The draft genome and transcriptome sequences of the wild potato species S. commersonii demonstrate the usefulness of genome sequences from wild relatives for elucidating evolutionary mechanisms contributing to Solanum species diversity and understanding changes in response to cold.
Abstract
Here, we report the draft genome sequence of Solanum commersonii, which consists of ∼830 megabases with an N50 of 44,303 bp anchored to 12 chromosomes, using the potato (Solanum tuberosum) genome sequence as a reference. Compared with potato, S. commersonii shows a striking reduction in heterozygosity (1.5% versus 53 to 59%), and differences in genome sizes were mainly due to variations in intergenic sequence length. Gene annotation by ab initio prediction supported by RNA-seq data produced a catalog of 1703 predicted microRNAs, 18,882 long noncoding RNAs of which 20% are shown to target cold-responsive genes, and 39,290 protein-coding genes with a significant repertoire of nonredundant nucleotide binding site-encoding genes and 126 cold-related genes that are lacking in S. tuberosum. Phylogenetic analyses indicate that domesticated potato and S. commersonii lineages diverged ∼2.3 million years ago. Three duplication periods corresponding to genome enrichment for particular gene families related to response to salt stress, water transport, growth, and defense response were discovered. The draft genome sequence of S. commersonii substantially increases our understanding of the domesticated germplasm, facilitating translation of acquired knowledge into advances in crop stability in light of global climate and environmental changes.
INTRODUCTION
The genus Solanum ranks among the largest of plant genera and includes several cultivated crops of regional or worldwide significance including potato (Solanum tuberosum) and tomato (Solanum lycopersicum). Potato is the most important non-cereal food crop worldwide, offering higher yields in calories per acre than any grain (Bradeen and Haynes, 2011). Unfortunately, potato is also host to a wide variety of pathogens of large economic impact, including fungi, oomycetes, bacteria, viruses, and nematodes (Stevenson et al., 2001). In addition, abiotic stress factors such as cold, heat, drought, and salinity have a significant effect on cultivated potato, affecting yield, tuber quality, and market value (Wang-Pruski and Schofield, 2012). To improve resistance to these adverse environmental factors, potato breeders can exploit the ∼200 tuber-bearing Solanum species native to South, Central, and North America. Although these genetic resources provide great potential to breed improved cultivars (Huamán et al., 2000), linkage drag typically limits the use of wild potato species since many exotic genes imparting undesirable traits (e.g., high alkaloid content, long stolons, etc.) can be cotransferred with desirable genes (Bradshaw and Ramsay, 2005).
Solanum commersonii is a tuber-bearing wild potato species native to Central and South America. The French taxonomist Michel-Felix Dunal named this species in honor of Philibert Commerson (1727–1773), who collected the type specimen (No. 47) in 1767 at Montevideo, Uruguay. This was probably the first wild potato to be collected on a scientific expedition (Hawkes, 1990). Analyses of chloroplast genome restriction sites and nitrate reductase gene sequence confirmed that S. commersonii is phylogenetically distinct from cultivated potato (Rodríguez and Spooner, 2009). Consistent with these analyses, S. commersonii and S. tuberosum are sexually incompatible and have been assigned different endosperm balance numbers (EBNs) (Johnston et al., 1980), with S. commersonii reported as 1 EBN and S. tuberosum reported as 4 EBN. Despite being genetically isolated from cultivated potato, S. commersonii has garnered significant research interest. It possesses several resistance traits not found in cultivated potato, including resistance to root knot nematode, soft rot and blackleg, bacterial and verticillium wilt, Potato virus X, tobacco etch virus, common scab, and late blight (Hanneman and Bamberg, 1986; Hawkes, 1990; Micheletto et al., 2000). Particularly attractive is its freezing tolerance and capacity to cold acclimate (i.e., ability to increase cold tolerance after exposure to low, nonfreezing temperatures). By contrast, the cultivated potato is classified as sensitive to low temperatures and is unable to cold acclimate (Palta and Simon, 1993). Both frost tolerance and cold acclimation capacity are important breeding traits, since temperatures below 0°C are a major cause of yield losses in several production regions. Using various strategies, breeders have successfully overcome sexual isolation barriers to introgress S. commersonii genetic material into cultivated potato (Cardi et al., 1993; Bamberg et al., 1994; Carputo et al., 1997). Despite such efforts, very little progress has been made in the release of new varieties originating from crosses involving S. commersonii. This is at least partially due to the lack of genomics resources for S. commersonii and other potato wild relatives. The genome sequence of the cultivated potato was published in 2011 (Potato Genome Sequencing Consortium, 2011), ushering in a new era of potato functional and comparative genomics. By contrast, no genome sequence of a wild potato species has been published to date. Comparative sequencing of cultivated potato and wild relatives will increase the use of wild species for crop breeding and improvement.
Here, we describe draft genome and transcriptome sequences of the wild potato species S. commersonii. We used Illumina technology to sequence and de novo assemble the genome of the cold-tolerant accession PI 243503, obtaining roughly 105x coverage. The genome sequence consists of ∼830 Mb, of which 44.5% comprises transposable elements. Gene annotation by ab initio prediction supported by RNA-seq data led to the identification of 39,290 protein-coding genes with a significant repertoire of nonredundant nucleotide binding site-encoding genes and 126 cold-related genes, which lack orthologs in S. tuberosum. Phylogenetic analyses offered new insights into recent duplications and divergence between S. commersonii and the domesticated potato. Overall, the draft genome sequence of S. commersonii provides an essential reference for studying Solanum diversity via resequencing of additional wild species genomes.
RESULTS
Genome Sequencing and Assembling
To obtain a whole-genome shotgun sequence assembly of S. commersonii clone cmm1t, we produced size-selected sequencing paired-end and mate pair libraries based on six insert sizes ranging from 400 bp to 10 kb (Supplemental Table 1). A total of 145.93 Gb of sequence reads was produced. After filtering low-quality sequences, the remaining 88 Gb were assembled into 278,460 contigs with an N50 contig length of 6506 bp (Table 1). All contigs were further assembled into 64,665 scaffolds (>1 kb), of which 4833 containing 50% of the assembly were 44.3 kb or larger (N50 = 44,298 kb; Table 1; Supplemental Table 2). Employing an interactive mapping approach using the potato genome as a reference, the S. commersonii scaffolds were anchored onto each chromosome, resulting in 12 pseudomolecules representing the S. commersonii scaffolds linked and ordered according to homology with S. tuberosum (Supplemental Figure 1). The S. commersonii genome size was measured at ∼830 Mb by flow cytometry (Supplemental Figure 2), consistent with genome size estimation (838 Mb) via 23-nucleotide depth distribution (Supplemental Figure 3). The sum of the Illumina sequences obtained represented ∼105x coverage (filtered reads) of the S. commersonii nuclear genome. Gaps within scaffolds ranged in length from 1 to 8369 bp, with a median length of 213 bp (Supplemental Figure 4). The GC content within S. commersonii coding DNA sequence was 34.5% (Supplemental Table 3). To assess the proportion of the gene space captured in this draft genome assembly, we aligned 248 sequences from the nonredundant core eukaryotic genes (CEGs) to the genome assembly. In total, 243 (98%) CEGs homologs were found in the S. commersonii genome, suggesting that the assembly captured a large majority of the gene space (Supplemental Figure 5).
Table 1. Metrics of S. commersonii Genome Assembly.
| Genome Assembly Statistics | |
|---|---|
| N50 index (contigs), number | 27,829 |
| N50 length (contigs), bp | 6,506 |
| Contig (>100 bp), number | 278,460 |
| Large (>500 bp) contig, number | 226,195 |
| Longest contig (bp) | 170,543 |
| Average contig length (bp) | 2,932 |
| N50 index (scaffolds), number | 4,833 |
| N50 length (scaffolds), bp | 44,298 |
| Longest scaffold (bp) | 458,668 |
| Average scaffold length (bp) | 13,543 |
Genomic Variations
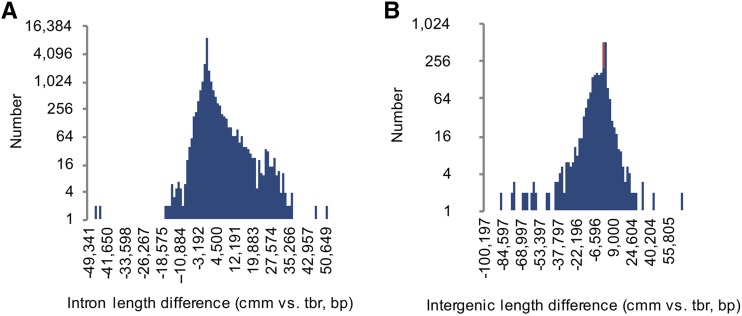
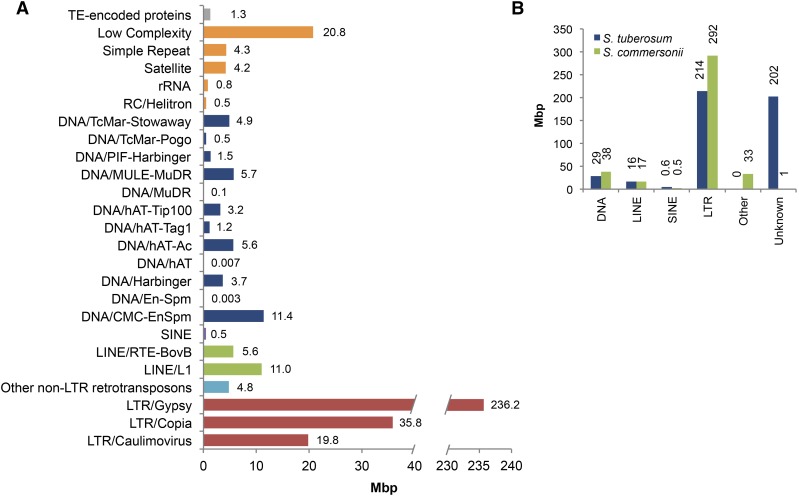
Compared with the cultivated potato, the S. commersonii genome showed a lower level of heterozygosity (Hirsch et al., 2013). A total of 9,894,571 reliable single-nucleotide polymorphisms (SNPs) were identified among 662,040,919 reliable genome bases (Supplemental Table 4), yielding a SNP frequency of 1.49%. We evaluated the structural and functional effect of SNPs. Of all SNPs, 92% had a distance of <50 bp to the nearest neighboring SNP. Overall 12,412 genes encompassed SNPs, of which 11,608 had a SNP rate of <1% (Supplemental Figure 6). With regard to functional annotation, most of the identified SNPs (84%) were located in intergenic regions (Supplemental Table 5). The 12,412 SNP-containing genes displayed overrepresentation of some major functional categories, including macromolecule metabolic processes, response to stimuli, carbohydrate derivative binding, localization, and ion binding (Supplemental Data Set 1). The genome size difference between S. commersonii (830 Mb) and the doubled monoploid clone DM1-3 516 R44 of S. tuberosum (838 Mb) was mainly due to differences in intergenic sequence length (Figure 1). The results of microsynteny analyses revealed greater frequency of SNPs and insertion-deletion events (indels) spanning intergenic regions, consistent with this observation. Roughly 383 Mb of repetitive sequences were identified, accounting for 44.5% of the current assembly of the S. commersonii genome (Figure 2A). Analysis of kmer distribution in unassembled reads estimated 51.3% of the genome as nonrepetitive (Liu et al., 2012), and graph-based clustering with RepeatExplorer (Novák et al., 2013) detected a fraction of repeated sequences equal to ∼36% of the total genome. Although these data are not conclusive, they suggest that, compared with potato (Potato Genome Sequencing Consortium, 2011), S. commersonii might have a lower amount of repetitive DNA (44.5% versus 55%), which might predict different genome dynamics in these two species since their separation from a common ancestor. The repetitive fraction of the S. commersonii genome assembly is dominated by long terminal repeat-retrotranspons (LTR-RTs) (∼34%) with lower levels of several other repeat types (Figure 2B). Characterization of SINE families allowed annotating 1925 SINEs with significant similarity to families previously described in S. tuberosum and in other Solanaceae (Wenke et al., 2011; Seibt et al., 2012) (Supplemental Table 6).
Figure 1.
Influence of Introns and Intergenic Regions on Genome Size Variation.
(A) Number of orthologous genes between S. commersonii (cmm) and S. tuberosum (tbr) showing differences in intron size.
(B) Number of orthologous regions between cmm and tbr showing intergenic size differences.
Figure 2.
Repetitive Sequence Annotation in the Draft Genome of S. commersonii.
(A) Classification of repetitive sequences in S. commersonii.
(B) Comparison of transposable element lengths between S. commersonii and S. tuberosum.
Gene Annotation
Gene prediction was performed by combining results obtained from ab initio prediction, homology searches, and experimental support (cmm ESTs). The de novo assembled transcriptome encompassed ∼96% of all predicted S. commersonii genes (Supplemental Table 7 and Supplemental Figure 7). Results for the functional annotation of the S. commersonii transcripts are reported in Supplemental Figures 8A to 8C. Fewer genes (37,662; annotation evidence distance ≤ 0.5) were predicted in S. commersonii than in potato (∼39,000), but the wild species has more predicted genes than tomato (34,727). Of predicted S. commersonii genes, 30,477 predicted protein-coding genes had significant BLAST similarity to protein-coding genes from other organisms in the nonredundant NCBI database. Nearly 20,500 S. commersonii genes were assigned to Gene Ontology (GO) terms, and more than 4900 proteins were annotated with a four-digit EC number. These data implied that more than 24% of the predicted proteome of S. commersonii has enzymatic function. A large number of transcripts (20,994) with no apparent coding capacity were predicted in S. commersonii (Supplemental Figure 8D). These noncoding RNAs (ncRNAs) comprised a diverse group of transcripts, including 22 tRNAs, 40 rRNAs, 18,882 long noncoding RNAs (lncRNAs), and 1703 putative microRNA (miRNA) precursors. Among the latter, 360 were predicted to fold into a secondary structure leading to the typical miRNA/miRNA* double-stranded RNA duplexes (Supplemental Data Set 2). In addition, 47 of these transcripts showed similarity to known mature miRNAs (Supplemental Tables 8 and 9). A key step toward understanding the biological functions of the predicted miRNAs was achieved through the identification of 4437 target sites. According to GO term classification, 22% (976) of the target genes are involved in cold response (Supplemental Table 8) and 10 are potential regulators of transcripts annotated as responsive to cold (Supplemental Table 10).
Phylogenetic Analysis and Genome Evolution
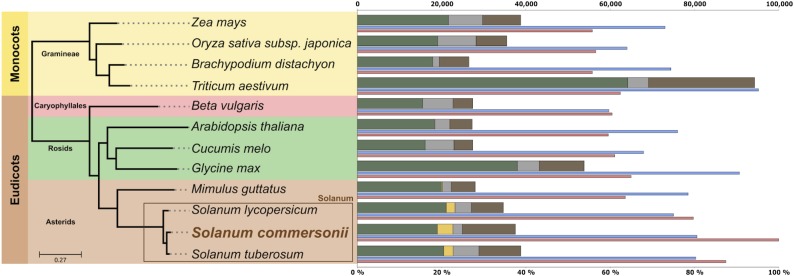
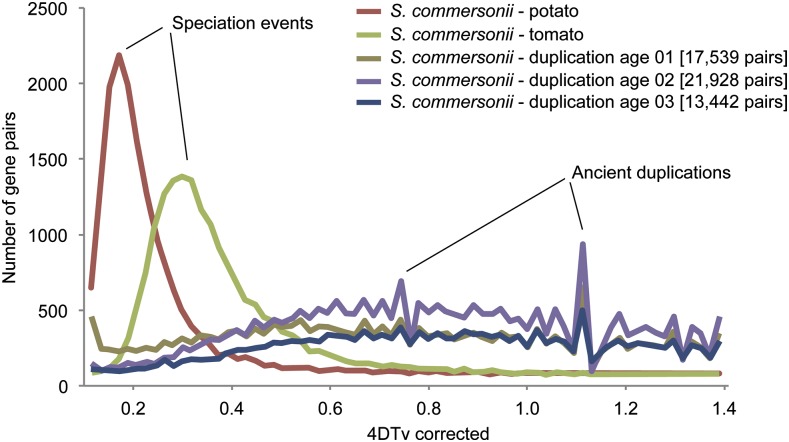
To gain insight into the evolution of the S. commersonii genome, we compared its virtual proteome with predicted proteins of 11 other fully sequenced plant genomes (Supplemental Table 11), including S. tuberosum and S. lycopersicum (Figure 3). The resulting 35,182 phylogenetic trees, available through PhylomeDB (Huerta-Cepas et al., 2014), were scanned to predict phylogeny-based orthology and paralogy relationships (Gabaldón, 2008), detect and date duplication events (Huerta-Cepas and Gabaldón, 2011), and transfer annotations to S. commersonii genes from their functionally characterized one-to-one orthologs (Huerta-Cepas and Gabaldón, 2011). Roughly 17,300 (44%) and 16,821 (42%) S. commersonii genes showed one-to-one orthology with genes from S. tuberosum and S. lycopersicum, respectively, but only 7058 (18%) with genes from the more distantly related asterid Mimulus guttatus (Supplemental Table 12). Out of 35,182 phylogenies obtained, 9445 (24%), 7316 (21%), and 14,061 (40%) showed at least one duplication event at the S. commersonii, potato ancestor, and Solanum ancestor nodes, respectively, compared with only 1814 trees (5%) showing a duplication at the base of asterids (Supplemental Table 13). The overall average number of duplications per branch (duplication density) was 0.66, 0.93, and 0.94 for S. commersonii, potato ancestor, and Solanum ancestor, respectively, whereas we found a low rate of 0.066 for the common ancestor of asterids (Supplemental Table 13). To gain further insight into the divergence between S. commersonii and the domesticated potato, we measured transversions at 4-fold degenerate sites (4DTv) for orthologous gene pairs between S. commersonii and either the domesticated potato or tomato (Figure 4) and between paralogous gene pairs diverged from duplications at each of the three relevant duplication periods investigated. Substitution rates at orthologous sites between S. commersonii and tomato peaked at 0.225, whereas those between S. commersonii and S. tuberosum at 0.077. Assuming a divergence time between tomato and potato of 7.3 million years (Potato Genome Sequencing Consortium, 2011), and a constant mutation rate between the three lineages, this renders an estimate of ∼2.3 million years for the separation of domesticated potato and S. commersonii lineages. The analysis of the paralogous pairs revealed at all three relative ages showed a similar pattern, with the two most prominent peaks largely preceding the divergence of S. commersonii and tomato. Paralogous genes mapped to the S. commersonii-specific duplication (age 1) did show an additional, younger peak at 4DTv values. We assessed the genomic organization of these recent duplicates and found that most of them were present in tandem (314) or at least closely associated in the same contig (141). Finally, we assessed functional enrichment among gene families duplicated at each of these three periods (Supplemental Data Set 3). Response to salt stress and water transport were terms found to be enriched exclusively among S. commersonii specific duplications. The ancestral potato duplication was enriched in terms related to cadmium, metal ion binding, or synthesis of terpenes, whereas terms related to nitrogen starvation, response to ethylene, response to gamma radiation, and maltose metabolism were enriched in the duplications preceding the common Solanum ancestor. All three duplication periods shared enrichments in defense response and growth. Transposon-related terms were enriched in the largest expanded families in S. commersonii, indicating active expansion of transposons (Supplemental Table 14).
Figure 3.
Phylogenetic Relationships of 12 Sequenced Plant Species and Comparative Protein Sequence Analysis.
Species tree based on maximum likelihood analysis of a concatenated alignment of 454 widespread single-copy protein sequences. Different background colors indicate taxonomic groupings within the species used to make the tree. Inset square highlights the species belonging to the genus Solanum. Bars represent the total number of genes for each species (scale on the top). Bars are divided to indicate different types of homology relationships: dark green, widespread genes that are found in at least 11 of the 12 species; orange, widespread but asterid-specific genes that are found in at least three of the four asterid species; gray, species-specific genes with no (detectable) homologs in other species; brown, genes without a clear homology pattern. The thin purple line under each bar represents the percentage of genes with at least one paralog in a given species. The thin dark-gray line represents the percentage of S. commersonii genes that have homologs in a given species.
Figure 4.
Major Evolutionary Events Related to the Evolution of S. commersonii as Revealed by the 4DTv Analysis.
Two speciation events at the split of ancestral potatoes and tomatoes (cyan line peak) and the split between S. tuberosum and S. commersonii (magenta line peak). No recent whole-genome duplication event was detected specifically for S. commersonii. Previously proposed ancient whole-genome duplication events were confirmed using paralogous genes mapped to three different relative evolutionary time points.
Pathogen-Receptor Gene Annotation
A catalog of 942 and 1406 nonredundant pathogen recognition proteins was created from the S. commersonii and S. tuberosum proteomes, respectively. We classified the corresponding genes into various structural categories based on the arrangement of encoded domains (Table 2). In S. commersonii, 286 coiled-coil-nucleotide binding site (NBS)-leucine-rich repeat (LRR) (CNL), 71 NBS, 143 Toll/interleukin-1 receptor (TIR)-NBS, and 37 TIR genes were found. More than 250 receptor-like kinases and 280 receptor-like proteins (RLPs) were also recorded. In comparison, in S. tuberosum, 506 CNL, 199 NBS, 199 TIR-NBS, 36 TIR, 313 receptor-like kinase, and 237 RLP genes were identified. The S. tuberosum genome also includes 14 TIR-LRR genes. Previously, using similar approaches, we cataloged the pathogen recognition proteins from tomato (Andolfo et al., 2014a, 2014b). While the S. tuberosum genome contains nearly twice as many CNL genes as S. commersonii (506 versus 286, respectively), the tomato genome contains roughly half as many CNL genes as S. commersonii (81 versus 186, respectively). By contrast, S. commersonii and S. tuberosum encode a larger complement of TIR-NBS-LRR and RLP proteins than does tomato. These findings suggest that the pathogen receptor gene repertoire in each Solanum species is uniquely shaped based on pathogen pressures and life history.
Table 2. Numbers of S. commersonii and S. tuberosum Genes Encoding Proteins with Domains Similar to Those Found in Plant Pathogen Receptor Proteins.
| Family ID | Domain | S. tuberosum, Number | S. commersonii, Number |
|---|---|---|---|
| Canonical cytoplasmic R genes | |||
| CNL or NL | CC-NBS-LRR | 194 | 186 |
| TNL | TIR-NBS-LRR | 46 | 36 |
| Single domains or incomplete structures | |||
| NL | NBS-LRR | 165 | 98 |
| N | NBS | 199 | 71 |
| T | TIR | 36 | 10 |
| L | LRR | 199 | 144 |
| TN | TIR-NBS | 14 | 12 |
| TL | TIR-LRR | 2 | 1 |
| Canonical transmembrane domains | |||
| RLK | Receptor like kinase | 313 | 252 |
| RLP | Receptor like protein | 237 | 180 |
| New combinations of domains or new structures | |||
| MN | Metallophos-NBS | 1 | 0 |
| GN | Glutaredox-NBS | 1 | 0 |
| RPW8N | RPW8-NBS | 1 | 0 |
| RPW8NL | RPW8-NBS-LRR | 1 | 0 |
| TPP2 | TIR-PP2 | 4 | 1 |
| ANL | Aldolase-NBS-LRR | 1 | 0 |
| RLP-M | RLP-Malectin | 4 | 0 |
| RLP-U | RLP-Ubiquitin | 4 | 0 |
| RLK-UPP | RLK-UPP | 1 | 1 |
| CelNL | Cellulose_synthase-NBS-LRR | 0 | 1 |
| PN | Peroxidase-NBS | 0 | 2 |
| PhN | Phage_GPO-NBS | 0 | 1 |
| HN | Homoserine-NBS | 0 | 4 |
| AL | Aldolase-LRR | 0 | 1 |
| HL | Hydrolase-LRR | 0 | 1 |
| LL | Lipase-LRR | 0 | 1 |
| PL | Peptidase-LRR | 0 | 1 |
| YL | YDG-LRR | 0 | 1 |
| ML | Malectin-LRR | 0 | 6 |
| RLK-L | RLK-Lipase | 0 | 1 |
| RLK-M | RLK-Malectin | 0 | 6 |
| RLK-P | RLK-PPR | 0 | 1 |
| SN | SBF-NBS | 0 | 1 |
| PT | PthA_Avr-TIR | 0 | 1 |
TIR, protein domain with homology to Drosophila TIR; eLRR, extracellular LRR; CC, coiled-coil motif; NB-ARC, nucleotide binding (NB) domain; RLP, an eLRR protein with a short cytoplasmic domain lacking homology to a protein kinase domain; RLK, receptor-like kinase, an eLRR plasma membrane-spanning protein with a cytoplasmic protein kinase domain; Gnk2, Ginkbilobin-2, an antifungal protein found in the endosperm of ginkgo seeds; TNL, TIR-NB-LRR protein, an R protein containing a central NB-ARC domain fused to an N-terminal TIR domain and a C-terminal LRR domain; CNL, an R protein containing a central NB-ARC domain fused to an N-terminal non-TIR domain and a C-terminal LRR domain; R1dom, the region CC of the protein, characteristic of the resistance protein R1; RPW8dom, the RPW8 domain found in several broad-spectrum mildew resistance proteins from Arabidopsis and other dicots.
Syntenic relationships between pathogen receptor genes in S. commersonii and S. tuberosum were further explored by comparative analysis of three loci involved in Phytophthora infestans resistance, Rpi-blb2 (van der Vossen et al., 2005), Tm-2 (Lanfermeijer et al., 2003), and R1 (Ballvora et al., 2002). All are members of the CNL superfamily and all exist within clusters of related gene copies. In S. tuberosum, Rpi-blb2 is part of a 15-gene cluster (van der Vossen et al., 2005), but in S. commersonii, only four corresponding gene copies were present. Similarly, in S. tuberosum, the Tm-2 cluster comprises four gene copies (Lanfermeijer et al., 2003), while in S. commersonii, only two genes were annotated. For R1, there was clear variation between the species in terms of the physical cluster size and number of genes included. The S. tuberosum R1 cluster was longer than that of S. commersonii (300 kb versus 37 kb, respectively) and comprised more gene copies. The S. commersonii genome contains three R1 genes with clear orthologous relationships to genes in S. tuberosum. Specifically, S. commersonii R1B-23-like, R1C-3-like, and R1A-4-like correspond to S. tuberosum R1B-23, R1C-3, and R1A-4, respectively. These three S. commersonii R1 orthologous pairs exhibited an average nucleotide identity of 93% but were arranged in reverse order in the two species (Supplemental Figure 9). Furthermore, additional unrelated genes found in the S. tuberosum R1B-23 to R1C-3 interval and R1C-3 and R1A-4 interval were completely absent in the S. commersonii R1 gene cluster. Comparison of S. commersonii and S. tuberosum orthologous R1 gene copies revealed further substantial structural variation in some cases. In particular, while the coding sequence lengths of R1B-23 and R1A-4 were similar for S. tuberosum and S. commersonii genes, S. tuberosum R1C-3 was only about one-third the size of S. commersonii R1C-3 (4207 bp versus 12,386 bp, respectively). Differences in the number of exons and introns between R1 orthologs were also found (Supplemental Figure 9).
Cold-Responsive Gene Analysis
A total of 5853 and 8666 predicted protein sequences similar to Arabidopsis thaliana proteins annotated as responsive to cold were identified in S. commersonii and S. tuberosum, respectively (Supplemental Figure 10). In S. commersonii, 1451 proteins were homologous to Arabidopsis sequences annotated with the GO term cold acclimation (hereinafter CA-like), 257 with the term cellular response to cold (hereinafter CRC-like), and 4145 with the term response to cold (hereinafter RC-like). In S. tuberosum, 2199 were in the CA-like group, 362 in the CRC-like group, and 6105 in the RC-like group. Enriched GO term categories were found in both species (Supplemental Figure 10A and Supplemental Data Sets 4 and 5). Roughly 2860 genes were in these categories in S. commersonii (707, 85, and 2072 belonging to the CA-like, CRC-like, and RC-like groups, respectively). By contrast only 532, 181, and 1539 genes were assigned to those categories in S. tuberosum (Supplemental Figures 10B and 10C). GO annotation also revealed that 34 CA-like, CRC-like, and RC-like categories encompassed a large number (1546) of cold-responsive genes harboring a SNP (12.4% of total) (Supplemental Figure 10D and Supplemental Data Set 6). In addition, out of 126 unique annotated S. commersonii genes involved in response to cold, 32 belonged to CA-like, 4 to CRTC-like, and 90 to RTC-like GO terms (Supplemental Figure 10E).
To identify genes involved in freezing and cold acclimation responses, the transcript expression profiles of frost stress acclimated (AC) and nonacclimated (NAC) plants were compared with that of non-frost-stressed plants grown at 24°C. We identified 855 differentially expressed genes: 720 under AC conditions and 784 under NAC conditions. Venn diagram analysis indicated that 71 genes were differently expressed (mostly upregulated) exclusively under AC conditions and 135 only under NAC conditions. Roughly 650 genes were found responsive to both conditions (Supplemental Figure 11 and Supplemental Data Set 7). Different functional categories appeared to be enriched under either AC or NAC conditions (Supplemental Table 15). Among NAC differentially expressed genes, the most significantly enriched groups were those involved in cytoplasmic part (GO:0044444), organelle metabolisms (GO:0044422 and GO:0043226), and in phytosteroid and brassinosteroid metabolic processes (GO:0016128 and GO:0016131, respectively). For AC differentially expressed genes, the most represented GO terms were in response to metal and cadmium ions (GO:0010038 and GO:0046686) symplast (GO:0055044) and vacuolar part (GO:0044437). Protein kinases and phosphatases with altered expression under NAC and AC conditions were among the most differentially expressed groups (Supplemental Data Set 7). In addition, proteins involved in the cold response machinery, such as antioxidant cascades, secondary metabolism, cell wall polysaccharide remodeling, starch metabolism, and protein folding (heat shock protein 70 [HSP70]), were found (Supplemental Data Set 6). Out of 855 cold differentially expressed genes, 56 (6.5%) were annotated as transcription factors (TFs) with known DNA binding domains (Supplemental Figure 12). Thirty-eight TFs were differentially expressed under both AC and NAC conditions, 15 only under NAC, and 3 exclusively under AC. The apetala 2/ethylene-response element binding factor (AP2/ERF) domain was the most represented TF family, accounting for 17 differently expressed TFs.
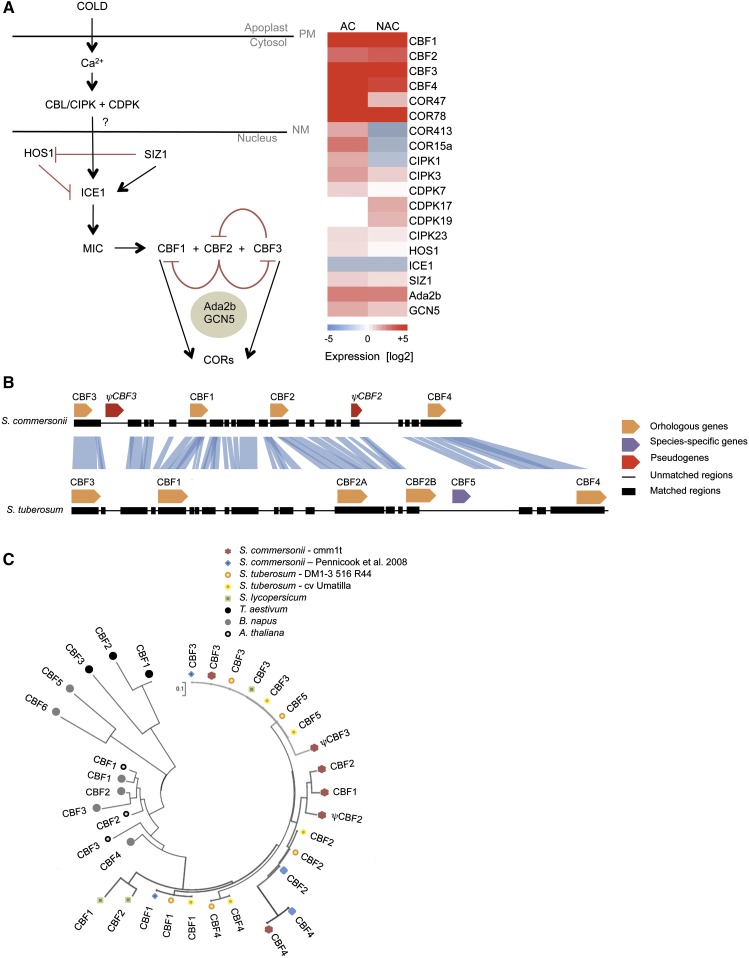
A set of 19 genes encoding cold-sensing and signaling proteins was specifically analyzed under both NAC and AC conditions (Figure 5A). The cold acclimation pathway is initiated when plants sense low temperatures through membrane rigidification, triggering a surge of Ca2+ into the cytosol. Plants possess groups of Ca2+ sensors, including CDPKs (Ca2+-dependent protein kinases), CBL (calcineurin B-like protein), and CIPKs (CBL interacting protein kinase). In S. commersonii, the expression of CDPK7, -17, and -19, as well as CIPK1, -3, and -23, was profiled under both AC and NAC conditions. Transcription of CDPK7, CIPK3, and CIPK23 was activated under NAC and AC conditions. By contrast, transcription of CIPK1 was suppressed under NAC and transcription of CDPK17 and -19 was not affected by acclimation.
Figure 5.
Cold-Responsive Gene Expression Analysis.
(A) Cold-sensing and signaling pathway and gene expression heat map. Expression levels are indicated by shades of blue (downregulation) and red (upregulation), whereas white indicates no differences between control and stressed plants (for AC or NAC plants). PM, plasma membrane; NM, nuclear membrane.
(B) Structural organization of the S. tuberosum and S. commersonii CBF regions.
(C) Similarity tree of the Solanum CBFs in relation to the Arabidopsis, Brassica, and Triticum polypeptides having the CBF signature sequences. The bar indicates branch length scale.
Inside the nucleus, the low-temperature signal transduction pathway triggers the expression of C-repeat binding factor (CBF) genes and their upstream regulators, namely, ICE1 (inducer of CBF expression), a positive regulator of CBF3, HOS1 (high expression of osmotically sensitive), a negative regulator of ICE1, and SIZ1, a SUMO E3 ligase, which mediates sumoylation (SUMO conjugation) of ICE1. In S. commersonii, we found ICE1 transcription was suppressed under both conditions tested, whereas HOS1 and SIZ1 expression was consistently downregulated under both NAC and AC. In light of their prominent role in plant cold acclimation, we also examined S. commersonii CBF gene structural organization and surveyed gene expression patterns under AC and NAC conditions. In total, we identified four S. commersonii CBFs (CBF1, -2, -3, and -4) and pseudogenes of CBF2 (ψCBF2) and CBF3 (ψCBF3). S. commersonii CBFs were collinear with S. tuberosum CBFs, although an S. tuberosum CBF5 ortholog was missing in S. commersonii. Structural variant analysis revealed the presence of few insertions and/or deletions (indels) within the coding sequences analyzed. By contrast, low conservation of CBF2 and CBF4 upstream regions was observed (Figure 5B). The S. commersonii ψCBF2 possessed only portions of a coding sequence with numerous nonsense codons in all reading frames (Supplemental Figure 13A) (Pennycooke et al., 2008). The duplicated S. commersonii CBF3 encoded the amino acid block ASP-ALA-SER-TRY-ARG (hereafter DASWR) positioned immediately downstream from the 60-amino-acid AP2/ERF DNA binding domain in the CBF protein. However, it lacked the PKKPAGR sequence positioned upstream of the domain (Supplemental Figure 13B). Phylogenetic analysis showed that S. commersonii CBF2 and CBF3 grouped with S. tuberosum CBF sequences (Figure 5C), supporting robust orthology relationships. By contrast, S. commersonii CBF1 was independent of the Solanum CBF1 clade, probably due to poorly conserved sequences flanking the AP2/ERF domain (Supplemental Figure 13C). We wondered whether the S. commersonii CBF transcripts accumulated differently under NAC and AC conditions. Interestingly, we found that all S. commersonii CBF genes were highly responsive, regardless of whether the plant experienced acclimation, with CBF1 and CBF3 being most actively responsive (Figure 5A).
CBFs can activate expression of a battery of downstream target genes, also called CBF regulon genes, by binding CRT/DRE elements in target promoter regions. Among these, cold responsive (COR) genes act in concert to enhance freezing tolerance. In S. commersonii, COR genes responded differently to the cold stimulus depending on whether the plants were first acclimated. In particular, COR47 and COR78 were upregulated under both AC and NAC conditions. By contrast, COR15a and COR413 were upregulated under AC but downregulated under NAC. In Arabidopsis, different components of the histone acetyltransferase complex were described to interact with CBF1 in vitro and are needed for CBF1 function (Stockinger et al., 2001). Therefore we checked the expression of S. commersonii ADA2b and GCN5, two components of histone acetyltransferase complex. Our data showed that both Ada2b and GCN5 were transcribed whether or not acclimation occurred (Figure 5A).
DISCUSSION
Genome Sequencing, Assembly, and Gene Annotation
In this work, we de novo sequenced the genome of stress-tolerant S. commersonii as an integral step toward deciphering the genetic bases of agricultural traits that can be improved using genes from this wild germplasm donor. The resulting S. commersonii genome assembly is comparable in length to the reference S. tuberosum genome, but divergence between the two sequences is demonstrated by the presence of SNPs and indels affecting target intergenic regions. These rearranged sites, together with genome-wide analyses of SNPs and indels, will shed light on selection processes shaping intergenic spaces and will likely facilitate the identification of polymorphic markers. In addition, the distribution of SNPs across S. commersonii genes indicated high variability in genes related to specific biological processes such as macromolecule metabolic processes, response to stimuli, carbohydrate derivative binding, localization, and ion binding.
Our data highlighted a striking difference between S. commersonii heterozygosity (1.5%) and that of the common potato (53 to 59%; Hirsch et al., 2013). Although in nature S. commersonii is an obligate sexually reproducing allogamous species, ex situ maintenance through randomly intercrossing seedlings of accession PI 243503 could have artificially affected its diversity level, causing a reduction in the level of heterozygosity compared with spontaneous populations. By contrast, the high level of potato heterozygosity likely reflects both its vegetative propagation (that fixes heterozygosity) and progress made in the past ∼150 years of concerted potato breeding to maximize heterosis. Since the magnitude of the difference in heterozygosity in S. commersonii relative to S. tuberosum was considerable, fundamental questions arise concerning the genetic constitution and maintenance of wild relatives of potato with regard to gene flow and population structure (Camadro et al., 2012; Hirsch et al., 2013). From a practical perspective, it seems that the use of species-wide diversity rather than individual accessions would be much more desirable to broaden the narrow genetic base of and increase allelic diversity in cultivated potato.
We also found differences between S. commersonii and other solanaceous species in terms of repetitive sequences. Transposable elements (TEs) are major components of all plant genomes studied, shaping genome structure and organization. In a comprehensive review of the first 50 sequenced plant genomes, Michael and Jackson (2013) reported that genome repetitive content ranged from 3% (Utricularia gibba) to 85% (maize [Zea mays]). TEs strongly affect gene expression levels and transcript splicing and consequently may impact plant phenotypes. Compared with potato (55%) and tomato (63%), S. commersonii showed a lower amount of repetitive DNA (∼383 Mb, accounting for 44.5% of the current assembly). As in other solanaceous species, there were many more Ty3-gypsy type than Ty1-copia type LTR-RTs identified in S. commersonii, suggesting that the former elements have been somewhat more successful in colonizing and persisting in Solanaceae genomes. Moreover, the ratio of Ty3-gypsy:Ty1-copia might also be driven by variation in the efficacy of illegitimate recombination and/or unequal homologous recombination in removing LTR-RTs from the genomes, as reported in Arabidopsis, maize, barley (Hordeum vulgare), and rice (Oryza sativa; reviewed in Bennetzen, 2007). LTR-RTs play a substantial role in genome size variation, and the lower frequency of TEs in S. commersonii may contribute to its smaller genome size as well as underline the occurrence of different evolutionary dynamics in individual solanaceous species genomes since their separation from a common ancestor. Vitte and Bennetzen (2006) suggested that the proportion of TEs in different genomes might be influenced by destabilization of epigenetic regulation.
In our study, we targeted different tissues to best represent the S. commersonii transcript repertoire. The de novo assembly of the transcriptome from leaf, flower, stolon, and tuber tissues allowed identification of 37,662 genes. Even though the number of genes found in S. commersonii was similar to that reported for S. tuberosum, the number of transcripts differed greatly between the two species. This might highlight the presence of more prominent alternative splicing activities in potato than in S. commersonii. This is consistent with observations by the Potato Genome Sequencing Consortium (2011) that ∼25% of potato genes encoded two or more isoforms, indicative of more functional variation than is represented by the gene set alone. We also identified ∼21,000 S. commersonii ncRNAs. Emerging evidence has revealed that ncRNAs are major products of the plant transcriptome (Rymarquis et al., 2008). They may have significant regulatory importance, especially during stress situations (Matsui et al., 2013). In this study, the perfect or near-perfect match to target sites allowed effective in silico prediction of the target sequences (Rhoades et al., 2002) and revealed that 10 miRNA targeted cold-responsive genes. Since manipulation of miRNA/siRNA-guided gene regulation may enable engineering of plants for improved stress tolerance, detailed analyses on miRNA-guided stress responsive gene regulation in S. commersonii may lead to new insights for efficient exploitation of this germplasm.
Phylogenomic Analysis across Plant Species
To assess evolutionary relationships between S. commersonii and other sequenced plant genomes, we undertook a comprehensive phylogenomic approach. This involved reconstruction of the complete collection of evolutionary histories of all S. commersonii protein-coding genes across a phylogeny of 12 sequenced plants (i.e., the phylome) and animals (Huerta-Cepas et al., 2010; Chipman et al., 2014). The usefulness of this approach in the annotation of newly sequenced genomes has been demonstrated in other plants (Garcia-Mas et al., 2012; Dohm et al., 2014). In total, 17,297 (44%) S. commersonii genes showed a one-to-one orthologous relationship with S. tuberosum genes and 16,821 (42%) with S. lycopersicum genes, but only 7058 (18%) with genes from the more distantly related asterid M. guttatus (Supplemental Table 12). This scenario, in which most orthology relationships are of the type one-to-many, many-to-many, or many-to-one, likely results from the past genome duplication shared by the three Solanum species (Tomato Genome Consortium, 2012), followed by differential loss of paralogous genes in each of the species. The overall average number of duplications per node (duplication density) was 0.66, 0.93, and 0.94 for S. commersonii, potato ancestor, and Solanum ancestor, respectively, whereas we found a low rate of 0.066 for the common ancestor of asterids. Collectively, these numbers suggest multiple rounds of duplications, at least at the lineage preceding the separation of the Solanum ancestor from the other asterid included in this analysis (M. guttatus). These two rounds of ancestral duplications were previously suggested by comparison of the domesticated potato and tomato genomes (Potato Genome Sequencing Consortium, 2011; Tomato Genome Consortium, 2012). Our study showed that these ancestral duplications were also shared with the wild species lineage represented by S. commersonii, as would be predicted by commonly accepted Solanum phylogeny. The analysis of paralogous gene pairs revealed that most duplicated genes diverged before the separation of Solanum species. These results are in agreement with the topological dating of duplications, which strongly suggest that these major duplications predate the divergence of the Solanum species and that most paralogous pairs dated as potato-specific or S. commersonii-specific result from differential retention of duplicated pairs in each of the investigated lineages. We assessed the genomic organization of these recent duplicates and found that most were present in tandem (314) or were closely associated along the same contig (141). Thus, results obtained are not compatible with a recent, specific genome duplication in S. commersonii but rather with differential retention of paralogs from ancient duplications and additional lineage-specific segmental duplications that blurred the syntenic and one-to-one correspondence between S. commersonii and S. tuberosum. This in turn may underlie the sexual incompatibility between these two related species.
Pathogen-Receptor Genes
Candidate S. commersonii disease resistance (R) genes were cataloged and compared with the R gene complement comprising the cultivated potato and tomato genomes. Our data revealed that S. commersonii contains fewer R gene candidates than S. tuberosum, but more than tomato. Polyploidization, genome size variation, natural selection, artificial selection including domestication, breeding, and cultivation, and gene family interactions have probably influenced pathogen recognition gene evolution in Solanum (Andolfo et al., 2014b). Differences in copy numbers of specific R gene families are important sources of genetic variation and are likely to play a role in phenotypic diversity and adaptation in different species (Peele et al., 2014). Our analyses revealed that different R locus arrangements emerged in different species after their separation from a common Solanum ancestor. Indeed, the size of the R1 locus varied 10-fold among the genomes analyzed. Previous comparative analysis of the R1 locus revealed highly conserved collinear regions that flank sequences showing high variability and tandem duplicated genes (namely, R1 homologs and F-box-containing genes) (Ballvora et al., 2007). Solanum R gene architecture seems to be shaped by the interplay of large-scale gene organization that determines global conservation of locus order genome wide and extensive local genome rearrangements mediated by tandem duplication, transposons, and other shuffling elements that determine distinct local arrangements (Zhang et al., 2014). Extant local arrangement of Solanum R genes within a genome may be indicative of biological and environmental factors influencing genotype adaptation and may have significant influence on phenotypic resistance diversity. More detailed comparative analyses of R genes within and among Solanum species are warranted.
Nonacclimated and Cold-Acclimated Gene Expression and Regulation
Previous studies provided evidence that, among potato species, S. commersonii is the most tolerant to low temperatures, with the best capacity to cold acclimate (Palta and Simon, 1993). The molecular basis of nonacclimated tolerance in potato is poorly understood, although it has been reported that it may be genetically determined by loci independent of acclimated tolerance (Stone et al., 1993). We therefore analyzed the cold-responsive transcriptome of S. commersonii to shed further light on processes that might explain its freezing tolerance and cold acclimation capacity. Overall, whole-genome expression data highlighted an extensive reorganization of the transcriptome under cold stress, with enhanced expression of genes affecting ROS scavenging enzymes (e.g., superoxide dismutase, SOD; catalase, CAT; ascorbate peroxidase, APX), those involved in cell repair (such as HSPs and dehydrins [DHNs]), and those encoding proteins that may function as osmoprotectants. Among the latter, we found a significant upregulation of S. commersonii galactinol synthase (GOLS1) under both NAC and AC conditions. Previously, overexpression of GOLS1 from Medicago falcata or Boea hygrometrica promoted the biosynthesis of increased amounts of raffinose family oligosaccharides, such as galactinol, raffinose, and stachyose, and resulted in elevated tolerance to low temperatures in transgenic tobacco (Nicotiana tabacum) plants (Wang et al., 2009; Zhuo et al., 2013). Given that, we hypothesize that high expression of S. commersonii GOLS1 in conjunction with the increased activity of the aforementioned cold-associated and -inducible proteins might contribute to S. commersonii frost tolerance.
It is notable that several genes were responsive to cold relative to control conditions, but with contrasting kinetics under AC versus NAC. For instance, BRASSINOSTEROID-SIGNALING KINASE1 (BSK1) was activated under AC and suppressed under NAC. Conversely, one MYB and one bHLH TF were cold induced under NAC and repressed under AC. As MYB and bHLH proteins often interact with each other to control transcription (Ramsay and Glover, 2005), this differential expression of MYB and bHLH TFs suggests that the regulation of some cold-responsive genes may be achieved by modulating the ratio of these partners. TFs were mostly upregulated under both conditions, as was observed in Arabidopsis (Lee et al., 2005). This is consistent with overall upregulation, rather than repression, of gene transcription following cold stress. Specifically, we identified 25 TFs correlating positively with acclimated and nonacclimated tolerance, and only 11 that showed negative correlations. Among the negatively correlating TFs was a Cys-2/His-2-type (C2H2) zinc-finger protein (Sakamoto et al., 2004). C2H2 zinc-finger-type TFs have been found to work downstream of DREB1/CBF and to be responsible for stress tolerance in plants (Sakamoto et al., 2004).
Our comparison of cold-responsive gene expression profiles between AC- and NAC-stressed plants highlighted remarkable features of some of those known to be critical in cold sensing and signaling pathways. Two genes for calcium-dependent protein kinases, CDPK17 and CDPK 19 (CPK8), were differentially expressed. Interestingly, CDPK19 was upregulated only under NAC conditions, whereas CDPK17 expression required acclimation. Neither gene has been previously implicated in cold stress response. Thus, the induction of CDPK19 (CPK8) and CDPK17 in S. commersonii suggests possible independent roles in response to freezing and cold acclimation, respectively. The structural organization and transcriptional activity of the S. commersonii genes for CBFs (C-repeat binding factors) also revealed intriguing features. Our cross-species comparisons indicated that the CBFs underwent rapid expansion via duplication processes. In S. commersonii, we found two pseudogenes, ψCBF2 and ψCBF3. Both pseudogenes contain premature stop codons. High identity (100%) between S. commersonii CBF3 paralogs suggests that gene duplication occurred in the last ∼2.3 million years, after the divergence between potato and S. commersonii from their most recent common ancestor. In particular, the paucity of sequence change indicates that, after the divergence of the two species lineages, there were likely strong constraints on CBF3 that conserved protein sequence. A different situation was found for S. commersonii ψCBF2, which shares only 80% of identity with the functional S. commersonii CBF2. This suggests that the gene duplication occurred prior to divergence of the S. tuberosum and S. commersonii lineages from their most recent common ancestor, with the duplicated copy subsequently undergoing rearrangements as observed also in other duplicated genes (Lynch and Force, 2000). Phylogenetic analysis highlighted a common origin of CBFs in Solanum species with respect to other plants from temperate regions that can cold acclimate, such as Arabidopsis, wheat (Triticum aestivum) and Brassica napus (Jaglo et al., 2001). This suggests homogenization mechanisms exist in Solanum, as previously reported (Pennycooke et al., 2008). Despite observed orthology for most of the CBF1 gene family, S. commersonii CBF1 clustered apart from other CBF1 sequences. This might be the result of strong selection pressure toward functional diversification of S. commersonii CBF1. Taken together, our data are consistent with a hypothesis of rapid evolution of CBFs within the genus Solanum (Carvallo et al., 2011). We hypothesize that a duplication event occurred after the S. tuberosum-S. commersonii divergence and may have led to a different functionalization of the S. commersonii CBF3, resulting in enhanced cold response capability in S. commersonii. To more deeply investigate the role of CBFs in S. commersonii, transcript levels were monitored both under AC and NAC. Our data showed that all S. commersonii CBFs were upregulated relative to controls under all tested conditions. This is in contrast with previous reports that CBF1, but not related CBFs, were responsive to low temperatures in both S. commersonii and S. tuberosum (Pennycooke et al., 2008; Carvallo et al., 2011). Our observations parallel patterns observed in tomato species. In cold-sensitive cultivated tomato, only CBF1 was upregulated in response to cold treatments, whereas in the cold-tolerant wild tomato species Solanum peruvianum, all three CBF genes were cold responsive (Mboup et al., 2012). High expression of S. commersonii CBF genes and genes regulated by CBF proteins (e.g., COR genes) may be directly responsible for enhanced cold tolerance and acclimation ability in this species.
In conclusion, we report the genome sequence for a wild relative of the cultivated potato S. tuberosum. The genome sequence of S. commersonii is a valuable resource for studying Solanum diversity and improving the cultivated potato. Among significant findings, we identified new cold-regulated genes. The information we generated provides a foundation for further experiments to explore the network of gene regulation required for cold tolerance and acclimation and to determine the function of cold-responsive genes through molecular and cellular approaches. Future challenges include translation of this new knowledge into advances in crop improvement in response to global climate and environment changes. In addition, our work demonstrated the utility of wild crop relative genome sequences in elucidating evolutionary mechanisms contributing to Solanum species diversity. Further sequencing and analysis of crop wild relative genomes will prove crucial to utilization of wild germplasm for improvement of specific traits in crop plants with narrow genetic bases.
METHODS
Genetic Background of Sequenced Material
We sequenced the genome of clone cmm1t of Solanum commersonii. It derived from a single seed from accession PI243503 obtained from the Inter-Regional Potato Introduction Station, Sturgeon Bay, Wisconsin (Supplemental Figure 2). This clone has been widely characterized and used in our breeding program as a source of resistance genes to biotic and abiotic stresses (Carputo et al., 1997, 2007, 2009, 2013).
Genome Sequencing and Assembly
Genomic DNA isolated from leaf material was sequenced on the Illumina HiSeq1000 sequencing platform and assembled using SOAPdenovo (Luo et al., 2012). Gaps were closed using GapCloser v1.12 (a SOAP suite tool), and sequences shorter than 1000 bp bases were discarded from the final assembly. The gene space of the assembled genome was assessed by aligning CEGs to the assembly using BLAST with a 65% identity threshold. Reads were aligned to the assembled genome using SOAPaligner v2.21 (a SOAP suite tool) with standard parameters but “-r 0” parameter. We called the SNPs by aligning and comparing S. commersonii reads to the assembled S. commersonii genome using SOAPsnp v1.03 (a SOAP suite tool) with “-u” and “-n” options enabled to give better accuracy for heterozygous SNP detection. Heterozygosity was then calculated by estimating the number of heterozygous calls over the total of the callable bases (Zheng et al., 2011; Varshney et al., 2012). We estimated the genome size of S. commersonii using flow cytometry. For more details, see Supplemental Methods.
Repeats, Protein-Coding Genes, and lncRNA Annotation
Annotation of repeats and protein coding genes was performed using the MAKER pipeline (v2.27) (Cantarel et al., 2008) with Illumina mRNA-seq reads obtained from root, stolon, tuber, leaf, and flower tissues and other cDNA read data as supporting evidence. In particular, repeats were annotated with RepeatMasker (v. 3.2.8) using the “Solanaceae” repeats database and with Repeatrunner using a database of TE-encoded proteins included into MAKER installation to help mask repeats that have diverged over time. The repeated fraction was also evaluated by graph-based clustering of repetitive elements in unassembled reads using the RepeatExplorer Web server (Novák et al., 2013) and by analysis of kmer content using Jellyfish and GCE software (Liu et al., 2012). Putative SINEs were detected using the SINE-Finder tool and were used to search against published SINE sequences of Solanum tuberosum and other Solanaceae using FASTA (Wenke et al., 2011; ftp://ftp.ebi.ac.uk/pub/software/unix/fasta/fasta36/). Members of each family detected in S. commersonii were aligned with MUSCLE (Edgar, 2004), and consensus sequences were calculated. Ab initio prediction of protein coding genes was performed using AUGUSTUS (Stanke and Waack, 2003) and GeneMark (Lukashin and Borodovsky, 1998). The OrthoMCL pipeline (Li et al., 2003) was used to identify and estimate the number of paralogous and orthologous gene clusters between S. commersonii, S. tuberosum, and Solanum lycopersicum. lncRNAs were identified using the approach described by Boerner and McGinnis (2012). To distinguish lncRNA from precursors of other ncRNA, the set of lncRNAs was first analyzed with cmscan (e-value 0.01) from Infernal 1.1 against the database of covariate models of Rfam 11.0. Noncoding transcripts were BLAST searched as well against a database of plant mature miRNA sequences in miRBase (http://www.mirbase.org/) to identify homologous miRNAs. For more details, see Supplemental Methods.
R Gene Analysis
Matrix-R was used to screen the proteomes of S. commersonii and S. tuberosum (37,662 and 39,031 proteins, respectively). The set of predicted proteins identified via HMM profiling was further analyzed using Interproscan software version 5.0 (http://www.ebi.ac.uk/Tools/pfa/iprscan5/) to verify the presence of conserved domains and motifs characteristic of R proteins. To identify S. tuberosum R1 orthologs in S. commersonii, we used the data from the orthology relationships. Then, selected homologous sequences were aligned further analyzed using Interproscan software version 5.0 (http://www.ebi.ac.uk/Tools/pfa/iprscan5/) to verify the presence of conserved domains and motifs characteristic of R1 proteins. The CBF protein sequences were aligned using Geneious R6 (Biomatters). The CBF evolutionary history was inferred by the maximum likelihood method using MEGA 6.06 (Tamura et al., 2011). For more details, see Supplemental Methods.
Cold-Responsive Gene Analysis
Twelve clonally propagated plants from cmm1t were cultured in a growth chamber under cool white fluorescent lamps (350 to 400 μmol m−2 s−1) at 24°C and then exposed to −2°C for 6 h to test their tolerance to low temperature under NAC conditions. To evaluate cold tolerance following AC, six plants were first transferred from a 24°C growth chamber to a cold room (4°C) under cool white fluorescent lamps (100 μmol m−2 s−1) for 2 weeks and then exposed to −2°C for 6 h. RNA expression analysis was performed on a Combimatrix S. tuberosum chip produced by the Plant Functional Genomics Center at the University of Verona. The chip contained 27,234 nonredundant 35-40-mer oligo probes in triplicate. For more details, see Supplemental Methods.
Phylogenetic Analysis, Genome Evolution, and Functional Annotation
The longest protein sequence associated with each predicated S. commersonii gene was used for a Smith-Waterman search against the protein sets of nine other plant species. Alignments were generated and quality-filtered, and phylogenetic trees were calculated for each S. commersonii sequence. A species tree was generated from a super tree of all trees and by multigene phylogenetic analysis of high-confidence 1:1 orthologs. We computed transversion rates at 4-fold degenerate sites (4DTv) as a conservative genetic distance to estimate recent major evolutionary events. To assess divergence between species, individual gene trees in the S. commersonii phylome were scanned to detect one-to-one orthologs between S. commersonii and S. tuberosum and between S. commersonii and S. lycopersicum. To estimate age of duplication waves, we used paralogous gene pairs assigned to the three relevant evolutionary time points. Protein-coding gene predictions were functionally annotated based on protein signatures and orthology relationships. For more details, see Supplemental Methods.
Accession Numbers
Illumina genome sequences have been deposited in the Sequence Read Archive under study SRP050408, and RNA-seq sequences have been deposited under study SRP050412. The Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under accession number GCHT00000000. The version described in this article is the first version, GCHT01000000. This Whole-Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under accession number JXZD00000000. The version described in this article is version JXZD01000000. Microarray data of AC and NAC experiments are downloadable from http://ddlab.sci.univr.it/files/scommersonii/results_CNAvsTA_sig.xls and http://ddlab.sci.univr.it/files/scommersonii/results_TNAvsCNA_sig.xls, respectively. Phylogenetic trees and alignments are available through PhylomeDB (http://www.phylomedb.org). Accession numbers and alignment of CBFs are reported in Supplemental Data Set 9.
Supplemental Data
Supplemental Figure 1. Ideograms of the 12 pseudochromosomes of S. commersonii.
Supplemental Figure 2. Phenotype and cytogenetic analysis of S. commersonii.
Supplemental Figure 3. Distribution of Illumina 23-kmer frequency for S. commersonii.
Supplemental Figure 4. Distribution of gap length within the scaffold assembly of S. commersonii.
Supplemental Figure 5. Percentage of core eukaryotic genes mapping on S. commersonii draft genome.
Supplemental Figure 6. SNP spacing in the S. commersonii genome.
Supplemental Figure 7. Proportion of transcriptome mapping to S. commersonii genome assembly.
Supplemental Figure 8. Functional annotation of S. commersonii transcriptome.
Supplemental Figure 9. R1 cluster in S. commersonii and S. tuberosum.
Supplemental Figure 10. Cold-responsive gene annotation analysis.
Supplemental Figure 11. Common and differentially expressed genes between AC and NAC conditions.
Supplemental Figure 12. Transcriptional factors with known DNA binding domains.
Supplemental Figure 13. Comparison between CBF2, CBF3, and CBF1 protein sequences of S. commersonii (clone cmm1t) and the orthologous sequences of S. commersonii (NCBI; Pennycook et al., 2009), S. tuberosum DM1-3 516 R44, S. tuberosum cv Umatilla, and S. lycopersicum.
Supplemental Figure 14. Real-time qPCR on four target genes in NAC and AC conditions.
Supplemental Table 1. Summary of sequence read statistics of the mate pair and paired-end libraries used in WGS sequencing.
Supplemental Table 2. Summary of the S. commersonii genome assembly.
Supplemental Table 3. CG content in S. commersonii genome.
Supplemental Table 4. Heterozygosity in S. commersonii genome.
Supplemental Table 5. Annotation of SNPs detected in S. commersonii.
Supplemental Table 6. SINE families in S. commersonii.
Supplemental Table 7. De novo assembled transcripts.
Supplemental Table 8. MicroRNA statistics.
Supplemental Table 9. Putative miRNA precursors showing miRNA/miRNA* duplexes and similarity to know miRNAs.
Supplemental Table 10. Transcripts annotated as responsive to cold stress and of their potential miRNA regulators.
Supplemental Table 11. Overview of the species used for the comparative genomics analyses.
Supplemental Table 12. Detected one-to-one orthologs between a given species and S. commersonii.
Supplemental Table 13. Number of duplication events detected in single gene trees according to their relative ages.
Supplemental Table 14. Functional enrichment analysis results after removing redundancy for the 10 biggest clusters of specifically expanded clusters of proteins in S. commersonii with statistically significant enriched functional terms.
Supplemental Table 15. Enrichment of functional categories among differentially expressed genes in nonacclimated and acclimated conditions.
Supplemental Table 16. Number of nonredundant protein families annotated with the Gene Ontology term cold acclimation, cellular response to cold, and proteins as response to cold and related number of proteins in Arabidopsis and S. tuberosum.
Supplemental Methods. Detailed information of the experimental and computational methods used.
Supplemental Data Set 1. Enriched GO terms for genes containing SNPs.
Supplemental Data Set 2. Putative miRNA precursors showing miRNA/miRNA* duplexes.
Supplemental Data Set 3. Functional enrichments for proteins duplicated at different points across evolution.
Supplemental Data Set 4. Enriched Gene Ontology term categories found in S. commersonii.
Supplemental Data Set 5. Enriched Gene Ontology term categories found in S. tuberosum.
Supplemental Data Set 6. S. commersonii cold-responsive genes containing SNPs.
Supplemental Data Set 7. Differentially expressed genes in acclimated and nonacclimated conditions.
Supplemental Data Set 8. Hidden Markov models for proteins annotated with Gene Ontology terms CA (cold acclimation), CRTC (cellular response to cold), and RC (response to cold).
Supplemental Data Set 9. CBF sequence alignment
Supplementary Material
Acknowledgments
This article is dedicated to Luigi Monti for his continuous support and for his ongoing exemplary career in plant genetics and breeding. This work was supported by the Ministry of University and Research (GenHORT Project PON02_00395_3215002) and by a grant from Campania Region (CARINA project POR_FSE 2007/2013). Research of the group of T.G. is also supported by the Spanish Ministry of Economy and Competitiveness (BIO2012-37161), by a grant from the Qatar National Research Fund (NPRP 5-298-3-086), and by a grant from the European Research Council under the European Union's Seventh Framework Programme (FP/2007-2013)/ERC (Grant Agreement ERC-2012-StG-310325). We thank M. Iovene for the cytological analysis, S. Lucretti for the cytometric assays, T. Cardi for producing clone cmm1t, and A. Di Matteo for RNA hybridizations. We also thank N. D’Agostino for kindly reviewing the article.
AUTHOR CONTRIBUTIONS
R.A. designed the research, performed experiments, analyzed data, and wrote the article. F.C. and V.G. performed research, contributed new computational tools, analyzed the data, and wrote the article. M.R.E. analyzed data and wrote the article. F.T., L.X., A.D.M., C.A., and S.C.G. contributed new computational tools. A.F. performed research and reviewed the article W.S., R.A.C., and T.G. contributed new computational tools, analyzed the data, and wrote the article. M.D., M.I., L.F., and J.M.B. reviewed the article. D.C. designed the research and wrote the article. All authors read and approved the article.
Glossary
- EBN
endosperm balance number
- CEG
core eukaryotic gene
- SNP
single-nucleotide polymorphism
- LTR-RT
long terminal repeat-retrotranspon
- GO
Gene Ontology
- ncRNA
noncoding RNA
- lncRNA
long noncoding RNA
- miRNA
microRNA
- AC
acclimated
- NAC
nonacclimated
- TF
transcription factor
- TE
transposable element
References
- Andolfo G., Sanseverino W., Aversano R., Frusciante L., Ercolano M.R. (2014a). Genome-wide identification and analysis of candidate genes for disease resistance in tomato. Mol. Breed. 33: 227–233. [Google Scholar]
- Andolfo G., Jupe F., Witek K., Etherington G.J., Ercolano M.R., Jones J.D.G. (2014b). Defining the full tomato NB-LRR resistance gene repertoire using genomic and cDNA RenSeq. BMC Plant Biol. 14: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballvora A., Ercolano M.R., Weiss J., Meksem K., Bormann C.A., Oberhagemann P., Salamini F., Gebhardt C. (2002). The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J. 30: 361–371. [DOI] [PubMed] [Google Scholar]
- Ballvora A., Jöcker A., Viehöver P., Ishihara H., Paal J., Meksem K., Bruggmann R., Schoof H., Weisshaar B., Gebhardt C. (2007). Comparative sequence analysis of Solanum and Arabidopsis in a hot spot for pathogen resistance on potato chromosome V reveals a patchwork of conserved and rapidly evolving genome segments. BMC Genomics 8: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamberg, J.B., Martin, M.W., and Schartner, J.J. (1994). Elite Selections of Tuber-Bearing Solanum Species Germplasm: Inter-Regional Potato Introduction Station, NRSP-6. (Madison, WI: University of Wisconsin Press). [Google Scholar]
- Bennetzen J.L. (2007). Patterns in grass genome evolution. Curr. Opin. Plant Biol. 10: 176–181. [DOI] [PubMed] [Google Scholar]
- Boerner S., McGinnis K.M. (2012). Computational identification and functional predictions of long noncoding RNA in Zea mays. PLoS ONE 7: e43047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradeen J.M., Haynes K.G. (2011). Introduction to potato. In Genetics, Genomics and Breeding of Potato, Bradeen J.M., Kole C., eds (Enfield, NH: CRC Press/Science Publishers; ), pp. 1–19. [Google Scholar]
- Bradshaw J.E., Ramsay G. (2005). Utilisation of the commonwealth potato collection in potato breeding. Euphytica 146: 9–19. [Google Scholar]
- Camadro E.L., Erazzú L.E., Maune J.F., Bedogni M.C. (2012). A genetic approach to the species problem in wild potato. Plant Biol. (Stuttg.) 14: 543–554. [DOI] [PubMed] [Google Scholar]
- Cantarel B.L., Korf I., Robb S.M.C., Parra G., Ross E., Moore B., Holt C., Sánchez Alvarado A., Yandell M. (2008). MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18: 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardi T., D’Ambrosio E., Consoli D., Puite K.J., Ramulu K.S. (1993). Production of somatic hybrids between frost-tolerant Solanum commersonii and S. tuberosum: characterization of hybrid plants. Theor. Appl. Genet. 87: 193–200. [DOI] [PubMed] [Google Scholar]
- Carputo D., Alioto D., Aversano R., Garramone R., Miraglia V., Villano C., Frusciante L. (2013). Genetic diversity among potato species as revealed by phenotypic resistances and SSR markers. Plant Genet. Resour. C. 11: 131–139. [Google Scholar]
- Carputo D., Aversano R., Barone A., Di Matteo A., Iorizzo M., Sigillo L., Zoina A., Frusciante L. (2009). Resistance to Ralstonia solanacearum of sexual hybrids between Solanum commersonii and S. tuberosum. Am. J. Potato Res. 86: 196–202. [Google Scholar]
- Carputo D., Barone A., Cardi T., Sebastiano A., Frusciante L., Peloquin S.J. (1997). Endosperm balance number manipulation for direct in vivo germplasm introgression to potato from a sexually isolated relative (Solanum commersonii Dun.). Proc. Natl. Acad. Sci. USA 94: 12013–12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carputo D., Castaldi L., Caruso I., Aversano R., Monti L., Frusciante L. (2007). Resistance to frost and tuber soft rot in near-pentaploid Solanum tuberosum-S.commersonii hybrids. Breed. Sci. 57: 145–151. [Google Scholar]
- Carvallo M.A., Pino M.-T., Jeknic Z., Zou C., Doherty C.J., Shiu S.-H., Chen T.H.H., Thomashow M.F. (2011). A comparison of the low temperature transcriptomes and CBF regulons of three plant species that differ in freezing tolerance: Solanum commersonii, Solanum tuberosum, and Arabidopsis thaliana. J. Exp. Bot. 62: 3807–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman A.D., et al. (2014). The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 12: e1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm J.C., et al. (2014). The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505: 546–549. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T. (2008). Large-scale assignment of orthology: back to phylogenetics? Genome Biol. 9: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mas J., et al. (2012). The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. USA 109: 11872–11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanneman R.E., Bamberg J.B. (1986). Inventory of Tuber-Bearing Solanum Species. 1st ed. (Sturgeon Bay, WI: Potato Introduction Station; ). [Google Scholar]
- Hawkes J.G. (1990). The Potato: Evolution, Biodiversity and Genetic Resources. (Washington, D.C.: John Wiley & Sons; ). [Google Scholar]
- Hirsch, C.N., Hirsch, C.D., Felcher, K., Coombs, J., Zarka, D., Van Deynze, A., De Jong, W., Veilleux, R.E., Jansky, S.H., Bethke, P., Douches, D.S., and Buell, C.R. (2013). Retrospective view of North American potato (Solanum tuberosum L.) breeding in the 20th and 21st centuries. G3 (Bethesda) 3: 1003–1013. [DOI] [PMC free article] [PubMed]
- Huamán Z., Hoekstra R., Bamberg J.B. (2000). The Inter-genebank potato database and the dimensions of available wild potato germplasm. Am. J. Potato Res. 77: 353–362. [Google Scholar]
- Huerta-Cepas J., Marcet-Houben M., Pignatelli M., Moya A., Gabaldón T. (2010). The pea aphid phylome: a complete catalogue of evolutionary histories and arthropod orthology and paralogy relationships for Acyrthosiphon pisum genes. Insect Mol. Biol. 19 (suppl. 2): 13–21. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J., Gabaldón T. (2011). Assigning duplication events to relative temporal scales in genome-wide studies. Bioinformatics 27: 38–45. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J., Capella-Gutiérrez S., Pryszcz L.P., Marcet-Houben M., Gabaldón T. (2014). PhylomeDB v4: zooming into the plurality of evolutionary histories of a genome. Nucleic Acids Res. 42: D897–D902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo K.R., Kleff S., Amundsen K.L., Zhang X., Haake V., Zhang J.Z., Deits T., Thomashow M.F. (2001). Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 127: 910–917. [PMC free article] [PubMed] [Google Scholar]
- Johnston S.A., den Nijs T.P., Peloquin S.J., Hanneman R.E. Jr. (1980). The significance of genic balance to endosperm development in interspecific crosses. Theor. Appl. Genet. 57: 5–9. [DOI] [PubMed] [Google Scholar]
- Lanfermeijer F.C., Dijkhuis J., Sturre M.J.G., de Haan P., Hille J. (2003). Cloning and characterization of the durable tomato mosaic virus resistance gene Tm-2(2) from Lycopersicon esculentum. Plant Mol. Biol. 52: 1037–1049. [DOI] [PubMed] [Google Scholar]
- Lee B.-H., Henderson D.A., Zhu J.-K. (2005). The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17: 3155–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C.J. Jr., Roos D.S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Shi Y., Yuan Y., Hu X., Zhang H., Li N., Li Z., Chen Y., Mu D., and Fan W. (2012). Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects, http://arxiv.org/abs/1308.2012.
- Lukashin A.V., Borodovsky M. (1998). GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26: 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Force A. (2000). The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A., Nguyen A.H., Nakaminami K., Seki M. (2013). Arabidopsis non-coding RNA regulation in abiotic stress responses. Int. J. Mol. Sci. 14: 22642–22654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mboup M., Fischer I., Lainer H., Stephan W. (2012). Trans-species polymorphism and allele-specific expression in the CBF gene family of wild tomatoes. Mol. Biol. Evol. 29: 3641–3652. [DOI] [PubMed] [Google Scholar]
- Michael T.P., Jackson S. (2013). The first 50 plant genomes. The Plant Genome 6: 1–7. [Google Scholar]
- Micheletto S., Boland R., Huarte M. (2000). Argentinian wild diploid Solanum species as sources of quantitative late blight resistance. Theor. Appl. Genet. 101: 902–906. [Google Scholar]
- Novák P., Neumann P., Pech J., Steinhaisl J., Macas J. (2013). RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29: 792–793. [DOI] [PubMed] [Google Scholar]
- Palta J.P., Simon G. (1993). Breeding potential for improvement of freezing stress resistance: genetic separation of freezing tolerance, freezing avoidance, and capacity to cold acclimate. In Advances in Plant Cold Hardiness, Li P.H., ed (Boca Raton, FL: CRC Press; ), pp. 299–310. [Google Scholar]
- Peele H.M., Guan N., Fogelqvist J., Dixelius C. (2014). Loss and retention of resistance genes in five species of the Brassicaceae family. BMC Plant Biol. 14: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennycooke J.C., Cheng H., Roberts S.M., Yang Q., Rhee S.Y., Stockinger E.J. (2008). The low temperature-responsive, Solanum CBF1 genes maintain high identity in their upstream regions in a genomic environment undergoing gene duplications, deletions, and rearrangements. Plant Mol. Biol. 67: 483–497. [DOI] [PubMed] [Google Scholar]
- Potato Genome Sequencing Consortium; Xu X., et al. (2011). Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195. [DOI] [PubMed] [Google Scholar]
- Ramsay N.A., Glover B.J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10: 63–70. [DOI] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. (2002). Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Rodríguez F., Spooner D.M. (2009). Nitrate reductase phylogeny of potato (Solanum sect. Petota) genomes with emphasis on the origins of the polyploid species. Syst. Bot. 34: 207–219. [Google Scholar]
- Rymarquis L.A., Kastenmayer J.P., Hüttenhofer A.G., Green P.J. (2008). Diamonds in the rough: mRNA-like non-coding RNAs. Trends Plant Sci. 13: 329–334. [DOI] [PubMed] [Google Scholar]
- Sakamoto H., Maruyama K., Sakuma Y., Meshi T., Iwabuchi M., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 136: 2734–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibt K.M., Wenke T., Wollrab C., Junghans H., Muders K., Dehmer K.J., Diekmann K., Schmidt T. (2012). Development and application of SINE-based markers for genotyping of potato varieties. Theor. Appl. Genet. 125: 185–196. [DOI] [PubMed] [Google Scholar]
- Stanke M., Waack S. (2003). Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19 (suppl. 2): ii215–ii225. [DOI] [PubMed] [Google Scholar]
- Stevenson W.R., Loria R., Franc G.D., Weingartner D.P. (2001). Compendium of Potato Diseases, 2nd ed. (St. Paul, MN: American Phytopathological Society Press; ). [Google Scholar]
- Stockinger E.J., Mao Y., Regier M.K., Triezenberg S.J., Thomashow M.F. (2001). Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29: 1524–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J.M., Palta J.P., Bamberg J.B., Weiss L.S., Harbage J.F. (1993). Inheritance of freezing resistance in tuber-bearing Solanum species: evidence for independent genetic control of nonacclimated freezing tolerance and cold acclimation capacity. Proc. Natl. Acad. Sci. USA 90: 7869–7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vossen E.A.G., Gros J., Sikkema A., Muskens M., Wouters D., Wolters P., Pereira A., Allefs S. (2005). The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J. 44: 208–222. [DOI] [PubMed] [Google Scholar]
- Varshney R.K., et al. (2012). Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 30: 83–89. [DOI] [PubMed] [Google Scholar]
- Vitte C., Bennetzen J.L. (2006). Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc. Natl. Acad. Sci. USA 103: 17638–17643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhu Y., Wang L., Liu X., Liu Y., Phillips J., Deng X. (2009). A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter. Planta 230: 1155–1166. [DOI] [PubMed] [Google Scholar]
- Wang-Pruski G., and Schofield A. (2012). Potato: improving crop productivity and abiotic stress tolerance. In Improving Crop Resistance to Abiotic Stress, Tuteja N., Gill S.S., Tiburcio F.A., Tuteja R., eds (Weinheim, Germany: Wiley-VCH Verlag; ), pp. 1121–1153. [Google Scholar]
- Wenke T., Döbel T., Sörensen T.R., Junghans H., Weisshaar B., Schmidt T. (2011). Targeted identification of short interspersed nuclear element families shows their widespread existence and extreme heterogeneity in plant genomes. Plant Cell 23: 3117–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Murat F., Pont C., Langin T., Salse J. (2014). Paleo-evolutionary plasticity of plant disease resistance genes. BMC Genomics 15: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L.-Y., Guo X.-S., He B., Sun L.-J., Peng Y., Dong S.-S., Liu T.-F., Jiang S., Ramachandran S., Liu C.-M., Jing H.-C. (2011). Genome-wide patterns of genetic variation in sweet and grain sorghum (Sorghum bicolor). Genome Biol. 12: R114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo C., Wang T., Lu S., Zhao Y., Li X., Guo Z. (2013). A cold responsive galactinol synthase gene from Medicago falcata (MfGolS1) is induced by myo-inositol and confers multiple tolerances to abiotic stresses. Physiol. Plant. 149: 67–78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.