Tissue boundary establishment and cell fate specification require BIRD proteins to regulate SHORT-ROOT signaling and transcriptional activities, highlighting the developmental plasticity in plants.
Abstract
Plant cells cannot rearrange their positions; therefore, sharp tissue boundaries must be accurately programmed. Movement of the cell fate regulator SHORT-ROOT from the stele to the ground tissue has been associated with transferring positional information across tissue boundaries. The zinc finger BIRD protein JACKDAW has been shown to constrain SHORT-ROOT movement to a single layer, and other BIRD family proteins were postulated to counteract JACKDAW’s role in restricting SHORT-ROOT action range. Here, we report that regulation of SHORT-ROOT movement requires additional BIRD proteins whose action is critical for the establishment and maintenance of the boundary between stele and ground tissue. We show that BIRD proteins act in concert and not in opposition. The exploitation of asymmetric redundancies allows the separation of two BIRD functions: constraining SHORT-ROOT spread through nuclear retention and transcriptional regulation of key downstream SHORT-ROOT targets, including SCARECROW and CYCLIND6. Our data indicate that BIRD proteins promote formative divisions and tissue specification in the Arabidopsis thaliana root meristem ground tissue by tethering and regulating transcriptional competence of SHORT-ROOT complexes. As a result, a tissue boundary is not “locked in” after initial patterning like in many animal systems, but possesses considerable developmental plasticity due to continuous reliance on mobile transcription factors.
INTRODUCTION
In multicellular organisms, a precise organization of different tissues and distinct cell types within tissues is critical for proper establishment and maintenance of a functional body. This structural organization relies on the formation of sharp borders between distinct cell populations, as cells with distinct functions must be kept physically separated. Such spatial patterning is achieved in part through intercellular signaling that induces specific tissues or cell types at boundary positions.
Plant cells are encased in rigid cell walls, which prevent their rearrangement during pattern formation. Therefore, cells must coordinate growth and development by interpreting a multitude of signals from their neighbors. Intercellular signaling through mobile transcriptional regulators has been shown to be essential for plant growth and development (Lucas et al., 1995; Nakajima et al., 2001; Kim et al., 2003; Kurata et al., 2005; Gallagher and Benfey, 2009). In the Arabidopsis thaliana root meristem, the cell fate determinant SHORT-ROOT (SHR) is produced in the stele and moves one cell layer outward to instruct ground tissue development (Helariutta et al., 2000; Nakajima et al., 2001; Sena et al., 2004; Gallagher et al., 2004; Gallagher and Benfey, 2009). After movement from the stele, SHR binds to its target SCR (SCARECROW) and promotes the asymmetric cell division (ACD) of the cortex-endodermis initial/daughter (CEI/CEID) as a bipartite SCR-SHR complex to generate the ground tissue (GT) consisting of two layers: cortex and endodermis (Di Laurenzio et al., 1996; Helariutta et al., 2000; Cui et al., 2007). The spatiotemporal distribution of ACDs at the CEI/CEID is regulated by a bistable circuit integrating cues provided by the radial movement of SHR and longitudinal auxin distribution patterns (Cruz-Ramírez et al., 2012). The establishment of the auxin gradient along the longitudinal axis has been extensively studied (Grieneisen et al., 2007; Santuari et al., 2011; Band et al., 2014), and deciphering SHR movement mechanisms is of equal importance to understand tissue boundary formation.
Several molecular factors that contribute to the generation of the SHR protein distribution are emerging. The HEAT domain protein SHR INTERACTING EMBRYONIC LETHAL was suggested to facilitate SHR movement through an endosome- and microtubule-dependent process (Koizumi et al., 2011; Wu and Gallagher, 2013). In addition, callose accumulation at plasmodesmata, symplastic channels that allow passage of hormones, proteins, and RNAs (Du et al., 2007; Schlereth et al., 2010; Matsuzaki et al., 2010), results in plasmodesmata closure and reduces SHR intercellular trafficking (Vatén et al., 2011). Furthermore, nuclear targeting of SHR by fusing a nuclear localization signal or by expressing SCR in the vasculature blocks SHR movement (Gallagher et al., 2004; Koizumi et al., 2012), suggesting that nuclear retention determines the range of SHR movement. Finally, JACKDAW (JKD) was identified as a factor that constrains SHR movement to a single cell layer and regulates the action range of SHR and SCR, while the JKD homolog MAGPIE (MGP) promotes SHR-dependent ACD (Welch et al., 2007). Both proteins bind to and are transcriptionally regulated by the SCR-SHR complex (Levesque et al., 2006; Cui et al., 2007, 2012) but the available data suggested that they had opposite roles in GT patterning (Welch et al., 2007).
Here, we report that JKD and its close homolog BALDIBIS (BIB) constrain SHR movement through nuclear retention in Arabidopsis. We show that JKD and BIB activate SCR expression and that the SHR-SCR complex requires JKD and BIB in transcriptional assays. We also show that JKD and BIB restrict CYCLIND6 (CYCD6) expression to the CEI/CEID. In addition, we demonstrate that two other homologs, MGP and NUTCRACKER (NUC), are required for periclinal cell divisions generating the two GT layers and, together with SCR, are necessary for endodermal fate specification in conjunction with JKD and not in opposition. Our findings illustrate a dual function of these proteins in maintaining sharp tissue boundaries within the root meristem and highlight a mechanism that can provide a high degree of developmental plasticity in patterning.
RESULTS
BIB and JKD Act Redundantly to Restrict SHR Movement and ACD in the Ground Tissue
In jkd mutants, SHR moves outward one layer beyond the endodermis and induces one additional endodermal file originating from the cortex (Welch et al., 2007). This phenotype is subtle and suggests that other factors or pathways may act in parallel to restrict SHR movement. JKD is a member of a plant-specific INDETERMINATE DOMAIN C2H2 zinc finger protein family (Colasanti et al., 1998; Kozaki et al., 2004; Welch et al., 2007); members of this family are referred to as BIRD proteins hereafter. BIRD proteins share high sequence similarity within Arabidopsis (Supplemental Data Set 1 and Supplemental Methods) and between species (Englbrecht et al., 2004; Colasanti et al., 2006), suggesting plausible functional redundancy. To identify potentially redundant partners of JKD, we first examined the expression patterns of its closest homologs BULBUL (BLB) and BIB (Supplemental Figure 1A). Promoter and protein fusions indicated that BLB was not expressed in roots, whereas BIB expression was similar to that of JKD (Figures 1A to 1D), with high levels in the cortex, endodermis, and quiescent center (QC). BIB could also be detected in vascular initials (Figure 1D). Therefore, we reasoned that BIB might act redundantly with JKD.
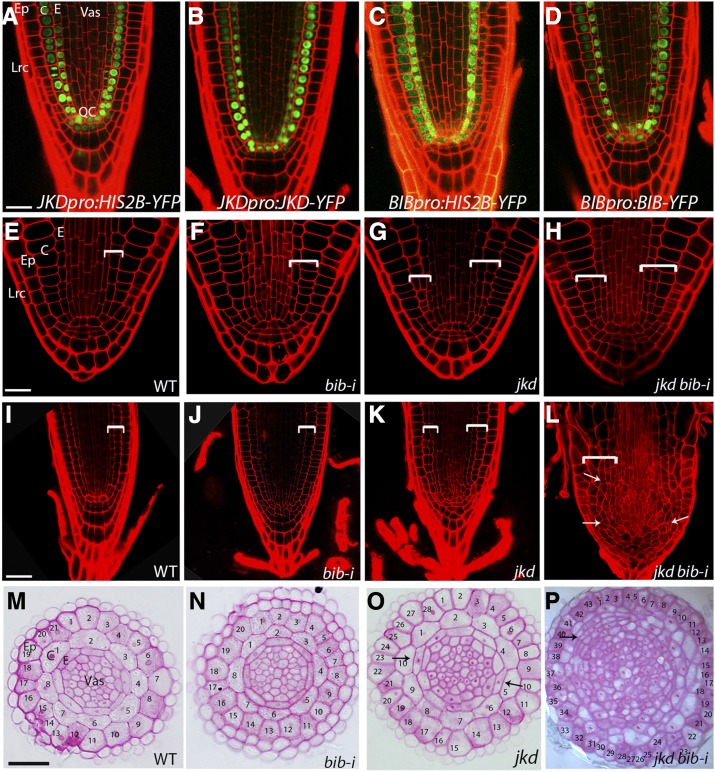
Figure 1.
JKD and BIB Act Redundantly to Restrict Cell Divisions in the Ground Tissue.
(A) to (D) Median longitudinal confocal sections of 4-d-old root expressing JKDpro:H2B-YFP (A), JKDpro:JKD-YFP (B), BIBpro:H2B-YFP (C), and BIBpro:BIB-YFP (D). Bar = 20 μm.
(E) to (H) Median longitudinal confocal sections of mature embryos of the wild type (E), bib-i (F), jkd (G), and jkd bib-i (H). Brackets indicate GT. Bar = 20 μm.
(I) to (L) Median longitudinal confocal sections of 7-d-old roots of the wild type (I), bib-i (J), jkd (K), and jkd bib-i (L). Brackets indicate GT; arrows point to examples of ectopic and disoriented divisions in jkd bib-i. Bar = 50 μm.
(M) to (P) Transverse sections of 5-d-old root meristem of the wild type (M), bib-i (N), jkd (O), and jkd bib-i (P). Note the increased cell number, disorganization of the circumferential pattern, and the expansion of the vasculature in jkd bib-i. Arrows point to examples of ectopic divisions derived from the cortex. Bar = 50 μm.
Ep, epidermis; C, cortex; E, endodermis; Vas, vasculature; Lrc, lateral root cap.
BIB knockouts were not available, so we generated RNA interference (RNAi) (bib-i) and artificial microRNA constructs, specifically targeting BIB and not its close homologs (Supplemental Figure 1). Both methods resulted in similar phenotypes, so we focused our analysis on RNAi-based bib-i lines.
Whereas wild-type and bib-i mature embryos did not reveal abnormal divisions at the root pole, 80% of jkd embryos and 95% of jkd bib-i embryos displayed one additional layer resulting from ectopic divisions in the GT region (Figures 1E to 1H). Cortical cells divided more frequently in jkd bib-i compared with jkd (47 and 22%, respectively, n = 25; measured by counting the number of cells in the cortex showing ectopic divisions). After germination, roots of bib-i displayed aberrant divisions in the QC (Figure 1J); however, its radial cellular organization was similar to the wild type with single layers of endodermis, cortex, and epidermis encircling the vasculature (Figures 1I, 1J, 1M, and 1N; Dolan et al., 1993). In jkd mutant roots, at least one extra GT cell file was evident (Figure 1K) with increased numbers of cells in the circumference within each layer (Figure 1O; Welch et al., 2007). More extensive divisions took place in jkd bib-i, resulting in roots with wider meristems (Figure 1L). Within these roots, additional layers were observed between the central stele and the epidermis (Figure 1P). Besides an increased number of cell layers, jkd bib-i roots possessed an increased cell number per layer and lacked clear morphological tissue distinctions (Figure 1P, Table 1).
Table 1. Cell Numbers in the Circumference in Root Sections of Wild-Type, jkd, and jkd bib-i Homozygotes.
| Genotype | Epidermis | Cortex | Endodermis | |
|---|---|---|---|---|
| Wild type | n = 13 | 22.9 ± 1.9 | 8 ± 0 | 8 ± 0 |
| bib-i | n = 11 | 22.5 ± 1.2 | 8 ± 0 | 8 ± 0 |
| jkd | n = 12 | 27.5 ± 1.6 | 9.2 ± 1.0 | 9.3 ± 1.1 |
| Layer 1 | Layer 2 | Layer 3 | ||
| jkd bib-i | n = 12 | 40.0 ± 2.9 | 26.3 ± 1.8 | 21.3 ± 1.6 |
In jkd bib-i, cells were counted in the outermost layer at the corresponding position of the epidermis (layer 1) and two layers inward (layers 2 and 3). n represents the number of roots counted; numbers are average and sd.
The increased number of layers in jkd bib-i roots reminded us of reported effects of ectopic SHR expression, which include extra layers displaying endodermal identity (Helariutta et al., 2000; Sena et al., 2004). Using the Casparian strip as a morphological marker for endodermal identity, we found that jkd bib-i roots possessed only one cell layer bearing endodermal features, located directly in contact with the epidermis, as recognized by emerging root hairs (Figures 2C and 2D). Occasionally Casparian strip features were present in the epidermis of the double mutant (Figure 2C). These data indicated that the extra divisions occurring in jkd bib-i did not produce supernumerary endodermal layers and thus was different from SHR overexpression phenotype.
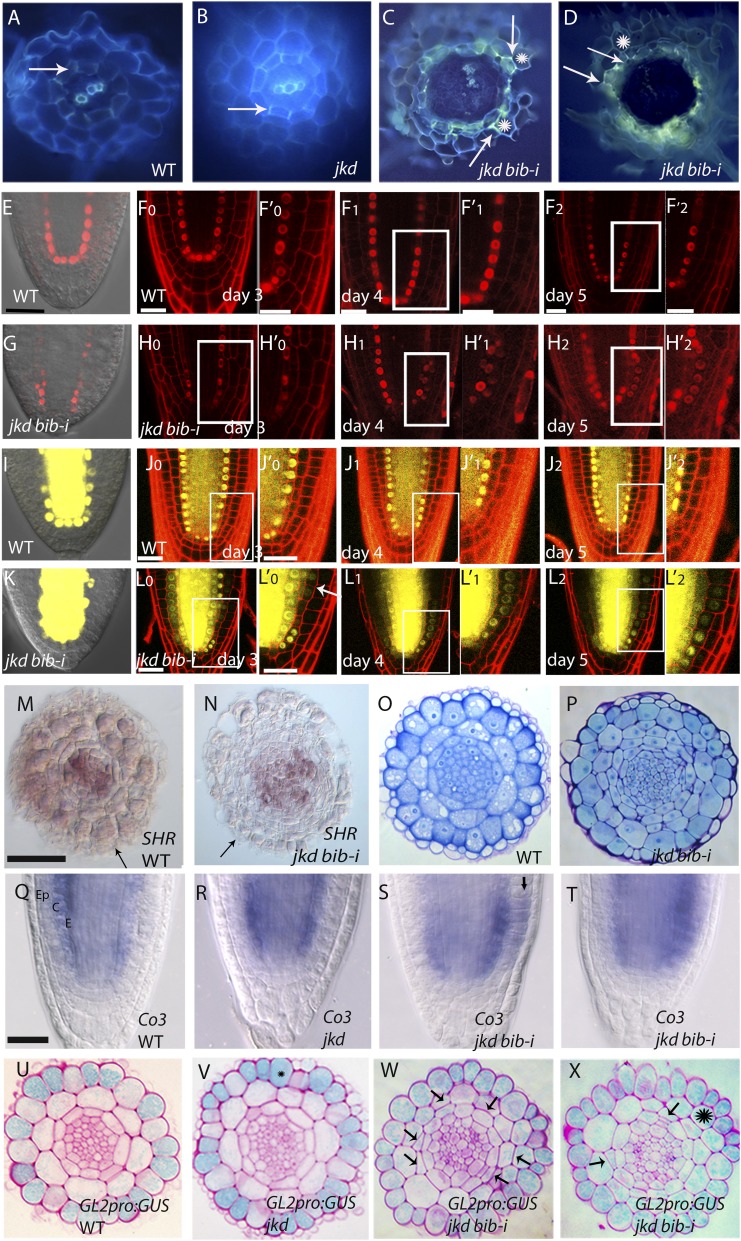
Figure 2.
JKD and BIB Determine Tissue Boundaries in the Arabidopsis Root Meristem.
(A) to (D) Freehand cross sections of 7-d-old roots stained with berberine hemisulfate and aniline blue, showing Casparian strips in the wild type (A), jkd (B), and jkd bib-i ([C] and [D]). Arrows mark Casparian strips, and asterisks mark epidermis bearing root hair ([C] and [D]).
(E) to (F’2) SCR:proSCR-mRFP localization in the wild-type embryo (E) and root at 3, 4, and 5 dag ([F0] to [F2]; [F’0], [F’1], and [F’2] are insets from [F0], [F1], and [F2]).
(G) to (H’2) SCRpro:SCR-mRFP localization in jkd bib-i embryo (G) and root at 3, 4, and 5 dag ([H0] to [H2]; [H’0], [H’1], and [H’2] are insets from [H0], [H1], and [H2]). For both the wild type and jkd bib-i, the same root was observed for three consecutive days. Bars = 20 μm.
(I) to (J’2) SHR:proSHR-YFP localization in the wild-type embryo (I) and root at 3, 4, and 5 dag ([J0] to [J2]; [J’0], [J’1], and [J’2] are insets from [J0], [J1], and [J2]).
(K) to (L’2) SHR:proSHR-YFP localization in jkd bib-i embryo (K) and root at 3, 4, and 5 dag ([L0] to [L2]; [L’0], [L’1], and [L’2] are insets from [L0], [L1], and [L2]). For both the wild type and jkd bib-i, the same root was observed for three consecutive days. Bars = 20 μm.
(M) and (N) Expansion of the vascular tissue in jkd bib-i. In situ hybridization shows SHR mRNA localization in transverse sections of 5-d-old root meristem in the wild type (M) and jkd bib-i (N); note the expanded SHR domain in jkd bib-i compared with the wild type. Arrows point to residual lateral root cap. Bar = 100 μm.
(O) and (P) Cross sections of 5-d-old roots stained with Toluidine blue showing more cells in the vasculature in jkd bib-i (P) compared with the wild type (O). Bar = 50 μm.
(Q) to (T) mRNA localization by whole-mount in situ hybridization of cortex marker Co3 in 3-d-old seedlings in the wild type (Q), jkd (R), and jkd bib-i ([S] and [T]). Arrow points to epidermal expansion of Co3 expression.
(U) to (X) Expression pattern of GL2pro:GUS in transverse root sections of 4-d-old wild type (U), jkd (V), and jkd bib-i ([W] and [X]). Unstained cells develop hairs, while blue-stained cells remain hairless. Arrows point to ectopic divisions. Asterisk indicates cortical cell expressing GL2 gene.
We next monitored SCR expression in jkd bib-i using a SCRpro:SCR-mRFP protein fusion. In the wild type, SCR is strongly expressed in the QC and endodermis (Supplemental Figure 2A). In the bib-i mutant, SCR expression was similar to the wild type but weaker in the QC (Supplemental Figure 2B). In jkd, as previously reported, SCR could be detected in the endodermis and at most in one additional GT layer and was absent from the QC (Supplemental Figure 2C; Welch et al., 2007). In jkd bib-i root meristems, only a subset of layers expressed SCR. Shootwards in the meristem, strong SCR-mRFP localization was observed in two layers (Supplemental Figure 2D). However, in the rootward meristematic region, where extensive divisions created additional layers, SCR expression decreased dramatically in its native stele-adjacent domain and was detected in cells located at, or internal to, the epidermal layer (Supplemental Figure 2D).
In jkd bib-i, only one layer occupying either the epidermal position or adjacent to it displayed endodermal features, while the inner layers did not. We monitored SCR expression dynamics in jkd bib-i to assess whether a shift in the SCR domain correlated with the occurrence of the divisions. In mature embryos of both the wild type and jkd bib-i, SCR expression was restricted to the stele-adjacent layer at the root pole although jkd bib-i roots occasionally revealed expression one layer further out (Figures 2E and 2G). In a time-lapse experiment, SCR showed predominant expression in the endodermis in the wild-type roots from 3 to 5 d after germination (dag; Figures 2F0 to 2F’2). Similarly, we observed strong SCR expression in the endodermis in jkd bib-i roots and weak expression in the adjacent outer layer at 3 dag (Figures 2H0 and 2H’0). However, on day 4, the additional divisions became more apparent in jkd bib-i and are correlated with an increase of SCR levels in the new cells (Figures 2H1 and 2H’1). On day 5, SCR levels were more elevated in the epidermal layer, while a decrease of expression was observed in the original SCR-expressing domain (Figures 2H2 and 2H’2), consistent with SCR expression in 7-d-old jkd bib-i roots (Supplemental Figure 2D). These data indicate that the SCR expression pattern observed in later stages in jkd bib-i roots results from cell fate respecifications, where outer cell layers acquire endodermal fate, while inner layers gradually lose it, thereby maintaining a one-layered, outward shifting “endodermis.”
In jkd mutants, a low level of SHR moves one extra layer outward from the endodermis, correlated with ectopic divisions in the cortex (Supplemental Figure 2G; Welch et al., 2007). We investigated whether SHR movement was enhanced further in jkd bib-i. In bib-i single mutants, SHR localization did not differ from the wild type; the protein was nuclear and cytoplasmic in vascular tissue, while it was retained in the nuclei in the endodermis (Supplemental Figures 2E and 2F). In jkd bib-i, however, SHR protein not only accumulated in the expanded inner vascular tissue, where it retained its cytoplasmic and nuclear localization, but was also detected in all surrounding cell layers, including epidermis (Supplemental Figure 2G).
To assess whether the ectopic divisions occurring in jkd bib-i correlated with enhanced SHR mobility and reduction of nuclear localization, we monitored SHR dynamics in a time-lapse experiment similar to the previous description for SCR. In jkd bib-i mature embryos, we did not observe SHR in the outer layers (Figures 2I and 2K). We then tracked SHR expression in the same root and found that wild-type root exhibited nuclear SHR localization in the endodermis from 3 to 5 dag (Figures 2J0 to 2J’2). In jkd bib-i at 3 dag, nuclear and cytoplasmic SHR could be detected in cells located at the cortex position spanning the entire meristem (Figures 2L0 and 2L’0); in the same lineage, we also detected early divisions, with nuclear and cytoplasmic SHR in the vasculature-facing inner cells, while in the epidermis-facing outer cells, SHR expression was predominantly nuclear (Figure 2L’0, arrow). At 4 and 5 dag, SHR could be detected in cells at the epidermal position and occasionally in lateral root cap cells (Figures 2L1 to 2L’2).
These findings clearly show that in jkd bib-i, inefficient SHR nuclear retention correlates with its spread outside its transcription domains. Consistent with this, we did not observe SHR mRNA in the outer layers (Figures 2M and 2N), confirming that this spreading of SHR protein was a result of enhanced SHR mobility.
Previous data indicated that the extra cortical division in the jkd mutant leads to a fate shift of the original endodermis to pericycle, evidenced by this layer producing lateral roots (Welch et al., 2007). We thus questioned whether the overproliferating inner cells in jkd bib-i resulted in an increase of the vasculature. Toluidine blue and basic fuchsin staining revealed that the central domain in jkd bib-i contained more vascular cell files (Figure 2P; Supplemental Figure 3) when compared with the wild type and jkd (Figures 1O and 2O; Supplemental Figure 3).
In mature embryos, additional divisions in jkd and jkd bib-i originated from the cortex, so we asked whether the newly formed layers in roots carried cortex identity. In the wild type, the Co3 promoter is highly expressed in the cortex, weakly in the endodermis, and excluded from the QC (ten Hove et al., 2010). Consistent with this reported promoter activity, in situ hybridization detected Co3 transcripts in the cortex of the wild type (Figure 2Q; Supplemental Figure 4A). In jkd, expansion of Co3 expression coincided with the additional ground tissue layer (Figure 2R; Supplemental Figure 4B). In jkd bib-i, Co3 expression expanded radially and could also be detected in the epidermis already at early stages (Figures 2S and 2T) and in the QC at later stages when extensive division has taken place (Supplemental Figures 4C and 4D).
Subsequently, we assessed whether the epidermal layer was also affected by the extensive divisions. In Arabidopsis, the epidermal layer comprises two cell types: cells directly in contact with a single cortex cell (N position) will adopt a non-hair fate, while cells located at the cleft between two cortical cells (H position) will generate root hairs. We checked the expression of the epidermal expressed homeobox gene GLABRA2 (GL2), specifically marking non-hair cells (Lee and Schiefelbein, 1999), by monitoring its promoter activity fused to the β-glucuronidase (GUS) reporter (GL2pro:GUS). In the wild type, GL2 was expressed in epidermal cells located at the N position (Lee and Schiefelbein, 1999; Figure 2U), whereas in jkd, ectopic GL2 expression could also be observed in cells located at the H position (Figure 2V) in agreement with a previous report that JKD affects epidermal fate specification (Hassan et al., 2010). In jkd bib-i, GL2 expression was observed in all epidermal cells, suggesting that BIB might also contribute to the JKD function in epidermal fate specification (Figure 2W). Interestingly, GL2 expression also extended inwards to the subepidermal layer (Figure 2X).
Collectively, our data indicate that tissue boundaries between layers in jkd bib-i mutant roots are destabilized, creating reprogrammed fates that correlate with SHR outspread.
BIB and JKD Cooperate with SCR to Retain SHR in the Nucleus
We explored the molecular basis by which BIB and JKD could restrict SHR movement. JKD can directly interact with both SCR and SHR (Welch et al., 2007; Supplemental Figure 5). We assessed whether BIB, similar to JKD, could interact with both SCR and SHR in yeast two-hybrid and bimolecular fluorescence complementation (BiFC) assays and found this to be the case (Supplemental Figure 5). In addition, BIB could directly bind to JKD in BiFC assays (Supplemental Figure 5B). These data are in agreement with previously reported findings that BIRD proteins are capable of binding among themselves and with SCR and SHR (Welch et al., 2007; Supplemental Figure 5).
Based on the observed SHR spread in jkd bib-i, and the potential for the BIRD proteins to bind SHR and SCR, we reasoned that BIRD proteins may restrict SHR movement by a nuclear retention mechanism, where these nuclear proteins bind SHR and prevent its nuclear exit. In support of this idea, we observed SHR-YFP in both the nuclei and cytoplasm in Arabidopsis protoplasts and Nicotiana benthamiana epidermal cells (Supplemental Figures 5B, 6A, and 6A’), but in the presence of SCR, BIB, or JKD, SHR localization became largely nuclear (Supplemental Figures 6B to 6E”), indicating a role of BIRD proteins in SHR nuclear retention. In agreement, in BiFC assays of SHR with BIRD proteins and SCR, the reconstituted YFP fluorescence signal was predominantly nuclear, indicating that SCR-SHR or BIRD-SHR protein complexes were largely nuclear (Supplemental Figure 5B).
To validate the observed effects on SHR nuclear retention in plants, we misexpressed JKD, BIB, and SCR using the promoter of the WOODENLEG (WOLpro) gene, which is normally expressed in the procambial region of the vascular tissue (Bonke et al., 2003; Vatén et al., 2011), where SHR is both nuclear and cytoplasmic (Figures 3A and 3A’). Expression of SCR in the vasculature directed nuclear SHR retention, consistent with a previous report (Koizumi et al., 2012; Figures 3B and 3B’). When BIB and JKD were ectopically expressed together or individually in the vasculature, they were also able to efficiently retain SHR in the nuclei (Figures 3C to 3E’’’, compare with 3A and 3A’). To quantify the effect of BIRD proteins in retaining nuclear SHR, we measured nuclear and cytoplasmic SHR-YFP fluorescence intensity in the vasculature with ectopic SCR or BIRD proteins expression and found that in the roots, the presence of BIRD proteins enhanced SHR nuclear portion to similar levels as SCR (Supplemental Figure 6F).
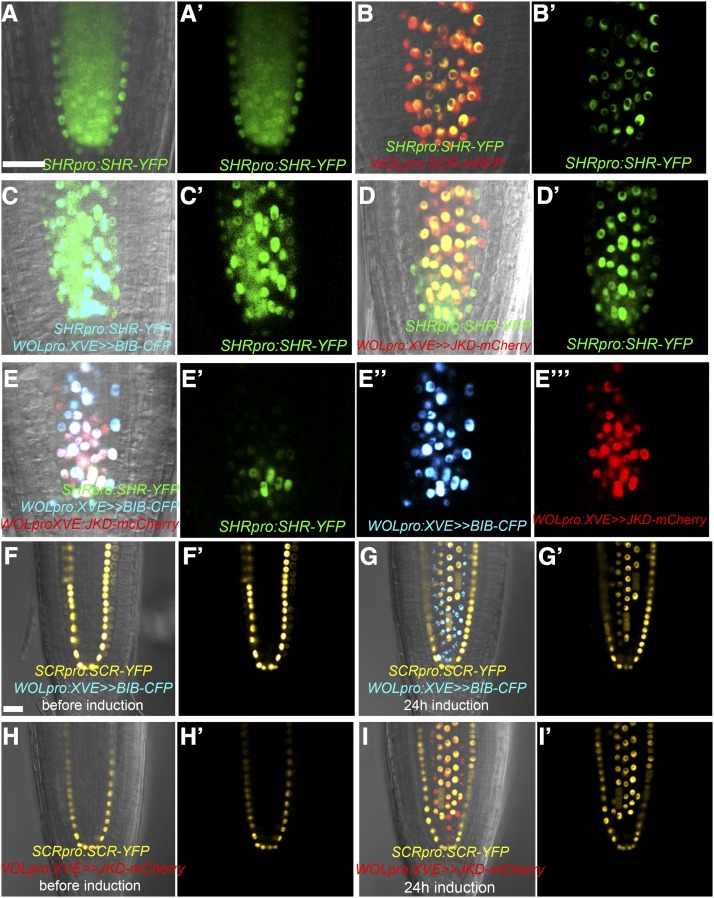
Figure 3.
JKD, BIB, and SCR Promote SHR Nuclear Retention in Planta.
(A) to (E’”) BIRD proteins and SCR are able to retain SHR in the nuclei when expressed in vasculature under WOLpro. Three-day-old roots expressing SHRpro:SHR-YFP alone ([A] and [A’]) or in combination with WOLpro:SCR-mRFP ([B] and [B’]), WOLpro:XVE>>BIB-CFP ([C] and [C’]), WOLpro:XVE>>JKD-mCherry ([D] and [D’]), and both WOLpro:XVE>>JKD-mCherry and WOLpro:XVE>>BIB-CFP ([E] to [E”’]). Bar = 20 μm.
(F) to (I’) Confocal images of 3- to 4-d-old roots containing SCRpro:SCR-YFP and WOLpro:XVE>>BIB-CFP or WOLpro:XVE>>JKD-mCherry before induction ([F], [F’], [H], and [H’]) and 24 h after estradiol induction ([G], [G’], [I], and [I’]). Note ectopic SCR expression in the vasculature. Bar = 20 μm.
As JKD is required to activate SCR expression (Ogasawara et al., 2011), we tested whether SHR nuclear localization mediated by JKD and BIB operated in part through induction of SCR. We first analyzed SCR expression in lines misexpressing JKD and BIB in the vascular domain and found that SCR was ectopically induced in the vascular tissue (Figures 3F to 3I’). This was in line with promoter assays using a dual-luciferase system where both JKD and BIB were able to significantly induce SCR promoter activity in Arabidopsis protoplasts (Supplemental Figure 7). In contrast, SCR and SHR were not sufficient to activate the SCR promoter effectively (Supplemental Figure 7). Additionally, we misexpressed JKD and BIB in the vasculature of scr mutants and found that JKD and BIB were not sufficient to fully retain SHR in the nuclei in the absence of functional SCR (Figure 4). Quantifications of nuclear SHR in scr background confirmed that ectopic JKD and BIB on their own are able to promote SHR nuclear retention to a certain extent and that their action is more efficient in presence of functional SCR (Supplemental Figure 6F).
Figure 4.
JKD and BIB Require SCR to Promote Full SHR Nuclear Retention in Planta.
(A) and (A’) Confocal images of roots expressing SHRpro:SHR-YFP in the wild type.
(B) to (E”’) Confocal images of 3-d-old scr-3 roots expressing SHRpro: SHR-YFP ([B] and [B’]) and coexpressing WOLpro:XVE>>-BIB-CFP ([C] to [C”]), WOLpro:XVE>>JKD-mCherry ([D] to [D”]), and both WOLpro:XVE>>BIB-CFP and WOLpro:XVE>>JKD-mCherry ([E] to [E”’]). Note that in vasculature, JKD and BIB are not able to fully retain SHR to the nuclei in the absence of SCR. Bar = 20 μm.
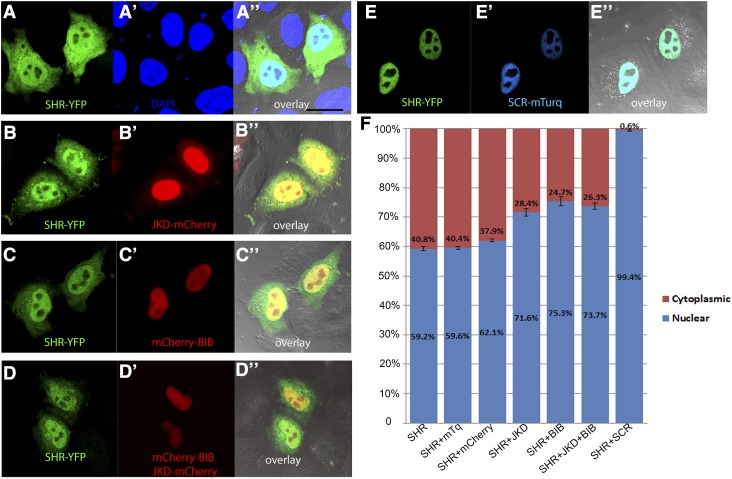
Besides being SHR targets, JKD and SCR also regulate each other’s expression (this study; Welch et al., 2007). To reveal a direct nuclear retention effect of SCR and BIRD proteins on SHR without the background effects of endogenous proteins involved in the pathway, we used HeLa cells as a heterologous system to prevent any plant-specific downstream regulation induced by SHR, SCR, or the tested BIRD proteins. When SHR-YFP alone was expressed in this system, it localized to both nuclei and cytoplasm (Figures 5A to 5A’’ and 5F) similar to its localization in the plant vasculature. When coexpressed with BIB and/or JKD, SHR-YFP nuclear localization was enhanced (Figures 5B to 5D”and 5F), but not to the same level as in plants (Supplemental Figure 6F). Interestingly, when SCR was added, SHR-YFP localization became fully nuclear (Figures 5E” and 5F). We conclude that BIB, JKD, and SCR all individually promote SHR nuclear targeting, with SCR showing the strongest effect.
Figure 5.
JKD, BIB, and SCR Promote SHR Nuclear Retention in HeLa Cells.
(A) to (E”) BIRD proteins and SCR are able to retain SHR in the nuclei of HeLa cells. After transfection, cells were stained with DAPI for nuclear visualization and protein colocalization. Images showing cells transfected with SHR-YFP only ([A] to [A”]), in combination with JKD-mCherry ([B] to [B”]), mCherry-BIB ([C] to [C”]), JKDm-Cherry + mCherry-BIB ([D] to [D”]), and SCR-mTurquoise (SCR-mTurq) ([E] to [E”]). Bar = 20 μm.
(F) Level of SHR nuclear retention in presence of BIRD proteins and SCR is indicated in percentages. Free mTurquoise (mTq) and mCherry were used as controls. Error bars represent se of mean; n > 15.
Together, our data suggest that JKD and BIB restrict SHR movement through SHR nuclear retention mediated by formation of nuclear complexes. In addition, JKD and BIB can promote SHR nuclear retention through activating SCR transcription.
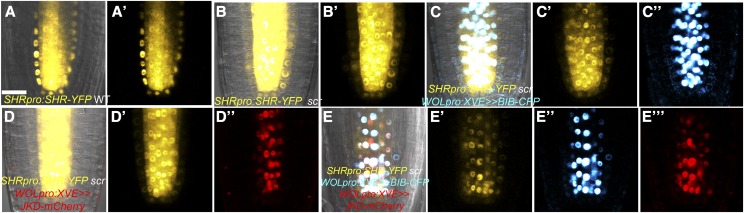
MGP and NUC Promote Formative Divisions and Endodermal Cell Fate in the Ground Tissue
MGP and NUC were previously described as direct SCR and SHR transcriptional targets (Levesque et al., 2006; Cui et al., 2007). Both proteins are expressed in the cortex, endodermis, and a subset of the vasculature and are predominantly excluded from the QC (Figures 6A and 6B). Since the additional GT divisions in jkd mutants were suppressed in jkd mgp-i (Welch et al., 2007), we wondered whether this suppression was enhanced in jkd mgp-i nuc-i. To assess NUC function in the root meristem, we generated RNAi to specifically reduce NUC levels (nuc-i; Supplemental Figure 1). We did not observe significant differences in meristem size in roots of double mgp-i nuc-i RNAi lines and triple jkd mgp-i nuc-i when compared with the wild type (Supplemental Figure 8A). Interestingly, jkd mgp-i nuc-i triple mutant roots contained patches of undivided GT, indicating that cortex and endodermis layers were not fully separated (Figures 6F and 6J). These cells maintained endodermal characteristics as revealed by the presence of Casparian strips (Figure 6N). This phenotype is reminiscent of scr mutants where an undivided layer maintains endodermal and cortical fate (Figures 6G, 6K, and 6O; Di Laurenzio et al., 1996; Heidstra et al., 2004).
Figure 6.
MGP, NUC, and JKD Cooperate with SCR to Specify Endodermal Fate.
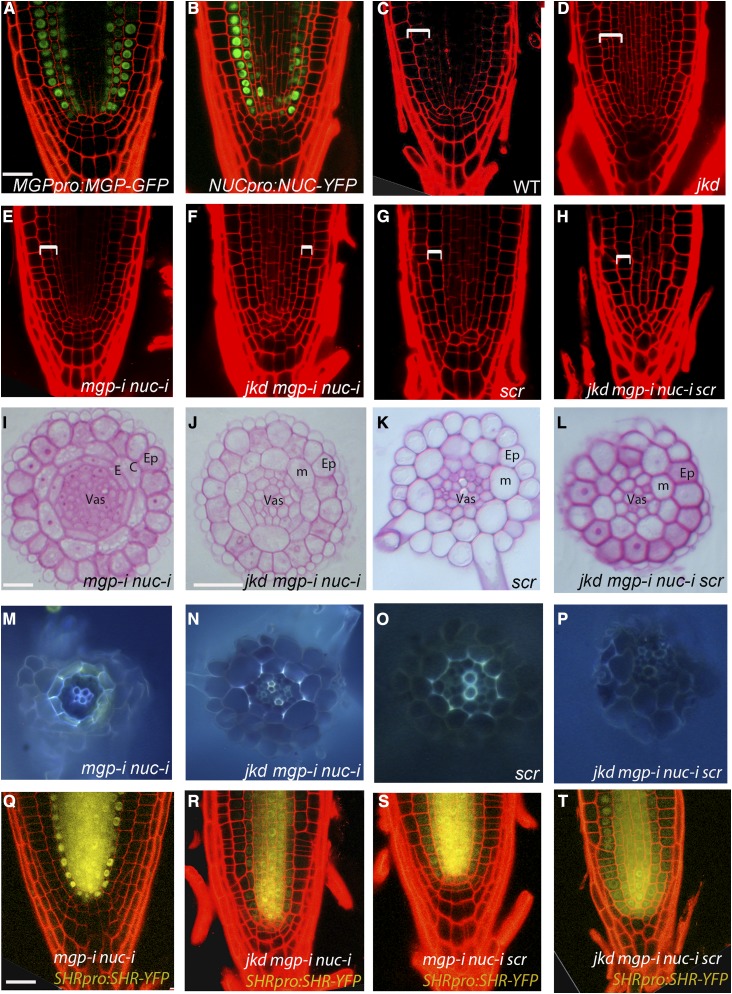
(A) and (B) Median longitudinal confocal images of 4-d-old roots expressing MGPpro:MGP-GFP (A) and NUCpro:NUC-YFP (B) showing expression in cortex, endodermis, and pericycle.
(C) to (H) Median longitudinal confocal sections of 4-d-old roots, with mgp-i nuc-i (E) exhibiting normal GT similar to the wild type (C), jkd roots showing divisions in the cortex layer (D), and jkd mgp-i nuc-i, scr, and jkd mgp-i nuc-i scr displaying one monolayer of ground tissue ([F] to [H]). Brackets indicate GT. Bar = 20 μm.
(I) to (L) Transverse sections of 4-d-old roots showing wild-type ground tissue layers in mgp-i nuc-i (I) and a reduction of ground tissue layers in jkd mgp-i nuc-i (J) and jkd mgp-i nuc-i scr (L) similar to scr (K). Bar = 50 μm.
Ep, epidermis; C, cortex; E, endodermis; Vas, vasculature; m, mutant GT layer.
(M) to (P) Freehand sections of 4-d-old roots stained with berberine hemisulfate and aniline blue to visualize Casparian strips in mgp-i nuc-i (M), jkd mgp-i nuc-i (N), scr (O), and jkd mgp-i nuc-i scr (P). Note absence of Casparian strips in jkd mgp-i nuc-i scr.
(Q) to (T) Confocal images of 4-d-old roots expressing SHRpro:SHR-YFP in mgp-i nuc-i (Q), jkd mgp-i nuc-i (R), mgp-i nuc-i scr (S), and jkd mgp-i nuc-i scr (T). Note SHR spreads to epidermis in jkd mgp-i nuc-i scr. Bar = 20 μm.
In contrast, the undivided GT layer in shr mutants lacks endodermal characteristics (Helariutta et al., 2000). To assess whether endodermal fate specification relies on the SHR targets MGP, NUC, SCR, and JKD, we generated a quadruple mutant line jkd mgp-i nuc-i scr. Similar to shr, these plants displayed shorter root meristems and no Casparian strip (Supplemental Figure 8A; Figures 6H, 6L, and 6P), suggesting a loss of endodermis. We then checked whether the remaining GT layer still retained cortex characteristics by in situ hybridization using the cortex specific gene Co3 (ten Hove et al., 2010) and revealed that the mutant GT layer of jkd mgp-i nuc-i scr still expressed a cortex marker (Supplemental Figures 8B and 8C), suggesting that SHR specification of endodermis requires these four targets.
Given the dual function of BIRD genes as transcriptional activators and SHR movement regulators, the lack of endodermal specification could be a result of SHR target removal or inefficient SHR movement. To test these hypotheses, we monitored SHR protein in the jkd mgp-i nuc-i scr mutant (Figures 6Q to 6T) and found that SHR movement was not hindered but rather enhanced, reaching the epidermis (Figure 6T). This effect was similar to that observed in jkd bib-i. Interestingly, jkd bib-i failed to induce divisions when combined with scr or shr mutants (Figures 7A to 7D), indicating that ectopic divisions in jkd bib-i depend on SCR and SHR activity. To test whether such dependency requires MGP and NUC, we generated jkd bib-i mgp-i nuc-i. The quadruple mutant failed to generate supernumerary layers, and patches of undivided cells were observed in the GT (Figures 7E and 7F).
Figure 7.
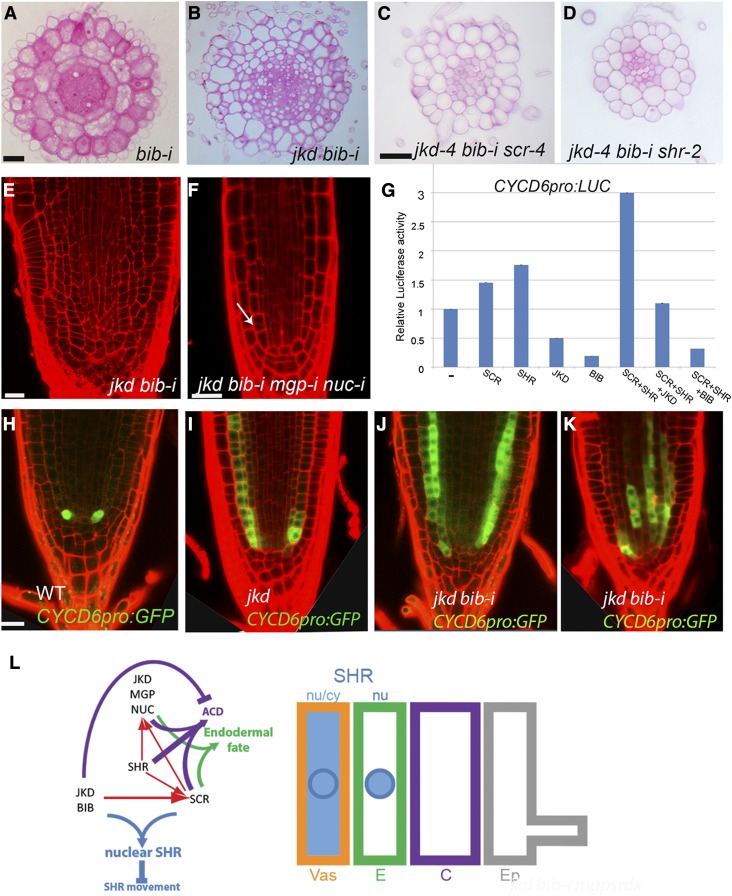
BIRD Proteins Regulate Formative Divisions through Modulation of Transcription.
(A) to (D) Root transverse sections of 4-d-old roots showing normal ground tissue layers in bib-i (A), multiple layers in jkd bib-i (B), and reduction of layers in both jkd bib-i scr (C) and jkd bib-i shr (D). Bars = 20 μm.
(E) and (F) Confocal images of roots showing reduction of ground tissue divisions in jkd bib-i mgp-i nuc-i (F) compared with jkd bib-i (E). Arrow points to GT separation. Bars = 20 μm.
(G) BIB and JKD reduce CYCD6 promoter activity, as measured by dual luciferase assay using protoplasts transiently cotransformed with firefly luciferase under CYCD6 promoter (CYCD6pro) and effectors plasmids carrying SCR, SHR, BIB, or JKD driven by the cauliflower mosaic virus 35S promoter. Values represent average of three replicates. The error bars represent sd. Each experiment was repeated at least three times with three technical replicates.
(H) to (K) Confocal images CYCD6pro:GFP expression in 5-d-old roots of the wild type (H), jkd (I), and jkd bib-i ([J] and [K]). Induced expression of CYCD6pro:GFP correlates with ectopic divisions in jkd and jkd bib-i. Bar = 20 μm.
(L) Model illustrating BIRD action on SHR movement range and root radial pattern specification. In the wild type, JKD, BIB, and SCR promote SHR nuclear retention (thick blue arrow) and restrict SHR movement (thick inhibition sign) in part through SCR activation (red arrow), which in turn together with SHR promotes JKD, NUC, and MGP expression. Together, they contribute to asymmetric cell division (thick purple arrow) and endodermal fate specification (thick green arrow). JKD and BIB restrict ACD and promote normal boundary specification leading to one layer from each tissue.
Taken together, our data suggest that BIRD proteins are collectively required for formative divisions in the GT. Endodermal fate is generated by combined transcriptional activity of SHR targets in the GT, and these transcriptional regulations can be uncoupled from the regulation of SHR movement in jkd bib-i background.
JKD and BIB Confine CYCD6 Expression to CEI/CEID
It has been shown that the SCR-SHR protein complex activates CYCD6 transcription and that ectopic CYCD6 expression is sufficient to induce formative divisions in the GT (Sozzani et al., 2010; Cruz-Ramírez et al., 2012). We thus asked whether divisions in jkd and jkd bib-i were associated with ectopic CYCD6 expression.
In wild-type roots, CYCD6 expression is predominantly constrained to the CEI/CEID (Figure 7H; Sozzani et al., 2010). In both jkd and jkd bib-i roots, CYCD6 expression expanded along the cortex and endodermis and occasionally could be found in the vasculature, correlating with the occurrence of extra divisions (Figures 7I to 7K).
To assess whether JKD and BIB can repress CYCD6 transcription, we tested the effect of JKD and BIB on the CYCD6 promoter in a transient protoplast dual-luciferase system. The SCR-SHR protein complex was able to activate the CYCD6 promoter in this assay. However, JKD and BIB countered SCR-SHR action: Addition of JKD to SCR-SHR reduced CYCD6 promoter activity back to basal levels, while BIB was able to repress it beyond basal levels (Figure 7G). Taken together, the genetic and molecular data suggest that JKD and BIB constrain CYCD6 expression to CEI/CEID by repressing its transcription in GT cells, showing an extra layer of regulation in a highly robust process.
DISCUSSION
Our work highlights the dual functions of BIRD proteins in setting up and maintaining boundaries between tissues in the root meristem, by being critical regulators of SHR movement, formative divisions, and endodermal fate specification. To achieve their function, BIRD proteins integrate two processes, nuclear retention and transcriptional regulation, as means to promote correct patterning. Our data suggest a mechanism by which the range of SHR action is restricted to one cell layer by a combined activity of BIRD proteins and SCR. We also pinpoint SHR targets required for endodermal fate specification. In addition, we show that BIRD proteins are required for CYCD6 restriction to the CEI/CEID. Furthermore, our transcription assays indicate that SCR and SHR are not sufficient to activate key downstream target genes, such as SCR, but require tethering by BIRD proteins, which have been demonstrated to bind DNA (Kozaki et al., 2004). Consistent with this notion, we also show that BIRD proteins on their own have the potential to act both as activators and repressors depending on the target promoters (Figure 7L; Supplemental Figure 9).
We present strong evidence that regulation of SHR movement mediated by BIRD proteins is important for setting up tissue boundaries, as the jkd bib-i mutant displays multiple cell layers in which such boundaries are no longer stable but shift (Supplemental Figure 9). A one-cell boundary shift was already observed in jkd mutants (Welch et al., 2007), but our current data highlight extreme boundary instability and underscore the potential of root cell files to continuously assess positional information and redirect cell fates. The boundary destabilization phenotype does not resemble the effect caused by ectopically expressing SHR under the SCR promoter, where all the multiple cell files exhibited endodermal fate. One major difference between the two phenotypes is that the first is a direct consequence of an enhanced SHR motility and spontaneous cell fate respecification, while the latter is a direct consequence of an hyperactive positive feedback loop of SHR on the SCR promoter (Nakajima et al., 2001; Sena et al., 2004).
This study shows that BIRD proteins employ nuclear retention as a mechanism to regulate SHR movement. As JKD and BIB form nuclear complexes with both SCR and SHR, it is plausible that BIRD proteins and SCR cooperate in the endodermis to prevent movement of SHR beyond one cell layer, in agreement with previously described data on the role of SCR (Gallagher et al., 2004; Cui et al., 2007; Koizumi et al., 2012).
In a heterologous system, JKD and BIB promoted SHR nuclear retention independently from SCR. However, in plants, this activity also involves SCR. Since both JKD and BIB were able to enhance SCR promoter activity in a protoplast system and induce SCR protein expression in the vasculature when expressed under the WOL promoter, it is likely that the in vivo role of the BIRD proteins in constraining SHR activity requires both nuclear retention and transcriptional activation of SCR (Supplemental Figure 9) to reinforce SCR action in this process. Finally, we show that SHR targets (SCR and BIRD genes studied here) cooperate to regulate GT divisions and endodermal fate specification. Reduction of MGP and NUC activities in jkd background not only suppressed the extra GT divisions in jkd but also prevented periclinal divisions generating the two GT layers, indicating that BIRD proteins act similarly to SCR in promoting periclinal divisions in the GT. However, the resulting GT monolayer retained endodermal fate, which is only removed when scr is additionally mutated even in the presence of SHR. These data show that BIRD proteins and SCR are essential SHR targets for endodermal fate. Interestingly, in jkd mgp-i nuc-i scr, SHR spreads to the epidermis and is both cytoplasmic and nuclear in all cells. Therefore, nuclear retention capacity is bestowed on SHR by a combination of BIRD proteins and SCR.
ACDs in the GT were shown to rely on a bistable circuit involving SCR-SHR interaction with RETINOBLASTOMA-RELATED (RBR) protein, which is in turn regulated by CYCD6 (Cruz-Ramírez et al., 2012). Expression of CYCD6, a direct SCR-SHR target, is usually constrained to the CEI/CEID (Sozzani et al., 2010; Cruz-Ramírez et al., 2012). In both jkd and jkd bib-i, CYCD6 expression expanded and correlated with ectopic divisions. This indicates that BIRD proteins can act not only as transcriptional activators, as shown by their action on SCR expression, but also as repressors. Alternatively, BIRD proteins might either activate a repressor to compete with the activation by SHR and SCR or bind to SCR-SHR complex to prevent it from activating CYCD6. Our data indicate that BIRD proteins can differentially regulate target genes. A recent study supports dual roles of BIRD proteins in differential transcriptional regulation of gibberellin-mediated downstream gene expression (Yoshida et al., 2014). A next step is to determine possible binding of BIRD proteins to these promoters, dissect which motifs in the sequences specify these activities, and test their relevance for GT patterning. Another question arising from our results is how BIRD proteins specifically restrict CYCD6 expression to the CEI/CEID, as BIRD expression domains are not excluded from the CEI/CEID. Taking into account that some BIRD proteins are part of the SCR-SHR complex and are also transcriptionally regulated by RBR (Wildwater et al., 2005; Cruz-Ramírez et al., 2012), further research is needed to address how BIRD proteins are involved in the regulation of the bistable circuit described by Cruz-Ramírez et al. (2012). As BIRD proteins can interact with both SCR and SHR and among each other, it will be interesting to map these interactions at cellular resolution to correlate the occurrence of specific complexes with cell type-specific activities.
Finally, our study indicates that SHR protein activity is regulated by dynamic interactions with BIRD proteins and SCR for the correct establishment of tissue boundaries. The continuous activity of this system may illustrate a major difference between plants and many animals. We show that, in plants, tissue boundaries can undergo major rearrangements under certain circumstances, indicating that plants maintain plasticity at adult stages, whereas in many animal systems, boundaries are locked after an initial patterning and cell movement phase.
METHODS
Plant Growth and Transformation
Growth conditions of Arabidopsis thaliana plants were described by Sabatini et al. (1999). Mutants and transgenic lines used in this study are as follows: jkd-4 (Columbia-0 [Col-0]) and mgp-i as described by Welch et al. (2007); scr-3 as described by Fukaki et al. (1996); scr-4 (Wassilewskija) as described by Fukaki et al. (1998); shr-2 as described by Nakajima et al. (2001).
For bib-i lines, an RNAi clone was obtained from pAGRIKOLA (www.agrikola.org) and transformed into Col-0. For nuc-i lines, a 300-bp fragment of NUC coding sequence was amplified and subcloned into pDONR221 in a Gateway reaction. The resulting entry clone was then recombined into a pHellsgate vector. Plants were transformed by floral dip as described by Clough and Bent (1998), and primary transformants were selected on Basta for bib-i and kanamycin for nuc-i. For every transgenic construct, at least 20 transformants were analyzed and 10 lines were selected to check RNA levels. Lines with more than 50% reduced transcripts were chosen for phenotypic analysis and follow-up experiments.
Double jkd bib-i was generated by crossing bib-i lines into jkd-4 followed by genotyping for jkd-4 as described by Welch et al. (2007). Information on the genotyping protocol for bib-i can be found at www.agrikola.org. mgp-i nuc-i and jkd mgp-i nuc-i were generated by transforming the pHellsgate vector containing the NUC RNAi construct into either mgp-i or jkd mgp-i. jkd bib-i scr, jkd bib-i shr, jkd bib-i mgp-i nuc-i, and jkd mgp-i nuc-i scr were generated by crossing. Homozygous lines were selected by genotyping.
Constructs for Cells and Plant Transformation
For mammalian constructs, JKD, BIB, SCR, and SHR coding sequences (CDSs) were amplified using primers described in Supplemental Table 1 and subcloned by ligation into mammalian vector containing mTurquoise, SYFP2, or mCherry under the constitutive cytomegalovirus promoter (Kremers et al., 2006; Goedhart et al., 2007, 2010). The resulting clones are listed in Supplemental Table 2. For constructs tested as effectors after protoplast transformation, CDSs of SCFP3A, SYFP2, mCherry, and mRFP were recombined in a multiple gateway reaction using the cauliflower mosaic virus 35S promoter combined with SCR, SHR, BIB, or JKD (Supplemental Table 3). Information on primers and transgenic constructs can be found in Supplemental Tables 4 and 5. Clones were generated using the Multigateway system (Invitrogen) and introduced to plants by floral dip (Clough and Bent, 1998).
Quantitative RT-PCR Analysis
Total RNA was extracted from 4-d-old roots (Spectrum Plant Total RNA Kit; Sigma-Aldrich). DNase treatment and cDNA synthesis were performed according to the manufacturer’s description (Fermentas). Quantitative RT-PCR was performed using SYBR Green Mastermix (Applied Biosystems). Results were normalized against ACTIN expression. Quantitative PCR primers are described in Supplemental Table 4.
Each experiment was repeated for three biological replicates.
Transient Transfection Assays
HeLa cell culture and transfection were as described by Jiang et al. (2014), and constructs were transfected using FuGENE 6 (Promega). Cells were fixed with formaldehyde and mounted in VectaShield mounting media with 4′,6-diamidino-2-phenylindole.
Four-week-old Nicotiana benthamiana plants were used for infiltration by Agrobacterium tumefaciens containing different constructs as described by Liu et al. (2010). The infiltrated region of the leaf was then mounted in water and checked for expression.
Promoter Luciferase Activity in Protoplast
CYCD6 and SCR promoters were amplified using primers described in Supplemental Table 4, cloned into pDONR221, and transferred to pUGW35 (Nakagawa et al., 2007) containing a minimal 35S promoter and the firefly luciferase (LUC) reporter.
Protoplasts were prepared from Arabidopsis mesophyll cells as described by Yoo et al. (2007). A suspension of protoplasts (100 μL, ∼105 protoplasts per mL) was cotransfected with 5 μg of CYCD6pro:LUC or SCRpro:LUC and the effector plasmids (Supplemental Table 3). One microgram of 35Spro:Renilla-LUC was used as internal control for transfection efficiency. Protoplasts were incubated overnight. Cells were checked for the expression of the effectors by confocal laser microscopy prior to measurement. LUC activities were measured using the dual-luciferase reporter assay system in a Glomax 96 microplate luminometer (Promega). LUC activity was normalized using Renilla luciferase, and the relative ratio was determined by comparing this to the obtained with the promoter activity without effectors.
Time-Lapse Analysis
SCRpro:SCR-mRFP and SHRpro:SHR-YFP roots were initially visualized 3 dag and followed daily up to 5 dag.
Fluorescence Microscopy
All confocal images were done using a Zeiss LSM 710 confocal laser scanning microscope (Carl Zeiss) with a C-Apochromat 40×/1.20 W Korr water-immersion objective. Cyan fluorescence was detected at 465 to 500 nm with 458-nm excitation and a 458/514 beam splitter, yellow fluorescence was detected at 520 to 560 nm with 514-nm excitation and a 458/514 beam splitter, and red fluorescence was detected at 600 to 660 nm with 543-nm excitation with a 488/543/633 beam splitter. Images were processed using Zeiss ZEN software and Adobe Photoshop CS6.
For mature embryo, seeds were incubated overnight in water before dissection and staining with periodic acid/Schiff as described by Truernit et al. (2008). Samples were mounted in Hoyer solution and analyzed.
For roots, samples were mounted in 10 µM propidium iodide. Casparian strip staining was performed in hand-sectioned roots as described by Scheres et al. (1995). Images were acquired using an Olympus fluorescence microscope. Fuchsin staining was done as described by Mähönen et al. (2000).
For HeLa cells, images were acquired as described above. For nuclear-cytoplasmic ratio quantification, three regions of interest in the nucleus excluding nucleoli and three regions of interest in the cytoplasm of each cell were measured with background subtraction using ImageJ with Multi Measure plug-in (http://www.optinav.com/Multi-Measure.htm).
Light Microscopy
Roots sections were performed as described by Scheres et al. (1995). Sections were then stained either with Rutenium red or Toluidine blue and photographed using a Normaski microscope (Axio Imager; Carl Zeiss).
Histochemical staining of plant material containing the GL2pro:GUS reporter gene was performed as described by Hassan et al. (2010). For in situ hybridization in sections using SHR probe, sectioning and preparation of probes were done as described by Mähönen et al. (2000). For cortex-specific expression, whole-mount in situ hybridizations were performed as described by Hejátko et al. (2006). Primers used to generate cortex probe are described in Supplemental Table 4.
Yeast Two-Hybrid Assay
Yeast two-hybrid interactions were analyzed using the ProQuest Two-Hybrid System (Invitrogen Life Technologies). The CDSs of BIB and NUC were amplified using Phusion High-Fidelity DNA Polymerase (Thermo Scientific) and Gateway-compatible primers (Supplemental Table 4). PCR products were cloned into pDONR221 with a Gateway BP II kit (Invitrogen) and sequences were verified. The resulting clones were used in a Gateway (Invitrogen) LR reactions, in combination with the destination yeast expression vectors pDEST22 (Gal4 AD) and pDEST32 (Gal4 BD), and were then checked by restriction analysis and sequencing. A nonautoactivating form of SHR was generated by eliminating acidic amino acids 58 to 64 as described by Koizumi et al. (2011). The resulting protein was fused to pDEST32 BD vector. Constructs for JKD, MGP, SCR, and SHR CDS were described by Welch et al. (2007). SCR and SHR were used as bait and interactions were selected using dropout medium supplemented by 5 mM 3-amino-1,2,4-triazole as described by Welch et al. (2007).
BiFC Assay
For BiFC analysis, we subcloned BIB and NUC CDS in vectors pARC233 and pARC234 by single Gateway LR reactions to generate N-terminal fusions to the two YFP fragments as described by Welch et al. (2007). JKD, MGP, SCR, and SHR CDSs were described by Welch et al. (2007). Arabidopsis Col-0 mesophyll protoplasts were transfected according to Yoo et al. (2007). YFP fluorescence was visualized using a Zeiss LSM 710 confocal laser scanning microscope, and images were processed with the confocal microscope Zeiss ZEN software and Adobe Photoshop CS6. Results from at least three independent experiments and more than 20 protoplast cells were visualized.
Image Processing
Images were processed using Adobe Photoshop CS6 as follows. For Figure 1, images were rotated to vertical position and background filled with black bucket paint in I and J. Contrast was adjusted in I and L to enhance red color. The color balance tool was used to equalize background in M to P. For Figure 2G, images were rotated to vertical position and the generated empty space was filled in with bucket paint tool. For Figure 6, levels were enhanced in T. For Figure 7, images were rotated to vertical position and background filled with black bucket paint in I and K.
Contrast was enhanced in Supplemental Figure 5 and brightness was enhanced in Figures 2J0 to 2J’2 to improve the quality of the signal in each panel.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: JKD, AT5G03150; BIB, AT3G45260; MGP, AT1G03840; NUC, AT5G44160; SCR, AT3G54220; and SHR, AT4G37650.
Supplemental Data
Supplemental Figure 1. Phylogenetic analysis of BIRD proteins using the conserved zinc finger domains.
Supplemental Figure 2. Outward shift of SCR expression domain in jkd bib-i correlates with SHR spread.
Supplemental Figure 3. Increased cell number of vasculature in jkd bib-i.
Supplemental Figure 4. JKD and BIB constrain cortex marker to one single layer.
Supplemental Figure 5. BIB and JKD proteins interact with SCR, SHR, and among themselves.
Supplemental Figure 6. BIRD proteins contribute to SHR nuclear retention together with SCR.
Supplemental Figure 7. JKD and BIB activate SCR expression more efficiently than SCR-SHR complex.
Supplemental Figure 8. JKD, MGP, NUC, and SCR genes regulate root meristem size but do not influence cortex cell fate.
Supplemental Figure 9. Model illustrating BIRD action on SHR movement range and root radial pattern specification.
Supplemental Table 1. Primers used to generate constructs for transformation in HeLa cells.
Supplemental Table 2. Constructs used for HeLa cell transformation.
Supplemental Table 3. Effectors and promoter constructs used for luciferase activity assays in protoplast.
Supplemental Table 4. Primer sequences used in this study.
Supplemental Table 5. Transgenic lines used in this study.
Supplemental Data Set 1. Sequence alignment of BIRD proteins using C2H2 conserved domain used to produce phylogenetic tree shown in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Philip Benfey, Renze Heidstra, Stephen Grigg, and Sara Díaz-Triviño for discussions and critical reading of the article. This work was supported by an NWO VIDI grant to I.B. and Y.L. Y.L. was further supported by an ERC Advanced Grant SysArc and NWO Spinoza Grant to B.S.
AUTHOR CONTRIBUTIONS
Y.L., I.B., and B.S. conceived the study and designed the experiments. Y.L. and I.B. generated lines and performed the experiments. Y.L., I.B., W.S., B.C., A.C.-R., and A.P.M. generated constructs. B.P.B. and A.A. provided mammalian cell lines for transfection. G.S.P. performed the phylogenetic analysis. Y.L., I.B., and B.S. wrote the article. All authors revised the article.
Glossary
- ACD
asymmetric cell division
- GT
ground tissue
- CEI/CEID
cortex-endodermis initial/daughter
- QC
quiescent center
- RNAi
RNA interference
- dag
days after germination
- BiFC
bimolecular fluorescence complementation
- Col-0
Columbia-0
- CDS
coding sequence
References
- Band L.R., et al. (2014). Systems analysis of auxin transport in the Arabidopsis root apex. Plant Cell 26: 862–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonke M., Thitamadee S., Mähönen A.P., Hauser M.-T., Helariutta Y. (2003). APL regulates vascular tissue identity in Arabidopsis. Nature 426: 181–186. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Colasanti J., Yuan Z., Sundaresan V. (1998). The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603. [DOI] [PubMed] [Google Scholar]
- Colasanti J., Tremblay R., Wong A.Y.M., Coneva V., Kozaki A., Mable B.K. (2006). The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A., et al. (2012). A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150: 1002–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Levesque M.P., Vernoux T., Jung J.W., Paquette A.J., Gallagher K.L., Wang J.Y., Blilou I., Scheres B., Benfey P.N. (2007). An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425. [DOI] [PubMed] [Google Scholar]
- Cui H., Hao Y., Kong D. (2012). SCARECROW has a SHORT-ROOT-independent role in modulating the sugar response. Plant Physiol. 158: 1769–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka-Diller J., Malamy J.E., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433. [DOI] [PubMed] [Google Scholar]
- Dolan L., Janmaat K., Willemsen V., Linstead P., Poethig S., Roberts K., Scheres B. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84. [DOI] [PubMed] [Google Scholar]
- Du T.-G., Schmid M., Jansen R.-P. (2007). Why cells move messages: the biological functions of mRNA localization. Semin. Cell Dev. Biol. 18: 171–177. [DOI] [PubMed] [Google Scholar]
- Englbrecht C.C., Schoof H., Böhm S. (2004). Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H., Fujisawa H., Tasaka M. (1996). SGR1, SGR2, SGR3: novel genetic loci involved in shoot gravitropism in Arabidopsis thaliana. Plant Physiol. 110: 945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H., Wysocka-Diller J., Kato T., Fujisawa H., Benfey P.N., Tasaka M. (1998). Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J. 14: 425–430. [DOI] [PubMed] [Google Scholar]
- Gallagher K.L., Benfey P.N. (2009). Both the conserved GRAS domain and nuclear localization are required for SHORT-ROOT movement. Plant J. 57: 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher K.L., Paquette A.J., Nakajima K., Benfey P.N. (2004). Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol. 14: 1847–1851. [DOI] [PubMed] [Google Scholar]
- Goedhart J., Vermeer J.E.M., Adjobo-Hermans M.J.W., van Weeren L., Gadella T.W.J. Jr. (2007). Sensitive detection of p65 homodimers using red-shifted and fluorescent protein-based FRET couples. PLoS ONE 2: e1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart J., van Weeren L., Hink M.A., Vischer N.O.E., Jalink K., Gadella T.W.J. Jr. (2010). Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nat. Methods 7: 137–139. [DOI] [PubMed] [Google Scholar]
- Grieneisen V.A., Xu J., Marée A.F.M., Hogeweg P., Scheres B. (2007). Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013. [DOI] [PubMed] [Google Scholar]
- Hassan H., Scheres B., Blilou I. (2010). JACKDAW controls epidermal patterning in the Arabidopsis root meristem through a non-cell-autonomous mechanism. Development 137: 1523–1529. [DOI] [PubMed] [Google Scholar]
- Heidstra R., Welch D., Scheres B. (2004). Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 18: 1964–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejátko J., Blilou I., Brewer P.B., Friml J., Scheres B., Benková E. (2006). In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat. Protoc. 1: 1939–1946. [DOI] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M.T., Benfey P.N. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567. [DOI] [PubMed] [Google Scholar]
- Jiang K., Hua S., Mohan R., Grigoriev I., Yau K.W., Liu Q., Katrukha E.A., Altelaar A.F.M., Heck A.J.R., Hoogenraad C.C., Akhmanova A. (2014). Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev. Cell 28: 295–309. [DOI] [PubMed] [Google Scholar]
- Kim J.-Y., Yuan Z., Jackson D. (2003). Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 130: 4351–4362. [DOI] [PubMed] [Google Scholar]
- Koizumi K., Wu S., MacRae-Crerar A., Gallagher K.L. (2011). An essential protein that interacts with endosomes and promotes movement of the SHORT-ROOT transcription factor. Curr. Biol. 21: 1559–1564. [DOI] [PubMed] [Google Scholar]
- Koizumi K., Hayashi T., Gallagher K.L. (2012). SCARECROW reinforces SHORT-ROOT signaling and inhibits periclinal cell divisions in the ground tissue by maintaining SHR at high levels in the endodermis. Plant Signal. Behav. 7: 1573–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki A., Hake S., Colasanti J. (2004). The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res. 32: 1710–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremers G.-J., Goedhart J., van Munster E.B., Gadella T.W.J. Jr. (2006). Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry 45: 6570–6580. [DOI] [PubMed] [Google Scholar]
- Kurata T., et al. (2005). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132: 5387–5398. [DOI] [PubMed] [Google Scholar]
- Lee M.M., Schiefelbein J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483. [DOI] [PubMed] [Google Scholar]
- Levesque M.P., Vernoux T., Busch W., Cui H., Wang J.Y., Blilou I., Hassan H., Nakajima K., Matsumoto N., Lohmann J.U., Scheres B., Benfey P.N. (2006). Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903. [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Bouché-Pillon S., Jackson D.P., Nguyen L., Baker L., Ding B., Hake S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270: 1980–1983. [DOI] [PubMed] [Google Scholar]
- Mähönen A.P., Bonke M., Kauppinen L., Riikonen M., Benfey P.N., Helariutta Y. (2000). A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 14: 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y., Ogawa-Ohnishi M., Mori A., Matsubayashi Y. (2010). Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329: 1065–1067. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P.N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311. [DOI] [PubMed] [Google Scholar]
- Ogasawara H., Kaimi R., Colasanti J., Kozaki A. (2011). Activity of transcription factor JACKDAW is essential for SHR/SCR-dependent activation of SCARECROW and MAGPIE and is modulated by reciprocal interactions with MAGPIE, SCARECROW and SHORT ROOT. Plant Mol. Biol. 77: 489–499. [DOI] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472. [DOI] [PubMed] [Google Scholar]
- Santuari L., Scacchi E., Rodriguez-Villalon A., Salinas P., Dohmann E.M.N., Brunoud G., Vernoux T., Smith R.S., Hardtke C.S. (2011). Positional information by differential endocytosis splits auxin response to drive Arabidopsis root meristem growth. Curr. Biol. 21: 1918–1923. [DOI] [PubMed] [Google Scholar]
- Scheres B., Laurenzio L.D., Willemsen V., Hauser M.T., Janmaat K., Weisbeek P., Benfey P.N. (1995). Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121: 53–62. [Google Scholar]
- Schlereth A., Möller B., Liu W., Kientz M., Flipse J., Rademacher E.H., Schmid M., Jürgens G., Weijers D. (2010). MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916. [DOI] [PubMed] [Google Scholar]
- Sena G., Jung J.W., Benfey P.N. (2004). A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development 131: 2817–2826. [DOI] [PubMed] [Google Scholar]
- Sozzani R., Cui H., Moreno-Risueno M.A., Busch W., Van Norman J.M., Vernoux T., Brady S.M., Dewitte W., Murray J.A.H., Benfey P.N. (2010). Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hove C.A., Willemsen V., de Vries W.J., van Dijken A., Scheres B., Heidstra R. (2010). SCHIZORIZA encodes a nuclear factor regulating asymmetry of stem cell divisions in the Arabidopsis root. Curr. Biol. 20: 452–457. [DOI] [PubMed] [Google Scholar]
- Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., Barthélémy J., Palauqui J.-C. (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatén A., et al. (2011). Callose biosynthesis regulates symplastic trafficking during root development. Dev. Cell 21: 1144–1155. [DOI] [PubMed] [Google Scholar]
- Welch D., Hassan H., Blilou I., Immink R., Heidstra R., Scheres B. (2007). Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 21: 2196–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildwater M., Campilho A., Perez-Perez J.M., Heidstra R., Blilou I., Korthout H., Chatterjee J., Mariconti L., Gruissem W., Scheres B. (2005). The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349. [DOI] [PubMed] [Google Scholar]
- Wu S., Gallagher K.L. (2013). Intact microtubules are required for the intercellular movement of the SHORT-ROOT transcription factor. Plant J. 74: 148–159. [DOI] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoshida H., et al. (2014). DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc. Natl. Acad. Sci. USA 111: 7861–7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.