A kinase complex regulates phosphate transporter trafficking in response to phosphate, inhibiting interaction with the factor regulating exit from the endoplasmic reticulum to the plasma membrane.
Abstract
Phosphate transporters (PTs) mediate phosphorus uptake and are regulated at the transcriptional and posttranslational levels. In one key mechanism of posttranslational regulation, phosphorylation of PTs affects their trafficking from the endoplasmic reticulum (ER) to the plasma membrane. However, the kinase(s) mediating PT phosphorylation and the mechanism leading to ER retention of phosphorylated PTs remain unclear. In this study, we identified a rice (Oryza sativa) kinase subunit, CK2β3, which interacts with PT2 and PT8 in a yeast two-hybrid screen. Also, the CK2α3/β3 holoenzyme phosphorylates PT8 under phosphate-sufficient conditions. This phosphorylation inhibited the interaction of PT8 with PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1, a key cofactor regulating the exit of PTs from the ER to the plasma membrane. Additionally, phosphorus starvation promoted CK2β3 degradation, relieving the negative regulation of PT phosphorus-insufficient conditions. In accordance, transgenic expression of a nonphosphorylatable version of OsPT8 resulted in elevated levels of that protein at the plasma membrane and enhanced phosphorus accumulation and plant growth under various phosphorus regimes. Taken together, these results indicate that CK2α3/β3 negatively regulates PTs and phosphorus status regulates CK2α3/β3.
INTRODUCTION
Phosphorus (P), as a component of key macromolecules, including nucleic acids (the largest organic P pool in plants) and phospholipids (important elements of membranes), is one of the essential mineral nutrients required by all organisms. P also influences photosynthesis, respiration, and the regulation of a wide array of cellular processes (Raghothama, 1999). Plants preferentially absorb P in the form of inorganic phosphate (Pi), which is poorly mobile in soils and often not readily available to plants. Thus, the level of available Pi is a major determinant of plant growth and productivity in many soils (Veneklaas et al., 2012; Zhang et al., 2014). However, plants have evolved an array of adaptive responses, involving developmental, physiological, and biochemical changes, to cope with growth in Pi-depleted soils. These responses are oriented toward improving Pi acquisition and use efficiency and to protecting plants from the stress caused by Pi starvation (Raghothama, 1999; Zhang et al., 2014).
The uptake of Pi is mediated by plasma membrane (PM)-localized Pi transporters (PTs) belonging to the PHT1 family, which shares similarity with the yeast high-affinity phosphate transporter PHO84 (Nussaume et al., 2011). Nine PHT1 genes have been identified in Arabidopsis thaliana and 13 in rice (Oryza sativa) (Raghothama, 1999; Goff et al., 2002). Gene expression analysis showed that most PHT1 family members are temporally and spatially upregulated in response to Pi starvation (Karthikeyan et al., 2002; Mudge et al., 2002; Misson et al., 2004; Shin et al., 2004). Their expression patterns extend beyond the root, in line with their roles in both Pi uptake and translocation: crucial functions for maintaining the internal P homeostasis in plants. For instance, in rice, the low-affinity phosphate transporter PT2 plays an important role in Pi translocation from root to shoot (Ai et al., 2009), whereas the high-affinity transporter PT8 plays a role in uptake, with half of the Pi uptake capacity dependent on PT8 (Chen et al., 2011; Jia et al., 2011). Therefore, increasing Pi transporter activity is one of the most important plant responses to Pi starvation (Raghothama, 1999; Nussaume et al., 2011).
In addition to transcriptional control, the activity of PTs, as PM-localized proteins, is also regulated posttranscriptionally. PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (PHF1) plays a major role in regulating the trafficking of PTs from the endoplasmic reticulum (ER) to the PM (González et al., 2005; Chen et al., 2011). In addition, posttranslational control of PHT1 family members at the level of protein stability involves both PHOSPHATE2 (PHO2) and NITROGEN LIMITATION ADAPTATION (NLA), two components of the ubiquitin-mediated degradation pathway (Huang et al., 2013; Lin et al., 2013; Park et al., 2014). Superimposed on these mechanisms, Pi transporters may undergo phosphorylation that negatively regulates the exit of PHT1 members from the ER (Nühse et al., 2004; Hem et al., 2007; Bayle et al., 2011). However, the nature of the kinase(s) involved in the control of PHT1 activity and the underlying molecular mechanism(s) remain unclear.
CK2, formerly known as casein kinase II, is a heterotetramer composed of two catalytic and two regulatory subunits, CK2α and CK2β, respectively (Niefind et al., 2001). The human genome has two genes encoding catalytic α- or α′-subunits and only one for the regulatory β-subunit (Wirkner et al., 1998; Ackermann et al., 2005), whereas plant genomes frequently have multigene families that encode each subunit (Espunya et al., 2005; Salinas et al., 2006). For example, four and two genes in rice encode CK2α and CK2β, respectively. CK2 is involved in a number of biological processes in mammals (Pinna, 2002; Meggio and Pinna, 2003), and plant CK2s also have substrates in several developmental, environmental, and hormonal response pathways (Mulekar and Huq, 2014). However, a biological role for CK2 in plant Pi homeostasis remains to be explored.
In this study, using a yeast two-hybrid screen, we identified a CK2 regulatory subunit from rice, CK2β3, which interacts with PT2 and PT8. We found that CK2α3/β3 holoenzyme phosphorylated PT8 under Pi-sufficient conditions, providing evidence of a role for CK2 kinase in Pi homeostasis. The CK2-mediated phosphorylation in turn prevented PT8 interaction with PHF1, providing a mechanism by which PT phosphorylation results in ER retention. Interestingly, phosphorylation of the CK2β3 subunit appeared to affect CK2β3 stability. In line with a negative regulatory role of PT phosphorylation, plants expressing a nonphosphorylatable mutant version of PT8 displayed increased PT8 levels at the PM and higher Pi uptake ability compared with plants expressing wild-type PT8. As the CK2 phosphorylation site in PTs is conserved among plant species, our findings suggest a potential strategy to engineer crop plants for increased Pi-acquisition efficiency.
RESULTS
The Rice CK2α3/β3 Complex Interacts with PTs
Previous phosphoproteomic studies have identified phosphorylation of PTs (Nühse et al., 2004), and posttranscriptional regulation has been reported to be involved in the exit of PHT1 from the ER in plants (Bayle et al., 2011). To identify kinases potentially acting on PTs, we fused PT8 (Os10g0444700), a rice high-affinity Pi transporter (Chen et al., 2011; Jia et al., 2011), to the C terminus of ubiquitin. We used the resulting chimeric bait vector PT8-Cub to screen a rice root cDNA library and identified multiple independent cDNAs encoding a putative protein kinase, CK2 (formerly known as casein kinase II) β-subunit (Os07g0495100; Rohila et al., 2006; Ding et al., 2009).
To confirm the initial library screening, we used PT8 and PT2 (Os03g0150800), the principal low-affinity PT for Pi translocation in rice roots (Ai et al., 2009), as bait to test for interaction with the CK2β subunit. Additionally, given that CK2 exists as a tetramer of two catalytic α-subunits, α2 (Os07g0114400) and α3 (Os03g0207300), and two regulatory β-subunits, β1 (Os10g0564900) and β3 (Os07g0495100) (Meggio and Pinna, 2003; Bibby and Litchfield, 2005; Rohila et al., 2006), the other three subunits were also used as prey for interaction analysis with PTs in both yeast and plants.
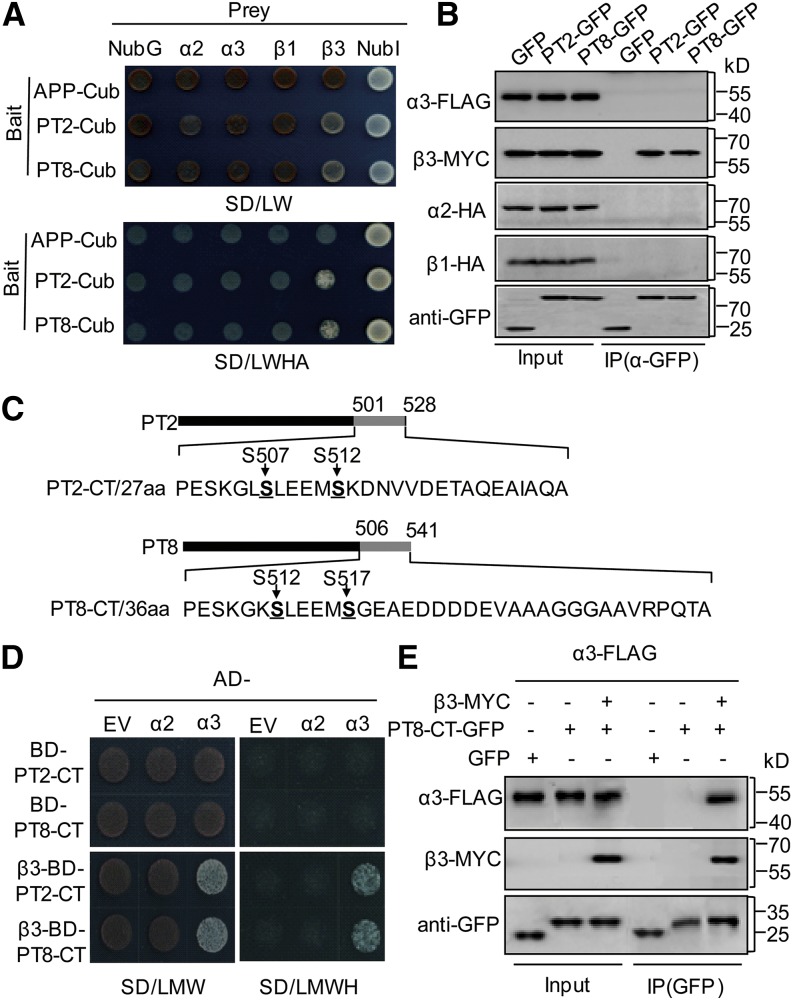
Split-ubiquitin yeast two-hybrid assays indicated that among these CK2 components, only β3 interacted with PT2 and PT8 in yeast cells (Figure 1A). To further validate this two-hybrid interaction between PT and the CK2 components, we conducted coimmunoprecipitation (co-IP) assays using Nicotiana benthamiana leaves infiltrated with either green fluorescent protein (GFP) or PT2/PT8-GFP together with the four CK2 components. β3-MYC was observed in PT2/PT8-GFP immunoprecipitates but not in the GFP control reactions (Figure 1B), whereas no physical interaction signal could be detected between PT2/PT8-GFP and the other three subunits of CK2 kinase in planta.
Figure 1.
Rice CK2β3 Directly Interacts with PT and Is Necessary for the CK2α3 Interaction with PT.
(A) Split-ubiquitin yeast two-hybrid analysis. Cub, C-terminal ubiquitin (the bait APP-Cub served as a control that could interact with NubI but not with NubG and NubG-CK2 subunits); NubI and NubG, the wild type and the mutated N-terminal fragment of ubiquitin; SD/LW, -Leu-Trp; SD/LWHA, -Leu-Trp-His-Ade.
(B) In vivo co-IP of PT2/PT8 and four CK2 subunits. An α-GFP affinity matrix was used for immunoprecipitation, and the immunoblots were developed using tag-specific antibodies.
(C) Schematic representation of the C-terminal (CT) structure of PT2 and PT8 used in the yeast three-hybrid and co-IP assays. Amino acids 501 to 528 of PT2-CT and 506 to 541 of PT8-CT, containing Ser-512 and Ser-517, respectively, are indicated.
(D) Yeast three-hybrid assay. CK2α2 and CK2α3 were used as prey (AD-α2/α3); pBridge plasmids containing PT2/PT8-CT and additional CK2β3 cloned into MSCII (β3-BD-PT2/8-CT) were used as bait and were cotransformed into the AH109 yeast strain. EV, empty vector; SD/LMW, -Leu-Met-Trp; SD/LMWH, -Leu-Met-Trp-His.
(E) co-IP of PT8-CT with CK2α3 and CK2β3 in planta.
The CK2β subunit often serves as an anchor element to bind its targets and interact with the catalytic α-subunits to form a holoenzyme (Ganley et al., 2001; Pinna, 2002). Accordingly, we tested for interactions of α-subunits with the PTs in the presence of CK2β3. The hydrophilic C termini (CT) of PT2/PT8, including the putative phosphorylation sites (Figure 1C; Bayle et al., 2011; Chen et al., 2011), were used in a yeast three-hybrid assay with two different CK2α (α2/α3) catalytic subunits in the presence or absence of CK2β3. CK2β3 was found to be necessary for the interaction between CK2α3 and PT2/PT8-CT in these assays (Figure 1D), suggesting that CK2α3, CK2β3, and the C termini of PT2/PT8 form a complex in yeast. In vivo co-IP assays confirmed this observation, as CK2β3 and CK2α3 were pulled down together with PT2/PT8-CT in plant cells (Figure 1E). The specificity of the CK2α subunit was confirmed, as even in the presence of CK2β3, no physical interaction between PT2/PT8-CT and CK2α2 was observed. Altogether, these results showed that the CK2α3/β3 complex interacted with PTs.
CK2α3/β3-Mediated Phosphorylation of PTs under Pi-Sufficient Conditions
Given our observation that the CK2α3/β3 complex interacted with PTs and the fact that phosphorylation of the PHT1 C terminus is known to regulate its exit from the ER (Bayle et al., 2011), we predicted that CK2α3/β3 might localize to the ER and phosphorylate PTs. To test this hypothesis, we examined the subcellular localization of CK2α3 and CK2β3. A z-stack image of confocal sections showed that both subunits localized in the ER (Supplemental Figure 1) as well as in the nucleus, which is consistent with a previously reported site of CK2 localization (Salinas et al., 2006).
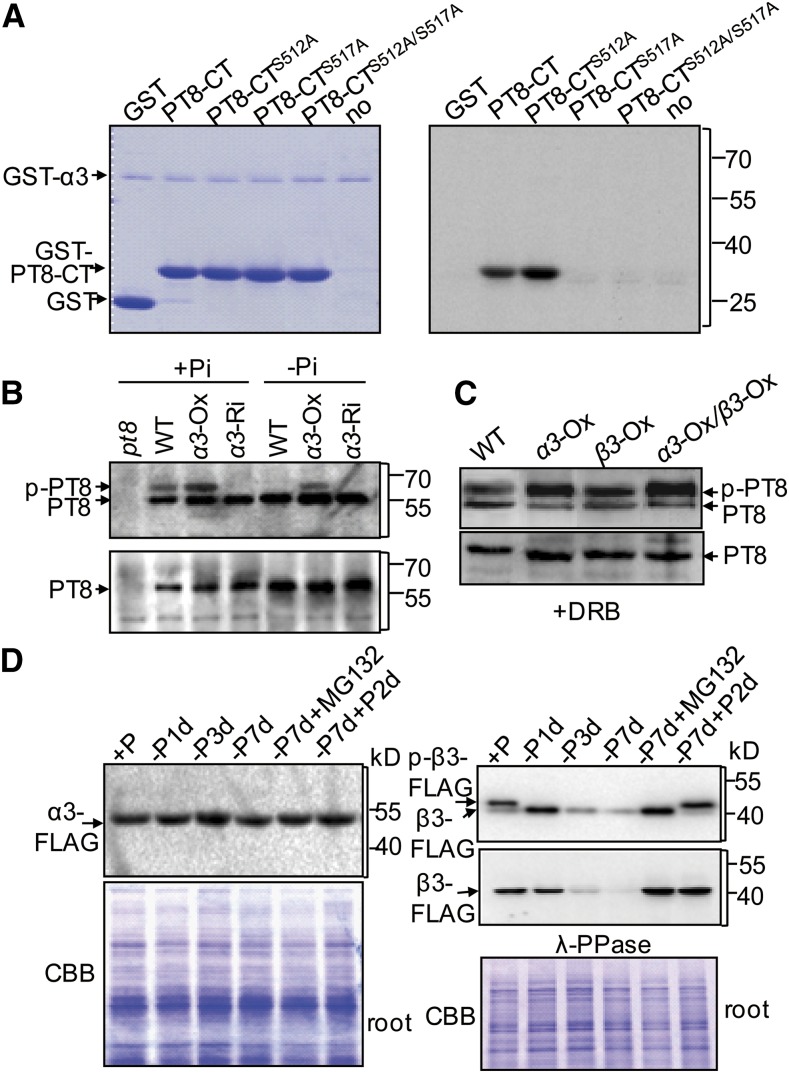
To determine whether CK2α3/β3 can phosphorylate PT in the ER, we performed in vitro phosphorylation assays using recombinant GST-α3, GST-β3, and GST-PT8-CT proteins. These assays showed that PT8-CT was phosphorylated in vitro by the catalytic subunit CK2α3 (Figure 2A) but not by the regulatory subunit CK2β3 (Supplemental Figure 2). This is consistent with the previous finding that both the isolated CK2 catalytic α-subunits and the holoenzyme have constitutive activity (Pinna, 2002; Sarno et al., 2002). To test whether the two conservative serine residues were target sites for CK2, we mutated PT8-CT peptides, replacing Ser-512 or Ser-517 by alanine (in peptides designated PT8-CTS512A and PT8-CTS517A, respectively, mimicking nonphosphorylatable forms of PT8-CT). The mutation of Ser-517 (but not Ser-512) prevented the phosphorylation of PT8-CT (Figure 2A), indicating that Ser-517 of PT8 is the site of phosphorylation by CK2α3.
Figure 2.
CK2α3-Mediated Phosphorylation of PT8 Is Affected by Pi Supply.
(A) Efficient phosphorylation of the hydrophilic C termini (CT) of PT8 by CK2α3 in vitro. Control reactions with GST (first lane) and no substrate (last lane) were performed side by side.
(B) Phosphorylation of PT8 by CK2α3 in vivo. Wild type, CK2α3 overexpression (α3-Ox), and CK2α3 knockdown (α3-Ri) plants (top) were used. The sensitivity of these bands to λ-PPase (bottom) was also tested.
(C) Effect of the CK2-specific inhibitor DRB (50 μM) on PT8 (p-PT8) phosphorylation by CK2α3 and CK2β3.
The immunoblots in (B) and (C) were developed with anti-PT8 in PhosTag SDS-PAGE.
(D) Pi supply-dependent phosphorylation and λ-PPase sensitivity of CK2β3. Ten-day-old transgenic plants were subjected to Pi starvation for different times as indicated, to Pi resupply after Pi starvation, and to Pi starvation in the presence of MG132 (10 μM). Proteins from the roots of those plants were probed by immunoblotting using an anti-FLAG antibody. The expression of α3-FLAG (left panel), phosphorylation of β3-FLAG (right panel, top blot), and sensitivity to λ-PPase treatment (right panel, bottom blot) are shown. Coomassie brilliant blue (CBB) staining was used as a loading control.
To confirm the phosphorylation of PT8 by CK2α3/β3 holoenzyme in vivo, we then performed PT8 phosphorylation assays using solubilized microsomal extracts from rice roots. A PT8-specific antibody (Supplemental Figure 3) was used to detect the phosphorylation status of PT8 via PhosTag SDS-PAGE (Figure 2B). In wild-type roots grown under +P (200 μM Pi), two bands were detected on the PhosTag immunoblots. The lower mobility band was determined to be phosphorylated PT8, as it was sensitive to λ-phosphatase (λ-PPase) treatment (Figure 2B). Furthermore, extracts from the roots of CK2α3 overexpressor (α3-OX) and CK2α3 knockdown (α3-Ri) plants displayed enhanced and reduced PT8 phosphorylation, respectively. This validated our in vitro experiments showing that CK2α3 could function as the CK2 catalytic subunit involved in PT phosphorylation.
We also transferred the two subunits into rice protoplasts together to test additive effects of CK2β3, and the results showed that the phosphorylation of PT8 by CK2α3 was enhanced in the presence of CK2β3 (Figure 2C). Phosphorylation by CK2 in these bands was further confirmed by their sensitivity to 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB), a CK2-specific inhibitor. It is noteworthy that CK2β3 did not provide catalytic activity, but it did increase holoenzyme function. Additionally, in vivo phosphorylation analysis of plants overexpressing CK2β3 (β3-OX) alone showed that increasing CK2β3 also enhanced PT8 phosphorylation in transgenic rice (Supplemental Figure 4), which is presumably dependent on endogenous expression of CK2α3. By contrast, downregulation of CK2β3 resulted in reduced PT8 phosphorylation, in agreement with in vivo CK2 function depending on the substrate-anchoring, regulatory subunit as well as the catalytic subunit.
Given that phosphorylation of PT8 was barely detectable in wild-type plants grown under −P (0 μM Pi) conditions (Figure 2B), the activity of CK2α3/β3 on PT might be regulated by Pi status. To test this hypothesis, proteins were extracted from roots and shoots of transgenic 35S-CK2α3-FLAG and 35S-CK2β3-FLAG plants grown under +P and −P conditions. PhosTag immunoblots, using tag-specific antibody, showed no change of CK2α3 protein levels under +P and −P conditions (Figure 2D; Supplemental Figure 5), whereas phosphorylation of CK2β3 was substantially reduced after Pi depletion, and CK2β3 protein accumulation gradually decreased under Pi starvation conditions in a time-dependent manner. After Pi recovery, phosphorylation level and protein abundance of CK2β3 rapidly recovered in both roots and shoots (Figure 2D; Supplemental Figure 5). Treatment with MG132 (10 μM), a 26S proteasome inhibitor, blocked the CK2β3 decline but not the reduction in phosphorylation (Figure 2D), indicative of protein degradation dependent on the ubiquitin/26S proteasome pathway. CK2β subunits that fail to form complexes with catalytic subunits are rapidly degraded (Zhang et al., 2002). In this context, the reduced accumulation of CK2β3 we observed under Pi starvation suggested that unphosphorylated CK2β3 may display decreased binding affinity with CK2α3 to form a stable holoenzyme. In vitro pull-down assays supported this hypothesis (Supplemental Figure 6). Together, all these results revealed that phosphorylation of PTs by CK2 was modulated in response to cellular Pi concentration.
CK2α3/β3 Affects the ER Exit of PT
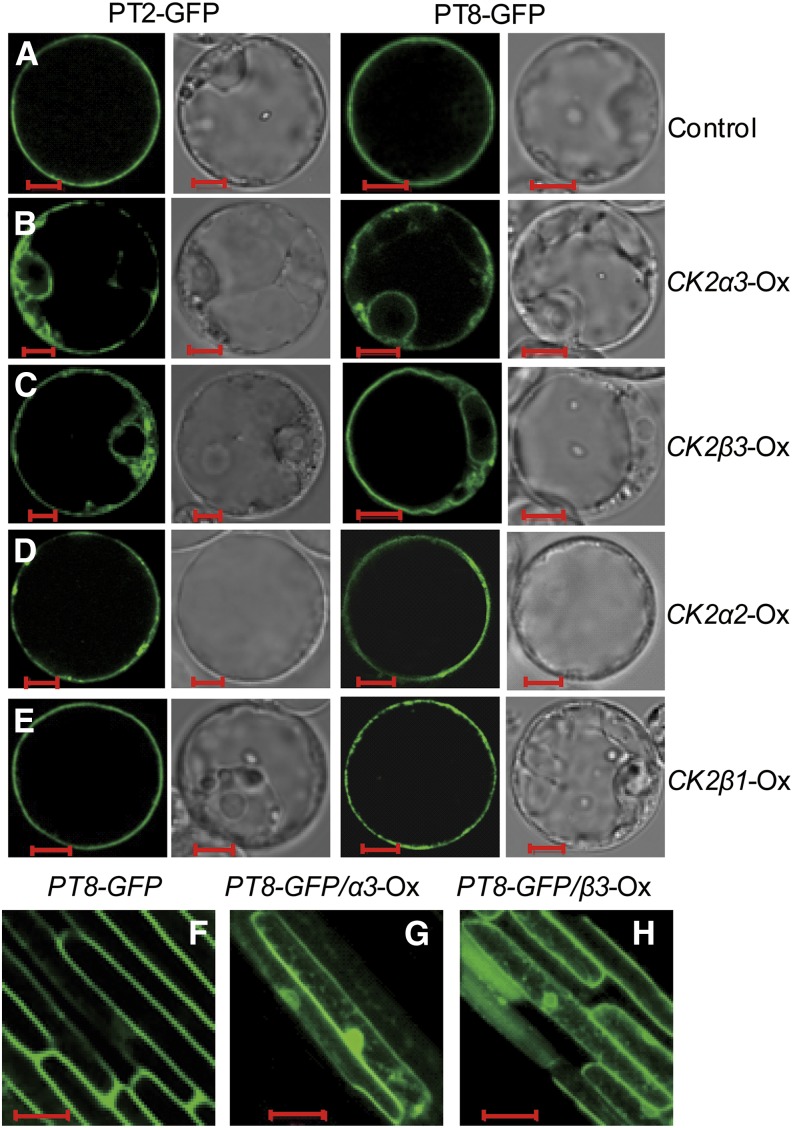
Transgenic lines expressing the phospho-mimicking PHT1;1-GFP (PHT1;1S514D-GFP) exhibited an ER retention pattern in Arabidopsis (Bayle et al., 2011). As rice CK2α3/β3 could phosphorylate the homologous serine site in PT8 (Figures 2A and 2C; Chen et al., 2011), this CK2 could function to modulate the ER exit of PTs. To test this possibility, we examined the subcellular localizations of GFP-tagged PT2/OsPT8. When transiently expressed in rice protoplasts, PT2/PT8-GFP proteins were mainly detected at the PM, as reported previously (Chen et al., 2011). Overexpression of CK2α3 or CK2β3 led to ER retention of PT2/PT8 (Figures 3B and 3C), whereas overexpression of other CK2 components (CK2α2 or CK2β1) had no effect on ER exit of the PTs (Figures 3D and 3E). This suggested a specific regulatory role for CK2α3/β3 in PT trafficking to the PM. Overexpression of CK2α3 or OsCK2β3 alone was sufficient to affect the ER exit of PTs, a result in accordance with the in vivo phosphorylation assay (Figure 2B; Supplemental Figure 4).
Figure 3.
CK2α3 and CK2β3 Affect ER Exit of PT.
(A) Subcellular localization of the PT2-GFP and PT8-GFP fusion proteins. Constructs of 35S-PT2/8-GFP were expressed in rice protoplast cells (control).
(B) to (E) Expression patterns of PT2-GFP and PT8-GFP fusion proteins in cells cotransformed with constructs overexpressing the four CK2 subunits.
(F) to (H) Subcellular localization of PT8-GFP in the epidermal cells of rice roots. Results from 7-d-old transgenic plants transformed with the PT8p-PT8-GFP construct alone (F) or simultaneously with PT8p-PT8-GFP and CK2α3 overexpression (G) or CK2β3 overexpression (H) are shown.
Bars in (A) to (E) = 5 μm; bars in (F) to (H) = 20 μm.
To further examine the effects of CK2α3/β3 on the subcellular localization of PTs in planta, a fully functional PT8-GFP driven by its native promoter (Supplemental Figures 7 and 8) was introgressed into the CK2α3/β3 overexpression transgenic plants (Oxα3/Oxβ3; Supplemental Figure 9). In the wild-type background, PT8-GFP displayed a PM localization pattern in root epidermis cells (Figure 3F) similar to that of PHT1;1 in Arabidopsis (Bayle et al., 2011). By contrast, cells coexpressing CK2α3/β3 showed a fluorescence pattern indicative of PT8-GFP retention in the ER (Figures 3G and 3H). These results indicated that the CK2α3/β3 holoenzyme regulates PT exit from the ER to the PM through its phosphorylation.
Phosphorylation of PT by CK2 Impairs Its Interaction with PHF1
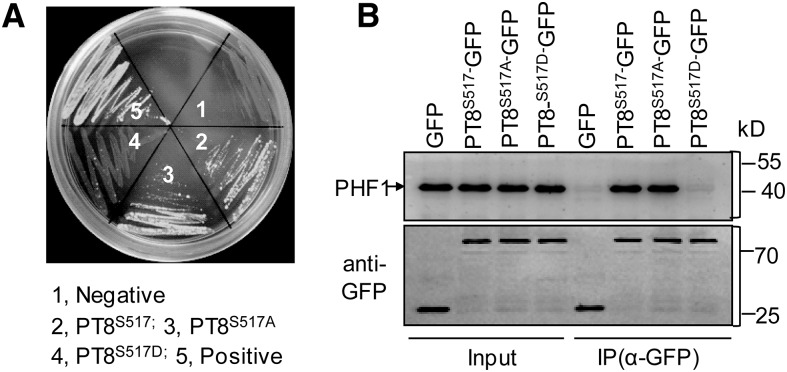
Phosphorylation of the PT C-terminal domain by CK2 resulted in ER retention of PT, mimicking the effect of mutations in the ER-exit cofactor PHF1 (González et al., 2005; Chen et al., 2011). This suggested that phosphorylation of PT by CK2 might affect its interaction with PHF1. To check this, split-ubiquitin yeast two-hybrid analyses were performed between PHF1 and PT8 as well as mutated versions of PT8 in which Ser-517 was replaced by Ala-517 or Asp-517 (designated PT8S517A or PT8S517D), representing nonphosphorylatable or phospho-mimicking forms of PT8, respectively. Notably, PT8S517D (phospho-mimicking) did not interact with PHF1 in yeast cells, whereas an interaction between PHF1 and wild-type PT8 or nonphosphorylatable PT8S517A (Figure 4A) was observed.
Figure 4.
Effect of the Mutation of Ser-517 of PT8 on Interaction with PHF1.
(A) Yeast split-ubiquitination assay among PHF1 and different versions of PT8. Coexpression of PHF1-Cub with NubI or NubG was used as a positive or negative control, respectively.
(B) co-IP of GFP, wild-type/mutated versions of GFP-PT8, and PHF1 from infiltrated N. benthamiana leaves. Precipitates were immunoblotted with GFP (bottom panel) and PHF1 (top panel) antibodies (Supplemental Figure 10). The first lane shows control immunoprecipitations using GFP empty vector.
To further validate our prediction in planta, co-IP assays were performed using solubilized microsomal extracts. They were prepared from N. benthamiana leaves coexpressing PHF1 together with PT8-GFP and its two mutated versions. PHF1 coimmunoprecipitated with PT8-GFP and PT8S517A-GFP but not with PT8S517D-GFP or the GFP control (Figure 4B), suggesting that phosphorylation of PT8 inhibits its interaction with PHF1.
In addition, we found that PHF1 protein abundance increased under the −P condition (0 µM Pi) in a time-dependent manner. This increase was reversed upon Pi resupply (Supplemental Figure 10). The response of PHF1 to Pi starvation was consistent with a function in assisting the exit of more dephosphorylated PT out of the ER under −P. Taken together, these results supported a model in which reduced CK2 and increased PHF1 coordinately enhance the exit from the ER of PTs under Pi starvation conditions.
CK2α3/β3 Influences Pi Uptake Ability in Rice
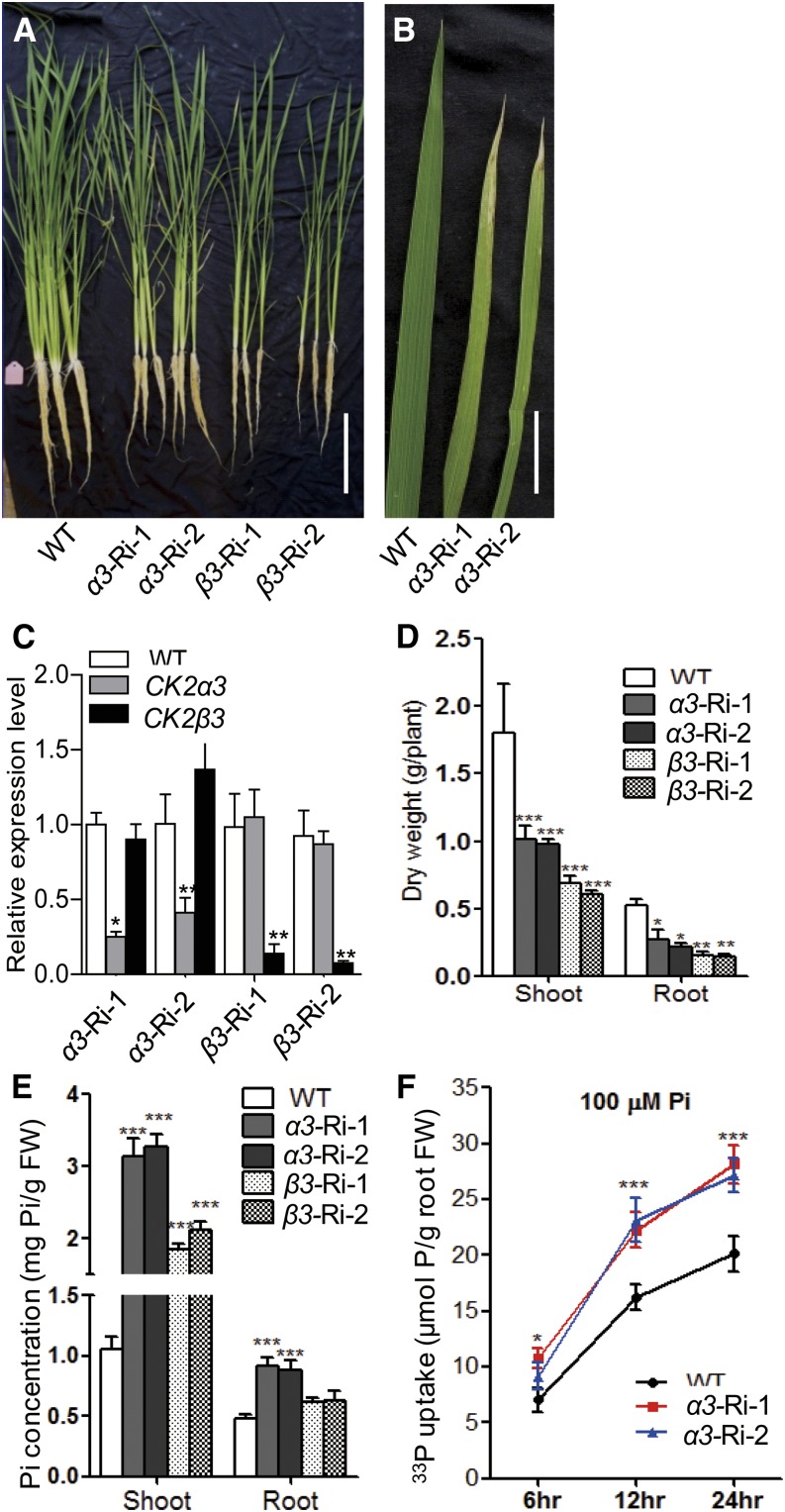
As CK2α3/β3 modulated the ER exit of PTs, it would be expected to also influence Pi uptake. Due to a lack of CK2α3 and CK2β3 T-DNA mutants, we used an RNA interference strategy (Ri) to develop transgenic plants with knockdown of CK2α3 or CK2β3 (Figure 5). Two independent lines of these knockdown plants were grown in +P hydroponic culture (200 μM Pi) for 30 d and used for Pi concentration measurements. As shown in Figures 5A to 5D, knockdown of CK2α3 or CK2β3 (α3-Ri or β3-Ri, respectively) led to necrotic symptoms on older leaf tips (Figure 5B) and retarded growth. α3-Ri and β3-Ri also showed significantly higher Pi concentrations in shoots and roots than that in the wild type (Figure 5E). We further used two independent α3-Ri lines to investigate their Pi uptake ability. Over a 24-h time course, uptake of 33P-labeled Pi by α3-Ri plants was above 30% higher than in the wild type (Figure 5F).
Figure 5.
Phenotypes and Pi Uptake Ability of Transgenic Plants with Knockdown of CK2α3 or CK2β3.
(A) and (B) Phenotypes of the wild type (cv Nipponbare) and two independent knockdown lines of CK2α3 or CK2β3 (α3-Ri-1/2 or β3-Ri-1/2, respectively). The plants were grown for 30 d with 200 μM Pi. Bars = 10 cm (A) and 2 cm (B).
(C) Relative expression of CK2α3/β3 in the transgenic plants.
(D) Dry weight of shoots and roots of the wild-type and transgenic plants shown in (A).
(E) Cellular Pi concentration of shoots and roots of the plants shown in (A).
(F) 33P-uptake ability of α3-Ri and wild-type plants. Twenty-day-old plants were grown in a Pi-deficient solution for 3 d and then supplied with 200 μM 33P-labeled Pi for 6, 12, and 24 h.
Error bars represent sd (n = 3 for [C], [E], and [F]; n = 5 for [D]). Values significantly different from the wild type are indicated (*P < 0.05, **P < 0.01, and ***P < 0.001; Student’s t test). FW, fresh weight.
Analysis of α2 and β1 loss-of-function T-DNA insertional mutants (Supplemental Figures 11 and 12) did not reveal significant differences in Pi concentration from the wild-type control (Supplemental Figures 11E and 12E), indicating that there is specificity among CK2 subunits in the regulation of PT intracellular trafficking (Figures 3A to 3E). By contrast, the α2 and β1 mutants showed retarded growth similar to that of α3-Ri and β3-Ri (knockdown lines of CK2α3/β3) (Figure 5A). In this context, whether the growth retardation observed for α3-Ri or β3-Ri is merely a direct consequence of Pi toxicity or reflects the multifaceted role of these subunits cannot be answered at present and merits further investigation.
Manipulation of PT Phosphorylation Status Affects Its PM Abundance and Pi Uptake Ability
One expectation of the regulatory steps affecting PT exit from the ER is that PT accumulation at the PM would be dependent on Pi availability. We tested this hypothesis by examining PT8 accumulation at the PM of plants under −P for different time spans. We also checked the PT8 level at the PM after Pi refeeding to Pi-starved plants; in these assays, roots and shoots were analyzed separately (Supplemental Figure 13A). PT8 accumulated with increasing time of Pi-starvation stress. Additionally, PT8 abundance at the PM declined rapidly after Pi restoration, but the decrease of PT8 differed between roots and shoots. We found that this tissue-specific effect of Pi refeeding on PT8 accumulation at the PM was associated with a higher phosphorylation level of PT8 in shoots (Supplemental Figure 13B).
These findings, as well as the result showing that Riα3 displayed enhanced Pi-uptake ability (Figure 5F) and the observation that phosphorylation of PT8 inhibited its interaction with PHF1, suggested that the nonphosphorylatable PTs should be more abundant at the PM as a result of enhanced ER exit, leading to an increase in Pi uptake ability, as reported previously for plants overexpressing PHF1 (Chen et al., 2011).
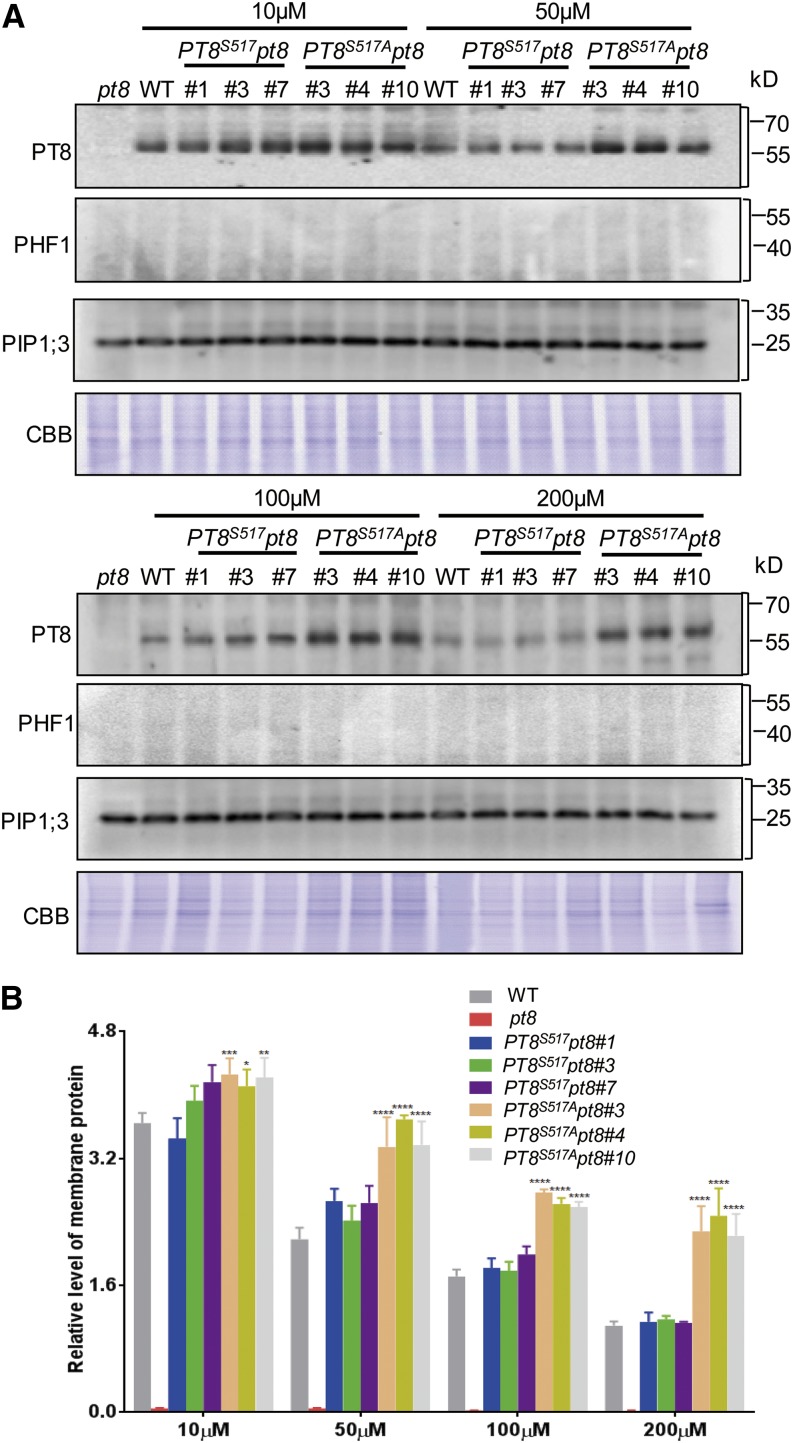
To test this notion, transgenic lines were generated with different versions of PT8. The PT8 antibody was then used to detect protein levels in PM-enriched fractions extracted from roots of the wild type, the pt8 mutant (Supplemental Figure 7), and complemented pt8 plants carrying a single copy of the wild-type PT8 (PT8S517) or the nonphosphorylatable PT8 (PT8S517A), designated PT8S517 pt8 and PT8S517A pt8, respectively (Figure 6; Supplemental Figure 14). Immunoblot analysis with the PHF1-specific antibody (ER marker; Supplemental Figure 10A) and the PM marker antibody PIP1;3 (Liu et al., 2013) demonstrated the purity and specificity of the PM-enriched fraction. These assays showed that under a wide range of Pi regimes (from 10 to 200 μM), the PT8 level in the PM fraction of PT8S517A pt8 plants was significantly higher than those in wild-type and PT8S517 pt8 plants, thus confirming that dephosphorylation of PTs favored their targeting efficiency to the PM.
Figure 6.
Increased Abundance of Nonphosphorylatable PT8 at the PM.
(A) PT8 protein levels in PM-enriched protein fractions in roots. Proteins extracted from seedlings (15 d old) of the wild type, the pt8 mutant, and transgenic pt8 mutant plants expressing PT8 (PT8S517) or the nonphosphorylatable PT8S517A mutant from the PT8 promoter were examined by immunoblotting using anti-PT8 antibody (Supplemental Figure 3) after growth at different Pi concentrations (10, 50, 100, and 200 μM). Immunoblots were also probed with antibodies against rice PM PIP1;3 and with anti-PHF1 antibody as controls for PM enrichment and ER contamination. Coomassie brilliant blue (CBB) staining was used as a loading control.
(B) Quantification of the results shown in (A). Relative PT protein (fold) is the ratio of the respective signal under the given Pi level to the loading control (protein gel blotting bands with anti-PIP1;3 antibody). Asterisks indicate statistically significant changes of PT8 signal: *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.0001.
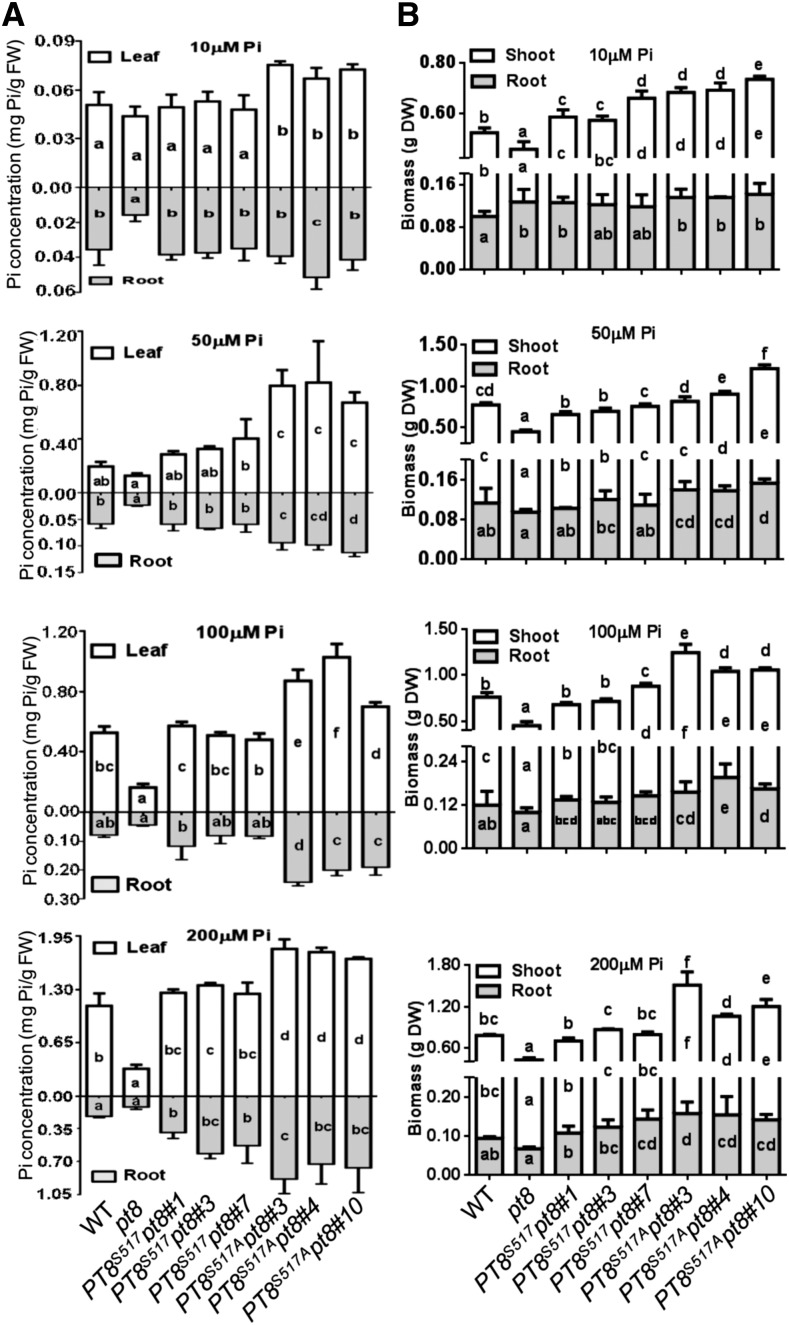
We also examined Pi accumulation and biomass of the genotypes described above grown under different Pi regimes. In general, Pi levels and biomass of shoots and roots in the three PT8S517A pt8 transgenic lines were significantly higher than those of the wild type (Figure 7). Taken together, these studies demonstrated that nonphosphorylatable PT8 (PT8S517A) exhibited higher abundance at the PM than wild-type PT8 (PT8S517) and resulted in enhanced Pi uptake and growth.
Figure 7.
The Nonphosphorylatable PT8 Increases Pi Uptake Ability.
(A) Pi concentrations of the plants shown in Supplemental Figure 14. Wild-type plants, pt8 mutants, and transgenic pt8 mutant plants expressing PT8 (PT8S517) or the nonphosphorylatable PT8S517A mutant from the PT8 promoter were grown for 30 d in solution culture with 10, 50, 100, and 200 μM Pi (n = 3).
(B) Biomass (dry weight [DW]) of the wild-type and related plants shown in Supplemental Figure 14 (n = 5).
Different letters indicate the statistical significance of variations analyzed by Duncan’s multiple range test (P < 0.05): letters inside the bars indicate the significance of variations in each part of individual lines, and letters above the bars indicate the significance of variations for overall plants. Error bars represent sd. FW, fresh weight.
DISCUSSION
PTs play a key role in Pi uptake and translocation. Multiple lines of evidence indicate that PT proteins undergo several posttranslational modifications in addition to transcriptional regulation (Bayle et al., 2011; Huang et al., 2013; Lin et al., 2013; Park et al., 2014). One key regulatory step is ER exit of PTs to the PM, which is regulated positively by PHF1 and negatively by phosphorylation. In this study, we found that the CK2 kinase is responsible for PT phosphorylation and that PT phosphorylation impairs the interaction of PT with PHF1. In addition, we also uncovered the mechanism regulating CK2 activity that is dependent on Pi supply conditions.
Rice CK2α3/β3 Functions as a Holoenzyme to Phosphorylate Pi Transporters
Protein kinase CK2 is a ubiquitous Ser/Thr kinase that is evolutionarily conserved in all eukaryotes (Pinna, 2002; Litchfield, 2003; Meggio and Pinna, 2003; Mulekar and Huq, 2014). CK2 has been studied extensively in mammalian systems, where it can phosphorylate more than 300 known substrates, including transmembrane proteins (Ganley et al., 2001; Litchfield, 2003; Meggio and Pinna, 2003; Fu et al., 2013). Characterization of CK2 in many plant species also demonstrates that this kinase is involved in diverse pathways (Plana et al., 1991; Espunya et al., 1999; Ogiso et al., 2010; Mulekar et al., 2012).
Our results reveal a role of CK2 in plant Pi homeostasis. We found that CK2 binds to high- and low-affinity PTs in rice through the regulatory subunit CK2β3 (Figures 1A to 1C), and CK2-mediated phosphorylation of PT inhibits the ER exit of PT protein, leading to reduced phosphate uptake ability in rice. These findings expand our understanding of CK2 function in plant growth and development and provide evidence for specific protein kinase effects on transmembrane proteins in plants as in animals (Ganley et al., 2001; Scott et al., 2006; Lambert and Hansen, 2011; Fu et al., 2013).
Genes encoding CK2 subunits show a higher degree of redundancy in plants compared with animals (Espunya et al., 2005; Ogiso et al., 2010; Mulekar et al., 2012). Here, we found that the duplication of CK2 subunits was accompanied by functional divergence, as only CK2α3/β3 was active in PT phosphorylation. In accordance, only overexpression of CK2α3/β3 inhibited the ER exit of PTs (Figures 3A to 3E). Consistent with this, rice transgenic lines with knockdown of CK2α3 or CK2β3 exhibited higher Pi concentration (Figure 5E; Supplemental Figures 11E and 12E).
Our results also reinforce the general notion that the noncatalytic CK2β subunit can function as an “anchoring element” for the substrate, rather than being responsible for the activation or inhibition of catalytic activity of the CK2α3 subunit (Pinna, 2002). Indeed, we have shown that the CK2β3 subunit is necessary to allow an interaction between CK2α3 and PT8 in vivo (Figure 1).
A minimum consensus sequence (Ser-Xaa-Xaa-acidic) for the CK2 target site has been defined (Pinna, 2002; Mulekar and Huq, 2014), and such a sequence also exists in the C-terminal domain of rice PT2 and PT8 (Figure 1C). Additionally, substrate specificity of different subunits could be achieved through their localization to distinct subcellular compartments. Here, we found that in rice, CK2α3 and CK2β3 are targeted to the ER compartment (Supplemental Figure 1), where PT exit from the ER was regulated by phosphorylation. Together, these findings indicate that CK2α3/β3 functions as a holoenzyme to phosphorylate PT in its C-terminal domain.
Phosphorylation of PTs Is Dependent on Pi Supply
It is notable that PT phosphorylation by CK2 is responsive to cellular Pi status (Figure 2B). Regulation of CK2 kinase activity has been considered puzzling, since neither of the typical regulatory mechanisms (control by second messenger molecules and switching off of catalytic activity by an interaction partner) for eukaryotic protein kinases held true for the enzyme (Allende and Allende, 1995). Recent crystal structure analysis of CK2 in Drosophila melanogaster supports a “regulation-by-aggregation” model in which CK2 tetramers are arranged as approximately linear aggregates with completely blocked active sites (Schnitzler et al., 2014). By contrast, according to our results, Pi deficiency in rice causes downregulation of the phosphorylation and protein level of CK2β3 (Figure 2E; Supplemental Figure 5), excluding the possibility of an aggregation mechanism analogous to that in Drosophila CK2.
In animals, the CK2β subunit is regulated by protein degradation through a YRQALDMILD “cyclin destruction” consensus sequence (Allende and Allende, 1995). In addition, phospho-mimicking in the human CK2β subunit enhances its stability (Zhang et al., 2002). In line with this result, we found that rice CK2β3 contains a related “destruction” box (YDYALDLILD) in the analogous position (amino acids 139 to 148), and abundance of CK2β3 was reduced when it was dephosphorylated in Pi-starved plants. Our findings support that phosphorylation of the CK2 regulatory subunit may result in enhanced activity of the holoenzyme, in agreement with a previous finding that the CK2β subunit is involved in the assembly of holoenzyme as a possible checkpoint for enzyme activity (Allende and Allende, 1998). Further investigation is required to identify the detailed mechanism involved in modulating the phosphorylation and degradation of CK2β3 under Pi starvation.
In addition to its crucial role in the regulation of CK2 activity, Pi supply also affects the accumulation of PHO2 and NLA (Lin et al., 2013; Park et al., 2014), which are involved in PT ubiquitination (and subsequent degradation). Thus, CK2, PHO2, and NLA generate a negative feedback loop for the regulation of PT activity according to plant Pi status. Such a negative loop is paralleled by a similar feedback mechanism for transcriptional control generated between the PHR1 master regulator of Pi starvation responses and several SPX proteins (Lv et al., 2014; Puga et al., 2014; Wang et al., 2014).
A CK2-Integrated Model for PT ER Exit
In this study, we found that phosphorylation of PT prevents its interaction with PHF1 (Figure 4), an integral membrane protein located in the ER that facilitates the ER trafficking of PHT1 family members. Together with the observation that PHF1 functions upstream of COPII vesicle formation (Bayle et al., 2011), our data make a critical contribution to our knowledge of the regulation of PT trafficking from the ER by identifying the kinase involved in the phosphorylation process, CK2, and uncovering its functional connection to the ER-exit facilitation function of PHF1.
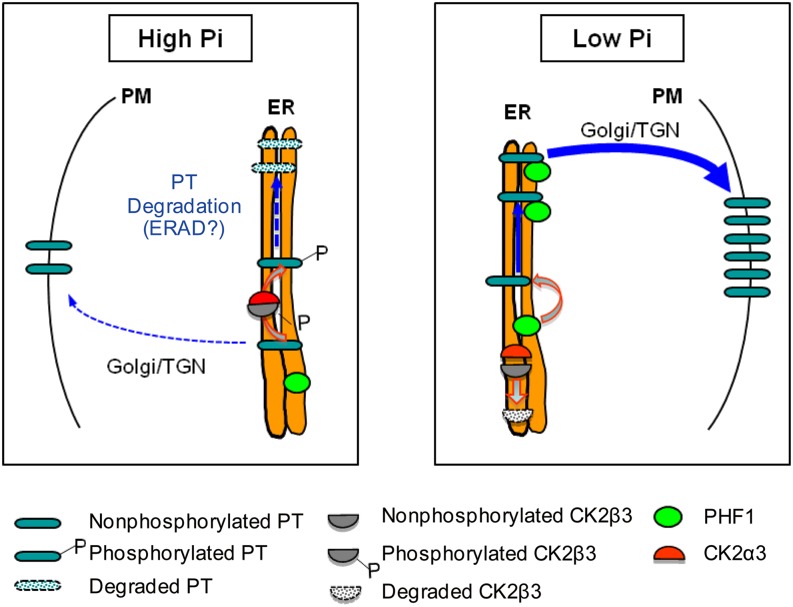
A model summarizing the role of CK2 phosphorylation is provided in Figure 8. It shows that low-Pi stress leads to a reduction of the phosphorylation and abundance of CK2β3, which limits CK2α3 access to PTs for phosphorylation. Therefore, the nonphosphorylated PTs can interact with PHF1 for efficient trafficking from the ER to the PM. By contrast, under Pi-sufficient conditions, phosphorylated CK2β3 is stable and, together with CK2α3, forms an active holoenzyme that phosphorylates PTs to inhibit the interaction between PTs and PHF1, resulting in ER retention of PTs.
Figure 8.
Model for CK2α3β3 Regulation of the ER Exit of PTs.
Under high Pi, phosphorylated CK2β3 interacts with CK2α3. This forms an active holoenzyme that phosphorylates PTs, thereby inhibiting the interaction between PTs and PHF1, resulting in ER retention of PTs. Under low Pi, CK2β3 is dephosphorylated and degraded. PHF1 interacts with nonphosphorylated PT, facilitating trafficking of PT through the ER and promoting its subsequent accumulation at the PM. The thickness of the arrows represents the intensity of the effects under different conditions. The pathway indicated by the dashed lines requires more evidence. See text for more details. ERAD, ER-associated protein degradation; TGN, trans-Golgi network.
The ER is the site for the maturation of secretory and membrane proteins, and improperly folded or excess proteins are frequently retained in the ER and subjected to ER-associated protein degradation (Arvan et al., 2002; Hegde and Ploegh, 2010). Recently, interplay between protein phosphorylation and ubiquitination in ER-associated protein degradation was demonstrated (Wang et al., 2012). Accordingly, it is possible that CK2-mediated phosphorylation of PT under high Pi may lead to ER retention and subsequent ER-associated degradation of PTs.
In summary, there is a regulatory step at the ER whereby PHF1 enables the exit of PTs and phosphorylation by CK2 inhibits it by impeding PT-PHF1 interaction in addition to transcriptional control increasing PT levels in response to low Pi (Karthikeyan et al., 2002; Mudge et al., 2002). The activity of this kinase is affected by Pi supply, because reduced phosphorylation of CK2β3 results in its decreased stability and interaction capacity with CK2α3. Thus, superimposing both regulatory mechanisms, transcriptional and posttranslational, should allow a sharper response to changes in Pi availability for better maintenance of P homeostasis in plants. Conversely, we show that relieving phosphorylation control by expressing a nonphosphorylatable version of PT8 enhances Pi acquisition efficiency and plant growth, providing a possible strategy for improving Pi acquisition efficiency in crops, especially under P-limited conditions.
METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa cv Nipponbare) as the wild type and transgenic plants were grown hydroponically in full-strength Kimura nutrient solution with high or low Pi in a greenhouse with a 12-h-day (30°C)/12-h-night (22°C) photoperiod, ∼200 μmol m−2 s−1 photon density, and ∼60% humidity. For the +P and −P conditions, the concentration of NaH2PO4 was adjusted to 200 or 0 μM, respectively, unless specified otherwise. Nicotiana benthamiana plants were cultivated in growth chambers as described before (Lv et al., 2014).
Rice Root cDNA Library Construction and Split-Ubiquitin Membrane Yeast Two-Hybrid Screening System
Total RNA was prepared using the RNeasy Plant Mini kit (Qiagen) from the roots of 14-d-old seedlings grown in a normal hydroponic solution. Isolated RNA was treated with RNase-free DNase (Qiagen) for DUALhunter library construction (Dualsystems Biotech). First-strand cDNA generated by reverse transcription was normalized and confirmed by quantitative PCR using two marker genes (ACTIN and GAPDH). Then, the normalized first-strand cDNA was size-selected and split into two size pools to optimize the representation of long and short fragments. The second-strand cDNA was generated separately for both size pools and directionally integrated into prey vector pPR3-N between two variable Sfi sites. Ultimately, a normalized rice root cDNA library with 2.9 × 106 independent clones was obtained. To minimize background arising from large-scale screening of the library, the cDNA library was transferred into the integrated yeast cell lines mentioned above, and selection was performed on leucine-tryptophan-histidine-adenine dropout plates (Clontech) with 7.5 mM 3-aminotriazole (Sigma-Aldrich), a competitive inhibitor of the imidazoleglycerolphosphate dehydratase involved in histidine biosynthesis. The screening was done according to the manufacturer’s protocol (DUALhunter starter kit; Dualsystems Biotech).
Yeast Split-Ubiquitination Assay
Split-ubiquitin yeast two-hybrid assays were performed following the instructions provided with the DUALmembrane pairwise interaction kit (Dualsystems Biotech). The coding regions of PT2/PT8 and CK2α2/α3/β1/β3 were cloned in frame in the vectors pBT3-STE and pPR3-N, respectively. The coding regions of PHF1 and PT8/PT8S51/PT8S517A/PT8S517D were cloned in frame into the vectors pBT3-N and pPR3-STE, respectively. Yeast strain NMY51 cells were cotransformed with these two constructs and plated onto synthetic medium lacking Leu and Trp. The protein-protein interactions were assessed by the growth of yeast colonies on synthetic medium lacking Leu, Trp, His, and Ade. The bait APP-Cub expressing the type I integral membrane protein APP (for amyloid A4 precursor protein) was used as the control that could interact with NubI but not with NubG.
Membrane Protein Isolation
Membrane proteins were isolated from rice seedlings (the tissue used is indicated in the respective experiments), infiltrated N. benthamiana leaves, or harvested protoplasts as follows. Tissues or suspension cells were first mechanically disrupted and suspended in homogenization buffer containing 50 mM Tris, 500 mM sucrose, 10% glycerol, 20 mM EDTA-Na2, 20 mM EGTA, 50 mM NaF, 5 mM β-glycerophosphate, 1 mM phenanthroline, 0.6% polyvinylpyrrolidone (average molecular weight from 30,000 to 40,000), and 10 mM ascorbic acid, with freshly added 1 mM leupeptin, 5 mM DTT, and 1 mM sodium orthovanadate. The resulting homogenate was filtered through a nylon cloth (100-μm diameter) and then centrifuged at 26,000g for 25 min. The resulting supernatant was filtered through two successive filters, 63- and 34-μm diameter. The microsomal pellet was obtained from the supernatant by centrifugation at 84,000g for 25 min. PM proteins were isolated from total microsomal proteins by two-phase partitioning as described previously (Szabó-Nagy et al., 1989). Total microsomal protein and PM protein-enriched fractions were loaded for immunoblot analysis on general gels or by PhosTag SDS-PAGE.
co-IP Assays
Microsomal fractions were prepared from coinfiltrated N. benthamiana leaves as described above. Samples were solubilized by dilution with 5 volumes of PBS containing 1.5% Triton X-100 and incubated for 1 h at 4°C. This was followed by centrifugation at 20,000g for 1 h and incubation with anti-GFP magnetic beads (Chromotek) for 2 h at 4°C. Beads were washed five times with washing buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.2% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail [Roche]). Following SDS-PAGE and transfer to nitrocellulose, GFP, PT8-CT-GFP, PT2-GFP, PT8-GFP, PT8S517A-GFP, and PT8S517D-GFP proteins and CK2 subunits were detected by immunoblotting with tag-specific antibodies (Sigma-Aldrich). PHF1 was detected using PHF1-specific antibody (see below).
Yeast Three-Hybrid Assays
The cDNA fragments encoding PT2/8-CT and CK2β3 were inserted into the pBridge vector (Clontech) to generate fusions with the GAL4 DNA-binding domain and Met promoter, respectively. CK2α2/α3 was inserted into the pGADT7 vector (Clontech) to generate pGAD-α2/α3 to serve as prey in yeast three-hybrid assays. The resulting constructs were cotransformed into yeast strain AH109 and selected on dropout medium lacking Leu, Met, and Trp or Leu, Met, Trp, and His.
Recombinant Protein Expression
Fragments encoding mature CK2α3/β3 and PT8-CT as well as alleles of PT8-CT were cloned into expression vector pGEX-4T-1 (GE Healthcare). The recombinant vectors were identified by sequencing. Recombinant plasmids were transformed into Escherichia coli TransB (DE3) (Transgen) and then treated with 0.05 mM isopropyl-β-d-thiogalactoside for 12 h under 22°C to induce the expression of fusion proteins. Expressed proteins were purified using GST-affinity chromatography on immobilized glutathione followed by competitive elution with excess reduced glutathione according to the manufacturer’s instructions (GE Healthcare).
In Vitro Phosphorylation Assays
In vitro kinase assays in solution were performed essentially as described previously (Meng et al., 2012) with a few modifications. Kinase subunits and substrate proteins were mixed with 1× kinase buffer (100 mM Tris-HCl, pH 8.0, 5 mM DTT, 5 mM EGTA, and 5 mM MgCl2; New England Biolabs) and 1× ATP solution (100 μM ATP and 1 μCi of [γ-32P]ATP; Perkin-Elmer) in a total volume of 50 μL. Equal amounts of GST-tagged PT8-CT and its Ser-to-Ala mutant proteins were incubated with CK2α3 in the presence of [γ-32P]ATP. The reactions were incubated at 30°C for 30 min and then stopped by adding 5× loading buffer and boiling for 5 min. Products were separated by electrophoresis through 12% acrylamide gels, and the gels were stained, dried, and then visualized by exposure to x-ray film.
In Vivo Phosphorylation Assays
Rice seedlings and CK2 overexpression/knockdown transgenic plants were grown for 7 d in full-strength Kimura nutrient solution with high or low Pi, and then the roots of these seedlings were harvested for microsomal fraction extraction. Membrane fractions were subjected to λ-PPase treatment as described previously (Michniewicz et al., 2007) with a few modifications. Treatment was performed with λ-PPase (Sigma-Aldrich) in a total volume of 50 μL, and samples were incubated at 30°C for 30 min. The reactions were stopped by adding 5× SDS loading buffer (Sangon) and boiled. For the in vivo phosphorylation assays performed in protoplasts, the membrane proteins were extracted from protoplasts developed from wild-type callus and callus harboring overexpressed CK2α3 or CK2β3, and simultaneously overexpressed CK2α3 and CK2β3, in the absence and presence of DRB inhibitor. Cells were treated with 50 μM DRB for 12 h. For the control, an equal volume of DMSO, which was the solvent used to prepare the inhibitor stock solutions, was added. Samples were separated on 10% PhosTag acrylamide gels and probed with PT8-specific antibody (1:2000). The second antibody, goat anti-rabbit IgG peroxidase antibody (Sigma-Aldrich), was used at 1:10,000. Detection was performed with the enhanced chemiluminescence reagent (Pierce).
Pull-Down Assays
β3-FLAG was synthesized by N. benthamiana leaves infiltrated with Agrobacterium tumefaciens. For in vitro binding, 20 μL of total N. benthamiana protein was added to 600 μL of binding buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 2 mM Na3VO4, 25 mM β-glycerophosphate, 10 mM NaF, 0.05 to 0.1% Tween 20, 1× Roche protease inhibitor, and 1 mM phenylmethylsulfonyl fluoride), followed by 50 μL of glutathione-agarose beads with bound GST-α3 or its alleles, and was incubated at 4°C for 3 h. The beads were washed with binding buffer three times. Bound proteins were eluted with 5× SDS loading buffer and resolved by 12% SDS-PAGE. Individual bands were detected by immunoblotting with tag-specific antibodies. Commercial antibodies were purchased from Sigma-Aldrich (anti-FLAG M2; 1:3000 protein gel blotting) and GE Healthcare (anti-GST; 1:5000 protein gel blotting).
Subcellular Localization of PT2/8 Proteins in Rice Protoplast Cells
Isolation of rice protoplasts and protoplast transient transformation were conducted as described previously (Chen et al., 2011). Observations were made on a Zeiss Axiovert LSM 710 laser scanning microscope. Protoplasts were observed under a 63× objective. For excitation of fluorescence proteins and mCherry, the following lines of an argon ion laser were used: 488 nm for GFP and 543 nm for mCherry. Fluorescence was detected at 493 to 542 nm for GFP and 578 to 625 nm for mCherry.
Construction of Vectors and Generation of Transgenic Plants
The nonphosphorylatable (S517A) mutation in PT8 and wild-type PT8 were generated with primers using the PT8S517A-pPR3-STE plasmid as template; released fragments were inserted into pCAMBIA1300 vector to generate pCAMBIA1300-PT8/PT8S517A. Plasmids bearing PT8p-OsPT8/PT8S517A, in which the genes were under the control of the native promoter, were derived from pCAMBIA1300-PT8/PT8S517A by replacing the cauliflower mosaic virus 35S promoter with 2679 bp of genome sequence upstream of the ATG of PT8. Plasmids encoding PT8-GFP under the control of its native promoter were similarly derived from pCAMBIA1300-PT8-GFP by replacing the cauliflower mosaic virus 35S promoter with 2679 bp of genome sequence upstream of the ATG of PT8. For the RNA interference construct, fragments 179 to 430 bp for CK2α3 and 517 to 763 bp for CK2β3 were cloned in both orientations in pCAMBIA35S-1300 vector and separated by the second intron of NIR1 of maize (Zea mays) to form a hairpin structure. The binary vectors and the 35S promoter-driven CK2α3/β3 vectors (see above) were introduced into Agrobacterium strain EHA105 and transformed into wild-type rice (cv Nipponbare) according to the method described previously (Chen et al., 2011).
RNA Isolation, RT-PCR, and Quantitative RT-PCR Analysis
Total RNA from rice samples was isolated using TRIzol reagent (Invitrogen) followed by treatment with DNase I (Qiagen) before quantitative RT-PCR (qRT-PCR) to eliminate genomic DNA contamination. cDNA was synthesized from 5 μg of total RNA using SuperScript III (Invitrogen). qRT-PCR was performed using the SYBR Green I Master (Roche) on the LightCycler 480 Real-Time PCR system (Roche) according to the manufacturer’s instructions. Relative expression levels were normalized to that of the housekeeping gene ACTIN. The primers used for RT-PCR and qRT-PCR are listed in Supplemental Table 1.
Determination of Cellular Pi Concentration and Pi Uptake Ability
Cellular Pi concentration was determined as described previously (Chen et al., 2011). 33P uptake analysis was conducted as described previously (Wu et al., 2011).
Development of PHF1 and PT8 Polyclonal Antibodies
Polyclonal rabbit PHF1 antibody was raised against a C-terminal fragment of rice PHF1 corresponding to amino acid residues 375 to 387 (C-KESPPVPEDQNPW-COOH) and affinity purified by Abmart. For an antibody against rice PT8, the synthetic peptide C-VLQVEIQEEQDKLEQMVT-COOH (positions 264 to 281 of rice PT8) was used to immunize rabbits. The obtained antiserum was purified through a peptide affinity column before use.
Accession Numbers
The Michigan State University Rice Genome Annotation Project Database accession numbers for the genes studied in this work are Os07g0187700 (PHF1), Os03g0150800 (PT2), Os10g0444700 (PT8), Os07g0114400 (CK2α2), Os03g0207300 (CK2α3), Os10g0564900 (CK2β1), and Os07g0495100 (CK2β3). NCBI accession numbers for the proteins are as follows: PHF1, NP_001059077; PT2, NP_001048979; PT8, NP_001064708; CK2α2, NP_001058752; CK2α3, NP_001049325; CK2β1, NP_001065415; and CK2β3, NP_001059693.
Supplemental Data
Supplemental Figure 1. Subcellular Localization of CK2α3/β3.
Supplemental Figure 2. CK2β3 Is Unable to Phosphorylate the C-Terminal Ser-517 Residue of PT8.
Supplemental Figure 3. Specificity of the Rice PT8 Polyclonal Antibody.
Supplemental Figure 4. Effect of CK2β3 Overexpression on PT8 Phosphorylation in Vivo.
Supplemental Figure 5. Immunoblotting Analysis for CK2α3/β3 Protein Level in Plants Grown under Pi-Sufficient and Pi-Starvation Conditions.
Supplemental Figure 6. Cellular Pi Sensitivity of the Interaction between CK2β3 and CK2α3.
Supplemental Figure 7. Isolation of pt8 Mutants in Nipponbare Background.
Supplemental Figure 8. Functional PT8-GFP Rescued pt8 Mutant.
Supplemental Figure 9. Phenotype and Pi Concentration of Transgenic Plants Overexpressing CK2α3/β3.
Supplemental Figure 10. Specificity of the PHF1 Polyclonal Antibody.
Supplemental Figure 11. Phenotype and Pi Content Analysis of α2 T-DNA Insertional Mutant.
Supplemental Figure 12. Phenotype and Pi Analysis of β1 T-DNA Insertional Mutant.
Supplemental Figure 13. PT8 Protein Accumulation and Phosphorylation Levels Differ in Roots and Shoots.
Supplemental Figure 14. DNA Gel Blot Analysis and Plant Phenotype of Independent Transgenic Lines of the pt8 Mutant Transformed with PT8S517 and PT8S517A Expressing Constructs.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Mingxiu Chen for helping develop the transgenic plants. This work was supported by the National Basic Research and Development Program of China (Grant 2011CB100303), the Ministry of Science and Technology of China (Grant 2012AA10A302), the Ministry of Agriculture of China (Grants 2011ZX08001-005, 2013ZX08001-005, and 2014ZX08001005), the Ministry of Education and Bureau of Foreign Experts of China (Grant B14027), the National Natural Science Foundation of China (Grant 31322048), and a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grant 22119002 to J.F.M.).
AUTHOR CONTRIBUTIONS
P.W. conceived the project. P.W., S.Q.Z., and J.P.-A. designed the experiments. J.Y.C. developed the assay and performed the experiments together with Y.F.W., F.W., J.Y., and M.X.G. Z.C.W., Y.L., C.Y.L., and Y.Y.L. analyzed the nutrients. N.Y. and J.F.M. developed the OsPT8 antibody. P.W. wrote the article with improvement from J.P.-A., S.Q.Z., L.N., K.K.Y., and J.F.M.
Glossary
- PT
Pi transporter
- PM
plasma membrane
- ER
endoplasmic reticulum
- co-IP
coimmunoprecipitation
- λ-PPase
λ-phosphatase
- DRB
5,6-dichloro-1-β-d-ribofuranosylbenzimidazole
- qRT-PCR
quantitative RT-PCR
Footnotes
Articles can be viewed online without a subscription.
References
- Ackermann K., Neidhart T., Gerber J., Waxmann A., Pyerin W. (2005). The catalytic subunit alpha′ gene of human protein kinase CK2 (CSNK2A2): Genomic organization, promoter identification and determination of Ets1 as a key regulator. Mol. Cell. Biochem. 274: 91–101. [DOI] [PubMed] [Google Scholar]
- Ai P., Sun S., Zhao J., Fan X., Xin W., Guo Q., Yu L., Shen Q., Wu P., Miller A.J., Xu G. (2009). Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 57: 798–809. [DOI] [PubMed] [Google Scholar]
- Allende C.C., Allende J.E. (1998). Promiscuous subunit interactions: A possible mechanism for the regulation of protein kinase CK2. J. Cell. Biochem. Suppl. 30-31: 129–136. [PubMed] [Google Scholar]
- Allende J.E., Allende C.C. (1995). Protein kinases. 4. Protein kinase CK2: An enzyme with multiple substrates and a puzzling regulation. FASEB J. 9: 313–323. [DOI] [PubMed] [Google Scholar]
- Arvan P., Zhao X., Ramos-Castaneda J., Chang A. (2002). Secretory pathway quality control operating in Golgi, plasmalemmal, and endosomal systems. Traffic 3: 771–780. [DOI] [PubMed] [Google Scholar]
- Bayle V., Arrighi J.F., Creff A., Nespoulous C., Vialaret J., Rossignol M., Gonzalez E., Paz-Ares J., Nussaume L. (2011). Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23: 1523–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby A.C., Litchfield D.W. (2005). The multiple personalities of the regulatory subunit of protein kinase CK2: CK2 dependent and CK2 independent roles reveal a secret identity for CK2beta. Int. J. Biol. Sci. 1: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Liu Y., Ni J., Wang Y., Bai Y., Shi J., Gan J., Wu Z., Wu P. (2011). OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiol. 157: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., et al. (2009). A rice kinase-protein interaction map. Plant Physiol. 149: 1478–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espunya M.C., Combettes B., Dot J., Chaubet-Gigot N., Martínez M.C. (1999). Cell-cycle modulation of CK2 activity in tobacco BY-2 cells. Plant J. 19: 655–666. [DOI] [PubMed] [Google Scholar]
- Espunya M.C., López-Giráldez T., Hernan I., Carballo M., Martínez M.C. (2005). Differential expression of genes encoding protein kinase CK2 subunits in the plant cell cycle. J. Exp. Bot. 56: 3183–3192. [DOI] [PubMed] [Google Scholar]
- Fu Y., Westenbroek R.E., Scheuer T., Catterall W.A. (2013). Phosphorylation sites required for regulation of cardiac calcium channels in the fight-or-flight response. Proc. Natl. Acad. Sci. USA 110: 19621–19626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley I.G., Walker S.J., Manifava M., Li D., Brown H.A., Ktistakis N.T. (2001). Interaction of phospholipase D1 with a casein-kinase-2-like serine kinase. Biochem. J. 354: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100. [DOI] [PubMed] [Google Scholar]
- González E., Solano R., Rubio V., Leyva A., Paz-Ares J. (2005). PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17: 3500–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde R.S., Ploegh H.L. (2010). Quality and quantity control at the endoplasmic reticulum. Curr. Opin. Cell Biol. 22: 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hem S., Rofidal V., Sommerer N., Rossignol M. (2007). Novel subsets of the Arabidopsis plasmalemma phosphoproteome identify phosphorylation sites in secondary active transporters. Biochem. Biophys. Res. Commun. 363: 375–380. [DOI] [PubMed] [Google Scholar]
- Huang T.K., et al. (2013). Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25: 4044–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Ren H., Gu M., Zhao J., Sun S., Zhang X., Chen J., Wu P., Xu G. (2011). The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol. 156: 1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan A.S., Varadarajan D.K., Mukatira U.T., D’Urzo M.P., Damsz B., Raghothama K.G. (2002). Regulated expression of Arabidopsis phosphate transporters. Plant Physiol. 130: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert I.H., Hansen D.B. (2011). Regulation of taurine transport systems by protein kinase CK2 in mammalian cells. Cell. Physiol. Biochem. 28: 1099–1110. [DOI] [PubMed] [Google Scholar]
- Lin W.Y., Huang T.K., Chiou T.J. (2013). Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 25: 4061–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchfield D.W. (2003). Protein kinase CK2: Structure, regulation and role in cellular decisions of life and death. Biochem. J. 369: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Shang-Guan K., Zhang B., Liu X., Yan M., Zhang L., Shi Y., Zhang M., Qian Q., Li J., Zhou Y. (2013). Brittle Culm1, a COBRA-like protein, functions in cellulose assembly through binding cellulose microfibrils. PLoS Genet. 9: e1003704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q., Zhong Y., Wang Y., Wang Z., Zhang L., Shi J., Wu Z., Liu Y., Mao C., Yi K., Wu P. (2014). SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26: 1586–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meggio F., Pinna L.A. (2003). One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17: 349–368. [DOI] [PubMed] [Google Scholar]
- Meng X., Wang H., He Y., Liu Y., Walker J.C., Torii K.U., Zhang S. (2012). A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24: 4948–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056. [DOI] [PubMed] [Google Scholar]
- Misson J., Thibaud M.C., Bechtold N., Raghothama K., Nussaume L. (2004). Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol. Biol. 55: 727–741. [DOI] [PubMed] [Google Scholar]
- Mudge S.R., Rae A.L., Diatloff E., Smith F.W. (2002). Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 31: 341–353. [DOI] [PubMed] [Google Scholar]
- Mulekar J.J., Huq E. (2014). Expanding roles of protein kinase CK2 in regulating plant growth and development. J. Exp. Bot. 65: 2883–2893. [DOI] [PubMed] [Google Scholar]
- Mulekar J.J., Bu Q., Chen F., Huq E. (2012). Casein kinase II α subunits affect multiple developmental and stress-responsive pathways in Arabidopsis. Plant J. 69: 343–354. [DOI] [PubMed] [Google Scholar]
- Niefind K., Guerra B., Ermakowa I., Issinger O.G. (2001). Crystal structure of human protein kinase CK2: Insights into basic properties of the CK2 holoenzyme. EMBO J. 20: 5320–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nühse T.S., Stensballe A., Jensen O.N., Peck S.C. (2004). Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16: 2394–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussaume L., Kanno S., Javot H., Marin E., Pochon N., Ayadi A., Nakanishi T.M., Thibaud M.C. (2011). Phosphate import in plants: Focus on the PHT1 transporters. Front. Plant Sci. 2: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiso E., Takahashi Y., Sasaki T., Yano M., Izawa T. (2010). The role of casein kinase II in flowering time regulation has diversified during evolution. Plant Physiol. 152: 808–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B.S., Seo J.S., Chua N.H. (2014). NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26: 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinna L.A. (2002). Protein kinase CK2: A challenge to canons. J. Cell Sci. 115: 3873–3878. [DOI] [PubMed] [Google Scholar]
- Plana M., Itarte E., Eritja R., Goday A., Pagès M., Martínez M.C. (1991). Phosphorylation of maize RAB-17 protein by casein kinase 2. J. Biol. Chem. 266: 22510–22514. [PubMed] [Google Scholar]
- Puga M.I., et al. (2014). SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 14947–14952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama K.G. (1999). Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 665–693. [DOI] [PubMed] [Google Scholar]
- Rohila J.S., et al. (2006). Protein-protein interactions of tandem affinity purification-tagged protein kinases in rice. Plant J. 46: 1–13. [DOI] [PubMed] [Google Scholar]
- Salinas P., Fuentes D., Vidal E., Jordana X., Echeverria M., Holuigue L. (2006). An extensive survey of CK2 alpha and beta subunits in Arabidopsis: Multiple isoforms exhibit differential subcellular localization. Plant Cell Physiol. 47: 1295–1308. [DOI] [PubMed] [Google Scholar]
- Sarno S., Ghisellini P., Pinna L.A. (2002). Unique activation mechanism of protein kinase CK2. The N-terminal segment is essential for constitutive activity of the catalytic subunit but not of the holoenzyme. J. Biol. Chem. 277: 22509–22514. [DOI] [PubMed] [Google Scholar]
- Schnitzler A., Olsen B.B., Issinger O.G., Niefind K. (2014). The protein kinase CK2(Andante) holoenzyme structure supports proposed models of autoregulation and trans-autophosphorylation. J. Mol. Biol. 426: 1871–1882. [DOI] [PubMed] [Google Scholar]
- Scott G.K., Fei H., Thomas L., Medigeshi G.R., Thomas G. (2006). A PACS-1, GGA3 and CK2 complex regulates CI-MPR trafficking. EMBO J. 25: 4423–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Shin H.S., Dewbre G.R., Harrison M.J. (2004). Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 39: 629–642. [DOI] [PubMed] [Google Scholar]
- Szabó-Nagy A., Bérczi A., Erdei L. (1989). Plasma membrane purification from roots of sunflower by phase partitioning. Acta Biochim. Biophys. Hung. 24: 203–211. [PubMed] [Google Scholar]
- Veneklaas E.J., Lambers H., Bragg J., Finnegan P.M., Lovelock C.E., Plaxton W.C., Price C.A., Scheible W.R., Shane M.W., White P.J., Raven J.A. (2012). Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol. 195: 306–320. [DOI] [PubMed] [Google Scholar]
- Wang Y., Guan S., Acharya P., Liu Y., Thirumaran R.K., Brandman R., Schuetz E.G., Burlingame A.L., Correia M.A. (2012). Multisite phosphorylation of human liver cytochrome P450 3A4 enhances its gp78- and CHIP-mediated ubiquitination: A pivotal role of its Ser-478 residue in the gp78-catalyzed reaction. Mol. Cell. Proteomics 11: 010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., et al. (2014). Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. USA 111: 14953–14958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirkner U., Voss H., Ansorge W., Pyerin W. (1998). Genomic organization and promoter identification of the human protein kinase CK2 catalytic subunit alpha (CSNK2A1). Genomics 48: 71–78. [DOI] [PubMed] [Google Scholar]
- Wu Z., Ren H., McGrath S.P., Wu P., Zhao F.J. (2011). Investigating the contribution of the phosphate transport pathway to arsenic accumulation in rice. Plant Physiol. 157: 498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Vilk G., Canton D.A., Litchfield D.W. (2002). Phosphorylation regulates the stability of the regulatory CK2beta subunit. Oncogene 21: 3754–3764. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Liao H., Lucas W.J. (2014). Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J. Integr. Plant Biol. 56: 192–220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.