Inhibition of sterol biosynthesis impairs the localization of GPI-anchored proteins at plasmodesmata and alters callose-mediated cell-to-cell connectivity.
Abstract
Plasmodesmata (PD) are nano-sized membrane-lined channels controlling intercellular communication in plants. Although progress has been made in identifying PD proteins, the role played by major membrane constituents, such as the lipids, in defining specialized membrane domains in PD remains unknown. Through a rigorous isolation of “native” PD membrane fractions and comparative mass spectrometry-based analysis, we demonstrate that lipids are laterally segregated along the plasma membrane (PM) at the PD cell-to-cell junction in Arabidopsis thaliana. Remarkably, our results show that PD membranes display enrichment in sterols and sphingolipids with very long chain saturated fatty acids when compared with the bulk of the PM. Intriguingly, this lipid profile is reminiscent of detergent-insoluble membrane microdomains, although our approach is valuably detergent-free. Modulation of the overall sterol composition of young dividing cells reversibly impaired the PD localization of the glycosylphosphatidylinositol-anchored proteins Plasmodesmata Callose Binding 1 and the β-1,3-glucanase PdBG2 and altered callose-mediated PD permeability. Altogether, this study not only provides a comprehensive analysis of the lipid constituents of PD but also identifies a role for sterols in modulating cell-to-cell connectivity, possibly by establishing and maintaining the positional specificity of callose-modifying glycosylphosphatidylinositol proteins at PD. Our work emphasizes the importance of lipids in defining PD membranes.
INTRODUCTION
To compartmentalize and coordinate biological processes, eukaryotic organisms have developed the propensity to laterally segregate their membrane constituents, thereby acquiring the ability to organize domains with specialized function within membrane bilayers (Simons and Sampaio, 2011; Spira et al., 2012; Jarsch et al., 2014). Not only proteins but also lipids are laterally organized within the plane of the membrane and contribute to the formation, dynamics, and function of microdomains (Lingwood and Simons, 2010; Sonnino and Prinetti, 2010; Schäfer et al., 2011; van den Bogaart et al., 2011; Contreras et al., 2012). In plants, a compelling example of membrane functional subcompartmentalization is illustrated by the specialization of the endoplasmic reticulum (ER) and the plasma membrane (PM) at sites of cell-to-cell junctions, called plasmodesmata (PD) (Maule, 2008; Tilsner et al., 2011). PD span the thick cell wall of virtually all plant cells, establishing both cytoplasmic and membrane continuity throughout the entire plant body. In recent years, PD have emerged as key elements of the cell-to-cell communication machinery and, as such, have been implicated in processes guaranteeing the collaborative functioning of the cells, the cooperative responses to exogenous and endogenous stimuli, and controlled developmental events (Chitwood and Timmermans, 2010; Dunoyer et al., 2010; Vatén et al., 2011; Xu et al., 2011, 2012; Burch-Smith and Zambryski, 2012; Furuta et al., 2012; Koizumi et al., 2012; Maule et al., 2012; Wu and Gallagher, 2012; Faulkner et al., 2013; Stahl et al., 2013; Tilsner et al., 2013; Wang et al., 2013; Han et al., 2014; Vaddepalli et al., 2014).
Despite morphological variability in plant tissues, a general unity of organization is observed at PD, attributable to their common function. They are lined by the PM and contain a cylindrical inner component, called the desmotubule, which is derived from the ER and positioned longitudinally in the center of the pore (Tilney et al., 1991; Ding et al., 1992). The space between the desmotubule and the PM provides a cytoplasmic conduit for cell-to-cell transport. Currently accepted models postulate that functionality at PD mostly hinges on the specification of both the PM and the ER membrane domains lining the channels (reviewed in Tilsner et al., 2011). Accordingly, substantial progress has been made over the last few years in identifying the PD-associated proteins (Reichelt et al., 1999; Faulkner et al., 2005, 2009, 2013; Sagi et al., 2005; Thomas et al., 2008; Faulkner and Maule, 2011; Fernandez-Calvino et al., 2011; Jo et al., 2011; Lee et al., 2011; Deeks et al., 2012; Salmon and Bayer, 2012; Ueki and Citovsky, 2014; Vaddepalli et al., 2014). Of particular interest are PD proteins involved in the regulation of callose deposition. Callose, a β-1,3-glucan, has emerged over the years as an important dynamic regulator of PD permeability, modulating key developmental and environmentally induced processes in plants (Levy et al., 2007; Simpson et al., 2009; Guseman et al., 2010; Rinne et al., 2011; Vatén et al., 2011; Zavaliev et al., 2011; Benitez-Alfonso et al., 2013; De Storme and Geelen, 2014; Han et al., 2014; Ueki and Citovsky, 2014). Enzymes regulating callose turnover are central to these processes, and recent work has led to the identification of several PD-localized β-1,3-glucanases that are involved in callose degradation and implicated in symplastic connectivity during lateral root organogenesis, virus movement, or chilling-induced release from dormancy (Iglesias and Meins, 2000; Levy et al., 2007; Rinne et al., 2011; Benitez-Alfonso et al., 2013). Although not bearing any apparent enzymatic activity, the PD-localized callose binding protein family (PDCB) has also been shown to regulate callose deposition at the neck of PD and, as such, has emerged as an important regulator of cell-to-cell communication (Simpson et al., 2009; Maule et al., 2013). Interestingly, PDCB proteins and most PD-located β-1,3-glucanases are glycosylphosphatidylinositol (GPI)-anchored proteins (Levy et al., 2007; Simpson et al., 2009; Fernandez-Calvino et al., 2011; Benitez-Alfonso et al., 2013), originating from a lipid posttranslational modification, which have been reported to often cluster in a sphingolipid- and sterol-rich membrane environment (Sangiorgio et al., 2004; Sharma et al., 2004; Borner et al., 2005; Kierszniowska et al., 2009). Lipid clustering at PD has also been suggested with the remorin protein, which localizes to sterol and sphingolipid nanodomains of the PM as well as at PD in Solanaceae (Raffaele et al., 2009; Mongrand et al., 2010; Tilsner et al., 2011). In addition, there is evidence that the specialized domain of the plasma membrane lining plasmodesmata (PD-PM) may house tetraspanin proteins, which have been reported in mammals to directly bind sterols (Bayer et al., 2006; Fernandez-Calvino et al., 2011). Sterol binding would impact their ability to associate with one another to build up an interacting network or “tetraspanin web” central to their function (Charrin et al., 2002, 2003; Silvie et al., 2006; Rubinstein, 2011).
Altogether, these data suggest that lipids are likely to be key elements of PD specialized membrane domains and may well contribute to proper functionality at PD channels (Delage and Zurzolo, 2013). However, to date, the lipid constituents of PD have not been identified, although research on biological membranes has unequivocally demonstrated that lipids can form functional units capable of modulating membrane organization, protein partitioning, and cellular functions (Lingwood and Simons, 2010; Sonnino and Prinetti, 2010; Munnik and Nielsen, 2011; Bigay and Antonny, 2012; Barrera et al., 2013; Li et al., 2014). Furthermore, the potential for membrane lipids to contribute to the regulation of intercellular communication in plants has never been investigated.
In this work, we focused our attention on the PD-PM with the aim of characterizing its lipid composition and establishing whether it differs from the bulk of the PM. We used Arabidopsis thaliana cultured cells and primary roots, which contain numerous primary PD on division walls, as model systems (Zhu, 1998; Bayer et al., 2004). We first showed that a strong lateral segregation of membrane constituents exists at the PD-PM of primary PD, illustrated by the exclusion of major PM proteins. We next investigated whether this lateral heterogeneity was also true for lipids. To this end, we isolated PD-enriched membrane fractions from Arabidopsis suspension cells and analyzed their lipid composition. Importantly, our isolation procedure allows access to the PD-PM domain without the requirement of detergent-based isolation methods criticized for not properly reflecting membrane compartmentalization in vivo. Our comparative lipidomic analysis showed that the PD-PM domain is distinct from the cellular PM and is characterized by an enrichment of sterols and complex sphingolipids with saturated very long chain fatty acids (VLCFAs) and a concomitant depletion of glycerolipids. Within the glycerolipid class, phospholipids with a higher saturation degree were detected. Remarkably, employing pharmacological approaches, we were able to show that the modification of the membrane pool of sterols strongly interferes with the ability of PDCB1 and the β-1,3-glucanase PdBG2, which are both GPI-anchored proteins, to associate with primary PD in the root. Alteration of PD targeting under sterol inhibitor treatment was correlated with a modification of both callose deposition and PD permeability. We propose that specific lipids and, in particular, sterols are key components of PD specialized membrane domains contributing to proper functionality at primary PD, possibly by participating in the lateral segregation of GPI-anchored proteins at sites of cell-to-cell junction.
RESULTS
Several PM Membrane Proteins Are Excluded from PD
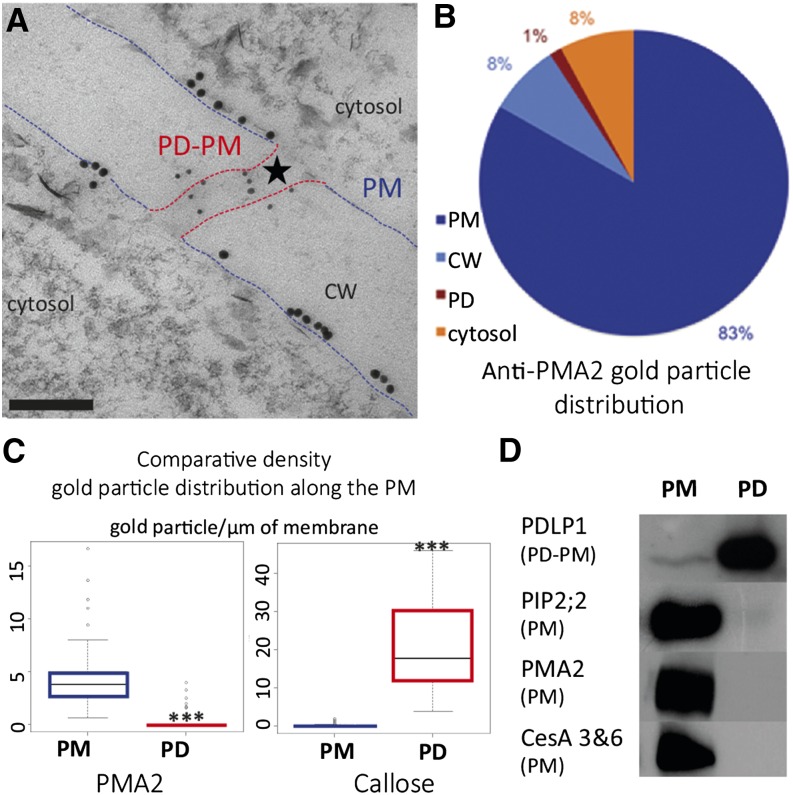
Despite its continuity from cell to cell, the PM is unlikely to remain uniform across PD cell boundaries. It is already known that some PM proteins concentrate at PD, but whether other PM proteins are excluded from PD remains elusive. To illustrate the existence of a locally confined membrane domain at PD, we analyzed the spatial distribution of several PM intrinsic markers in Arabidopsis cultured cells. First, we employed an immunogold labeling approach using antibodies directed against the PM proton pump ATPase (PMA2) (Morsomme et al., 1998). PMA2 was mainly detected along the cell periphery tightly associated with the PM (Supplemental Figures 1A and 1B). Quantification of gold particle distribution along division walls showed a strong preponderance for PMA2 labeling associated with the PM outside PD (83% of the total gold particles for the PM versus 1% for PD; Figures 1A and 1B). By contrast, callose, a PD marker, was almost exclusively associated with the channels (86% of the total gold particles versus 3% for the PM; Supplemental Figures 1C and 1D). Given that the relative proportion of the PD-PM domain is small compared with the total PM surface, we subsequently analyzed the gold particle density for each membrane segment (Figure 1C). The density of PMA2 within PD is lower (∼9-fold) than that at the PM outside the channels (0.56 versus 5.16 gold particles/μm membrane). Furthermore, a specific association of callose with PD channels but not with the PM was evident (0.48 against 19.52 gold particles/μm membrane). Although PMA2 can be associated with the PM close to the PD entrance (Figure 1A), our results indicate that PMA2 is largely excluded from PD.
Figure 1.
Exclusion of Major PM Proteins from PD Channels.
(A) Immunolocalization of PMA2 (10-nm gold particles) and callose (5-nm gold particles) at the division cell wall (CW) in cryosubstituted Arabidopsis suspension cells observed by transmission electron microscopy. While callose localizes within PD (star), the PMA2 protein associates with the PM outside PD (lined in blue) and remains excluded from the PD channel (lined in red). Bar = 100 nm.
(B) Quantification of PMA2 gold particle distribution at the PM, cell wall, PD, and cytosol (n = 1073 gold particles from 55 micrographs).
(C) Box-plot representation of the quantification of callose and PMA2 gold particles per µm of membrane (n = 55 micrographs containing 65 PD; total number of gold particles for the PM: PMA2 = 890 and callose = 27; for the PD-PM: PMA2 = 12 and callose = 367). Asterisks indicate significant differences (P < 0.001) between samples by Wilcoxon test. PMA2 density is 9 times lower at the PD-PM when compared with the cellular PM (0.56 and 5.16 gold particles per µm of membrane, respectively). By comparison, callose concentrates at PD compared with the PM (19.52 and 0.48 gold particles per µm of membrane, respectively).
(D) Immunoblot analysis of purified PM- and PD-enriched membrane fractions (7 μg of protein was loaded in each lane). Note that while the PD-PM marker, PDLP1, is enriched in the PD fraction, some major integral PM proteins, namely the aquaporin PIP2;2, the proton pump ATPase PMA2, and the cellulose synthase subunits CesA3 and CesA6, are excluded from PD.
To investigate whether this exclusion was specific for PMA2, we further assessed the distribution of two other PM intrinsic markers, the aquaporin PIP2;2 and the cellulose synthase subunits CesA3 and CesA6, by immunoblot analysis. For that we used PM- and PD-enriched membrane fractions purified from Arabidopsis cultured cells (Figure 1D). The results confirmed the exclusion of PMA2 from the PD channels. Similar to PMA2, CesA3, CesA6, and PIP2;2 were enriched in the PM fraction but absent from the PD-enriched membrane fraction. Immunoblot analysis using antibodies directed against PDLP1, a membrane-bound PD marker (Thomas et al., 2008; Fernandez-Calvino et al., 2011), confirmed the presence of PD-derived membranes in the PD fraction (Figure 1D).
Altogether, these results indicate that a conspicuous lateral segregation of the PM constituents operates at the PD, excluding some major PM proteins from the channels. Despite its continuity with the PM, the PD-PM appears as a highly specialized membrane domain with a protein composition distinct from the PM.
Detergent-Free Isolation of PD Membranes Provides Access to Lipidomic Analysis
Although previous studies and our own results showed that PM proteins laterally segregate across the PD cell boundary, no study has so far addressed the question of whether lipids, as functionally important membrane components, also segregate at PD. To address this consequential issue, we adapted a detergent-free protocol that generates PD-derived membranes from Arabidopsis suspension cells with the aim of analyzing their lipid composition (Fernandez-Calvino et al., 2011; Grison et al., 2015). Getting access to reliable data on PD lipid constituents is greatly contingent on the level of purity of PD-enriched membrane fractions. Therefore, we first ensured that virtually no contaminants from other membranes were present in our final extract.
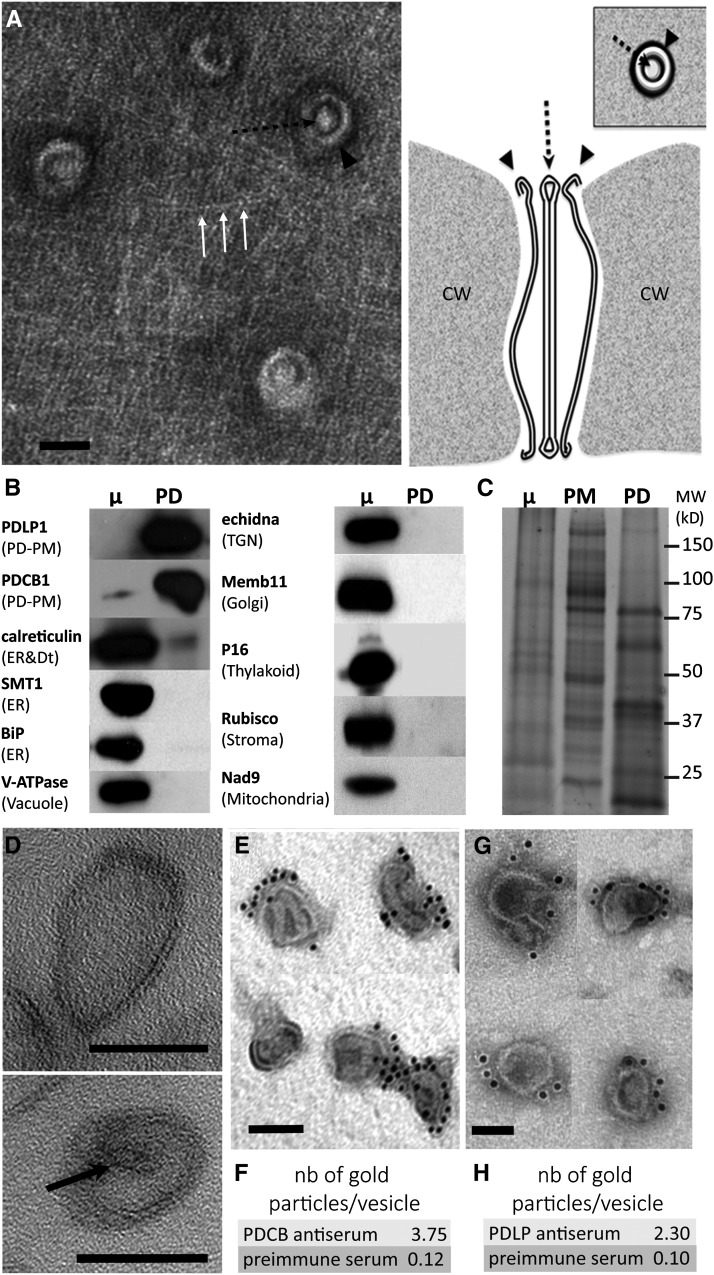
The purification procedure consists of two steps. First, wall fragments containing PD are isolated by means of mechanical disruption. Second, PD channels are “released” from the extracellular matrix through the action of cell wall-degrading enzymes. Electron microscopy analyses of the cell wall intermediate fraction indicated that both the PM and the ER membranes were disrupted close to the PD entrance during the purification process, revealing the concentric arrangement of the desmotubule and the PD-PM as well as cellulose fibers around the PD pore entrances (Figure 2A). This implies that no segment of the PM or the ER, continuous with PD in intact cells, is likely to persist after wall isolation. The absence of PM contamination of the final PD-enriched membrane fraction was validated by our immunoblot analysis (Figure 1D). Indeed, PMA2 is absent from the PD extract despite its association with the PM close to the PD entrance (Figure 1A). We complemented the purity assessment by testing for the presence of proteins representative of the ER (sterol methyltransferase [SMT1] and immunoglobulin binding protein [BiP]). Similar to the PM marker tested, neither SMT1 nor BiP was detected in the final PD-enriched membrane fraction (Figure 2B). Likewise, markers from other membranous compartments, such as the trans-Golgi network (ECHIDNA), Golgi (membrine11), vacuole (V-ATPase; ε-subunit of tonoplast H+-ATPase), chloroplast (P16), mitochondria (Nad9), and chloroplast stroma (Rubisco), were also absent from our PD extracts, while PD-PM intrinsic membrane proteins such as PDCB1 and PDLP1 were enriched (Figure 2B). We also showed that the PD-enriched membrane fraction displayed a distinct protein profile when compared with the microsomal or PM samples (Figure 2C). To confirm the enrichment of PD-derived membranes, we conducted quantitative proteomic analysis between the PD fraction and purified cellular PM and specifically looked at known PD and PM protein markers. The results confirmed the substantial enrichment of known PD-associated proteins such as tetraspanin 3, receptor-like kinase 3 (Fernandez-Calvino et al., 2011), several β-1,3-glucanases (Levy et al., 2007; Benitez-Alfonso et al., 2013), callose synthase 12 (Vatén et al., 2011), and members of the PDLP and PDCB families (Thomas et al., 2008; Simpson et al., 2009) (Supplemental Figure 2 and Supplemental Table 1).
Figure 2.
The PD-Enriched Membrane Fraction Predominantly Contains PD-PM-Derived Vesicles and Is Not Contaminated by Other Membrane Compartments.
(A) Left, surface view electron micrograph of a purified wall fragment containing embedded PD revealing the concentric arrangement of the PD-PM (black arrowhead), the cytoplasmic sleeve, and the central desmotubule (black dashed arrow). Cellulose fibers are visible around PD (white arrows). Bar = 50 nm. Right, interpretative diagram of the structure of PD (longitudinal section and surface view) in purified walls as shown in the left panel. CW, cell wall.
(B) Immunoblot analysis showing that the PD fraction is enriched in PD-PM markers (PDLP1 and PDCB1) while deprived of markers from the ER (SMT1 and BiP), vacuole (V-ATPase), trans-Golgi network (TGN; echidna), Golgi (membrine11), thylakoid (P16), soluble proteins/stroma (Rubisco), and mitochondria (Nad9). The desmotubule-associated ER protein calreticulin is detected in the PD fraction but not enriched compared with the microsomal (μ) protein extract. The same amount of protein was loaded in each lane (7 μg).
(C) The protein profile of the PD-enriched membrane fraction is distinct from that of the PM and microsomal (μ) extracts. The same amount of protein was loaded in each lane (20 μg).
(D) Electron micrograph of the PD-enriched membrane fraction showing that the final extract is composed of membrane vesicles ∼50 to 150 nm in diameter. Note that internal desmotubule-like structures are also detected (arrow). Bars = 50 nm.
(E) to (H) Electron micrographs of PDCB1 (E) and PDLP1 (G) immunogold labeling of the PD fraction. The density of gold particles per vesicle was calculated ([F] and [H]) for both preimmune antisera (n = 218 vesicles for PDCB1; n = 97 vesicles for PDLP1 [not shown]) and specific antisera (n = 163 vesicles for PDCB1; n = 75 vesicles for PDLP1). Bars = 50 nm.
Altogether, our data indicate that the level of purity required to conduct a reliable and accurate lipidomic analysis of the PD-enriched membrane fraction has been reached. In particular, we show that the cellular membrane compartments continuous with the PD pores, namely the ER and the PM, are excised during the purification process and do not significantly contaminate the final PD fraction.
The PD-Enriched Membrane Fraction Is Predominantly Composed of the PD-PM Domain
While our immunoblot analysis showed enrichment of PD-PM intrinsic membrane proteins in the final PD-enriched membrane fraction (i.e., PDCB1 and PDLP1), we also detected calreticulin, an ER resident protein that associates with PD, most likely with the desmotubule (Baluška et al., 1999, 2001; Bayer et al., 2004; Chen et al., 2005) (Figure 2B). Although calreticulin was not enriched in the PD-enriched membrane fraction, its presence suggested that the final extract contained a portion of the desmotubule. To better characterize the PD-enriched membrane fraction, thin sections of chemically fixed material were observed by transmission electron microscopy (Figure 2D). The archetypal channel-like structure was not identifiable, indicating that PD ultrastructural organization and shape are unlikely to be maintained during the cell wall digestion process. The PD-enriched membrane fraction appeared to be mainly composed of vesicles from 50 to 150 nm in diameter, which sometimes contained a central rod-like component possibly corresponding to the desmotubule (Figure 2D, arrow). To ensure that the vesicles observed were indeed PD-derived membranes, we performed anti-PDCB1 and anti-PDLP1 immunogold labeling directly on the PD fraction, which was then negatively stained (Figures 2E to 2H; Supplemental Figure 3). Gold particles associated with antibodies to PDCB1 and PDLP1 were tightly associated with the membrane vesicles observed in the PD-enriched membrane fraction, suggesting that these vesicles are indeed derived from the PD-PM domain. By contrast, PMA2 did not label the membrane vesicles of the PD fraction, whereas strong labeling was observed for PM-derived membrane vesicles (Supplemental Figure 3).
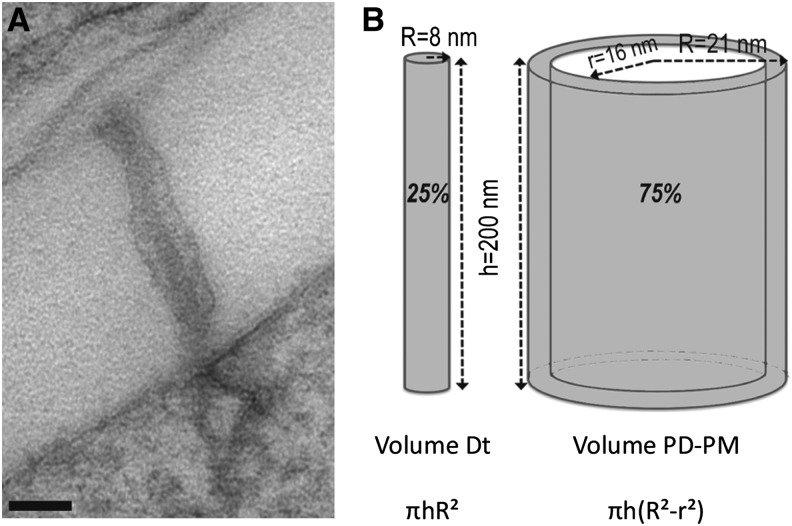
Altogether, our data suggest that, although enriched in PD-PM protein markers, the PD fraction likely contains part of the desmotubule. Therefore, before conducting the lipid analysis, we estimated the relative proportion of the two domains (PD-PM versus desmotubule) in terms of membrane volume. Thin sections of cryofixed freeze-substituted Arabidopsis cultured cells were examined, and the dimensions of the PD channels were estimated (Figure 3). On the basis of the thickness of division walls, we established that the length of the channels varied from 150 to 300 nm, whereas the PD diameter varied from 35 to 50 nm with an average diameter of 42 nm (n = 30). The thickness of the PD-PM and the diameter of the desmotubule were constant and estimated to be 5 and 16 nm, respectively. Based on these measurements, we reckoned that the volume of the desmotubule represented on average 25% of the PD membrane volume versus 75% for the PD-PM (the desmotubule was considered as a solid rod devoid of ER lumen and the PD-PM as a hollow cylinder) (Figure 3B). Therefore, we concluded that lipid analysis of the PD-enriched membrane fraction will predominantly reveal that of the PD-PM domain.
Figure 3.
Estimation of the Volume of the PD-PM and Desmotubule Domains in Arabidopsis Suspension Cells.
(A) Electron micrograph of cryofixed Arabidopsis cultured cells from which PD dimensions and membrane volumes have been calculated. Bar = 50 nm.
(B) From micrographs (n = 30), we determined that the average length and diameter of the PD channels were ∼200 nm and 42 nm, respectively. The thickness of the PD-PM was 5 nm, and the diameter of the desmotubule (Dt) was 16 nm. Considering the desmotubule as a solid cylinder with no ER lumen and the PD-PM as a hollow cylinder, we calculated that the PD-PM domain represented ∼75% of the total PD membrane volume versus 25% for the desmotubule.
PD Membranes Contain Phospholipids with a Higher Saturation Degree When Compared with the PM
Having validated our PD purification procedure for lipidomic analysis, we next compared the lipid profile of the PD-enriched membrane fraction with that of purified PM. The purity of the PM fraction was assessed by immunoblot analysis (Supplemental Figure 4). The two membrane fractions, PD and PM, were isolated from Arabidopsis suspension cells (see Methods for details). Early during the purification of the PM, a high-speed centrifugation is required to eliminate cell debris, which contain wall fragments and PD. The PM is then recovered from the microsomal fraction using a two-phase polyethylene glycol/dextran system, which separates the PM from other membranes based on differential surface charge density. By contrast, during PD purification, the later microsomal fraction (hence the PM) is discarded, whereas wall fragments are collected prior to cellulase treatment for PD membrane recovery. Consistently, our results show that the two membrane fractions have distinct protein profiles (Figures 1D and 2C; Supplemental Figure 2). The presence, in low amount, of PDLP1 in the PM fraction (Figure 1D), however, may indicate that residual PD copurify with the PM, although in negligible amounts (which is supported by the much larger size of the bulk PM compared with the PD-PM domain). Alternatively, PDLP1 may distribute between both membrane compartments but is strongly enriched at PD.
We measured the enrichment or depletion of lipid classes by calculating the ratios of the different lipid species. As a control, we also ensured that the cellulase solution used during the PD purification procedure did not contain any lipids and that no lipase activity could be detected (Supplemental Figure 5).
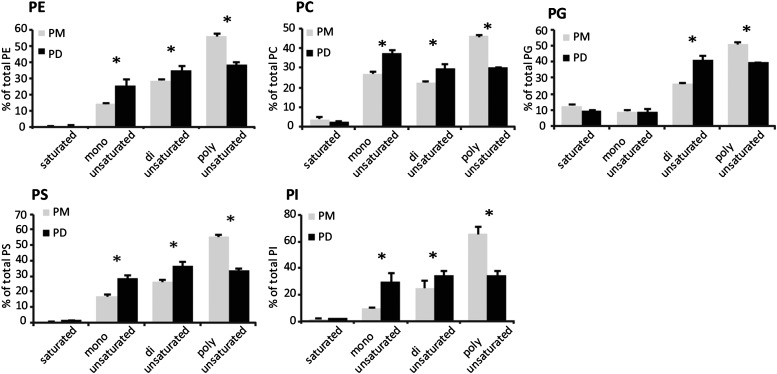
We first separated polar lipids by high-performance thin layer chromatography (HP-TLC) and determined their relative abundance by densitometry scanning. The major phospholipids in PD and PM were phosphatidylethanolamine (PE; ∼45%) and phosphatidylcholine (PC; ∼20%) (Supplemental Figure 6). Minor phospholipids (below 10%) were phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidic acid, and phosphatidylglycerol (PG). PE and PG comigrate on HP-TLC analysis. Therefore, the PE:PG ratios were established by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) to be 29 and 30 for the PD and PM fractions, respectively, confirming that PG is indeed a minor species. We also detected the galactolipid digalactosyldiacylglycerol (∼20%), which was described previously as a PM lipid component (Andersson et al., 2003; Mongrand et al., 2004). To get more information about the phospholipid molecular species, we next used LC-MS/MS. This analysis revealed that phospholipids associated with the PD and PM fractions (PE, PC, PS, PG, and PI) were primarily composed of the molecular species C34 and C36 containing C16 and C18 long-chain fatty acids (Supplemental Figure 7). Interestingly, we found that all phospholipid species analyzed showed a higher degree of saturation in the PD extract when compared with the PM, with a significant increase of monounsaturated (except for PG) and diunsaturated species and a concomitant reduction of polyunsaturated species (Figure 4; Supplemental Figure 8).
Figure 4.
The Saturation Degree of PD-Associated Phospholipid Species Is Elevated Compared with PM.
The distribution of phospholipid molecular species (saturated, monounsaturated, diunsaturated, and polyunsaturated) was analyzed by LC-MS/MS for each phospholipid class. The quantification of saturated, monounsaturated, diunsaturated, and polyunsaturated species takes into account the two acyl chains esterified on the phospholipid glycerol backbone. For each class, values were normalized to the total of molecular species. Asterisks indicate significant differences (P < 0.05) between two samples by ANOVA of the Kruskal-Wallis test. n = 3 for PD and n = 4 for PM. Error bars indicate sd.
Analysis of Total Fatty Acids Revealed That Complex Sphingolipids Accumulate in the PD-Enriched Membrane Fraction When Compared with the PM per se
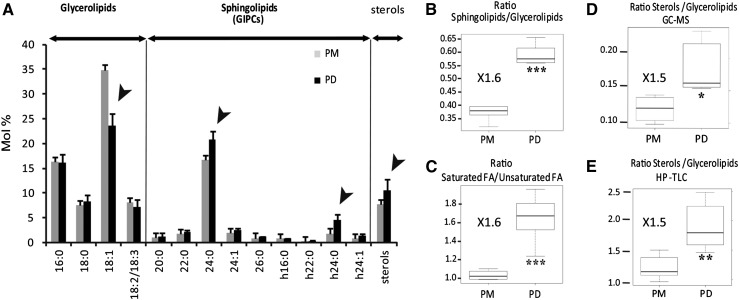
We next assessed the relative proportion of the glycerolipid and sphingolipid classes by analyzing the total fatty acid profile of the PM and the PD-enriched membrane fractions (Figure 5A). As a general rule, sphingolipids tend to contain VLCFAs (20 to 24 carbon atoms long) as well as 2-hydroxylated fatty acids that are linked to the sphingobase via an amide bond, whereas glycerolipids are composed of long-chain fatty acids (16 to 18 carbon atoms long), which are esterified to a glycerol backbone. The two most abundant sphingolipid classes found in plant membranes are the glucosylceramides and the glycosylinositolphosphoceramides (GIPCs), a group of highly glycosylated sphingolipids. GIPCs were the main sphingolipid classes detected in the lipid extract from PD and PM (Supplemental Figure 9). GIPCs are organized into distinct series depending on the number and type of monosaccharides bound to the inositol group (Buré et al., 2011). GIPC from series A, bearing hexose-hexuronic acid-inositol, was the most prevalent species in PD and PM (∼70%; Supplemental Figure 9B). In Arabidopsis Landsberg erecta-0 suspension cells, GIPC species contain two major VLCFAs, 24:0 (50%) and 24:0 2-hydroxylated (h24:0; 20%), whereas minor species contain 22:0, 24:1, and h24:1 (Buré et al., 2011). Therefore, we used the VLCFA content as a marker for the presence of GIPCs. The complete fatty acid profile was quantified by gas chromatography-mass spectrometry (GC-MS) analysis of fatty acid methyl esters (FAMEs). These measurements revealed that the main VLCFAs detected in both the PM and the PD-enriched membrane fractions were 24:0 and h24:0, characteristic of GIPC species (Figure 5A). The relative proportion of VLCFA species was similar to the one reported for total GIPCs in Arabidopsis Landsberg erecta-0 suspension cells (Buré et al., 2011). A survey of the FAME profile indicated an augmentation in GIPC species containing 24:0 and h24:0 in the PD extract. To assess the relative enrichment of sphingolipids (GIPCs) versus glycerolipids, we next calculated the ratio between VLCFAs and the long-chain fatty acids (C16-C18). Interestingly, PD membranes showed a significant 1.6-fold enrichment of VLCFAs (sphingolipids) versus C16-C18 fatty acids (glycerolipids) compared with the PM per se (Figure 5B). We also observed that the PD-enriched membrane fraction contained a higher proportion of saturated fatty acids over unsaturated fatty acids (1.6-fold enrichment; Figure 5C), consistent with the LC-MS data on phospholipids (Figure 4).
Figure 5.
The PD-PM Domain Is Depleted in Unsaturated Fatty Acids and Enriched in Complex Sphingolipids and Sterols When Compared with the PM.
(A) Total fatty acid profiles (FAMEs) quantified by GC-MS. Sphingolipid (in particular GIPC) species are characterized by VLCFAs (20:0 to 26:0) and hydroxylated fatty acids (h16:0 to h24:1), with the main GIPC-derived acyl groups being 24:0 and h24:0. Long-chain fatty acids (16:0 to 18:3) are mainly derived from glycerolipids. Results are expressed in mol % of a given fatty acid compared with the sum of total fatty acids. Arrowheads indicate changes in fatty acid composition between the PD and PM fractions. Error bars indicate sd.
(B) Ratio between sphingolipids (20:0 to h24:1 fatty acids) versus glycerolipids (16:0 to 18:3 fatty acids), showing a significant enrichment (1.6-fold) of sphingolipids in PD compared with the PM, according to the GC-MS measurement.
(C) Ratio between saturated versus unsaturated fatty acids (GC-MS analysis), showing a significant enrichment (1.6-fold) of saturated fatty acids in PD compared with the PM.
(D) Ratio between sterols versus glycerolipids (GC-MS analysis), showing a significant enrichment (1.5-fold) of sterols in PD compared with the PM.
(E) HP-TLC analysis confirmed the sterol enrichment relative to glycerolipids in the PD fraction compared with the PM fraction.
Asterisks indicate significant differences (*P < 0.05, **P < 0.01, ***P < 0.001) by Wilcoxon test. n = 6 for PD and n = 5 for PM.
The PD-Enriched Membrane Fraction Shows an Enrichment in Sterols versus Glycerolipids When Compared with the PM per se
Plant sterol lipids are composed of free sterols, the most prevalent ones being sitosterol, stigmasterol, campesterol, and cholesterol, as well as conjugated sterols. Conjugated sterols are divided into different classes: the sterol glucosides (SG), which contain a sugar head group linked to the steroid backbone, and the acylated sterol glucosides (ASG), which contain an acyl chain esterified to the sugar head group. Sterol esters are not found in membranes but localize to oil bodies. The sterol profiles of the PM and PD-enriched membrane fractions were determined using high-resolution quadrupole time-of-flight mass spectrometry (Q-TOF-MS/MS) (Supplemental Figure 10). In both PD and PM membrane fractions, free sterols constituted the major sterol lipid class (∼80%), whereas the conjugated sterols, SG and ASG, were present in lower amounts. Sitosterol was the main sterol in free sterols and conjugated sterol classes in both PD and PM fractions (Supplemental Figure 10A).
We then examined whether the relative proportion of sterols, compared with the total pool of glycerolipids, differed between the PD-PM domain (i.e., the PD-enriched membrane fraction) and the cellular PM. Individual PD and PM samples were split into two, and the amounts of total sterols (steroid backbone) as well as total FAMEs were determined in parallel by GC-MS. The ratio of sterols to glycerolipids (i.e., sterol versus C16-C18 FAMEs) was found to be significantly higher in PD when compared with the PM (1.5-fold; Figure 5D). To confirm these results, individual membrane samples were separated in parallel by HP-TLC into neutral and polar lipids, quantified by densitometric scanning, and the ratio between sterols and glycerolipids was established. Our results confirmed that the PD-enriched membrane fraction displayed a significant enrichment in sterols (∼1.5-fold; Figure 5E). Altogether, our results showed that the PD-PM is enriched in sterols with a concomitant reduction in glycerolipids. Interestingly, we found that within the sterol pool, the proportion of the different sterol classes and molecular species was fairly similar between the PD and the PM fractions, indicating that sterol enrichment at PD does not involve strong structural selectivity (Supplemental Figure 10).
The Localization of PDCB1 and PdBG2 GPI-Anchored Proteins to Root Tip Primary PD Is Altered upon the Modulation of Sterol Composition
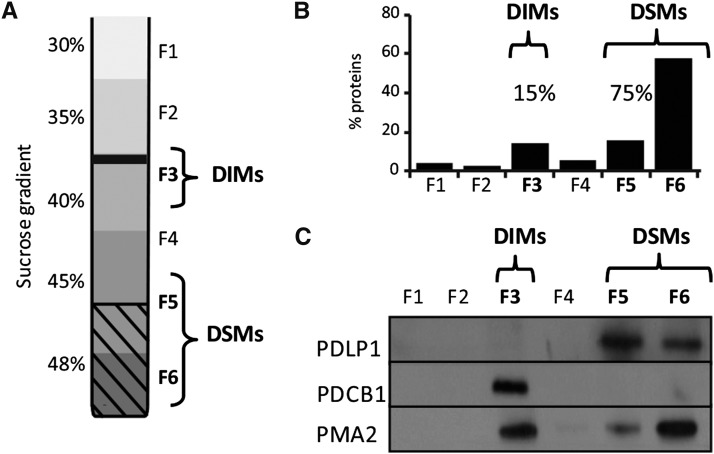
Our results show that specific lipids, namely sterols and sphingolipids, are locally enriched at the specialized PM domain across PD cell boundaries. Notably, several of the few PD proteins identified so far are GPI-anchored proteins, lipid-tethered polypeptides that cosegregate with the sphingolipid- and sterol-rich membrane environment in Arabidopsis (Borner et al., 2005; Levy et al., 2007; Kierszniowska et al., 2009; Simpson et al., 2009; Benitez-Alfonso et al., 2013; Faulkner et al., 2013). Focusing on the GPI-anchored protein PDCB1, which associates with the neck region of PD and has been implicated in the regulation of PD permeability (Simpson et al., 2009; Maule et al., 2013), we first tested whether PDCB1 displays an affinity for sphingolipid- and sterol-rich membrane environments. We isolated sphingolipid- and sterol-rich microdomains, referred to as detergent-insoluble membranes (DIMs) (Mongrand et al., 2004; Borner et al., 2005), exploiting their resistance to solubilization with mild nonionic detergents at low temperature. As expected, the low buoyant density DIM fraction represented only 15% of total proteins (fraction 3; Figure 6B) under our experimental conditions. The results showed that PDCB1 associates exclusively with DIMs (fraction F3; Figure 6C), whereas PDLP1, a type 1 integral membrane protein, which also localizes at PD but does not bear a GPI anchor (Thomas et al., 2008), associated instead with the detergent-soluble membranes (DSMs) (fractions 5 and 6; Figure 6C). The conspicuous partitioning of PDCB1 and PDLP1 in DIMs versus DSMs contrasted with PMA2, which associated with the two fractions in proportions of ∼40 and 60% for DIMs and DSMs, respectively. As the DIMs only represent 15% of total proteins, PMA2 showed enrichment in the DIM fraction by a factor of 3.7. These results are consistent with previous studies mostly showing a preferential association of PM-associated H+-ATPases with DIM fractions isolated from plant tissues (Mongrand et al., 2004; Borner et al., 2005; Morel et al., 2006; Raffaele et al., 2009; Srivastava et al., 2013). From these data, we concluded that PDCB1 and PDLP1 may reside in different membrane lipid environments and that the GPI-anchored protein PDCB1 is likely to reside in locally confined membrane environments enriched in sterols and sphingolipids.
Figure 6.
The GPI-Anchored PD Protein, PDCB1, Associates with the DIM Fraction, while the PD Protein, PDLP1, Does Not.
(A) Isolation of DIMs from Arabidopsis suspension cells. Six fractions of equal volume were collected from the top to the bottom of the sucrose gradient (F1 to F6).
(B) Protein quantification of collected fractions F1 to F6 shows that the DIM fraction (F3) contains ∼15% of total proteins, while the DSM fractions (F5 and F6) contain 75% of total proteins.
(C) Immunoblot of fractions F1 to F6. The GPI-anchored protein PDCB1 is highly enriched in the DIM fraction (F3), while PDLP1 associates with the DSM fractions (F5 and F6).
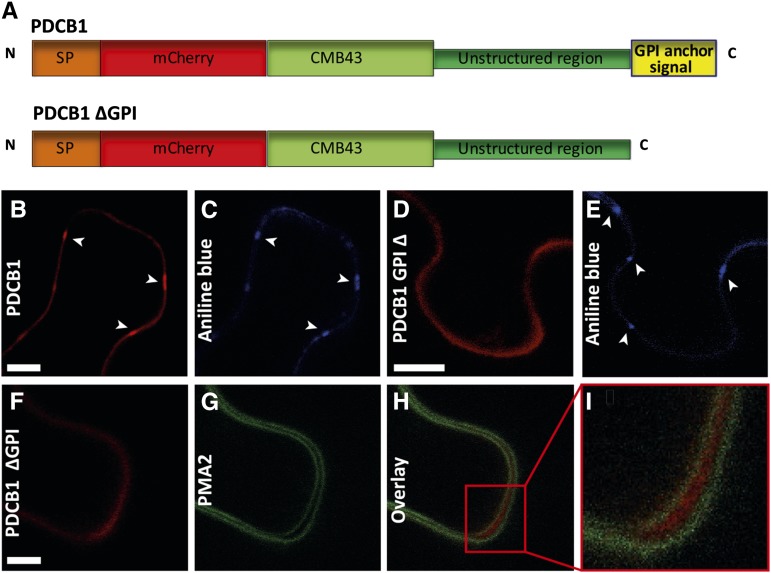
In mammalian cells, GPI-anchored proteins have been shown to organize in nanoscale cholesterol-sensitive clusters (Varma and Mayor, 1998; Sharma et al., 2004; Goswami et al., 2008; van Zanten et al., 2009). First, we tested whether GPI anchoring was a determinant for PD targeting. The GPI attachment signal (GPI signal peptide) is located at the very C terminus of the protein sequence and consists of a hydrophobic domain, an Ω cleavage site, and a short stretch of hydrophilic amino acids (Galian et al., 2012). This signal sequence is recognized in the ER lumen by a multisubunit transamidase, which concomitantly cleaves the C-terminal GPI signal peptide at the Ω site and adds the GPI moiety. The C-terminal GPI sequence signal of PDCB1-mCherry-tagged protein was deleted (PDCB1∆GPI), while the protein still contained its callose binding module (CBM43) (Figure 7A). Transient expression in Nicotiana benthamiana leaves showed that, upon GPI deletion, PDCB1 did not cluster at PD anymore (Figures 7D and 7E) like control full-length construct (Figures 7B and 7C). Instead, PDCB1∆GPI was secreted into the extracellular matrix, as shown by coexpression with the PM protein PMA2-GFP (Figures 7F to 7I). These data indicate that the GPI lipid moiety of PDCB1 is required for PD targeting.
Figure 7.
The GPI Anchor of PDCB1 Is Required for PD Targeting.
(A) Structural organization of PDCB1.mCherry and PDCB1.mCherry∆GPI. Chimeric constructs consist of the signal peptide of PDCB1, followed by the coding sequence of mCherry fused to the callose binding domain CBM43, unstructured region, and C-terminal GPI anchor signal. For PDCB1.mCherry∆GPI, the C-terminal GPI anchor signal was deleted.
(B) and (C) PDCB1.mCherry is targeted to PD (arrowheads) when transiently expressed in N. benthamiana leaves, as revealed by costaining with aniline blue ([B] and [C], respectively). Bar = 10 μm.
(D) and (E) PD targeting (arrowheads) was lost upon deletion of the PDCB1 C-terminal GPI anchor signal ([D] and [E]). Bar = 10 μm.
(F) to (I) Coexpression of PDCB1.mCherry∆GPI (F) and the PM-located protein PMA2.GFP (G) demonstrates that PDCB1.mCherry∆GPI is secreted into the extracellular space ([H] and [I]). (I) shows a higher magnification image of the box in (H). The epidermal cells were slightly plasmolyzed with 0.4 M mannitol to distinguish wall versus PM association. Bar = 5 μm.
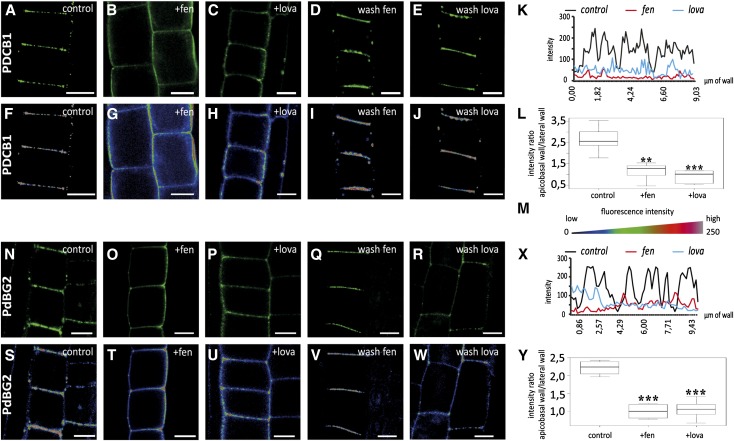
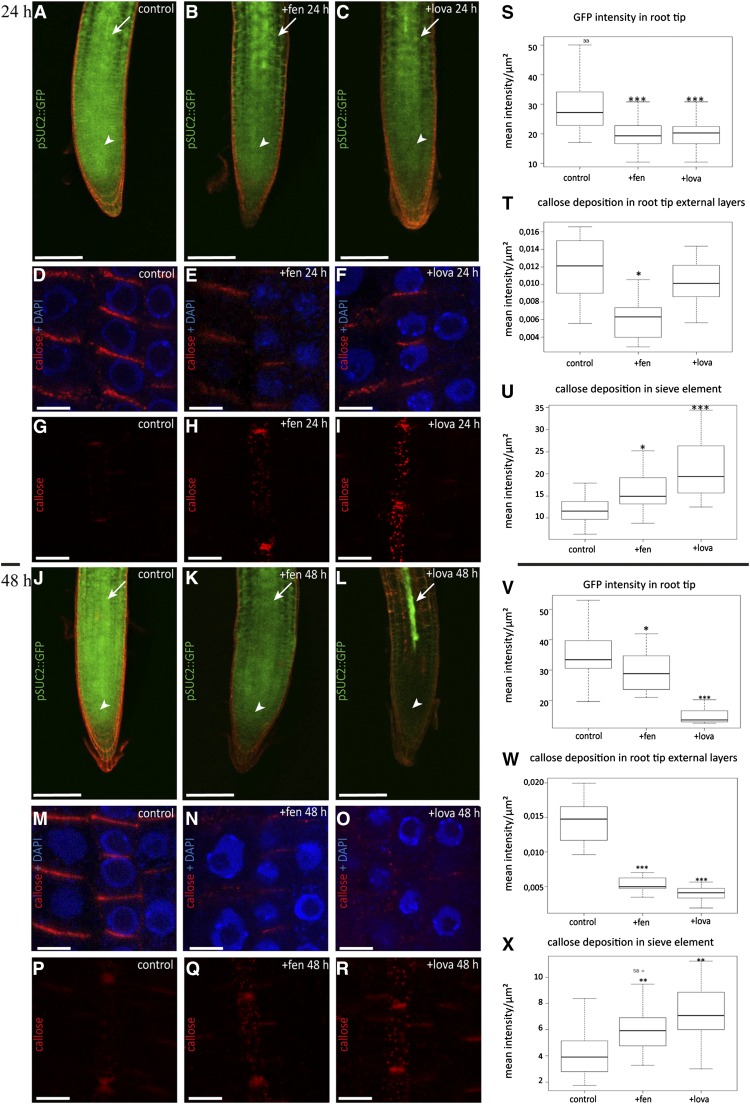
To address the question of whether PDCB1 localization at primary PD is dependent on sterol composition, we used yellow fluorescent protein (YFP)-PDCB1 lines and drugs specifically inhibiting sterol biosynthesis. We selected two different inhibitors, fenpropimorph (fen) and lovastatin (lova), which act at different steps along the biosynthetic pathway and have been well characterized in Arabidopsis roots (Vögeli and Chappell, 1991; Hartmann et al., 2002; He et al., 2003). We focused on epidermal cells of the root meristematic zone, which contain actively dividing cells with mainly primary PD like cultured cells. To minimize undesired secondary effects, seedlings were first grown on normal medium for 5 d before being transferred for 24 h to medium containing fen or lova. In control plants, the PDCB1 fluorescent signal was most prominent at the apical-basal division plane of epidermal root cells, where abundant primary PD are established during cytokinesis (Zhu, 1998; Simpson et al., 2009) (Figures 8A and 8F). At apical-basal walls, the YFP-PDCB1 fluorescence pattern was clearly punctuated, each spot of fluorescence corresponding to a PD or group of PD (Figures 8A and 8F; see Figure 8K for plot intensity along division walls). After 24 h of treatment with either fen or lova, the spotty PD fluorescence pattern had almost entirely vanished (Figures 8B, 8C, 8G, and 8H; see Figure 8K for plot intensity along division walls). Instead, YFP-PDCB1 was evenly distributed not only along the apical-basal membrane but also along the entire lateral membranes. This shift of YFP-PDCB1 localization from apical-basal to lateral was quantified by measuring the fluorescence intensity ratio between lateral and apical-basal membranes. Our results confirmed the mislocalization of PDCB1 upon both fen and lova treatment (Figure 8L). We next tested whether the effect of fen and lova treatment on PDCB1 targeting was reversible. When fen- or lova-treated seedlings were transferred back to Murashige and Skoog (MS) control medium for 24 h, the PDCB1 localization pattern was similar to that of control plants (Figures 8D, 8E, 8I, and 8J). The latter result suggests that mislocalization of PDCB1 under short sterol inhibition treatment is not due to a strong defect in PD formation. However, to verify this point, we performed transmission electron microscopy experiments on seedlings treated with fen for 24 h. We observed no obvious differences between PD in fen-treated and control plants, suggesting that PD formation and ultrastructure were not significantly compromised after 24 h of treatment (Supplemental Figure 11). Altogether, our results showed that targeting of the GPI-anchored PDCB1 to the laterally restricted PD-PM domain of young PD is sterol-dependent.
Figure 8.
Sterol Composition Defines the PD Localization of the GPI-Anchored Proteins PDCB1 and PdBG2 in Arabidopsis Root Tips.
Localization of YFP-PDCB1 ([A] to [L]) and mCitrine-PdBG2 ([N] to [Y]) GPI-anchored proteins in epidermal cells of Arabidopsis roots tips, visualized by confocal microscopy. (F) to (J) and (S) to (W) show quantitative color-coded heat-map micrographs of (A) to (E) and (N) to (R), respectively. (M) shows a color-coding bar for heat-map images. (A), (F), (N), and (S) Untreated control cells. (B), (C), (G), (H), (O), (P), (T), and (U) show cells from seedlings grown on normal agar plates for 5 d and then transferred to 50 μg/mL fen (+fen) agar plates ([B], [G], [O], and [T]) or 1 μM lova (+lova) agar plates ([C], [H], [P], and [U]) for 24 h. Under fen or lova treatment, GPI-anchored proteins do not accumulate at PD anymore, but instead label the whole cell periphery. (D), (E), (I), (J), (Q), (R), (V), and (W) show cells from seedlings grown on normal agar plates for 5 d, transferred to 50 μg/mL fen (+fen) agar plates ([D], [I], [Q], and [V]) or 1 μM lova (+lova) agar plates ([E], [J], [R], and [W]) for 24 h, and then transferred back to normal agar plates for 24 h (wash treatment). Note that GPI-anchored proteins are now restricted to the division plane as in untreated control experiments ([A], [F], [N], and [S]). (L) and (Y) show the quantification of fluorescence intensity ratios for apical-basal membranes versus lateral membranes in untreated control (corresponding panels [A], [F], [N], and [S]), fen (corresponding panels [B], [G], [O], and [T]), and lova (corresponding panels [C], [H], [P], and [U]) treatments for YFP-PDCB1-expressing (L) and mCitrine-PdBG2-expressing (Y) lines. n = 10 roots. Six cells per root were analyzed. Asterisks indicate significant differences (*P < 0.05, **P < 0.01, ***P < 0.001) by Wilcoxon test. (K) and (X) show representative intensity plots along division walls of control and treated plants. In control conditions, the PD association of YFP-PDCB1 (K) and mCitrine-PdBG2 (X) is illustrated by peaks of high fluorescence intensity along the apical-basal walls (black lines). After either fen or lova treatment, these peaks of high intensity disappeared, indicating that PD association was lost (red and blue lines, respectively).
To confirm these results, we next analyzed another PD-PM-localized GPI-anchored protein, the β-1,3-glucanase PdBG2, which, like PDCB1, has been shown to regulate callose deposition at PD in roots (Benitez-Alfonso et al., 2013; Maule et al., 2013). Consistently, we found that PdBG2 localization at PD in Arabidopsis root tips was also dependent on the presence of sterols. Reminiscent of PDCB1, PdBG2 localization was found to be punctuated at apical-basal membranes of meristematic root cells (Figures 8N, 8S, and 8X). After 24 h of treatment with either fen or lova, the spotty mCitrine-PdBG2 fluorescence at PD was weaker and the fluorescence was redistributed evenly at the whole cell periphery (Figures 8O, 8P, 8T, and 8U; see Figure 8X for plot intensity along division walls). As for PDCB1, fluorescence quantification confirmed the mislocalization defect of PdBG2 upon fen or lova treatment (Figure 8Y).
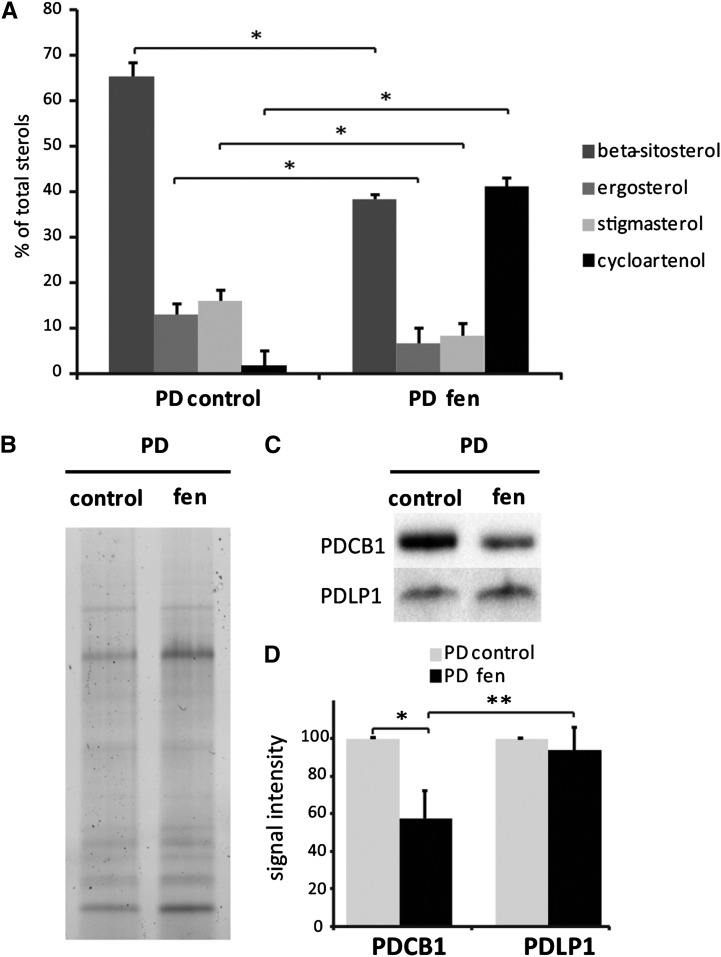
We next exposed Arabidopsis suspension cells to sterol inhibitor treatment with the aim of correlating PD targeting and modification of the sterol pool at the PD-PM specifically. After 24 h of treatment with either fen or lova using the same concentrations as before (i.e., 50 μg/mL and 1 μM, respectively), we observed little change in the sterol pool at the cellular level (Supplemental Figure 12C). By contrast, Arabidopsis seedlings showed a reduction of ∼20% after fen or lova treatment (Supplemental Figure 12B). These results suggest that depending on the tissue type and growth condition, the effect of the drugs varies. Therefore, we tried different exposure times and drug concentrations with the sterol inhibitor fen. Fen is expected to induce milder effects than lova, as it changes the sterol pool qualitatively by increasing the amount of intermediate cyclopropyl sterols but does not change the total amount of sterol species (He et al., 2003). When Arabidopsis cells were treated for 48 h with 250 µg/mL fen, the cellular pool of sterols was strongly altered (Supplemental Figure 12C). Similarly, purified PD showed a reduction of sterols by 40% with a concomitant increase of cyclopropyl sterols in treated versus control samples (Figure 9A). The modification of the PD-PM sterol composition was correlated with a significant reduction of PDCB1 association with the channels (∼50%), whereas PDLP1 remained relatively stable (Figures 9B to 9D).
Figure 9.
Modification of the Sterol Pool of the PD-PM Domain Correlates with a Reduction of PDCB1 Association with PD Channels.
(A) Quantification by GC-MS of the sterol lipids in PD-enriched membrane fractions purified from control and fen-treated liquid cultured Arabidopsis cells (48 h, 250 μg/mL). Sterol inhibitor treatment leads to a strong decrease of total sterols and a concomitant increase of intermediate cyclopropyl sterols. n = 3 for PD and PD + fen samples. Error bars indicate sd.
(B) to (D) Immunoblot analysis of control and fen-treated PD fractions showing that, upon modification of the sterol pool, the PDCB1 association with PD channels is reduced, whereas the PDLP1 signal remains stable. (B) shows SDS-PAGE protein profiles of the PD- and PD-fen-treated fractions used for blotting shown in (C). (C) shows representative immunoblots for PDCB1 and PDLP1. (D) shows the quantification from immunoblot analyses of the PDCB1 and PDLP1 association with PD using the Bio-Rad Chemidoc MP system and Image Lab software.
Asterisks indicate significant differences (*P < 0.05, **P < 0.01) using the Wilcoxon test. n = 3 for PD and n = 6 PD + fen samples. Error bars indicate sd.
Callose-Mediated PD Permeability Is Altered upon Sterol Inhibition
Both PDCB1 and PdBG2 play determinant roles in regulating callose deposition at PD and, as a consequence, in the fine-tuning of intercellular trafficking (Simpson et al., 2009; Benitez-Alfonso et al., 2013; Maule et al., 2013). Therefore, we investigated whether a modulation of the sterol pool would also alter symplastic connectivity.
To assess any potential changes in PD permeability under sterol inhibition, we used Arabidopsis plants expressing green fluorescent protein (GFP) under the control of the phloem-specific SUCROSE-H SYMPORTER2 gene promoter (ProSUC2) (Imlau et al., 1999). Under control conditions, GFP diffused from the phloem into surrounding tissues, including the root meristem (Figure 10A). After 24 h of sterol inhibitor treatment, we observed a significant reduction of GFP intensity in the root tip of both fen- and lova-treated plants (Figures 10A to 10C and 10S), whereas GFP intensity in the phloem was either stable or slightly increased (Supplemental Figure 13B). These results suggest restricted GFP diffusion from the vasculature to surrounding tissues via PD. To better understand the effect of sterol inhibition on cell-to-cell connectivity, we then examined the level of callose at PD in the root meristem and stele. To this end, Arabidopsis roots were chemically fixed and immunofluorescence-labeled using callose-specific antibodies. Our results showed that a significant increase of callose signal was detected in the phloem tissue of 24-h fen- and lova-treated seedlings (Figures 10G to 10I and 10U). In particular, the apical-basal and lateral walls of protophloem sieve elements were intensely stained compared with controls (Figures 10G to 10I). Therefore, we concluded that occlusion caused by callose deposition in the phloem was likely responsible for reducing GFP symplastic unloading from the vasculature to the surrounding meristematic tissues upon sterol depletion. Surprisingly, however, increased callose deposition was specific to the root vasculature. Surrounding tissues either showed a minor decrease of callose deposits at the apical-basal walls (fen treatment) or no significant changes (lova treatment) when compared with control plants (Figures 10E and 10F; see Figure 10T for quantification). At 48 h, this tendency was accentuated, and callose deposition was dramatically reduced in root tip tissues for both fen- and lova-treated plants (Figures 10M to 10O; see Figure 10W for quantification). In phloem sieve elements, we measured an increase of callose deposition similar to what we observed in 24-h treated plants (Figures 10P to 10R; see Figure 10X for quantification) and a concomitant reduction of GFP unloading from the phloem to surrounding tissues (Figures 10J to 10L; see Figure 10V for quantification). Taken together, these results suggest that modification of the sterol pool interferes with callose production/degradation at PD and, as a consequence, impairs intercellular trafficking. Moreover, our data indicate that callose deposition is differentially regulated in protophloem compared with other root tissues.
Figure 10.
Altered Sterol Composition Affects Both Symplastic Phloem Unloading and Callose Deposition at PD in Arabidopsis Roots.
(A) to (C), (J) to (L), (S), and (V) GFP symplastic unloading from the phloem to surrounding tissues is reduced in Arabidopsis roots grown on fen ([B] and [K]) or lova ([C] and [L]) for 24 h ([A] to [C]) and 48 h ([J] to [L]). Control seedlings expressing ProSUC2:GFP show diffusion out of the phloem (white arrows) into surrounding tissues including the root meristem (white arrowheads) ([A] and [J]). After fen ([B] and [K]) or lova ([C] and [L]) treatment, GFP transport from companion cells was altered, as indicated by the apparent reduction of the GFP intensity in the root tip and cells surrounding the vasculature. Quantification of fluorescence intensity in the root meristem in control, fen-, and lova-treated plants for 24 and 48 h confirmed that sterol inhibition significantly reduces GFP symplastic unloading from the companion cells to surrounding tissues ([S] and [V], respectively). White arrows indicate the root vasculature where ProSUC2:GFP is expressed, and white arrowheads indicate the GFP unloading zone at the root tip. Bars = 200 μm.
(D) to (I), (M) to (R), (T) to (U), (W), and (X) Callose immunofluorescence (red) in Arabidopsis seedlings treated with fen ([E], [H], [N], and [Q]) or lova ([F], [I], [O], and [R]) for 24 and 48 h. DAPI staining of DNA (blue) was done to highlight the cellular organization of root tissues. Sterol inhibition treatment induced a significant accumulation of callose deposition in sieve elements at both 24 and 48 h, as confirmed by fluorescence quantification ([H] and [U], fen, 24 h; [Q] and [X], fen, 48 h). In external cell layers, callose signal was reduced significantly under 24 h of fen but not lova ([E] and [F], respectively; [T] for fluorescence quantification). However, at 48 h, a strong decrease in callose accumulation was observed in both fen- and lova-treated plants ([N] and [O], respectively; [W] for fluorescence quantification). Bars = 5 μm.
Callose immunolabeling and ProSUC2:GFP experiments were performed at least three independent times using a total of 20 seedlings per condition. Asterisks indicate significant differences (*P < 0.05, **P < 0.01, ***P < 0.001) by Wilcoxon test.
In conclusion, our results show that the PD-PM is enriched in sterols and sphingolipids compared with glycerolipids and that the modulation of sterol biosynthesis results in mislocalization of the PD-localized GPI-anchored proteins PDCB1 and PdBG2 at primary PD and in an alteration of PD permeability due to a defect in callose deposition.
DISCUSSION
PD Are Defined by Specialized Domains of the PM Displaying a Unique Lipid Composition
A striking feature of PD organization is the apparent continuity of the PM from cell to cell through the cylindrical pores. However, to guarantee the compartmentalization of PD-related function, the PM is unlikely to continue homogeneously through the PD channel. Specialization of the PM along the PD channel is illustrated by the local enrichment of a specific set of membrane-associated proteins (Levy et al., 2007; Thomas et al., 2008; Simpson et al., 2009; Fernandez-Calvino et al., 2011; Jo et al., 2011; Benitez-Alfonso et al., 2013; Faulkner et al., 2013). Interestingly, the results from our study additionally show that integral membrane proteins, such as the H+-ATPase PMA2, the cellulose synthase catalytic subunits CesA3 and CesA6, and the aquaporin PIP2;2, remain largely absent from PD. Our results are consistent with an earlier study showing that the PM proton pump H+-ATPase is not associated with PD in young pulvini of Mimosa pudica (Fleurat-Lessard et al., 1995). Hence, lateral heterogeneity of the PM at the site of PD intercellular junction is not only illustrated by the clustering of a given set of proteins but also by the exclusion of some proteins predominant in the PM. As for proteins, lipid segregation along the PD-PM has also been suggested to be fundamental for both PD function and structure (Raffaele et al., 2009; Mongrand et al., 2010; Tilsner et al., 2011; Delage and Zurzolo, 2013; Faulkner, 2013). However, until now, the lipid constituents of PD specialized membranes had remained unknown.
In this study, we used highly purified PD-derived membranes to perform comparative lipid analysis. Importantly the purification protocol employed does not require the use of detergent, which has been criticized for leading to the artificial aggregation of sterols and sphingolipids (Tanner et al., 2011). We convincingly demonstrated that our procedure allows the isolation of a fraction highly enriched in the PD-PM domains without notable contamination by the cellular PM or other membrane compartments. Our results showed that not only proteins but also the lipids of the PD-PM domain differ from the cellular PM. In particular, our data indicate that the PM domain lining the primary PD is enriched in sterols, highly glycosylated sphingolipids harboring VLCFAs (GIPCs), and phospholipids containing fatty acids with a reduced degree of desaturation. Although our purified PD-enriched membrane fraction probably contains remains of the desmotubule, the enrichment in sphingolipids and sterols is unlikely to be derived from the desmotubule membranes. First, we estimated that the PD-enriched membrane fraction is highly enriched in the specialized PD-PM domains (Figures 2 and 3). Second, it is known that the final steps of the GIPC biosynthetic pathway are localized at the Golgi apparatus (Pata et al., 2010). Thus, desmotubule membranes, which are derived from the ER, are unlikely to contain these complex sphingolipids (GIPCs). Similarly, sterols are enriched along the secretory pathway and usually depleted in the ER (Hartmann and Benveniste, 1987). This implies that the enrichment factors (∼1.5-fold) for GIPCs and sterols versus glycerolipids very likely indicate that these lipids predominantly localize to the PD-PM domain. The local confinement of sterols, GIPCs, and phospholipids with a higher saturation degree raises the question of how a specific PD-PM domain with a particular lipid composition is established and maintained at PD and suggests the existence of a diffusion barrier at the PD-PM across PD channels. Consistently, pioneering work by Grabski et al. (1993) has shown that lipid analogs were indeed unable to diffuse from cell to cell through the PM, whereas ER-located lipids were able to move through the PD channels.
Clustering of Sterols and Sphingolipids Enriched Lipid Raft-Like Domains at PD?
The existence of a locally confined sphingolipid and sterol concentration at PD raises the important question of lateral compartmentalization within the PM. In model membranes, autoassociation between sterols and sphingolipids results in domain formation observable with phase-partitioning membrane probes (Simons and Vaz, 2004). In biological membranes, the “lipid raft” hypothesis proposes a mechanism for membrane nanodomain formation based on the preferential association between sterols and sphingolipids creating a liquid-ordered phase within the membrane (Pike, 2006; Simons and Sampaio, 2011). Nonetheless, the very existence of lipid nanodomains remains controversial, owing to the lack of suitable detection techniques, especially in plants. To some extent, this is due to the fact that isolating nanoscale domains for biochemical characterization is technically challenging, especially since the characterization of lipid domains has relied largely on the use of DIM fractions (Mongrand et al., 2004; Lefebvre et al., 2007; Kierszniowska et al., 2009; Keinath et al., 2010). The biological relevance of this fraction and its accurate correspondence to lipid domains in vivo have been rightly questioned (Schuck et al., 2003; Tanner et al., 2011; Malinsky et al., 2013). Appreciably, our PD isolation procedure is detergent-free, and our analysis of PD lipid composition still revealed a similar pattern to that reported previously for DIMs (i.e., an enrichment of sphingolipids and sterols versus glycerolipids and a higher proportion of saturated fatty acyl chain-containing lipids). Compared with DIM fractions, however, the ∼1.5-fold enrichment in sterols and sphingolipids in the PD-PM in comparison with the bulk PM is moderate. In DIM fractions, enrichment levels of 4- to 6.7-fold have been measured for sphingolipids and from 1.7- to 4-fold for sterols (Mongrand et al., 2004; Borner et al., 2005; Lefebvre et al., 2007). Direct comparison is difficult, however, especially considering that cold detergent treatment of membrane samples may lead, at least to some extent, to the artificial aggregation of sterols and sphingolipids (Heerklotz, 2002). Alternatively, the PD-PM may itself be subcompartmentalized, and sterols/GIPC nanodomains may not span the entire length of the membrane. In line with the scenario that PD are defined by specific lipid domains of the PM, it was recently shown that the remorin protein, which associates with DIMs and clusters in sterol-dependent nanoscale domains of the PM in vivo, also localizes to PD (Raffaele et al., 2009; Gui et al., 2014). Moreover, recent data suggest that PD contain tetraspanin-enriched microdomains (TEMs) (Fernandez-Calvino et al., 2011). Tetraspanins are transmembrane proteins that have the ability to associate with one another and recruit specific proteins to build up TEMs, which, similar to lipid rafts in mammalian cells, play a role in the membrane compartmentalization of biological processes such as cell adhesion, signaling, and intracellular trafficking (Stipp et al., 2003; Yunta and Lazo, 2003; Rubinstein, 2011). Importantly, tetraspanins have been reported to directly bind cholesterol in mammalian cells through their palmitate moieties (Charrin et al., 2003). Our own proteomic analysis revealed that several tetraspanins are indeed enriched in the PD-enriched membrane fraction (Supplemental Table 1). Interestingly, all members display several putative S-acylation sites (Supplemental Figure 14). Future work will be necessary to determine whether cysteine palmitoylation is important for direct interaction between sterols and plant tetraspanins and for PD targeting. However, our data showing an enrichment of sterols at PD are consistent with the presence of TEMs at PD. Altogether, our results strongly support the notion that the PD membrane clusters sterol- and sphingolipid-enriched domains.
Modification of the Sterol Pool Affects the Clustering of GPI-Anchored Proteins at Primary PD and Modifies Callose-Mediated PD Permeability in Arabidopsis Roots
Our results show a local enrichment of sterols at PD when compared with PM. A crucial question was to determine whether the sterol profile is important for proper PD functionality and the spatial organization of PD membranes with the recruitment of a specific set of proteins. GPI-anchored proteins, which are tethered to the outer leaflet of the PM through two saturated fatty acids, have been shown to preferentially associate with nanodomains in a cholesterol-dependent manner (Mayor and Riezman, 2004; Sangiorgio et al., 2004; Sharma et al., 2004; Borner et al., 2005; Kierszniowska et al., 2009; Levental et al., 2010). Strikingly, of the few PD-PM-associated proteins identified so far, a substantial number harbor a GPI anchor (Levy et al., 2007; Simpson et al., 2009; Benitez-Alfonso et al., 2013; Faulkner et al., 2013). Among those are the callose binding protein PDCB1 and the β-1,3-glucanase PdBG2 (Simpson et al., 2009; Benitez-Alfonso et al., 2013; Maule et al., 2013). In line with our comparative lipidomic analysis, we show that the GPI-anchored protein PDCB1 associates with DIMs, suggesting a preferential location of PDCB1 in sterol-rich membrane environments. Additionally, several PD-associated GPI-anchored proteins, including PDCB5 and several members of the β-1,3-glucanase family, have been shown by others to be enriched in DIMs in a sterol-dependent manner (see Supplemental Table 2 for a listing of GPI-anchored PD proteins found in DIMs in the literature). Consistently, we showed that clustering of both PDCB1 and PdBG2 at PD is strongly abolished in young epidermal cells of the root tip after alteration of the sterol composition with two different sterol biosynthesis inhibitors. Significant reduction of the PDCB1, but not PDLP1, association with primary PD was correlated with a modification of the sterol pool of the PD-PM domains in Arabidopsis cultured cells. Altogether, our results suggest that, upon modification of sterols, the PD-PM domain is not recognized anymore as a subcellular targeting address for GPI-anchored proteins in young tissues.
Little information exists regarding how GPI proteins specifically associate with PD channels. In addition to the lipid moiety, both PDCB1 and PdBG2 contain a callose binding module (Carbohydrate Binding Module Family 43 [CBM43]), which may participate in PD targeting (Simpson et al., 2009; Benitez-Alfonso et al., 2013). To establish whether PD association is determined by GPI anchoring, we generated a PDCB1 mutant deleted in the C-terminal GPI signal but still containing the CBM43 callose binding motif. Our results indicate that the GPI anchor is indeed necessary for PD association, whereas the callose binding motif is not sufficient by itself to drive clustering of the protein at PD. Callose binding may nevertheless contribute to the stabilization of the GPI-anchored proteins at PD once the protein is associated with the outer leaflet of the PD-PM domains. Altogether, our data suggest that PDCB1 and PdBG2 are recruited to primary PD, most likely from the surrounding PM, through their GPI anchor in a sterol-dependent manner.
Callose deposition at the neck region of PD channels is a key regulatory process involved in the fine-tuning of symplastic connectivity during both developmental processes and biotic/abiotic responses (Iglesias and Meins, 2000; Ruan et al., 2004; Levy et al., 2007; Guseman et al., 2010; Hofmann et al., 2010; Rinne et al., 2011; Vatén et al., 2011; Benitez-Alfonso et al., 2013; Han et al., 2014). Both PdBG2 and PDCB1 have been shown to play an important role in the regulation of callose deposition at PD and in symplastic connectivity (Simpson et al., 2009; Benitez-Alfonso et al., 2013). In line with the mislocalization of the two callose-modifying GPI-anchored proteins, we demonstrated that sterol inhibition induces defective cell-to-cell permeability due to altered callose deposition at PD. Interestingly, our data indicate that callose accumulation is differentially regulated in the highly specialized protophloem sieve elements and other tissues of the root. This likely indicates that the action of structural sterols in the modulation of callose deposition will be a function of the developmental stage and tissue types considered. Callose levels at PD are regulated through a complex network involving callose-modifying proteins such as the β-1,3-glucanases, PDCB, and callose synthase complexes as well as additional factors such as the production of reactive oxygen species and Ca2+ (Benitez-Alfonso et al., 2011; Zavaliev et al., 2011). Therefore, further work will be necessary to fully understand the impact of sterols on PD functionality. Nevertheless, our work suggests that one mode of action of sterols, in relation to PD function, is to participate in the local recruitment of callose-modifying GPI-anchored proteins into PD-specific lipid nanodomains.
Both PDCB1 and the β-1,3-glucanase PdBG2 are likely to be preferentially targeted to the neck region of PD, where callose is deposited. In agreement with this scenario, PDCB1 was immunolocalized at the entrance of the PD channels while PDLP1, which is not GPI-anchored, was mainly found associated along the length of the PM in the pores (Maule et al., 2011). Interestingly, our data indicate that PDLP1 and PDCB1 display a radically different affinity for DIMs and respond differently upon sterol inhibition. This most likely indicates that the two proteins are present in different membrane environments, PDCB1 being preferentially present in sterol-rich membrane environments. Interestingly, many of the PD-localized β-1,3-glucanases identified so far harbor a GPI anchor (Supplemental Table 2). Similar to PDCB1 (and other members of the PDCB family), GPI-anchored β-1,3-glucanases are likely to be specifically targeted to the neck region of the PD, consistent with their physical association with callose. Altogether, our results suggest that sterol- and sphingolipid-enriched domains may be confined to the entrance of the PD pore and concentrate functional elements involved in the regulation of callose deposition and modulation of the PD aperture. It should be mentioned, however, that in our hands immunogold labeling for callose in Arabidopsis cultured cells did not show strong enrichment of the β-1,3-glucan at the neck regions when compared with the central part (Supplemental Figure 1E). This may indicate that callose is evenly distributed along the entire length of the channels in primary PD. However, interpretation of these results is difficult given that young dividing cells mostly consist of relatively thin walls, meaning that callose clustering at the neck of the channels may be difficult to appraise.
Overall, our study shows that specific lipids are locally enriched at the PM across cell boundaries and likely act to functionally define the PD-PM domain. Moreover, we highlight a potential role for sterols in establishing and maintaining positional specificity at the primary PD as well as regulating callose-mediated PD permeability. However, further work will be needed to fully understand the mode of action of sterols on PD structure and function. Our study was limited to the primary PD of young meristematic tissues. An exciting new path of investigation will be to establish whether developmental and/or biotic/abiotic stresses trigger dynamic changes in the PD lipidome to adjust cell-to-cell connectivity.
Altogether, we believe that our results pave new paths toward a better understanding of how lipids act as functional units of PD-specialized membranes.
METHODS
Plant Material and Growth Conditions
Suspension cell cultures of Arabidopsis thaliana (ecotype Landsberg erecta) were grown as described (Bayer et al., 2004) except that cells were subcultured once instead of twice per week (20 mL into 200 mL of fresh medium).
The following transgenic Arabidopsis lines expressing fluorescent proteins in the Columbia-0 background were used: p35s:YFP-PDCB1 (Simpson et al., 2009), p35s:mCit:pdBG2 (Benitez-Alfonso et al., 2013), and ProSUC2:GFP (Imlau et al., 1999). Seedlings were grown on agar (8 g/L) plates containing MS salts (4.4 g/L) supplemented with vitamins and MES (0.5 g/L) at pH 5.8 in a growth chamber at 24°C in long-day light conditions (irradiance of 5 μE m−2 s−1).
Inhibitor Treatments
For inhibitor experiments, 5-d-old seedlings were transferred to MS agar plates containing 50 μg/mL fen (stock solution 50 μg/μL in DMSO) or 1 μM lova (stock solution 1 mM in DMSO). Control plates contained an equal amount of 0.1% DMSO solvent. Seedlings were observed by confocal microscopy (GFP-PDCB1, mCitrine-PdBG2, and ProSUC2:GFP) or whole-mount fixed for callose immunolabeling 1:200 (Boutté and Grebe, 2014) after 24 or 48 h of treatment. Washout experiments were performed by transferring 24-h treated seedlings onto MS agar plates containing 0.1% DMSO for another 24 h before confocal observation.
Cell Wall, PD, PM, and DIM Purification
Cell wall fractions were prepared from 6-d-old Arabidopsis suspension cultured cells as described by Bayer et al. (2004), except that cell disruption through the N2 cell disruption vessel was repeated five consecutive times instead of three. The PD-enriched membrane fraction was obtained from purified wall fragments as described (Fernandez-Calvino et al., 2011; Grison et al., 2015). Briefly, purified cell walls were digested with 0.7% (w/v) cellulase R10 (Karlan) in digestion buffer (10 mM MES, pH 5.5, and 4.4% mannitol) containing 1 μM phenylmethylsulfonyl fluoride and complete protease inhibitor cocktail (Roche Diagnostic) for 1.5 h at 37°C with 50 to 100 rpm shaking. After centrifugation at 5850g for 5 min at 4°C, the supernatant and pellet fractions were collected separately. The pellet was washed in an excess of digestion buffer and centrifuged again, and the two supernatants were combined before centrifugation at 110,000g for 40 min at 4°C. The resulting pellet, containing PD-derived membranes, was washed in an excess volume of cold Tris-buffered saline (TBS; 20 mM Tris-HCl, 0.14 M NaCl, and 2.5 mM KCl, pH 7.4). Finally, the pellet was resuspended in cold 1× TBS containing protease inhibitors (Roche Diagnostic). Approximately 600 mL of cultured cells was used to obtain 100 μg of PD-enriched membrane fraction. Protein amount was determined with a bicinchoninic acid protein assay using BSA as standard. For the purification of PD-enriched membrane fractions from cells treated with sterol inhibitor, fen was added to 5-d-old liquid cultured Arabidopsis cells at a final concentration of 250 μg/mL (stock solution 500 μg/μL in DMSO) and incubated as described previously for 48 h before purification.
The PM was purified by a two-phase partitioning system according to Marmagne et al. (2006) with some modifications. Briefly, vacuum-filtered 6-d-old suspension cells (∼80 g) were homogenized in a Waring blender with 3 volumes of 0.5 M sucrose, 50 mM Tris-MES, pH 8, 40 mM EDTA, 20 mM DTT, and 1 mM phenylmethylsulfonyl fluoride. All buffers were ice cold, and centrifugation steps were performed at 4°C. The homogenate was centrifuged for 20 min at 13,000g, and the resulting supernatant was centrifuged for 1 h at 100,000g. This microsomal pellet was resuspended in 300 mM sucrose, 3 mM KCl, and 5 mM KH2PO4/K2HPO4, pH 7.8, buffer (15 g final) and then gently mixed with 50 g of an aqueous polymer phase mixture (6.4% [w/w] polyethylene glycol 3350, 6.4% [w/w] Dextran T-500 in 300 mM sucrose, 3 mM KCl, and 5 mM KH2PO4/K2HPO4, pH 7.8). After a short low-speed centrifugation (∼1000g), the upper phase was recovered. Phase partitioning was performed one more time. The final upper phase was recovered and centrifuged at 100,000g for 1 h. The pellet was washed once with excess 1× TBS buffer to remove residual polyethylene glycol-dextran. The final pellet was resuspended in TBS containing protease inhibitors (Roche Diagnostic). Please note that isolated PM does not contain PD, which are discarded with wall debris by low-speed centrifugation early in the purification process.
For DIM purification, PM preparations (with one phase partitioning only) were further submitted to Triton X-100 treatment (final concentration 1% [v/v]) with a detergent-to-PM protein mass ratio of 10 (w/w) at 4°C for 30 min. Treated membranes were brought to a final concentration of 48% (w/w) sucrose, overlaid with successive 2-mL steps of 45, 40, 35, and 30% (w/w) sucrose in 1× TBS buffer, and then centrifuged for 16 h at 200,000g. DIMs were visible at the interface between the 35 and 40% layers as an opaque band. Fractions (1.8 mL) were recovered along the sucrose gradient, and one-tenth was used for protein concentration using the bicinchoninic acid protein assay to avoid Triton X-100 interference. Proteins from the different fractions were precipitated in 10% cold trichloroacetic acid for 30 min at 48°C. After centrifugation, the pellet was first washed with 10% trichloroacetic acid in water to remove residual sucrose and finally with cold acetone before being resuspended in Laemmli buffer for SDS-PAGE.
Immunoblot Analysis
Proteins from the PD, PM, microsomal, and DIM fractions were solubilized with 1× Laemmli buffer (Laemmli, 1970) for 30 min at 50°C. Proteins were separated by 7 or 12% SDS-PAGE, blotted to nitrocellulose, and analyzed with antiserum specific for PDLP1 (Fernandez-Calvino et al., 2011) at a dilution of 1:1250, PDCB1 (Simpson et al., 2009) at a dilution of 1:1000, PMA2 (Morsomme et al., 1998) at a dilution of 1:16,000, PIP2;2 (Santoni et al., 2003) at a dilution of 1:5000, CesA3 and CesA6 (Desprez et al., 2007) at a dilution of 1:500, calreticulin (Baluška et al., 1999) at a dilution of 1:5000, SMT1 (Boutté et al., 2010) at a dilution of 1:50, ECHIDNA (Gendre et al., 2011) at a dilution of 1:1000, V-ATPase (Agrisera) at a dilution of 1:1400, BiP (Höfte and Chrispeels, 1992) at a dilution of 1:8000, P16 (Vallon et al., 1986) at a dilution of 1:20,000, membrine11 (provided by L. Maneta-Peret and P. Moreau) at a dilution of 1:400, and Nad9 at a dilution of 1:5000. Specific binding was visualized by chemiluminescence standard techniques. For quantification, blot exposure to reveal the PDCB1 or PDLP1 signal was kept to a minimum to avoid saturation. Blot imaging and quantification were done using the Bio-Rad Chemidoc MP system, which enables quantitative chemiluminescence measurement, and Image Lab software.
Electron Microscopy
For transmission electron microscopy, 6-d-old Arabidopsis suspension cells or 5-d-old seedlings treated for 24 h with 50 μg/mL fen or DMSO for a control were high-pressure frozen with the Leica EM-PACT-1 system. Tissues were inserted into a flat copper carrier, fast-frozen, and cryosubstituted into the Leica AFS2 device. The different freeze-substitution steps were as follows: 72 h at −90°C in acetone solution containing 0.5% glutaraldehyde, 2% osmium tetroxide, and 0.1% uranyl acetate, and the temperature was then raised to 4°C. Samples were washed three times for 20 min in 100% acetone before embedding in Spurr’s resin, which was performed progressively (2 h in 25% Spurr’s resin in acetone, 2 h in 50% Spurr’s resin in acetone, overnight in 75% Spurr’s resin in acetone, and two times for 2 h in 100% Spurr’s resin). Polymerization was performed at 60°C for 24 h.
For immunogold labeling of Arabidopsis suspension cells, the different freeze-substitution steps were as follows: 72 h at −90°C in acetone solution containing 0.5% glutaraldehyde and 0.1% uranyl acetate. The temperature was then raised to −50°C, and after 12 h at −50°C, samples underwent three 20-min washes in 100% acetone and then three 20-min washes in 100% ethanol. The embedding in HM20 was performed at −50°C as follows: 2 h in 25% HM20 in acetone, 2 h in 50% HM20 in acetone, overnight in 75% HM20 in acetone, and two times for 2 h in 100% HM20. Polymerization was performed under UV light for 48 h at −50°C and then for 48 h at 20°C. Immunogold labeling was performed as described previously (Kang et al., 2011) on 70-nm sections with the following antibodies: monoclonal anti-β-1,3-glucan antibody (Biosupplies; Supplemental Figure 1) diluted 1:20 and polyclonal antiserum against the ATPase PMA2 (Morsomme et al., 1998) diluted 1:300. Antibody binding was detected with the following secondary antibodies: 5-nm diameter gold-conjugated goat anti-mouse antibodies (for callose) diluted 1:30 and 10-nm diameter gold-conjugated goat anti-rabbit antibodies (for PMA2) diluted 1:30. The quantification of PMA2- and callose-associated gold particle distribution in Figure 1C was performed using Arabidopsis ultrathin sections that were colabeled with anti-PMA2 and anti-callose antibodies. A total of 55 micrographs containing 65 PD were examined. Gold particles specifically associated with the PM or PD were counted (together with the length of the membrane segment considered). Statistical analysis was performed with the software R using the nonparametrical Wilcoxon test.
For transmission electron microscopy observation of the PD fraction, the membranes were concentrated in 2% low-melting agarose. Agarose cubes of 5 mm were chemically fixed for 2 h in 0.1 M sodium-cacodylate buffer, pH 7.4, containing 2% glutaraldehyde before being postfixed in the same buffer containing 1% osmium tetroxide. During dehydration, water was progressively replaced by acetone. Spurr’s resin was used for stepwise embedding.
For immunogold labeling of the PD and PM fractions, 5 μL of purified membranes, at a protein concentration of 0.1 μg/μL, was pipetted onto plastic- and carbon-coated grids. Excess liquid was gently removed, and the grids were incubated with 10 μL of PBS blocking buffer (0.15 M NaCl, 7.5 mM Na2HPO4, and 0.25 mM NaH2PO4 containing 5% BSA, 5% normal goat serum, and 0.1% cold waterfish skin) for 1 h and then washed once with 1× PBS containing 0.05% Tween 20, before being incubated for 90 min with the primary antibodies diluted in PBS containing 0.1% acetylated BSA. Dilutions of 1:100, 1:20, and 1:1000 were used for PDCB1 (Simpson et al., 2009), PDLP1 (Thomas et al., 2008), and PMA2 (Morsomme et al., 1998) antisera and for the preimmune sera. After six washes, for 5 min each with PBS containing 0.05% Tween 20, antibody binding was detected using the following 10-nm gold-conjugated goat anti-rabbit antibodies diluted at 1:30. After 1 h of incubation at room temperature, the grids were washed six times with PBS containing 0.05% Tween 20 and three times with 0.2 µm filtered water and then negatively stained with 2% (w/v) uranyl acetate.
For negative staining of wall fragments, 5 μL of diluted wall suspension, prewashed with 0.2 µm filtered water, was mixed with an equal volume of 2% (w/v) uranyl acetate in a droplet on a pyroxylin-filmed electron microscope grid (400 mesh). After a few seconds, the grid was blotted to remove excess stain and then allowed to air dry.
Sections and negatively stained samples were viewed with the MET Philips CM10 100-kV device.
Construction of the PDCB1.mCherry GPI Mutant
PCR mutagenesis of PDCB1 (At5g61130) was used to delete the C-terminal GPI anchor signal. The sequence of the GPI anchor signal at the C-terminal end of the protein was determined using bioinformatic prediction programs (Big PI [http://mendel.imp.ac.at/gpi/gpi_server.html] and GPI-SOM [http://gpi.unibe.ch]) as well as experimental data (Elortza et al., 2003).
The PDCB1∆GPI.mCherry sequence was amplified by PCR using Q5 DNA polymerase (New England Biolabs) from a pDONR221 vector containing the N-terminal secretion signal of PDCB1 fused to the coding sequence of mCherry translationally fused to the rest of the coding sequence of PDCB1 (a gift from Yoselin Benitez-Alfonso). The primer sequences used for cloning are as follows: FW primer (3′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCCGAATCATGGCTGCTCTGGTGCTTTCAC-5′) and Rv primer (3′-GGGGACCACTTTGTACAAGAAAGCTGGGTATTATGTCGTGTAATCCGGGTTAATCCC-5′). The resulting DNA fragments were purified and transferred by recombination into the entry vector pDONR221 using BP Clonase II (Invitrogen Gateway Technology) following the manufacturer’s conditions. The sequence of the resulting pDONR clone was verified by automated sequencing. The coding sequence for PDCB1∆GPI.mCherry was then transferred to the Gateway binary destination vector pK2GW7 using LR Clonase II for expression in plants under the control of the 35S promoter. The constructs were transformed into Agrobacterium tumefaciens GV3101 to be used for transient expression in Nicotiana benthamiana.
Confocal Laser Scanning Microscopy
Live imaging was performed using a Leica SP8 confocal laser scanning microscopy system equipped with a white laser and hybrid detectors. For transient expression in N. benthamiana, leaves of a 3-week-old plant were pressure-infiltrated with the appropriate bacterial culture, and fluorescence was visualized by confocal microscopy at 3 d after inoculation.
Arabidopsis seedlings or agroinfiltrated N. benthamiana leaves were gently transferred between a glass slide and a cover slip in a drop of water. GFP, YFP, and mCitrine fluorescence were observed with similar settings (i.e., excitation wavelengths of 488 nm and emission wavelengths of 500 to 555 nm).
To quantify fluorescence intensities (ProSUC2:GFP, YFP-PDCB1, mCitrine-PdBG2, and callose immunofluorescence), experiments were performed using strictly identical confocal acquisition parameters (laser power, gain, zoom factor, and resolution) between samples treated with sterol inhibitors and controls.
For quantifying YFP-PDCB1 and mCitrine-PdBG2 fluorescence, intensity values at the apical-basal and lateral walls were measured by manually outlining the region of interest (ROI) of a 3-μm × 10-μm rectangle and calculating the mean gray value (sum of gray values of all the pixels in the selection divided by the ROI surface) for each ROI using Leica LAS AF Lite (http://leica-las-af-lite.software.informer.com). The ratio of the fluorescence intensities between apical-basal and lateral walls was then calculated. For each experiment, six cells from each of 10 seedlings were measured (n = 60). Statistical analysis was done using the R software version 2.15.1 (http://www.r-project.org). Intensity plot measurement along transverse walls was done using the image-processing software ImageJ version 1.47 (http://rsb.info.nih.gov/ij).
For quantification of ProSUC2:GFP, the intensity values were measured by manually outlining three ROIs in both the vasculature and the root meristem by calculating the mean gray value. For each experiment, 25 plants were processed per condition.
For quantification of callose immunofluorescence, the intensity values at the apical-basal walls of sieve elements were measured by manually outlining the ROI (i.e., apical-basal walls) and calculating the mean gray value as described above. For external cell layers, callose immunofluorescence intensity values were measured by manually outlining the ROI (20-μm × 20-μm square) and calculating the mean gray value as described above. For each experiment, a minimum of six measurements was performed for each of the 20 plants per condition.
Lipid Analysis
For HP-TLC (Merck 60 F254) analysis of polar and neutral lipids, lipids were extracted and purified from the different fractions according to Folch et al. (1957). Briefly, the lipids from 80 to 200 µg (protein equivalent) of PD and PM fractions were extracted with chloroform:methanol (2:1, v/v) for 20 min at room temperature. Polar contaminants such as proteins or nucleic acid were removed by adding 1 volume of 0.09% NaCl solution. After phase separation, the lower organic phase, which contains lipids, was harvested and the solvent was evaporated under a gentle flow of N2 gas. The lipids were then resuspended in 100 μL of chloroform:methanol (2:1, v/v) solution and applied onto a silica-coated chromatography plate with lipid standards. To resolve conjugated sterols and glucosylceramide, the Hillig solvent (chloroform:methanol, 85:15, v/v) was used (Hillig et al., 2003). For polar lipids, a Vitiello-Zanetta solvent mixture (Vitiello and Zanetta, 1978) was used. Lipids were then quantified by densitometry scanning after coloration with copper sulfate (Macala et al., 1983).
For HP-TLC analysis of GIPCs, lipids were extracted using a more polar solvent (methanol:water:chloroform, 60:8:30) and then separated by HP-TLC using Kaul and Lester solvent (Kaul and Lester, 1978). Silica plates were preactivated with methanol:ammonium acetate 0.2 M final concentration and dried for 10 min at 110°C before use. GIPCs were identified by running purified standards extracted from Arabidopsis (Buré et al., 2011). Lipids were then quantified by densitometry scanning after coloration by copper sulfate (Macala et al., 1983).
For the analysis of total fatty acids by GC-MS, ∼150 μg (protein equivalent) of PD and PM membranes was spun for 30 min at 110,000g at 4°C to eliminate traces of water. Transmethylation of fatty acids was performed overnight in 1 mL of methanol:H2SO4 solution (100:2.5, v/v) containing the internal standards C17:0 and h14:0 (5 μg/mL). After cooling, 1 mL of hexane:2.5% NaCl (1:1, v/v) was added, and the upper hexane phase containing FAMEs was recovered and buffered with 1 mL of Tris buffer (100 mM Tris‐HCl, pH 8, and 0.9% NaCl). The upper hexane phase was harvested, and the solvent was evaporated by a gentle flow of N2 gas. Prior to analysis by GC-MS, free hydroxyl groups were trimethylsilylated by N,O‐bis(trimethylsilyl) trifluoroacetamide (BSTFA)-trimethylchlorosilane for 15 min at 100°C. After complete evaporation of BSTFA under N2 gas, FAMEs were dissolved in 100 μL of hexane and analyzed by GC-MS. An HP-5MS capillary column (5% phenyl-methyl-siloxane, 30-m, 250-mm, and 0.25-μm film thickness; Agilent) was used with helium carrier gas at 2 mL/min; injection was done in splitless mode; injector and mass spectrometry detector temperatures were set to 250°C; the oven temperature was held at 50°C for 1 min, then programmed with a 25°C/min ramp to 150°C (2-min hold) and a 10°C/min ramp to 320°C (6-min hold). Quantification of nonhydroxylated and hydroxylated fatty acids was based upon peak areas that were derived from the total ion current.
For the analysis of phospholipids by LC-MS/MS, phospholipid extracts (chloroform:methanol extraction as described for HP-TLC analysis of polar lipids) were dissolved in 50 μL of eluent A (isopropanol:CH3OH:water + 0.2% formic acid + 0.028% NH3) containing synthetic internal lipid standards (PE, 17:0/17:0; PS, 17:0/17:0; PC, 17:0/17:0; PI, 17:0/14:1; and PG, 17:0/17:0 from Avanti Polar Lipids). LC-MS/MS (multiple reaction monitoring mode) analyses were performed with a model QTRAP 5500 (ABSciex) mass spectrometer coupled to a liquid chromatography system (Ultimate 3000; Dionex). Analyses were performed in the negative (PE, PS, PI, and PG) and positive (PC) modes with fast polarity switching (50 ms); nitrogen was used for the curtain gas (set to 15), gas 1 (set to 20), and gas 2 (set to 0). Needle voltage was at −4500 or +5500 V without needle heating; the declustering potential was adjusted between −180 and −85 V or set at +40 V. The collision gas was also nitrogen; collision energy varied from −48 to −62 eV and +47 eV on a compound-dependent basis. The dwell time was set to 3 ms. Reverse-phase separations were performed at 50°C on a Luna C8 150×1 mm column with 100-Å pore size and 5-μm particles (Phenomenex). The gradient elution program was as follows: 0 min, 30% eluent B (isopropanol + 0.2% formic acid + 0.028% NH3); 5 min, 50% eluent B; 30 min, 80% eluent B; 31 to 41 min, 95% eluent B; 42 to 52 min, 30% eluent B. The flow rate was set at 40 μL/min, and 3-μL sample volumes were injected. The areas of LC peaks were determined using MultiQuant software (version 2.1; ABSciex) for phospholipid quantification.
For determination of the ratios of sterols versus glycerolipids by HP-TLC, 300 µg of the PD and PM samples (n = 5) was split into two aliquots and analyzed in parallel by HP-TLC for neutral and polar lipids as described above. Quantification of the sterol lipid classes (free sterols, ASG, and SG) and the glycerolipid classes (digalactosyldiacylglycerol, PC, PS, phosphatidic acid, and PE:PG) was done by densitometry scanning after Macala staining (Macala et al., 1983).
For determination of the ratios of sterols versus glycerolipids by GC-MS, PD and PM samples (n = 6 for PD and n = 3 for PM; ∼300 μg) were split into two aliquots and analyzed in parallel for total FAMEs as described above and for sterols. For sterol analysis, the lipids were first extracted in chloroform:methanol (2:1, v/v) containing the internal standard α‐cholestanol (25 μg/mL). After 1 h at room temperature under gentle shaking, 1 mL of 0.9% NaCl was added and the organic phase was recovered. A saponification step was performed, after total evaporation of the solvent, by adding 1 mL of ethanol and 100 μL of 11 n KOH and incubating it for 1 h at 80°C. After the addition of 1 mL of hexane and 2 mL of water, the sterol-containing upper phase was recovered and the solvent was evaporated under an N2 gas stream. Sterols were derivatized by BSTFA as described for FAMEs and resuspended in 100 μL of hexane before analysis by GC-MS (see FAME analysis).
For quantification of sterol lipids by Q-TOF-MS/MS, sterol lipids were analyzed by direct nanospray infusion Q-TOF-MS/MS after collision-induced dissociation as described previously (Wewer et al., 2011). The measurements were performed on an Agilent 6530 Q-TOF-LC/MS instrument equipped with a nanospray infusion ion source (HPLC/chip MS 1200 with infusion chip; Agilent). Lipids were extracted from the PM and PD fractions (n = 3) with 2 volumes of chloroform:methanol:formic acid (1:1:0.1) and 1 volume of 1 M KCl/0.2 M H3PO4. Lipid extracts were purified by solid-phase extraction on silica columns (100 mg; Phenomenex). Free sterols were eluted with chloroform, and SG and ASG were eluted with acetone/2-propanol (1:1, v/v). Free sterols were derivatized with N-chlorobetainylchloride. Betainylated sterols, SG, and ASG were dried and dissolved in methanol:chloroform:300 mM ammonium acetate (665:300:35, v/v/v) for Q-TOF-MS/MS analysis (Welti et al., 2002).
Statistical analysis of the data was done using the R software package. When fewer than 20 or 5 samples were used, the nonparametrical Wilcoxon test or ANOVA of the Kruskal-Wallis test was employed, respectively.
Proteomic Analysis
Sample Preparation and Protein Digestion
Ten micrograms of each protein sample was solubilized in Laemmli buffer and deposited onto an SDS-PAGE gel for concentration and cleaning purposes. Separation was stopped after proteins entered the resolving gel. After colloidal blue staining, bands were excised from the SDS-PAGE gel and subsequently cut in 1-mm × 1-mm gel pieces. Gel pieces were destained in 25 mM ammonium bicarbonate and 50% acetonitrile (ACN), rinsed twice in ultrapure water, and shrunk in ACN for 10 min. After ACN removal, gel pieces were dried at room temperature, covered with trypsin solution (10 ng/μL in 40 mM NH4HCO3 and 10% ACN), rehydrated at 4°C for 10 min, and finally incubated overnight at 37°C. Gel pieces were then incubated for 15 min in 40 mM NH4HCO3 and 10% ACN at room temperature. The supernatant was collected, and a water:ACN:HCOOH (47.5:47.5:5) extraction solution was added to gel slices for 15 min. The extraction step was repeated twice. Supernatants were dried in a vacuum centrifuge and resuspended in 100 μL of 0.1% HCOOH.
Nano-LC-MS/MS Analysis
The peptide mixture was analyzed with the Ultimate 3000 nanoLC system (Dionex) coupled to the LTQ Orbitrap XL mass spectrometer (ThermoFinnigan). Ten microliters of peptide digest was loaded onto a 300-µm (i.d.) × 5-mm C18 PepMap trap column (LC Packings) at a flow rate of 20 µL/min. The peptides were eluted from the trap column onto an analytical 75-mm (i.d.) × 15-cm C18 PepMap column (LC Packings) with a 2 to 40% linear gradient of solvent B in 108 min (solvent A was 0.1% formic acid in 5% ACN, and solvent B was 0.1% formic acid in 80% ACN). The separation flow rate was set at 200 nL/min. The mass spectrometer operated in positive ion mode at a 1.8-kV needle voltage. Data were acquired using Xcalibur 2.0.7 software in data-dependent mode. Mass spectrometry scans (m/z 300 to 2000) were recorded in the Orbitrap cell at a resolution of 60,000 (at m/z 400) and an AGC target of 5 × 105 ions collected within 500 ms. Dynamic exclusion was set to 30 s, and the top six ions were selected from fragmentation in collision-induced dissociation mode. Tandem mass spectrometry scans with a target value of 104 ions were collected in the linear ion trap with a maximum fill time of 200 ms. Additionally, only +2 and +3 charged ions were selected for fragmentation. Others settings were as follows: spray voltage, 1.8 kV; no sheath or auxiliary gas flow; heated capillary temperature, 200°C; normalized collision energy of 0.25; and isolation width of 3 m/z.
Database Search and Results Processing
Data were searched by SEQUEST through Proteome Discoverer 1.4 (Thermo Fisher Scientific) against the 10th version of TAIR (32,785 entries). Spectra from peptides higher than 5000 D or lower than 350 D were rejected. The search parameters were as follows: the mass accuracy of the monoisotopic peptide precursor and peptide fragments was set to 10 ppm and 0.6 D, respectively. Only b and y ions were considered for mass calculation. Oxidation of methionines (+16 D) was considered a variable modification, and carbamidomethylation of cysteines (+57 D) was considered a fixed modification. Two missed trypsin cleavages were allowed. Peptide validation was performed using the Percolator algorithm (Käll et al., 2007), and only “high-confidence” peptides were retained, corresponding to a 1% false positive rate at the peptide level.
Label-Free Quantitative Data Analysis
Raw LC-MS/MS data were imported in Progenesis LC-MS 4.1 (Nonlinear Dynamics). Data processing includes the following steps: (1) features detection; (2) features alignment across the six samples; (3) volume integration for two to six charge-state ions; (4) import of sequence information; and (5) calculation of protein abundance (sum of the volume of corresponding peptides). Only nonconflicting features and unique peptides were considered for calculation at the protein level.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the accession numbers provided in Supplemental Tables 1 and 2.
Supplemental Data
Supplemental Figure 1. Immunogold labeling of PMA2 and callose in cryo-substituted Arabidopsis suspension cells.
Supplemental Figure 2. Quantitative proteomic analysis reveals that PD-associated proteins are highly enriched in the PD fraction compared with the bulk PM.
Supplemental Figure 3. Immunogold labeling of PMA2 on purified PD and PM fractions.
Supplemental Figure 4. Purity assessment of the PM fraction by immunoblot analyses.
Supplemental Figure 5. The cellulase solution used for PD isolation does not contain lipids or lipase activity.
Supplemental Figure 6. Quantification by HP-TLC of the different glycerolipid classes from isolated PD and PM fractions from Arabidopsis suspension cells.
Supplemental Figure 7. Quantification by LC-MS/MS of the phospholipid molecular species associated with the PD and PM fractions from Arabidopsis suspension cells.
Supplemental Figure 8. Molecular composition of phospholipid species identified by LC-MS/MS.
Supplemental Figure 9. The complex sphingolipids GIPCs constitute the major sphingolipid classes found in isolated PD and PM fractions from Arabidopsis suspension cells.
Supplemental Figure 10. The molecular species distribution of sterol lipid classes in isolated PD and PM fractions is similar. Sterol molecular species were determined by Q-TOF MS/MS analyses.
Supplemental Figure 11. Electron micrograph of cryofixed Arabidopsis roots treated with fenpropimorph for 24 h.
Supplemental Figure 12. Sterol composition of lova and fen treated Arabidopsis seedlings and cultured cells.
Supplemental Figure 13. proSUC2:GFP intensity measurement in the vasculature of Arabidopsis roots treated with fen or lova for 24 h and 48 h.
Supplemental Figure 14. List of the tetraspanins identified in the PD proteome and their putative S-acylation sites.
Supplemental Table 1. List of PD- and PM-associated proteins identified by proteomic analysis.
Supplemental Table 2. List of PD GPI-anchored proteins and their occurrence in DIM.
Supplementary Material
Acknowledgments
We thank Patrick Moreau for critical review of the article prior to submission. We also thank Sébastien Marais, Philippe Legros, Fabrice Cordelieres, and Christel Poujol from the Bordeaux Imaging Center (http://www.bic.ubordeaux2.fr/) for providing technical support on the confocal microscope and data quantification; Christine Faulkner (John Innes Centre) for providing the p35S:YFP:PDCB1 plasmid; Martine Gonneau for providing the antibodies against CesA3 and CesA6; and Véronique Santoni for providing the antibodies against PIP2;2. The lipidomic analyses were performed at the Functional Genomic Center of Bordeaux, Metabolome/Lipidome platform (http://www.biomemb.cnrs.fr/page_8ENG.html), funded by Grant MetaboHUB-ANR-11-INBS-0010. The mass spectrometry-based analysis of sterols was performed at the Institute of Molecular Physiology and Biotechnology of Plants, University of Bonn. Electron and fluorescence microscopy analyses were performed at the Bordeaux Imaging Centre. This work was supported by an SFR BIE grant 2010 and the Agence Nationale de la Recherche CONNECT (Grant ANR-14-CE19-0006-01 to E.M.B.).
AUTHOR CONTRIBUTIONS
E.M.B., M.S.G., and S.M. designed the research. E.M.B., H.N., L.B., L.F., M.S.G., S.C., V.G., V.W., Y.B.-A., and W.N. performed research. E.M.B., L.B., L.F., M.S.G., P.D., S.M., W.N., Y.B.-A., and Y.B. analyzed data. E.M.B., M.S.G., S.M., and Y.B. wrote the article.
Glossary
- ER
endoplasmic reticulum
- PM
plasma membrane
- PD
plasmodesmata
- GPI
glycosylphosphatidylinositol
- PD-PM
specialized domain of the plasma membrane lining plasmodesmata
- HP-TLC
high-performance thin layer chromatography
- PE
phosphatidylethanolamine
- PC
phosphatidylcholine
- PS
phosphatidylserine
- PI
phosphatidylinositol
- PG
phosphatidylglycerol
- LC-MS/MS
liquid chromatography coupled to tandem mass spectrometry
- VLCFA
very long chain fatty acid
- GIPC
glycosylinositolphosphoceramide
- GC-MS
gas chromatography-mass spectrometry
- FAME
fatty acid methyl ester
- SG
sterol glucosides
- ASG
acylated sterol glucosides
- Q-TOF-MS/MS
quadrupole time-of-flight mass spectrometry
- DIM
detergent-insoluble membrane
- DSM
detergent-soluble membrane
- fen
fenpropimorph
- lova
lovastatin
- MS
Murashige and Skoog
- TEM
tetraspanin-enriched microdomain
- TBS
Tris-buffered saline
- ROI
region of interest
- BSTFA
N,O‐bis(trimethylsilyl) trifluoroacetamide
- ACN
acetonitrile
References
- Andersson M.X., Stridh M.H., Larsson K.E., Liljenberg C., Sandelius A.S. (2003). Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett. 537: 128–132. [DOI] [PubMed] [Google Scholar]
- Baluška F., Cvrcková F., Kendrick-Jones J., Volkmann D. (2001). Sink plasmodesmata as gateways for phloem unloading. Myosin VIII and calreticulin as molecular determinants of sink strength? Plant Physiol. 126: 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluška F., Samaj J., Napier R., Volkmann D. (1999). Maize calreticulin localizes preferentially to plasmodesmata in root apex. Plant J. 19: 481–488. [DOI] [PubMed] [Google Scholar]
- Barrera N.P., Zhou M., Robinson C.V. (2013). The role of lipids in defining membrane protein interactions: Insights from mass spectrometry. Trends Cell Biol. 23: 1–8. [DOI] [PubMed] [Google Scholar]
- Bayer E., Thomas C.L., Maule A.J. (2004). Plasmodesmata in Arabidopsis thaliana suspension cells. Protoplasma 223: 93–102. [DOI] [PubMed] [Google Scholar]
- Bayer E.M., Bottrill A.R., Walshaw J., Vigouroux M., Naldrett M.J., Thomas C.L., Maule A.J. (2006). Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics 6: 301–311. [DOI] [PubMed] [Google Scholar]
- Benitez-Alfonso Y., Faulkner C., Pendle A., Miyashima S., Helariutta Y., Maule A. (2013). Symplastic intercellular connectivity regulates lateral root patterning. Dev. Cell 26: 136–147. [DOI] [PubMed] [Google Scholar]
- Benitez-Alfonso Y., Jackson D., Maule A. (2011). Redox regulation of intercellular transport. Protoplasma 248: 131–140. [DOI] [PubMed] [Google Scholar]
- Bigay J., Antonny B. (2012). Curvature, lipid packing, and electrostatics of membrane organelles: Defining cellular territories in determining specificity. Dev. Cell 23: 886–895. [DOI] [PubMed] [Google Scholar]
- Borner G.H., Sherrier D.J., Weimar T., Michaelson L.V., Hawkins N.D., Macaskill A., Napier J.A., Beale M.H., Lilley K.S., Dupree P. (2005). Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137: 104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutté Y., Grebe M. (2014). Immunocytochemical fluorescent in situ visualization of proteins in Arabidopsis. Methods Mol. Biol. 1062: 453–472. [DOI] [PubMed] [Google Scholar]
- Boutté Y., Frescatada-Rosa M., Men S., Chow C.M., Ebine K., Gustavsson A., Johansson L., Ueda T., Moore I., Jürgens G., Grebe M. (2010). Endocytosis restricts Arabidopsis KNOLLE syntaxin to the cell division plane during late cytokinesis. EMBO J. 29: 546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith T.M., Zambryski P.C. (2012). Plasmodesmata paradigm shift: regulation from without versus within. Annu. Rev. Plant Biol. 63: 239–260. [DOI] [PubMed] [Google Scholar]
- Buré C., Cacas J.L., Wang F., Gaudin K., Domergue F., Mongrand S., Schmitter J.M. (2011). Fast screening of highly glycosylated plant sphingolipids by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 25: 3131–3145. [DOI] [PubMed] [Google Scholar]
- Charrin S., Manié S., Oualid M., Billard M., Boucheix C., Rubinstein E. (2002). Differential stability of tetraspanin/tetraspanin interactions: Role of palmitoylation. FEBS Lett. 516: 139–144. [DOI] [PubMed] [Google Scholar]
- Charrin S., Manié S., Thiele C., Billard M., Gerlier D., Boucheix C., Rubinstein E. (2003). A physical and functional link between cholesterol and tetraspanins. Eur. J. Immunol. 33: 2479–2489. [DOI] [PubMed] [Google Scholar]
- Chen M.H., Tian G.W., Gafni Y., Citovsky V. (2005). Effects of calreticulin on viral cell-to-cell movement. Plant Physiol. 138: 1866–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Timmermans M.C. (2010). Small RNAs are on the move. Nature 467: 415–419. [DOI] [PubMed] [Google Scholar]
- Contreras F.X., et al. (2012). Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature 481: 525–529. [DOI] [PubMed] [Google Scholar]
- Deeks M.J., Calcutt J.R., Ingle E.K., Hawkins T.J., Chapman S., Richardson A.C., Mentlak D.A., Dixon M.R., Cartwright F., Smertenko A.P., Oparka K., Hussey P.J. (2012). A superfamily of actin-binding proteins at the actin-membrane nexus of higher plants. Curr. Biol. 22: 1595–1600. [DOI] [PubMed] [Google Scholar]
- Delage E., Zurzolo C. (2013). Exploring the role of lipids in intercellular conduits: Breakthroughs in the pipeline. Front. Plant Sci. 4: 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T., Juraniec M., Crowell E.F., Jouy H., Pochylova Z., Parcy F., Höfte H., Gonneau M., Vernhettes S. (2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 15572–15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N., Geelen D. (2014). Callose homeostasis at plasmodesmata: molecular regulators and developmental relevance. Front. Plant Sci. 5: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Turgeon R., Parthasarathy M.V. (1992). Substructure of freeze substituted plasmodesmata. Protoplasma 169: 28–41. [Google Scholar]
- Dunoyer P., Schott G., Himber C., Meyer D., Takeda A., Carrington J.C., Voinnet O. (2010). Small RNA duplexes function as mobile silencing signals between plant cells. Science 328: 912–916. [DOI] [PubMed] [Google Scholar]
- Elortza F., Nühse T.S., Foster L.J., Stensballe A., Peck S.C., Jensen O.N. (2003). Proteomic analysis of glycosylphosphatidylinositol-anchored membrane proteins. Mol. Cell. Proteomics 2: 1261–1270. [DOI] [PubMed] [Google Scholar]
- Faulkner C. (2013). Receptor-mediated signaling at plasmodesmata. Front. Plant Sci. 4: 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C., Maule A. (2011). Opportunities and successes in the search for plasmodesmal proteins. Protoplasma 248: 27–38. [DOI] [PubMed] [Google Scholar]
- Faulkner C., Petutschnig E., Benitez-Alfonso Y., Beck M., Robatzek S., Lipka V., Maule A.J. (2013). LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc. Natl. Acad. Sci. USA 110: 9166–9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C.R., Blackman L.M., Collings D.A., Cordwell S.J., Overall R.L. (2009). Anti-tropomyosin antibodies co-localise with actin microfilaments and label plasmodesmata. Eur. J. Cell Biol. 88: 357–369. [DOI] [PubMed] [Google Scholar]
- Faulkner C.R., Blackman L.M., Cordwell S.J., Overall R.L. (2005). Proteomic identification of putative plasmodesmatal proteins from Chara corallina. Proteomics 5: 2866–2875. [DOI] [PubMed] [Google Scholar]
- Fernandez-Calvino L., Faulkner C., Walshaw J., Saalbach G., Bayer E., Benitez-Alfonso Y., Maule A. (2011). Arabidopsis plasmodesmal proteome. PLoS ONE 6: e18880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleurat-Lessard P., Bouche-Pillon S., Leloup C., Lucas W.J., Serrano R., Bonnemain J.L. (1995). Absence of plasma membrane H+-ATPase in plasmodesmata located in pit-fields of the young reactive pulvinus of Mimosa pudica L. Protoplasma 188: 180–185. [Google Scholar]
- Folch J., Lees M., Sloane Stanley G.H. (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- Furuta K., Lichtenberger R., Helariutta Y. (2012). The role of mobile small RNA species during root growth and development. Curr. Opin. Cell Biol. 24: 211–216. [DOI] [PubMed] [Google Scholar]
- Galian C., Björkholm P., Bulleid N., von Heijne G. (2012). Efficient glycosylphosphatidylinositol (GPI) modification of membrane proteins requires a C-terminal anchoring signal of marginal hydrophobicity. J. Biol. Chem. 287: 16399–16409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendre D., Oh J., Boutté Y., Best J.G., Samuels L., Nilsson R., Uemura T., Marchant A., Bennett M.J., Grebe M., Bhalerao R.P. (2011). Conserved Arabidopsis ECHIDNA protein mediates trans-Golgi-network trafficking and cell elongation. Proc. Natl. Acad. Sci. USA 108: 8048–8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami D., Gowrishankar K., Bilgrami S., Ghosh S., Raghupathy R., Chadda R., Vishwakarma R., Rao M., Mayor S. (2008). Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell 135: 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabski S., De Feijter A.W., Schindler M. (1993). Endoplasmic reticulum forms a dynamic continuum for lipid diffusion between contiguous soybean root cells. Plant Cell 5: 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grison M.S., Fernandez-Calvino L., Mongrand S., Bayer E.M. (2015). Isolation of plasmodesmata from Arabidopsis suspension culture cells. Methods Mol. Biol. 1217: 83–93. [DOI] [PubMed] [Google Scholar]
- Gui J., Liu C., Shen J., Li L. (2014). Grain setting defect1, encoding a remorin protein, affects the grain setting in rice through regulating plasmodesmatal conductance. Plant Physiol. 166: 1463–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseman J.M., Lee J.S., Bogenschutz N.L., Peterson K.M., Virata R.E., Xie B., Kanaoka M.M., Hong Z., Torii K.U. (2010). Dysregulation of cell-to-cell connectivity and stomatal patterning by loss-of-function mutation in Arabidopsis chorus (glucan synthase-like 8). Development 137: 1731–1741. [DOI] [PubMed] [Google Scholar]
- Han X., Hyun T.K., Zhang M., Kumar R., Koh E.J., Kang B.H., Lucas W.J., Kim J.Y. (2014). Auxin-callose-mediated plasmodesmal gating is essential for tropic auxin gradient formation and signaling. Dev. Cell 28: 132–146. [DOI] [PubMed] [Google Scholar]
- Hartmann M.A., Benveniste P. (1987). Plant membrane sterols: Isolation, identification and biosynthesis. Methods Enzymol. 148: 632–650. [Google Scholar]
- Hartmann M.A., Perret A.M., Carde J.P., Cassagne C., Moreau P. (2002). Inhibition of the sterol pathway in leek seedlings impairs phosphatidylserine and glucosylceramide synthesis but triggers an accumulation of triacylglycerols. Biochim. Biophys. Acta 1583: 285–296. [DOI] [PubMed] [Google Scholar]
- He J.X., Fujioka S., Li T.C., Kang S.G., Seto H., Takatsuto S., Yoshida S., Jang J.C. (2003). Sterols regulate development and gene expression in Arabidopsis. Plant Physiol. 131: 1258–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerklotz H. (2002). Triton promotes domain formation in lipid raft mixtures. Biophys. J. 83: 2693–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillig I., Leipelt M., Ott C., Zähringer U., Warnecke D., Heinz E. (2003). Formation of glucosylceramide and sterol glucoside by a UDP-glucose-dependent glucosylceramide synthase from cotton expressed in Pichia pastoris. FEBS Lett. 553: 365–369. [DOI] [PubMed] [Google Scholar]
- Hofmann J., Youssef-Banora M., de Almeida-Engler J., Grundler F.M. (2010). The role of callose deposition along plasmodesmata in nematode feeding sites. Mol. Plant Microbe Interact. 23: 549–557. [DOI] [PubMed] [Google Scholar]
- Höfte H., Chrispeels M.J. (1992). Protein sorting to the vacuolar membrane. Plant Cell 4: 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias V.A., Meins F. Jr. (2000). Movement of plant viruses is delayed in a beta-1,3-glucanase-deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J. 21: 157–166. [DOI] [PubMed] [Google Scholar]
- Imlau A., Truernit E., Sauer N. (1999). Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11: 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarsch I.K., Konrad S.S., Stratil T.F., Urbanus S.L., Szymanski W., Braun P., Braun K.H., Ott T. (2014). Plasma membranes are subcompartmentalized into a plethora of coexisting and diverse microdomains in Arabidopsis and Nicotiana benthamiana. Plant Cell 26: 1698–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo Y., Cho W.K., Rim Y., Moon J., Chen X.Y., Chu H., Kim C.Y., Park Z.Y., Lucas W.J., Kim J.Y. (2011). Plasmodesmal receptor-like kinases identified through analysis of rice cell wall extracted proteins. Protoplasma 248: 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L., Canterbury J.D., Weston J., Noble W.S., MacCoss M.J. (2007). Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4: 923–925. [DOI] [PubMed] [Google Scholar]
- Kang B.H., Nielsen E., Preuss M.L., Mastronarde D., Staehelin L.A. (2011). Electron tomography of RabA4b- and PI-4Kβ1-labeled trans Golgi network compartments in Arabidopsis. Traffic 12: 313–329. [DOI] [PubMed] [Google Scholar]
- Kaul K., Lester R.L. (1978). Isolation of six novel phosphoinositol-containing sphingolipids from tobacco leaves. Biochemistry 17: 3569–3575. [DOI] [PubMed] [Google Scholar]
- Keinath N.F., Kierszniowska S., Lorek J., Bourdais G., Kessler S.A., Shimosato-Asano H., Grossniklaus U., Schulze W.X., Robatzek S., Panstruga R. (2010). PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 285: 39140–39149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszniowska S., Seiwert B., Schulze W.X. (2009). Definition of Arabidopsis sterol-rich membrane microdomains by differential treatment with methyl-beta-cyclodextrin and quantitative proteomics. Mol. Cell. Proteomics 8: 612–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi K., Hayashi T., Wu S., Gallagher K.L. (2012). The SHORT-ROOT protein acts as a mobile, dose-dependent signal in patterning the ground tissue. Proc. Natl. Acad. Sci. USA 109: 13010–13015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Wang X., Cui W., Sager R., Modla S., Czymmek K., Zybaliov B., van Wijk K., Zhang C., Lu H., Lakshmanan V. (2011). A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 23: 3353–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B., Furt F., Hartmann M.A., Michaelson L.V., Carde J.P., Sargueil-Boiron F., Rossignol M., Napier J.A., Cullimore J., Bessoule J.J., Mongrand S. (2007). Characterization of lipid rafts from Medicago truncatula root plasma membranes: a proteomic study reveals the presence of a raft-associated redox system. Plant Physiol. 144: 402–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental I., Grzybek M., Simons K. (2010). Greasing their way: Lipid modifications determine protein association with membrane rafts. Biochemistry 49: 6305–6316. [DOI] [PubMed] [Google Scholar]
- Levy A., Erlanger M., Rosenthal M., Epel B.L. (2007). A plasmodesmata-associated beta-1,3-glucanase in Arabidopsis. Plant J. 49: 669–682. [DOI] [PubMed] [Google Scholar]
- Li L., Shi X., Guo X., Li H., Xu C. (2014). Ionic protein-lipid interaction at the plasma membrane: What can the charge do? Trends Biochem. Sci. 39: 130–140. [DOI] [PubMed] [Google Scholar]
- Lingwood D., Simons K. (2010). Lipid rafts as a membrane-organizing principle. Science 327: 46–50. [DOI] [PubMed] [Google Scholar]
- Macala L.J., Yu R.K., Ando S. (1983). Analysis of brain lipids by high performance thin-layer chromatography and densitometry. J. Lipid Res. 24: 1243–1250. [PubMed] [Google Scholar]
- Malinsky J., Opekarová M., Grossmann G., Tanner W. (2013). Membrane microdomains, rafts, and detergent-resistant membranes in plants and fungi. Annu. Rev. Plant Biol. 64: 501–529. [DOI] [PubMed] [Google Scholar]
- Marmagne A., Salvi D., Rolland N., Ephritikhine G., Joyard J., Barbier-Brygoo H. (2006). Purification and fractionation of membranes for proteomic analyses. Methods Mol. Biol. 323: 403–420. [DOI] [PubMed] [Google Scholar]
- Maule A., Faulkner C., Benitez-Alfonso Y. (2012). Plasmodesmata “in communicado.” Front. Plant Sci. 3: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule A.J. (2008). Plasmodesmata: Structure, function and biogenesis. Curr. Opin. Plant Biol. 11: 680–686. [DOI] [PubMed] [Google Scholar]
- Maule A.J., Benitez-Alfonso Y., Faulkner C. (2011). Plasmodesmata—Membrane tunnels with attitude. Curr. Opin. Plant Biol. 14: 683–690. [DOI] [PubMed] [Google Scholar]
- Maule A.J., Gaudioso-Pedraza R., Benitez-Alfonso Y. (2013). Callose deposition and symplastic connectivity are regulated prior to lateral root emergence. Commun. Integr. Biol. 6: e26531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S., Riezman H. (2004). Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 5: 110–120. [DOI] [PubMed] [Google Scholar]
- Mongrand S., Morel J., Laroche J., Claverol S., Carde J.P., Hartmann M.A., Bonneu M., Simon-Plas F., Lessire R., Bessoule J.J. (2004). Lipid rafts in higher plant cells: Purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 279: 36277–36286. [DOI] [PubMed] [Google Scholar]
- Mongrand S., Stanislas T., Bayer E.M., Lherminier J., Simon-Plas F. (2010). Membrane rafts in plant cells. Trends Plant Sci. 15: 656–663. [DOI] [PubMed] [Google Scholar]
- Morel J., Claverol S., Mongrand S., Furt F., Fromentin J., Bessoule J.J., Blein J.-P., Simon-Plas F. (2006). Proteomics of plant detergent-resistant membranes. Mol. Cell. Proteomics 5: 1396–1411. [DOI] [PubMed] [Google Scholar]
- Morsomme P., Dambly S., Maudoux O., Boutry M. (1998). Single point mutations distributed in 10 soluble and membrane regions of the Nicotiana plumbaginifolia plasma membrane PMA2 H+-ATPase activate the enzyme and modify the structure of the C-terminal region. J. Biol. Chem. 273: 34837–34842. [DOI] [PubMed] [Google Scholar]
- Munnik T., Nielsen E. (2011). Green light for polyphosphoinositide signals in plants. Curr. Opin. Plant Biol. 14: 489–497. [DOI] [PubMed] [Google Scholar]
- Pata M.O., Hannun Y.A., Ng C.K. (2010). Plant sphingolipids: Decoding the enigma of the sphinx. New Phytol. 185: 611–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike L.J. (2006). Rafts defined: A report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 47: 1597–1598. [DOI] [PubMed] [Google Scholar]
- Raffaele S., et al. (2009). Remorin, a Solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21: 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt S., Knight A.E., Hodge T.P., Baluska F., Samaj J., Volkmann D., Kendrick-Jones J. (1999). Characterization of the unconventional myosin VIII in plant cells and its localization at the post-cytokinetic cell wall. Plant J. 19: 555–567. [DOI] [PubMed] [Google Scholar]
- Rinne P.L., Welling A., Vahala J., Ripel L., Ruonala R., Kangasjärvi J., van der Schoot C. (2011). Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-β-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell 23: 130–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y.L., Xu S.M., White R., Furbank R.T. (2004). Genotypic and developmental evidence for the role of plasmodesmatal regulation in cotton fiber elongation mediated by callose turnover. Plant Physiol. 136: 4104–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein E. (2011). The complexity of tetraspanins. Biochem. Soc. Trans. 39: 501–505. [DOI] [PubMed] [Google Scholar]
- Sagi G., Katz A., Guenoune-Gelbart D., Epel B.L. (2005). Class 1 reversibly glycosylated polypeptides are plasmodesmal-associated proteins delivered to plasmodesmata via the Golgi apparatus. Plant Cell 17: 1788–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon M.S., Bayer E.M. (2012). Dissecting plasmodesmata molecular composition by mass spectrometry-based proteomics. Front. Plant Sci. 3: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangiorgio V., Pitto M., Palestini P., Masserini M. (2004). GPI-anchored proteins and lipid rafts. Ital. J. Biochem. 53: 98–111. [PubMed] [Google Scholar]
- Santoni V., Vinh J., Pflieger D., Sommerer N., Maurel C. (2003). A proteomic study reveals novel insights into the diversity of aquaporin forms expressed in the plasma membrane of plant roots. Biochem. J. 373: 289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer L.V., de Jong D.H., Holt A., Rzepiela A.J., de Vries A.H., Poolman B., Killian J.A., Marrink S.J. (2011). Lipid packing drives the segregation of transmembrane helices into disordered lipid domains in model membranes. Proc. Natl. Acad. Sci. USA 108: 1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S., Honsho M., Ekroos K., Shevchenko A., Simons K. (2003). Resistance of cell membranes to different detergents. Proc. Natl. Acad. Sci. USA 100: 5795–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Varma R., Sarasij R.C., Ira, Gousset K., Krishnamoorthy G., Rao M., Mayor S. (2004). Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell 116: 577–589. [DOI] [PubMed] [Google Scholar]
- Silvie O., Charrin S., Billard M., Franetich J.F., Clark K.L., van Gemert G.J., Sauerwein R.W., Dautry F., Boucheix C., Mazier D., Rubinstein E. (2006). Cholesterol contributes to the organization of tetraspanin-enriched microdomains and to CD81-dependent infection by malaria sporozoites. J. Cell Sci. 119: 1992–2002. [DOI] [PubMed] [Google Scholar]
- Simons K., Sampaio J.L. (2011). Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 3: a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K., Vaz W.L. (2004). Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct. 33: 269–295. [DOI] [PubMed] [Google Scholar]
- Simpson C., Thomas C., Findlay K., Bayer E., Maule A.J. (2009). An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21: 581–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnino S., Prinetti A. (2010). Lipids and membrane lateral organization. Front Physiol 1: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spira F., Mueller N.S., Beck G., von Olshausen P., Beig J., Wedlich-Söldner R. (2012). Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat. Cell Biol. 14: 640–648. [DOI] [PubMed] [Google Scholar]
- Srivastava V., Malm E., Sundqvist G., Bulone V. (2013). Quantitative proteomics reveals that plasma membrane microdomains from poplar cell suspension cultures are enriched in markers of signal transduction, molecular transport, and callose biosynthesis. Mol. Cell. Proteomics 12: 3874–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y., et al. (2013). Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr. Biol. 23: 362–371. [DOI] [PubMed] [Google Scholar]
- Stipp C.S., Kolesnikova T.V., Hemler M.E. (2003). Functional domains in tetraspanin proteins. Trends Biochem. Sci. 28: 106–112. [DOI] [PubMed] [Google Scholar]
- Tanner W., Malinsky J., Opekarová M. (2011). In plant and animal cells, detergent-resistant membranes do not define functional membrane rafts. Plant Cell 23: 1191–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C.L., Bayer E.M., Ritzenthaler C., Fernandez-Calvino L., Maule A.J. (2008). Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 6: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney L.G., Cooke T.J., Connelly P.S., Tilney M.S. (1991). The structure of plasmodesmata as revealed by plasmolysis, detergent extraction, and protease digestion. J. Cell Biol. 112: 739–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J., Amari K., Torrance L. (2011). Plasmodesmata viewed as specialised membrane adhesion sites. Protoplasma 248: 39–60. [DOI] [PubMed] [Google Scholar]
- Tilsner J., Linnik O., Louveaux M., Roberts I.M., Chapman S.N., Oparka K.J. (2013). Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 201: 981–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki S., Citovsky V. (2014). Plasmodesmata-associated proteins: Can we see the whole elephant? Plant Signal. Behav. 9: e27899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaddepalli P., Herrmann A., Fulton L., Oelschner M., Hillmer S., Stratil T.F., Fastner A., Hammes U.Z., Ott T., Robinson D.G., Schneitz K. (2014). The C2-domain protein QUIRKY and the receptor-like kinase STRUBBELIG localize to plasmodesmata and mediate tissue morphogenesis in Arabidopsis thaliana. Development 141: 4139–4148. [DOI] [PubMed] [Google Scholar]
- Vallon O., Wollman F.A., Olive J. (1986). Lateral distribution of the main protein complexes of the photosynthetic apparatus in Chlamydomonas reinhardttii and in spinach: An immunocytochemical study using intact thylakoid membranes and PS II enriched membrane preparation. Photobiochem. Photobiophys. 12: 203–220. [Google Scholar]
- van den Bogaart G., Meyenberg K., Risselada H.J., Amin H., Willig K.I., Hubrich B.E., Dier M., Hell S.W., Grubmüller H., Diederichsen U., Jahn R. (2011). Membrane protein sequestering by ionic protein-lipid interactions. Nature 479: 552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten T.S., Cambi A., Koopman M., Joosten B., Figdor C.G., Garcia-Parajo M.F. (2009). Hotspots of GPI-anchored proteins and integrin nanoclusters function as nucleation sites for cell adhesion. Proc. Natl. Acad. Sci. USA 106: 18557–18562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma R., Mayor S. (1998). GPI-anchored proteins are organized in submicron domains at the cell surface. Nature 394: 798–801. [DOI] [PubMed] [Google Scholar]
- Vatén A., et al. (2011). Callose biosynthesis regulates symplastic trafficking during root development. Dev. Cell 21: 1144–1155. [DOI] [PubMed] [Google Scholar]
- Vitiello F., Zanetta J.P. (1978). Thin-layer chromatography of phospholipids. J. Chromatogr. A 166: 637–640. [DOI] [PubMed] [Google Scholar]
- Vögeli U., Chappell J. (1991). Inhibition of a plant sesquiterpene cyclase by mevinolin. Arch. Biochem. Biophys. 288: 157–162. [DOI] [PubMed] [Google Scholar]
- Wang X., Sager R., Cui W., Zhang C., Lu H., Lee J.Y. (2013). Salicylic acid regulates plasmodesmata closure during innate immune responses in Arabidopsis. Plant Cell 25: 2315–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welti R., Li W., Li M., Sang Y., Biesiada H., Zhou H.E., Rajashekar C.B., Williams T.D., Wang X. (2002). Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 277: 31994–32002. [DOI] [PubMed] [Google Scholar]
- Wewer V., Dombrink I., vom Dorp K., Dörmann P. (2011). Quantification of sterol lipids in plants by quadrupole time-of-flight mass spectrometry. J. Lipid Res. 52: 1039–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Gallagher K.L. (2012). Transcription factors on the move. Curr. Opin. Plant Biol. 15: 645–651. [DOI] [PubMed] [Google Scholar]
- Xu M., Cho E., Burch-Smith T.M., Zambryski P.C. (2012). Plasmodesmata formation and cell-to-cell transport are reduced in decreased size exclusion limit 1 during embryogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 5098–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.M., Wang J., Xuan Z., Goldshmidt A., Borrill P.G., Hariharan N., Kim J.Y., Jackson D. (2011). Chaperonins facilitate KNOTTED1 cell-to-cell trafficking and stem cell function. Science 333: 1141–1144. [DOI] [PubMed] [Google Scholar]
- Yunta M., Lazo P.A. (2003). Tetraspanin proteins as organisers of membrane microdomains and signalling complexes. Cell. Signal. 15: 559–564. [DOI] [PubMed] [Google Scholar]
- Zavaliev R., Ueki S., Epel B.L., Citovsky V. (2011). Biology of callose (β-1,3-glucan) turnover at plasmodesmata. Protoplasma 248: 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T., Lucas W.J., Rost T.L. (1998). Directional cell to cell communication in the Arabidopsis root apical meristem. I. An ultrastructural and functional analysis. Protoplasma 180: 169–184. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.