Root-to-shoot transportation of the essential nutrient sulfate requires a unique molecular mechanism that controls the expression of a sulfate transporter in root vascular tissues.
Abstract
Under sulfur deficiency (−S), plants induce expression of the sulfate transport systems in roots to increase uptake and root-to-shoot transport of sulfate. The low-affinity sulfate transporter SULTR2;1 is predominantly expressed in xylem parenchyma and pericycle cells in Arabidopsis thaliana roots under –S. The mechanisms underlying –S-inducible expression of SULTR2;1 in roots have remained unclear, despite the possible significance of SULTR2;1 for acclimation to low-sulfur conditions. In this investigation, examination of deletions and base substitutions in the 3′-intergenic region of SULTR2;1 revealed novel sulfur-responsive elements, SURE21A (5′-CAATGTATC-3′) and SURE21B (5′-CTAGTAC-3′), located downstream of the SULTR2;1 3′-untranslated region. SURE21A and SULTR21B effectively induced reporter gene expression from fusion constructs under –S in combination with minimal promoters or promoters not inducible by –S, suggesting their versatility in controlling transcription. T-DNA insertions near SURE21A and SULTR21B abolished −S-inducible expression of SULTR2;1 in roots and reduced the uptake and root-to-shoot transport of sulfate. In addition, these mutations partially suppressed SULTR2;1 expression in shoots, without changing its –S-responsive expression. These findings indicate that SULTR2;1 contributes to the increase in uptake and internal translocation of sulfate driven by gene expression induced under the control of sulfur-responsive elements in the 3′-nontranscribed intergenic region of SULTR2;1.
INTRODUCTION
Sulfur is an essential element required for plant growth. Sulfate is the major form of sulfur that plants can use for synthesizing sulfur-containing compounds, such as the amino acids cysteine and methionine, proteins, lipids, coenzymes, and various secondary metabolites (Leustek et al., 2000; Saito, 2004). The essentiality of these compounds clearly indicates the importance of sulfate uptake, distribution, and metabolism. Following uptake of sulfate from the soil, sulfate moves horizontally through the apoplast and symplast and is loaded into xylem to be transported to aerial parts of the plant. Sulfate transporters mediate the uptake and internal mobilization of sulfate.
The Arabidopsis thaliana genome encodes 12 sulfate transporters (SULTRs), classified into four groups (SULTR1, SULTR2, SULTR3, and SULTR4) based on the similarity in their protein sequences (Takahashi et al., 2012). Their biochemical properties, tissue localization, and functions in plants have been studied extensively (Davidian and Kopriva, 2010; Takahashi et al., 2011). The group 1 sulfate transporters consist of the high-affinity transporters SULTR1;1, SULTR1;2, and SULTR1;3. SULTR1;1 and SULTR1;2 are expressed in the epidermis and cortex of roots and facilitate the initial uptake of sulfate from the soil (Takahashi et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002). SULTR1;3 is localized in the phloem and mediates source-to-sink translocation of sulfate (Yoshimoto et al., 2003). The group 4 sulfate transporters SULTR4;1 and SULTR4;2 are localized to the tonoplast and are involved in remobilization of vacuolar sulfate pool (Kataoka et al., 2004a).
When the soil concentration of sulfate declines, plants increase the capacities of sulfate transport systems in roots. In Arabidopsis, −S induces the expression of several sulfate transporters, including SULTR1;1, SULTR1;2, SULTR1;3, SULTR4;1, and SULTR4;2 (Takahashi et al., 2000; Vidmar et al., 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002; 2007; Kataoka et al., 2004a). These sulfate transporters are essential for the initial uptake and vascular translocation of sulfate and release of vacuolar sulfate to support efficient utilization of sulfate pools. Our recent studies have indicated several molecular mechanisms required for the –S-responsive gene expression of Arabidopsis SULTRs (Maruyama-Nakashita et al., 2005, 2006). A sulfur-responsive cis-acting element, comprising 16 bp of a DNA sequence named the sulfur-responsive element (SURE) occurs in the 5′-region of SULTR1;1 and induces its gene expression in response to –S (Maruyama-Nakashita et al., 2005). An EIL-family transcription factor, SLIM1, has been identified as a transcriptional regulator controlling the main pathways of sulfate uptake and metabolism, including SULTR1;1 and SULTR1;2, under –S conditions in Arabidopsis (Maruyama-Nakashita et al., 2006).
In contrast to the group 1 and 4 sulfate transporters, the physiological functions of group 2 and 3 sulfate transporters are not well understood. SULTR2;1, a group 2 sulfate transporter, exhibits a low-affinity sulfate transport activity in yeast and is expressed in the xylem and phloem parenchyma cells of leaves and xylem parenchyma and pericycle cells of roots in Arabidopsis (Takahashi et al., 1997, 2000). Based on its tissue-specific localization, SULTR2;1 has been suggested to mediate the uptake of sulfate from the apoplast within the vascular bundle. Furthermore, it has been considered to function as a component of a sulfate transport system that possibly involves a functional interplay with SULTR3;5 mediating the root-to-shoot transport of sulfate in Arabidopsis (Kataoka et al., 2004b). Simultaneous expression of SULTR2;1 and SULTR3;5 in yeast enhances sulfate uptake capacity compared with the expression of either SULTR2;1 or SULTR3;5 (Kataoka et al., 2004b). The synergistic contribution of SULTR2;1 and SULTR3;5 to root-to-shoot transport of sulfate is suggested based on the overlap of tissue-specific gene expression in the xylem parenchyma and pericycle cells of Arabidopsis roots (Takahashi et al., 1997, 2000; Kataoka et al., 2004b). Because the coexpression of SULTR2;1 with SULTR3;5 increased sulfate uptake activity in yeast, the inducible expression of SULTR2;1 in roots has been suggested to act as a key factor in increasing root-to-shoot transport of sulfate under –S conditions (Kataoka et al., 2004b).
Expression of SULTR2;1 shows complicated responses to –S conditions. The transcript level of SULTR2;1 is highly upregulated in response to −S in roots but is decreased in shoots (Takahashi et al., 2000). The repression of SULTR2;1 in shoots involves microRNA-395 (miR395), which is induced by −S in a SLIM1-dependent manner in phloem (Kawashima et al., 2009) and targets SULTR2;1 mRNA (Jones-Rhoades and Bartel, 2004; Allen et al., 2005; Kawashima et al., 2009). However, this posttranscriptional regulatory mechanism contrasts with the situation in roots, where the SULTR2;1 mRNA level increases significantly under –S conditions; nevertheless, miR395 accumulates to high levels in roots, as in shoots (Kawashima et al., 2009). This disagreement between miR395 and SULTR2;1 accumulations has been suggested to be due to their cell-type-specific expression in root vascular tissues; i.e., the expression of miR395 is restricted in the phloem companion cells, which leaves the target SULTR2;1 mRNA to remain intact and accumulate in xylem parenchyma and pericycle cells (Kawashima et al., 2009). Thus, an alternative regulatory mechanism independent of SLIM1 and miR395 must underlie the –S-responsive induction of SULTR2;1 expression in roots, particularly in cell types where an induced expression of sulfate transporters (i.e., SULTR2;1) can increase the flux of sulfate loaded to the xylem stream and transferred from roots to aerial organs. This study reveals the presence of cis-acting regulatory elements in the 3′-nontranscribed intergenic region for the –S-responsive transcriptional regulation of SULTR2;1 in Arabidopsis. Molecular dissection of regulatory elements and disruption of their function in T-DNA knockout lines demonstrate the significance of this mechanism for the –S-responsive induction of SULTR2;1 in roots.
RESULTS
The 3′-Downstream Intergenic Region of SULTR2;1 Controls –S-Inducible Expression in Roots
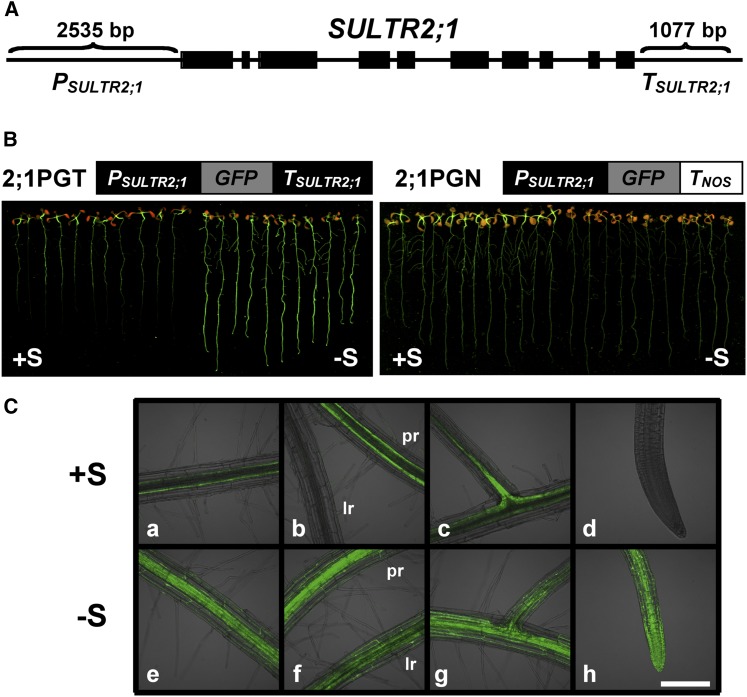
To identify the cis-acting molecular mechanisms controlling –S-inducible expression of SULTR2;1 in Arabidopsis roots, regulatory functions of 5′-upstream and 3′-downstream intergenic regions flanking SULTR2;1 (Figure 1A) were investigated by expressing their green fluorescent protein (GFP) fusion genes in transgenic Arabidopsis. Fusion genes containing the fragments, a 2535-bp 5′-upstream region of SULTR2;1 (PSULTR2;1), GFP coding region, and either the 1077-bp 3′-downstream region of SULTR2;1 (TSULTR2;1) or the nopaline synthase gene terminator (TNOS) were constructed for generating transgenic plants (Figures 1A and 1B). The fusion gene constructs, PSULTR2;1:GFP:TSULTR2;1 (2;1PGT) and PSULTR2;1:GFP:TNOS (2;1PGN), were introduced into Arabidopsis plants (ecotype Columbia-0 [Col-0]) by Agrobacterium tumefaciens infection. T2 progenies of these transgenic Arabidopsis plants were grown on culture media containing 1500 μM sulfate (+S) or 15 μM sulfate (–S) for 10 d, and the GFP accumulation in plants was visualized (Figure 1B). The fluorescent images indicated significant accumulation of GFP in the root tissues of 2;1PGT transgenic lines under –S conditions (Figure 1B). In contrast, GFP expression was slightly reduced in the root tissues of 2;1PGN transgenic lines under –S (Figure 1B). These results suggested that –S-inducible transcript accumulation of SULTR2;1 was controlled in the roots by the 3′-downstream region rather than the 5′-upstream region of SULTR2;1.
Figure 1.
The –S-Inducible Expression of SULTR2;1 in Roots Is Controlled by the 3′-Downstream Region.
(A) Genomic structure of SULTR2;1. Thick black bars represent exons. PSULTR2;1 (2535-bp upstream sequence from the translational initiation codon of SULTR2;1) and TSULTR2;1 (1077-bp downstream sequence from the translational termination codon of SULTR2;1) indicate the intergenic regions used for making GFP fusion constructs.
(B) GFP accumulation in PSULTR2;1:GFP:TSULTR2;1 (2;1PGT) and PSULTR2;1:GFP:TNOS (2;1PGN) plants grown under +S and –S conditions. Fusion gene constructs used for transformation of Arabidopsis are shown above the fluorescence images. TNOS indicates the terminator sequence of nopaline synthase. 2;1PGT and 2;1PGN plants were grown for 10 d with 1500 μM (+S) or 15 μM of sulfate (–S). Fluorescence of GFP was visualized under an image analyzer as described in Methods.
(C) Tissue distribution of GFP fluorescence in 2;1PGT plant roots grown under +S or –S conditions. Fluorescence images in basal region of roots (a and e), primary roots and lateral roots (b and f), lateral root initiation zone (c and g), and root tip (d and h) of 2;1PGT plants grown under +S (a to d) and –S (e to h) conditions were visualized using laser confocal microscopy as described in Methods. pr, primary root; lr, lateral root. Bar = 200 μm.
Previous studies based on in situ hybridization and reporter analysis using a 2990-bp 5′-upstream region of SULTR2;1 and a uidA gene indicated predominant localization of SULTR2;1 gene expression in the vascular tissues in both leaves and roots (Takahashi et al., 1997, 2000). The tissue localizations of GFP expression in 2;1PGT plants under +S in this study were basically identical to these previously reported results. GFP accumulated in the vascular tissues of roots, especially in basal region of primary roots and lateral root initiation zones under +S (Figure 1C, a to d), and these GFP signals significantly increased under –S (Figure 1C, e to h). In addition, weaker GFP fluorescence was detected in both cortex and epidermis in the roots of 2;1PGT plants grown under –S (Figure 1C, e to h).
Identification of Sulfur-Responsive Elements in the 3′-Downstream Intergenic Region of SULTR2;1
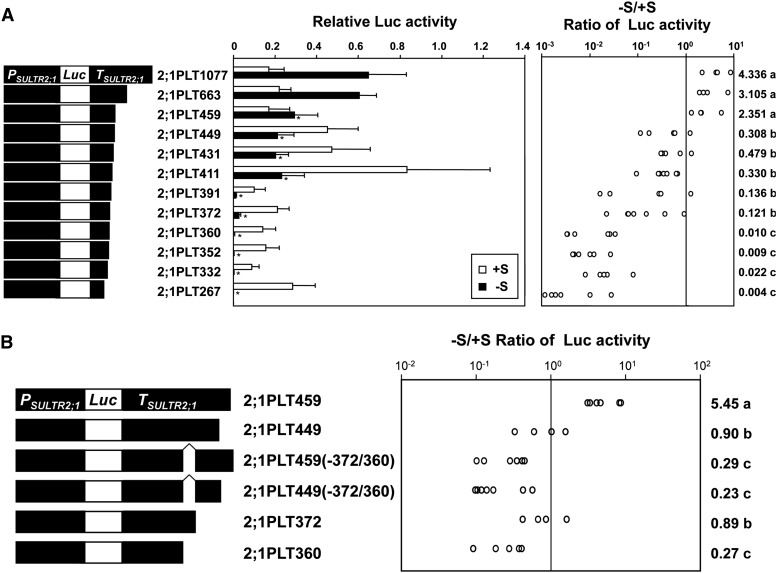
To determine the sulfur-responsive element in the 3′-downstream region of SULTR2;1, a series of 3′-truncated fragments were fused to the luciferase (Luc) gene, and these fusion genes were introduced into Arabidopsis for monitoring Luc reporter activities in the transgenic plants (Figure 2). Twelve independent fragments of the 3′-region of SULTR2;1 truncated at positions +1077, +663, +459, +449, +431, +411, +391, +372, +360, +352, +332, and +267 from the end of the SULTR2;1 coding region were prepared and fused to the Luc gene placed downstream of PSULTR2;1. These Luc fusion constructs were designated 2;1PLT1077, 2;1PLT663, 2;1PLT459, 2;1PLT449, 2;1PLT431, 2;1PLT411, 2;1PLT391, 2;1PLT372, 2;1PLT360, 2;1PLT352, 2;1PLT332, and 2;1PLT267, respectively. Luc activities in roots were determined using T2 progeny of the transgenic Arabidopsis plants grown under +S and –S conditions (Figure 2).
Figure 2.
Deletion Analysis of SULTR2;1 3′-Downstream Region.
(A) Effects of the 3′-deletions of the SULTR2;1 downstream region on Luc reporter activity under +S and –S conditions. Schematic representation of the 3′-deletions of the SULTR2;1 downstream region fused to the Luc gene (left), relative Luc activities (middle), and the –S/+S ratios of Luc activities (right) are described. Error bars in the middle panel denote the se of the mean. Asterisks indicate significant differences (Student’s t test; P < 0.05) between 2;1PLT1077 and other lines. The –S/+S ratios of Luc activities in each independent transgenic line were calculated and are shown as open circles. The geometric means of the –S/+S ratios are shown on the right with lowercase letters representing the significant differences (P < 0.05).
(B) Deletion analysis of the region between +372 and +361 from the translational termination codon of SULTR2;1. The deletion constructs of the SULTR2;1 downstream region (left) and the –S/+S ratios of Luc activities (right) are described as in (A).
T2 progenies of five to seven independent transgenic lines from each construct were grown for 10 d on agar medium containing 1500 μM (+S, open bars in [A]) or 15 μM (–S, closed bars in [A]) of sulfate. Luc activities of root tissues from 25 plantlets were assayed as described in the Methods.
Plants expressing 2;1PLT1077, 2;1PLT663, and 2;1PLT459 showed significantly higher levels of Luc activities under –S compared with those on +S conditions. The Luc activity was 2.4- to 4.3-fold higher in –S than in +S (Figure 2A). However, the Luc activities were lower under –S relative to +S in other transgenic plants, 2;1PLT449, 2;1PLT431, 2;1PLT411, 2;1PLT391, 2;1PLT372, 2;1PLT360, 2;1PLT352, 2;1PLT332, and 2;1PLT267 (Figure 2A). These transgenic lines showing reduction in –S responses were further classified into two groups based on their –S/+S ratios of Luc activities. In one group, from 2;1PLT449 to 2;1PLT372, the –S/+S ratios of Luc activities were relatively high (from 0.12 to 0.48), whereas in the second group, from 2;1PLT360 to 2;1PLT267, the –S/+S ratios of Luc activity were low (from 0.009 to 0.022). These results indicated the necessity of the 12-bp region between +372 and +360 and the 10-bp region between +459 and +449 for either inducing Luc reporter gene expression in roots in response to –S or repressing it on +S (Figure 2A). Removing the +372 to +360 region from 2;1PLT459 and 2;1PLT449 resulted in significantly lower –S/+S ratios of Luc activities in comparison to 2;1PLT372 and 2;1PLT360, which clearly indicated the importance of the nucleotide sequence from +372 to +360 in driving –S-responsive induction of gene expression in roots [Figure 2B; fusion constructs 2;1PLT459(-372/360) and 2;1PLT449(-372/360)]. In contrast, a moderate increment of Luc activity was observed in 2;1PLT449, 2;1PLT431, and 2;1PLT411 compared with 2;1PLT459 plants when they were grown on +S, indicating that the nucleotide sequence between +459 and +449 was involved in repression of Luc gene expression under +S and, thus, the absence of this region released the repression in the deletion lines (Figure 2A). Besides these two specific regions conferring significant +/–S responses, two additional regions from +460 to +663 and from +392 to +411 were found to affect Luc expression (Figure 2A). Deletion of a region from +460 to +663 reduced the Luc activity on –S; however, the –S/+S ratio of Luc activity was not altered to an extent that could be supported as statistically significant. Deletion of a region from +392 to +411 diminished the Luc activity on both +S and –S with a stronger reduction observed on –S; however, the resultant decrease in the –S/+S ratio of Luc activity was not statistically significant (Figure 2A).
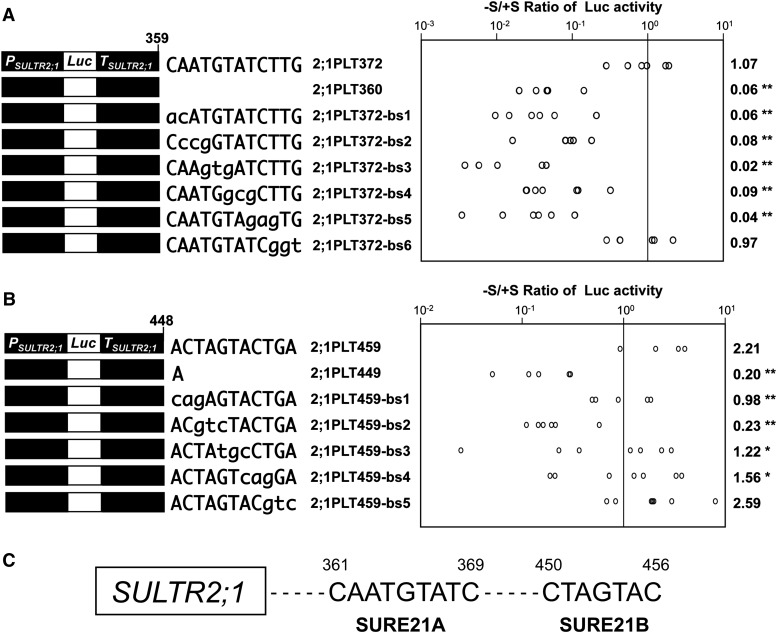
To determine the core element within the sulfur-responsive region between +372 and +360 of the SULTR2;1 3′-downstream sequence, base substitution analysis was performed in transgenic Arabidopsis (Figure 3A). Six independent constructs, 2;1PLT372-bs1, 2;1PLT372-bs2, 2;1PLT372-bs3, 2;1PLT372-bs4, 2;1PLT372-bs5, and 2;1PLT372-bs6, representing successive 3-bp substitutions of nucleotide sequences from +372 to +360 in 2;1PLT372 were prepared and introduced into Arabidopsis. The T2 progeny of transgenic lines was grown for 10 d on +S and –S medium, and Luc activities in roots were determined in independent lines prepared for each construct (Figure 3A). Plants expressing 2;1PLT372-bs1, 2;1PLT372-bs2, 2;1PLT372-bs3, 2;1PLT372-bs4, and 2;1PLT372-bs5 showed –S/+S ratios of Luc activities from 0.02 to 0.09, comparable to the value in 2;1PLT360. In contrast, 2;1PLT372-bs6 plants showed an –S/+S ratio of 0.97, which was fairly consistent with the value in 2;1PLT372 (Figure 3A). These results indicated that the 9-bp sequence between +361 and +369, 5′-CAATGTATC-3′ (Figure 3A), was indispensable for driving –S-inducible expression of SULTR2;1 in roots.
Figure 3.
Base Substitution Analysis of the –S-Responsive Segments in the SULTR2;1 3′-Downstream Region.
(A) Base-substituted constructs from +372 to +361 of the SULTR2;1 downstream region (left) and the –S/+S ratios of Luc activities (right).
(B) Base-substituted constructs from +449 to +459 of the SULTR2;1 downstream region (left) and the –S/+S ratios of Luc activities (right).
In (A) and (B), T2 progenies of five to seven independent transgenic lines from each construct were grown and analyzed as described in Figure 2. The –S/+S ratios of Luc activities in each independent transgenic line were calculated and are shown as open circles. The geometric means of the –S/+S ratios are shown on the right with asterisks representing the significant differences (*0.05 ≤ P < 0.1 and **P < 0.05). Statistical analysis was performed using Student’s t test compared with 2;1PLT372 (A) or 2;1PLT459 (B).
(C) Sulfur-responsive elements determined in the 3′-downstream region of SULTR2;1. Two regions required for the –S-inducible expression of SULTR2;1, 5′-CAATGTATC-3′ between +361 and +369, and 5′-CTAGTAC-3′ between +450 and +456, were designated SURE21A and SURE21B, respectively.
A similar base substitution analysis was performed with the +459 to +450 region as well (Figure 3B). Five independent constructs, 2;1PLT459-bs1, 2;1PLT459-bs2, 2;1PLT459-bs3, 2;1PLT459-bs4, and 2;1PLT459-bs5, representing 3-bp successive substitutions of nucleotide sequences from +449 to +459 region in the 2;1PLT459, were prepared and used for generating stable Arabidopsis transformants. The Luc activities in root tissues were determined in 10-d-old transgenic plants grown under +S and –S conditions (Figure 3B). Plants containing 2;1PLT459-bs5 showed almost the same level of induction of Luc activity as in 2;1PLT459. In contrast, other transgenic plants, 2;1PLT459-bs1, 2;1PLT459-bs2, 2;1PLT459-bs3, and 2;1PLT459-bs4, showed intermediate –S/+S ratios ranging from 0.23 to 1.56, but still lower than the values in 2;1PLT459 and 2;1PLT459-bs5. These results indicated that the 7-bp sequence between +450 and +456, 5′-CTAGTAC-3′ (Figure 3B), was necessary for the +S repression (Figure 2A) and consequently required for driving the –S-inducible gene expression of SULTR2;1 in roots.
From these results, we designated the 9-bp sequence 5′-CAATGTATC-3′ between +361 and +369 as SURE21A and the 7-bp sequence 5′-CTAGTAC-3′ between +450 and +456 as SURE21B, representing the SURE of SULTR2;1 (Figure 3C).
The SURE21A and SURE21B Enhancers Induce Gene Expression in Response to Sulfur Limitation
The 3′-untranslated region (UTR) controls mRNA stability and translational efficiency in plants (Gutiérrez et al., 1999; Fabian et al., 2010). The 3′UTR of SULTR2;1 is reported to be 302 bp in length at the maximum length, according to the sequences deposited in the public database (TAIR; http://www.arabidopsis.org). Therefore, SURE21A and SURE21B are predicted to be located 60 to 150 bp downstream of the 3′UTR. To investigate whether the lack of sulfur nutrition alters the length of 3′UTR to incorporate SURE21A and SURE21B into the SULTR2;1 transcripts, and whether the presence of these elements affects transcriptional termination in response to sulfur availabilities, the 3′-end nucleotide sequences of the Luc-TSULTR2;1 fusion transcripts were determined in 2;1PLT1077 and 2;1PLT360 plants grown under +S and –S conditions. In all 20 independent clones tested in each condition, the Luc-TSULTR2;1 transcripts carried a 241-bp SULTR2;1 3′UTR, which was identical to the native SULTR2;1 transcripts found in wild-type plants (Supplemental Figure 1). These results showing the consistency in the length of the 3′UTR suggest that SURE21A and SURE21B are not likely to be associated with posttranscriptional mechanisms because these elements are located in the nontranscribed region and because they are neither incorporated into the 3′UTR nor do they alter the position of transcriptional termination in shoots and roots in response to sulfur nutrition.
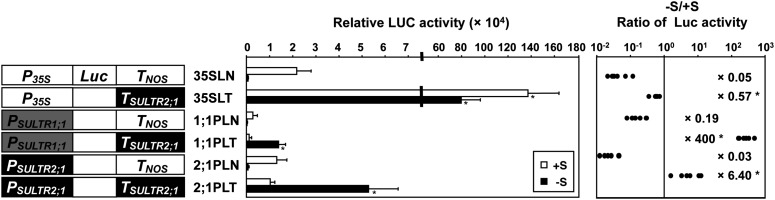
To investigate the molecular mechanism of SURE21A and SURE21B controlling the –S-inducible transcript accumulation of SULTR2;1 in roots, the effect of the SULTR2;1 3′-downstream intergenic region on Luc reporter gene expression was tested with combinations of several different promoters (Figure 4). The 2535-bp 5′-upstream region of SULTR2;1 (PSULTR2;1), the 2453-bp 5′-upstream region of SULTR1;1 (PSULTR1;1) lacking the sulfur-responsive region (Maruyama-Nakashita et al., 2005), and the cauliflower mosaic virus (CaMV) 35S promoter (P35S) were used as promoters with the SULTR2;1 3′-downstream intergenic region (TSULTR2;1). In addition to PSULTR2;1:Luc:TSULTR2;1 (2;1PLT, named 2;1PLT1077 in Figure 2) and PSULTR1;1:Luc:TNOS (1;1PLN; Maruyama-Nakashita et al., 2005) used in previous experiments, fusion gene constructs PSULTR2;1:Luc:TNOS (2;1PLN), PSULTR1;1:Luc:TSULTR2;1 (1;1PLT), P35S:Luc:TSULTR2;1 (35SLT), and P35S:Luc:TNOS (35SLN) were created and stably transformed into Arabidopsis plants by Agrobacterium infection. These transgenic plants were grown on +S and –S media for 10 d, and the Luc activities in root tissues were analyzed in independent lines prepared for each construct (Figure 4). In 2;1PLT plants, Luc activity was significantly higher (6.4-fold) under –S compared with +S. In contrast, in 2;1PLN plants, Luc activity was significantly lower (0.03-fold of +S) when grown under –S (Figure 4). These results were consistent with the observations when GFP was used as a reporter (Figure 1).
Figure 4.
The SULTR2;1 3′-Region Controls –S-Inducible Expression Independent of the Promoter.
Schematic representation of the promoter-Luc constructs with or without SULTR2;1 3′-region (left), average values of relative Luc activities (middle), and the –S/+S ratios of Luc activities (right) are described. T2 progenies of five to six independent transgenic lines from each construct were grown and analyzed as described in Figure 2. Error bars in the middle panel denote se. Asterisks in the middle panel indicate statistically significant differences (P < 0.05) between the plants harboring TSULTR2;1 and TNOS in the downstream of the same promoter. The –S/+S ratios of Luc activities in each independent transgenic line were calculated and are shown as closed circles. The geometric means of the –S/+S ratios are shown in the right graph with asterisks representing the significant differences (P < 0.01). P35S, CaMV 35S promoter; PSULTR1;1, 2453-bp upstream sequence from the translation initiation codon of SULTR1;1. Statistical analysis was performed using Student’s t test.
Analysis of 1;1PLT, 1;1PLN, 35SLT, and 35SLN plants revealed the ability of TSULTR2;1 to induce Luc expression in response to –S (Figure 4). The Luc activity was 400-fold higher under –S compared with +S in 1;1PLT plants, whereas it significantly decreased under –S (0.19-fold of +S) in 1;1PLN plants (Figure 4). The cis-acting element for the –S-inducible expression of SULTR1;1 is located between the 5′-upstream positions –2777 and –2762 of SULTR1;1 (Maruyama-Nakashita et al., 2005). Since this element is absent from the 5′-region of SULTR1;1 used in this experiment, the –S-responsive increase in the Luc activity observed in 1;1PLT plants is suggested to be dependent on the function of TSULTR2;1. The positive effect of TSULTR2;1 was also observed in a comparison between 35SLT and 35SLN plants. The 35SLT plants showed 84- and 1832-fold enhancement of Luc activities compared with the 35SLN plants under +S and –S conditions (Figure 4). The –S/+S ratios of Luc activities were 0.57 in 35SLT plants and 0.05 in 35SLN plants, indicating the presence of the –S-responsive region of TSULTR2;1 gaining strong ability to increase the reporter gene expression driven by P35S (Figure 4). These results suggest that the SULTR2;1 3′-region (TSULTR2;1) positively controls reporter gene expression by acting as an enhancer to modulate the activity of the basic transcription machinery acting on the SULTR2;1, SULTR1;1, and CaMV 35S promoters.
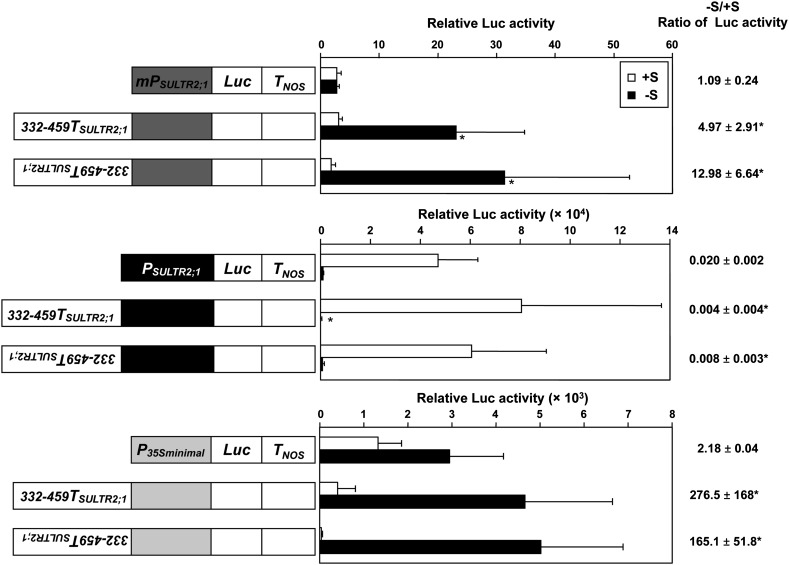
We further tested whether the transposition of SURE21A and SURE21B to the vicinity of the promoter region and their placement in inverted orientations could still induce gene expression in response to –S (Figure 5). The –S-responsive 3′-region of SULTR2;1 (from positions +332 to +459; 332-459TSULTR2;1) was relocated from the original position and placed in front of either the 2535-bp 5′-upstream region of SULTR2;1 (PSULTR2;1), the minimal promoter of SULTR2;1 (mPSULTR2;1), or a minimal 35S promoter (Benfey et al., 1989; P35S minimal) in the sense or antisense orientation. These synthetic constructs were fused to the Luc reporter gene and introduced into Arabidopsis plants. The luciferase activities in roots were measured in 10-d-old transgenic plants grown under +S and –S (Figure 5). When sense and antisense 332-459TSULTR2;1 fragments were placed in front of mPSULTR2;1 or P35S minimal, the Luc activities of plants on –S were significantly higher than those on +S. In contrast, the Luc activities were not induced under –S when sense and antisense 332-459TSULTR2;1 were fused in front of the 2535-bp 5′-upstream region of SULTR2;1 (PSULTR2;1).
Figure 5.
Transposition of SULTR2;1 3′-Region and the Complementary Sequence.
Schematic representation of the gene constructs of SULTR2;1 3′-region and the complementary sequence fused to the 5′-upstream region of SULTR2;1 (PSULTR2;1), minimal promoter of SULTR2;1 (mPSULTR2;1) or CaMV 35S minimal promoter (P35S minimal) and Luc (left), relative Luc activities (middle), and the geometric means ± se of the –S/+S ratios of Luc activities (right) are described. T2 progenies of five to seven independent transgenic lines from each construct were grown and analyzed. Error bars in the middle panel denote se. Significant differences (P < 0.05) between the transgenic plants harboring the SULTR2;1 3′-region and the corresponding control are shown as asterisks. Statistical analysis was performed using Student’s t test. PSULTR2;1 and mPSULTR2;1 represent the 2535- and 142-bp upstream sequences from the translational initiation codon of SULTR2;1. 332-459TSULTR2;1 indicates the 127-bp sequence between +332 and +459 downstream of the translational termination codon of SULTR2;1. The inverted description of 332-459TSULTR2;1 indicates the complementary sequence of 332-459TSULTR2;1.
3′-Downstream Intergenic Region of SULTR2;1 Induces Tissue-Specific Expression
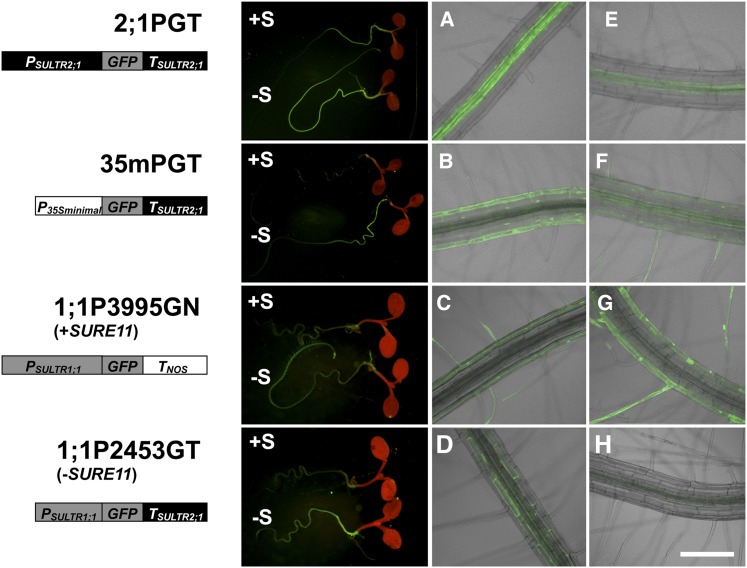
With regard to tissue specificity, the 1077-bp 3′-downstream intergenic region of SULTR2;1 (TSULTR2;1) induced GFP accumulation under –S not only in vascular tissues but also in cortex and epidermis in 2;1PGT plants (Figure 1C). We further analyzed the effect of the TSULTR2;1 on tissue-specific distribution of GFP expression in combination with other –S-noninducible promoters in transgenic Arabidopsis to demonstrate whether the elements in the 3′-region alone were able to influence gene expression in other root cell types while still retaining the –S inducibility (Figure 6). For this purpose, the SULTR2;1 3′-region (TSULTR2;1) was fused to GFP reporter and placed downstream of either P35S minimal (Benfey et al., 1989) or 2453-bp 5′-upstream region of SULTR1;1 promoter [PSULTR1;1(2453)] to generate P35S minimal:GFP:TSULTR2;1 (35mPGT) and PSULTR1;1(2453):GFP:TSULTR2;1 (1;1P2453GT). These fusion gene constructs were introduced into Arabidopsis plants by Agrobacterium infection, and independent transgenic lines were obtained for microscopic analysis. 2;1PGT (Figure 1) and PSULTR1;1(3995):GFP:TNOS (1;1P3995GN; Maruyama-Nakashita et al., 2004a) were used as positive control lines to monitor the –S-inducible expression in roots. 35mPGT, 1;1P2453GT, and the control lines were grown on +S and –S media for 9 d, and the GFP in root tissues was observed by fluorescence microscopy (Figure 6).
Figure 6.
Tissue Specificity of –S-Inducible Expression Driven by the SULTR2;1 3′-Region.
Schematic representation of the promoter-GFP constructs with or without the SULTR2;1 3′-region (left), GFP accumulation in transgenic lines grown under +S or –S condition (middle), and the close-up images of basal ([A] to [D]) and apical ([E] to [H]) regions of root tissues grown under –S condition (right) are shown. T2 transformants carrying each construct were grown for 10 d on +S and –S medium. P35S minimal, CaMV 35S minimal promoter; PSULTR1;1, 5′-upstream sequence from the translation initiation codon of SULTR1;1. The –S-responsive element, SURE11, is present in the 5′-upstream sequence of SULTR1;1 in 1;1P3995GN and absent from 1;1P2453GT, as indicated in the diagram. Bar = 200 μm.
In 2;1PGT plants, GFP accumulation was more significant in the basal region than in apical region of roots in both +S and –S conditions (Figure 6). The –S-induced GFP accumulation in 2;1PGT roots was mainly observed in vascular tissues, but minor fluorescence was also detected in the cortex and epidermis (Figures 1C and 6A). In 35mPGT plants, GFP fluorescence was not detected under +S conditions. However, under –S conditions, 35mPGT plants showed strong GFP accumulation in roots, and GFP levels were much higher in the basal region than in apical region of roots (Figure 6B). GFP fluorescence was mainly distributed in the cortex and epidermis including root hairs, and minor fluorescence was detected in vascular tissues (Figures 6B and 6F).
SULTR1;1 is a sulfate transporter that facilitates sulfate uptake in roots, and it is localized in root epidermis including root hairs (Takahashi et al., 2000; Vidmar et al., 2000; Yoshimoto et al., 2007). The –S-response of SULTR1;1 is dependent on the sulfur-responsive cis-acting element (SURE11) located at positions –2777 to –2762 of the 5′-upstream region of SULTR1;1 (Maruyama-Nakashita et al., 2005). In 1;1P3995GN plants carrying the transgene construct with SURE11, GFP fluorescence was greatly induced under –S conditions mainly in the epidermal cells of the apical zone with a high density of root hairs observed near the root tip (Figures 6C and 6G; Maruyama-Nakashita et al., 2004a). In contrast, in 1;1P2453GT plants carrying the construct missing SURE11 from the promoter region but containing SURE21A and SURE21B in the 3′-region, GFP was predominantly localized in the basal part of roots under –S, mainly in the cortex (Figure 6D) with slightly moderate but significant levels of signals observed in vascular tissues (Figures 6D and 6H), suggesting that –S-induced GFP accumulations in these cell types and tissues were due to the presence of TSULTR2;1.
These results indicated that TSULTR2;1 was able to induce gene expression preferentially in the basal region of roots. Although the precise distributions of GFP fluorescence through the radial axis of roots were not identical among the lines made with different fusion constructs, the induction of GFP accumulation was dependent on TSULTR2;1 and was observed in the basal regions of roots in all cases tested in this study (Figures 6A, 6B, and 6D). Therefore, it is reasonable to assume that TSULTR2;1 is able to induce gene expression in response to –S regardless of the promoter sequences of SULTR2;1 and SULTR1;1 determining tissue specificities (Figures 4 and 6). Furthermore, results obtained with the 35mPGT plants suggest that TSULTR2;1 has enhancer activity to induce gene expression in response to –S specifically in roots.
Induction of SULTR2;1 in Roots Contributes to Both Sulfate Uptake and Internal Translocation
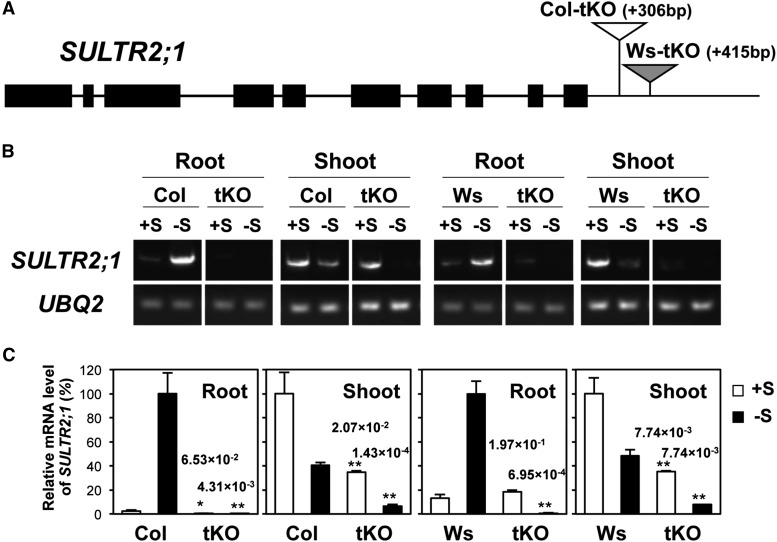
To elucidate the physiological meaning of –S-inducible SULTR2;1 expression in roots, Arabidopsis mutant lines containing T-DNA insertions between the end of SULTR2;1 3′UTR and SURE21A or SURE21B in the 3′-nontranscribed intergenic region were searched in the database, and SAIL_363_C06 in Col-0 background and FLAG_373B04 in Wassilewskija (Ws) background (Samson et al., 2002; Sessions et al., 2002; T-DNA Express, http://signal.salk.edu/cgi-bin/tdnaexpress) were identified as candidates. These T-DNA insertion lines were selected as homozygous lines by PCR-based screening, and the T-DNA insertions of 306 and 415 bp downstream of SULTR2;1 in SAIL_363_C06 and FLAG_373B04, respectively, were confirmed by sequencing. These lines were named tKO (Figure 7A) as they contained T-DNA insertions in the 3′-nontranscribed region.
Figure 7.
T-DNA Insertion between Stop Codon and SURE21A/SURE21B Disrupts –S-Inducible Expression of SULTR2;1 in Roots.
(A) Structure of SULTR2;1 genomic region and T-DNA insertion sites (triangles) in the knockout (tKO) mutants. Thick black bars represent exons. In the tKO mutants in Col-0 and Ws backgrounds, T-DNAs are inserted in positions +306 and +415 bp from the translational termination codon of SULTR2;1, as indicated by white and gray triangles, respectively.
(B) Gel images of real-time RT-PCR products showing SULTR2;1 and UBQ2 (UBIQUITIN2) mRNA accumulations. The wild type (Col-0 and Ws) and tKO mutants were grown for 10 d on +S and –S media.
(C) Relative transcript levels of SULTR2;1 quantified by real-time RT-PCR. UBQ2 transcript levels were used for normalization. SULTR2;1 transcript levels in wild-type roots under –S and wild-type shoots under +S are shown as 100%. Error bars denote se (n = 3). P values of the Student’s t tests of the comparisons between the wild-type and tKO plants on either +S or –S conditions are indicated above the tKO data with asterisks (*0.05 ≤ P < 0.1 and **P < 0.05).
The tKO and the wild-type plants (Col-0 and Ws) were grown under +S and –S conditions, and the SULTR2;1 transcript levels were analyzed by quantitative RT-PCR (Figures 7B and 7C). In wild-type plants, the transcript levels of SULTR2;1 were higher in roots grown on –S compared with those on +S, while they were reversed in shoots, i.e., lower on –S compared with +S, as reported previously (Takahashi et al., 2000; Kataoka et al., 2004b). In roots grown under +S conditions, the transcript levels of SULTR2;1 were not significantly different between the wild type and tKO. However, in roots grown under –S conditions, the SULTR2;1 transcript accumulation was observed only in the wild type but not in the tKO (Figure 7C). Furthermore, the transcript levels of SULTR2;1 in the tKO roots were significantly lower on –S compared with those on +S. These data demonstrated the essential roles of SURE21A and SURE21B in the induction and maintenance of SULTR2;1 expression in roots grown under –S conditions.
In shoots, by contrast, SULTR2;1 was partially suppressed in the tKO lines under both +S and –S conditions. The –S-responsive repression of SULTR2;1 transcript was reproducible in the tKO lines as those in the wild-type plants, although the transcript levels of SULTR2;1 were consistently lower in tKO than in the wild-type plants under both +S and –S conditions (Figure 7C). Our results further suggested that SULTR2;1 repression in shoots was more significant in tKO, as indicated by the +S/–S ratios of SULTR2;1 transcript levels exhibiting an increase in the magnitude of repression from 2.46 in Col-0 to 5.23 in Col-tKO or from 2.07 in Ws to 4.58 in Ws-tKO. To investigate whether the T-DNA insertions in tKOs have any significant effects on the SULTR2;1 transcript terminations as those were confirmed in 2;1PLT1077 and 2;1PLT360, the 3′-end nucleotide sequences of the SULTR2;1 transcripts were determined in tKO and wild-type plants grown under +S and –S conditions. The length of 3′UTR was 241 bp, which was mostly consistent and conserved in shoots and roots of the wild type and tKOs under both +S and –S growth conditions, with a few exceptions showing a length of 254 bp in roots of –S-grown tKOs (Supplemental Figure 1).
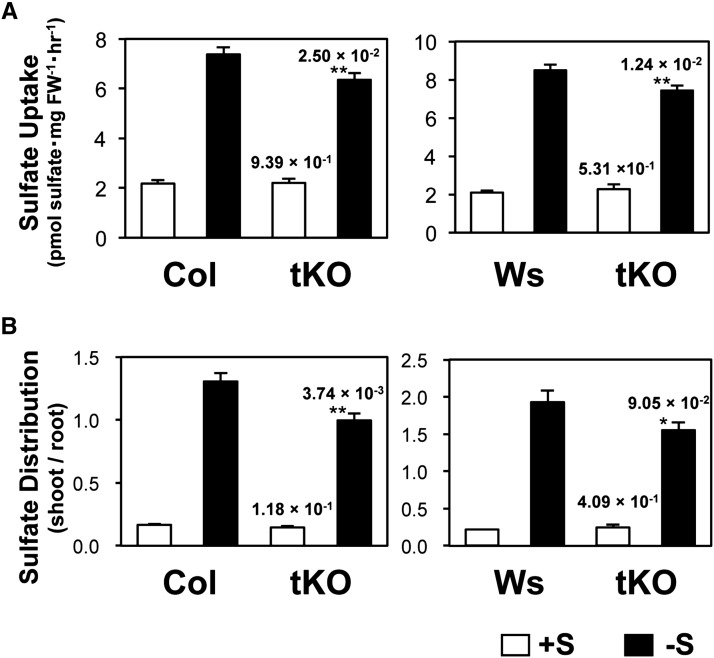
We further analyzed sulfate uptake and translocation in tKO plants to assess the contribution of the –S-inducible expression of SULTR2;1 in roots (Figure 8). Ten-day-old plants grown under +S and –S conditions were incubated for 1 h with +S medium containing the [35S] sulfate. Sulfate uptake activity was calculated by dividing the total [35S] radioisotope accumulated in the seedlings with the fresh weight of the root (Figure 8A). The sulfate translocation activity was calculated as a shoot-to-root ratio of distribution of [35S] sulfate in the seedlings (Figure 8B). Under +S conditions, both sulfate uptake and translocation activity did not differ between wild-type and tKO plants (Figure 8). These results indicated that the decrease in SULTR2;1 transcripts in the foliar part of tKO did not modulate the sulfate uptake activity and sulfate distribution in 10-d-old plants. In contrast, sulfate uptake and translocation activity were significantly lower in tKO than in the wild-type plants grown under –S conditions (Figure 8). These results provided evidence that induction of SULTR2;1 expression in roots through the function of the 3′-nontranscribed region was important for controlling both the sulfate uptake and translocation under –S conditions.
Figure 8.
Sulfate Uptake Activity and Sulfate Distribution in Wild-Type and tKO Plants Grown under +S and –S Conditions.
(A) Sulfate uptake activity of the wild type and tKO mutant. Ten-day-old plants grown under +S and –S conditions were used for the analysis. The absolute values of [35S] sulfate uptake rates are indicated.
(B) Sulfate distribution in the wild type and tKO mutant shown as shoot/root ratios of [35S] sulfate accumulations. Error bars denote se (n = 8). P values of the Student’s t tests of the comparisons between the wild-type and tKO plants on either +S or –S conditions are indicated above the tKO data with asterisks (*0.05 ≤ P < 0.1 and **P < 0.05).
DISCUSSION
The 3′-Nontranscribed Intergenic Region Is Responsible for the Induction of SULTR2;1 Gene Expression in Arabidopsis Roots under Sulfur-Limited Conditions
An increase of SULTR2;1 transcript levels in Arabidopsis roots is a typical –S response (Takahashi et al., 2000; Hirai et al., 2003; Maruyama-Nakashita et al., 2003, 2005; Nikiforova et al., 2003; Iyer-Pascuzzi et al., 2011). However, unlike the other –S-inducible isoforms of the SULTR family members, SULTR2;1 does not require the function of SLIM1 transcription factor to induce its expression in roots under S-limited conditions (Maruyama-Nakashita et al., 2006). The APS reductase gene family is also induced by –S in a SLIM1-independent manner; however, the molecular mechanisms underlying their –S-responsive expression have not been elucidated (Maruyama-Nakashita et al., 2006; Davidian and Kopriva, 2010). In this study, we analyzed the regulation of SULTR2;1 with a particular focus on the –S induction of its transcript accumulation in Arabidopsis roots and identified the sulfur-responsive cis-acting elements, SURE21A and SURE21B, located at the positions +361/+369 and +450/+456, respectively, in the 3′-nontranscribed intergenic region (Figures 1 to 3).
The distinct function of both these elements in the –S-inducible expression of SULTR2;1 is suggested by the deletion study (Figure 2). Deletion of +450/+459 region of SULTR2;1 significantly enhances the Luc activity under +S, indicating that SURE21B, 5′-CTAGTAC-3′, contributes to the repression of SULTR2;1 expression under +S. In contrast, deletion of +361/+372 region of SULTR2;1 significantly diminishes the Luc activity under –S, indicating that SURE21A, 5′-CAATGTATC-3′, contributes to the induction of SULTR2;1 gene expression under –S. These results suggest that the –S-dependent upregulation of SULTR2;1 occurs due to a combinatorial effect of two elements operating in favor of releasing the repression and stimulating the induction of gene expression, allowing a strict control of SULTR2;1 expression possibly under two different signals derived from changes in the sulfur status. In addition to these two core elements, +460/+663 and +392/+411 regions are suggested to be partly required for maintenance of SULTR2;1 transcript levels although their roles in controlling the +/–S responses appear subsidiary (Figure 2).
Several lines of evidence indicate that SULTR2;1 transcripts accumulate in roots in response to –S due to a transcriptional mechanism, despite the presence of two cis-acting elements, SURE21A and SURE21B, identified in the 3′-downstream intergenic region. Because of this unusual location of cis-acting elements, posttranscriptional regulation of mRNA stability associated with the nucleotide sequences of 3′UTR was suspected as a mechanism underlying SULTR2;1 transcript modulation. However, since SURE21A and SURE21B are located downstream of the 3′UTR, and since the length of the 3′UTRs of SULTR2;1 mRNA in tKO or Luc-fusion mRNA in reporter lines were unchanged regardless of sulfur conditions or of the presence of these –S-responsive elements (Supplemental Figure 1), a posttranscriptional regulation of the SULTR2;1 mRNA stability through the function of 3′UTR seems an unlikely scenario. Our results rather strongly support the involvement of SURE21A and SURE21B as enhancers involved in transcriptional mechanisms. SURE21A and SURE21B effectively enhanced reporter gene expression under –S when they were relocated to the vicinity of promoter sequence in the 5′-upstream region (Figure 5). In addition, they enhanced reporter gene expression in combination with several different promoters lacking the ability to respond to –S (Figures 4 and 6). The SURE21A/SURE21B-dependent activation of the reporter activity is detected under –S even in combination with the 35S minimal promoter (Figures 5 and 6). These results indicate that SURE21A and SURE21B function sufficiently as cis-acting elements that may act on the basic transcription machinery, possibly by direct or indirect interactions with the preinitiation complex of general transcription factors. Nevertheless their placement at a distance from the reporter gene toward the 5′ direction appears to limit their proper function. The T-DNA insertions between the SULTR2;1 coding sequence and SURE21 block –S induction of SULTR2;1 in roots (Figure 7), suggesting that distances toward the 3′-direction also affect their function as enhancers.
Several cis-acting elements or regions responsive to –S, such as SURE11, UPE-box, –S-responsive region of NIT3, and β-subunit gene promoter of β-conglycinin, have been previously reported (Awazuhara et al., 2002; Kutz et al., 2002; Maruyama-Nakashita et al., 2005; Davidian and Kopriva, 2010; Wawrzyńska et al., 2010). Neither SURE21A nor SURE21B shows any sequence similarity to these –S-responsive sequences. We further searched for SURE21A and SURE21B in the 500-bp 5′-upstream and 3′-downstream sequences of the 469 –S-regulated genes previously selected from microarray analysis (Table 1; Maruyama-Nakashita et al., 2006). SURE21A (5′-CAATGTATC-3′) and SURE21B (5′-CTAGTAC-3′) or their complementary sequences were found in 11 and 25 genes, respectively (Table 1). Among them, four of the –S-upregulated genes, SULTR2;1 (AT5G10180), γ-glutamyl cyclotransferase 2;1 (GGCT2;1; AT5G26220; Paulose et al., 2013), LSU1 (AT3G49580; Wawrzyńska et al., 2010; Lewandowska et al., 2010), and nuclear transport factor 2 (NTF2) family protein (AT3G09250), contained both SURE21A and SURE21B. In contrast, none of the –S-downregulated genes contained sequences that matched with both the cis-acting elements. GGCT2;1 and LSU1 carried SURE21A or its complementary sequence in their upstream region and the complementary sequence of SURE21B in their downstream region. Other than SULTR2;1, only the NTF2 family protein carried both sequences in their downstream region, but they were found on the complementary strand. These results indicate that SURE21A and SURE21B may be present as cis-acting elements in –S-upregulated genes, but their combinations and presence in the 3′-region may be specific to SULTR2;1 and NTF2.
Table 1. SURE21A (CAATGTATC), SURE21B (CTAGTAC), and the Complementary Sequences, GATACATTG and GTACTAG, in the Upstream or Downstream Regions of –S-Responsive Genes.
| −S Response | Sequence | Affymetrix ID | AGI Code | Gene Name | Position |
|---|---|---|---|---|---|
| Upregulated | SURE21A | 250475_at | AT5G10180 | Sulfate transporter, SULTR2;1 | +361 |
| 5′-CAATGTATC-3′ | 254113_at | AT4G24900 | Titan-like, TTL (nuclear C2H2 domain-containing protein) | +322 | |
| 258151_at | AT3G18080 | B-S Glucosidase 44, BGLU44 | +4 | ||
| 246884_at | AT5G26220 | γ-Glutamyl cyclotransferase 2;1 (GGCT2;1) | -305 | ||
| 5′-GATACATTG-3′ | 259039_at | AT3G09250 | Nuclear transport factor 2 (NTF2) family protein | +72 | |
| 252269_at | AT3G49580 | Response to low sulfur 1, LSU1 | -131 | ||
| 249752_at | AT5G24660 | Response to low sulfur 2, LSU2 | −159 | ||
| SURE21B | 266709_at | AT2G03120 | Signal peptide peptidase family protein, ATSPP | +476 | |
| 5′-CTAGTAC-3′ | 250475_at | AT5G10180 | Sulfate transporter, SULTR2;1 | +450 | |
| 251928_at | AT3G53980 | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein | +263 | ||
| 260064_at | AT1G73730 | Sulfur limitation 1/EIL3 | +149 | ||
| 5′-GTACTAG-3′ | 246884_at | AT5G26220 | γ-Glutamyl cyclotransferase 2;1 (GGCT2;1) | +221 | |
| 252269_at | AT3G49580 | Response to low sulfur 1, LSU1 | +190 | ||
| 260196_at | AT1G67570 | Protein of unknown function (DUF3537) | +188 | ||
| 259039_at | AT3G09250 | Nuclear transport factor 2 (NTF2) family protein | +184 | ||
| 263297_at | AT2G15310 | ADP-ribosylation factor B1A, ATARFB1A | +65 | ||
| 259476_at | AT1G19000 | Homeodomain-like superfamily protein | +61 | ||
| Downregulated | SURE21A | 255773_at | AT1G18590 | Sulfotransferase, SOT17 | +476 |
| 5′-CAATGTATC-3′ | 252612_at | AT3G45160 | Putative membrane lipoprotein | +433 | |
| 260902_at | AT1G21440 | Phosphoenolpyruvate carboxylase family protein | +38 | ||
| 5′-GATACATTG-3′ | 263477_at | AT2G31790 | UDP-glycosyltransferase superfamily protein | +204 | |
| SURE21B | 260745_at | AT1G78370 | Glutathione S-transferase, ATGSTU20 | +240 | |
| 5′-CTAGTAC-3′ | 255711_at | AT4G00090 | Transducin/WD40 repeat-like superfamily protein | +185 | |
| 250226_at | AT5G13780 | Acyl-CoA N-acyltransferase (NAT) superfamily protein | +52 | ||
| 261536_at | AT1G01790 | K+ efflux antiporter, KEA1 | −85 | ||
| 262774_at | AT1G13230 | Leucine-rich repeat protein pii-2 | −402 | ||
| 247193_at | AT5G65380 | MATE efflux family protein | −488 | ||
| 5′-GTACTAG-3′ | 250226_at | AT5G13780 | Acyl-CoA N-acyltransferase (NAT) superfamily protein | +362 | |
| 260557_at | AT2G43610 | Glycoside hydrolase family 19, similar to chitinase | +362 | ||
| 250032_at | AT5G18170 | Glutamate dehydrogenase 1, GDH1 | +135 | ||
| 250438_at | AT5G10580 | Protein of unknown function | +111 | ||
| 259264_at | AT3G01260 | Aldose 1-epimerase family protein | +105 | ||
| 258977_s_at | AT3G02020 | Aspartate kinase 3, AK3 | +31 | ||
| 262064_at | AT1G56070 | Low expression of osmotically Responsive genes 1, LOS1 | +15 | ||
| 249025_at | AT5G44720 | Molybdenum cofactor sulfurase family protein | −347 | ||
| 259813_at | AT1G49860 | Glutathione S-transferase 14, ATGSTF14 | −362 |
Positions of SURE21A and SURE21B between the 1- to 500-bp upstream or downstream regions of −S-responsive genes are indicated. Genes having both SURE21A and SURE21B are shown in bold.
Induction of SULTR2;1 Expression in Roots Contributes to –S-Induced Root-to-Shoot Transport and Uptake of Sulfate
SULTR2;1 is considered to be responsible for retrieval of apoplasmic sulfate in root vasculature (Takahashi et al., 1997, 2000; Kataoka et al., 2004b). It is suggested that efficient retrieval of sulfate into xylem parenchyma cells would help the loading of sulfate to the xylem stream under –S. A recent study using a sultr2;1-null knockout line suggests that the disruption of SULTR2;1 diminishes root-to-shoot transport of sulfate in plants grown under +S but not under –S (Kawashima et al., 2011). In contrast, a decrease in the root-to-shoot translocation of sulfate specifically under –S conditions was observed in the mutants containing T-DNA insertions in the 3′-nontranscribed region of SULTR2;1 (tKO) in this study (Figure 8). Since the major impact of the T-DNA insertion mutations on SULTR2;1 expression in tKO lines is observed in roots under –S conditions (Figure 7), the phenotype of root-to-shoot sulfate transport is suggested to be relevant to the function of SULTR2;1 in roots. The results presented in this study therefore provide a strong evidence for the root-specific function of SULTR2;1, increasing the root-to-shoot transport of sulfate under –S. Our results support the previously suggested roles of SULTR2;1 in Arabidopsis roots (Takahashi et al., 2000; Kataoka et al., 2004b) and explain the relevance of its –S-responsive induction of gene expression led by a transcriptional control mechanism requiring the cis-acting elements, SURE21A and SURE21B.
Results shown in this study further indicate the partial contribution of SULTR2;1 to the uptake of sulfate in roots (Figure 8). Previous studies indicate that the –S-inducible sulfate uptake activity is mainly attributed to the function of the two sulfate transporter genes, SULTR1;1 and SULTR1;2, in Arabidopsis (Takahashi et al., 2000; Vidmar et al., 2000; Shibagaki et al., 2002; Maruyama-Nakashita et al., 2003; Yoshimoto et al., 2002, 2007). Gene expression studies using promoter-reporter constructs further indicate a strong regulation of both SULTR1;1 and SULTR1;2 dependent on their –S-responsive promoters (Maruyama-Nakashita et al., 2004a, 2004b, 2005). Thus, SULTR1;1 and SULTR1;2 are considered as major components of the –S-inducible sulfate uptake activity in Arabidopsis roots. However, the double T-DNA knockout plant of SULTR1;1 and SULTR1;2 still can increase sulfate uptake activity in response to –S (Yoshimoto et al., 2007), suggesting that another transport system in roots induced by –S remains unidentified. Reduction of sulfate uptake activity shown by T-DNA insertions disrupting SURE21A and SURE21B functions suggests that SULTR2;1 is a potential candidate for this alternative –S-inducible component (Figure 8). The localization of SULTR2;1 gene expression in cortex cells (Figures 1 and 6) in addition to xylem parenchyma and pericycle cells (Takahashi et al., 1997, 2000) also supports the functionality of SULTR2;1 mediating sulfate uptake in roots.
The SULTR2;1 3′-Nontranscribed Region Is Also Effective for the Maintenance of Basal Levels of Gene Expression
Besides controlling –S responses, the 3′-nontranscribed region of SULTR2;1 also seems to be responsible for maintaining the basal levels of gene expression in both roots and shoots (Figures 2, 4, and 7). Luc activities of 2;1PLT391 roots were 17- and 40-fold lower than those of 2;1PLT411 roots under +S and –S, respectively (Figure 2). This suggests that the nucleotide sequence between +392 and +411, 5′-TATTTGGTGAATCAAATTAG-3′, contains information required for the maintenance of SULTR2;1 basal expression levels in roots independent of the functions of SURE21A and SURE21B in controlling the –S responses. The Luc activity driven by CaMV 35S promoter with SULTR2;1 3′-region (35SLT) is 26-fold higher than with NOS terminator (35SLN) even under +S (Figure 4). In contrast, the Luc activity driven by SULTR1;1 promoter lacking SURE11 but with SULTR2;1 3′-region (1;1PLT) is 2.7-fold lower than with the NOS terminator (1;1PLN) under +S (Figure 4). These results indicate that SULTR2;1 3′-region may enhance expression levels of genes located nearby, although the effect seems restricted by the promoter sequences (Figures 2 and 4).
The effect of the SULTR2;1 3′-region on basal gene expression is also suggested by comparisons between tKOs and wild types in both Col-0 and Ws backgrounds. SULTR2;1 transcript levels in shoots were 3- and 6-fold lower in tKOs than in wild types under +S and –S, respectively (Figure 7). On the other hand, SULTR2;1 transcript levels in roots did not significantly differ between tKO and wild-type plants under +S. These results indicate that T-DNA insertions near SURE21A and SURE21B in the SULTR2;1 3′-region may have additional negative impacts on SULTR2;1 expression in shoots under both +S and –S conditions. Recent studies using the sultr2;1 null-KO plants suggest that SULTR2;1 contributes to the translocation of sulfate or sulfur-containing metabolites from old to young leaves possibly through phloem (Liang et al., 2010; Kawashima et al., 2011). Partial suppression of SULTR2;1 expression due to T-DNA insertions in the 3′-region may also affect the distribution of sulfur in the aerial tissues of tKO. How this gene expression mechanism driven by the SULTR2;1 3′-region can be controlled in parallel with miR395 regulating the SULTR2;1 transcript levels in shoots remains to be investigated.
Prediction of SURE21A and SURE21B Binding Proteins
Unraveling the molecular machinery of –S-inducible gene expression of SULTR2;1 will likely involve the identification of trans-acting factors binding to SURE21A and SURE21B. The trans-factors expected to be involved in this mechanism would be repressed or activated under –S by binding to or dissociating from the cis-elements, thereby influencing the basic machinery of transcription, and ultimately would enhance expression of SULTR2;1. SURE21 consists of two elements, SURE21A and SURE21B, which are located 80 bp apart, suggesting that the two kinds of DNA binding proteins may bind to each element and cooperatively stimulate SULTR2;1 transcription under –S.
Database searches on PLACE (Higo et al., 1999) revealed sequence similarity between SURE21A and SURE21B and several cis-acting elements reported in plants. SURE21A, 5′-CAATGTATC-3′, includes the CAAT and GATA motifs. CAAT has been reported as a tissue-specific sequence of a pea (Pisum sativum) legumin gene that is functional in tobacco (Nicotiana tabacum; Shirsat et al., 1989); however, its binding protein has not been found yet. The plant GATA motif was first identified in the light-responsive Cab promoters in tobacco (Lam and Chua, 1989). Subsequently, it was also found in the promoter of spinach (Spinacia oleracea) nitrite reductase gene, whose expression is controlled by the availability of ammonium (Rastogi et al., 1997). The Arabidopsis genome has 29 GATA family transcription factors, classified into four subfamilies that are characterized by a single zinc finger domain with 18 or 20 residues in the zinc finger loop (Reyes et al., 2004). Several GATA transcription factors are reported to have biological functions in plant growth and development, including cell elongation, cell division, chloroplast development through chlorophyll synthesis, and gibberellin signaling (Shikata et al., 2004; Bi et al., 2005; Richter et al., 2010; Hudson et al., 2011; Chiang et al., 2012). SURE21B, 5′-CTAGTAC-3′, includes YATC and GTAC motifs. YATC has been reported as a cis-regulatory element for mesophyll-specific expression of phosphoenolpyruvate carboxylase in the C4 plant Flaveria trinevia (Gowik et al., 2004). However, its binding protein has not been reported. Arabidopsis also has 16 members of the SQUAMOSA-promoter binding protein-like (SPL) family of GTAC binding transcription factors (Cardon et al., 1999; Guo et al., 2008; Yang et al., 2008). SPL is a plant-specific transcription factor, with a highly conserved region of 76 amino acids named the SBP-box, that is responsible for both the interaction with DNA and for nuclear import (Klein et al., 1996; Birkenbihl et al., 2005; Yang et al., 2008). The SPL family members play diverse roles in developmental processes and stress responses in plants (Yamasaki et al., 2009; Preston and Hileman, 2013).
Transcript levels of Arabidopsis GATA and SPL family transcription factors in +S and –S roots are summarized in Supplemental Tables 1 and 2. GATA4, GATA18, SPL2, and SPL15 are repressed under –S, but none of them is induced under the same condition (Supplemental Tables 1 and 2). Our deletion study suggested that SURE21A including a GATA motif is involved in the induction of transcription in response to –S (Figure 2). If any of the GATA transcription factors function as a repressor, decreased levels of GATA4 and GATA18 may trigger an increase in SULTR2;1 expression under –S. The deletion study also suggested that SURE21B, which includes a GTAC motif, is involved in the repression of transcription under +S (Figure 2). None of the SPL transcription factors has been reported as a transcriptional repressor; however, it can be hypothesized that the binding of these SPL proteins inhibits SULTR2;1 expression under +S, while their repression leads to an induction of SULTR2;1 expression under –S. A number of questions and possibilities still remain as to whether and how the binding of these two transcription factors to SURE21A and SURE21B and their interactions with each other would induce SULTR2;1 expression. The existence of novel proteins that bind to SURE21A and SURE21B would be another possibility. Identification of other SURE21A and SURE21B binding proteins and clarification of their functions need to be investigated for understanding of this transcriptional control machinery.
In summary, in this study, we identified two novel sulfur-responsive elements, SURE21A and SURE21B, located in the 3′-nontranscribed region as cis-acting elements indispensable for inducing SULTR2;1 expression in roots in response to –S. The function of SULTR2;1 in the uptake and translocation of sulfate under –S in the roots was revealed from the experiments using tKO mutants containing T-DNA insertions in the 3′-nontranscribed region of SULTR2;1. This work provides evidence for a molecular mechanism underlying –S-induced expression of SULTR2;1 in Arabidopsis roots and explains the physiological meaning of the induction of SULTR2;1 expression with relevance to the presence of the two possible enhancers, SURE21A and SURE21B. The ability of the SULTR2;1 3′-nontranscribed region to control the –S-responsive and the tissue-specific gene expressions of SULTR2;1 demonstrates another regulatory mechanism of sulfate uptake and translocation in plants. The mechanism demonstrated in this study represents a part of the transcriptional machinery, which is under control of signal transduction pathways operating in response to changes in sulfur conditions, but is distinct from the SLIM1-dependent regulatory pathway. It is suggested that SURE21A/SURE21B-dependent transcriptional induction and miR395-dependent posttranscriptional regulation allow fine-tuning of the SULTR2;1 transcript levels in roots. These multiple mechanisms are probably controlled by signals carrying spatio-temporal and conditional information on sulfur status to make timely and flexible responses to deficiency of such an essential macronutrient as sulfur, which needs to be acquired from the soil environment by the expression of functional sulfate transporters in roots.
METHODS
Plant Growth
Arabidopsis thaliana plants were grown at 22°C under 16-h-light/8-h-dark long-day cycles. Plants were grown on mineral nutrient media (Hirai et al., 1995) containing 1% sucrose. For preparation of agar medium, agar was washed twice with 1 liter of deionized water and vacuum filtered to remove sulfate. +S agar medium was supplied with 1500 μM MgSO4. –S agar medium was supplied with 15 μM MgSO4, and Mg concentration was adjusted to 1500 μM by adding MgCl2.
Transgenic Plants
The chimeric constructs are shown schematically in Figures 1 to 6. The primers used are listed in Supplemental Table 3.
The synthesis of PSULTR2;1:GFP:TSULTR2;1, PSULTR2;1:Luc:TSULTR2;1, PSULTR2;1:GFP:TNOS, and PSULTR2;1:Luc:TNOS fusion gene constructs (Figures 1, 2, 5, and 6) was performed as follows: The SULTR2;1 5′region (PSULTR2;1) 2535 bp upstream of the translational initiation site of SULTR2;1 was amplified from genomic DNA of Arabidopsis (ecotype Col-0) by PCR using Pfu turbo DNA polymerase (Stratagene-Agilent) and SULTR2;1PFSpe and SULTR2;1PRBam primers. The amplified PCR fragment containing the SpeI and BamHI sites on the ends was cloned into pCR-BluntII-TOPO (Invitrogen) and sequenced. Similarly, the SULTR2;1 3′-region (TSULTR2;1) 1077 bp downstream of the translational termination site of SULTR2;1 was amplified using SULTR2;1TerFNot and SULTR2;1TerREco primers, cloned, and sequenced. Then, TSULTR2;1 as the NotI-EcoRI-ended fragment was cloned between the NotI and EcoRI sites of pTH2 or pTHLuc, replacing the nopaline synthase terminator region (TNOS) in these vectors. pTHLuc is a plasmid made by substitution of the NcoI-NotI GFP region in pTH2 (Chiu et al., 1996) by the Luc coding sequence (Promega). From these plasmids, either in pCR-BluntII-TOPO, pTH2, or pTHLuc vector backgrounds, PSULTR2;1 as the SpeI-BamHI fragment, and GFP:TSULTR2;1, Luc:TSULTR2;1, GFP:TNOS, and Luc:TNOS as the BamHI-EcoRI fragments were isolated and cloned between the XbaI and EcoRI sites of pBI101 (Clontech), replacing the GUS:TNOS region in this binary vector to obtain the promoter:reporter:terminator fusion constructs.
For the synthesis of P35S:Luc:TSULTR2;1 and P35S:Luc:TNOS fusion gene constructs (Figure 4), Luc:TSULTR2;1 or Luc:TNOS as the BamHI-EcoRI fragments were cloned between the BamHI and EcoRI sites of pBI121 (Clontech), replacing the GUS-TNOS region. For the synthesis of PSULTR1;1:Luc:TSULTR2;1 and PSULTR1;1:Luc:TNOS fusion gene constructs (Figure 4), the SULTR1;1 5′region (PSULTR1;1) 2453 bp upstream of the translation initiation site of SULTR1;1, which lacks the sulfur-responsive element of the SULTR1;1 promoter (SURE11; Maruyama-Nakashita et al., 2005), was obtained as the SalI-BamHI fragment, ligated with the Luc:TSULTR2;1 or Luc:TNOS BamHI-EcoRI fragments, and cloned between the SalI and EcoRI sites of pBI101, replacing the GUS:TNOS region.
For the deletion analysis of TSULTR2;1 (Figure 2A), the regions starting from the translational termination site of SULTR2;1 and ending at the positions +663, +459, +449, +431, +411, +391, +372, +360, +352, +332, and +267 were amplified from the plasmid containing the 1077-bp TSULTR2;1 fragment by PCR using Pfu turbo DNA polymerase. PCR was performed using SULTR2;1TerFNot as a forward primer and SULTR2;1TerREco663, 459, 449, 431, 411, 391, 372, 360, 352, 332, or 267 as reverse primers. The amplified PCR fragments containing the NotI and EcoRI sites on ends were cloned and sequenced. These TSULTR2;1 deletion fragments were then cloned between the NotI and EcoRI sites of pTHLuc to be fused to Luc using NotI. The Luc:TSULTR2;1 fusions including TSULTR2;1 deletions were isolated as the BamHI-EcoRI-ended fragments and cloned into the XbaI-EcoRI sites of pBI101 together with the PSULTR2;1 SpeI-BamHI fragment.
The fragments having deletions of +361/+372 region with or without +449/+459 region of TSULTR2;1 (Figure 2B) were made by overlap extension and amplified by two-step PCR. In the first PCR, a segment from the translational termination site of SULTR2;1 to the position +360 and two other segments from +373 to either +449 or +459 were amplified from the plasmid containing the 1077-bp TSULTR2;1 fragment using high-fidelity KOD-Plus DNA polymerase (Toyobo) and the primer sets, SULTR2;1TerFNot and 2;1T(360/372)R, and 2;1T(360/372)F and either 2;1TerREco449 or 2;1TerREco459. The resultant fragments were mixed to make overlaps and used as templates for the second PCR, which was performed using SULTR2;1TerFNot and either 2;1TerREco449 or 2;1TerREco459.
The mutated fragments of the +449/+459 region (Figure 3) were amplified from the plasmid containing the 459-bp TSULTR2;1 fragment using 2;1TerFNot as the forward primer and 2;1T459Rbs-1, -2, -3, -4, or -5 as reverse primers. The mutated fragments of the +361/+372 region (Figure 3) were made by overlap extension and amplified by a two-step PCR. In the first PCR, the regions from the translation termination site of SULTR2;1 to the mutated position, and the regions from mutated position to +459 were amplified from the plasmid containing the 459-bp TSULTR2;1 fragment, using the primer pairs between the forward primer 2;1TerFNot and either of the reverse primers 2;1T(360-372)Rbs-1, -2, -3, -4, -5, or -6, and the primer pairs between either of the forward primers 2;1T(360-372)Fbs-1, -2, -3, -4, -5, or -6 and the reverse primer 2;1TerREco459. The resultant fragments containing the same mutations were mixed to make overlaps and used as templates for the second PCR, which was performed using SULTR2;1TerFNot and 2;1TerREco459. The amplified PCR fragments containing the NotI and EcoRI sites on ends were cloned into pCR-BluntII-TOPO and sequenced. The NotI-EcoRI-ended fragments with mutations were cloned between the NotI and EcoRI sites of pTHLuc to be fused to Luc using NotI. From these plasmids, the Luc:TSULTR2;1 fusions with mutations were isolated as BamHI-EcoRI fragments and cloned between the XbaI and EcoRI sites of pBI101, together with the PSULTR2;1 SpeI-BamHI fragment.
Construction of 332-459TSULTR2;1(sense/antisense):PSULTR2;1:Luc:TNOS, 332-459TSULTR2;1(sense/antisense):mPSULTR2;1:Luc:TNOS, and 332-459TSULTR2;1(sense/antisense):P35S minimal:Luc:TNOS fusion genes was performed as follows: The +332 to +459 region of TSULTR2;1 in sense or antisense orientations [332-459TSULTR2;1(sense) or 332-459TSULTR2;1(antisense)] were created by annealing the two sets of complementary synthetic oligomers, 2;1TerFHindXba(sense) and 2;1TerRHindXba(sense), and 2;1TerFHindXba(antisense) and 2;1TerRHindXba(antisense), to have HindIII and XbaI sites on ends. As mPSULTR2;1, a DNA fragment covering 142 bp upstream of the translation start site of SULTR2;1 was created by annealing the two complementary synthetic oligomers, 2;1miniProFXbaBam and 2;1miniProRXbaBam. As P35S minimal, the –46/0 region of the CaMV 35S promoter was created by annealing the two complementary synthetic oligomers, 35SminiF-Xba and 35SminiR-Bam. These minimal promoter fragments were cloned between the XbaI and BamHI sites of pBI101-Luc (Maruyama-Nakashita et al., 2005). The resultant plasmid containing the mPSULTR2;1:Luc:TNOS or P35S minimal:Luc:TNOS fusion gene was used to clone the HindIII-XbaI-ended 332-459TSULTR2;1(sense) or 332-459TSULTR2;1(antisense) fragments in front of mPSULTR2;1 or P35S minimal. PSULTR2;1:Luc:TNOS fusion construct made in pBI101 was also used for cloning these sense or antisense fragments in front of the 2535-bp PSULTR2;1 fragment.
For the construction of P35S minimal:GFP:TSULTR2;1 (35mPGT in Figure 6), the –46/0 region of the CaMV 35S promoter was created by annealing the two complementary synthetic oligomers 35SminiF-Sal and 35SminiR-Bam, and the resultant fragment was cloned between the SalI and BamHI site in front of the GFP:TSULTR2;1 fusion construct made in pBI101. For the construction of PSULTR1;1(2453):GFP:TSULTR2;1 fusion gene (1;1P2453GT in Figure 6), the 2453-bp SULTR1;1 5′-region was isolated as a SalI-BamHI fragment from the plasmid created previously (Maruyama-Nakashita et al., 2004a) and cloned between the SalI and BamHI site in front of the GFP:TSULTR2;1 fusion construct made in pBI101. PSULTR1;1(3995):GFP:TNOS was described previously (Maruyama-Nakashita et al., 2004a; 1;1P3995GT in Figure 6).
The resultant binary plasmids were transferred to Agrobacterium tumefaciens GV3101 (pMP90) (Koncz and Schell, 1986) by freeze and thaw methods. Arabidopsis plants were transformed according to the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on GM media (Valvekens et al., 1988) containing 50 mg L−1 kanamycin sulfate. Kanamycin-resistant T2 progenies were used for the analysis.
Imaging of GFP Expression
Expression of GFP in whole intact Arabidopsis seedlings was visualized using a FluorImager 595 image analyzer under 488-nm laser excitation (Molecular Dynamics-GE Healthcare). Fluorescence of GFP was detected using a 515- to 545-nm band-pass filter. Autofluorescence derived from chlorophyll was scanned in parallel using a 610-nm long-pass filter.
Tissue-specific expression of GFP in transgenic plants was visualized using a FluoView 500 confocal laser scanning microscopy system equipped with a 505- to 525-nm band-pass filter (Olympus).
Luciferase Assay
Luciferase activity was determined using a Mithras LB 940 multilabel reader (Berthold Technologies) following the manufacturer’s instructions. Protein extraction and luciferase assay were performed using the Luciferase Assay System (Promega) as described previously (Maruyama-Nakashita et al., 2005). The luciferase activities were expressed as relative luminescence units per milligram of protein. Protein concentration was determined with a Bio-Rad protein assay kit according to Bradford (1976) using BSA as a standard protein.
Prediction of SURE21A and SURE21B in Up- and Downstream Sequences of –S-Regulated Genes
Subio Platform programs were used for data analysis. To identify the SURE21A (5′-CAATGTATC-3′) and SURE21B (5′-CTAGTAC-3′), and their complementarily sequences in upstream and downstream sequences of –S-regulated genes, the list of 469 –S-regulated genes selected from microarray analysis was imported (Maruyama-Nakashita et al., 2006). Among the 469 genes, –S-upregulated and -downregulated genes were separated by their fold change of mRNA levels in –S compared with +S. A group of 238 genes with –S/+S fold-change values of >1 was categorized as –S-upregulated, and the remaining 231 genes with –S/+S fold-change values of <1 were categorized as –S-downregulated genes. The presence and the positions of SURE21A and SURE21B were searched using Genomic Analysis Plug-In (Subio).
Isolation of T-DNA Insertion Mutants for 3′-Nontranscribed Region of SULTR2;1
Homozygous knockout mutants for SAIL_363_C06 (Sessions et al., 2002; Col-0 background) and FLAG_373B04 (Samson et al., 2002; Ws background) containing T-DNA insertions in the 3′-nontranscribed region of SULTR2;1 were obtained by a screening based on PCR analysis. Confirmation of the T-DNA insertion in SAIL_363_C06 was done by PCR on genomic DNA using T-DNA left border primer SAIL-LB3 (Sessions et al., 2002) and SULTR2;1-specific primers, SULTR2;1TerFNot or 2;1T656R. The T-DNA insertion in FLAG_373B04 was confirmed by PCR on genomic DNA using T-DNA left border primer FLAG-LB4 (Samson et al., 2002) and SULTR2;1-specific primers. The primers used are listed in Supplemental Table 4.
Real-Time RT-PCR Analysis of SULTR2;1
RNA preparation and reverse transcription were performed as reported previously (Maruyama-Nakashita et al., 2004a). Real-time PCR was performed using SYBR Green Perfect real-time kit (Takara Bio) and Thermal Cycler Dice real-time system (Takara Bio) using the gene-specific primers for SULTR2;1 (SULTR2;1-1648F, 5′-ATGACGGTTAAGACTCCCGGA-3′, and SULTR2;1-1750R, 5′-CGACCCATCCCATAATCCTTT-3′) and for UBIQUITIN2 (UBQ2-144F, 5′-CCAAGATCCAGGACAAAGAAGGA-3′, and UBQ2-372R, 5′-TGGAGACGAGCATAACACTTGC-3′). The relative mRNA levels were calculated using UBIQUITIN2 as an internal standard.
3′-Rapid Amplification of cDNA Ends
RNA preparation was performed as reported previously (Maruyama-Nakashita et al., 2004a). Reverse transcription and RT-PCR was performed using Smart RACE cDNA amplification kit according to the user’s manual (Clontech-Takara Bio). Following the reverse transcription using 3′-RACE CDS primer, the first PCR was performed using Universal Primer A mix (Short) and the gene-specific primers SULTR2;1-3′RACE(1781F), 5′-GCAATGCCAAGAGAAAGATCCTCTTTGTAGT-3′, for amplification of the 3′UTR of SULTR2;1 in Col-0, tKO-Col, Ws, and tKO-Ws, and Luc-3′RACE-1: 5′-CATCTTCGACGCAGGTGTCGCAGGT-3′, for amplification of 3′UTR of Luc in 2;1PLT1077 and 2;1PLT360 plants. The reaction mixtures of the first PCR were used as the templates for the nested PCR. The nested PCR were performed using Nested Universal Primer A and the gene-specific primers SULTR2;1-3′RACE(1938F), 5′-CAAGCTGAATCAAGCAAAGTTCGTCGACAGA-3′, for amplification of 3′UTR of SULTR2;1, and Luc-3′RACE-2: 5′-GAGATCGTGGATTACGTCGCCAGT-3′, for amplification of 3′UTR of Luc. The amplified fragments were cloned into pGEM-T Easy Vectors (Promega) and sequenced.
Sulfate Uptake
Plants were grown vertically for 10 d on +S (1500 μM sulfate) or –S (15 μM sulfate) media. The roots were submerged in +S nutrient solution containing 15 μM [35S] sodium sulfate (Amersham Biosciences) and incubated for 1 h. Washing and measurement were performed as described previously (Kataoka et al., 2004b; Maruyama-Nakashita et al., 2004b).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers and AGI numbers: SULTR2;1 (At5g10180), SULTR1;1 (At4g08620), and UBQ2 (At2g36170).
Supplemental Data
Supplemental Figure 1. Length of 3′-Untranslated Region Determined by 3′-RACE.
Supplemental Table 1. Transcript Levels of GATA Transcription Factor Genes in Plant Roots under Sulfur-Sufficient and -Deficient Conditions.
Supplemental Table 2. Transcript Levels of SPL Transcription Factor Genes in Plant Roots under Sulfur-Sufficient and -Deficient Conditions.
Supplemental Table 3. Oligonucleotides Used for the Construction of Plant Transformation Vectors.
Supplemental Table 4. Primers Used for the Isolation of T-DNA Insertion Mutants.
Supplementary Material
Acknowledgments
We gratefully acknowledge the ABRC, the Salk Institute Genomic Analysis Laboratory, and the Institut National de la Recherche Agronomique for providing the T-DNA insertion lines of SULTR2;1 3′-region. We thank Yukiko Okuo, Akiko Hayashi, Atsumi Onoda, Kazue Nakabayashi, Chieko Komori, Chie Uchiyama, Keiko Nojima, and Yoko Suzuki for technical support; Tatsuhiko Kataoka and Naoko Yoshimoto for advice on the sulfate uptake assay; and Yasuo Niwa (University of Shizuoka, Japan) for providing the sGFP(S65T) vector. This work was supported by JSPS KAKENHI Grant 24380040 and MEXT KAKENHI Grant 23119516 (to A.M.-N.). H.T. is supported by the National Science Foundation (MCB-1244300) and AgBioResearch.
AUTHOR CONTRIBUTIONS
A.M.-N. designed the research. A.M.-N., A.W.-T., and E.I. performed research. A.M.-N., A.W.-T., E.I., T.Y., K.S., and H.T. analyzed data. A.M.-N and H.T. wrote the article.
Glossary
- Col-0
Columbia-0
- UTR
untranslated region
- CaMV
cauliflower mosaic virus
- Ws
Wassilewskija
References
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. [DOI] [PubMed] [Google Scholar]
- Awazuhara M., Kim H., Goto D.B., Matsui A., Hayashi H., Chino M., Kim S.-G., Naito S., Fujiwara T. (2002). A 235-bp region from a nutritionally regulated soybean seed-specific gene promoter can confer its sulfur and nitrogen response to a constitutive promoter in aerial tissues of Arabidopsis thaliana. Plant Sci. 163: 75–82. [Google Scholar]
- Benfey P.N., Ren L., Chua N.H. (1989). The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J. 8: 2195–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y.M., Zhang Y., Signorelli T., Zhao R., Zhu T., Rothstein S. (2005). Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 44: 680–692. [DOI] [PubMed] [Google Scholar]
- Birkenbihl R.P., Jach G., Saedler H., Huijser P. (2005). Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 352: 585–596. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Cardon G., Höhmann S., Klein J., Nettesheim K., Saedler H., Huijser P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237: 91–104. [DOI] [PubMed] [Google Scholar]
- Chiang Y.H., Zubo Y.O., Tapken W., Kim H.J., Lavanway A.M., Howard L., Pilon M., Kieber J.J., Schaller G.E. (2012). Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 160: 332–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6: 325–330. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Davidian J.C., Kopriva S. (2010). Regulation of sulfate uptake and assimilation—the same or not the same? Mol. Plant 3: 314–325. [DOI] [PubMed] [Google Scholar]
- Fabian M.R., Sonenberg N., Filipowicz W. (2010). Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79: 351–379. [DOI] [PubMed] [Google Scholar]
- Gowik U., Burscheidt J., Akyildiz M., Schlue U., Koczor M., Streubel M., Westhoff P. (2004). cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell 16: 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez R.A., MacIntosh G.C., Green P.J. (1999). Current perspectives on mRNA stability in plants: multiple levels and mechanisms of control. Trends Plant Sci. 4: 429–438. [DOI] [PubMed] [Google Scholar]
- Guo A.-Y., Zhu Q.-H., Gu X., Ge S., Yang J., Luo J. (2008). Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 418: 1–8. [DOI] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson D., Guevara D., Yaish M.W., Hannam C., Long N., Clarke J.D., Bi Y.-M., Rothstein S.J. (2011). GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis. PLoS ONE 6: e26765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M.Y., Fujiwara T., Awazuhara M., Kimura T., Noji M., Saito K. (2003). Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J. 33: 651–663. [DOI] [PubMed] [Google Scholar]
- Hirai M.Y., Fujiwara T., Chino M., Naito S. (1995). Effects of sulfate concentrations on the expression of a soybean seed storage protein gene and its reversibility in transgenic Arabidopsis thaliana. Plant Cell Physiol. 36: 1331–1339. [PubMed] [Google Scholar]
- Iyer-Pascuzzi A.S., Jackson T., Cui H., Petricka J.J., Busch W., Tsukagoshi H., Benfey P.N. (2011). Cell identity regulators link development and stress responses in the Arabidopsis root. Dev. Cell 21: 770–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14: 787–799. [DOI] [PubMed] [Google Scholar]
- Kataoka T., Watanabe-Takahashi A., Hayashi N., Ohnishi M., Mimura T., Buchner P., Hawkesford M.J., Yamaya T., Takahashi H. (2004a). Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 16: 2693–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T., Hayashi N., Yamaya T., Takahashi H. (2004b). Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol. 136: 4198–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima C.G., Yoshimoto N., Maruyama-Nakashita A., Tsuchiya Y.N., Saito K., Takahashi H., Dalmay T. (2009). Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J. 57: 313–321. [DOI] [PubMed] [Google Scholar]
- Kawashima C.G., et al. (2011). Interplay of SLIM1 and miR395 in the regulation of sulfate assimilation in Arabidopsis. Plant J. 66: 863–876. [DOI] [PubMed] [Google Scholar]
- Klein J., Saedler H., Huijser P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 250: 7–16. [DOI] [PubMed] [Google Scholar]
- Koncz C., Schell J. (1986). The promoter of T-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel types of Agrobacterium binary vector. Mol. Gen. Genet. 204: 383–396. [Google Scholar]
- Kutz A., Müller A., Hennig P., Kaiser W.M., Piotrowski M., Weiler E.W. (2002). A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J. 30: 95–106. [DOI] [PubMed] [Google Scholar]
- Lam E., Chua N.H. (1989). ASF-2: a factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in Cab promoters. Plant Cell 1: 1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T., Martin M.N., Bick J.A., Davies J.P. (2000). Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51: 141–165. [DOI] [PubMed] [Google Scholar]
- Lewandowska M., Wawrzynska A., Moniuszko G., Lukomska J., Zientara K., Piecho M., Hodurek P., Zhukov I., Liszewska F., Nikiforova V., Sirko A. (2010). A contribution to identification of novel regulators of plant response to sulfur deficiency: characteristics of a tobacco gene UP9C, its protein product and the effects of UP9C silencing. Mol. Plant 3: 347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Yang F., Yu D. (2010). MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 62: 1046–1057. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Nakamura Y., Watanabe-Takahashi A., Inoue E., Yamaya T., Takahashi H. (2005). Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J. 42: 305–314. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Nakamura Y., Tohge T., Saito K., Takahashi H. (2006). Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18: 3235–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Nakamura Y., Watanabe-Takahashi A., Yamaya T., Takahashi H. (2004a). Induction of SULTR1;1 sulfate transporter in Arabidopsis roots involves protein phosphorylation/dephosphorylation circuit for transcriptional regulation. Plant Cell Physiol. 45: 340–345. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Inoue E., Watanabe-Takahashi A., Yamaya T., Takahashi H. (2003). Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol. 132: 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Nakamura Y., Yamaya T., Takahashi H. (2004b). A novel regulatory pathway of sulfate uptake in Arabidopsis roots: implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J. 38: 779–789. [DOI] [PubMed] [Google Scholar]
- Nikiforova V., Freitag J., Kempa S., Adamik M., Hesse H., Hoefgen R. (2003). Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J. 33: 633–650. [DOI] [PubMed] [Google Scholar]
- Paulose B., Chhikara S., Coomey J., Jung H.I., Vatamaniuk O., Dhankher O.P. (2013). A γ-glutamyl cyclotransferase protects Arabidopsis plants from heavy metal toxicity by recycling glutamate to maintain glutathione homeostasis. Plant Cell 25: 4580–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J.C., Hileman L.C. (2013). Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front. Plant Sci. 4: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi R., Bate N.J., Sivasankar S., Rothstein S.J. (1997). Footprinting of the spinach nitrite reductase gene promoter reveals the preservation of nitrate regulatory elements between fungi and higher plants. Plant Mol. Biol. 34: 465–476. [DOI] [PubMed] [Google Scholar]
- Reyes J.C., Muro-Pastor M.I., Florencio F.J. (2004). The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 134: 1718–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R., Behringer C., Müller I.K., Schwechheimer C. (2010). The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev. 24: 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K. (2004). Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol. 136: 2443–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson F., Brunaud V., Balzergue S., Dubreucq B., Lepiniec L., Pelletier G., Caboche M., Lecharny A. (2002). FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30: 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibagaki N., Rose A., McDermott J.P., Fujiwara T., Hayashi H., Yoneyama T., Davies J.P. (2002). Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J. 29: 475–486. [DOI] [PubMed] [Google Scholar]
- Shikata M., Matsuda Y., Ando K., Nishii A., Takemura M., Yokota A., Kohchi T. (2004). Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J. Exp. Bot. 55: 631–639. [DOI] [PubMed] [Google Scholar]
- Shirsat A., Wilford N., Croy R., Boulter D. (1989). Sequences responsible for the tissue specific promoter activity of a pea legumin gene in tobacco. Mol. Gen. Genet. 215: 326–331. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Buchner P., Yoshimoto N., Hawkesford M.J., Shiu S.H. (2012). Evolutionary relationships and functional diversity of plant sulfate transporters. Front. Plant Sci. 2: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Kopriva S., Giordano M., Saito K., Hell R. (2011). Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 62: 157–184. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Watanabe-Takahashi A., Smith F.W., Blake-Kalff M., Hawkesford M.J., Saito K. (2000). The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J. 23: 171–182. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Yamazaki M., Sasakura N., Watanabe A., Leustek T., Engler J.A., Engler G., Van Montagu M., Saito K. (1997). Regulation of sulfur assimilation in higher plants: a sulfate transporter induced in sulfate-starved roots plays a central role in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94: 11102–11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D., Montagu M.V., Van Lijsebettens M. (1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85: 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidmar J.J., Tagmount A., Cathala N., Touraine B., Davidian J.E. (2000). Cloning and characterization of a root specific high-affinity sulfate transporter from Arabidopsis thaliana. FEBS Lett. 475: 65–69. [DOI] [PubMed] [Google Scholar]
- Wawrzyńska A., Lewandowska M., Sirko A. (2010). Nicotiana tabacum EIL2 directly regulates expression of at least one tobacco gene induced by sulphur starvation. J. Exp. Bot. 61: 889–900. [DOI] [PubMed] [Google Scholar]
- Yamasaki H., Hayashi M., Fukazawa M., Kobayashi Y., Shikanai T. (2009). SQUAMOSA promoter binding protein–like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21: 347–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Wang X., Gu S., Hu Z., Xu H., Xu C. (2008). Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 407: 1–11. [DOI] [PubMed] [Google Scholar]
- Yoshimoto N., Takahashi H., Smith F.W., Yamaya T., Saito K. (2002). Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J. 29: 465–473. [DOI] [PubMed] [Google Scholar]
- Yoshimoto N., Inoue E., Saito K., Yamaya T., Takahashi H. (2003). Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol. 131: 1511–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto N., Inoue E., Watanabe-Takahashi A., Saito K., Takahashi H. (2007). Posttranscriptional regulation of high-affinity sulfate transporters in Arabidopsis by sulfur nutrition. Plant Physiol. 145: 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.