The identification of Opaque2 DNA binding targets clarified its direct target genes and revealed its broad transcriptional regulation during endosperm development.
Abstract
Opaque2 (O2) is a transcription factor that plays important roles during maize endosperm development. Mutation of the O2 gene improves the nutritional value of maize seeds but also confers pleiotropic effects that result in reduced agronomic quality. To reveal the transcriptional regulatory framework of O2, we studied the transcriptome of o2 mutants using RNA sequencing (RNA-Seq) and determined O2 DNA binding targets using chromatin immunoprecipitation coupled to high-throughput sequencing (ChIP-Seq). The RNA-Seq analysis revealed 1605 differentially expressed genes (DEGs) and 383 differentially expressed long, noncoding RNAs. The DEGs cover a wide range of functions related to nutrient reservoir activity, nitrogen metabolism, stress resistance, etc. ChIP-Seq analysis detected 1686 O2 DNA binding sites distributed over 1143 genes. Overlay of the RNA-Seq and ChIP-Seq results revealed 35 O2-modulated target genes. We identified four O2 binding motifs; among them, TGACGTGG appears to be the most conserved and strongest. We confirmed that, except for the 16- and 18-kD zeins, O2 directly regulates expression of all other zeins. O2 directly regulates two transcription factors, genes linked to carbon and amino acid metabolism and abiotic stress resistance. We built a hierarchical regulatory model for O2 that provides an understanding of its pleiotropic biological effects.
INTRODUCTION
opaque2 (o2) is a nonlethal maize (Zea mays) seed mutant that has been used as a classic genetic marker for chromosome 7 since the early part of the last century (Emerson et al., 1935). o2 kernels contain over 70% higher lysine contents than wild-type maize kernels (Mertz et al., 1964), thereby significantly improving the nutritional value of the grain. Therefore, o2 has been the subject of intense research over the past several decades. However, due to pleiotropic effects that reduce its agronomic quality (Lambert et al., 1969; Nass and Crane, 1970; Loesch et al., 1976), the o2 mutant was not widely adopted for breeding high nutrition maize lines.
The O2 gene was cloned by transposon tagging (Schmidt et al., 1987) and shown to encode a DNA binding protein belonging to the bZIP class of transcription factors (Schmidt et al., 1990). The 22-kD α-zein storage protein subfamily is the best characterized target of O2 regulation. O2 activates transcription by binding conserved DNA motifs in its target genes, most of which contain an ACGT core sequence named the O2-box (Schmidt et al., 1992; Muth et al., 1996). O2 recognizes and interacts with three clustered binding sites (Z1-Z3) in the 22-kD α-zein gene promoter (Muth et al., 1996) at the ACGT core sequence in the Z3 site. O2 activates expression of the 14-kD β-zein gene by binding to an ACGT-containing cis-element (Cord Neto et al., 1995). In addition to the 22- and 14-kD zeins, other zeins, such as the 19- and 10-kD zeins, show decreased expression in the o2 mutant (Hunter et al., 2002). Although 27-kD γ-zein genes are not downregulated by this mutation, an O2-like box containing the ACGT core sequence is found in the promoter region (Wu and Messing, 2012), but it remains unclear whether O2 also directly regulates these zein genes.
Previous studies indicated that O2 directly regulates a number of genes encoding nonzein proteins, including b-32 (Di Fonzo et al., 1986), which encodes a ribosome inactivating protein involved in pathogen defense. O2 is capable of activating b-32 gene expression by interacting with five binding sites (B1-B5), but only the B5 site contains the ACGT core sequence (Lohmer et al., 1991). O2 also regulates cyPPDK1, which encodes an endosperm-specific cytoplasmic pyruvate orthophosphate dikinase (PPDK) that is required for carbon partitioning (Sheen, 1991; Maddaloni et al., 1996). cyPPDK1 has an ACGT binding site (O2 BS-2) in the promoter region. LKR/SDH, which encodes a bifunctional enzyme involved in Lys catabolism (Kemper et al., 1999), is also regulated by O2. Gene expression profiling analysis of o2 mutants revealed that O2 regulates transcription of many genes involved in a variety of pathways (Frizzi et al., 2010; Hartings et al., 2011; Hunter et al., 2002; Jia et al., 2007; Jia et al., 2013), which is consistent with the described pleiotropic effects of the mutant.
In this study, we used RNA sequencing (RNA-Seq) to identify protein-coding and noncoding RNAs that are differentially expressed in wild-type and o2 endosperms to build a comprehensive and genome-wide regulatory framework for O2. These data indicate O2 significantly affects expression of 1605 genes and 384 long, noncoding RNAs (lncRNAs). We conducted chromatin immunoprecipitation using an O2-specific antibody coupled with high-throughput sequencing (ChIP-Seq) to determine which of the differentially expressed genes are direct targets of O2. The additional target genes were confirmed and functionally characterized by experiments.
RESULTS
RNA-Seq Identifies O2-Regulated Protein-Coding and Noncoding RNAs
To establish the regulatory function of the O2 gene, RNA-Seq analysis was performed with total RNA extracts obtained from wild-type and o2 endosperms 15 d after pollination (DAP; see Methods; Supplemental Figure 1). High-throughput sequencing using the Illumina Hisequation 2500 platform resulted in generation of ∼110 million raw reads for each sample. Approximately 90 million of these reads were of high quality, of which ∼75 million aligned well to the maize genome sequence (Schnable et al., 2009). Approximately 55 million of the aligned reads were unique, yielding an effective reads ratio (unique aligned reads/sequenced raw reads) of ∼50%. The unique aligned reads were used to calculate the relative abundance of transcripts, reported here as the expected number of fragments per kilobase of the transcript sequence per million base pairs sequenced (Trapnell et al., 2010).
RNA molecules that are not templates for protein synthesis are designated noncoding RNAs (ncRNAs) (Ernst and Morton, 2013; Novikova et al., 2013), and ncRNAs larger than 200 bases are defined as lncRNAs (Cesana et al., 2011). A number of in vivo and in vitro experiments have demonstrated important biological roles for lncRNAs (Eddy, 2001; Mattick, 2004; Mattick and Makunin, 2006), but a comparatively small number of lncRNAs have been characterized in plants (Swiezewski et al., 2009). Transcripts in the RNA-Seq calculations included protein-coding genes and ncRNAs, including several potential lncRNAs that were identified based on the PhyloCSF platform (http://compbio.mit.edu/PhyloCSF; Lin et al., 2011). Potential trans-targets of the lncRNAs were screened using an RNAplex program that identifies similar sequences required for short, highly stable RNA-RNA interactions (Tafer and Hofacker, 2008; Wang and Chang, 2011).
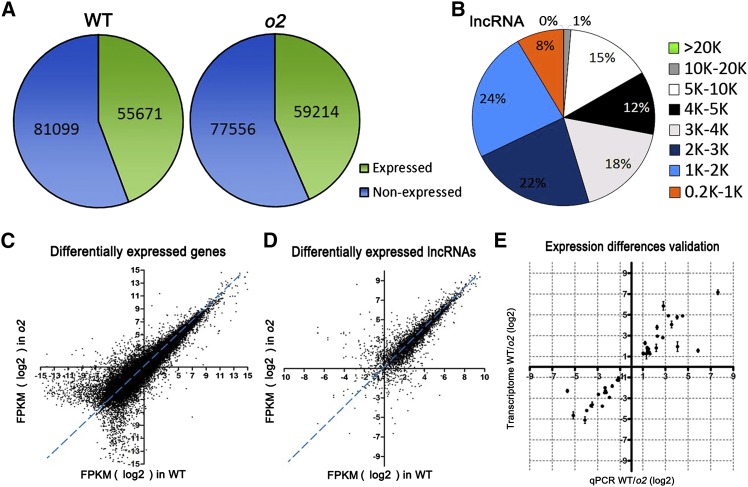
First, we examined the number of genes that are significantly expressed in both 15 DAP wild-type and o2 endosperms. We found 55,671 of the total 136,770 maize gene models transcribed in the wild-type endosperm. In o2, transcripts for 59,214 genes were detected (Figure 1A). There are 52,601 genes transcribed in both wild-type and o2 15 DAP endosperm, leaving 3070 genes that are only expressed in the 15 DAP wild-type endosperm and 6613 genes that are only expressed in the o2 endosperm. For lncRNAs, 5154 transcripts were detected in the 15 DAP wild-type endosperm, and 5163 lncRNAs were detected in the 15 DAP o2 endosperm. The size of the lncRNAs was distributed from 0.2 to 20 kb, with most lengths significantly concentrated at 1 to 2 kb (Figure 1B).
Figure 1.
Genome-Wide Transcript Profile of 15 DAP Wild-Type and o2 Endosperm Determined by RNA-Seq Analysis.
(A) Number of significantly expressed genes in 15 DAP wild-type and o2 endosperm.
(B) Length distribution of RNA-Seq-identified potential lncRNAs.
(C) Comparison of gene expression in 15 DAP wild-type and o2 endosperm.
(D) Comparison of lncRNA expression in 15 DAP wild-type and o2 endosperm.
(E) Validation of expression differences observed by RNA-Seq through qRT-PCR of 40 DEGs. Values are the means with se (n = three individuals). R2 = 0.66.
Next, we determined how many genes that O2 affects O2 by comparing the relative abundance of the transcripts in the wild type in relation to o2 endosperm. Significantly differentially expressed genes (DEGs) were identified as those with a differential expression threshold above a P value < 0.05. Based on this criterion, we determined O2 affects steady state mRNA levels of 1605 genes: 767 show higher levels in the wild type, and 838 show greater accumulations in endosperm lacking O2 function (Figure 1C).
A total of 383 lncRNAs are differently expressed in the wild type and o2: 125 are downregulated and 258 are upregulated in o2 (Figure 1D). To validate the expression differences between wild-type and o2 endosperm observed by RNA-Seq, we performed quantitative RT-PCR (qRT-PCR) on 40 differentially expressed genes (Supplemental Table 1). The results showed similar degrees of mRNA accumulation differences identified by RNA-Seq for 38 of the 40 transcripts (Figure 1E). This demonstrated that the gene expression differences determined by RNA-Seq are reliable.
A Mutation in O2 Creates Extensive Changes in Gene Expression Patterns
Of the 1605 genes that showed significantly altered expression between wild-type and o2 endosperm, 37% could be functionally annotated (annotations were found using BLASTN and BLASTX analyses against the GenBank database [http://www.ncbi.nlm.nih.gov/]). The most significantly related cellular components, biological processes, and molecular functions of the 607 functionally annotated DEGs were characterized using the Gene Ontology (GO) database (http://bioinfo.cau.edu.cn/agriGO/), and this classification is illustrated in Figure 2 and in Supplemental Data Set 1.
Figure 2.
GO Classification for Genes with Altered Expression in o2.
The most significantly related cellular components, biological processes, and molecular functions of the 607 functional annotated DEGs. The significance and number of genes classified within each GO term is shown.
DEGs classified as cellular components corresponded to only one GO term, the ribosome (GO: 0005840, P value = 1.17e-59), including large ribosomal subunit proteins, e.g., eRPL7 (GRMZM2G100225) and eRPL24 (GRMZM2G142640); and small ribosomal subunit proteins, e.g., eRPS24 (GRMZM2G091383) and eRPS27 (GRMZM2G132121). Transcription of all these genes was upregulated in the o2 endosperm.
For DEGs involved in biological processes, o2 has a strong effect on translational elongation (GO: 0006414, P value = 2e-06), cellular nitrogen compound metabolic processes (GO: 0034641, P value = 0.0429), and protein folding (GO: 0006457, P value = 0.0429). Twelve increased DEGs are related to translational elongation, including eEF1As (e.g., GRMZM2G153541) and 60S acidic ribosomal proteins such as LP0 (GRMZM2G066460) and LP2 (GRMZM2G119809). Thirty-one DEGs related to cellular nitrogen compound metabolic processes are significantly associated with amino acid metabolism: serine acetyltransferase (SAT; GRMZM2G048740), which is involved in cysteine biosynthesis, was upregulated by 3.67-fold; the previously identified O2-regulated gene LKR/SDH (GRMZM2G181362), which is involved in Lys catabolism, was downregulated by 4.57-fold; phosphoserine phosphatase (PSPH; GRMZM2G165383), a key enzyme in serine biosynthesis is upregulated; and proline oxidases (GRMZM2G117956 and GRMZM2G053720) have reduced expression. Transcription of other putative amino acid biosynthesis-related genes and the acyl carrier protein (ACP; GRMZM2G091715) involved in fatty acid synthesis were also reduced by o2. Nineteen genes related to protein folding, most of which encode chaperones such as heat shock protein 70 (HSP70; GRMZM2G001500), HSP90 (GRMZM5G813217), and protein disulfide isomerases (PDIs; GRMZM2G091481) are upregulated.
Nutrient reservoir activity (GO: 0045735, P value = 2.29e-17) and endopeptidase inhibitor activity (GO: 0004866, P value = 0.00262) are two subcategories of the molecular functions category that are markedly affected in o2 endosperm. Thirty DEGs related to nutrient reservoir activity, including the genes encoding the zein storage proteins (22-kD α-zeins, 19-kD α-zeins, 14-kD β-zein, 27-kD γ-zein, and 50-kD γ-zein), are reduced in o2. In contrast to zein proteins, transcripts of nonzein seed proteins (e.g., Globulin-2, GRMZM2G026703) typically are preferentially expressed in the embryo and increased 35.27-fold in o2. The nine DEGs related to increased endopeptidase inhibitor activity corresponded to different types of protease inhibitors and stress-related genes.
ChIP-Seq Identifies Genes Directly Targeted by O2
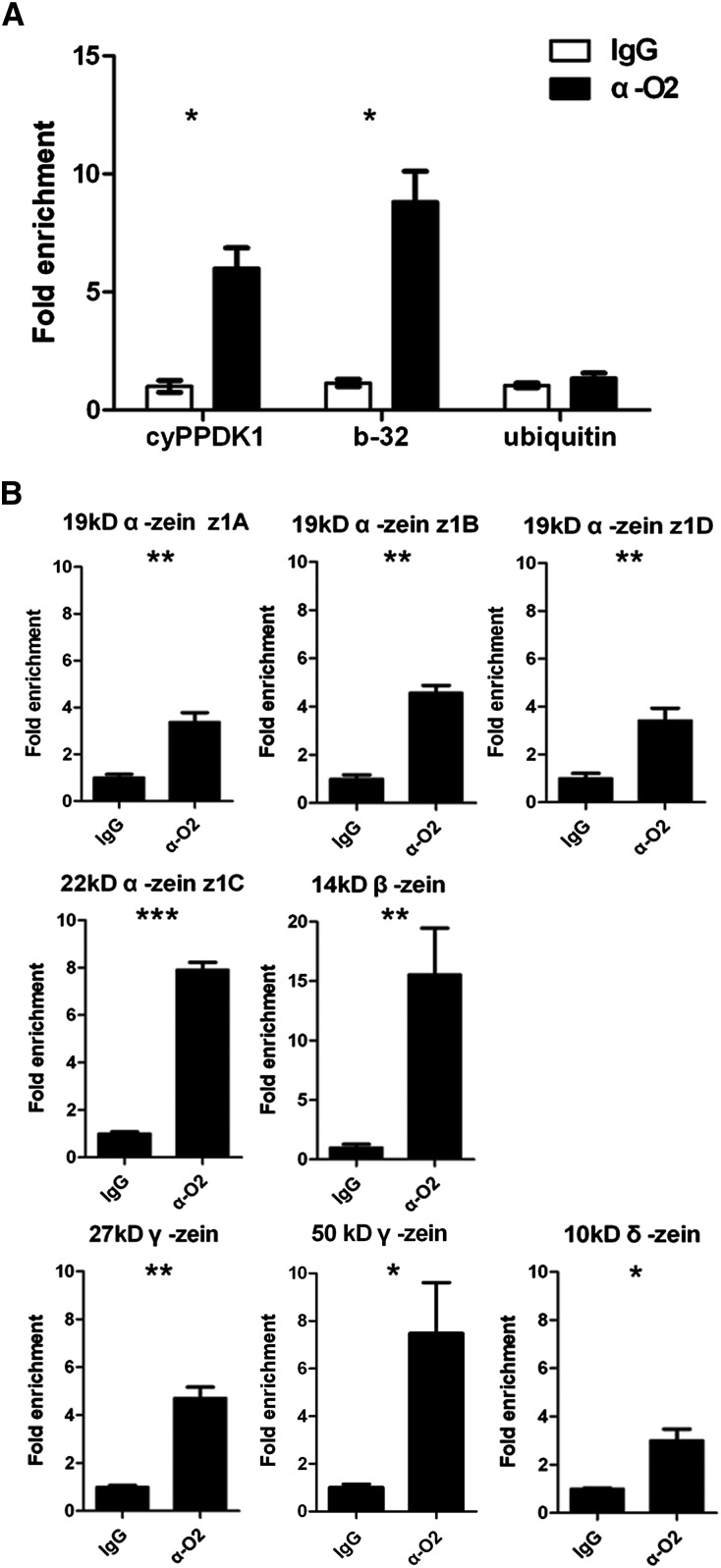
To further understand which DEGs between the wild type and o2 are directly regulated by O2, we identified O2 target genes by investigating the global in vivo DNA binding profile of O2. A ChIP-Seq assay was performed using O2-specific (see Methods) and control antibodies (IgG) on chromatin extracted from the same 15 DAP wild-type endosperm sample used in the RNA-Seq analysis. We modified the conventional chromatin immunoprecipitation (ChIP) method to overcome complications due to the high starch granule content of maize endosperm extracts (see Methods). The O2-specific antibody recognizes the full-length O2 protein and was used in a previous study (Zhang et al., 2012). In ChIP experiments, we determined the specificity of the O2 antibody using immunoblot analyses on the input, post binding input, and elution samples (Supplemental Figure 2). For quality control, chromatin immunoprecipitation with quantitative PCR (ChIP-qPCR) was performed on the ChIPed DNA of positive controls, including the 22-kD α-zein gene, the b-32 gene, and the cyPPDK1 gene, which were all previously shown to be directly regulated by O2 (Lohmer et al., 1991; Maddaloni et al., 1996; Muth et al., 1996). ChIP-qPCR results showed the core promoter fragments of the three genes were greatly enriched in the ChIPed DNA. In contrast, promoter fragments from a ubiquitin gene, the negative control, were not enriched (see Figures 5A and 5B).
Figure 5.
ChIP-qPCR Assays Confirming in Vivo Binding Activity of O2 to the Promoter Region of Zeins.
(A) Anti-O2 antibody was used to precipitate chromatin prepared from 15 DAP wild-type endosperm. IgG was used as an antibody control. Primers from b-32, cyPPDK1, or ubiquitin were used to detect the corresponding promoter fragments in ChIP products for positive and negative control analysis. The fold changes were calculated based on the relative enrichment in anti-O2 compared with anti-IgG. Values are the means with se (n = three individuals; *P < 0.05, Student’s t test).
(B) Primers from the 19-kD α-zein z1A (GRMZM2G059620), the 19-kD α-zein z1B (AF546188.1_FG001), the 19-kD α-zein z1D (AF546187.1_FG001), the 22-kD α-zein gene (GRMZM2G346897), the 14-kD β-zein gene, the 27-kD γ-zein gene, the 50-kD γ-zein gene, and the 10-kD δ-zein gene were used to detect the corresponding promoter fragments in ChIP products by detecting direct binding of O2 to the promoter region of the genes of different zein classes. The fold changes were calculated based on the relative enrichment in the anti-O2 treatment compared with anti-IgG treatment. Values are the means with se (n = three individuals; *P < 0.05, **P < 0.01, and ***P < 0.001, Student’s t test).
ChIP-Seq using the Illumina platform (100-bp-long pair-end reads) produced a total of ∼25 million reads for each sample, of which ∼15 million reads uniquely mapped to the maize genome sequence (Schnable et al., 2009) with an effective reads ratio of ∼60%. O2 binding sites were predicted by Model Based Analysis for ChIP-Seq data (MACS2) (Zhang et al., 2008; q-value < 0.05, based on a Poisson distribution comparing the O2 and IgG ChIP-Seq samples), and 1686 peaks were reported.
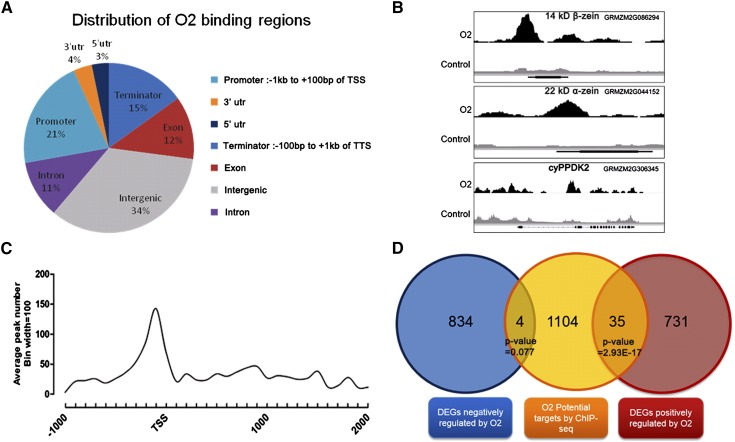
First, we analyzed whether the peaks in the O2 ChIP-Seq experiment were distributed in genic regions, i.e., the region containing the genes as well as 1 kb upstream from the transcription start site (TSS) to 1 kb downstream of the transcription termination site (TTS). Based on these analyses, 66% of O2 binding sites are located in the genic regions of 1143 genes (Figure 3A). Of these, 21% are located in the promoter regions (−1 kb to +100 bp of the TSS), 3% are located in the 5′-untranslated regions (untranslated regions), 11% are located in intron regions, 12% are located in exon regions, 4% are located in 3′-untranslated regions, and 15% are located in the terminator regions (−100 bp to +1 kb of the TTS). These results suggest gene promoter regions are significantly enriched with O2 binding sites. Our positive controls, the 22-kD α-zein, 14-kD β-zein, and cyPPDK1 genes (Cord Neto et al., 1995; Maddaloni et al., 1996; Muth et al., 1996), were detected among the 1143 ChIP-Seq genes with O2 binding sites (Figure 3B). Surprisingly, an O2 binding site in the b-32 gene (Lohmer et al., 1991) was not detected by this experiment, despite the ChIP-qPCR data showing enrichment of the core promoter fragment of b-32 in ChIP product. Sequence analysis indicated highly repetitive DNA sequences in the b-32 promoter region, which may explain our inability to detect it. To investigate the detailed O2 binding profile of the promoter regions, the distance between each peak and its nearest gene’s TSS was calculated. The O2 binding sites are significantly concentrated in the 300 bp immediately upstream of the TSSs in the core promoter regions (Figure 3C). These distribution patterns of O2 binding sites in the ChIP-Seq experiment are consistent with the fact that O2 functions as a transcription factor.
Figure 3.
Summary of O2 ChIP-Seq Results.
(A) Distribution of O2 binding regions in the maize genome.
(B) O2 binding peaks for the positive control genes are shown in the Integrated Genome Browser. For each gene model, the T01 splicing variant is shown. Large boxes indicate exons. Aligned reads are indicated in black (O2) or gray (IgG).
(C) Distribution of O2 binding peaks per 100-bp bin corresponding to the −1000- to +2000-bp region flanking the TSS.
(D) Venn diagram representing a comparison between RNA-Seq and ChIP-Seq results and P value by χ2 test is shown under the gene number of each intersection.
The RNA-Seq revealed 1605 DEGs between wild-type and o2 endosperm, and ChIP-Seq identified 39 of these genes as putative targets directly modulated by O2. Thirty-five of these genes were downregulated in o2, whereas four were upregulated (Figure 3D). There is significant overlap (P value = 2.93E-17, χ2 test) between the downregulated DEGs and the ChIP-Seq data, supporting previous results showing that O2 acts as a transcriptional activator (Schmidt et al., 1992). The 35 downregulated genes with O2 binding sites that are potential O2 direct targets include two 22-kD α-zein genes, one 19-kD α-zein z1A subfamily gene, the 14-kD β-zein gene, the 27-kD γ-zein gene, the 50-kD γ-zein gene, other transcription factors, and genes with predicted functions (Table 1). However, while the RNA-Seq revealed 384 differentially expressed lncRNAs between the wild-type and o2 endosperm, ChIP-Seq identified no lncRNA with an O2 binding site that could be putative O2 targets.
Table 1. Downregulated DEGs with O2 Binding Site Detected.
| Gene | Description | Fold Change | P Value | Peak |
|---|---|---|---|---|
| GRMZM2G160739 | 22-kD α-zein | −37.46 | 1.25E-231 | o2_uniq_peak_783 |
| GRMZM2G044152 | 22-kD α-zein | −40.31 | 6.84E-70 | o2_uniq_peak_782 |
| AF546188.1_FG003 | 19-kD α-zein z1B | −15.36 | 2.45E-07 | o2_uniq_peak_1301 |
| GRMZM2G086294 | 14-kD β-zein | −6.06 | 0.00E+00 | o2_uniq_peak_1195 |
| GRMZM2G138727 | 27-kD γ-zein | −1.32 | 2.22E-10 | o2_uniq_peak_1373 |
| GRMZM2G138689 | 50-kD γ-zein | −1.15 | 3.30E-06 | o2_uniq_peak_1372 |
| GRMZM2G011932 | Transcription factor GBF1 | −3.22 | 1.25E-02 | o2_uniq_peak_1281 |
| GRMZM2G091201 | Myb-like transcription factor | −6.79 | 2.42E-06 | o2_uniq_peak_1260 |
| GRMZM2G384528 | Transcription factor NFYB/HAP3 | −4.79 | 1.18E-02 | o2_uniq_peak_1418 |
| GRMZM2G306345 | cyPPDK1 | −2.64 | 1.21E-68 | o2_uniq_peak_1256 |
| GRMZM2G097457 | cyPPDK2 | −1.28 | 9.94E-16 | o2_uniq_peak_1490 |
| GRMZM2G117956 | Proline oxidase | −3.93 | 3.71E-03 | o2_uniq_peak_1645 |
| GRMZM2G053720 | Proline oxidase | −1.96 | 1.03E-02 | o2_uniq_peak_81 |
| GRMZM2G442658 | Alcohol dehydrogenase 1 | −1.53 | 6.90E-14 | o2_uniq_peak_237 |
| GRMZM2G378106 | Indole-3-acetic acid amido synthetase | −1.63 | 3.52E-02 | o2_uniq_peak_537 |
| GRMZM2G312877 | Lactoylglutathione lyase | −2.01 | 3.55E-05 | o2_uniq_peak_1478 |
| GRMZM2G028110 | Lactoylglutathione lyase | −2.53 | 2.18E-03 | o2_uniq_peak_1239 |
| GRMZM2G130987 | Translocon Sec61/SecY | −1.27 | 1.30E-03 | o2_uniq_peak_33 |
| GRMZM2G341934 | Peroxidase | −1.97 | 2.13E-02 | o2_uniq_peak_13 |
| GRMZM2G152616 | β-Lactamase-inhibitor protein II | −1.53 | 3.89E-02 | o2_uniq_peak_885 |
| GRMZM2G105954 | Cytochrome P450 | −1.51 | 4.27E-02 | o2_uniq_peak_830 |
| GRMZM5G847052 | Unknown | −1.74E+07 | 3.51E-51 | o2_uniq_peak_1361 |
| GRMZM2G438538 | Unknown | −1.10 | 2.61E-08 | o2_uniq_peak_1373 |
| GRMZM5G896196 | Unknown | −4.45 | 1.24E-06 | o2_uniq_peak_1568 |
| GRMZM5G811968 | Unknown | −2.53 | 1.26E-04 | o2_uniq_peak_81 |
| GRMZM5G895499 | Unknown | −1.81 | 1.09E-03 | o2_uniq_peak_702 |
| GRMZM5G894976 | Unknown | −4.04 | 1.31E-03 | o2_uniq_peak_1587 |
| GRMZM2G012031 | Unknown | −1.86 | 1.53E-03 | o2_uniq_peak_1289 |
| GRMZM2G054233 | Unknown | −2.24 | 9.38E-03 | o2_uniq_peak_959 |
| GRMZM2G163304 | Unknown | −21.57 | 1.17E-02 | o2_uniq_peak_973 |
| GRMZM5G817886 | Unknown | −1.58 | 1.19E-02 | o2_uniq_peak_384 |
| GRMZM2G371606 | Unknown | −1.99 | 1.44E-02 | o2_uniq_peak_762 |
| GRMZM2G371612 | Unknown | −4.79 | 2.12E-02 | o2_uniq_peak_762 |
| GRMZM2G446225 | Unknown | −1.47 | 2.14E-02 | o2_uniq_peak_730 |
| GRMZM2G404599 | Unknown | −28.13 | 2.14E-02 | o2_uniq_peak_1584 |
Binding Motif Analysis Reveals Novel O2 cis-Elements
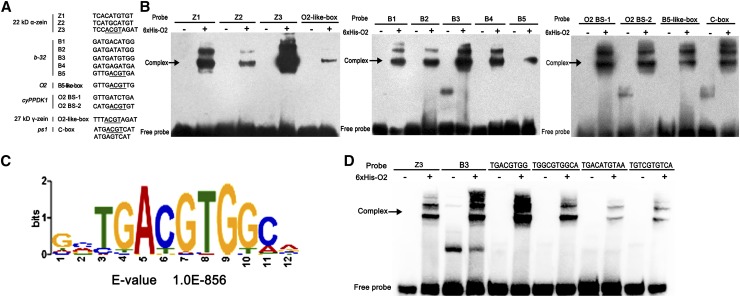
To confirm O2 was binding previously published cis-elements, an electrophoretic mobility shift assay (EMSA) was performed using a purified His-fusion recombinant O2 protein and a labeled DNA probe containing the O2 binding sites within a 50-bp sequence derived from O2 target genes. The cis-elements studied included the Z1 (TCACATGTGT), Z2 (TCATGCATGT) and Z3 (TCCACGTAGAT) sites from the 22-kD α-zein gene promoter (Muth et al., 1996); the B1 (GATGACATGG), B2 (GATGATATGG), B3 (GATGATGTGG), B4 (GATGAGATGA), and B5 (GTTGACGTGA) from the b-32 gene (Lohmer et al., 1991); the O2 BS-1 (GTTGATCTGA) and O2 BS-2 (CATGACGTGT) from the cyPPDK1 gene (Maddaloni et al., 1996); the O2-like-box (TTTACGTAGAT) from the 27-kD γ-zein gene promoter (Wu and Messing, 2012); the C-box and odd C-box (ATGACGTCAT or ATGAGTCAT) from the pea (Pisum sativum) lectin (ps1) gene (de Pater et al., 1994; Cord Neto et al., 1995); and the B5-like-box (GTTGACGTTG) from the O2 gene (Lohmer et al., 1991; Rossi et al., 1997). As shown in Figure 4B, without the O2 protein no bands were observed, except for the free probe. After adding the O2 protein, high molecular weight complexes were formed for all 13 cis-elements, as demonstrated by the appearance of the upshifted band.
Figure 4.
O2 Binding Sites.
(A) The specific sequence and origin of 13 previously reported O2 binding sites. The ACGT core is underlined.
(B) EMSAs with 6XHis-O2 fusion protein using probes derived from O2 target gene promoters containing 13 previously reported O2 binding sites (Supplemental Table 2). Arrows on the left indicate the formation of O2 and O2 binding-element complexes.
(C) O2 binding motif identified by MEME-ChIP in 1-kb flanking sequences around the genic peak summits and the density plot of this motif around the summits of peaks.
(D) EMSAs with probe derived from the O2 target gene promoters containing Z3, B3, and four O2 binding sites identified in this study (Supplemental Table 2). Arrows on the left indicate the formation of O2 and O2 binding element complexes.
To explore novel O2 binding motifs, the 1-kb flanking sequences around all of the genic peak summits were applied to the motif discovery tool MEME-ChIP (http://meme.nbcr.net; Machanick and Bailey, 2011), and the motif TGACGTGG with an ACGT core was identified as a statistically defined motif (E-value = 1.0E-856; Figure 4C). The previously published 13 cis-elements were also submitted to the MEME platform, and the same TGACGTGG motif was generated. Therefore, this motif was used for further analysis. By examining the density plot of this newly identified motif within all peaks, the TGACGTGG motif was mostly enriched from −100 to 100 bp around the peak maxima (Figure 4C). We performed cis-element screening using the TGACGTGG motif and the 13 previously published cis-elements on the 1-kb flanking sequences around the peak maxima of the 35 potential O2 direct target genes (Solovyev et al., 2010). The TGACGTGG motif was detected in 13 of the 35 genes, including the 14-kD β-zein gene. However, the 13 cis-elements, the TGACGTGG motif, and the ACGT core were not detected in the following O2 target genes: a Myb-like transcription factor (GRMZM2G091201), the 50-kD γ-zein gene (GRMZM2G138689), the cyPPDK2 (GRMZM2G097457), and peroxidase (GRMZM2G341934). Specific binding motifs (TGGCGTGGCA, TGACATGTAA, and TGTCGTGTCA) were calculated for the Myb-like transcription factor, the 50-kD γ-zein gene, and cyPPDK2, respectively, using the MEME platform. To investigate O2 binding on these four cis-elements, EMSA was performed with a labeled DNA probe with a 50-bp sequence derived from the 14-kD β-zein gene, the Myb-like transcription factor, the 50-kD γ-zein gene, and cyPPDK2 containing TGACGTGG, TGGCGTGGCA, TGACATGTAA, and TGTCGTGTCA, respectively. As shown in Figure 4D, O2 binding to TGACGTGG was even stronger than previously reported for the strong O2 binding sites Z3 and B3. High molecular weight complexes were also formed for the three other newly identified cis-elements upon addition of the O2 fusion protein, as observed by the shifted band. The 1-kb flanking sequences around all of the intergenic peak maxima were also applied to MEME-ChIP, and the same TGACGTGG core sequence was generated (E-value = 2.1E-1867; Supplemental Figure 2). It remains to be explored whether the binding of O2 to the intergenic regions could be involved in transcriptional regulation of distant genes or take part in other biological processes.
O2 Directly Regulates Different Zein Gene Classes
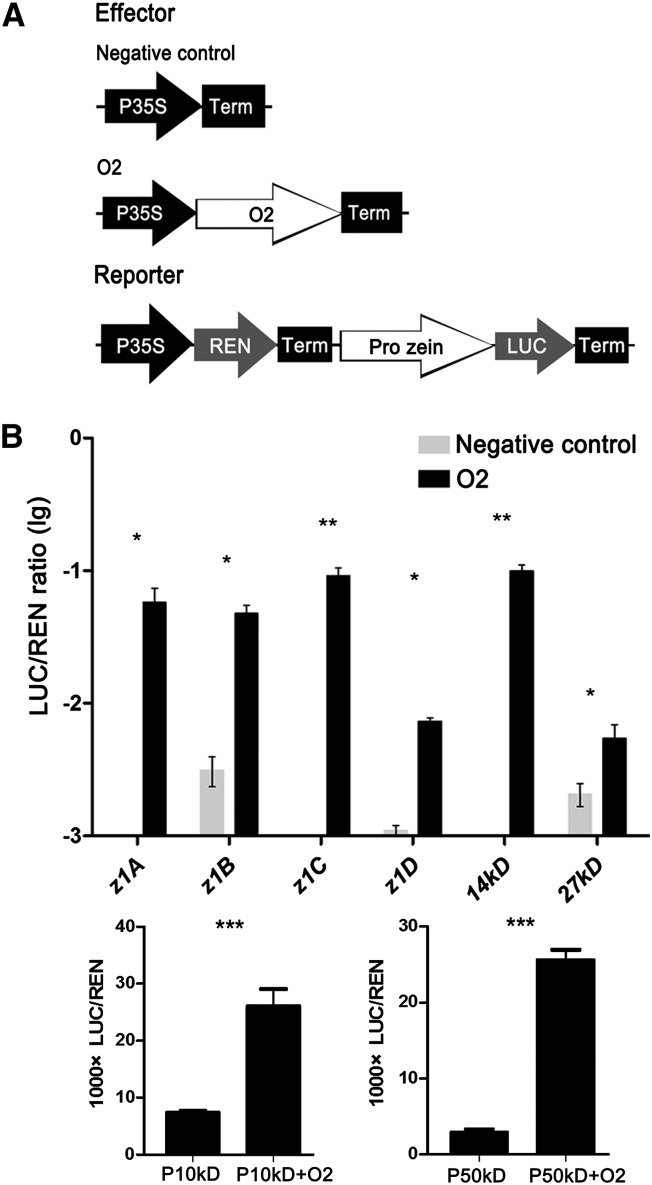
According to the RNA-Seq and ChIP-Seq results, among the zein genes, two 22-kD α-zein genes, one 19-kD α-zein z1A subfamily gene, the 14-kD β-zein gene, the 27-kD γ-zein gene, and the 50-kD γ-zein gene are potential O2 targets, whereas the 16-kD γ-zein gene and the 18-kD δ-zein gene are not directly regulated by O2. Because α-zein gene sequences share high similarity, some O2 target α-zeins might be absent in our ChIP-Seq results. Additionally, because the 10-kD δ-zein has no gene model information, it might not be detected by our ChIP-Seq analysis. Consequently, ChIP-qPCR was performed to assess the capacity of O2 to directly regulate the different classes of zein genes with O2 binding sites detected in the ChIP-Seq experiment (Figure 5B). Real-time PCR showed that the 100-bp promoter fragments located within the −300-bp region of the TSS of several randomly selected genes, including the 22-kD α-zein gene (GRMZM2G346897), the 19-kD α-zein z1A (GRMZM2G059620), z1B (AF546188.1_FG001), and z1D (AF546187.1_FG001) subfamily genes, the 14-kD β-zein gene, the 27-kD γ-zein gene, the 50-kD γ-zein gene, and the 10-kD δ-zein gene, showed significant enrichment in ChIP products.
To understand whether O2 functions as a transcriptional activator of the different zeins, we performed a dual-luciferase transient transcriptional activity assay in onion (Allium cepa) epidermal cells with O2 driven by the cauliflower mosaic virus (CaMV) 35S promoter as an effecter and LUC (the firefly luciferase-coding gene driven by the different −500- to −1-bp zein promoters, respectively) as the reporter (Figure 6A). The results showed that O2 specifically induced the expression of LUC with the 22-kD α-zein promoter, the 19-kD α-zein z1A, z1B, and z1D promoters, the 14-kD β-zein promoter, the 27-kD γ-zein promoter, the 50-kD γ-zein promoter, and the 10-kD δ-zein promoter (Figure 6B).
Figure 6.
O2 Functions as a Transcriptional Activator of Different Zein Genes.
(A) The 35S:REN-Pro zein:LUC reporter constructs were transiently expressed in onion epidermal cells together with control vector or 35S:O2 effector, respectively.
(B) The expression level of Renilla (REN) was used as an internal control. The LUC/REN ratio represents the relative activity of zein promoters. Data are values of three independent experiments. Significant differences from the corresponding control values (using Student’s t test [n = 3]): *P < 0.05, **P < 0.01, and ***P < 0.001.
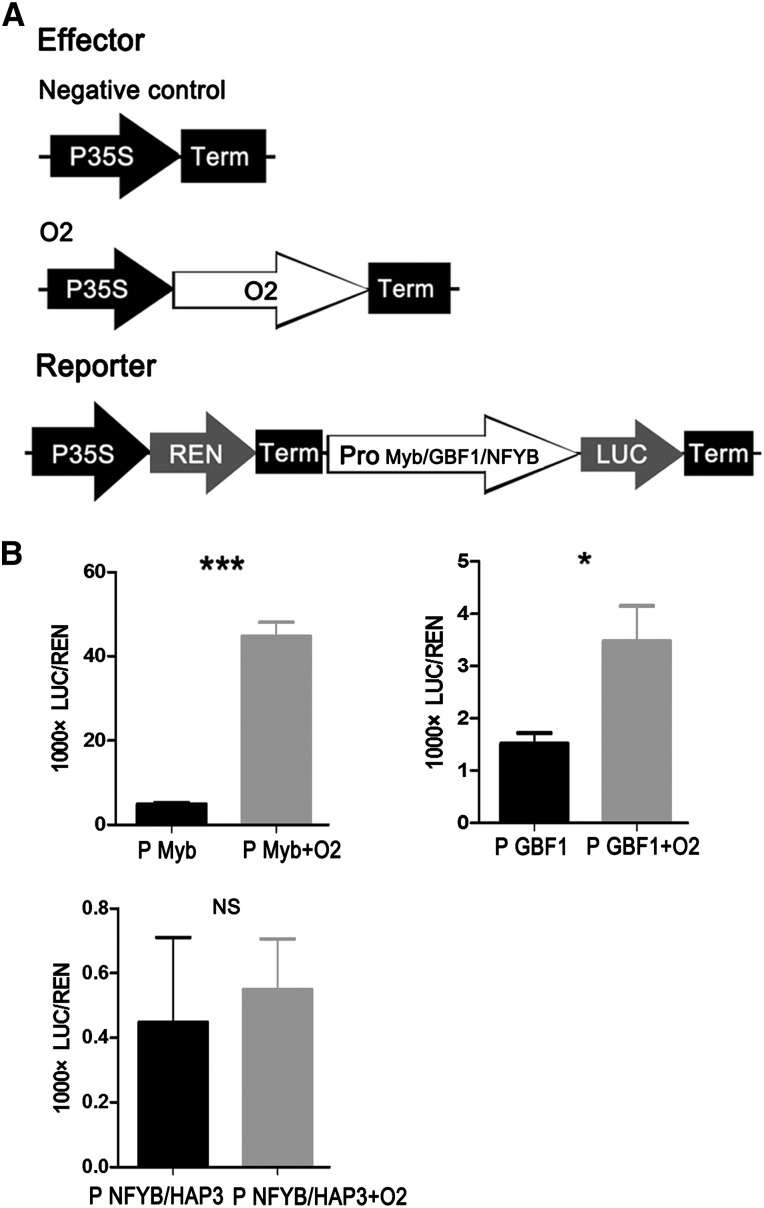
O2 Directly Regulates Other Transcriptional Factors
The altered expression of many genes in our study is consistent with previously reported transcript profile analysis of the o2 mutant and cannot be explained by the direct transcriptional activation of the O2 protein alone. Therefore, there might be hierarchical transcriptional regulation within the genome-wide regulatory framework of O2. To investigate this, we analyzed transcription factors within the 35 potential O2 direct target genes (Table 1), including the Myb-like transcription factor (GRMZM2G091201), the bZIP G-box binding factor 1 (GBF1; GRMZM2G011932), and a NFYB/HAP3 transcription factor (GRMZM2G384528). We performed dual-luciferase transient transcriptional activity assays to investigate whether O2 could function as an activator of these genes. Again, O2 expression was driven by the CaMV 35S promoter, and LUC controlled by the promoter sequence of the transcription factors was used as the reporter (Figure 7A). The results showed that LUC expression driven by the Myb-like transcription factor and GBF1 promoters was prominently induced by O2 (Figure 7B). This result indicates the Myb-like transcription factor and GBF1 are O2 target genes.
Figure 7.
O2 Functions as a Transcriptional Activator of Two Transcriptional Factors.
(A) The 35S:REN-Pro Myb TF:LUC, 35S:REN-Pro GBF1:LUC, and 35S:REN-Pro NFYB/HAP3 TF:LUC reporter constructs were transiently expressed in onion epidermal cells, together with control vector or 35S:O2 effector, respectively.
(B) The LUC/REN ratio represents the relative activity of the promoters. Data are values of three independent experiments. Significant differences from the corresponding control values (using Student’s t test [n = 3]): *P < 0.05 and ***P < 0.001.
O2 Regulates Genes Related to Nitrogen Metabolism
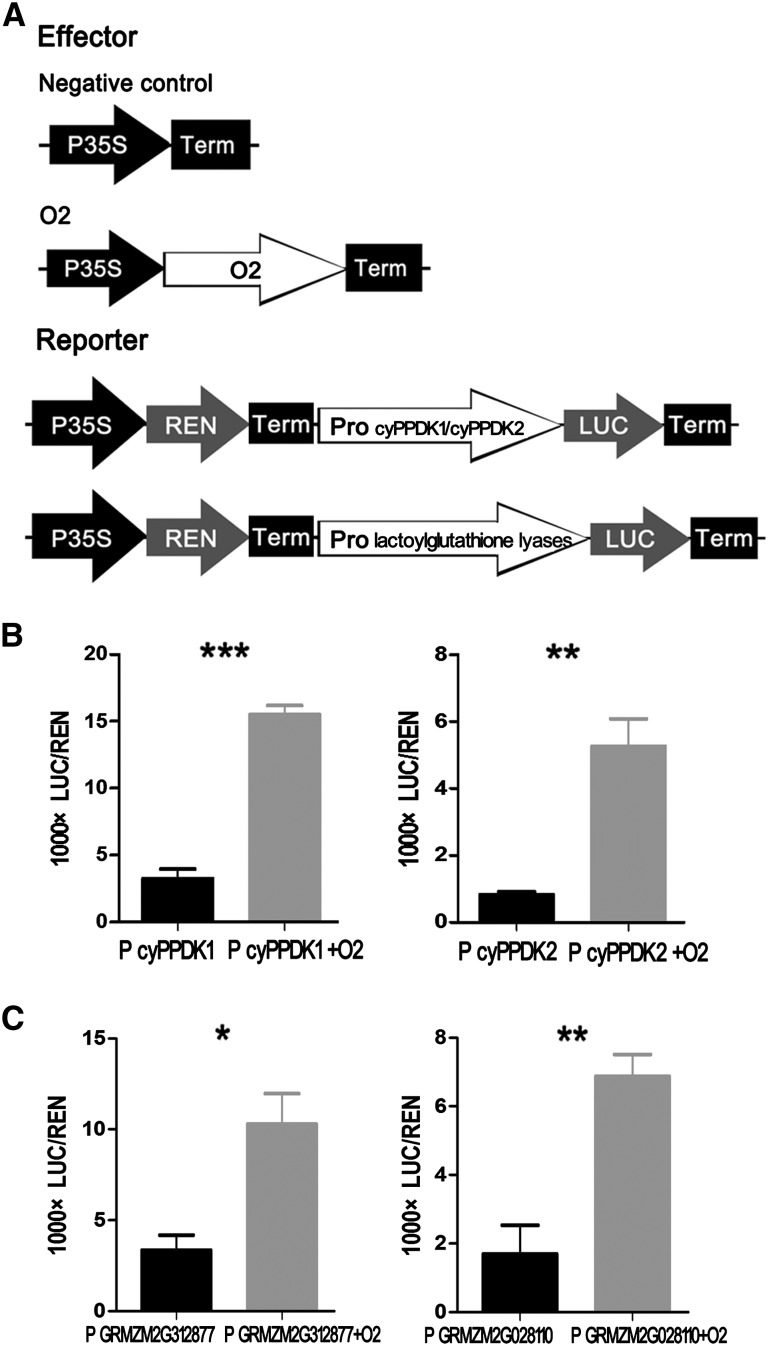
Cellular nitrogen compound metabolic processes (GO: 0034641) are prominently altered in o2 (Figure 2). For example, cyPPDK1 is a well-documented O2 target gene involved in nitrogen metabolism, and cyPPDK2 was also detected among the 35 potential O2 direct target genes (Table 1). Both cyPPDK1 and cyPPDK2 enzymes are major regulators of the glycolytic pathway and could be linked to carbon and amino acid metabolism by converting pyruvate to phosphoenol pyruvate (Wang and Larkins, 2001). We performed dual-luciferase transient transcriptional activity assays with O2 protein and LUC driven by the promoter sequences of cyPPDK1 and cyPPDK2 as reporters (Figure 8A), and both promoters were significantly induced by O2 (Figure 8B). Proline oxidase is involved in proline metabolism (Huang and Cavalieri, 1979), and two proline oxidase genes were detected as O2 targets according to the ChIP-Seq data (Table 1), implicating genes involved with carbon and amino acid metabolism.
Figure 8.
O2 Functions as a Transcriptional Activator of cyPPDK1, cyPPDK2, and Two Lactoylglutathione Lyases.
(A) The 35S:REN-Pro cyPPDK1:LUC, 35S:REN-Pro cyPPDK2:LUC, 35S:REN-Pro GRMZM2G312877:LUC, and 35S:REN-Pro GRMZM2G028110:LUC reporter constructs were transiently expressed in onion epidermal cells together with control vector or 35S:O2 effecter.
(B) The LUC/REN ratio represents the relative activity of the cyPPDK1 and cyPPDK2 promoters. Data are values of three independent experiments. Significant differences from the corresponding control values (using Student’s t test [n = 3]): **P < 0.01 and ***P < 0.001.
(C) The LUC/REN ratio represents the relative activity of two lactoylglutathione lyase promoters. Data are values of three independent experiments. Significant differences from the corresponding control values (using Student’s t test [n = 3]): *P < 0.05 and **P < 0.01.
O2 Regulates Genes Involved in Stress Resistance
b-32 is a well-known O2 target that plays a role in defense against fungal pathogenesis (Ferreira et al., 2007). Endopeptidase inhibitor activity (GO: 0004866), a subcategory of genes that contribute to stress resistance was also prominently altered in the o2 mutant (Figure 2). Lactoylglutathione lyase (LGL; also known as glyoxalase I) is the first enzyme of the glyoxalase system and detoxifies methylglyoxal, a cytotoxic compound that increases rapidly under stressful conditions. Consequently, LGLs play an important role in cellular detoxification and tolerance to abiotic stresses (Wu et al., 2013). Two LGLs (GRMZM2G312877 and GRMZM2G028110) were detected as O2 target genes according to the ChIP-Seq results (Table 1), and dual-luciferase transient transcriptional activity assays (Figure 8A) indicated that both LGLs were directly regulated by O2 (Figure 8C).
DISCUSSION
The o2 mutation is known to have many pleiotropic effects on maize endosperm development. Some of these are understood, but many are not. The best known consequence of o2 is a starchy endosperm texture and high lysine content. The starchy endosperm phenotype appears to be due to a substantial reduction of zein protein body size and number, as a consequence of reduced zein synthesis. This disrupts embedding of starch granules in the protein matrix of the mature endosperm (Mertz et al., 1964). The high lysine phenotype of o2 is mostly attributed to a reduction in zeins, which lack lysine residues. As a result of reduced zein synthesis, there is a compensatory increase in lysine-containing nonzein endosperm proteins. These changes in o2 protein composition result in nearly twice the lysine content of wild-type maize, which improve nutritional value (Mertz et al., 1964). The increased amount of lysine in o2 endosperm is also due in part to reduced expression of LKR/SDH, a key enzyme responsible for lysine catabolism (Kemper et al., 1999; Arruda et al., 2000; Stepansky et al., 2006). In addition, a lysine-sensitive aspartate kinase isoenzyme of the aspartate biosynthetic pathway is inhibited by the o2 mutation (Azevedo and Arruda, 2010).
Cloning of the O2 gene provided some insight into the pleiotropic effects of the o2 mutation. O2 encodes a bZIP transcription factor (Schmidt et al., 1990) that activates expression of 22-kD α-zeins (Muth et al., 1996) and the 14-kD β-zein (Cord Neto et al., 1995). The first characterized nonzein target of O2 is the b-32 gene (Di Fonzo et al., 1986), which is associated with the defense against fungal pathogens (Ferreira et al., 2007). The cyPPDK1 gene, which is involved in carbon and amino acid metabolism, was also identified as directly regulated by O2 (Sheen, 1991; Maddaloni et al., 1996). The LKR/SDH mRNA is reduced by >90% in o2; consequently, this gene was predicted to be directly regulated by O2 (Kemper et al., 1999; Prioul et al., 2008). O2 itself is believed to be positively autoregulated by its gene product, based on a B5-like box in its promoter region (Lohmer et al., 1991; Rossi et al., 1997). Identification of this diverse group of O2-regulated genes provided insight into the pleiotropic phenotypic effects of the o2 mutation.
To provide a more detailed understanding of O2-regulated gene expression, profiling studies based on microarray analysis were performed that revealed multiple DEGs in o2 endosperm (Hunter et al., 2002; Jia et al., 2007; Frizzi et al., 2010; Hartings et al., 2011; Jia et al., 2013). These studies showed that, along with 22-kD α-zeins and the 14-kD β-zein, a number of other zein genes, e.g., 19-kD α-zeins and the 10-kD δ-zein, also showed decreased transcription. There is simultaneous upregulation of several nonzein seed storage protein genes (Globulin S and α-Globulin) (Hunter et al., 2002), translation-related proteins (eEF1A and some ribosomal proteins) (Hunter et al., 2002), and lysine-rich proteins (sorbitol dehydrogenase and glyceraldehyde 3-phosphate dehydrogenase) (Jia et al., 2013) in o2; some stress-responsive and defense proteins are strongly regulated (HSPs, alcohol dehydrogenases, and protease inhibitors) (Hunter et al., 2002; Jia et al., 2013).
In this study, we greatly expanded our understanding of O2 regulation through comprehensive, genome-wide gene expression profiling of o2 using RNA-Seq. This experiment led to the identification of 1605 DEGs in o2 compared with the wild type (Figure 1) and confirmed many DEGs identified in previous studies, including zein genes, amino acid metabolism-related genes, stress response-related genes, and translational elongation-related genes (Hunter et al., 2002; Jia et al., 2013; Supplemental Data Set 1). In addition, we identified a number of additional DEGs. For example, expression of additional zein genes, such as the 50-kD γ-zein gene, was found to be prominently reduced in o2 (Supplemental Data Set 1). Genes involved in amino acid metabolism, including two proline oxidase genes, were significantly downregulated, and the SAT gene was upregulated in o2 (Supplemental Data Set 1). Translation-related proteins, e.g., eEF1B and many ribosomal proteins, were prominently induced in o2 (Supplemental Data Set 1). PDI, which encodes an endoplasmic reticulum enzyme involved in the oxidation of cysteine to form disulfide bonds (Tsai et al., 2001), showed increased transcript accumulation (Supplemental Data Set 1). We also identified 383 lncRNAs as DEGs in o2 (Supplemental Data Set 1), although their function was not investigated.
To identify which DEGs are directly regulated by O2, we performed ChIP-Seq with an O2-specific antibody. This analysis revealed 1686 O2 binding sites. O2 binds target genes by recognizing various cis-elements (Figure 4) not only through previously identified O2 binding motifs, but also four more motifs. TGACGTGG was determined to be the most conserved O2 binding motif, even stronger than the previously reported strongest binding sites (Z3 and B5). This sequence is found in the 14-kD β-zein gene promoter and explains the high expression level of this zein gene. Another O2 binding cis-element, TGACATGTAA, was detected in 50-kD γ-zein promoter (Figure 4).
Thus, we discovered that O2 not only directly binds to the promoters of known targets (22-kD α-zein and 14-kD β-zein genes), but also to many other zein genes. The promoter regions of 19-kD α-zein (the z1A, z1B, z1D subfamily) genes directly interact with O2 (Table 1, Figure 5). Transactivation of O2 on the promoters of z1A, z1B, and z1D zein genes was confirmed by dual-luciferase transactivation assay. The 27-kD γ-zein gene was hypothesized to be an O2 direct target (Wu and Messing, 2012). The 16- and 27-kD γ-zein genes originated from a common progenitor (Xu and Messing, 2008), but the promoter sequence of 16-kD γ-zein is quite different from that in other zeins (Wu and Messing, 2012). There are O2 binding sites in 27-kD γ-zein and 50-kD γ-zein genes (Figure 5). Strong transcriptional activation by O2 for the 50-kD γ-zein and weak transcriptional activation for the 27-kD γ-zein was detected (Figure 6), indicating that O2 directly regulates the γ-zein genes. The 10- and 18-kD δ-zein genes also originated from a common progenitor and share high sequence similarity (Xu and Messing, 2008). Direct binding of O2 to the promoter region of the 10-kD δ-zein was detected (Figure 5), but not to the promoter region of the 18-kD δ-zein, although the transactivation assay confirmed that O2 regulates the 10-kD δ-zein. Therefore, O2 directly regulates most of the zein classes, except for the 16- and 18-kD zeins. But the transactivation effect of O2 on different zein genes is variable. In the case of α- and β-zein genes, O2 appears to be a prominent transcriptional activator. However, for the 10 kD δ-zein, and the 27 kD and 50 kD γ-zein genes, O2 seem to act more as a co-factor with some unknown transcriptional activator(s). It was reported that O2 interacts with the prolamin-box binding factor (PBF1) and may cooperatively activate expression of the 22-kD α-zein genes (Vicente-Carbajosa et al., 1997). PBF1 was also found to regulate the 27-kD γ-zein gene (Wu and Messing, 2012).
Among nonzein gene O2 targets, we confirmed that the previously characterized b-32 and cyPPDK1 genes are O2 regulated (Table 1). However, our data indicate the LKR/SDH gene is not a direct O2 target because no O2 binding site was detected in its genic sequence. We also demonstrated that O2 does not directly bind its own sequence. We also report that cyPPDK2 and proline oxidases (nitrogen compound metabolism related genes) are O2 direct targets (Table 1, Figure 8). Direct activation by O2 on PPDK genes is likely critical in controlling the balance between C and N metabolism. We also identified LGLs (stress resistance-related genes) as O2 target genes (Table 1, Figure 8). LGLs respond to salt and oxidative stresses (Wu et al., 2013), suggesting an important role in cellular detoxification and tolerance to abiotic stresses. The identification of LGLs as O2 directly regulated genes is consistent with their functions, which are associated with alterations in nutrient reservoir activity, nitrogen compound metabolism, and endopeptidase inhibitor activity (Figure 2). Despite the broad range of functions associated with O2 target genes, they cannot explain all of the phenotypic alterations of the o2 mutation.
We also identified the genes encoding GBF1 and the Myb transcriptional factor (GRMZM2G091201) as O2 direct targets. Maize GBF1 is a bZIP protein that binds to G-box elements and was previously reported to be one of the factors involved in activation of Alcohol dehydrogenase 1 (Adh1; de Vetten and Ferl, 1995). Expression of ADHs was consistently shown to be downregulated in o2 (Hunter et al., 2002), and Adh1 was detected in our O2 direct target profiling results (Table 1). These results indicate O2 and GBF1 might function collaboratively to regulate Adh1 gene expression. Transcriptional modulation by O2 involves interaction with ADA2 and GCN5 (Bhat et al., 2003, 2004). The activity of O2 is also affected by interaction with a Taxilin, leading to a change in subcellular distribution (Zhang et al., 2012). Through the direct regulation of other transcriptional factors, or direct interaction with other proteins, the transcriptional regulatory function of O2 could be broadened and amplified.
None of the differentially expressed lncRNAs were identified by ChIP-Seq as putative O2 direct targets, indicating that the differentially expressed lncRNAs might be the targets of downstream transcription factors. The metabolic changes in o2 might also trigger the transcriptional activation of noncoding regions of the maize genome. The biological roles of lncRNAs include transcription and translation regulation, RNA modification, and epigenetic modification of chromatin structure (Mattick, 2004; Mattick and Makunin, 2006; Eddy, 2001). Differences in expression levels of some DEGs could also be explained by hierarchical control of O2-regulated lncRNAs.
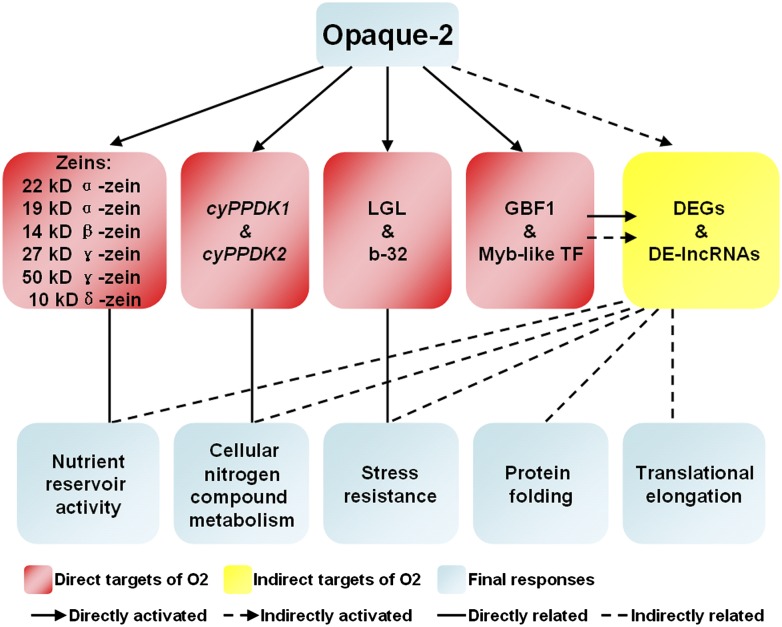
In summary, our results demonstrate that O2 is a central regulator of multiple metabolic pathways related to anabolic functions during maize endosperm development. It acts in both direct and indirect ways to affect the patterns of gene transcription. We built a hierarchical regulatory model describing O2 and O2-targeted transcriptional factors to explain the pleiotropic biological effects of the O2 mutation. This is illustrated in Figure 9, which shows a genome-wide transcriptional regulatory network affected by O2. Our data suggest that O2 not only directly activates genes associated with a variety of nutrient storage genes and enzymes involved in carbon and amino acid metabolism pathways, but also controls transcription factors that are, in turn, regulating various other aspects of plant metabolism. Quality protein maize (QPM), a modified version of the o2 mutant, has provided important insights that direct future approaches for the production of high-yield, high-lysine maize (Azevedo and Arruda, 2010). However, the nature of genetic modifiers in QPM is poorly understood, and a better understanding of O2 regulatory pathways will provide insight into the pleiotropic effects of the o2 mutation and approaches for developing more nutritious maize varieties, such as QPM, in the future.
Figure 9.
A Hierarchical Genome-Wide Transcriptional Regulatory Framework for O2 Involving O2 Targeted Transcription Factors.
O2 not only directly activates genes associated with nutrient storage and enzymes involved in carbon and amino acid metabolism but also controls transcription factors that regulate various other aspects of plant metabolism.
METHODS
Plant Materials
The o2 mutant stock (701D) was obtained from the Maize Genetics Cooperation Stock Center and then backcrossed into the W22 genetic background for six generations (BC6). Maize (Zea mays) plants were cultivated in the field at the campus of Shanghai University. After six backcross generations followed by two more generations of selfing, lines of homozygous o2 mutant (o2/o2) or wild type (+/+) were obtained. Kernels at 15 DAP were collected from selfing homozygous o2 or wild-type ears, as experimental materials. The seeds were frozen in liquid nitrogen and stored at −80°C. Mature seeds were also harvested for phenotype observation (Supplemental Figure 1).
RNA-Seq Analysis
Total RNA (10 μg) was extracted from endosperm of o2 and wild-type kernels harvested at 15 DAP and, three o2 or wild-type biological samples were pooled together. The RNA-Seq library was prepared according to Illumina standard instruction (TruSeq Stranded RNA LT Guide). Library DNA was checked for concentration and size distribution in an Agilent 2100 bioanalyzer before sequencing with an Illumina HiSequation 2500 system according to the manufacturer’s instructions (HiSequation 2500 User Guide). Reads were aligned to the maize B73 genome using TopHat 2.0.6 (Langmead et al., 2009). Data were normalized as fragments per kilobase of exon per million fragments mapped, for the sensitivity of RNA-Seq depends on the transcript length. Significant DEGs were identified as those with a P value of differential expression above the threshold (P < 0.05). lncRNAs were identified based on the PhyloCSF platform (http://compbio.mit.edu/PhyloCSF; Lin et al., 2011).
RNA Extraction and Real-Time PCR Analysis
Total RNA was extracted from 15 DAP endosperm of o2 and wild-type kernels harvested at with TRIzol reagent (Tiangen), removing DNA by treatment with RNase free DNase I (Takara). RNA was then reverse transcribed to cDNA using ReverTra Ace reverse transcriptase (Toyobo). Quantitative real-time PCR of the randomly selected 40 DEGs was performed using a Bio-Rad iCycler IQ real-time PCR detection system (Bio-Rad) with SYBR Green Real-Time PCR Master Mix (ABI) according to the standard protocol. Specific primers of 40 DEGs were designed (Supplemental Table 3), and the experiments were performed using two independent RNA samples sets with ubiquitin as the internal control gene. Each RNA sample was extracted from a pool of three kernels, and three technical replicates were analyzed. Relative quantifiable differences in gene expression were analyzed as described previously (Livak and Schmittgen, 2001).
Polyclonal Antibody
For O2 antibody production, a full-length O2 cDNA was cloned into the BglII and HindIII sites of the pET-32a+ vector (Novagen) and the construct transformed into Rosetta (DE3). Cells were grown at 37°C and induced by the addition of isopropylthio-β-galactoside to a final concentration of 1 mM when the optical density at 600 nm was 0.6. The 6xHis fusion recombinant O2 protein was purified with the ÄKTA purification system using a 1-mL HisTrapTM FF crude column (GE Healthcare). Protein analysis and purification followed established procedures. Antibodies were produced in rabbits according to standard protocols of Shanghai ImmunoGen Biological Technology.
ChIP
ChIP with O2 antibody was performed as previously described (Kaufmann et al., 2010) with modifications to accommodate the high concentration of starch granules in maize endosperm. Briefly, 5 g of dissected starchy 15 DAP endosperms was cross-linked in MC buffer (25 mL) with 1% formaldehyde on ice under vacuum; the vacuum was released after 15 min and reapplied for another 14 min. Fixation was stopped by adding glycine to a final concentration of 0.125 M, and the fixed sample was washed thrice with MC buffer and ground to a powder in liquid nitrogen, followed by nuclear isolation by adding the frozen powder to 30 mL of M1 buffer. The homogenate was filtered through four layers of Miracloth prior to nuclei isolation. Nuclear-enriched extracts were resuspended in 5 mL lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride, and 10 mM sodium butyrate) containing a plant proteinase inhibitor cocktail (Sigma-Aldrich), followed by sonication for 5 min on medium power in 1.5 mL sonic buffer using a Bioruptor UCD-200 (Diangenode) and centrifugation at 500g to remove starch granules. The chromatin solution was sonicated for 5 min on medium power five times to create ∼300-bp average fragment sizes, as estimated by agarose gel electrophoresis. Antibody against O2 and the IgG (Sigma-Aldrich) control were used for immunoprecipitation. The precipitated DNA was recovered using a QIAquick PCR purification kit (Qiagen) and analyzed by real-time quantitative PCR using the appropriate DNA primers (Supplemental Table 3) and SYBR Green Real-Time PCR Master Mix (Applied Biosystems). Equal amounts of starchy endosperm tissue and ChIP products were used for the quantitative real-time PCR reaction. ChIP values were normalized to their respective DNA input values, and the fold changes in concentration were calculated based on the relative enrichment in anti-O2 compared with anti-IgG immunoprecipitates. The means and standard deviations were calculated from three biological replicates.
Analysis of ChIP-Seq Data
ChIP DNA samples were subjected to end repair and A-base addition, followed by ligation with adapters. Barcode sequences of adapters correspond to the following: AGTCAA, ChIP of O2; AGTTCC, ChIP of IgG. Illumina Genome Analyzer (GA) sequencing was performed by the OSUCCC Nucleic Acid Shared Resource-Illumina GA II Core. We defined target genes as those that contain ChIP-Seq peaks located within transcribed regions of genes, in introns or 1 kb upstream of the TSS, or 1 kb downstream of the TTS. ChIP-Seq reads were aligned to the maize genome (Schnable et al., 2009) using Bowtie2 (Langmead et al., 2009). Bowtie 2 supports gapped and paired-end alignment modes. We ran Bowtie version 2.2.3 with default parameters and reported only unique alignments. ChIP-Seq peaks were detected by MACS2 (Zhang et al., 2008). We used MACS version 2.0.10 with default parameters, as duplicates were allowed and q-value < 0.05.
EMSA
The full-length O2 cDNA was amplified with gene-specific primers and fused into the BamHI and NotI sites of the expression vector pET-32a+. The construct was transformed into Escherichia coli BL21 (DE3) cells, grown at 37°C, and induced by the addition of isopropylthio-β-galactoside to a final concentration of 1 mM when the optical density at 600 nm was 0.6. The pET-32a-O2-expressed 6xHis-O2 fusion protein was purified with HisPur Ni-NTA Resin (Thermo Fisher) and used for EMSA. Oligonucleotide probes (Supplemental Table 2) were synthesized and labeled according to the standard protocol of Shanghai Invitrogen Technology. We used standard reaction mixtures for EMSA containing 20 ng of purified 6xHis-O2 fusion protein, 5 ng of biotin-labeled annealed oligonucleotides, 2 μL of 10× binding buffer (100 mM Tris, 500 mM KCl, and 10 mM DTT, pH 7.5), 1 μL of 50% (v/v) glycerol, 1 μL of 100 mM MgCl2, 1 μL of 1 mg/mL poly(dI-dC), 1 μL of 1% (v/v) Nonidet P-40, and double-distilled water to a final volume of 20 μL. The reactions were incubated at 25°C for 20 min, electrophoresed in 6% (w/v) polyacrylamide gels, and then transferred to N+ nylon membranes (Millipore) in 0.5× Trisborate/ EDTA buffer at 380 mA at 4°C for 30 min. Biotin-labeled DNA was detected using the LightShift Chemiluminescent EMSA kit (Thermo Fisher). Bands were visualized by autoradiography.
Transient Assays for in Vivo Activation Activity
To generate the Pro zein:LUC reporters and Pro TF:LUC reporters for the dual-luciferase assays, the −500 bp from TSS promoter region of different zein genes, transcription factors, and other potential targets were inserted into the XhoI and NotI, SmaI and NotI, HindIII and NotI, or BamHI and NotI sites of pGreenII0800-LUC. To generate the CaMV 35S promoter-driven O2 effector and CaMV 35S promoter-driven GBF1 effector, full-length coding sequence of the O2 gene and GBF1 genes was inserted into the BamHI and HindIII sites of pUC18-35S. Transient dual-luciferase assays in onion (Allium cepa) epidermal cells were performed and checked using dual-luciferase assay reagents (Promega). Briefly, 1 μg of DNA was used to coat 0.3 mg of 1.0-μm-diameter tungsten particles. The onion epidermal cells were bombarded using the PDS-1000 system (Bio-Rad) at 1100 p.s.i. helium pressure twice and then the bombarded samples were incubated for at least 8 h in the dark at 25°C. Sample discs (1 to 2 cm in diameter) were ground in liquid nitrogen and homogenized in 100 μL passive lysis buffer from the dual-luciferase assay reagents (Promega). The crude extract (20 μL) was mixed with 100 μL of luciferase assay buffer and the firefly luciferase (LUC) activity measured using the Tecan M200 system. One hundred microliters of Stop and Glow Buffer was then added to the reaction and Renilla luciferase (REN) activity was measured. For this analysis, the ratio between LUC and REN activities was measured three times.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Zm O2, NM_001111884, GRMZM2G015534; Zm GBF1, NM_001111894, GRMZM2G011932; Myb-like transcription factor, NM_001158143, GRMZM2G091201; and lactoylglutathione lyases, NM_001152619, GRMZM2G312877, and NM_001159147, GRMZM2G028110. RNA-Seq and ChIP-Seq data are available from the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the series entries GSE61830 and GSE63991, respectively.
Supplemental Data
Supplemental Figure 1. Phenotype of Mature Wild-Type and o2 Kernels.
Supplemental Figure 2. Determination of the O2 Antibody Specificity in Input Sample, Post Bind Input Sample, and Elution Sample of the ChIP Experiment by Immunoblot Analyses.
Supplemental Figure 3. O2 Binding Motif Identified by MEME-ChIP on 1-kb Flanking Sequences around the Intergenic Peak Summits and Its Density Plot around the Summits of Peaks.
Supplemental Table 1. mRNA Accumulation Differences Identified by RNA-Seq and qRT-PCR for 38 out of the 40 Selected DEGs.
Supplemental Table 2. EMSA Probes as Specific Sequences Derived from Potential O2 Direct Targets Containing O2 Binding Sites.
Supplemental Table 3. Primers Used in These Experiments.
Supplemental Data Set 1. Gene Ontology Classification of DEGs with Functional Annotation.
Supplementary Material
Acknowledgments
This work was supported by The National Science Fund for Distinguished Young Scholars of China (31425019), The Ministry of Science and Technology of China (2012AA10A305 and 2014CB138204), and The National Natural Sciences Foundation of China (31271733). We thank Lin Xu (Shanghai Institute of Plant Physiology and Ecology, SIBS) for technical support on ChIP experiments and Brian A. Larkins (University of Nebraska, Lincoln) for critical reading of the article.
AUTHOR CONTRIBUTIONS
R.S., W.Q., C.L., and Z.Q. designed the experiment. C.L., Z.Q., Q.W., Y.Y., X.Y., and H.X. performed the experiments. C.L., Z.Q., W.Q., Y.T., B.M., Y.L., H.Z., and R.S. analyzed the data. W.Q. and R.S. wrote the article.
Glossary
- RNA-Seq
RNA sequencing
- lncRNA
long, noncoding RNA
- ChIP-Seq
chromatin immunoprecipitation coupled to high-throughput sequencing
- DAP
days after pollination
- ncRNA
noncoding RNA
- DEG
differentially expressed gene
- qRT-PCR
quantitative RT-PCR
- GO
Gene Ontology
- ChIP-qPCR
chromatin immunoprecipitation with quantitative PCR
- TSS
transcription start site
- TTS
transcription termination site
- EMSA
electrophoretic mobility shift assay
- CaMV
cauliflower mosaic virus
- LGL
lactoylglutathione lyase
- QPM
quality protein maize
- ChIP
chromatin immunoprecipitation
Footnotes
Articles can be viewed online without a subscription.
References
- Arruda P., Kemper E.L., Papes F., Leite A. (2000). Regulation of lysine catabolism in higher plants. Trends Plant Sci. 5: 324–330. [DOI] [PubMed] [Google Scholar]
- Azevedo R.A., Arruda P. (2010). High-lysine maize: the key discoveries that have made it possible. Amino Acids 39: 979–989. [DOI] [PubMed] [Google Scholar]
- Bhat R.A., Borst J.W., Riehl M., Thompson R.D. (2004). Interaction of maize Opaque-2 and the transcriptional co-activators GCN5 and ADA2, in the modulation of transcriptional activity. Plant Mol. Biol. 55: 239–252. [DOI] [PubMed] [Google Scholar]
- Bhat R.A., Riehl M., Santandrea G., Velasco R., Slocombe S., Donn G., Steinbiss H.H., Thompson R.D., Becker H.A. (2003). Alteration of GCN5 levels in maize reveals dynamic responses to manipulating histone acetylation. Plant J. 33: 455–469. [DOI] [PubMed] [Google Scholar]
- Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147: 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cord Neto G., Yunes J.A., da Silva M.J., Vettore A.L., Arruda P., Leite A. (1995). The involvement of Opaque 2 on beta-prolamin gene regulation in maize and Coix suggests a more general role for this transcriptional activator. Plant Mol. Biol. 27: 1015–1029. [DOI] [PubMed] [Google Scholar]
- de Pater S., Katagiri F., Kijne J., Chua N.H. (1994). bZIP proteins bind to a palindromic sequence without an ACGT core located in a seed-specific element of the pea lectin promoter. Plant J. 6: 133–140. [DOI] [PubMed] [Google Scholar]
- de Vetten N.C., Ferl R.J. (1995). Characterization of a maize G-box binding factor that is induced by hypoxia. Plant J. 7: 589–601. [DOI] [PubMed] [Google Scholar]
- Di Fonzo N., Manzocchi L., Salamini F., Soave C. (1986). Purification and properties of an endospermic protein of maize associated with the Opaque-2 and Opaque-6 genes. Planta 167: 587–594. [DOI] [PubMed] [Google Scholar]
- Eddy S.R. (2001). Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2: 919–929. [DOI] [PubMed] [Google Scholar]
- Emerson R.A., Beadle G.W., Fraser A.C. (1935). A summary of linkage studies in maize. Cornell Univ. Agric. Exp. Stn. Memoir 180: 1–83. [Google Scholar]
- Ernst C., Morton C.C. (2013). Identification and function of long non-coding RNA. Front. Cell. Neurosci. 7: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R.B., Monteiro S., Freitas R., Santos C.N., Chen Z., Batista L.M., Duarte J., Borges A., Teixeira A.R. (2007). The role of plant defence proteins in fungal pathogenesis. Mol. Plant Pathol. 8: 677–700. [DOI] [PubMed] [Google Scholar]
- Frizzi A., Caldo R.A., Morrell J.A., Wang M., Lutfiyya L.L., Brown W.E., Malvar T.M., Huang S. (2010). Compositional and transcriptional analyses of reduced zein kernels derived from the opaque2 mutation and RNAi suppression. Plant Mol. Biol. 73: 569–585. [DOI] [PubMed] [Google Scholar]
- Hartings H., Lauria M., Lazzaroni N., Pirona R., Motto M. (2011). The Zea mays mutants opaque-2 and opaque-7 disclose extensive changes in endosperm metabolism as revealed by protein, amino acid, and transcriptome-wide analyses. BMC Genomics 12: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.H., Cavalieri A.J. (1979). Proline oxidase and water stress-induced proline accumulation in spinach leaves. Plant Physiol. 63: 531–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter B.G., Beatty M.K., Singletary G.W., Hamaker B.R., Dilkes B.P., Larkins B.A., Jung R. (2002). Maize opaque endosperm mutations create extensive changes in patterns of gene expression. Plant Cell 14: 2591–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Nettleton D., Peterson J.M., Vazquez-Carrillo G., Jannink J., Scott M.P. (2007). Comparison of transcript profiles in wild-type and o2 maize endosperm in different genetic backgrounds. Crop Sci. 47: S45–S59. [Google Scholar]
- Jia M., Wu H., Clay K.L., Jung R., Larkins B.A., Gibbon B.C. (2013). Identification and characterization of lysine-rich proteins and starch biosynthesis genes in the opaque2 mutant by transcriptional and proteomic analysis. BMC Plant Biol. 13: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Østerås M., Farinelli L., Krajewski P., Angenent G.C. (2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5: 457–472. [DOI] [PubMed] [Google Scholar]
- Kemper E.L., Neto G.C., Papes F., Moraes K.C.M., Leite A., Arruda P. (1999). The role of opaque2 in the control of lysine-degrading activities in developing maize endosperm. Plant Cell 11: 1981–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert R.J., Alexander D.E., Dudley J.W. (1969). Relative performance of normal and modifi ed protein (opaque2) maize hybrids. Crop Sci. 9: 187–190. [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.F., Jungreis I., Kellis M. (2011). PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27: i275–i282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Loesch J., Foley D.C., Cox D.F. (1976). Comparative resistance of opaque-2 and normal inbred lines of maize to ear-rotting pathogens. Crop Sci. 16: 841–842. [Google Scholar]
- Lohmer S., Maddaloni M., Motto M., Di Fonzo N., Hartings H., Salamini F., Thompson R.D. (1991). The maize regulatory locus Opaque-2 encodes a DNA-binding protein which activates the transcription of the b-32 gene. EMBO J. 10: 617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P., Bailey T.L. (2011). MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddaloni M., Donini G., Balconi C., Rizzi E., Gallusci P., Forlani F., Lohmer S., Thompson R., Salamini F., Motto M. (1996). The transcriptional activator Opaque-2 controls the expression of a cytosolic form of pyruvate orthophosphate dikinase-1 in maize endosperms. Mol. Gen. Genet. 250: 647–654. [DOI] [PubMed] [Google Scholar]
- Mattick J.S. (2004). RNA regulation: a new genetics? Nat. Rev. Genet. 5: 316–323. [DOI] [PubMed] [Google Scholar]
- Mattick J.S., Makunin I.V. (2006). Non-coding RNA. Hum. Mol. Genet. 15: R17–R29. [DOI] [PubMed] [Google Scholar]
- Mertz E.T., Bates L.S., Nelson O.E. (1964). Mutant gene that changes protein composition and increase lysine content of maize endosperm. Science 145: 279–280. [DOI] [PubMed] [Google Scholar]
- Muth J.R., Müller M., Lohmer S., Salamini F., Thompson R.D. (1996). The role of multiple binding sites in the activation of zein gene expression by Opaque-2. Mol. Gen. Genet. 252: 723–732. [DOI] [PubMed] [Google Scholar]
- Nass H.G., Crane P.L. (1970). Effect of endosperm mutants on germination and early seedling growth rate in maize (Zea mays L.). Crop Sci. 10: 139–140. [Google Scholar]
- Novikova I.V., Hennelly S.P., Tung C.S., Sanbonmatsu K.Y. (2013). Rise of the RNA machines: exploring the structure of long non-coding RNAs. J. Mol. Biol. 425: 3731–3746. [DOI] [PubMed] [Google Scholar]
- Prioul J.L., Méchin V., Damerval C. (2008). Molecular and biochemical mechanisms in maize endosperm development: the role of pyruvate-Pi-dikinase and Opaque-2 in the control of C/N ratio. C. R. Biol. 331: 772–779. [DOI] [PubMed] [Google Scholar]
- Rossi V., Motto M., Pellegrini L. (1997). Analysis of the methylation pattern of the maize opaque-2 (O2) promoter and in vitro binding studies indicate that the O2 B-Zip protein and other endosperm factors can bind to methylated target sequences. J. Biol. Chem. 272: 13758–13765. [DOI] [PubMed] [Google Scholar]
- Schmidt R.J., Burr F.A., Burr B. (1987). Transposon tagging and molecular analysis of the maize regulatory locus opaque-2. Science 238: 960–963. [DOI] [PubMed] [Google Scholar]
- Schmidt R.J., Burr F.A., Aukerman M.J., Burr B. (1990). Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif that binds to zein DNA. Proc. Natl. Acad. Sci. USA 87: 46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R.J., Ketudat M., Aukerman M.J., Hoschek G. (1992). Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell 4: 689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Sheen J. (1991). Molecular mechanisms underlying the differential expression of maize pyruvate, orthophosphate dikinase genes. Plant Cell 3: 225–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev V.V., Shahmuradov I.A., Salamov A.A. (2010). Identification of promoter regions and regulatory sites. Methods Mol. Biol. 674: 57–83. [DOI] [PubMed] [Google Scholar]
- Stepansky A., Less H., Angelovici R., Aharon R., Zhu X., Galili G. (2006). Lysine catabolism, an effective versatile regulator of lysine level in plants. Amino Acids 30: 121–125. [DOI] [PubMed] [Google Scholar]
- Swiezewski S., Liu F., Magusin A., Dean C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802. [DOI] [PubMed] [Google Scholar]
- Tafer H., Hofacker I.L. (2008). RNAplex: a fast tool for RNA-RNA interaction search. Bioinformatics 24: 2657–2663. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B., Rodighiero C., Lencer W.I., Rapoport T.A. (2001). Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 104: 937–948. [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa J., Moose S.P., Parsons R.L., Schmidt R.J. (1997). A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc. Natl. Acad. Sci. USA 94: 7685–7690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.C., Chang H.Y. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell 43: 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Larkins B.A. (2001). Genetic analysis of amino acid accumulation in opaque-2 maize endosperm. Plant Physiol. 125: 1766–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Ma C., Pan Y., Gong S., Zhao C., Chen S., Li H. (2013). Sugar beet M14 glyoxalase I gene can enhance plant tolerance to abiotic stresses. J. Plant Res. 126: 415–425. [DOI] [PubMed] [Google Scholar]
- Wu Y., Messing J. (2012). Rapid divergence of prolamin gene promoters of maize after gene amplification and dispersal. Genetics 192: 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.H., Messing J. (2008). Organization of the prolamin gene family provides insight into the evolution of the maize genome and gene duplications in grass species. Proc. Natl. Acad. Sci. USA 105: 14330–14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Qiao Z., Liang Z., Mei B., Xu Z., Song R. (2012). Zea mays Taxilin protein negatively regulates opaque-2 transcriptional activity by causing a change in its sub-cellular distribution. PLoS ONE 7: e43822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.