The See1 effector acts organ specifically in U. maydis-triggered DNA synthesis during tumor formation, interfering with the phosphorylation of SGT1, a central regulator of immunity and cell cycle control.
Abstract
The biotrophic smut fungus Ustilago maydis infects all aerial organs of maize (Zea mays) and induces tumors in the plant tissues. U. maydis deploys many effector proteins to manipulate its host. Previously, deletion analysis demonstrated that several effectors have important functions in inducing tumor expansion specifically in maize leaves. Here, we present the functional characterization of the effector See1 (Seedling efficient effector1). See1 is required for the reactivation of plant DNA synthesis, which is crucial for tumor progression in leaf cells. By contrast, See1 does not affect tumor formation in immature tassel floral tissues, where maize cell proliferation occurs independent of fungal infection. See1 interacts with a maize homolog of SGT1 (Suppressor of G2 allele of skp1), a factor acting in cell cycle progression in yeast (Saccharomyces cerevisiae) and an important component of plant and human innate immunity. See1 interferes with the MAPK-triggered phosphorylation of maize SGT1 at a monocot-specific phosphorylation site. We propose that See1 interferes with SGT1 activity, resulting in both modulation of immune responses and reactivation of DNA synthesis in leaf cells. This identifies See1 as a fungal effector that directly and specifically contributes to the formation of leaf tumors in maize.
INTRODUCTION
To establish a successful infection and dampen plant defense responses during colonization, plant pathogens secrete proteins and other molecules, collectively termed effectors, to various host compartments (Jones and Dangl, 2006). Effectors are key to the alterations of host structures and functions during infection (Hogenhout et al., 2009). They act either in the intercellular space to handle the primary defense response or inside the host cell to execute functions such as reprogramming of the host to favor infection (Doehlemann et al., 2014).
The basidiomycetous plant pathogens are highly specialized colonizers that develop biotrophic interactions. Members of the Ustilaginales, a major order of this class, invade mainly monocots, including all major cereal crops. The infection normally occurs in seedlings, often immediately after mating of the compatible sporidia to form a dikaryotic filament (Kämper et al., 2006; Brefort et al., 2009). Fungal hyphae growing both intracellularly and intercellularly colonize the host systemically and grow toward the shoot apical meristem without inducing visible disease symptoms. Disease symptoms become evident upon the floral transition, and the fungus completes sporulation within the infected inflorescence, liberating a sooty mass of black teliospores (Brefort et al., 2009). Among Ustilaginales, Ustilago maydis, a model organism for biotrophic fungi (Kämper et al., 2006; Ökmen and Doehlemann, 2014), has the unique ability to colonize all the aerial organs of its host plant maize (Zea mays) and to induce the formation of plant tumors locally at sites of infection. The fungus penetrates the epidermal cells and then the subepidermal cells, forming an interaction zone called the biotrophic interface in which the hyphae are encapsulated by the host plasma membrane. After successful establishment in leaves, the fungus grows in the mesophyll and the living cells of the vasculature (Ökmen and Doehlemann, 2014). Proliferation of host and fungal cells results in tumors, which are supported by the comprehensive reprogramming of both plant signaling and metabolism early in infection (Doehlemann et al., 2008; Horst et al., 2010) and alteration of the pace and pattern of host cell division.
The U. maydis genome encodes ∼550 proteins that are predicted to be secreted and likely function as effectors (Mueller et al., 2008; Djamei and Kahmann, 2012). Many potential effector genes are arranged in clusters, and examination of deletion mutants revealed the importance of these genes in virulence (Kämper et al., 2006; Brefort et al., 2014). So far, only a few effector genes of U. maydis have been functionally characterized. Pep1 (Protein essential for penetration1) is involved in penetration and the establishment of initial compatibility by targeting and inhibiting the activity of the plant peroxidase POX12 (Doehlemann et al., 2009; Hemetsberger et al., 2012). Pit2 (Protein involved in tumors2), a protein essential for tissue colonization and plant defense suppression, inhibits apoplastic cysteine proteases (Doehlemann et al., 2011; Mueller et al., 2013). In addition, two translocated U. maydis effectors have been analyzed. The U. maydis chorismate mutase Cmu1 rechannels chorismate metabolism in the plant cell cytoplasm to prevent the synthesis of salicylic acid, a major defense signal (Djamei et al., 2011). The effector Tin2 (Tumor inducing2), which is part of the largest cluster of effectors in U. maydis (Brefort et al., 2014), masks a ubiquitin-proteasome degradation motif in TTK1, a maize protein kinase that regulates the anthocyanin biosynthetic pathway. Tin2 protects the active kinase against ubiquitination and thereby promotes the production of anthocyanin in infected tissue and suppresses lignin biosynthesis, a defense pathway (Tanaka et al., 2014).
U. maydis infects all maize aerial organs and thus interacts with different, developmentally distinct immature host tissues (Walbot and Skibbe, 2010). In a previous study, organ-specific transcriptomes of both the host and the pathogen were documented in seedlings, adult leaves, and tassels (Skibbe et al., 2010). It was hypothesized that effectors in U. maydis act in an organ-specific manner, a new concept now extended to anthers within the tassels (Gao et al., 2013). A recent study showed that individual effector genes of U. maydis act in specific plant organs and that deletion of one organ-specific effector does not hamper virulence in a nontarget organ (Schilling et al., 2014). To date, however, the functional basis of organ-specific effectors remains elusive.
Effectors may be recognized by plant receptor proteins, which in turn induce defense responses. Several plant receptor proteins function with the help of chaperones and cochaperones, including HSP90 (heat shock protein 90), RAR1 (required for Mla12 resistance), and SGT1 (suppressor of G2 allele of skp1) (Shirasu, 2009; Zhang et al., 2010). SGT1 was originally identified in Saccharomyces cerevisiae as an essential cell cycle protein that interacts with Skp1p, a component of the conserved eukaryotic Skp1/Cullin/F-box (SCF) E3 ubiquitin ligase. In yeast, Sgt1p is required for progression through the G1/S and G2/M checkpoints (Kitagawa et al., 1999) and is highly conserved, as its orthologs in both animal and plant kingdoms retain the cell cycle functions (Bhavsar et al., 2013). Maturation of SGT1 as a signaling molecule depends on phosphorylation by an upstream MAPK (Hoser et al., 2013).
In this study, we present the functional characterization of the U. maydis organ-specific effector See1 (Seedling efficient effector1; Um02239), which is specifically required during tumor formation in seedling leaves. See1 is translocated by the fungus into the plant cell cytoplasm and nucleus, where it interacts with the maize homolog of SGT1 and interferes with the MAPK-induced phosphorylation of SGT1. See1 participates in U. maydis-triggered reactivation of plant DNA synthesis in maize leaves and contributes to vegetative tumor formation.
RESULTS
See1 Is Required for the Induction of Leaf Tumors
After infection, U. maydis hyphae mainly grow intracellularly. About 4 d postinfection (DPI), small tumors are visible and the fungus proliferates massively both intracellularly and intercellularly. In mature tumors at 10 to 14 DPI, U. maydis forms masses of melanized teliospores (Doehlemann et al., 2008). Unlike other smut fungi of monocots, U. maydis causes these symptoms in both maize inflorescences and aerial vegetative tissues, such as seedling leaves (Skibbe et al., 2010). A previous analysis of U. maydis effector candidates with organ-specific expression patterns identified seven genes whose deletion resulted in a leaf-specific reduction of tumor formation (Schilling et al., 2014), and here we investigate one of these genes (um02239, now termed see1) for its specific role.
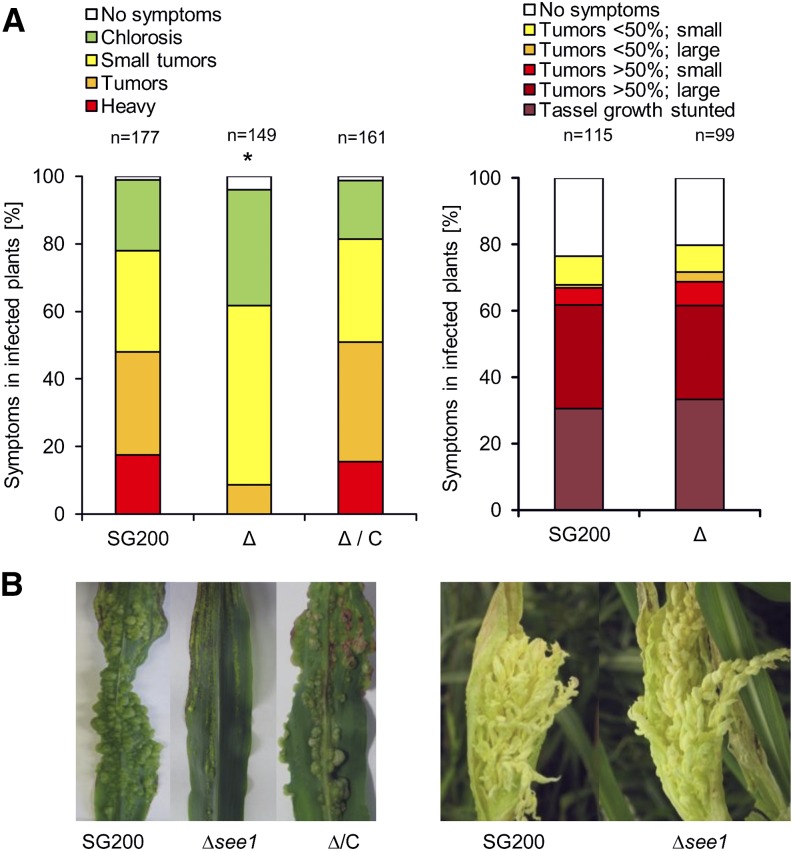
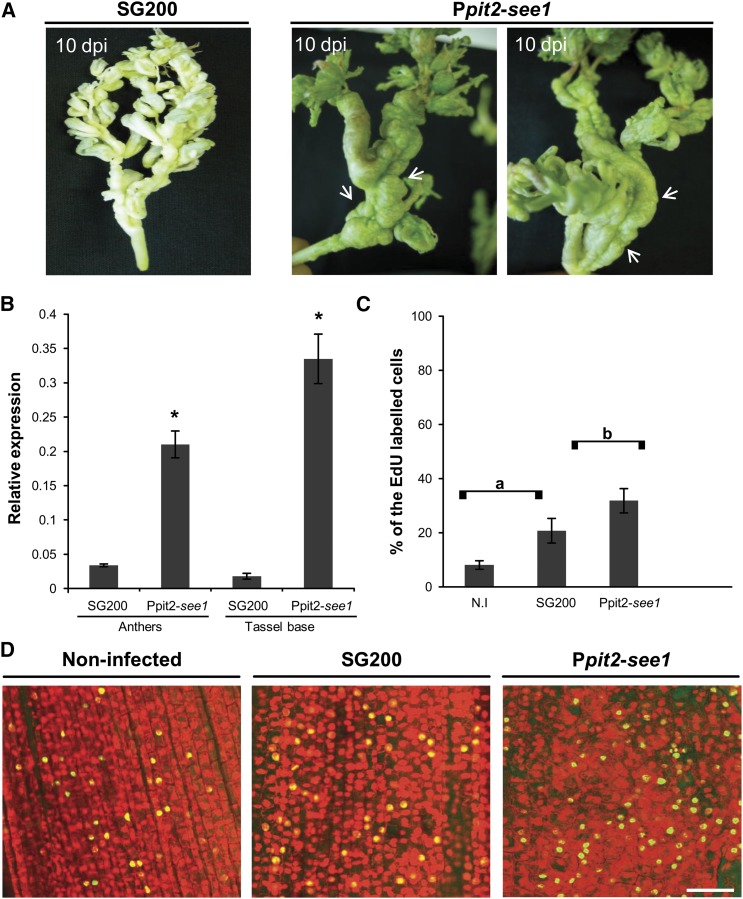
Deletion mutants of see1 (SG200∆see1) mainly formed tumors of 1 to 4 mm in diameter on seedling leaves at 12 DPI; these symptoms represent about half of the total tumors formed. Tumors of >6 to 20 mm occurred frequently in wild-type infections, representing around 28% of the total tumors, but they occurred much less frequently and were reduced in size in SG200∆see1 infections, representing only 9% of tumors. Heavy tumors, which cause altered leaf shape or even stunted growth of infected seedlings, were not observed after infection by SG200∆see1 (Figure 1A; Supplemental Figure 1). The SG200∆see1 mutant induces normal tumors in maize tassels, indistinguishable from the virulent progenitor strain SG200 (Figures 1A and 1B). Teliospores dissected from these tassel tumors were normal in shape and fully viable (Supplemental Figure 2). Similarly, in the maize ear, tumor formation was comparable to that in the wild type, supporting a strictly leaf-specific role of See1 in tumor induction (Supplemental Figure 3A). Confocal microscopy showed that SG200∆see1 hyphae initially colonize similarly to the progenitor strain SG200. At 3 DPI, when wild-type fungal hyphae reach the leaf mesophyll and are interspersed within the vasculature, mutant hyphae clustered at collapsed, highly fluorescent mesophyll cells (Supplemental Figure 4A). In addition, mutant hyphae failed to traverse from an infected cell into uninfected neighboring cells; this was particularly observed in bundle-sheath cells (Supplemental Figure 4B). Reintroduction of the see1 gene into the U. maydis ip locus fully restored virulence, demonstrating functional complementation of see1 and confirming that the observed growth defects reflected the absence of See1 (Figures 1A and 1B).
Figure 1.
Organ-Specific Phenotype of See1 Demonstrating Its Role in Leaves.
(A) Disease symptoms caused by SG200∆see1 in comparison with the wild-type progenitor strain SG200 in leaves and tassels. The mutant shows a significant reduction in leaf virulence. Maize seedling leaves were scored at 12 DPI. Disease symptoms in maize tassels were scored at 14 DPI as described by Schilling et al. (2014). SG200, the virulent U. maydis progenitor strain; Δ, deletion mutant for see1; Δ/C, genetic complementation of the deletion strain. The experiment was performed in three independent biological replicates. n = number of plants infected. *P ≤ 0.001.
(B) Symptoms caused by U. maydis strain SG200 in comparison with the SG200∆see1 mutant and the complemented strain in leaves and tassels. The leaf photograph shows typical disease symptoms at 12 DPI; the tassel photograph is at 14 DPI. Similar to SG200, the mutant caused disease symptoms in tassels, but leaf tumors were significantly reduced.
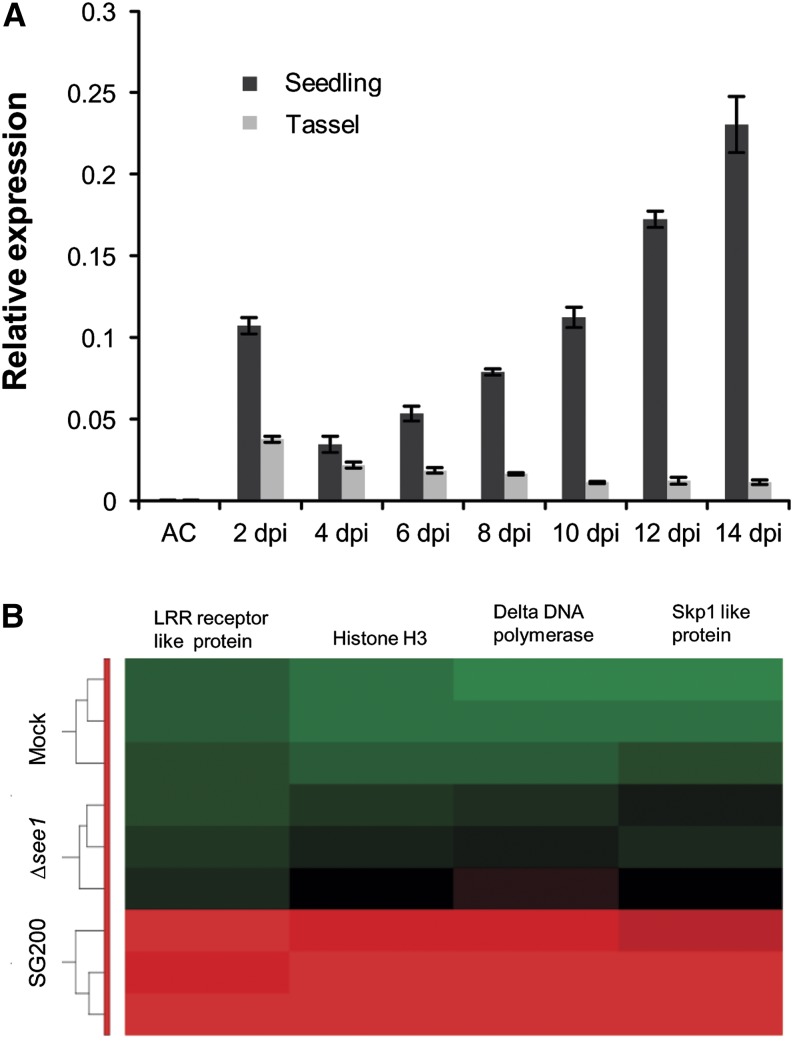
Transcription of see1 specifically increased during biotrophic growth of U. maydis (Figure 2A). Comparison of the temporal and spatial profiles of see1 expression during successive stages of tumor progression showed that see1 expression is constitutive and then upregulated at the later stages of tumor expansion in maize leaves but not in tassels (Figure 2A). In maize ear tumors, see1 transcript abundance was low as in tassels at 12 DPI. At this time point, see1 expression was >50-fold induced in leaves compared with the floral organs (Supplemental Figure 3B). To gain comprehensive insight into host processes affected by see1 deletion, Agilent microarrays were used to profile the transcriptome of maize leaves at 6 DPI by SG200, SG200∆see1, and mock control infections. RNA of infected tissue was prepared from three biological replicates, analyzed by hybridization, and subjected to data normalization and statistical analysis (see Methods for details). The abundances of 10,952 maize transcripts were altered in response to infection with wild-type U. maydis; by contrast, only 773 transcripts were altered in response to infection with SG200∆see1 (Supplemental Figure 5 and Supplemental Data Set 1). Hierarchical clustering of the SG200-induced maize genes visualized the reduced transcriptional response of maize leaves to the see1 deletion mutant (Supplemental Figure 5). A direct comparison of SG200 with SG200∆see1 showed that 549 genes were significantly induced (>2-fold) in SG200 compared with SG200∆see1 at 6 DPI, while only two genes were repressed (Supplemental Data Set 2). The transcripts induced by infection with wild-type pathogen were enriched for genes involved in DNA modification (i.e., histones), DNA replication, and DNA damage repair as well as genes associated with the cell cycle (Supplemental Data Set 2). Gene Ontology (GO) analysis showed that 71 of the 549 SG200-induced genes are associated with DNA metabolism and cell cycle regulation (Supplemental Data Set 2 and Supplemental Table 1 list the top 30 GO terms that are associated with DNA metabolism and cell cycle regulation). As shown in Figure 2B, DNA replicase Δ (TC280511), which is involved in S-phase DNA replication, was induced 690-fold in wild-type infections compared with SG200∆see1. DNA histone H3 (TC298222), which is required to generate nucleosomes, was induced 862-fold in wild-type infections. Maize Skp1 (TC293032) was induced 875-fold in SG200 infections versus SG200∆see1. Also, a Leu-rich repeat receptor-like protein responsible for protein phosphorylation and regulation of cell division (TC307447) increased 230-fold in wild-type infections (Supplemental Data Set 2). Together, these data suggest that SG200∆see1 fails to induce leaf tumor growth at the level of host cell DNA synthesis and cell proliferation, processes that are hallmarks of maize responses to infection (Doehlemann et al., 2008).
Figure 2.
Gene Expression during Maize Colonization with SG200 and SG200∆see1.
(A) RT-qPCR expression profiling of the see1 gene during the biotrophic phase of U. maydis growth in seedling and tassel tissues. Expression levels are shown relative to mean expression of ppi transcripts. Gene expression was analyzed in axenic culture (AC), seedling, and tassel tissues at consecutive time points from 2 to 14 DPI. The experiment was performed in three independent biological replicates.
(B) Transcriptional regulation of the key genes involved in the process of DNA synthesis and histone modification between wild-type SG200- and SG200Δsee1 (mutant)-infected seedlings at 6 DPI. Hierarchical clustering was performed by the Partek Genomics Suite version 6.12 to visualize the expression of maize genes transcriptionally regulated at 6 DPI by U. maydis strain SG200 (bottom), infection by SG200Δsee1 (middle), and mock inoculation (top). The x axis depicts clustering of the microarray samples for each of the three biological replicates for each treatment. The y axis shows clustering of the regulated maize transcripts based on the similarity of their expression patterns. red, upregulated genes; green, downregulated genes; black, not significantly altered. LRR, Leu-rich repeat.
See1 Is Required for U. maydis-Induced Plant DNA Synthesis during Leaf Tumor Formation
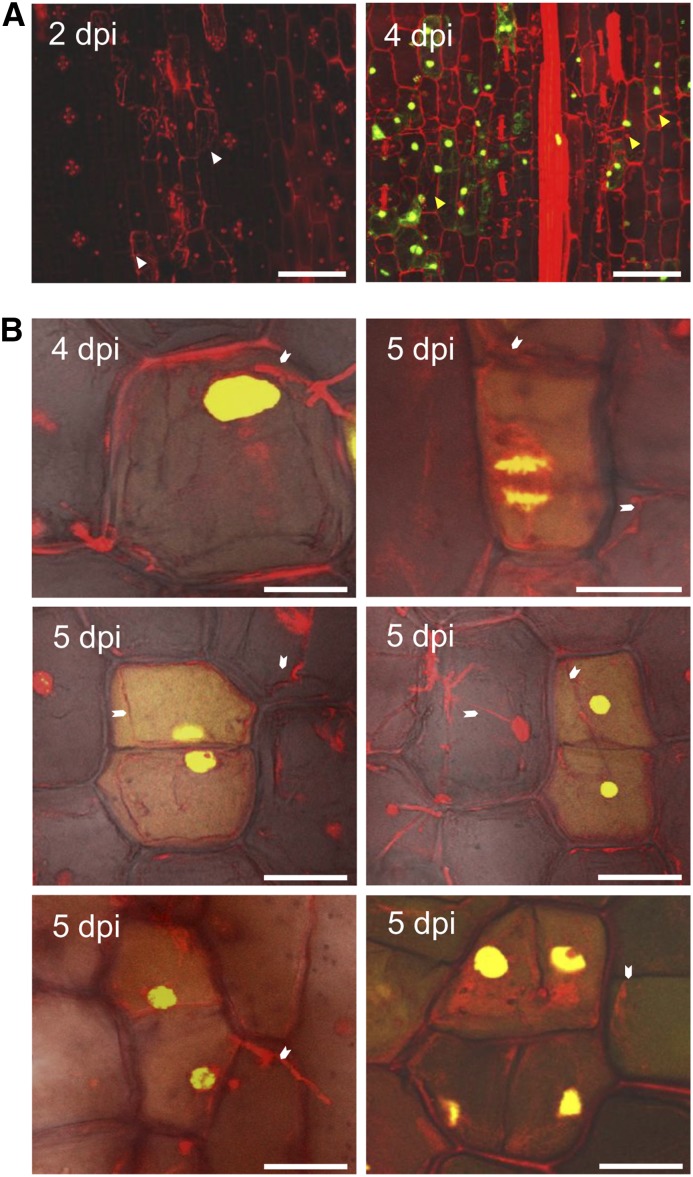
Because see1 expression is prominent during tumor enlargement and our initial observations indicated that SG200∆see1 hyphae were mainly restricted within mesophyll and bundle-sheath cells, we performed a more thorough confocal microscopy investigation of leaf infections. U. maydis-induced tumor growth reflects host proliferation, then cell expansion; thus, DNA synthesis is a prerequisite for growth. To monitor DNA synthesis in planta, we treated uninfected and infected leaves with 5-ethynyl-2-deoxyuridine (EdU) at several time points over a period of 5 h and then harvested samples. Incorporation of EdU was visualized by attaching a fluorescent tag (AF-488). Maize nuclei were stained with propidium iodide (PI) following a procedure described previously for maize anthers (Kelliher and Walbot, 2011). In maize leaves at 2 DPI, EdU treatment did not result in any detectable labeling. We observed this in maize leaves colonized with U. maydis and in uninfected maize leaves (Figure 3A), suggesting that no or only rare, sporadic maize DNA synthesis occurs in seedling leaf blades during the early phase of infection. We conclude that in the infected zones, the host cells were already postmitotic. By 4 DPI, when the first macroscopic symptoms appear in wild-type infections, EdU incorporation into leaf DNA was widespread (Figure 3A). Leaf cells invaded by fungal hyphae synthesized new DNA, and this coincided with the induction of mitosis, which could be visualized at different stages of cell division and as contiguous pairs of similarly labeled cells (Figure 3B). Such invaded cells also underwent multiple division events over several days (Figure 3B).
Figure 3.
U. maydis Induces DNA Synthesis in Infected Maize Seedlings.
(A) Maize seedlings were infected by U. maydis wild-type strain SG200, and then tissue was incubated in EdU to visualize in vivo DNA synthesis in the host cells. Samples were imaged at 2 and 4 DPI by confocal microscopy. Left, at 2 DPI, the fungal proliferation was observed subepidermally; host cells adjacent to fungal hyphae were considered to be colonized cells (white arrowheads). No EdU incorporation was observed. Right, at 4 DPI, numerous colonized cells showed EdU labeling (green stain), indicating the onset of DNA synthesis in host cells (yellow arrowheads). Bars = 75 μm.
(B) Cell division events were observed in maize seedlings infected by U. maydis wild-type strain SG200 at 4 and 5 DPI. EdU incorporation into a cell will result in equally labeled contiguous daughter cells after cell division. Such equally labeled cell pairs were readily observed in SG200-infected seedling leaf tissue. The white arrowheads point to fungal hyphae associated with maize cells undergoing cell division. It is inferred that reactivation of the cell cycle and rapid divisions are responsible for tumor formation. Bars = 25 μm.
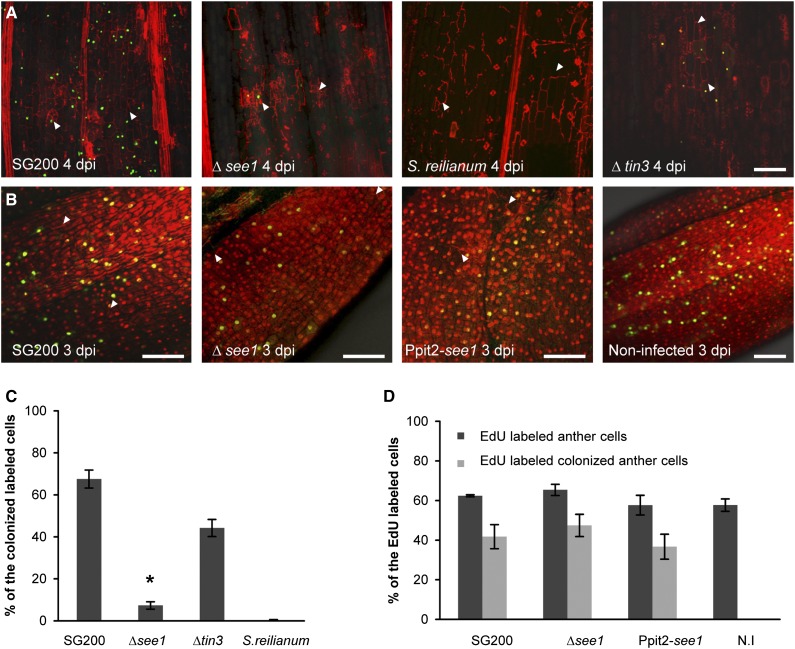
As an additional negative control for the EdU labeling, we injected 5 mM hydroxyurea, a DNA synthesis inhibitor, into seedling leaves infected with wild-type SG200 or SG200Δsee1 1 d before labeling them with EdU. Pretreatment with hydroxyurea eliminated EdU incorporation in all samples; this was also true in SG200-infected cells that already initiated division before treatment. Hydroxyurea appears to block DNA synthesis completely and validates the specificity of the EdU labeling assay (Supplemental Figure 6). Quantification of EdU labeling showed that 67.5% ± 4.2% of the maize cells colonized by SG200 incorporated EdU at 4 DPI (Figures 4A and 4C). Labeling was initiated from 3 to 4 DPI, while no DNA synthesis was observed in uninfected leaves of the same age. Therefore, U. maydis reactivated DNA synthesis and cell division in maize leaves at the onset of tumor induction. By contrast, SG200∆see1-infected leaf samples showed only 7.3% ± 1.7% EdU-positive cells at 4 DPI (Figures 4A and 4C; Supplemental Figure 7). The SG200∆see1 deletion mutant fails to trigger DNA synthesis and cell division to support the formation of large tumors.
Figure 4.
See1 Requirement for Host Cell Cycle Release in Leaf Tumor Formation.
(A) In vivo DNA synthesis in seedling tissue infected with SG200∆see1 in comparison with wild-type SG200. Samples infected with S. reilianum and SG200∆tin3, which has a similar phenotype to SG200∆see1 with respect to tumor size, were used as controls. Fungal hyphae and plant cell walls were visualized by PI staining (red), and the EdU-labeled host cell nuclei are visualized by AF488 staining (green). Fungal hyphae are shown by the white arrowheads. Bar = 100 μm.
(B) DNA synthesis in anther tissue infected with SG200∆see1 in comparison with wild-type SG200. Samples infected with the strain overexpressing See1 and uninfected anthers served as controls (right panel). Nuclei were visualized by PI staining (red), and EdU-labeled host cell nuclei are visualized by AF488 staining (green). Fungal hyphae are marked by white arrowheads. Bars = 100 μm.
(C) Quantification of the EdU-labeled seedling leaf cells in the in vivo DNA synthesis assay comparing infections with wild-type SG200, SG200∆see1, SG200∆tin3, and S. reilianum. Error bars show se. *P ≤ 0.001.
(D) Quantification of the EdU-labeled nuclei relative to total anther nuclei per image examined after infection with wild-type SG200, SG200∆see1, See1-overexpressing strain Ppit2-see1, and noninfected (N.I) tissue. Within the population of EdU-positive cells, the number colonized by fungal hyphae was also quantified in the infected samples. Error bars show se.
One might argue that the reduction in host cell DNA synthesis could be a general consequence of reduced virulence (i.e., an impaired biotrophic interaction); therefore, it would not be functionally linked with the action of See1. To test this, we included two additional strains. First, leaves were infected with wild-type strains of the maize head smut fungus Sporisorium reilianum, a close relative of U. maydis. S. reilianum also establishes a biotrophic interaction with maize, but it causes visible symptoms only in the inflorescences, never in leaves (Schirawski et al., 2010). Strikingly, at 4 DPI, the leaves infected with S. reilianum lacked detectable DNA synthesis, although dense tissue colonization was observed (Figure 4A). This observation indicates that biotrophic colonization of maize smut per se does not induce host DNA synthesis. Reactivation of DNA synthesis is specific to tumor formation. Next, a U. maydis deletion mutant lacking the secreted effector Tin3 was tested. Deletion of tin3 results in small leaf tumors similar to SG200∆see1 strains (Brefort et al., 2014). Despite its severely reduced virulence, the SG200∆tin3 deletion mutant activated EdU labeling in 44.22% ± 4.0% of colonized leaf cells at 4 DPI (Figures 4A and 4C). Therefore, there is more than one cause of impaired tumor induction, separating See1 from other mutants that lack large tumors but retain the ability to reactivate widespread host DNA synthesis. From these results, we conclude that the inability of the SG200∆see1 mutant to reactivate maize cell DNA synthesis and proliferation for tumor formation is not an indirect consequence of reduced tumor size but reflects a required activity of the See1 effector.
Tumor Formation in Anthers Does Not Involve U. maydis-Induced DNA Synthesis
The reproductive spikelets each contain two florets with three anthers and arise within the tassel inflorescence; the nonreproductive floral tissues such as the glumes, palea, and lemma of each spikelet, as well as the tassel stem, are readily infected and transformed to tumors by U. maydis. Interestingly, the fungus is only effective in causing anther tumors during the period of rapid anther growth by cell division prior to meiosis (Walbot and Skibbe, 2010). The intrinsic anther developmental program of rapid proliferation is reprogrammed into a tumor pathway, with different cell types affected depending on when fungal hyphae invade the cells (Gao et al., 2013). From this observation, Gao et al. (2013) concluded that tumor formation in anthers mainly occurs by restructuring of the usual sequential events in cell fate specification. In line with this hypothesis, we did not observe significant differences in EdU-labeled cells in uninfected tissue compared with U. maydis-infected anther tissue. Uninfected anthers as well as SG200- and SG200∆see1-infected premeiotic anthers contained ∼60% cells labeled with EdU in a 5-h treatment (Figures 4B and 4D). This is consistent with the previous report that, during the rapid proliferation period of anthers, EdU incorporation was found in the majority of cells after a 4-h labeling (Kelliher and Walbot, 2011). Therefore, in contrast with leaves, U. maydis does not alter anther cell DNA synthesis. In line with this, ∼42% of the EdU-positive anther cells were colonized by U. maydis at 4 DPI, with no significant difference between SG200 and SG200∆see1. We conclude that See1 is not involved in modulating host DNA synthesis and cell division during colonization and tumor induction in anthers and that it is dispensable for tumor formation in anthers.
See1 Actively Contributes to Tumor Formation and Maize DNA Synthesis
To test whether See1 actively contributes to tumor formation and DNA synthesis, a U. maydis strain was generated that constitutively expresses see1 during the entire infection process, independently of the colonized tissue. See1 was expressed from the promoter of pit2, which is one of the U. maydis genes with the strongest in planta transcription (Skibbe et al., 2010; Doehlemann et al., 2011). In leaf infections, the Ppit2-driven overexpression of see1 RNA (verified by reverse transcription and quantitative PCR [RT-qPCR]) and protein (confirmed by immunoblot analysis) did not result in a phenotype significantly different from the wild type (Supplemental Figures 8 and 9). Therefore, ectopic overexpression of see1 did not augment the virulence potential of U. maydis in leaves. Interestingly, the see1-overexpressing strain caused an unexpected tassel phenotype, although tassel tumors caused by wild-type U. maydis are largely restricted to the floral tissues, particularly the anthers. By contrast, Ppit2-driven see1 expression caused extensive tumor formation in the vegetative tassel base (the terminal node and internode) (Figure 5A). This effect, which resulted in bizarre alterations of tassel architecture, was observed in ∼38% of the infected plants (wild type, 8%) (Supplemental Figures 8C and 10B). Additionally, ∼20% of infected tassels including the spikelets became green 10 DPI with the see1-overexpressing strain (Supplemental Figures 8C and 10C).
Figure 5.
Overexpression of see1 Results in Tumor Proliferation in Vegetative Tassel Parts.
(A) Tassel base abnormality occurs much more frequently with constitutive overexpression of see1 in comparison with the wild-type strain SG200. Tumor formation in the tassel base is indicated by the white arrows.
(B) Quantification of see1 gene expression in tassels infected with the overexpressing strain Ppit2-see1 in comparison with plants infected with the wild-type SG200 strain. Error bars show se. *P ≤ 0.001.
(C) Quantification of EdU-labeled tassel base cells in the in vivo DNA synthesis assay after infection with the See1-overexpressing strain Ppit2-see1 in comparison with either the wild-type SG200 infected or noninfected (N.I) tassels. There was a significant difference in the number of EdU-labeled nuclei in the abnormal tassel base region as compared with the wild-type SG200 infected or noninfected tissue. Error bars show se. Comparisons a and b, P ≤ 0.05.
(D) Detection of in vivo DNA synthesis in the tassel base colonized by See1-overexpressing strain Ppit2-see1 in comparison with tissue colonized by wild-type strain SG200 and noninfected tissue. The total nuclei were visualized by PI staining (red), and the EdU-labeled cell nuclei were visualized by AF488 staining (green). Bar = 50 μm.
Tissue infected by the see1-overexpressing strain was also used to quantify EdU-labeled cells in anthers as well as the vegetative tassel base. In anthers, overexpression of see1 did not cause any significant differences in EdU labeling compared with SG200, SG200∆see1, and noninfected samples (Figures 4B and 4D), further evidence that See1 is not involved in tumor formation in anthers. In the terminal node/internodes, however, the frequency of EdU-labeled cells was significantly increased by see1 overexpression as compared with SG200 (Ppit2-see1, 31.8% ± 4.5%; SG200, 20.7% ± 4.5%; mock, 8.1% ± 1.6%). Therefore, the abnormal phenotype is a direct consequence of the excessive cell division resulting from see1 overexpression (Figures 5C and 5D). In summary, we conclude that See1 is required to stimulate U. maydis-induced tumor formation by promoting host DNA synthesis in vegetative tissue but not in maize anthers.
See1 Localizes to Maize Cytoplasm and Nuclei
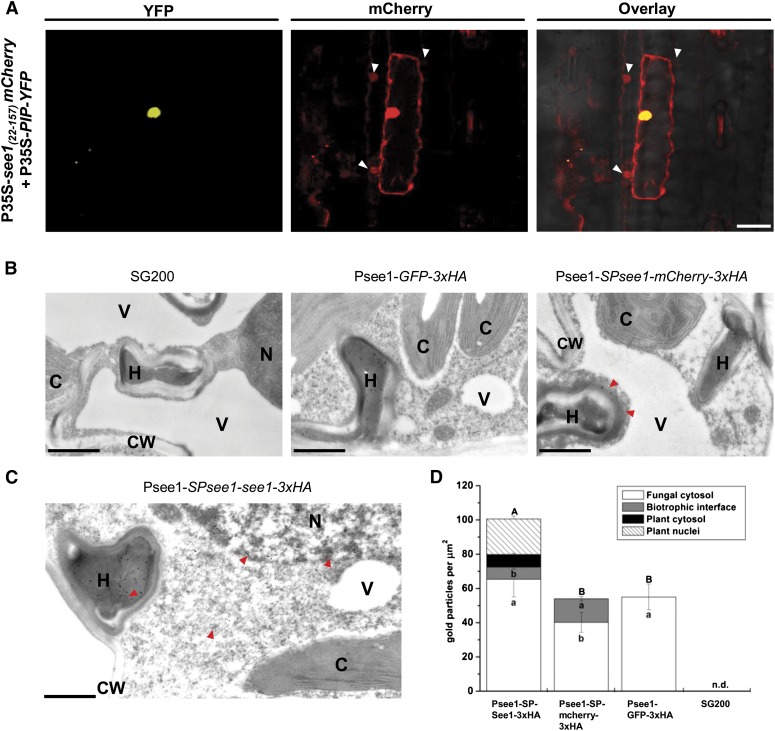
Live cell imaging and immunolabeling using transmission electron microscopy (TEM) were used to localize See1 in planta. An mCherry-tagged version of See1 lacking its N-terminal secretion signal (35S promoter:See122–157-mCherry) was transiently expressed in maize leaves by particle bombardment (Figure 6A). See122–157-mCherry localized to both the maize cytoplasm and nuclei (Figure 6A). As a transformation control, the nuclear marker protein PCNA-interacting protein (PIP426–593)-yellow fluorescent protein (YFP) was coexpressed and localized exclusively to the nuclei. As a localization control, mCherry expressed alone and Pit2-mCherry (Pit226–118-mCherry) also showed the same localization pattern to the cytoplasm and nucleus when transiently expressed in maize epidermal cells (Supplemental Figures 11A and 11B). Interestingly, the See122–157-mCherry signal spread to the nuclei of cells surrounding individual transformed cells (Figure 6A; Supplemental Figure 11C). Fluorescence signal movement was not observed for mCherry or for the Pit2-mCherry fusion protein (Supplemental Figures 11A and 11B). These results indicate that See1 may traffic in planta.
Figure 6.
See1 Localizes to the Plant Cell Cytoplasm and Nucleus.
(A) Confocal microscopy of 35S-see122–157-mCherry transiently expressed in maize epidermal cells. Left panel, transformation with the PIP-YFP control results in fluorescence that is specifically localized to the nucleus. Right panel, See1-mCherry is localized to the cytoplasm and nucleus and is transferred to the adjacent neighboring cells, which are shown by the white arrowheads in the mCherry and overlay channels. Bar = 25 μm.
(B) Controls for the TEM micrographs showing immunogold labeling of See1-3xHA in leaves of U. maydis-infected maize. No gold particles were bound to wild-type infected tissue specimens (left panel). Gold labeling was restricted to fungal hyphae in GFP-3xHA samples, as GFP was not secreted by the fungus (middle panel). Gold particles bound to the secreted mCherry control could be found in hyphae and at the biotrophic interface (red arrowheads) but not inside the plant cells, despite proximity to hyphae. Psee1-SPsee1-mCherry-3xHA expression demonstrates that mCherry is secreted by the fungus but not taken up by the plant (right panel). Bars = 1 μm.
(C) Immunogold labeling of See1 could be found in hyphae (H), the cytosol, and nuclei (N), as shown by the red arrowheads, but not in chloroplasts (C), vacuoles (V), or the cell wall (CW) when the See1 effector was tagged with 3xHA in the strain Psee1-SPsee1-See1-3xHA. Bar = 1 μm.
(D) Graph depicts the spatial distribution of gold particles bound to See1-3xHA in different cell compartments of leaves from Psee1-See1-3x-HA along with the secretory (mCherry-3xHA), nonsecreted (GFP-3xHA), and SG200 wild-type controls. Means are shown with se for the number of gold particles per μm2 in the individual cell compartments of three independent transverse sections. Lowercase letters indicate significant differences (P < 0.05) between the individual cell compartments, whereas uppercase letters indicate significant differences between the total sum of labeling signal for all analyzed cell compartments. Data were analyzed with the Kruskal-Wallis test followed by post-hoc comparison according to Conover (1999). n.d., not detected, for all analyzed cell compartments.
In addition to the heterologous expression of See1 in maize cells, the effector was localized upon natural delivery (i.e., when it was secreted from infectious U. maydis hyphae). A C-terminal 3xHA-tagged version of the effector was expressed under the control of the native promoter in the SG200∆see1 deletion strain. Immunoblot analysis after immunoprecipitation of See1-3xHA from infected plant tissue verified the expression and stability of the fusion protein (Supplemental Figure 12). For the immunolocalization of See1-3xHA, plants infected with U. maydis strain SG200 served as negative controls. Two additional controls for immunolabeling were employed: strain SG200 Psee1-GFP-3xHA expresses cytoplasmic green fluorescent protein (GFP) driven by the see1 promoter, and strain SG200 Psee1-SPsee1-mCherry-3xHA expresses a secreted mCherry under the control of the see1 promoter. Maize leaves were inoculated, and samples were harvested at 6 DPI for immunogold detection of the 3xHA tag. In the TEM images, no gold labeling was seen in plant tissue infected with the parental strain SG200, indicating the absence of nonspecific background labeling (Figure 6B, left panel). The nonsecreted GFP-3xHA was detected exclusively inside fungal hyphae at established biotrophic interfaces (Figure 6B, middle panel). The secreted mCherry-3xHA control showed labeling mainly in the biotrophic zone surrounding fungal cells as well as inside fungal hyphae (Figure 6B, right panel). By contrast, See1-3xHA was detected in the fungal hyphae, at the biotrophic interface, in plant cytoplasm, and prominently inside plant cell nuclei (Figure 6C; Supplemental Figure 13). The distribution of gold particles was quantified for all constructs. While all the encoded proteins were found inside fungal cells, only the two proteins with N-terminal secretion signals were found in the biotrophic interface. Only See1-3xHA was quantitatively detected inside host cells, with ∼20% of particles localizing to maize nuclei (Figure 6D). These results conclusively demonstrate the translocation of See1 from biotrophic fungal hyphae into maize cytoplasm and nuclei.
See1 Interacts with a Maize Homolog of SGT1
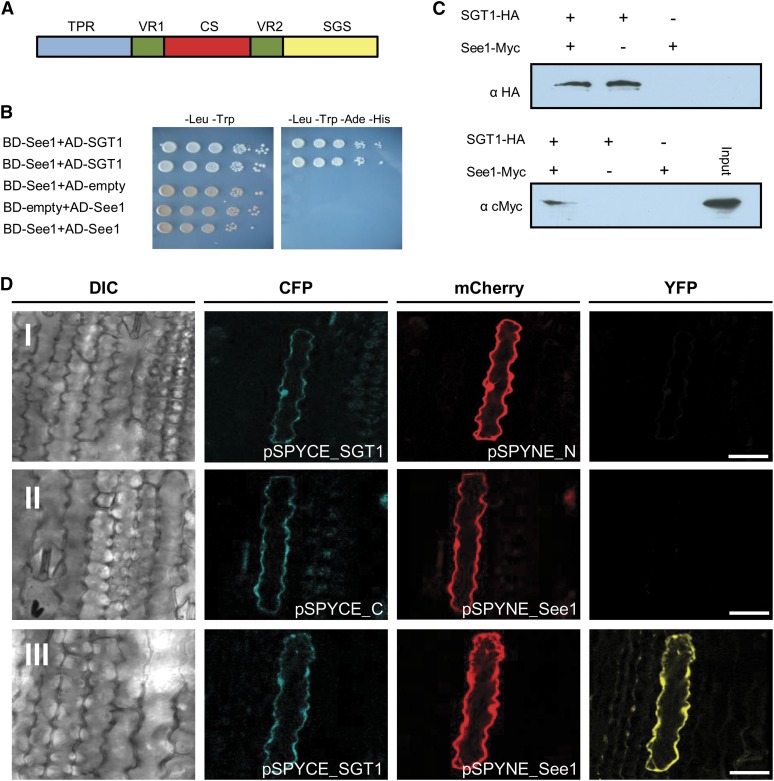
To identify proteins interacting with See1, we performed a yeast two-hybrid screen using a normalized cDNA library of U. maydis-infected maize leaves and tassels. From 60 clones that were isolated after plating on high-stringency selection medium, sequences corresponding to a maize homolog of SGT1 were identified (Figures 7A and 7B). SGT1, a known regulator of cell cycle progression in yeast and an important factor in plant host and nonhost resistance, has three functional domains: the tetratricopeptide repeat (TPR) domain, the Chord SGT1 (CS) domain, and an SGT1-specific (SGS) domain (Figure 7A) (Kitagawa et al., 1999; Peart et al., 2002). There are also two variable protein regions that are species-specific (Figure 7A). To test whether the identified maize protein exhibits SGT1 functions, temperature-sensitive mutants of S. cerevisiae, YKK57 (sgt1-5) and YKK65 (sgt1-3) (Kitagawa et al., 1999), were used for complementation experiments. At 37°C, both mutants are restricted in cell cycle phases, arrested at G1 and G2, respectively. Expression of full-length maize SGT1 under the control of the GAL4 yeast promoter complemented the growth defect of S. cerevisiae strain YKK57 (sgt1-5), indicating the functionality of the identified maize homolog. Expression of maize SGT1 in S. cerevisiae strain YKK65 (sgt1-3), which is defective at G2, showed normal growth at permissive temperature and partial complementation at 37°C (Supplemental Figure 14).
Figure 7.
See1 Interacts with the Cell Cycle and the Immune Response Modulator SGT1.
(A) Domain structure of maize SGT1 depicting three important domains: TPR, CS, and SGS. The two variable regions (VR1 and VR2) in the protein are species-specific.
(B) Yeast two-hybrid experiment to test for the interaction of See1 and maize SGT1. The drop assay was done by serial dilutions (see Methods), and strains were tested on low- and high-stringency plates to check for the specificity of the interaction. Results were documented after 4 d.
(C) Coimmunoprecipitation shows the interaction of See1 and SGT1 fusion proteins isolated from transiently expressing N. benthamiana cells. SGT1 was tag purified, and See1 was pulled down. In the absence of SGT1, no See1 signal was detected.
(D) In vivo interaction of See1 with SGT1. Confocal images show maize epidermal cells expressing BiFC constructs. Row I shows a plant cell coexpressing pSPYCE-SGT1 and pSPYNE-mCherry. Blue and red channels show cytoplasmic colocalization of the respective signals. No complementation of fluorescence is observed in the YFP channel. Row II shows the coexpression of pSPYCE-CFP and pSPYNE-See1. Blue and red channels show cytoplasmic colocalization of the respective signals. No complementation of fluorescence is observed in the YFP channel. Row III shows a cell coexpressing pSPYCE-SGT1 and pSPYNE-See1. Both signals colocalize in the nucleus and cytoplasm. The YFP channel exhibits YFP fluorescence reflecting the direct interaction of See1 and SGT1. DIC, differential interference contrast. Bars = 25 μm.
To verify the See1-SGT1 interaction in planta, both proteins were transiently expressed in Nicotiana benthamiana. As expression controls, P35S-See1-Myc and P35S-SGT1-HA were separately expressed in N. benthamiana leaves (Figure 7C). Using anti-HA matrix, See1-Myc was coimmunoprecipitated by the HA-tagged SGT1 but not in the absence of SGT1 (Figure 7C), confirming the See1-SGT1 interaction in planta. To localize the See1-SGT1 interaction in plant cells, bimolecular fluorescence complementation (BiFC) was employed, using an enhanced split-YFP system (Hemetsberger et al., 2012). An mCherry tag was fused to the C terminus of the N-terminal part of YFP (pSPYNE_N). Similarly, a cyan fluorescent protein (CFP) tag was added to the C-terminal part of YFP (pSPYCE_C). Via ballistic transformation of maize epidermal cells, both constructs were transiently expressed under the control of the 35S promoter. Cells expressing both pSPYCE_C and pSPYNE_N fused to SGT1 and See1, respectively, were designated as pSPYNE-P35S-see1-mCherry-N_YFP-Myc and pSPYCE-SGT1-CFP-CYFP-HA. The cells exhibited cytoplasmic and nuclear fluorescence signals for both mCherry and CFP, indicating expression of the fusion proteins. Expression of pSPYNE_N-mCherry with pSPYCE_SGT1 did not result in any detectable YFP signal, demonstrating that no unspecific protein dimerization occurred (Figure 7D). Similarly, no YFP fluorescence was detected when pSPYCE-CFP was coexpressed with pSPYNE-mCherry fused to see1 (pSPYNE_see1) (Figure 7D). By contrast, cells that coexpressed pSPYNE_see1 and pSPYCE_SGT1 showed a complementation of YFP fluorescence (Figure 7D), indicating an interaction of See1 and SGT1 in the cytoplasm and nucleus of maize cells. Altogether, we demonstrate that See1 interacts with SGT1 in the cytoplasm and nucleus of maize cells.
See1 Interferes with the Phosphorylation of SGT1
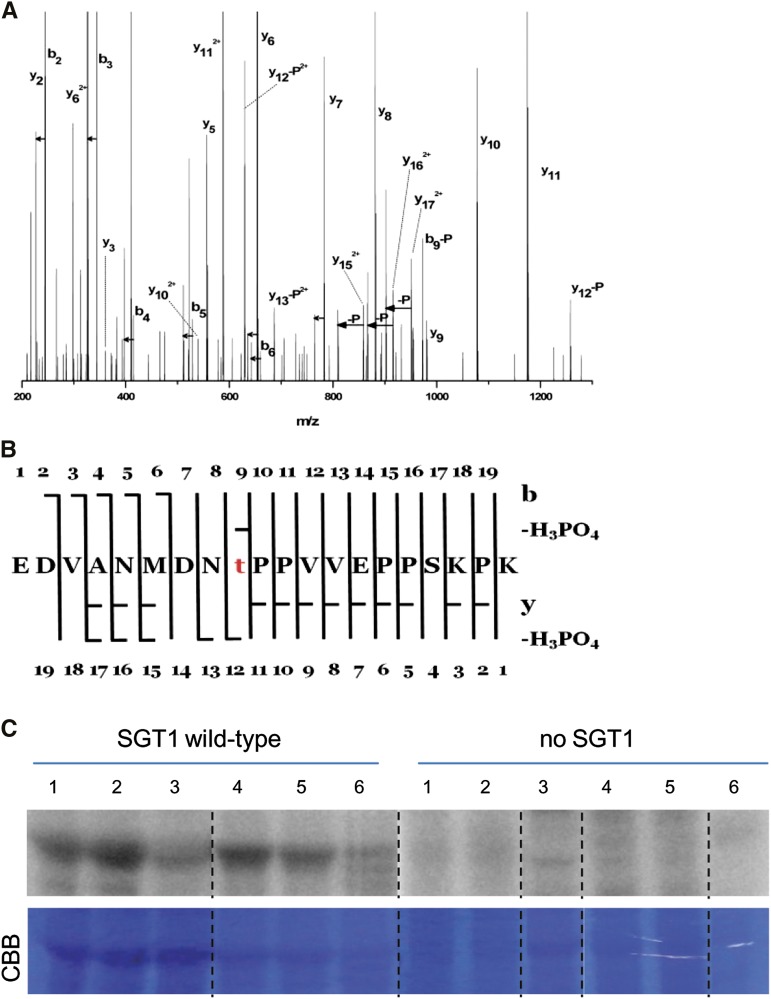
A major question arising from the See1 interaction with SGT1 concerns how the effector interferes with SGT1 function at the molecular level. Recent work showed that activation of SGT1 signaling activity requires the phosphorylation of SGT1 by salicylic acid-induced protein kinase (SIPK), a MAPK activated in response to pathogen assault (Hoser et al., 2013). SIPK-mediated phosphorylation of SGT1 was concluded to trigger enhanced nuclear compartmentalization of SGT1, thereby possibly activating defense-related signaling via the modulation of transcription. Based on these observations, we tested whether See1 interferes with the phosphorylation of SGT1. To facilitate the analysis of the in planta phosphorylation of SGT1 in the presence of See1, we used the well-established Agrobacterium tumefaciens expression system in N. benthamiana. N. benthamiana SGT1, which shares 65% identity to maize SGT1 (Supplemental Figure 15), was previously shown to be activated by Nt-SIPK, which, in turn, shares 84% to 86% identity to a set of five putative MAPK proteins of maize (Supplemental Figure 16). Agrobacterium strains carrying P35S-Zm-SGT1-StrepII, a gene encoding constitutively active Nt-MEK2 (Nt-MEK2DD) under the control of a dexamethasone (DEX)-inducible promoter, P35S-SIPK, and Dex-see1-HA or pTA7001 empty constructs were coinfiltrated into N. benthamiana leaves and maintained for 2 d to ensure expression of the constitutive promoter constructs. Subsequently, Nt-MEK2DD and See1 expression were induced by DEX treatment. Leaf samples were collected 5 h after the induction, and SGT1 was affinity-purified via its Strep tag II. Expression of all heterologous proteins has been verified by immune detection (Supplemental Figure 17). We detected two sites of phosphorylation in Zm-SGT1. Constitutive phosphorylation that was independent from See1 as well as from SIPK was detected for residue Thr-262 (Supplemental Table 5 and Supplemental Data Set 3). This residue is situated in the second variable region of Zm-SGT1 and only conserved in maize and sorghum (Sorghum bicolor) (Supplemental Figure 18). Surprisingly, upon SIPK activation and in the absence of See1, there was a high abundance of a Zm-SGT1 phosphopeptide with phosphorylation at Thr-150 (Figures 8A and 8B) in all three independently performed biological replicates. This phosphorylated position is situated in the variable region of Zm-SGT1, represents a putative MAPK target site, and is conserved in monocots including maize, rice (Oryza sativa), and sorghum (Supplemental Figure 18). Strikingly, in See1 coinfiltrated samples, no phosphorylation at Thr-150 could be detected, reflecting the interference of See1 with SIPK1-induced phosphorylation of SGT1 (Supplemental Table 5 and Supplemental Data Set 3). As a negative control, we coexpressed an inactive form of Nt-MEK2 (Nt-MEK2KR) with SGT1. In this case, SIPK was not included because its transient expression results in slightly increased SIPK activity and the associated activation of defense responses (Zhang and Liu, 2001). Under these conditions, coexpression of Nt-MEK2KR and SGT1 did not result in SGT1 phosphorylation at Thr-150. Therefore, we conclude that See1 interferes with the Zm-SGT1 phosphorylation at Thr-150, thereby delaying or preventing its activity.
Figure 8.
In Planta Phosphorylation of Maize SGT1.
(A) Fragmentation spectrum assigned to the phosphorylated form of the peptide EDVANMDNTPPVVEPPSKPK (Mascot score 126). Loss of H3PO4 is denoted by –P, loss of water is marked by short horizontal arrows, whereas a longer arrow symbolizes pairs of detected signals corresponding to yn and yn-H3PO4. The majority of signals of the tandem mass spectrometry spectra are assigned to the above species. The presence of several yn>11-H3PO4 and b9-H3PO4 ions accompanied by y15, y16, and y17 pinpoints threonine at position 9 as the unequivocal phosphorylation site within the peptide.
(B) Peptide sequence with assigned y, b, y-H2O, b-H2O, y-H3PO4, and b-H3PO4 ions present.
(C) Recombinant maize SGT1 produced in E. coli was incubated in the buffer containing [γ-32P]ATP, and total proteins were extracted from maize seedlings or tassels infected by various U. maydis strains. The samples were fractionated by SDS-PAGE and analyzed with a phosphor imager. Columns 1 to 3, extracts from seedling leaves 6 DPI with U. maydis wild-type SG200 (1), SG200∆see1 (2), or mock-inoculated (3). Columns 4 to 6, extracts from tassel base 9 DPI with U. maydis wild-type SG200 (4), U. maydis-overexpressing Ppit2:see1 (single-copy integration; 5), or U. maydis-overexpressing Ppit2:see1 (multiple-copy integration; 6). Representative data of four independent biological replicates are shown. CBB, Coomassie Brilliant Blue.
To corroborate the result that SIPK could phosphorylate Zm-SGT1, purified recombinant proteins Zm-SGT1, SIPK, and MEK2DD were coincubated in the presence of radioactive ATP in vitro. As shown in Supplemental Figure 19, the SIPK activated by MEK2DD was able to phosphorylate Zm-SGT1 (lane 1). In the negative control, which did not contain SIPK, we did not observe any phosphorylation of Zm-SGT1, and the signal detected corresponded to MEK2DD (Supplemental Figure 19, lane 2). As a positive control, a nonspecific substrate, myelin basic protein (Supplemental Figure 19, lane 3), was used; this protein was intensively phosphorylated in our assay. Collectively, from these results, we conclude that SIPK from tobacco (Nicotiana tabacum) can phosphorylate Zm-SGT1 in planta or in vitro and, therefore, could be used for transient assays to study the See1-SGT1 interaction.
Phosphorylation can alter protein subcellular localization and stability (Cohen, 2000). A truncated Arabidopsis thaliana SGT1b lacking a MAPK phosphorylation site was detected only in the nucleus-depleted fraction in leaf extracts (Noël et al., 2007), while a phosphomimic variant of At-SGT1b exhibited enhanced nuclear localization (Hoser et al., 2013). To test whether the phosphorylation of Zm-SGT1 altered its subcellular localization, constructs encoding YFP fused C-terminally to the wild-type Zm-SGT1 or to its phosphovariants were transiently expressed in N. benthamiana epidermal cells via particle bombardment. Wild-type Zm-SGT1 localized to the nucleus in ∼33% of the transformed cells, and its relative nuclear fraction in all cells tested was 39.4% (Supplemental Figure 20). Upon bombardment with constructs encoding Zm-SGT1 fused to a strong nuclear localization signal derived from SV40 large T antigen or protein serine kinase protein-derived nuclear export signal, the nuclear fraction was changed to 94.3% and 15% of the transformed cells, respectively (Wen et al., 1995). Control-like subcellular distribution could be restored by mutation of either the nuclear localization signal or the nuclear export signal (Supplemental Figure 20), demonstrating the suitability of the bombardment assay to assess protein localization. The phosphonull (Zm-SGT1AP) variant localized to the nucleus in around 20% of the transformed cells, and the nuclear fraction was 33.5% (Supplemental Figure 20). By contrast, the phosphomimic variant of Zm-SGT1 (Zm-SGT1DP) was observed in the nucleus of ∼40% of transformed cells, with an average nuclear fraction of 46.2%. These data suggest that Zm-SGT1 phosphorylation status affects nuclear import or export. Because the differences are subtle compared with the wild type, we conclude that the disturbance in localization of the Zm-SGT1 is not the primary virulence function of See1.
The See1-SGT1 Interaction Has Functional Relevance
To further investigate the role of See1 in targeting SGT1, we checked if the identified phosphorylation at Thr-150 in Zm-SGT1 is required for SGT1 function. To address this point, we first performed a yeast complementation assay using sgt1 cell cycle mutants to test for the complementation ability of phosphomimic and phosphonull mutants of Zm-SGT1 at this position. This assay did not show any differences in the complementation ability compared with the wild-type SGT1 (Supplemental Figure 21). This suggests that the phosphorylation site we identified in planta is not required for the cell cycle-related SGT1 function in yeast. Because the relevant phosphorylation site is specific to monocot SGT1 homologs and not present in yeast, one could not expect a function of this residue in yeast.
To elucidate the relevance of SGT1 phosphorylation in tumor formation, an alternative approach was employed. Specifically, this aimed to identify the phosphorylation status of SGT1 upon U. maydis infection and to further test the phosphorylation state in overexpression strains as compared with the U. maydis wild-type SG200. To this end, maize leaves were infected with the wild-type U. maydis strain SG200 and SG200Δsee1. In addition, tassels were infected with strains overexpressing See1 (which caused the abnormal tassel base phenotype) to collect the vegetative tassel base tumors. An in vivo detection of SGT1 phosphorylation was performed by preparing extracts from infected tissues, which served as a kinase source in the assay. Supplemental recombinant Zm-SGT1 in maize leaf or tassel base extracts was monitored for its phosphorylation state by autoradiography. It is clear that Zm-SGT1 is phosphorylated after incubation with the extracts from U. maydis-infected leaves as compared with the uninfected control (Figure 8C). Moreover, incubation with the extracts from leaves infected with SG200Δsee1 showed increased phosphorylation of SGT1 compared with the extracts from wild-type SG200-infected tissue. With U. maydis overexpression of See1 (by single and multiple integration of overexpression constructs), tassel bases showed quantitative reduction of SGT1 phosphorylation (Figure 8C). When using Zm-SGT1 protein with the T150A (SGT1AP) mutation in the same assay, residual phosphorylation was observed mainly in the tassel tissue (Supplemental Figure 22). This signal did not correlate with See1 expression and, therefore, most likely represents the previously observed constitutive phosphorylation of residue Thr-262 (Supplemental Table 5 and Supplemental Data Set 3). Together, these findings demonstrate that Thr-150 phosphorylation of maize SGT1 is triggered in response to U. maydis infection and that this is modulated by the See1 effector.
DISCUSSION
The secreted effector protein See1 is an organ-specific U. maydis virulence factor that promotes tumor formation in maize vegetative tissues. The expression profile of see1 shows strong organ specificity, supporting the specific requirement for this effector in supporting tumors in maize leaves but not in floral tissues. See1 is translocated from biotrophic hyphae to the maize cell cytoplasm and nucleus, where it interacts with maize SGT1 and interferes with its MAPK-induced phosphorylation. In leaf zones with postmitotic differentiated cells, U. maydis requires See1 to reactivate host cell division, a prerequisite for tumor formation. By contrast, anther tumor induction does not require de novo activation of plant cell proliferation, because in this highly proliferating tissue, the tumors result from redirecting host cell division and cell expansion into a tumor pathway (Gao et al., 2013).
During maize leaf development, most cell divisions occur in a narrow zone at the base of the blade adjacent to the ligule, with only sporadic divisions in the differentiating leaves. Tumors formed in U. maydis infections result from the profuse and rapid cell division in the subepidermal leaf cells. In uninfected plants, no such activity was visible in the corresponding leaf area (which is neither part of the apical meristem nor the basal region of the leaf) (Li et al., 2010). Confocal microscopy of leaves infected by SG200∆see1 showed that it successfully penetrated the host tissue and established itself in the initial stages of colonization. During the later stages, when the fungi reached the mesophyll cell layer, the mutant displayed defects in passing from cell to cell, with entrapment of the fungal hyphae in the mesophyll cells or the adjacent vascular cells (Supplemental Figure 4). This stage of infection at 4 DPI coincides with the normal appearance of heavy EdU labeling, indicative of reinitiation of the cell cycle in leaves infected with the wild-type fungi, as monitored by DNA precursor incorporation (Gratzner, 1982; Salic and Mitchison, 2008). EdU labeling of seedling tissue colonized by U. maydis showed that several division events had occurred in contiguous cells by 4 to 5 DPI, indicating that reactivation of host cell division occurs after initial fungal establishment and is followed by sustained proliferation of maize leaf cells.
During the initial 2 d of anther colonization, U. maydis is present on the epidermis (Gao et al., 2013). At later time points, the fungus is subepidermal and alters cell fate specification events, ongoing cell division patterns, and cell expansion depending upon developmental stage and cell type. The fungus mainly induces ectopic periclinal divisions in anther somatic cells, generating an extra cell layer resulting in disrupted anther lobe architecture. Frequent anticlinal and periclinal divisions are also observed in the middle layers of infected anther, which otherwise undergo only a few anticlinal divisions after their birth, prior to programmed cell death. Hence, in floral tissues, U. maydis reprograms cell fate but does not act as an oncogenic agent (Gao et al., 2013). Constitutive overexpression of see1 led to tumor formation in the vegetative parts of the tassel base, which in wild-type infections were not tumorous under the tested conditions. This suggests that See1 specifically acts in vegetative tissues. The phenomenon might be of significance to the fungus in nature, where floral tumors are more frequent in occurrence. See1 and other leaf-specific effectors might be of importance for host adaptation and the evolution of U. maydis, as they promote the formation of tumors in vegetative parts, an important factor in colonizing perennial grasses or exploiting seedlings. Infection of seedlings and immature plants allows the fungus to complete its short, 2-week life cycle multiple times during plant vegetative growth, because this infection style is independent from the development of plant inflorescences.
Where does the See1 effector act in leaf cells? Transient expression assays showed that See1 localizes to the cytoplasm and nucleus of maize cells. Interestingly, the effector protein also moved to cells neighboring a transformed cell. The specific translocation of See1-mCherry between neighboring maize cells suggests that there may be a second route for fungal effectors to enter maize cells. The U. maydis effectors Cmu1 and Tin2 are both translocated from biotrophic hyphae into the host cell (Djamei et al., 2011; Tanaka et al., 2014). We speculate that subsequent cell-to-cell movement may reflect a general feature of effectors taken up by plant cells. These proteins could act to stimulate the surrounding cells not yet in contact with fungal hyphae to promote fungal proliferation. For See1, the independent approach of immunolabeling clearly confirmed that the effector is translocated to the plant cell cytoplasm and nucleus. Movement of See1 to the neighboring cells was not observed in the TEM immunogold assay. This discrepancy might result from different expression levels in the two approaches; in the immunolabeling, See1 was expressed by fungal hyphae from its native promoter, but plant-derived expression was driven by the 35S promoter. Nevertheless, based on our data, we cannot exclude that the cell-to-cell movement observed in the transient assay is an experimental artifact of the exogenous expression of the effector rather than a feature of normal infections. On the other hand, the phenomenon of cell-to-cell movement of translocated effectors has also been described for Magnaporthe oryzae effectors in rice cells (Khang et al., 2010). This movement between cells was proposed to occur via the plasmodesmata, which are coopted by hyphae for cell-to-cell movement (Kankanala et al., 2007; Djamei et al., 2011).
See1 interacts with maize SGT1 in a yeast two-hybrid screen. Maize SGT1 is present in both the cytoplasm and nucleus of plant cells; hence, localization is consistent with the dual localization of See1. The interaction of See1 and SGT1 was confirmed independently by in planta coimmunoprecipitation. In addition, BiFC data showed that this interaction takes place in both the cytoplasm and nucleus of maize cells. This evidence is congruent with the electron microscopy analyses that show the same localization pattern of the See1 effector. Mass spectrometry analysis showed that the interaction with See1 results in an inhibition of SGT1 MAPK-triggered phosphorylation at Thr-150. Why would See1 interfere with the phosphorylation of SGT1 during leaf infection? Hijacking of SGT1 may contribute to the deactivation of immune responses. The literature provides a large body of data on SGT1. From all the available evidence concerning plant SGT1, this protein is widely seen to be active in vegetative leaf tissues (Noël et al., 2007). In Arabidopsis, the SGT1b isoform was found to be required for the SCF-mediated auxin response in seedling roots (Gray et al., 2003). Arabidopsis SGT1 has a regulatory role in early R gene-mediated plant defenses (Austin et al., 2002) and was shown to be involved in forming a cochaperone complex with HSP90; this complex functions in sensing immune responses of the host receptor proteins (Shirasu, 2009). Among the three domains of SGT1, namely the TPR, CS, and SGS domains, the CS domain resembles the α-crystalline domain of the cochaperone HSP20 (Dubacq et al., 2002; Garcia-Ranea et al., 2002). The other components of the immune regulatory cochaperone complex, RAR1 and HSP90, have been shown to interact with the CS domain of SGT1 (Azevedo et al., 2002; Takahashi et al., 2003).
The host SGT1/RAR1/HSP90 complex is a target of several bacterial effector proteins. The Pseudomonas syringae effector AvrB interacts weakly with Arabidopsis SGT1b (Cui et al., 2010). Another P. syringae effector, AvrPtoB, showed a genetic interaction with SGT1 and RAR1, requiring these cochaperones to suppress plant immunity (Hann and Rathjen, 2007). Additionally, the P. syringae effector HopI1 interacts with HSP70 (Jelenska et al., 2010), which is an active component initiating signaling by interaction with the SGT1/RAR1 complex. Recently, effector proteins from Salmonella enterica and Xanthomonas campestris have been shown to interact with SGT1 (Bhavsar et al., 2013; Kim et al., 2014). Consistent with the previous findings, these effectors bind to SGT1 at the CS domain, confirming the importance of this domain for SGT1 regulation during immune defenses. SGT1-mediated pathways may vary by plant species and are also specific to a particular pathogen (Wang et al., 2010). SGT1 is known to be used by some fungal pathogens in promoting disease symptoms. Like X. campestris, the necrotrophic fungus Botrytis cinerea uses SGT1 to initiate the hypersensitive response-mediated cell death pathway for the necrotrophic lifestyle (El Oirdi and Bouarab, 2007). Fusarium culmorum is known to require SGT1b to cause full disease symptoms in buds and flowers of Arabidopsis (Cuzick et al., 2009), the only prior report that described SGT1 in floral tissues. These observations suggest that searching for a tissue-specific role of SGT1 protein in Arabidopsis flowers may be fruitful.
We propose that SGT1 represents a conserved hub targeted by several effectors from bacterial as well as fungal pathogens, utilizing it according to the need of the pathogen lifestyle. This supports the early study of evolutionarily different effectors targeting a common host defense protein (Song et al., 2009) and is also consistent with the model of evolutionarily different virulence effectors targeting conserved hubs in a plant immune system network (Mukhtar et al., 2011). Work to date demonstrated that SGT1 is involved in resistance during biotrophic interactions. The results reported here indicate that U. maydis alters SGT1 function for symptom development. The precise steps following the interference of See1 with the posttranslational modification of SGT1, resulting in the reactivation of maize DNA synthesis and ultimately in tumor formation, remain to be elucidated biochemically. Therefore, it will be of prime interest to work out the detailed molecular mechanism of the See1-SGT1 interaction and to dissect the downstream signaling network (i.e., to identify and characterize proteins that interact with and/or are affected by SGT1 and to pinpoint residues in See1 required to interfere with SGT1).
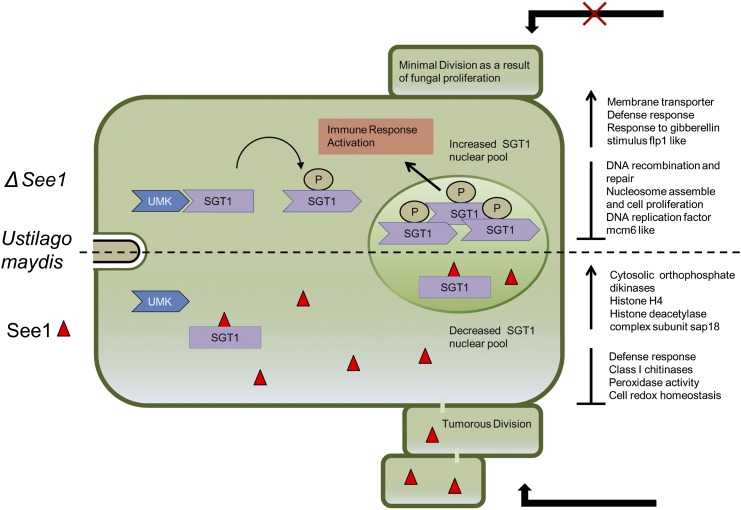
Transcriptome analysis of U. maydis wild-type and SG200∆see1-infected maize leaves at 6 DPI showed that genes involved in DNA binding, replication (including Skp1), as well as repair mechanisms characterize normal infections but are not induced in the mutant interaction. We hypothesize that this reflects the reduced activation of cell division and, consequently, limited tumor expansion in the mutant-infected leaves. Recent work by Bao et al. (2013) found an unexpected link between cell cycle progression and plant immunity, suggesting that cell cycle misregulation impacts the expression of R genes. Also, DNA repair proteins have been shown to be directly involved in the regulation of gene expression during plant defense responses (Song et al., 2011). The DNA damage response is an intrinsic component of the plant immune response and, in turn, enhances salicylic acid-mediated defense gene expression (Yan et al., 2013). Among the Ustilaginales, which cause characteristic floral symptoms in the immature, proliferative host floral organs, U. maydis is the only species that causes local tumors in vegetative tissue. Seedling-specific effectors such as See1 promote the generation of a mitotically active sink tissue within vegetative organs. By regulating SGT1, See1 may not only shut down defense signaling but also activate the host cell cycle, a prerequisite for tumor development. Hence, a combination of immune suppression and nutrient rechanneling, particularly facilitating the acquisition of sucrose, could trigger uncontrolled cell proliferation, ultimately resulting in plant tumors. Based on the results obtained in this study, our hypothesis on how the See1-SGT1 interaction could affect the formation of leaf tumors is shown in a tentative model (Figure 9). Detailed understanding of such processes should shed light on colonization biology in various biotrophic plant pathogens and on intrinsic host mechanisms that operate to prevent cell proliferation in differentiated organs.
Figure 9.
Tentative Model of the Role of the See1-SGT1 Interaction during U. maydis Tumor Formation.
The SGT1 protein is known to occur in the cytoplasmic and nuclear pools (Hoser et al., 2013). In U. maydis wild-type infections, activated unidentified maize kinase (UMK) triggers the phosphorylation of SGT1 at a monocot-specific target site. The See1 effector binds to SGT1, interferes with its phosphorylation status, and thereby disturbs the subcellular distribution (i.e., transport into the nucleus). This misbalancing of SGT1 phosphorylation and distribution contributes to the induction of cell cycle genes, leading to the induction of tumorous division.
METHODS
Growth Conditions and Virulence Assays
Maize (Zea mays cv Early Golden Bantam [Olds Seeds] and cv Gaspe Flint [maintained by self-pollination]) plants were grown in a temperature-controlled greenhouse (14-h/10-h light/dark cycle, 28/20°C). Gaspe Flint plants were mainly used for tassel infection and for the overexpression of See1, as they have an early floral switch (15 d) and are suitable for early meristematic tassel infections. Both cultivars of maize exhibit similar infection symptoms. Both varieties were grown in T-type soil (Frühstorfer Pikiererde). Ustilago maydis strains were grown in YEPSL (0.4% yeast extract, 0.4% peptone, and 2% sucrose) at 28°C with shaking at 200 rpm to an OD600 of 0.6 to 0.8. Cells were centrifuged at 900g for 5 min, resuspended in water to OD600 of 1.0, and injected into stems of 7-d-old maize seedlings with a syringe, as described previously (Kämper et al., 2006). All infection assays were performed in three independent infection trials with multiple seedlings. Disease symptoms were scored at 12 DPI using a previously developed scoring scheme (Kämper et al., 2006). Tassel infections were done after 15 d of seed sowing in Gaspe Flint and after 4 weeks in Early Golden Bantam, as described previously (Walbot and Skibbe, 2010). Disease symptoms in the tassels were scored 14 DPI following the criteria described in a previous study (Schilling et al., 2014). Nicotiana benthamiana plants (BN3) were grown in a phytochamber (Vötsch) under controlled environmental conditions (21°C, 16 h of light, 8 h of dark) as described previously (Talarczyk et al., 2002).
RNA Extraction and RT-qPCR
Expression of the candidate gene was analyzed by RT-qPCR. Total RNA was isolated from infected samples (leaf, tassel base, or anthers). The infected plant material was collected at successive time points from 2 to 14 DPI. To confirm the high expression of see1 at 9 DPI, seedlings, anthers, and tassel base were collected from plants infected with U. maydis strain Ppit2-see1 and the SG200 control. The samples were taken in three independently conducted experiments. RNA was isolated using Trizol reagent (Invitrogen) and purified using an RNeasy kit (Qiagen). For cDNA synthesis, the SuperScript III first-strand synthesis SuperMix kit (Invitrogen) was used to reverse transcribe 1 μg of total RNA with oligo(dT) primer. The RT-qPCR analysis was performed using an iCycler machine (Bio-Rad) in combination with iQ SYBR Green Supermix (Bio-Rad). Cycling conditions were 2 min at 95°C followed by 45 cycles of 30 s at 95°C, 30 s at 61°C, and 30 s at 72°C. Gene expression levels were calculated relative to the peptidylprolyl isomerase gene (ppi) of U. maydis for quantifying see1 expression (van der Linde et al., 2012). Error bars in all figures that show RT-qPCR data represent the sd that was calculated from the original cycle threshold values of three independent biological replicates. Primer sequences used for RT-qPCR are listed in Supplemental Table 2.
EdU-Based DNA Synthesis and Cell Proliferation Assay
Published protocols were used for U. maydis seedling infections (Kämper et al., 2006) and tassel infections as described (Walbot and Skibbe, 2010). At 4 DPI, the third infected leaf where the first infection symptoms appeared was used for the EdU assay and incubated for 5 h with 10 μM EdU (Invitrogen) in small chambers designed for labeling physiologically active leaves. For the tassels at 3 DPI, the immature mitotically active tassel was bathed with 1 mL of 20 μM EdU after delivery with a 26-gauge hypodermic needle through the whorl of leaves surrounding the inflorescence apex ∼17 d after germination in the Gaspe Flint maize variety. EdU seeps into spikelets through the small air spaces between the external organs of the spikelet and florets and reaches anthers over the labeling time of 5 h. After the labeling procedure, the area in seedlings below the infection holes was detached and fixed in 100% (v/v) ethanol. For the tassel tissue, around 150 anthers from different parts of the tassel were dissected to ensure random sampling with equal probability of labeled anthers and fixed in 100% (v/v) ethanol 5 h after labeling, as done for seedlings. The EdU staining procedure was done as described previously (Kelliher and Walbot, 2011). The samples were washed once with fresh 100% (v/v) ethanol followed by two washes in PBS, pH 7.4, plus 2% (w/v) BSA, then the samples were transferred to permeabilization solution (PBS +1% [v/v] Triton X-100) at room temperature for 20 min with rocking. After permeabilization, samples were washed twice in PBS plus 2% (w/v) BSA and then directly incubated for 30 min at room temperature with EdU Click-IT cocktail for detection (Invitrogen) and 20 μg/mL PI (Molecular Probes) directly added to the staining solution. The addition of EdU detection solution was done according to the manufacturer’s instructions. The samples were then washed twice in PBS, pH 7.4, plus 2% (w/v) BSA, transferred to PBS, pH 7.4, and kept at 4°C in the dark for several days before imaging. Triton X-100 treatment results in a nuclear stain and is compatible with EdU costaining. Moreover, it prevents the cell shrinkage and quick penetration of the fixer and allows better preservation of mitotic stages (Kotogány et al., 2010).
Yeast Transformation and Two-Hybrid Interaction Assay
To identify host interactors of See1, See1 lacking the signal peptide was expressed from pGBKT7-See122–157 and screened against a maize cDNA library in pGADT7. Preliminary testing indicated that See1 was nontoxic to yeast (Saccharomyces cerevisiae) and did not autoactivate. The yeast two-hybrid library analyses were done using a normalized cDNA library of infected maize tissue containing seedling and tassel samples. The strain AH109 (Clontech) was used for all yeast assays unless otherwise mentioned. Yeast transformation was done as described in the DUAL membrane starter kit manual (Dualsystems Biotech). The yeast two-hybrid screen was performed following the instructions of the Matchmaker yeast two-hybrid manual (Clontech) using 1 mg of bait DNA (pGBKT7-See1) and 0.5 mg of library DNA. All resulting yeast clones were tested by immunodetection for expression of the respective proteins. To perform a yeast dilution assay, 3 mL of selective medium (SD-Leu-Trp) was inoculated with a single colony of the respective yeast strain and incubated overnight at 28°C. OD600 was adjusted to 0.2, and the cells were grown to an OD600 of 0.6 to 0.8. Next, 1 mL of yeast culture was centrifuged for 10 min at 3500g, and the pellet was washed twice with 1 mL of sterile water and finally resuspended in 500 μL of sterile water. OD600 was adjusted to 1.0 with sterile water, and 5 μL of this suspension, as well as 1:10, 1:100, and 1:1000 dilutions, were applied on SD-Leu-Trp plates (low stringency) as a growth control and SD-Leu-Trp-Ade-His plates (high stringency) to test for protein-protein interaction. Growth was scored after 4 to 5 d of incubation at 28°C.
For the yeast complementation assay of the yeast sgt1 mutants, yeast strains YKK 57 (sgt1-5) and YKK65 (sgt1-3) were transformed with constructs to express maize SGT1 and the phosphovariants of SGT1 (cloned into pGREG536 vector under the control of the Gal1 promoter). The transformation was done as described previously, and the transformants were selected on SC-Ura-2% (w/v) glucose plates. The strains were then shifted to SC-Ura-2% (w/v) galactose and incubated for 4 d to test their ability to complement the temperature-sensitive sgt1-5 and sgt1-3 growth defects. Also, the transformants selected were serially diluted 5-fold for a drop assay and incubated at 25 and 37°C for 4 d to check for complementation.
Microscopy
Confocal images were taken on a TCS-SP5 confocal microscope (Leica) as described previously (Doehlemann et al., 2009). Details of the AF488 WGA and PI microscopy are given in Supplemental Method 1. Fluorescence of AF488 alone or coupled to EdU was elicited at 488 nm and detected at 495 to 540 nm. PI was excited at 561 nm and detected at 570 to 640 nm. YFP was excited at 514 nm and detected at 520 to 540 nm, mCherry fluorescence was excited at 561 nm and detected at 590 to 630 nm, and cell wall autofluorescence used excitation of 405 nm and detection at 435 to 480 nm.
Preparation of samples for TEM and immunogold labeling was performed as described previously (Heyneke et al., 2013). Small samples (∼1.5 mm2) from at least 12 different leaves were cut on a modeling wax plate in a drop of 2.5% (w/v) paraformaldehyde and 0.5% (v/v) glutaraldehyde in 0.06 M Sørensen phosphate buffer, pH 7.2. Samples were then fixed for 90 min in the same fixing solution. Samples were rinsed in buffer four times for 15 min and dehydrated in increasing concentrations of acetone (50, 70, and 90% [v/v]) for 20 min at each step. Subsequently, specimens were gradually infiltrated with increasing concentrations of LR-White resin (30, 60, and 100% [w/v]; London Resin) mixed with 90% (v/v) acetone for a minimum of 3 h per step. Samples were finally embedded in pure, fresh LR-White resin and polymerized at 50°C for 48 h in small plastic containers under anaerobic conditions. Ultrathin sections (80 nm) were cut with a Reichert Ultracut S ultramicrotome (Leica Microsystems).
Immunogold labeling of See1-3xHA was done with ultrathin sections on coated nickel grids with the automated immunogold labeling system Leica EM IGL (Leica Microsystems). The ideal dilutions and incubation times of the primary monoclonal anti-HA antibody (produced in mouse by Sigma-Aldrich) and secondary antibodies (goat anti-mouse antibodies from British BioCell International) were determined in preliminary studies by evaluating the labeling density after a series of labeling experiments. The final dilutions of primary and secondary antibodies used in this study showed a minimum background labeling outside the sample with a maximum specific labeling in the sample. The sections were blocked for 20 min with 2% (w/v) BSA (Sigma-Aldrich) in PBS, pH 7.2, and then treated with the primary antibody against See1 3xHA diluted 1:2000 in PBS containing 1% (w/v) BSA. After section washing with PBS containing 1% (w/v) BSA three times for 5 min, each grid was treated with 10-nm gold-conjugated secondary antibodies (goat anti-mouse IgG) diluted 1:100 in PBS containing 1% (w/v) BSA for 90 min at room temperature. After a short wash in PBS (three times for 5 min) and distilled water (two times for 5 min), labeled grids were poststained with 2% uranyl-acetate aqueous solution for 15 s and then investigated with a Philips CM10 TEM apparatus. Micrographs of randomly photographed immunogold-labeled sections were digitized, and gold particles were counted automatically using the software package Cell D with the particle analysis tool (Olympus, Life and Material Science Europa) in different visually identified and manually traced cell structures. The obtained data were statistically evaluated using Statistica (Stat-Soft Europe).
Coimmunoprecipitation of See1 and SGT1 in N. benthamiana
pSPYNE-P35S-See1-mCherry-N_YFP-Myc and pSPYCE-SGT1-CFP-C_YFP-HA were heterologously expressed in N. benthamiana. As expression controls, the constructs were separately expressed with the appropriate empty vector. For all experiments, Agrobacterium tumefaciens GV3101 was transformed as described previously (Flowers and Vaillancourt, 2005). The transformants were infiltrated into N. benthamiana leaves (3 to 4 weeks old) according to Sparkes et al. (2006). Four days after infiltration, the leaves were harvested and ground in liquid nitrogen. The ground powder was mixed with buffer (50 mM Tris-HCl and 150 mM NaCl, pH 7.0). The resulting leaf extract was centrifuged at 3500 rpm at 4°C and subsequently sterile-filtered. The protein concentration of the extract was determined with the Roti-Quant Protein quantification assay (Carl Roth). To 1 mL of leaf extract containing 2 mg/mL protein, 50 μL of anti-HA Affinity Matrix (Roche Diagnostics) was added, and samples were incubated overnight at 4°C on a rotation wheel. The samples were then centrifuged through Pierce SpinColumns (Thermo Scientific) and washed once with buffer (50 mM Tris-HCl and 150 mM NaCl, pH 7.0), and protein was finally eluted by boiling samples in 2× SDS loading buffer for 5 min. Appropriate amounts of the eluted proteins were separated by SDS-PAGE, followed by transfer to a nitrocellulose membrane. After electroblotting, the membrane was saturated with 5% (w/v) nonfat dry milk in TBS-T (50 mM Tris-HCl, 150 mM NaCl, pH 7.6, and 0.1% [v/v] Tween 20) for 1 h at room temperature. After blocking, the membrane was washed three times with TBS-T followed by incubation with the primary antibody (anti-HA antibody, 1:10,000; anti-c-Myc antibody, 1:5000; Sigma-Aldrich) overnight at 4°C. Membranes were washed three times prior to incubation for 1 h with horseradish peroxidase-conjugated secondary antibody (anti-mouse antibody, 1:5000; Cell Signaling). Signals were detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific).
In Planta Phosphorylation Assay
The constructs encoding maize SGT1 in the presence or absence of See1 and also in addition to the earlier mentioned controls (described in Results) were infiltrated into 4-week-old N. benthamiana leaves as described earlier. Expression of the Nb-MEK2 variants was induced with 30 μM DEX 40 to 48 h later (Yang et al., 2001). Treated leaves were collected ∼5 h after DEX infiltration. Ground leaf material was thawed in 10 mL of Ex-strep buffer (100 mM Tris-HCl, pH 8.0, 5 mM EGTA, 5 mM EDTA, 150 mM NaCl, 10 mM DTT, 0.5 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, 5 μg/mL antipain, 5 μg/mL leupeptin, 50 mM NaF, 1% [v/v] Phosphatase Inhibitor Cocktail 1 [Sigma-Aldrich], 0.5% [v/v] Triton X-100, and 100 μg/mL avidin) as described previously (Witte et al., 2004). The slurry was centrifuged for 10 min at 4°C (15,000g), the supernatant was filtered through Miracloth, and 0.5 mL of StrepTactin Sepharose (IBA) was added. Binding was performed by incubation of this suspension on a rotator for 1 h at 4°C. The slurry was transferred into a Poly-Prep column (Bio-Rad), and the flow-through was discarded. The resin was washed twice with 10 mL of W-buffer (100 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 1 mM EDTA). Four times 250 μL of E-buffer (100 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, and 2.5 mM desthiobiotin) was added, and eluates were collected. The samples were concentrated on a Microcon YM-10 (Millipore) for 30 min at 4°C (13,000g) to a volume of 20 μL and resolved by SDS-PAGE.
Recombinant Protein Preparation
The full-length cDNA of maize SGT1 was cloned into the pET-15b vector. The full-length cDNA of SIPK was cloned into the pGEX-6P-1 vector. The full-length cDNA of tobacco (Nicotiana tabacum) MEK2DD was cloned into the pGEX-6P-2 vector. Escherichia coli (BL21) cells were induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside for Zm-SGT1 at 28°C for 4 h and with 0.25 mM isopropyl β-d-1-thiogalactopyranoside for SIPK and MEK2DD at 18°C for 4 h. His-tagged recombinant protein was purified using Ni-NTA resin (Qiagen). Glutathione S-transferase-tagged recombinant proteins were purified using the Glutathione Agarose resin (Sigma-Aldrich).
Targeted Site-Directed Mutagenesis of Maize SGT1
The targeted exchange of one or more bases in the plasmids was performed by PCR using the QuikChange Multi Site-Directed Mutagenesis kit (Stratagene). The protocol used for this mutation and base substitution was according to the manufacturer’s instructions. Up to three oligonucleotides were designed for the amplification of the entire plasmid, with one oligonucleotide containing the corresponding mutation(s) in the primer sequence.
In Vitro Phosphorylation Assays
The purified recombinant 6xHis-Zm-SGT1 or 6xHis-Zm-SGT1AP (5 μg each) was incubated at 30°C for 30 min with SIPK and MEK2DD (2 μg each) in the reaction buffer (20 mM Tris-HCl, pH 7.5, 40 mM MgCl2, and 10 mM EGTA) or with crude plant protein extracts from the infected maize samples of seedling leaves and tassel base with U. maydis wild-type SG200, SG200Δsee1, or the overexpression strain with See1 (approximately 20 μg each) in the reaction buffer (40 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 1 mM DTT, and 0.1% [v/v] Triton X-100). Both buffers contained 50 μM ATP and 1.5 μCi of [γ-32P]ATP. The reaction was terminated by adding 3× Laemmli sample buffer.
Mass Spectrometry Analysis
Gel bands containing the proteins of interest were subjected to a standard proteomic procedure as described in Supplemental Method 2. Briefly, reduced and alkylated proteins were subjected to trypsin digestion. The resulting peptides were eluted from the gel, and phosphopeptide enrichment was performed on SwellGel Gallium-Chelated Discs (Phosphopeptide Isolation Kit; Thermo Scientific). Liquid chromatography-mass spectrometry analyses of the peptide mixtures were performed on the Orbitrap Velos spectrometer (Thermo Scientific). The Mascot program was used for database searches, and tandem mass spectrometry spectra of phosphorylated peptides were also curated manually.
Microarray Analysis
For the microarray experiments, maize plants (cv Early Golden Bantam) were grown in a phytochamber with the same climatic conditions as described previously. Plants were inoculated in three independent biological replicates with water (mock), SG200, and SG200Δsee1 as described previously in virulence assays for seedling infections (Kämper et al., 2006; Doehlemann et al., 2008). Infected or mock-inoculated tissue from 15 plants per treatment was harvested at 6 DPI by excising a section of the third leaf between 1 and 3 cm below the injection holes. For RNA extraction, material from 15 plants per treatment was pooled and ground to powder in liquid nitrogen, and RNA was extracted with Trizol reagent (Invitrogen). RNA was purified using the RNeasy kit (Qiagen). Agilent 4x44k maize genome arrays were used in three biological replicates, using the standard Agilent One Color Microarray-based gene expression analysis low-input quick Amp labeling protocol. Expression data were submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession number GSE63077). For further experimental details, see Supplemental Method 3.
Plasmid Constructs and Nucleic Acid Construction
For plasmid construction, standard molecular cloning strategies and techniques were applied (Sambrook et al., 1989). All plasmids generated and used in this study are listed in Supplemental Table 3. Oligonucleotides that were used for PCR are shown in Supplemental Table 2. All restriction enzymes used in this study were purchased from New England Biolabs. For a detailed description of plasmid constructs, see Supplemental Method 4.
The isolation of genomic U. maydis DNA was performed as described previously (Schulz et al., 1990). All U. maydis strains (Supplemental Table 4) are derived from SG200 and were generated by insertion of p123 derivatives into the ip locus as described (Loubradou et al., 2001). Isolated U. maydis transformants were tested by DNA gel blot hybridization to assess single or multiple integration events in the ip locus. For all the cloning work, PCR was performed using Phusion High-Fidelity (New England Biolabs). The PCR products of the different genes were cleaned up before digestion using the Wizard SV Gel and PCR Clean-Up System (Promega). The vectors were transformed into either DH5α or Top10 cells (Invitrogen) and then plated on YT-agar plates containing a specific selection marker. Plasmids were extracted using the QIAprep system (Qiagen). All plasmid constructs containing amplified gene fragments were sequenced.
Bioinformatics Tools Applied in This Study
Signal peptide prediction was performed with the online program SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/). Protein conserved domain search was performed with the online program Pfam (http://pfam.sanger.ac.uk/search/). The mass spectrometry data were analyzed using Mascot Distiller software (version 2.1.1; Matrix Science) and compared with the NCBInr database using the Mascot database search engine (version 2.1; Matrix Science), as described in detail in Supplemental Methods.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: U. maydis see1, XP_758386.1; U. maydis pit2, XP_757522.1; U. maydis ppi, XM_754780.1; maize SGT1.1 of cv Early Golden Bantam, KP789376; maize SGT1.2, ACF84496.1; N. benthamiana SGT1 AY, AY899199.1; N. benthamiana SGT1 AF, AF494083.1; tobacco SIPK, AF165186; tobacco MEK2, AF325168; maize MAPK kinase 2, NP_001104843.1; maize putative MAPK family protein, AFW85791.1; maize putative MAPK MPK6, ACG37232.1; maize ABA stimulation MAPK, NP_001152745.1; maize unknown kinase, ACF85409.1.
Supplemental Data
Supplemental Figure 1. Scoring Scheme for Leaf Tumors Based on Symptoms Observed at 12 DPI.
Supplemental Figure 2. SG200∆see1 Mutant Completes Its Entire Life Cycle.
Supplemental Figure 3. Formation of Ear Tumors in SG200∆see1 Mutant Demonstrates the Independence of Floral Tumors on See1 Activity.
Supplemental Figure 4. Growth of SG200∆see1 Mutant Is Arrested in the Mesophyll and Vascular Cell Layers of Leaves.
Supplemental Figure 5. Gene Expression during Maize Colonization with SG200 and SG200∆see1.
Supplemental Figure 6. Specificity of the EdU Labeling Assay.
Supplemental Figure 7. See1 Requirement in Leaf Tumor Formation.
Supplemental Figure 8. Constitutive Overexpression of See1 Results in Tumors on the Vegetative Parts of Tassels.
Supplemental Figure 9. Stable Expression of the Ppit2 overexpressed See1-HA Protein in Infected Seedlings and Tassel Base.
Supplemental Figure 10. Overexpression of See1 Results in Two Abnormalities in Tassels: Tumors at the Tassel Base and Greenish Tassels.
Supplemental Figure 11. See1 Is Transferred to Cells Neighboring to the Transformed Cell.
Supplemental Figure 12. Stable Expression of the See1-3xHA Protein in Infected Tissues.
Supplemental Figure 13. See1 Is Translocated to the Plant Cell Cytoplasm and Nucleus.
Supplemental Figure 14. Complementation of Two Yeast sgt1 Cell Cycle Temperature Sensitive Mutants with Maize SGT1.
Supplemental Figure 15. The Deduced SGT1 Protein Sequences of Z. mays and N. benthamiana Showing a Multiple Alignment.
Supplemental Figure 16. The Deduced SIPK Protein Sequences of N. tabacum and a Set of Five Putative Z. mays SIPK Showing a Multiple Alignment.
Supplemental Figure 17. Expression Level of the Nt-MEK2, Nb-Zm-SGT1, and Um-See1 Transiently Expressed in N. benthamiana Leaves to Assess Zm-SGT1 Phosphorylation in Planta.
Supplemental Figure 18. The Deduced SGT1 Protein Sequences with Marked Phosphorylation Sites within Zm-SGT1.
Supplemental Figure 19. Salicylic Acid Induced Protein Kinase Phosphorylates Zm-SGT1 in Vitro.
Supplemental Figure 20. Subcellular Localization of the Phosphovariants of Maize SGT1.
Supplemental Figure 21. Complementation of Two Yeast sgt1 Cell Cycle Temperature-Sensitive Mutants with Maize SGT1 Wild Type, Phosphomimic, and Phosphonull.
Supplemental Figure 22. In Planta Phosphorylation of Maize SGT1AP.
Supplemental Table 1. Differentially Expressed Top GO Terms Related to the DNA Synthesis and Cell Differentiation.
Supplemental Table 2. Oligonucleotides Used in This Study.
Supplemental Table 3. Plasmids Used in This Study.
Supplemental Table 4. U. maydis Strains Used in This Study.
Supplemental Table 5. Peptides Identified by the Mass Spectrometry.
Supplemental Data Set 1. Comparison of Differentially Expressed Genes at 6 dpi In Maize Leaves as Comparison of Mock Infected versus the Wild Type-Infected SG200 and Mock-Infected versus SG200∆see1 Mutant Infected at 6 dpi.
Supplemental Data Set 2. Comparison of Differentially Expressed Genes at 6 dpi In Maize Leaves Infected with SG200 and SG200∆see1 Mutant.
Supplemental Data Set 3. Full Mascot Search Results of Zm-SGT1 Mass Spectrometry Analysis.
Supplementary Material
Acknowledgments
We thank Urs Lahrmann for support with the microarray experiment, Christoph Hemetsberger and Malgorzata Lichocka for help with confocal microscopy, and Daniela Assmann for expert technical support. We thank Nam Hai Chua from The Rockefeller University for the DEX-inducible plasmid pTA7001 and Katsumi Kitagawa for the sgt1 yeast strains. We thank Michal Dadlez (Institute of Biochemistry and Biophysics) for mass spectrometry analyses. This work was supported by the Max Planck Society (www.mpg.de), the German Research Foundation (www.dfg.de; Grant DO 1421/3-1), and the Cluster of Excellence on Plant Science. A.R. was supported by a fellowship from the German Academic Exchange Service (www.daad.de) and a Short Term Scientific Mission grant from the COST FA 1208 program (http://www.cost-sustain.org). R.H. was supported by the National Science Centre, Poland (Grant 2013/11/B/NZ9/01970).
AUTHOR CONTRIBUTIONS
A.R., R.H., M.K., and G.D. conceived and designed the experiments. A.R., L.S., R.H., and B.Z. performed the research. A.R., L.S., R.H., B.Z., M.K., V.W., and G.D. analyzed data. A.R. and G.D. wrote the article. M.K. and V.W. edited the article.
Glossary
- DPI
days postinfection
- GO
Gene Ontology
- EdU
5-ethynyl-2-deoxyuridine
- PI
propidium iodide
- RT-qPCR
reverse transcription and quantitative PCR
- TEM
transmission electron microscopy
- BiFC
bimolecular fluorescence complementation
- DEX
dexamethasone
References
- Austin M.J., Muskett P., Kahn K., Feys B.J., Jones J.D., Parker J.E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295: 2077–2080. [DOI] [PubMed] [Google Scholar]
- Azevedo C., Sadanandom A., Kitagawa K., Freialdenhoven A., Shirasu K., Schulze-Lefert P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295: 2073–2076. [DOI] [PubMed] [Google Scholar]
- Bao Z., Yang H., Hua J. (2013). Perturbation of cell cycle regulation triggers plant immune response via activation of disease resistance genes. Proc. Natl. Acad. Sci. USA 110: 2407–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar A.P., et al. (2013). The Salmonella type III effector SspH2 specifically exploits the NLR co-chaperone activity of SGT1 to subvert immunity. PLoS Pathog. 9: e1003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brefort T., Doehlemann G., Mendoza-Mendoza A., Reissmann S., Djamei A., Kahmann R. (2009). Ustilago maydis as a pathogen. Annu. Rev. Phytopathol. 47: 423–445. [DOI] [PubMed] [Google Scholar]
- Brefort T., Tanaka S., Neidig N., Doehlemann G., Vincon V., Kahmann R. (2014). Characterization of the largest effector gene cluster of Ustilago maydis. PLoS Pathog. 10: e1003866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. (2000). The regulation of protein function by multisite phosphorylation—A 25 year update. Trends Biochem. Sci. 25: 596–601. [DOI] [PubMed] [Google Scholar]
- Conover W.J. (1999). Practical Nonparametric Statistics, 3rd ed. (New York: John Wiley & Sons; ). [Google Scholar]
- Cui H., Wang Y., Xue L., Chu J., Yan C., Fu J., Chen M., Innes R.W., Zhou J.M. (2010). Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe 7: 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick A., Maguire K., Hammond-Kosack K.E. (2009). Lack of the plant signalling component SGT1b enhances disease resistance to Fusarium culmorum in Arabidopsis buds and flowers. New Phytol. 181: 901–912. [DOI] [PubMed] [Google Scholar]
- Djamei A., Kahmann R. (2012). Ustilago maydis: Dissecting the molecular interface between pathogen and plant. PLoS Pathog. 8: e1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djamei A., et al. (2011). Metabolic priming by a secreted fungal effector. Nature 478: 395–398. [DOI] [PubMed] [Google Scholar]
- Doehlemann G., Reissmann S., Assmann D., Fleckenstein M., Kahmann R. (2011). Two linked genes encoding a secreted effector and a membrane protein are essential for Ustilago maydis-induced tumour formation. Mol. Microbiol. 81: 751–766. [DOI] [PubMed] [Google Scholar]
- Doehlemann G., Requena N., Schaefer P., Brunner F., O’Connell R., Parker J.E. (2014). Reprogramming of plant cells by filamentous plant-colonizing microbes. New Phytol. 204: 803–814. [DOI] [PubMed] [Google Scholar]
- Doehlemann G., van der Linde K., Assmann D., Schwammbach D., Hof A., Mohanty A., Jackson D., Kahmann R. (2009). Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog. 5: e1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G., Wahl R., Horst R.J., Voll L.M., Usadel B., Poree F., Stitt M., Pons-Kühnemann J., Sonnewald U., Kahmann R., Kämper J. (2008). Reprogramming a maize plant: Transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J. 56: 181–195. [DOI] [PubMed] [Google Scholar]
- Dubacq C., Guerois R., Courbeyrette R., Kitagawa K., Mann C. (2002). Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Eukaryot. Cell 1: 568–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Oirdi M., Bouarab K. (2007). Plant signalling components EDS1 and SGT1 enhance disease caused by the necrotrophic pathogen Botrytis cinerea. New Phytol. 175: 131–139. [DOI] [PubMed] [Google Scholar]
- Flowers J.L., Vaillancourt L.J. (2005). Parameters affecting the efficiency of Agrobacterium tumefaciens-mediated transformation of Colletotrichum graminicola. Curr. Genet. 48: 380–388. [DOI] [PubMed] [Google Scholar]
- Gao L., Kelliher T., Nguyen L., Walbot V. (2013). Ustilago maydis reprograms cell proliferation in maize anthers. Plant J. 75: 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ranea J.A., Mirey G., Camonis J., Valencia A. (2002). p23 and HSP20/alpha-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett. 529: 162–167. [DOI] [PubMed] [Google Scholar]
- Gratzner H.G. (1982). Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: A new reagent for detection of DNA replication. Science 218: 474–475. [DOI] [PubMed] [Google Scholar]
- Gray W.M., Muskett P.R., Chuang H.W., Parker J.E. (2003). Arabidopsis SGT1b is required for SCFTIR1-mediated auxin response. Plant Cell 15: 1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann D.R., Rathjen J.P. (2007). Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana. Plant J. 49: 607–618. [DOI] [PubMed] [Google Scholar]
- Hemetsberger C., Herrberger C., Zechmann B., Hillmer M., Doehlemann G. (2012). The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLoS Pathog. 8: e1002684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyneke E., Luschin-Ebengreuth N., Krajcer I., Wolkinger V., Müller M., Zechmann B. (2013). Dynamic compartment specific changes in glutathione and ascorbate levels in Arabidopsis plants exposed to different light intensities. BMC Plant Biol. 13: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenhout S.A., Van der Hoorn R.A.L., Terauchi R., Kamoun S. (2009). Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact. 22: 115–122. [DOI] [PubMed] [Google Scholar]
- Horst R.J., Doehlemann G., Wahl R., Hofmann J., Schmiedl A., Kahmann R., Kämper J., Sonnewald U., Voll L.M. (2010). Ustilago maydis infection strongly alters organic nitrogen allocation in maize and stimulates productivity of systemic source leaves. Plant Physiol. 152: 293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoser R., Zurczak M., Lichocka M., Zuzga S., Dadlez M., Samuel M.A., Ellis B.E., Stuttmann J., Parker J.E., Hennig J., Krzymowska M. (2013). Nucleocytoplasmic partitioning of tobacco N receptor is modulated by SGT1. New Phytol. 200: 158–171. [DOI] [PubMed] [Google Scholar]
- Jelenska J., van Hal J.A., Greenberg J.T. (2010). Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. Proc. Natl. Acad. Sci. USA 107: 13177–13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kämper J., et al. (2006). Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444: 97–101. [DOI] [PubMed] [Google Scholar]
- Kankanala P., Czymmek K., Valent B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19: 706–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T., Walbot V. (2011). Emergence and patterning of the five cell types of the Zea mays anther locule. Dev. Biol. 350: 32–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khang C.H., Berruyer R., Giraldo M.C., Kankanala P., Park S.Y., Czymmek K., Kang S., Valent B. (2010). Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22: 1388–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.H., Kim D.S., Chung E.H., Hwang B.K. (2014). Pepper suppressor of the G2 allele of skp1 interacts with the receptor-like cytoplasmic kinase1 and type III effector AvrBsT and promotes the hypersensitive cell death response in a phosphorylation-dependent manner. Plant Physiol. 165: 76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K., Skowyra D., Elledge S.J., Harper J.W., Hieter P. (1999). SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 4: 21–33. [DOI] [PubMed] [Google Scholar]
- Kotogány E., Dudits D., Horváth G.V., Ayaydin F. (2010). A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods 6: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., et al. (2010). The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42: 1060–1067. [DOI] [PubMed] [Google Scholar]
- Loubradou G., Brachmann A., Feldbrügge M., Kahmann R. (2001). A homologue of the transcriptional repressor Ssn6p antagonizes cAMP signalling in Ustilago maydis. Mol. Microbiol. 40: 719–730. [DOI] [PubMed] [Google Scholar]
- Mueller A.N., Ziemann S., Treitschke S., Aßmann D., Doehlemann G. (2013). Compatibility in the Ustilago maydis-maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. PLoS Pathog. 9: e1003177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller O., Kahmann R., Aguilar G., Trejo-Aguilar B., Wu A., de Vries R.P. (2008). The secretome of the maize pathogen Ustilago maydis. Fungal Genet. Biol. 45 (suppl. 1): S63–S70. [DOI] [PubMed] [Google Scholar]
- Mukhtar M.S., et al. (2011). Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333: 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël L.D., Cagna G., Stuttmann J., Wirthmüller L., Betsuyaku S., Witte C.P., Bhat R., Pochon N., Colby T., Parker J.E. (2007). Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell 19: 4061–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ökmen B., Doehlemann G. (2014). Inside plant: Biotrophic strategies to modulate host immunity and metabolism. Curr. Opin. Plant Biol. 20: 19–25. [DOI] [PubMed] [Google Scholar]
- Peart J.R., et al. (2002). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99: 10865–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A., Mitchison T.J. (2008). A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 105: 2415–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Schilling L., Matei A., Redkar A., Walbot V., Doehlemann G. (2014). Virulence of the maize smut Ustilago maydis is shaped by organ-specific effectors. Mol. Plant Pathol. 15: 780–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirawski J., et al. (2010). Pathogenicity determinants in smut fungi revealed by genome comparison. Science 330: 1546–1548. [DOI] [PubMed] [Google Scholar]
- Schulz B., Banuett F., Dahl M., Schlesinger R., Schäfer W., Martin T., Herskowitz I., Kahmann R. (1990). The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60: 295–306. [DOI] [PubMed] [Google Scholar]
- Shirasu K. (2009). The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 60: 139–164. [DOI] [PubMed] [Google Scholar]
- Skibbe D.S., Doehlemann G., Fernandes J., Walbot V. (2010). Maize tumors caused by Ustilago maydis require organ-specific genes in host and pathogen. Science 328: 89–92. [DOI] [PubMed] [Google Scholar]
- Song J., Durrant W.E., Wang S., Yan S., Tan E.H., Dong X. (2011). DNA repair proteins are directly involved in regulation of gene expression during plant immune response. Cell Host Microbe 9: 115–124. [DOI] [PubMed] [Google Scholar]
- Song J., Win J., Tian M., Schornack S., Kaschani F., Ilyas M., van der Hoorn R.A., Kamoun S. (2009). Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Natl. Acad. Sci. USA 106: 1654–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025. [DOI] [PubMed] [Google Scholar]
- Takahashi A., Casais C., Ichimura K., Shirasu K. (2003). HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 11777–11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talarczyk A., Krzymowska M., Borucki W., Hennig J. (2002). Effect of yeast CTA1 gene expression on response of tobacco plants to tobacco mosaic virus infection. Plant Physiol. 129: 1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Brefort T., Neidig N., Djamei A., Kahnt J., Vermerris W., Koenig S., Feussner K., Feussner I., Kahmann R. (2014). A secreted Ustilago maydis effector promotes virulence by targeting anthocyanin biosynthesis in maize. eLife 3: e01355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linde K., Hemetsberger C., Kastner C., Kaschani F., van der Hoorn R.A., Kumlehn J., Doehlemann G. (2012). A maize cystatin suppresses host immunity by inhibiting apoplastic cysteine proteases. Plant Cell 24: 1285–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V., Skibbe D.S. (2010). Maize host requirements for Ustilago maydis tumor induction. Sex. Plant Reprod. 23: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Uppalapati S.R., Zhu X., Dinesh-Kumar S.P., Mysore K.S. (2010). SGT1 positively regulates the process of plant cell death during both compatible and incompatible plant-pathogen interactions. Mol. Plant Pathol. 11: 597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., Meinkoth J.L., Tsien R.Y., Taylor S.S. (1995). Identification of a signal for rapid export of proteins from the nucleus. Cell 82: 463–473. [DOI] [PubMed] [Google Scholar]
- Witte C.P., Noël L.D., Gielbert J., Parker J.E., Romeis T. (2004). Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol. Biol. 55: 135–147. [DOI] [PubMed] [Google Scholar]
- Yan S., Wang W., Marqués J., Mohan R., Saleh A., Durrant W.E., Song J., Dong X. (2013). Salicylic acid activates DNA damage responses to potentiate plant immunity. Mol. Cell 52: 602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K.Y., Liu Y., Zhang S. (2001). Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. USA 98: 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Kadota Y., Prodromou C., Shirasu K., Pearl L.H. (2010). Structural basis for assembly of Hsp90-Sgt1-CHORD protein complexes: Implications for chaperoning of NLR innate immunity receptors. Mol. Cell 39: 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Liu Y. (2001). Activation of salicylic acid-induced protein kinase, a mitogen-activated protein kinase, induces multiple defense responses in tobacco. Plant Cell 13: 1877–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.