A transcriptional regulator physically interacts with and targets an evening complex component for degradation, enabling increased expression of dark-associated growth-promoting transcription factors.
Abstract
Hypocotyl elongation is a highly coordinated physiological response regulated by myriad internal and external cues. Here, we show that BBX19, a transcriptional regulator with two B-box motifs, is a positive regulator of growth; diminished BBX19 expression by RNA interference reduces hypocotyl length, and its constitutive expression promotes growth. This function of BBX19 is dependent on the E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), EARLY FLOWERING3 (ELF3), and PHYTOCHROME-INTERACTING FACTOR4 (PIF4) and PIF5. BBX19 is nucleus-colocalized and interacts physically with COP1 and ELF3, a component of the evening complex that represses the expression of PIF4 and PIF5. Moreover, ELF3 protein abundance inversely correlates with BBX19 expression levels in a COP1-dependent manner. By contrast, PIF expression, coinciding with the initiation of hypocotyl growth in the early evening, is positively correlated with the BBX19 transcript abundance. These results collectively establish BBX19 as an adaptor that binds to and recruits ELF3 for degradation by COP1 and, as such, dynamically gates the formation of the evening complex, resulting in derepression of PIF4/5. This finding refines our perspective on how plants grow by providing a molecular link between COP1, ELF3, and PIF4/5 as an underlying mechanism of photomorphogenic development in Arabidopsis thaliana.
INTRODUCTION
The B-BOX family of proteins (BBX) is a functionally diverse family of 32 zinc-finger transcription factors in Arabidopsis thaliana characterized by the presence of one or two B-box motifs at the N-terminal domain, alone or in combination with a C-terminal CCT (for CONSTANS, CO like, and TOC) domain (Khanna et al., 2009; Huang et al., 2012; Gangappa and Botto, 2014). The B-box motif is critical for both transcriptional regulation and heterodimeric protein formation, and the CCT domain is implicated in transcriptional regulation and, in some instances, nuclear protein import (Crocco and Botto, 2013; Gangappa and Botto, 2014). Eight of the BBX proteins belong to structural group IV (BBX18 to BBX25), by virtue of containing two tandem repeat B-box motifs (Box1 and Box2) in the N terminus and lacking the CCT domain (Kumagai et al., 2008; Khanna et al., 2009; Gangappa and Botto, 2014). Members of group IV are considered to be evolutionarily conserved functional proteins associated with photomorphogenesis and stress responses (Yamawaki et al., 2011). Among the group members, BBX18, BBX19, BBX24, and BBX25 are implicated as negative regulators of photomorphogenesis, whereas BBX20, BBX21, and BBX22 are known to promote photomorphogenesis (Datta et al., 2006, 2007, 2008; Indorf et al., 2007; Kumagai et al., 2008; Chang et al., 2011; Wang et al., 2011; Yan et al., 2011; Fan et al., 2012; Jiang et al., 2012; Gangappa et al., 2013). BBX25 and its homolog BBX24 function through their direct interaction with ELONGATED HYPOCOTYL5 (HY5), a basic domain/leucine zipper transcription factor, and with CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), additively enhancing COP1 and suppressing HY5 functions (Holm and Deng, 1999; Hardtke et al., 2000; Lau and Deng, 2012; Gangappa et al., 2013). HY5 plays a central role in the regulation of seedling photomorphogenesis, and COP1, an E3 ubiquitin ligase, is a crucial mediator of ubiquitination and subsequent targeted degradation of positive regulators of light signal transduction, such as HY5 and BBX22, in dark conditions (Holm and Deng, 1999; Hardtke et al., 2000; Yu et al., 2008; Lau and Deng, 2012). The functioning of promoters of the photomorphogenic response, BBX21 and BBX22, is reported to be through their recruitment by COP1 to nuclear speckles (nuclear bodies) and the enhancement of HY5 activity by their physical interaction via the B-box motifs (Datta et al., 2007, 2008; Chang et al., 2008).
Hypocotyl elongation is a physiological response controlled by myriad internal and external cues, including the convergence of light signaling and the circadian clock, with maximal growth occurring at the end of night under diurnal conditions (Nozue et al., 2007; Nusinow et al., 2011). One regulator of growth is a protein complex, known as the evening complex, comprising EARLY FLOWERING3 (ELF3), ELF4, and the transcription factor LUXARRHYTHMO (LUX) (Nusinow et al., 2011). ELF3 and ELF4 are nuclear proteins that regulate circadian rhythms, and LUX is a single MYB domain-containing SHAQYF-type GARP transcription factor initially identified in a genetic screen for long-hypocotyl mutants (Zagotta et al., 1996; Hicks et al., 2001; Doyle et al., 2002; Khanna et al., 2003). Early in the night, the evening complex represses the expression of two growth-promoting basic helix-loop-helix transcription factors, PHYTOCHROME-INTERACTING FACTOR4 (PIF4) and PIF5, via binding to their promoters (Leivar et al., 2009; Leivar and Quail, 2011; Nusinow et al., 2011; Zhang et al., 2013). Later in the night, derepression of PIF4/5 expression promotes hypocotyl elongation, but at dawn, when PIF4/5 proteins are targeted for degradation through their interaction with the light-activated form of phytochrome B, hypocotyl growth concludes (Huq and Quail, 2002; Khanna et al., 2004). As such, PIF4 and PIF5 are considered to be integrators of the light and circadian clock signaling networks (Nozue et al., 2007).
Recently, we established that the small metabolite methylerythritol cyclodiphosphate (MEcPP), a plastidial precursor of isoprenoids produced by the methylerythritol phosphate pathway, also functions as a stress-specific retrograde signal, relaying environmental perturbations sensed by plastids back to the nucleus (Xiao et al., 2012). We have also demonstrated that light-grown ceh1 (for constitutively expressing HPL) mutant plants accumulating high MEcPP levels are severely dwarfed (Xiao et al., 2012). Microarray analyses in conjunction with quantitative PCR identified a number of genes with robustly altered transcript levels in ceh1, among them BBX19, whose expression level is reduced by half (Xiao et al., 2012; Wang et al., 2014). The altered expression level of BBX19 and the severe dwarfism of ceh1 in conjunction with the reported function of BBX19 as a negative regulator of photomorphogenesis prompted us to explore the mode of action of this transcriptional regulator in plant photomorphogenic development.
Here, we demonstrate that BBX19 is a suppressor of photomorphogenesis by physically interacting with and targeting ELF3 for degradation by COP1 in Arabidopsis, thereby depleting/reducing the ELF3 pool required for evening complex formation and, in turn, enabling increased expression of the dark-associated growth-promoting transcription factors PIF4/5. Furthermore, we ascertain the significance of the integrity of the BBX19-Box1 motif in the protein-protein interaction and, by extension, in BBX19 suppression of photomorphogenesis.
Taken together, this report establishes BBX19 as a circadian gatekeeper of the evening complex and, as such, an integrator of clock and light signaling inputs necessary for growth promotion.
RESULTS
BBX19 Is a Suppressor of Photomorphogenesis
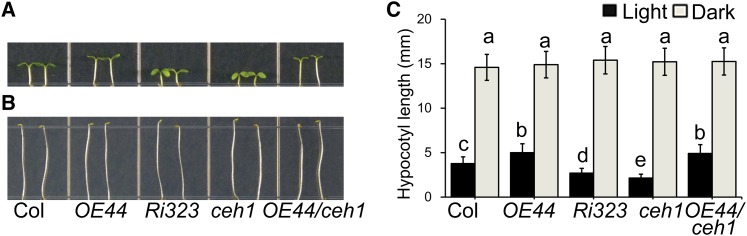
We have previously established that mutant Arabidopsis plants (ceh1) accumulating high levels of the stress-specific retrograde signal MEcPP are dwarfed and have reduced BBX19 transcript levels (Xiao et al., 2012; Wang et al., 2014). The reported role of BBX19 in promoting hypocotyl growth (Kumagai et al., 2008) prompted us to examine its function in ceh1 as compared with the Columbia-0 (Col-0) wild-type plants. Initially, we generated BBX19-overexpressing (CaMV35S:BBX19-GFP; designated OE44 and OE56) and RNA interference (RNAi; designated Ri323 and Ri513) lines in the Col-0 wild-type background (Wang et al., 2014). Generation of RNAi lines was required because, in agreement with the previous report (Khanna et al., 2006), the presence of notable BBX19 transcript levels in the available T-DNA insertion lines renders these lines functionally similar to wild-type plants (Wang et al., 2014). Dark-grown seedlings of the aforementioned genotypes exhibit similar hypocotyl length, whereas those grown under long-day (LD) photoperiod conditions display a positive association between hypocotyl length and BBX19 expression levels (Figures 1A to 1C), indicating the regulatory role of BBX19 in photomorphogenesis. To verify this regulatory role, we extended our studies and examined the hypocotyl length of these genotypes grown under short-day conditions as well as under continuous monochromatic wavelengths, including blue, red, and far-red light, along with continuous white light as the control (Supplemental Figures 1A to 1D). Collectively, these data clearly identify BBX19 as a negative regulator of photomorphogenesis, since overexpressor lines (OE44 and OE56) as opposed to RNAi lines (Ri323 and Ri513) exhibit a longer hypocotyl than the wild-type control under all the light conditions examined (Supplemental Figures 1A to 1D).
Figure 1.
BBX19 Is a Suppressor of Photomorphogenesis.
(A) and (B) Hypocotyl phenotypes of 5-d-old LD-grown (A) and 3-d-old dark-grown (B) wild-type Col-0, BBX19-OE (OE44), BBX19 RNAi line (Ri323), ceh1, and OE44/ceh1 seedlings.
(C) Measurements of hypocotyl length of the same genotypes grown under LD (black boxes) and dark (gray boxes) conditions. Data are means ± sd; n ≥ 90. Letters above the bars indicate significant differences as determined by Tukey’s HSD method (P < 0.05).
Because of the similar trend in responsiveness of hypocotyl growth to BBX19 expression levels, the subsequent studies were performed with OE44 and Ri323 as representative transgenic lines with opposing levels of BBX19 expression. Next, we generated BBX19-overexpressing ceh1 lines (OE44/ceh1) by performing crosses between the two corresponding parent lines. We compared the hypocotyl lengths of various genotypes and found them to be indistinguishable when grown in the dark, while under LD conditions, BBX19-overexpressing seedlings displayed similar and longer hypocotyls in both wild-type and ceh1 mutant backgrounds (Figures 1A to 1C). Specifically, as reported previously (Khanna et al., 2006), overexpression of BBX19 not only resulted in a longer hypocotyl in the OE44 line as compared with the wild type but also fully compensated the ceh1 reduced hypocotyl length in OE44/ceh1 plants to that of the overexpressor parent line (Figures 1A and 1C). This result provides a direct link between the retrograde signal MEcPP and the regulation of photomorphogenic growth through the adjustment of BBX19 expression levels. Moreover, the hyposensitive photomorphogenic response of BBX19 overexpression lines versus the hypersensitive response of the RNAi lines plus the full recovery of ceh1 photomorphogenic phenotypes in OE44/ceh1 lines to the OE44 levels establish BBX19 as a negative regulator of photomorphogenesis.
BBX19 Is a COP1-Dependent and HY5-Independent Suppressor of Photomorphogenesis
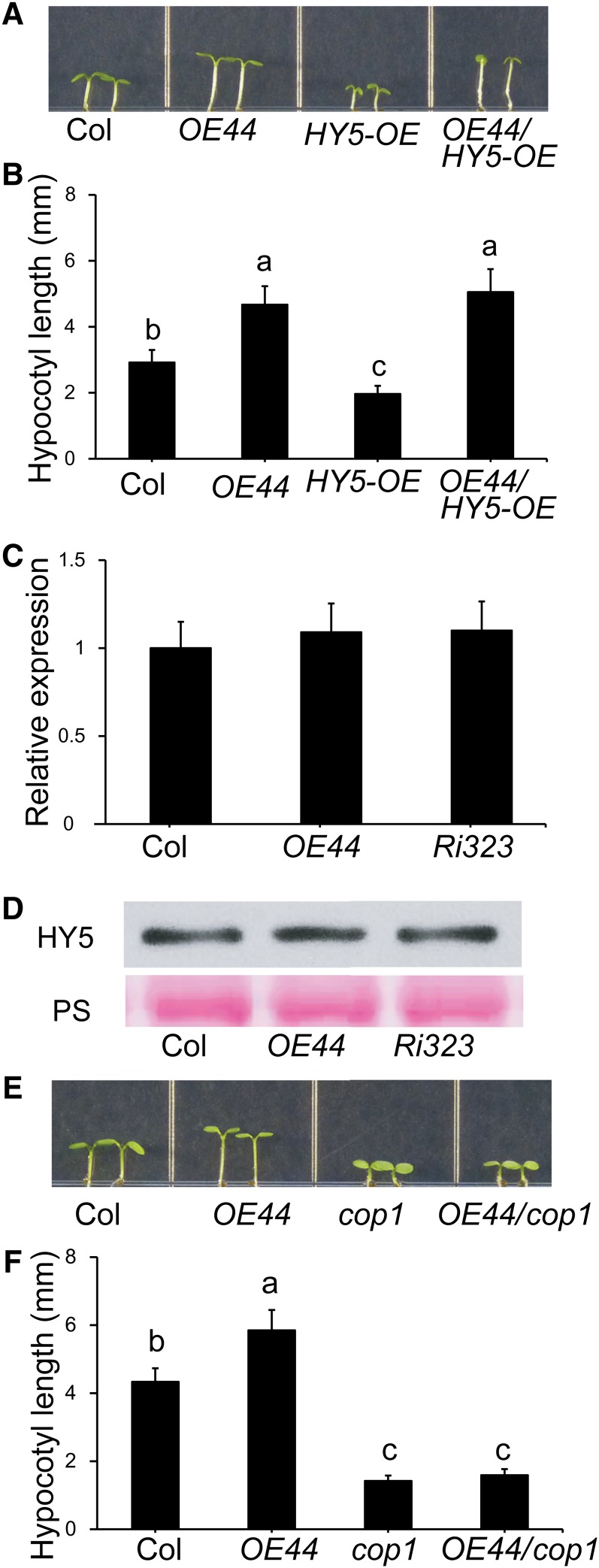
Next, we questioned whether, similar to the other reported members of group IV proteins (Datta et al., 2007, 2008; Chang et al., 2011), BBX19-mediated photomorphogenic development in Arabidopsis is genetically dependent on the two central regulators of photomorphogenesis, namely COP1 and HY5 (Ang et al., 1998; Lau and Deng, 2012). Thus, we generated HY5-overexpressing parent lines (HY5-OE) followed by the incorporation of OE44 into this background and the generation of OE44/HY5-OE plants. Examination of the hypocotyl length of LD-grown seedlings from these various genotypes showed the predicted photomorphogenic short-hypocotyl phenotype in HY5-OE lines (Ang et al., 1998) and the reversal of this phenotype in BBX19-overexpressing lines (OE44/HY5-OE), suggesting the functional independence of BBX19 from HY5 (Figures 2A and 2B).
Figure 2.
BBX19 Is a COP1-Dependent and HY5-Independent Suppressor of Photomorphogenesis.
(A) and (B) Hypocotyl phenotypes of 5-d-old Col-0, OE44, HY5-OE, and OE44/HY5-OE seedlings grown under LD conditions (A) and measurements of their hypocotyl lengths (B).
(C) and (D) Relative expression levels (C) and the corresponding protein levels (D) of HY5 examined by protein gel blot analyses using HY5-specific antibody in Col-0, OE44, and Ri323 lines. The Ponceau S (PS) staining shows equal protein loadings. Total RNA was extracted from these genotypes and subjected to quantitative PCR analysis. The transcript levels of HY5 were normalized to At4g34270 (T1P41-like family protein) and At4g26410 (M3E9) measured in the same samples. Data are mean fold differences ± sd of three biological replicates each with three technical repeats.
(E) and (F) Hypocotyl phenotypes of 5-d-old Col-0, OE44, cop1, and OE44/cop1 seedlings grown under LD conditions (E) and measurements of their hypocotyl lengths (F).
For (B) and (F), data are means ± sd; n ≥ 90. Letters above the bars indicate significant differences as determined by Tukey’s HSD method (P < 0.05).
However, the above genetic approaches could not eliminate the possibility that the observed phenotype is the result of the decreased levels of expression and/or degradation of the HY5 protein in response to the alteration of BBX19 expression levels in OE44 plants. To address these possibilities, we first examined HY5 expression levels in wild-type, OE44, and Ri323 lines (Figure 2C). Similar relative expression levels of HY5 in these genotypes clearly indicate that overexpression or suppression of BBX19 does not alter the expression levels of HY5. Next, we employed these genotypes and examined HY5 protein levels using a HY5-specific antibody (Figure 2D). These data also display similar HY5 protein levels in wild-type, OE44, and Ri323 lines. These findings collectively confirm the HY5-independent function of BBX19 in mediating the observed photomorphogenic development.
To examine the functional dependency of BBX19 on COP1, we also introduced OE44 into the existing cop1-4 mutant line (McNellis et al., 1994), which, for simplicity herein, is referred to as cop1, and generated OE44/cop1 lines and subsequently performed similar studies to those performed with HY5 lines (Figures 2E and 2F). These studies established that, in contrast with the wild-type background and HY5-overexpressing lines, overexpression of BBX19 failed to rescue the photomorphogenic short-hypocotyl phenotype of cop1 (Figures 2E and 2F), a clear indication of a COP1-dependent function of BBX19.
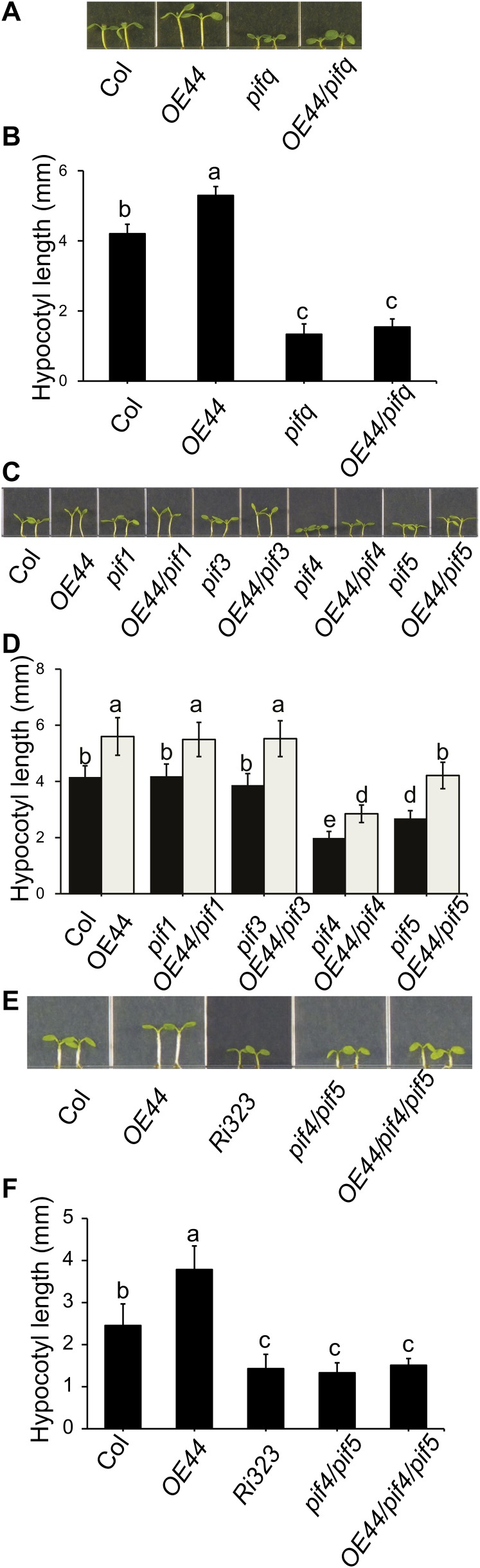
BBX19 Suppression of Photomorphogenesis Is Mediated by PIF4/5
The phytochrome-interacting basic helix-loop-helix transcription factors (PIFs) are responsible for the active repression of deetiolation of dark-grown seedlings, which is reversed in light by their proteolytic degradation (Leivar and Quail, 2011). The hypocotyl growth phenotype associated with enhanced BBX19 expression levels, together with the reported involvement of BBX23 in one of the PIF3 signaling branches inhibiting photomorphogenesis in the dark (Soy et al., 2012), prompted us to genetically examine the potential association of BBX19 function with PIF-regulated signaling cascades. Toward this goal, we initially introduced OE44 into the pif quadruple mutant pif1-1/pif3-3/pif4-2/pif5-3 (pifq) deficient in the functions of all these PIFs (Leivar et al., 2008). Next, we examined the hypocotyl length of LD-grown seedlings and determined that, in contrast with the wild-type background, BBX19 overexpression in the pifq mutant background failed to rescue the shortened hypocotyl of the pifq mutant lines (Figures 3A and 3B). This indicates that the BBX19 suppression of photomorphogenesis is mediated through PIFs. To identify the responsible PIF member(s), we introduced the OE44 line into pif single mutants and examined the hypocotyl length of seedlings homozygous for both loci (Figures 3C and 3D). This result clearly identifies PIF4 and PIF5 as the only PIFs examined required for hypocotyl growth in the wild-type background and, as such, necessary for the full functionality of BBX19 in mediating hypocotyl elongation, albeit to different degrees. Specifically, hypocotyls of OE44/pif5 seedlings are longer than those of OE44/pif4 (Figures 3C and 3D). This difference in the functional effectiveness of PIF4 and PIF5 in regulating the photomorphogenic growth response led us to examine the hypocotyl length of pif4/pif5 lines as compared with the progeny of this double mutant crossed to OE44 (OE44/pif4/pif5) and to extend this comparison to the Col-0, Ri323, and OE44 genotypes. Similarly reduced hypocotyl lengths in Ri323, pif4/5 parent, and OE44/pif4/pif5 seedlings, as compared with either OE44 or Col-0 plants (Figures 3E and 3F), lend strong support to BBX19’s functional dependence on both PIF4 and PIF5.
Figure 3.
BBX19 Suppression of Photomorphogenesis Is Mediated by PIF4/5.
(A) and (C) Phenotypes of 5-d-old seedlings from various genotypes (Col-0, OE44, pifq, OE44/pifq, and single pif mutant/OE44 combinations) grown under LD conditions.
(B) and (D) Hypocotyl lengths of the genotypes in (A) and (C), respectively.
(E) Hypocotyl phenotypes of Col-0, BBX19-overexpressing (OE44), RNAi (Ri323), pif4/5 double mutant, and OE44/pif4/5 seedlings grown under LD conditions.
(F) Measurements of hypocotyl lengths of the genotypes in (E).
For (B), (D), and (F), data are means ± sd; n ≥ 90. Letters above the bars indicate significant differences as determined by Tukey’s HSD method (P < 0.05).
BBX19 Suppression of Photomorphogenesis Is Dependent on ELF3
The established role of the evening complex in repressing the expression of PIF4 and PIF5 and, consequently, hypocotyl growth (Nusinow et al., 2011) alerted us to a potential involvement of ELF3 in the PIF4/PIF5-dependent function of BBX19 in mediating hypocotyl elongation. To examine this possibility genetically, we generated ELF3-overexpressing lines (ELF3-OE), followed by incorporating OE44 into this background (OE44/ELF3-OE). As anticipated, hypocotyls of ELF3-OE seedlings grown under LD conditions were shorter than those of Col-0 and OE44 seedlings, but they were equally long compared with those of Ri323 lines and OE44/ELF-OE lines (Figures 4A and 4B). This lack of reversion and/or compensation of the ELF3-OE reduced hypocotyl length by overexpression of BBX19 (OE44/ELF3-OE) lines implies a functional dependency of BBX19 on ELF3.
Figure 4.
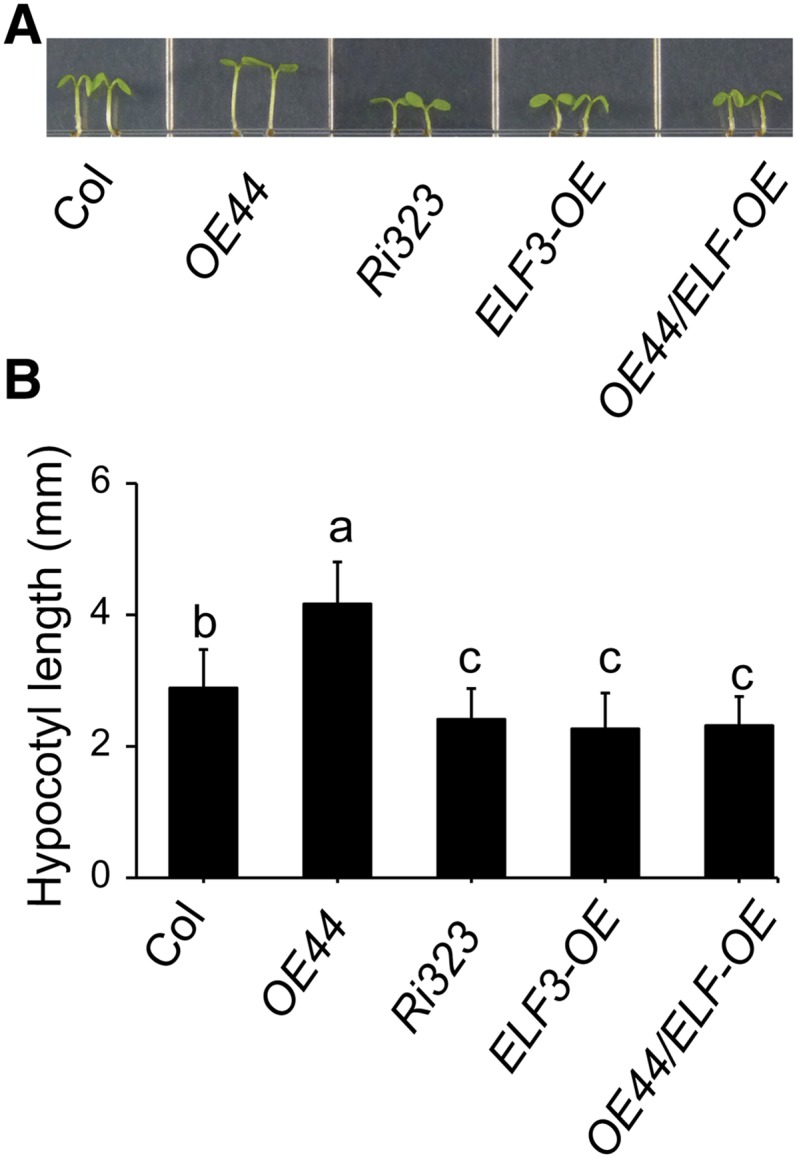
BBX19 Suppression of Photomorphogenesis Is Dependent on ELF3.
(A) Hypocotyl phenotypes of Col-0, OE44, Ri323, ELF3-OE, and OE44/ELF3-OE seedlings grown under LD conditions.
(B) Hypocotyl length measurements of these genotypes. Data are means ± sd; n ≥ 90. Letters above the bars indicate significant differences as determined by Tukey’s HSD method (P < 0.05).
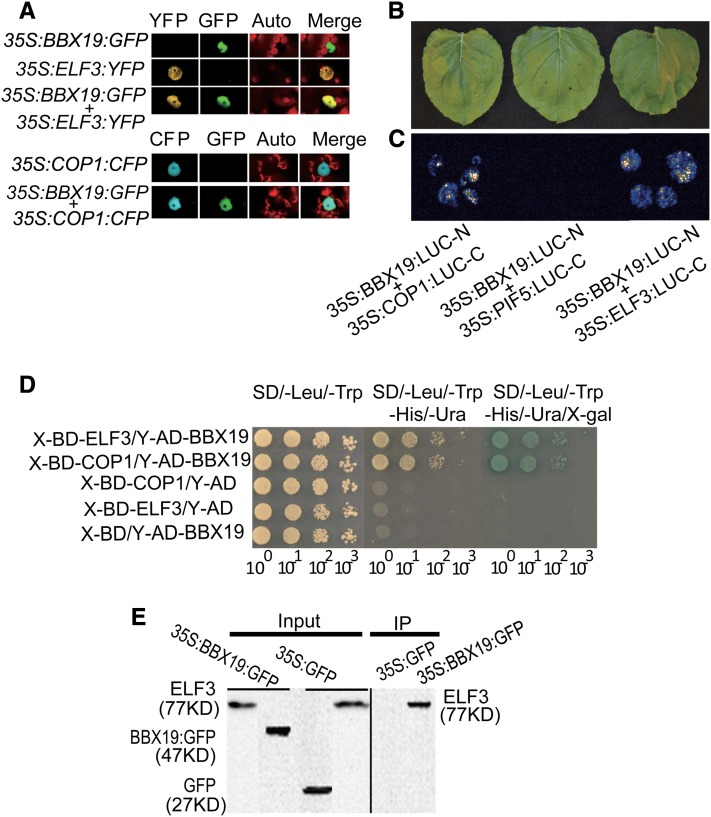
BBX19 Is Nucleus-Colocalized with and Interacts Physically with ELF3 and COP1
As a prelude to gaining insight into the mechanism(s) underlying the dependency of BBX19 on COP1 and ELF3 for photomorphogenic development of Arabidopsis, we performed colocalization assays in Nicotiana benthamiana (wild tobacco) leaves using 35S:BBX19:GFP (green fluorescent protein), 35S:ELF3:YFP (yellow fluorescent protein), and 35S:COP1:CFP (cyan fluorescent protein) constructs individually or in the combinations shown (Figure 5A). These data show nuclear localization of these proteins singularly in addition to the colocalization of BBX19 with ELF3 and BBX19 with COP1. In view of this colocalization, we tested for a potential physical interaction between BBX19 and COP1 and between BBX19 and ELF3 using a combination of N. benthamiana transient expression and yeast two-hybrid assays in yeast (Saccharomyces cerevisiae) (Figures 5B to 5D). To perform split-luciferase assays, we generated fusion constructs of BBX19 to the N-terminal fragment of luciferase (LUC) and fusion constructs of COP1 and ELF3 individually to the C-terminal fragment of LUC. Subsequently, the constructs in the combinations shown (Figures 5B and 5C) were used in agroinfiltration-based transient assays in N. benthamiana leaves. Both the yeast two-hybrid and split-luciferase assays confirmed a direct protein-protein interaction of BBX19 with both COP1 and ELF3 but not with the other proteins tested. In addition, we examined the interaction of BBX19 with ELF3 in vivo by coimmunoprecipitation using a GFP-specific antibody for immunoprecipitation of transgenic Arabidopsis plants constitutively expressing BBX19 fused to GFP (35S:BBX19:GFP), followed by protein gel blot analyses using the reported ELF3-specific antibody (Nusinow et al., 2011). The clear and specific presence of an ELF3-reacting band in the immunoprecipitation fraction of 35S:BBX19:GFP lines, but not in 35S:GFP plant proteins, verified the in vivo interaction of BBX19 with the ELF3 protein (Figure 5E).
Figure 5.
BBX19 Is Nucleus-Colocalized with and Interacts Physically with ELF3 and COP1.
(A) Representatives of transient assays in N. benthamiana expressing 35S:ELF3-YFP, 35S:COP1-CFP, or 35S:BBX19-GFP individually or in a combination, displaying nuclear colocalization of BBX19 and COP1 as well as BBX19 and ELF3.
(B) and (C) Representatives of split-luciferase complementation assays in N. benthamiana displayed by bright-field (B) and dark-field (C) imaging of leaves expressing BBX19/COP1 and BBX19/ELF3 each fused to N- and C-terminal fragments of luciferase. The fusion construct with PIF5 was used as a negative control.
(D) Yeast two-hybrid assays display the BBX19 physical interaction with ELF3 and with COP1. Diploid yeast cells containing both prey construct Y-AD-BBX19 and bait constructs X-BD-COP1 and X-BD-ELF3 were obtained by mating. Overnight cultures were normalized to an OD600 of 1, serially diluted as indicated, and spotted onto nonselective medium lacking Leu and Trp (left panel) and selective medium lacking Leu, Trp, His, and Ura (middle panel) and with X-Gal (right panel). Negative controls contained empty bait and/or prey vectors.
(E) In vivo interaction between BBX19 and ELF3 determined by coimmunoprecipitation assay. Protein samples obtained from 35S:BBX19:GFP and 35S:GFP seedlings were immunoprecipitated (IP) with GFP antibody, and ELF3 (77 kD) was detected by immunoblotting using α-ELF3 antibody.
The Integrity of the Box1 Motif Is Indispensable for BBX19 Interaction with COP1 and ELF3 and BBX19-Associated Suppression of Photomorphogenesis
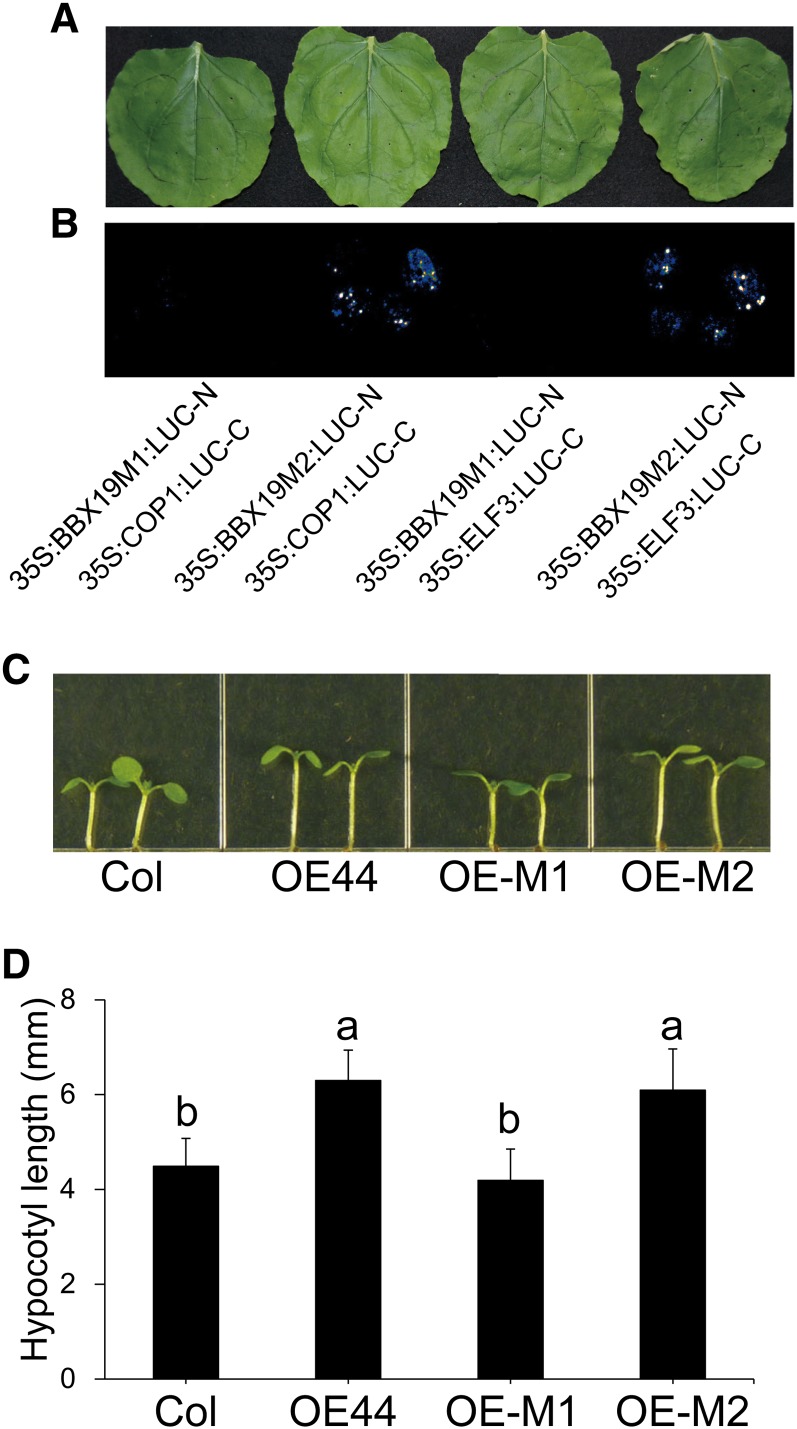
Previously, we established the indispensable role of BBX19-Box1 in enabling the interaction with the regulator of flowering CONSTANS (CO) and, by extension, in the regulation of flowering time (Wang et al., 2014). Specifically, we showed that substitution of the conserved cysteine-25 to serine in the BBX19-Box1 motif (BBX19-M1) abolishes the BBX19/CO interaction and, as the result, eliminates the BBX19-associated regulation of flowering time. By contrast, however, the analogous Cys-76-to-Ser substitution in the Box2 motif (BBX19-M2) was ineffective (Wang et al., 2014). Based on the earlier findings, we questioned whether Box1 plays a parallel role in the suppression of photomorphogenesis. To examine this possibility, we performed split-luciferase complementation assays using the previously reported Box1 (BBX19-M1) or Box2 (BBX19-M2) construct fused to the N-terminal fragment of LUC. These constructs were coinfiltrated with individual 35S:COP1:LUC-C and 35S:ELF3:LUC-C fusion constructs into N. benthamiana leaves (Figures 6A and 6B). Similar to the previous findings (Wang et al., 2014), the data presented here clearly display the indispensable role of BBX19-Box1 integrity in enabling BBX19/COP1 and BBX19/ELF3 interactions, while BBX19-Box2 remained ineffective.
Figure 6.
Box1 Is Indispensable for BBX19 Interaction with COP1 and ELF3 and Associated Suppression of Photomorphogenesis.
(A) and (B) Representatives of split-luciferase complementation assays in N. benthamiana displayed by bright-field (A) and dark-field (B) imaging of leaves expressing BBX19M1/ or BBX19M2/COP1 and BBX19M1/ or BBX19M2/ELF3 each fused to N- and C-terminal fragments of luciferase.
(C) Representative hypocotyl lengths of Col-0 and lines overexpressing intact BBX19 (OE-44), mutated Box1 (OE-M1), and mutated Box2 (OE-M2).
(D) Hypocotyl length measurements of these genotypes. Data are means ± sd; n ≥ 30. Letters above the bars indicate significant differences as determined by Tukey’s HSD method (P < 0.05).
Because of the critical role of Box1 in the BBX19/COP1 and BBX19/ELF3 interactions, we next examined the role of this motif, along with the Box2 motif as the control, in the regulation of photomorphogenesis. Thus, we examined the hypocotyl length of the previously reported transgenic lines overexpressing BBX19-M1 (OE-M1) or BBX19-M2 (OE-M2) grown under LD conditions (Figures 6C and 6D). The data clearly show a reversion of OE-M1 hypocotyl length to that of the wild type, whereas OE-M2 plants display long hypocotyls similar to those of OE44 lines.
Collectively, these data confirm the significance of BBX19-Box1 integrity in facilitating the protein-protein interactions and, consequently, the functionality of BBX19 as a suppressor of photomorphogenesis.
BBX19 Promotes COP1-Mediated Degradation of ELF3
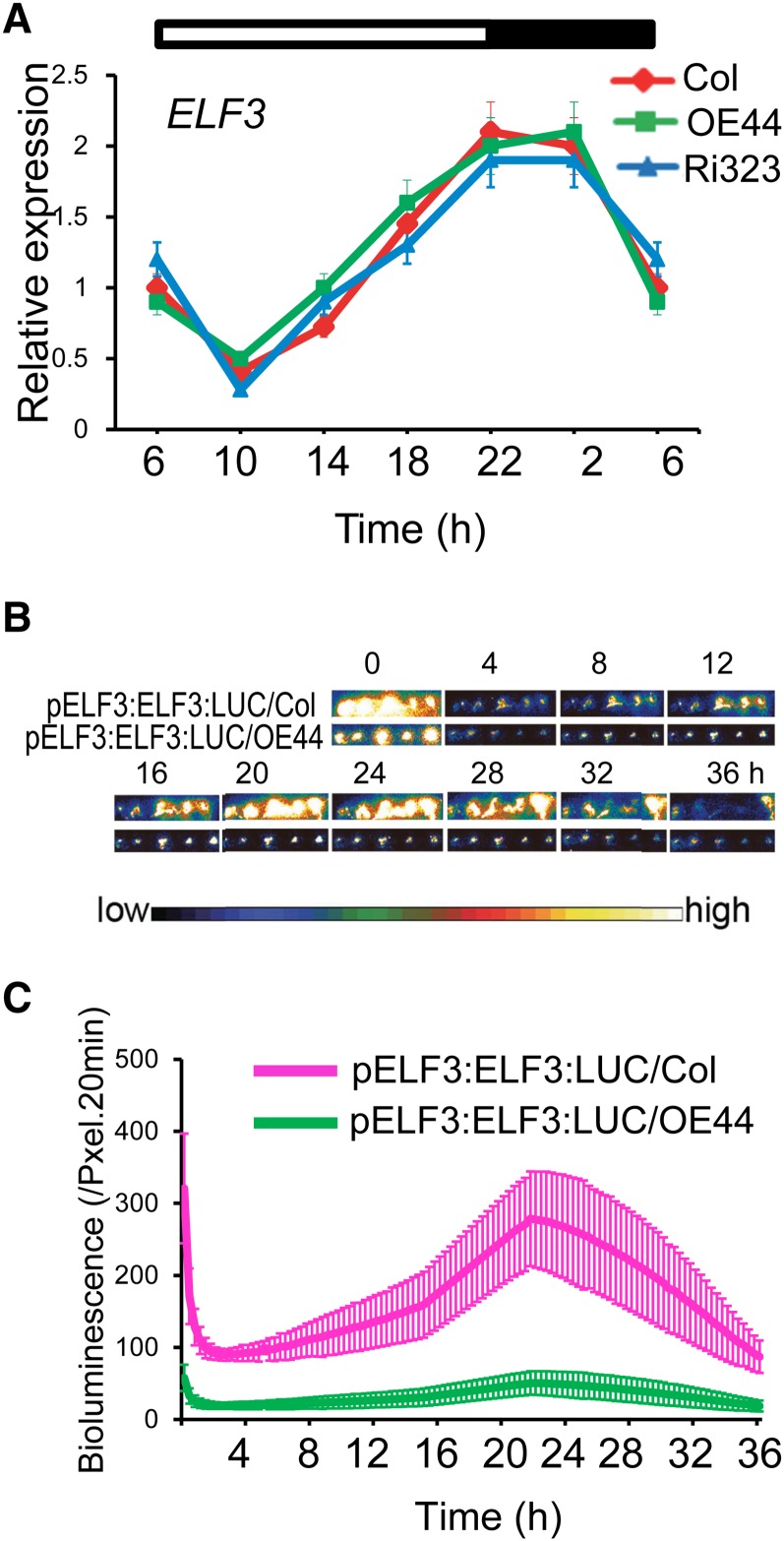
The in vivo interaction of BBX19 with ELF3 (Figures 5B and 5E) led us to question whether BBX19 might also alter either the diurnal rhythm of expression or the protein stability of ELF3. To address these possibilities, we first examined the expression rhythm and levels of ELF3 in wild-type, OE44, and Ri323 lines over a 24-h period starting at the onset of light at 6 am (Figure 7A). These data clearly established that variation in BBX19 expression levels alter neither the rhythm nor the level of ELF3 expression. We next questioned whether circadian-controlled ELF3 protein levels might be altered by BBX19. Toward this goal, we performed transient assays in wild-type and OE44 Arabidopsis seedlings infiltrated with the pELF3:ELF3:LUC construct (Figures 7B and 7C). Visual assessment and quantitative luciferase activity analyses, as a measure of ELF3 protein levels, over a 36-h time course clearly illustrate the concordance of the circadian rhythm of the ELF3 protein with ELF3 transcript in wild-type plants (Figures 7A to 7C). By contrast, however, the strongly diminished levels and circadian rhythm of ELF3 protein in the OE44 background do not match the unaltered expression levels and pattern of ELF3 in these lines (Figures 7A to 7C). This finding supports the notion of BBX19-mediated destabilization of the ELF3 protein.
Figure 7.
BBX19 Does Not Alter the Rhythm or Levels of Expression but Modifies ELF3 Protein Stability.
(A) Levels and diurnal rhythms of ELF3 expression in Col-0, OE44, and Ri323 lines.
(B) and (C) Diurnal rhythms of ELF3 protein levels displayed by LUC activity in Col-0 and OE44 seedlings transiently expressing pELF3:ELF3:LUC. Representative dark-field images of seedlings (B) and the quantification of LUC activity using Andor Solis image analysis software (C) of corresponding seedlings from the two backgrounds are shown. The color-coded bar displays the intensity of LUC activity.
Data are means ± se (n ≥ 60).
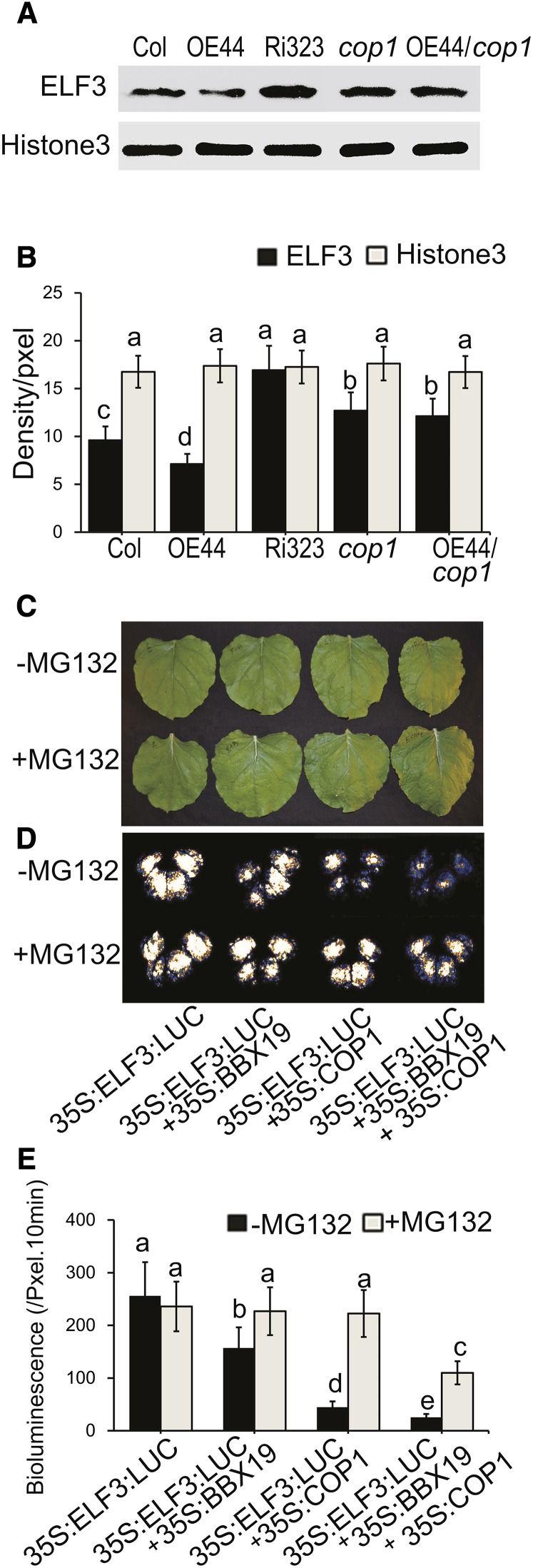
The coordinated function of COP1 and ELF3 in the regulation of flowering time and circadian rhythm, through COP1 interaction with and degradation of ELF3, is well established (Yu et al., 2008). The knowledge of this coordinated function, together with our data confirming BBX19 physical interaction with and functional dependency on both ELF3 and COP1 (Figures 2E and 2F, 4A and 4B, and 6A to 6D), led us to question whether BBX19 expression levels might alter the stability of the ELF3 protein in a COP1-dependent manner. Thus, we employed an ELF3-specific antibody to examine ELF3 protein levels in various genotypes (wild type, OE44, Ri323, cop1, and OE/cop1) by protein gel blot analyses using nucleus-enriched proteins extracted from plants collected at the peak (6 pm) of evening complex formation (Nusinow et al., 2011) (Figures 8A and 8B). We observed uniform levels of control protein (Histone3) in these genotypes, in contrast with ELF3, whose level was inversely correlated with BBX19 expression levels. Specifically, the highest levels of ELF3 are found in Ri323 lines and the lowest level in OE44 plants as compared with the Col-0 plants (Figure 8B). However, similar levels of ELF3 protein are present in all the cop1 mutant backgrounds, including BBX19-overexpressing cop1 lines (OE44/cop1) (Figures 8A and 8B). Taken together, our data imply that the BBX19 physical interaction with ELF3 is a crucial recruiting mechanism that facilitates the COP1-mediated degradation of ELF3.
Figure 8.
BBX19 Promotes COP1-Dependent Degradation of ELF3.
(A) ELF3 protein levels at a selected time point (6 pm) in wild-type Col-0, OE44, Ri323, cop1, and OE44/cop1 seedlings grown under LD conditions. Each lane was loaded equally with enriched nuclear protein from various genotypes, followed by protein gel blot analyses using antibodies against ELF3 and Histone3 as the loading control.
(B) Quantification of protein levels measuring density/pixel.
(C) and (D) Representatives of transient expression assays in N. benthamiana displayed by bright-field (C) and dark-field (D) imaging of leaves expressing 35S:ELF3:LUC alone, or together with 35S:BBX19, or with 35S:COP1, or with 35S:BBX19 and 35S:COP1 combined, in the presence or absence of the 26S proteasome inhibitor MG132.
(E) Intensity of LUC bioluminescence in control (black bars) and MG132-treated (gray bars) leaves quantified using Andor Solis image analysis software.
For (B) and (E), data are means ± sd (n = 20). Letters above the bars indicate significant differences as determined by Tukey’s HSD method (P < 0.05).
To examine the function of BBX19 as a facilitator of COP-mediated degradation of ELF3, we examined the stability of ELF3 using coinfiltration assays in N. benthamiana leaves. Leaves were infiltrated with various constructs, including 35S:ELF3:LUC alone or in combination with either 35S:BBX19 or 35S:COP1, or together with both 35S:BBX19 and 35S:COP1, in the presence or absence of the 26S proteasome inhibitor, MG132. Collectively, the visual and quantitative luciferase activity analyses by a CCD camera show reduced LUC activity levels as a measure of ELF3 degradation in the presence of either BBX19 or COP1, albeit to a greater extent with COP1 than with BBX19 (Figures 8C to 8E). This decrease in ELF3 stability when coinfiltrated with either BBX19 or COP1 individually is potentially accomplished by the endogenous levels of the N. benthamiana version of the component required to destabilize the protein. Importantly, MG132 abolished the individual or combinatorial destabilizing effects of BBX19 and COP1 on the ELF3 protein (Figures 8C to 8E). In the absence of MG132, degradation of ELF3, however, is strongest in the combinatorial presence of COP1 and BBX19 (Figures 8D and 8E). Interestingly, addition of the proteasome inhibitor MG132 is only able to partially prevent the degradation of ELF3 in the presence of both BBX19 and COP1, potentially due to inadequate uptake of the inhibitor necessary to hinder the abundant presence and enhanced function of COP1 on BBX19-recruited ELF3.
Taken together, these results strongly support the function of BBX19 as an adaptor that binds to, and recruits, ELF3 for degradation by COP1.
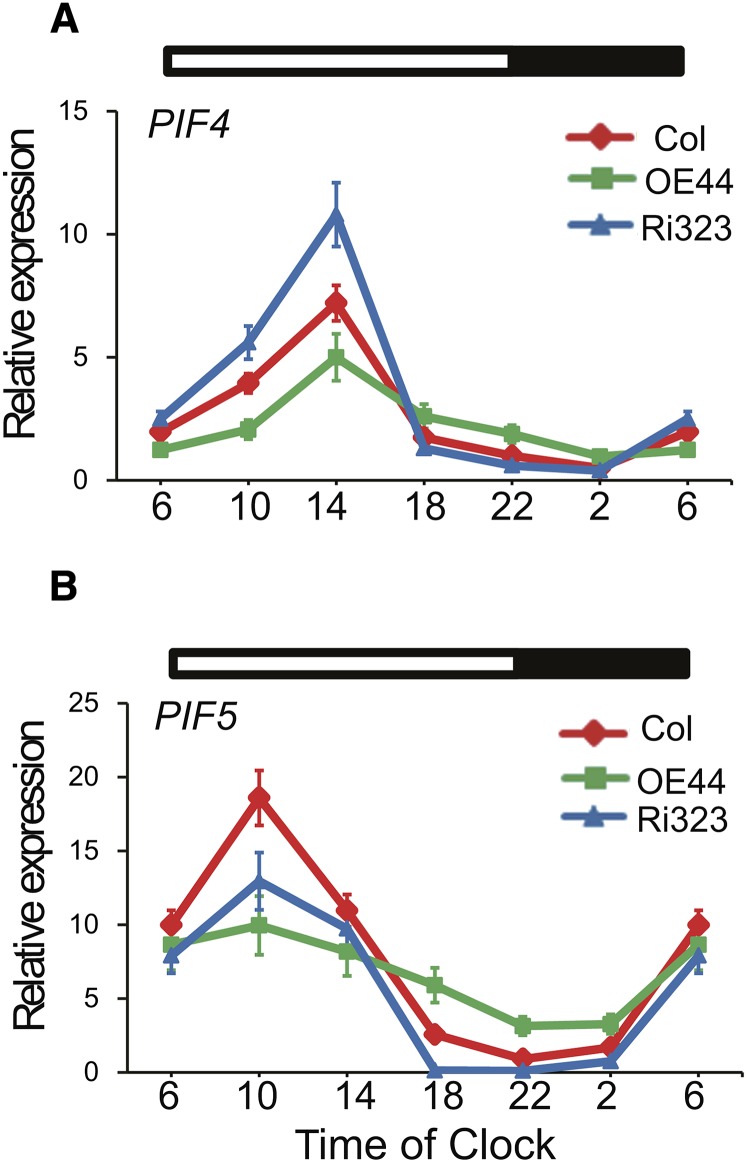
BBX19 Is a Modulator of PIF4 and PIF5 Expression Levels in Early Evening
In view of the BBX19-mediated degradation of a component of the evening complex, ELF3, by COP1, and the PIF4/5-dependent phenotype of the BBX19 overexpressor (Figures 3E, 3F, and 8C to 8E), we questioned whether BBX19 expression levels might influence the amplitude and/or phase of PIF4 and PIF5 rhythmic accumulation in the wild-type, Ri323, and OE44 lines. Thus, we measured the transcript levels of PIF4 and PIF5 every 4 h in a 24-h cycle starting at the onset of light at 6 am in these genotypes. The results show that BBX19 transcript levels influence the amplitude, but not the diurnal rhythms, of PIF4 and PIF5 expression (Figures 9A and 9B). However, a definable positive correlation between changes in PIF4 and PIF5 transcript amplitudes and BBX19 expression levels is observed only between 6 pm and 2 am, the period that overlaps with the repression of PIF4/PIF5 expression by the early evening complex (Nusinow et al., 2011). This suggests that BBX19 modulation of PIF4 and PIF5 transcript levels is restricted in the evening through gating the formation of the evening complex by binding to and recruiting ELF3 for degradation by COP1.
Figure 9.
BBX19 Modulation of PIF4/5 Expression Levels.
Diurnal rhythms of PIF4 (A) and PIF5 (B) expression are shown in Col-0, OE44, and Ri323 lines. Total RNA extracted from these genotypes was subjected to quantitative PCR analysis. The transcript levels of PIF4/5 were normalized to At4g34270 (T1P41-like family protein) and At4g26410 (M3E9) measured in the same samples. The rectangles above the graphs indicate the light (white) and dark (black) conditions. Data are mean fold differences ± sd of three biological replicates each with three technical repeats.
DISCUSSION
Delicate and synchronized responses of plants to a complex range of internal and environmental signals are crucial for coordination of the physiological and metabolic processes required for growth. The complexities of this coordinated regulation have impeded efforts to identify the full complements of growth governing regulatory networks.
Here, we establish BBX19 as a suppressor of seedling photomorphogenesis, functioning through a mechanism of action distinct from those of the other associated and evolutionarily conserved group IV members of the BBX family, which are known to be involved in photomorphogenic processes (Yamawaki et al., 2011). Specifically, we show that BBX19 functions independently of HY5 in regulating seedling growth. This mode of action contrasts with those other members of group IV that interact with HY5, albeit in the opposing direction, with BBX21 and BBX22 acting as transcriptional coactivators and BBX24 and BBX25 functioning as corepressors of HY5 action in photomorphogenesis (Datta et al., 2007, 2008; Chang et al., 2011; Gangappa et al., 2013; Gangappa and Botto, 2014).
The diminished levels and circadian rhythm of the ELF3 protein in OE44, in contrast with those of wild-type plants, established BBX19 as a destabilizing agent of ELF3. This notion is further strengthened by the nuclear colocalization and physical interaction of BBX19 specifically mediated by the BBX19-Box1 motif with ELF3 and COP1. Moreover, this physical interaction enhances the COP1-mediated ubiquitination of ELF3 as opposed to BBX20, BBX21, and BBX22, which repress COP1 function (Gangappa and Botto, 2014).
The genetic, molecular, and biochemical approaches employed here identify BBX19 as an adaptor that binds to and recruits ELF3 for COP1-mediated ubiquitination and, in turn, triggers the degradation and hence depletion of ELF3 for the formation of the evening complex (Nusinow et al., 2011). In the absence and/or reduced levels of the evening complex, two growth-promoting transcription factors, PIF4 and PIF5, continue to be expressed accordingly, thereby permitting hypocotyl elongation.
In summary, as depicted in the proposed model (Figures 10A and 10B), the mechanism of BBX19 action as a suppressor of photomorphogenesis is through hindering the formation of the evening complex by physically interacting with and targeting ELF3 for degradation by COP1 and, as such, lifting and/or reducing the transcriptional repression of PIF4 and PIF5, leading to enhanced hypocotyl growth. This defines BBX19 as a gatekeeper of evening complex formation and, consequently, a modulator of PIF4 and PIF5 expression and, as such, an integrator of clock and light signaling inputs regulating growth.
Figure 10.
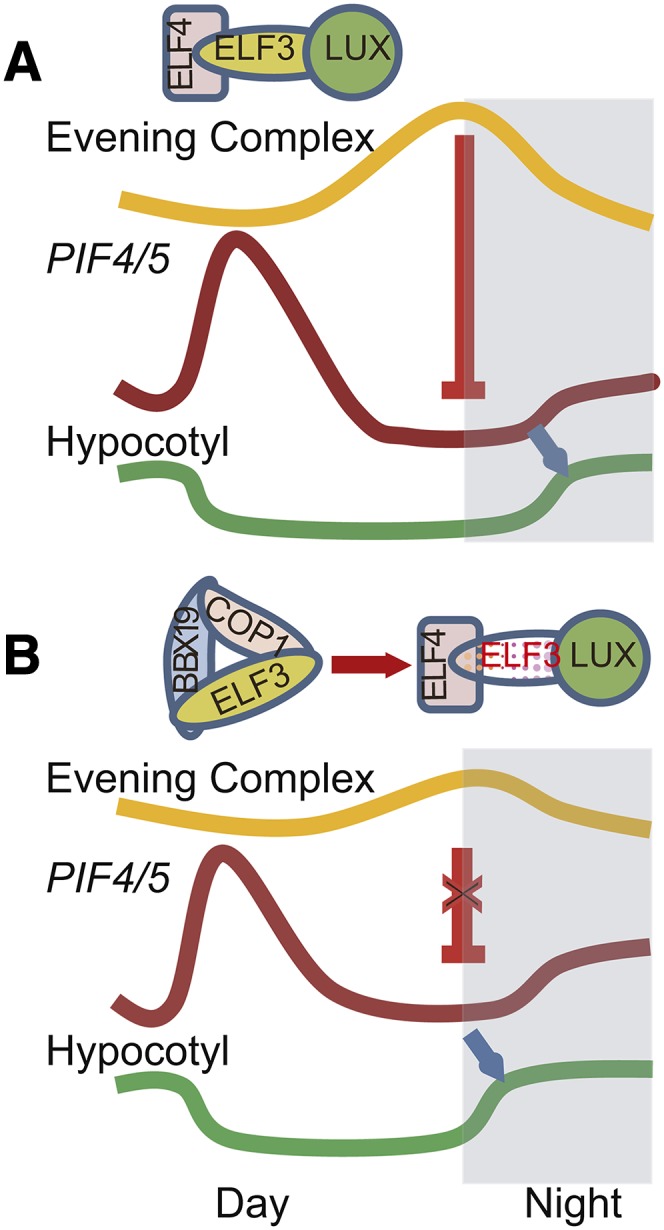
Simplified Schematic Model Representing BBX19 as a Gatekeeper of Evening Complex Formation through Recruitment and COP1-Mediated Degradation of ELF3 Leading to Hypocotyl Elongation.
(A) Model of evening complex (ELF4/ELF3/LUX) repression of PIF4/5 expression and inhibition of hypocotyl elongation modified from Nusinow et al. (2011).
(B) Model of BBX19 depletion/reduction of the ELF3 pool for evening complex formation through recruitment and COP1-dependent degradation of ELF3, enabling PIF4 and PIF5 expression and, consequently, hypocotyl growth.
METHODS
Plant Materials and Growth Conditions
All transgenic lines were generated in Arabidopsis thaliana ecotype Col-0. The PIF quadruple mutant pifq and single mutants pif1-1, pif3-3, pif4-2, and pif5-3 were gifts from Peter Quail (University of California, Berkeley), and cop1-4 was kindly provided by Wei Hu (University of California, Davis). Homozygous plants from all mutants were identified through PCR-based genotyping (primers are listed in Supplemental Table 1).
Arabidopsis plants were grown at 22°C either under LD (16-h-light/8-h-dark cycle) or short-day (8-h-light/16-h-dark cycle) conditions. Plants were also grown under continuous white light or monochromatic wavelengths at various intensities: 100 μmol m−2 s−1 for white light, 50 μmol m−2 s−1 for red light, 10 μmol m−2 s−1 for blue light, and 5 μmol m−2 s−1 for far-red light. Dark-grown seedlings were kept for 3 d on foil-covered plates.
Surface-sterilized seeds were planted on half-strength Murashige and Skoog medium (2.16 g/L Murashige and Skoog salts, pH 5.75, and 8 g/L agar) and imbibed for 3 d at 4°C to synchronize germination. For the time-course expression analysis, leaves of 12-d-old seedlings grown on plates were collected every 4 h over 24 h.
Treatment of Nicotiana benthamiana (wild tobacco) leaves with the proteasome inhibitor MG132 was performed by spraying the leaves with either 50 μM MG132 in 0.5% DMSO (Sigma-Aldrich; M7449) or 0.5% DMSO as the control 12 h prior to imaging with a CCD camera.
Construction of Plasmids and Generation of Transgenic Plants
To overexpress BBX19, HY5, and ELF3, the respective coding regions were amplified from cDNA of Arabidopsis (Col-0) using the listed primers (Supplemental Table 1), followed by cloning them into pENTR-D-TOPO vector (Invitrogen) and subsequent subcloning into Gateway vector pFAST-R05. HY5 and ELF3 constructs were generated using pENTR vectors and recombination with pEN-L4-p35S-R1 and pEN-R2-LUC-L3 using multiple LR reaction and pB7m34GW as the destination vector. The RNAi lines of BBX19 were generated as described previously (Wang et al., 2014). Protein-protein interaction analyses were performed with a split-luciferase construct prepared as described previously (Chen et al., 2008).
Hypocotyl Measurement and Expression Analysis
Hypocotyl length measurements were aided by the ImageJ software. Expression analysis was performed by isolating total RNA from rosette leaves by TRIzol extraction (Life Technologies) and treating it with DNase to control for DNA contamination. One microgram of RNA was reverse transcribed using SuperScript III (Invitrogen). Quantitative PCR was conducted using 6 μL of SYBR Green mix (Bio-Rad; iTaq Universal SYBR Green Supermix), 0.92 μL of 5 ng/μL cDNA, 4.6 μL of water, and 0.24 μL of each primer (10 mM). The Bio-Rad iCycler iQ multicolor real-time detection system was used with a two-step reaction condition (extension temperature was primer-specific but was typically at 60°C), followed by a melt curve encompassing 80 steps of 0.5°C from 60°C to 100°C. Gene-specific primers were designed using the QuantPrime q-PCR primer design tool (http://www.quantprime.de/) and are listed in Supplemental Table 1. The iQ5 Optical System Software version 2.1 (Bio-Rad) was used to calculate relative RNA levels normalized with At4g34270 and At4g26410 used as reference genes for the internal controls as described previously (Walley et al., 2007). Each experiment was performed with three biological replicates and three technical replicates for each.
N. benthamiana and Arabidopsis Transient Transformation
Agrobacterium tumefaciens GV3101 infiltration assays using N. benthamiana leaves were performed as described previously (Wang et al., 2014). Degradation levels were determined by luciferase activity using a CCD camera (Andor Technology) as described previously (Wang et al., 2014). To prevent potential gene silencing, an Agrobacterium strain carrying the p19 suppressor from Tomato bushy stunt virus was coinfiltrated with all the other constructs as described previously (Wang et al., 2014).
Arabidopsis transient assays were performed using 5-d-old seedlings infiltrated with Agrobacterium GV3101 carrying pELF3:ELF3:LUC according to a previously described procedure (Wu et al., 2014). Luciferase activity measurements were performed for 36 h with a CCD camera.
Yeast Two-Hybrid Analysis
A yeast mating-based two-hybrid system was kindly provided by Siobhan Brady (University of California, Davis). According to the protocol described previously (Walhout and Vidal, 2001), full-length cDNAs of BBX19, ELF3, and COP1 were cloned into the pENTR/D-TOPO vector (Invitrogen) for sequencing and subsequently subcloned into the Gateway vector Y-AD or X-BD using an LR reaction to generate the prey or bait plasmid. The plasmids were transformed into the MaV103 and MaV203 yeast strains, respectively. Yeast mating and selection were performed as described previously (Wang et al., 2014).
Immunoprecipitation and Protein Gel Blotting
Plant nuclei were isolated, and nuclear proteins were extracted using the CelLytic PN Isolation/Extraction Kit (Sigma-Aldrich) following the manufacturer’s instructions. For immunoprecipitation, α-GFP antibody (Genescript) was cross-linked to proteins, which were then precipitated using protein A beads (Qiagen) and subsequently analyzed by protein gel blot using α-ELF3 (1:750) as the primary antibody and horseradish peroxidase-conjugated α-rabbit as the secondary antibody (1:5000; Pierce).
Western analyses for HY5 measurements were performed with total protein extract obtained from 10-d-old seedlings grown under LD conditions. The specific α-HY5 antibody was provided by Xing-wang Deng.
Luciferase Imaging
Luciferase imaging was performed as described previously (Walley et al., 2007).
Statistical Analyses
To determine statistical significance, we employed Tukey’s honestly significant difference (HSD) test. The difference was considered significant at P < 0.05.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BBX19 (At4g38960.1), ELF3 (At2g25930.1), HY5 (At5g11260.1), COP1 (At2g32950.1), PIF4 (At2g43010.1), and PIF5 (At3g59060.1).
Supplemental Data
Supplemental Figure 1. BBX19 is a negative regulator of photomorphogenesis.
Supplemental Table 1. List of primers used.
Supplementary Material
Acknowledgments
We thank Marta Bjornson and Jinzheng Wang for their critical reading of the article and Geoff Benn for critical comments on the article and assistance with performing statistical analyses. We also thank Steve Kay for providing the ELF3 antibody, Peter Quail for providing various pif mutant seeds, Wei Hu for the cop1 mutant seeds, and Siobhan Brady for providing the yeast mating-based two-hybrid system. This work was supported by the National Institutes of Health (Grant R01GM107311 to K.D.) and the National Science Foundation (Grants IOS-1036491 and IOS-1352478 to K.D.).
AUTHOR CONTRIBUTIONS
C.-Q.W. planned and executed experimental procedures. M.K.S. executed experiments. J.J. performed experiments. K.D. provided guidance and prepared the article.
Glossary
- MEcPP
methylerythritol cyclodiphosphate
- Col-0
Columbia-0
- RNAi
RNA interference
- LD
long-day
- HSD
honestly significant difference
References
- Ang L.H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213–222. [DOI] [PubMed] [Google Scholar]
- Chang C.S., Li Y.H., Chen L.T., Chen W.C., Hsieh W.P., Shin J., Jane W.N., Chou S.J., Choi G., Hu J.M., Somerville S., Wu S.H. (2008). LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 54: 205–219. [DOI] [PubMed] [Google Scholar]
- Chang C.S., Maloof J.N., Wu S.H. (2011). COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol. 156: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocco C.D., Botto J.F. (2013). BBX proteins in green plants: Insights into their evolution, structure, feature and functional diversification. Gene 531: 44–52. [DOI] [PubMed] [Google Scholar]
- Datta S., Hettiarachchi C., Johansson H., Holm M. (2007). SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19: 3242–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Hettiarachchi G.H., Deng X.W., Holm M. (2006). Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18: 70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Johansson H., Hettiarachchi C., Irigoyen M.L., Desai M., Rubio V., Holm M. (2008). LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20: 2324–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M.R., Davis S.J., Bastow R.M., McWatters H.G., Kozma-Bognár L., Nagy F., Millar A.J., Amasino R.M. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77. [DOI] [PubMed] [Google Scholar]
- Fan X.Y., Sun Y., Cao D.M., Bai M.Y., Luo X.M., Yang H.J., Wei C.Q., Zhu S.W., Sun Y., Chong K., Wang Z.Y. (2012). BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol. Plant 5: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa S.N., Botto J.F. (2014). The BBX family of plant transcription factors. Trends Plant Sci. 19: 460–470. [DOI] [PubMed] [Google Scholar]
- Gangappa S.N., Crocco C.D., Johansson H., Datta S., Hettiarachchi C., Holm M., Botto J.F. (2013). The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 25: 1243–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C.S., Gohda K., Osterlund M.T., Oyama T., Okada K., Deng X.W. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19: 4997–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks K.A., Albertson T.M., Wagner D.R. (2001). EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13: 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M., Deng X.W. (1999). Structural organization and interactions of COP1, a light-regulated developmental switch. Plant Mol. Biol. 41: 151–158. [DOI] [PubMed] [Google Scholar]
- Huang J., Zhao X., Weng X., Wang L., Xie W. (2012). The rice B-box zinc finger gene family: Genomic identification, characterization, expression profiling and diurnal analysis. PLoS ONE 7: e48242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Quail P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21: 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indorf M., Cordero J., Neuhaus G., Rodriguez-Franco M. (2007). Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant J. 51: 563–574. [DOI] [PubMed] [Google Scholar]
- Jiang L., Wang Y., Li Q.F., Björn L.O., He J.X., Li S.S. (2012). Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res. 22: 1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Huq E., Kikis E.A., Al-Sady B., Lanzatella C., Quail P.H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Kikis E.A., Quail P.H. (2003). EARLY FLOWERING 4 functions in phytochrome B-regulated seedling de-etiolation. Plant Physiol. 133: 1530–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Kronmiller B., Maszle D.R., Coupland G., Holm M., Mizuno T., Wu S.H. (2009). The Arabidopsis B-box zinc finger family. Plant Cell 21: 3416–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Shen Y., Toledo-Ortiz G., Kikis E.A., Johannesson H., Hwang Y.S., Quail P.H. (2006). Functional profiling reveals that only a small number of phytochrome-regulated early-response genes in Arabidopsis are necessary for optimal deetiolation. Plant Cell 18: 2157–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai T., Ito S., Nakamichi N., Niwa Y., Murakami M., Yamashino T., Mizuno T. (2008). The common function of a novel subfamily of B-box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 72: 1539–1549. [DOI] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17: 584–593. [DOI] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Monte E., Calderon R.H., Liu T.L., Quail P.H. (2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Miséra S., Deng X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K., Covington M.F., Duek P.D., Lorrain S., Fankhauser C., Harmer S.L., Maloof J.N. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361. [DOI] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J., Leivar P., Gonzalez-Schain N., Sentandreu M., Prat S., Quail P.H., Monte E. (2012). Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J. 71: 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout A.J., Vidal M. (2001). High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24: 297–306. [DOI] [PubMed] [Google Scholar]
- Walley J., Coughlan S., Hudson M.E., Covington M.F., Kaspi R., Banu G., Harmer S.L., Dehesh K. (2007). Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet. e172. [DOI] [PMC free article] [PubMed]
- Wang C.Q., Guthrie C., Sarmast M.K., Dehesh K. (2014). BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell 26: 3589–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zeng J., Deng K., Tu X., Zhao X., Tang D., Liu X. (2011). DBB1a, involved in gibberellin homeostasis, functions as a negative regulator of blue light-mediated hypocotyl elongation in Arabidopsis. Planta 233: 13–23. [DOI] [PubMed] [Google Scholar]
- Wu H.Y., Liu K.H., Wang Y.C., Wu J.F., Chiu W.L., Chen C.Y., Wu S.H., Sheen J., Lai E.M. (2014). AGROBEST: An efficient Agrobacterium-mediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant Methods 10: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Savchenko T., Baidoo E.E., Chehab W.E., Hayden D.M., Tolstikov V., Corwin J.A., Kliebenstein D.J., Keasling J.D., Dehesh K. (2012). Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149: 1525–1535. [DOI] [PubMed] [Google Scholar]
- Yamawaki S., Yamashino T., Nakamichi N., Nakanishi H., Mizuno T. (2011). Light-responsive double B-box containing transcription factors are conserved in Physcomitrella patens. Biosci. Biotechnol. Biochem. 75: 2037–2041. [DOI] [PubMed] [Google Scholar]
- Yan H., Marquardt K., Indorf M., Jutt D., Kircher S., Neuhaus G., Rodríguez-Franco M. (2011). Nuclear localization and interaction with COP1 are required for STO/BBX24 function during photomorphogenesis. Plant Physiol. 156: 1772–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.W., et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta M.T., Hicks K.A., Jacobs C.I., Young J.C., Hangarter R.P., Meeks-Wagner D.R. (1996). The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 10: 691–702. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.