Three nucleotide sugar transporters were shown to transport UDP-Xyl in vitro, demonstrating the existence of plant nucleotide sugar transporters with specificity for UDP-Xyl. UXT1 provides UDP-Xyl for cell wall biosynthesis.
Abstract
Most glycosylation reactions require activated glycosyl donors in the form of nucleotide sugars to drive processes such as posttranslational modifications and polysaccharide biosynthesis. Most plant cell wall polysaccharides are biosynthesized in the Golgi apparatus from cytosolic-derived nucleotide sugars, which are actively transferred into the Golgi lumen by nucleotide sugar transporters (NSTs). An exception is UDP-xylose, which is biosynthesized in both the cytosol and the Golgi lumen by a family of UDP-xylose synthases. The NST-based transport of UDP-xylose into the Golgi lumen would appear to be redundant. However, employing a recently developed approach, we identified three UDP-xylose transporters in the Arabidopsis thaliana NST family and designated them UDP-XYLOSE TRANSPORTER1 (UXT1) to UXT3. All three transporters localize to the Golgi apparatus, and UXT1 also localizes to the endoplasmic reticulum. Mutants in UXT1 exhibit ∼30% reduction in xylose in stem cell walls. These findings support the importance of the cytosolic UDP-xylose pool and UDP-xylose transporters in cell wall biosynthesis.
INTRODUCTION
Plant cell walls are composed of various polysaccharides and with the exception of cellulose and callose, these cell wall polysaccharides are biosynthesized in the lumen of the Golgi apparatus by families of glycosyltransferases (Scheible and Pauly, 2004; Liepman et al., 2010). The nucleotide sugar substrates essential for the biosynthesis of these polysaccharides are predominantly made in the cytosol. To overcome the subcellular partitioning of substrates and enzymes, nucleotide sugar transporters (NSTs) have evolved to allow the transport of nucleotide sugars from the cytosol into the Golgi and endoplasmic reticulum (ER) lumen. NSTs belong to the NST/triose phosphate translocator (TPT) superfamily, and the fact that they are present in all eukaryotes testifies to their biological significance (Knappe et al., 2003). Phylogenetic analyses have identified more than 50 members in Arabidopsis thaliana that are distributed in six clades (Rautengarten et al., 2014). However, functional characterization of members of the NST family at the molecular level has progressed slowly. In the past decade, only a few NSTs have been characterized, thus far accounting for the transport of GDP-mannose (GDP-Man), UDP-galactose (UDP-Gal), UDP-glucose (UDP-Glc), and CMP-sialic acid, although sialic acid has not been found in plants (Baldwin et al., 2001; Norambuena et al., 2002, 2005; Handford et al., 2004, 2012; Bakker et al., 2005, 2008; Rollwitz et al., 2006; Zhang et al., 2011; Mortimer et al., 2013). Recently, we developed a biochemical approach that allows the rapid and reliable determination of NST activities and led to the identification and characterization of the Arabidopsis bifunctional UDP-rhamnose (UDP-Rha)/UDP-Gal transporter (URGT) clade (Rautengarten et al., 2014).
Xyl is a key component of various plant cell wall polymers, including xylan and xyloglucan, which are two of the most abundant cell wall polysaccharides in plants (Ebringerová and Heinze, 2000; Scheller and Ulvskov, 2010). While glucuronoxylan is a major hemicellulose in secondary cell walls, xyloglucan is the major component of the hemicellullosic fraction of primary walls of dicot plants. Minor amounts of Xyl can also be found in pectic polysaccharides, such as rhamnogalacturonan-II and xylogalacturonan (Jensen et al., 2008; Atmodjo et al., 2013), glycoproteins (Strasser et al., 2000), and diverse metabolites. Xylans in vascular plants are mainly composed of a backbone of β-(1,4)-linked xylopyranosyl residues, which may be decorated at O-2 or O-3 with arabinofuranosyl residues or at O-2 with glucuronosyl and 4-O-methylglucuronosyl residues to form arabinoxylan found in grasses and glucuronoxylan, the main xylan found in dicots (Tan et al., 2013; Rennie and Scheller, 2014). UDP-Xyl, the activated sugar donor for xylosyltransferases, is biosynthesized via decarboxylation of UDP-glucuronic acid by UDP-XYLOSE SYNTHASE (UXS) (Harper and Bar-Peled, 2002). While most nucleotide sugars are made in the cytosol and require transport into the Golgi lumen, in plants, members of the UXS family have been localized to both the Golgi and cytosolic fractions (Harper and Bar-Peled, 2002; Pattathil et al., 2005). Furthermore, UDP-Xyl can also be biosynthesized in the cytosol by a related gene family that also produces UDP-apiose, namely, UDP-API SYNTHASE/UDP-XYL SYNTHASE (Mølhøj et al., 2003). Thus, in plants there seem to be three distinct pathways for UDP-Xyl biosynthesis (Bar-Peled and O’Neill, 2011).
The biosynthesis of UDP-Xyl in the Golgi lumen would argue against the requirement for a UDP-Xyl-specific transporter residing in the Golgi membrane for the biosynthesis of Xyl-containing polysaccharides and glycoproteins. Nevertheless, we obtained evidence that Golgi-localized UDP-Xyl specific transporters exist in Arabidopsis and that at least one Golgi-localized UDP-Xyl transporter is necessary for proper biosynthesis of Xyl-containing cell wall polysaccharides. We designated the three members of this gene family as UDP-XYL TRANSPORTER1 (UXT1) to UXT3.
RESULTS
The NST-KT Clade of the Arabidopsis NST/TPT Family
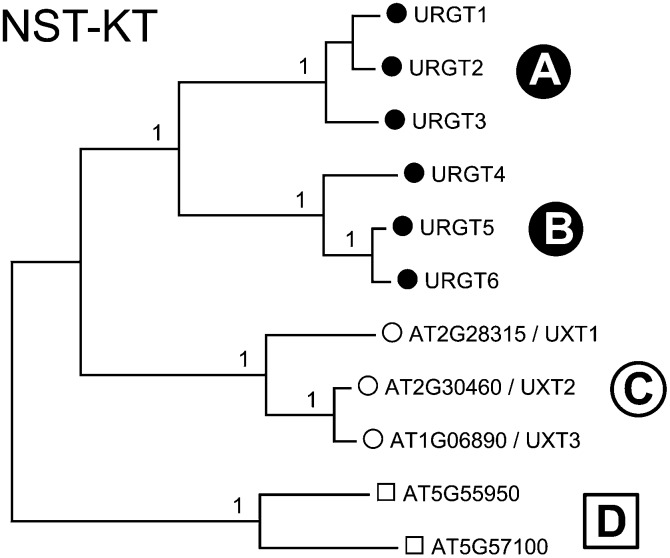
The previously classified NST-KT subfamily is characterized by a highly conserved lysine/threonine (KT) motif (Knappe et al., 2003) and forms clade I of the NST/TPT family (Rautengarten et al., 2014) encompassing 11 putative NSTs (Figure 1). Collectively, they share 25 to 93% identity in their amino acid sequences and include the recently identified URGTs that form subclades (A) and (B) of the NST-KT subfamily (Rautengarten et al., 2014). Another subclade (C) contains three additional members, namely, UXT1 (AT2G28315), UXT2 (AT2G30460), and UXT3 (AT1G06890). UXT1 shares 66% and 67% identity with UXT2 and UXT3, respectively. UXT2 and UXT3 share 92% identity in their amino acid sequences. Members of the uncharacterized subclade (D) share 25 to 28% identity in their amino acid sequences when compared with all other members of the NST-KT family (Rautengarten et al., 2014).
Figure 1.
Phylogenetic Tree of the Arabidopsis NST-KT Subfamily.
The full-length amino acid sequences of the Arabidopsis NST-KT subfamily (Knappe et al., 2003) were aligned with Clustal Omega and the tree generated using MEGA6. Numbers at the nodes indicate bootstrap values calculated for 1000 replicates. Subclades were assigned as previously reported (Rautengarten et al., 2014).
UXTs Are Ubiquitously Expressed and Localized to the Golgi Apparatus
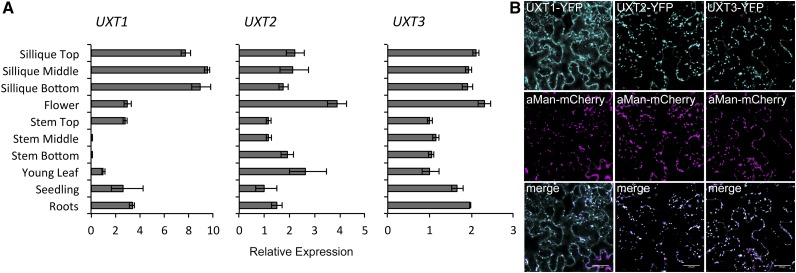
Publicly available microarray expression data comprising the AtGenExpress Developmental Data Set (Schmid et al., 2005) have shown ubiquitous expression for UXT2 and UXT3 throughout plant development, with UXT3 showing highest expression in pollen and flowers. Since UXT1 is not present on the Affymetrix ATH1 array, we assessed the relative expression levels using quantitative RT-PCR (Figure 2A). Expression data obtained by quantitative RT-PCR for UXT2 and UXT3 are consistent with the microarray expression data, confirming ubiquitous but relatively low expression for both genes. UXT1 is more highly expressed in most tissues analyzed, with some variation in expression, especially in the stem tissue. To determine the subcellular localization of the UXTs, we generated C-terminal yellow fluorescent protein (YFP) fusions of the coding sequences and expressed them transiently in Nicotiana benthamiana leaves. All three UXTs localized to Golgi-like punctate structures and colocalized with the Golgi-marker α-mannosidase I, supporting their function as Golgi NSTs (Figure 2B). In contrast to UXT2 and UXT3, UXT1 also appeared to be localized to the ER and colocalized with the ER marker ER-ck (Nelson et al., 2007) (Supplemental Figure 1).
Figure 2.
Expression Pattern and Subcellular Localizations of UXTs.
(A) Quantitative RT-PCR of UXT expression in Arabidopsis organs and developmental stages. The three UXTs are expressed throughout the plant with UXT1 showing the highest expression levels. Expression levels are mean ± sd (n = 3) technical replicates relative to the reference genes.
(B) Subcellular localization of the UXTs. C-terminal YFP fusions were transiently coexpressed with the α-mannosidase I Golgi marker in N. benthamiana leaves. All three UXTs colocalize with the Golgi marker. UXT1 also colocalizes with an ER marker (Supplemental Figure 1). Bar = 25 μm.
Determining the in Vitro Functions of the Arabidopsis UXTs
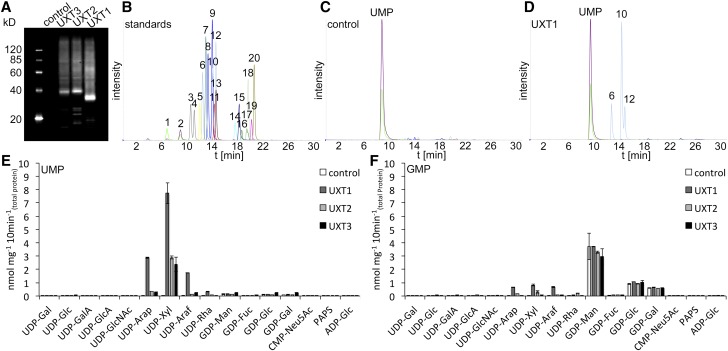
To assess the function of the UXTs, each was heterologously expressed in Saccharomyces cerevisiae (yeast) and microsomal proteins were prepared. Immunoblot analysis confirmed the presence of the specific UXT proteins in yeast microsomal extracts (Figure 3A). Subsequently, microsomal proteins were reconstituted into liposomes for transport assays. Proteoliposomes preloaded with either UMP, GMP, CMP, or AMP were incubated with a mixture of 16 nucleotides/nucleotide sugars (Figure 3B). Nontransported substrates were removed by gel filtration, and the content of the liposomes was analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The LC-MS/MS analysis of nucleotide sugars after transport by UXTs could be readily assessed when compared with the empty vector control (Figures 3C and 3D). All three UXTs had the capacity to transport UDP-Xyl as well as minor amounts of UDP-arabinopyranose (UDP-Arap) in vitro when the proteoliposomes were preloaded with UMP (Figure 3E). By contrast, when proteoliposomes were preloaded with GMP, only transport of GDP-sugars was observed (Figure 3F), resulting from endogenous activity present in yeast microsomal preparations, since the incorporation levels were similar to those observed in control reactions (yeast transformed with the empty vector). No significant transport activities were observed when proteoliposomes were preloaded with AMP or CMP.
Figure 3.
LC-MS/MS Analysis of NST Activities of UXTs.
(A) Immunoblot analysis of UXT expression in yeast microsomal protein extracts (2.5 μg), including the empty vector control.
(B) Separation of a 20 nucleotide/nucleotide sugar mix: 1, CMP; 2, UMP; 3, UDP-GalA; 4, UDP-glucuronic acid; 5, CMP-sialic acid; 6, UDP-Arap; 7, UDP-Rha; 8, UDP-Gal; 9, UDP-Glc; 10, UDP-Xyl; 11, UDP-GlcNAc/GalNAc; 12, UDP-Araf; 13, adenosine 3′-phosphate 5′ phosphosulfate; 14, GMP; 15, AMP; 16, GDP-Man; 17, GDP-Gal; 18, GDP-Glc, 19, GDP-Fuc; and 20, ADP-Glc.
(C) and (D) Reconstitution of empty vector control (C) and UXT1 (D) into liposomes and analysis by LC-MS/MS after simultaneous incubation with 16 nucleotide sugar substrates.
(E) and (F) Quantification of nucleotide sugar uptake of proteoliposomes containing UXT1 and preloaded with UMP (E) and GMP (F). Data represent the mean and sd of n = 2 independent experiments.
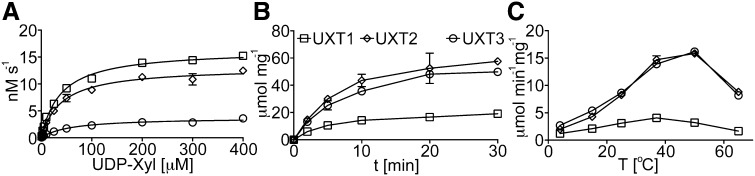
The UXT-mediated transport of UDP-Xyl was saturable in a concentration and time-dependent manner (Figures 4A and 4B). To determine Kcat, we measured the amount of UXT protein in the proteoliposomes using multiple reaction monitoring (MRM) mass spectrometry as explained in Methods and Supplemental Table 1. The analysis of the UXTs revealed apparent Km values for UDP-Xyl in the range of 40 to 60 μM with turnover rates of 3 to 12 s−1 (Table 1). As previously determined, the UDP-Xyl content in various Arabidopsis organs is in the range of 40 to 120 pmol mg−1 dry weight (Rautengarten et al., 2014). Considering the volume of the central vacuole in plants, these measurements indicate that the cellular levels of UDP-Xyl are in the micromolar range. Thus, we estimate that the affinity constants (Km) for all three UXTs are within physiological range. By contrast, estimations of the Km for UDP-Arap, which was transported by all three UXTs to a lower extent, revealed values of ≥200 μM, which would be inconsistent with endogenous concentrations. UDP-Arap concentrations in Arabidopsis organs are very similar to UDP-Xyl concentrations (Rautengarten et al., 2014); hence, the high affinity constants or Km values indicate that the UXTs are most likely not significantly involved in UDP-Arap transport in vivo. The temperature optimums for transport of UDP-Xyl for the three UXTs ranged from 37 to 55°C (Figure 4C).
Figure 4.
Time- and Temperature-Dependent Transport Activities of the Arabidopsis UXTs.
(A) Proteoliposomes, preloaded with UMP, were incubated with UDP-Xyl at varying concentration (0.5 to 400 μM) for 2 min at 25°C.
(B) and (C) Proteoliposomes were incubated with UDP-Xyl at a concentration of 50 μM for the indicated time points at 25°C, or varying temperatures. Values are normalized to the actual NST content present in the proteoliposome preparations. Data represent the mean and sd of n = 2 independent experiments.
Table 1. Kinetic Parameters of UDP-Xyl Transport into Proteoliposomes.
| Parameter | UXT1 | UXT2 | UXT3 |
|---|---|---|---|
| Km (μM) | 39 (3) | 40 (4) | 58 (9) |
| Vmax (nM s−1) | 16 (0) | 13 (0) | 4 (0) |
| Kcat (s−1) | 3.6 | 10.3 | 11.7 |
For each UXT, 20 data points with varying substrate concentrations (0.5 to 400 μM) were acquired. Standard errors are in parentheses.
The Role of the UXTs in Planta
To evaluate the in vivo function of the UXTs, we obtained homozygous T-DNA lines. Two independent lines were acquired for UXT1, and single insertion lines were identified for UXT2 and UXT3. Finally, a double knockout was generated between uxt2 and uxt3, and the absence of respective full-length transcripts was confirmed by PCR (Supplemental Figure 2).
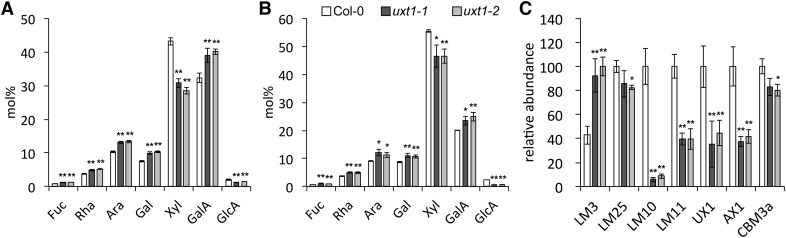
To assess if mutations in the UXTs affect the biosynthesis of specific polysaccharides, we prepared alcohol insoluble residue (AIR) from leaves, flowers, and young (upper) and mature (lower) inflorescence stem tissue from 6- to 8-week-old plants and analyzed the monosaccharide composition (Supplemental Table 2). Flowers and leaves showed no significant differences in the monosaccharide composition between any of the mutants and Columbia-0 (Col-0; P > 0.05, ANOVA and Duncan’s test for multiple comparisons). Stem data showed a significant difference in the monosaccharide composition of uxt1-1 and uxt1-2 compared with Col-0 (P < 0.05), whereas the other mutants did not show a difference from Col-0. In uxt1-1 and uxt1-2, only Xyl and glucuronic acid (GlcA) were significantly decreased in stems compared with Col-0 (Figures 5A and 5B). The Xyl content in mature inflorescence stems from the uxt1 mutants was decreased by 16% (Figure 5B), whereas a reduction of 34% was observed in young stem tissue (Figure 5A). These data confirm the importance of UXT1 for the biosynthesis of Xyl-containing cell wall polymers. In addition, a significant reduction in cell wall GlcA content of ∼25 to 37% was observed in mature and young parts of the inflorescence stems from uxt1 mutants. Since the monosaccharide compositions are relative measurements, the decrease in Xyl and GlcA was accompanied by an apparent increase in other monosaccharides. However, the ratio between the other sugars was not significantly changed in any of the samples, indicating that a loss of function of UXT1 had no direct effect on any sugars besides Xyl and GlcA. Notably, while GlcA content was decreased in the mutant, there was no change in the 4-O-methyl ether (MeGlcA) content, i.e., the ratio between the methylated and nonmethylated form of GlcA was much higher in the mutant than in the wild type (Supplemental Figure 3). However, even though there was a significant reduction of Xyl and GlcA in the uxt1 mutants, the plants did not exhibit a morphological phenotype compared with wild-type plants.
Figure 5.
Cell Wall Analysis of UXT1 Mutants.
(A) and (B) Monosaccharide composition of total cell wall extracts from upper (A) and lower (B) parts of the inflorescence stem. Data represent 10 pooled individuals and mean and sd from six technical replicates. Values are expressed in mol %.
(C) CoMPP analysis with relative abundance of cell wall glycan epitopes. Mean and sd for spot signals (MSS) were obtained by probing microarrays with antibodies (x axis) from four technical replicates and the highest MSS set to 100 and all other values adjusted accordingly. For all data, values significantly different from the wild type are marked with asterisks (*P < 0.05 and **P < 0.01; t test).
Cell Wall Profiling of the uxt1 Mutants
To analyze the changes in the cell wall composition of uxt1 mutants in more detail, we performed comprehensive microarray polymer profiling (CoMPP) analysis on mature (lower) stem material. Cell wall matrix polymers were extracted, spotted onto nitrocellulose membranes to generate microarrays, and probed with a number of different antibodies with specificity for epitopes borne on cell wall polymers (Figure 5C). Since we observed a significant decrease in cell wall xylose in the uxt1 mutants, we focused specifically on antibodies recognizing xylan structures, including LM10/LM11 (which bind unsubstituted xylans), UX1 (which recognizes GlcA or MeGlcA substitutions on xylan), and AX1 [which was produced against arabinose-substituted β-(1,4)-xylan from wheat (Triticum aestivum)]. Clear differences were observed between the uxt1-1 and uxt1-2 mutants and the wild type (Figure 5C). Collectively, these probes revealed an apparent reduction in the xylan content and, more specifically, as indicated in the results for the UX1 antibody, in the glucuronoxylan content in UXT1 mutants when compared with the wild type (Figure 5C). Concomitant with the reduction in xylan content, the results from the CoMPP analysis indicate that other polymers, such as cellulose (recognized by the carbohydrate bonding module CBM3a) and xyloglucan (recognized by antibody LM25), were only slightly affected in UXT1 mutants. By contrast, results obtained with the LM3 antibody, which recognizes glycan moieties on extensins, indicated that these proteoglycans are enriched in UXT1 mutants. In addition, oligosaccharide mass profiling (OLIMP) revealed no significant differences in the xyloglucan structure in uxt1 cell wall preparations compared with the wild type (Table 2).
Table 2. Xyloglucan Structure Determined by Oligosaccharide Mass Profiling.
| Tissue | Plant Line | GXXG (953 m/z) | XXXG (1085 m/z) | XXLG/XLXG (1247 m/z) | XXLG/XLXG +OAc (1289 m/z) | XXFG (1393 m/z) | XXFG +OAc (1435 m/z) | XLFG (1555 m/z) | XLFG +OAc (1597 m/z) |
|---|---|---|---|---|---|---|---|---|---|
| Lower | Col-0 | 2.0 ± 0.2 | 32.5 ± 2.8 | 9.3 ± 0.7 | 1.8 ± 0.2 | 15.0 ± 0.9 | 14.1 ± 0.8 | 13.7 ± 1.6 | 11.5 ± 0.5 |
| Stem | uxt1-1 | 2.1 ± 0.3 | 33.2 ± 1.8 | 8.6 ± 0.6 | 1.4 ± 0.1 | 16.1 ± 0.5 | 14.1 ± 0.4 | 14.3 ± 1.1 | 10.3 ± 1.0 |
| uxt1-2 | 2.1 ± 0.1 | 35.0 ± 1.3 | 7.8 ± 0.2 | 1.4 ± 0.1 | 15.2 ± 0.2 | 14.4 ± 0.4 | 13.3 ± 0.3 | 10.9 ± 0.9 | |
| Upper | Col-0 | 3.2 ± 0.5 | 37.5 ± 0.9 | 13.0 ± 0.2 | 1.0 ± 0.1 | 19.0 ± 1.3 | 7.3 ± 0.3 | 15.5 ± 0.3 | 3.5 ± 0.3 |
| Stem | uxt1-1 | 2.7 ± 0.1 | 33.8 ± 0.7 | 12.1 ± 0.7 | 1.2 ± 0.1 | 20.7 ± 0.9 | 8.6 ± 1.1 | 16.1 ± 0.8 | 4.8 ± 0.2 |
| uxt1-2 | 2.9 ± 0.2 | 36.1 ± 1.4 | 12.4 ± 0.7 | 1.4 ± 0.1 | 20.1 ± 1.2 | 7.7 ± 0.5 | 15.0 ± 0.4 | 4.4 ± 0.3 |
Relative abundance of xyloglucan oligosaccharides in the upper and lower stem walls was determined by OLIMP. No significant differences (P < 0.05) were observed between the wild type and mutants. Data represent the mean (±se) of n = 3 independent biological replicates. The mass-to-charge ratio (m/z) is provided for each oligosaccharide; for nomenclature of the oligosaccharides, see Fry et al. (1993).
In Arabidopsis, xylan is comprised of domains that differ in their pattern of MeGlcA substitution and is generated by two glucuronosyltransferases, GUX1 and GUX2. The GUX1 enzyme is responsible for the addition of GlcA only to evenly spaced Xyl residues, whereas GUX2 decorates both even and odd spaced Xyl residues (Bromley et al., 2013). To determine if mutations in UXT1 preferentially affect one of the domains, we digested xylan from the uxt1 mutants and the wild type with the glucuronoxylanase C (XynC) and analyzed the released oligosaccharides by high-performance anion exchange chromatography (HPAEC; Supplemental Figure 4). The xylooligosaccharide profile from the uxt1 mutants is similar to the profile observed for the wild type, and differences detected in the chromatograms are consistent with the increased ratio of the methylated and nonmethylated form of GlcA in the mutant compared with the wild type (Supplemental Figure 3). However, the pattern of xylan substitution is clearly different from those observed for the gux1 and gux2 mutants, thus indicating that UXT1 does not play a role in defining the domain pattern of decoration. Compared with the wild type, the profile from the uxt1 mutants showed a slight shift toward shorter oligosaccharides, indicating a higher degree of substitution (Supplemental Figure 3).
Since UXT1 localizes to both the Golgi apparatus and the ER, it is possible that UXT1 could also play a role in protein glycosylation. To investigate this in more detail, we analyzed total protein from UXT mutants with antibodies against plant N-glycan xylosyl and fucosyl epitopes. However, no obvious differences in the N-glycan xylosylation pattern of the UXT mutants compared with the wild type could be observed. The same was true for the fucosylation pattern, which we analyzed as a control (Supplemental Figure 5). Hence, our results indicate that the UXTs do not have an obvious effect on protein N-glycosylation.
DISCUSSION
The majority of nucleotide sugars are actively transported into the Golgi and ER lumen by NSTs (Bar-Peled and O’Neill, 2011). Here, we present evidence for the existence of a Golgi UDP-Xyl transporter family in plants. We identified three previously uncharacterized Arabidopsis NSTs comprising the NST-KT family subclade C. All three members are capable of transporting UDP-Xyl and to a lower extent UDP-Arap in vitro (Figure 4). However, estimations of the Km values for UDP-Arap indicate that these UXTs are unlikely to transport UDP-Arap in vivo, whereas determined Km values for UDP-Xyl transport are within the physiological range (Rautengarten et al., 2014).
Analysis of the subcellular localization of the UXTs showed that all three are located in the Golgi apparatus and that UXT1 also localized to the ER, which could indicate a distinct functional role for UXT1. However, we are not aware of any xylosyltransferase reaction that is known to take place in the ER. Although protein N-glycosylation is initiated in the ER, the xylosylation has been shown to occur in the Golgi apparatus (Fitchette-lainé et al., 1994; Egelund et al., 2006). Therefore, the possible biological significance of the partial localization of UXT1 to the ER remains unclear. By contrast, the localization to the Golgi apparatus of all three transporters is consistent with the biosynthesis of Xyl-containing glycans in this compartment. The three UXTs are expressed throughout plant development. Compared with UXT1, UXT2 and UXT3 have lower overall expression levels in the tissues analyzed. The UXT1 transcript varies more substantially, especially in mature stem material where relative expression was lowest but still detectable.
To determine the functions of the UXTs in planta, we identified and analyzed loss-of-function mutants in all three transporters. Neither the uxt2 nor the uxt3 mutants exhibited any changes in cell wall monosaccharide composition. This could be due to the lower relative levels of expression for these two genes when compared with UXT1. Similarly, the uxt2 uxt3 double mutants did not show any discernable phenotype, indicating possible minor roles for these genes. Due to the genetic linkage between UXT1 and UXT2, we have not been able to generate a triple homozygous mutant line.
Only UXT1 mutants showed a significant decrease in cell wall-derived Xyl. This decrease was exclusive to stem material and was more pronounced in material isolated from younger parts of inflorescence stems. This difference based on stem maturity can be explained by the fact that in the developing stem, active cell wall biosynthesis and xylan production is occurring, whereas in mature stem tissue the function of the UDP-Xyl transporter may no longer be needed. Together with the decrease in Xyl in stems, we also observed a proportional reduction in GlcA content. Most of the GlcA in stem cell walls originates from glucuronoxylan; therefore, the proportional reduction in GlcA content is consistent with the suggestion that UXT1 functions predominantly in the biosynthesis of glucuronoxylans. However, the methylated GlcA content remained unchanged in uxt1 mutant plants when compared with the wild type (Supplemental Figure 3). This observation is consistent with previously published data on xylan mutants, such as irx8, irx9, and fra8, in which the ratio of GlcA to MeGlcA is lower and methylated GlcA predominates (Liepman et al., 2010). Xylooligosaccharide profiling indicated that UXT1 affects both GUX1-dependent and GUX2-dependent xylan domains.
A more detailed characterization of uxt1 mutants using CoMPP and OLIMP techniques also indicated the importance of UXT1 in glucuronoxylan biosynthesis and revealed that it has little effect on xyloglucan biosynthesis. The latter could be explained by a lower requirement for UDP-Xyl for decoration of matrix polysaccharides, such as xyloglucan and xylogalacturonan, or could hint at a role for the endogenously biosynthesized luminal pool of UDP-Xyl. These findings suggest that UXT1 may be involved in protein interactions with specific glycosyltransferases and associated enzymes involved in biosynthesis of xylan.
The lack of an obvious mutant phenotype for UXT2 and UXT3 could indicate that they are not specifically involved in a functional protein complex and may play a generic role in the delivery of cytosolic-derived UDP-Xyl into the Golgi lumen. Interestingly, the URGT1 transporter of UDP-Rha/UDP-Gal also showed a differential role in biosynthesis of different polysaccharides in vivo (Rautengarten et al., 2014). Loss-of-function mutants and overexpressors of URGT1 showed large changes in content of pectic galactan but no change in the galactose substitutions of xyloglucan. While these observations could suggest substrate channeling in the case of URGT1 and UXT1, it is also possible that differences in Km values for different glycosyltransferases or different sub-Golgi localizations can explain the apparent specificity.
Both β-(1,4)-xylan synthase and β-(1,4)-galactan synthase are enzymes that make a homopolymer and are nonprocessive enzymes in vitro, i.e., the product profiles indicate that the product is released from the enzyme after each round of catalysis (York and O’Neill, 2008; Liwanag et al., 2012; Jensen et al., 2014; Urbanowicz et al., 2014). This is in contrast with processive enzymes, such as cellulose synthase, where the product remains associated with the enzyme. Biosynthesis of β-(1,4)-xylan (this study) and β-(1,4)-galactan (Rautengarten et al., 2014) have been found to be affected by specific nucleotide transporters, and we speculate that the functional association with a transporter is a mechanism that allows the synthases to maintain a degree of processivity and operate efficiently in vivo.
In mammalian cells, UXS enzymes are located only in the Golgi lumen and UDP-Xyl transport is therefore apparently not required (Ashikov et al., 2005). However, a mutant in the UXS enzyme in Chinese hamster ovary cells could be complemented by a cytoplasmic isoform of UXS from Arabidopsis, showing that in these cells the route for delivery of UDP-Xyl is not important for the function of the xylosyltransferases (Bakker et al., 2009). Since UDP-Xyl in plants is biosynthesized both in the cytoplasm and in the Golgi lumen by UXS enzymes, it seemed highly likely that plant UDP-Xyl transport into the Golgi would be a redundant process. However, our results show that, at least for xylan biosynthesis, the transport of UDP-Xyl is important and the Golgi-localized UXS enzymes cannot deliver sufficient UDP-Xyl for proper xylan biosynthesis.
Conclusions
We identified three Golgi-localized nucleotide sugar transporters that are able to transport UDP-Xyl in vitro. This demonstrates the existence of NSTs with specificity for UDP-Xyl in plants. uxt1 mutant plants showed a significant decrease in total cell wall Xyl content in stems, thus confirming a role for UXT1 in providing UDP-Xyl for cell wall biosynthesis.
METHODS
Nucleotide and Nucleotide Sugar Standards
Nucleotide and nucleotide sugar standards were obtained from the following sources: UDP-α-d-xylose, UDP-β-l-arabinopyranose, and UDP-α-d-galacturonic acid (Carbosource Services, Complex Carbohydrate Research Center, Athens, GA); UMP, GMP, CMP, AMP, UDP-α-d-glucuronic acid, UDP-α-d-glucose, UDP-α-d-galactose, UDP-N-acetyl-α-d-glucosamine, UDP-N-acetyl-α-d-galactosamine, GDP-α-d-mannose, GDP-β-l-fucose, GDP-α-d-glucose, adenosine 3′-phosphate 5′ phosphosulfate, CMP-N-acetylneuraminic acid, and ADP-α-d-glucose (Sigma-Aldrich); and UDP-β-l-arabinofuranose (Peptides International). GDP-α-l-galactose was enzymatically synthesized according to Major et al. (2005) and HPLC purified using a linear ammonium formate gradient (Rautengarten et al., 2011). UDP-β-l-rhamnose was enzymatically synthesized by a two-step reaction using UDP-Glc as substrate as previously described (Rautengarten et al., 2014).
Sequence Analysis
Amino acid sequences were retrieved from The Arabidopsis Information Resource (Lamesch et al., 2012). Deduced amino acid sequences were aligned using the Clustal Omega program (Sievers et al., 2011) using default parameters (Supplemental Data Set 1). Phylogenetic trees were created using the neighbor-joining statistical method and applying the bootstrap method with 1000 replications and visualized using the Molecular Evolutionary Genetics Analysis (MEGA) version 6.0 application (Tamura et al., 2013).
Heterologous Expression, Reconstitution, and in Vitro Assay of Transport Activities
Heterologous expression in Saccharomyces cerevisiae (strain INVSc1: MATa his3D1 leu2 trp1-289 ura3-52 MAT his3D1 leu2 trp1-289 ura3-52; Life Technologies), reconstitution of microsomal proteins, and subsequent transport activity assay were performed as previously described (Rautengarten et al., 2014). Kinetic parameters were calculated by nonlinear regression using the Prism6 application (GraphPad Software). PAGE and immunoblot analyses were done as previously described (Rautengarten et al., 2011) using 2.5 μg yeast microsomal protein. Filters were probed using the anti-V5 antibody (Life Technologies).
Chromatographic Separation and Detection of Nucleotide Sugars by Mass Spectrometry
LC-MS/MS was performed using porous graphitic carbon as the stationary phase on an 1100 series HPLC system (Agilent Technologies) and a 4000 QTRAP LC-MS/MS system (AB Sciex) equipped with a TurboIonSpray ion source using methods previously described (Ito et al., 2014; Rautengarten et al., 2014)
Absolute Quantification of Reconstituted NSTs by MRM Mass Spectrometry
The yeast expression vector pYES-DEST52 contains an in-frame V5-tag and 6xHis-tag epitope at the 3′ end of the cloning site. The expressed UXT proteins all yield a common tryptic peptide, namely, R.SRGPFEGKPIPNPLLGLDSTR.T, as previously described (Rautengarten et al., 2014). A synthesized peptide was used to determine optimal parameters for MRM analysis with the following parameters: dwell (25 ms), fragmentor (130 V), collision energy (11.1 V), and cell accelerator voltage (7 V). Analysis of samples and standard curves were conducted on a 6460 Triple Quad LC/MS system equipped with a Jet Stream ESI source (Agilent Technologies). The system was operated in positive ion mode using the MRM scan type with both MS1 and MS2 resolutions set to unit. The following mass spectrometer parameters were applied: gas temperature (350°C), gas flow (10 L/min), nebulizer (45 p.s.i.), sheath gas temperature (400°C), sheath gas flow (11 L/min), capillary (5000 V), and MS1/MS2 heater (100°C). A total of 5 μg of trypsin-digested (1:10 [w/w]) proteoliposome was loaded onto an Ascentis Express Peptide ES-C18 (5 cm × 2.1 mm, 2.7 µm) column (Sigma-Aldrich) using a 1290 series HPLC (Agilent Technologies) at a flow rate of 0.4 mL/min as follows: 95% Buffer A (99.9% water and 0.1% formic acid) and 5% Buffer B (99.9% acetonitrile and 0.1% formic acid) for 0.2 min, followed by an increase to 35% Buffer B over 5.5 min, then 90% Buffer B in 0.3 min, where it was held for 2 min. The buffer composition was ramped back to 5% Buffer B over 5 min, giving a total runtime of 13 min. The column temperature was maintained at 60°C. Data were acquired using MassHunter Workstation Software Version B.06.00 Build 6.0.6025.4 SP4 (Agilent Technologies). The raw data were imported into Skyline (v2.5.0.6157) (MacLean et al., 2010) and transition peaks manually inspected for retention time and adjusted accordingly. The abundance of the expressed UXTs in a sample was calculated by integrating the total signal peak area (total area) from Skyline for the two transitions on the predominant 563.560 [M+4H]4+ precursor ion, namely, L [y7] 761.452 [M+H]1+ and G [y6] 648.3311 [M+H]1+, and calculating total moles in the sample against a standard curve for the synthesized peptide. The standard curve was created by linear regression using a range of abundances (0.5 to 10 pmol), which were interspersed as separate runs during sample analysis. The UXTs represent from 0.01 to 0.1% of total protein of the reconstituted proteoliposomes with errors representing the sd of two technical replicates (Supplemental Table 1). Values were used for enzyme kinetic calculations.
Plant Material and Growth Conditions
Arabidopsis thaliana Col-0 was obtained from the ABRC (http://abrc.osu.edu/). T-DNA insertion mutants for UXT1 (uxt1-1, SAIL_147_F11; uxt1-2, SALK_086773), UXT2 (uxt2-1, SALK_078576), and UXT3 (uxt3-1, SALK_013372) were obtained from the SIGnAL Salk collection (http://signal.salk.edu/). Plants were germinated and grown on soil (PRO-MIX; Premier Horticulture) in an Arabidopsis growth chamber (Percival-Scientific) under short-day light conditions (10 h of fluorescent light [120 μmol m−2 s−1] at 22°C and 60% RH/14 h of dark at 22°C and 60% RH). After 4 weeks, plants were transferred to long-day conditions (16 h of fluorescent light [120 μmol m−2 s−1] at 22°C and 60% RH/8 h of dark at 22°C and 60% RH).
Cloning Procedures
Coding sequences for Arabidopsis UXTs without native stop codon were PCR amplified using the primer pairs listed in Supplemental Table 3. PCR products were introduced into the pENTR/SD/D-TOPO cloning vector (Life Technologies) according to the manufacturer’s protocol and confirmed by sequencing. To obtain C-terminal YFP fusions, the constructs were introduced into the 35S promoter carrying pEarleyGate101 plant transformation vector (Earley et al., 2006) using the LR Clonase II reaction (Life Technologies) following the manufacturer’s protocol. For yeast expression, the constructs were introduced into the yeast expression vector pYES-DEST52 (Life Technologies) using the LR Clonase II reaction (Life Technologies).
Subcellular Localization and Microscopy
Nicotiana benthamiana plants were grown on soil (PRO-MIX) in a growth chamber (Percival-Scientific) using the following conditions: 24°C day/night temperature, 60% humidity, and 16-h-light/8-h-dark cycles. Four-week-old leaves were coinfiltrated with Agrobacterium tumefaciens strain GV3101 pmp90 carrying the C-terminal YFP fusion constructs (OD600 = 0.15) and the α-mannosidase-mCherry marker (OD600 = 0.01) (Nelson et al., 2007) using the previously described method (Jensen et al., 2008). Visualization by confocal laser scanning microscopy was performed as previously described (Rautengarten et al., 2012).
Determination of Monosaccharide Composition
AIR was prepared as described earlier (Harholt et al., 2006). Samples were hydrolyzed in 2 n trifluoroacetic acid for 1 h at 120°C. HPAEC with pulsed amperometric detection was performed as described (ØBro et al., 2004) on an ICS 3000 (Dionex) using a CarboPac PA20 anion exchange column (3 × 150 mm; Dionex).
PCR Characterization of Mutants
Homozygous T-DNA insertion lines were verified by PCR to confirm the presence of the insert using the primers listed in Supplemental Table 3. Subsequently, absence of the transcript was verified by RT-PCR using the primers listed in Supplemental Table 3. Arabidopsis ACTIN-2 (At3g18780) was used as a control for equal loading.
RT-PCR
Plant RNA was extracted using the RNEasy RNA Plant Kit (Qiagen) according to the manufacturer’s protocol, and 0.5 to 1 μg was reverse transcribed using SuperScript II reverse transcriptase and d(T)15 oligomers (Life Technologies) according to the manufacturer’s protocol. UXT1-3 expression in different organs was analyzed by quantitative RT-PCR using SYBR Select Master Mix (Applied Biosystems) on a StepOnePlus Real-Time PCR system (Applied Biosystems) according to the conditions described earlier (Czechowski et al., 2005) using StepOne 2.0 software (Applied Biosystems). The UXT genes were amplified using the primers listed in Supplemental Table 3. As references, primers for UBQ10 (At4g05320), PP2A (At1g13320), and a SAND family member (MON1, At2g28390) were used (Supplemental Table 3). Expression levels were calculated using the comparative CT method, which involves normalizing against the geometric mean of the three housekeeping genes (UBI10, PP2A and SAND family) for each tissue type (Schmittgen and Livak, 2008).
Xylan Oligosaccharide Profiling
Xylan was digested with endoglucuronoxylanase GH30 (XynC) from Bacillus subtilis (St John et al., 2006) as previously described (Bromley et al., 2013). Profiling of the released oligosaccharides by HPAEC was performed using the conditions previously described (Chiniquy et al., 2012). Cell wall preparations from the gux1 and gux2 mutants (Oikawa et al., 2010; Bromley et al., 2013) were analyzed for comparison.
Protein Extraction and Immunoblotting
Inflorescence stems from 6-week-old plants were ground in extraction buffer (10 mM Tris, pH 8, 150 mM NaCl, 2% Triton, 1 mM PMSF, protease inhibitor, and 10 mM CaCl2) incubated for 1 h at 4°C under constant shaking, and centrifuged for 30 min at 20,800g at 4°C to remove cell debris. Subsequently, protein was precipitated with 20% trichloroacetic acid, incubated on ice, and spun down. After removal of the supernatant, the samples were washed twice with ice-cold acetone, dried, and suspended in the appropriate buffer. Samples were separated by SDS-PAGE, and glycosylation was detected by immunoblotting using antibodies raised against β-(1,2)-Xyl and α-(1,3)-Fuc (Agrisera). Detection was performed with an ECL Plus Western Blotting Detection System (GE Healthcare).
OLIMP
Mass profiling of xyloglucan oligosaccharides derived from various stem tissues was performed as previously described (Lerouxel et al., 2002). The cell wall material was digested with a xyloglucans-specific endoglucanase (Pauly et al., 1999), and the resulting solubilized xyloglucan oligosaccharides were detected using an Axima matrix-assisted laser desorption/ionization time-of-flight system (Shimadzu) set in linear positive mode with an acceleration voltage of 20,000 V.
CoMPP
CoMPP was undertaken essentially according to previously described work (Moller et al., 2007). AIR samples were extracted from mature stems of pooled material (approximately eight individuals) from 8-week-old uxt1-1, uxt1-2, and wild-type plants. A total of 4 mg AIR was subsequently extracted sequentially with CDTA and then NaOH solutions to obtain pectin-rich and hemicellulose-rich extracts, respectively. These extracts were spotted onto membranes and probed with monoclonal antibodies and carbohydrate binding modules (CBMs) that recognize specific cell wall epitopes, namely, LM3, extensins; LM25, xyloglucan; LM10, β-(1-4)-d-xylan; LM11, (1-4)-β-d-xylan; UX1, glucuronoxylan; AX1, arabinose substituted β-(1-4)-d-xylan; and CBM3a, cellulose (Guillon et al., 2004; McCartney et al., 2005; Blake et al., 2006; Koutaniemi et al., 2012; Pedersen et al., 2012).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: UXT1 (AT2G28315), UXT2 (AT2G30460), UXT3 (AT1G06890), ACTIN-2 (At3g18780), UBQ10 (At4g05320), PP2A (At1g13320), and MON1 (At2g28390).
Supplemental Data
Supplemental Figure 1. Subcellular localization of UXT1 with an ER marker.
Supplemental Figure 2. Assessment of UXT transcripts by RT-PCR in the of uxt mutant backgrounds.
Supplemental Figure 3. The 4-O-Methyl-d-glucuronic acid content of pooled stem material.
Supplemental Figure 4. Xylan profiling of the uxt1 mutants.
Supplemental Figure 5. Immunoblot analysis of N-glycosylation in the uxt mutants.
Supplemental Table 1. Calculations of UXT protein contents in reconstituted proteoliposomes used for transport assays.
Supplemental Table 2. Monosaccharide composition of UXT mutant cell wall preparations derived from different Arabidopsis organs.
Supplemental Table 3. Primer list.
Supplemental Data Set 1. Text file of the alignment used for the phylogenetic analysis shown in Figure 1.
Supplementary Material
Acknowledgments
This work was supported by the U. S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through Contract DE-AC02-05CH11231 between the Lawrence Berkeley National Laboratory and the U.S. Department of Energy. J.L.H. is supported by an Australian Research Council Future Fellowship (FT130101165). Part of the work was supported by the Danish Strategic Research Council (Set4Future 11-116795). The substrates obtained from Carbosource Services (Athens, GA) were supported in part by NSF-RCN Grant 0090281. We thank James F. Preston (University of Florida) for the generous gift of xylanase XynC and Breeanna Urbanowicz (University of Georgia) for providing a 4-O-Me-GlcA standard. We also thank Devon Birdseye and Mi Yeon Lee for assistance with plant growth and maintenance.
AUTHOR CONTRIBUTIONS
B.E., C.R., P.D.A., C.J.P., J.L.H., and H.V.S. designed the study and directed its implementation. B.E., C.R., X.G., G.X., T.H., L.J.G.C., and S.S. conducted the experiments. B.E., C.R., J.L.H., A.M.S.-M., M.P., W.G.T.W., and H.V.S. analyzed the data. B.E., C.R., J.L.H., and H.V.S. wrote the article.
Glossary
- NST
nucleotide sugar transporter
- ER
endoplasmic reticulum
- TPT
triose phosphate translocator
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- UDP-Arap
UDP-arabinopyranose
- MRM
multiple reaction monitoring
- AIR
alcohol insoluble residue
- Col-0
Columbia-0
- GlcA
glucuronic acid
- CoMPP
comprehensive microarray polymer profiling
- MeGlcA
4-O-methyl ether
- OLIMP
oligosaccharide mass profiling
- XynC
glucuronoxylanase C
- HPAEC
high-performance anion exchange chromatography
Footnotes
Articles can be viewed online without a subscription.
References
- Ashikov A., Routier F., Fuhlrott J., Helmus Y., Wild M., Gerardy-Schahn R., Bakker H. (2005). The human solute carrier gene SLC35B4 encodes a bifunctional nucleotide sugar transporter with specificity for UDP-xylose and UDP-N-acetylglucosamine. J. Biol. Chem. 280: 27230–27235. [DOI] [PubMed] [Google Scholar]
- Atmodjo M.A., Hao Z., Mohnen D. (2013). Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 64: 747–779. [DOI] [PubMed] [Google Scholar]
- Bakker H., Routier F., Ashikov A., Neumann D., Bosch D., Gerardy-Schahn R. (2008). A CMP-sialic acid transporter cloned from Arabidopsis thaliana. Carbohydr. Res. 343: 2148–2152. [DOI] [PubMed] [Google Scholar]
- Bakker H., Routier F., Oelmann S., Jordi W., Lommen A., Gerardy-Schahn R., Bosch D. (2005). Molecular cloning of two Arabidopsis UDP-galactose transporters by complementation of a deficient Chinese hamster ovary cell line. Glycobiology 15: 193–201. [DOI] [PubMed] [Google Scholar]
- Bakker H., Oka T., Ashikov A., Yadav A., Berger M., Rana N.A., Bai X., Jigami Y., Haltiwanger R.S., Esko J.D., Gerardy-Schahn R. (2009). Functional UDP-xylose transport across the endoplasmic reticulum/Golgi membrane in a Chinese hamster ovary cell mutant defective in UDP-xylose Synthase. J. Biol. Chem. 284: 2576–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin T.C., Handford M.G., Yuseff M.I., Orellana A., Dupree P. (2001). Identification and characterization of GONST1, a Golgi-localized GDP-mannose transporter in Arabidopsis. Plant Cell 13: 2283–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled M., O’Neill M.A. (2011). Plant nucleotide sugar formation, interconversion, and salvage by sugar recycling. Annu. Rev. Plant Biol. 62: 127–155. [DOI] [PubMed] [Google Scholar]
- Blake A.W., McCartney L., Flint J.E., Bolam D.N., Boraston A.B., Gilbert H.J., Knox J.P. (2006). Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J. Biol. Chem. 281: 29321–29329. [DOI] [PubMed] [Google Scholar]
- Bromley J.R., Busse-Wicher M., Tryfona T., Mortimer J.C., Zhang Z., Brown D.M., Dupree P. (2013). GUX1 and GUX2 glucuronyltransferases decorate distinct domains of glucuronoxylan with different substitution patterns. Plant J. 74: 423–434. [DOI] [PubMed] [Google Scholar]
- Chiniquy D., et al. (2012). XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proc. Natl. Acad. Sci. USA 109: 17117–17122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Ebringerová A., Heinze T. (2000). Xylan and xylan derivatives: biopolymers with valuable properties, 1 Naturally occurring xylans structures, procedures and properties. Macromol. Rapid Commun. 21: 542–556. [Google Scholar]
- Egelund J., Petersen B.L., Motawia M.S., Damager I., Faik A., Olsen C.E., Ishii T., Clausen H., Ulvskov P., Geshi N. (2006). Arabidopsis thaliana RGXT1 and RGXT2 encode Golgi-localized (1,3)-alpha-D-xylosyltransferases involved in the synthesis of pectic rhamnogalacturonan-II. Plant Cell 18: 2593–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitchette-lainé A.C., Gomord V., Chekkafi A., Faye L. (1994). Distribution of xylosylation and fucosylation in the plant Golgi-apparatus. Plant J. 5: 673–682. [Google Scholar]
- Fry S.C., et al. (1993). An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol. Plant. 89: 1–3. [Google Scholar]
- Guillon F., Tranquet O., Quillien L., Utille J.P., Ortiz J.J.O., Saulnier L. (2004). Generation of polyclonal and monoclonal antibodies against arabinoxylans and their use for immunocytochemical location of arabinoxylans in cell walls of endosperm of wheat. J. Cereal Sci. 40: 167–182. [Google Scholar]
- Handford M., Rodríguez-Furlán C., Marchant L., Segura M., Gómez D., Alvarez-Buylla E., Xiong G.Y., Pauly M., Orellana A. (2012). Arabidopsis thaliana AtUTr7 encodes a golgi-localized UDP-glucose/UDP-galactose transporter that affects lateral root emergence. Mol. Plant 5: 1263–1280. [DOI] [PubMed] [Google Scholar]
- Handford M.G., Sicilia F., Brandizzi F., Chung J.H., Dupree P. (2004). Arabidopsis thaliana expresses multiple Golgi-localised nucleotide-sugar transporters related to GONST1. Mol. Genet. Genomics 272: 397–410. [DOI] [PubMed] [Google Scholar]
- Harholt J., Jensen J.K., Sørensen S.O., Orfila C., Pauly M., Scheller H.V. (2006). ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol. 140: 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper A.D., Bar-Peled M. (2002). Biosynthesis of UDP-xylose. Cloning and characterization of a novel Arabidopsis gene family, UXS, encoding soluble and putative membrane-bound UDP-glucuronic acid decarboxylase isoforms. Plant Physiol. 130: 2188–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J., Herter T., Baidoo E.E., Lao J., Vega-Sánchez M.E., Michelle Smith-Moritz A., Adams P.D., Keasling J.D., Usadel B., Petzold C.J., Heazlewood J.L. (2014). Analysis of plant nucleotide sugars by hydrophilic interaction liquid chromatography and tandem mass spectrometry. Anal. Biochem. 448: 14–22. [DOI] [PubMed] [Google Scholar]
- Jensen J.K., Johnson N.R., Wilkerson C.G. (2014). Arabidopsis thaliana IRX10 and two related proteins from psyllium and Physcomitrella patens are xylan xylosyltransferases. Plant J. 80: 207–215. [DOI] [PubMed] [Google Scholar]
- Jensen J.K., et al. (2008). Identification of a xylogalacturonan xylosyltransferase involved in pectin biosynthesis in Arabidopsis. Plant Cell 20: 1289–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe S., Flügge U.I., Fischer K. (2003). Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol. 131: 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutaniemi S., Guillon F., Tranquet O., Bouchet B., Tuomainen P., Virkki L., Petersen H.L., Willats W.G., Saulnier L., Tenkanen M. (2012). Substituent-specific antibody against glucuronoxylan reveals close association of glucuronic acid and acetyl substituents and distinct labeling patterns in tree species. Planta 236: 739–751. [DOI] [PubMed] [Google Scholar]
- Lamesch P., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40: D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouxel O., Choo T.S., Séveno M., Usadel B., Faye L., Lerouge P., Pauly M. (2002). Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiol. 130: 1754–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman A.H., Wightman R., Geshi N., Turner S.R., Scheller H.V. (2010). Arabidopsis - a powerful model system for plant cell wall research. Plant J. 61: 1107–1121. [DOI] [PubMed] [Google Scholar]
- Liwanag A.J., Ebert B., Verhertbruggen Y., Rennie E.A., Rautengarten C., Oikawa A., Andersen M.C., Clausen M.H., Scheller H.V. (2012). Pectin biosynthesis: GALS1 in Arabidopsis thaliana is a β-1,4-galactan β-1,4-galactosyltransferase. Plant Cell 24: 5024–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. (2010). Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26: 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major L.L., Wolucka B.A., Naismith J.H. (2005). Structure and function of GDP-mannose-3′,5′-epimerase: an enzyme which performs three chemical reactions at the same active site. J. Am. Chem. Soc. 127: 18309–18320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney L., Marcus S.E., Knox J.P. (2005). Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J. Histochem. Cytochem. 53: 543–546. [DOI] [PubMed] [Google Scholar]
- Mølhøj M., Verma R., Reiter W.D. (2003). The biosynthesis of the branched-chain sugar d-apiose in plants: functional cloning and characterization of a UDP-d-apiose/UDP-d-xylose synthase from Arabidopsis. Plant J. 35: 693–703. [DOI] [PubMed] [Google Scholar]
- Moller I., Sørensen I., Bernal A.J., Blaukopf C., Lee K., Øbro J., Pettolino F., Roberts A., Mikkelsen J.D., Knox J.P., Bacic A., Willats W.G. (2007). High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J. 50: 1118–1128. [DOI] [PubMed] [Google Scholar]
- Mortimer J.C., et al. (2013). Abnormal glycosphingolipid mannosylation triggers salicylic acid-mediated responses in Arabidopsis. Plant Cell 25: 1881–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Norambuena L., Marchant L., Berninsone P., Hirschberg C.B., Silva H., Orellana A. (2002). Transport of UDP-galactose in plants. Identification and functional characterization of AtUTr1, an Arabidopsis thaliana UDP-galactos/UDP-glucose transporter. J. Biol. Chem. 277: 32923–32929. [DOI] [PubMed] [Google Scholar]
- Norambuena L., Nilo R., Handford M., Reyes F., Marchant L., Meisel L., Orellana A. (2005). AtUTr2 is an Arabidopsis thaliana nucleotide sugar transporter located in the Golgi apparatus capable of transporting UDP-galactose. Planta 222: 521–529. [DOI] [PubMed] [Google Scholar]
- ØBro J., Harholt J., Scheller H.V., Orfila C. (2004). Rhamnogalacturonan I in Solanum tuberosum tubers contains complex arabinogalactan structures. Phytochemistry 65: 1429–1438. [DOI] [PubMed] [Google Scholar]
- Oikawa A., Joshi H.J., Rennie E.A., Ebert B., Manisseri C., Heazlewood J.L., Scheller H.V. (2010). An integrative approach to the identification of Arabidopsis and rice genes involved in xylan and secondary wall development. PLoS ONE 5: e15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S., Harper A.D., Bar-Peled M. (2005). Biosynthesis of UDP-xylose: characterization of membrane-bound AtUxs2. Planta 221: 538–548. [DOI] [PubMed] [Google Scholar]
- Pauly M., Andersen L.N., Kauppinen S., Kofod L.V., York W.S., Albersheim P., Darvill A. (1999). A xyloglucan-specific endo-β-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9: 93–100. [DOI] [PubMed] [Google Scholar]
- Pedersen H.L., et al. (2012). Versatile high resolution oligosaccharide microarrays for plant glycobiology and cell wall research. J. Biol. Chem. 287: 39429–39438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C., Ebert B., Herter T., Petzold C.J., Ishii T., Mukhopadhyay A., Usadel B., Scheller H.V. (2011). The interconversion of UDP-arabinopyranose and UDP-arabinofuranose is indispensable for plant development in Arabidopsis. Plant Cell 23: 1373–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C., Ebert B., Ouellet M., Nafisi M., Baidoo E.E., Benke P., Stranne M., Mukhopadhyay A., Keasling J.D., Sakuragi Y., Scheller H.V. (2012). Arabidopsis Deficient in Cutin Ferulate encodes a transferase required for feruloylation of ω-hydroxy fatty acids in cutin polyester. Plant Physiol. 158: 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C., et al. (2014). The Golgi localized bifunctional UDP-rhamnose/UDP-galactose transporter family of Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 11563–11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie E.A., Scheller H.V. (2014). Xylan biosynthesis. Curr. Opin. Biotechnol. 26: 100–107. [DOI] [PubMed] [Google Scholar]
- Rollwitz I., Santaella M., Hille D., Flügge U.I., Fischer K. (2006). Characterization of AtNST-KT1, a novel UDP-galactose transporter from Arabidopsis thaliana. FEBS Lett. 580: 4246–4251. [DOI] [PubMed] [Google Scholar]
- Scheible W.R., Pauly M. (2004). Glycosyltransferases and cell wall biosynthesis: novel players and insights. Curr. Opin. Plant Biol. 7: 285–295. [DOI] [PubMed] [Google Scholar]
- Scheller H.V., Ulvskov P. (2010). Hemicelluloses. Annu. Rev. Plant Biol. 61: 263–289. [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506. [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John F.J., Rice J.D., Preston J.F. (2006). Characterization of XynC from Bacillus subtilis subsp. subtilis strain 168 and analysis of its role in depolymerization of glucuronoxylan. J. Bacteriol. 188: 8617–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R., Mucha J., Mach L., Altmann F., Wilson I.B., Glössl J., Steinkellner H. (2000). Molecular cloning and functional expression of β1, 2-xylosyltransferase cDNA from Arabidopsis thaliana. FEBS Lett. 472: 105–108. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., et al. (2013). An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25: 270–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz B.R., Peña M.J., Moniz H.A., Moremen K.W., York W.S. (2014). Two Arabidopsis proteins synthesize acetylated xylan in vitro. Plant J. 80: 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York W.S., O’Neill M.A. (2008). Biochemical control of xylan biosynthesis - which end is up? Curr. Opin. Plant Biol. 11: 258–265. [DOI] [PubMed] [Google Scholar]
- Zhang B., Liu X., Qian Q., Liu L., Dong G., Xiong G., Zeng D., Zhou Y. (2011). Golgi nucleotide sugar transporter modulates cell wall biosynthesis and plant growth in rice. Proc. Natl. Acad. Sci. USA 108: 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.