A homeostatic mechanism in Arabidopsis allows the lipid composition of the endoplasmic reticulum to directly control its rate of biogenesis.
Abstract
Regulation of membrane lipid biosynthesis is critical for cell function. We previously reported that disruption of PHOSPHATIDIC ACID PHOSPHOHYDROLASE1 (PAH1) and PAH2 stimulates net phosphatidylcholine (PC) biosynthesis and proliferation of the endoplasmic reticulum (ER) in Arabidopsis thaliana. Here, we show that this response is caused specifically by a reduction in the catalytic activity of the protein and positively correlates with an accumulation of its substrate, phosphatidic acid (PA). The accumulation of PC in pah1 pah2 is suppressed by disruption of CTP:PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE1 (CCT1), which encodes a key enzyme in the nucleotide pathway for PC biosynthesis. The activity of recombinant CCT1 is stimulated by lipid vesicles containing PA. Truncation of CCT1, to remove the predicted C-terminal amphipathic lipid binding domain, produced a constitutively active enzyme. Overexpression of native CCT1 in Arabidopsis has no significant effect on PC biosynthesis or ER morphology, but overexpression of the truncated constitutively active version largely replicates the pah1 pah2 phenotype. Our data establish that membrane homeostasis is regulated by lipid composition in Arabidopsis and reveal a mechanism through which the abundance of PA, mediated by PAH activity, modulates CCT activity to govern PC content.
INTRODUCTION
Phospholipids are a major component of most eukaryotic cell membranes, and their amphipathic properties are instrumental to the formation of lipid bilayers. In plants, as in other eukaryotes, both the quantity and composition of phospholipids must be tightly regulated to allow membranes to maintain their structure and function (Ohlrogge and Browse, 1995). Research performed in mammalian and yeast (Saccharomyces cerevisiae) cells has revealed intricate lipid sensing and signaling pathways that play a critical role in membrane homeostasis and govern phospholipid metabolism at both the transcriptional and posttranslational levels (Rawson, 2003; Jackowski and Fagone, 2005; Kent, 2005; Carman and Henry, 2007; Sugimoto et al., 2008; Carman and Han, 2009). Equivalent mechanisms must also exist in plants (Wang and Moore, 1990; Xu et al., 2005), but they remain comparatively poorly defined at the molecular level. Understanding these regulatory mechanisms is important given that membrane lipids play a central role in so many aspects of plant biology (Ohlrogge and Browse, 1995).
The Mg(2+)-dependent form of the enzyme phosphatidic acid phosphatase (PAP; EC 3.1.3.4) has recently emerged as an important player in eukaryotic lipid homeostasis with both structural and regulatory roles (Finck et al., 2006; Carman and Henry, 2007; Donkor et al., 2009; Pascual and Carman, 2013). Yeast contains a single PAP protein called Pah1p (Han et al., 2006), while mammals possess a small family of proteins termed lipin1, 2, and 3 (Péterfy et al., 2001). PAP catalyzes an essential step in the Kennedy pathway that is responsible for glycerolipid assembly. It converts phosphatidic acid (PA) to diacylglycerol (DG), which then serves as a substrate for both phospholipid and triacylglycerol (TG) biosynthesis (Carman and Henry, 2007). However, PAP also participates in signaling pathways that govern the expression of multiple genes associated with membrane and storage lipid metabolism (Finck et al., 2006; Carman and Henry, 2007; Donkor et al., 2009; Pascual and Carman, 2013).
The signaling function of PAP has been best characterized in yeast, where it is dependent on the protein’s catalytic activity (Pascual and Carman, 2013). Pah1p is an amphitropic enzyme that can interconvert between a soluble inactive form that is phosphorylated and a membrane-bound active form that is dephosphorylated (Choi et al., 2011). Recruitment of the phosphorylated enzyme to the nuclear-endoplasmic reticulum (ER) membrane is dependent on the nuclear envelope morphology 1–sporulation 7 (Nem1p-Spo7p) protein phosphatase complex, which dephosphorylates Pah1p and allows for its association with the membrane through a process mediated by a short N-terminal amphipathic helix (Karanasios et al., 2010; Choi et al., 2011). Pah1p is phosphorylated by several protein kinases, including the cyclin-dependent kinases cell division cycle 28 (Cdc28p) and phosphate metabolism 85 (Pho85p) (Choi et al., 2011, 2012). Crosstalk between these two kinases allows for regulation of a host of cellular processes in response to environmental stimuli (Enserink and Kolodner, 2010). Pah1p is also a substrate for protein kinase A (Su et al., 2012) and protein kinase C (Su et al., 2014). The former is a principal mediator of metabolic status signals transmitted through the cyclic adenosine monophosphate pathway in yeast (Thevelein, 1994).
A reduction in Pah1p activity (as found in the pah1Δ mutant) leads to elevated levels of its substrate PA, massive expansion of the nuclear-ER membrane, and abnormally high expression of key phospholipid biosynthetic genes (Santos-Rosa et al., 2005; Han et al., 2006, 2007). Conversely, overexpression of PAH1 leads to repression of phospholipid biosynthetic gene expression, and this response can be abolished by specific mutation of the Pah1p catalytic site (O’Hara et al., 2006; Han et al., 2007). Many yeast genes that encode phospholipid biosynthetic enzymes contain upstream activating sequence inositol-responsive (UASINO) elements in their promoters (Carman and Henry, 2007). Their expression is enhanced by interaction of the inositol requiring 2–inositol requiring 4 (Ino2p-Ino4p) transcription factor complex with UASINO elements, whereas it is blocked by interaction of the repressor protein overproducer of inositol 1 (Opi1p) with Ino2p (Carman and Henry, 2007). PA levels influence the Opi1p-mediated repression of gene expression because PA tethers Opi1p at the nuclear-ER membrane, together with a vesicle-associated protein homolog called suppressor of choline sensitivity 2 (Loewen et al., 2004). A reduction in PA concentration promotes translocation of Opi1p from the nuclear-ER membrane into the nucleus, where it interacts with Ino2p to repress gene expression and reduce the rate of phospholipid biosynthesis (Loewen et al., 2004).
PAP has been characterized in many eukaryotes, including plants (Pascual and Carman, 2013). Arabidopsis thaliana contains two genes, called PAH1 and PAH2, that encode proteins with PAP activity (Nakamura et al., 2009; Eastmond et al., 2010), and PAH1 has been shown to complement the yeast pah1Δ mutant (Mietkiewska et al., 2011). We have previously shown that these genes also act redundantly to repress phospholipid biosynthesis at the ER (Eastmond et al., 2010). Leaves from pah1 pah2 double mutants contain almost twice as much phospholipid as the wild type (Eastmond et al., 2010; Wang et al., 2014) and exhibit gross changes in ER morphology, which are consistent with massive endomembrane proliferation (Eastmond et al., 2010). An increase in the net rate of phosphatidylcholine (PC) biosynthesis (Eastmond et al., 2010; Wang et al., 2014) and in gene expression and enzyme activities associated with this pathway are also evident in pah1 pah2 (Eastmond et al., 2010).
The aim of this study was to investigate the mechanism by which PAP represses phospholipid biosynthesis and ER membrane biogenesis in Arabidopsis. Using catalytic site mutants, we found that regulation of these processes relies on PAP activity, suggesting that they are likely to be mediated by local changes in the level of PA, as has been shown previously in yeast (O’Hara et al., 2006; Han et al., 2007). However, our analysis of gene expression in Arabidopsis suggested that transcriptional responses to a reduction in PAP activity are very limited and cannot account for the elevated rate of PC biosynthesis and ER membrane proliferation observed in all tissues. Instead, our data support a biochemical regulatory mechanism in which PA accumulation in the ER membrane directly activates the enzyme CYTIDINE 5′-TRIPHOSPHATE (CTP):PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE (CCT), which is believed to catalyze the rate-limiting step of the nucleotide pathway of PC biosynthesis (Kinney et al., 1987; Tasseva et al., 2004). We found that relieving CCT1 of its sensitivity to PA through deletion of its putative lipid binding domain (Wang and Kent, 1995) produced a constitutively active enzyme, which can directly trigger PC overproduction and ER membrane proliferation in Arabidopsis, thereby phenocopying the pah1 pah2 mutant.
RESULTS
PAH1 Is Inactivated by Replacing Aspartic Acid Residues in the Catalytic Motif
Yeast Pah1p and mammalian lipin1 both regulate cellular lipid metabolism, but their mechanisms of action have been reported to differ. In yeast, Pah1p appears to function almost exclusively through its catalytic activity to modulate the levels of PA (O’Hara et al., 2006; Han et al., 2007), which in turn influence gene expression (Loewen et al., 2004). However, in mammals, lipin1 can also regulate gene expression independently of its catalytic function by migrating to the nucleus, where it functions as a transcriptional coactivator by interacting with members of the peroxisome proliferator-activated receptor family (Finck et al., 2006; Donkor et al., 2009). Interestingly, Mietkiewska et al. (2011) have reported that Arabidopsis PAH1 can also translocate to the nucleus when expressed in yeast supplemented with oleic acid. In both mammalian and yeast systems, mutants in the active site of the proteins have been used as a tool to distinguish catalytic from noncatalytic signaling functions (Finck et al., 2006; O’Hara et al., 2006). To investigate how PAP regulates phospholipid biosynthesis and ER biogenesis in plants, we attempted to complement the Arabidopsis pah1 pah2 mutant with wild-type PAH1, expressed under the control of the constitutive CaMV (cauliflower mosaic virus) 35S promoter, and with a mutated form (PAH1Δcat) containing D707E and D709E substitutions within the conserved catalytic motif DI/VDGT (Finck et al., 2006; Han et al., 2007; Mietkiewska et al., 2011).
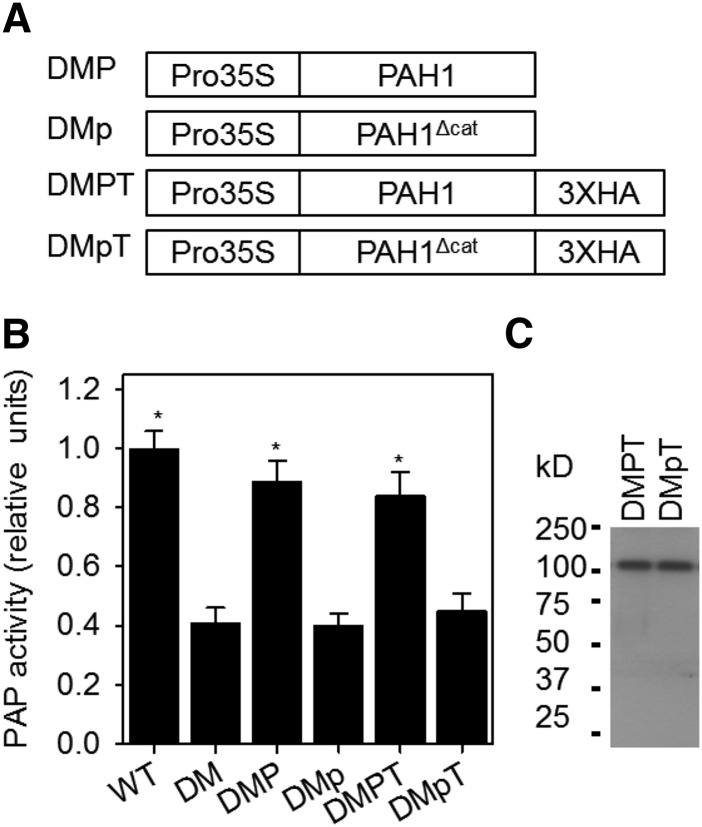
To simplify our analysis, we chose to use roots rather than leaves as an experimental system because phospholipids are the predominant class in this tissue, constituting ∼90% of total lipid content (Ohlrogge and Browse, 1995). Due to the increase in phospholipid production in pah1 pah2 roots, they exhibit an ∼1.8-fold increase in total fatty acid (FA) content (Eastmond et al., 2010). PAP activity in root extracts from pah1 pah2 was reduced to ∼40% of that in the wild type (Figure 1A). Nakamura et al. (2009) previously reported a similar reduction in pah1 pah2 leaves and proposed that the residual activity is due to the presence of other phospholipid phosphatases. Expression of both PAH1 and PAH1-HA (hemagglutinin tag) in pah1 pah2 restored PAP activity to near wild-type levels, but expression of PAH1Δcat and PAH1Δcat-HA failed to restore PAP activity, which did not differ significantly from that in pah1 pah2 (P > 0.05) (Figure 1A). Immunoblotting confirmed that lines expressing PAH1-HA and PAH1Δcat-HA produced similar amounts of fusion proteins (Figure 1B). These data suggest that in pah1 pah2 lines expressing PAH1Δcat, the protein is present but it is not catalytically functional. Therefore, the lines can be used as a tool to investigate the relationship between the activity of the protein and its influence over phospholipid biosynthesis and ER biogenesis.
Figure 1.
Effect of Mutation in the Catalytic Site of PAH1 on the Activity of the Protein in Vivo.
A diagram of the gene constructs transformed into pah1 pah2 (A), PAP activity (B), and protein abundance (C). Activity values are the mean ± se (n = 5) of measurements on crude extracts from roots of 4-week-old plants and are normalized on a per unit FW basis. Protein abundance was determined by immunoblotting using anti-HA antibodies. DM, pah1 pah2 double mutant; DMP, DM expressing wild-type PAH1; DMp, DM expressing mutated PAH1Δcat containing amino acid substitutions D707E and D709E in the conserved catalytic motif DI/VDGT; DMPT and DMpT, C-terminal HA tagged versions of DMP and DMp, respectively. Asterisk denotes a significant difference from DM (P > 0.05).
PAP Activity Represses PC Biosynthesis
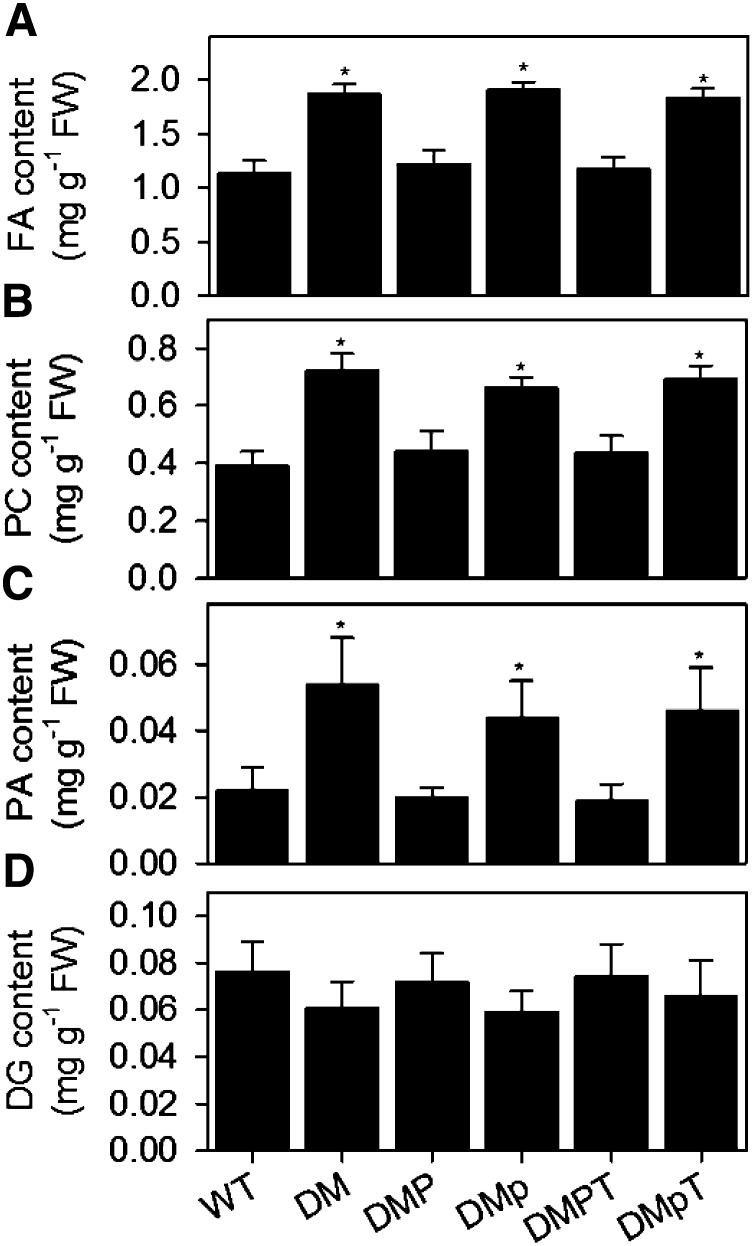
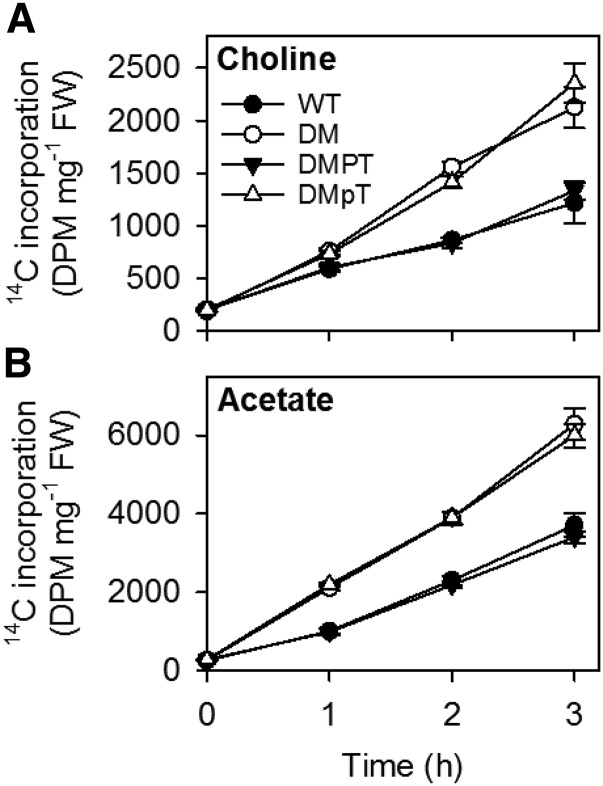
To investigate whether expression of the inactive form of PAH1 (PAH1Δcat) can repress PC production in pah1 pah2, lipid measurements were performed on roots (Figure 2). The total FA content and PC content was increased by ∼80% in pah1 pah2, when compared with the wild type (Figures 2A and 2B). Expression of PAH1 in pah1 pah2 largely suppressed these increases, but expression of PAH1Δcat led to no significant differences (P > 0.05) from pah1 pah2 alone (Figures 2A and 2B). Measurements of the precursor and product of PAP (PA and DG, respectively) showed that reduced PAP activity was associated with PA accumulation, but DG content was only marginally depleted (Figures 2C and 2D). Radiolabel feeding experiments using [1-14C]acetate and [methyl-14C]choline as substrates also showed that the rates of incorporation into total lipids and PC were ∼2-fold greater in pah1 pah2 roots than in the wild type on a fresh weight basis (Figures 3A and 3B). Expression of PAH1 in pah1 pah2 reduced the rates of incorporation into both products, whereas expression of PAH1Δcat did not (Figures 3A and 3B).
Figure 2.
Effect of Expression of Catalytically Active and Inactive PAH1 on Lipid Content in pah1 pah2.
The total fatty acid content (A) PC content (B), PA content (C), and DG content (C) was measured in roots from 4-week-old plants. Values are the mean ± se (n = 5). DM, pah1 pah2 double mutant; DMP, DM expressing wild-type PAH1; DMp, DM expressing mutated PAH1Δcat containing amino acid substitutions D707E and D709E in the conserved catalytic motif DI/VDGT; DMPT and DMpT, C-terminal HA-tagged versions of DMP and DMp, respectively. Asterisk denotes a significant difference from the wild type (P < 0.05).
Figure 3.
Effect of Expression of Catalytically Active and Inactive PAH1 on Incorporation of 14C-Labeled Precursors into Lipids in pah1 pah2.
Roots of 4-week-old plants were incubated with [methyl-14C]choline (A) and [1-14C]acetate (B), and the incorporation of 14C into lipids was determined over the course of 3 h. Values are the mean ± se (n = 5). DM, pah1 pah2 double mutant; DMPT, DM expressing wild-type PAH1 with a C-terminal HA tag; DMpT, DM expressing mutated PAH1Δcat containing amino acid substitutions D707E and D709E in the conserved catalytic motif DI/VDGT. At 1, 2, and 3 h, the values for DM and DMpT are significantly different from the wild type (P < 0.05).
PAP Activity Represses ER Proliferation
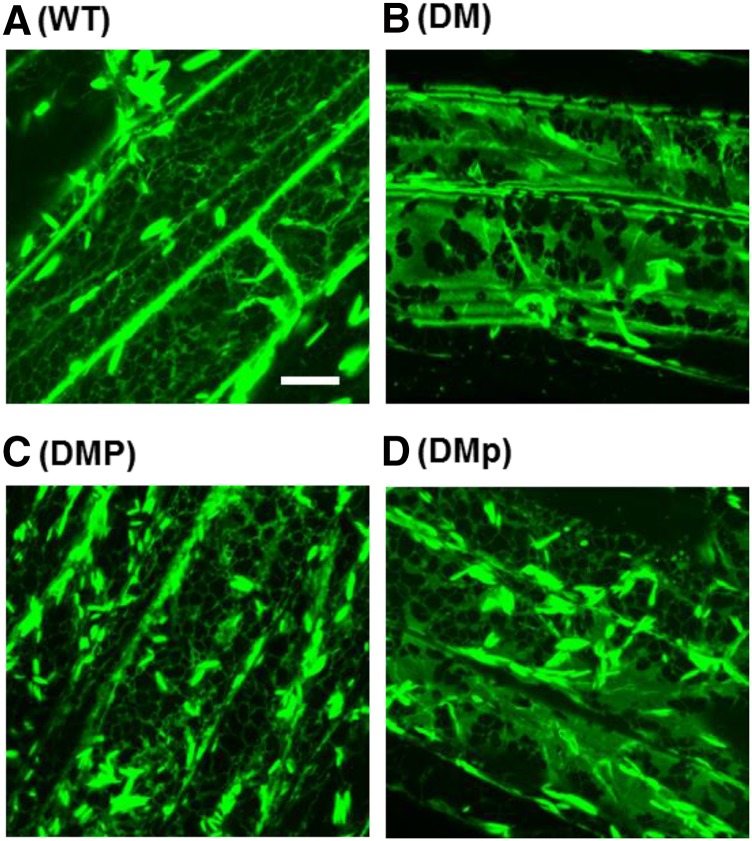
To confirm that PAP regulation of PC production is coupled to ER proliferation, red fluorescent protein (RFP) tagged with the C-terminal ER retention signal sequence HDEL was introduced into pah1 pah2 lines expressing PAH1 and PAH1Δcat (Eastmond et al., 2010). Laser-scanning confocal microscopy showed that ER morphology was normal in root epidermal cells of wild-type plants expressing RFP-HDEL and perturbed in pah1 pah2 plants expressing RFP-HDEL (Figure 4). Note that in all images, cigar-shaped ER bodies were observed, and these structures are well documented in Arabidopsis roots expressing ER marker proteins (Matsushima et al., 2003). The ER is usually connected to the nuclear envelope in plant cells and comprises sheet-like regions (cisternae) and an extensive cortical network of tubules, predominantly connected by three-way junctions (Staehelin, 1997). In pah1 pah2, it appears that the network of tubules, which usually makes up the majority of the ER, is partially replaced by ER sheets (Eastmond et al., 2010; Figure 4). Expression of PAH1 in pah1 pah2 reverted ER morphology to the wild type, but the ER remained abnormal in pah1 pah2 lines expressing PAH1Δcat (Figure 4). Therefore, PAP activity determines gross ER morphology, as well as phospholipid production.
Figure 4.
Effect of Expression of Catalytically Active and Inactive PAH1 on ER Morphology in pah1 pah2.
Laser-scanning confocal microscopy images of root epidermal cells from various genotypes expressing the ER lumen marker RFP-HDEL under the control of the CaMV 35S promoter. DM, pah1 pah2 double mutant; DMP, DM expressing wild-type PAH1; DMp, DM expressing mutated PAH1Δcat containing amino acid substitutions D707E and D709E in the conserved catalytic motif DI/VDGT. Bar = 10 μm.
PEAMT1 and CCT1 Are Required for Enhanced PC Production in pah1 pah2
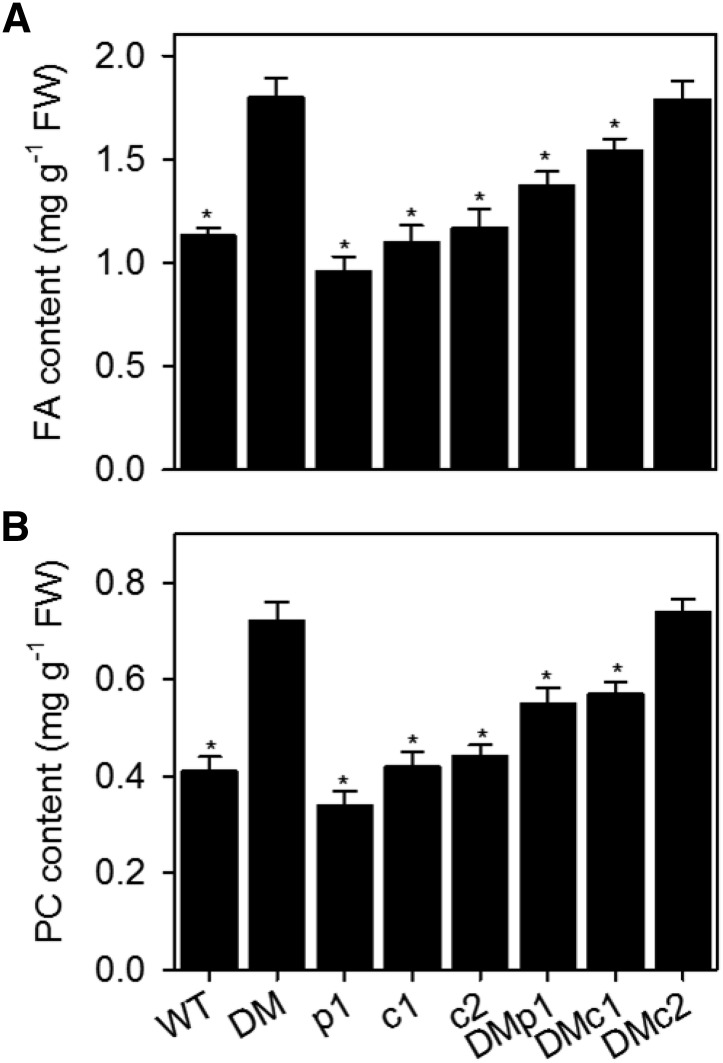
Our data suggest that repression of PC production by PAP in Arabidopsis is dependent on the catalytic activity of the protein and correlates positively with the accumulation of PA, as has been shown in yeast (O’Hara et al., 2006; Han et al., 2007). In yeast, PA acts by stimulating the expression of key genes encoding enzymes of the methylation pathway that biosynthesizes PC from phosphatidylethanolamine (O’Hara et al., 2006; Han et al., 2007). However, the pathways for PC biosynthesis in yeast and Arabidopsis are markedly different. In Arabidopsis, choline is biosynthesized by the sequential methylation of ethanolamine, predominantly in its phosphorylated form, to produce phosphocholine (P-Cho), which is then converted to PC by the nucleotide pathway using CTP phosphocholine cydylyltransferase and aminoalcohol phosphotransferase (Keogh et al., 2009). Our previous study suggested that PHOSPHOETHANOLAMINE N-METHYLTRANSFERASE1 (PEAMT1/XIPOTL1) and CCT are the most likely regulatory targets of PAP action in Arabidopsis leaves (Eastmond et al., 2010). To investigate whether these proteins are necessary for the derepression of PC biosynthesis that occurs in pah1 pah2 roots, T-DNA insertion mutants in each gene (peamt1, cct1, and cct2) were obtained (Cruz-Ramírez et al., 2004; Inatsugi et al., 2009), and triple mutants were constructed with pah1 pah2. Disruption of PEAMT1 or CCT1 partially suppressed the increase in FA and PC content observed in pah1 pah2 roots, while disruption of CCT2 had no significant effect (Figure 5). PEAMT1 and CCT1 could therefore play a role in PAP-mediated regulation of PC biosynthesis in Arabidopsis.
Figure 5.
Effect of PEAMT1, CCT1, and CCT2 Disruption on Lipid Content in pah1 pah2.
The total fatty acid content (A) and PC content (B) was measured in roots of 4-week-old plants. Values are the mean ± se (n = 5). DM, pah1 pah2 double mutant; p1, peamt1; c1, cct1; c2, cct2; DMp1, pah1 pah2 peamt1; DMc1, pah1 pah2 cct1; DMc2, pah1 pah2 cct2. Asterisk denotes a significant difference from DM (P > 0.05).
Transcriptional Induction of PEAMT1 Is Not Necessary for Enhanced PC Production in pah1 pah2
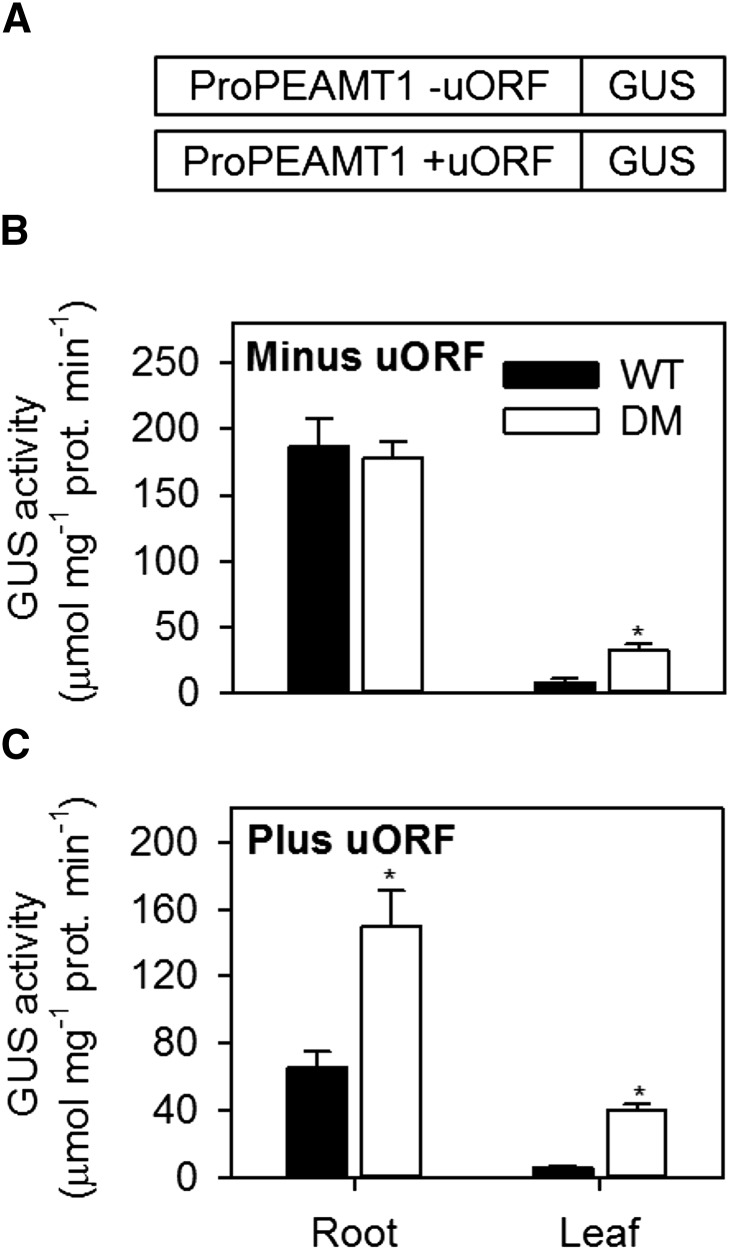
Our previous survey of phospholipid biosynthetic gene expression in leaves suggested that PEAMT1 transcript abundance was substantially increased in pah1 pah2 (Eastmond et al., 2010). PEAMT1 catalyzes the first committed step in choline biosynthesis in plants and is subject to transcriptional, translational, and biochemical regulation (Tabuchi et al., 2006; BeGora et al., 2010; Eastmond et al., 2010; Alatorre-Cobos et al., 2012). Of particular interest is the 5′ untranslated region (5′UTR) of PEAMT1, which contains an upstream open reading frame (uORF). This uORF enables translational repression of PEAMT1 by P-Cho without significantly affecting its steady state transcript abundance (Tabuchi et al., 2006; Alatorre-Cobos et al., 2012). We previously investigated PEAMT1 regulation in leaves using transient expression from two ProPEAMT1:β-glucuronidase (GUS) constructs: one containing an intact 5′UTR and the other a 5′UTR carrying a mutation in the initiation codon of the uORF that prevents repression of GUS translation by P-Cho. Our data suggested that PEAMT1 is subject to transcriptional regulation by PAP as well as translational regulation by P-Cho (Eastmond et al., 2010).
To study the regulation of PEAMT1 in greater detail, stable homozygous transgenic lines were created containing each of the two ProPEAMT1:GUS constructs. The constructs were first transformed into the wild type and homozygous single insertion site lines were identified. These T-DNAs were then crossed into the pah1 pah2 background so that a direct comparison could be made between the two genotypes. Analysis of GUS by histochemical staining suggested that PEAMT1 promoter activity is strongest in wild-type roots, both in the presence and absence of the uORF, whereas staining was only very weak in leaves and was largely confined to the veins (Supplemental Figure 1). The spatial pattern of GUS expression was also very similar for the same ProPEAMT1:GUS T-DNA insertions in the pah1 pah2 background (Supplemental Figure 1). To obtain quantitative data, GUS assays were performed on whole root and leaf extracts (Figure 6). These data showed that promoter activity is generally >20-fold higher in wild-type roots than in leaves of the same genotype (Figure 6B). In the absence of a functional uORF, GUS activity was enhanced in pah1 pah2 leaves but was not significantly upregulated (P > 0.05) in roots (Figure 6B). A comparison of GUS activity between genotypes harboring each of the two ProPEAMT1:GUS constructs also suggests that uORF-dependent repression of PEAMT1 translation occurs in both pah1 pah2 roots and leaves (Figure 6). On the basis of these data, the stimulation of PC production observed in root tissues of pah1 pah2 cannot be explained by changes in PEAMT1 gene expression but could rely on derepression of translation in response to increased utilization of P-Cho (Tabuchi et al., 2006; Eastmond et al., 2010; Alatorre-Cobos et al., 2012).
Figure 6.
ProPEAMT1:GUS Expression in Roots and Leaves of Wild-Type and pah1 pah2 Plants.
GUS assays were performed on roots and leaves of 4-week-old wild-type and DM (pah1 pah2) plants containing ProPEAMT1:GUS constructs (A) without (B) or with (C) a functional uORF in the 5′UTR. Single insertion site transgenic lines were first created by transforming the wild type, and the T-DNAs were then introduced into the pah1 pah2 background by crossing. Activities are the mean ± se of measurements on tissue from four separate plants. Asterisk denotes a significant difference from the wild type (P < 0.05).
The pah1 pah2 Mutant Exhibits Very Limited Changes in Gene Expression
Given that PEAMT1 does not appear to be transcriptionally upregulated in the roots of pah1 pah2, we performed a wider analysis of gene expression in this tissue. Real-time PCR analysis in pah1 pah2 leaves previously revealed only modest upregulation of other genes associated with phospholipid biosynthesis (Eastmond et al., 2010). Analysis of the same 13 genes in pah1 pah2 roots showed that in this tissue none were more than 2-fold upregulated (Supplemental Table 1). Therefore, a global survey of gene expression in whole 2-week-old plants was performed using the Affymetrix Ath1 chip to assess the wider impact that disruption of PAP activity might have on transcript abundance (Supplemental Data Set 1). Using Extraction of Differential Gene Expression (EDGE) software (Leek et al., 2006), only 14 genes were found with significant changes in expression (P < 0.01) that were also more than 2-fold up- or downregulated. The most strongly upregulated gene in this list was PEAMT1 (∼3-fold), which was also the only gene listed that is known to be associated with phospholipid biosynthesis. These data suggest that the global transcriptional response to disruption of PAP in Arabidopsis is very limited, despite the gross changes observed in lipid content and ER membrane morphology (Eastmond et al., 2010).
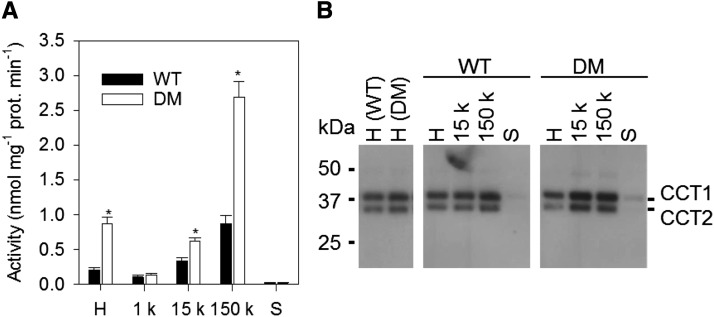
Microsomal CCT Activity Increases in pah1 pah2 but Protein Abundance and Localization Are Unchanged
We have previously shown that the enhanced production of PC in pah1 pah2 is accompanied by a >2-fold increase in CCT activity in crude leaf extracts and that radiolabeling experiments suggest this enzyme (which uses P-Cho as substrate) may also be rate-limiting for PC biosynthesis (Kinney et al., 1987; Tasseva et al., 2004; Eastmond et al., 2010). To investigate whether posttranscriptional regulation of CCT might explain the pah1 pah2 phenotype, we measured CCT activity and protein abundance in subcellular fractions of wild-type and pah1 pah2 root extracts. Analysis of a crude homogenate confirmed that total CCT activity is >2-fold higher in pah1 pah2 (Figure 7A). However, immunoblotting showed that CCT1 and CCT2 protein abundance is similar in wild-type and pah1 pah2 roots (Figure 7B). Fractionation experiments performed on the homogenates (Inatsugi et al., 2009) showed that the majority of CCT activity is associated with microsomal membranes (150,000g pellet) in both wild-type and pah1 pah2 tissues and that the activity in this fraction also increases ∼3-fold in pah1 pah2 (Figure 7A). CCT1 and CCT2 proteins were also readily detected in the microsomal membrane fraction from the wild type and pah1 pah2 but they were only present at trace levels in the soluble (150,000g supernatant) fraction (Figure 7B). These data suggest that the increase in CCT activity observed in pah1 pah2 occurs in the microsomal membrane fraction and is due to activation of the protein rather than to an increase in either total protein abundance or the degree of membrane association.
Figure 7.
Subcellular Distribution of CCT in Roots of Wild-Type and pah1 pah2 Plants.
Specific activity of CCT (A) and protein abundance (B) in subcellular fractions. H, crude homogenate; 1 k, 1000g pellet; 15 k, 15,000g pellet; 150 k, 150,000g pellet; S, supernatant. Values are the mean ± se of measurements on four separate fractionations. CCT proteins were detected by immunoblotting using anti-CCT1 antibodies. Asterisk denotes a significant difference from the wild type (P < 0.05).
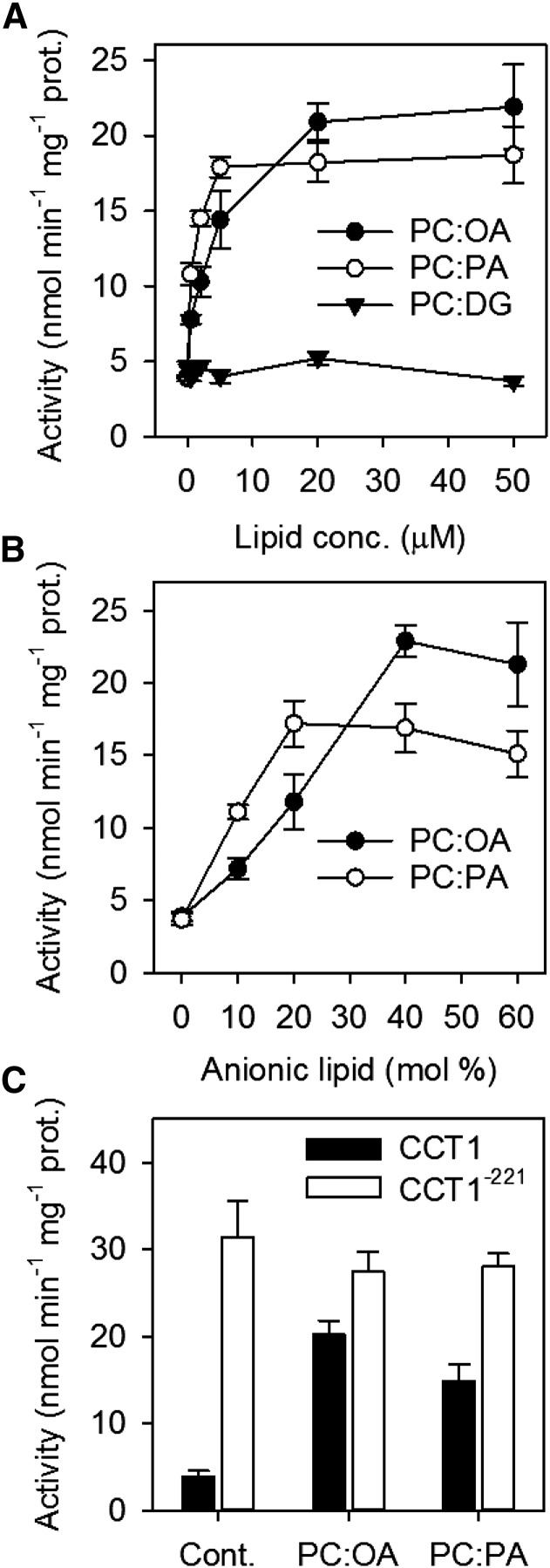
CCT1 Is Activated in Vitro by Lipids and Is Highly Sensitive to PA
In mammals, allosteric regulation of CCTα plays a key role in controlling PC biosynthesis (Cornell and Northwood, 2000). CCTα is an amphitropic enzyme that can interconvert between a soluble inactive form and a membrane-bound active form. When CCTα is membrane bound, its activity is stimulated by the presence of lipids such as free fatty acids and PA, and also by DG (Cornell and Northwood, 2000). In Arabidopsis, the bulk of CCT1 and CCT2 activity and protein appears to be membrane associated (Inatsugi et al., 2009; Figure 7), and several studies suggest that CCT activity in plants can be activated by lipids (Kinney and Moore, 1987; Wang and Moore, 1990).
To investigate whether Arabidopsis CCT1 activity is regulated by lipids, the protein was expressed in the yeast pct1Δ mutant, which is deficient in CCT activity (Nishida et al., 1996; Choi et al., 1997). In vitro enzyme assays were performed on fractionated cell extracts (Nishida et al., 1996). The majority of CCT activity (∼80%) was membrane associated (150,000g pellet) but a significant proportion was soluble (Nishida et al., 1996). The addition of lipid vesicles containing a 1:1 ratio of PC to either oleic acid or PA stimulated the activity in the soluble (150,000g supernatant) fraction >3-fold, with a maximum activity attained at ∼20 and ∼5 μM, respectively (Figure 8A). By contrast, vesicles containing DG did not stimulate this activity (Figure 8A). When an optimum lipid vesicle concentration was maintained, varying the molar ratio of PC to either oleic acid or PA affected enzyme stimulation. Vesicles containing PC alone had no effect, but activity was progressively stimulated with increasing oleic acid and PA content up to ∼40 and 20 mol %, respectively (Figure 8B). The kinetic parameters Vmax and Km were also determined for CCT1 in the presence and absence of lipids vesicles containing a 1:1 ratio of PC to either oleic acid or PA. The Vmax values in the presence of oleic acid and PA were ∼24 and ∼18 nmol min−1 mg−1 protein, respectively, while the Vmax value in their absence was ∼4 nmol min−1 mg−1 protein (Supplemental Table 2). The Km values for P-Cho and CTP were ∼0.6 and ∼0.2 mM, respectively, and were not appreciably affected by the presence of either oleic acid or PA (Supplemental Table 2). The activation of CCT1 by both oleic acid and PA is therefore accompanied by an increase in the Vmax value, which has also been observed for CCT from several other organisms (Friesen et al., 1999, 2001a, 2001b). Given that suppression of PAP catalytic function leads to accumulation of the substrate PA (Eastmond et al., 2010; Figure 2C), it is plausible that direct activation of CCT1 by membranes enriched with PA (and possibly other anionic lipids derived from this metabolite) is the major mechanism that stimulates PC production in pah1 pah2 tissues.
Figure 8.
Effect of Lipids on the Activity of Recombinant CCT1.
Influence of lipid vesicle concentration (A), composition (B), and protein C-terminal truncation (C) on CCT activity from the 100,000g supernatant of yeast pct1Δ cell expressing CCT1. Vesicles with a 1:1 ratio of PC to oleic acid (OA), PA, or DG were used in (A) and (C). Values are the mean ± se of measurements for four separate assays.
Truncation of the C Terminus of CCT1 Leads to Constitutive Activity
In mammalian CCTα, removal of the amphipathic C-terminal lipid binding domain (“M”) prevents membrane interaction and causes the enzyme to become lipid insensitive and constitutively active (Wang and Kent, 1995; Friesen et al., 1999). Arabidopsis CCT1 and CCT2 contain a shorter predicted amphipathic α-helical region that may play a similar role, although this hypothesis has not been tested experimentally (Braker et al., 2009; Inatsugi et al., 2009). To investigate whether removal of this region from CCT1 affects the protein’s activity, a truncated version of CCT1 containing only the first 221 amino acid residues (CCT1−211) was expressed in the yeast pct1Δ mutant. In fractionated cell extracts, the majority of CCT activity (>90%) was found to be soluble (150,000g supernatant). This supports the hypothesis that the C-terminal portion of CCT1 is involved in membrane binding (Braker et al., 2009; Inatsugi et al., 2009). The addition of lipids vesicles containing a 1:1 ratio of PC to either oleic acid or PA did not lead to a further stimulation of activity in this fraction (Figure 8C), suggesting that CCT1−211 is constitutively active and lipid insensitive.
Expression of Truncated CCT1−211 in Arabidopsis Phenocopies pah1 pah2
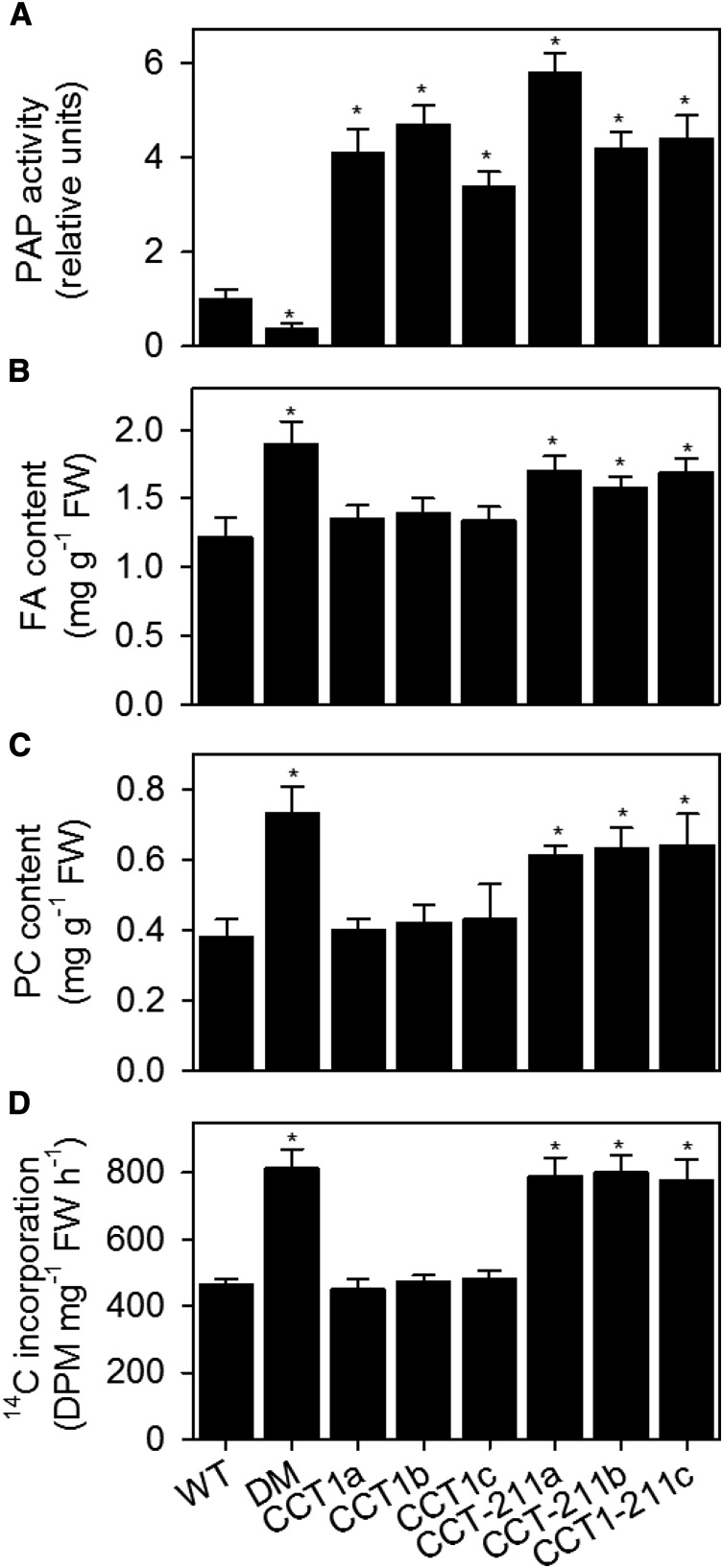
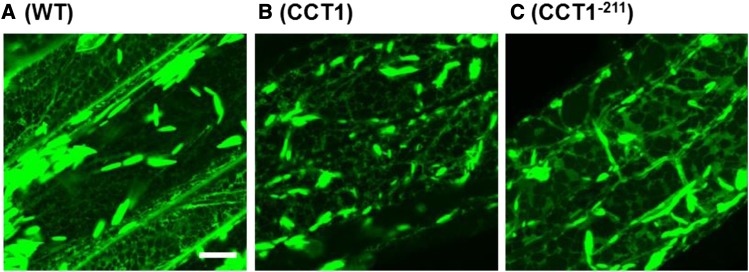
In mammalian cells, expression of CCTα can lead to a small stimulation of PC biosynthesis (Walkey et al., 1994), but this effect becomes far more pronounced in cells harboring the constitutively active form of the protein that lacks the M domain (Wang and Kent, 1995). Therefore, to determine whether CCT activation is sufficient to account for the increased production of PC observed in pah1 pah2, CCT1 and CCT1−211 were expressed under the control of the constitutive CaMV 35S promoter in wild-type Arabidopsis plants. Three independent homozygous transgenic lines were isolated for each construct. Enzyme assays performed on crude root homogenates confirmed that CCT activity was increased between 3- and 6-fold in all lines (Figure 9A). The total FA content, PC content, and the initial rate of [methyl-14C]choline incorporation into PC were measured in the roots of each line. The FA content was increased by 30 to 50% in plants expressing CCT1−211, when compared with the wild type (Figure 9B). By contrast, expression of CCT1 did not lead to a significant (P > 0.05) increase in root FA content (B). The increase in FA content in plants expressing CCT1−211 was also accompanied by a higher PC content and an enhanced rate of [methyl-14C]choline incorporation into PC (Figures 9C and 9D). Finally, to determine whether expression of CCT1−211 also causes ER proliferation, the ER lumen marker RFP-HDEL was introduced into one line of each genotype. Laser-scanning confocal microscopy showed that ER morphology in root epidermal cells was perturbed (Figure 10) and had a similar morphology to that observed in pah1 pah2 (Figure 4). These data support the hypothesis that CCT1 activity modulates the production of PC in response to the relative abundance of PA (and other anionic lipids) in the ER.
Figure 9.
Effect of CCT1 Overexpression on PC Content.
The PAP activity (A), total fatty acid content (B), PC content (C), and rate of [methyl-14C]choline incorporation into lipids (D) were measured in roots from 4-week-old plants and are expressed on a per unit FW basis. Values are the mean ± se (n = 5). DM, pah1 pah2; CCT1a, b, and c, independent homozygous lines expressing CCT1 under the CaMV 35S promoter; CCT1−211a, b, and c, independent homozygous lines expressing C-terminal truncated CCT1 under the CaMV 35S promoter. Values are the mean ± se of measurements on four separate samples. Asterisk denotes a significant difference from the wild type (P < 0.05).
Figure 10.
Effect of CCT1 Overexpression on ER Morphology.
Laser-scanning confocal microscopy images of root epidermal cells from various genotypes expressing the ER lumen marker RFP-HDEL under the control of the CaMV 35S promoter. DM, pah1 pah2; CCT1 and CCT1−211, homozygous lines (a in Figure 9) expressing CCT1 and C-terminal truncated CCT1, respectively, under the control of the CaMV 35S promoter. Bar = 10 μm.
DISCUSSION
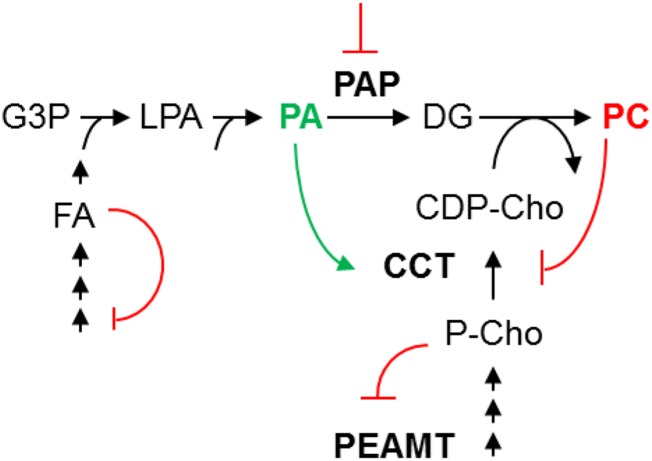
In this study, we show that direct PA activation of CCT activity is the primary mechanism that accounts for the pronounced increase in PC production and massive ER membrane proliferation observed in the Arabidopsis pah1 pah2 mutant (Eastmond et al., 2010). In vitro stimulation of plant CCT activity by lipids has been reported previously (Kinney and Moore, 1987; Wang and Moore, 1990; Parkin and Rolph, 1999), although it has not been observed in all studies (Price-Jones and Harwood, 1983, 1986). Here, we provide in vivo evidence that CCT does indeed function as a metabolic sensor in plant cells, monitoring membrane lipid composition and adjusting the rate of PC biosynthesis accordingly (Figure 11). This role appears to be analogous to that of CCTα in mammals (Cornell and Northwood, 2000). CCTα is an amphitropic enzyme that can interconvert between a soluble inactive form and a membrane-bound active form. The membrane binding domain of CCT is a long amphipathic α helix that responds to changes in the physical properties of membranes. Binding of this domain to membranes activates CCT by relieving an inhibitory constraint in the catalytic domain (Weinhold et al., 1991; Yang et al., 1995). The precise mechanism by which membrane lipid composition modulates CCTα activity is still not fully understood (Huang et al., 2013; Lee et al., 2014), but a relatively high abundance of anionic lipids, such as PA, are known to act as a stimulant (Arnold and Cornell, 1996; Cornell and Northwood, 2000).
Figure 11.
A Simplified Model Explaining the Proposed Biochemical Regulation of PC Production by PAP Activity in Arabidopsis Roots.
A reduction in PAP activity leads to an increase in the precursor (PA) pool in the ER. This enhances the ratio of PA to PC and stimulates CCT1 activity. Increased production of CDP-Cho by CCT1 promotes PC biosynthesis from DG by aminoalcohol phosphotransferase. Increased consumption of P-Cho by CCT1 also leads to enhanced P-Cho production through derepression of PEAMT1 translation and activity (Tabuchi et al., 2006; Alatorre-Cobos et al., 2012). Increased demand for fatty acids for glycerolipid biosynthesis also leads to derepression of fatty acid biosynthesis (Andre et al., 2012).
Although both mammalian CCTα and Arabidopsis CCT1 activities respond to lipids, there appear to be differences in both their structure and function. Both CCT1 and CCT2 are predicted to have shorter amphipathic α-helix domains than CCTα, and they also lack the phosphorylation domain, which in CCTα can block membrane binding (Braker et al., 2009; Inatsugi et al., 2009). Indeed, both CCT1 and CCT2 appear to maintain a tight membrane association in Arabidopsis (Inatsugi et al., 2009; Figure 7). In terms of their lipid sensitivity, CCTα is also activated by DG (Johnson et al., 1992), while CCT1 is insensitive (Figure 8). CCT1 also responds to far lower concentrations of lipid activators than does CCTα (Johnson et al., 1992). The differences in lipid responsiveness exhibited by CCT1 may be important for PAP-mediated regulation of PC biosynthesis in plants. Interestingly, biochemical studies on yeast PCT1 have previously shown that it is also highly sensitive to anionic lipids and fatty acids but insensitive to DG (Johnson et al., 1992; Friesen et al., 2001b). In yeast, the regulation of PC biosynthesis by Pah1p has been characterized predominantly as a transcriptional response of certain genes that encode phospholipid biosynthetic enzymes, to changes in PA levels (Carman and Henry, 2007). The target genes contain UASINO elements in their promoters and include those encoding key enzymes of the methylation pathway that biosynthesizes PC from phosphatidylethanolamine (O’Hara et al., 2006; Han et al., 2007). However, yeast PCT1, which encodes an enzyme of the nucleotide pathway, does not contain UASINO elements in its promoter and it does not appear to be differentially expressed in response to PA (Jesch et al., 2005; Carman and Henry, 2007). Given our evidence that biochemical regulation of CCT1 by PA plays a key role in controlling PC biosynthesis in Arabidopsis, it is possible that biochemical regulation of CCT1 might also contribute to Pah1p regulation of PC biosynthesis in yeast, despite the fact that the methylation pathway is usually predominant. In Caenorhabditis elegans, the downregulation of LIPIN-1 also leads to changes in ER morphology that are similar to those observed in Arabidopsis pah1 pah2 (Golden et al., 2009; Gorjánácz and Mattaj, 2009), and C. elegans CCT has also been shown to be highly sensitive to activation by PA (Friesen et al., 2001a).
Removal of the amphipathic α-helix lipid binding domains of CCT proteins from both mammals and C. elegans has been shown to abolish their sensitivity to lipids (Wang and Kent, 1995; Friesen et al., 1999, 2001a). Expression of a constitutively active form of CCT in mammalian cells stimulates PC biosynthesis, but no net increase in PC content is detected under these circumstances because PC turnover is enhanced to compensate (Wang and Kent, 1995). By contrast, PC content does increase substantially in Arabidopsis roots expressing a constitutively active form of CCT, suggesting that in this case PC turnover does not fully compensate (Figure 10). In pah1 pah2, net PC accumulation also occurs (Eastmond et al., 2010; Wang et al., 2014), despite evidence for an increased rate of turnover (Wang et al., 2014). How PC turnover is regulated in plants is still relatively poorly understood, and there are several possible pathways involving phospholipases A, C, or D or the reverse activity of lyso-PC acyltransferase (LPCAT). Wang et al. (2014) have recently shown that both LPCAT activity and the in vivo rate of PC deacylation are increased in pah1 pah2 mutant leaves. PC may also be used as a substrate for TG biosynthesis by PHOSPHOLIPID:DIACYLGLYCEROL ACYLTRANSFERASE1 (PDAT1), and recent work has shown that TG formed from PC in leaves is subsequently hydrolyzed by the lipase SUGAR-DEPENDENT1 and the fatty acids are β-oxidized (Fan et al., 2013, 2014).
Achieving the net increase in PC content observed in the Arabidopsis pah1 pah2 mutant, as well as in plants expressing a constitutively active form of CCT, requires enhanced provision of key metabolic precursors (Figure 11). In the case of fatty acids, recent studies have shown that creating a stronger metabolic sink in Arabidopsis leaves can enhance the rate of biosynthesis (Fan et al., 2013, 2014). Furthermore, acetyl-CoA carboxylase, which is considered to be the rate-limiting enzyme of fatty acid biosynthesis, is subject to feedback inhibition by oleoyl-acyl carrier protein, providing a possible mechanism to explain this effect (Andre et al., 2012). It may seem counterintuitive that the reduction in PAP activity in the pah1 pah2 mutant can give rise to an increase in net PC biosynthesis since the product of this reaction (DG) is a direct precursor (Figure 11). However, pah1 pah2 still contains a substantial amount of PAP activity that must be accounted for by other proteins (Nakamura et al., 2009). This is the case in the yeast pah1Δ mutant that also exhibits an increase in net PC biosynthesis, despite a reduction in PAP activity (Santos-Rosa et al., 2005; Han et al., 2006, 2007). It is believed that flux through the nucleotide pathway for PC biosynthesis is most limited by the provision of CDP-Cho by CCT rather than by the availability of DG, at least within the normal range of flux (Kinney et al., 1987; Tasseva et al., 2004). This is also consistent with the data presented in this study, which suggests that CCT activity as a major determinant of the net rate of PC biosynthesis in Arabidopsis.
CCT support of PC biosynthesis in Arabidopsis also requires the biosynthesis of P-Cho from ethanolamine catalyzed by PEAMT (Figure 11). Both PEAMT1 translation and PEAMT enzyme activity are inhibited by P-Cho (Tabuchi et al., 2006; BeGora et al., 2010; Alatorre-Cobos et al., 2012), providing a mechanism to balance substrate supply with the demand by CCT. We previously reported that PEAMT1 is also transcriptionally upregulated in pah1 pah2 leaves (Eastmond et al., 2010). Analysis of stable ProPEAMT1:GUS reporter lines in this study supports this finding, but also shows that PEAMT1 is relatively weakly expressed in leaves, where it is largely restricted to the veins. PEAMT1 is far more strongly expressed in roots, and in this tissue disruption of PAP does not lead to transcriptional induction of the gene. Hence, changes in PEAMT1 expression cannot account for the increased net rate of PC biosynthesis that is observed in both leaves and roots of pah1 pah2 (Eastmond et al., 2010). Since PEAMT1 is expressed at a low level in leaves, it may not be responsible for supplying the bulk of the P-Cho for PC biosynthesis in this tissue. Other PEAMT genes may contribute to the biosynthesis of P-Cho in leaves (BeGora et al., 2010). However, there is also evidence that P-Cho is transported in the xylem from the root (Gout et al., 1990).
Analysis of the transcript abundance of a set of genes associated with PC biosynthesis in Arabidopsis roots suggested that there are very limited changes in expression in pah1 pah2. In our previous study, we found that of these same genes only PEAMT1 was prominently upregulated in leaves (Eastmond et al., 2010). To investigate what effect PAP disruption has on global gene expression, we also performed microarray experiments on whole 2-week-old plants of pah1 pah2 using the Affymetrix Ath1 chip (Supplemental Data Set 1). Remarkably, given the pronounced changes in lipid composition and cell ultrastructure present in pah1 pah2 (Eastmond et al., 2010), very few genes were more than 2-fold up- or downregulated. PEAMT1 was the only differentially expressed gene with a known role in phospholipid biosynthesis. Wang et al. (2014) have recently shown that the transcript abundance of LPCAT1 and LPCAT2 increase ∼2-fold in pah1 pah2 leaves, as measured using real-time PCR, while those of PDAT1 and seven PLA2 genes do not change significantly. No significant upregulation of LPCAT1 and LPCAT2 was detected in our microarray experiment. However, this discrepancy is very likely due to the relatively low level of induction of these genes combined with the difference in methodologies and tissues used in each study. In general, the data suggest that transcription plays a very limited role in the deregulation of PC biosynthesis caused by PAP disruption in Arabidopsis. This appears to contrast with yeast where the pah1Δ mutant exhibits wide-scale transcriptional reprogramming of fatty acid and phospholipid metabolism (Carman and Henry, 2007; Pascual and Carman, 2013). Nevertheless the mechanism by which PEAMT1 is induced in pah1 pah2 leaves still warrants further study since this enzyme is specific to plants (Keogh et al., 2009). Our microarray data suggest that the next most strongly upregulated gene in pah1 pah2 after PEAMT1 is one or more of five highly homologous auxin response factors situated within a cluster on the top arm of chromosome 1. It is unclear which gene (or genes) within this group are differentially expressed because the probe lacks specificity. The precise function of these auxin response factor family members is also unknown (Okushima et al., 2005), but it is possible to speculate that one or more of these transcription factors might regulate PEAMT1.
In conclusion, our data establish that membrane homeostasis is regulated by lipid composition in Arabidopsis and reveal a mechanism through which the relative abundance of PA (and potentially other anionic lipids), determined by PAP activity, modulates CCT1 activity to determine PC content. This signaling pathway appears to be mechanistically different from that so far described in yeast because it uses predominantly biochemical rather than transcriptional regulation (Carman and Henry, 2007; Pascual and Carman, 2013). However, the overall effect is very similar in that the rate of phospholipid biosynthesis and ER membrane biogenesis can be regulated by the activity of PAP. In yeast, Pah1p activity (and membrane association) is regulated via phosphorylation by several protein kinases (i.e., Cdc28p, Pho85p, and protein kinase A and C) and dephosphorylation by the Nem1p-Spo7p phosphatase complex situated on the nuclear-ER membrane (Santos-Rosa et al., 2005; O’Hara et al., 2006). The phosphorylation status of Pah1p therefore coordinates phospholipid biosynthetic gene expression and growth of the nuclear-ER membrane with multiple developmental and environmental signals (O’Hara et al., 2006; Han et al., 2007; Pascual and Carman 2013). It will be interesting to determine whether Arabidopsis PAP proteins are regulated in a similar manner and in particular whether there might be functional coupling to the cell cycle as discovered in yeast (O’Hara et al., 2006; Han et al., 2007), perhaps via the Arabidopsis Cdc28p homolog CDKA;1.
METHODS
Plant Material and Growth Conditions
Wild-type Arabidopsis thaliana ecotype Columbia-0 and the pah1, pah2, peamt1, cct1, and cct2 mutants (SALK_042970, SALK_047457, SAIL_28_H12, GABI_349C03, and SAIL_71_H04) were obtained from the European Arabidopsis Stock Centre (University of Nottingham, UK). Seeds were surface sterilized, applied to agar plates containing half-strength MS salts (Sigma-Aldrich), and imbibed in the dark for 4 d at 4°C. The plates were then placed vertically in a growth chamber set to 21°C 16 h light/18°C 8 h dark; PPFD = 150 μmol m−2 s−1. T-DNA mutants were genotyped by performing PCR on genomic DNA (Eastmond et al., 2010) using primer pairs designed by T-DNA Express (http://signal.salk.edu/tdnaprimers.2.html).
Lipid Analysis
The total fatty acid content of root tissue was measured by gas chromatography analysis after combined digestion and fatty acid methyl ester formation using the method described by Browse et al. (1986). Lipids were extracted from 0.2 g fresh weight (FW) of root tissue according to the method of Dörmann et al. (1995). The extracts were applied to silica gel 60A plates (Merck) and analyzed by thin-layer chromatography (TLC). PC and PA were separated by two-dimensional TLC using chloroform/methanol/water (75:25:2.5, by volume), followed by chloroform/methanol/acetic acid/water (80:9:12:2, by volume). DG was separated by one-dimensional TLC using hexane/diethyl ether/acetic acid (70:30:1, by volume). Lipids were visualized under UV light having been sprayed with primulin solution (0.01% [w/v] in 60% [v/v] acetone). The PC, PA, and DG were identified by cochromatography with commercial standards (Sigma-Aldrich), and their fatty acid content was determined by gas chromatography analysis (Browse et al., 1986). To allow quantification, the lipid extracts were spiked with 15:0 TG, the TG band was recovered from TLC plates, and the absolute quantity of each lipid class was calculated based on the final recovery of 15:0.
Reporter Protein Analysis
Histochemical staining and quantitative assays for GUS were performed as described by Jefferson et al. (1987). For GUS imaging, the tissue was destained in ethanol, mounted in 50% (v/v) glycerol, and analyzed using the Zeiss Axiophot upright microscope. Images were acquired using a Q-Imaging Retiga digital camera and Metamorph software. For confocal laser microscopy, the material was mounted on a slide in water and imaged with a Zeiss LSM 780 using a 40× or 63× objective and an excitation wavelength of 561 nm and detected using an emission band of 570 to 650 nm. Protein extraction, quantification, SDS-PAGE, and immunoblotting were performed as described previously (Eastmond, 2004) except that anti-HA (or CCT) and anti-IgG-HRP (Invitrogen) were used as primary and secondary antibodies at 1 in 1000 and 1 in 10,000 dilutions, respectively, and HRP was detected using an Enhanced Chemiluminescence kit (Perkin-Elmer). The affinity-purified antibody used to detect CCT was raised against the synthetic oligopeptide CVGRFKETQRTEGIS by Eurogentec.
Radiolabel Assays
For radiolabel feeding experiments, roots (0.2 g FW) were cut into fragments using a razor blade and placed in vials containing 0.5 mL of half-strength MS salts supplemented with 1 mM [methyl-14C]choline chloride or [1-14C]acetate. The tissue was vacuum infiltrated for 5 min. Following incubation at 22°C with gentle agitation for up to 3 h, the tissue was gently blotted dry and frozen in liquid nitrogen. The extraction of lipids and analysis of labeled PC was conducted according to the methods described by Tasseva et al. (2004). PAP assays were performed on crude homogenates from roots according to the method described by Nakamura et al. (2009). CCT assays were performed on Arabidopsis and yeast extracts following the method described by Inatsugi et al. (2002) using [14C]phosphocholine substrate prepared from [methyl-14C]choline chloride as described by Cornell (1989). Subcellular fractionation of extracts from Arabidopsis roots was performed according to the protocol described by Inatsugi et al. (2009). To prepare lipid vesicles for enzyme assays, lipids dissolved in chloroform or chloroform/methanol were evaporated to dryness under a stream of nitrogen and suspended in 20 mM Tris/HCl (pH 7.5) with a probe sonicator. The rate of enzyme the reaction was confirmed to be linear with time for up to 30 min and also to be proportional to the amount of protein included in the assay.
Gene Expression Analysis
DNase-treated total RNA was isolated from Arabidopsis roots using the RNeasy kit from Qiagen. The synthesis of single-stranded cDNA was performed using SuperScript II RNase H- reverse transcriptase from Invitrogen. Quantitative real-time PCR was performed with an ICycler (Bio-Rad) using qPCR Mastermix Plus (Eurogentec), according to the manufacturer’s instructions, and the data were analyzed with the ICycler IQ5 software. Details of the methodology and primer sequences were reported by Eastmond et al. (2010). Microarray analysis was performed at the University of Warwick Genomics Facility using the Affymetrix Ath1 chip, according to the manufacturer’s protocols. Raw data were normalized by MAS5 (Affymetrix) to a target signal of 500.
Creation of DNA Constructs and Arabidopsis Transformation
Sequences encoding PAH1, CCT1, and CCT1−211 were amplified by PCR from Arabidopsis cDNA, and the products were cloned into the entry vector pDONR207 using the Gateway BP clonase enzyme mix from Invitrogen. PAH1Δcat was then created from the PAH1 template using the Quikchange Lightning site-directed mutagenesis kit from Stratagene. The primers used for cloning and mutagenesis are listed in Supplemental Table 3. The various gene cassettes were cloned into the destination vectors pEG100 and pEG101 (Earley et al., 2006) using the Gateway LR Clonase enzyme mix (Invitrogen). All plasmids were transformed into Agrobacterium tumefaciens strain GV3101 by heat shock and into Arabidopsis by the floral dip method (Clough and Bent, 1998). Transformants containing T-DNA insertions were selected via herbicide resistance.
Heterologous Expression of CCT1
For the heterologous expression of CCT1 and CCT−211 in yeast under a GAL1 promoter, the gene cassettes in the entry vector pDONR207 were cloned into the destination vector pYES-DEST52, using the Gateway LR clonase enzyme mix (Invitrogen). The plasmids were then transformed into yeast pct1Δ mutant (Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YGR202c::kanMX4) cells using the Invitrogen S. c. EasyComp kit. Protein expression was induced and yeast cells were grown to early-stationary phase as described in the Invitrogen pYES2 user manual. Yeast cells were harvested and washed with isolation buffer (50 mM Tris-HCl, pH 7.5, 300 mM sucrose, 2 mM DTT, and 0.5 mM phenylmethylsulfonyl fluoride), pelleted by centrifugation, and resuspended in isolation buffer. Cells were broken using three 60-s pulses with a bead mill using 0.5-mm glass beads. The homogenate was collected and briefly centrifuged to remove unbroken cells. Membrane and soluble fractions were obtained using 100,000g centrifugation of the 15,000g supernatant derived from the crude homogenate (Mietkiewska et al., 2011). The soluble fraction was concentrated by ultrafiltration.
Statistical Analysis
One-way ANOVA was used to assess the differences between genotypes or treatments. Following significant (P < 0.05) F-test results, means of interest were compared using the appropriate least significant difference value at the 5% (P = 0.05) level of significance, on the corresponding degrees of freedom. The GenStat statistical system (2011, 14th edition; VSN International) was used for these analyses. For analysis of microarray data, P and Q values for the comparison of genotypes were calculated using EDGE software (http://www.genomine.org/edge/), and a P = 0.1 cutoff was used to select the genes that were differentially regulated (Liu and Howell, 2010).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: PAH1, At3g09560; PAH2, At5g42870; PEAMT1, At3g18000; CCT1, At2g32260; and CCT2, At4g15130. The raw microarray data are deposited as accession E-MTAB-3253 at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/).
Supplemental Data
Supplemental Figure 1. Histochemical staining of ProPEAMT1:GUS plants.
Supplemental Table 1. Analysis of gene expression using real-time RT-PCR for genes associated with phospholipid biosynthesis in pah1 pah2 roots.
Supplemental Table 2. Kinetic parameters for CCT1 in the presence and absence of lipids.
Supplemental Table 3. Primers used for cloning.
Supplemental Data Set 1. ATH1 microarray data from 2-week-old plants of pah1 pah2.
Supplementary Material
Acknowledgments
We thank Ilaria Lavagi for assisting with this research. The work was funded by the UK Biotechnology and Biological Sciences Research Council through Grants BB/G009724/1 and BBS/E/C/00005207.
AUTHOR CONTRIBUTIONS
P.J.E. designed the research and wrote the article. C.P.C. and N.A. performed most of the research and data analysis. F.M.B., S.K., and P.J.E. performed some aspects of the research and data analysis.
Glossary
- PAP
phosphatidic acid phosphatase
- DG
diacylglycerol
- PA
phosphatidic acid
- TG
triacylglycerol
- ER
endoplasmic reticulum
- UASINO
upstream activating sequence inositol-responsive
- PC
phosphatidylcholine
- CaMV
cauliflower mosaic virus
- FA
fatty acid
- P-Cho
phosphocholine
- 5′UTR
5′ untranslated region
- uORF
upstream open reading frame
- FW
fresh weight
- TLC
thin-layer chromatography
Footnotes
Articles can be viewed online without a subscription.
References
- Alatorre-Cobos F., Cruz-Ramírez A., Hayden C.A., Pérez-Torres C.A., Chauvin A.L., Ibarra-Laclette E., Alva-Cortés E., Jorgensen R.A., Herrera-Estrella L. (2012). Translational regulation of Arabidopsis XIPOTL1 is modulated by phosphocholine levels via the phylogenetically conserved upstream open reading frame 30. J. Exp. Bot. 63: 5203–5221. [DOI] [PubMed] [Google Scholar]
- Andre C., Haslam R.P., Shanklin J. (2012). Feedback regulation of plastidic acetyl-CoA carboxylase by 18:1-acyl carrier protein in Brassica napus. Proc. Natl. Acad. Sci. USA 109: 10107–10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold R.S., Cornell R.B. (1996). Lipid regulation of CTP: phosphocholine cytidylyltransferase: electrostatic, hydrophobic, and synergistic interactions of anionic phospholipids and diacylglycerol. Biochemistry 35: 9917–9924. [DOI] [PubMed] [Google Scholar]
- BeGora M.D., Macleod M.J., McCarry B.E., Summers P.S., Weretilnyk E.A. (2010). Identification of phosphomethylethanolamine N-methyltransferase from Arabidopsis and its role in choline and phospholipid metabolism. J. Biol. Chem. 285: 29147–29155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braker J.D., Hodel K.J., Mullins D.R., Friesen J.A. (2009). Identification of hydrophobic amino acids required for lipid activation of C. elegans CTP:phosphocholine cytidylyltransferase. Arch. Biochem. Biophys. 492: 10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J., McCourt P.J., Somerville C.R. (1986). Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal. Biochem. 152: 141–145. [DOI] [PubMed] [Google Scholar]
- Carman G.M., Han G.S. (2009). Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J. Biol. Chem. 284: 2593–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G.M., Henry S.A. (2007). Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 282: 37293–37297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.-B., Lee K.W., Cho S.H. (1997). Cloning of CTP:phosphocholine cytidylyltransferase cDNA from Arabidopsis thaliana. Mol. Cells 7: 58–63. [PubMed] [Google Scholar]
- Choi H.S., Su W.M., Han G.S., Plote D., Xu Z., Carman G.M. (2012). Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J. Biol. Chem. 287: 11290–11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.S., Su W.M., Morgan J.M., Han G.S., Xu Z., Karanasios E., Siniossoglou S., Carman G.M. (2011). Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of SER(602), THR(723), AND SER(744) as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J. Biol. Chem. 286: 1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cornell R. (1989). Chemical cross-linking reveals a dimeric structure for CTP:phosphocholine cytidylyltransferase. J. Biol. Chem. 264: 9077–9082. [PubMed] [Google Scholar]
- Cornell R.B., Northwood I.C. (2000). Regulation of CTP:phosphocholine cytidylyltransferase by amphitropism and relocalization. Trends Biochem. Sci. 25: 441–447. [DOI] [PubMed] [Google Scholar]
- Cruz-Ramírez A., López-Bucio J., Ramírez-Pimentel G., Zurita-Silva A., Sánchez-Calderon L., Ramírez-Chávez E., González-Ortega E., Herrera-Estrella L. (2004). The xipotl mutant of Arabidopsis reveals a critical role for phospholipid metabolism in root system development and epidermal cell integrity. Plant Cell 16: 2020–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkor J., Zhang P., Wong S., O’Loughlin L., Dewald J., Kok B.P., Brindley D.N., Reue K. (2009). A conserved serine residue is required for the phosphatidate phosphatase activity but not the transcriptional coactivator functions of lipin-1 and lipin-2. J. Biol. Chem. 284: 29968–29978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann P., Hoffmann-Benning S., Balbo I., Benning C. (1995). Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7: 1801–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Eastmond P.J. (2004). Cloning and characterization of the acid lipase from castor beans. J. Biol. Chem. 279: 45540–45545. [DOI] [PubMed] [Google Scholar]
- Eastmond P.J., Quettier A.-L., Kroon J.T., Craddock C., Adams N., Slabas A.R. (2010). Phosphatidic acid phosphohydrolase 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell 22: 2796–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink J.M., Kolodner R.D. (2010). An overview of Cdk1-controlled targets and processes. Cell Div. 5: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Yan C., Roston R., Shanklin J., Xu C. (2014). Arabidopsis lipins, PDAT1 acyltransferase, and SDP1 triacylglycerol lipase synergistically direct fatty acids toward β-oxidation, thereby maintaining membrane lipid homeostasis. Plant Cell 26: 4119–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Yan C., Zhang X., Xu C. (2013). Dual role for phospholipid:diacylglycerol acyltransferase: enhancing fatty acid synthesis and diverting fatty acids from membrane lipids to triacylglycerol in Arabidopsis leaves. Plant Cell 25: 3506–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck B.N., Gropler M.C., Chen Z., Leone T.C., Croce M.A., Harris T.E., Lawrence J.C. Jr., Kelly D.P. (2006). Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 4: 199–210. [DOI] [PubMed] [Google Scholar]
- Friesen J.A., Campbell H.A., Kent C. (1999). Enzymatic and cellular characterization of a catalytic fragment of CTP:phosphocholine cytidylyltransferase α. J. Biol. Chem. 274: 13384–13389. [DOI] [PubMed] [Google Scholar]
- Friesen J.A., Liu M.F., Kent C. (2001a). Cloning and characterization of a lipid-activated CTP:phosphocholine cytidylyltransferase from Caenorhabditis elegans: identification of a 21-residue segment critical for lipid activation. Biochim. Biophys. Acta 1533: 86–98. [DOI] [PubMed] [Google Scholar]
- Friesen J.A., Park Y.S., Kent C. (2001b). Purification and kinetic characterization of CTP:phosphocholine cytidylyltransferase from Saccharomyces cerevisiae. Protein Expr. Purif. 21: 141–148. [DOI] [PubMed] [Google Scholar]
- Golden A., Liu J., Cohen-Fix O. (2009). Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J. Cell Sci. 122: 1970–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjánácz M., Mattaj I.W. (2009). Lipin is required for efficient breakdown of the nuclear envelope in Caenorhabditis elegans. J. Cell Sci. 122: 1963–1969. [DOI] [PubMed] [Google Scholar]
- Gout E., Bligny R., Roby C., Douce R. (1990). Transport of phosphocholine in higher plant cells: 31P nuclear magnetic resonance studies. Proc. Natl. Acad. Sci. USA 87: 4280–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.S., Siniossoglou S., Carman G.M. (2007). The cellular functions of the yeast lipin homolog PAH1p are dependent on its phosphatidate phosphatase activity. J. Biol. Chem. 282: 37026–37035. [DOI] [PubMed] [Google Scholar]
- Han G.S., Wu W.I., Carman G.M. (2006). The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281: 9210–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.K., Taneva S.G., Lee J., Silva L.P., Schriemer D.C., Cornell R.B. (2013). The membrane-binding domain of an amphitropic enzyme suppresses catalysis by contact with an amphipathic helix flanking its active site. J. Mol. Biol. 425: 1546–1564. [DOI] [PubMed] [Google Scholar]
- Inatsugi R., Nakamura M., Nishida I. (2002). Phosphatidylcholine biosynthesis at low temperature: differential expression of CTP:phosphorylcholine cytidylyltransferase isogenes in Arabidopsis thaliana. Plant Cell Physiol. 43: 1342–1350. [DOI] [PubMed] [Google Scholar]
- Inatsugi R., Kawai H., Yamaoka Y., Yu Y., Sekiguchi A., Nakamura M., Nishida I. (2009). Isozyme-specific modes of activation of CTP:phosphorylcholine cytidylyltransferase in Arabidopsis thaliana at low temperature. Plant Cell Physiol. 50: 1727–1735. [DOI] [PubMed] [Google Scholar]
- Jackowski S., Fagone P. (2005). CTP:phosphocholine cytidylyltransferase: paving the way from gene to membrane. J. Biol. Chem. 280: 853–856. [DOI] [PubMed] [Google Scholar]
- Jesch S.A., Zhao X., Wells M.T., Henry S.A. (2005). Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J. Biol. Chem. 280: 9106–9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.E., Kalmar G.B., Sohal P.S., Walkey C.J., Yamashita S., Cornell R.B. (1992). Comparison of the lipid regulation of yeast and rat CTP:phosphocholine cytidylyltransferase expressed in COS cells. Biochem. J. 285: 815–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent C. (2005). Regulatory enzymes of phosphatidylcholine biosynthesis: a personal perspective. Biochim. Biophys. Acta 1733: 53–66. [DOI] [PubMed] [Google Scholar]
- Keogh M.R., Courtney P.D., Kinney A.J., Dewey R.E. (2009). Functional characterization of phospholipid N-methyltransferases from Arabidopsis and soybean. J. Biol. Chem. 284: 15439–15447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney A.J., Clarkson D.T., Loughman B.C. (1987). The regulation of phosphatidylcholine biosynthesis in rye (Secale cereale) roots. Stimulation of the nucleotide pathway by low temperature. Biochem. J. 242: 755–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney A.J., Moore T.S. Jr. (1987). Phosphatidylcholine synthesis in castor bean endosperm: the localization and control of CTP: choline-phosphate cytidylyltransferase activity. Arch. Biochem. Biophys. 259: 15–21. [DOI] [PubMed] [Google Scholar]
- Karanasios E., Han G.S., Xu Z., Carman G.M., Siniossoglou S. (2010). A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc. Natl. Acad. Sci. USA 107: 17539–17544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Taneva S.G., Holland B.W., Tieleman D.P., Cornell R.B. (2014). Structural basis for autoinhibition of CTP:phosphocholine cytidylyltransferase (CCT), the regulatory enzyme in phosphatidylcholine synthesis, by its membrane-binding amphipathic helix. J. Biol. Chem. 289: 1742–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T., Monsen E., Dabney A.R., Storey J.D. (2006). EDGE: extraction and analysis of differential gene expression. Bioinformatics 22: 507–508. [DOI] [PubMed] [Google Scholar]
- Liu J.X., Howell S.H. (2010). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22: 782–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen C.J., Gaspar M.L., Jesch S.A., Delon C., Ktistakis N.T., Henry S.A., Levine T.P. (2004). Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304: 1644–1647. [DOI] [PubMed] [Google Scholar]
- Matsushima R., Hayashi Y., Yamada K., Shimada T., Nishimura M., Hara-Nishimura I. (2003). The ER body, a novel endoplasmic reticulum-derived structure in Arabidopsis. Plant Cell Physiol. 44: 661–666. [DOI] [PubMed] [Google Scholar]
- Mietkiewska E., Siloto R.M., Dewald J., Shah S., Brindley D.N., Weselake R.J. (2011). Lipins from plants are phosphatidate phosphatases that restore lipid synthesis in a pah1Δ mutant strain of Saccharomyces cerevisiae. FEBS J. 278: 764–775. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Koizumi R., Shui G., Shimojima M., Wenk M.R., Ito T., Ohta H. (2009). Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc. Natl. Acad. Sci. USA 106: 20978–20983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida I., Swinhoe R., Slabas A.R., Murata N. (1996). Cloning of Brassica napus CTP: phosphocholine cytidylyltransferase cDNAs by complementation in a yeast cct mutant. Plant Mol. Biol. 31: 205–211. [DOI] [PubMed] [Google Scholar]
- O’Hara L., Han G.S., Peak-Chew S., Grimsey N., Carman G.M., Siniossoglou S. (2006). Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem. 281: 34537–34548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J., Browse J. (1995). Lipid biosynthesis. Plant Cell 7: 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin E.T., Rolph C.E. (1999). Modulation of phosphatidylcholine biosynthesis in celery by exogenous fatty acids. Phytochemistry 50: 47–51. [DOI] [PubMed] [Google Scholar]
- Pascual F., Carman G.M. (2013). Phosphatidate phosphatase, a key regulator of lipid homeostasis. Biochim. Biophys. Acta 1831: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péterfy M., Phan J., Xu P., Reue K. (2001). Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27: 121–124. [DOI] [PubMed] [Google Scholar]
- Price-Jones M.J., Harwood J.L. (1983). Hormonal regulation of phosphatidylcholine synthesis in plants. The inhibition of cytidylyltransferase activity by indol-3-ylacetic acid. Biochem. J. 216: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price-Jones M.J., Harwood J.L. (1986). The control of CTP:choline-phosphate cytidylyltransferase activity in pea (Pisum sativum L.). Biochem. J. 240: 837–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson R.B. (2003). The SREBP pathway—insights from Insigs and insects. Nat. Rev. Mol. Cell Biol. 4: 631–640. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., Siniossoglou S. (2005). The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24: 1931–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin L.A. (1997). The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J. 11: 1151–1165. [DOI] [PubMed] [Google Scholar]
- Su W.M., Han G.S., Carman G.M. (2014). Cross-talk phosphorylations by protein kinase C and Pho85p-Pho80p protein kinase regulate Pah1p phosphatidate phosphatase abundance in Saccharomyces cerevisiae. J. Biol. Chem. 289: 18818–18830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W.M., Han G.S., Casciano J., Carman G.M. (2012). Protein kinase A-mediated phosphorylation of Pah1p phosphatidate phosphatase functions in conjunction with the Pho85p-Pho80p and Cdc28p-cyclin B kinases to regulate lipid synthesis in yeast. J. Biol. Chem. 287: 33364–33376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto H., Banchio C., Vance D.E. (2008). Transcriptional regulation of phosphatidylcholine biosynthesis. Prog. Lipid Res. 47: 204–220. [DOI] [PubMed] [Google Scholar]
- Tabuchi T., Okada T., Azuma T., Nanmori T., Yasuda T. (2006). Posttranscriptional regulation by the upstream open reading frame of the phosphoethanolamine N-methyltransferase gene. Biosci. Biotechnol. Biochem. 70: 2330–2334. [DOI] [PubMed] [Google Scholar]
- Tasseva G., Richard L., Zachowski A. (2004). Regulation of phosphatidylcholine biosynthesis under salt stress involves choline kinases in Arabidopsis thaliana. FEBS Lett. 566: 115–120. [DOI] [PubMed] [Google Scholar]
- Thevelein J.M. (1994). Signal transduction in yeast. Yeast 10: 1753–1790. [DOI] [PubMed] [Google Scholar]
- Walkey C.J., Kalmar G.B., Cornell R.B. (1994). Overexpression of rat liver CTP:phosphocholine cytidylyltransferase accelerates phosphatidylcholine synthesis and degradation. J. Biol. Chem. 269: 5742–5749. [PubMed] [Google Scholar]
- Wang L., Kazachkov M., Shen W., Bai M., Wu H., Zou J. (2014). Deciphering the roles of Arabidopsis LPCAT and PAH in phosphatidylcholine homeostasis and pathway coordination for chloroplast lipid synthesis. Plant J. 80: 965–976. [DOI] [PubMed] [Google Scholar]
- Wang Y., Kent C. (1995). Identification of an inhibitory domain of CTP:phosphocholine cytidylyltransferase. J. Biol. Chem. 270: 18948–18952. [DOI] [PubMed] [Google Scholar]
- Wang X., Moore T.S. (1990). Phosphatidylcholine biosynthesis in castor bean endosperm : purification and properties of cytidine 5′-triphosphate:choline-phosphate cytidylyltransferase. Plant Physiol. 93: 250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold P.A., Charles L.G., Feldman D.A. (1991). Microsomal CTP:choline phosphate cytidylyltransferase: kinetic mechanism of fatty acid stimulation. Biochim. Biophys. Acta 1086: 57–62. [DOI] [PubMed] [Google Scholar]
- Xu C., Fan J., Froehlich J.E., Awai K., Benning C. (2005). Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. Plant Cell 17: 3094–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Boggs K.P., Jackowski S. (1995). The association of lipid activators with the amphipathic helical domain of CTP:phosphocholine cytidylyltransferase accelerates catalysis by increasing the affinity of the enzyme for CTP. J. Biol. Chem. 270: 23951–23957. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.