Phosphatidylinositol 4-phosphate binds specifically to PDV1 and PDV2, the components of the chloroplast division apparatus, and plays an important role as a negative regulator of chloroplast division.
Abstract
Chloroplast division is performed by the constriction of envelope membranes at the division site. Although constriction of a ring-like protein complex has been shown to be involved in chloroplast division, it remains unknown how membrane lipids participate in the process. Here, we show that phosphoinositides with unknown function in envelope membranes are involved in the regulation of chloroplast division in Arabidopsis thaliana. PLASTID DIVISION1 (PDV1) and PDV2 proteins interacted specifically with phosphatidylinositol 4-phosphate (PI4P). Inhibition of phosphatidylinositol 4-kinase (PI4K) decreased the level of PI4P in chloroplasts and accelerated chloroplast division. Knockout of PI4Kβ2 expression or downregulation of PI4Kα1 expression resulted in decreased levels of PI4P in chloroplasts and increased chloroplast numbers. PI4Kα1 is the main contributor to PI4P synthesis in chloroplasts, and the effect of PI4K inhibition was largely abolished in the pdv1 mutant. Overexpression of DYNAMIN-RELATED PROTEIN5B (DRP5B), another component of the chloroplast division machinery, which is recruited to chloroplasts by PDV1 and PDV2, enhanced the effect of PI4K inhibition, whereas overexpression of PDV1 and PDV2 had additive effects. The amount of DRP5B that associated with chloroplasts increased upon PI4K inhibition. These findings suggest that PI4P is a regulator of chloroplast division in a PDV1- and DRP5B-dependent manner.
INTRODUCTION
Chloroplasts arose from the integration of a cyanobacterial endosymbiont into primitive eukaryotic cells more than 1 billion years ago. Reminiscent of their free-living ancestor, chloroplasts multiply by division (Keeling, 2010), which is initiated by stromal FtsZ (originally named prokaryotic cell division gene, Filamenting temperature-sensitive Z) ring formation at the division site (Vitha et al., 2001; Kuroiwa et al., 2002). ACCUMULATION AND REPLICATION OF CHLOROPLAST6 (ARC6) and PARALOG OF ARC6 (PARC6) inner envelope-spanning proteins regulate FtsZ polymerization and mediate the interactions between the stromal and cytosolic division machinery (Vitha et al., 2003; Glynn et al., 2009). FtsZ and ARC6 are derived from the cell division machinery of the cyanobacterial endosymbiont (Osteryoung and Vierling, 1995; Vitha et al., 2003), and PARC6 is derived from ARC6 by gene duplication and differentiation (Glynn et al., 2009). After stromal FtsZ ring formation, the outer envelope-spanning proteins PLASTID DIVISION1 (PDV1) and PDV2 are recruited to the division site through direct interactions with PARC6 and ARC6 (Glynn et al., 2008, 2009). Finally, DYNAMIN-RELATED PROTEIN5B (DRP5B; also known as ARC5), a member of the dynamin family of self-assembling proteins, is recruited by PDV1 and PDV2 to chloroplasts to complete chloroplast division (Miyagishima et al., 2006). DRP5B is specific to plants and algae and is thought to have evolved from a dynamin-related protein involved in eukaryotic cytokinesis (Miyagishima et al., 2008).
PDV1 and PDV2 are specific to land plants (Miyagishima et al., 2006; Glynn et al., 2008), in which the levels of PDV proteins determine the rate of chloroplast division (Okazaki et al., 2009). The levels of PDV proteins, but not other division components, decrease during leaf development, with a concomitant decline in the rate of chloroplast division. The amount of PDV2 is increased by cytokinin treatment or overexpression of the cytokinin-responsive transcription factor CRF2, with a corresponding increase in the chloroplast division rate. The regulation of PDV2 level by cytokinin enables land plant cells to change chloroplast size and number in coordination with cell differentiation (Okazaki et al., 2009). Although PDV proteins have been shown to have a key function in land plants, the mechanisms that regulate the size and number of chloroplasts are largely unknown. In addition, several other proteins were reported to affect chloroplast division, but most of them showed no direct interactions with the chloroplast division machinery or the mechanisms of regulation were not clear (Basak and Møller, 2013). FHY3, a key regulator of far-red light signaling, and its homolog FRS4 were recently reported to bind the promoter region of DRP5B and activate its expression (Ouyang et al., 2011; Gao et al., 2013). These results showed that transcription factors are also involved in the regulation of chloroplast division, although a relationship between far-red light signaling and chloroplast division was not demonstrated. Mutations in genes involved in lipid synthesis and/or trafficking also affect chloroplast division, suggesting that an inadequate lipid supply and/or alteration of lipid composition limits membrane proliferation during chloroplast division (Wu and Xue, 2010; Ajjawi et al., 2011; Fan and Xu, 2011; Nobusawa and Umeda, 2012). These findings revealed relationships between fundamental cellular processes and chloroplast division, although the mechanism of regulation remains unclear.
Phosphoinositides, the phosphorylated derivatives of phosphatidylinositol (PI), modulate fundamental cellular processes. The inositol ring of PI can be phosphorylated at position 3, 4, or 5 in all possible combinations, and seven phosphorylated derivatives have been detected (Boss and Im, 2012). Phosphatidylinositol 4-phosphate (PI4P) is the most abundant PI molecule in plant cells (Krinke et al., 2007; Munnik and Vermeer, 2010). In addition to being important for signaling as a direct substrate for phospholipase C (Gonorazky et al., 2010) or as a precursor for phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] (Berridge and Irvine, 1989), PI4P itself acts as a signaling lipid (Jung et al., 2002; Preuss et al., 2006; Chapman and Goring, 2011). PI4P is synthesized by phosphatidylinositol 4-kinase (PI4K), and PI4K activities are present in many cellular compartments, including the plasma membrane (Sommarin and Sandelius, 1988), cytosol (Okpodu et al., 1995), cytoskeleton (Xu et al., 1992), and nucleus (Hendrix et al., 1989). The existence of PI kinase activity was reported in chloroplast envelope membranes, and phosphatidylinositol 3-phosphate (PI3P) and PI4P were identified as its products (Bovet et al., 2001). Although the presence of phosphoinositides in chloroplast membranes suggests that signal transduction pathways and/or protein regulation occur in the envelopes, little is known about the functions of the PI phosphates and what genes are involved in their synthesis.
Here, we show that PI4P negatively regulates chloroplast division. PI4P bound specifically to PDV1 and PDV2, and the inhibition of PI4K decreased the level of PI4P in chloroplasts and accelerated chloroplast division. The regulation of chloroplast division by PI4P was mediated primarily by PDV1. A decrease in PI4P caused increased recruitment of DRP5B to chloroplasts and an increased rate of chloroplast division. Our results demonstrate the existence of a lipid signaling pathway involved in chloroplast division and identify PI4P in the chloroplast envelope as a regulator of chloroplast division.
RESULTS
PDV1, PDV2, and DRP5B Exhibit Specific Interactions with Lipids
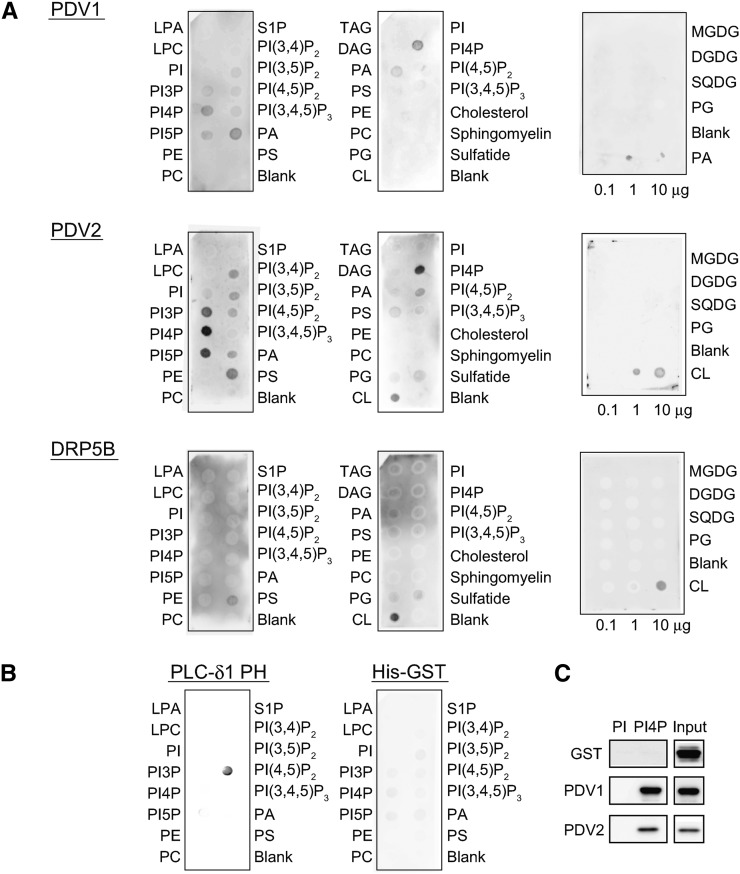
DRP5B belongs to the dynamin protein superfamily and has a pleckstrin homology (PH) domain (Hong et al., 2003). The PH domains of dynamins, which often bind specific lipids such as PI(4,5)P2 or protein ligands, regulate the GTPase activity of their own GTPase domains (Zheng et al., 1996; Barylko et al., 1998). The PH domain of DRP5B was reported to interact with PDV1 and PDV2 (Holtsmark et al., 2013), but whether DRP5B binds to lipids is unknown. PDV1 and PDV2 are integral outer envelope proteins (Miyagishima et al., 2006), so they may exhibit specific interactions with lipids. To examine whether PDV1, PDV2, and DRP5B bind specifically to different lipids, we performed lipid-protein interaction assays. Commercial membranes spotted with different phospholipids and a membrane spotted with chloroplast lipids (Figure 1A) were used. PDV1 bound to PI4P, phosphatidylinositol 5-phosphate, and phosphatidic acid (PA), whereas PDV2 bound to PI4P with high affinity. Weak signals were detected between PDV2 and PI3P, phosphatidylinositol 5-phosphate, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, phosphatidylserine (PS), and cardiolipin (CL) (Figure 1A). DRP5B bound to PS and CL. None of the proteins bound to chloroplast lipids such as monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), sulfoquinovosyldiacylglycerol (SQDG), and phosphatidylglycerol (PG) (Figure 1A). The PH domain of phospholipase Cδ (PLCδ1-PH) used as a positive control interacted specifically with PI(4,5)P2, as reported previously (Kavran et al., 1998), and His-tagged GST alone used as a negative control bound to lipids very weakly (Figure 1B). To confirm that PDV1 and PDV2 bind to PI4P, liposome pull-down assays were also performed. As shown in Figure 1C, PDV1 and PDV2, but not GST, bound to PI4P. Since PI4P is present in chloroplast membranes (Bovet et al., 2001), PDV1 and PDV2 are probably able to interact with PI4P in vivo. Chloroplast membranes contain neither PS nor CL (Douce and Joyard, 1990); thus, they would not be targets for DRP5B binding on the chloroplast membrane. Because DRP5B localizes to peroxisomes (Zhang and Hu, 2010) as well as chloroplasts, and peroxisome membranes probably contain PS, DRP5B may bind to PS on peroxisome membranes. In this study, we focused on the functions of PI4P, which interacts with PDV1 and PDV2, in chloroplast division.
Figure 1.
Binding of Recombinant PDV1, PDV2, and DRP5B Proteins to Lipids.
(A) Membrane binding assays with lipid-spotted membranes. Each spot on the left and middle membranes contained 100 pmol of lipids. The right membranes contained chloroplast lipids, MGDG, DGDG, SQDG, and PG. The chloroplast lipid membranes also contained CL or PA as the control. The membranes were incubated with 0.5 μg/mL GST-PDV1, His-PDV2, or GST-DRP5B. Abbreviations are as follows: LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; S1P, sphingosine 1-phosphate; PI(3,4,5)P3, phosphatidylinositol 3,4,5-trisphosphate; TAG, triacylglycerol; DAG, 1,2-diacylglycerol.
(B) Membrane binding assays with control proteins. PIP Strips were incubated with 0.5 μg/mL His-GST or PLCδ1-PH protein. His-GST and PLCδ1-PH were used as negative and positive controls, respectively.
(C) Liposome pull-down assays with PolyPIPosomes containing PI or PI4P. Ten micrograms of His-GST, GST-PDV1, or His-PDV2 was incubated with 10 μmol of biotinylated liposomes containing 5 mol % of PI or PI4P. Proteins sedimented with liposomes were separated by SDS-PAGE and detected with anti-His antibody. Input denotes 5% of total protein in each assay.
Inhibition of PI4K Increases the Rate of Chloroplast Division in Arabidopsis thaliana
To examine the relationship between chloroplast division and PI4P, we performed PI4K inhibition assays. Wortmannin (WM) inhibits phosphatidylinositol 3-kinase (PI3K) at nanomolar concentrations and inhibits both PI3K and type III PI4K at micromolar concentrations (Matsuoka et al., 1995; Westergren et al., 1999). PI4K and PI3K phosphorylate specific positions in the inositol ring of PI and produce PI4P and PI3P, respectively. Phenylarsine oxide (PAO) and LY294002 are used as specific inhibitors of PI4K and PI3K, respectively (Walker et al., 2000; Vermeer et al., 2009).
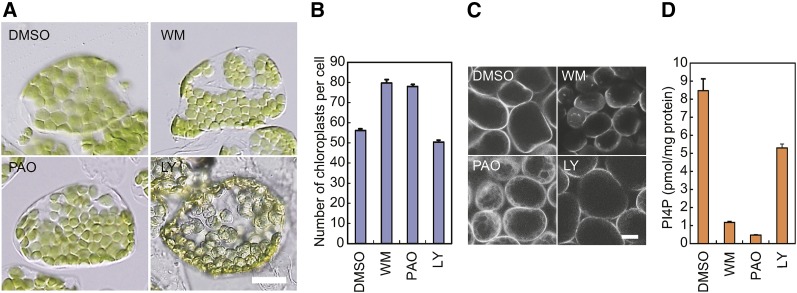
Wild-type seedlings were grown on medium containing 200 μM WM, 25 μM PAO, or 50 μM LY294002 (Figures 2A and 2B). All three inhibitors impaired plant growth. When 4-d-old seedlings were treated with inhibitors for 3 d, fresh weight per seedling was 0.820 ± 0.0234 mg for WM, 1.11 ± 0.0398 mg for PAO, and 1.02 ± 0.0197 mg for LY294002, whereas it was 2.13 ± 0.0838 mg for DMSO used as a control. Even though plant growth was inhibited, the sizes of mesophyll cells were similar between inhibitor-treated and untreated plants when inhibitors were used at the above concentrations. We then compared the size and number of chloroplasts between inhibitor-treated and untreated seedlings. Chloroplasts in the leaf cells of plants treated with 200 μM WM or 25 μM PAO were smaller and more numerous than those in plants grown without inhibitors (Figures 2A and 2B). WM treatment at 50 and 100 μM also increased the number of chloroplasts, but the increase was more significant at 200 μM WM (Supplemental Figure 1). Chloroplasts treated with 50 μM LY294002 were pale and deformed, but the size and number of chloroplasts were similar to those in the untreated plants (Figures 2A and 2B).
Figure 2.
PI4K Inhibition Increases Chloroplast Number, Decreases Chloroplast Size, and Decreases the Level of PI4P.
(A) Chloroplasts of wild-type plants treated with inhibitors of PI4K and/or PI3K. Chloroplasts in a single mesophyll cell are shown. Wild-type 4-d-old seedlings were transferred onto agar plates with 200 μM WM, 25 μM PAO, or 50 μM LY294002 (LY) or onto plates lacking inhibitors (DMSO) and grown for 2 weeks. Bar = 10 μm.
(B) Statistical comparison of the number of chloroplasts per mesophyll cell in inhibitor-treated plants. Error bars represent se (n = 50 cells).
(C) Intracellular localization of the PI4P biosensor, eYFP-PHFAPP1, in the cells of transgenic plants treated with PIK inhibitors. Transgenic plants overexpressing eYFP-PHFAPP1 were treated with WM, PAO, or LY294002, or without inhibitors (DMSO), for 3 d. Images of YFP fluorescence were taken using a confocal laser-scanning microscope. Bar = 20 μm.
(D) PI4P levels in the isolated intact chloroplasts prepared from plants treated with PIK inhibitors. Wild-type plants were treated with WM, PAO, or LY294002, or without inhibitors (DMSO), for 3 d. Intact chloroplasts were isolated, and the amounts of PI4P were measured using a PI(4)P Mass Strip Kit. Error bars represent se (n = 3).
To confirm that the treatments with WM or PAO diminished cellular PI4P, transgenic plants overexpressing the PI4P biosensor, eYFP-PHFAPP1, were treated with WM, PAO, or LY294002 (Figure 2C). PHFAPP1 is a lipid binding domain that specifically binds PI4P (Vermeer et al., 2009). The fluorescence of eYFP-PHFAPP1 was localized to the plasma membrane in the cells of plants treated with LY294002 or DMSO (Figure 2C), indicating that PI4P was mainly present in the plasma membranes of these plants. By contrast, the fluorescence of eYFP-PHFAPP1 was localized mainly in the cytoplasm of PAO-treated cells (Figure 2C), as reported previously (Vermeer et al., 2009), indicating that PI4P-free eYFP-PHFAPP1 was present in the cytoplasm of PAO-treated plants. This finding suggests that the level of PI4P decreased in the cells of PAO-treated plants. In plants treated with WM, eYFP-PHFAPP1 appeared to be partially localized in the cytoplasm.
We measured PI4P levels in the isolated intact chloroplasts prepared from plants treated with WM, PAO, LY294002, or DMSO. When wild-type plants were treated with WM or PAO, PI4P levels in the chloroplasts decreased dramatically compared with that in the chloroplasts of DMSO-treated plants (Figure 2D). On the other hand, the amount of PI4P in the chloroplasts of LY294002-treated plants was only slightly decreased compared with that in the chloroplasts from the DMSO-treated plants.
These results suggest that the level of PI4P was lowered in chloroplasts by treatment with WM or PAO, with an accompanying positive effect on the rate of chloroplast division.
PI4Kα1 Contributes Mainly to the Regulation of Chloroplast Division
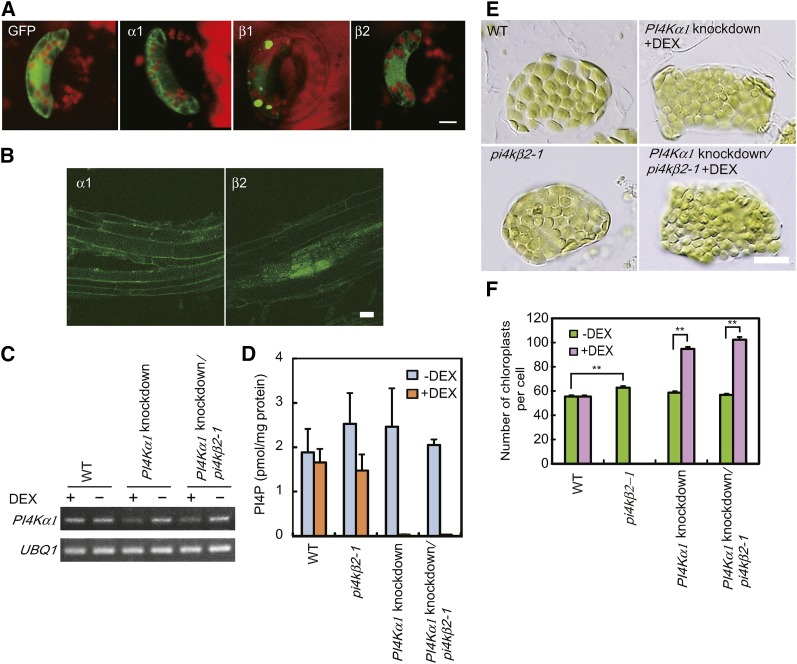
Four PI4K genes, PI4Kα1, PI4Kα2, PI4Kβ1, and PI4Kβ2, have been identified in Arabidopsis (Müller-Röber and Pical, 2002). To identify the PI4Ks involved in production of PI4P in the outer envelope membrane of chloroplasts, green fluorescent protein (GFP) fusion proteins were transiently expressed under the control of the cauliflower mosaic virus 35S promoter (35S) in guard cells of broad bean (Vicia faba). Because we were not able to obtain the PI4Kα2 transcripts encoding an active PI4K catalytic domain, we used PI4Kα1-GFP, PI4Kβ1-GFP, and PI4Kβ2-GFP fusion constructs. PI4Kβ1-GFP fluorescence exhibited a dot-like pattern (Figure 3A). PI4Kβ1 was shown previously to be localized to the trans-Golgi network (TGN; Kang et al., 2011). PI4Kβ1 interacts with RabA4b, a Rab GTPase specifically localized to the TGN, through a novel homology domain specific to eukaryotic type IIIβ PI4K (Preuss et al., 2006). The dot-like pattern of fluorescence may indicate that PI4Kβ1-GFP is localized to the TGN. By contrast, diffused fluorescence was observed throughout the cytosol in cells expressing PI4Kα1-GFP and PI4Kβ2-GFP (Figure 3A).
Figure 3.
Intracellular Localization of PI4K-GFP, Increased Chloroplast Number, Decreased Chloroplast Size, and Decreased Level of PI4P in PI4Kα1 Knockdown and/or PI4Kβ2 Knockout Plants.
(A) GFP, PI4Kα1-GFP, PI4Kβ1-GFP, and PI4Kβ2-GFP were transiently overexpressed in guard cells of broad bean. Images of GFP and chlorophyll fluorescence were taken using a confocal laser-scanning microscope. Bar = 10 μm.
(B) GFP-PI4Kα1 and GFP-PI4Kβ2 were expressed under the control of their respective promoters in Arabidopsis. Root cells of transgenic plants are shown. Bar = 20 μm.
(C) PI4Kα1 expression was diminished by PI4Kα1 expression knockdown in transgenic plants. Expression of amiRNAs with high specificity to PI4Kα1 was induced by treatment with (+) DEX in wild-type or pi4kβ2-1 mutant backgrounds. Total RNA extracted from whole plants was used for RT-PCR. UBQ1 was used as an internal control. Signal intensities were estimated based on ethidium bromide staining. Two biological replicates showed equivalent results.
(D) PI4P levels in the isolated intact chloroplasts decreased upon knockdown of PI4Kα1. Wild-type, pi4kβ2-1 mutant, and PI4Kα1 knockdown plants were grown for 4 d on MS agar plates and then transferred onto agar plates with or without 5 μM DEX and grown for 1 week. Intact chloroplasts were isolated, and the amounts of PI4P were measured using a PI(4)P Mass Strip Kit. Error bars represent se (n = 3).
(E) Chloroplasts in mesophyll cells of wild-type, pi4kβ2-1 mutant, and PI4Kα1 knockdown plants. Chloroplasts in a single mesophyll cell are shown. Four-day-old seedlings were transferred to agar plates with or without 5 μM DEX and grown for 2 weeks. Bar = 10 μm.
(F) Statistical comparison of the number of chloroplasts per mesophyll cell in DEX-treated or untreated wild-type, pi4kβ2-1 mutant, and PI4Kα1 knockdown plants. Error bars represent se (n = 50 cells). **P < 0.0001 by Student’s t test.
To further examine the localization of PI4Kα1 and PI4Kβ2, we also expressed GFP-PI4Kα1 and GFP-PI4Kβ2 under the control of their respective promoters in Arabidopsis. When GFP-PI4Kα1 was expressed in PI4Kα1/pi4kα1 heterozygous plants, pi4kα1 homozygous plants carrying a GFP-PI4Kα1 gene were obtained. Because null mutations of PI4Kα1 are lethal (Delage et al., 2012), we conclude that the GFP-PI4Kα1 transgene complemented the pi4kα1 mutation and that GFP-PI4Kα1 is functional. GFP-PI4Kα1 and GFP-PI4Kβ2 signals were detected in leaf and root cells, but the signals of GFP-PI4Kα1 and GFP-PI4Kβ2 in plants expressing GFP-PI4Kα1 and GFP-PI4Kβ2 driven by their own promoters were too weak to determine the intracellular localization, especially in mesophyll cells, because of the high level of background chlorophyll fluorescence. Strong signals were detected in the anthers of GFP-PI4Kα1- or GFP-PI4Kβ2-expressing plants and in the stigma of GFP-PI4Kα1 plants (Supplemental Figure 2). As shown in Figure 3B, fluorescence was observed in the plasma membrane and throughout the cytosol in root cells of GFP-PI4Kα1- or GFP-PI4Kβ2-expressing plants. Bright speckles in the cytosol were also observed in some cells of both plants. PI4Kα1 has a PH domain, which binds specifically to PI4P, and the GFP fusion protein is localized in perinuclear membranes when it is expressed in Sf9 insect cells (Stevenson-Paulik et al., 2003). Our results suggest that PI4Kα1 localizes in membranes and the cytosol, as does PI4KIIIα in mammalian cells (Kakuk et al., 2006). The localization of PI4Kβ2 has not been demonstrated, although it was reported that the novel homology domain of PI4Kβ2 interacts with RabA4b as does that of PI4Kβ1 (Preuss et al., 2006). Therefore, the speckles in the cytosols of GFP-PI4Kβ2-expressing plants may indicate that GFP-PI4Kβ2 is localized to the TGN by RabA4b recruitment as does PI4Kβ1. However, the finding that PI4Kβ2-GFP signal was also detected in the plasma membrane and throughout the cytosol suggests that a part of PI4Kβ2 is not recruited by RabA4b, in contrast with the complete colocalization of PI4Kβ1 and RabA4b (Preuss et al., 2006). These results suggest that PI4Kα1 and PI4Kβ2 in the cytosol may convert PI to PI4P in the chloroplast outer envelope.
We examined whether PI4Kα1 or PI4Kβ2 is involved in the regulation of chloroplast division using the previously characterized pi4kβ2-1 (SALK_098069) null mutant (Preuss et al., 2006). The number of chloroplasts was slightly but significantly higher in pi4kβ2-1 compared with the wild type (Figures 3E and 3F; Student’s t test, one-tailed, P < 0.0001). Because plants homozygous for a T-DNA insertion in PI4Kα1 could not be produced due to embryonic lethality (Delage et al., 2012), we generated transgenic plants in which PI4Kα1 expression was knocked down. Artificial microRNAs (amiRNAs; Schwab et al., 2006) with high specificity for PI4Kα1 were expressed in wild-type and pi4kβ2-1 mutant plants under the control of dexamethasone (DEX)-inducible promoters (Aoyama and Chua, 1997) to downregulate PI4Kα1 expression by RNA interference (Figure 3C). When PI4Kα1 expression was transiently knocked down (Figure 3C), the levels of PI4P decreased in chloroplasts (Figure 3D), the number of chloroplasts increased, and their size was diminished compared with noninduced plants (Figures 3E and 3F). These results indicate that PI4Kα1 and PI4Kβ2 negatively regulate the rate of chloroplast division and that PI4Kα1 is the main contributor to the regulation.
The Effects of PI4K Inhibition Are Decreased by Deletion of PDV1 and Enhanced by Overexpression of DRP5B
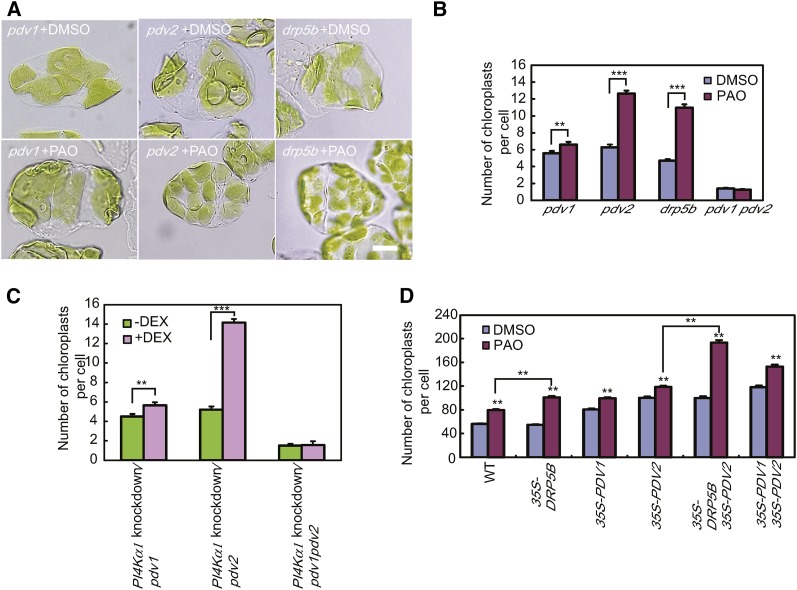
Because PDV1 and PDV2 bound PI4P, and DRP5B interacts with PDV1 and PDV2 (Holtsmark et al., 2013), we examined whether PDV1, PDV2, and DRP5B are involved in the regulation of chloroplast division by PI4P. We treated the pdv1, pdv2, and drp5b mutants and a pdv1 pdv2 double mutant with PAO (Figures 4A and 4B). The number of chloroplasts was 2-fold higher in pdv2 and drp5b mutant plants grown with 25 μM PAO than without PAO. By contrast, PAO treatment had much less of an effect on chloroplast number in the pdv1 mutant, although the difference between PAO-treated and untreated plants was significant (Student’s t test, one-tailed, P < 0.01). The number of chloroplasts was similar in PAO-treated and untreated pdv1 pdv2 double mutant plants. Knockdown of PI4Kα1 in the pdv1, pdv2, and pdv1 pdv2 mutants caused similar results (Figure 4C). These results indicate that PDV1 has a major role in the regulation of chloroplast division mediated by PI4P.
Figure 4.
Effects of PAO Treatment on the Chloroplast Division Rate in pdv1, pdv2, pdv1 pdv2, and drp5b Mutants and PDV1-, PDV2-, and/or DRP5B-Overexpressing Plants.
(A) Chloroplasts of pdv1, pdv2, pdv1 pdv2, and drp5b mutant plants treated or not with 25 μM PAO. Bar = 10 μm.
(B) Statistical comparison of the number of chloroplasts per mesophyll cell of mutant plants treated or not with 25 μM PAO. Error bars represent se (n = 50 cells). **P < 0.01 and ***P < 0.0001 by Student’s t test.
(C) Statistical comparison of the number of chloroplasts per mesophyll cell in PI4Kα1 knockdown/pdv1, PI4Kα1 knockdown/pdv2, and PI4Kα1 knockdown/pdv1 pdv2 mutants. Four-day-old seedlings were transferred onto agar plates with or without 5 μM DEX and grown for 2 weeks. Error bars represent se (n = 50 cells). **P < 0.01 and ***P < 0.0001 by Student’s t test.
(D) Statistical comparison of the number of chloroplasts per mesophyll cell in 25 μM PAO-treated and untreated plants overexpressing PDV1, PDV2, and/or DRP5B. Error bars represent se (n = 50 cells). **P < 0.0001 by Student’s t test.
We also treated plants overexpressing PDV1, PDV2, or DRP5B with PAO. Overexpression of PDV1 and/or PDV2 resulted in an increased number of chloroplasts in the transgenic plants compared with wild-type plants, as reported previously (Okazaki et al., 2009). PAO treatment caused an additional increase in chloroplast numbers in the overexpressing lines (Figure 4D). By contrast, the numbers of chloroplasts in DRP5B-overexpressing plants and wild-type plants were similar, as reported previously (Okazaki et al., 2009), but the effect of PAO treatment was enhanced by the overexpression of DRP5B (Figure 4D). The number of chloroplasts was 1.85 times higher in 35S-DRP5B plants treated with PAO than in untreated 35S-DRP5B plants, but it was only 1.39 times higher in wild-type plants treated with PAO than in untreated wild-type plants (Student’s t-test, one-tailed, P < 0.0001). Chloroplast number increased 1.93-fold in 35S-PDV2 35S-DRP5B plants by PAO treatment but increased only 1.18-fold in 35S-PDV2 plants treated with PAO (Student’s t test, one-tailed, P < 0.0001). These results indicate that DRP5B is also involved in the regulation of chloroplast division mediated by PI4P, although DRP5B is not indispensable to the regulation.
PAO Treatment Increases the Amount of DRP5B Associated with the Chloroplast Surface
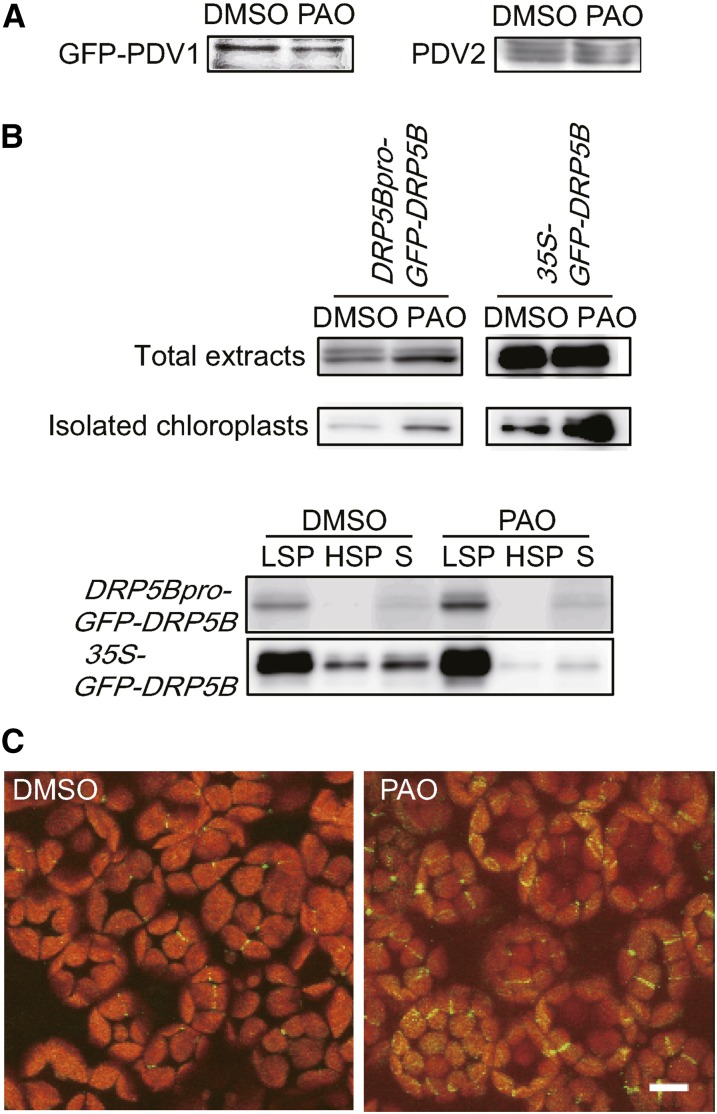
PDV protein levels were shown previously to determine the rate of chloroplast division (Okazaki et al., 2009). Elevated levels of PDV proteins increase the number of chloroplasts and decreased PDV levels have the opposite effect, suggesting that PI4P might regulate the levels of PDV proteins. Therefore, we compared the levels of PDV1 and PDV2 in PAO-treated and untreated plants (Figure 5A). GFP-PDV1 was expressed under the control of the PDV1 promoter in the homozygous pdv1 knockout mutant, and the expressed protein was detected using an anti-GFP antibody. PDV2 expression was also detected in the same samples using an anti-PDV2 antibody. Immunoblot analyses showed that the levels of PDV1 and PDV2 were similar in the PAO-treated and untreated plants (Figure 5A). These results suggest that the increase in chloroplast number in the cells of PAO-treated plants was not caused by an increase in PDV protein levels. We also compared the levels of DRP5B in PAO-treated and untreated plants (Figure 5B). GFP-DRP5B was expressed under the control of the DRP5B promoter in the homozygous drp5b knockout mutant. The level of DRP5B increased in the total extracts and isolated intact chloroplasts of PAO-treated drp5b plants (Figure 5B). When total extracts of GFP-DRP5B-expressing plants were fractionated by centrifugation, GFP-DRP5B was detected primarily in the low-speed pellet (LSP), consisting mostly of chloroplasts, and partly in the soluble fraction (Figure 5B). PAO treatment significantly increased GFP-DRP5B levels in the LSP fractions of drp5b plants expressing GFP-DRP5B under the control of either the DRP5B or 35S promoter. In addition, the level of DRP5B in the soluble fraction of GFP-DRP5B-overexpressing drp5b plants treated with PAO decreased relative to untreated overexpressing plants, even though the total DRP5B level increased slightly (Figure 5B). Overexpression of PDV2 had no effect on the level or localization of DRP5B (Supplemental Figure 3A).
Figure 5.
PAO Treatment Increases DRP5B Expression and Enhances DRP5B Recruitment to Chloroplasts.
(A) Levels of GFP-PDV1 and PDV2 in pdv1 plants expressing GFP-PDV1 under the control of the PDV1 promoter. Plants were treated with DMSO or 25 μM PAO for 3 d. Total extracts containing 50 μg of protein were loaded for analysis of GFP-PDV1 and PDV2. GFP-PDV1 and PDV2 were detected with anti-GFP and anti-PDV2 antibodies, respectively.
(B) Levels of GFP-DRP5B in drp5b plants expressing GFP-DRP5B under the control of the DRP5B promoter (DRP5Bpro-GFP-DRP5B) and the cauliflower mosaic virus 35S promoter (35S-GFP-DRP5B). In total extracts, 50 and 2.5 μg of proteins extracted from whole plants were loaded for analysis of DRP5Bpro-GFP-DRP5B and 35S-GFP-DRP5B, respectively. In isolated chloroplasts, 2 and 0.2 μg of proteins were loaded for analysis of DRP5Bpro-GFP-DRP5B and 35S-GFP-DRP5B, respectively. GFP-DRP5B was detected with an anti-GFP antibody. In fractionated samples, homogenates prepared from 30 to 50 mg of leaves were fractionated into LSP, high-speed pellet (HSP), and supernatant (S) fractions by centrifugation. Two biological replicates showed equivalent results.
(C) DRP5Bpro-GFP-DRP5B drp5b plants were treated with DMSO (left panel) or 25 μM PAO (right panel) for 3 d. Images of GFP and chlorophyll fluorescence were taken using a confocal laser-scanning microscope. Bar = 10 μm.
To further examine the localization of DRP5B, we observed DRP5Bpro-GFP-DRP5B drp5b plants treated with DMSO or PAO using fluorescence microscopy. GFP-DRP5B signals were observed with higher frequency at the midpoint of chloroplasts in the cells of DRP5Bpro-GFP-DRP5B plants treated with PAO than in untreated plants (Figure 5C). In several chloroplasts of PAO-treated plants, GFP-DRP5B fluorescence was visible as small puncta distributed over the chloroplast envelope (Figure 5C). GFP-DRP5B signals associated with the chloroplast surface also increased in the cells of 35S-GFP-DRP5B plants treated with PAO compared with untreated plants (Supplemental Figure 3B). Large aggregations were often observed in the cytosol of untreated 35S-GFP-DRP5B plants; these aggregations were diminished in the cells of PAO-treated plants. These results indicate that PI4P affects the localization of DRP5B, which is recruited to chloroplasts by PDV1 and PDV2 (Miyagishima et al., 2006; Glynn et al., 2008, 2009; Holtsmark et al., 2013), and our results highlight the importance of PDV1 for PI4P regulation (Figures 4A and 4B). We conclude that PI4P regulates the recruitment of DRP5B to chloroplasts by PDV1.
DISCUSSION
In this study, we showed that PI4K inhibition increased the rate of chloroplast division. Two kinds of PI4K inhibitor treatments and downregulation of PI4K expression similarly increase the number of chloroplasts, indicating that PI4P is a negative regulator of chloroplast division. Inhibition of PI4K caused an increase of chloroplast division in parallel with an increase in the amount of DRP5B localized on the surface of chloroplasts (Figures 5B and 5C). These results suggest that the binding of PI4P to PDV1 changes the interaction between PDV1 and DRP5B. Decrease of PI4P in envelope membranes probably causes increases in the binding affinity of PDV1 for DRP5B or inhibits the dissociation of DRP5B from chloroplasts. As a result, the recruitment of DRP5B to chloroplasts increases and may enhance the rate of chloroplast division. In addition, the total protein level of DRP5B was increased by the PAO treatment (Figure 5B). The increase in DRP5B recruited to chloroplasts suggests that the expression of DRP5B is increased or its degradation is reduced.
Although our results show that DRP5B is involved in the regulation of chloroplast division mediated by PI4P, the increase in chloroplast number in PAO-treated drp5b mutant plants indicates that DRP5B-independent regulation also exists. The pdv1 pdv2 double mutation abolished the response to changes in PI4P levels, indicating that PDVs are involved in the DRP5B-independent regulation as well as in the DRP5B-dependent regulation. The pdv1 pdv2 double mutant was shown previously to exhibit a more severe phenotype than the drp5b mutant (Miyagishima et al., 2006; Figure 4B). We suggest that proteins other than DRP5B are involved in chloroplast division and its regulation by PI4P and PDV. One possibility is that other members of the DRP superfamily play a role, as in peroxisomal or mitochondrial division (Zhang and Hu, 2009, 2010; Aung and Hu, 2012). Identification and characterization of such proteins will provide insights into chloroplast division and its regulation.
Previous studies showed that PDV1 and PDV2 are homologs and have partial functional redundancy (Miyagishima et al., 2006; Okazaki et al., 2009). Our results indicate several differences between PDV1 and PDV2. Lipid-protein interaction assays revealed that the lipid binding specificity of PDV1 differed from that of PDV2, although both PDV1 and PDV2 bound to PI4P (Figure 1A), suggesting that the lipid binding regions of PDV1 are distinct from those of PDV2. PDV1 exhibited relatively weak signals in lipid-protein interaction assays but strong signals in liposome pull-down assays, which are thought to be more similar to the in vivo condition. On the other hand, PDV2 showed specific interaction with lipids in both lipid-protein interaction assays and liposome pull-down assays (Figures 1A and 1C). This finding also suggests that the lipid recognition site of PDV1 is structurally different from that of PDV2. Furthermore, the regulation of chloroplast division by PI4P is almost entirely via PDV1. It can be assumed that the binding of PI4P to PDV1 causes a structural change of PDV1 and influences the interaction between PDV1 and DRP5B. Further biochemical investigations will be required to understand why the binding affinity of PDV1 with PI4P is different from that of PDV2 and how the binding of PDV1 with PI4P influences the recruitment of DRP5B to the chloroplast envelope.
The binding of PI4P to PDV2 probably causes a minor change or a structural change that does not affect the interaction between PDV2 and DRP5B. However, the number of chloroplasts increased slightly but significantly in PAO-treated pdv1 mutant plants compared with untreated plants, but PAO treatment did not increase chloroplast numbers in the pdv1 pdv2 double mutant (Figure 4B). The results of lipid-protein interaction assays and liposome pull-down assays suggest that PI4P interacts with PDV2 as well as PDV1 on chloroplast envelopes (Figures 1A and 1C). These results suggest that PDV2 may contribute the remainder of the overall response to alterations in PI4P levels in the pdv1 mutant.
PDV1 and PDV2 specifically bound PI4P (Figures 1A and 1C). Lipid binding specificities of PDV proteins must be important for the regulation by PI4P. Major lipid constituents of chloroplast envelopes, such as MGDG, DGDG, SQDG, PG, and phosphatidylcholine, did not interact with PDV1 and PDV2 (Figure 1A). PI4P is a minor lipid, but it is involved in major biological processes and regulates the activities of many proteins. PDV1 and PDV2 are key components in the regulation of chloroplast division (Okazaki et al., 2009); thus, the regulation of PDV1 activity by PI4P links chloroplast division to developmental processes and environmental responses. Although PI and its phosphorylated derivatives were detected in chloroplasts (Douce and Joyard, 1990; Bovet et al., 2001), their functions in chloroplasts have not been clarified. In this study, we clarified that PI4P negatively regulates chloroplast division and the function of phosphoinositides in chloroplasts.
Our results indicate that PI4Kα1 is the main contributor to the regulation of chloroplast division by changing the level of PI4P in chloroplasts (Figures 3E and 3F). Phosphorylation of PI in chloroplasts was reported to be sensitive to WM (Bovet et al., 2001), which was consistent with a previous study showing that PI4Kα1 was inhibited by WM (Stevenson-Paulik et al., 2003). Although we demonstrated the relationship between PI4Kα1 and chloroplast division, the functions of PI4Kα1 have not been thoroughly studied because null mutations of the PI4Kα1 gene are lethal (Delage et al., 2012). Further investigations into PI4Kα1 function will provide important insights into the phosphoinositide signaling pathways in the chloroplast envelope.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana (Columbia-0) was used as the wild-type control and for plant transformation in this study. The pdv1 and pdv2 mutants used in this study were pdv1-1 (contains a premature stop codon close to the start codon) and pdv2-1 (T-DNA insertion in the first intron), respectively (Miyagishima et al., 2006). pdv1 mutant plants transformed with ProPDV1-GFP-PDV1, drp5b mutant plants transformed with ProDRP5B -GFP-DRP5B, and wild-type plants transformed with Pro35S-GFP-DRP5B, Pro35S-PDV1, and Pro35S-PDV2 constructs were generated as described previously (Okazaki et al., 2009). ProDRP5B-GFP-DRP5B and Pro35S-DRP5B plants were crossed with Pro35S-PDV2 plants to produce plants overexpressing PDV2 and both DRP5B and PDV2. Arabidopsis seeds were surface-sterilized, sown on Murashige and Skoog (MS) plates, and stratified at 4°C for 48 h in darkness before germination. Plants were grown in controlled-environment chambers with continuous light (100 µmol m−2 s−1) at 23°C. For treatments with PI3K and PI4K inhibitors, 4-d-old seedlings were transferred onto MS plates supplemented with 200 μM WM (Wako), 25 μM PAO (Wako), 50 μM LY294002 (Wako), or without inhibitors and grown for 2 weeks after the transfer. For transient knockdown of PI4Kα1 expression, 4-d-old seedlings of transgenic plants expressing amiRNAs were transferred onto MS plates supplemented with 5 μM DEX.
Expression and Purification of GST- and His-Tagged Proteins
The coding regions of DRP5B, PDV1, and PDV2 were amplified by RT-PCR with the following primer sets: 5′-GGATCCGATGGCGGAAGTATCAGCAAAA-3′ and 5′-GAATTCCCATGCTGCACCGAAGGAGCCTT-3′ for DRP5B (the BamHI and EcoRI sites are underlined, respectively); 5′-AAAGGATCCGATGGGAGAAATGGAGATCGAA-3′ and 5′-TTTGAATTCCCACCACGAGCCATCATTACGTC-3′ for PDV1 (the BamHI and EcoRI sites are underlined, respectively); and 5′-CACCATGGAAGACGAAGAAGGCATC-3′ and 5′-TCAACCGTATCCGTAAGTTAC-3′ for PDV2. Amplified cDNA fragments for DRP5B and PDV1 were digested with BamHI and EcoRI and cloned into the pET-49b(+) vector (Novagen). Amplified cDNA fragments of PDV2 were cloned into the pET100/D-TOPO vector (Invitrogen) according to the manufacturer’s instructions. The resultant constructs were used for the transformation of Escherichia coli BL21 cells, and the obtained transformants were used for the expression of recombinant proteins. The expressed recombinant proteins were purified using GSTrap FF or HisTrap FF columns according to the manufacturer’s protocols (GE Healthcare).
Lipid-Protein Interaction Assay
PIP Strips and Membrane Lipid Strips were bought from Echelon Biosciences. PA and CL were purchased from Sigma-Aldrich. MGDG, DGDG, SQDG, and PG were purified from the cyanobacterium Synechocystis sp PCC 6803. Lipids were spotted onto Hybond-ECL membranes (GE Healthcare). The strips were first blocked with 3% fatty acid-free BSA in PBS (10 mM phosphate and 150 mM NaCl, pH 7.4) for 1 h and incubated overnight at 4°C in blocking buffer containing 0.5 μg/mL GST-PDV1, His-PDV2, GST-DRP5B, His-GST, or PLCδ1-PH protein. To detect the proteins, an anti-penta-His mouse monoclonal antibody (Qiagen) and an anti-GST mouse monoclonal antibody (Sigma-Aldrich) were used at 1:2000 and 1:1000 dilution, respectively.
Liposome Pull-Down Assay
PolyPIPosomes (Echelon Biosciences) contain phosphatidylcholine (65 mol %), phosphatidylethanolamine (29 mol %), biotin-phosphatidylethanolamine (1 mol %), and PI or PI4P (5 mol %). PolyPIPosomes (10 μmol) were diluted in 1 mL of binding buffer (20 mM HEPES-KOH, pH 7.5, 150 mM KCl, and 0.05% Nonidet P-40) and incubated overnight at 4°C with 10 μg of His-GST, GST-PDV1, or His-PDV2 protein. Samples were centrifuged at 15,000g for 10 min at 4°C, and pellets were washed in 1 mL of binding buffer three times. The bound proteins were resolved by SDS-PAGE. Immunoblotting assays were performed as described previously (Nakanishi et al., 2009). An anti-His antibody was used at a dilution of 1:2000.
Measurement of PI4P Levels
For treatments with PI3K and PI4K inhibitors, wild-type plants were grown for 4 d on MS agar plates and then transferred onto agar plates containing 200 μM WM, 25 μM PAO, or 50 μM LY294002 or onto plates lacking inhibitors (DMSO) and grown for 3 d. For transient knockdown of PI4Kα1 gene expression, 4-d-old seedlings were transferred onto MS plates with or without 5 μM DEX and grown for 1 week. Intact chloroplasts were isolated as described previously (Nakanishi et al., 2009) with the following modifications. Seedlings (0.5 to 1 g) were homogenized in 6 mL of grinding buffer (0.33 M sorbitol, 30 mM HEPES-KOH, pH 7.5, and 2 mM EDTA) containing protease inhibitor cocktail (Nakarai). The homogenate was filtered through a Miracloth (Calbiochem) and then centrifuged at 2500g for 15 min at 4°C. The obtained pellet was resuspended in grinding buffer, and Percoll was added to a final concentration of 20% (v/v). The suspension was overlaid onto a stepwise Percoll gradient (40 and 80% [v/v]) and then centrifuged at 3200g for 30 min at 4°C. A green band corresponding to intact chloroplasts at the interface between 40 and 80% Percoll was collected, washed twice with grinding buffer by centrifugation at 700g for 5 min at 4°C, and used as intact chloroplasts. Lipids were extracted from the isolated chloroplasts according to the manufacturer’s protocols, and the level of PI4P was measured with a PI(4)P Mass Strip Kit (Echelon Biosciences).
Transient Overexpression of PI4K in Broad Bean Cells
For transient overexpression of PI4Kα1-GFP, PI4Kβ1-GFP, and PI4Kβ2-GFP, cDNA fragments containing respective open reading frame sequences were amplified by RT-PCR with the following primer sets: 5′-ACGCGTCGACATGGAGGCACTGACGGAGCT-3′ and 5′-TCGCGTCGACCTTCTCGATGCCTTGTTGCA-3′ for PI4Kα1; 5′-ACGCGTCGACATGCCGATGGGACGCTT-3′ and 5′-TCGCGTCGACCAATATTCCATTTAAGACC-3′ for PI4Kβ1; and 5′-ACGCGTCGACATGCAGATGGCACAGTT-3′ and 5′-TCGCGTCGACTCGTATTCCATTCAACAC-3′ for PI4Kβ2 (the SalI sites are underlined). The amplified cDNA fragments were cloned into a pGEM-T Easy vector (Promega). The subcloned fragments were digested with SalI and ligated into the SalI site (downstream of the cauliflower mosaic virus 35S promoter) of the pTH2 vector (Niwa, 2003). The plasmids were introduced into leaves of broad bean (Vicia faba) using a particle bombardment device (PDS-1000/He Biolistic Particle Delivery System; Bio-Rad) according to the manufacturer’s instructions. Conditions were 1350 p.s.i. of helium gas pressure, a distance of 9 cm between the macrocarrier and the sample, and a decompression vacuum of 28 inches of Hg. Tungsten particles (1 μm) were used as plasmid DNA carriers. The bombarded samples were incubated overnight at room temperature in darkness. The epidermis was stripped from the bombarded leaves after the incubation and used for the observation of red fluorescence from chlorophyll and green fluorescence from GFP using a confocal laser-scanning microscope.
Generation of Transgenic Arabidopsis Plants Expressing GFP-PI4Kα1, GFP-PI4Kβ2, or amiRNAs
For the expression of GFP-PI4Kα1 and GFP-PI4Kβ2 by their respective promoters, we amplified the PI4Kα1 and PI4Kβ2 open reading frame sequences using the primers 5′-AGGAGGAGGTACCATGGAGGCACTGACGGA-3′ (the KpnI site is underlined, and overlaps are indicated by boldface letters) and 5′-AAAGCGGCCGCTTACTTCTCGATGCCTTGT-3′ (the NotI site is underlined) for PI4Kα1 and 5′-AGGAGGAGGTACCATGCCGATGGGACGCTT-3′ (the KpnI site is underlined, and overlaps are indicated by boldface letters) and 5′-AAAGCGGCCGCTCACAATATTCCATTTAAGACC-3′ (the NotI site is underlined) for PI4Kβ2, and the GFP sequence was amplified by primers 5′-AAAGGATCCATGGTGAGCAAGGGCGAGGAG-3′ and 5′-TTTGGTACCTCCTCCTCCCTTGTA-3′ (the BamHI sites are underlined). Amplified PI4Kα1 or PI4Kβ2 fragments were mixed with the GFP fragment and fused by overlap-extension PCR using primers 5′-AAAGGATCCATGGTGAGCAAGGGCGAGGAG-3′ (the BamHI site is underlined) and 5′-AAAGCGGCCGCTTACTTCTCGATGCCTTGT-3′ or 5′-AAAGCGGCCGCTCACAATATTCCATTTAAGACC-3′ (the NotI sites are underlined). The fused fragment was cloned into pGEM-T Easy. The 1.2-kb 5′ upstream sequences of PI4Kα1 and GFP-PI4Kβ2, including the start codon, were amplified by primers 5′-AAATCTAGAGCGGCCGCGTGAGAGAAATATGCAATTGC-3′ (the XbaI and NotI sites are underlined) and 5′-TTTTCTAGAAATGTCACAAAGCTCCGTCAG-3′ (the XbaI site is underlined) for PI4Kα1 and 5′-AAATCTAGAGCGGCCGCAACCGTCGGTGTTCCTCGTAA-3′ (the XbaI and NotI sites are underlined) and 5′-TTTTCTAGAAACCAATGAAAGAAACTGTGC-3′ (the XbaI site is underlined) for PI4Kβ2. Promoter fragments were digested with XbaI and cloned upstream of GFP in the plasmid constructs. The resulting ProPI4Kα1-GFP-PI4Kα1 and ProPI4Kβ2-GFP-PI4Kβ2 fusion constructs were excised with NotI and then transferred into pMLBART (Vitha et al., 2001; conferring resistance to glufosinate ammonium).
For transient knockdown of PI4Kα1 expression, transgenic plants that expressed amiRNAs (Schwab et al., 2006) under the control of DEX-inducible promoters (Aoyama and Chua, 1997) were generated. PI4Kα1 amiRNAs were designed using Web MicroRNA Designer (http://wmd.weigelworld.org) software. A miR319a sequence was amplified by RT-PCR, and the target sequences were replaced by overlap-extension PCR. The miR319a sequence was amplified using the primer set 5′-CTCGAGCAAACACACGCTCGGACGCAT-3′ (the XhoI site is underlined) and 5′-CATGGCGATGCCTTAAATAAAGA-3′ and cloned into a pGEM-T Easy vector. The cloned miR319a sequence was further amplified by PCR with the following three primer sets: 5′-GTTTTCCCAGTCACGAC-3′ and 5′-GAAAAACTAACCAGACATGATGTTCTACATATATATTCCT-3′, 5′-GAACGTCATGTCTGGATAGTTTATCAAAGAGAATCAATGA-3′ and 5′-GAACATCATGTCTGGTTAGTTTTTCACAGGTCGTGATATG-3′, and 5′-GATAAACTATCCAGACATGACGTTCTCTCTTTTGTATTCC-3′ and 5′-CAGGAAACAGCTATGAC-3′ (overlaps are indicated by boldface letters). The three amplified DNA fragments were mixed and fused by PCR using the primer set 5′-GTTTTCCCAGTCACGAC-3′ and 5′-CAGGAAACAGCTATGAC-3′. The amplified DNA fragment was digested with SpeI and XhoI and cloned into a pTA7002 vector (Aoyama and Chua, 1997).
All constructs were transferred to Agrobacterium tumefaciens GV3101 and introduced into the wild type and pi4kβ2 (SALK_098069; Preuss et al., 2006) homozygous mutant or pi4kα1 (GK_502D11) heterozygous mutant plants. T1 plants were selected for resistance to glufosinate ammonium or hygromycin B. A PI4Kα1 knockdown plant was crossed with the pdv1 pdv2 mutant to produce PI4Kα1 knockdown/pdv1, PI4Kα1 knockdown/pdv2, and PI4Kα1 knockdown/pdv1 pdv2 mutants.
Downregulation of PI4Kα1 was confirmed by RT-PCR analysis. Total RNA was extracted using an RNeasy Mini Kit (Qiagen). Four-day-old seedlings were transferred onto MS plates with or without 5 μM DEX and grown for 1 week. RNA (0.8 μg) was reverse-transcribed using the oligo(dT) (20 nucleotides) primer. The resulting cDNA was used as a template for PCR. Before comparing the expression levels, we confirmed that the amplification was in the linear range by comparing different cycles of amplification by ethidium bromide staining. PCR was performed using the primer set 5′-TGGCGTAAAATGAAGGCCTGT-3′ and 5′-TCCGGCTTCTTAACAGCAACA-3′. As a control, UBQ1 cDNA was amplified using the primers 5′-GGCCAAGATCCAAGACAAAG-3′ and 5′-GTTGACAGCTCTTGGGTGAA-3′. The number of PCR cycles was 30 for PI4Kα1 and 25 for UBQ1.
Microscopic Observation
Observations of chloroplasts in Arabidopsis leaf cells were performed as described previously (Okazaki et al., 2009). Samples were observed using Nomarski optics. Images of GFP and chlorophyll fluorescence were taken using a confocal laser-scanning microscope.
Antibodies and Immunoblotting
Four-day-old seedlings were transferred onto MS plates with or without 25 μM PAO and grown for 3 d. Plants were ground with a mortar and pestle on ice and homogenized in extraction buffer (50 mM Tris, pH 7.5, 2 mM MgCl2, 5 mM EDTA, and a protease inhibitor mixture [cOmplete ULTRA; Roche Applied Science]). The homogenate was filtered through Miracloth (Calbiochem). The protein concentration of the homogenate was determined, and then the homogenate was subjected to fractionation and immunoblotting. The total extract was centrifuged at 20,000g for 20 min at 4°C to sediment an LSP. The supernatant fraction was further centrifuged at 100,000g for 1 h at 4°C to obtain a high-speed microsomal pellet and a supernatant fraction. Isolation of intact chloroplasts was performed as described in Measurement of PI4P Levels above.
Immunoblotting assays were performed as described previously (Nakanishi et al., 2009). An anti-PDV2 antibody (Okazaki et al., 2009) and an anti-GFP mouse monoclonal antibody (JL-8; Invitrogen) were used at dilutions of 1:20,000 and 1:1000, respectively.
Accession Numbers
Sequence data from this work can be found in the Arabidopsis Genome Initiative or GenBank databases under the following accession numbers and GI numbers: PDV1 (At5g53280), PDV2 (At2g16070), DRP5B (At3g19720), PI4Kα1 (At1g49340), PI4Kβ1 (At5g64070), and PI4Kβ2 (At5g09350).
Supplemental Data
Supplemental Figure 1. WM Treatment Increases Chloroplast Number.
Supplemental Figure 2. Intracellular Localization of GFP-PI4Kα1 and GFP-PI4Kβ2 in Arabidopsis.
Supplemental Figure 3. Phenylarsine Oxide (PAO) Treatment Increases DRP5B Expression and Enhances DRP5B Recruitment to Chloroplasts.
Supplementary Material
Acknowledgments
We thank J.E.M. Vermeer for providing us with EYFP-FAPP1 plants. This work was supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (to K.O.).
AUTHOR CONTRIBUTIONS
K.O. designed and performed experiments and wrote the article. S.M. and H.W. designed experiments. All authors discussed the results and collectively edited the article.
Glossary
- PI
phosphatidylinositol
- PI4P
phosphatidylinositol 4-phosphate
- PI(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- PI3P
phosphatidylinositol 3-phosphate
- PI4K
phosphatidylinositol 4-kinase
- PH
pleckstrin homology
- PS
phosphatidylserine
- CL
cardiolipin
- MGDG
monogalactosyldiacylglycerol
- DGDG
digalactosyldiacylglycerol
- SQDG
sulfoquinovosyldiacylglycerol
- PA
phosphatidic acid
- PG
phosphatidylglycerol
- WM
wortmannin
- PI3K
phosphatidylinositol 3-kinase
- PAO
phenylarsine oxide
- TGN
trans-Golgi network
- amiRNA
artificial microRNA
- DEX
dexamethasone
- LSP
low-speed pellet
- MS
Murashige and Skoog
Footnotes
Articles can be viewed online without a subscription.
References
- Ajjawi I., Coku A., Froehlich J.E., Yang Y., Osteryoung K.W., Benning C., Last R.L. (2011). A J-like protein influences fatty acid composition of chloroplast lipids in Arabidopsis. PLoS ONE 6: e25368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612. [DOI] [PubMed] [Google Scholar]
- Aung K., Hu J. (2012). Differential roles of Arabidopsis dynamin-related proteins DRP3A, DRP3B, and DRP5B in organelle division. J. Integr. Plant Biol. 54: 921–931. [DOI] [PubMed] [Google Scholar]
- Barylko B., Binns D., Lin K.-M., Atkinson M.A.L., Jameson D.M., Yin H.L., Albanesi J.P. (1998). Synergistic activation of dynamin GTPase by Grb2 and phosphoinositides. J. Biol. Chem. 273: 3791–3797. [DOI] [PubMed] [Google Scholar]
- Basak I., Møller S.G. (2013). Emerging facets of plastid division regulation. Planta 237: 389–398. [DOI] [PubMed] [Google Scholar]
- Berridge M.J., Irvine R.F. (1989). Inositol phosphates and cell signalling. Nature 341: 197–205. [DOI] [PubMed] [Google Scholar]
- Boss W.F., Im Y.J. (2012). Phosphoinositide signaling. Annu. Rev. Plant Biol. 63: 409–429. [DOI] [PubMed] [Google Scholar]
- Bovet L., Müller M.O., Siegenthaler P.A. (2001). Three distinct lipid kinase activities are present in spinach chloroplast envelope membranes: Phosphatidylinositol phosphorylation is sensitive to wortmannin and not dependent on chloroplast ATP. Biochem. Biophys. Res. Commun. 289: 269–275. [DOI] [PubMed] [Google Scholar]
- Chapman L.A., Goring D.R. (2011). Misregulation of phosphoinositides in Arabidopsis thaliana decreases pollen hydration and maternal fertility. Sex. Plant Reprod. 24: 319–326. [DOI] [PubMed] [Google Scholar]
- Delage E., Ruelland E., Guillas I., Zachowski A., Puyaubert J. (2012). Arabidopsis type-III phosphatidylinositol 4-kinases β1 and β2 are upstream of the phospholipase C pathway triggered by cold exposure. Plant Cell Physiol. 53: 565–576. [DOI] [PubMed] [Google Scholar]
- Douce R., Joyard J. (1990). Biochemistry and function of the plastid envelope. Annu. Rev. Cell Biol. 6: 173–216. [DOI] [PubMed] [Google Scholar]
- Fan J., Xu C. (2011). Genetic analysis of Arabidopsis mutants impaired in plastid lipid import reveals a role of membrane lipids in chloroplast division. Plant Signal. Behav. 6: 458–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Liu H., An C., Shi Y., Liu X., Yuan W., Zhang B., Yang J., Yu C., Gao H. (2013). Arabidopsis FRS4/CPD25 and FHY3/CPD45 work cooperatively to promote the expression of the chloroplast division gene ARC5 and chloroplast division. Plant J. 75: 795–807. [DOI] [PubMed] [Google Scholar]
- Glynn J.M., Froehlich J.E., Osteryoung K.W. (2008). Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 20: 2460–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn J.M., Yang Y., Vitha S., Schmitz A.J., Hemmes M., Miyagishima S.Y., Osteryoung K.W. (2009). PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J. 59: 700–711. [DOI] [PubMed] [Google Scholar]
- Gonorazky G., Laxalt A.M., de la Canal L. (2010). Involvement of phospholipase C in the responses triggered by extracellular phosphatidylinositol 4-phosphate. J. Plant Physiol. 167: 411–415. [DOI] [PubMed] [Google Scholar]
- Hendrix W., Assefa H., Boss W.F. (1989). The polyphosphoinositides, phosphatidylinositol monophosphate and phosphatidylinositol bisphosphate are present in nuclei isolated from carrot protoplasts. Protoplasma 151: 62–72. [Google Scholar]
- Holtsmark I., Lee S., Lunde K.A., Auestad K., Maple-Grødem J., Møller S.G. (2013). Plastid division control: The PDV proteins regulate DRP5B dynamin activity. Plant Mol. Biol. 82: 255–266. [DOI] [PubMed] [Google Scholar]
- Hong Z., Bednarek S.Y., Blumwald E., Hwang I., Jurgens G., Menzel D., Osteryoung K.W., Raikhel N.V., Shinozaki K., Tsutsumi N., Verma D.P.S. (2003). A unified nomenclature for Arabidopsis dynamin-related large GTPases based on homology and possible functions. Plant Mol. Biol. 53: 261–265. [DOI] [PubMed] [Google Scholar]
- Jung J.Y., Kim Y.W., Kwak J.M., Hwang J.U., Young J., Schroeder J.I., Hwang I., Lee Y. (2002). Phosphatidylinositol 3- and 4-phosphate are required for normal stomatal movements. Plant Cell 14: 2399–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuk A., Friedländer E., Vereb G. Jr., Kása A., Balla A., Balla T., Heilmeyer L.M.G. Jr., Gergely P., Vereb G. (2006). Nucleolar localization of phosphatidylinositol 4-kinase PI4K230 in various mammalian cells. Cytometry A 69: 1174–1183. [DOI] [PubMed] [Google Scholar]
- Kang B.H., Nielsen E., Preuss M.L., Mastronarde D., Staehelin L.A. (2011). Electron tomography of RabA4b- and PI-4Kβ1-labeled trans Golgi network compartments in Arabidopsis. Traffic 12: 313–329. [DOI] [PubMed] [Google Scholar]
- Kavran J.M., Klein D.E., Lee A., Falasca M., Isakoff S.J., Skolnik E.Y., Lemmon M.A. (1998). Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J. Biol. Chem. 273: 30497–30508. [DOI] [PubMed] [Google Scholar]
- Keeling P.J. (2010). The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 729–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krinke O., Ruelland E., Valentová O., Vergnolle C., Renou J.P., Taconnat L., Flemr M., Burketová L., Zachowski A. (2007). Phosphatidylinositol 4-kinase activation is an early response to salicylic acid in Arabidopsis suspension cells. Plant Physiol. 144: 1347–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa H., Mori T., Takahara M., Miyagishima S.Y., Kuroiwa T. (2002). Chloroplast division machinery as revealed by immunofluorescence and electron microscopy. Planta 215: 185–190. [DOI] [PubMed] [Google Scholar]
- Matsuoka K., Bassham D.C., Raikhel N.V., Nakamura K. (1995). Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J. Cell Biol. 130: 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S.Y., Froehlich J.E., Osteryoung K.W. (2006). PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18: 2517–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S.Y., Kuwayama H., Urushihara H., Nakanishi H. (2008). Evolutionary linkage between eukaryotic cytokinesis and chloroplast division by dynamin proteins. Proc. Natl. Acad. Sci. USA 105: 15202–15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Röber B., Pical C. (2002). Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol. 130: 22–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T., Vermeer J.E.M. (2010). Osmotic stress-induced phosphoinositide and inositol phosphate signalling in plants. Plant Cell Environ. 33: 655–669. [DOI] [PubMed] [Google Scholar]
- Nakanishi H., Suzuki K., Kabeya Y., Miyagishima S.Y. (2009). Plant-specific protein MCD1 determines the site of chloroplast division in concert with bacteria-derived MinD. Curr. Biol. 19: 151–156. [DOI] [PubMed] [Google Scholar]
- Niwa Y. (2003). A synthetic green fluorescent protein gene for plant biotechnology. Plant Biotechnol. 20: 1–11. [Google Scholar]
- Nobusawa T., Umeda M. (2012). Very-long-chain fatty acids have an essential role in plastid division by controlling Z-ring formation in Arabidopsis thaliana. Genes Cells 17: 709–719. [DOI] [PubMed] [Google Scholar]
- Okazaki K., Kabeya Y., Suzuki K., Mori T., Ichikawa T., Matsui M., Nakanishi H., Miyagishima S.Y. (2009). The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell 21: 1769–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okpodu C.M., Gross W., Burkhart W., Boss W.F. (1995). Purification and characterization of a soluble phosphatidylinositol 4-kinase from carrot suspension culture cells. Plant Physiol. 107: 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W., Vierling E. (1995). Conserved cell and organelle division. Nature 376: 473–474. [DOI] [PubMed] [Google Scholar]
- Ouyang X., et al. (2011). Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23: 2514–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss M.L., Schmitz A.J., Thole J.M., Bonner H.K.S., Otegui M.S., Nielsen E. (2006). A role for the RabA4b effector protein PI-4Kbeta1 in polarized expansion of root hair cells in Arabidopsis thaliana. J. Cell Biol. 172: 991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommarin M., Sandelius A.S. (1988). Phosphatidylinositol and phosphatidylinositolphosphate kinases in plant plasma membranes. Biochim. Biophys. Acta 958: 268–278. [Google Scholar]
- Stevenson-Paulik J., Love J., Boss W.F. (2003). Differential regulation of two Arabidopsis type III phosphatidylinositol 4-kinase isoforms. A regulatory role for the pleckstrin homology domain. Plant Physiol. 132: 1053–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer J.E.M., Thole J.M., Goedhart J., Nielsen E., Munnik T., Gadella T.W.J. Jr (2009). Imaging phosphatidylinositol 4-phosphate dynamics in living plant cells. Plant J. 57: 356–372. [DOI] [PubMed] [Google Scholar]
- Vitha S., Froehlich J.E., Koksharova O., Pyke K.A., van Erp H., Osteryoung K.W. (2003). ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15: 1918–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S., McAndrew R.S., Osteryoung K.W. (2001). FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 153: 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E.H., Pacold M.E., Perisic O., Stephens L., Hawkins P.T., Wymann M.P., Williams R.L. (2000). Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol. Cell 6: 909–919. [DOI] [PubMed] [Google Scholar]
- Westergren T., Ekblad L., Jergil B., Sommarin M. (1999). Phosphatidylinositol 4-kinase associated with spinach plasma membranes. Isolation and characterization of two distinct forms. Plant Physiol. 121: 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.Z., Xue H.W. (2010). Arabidopsis β-ketoacyl-[acyl carrier protein] synthase I is crucial for fatty acid synthesis and plays a role in chloroplast division and embryo development. Plant Cell 22: 3726–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Lloyd C.W., Staiger C.J., Drøbak B.K. (1992). Association of phosphatidylinositol 4-kinase with the plant cytoskeleton. Plant Cell 4: 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Hu J. (2009). Two small protein families, DYNAMIN-RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis. Plant J. 57: 146–159. [DOI] [PubMed] [Google Scholar]
- Zhang X., Hu J. (2010). The Arabidopsis chloroplast division protein DYNAMIN-RELATED PROTEIN5B also mediates peroxisome division. Plant Cell 22: 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Cahill S.M., Lemmon M.A., Fushman D., Schlessinger J., Cowburn D. (1996). Identification of the binding site for acidic phospholipids on the pH domain of dynamin: Implications for stimulation of GTPase activity. J. Mol. Biol. 255: 14–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.