A genome-wide analysis and interspecific comparison of MIRNA genes revealed the patterns of MIRNA gene evolution subsequent to the recent whole-genome duplication event in soybean.
Abstract
The evolutionary dynamics of duplicated protein-encoding genes (PEGs) is well documented. However, the evolutionary patterns and consequences of duplicated MIRNAs and the potential influence on the evolution of their PEG targets are poorly understood. Here, we demonstrate the evolution of plant MIRNAs subsequent to a recent whole-genome duplication. Overall, the retention of MIRNA duplicates was correlated to the retention of adjacent PEG duplicates, and the retained MIRNA duplicates exhibited a higher level of interspecific preservation of orthologs than singletons, suggesting that the retention of MIRNA duplicates is related to their functional constraints and local genomic stability. Nevertheless, duplication status, rather than local genic collinearity, was the primary determinant of levels of nucleotide divergence of MIRNAs. In addition, the retention of duplicated MIRNAs appears to be associated with the retention of their corresponding duplicated PEG targets. Furthermore, we characterized the evolutionary novelty of a legume-specific microRNA (miRNA) family, which resulted from rounds of genomic duplication, and consequent dynamic evolution of its NB-LRR targets, an important gene family with primary roles in plant-pathogen interactions. Together, these observations depict evolutionary patterns and novelty of MIRNAs in the context of genomic duplication and evolutionary interplay between MIRNAs and their PEG targets mediated by miRNAs.
INTRODUCTION
Genomic duplications, such as whole-genome duplication (WGD), segmental duplication, and tandem duplication are recognized as essential sources of genetic material for the origin of evolutionary novelties (Ohno, 1970). WGD, caused by polyploidization, is particularly common in the plant kingdom (Soltis and Soltis, 1999; Otto and Whitton, 2000) and largely responsible for reproductive isolation and sympatric speciation (McGrath et al., 2014). Tandem duplication frequently occurs in higher eukaryotes and is often revealed by the prevalence of clusters of genes and their copy number variation among related species (Rizzon et al., 2006; Fan et al., 2008). Recent studies demonstrated that a large number of genes or gene fragments could be captured and moved by transposable elements (TEs) (Jiang et al., 2004; Lai et al., 2005), as a mechanism for the creation and amplification of new genes. In addition, TEs themselves could be domesticated into new forms with novel functions that regulate the activity of protein-encoding genes (PEGs) in the host genomes (Vogt et al., 2013), representing another TE-mediated mechanism for gene creation. Among these mechanisms, WGD appears to have the most significant impact on genome evolution and innovation, as it creates a large set of genes simultaneously in a genome.
Theoretically, functional redundancy of duplicated genes would subject one copy to a period (e.g., perhaps a few million years) of relaxed selection after duplication, resulting in accumulation of deleterious mutations or ultimate elimination of one copy of the gene (Ohno, 1970). This process is evident from the presence of a large number of pseudogenes and singletons in many organisms that have undergone WGD (Schnable et al., 2009, 2011; Schmutz et al., 2010; Moghe et al., 2014). However, tens of thousands of duplicated gene pairs are generally retained in paleopolyploid genomes after tens of millions of years of natural selection (Schnable et al., 2009; Schmutz et al., 2010), indicating their functional importance. These duplicated genes may both have been retained with conserved ancestral functions (conservation), one copy with ancestral functions and the other with novel functions (neofunctionalization), both copies with deleterious mutations but together maintaining the ancestral functions (subfunctionalization), or both copies with functions diverged from each other and from the ancestral gene (specialization) (Ohno, 1970; Force et al., 1999; Stoltzfus, 1999; He and Zhang, 2005). Despite the widespread acceptance of these evolutionary models, with empirical examples for each (Assis and Bachtrog, 2013), more and more studies suggest that rewiring of the regulatory networks following WGD might have had a significant impact on the evolutionary consequences of duplicated genes rather than functional divergence of duplicated proteins (De Smet and Van de Peer, 2012). This proposition seems to be well supported by the observation that the majority of duplicated genes exhibit distinct patterns of expression between the two members of each pair, as revealed by genome-wide profiling of plant transcriptomes across different tissues and development stages (Danilevskaya et al., 2003; Du et al., 2012; Roulin et al., 2012).
MicroRNAs (miRNAs) are important regulators of PEGs and are involved in many biological processes, such as development, differentiation, growth, and immune responses (Carrington and Ambros, 2003; Bartel, 2004; Carthew and Sontheimer, 2009). In plants, miRNAs are mainly 21- or 22-nucleotides long and processed by DICER-LIKE1 from single-stranded MIRNA precursors with stem-loop secondary structures (Voinnet, 2009). In general, miRNAs reduce levels of specific proteins through direct cleavage of mRNAs or translational repression (Khraiwesh et al., 2010). Recent work demonstrated that some miRNAs can trigger the biogenesis of secondary small interfering RNAs (siRNAs), termed trans-acting siRNAs or phased siRNAs (phasiRNAs) from the transcripts of genic or intergenic sequences (termed TAS or PHAS loci) through the RDR6/DCL4 pathway (Peragine et al., 2004; Vazquez et al., 2004; Allen et al., 2005; Yoshikawa et al., 2005; Zhai et al., 2011). These trans-acting siRNAs or phasiRNAs are generally clustered in a 21-nucleotide phased register starting at the initiation cleavage sites (Chen et al., 2007; Howell et al., 2007). Similar to miRNAs, phasiRNAs function in homology-dependent manner to suppress the expression of their target genes in cis or in trans (Zhai et al., 2011; Li et al., 2012; Shivaprasad et al., 2012). It is believed that the effects of miRNAs on gene regulation could be amplified by production of phasiRNAs (Fei et al., 2013). To date, numerous miRNAs have been identified in plants, and many of these mRNAs are evolutionarily and functionally conserved across species (Fahlgren et al., 2010; Fei et al., 2013). On the other hand, a few studies have illustrated the rapid divergence of miRNAs and their target genes (Allen et al., 2004; Smith et al., 2015). Nevertheless, few genome-wide studies have attempted to investigate the nature of MIRNA duplication, or the evolutionary patterns, consequences, and novelty of duplicated MIRNAs, the evolutionary mechanisms for creation of novel miRNAs and potential coevolution of miRNAs and their target PEGs in the context of WGD events.
Soybean (Glycine max) is one of the most economically important leguminous crops, domesticated from its wild progenitor species, Glycine soja, ∼5000 years ago (Carter et al., 2004). It is proposed that the Glycine lineage has undergone at least two rounds of WGD after its divergence from the lineage that gave rise to Arabidopsis thaliana. One WGD occurred ∼59 million years ago (MYA) and a second ∼13 MYA (Schmutz et al., 2010), after the split of the Glycine lineage from its close relative, common bean (Phaseolus vulgaris), ∼19 MYA (Lavin et al., 2005; McClean et al., 2010; Schmutz et al., 2014). With the availability of the reference genome sequences from both soybean and common bean (Schmutz et al., 2010, 2014), and two clearly defined duplication events before and after their split, these two leguminous crops have become an important model system suitable for investigation of the nature, timing, and evolutionary dynamics and consequences of recurrent WGD events.
Here, we performed a comprehensive, genome-wide study on soybean MIRNAs to examine the evolutionary rates of duplicated MIRNAs versus singletons, the relative retention of duplicated MIRNAs after WGD, amplification of MIRNAs by tandem duplication, and TE-mediated creation of MIRNAs in the context of the recent WGD. We then investigated intraspecific and interspecific preservation/conservations of MIRNAs including both MIRNA duplicates and singletons by comparing homologous/orthologous genomic regions from the two species. Finally, we analyzed the potential interplay between MIRNAs and their PEG targets and the coevolution of these sequences in the paleopolyploid genome. These analyses not only depict the evolutionary patterns and consequences of duplicated MIRNAs in a complex paleopolyploid genome, but also illustrate the evolutionary novelty of duplicated PEGs and MIRNAs mediated by miRNAs. In addition to these observations, we annotated 186 MIRNAs that produce 122 unique mature miRNAs in the newly available reference genome of common bean (Schmutz et al., 2014).
RESULTS
Distribution of MIRNAs in Terms of Their Genomic Environments in Soybean
A total of 638 nonredundant MIRNAs (i.e., precursor genes of mature miRNAs) were previously annotated based on small RNA libraries generated from various tissues, including seeds, roots, stems, leaves, flowers, and developing nodules at various stages (Song et al., 2011; Zhai et al., 2011; Arikit et al., 2014; Zhao et al., 2015). However, some of these miRNAs were recently reannotated as siRNA-like miRNAs; thus, the predicted MIRNAs producing these more dubious “miRNAs” were excluded from our analyses. In total, 454 MIRNAs showing the typical features of MIRNA precursors with capability to generate miRNAs in at least one of these many tissues were considered as genuine MIRNAs. This consideration is consistent with the criteria generally used for annotation of MIRNAs and miRNAs in plants (Meyers et al., 2008). A previous study predicted 28,281 MIRNA-like elements in soybean based on their hairpin structures (Zhou et al., 2013), but nearly 98% of those actual “hairpins” were not able to produce miRNAs at a detectable level in any of these small RNA libraries, and the majority of those are portions of “dead” TEs belonging to a limited number of large TE families (Du et al., 2010). Therefore, such so-called MIRNA-like elements, solely predicted based on hairpin structures, were also excluded from our analyses, unless their homologs generated by the recent WGD or orthologs in common bean were found to be able to produce miRNAs. In such a case, the unexpressed MIRNA homologs or orthologs were referred to as pseudo-MIRNAs or pMIRNAs.
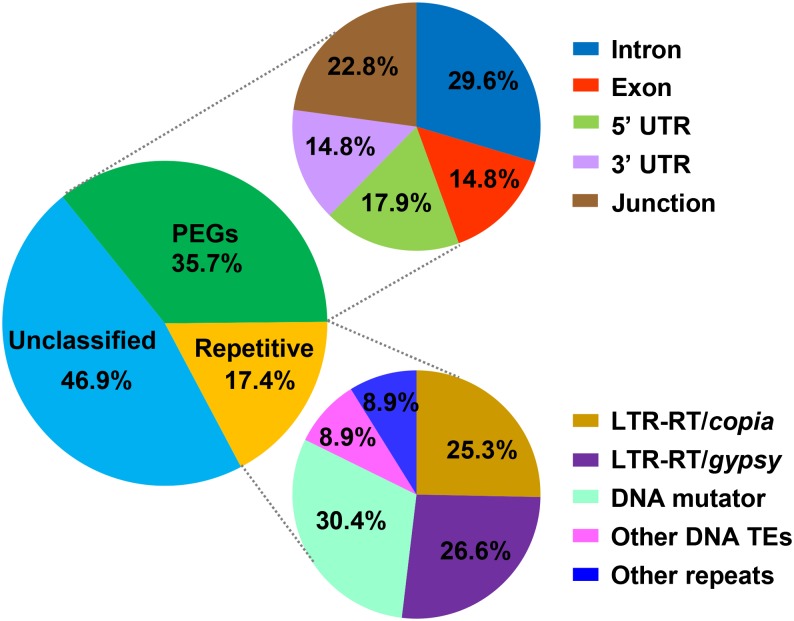
To shed light on how the expression of MIRNAs might be affected by their surrounding sequences, we analyzed the distribution of the 454 MIRNAs in the context of three categories of genomic components: PEGs, repetitive sequences, and unclassified sequences in the soybean reference genome (version 1.1, www.soybase.org). Of these MIRNAs, 162 (35.7%) were located within PEGs, 213 (46.9%) were harbored by unclassified sequences, and 79 (17.4%) were found within repetitive sequences, primarily TEs (Figure 1; Supplemental Data Sets 1 and 2). Of the 162 MIRNAs within PEGs, 48 (29.6%) were within introns, 53 (32.7%) were within untranslated regions (UTRs), 24 (14.8%) were within exons, and 37 (22.8%) spanned the boundaries of different genic components (Figure 1; Supplemental Data Set 1). Approximately 84.6% of these MIRNA genes showed the same transcriptional orientations as their host PEGs, while the remaining 15.4% were predicted to transcribe in the orientations opposite to their host genes’ transcriptional orientations (Supplemental Table 1). The direct transcripts from MIRNAs are generally processed to produce mature miRNAs and thus have been well characterized. Interestingly, the transcripts from 26 of the 48 MIRNAs within introns were detected in 28 mRNA libraries described previously (Shen et al., 2014), as were the splicing patterns of their host PEGs detected in the same libraries that remove these intronic sequences (Supplemental Table 2). Because some of these putative MIRNA transcripts cover the intron-exon junctions of their host PEGs, and some are arranged in opposite orientations to the hosting PEG’s transcripts, the coexistence of transcripts from these intronic MIRNAs and respective host PEGs in the same libraries would suggest separate and independent transcription of the PEGs and respective MIRNAs. However, we could not rule out the possibility of alternative splicing or antisense transcription of these PEGs. Of the 79 MIRNAs found in repetitive sequences, 41 (51.9%) were in long terminal repeat (LTR)-retrotransposon sequences, 31 (39.2%) were in DNA transposon sequences, and seven (8.9%) in other repeats (Figure 1). When only the 17 MIRNA-containing intact TEs with clearly defined boundaries were counted, 10, 3, 3, and 1 were Mutator elements, CACTAs, LTR-retrotransposons, and a Helitron, respectively (Supplemental Figure 1 and Supplemental Data Set 2). No two of these TEs were found to contain the same MIRNA, regardless if they belong to a same TE family (Supplemental Data Set 2), suggesting that, instead of TE amplification, mutations within existing TEs were responsible for the birth of these TE-related MIRNAs, and such mutations may have led to direct or indirect silencing of those TEs.
Figure 1.
Genomic Locations of MIRNAs in the Soybean Genome.
The left pie chart indicates the physical locations of the 454 MIRNAs in the soybean genome. The two pie charts at right indicate the detailed classifications of MIRNAs mapped to PEGs and repetitive DNA. “Junction” (top, right) indicates MIRNAs span the boundaries of different genomic components.
Retention of MIRNA Duplicates versus Retention of PEG Duplicates in Soybean
Based on the newly annotated set of PEGs in the soybean reference genome (version 1.1, www.soybase.org) and reanalysis of the duplicated genomic blocks retained after the recent WGD event (Schmutz et al., 2010; Du et al., 2012), 16,996 singletons and 17,627 PEG pairs that correspond to 35,254 duplicates were defined (Supplemental Data Set 3). As per the assumption that a singleton is the product of deletion of one of two members of a duplicated gene pair (Schmutz et al., 2010), it is estimated that, maximally, 16,996 PEGs were eliminated after the recent soybean WGD event (Supplemental Data Set 3), suggesting rapid fragmentation of the duplicated genomes. To understand the evolutionary process and consequences of MIRNAs after the recent WGD, we first analyzed the large genomic regions containing each of the 454 MIRNAs and their flanking PEGs and were able to classify these MIRNAs into 234 MIRNA singletons and 220 MIRNA duplicates that correspond to 110 duplicated MIRNA pairs. The ratio of MIRNA singletons to MIRNA duplicates is ∼1.06:1, significantly higher than the ratio (0.48:1) of the PEG singletons to PEG duplicates generated by the same WGD event (P < 0.01, χ2 test).
Further comparison of genomic regions harboring these MIRNAs with respective putative duplicated regions generated by the recent WGD (Schmutz et al., 2010; Du et al., 2012) identified 39 MIRNA sequences that are homoeologous to 39 of the 234 singletons defined above (Supplemental Data Set 4). Of these 39 homoeologs, 30 contains point mutations and two contain small insertions/deletions (indels) in the mature miRNA regions compared with their duplicates, and the remaining seven were more diverged from their duplicates. None of these 39 homoeologs were able to generate miRNAs based on the small RNA libraries that we examined; thus, they were considered as pseudo-MIRNAs or pMIRNAs. Because these 39 pMIRNAs were physically retained in pairs with their respective expressed MIRNA duplicates in homoeologous regions, these pMIRNA-MIRNA pairs were considered as retained duplicates. Under this specific consideration for categorization of duplicated MIRNA pairs retained after WGD, the ratio (0.65:1) of MIRNA singletons to MIRNA duplicates remained to be significantly higher than the ratio (0.48:1) of the PEG singletons to the PEG duplicates (P < 0.01, χ2 test).
The relative proportion of MIRNA singletons versus MIRNA duplicates located in the three categories of genomic components were further analyzed and compared (Table 1; Supplemental Table 3). Of the 226 MIRNAs located in unclassified sequences, 56 are singletons and 170 are duplicates, corresponding to a singleton-to-duplicates ratio of 1:3.0, which is significantly lower than the ratio (1:2.1) of the PEG singletons to the PEG duplicates in the whole soybean genome (P < 0.01, χ2 test). Of the 177 MIRNAs located in genic sequences, 71 are singletons and 106 are duplicates, corresponding to a singleton-to-duplicate ratio of 1:1.5, which is significantly higher than the ratio of the PEG singletons to the PEG duplicates in the whole soybean genome (P < 0.05, χ2 test). It is also notable that the ratios of singletons to duplicates in genic sequences vary greatly among different genic components, ranging from 2.1:1 in introns as the highest to 1:4.8 in exons as the lowest. Such variation appears to be the outcome of different levels of selection applied to different portions of the PEGs. Of the 90 MIRNAs in repetitive DNA, primarily composed of TE sequences, 68 are singletons and 22 are duplicates, corresponding to a singleton-to-duplicate ratio of 3.1:1.
Table 1. Description of Soybean MIRNAs with Orthologs and Homologs in Common Bean.
| Duplicates |
Singletons |
||||||
|---|---|---|---|---|---|---|---|
| Features | Totala | MIRNAs | Orthologsb | Homologsc | MIRNAs | Orthologsb | Homologsc |
| Genic | |||||||
| Intron | 55 | 18 | 5 | 2 | 37 | 1 | 6 |
| Exon | 29 | 24 | 15 | 2 | 5 | 0 | 2 |
| 5′ UTR | 29 | 21 | 15 | 1 | 9 | 5 | 0 |
| 3′ UTR | 24 | 16 | 13 | 1 | 7 | 1 | 0 |
| Junction | 40 | 27 | 24 | 1 | 13 | 4 | 2 |
| Subtotal | 177 | 106 | 72 | 7 | 71 | 11 | 10 |
| Unclassified | |||||||
| Subtotal | 226 | 170 | 114 | 19 | 56 | 14 | 10 |
| Repetitive DNA | |||||||
| Subtotal | 90 | 22 | 2 | 0 | 68 | 3 | 2 |
| Total (%) | 493 | 298 | 188 | 26 | 195 | 28 | 22 |
Total number of MIRNAs, including 39 pseudo-MIRNAs identified in soybean.
Number of orthologous MIRNAs of soybean shared by common bean.
Number of homologous MIRNAs of soybean shared by common bean.
Different from the PEG singletons, which may be the products of deletion of one of two members of a duplicated PEG pair, the overall overrepresentation of MIRNA singletons was primarily caused by a combination of deletion of one of the two members of duplicated MIRNA pairs, asymmetric tandem duplication of two members of a duplicated MIRNA pair, and sequence variation within amplified TEs. Of the 195 singletons, 23 (11.8%) were involved in asymmetric tandem duplication, which could be the products of either uneven deletion of tandemly duplicated MIRNA homologs that were formed prior to the WGD event, or uneven tandem duplication of two members of a duplicated MIRNA pair produced by the WGD, or both, and 62 (31.8%) were found in repetitive DNA primarily composed of TEs (Figure 1), and the remaining 110 (56.4%) were considered as the products of deletion per the same assumption for the PEG singletons. Although shared TEs at homologous loci are rarely identified in soybean (Du et al., 2010; Schmutz et al., 2010), the majority of these MIRNA singletons could be born from TEs amplified after the recent WGD event, as shown in Supplemental Figure 2. The presence of 22 MIRNA duplicates in TE sequences indicates that is also possible that some MIRNA singletons derived from TEs were generated by deletion after the WGD event.
When both MIRNA duplicates and singletons associated with tandem duplication plus those located in repetitive DNA were excluded, the ratio of MIRNA singletons to duplicates is 1:1.8, significantly higher than the ratio (1:2.1) of PEG singletons to duplicates (P < 0.01, χ2 test), excluding PEG duplicates and singletons associated with tandem duplications, in the soybean genome (Supplemental Table 4). This observation may reflect an overall lower retention rate for duplicated MIRNAs than that of the duplicated PEGs in the soybean genome.
MIRNA Duplicates Are Evolutionarily More Conserved Than MIRNA Singletons: A Scenario of Interspecific Preservation
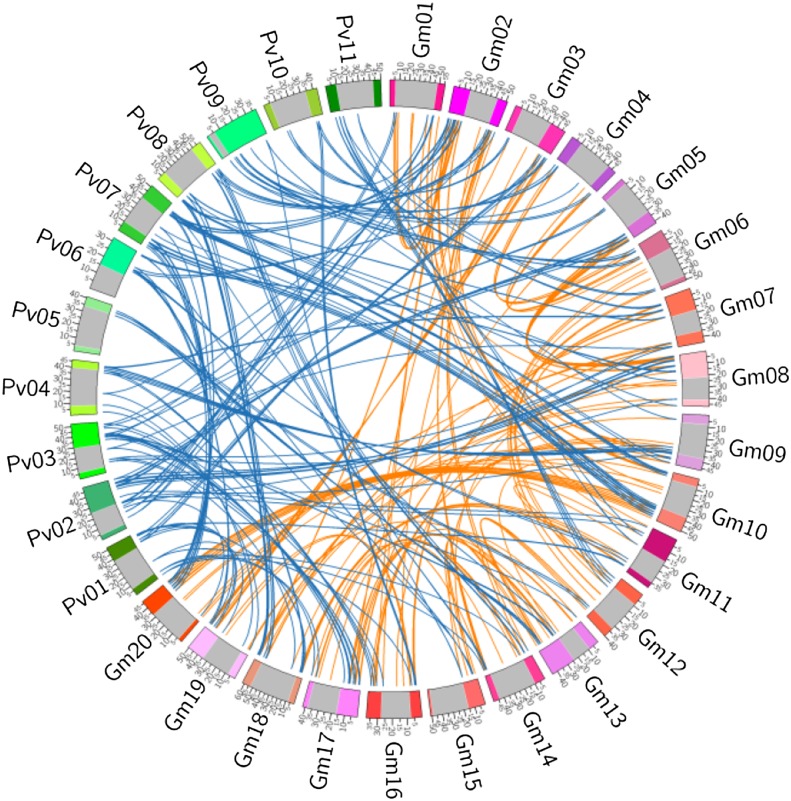
To further understand the nature and evolutionary process of genomic fractionation shaping the distribution and relative abundance of MIRNA singletons versus duplicates in soybean, we identified the genomic regions in the sequenced common bean reference genome (Schmutz et al., 2014) that are putatively orthologous to the soybean genomic regions harboring the 149 pairs of MIRNA homoeologs and the 195 singletons, as well as all homologs of these soybean MIRNAs in the common bean genome (Figure 2; Supplemental Data Sets 5 and 6). Subsequently, all genomic regions containing these putative orthologs or homologs were compared between the two species. Of the 298 MIRNA duplicates, 188 (63.1%) were found to have putative orthologs in common bean, 26 (8.7%) were found to have homologs in common bean without a detectable orthologous relationship between regions surrounding these homologs, and the remaining 84 (28.2%) did not have any detectable homologs in common bean. By contrast, of the 195 singletons, 28 (14.4%) were found to have detectable putative orthologs in common bean, 22 (11.3%) had homologs in common bean without a detectable orthologous relationship between regions surrounding these homologs, and the remaining 145 (74.4%) did not have any detectable homologs in common bean (Table 1). These observations suggest that the WGD MIRNA pairs are evolutionarily more conserved than the MIRNA singletons in soybean in terms of the preservation of their orthologs in common bean.
Figure 2.
Distribution of Duplicated MIRNA Pairs in Soybean and Orthologous MIRNAs in Common Bean.
The 11 chromosomes in common bean (Pv01 to Pv11) and 20 chromosomes in soybean (Gm01 to Gm20) are shown in a circle. Orange links indicate the MIRNAs derived from WGD events, and dark-blue links indicate the orthologous MIRNAs of soybean and common bean. The gray regions of each chromosome represent pericentromeric regions, and the colored green regions of the 11 chromosomes in common bean and pink regions of the 20 chromosomes in soybean represent chromosomal arms.
We also compared these MIRNA orthologs and homologs between the two species, based on genomic components in which these MIRNAs are located. Of the 106 MIRNA duplicates and 71 singletons located within genes, 72 (67.9%) and 11 (15.5%) have putative orthologs in common bean, respectively. Of the 170 MIRNA duplicates and 56 singletons located within unclassified sequences, 114 (67.1%) and 14 (25%) have putative orthologs in common bean. Of the 22 MIRNA duplicates and 68 singletons associated with repetitive DNA, only 2 (9.1%) and 3 (4.4%) have putative orthologs in common bean. Such a low level of preservation of MIRNA genes associated with repetitive DNA, mostly TE sequences, may be explained by rapid movement and divergence of TEs (Du et al., 2010) if inserted in the soybean before the recent WGD or recent birth of MIRNAs within younger TEs amplified after the WGD event. When the MIRNAs in repetitive DNA were excluded, the frequencies of preservation of the MIRNA orthologs in common bean observed for the soybean MIRNA duplicates were significantly higher than those observed for the soybean MIRNA singletons (Table 1; P < 0.01, χ2 test).
Effects of Local Genomic Stability on Retention of MIRNA Duplicates in Soybean
Previous studies demonstrated that the frequency of small deletions of genomic DNA was associated with local genomic features, such as rates of genetic recombination (Gaut et al., 2007; Tian et al., 2009); we thus wondered whether the stability of local genomic regions surrounding MIRNAs had local effects on the retention of MIRNA duplicates. The ratio of MIRNA singletons to MIRNA duplicates, excluding those associated with tandem duplication and those located in repetitive DNA, was compared with the ratios of singletons to duplicates for two individual PEGs flanking each of these MIRNA loci. As shown in Supplemental Table 4, the ratio of MIRNA singletons to MIRNA duplicates (0.56:1) did not show a significant difference from the ratio of PEG singletons to PEG duplicates for the first and second genes (0.65:1 and 0.54:1, respectively) flanking each of these MIRNAs.
We then paid a particular attention to the duplication status of the first PEG genes adjacent to individual MIRNAs (those immediately flanking the MIRNAs), excluding those involved in tandem duplication and associated with TEs. Of the 396 PEGs flanking the 198 duplicated MIRNA genes, 253 (63.9%) were duplicated genes and 143 (36.1%) were singletons. By contrast, of the 220 PEGs flanking the 110 MIRNA singletons, 120 (54.5%) were duplicated genes and 100 (45.4%) are singletons. The ratio of PEG singletons to PEG duplicates flanking the MIRNA duplicates is significantly lower than the ratio of PEG singletons to PEG duplicates flanking the MIRNA singletons (P < 0.001, χ2 test; Supplemental Figure 3). Together, these observations indicate that the MIRNA duplicates tend to be physically linked with the PEG duplicates in soybean and suggest that, in addition to the functional constraints of MIRNAs, the retention of MIRNA duplicates was affected by the stability of local genomic regions.
Asymmetric Evolutionary Rates for MIRNAs: TE-Related versus Non-TE-Related, Duplicates versus Singletons, and Conserved versus Nonconserved
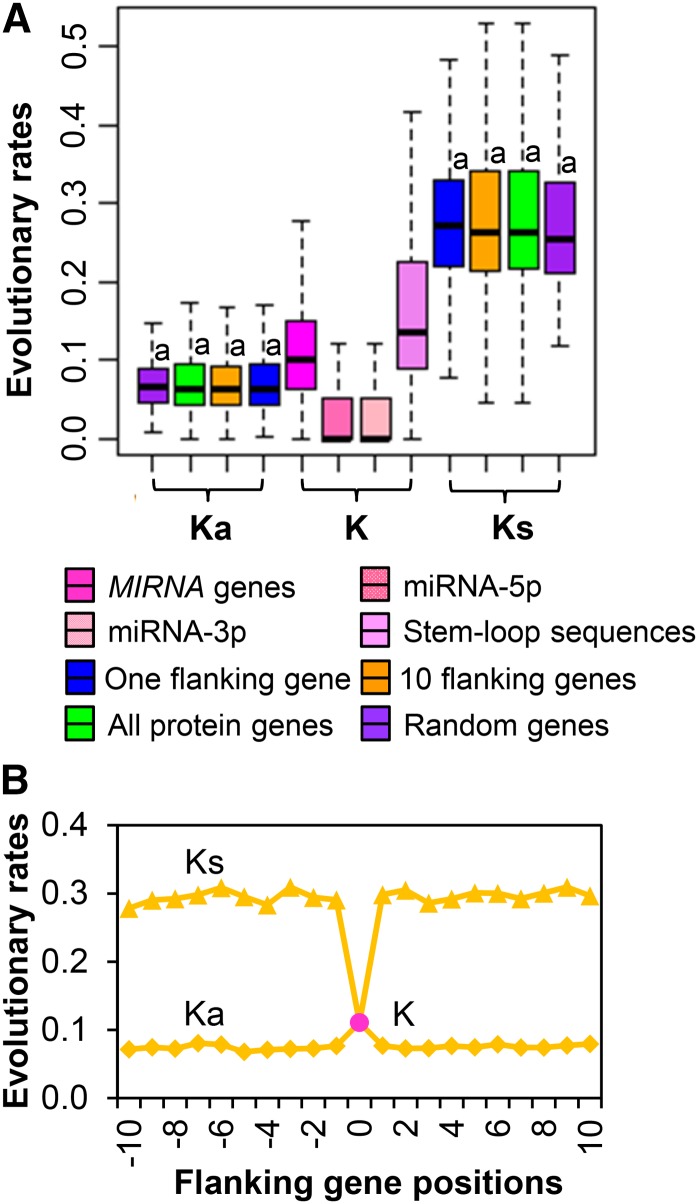
The evolutionary rates of MIRNAs and PEGs in soybean were estimated by intraspecific and interspecific sequence comparisons. Intraspecific comparison was conducted with sequences generated from seven deeply sequenced and de novo assembled genomes of G. soja (Li et al., 2014), the progenitor species of cultivated soybean. These genomes were estimated to have diverged from each other for 0.7 to 1.1 million years and are quite representative of the natural G. soja population (Li et al., 2014). Interspecific comparison was performed with orthologous sequences between soybean and common bean (Schmutz et al., 2014), reflecting levels of nucleotide divergence obtained up to the past 19 million years (Lavin et al., 2005; McClean et al., 2010; Schmutz et al., 2014), although the birth and loss of a particular MIRNA may have occurred more recently. In the intraspecific comparison, the pattern of nucleotide divergence (K) of MIRNAs was different among the three categories in comparison with synonymous substitution (Ks) and nonsynonymous substitution (Ka) of PEG genes (Supplemental Figure 4). In the interspecific comparison, the level of nucleotide divergence (K) of MIRNAs was found to be significantly smaller than the level of synonymous substitution (Ks) of PEG genes (P < 0.01, χ2 test) but significantly higher than the level of nonsynonymous substitution (Ka) for both genic (P < 0.01, χ2 test) and unclassified (P < 0.01, χ2 test) categories, although the levels of difference are different (Figure 3; Supplemental Figure 4). As expected, the portions of miRNA-5p and miRNA-3p are highly conserved and evolved at the lowest pace in comparison with the coding sequences of PEGs. No significant difference of the levels of nucleotide divergence was observed between the miRNA-5p and miRNA-3p sequences (Supplemental Table 5).
Figure 3.
Comparison of Evolutionary Rates between MIRNAs and PEGs in Soybean.
(A) Comparison of evolutionary rates of MIRNAs with different sets of PEGs. The statistical analysis was conducted between each set of MIRNAs and PEGs by Student’s t test. The “a” above each column indicates P < 0.01.
(B) Comparison of evolutionary rates of MIRNAs with the up- and downstream 10 flanking genes related to the MIRNAs. The pink circle indicates MIRNAs.
Ka, Ks, and K were calculated by pairwise comparison of the orthologous MIRNAs or PEGs between soybean and common bean.
Pairwise comparisons of K among the three categories of MIRNA duplicates and among the three categories of MIRNA singletons were conducted separately using the intraspecific comparative sequence data. For both MIRNA duplicates and MIRNA singletons, those related to TEs exhibited significantly higher K than those located in genes and unclassified sequences. No significant difference of K was observed between MIRNAs located in genes and those in unclassified sequences (Figure 4).
Figure 4.
Pairwise Comparison of Evolutionary Rates among the Three Categories of MIRNA Duplicates and MIRNA Singletons.
Evolutionary rates were calculated by pairwise comparisons between orthologous genes among seven pan genomes of G. soja. The statistical analysis was conducted by Student’s t test. The “a” above two columns indicates P < 0.01.
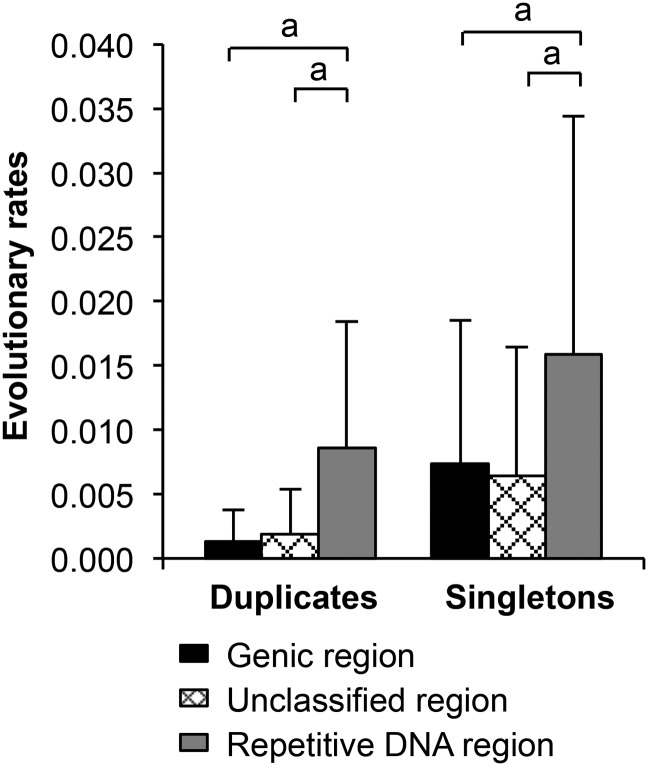
We also compared K between singletons and duplicates under the three categories of MIRNAs using the intraspecific comparative sequence data. For the MIRNAs located in genes, unclassified sequences, and repetitive sequences, significantly higher K values were observed for singletons than duplicates (Table 2). To understand potential effects of tandem duplication on nucleotide divergence, we further compared MIRNA singletons and duplicates associated with tandem duplication and those that are not associated with tandem duplication. As shown in Supplemental Table 6, interestingly, if tandem duplication was not involved, the K for MIRNA duplicates was significantly smaller than that for MIRNA singletons (P < 0.001, χ2 test). No significant difference of K was observed between MIRNA duplicates involved in tandem duplication and MIRNA duplicates uninvolved in tandem duplication but a significantly higher K for MIRNA singletons uninvolved in tandem duplication than that for MIRNA singletons involved in tandem duplication (P < 0.001, χ2 test).
Table 2. Comparison of Evolutionary Rates between MIRNA Duplicates and MIRNA Singletons.
| Comparison | Duplicates | Singletons | Pa |
|---|---|---|---|
| Soybean vs. common bean | |||
| Overall | 0.1063 ± 0.0628 | 0.1404 ± 0.0547 | 0.0070 |
| Genic region | 0.0971 ± 0.0679 | 0.1454 ± 0.0521 | 0.0276 |
| Unclassified region | 0.1046 ± 0.0565 | 0.1280 ± 0.0511 | 0.1426 |
| Among seven soybean pan genomes | |||
| Overall | 0.0022 ± 0.0044 | 0.0100 ± 0.0144 | <0.0001 |
| Genic region | 0.0013 ± 0.0025 | 0.0074 ± 0.0111 | <0.0001 |
| Unclassified region | 0.0019 ± 0.0035 | 0.0064 ± 0.0100 | 0.0020 |
| Repetitive DNA region | 0.0086 ± 0.0098 | 0.0159 ± 0.0185 | 0.0234 |
Student's t test.
Finally, K values between interspecifically conserved and nonconserved MIRNAs, under the categories of MIRNA duplicates and singletons, were compared using the intraspecific comparative sequence data. In both genic and unclassified categories, nonconserved MIRNA singletons consistently showed significantly higher K than conserved MIRNA singletons, but no significant differences were detected between conserved duplicates and nonconserved duplicates (Table 3). These observations suggest that the status of duplication, including WGD and tandem duplication, of non-TE-related MIRNAs is the main determinant of their evolutionary rates.
Table 3. Comparison of Evolutionary Rates between Conserved and Nonconserved MIRNAs with Genomic Sequences from the Seven G. soja Accessions Used for Construction of a G. soja Pan Genome.
| Comparison | Conserved | Nonconserved | Pa |
|---|---|---|---|
| Duplicates | |||
| Overallb | 0.0017 ± 0.0035 | 0.0024 ± 0.0038 | 0.1265 |
| Genic region | 0.0013 ± 0.0028 | 0.0015 ± 0.0022 | 0.7824 |
| Unclassified region | 0.0019 ± 0.0040 | 0.0019 ± 0.0025 | 0.9083 |
| Singletons | |||
| Overallb | 0.0016 ± 0.0028 | 0.0083 ± 0.0114 | <0.0001 |
| Genic region | 0.0005 ± 0.0012 | 0.0087 ± 0.0117 | <0.0001 |
| Unclassified region | 0.0025 ± 0.0034 | 0.0078 ± 0.0111 | 0.0099 |
Student's t test.
Does not contain MIRNAs located in repetitive DNA region.
Functional Divergence of MIRNA Duplicates Reflected by Variations in miRNA Sequence and Abundance
Using soybean small RNA libraries previously reported (Zhao et al., 2015), we evaluated average levels of accumulation of the mature miRNAs for the MIRNA precursors analyzed in this study (Table 4; Supplemental Table 7). Among the three categories of miRNAs, those located in genic regions showed the highest level of abundance, and those that arose from TEs showed the lowest levels of abundance. Overall, miRNA duplicates located in genic regions showed higher levels of abundance than miRNA singletons located in genic regions, but such a difference was rather modest in comparisons between miRNA duplicates and singletons located in unclassified sequences. Because, generally, a small number of miRNAs are predominant in a particular library (Arikit et al., 2014; Zhao et al., 2015), as indicated by the sd of the evaluated expression levels (Table 4), the accumulation levels measured in this set of small RNA data could be somewhat biased. Nevertheless, the distinction was clear for the overall accumulation patterns among the three categories of miRNAs and between the miRNA duplicates and singletons.
Table 4. Comparison of miRNA Expression Levels between MIRNA Duplicates and MIRNA Singletons.
| Comparison | Duplicates (TPM)a | Singletons (TPM)a | Pb |
|---|---|---|---|
| Overall | 10,582 ± 54,134 | 1,131 ± 7,800 | 0.0790 |
| Genic region | 19,437 ± 78,735 | 412 ± 1,311 | 0.1206 |
| Unclassified region | 4,889 ± 26,484 | 2,882 ± 13,956 | 0.6341 |
| Repetitive DNA region | 263 ± 507 | 290 ± 857 | 0.9410 |
Transcripts per million mapped reads.
Student's t test.
In an attempt to understand the evolutionary consequences of duplicated MIRNAs, we compared the 94 duplicated MIRNA pairs and 28 MIRNA singletons in soybean with their respective orthologs in common bean. As shown in Table 5, of the 94 soybean MIRNA orthologous pairs, 67 (71.3%) each produced an identical miRNA by the two duplicated members and by their ortholog in common bean (dubbed type I: S1=S2=C), seven (7.4%) each produced an identical miRNA, which was diverged from the miRNA produced by an respective ortholog in common bean (dubbed type II: S1=S2∼C); eight (8.5%) each produced two diverged miRNAs, one of which was identical to the miRNA produced by an respective ortholog in common bean (type III: S1∼S2=C); 10 (10.6%) each produced two diverged miRNAs, neither of which was identical to the miRNA produced by an respective ortholog in common bean (type IV: S1∼S2∼C), and two (9.6%) each produced two diverged miRNAs, but the respective ortholog in common bean was a pMIRNA (type V: S1∼S2?C). By contrast, of the 28 MIRNA singletons in soybean and respective orthologs in common bean, 19 (67.9%) each produced an identical miRNA in both species (type VI: S=C), seven (25%) each produced diverged miRNAs (type VII: S∼C), and two (7.1%) were pMIRNAs in common bean (type VIII: S?C).
Table 5. Comparison of the Orthologous MIRNAs in Soybean and Common Bean.
| Conservation or Divergence of MIRNAs | Types | Numbers |
|---|---|---|
| Category of duplicates | Pairs of duplicates | |
| miRNAs from two MIRNA duplicates in soybean and their ortholog in common bean are same | S1=S2=C | 67 |
| miRNAs from two MIRNA duplicates in soybean are same, but different from their counterpart in common bean | S1=S2∼C | 7 |
| miRNAs from one of the two MIRNA duplicates in soybean is same as its counterpart in common bean | S1∼S2=C | 8 |
| miRNAs from two MIRNA duplicates in soybean and their ortholog in common bean are all different | S1∼S2∼C | 10 |
| miRNAs from two MIRNA duplicates in soybean are different and no counterparts were detected in common bean | S1∼S2?C | 2 |
| Category of singletons | Singletons | |
| miRNAs from the MIRNA orthologs in soybean and common bean are same | S=C | 19 |
| miRNAs from the MIRNA orthologs in soybean and common bean are different | S∼C | 7 |
| miRNAs from MIRNAs in soybean were not detected in common bean | S?C | 2 |
It is notable that the majority (90.5%) of duplicated MIRNA pairs, whose two members generate identical miRNAs in soybean, have orthologs producing identical miRNAs in common bean. By contrast, of the 20 duplicated MIRNA pairs whose two members produce diverged miRNAs, only eight (40%) have orthologs producing miRNAs identical to one of the two miRNAs in soybean. These observations suggest that homologous MIRNAs producing identical miRNAs in soybean tend to have orthologs that produce identical miRNAs in common bean.
Comparative analysis of the MIRNA orthologs between soybean and common bean revealed potential gain- or loss-of-function mutations that have occurred in one of the two duplicated MIRNA homologs in soybean. As exemplified in Supplemental Figure 5, an insertion of a single nucleotide “G” in one of a duplicated pMIRNA structures appears to have occurred after the recent soybean WGD event in soybean, which may have created a gain-of-function MIRNA that produces miR169 in soybean given the fact that neither its homoeolog in soybean nor its ortholog in common bean without the insertion of “G” apparently enable to produce miRNA products. Another example is a single point mutation that appears to have occurred in one of the two duplicated MIR5778 precursor genes after the recent WGD event, which may have led to the formation of a pMIRNA in soybean. In addition to expressional gain and loss of duplicated MIRNAs, changes in accumulation levels of duplicated MIRNAs were detected by analysis of the relative abundance of distinguishable miRNAs produced by the paired MIRNAs (Supplemental Table 8).
The PEG Targets of MIRNA Duplicates Are More Preferentially Retained as Duplicates Than the PEG Targets of MIRNA Singletons
To understand whether the fractionation of duplicated MIRNAs may have shaped the pattern of retention and elimination of their miRNA targets following the recent WGD event in soybean, we selected and analyzed 289 miRNA targets that have been validated using three parallel analysis of RNA ends (PARE) libraries (Song et al., 2011; Shamimuzzaman and Vodkin, 2012; Hu et al., 2013; Arikit et al., 2014; Supplemental Figure 6). It was found that the 289 PEGs were targeted by 155 miRNAs generated from 265 MIRNAs, including 186 MIRNA duplicates and 79 MIRNA singletons. Among these 186 duplicated MIRNAs and 79 MIRNA singletons, 123 and 47 were predicted to be able to generate 71 and 43 miRNAs, respectively, to target two sets of nonoverlapping genes. It was predicted that the miRNAs from the 123 duplicated MIRNAs targeted 111 PEG duplicates and 31 singletons, while the miRNAs from the 47 MIRNA singletons targeted 39 PEG duplicates and 20 singletons. Statistically, the ratio (1:3.6) of PEG singletons versus PEG duplicates targeted by miRNAs from the duplicated MIRNAs was significantly higher than the ratio (1:2.0) of PEG singletons versus PEG duplicates targeted by miRNAs from the MIRNA singletons (P < 0.01, χ2 test).
The PEG Targets of Conserved MIRNAs Are More Preferentially Retained as Duplicates Than the PEG Targets of Nonconserved MIRNAs
To understand whether the intra- and interspecific conservation of MIRNAs is associated with conservation of the corresponding miRNA targets, we further analyzed the 289 genes targeted by 155 miRNAs generated by 265 MIRNAs, as described above (Supplemental Figure 7). These 265 MIRNAs were grouped into two categories: conserved MIRNAs and nonconserved MIRNAs. The former refer to the MIRNAs with orthologs in common bean and the latter refer to the MIRNAs without orthologs in common bean. Of the 265 MIRNAs, 163 are conserved and 102 are nonconserved between the two species. When predicted targets of miRNAs generated by both conserved and nonconserved MIRNAs were excluded, there remained 105 conserved MIRNAs and 70 nonconserved MIRNAs, which were predicted to be able to generate 59 and 56 miRNAs, respectively, to target two sets of nonoverlapping genes. The miRNAs from the 105 duplicated MIRNAs targeted 95 duplicated genes versus 21 singletons, while the miRNAs from the 70 MIRNA singletons targeted 53 duplicated genes and 34 singletons. Statistically, the ratio (1:4.5) of singletons versus duplicated genes targeted by miRNAs from the duplicated MIRNAs was significantly lower than the ratio (1:1.6) of singletons versus duplicated genes targeted by miRNAs from the MIRNA singletons (P < 0.001, χ2 test).
The MIR1510 Family Was Derived from an Ancient Duplication in Legumes and Targets Numerous NB-LRR Genes
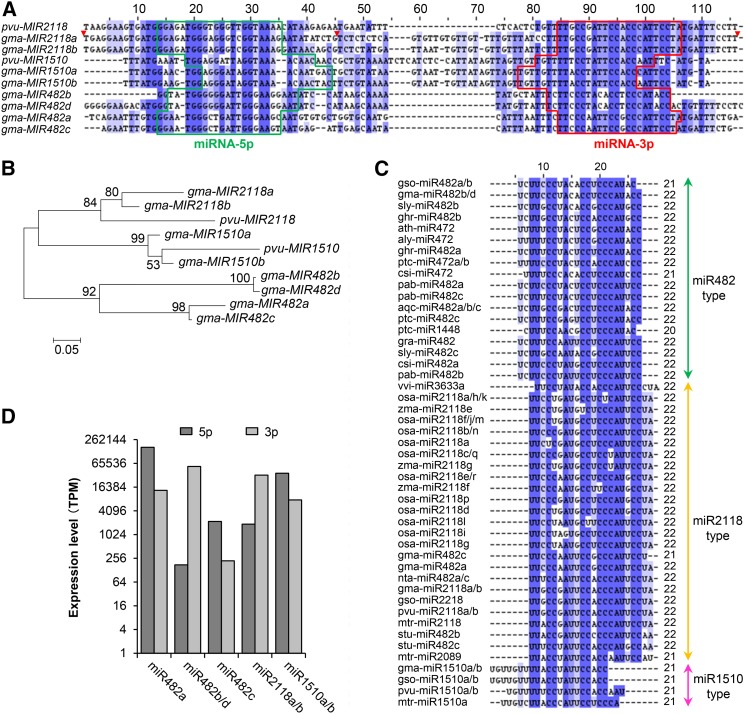
Plant nucleotide binding leucine-rich repeat proteins (encoded by NB-LRR genes) are generally grouped into two subclasses: the Toll/Interleukin-1 receptors-like group (TIR-NB-LRRs [TNLs]) and a coiled-coil domain containing group (CC-NB-LRRs [CNLs]) (Meyers et al., 1999; Dangl and Jones, 2001; Jones and Dangl, 2006). Both classes are targeted by miRNAs, typically generating phasiRNAs, which could reduce the levels of the transcripts of their targets in cis and in trans (Fei et al., 2013). A MIRNA superfamily composed of the MIR482 family and the MIR2118 family target NB-LRRs at the encoded and conserved P-loop motif and is highly conserved among divergent plant species including Arabidopsis, tomato (Solanum lycopersicum), Medicago truncatula, soybean, and even the grasses (Lu et al., 2006; Subramanian et al., 2008; Szittya et al., 2008; Zhai et al., 2011; Shivaprasad et al., 2012). In addition, a third, related family, MIR1510, was recently identified in legumes such as M. truncatula and soybean (Subramanian et al., 2008; Szittya et al., 2008; Zhai et al., 2011; Arikit et al., 2014; Zhao et al., 2015), indicating the existence of this MIRNA family prior to the divergence of these two species from a common ancestor ∼50 MYA (Bertioli et al., 2009; Severin et al., 2011). As expected, MIR1510 was also found in common bean (Figures 5A and 5B).
Figure 5.
MIR1510, MIR482, and MIR2118 Share a Common Origin.
(A) Sequence alignment of MIR1510, MIR482, and MIR2118 family members. The unaligned or poorly aligned regions (red triangles) of gma-MIR2118a were removed for better view. pvu-MIR2118 and pvu-MIR1510 indicate the orthologous copies from the common bean genome.
(B) Phylogenetic relationship of MIRNA precursors. Bootstrap values were calculated from 1000 replicates. Sequence alignment is shown in Supplemental Data Set 8.
(C) Conservation profile of miR1510 with miR482 and miR2118 in diverse plant species. The mature miRNA sequences were retrieved from miRBase.
(D) The expression values of miR482, miR2118, and miR1510 in the soybean small RNA libraries. TPM, transcripts per million mapped reads.
To elucidate the evolutionary origin of MIR1510, we extracted the MIRNA sequences of all MIRNAs belonging to the MIR482/MIR2118/MIR1510 superfamily from soybean and common bean, compared the genomic regions harboring these MIRNAs between the two species, and generated a phylogeny of these MIRNAs. Our results suggest that the two MIR2118s (i.e., gma-MIR2118a and gma-MIR2118b) and the two MIR1510s (i.e., gma-MIR1510a and gma-MIR1510b) are homoeologous pairs generated by the recent WGD, which occurred ∼13 MYA, orthologous to the common bean pvu-MIR2118 and pvu-MIR1510, respectively (Figures 5A and 5B; Supplemental Figures 8 and 9). The results also suggest that MIR2118 and the MIR1510 were generated by a duplication seemingly predating the split of soybean and common bean from a common ancestor. Because MIR1510 was also found in M. truncatula (Figure 5C; Zhai et al., 2011), it is thus reasonable to deduce that the duplication of the MIR2118 and MIR1510 lineages occurred prior to the divergence of soybean/common bean and M. truncatula (Bertioli et al., 2009; Severin et al., 2011). However, because the genomic regions harboring MIR2118 and MIR1510 did not show clear syntenic relationships in either soybean/common bean or M. truncatula, while the MIR2118 region in common bean is syntenic to a genomic region without MIR1510 (Schmutz et al., 2014), suggesting that the duplication event producing these two MIRNAs may not be the WGD shared by the soybean/common bean and M. truncatula lineages proposed to have occurred ∼50 to 60 MYA. Given that MIR2118 is shared by dicots and monocots (Figure 5C; Supplemental Figure 9), while MIR1510 is apparently specific to legumes, the MIR1510 group is perhaps a variant specifically formed in the legume lineage, representing an evolutionary novelty mediated by segmental or single-gene duplication. It is also interesting to mention that MIR482 variants were not present in the orthologous regions (Supplemental Figure 10) or any other regions in the common bean genome (Schmutz et al., 2014), but they present in two pairs of homoeologous regions in soybean that were derived from the older WGD event (Figure 5C), suggesting that the original counterpart(s) of soybean MIR482b/d and MIR482a/c in common bean must have both been eliminated. For all members of the MIR482/MIR2118/MIR1510 superfamily in soybean, high levels of abundance of both miRNA-5p and miRNA-3p were detected, with the former predicted to target a variety of PEGs and the later mainly regulating NB-LRR gene family (Figure 5D).
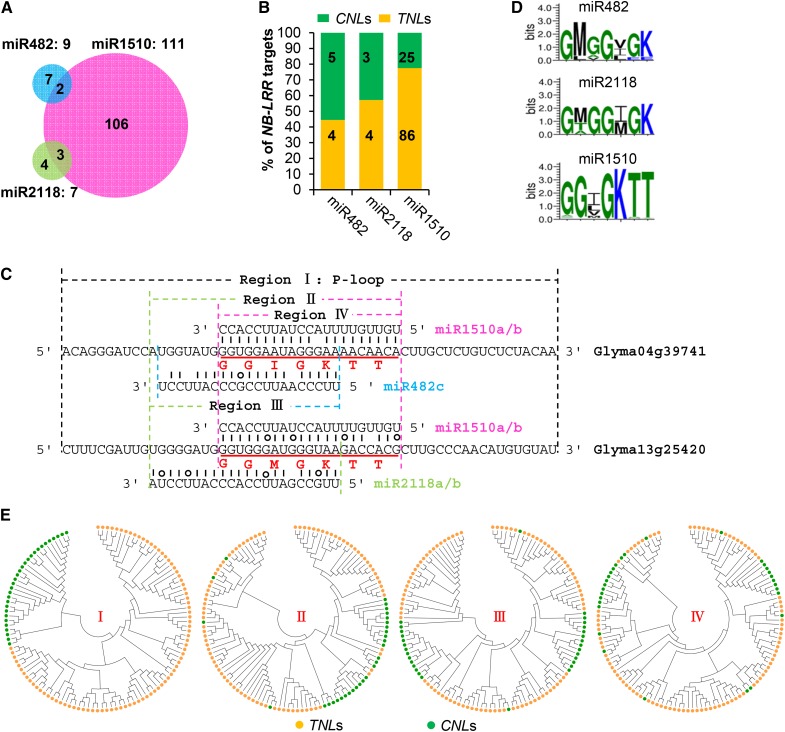
More interestingly, miR1510, which is 21 nucleotides in length, was predicted to target 111 NB-LRRs, including 86 TNLs and 25 CNLs in soybean. By contrast, the highly conserved 22-nucleotide miR2118 and 22-nucleotide miR482 were predicted to only target seven and nine NB-LRRs, respectively, with three and two predicted targets overlapped with the predicted targets of miR1510 (Figures 6A and 6B; Supplemental Figure 11 and Supplemental Data Set 7). A careful examination of the precursor sequences of the three MIRNAs reveals a shift of eight nucleotides resulting from natural variation in the stem of the predicted MIR1510 (Figure 6C; Supplemental Figure 12). This 8-nucleotide shift better aligns miR1510 with the highly conserved nucleotides that encode the core of the P-loop and appears to be responsible for the formation of MIR1510, including both miR1510-5p and miR1510-3p.
Figure 6.
Predicted Targets of miR482, miR2118, and miR1510 in Soybean.
(A) Comparison of the predicted NB-LRR targets of miR482, miR2118, and miR1510 at or under a penalty score of 5.
(B) Different classes of NB-LRR targets. The numbers embedded in the regions of each column represent the counts of NB-LRR targets of the corresponding class. CNLs, CC-NB-LRRs; TNLs, TIR-NB-LRRs.
(C) Two randomly selected NB-LRR genes targeted by miR482, miR2118, and miR1510.
(D) Consensus coding sequences of the target regions.
(E) Phylogenetic analysis of NB-LRR targets of miR1510, miR482, and miR2118 in soybean. The first phylogenetic tree was constructed using the nucleotide sequences of the conserved P-loop domains described by Meyers et al. (2003) (Region I in [C]). The second, third, and fourth trees were constructed using the nucleotide sequences of Regions II, III, and IV in (C), respectively. Sequence alignments are shown in Supplemental Data Sets 9 to 12.
The 21-nucleotide miRNA1510 showed canonical characteristics of the 22-nucleotide miRNA triggers associated with ARGONAUTE1 (AGO1) for initiation of PHAS loci (Figure 5C), such as “U” in the 5′ position, a common feature of miRNAs loading to AGO1, “A” in position 10, and “C” in the 3′ position (Chen et al., 2010). In addition, a previous analysis of PARE data and soybean miRNAs demonstrated that miR1510 directs cleavage and triggers biogenesis of phasiRNAs from at least 20 NB-LRRs (Arikit et al., 2014). Therefore, miR1510 is clearly a trigger for producing phasiRNAs, although the miR1510-5p:miR1510-3p duplex does not show any asymmetrically positioned bulged base(s) (Supplemental Figure 8). Such bulged bases, rather than miRNA-5p or miRNA-3p length, have been shown to be a critical factor for some 22-nucleotide miRNAs to trigger the production of secondary siRNAs (Manavella et al., 2012). The PARE data and small RNA data previously generated by our studies in soybean (Song et al., 2011; Shamimuzzaman and Vodkin, 2012; Hu et al., 2013; Arikit et al., 2014; Zhao et al., 2015) reveal that miR1510 targets the encoded, core P-loop motif of NB-LRRs, a sequence highly conserved among the 111 predicted targets in soybean (Figures 6C and 6D; Supplemental Figure 13). By contrast, the target sites of miR482 and miR2118 in NB-LRR are more diverged, to a level at which only a few NB-LRR copies could be targeted by these two interspecifically highly conserved miRNAs; as a result, NB-LRRs in soybean are primarily targeted by miR1510. In soybean, miR1510 functions as the primary miRNA trigger of phasiRNAs from NB-LRRs despite its 21-nucleotide length.
Evolution of NB-LRR Genes Appears to Be Affected by the Evolution of the miRNA Triggers of PhasiRNAs
To understand the potential evolutionary interplay between NB-LRR genes and their miRNA trigger of phasiRNAs, particularly after the birth of miR1510, we performed phylogenetic analysis of the putative NB-LRR targets of these three MIRNA families using sequences from multiple region/subregions of the P-loop domain, including the full length of the P-loop, the region covered by the target sites of miR482/miR2118 and miR1510, the miR482/miR2118 target site, and the miR1510 target site (Figure 6C). When the full length of the P-loop was used in phylogenetic analysis, the TNLs and CNLs were exclusively grouped into two distinct and distant clades (Figure 6E; Supplemental Figure 11). When the three subregions within the P-loop were used, proportions of CNLs were grouped into clades dominated by TNLs, and some CNLs were even found to be the ones closet to some of the TNLs within the same clades (Figures 6E). These observations suggest that sequence homogenization or recombination involving these miRNA target sites may have occurred between some of the TNLs and CNLs in soybean, leading to the formation of the CNL variants that can be targeted by miR1510.
DISCUSSION
Although miRNA-mediated gene regulation that affects various biological pathways is recognized as a widespread phenomenon in the plant kingdom (Carrington and Ambros, 2003; Bartel, 2004; Carthew and Sontheimer, 2009), our understanding of MIRNA evolution, particularly after WGD events, has been limited. On the one hand, some plant miRNA families are highly conserved across species (Zhang et al., 2006; Fahlgren et al., 2010), but on the other hand, more miRNA families are not shared than are shared across phylogenetically distant species (Fahlgren et al., 2010; Cuperus et al., 2011; Montes et al., 2014). Furthermore, these studies generally focused on miRNAs instead of their precursors. Thus, our view of the evolutionary conversation and divergence of MIRNAs across plants has remained blurry, including the evolutionary processes and consequences of duplicated MIRNAs in the complex paleopolyploid genomes. In this study, we illustrated the origin, distribution, duplication types and status, evolutionary rates, subgenomic and interspecific conservation and divergence, evolutionary novelty of MIRNAs, and miRNA-mediated coevolution between MIRNAs and target PEGs in the paleopolyploid soybean by genome-wide comparison with orthologous regions in common bean, thereby providing novel insights into the nature, patterns, and processes of MIRNA evolution primarily triggered by a WGD event.
Rapid Birth and Purge of MIRNA Genes
We observed a significantly higher ratio of MIRNA singletons to MIRNA duplicates than the ratio of PEG singletons to PEG duplicates in soybean. Nearly half of these MIRNA singletons appear to have arisen by gain-of-function changes from repetitive sequences that are predominantly composed of TEs (Figure 1, Table 1). Among these TEs, >80% are truncated fragments, remnants of ancient TEs (Supplemental Data Set 2). Actually, a proportion of TE sequences harboring MIRNAs are present even at homoeologous loci in soybean or orthologous loci between soybean and common bean (Table 1), suggesting that their insertions predate the recent WGD event that occurred in soybean ∼13 MYA or the split of the two species ∼19 MYA. Nevertheless, the ages of TEs do not always or may not reflect the dates when the MIRNAs came into existence. Indeed, many of the TE-related MIRNAs appear to have been formed very recently, given the fact that none of these MIRNAs are shared by any other TEs belonging to a same family. For example, only three MIRNA-containing TEs were found to be intact LTR-retrotransposons, which were dated to 1.3, 1.6, and 2.8 MYA, but none of any other intact LTR-retrotransposons belonging to these families, regardless of their ages (Supplemental Figure 2; Du et al., 2010), were detected to harbor MIRNAs, suggesting that the TE-derived MIRNAs were formed independently without further proliferation via their host elements. It is unclear whether these host elements were “dead” before the MIRNA formation or whether their activities were suppressed upon the formation of the MIRNAs.
We would like to point out that the MIRNAs within TE sequences shared by soybean and common beans at orthologous regions were presumably formed before the split of the two species, if one believes that independent formations of an MIRNA at an orthologous locus between the two species would rarely occur. If this is true, then the orthologous TE-derived MIRNAs could have been under some level of functional constraints during independent evolution of soybean and common bean, resulting in their preservation in both species. Under this assumption, it is also reasonable to deduce that some MIRNA singletons in soybean, particularly those shared by common bean at orthologous loci, are more likely to be formed by deletion or decay of the other MIRNA copies derived from the WGD, rather than birth of new MIRNAs after the WGD event. Nevertheless, the relative abundances of the predicted miRNAs from the majority of these TE-related MIRNAs are relatively low; thus, this category of MIRNAs remains to be further validated.
We demonstrated different retention rates for MIRNA duplicates among those located in different portions of PEGs, including introns, exons, UTRs, and junctions of different genic components, and between those located in genic regions and harbored by unclassified sequences (Table 1). An extremely low retention rate (2.1:1) for MIRNA duplicates was observed in introns compared with other genic portions. Because these introns did not contain any detectable TE sequences, such a low retention rate is more likely to be the outcome of a faster rate of purge of MIRNAs from introns, which generally have limited impact on gene functions and are thus more easily retained. Of course, we could not fully rule out the possibility that some of the MIRNA singletons could be the products of insertions by transposition or other mechanisms. The average retention rate for MIRNAs in unclassified regions was found to be even higher than that for MIRNAs in genic regions. If intronic sequences are nearly neutral, the detected higher retention rate for MIRNAs in unclassified sequences may indicate that these unclassified sequences were under some level of functional constraint.
Duplication Status Is a Key Indicator of Interspecific Conservation of MIRNAs
Comparative analysis of MIRNAs and their flanking regions between soybean and common bean revealed a striking distinction between the soybean MIRNA duplicates and MIRNA singletons in the relative preservation of their orthologs in common bean (Tables 1 and 5). When TE-related MIRNAs were excluded, ∼67% of MIRNA duplicates in soybean were found to have orthologs in common bean, while only ∼20% of MIRNA singletons in soybean were found to have orthologs in common bean. In general, when both members of a duplicated MIRNA pair are retained at homoeologous sites of duplicated regions, it is believed that neither of the two members was involved in local genomic rearrangements. By contrast, a singleton could be explained solely by a local deletion/insertion, translocation, or tandem duplication event without DNA removal from the genome. The possibility for formation of some MIRNA singletons without DNA removal from the genome appears to be echoed by the existence of homologous sequences of 22 soybean MIRNAs that do not have orthologs in common bean. However, only seven of the 48 MIRNA homologs in soybean were found to be MIRNAs in common bean, versus 118 of the 122 MIRNA orthologs were identified to be MIRNAs in common bean (Tables 1 and 5; Supplemental Data Set 5), suggesting that it is very unlikely that these common bean homologs are the relocated copies of original orthologs of soybean. Moreover, ∼74% of the soybean MIRNA singletons do not even have homologous sequences in common bean, versus only ∼28% of the soybean MIRNA duplicates lacking homologous sequences in common bean. Together, these lines of observations suggest that the majority of the singletons in soybean, particularly those unrelated to TEs, were formed by rapid decay or removal of their respective duplicated copies from the soybean genome. If true, then the lower rate of preservation of MIRNA singleton orthologs than those of MIRNA duplicates in common bean would indicate that overall the MIRNA duplicates retained in soybean have been under stronger functional constraints than MIRNA singletons. This would be also true for the orthologs of the soybean MIRNA duplicates in common bean.
Effects of Local Genomic Stability on Formation of MIRNA Singletons Are Detectable at Small Scales
The effects of local genomic features, such as gene density, proportion of TEs, rates of recombination, and their interplay, on removal of both TE sequences and genic sequences have been described (Wicker et al., 2007; Tian et al., 2009; Du et al., 2012). In addition, analyses of the distribution of PEG singletons versus duplicates revealed that a majority of singletons in paleopolyploid genomes (maize and soybean) were dispersed in duplicated blocks, rather than clustered (Langham et al., 2004; Ma et al., 2005; Wicker et al., 2007). This suggests that these PEG singletons were likely formed by independent events that removed single genes, perhaps primarily through accumulation of small deletions generated by illegitimate recombination (Devos et al., 2002; Ma et al., 2004; Wicker et al., 2007). In this study, we found that the duplication status of MIRNAs is associated with the duplication status of their flanking PEGs, suggesting that local genomic features do have effects, at a very fine scale, on formation of small genomic deletions that remove both PEG and adjacent MIRNA singletons.
Interplays among Duplication Status, Evolutionary Rates, Relative Abundance, and Functionality of MIRNAs
PEG duplicates evolve significantly slower than singletons in eukaryotes (Davis and Petrov, 2004; Jordan et al., 2004; Yang and Gaut, 2011; Du et al., 2012). Similar to PEGs, MIRNA duplicates were found to evolve significantly slower than MIRNA singletons in the past million years or so, regardless of whether these MIRNAs were located in genic or unclassified regions (Table 2). The miRNA-5p and miRNA-3p are the portions of MIRNAs that have undergone strongest selection (Figure 5; Supplemental Table 5). The relatively low rates of nucleotide divergence for MIRNA duplicates, consistent with the relatively high rates of preservation for MIRNA duplicates indicate a relatively strong functional constraint on these retained MIRNA duplicates, which explains why the majority of duplicated MIRNA pairs, whose two members generate identical miRNAs in soybean, have orthologous MIRNAs in common bean (Table 5; Supplemental Data Set 5). Indeed, duplicated status, rather than maintenance of orthologous relationship, appears to the primary force driving MIRNA divergence, based on the observations that nonconserved MIRNA singletons evolve faster than conserved MIRNA singletons, but the conserved duplicates and nonconserved duplicates did not show differences in the overall rates of nucleotide divergence (Table 3), and MIRNA duplicates showed relatively higher expression than singletons (Table 4). The relative slow evolution and strong functional constraint of both PEG and MIRNA duplicates and preferential retention of PEG targets of conserved MIRNAs appear to support the gene balance theory, which predicts that maintaining proper balance in the concentrations of protein subunits in a macromolecular complex and members of regulatory networks and highly connected portions of signaling duplicated networks is vital to maintain normal function and that an imbalance may lead to either decreased fitness or lethality (Birchler and Veitia, 2007; Freeling, 2008; Veitia et al., 2008; Edger and Pires, 2009).
The relatively low rates of nucleotide divergence are not limited to duplicated MIRNAs generated by WGD; MIRNA duplicates generated by tandem duplication also showed slower evolution than singletons associated with tandem duplication events (Supplemental Table 6). Interestingly, no significant difference in evolutionary rates was observed between MIRNA duplicates with both homologs generated by WGD and with paralogs produced by tandem duplication retained in the genome versus MIRNA duplicates without paralogs in the genome, suggesting that the dosage effects of MIRNA duplicates may be negligible.
What are the evolutionary consequences of MIRNA duplicates? Clearly, 39 p-MIRNAs that are homoeologous to 39 expressed MIRNAs, but not expressed in any tissues, would be considered as the outcomes of nonfunctionalization, whereas the 69 duplicated MIRNA pairs each with two members producing identical miRNAs likely maintained the same original functions, unless they are differentially regulated to accumulate in different tissues or at different developmental stages. However, because of their slower evolution and higher interspecific preservation, and because the majority of these duplicates have common orthologs producing identical miRNAs (i.e., S1=S2=C; Table 5), these duplicates are more likely to be functionally conserved. Variations in sequence divergence and levels of expression among homoeologous and orthologous MIRNAs, e.g., “S1~S2=C” and “S1~S2~C”, could be the outcomes of neofunctionalization, subfunctionalization, or speciation. Further in-depth structural and functional analyses of specific miRNA families may help to exemplify these evolutionary scenarios.
Coevolution of MIRNAs and PEGs through miRNA-Mediated Regulation
Because multiple MIRNAs could produce identical miRNAs, and an individual miRNA could regulate expression of multiple PEG targets, in many cases, the evolution of MIRNAs on the evolution of their PEG targets cannot be elucidated. This is particularly true for many MIRNA duplicates producing identical miRNAs. Nevertheless, we detected propensities for miRNA-mediated MIRNA-PEG interactions, which are reflected by two tendencies for retention of duplicates: (1) PEG targets of MIRNA duplicates tend to be retained as duplicates rather than PEG targets of MIRNA singletons, and (2) PEG targets of conserved MIRNAs tend to be retained as duplicates rather than PEG targets of nonconserved MIRNAs. Given that both MIRNA duplicates and PEG duplicates have undergone stronger functional constraints than MIRNA singletons and PEG singletons, such tendencies would be indicative of coevolution between MIRNAs and their PEG targets.
The coevolution between MIRNAs and PEG targets was exemplified by structural and evolutionary analysis of NB-LRRs targeted by legume-specific miR1510, derived from the miR2118/miR482 superfamily (Figure 5). In tomato, CNLs comprise the major type of NB-LRRs and are targeted by miR482 (Shivaprasad et al., 2012). By contrast, TNLs are the main targets of miR1510 in soybean (Figures 6A and 6B), although CNLs remain more predominant than TNLs (Zhao et al., 2015). This distinction in the relative proportion of CNLs versus TNLs appears to be associated with the shift of an 8-nucleotide motif between miR482 and miR1510 (Figure 6C; Supplemental Figure 12). More intriguingly, chimeric structures seemingly caused by intergenic sequence exchanges between TNLs and CNLs or sequence homogenization biased toward the enrichment of miR1510 target site within the P-loop domains in the soybean genome was revealed by phylogenetic analysis (Figure 6E). Based on these observations, we propose that after the emergence of miR1510, miR1510 took over the targeting of some NB-LRRs from miR2118 as a result of target site mutations. Although the dynamic evolutionary history of NB-LRRs in soybean remains to be more fully elucidated, our study provides an appealing example of miRNA-mediated coevolution between an MIRNA and its PEG targets, showing how such coevolution has shaped the composition of NB-LRRs, an important gene family with primary roles in plant-pathogen interactions.
METHODS
Detection of Mature miRNA, MIRNA Gene, and PEG Expression
Small RNAs generated from the susceptible cultivar Williams and its nine NILs (Lin et al., 2013; Zhao et al., 2015) were used to estimate relative abundance (or levels of accumulation) of mature miRNAs. To detect the transcripts of MIRNAs embedded in the host PEGs and splicing sites of the host PEGs, RNA-seq data from 28 samples of soybean (Glycine max) tissues at different developmental stages previously generated (Shen et al., 2014) were retrieved and reanalyzed. The RNA-seq reads were uniquely mapped to the soybean reference genome allowing mismatch of a single nucleotide per paired reads using TopHat2 (Trapnell et al., 2009). The MIRNA transcripts and the splicing sites of the host genes with matched full-length RNA-seq reads were considered as coexistence of the MIRNA transcripts and normal transcripts of the PEG harboring the MIRNAs.
Identification of WGD MIRNA Pairs and Singletons
All known MIRNAs collected from miRBase (Kozomara and Griffiths-Jones, 2011) were further examined to remove the ones that were reannotated as siRNA-like miRNAs (Arikit et al., 2014). The remaining MIRNAs after manual inspection and the novel MIRNA genes previously identified by our team (Zhao et al., 2015) were included in our analyses. To characterize duplicated MIRNA pairs and MIRNA singletons, homoeologous PEG pairs and singletons were identified using the new version of soybean gene annotation (G. max v1.1) following the methods previously described (Du et al., 2012). The duplicated MIRNA pairs were defined as: (1) pairs of MIRNA sequences with high similarity (identity >80% and matched length >80%) and (2) pairs of MIRNA sequences flanked by upstream and downstream homoeologous PEG pairs in the duplicated genomic blocks as previously defined (Schmutz et al., 2010; Du et al., 2012).
Determination of Soybean MIRNA Orthologous Genes and Identification of Novel miRNAs in the Common Bean Genome
To identify orthologous MIRNA genes in the common bean (Phaseolus vulgaris) genome, homologous protein gene synteny between soybean and common bean were determined using MCScanX (Schmutz et al., 2010, 2014; Wang et al., 2012). Soybean MIRNA precursor sequences were used as queries to search again common bean reference genome using BLASTN with default parameters (Altschul et al., 1997). The candidate MIRNA gene hits to the corresponding pseudomolecules were further checked for the flanking gene synteny between soybean and common bean. MIRNA genes in the same syntenic region were considered as interspecific orthologs. The WGD MIRNA pairs in soybean and interspecific MIRNA orthologs were view using the Circos software (Krzywinski et al., 2009).
A combination of de novo prediction following a pipeline previously described (Zhai et al., 2011; Zhao et al., 2015) and ortholog comparison was used to identify new miRNAs in the assembled common bean genome (Schmutz et al., 2014). Publicly available small RNA sequencing data from root, flower, nodule, and developing seed were downloaded, trimmed, and mapped to the common bean genome using Bowtie (Langmead et al., 2009; Zhai et al., 2011) with the parameters only allowing perfect matches. CentroidFold with the CONTRAfold engine (Sato et al., 2009) was used to predict the secondary structures of MIRNA precursors.
Identification of MIRNAs Associated with Tandem Duplication
A total of 493 MIRNA precursors were used for identification of tandem duplicated MIRNAs. An all-against-all BLASTN search was performed using default parameters. The candidate BLAST hits were kept for manual inspection. Tandemly duplicated MIRNA genes were defined as genes in the gene pairs that (1) belong to the same MIRNA gene family and (2) are separated by less than one protein spacer gene.
Analysis of Sequence Divergence
Homologous sequences were aligned using MUSCLE program (Edgar, 2004) or ClustalW (Thompson et al., 1994) using default parameters with manual refinement and viewed by Jalview. The Ka and Ks of PEGs and nucleotide sequence divergence K of MIRNA genes were estimated using the yn00 and baseml modules in the PAML software, respectively (Yang 2007). Ka, Ks, and K were calculated by comparison of orthologs of protein and MIRNA genes between soybean and common bean and orthologs within seven highly diverged Glycine soja accessions (Li et al., 2014).
Phylogenetic Analysis of MIRNA Precursors and NB-LRR Targets
Potential targets of miR482, miR2118, and miR1510 were predicted using TargetFinder 1.6 (http://carringtonlab.org/resources/targetfinder) with the parameters of penalty score ≤ 5. The MIRNA precursor sequences and the target region or/and flanking sequences of 122 predicted NB-LRR genes were used to constructed the phylogenetic trees using the neighbor-joining maximum composite likelihood model integrated in MEGA4 (Tamura et al., 2007). Bootstrap values were calculated from 1000 replicates.
Statistical Analysis
The significance of difference of sequence divergence level, between MIRNA genes and different sets of PEGs, between conserved and nonconserved MIRNA genes, between MIRNA duplicates and singletons was estimated by Student’s t test or Student’s paired t test in the SAS software. The significance of different ratios of MIRNA singletons to MIRNA duplicates was evaluated by χ2 test.
Supplemental Data
Supplemental Figure 1. Schematic Description of MIRNA Genes Colocated with Intact Transposable Elements.
Supplemental Figure 2. Age Distribution of the Elements of Three LTR-RT Families Gmr21, Gmr42, and Gmr124.
Supplemental Figure 3. Comparison of the Duplicate Status of Flanking Genes of MIRNA Duplicates and MIRNA Singletons.
Supplemental Figure 4. Comparison of Evolutionary Rates between Different Categories of MIRNA Genes and PEGs in Soybean.
Supplemental Figure 5. Two Representative miRNAs May Have Gained Function via Mutation or Indels.
Supplemental Figure 6. Comparisons among MIRNA Duplicates and MIRNA Singletons and among Their Target Genes.
Supplemental Figure 7. Comparisons among Conserved and Nonconserved MIRNAs and among Their Target Genes.
Supplemental Figure 8. Predicted Stem-Loop Structures of Transcripts from MIR482, MIR2118, and MIR1510 in Soybean.
Supplemental Figure 9. Phylogenetic Relationship of MIRNAs Related to MIR482, MIR2118, and MIR1510. Sequence Alignment Is Shown in Supplemental Data Set 13.
Supplemental Figure 10. Orthologous Relationships of MIR1510, MIR2118, and MIR482 between Soybean and Common Bean.
Supplemental Figure 11. Phylogenetic Analysis of NB-LRR Genes in Soybean.
Supplemental Figure 12. Comparative Analysis of the miRNA-5p and miRNA-3p Regions of MIR1510 and MIR2118.
Supplemental Figure 13. Genomic View of Phased Secondary siRNAs Initiated from miR1510 Target Sites in an NB-LRR Gene in Soybean.
Supplemental Table 1. Orientations of MIRNA Genes Co-located with PEGs.
Supplemental Table 2. Description of MIRNAs with RNA-seq Reads Matched in 28 Soybean Tissues.
Supplemental Table 3. Description of Soybean MIRNA Genes with Orthologs and Homologs in Common Bean.
Supplemental Table 4. Statistics Analysis of Differential Retention of MIRNAs and PEGs in Soybean.
Supplemental Table 5. Comparison of Evolutionary Rates among Different Regions of MIRNA Hairpins.
Supplemental Table 6. Comparison of Evolutionary Rates of MIRNA Genes Involved and Not Involved in Tandem Duplication.
Supplemental Table 7. Comparisons of the miRNA Expression Levels of MIRNA Genes Located in Different Regions.
Supplemental Table 8. Comparison of the Expression Values of the 11 Duplicated miRNAs with Mutations.
Supplemental Data Set 1. Summary of MIRNAs Colocated with PEGs.
Supplemental Data Set 2. Summary of MIRNAs Colocated with Repetitive Sequences.
Supplemental Data Set 3. Summary of the Whole-Genome Duplicated PEGs in the Soybean Genome.
Supplemental Data Set 4. Summary of Duplicated MIRNA Pairs Identified in the Soybean Genome.
Supplemental Data Set 5. Mature miRNAs and MIRNAs Identified in the Common Bean Genome by Comparison with the MIRNA orthologs in Soybean and by de Novo Prediction.
Supplemental Data Set 6. Identification of Soybean Orthologous MIRNAs in Common Bean.
Supplemental Data Set 7. The Predicted NB-LRR Targets of miR482, miR2118, and miR1510 in the Soybean Genome.
Supplemental Data Set 8. Sequence Alignment of MIRNA MIR1510, MIR482, and MIR2118 Corresponding to the Phylogenetic Analysis in Figure 5B.
Supplemental Data Set 9. Sequence Alignment of the Full Length of the P-loop Domain Corresponding to the Phylogenetic Analysis in Figure 6E.
Supplemental Data Set 10. Sequence Alignment of the Region Covered by the Target Sites of miR482/miR2118/miR1510 Corresponding to the Phylogenetic Analysis in Figure 6E.
Supplemental Data Set 11. Sequence Alignment of the Region Covered by the miR482/2118 Target Sites Corresponding to the Phylogenetic Analysis in Figure 6E.
Supplemental Data Set 12. Sequence Alignment of the Region Covered by the miR1510 Target Sites Corresponding to the Phylogenetic Analysis in Figure 6E.
Supplemental Data Set 13. Sequence Alignment of MIRNA Genes Related to MIR482, MIR2118, and MIR1510 Corresponding to the Phylogenetic Analysis in Supplemental Figure 9.
Supplemental Data Set 14. Sequence Alignment of NB-LRR Genes Corresponding to the Phylogenetic Analysis in Supplemental Figure 11.
Supplementary Material
Acknowledgments
We thank Phillip SanMiguel for assistance in small RNA sequencing, Doug Yatcilla for software installation and testing, and Damon Lisch for helpful discussion and comments. This work was mainly supported by soybean check-off funds from the Indiana Soybean Alliance (Grant 205267) and partially supported by funds from the National Natural Science Foundation of China (Grant 31371647) and the Taishan Scholarship and High Level Talents Foundation of QAU (Grant 631304). Work on soybean small RNAs in the Meyers lab is supported by funding from the United Soybean Board and the North Central Soybean Research Program.
AUTHOR CONTRIBUTIONS
M.Z. performed the research and analyzed the data. B.C.M., C.C., and W.X. contributed analytic tools and methods. M.Z. and J.M. designed the research. M.Z., B.C.M., and J.M. wrote the article.
Glossary
- WGD
whole-genome duplication
- TE
transposable element
- PEG
protein-encoding gene
- miRNA
microRNA
- siRNA
small interfering RNA
- phasiRNA
phased siRNA
- MYA
million years ago
- UTR
untranslated region
- LTR
long terminal repeat
- PARE
parallel analysis of RNA ends
References
- Allen E., Xie Z., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C. (2004). Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36: 1282–1290. [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikit S., Xia R., Kakrana A., Huang K., Zhai J., Yan Z., Valdés-López O., Prince S., Musket T.A., Nguyen H.T., Stacey G., Meyers B.C. (2014). An atlas of soybean small RNAs identifies phased siRNAs form hundreds of coding genes. Plant Cell 26: 4584–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R., Bachtrog D. (2013). Neofunctionalization of young duplicate genes in Drosophila. Proc. Natl. Acad. Sci. USA 110: 17409–17414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bertioli D.J., et al. (2009). An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes. BMC Genomics 10: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Veitia R.A. (2007). The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19: 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington J.C., Ambros V. (2003). Role of microRNAs in plant and animal development. Science 301: 336–338. [DOI] [PubMed] [Google Scholar]
- Carter, T.E., Nelson, R.L., Sneller, C.H., and Cui, Z. (2004). Genetic diversity in soybean. In Soybeans: Improvement, Production, and Uses, 3rd ed, H.R. Boerma and J.E. Specht, eds (Madison, WI: American Society of Aronomy), pp. 303–416. [Google Scholar]
- Carthew R.W., Sontheimer E.J. (2009). Origins and Mechanisms of miRNAs and siRNAs. Cell 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Chen L.T., Patel K., Li Y.H., Baulcombe D.C., Wu S.H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 107: 15269–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Li Y.H., Wu S.H. (2007). Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 3318–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Fahlgren N., Carrington J.C. (2011). Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J.L., Jones J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Danilevskaya O.N., Hermon P., Hantke S., Muszynski M.G., Kollipara K., Ananiev E.V. (2003). Duplicated fie genes in maize: expression pattern and imprinting suggest distinct functions. Plant Cell 15: 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J.C., Petrov D.A. (2004). Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2: E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet R., Van de Peer Y. (2012). Redundancy and rewiring of genetic networks following genome-wide duplication events. Curr. Opin. Plant Biol. 15: 168–176. [DOI] [PubMed] [Google Scholar]
- Devos K.M., Brown J.K.M., Bennetzen J.L. (2002). Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12: 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Grant D., Tian Z., Nelson R.T., Zhu L., Shoemaker R.C., Ma J. (2010). SoyTEdb: a comprehensive database of transposable elements in the soybean genome. BMC Genomics 11: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Tian Z., Sui Y., Zhao M., Song Q., Cannon S.B., Cregan P., Ma J. (2012). Pericentromeric effects shape the patterns of divergence, retention, and expression of duplicated genes in the paleopolyploid soybean. Plant Cell 24: 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger P.P., Pires J.C. (2009). Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 17: 699–717. [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Jogdeo S., Kasschau K.D., Sullivan C.M., Chapman E.J., Laubinger S., Smith L.M., Dasenko M., Givan S.A., Weigel D., Carrington J.C. (2010). MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 22: 1074–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C., Chen Y., Long M. (2008). Recurrent tandem gene duplication gave rise to functionally divergent genes in Drosophila. Mol. Biol. Evol. 25: 1451–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q., Xia R., Meyers B.C. (2013). Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25: 2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. (2008). The evolutionary position of subfunctionalization, downgraded. Genome Dyn. 4: 25–40. [DOI] [PubMed] [Google Scholar]
- Gaut B.S., Wright S.I., Rizzon C., Dvorak J., Anderson L.K. (2007). Recombination: an underappreciated factor in the evolution of plant genomes. Nat. Rev. Genet. 8: 77–84. [DOI] [PubMed] [Google Scholar]
- He X., Zhang J. (2005). Gene complexity and gene duplicability. Curr. Biol. 15: 1016–1021. [DOI] [PubMed] [Google Scholar]
- Howell M.D., Fahlgren N., Chapman E.J., Cumbie J.S., Sullivan C.M., Givan S.A., Kasschau K.D., Carrington J.C. (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Jiang Q., Ni Z., Chen R., Xu S., Zhang H. (2013). Analyses of a Glycine max degradome library identify microRNA targets and microRNAs that trigger secondary siRNA biogenesis. J. Integr. Plant Biol. 55: 160–176. [DOI] [PubMed] [Google Scholar]
- Jiang N., Bao Z., Zhang X., Eddy S.R., Wessler S.R. (2004). Pack-MULE transposable elements mediate gene evolution in plants. Nature 431: 569–573. [DOI] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Jordan I.K., Wolf Y.I., Koonin E.V. (2004). Duplicated genes evolve slower than singletons despite the initial rate increase. BMC Evol. Biol. 4: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B., Arif M.A., Seumel G.I., Ossowski S., Weigel D., Reski R., Frank W. (2010). Transcriptional control of gene expression by microRNAs. Cell 140: 111–122. [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2011). miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39: D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J., Li Y., Messing J., Dooner H.K. (2005). Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc. Natl. Acad. Sci. USA 102: 9068–9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langham R.J., Walsh J., Dunn M., Ko C., Goff S.A., Freeling M. (2004). Genomic duplication, fractionation and the origin of regulatory novelty. Genetics 166: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M., Herendeen P.S., Wojciechowski M.F. (2005). Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol. 54: 575–594. [DOI] [PubMed] [Google Scholar]
- Li F., Pignatta D., Bendix C., Brunkard J.O., Cohn M.M., Tung J., Sun H., Kumar P., Baker B. (2012). MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 109: 1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.H., et al. (2014). De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 32: 1045–1052. [DOI] [PubMed] [Google Scholar]
- Lin F., Zhao M., Ping J., Johnson A., Zhang B., Abney T.S., Hughes T.J., Ma J. (2013). Molecular mapping of two genes conferring resistance to Phytophthora sojae in a soybean landrace PI 567139B. Theor. Appl. Genet. 126: 2177–2185. [DOI] [PubMed] [Google Scholar]
- Lu C., Kulkarni K., Souret F.F., MuthuValliappan R., Tej S.S., Poethig R.S., Henderson I.R., Jacobsen S.E., Wang W., Green P.J., Meyers B.C. (2006). MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 16: 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Devos K.M., Bennetzen J.L. (2004). Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14: 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., SanMiguel P., Lai J., Messing J., Bennetzen J.L. (2005). DNA rearrangement in orthologous Orp regions of the maize, rice and sorghum genomes. Genetics 170: 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella P.A., Koenig D., Weigel D. (2012). Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. USA 109: 2461–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClean P.E., Mamidi S., McConnell M., Chikara S., Lee R. (2010). Synteny mapping between common bean and soybean reveals extensive blocks of shared loci. BMC Genomics 11: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath C.L., Gout J.F., Doak T.G., Yanagi A., Lynch M. (2014). Insights into three whole-genome duplications gleaned from the Paramecium caudatum genome sequence. Genetics 197: 1417–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., Dickerman A.W., Michelmore R.W., Sivaramakrishnan S., Sobral B.W., Young N.D. (1999). Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20: 317–332. [DOI] [PubMed] [Google Scholar]
- Meyers B.C., Kozik A., Griego A., Kuang H., Michelmore R.W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., et al. (2008). Criteria for annotation of plant microRNAs. Plant Cell 20: 3186–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghe G.D., Hufnagel D.E., Tang H., Xiao Y., Dworkin I., Town C.D., Conner J.K., Shiu S.H. (2014). Consequences of whole-genome triplication as revealed by comparative genomic analyses of the wild radish Raphanus raphanistrum and three other Brassicaceae species. Plant Cell 26: 1925–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes R.A., de Fátima Rosas-Cárdenas F., De Paoli E., Accerbi M., Rymarquis L.A., Mahalingam G., Marsch-Martínez N., Meyers B.C., Green P.J., de Folter S. (2014). Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat. Commun. 5: 3722. [DOI] [PubMed] [Google Scholar]
- Ohno S. (1970). Evolution by Gene Duplication. (New York: Springer-Verlag; ). [Google Scholar]
- Otto S.P., Whitton J. (2000). Polyploid incidence and evolution. Annu. Rev. Genet. 34: 401–437. [DOI] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18: 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzon C., Ponger L., Gaut B.S. (2006). Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLOS Comput. Biol. 2: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulin A., Auer P.L., Libault M., Schlueter J., Farmer A., May G., Stacey G., Doerge R.W., Jackson S.A. (2012). The fate of duplicated genes in a polyploid plant genome. Plant J. 73: 143–153. [DOI] [PubMed] [Google Scholar]
- Sato K., Hamada M., Asai K., Mituyama T. (2009). CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res. 37: W277–W280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D.E., Soltis P.S. (1999). Polyploidy: recurrent formation and genome evolution. Trends Ecol. Evol. (Amst.) 14: 348–352. [DOI] [PubMed] [Google Scholar]
- Schmutz J., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183. [DOI] [PubMed] [Google Scholar]
- Schmutz J., et al. (2014). A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 46: 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J.C., Springer N.M., Freeling M. (2011). Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108: 4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Severin A.J., Cannon S.B., Graham M.M., Grant D., Shoemaker R.C. (2011). Changes in twelve homoeologous genomic regions in soybean following three rounds of polyploidy. Plant Cell 23: 3129–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamimuzzaman M., Vodkin L. (2012). Identification of soybean seed developmental stage-specific and tissue-specific miRNA targets by degradome sequencing. BMC Genomics 13: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Zhou Z., Wang Z., Li W., Fang C., Wu M., Ma Y., Liu T., Kong L.A., Peng D.L., Tian Z. (2014). Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 26: 996–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad P.V., Chen H.M., Patel K., Bond D.M., Santos B.A.C.M., Baulcombe D.C. (2012). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L.M., Burbano H.A., Wang X., Fitz J., Wang G., Ural-Blimke Y., Weigel D. (2015). Rapid divergence and high diversity of miRNAs and miRNA targets in the Camelineae. Plant J. 81: 597–610. [DOI] [PubMed] [Google Scholar]
- Song Q.X., Liu Y.F., Hu X.Y., Zhang W.K., Ma B., Chen S.Y., Zhang J.S. (2011). Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 11: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus A. (1999). On the possibility of constructive neutral evolution. J. Mol. Evol. 49: 169–181. [DOI] [PubMed] [Google Scholar]
- Subramanian S., Fu Y., Sunkar R., Barbazuk W.B., Zhu J.K., Yu O. (2008). Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics 9: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G., Moxon S., Santos D.M., Jing R., Fevereiro M.P., Moulton V., Dalmay T. (2008). High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics 9: 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z., Rizzon C., Du J., Zhu L., Bennetzen J.L., Jackson S.A., Gaut B.S., Ma J. (2009). Do genetic recombination and gene density shape the pattern of DNA elimination in rice long terminal repeat retrotransposons? Genome Res. 19: 2221–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crété P. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16: 69–79. [DOI] [PubMed] [Google Scholar]
- Veitia R.A., Bottani S., Birchler J.A. (2008). Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 24: 390–397. [DOI] [PubMed] [Google Scholar]
- Vogt A., Goldman A.D., Mochizuki K., Landweber L.F. (2013). Transposon domestication versus mutualism in ciliate genome rearrangements. PLoS Genet. 9: e1003659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687. [DOI] [PubMed] [Google Scholar]
- Wang Y., Tang H., Debarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H., Kissinger J.C., Paterson A.H. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T., Yahiaoui N., Keller B. (2007). Illegitimate recombination is a major evolutionary mechanism for initiating size variation in plant resistance genes. Plant J. 51: 631–641. [DOI] [PubMed] [Google Scholar]
- Yang L., Gaut B.S. (2011). Factors that contribute to variation in evolutionary rate among Arabidopsis genes. Mol. Biol. Evol. 28: 2359–2369. [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25: 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Pan X., Cannon C.H., Cobb G.P., Anderson T.A. (2006). Conservation and divergence of plant microRNA genes. Plant J. 46: 243–259. [DOI] [PubMed] [Google Scholar]
- Zhao, M., Cai, C., Zhai, J., Lin, F., Li, L., Shreve, J., Thimmapuram, J., Hughes, T.J., Meyers, B.C., and Ma, J. (2015). Coordination of microRNAs, phasiRNAs, and NB-LRR genes in response to a plant pathogen: Insights from analyses of a set of soybean Rps gene near-isogenic lines. Plant Gen., 10.3835/plantgenome2014.09.0044. [DOI] [PubMed]
- Zhou Z., Wang Z., Li W., Fang C., Shen Y., Li C., Wu Y., Tian Z. (2013). Comprehensive analyses of microRNA gene evolution in paleopolyploid soybean genome. Plant J. 76: 332–344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.