynamic mobility shift upon flg22 treatment as detected by immunoblotting (Figure 2A). The mobility shift of ASR3 protein could be detected as early as 3 min, peaked at 15 min, gradually decreased to 30 min, and then returned to the unshifted form at 2 h of flg22 treatment. The flg22-induced ASR3 mobility shift was also detected in ASR3-HA transgenic plants (Figure 2B). In addition to flg22, other MAMPs, including elf18, chitin, and LPS, also induced ASR3 mobility shift, although to a lesser extent (Figure 2C). By contrast, ectopic expression of bacterial effector AvrRpt2 or AvrRps4 did not induce a detectable mobility shift of ASR3 in protoplasts (Figure 2D). In addition, abiotic stresses, such as treatment with low temperature (4°C) or excessive heat (42°C), did not induce a demonstrable ASR3 mobility shift (Figure 2E). Thus, the mobility shift of ASR3 seems to be specifically induced in plant PTI signaling.
Figure 2.
flg22 Perception Induces ASR3 Phosphorylation.
(A) ASR3 displays a mobility shift upon flg22 treatment. Protoplasts were transfected with HA-tagged ASR3 for 12 h and treated with 100 nM flg22 for the indicated amounts of time. RBC, Rubisco.
(B) The flg22 treatment induces ASR3 mobility shift in 35S:ASR3-HA transgenic plants. The leaves from 4-week-old ASR3-HA transgenic plants were hand-inoculated with 100 nM flg22 or water for 30 min. Control “-” denotes the leaves without inoculation.
(C) Multiple MAMPs trigger ASR3 mobility shift. Protoplasts were transfected with ASR3 for 12 h and treated with 100 nM flg22, 100 nM elf18, 50 μg/mL chitin, or 5 μg/mL LPS for 15 min.
(D) AvrRpt2 or AvrRps4 does not induce ASR3 mobility shift. Protoplasts were cotransfected with ASR3-FLAG and AvrRpt2-HA or AvrRps4-HA.
(E) Cold or heat treatment does not induce ASR3 mobility shift. Protoplasts were transfected with ASR3-HA for 8 h at 23°C and incubated for additional 2 h at 23, 4, or 42°C.
(F) The flg22-induced ASR3 mobility shift depends on functional FLS2/BAK1 receptor complex. Protoplasts isolated from fls2 or bak1-4 mutants were cotransfected with ASR3-HA and empty vector control (Ctrl), FLAG-tagged FLS2, FLS2 kinase mutant (FLS2km), or BAK1.
(G) Kinase inhibitor K252a blocks flg22-induced ASR3 mobility shift. K252a was applied 30 min before 100 nM flg22 treatment. The controls were nontreatment (-) or solvent (DMSO).
(H) λPP removes the flg22-induced mobility shift of ASR3. Protein extracts from protoplasts transfected with ASR3-HA or BIK1-HA were treated with λPP following the standard protocol. NaF, a phosphatase inhibitor, compromised λPP phosphatase activity.
(I) CIP treatment abolishes the mobility shift of BIK1 but not ASR3. Protoplasts were transfected with ASR3-HA or BIK1-HA and treated with or without 100 nM flg22 for 30 min.
The above experiments were repeated three times with similar results.
The flg22-induced ASR3 mobility shift was not observed in fls2 and bak1 mutant protoplasts (Figure 2F). Importantly, expression of FLS2 in fls2 mutant protoplasts or BAK1 in bak1 mutant protoplasts restored the flg22-induced ASR3 mobility shift, suggesting the requirement of a functional flagellin receptor complex (Figure 2F). By contrast, expression of the FLS2 kinase-inactive mutant, FLS2Km, failed to complement the ASR3 mobility shift in the fls2 mutant, indicating that the FLS2 kinase activity is required for the ASR3 mobility shift (Figure 2F). Consistent with those results, the ASR3 mobility shift was blocked in the presence of a general kinase inhibitor K252a (Figure 2G). Since K252a likely interferes with multiple phosphorylation steps in FLS2 signaling, we used phosphatase treatments of ASR3 proteins from flg22-induced samples to examine whether the mobility shift was caused by phosphorylation. Treatment of ASR3 proteins with lambda protein phosphatase (λPP), a Mn2+-dependent protein phosphatase with activity toward phosphorylated serine, threonine, and tyrosine residues, was able to completely remove the flg22-induced mobility shift of ASR3. NaF, a phosphatase inhibitor, compromised λPP phosphatase activity (Figure 2H). Treatment with calf intestinal alkaline phosphatase (CIP), a phosphatase that preferentially alters phosphotyrosine residues, did not significantly affect the flg22-induced ASR3 mobility shift, although it completely restored the mobility of phosphorylated BIK1 to that of the unmodified form (Figure 2I) (Lu et al., 2010). This is consistent with the reports that BIK1 possesses tyrosine phosphorylation activity (Xu et al., 2013; Lin et al., 2014). These results imply that ASR3 undergoes phosphorylation, likely on serine and/or threonine residues, upon flg22 perception.
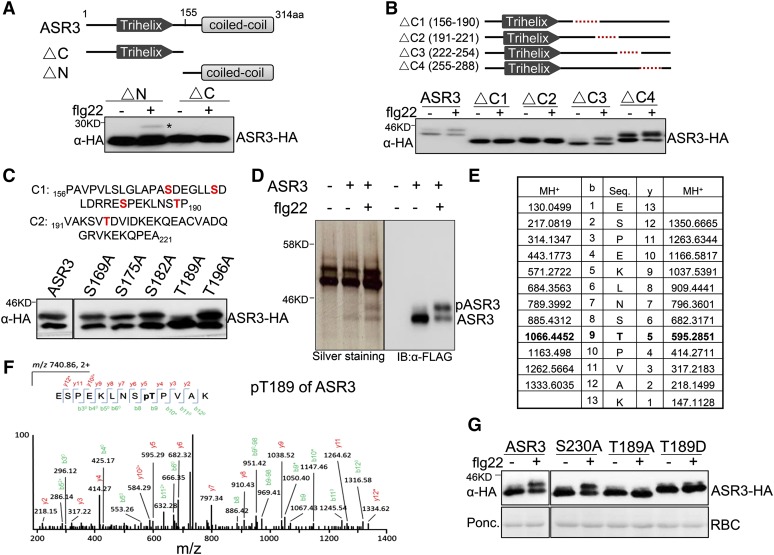
Flg22 Induces in Vivo ASR3 Phosphorylation at Thr-189
To identify the flg22-induced in vivo ASR3 phosphorylation site, we performed a series of deletion/mutation and mass spectrometry analyses of ASR3. The N-terminal deletion (ΔN), but not the C-terminal deletion (ΔC), still exhibited a mobility shift upon flg22 treatment (Figure 3A), implying that the phosphorylation underlying the mobility shift occurs at the C terminus of ASR3. To map the phosphorylated residues, we generated a series of truncation mutants each with ∼35 amino acids deleted, which in total span the entire C-terminal half of ASR3 (Figure 3B). Interestingly, ΔC1 (156 to 190 amino acids) or ΔC2 (191 to 221 amino acids), but not ΔC3 (222 to 254 amino acids) nor ΔC4 (255 to 288 amino acids), completely blocked the flg22-induced mobility shift (Figure 3B). There are multiple serine (S) and threonine (T) residues in the C1 and C2 regions. We further mutated some of the individual serine or threonine residues to alanine (A) in these regions. Remarkably, the T189A mutant, but not the S169A, S175A, S182A, or T196A mutant, blocked the flg22-induced ASR3 mobility shift (Figure 3C). Deletion of the C2 (191 to 221) region may impose a conformational change on the nearby Thr-189 residue, there by abolishing flg22-induced mobility shift. Taken together, the data suggest that Thr-189 is an important site of ASR3 phosphorylation in response to flg22 treatment.
Figure 3.
The flg22-Induced ASR3 Phosphorylation Occurs at Thr-189.
(A) The flg22-induced ASR3 phosphorylation occurs at its C-terminal half. The top panel shows schematic diagrams of deletion mutants with the putative trihelix and coiled-coil motifs labeled. The deletion was made at the 155-amino acid position of ASR3.
(B) C1 and C2 regions are required for flg22-induced ASR3 phosphorylation. The top panel shows schematic diagrams with the dashed red line indicating the deleted sequence. Protoplasts were transfected with different ASR3 truncation variants for 12 h and treated with 100 nM flg22 for 15 min.
(C) ASR3T189A mutation abolishes flg22-induced ASR3 phosphorylation. Protoplasts were transfected with ASR3 point mutation variants and treated with 100 nM flg22 for 15 min. The amino acid sequences and potential phosphorylation residues (in red) in C1 and C2 regions are listed on the top.
(D) Silver staining and immunoblot analysis of immunoprecipitated ASR3-FLAG from Arabidopsis protoplasts. Protoplasts expressing ASR3-FLAG were treated with or without 100 nM flg22 for 15 min. Protein lysis was subjected to immunoprecipitation with α-FLAG antibody followed by SDS-PAGE sliver staining or immunoblotting with an α-FLAG-HRP antibody.
(E) and (F) LC-MS/MS analysis showing that ASR3 Thr-189 is phosphorylated. The sequence of a doubly charged peptide ion at m/z 740.86 matches ESPEKLNSpTPVAK of ASR3.
(G) Thr-189 is an essential phosphorylation site induced by flg22 treatment. Different ASR3 mutants were expressed in protoplasts, treated with flg22, and detected by immunoblotting with an α-HA antibody.
The above assays, except the mass spectrometry analysis, were repeated at least three times with similar results.
Furthermore, we performed a liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis with the FLAG epitope-tagged ASR3 protein expressed in Arabidopsis protoplasts with or without flg22 treatment (Figure 3D). The ASR3 proteins were immunoprecipitated with α-FLAG agarose and subjected to SDS-PAGE and silver staining analysis. Compared with the vector control-transfected protoplasts, a discrete band with a molecular mass of ∼35 kD was observed in the ASR3-transfected protoplast samples (Figure 3D). Another band with a molecular mass of ∼38 kD could be detected upon flg22 treatment. Immunoblot analysis using an α-FLAG antibody with a small aliquot of the same protein samples as used for silver staining confirmed that these bands were likely unphosphorylated and phosphorylated ASR3. We enriched the phosphorylated peptides and analyzed them by LC-MS/MS analysis. The LC-MS/MS analysis of the upper band derived from flg22-treated samples revealed that 13 peptides contained Thr-189 as the phosphorylation site (Figures 3E and 3F; Supplemental Table 1). The data in large part support our deletion and mutation analysis findings that the ASR3T189A mutant no longer exhibited the mobility shift upon flg22 treatment. There were three peptides containing Ser-230 as a high confidence phosphorylation site (Supplemental Table 1). However, the ASR3S230A mutation did not block the flg22-induced ASR3 mobility shift (Figure 3G). In addition, the ASR3 Thr-189 phospho-mimetic mutant (ASR3T189D) with a substitution of aspartic acid (D) showed a constitutive mobility shift in the absence of flg22 treatment (Figure 3G). Taken together, the data indicate that Thr-189 is a major phosphorylation residue of ASR3 induced by flg22 treatment.
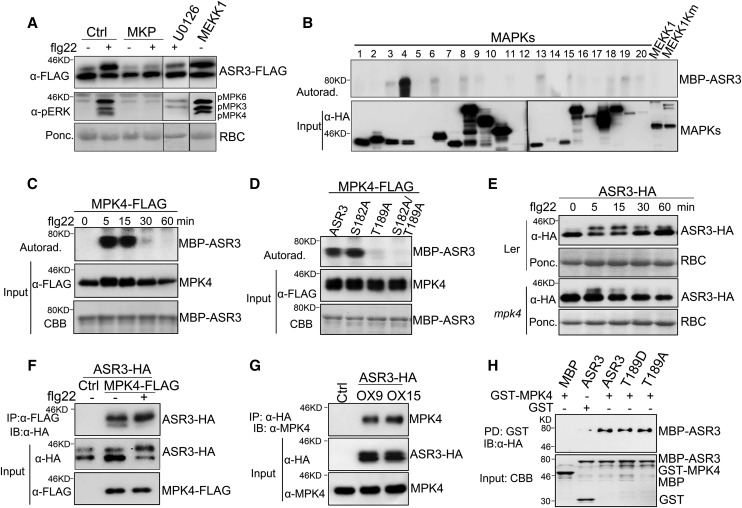
ASR3 Is a Substrate of MPK4
We further examined the signaling events that potentially regulate ASR3 phosphorylation. Various chemical inhibitors, which could specifically interfere with distinct early defense responses following receptor complex activation, including calcium influx, ROS burst, or MAPK activation, were used. Treatment with Ca2+ channel inhibitors lanthanum chloride (LaCl3), gallium chloride (GaCl3), or ruthenium red (RR) or the NADPH oxidase inhibitor diphenylene iodonium (DPI) did not affect flg22-induced ASR3 phosphorylation (Supplemental Figure 2). However, treatment with MAPK pathway inhibitor U0126 markedly reduced flg22-triggered ASR3 phosphorylation. In addition, coexpression of the MAPK phosphatase MKP almost completely abolished the ASR3 mobility shift (Figure 4A). These data suggest that the MAPK cascade(s) is required for flg22-triggered ASR3 phosphorylation. Ectopic expression of the full-length MEKK1, the most upstream kinase in the flg22-activated MAPK cascade, was sufficient to induce ASR3 phosphorylation in the absence of flg22 treatment (Figure 4A). Notably, the flg22-induced phosphorylation residue Thr-189 is typical of MAPK phosphorylation sites in that it is followed by a proline (P) residue. Thus, the data suggest that ASR3 may function as a direct target of certain MAPKs in immune signaling. The Arabidopsis genome encodes 20 MPK genes. To discern which MAPK(s) could phosphorylate ASR3, we screened individual Arabidopsis MAPKs for the ability to phosphorylate ASR3. The HA-tagged MAPKs were expressed in protoplasts, activated by flg22 treatment, and immunoprecipitated for an in vitro kinase assay using ASR3 protein fused to maltose binding protein (MBP) as a substrate. Importantly, flg22-activated MPK4 strongly phosphorylated ASR3 in vitro (Figure 4B).
Figure 4.
MPK4 Phosphorylates and Interacts with ASR3.
(A) MAPK-dependent ASR3 phosphorylation. Protoplasts were coexpressed with ASR3-FLAG and MAPK phosphatase MKP or MEKK1. MEK/MKK inhibitor U0126 was added to protoplasts 30 min before flg22 treatment. MAPK activation is shown by immunoblotting with α-pERK antibody (middle panel).
(B) flg22-activated MPK4 phosphorylates ASR3. The individual MAPKs were expressed in protoplasts, activated by flg22 treatment, immunoprecipitated with α-HA antibody, and subjected to an in vitro kinase assay using MBP-ASR3 as a substrate in the presence of [γ-32P]ATP. Proteins were separated with SDS-PAGE and analyzed by autoradiography (upper panel), and the MAPK expression is shown by immunoblotting (bottom panel).
(C) Time course of flg22-activated MPK4 phosphorylation on ASR3. The experiment was performed as in (B) with 100 nM flg22 treatment for the indicated time. MBP-ASR3 is shown by Coomassie blue staining (CBB).
(D) Thr-189 is required for MPK4-mediated ASR3 phosphorylation. The experiment was performed as in (B) with different MBP-ASR3 mutants as substrates.
(E) The mpk4 mutant abolishes flg22-induced ASR3 phosphorylation. Protoplasts were isolated from Ler and the mpk4 mutant (in Ler background), transfected with ASR3-HA, and treated with 100 nM flg22 for the indicated time.
(F) ASR3 associates with MPK4 in Arabidopsis protoplasts. Protoplasts were cotransfected with ASR3-HA and MPK4-FLAG or an empty vector control (Ctrl). co-IP was performed with α-FLAG antibody (IP: α-FLAG), and the proteins were analyzed using immunoblots with α-HA antibody (IB: α-HA).
(G) ASR3 associates with MPK4 in 35S:ASR3-HA transgenic plants. Ten-day-old seedlings from two independent transgenic lines (OX9 and OX15) were used for co-IP, and transgenic plants carrying an empty vector were used as the control (Ctrl). Co-IP assay was performed with α-HA antibody, and the proteins were analyzed using immunoblots with α-MPK4 antibody (top). The input of ASR3-HA and MPK4 proteins is shown by immunoblots (middle and bottom).
(H) ASR3 directly interacts with MPK4 with an in vitro pull-down assay. GST or GST-MPK4 immobilized on glutathione Sepharose beads was incubated with MBP, MBP-ASR3, MBP-ASR3T189D, or MBP-ASR3T189A proteins. The beads were washed and pelleted for immunoblot analysis with α-HA antibody. PD, pull-down.
The above experiments were repeated three times with similar results.
A time-course study suggested that MPK4 exhibited the highest phosphorylation activity toward ASR3 at 5 to 15 min of flg22 treatment (Figure 4C). MPK4 was unable to phosphorylate ASR3T189A, which abolished the flg22-induced ASR3 mobility shift (Figure 4D). By contrast, mutation of another putative MAPK phosphorylation site (S182A) did not impair the phosphorylation of ASR3 by MPK4 (Figure 4D). ASR3S182 was also not detected as a confident phosphorylation site by LC-MS/MS analysis (Supplemental Table 1). These data suggest that MPK4 directly phosphorylates ASR3 on the Thr-189 residue during FLS2 signaling. Importantly, flg22-induced ASR3 phosphorylation was largely abolished in the mpk4 mutant compared with wild-type (Landsberg erecta [Ler]) plants, providing genetic evidence of the involvement of MPK4 in flg22-induced ASR3 phosphorylation (Figure 4E).
In addition, MPK4 coimmunoprecipitated with ASR3 when coexpressed in protoplasts (Figure 4F). The association was also confirmed in ASR3-HA transgenic plants. Following immunoprecipitation with the α-HA antibody, endogenous MPK4 detected by the α-MPK4 antibody was observed in ASR3-HA transgenic plants but not in the empty vector control transgenic plants (Figure 4G). To test whether MPK4 directly interacts with ASR3, an in vitro pull-down assay was performed with glutathione S-transferase (GST)-tagged MPK4 immobilized on glutathione Sepharose beads as bait against MBP-ASR3 fusion protein with an HA epitope tag. As shown in Figure 4H, MBP-ASR3 could be pulled down by GST-MPK4, but not by GST alone. It appears that ASR3T189A did not affect MPK4 and ASR3 interaction (Figure 4H). Taken together, the data indicate that ASR3 directly interacts with MPK4 and is phosphorylated by MPK4 mainly on the Thr-189 residue upon flg22 perception.
ASR3 Is a Transcriptional Repressor
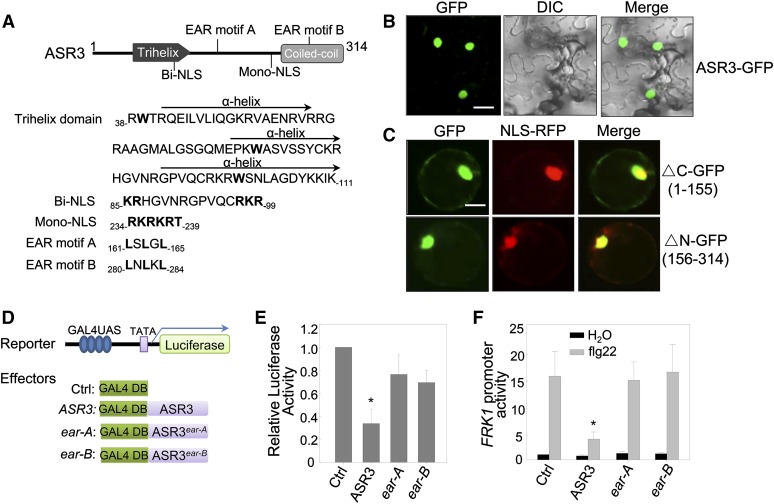
The putative trihelix DNA binding domain of ASR3 is located at its N terminus with three amphipathic α-helices and the conserved tryptophan (W) residues. The ASR3 C terminus is predicted to form a coiled-coil, which is loosely conserved within the clade (Figure 5A). Consistent with its potential function as a transcription factor, fluorescence signals derived from green fluorescent protein (GFP) fusion of ASR3 were observed mainly in the nucleus in Nicotiana benthamiana transient assays (Figure 5B). Similarly, stable Arabidopsis transgenic plants carrying ASR3-GFP under the control of the Cauliflower mosaic virus 35S promoter showed strong fluorescence signals in the nucleus (Supplemental Figure 3). There are two predicted nuclear localization signals (NLSs) in ASR3, a bipartite NLS at the N terminus and a monopartite NLS at the C terminus (Figure 5A). Apparently, either NLS is sufficient to mediate ASR3 nuclear localization since both ASR3-ΔC and ASR3-ΔN were mainly localized in the nucleus when transiently expressed in Arabidopsis protoplasts (Figure 5C).
Figure 5.
ASR3 Is a Transcriptional Repressor.
(A) ASR3 is a putative trihelix transcription factor. The structure of ASR3 labeled with the putative protein motifs is shown on top, and the amino acid sequence with the starting and ending positions for each motif is shown below. The trihelix domain contains three amphipathic α-helices with the conserved Trp residues in bold. Bi-NLS and Mono-NLS stand for bipartite NLS and monopartite NLS, respectively. The bold letters in the sequence indicate the conserved sites in the motifs.
(B) ASR3-GFP localizes in the nucleus. N. benthamiana leaves were infiltrated with Agrobacterium tumefaciens carrying 35S:ASR3-GFP, and the images were taken 2 dpi with a confocal microscope. Bar = 20 μm.
(C) Both N-terminal and C-terminal halves of ASR3 localize in the nucleus. The GFP fusion of N-terminal half (△C) or C-terminal half (△N) was coexpressed with NLS-RFP in Arabidopsis protoplasts for 12 h and the images were taken with a confocal microscope. NLS-RFP is a control for nuclear localization. Bar = 10 μm.
(D) Schematic diagram of the effector and reporter constructs used in the transactivation assay. The reporter construct contains four copies of GAL4-UAS, a minimal 35S promoter (TATA), and a luciferase reporter gene. The effector constructs contain GAL4 DNA binding domain alone (Ctrl) or with ASR3 wild type or ear mutants under the control of 35S promoter.
(E) Relative transcriptional activity of ASR3 and ear mutants. Protoplasts were cotransfected with the reporter construct and different effector constructs.
(F) Overexpression of ASR3 in Arabidopsis protoplasts suppresses flg22-induced FRK1 promoter activity. Protoplasts were cotransfected with pFRK1:LUC and ASR3, ASR3ear-A, ASR3ear-B, or an empty vector control. In (E) and (F), UBQ10-GUS was included in all transfections and served as an internal transfection control. The luciferase activity was normalized with GUS activity. The data are shown as mean ± sd from three independent repeats. The asterisk indicates a significant difference with a Student’s t test (P < 0.05) when compared with control.
The above experiments were repeated three times with similar results.
We also determined the transcriptional activity of ASR3 with an effector construct containing 35S promoter-driven yeast transcriptional activator GAL4 DNA binding domain fused with ASR3 and a reporter construct containing the GAL4 upstream activation sequence (UAS) and the 35S minimal promoter-driven a luciferase reporter gene (Figure 5D). Transactivation assays were performed by coexpression of the effector and the reporter constructs in Arabidopsis protoplasts. Surprisingly, compared with the empty vector control, expression of ASR3 resulted in ∼3-fold reduction of luciferase activity, suggesting that ASR3 functions as a transcriptional repressor (Figure 5E). In line with this observation, we identified two EAR motifs in ASR3 (Figure 5A). The EAR motif with the consensus sequence of either LxLxL or DLNxxP has been reported to constitute a predominant form of transcriptional repression motif in plants (Kagale et al., 2010). Mutations in the conserved leucine (L) residues in the EAR motifs (ear-A, L161A/L163A/L165A, or ear-B, L280A/L282A/L284A) impaired ASR3 transcriptional repressor activity (Figure 5E), indicating that its repressor activity is largely conferred by the EAR motifs. Consistent with these results, expression of ASR3 in protoplasts suppressed flg22-induced FRK1 promoter activity and this suppression activity depended on the EAR motifs (Figure 5F). Together, the data indicate that ASR3 functions as a transcriptional repressor to suppress certain flg22-induced immune gene expression.
ASR3 Forms a Homodimer
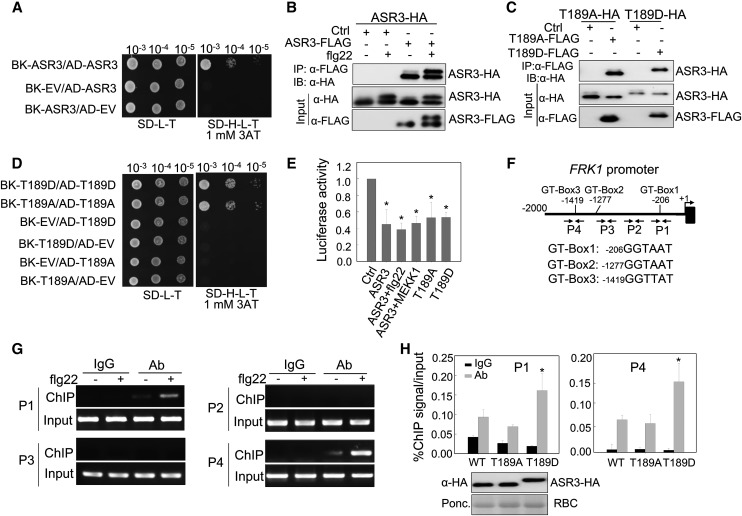
The C-terminal coiled-coil domain of the trihelix transcription factors is predicted to be involved in protein dimerization (Kaplan-Levy et al., 2012; Qin et al., 2014) (Figure 5A). We tested the potential homodimerization of ASR3 and the involvement of the coiled-coil domain by yeast two-hybrid (Y2H) and coimmunoprecipitation (co-IP) assays (Figures 6A and 6B). ASR3 interacted with itself in the Y2H assay (Figure 6A). In addition, ASR3-FLAG could immunoprecipitate ASR3-HA when transiently expressed in Arabidopsis protoplasts (Figure 6B). It appears that flg22-induced ASR3 phosphorylation did not affect ASR3 homodimerization (Figure 6B). Consistent with this observation, both ASR3T189A (phospho-inactive mutant) and ASR3T189D (phospho-mimetic mutant) could interact with not only wild-type ASR3 (Supplemental Figures 4A and 4B) but also with themselves (Figures 6C and 6D) in co-IP and Y2H assays. In line with the role of C-terminal coiled-coil domain in protein dimerization, ASR3△C, but not ASR3△N, lost the interaction with full-length ASR3 (Supplemental Figure 4C). Furthermore, the △C4 (255 to 288) truncation mutant, of which the coiled-coil domain was deleted, blocked ASR3 homodimerization (Supplemental Figure 4C). These data indicate that ASR3 forms a homodimer that is likely mediated by the C-terminal coiled-coil domain.
Figure 6.
Phosphorylation of ASR3 by MPK4 Enhances Its DNA Binding Activity.
(A) ASR3 forms a homodimer in Y2H assays. The interaction between pAD-ASR3 and pBK-ASR3 was tested on SD-H-L-T supplemented with 1 mM 3-amino-1,2,4-triazole (3AT). EV indicates the empty vectors for either pGADT7 or pGBKT7. Serial dilutions of the yeast colonies were plated.
(B) ASR3 forms a homodimer in vivo. Co-IP was performed with Arabidopsis protoplasts coexpressing ASR3-HA and ASR3-FLAG or an empty vector control (Ctrl).
(C) Neither ASR3T189A nor ASR3T189D exhibits altered homodimerization in co-IP assays.
(D) Neither ASR3T189A nor ASR3T189D exhibits altered homodimerization in Y2H assays.
(E) The effect of phosphorylation on ASR3 transcriptional regulation activity. All the transfections included the UAS-LUC (reporter construct), UBQ10-GUS (internal transfection control), and different ASR3 effector constructs. One sample was treated with 100 nM flg22 for 4 h, and one sample was cotransfected with MEKK1. The asterisk indicates a significant difference with a Student’s t test (P < 0.05) when compared with control.
(F) Schematic diagram of the FRK1 promoter with the positions of putative GT-boxes and PCR primers for the ChIP assays. The sequence of each GT-box is shown with the starting nucleotide position.
(G) ASR3 binds to the endogenous FRK1 promoter in vivo based on ChIP assays. Twelve-day-old seedlings from 35S:ASR3-HA transgenic plants were used for chromatin isolation. ASR3-chromatin complex was immunoprecipitated with α-HA antibody (with mouse IgG as a negative control) and subjected to PCR analysis with primers as indicated in Figure 6F. Sheared DNA before immunoprecipitation served as the input control.
(H) Phosphorylation of ASR3 enhances its DNA binding activity. Twelve-day-old seedlings from wild-type ASR3-HA, ASR3T189D-HA, and ASR3T189A-HA transgenic plants were used for chromatin isolation. ChIP- and input-DNA samples were quantified by PCR using P1 and P4 primers. The ChIP results are presented as the percentage of input DNA. Error bars indicate sd (n = 3). The asterisk indicates a significant difference with a Student’s t test (P < 0.05) when compared with wild-type ASR3. The protein expression level of different ASR3 variants in transgenic plants is shown by immunoblot on the bottom.
Phosphorylation of ASR3 by MPK4 Enhances Its DNA Binding Activity
Our data suggest that flg22-activated MPK4 could directly phosphorylate ASR3 and that ASR3 possesses transcriptional repressor activity. Next, we determined whether MPK4-mediated ASR3 phosphorylation affects its repressor activity and/or DNA binding activity. As shown in Figure 6E, flg22 treatment or activation of ASR3 by MEKK1 did not affect its transcriptional repressor activity in the GAL4-UAS-based protoplast transactivation assay. In addition, the T189A and T189D mutants behaved similarly as wild-type ASR3 in terms of transcriptional repressor activity (Figure 6E). Apparently, phosphorylation of ASR3 may not regulate its transcriptional repressor activity.
It has been reported that trihelix transcription factors bind to the GT-like motif [GGT(A/T)(A/T)(A/T)] of target genes to regulate transcription (Kaplan-Levy et al., 2012). The FRK1 promoter region (2 kb upstream of the translational start site) contains three putative GT-like motifs (Figure 6F). We tested whether ASR3 is able to bind to any of these motifs in the FRK1 promoter by a chromatin immunoprecipitation (ChIP)-PCR assay with four pairs of primers amplifying different regions of the FRK1 promoter in transgenic plants expressing 35S:ASR3-HA. ASR3 was able to bind to the P1 and P4 regions of the FRK1 promoter. Interestingly, the binding of ASR3 to these regions was enhanced upon flg22 treatment (Figure 6G). Furthermore, a ChIP-PCR assay with transgenic plants carrying 35S promoter-driven wild-type ASR3-HA, ASR3T189A-HA, or ASR3T189D-HA indicated that the phospho-mimetic form (ASR3T189D) displayed higher DNA binding affinity than the wild-type ASR3 and phospho-inactive form (ASR3T189A) (Figure 6H). The data support that flg22-induced ASR3 phosphorylation enhanced its binding to the FRK1 promoter.
Overexpression of ASR3 Compromises Disease Resistance to Virulent Bacterial Pathogens
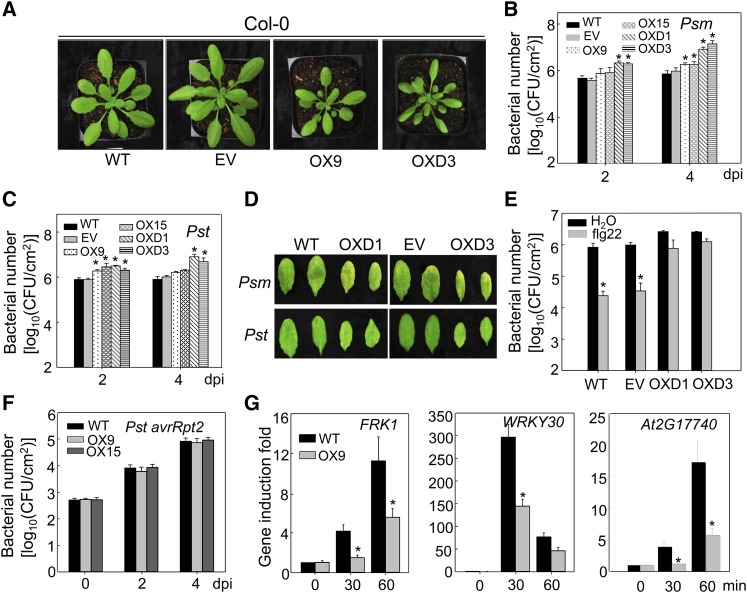
We further determined the disease phenotype of transgenic plants carrying ASR3T189D with a C-terminal HA epitope tag under the control of the constitutive 35S promoter. We also generated transgenic plants carrying wild-type ASR3-HA under the control of the 35S promoter. Multiple lines of each construct were obtained and two lines with comparable transcript and protein expression levels for each were chosen for plant defense response assays (Supplemental Figures 5A and 5B). OX9 and OX15 were the representative lines for 35S:ASR3-HA, whereas OXD1 and OXD3 were the representatives for 35S:ASR3T189D-HA transgenic plants. We observed that transgenic plants overexpressing wild-type ASR3 or ASR3T189D were smaller in size than wild-type Columbia-0 (Col-0) or transgenic plants carrying an empty vector (Figure 7A). The transgenic plants overexpressing wild-type ASR3 displayed slightly but statistically significant enhanced susceptibility to infections by virulent bacterial pathogens Psm and Pst as measured by in planta bacterial multiplication (Figures 7B and 7C). The enhanced susceptibility was more evident in plants overexpressing ASR3T189D with over 10-fold more bacterial population in transgenic plants than that in wild-type plants 4 dpi (Figures 7B and 7C). The disease symptoms were also more severe in the transgenic plants than in the wild type (Figure 7D). The flg22 treatment primed plant resistance against Pst infection in wild-type plants. However, flg22-induced resistance was blocked in ASR3T189D overexpression plants (Figure 7E). Consistent with the asr3-1 mutant, overexpression lines showed unaltered disease resistance to the avirulent pathogen Pst avrRpt2 (Figure 7F). The data further indicate that ASR3 plays a negative role in plant PTI defense and that phosphorylation at Thr-189 is important for its function. Consistent with this, the transgenic plants overexpressing ASR3 displayed reduced induction of immune-responsive genes FRK1, WRKY30, and At2G17740 upon flg22 treatment (Figure 7G).
Figure 7.
Overexpression of ASR3 Compromises Disease Resistance to Virulent Bacterial Pathogens.
(A) Morphological phenotype of wild-type, the empty vector control (EV), 35S:ASR3-HA (OX9), and 35S:ASR3T189D-HA (OXD3) transgenic lines. Four-week-old soil-grown plants are shown.
(B) and (C) Bacterial multiplication of Psm ES4326 (B) or Pst DC3000 (C) in wild-type, empty vector control, 35S:ASR3-HA (OX9 and OX15), and 35S:ASR3T189D-HA (OXD1 and OXD3) transgenic plants at 2 and 4 dpi. Leaves from 4-week-old plants were hand-inoculated with Psm or Pst at 5 × 105 cfu/mL, and bacterial counting was performed at the indicated time points.
(D) The disease symptoms upon Psm or Pst infection. The pictures were taken at 4 dpi.
(E) Compromised flg22-mediated immunity to Pst infection in ASR3T189D overexpression lines. Leaves from 4-week-old plants were hand-inoculated with water or 100 nM flg22, and 24 h later hand-inoculated with Pst at 5 × 105 cfu/mL. Bacterial counting was performed at 3 dpi.
(F) Bacterial growth of Pst avrRpt2. The bacteria at 5 × 105 cfu/mL were hand-inoculated into leaves of 4-week-old plants.
(G) Reduced immune gene expression in ASR3 overexpression lines. Ten-day-old seedlings were treated with 100 nM flg22 for 30 and 60 min for qRT-PCR analysis. Gene expression level was normalized with internal control UBQ10.
The data in (B), (C), and (E) to (G) are shown as means ± sd from three biological repeats. The asterisk indicates a significant difference with a Student’s t test (P < 0.05) when compared with the wild-type or control treatment. The above experiments were repeated three times with similar results.
ASR3 Globally Regulates flg22-Induced Immune Genes
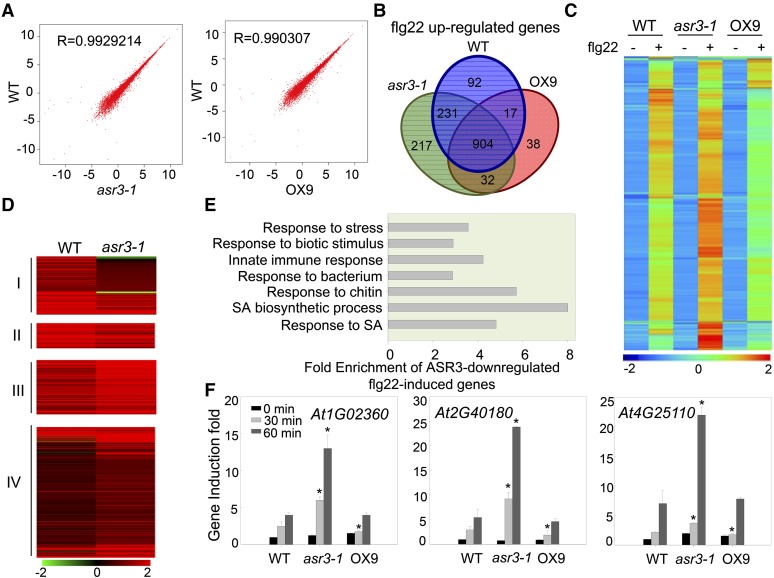
To further identify the ASR3-regulated flg22-induced genes, we performed RNA sequencing (RNA-seq) analysis with 10-d-old seedlings of wild-type, asr3-1, and 35S:ASR3-HA transgenic line OX9 with or without 100 nM flg22 treatment for 30 min. Samples from four independent biological repeats were collected and RNAs from two repeats were pooled for RNA-seq. The correlation coefficient (R) for the expression profiles of all transcripts between the wild type and asr3-1, and between the wild type and OX9 without flg22 treatment was close to linear (0.99), suggesting that ASR3 does not affect general gene transcription (Figure 8A). Among 23,317 detectable transcripts, 48 genes showed differential expression (fold change ≥ 2, P value < 0.05), with 31 showing enhanced and 17 reduced expression in the asr3-1 mutant compared with wild-type plants without treatment (Supplemental Data Set 1A). Compared with no treatment, flg22 treatment induced 1244, 1384, and 991 genes (fold change ≥ 2, P value < 0.05) in the wild type, asr3-1, and OX9, respectively, with 904 genes induced in all three genotypes (Figure 8B; Supplemental Data Set 1B). Hierarchical clustering analysis with 1531 genes induced by flg22 treatment in any of the three genotypes suggested that the asr3-1 mutant displayed an overall enhanced flg22 response, whereas OX9 displayed a reduced response compared with wild-type plants (Figure 8C). We further analyzed the differential flg22-induced genes in the wild type and the asr3-1 mutant, which were defined as ASR3-dependent flg22-induced genes and classified them into four groups: Group I, 109 genes as ASR3-required flg22-induced genes (genes induced in the wild type but not in asr3-1); Group II, 43 genes as ASR3-potentiated flg22-induced genes (genes induced in both the wild type and asr3-1 with at least 1.5-fold higher induction in the wild type than asr3-1); Group III, 89 genes as ASR3-attenuated flg22-induced genes (genes induced in both the wild type and asr3-1 with at least 1.5-fold higher induction in asr3-1 than the wild type); and Group IV, 249 genes as ASR3-suppressed flg22-induced genes (genes induced in asr3-1 but not in the wild type) (Figure 8D; Supplemental Data Set 1C). Importantly, 338 out of 490 (69%) ASR3-dependent flg22-induced genes showed enhanced flg22-induction in asr3-1 compared with wild-type plants. Notably, 227 out of 338 (67%) ASR3 negatively regulated flg22-induced genes displayed reduced flg22 induction in OX9 plants compared with that in the wild type (Supplemental Data Set 1D). Enrichment analysis of Gene Ontology (GO) categories indicates that genes associated with response to stress, response to biotic stimulus, immune system response, and response to salicylic acid were significantly enriched (P value < 0.01) among Group III and IV genes (Figure 8E; Supplemental Data Set 1E). The elevated expression of several flg22-induced genes, including At1G02360 (chitinase family gene), At2G40180 (PP2C family phosphatase gene), and At4G25110 (type I metacaspase gene), in asr3-1 was confirmed with quantitative RT-PCR (qRT-PCR) analysis (Figure 8F). There were 133 genes identified as flg22 downregulated genes (fold change ≥ 2, P value < 0.05) in either the wild type, asr3-1, or OX9 (Supplemental Data Set 1F). Compared with the wild type (25 genes), OX9 (69 genes) had more downregulated genes and enhanced fold change of downregulation, which is consistent with the idea that ASR3 functions as a transcriptional repressor. Thus, ASR3 appears to regulate both flg22-induced and flg22-reduced genes. Taken together, the global gene expression data suggest that ASR3 plays a negative role in regulating a large subset of flg22-regulated genes.
Figure 8.
ASR3 Globally Regulates flg22-Induced Gene Expression.
(A) Scatter plots of whole-genome transcript fragments per kilobase of transcript per million mapped reads (FPKM) in Col-0 (wild type) versus asr3-1 mutant (left) or OX9 transgenic plants (right). Gene expression levels were detected in 10-d-old seedlings without treatment. The y axis indicates gene expression in the wild type, and the x axis indicates gene expression in asr3-1 or OX9 transgenic plants.
(B) Venn diagram of flg22-induced genes (fold change ≥ 2 and P value < 0.05; 30 min after 100 nM flg22 treatment) in wild-type, asr3-1, or OX9 transgenic plants.
(C) Heat map of flg22-induced genes in wild-type, asr3-1, or OX9 transgenic plants. The original FPKM values were subjected to data adjustment by normalized genes/rows and hierarchical clustering was generated with the average linkage method using MeV4.0. Red color indicates relatively high expression, and blue indicates relatively low expression. A list of flg22-induced genes is shown in Supplemental Data Set 1B.
(D) Clustering display of ASR3-dependent flg22 upregulated genes in wild-type and asr3-1 mutant plants. The four clusters are defined in the text. The flg22 induction fold of individual genes with the log2-transformed values was used for hierarchical clustering analysis with the average linkage method using MeV4.0. Red color indicates upregulation and green indicates downregulation with flg22 treatment. The gene list for this analysis is shown in Supplemental Data Set 1C.
(E) Enrichment of genes with GO terms related to defense response for Group III and IV genes. The fold enrichment was calculated based on the frequency of genes annotated to the term compared with their frequency in the genome.
(F) qRT-PCR analysis of ASR3-regulated genes. At1G02360 encodes a chitinase family protein, At2G40180 encodes a PP2C family phosphatase, and At4G25110 encodes a type I metacaspase. Gene expression was normalized to internal control UBQ10. The data are shown as means ± sd from three biological replicates with a Student’s t test. Asterisk indicates a significant difference with P < 0.05 when compared with the wild type.
DISCUSSION
Proper transcriptional reprogramming of immune-related genes is crucial for organisms to achieve efficient defense responses against pathogen infections. Although more than 1000 genes are activated by MAMP treatments, the regulation of immune-related gene expression remains largely unknown. In this study, we report that a putative trihelix transcription factor, ASR3, plays a negative role in regulating immune-related gene expression and defense in FLS2 signaling. ASR3 functions as a transcriptional repressor via its EAR motifs. ASR3 directly interacted with MPK4 in vivo and in vitro. Upon flg22 perception, MPK4 rapidly phosphorylated ASR3 primarily on the Thr-189 residue, which enhanced ASR3 DNA binding activity toward the promoters of target genes. FRK1, a PTI marker gene, is a direct target of ASR3 and its induction was suppressed by overexpression of ASR3. The asr3 knockout mutant showed enhanced disease resistance to virulent P. syringae strains accompanied with elevated immune-related gene induction. Thus, the data revealed that ASR3, a new MPK4 substrate, functions as a transcriptional repressor to downregulate expression of certain immune-related genes and negatively regulate PTI responses.
ASR3 was annotated as an unknown function protein containing a putative MYB-like DNA binding domain by TAIR10 (http://www.arabidopsis.org/) and does not bear significant similarity to any other proteins. ASR3 was not classified as a member of the MYB or MYB-like gene family, as the individual helix motif of its DNA binding domain is significantly longer than the classical MYB or MYB-like domain and the target sequences are also different. It was recently shown that its DNA binding domain bears features of the trihelix DNA binding motif (Kaplan-Levy et al., 2012; Qin et al., 2014). In addition, ASR3 contains a conserved coiled-coil motif at its C terminus (Figure 5A). Based on these features and phylogenetic analysis, ASR3 was classified as a member of the SH4 clade in the trihelix transcription factor family (Kaplan-Levy et al., 2012). The Arabidopsis genome contains four SH4-related genes, and none of them has been assigned a function. ASR3 does not bear high sequence similarity to other Arabidopsis SH4-related proteins, with sequence identity of 22.6% to At4G31270, 20.2% to At2G35640, and 19.5% to At1G31310 at the amino acid level. Trihelix transcription factors appear to be specific to land plants and do not exist in algae, insects, and animals. In Arabidopsis, there are 30 members in this gene family. Compared with other transcription factor families, trihelix family transcription factors remain poorly characterized and most of them have not been assigned a function. Several characterized trihelix transcription factors have been reported to be involved in light response, plant development, and abiotic stress responses (Kaplan-Levy et al., 2012). The founding members of trihelix transcription factors, GT factors (GT-1 and GT-2), bind to the GT elements in the promoters of light-induced genes (Dehesh et al., 1990; Hiratsuka et al., 1994). Whether and how trihelix transcription factors function in plant biotic stresses was not clear. It has been reported that GT-3b, a GT-1 clade of the trihelix transcription factor, is transcriptionally induced 30 min after P. syringae infection, although the biological function of this was unclear (Park et al., 2004). Here, we provide genetic evidence that ASR3, a SH-4 clade trihelix transcription factor, negatively regulates plant immune gene expression and defense. In contrast to GT-1 and GT-2, which function as transcriptional activators (Ni et al., 1996), ASR3 is a transcriptional repressor through its EAR motifs.
Transcription factors are often transcriptionally and/or posttranslationally regulated in response to internal and external stimuli. Transcripts of WRKY transcription factors are quickly activated upon pathogen infections (Dong et al., 2003). By contrast, transcripts of ASR3 do not appear to change significantly upon flg22 treatment. Similarly, expression of GT-1 and GT-2 is constitutive and is not affected by light signals (Ni et al., 1996). It has been speculated that GT-1 and GT-2 are likely regulated by posttranslational modification in response to light. Indeed, in vitro phosphorylation of GT-1 by mammalian calcium/calmodulin kinase II increases its DNA binding activity, but the biological significance of this and the corresponding plant kinase remain unknown (Maréchal et al., 1999). We found that MPK4 directly phosphorylated ASR3 and that the phosphorylation enhanced its DNA binding activity to the promoters of target genes. There are several putative GT-like elements in the promoter region of FRK1. We show that flg22 treatment or the phospho-mimetic form ASR3T189D enhanced ASR3 binding to some of the GT-like elements in the FRK1 promoter. It appears that different mechanisms underlie the phosphorylation-induced enhancement of DNA binding activity of ASR3 and GT-1. The phosphorylation site of GT-1 is located in the DNA binding domain and structural modeling suggests that the phosphorylated side chain of GT-1 is involved in direct interaction with bases of the DNA (Maréchal et al., 1999; Nagata et al., 2010). However, the MPK4-mediated phosphorylation occurs outside of the DNA binding domain of ASR3. The mechanistic details of how phosphorylation enhances the DNA binding activity of ASR3 await further elucidation.
The MEKK1-MKK1/MKK2-MPK4 cascade is considered to be a negative regulator in plant innate immunity (Gao et al., 2008). Recent functional study of a constitutively active (CA) form of MPK4 further supports this hypothesis. CA-MPK4 transgenic plants show compromised disease resistance to virulent bacterium Pst DC3000 and nonpathogenic mutant hrcC infection (Berriri et al., 2012). Activation of the MPK4 pathway has been hypothesized to antagonize the positive MKK4/MKK5-MPK3/MPK6 pathway and balance the strength of the defense response (Rodriguez et al., 2010). MPK4 phosphorylates and interacts with a VQ motif-containing protein, MKS1 (MPK4 SUBSTRATE1), which likely serves as a scaffold protein to form the MPK4-MKS1-WRKY33 complex (Andreasson et al., 2005). Upon pathogen signal perception, phosphorylation of MKS1 by MPK4 results in the complex disassembly, thereby releasing WRKY33 to bind to the promoters of its target genes, including PAD3, which encodes an enzyme required for the synthesis of phytoalexin camalexin (Qiu et al., 2008). Consistent with this, the wrky33 mutant displayed reduced MAMP- or pathogen-induced PAD3 expression and camalexin production. However, the mks1 and mpk4 mutants have no detectable change of camalexin levels (Qiu et al., 2008). Unlike the MPK4-MKS1-WRKY33 complex, MPK4 and ASR3 interact constitutively and the phosphorylation status or pathogen signal perception does not appear to exert a demonstrable change in complex formation (Figure 4F). In addition, the mks1 mutant compromised plant basal defense to Pst DC3000 infection (Petersen et al., 2010), whereas the asr3 mutant enhanced plant resistance to virulent bacterial infections. Apparently, ASR3 functions in parallel with MKS1-WRKY33 downstream of MPK4 to regulate plant defense genes.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana accessions Col-0 and Ler and mutants fls2 (Salk_141277), bak1-4 (Salk_116202), mpk4 (CS5205, in Ler background), asr3-1 (SALK_112571C), and asr3-2 (SALK_047951C) were obtained from the ABRC. Transgenic plants pASR3:ASR3-HA in the background of the asr3-1 mutant, p35S:ASR3-HA, p35S:ASR3T189A-HA, p35S:ASR3T189D-HA, and p35S:ASR3-GFP in the background of Col-0 were generated in the studies. Plants were grown in soil (Metro Mix 366) in a growth room at 23°C, 45% humidity, and 75 μE m−2 s−1 light with a 12-h-light/12-h-dark photoperiod. Four-week-old plants were used for protoplast isolation, pathogen infection, and ROS production assays. Seedlings were germinated on half-strength Murashige and Skoog plates containing 1% sucrose and 0.8% agar, grown under the same conditions as above for 10 d, transferred to a six-well tissue culture plate with 2 mL water overnight, and then treated with flg22 for the indicated amounts of time for MAPK and qRT-PCR assays.
Plasmid Construction, Protoplast Transient Assays, and Generation of Transgenic Plants
The ASR3 gene was amplified from Col-0 cDNA with primers containing BamHI and NcoI at N terminus and StuI at the C terminus (Supplemental Table 2) and introduced into the plant expression vector pHBT with an HA, FLAG, or GFP epitope-tag at the C terminus. The clone was sequenced to cover the entire ASR3 gene. ΔC, ΔN, ΔC1, ΔC2, ΔC3, and ΔC4 were cloned using the above construct pHBT-ASR3-HA as the template and primers as listed in Supplemental Table 2. The point mutations of ASR3S169A, ASR3S175A, ASR3S182A, ASR3T189A, ASR3S182A/T189A, ASR3T189D, ASR3T196A, and ASR3S230A were generated by site-directed mutagenesis kit with primers as listed in Supplemental Table 2. For Escherichia coli fusion protein constructs, ASR3, ASR3S182A, ASR3T189A, ASR3S182A/T189A, and ASR3T189D were subcloned into a modified pMAL-c2 vector (NEB) with BamHI and StuI digestion. To construct the pCB302-35S:ASR3-GFP binary vector for Agrobacterium tumefaciens-mediated transient expression assay in Nicotiana benthamiana and Arabidopsis transformation, the ASR3-GFP fragment was released from pHBT-35S:ASR3-GFP using NcoI and PstI digestion and ligated into pCB302 binary vector. To construct the pCB302-pASR3:ASR3-HA binary vector, the native promoter of ASR3 (2.1 kb upstream of the start codon) was amplified from Col-0 genomic DNA with primers containing XhoI at the N terminus and BamHI at the C terminus and introduced into the vector pHBT-ASR3-HA. The pASR3:ASR3 fragment was released via XhoI and StuI digestion and ligated into the pCB302 binary vector with an HA-tag at the ASR3 C terminus. For Y2H assay, ASR3, ASR3T189A, and ASR3T189D were subcloned into a modified pGBKT7 vector and a modified pGADT7 vector (Clontech) with BamHI and StuI digestion. For transcriptional activity assays, different ASR3 variants were subcloned into the effector vector containing 35S promoter-driven GAL4 DNA binding domain with BamHI and StuI digestion. MEKK1, MEKK1Km, and MPK clones in protoplast expression vector and protoplast transient assays were reported previously (He et al., 2006). For transgenic plant generation, a standard protocol for Agrobacterium-mediated floral dip method was used. The transgenic plants were selected by glufosinate-ammonium (50 μg/mL). Equal numbers of leaves from multiple transgenic plants were ground in 4× SDS loading buffer, boiled samples were subjected to SDS-PAGE gel separation, and proteins were detected by immunoblotting with an α-HA antibody (Roche; 12013819001). Two lines with single insertions and similar protein expression levels were chosen for further assays.
Elicitor and Chemical Inhibitor Treatments
The flagellin peptide flg22, EF-Tu peptide elf18, chitin, and LPS (Supplemental Table 3) were used in the concentration as indicated. Chemical inhibitors LaCl3, GaCl3, and RR were purchased from Sigma-Aldrich and K-252a, DPI, and U0126 were purchased from A.G. Scientific. Chemical inhibitors used at a final concentration of 1 μM for K-252a, 5 μM for DPI, 0.5 to ∼1 mM for LaCl3, 0.5 to ∼1 mM for GaCl3, 0.1 to ∼0.2 mM for RR, and 5 μM for U0126. Different chemical inhibitors were added to protoplasts 1 h before flg22 treatment. CIP and λPP were purchased from New England Biolabs, and the treatments were performed following the manufacturer’s instructions. A detailed summary on the chemical inhibitor usage is included in Supplemental Table 3.
MAPK Assays
Ten-day-old seedlings were ground in lysis buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% glycerol, and 1% Triton X-100), and supernatant was collected after centrifugation. Protein samples with 1× SDS buffer were separated in 10% SDS-PAGE gel to detect pMPK3, pMPK6, and pMPK4 by immunoblotting with α-pERK1/2 antibody (Cell Signaling).
LC-MS/MS Analysis
To obtain samples for mass spectrometry analysis, FLAG-tagged ASR3 was expressed in protoplasts for 12 h and treated with or without flg22 for 15 min. Protoplasts were lysed with buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10% glycerol, 1% Triton X-100, and protease inhibitor cocktail) and immunoprecipitated with α-FLAG Agarose (Sigma-Aldrich). The immunoprecipitated ASR3 was separated in 10% SDS-PAGE gel followed by silver staining. A small aliquot of immunoprecipitated ASR3 was subjected for immunoblot using α-FLAG antibody. The corresponding bands were sliced and subjected for in-gel digestion with trypsin. The phospho-peptides were enriched and analyzed using a LTQ Orbitrap XL LC-MS/MS system (Thermo Scientific) as previously described (Gao et al., 2013). The MS/MS spectra were analyzed with Mascot (Matrix Science; version 2.2.2), and the identified phosphorylated peptides were manually inspected to ensure confidence in phosphorylation site assignment.
RNA Isolation and qRT-PCR Analysis
Total RNA from 10-d-old seedlings was extracted by TRIzol reagent (Invitrogen) and quantified with NanoDrop (Thermo Scientific). RNA was then reverse transcribed to synthesize first-strand cDNA with M-MuLV Reverse Transcriptase and oligo(dT) primer following RNase-free DNase I (New England Biolabs) treatment. qRT-PCR analysis was performed using iTaq SYBR green Supermix (Bio-Rad) with an ABI GeneAmp PCR System 9700 following standard protocol. The expression of each gene was normalized to the expression of UBQ10.
RNA-seq and Data Analysis
Two independent repeats were performed for RNA-seq analysis. For each repeat, equal amounts of RNA from two biological replicates was pooled for RNA-seq library construction. RNA-seq library preparation and sequencing were performed on an Illumina HiSequation 2500 platform with 100-nucleotide single-end reads at Texas AgriLife Genomics and Bioinformatics Service (College Station, TX). Approximately 15 million reads were obtained for each sample, which corresponds to 30× coverage of the Arabidopsis transcriptome. RNA-seq reads with low sequencing quality or reads with sequencing adaptors were filtered from the raw data. The resulting clean reads were then aligned to the Arabidopsis reference genome (TAIR10) using TopHat (Trapnell et al., 2009) with default parameters. A GFF (general feature format) formatted gene model annotation file was provided for reads alignment. Following the alignments, Cuffdiff (Trapnell et al., 2010) was used to calculate the number of fragments per kilobase of exon per million fragments mapped and to find the significant differential gene expression. Genes with expression fold change ≥ 2 and P value < 0.05 were considered as significantly different between samples with and without flg22 treatment. The differentially expressed genes were chosen for the hierarchical clustering analysis. A clustering heat map was generated using the Mev software (Howe et al., 2011). GO term enrichment in each gene list was identified using GO::Term Finder (Boyle et al., 2004) with the latest Arabidopsis GO term annotations. The cutoff for significant enrichment is P value < 0.01 and calculation false discovery rate < 0.5. The fold enrichment was calculated based on the frequency of genes annotated to the term compared with their frequency in the genome.
ROS Assay
Around 25 leaves of 5-week-old soil-grown Arabidopsis plants for each genotype were excised into leaf discs (5-mm diameter) and then cut into leaf strips, followed by an overnight incubation with water in 96-well plates to eliminate the wounding effect. ROS burst was determined by a luminol-based assay. Leaf strips were soaked with solution containing 50 μM luminol and 10 μg/mL horseradish peroxidase (Sigma-Aldrich) supplemented with 100 nM flg22. The measurement was performed immediately after adding the solution with a Multilabel Plate Reader (Perkin-Elmer; Victor X3) for a period of 30 min. The values for ROS production from each line were indicated as means of relative light units.
In Vivo Co-IP
Arabidopsis protoplasts were transfected with a pair of constructs tested (empty vector as control) and incubated for 12 h. Samples were collected by centrifugation and lysed with co-IP buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Triton X-100, and protease inhibitor cocktail) by vortexing. For p35S:ASR3-HA transgenic plants, 2-week-old seedlings were homogenized by mortar and pestle with liquid nitrogen, and the fine powders were transferred into co-IP buffer for lysis. For the co-IP assay, protein extract was preincubated with protein-G-agarose beads for 1 h at 4°C with gentle shaking. Immunoprecipitation was performed with an α-HA or α-FLAG antibody for 2 h and then with protein-G-agarose beads for another 2 h at 4°C. The beads were collected and washed three times with washing buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, and 0.1% Triton X-100). The immunoprecipitated and input proteins were analyzed by immunoblot with indicated antibodies.
In Vitro Pull-Down Assay
Fusion proteins were expressed from bacterial protein expression vector in E. coli BL21 strain using Lysogeny broth medium supplemented with 0.25 mM isopropyl β-d-1-thiogalactopyranoside. GST and GST-MPK4 were purified with Pierce glutathione agarose (Thermo Scientific), and MBP, MBP-ASR3, MBP-ASR3T189A, and MBP-ASR3T189D proteins were purified using amylose resin (New England Biolabs) according to the standard protocols from company. MBP fusion proteins (tagged with HA) as preys were preincubated with 5 μL prewashed glutathione agarose in 300 μL incubation buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1mM EDTA, and 0.5% Triton X-100) for 0.5 h at 4°C. After centrifugation, the supernatant was collected and incubated with prewashed GST or GST-MPK4 beads for another 1 h. The beads were collected and washed three times with washing buffer (20 mM Tris-HCl, pH 7.5, 300 mM NaCl, 0.1 mM EDTA, and 0.1% Triton X-100). The pull-down proteins were detected with an α-HA antibody by immunoblot.
Subcellular Localization
Agrobacterium strain GV3101 containing pCB302-35S:ASR3-GFP vector was cultured at 28°C overnight. Bacteria were harvested by centrifugation and resuspended with buffer (10 mM MES, pH 5.7, 10 mM MgCl2, and 200 μM acetosyringone) at OD600 = 0.75. Leaves of 3-week-old soil-grown N. benthamiana were infiltrated with Agrobacterium cultures. Fluorescence signals were detected 2 dpi. Arabidopsis transgenic plants expressing ASR3-GFP were generated by Agrobacterium-mediated floral dipping transformation. For transient protoplast expression, protoplasts were cotransfected with GFP-tagged ASR3-ΔN or ASR3-ΔC vector and a nuclear-localized red fluorescence protein (NLS-RFP), and signals were observed 12 h after transfection. Fluorescence images were taken with Nikon-A1 confocal laser microscope systems and images were processed using NIS-Elements microscope imaging software. The excitation laser of 488 and 561 nm was used for imaging GFP and RFP signals, respectively.
Transcriptional Activity Assay and FRK1 Reporter Assay
Transcriptional activity assay was performed by coexpression of the effector and the reporter constructs in Arabidopsis protoplasts. The effector vector containing GAL4 binding domain was used as the transcriptional activity control, and UBQ10-GUS was included for all the samples as the internal transfection efficiency control. For FRK1 reporter assay, protoplasts were cotransfected with empty vector, 35S:ASR3-HA, 35S:ASR3ear-A-HA, or 35S:ASR3ear-B-HA and pFRK1:LUC for 4 h and then treated with 100 nM flg22 for another 4 h. The cells were collected and resuspended with cell lysis buffer (25 mM Tris-phosphate, pH 7.8, 2 mM 1,2-diaminocyclohexane-N,N,N’,N’-tetraacetic acid, 10% glycerol, 1% Triton X-100, and 2 mM DTT). The luciferase activity was detected by Glomax Multi-Detection System (Promega) with the luciferase assay substrate (Promega). For the GUS activity, methylumbelliferyl-β-d-glucuronide was mixed with the lysed cells, and the fluorescence signals were analyzed with a Multilabel Plate Reader (Perkin-Elmer; Victor X3).
Y2H Assay
The different combinations of ASR3 variants in pGADT7 and pGBKT7 as indicated in the figures were cotransformed into yeast AH109 strain. Polyethylene glycol/LiAc-mediated yeast transformation was performed according to the protocol of Yeastmaker Yeast Transformation System 2 (Clontech). Protein-protein interaction was tested by growing yeast colonies on the synthetic defined (SD) medium without histidine, leucine, and tryptophan (SD-H-L-T) and supplemented with 1 mM 3-amino-1,2,4-triazole.
ChIP Assay
Two-week-old seedlings from T3 homozygous transgenic lines of 35S:ASR3-HA, 35S:ASR3T189A-HA, and 35S:ASR3T189D-HA were used for ChIP assay following the protocol described previously (Gao et al., 2013). Plant tissues were cross-linked with 1% formaldehyde under vacuum and quenched by glycine. Fixed samples were ground in a mortar with liquid nitrogen, and nuclei were extracted with freshly prepared buffer (15 mM Tris-HCl, pH 7.0, 0.25 M sucrose, 5 mM MgCl2, 15 mM NaCl, 1 mM CaCl2, 0.5% Triton X-100, and protease inhibitor cocktail). Chromatin was sheared into ∼500-bp fragments by sonication, six times with Output 2, Duty cycle 3 (Brason Sonifier 250). Immunoprecipitation was performed with α-HA antibody and protein G-agarose (Roche). Immunoprecipitated DNA was precipitated with ethanol following proteinase K digestion. PCR amplification was performed with four pairs of primers amplifying different regions of FRK1 promoter (Supplemental Table 2).
Bacterial Pathogen Infection Assay
Pst DC3000, Psm ES4326, or Pst DC3000 avrRpt2 strains were cultured for overnight at 28°C in the King’s B medium with appropriate antibiotics. Bacteria were harvested by centrifugation, washed with double distilled water, and adjusted to the density of 5 × 105 colony-forming units (cfu)/mL with 10 mM MgCl2. Leaves of 4-week-old soil-grown plants were hand-infiltrated with bacterial suspension using a needleless syringe. For flg22 protection assay, leaves were preinoculated with 100 nM flg22 or double distilled water as control 24 h before bacterial pathogen infiltration. To measure in planta bacterial growth, six leaf discs separated as three repeats were ground and serial dilutions were plated on medium (1% tryptone, 1% sucrose, 0.1% glutamatic acid, and 1.5% agar) with the corresponding antibiotics. Bacterial colony forming units were counted at 0, 2, and 4 dpi.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ASR3 (AT2G33550), FLS2 (AT5G46330), BAK1 (AT4G33430), MEKK1 (AT4G08500), MPK1 (AT1G10210), MPK2 (AT1G59580), MPK3 (AT3G45640), MPK4 (AT4G01370), MPK5 (AT4G11330), MPK6 (AT2G43790), MPK7 (AT2G18170), MPK8 (AT1G18150), MPK9 (AT3G18040), MPK10 (AT3G59790), MPK11 (AT1G01560), MPK12(AT2G46070), MPK13 (AT1G07880), MPK14 (AT4G36450), MPK15 (AT1G73670), MPK16 (AT5G19010), MPK17(AT2G01450), MPK18(AT1G53510), MPK19 (AT3G14720), MPK20 (AT2G42880), FRK1(AT2G19190), PP2C (AT1G07160), WRKY30 (AT5G24110), UBQ1 (AT3G52590), and UBQ10 (AT4G05320). The RNA-seq data were deposited in the Gene Expression Omnibus database (GSE63603) at the National Center for Biotechnology Information.
Supplemental Data
Supplemental Figure 1. Characterization of asr3 T-DNA Knockout Lines and Complementation Transgenic Lines.
Supplemental Figure 2. The flg22-Induced ASR3 Phosphorylation Is Not Affected by Ca2+ Channel Inhibitor or NADPH Oxidase Inhibitor Treatment.
Supplemental Figure 3. ASR3 Localizes to the Nucleus in 35S:ASR3-GFP Transgenic Plants.
Supplemental Figure 4. The Coiled-Coil Domain of ASR3 Is Required for ASR3 Homodimerization.
Supplemental Figure 5. Characterization of 35S:ASR3-HA and 35S:ASR3T189D-HA Transgenic Lines.
Supplemental Table 1. LC-MS/MS Analysis of in Vivo Phosphorylation of ASR3 upon flg22 Treatment.
Supplemental Table 2. Primers Used in This Study.
Supplemental Table 3. Elicitors and Chemical Inhibitors Used in This Study.
Supplemental Data Set 1A. List of ASR3-Regulated Genes without Treatment.
Supplemental Data Set 1B. List of flg22-Induced Genes in Either the Wild Type, asr3-1, or OX9.
Supplemental Data Set 1C. Four Groups of ASR3-Dependent flg22-Induced Genes.
Supplemental Data Set 1D. The flg22-Induced Genes with Enhanced Induction in asr3-1 Mutant and Reduced Induction in OX9 Plants Compared with Wild-Type Plants.
Supplemental Data Set 1E. GO Biological Process Enrichment of ASR3-Dependent flg22-Induced Genes.
Supplemental Data Set 1F. List of flg22-Reduced Genes in Either the Wild Type, asr3-1, or OX9.
Supplementary Material
Acknowledgments
We thank Salk Institute and ABRC for the Arabidopsis T-DNA insertion lines, Marie Boudsocq for the transactivation assay vectors, Ning Zhu from the University of Florida for in-gel digestion and phospho-peptide enrichment, Xiangzong Meng and Kevin Cox for critical reading of the article, and members of the laboratories of L.S. and P.H. for comments and suggestions of the experiments. The work was supported by National Institutes of Health (NIH) (R01GM092893) and the National Science Foundation (IOS-1252539) to P.H. and NIH (R01GM097247) and the Robert A. Welch foundation (A-1795) to L.S. The next-generation sequencing was supported by Texas AgriLife Genomics Seed Grant. The authors have declared no conflict of interests.
AUTHOR CONTRIBUTIONS
B.L., P.H., and L.S. conceived and designed the experiments. B.L., S.J., X.Y., C.C., and S.C. performed the experiments. B.L., S.J., Y.C., J.S.Y., D.J., P.H., and L.S. analyzed the data. B.L., S.J., P.H., and L.S. wrote the article.
Glossary
- MAMP
microbe-associated molecular pattern
- PRR
pattern recognition receptor
- PTI
pattern-triggered immunity
- ROS
reactive oxygen species
- LPS
lipopolysaccharide
- LRR-RLK
leucine-rich repeat receptor-like kinase
- MAPK
mitogen-activated protein kinase
- Pst
Pseudomonas syringae pv tomato
- dpi
days postinoculation
- Psm
P. syringae pv maculicola
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- RR
ruthenium red
- DPI
diphenylene iodonium
- Ler
Landsberg erecta
- NLS
nuclear localization signal
- co-IP
coimmunoprecipitation
- Y2H
yeast two-hybrid
- Col-0
Columbia-0
- RNA-seq
RNA sequencing
- GO
Gene Ontology
- qRT-PCR
quantitative RT-PCR
- ChIP
chromatin immunoprecipitation
- cfu
colony-forming units
References
- Andreasson E., et al. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24: 2579–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. [DOI] [PubMed] [Google Scholar]
- Berriri S., Garcia A.V., Frei dit Frey N., Rozhon W., Pateyron S., Leonhardt N., Montillet J.L., Leung J., Hirt H., Colcombet J. (2012). Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell 24: 4281–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G. (2004). GO:TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D.G., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500. [DOI] [PubMed] [Google Scholar]
- Dehesh K., Bruce W.B., Quail P.H. (1990). A trans-acting factor that binds to a GT-motif in a phytochrome gene promoter. Science 250: 1397–1399. [DOI] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Dong J., Chen C., Chen Z. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51: 21–37. [DOI] [PubMed] [Google Scholar]
- Dou D., Zhou J.M. (2012). Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12: 484–495. [DOI] [PubMed] [Google Scholar]
- Elmore J.M., Lin Z.J., Coaker G. (2011). Plant NB-LRR signaling: upstreams and downstreams. Curr. Opin. Plant Biol. 14: 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B., Liu C., de Oliveira M.V., Intorne A.C., Li B., Babilonia K., de Souza Filho G.A., Shan L., He P. (2015). Protein poly(ADP-ribosyl)ation regulates Arabidopsis immune gene expression and defense responses. PLoS Genet. 11: e1004936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., Zhang Y. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18: 1190–1198. [DOI] [PubMed] [Google Scholar]
- Gao X., Chen X., Lin W., Chen S., Lu D., Niu Y., Li L., Cheng C., McCormack M., Sheen J., Shan L., He P. (2013). Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 9: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann W., Bhattacharjee S. (2012). Effector-triggered immunity signaling: from gene-for-gene pathways to protein-protein interaction networks. Mol. Plant Microbe Interact. 25: 862–868. [DOI] [PubMed] [Google Scholar]
- Halter T., et al. (2014). The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 24: 134–143. [DOI] [PubMed] [Google Scholar]
- He P., Shan L., Lin N.C., Martin G.B., Kemmerling B., Nürnberger T., Sheen J. (2006). Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125: 563–575. [DOI] [PubMed] [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M.E., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka K., Wu X., Fukuzawa H., Chua N.H. (1994). Molecular dissection of GT-1 from Arabidopsis. Plant Cell 6: 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe E.A., Sinha R., Schlauch D., Quackenbush J. (2011). RNA-Seq analysis in MeV. Bioinformatics 27: 3209–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J.D., Shirasu K., Menke F., Jones A., Zipfel C. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54: 43–55. [DOI] [PubMed] [Google Scholar]
- Kagale S., Links M.G., Rozwadowski K. (2010). Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 152: 1109–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan-Levy R.N., Brewer P.B., Quon T., Smyth D.R. (2012). The trihelix family of transcription factors—light, stress and development. Trends Plant Sci. 17: 163–171. [DOI] [PubMed] [Google Scholar]
- Li C., Zhou A., Sang T. (2006). Rice domestication by reducing shattering. Science 311: 1936–1939. [DOI] [PubMed] [Google Scholar]
- Li F., et al. (2014a). Modulation of RNA polymerase II phosphorylation downstream of pathogen perception orchestrates plant immunity. Cell Host Microbe 16: 748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., Cai G., Gao L., Zhang X., Wang Y., Chen S., Zhou J.M. (2014b). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338. [DOI] [PubMed] [Google Scholar]
- Lin W., Li B., Lu D., Chen S., Zhu N., He P., Shan L. (2014). Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc. Natl. Acad. Sci. USA 111: 3632–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Griffith M.E., Li X., Zhu Z., Tan L., Fu Y., Zhang W., Wang X., Xie D., Sun C. (2007). Origin of seed shattering in rice (Oryza sativa L.). Planta 226: 11–20. [DOI] [PubMed] [Google Scholar]
- Lu D., Wu S., Gao X., Zhang Y., Shan L., He P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho A.P., Zipfel C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54: 263–272. [DOI] [PubMed] [Google Scholar]
- Mao G., Meng X., Liu Y., Zheng Z., Chen Z., Zhang S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal E., Hiratsuka K., Delgado J., Nairn A., Qin J., Chait B.T., Chua N.H. (1999). Modulation of GT-1 DNA-binding activity by calcium-dependent phosphorylation. Plant Mol. Biol. 40: 373–386. [DOI] [PubMed] [Google Scholar]
- Meng X., Zhang S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51: 245–266. [DOI] [PubMed] [Google Scholar]
- Meng X., Xu J., He Y., Yang K.Y., Mordorski B., Liu Y., Zhang S. (2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25: 1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J., Matschi S., Shorinola O., Rovenich H., Matei A., Segonzac C., Malinovsky F.G., Rathjen J.P., MacLean D., Romeis T., Zipfel C. (2014). The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16: 605–615. [DOI] [PubMed] [Google Scholar]
- Nagata T., Niyada E., Fujimoto N., Nagasaki Y., Noto K., Miyanoiri Y., Murata J., Hiratsuka K., Katahira M. (2010). Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: mechanism for an increase in DNA-binding affinity through phosphorylation. Proteins 78: 3033–3047. [DOI] [PubMed] [Google Scholar]
- Ni M., Dehesh K., Tepperman J.M., Quail P.H. (1996). GT-2: in vivo transcriptional activation activity and definition of novel twin DNA binding domains with reciprocal target sequence selectivity. Plant Cell 8: 1041–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.C., et al. (2004). Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 135: 2150–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen K., Qiu J.L., Lütje J., Fiil B.K., Hansen S., Mundy J., Petersen M. (2010). Arabidopsis MKS1 is involved in basal immunity and requires an intact N-terminal domain for proper function. PLoS ONE 5: e14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A., Schikora A., Hirt H. (2009). MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12: 421–426. [DOI] [PubMed] [Google Scholar]
- Qi D., Innes R.W. (2013). Recent advances in plant NLR structure, function, localization, and signaling. Front. Immunol. 4: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, Y., Ma, X., Yu, G., Wang, Q., Wang, L., Kong, L., Kim, W., and Wang, H.W. (2014). Evolutionary history of trihelix family and their functional diversification. DNA Res. 21: 499–510. [DOI] [PMC free article] [PubMed]
- Qiu J.L., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27: 2214–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649. [DOI] [PubMed] [Google Scholar]
- Schwessinger B., Ronald P.C. (2012). Plant innate immunity: perception of conserved microbial signatures. Annu. Rev. Plant Biol. 63: 451–482. [DOI] [PubMed] [Google Scholar]
- Segonzac C., Macho A.P., Sanmartín M., Ntoukakis V., Sánchez-Serrano J.J., Zipfel C. (2014). Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J. 33: 2069–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li L., Macho A.P., Han Z., Hu Z., Zipfel C., Zhou J.M., Chai J. (2013). Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342: 624–628. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X.F., He S.Y. (2013). Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51: 473–498. [DOI] [PubMed] [Google Scholar]
- Xu J., Wei X., Yan L., Liu D., Ma Y., Guo Y., Peng C., Zhou H., Yang C., Lou Z., Shui W. (2013). Identification and functional analysis of phosphorylation residues of the Arabidopsis BOTRYTIS-INDUCED KINASE1. Protein Cell 4: 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.