Expression of circadian clock genes in rice, individually responding to given field conditions, coordinately encode punctual internal time irrespective of weather, daylength, or developmental stage.
Abstract
Plant circadian clocks that oscillate autonomously with a roughly 24-h period are entrained by fluctuating light and temperature and globally regulate downstream genes in the field. However, it remains unknown how punctual internal time produced by the circadian clock in the field is and how it is affected by environmental fluctuations due to weather or daylength. Using hundreds of samples of field-grown rice (Oryza sativa) leaves, we developed a statistical model for the expression of circadian clock-related genes integrating diurnally entrained circadian clock with phase setting by light, both responses to light and temperature gated by the circadian clock. We show that expression of individual genes was strongly affected by temperature. However, internal time estimated from expression of multiple genes, which may reflect transcriptional regulation of downstream genes, is punctual to 22 min and not affected by weather, daylength, or plant developmental age in the field. We also revealed perturbed progression of internal time under controlled environment or in a mutant of the circadian clock gene GIGANTEA. Thus, we demonstrated that the circadian clock is a regulatory network of multiple genes that retains accurate physical time of day by integrating the perturbations on individual genes under fluctuating environments in the field.
INTRODUCTION
Plants gain energy from sunlight via photosynthesis in the daytime and risk injury or death from photoinhibition or drought. Considering the large variations in intensity and limited hours of sunlight as the sole source of energy, preparation for diurnal cycles of solar radiation is critical. Circadian clocks allow plants to anticipate daily changes in the environment that affect their growth and fitness (Dodd et al., 2005; Yerushalmi and Green, 2009). Plants also adjust the timing of flowering and fruit production based on photoperiodism, a function recognizing daylength. The circadian clock contributes to the photoperiodic control of flowering by producing internal time, which may not be directly affected by a rapidly changing external environment. It has been implied that gating, which is internal time-dependent sensitivity to light signals, can confer diurnal regulation of the expression of genes involved in photoperiodic flowering. For example, two flowering time genes in rice (Oryza sativa), a short-day plant, Early heading date 1 (Ehd1) and Grain-number, plant height, and heading-date 7, are acutely induced by light signals, with the gating conferred by the circadian clock, of which the rice ortholog of the Arabidopsis thaliana gene GIGANTEA (GI) is a component (Itoh et al., 2010; Izawa et al., 2011; Itoh and Izawa, 2013). GI has been shown to be a key controller of the global transcriptome in rice under fluctuating field conditions. In Arabidopsis, flowering is induced under long-day conditions via CONSTANS (CO), which is transcriptionally activated by GI (Suárez-López et al., 2001). Expression of a rice ortholog of CO, Heading date 1 (Hd1), is also regulated by GI, but the Hd1 regulation of flowering is bifunctional; Hd1 represses flowering under short-day conditions and induces it under long-day conditions (Yano et al., 2000; Izawa et al., 2011). Both Ehd1 and Hd1 regulate rice florigen genes, Hd3a and Rice FT-like 1 (Itoh and Izawa, 2013). In addition, it has been shown that Hd3a expression depends on the subtle differences in daylength of <30 min under the laboratory conditions (Itoh et al., 2010). Thus, daylength measurement might depend on punctuality (small error) and precision (small variance of error) of the circadian clock relative to physical time. Therefore, it is important to know the punctuality and precision of the circadian clock in the field to elucidate the photoperiodic control of flowering in rice under natural conditions.
In Arabidopsis, interlocked transcriptional negative-feedback loops, mainly consisting of CIRCADIAN CLOCK ASSOCIATED1 (CCA1), LONG ELONGATED HYPOCOTYL (LHY), TIMING OF CAB EXPRESSION1 (TOC1), the PSEUDO RESPONSE REGULATOR (PRR) gene family, and CCA1 HIKING EXPEDITION, constitute the core autonomous oscillator of ∼24-h period of the plant circadian clock (Nagel and Kay, 2012; Pokhilko et al., 2013).
The circadian clock synchronizes to the light/dark diurnal cycles of the external environment; this synchronization is termed entrainment. In Arabidopsis, light stimuli for entrainment are perceived by photoreceptors, such as phytochromes, cryptochromes, ZEITLUPE (ZTL), and FLAVIN BINDING KELCH F-BOX1 (FKF1), and are transduced to the core oscillator via At-GI, At-PRR5, At-PRR7, and At-PRR9 (Franklin et al., 2014). To elucidate the mechanisms of light entrainment, phase response curves (PRCs) to light pulses have been obtained for various organisms (Pfeuty et al., 2011). The PRCs for both Arabidopsis (Covington et al., 2001) and rice (Sugiyama et al., 2001) show some advance in the morning and some delay in the evening for luciferase promoter reporters regulated by the circadian clock. It has been proposed that the circadian clocks are entrained to solar time, in which a day is defined by the interval between two successive culminations (Pittendrigh and Daan, 1976; Geier et al., 2005; Yeang, 2013).
The circadian clocks are also entrained by hot/cold diurnal cycles, where the temperature signals are transduced by At-PRR7, At-PRR9, EARLY FLOWERING3 (ELF3), and At-ZTL in Arabidopsis (Franklin et al., 2014). Thus, signaling pathways leading to the entrainment of the circadian clock by light and temperature have common components (Franklin et al., 2014), while the specific molecular components for temperature entrainment are not yet known.
In Arabidopsis, many genetic components have been identified by screening for mutants with changes in periods and amplitudes of oscillation of circadian clock-regulated genes under constant light or temperature conditions after entrainment by light/dark or hot/cold diurnal cycles. However, the amplitudes of rhythmic gene expression are often strongly reduced under constant conditions, implying that the observed characteristics may be different from those in the field. Thus, molecular functions of the clock components should be examined also under diurnal environmental cycles (Izawa, 2012), where plants evolved. For example, hypocotyl growth of Arabidopsis is regulated by light and phytohormone signaling gated by the circadian clock and shows different peaking time under an 8-h-light/16-h-dark cycle from those under continuous light; the circadian clock components At-CCA1, At-LHY, At-ELF3, At-ELF4, and At-PHYTOCLOCK1 (PCL1) contribute to the gating of light and phytohormone signaling (Nozue et al., 2007; Michael et al., 2008; Nusinow et al., 2011). Light signals are transmitted via At-PHYTOCHROME B and PHYTOCHROME-INTERACTING FACTOR4 and 5 (Nozue et al., 2007). The diurnal variation in hypocotyl growth is regulated at least partially through repression of phytohormone signaling by both the circadian clock and light (Michael et al., 2008). However, the circadian clock is markedly robust against genetic perturbations under environmental diurnal cycles in Arabidopsis. A few mutant lines, such as elf3 (Hicks et al., 1996) and lhy cca1 (Alabadí et al., 2002; Mizoguchi et al., 2002) and double mutants of At-PRR7 and At-PRR9, At-PRR5, or At-TOC1 (Yamashino et al., 2008) exhibit significant changes in circadian oscillation of expression of key rhythmic genes under diurnal cycles of light or temperature.
Furthermore, experiments under controlled diurnal cycles of light or temperature could be insufficient to understand molecular functions of the circadian clock components in an evolutionary context because plants may evolve to use correlations among internal time produced by the circadian clock, light, and temperature in the field. Under natural conditions, solar radiation and ambient temperature generally increase and decrease gradually and peak around noon. Cloudy and rainy weather causes rapid fluctuations of diurnal patterns within the range of diurnal and seasonal trends. The onset and peak of solar radiation precedes those of the ambient temperature on both daily and seasonal time scales. Such correlations between external environments can be used by genetic regulatory networks. For instance, Escherichia coli can use high temperature as a sign of low oxygen concentration because it evolved under conditions of a negative correlation between temperature and oxygen resulting from its migration between mammal intestines and the outside world (Tagkopoulos et al., 2008). Plants might also use correlation between environmental stimuli to achieve proper function of their circadian clocks. However, correlations among external environments or between the phase of the circadian clock and external environments are disrupted in the controlled environments; thus, experiments under natural environments are necessary.
To evaluate the state of the circadian clock (internal time) from expression of multiple genes, the molecular-timetable method was developed for mammals (Ueda et al., 2004) and applied to plants as well (Kerwin et al., 2011). In this method, diurnal expression patterns of multiple circadian-regulated genes are modeled as cosine curves of various peak phases. The internal time of a sample is evaluated by searching a phase when the expression of the multiple genes coincides with the model output. However, the method has a drawback in that it is difficult to use genes with diurnal expression patterns other than a cosine curve even if their rhythmicity is strong, and major components of the circadian clock such as PRR95, PRR73, and GI often show diurnal expression patterns deviating from cosine curves (Izawa et al., 2011). Thus, to evaluate internal time from components of the circadian clock, it is necessary to develop a method applicable to genes of any diurnal expression pattern.
Using time sequences of transcriptome data, we previously performed genome-wide statistical modeling to dissect the effects of external environmental factors on expression of individual genes (Nagano et al., 2012). Although this modeling enabled the selection of only one factor (among six distinct environmental factors) that contributed most to the expression of each gene, it could be flawed in cases where gene expression is simultaneously affected by multiple environmental stimuli such as light and temperature.
The outline of this study (Supplemental Figure 1) is as follows. We built a new statistical model of individual gene expression integrating the effects of a diurnally entrained circadian clock with a fixed amplitude and phase entrained by solar radiation, two distinct environmental factors, solar radiation and ambient temperature gated by the circadian clock, and development and growth depending on accumulating responses to ambient temperature. We revealed that temperature strongly affect expression of circadian clock genes. By virtue of this new model, we estimated the expression dynamics of 25 circadian clock-related genes in the leaves of field-grown rice at 10-min intervals over an entire crop season. We then estimated internal time produced by the circadian clock based on the relationship between physical time of day and expression of multiple genes using a newly developed Bayesian method that enables us to utilize genes of any diurnal expression pattern and quantified the punctuality and precision of internal time versus physical time of day. Although the expression of individual circadian clock-related genes was strongly affected by fluctuations of light and temperature, we found that the expression of a group of 16 circadian clock-related genes encoded information on physical time of day with a punctuality of 22 min under fluctuating field environments through the entire crop season. We revealed that this monitoring of internal time could be a good indicator to evaluate the behavior of the circadian clock even under controlled environments. We also found some downstream genes regulated by both a normal circadian clock in the wild type and the perturbed circadian clock in a circadian clock mutant, osgi, and identified distinct regulation of downstream genes by distinct groups of circadian clock genes in both the wild type and osgi.
RESULTS
Modeling the Expression of Individual Genes Regulated by Fluctuating Light and Temperature Stimuli
To estimate the expression of individual genes, we developed a statistical model, which combined five factors: (1) circadian clock, i.e., an internal clock with fixed amplitude and phase entrained by solar radiation (Roenneberg et al., 2010); (2) gated light response, i.e., a response the sensitivity of which to solar radiation is regulated by the circadian clock; (3) gated temperature response; (4) development, the developmental stage defined as an accumulating response to ambient temperature; and (5) genotype (Figure 1A). It is of note that, here, we adopted 24 h as the free-running period in darkness in this model since we would like to focus on phase setting of circadian clocks by light under natural diurnal conditions. We denote model outputs corresponding to the observed data used for training the model as “estimation” and those corresponding to validation data not used for the training as “prediction.” To prevent overfitting to training data, which results in poor prediction performance for validation data, we removed unnecessary terms and initial values for each gene based on a likelihood ratio test (Supplemental Figure 2). In comparison with our previous model (Nagano et al., 2012), this model required higher computational cost for parameter optimization. Thus, we had to focus on only dozens of genes involved in a single biological trait and selected the circadian clock as the single target trait in this work. We first chose microarray probes for 25 rice orthologs of Arabidopsis genes for circadian clock components (Supplemental Figure 3) (Murakami et al., 2007) and light receptors (denoted hereafter as “clock-related genes”; see Supplemental Tables 1 to 3 for genes analyzed in each figure and table). Here, the genetic analysis of the osgi mutant (Izawa et al., 2011) led us to hypothesize that these orthologs contain major circadian clock components in rice. As training data, we used previously obtained transcriptome from 461 leaf samples of rice grown in a paddy field in Tsukuba, Japan, collected under various conditions (time of day, weather, and developmental stage) from June to September 2008 (the same data as used in Nagano et al., 2012). See Supplemental Tables 4 to 6 for transcriptome data sets analyzed in each figure and table. Estimation performance of the model was evaluated using adjusted R2, which is the fraction of variance explained by the model considering the number of parameters in the model. Most new models fitted the training data well (adjusted R2 > 0.5 for 21 out of the 25 clock-related genes; Figure 1B), as shown for GI (Figure 1C; Supplemental Figure 4A) and LHY (Supplemental Figure 5A). Furthermore, prediction performance was evaluated for validation data (108 samples in 2009; these data were also used in Nagano et al., 2012) using R2, which is the fraction of variance explained by the model without considering the number of parameters, and was found to be >0.5 for 16 out of the 25 genes (Figure 1B; Supplemental Figures 4B and 5B). Models for some genes such as ELF4_chr.3 showed negative R2, which indicates worse prediction performance than a model composed of just a constant value, that is, the average of the observed gene expression. This worse performance may be because the variations in observed expression of such genes were small and, thus, contribution of random noise in gene expression became relatively large. As expected from our previous report (Nagano et al., 2012), prediction performance for samples under controlled light/dark environment (26 samples) was also worse than the estimation performance for most of the genes even using this new model (Supplemental Figure 6A). However, when compared with our previous models (Nagano et al., 2012), the estimation (Supplemental Figure 6B) and prediction (Supplemental Figure 6C) performance for most of the 25 genes for the data from the fields were improved.
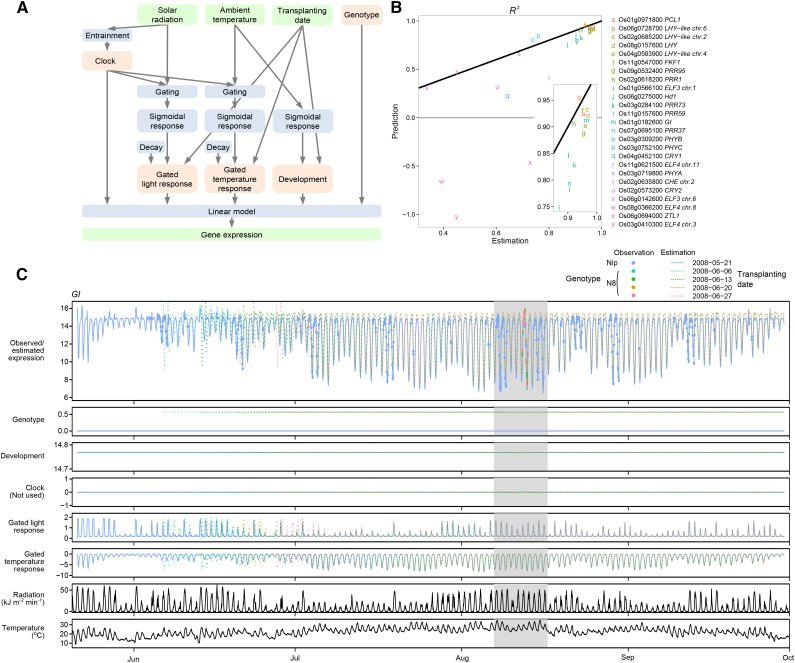
Figure 1.
Structure and Performance of the Gene Expression Model.
(A) Structure of the model. Observed variables (except for genotype) are in green, response processes in blue, and terms in pink.
(B) Estimation and prediction performance of the model for clock-related genes. R2, the fraction of variance explained by the model relative to variance of the data, was used as an index of fit for each gene model. Estimation performance was obtained with the same training samples (n = 461) as in (C). The black line indicates points where R2 values for estimation and prediction are equal. Prediction performance was obtained from validation samples (n = 108) collected in a different year. The genes are listed in order of rhythmicity evaluated as mutual information between the estimated gene expression and time of day (Supplemental Figure 3).
(C) The GI model as an example of estimated gene expression, inputs to the linear model, and observed gene expression. The shaded time interval is expanded in Supplemental Figure 4A. Nip, ‘Nipponbare’; N8, ‘Norin 8.’
Characteristics of the Response of Circadian Clock-Related Genes to Environmental Stimuli in the Field
We analyzed response of the gene expression to environmental stimuli from composition of terms and behavior of each term in the model. For example, in GI, the troughs of diurnal oscillation were often high in June and September compared with July to August, which is mostly explained by the gated temperature response (Figure 1C). The gated temperature response showed negative response to temperature, with the gate opening from midnight to morning (Supplemental Figure 4A). Therefore, the variations in trough expression level were likely to be determined by variation of temperature from midnight to morning. By contrast, the contribution of gated light response, development, and genotype were little, as reflected in the range of the responses and that clock term was removed. Compared with GI, seasonal variations of trough expression level for LHY were smaller, which might be determined by the contribution of circadian clock, which has an invariant amplitude, and by less contribution of gated temperature response (Supplemental Figure 5A).
Generally, the gated temperature response term remained for all 16 genes with high prediction performance (R2 > 0.5). The term corresponds to a response to ambient temperature, where the sensitivity is regulated temporally by the circadian clock. This term also contributed most to the variation of 11 genes among them (Figure 2A). These results are consistent with our previous work, where the gated temperature response affected the largest number of genes when we evaluated light, temperature, and other environmental factors separately for the same data (Nagano et al., 2012).
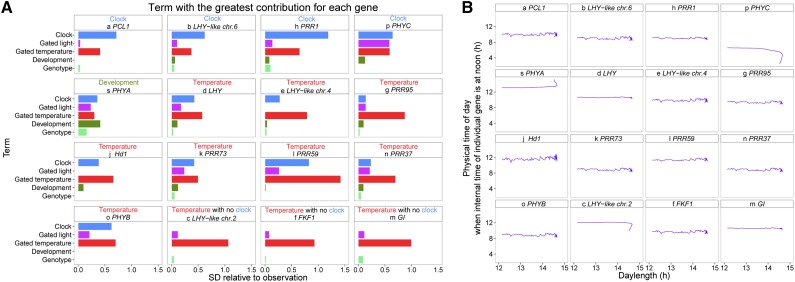
Figure 2.
Analysis of the Expression Model for the Genes with High Prediction Performance.
(A) Contribution of each term to the estimation. The sd of each term relative to that of the observation is shown. Letters preceding gene names correspond to those in Figure 1B. Only the 16 genes with the highest prediction performance (R2 for prediction >0.5; Figure 1B) are shown. The model of ELF3_chr.1 (data not shown) was considered unrealistic because the sds of the gated light response and development terms were >10 times that of the observation.
(B) Relationship between daylength and physical time of day when internal time of individual genes is at noon. Points for each day are connected along the lapse of date. Daylength was based on the astronomical estimate at the longitude, latitude, and altitude where the samples were taken (National Astronomical Observatory of Japan, http://eco.mtk.nao.ac.jp/cgi-bin/koyomi/koyomix_en.cgi). The internal time that corresponded to noon of physical time of day for most genes was earlier than 12:00 h because sensitivity to solar radiation is higher for the progress in the morning than that for the delay in the evening in those models, as indicated by asymmetry in response q > 1 (Supplemental Table 10; see Equation 5 in Methods). With the higher sensitivity for progress in the morning, the progress and the delay are counterbalanced when noon of internal time is set at earlier than 12:00 h of physical time.
We also estimated internal time of individual gene for the circadian clock entrained to solar radiation in the model for the 16 clock-related genes for which the model showed high prediction performance. The models for some genes lack the explicit circadian clock term. For example, the models for LHY-like chr.2, FKF1, and GI have the gated temperature term, but not the circadian clock term (Figure 2A). These results suggest that these genes are affected mainly by temperature fluctuations but not by direct regulation by the circadian clock. However, the gated temperature response or the gated light response term intrinsically includes the circadian clock, which gates the light or temperature response (see Figure 1A for the model structure). Thus, we were able to monitor the variation of the internal time of individual gene (defined by the state variable φ) relative to physical time of day for all 16 genes. The entrainment of the circadian clock is implemented in the model as change in progress rate of the internal time of individual gene in response to solar radiation. The period of the circadian clock in darkness was set to be 24 h, and we did not consider constant shortening or lengthening of period to 24 h by light (see Methods). We found that the seasonal variation in physical time of day at subjective noon (when internal time of individual gene is at noon) was <30 min and was not affected by seasonal changes in daylength of >3 h during the entire crop season in 2008 for most of the 16 genes (Figure 2B). Even if we consider the threshold for light detection by rice leaves in the field (Nagano et al., 2012), the seasonal variation of subjective noon cannot be relevant to seasonal changes in daylength.
In this model, the sensitivity to solar radiation, which depends on the internal time of individual gene and affects the entrainment of circadian clocks, is represented by circadian integrated response characteristic (CIRC; Roenneberg et al., 2010), which is determined according to the phase response curve obtained from phase progress or delay of the circadian clock in response to light of short duration. CIRC switches from negative to positive at midnight of internal time of an individual gene and from positive to negative at noon of internal time of an individual gene. When the model is provided with a specific range of parameters and diurnal cycle of light, the internal time of individual genes in the model is autonomously entrained so that the integral of positive and negative phase responses to light is counterbalanced according to the CIRC every day. Furthermore, asymmetry of positive and negative sensitivity and dead zone (no response) around noon is also incorporated in CIRC, which enables various types of entrainment to the fluctuating daily environments (Pittendrigh and Daan, 1976; Geier et al., 2005).
The small seasonal variation of internal time of individual genes was also seen in gate on radiation, gate on temperature, and CIRC. Considering the case of GI as a typical example, <20% daily change (100 to ∼120%) in progress rate of internal time of GI was observed around dawn during the crop season, resulting in ∼30 min range of advance (1.2 to ∼1.7 h) in the internal time of GI relative to physical time (Supplemental Figure 7A). The progress rate of internal time and the advance in internal time of individual genes is a response to radiation according to the CIRC. The small day-to-day variation against large fluctuations of radiation under various weather conditions (Supplemental Figure 7A) can be attributed to the low sensitivity of CIRC to radiation in the daytime (Supplemental Figure 7B). The small seasonal variations of internal time of individual genes against changes in seasonal daylength were attributed to the switch from positive to negative CIRC in the daytime (Supplemental Figures 7A and 7B).
Bayesian Inference Decoding Internal Time on the Basis of Expression of Circadian Clock-Related Genes in the Field
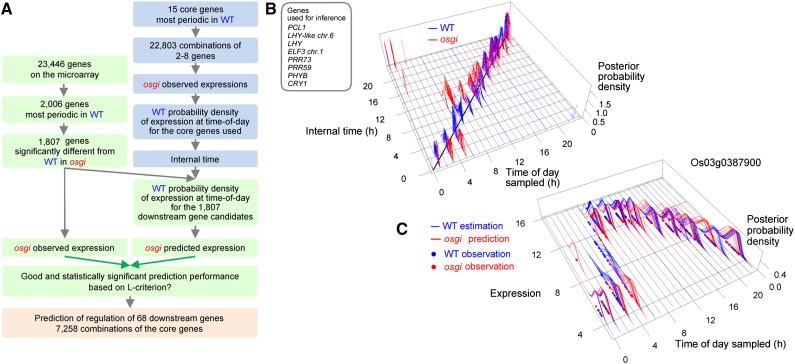
The internal time of individual genes is estimated from expression of a single gene with a model of the circadian clock. The parameters in the model of the circadian clock were estimated separately for each gene, which might have produced an unlikely consequence: different phase setting of circadian clock for each gene. Since the circadian clock is a network of regulation among multiple genes, we would be able to decode internal time from the expression of multiple genes. Therefore, we next estimated internal time from multiple genes composing the circadian clock based on the relationship between physical time of day and expression and quantified the punctuality and precision of internal time versus physical time of day. Because of rapid fluctuations in solar radiation and ambient temperature, data from several hundred samples cannot include all of temporal gene expression during the entire crop season. However, our new model with high predictive performance enables us to interpolate temporal gene expression accurately (i.e., with small error) with high time resolution, once we have corresponding data for both solar radiation and ambient temperature. Therefore, we estimated temporal gene expression at 10-min intervals based on the improved model using temporal data on solar radiation and ambient temperature from May to September 2008 and data for transplanting date in 2008. We then calculated the relationships between time of day and the expression of each gene as empirical probability distributions for the 25 clock-related genes as two dimensional heat maps (Figure 3A; Supplemental Figure 1), which should reflect the effect of all fluctuations in solar radiation and ambient temperature during the entire crop season on expression of each gene.
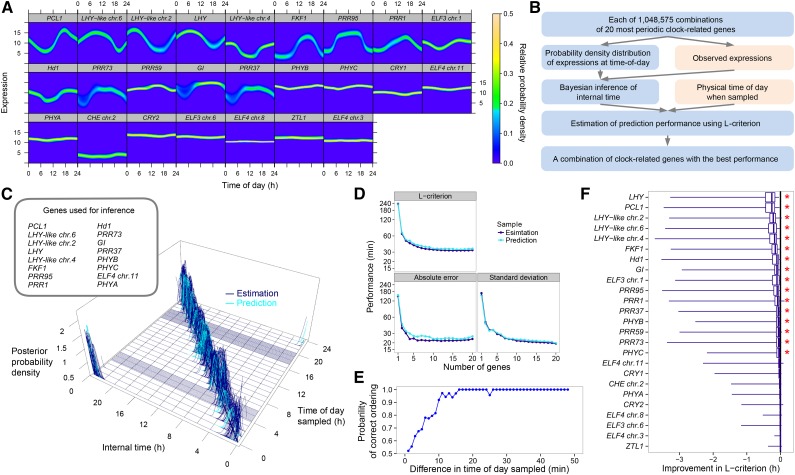
Figure 3.
Inference of Internal Time from the Expression Data of Circadian Clock-Related Genes.
(A) Probability density distributions between physical time of day and expression of the 25 clock genes based on the variation during an entire crop season in 2008. The genes are listed in order of rhythmicity (Supplemental Figure 3).
(B) Time inference process, evaluation, and optimization of performance. Observed variables are in pink and the inference processes in blue. See Supplemental Figure 8A.
(C) Estimation and prediction of internal time using the gene combination with the best estimation performance (i.e., the lowest mean absolute error in estimation). Posterior probability density of internal time is plotted against time of day sampled. Each blue (training sample for estimation; n = 461) or turquoise (validation sample for prediction, n = 125) line corresponds to a single sample. Among the training samples, samples obtained at 10-min intervals at 04:00 to 06:00 and 17:00 to 20:00 are included. Ranges of internal time with zero posterior probability density for those samples are represented as areas with dense blue lines at the bottom of the 3D space. Thick black diagonal line at the bottom of the 3D space indicates the correspondence between internal time and time of day sampled.
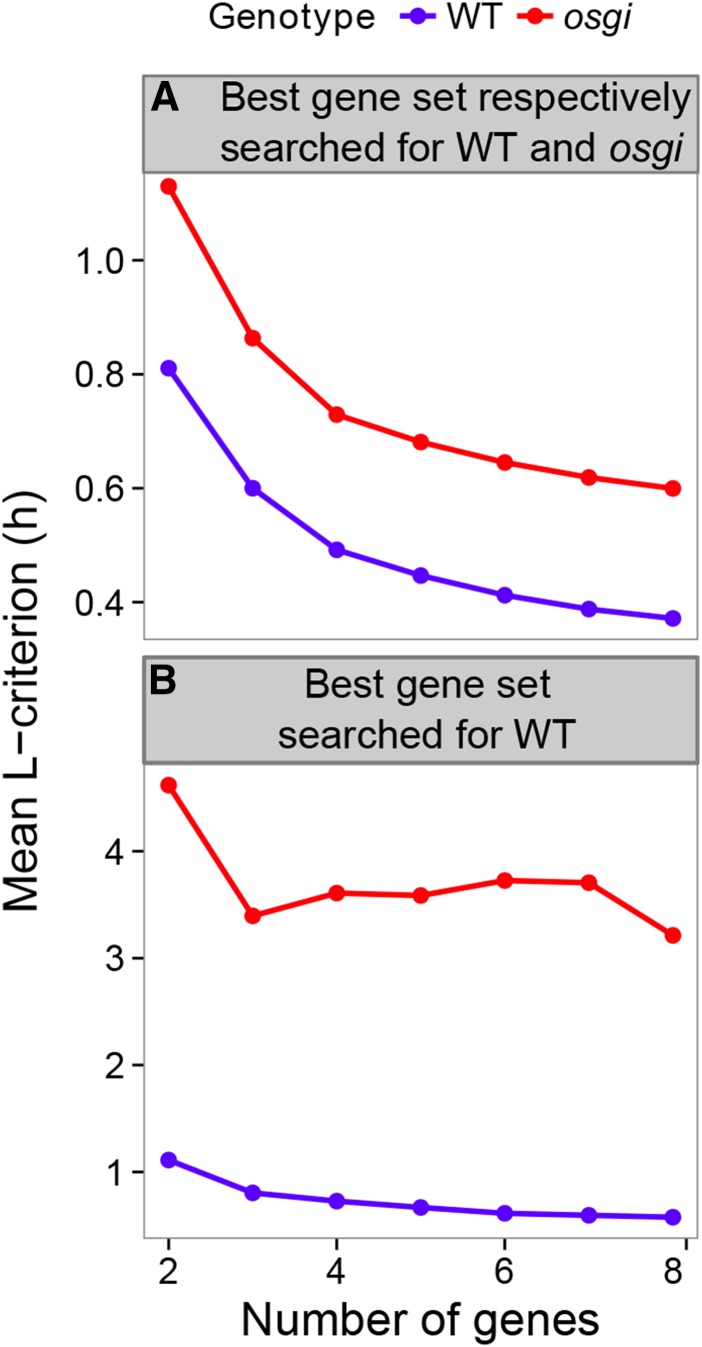
(D) Performance of the gene combination with the least L-criterion for each number of genes per combination.
(E) Probability of correct prediction of the sampling order for all possible pairs from 48 sequential samples collected at 1-min (or 2-min) intervals (based on the data in Supplemental Figure 8E).
(F) Contribution of each gene to the estimation performance. Improvement in the L-criterion by inclusion of the gene in the gene combinations is shown. Right ends of the horizontal bars, maximum values; right ends of the boxes, 75% quantile; vertical bars in the boxes, median; left ends of the boxes, 25% quantile; left ends of the horizontal bars, minimum values. Genes are listed in decreasing order of median improvement. Asterisks indicate significant improvement (P < 0.05 by the Wilcoxon signed-rank test with random permutation and Bonferroni correction; Supplemental Table 7).
We then performed Bayesian inference of time of day on the basis of gene expression by calculating the posterior probabilities of time of day having the observed expression. In this way, we were able to narrow down the range of times of day by calculating the product of the posterior probabilities of time of day for multiple genes. An example of such an approach for a pair of genes (PCL1 and GI) is shown in Supplemental Figure 8A. Apparently, the performance of time inference varied among the gene combinations used. Thus, we first selected the 20 genes for which expression was most dependent on physical time of day from among the 25 clock-related genes (Supplemental Figure 3) to reduce the number of gene combinations to a practical range. We then searched for a gene combination (2 to 20 genes) with the best performance from among 1,048,575 all possible combinations of the genes (Figure 3B). The expected value of the posterior distribution of time of day (the mean of time of day weighted with the posterior probabilities) is denoted here as “internal time.” We denote internal time obtained from the data used for computing relationships between time of day and the gene expression as “estimation” and internal time obtained from other data as “prediction.” “Error” denotes the difference between internal time and physical time of day sampled. The estimation (or prediction) performance was evaluated using the L-criterion, which was originally devised as an index of predictive performance estimated solely from the training data (Laud and Ibrahim, 1995). The gene combination with the best predictive performance contained 16 genes (Figures 3C and 3D); the mean of absolute values of error was 22 min for estimation from the training data (461 samples collected in 2008) and 24 min for prediction from the validation data (125 samples collected in 2009 and 2010). The means of raw values of estimation and prediction error were 5 ± 28 min and 8 ± 26 min (circular mean ± sd; Batschelet, 1965; Zar, 1999), respectively (Supplemental Figure 8B). No systematic and significant error was observed against solar radiation, ambient temperature, or daylength when sampled or developmental stage measured as days after transplanting (Supplemental Figure 8C). These results demonstrate that the internal time inferred from multiple gene expression relative to physical time of day is progressing punctually and thus constantly even in the field.
When the number of clock-related genes in a gene combination was limited to five, 1644 combinations showed a prediction error of <30 min (Supplemental Data Set 1). Even with only two genes, the best combination (PCL1 and GI; Supplemental Figure 8D) showed an average error of 38 min for estimation and 44 min for prediction (Figure 3D). These results strongly suggest that certain downstream genes can exhibit diurnal expression at punctual and precise timing when they are directly regulated by such combinations of clock-related genes.
To evaluate the resolution of two distinct time points in sequential sampling, we determined the internal time from independent leaf samples obtained at 1-min intervals (including a pair of samples collected with a 2-min interval) for a 49-min period in 2008 (48 samples in total, data in this study; Supplemental Figure 8E) and examined whether the predicted sampling order according to the internal time was correct among all possible pairs of 48 samples (Supplemental Figure 8F). The sampling order was correctly predicted with >95% probability for intervals between sample collection of at least 15 min (Figure 3E). This indicates that the internal time conferred by the expression of clock-related genes progresses to a significantly distinguishable state on a time scale of 15 min even under fluctuating external environments in the field.
To reveal the contribution of each clock-related gene to this time inference using temporal gene expression dynamics of circadian clock-related genes during the crop season with high time resolution, the improvement in estimation performance conferred by each gene was evaluated by using the L-criterion for combinations of two to eight genes from among the 25 clock-related genes (Figure 3F; see Methods for the details). Sixteen of these genes significantly (P < 0.05 with Bonferroni correction) improved the L-criterion based on the Wilcoxon signed-rank test, where the null distribution was calculated for each gene by random permutations (Figure 3F; Supplemental Table 7). As expected, genes whose expression were more dependent on time of day, such as PCL1 (Supplemental Figure 3), contributed most to the median improvement in the L-criterion. Genes with large seasonal (or day-to-day) variation of expression from midnight to dawn, as seen in the broad ranges of expression with non-zero relative probability density in the heat maps, such as GI, PRR37, and PRR73 (Figure 3A), also contributed significantly. The expression dynamics of these genes during daytime may contribute to stable time progression of the circadian clocks during daytime and thus confer the temporally proper regulation of downstream genes. The variation in expression observed from midnight to dawn may result from the variation in temperature (Figure 2A), according to which the downstream genes may be temporally regulated.
Progression of Internal Time under a Controlled Environment
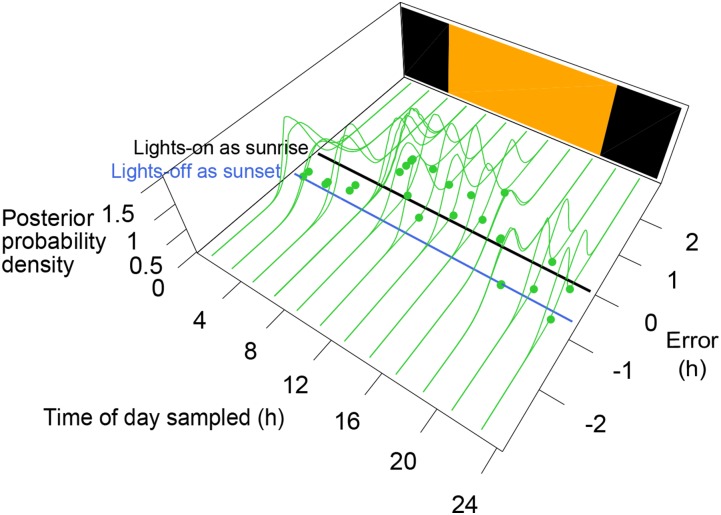
The progression of internal time is likely to be different under controlled environment (in a growth chamber) versus field conditions since light intensity and ambient temperature gradually increase from sunrise and then decrease to sunset with daily fluctuations in the fields (the lowermost panel of Supplemental Figure 7A), whereas light intensity is almost constant during the light period and ambient temperature rapidly increases and decreases at the beginning and the end of the light period, respectively, under the controlled environment. We determined the internal time under a controlled light/dark environment, using a combination of 15 genes with the best estimation performance for the training data (see Methods). The mean error throughout a day between the controlled and field environments became smallest (∼3.5 min) when the time of lights-on was set to mean of sunrise time during the crop season for the training data (zero error in this case is shown as a black line in Figure 4). When the time of the lights-off was set to sunset time (zero error in this case is shown as a blue line in Figure 4), the mean error in a day became 45.1 min. This indicates that state of the circadian clock at lights-on in the growth chamber is similar to that at sunrise for the growth condition applied. Even when setting the time of lights-on to sunrise time, a significant time advance of ∼2 h after lights-on and a time delay just before lights-off were observed under the controlled environment in comparison with the field environment (Figure 4; Supplemental Figure 8G). Light intensity around lights-on and lights-off is likely to be stronger for the controlled environment than that around sunrise or sunset in the field because the light intensity acutely increases to maximum and decreases to darkness under the controlled environment. Therefore, the progress after lights-on and the delay before lights-off observed in gene expression under the controlled environment are consistent with CIRC assumed in the gene expression model, which would cause an advance in the morning and a delay in the evening in response to more light intensity in the controlled environment around lights-on and lights-off. The fact that such changes in time progression were not observed in rice grown in the field (Figure 3C) indicates that global diurnal gene expression regulated by the circadian clock differs under controlled and field environments.
Figure 4.
Time Progression under a Controlled Diurnal Environment.
Prediction errors of internal time were calculated relative to physical time of day sampled. Green lines denote posterior distributions of errors, of which expectations are denoted by green points. Time of day sampled under the controlled environment was set so that lights-on time under the controlled environment corresponded to the average time of sunrise during the crop season. We also tried to set time of day so that the lights-off time under the controlled environment corresponded to the average time of sunset. Black and blue lines indicate the zero error line for the former and the latter setting, respectively. Orange and black rectangles indicate the light and dark periods, respectively.
Perturbed Progression of Internal Time in a Circadian Clock Mutant in the Field and Prediction of Downstream Genes
In the rice circadian clock mutant osgi, diurnal expression patterns of many clock-related genes were perturbed (Supplemental Figure 9A), in line with a previous report (Izawa et al., 2011), and time of day information was lost (Supplemental Figure 9B). When probability density of expression along time of day was calculated by a smoothing spline model (see Methods for details) using 212 pairs of osgi and wild-type samples obtained in the fields in 2008 and 2009 (Supplemental Table 5) and gene combinations were optimized separately for osgi and the wild type (Supplemental Figure 10A), the punctuality of time inference in osgi was found to become worse in comparison with that of the wild type (Figure 5A). When probability density of expression was calculated and gene combination was optimized only for the wild type (Supplemental Figure 10B), time progression in osgi resulted in more severely impaired predictive performance (Figure 5B). The pattern of altered time progression in osgi based on probability density of expression along time of day for the wild type depended on the gene combinations used for monitoring of internal time (Supplemental Figures 10C to 10E).
Figure 5.
Punctuality of Internal Time for the osgi Mutant.
(A) Punctuality of internal time encoded in a combination of clock-related genes showing the best performance for the osgi mutant and the wild type, respectively (see Supplemental Figure 10A for a scheme).
(B) Punctuality of internal time encoded in a combination of clock-related genes with the best performance for the wild type (see Supplemental Figure 10B for a scheme).
These results led us to an idea that transcriptome data containing both a certain genetic perturbation such as osgi mutation and environmental fluctuations in the fields may encode information of some regulatory genes linked with regulation of downstream genes. We first tried to extract such information from only transcriptome data of the wild type with environmental fluctuations in the fields but did not identify any such significant relationships (data not shown). We then tried to identify the downstream genes that are still under the control of a deficient circadian clock by osgi in order to reveal some structures in the gene network of the rice circadian clock from transcriptome data at the first step because circadian clocks are often robust to genetic perturbations (Baggs et al., 2009). We first chose 15 genes with the most marked rhythmicity to reduce the computational cost (denoted hereafter “core genes”; see Supplemental Tables 1 to 3). Then we searched for their downstream genes (among all the genes on the microarray platform), whose expression patterns in osgi were predicted based on the observed relationship between expression of downstream genes in the wild type and physical time, assigning the perturbed progression of internal time inferred from any combinations of the core genes in osgi as physical time (Figure 6A). Among the 1807 genes that were fairly rhythmic in the wild type (Supplemental Figure 9B) and further showed significantly different expression between osgi and the wild type, we were able to predict the expression of at least 68 downstream genes (Supplemental Table 8) significantly from internal time inferred from 7258 distinct combinations of the core genes among all gene combinations of two to eight genes out of 15 core genes (see Methods for details). For the downstream gene with the best prediction performance (Os03g0387900), progression of internal time was delayed in osgi relative to the wild type from midnight to dawn but jumped at 6 to 7 am to the internal time corresponding to noon and converged to the internal time of the wild type by noon (Figure 6B). For this best gene, on the basis of the progression of internal time, we were able to accurately predict a delay in the acute increase of gene expression at dawn and occasional outlier expression after midnight in osgi relative to the wild type (Figure 6C). We also predicted gene expression of downstream genes in the wild type (Supplemental Figure 11A) based on the relationship between gene expression in osgi and physical time, assigning perturbed internal time for the wild type inferred from the combination of core genes that gave the most accurate internal time for osgi as physical time (Figure 6A; Supplemental Figure 11B). Since this resulted in poorer performance, we concluded that osgi-based internal time was not appropriate to search for such downstream genes.
Figure 6.
Identification of Downstream Genes of the Circadian Clock Using Perturbed Progression in the osgi Mutant.
(A) A scheme to find downstream genes regulated by core genes both in the wild type and osgi (Supplemental Table 8). Processes for time inference from expression of the core genes are shown in blue and those for prediction of downstream gene expression are in green. The gene expression in osgi was predicted by plugging in internal time in osgi (determined by a combination of core genes in osgi) for physical time and inferring gene expression based on the relationship between the physical time and expression of a downstream gene in the wild type.
(B) Time progression in osgi that most accurately explains the expression of downstream genes. The seven core genes were used for inference of internal time.
(C) A downstream gene with the best prediction performance (Os03g0387900). Note that we can predict a delay in the acute increase at dawn and occasional outlier expression after midnight in osgi relative to the wild type. The corresponding internal time for osgi is delayed from midnight to dawn with occasional severity but jumps in the morning to catch up with the wild type by noon as shown in (B).
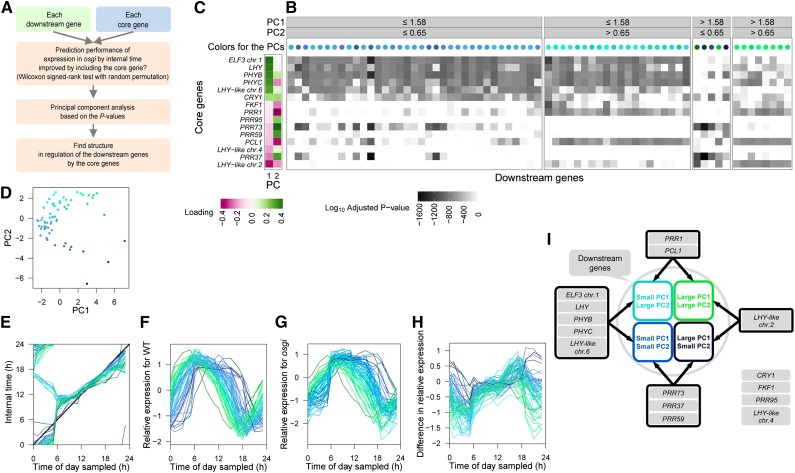
To quantify the contribution of each core gene to the prediction of expression of downstream genes in osgi (Figure 7A), we next calculated statistical significance of the contribution based on the Wilcoxon signed-rank test with random permutation (Figure 7B). This contribution implies that the core gene is likely to regulate each downstream gene together with OsGI and other core genes in the combination. Using principal component (PC) analysis of the obtained P values (Figures 7B to 7D), we classified the downstream genes by the core genes that likely to regulate them. We then found that downstream gene regulation by a group of core genes including ELF3_chr.1 is mutually exclusive with regulation by LHY-like_chr.2, whereas regulation by PRR73, PRR37, and PRR59 is mutually exclusive with that by PRR1 and PCL1 (Figures 7C and 7I). As expected, variations in the PC scores (Figure 7D), which should reflect variations of core genes regulating each downstream gene, corresponded to variations in progression of internal time in osgi (Figure 7E), variations of phase, and the diurnal pattern of the relative expression in the wild type (Figure 7F), that in osgi (Figure 7G), and the difference between osgi and the wild type (Figure 7H). Thus, a significant relationship between the expression dynamics of the core genes and that of the downstream genes, identified on the basis of genetic perturbations in osgi and environmental fluctuations, shows possible regulation of the specific downstream genes by those groups of core genes (Figure 7I). However, it is necessary to evaluate these relationships by further genetic analysis in the future.
Figure 7.
Structure of Regulation by the Core Genes Revealed by the Perturbed Time Progression in osgi.
(A) A scheme for finding the structure of regulation.
(B) Contribution of 15 core genes to the prediction of expression of downstream genes. Bonferroni-corrected log10 P values for each core gene determined by the Wilcoxon signed-rank test with random permutation are shown as a gray-scaled heat map. PC analysis was performed using the log10 P values for 15 core genes with each downstream gene. The scores of each downstream gene on PCs 1 and 2 are indicated by the colors of the circles above the heat map (as also shown in [D]). Small PC1 values are expressed as blue and large PC2 values green. The same labeling of colors are used to distinguish the 68 downstream genes in (E) to (I).
(C) Loading of the log10 P values of each core gene on PC1 and PC2 scores of the downstream genes.
(D) Scores of PC1 and PC2.
(E) Progression of internal time with the best prediction performance for each downstream gene. The osgi time progression for a downstream gene is shown as a polygonal line connecting the means of expectation of the internal time for each time of day sampled. The thick black line shows time progression along physical time of day.
(F) and (G) Relative expression of predicted downstream genes in the wild type (F) and osgi (G). Log2 expression levels are standardized so that the mean is 0 and sd is 1 for the wild type.
(H) Difference in the relative expression between the wild type and osgi.
(I) Significant relationships between groups of the core genes and downstream gene clusters based on PC analysis.
DISCUSSION
We demonstrated that the progression of the plant circadian clock as a whole system corresponds to physical time of day and was not affected much by fluctuating external environments in the field during the entire crop season, regardless of whether estimation methodology was based on expression of individual genes (Figure 2B) or multiple genes (Figures 3C and 3D). While the time resolution of the circadian clock state was found to be 15 min in the fields (Figure 3E), the progress rate of the circadian clock is also constant (Figure 3C; Supplemental Figure 7). However, the expression of most individual clock-related genes was strongly affected by both solar radiation and ambient temperature (Figure 2A). It may seem contradictory that the external environment strongly affected the expression of each clock-related gene, but did not affect the internal time of individual genes or the internal time estimated from the expression of multiple genes. Since a significant contribution of the genes even with drastic variations in expression from midnight till dawn and almost constant expression in daytime (such as GI, PRR73, and PRR37; Figure 3A) to the internal time (Figure 3F) was observed, the expression patterns of such circadian clock genes can retain partial, but critical information on physical time of day. Thus, the plant circadian clock as a network of regulations among genes may integrate perturbations from various factors such as daily fluctuations of the external environments, developmental stage, and intrinsic stochasticity of molecular processes to keep the progression of internal time constant even under fluctuating environments. Compared with studies using Arabidopsis, there is a lack of genetic evidence for the molecular mechanisms of circadian clocks in rice. However, we were able to demonstrate that the entire network of transcriptional regulation of circadian clock-related genes is punctually regulated under fluctuating field conditions in rice by a physiological approach using statistical modeling and inference.
We also found that the internal time under a controlled environment showed an advance in the morning and a delay in the evening compared with that in the field (Figure 4). Such difference between field and controlled conditions is implied in the PRCs obtained by light pulses in many organisms (Pfeuty et al., 2011), including rice (Sugiyama et al., 2001). The difference in transcriptomes between controlled and field environments should be taken into account, especially when knowledge on a trait and its underlying mechanisms is limited to that under such controlled environments. Running our model for lab conditions will be more informative to assess the data from the labs when enough of such data has been obtained.
We further revealed that time progression was severely disturbed in the osgi mutant in the field. The characteristic changes, such as the jump in time progression around dawn in osgi, remain largely unexplored; our approach would be useful in elucidating details of the action of each circadian clock gene under diurnal conditions. We also demonstrated a statistical analysis to find possible downstream genes in the genetic network with GI as a hub gene and to assess the specific relationship between each downstream gene and several core genes of the circadian clock.
The punctuality of internal time in this work outperformed the body time estimates based on the mouse liver transcriptome (Ueda et al., 2004), mouse blood metabolome (Minami et al., 2009), and human blood metabolome (Kasukawa et al., 2012) under controlled light/dark conditions (mean estimation error of 1.0, 1.0, and ∼3 h, respectively). This may result from more effective extraction of information on the internal time from gene expression by our methodology compared with the previous attempts. Otherwise, more accurate information may be encoded in gene expression of plants than in that of mammals.
The punctual and precise internal time enables prediction of strong light and high temperature in the daytime, which correlate to the internal time with some delay in a day, possibly resulting in effective prevention of photoinhibition and drought damage, although these effects have not been experimentally confirmed. Instead, it is very likely that the punctuality and precision of internal time can contribute to the photoperiodic control of flowering time in rice. It has been shown that expression of both Ehd1 and its downstream florigen gene shows a binary response to a change in daylength from 13.5 to 13.0 h (Itoh et al., 2010). This detection of a ≤30-min difference in daylength is easily achievable because even distinct plants grown in the same field can distinguish a 15-min difference at the transcriptional level (Figure 3E). According to the external coincidence model, which is supported by several lines of strong molecular evidence in Arabidopsis and rice (Itoh and Izawa, 2013; Yeang, 2013), the interaction of the external light signal and the circadian clock is crucial for accurate recognition of daylength changes (Pittendrigh and Minis, 1964). The punctual time progression of the circadian clock in the field confirmed in this work strongly supports this model. However, we cannot exclude that a distinct combination of circadian clock-related genes may produce two distinct internal time progressions, which are differently affected by subtle changes in seasonal daylength; this would support the internal coincidence model (Pittendrigh, 1972), another model of photoperiodic flowering.
Our approach can be used to determine the endogenous “time of day” state in plants grown in the field at different latitudes and longitudes and in plants grown under controlled environments such as those in plant factories. The detection of internal time may be used for the diagnosis of agronomic trait-related gene expression in a time of day-dependent manner.
In conclusion, the circadian clock as a regulatory network of multiple genes retains accurate and precise information related to physical time of day under fluctuating field environments by integrating perturbations from external and internal factors and stochastic noise in gene expression. In-depth analyses using statistical modeling of the field transcriptome combined with circadian clock-related experiments will enable us to further elucidate the molecular mechanisms conferring the punctuality and precision of the circadian clock under fluctuating field environments.
METHODS
All the analyses were performed using R, a language and environment for statistical computing (R Core Team, 2013).
Plant Materials and Microarray Analyses
Rice plants (Oryza sativa cv Nipponbare, cv Norin 8, osgi mutant), growth conditions, and acquisition of microarray data were previously described (Izawa et al., 2011; Sato et al., 2011; Nagano et al., 2012) except those for sampling at 1 min (2 min in part) intervals and those under a controlled environment. See Supplemental Tables 4 to 6 for transcriptome data sets analyzed in each figure and table. For 1-min (2-min) sampling (Figure 3E; Supplemental Figures 8E and 8F), the youngest fully expanded leaf from one rice plant (O. sativa cv ‘Norin 8’) was collected and microarray analysis was performed as described (Nagano et al., 2012); the data have been deposited in Gene Expression Omnibus (GEO) as GSE52120. For gene expression modeling (Figures 1 and 2; Supplemental Figures 3 to 7 and Supplemental Tables 9 to 12), training data for estimation have been deposited in GEO as GSE21397, GSE36040, GSE36042, GSE36043, GSE36044, and GSE18685 (wild type data only) and validation data for prediction as GSE36777 (wild type data only). For time inference in the wild type (Figures 3C and 3D; Supplemental Figures 8B to 8D and Supplemental Table 7), data deposited as GSE39520 were also used for validation. For analyses of osgi versus the wild type (Figures 5 to 7; Supplemental Figures 9 and 10 and Supplemental Table 8), paired data with the same sampling time have been deposited as GSE18685 and GSE36777. For experiments performed under a controlled environment (Figure 4; Supplemental Figures 6A and 8G), rice plants (O. sativa ‘Koshihikari’; n = 26) were grown in a growth chamber under metal halide lamps (photosynthetic photon flux density of 450 μmol m−2 s−1 or 5.88 kJ m−2 min−1) with a light period from 08:00 until 22:30 at 28°C (24°C during the dark period). For 24-h time series, the youngest fully expanded leaves were collected at 2-h intervals 28 and 49 d after sowing, and microarray analysis was performed as described (Nagano et al., 2012). Because the microarray platform for the data collected under the controlled environment was different from that used for the field samples, only probes common to both platforms were considered. To reduce the effects of different numbers of replicated probes for the same sequence in the two platforms, we averaged signals of the replicated probes after transformation to log2 scale and assigned the same value to all replicated probes in the microarray platform used for field samples. The data were deposited as GSE54525.
Gene expression data for each probe processed by the Agilent protocol were transformed to log2 scale. Then q-spline normalization (Workman et al., 2002) was applied to distinct arrays. Probes were annotated on the basis of RAP build 5 (Sakai et al., 2013); RAP build 3 (Ohyanagi et al., 2006) was used for Hd1. For each probe on the Agilent array, rice coding sequences were searched using BLASTn for hits of >51 bp with a ratio between the E-values of the best hit and the second best hit of at least 10−3. If both criteria were met, the probe was annotated as corresponding to the locus that included the hit. Expression values of probes annotated to the same locus were averaged.
To evaluate rhythmicity of gene expression, we calculated mutual information between time of day and gene expression. To calculate mutual information, time of day and gene expression was discretized to bins with equal frequency and then empirical mutual information was calculated using R package “infotheo” (Meyer P.E; http://www.r-project.org/). Among the 25 clock-related genes, Hd1 is often considered as a flowering-time gene in rice but was included owing to its high mutual information with time of day in this study. See Supplemental Tables 1 to 3 for genes analyzed in each figure and table.
Meteorological Data
Data from the nearest meteorological station belonging to the Japan Meteorological Agency (Tateno, Tsukuba, 36°03′N, 140°08′E, altitude 25.2 m above sea level) were obtained. Global solar radiation (kJ m−2 min−1) and ambient temperature (°C) were transformed to relative scales (0 to 1) by dividing by 100 and 50, respectively, and averaged over each 1-h interval. When data for finer time intervals were necessary to solve ordinary differential equations, linear interpolation was applied.
The Model of Gene Expression Responding to Two Environmental Variables
The model considers two environmental stimuli (solar radiation and ambient temperature) equally. In addition, the major changes from the previous model (Nagano et al., 2012) are (1) autonomous entrainment of a circadian clock to solar radiation, (2) sigmoidal responses to environmental changes with continuous variation of response characteristics from linear to binary, (3) a development term reflecting the accumulating effect of ambient temperature, and (4) removal of the interaction terms.
Gene expression (y) was approximated by a linear model composed of clock (C), gated light response (L), gated temperature response (T), development (D), and genotype (G) terms (Figure 1A, Equation 1), where the coefficient of each term is represented by β followed by a specific subscript:
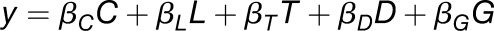 |
(1) |
The genotype term (G) indicates whether the cultivar was ‘Nipponbare’ (0) or ‘Norin 8’ (1); other terms describe characteristic responses to the environment by the following ordinary differential equations (ODEs). Supplemental Tables 9 to 12 contain the search ranges of the parameters. All parameters and variables except for observed solar radiation XL(t) and the ambient temperature XT(t) are gene specific.
The clock term (C) describes autonomous entrainment of the circadian clock to solar radiation by the following differential equations, where the state variable φ indicates internal time of individual gene:
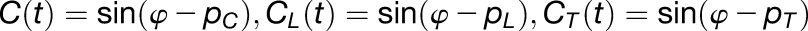 |
(2) |
Equation 2 describes the output of the circadian clock (C, CL, and CT) with different phases (pC, pL, and pT) for the clock term, a gate for the gated light response term, and a gate for the gated temperature response term, respectively.
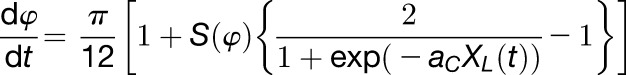 |
(3) |
Equation 3 describes the progress of the internal time of individual gene (φ) with a saturation response with gain aC to observed solar radiation (XL(t)), gated by a CIRC (S(φ); Roenneberg et al., 2010). When in darkness (XL(t)=0), progress rate is constant and equals to π/12, which corresponds to a 24-h period.
The maximum and minimum progress rates are twice as fast as physical time and 0, respectively.
 |
(4) |
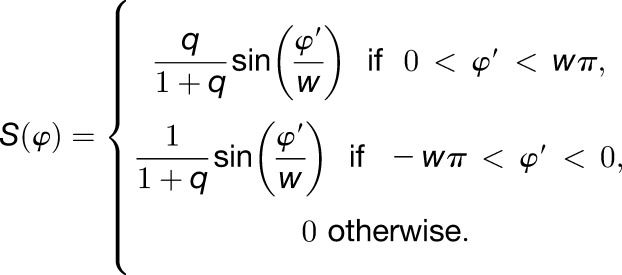 |
(5) |
Equation 4 transforms φ to φ’ confined within range of (-π, π], and Equation 5 gives CIRC S(φ), where q is the asymmetry of sensitivity between progress and delay and w (0.1 ≤ w ≤ 1) is the width of the responsive time interval. The mean period of the circadian clock without light stimulus was estimated as 25.7 ± 0.6 h for rice (Sugiyama et al., 2001). However, the period without light stimulus does not critically affect the response to changing daylength, compared with the width of the responsive time interval and the asymmetry of sensitivity between progress and delay (Pittendrigh and Daan, 1976; Geier et al., 2005). Therefore, in this model, the period without light stimulus was fixed to 24 h; at this value, the effect of the period on tracking noon, dusk, and dawn is neutral.
The gated radiation response term (Equation 6) and the gated temperature response term (Equation 7) have a common structure with different inputs (solar radiation and ambient temperature):
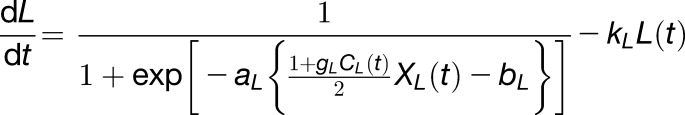 |
(6) |
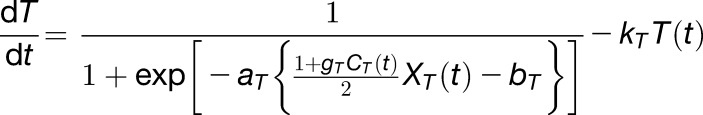 |
(7) |
The first terms of Equations 6 and 7 describe sigmoidal responses to observed solar radiation (XL(t)) and observed ambient temperature (XT(t)), gated by the outputs of the circadian clock (CL(t) and CT(t)) with amplitudes gL and gT, respectively (0<(1+ gL CL(t))/2 <1, 0<(1+ gT CT(t))/2 <1). The sigmoidal response can describe a saturation response with gains aL and aT, respectively, and intercepts bL and bT. The intercepts can guarantee some constant response rates to L and T, respectively. The second terms describe the decay of the gated responses with rates kL and kT, respectively.
The development term (Equation 8) is a simplified version of the gated temperature term (Equation 7), where the decay and the gate are removed. A sigmoidal response with gain aD and intercept bD confers a monotonic accumulation of D, which is affected by fluctuations of observed ambient temperature (XT(t)):
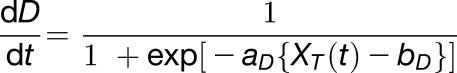 |
(8) |
The initial values of the state variables (φ, L, T, and D) of the ODEs were defined as values at noon of the transplanting date. The initial value of the internal time of individual gene (φ; Equation 3) was set to 0 because the autonomous entrainment became stable within 10 d for most of the genes examined. For other ODEs, the initial values were included in the parameters.
Solution of the Differential Equations and Optimization of the Model Parameters and Structure
To solve the system of ODEs, we used lsoda, a solver for ODEs implemented in the R package deSolve (Soetaert et al., 2010), with the maximum time step set to 10 min and relative and absolute tolerances for expression of 10−6. To fit the model to the training data, a parameter set with the least sum of squared residuals of gene expression was searched with particle swarm optimization (Kennedy and Eberhart, 1995) for nonlinear parameters and QR decomposition for coefficients of the linear model. Particle swarm optimization was implemented in the R package pso (C. Bendtsen; http://www.r-project.org/) and QR decomposition as the “lm” function in R. Particle swarm optimization was performed with the number of particles set to 10 times that of nonlinear parameters and a maximum of 5000 iterations. It took 15 to 30 h on a node with 16 cores (two CPUs of Intel Xeon E5-2670) for the parameter optimization of a model with specific combination of terms for a single gene. Unnecessary inputs to the linear model and the initial values were removed based on a likelihood-ratio test (Supplemental Figure 2), which is an F-test based on sum of squared errors, number of parameters, and number of observations for the models. See Supplemental Tables 9 to 12 for the estimated parameters of the final model.
Probabilities of Expression as a Function of Physical Time of Day
We used the new model to calculate daily gene expression at 10-min intervals for a sampling period that included 357 Nipponbare samples collected during the crop season in 2008 (Figure 3A). We used only the Nipponbare data because the transplanting date was earlier than that of ‘Norin 8’ and the samples covered the longest time period during the crop season. To obtain probabilities of expression level dependent on time of day during the crop season in 2008, first we calculated probabilities for each day at each grid point of 10 min intervals for time of day and 0.2 intervals for expression level. The dependence of probability on expression level for time of day was modeled as normal distribution, with means at the estimated gene expression and constant variance. The variance was estimated from residuals at time points when gene expression was observed. To obtain probabilities of expression level during the entire crop season, we averaged the probabilities for each day at each grid point.
To compare estimation optimized separately for osgi and the wild type (Figure 5A), we obtained a smoothing spline model (rather than the model of environmental response) separately for the wild type (cv ‘Norin 8’) and osgi from the paired samples to estimate the expression levels at a given time of day irrespective of sampling date. Cubic splines with a cyclic border (Wood, 2011) were used as the smoothing spline. The estimations of expression levels were obtained at each grid point of time of day. The probabilities at each grid point of expression level and time of day were then calculated as described above.
To estimate the probabilities of expression of downstream genes in osgi as a function of time of day irrespective of sampling date, we also used smoothing splines as described above. Only the data for the wild type (cv ‘Norin 8’) from the paired samples of ‘Norin 8’ and osgi were used to estimate the probabilities.
Bayesian Inference of Time of Day
Posterior probabilities of time of day conditional on expression level of genes used for inference were obtained based on Bayes’ rule, assuming uniform prior probabilities, as:
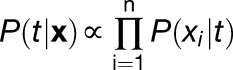 |
(9) |
where xi is expression of one of n genes used for the inference, P(xi | t) is the probability of expression as a function of time of day, and x is a vector of expression levels of all genes in a gene combination used for the inference. First, probabilities of time of day for the observed expression level were calculated at 2.5-min intervals, using a linear interpolation of the relative probabilities (Supplemental Figure 8A). The probabilities were then collected for all the genes used for the inference, and the probability product of them at each grid point of time of day (right hand side of Equation 9) was calculated. To obtain posterior probability (left-hand side of Equation 9), the probability product, which is in a relative scale, was normalized so that integration over 24 h by the trapezoidal rule gave a value of 1 to meet the definition of probability.
The sd for the circular variable s in an hour (Batschelet, 1965; Zar, 1999) was calculated as show in Equation 10:
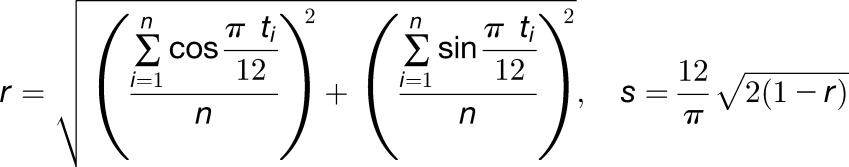 |
(10) |
where ti is the inferred internal time (h) for the i th sample out of n samples.
The L-criterion, which is an index of predictive performance estimated solely from the training data, is defined as the length of a vector composed of the error (otherwise, the difference between observed data and mean of the posterior distribution) and sd of the posterior distribution (Laud and Ibrahim, 1995).
For inference of internal time for samples collected under the controlled environment (Figure 4; Supplemental Figure 8G), one of the probes detecting OsLHY-like_chr.4 (Os04g0583900) was absent in the microarray platform. Thus, OsLHY-like_chr.4 was removed from the 20 most rhythmic clock-related genes, and a combination of genes with the least L-criterion for estimation of the training data was searched separately. As a result, a combination of 15 genes was obtained, where just OsLHY-like_chr.4 was absent from the 16 genes (Figure 3C) searched from all 20 most rhythmic clock-related genes.
Wilcoxon Signed-Rank Test with Random Permutation
The tests for the contribution of clock-related genes to the time inference performance (Figure 3F; Supplemental Table 7) and to the prediction performance of gene expression in osgi for each downstream gene (Figure 7B; Supplemental Table 8) were based on the estimation (or prediction) performances evaluated using the L-criterion. The null hypothesis was that the L-criterion of a gene combination with the tested gene is not less than that of the corresponding gene combination without the tested gene. The null distributions of the W-statistic in the Wilcoxon signed-rank test for all tested gene combinations were generated on the basis of 10,000 (Figure 3F; Supplemental Table 7) or 100,000 (Figure 7B; Supplemental Table 8) paired samples, each consisting of a gene combination without the tested gene and the corresponding combination with a randomly added gene. These null distributions were approximated to a normal distribution, and the cumulative probability of the lower tail was calculated as the P value. None of the null distributions significantly deviated from normality (P > 0.05, Kolmogorov-Smirnov test) in the test of the time inference performance (Supplemental Table 7). For the test of the prediction performance of gene expression in osgi, one combination consisting of a core circadian clock gene and a downstream gene out of 1020 combinations was deviated from normality (P < 0.05, Kolmogorov-Smirnov test), which was not significant after Holm’s correction for multiple comparison.
Search for Downstream Genes
Downstream genes that follow the perturbed progression of internal time in osgi were searched on the basis of the prediction performance of gene expression in osgi. The gene expression in osgi was predicted by plugging in internal time in osgi for physical time and inferring gene expression based on the relationship between physical time and expression in the wild type (Figure 6A). The internal time for each osgi sample was obtained for 22,803 gene combinations (all combination of two to eight genes out of 15 core genes). Predictions of gene expression in osgi were obtained for the 1807 genes that were most rhythmic in the wild type (Supplemental Figure 9B) and that showed significantly different expression between osgi and the wild type. For each of 41,205,021 combinations (22,803 combinations of core genes × 1807 genes), the predictions of gene expression in osgi were obtained as a probability distribution, and the prediction performance was evaluated with the L-criterion for gene expression in osgi (Figure 6A).
To identify a relationship between the core genes and a rhythmic gene statistically, we first scaled L-criteria for predicted gene expression in osgi divided by the sd of the observed gene expression, searched the genes with the average scaled L-criteria for 13 time points of <0.5, and found 68 genes out of the 1807 rhythmic genes. Since these 68 genes were well predicted, these genes were considered to be strictly regulated by the circadian clocks in both the wild type and osgi (even when the circadian clocks were impaired in osgi).
We further assessed statistical significance of the relationship by testing whether the prediction performance of gene expression in osgi occurred by chance using a random permutation test for the L-criterion, where the null hypothesis was that the L-criterion was not less than that for the prediction of gene expression in osgi for random permutation of observed gene expression and physical time of day sampled. The null distribution of the L-criterion was obtained from 10,000 samples of randomly permuted observed expression data of the downstream gene. The null distributions were approximated by a normal distribution, and the cumulative probability of the lower tail was calculated as the P value. None of the null distributions deviated significantly from normality (P > 0.05, Kolmogorov-Smirnov test). The Bonferroni correction for the 41,205,021 tests was applied to the P values. The values for all 68 downstream genes were found to be significant (P < 10−30), where those L-criterion were unlikely to be obtained by chance.
Principal Component Analysis
To reveal the structure of regulation of the downstream genes by the 15 core genes, we performed PC analysis of the log10 P values of the Wilcoxon signed-rank test with random permutation. For all 68 downstream genes, 15 log10 P values of the Wilcoxon signed-rank test for each core gene were calculated. For each core gene, the log10 P values were first centered and scaled to have an average of 0 and sd = 1.0. Using 15 set of the standardized log10 P values, PC analysis classified all downstream genes using the R function “prcomp.” The core genes with an absolute loading of >0.3 were considered to contribute to the PCs. The cumulative proportion of variance explained by PC1 and PC2 was 0.52. The PC1 and PC2 scores of a downstream gene were determined from a vector of the normalized log10 P values of the core genes projected onto a pair of orthogonal axes that give the best and second best explanations for the variation of the normalized log10 P values. Thus, the PC scores should correspond to a classification of the core genes contributing to the prediction of expression for each downstream gene in osgi. The loadings of PC analysis are coefficients of the normalized log10 P values of each core gene used to calculate the PC scores of the downstream genes and indicate their contribution to the classification by each PC axis and its direction.
Accession Numbers
Sequence data from this article can be found GEO under the following accession numbers: GSE52120, GSE21397, GSE36040, GSE36042, GSE36043, GSE36044, GSE18685 (wild type data only), GSE36777 (wild type data only), GSE39520, GSE18685, GSE36777, and GSE54525.
Supplemental Data
Supplemental Figure 1. Major Findings and Outline of Methods in This Study.
Supplemental Figure 2. Model Improvement by Reducing the Number of Parameters.
Supplemental Figure 3. Rhythmicity of Gene Expression.
Supplemental Figure 4. A Model for GI.
Supplemental Figure 5. A Model for LHY.
Supplemental Figure 6. Comparison of Model Performance with Data Sets under Controlled Environment and with Previous Models.
Supplemental Figure 7. Entrainment of the Circadian Clock to Solar Radiation in the Field.
Supplemental Figure 8. Processes and Results of Bayesian Inference of Internal Time.
Supplemental Figure 9. Comparison of Diurnal Gene Expression and Rhythmicity between the osgi Mutant and the Wild Type.
Supplemental Figure 10. Evaluations of Time Inference Based on Gene Expression in osgi and the Wild Type.
Supplemental Figure 11. Prediction of Wild-Type Gene Expression of a Downstream Gene from a Combination of Core Genes That Gives the Most Accurate Internal Time in osgi.
Supplemental Table 1. Circadian Clock-Related Genes Analyzed in This Study and Corresponding Figures.
Supplemental Table 2. Circadian Clock-Related Genes Analyzed in This Study and Corresponding Supplemental Figures.
Supplemental Table 3. Circadian Clock-Related Genes Analyzed in This Study and Corresponding Supplemental Tables and Data.
Supplemental Table 4. Transcriptome Data Set Analyzed in This Study and Corresponding Figures.
Supplemental Table 5. Transcriptome Data Set Analyzed in This Study and Corresponding Supplemental Figures.
Supplemental Table 6. Transcriptome Data Set Analyzed in This Study and Corresponding Supplemental Tables and Data.
Supplemental Table 7. Contributions of the Genes to the Time Inference.
Supplemental Table 8. Downstream Genes Found, Their Prediction Performance, and Principal Component Scores.
Supplemental Table 9. Coefficients for the Terms in the Linear Model.
Supplemental Table 10. Parameters for the Development and Clock Terms.
Supplemental Table 11. Parameters for the Gated Light Response Term.
Supplemental Table 12. Parameters for the Gated Temperature Response Term.
Supplemental Data Set 1. Combinations of up to Five Clock-Related Genes with a Prediction Error for Internal Time of Less Than 30 min.
Supplementary Material
Acknowledgments
We thank A.J. Nagano, R. Motoyama, N. Tanabe, Y. Sato, and Y. Nagamura for preparation of the raw microarray data published and registered in GEO, A.J. Nagano and S. Ishida for evaluation of the previous model by A.J. Nagano, M. Fukata for design of the microarray platform and registration of new microarray data with GEO, and J. Sese and A.J. Nagano for comments on the article. All microarray analyses were performed at the Open Laboratory at the National Institute of Agrobiological Sciences. We used the high-performance cluster computing system of AFFRIT, Ministry of Agriculture, Forestry, and Fisheries (MAFF), Japan. This work was supported by grants from MAFF, Japan (Genomics for Agricultural Innovation, RTR-0004; Genomics-based Technology for Agricultural Improvement, PFT-1001) to T.I.
AUTHOR CONTRIBUTIONS
T.I. conceived this plan and organized the entire research. T.I. and J.M. designed the research. T.I. and Y.K. performed the experiments. J.M. conceived and performed the data analyses. J.M. and T.I. wrote the article and Y.K. revised it.
Glossary
- PRC
phase response curve
- CIRC
circadian integrated response characteristic
- PC
principal component
- GEO
Gene Expression Omnibus
- ODE
ordinary differential equation
Footnotes
Articles can be viewed online without a subscription.
References
- Alabadí D., Yanovsky M.J., Más P., Harmer S.L., Kay S.A. (2002). Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 12: 757–761. [DOI] [PubMed] [Google Scholar]
- Baggs J.E., Price T.S., DiTacchio L., Panda S., Fitzgerald G.A., Hogenesch J.B. (2009). Network features of the mammalian circadian clock. PLoS Biol. 7: e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batschelet E. (1965). Statistical Methods for the Analysis of Problems in Animal Orientation and Certain Biological Rhythms. (Washington, DC: American Institute of Biological Sciences; ). [Google Scholar]
- Covington M.F., Panda S., Liu X.L., Strayer C.A., Wagner D.R., Kay S.A. (2001). ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13: 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A.N., Salathia N., Hall A., Kévei E., Tóth R., Nagy F., Hibberd J.M., Millar A.J., Webb A.A.R. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633. [DOI] [PubMed] [Google Scholar]
- Franklin K.A., Toledo-Ortiz G., Pyott D.E., Halliday K.J. (2014). Interaction of light and temperature signalling. J. Exp. Bot. 65: 2859–2871. [DOI] [PubMed] [Google Scholar]
- Geier F., Becker-Weimann S., Kramer A., Herzel H. (2005). Entrainment in a model of the mammalian circadian oscillator. J. Biol. Rhythms 20: 83–93. [DOI] [PubMed] [Google Scholar]
- Hicks K.A., Millar A.J., Carré I.A., Somers D.E., Straume M., Meeks-Wagner D.R., Kay S.A. (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792. [DOI] [PubMed] [Google Scholar]
- Itoh H., Izawa T. (2013). The coincidence of critical day length recognition for florigen gene expression and floral transition under long-day conditions in rice. Mol. Plant 6: 635–649. [DOI] [PubMed] [Google Scholar]
- Itoh H., Nonoue Y., Yano M., Izawa T. (2010). A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 42: 635–638. [DOI] [PubMed] [Google Scholar]
- Izawa T. (2012). Physiological significance of the plant circadian clock in natural field conditions. Plant Cell Environ. 35: 1729–1741. [DOI] [PubMed] [Google Scholar]
- Izawa T., Mihara M., Suzuki Y., Gupta M., Itoh H., Nagano A.J., Motoyama R., Sawada Y., Yano M., Hirai M.Y., Makino A., Nagamura Y. (2011). Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell 23: 1741–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasukawa T., Sugimoto M., Hida A., Minami Y., Mori M., Honma S., Honma K., Mishima K., Soga T., Ueda H.R. (2012). Human blood metabolite timetable indicates internal body time. Proc. Natl. Acad. Sci. USA 109: 15036–15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, J., and Eberhart, R. (1995). Particle swarm optimization. Proc. IEEE Internat. Conf. Neural Networks 4: 1942–1948. [Google Scholar]
- Kerwin R.E., Jimenez-Gomez J.M., Fulop D., Harmer S.L., Maloof J.N., Kliebenstein D.J. (2011). Network quantitative trait loci mapping of circadian clock outputs identifies metabolic pathway-to-clock linkages in Arabidopsis. Plant Cell 23: 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laud P., Ibrahim J. (1995). Predictive model selection. J. R. Stat. Soc. B 57: 247–262. [Google Scholar]
- Michael T.P., Breton G., Hazen S.P., Priest H., Mockler T.C., Kay S.A., Chory J. (2008). A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 6: e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami Y., Kasukawa T., Kakazu Y., Iigo M., Sugimoto M., Ikeda S., Yasui A., van der Horst G.T.J., Soga T., Ueda H.R. (2009). Measurement of internal body time by blood metabolomics. Proc. Natl. Acad. Sci. USA 106: 9890–9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T., Wheatley K., Hanzawa Y., Wright L., Mizoguchi M., Song H.-R., Carré I.A., Coupland G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2: 629–641. [DOI] [PubMed] [Google Scholar]
- Murakami M., Tago Y., Yamashino T., Mizuno T. (2007). Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48: 110–121. [DOI] [PubMed] [Google Scholar]
- Nagano A.J., Sato Y., Mihara M., Antonio B.A., Motoyama R., Itoh H., Nagamura Y., Izawa T. (2012). Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell 151: 1358–1369. [DOI] [PubMed] [Google Scholar]
- Nagel D.H., Kay S.A. (2012). Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 22: R648–R657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K., Covington M.F., Duek P.D., Lorrain S., Fankhauser C., Harmer S.L., Maloof J.N. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361. [DOI] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyanagi H., et al. (2006). The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res. 34: D741–D744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeuty B., Thommen Q., Lefranc M. (2011). Robust entrainment of circadian oscillators requires specific phase response curves. Biophys. J. 100: 2557–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh C., Daan S. (1976). A functional analysis of circadian pacemakers in nocturnal rodents, IV. Entrainment: pacemaker as clock. J. Comp. Physiol. 106: 291–331. [Google Scholar]
- Pittendrigh C.S. (1972). Circadian surfaces and the diversity of possible roles of circadian organization in photoperiodic induction. Proc. Natl. Acad. Sci. USA 69: 2734–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh C.S., Minis D.H. (1964). The entrainment of circadian oscillations by light and their role as photoperiodic clocks. Am. Nat. 98: 261–294.. [Google Scholar]
- Pokhilko A., Mas P., Millar A.J. (2013). Modelling the widespread effects of TOC1 signalling on the plant circadian clock and its outputs. BMC Syst. Biol. 7: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2013). R: A language and environment for statistical computing. (Vienna, Austria: R Foundation for Statistical Computing; ). [Google Scholar]
- Roenneberg T., Rémi J., Merrow M. (2010). Modeling a circadian surface. J. Biol. Rhythms 25: 340–349. [DOI] [PubMed] [Google Scholar]
- Sakai H., et al. (2013). Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 54: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Antonio B., Namiki N., Motoyama R., Sugimoto K., Takehisa H., Minami H., Kamatsuki K., Kusaba M., Hirochika H., Nagamura Y. (2011). Field transcriptome revealed critical developmental and physiological transitions involved in the expression of growth potential in japonica rice. BMC Plant Biol. 11: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soetaert K., Petzoldt T., Setzer R.W. (2010). Solving differential equations in R: Package deSolve. J. Stat. Softw. 33: 1–25.20808728 [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120. [DOI] [PubMed] [Google Scholar]
- Sugiyama N., Izawa T., Oikawa T., Shimamoto K. (2001). Light regulation of circadian clock-controlled gene expression in rice. Plant J. 26: 607–615. [DOI] [PubMed] [Google Scholar]
- Tagkopoulos I., Liu Y.-C., Tavazoie S. (2008). Predictive behavior within microbial genetic networks. Science 320: 1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H.R., Chen W., Minami Y., Honma S., Honma K., Iino M., Hashimoto S. (2004). Molecular-timetable methods for detection of body time and rhythm disorders from single-time-point genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 101: 11227–11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood S.N. (2011). Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J. R. Stat. Soc. B 73: 3–36. [Google Scholar]
- Workman C., Jensen L.J., Jarmer H., Berka R., Gautier L., Nielser H.B., Saxild H.H., Nielsen C., Brunak S., Knudsen S. (2002). A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 3: h0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T., Ito S., Niwa Y., Kunihiro A., Nakamichi N., Mizuno T. (2008). Involvement of Arabidopsis clock-associated pseudo-response regulators in diurnal oscillations of gene expression in the presence of environmental time cues. Plant Cell Physiol. 49: 1839–1850. [DOI] [PubMed] [Google Scholar]
- Yano M., Katayose Y., Ashikari M., Yamanouchi U., Monna L., Fuse T., Baba T., Yamamoto K., Umehara Y., Nagamura Y., Sasaki T. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeang H.-Y. (2013). Solar rhythm in the regulation of photoperiodic flowering of long-day and short-day plants. J. Exp. Bot. 64: 2643–2652. [DOI] [PubMed] [Google Scholar]
- Yerushalmi S., Green R.M. (2009). Evidence for the adaptive significance of circadian rhythms. Ecol. Lett. 12: 970–981. [DOI] [PubMed] [Google Scholar]
- Zar J.H. (1999). Biostatistical Analysis. (Upper Saddle River, NJ: Prentice-Hall; ). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.