ATG3 interacts with GAPCs, which negatively regulate autophagy and play an essential role in effector-triggered immunity and basal defense. The interaction between GAPCs and ATG3 is regulated by ROS.
Abstract
Autophagy as a conserved catabolic pathway can respond to reactive oxygen species (ROS) and plays an important role in degrading oxidized proteins in plants under various stress conditions. However, how ROS regulates autophagy in response to oxidative stresses is largely unknown. Here, we show that autophagy-related protein 3 (ATG3) interacts with the cytosolic glyceraldehyde-3-phosphate dehydrogenases (GAPCs) to regulate autophagy in Nicotiana benthamiana plants. We found that oxidative stress inhibits the interaction of ATG3 with GAPCs. Silencing of GAPCs significantly activates ATG3-dependent autophagy, while overexpression of GAPCs suppresses autophagy in N. benthamiana plants. Moreover, silencing of GAPCs enhances N gene-mediated cell death and plant resistance against both incompatible pathogens Tobacco mosaic virus and Pseudomonas syringae pv tomato DC3000, as well as compatible pathogen P. syringae pv tabaci. These results indicate that GAPCs have multiple functions in the regulation of autophagy, hypersensitive response, and plant innate immunity.
INTRODUCTION
Autophagy is an evolutionarily conserved vacuole/lysosome-dependent cellular process responsible for the bulk degradation of cytoplasmic components. In plants, autophagy can be induced by both abiotic and biotic stresses, such as starvation, oxidative stress, drought, salt, and pathogen invasion (Bassham, 2007; Han et al., 2011; Hayward and Dinesh-Kumar, 2011; Liu and Bassham, 2012). Generally, autophagy can be divided into three main types, including macroautophagy, chaperone-mediated autophagy, and microautophagy (Xie and Klionsky, 2007; Reggiori and Klionsky, 2013). Macroautophagy, hereafter referred as autophagy, mainly describes a biological process in which a double membrane-bound autophagosome engulfs cellular components to fuse with vacuole or lysosome for the final degradation. More than 30 autophagy-related genes have been identified in yeast, most of which have homologs in plants. These gene products can be divided into several functional groups: the ATG1-ATG13 kinase complex, ATG9 and ATG9-associated proteins, a phosphatidylinositol 3-kinase complex, and two ubiquitin-like conjugation systems (Xie and Klionsky, 2007). Recent studies show that plant autophagy participates in multiple physiological processes, such as nutrient recycling, development, senescence, abiotic stress responses, and pathogen defense (Doelling et al., 2002; Hanaoka et al., 2002; Liu et al., 2005, 2009, 2012; Bassham, 2007; Han et al., 2011). ATG3 is an E2-like conjugating enzyme that can catalyze the ATG8-PE conjugation formation during autophagy process (Ichimura et al., 2000).
Reactive oxygen species (ROS) are key cellular signals for plant response to multiple stresses, such as drought, UV irradiation, high-light intensity, wounding, ozone, low and high temperatures, and pathogens. H2O2 is the major and most stable type of ROS. In mammals, autophagy can be induced by exogenous H2O2 (Zhang et al., 2009) or internal ROS generated mainly from mitochondria (Chen et al., 2007; Scherz-Shouval et al., 2007; Moore, 2008; Azad et al., 2009). However, autophagy can serve to reduce ROS levels and reduce oxidative damage (Kaushik and Cuervo, 2006; Scherz-Shouval and Elazar, 2007; Gurusamy et al., 2009; Jain et al., 2010). In plants, ROS is reported to induce autophagy, while downregulation of ATG18 in Arabidopsis thaliana leads to the accumulation of oxidized proteins and subsequent increased sensitivity to oxidative stress (Xiong et al., 2007a, 2007b). In rice (Oryza sativa) atg10b mutant seedlings, the amount of oxidized protein induced by methyl viologen (MV) treatment is also increased (Shin et al., 2009). Knockout of either ATG5 or ATG2 in Arabidopsis causes a dramatic accumulation of H2O2 (Yoshimoto et al., 2009). Taken together, these observations suggest autophagy functions to alleviate oxidative stress in plants. However, how autophagy is activated by ROS signaling is still unclear.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a key enzyme in the glycolytic pathway. It catalyzes the major step of the conversion of glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate, linking the energy-consuming steps of the pathway with the energy-producing steps and providing intermediates for cellular metabolism (Plaxton, 1996). In addition, “moonlighting” functions of GAPDHs have been increasingly discovered outside of glycolysis. In animal cells, GAPDH participates in multiple nonmetabolic processes, including apoptosis activation, S phase-dependent histone H2B transcription, DNA replication, DNA repair, and immune response to various diseases (Sirover, 1997; Zheng et al., 2003; Hara et al., 2005; Bae et al., 2006; Harada et al., 2007). GAPDH is also involved in NF-κB-dependent host innate immune responses, while pathogen effector NieB mediates O-GlcNAcylation of GAPDH to disrupt the TRAF2-GAPDH interaction and further suppresses TRAF2 polyubiquitination and NF-κB activation (Gao et al., 2013). In plants, GAPDHs have been implicated in embryo development, pollen development, root growth, lipid metabolism, seed oil accumulation, and abscisic acid signal transduction (Rius et al., 2006, 2008; Muñoz-Bertomeu et al., 2009, 2010, 2011; Kim et al., 2013; Guo et al., 2014). In Arabidopsis, cytosolic glyceraldehyde-3-phosphate dehydrogenase (GAPC) is thought to function in oxidation signaling, and cadmium treatment causes inactivation of GAPC enzyme activity and relocation of GAPC from cytoplasm to nuclei (Vescovi et al., 2013). GAPC1 has been suggested to interact directly with H2O2 and to be a potential modification target involved in ROS response in Arabidopsis (Hancock et al., 2005; Holtgrefe et al., 2008). Furthermore, GAPCs interact with the plasma membrane-associated phospholipase D to transduce H2O2 signals in the Arabidopsis response to stress (Guo et al., 2012).
ATG3 is not only a key autophagy component but also a regulator of plant immunity-related cell death (Liu et al., 2005). To study how ATG3 is involved in these processes, we used a mass spectrometry-based strategy to screen for potential ATG3 binding proteins. Here, we report that GAPCs interacts with ATG3 to regulate autophagy and programmed cell death during innate immunity responses in N. benthamiana.
RESULTS
Identification of Nb-GAPC as Nb-ATG3-Interacting Partners
A cassette driven by the cauliflower mosaic virus (CaMV) 35S promoter was generated to transiently express the NbATG3-GFP (green fluorescent protein) fusion protein in the leaves of Nicotiana benthamiana. Total crude proteins were extracted from agroinfiltrated leaf tissues and followed by a coimmunoprecipitation assay using anti-GFP agarose beads. The putative ATG3 interacting proteins were then separated by SDS-PAGE gels followed by silver staining. Protein extracts from leaves in which only free GFP was expressed served as negative control. The differentially displayed gel bands were excised, in-gel digested with trypsin, and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. When the results were searched against databases, Nicotiana tabacum cytosolic glyceraldehyde-3-phosphate dehydrogenase was found with high scores. We next searched the N. benthamiana genome database using N. tabacum GAPC and found three GAPC counterparts, which are named Nb-GAPC1, Nb-GAPC2, and Nb-GAPC3, respectively. Nb-GAPC1 shares an amino acid identity of 95.0% with Nb-GAPC2 and 94.7% with Nb-GAPC3, while identity between Nb-GAPC2 and Nb-GAPC3 proteins is up to 97.6% (Supplemental Figure 1). Because N. benthamiana is allotetraploid, Nb-GAPC2 and Nb-GAPC3 may be the alleles of one gene from different ancestry. Given this high identity, we focused on Nb-GAPC1 and Nb-GAPC2 for further analyses.
Nb-GAPCs and Nb-ATG3 Mainly Localize to Cytoplasm
GAPC is an isoform of GAPDH, which mainly localizes in the cytoplasm. To determine the subcellular localization of Nb-GAPCs and Nb-ATG3, yellow fluorescent protein (YFP)-NbGAPCs and GFP-NbATG3 fusion proteins were transiently expressed in N. benthamiana by agroinfiltration. All three GAPCs were predominantly present in the cytoplasm and hardly visible in nucleus under normal physiological conditions. A similar cellular pattern of the GFP-NbATG3 fluorescence was also found (Supplemental Figure 2), indicating that GAPCs and ATG3 are mainly localized to cytoplasm.
GAPCs Interacts with ATG3 in Vivo and in Vitro
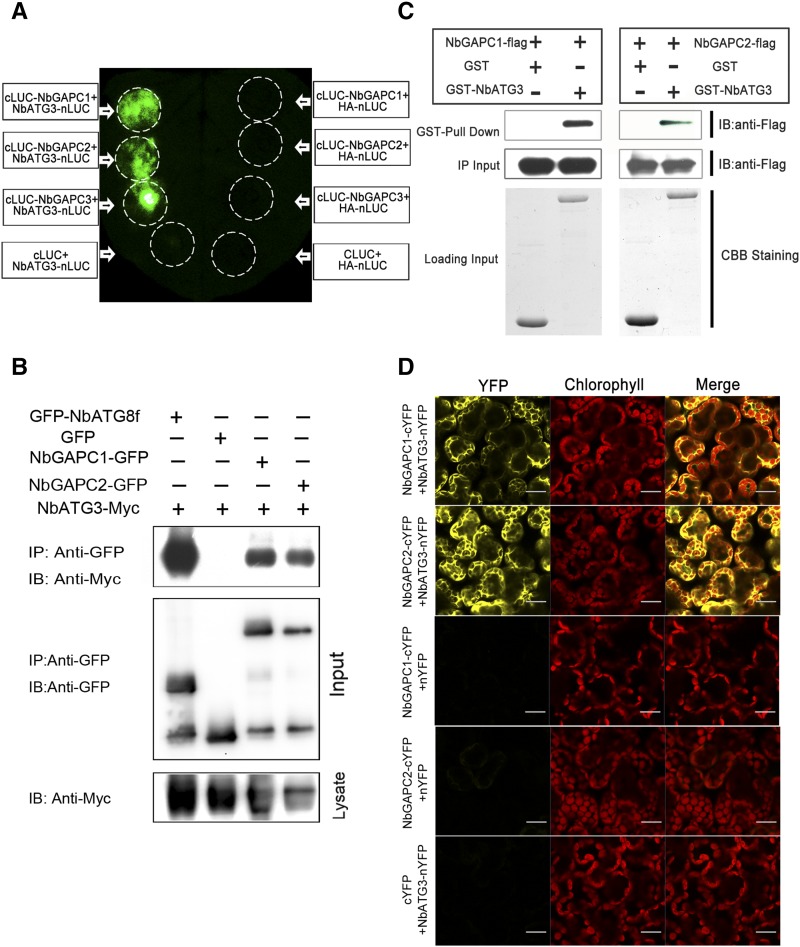
Since GAPC was identified by the affinity purification coupled to mass spectrometry, it was essential to verify that the interaction between GAPCs and ATG3 occurs in plants. To test this, we used a firefly luciferase complementation imaging (LCI) assay (Chen et al., 2008). The three GAPC genes were each fused to the C-terminal domain of luciferase (cLUC), while ATG3 was fused to the N-terminal domain of luciferase (nLUC) (Figure 1A). NbATG3-nLUC was coexpressed with NbGAPC1-cLUC, NbGAPC2-cLUC, and NbGAPC3-cLUC, respectively, in N. benthamiana. Chemical signals were detected when ATG3 was combined with all three GAPCs, due to the reconstitution of the luciferase activity by ATG3-GAPC interactions. However, no such interactions were detected in negative controls (Figure 1A).
Figure 1.
Nb-ATG3 Interacts with Nb-GAPCs in Vivo and in Vitro.
(A) LCI assays showed the interaction of Nb-ATG3 with Nb-GAPCs in plants. Image shown is luminescence of N. benthamiana leaf that was agroinfiltrated with NbATG3-nLUC (left) or negative control HA-nLUC (right) with the cLUC-tagged GAPC1, GAPC2, GAPC3, and a negative control cLUC. The experiments were repeated three times with similar results.
(B) Nb-ATG3 coimmunoprecipitated with Nb-ATG8f, Nb-GAPC1, and Nb-GAPC2. NbATG3-Myc was coexpressed with GFP-NbATG8f, NbGAPC1-GFP, or NbGAPC2-GFP in N. benthamiana leaves by agroinfiltration. NbATG3-Myc coexpressed with GFP was introduced as a negative control. At 60 hpi, leaf lysates were immunoprecipitated with anti-GFP beads and then the immunoprecipitates were assessed by immunoblotting (IB) using anti-Myc (upper panel) and anti-GFP antibodies (middle panel). In addition to immunoblotting for co-IP, presence of NbATG3-Myc in each treatment of the cell lysates was also analyzed (lower panel).
(C) GST pull-down assay for detection of in vitro interaction of ATG3 with GAPCs. The total soluble proteins of E. coli expressing NbGAPC1-3×FLAG-6×His or NbGAPC2-3×FLAG-6×His were incubated with GST-NbATG3 or GST immobilized on glutathione-Sepharose beads. Beads were washed and proteins analyzed by immunoblot assays using an anti-Flag antibody (upper panel). The middle panel shows inputs of Flag-tagged proteins in pull-down assays. Equal aliquots of glutathione beads loaded with GST-NbATG3 or GST were separated by SDS-PAGE and stained with Coomassie blue (CBB).
(D) BiFC assays to show the interaction of ATG3 with GAPCs in plants. NbATG3-nYFP or nYFP was coexpressed transiently with NbGAPC1-cYFP, NbGAPC2-cYFP, or cYFP in N. benthamiana leaves, and fluorescence was detected for mesophyll cells. Combinations of NbATG3-nYFP with NbGAPC1-cYFP or NbGAPC2-cYFP, but not other combinations, gave YFP fluorescence signals (left). Yellow color indicates positive interaction signal, while red color indicates the signals from chloroplasts (middle). The experiments were repeated three times with similar results. Bars = 20 μm.
We further confirmed the interaction of ATG3 with GAPCs using coimmunoprecipitation (co-IP) assays. Myc-tagged ATG3 (NbATG3-Myc) was transiently coexpressed with GFP, GFP-tagged GAPC1 (NbGAPC1-GFP), GAPC2 (NbGAPC2-GFP), or ATG8f (GFP-NbATG8f) in N. benthamiana. Leaf tissues were then collected at 60 h postinfiltration (hpi). Total protein extracts were immunoprecipitated using anti-GFP antibody coupled to agarose beads, and the resulting precipitates were analyzed by immunoblot using anti-Myc antibodies. We observed that ATG3 coimmunoprecipitated with NbGAPC1-GFP, NbGAPC2-GFP, and positive control GFP-NbATG8f, but not with GFP alone (Figure 1B). These experiments, along with our LCI assays, demonstrate that ATG3 interacts with GAPC1 and GAPC2 in N. benthamiana cells.
We also tested whether ATG3 interacted directly with GAPCs by in vitro glutathione S-transferase (GST) pull-down assays. GST-tagged ATG3 (GST-NbATG3) and 3×Flag-6×His double tagged GAPCs (NbGAPC1-3×Flag-6×His or NbGAPC2-3×Flag-6×His) proteins were expressed in Escherichia coli. NbGAPC1-3×Flag-6×His or NbGAPC2-3×Flag-6×His were incubated with glutathione-agarose purified GST-NbATG3 or GST control alone. The pull-down products were separated by SDS-PAGE and subjected to immunoblot assays with anti-Flag antibody. NbGAPC1-3×Flag-6×His or NbGAPC2-3×Flag-6×His was only detected upon incubation with GST-NbATG3, but not with GST protein alone, suggesting that GAPCs directly interact with ATG3 (Figure 1C).
Subcellular localization of the ATG3-GAPC interactions was investigated using bimolecular fluorescence complementation (BiFC) assays (Burch-Smith et al., 2007). GAPC1 and GAPC2 were tagged with the C-terminal half of YFP (cYFP) to generate NbGAPC1-cYFP and NbGAPC2-cYFP, and ATG3 was in-frame fused with the N-terminal half of YFP (nYFP) to generate NbATG3-nYFP. NbATG3-nYFP was transiently coexpressed in N. benthamiana with either NbGAPC1-cYFP or NbGAPC2-cYFP. A positive interaction signal indicated by yellow fluorescence was observed in cytoplasm only when NbATG3-nYFP was combined with NbGAPC1-cYFP or NbGAPC2-cYFP, but not with cYFP alone. No yellow fluorescence was observed when either NbGAPC1-cYFP or NbGAPC2-cYFP was combined with free nYFP alone (Figure 1D). Using BiFC assays, we further confirmed in vivo ATG3-GAPC interactions by expressing ATG3 or GAPCs driven by the native promoter of corresponding gene and found that ATG3 still specifically interacted with GAPC1 or GAPC2 in cytoplasm (Supplemental Figure 3).
Taken together, our results demonstrate that GAPCs interact with ATG3 in vitro and that their in planta interactions occur in cytoplasm.
ROS Affects the Interaction between GAPCs and ATG3 in Plants
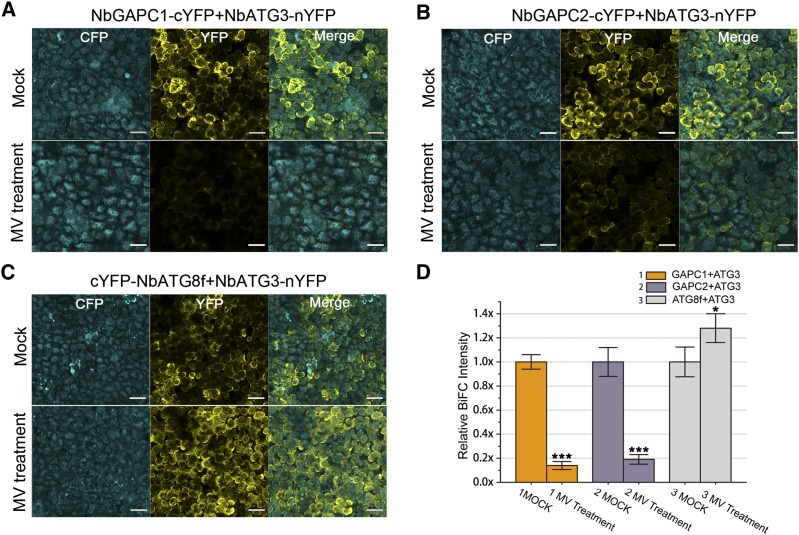
GAPC is a direct regulation target of H2O2 and acts as an H2O2 signaling transducer in plants (Hancock et al., 2005; Guo et al., 2012). We therefore investigated whether ROS could influence the GAPC-ATG3 interactions in N. benthamiana. Because it has been reported that H2O2 diffusion across cell membrane is limited (Bienert et al., 2006) and that exogenous application of H2O2 does not alter the endogenous H2O2 level of plants (Yu et al., 2003), we used MV, a well-known ROS inducer in plants, to generate ROS and then used BiFC to scrutinize the effect of ROS on the GAPC-ATG3 interaction. MV treatment effectively led to production of superoxide anion and H2O2 as indicated by nitroblue tetrazolium and 3,3′-diaminobenzidine tetrahydrochloride (DAB) staining, respectively (Supplemental Figures 4 and 5). We coexpressed NbGAPC1-cYFP, NbGAPC2-cYFP, or cYFP-NbATG8f (as positive control) with NbATG3-nYFP, along with cyan fluorescent protein (CFP) as an internal control, in N. benthamiana leaves and then treated them with 20 μM MV or water (mock). Here, CFP served as an internal normalization for reconstituted YFP fluorescence to eliminate the differences in protein expression of different combinations. As expected, MV led to increased ATG3-ATG8f interaction. However, MV treatment weakened the interaction of ATG3 with either GAPC1 or GAPC2, measured by YFP/CFP ratio using the confocal laser imaging system. We found that MV decreased GAPC-ATG3 interactions by ∼80% compared with the mock treatment of water, while the ATG8f-ATG3 interaction was enhanced by ∼28% (Figure 2).
Figure 2.
BiFC Assays Show That MV Treatments Weaken ATG3-GAPC Interactions.
(A) to (C) NbATG3-nYFP, along with CFP, was coexpressed transiently with NbGAPC1-cYFP (A), NbGAPC2-cYFP (B), or cYFP-NbATG8f ([C]; as a control) in N. benthamiana leaves, followed by mock (water) treatment (upper panel) or treatment with 20 μM MV (lower panel). Yellow color indicates the YFP signal. Cyan color indicates the signal of CFP alone, which was used as an internal control. For a view of a large area, a 10× objective lens was used for confocal image. Bars =50 μm.
(D) BiFC intensity was quantified by YFP/CFP ratio. Relative BiFC intensity was normalized to the control. Values represent means ± se from three independent experiments. Student’s t test was used to determine significant differences from mock (***P < 0.001 and *P < 0.05, Student’s t test).
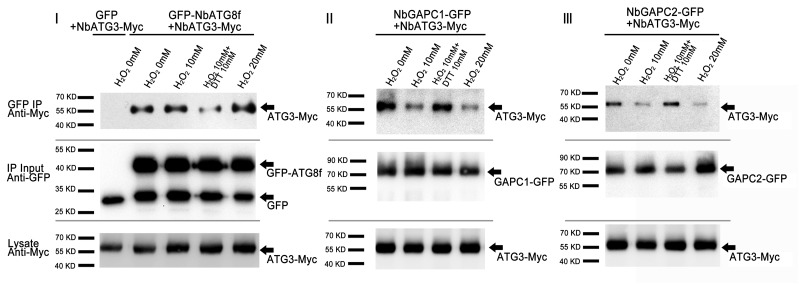
We confirmed the effect of ROS on GAPC-ATG3 interaction using co-IP. For this purpose, we coexpressed NbGAPC1-GFP, NbGAPC2-GFP, or GFP-NbATG8f (as positive control) with NbATG3-Myc. Total protein was extracted separately from the agroinfiltrated leaf tissues, treated with H2O2 with or without reducing reagent DTT, and then immunoprecipitated using anti-GFP beads. The resulting precipitates were analyzed by immunoblot using anti-Myc antibodies. We found that less NbGAPC1-GFP or NbGAPC2-GFP, but more GFP-NbATG8f, was coimmunoprecipitated with NbATG3 in the presence of H2O2 in a dose-dependent manner. However, the addition of DTT, a scavenger of ROS, reversed the effect of H2O2 on the co-IP of NbATG3-Myc with either NbGAPC1-GFP or NbGAPC2-GFP (Figure 3), consistent with previous reports of a direct role for H2O2 in regulating GAPC function (Hancock et al., 2005; Guo et al., 2012).
Figure 3.
Co-IP Assays Show That H2O2 Treatments Weaken ATG3-GAPC Interactions.
Total proteins from N. benthamiana leaves coexpressing NbATG3-Myc with GFP-NbATG8f (I), NbGAPC1-GFP (II), or NbGAPC2-GFP (III) were immunoprecipitated with anti-GFP beads, followed by immunoblotting using anti-Myc (upper panel) and anti-GFP antibodies (middle panel) in different redox environments. Different interaction levels were observed in the water, 10 mM H2O2, 10 mM H2O2 with 10 mM DTT, and 20 mM H2O2 treatments as indicated. DTT was added before the addition of H2O2 when both were applied. The middle panel shows the input of immunoprecipitates using anti-GFP antibodies. The presence of NbATG3-Myc in lysate was also analyzed (bottom panel).
Taken together, these results clearly show that ROS inhibits the interaction between GAPCs and ATG3 in plants.
Silencing of GAPCs Activates Autophagy
To investigate the role of three Nb-GAPC genes (GAPC1, 2, and 3) in autophagy, we silenced individual or multiple GAPCs in N. benthamiana using the Tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) system (Liu et al., 2002b). Real-time RT-PCR indicated that agroinfiltration of VIGS vectors targeting the 3′-untranslated region (UTR) of each GAPC gene caused specific RNA silencing, while cosuppression of GAPC1-3 was triggered by the VIGS vector carrying a tandem sequence of UTRs from GAPC1-3 (Supplemental Figure 6).
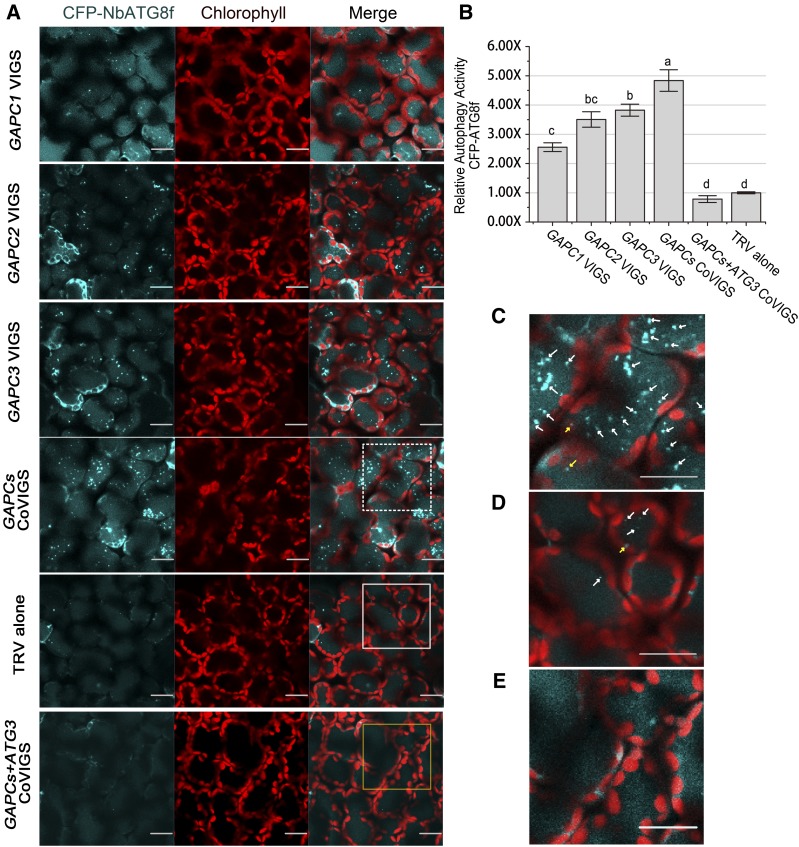
We then examined the effect of three GAPC genes on autophagy using CFP-tagged ATG8f (CFP-NbATG8f), which allows visualization of autophagosomes in the cytoplasm as well as autophagic bodies in the vacuole (Wang et al., 2013). In individually silenced and cosilenced plants, numerous CFP-labeled autophagic bodies accumulated (Figures 4A and 4C). However, only a few CFP-labeled autophagic bodies were seen in the TRV controls (Figures 4A and 4D). The relative autophagy level in GAPCs-silenced plants was 3- to 5-fold of that in the TRV controls (Figure 4B).
Figure 4.
Silencing of GAPCs Activates Autophagy.
(A) Representative confocal images of dynamic autophagic activity revealed by specific autophagy marker CFP-NbATG8f in GAPC-silenced plants (GAPC-CoVIGS) and control plants. Autophagosomes and autophagic bodies are revealed as CFP-positive puncta in mesophyll cells. CFP-NbATG8f fusion proteins are in cyan, and chloroplasts are in red. Bars = 20 μm.
(B) Relative autophagic activity in GAPC-silenced plants was normalized to that of TRV control plants, which was set to 1.0. Quantification of the CFP-NbATG8-labeled autophagic puncta per cell was performed. More than 150 mesophyll cells for each treatment were used for the quantification. Values represent means ± se from three independent experiments. Different letters indicate significant differences (ANOVA, P < 0.05).
(C) to (E) Magnification of the mesophyll cells in (A) surrounded by a dashed line (C), a solid line (D), and a yellow line (E). Autophagosomes labeled by CFP-NbATG8f are indicated by arrows. Yellow arrows indicate autophagosomes in the cytoplasm. White arrows indicate autophagic bodies in the vacuole. Bars = 20 μm.
We further investigated the effect of GAPC1-3 on autophagy by monodansylcadaverine (MDC) staining, a method frequently used to detect autophagic activity in plants and other organisms (Mitou et al., 2009). Here, a large number of MDC-stained structures were seen in plants where GAPC1-3 were either individually silenced or cosilenced, but not in TRV controls (Supplemental Figures 7A to 7D). Consistent with findings from direct visualization of autophagosomes (Figure 4), the relative autophagy level in GAPCs-silenced plants was 3 to ∼6 times more than that in the TRV controls, and cosilencing of GAPC1-3 caused the most autophagy activation (Supplemental Figure 7B).
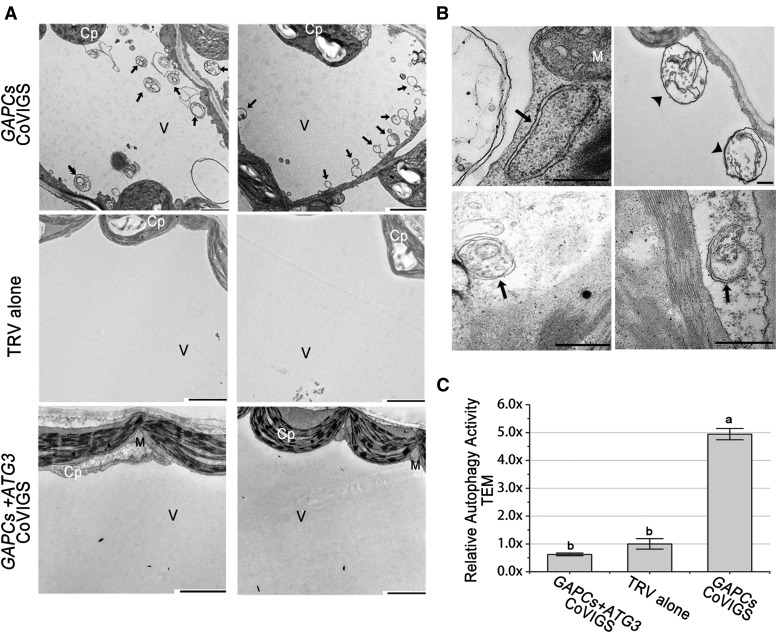
We tested whether induction of autophagy by GAPCs silencing is dependent on ATG3 function through three different experimental approaches, including using the CFP-NbATG8f marker (Figure 4), MDC staining (Supplemental Figure 7), and transmission electron microscopy (TEM) observation (Figure 5). We used VIGS to effectively silence GAPCs and ATG3 (Supplemental Figure 6). We observed a number of autophagic bodies in the GAPCs-silenced plants, but not in GAPCs/ATG3 cosilenced plants or TRV controls (Figures 4 and 5; Supplemental Figure 7).
Figure 5.
Representative TEM Images of Autophagic Structures.
(A) Representative ultrastructure of autophagic bodies (arrows) observed in the vacuoles of mesophyll cells of GAPC-silenced plants (top panels), TRV alone control (middle panels), or plants cosilencing GAPC1-3 and ATG3 (bottom panels). Cp, chloroplast; M, mitochondrion; V, vacuole. Bars = 2 μm.
(B) Representative ultrastructure of autophagosomes observed in the cytoplasm of mesophyll cells. The black arrows show the classic double-membrane autophagosomes, and the black arrowheads indicate the autophagic bodies inside the vacuole. Bars = 500 nm.
(C) Relative autophagic activity in GAPC-silenced plants was normalized to that of TRV control plants, which was set to 1.0. Approximately 20 cells were used to quantify autophagic structures in each treatment. Values represent means ± se from three independent experiments. Different letters indicate significant differences (ANOVA, P < 0.05).
Taken together, our results reveal that silencing of GAPCs can activate ATG3-dependent autophagy.
Expression of GAPCs Inhibits ATG3-Induced Autophagy
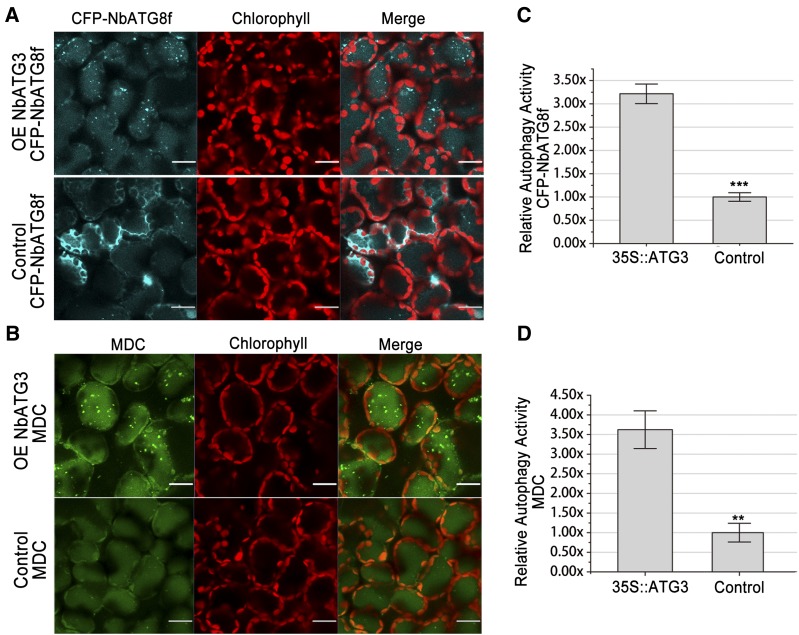
Expression of some ATG genes could activate autophagy (Scott et al., 2007; Pyo et al., 2013). However, the role of Nb-ATG3 in activation of autophagy remains to be elucidated. To achieve this, we expressed ATG3 in N. benthamiana and studied its effect on autophagy using CFP-NbATG8f marker and MDC staining (Figure 6). We noticed few autophagic bodies in leaf tissues agroinfiltrated with empty vector control. However, we found a significantly high level of autophagic body accumulation in leaves where ATG3 was transiently overexpressed (Figure 6). In addition, overexpression of Myc-tagged ATG3 (NbATG3-Myc) also activated autophagy (Figure 7).
Figure 6.
Overexpression of ATG3 Induces Autophagy.
(A) and (B) Representative confocal images of autophagic activity revealed by autophagy marker CFP-NbATG8f (A) or MDC staining (B) in cells overexpressing ATG3 (upper panel) and nLUC control (lower panel). CFP-NbATG8f-labeled punctate autophagosomes and autophagic bodies inside vacuoles (cyan), MDC-labeled punctate autophagosomes and autophagic bodies inside vacuoles (green), autofluorescence from chloroplasts (red), and merged images (merge). Bars = 20 μm.
(C) and (D) Relative autophagic activity revealed by autophagy marker CFP-NbATG8f (C) or MDC staining (D) in leaves overexpressing ATG3 was normalized to that of leaves overexpressing nLUC. Quantification of the CFP-NbATG8-labeled autophagic puncta or MDC-stained structures per cell was performed to calculate the autophagic activity using more than 150 mesophyll cells for each treatment. Values represent means ± se from three independent experiments (***P <0.001 and ** P < 0.01, Student’s t test).
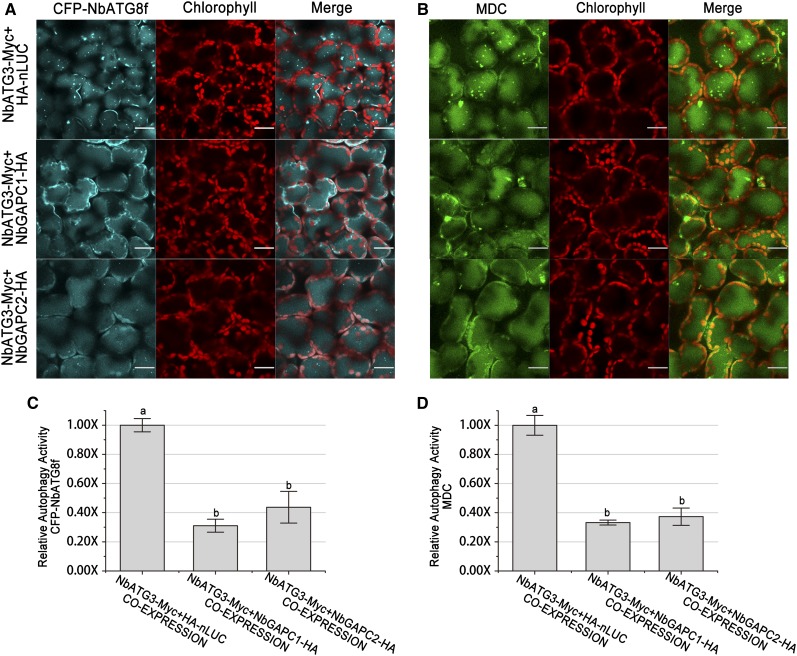
Figure 7.
Visualization of Autophagy Level in ATG3/GAPC Co-Overexpressing Leaves.
(A) and (B) Representative images of autophagic activity revealed by autophagy marker CFP-NbATG8f (A) or MDC staining (B) in leaves coexpressing NbATG3-Myc with HA-nLUC (upper), NbGAPC1-HA (middle), or NbGAPC2-HA (bottom). CFP-NbATG8f fusion proteins are in cyan, MDC are in green, and chloroplasts are in red. Bars = 20 μm.
(C) and (D) Relative autophagic activity revealed by autophagy marker CFP-NbATG8f (C) or MDC staining (D) in leaves coexpressing NbATG3-Myc with NbGAPC1-HA or NbGAPC2-HA was normalized to that of leaves coexpressing NbATG3-Myc and HA-nLUC. Quantification of the CFP-NbATG8-labeled autophagic puncta per cell (C) and MDC positive structures per cell (D) were performed to calculate the autophagic activity. More than 150 mesophyll cells for each treatment were used for the quantification. Values represent means ± se from three independent experiments. Different letters indicate significant differences (ANOVA, P < 0.05).
To further confirm the involvement of GAPCs in ATG3-related autophagy, we coexpressed ATG3 along with GAPC1, GAPC2, or nLUC. Expression of ATG3, GAPC1, or GAPC2 in agroinfiltrated leaf tissues was verified by immunoblots (Supplemental Figure 8A). Overexpression of ATG3 together with nLUC control was able to induce obvious autophagic body accumulation. However, ATG3 induced much less autophagy if GAPCs were coproduced in the same plant tissues (Figure 7). Compared with the HA-nLUC negative control, GAPC1 or GAPC2 caused 2- to ∼3-fold reduction of the ATG3-induced autophagic body accumulation as assayed by both CFP-NbATG8f marker and MDC staining methods (Figure 7). These results reveal that overexpression of GAPCs leads to suppression of ATG3-induced autophagy.
GAPCs Inhibit ROS-Induced Autophagy in Plants
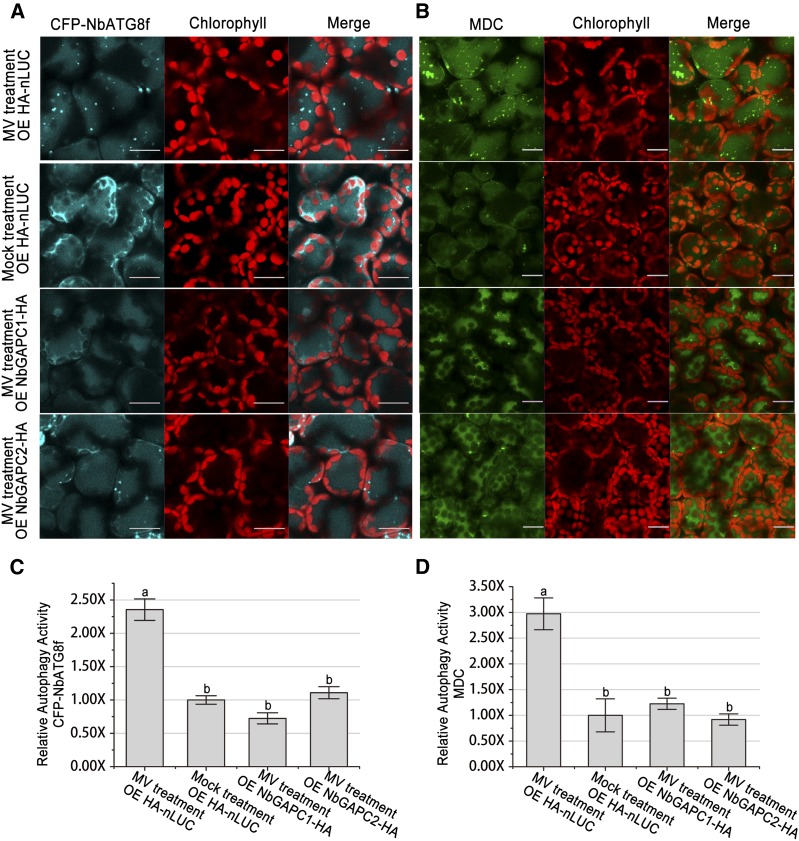
ROS affects the interaction between GAPCs and ATG3 (Figure 2), indicating that GAPCs could influence ROS-mediated autophagy. To test this idea, we used the ROS inducer MV to trigger autophagy (Xiong et al., 2007b) and investigated whether GAPCs indeed affect MV-induced autophagy. GAPC1, GAPC2, or nLUC was expressed in leaves of N. benthamiana, followed by treatment of 10 μM MV or water under a 48-h period of constant light. Immunoblot assays showed that all transgenes were expressed in the agroinfiltrated leaf tissues (Supplemental Figure 8B). As expected, autophagy was induced significantly in nLUC/MV, but not in nLUC/H2O combinations, with a 2.36- and 2.97-fold difference as indicated by CFP-NbATG8f marker and MDC staining, respectively (Figure 8). Interestingly, overexpression of either GAPC1 or GAPC2 inhibited MV-induced autophagy, and the level of MV-induced autophagy was reduced by 50 to 70% compared with that of nLUC controls (Figure 8), demonstrating that GAPCs are able to inhibit ROS-induced autophagy in plants.
Figure 8.
Overexpression of GAPCs Inhibits MV-Induced Autophagy.
(A) and (B) Representative images of autophagic activity revealed by autophagy marker CFP-NbATG8f (A) or MDC staining (B) in leaves expressing NbGAPC1-HA, NbGAPC2-HA, or HA-nLUC, followed by treatment with 10 μM MV or mock (water). Numerous CFP-NbATG8f-labeled positive puncta or MDC-stained structures were observed in leaves expressing HA-nLUC followed by MV treatment, but not in other leaves. CFP-NbATG8f fusion proteins are in cyan, MDC is in green, and chloroplasts are in red. Bars = 20 μm.
(C) and (D) Relative autophagic activity revealed by autophagy marker CFP-NbATG8f (C) or MDC staining (D) in leaves expressing NbGAPC1-HA, NbGAPC2-HA, or HA-nLUC, followed by treatment with 10 μM MV was normalized to that of leaves expressing HA-nLUC followed by mock treatment. Quantification of the CFP-NbATG8-labeled autophagic puncta per cell (C) and MDC-positive structures per cell (D) was performed to calculate the autophagic activity. More than 150 mesophyll cells for each treatment were used for the quantification. Values represent means ± se from three independent experiments. Different letters indicate significant differences (ANOVA, P < 0.05).
GAPCs Are Involved in Plant Immunity
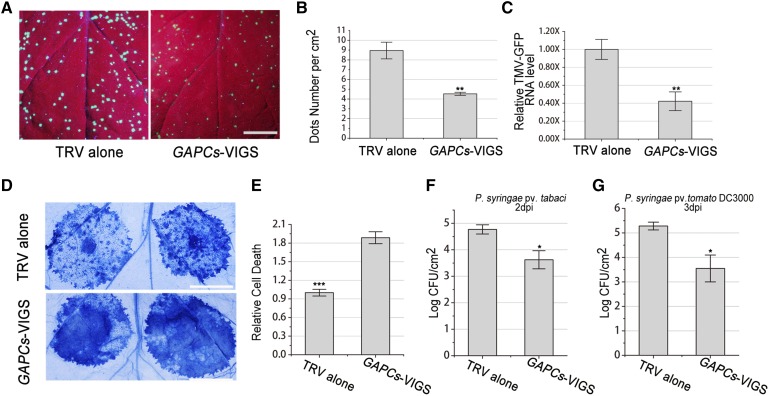
Autophagy plays an essential role in plant immunity (Liu et al., 2005). This led us to hypothesize that GAPCs may also affect innate immunity. We therefore challenged the GAPC-silenced N gene-containing N. benthamiana (designated NN) plants with GFP-tagged Tobacco mosaic virus (TMV-GFP). At 3 d postinfection (dpi), the number of TMV-GFP local infection loci showing GFP fluorescence, visualized under long-wavelength UV light in the GAPC-silenced NN, was reduced by ∼50% compared with that in TRV control plants (Figures 9A and 9B). Real-time PCR showed that TMV-GFP RNA level in GAPC-silenced NN plants was only 40% of the control plants (Figure 9C). Taken together, these findings reveal that VIGS of GAPCs reduced TMV viral RNA accumulation and NN susceptibility to TMV infection. Furthermore, we analyzed the effect of silencing of GAPCs on N-mediated hypersensitive response (HR) induced by N gene elicitor protein, TMV-P50. At 48 hpi, HR cell death in GAPC-silenced NN plants was enhanced compared with control NN plants indicated by trypan blue staining (Figures 9D and 9E). These results suggested that GAPCs negatively regulate N gene-mediated resistance against TMV.
Figure 9.
Silencing of GAPCs Enhances Resistance and Hypersensitive Response.
(A) to (C) Silencing of Nb-GAPCs (GAPCs-VIGS) enhanced N gene-mediated resistance against TMV.
(A) Representative photographs of TMV-GFP-inoculated leaves were taken at 3 dpi under UV light to assess TMV infection in nonsilenced TRV control (left) or GAPC-silenced plants (right). Bars = 1 cm.
(B) Average TMV-GFP infection foci at 3 dpi are shown. All values in bar graphs represent means with sd (**P < 0.01, Student’s t test, n = 3). The experiments were repeated three times with similar results.
(C) Real-time RT-PCR confirmed that silencing of GAPCs reduced TMV-GFP RNA levels in local inoculated leaves at 3 dpi.
(D) Silencing of GAPCs accelerated TMV-p50-induced hypersensitive response cell death in NN plants. TMV-p50 was expressed by agroinfiltration in nonsilenced TRV control (upper) or GAPCs-silenced plants (lower). Representative photographs were taken at 2 dpi after trypan blue staining. Bars = 1 cm.
(E) Quantitative representation of TMV-p50-induced HR programmed cell death. For quantification of death intensity, images were converted to gray scale, and mean gray value of inoculation area minus noninoculation area was calculated using ImageJ. Values represent means ± se (***P < 0.001, Student’s t test, n = 12). The experiments were repeated three times with similar results.
(F) Silencing of GAPCs inhibited the growth of compatible pathogen P. syringae pv tabaci. Control plants (TRV alone) and GAPC-silenced plants (GAPCs-VIGS) were infiltrated with P. syringae pv tabaci at 103 cfu/mL, and bacterial populations were quantified at 2 dpi (*P < 0.05, Student’s t test, n = 5). The experiments were repeated three times with similar results.
(G) Silencing of GAPCs inhibited the growth of incompatible pathogen P. syringae pv tomato DC3000. Control plants (TRV alone) and GAPCs-silenced plants (GAPCs-VIGS) were infiltrated with P. syringae pv tomato DC3000 at 103 cfu/mL, and bacterial populations were quantified at 3 dpi (*P < 0.05, Student’s t test, n = 3). The experiments were repeated twice with similar results.
Furthermore, we investigated whether GAPCs would have a general role in plant immunity against bacterial pathogens. In this regard, we tested whether GAPCs silencing has an effect on a compatible pathogen, Pseudomonas syringae pv tabaci, as well as an incompatible pathogen, P. syringae pv tomato (Pst) strain DC3000, in N. benthamiana plants. We found that the growth of P. syringae pv tabaci was reduced ∼14-fold at 2 dpi in GAPC-silenced plants compared with that in control plants (Figure 9F). We also found that the growth of Pst DC3000 was reduced ∼54-fold at 3 dpi in GAPC-silenced plants compared with that in control plants (Figure 9G). However, we did not find an obvious effect of GAPCs silencing on HR cell death induced by Pst DC3000 (Supplemental Figure 9).
Taken together, these results suggest that GAPCs function in both R gene-mediated defense and basal defense.
DISCUSSION
In this study, we found that N. benthamiana GAPC proteins, herein established as ATG3-interacting proteins, negatively regulate autophagy. Intriguingly, the interaction of GAPCs with ATG3 is regulated by ROS. Furthermore, we found that N. benthamiana GAPCs play an essential role in both effector-triggered immunity and basal defense.
Role of GAPC-ATG3 Interaction in Autophagy
ATG3 is a key autophagy component that participates in the ATG8-PE conjugation, an important step of autophagy in yeast and mammals. In plants, silencing of ATG3 also compromises autophagy (Wang et al., 2013). Human ATG3 interacts with ATG12 and its overexpression increases ATG5-ATG12 conjugation (Tanida et al., 2002). In addition, overexpression of ATG5 in mice activates autophagy and extends lifespan (Pyo et al., 2013). Here, we further found that overexpression of ATG3 induced autophagy in plants (Figure 6). Plant ATG3 may induce autophagy by increasing ATG8/LC3 lipidation and Atg5-Atg12 conjugation, similar to mammalian ATG3. Furthermore, a similar level of autophagy (∼3.2 times) was induced by overexpression of wild-type ATG3 and C-terminal Myc-tagged ATG3 (Figure 7), suggesting that C-terminal tagged ATG3 functions properly.
Autophagy is involved in a broad range of physiological processes, and its biological significance is explained by its crosstalk with various cellular pathways through interactions among key gene components. The role of ATG3 in autophagy can be regulated by its interacting partners. In yeast, Atg3 can interact with Esa1 and Rpd3 to serve as an acetylation substrate to regulate autophagy (Yi et al., 2012). In mammals, FNBP1L/ATG3 interaction is essential for the antibacterial autophagy against the intracellular pathogen Salmonella enterica serovar Typhimurium (Huett et al., 2009). The protein vFLIP encoded by Human herpesvirus 8 (HHV-8), a viral counterpart of cellular FLIP, interacts with ATG3 to repress the cellular antiviral autophagic mechanism (Lee et al., 2009). However, in plants no ATG3 binding protein has been experimentally documented. In this study, we report that Nb-GAPCs can interact with Nb-ATG3 and negatively regulate autophagy.
Nb-GAPC-silenced plants did not show obvious developmental defects, consistent with the finding that single and double GAPC-knockout Arabidopsis lines grow normally (Guo et al., 2012, 2014). However, silencing of Nb-GAPCs activates ATG3-dependent autophagy (Figures 4 and 5; Supplemental Figure 7). Furthermore, overexpression of Nb-GAPCs inhibits autophagy induced by Nb-ATG3 (Figure 7). This suggests that GAPCs negatively regulate autophagy through direct interactions with ATG3. Interestingly, silencing of single GAPC genes activates autophagy (Figure 4), despite the fact that all GAPCs are localized in cytoplasm and share high sequence conservation. ATG3 interacts with all GAPCs (Figure 1), while overexpression of either GAPC1 or GAPC2 inhibits autophagy (Figures 7 and 8), suggesting that the multiple cytoplastic GAPDH isoforms play a similar role in the regulation of autophagy. This regulation process could occur in a sensitive and accurate manner and involve the total amount of GAPCs, and any interference with the balance of GAPCs pools may lead to autophagy activation. In addition, simultaneous silencing of GAPC1-3 caused the strongest activation of autophagy, (Figure 4; Supplemental Figure 7). These data indicate that the three GAPC proteins may be functionally synergistic, to some extent, in the control of plant autophagy.
GAPDH has also been reported to function in oxidative stress responses besides its biochemical housekeeping function in glycolysis. In mammalian cells, GAPDH acts as a key mediator of many oxidative stress responses, involving GAPDH nuclear translocation and induction of cancer cell senescence (Hwang et al., 2009; Nicholls et al., 2012). Interestingly, GAPDH interacts with mammalian RNA binding protein p54nrb (54-kD nuclear RNA binding protein) in the cytosol in a manner dependent on the dose of H2O2 to exert a potential role in transcription or replication (Hwang et al., 2009). In plants, GAPCs are identified as the direct inhibition targets of H2O2 (Hancock et al., 2005; Holtgrefe et al., 2008; Zaffagnini et al., 2013). GAPCs could also respond to ROS signals by regulating their interacting partner proteins. In Arabidopsis, H2O2 promotes the interaction of GAPCs with phospholipase D to transduce H2O2 signals in the response to ROS and water stress (Guo et al., 2012). Here, we found that both MV and H2O2 weakened the interaction between ATG3 and GAPCs, but enhanced the ATG3-ATG8 interaction in N. benthamiana (Figures 2 and 3). It is possible that ROS regulates cellular functions by altering the interaction of GAPC-ATG3 at different redox status.
ROS, such as H2O2, as well as ROS inducers, including MV, can induce autophagy in mammalian cells and Arabidopsis plants (Kunchithapautham and Rohrer, 2007; Scherz-Shouval et al., 2007; Xiong et al., 2007b; Chen et al., 2009; Pérez-Pérez et al., 2012). In this study, we found that MV can induce autophagy in N. benthamiana plants (Figure 8). In Arabidopsis, cells exploit autophagy to remove ROS-damaged proteins (Xiong et al., 2007b). However, under oxidative stress, how autophagy is induced to eliminate the oxidized proteins remains unclear. It is possible that ROS stress induces autophagy by specifically changing the status of GAPCs. Indeed, overexpression of GAPCs inhibits MV-induced autophagy (Figure 8). H2O2 and MV weaken the interaction of ATG3 with GAPCs (Figures 2 and 3), while MV treatment did not have an effect on expression of ATG3 during the 3-d time analyzed (Supplemental Figure 10). Under oxidative stress, there may be more free ATG3 proteins available for use in autophagy.
Roles of GAPCs in Plant Defense
In mammals, GAPDH is a key cell death regulator. S-nitrosylated GAPDH binds Siah1 to induce nuclear translocation and initiate apoptotic cell death (Hara et al., 2005). Overexpression of GAPDH inhibits caspase-independent cell death by stimulating glycolysis activity and autophagy (Colell et al., 2007). GAPDH also functions as a pro-survival factor in other cellular processes, such as DNA repair, cell cycle progression, and mRNA binding and stability (Chambers et al., 1997; Carujo et al., 2006; Azam et al., 2008; Colell et al., 2009; Lavallard et al., 2009). Recently, a chloroplast-located isoform GAPDH-A has been reported to recruit a movement protein of Red clover necrotic mosaic virus to facilitate viral cell-to-cell movement (Kaido et al., 2014). Here, we report that GAPCs negatively regulate immunity-associated cell death and defense in plants. Silencing of GAPCs reduced TMV accumulation in N gene-containing plants and enhanced N gene-mediated HR cell death (Figures 9A to 9E). Enhanced N gene-mediated HR cell death in GAPC-silenced plants may be caused by enhanced autophagy, since autophagy is reported to participate in the execution of immunity-related cell death within the infection site (Hofius et al., 2009; Munch et al., 2014) and excess autophagy can cause cell death after pathogen treatments (Kwon et al., 2013). Reduced TMV accumulation in GAPC-silenced plants seems to be R gene dependent and associated with the enhanced autophagy. This is supported by that no effect of silencing of GAPCs on TMV infection was observed in wild-type N. benthamiana plants (data not shown) and that disruption of autophagy caused more TMV accumulation in N gene-containing plants (Liu et al., 2005). Furthermore, silencing of GAPCs enhanced plant resistance against both compatible pathogen P. syringae pv tabaci and incompatible pathogen Pst DC3000 (Figures 9F and 9G). Given that autophagy was constitutively activated when GAPCs were silenced in plants, autophagy could positively contribute to plant basal defense and N gene-mediated cell death through negative regulation of GAPCs. ROS and autophagy have been associated with plant defense and cell death, and GAPCs are also shown to be involved in the regulation of ROS-mediated autophagy and defense in this study. The mechanistic link between the enhanced defense and ROS-induced autophagy awaits further investigation. Considering that silencing of GAPCs activates autophagy but has no effect on HR cell death induced by Pst DC3000 in N. benthamiana plants (Supplemental Figure 9), it is also possible that GAPCs have a direct function in plant immunity, independent of their function in autophagy.
METHODS
Plant Materials
Wild-type Nicotiana benthamiana NN transgenic plants were as described (Liu et al., 2002b). N. benthamiana plants were grown in pots placed in growth rooms at 25°C under a 16-h-light/8-h-dark cycle.
Plasmid Constructs
Full-length Nb-ATG3 cDNA was PCR amplified and cloned into NdeI-XhoI-digested pGEX4T-1 vector to express GST-tagged fusion proteins in Escherichia coli. Full-length cDNA of Nb-GAPC1 and Nb-GAPC2 was individually cloned into BamHI-XhoI-digested pET28a to express the double-tagged NbGAPC1-3×FLAG-6×His and NbGAPC2-3×FLAG-6×His in E. coli.
DNA fragments for NbATG3-GFP, NbATG3-nLUC, NbATG3-nYFP, NbATG3-Myc, GFP-NbGAPC3, NbGAPC1-GFP, NbGAPC1-cYFP, NbGAPC1-HA, cLUC-NbGAPC1, YFP-NbGAPC1, NbGAPC2-GFP, NbGAPC2-cYFP, NbGAPC2-HA, cLUC-NbGAPC2, YFP-NbGAPC2, cLUC-NbGAPC3, cYFP-NbATG8f, and GFP-NbATG8f were obtained by overlapping PCR. The resulting PCR products and full-length Nb-ATG3 cDNA were cloned between the duplicated CaMV 35S promoter and the NOS terminator of pJG045, a pCAMBIA1300-based T-DNA vector (Du et al., 2013). HA-nLUC and cLUC driven by CaMV 35S promoter were described previously (Du et al., 2013).
Vectors pTRV1 (Liu et al., 2002a) and pTRV2-LIC (Dong et al., 2007) were described previously. pTRV2-NbGAPC1, pTRV2-NbGAPC2, and pTRV2-NbGAPC3 were generated by cloning PCR products of 3′-UTR fragments of Nb-GAPC1, Nb-GAPC2, and Nb-GAPC3 into pTRV2-LIC for VIGS. Two fusion DNA fragments NbGAPC1-3 and NbGAPC1-3+NbATG3 were obtained by overlapping PCR and cloned into pTRV2-LIC to generate pTRV2-GAPCs for cosilencing GAPC1-3 and pTRV2-NbGAPCs+NbATG3 for cosilencing GAPC1-3 together with ATG3.
About 2 kb of DNA upstream of the start codon of Nb-ATG3, Nb-GAPC1, or Nb-GAPC2 was used as the native promoter of each corresponding gene and named ATG3PRO, GAPC1PRO, and GAPC2PRO. These promoters were PCR amplified from N. benthamiana genomic DNA to replace the CaMV 35S promoter in BiFC vectors containing NbATG3-nYFP, NbGAPC1-cYFP, NbGAPC2-cYFP, nYFP, or cYFP to generate ATG3PRO:ATG3-nYFP, ATG3PRO:nYFP, GAPC1PRO:GAPC1-cYFP, GAPC1PRO:cYFP, GAPC2PRO:GAPC2-cYFP, and GAPC2PRO:cYFP.
All constructs were confirmed by DNA sequencing. Primers used for plasmid construction in this study are listed in Supplemental Table 1.
Mass Spectrometry Analysis
Total protein was extracted from 1 g of N. benthamiana leaves agroinfiltrated with ATG3-GFP or GFP, driven by CaMV 35S promoter, with extraction buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 10% [v/v] glycerol, 0.5% [v/v] Nonidet P-40, and 1× protease inhibitor cocktail [Roche]). The total protein extracts were concentrated and incubated with GFP-Trap_A beads (ChromoTek) for 3 h at 4°C with gentle rocking. After washing, the purified GFP-tagged ATG3 proteins were denatured at 98°C and separated by SDS-PAGE. The differentially displayed gel bands were excised, reduced with 10 mM DTT, and alkylated with 55 mM iodoacetamide (Calbiochem). Then, in-gel digestion was performed with trypsin (Promega) in 50 mM ammonium bicarbonate at 37°C overnight. The peptides were extracted twice with 1% (v/v) trifluoroacetic acid in 50% (v/v) acetonitrile aqueous solution for 30 min.
For LC-MS/MS analysis, peptides were separated by a 60-min gradient elution at a flow rate 0.30 μL/min with a Thermo-Dionex Ultimate 3000 HPLC system, which was directly interfaced with a Thermo LTQ-Orbitrap Velos pro mass spectrometer. The analytical column was a fused silica capillary column (Zorbax 300SB-C18, 3.5 μm 150 mm × 75 μm; Agilent). Mobile phase A consisted of 0.1% formic acid, and mobile phase B consisted of 80% acetonitrile and 0.08% formic acid. The LTQ-Orbitrap mass spectrometer was operated in the data-dependent acquisition mode using Xcalibur 2.0.7 software with a single full-scan mass spectrum in the Orbitrap (400 to 1500 m/z; 30,000 resolution) followed by 20 data-dependent MS/MS scans in an ion trap at 35% normalized collision energy. MS/MS spectra from each LC-MS/MS run were searched against the selected database using the Proteome Discoverer (version 1.4) searching algorithm.
LCI Assays
LCI assays were performed as described (Du et al., 2013). All combinations tested were agroinfiltrated into leaves of N. benthamiana. The leaves were detached 60 hpi, sprayed with 1 mM luciferin, and observed under a low-light cooled CCD imaging apparatus (iXon; Andor Technology). The pictures were taken 5 min after exposure.
Protein Analyses and Co-IP
For protein analysis, total proteins were extracted from N. benthamiana leaves with a ratio of 1:1 of 2× Laemmli buffer. After boiling for 10 min, protein extracts were separated by SDS-PAGE for immunoblot analysis using indicated antibodies. For co-IP assays, total proteins from N. benthamiana leaves (∼1 g leaf tissues for each sample) were extracted in ice-cold immunoprecipitation buffer (10% [v/v] glycerol, 25 mM Tris, pH 7.5, 150 mM NaCl, 1× protease inhibitor cocktail [Roche], and 0.15% [v/v] Nonidet P-40). Protein extracts were incubated with GFP-Trap_A (ChromoTek) beads for 3 h at 4°C. The precipitations were washed four times with ice-cold immunoprecipitation buffer at 4°C and were analyzed by immunoblot using anti-Myc (Abmart), anti-HA (Cell Signaling Technology), or anti-GFP (ChromoTek) antibodies.
GST Pull-Down Assays
The GST pull-down assays were performed as described previously (Zhao et al., 2013). GST-NbATG3 and NbGAPC1-3×Flag-6×His or NbGAPC2-3×Flag-6×His fusion proteins were produced in BL21 (DE3) cells (Stratagene). GST-NbATG3 was purified using glutathione-Sepharose beads (GE Healthcare) according to the manufacturer’s instructions. About 1 mg of purified GST fusion proteins or GST was incubated for 3 h with NbGAPC1-3×Flag-6×His or NbGAPC2-3×Flag-6×His at 4°C in 1 mL of buffer A (100 mM NaCl, 50 mM Tris-Cl, pH 7.5, 0.1 mM EDTA, 0.1 mM EGTA, 0.2% Triton X-100, 0.1% β-mercaptoethanol, 1 mM PMSF, and complete protease inhibitor). The beads were washed three times with ice-cold buffer B (100 mM NaCl, 50 mM HEPES, pH 7.5, 0.1 mM EDTA, 1 mM PMSF, and complete protease inhibitor) at 4°C. The washed beads were boiled in 1× SDS sample buffer, and proteins were separated by SDS-PAGE for immunoblot assays using the anti-FLAG antibody.
Confocal Microscopy and TEM
Confocal imaging was performed using an inverted Zeiss LSM 710 three-channel laser scanning microscope (Carl-Zeiss). For MDC staining, the leaves were infiltrated with 20 μM E-64d (Sigma-Aldrich) for 8 h. Leaves were immediately vacuum infiltrated with 50 μM MDC (Sigma-Aldrich) for 10 min, followed by two washes with PBS buffer before observation. MDC-incorporated structures were excited by a wavelength of 405 nm and detected at 410 to 585 nm; chloroplast autofluorescence was excited at 405 nm and detected at 635 to 708 nm.
Agrobacterium tumefaciens harboring autophagy marker CFP-NbATG8f was infiltrated into leaves for a 60-h expression, and the samples underwent additional infiltration with the protease inhibitor 20 μM E-64d for 8 h before being monitored by a Zeiss LSM 710 three-channel microscope with an excitation light of 405 nm, and the emission was captured at 454 to 581 nm.
TEM observation was performed as described previously (Wang et al., 2013). Leaves pretreated with 20 μM E-64d for 8 h were cut into small pieces (∼1 × 2mm) and infiltrated with 2.5% glutaraldehyde fixation in 0.1 M PBS buffer for fixation. The samples were then postfixed in OsO4, dehydrated in ethanol, and embedded in Epon 81. The sections (∼70 nm) were cut with a diamond knife on an ultramicrotome (Leica EM UC6). The ultrathin sections were collected on 3-mm copper (mesh) grids and stained with uranyl acetate and lead citrate before final examination on an electron microscope (Hitachi H-7650).
VIGS Assay, Virus Infection, and GFP Imaging
For VIGS assays, pTRV1 and pTRV2 or its derivatives were introduced into Agrobacterium strain GV3101. VIGS assays were performed as described (Liu et al., 2005). GFP-tagged TMV (TMV-GFP) sap was prepared from the infiltrated leaves of N. benthamiana plants infiltrated with Agrobacterium containing TMV-GFP plasmid pSPDK661 (Liu et al., 2002a). GFP was imaged under long-wavelength UV light, and photographs were taken using a Canon 650D camera.
RT-PCR and Real-Time RT-PCR
Total RNA was extracted from leaves of N. benthamiana plants using TRNzol-A+ solution (Tiangen Biotech) and treated with RNase-free DNase I (Sigma-Aldrich) to remove potential DNA contamination. First-strand cDNA was synthesized using 2 μg of total RNA, oligo(dT)15, or TMV-specific primers and M-MuLV reverse transcriptase according to the manufacturer’s protocol (MBI). Real-time RT-PCR was performed as described (Wang et al., 2013). Primers used for RT-PCR are listed in Supplemental Table 2.
Bacterial Strain Growth
For pathogen growth assays, the compatible pathogen Pseudomonas syringae pv tabaci and incompatible pathogen Pst DC3000 were resuspended in 10 mM MgCl2 (10,000-fold dilution from OD600 = 0.1, ∼3.75 × 103 colony-forming units [cfu]/mL) and infiltrated into N. benthamiana leaves. Four discs of 6-mm diameter were collected per sample at the indicated time point. Leaf samples were ground, subjected to serial dilution, plated on King’s B agar medium supplemented with appropriate antibiotics, and incubated at 28°C for 2 d for bacterial colony counting, as previously described (Rojas et al., 2012). For Pst DC3000-induced HR cell death assays, the leaves of GAPC-silenced and control N. benthamiana plants were inoculated with Pst DC3000 at 107 cfu/mL, and HR was observed at 15 hpi.
Trypan Blue Staining
Lactophenol-trypan blue staining and destaining with chloral hydrate were performed as described earlier (Kamoun et al., 1998).
DAB Staining
DAB (Sigma-Aldrich) staining was performed using a previously described method (Daudi et al., 2012).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Nb-GAPC1 (KM986323), Nb-GAPC2 (KM986324), and Nb-GAPC3 (KM986325).
Supplemental Data
Supplemental Figure 1. Alignment of the Nb-GAPC amino acid sequences with their Arabidopsis homologs.
Supplemental Figure 2. Subcellular localization of YFP-NbGAPCs and GFP-NbATG3 in N. benthamiana epidermal cells.
Supplemental Figure 3. BiFC assays show ATG3-GAPC interactions when they are expressed under control of their native promoters.
Supplemental Figure 4. NBT staining indicates that MV concentration is positively correlated with superoxide anion accumulation.
Supplemental Figure 5. DAB staining indicates that MV concentration is positively correlated with H2O2 accumulation.
Supplemental Figure 6. Real-time RT-PCR to confirm VIGS efficiency.
Supplemental Figure 7. MDC staining indicates that silencing of GAPCs activates autophagy.
Supplemental Figure 8. Protein expression tested by immunoblot.
Supplemental Figure 9. Silencing of GAPCs does not have an obvious effect on HR cell death induced by nonhost pathogen Pst DC3000.
Supplemental Figure 1. MV application does not affect mRNA level of ATG3.
Supplemental Table 1. Primers used in generating constructs.
Supplemental Table 2. Primers used in RT-PCR.
Supplementary Material
Acknowledgments
We thank Haiteng Deng for help with mass spectrometry and Jian-Min Zhou for providing P. syringae strains. This work was supported by the National Basic Research Program of China (2011CB910100 and 2014CB138400), the National Natural Science Foundation of China (31421001, 31470254, 31270182, and 31370180), and the National Transgenic Program of China (2014ZX08009-003 and 2014ZX0800104B).
AUTHOR CONTRIBUTIONS
Y.L., S.H., and Y.W. initiated the project and designed the experiments. Y.L. and S.H. wrote the article. S.H., X.Z., Y.W. Q.J., J.Z., and F.B. performed the research. Y.L., S.H., Y.W., and X.Z. analyzed the data. Y.H. discussed and revised the article.
Glossary
- ROS
reactive oxygen species
- MV
methyl viologen
- GAPC
cytosolic glyceraldehyde-3-phosphate dehydrogenase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- CaMV
cauliflower mosaic virus
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- LCI
luciferase complementation imaging
- co-IP
coimmunoprecipitation
- hpi
hours postinfiltration
- BiFC
bimolecular fluorescence complementation
- DAB
3,3′-diaminobenzidine
- TRV
Tobacco rattle virus
- VIGS
virus-induced gene silencing
- UTR
untranslated region
- MDC
monodansylcadaverine
- TEM
transmission electron microscopy
- TMV
Tobacco mosaic virus
- dpi
days postinfection
- HR
hypersensitive response
- Pst
Pseudomonas syringae pv tomato
- cfu
colony-forming units
References
- Azad M.B., Chen Y., Gibson S.B. (2009). Regulation of autophagy by reactive oxygen species (ROS): implications for cancer progression and treatment. Antioxid. Redox Signal. 11: 777–790. [DOI] [PubMed] [Google Scholar]
- Azam S., Jouvet N., Jilani A., Vongsamphanh R., Yang X., Yang S., Ramotar D. (2008). Human glyceraldehyde-3-phosphate dehydrogenase plays a direct role in reactivating oxidized forms of the DNA repair enzyme APE1. J. Biol. Chem. 283: 30632–30641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae B.I., Hara M.R., Cascio M.B., Wellington C.L., Hayden M.R., Ross C.A., Ha H.C., Li X.J., Snyder S.H., Sawa A. (2006). Mutant huntingtin: nuclear translocation and cytotoxicity mediated by GAPDH. Proc. Natl. Acad. Sci. USA 103: 3405–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D.C. (2007). Plant autophagy—more than a starvation response. Curr. Opin. Plant Biol. 10: 587–593. [DOI] [PubMed] [Google Scholar]
- Bienert G.P., Schjoerring J.K., Jahn T.P. (2006). Membrane transport of hydrogen peroxide. Biochim. Biophys. Acta 1758: 994–1003. [DOI] [PubMed] [Google Scholar]
- Burch-Smith T.M., Schiff M., Caplan J.L., Tsao J., Czymmek K., Dinesh-Kumar S.P. (2007). A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 5: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carujo S., Estanyol J.M., Ejarque A., Agell N., Bachs O., Pujol M.J. (2006). Glyceraldehyde 3-phosphate dehydrogenase is a SET-binding protein and regulates cyclin B-cdk1 activity. Oncogene 25: 4033–4042. [DOI] [PubMed] [Google Scholar]
- Chambers S.K., Kacinski B.M., Ivins C.M., Carcangiu M.L. (1997). Overexpression of epithelial macrophage colony-stimulating factor (CSF-1) and CSF-1 receptor: a poor prognostic factor in epithelial ovarian cancer, contrasted with a protective effect of stromal CSF-1. Clin. Cancer Res. 3: 999–1007. [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Azad M.B., Gibson S.B. (2009). Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 16: 1040–1052. [DOI] [PubMed] [Google Scholar]
- Chen Y., McMillan-Ward E., Kong J., Israels S.J., Gibson S.B. (2007). Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J. Cell Sci. 120: 4155–4166. [DOI] [PubMed] [Google Scholar]
- Colell A., Green D.R., Ricci J.E. (2009). Novel roles for GAPDH in cell death and carcinogenesis. Cell Death Differ. 16: 1573–1581. [DOI] [PubMed] [Google Scholar]
- Colell A., et al. (2007). GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell 129: 983–997. [DOI] [PubMed] [Google Scholar]
- Daudi A., Cheng Z., O’Brien J.A., Mammarella N., Khan S., Ausubel F.M., Bolwell G.P. (2012). The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell 24: 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277: 33105–33114. [DOI] [PubMed] [Google Scholar]
- Dong Y., Burch-Smith T.M., Liu Y., Mamillapalli P., Dinesh-Kumar S.P. (2007). A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and -2 in floral development. Plant Physiol. 145: 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Zhao J., Chen T., Liu Q., Zhang H., Wang Y., Hong Y., Xiao F., Zhang L., Shen Q., Liu Y. (2013). Type I J-domain NbMIP1 proteins are required for both Tobacco mosaic virus infection and plant innate immunity. PLoS Pathog. 9: e1003659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Wang X., Pham T.H., Feuerbacher L.A., Lubos M.L., Huang M., Olsen R., Mushegian A., Slawson C., Hardwidge P.R. (2013). NleB, a bacterial effector with glycosyltransferase activity, targets GAPDH function to inhibit NF-κB activation. Cell Host Microbe 13: 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Ma F., Wei F., Fanella B., Allen D.K., Wang X. (2014). Cytosolic phosphorylating glyceraldehyde-3-phosphate dehydrogenases affect Arabidopsis cellular metabolism and promote seed oil accumulation. Plant Cell 26: 3023–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Devaiah S.P., Narasimhan R., Pan X., Zhang Y., Zhang W., Wang X. (2012). Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell 24: 2200–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurusamy N., Lekli I., Gorbunov N.V., Gherghiceanu M., Popescu L.M., Das D.K. (2009). Cardioprotection by adaptation to ischaemia augments autophagy in association with BAG-1 protein. J. Cell. Mol. Med. 13: 373–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, S., Yu, B., Wang, Y., and Liu, Y. (2011). Role of plant autophagy in stress response. Protein & Cell 2: 784–791. [DOI] [PMC free article] [PubMed]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129: 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J.T., Henson D., Nyirenda M., Desikan R., Harrison J., Lewis M., Hughes J., Neill S.J. (2005). Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol. Biochem. 43: 828–835. [DOI] [PubMed] [Google Scholar]
- Hara M.R., et al. (2005). S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7: 665–674. [DOI] [PubMed] [Google Scholar]
- Harada N., Yasunaga R., Higashimura Y., Yamaji R., Fujimoto K., Moss J., Inui H., Nakano Y. (2007). Glyceraldehyde-3-phosphate dehydrogenase enhances transcriptional activity of androgen receptor in prostate cancer cells. J. Biol. Chem. 282: 22651–22661. [DOI] [PubMed] [Google Scholar]
- Hayward A.P., Dinesh-Kumar S.P. (2011). What can plant autophagy do for an innate immune response? Annu. Rev. Phytopathol. 49: 557–576. [DOI] [PubMed] [Google Scholar]
- Hofius D., Schultz-Larsen T., Joensen J., Tsitsigiannis D.I., Petersen N.H., Mattsson O., Jørgensen L.B., Jones J.D., Mundy J., Petersen M. (2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783. [DOI] [PubMed] [Google Scholar]
- Holtgrefe S., Gohlke J., Starmann J., Druce S., Klocke S., Altmann B., Wojtera J., Lindermayr C., Scheibe R. (2008). Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol. Plant. 133: 211–228. [DOI] [PubMed] [Google Scholar]
- Huett A., Ng A., Cao Z., Kuballa P., Komatsu M., Daly M.J., Podolsky D.K., Xavier R.J. (2009). A novel hybrid yeast-human network analysis reveals an essential role for FNBP1L in antibacterial autophagy. J. Immunol. 182: 4917–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang N.R., Yim S.H., Kim Y.M., Jeong J., Song E.J., Lee Y., Lee J.H., Choi S., Lee K.J. (2009). Oxidative modifications of glyceraldehyde-3-phosphate dehydrogenase play a key role in its multiple cellular functions. Biochem. J. 423: 253–264. [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492. [DOI] [PubMed] [Google Scholar]
- Jain A., Lamark T., Sjøttem E., Larsen K.B., Awuh J.A., Øvervatn A., McMahon M., Hayes J.D., Johansen T. (2010). p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 285: 22576–22591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaido M., Abe K., Mine A., Hyodo K., Taniguchi T., Taniguchi H., Mise K., Okuno T. (2014). GAPDH—a recruits a plant virus movement protein to cortical virus replication complexes to facilitate viral cell-to-cell movement. PLoS Pathog. 10: e1004505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S., van West P., Vleeshouwers V.G., de Groot K.E., Govers F. (1998). Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10: 1413–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S., Cuervo A.M. (2006). Autophagy as a cell-repair mechanism: activation of chaperone-mediated autophagy during oxidative stress. Mol. Aspects Med. 27: 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-C., Guo L., Wang X. (2013). Phosphatidic acid binds to cytosolic glyceraldehyde-3-phosphate dehydrogenase and promotes its cleavage in Arabidopsis. J. Biol. Chem. 288: 11834–11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunchithapautham K., Rohrer B. (2007). Apoptosis and autophagy in photoreceptors exposed to oxidative stress. Autophagy 3: 433–441. [DOI] [PubMed] [Google Scholar]
- Kwon S.I., Cho H.J., Kim S.R., Park O.K. (2013). The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol. 161: 1722–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavallard V.J., Pradelli L.A., Paul A., Bénéteau M., Jacquel A., Auberger P., Ricci J.-E. (2009). Modulation of caspase-independent cell death leads to resensitization of imatinib mesylate-resistant cells. Cancer Res. 69: 3013–3020. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Li Q., Lee J.Y., Lee S.H., Jeong J.H., Lee H.R., Chang H., Zhou F.C., Gao S.J., Liang C., Jung J.U. (2009). FLIP-mediated autophagy regulation in cell death control. Nat. Cell Biol. 11: 1355–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C. (2012). Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63: 215–237. [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Marathe R., Dinesh-Kumar S.P. (2002a). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30: 415–429. [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Serino G., Deng X.W., Dinesh-Kumar S.P. (2002b). Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell 14: 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Czymmek K., Tallóczy Z., Levine B., Dinesh-Kumar S.P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577. [DOI] [PubMed] [Google Scholar]
- Liu Y., Burgos J.S., Deng Y., Srivastava R., Howell S.H., Bassham D.C. (2012). Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24: 4635–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xiong Y., Bassham D.C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5: 954–963. [DOI] [PubMed] [Google Scholar]
- Mitou G., Budak H., Gozuacik D. (2009). Techniques to study autophagy in plants. Int. J. Plant Genomics 2009: 451357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.N. (2008). Autophagy as a second level protective process in conferring resistance to environmentally-induced oxidative stress. Autophagy 4: 254–256. [DOI] [PubMed] [Google Scholar]
- Munch D., Rodriguez E., Bressendorff S., Park O.K., Hofius D., Petersen M. (2014). Autophagy deficiency leads to accumulation of ubiquitinated proteins, ER stress, and cell death in Arabidopsis. Autophagy 10: 1579–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J., Bermúdez M.A., Segura J., Ros R. (2011). Arabidopsis plants deficient in plastidial glyceraldehyde-3-phosphate dehydrogenase show alterations in abscisic acid (ABA) signal transduction: interaction between ABA and primary metabolism. J. Exp. Bot. 62: 1229–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J., Cascales-Miñana B., Mulet J.M., Baroja-Fernández E., Pozueta-Romero J., Kuhn J.M., Segura J., Ros R. (2009). Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol. 151: 541–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J., Cascales-Miñana B., Irles-Segura A., Mateu I., Nunes-Nesi A., Fernie A.R., Segura J., Ros R. (2010). The plastidial glyceraldehyde-3-phosphate dehydrogenase is critical for viable pollen development in Arabidopsis. Plant Physiol. 152: 1830–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls C., Pinto A.R., Li H., Li L., Wang L., Simpson R., Liu J.P. (2012). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) induces cancer cell senescence by interacting with telomerase RNA component. Proc. Natl. Acad. Sci. USA 109: 13308–13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez M.E., Lemaire S.D., Crespo J.L. (2012). Reactive oxygen species and autophagy in plants and algae. Plant Physiol. 160: 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton W.C. (1996). The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 185–214. [DOI] [PubMed] [Google Scholar]
- Pyo J.-O., Yoo S.-M., Ahn H.-H., Nah J., Hong S.-H., Kam T.-I., Jung S., Jung Y.-K. (2013). Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat. Commun. 4: 2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D.J. (2013). Autophagic processes in yeast: mechanism, machinery and regulation. Genetics 194: 341–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rius S.P., Casati P., Iglesias A.A., Gomez-Casati D.F. (2006). Characterization of an Arabidopsis thaliana mutant lacking a cytosolic non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase. Plant Mol. Biol. 61: 945–957. [DOI] [PubMed] [Google Scholar]
- Rius S.P., Casati P., Iglesias A.A., Gomez-Casati D.F. (2008). Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiol. 148: 1655–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas C.M., Senthil-Kumar M., Wang K., Ryu C.M., Kaundal A., Mysore K.S. (2012). Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. Plant Cell 24: 336–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R., Elazar Z. (2007). ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 17: 422–427. [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. (2007). Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26: 1749–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R.C., Juhász G., Neufeld T.P. (2007). Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 17: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.-H., Yoshimoto K., Ohsumi Y., Jeon J.S., An G. (2009). OsATG10b, an autophagosome component, is needed for cell survival against oxidative stresses in rice. Mol. Cells 27: 67–74. [DOI] [PubMed] [Google Scholar]
- Sirover M.A. (1997). Role of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in normal cell function and in cell pathology. J. Cell. Biochem. 66: 133–140. [PubMed] [Google Scholar]
- Tanida I., Tanida-Miyake E., Komatsu M., Ueno T., Kominami E. (2002). Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J. Biol. Chem. 277: 13739–13744. [DOI] [PubMed] [Google Scholar]
- Vescovi M., Zaffagnini M., Festa M., Trost P., Lo Schiavo F., Costa A. (2013). Nuclear accumulation of cytosolic glyceraldehyde-3-phosphate dehydrogenase in cadmium-stressed Arabidopsis roots. Plant Physiol. 162: 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yu B., Zhao J., Guo J., Li Y., Han S., Huang L., Du Y., Hong Y., Tang D., Liu Y. (2013). Autophagy contributes to leaf starch degradation. Plant Cell 25: 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Klionsky D.J. (2007). Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9: 1102–1109. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C. (2007a). Disruption of autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy 3: 257–258. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Nguyen P.Q., Bassham D.C. (2007b). Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 143: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C., et al. (2012). Function and molecular mechanism of acetylation in autophagy regulation. Science 336: 474–477. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.-W., Murphy T.M., Lin C.-H. (2003). Hydrogen peroxide-induced chilling tolerance in mung beans mediated through ABA-independent glutathione accumulation. Funct. Plant Biol. 30: 955–963. [DOI] [PubMed] [Google Scholar]
- Zaffagnini M., Fermani S., Costa A., Lemaire S.D., Trost P. (2013). Plant cytoplasmic GAPDH: redox post-translational modifications and moonlighting properties. Front. Plant Sci. 4: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kong X., Kang J., Su J., Li Y., Zhong J., Sun L. (2009). Oxidative stress induces parallel autophagy and mitochondria dysfunction in human glioma U251 cells. Toxicol. Sci. 110: 376–388. [DOI] [PubMed] [Google Scholar]
- Zhao J., Liu Q., Zhang H., Jia Q., Hong Y., Liu Y. (2013). The rubisco small subunit is involved in tobamovirus movement and Tm-2²-mediated extreme resistance. Plant Physiol. 161: 374–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Roeder R.G., Luo Y. (2003). S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114: 255–266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.