Arabidopsis protein NCA1 interacts with catalases in the cytosol and increases catalase activity through maintaining catalase folding state, which is required for stress responses.
Abstract
Catalases are key regulators of reactive oxygen species homeostasis in plant cells. However, the regulation of catalase activity is not well understood. In this study, we isolated an Arabidopsis thaliana mutant, no catalase activity1-3 (nca1-3) that is hypersensitive to many abiotic stress treatments. The mutated gene was identified by map-based cloning as NCA1, which encodes a protein containing an N-terminal RING-finger domain and a C-terminal tetratricopeptide repeat-like helical domain. NCA1 interacts with and increases catalase activity maximally in a 240-kD complex in planta. In vitro, NCA1 interacts with CATALASE2 (CAT2) in a 1:1 molar ratio, and the NCA1 C terminus is essential for this interaction. CAT2 activity increased 10-fold in the presence of NCA1, and zinc ion binding of the NCA1 N terminus is required for this increase. NCA1 has chaperone protein activity that may maintain the folding of catalase in a functional state. NCA1 is a cytosol-located protein. Expression of NCA1 in the mitochondrion of the nca1-3 mutant does not rescue the abiotic stress phenotypes of the mutant, while expression in the cytosol or peroxisome does. Our results suggest that NCA1 is essential for catalase activity.
INTRODUCTION
Due to their sessile nature, plants face numerous environmental changes during their life cycle. Soil alkalization is a major problem that affects plant development and crop productivity. Apoplastic pH is weakly acidic (about pH 5.8) and apoplastic buffering capacity is low. When plants are exposed to alkali stress, apoplastic pH increases, leading to impaired cell wall extension and reduced plant growth (Gout et al., 1992; Rayle and Cleland, 1992; Rober-Kleber et al., 2003; Lager et al., 2010; Yang et al., 2010). Higher apoplastic pH also impairs root hair formation and reduces water and ion uptake (Palmgren, 2001). Each plant cell organelle has an optimal pH required for the function of its specialized enzymes (Werdan et al., 1975; Whitten et al., 2005; Casey et al., 2010), and high external pH also impairs pH homeostasis in these organelles. The result is that many plant biological processes, including photosynthesis and photorespiration (Heldt et al., 1973; Servaites, 1977; Song et al., 2004), are affected.
Alkaline and other abiotic stresses, including salt, drought, and low/high temperature, induce the production of reactive oxygen species (ROS) (Foreman et al., 2003; Mittler et al., 2004; Selivanov et al., 2008; Munné-Bosch et al., 2013; Yao et al., 2013) in chloroplasts, mitochondria, and peroxisomes, as well as at the plasma membrane. Accumulation of ROS in cellular compartments results in oxidative stress and affects organelle integrity. However, ROS also functions as a signal molecule triggering pathways for plant growth, development, and response to stress. Therefore, tight control of ROS homeostasis is critical (Suzuki et al., 2012). ROS include singlet oxygen (1O2), superoxide anion (O2−), hydrogen peroxide (H2O2), and hydroxyl radicals (HO⋅; likely responsible for oxidative damage during many stresses). Transformation among these species is a common phenomenon in the cell. Accumulation of ROS is eliminated by antioxidants, antioxidative enzymes, and scavenging enzymes like catalase (CAT), ascorbate peroxidase (APX), and superoxide dismutase, leading to the regulation of ROS homeostasis (Alscher et al., 1997; Mhamdi et al., 2010a).
The Arabidopsis genome encodes three catalase proteins, which consist of 492 amino acids and share high sequence similarity. While catalase gene expression and enzyme activity have been found in most plant tissues, expression and activity are higher in aerial tissues than in the root. These results suggest that catalases play important roles in plant antioxidative and detoxification processes that are closely correlated with ROS generation during photosynthesis and photorespiration (Mhamdi et al., 2012). In contrast to other peroxidases (PRXs), which require a cofactor to catalyze the reaction and for which water is the only product, catalases catalyze a dismutation reaction to produce both H2O and O2. The affinity (Km) of catalases for H2O2 is in the range 40 to 600 mM (Chelikani et al., 2004), which is much lower than those of APX and PRX (Km below 100 µM), suggesting that catalases may function in the plant cell mainly when local H2O2 concentrations reach a high level.
While evidence exists for catalases in different cell compartments, they predominantly localize to the peroxisomes. To achieve higher activity, monofunctional catalases bind heme as a cofactor and form homotetramers in the peroxisomes. In Arabidopsis thaliana, genetic studies show that knockout of CAT3 only slightly reduces catalase activity; deletion of CAT2 reduces catalase activity by 80%, while deletion of CAT1 has no effect on this activity (Mhamdi et al., 2010a). Consistent with these findings, no obvious phenotype is detected in Arabidopsis cat1 and cat3 knockout mutants. The cat2 mutant shows defects in many processes, including photorespiration and pathogenesis, salicylic acid-dependent hypersensitive response-like lesion formation (Chaouch and Noctor, 2010), autophagy-dependent cell death (Hackenberg et al., 2013), sensitivity to the inhibitor 3-amino-1,2,4-triazole (3-AT), and gene expression during both abiotic and biotic stresses, including hypersensitive response, cold, heat, and drought (Vanderauwera et al., 2005).
Several proteins have been reported to interact with catalase and affect its activity, including calmodulin (Yang and Poovaiah, 2002), nucleoside diphosphate kinase 1 (Fukamatsu et al., 2003), Salt Overly Sensitive2 (Verslues et al., 2007), triple gene block protein 1 (Mathioudakis et al., 2013), NCA1 (Hackenberg et al., 2013), and LESION SIMULATING DISEASE1 (LSD1) (Li et al., 2013). However, it is not clear if or how these proteins directly regulate catalase activity.
Here, we found that NCA1 is required for catalase activity. In a screen for Arabidopsis mutants that are sensitive to high pH, we identified a mutant altered in NCA1, a cytosol-localized protein. We found that the tetratricopeptide repeat (TPR) domain in the NCA1 C terminus mediates interaction with CAT2 and that the binding of a zinc ion in the N-terminal RING-finger domain of NCA1 is essential for the full function of CAT2. Our findings reveal a mechanism of maintaining catalase activity both in cytosol and in peroxisomes.
RESULTS
Genetic Cloning of NCA1
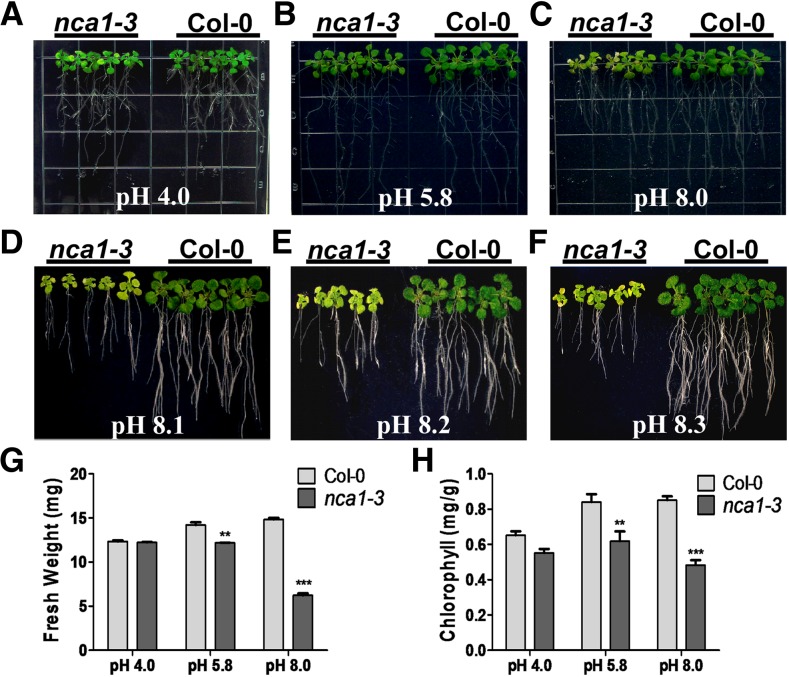
To identify genes involved in the response to alkaline stress in Arabidopsis, we generated a T-DNA insertion (Agrobacterium tumefaciens-transferred DNA) pool (Zhao et al., 2011) and used it to screen for mutants whose growth was affected by high external pH. Several alkaline-sensitive mutants were recovered, and one of them, subsequently named nca1-3 based on cloning results (see below), was hypersensitive to high external pH. When Columbia-0 (Col-0) and nca1-3 seeds were germinated on Murashige and Skoog (MS) medium at pH 5.8, the growth of 5-d-old nca1-3 seedlings was similar to that of Col-0 seedlings (Supplemental Figure 1). When these seedlings were transferred to medium at pH 8.0, or above, shoot growth of the nca1-3 mutant was reduced relative to the growth of Col-0 after 10 d of growth at pH 5.8, as were seedling fresh weight and chlorophyll content (Figure 1). The mutant and wild-type plants displayed no significant differences when grown on MS medium at pH 4.0 (Figures 1A, 1G, and 1H). On medium at pH 8.0 or above, root elongation in nca1-3 was dramatically reduced compared with that of Col-0 (Figures 1C to 1H).
Figure 1.
The Arabidopsis nca1-3 Mutant Is Hypersensitive to High-External pH.
(A) to (F) Screening of mutants with reduced growth at high-external pH. Five-day-old Col-0 and nca1-3 mutant seedlings germinated and grown on MS medium at pH 5.8 were transferred to MS medium at pH 4.0, 5.8, 8.0, 8.1, 8.2, or 8.3. Photographs were taken 10 d after transfer.
(G) Fresh weight of Col-0 and nca1-3 seedlings germinated and grown on MS medium at pH 5.8 and then transferred to MS medium at pH 4.0, 5.8, or 8.0. Data represent means ± sd of at least three replicate experiments. Statistical significance compared with Col-0 values is based on a Student’s t test (**P < 0.01 and ***P < 0.001).
(H) Chlorophyll content of Col-0 and nca1-3 seedlings germinated and grown on MS medium at pH 5.8 and then transferred to MS medium at pH 4.0, 5.8, or 8.0. Data represent means ± sd of at least three replicate experiments. Statistical significance compared with Col-0 values is based on a Student’s t test (**P < 0.01 and ***P < 0.001).
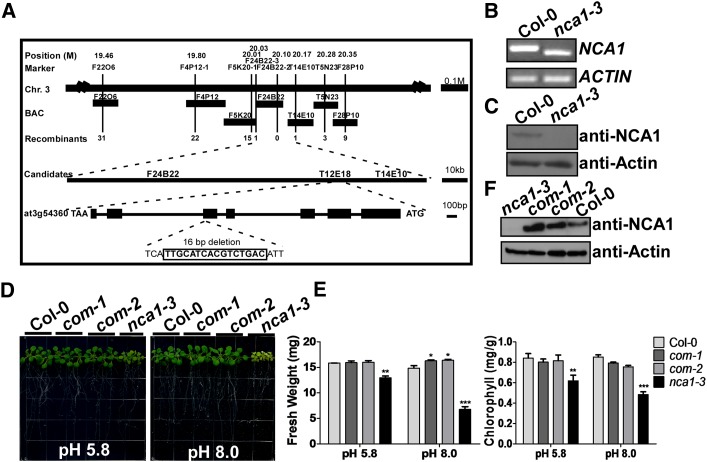
Plasmid rescue and thermal asymmetric interlaced PCR were attempted to clone the mutated gene; however, the T-DNA insertion could not be identified. Using simple sequence length polymorphism markers, we mapped the mutated locus to chromosome III between the F24B22 and T14E10 BAC clones (TAIR) within ∼140 kb. All coding sequences in this region were then sequenced, and a 16-bp deletion was identified in exon 5 in At3g54360, a gene that had been identified as NCA1 and shown to be involved in regulating autophagy-dependent cell death (Hackenberg et al., 2013). As a result, we named the mutant nca1-3. The deletion was located 960 bp downstream of the NCA1 translation initiation codon and introduced a frame shift and a premature stop codon in the NCA1 coding sequence (Figure 2A). NCA1 encodes a protein containing a RING-finger domain and two TPR motifs. To determine if expression of NCA1 is altered in the mutant, total RNA was extracted from 10-d-old Col-0 and nca1-3 seedlings and analyzed using RT-PCR. A shorter NCA1 transcript was detected in nca1-3 compared with that in Col-0 (Figure 2B). To determine if NCA1 protein level is changed in the mutant, NCA1-specific antibodies were generated by immunizing mice with Escherichia coli-expressed NCA1. Total protein was extracted from Col-0 and nca1-3 and analyzed using immunoblot assays. Strong cross-reaction was detected in Col-0; no cross-reaction was observed in the nca1-3 mutant. As a control, Actin was monitored and detected in both nca1-3 and Col-0 (Figure 2C).
Figure 2.
Cloning of NCA1.
(A) Map-based cloning of the mutation in nca1-3. The chromosomal region containing the mutation is represented on the top line with the simple sequence length polymorphism markers indicated above and the corresponding BAC clones indicated below. The number of recombinants between these markers and the mutation in 1850 samples is included. The region covered by BAC clones F24B22 to T14E10 is labeled as “Candidates.” The gene structure is indicated with exons represented by boxes and introns represented by lines. The 16-bp deletion at the end of the fifth exon generates a new stop codon. Size markers are shown on the right.
(B) Analysis of NCA1 expression by RT-PCR. NCA1 expression in Col-0 and the nca1-3 mutant is shown. ACTIN was used as loading control. One representative analysis of three replicates experiments is shown.
(C) Analysis of NCA1 protein level in Col-0 and the nca1-3 mutant. Total protein was extracted from 10-d-old seedlings and probed with anti-NCA1 and antiactin (used as a loading control) antibodies. One representative analysis of three replicates experiments is shown.
(D) Complementation of the nca1-3 mutant with the full-length NCA1 genomic sequence. Five-day-old seedlings of Col-0, nca1-3, and two independent complemented lines (com-1 and com-2) from seeds germinated on MS at pH 5.8 were transferred to MS medium at pH 5.8 or 8.0. Photographs were taken 10 d after transfer.
(E) Fresh weight and chlorophyll content of Col-0, nca1-3, com-1, and com-2 seedlings. Data represent means ± sd of at least three replicate experiments. Statistical significance compared with Col-0 values is based on a Student’s t test (**P < 0.01 and ***P < 0.001).
(F) Analysis of NCA1 protein in Col-0, nca1-3, com-1, and com-2 seedlings. Protein was extracted from 10-d-old seedlings and probed with anti-NCA1 and antiactin (used as a loading control) antibodies. One representative analysis of three replicates experiments is shown.
To complement the nca1-3 alkaline-sensitive phenotype, a 6000-bp genomic DNA fragment (corresponding to the sequence from 2000 bp upstream of the NCA1 translation start codon to 500 bp downstream of the stop codon) was cloned into the pCAMBIA1300 vector. The resulting construct was transformed into the nca1-3 mutant. Two transgenic T3 lines grown in media at pH 8.0 demonstrated that the transgene rescued the mutant phenotype to the level of wild-type alkaline tolerance (Figures 2D and 2E). The protein levels of NCA1 in the transgenic lines were slightly higher than levels in Col-0 (Figure 2F).
NCA1 Gene Expression and Protein Localization
NCA1 expression was determined using real-time RT-PCR on total RNA extracted from roots, stems, leaves, flowers, and siliques of Col-0 plants. NCA1 was expressed in all of these tissues, with highest expression in rosette leaves and siliques (Supplemental Figure 2A). The 2000-bp promoter of NCA1 was fused with the β-GLUCURONIDASE (GUS) reporter gene, and the resulting constructs were introduced into Arabidopsis Col-0 using Agrobacterium-mediated floral transformation. Consistent with the RT-PCR results, GUS expression driven by the NCA1 promoter was detected in young seedlings, roots, stems, leaves, flowers, and siliques (Supplemental Figure 2B).
To determine the subcellular localization of the NCA1 protein, we fused green fluorescent protein (GFP) to the C terminus of NCA1 using a construct in which expression was driven by the cauliflower mosaic virus 35S promoter. The resulting construct was transformed into Col-0 protoplasts, and the NCA1-GFP signal was observed in the cytosol. The control, GFP alone, was observed in both the cytosol and nucleus (Supplemental Figure 2C).
The nca1-3 Mutant Is Sensitive to Multiple Stresses
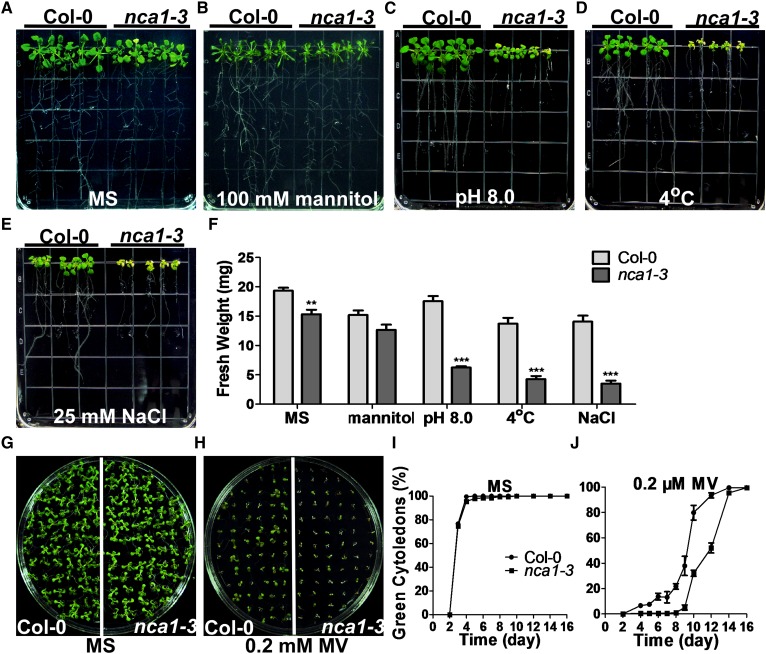
To determine whether NCA1 is involved only in the response of Arabidopsis to high pH, we transferred 5-d-old Col-0 and nca1-3 seedlings from seeds germinated on MS medium at pH 5.8 to MS medium at pH 5.8 or pH 8.0, or pH 5.8 containing 25 mM NaCl or 100 mM mannitol, or treated at 4°C for 3 d. While the nca1-3 mutant was insensitive to mannitol treatment, it was hypersensitive to NaCl and cold treatment relative to Col-0 with reduced root and shoot growth and bleached leaves (Figures 3A to 3F). When seeds of the nca1-3 mutant and Col-0 were germinated on MS medium (pH 5.8) containing 0.2 μM methyl viologen (MV), the nca1-3 mutant was also hypersensitive, with reduced cotyledon greenness relative to Col-0 (Figures 3G to 3J). These results suggest that NCA1 functions in multiple stress responses.
Figure 3.
nca1-3 Is Sensitive to Multiple Stresses.
(A) Analysis of Col-0 and nca1-3 on MS medium. Five-day-old seedlings of Col-0 and nca1-3 from seeds germinated on MS medium at pH 5.8 were transferred to fresh MS medium at pH 5.8. Photographs were taken 10 d after transfer.
(B) Analysis of mannitol sensitivity in Col-0 and nca1-3. Five-day-old seedlings of Col-0 and nca1-3 from seeds germinated on MS medium at pH 5.8 were transferred to MS medium at pH 5.8 containing 100 mM mannitol. Photographs were taken 10 d after transfer.
(C) Analysis of sensitivity to high external pH in Col-0 and nca1-3. Five-day-old seedlings of Col-0 and nca1-3 from seeds germinated on MS medium at pH 5.8 were transferred to MS medium at pH 8.0. Photographs were taken 10 d after transfer.
(D) Analysis of cold stress sensitivity in Col-0 and nca1-3. Five-day-old seedlings of Col-0 and nca1-3 from seeds germinated on MS at pH 5.8 medium were transferred to MS medium at pH 5.8 and moved to a light chamber after being treated at 4°C for 3 d. Photographs were taken 13 d after transfer.
(E) Analysis of salt stress sensitivity in Col-0 and nca1-3. Five-day-old seedlings of Col-0 and nca1-3 from seeds germinated on MS medium at pH 5.8 were transferred to MS medium containing 25 mM NaCl. Photographs were taken 10 d after transfer.
(F) Fresh weight of the Col-0 and nca1-3 seedlings shown in (A) to (E). Data represent means ± sd of at least three replicate experiments. Statistical significance compared with Col-0 is based on a Student’s t test (**P < 0.01 and ***P < 0.001).
(G) Analysis of MV sensitivity in Col-0 and nca1-3. Col-0 and nca1-3 seeds were sown on MS medium at pH 5.8 and germinated in continuous light after stratification at 4°C. Photographs were taken 10 d after seeds were sown.
(H) Analysis of MV sensitivity in Col-0 and nca1-3. Col-0 and nca1-3 seeds were sown on MS medium at pH 5.8 with 0.2 µM MV and germinated in continuous light after stratification at 4°C. Photographs were taken 10 d after seeds were sown.
(I) Percentage of green cotyledons in Col-0 and nca1-3 grown on MS medium were scored at the indicated times and represent an average of 100 seeds from at least three independent experiments ±sd.
(J) Percentage of green cotyledons in Col-0 and nca1-3 grown on MS medium with 0.2 µM MV were scored at the indicated times and represent an average of 100 seeds from at least three independent experiments ±sd.
NCA1 Interacts with Catalases
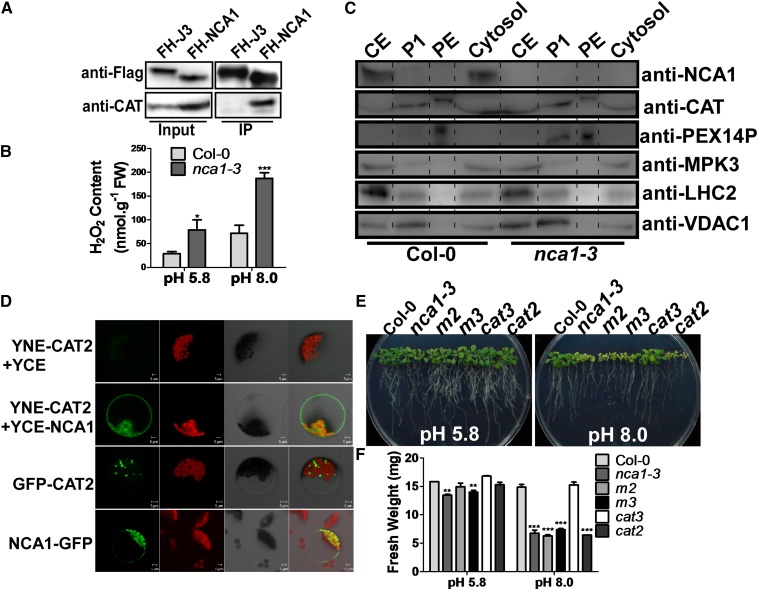
The hypersensitivity of the nca1-3 mutant to multiple abiotic stresses suggests that NCA1 is part of a conserved mechanism that is required for plant stress responses. To determine how NCA1 functions in these responses, we generated Pro35S:3×Flag-HA-NCA1 (FH-NCA1) transgenic plants in the nca1-3 background to identify interacting proteins; this construct rescued the nca1-3 mutant phenotypes (Supplemental Figure 3). Total protein was extracted from 10-d-old T3 homozygous plants, and Flag-HA-NCA1 was immunoprecipitated with anti-Flag antibody-conjugated agarose. The Flag epitope was then cleaved off and anti-HA-conjugated agarose was used for HA-NCA1 immunoprecipitation from the resulting supernatant. Agarose-bound HA-NCA1 was eluted from the HA agarose using an HA peptide. After this two-step purification, NCA1 and its interacting proteins were enriched and analyzed by mass spectrometry. CAT1, 2, and 3 were identified as putative NCA1-interacting proteins.
To confirm the interaction between NCA1 and the catalases, CAT antibodies were generated by immunizing mice with E. coli-expressed CAT3. To determine the specificity of the anti-CAT antibodies, Pro35S:6Myc-NCA1, Pro35S:6Myc-CAT2, and Pro35S:6Myc-CAT3 transgenic plants were generated, and the cat2 (SALK_076998; Buesoet al., 2007; Queval et al., 2007) and cat3 T-DNA insertion lines (SALK_092911; Mhamdi et al., 2010a) (reported to be knockout lines) were obtained. In both cat2 and cat3 mutants, catalase protein was detected using anti-CAT antibodies (Supplemental Figure 4A). In the Pro35S:6Myc-CAT2 transgenic line, both catalase and 6Myc-CAT2 bands were identified using anti-CAT antibodies (Supplemental Figure 4B), suggesting that this antibody recognizes at least both CAT2 and CAT3. Flag-HA-NCA1 and Flag-HA-J3 (Yang et al., 2010; a cytosolic protein used as a control) were immunoprecipitated with anti-flag antibody-conjugated agarose from their respective transgenic plants. NCA1 and J3 loading was determined with anti-Flag antibody, and catalases were detected in the immunoprecipitated products using anti-CAT antibodies. Catalase proteins were pulled down by NCA1 but not by J3 (Figure 4A). Consistent with this result, H2O2 content was higher in the nca1-3 mutant than in Col-0 under control conditions, and this difference increased significantly when the plants were grown at high pH (8.0) for 10 d (Figure 4B).
Figure 4.
NCA1 Interacts with Catalases in the Cytosol.
(A) Analysis of the interaction between NCA1 and catalases in vivo. The Pro35S:3×Flag-HA-NCA1 and Pro35S:3×Flag-HA-J3 plasmids were introduced into Col-0. Flag-HA-NCA1 or Flag-HA-J3 was immunoprecipitated with anti-Flag agarose and analyzed by immunoblot analysis with anti-Flag antibody to detect Flag-HA-NCA1 or Flag-HA-J3 and with anti-CAT antibody to detect catalases in the precipitates. J3 is a cytosolic protein used as a control.
(B) Analysis of H2O2 content in Col-0 and nca1-3. Five-day-old seedlings from seeds germinated on MS medium at pH 5.8 were transferred to MS medium at pH 5.8 or 8.0 for 10 d. H2O2 content was monitored with an Amplex Red H2O2/peroxidase assay kit. Data represent means ± sd of at least three replicate experiments. Statistical significance is based on a Student’s t test in comparison to Col-0 values (*P < 0.05 and ***P < 0.001).
(C) Analysis of NCA1 and catalase localization in Col-0 seedlings. Peroxisomes and cytosol were separated by density gradient centrifugation and protein was detected by immunoblot analysis with anti-NCA1 and anticatalase antibodies. Anti-MPK3 antibody was used as a cytosolic marker, anti-PEX14p antibody as a peroxisomal marker, anti-LHC2 as a chloroplast marker, and anti-VDAC1 as a mitochondria marker. CE, crude extract; P1, first purified peroxisomes; PE, high purity peroxisomes.
(D) NCA1 interacts with CAT2 in the cytosol. Plasmids were introduced into Arabidopsis protoplasts and fluorescence was detected by confocal microscopy. Top row, pSPYNE-CAT and pSPYCE vectors were cotransformed as a negative control. Second row, pSPYNE-CAT and pSPYCE-NCA1 were cotransformed. Third row, GFP-CAT2 was introduced. Bottom row, NCA1-GFP was introduced. In each row, the first panel, YFP/GFP signal; the second panel, autofluorescence; the third panel, bright-field image; and the fourth panel, merged image.
(E) Analysis of alkaline sensitivity in Col-0, nca1-3, cat2, cat3, m2 (cat2 cat3), and m3 (nca1-3 cat2 cat3) mutant seedlings. Five-day-old seedlings of Col-0, nca1-3, cat2, cat3, m2, and m3 from seeds germinated on MS medium at pH 5.8 were transferred to MS medium at pH 5.8 or 8.0. Photographs were taken 10 d after transfer.
(F) Fresh weight of Col-0, nca1-3, cat2, cat3, m2, and m3 seedlings in (E). Data represent means ± sd of at least three replicate experiments. Statistical significance is based on a Student’s t test in comparison to Col-0 values (**P < 0.01 and ***P < 0.001).
Because NCA1 localizes in the cytosol while the major functional site of catalases is reported to be the peroxisomes, additional biochemical and genetic assays were done to monitor the interaction between NCA1 and the catalases. First, a combination of Percoll and sucrose density gradient centrifugation was used to investigate colocalization of these two proteins. Ten-day-old Arabidopsis Col-0 or nca1-3 seedlings were harvested to isolate peroxisomes as described by Reumann et al. (2007). The crude extract, first-purified peroxisomes (P1), high purity peroxisomes (PEs), and the cytosolic fraction were analyzed using immunoblots with anti-NCA1, -CAT, -PEX14p (a peroxisomal membrane marker), -MPK3 (a cytosolic marker), -LHC2 (a chloroplast marker), and -VDAC1 (a mitochondrial marker) antibodies (Figure 4C). NCA1 was observed in the cytosol, and CATs were observed in both the cytosol and peroxisomes, with higher expression in the peroxisomes. Both NCA1 and CATs colocalized with the MPK3 cytosolic marker. PEX14p was observed in P1 and enriched in PE where CATs were also found, while no signals for LHC2 or VDAC1 were detected in PE (Figure 4C). These results indicate that NCA1 was undetectable in high purity peroxisomes and that NCA1 colocalizes with CATs in the cytosol.
To further confirm the cytosolic NCA1-CAT interaction, bimolecular fluorescence complementation was performed. The coding sequences of CAT2 and NCA1 were amplified and cloned into the pSPYNE(R)173 and pSPYCE(M) vectors to generate YNE-CAT2 and YCE-NCA1 constructs. These plasmids were introduced into Arabidopsis protoplasts by polyethylene glycol-mediated transformation. After incubation for 18 h, the interaction between NCA1 and CAT2 was detected by confocal microscopy. A strong florescence signal was detected in the cytosol (Figure 4D), providing additional evidence that NCA1 colocalizes and interacts with CAT2 in cytosol.
To confirm that NCA1 genetically interacts with the catalases, the cat2 and cat3 T-DNA insertion lines (SALK_076998 and _092911, respectively) and the nca1-3 mutant were used to generate the cat2 cat3 (m2) and nca1-3 cat2 double mutants and the nca1-3 cat2 cat3 (m3) triple mutant. To determine the sensitivity of these mutants to high pH (8.0), 5-d-old seedlings of Col-0, nca1-3, the cat2 and cat3 single mutants, the cat2 cat3 (m2) double, and the nca1-3 cat2 cat3 (m3) triple mutant were transferred to MS medium at pH 5.8 or 8.0. On medium at pH 5.8, the phenotype of the nca1-3 cat2 cat3 (m3) triple mutant was similar to that of the nca1-3 mutant in which the fresh weight was lower than in Col-0 (Figure 4E). On medium at pH 8.0, the growth of Col-0 and the cat3 single mutant was similar, while the other mutants were very sensitive to the high pH (Figure 4E).
cat2 is a photorespiratory-defective mutant, and its phenotype can be readily rescued by high external CO2 concentration (Mhamdi et al., 2010b). When grown under an irradiance of 90 μmol m−2 s−1, both cat2 and nca1-3 seedlings showed severely decreased growth compared with Col-0, and this growth reduction was further enhanced by high external pH treatment (Supplemental Figures 5A and 5B). When plants were grown with high levels of CO2 (3000 ppm), nca1-3, cat2, and Col-0 showed no difference in appearance or biomass at 90 μmol m−2 s−1 light onx pH 5.8 medium. Under the same light and high CO2 conditions, when the pH was increased to 8.0, the growth of cat2 was similar to that of Col-0, and the growth of nca1-3 was also largely rescued; however, it was still impaired by the pH 8.0 treatment (Supplemental Figures 5C and 5D). This phenotype was completely rescued in the NCA1 complemented transgenic plants (Supplemental Figures 5E to 5H).
To identify phenotypic changes when nca1-3 plants were grown in soil, Col-0 and the nca1-3 and cat2 mutants were treated with an irradiance of 30 or 100 μmol m−2 s−1 in soil. No significant difference was detected between Col-0 and cat2 plants in rosette biomass at an irradiance of 30 μmol m−2 s−1 (16 h) for 3 weeks; however, nca1-3 plants were slightly smaller than Col-0 and cat2. Consistent with previously published results (Queval, et al., 2007), at an irradiance of 100 μmol m−2 s−1 (16 h), cat2 plants were smaller; this phenomenon was also observed in nca1-3 plants (Supplemental Figures 5I and 5J). These data suggest that CAT2 may not be the only target of NCA1 and that other catalases or proteins may also be affected by NCA1 in response to abiotic stresses or high light stress.
It has been reported that hydroxyurea (HU) directly inhibits catalase-mediated H2O2 decomposition and that the cat2 mutant is more tolerant to HU than Col-0 (Juul et al., 2010). Five-day-old seedlings of Col-0, nca1-3, cat2, and the nca1-3 cat2 double mutant were transferred to MS medium (pH 5.8) without or with 3 mM HU. In the absence of 3 mM HU, the nca1-3 single and nca1-3 cat2 double mutant displayed a weak growth phenotype compared with Col-0 and cat2 (Supplemental Figures 5K and 5L). In the presence of 3 mM HU, consistent with published results (Juul et al., 2010), the cat2 mutant was more tolerant to HU than Col-0, and the nca1-3 single and nca1-3 cat2 double mutant showed a tolerance level similar to the cat2 mutant (Supplemental Figures 5K and 5L). These results suggest that NCA1 genetically interacts with CAT2.
NCA1 Increases CAT Activity in Vivo
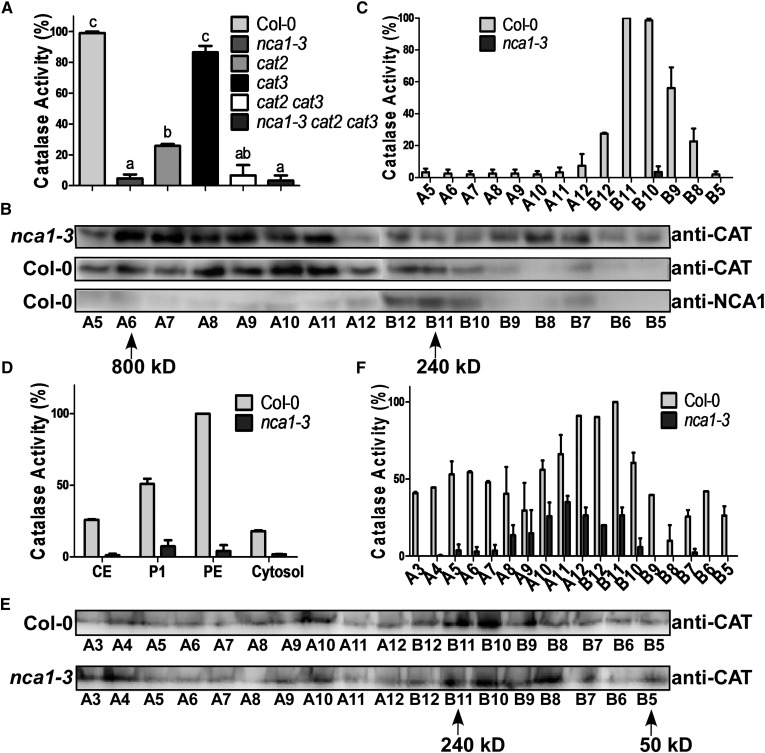
NCA1 interacts with three catalases in Arabidopsis and the nca1-3 cat2 cat3 triple mutant shows a phenotype similar to nca1-3, suggesting that NCA1 plays an important role in regulating catalase activity. To examine if the activity of CATs is affected by NCA1, total protein was extracted from 10-d-old seedlings of Col-0 and the nca1-3, cat2, cat3, cat2 cat3, and nca1-3 cat2 cat3 mutants grown on MS medium and CAT activity was analyzed using a Catalase Activity Assay Kit (Beyotime). One unit of catalase activity is defined as the quantity of enzyme catalyzing the decomposition of 1 μmol H2O2 per minute. CAT activity was dramatically reduced in nca1-3 to one-tenth of the level in Col-0 (Figure 5A), suggesting that NCA1 is required for activating catalase in Arabidopsis. Consistent with published results, deleting CAT2 but not CAT3 significantly reduced catalase activity in Arabidopsis (Figure 5A). The cat2 cat3 double mutant and the nca1-3 cat2 cat3 triple mutant displayed catalase activity similar to activity in the nca1-3 mutant (Figure 5A).
Figure 5.
NCA1 Increases CAT Activity in Vivo.
(A) Analysis of catalase activity in total protein extracted from Col-0, nca1-3, cat2, cat3, cat2 cat3, and the nca1-3 cat2 cat3 mutant. Catalase activity is indicated in units/mg protein, and all activities were calculated relative to Col-0 activity (80 units/mg). Data represent means ± sd of at least three replicate experiments. Statistical significance is based on a Student’s t test. Significant differences (P < 0.05) are indicated by different lowercase letters.
(B) Gel filtration analysis of catalases and NCA1 in nca1-3 and Col-0 seedlings. Total protein was extracted from 10-d-old seedlings and loaded onto a Superdex-200 10/300GL column (GE). The samples were collected every 500 μL in order of A1 (tube number) to A12 then B12 to B1 and examined by immunoblot analysis with anti-CAT and anti-NCA1 antibodies. Arrows, proteins of molecular mass of 800 and 240 kD.
(C) Catalase activity in the Col-0 and nca1-3 gel filtration products. Catalase activity is indicated in units/mg protein, and all activities were calculated relative to Col-0 B11 activity (350 units/mg). Data represent means ± sd of at least three replicate experiments.
(D) Catalase activity of the density gradient products shown in Figure 4C. Catalase activity is indicated in units/mg protein, and all activities were calculated relative to Col-0 PE activity (700 units/mg). Data represent means ± sd of at least three replicate experiments.
(E) Gel filtration analysis of peroxisomal catalases. PE proteins from the density gradients shown in Figure 4C were loaded onto a Superdex-200 10/300GL column (GE). Samples were collected every 500 μL (from A1 to A12 and then B12 to B1) and proteins examined by immunoblot analysis with anti-CAT antibodies. Arrows, proteins of molecular mass of 240 and 50 kD.
(F) Catalase activity of the gel filtration products shown in Figure 5E. Catalase activity is indicated in units/mL protein, and all activities were calculated relative to Col-0 B11 activity (170 units/mL). Data represent means ± sd of at least three replicate experiments.
To determine if NCA1 forms a complex with catalases that is required for their activity, total protein from Col-0 and nca1-3 was extracted and loaded onto a Superdex-200 gel filtration column. The samples were collected every 500 μL and examined by immunoblot analysis with anti-CAT or anti-NCA1 antibodies (Figure 5B). In Col-0, the major NCA1 protein eluted in a peak corresponding to a species with a molecular mass of 240 kD; however, catalase proteins dispersed from 800 to 200 kD and mainly existed in the higher molecular mass region (Figure 5B). In the nca1-3 mutant, catalase protein also localized to the lower molecular mass region. We subsequently tested the catalase activity of these samples. In Col-0, the highest catalase activity appeared at the 240 kD region that colocalized with NCA1 (Figure 5C). In the nca1-3 mutant, catalase activity was significantly reduced in all of the gel filtration samples. These results suggest that NCA1 is required for catalase activation and may be involved in formation of a functional complex.
Because NCA1 and CATs colocalize and interact in the cytosol and NCA1 is required for increasing catalase activity, Percoll-sucrose density gradient fractions (as in Figure 4C) were used in catalase activity assays to determine where NCA1 increases the catalase activity. Catalase activities in all fractions (crude extract, P1, PE, and the cytosol) were much higher in Col-0 than in nca1-3 (Figure 5D). These results suggest that NCA1 is essential for catalase activation in the plant.
When catalases form tetramers in plants, higher enzyme activity results (Mhamdi et al., 2012). To determine if NCA1 is required for the formation of tetramers in peroxisomes, the PE fractions from the wild type or nca1-3 were loaded onto a Superdex-200 gel filtration column. Samples were collected every 500 μL and examined by immunoblot analysis with anti-CAT antibodies. Peroxisomal catalases in both Col-0 and nca1-3 were enriched in the fractions with a molecular mass around 240 kD, corresponding to CAT tetramers (∼220 kD) (Figure 5E). When catalase activity in these samples was tested, the fractions containing more peroxisomal CAT tetramers had higher catalase activity (Figure 5F). The activity of these peroxisomal catalases was significantly higher in Col-0 than in nca1-3. These results suggest that NCA1 is not required for CAT tetramer formation.
It has been reported that catalase activity is regulated by environmental stimuli (Gossett et al., 1996; Dat et al., 1998; Du et al., 2008; Distelbarth et al., 2013). To determine if NCA1-catalase interaction is affected by abiotic stress, Pro35S:3×Flag-HA-NCA1 (FH-NCA1) transgenic plants were treated with liquid MS medium at pH 5.8 or 8.0, or with MS medium (pH 5.8) containing 20 mM H2O2, 150 mM NaCl, or 20 mM 3-AT for 3 h. FH-NCA1 was immunoprecipitated with anti-Flag antibody-conjugated agarose. FH-NCA1 in the input was used to adjust protein levels for pull-downs with anti-Flag antibody. No significant difference in CAT levels was detected in the NCA1 pull-down products among these treatments (Supplemental Figure 6A). When catalase activity was determined by detecting the decrease in the absorbance of H2O2 at 240 nm, no difference in catalase activity was observed in pH 8.0-treated samples, while, as previously reported, H2O2 and NaCl treatments induced catalase activity (Supplemental Figure 6B) (Li and Yi, 2012). As a control, 3-AT (a potent inhibitor of catalases; Gechev et al., 2002) treatment caused a significant decrease in catalase activity (Supplemental Figure 6B). Ten-day-old Col-0 seedlings grown on MS medium were treated with liquid MS medium at pH 5.8 or 8.0, or with liquid MS medium containing 20 mM H2O2, 150 mM NaCl, or 20 mM 3-AT for 3 h. Total RNA was extracted and the expression of NCA1 and CAT2 was measured using real-time RT-PCR. The expression of CAT2 was induced by all four treatments; however, NCA1 expression was induced by H2O2 or NaCl treatment but was not affected by pH 8.0 or 3-AT treatment (Supplemental Figure 6C). These results suggest that the interaction between NCA1 and catalases is not induced by stress and that the role of NCA1 in increasing catalase activity is more widespread and fundamental.
NCA1 Has Chaperone Protein Activity
To determine the mechanism underlying NCA1 increasing catalase activity, we examined the localization of the catalases in Col-0 and the nca1-3 mutant. We fused the coding regions of CAT2 and CAT3 individually to GFP and cloned them into pCAMBIA 1205. The resulting constructs were transformed individually into Col-0 and nca1-3, and the transgenic plants were used to visualize the subcellular localization of CAT2 and CAT3. The GFP signal was predominately observed in the peroxisomes in both Col-0 and nca1-3 (Supplemental Figure 7), suggesting that the peroxisome localization of catalases was not altered in nca1-3.
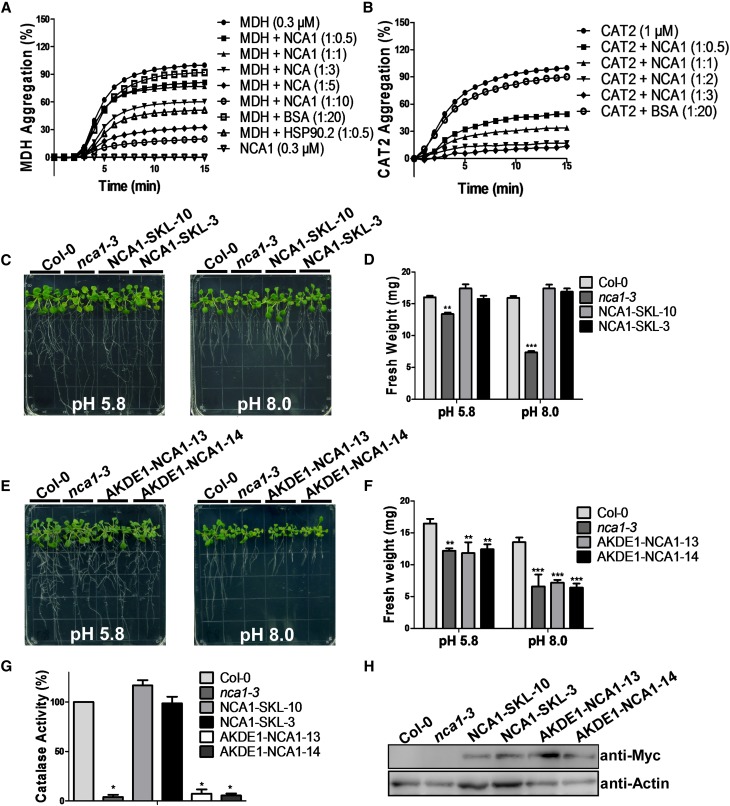
Because NCA1 is required for basic catalase activity but not subcellular localization, we hypothesized that NCA1 may help to maintain catalase folding in a functional structure. To determine if NCA1 has holdase chaperone activity that effectively maintains nascent chains in a folding-competent conformation, the holdase activity of His-NCA1 was assayed by measuring its capacity to suppress thermal aggregation of Arabidopsis malate dehydrogenase (MDH, EC 1.1.1.37; a model substrate for holdase assay, as described in Cha et al., 2009). MDH aggregation was monitored by measuring absorbance at 340 nm without or with NCA1 under thermal denaturing conditions at 45°C. Heat-induced aggregation of MDH was suppressed by NCA1 in a dose-dependent manner (Figure 6A). We next tested the holdase activity of NCA1 using a physiological substrate, CAT2. As was seen for MDH, NCA1 repressed the denaturation of CAT2 during thermal stress (Figure 6B). These results suggest that NCA1 functions as a molecular chaperone and interacts with and maintains CAT2 in an optimal structure.
Figure 6.
NCA1 Has Chaperone Activity.
(A) Holdase chaperone activity of NCA1 with the model substrate malate dehydrogenase. MDH (0.3 μM) was incubated without or with NCA1 at 45°C for the indicated time. Arabidopsis HSP90.2 was used as a positive control and BSA as negative control.
(B) Holdase chaperone activity of NCA1 with CAT2. CAT2 (1 μM) was incubated without or with NCA1 at 45°C for the indicated time. BSA was used as negative control.
(C) Analysis of alkaline sensitivity in Col-0, nca1-3, and T3 transgenic Pro35S:6×Myc-GFP-NCA1-SKL seedlings. Five-day-old seedlings of Col-0, nca1-3, NCA1-SKL-10, and NCA1-SKL-3 from seeds germinated on MS medium at pH 5.8 were transferred to MS medium at pH 5.8 or 8.0. Photographs were taken 10 d after transfer.
(D) Fresh weight of Col-0, nca1-3, NCA1-SKL-10, and NCA1-SKL-3 seedlings in (C). Data represent means ± sd of at least three replicate experiments. Statistical significance is based on a Student’s t test in comparison to Col-0 values (**P < 0.01 and ***P < 0.001).
(E) Analysis of alkaline sensitivity in Col-0, nca1-3, and T3 transgenic Pro35S:6×Myc-AKDE1 -NCA1 seedlings. Five-day-old seedlings of Col-0, nca1-3, AKDE1-NCA1-13, and AKDE1-NCA1-14 from seeds germinated on MS medium at pH 5.8 were transferred to MS medium at pH 5.8 or 8.0. Photographs were taken 10 d after transfer.
(F) Fresh weight of Col-0, nca1-3, NCA1-SKL-10, and NCA1-SKL-3 seedlings in (E). Data represent means ± sd of at least three replicate experiments (**P < 0.01 and ***P < 0.001).
(G) Catalase activity in total protein of Col-0, nca1-3, NCA1-SKL-10, NCA1-SKL-3, AKDE1-NCA1-13, and AKDE1-NCA1-14 seedlings. Catalase activity is indicated in units/mg protein, and all activities were calculated relative to Col-0 activity (80 units/mg). Data represent means ± sd of at least three replicate experiments. Statistical significance is based on a Student’s t test. Significant differences (P < 0.05) are indicated by different lowercase letters.
(H) Analysis of Myc-NCA1 protein in the Col-0, nca1-3, NCA1-SKL-10, NCA1-SKL-3, AKDE1-NCA1-13, and AKDE1-NCA1-14 seedlings. Protein was extracted from 10-d-old seedlings and probed with anti-Myc and antiactin (used as a loading control) antibodies. One representative analysis of three replicates experiments is shown.
Because NCA1 localizes mainly to the cytosol while catalases localize mainly in the peroxisome, and because NCA1 does not affect catalase peroxisomal localization, it is possible that NCA1 plays a role in maintaining structure of cytosol-localized catalases before they are transported into peroxisomes/organelles. If this is the case, expression of NCA1 in organelles, such as the peroxisome where catalases function, should partially rescue the nca1-3 phenotype. To test this hypothesis, sequence encoding a Ser-Lys-Leu tripeptide (SKL; targeting sequence that confers specific sorting to peroxisomes) (Hayashi, et al., 1997) or AKDE1 (the first 66 amino acids of the mitochondrion-localized 2-oxoglutarate dehydrogenase E1 that directs protein into the mitochondrial matrix) (Mehlmer et al., 2012) fused to the C or N terminus of NCA1 was cloned into the pCAMBIA1307-MYC vector. The resulting constructs were transformed into nca1-3. Five-day-old T3 transgenic plants (Pro35S:6×MycNCA1-SKL and Pro35S:6×Myc-AKDE1-NCA1) germinated on pH 5.8 MS medium were transferred to MS medium at pH 5.8 or 8.0. The NCA1-SKL transgene rescued the mutant’s alkaline-sensitive phenotype and the catalase activity to the level of the wild type (Figures 6C, 6D, and 6G), while the AKDE1-NCA1 transgene failed to rescue the sensitivity and catalase activity of nca1-3 (Figures 6E to 6G). The protein levels of NCA1 in the transgenic lines were similar (Figure 6H). The fact that the peroxisome-targeted NCA1 rescued the phenotype of nca1-3 while the mitochondrion-targeted NCA1 did not indicates that NCA1 plays an important role in the increase of catalase activity.
NCA1 Increases CAT2 Activity in a 1:1 Molar Ratio
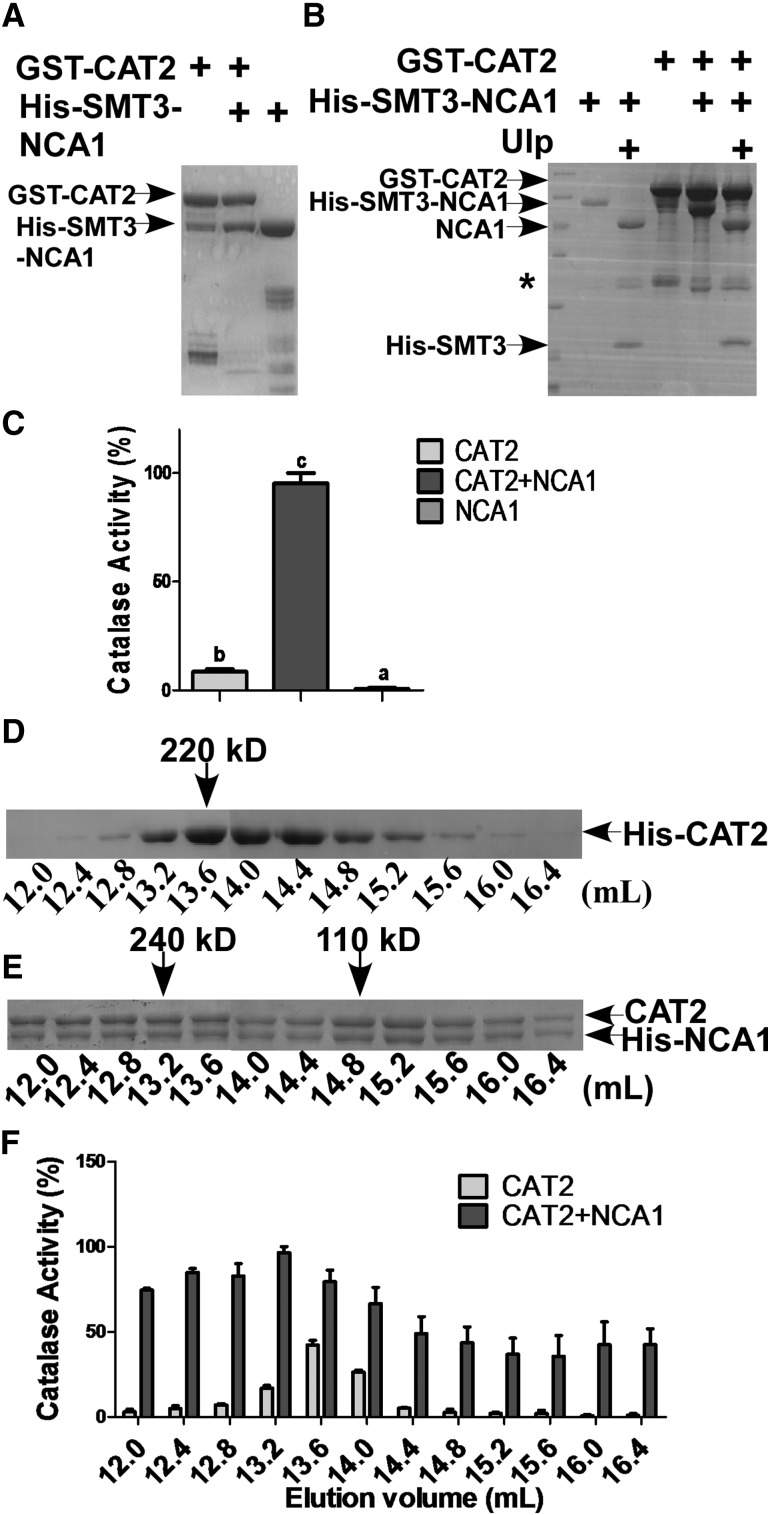
To determine the biochemical nature of NCA1 increasing catalase activity, we introduced GST-CAT2, His-Small Ubiquitin Like Modifier3 (SMT3)-NCA1, or the two in combination into E. coli BL21 cells. For subsequent experiments, we employed mainly CAT2, based on the 80% reduction in catalase activity in the cat2 mutant in Arabidopsis. Consistent with the holdase chaperone activity of NCA1, the GST-CAT2 and His-SMT3-NCA1 proteins were unstable in vitro and easily precipitated/aggregated and degraded (Figure 7A); however, they appeared to stabilize each other when coexpressed in E. coli (Figure 7A). CAT2 recombinant protein pulled down NCA1 in a nearly 1:1 molar ratio. When His-SMT3 was cleaved from His-SMT3-NCA1 using a Ubiquitin-like-specific protease (Ulp), NCA1 still pulled down and tightly bound to CAT2 in the same molar ratio (Figure 7B). We next tested the catalase activity of the recombinant proteins shown in Figure 7A. When CAT2 was pulled down by NCA1, the catalase activity was 10-fold higher than in CAT2 alone (Figure 7C), consistent with in vivo results (Figure 5C) and demonstrating that NCA1 directly increases CAT2 activity.
Figure 7.
NCA1 Increases CAT2 Activity in a 1:1 Molar Ratio.
(A) Analysis of the interaction between NCA1 and CAT2 in vitro. His-SMT3-NCA1 and GST-CAT2 were coexpressed in E. coli DE3 cells and proteins were purified with glutathione-agarose. The pull-down products were separated by SDS-PAGE.
(B) Analysis of the pull-down products in (A). Proteins were digested with SUMO (Ulp) protease and separated on SDS-PAGE. The asterisk indicates a nonspecific band.
(C) Analysis of recombinant protein catalase activity. Catalase activity is indicated in units/mg protein, and all activities were calculated relative to the activity of the coexpressed CAT2 and NCA1 sample (200 units/mg). Data represent means ± sd of at least three replicate experiments. Statistical significance is based on a Student’s t test. Significant differences (P < 0.05) are indicated by different lowercase letters.
(D) Gel filtration analysis of recombinant protein. Eluted His-CAT2 protein was loaded onto a Superdex-200 10/300GL column (GE), and samples were collected every 400 μL and separated on SDS-PAGE.
(E) Gel filtration analysis of recombinant protein. Coexpressed CAT2 and His-NCA1 protein was loaded onto a Superdex-200 10/300GL column (GE), and samples were collected every 400 μL and separated on SDS-PAGE.
(F) Catalase activity in the samples from (D) and (E). Catalase activity is indicated in units/mL protein, and all the activities were calculated relative to the activity of the fraction at 13.2 mL elution volume in which CAT2 and NCA1 were coexpressed (400 units/mL) (E). Data represent means ± sd of at least three replicate experiments.
To determine how NCA1 forms a complex with CAT2 and if it is required for activation of CAT2, CAT2 or the CAT2-NCA1 complex was loaded onto a Superdex-200 gel filtration column and samples were collected every 400 µL. Recombinant CAT2 protein eluted between 12.4 and 16 mL (corresponding to molecular masses of ∼300 and 70 kD) (Figure 7D). The calculated molecular mass of 6×His-tag CAT2 fusion protein is 58.75 kD, which means that recombinant CAT2 protein is present mainly as oligomers in solution. Consistent with previous reports (Kirkman and Gaetani, 1984; Frugoli et al., 1996), CAT2 proteins that eluted at 13.6 mL (corresponding to molecular mass of ∼220 kD, the size of a CAT2 tetramer) had the highest activity in our assay (Figure 7F). For the NCA1-CAT2 complex, the protein eluted at a broad range of molecular masses. The protein complex that eluted at 13.2 mL, corresponding to a molecular mass of ∼240 kD (Figure 7E), had the highest activity (Figure 7F), indicating that a heterotetrameric NCA1-CAT2 complex might form in solution. Surprisingly, we found that NCA1-CAT2 complex proteins with a higher molecular weight were still present in a molar ratio of 1:1 (Figure 7E). These results indicate that this molar ratio plays a critical role in increasing catalase activity.
NCA1 Interacts with CAT2 through the NCA1 C Terminus and Increase of CAT2 Activity Requires Zinc Binding of the NCA1 RING-Finger Domain
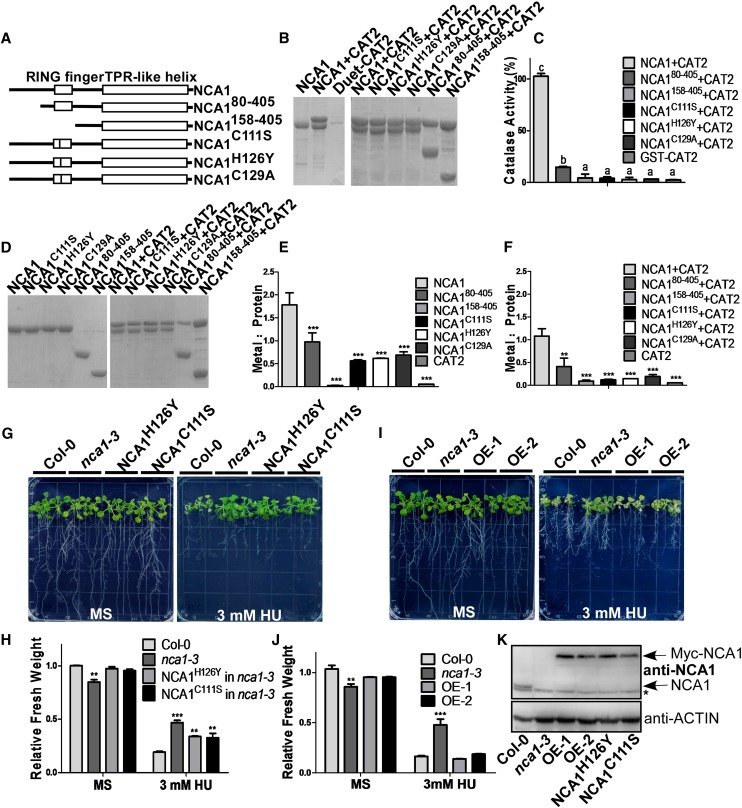
To investigate the mechanism of interaction between NCA1 and CAT2, we cloned sequences encoding various truncated versions of NCA1 into the pET28a vector for fusion with a His-tag (Figure 8A). We coexpressed full-length or truncated His-NCA1 proteins with untagged CAT2 in BL21 cells, purified His-NCA1, and separated the proteins using SDS-PAGE (Figure 8B). When the first 158 amino acids from the N terminus of NCA1 (including the entire RING-type zinc finger domain) were deleted, the truncated protein still interacted with CAT2 in a 1:1 molar ratio (Figure 8B). These results indicate that NCA1 interacts with CAT2 mainly through the NCA1 C-terminal TPR-like helical domain. However, the catalytic activity of CAT2 showed significant dependence on the integrity of the RING-type zinc finger domain when the NCA1-CAT2 complex was formed. In the presence of full-length NCA1, CAT2 showed enhanced catalytic activity compared with CAT2 alone. Deletion of 80 amino acids from the NCA1 N terminus resulted in decreased catalytic activity of CAT2, and catalytic activity was not increased when the RING-finger domain was deleted (Figure 8C). These results suggest that the RING-finger domain in NCA1 plays a key role in increasing CAT2 activity. To further demonstrate the importance of this domain, we mutated conserved cysteines (Cys-111 and Cys-129) or histidine (His-126) residues in the RING-finger domain of NCA1 to alanine, serine, or tyrosine. The mutated proteins were pulled down with CAT2 in a 1:1 molar ratio but did not increase CAT2 activity (Figures 8A to 8C).
Figure 8.
NCA1 Interacts with CAT2 through the CAT2 C Terminus, and Increase of CAT2 Activity Requires Zinc Binding of the NCA1 RING-Finger Domain.
(A) Schematic diagram of NCA1, NCA180-405, NCA1158-405, NCA1C111S, NCA1H126Y, and NCA1H129A.
(B) Analysis of the interaction between full-length, truncated, or mutated NCA1 and CAT2 in vitro. His-NCA1 and CAT2 were coexpressed in E. coli DE3 cells, and proteins were purified with Ni-NTA agarose. The pull-down products were separated on SDS-PAGE. CAT2 was expressed in DE3 cells and pulled down with Ni-NTA agarose as a negative control.
(C) Catalase activity of the coexpressed NCA1 and CAT2 recombinant proteins. Catalase activity is indicated in units/mg protein, and all activities were calculated relative to the activity of coexpressed CAT2 and NCA1 sample (200 units/mg). Data represent means ± sd of at least three replicate experiments.
(D) SDS-PAGE analysis of pull-down products of full-length, truncated, or mutated NCA1 without or with CAT2. Coomassie blue staining was performed after SDS-PAGE.
(E) Analysis of zinc content in full-length, truncated, or mutated NCA1 protein by ICP-AES. Data represent means ± sd of at least three replicate experiments.
(F) Analysis of zinc content in NCA1/CAT2 coexpressed protein by ICP-AES. Data represent means ± sd of at least three replicate experiments.
(G) Analysis of HU resistance in T3 transgenic nca1-3 plants expressing Pro35S:myc-NCA1C111S or Pro35S:myc-NCA1H126. Five-day-old seedlings from seeds germinated on MS medium at pH 5.8 were transferred to MS medium at pH 5.8 without or with 3 mM HU. Photographs were taken 10 d after transfer.
(H) Fresh weight of the Col-0, nca1-3, and nca1-3 seedlings expressing Pro35S:myc-NCA1C111S or Pro35S:myc-NCA1H126Y shown in (G). Data represent means ± sd of at least three replicate experiments. Statistical significance is based on a Student’s t test (**P < 0.01 and ***P < 0.001).
(I) Analysis of HU resistance in T3 transgenic nca1-3 plants expressing Pro35S:myc-NCA1 (OE-1 and OE-2). Five-day-old seedlings from seeds germinated on MS medium at pH 5.8 were transferred to MS medium at pH 5.8 without or with 3 mM HU. Photographs were taken 10 d after transfer.
(J) Fresh weight of Col-0, nca1-3, and nca1-3 seedlings expressing Pro35S:myc-NCA1 shown in (I). Data represent means ± sd of at least three replicate experiments. Statistical significance is based on a Student’s t test (**P < 0.01 and ***P < 0.001).
(K) Immunoblot analysis of NCA1 in total proteins from nca1-3, Col-0, and transgenic plants expressing Pro35S:myc-NCA1C111S, Pro35S:myc-NCA1H126Y, or Pro35S:myc-NCA1. ACTIN was used as a loading control. Three replicate experiments were performed.
The RING-finger domain in NCA1 can bind zinc ions (Kosarev et al., 2002); mutation or deletion of this domain results in decreased or in the absence of CAT2 activation. To determine if zinc ions play a key role in increasing CAT2 activity in the NCA1-CAT2 complex, inductively coupled plasma-atomic emission spectroscopy (ICP-AES) was performed to determine the content of zinc in purified full-length and truncated NCA1 with or without CAT2 (Figure 8D). In native NCA1, the zinc ion and protein molar ratio is ∼2:1, indicating two zinc ions are bound to each NCA1 through the RING-finger domain (Figure 8E). Point mutation of the conserved amino acids in the RING-finger domain of NCA1 (NCA1C111S, NCA1H126Y, or NCA1C129A) resulted in a reduced zinc to protein ratio, indicating the importance of the coordination between zinc and the RING-finger domain (Figure 8E). With the RING-finger domain removed, the zinc content of NCA1 and the NCA1-CAT2 complex dropped to a level that was barely beyond the buffer control (Figures 8E and 8F). Thus, the RING-finger domain is actively involved in binding zinc ions in the NCA1-CAT2 complex. Deletion of first 80 amino acids from the NCA1 N terminus also resulted in decreased zinc binding and catalytic activity of CAT2 (Figures 8C to 8F). Taken together, these results indicate that the integrity of the RING-finger domain affects both zinc ion content and NCA1-CAT2 catalytic activity, suggesting that zinc ions may be critical for increasing CAT2 activity in the NCA1-CAT2 complex.
To genetically confirm this conclusion, Pro35S:6×Myc-NCA1, Pro35S:6×Myc-NCA1C111S, and Pro35S:6×Myc-NCA1H126Y transgenic plants were generated in nca1-3, and 5-d-old T3 seedlings from seed germinated on MS medium were transferred to MS medium without or with 3 mM HU. More than three independent transgenic lines were used for the assay and showed similar phenotypes; one line from each mutant construct is shown in Figure 8. Neither NCA1C111S nor NCA1H126Y in the nca1-3 mutant rescued the HU-tolerant phenotype (Figure 8G). By contrast, NCA1 rescued the phenotype (Figure 8I), although the protein levels of NCA1C111S, NCA1H126Y, and NCA1 were similar in the transgenic plants (Figure 8K).
To further determine if the increase of catalase activity by the RING-finger domain of NCA1 is required for the tolerance of high pH, the seedlings from the HU-tolerance assay (Figure 8G) were transferred to MS medium at pH 5.8 or 8.0. None of the mutated NCA1 proteins (NCA1C111S, NCA1H126Y, and NCA1C129A) in the nca1-3 mutant rescued the high pH-sensitive phenotype (Supplemental Figures 8A and 8B). By contrast, NCA1 did rescue the phenotype (Supplemental Figures 8C and 8D). These results demonstrate that NCA1 interacts with CAT2 through its C terminus and increases CAT2 activity through its RING-finger domain and that this increase is critical for plant stress tolerance.
DISCUSSION
Catalase proteins are important antioxidative enzymes, and plant genomes encode three catalase genes that play an essential role during plant growth, development, and response to stress (reviewed in Mhamdi et al., 2012). In Arabidopsis, loss of CAT2 results in an 80% loss of catalase activity and mutant plants are hypersensitive to high-intensity light, H2O2, NaCl, and cold, but tolerant to LiCl and HU (Bueso et al., 2007; Juul et al., 2010). Because catalase genes are highly expressed in plants, overexpression of such genes normally does not increase plant catalase activity or affect plant growth and response to stress. However, a few studies have shown that introduction of bacterial catalases in plants increases plant catalase activity and results in transgenic plants that are resistant to oxidative stress, NaCl, and high light intensities (Mhamdi et al., 2012). In this study, by genetic screening for alkaline-stress defective Arabidopsis mutants, we discovered that NCA1 increases catalase activity. Among all the stresses tested, only mannitol did not lead to growth reduction in the nca1-3 mutant, which may be due to mannitol’s function as a hydroxyl radical scavenger (Shen, et al., 1997).
NCA1 was previously identified by screening for HU-resistant mutants (Hackenberg et al., 2013). Catalase protein levels and enzymatic activities were significantly decreased in nca1 mutants, suggesting that NCA1 is required for catalase activity mainly by mediating catalase protein level. NCA1 is a cytosol-localized protein and does not affect type 1 peroxisomal targeting signal (PTS1)-guided protein peroxisome import. Both nca1 and cat2 share similar phenotypes, including altered expression of oxidative stress marker genes, altered protein stability, and suppression of RPM1-triggered cell death and avrRpm1-induced autophagic degradation. These results suggested that NCA1 regulates autophagy-dependent cell death mainly through mediating catalase activity. However, the molecular and biochemical relationships between NCA1 and catalases were not identified previously.
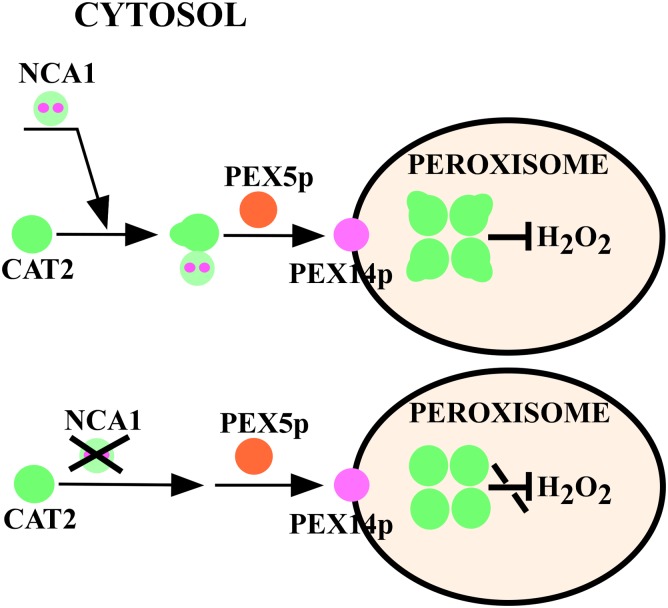
In our study, we demonstrate that NCA1 interacts with catalases through the NCA1 C-terminal TPR-like helical domain and increases catalase activity by stabilizing catalase proteins through the NCA1 N-terminal RING-type zinc finger domain and that the zinc ion binding is required for this increase. NCA1 exhibits holdase chaperone activity that may fold catalase to a functional structure. We propose a model of modulation of catalase (e.g., CAT2) by NCA1 in vivo (Figure 9). Nascent CAT2 in the cytosol interacts with NCA1 and form complexes in a 1:1 ratio. At this step, the interaction helps fold CAT2 into a functional structure. However, it is not required for the peroxisome import of catalases. After import into peroxisomes through plant peroxins, CAT2, with this optimal structure, forms a tetramer for full function.
Figure 9.
Model of NCA1 Modulation of Catalases in Vivo.
NCA1 binds to nascent CATs (e.g., CAT2) and forms complexes in a 1:1 ratio in the cytosol. The interaction maintains the optimal structure of CATs. After peroxin-mediated import into peroxisomes, CATs with this optimal structure form tetramers for maximal activity.
As reported previously, catalase activity is regulated by stresses including NaCl, drought, cold, H2O2, MV, high light, or heat shock (Gossett et al., 1996; Dat et al.,1998; Du et al., 2008; Distelbarth et al., 2013). However, it is likely that the interaction between NCA1 and catalases was not induced by alkaline stress or H2O2 and NaCl treatments. The affinity of catalase for H2O2 is very low, and its Km for H2O2 is above 40 mM in vivo (Mhamdi et al., 2010a). It is likely that NCA1 interacts with nascent catalases in the cytosol under normal conditions to help maintain catalase in a functional state that is ready to scavenge H2O2. High intracellular H2O2 levels could be a switch for activating catalase. Under certain conditions, when local concentrations of intracellular H2O2 increase to a high level, catalases function as H2O2 scavengers; however, under low concentration of H2O2, other enzymes, such as APX and PRX, with higher H2O2 affinities for catalase, perform this function. In nca1 mutants, the loss of NCA1 function leads to very low catalase activity that cannot remove high levels of H2O2, resulting in hypersensitivity to multiple stresses in the mutant. It is currently not understood how catalase interacts with and is released from NCA1; it is possible that post translational modification of catalase or NCA1 is required to release catalase and/or for further activating catalase during environmental stress.
Previous studies have shown that the TPR domain functions as a protein interaction scaffold for the formation of different protein complexes (Allan and Ratajczak, 2011). Peroxisome import of Arabidopsis catalases requires PEX5 and other PTS1 components. PEX5 contains seven TPR motifs in its C terminus. However, based on yeast two-hybrid analysis, it has been shown that catalases interact with the N terminus of PEX5 but not the C-terminal TPR domain (Horiguchi et al., 2001). In this study, CAT2 interacted with the C-terminal TPR domain of NCA1; this interaction likely occurred in cytosol, and NCA1 did not recruit catalases to the peroxisomes, indicating that TPR domain-containing proteins play roles in both recruiting catalase to peroxisomes and increasing catalase activity in the cytosol. When NCA1 was targeted to the mitochondrion in the nca1-3 mutant using the AKDE1 peptide, the mutant phenotype was not rescued. However, when NCA1 was targeted to the peroxisome in the nca1-3 mutant using the SKL peptide, the mutant phenotype was rescued. These results suggest that NCA1 plays a fundamental and essential role in increasing catalase activity, although we cannot presently exclude the possibility that NCA1-SKL was still in cytosol due to the NCA1-SKL leakage. In the presence of high CO2, the high external pH-sensitive phenotype of nca1-3 was largely rescued but still displayed a growth reduction compared with the wild type, while cat2 showed a similar sensitivity to that found in the wild type. These results suggest that catalases are major targets of NCA1 and that NCA1 also regulates other protein activities in response to alkaline stress. These results are supported by differential protein regulation that has been reported for the cat2 and nca1 mutants (Hackenberg et al., 2013).
CAT2 and NCA1 proteins were unstable in vitro and easily degraded. However, heterodimeric NCA1-CAT2 was more stable than the individual proteins, consistent with published results that indicate that NCA1 is required for stabilization of catalases in plants (Hackenberg et al., 2013). When we expressed GFP-CAT2 and GFP-CAT3 in Col-0 and the nca1-3 mutant, we found that NCA1 is not required for the peroxisomal localization of CAT2 and CAT3 and that the GFP signals did not significantly change in the nca1-3 mutant. It is possible that when GFP-labeled catalases were overexpressed in the nca1-3 knockout mutant, the GFP signal was not sensitive enough to respond to the reduced level of catalase or that nca1-1 and nca1-2, which are point mutations with an exchanged amino acid and truncated NCA1 protein, respectively, have more effect on the regulation of catalase stability.
As reported in many studies (Kirkman and Gaetani, 1984; Frugoli et al., 1996) and measured in this study, homotetrameric catalases have maximum activity, while monomeric catalases have almost no activity. NCA1 likely does not affect catalase tetramer formation in peroxisomes. The activity of monomeric CAT2 was greatly enhanced by NCA1 and heterodimeric NCA1-CAT2 had higher activity compared with the native homotetrameric CAT2. The higher molecular mass of the NCA1-CAT2 protein complex, which also had a molar ratio of 1:1, had even higher activity compared with the native homotetrameric CAT2. In vivo, NCA1 and catalases form complexes with the highest catalase activity at a molecular mass of ∼240 kD, which means that two molecules of NCA1 form a complex with two molecules of catalase. These results suggest that the NCA1 is required for increasing catalase activity.
NCA1 increased CAT2 activity through the NCA1 N-terminal RING-type zinc finger domain. Recently, LSD1, another zinc finger domain-containing protein, has been reported to interact with CAT1, CAT2, and CAT3 (Li et al., 2013). Catalase activity is reduced in the lsd1 mutant, although not as dramatically as in the nca1-3 mutant. The interaction between LSD1 and catalases requires the zinc finger domain of LSD1 (Li et al., 2013); however, it is not clear whether LSD1 directly activates catalases. The increase of CAT2 activity by NCA1 requires the zinc finger domain and the binding of zinc ions. For maximal activity, catalases form a tetramer in peroxisomes with a heme Fe in each unit (Nicholls, 2012). Crystal structure analysis will be required to dissect the roles of the zinc finger domain and zinc ions in the NCA1-mediated catalase activity increase and to determine the functional complex of these two proteins.
METHODS
Plant Growth and Treatments
Arabidopsis thaliana Col-0 was used as the wild type in all experiments. Homozygous cat2 (SALK_076998) and cat3 (SALK_092911) mutants were obtained from the ABRC.
Arabidopsis seeds were sterilized in a solution containing 20% sodium hypochlorite and 0.1% Triton X-100 for 10 min and washed five times with sterilized distilled water. Sterilized seeds were sown on MS medium with 0.3% Phytagel for germination or with 0.6% Phytagel for seedling growth and transferred to various treatments. Plates were kept in dark at 4°C for 3 d and transferred to the light for germination. Plants were grown under 16 h light/8 h dark or under continuous white light (light intensity of 30 μmol m–2 s–1) at 23°C.
To examine the tolerance of Arabidopsis to pH or HU (Sigma-Aldrich), seeds were sown on MS medium at pH 5.8 with 0.6% Phytagel and germinated. Five-day-old seedlings were transferred to MS medium at pH 8.0 or to MS medium (pH 5.8) with the indicated concentration of HU for vertical growth. At the indicated times, photos were taken and seedling weight or chlorophyll content was measured.
For germination assays, Arabidopsis seeds were sown on MS medium at pH 5.8 without or with the indicated concentration of MV (Sigma-Aldrich), kept in dark at 4°C for 3 d, and transferred to chambers with a light intensity of 30 μmol m–2 s–1. The percentage of green cotyledons was counted at the indicated times.
To assay a photorespiratory phenotype, 5-d-old seedlings were transferred to MS medium at pH 5.8 or 8.0 and grown under an irradiance of 90 μmol m−2 s−1 in air or in 3000 ppm CO2. To monitor phenotypes in older seedlings, seeds were sown in soil, germinated, and grown in a light chamber with irradiances of 30 μmol m–2 s–1 or 100 μmol m–2 s–1, 16 h/8 h.
Analysis of Chlorophyll Content
Arabidopsis seedlings were weighed and put into 1.5-mL Eppendorf tubes. Acetone (80%, 1 mL) was added and samples were incubated in the dark overnight at room temperature. The samples were centrifuged at 13,000g for 10 min, and the resulting supernatant was transferred into a 96-well plate and absorption measured at 663 and 645 nm with a microplate reader. The concentration of chlorophyll (total chlorophyll, chlorophyll a, and chlorophyll b) was calculated according to the following formula: chlorophyll a = 12.7A665 − 2.69A645; chlorophyll b = 22.9A645 − 4.68A663; and total chlorophyll = chlorophyll a + chlorophyll b.
Complementation Assays
The full-length NCA1 genomic DNA fragment (corresponding to the sequence from 2000 bp upstream of the NCA1 translation start codon to 500 bp downstream of the stop codon) was cloned into the pCAMBIA1300 vector between SacI and BamHI. The resulting construct was introduced into the nca1-3 mutant using Agrobacterium tumefaciens-mediated floral transformation (Clough and Bent, 1998) and T3 transgenic plants were used for analysis.
Plasmid Construction and Generation of Transgenic Plants
Full-length coding sequences of NCA1, NCA1C111S, NCA1H126Y, and NCA1C129A were cloned into the pCAMBIA1307-6×Myc or pCAMBIA1307-Flag-HA binary vectors between the AscI and PacI sites. Full-length coding regions of NCA1 and CAT2 were cloned into the AscI and PacI sites of the pET28a or pGEX-6p1 vectors. NCA180-405 and NCA1158-405 were cloned into the pET28a vector between the NdeI and AccI sites. Primers used for cloning are listed in Supplemental Table 1.
The resulting constructs were introduced into Arabidopsis using Agrobacterium-mediated floral transformation and verified by immunoblot analysis. T3 transgenic plants were used for analysis.
Real-Time and RT-PCR Analysis
Total RNA was extracted with Trizol reagent (Invitrogen) from 10-d-old seedlings grown on MS medium or root, stem, rosette, flower, or silique of 4-week-old plants grown in soil under a 16-h-light/8-h-dark photoperiod. Total RNA was treated with RNase-free DNase I (Takara) to remove genomic DNA. RNA was used for reverse transcription with M-MLV reverse transcriptase (Promega) according to the manufacturer’s instructions. cDNAs were used for quantitative real-time PCR or RT-PCR analysis with the primers listed in Supplemental Table 2. ACTIN was used as an internal control.
GUS Staining and Subcellular Localization
A 2000-bp genomic DNA fragment (1 to 2000 bp upstream of the translational start site) of NCA1 was amplified (see primers in Supplemental Table 1) and cloned into the EcoRI and BamHI sites of the pCAMBIA1381 binary vector. The resulting constructs were introduced into Col-0 plants using Agrobacterium-mediated floral transformation and T3 transgenic lines were used for GUS staining.
For subcellular localization, the coding regions of NCA1 and CAT3 (primers in Supplemental Table 1) were cloned into the pCAMBIA1205-GFP binary vector. NCA1 was introduced into the BamHI site and CAT3 was introduced into the AscI and PacI sites. Transgenic plants were generated using Agrobacterium-mediated floral transformation. T2 transgenic lines were used for GFP observation with a Zeiss confocal microscope.
Antibody Preparation and Immunoblot Analysis
Recombinant NCA1 and CAT3 proteins prepared from Escherichia coli were used as antigens for polyclonal antibody preparation in mouse. The immunoblots were probed with anti-NCA1 (1:1000) and anti-CAT (1:800) antibodies, and chemiluminescence signals were detected by autoradiography.
CAT Activity Assays
Ten-day-old Col-0, nca1-3, cat2, cat3, cat2 cat3, and nca1-3 cat2 cat3 seedlings were ground with 100 μL extraction buffer (10 mM Tris-HCl, pH 7.6, 150 mM NaCl, 2 mM EDTA, and 0.5% Nonidet P-40) and centrifuged at 10,000g for 10 min at 4°C. The supernatant was used as the crude extract and catalase activity was tested with the Catalase Activity Assay Kit (Beyotime). Protein concentration was determined using the Bradford protein assay. Crude extract (1 to 5 μL) was mixed with catalase testing buffer, and 50 mM H2O2 was used as the substrate. Reaction time was strictly controlled and stopped with addition of stop buffer. The mixture was then added into the working color solution and incubated for at least 15 min. Absorbance at 520 nm was measured and activity was calculated. Catalase activity was indicated in units/mL or units/mg. One unit of catalase activity is defined as the quantity of enzyme catalyzing the decomposition of 1 μmol H2O2 per minute.
Coimmunoprecipitation and Immunoblot Analysis
The NCA1 coding sequence was amplified and cloned into the SpeI site of the pCAMBIA1307- Flag-HA vector. Ten-day-old transgenic plants were ground in extraction buffer containing 10 mM Tris-HCl, pH 7.6, 150 mM NaCl, 2 mM EDTA, 0.5% (v/v) Nonidet P-40, and 2× protease inhibitor (Roche) and centrifuged at 10,000g for 15 min at 4°C. Supernatant (50 μL) was kept for analysis as input and the remaining supernatant was incubated with 20 μL anti-Flag antibody-conjugated agarose (Sigma-Aldrich) for 2 h at 4°C. After washing five times with extraction buffer, the products were analyzed by immunoblot analysis. The proteins were probed with anti-Flag (Sigma-Aldrich), and anti-CAT antibodies and chemiluminescence signals were detected with a chemiluminescence system (GE Healthcare).
Isolation of Intact Peroxisomes
Ten-day-old Arabidopsis seedlings were harvested and peroxisomes isolated as described by Reumann et al. (2007) with modification. Col-0 and nca1-3 seedlings (20 g) were ground individually in grinding buffer (170 mM Tricine-KOH, pH 7.5, 1.0 M sucrose, 1% [w/v] BSA, 2 mM EDTA, 5 mM DTT, 10 mM KCl, 1 mM MgCl2, and protease inhibitor cocktail). The products were filtered through two layers of Miracloth and centrifuged at 5,000g for 1 min at 4°C to obtain the crude extract. The supernatant was loaded onto Percoll density gradients prepared in TE buffer (20 mM Tricine-KOH, pH 7.5, and 1 mM EDTA) as described (Reumann et al., 2007) and centrifuged for 12 min at 13,000g (JA 25.5 rotor) followed by centrifugation for 20 min at 27,000g (JA 25.5 rotor). After centrifugation, peroxisomes are located at the bottom of the gradients. Fractions (∼2 mL) were collected from the top and bottom of the centrifugation tubes and washed in 36% (w/w) sucrose in TE buffer and centrifuged for 30 min at 39,000g (JA 25.5 rotor). The top fraction was labeled as the cytosol and the pellet as P1. Subsequently, P1 was loaded onto a sucrose density gradient (2 mL 41% [w/w], 2 mL 44% [w/w], 2 mL 46% [w/w], 3 mL 49% [w/w], 1 mL 51% [w/w], 1.5 mL 55% [w/w], and 1 mL 60% [w/w] in TE buffer) and centrifuged at 80,000g for 2 h with acceleration 7 and deceleration 7 (SW32Ti). After centrifugation, a white band appeared at the interface of 55 and 51% sucrose. The white band was collected and labeled PE. Protease inhibitor cocktail was added to the crude extract, cytosol, P1, and PE fractions. These fractions were analyzed using immunoblot analysis with anti-NCA1, anti-CAT, anti-MPK3 (Sigma-Aldrich), anti-PEX14p (Agrisera), anti-LHC2 (Agrisera), and anti-VDAC1 (Agrisera) antibodies. MPK3 was used as a marker for the cytosol, PEX14p as a peroxisomal marker, LHC2 as a chloroplast marker, and VDAC1 as a mitochondria marker.
Gel Filtration
Ten-day-old Arabidopsis seedlings were homogenized in buffer containing 10 mM Tris-HCl, pH 7.6, 150 mM NaCl, 2 mM EDTA, 0.5% Nonidet P-40, and 2× protease inhibitor (Roche) on ice and centrifuged at 10,000g for 10 min at 4°C to remove cell debris. The supernatant was subsequently centrifuged at 100,000g for 1 h at 4°C, and the resulting supernatant was filtered through a 0.45-µm filter (Millipore) before being loaded onto a Superdex-200 10/300GL column (GE Healthcare). Before loading the samples, the column was equilibrated with homogenization buffer without Nonidet P-40, and fractions (0.5 mL) were eluted with this buffer at a speed of 0.3 mL/minute. All fractions were analyzed via immunoblot analysis with anti-CAT and anti-NCA1 antibodies and catalase activity assays were performed. The molecular mass standards used for gel filtration size estimation were ferritin (440 kD), aldolase (158 kD), conalbumin (75 kD), and carbonic anhydrase (25 kD).
In Vitro Pull-Down Assay
Full-length coding sequences for NCA1 and CAT2 were introduced into the pET28a-His-SMT3 or pGEX-6p1-GST vectors for recombinant protein expression. NCA1 sequence was inserted into the pET28a vector at the BamHI site and CAT2 sequence was inserted into pGEX-6p1 between the AscI and PacI sites.
The pET28a-His-SMT3-NCA1 and pGEX-6p1-GST-CAT2 plasmids were cotransformed into E. coli DE3 cells and screened on Luria-Bertani plates containing both ampicillin and kanamycin. Colony PCR was performed to confirm the presence of both plasmids. Protein expression was induced at 16°C with 0.3 mM isopropyl β-d-1-thiogalactopyranoside overnight. The recombinant protein was purified with glutathione-agarose according to the manufacturer’s protocol. The resulting proteins were separated using SDS-PAGE and digested with Ulp to allow identification of the 55-kD band as His-SMT3-NCA1.
ICP-AES
Eluted recombinant proteins were used for ICP-AES analysis with a SPECTRO Arcos EOP inductively coupled plasma spectrometer (SPECTRO Analytical Instruments). The plasma power was set at 1.4 kW and nebulizer flow was set at 0.8 L/min.
Holdase Chaperone Assay
Recombinant His-NCA1 and His-CAT2 proteins were expressed as described previously and purified using Ni-NTA affinity chromatography. Arabidopsis MDH (EC 1.1.1.37) and HSP90.2 were prepared as described previously (Cha et al., 2009; Kim et al., 2011). The proteins were further dialyzed against 50 mM Tris-HCl, pH 8.0. Holdase chaperone activity of His-NCA1 was assayed by measuring its capacity to suppress thermal aggregation of MDH or His-CAT2, as described by Cha et al. (2009). MDH or CAT2 aggregation was monitored in reaction buffer (40 mM HEPES, pH 7.5) by measuring absorbance at 340 nm using a Beckman DU-800 spectrophotometer attached to a thermostatic cell holder assembly at 45°C. In each set, recombinant His-NCA1 was prepared independently.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At3g54360 (NCA1), At4g35090 (CAT2), and At1g20620 (CAT3).
Supplemental Data
Supplemental Figure 1. Growth of 5-d-Old nca1-3 Seedlings Is Similar to That in Col-0 Seedlings.
Supplemental Figure 2. NCA1 Gene Expression and Protein Localization.
Supplemental Figure 3. Flag-HA-NCA1 Rescues the nca1-3 Phenotype.
Supplemental Figure 4. Analysis of Anti-CAT Antibody Specificity.
Supplemental Figure 5. NCA1 Genetically Interacts with CAT2.
Supplemental Figure 6. Increase of Catalase Activity Is Not Affected by Alkaline Stress.
Supplemental Figure 7. NCA1 Does Not Affect the Subcellular Localization of Catalase.
Supplemental Figure 8. The RING-Finger Domain of NCA1 Is Required for Seedling Tolerance of High pH.
Supplemental Table 1. Primers for Plasmid Construction.
Supplemental Table 2. Primers for RT-PCR.
Supplementary Material
Acknowledgments
We thank Jianping Hu for providing the protocol to purify peroxisomes. This work was supported by the National Basic Research Program of China (Grants 2012CB114200 and 2015CB910203), the National Natural Science Foundation of China (Grant 31430012), the Foundation for Innovative Research Group of the National Natural Science Foundation of China (Grant 31421062), the U.S. Department of Energy/Energy Biosciences (Grant DE-FG02-04ER15616 to K.S.S.), the National Research Foundation of Korea (Grant 2013R1A2A1A01005170 to D.-J.Y.), and the Chinese Universities Scientific Fund (2013YJ002 to J. Li).
AUTHOR CONTRIBUTIONS
J. Li and J. Liu performed the research. J. Li, J. Liu, and Y.G. designed the research and analyzed the data. G.W. and G.L. contributed to the purification of recombinant proteins. J.-Y.C. and D.-J.Y. performed the research on analysis of holdase activity. Z.L. and J.G. contributed to the ICP-AES experiments. C.Z. screened the mutant. J. Li, J. Liu, Y.G., S.C., Y.Y., W.-Y.K., D.-J.Y., Z.C., and K.S.S. contributed to the discussion and wrote the article.
Glossary
- TPR
tetratricopeptide repeat
- ROS
reactive oxygen species
- CAT
catalase
- APX
ascorbate peroxidase
- PRX
peroxidase
- Col-0
Columbia-0
- MS
Murashige and Skoog
- MV
methyl viologen
- PE
high purity peroxisome
- HU
hydroxyurea
- 3-AT
3-amino-1,2,4-triazole
- ICP-AES
inductively coupled plasma-atomic emission spectroscopy
References
- Allan R.K., Ratajczak T. (2011). Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones 16: 353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alscher R.G., Donahue J.L., Cramer C.L. (1997). Reactive oxygen species and antioxidants: relationships in green cells. Physiol. Plant. 100: 224–233. [Google Scholar]
- Bueso E., Alejandro S., Carbonell P., Perez-Amador M.A., Fayos J., Bellés J.M., Rodriguez P.L., Serrano R. (2007). The lithium tolerance of the Arabidopsis cat2 mutant reveals a cross-talk between oxidative stress and ethylene. Plant J. 52: 1052–1065. [DOI] [PubMed] [Google Scholar]
- Casey J.R., Grinstein S., Orlowski J. (2010). Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 11: 50–61. [DOI] [PubMed] [Google Scholar]
- Cha J.-Y., Jung M.H., Ermawati N., Su’udi M., Rho G.-J., Han C.D., Lee K.H., Son D. (2009). Functional characterization of orchardgrass endoplasmic reticulum-resident Hsp90 (DgHsp90) as a chaperone and an ATPase. Plant Physiol. Biochem. 47: 859–866. [DOI] [PubMed] [Google Scholar]
- Chaouch S., Noctor G. (2010). Myo-inositol abolishes salicylic acid-dependent cell death and pathogen defence responses triggered by peroxisomal hydrogen peroxide. New Phytol. 188: 711–718. [DOI] [PubMed] [Google Scholar]
- Chelikani P., Fita I., Loewen P.C. (2004). Diversity of structures and properties among catalases. Cell. Mol. Life Sci. 61: 192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dat J.F., Lopez-Delgado H., Foyer C.H., Scott I.M. (1998). Parallel changes in H2O2 and catalase during thermotolerance induced by salicylic acid or heat acclimation in mustard seedlings. Plant Physiol. 116: 1351–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelbarth H., Nägele T., Heyer A.G. (2013). Responses of antioxidant enzymes to cold and high light are not correlated to freezing tolerance in natural accessions of Arabidopsis thaliana. Plant Biol (Stuttg) 15: 982–990. [DOI] [PubMed] [Google Scholar]
- Du Y.Y., Wang P.C., Chen J., Song C.P. (2008). Comprehensive functional analysis of the catalase gene family in Arabidopsis thaliana. J. Integr. Plant Biol. 50: 1318–1326. [DOI] [PubMed] [Google Scholar]
- Foreman J., Demidchik V., Bothwell J.H., Mylona P., Miedema H., Torres M.A., Linstead P., Costa S., Brownlee C., Jones J.D., Davies J.M., Dolan L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446. [DOI] [PubMed] [Google Scholar]
- Frugoli J.A., Zhong H.H., Nuccio M.L., McCourt P., McPeek M.A., Thomas T.L., McClung C.R. (1996). Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 112: 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukamatsu Y., Yabe N., Hasunuma K. (2003). Arabidopsis NDK1 is a component of ROS signaling by interacting with three catalases. Plant Cell Physiol. 44: 982–989. [DOI] [PubMed] [Google Scholar]
- Gechev T., Gadjev I., Van Breusegem F., Inzé D., Dukiandjiev S., Toneva V., Minkov I. (2002). Hydrogen peroxide protects tobacco from oxidative stress by inducing a set of antioxidant enzymes. Cell. Mol. Life Sci. 59: 708–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossett D.R., Banks S.W., Millhollon E.P., Lucas M.C. (1996). Antioxidant response to NaCl stress in a control and an NaCl-tolerant cotton cell line grown in the presence of paraquat, buthionine sulfoximine, and exogenous glutathione. Plant Physiol. 112: 803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout E., Bligny R., Douce R. (1992). Regulation of intracellular pH values in higher plant cells. Carbon-13 and phosphorus-31 nuclear magnetic resonance studies. J. Biol. Chem. 267: 13903–13909. [PubMed] [Google Scholar]
- Hackenberg T., et al. (2013). Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in Arabidopsis. Plant Cell 25: 4616–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Aoki M., Kondo M., Nishimura M. (1997). Changes in targeting efficiencies of proteins to plant microbodies caused by amino acid substitutions in the carboxy-terminal tripeptide. Plant Cell Physiol. 38: 759–768. [DOI] [PubMed] [Google Scholar]
- Heldt H.W., Werdan K., Milovancev M., Geller G. (1973). Alkalization of the chloroplast stroma caused by light-dependent proton flux into the thylakoid space. Biochim. Biophys. Acta 314: 224–241. [DOI] [PubMed] [Google Scholar]
- Horiguchi H., Yurimoto H., Goh T., Nakagawa T., Kato N., Sakai Y. (2001). Peroxisomal catalase in the methylotrophic yeast Candida boidinii: transport efficiency and metabolic significance. J. Bacteriol. 183: 6372–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juul T., Malolepszy A., Dybkaer K., Kidmose R., Rasmussen J.T., Andersen G.R., Johnsen H.E., Jørgensen J.-E., Andersen S.U. (2010). The in vivo toxicity of hydroxyurea depends on its direct target catalase. J. Biol. Chem. 285: 21411–21415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.S., Kim W.Y., Fujiwara S., Kim J., Cha J.-Y., Park J.H., Lee S.Y., Somers D.E. (2011). HSP90 functions in the circadian clock through stabilization of the client F-box protein ZEITLUPE. Proc. Natl. Acad. Sci. USA 108: 16843–16848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkman H.N., Gaetani G.F. (1984). Catalase: a tetrameric enzyme with four tightly bound molecules of NADPH. Proc. Natl. Acad. Sci. USA 81: 4343–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosarev P., Mayer K., Hardtke C.S. (2002). Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol. 3: RESEARCH0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lager I., Andréasson O., Dunbar T.L., Andreasson E., Escobar M.A., Rasmusson A.G. (2010). Changes in external pH rapidly alter plant gene expression and modulate auxin and elicitor responses. Plant Cell Environ. 33: 1513–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Yi H. (2012). Effect of sulfur dioxide on ROS production, gene expression and antioxidant enzyme activity in Arabidopsis plants. Plant Physiol. Biochem. 58: 46–53. [DOI] [PubMed] [Google Scholar]
- Li Y., Chen L., Mu J., Zuo J. (2013). LESION SIMULATING DISEASE1 interacts with catalases to regulate hypersensitive cell death in Arabidopsis. Plant Physiol. 163: 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathioudakis M.M., Veiga R.S., Canto T., Medina V., Mossialos D., Makris A.M., Livieratos I. (2013). Pepino mosaic virus triple gene block protein 1 (TGBp1) interacts with and increases tomato catalase 1 activity to enhance virus accumulation. Mol. Plant Pathol. 14: 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlmer N., Parvin N., Hurst C.H., Knight M.R., Teige M., Vothknecht U.C. (2012). A toolset of aequorin expression vectors for in planta studies of subcellular calcium concentrations in Arabidopsis thaliana. J. Exp. Bot. 63: 1751–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A., Hager J., Chaouch S., Queval G., Han Y., Taconnat L., Saindrenan P., Gouia H., Issakidis-Bourguet E., Renou J.P., Noctor G. (2010b). Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol. 153: 1144–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A., Noctor G., Baker A. (2012). Plant catalases: peroxisomal redox guardians. Arch. Biochem. Biophys. 525: 181–194. [DOI] [PubMed] [Google Scholar]
- Mhamdi A., Queval G., Chaouch S., Vanderauwera S., Van Breusegem F., Noctor G. (2010a). Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 61: 4197–4220. [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9: 490–498. [DOI] [PubMed] [Google Scholar]
- Munné-Bosch S., Queval G., Foyer C.H. (2013). The impact of global change factors on redox signaling underpinning stress tolerance. Plant Physiol. 161: 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls P. (2012). Classical catalase: ancient and modern. Arch. Biochem. Biophys. 525: 95–101. [DOI] [PubMed] [Google Scholar]
- Palmgren M.G. (2001). Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 817–845. [DOI] [PubMed] [Google Scholar]
- Queval G., Issakidis-Bourguet E., Hoeberichts F.A., Vandorpe M., Gakière B., Vanacker H., Miginiac-Maslow M., Van Breusegem F., Noctor G. (2007). Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52: 640–657. [DOI] [PubMed] [Google Scholar]
- Rayle D.L., Cleland R.E. (1992). The Acid Growth Theory of auxin-induced cell elongation is alive and well. Plant Physiol. 99: 1271–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Babujee L., Ma C., Wienkoop S., Siemsen T., Antonicelli G.E., Rasche N., Lüder F., Weckwerth W., Jahn O. (2007). Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19: 3170–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rober-Kleber N., Albrechtová J.T., Fleig S., Huck N., Michalke W., Wagner E., Speth V., Neuhaus G., Fischer-Iglesias C. (2003). Plasma membrane H+-ATPase is involved in auxin-mediated cell elongation during wheat embryo development. Plant Physiol. 131: 1302–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selivanov V.A., Zeak J.A., Roca J., Cascante M., Trucco M., Votyakova T.V. (2008). The role of external and matrix pH in mitochondrial reactive oxygen species generation. J. Biol. Chem. 283: 29292–29300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servaites J.C. (1977). pH dependence of photosynthesis and photorespiration in soybean leaf cells. Plant Physiol. 60: 693–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Jensen R.G., Bohnert H.J. (1997). Mannitol protects against oxidation by hydroxyl radicals. Plant Physiol. 115: 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C.-P., Guo Y., Qiu Q., Lambert G., Galbraith D.W., Jagendorf A., Zhu J.-K. (2004). A probable Na+(K+)/H+ exchanger on the chloroplast envelope functions in pH homeostasis and chloroplast development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 101: 10211–10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Koussevitzky S., Mittler R., Miller G. (2012). ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35: 259–270. [DOI] [PubMed] [Google Scholar]
- Vanderauwera S., Zimmermann P., Rombauts S., Vandenabeele S., Langebartels C., Gruissem W., Inzé D., Van Breusegem F. (2005). Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol. 139: 806–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues P.E., Batelli G., Grillo S., Agius F., Kim Y.-S., Zhu J., Agarwal M., Katiyar-Agarwal S., Zhu J.-K. (2007). Interaction of SOS2 with nucleoside diphosphate kinase 2 and catalases reveals a point of connection between salt stress and H2O2 signaling in Arabidopsis thaliana. Mol. Cell. Biol. 27: 7771–7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werdan K., Heldt H.W., Milovancev M. (1975). The role of pH in the regulation of carbon fixation in the chloroplast stroma. Studies on CO2 fixation in the light and dark. Biochim. Biophys. Acta 396: 276–292. [DOI] [PubMed] [Google Scholar]
- Whitten S.T., García-Moreno E.B., Hilser V.J. (2005). Local conformational fluctuations can modulate the coupling between proton binding and global structural transitions in proteins. Proc. Natl. Acad. Sci. USA 102: 4282–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Poovaiah B.W. (2002). Hydrogen peroxide homeostasis: activation of plant catalase by calcium/calmodulin. Proc. Natl. Acad. Sci. USA 99: 4097–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Qin Y., Xie C., Zhao F., Zhao J., Liu D., Chen S., Fuglsang A.T., Palmgren M.G., Schumaker K.S., Deng X.W., Guo Y. (2010). The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. Plant Cell 22: 1313–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T., Jin D., Liu Q., Gong Z. (2013). Abscisic acid suppresses the highly occurred somatic homologous recombination in Arabidopsis rfc1 mutant. J. Genet. Genomics 40: 465–471. [DOI] [PubMed] [Google Scholar]
- Zhao F., Chen L., Perl A., Chen S., Ma H. (2011). Proteomic changes in grape embryogenic callus in response to Agrobacterium tumefaciens-mediated transformation. Plant Sci. 181: 485–495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.