The inositol pyrophosphate InsP8 plays an important role in plant defenses against herbivorous insects and necrotrophic fungi and is a key cofactor of the jasmonate receptor complex.
Abstract
Diphosphorylated inositol polyphosphates, also referred to as inositol pyrophosphates, are important signaling molecules that regulate critical cellular activities in many eukaryotic organisms, such as membrane trafficking, telomere maintenance, ribosome biogenesis, and apoptosis. In mammals and fungi, two distinct classes of inositol phosphate kinases mediate biosynthesis of inositol pyrophosphates: Kcs1/IP6K- and Vip1/PPIP5K-like proteins. Here, we report that PPIP5K homologs are widely distributed in plants and that Arabidopsis thaliana VIH1 and VIH2 are functional PPIP5K enzymes. We show a specific induction of inositol pyrophosphate InsP8 by jasmonate and demonstrate that steady state and jasmonate-induced pools of InsP8 in Arabidopsis seedlings depend on VIH2. We identify a role of VIH2 in regulating jasmonate perception and plant defenses against herbivorous insects and necrotrophic fungi. In silico docking experiments and radioligand binding-based reconstitution assays show high-affinity binding of inositol pyrophosphates to the F-box protein COI1-JAZ jasmonate coreceptor complex and suggest that coincidence detection of jasmonate and InsP8 by COI1-JAZ is a critical component in jasmonate-regulated defenses.
INTRODUCTION
Inositol polyphosphates became an intense focus of research with the discovery that myo- inositol 1,4,5-trisphosphate (InsP3) mobilizes Ca2+ in a receptor-dependent fashion from intracellular stores in pancreatic cells (Streb et al., 1983). The stereochemistry of d-myo-inositol suggested that the inositol ring represents a 6-bit signaling scaffold with the potential to “encode” 64 unique signaling states (York, 2006). In plants, InsP3 has been associated with a wide range of cellular functions, such as guard cell physiology (Blatt et al., 1990; Gilroy et al., 1990; Burnette et al., 2003; Han et al., 2003), drought tolerance (Knight et al., 1997; Perera et al., 2008), heat shock responses (Liu et al., 2006), blue light perception (Chen et al., 2008), root gravitropism (Wang et al., 2009; Zhang et al., 2011), response to mechanical wounding (Mosblech et al., 2008), and pollen dormancy (Y. Wang et al., 2012). However, the role of InsP3 and other inositol phosphates in plant signaling remains controversial as no inositol phosphate receptor has been identified to date (Munnik and Vermeer, 2010; Munnik and Nielsen, 2011; Gillaspy, 2013).
Recent discoveries that InsP6 binds to the auxin receptor complex TIR1/IAA (Tan et al., 2007) and InsP5 binds to the jasmonate receptor complex COI1/JAZ (Sheard et al., 2010) suggest that inositol polyphosphates are involved in plant hormone perception. COI1 is the F-box component of a SKP1-CUL1-F-box protein (SCF) ubiquitin E3 ligase complex that recruits Jasmonate ZIM-domain (JAZ) transcriptional repressors upon binding to the bioactive jasmonic acid (JA) conjugate JA-Ile. This triggers polyubiquitylation and subsequent degradation of the JAZ repressors by the 26S proteasome (Chini et al., 2007; Thines et al., 2007; Katsir et al., 2008; Sheard et al., 2010). JAZ degradation relieves repression of MYC2 and other transcription factors, thus permitting the expression of jasmonate-responsive genes (Chini et al., 2007). Mass spectrometry and NMR analyses revealed that inositol-1,2,4,5,6-pentakisphosphate [Ins(1,2,4,5,6)P5] copurified with Arabidopsis SKP1 homolog (ASK1)-COI1 expressed in insect cells (Sheard et al., 2010). Although ligand binding based reconstitution assays suggested that Ins(1,2,4,5,6)P5 potentiates jasmonate receptor assembly in vitro, its physiological role remains unclear. Two studies that analyzed Arabidopsis thaliana backgrounds altered in inositol polyphosphate metabolism provide evidence that this class of molecules contributes to COI1 function during the plants wound response (Mosblech et al., 2008, 2011).
The discovery of diphosphoinositol polyphosphates, also referred to as inositol pyrophosphates, in amoebae, mammals, and yeast (Stephens et al., 1991; Menniti et al., 1993; Saiardi et al., 2000b) revealed an even higher complexity of this family of signaling molecules. In these organisms, inositol pyrophosphates regulate many cellular processes, including stress responses, membrane trafficking, telomere maintenance, ribosome biogenesis, cytoskeletal dynamics, insulin signaling, apoptosis, phosphate homeostasis, and neutrophil activation (Barker et al., 2009; Burton et al., 2009; Shears, 2009; Chakraborty et al., 2011; Wundenberg and Mayr, 2012). Two distinct classes of inositol pyrophosphate synthetases have been described: inositol hexakisphosphate kinases (also termed IP6Ks or Kcs1-like proteins) and diphosphoinositol pentakisphosphate kinases (PPIP5K or IP7K/Vip1-like proteins). IP6Ks phosphorylate the phosphate in the 5-position of InsP6 and Ins(1,3,4,5,6)P5 (InsP5) and can use the resulting inositol pyrophosphates as substrates to generate more complex molecules containing two or more additional pyrophosphate moieties (Saiardi et al., 2000a, 2001; Draskovic et al., 2008). In contrast, PPIP5Ks phosphorylate the phosphate in the 1-position of both 5-InsP7 and InsP6, leading to the formation of the InsP8 isomer 1,5(PP)2-InsP4 (1,5-InsP8) and the InsP7 isomer 1PP-InsP5 (1-InsP7), respectively (Mulugu et al., 2007; Lin et al., 2009; H. Wang et al., 2012).
The existence of inositol species more polar than InsP6 has been reported in Spirodela polyrrhiza (Flores and Smart, 2000), barley (Hordeum vulgare; Brearley and Hanke, 1996; Dorsch et al., 2003), guard cells of intact guard cells of potato (Solanum tuberosum; Lemtiri-Chlieh et al., 2000), and in extracts of Arabidopsis and maize (Zea mays; Desai et al., 2014). In the later study, the authors addressed a possible involvement of Arabidopsis Vip1 homologs in the synthesis of inositol pyrophosphates by performing yeast complementation assays (Desai et al., 2014). Based on these assays, the authors proposed a function of Arabidopsis Vip1 homologs as IP6K enzymes with a possible role in InsP7 biosynthesis. However, plants with altered Vip1 functions have not been investigated and the physiological role(s) of Vip1 proteins and inositol pyrophosphates in plants await clarification.
Here, we show that plant genomes of phylogenetically diverse taxa encode Vip1 homologs that appear to have evolved from a single common ancestor. We also provide evidence that InsP7 and InsP8 are readily detected in Arabidopsis extracts. The Vip1 homologs VIH1 and VIH2 are functional PPIP5Ks, and VIH2 is critical for InsP8 production in Arabidopsis seedlings. Our data further suggest that VIH2-dependent inositol pyrophosphates represent key cofactors of the COI1-JAZ receptor complex, thereby playing an important role in jasmonate perception and jasmonate-regulated defenses.
Note that in a previous published work (Desai et al., 2014), which was published during the preparation of this article, Arabidopsis homologs of yeast Vip1 were named AtVIP1 and AtVIP2 (corresponding to VIH2 and VIH1, respectively). Because VIP1 is already in use for an unrelated protein (Arabidopsis VirE2 Interacting Protein 1, encoded by At1g43700), we propose to use the gene symbol VIH (VIP1 homolog) registered at the TAIR database (see http://www.arabidopsis.org).
RESULTS
Vip1/PPIP5K Homologs Are Widespread in Plants
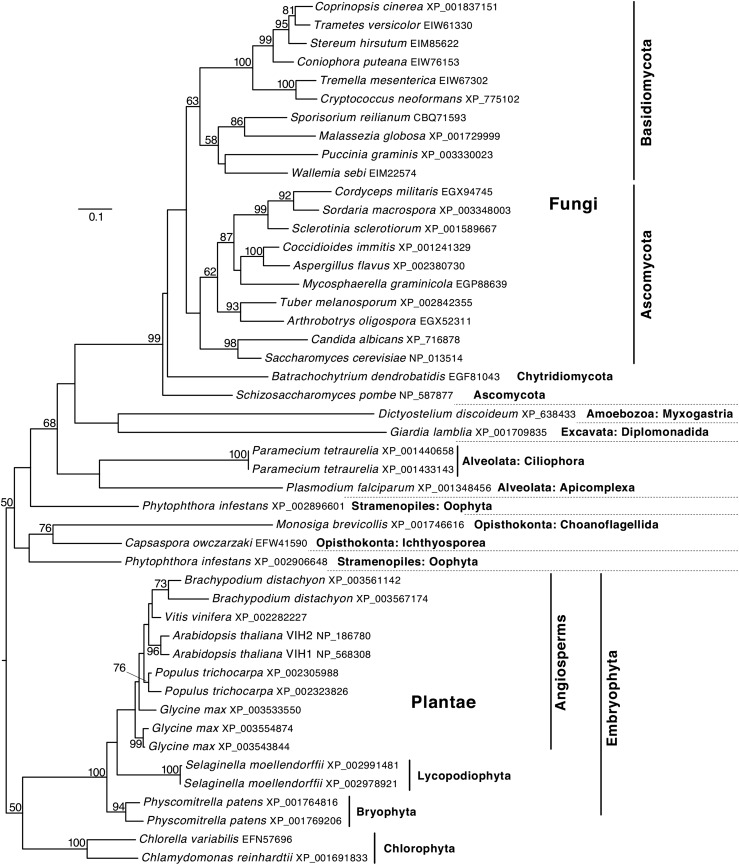
BLAST searching with the N-terminal ATP-grasp kinase domain of Vip1 as the query sequence allowed us to identify genes encoding putative Vip1/PPIP5K proteins in all available plant genomes, including diverse taxa such as green algae (Chlorophyta), mosses (Bryophyta), lycopods, and monocot and eudicot angiosperms, suggesting that PPIP5Ks play important basic functions in all plants. Phylogenetic analysis of the N-terminal ATP-grasp kinase domain of selected proteins, with a focus on plants and fungi, reflects major monophyletic groups as currently accepted (Keeling et al., 2009; Blackwell et al., 2012) (Figure 1). According to the maximum likelihood tree (Figure 1), all of the plant homologs are derived from a single ancestral gene, with subsequent gene duplications in the individual lineages.
Figure 1.
Vip1/PPIP5K Homologs Are Ubiquitous in Plants.
BLAST search analyses were performed employing the protein sequence of the Saccharomyces cerevisiae Vip1 ATP-grasp kinase domain (residues 1 to 535) as a search template. The tree was estimated from an alignment of ATP-grasp kinase domains of the encoded proteins using maximum likelihood. Branch support was calculated from 10,000 bootstrap replicates, and values below 50% are omitted. Branch lengths are given in terms of expected numbers of amino acid substitutions per site.
The N-Terminal ATP-Grasp Kinase Domain in Arabidopsis Vip1 Homologs Has a Two-Domain Architecture and Is Structurally Conserved
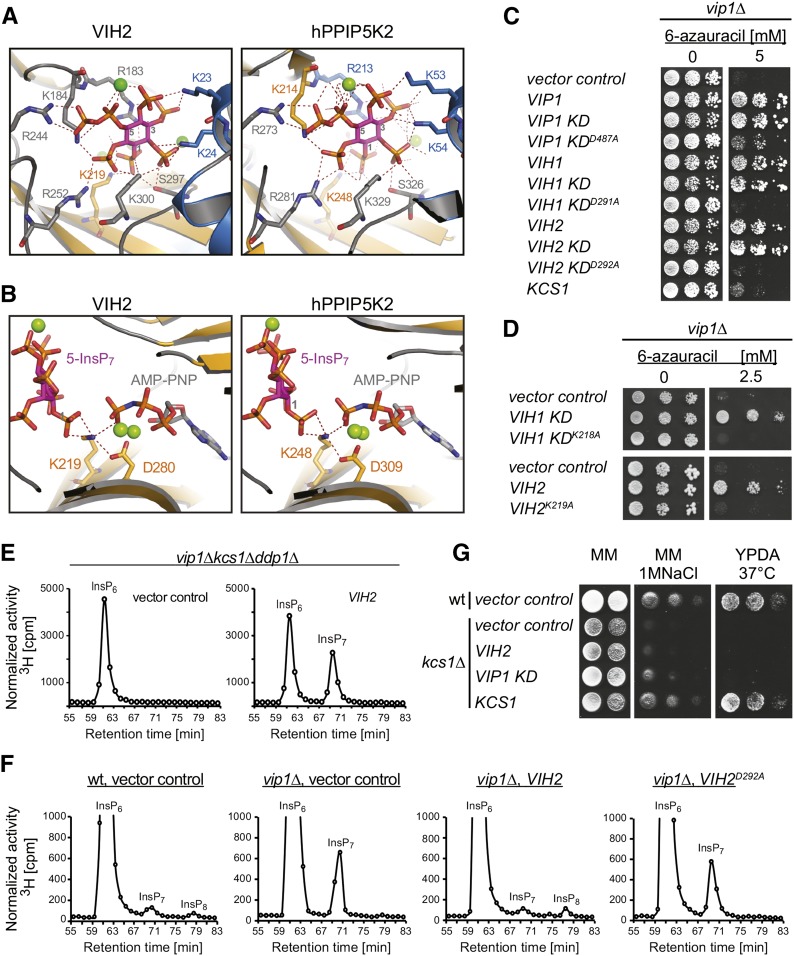
We named the Arabidopsis Vip1 homologs identified in our BLAST search VIH1 and VIH2 (Vip1 homolog), respectively. Protein sequence comparison suggests that both proteins possess a two-domain architecture conserved in members of the Vip1/PPIP5K family, with an N-terminal ATP-grasp kinase domain and a C-terminal phosphatase-like domain (Supplemental Figure 1A). A model of the VIH2 kinase domain based on the crystal structure of human diphosphoinositol pentakisphosphate kinase 2 (hPPIP5K2) predicts the nucleotide analog AMP-PNP to be coordinated between two sets of antiparallel β-sheets as it has been described for hPPIP5K2 (H. Wang et al., 2012) (Supplemental Figure 1B). InsP7 is coordinated exclusively by VIH2 lysine and arginine residues with the exception of one serine residue (as in hPPIP5K2), a hallmark of PPIP5K enzymes (H. Wang et al., 2012). Importantly, all protein-substrate interactions, including residues involved in the phosphotransfer reaction are conserved: VIH2 (hPPIP5K2) residues that coordinate the substrate are Lys-23 (Lys-53), Lys-24 (Lys-54), Arg-183 (Arg-213), Lys-184 (Lys-214), Lys-219 (Lys-248), Arg-244 (Arg-273), Arg-252 (Arg-281), Ser-297 (Ser-326), and Lys-300 (Lys-329) (Figures 2A and 2B) (H. Wang et al., 2012). These residues are also conserved in the VIH1 polypeptide (Supplemental Figure 1C). Collectively, these results suggest Arabidopsis VIH proteins execute Vip1/PPIP5K-like activities.
Figure 2.
VIH1 and VIH2 Are Functional Vip1-Type PPIP5 Kinases.
(A) and (B) Structural model of the VIH2 ATP-grasp kinase domain (left) and the hPPIP5K2 ATP-grasp kinase domain (PDB ID: 3T9D, right) depicting the 5-InsP7 binding sites and key catalytic residues. Residues coordinating substrate via polar contacts are shown as sticks, polar interactions are highlighted by dashed lines, α-helices are rendered in blue, β-sheets in orange, substrate (5-InsP7) is rendered in magenta, and Mg2+ ions are presented as green spheres. Three carbon atoms on the inositol ring are numbered. The ATP analog AMP-PNP (in [B]) is depicted with gray carbon and orange and red phosphate groups.
(C) and (D) Complementation of vip1Δ-associated growth defects in yeast by ectopic expression of inositol pyrophosphate synthetases. The vip1Δ yeast strain transformed with episomal pDR195(URA3) plasmids carrying either VIP1, VIH1, or VIH2, sequences encoding their respective ATP-grasp kinase domains (KD) or designated kinase domain mutants, or carrying KCS1 were spotted in 8-fold serial dilutions onto uracil-free minimal medium in presence or absence of 6-azauracil, as indicated. Rescue on medium supplemented with 6-azauracil (right) reports Vip1 activity.
(E) and (F) Normalized HPLC profiles of inositol phosphates of extracts from designated [3H] inositol-labeled yeast transformants. Extracts were resolved by Partisphere SAX HPLC and fractions collected each minute for subsequent determination of radioactivity as indicated. Changes in elution times in independent experiments were observed and can be explained by subtle changes in column properties or column change. Experiments were repeated three times with similar results.
(G) Complementation assays of kcs1Δ-associated growth defects on high salt by ectopic expression inositol pyrophosphate synthetases. Wild-type (wt) or kcs1Δ yeast transformants (both DDY1810 background) carrying designated plasmids were spotted in 8-fold serial dilutions onto solid minimal media (MM, uracil deficient CSM media with YNB and appropriate supplements) in presence or absence of NaCl and onto solid YPDA media incubated at 37°C.
VIH1 and VIH2 Complement vip1Δ- but Not kcs1Δ-Associated Defects in Yeast
To address the function of VIH more directly, we investigated consequences of heterologous VIH expression in yeast. A previously identified vip1Δ-associated growth defect on 6-azauracil in yeast (Osada et al., 2012) was rescued by Arabidopsis VIH1 and VIH2, suggesting that these proteins execute Vip1-like activities in vivo (Figure 2C). In contrast, ectopic overexpression of Kcs1 under similar conditions failed to cause growth complementation of the single vip1Δ strain (Figure 2C). Rescue of this yeast strain was also observed by expression of the N-terminal kinase domains of Vip1, VIH1, and VIH2, indicating that the 6-azauracil sensitivity of vip1Δ is caused by loss of Vip1 kinase activity and, importantly, that VIH1 and VIH2 possess a functional Vip1-like ATP-grasp kinase domain. This idea is supported by the finding that kinase catalytic dead mutant Vip1D487A (Mulugu et al., 2007) and the corresponding mutant proteins VIH1D291A and VIH2D292A failed to rescue vip1Δ-associated growth defects on 6-azauracil (Figure 2C). In the context of the hPPIP5K2 enzyme, another residue, Lys-248, interacts with both the 1-phosphate of 5-InsP7 and the γ-phosphate of ATP and therefore plays an essential function in the catalytic cycle of 5-InsP7 phosphorylation and the specificity of the phosphotransfer reaction (Figure 2B) (H. Wang et al., 2012). This residue is conserved in plant Vip1 homologs and our structural model suggests that the corresponding Arabidopsis VIH2 residue K219 interacts with substrate and cofactor in a similar manner (Figure 2B).
In agreement with this idea, mutant polypeptides VIH1-KDK218A and VIH2K219A are dysfunctional (Figure 2D). All proteins, including catalytic dead mutants, were correctly expressed (Supplemental Figures 2A and 2B). We further investigated consequences of VIH2 expression on inositol polyphosphate metabolism in different yeast strains. We expressed VIH2 in a vip1Δ kcs1Δ ddp1Δ triple mutant yeast strain, which lacks inositol pyrophosphates and is devoid of Ddp1 (diadenosine-and-diphosphoinositol-polyphosphate-phosphohydrolase)-dependent inositol pyrophosphatase activity, thus facilitating the detection of inositol pyrophosphates synthesized by ectopically expressed kinases (Safrany et al., 1998; Mulugu et al., 2007). VIH2 expression in this background resulted in a robust InsP7 peak that eluted at a retention time identical to the 1-InsP7 peak in transformants ectopically expressing Vip1 (Figure 2E; Supplemental Figure 2D), supporting recent observations by Desai et al. (2014). However, these results did not address whether VIH proteins have Vip1/PPIP5K or Kcs1/IP6K-like activities.
Therefore, we expressed VIH2 in single vip1Δ or kcs1Δ mutant backgrounds. Because of a preference of Vip1 to phosphorylate 5-InsP7 to 1,5-InsP8, vip1Δ yeast cells accumulate the nonmetabolized substrate 5-InsP7 (Azevedo et al., 2009; Onnebo and Saiardi, 2009; Padmanabhan et al., 2009) (Figure 2F). Levels of 1-InsP7 and 1,5-InsP8 are generally low in wild-type yeast due to the activity of Ddp1 (Safrany et al., 1998; Mulugu et al., 2007). As apparent from a robust rescue of (i.e., decrease in) 5-InsP7 levels, expression of VIH2 complemented vip1Δ-associated defects in inositol pyrophosphate homeostasis in a kinase-dependent manner (Figure 2F; Supplemental Figure 2C). We also investigated the consequences of ectopic expression of Kcs1, Vip1, and VIH2 in a kcs1Δ single mutant yeast strain. While ectopic expression of Kcs1 complemented a previously described growth defect of kcs1Δ cells on 1 M NaCl at 37°C, ectopic expression of Vip1 and VIH2 under similar conditions failed to do so. This is in agreement with the idea that VIH2 does not have Kcs1/IP6K-like activities (Figure 2G). We also found that overexpression of Vip1 kinase activity in kcs1Δ cells causes production of InsP7 and InsP8 (Supplemental Figure 2E). Based on previous in vitro studies (Losito et al., 2009), these species are likely to represent 1-InsP7 and 1,3-InsP8 or 1PPP-InsP7. Likewise, ectopic expression of VIH2 caused peaks with identical chromatographic mobilities (Supplemental Figure 2E). Collectively, these data suggest that VIH2 executes Vip1/PPIP5K but not Kcs1/IP6K-like activities in yeast.
Levels of the Inositol Pyrophosphate InsP8 Are Regulated by Methyl Jasmonate and Depend on VIH2
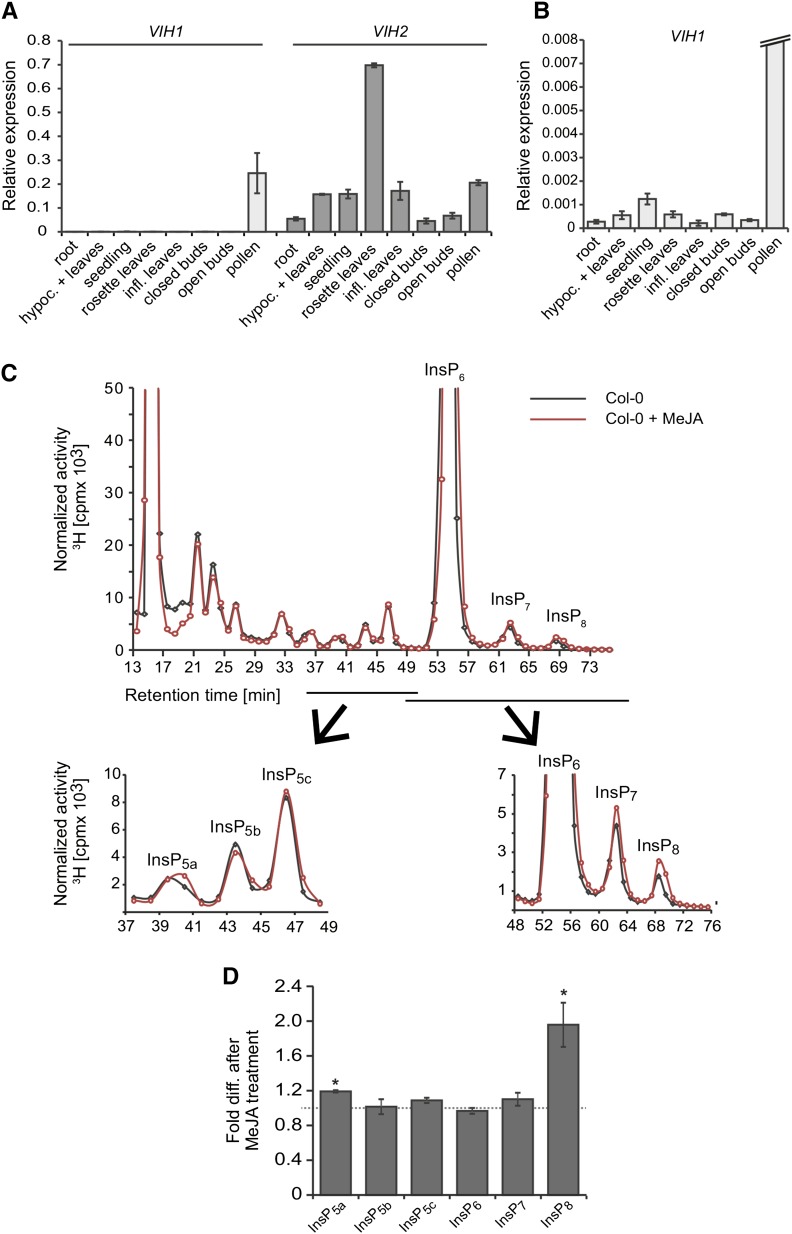
Quantitative PCR (qPCR) analyses showed expression of VIH1 to be restricted mainly to pollen (Figures 3A and 3B). VIH2 expression, on the other hand, was ubiquitous and especially strong in rosette leaves (Figure 3A), suggesting specialized functions of both isoforms. To investigate VIH2 functions in planta, we analyzed inositol polyphosphates in 3-week-old seedlings of wild-type (Columbia-0 [Col-0]) and vih2 mutant plants. HPLC runs of [3H]-inositol-labeled Col-0 plant extracts showed a similar profile as reported previously (Stevenson-Paulik et al., 2005), with a robust peak at a retention time identical to the [3H]-InsP6 standard (Figure 3C; Supplemental Figure 3A). However, in contrast to this previous report, we also detected two additional peaks more anionic than InsP6 that eluted at elution times expected for InsP7 and InsP8, respectively (Figure 3C), supporting recent findings by Desai et al. (2014).
Figure 3.
Expression Analyses Suggest Specialized Functions of VIH1 and VIH2, and Inositol Pyrophosphates Can Be Detected in Arabidopsis Extracts and Are Regulated by Jasmonate.
(A) and (B) qPCR analyses of VIH expression in Col-0 plants using cDNA prepared from RNA extracts of different plant tissues as indicated. Averages of triplicate reactions ± sd are shown. β-TUBULIN was used as reference gene. Transcript levels of VIH1 and VIH2 are presented relative to β-TUBULIN transcript. The experiment was repeated three times with similar results.
(C) and (D) MeJA increases InsP8 level. Normalized HPLC profiles (C) of 3-week old [3H] inositol-labeled Col-0 seedlings that were untreated (solid gray line) or treated for 4 h with 50 μM MeJA (solid red lines). Treated and nontreated plants were harvested simultaneously to avoid daytime-dependent differences in inositol polyphosphate homeostasis. The experiment was repeated with similar results, and representative results from one experiment are shown. For relative amounts of respective species (D), averages of fold differences after MeJA treatment of three independent experiments ± se are shown. Asterisks indicate statistical differences (Student’s t test; *P < 0.02).The isomeric identity of InsP5b is unknown in Arabidopsis seedlings. Based on chromatographic mobilities presented in a previous study on seedlings of Col-0 plants and ipk1-1 plants (Stevenson-Paulik et al., 2005), and comparison with chromatographic mobilities of inositol polyphosphates in the same ipk1-1 line on our HPLC (Supplemental Figures 8A and 8B), InsP5a represents Ins(1,3,4,5,6)P5 and InsP5c represents Ins(1,2,4,5,6)P5 or its enatiomer Ins(2,3,4,5,6)P5.
Inositol phosphates have been implicated in the wound response (Mosblech et al., 2008, 2011; Sheard et al., 2010), a process that is regulated by the oxylipin JA and related signaling molecules (collectively referred to as jasmonates), as well as by the stress hormone abscisic acid (ABA) (Vos et al., 2013). Therefore, we explored the role of jasmonates and ABA in the regulation of inositol pyrophosphates. Treatment of Col-0 seedlings with methyl jasmonate (MeJA) caused a 2-fold increase in InsP8, but only had subtle effects on other inositol polyphosphate species (as exemplified for various InsP5 species; Figures 3C and 3D). A time-course experiment with MeJA-treated plants showed an almost 2-fold increase in levels of InsP8 already after 15 min, which remained stable for the course of 3 h. In contrast, InsP7 levels were not affected by MeJA treatment (Supplemental Figures 3C and 3D). Very different effects were observed in ABA-treated plants: ABA induced increases in both InsP7 and InsP8 in a dose-dependent manner (Supplemental Figure 3E).
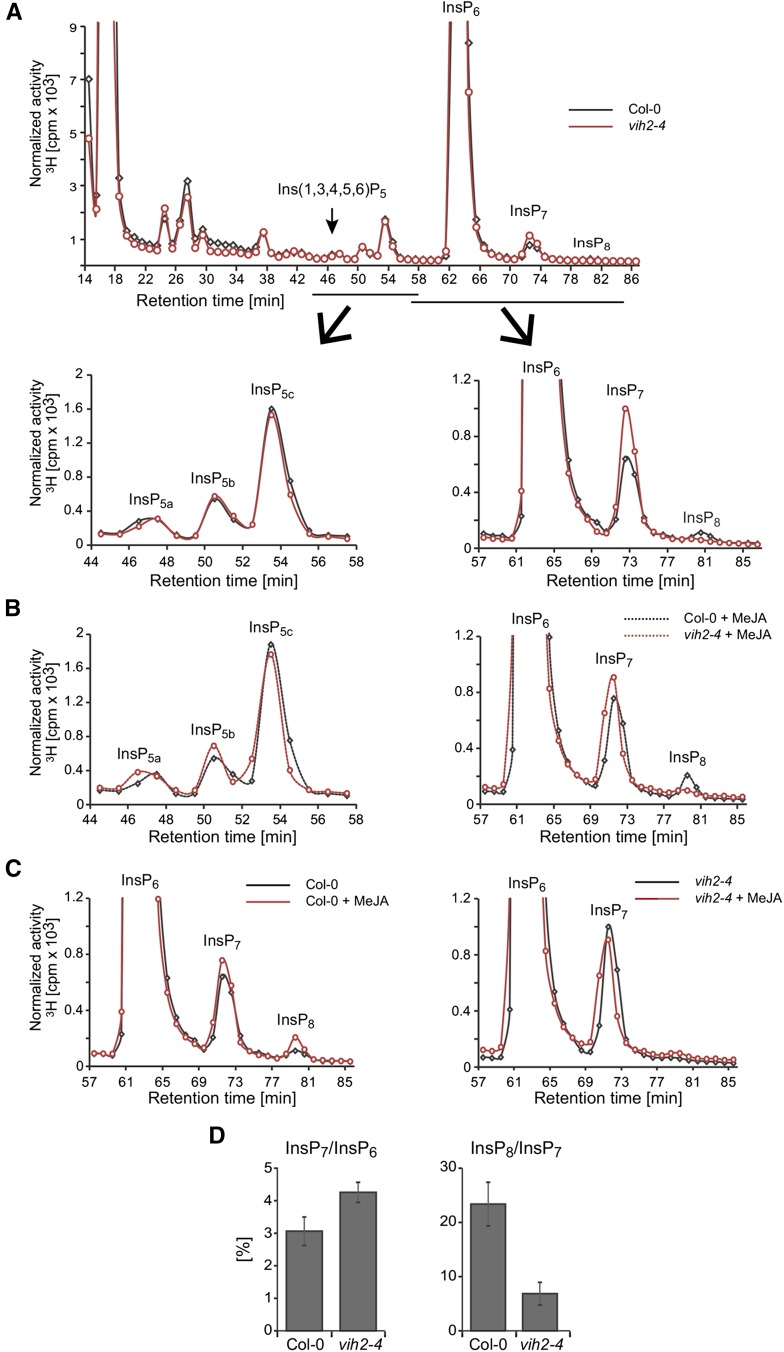
To investigate the potential role of VIH2 in inositol pyrophosphate homeostasis, we analyzed two independent T-DNA insertion lines (vih2-3 and vih2-4) that lack VIH2 transcript (Supplemental Figures 4A and 4B). Overall incorporation of [3H]-myo-inositol was not affected in these mutants (Supplemental Figure 4C). HPLC profiles of vih2 plants were similar to those of wild-type plants but showed a robust reduction of InsP8 with a concomitant increase in InsP7, suggesting that, like Vip1 in yeast, Arabidopsis VIH2 catalyzes the conversion of InsP7 to InsP8 (Figures 4A and 4D; Supplemental Figures 5A to 5C). Small residual levels of InsP8 in vih2 plants remained completely insensitive to MeJA, independent of the exposure time (30 min MeJA in Figures 4B and 4C; 4 h MeJA in Supplemental Figure 5D). In summary, these results show that in seedlings, InsP8 levels are upregulated by MeJA treatment and that both bulk/steady state and MeJA-induced pools of InsP8 depend on VIH2.
Figure 4.
Bulk Steady State and Jasmonate-Induced Pools of InsP8 in Arabidopsis Seedlings Depend on VIH2.
Normalized HPLC profiles ([A] to [C]) or relative amounts (D) of inositol phosphate species of 3-week-old [3H] inositol-labeled Col-0 (solid black line) and vih2-4 seedlings. In (B) and (C), plants were treated with 50 µM MeJA and harvested after 30 min together with nontreated plants. Extracts were resolved by Partisphere SAX HPLC and fractions collected each minute for subsequent determination of radioactivity. The experiment was repeated with similar results, and representative results from one experiment are shown. (B) is a zoom-in into the InsP5 (left) and InsP6-8 (right) regions of HPLC runs with extracts of MeJA-treated Col-0 and vih2 seedlings as indicated. For InsP5a-c isomer identities, see comment in Figure 3. (C) is a zoom-in into the InsP6-8 regions of HPLC runs with extracts of Col-0 (left) and vih2 (right) seedlings with or without MeJA treatment as indicated. For relative amounts (D), data are presented either as InsP7/InsP6 ratio (a measure of IP6K activity) or as InsP8/InsP7 ratio (a measure of PPIP5K activity). The data represent means ± se.
VIH2 Plays a Critical Role in Jasmonate-Regulated Defenses
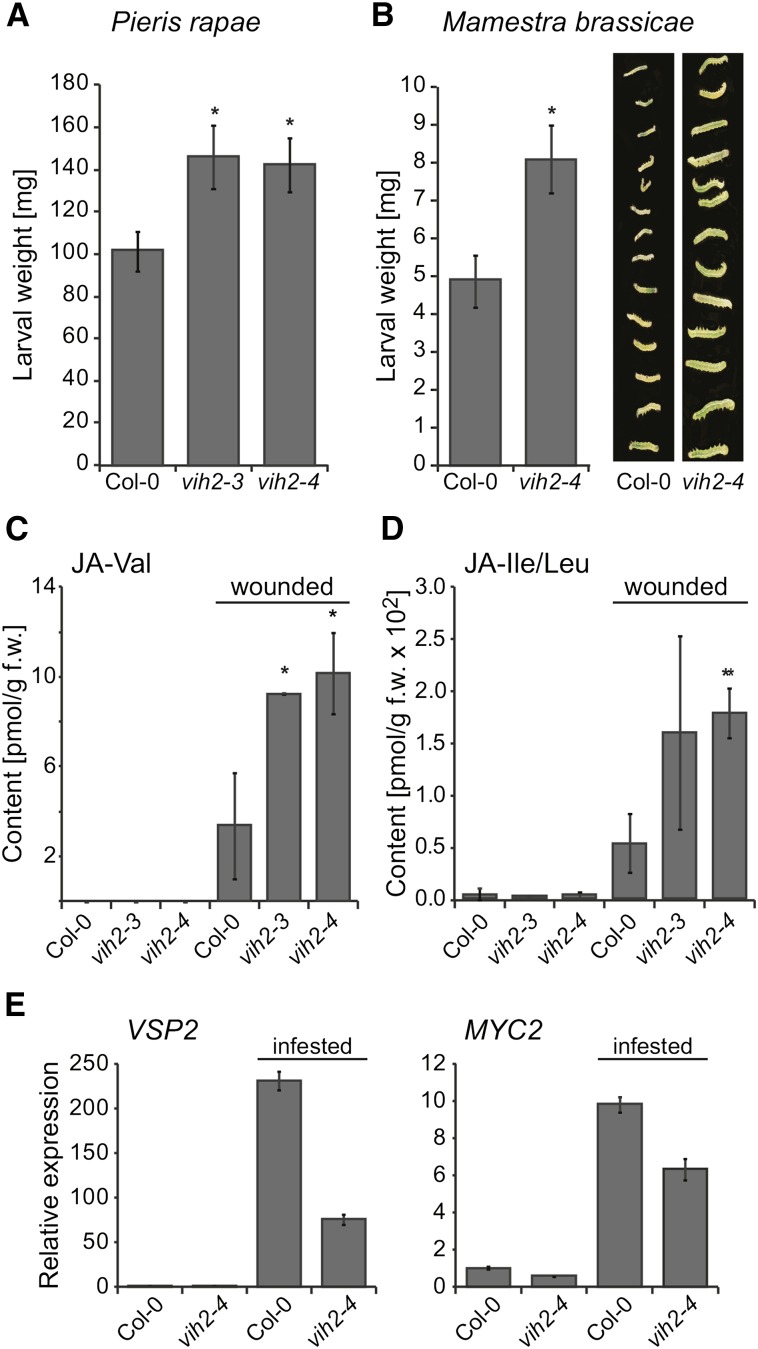
We examined the functional role of VIH2 in the defense against Brassicaceae specialist Pieris rapae (small white butterfly) and the generalist Mamestra brassicae (cabbage moth) by monitoring larval development in a no-choice setup in which larvae are contained and allowed to only graze on one specific genotype. Both P. rapae and M. brassicae larvae feeding on vih2 plants showed a significant weight increase compared with larvae feeding on Col-0 plants (Figures 5A and 5B), suggesting that VIH2 plays a role in activating defenses that interfere with insect herbivore development.
Figure 5.
Arabidopsis vih2 Lines Have Reduced Defenses against Larvae of Herbivorous Insects and Are Compromised in Jasmonate Perception.
(A) and (B) Larval development was monitored in a no choice setup. One caterpillar each (larval stage L1) of the Brassicaceae specialist P. rapae (A) or the generalist M. brassicae (B) was released onto a single 5-week-old plant (n = 20) of the designated genotype. Fresh weight of caterpillars was determined after 7 d (P. rapae) or 8 d (M. brassicae). The values represent means ± se. Asterisks indicate statistical differences (Student’s t test; *P < 0.02). Plant genotype-dependent size differences of M. brassicae larvae are also visualized by a photograph ([B], right panel). Experiments were repeated with similar results.
(C) and (D) Determination of bioactive conjugates JA-Val and JA-Ile/Leu. Conjugate levels were determined in rosette leaves of 4-week-old plants of designated genotypes under sterile conditions and 3 h after inflecting wounding by squeezing each leaf with forceps. Data represent means of three independent biological replica ± sd. Statistical significance is indicated by asterisks (Student’s t test; *P < 0.02 and **P < 0.005).
(E) qPCR analysis of JA-dependent genes. Gene expression was analyzed by qPCR analyses using RNA extracted from pooled leaves (n = 5) of 5-week-old plants of the designated genotype that were untreated or infested for 24 h by P. rapae larvae as indicated. PP2AA3 was used as a reference gene. The expression value of untreated Col-0 was set to 1. Shown are means ± se (n = 3). qPCR analyses were repeated with similar results.
To examine whether decreased herbivore resistance was caused by compromised jasmonate production or perception, we analyzed jasmonates and jasmonate-responsive gene expression. To our surprise, upon mechanical wounding, vih2 mutants exhibited increased levels of JA and bioactive conjugates such as JA-Leu/Ile and JA-Val compared with Col-0 (Figures 5C and 5D; Supplemental Figure 6A), an observation that is counterintuitive to decreased insect herbivore resistance in these plants. However, expression of VSP2, a marker gene of the MYC branch of JA signaling known to be induced by jasmonates and herbivores, was reduced in vih2 plants after infestation with P. rapae larvae relative to Col-0 plants (Figure 5E; Supplemental Figure 6B). Similar results were obtained for MYC2 expression (Figure 5E). These findings are in agreement with a reduced resistance of vih2 plants in the performance assays. The observation that MYC-branch marker gene expression in vih2 plants was reduced despite an increase in jasmonates suggests a defect in jasmonate perception. Supporting a defect in jasmonate-regulated defenses, vih2 plants were also found to be more susceptible to the necrotrophic fungi Botrytis cinerea and Alternaria brassicicola (Supplemental Figures 6C and 6D).
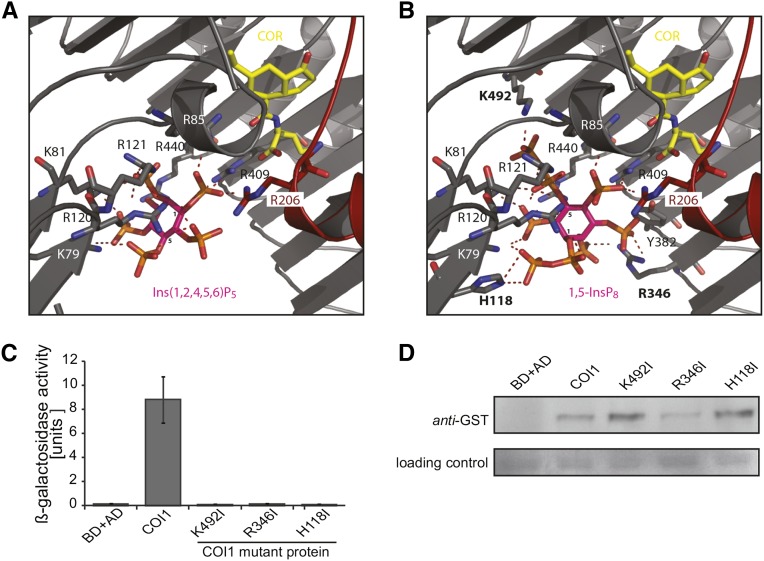
Inositol Pyrophosphates Bind to the ASK1-COI1-JAZ Jasmonate Receptor Complex
To further investigate the role of VIH2 in jasmonate perception, we performed molecular docking of Ins(1,2,4,5,6)P5, which copurified with ASK1-COI1 from insect cells (Sheard et al., 2010), and 1,5-InsP8 into the proposed inositol polyphosphate binding pocket of the ASK1-COI1-JAZ1-JA-Ile complex. Poses with the highest scores (shown in Figures 6A and 6B) predict that the concave surface of the COI1 solenoid fold surrounds and binds molecules at overlapping, yet distinct, sites. An intricate network of basic COI1 residues (Lys-79, Lys-81, Arg-85, Arg-120, Arg-121, Arg-409, and Arg-440) and JAZ1 residue Arg-206 are predicted to coordinate Ins(1,2,4,5,6)P5 and 1,5-InsP8 (Figures 6A and 6B). The 1,5-InsP8 molecule is predicted to be additionally stabilized by COI1 residues His-118, Arg-346, Tyr-382, and Lys-492, which coordinate the 1-β-phosphate, the 3-phosphate, the 3-phosphate, and the 5-β-phosphate of 1,5-InsP8, respectively. We investigated the involvement of three of these residues (His-118, Arg-346, and Lys-492; highlighted in Figure 6B) in COI1-JAZ1 interaction in a yeast two-hybrid system. These residues are positioned as an almost equilateral triangle (distance of respective coordinating groups: His-118, Lys-492, 13.01 Å; His-118-Arg-346, 13.06 Å; and Lys-492-Arg-346, 13.57 Å) and for geometrical reasons and assuming rigid ligand binding, not all three residues can interact with Ins(1,2,4,5,6)P5 or other InsP5 isomers (not containing diphosphobonds) simultaneously. Individual substitution of these amino acids (predicted to specifically coordinate 1,5-InsP8) by Ile completely abolished COI1-JAZ1 interaction, even though protein stability was not (His-118I and Lys-492I) or only mildly (Arg-346I) affected (Figures 6C and 6D). These results suggest a critical role of InsP8 rather than InsP5 isomers in COI1-JAZ1 complex formation.
Figure 6.
Structural Models of ASK1-COI1-JAZ1-Coronatine in Complex with Ins(1,2,4,5,6)P5 or 1,5-InsP8 and Functional Evaluation of Proposed 1,5-InsP8 Binding Mutants Suggest a Role of InsP8 in Jasmonate Receptor Complex Formation.
(A) and (B) COI1-JAZ structures containing Ins(1,2,4,5,6)P5 or 1,5-InsP8 as obtained from in silico docking experiments are shown. COI1 (gray ribbon), coronatine (COR) in yellow stick representation, and inositol polyphosphates (rendered as stick in magenta) are presented. Hydrogen bonds and salt bridge networks are depicted as dashed lines. Residues in bold were substituted by Ile for yeast two-hybrid studies.
(C) JAZ1 interaction with wild-type or mutant COI1 in yeast was evaluated in the presence of 50 μM coronatine by coexpression of pGBKT7-COI1 (and mutated versions as indicated) with pGADT7-JAZ1 in yeast strain Y187 (Clontech) and subsequent quantification of β-galactosidase-mediated hydrolysis of ortho-nitrophenyl-β-d-galactopyranoside. Values represent means of four independent biological replica ± sd.
(D) Stability of mutant COI1 protein. Immunoblots of soluble lysates prepared from tobacco (Nicotiana benthamiana) leaves expressing COI1 mutants (as designated) in translational fusion with N-terminal GST. Equal amounts of total protein were loaded, and COI1 was detected with antibodies against GST (Sigma-Aldrich). As a normalization control (lower panel), a representative unspecific band was chosen.
To investigate whether inositol pyrophosphate can bind directly to the COI1-JAZ1 complex, we performed binding assays of ASK1-COI1-JAZ1 with radiolabeled [3H]-InsP5, [3H]-InsP6, and [3H]-InsP7 purified and desalted from [3H]-myo-inositol-labeled seedlings as described in experimental procedures. Inositol polyphosphate binding was only observed in the presence of the structural JA-Ile analog coronatine, suggesting that inositol polyphosphates do not stimulate ASK1-COI1-JAZ1 complex formation in the absence of jasmonates (Figure 7A). Importantly, plant InsP7 bound more efficiently than InsP6, which bound more efficiently than Ins(1,3,4,5,6)P5, indicating that inositol pyrophosphates are superior ligands of the ASK1-COI1-JAZ1 complex compared with less anionic inositol polyphosphate species (Figures 7A and 7B).
Figure 7.
Plant Inositol Pyrophosphates Are Superior Ligands of the ASK1-COI1-JAZ1 Complex Compared with Less Anionic Inositol Polyphosphate Species.
Direct binding of [3H]-InsP5, [3H]-InsP6, and [3H]-InsP7 (purified and desalted from [3H]-myo-inositol labeled seedlings of the ipk1-1 mutant [InsP5] or Col-0 seedlings [InsP6 and InsP7]) to the ASK1/COI1/His8-MBP-JAZ1 jasmonate receptor complex or to individual components of the receptor complex (ASK1-COI1 or His8-MBP-JAZ1) was analyzed with or without 1 μM coronatine. A total activity of 2000 dpm was used for each [3H]-labeled inositol phosphate species. The average of recovered radiolabel with [3H]-InsP7 in (B) is set to 100%. Values show means ± se (n = 2 or 3) of radiolabel recovered by pull-down of His8-MBP-JAZ1 via metal affinity chromatography, and experiments were repeated with similar results.
DISCUSSION
Inositol pyrophosphates have gained recent attention as signaling molecules in amoeba, yeast, and mammalian cells (Mulugu et al., 2007; Shears, 2009; Chakraborty et al., 2011; Szijgyarto et al., 2011; Wundenberg and Mayr, 2012; Pöhlmann et al., 2014). Here, we describe the presence of InsP7 and InsP8 in the model plant Arabidopsis and show that VIH2 is a functional inositol pyrophosphate synthetase responsible for InsP8 production, playing a critical role in jasmonate-regulated defenses. The ubiquitous presence of Vip1/PPIP5K homologs in plants as suggested by our work supports and extends previous reports of the wide distribution of these enzymes in eukaryotic organisms and underlines the fundamental importance of inositol pyrophosphates in regulating cellular functions.
VIH Proteins Have Vip1/PPIP5K-Like Activities
A structural model of the VIH2 ATP-grasp kinase domain and complementation of defects in growth and inositol polyphosphate homeostasis of yeast vip1 mutants indicate that VIH1 and VIH2 execute Vip1/PPIP5K-like activities and are likely to pyrophosphorylate the 1-position of InsP6 and 5-InsP7 in yeast (Figure 2). A recent study by Desai et al. (2014) that was published while this article was in preparation also addressed the function of Arabidopsis Vip1 homologs. Similar to the experiment shown in Figure 2E, the authors show that expression of these proteins in a yeast vip1∆ kcs1∆ ddp1∆ triple mutant results in InsP7 production. Based on these results, the authors predicted a role of these enzymes as IP6K (InsP6 kinase) enzymes. Unfortunately, this experiment does not allow discrimination between Kcs1/IP6K and Vip1/PPIP5K activities because Vip1/PPIP5K enzymes are well known to efficiently use InsP6 as a substrate to produce (1-)InsP7 (Mulugu et al., 2007; Lin et al., 2009; Losito et al., 2009). We therefore investigated the ability of VIH proteins to complement single mutant kcs1∆ or vip1∆ yeast phenotypes and analyzed the role of VIH2 in planta. We show that VIH proteins rescue vip1∆-associated growth defects on 6-azauracil, whereas they fail to rescue kcs1∆-associated growth defects (Figures 2C, 2D, and 2G). Furthermore, we show that VIH2 complements the vip1∆-associated defect in InsP7 to InsP8 conversion (Figure 2F; Supplemental Figure 2C) and that vih2 lines are compromised in InsP8 synthesis and (similar to yeast vip1∆ mutants) accumulate InsP7 (Figures 4B and 4D; Supplemental Figure 5). Collectively, these data provide strong evidence that in vivo VIH proteins do not execute IP6K/Kcs1-like activities as suggested by Desai et al. (2014) but PPIP5K/Vip1-like activities.
The Isomer Identity of Plant Inositol Pyrophosphates Remains Unresolved
The expression pattern of VIH2 and the pronounced effect on seedling InsP8 production observable in vih2 plants indicate that VIH2 is the major enzyme synthesizing InsP8 in Arabidopsis (Figures 3 and 4; Supplemental Figures 5A and 5B). It remains unclear, however, whether plant InsP7 and InsP8 have the same isomer identity as in yeast. Anion exchange HPLC chromatography does not allow unambiguous discrimination between different inositol pyrophosphate isomers of same molecular mass. Two major observations challenge the idea that yeast and plant InsP7 and InsP8 isomers are identical. First, BLAST search analyses did not allow the identification of plant Kcs1/IP6K homologs, suggesting that an unknown enzyme activity is responsible for plant InsP7 production. Second, plant-purified InsP7 exhibits a higher binding affinity for the ASK1-COI1-JAZ1 jasmonate receptor complex than InsP6 or InsP(1,3,4,5,6)P5 (Figure 7B). In contrast, competition assays following a similar strategy recently employed to evaluate InsP7 binding to human casein kinase-2 (Rao et al., 2014) showed that available chemically synthesized InsP7 isomers have lower affinities than InsP6 (1-InsP7, 5-InsP7, and 6-InsP7) or a similar affinity (4-InsP7) for the jasmonate receptor complex (Supplemental Figures 7A to 7C). Future research will have to address purification of plant InsP7 in sufficient amounts for NMR analyses or crystallography to reveal plant InsP7 (and by extension InsP8) isomer identity. Equally important for future studies will be the identification of the protein(s) responsible for plant InsP7 synthesis.
Plant InsP8 Has a Role in Resistance against Insect Herbivores and Necrotrophic Fungal Pathogens
The finding that ABA induces InsP7 and InsP8 production (Supplemental Figure 3E), whereas MeJA induces production of primarily InsP8 and not InsP7 (Figures 3C, 3D, and 4C; Supplemental Figures 3C and 3D) identifies interesting interactions between jasmonate and ABA signaling that are distinct from those reported previously (Pieterse et al., 2012). Several lines of evidence suggest that plant InsP8 is responsible for the VIH2-dependent contribution to resistance against herbivores and necrotrophs: (1) vih2-mutant seedlings have robustly decreased levels of InsP8 and exhibit a decreased resistance against larvae of herbivorous insects and necrotrophs and have a defect in jasmonate perception (Figures 4 and 5; Supplemental Figures 5 and 6); (2) MeJA treatment causes a robust and specific increase in a VIH2-dependent pool of InsP8 (Figures 3 and 4; Supplemental Figures 3 and 5); (3) direct binding assays suggest that plant-derived inositol pyrophosphate (InsP7) binds more efficiently to the ASK1-COI1-JAZ1 complex than less anionic inositol polyphosphates such as InsP6 and Ins(1,3,4,5,6)P5 (Figure 7B); (4) the proposed inositol polyphosphate binding pocket of the ASK1-COI1-JAZ1 jasmonate receptor complex is large enough to accommodate a single InsP8 molecule (Figures 6A and 6B) and COI1 mutants designed to specifically prevent InsP8 binding failed to interact with JAZ1 in yeast (Figure 6C). Unfortunately, we failed to purify sufficient amounts of plant derived InsP8 to perform radioligand binding based reconstitution assays, which will be an important task for future work.
Our study does not rule out that other inositol polyphosphates may influence assembly of the jasmonate receptor complex. Ins(1,2,4,5,6)P5, which copurified with the ASK1-COI1 complex from insect cells (Sheard et al., 2010), is an interesting candidate in this regard. However, it is unclear whether plants synthesize Ins(1,2,4,5,6)P5 or its enantiomer Ins(2,3,4,5,6)P5 (Stevenson-Paulik et al., 2005; Hanke et al., 2012), and a physiological significance of either species in herbivore resistance has not been established so far. Another isomer, Ins(1,3,4,5,6)P5, that highly accumulates in ipk1-1 plants (Supplemental Figure 8) has been implicated in the increased herbivore resistance of this plant mutant (Mosblech et al., 2011). While it remains to be shown whether this InsP5 isomer has any function in ASK1-COI1-JAZ complex formation in the context of a wild-type plant, we can exclude the possibility that it plays a role in VIH2-dependent insect herbivore resistance because Ins(1,3,4,5,6)P5 levels did not change in vih2 lines (Figures 4A and 4B; Supplemental Figures 5A and 5B).
Coincidence Detection of Jasmonate and Inositol Phosphates by the Jasmonate Coreceptor
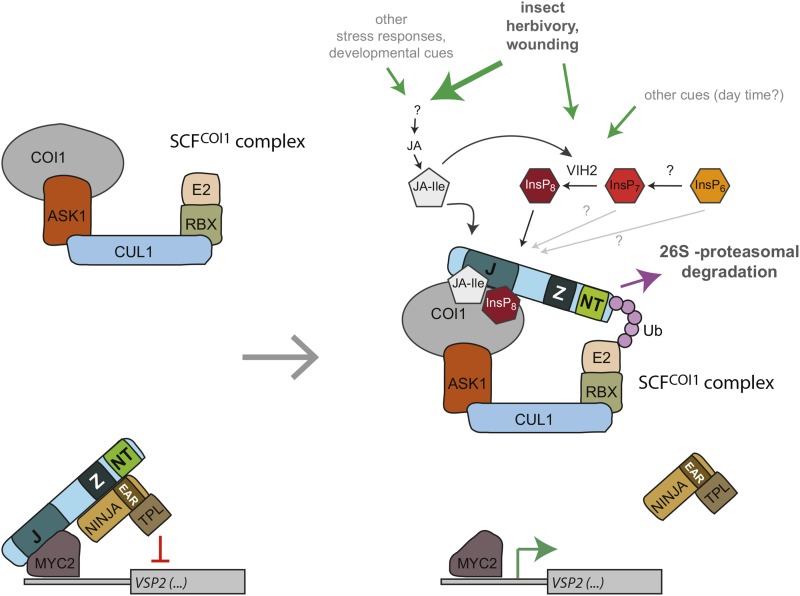
Direct binding assays with [3H]-Ins(1,3,4,5,6)P5, [3H]-InsP6, and [3H]-InsP7 (Figures 7A and 7B) indicate that inositol polyphosphate binding is not sufficient for ASK1-COI1-JAZ1 assembly but still requires the presence of coronatine (or by extension JA-Ile). Combined with the previous observation that coronatine fails to trigger the formation of the ASK1-COI1-JAZ complex in the absence of inositol polyphosphate (Sheard et al., 2010), these data suggest that only coincidence detection of both inositol polyphosphate or pyrophosphate and bioactive jasmonate allows complex formation and subsequent proteasomal degradation of JAZ repressor proteins to stimulate jasmonate responsive gene expression (Figure 8). We propose that coincidence detection of two unrelated molecules might prevent an uncontrolled accidental trigger of immune responses that are known to severely affect plant growth and development (Pieterse and Dicke, 2007; Howe and Jander, 2008). This idea is in agreement with a recent finding that COI1 protein levels are strictly regulated by a dynamic balance between SCFCOI1-mediated stabilization and 26S proteasome-mediated degradation (Yan et al., 2013).
Figure 8.
Model of the Role of VIH2 and InsP8 in the Wound Response.
Mechanical wounding or herbivore attack stimulate the synthesis of JA and bioactive JA conjugates such as JA-Ile. Increasing jasmonate levels trigger a fast VIH2-dependent increase in InsP8, which is most likely caused by posttranslational activation of the VIH2 protein. Both JA-Ile and InsP8 occupy designated binding pockets in COI1-ASK1 and might work as molecular glue to recruit the JAZ repressor protein. Subsequent polyubiquitylation of JAZ by the SCF ubiquitin E3 ligase complex causes proteasomal degradation of the JAZ repressor and allows expression of jasmonate/InsP8-responsive genes such as VSP2. The physiological role of other inositol polyphosphates on potentiating jasmonate dependent formation of the SCFCOI1 ubiquitin E3 ligase complex remains unclear.
In addition, coincidence detection could allow differentiated immune responses. It still remains a central unanswered question how plants achieve specificity in their response to herbivore attack. It has been shown, for instance, that transcriptional responses of Arabidopsis to caterpillar (P. rapae) and thrips (Frankliniella occidentalis) infestation is primarily via COI1-dependent gene regulation, but that expression patterns of these genes are specific to one or the other insect herbivore (De Vos et al., 2005). We speculate that a differentiation in COI1-dependent responses might be in part determined by the inositol pyrophosphate signature of a given tissue under herbivore attack. It is important to note that InsP6 also has previously been suggested to play an important role in the maintenance of basal resistance to plant pathogens. The reduction of InsP6 in potato and Arabidopsis was correlated with increased susceptibility toward different viral infections and also caused hypersensitivity to fungal and bacterial infections in Arabidopsis (Murphy et al., 2008). In future experiments, it will be important to study whether these effects are an immediate consequence of reduced InsP6 or whether they are caused by the reduction of InsP6-dependent inositol pyrophosphates. Independent of this outcome, breeding strategies and biotechnological approaches to reduce InsP6 will have to consider possible negative side effects in crop plants.
METHODS
BLAST Search and Phylogenetic Analyses
Sequence sampling focused on plants and fungi with some additions of protist species. BLAST search analyses (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were performed using the N-terminal part of Saccharomyces cerevisiae Vip1 (residues 1 to 535), which contains the entire ATP-grasp kinase domain (Mulugu et al., 2007). The amino acid sequences were aligned using MAFFT, version 6.927b (Katoh et al., 2005). Heterogeneous alignment regions were excluded prior to phylogenetic analyses using Gblocks (Castresana, 2000), with the minimum length of a block set to five, allowing gaps in up to 50% of the sequences at a given site, a minimum number of sequences of 24 for a conserved or a flanking position, and a maximum number of contiguous nonconserved positions of eight. A phylogenetic tree was estimated using maximum likelihood (Felsenstein, 1981) with RAxML version 7.3.2 (Stamatakis, 2006a). Fast bootstrap analyses (Felsenstein, 1985; Stamatakis et al., 2008) over 10,000 rounds were run on the Web-based bioportal facility (Kumar et al., 2009) (http://www.mn.uio.no/ibv/bioportal/) with eight parallel processors, using bootstrap trees as starting trees for heuristic searches, and employing the DAYHOFF model of amino acid substitution as inferred with the ProteinModelSelection perl script (http://www.exelixis-lab.org/), accounting for rate heterogeneity by using the CAT model (Stamatakis, 2006b). The final tree was optimized using the Gamma model of rate heterogeneity (Yang, 1993).
Plants and Growth Conditions
For T-DNA insertion lines, seeds of mutant lines of Arabidopsis thaliana (ecotype Col-0) were obtained from The European Arabidopsis Stock Centre (http://arabidopsis.info/). The T-DNA lines used in this study are as follows: vih2-3a (SAIL_165_F12), vih2-4 (GK-080A07), and ipk1-1 (SALK_065337C). Homozygous lines were identified by PCR using T-DNA left and right border primers and gene-specific sense or antisense primers (Supplemental Table 1). The isolated homozygous progeny of the vih2-3a line identified by PCR-based genotyping was found to have short root hairs compared with Col-0. However, the phenotype did not cosegregate with the vih2-3 allele in the F2 generation of a cross with the vih1-1 T-DNA line (SAIL_543_F08), suggesting that the original vih2-3a plant had an additional insertion or mutation causing the root hair phenotype. Therefore, vih2-3a was crossed with a VIH2 wild-type plant (Col-0 background) and F3 progeny homozygous for the vih2-3 allele (exhibiting normal root hairs) used for further analyses. All lines analyzed in this study, including Col-0 plants, were grown in parallel for two generations under identical conditions on soil (16 h light and 8 h dark, day/night temperature 22/18°C and 120 μmol−1 m−2 light intensity), and seeds of the respective last progenies were used for all analyses described in this article. For growth in sterile conditions, seeds were sterilized in 70% (v/v) ethanol and 0.05% (v/v) Triton X-100 for 30 min and washed twice in 90% (v/v) ethanol. Sterilized seeds were plated onto 0.5× MS, 1% sucrose, 0.7 to 0.8% phytagel stratified for 2 d at 4°C, and grown under conditions of 12 h light (23°C) and 12 h dark (21°C). To investigate the expression of distinct VIH2 domains in the respective T-DNA insertion lines, qPCR analyses were performed using the primers listed in Supplemental Table 2.
Performance and Disease Assays
Plants were grown at standard growth conditions in the greenhouse as described earlier (Verhage et al., 2011). Freshly hatched larvae (L1 stage) of the Brassicaceae specialist Pieris rapae (small cabbage white butterfly) or the generalist Mamestra brassicae (cabbage moth) were released onto fully expanded rosette leaves of 5-week-old plants of the designated genotype. Individual plants of the designated genotype were infested with a single caterpillar of either P. rapae or M. brassicae. The caterpillar-challenged plants were placed in a transparent plastic container sealed with insect-proof meshes allowing adequate gas exchange and light transmission. Fresh weight of caterpillars was measured after 7 d (P. rapae) or 8 d (M. brassicae) of feeding.
Botrytis cinerea and Alternaria brassicicola assays were performed as previously described (Kemmerling et al., 2007; Van Wees et al., 2013).
Chemicals
Coronatine, methyl jasmonate, and abscisic acid were from Sigma-Aldrich. 2-Nitrophenyl-β-d-galactopyranoside was from Applichem. InsP6 was from Sichem. 1-InsP7, 4-InsP7, 5-InsP7, 6-InsP7, and 1,5-InsP8 were synthesized as recently described (Capolicchio et al., 2013, 2014). [3H]-InsP6 and [3H]-InsP7 were extracted and purified from [3H]-myo-inositol-labeled Col-0 seedlings using a desalting protocol as described earlier (Azevedo et al., 2010). [3H]-Ins(1,3,4,5,6)P5 was purified from [3H]-myo-inositol-labeled ipk1-1 seedlings using the same desalting protocol. Standards were [3H]-InsP6 (Azevedo and Saiardi, 2006) and [3H]-5-InsP7 that was generated in vitro from [3H]-InsP6 and recombinant mammalian IP6K1 (Azevedo et al., 2010).
In Vitro Binding Assays
In vitro binding assays were performed with recombinant COI1-ASK1 and His8-MBP-JAZ in 1:2 molar ratios. Purified and desalted [3H]-InsP6 was added to the reaction buffer containing 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM imidazole, 10% (v/v) glycerol, 0.1% (v/v) Tween 20, and 5 mM 2-mercaptoethanol in a total volume of 0.5 mL. Unless mentioned otherwise, [3H]-InsP6 at a total activity of 4000 dpm and 1 μM coronatine was added to each reaction. The reaction was incubated at room temperature (22 to 24°C) for 90 min, then 30 μL of Ni-NTA resin was added with a further incubation at 4°C for 90 min. The resin was centrifuged for 5 min at 900g and washed three times with ice-cold reaction buffer. Proteins were eluted with 250 mM imidazole and recovered radioactivity analyzed by scintillation counting.
Extraction and HPLC Analyses of Inositol Phosphates
Inositol polyphosphates from yeast were extracted and analyzed as described (Azevedo and Saiardi, 2006). Extraction and measurement of inositol polyphosphates from Arabidopsis seedling were performed as follows. Seedlings were grown under sterile conditions in liquid 0.5× MS with 2% sucrose for 10 d and then transferred to sucrose-free low MS semi-liquid medium (0.25 MS and 0.3% Phytagel). Labeling was started at 2 weeks of age by addition of 40 μCi mL−1 of [3H]-myo-inositol (30 to 80 Ci mmol−1 and 1 mCi mL−1; Biotrend; ART-0261-5) for 2 mL liquid MS media containing 10 seedlings. After 6 d of labeling, leaves or seedlings were washed two times with ultrapure water before harvesting and freezing into liquid N2. Inositol polyphosphates were extracted as described previously (Azevedo and Saiardi, 2006) and resolved by strong anion exchange chromatography HPLC (using the partisphere SAX 4.6 × 125 mm column; Whatman) at a flow rate of 0.5 mL min−1 with the gradient of buffers A (1 mM EDTA) and B [1 mM EDTA and 1.3 M (NH4)2HPO4, pH 3.8, with H3PO4] following the standard protocol mentioned above. Fractions were collected each minute, mixed with scintillation cocktail (Perkin-Elmer; ULTIMA-FLO AP), and analyzed by scintillation counting. To account for differences in fresh weight and extraction efficiencies between samples, values shown are normalized activities based on the total activity of each sample. To avoid misleading results derived from unincorporated [3H]-myo-inositol in the HPLC run, “total” activities for normalization were calculated by counting fractions from 17 min (Figures 2F and 2G; Supplemental Figures 2C and 2D), 22 min (Figure 3C; Supplemental Figures 2E, 3B, and 5A), or from 18 min (Figure 4) until the end of runs. HPLC runs within an experimental set were normalized in the following way: If sample B had less “total” activity than sample A, the equation used for normalization was −[individual data point of sample A*(“total” InsP of B/“total” InsP of A)]. Results are presented as minute fractions (circle or diamond) connected by lines.
Total RNA Extraction and qPCR Analyses
Leaf samples (up to 100 mg) were harvested for total RNA extraction using the RNeasy Plant Mini Kit (Qiagen). A total of 1 μg RNA was used for cDNA preparation following DNaseI digest (Fermentas). The reverse transcription was done according to the manufacturer’s instructions (Roboklon; AMV Reverse Transcriptase Native). The qPCR was performed with the SYBR Green reaction mix (Bioline; Sensimix SYBR No-ROX kit) in a Bio-Rad CFX384 real-time system. Data were analyzed using the Bio-Rad CFX Manager 2.0 (admin) system. PP2AA3 or β-TUBULIN was used as a reference gene.
Yeast Two-Hybrid Assays
The full-length coding sequences of COI1 and JAZ1 were cloned into the yeast two-hybrid vector pGBKT7 and pGADT7, respectively, in fusion with N -terminal binding domain or activation domain. The yeast strain Y187 was transformed with individual wild-type or COI1 mutant constructs generated by site-directed mutagenesis (see above) together with JAZ1 construct following the standard yeast transformation protocol mentioned above. Yeast transformants were selected on solid CSM-Leu-Trp media after which single fresh colonies from independent transformants were grown overnight in liquid CSM-Leu-Trp media. JAZ1 interaction with wild-type or mutant COI1 in the presence of 50 μM coronatine was evaluated by quantification of β-galactosidase-mediated hydrolysis of ortho-nitrophenyl-β-d-galactopyranoside.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: VIH1 (At5g15070), VIH2 (At3g01310), MYC2 (At1g32640), VSP2 (At5g24770), PP2AA3 (AT1G13320), β-TUBULIN (AT5G62700), JAZ1 (At1g19180), COI1 (At2g39940), IPK1 (At5g42810), Saccharomyces cerevisiae VIP1 (YLR410W), S. cerevisiae KCS1 (YDR017C), and hPPIP5K2 (NM_001276277). Accession numbers for T-DNA insertion lines are as follows: vih2-3a (SAIL_165_F12), vih2-4 (GK-080A07), ipk1-1 (SALK_065337C), and vih1-1 T-DNA line (SAIL_543_F08).
Supplemental Data
Supplemental Figure 1. Two-domain Architecture, Structural Conservation of the ATP-Grasp Domain, and Conserved Binding Residues Suggest That VIH Proteins Are Functional PPIP5 Kinases.
Supplemental Figure 2. VIH Proteins Are Functional Inositol Pyrophosphate Synthetases in Yeast.
Supplemental Figure 3. A Time-Course Experiment Reveals a Fast and Specific Induction of InsP8 by MeJA.
Supplemental Figure 4. Genome Structure and Identification of vih2::T-DNA Insertion Lines.
Supplemental Figure 5. Bulk Steady State and Jasmonate-Induced Pools of InsP8 in Arabidopsis Seedlings Depend on VIH2.
Supplemental Figure 6. VIH2 Regulates Jasmonate Perception.
Supplemental Figure 7. Different Inositol Polyphosphates Exhibit Distinct Binding Affinities for the ASK1-COI1-JAZ Jasmonate Receptor Complex.
Supplemental Figure 8. Detection of Inositol Pyrophosphate in the ipk1-1 Mutant Line.
Supplemental Table 1. Primer List for PCR-Based Characterization of T-DNA Insertion Lines.
Supplemental Table 2. Primer List for qPCR Analyses.
Supplemental Table 3. Primer List for Generation of pDR195-Based Yeast Episomal Expression Vectors.
Supplemental Table 4. List of Primer Sequences Used for Site-Directed Mutagenesis.
Supplemental Table 5. Primers and Plasmids Used to Generate Yeast Knockout Strains.
Supplemental Table 6. Primers Used to Clone JAZ Homolog into the pET28- His8-MBP Bacterial Expression Vector
Supplemental Data Set 1. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis Shown in Figure 1.
Supplementary Material
Acknowledgments
We thank Tsuyoshi Nakagawa for Gateway binary vectors containing the bar gene, which was identified by Meiji Seika Kaisha, David Waugh for sharing the pDEST-HisMBP vector, Ana Pineda for providing M. brassicae larvae, Andreas Wachter and Martin Bayer for providing RNA, Birgit Kemmerling for providing fungal spores, and Vytas Bankaitis for the anti-Kes1 monoclonal antibody. We thank Elke Sauberzweig, Michael Fitz, and Hans van Pelt for excellent technical assistance and Junpei Takano, Sascha Laubinger, Nargis Parvin, and Kristina E. Ile for critical reading of previous versions of the article. This work was supported by Emmy Noether Grant SCHA 1274/2-1, Grants SCHA 1274/3-1 and SFB 1101/TP A05 from the Deutsche Forschungsgemeinschaft to G.S. Efforts of D.L. were also supported by the Deutscher Akademischer Austauschdienst. A.S. and C.A. are supported by the Medical Research Council (MRC) core support to the MRC/UCL Laboratory for Molecular Cell Biology University Unit (MC_U122680443). H.J.J. and S.C. are supported by the Swiss National Science Foundation (Grant PZ00P2_136816). N.Z. is a Howard Hughes Medical Institute Investigator and is supported by National Institutes of Health Grant R01CA107134. S.C.M.V.W. and M.S. were supported by Vidi Grant 11281 of the Dutch Technology Foundation STW. We dedicate this work to the memory of Laura B. Sheard. Her work greatly stimulated our interest in the regulation of jasmonate perception by inositol polyphosphates. ASK1-COI1 employed in this study was purified by her and her colleagues. Laura passed away in a tragic car accident in Seattle on November 13, 2011.
AUTHOR CONTRIBUTIONS
D.L., S.C.M.V.W., A.S., and G.S. designed the research. D.L., P.J., C.A., M.D., S.C., H.M., T.I., M.S., M.F., P.G., M.F.K.D.C, A.S., and G.S. performed the experiments. D.L., P.J., C.A., M.D., M.W., N.Z., I.F., H.J.J., S.C.M.V.W., A.S., and G.S. analyzed the data and revised the article. D.L. and G.S. wrote the article.
Glossary
- InsP3
myo- inositol 1,4,5-trisphosphate
- Ins(1,2,4,5,6)P5
inositol-1,2,4,5,6-pentakisphosphate
- JA
jasmonic acid
- qPCR
quantitative PCR
- Col-0
Columbia-0
- ABA
abscisic acid
- MeJA
methyl jasmonate
Footnotes
Articles can be viewed online without a subscription.
References
- Azevedo C., Saiardi A. (2006). Extraction and analysis of soluble inositol polyphosphates from yeast. Nat. Protoc. 1: 2416–2422. [DOI] [PubMed] [Google Scholar]
- Azevedo C., Burton A., Ruiz-Mateos E., Marsh M., Saiardi A. (2009). Inositol pyrophosphate mediated pyrophosphorylation of AP3B1 regulates HIV-1 Gag release. Proc. Natl. Acad. Sci. USA 106: 21161–21166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C., Burton A., Bennett M., Onnebo S.M., Saiardi A. (2010). Synthesis of InsP7 by the Inositol Hexakisphosphate Kinase 1 (IP6K1). Methods Mol. Biol. 645: 73–85. [DOI] [PubMed] [Google Scholar]
- Barker C.J., Illies C., Gaboardi G.C., Berggren P.O. (2009). Inositol pyrophosphates: structure, enzymology and function. Cell. Mol. Life Sci. 66: 3851–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell, M., Vilgalys, R., James, T.Y., and Taylor, J.W. (2012). Fungi. Eumycota: mushrooms, sac fungi, yeast, molds, rusts, smuts, etc. In The Tree of Life Web Project, http://tolweb.org/.
- Blatt M.R., Thiel G., Trentham D.R. (1990). Reversible inactivation of K+ channels of Vicia stomatal guard cells following the photolysis of caged inositol 1,4,5-trisphosphate. Nature 346: 766–769. [DOI] [PubMed] [Google Scholar]
- Brearley C.A., Hanke D.E. (1996). Inositol phosphates in barley (Hordeum vulgare L.) aleurone tissue are stereochemically similar to the products of breakdown of InsP6 in vitro by wheat-bran phytase. Biochem. J. 318: 279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette R.N., Gunesekera B.M., Gillaspy G.E. (2003). An Arabidopsis inositol 5-phosphatase gain-of-function alters abscisic acid signaling. Plant Physiol. 132: 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A., Hu X., Saiardi A. (2009). Are inositol pyrophosphates signalling molecules? J. Cell. Physiol. 220: 8–15. [DOI] [PubMed] [Google Scholar]
- Capolicchio S., Thakor D.T., Linden A., Jessen H.J. (2013). Synthesis of unsymmetric diphospho-inositol polyphosphates. Angew. Chem. Int. Ed. Engl. 52: 6912–6916. [DOI] [PubMed] [Google Scholar]
- Capolicchio S., Wang H., Thakor D.T., Shears S.B., Jessen H.J. (2014). Synthesis of densely phosphorylated bis-1,5-diphospho-myo-inositol tetrakisphosphate and its enantiomer by bidirectional P-anhydride formation. Angew. Chem. Int. Ed. Engl. 53: 9508–9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Chakraborty A., Kim S., Snyder S.H. (2011). Inositol pyrophosphates as mammalian cell signals. Sci. Signal. 4: re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Lin W.H., Wang Y., Luan S., Xue H.W. (2008). An inositol polyphosphate 5-phosphatase functions in PHOTOTROPIN1 signaling in Arabidopis by altering cytosolic Ca2+. Plant Cell 20: 353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- De Vos M., Van Oosten V.R., Van Poecke R.M., Van Pelt J.A., Pozo M.J., Mueller M.J., Buchala A.J., Métraux J.P., Van Loon L.C., Dicke M., Pieterse C.M. (2005). Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 18: 923–937. [DOI] [PubMed] [Google Scholar]
- Desai M., Rangarajan P., Donahue J.L., Williams S.P., Land E.S., Mandal M.K., Phillippy B.Q., Perera I.Y., Raboy V., Gillaspy G.E. (2014). Two inositol hexakisphosphate kinases drive inositol pyrophosphate synthesis in plants. Plant J. 80: 642–653. [DOI] [PubMed] [Google Scholar]
- Dorsch J.A., Cook A., Young K.A., Anderson J.M., Bauman A.T., Volkmann C.J., Murthy P.P., Raboy V. (2003). Seed phosphorus and inositol phosphate phenotype of barley low phytic acid genotypes. Phytochemistry 62: 691–706. [DOI] [PubMed] [Google Scholar]
- Draskovic P., Saiardi A., Bhandari R., Burton A., Ilc G., Kovacevic M., Snyder S.H., Podobnik M. (2008). Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem. Biol. 15: 274–286. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17: 368–376. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Flores S., Smart C.C. (2000). Abscisic acid-induced changes in inositol metabolism in Spirodela polyrrhiza. Planta 211: 823–832. [DOI] [PubMed] [Google Scholar]
- Gillaspy G.E. (2013). The role of phosphoinositides and inositol phosphates in plant cell signaling. Adv. Exp. Med. Biol. 991: 141–157. [DOI] [PubMed] [Google Scholar]
- Gilroy S., Read N.D., Trewavas A.J. (1990). Elevation of cytoplasmic calcium by caged calcium or caged inositol triphosphate initiates stomatal closure. Nature 346: 769–771. [DOI] [PubMed] [Google Scholar]
- Han S., Tang R., Anderson L.K., Woerner T.E., Pei Z.M. (2003). A cell surface receptor mediates extracellular Ca2+ sensing in guard cells. Nature 425: 196–200. [DOI] [PubMed] [Google Scholar]
- Hanke D.E., Parmar P.N., Caddick S.E., Green P., Brearley C.A. (2012). Synthesis of inositol phosphate ligands of plant hormone-receptor complexes: pathways of inositol hexakisphosphate turnover. Biochem. J. 444: 601–609. [DOI] [PubMed] [Google Scholar]
- Howe G.A., Jander G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59: 41–66. [DOI] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling, P., Leander, B.S., and Simpson, A. (2009). Eukaryotes. Eukaryota: Organisms with nucleated cells. In The Tree of Life Project, http://tolweb.org/Eukaryotes/3/2009.10.28.
- Kemmerling B., et al. (2007). The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr. Biol. 17: 1116–1122. [DOI] [PubMed] [Google Scholar]
- Knight H., Trewavas A.J., Knight M.R. (1997). Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 12: 1067–1078. [DOI] [PubMed] [Google Scholar]
- Kumar S., Skjaeveland A., Orr R.J.S., Enger P., Ruden T., Mevik B.H., Burki F., Botnen A., Shalchian-Tabrizi K. (2009). AIR: A batch-oriented web program package for construction of supermatrices ready for phylogenomic analyses. BMC Bioinformatics 10: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F., MacRobbie E.A., Brearley C.A. (2000). Inositol hexakisphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc. Natl. Acad. Sci. USA 97: 8687–8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Fridy P.C., Ribeiro A.A., Choi J.H., Barma D.K., Vogel G., Falck J.R., Shears S.B., York J.D., Mayr G.W. (2009). Structural analysis and detection of biological inositol pyrophosphates reveal that the family of VIP/diphosphoinositol pentakisphosphate kinases are 1/3-kinases. J. Biol. Chem. 284: 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.T., Gao F., Cui S.J., Han J.L., Sun D.Y., Zhou R.G. (2006). Primary evidence for involvement of IP3 in heat-shock signal transduction in Arabidopsis. Cell Res. 16: 394–400. [DOI] [PubMed] [Google Scholar]
- Losito O., Szijgyarto Z., Resnick A.C., Saiardi A. (2009). Inositol pyrophosphates and their unique metabolic complexity: analysis by gel electrophoresis. PLoS ONE 4: e5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menniti F.S., Miller R.N., Putney J.W. Jr., Shears S.B. (1993). Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J. Biol. Chem. 268: 3850–3856. [PubMed] [Google Scholar]
- Mosblech A., Thurow C., Gatz C., Feussner I., Heilmann I. (2011). Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 65: 949–957. [DOI] [PubMed] [Google Scholar]
- Mosblech A., König S., Stenzel I., Grzeganek P., Feussner I., Heilmann I. (2008). Phosphoinositide and inositolpolyphosphate signalling in defense responses of Arabidopsis thaliana challenged by mechanical wounding. Mol. Plant 1: 249–261. [DOI] [PubMed] [Google Scholar]
- Mulugu S., Bai W., Fridy P.C., Bastidas R.J., Otto J.C., Dollins D.E., Haystead T.A., Ribeiro A.A., York J.D. (2007). A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316: 106–109. [DOI] [PubMed] [Google Scholar]
- Munnik T., Vermeer J.E. (2010). Osmotic stress-induced phosphoinositide and inositol phosphate signalling in plants. Plant Cell Environ. 33: 655–669. [DOI] [PubMed] [Google Scholar]
- Munnik T., Nielsen E. (2011). Green light for polyphosphoinositide signals in plants. Curr. Opin. Plant Biol. 14: 489–497. [DOI] [PubMed] [Google Scholar]
- Murphy A.M., Otto B., Brearley C.A., Carr J.P., Hanke D.E. (2008). A role for inositol hexakisphosphate in the maintenance of basal resistance to plant pathogens. Plant J. 56: 638–652. [DOI] [PubMed] [Google Scholar]
- Onnebo S.M., Saiardi A. (2009). Inositol pyrophosphates modulate hydrogen peroxide signalling. Biochem. J. 423: 109–118. [DOI] [PubMed] [Google Scholar]
- Osada S., Kageyama K., Ohnishi Y., Nishikawa J., Nishihara T., Imagawa M. (2012). Inositol phosphate kinase Vip1p interacts with histone chaperone Asf1p in Saccharomyces cerevisiae. Mol. Biol. Rep. 39: 4989–4996. [DOI] [PubMed] [Google Scholar]
- Padmanabhan U., Dollins D.E., Fridy P.C., York J.D., Downes C.P. (2009). Characterization of a selective inhibitor of inositol hexakisphosphate kinases: use in defining biological roles and metabolic relationships of inositol pyrophosphates. J. Biol. Chem. 284: 10571–10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera I.Y., Hung C.Y., Moore C.D., Stevenson-Paulik J., Boss W.F. (2008). Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell 20: 2876–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C.M., Dicke M. (2007). Plant interactions with microbes and insects: from molecular mechanisms to ecology. Trends Plant Sci. 12: 564–569. [DOI] [PubMed] [Google Scholar]
- Pieterse C.M., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S.C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28: 489–521. [DOI] [PubMed] [Google Scholar]
- Pöhlmann J., Risse C., Seidel C., Pohlmann T., Jakopec V., Walla E., Ramrath P., Takeshita N., Baumann S., Feldbrügge M., Fischer R., Fleig U. (2014). The Vip1 inositol polyphosphate kinase family regulates polarized growth and modulates the microtubule cytoskeleton in fungi. PLoS Genet. 10: e1004586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F., et al. (2014). Inositol pyrophosphates mediate the DNA-PK/ATM-p53 cell death pathway by regulating CK2 phosphorylation of Tti1/Tel2. Mol. Cell 54: 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safrany S.T., Caffrey J.J., Yang X., Bembenek M.E., Moyer M.B., Burkhart W.A., Shears S.B. (1998). A novel context for the ‘MutT’ module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J. 17: 6599–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A., Caffrey J.J., Snyder S.H., Shears S.B. (2000a). The inositol hexakisphosphate kinase family. Catalytic flexibility and function in yeast vacuole biogenesis. J. Biol. Chem. 275: 24686–24692. [DOI] [PubMed] [Google Scholar]
- Saiardi A., Caffrey J.J., Snyder S.H., Shears S.B. (2000b). Inositol polyphosphate multikinase (ArgRIII) determines nuclear mRNA export in Saccharomyces cerevisiae. FEBS Lett. 468: 28–32. [DOI] [PubMed] [Google Scholar]
- Saiardi A., Nagata E., Luo H.R., Snowman A.M., Snyder S.H. (2001). Identification and characterization of a novel inositol hexakisphosphate kinase. J. Biol. Chem. 276: 39179–39185. [DOI] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears S.B. (2009). Diphosphoinositol polyphosphates: metabolic messengers? Mol. Pharmacol. 76: 236–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006a). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2006b). Phylogenetic models of rate heterogeneity: A high performance computing perspective. In 20th International Parallel and Distributed Processing Symposium (IPDPS), 10.1109/IPDPS.2006.1639535. [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. (2008). A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57: 758–771. [DOI] [PubMed] [Google Scholar]
- Stephens L.R., Hawkins P.T., Stanley A.F., Moore T., Poyner D.R., Morris P.J., Hanley M.R., Kay R.R., Irvine R.F. (1991). Myo-inositol pentakisphosphates. Structure, biological occurrence and phosphorylation to myo-inositol hexakisphosphate. Biochem. J. 275: 485–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson-Paulik J., Bastidas R.J., Chiou S.T., Frye R.A., York J.D. (2005). Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc. Natl. Acad. Sci. USA 102: 12612–12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb H., Irvine R.F., Berridge M.J., Schulz I. (1983). Release of Ca2+ from a nonmitochondrial intracellular store in pancreatic acinar cells by inositol-1,4,5-trisphosphate. Nature 306: 67–69. [DOI] [PubMed] [Google Scholar]
- Szijgyarto Z., Garedew A., Azevedo C., Saiardi A. (2011). Influence of inositol pyrophosphates on cellular energy dynamics. Science 334: 802–805. [DOI] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L.I., Sharon M., Zheng C., Robinson C.V., Estelle M., Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645. [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Van Wees S.C., Van Pelt J.A., Bakker P.A., Pieterse C.M. (2013). Bioassays for assessing jasmonate-dependent defenses triggered by pathogens, herbivorous insects, or beneficial rhizobacteria. Methods Mol. Biol. 1011: 35–49. [DOI] [PubMed] [Google Scholar]
- Verhage A., Vlaardingerbroek I., Raaymakers C., Van Dam N.M., Dicke M., Van Wees S.C., Pieterse C.M. (2011). Rewiring of the jasmonate signaling pathway in Arabidopsis during insect herbivory. Front. Plant Sci. 2: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos I.A., Verhage A., Schuurink R.C., Watt L.G., Pieterse C.M.J., Van Wees S.C.M. (2013). Onset of herbivore-induced resistance in systemic tissue primed for jasmonate-dependent defenses is activated by abscisic acid. Front. Plant Sci. 4: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Falck J.R., Hall T.M., Shears S.B. (2012). Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat. Chem. Biol. 8: 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chu Y.J., Xue H.W. (2012). Inositol polyphosphate 5-phosphatase-controlled Ins(1,4,5)P3/Ca2+ is crucial for maintaining pollen dormancy and regulating early germination of pollen. Development 139: 2221–2233. [DOI] [PubMed] [Google Scholar]
- Wang Y., Lin W.H., Chen X., Xue H.W. (2009). The role of Arabidopsis 5PTase13 in root gravitropism through modulation of vesicle trafficking. Cell Res. 19: 1191–1204. [DOI] [PubMed] [Google Scholar]
- Wundenberg T., Mayr G.W. (2012). Synthesis and biological actions of diphosphoinositol phosphates (inositol pyrophosphates), regulators of cell homeostasis. Biol. Chem. 393: 979–998. [DOI] [PubMed] [Google Scholar]
- Yan J., Li H., Li S., Yao R., Deng H., Xie Q., Xie D. (2013). The Arabidopsis F-box protein CORONATINE INSENSITIVE1 is stabilized by SCFCOI1 and degraded via the 26S proteasome pathway. Plant Cell 25: 486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (1993). Maximum-likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Mol. Biol. Evol. 10: 1396–1401. [DOI] [PubMed] [Google Scholar]
- York J.D. (2006). Regulation of nuclear processes by inositol polyphosphates. Biochim. Biophys. Acta 1761: 552–559. [DOI] [PubMed] [Google Scholar]
- Zhang J., et al. (2011). Inositol trisphosphate-induced Ca2+ signaling modulates auxin transport and PIN polarity. Dev. Cell 20: 855–866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.