A microRNA-auxin/indole acetic acid repressor module regulates the extent of cell competence, thereby affecting the duration of cell proliferation and the development of lateral organs.
Abstract
MicroRNAs function in a range of developmental processes. Here, we demonstrate that miR847 targets the mRNA of the auxin/indole acetic acid (Aux/IAA) repressor-encoding gene IAA28 for cleavage. The rapidly increased accumulation of miR847 in Arabidopsis thaliana coincided with reduced IAA28 mRNA levels upon auxin treatment. This induction of miR847 by auxin was abolished in auxin receptor tir1-1 and auxin-resistant axr1-3 mutants. Further analysis demonstrates that miR847 functions as a positive regulator of auxin-mediated lateral organ development by cleaving IAA28 mRNA. Importantly, the ectopic expression of miR847 increases the expression of cell cycle genes as well as the neoplastic activity of leaf cells, prolonging later-stage rosette leaf growth and producing leaves with serrated margins. Moreover, both miR847 and IAA28 mRNAs are specifically expressed in marginal meristems of rosette leaves and lateral root initiation sites. Our data indicate that auxin-dependent induction of miR847 positively regulates meristematic competence by clearing IAA28 mRNA to upregulate auxin signaling, thereby determining the duration of cell proliferation and lateral organ growth in Arabidopsis. IAA28 mRNA encodes an Aux/IAA repressor protein, which is degraded through the proteasome in response to auxin. Altered signal sensitization to IAA28 mRNA levels, together with targeted IAA28 degradation, ensures a robust signal derepression.
INTRODUCTION
MicroRNAs (miRNAs), a class of short (21 to 24 nucleotides) noncoding RNAs, are processed in the nucleus as duplexes (miRNA:miRNA*) from primary miRNA transcripts (or MIRNA) containing imperfect intramolecular stem loops. The mature guide-stranded miRNA mediates the sequence-specific, posttranscriptional repression of mRNA targets (Jones-Rhoades et al., 2006; Carthew and Sontheimer, 2009). Most plant miRNAs directly cleave their targets, although in one case, diminution of the target protein without a corresponding reduction in the target mRNA indicates that plant miRNAs can also inhibit productive translation (Aukerman and Sakai, 2003; Chen, 2004). A recent study revealed the endoplasmic reticulum as the site of miRNA-mediated translation repression in Arabidopsis thaliana (Li et al., 2013). Plant miRNAs function in a range of developmental processes, ensuring proper embryonic, vegetative, and floral development (Jones-Rhoades et al., 2006) as well as tolerance and responses to extrinsic stresses (Sunkar and Zhu, 2004; Sunkar et al., 2006; Kruszka et al., 2012).
In Arabidopsis, miR172, miR156, miR159, and miR824 play important roles in the floral transition by targeting transcripts of the Apetala2, Squamosa-promoter Binding Protein-Like, MYB transcription factor, and Agamous-Like16 genes (Park et al., 2002; Aukerman and Sakai, 2003; Achard et al., 2004; Chen, 2004; Schwab et al., 2005; Wang et al., 2009; Wu et al., 2009; Kim et al., 2012; Hu et al., 2014). miR165/166 and miR319 regulate leaf development and morphogenesis by targeting gene transcripts encoding HD-ZIP and TCP transcription factors, respectively (Reinhart et al., 2002; Rhoades et al., 2002; Palatnik et al., 2003; Tang et al., 2003; Juarez et al., 2004). miR160, miR167, and miR390 play roles in root and lateral root development, as well as embryo, leaf, and floral organ development, by targeting transcripts of Auxin Response Factor10 (ARF10)/ARF16/ARF17, ARF6/ARF8, and ARF2/ARF3/ARF4 genes, respectively (Mallory et al., 2005; Wang et al., 2005; Wu et al., 2006; Gutierrez et al., 2009; Liu et al., 2010; Marin et al., 2010; Yoon et al., 2010). While miR160 and miR167 directly target their ARF transcripts, miR390 targets Trans-Acting siRNA3 to trigger the biogenesis of trans-acting small interfering RNAs, which subsequently inhibit ARF2/ARF3/ARF4 to release the repression of lateral root growth (Marin et al., 2010; Yoon et al., 2010). One miRNA may regulate multiple aspects of plant growth and development by targeting different mRNAs or interacting with other miRNA/target and environmental factors. For instance, targets of miR164 include mRNAs encoding five members of the NAC domain transcription factor family, including NAC1, Cup-Shaped Cotyledon1 (CUC1), and CUC2. The expression of miR164 in root targets NAC1 mRNA for cleavage to downregulate auxin signals for lateral root development (Guo et al., 2005), whereas the expression of miR164 in inflorescences targets CUC1/2 to regulate the establishment and maintenance of the shoot apical and axillary meristems and floral boundary formation (Laufs et al., 2004; Mallory et al., 2004; Baker et al., 2005). A recent study shows that miR156 also functions antagonistically with miR171 in regulating trichome distribution in Arabidopsis (Xue et al., 2014).
Auxin signaling plays diverse and pivotal roles in many aspects of the growth and development of plants. ARFs are repressed by AUXIN/INDOLE ACETIC ACID (Aux/IAA) proteins, which are degraded through the proteasome mediated by the auxin receptor TRANSPORT INHIBITOR RESPONSE1 (TIR1) protein in response to auxin (Dharmasiri et al., 2005a, 2005b). The degradation of Aux/IAA releases the ARFs that bind to the auxin-responsive cis-acting element in early auxin response genes, including Aux/IAAs, small auxin-up RNAs, and a group of GH3 proteins identified as early auxin-responsive proteins (Hagen and Guilfoyle, 2002). Several GH3 family genes mediate auxin homeostasis by conjugating amino acids to indole-3-acetic acid (Staswick et al., 2005) and further regulate root development (Guilfoyle and Hagen, 2007). In addition to repression by Aux/IAAs, the expression of many ARFs is directly mediated by at least one miRNA, such as miR160, miR167, or miR390 (Wang et al., 2005; Gutierrez et al., 2009; Marin et al., 2010; Yoon et al., 2010). miR393 also suppresses auxin signaling by degrading TIR1 mRNA (Navarro et al., 2006). NAC1 acts downstream of TIR1 to transmit auxin signals (Xie et al., 2000), whereas miR164 induced by auxin downregulates auxin signals by clearing NAC1 mRNA (Guo et al., 2005). These findings suggest that many miRNAs are involved in the response of plant hormones, particularly auxin, in the regulation of plant growth and development. However, the networks involving miRNA, mRNA, and plant hormones remain elusive.
Despite considerable research surveying the miRNA repertoire of individual plant species and the thousands of miRNAs in a diversity of plant species that have been predicted in the past decades (miRBase databases; http://mirbase.org), experimental functional studies of plant miRNAs are rather limited; as described above, most of these studies were confined to the abundant, previously identified, conserved miRNAs (Rajagopalan et al., 2006). Only a few low-abundance miRNAs, such as miR395 and miR399, have been studied in plants subjected to abiotic stresses (Jones-Rhoades and Bartel, 2004; Fujii et al., 2005; Chiou et al., 2006).
We report here a functional study of a low-abundance miRNA, miR847, in Arabidopsis. We found that the accumulation levels of miR847, which was not detected under normal circumstances, increased with decreasing IAA28 mRNA levels upon auxin treatment. We confirmed the miR847-mediated cleavage of IAA28 mRNA in vivo. IAA28, an Aux/IAA repressor, represses the auxin-induced transcription of genes that promote lateral root initiation (Rogg et al., 2001). We found that both miR847 and IAA28 mRNAs have similar expression patterns and striking cell type specificity not only in the roots but also in the aerial portions of Arabidopsis. Analysis of transgenic plants overexpressing miR847 or IAA28 cDNA and the IAA28 knockout iaa28-ko mutant and the gain-of-function iaa28-1 mutant revealed that the miR847/IAA28 module regulates the extent of cell competence, thereby determining the duration of cell proliferation and the development of lateral organs, including lateral roots, rosette leaves, and axillary shoots in Arabidopsis.
RESULTS
Identification and Expression Profile of miR847
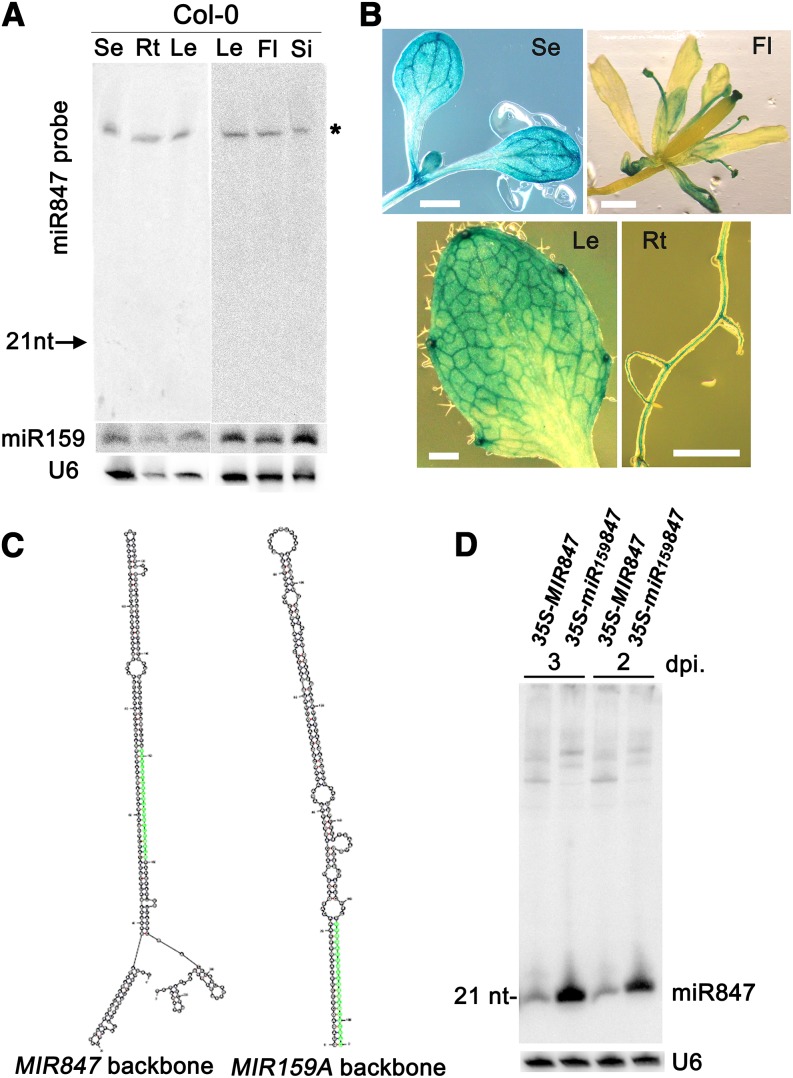
The function of miR847, which accumulates at low levels and was mainly isolated from Arabidopsis (Columbia-0 [Col-0] ecotype) seedlings (with 45 reads in a small RNA library containing 887,266 reads and 2, 2, and 5 reads in rosette leaves, flowers, and siliques, respectively) (Rajagopalan et al., 2006), was investigated. The accumulation of miR847 was undetectable in 8-d-old seedlings and roots and 2-week-old leaves, flowers, and siliques (Figure 1), whereas the accumulation of miR159 was readily detected (Figure 1A), consistent with the high cloning frequency of up to 6621 (for miR159a) and 982 (for miR159b) reads in the same small RNA library (Rajagopalan et al., 2006); these results confirm the low accumulation level of miR847. To determine the expression profile of the miR847-coding gene, we generated transgenic plants carrying a fusion of the precursor miR847 (MIR847) promoter and the β-glucuronidase gene (GUS). The expression of MIR847-GUS was readily detected in the shoot apex, leaf veins of the cotyledons, marginal meristems (leaf tooth) of the rosette leaves, sepals, stamen, and lateral root initiation sites (Figure 1B), indicating that MIR847 is expressed at higher levels but within specialized cells of different organs. We then examined the processing efficiency of mature miR847 production. The MIR847 backbone sequence was cloned into a construct under the control of the 35S promoter, producing 35S-MIR847. As a control, the miR159a precursor MIR159a, which is effective in expressing artificial miRNAs (Duan et al., 2008), was used as a backbone for the construction and expression of artificial miR847 under the control of the 35S promoter, producing 35S-miR159847 (Figure 1C). As shown in Figure 1D, miR847 was more highly produced from 35S-miR159847 than from 35S-MIR847 when they were transiently expressed in Nicotiana benthamiana via Agrobacterium tumefaciens infiltration, indicating the low processing efficiency of mature miRNA from the precursor-miR847 backbone sequence. This result might also explain the previous finding that only a close heterogeneous variant of the perfect miRNA* without the two-nucleotide 3′ overhangs typical of the Dicer-like products was sequenced (Rajagopalan et al., 2006). Nevertheless, both the striking cell type specificities and low processing efficiency indicate the low accumulation of miR847 under normal conditions.
Figure 1.
Expression Pattern of miR847.
(A) Examination of miR847 expression in various tissues of Arabidopsis. Small RNAs (60 µg, except 40 µg for roots) isolated from 8-d-old seedlings (Se), roots (Rt), and leaves (Le) and 2-week-old leaves, flowers (Fl), and siliques (Si) were loaded. LNA-modified oligonucleotide complementary to miR847 was used as a probe. No mature miR847 hybridization signal was detected. A band visible in the top blot (labeled with the asterisk) was possibly the miR847-coding precursor, MIR847, based on the size (230 bp) and detection by the miR847-specific sequence probe. The membrane was stripped and rehybridized with miR159-specific probe. The position of the miR159 hybridization signal in the original membrane is indicated by the arrow. U6 RNA hybridization is shown as a control.
(B) MIR847-GUS expression in 6-d-old seedling and 2-week-old leaf, root, and flower. Bars = 1 mm.
(C) Folding structure of MIR847 and MIR159A backbones. Green highlighted nucleotides indicate the locations of mature miR847 and miR159.
(D) Detection of miR847 by transient expression of 35S-MIR847 and 35S-miR159847 in N. benthamiana. Small RNAs (20 µg) isolated from N. benthamiana leaf at 2 and 3 d after infiltration (dpi) were used.
miR847-Mediated Cleavage of the Predicted Target IAA28 mRNA
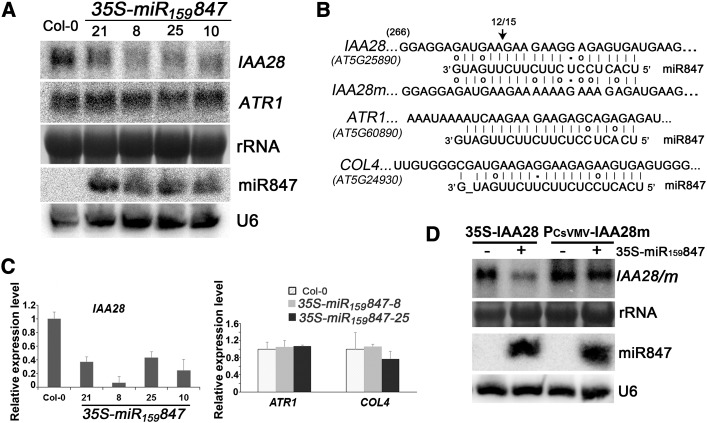
Putative targets for miR847 were predicted through computational approaches (Wu et al., 2012). Plenty of potential targets were predicted (Supplemental Data Set 1). Dozens of sequences of the best putative targets, which had scores of 2.2 or 2.5, were examined by 5′ rapid amplification of cDNA ends (RACE). No cleavage fragments corresponding to predicted targets were obtained, presumably due to the low expression of miR847. Therefore, we transformed the above 35S-miR159847 into wild-type Arabidopsis (Col-0). Figure 2 shows that the accumulation of miR847 was readily detected in all of the individual 35S-miR159847 transgenic lines (Figure 2A). A 5′ RACE assay was performed, and a specific cleavage site within the miR847 complementary regions was obtained for only one potential target, AT5G25890, encoding IAA28, but not for any other predicted targets (Figure 2B). The cleaved site was at nucleotide 278 of the IAA28 mRNA, which is located in, but not in the middle of, the miR847/IAA28 mRNA complementary region. Consistent with the results of 5′ RACE, the accumulation level of IAA28 was reduced consistently in 8-d-old seedlings and 25-d-old rosette leaves of different 35S-miR159847 lines, as determined by RNA gel blot and quantitative RT-PCR (qRT-PCR) analyses (Figures 2A and 2C). There was no obvious change in the accumulation levels of other putative targets of miR847 in any of the 35S-miR159847 lines tested (Figures 2A and 2C). To further confirm that the IAA28 mRNA was the target of miR847, we introduced four nucleotide mutations into the miR847/IAA28 mRNA complementary region of the IAA28 mRNA sequence without changing the encoded amino acids, and the mutant (designated IAA28m) was confirmed to be resistant to miR847-mediated cleavage in a transient plant system assay (Figure 2D). Our results indicate that IAA28 mRNA is a target of miR847.
Figure 2.
Identification of miR847 Targets.
(A) Overexpression of miR847 and effect of miR847 on IAA28 mRNA accumulation. Total RNAs (30 µg) or small RNAs (60 µg) from 8-d-old seedlings of 35S-miR159847 transgenic Arabidopsis and control Col-0 were used. IAA28 mRNA and ATR1 mRNA, two computational predicted targets of miR847, as well as miR847 are indicated. 28S rRNA and U6 were used as loading controls.
(B) Alignments of miR847 with IAA28 (wild type) and IAA28 mutant (IAA28m) and ATR1 and COL4 mRNAs, two of the computationally predicted targets. G-U base pairing is shown with dots, and mismatches are indicated with circles. The arrow indicates the cleavage site between nucleotides 277 and 278 of the IAA28 mRNA detected by 5′ RACE analysis. Fifteen clones were sequenced, and an identical 5′ end was detected in 12 clones. No cleavage site was detected in the other computationally predicted targets, including ATR1 and COL4 mRNAs.
(C) Expression of IAA28, ATR1, and COL4 mRNAs in 35S-miR159847 transgenic plants. Total RNAs isolated from leaves of 25-d-old plants were quantified by qRT-PCR and normalized with the corresponding input RNA and EIF1a (as the internal standard) levels. The value of IAA28 mRNA in Col-0 was arbitrarily designated as 1. Error bars represent sd for three replicates.
(D) Effect of miR847 on IAA28 and IAA28m transcripts in coinfiltration assays. Total RNAs (30 μg) or small RNAs (20 μg) isolated at 2 d after infiltration were used. 28S rRNA and U6 were used as loading controls. Similar results were obtained from two individual assays.
miR847-Mediated Reduction of IAA28 mRNA Levels upon Auxin Treatment
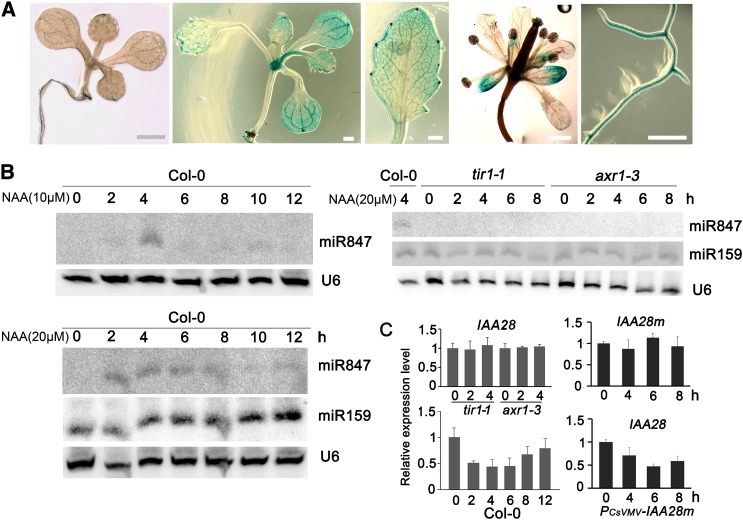
In a previous study, staining of 8-d-old transgenic Arabidopsis seedlings carrying a fusion of the IAA28 promoter and the GUS gene showed promoter-GUS expression limited to roots (Rogg et al., 2001). IAA28 mRNA was also concentrated in the roots and inflorescence stems, with minor levels in leaves and flowers (Rogg et al., 2001). To further examine whether the low level of IAA28 in aerial portions of Arabidopsis was due to cell type specificities, as found for miR847, we first examined the expression profile of IAA28 by generating IAA28-promoter-GUS (IAA28-GUS) transgenic Arabidopsis (Col-0). When 8-d-old seedlings were stained, IAA28-GUS was readily detected in roots but very weak in leaf veins of the cotyledons (Figure 3). However, staining of 12-d-old seedlings showed that IAA28-GUS was highly expressed in the shoot apex, leaf veins of the cotyledons, marginal meristems of the rosette leaves, sepals (but not stamens), and roots, especially at lateral root initiation sites (Figure 3A), revealing the striking cell type specificity and developmental regulation of IAA28 expression. These expression patterns are similar to that of MIR847, suggesting that IAA28 and miR847 function in the same types of cell.
Figure 3.
Expression Pattern of IAA28 and Its Reduction Coincident with the Induction of miR847 upon Auxin Treatment.
(A) IAA28-GUS expression in 8- or 12-d-old seedling, leaves, flower, and root. Bars = 1 mm.
(B) Induced expression of miR847 upon auxin treatment. Ten-day-old wild-type Col-0 and tir1-1 and axr1-3 mutant seedlings were treated with 10 or 20 µM NAA, and samples were collected at the times indicated at top. Eighty micrograms of small RNA samples was loaded for hybridization with miR847 LNA probe. miR159 and U6 hybridization was used as controls.
(C) Reduced expression of IAA28, but not IAA28m, upon auxin treatment. Total RNAs were isolated from 10-d-old Col-0, PCsVMV-IAA28m, and tir1 and axr1-3 mutant seedlings treated with 20 μM NAA at the indicated time points. The expression of IAA28 or IAA28m was quantified by qRT-PCR amplified with sequence-specific primers (Supplemental Data Set 2) and normalized with the corresponding input RNA and UBQ10 (as the internal standard) levels. The value of IAA28 or IAA28m mRNA at 0 h was arbitrarily designated as 1. Error bars represent sd for three replicates.
IAA28 is a negative regulator of auxin signaling, and a reduction of IAA28 mRNA was detected in response to auxin treatment (Rogg et al., 2001). To test whether the auxin-induced decrease in the IAA28 mRNA level were mediated by miR847, the expression of miR847 in response to auxin treatment was examined. Ten-day-old wild-type Col-0 seedlings were treated with 1-naphthalene acetic acid (NAA) at 10 μM; we consistently detected an increase in miR847, but not in miR159a, at 2 h, and it reached the highest level at 4 h after treatment (Figure 3B). With 20 μM NAA treatment, the increase in miR847 was maintained at high levels for several hours (detected from 2 to 8 h after treatment), consistent with the increased expression of GUS transcript in MIR847-GUS transgenic plants upon auxin treatment (Supplemental Figure 1). Coincidently, reduced IAA28 mRNA was detected from 2 h after auxin treatment, consistent with the detection of increased miR847 levels at 2 h (Figure 3C). Reduction of IAA28 mRNA was not due to the decreased transcription of IAA28 mRNA, because no change was detected in the expression of GUS in IAA28-GUS transgenic plants upon auxin treatment (Supplemental Figure 1). To confirm that IAA28 is a direct target of miR847, transgenic PCsVMV-IAA28m plants expressing the cleavage-resistant mutant IAA28m were created. Ten-day-old seedlings were treated with 20 µM NAA, and a reduction of IAA28 mRNA, but not the mutant IAA28m mRNA, by auxin treatment was observed (Figure 3C). Neither induction of miR847 nor reduction of IAA28 mRNA levels by auxin was observed in the auxin-insensitive mutants transport inhibitor response1 (tir1-1) and auxin resistant1 (axr1-3) (Figures 3B and 3C). Taken together, our data indicate that the increase in the miR847 level is auxin-dependent, and the reduction in the IAA28 mRNA level after auxin treatment resulted, at least in part, from miR847-mediated cleavage.
Transgenic Plants Overexpressing miR847 Promoted Lateral Root Initiation
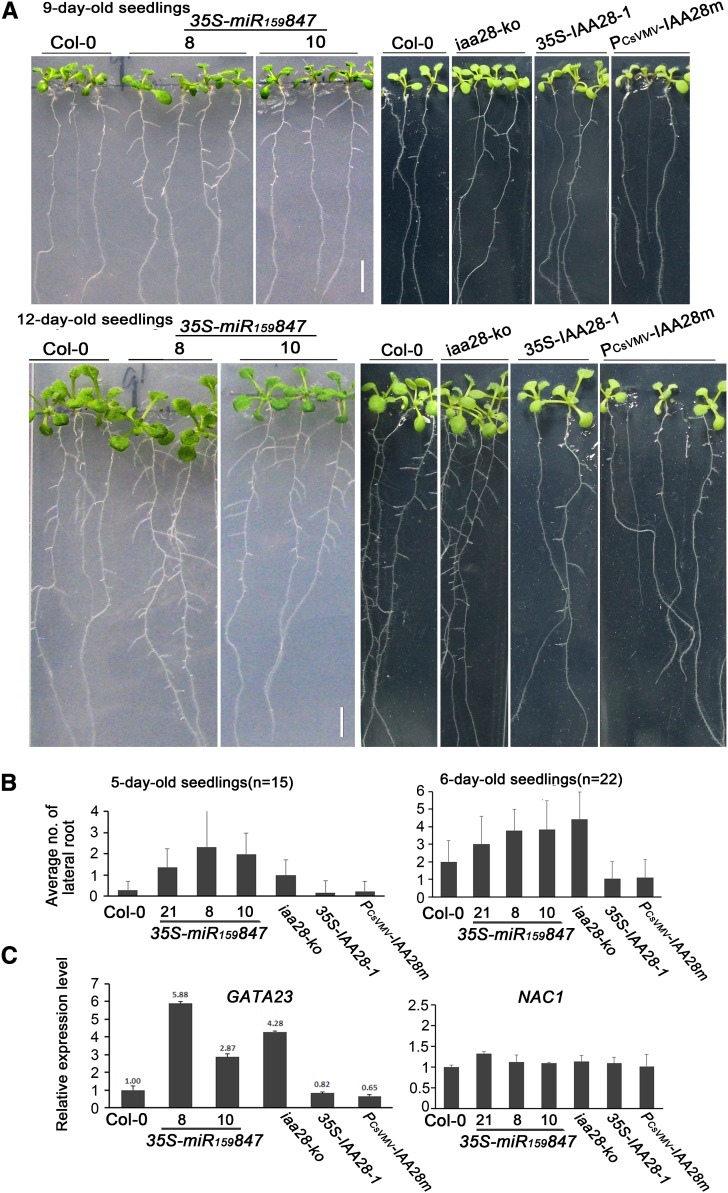
IAA28 belongs to the Aux/IAA family and functions as an important component of the auxin signaling pathway implicated in lateral root initiation (De Rybel et al., 2010). A gain-of-function mutant, iaa28-1, strongly suppresses lateral root development in Arabidopsis Wassilewskija (Ws) (Rogg et al., 2001), revealing the negative effect of IAA28 on lateral root initiation (De Rybel et al., 2010). Therefore, the effect of miR847-mediated degradation of IAA28 on lateral root development was examined in 35S-miR159847 transgenic plants, an IAA28 knockout line (SALK_129988C in Col-0; referred to iaa28-ko to distinguish it from the gain-of function iaa28-1 in Ws), and wild-type Col-0. The overexpression of miR847 and the knockout of IAA28 were hypothesized to result in more lateral roots. As expected, clear lateral roots were observed in 9-d-old seedlings of 35S-miR159847 and iaa28-ko plants but were rarely found in Col-0 seedlings (Figure 4). Statistical analysis of the lateral root count per primary root in 5- and 6-d-old seedlings of 35S-miR159847 lines, iaa28-ko, and Col-0 showed that the overexpression of miR847 or knockout of IAA28 produced 2- to 3-fold more lateral roots than were present in the Col-0 controls (Figure 4B). Earlier and more lateral root development was conspicuous in 9- and 12-d-old 35S-miR159847 and iaa28-ko seedlings (Figure 4A). By contrast, 9- and 12-d-old seedlings of 35S-IAA28-1 and PCsVMV-IAA28m expressing the natural or the cleavage-resistant mutant IAA28 mRNA (Figure 4A) developed fewer lateral roots compared with Col-0 control seedlings (Figure 4A), consistent with the opposite levels of accumulation of IAA28 mRNA in 35S-IAA28 transgenic and iaa28-ko mutant plants. Similarly, fewer lateral roots were observed in the gain-of-function iaa28-1 mutant compared with Ws control seedlings (Supplemental Figure 2). There was no significant difference in lateral root development between seedlings of 35S-IAA28 and PCsVMV-IAA28m, presumably because there was no substantial difference between wild-type and mutant IAA28 mRNAs at the low-abundance level of miR847 under normal conditions.
Figure 4.
Effect of the Overexpression of miR847 on Lateral Root Development.
(A) Early and sustained formation of lateral roots in the 35S-miR159847 transgenic lines. Col-0 and 35S-miR159847, iaa28-ko, 35S-IAA28-1, and PCsVMV-IAA28m seedlings were grown on MS medium with 3% sucrose. Seedlings were photographed at 9 and 12 DAG. Bars = 5 mm.
(B) Statistical histogram of the average number of lateral roots in 5- and 6-d-old seedlings of each genotype. Roots were observed with a binocular microscope to acquire numbers of lateral root initials identified by staining whole seedlings with a mixture of toluidine blue and basic fuchsin and then counted. The average number of lateral roots was obtained from 15 (n = 15) or 22 (n = 22) seedlings of each genotype. Error bars represent sd for the average number.
(C) Analysis of the expression of GATA23 and NAC1 mRNAs in 8-d-old roots. Total RNAs isolated from roots of each genotype were quantified by qRT-PCR and normalized with the corresponding input RNA and ACTIN2 (as the internal standard) levels. The values of GATA23 and NAC1 mRNAs in Col-0 were arbitrarily designated as 1. Error bars represent sd for three replicates.
Previous studies demonstrated that an IAA28-dependent auxin signaling mechanism in the basal meristem regulates the expression of the transcription factor gene GATA23, which determines lateral root founder cell identity (De Rybel et al., 2010). Therefore, we examined the expression of GATA23 mRNA in the above plants of various genotypes (the 35S-miR159847, iaa28-ko, 35S-IAA28-1, and PCsVMV-IAA28m lines). A qRT-PCR analysis demonstrated that the expression level of GATA23 mRNA was remarkably increased in the roots of 8-d-old 35S-miR159847 and iaa28-ko seedlings compared with the Col-0 control (Figure 4C). Plants of the 35S-miR159847 line (line 8), with a lower level of IAA28 mRNA (Figure 2C), accumulated higher levels of GATA23 mRNA (Figure 4C), consistent with greater lateral root formation (Figure 4A). In contrast, decreased levels of GATA23 mRNA were detected in the roots of 35S-IAA28-1 and PCsVMV-IAA28m seedlings (Figure 4C). To determine whether the production of more lateral roots in 35S-miR159847 plants possibly involved other factors that also affect lateral root development, the expression of transcription factor NAC1, which transduces auxin signals for lateral root emergence (Xie et al., 2000; Guo et al., 2005), was examined. A qRT-PCR analysis showed no difference in the expression levels of NAC1 mRNA between the roots of 8-d-old seedlings of the various genotypes and the Col-0 control (Figure 4C). Thus, our data indicate that greater lateral root development in 35S-miR159847 seedlings presumably results from the miR847-mediated reduction of IAA28 mRNA and subsequent activation of GATA23 to promote lateral root initiation.
Transgenic Plants Overexpressing miR847 Presented a Series of Abnormal Aerial Phenotypes Opposite to Those of Plants Overexpressing IAA28 mRNA
In addition to greater lateral root development in the 35S-miR159847 plants, we observed serial abnormal aerial phenotypes in these transgenic lines throughout plant growth. Because both miR847 and IAA28 presented striking cell type specificities in aerial tissues (Figures 1 and 3) and the aerial portions of the gain-of-function iaa28-1 mutant plants were also malformed (Rogg et al., 2001), we investigated the possible involvement of miR847 and IAA28 in the regulation of the development of the aerial part of Arabidopsis. We compared the aerial part phenotypes of the 35S-miR159847, iaa28-ko, and PCsVMV-IAA28m plants as well as the wild-type Col-0. In 6- and 8-d-old seedlings, Col-0 developed normal oval cotyledons, and round and/or slightly heart-shaped cotyledons were observed in the 35S-miR159847 and iaa28-ko seedlings, while the PCsVMV-IAA28m seedlings showed slightly longer oval cotyledons (Supplemental Figure 3). During the early development stage, 35S-miR159847 and iaa28-ko seedlings were generally smaller, with shorter petioles, smaller leaf size, and shorter hypocotyls, compared with the Col-0 control, while the PCsVMV-IAA28m seedlings presented an opposite phenotype, with longer petioles and hypocotyls (Supplemental Figure 3). Consistently, slightly longer petioles and hypocotyls in the gain-of-function mutant iaa28-1 were also observed compared with wild-type Ws (Supplemental Figure 3). Throughout plant growth, the morphology of the 35S-miR159847 plants appeared compact and leafy compared with that of Col-0 (Figure 5). At ∼20 d after germination (DAG), 7 to 8 true leaves developed in the Col-0 plants and 9 to 11 leaves were consistently observed in the 35S-miR159847and iaa28-ko plants, whereas 6 to 7 leaves were observed in the PCsVMV-IAA28m plants (Figures 5A and 5B). Inverse phenotypes were also observed in both the true leaf size and the morphology of the leaf margin between the 35S-miR159847, iaa28-ko, and PCsVMV-IAA28m plants. The early developed leaves (e.g., 3rd and 4th true leaves) of the 35S-miR159847 and iaa28-ko plants were shorter and wider, but those of PCsVMV-IAA28m were smaller than those of Col-0 (Figure 5C). Then, juvenile leaves (e.g., 6th to 10th true leaves) emerged that were larger in both 35S-miR159847 and iaa28-ko plants compared with those in Col-0 (Figure 5C). Some of the 35S-miR159847 plants had a notable serrated margin (Figure 5C). A strong serrated leaf margin was observed in 39 out of 63 primary 35S-miR159847 transformants (Figure 5D). Leaves in similar layers were slender with a smooth margin in PCsVMV-IAA28m plants (Figure 5C). The production of the floral organs was normally observed at 22 DAG in Col-0 but with a 3- to 5-d delay in the 35S-miR159847 plants (Figure 5E).
Figure 5.
Phenotypes of 35S-miR159847and PCsVMV-IAA28m Transgenic and iaa28-ko Mutant Plants.
(A) Developmental phenotypes of Col-0, 35S-miR159847-8, iaa28-ko mutant, and PCsVMV-IAA28m plants at 19 DAG. Bars = 10 mm.
(B) Statistical histogram of the average number of leaves in Col-0, 35S-miR159847 transgenic lines 8, 25, and 21, iaa28-ko mutant, and PCsVMV-IAA28m plants at 20 DAG. Sixteen plants of each genotype were counted. Error bars represent sd for the average number.
(C) Leaf morphologies of Col-0, 35S-miR159847 and PCsVMV-IAA28m transgenic lines, and iaa28-ko mutant plants at 21 and 25 DAG. Bars = 5 mm.
(D) Representative phenotypes of various 35S-miR159847 and PCsVMV-IAA28m transgenic lines and iaa28-ko mutant plants. Bars = 10 mm.
(E) Floral development in Col-0 and 35S-miR159847 transgenic lines at 25 DAG. Bar = 10 mm.
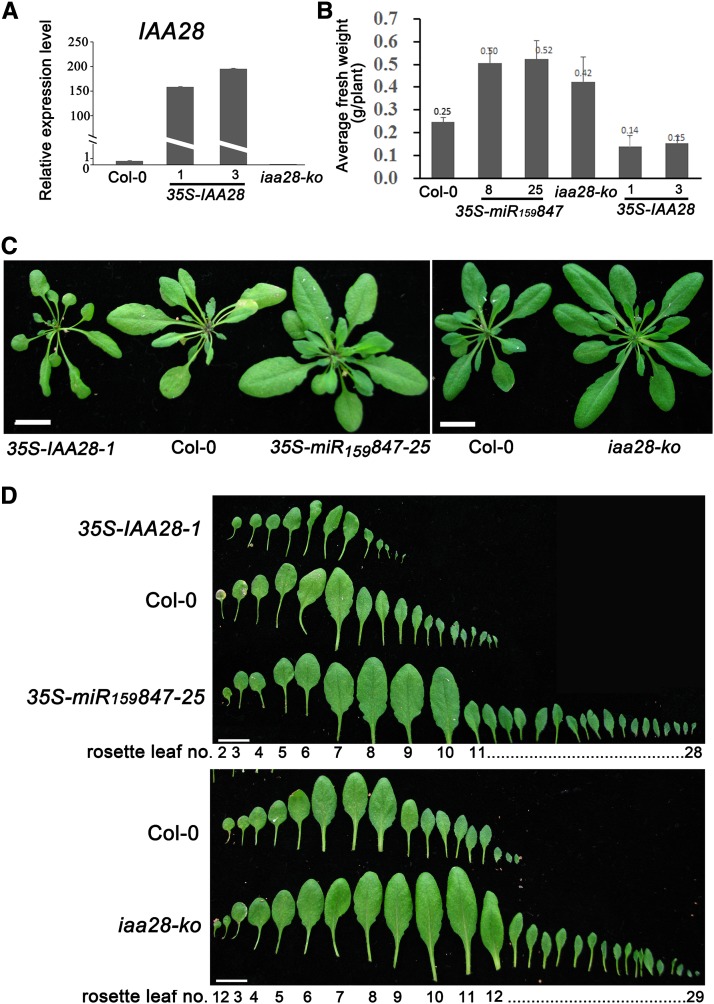
The total leaf number and average fresh weight per plant were also compared among the 35S-IAA28, 35S-miR159847 transgenic, iaa28-ko mutant, and Col-0 plants. Plants of these genotypes that were 33 d old were used (Figure 6). The 35S-IAA28 plants had fewer rosette leaves and lower average fresh weights compared with the Col-0 plants (Figures 6B and 6D). The 35S-miR159847 and iaa28-ko mutant plants produced more rosette leaves and had a remarkably increased average fresh weight compared with Col-0 (Figures 6B and 6D). The finding that the phenotypes of plants overexpressing miR847 resembled those of iaa28-ko plants and were opposite those of plants overexpressing IAA28 mRNA reveals that miR847-mediated degradation of IAA28 is involved in the regulation of aerial portion development.
Figure 6.
Plants Overexpressing the IAA28 mRNA Display the Opposite Phenotype to That of 35S-miR159847.
(A) The expression of IAA28 mRNA in Col-0, 35S-IAA28 transgenic, and iaa28-ko mutant plants at 33 DAG was quantified by qRT-PCR as described in Figure 2C.
(B) Average fresh weight of rosette leaves of Col-0, 35S-miR159847, iaa28-ko, and 35S-IAA28 transgenic plants at 33 DAG with inflorescences removed. Ten plants of each genotype were used. Error bars represent sd for the average number.
(C) and (D) Plants with inflorescences removed (C) and peeled off rosette leaves (D) of 35S-IAA28-1, Col-0, 35S-miR159847-25, and iaa28-ko at 33 DAG. Bars = 10 mm.
Notably, a previous study reported that only transgenic plants overexpressing the gain-of-function mutant iaa28-1 gene had severe defects, with an infertile dwarf phenotype, while overexpression of IAA28 caused no discernible morphological effects (Rogg et al., 2001). In our 35S-IAA28 plants, slender and fewer leaves and advanced inflorescence were observed, but none of the 35S-IAA28 lines had extreme phenotypes of infertile dwarfs throughout the experimental periods (longer than 2 months). IAA28 was previously shown to have the longest (∼80 min) half-life of the four canonical Aux/IAA proteins, as estimated using luciferase-fusing proteins (10 to 20 min for IAA1, IAA9, and IAA17) (Ramos et al., 2001; Dreher et al., 2006). We speculate that the different growth conditions and/or different product levels of the transgenic IAA28 mRNA, in addition to the dramatically different half-life of IAA28 compared with other Aux/IAAs (Dreher et al., 2006), might cause the phenotypic discrepancies of transgenic plants overexpressing wild-type IAA28 cDNA.
Overexpression of miR847 Downregulates the IAA28-Mediated Repression of Auxin Signaling in Aerial Portions
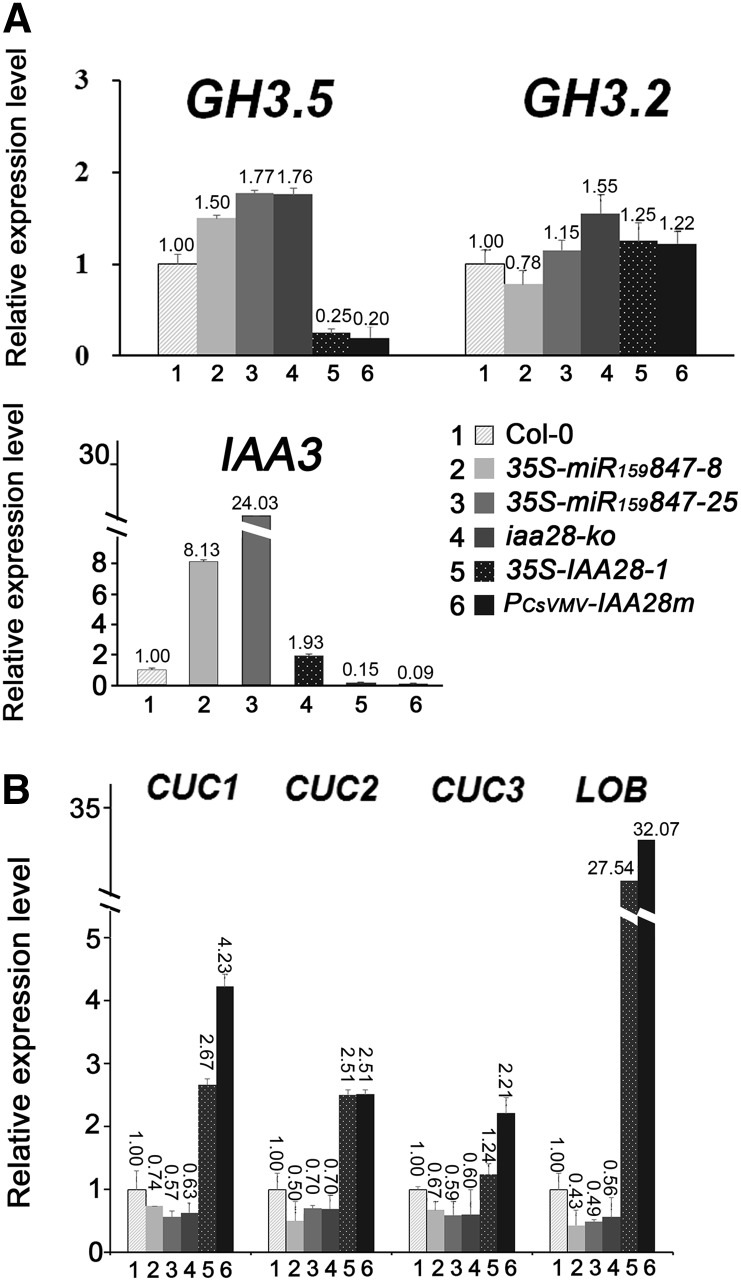
Because the miR847-mediated cleavage of IAA28 mRNA resulted in the derepression of auxin signaling, leading to lateral root formation (Figure 4), we hypothesized that the opposite abnormal phenotypes observed in the aerial portions of the 35S-IAA28 and 35S-miR159847 plants also resulted from the repression and derepression of auxin signaling in aerial portions of two genotypes. To determine which auxin regulation genes are possibly regulated by IAA28, several early auxin response genes of two major classes, GH3 and Aux/IAA, were tested using samples from the aerial portions of 13-d-old seedlings. The accumulation levels of GH3.5, an auxin-responsive GH3 gene also known as WES1 (At4g27260), which encodes IAA-amide synthetases that help maintain auxin homeostasis (Staswick et al., 2005), but not GH3.2, were reduced in the 35S-IAA28 and PCsVMV-IAA28m plants but increased in the 35S-miR159847 and iaa28-ko mutant plants compared with Col-0 (Figure 7), consistent with a previous report that a light-grown GH3.5 (WES1)-overexpressing wes1-D mutant developed short hypocotyls. By contrast, the knockout wes1 mutant hypocotyls were slightly longer than the control Col-0 hypocotyls (Park et al., 2007). The expression of IAA3, a primary auxin response gene, was also reduced in 35S-IAA28 and PCsVMV-IAA28m but increased in 35S-miR159847 and iaa28-ko mutants compared with Col-0 plants, although the increased level in iaa28-ko was not as high as in 35S-miR159847 (Figure 7A). This is consistent with the gain-of-function mutations in IAA3 producing short hypocotyls (Tian et al., 2002). Our results suggest that the short hypocotyl auxin phenotype in 35S-miR159847 plants (Figure 5) can probably be attributed to the increase in the expression of GH3.5 and IAA3 upon miR847-mediated downregulation of IAA28.
Figure 7.
RNA Analysis of Early Auxin-Responsive and Auxin Signaling-Repressive Genes in 35S-miR159847 Transgenic Plants.
The expression of early auxin response genes, GH3.5, GH3.2, and IAA3 (A), and auxin signaling-repressive genes, CUC1, CUC2, CUC3, and LOB (B), is shown in Col-0, 35S-miR159847 transgenic (lines 8 and 25), iaa28-ko mutant, and 35S-IAA28-1 and PCsVMV-IAA28m plants. Total RNAs isolated from aerial parts of 13-d-old seedlings were quantified by qRT-PCR and normalized with the corresponding input RNA and UBQ10 (as the internal standard) levels. The value of each mRNA in Col-0 was arbitrarily designated as 1. Error bars represent sd for three replicates.
The initiation of plant lateral organs from the shoot apical meristem (SAM) is associated with the formation of boundaries that separate the primordia from the surrounding tissue (Aida and Tasaka, 2006; Rast and Simon, 2008). We examined the CUC family genes, which are required for the activation of genes that maintain meristematic fate in the SAM and are negatively regulated by auxin-dependent signaling (Berleth et al., 2004; Laufs et al., 2004; Hibara et al., 2006; Rast and Simon, 2008), and LATERAL ORGAN BOUNDARIES (LOB) mRNA, which is expressed at the base of all lateral organs and mediates the response to auxin (Inukai et al., 2005; Okushima et al., 2007). In the aerial portions of 13-d-old seedlings, the accumulation of CUC1, CUC2, CUC3, and, especially, LOB mRNAs was reduced in 35S-miR159847 and iaa28-ko mutant plants but increased in the 35S-IAA28 and PCsVMV-IAA28m plants compared with Col-0 (Figure 7B). The induction was also prominent for LOB. The redundant functions of the CUC1, CUC2, and CUC3 genes (Mallory et al., 2004; Shani et al., 2006) in the regulation of embryonic shoot meristem formation and cotyledon boundary specification (Hibara et al., 2006) might account for the inconspicuous changes in transcript levels of CUC family genes and the slightly heart-shaped cotyledon phenotype in the 35S-miR159847 seedlings (Supplemental Figure 3). Nonetheless, our findings suggest that the miR847-mediated degradation of IAA28 increases auxin signaling, probably by decreasing the expression of LOB and CUC family genes to promote the initiation of leaf primordia and consequently increase the number of leaves.
Overexpression of miR847 Reduces IAA28 mRNA Levels to Prolong Growth and Cell Proliferation
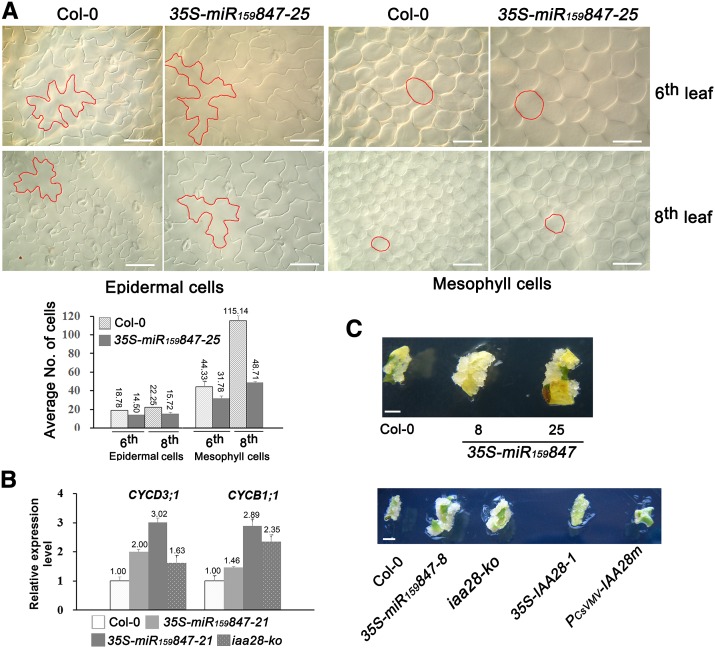
In addition to exhibiting an increased number of leaves, similar to the iaa28-ko mutant, 35S-miR159847 plants also had remarkably increased leaf sizes of the 8th to 10th rosette leaves compared with those of Col-0, in which the leaves from the 8- to 9-leaf stage generally stopped growing after flowering (Figure 6). To determine whether the larger leaves were due to cell expansion or increased cell numbers, the 6th and 8th leaves of 33-d-old 35S-miR159847-25 and Col-0 plants were selected for microscopic observation. In these two leaves, cells, especially the epidermal cells, in the 35S-miR159847-25 plants were likely turgid, because the cell margin was less wrinkled compared with that in Col-0 (Figure 8), leading to the slightly larger size of both epidermal and mesophyll cells in the 35S-miR159847-25 plants than in Col-0. Except for mesophyll cells in the 8th leaf, the number of cells in the same area of leaves was not much different between Col-0 and 35S-miR159847-25. The numbers of mesophyll cells in the same area in the 8th leaf of Col-0 was ∼2.3-fold more than that of 35S-miR159847-25 (Figure 8A), in agreement with the slightly larger size of mesophyll cells in the 35S-miR159847-25 plants than in Col-0. However, this slightly larger size of the cells could not explain the at least four times larger 8th, 9th, and 10th leaves in 35S-miR159847-25 plants compared with Col-0 (Figure 6D).
Figure 8.
Ectopic Expression of miR847 Prolongs Growth and Cell Proliferation.
(A) Morphologies of adaxial epidermal cells and mesophyll cells of sixth and eighth leaves of Col-0 and 35S-miR159847-25 plants at 33 DAG. Leaves were transparentized and observed using an HC/DMR microscope (Leica) equipped with Nomarski optics. One representative cell in each photograph is outlined in red. The average number of cells counted under the same magnification is indicated. Error bars represent sd for three views of each sample. Bars = 50 μm.
(B) The expression of CYCD3;1 and CYCB1;1 in 33-d-old rosette leaves of Col-0, 35S-miR159847 transgenic (lines 21 and 25), and iaa28-ko plants was quantified by qRT-PCR and normalized with the corresponding input RNA and AT3G33380 (as the internal standard) levels. Error bars represent sd for three replicates.
(C) Callus growth in leaf explants of Col-0, 35S-miR159847 transgenic (lines 8 and 25), iaa28-ko mutant, and 35S-IAA28-1 and PCsVMV-IAA28m transgenic plants. The explants were cultured on MS medium containing 100 ng/L 2,4-D and 100 ng/L 6-BA in the dark and photographed on day 7 without changing the medium. Bars = 2 mm.
Because both MIR847 and IAA28 were expressed in the marginal meristems (leaf tooth) of the rosette leaves (Figures 1 and 3), where cells maintain meristematic competence, we investigated the involvement of miR847 and IAA28 in cell proliferation. Two genes that are involved in the cell cycle were examined. CYCD3;1, a G1 cyclin gene (Hu et al., 2000; Dewitte et al., 2003), and CYCB1;1, which is expressed during the G2/M phase of the cell cycle (Ferreira et al., 1994a, 1994b; Donnelly et al., 1999), were analyzed in 33-d-old rosette leaves of the 35S-miR159847, iaa28-ko, and Col-0 plants. The upregulation of the two mRNAs was detected in 35S-miR159847 and iaa28-ko plants compared with Col-0 (Figure 8B), indicating that the ectopic expression of miR847 or knockout of IAA28 increased the expression of CYCD3;1 and CYCB1;1 and sustained cell meristematic competence to prolong later-stage rosette leaf growth. Consistently, the leaves of the 35S-miR159847 plants displayed strong serrations in the leaf blade, whereas smooth leaf blades were observed in the IAA28-overexpressed PCsVMV-IAA28m plants (Figure 5C), suggesting the important regulatory roles of miR847/IAA28 in marginal cell division during development. The opposite effects of miR847 and IAA28 on cell proliferation were further supported by the formation of callus during tissue culture. Leaf explants that were cultured on callus induction medium (with 2,4-D and 6-benzyl aminopurine [6-BA]) for 1 week produced larger calli for 35S-miR159847 and iaa28-ko than for the Col-0 plants; by contrast, the leaf explants of the 35S-IAA28-1 and PCsVMV-IAA28m plants produced almost no visible calli in 1 week (Figure 8C), and small calli from the 35S-IAA28-1 and PCsVMV-IAA28m plant leaf explants were observed after 2 weeks, consistent with the high activity of IAA28 in repressing cell division during the auxin response. Taken together, our results indicate that the overexpression of miR847 downregulates IAA28, leading to prolonged growth and cell proliferation.
Overexpression of a Cleavage-Resistant Form of IAA28 mRNA Rescues the Abnormal Developmental Morphologies of 35S-miR159847 Transgenic Plants
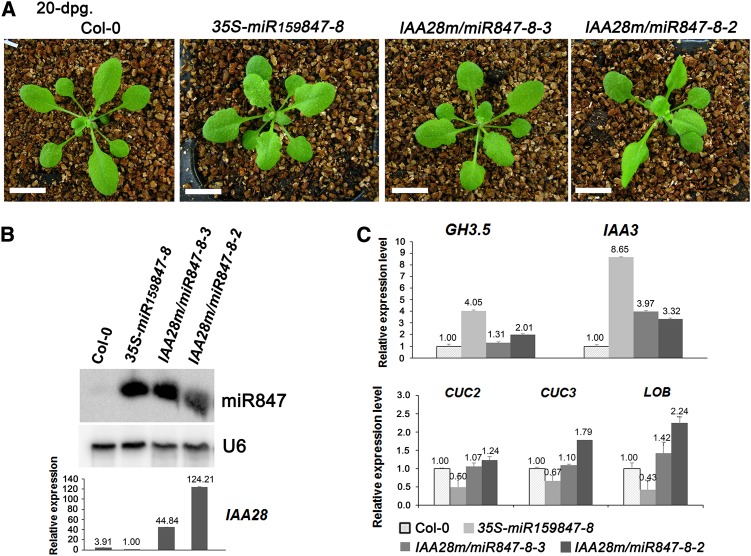
To further demonstrate that the overexpression of miR847, leading to the downregulation of IAA28, is responsible for the abnormal development morphologies, we introduced the cleavage-resistant form of IAA28m mRNA into 35S-miR159847 plants. The 35S-miR159847 plants of line 8, which showed a strong abnormal phenotype, were retransformed with PCsVMV-IAA28m, a construct harboring IAA28m under the control of the cassava vein mosaic virus (CsVMV) promoter, generating IAA28m/miR847-8 transgenic plants. If miR847 targets IAA28, but not other computationally predicted endogenous genes, for cleavage, overexpression of cleavage-resistant IAA28m in 35S-miR159847 plants would rescue its abnormal phenotypes. The accumulation of miR847 and the increased expression of IAA28m were confirmed in all retransformants (Figure 9). As expected, most of the IAA28m/miR847-8 primary transformants (five out of seven lines, including line 3) likely rescued the abnormal development morphologies of 35S-miR159847-8, producing morphologies such as the normal length of the leaf petioles and hypocotyls, normal ovate cotyledons (Supplemental Figure 3), and leaf numbers and sizes that are similar to those of wild-type Col-0 (Figure 9A); these rescued phenotypes are consistent with the restoration of the expression levels of most of the tested auxin response genes, such as GH3.5, CUC2, CUC3, and LOB mRNAs, compared with that in Col-0 (Figure 9C). The expression levels of IAA3 mRNA in the IAA28m/miR847-8 plants were also remarkably reduced compared with those of the 35S-miR159847-8 plants, but they were higher than those of Col-0 (Figure 9C), reflecting the complex roles of negative regulators in auxin signaling. Negative autoregulation has been demonstrated in an IAA3 gain-of-function mutant (Tian et al., 2002). The high level of IAA28m in the IAA28m/miR847-8 plants might account for this discrepancy. Indeed, two out of the seven IAA28m/miR847 lines (including line 2) with a high accumulation level of IAA28m mRNAs presented weak phenotypes similar to that of the 35S-IAA28-1 and PCsVMV-IAA28m transgenic plants (Figures 5 and 6), such as fewer leaves and slender and uneven leaf blades with downward curled leaf margins (Figure 9A). In agreement with the fewer-leaf phenotype, higher levels of CUC3 and LOB mRNAs were also detected in IAA28m/miR847-8 line 2 plants (Figure 9C). Nevertheless, the overexpression of the cleavage-resistant form of IAA28m in 35S-miR159847-8 mostly rescued its abnormal development morphologies in most of the tested auxin response genes, indicating that the overexpression of miR847 resulting in the cleavage of IAA28 mRNA contributed to the abnormal phenotypes of 35S-miR159847 plants and that miR847/IAA28 functions as an important module in the auxin signaling pathway, implicating miR847/IAA28 in the regulation of cell competence, thereby determining the duration of cell proliferation and lateral organ development in Arabidopsis.
Figure 9.
Phenotypes of Plants Overexpressing the Cleavage-Resistant Form of IAA28 mRNA in 35S-miR159847-8 Transgenic Lines.
(A) Phenotypes of Col-0, 35S-miR159847-8, and IAA28m/miR847-8 plants. Bars = 10 mm.
(B) Expression of miR847 and IAA28 mRNA in 10-d-old seedlings of Col-0, 35S-miR159847-8, and IAA28m/miR847-8 plants. RNA gel blotting and qRT-PCR were done as described in Figure 2. The value of IAA28 mRNA in 35S-miR159847-8 was arbitrarily designated as 1. Error bars represent sd for three replicates.
(C) Expression of GH3.5, IAA3, CUC2, CUC3, and LOB mRNAs in 10-d-old seedlings of Col-0, 35S-miR159847-8, and IAA28m/miR847-8 (lines 2 and 3) plants. qRT-PCR was done as described in Figure 7. The value of each mRNA in Col-0 was arbitrarily designated as 1. Error bars represent sd for three replicates.
DISCUSSION
miR847-Mediated IAA28 mRNA Cleavage in Vivo
In both plants and animals, some MIRNA families are highly conserved through hundreds of millions of years (Axtell and Bartel, 2005; Grimson et al., 2008; Fahlgren et al., 2010). The miR847 sequence was identified by deep sequencing in Arabidopsis and Arabidopsis lyrata, but not in Capsella rubella, a species closely related to Arabidopsis within the Brassicaceae family (Rajagopalan et al., 2006; Fahlgren et al., 2010; Ma et al., 2010). A. lyrata and C. rubella diverged from Arabidopsis ∼10 and ∼20 million years ago, respectively (Koch et al., 2000; Wright et al., 2002; Fahlgren et al., 2010). Thus, the miR847-coding gene (MIR847) should be an orthologous conserved locus in the Arabidopsis lineage and one of the newer MIRNA genes (Fahlgren et al., 2010), which are generally expressed from single genes at low levels or primarily in specific cells or growth conditions (Rajagopalan et al., 2006). Using coexpression in N. benthamiana, we demonstrate here that miR847 targets IAA28 mRNA for cleavage in vivo at the IAA28/miR847 complementary region and that the disruption of the base-pairing region compromised cleavage. In Arabidopsis, both miR847 and IAA28 mRNA exhibit similar expression patterns and striking cell type specificity. IAA28 mRNA cleavage might be developmentally regulated in response to auxin in Arabidopsis because the accumulation of miR847, which was undetectable under normal growth conditions, increased with decreasing IAA28 mRNA levels upon auxin treatment. IAA28 mRNA encodes an Aux/IAA repressor protein that is targeted for degradation through the proteasome. This study extends our knowledge of the sensitization of auxin signaling to IAA28 mRNA levels, which, together with targeted IAA28 degradation, ensures a robust signal derepression. This result is in agreement with the previous corollary that unstable mRNAs are less likely to encode stable proteins, such as NAC1, which is targeted by the SINAT5 E3 ligase for the 26S proteasome through polyubiquitination (Xie et al., 2002); its mRNA is also targeted by miR164-mediated cleavage (Guo et al., 2005).
miR847 Positively Regulates Lateral Root Development by Derepressing the IAA28-Dependent Auxin Signaling Cascade
The plant hormone auxin plays an important role in lateral root initiation. IAA28 is a negative regulator of auxin signaling during lateral root development (Rogg et al., 2001), which functions by regulating the expression of the transcription factor gene GATA23 in the basal meristem region (De Rybel et al., 2010). In plants overexpressing miR847 or in mutant plants with knocked out IAA28 gene expression, the downregulation of IAA28 mRNA produced more lateral roots, correlated with the increased expression of GATA23 mRNA, compared with the wild-type Col-0. The transcription factor NAC1 mRNA also positively transduces the auxin signal leading to lateral root emergence (Xie et al., 2000), and miR164 negatively regulates NAC1 mRNA and lateral root development in response to auxin treatment (Guo et al., 2005). However, increased auxin signaling through the miR847-mediated downregulation of IAA28 likely did not obviously affect miR164/NAC1 mRNA expression. Indeed, the 35S-miR159847-8 line, which highly expressed GATA23 mRNA, not only developed lateral roots earlier but also exhibited prolonged production of lateral roots in the basal meristem region compared with Col-0. By contrast, increased NAC1 mRNA levels promoted lateral root production, which was mostly observed near the hypocotyl/root junction (Guo et al., 2005). Thus, our data suggest that miR847 positively regulates lateral root development by derepressing the IAA28-dependent repression of GATA23 expression during the auxin response.
miR847 Positively Regulates Aerial Lateral Organ Development by Derepressing the IAA28-Dependent Auxin Signaling Cascade
In addition to greater lateral root development, overexpressing miR847 or knockout of IAA28 also altered the development of the aerial portion. We have provided several lines of evidence demonstrating that miR847 functions as a positive regulator of auxin-mediated lateral organ development in the aerial portion by cleaving IAA28 mRNA. (1) Plants overexpressing IAA28 mRNA produced long hypocotyls and fewer rosette leaves, resembling the gain-of-function iaa28-1 mutant. (2) Plants with high miR847 production accumulated lower levels of IAA28 mRNA and produced short hypocotyls and more rosette leaves, resembling the knockout iaa28-ko mutant. (3) Overexpressing cleavage-resistant IAA28m in plants with high levels of miR847 restored the normal growth of the hypocotyl and the number of rosette leaves. (4) In addition to the hypocotyl and leaf numbers, the leaf size and morphology were opposite between the plants overexpressing miR847 and those overexpressing IAA28 mRNA. In every case, there is a strict inverse correlation between changes in the miR847 levels and changes in both the IAA28 mRNA levels and leaf numbers and morphology.
The roles of auxin signaling mediators have been demonstrated in diverse development processes, including hypocotyl growth (Hagen and Guilfoyle, 2002). The Arabidopsis GH3.5 gene (WES1) encodes an auxin-conjugating enzyme that functions in hypocotyl growth (Park et al., 2007). Both GH3.5 and IAA3 expression decreased in the long-hypocotyl 35S-IAA28 plants but increased in 35S-miR159847 plants, which had short hypocotyls. IAA3 is a negative regulator of auxin signaling (Tian et al., 2002). Our findings, together with the observation that gain-of-function mutations in IAA3 cause short hypocotyls and do not affect IAA28 expression (Tian et al., 2002), suggest that IAA28 probably acts upstream of IAA3 and that a reduction in IAA28 expression stimulates an increase in the expression of other auxin signaling repressors, such as IAA3. However, increased IAA3 expression in 35S-miR159847 plants did not affect the expression of GH3.5 mRNA. Thus, our data suggest that an auxin signaling pathway, miR847-IAA28-GH3.5, and its regulation function in hypocotyl development in Arabidopsis.
The overexpression of miR847 or knockout of IAA28 in plants also produces more rosette leaves. The initiation of lateral organs from the SAM is associated with the formation of boundaries that separate the primordia from surrounding tissue (Aida and Tasaka, 2006; Rast and Simon, 2008). CUC2 and CUC3 mRNAs are expressed at the boundary between the shoot meristem and leaf primordia (Hibara et al., 2006). The region that produces leaf primordium in the SAM requires high auxin concentrations to restrict the expression of CUC genes, which are required for the activation of Shoot Meristemless, which maintains the meristematic fate in the SAM (Berleth et al., 2004; Laufs et al., 2004; Inukai et al., 2005; Shani et al., 2006; Okushima et al., 2007; Rast and Simon, 2008). LOB is also expressed in the boundary region at the base of all lateral organs and mediates responses to auxin (Husbands et al., 2007). The increased rosette leaf numbers in the 35S-miR159847 and iaa28-ko plants decreased the expression of CUC family and LOB genes, suggesting that the miR847-mediated downregulation of IAA28 results in auxin responsiveness in the SAM, promoting leaf primordia formation, consequently impeding the vegetative SAMs from being transformed into inflorescence meristems and generating floral meristems that produce floral organs. This notion explains the delayed florescence in 35S-miR159847 plants. By contrast, the 35S-IAA28 plants develop early inflorescences due to the more stable IAA28 protein depressing the auxin signaling pathway. In Arabidopsis, the redundant functions of the CUC1, CUC2, and CUC3 genes regulate embryonic shoot meristem formation and cotyledon boundary specification (Mallory et al., 2004; Shani et al., 2006). CUC3 and CUC2 are responsible for the axillary shoot formation and boundary specification of various postembryonic shoot organs, whereas the contribution of CUC1 and CUC2 during embryogenesis is greater than that of CUC3 (Hibara et al., 2006). There was no obvious developmental defect observed in embryogenesis in the 35S-miR159847 plants. Taken together, our data suggest that miR847-IAA28-mediated auxin signaling affects the formation of postembryonic shoot meristems, but not of the embryonic meristem, by regulating LOB and CUC family genes.
miR847 Positively Regulates Meristematic Competence and Cell Proliferation by Derepressing the IAA28-Dependent Auxin Signaling Cascade
The determination of organ size is a fundamental aspect of growth and development. The cell cycle genes CYCD3;1 and CYCB1;1 determine the leaf cell number and are regulated by auxin (Casimiro et al., 2001; Oakenfull et al., 2002; Dewitte et al., 2003). The finding that the expression of both CYCD3;1 and CYCB1;1 increases in 35S-miR159847 and iaa28-ko plants, together with the observation that both MIR847 and IAA28 are expressed in the marginal meristems of rosette leaves where cells maintain meristematic competence, prolonging the growth of 8th to 10th rosette leaves in 35S-miR159847 and iaa28-ko plants before floral organ development, indicate that miR847 plays a positive regulatory role in cell division-related leaf development. The transition from vegetative SAMs to inflorescence meristems and, finally, to floral organ development is mediated by a complex network of genetic pathways that regulate flowering in response to environmental and developmental signals (Liu et al., 2009). The regulation of auxin flux in response to endogenous cues in this transition is likely strong and restricts miR847-related regulation. The growth of rosette leaves after floral organ development was restricted in all plant genotypes, including wild-type Col-0, 35S-miR159847, and iaa28-ko mutant as well as 35S-IAA28 plants. Although more leaf primordia likely formed before inflorescence meristem formation in 35S-miR159847 and iaa28-ko plants, the growth of the late-stage rosette leaves after visible floral growth was also restricted. Nonetheless, the effect of miR847 on cell division-related leaf development before floral development is demonstrated. Our results, together with the fact that the 35S-IAA28 and PCsVMV-IAA28m plants present small leaves and that miR847 and IAA28 have opposite effects on cell proliferation during the formation of calli by tissue culture, suggest that miR847 positively regulates meristematic competence and cell proliferation by derepressing the IAA28-dependent auxin signaling cascade.
In summary, we identify a nonconserved miR847 that is not detected under normal conditions and is induced upon auxin treatment. IAA28 mRNA, encoding an Aux/IAA repressor protein, is targeted by miR847 for cleavage. The striking cell type specificity of both miR847 and IAA28 as well as the induction of miR847 coincide with decreased IAA28 mRNA levels upon auxin treatment, which was not observed in tir1-1 and axr1-3 mutants. These findings suggest that the auxin-dependent induction of miR847 is positively regulated in response to developmental switches to regulate the extent of cell competence, thereby determining the duration of cell proliferation and lateral organ growth through degradation of the transcript encoding the Aux/IAA28 repressor protein, which is concurrent with its degradation by the proteasomes and followed by the activation of relevant auxin-responsive molecular components.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotypes Col-0 and Ws were used as the wild-type lines. The tir1-1 and axr1-3 mutant seeds (gifts from Mark Estelle), all transgenic plants, and knockout mutant iaa28-ko (SALK_129988C) used in this study are in the Col-0 background, and the gain-of-function iaa28-1 mutant is in the Ws background. Arabidopsis plants were grown under long-day conditions (16 h of light/8 h of dark) at 22°C.
Cloning, Constructs, and Plant Transformation
For the MIR847-GUS and IAA28-GUS constructs, the 2480-bp upstream sequence of the MIR847 gene and the 2041-bp upstream sequence of the IAA28 gene were amplified, and PCR products were cloned into T-vectors (Tiangen) using T4 DNA Ligase (Takara). After confirmation by sequencing, MIR847 and IAA28 promoter sequences were individually subcloned into the pCAMBIA1300-221 binary vectors fused to the GUS reporter gene. For 35S-MIR847 and 35S-IAA28 constructs, the 259-bp genomic sequence of the MIR847 and IAA28 coding sequences was amplified, and the PCR product was cloned into T-vectors and confirmed by sequencing. MIR847 and IAA28 gene sequences were individually subcloned into the pCAMBIA1300-221 binary vector under the control of the 35S promoter. These plasmids, MIR847-GUS, IAA28-GUS, 35S-MIR847, and 35S-IAA28, were individually introduced into Col-0 plants using Agrobacterium tumefaciens EHA105 (Hood et al., 1993), and transformants were selected on medium containing hygromycin (30 mg/mL).
For the 35S-miR159847 construct, the miR847-containing artificial miRNA precursor was amplified with the Arabidopsis precursor miR159a sequence as a backbone as described previously (Niu et al., 2006). The forward and reverse primers contain the miR847* sequence (Supplemental Data Set 2) and the mature miR847 reverse complementary sequence, respectively. The resulting PCR fragment was cloned into T-vector and confirmed by sequencing, then subcloned into the pCAMBIA1300-221 binary vector under the control of the 35S promoter to generate 35S-miR159847 for transformation.
For the cleavage-resistant PCsVMV-IAA28m construct, IAA28 genomic sequence was amplified from the wild-type Col-0 genome. Oligonucleotide-directed mutagenesis was introduced into the IAA28 genomic fragment with two primers in opposite orientations, IAA28m5′ and IAA28m3′ (Supplemental Data Set 2), using the Site-Directed Mutagenesis Kit (New England Biolabs) according to the manual. After it was confirmed by sequencing, the IAA28m sequence was subcloned into the pCAMBIA1300-221 binary vector, which contains the CsVMV promoter (Li et al., 2010), giving PCsVMV-IAA28m, and retransformed to 35S-miR159847 line 8 plants using Agrobacterium EHA105. Transformants were selected on medium containing 30 mg/L hygromycin and 50 mg/L kanamycin.
All primer sequences are listed in Supplemental Data Set 2.
GUS Staining
Seedlings, leaves, and flowers of MIR847-GUS and IAA28-GUS transgenic plants were stained in a GUS staining solution [2 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, 100 mM Na3PO4 buffer, 2 mM each K3Fe(CN)6/K4Fe(CN)6, and 0.5% Triton X-100] and incubated at 37°C overnight. After GUS staining, chlorophyll was removed using 70% ethanol.
5′ RLM-RACE Assay
The 5′ RACE assay was performed by using the First Choice RLM-RACE kit (Ambion). Portions (2 µg) of total RNAs were collected for direct ligation to the 5′ RACE RNA adapter, and subsequent steps were performed according to the manufacturer’s directions. PCR fragments obtained from 5′ RACE were inserted into the pGEM-T vector (Tiangen), and individual clones were selected for DNA sequencing.
Agrobacterium Infiltration
The 35S-miR159847, PCsVMV-IAA28m, 35S-IAA28, and pCAMBIA1300-221 (control vector) constructs were transformed into the EHA105 strain of Agrobacterium by electroporation and selected on Luria-Bertani medium containing kanamycin at 50 mg/L and rifampicin at 10 mg/L. For Agrobacterium infiltration experiments, equal volumes of an Agrobacterium culture containing 35S-IAA28 or PCsVMV-IAA28m (OD600 = 0.5) with the 35S-miR159847 or pCAMBIA1300-221 (OD600 = 1.5) were mixed before infiltration and coinfiltrated into leaves of Nicotiana benthamiana.
NAA Treatment
Wild-type Col-0 and tir1-1 and axr1-3 mutants were cultured in Murashige and Skoog (MS) medium with 30% sucrose for 10 d, then placed in solutions with 10 or 20 µM NAA. Seedling samples were collected at various times for RNA analysis.
RNA Gel Blotting
Total RNA was extracted from plant tissue using the TRIzol reagent (Invitrogen). For low molecular weight RNA gel blots, 80 or 60 μg of total RNA was separated by electrophoresis on 17% PAGE gels and electrically transferred to nylon N+ membrane. [γ-32P]ATP-labeled specific oligonucleotide probe sequences were used. For high molecular weight RNA gel blots, 30 μg of total RNA was separated on 1.2% agarose gels containing 6% formaldehyde and transferred to nylon N+ membrane. DNA probes were labeled with [α-32P]dCTP using the Rediprime II system (Amersham).
RNA Isolation and qRT-PCR Analysis
Total RNA was extracted from Arabidopsis seedlings and rosette leaves using TRIzol reagent (Invitrogen). DNA residues of total RNA were removed using DNase I (Takara). Total RNA was then reverse transcribed into cDNA using SuperScript III reverse transcriptase (Promega). qRT-PCR analysis was performed with a 1000 series Thermal Cycling Platform (Bio-Rad) using GoTaq qPCR Master Mix (Promega). The primers used for qRT-PCR are listed in Supplemental Data Set 2. At least three biological replicates and three technical replicates within an experiment for each sample were performed.
Histological Analysis
Leaves and seedlings were fixed in 90% ethanol and 10% acetic acid for 1 h and gradually hydrated using serial dilutions of ethanol solutions (90, 70, 50, and 30%). The samples were washed with distilled water and then dipped in 16.1 M chloral hydrate in glycerol:water (1:2, v/v) solution for transparentizing. The transparentized tissues were observed using an HC/DMR microscope (Leica) equipped with Nomarski optics. The middle part of the leaf was used for microscopic observation.
Leaf Explant Culture
Rosette leaves of 2-week-old Col-0, 35S-miR159847, 35S-IAA28, and PCsVMV-IAA28m transgenic plants, and the iaa28-ko mutant were excised. The explants were cultured on callus induction medium in the dark at 22°C. The callus induction medium used was MS medium containing 100 ng/L 2,4-D and 100 ng/L 6-BA (Hu et al., 2000). Photograph was taken at 7 d without changing the medium.
Bioinformatics and Statistical Analysis
Putative targets for miR847 were predicted through computational approaches at http://omicslab.genetics.ac.cn/psRobot/ (Wu et al., 2012). Statistical analysis was performed using Excel at http://www.microsoft.com/. Data shown are means ± sd.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: AT1G07051 (MIR847A), AT5G25890 (IAA28), AT5G60890 (ATR1), AT3G62980 (TIR1), AT1G05180 (AXR1), AT5G26930 (GATA23), AT1G56010 (NAC1), AT4G27260 (GH3.5), AT4G37390 (GH3.2), AT1G04240 (IAA3), AT3G15170 (CUC1), AT5G53950 (CUC2), AT1G76420 (CUC3), AT5G63090 (LOB), AT4G34160 (CYCD3;1), AT4G37490 (CYCB1;1), and AT4G14560 (IAA1).
Supplemental Data
Supplemental Figure 1. Expression pattern of GUS transcript in IAA28-GUS and MIR847-GUS transgenic plants upon auxin treatment.
Supplemental Figure 2. Lateral root development in gain-of-function mutant iaa28-1 and wild type control Wassilewskija.
Supplemental Figure 3. Phenotypes of transgenic plants overexpressing miR847 or IAA28, and IAA28 lost-of-function and gain-of-function mutants.
Supplemental Data Set 1. Predicted putative targets for miR847 through computational approaches (Wu et al., 2012).
Supplemental Data Set 2. List of primers and DNA oligos used in this study.
Supplementary Material
Acknowledgments
We thank Bonnie Bartel for the iaa28-1 mutant seeds, Qi Xie for the iaa28-ko (SALK_129988C from the ABRC) mutant seeds and the wild-type Arabidopsis Ws seeds, and Oliver Yu for CsVMV promoter-containing plasmid. We thank Xiang-Feng He and Meng Liu for the 35S-miR159847 construct. This research was supported by the National Science Foundation of China (Grant 31030009) and the Ministry of Science and Technology (Grant 2011CB100703).
AUTHOR CONTRIBUTIONS
J.-J.W. and H.-S.G. designed the research. J.-J.W. performed the research. J.-J.W. and H.-S.G. wrote the article.
Glossary
- miRNA
microRNA
- RACE
rapid amplification of cDNA ends
- Col-0
Columbia-0
- qRT-PCR
quantitative RT-PCR
- NAA
1-naphthalene acetic acid
- Ws
Wassilewskija
- DAG
days after germination
- SAM
shoot apical meristem
- 6-BA
6-benzyl aminopurine
- CsVMV
cassava vein mosaic virus
- MS
Murashige and Skoog
References
- Achard P., Herr A., Baulcombe D.C., Harberd N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131: 3357–3365. [DOI] [PubMed] [Google Scholar]
- Aida M., Tasaka M. (2006). Genetic control of shoot organ boundaries. Curr. Opin. Plant Biol. 9: 72–77. [DOI] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Bartel D.P. (2005). Antiquity of microRNAs and their targets in land plants. Plant Cell 17: 1658–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C.C., Sieber P., Wellmer F., Meyerowitz E.M. (2005). The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 15: 303–315. [DOI] [PubMed] [Google Scholar]
- Berleth T., Krogan N.T., Scarpella E. (2004). Auxin signals—Turning genes on and turning cells around. Curr. Opin. Plant Biol. 7: 553–563. [DOI] [PubMed] [Google Scholar]
- Carthew R.W., Sontheimer E.J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I., Marchant A., Bhalerao R.P., Beeckman T., Dhooge S., Swarup R., Graham N., Inzé D., Sandberg G., Casero P.J., Bennett M. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T.J., Aung K., Lin S.I., Wu C.C., Chiang S.F., Su C.L. (2006). Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B., et al. (2010). A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 20: 1697–1706. [DOI] [PubMed] [Google Scholar]
- Dewitte W., Riou-Khamlichi C., Scofield S., Healy J.M., Jacqmard A., Kilby N.J., Murray J.A. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005a). The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., Ehrismann J.S., Jürgens G., Estelle M. (2005b). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9: 109–119. [DOI] [PubMed] [Google Scholar]
- Donnelly P.M., Bonetta D., Tsukaya H., Dengler R.E., Dengler N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215: 407–419. [DOI] [PubMed] [Google Scholar]
- Dreher K.A., Brown J., Saw R.E., Callis J. (2006). The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan C.G., Wang C.H., Fang R.X., Guo H.S. (2008). Artificial microRNAs highly accessible to targets confer efficient virus resistance in plants. J. Virol. 82: 11084–11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N., Jogdeo S., Kasschau K.D., Sullivan C.M., Chapman E.J., Laubinger S., Smith L.M., Dasenko M., Givan S.A., Weigel D., Carrington J.C. (2010). MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 22: 1074–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira P., Hemerly A., de Almeida Engler J., Bergounioux C., Burssens S., Van Montagu M., Engler G., Inzé D. (1994a). Three discrete classes of Arabidopsis cyclins are expressed during different intervals of the cell cycle. Proc. Natl. Acad. Sci. USA 91: 11313–11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira P.C., Hemerly A.S., Engler J.D., van Montagu M., Engler G., Inzé D. (1994b). Developmental expression of the Arabidopsis cyclin gene cyc1At. Plant Cell 6: 1763–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Chiou T.J., Lin S.I., Aung K., Zhu J.K. (2005). A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15: 2038–2043. [DOI] [PubMed] [Google Scholar]
- Grimson A., Srivastava M., Fahey B., Woodcroft B.J., Chiang H.R., King N., Degnan B.M., Rokhsar D.S., Bartel D.P. (2008). Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455: 1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T.J., Hagen G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10: 453–460. [DOI] [PubMed] [Google Scholar]
- Guo H.S., Xie Q., Fei J.F., Chua N.H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17: 1376–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L., Bussell J.D., Pacurar D.I., Schwambach J., Pacurar M., Bellini C. (2009). Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21: 3119–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G., Guilfoyle T. (2002). Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 49: 373–385. [PubMed] [Google Scholar]
- Hibara K., Karim M.R., Takada S., Taoka K., Furutani M., Aida M., Tasaka M. (2006). Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18: 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood E.E., Gelvin S.B., Melchers L.S., Hoekema A. (1993). New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2: 208–218. [Google Scholar]
- Hu J.Y., Zhou Y., He F., Dong X., Liu L.Y., Coupland G., Turck F., de Meaux J. (2014). miR824-regulated AGAMOUS-LIKE16 contributes to flowering time repression in Arabidopsis. Plant Cell 26: 2024–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Bao F., Li J. (2000). Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J. 24: 693–701. [DOI] [PubMed] [Google Scholar]
- Husbands A., Bell E.M., Shuai B., Smith H.M., Springer P.S. (2007). LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 35: 6663–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai Y., Sakamoto T., Ueguchi-Tanaka M., Shibata Y., Gomi K., Umemura I., Hasegawa Y., Ashikari M., Kitano H., Matsuoka M. (2005). Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17: 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14: 787–799. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B. (2006). MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 57: 19–53. [DOI] [PubMed] [Google Scholar]
- Juarez M.T., Kui J.S., Thomas J., Heller B.A., Timmermans M.C. (2004). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88. [DOI] [PubMed] [Google Scholar]
- Kim J.J., Lee J.H., Kim W., Jung H.S., Huijser P., Ahn J.H. (2012). The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol. 159: 461–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M.A., Haubold B., Mitchell-Olds T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17: 1483–1498. [DOI] [PubMed] [Google Scholar]
- Kruszka K., Pieczynski M., Windels D., Bielewicz D., Jarmolowski A., Szweykowska-Kulinska Z., Vazquez F. (2012). Role of microRNAs and other sRNAs of plants in their changing environments. J. Plant Physiol. 169: 1664–1672. [DOI] [PubMed] [Google Scholar]
- Laufs P., Peaucelle A., Morin H., Traas J. (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131: 4311–4322. [DOI] [PubMed] [Google Scholar]
- Li H., Deng Y., Wu T., Subramanian S., Yu O. (2010). Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol. 153: 1759–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., et al. (2013). MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153: 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Thong Z., Yu H. (2009). Coming into bloom: The specification of floral meristems. Development 136: 3379–3391. [DOI] [PubMed] [Google Scholar]
- Liu X., Huang J., Wang Y., Khanna K., Xie Z., Owen H.A., Zhao D. (2010). The role of floral organs in carpels, an Arabidopsis loss-of-function mutation in microRNA160a, in organogenesis and the mechanism regulating its expression. Plant J. 62: 416–428. [DOI] [PubMed] [Google Scholar]
- Ma Z., Coruh C., Axtell M.J. (2010). Arabidopsis lyrata small RNAs: Transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell 22: 1090–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Bartel D.P., Bartel B. (2005). MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17: 1360–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Dugas D.V., Bartel D.P., Bartel B. (2004). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14: 1035–1046. [DOI] [PubMed] [Google Scholar]
- Marin E., Jouannet V., Herz A., Lokerse A.S., Weijers D., Vaucheret H., Nussaume L., Crespi M.D., Maizel A. (2010). miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J.D. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439. [DOI] [PubMed] [Google Scholar]
- Niu Q.W., Lin S.S., Reyes J.L., Chen K.C., Wu H.W., Yeh S.D., Chua N.H. (2006). Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 24: 1420–1428. [DOI] [PubMed] [Google Scholar]
- Oakenfull E.A., Riou-Khamlichi C., Murray J.A.H. (2002). Plant D-type cyclins and the control of G1 progression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357: 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425: 257–263. [DOI] [PubMed] [Google Scholar]
- Park J.E., Seo P.J., Lee A.K., Jung J.H., Kim Y.S., Park C.M. (2007). An Arabidopsis GH3 gene, encoding an auxin-conjugating enzyme, mediates phytochrome B-regulated light signals in hypocotyl growth. Plant Cell Physiol. 48: 1236–1241. [DOI] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20: 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J.A., Zenser N., Leyser O., Callis J. (2001). Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13: 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast M.I., Simon R. (2008). The meristem-to-organ boundary: More than an extremity of anything. Curr. Opin. Genet. Dev. 18: 287–294. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. (2002). MicroRNAs in plants. Genes Dev. 16: 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. (2002). Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Rogg L.E., Lasswell J., Bartel B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13: 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527. [DOI] [PubMed] [Google Scholar]
- Shani E., Yanai O., Ori N. (2006). The role of hormones in shoot apical meristem function. Curr. Opin. Plant Biol. 9: 484–489. [DOI] [PubMed] [Google Scholar]
- Staswick P.E., Serban B., Rowe M., Tiryaki I., Maldonado M.T., Maldonado M.C., Suza W. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Zhu J.K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Kapoor A., Zhu J.K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18: 2051–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G., Reinhart B.J., Bartel D.P., Zamore P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q., Uhlir N.J., Reed J.W. (2002). Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14: 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749. [DOI] [PubMed] [Google Scholar]
- Wang J.W., Wang L.J., Mao Y.B., Cai W.J., Xue H.W., Chen X.Y. (2005). Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17: 2204–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S.I., Lauga B., Charlesworth D. (2002). Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol. Biol. Evol. 19: 1407–1420. [DOI] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.J., Ma Y.K., Chen T., Wang M., Wang X.J. (2012). PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 40: W22–W28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.F., Tian Q., Reed J.W. (2006). Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133: 4211–4218. [DOI] [PubMed] [Google Scholar]
- Xie Q., Frugis G., Colgan D., Chua N.H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14: 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Guo H.S., Dallman G., Fang S., Weissman A.M., Chua N.H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170. [DOI] [PubMed] [Google Scholar]
- Xue X.-Y., Zhao B., Chao L.-M., Chen D.-Y., Cui W.-R., Mao Y.-B., Wang L.-J., Chen X.-Y. (2014). Interaction between two timing microRNAs controls trichome distribution in Arabidopsis. PLoS Genet. 10: e1004266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon E.K., Yang J.H., Lim J., Kim S.H., Kim S.K., Lee W.S. (2010). Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res. 38: 1382–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.