A targeted search for jasmonate-induced metabolites in rice identified an isomer of the common amino acid (S)-α-tyrosine, (R)-β-tyrosine, which may contribute to the allelopathic potential of rice.
Abstract
Non-protein amino acids, often isomers of the standard 20 protein amino acids, have defense-related functions in many plant species. A targeted search for jasmonate-induced metabolites in cultivated rice (Oryza sativa) identified (R)-β-tyrosine, an isomer of the common amino acid (S)-α-tyrosine in the seeds, leaves, roots, and root exudates of the Nipponbare cultivar. Assays with 119 diverse cultivars showed a distinct presence/absence polymorphism, with β-tyrosine being most prevalent in temperate japonica cultivars. Genetic mapping identified a candidate gene on chromosome 12, which was confirmed to encode a tyrosine aminomutase (TAM1) by transient expression in Nicotiana benthamiana and in vitro enzyme assays. A point mutation in TAM1 eliminated β-tyrosine production in Nipponbare. Rice cultivars that do not produce β-tyrosine have a chromosome 12 deletion that encompasses TAM1. Although β-tyrosine accumulation was induced by the plant defense signaling molecule jasmonic acid, bioassays with hemipteran and lepidopteran herbivores showed no negative effects at physiologically relevant β-tyrosine concentrations. In contrast, root growth of Arabidopsis thaliana and other tested dicot plants was inhibited by concentrations as low as 1 μM. As β-tyrosine is exuded into hydroponic medium at higher concentrations, it may contribute to the allelopathic potential of rice.
INTRODUCTION
Plants use a large variety of physical and chemical defenses to protect themselves against pests, pathogens, and competitors in the environment. Many species produce non-protein amino acids, a diverse class of metabolites that can have toxic and deterrent properties (Fowden, 1981; Bell, 2003). Although some non-protein amino acids, e.g., homoserine and ornithine, are essential components of primary metabolism, most of the hundreds of known non-protein amino acids are secondary metabolites with sporadic distribution in the plant kingdom and likely defensive functions. Many non-protein amino acids are toxic analogs of the standard 20-protein amino acids. Incorporation of non-protein amino acids can result in enzyme inhibition or misfolding of proteins. In other instances, non-protein amino acids serve as substrates for the biosynthesis of larger defensive metabolites.
β-Amino acids, which have the amino group shifted from the α-carbon to the adjacent β-carbon, are relatively rare in biological systems. However, they have been found as components of nonribosomal peptides, bacterial antibiotics, anticancer drugs, and other natural products (Kudo et al., 2014). In particular, the β-tyrosine and β-phenylalanine have been investigated as building blocks for the biosynthesis of several medically relevant metabolites. β-Phenylalanine is a component of andrimid, an antibiotic from the bacterium Pantoea agglomerans (Ratnayake et al., 2011), as well as the anticancer drug taxol from yew trees (Taxus sp; Walker et al., 2004). β-Tyrosine is a pathway intermediate in the biosynthesis of several bacterial antibiotics: C-1027 from Streptomyces globisporus (Liu et al., 2002), myxovalargin from Myxococcus fulvus (Krug and Müller, 2009), chondramide C from Chondromyces crocatus (Rachid et al., 2007), kedarcidin from Streptoalloteichus sp (Van Lanen et al., 2005), and maduropeptin from Actinomadura madurae (Van Lanen et al., 2007).
β-Amino acids in microorganisms and plants can be formed from α-amino acids by the action of aminomutases, which catalyze the reversible exchange of an amine group and a hydrogen on adjacent carbons (Lohman and Shen, 2012). One class of aminomutases has structural similarity to phenylalanine aminolyase (PAL) and tyrosine aminolyase (TAL), which catalyze the deamination of l-phenylalanine and l-tyrosine, respectively, to form (E)-cinnamic acid and (E)-4-hydroxycinnamic acid. For instance, a PAL-like phenylalanine aminomutase (PAM) in Tsuga canadensis isomerizes (S)-α-phenylalanine to the (R)-β-isomer (Feng et al., 2011), and a TAL-like tyrosine aminomutase (TAM) from C. crocatus isomerizes (S)-α-tyrosine to (R)-β-tyrosine (Wanninayake and Walker, 2013). The aminolyases and the aminomutases of this enzyme family both use a 3,5-dihydro-5-methylidene-4H-imidazol-4-one (MIO) cofactor in the active site to catalyze the reaction, either removing an amino group or shifting it to an adjacent carbon atom.
Like other plants that have been investigated, rice (Oryza sativa) produces a large variety of defense-related secondary metabolites (Chen et al., 2014; Matsuda et al., 2015). Some of these metabolites have known allelopathic effects, i.e., they inhibit the growth of neighboring plants after being exuded into the soil or paddy water (Olofsdotter, 2001b). There is considerable natural variation in this phenotype, with ∼4% of tested rice cultivars being classified as strongly allelopathic (Belz, 2007). In some cases, specific allelopathic compounds, including the diterpenes momilactone A and B (Kato-Noguchi and Ino, 2003; Kato-Noguchi et al., 2010), phenolic acids (Chung et al., 2001; Rimando et al., 2001; Chung et al., 2002), and flavones (Kong et al., 2004), have been identified in rice exudates and/or extracts. Transposon knockout mutations of rice diterpene synthases reduced momilactone accumulation and compromised the allelopathic effects of rice root exudates on root growth of lettuce (Lactuca sativa) and barnyard grass (Echinochloa crus-galli) (Xu et al., 2012a). Similarly, reducing rice PAL expression using an RNA interference approach attenuated the allelopathic inhibition of barnyard grass root growth, suggesting the involvement of phenolic compounds (Fang et al., 2013). Given the multiple growth-inhibiting compounds that have been identified in rice exudates, it is likely that more than one biosynthetic pathway is required to generate strong allelopathic properties. Nevertheless, natural variation in the production of allelopathic compounds has the potential to be incorporated into breeding programs for production of rice cultivars that are naturally weed-suppressive (Olofsdotter, 2001a; Kong et al., 2011). Such breeding efforts will be facilitated by the identification of specific rice genes that catalyze the formation of allelopathic compounds.
Rice genome sequencing projects (International Rice Genome Sequencing Project, 2005; Xu et al., 2012b), metabolic diversity among cultivars, and new genetic mapping approaches facilitate the identification of genes involved in the biosynthesis of secondary metabolites. Genome-wide association studies demonstrate that, for many secondary metabolites, natural variation among rice cultivars is associated with a small number of quantitative trait loci (QTL) that have large effects on metabolite abundance (Chen et al., 2014; Matsuda et al., 2015). Such large-effect QTL, in combination with information about the types of enzymes that would be involved in the biosynthesis of particular metabolites, make it possible to progress rapidly from the identification of previously unknown rice metabolites to the characterization of genes and enzymes that are involved in their biosynthesis.
Here, we describe the identification of β-tyrosine, which is more commonly associated with the biosynthesis of medically relevant antibiotics in bacteria, in the vegetative tissue and seeds of rice, one of the world’s most important food crops. On the basis of natural variation among rice cultivars, we discovered a rice tyrosine aminomutase that converts the common protein amino acid tyrosine (α-tyrosine) into β-tyrosine. Consistent with a possible defensive function, we show that Pseudomonas syringae growth is inhibited by β-tyrosine, and seedlings of dicot species are much more sensitive to exogenous β-tyrosine than rice and other tested monocots.
RESULTS
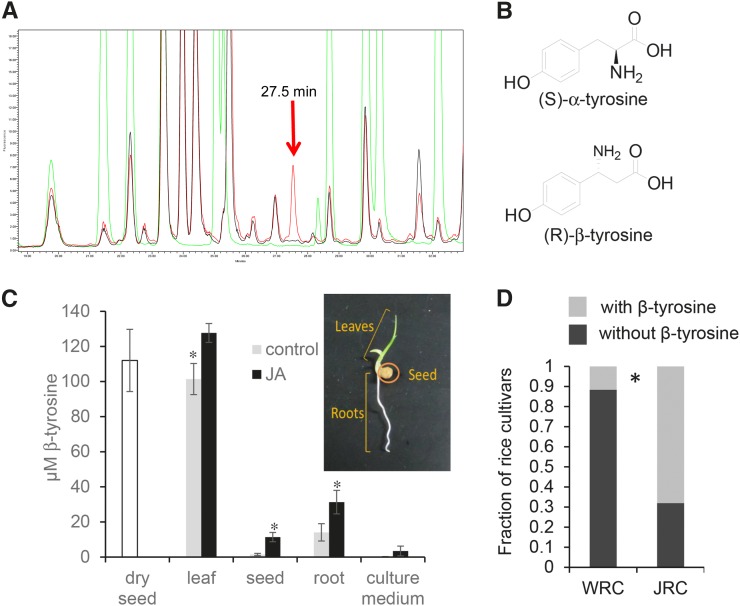
In a targeted effort to find previously unknown rice defensive metabolites, we conducted a screen for non-protein amino acids that are induced by jasmonic acid, a plant defense signaling molecule that is commonly associated with enhanced insect resistance. Liquid chromatography (LC) assays of rice cultivar Nipponbare seedlings identified a peak in the LC chromatogram that was distinct from the twenty protein amino acids (Figure 1A). The area of this unknown peak increased in seedlings that had been treated with jasmonic acid. Another tested rice cultivar, Kasalath, did not appear to contain the compound of interest (Figure 1A).
Figure 1.
Detection of β-Tyrosine in Rice.
(A) HPLC fluorescence detection chromatogram of derivatized rice foliar amino acids. Red, Nipponbare; black, Kasalath; green, amino acid standards (20 protein amino acids, meta-tyrosine, and norleucine). The arrow indicates the induced peak of interest.
(B) Structures of (S)-α-tyrosine and (R)-β-tyrosine.
(C) Relative abundance of (R)-β-tyrosine in dry rice seeds and rice seedlings, with and without jasmonic acid treatment. Mean ± se of n = 3. *P < 0.05, t test comparing control and jasmonic acid-treated samples.
(D) Cultivars that contain β-tyrosine are relatively more abundant in a collection of 50 Japanese rice cultivars (JRC) than in a worldwide collection of 69 rice cultivars (world rice collection [WRC]) from the NIAS Genebank (http://www.gene.affrc.go.jp). *P < 0.05, χ2 test. β-Tyrosine data for each of the 119 tested rice cultivars are in Supplemental Figure 4.
Gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-mass spectrometry assays of Nipponbare and Kasalath samples, as well as comparisons to authentic standards (Supplemental Figures 1 and 2) showed that the unknown peak in the chromatogram is β-tyrosine, an isomer of the common protein amino acid tyrosine (α-tyrosine) (Figure 1B). Liquid chromatography-tandem mass spectrometry comparison of the purified Nipponbare metabolite samples to authentic (R)-β-tyrosine and (S)-β-tyrosine showed that the endogenous rice compound is (R)-β-tyrosine (Supplemental Figure 3).
Although β-tyrosine was originally detected in Nipponbare leaves, subsequent assays showed that it is present in all parts of the rice seedlings, with the highest concentration in the leaves (Figure 1C). Jasmonic acid treatment of the seedlings increased β-tyrosine content in the leaves, roots, and imbibed seed material. Dry rice seeds from plants that had not been elicited with jasmonic acid also contained significant amounts of β-tyrosine. When rice seedlings were grown hydroponically, β-tyrosine was secreted into the medium.
Unlike Nipponbare, the Kasalath rice cultivar does not contain β-tyrosine, with or without jasmonic acid treatment. To investigate the prevalence of β-tyrosine in rice more broadly, we screened cultivars from a Japanese rice collection and a world rice collection that were available from the National Institute of Agrobiological Sciences (NIAS) Genebank (https://www.gene.affrc.go.jp/databases-core_collections_en.php). Among 50 tested Japanese rice cultivars, 32 contained β-tyrosine (Supplemental Figure 4). Including Nipponbare, eight out of 69 tested cultivars in the world rice collection contained β-tyrosine (Supplemental Figure 4B), significantly fewer than in the Japanese rice collection (Figure 1D). Based on their phylogenetic relationship, rice cultivars can be broadly classified into the indica, aus, aromatic, temperate japonica, and tropical japonica populations (Garris et al., 2005). β-Tyrosine production was significantly overrepresented in the japonica cultivars, with 36 of the 40 β-tyrosine-containing cultivars coming from the two japonica groups. The presence of β-tyrosine in the Korean cultivar Milyang 23 (Supplemental Figure 4B) may be explained by its mixed indica and japonica ancestry. Nipponbare, a temperate japonica cultivar, was somewhat unusual in having significantly increased β-tyrosine production in response to jasmonic acid treatment. Among the other 39 β-tyrosine-producing cultivars, only three exhibit a significant β-tyrosine increase in response to jasmonic acid (Supplemental Figure 4).
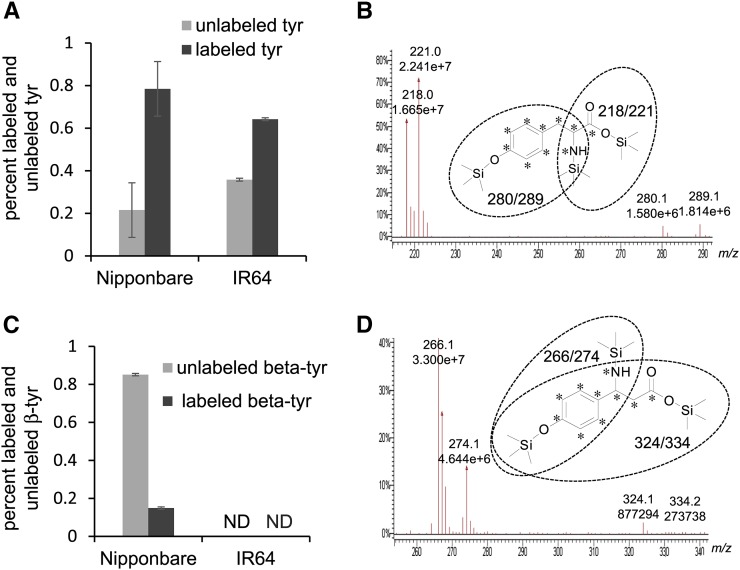
As β-tyrosine can be formed through isomerization of α-tyrosine by PAL-like enzymes in bacteria (Christenson et al., 2003; Krug and Müller, 2009; Wanninayake and Walker, 2013), we conducted stable isotope labeling experiments to determine whether this is also the case in rice. Exogenously added [13C9 15N]tyrosine was taken up efficiently via the petioles of detached leaves of both Nipponbare, which produces β-tyrosine, and IR64, which does not (Figures 2A and 2B). Nipponbare, but not IR64, accumulated [13C9 15N]β-tyrosine in response to the uptake of [13C9 15N]tyrosine from the medium (Figures 2C and 2D). All isotope-labeled carbon atoms and the nitrogen were conserved in the conversion of tyrosine to β-tyrosine by the rice seedlings, indicating TAM activity. More details of the chromatographic analysis that confirmed the synthesis of [13C9 15N]β-tyrosine are provided in Supplemental Figures 5 and 6. Together, these results show that Nipponbare, but not IR64, contains TAM enzymatic activity that converts tyrosine to β-tyrosine.
Figure 2.
Incorporation of [13C915N]Tyrosine into β-Tyrosine after Uptake via Leaf Petioles.
(A) Unlabeled and 13C915N-labeled tyrosine content in rice leaves that had taken up a 1 mM [U-13C9,15N]tyrosine solution via the petioles. Mean ± se of n = 3.
(B) GC-MS chromatograms of unlabeled (m/z 218 and 280) and 13C915N-labeled (m/z 221and 289) tyrosine fragments, derivatized with N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA).
(C) Unlabeled and 13C9,15N-labeled β-tyrosine content in rice leaves that had taken up a 1 mM [13C915N]tyrosine solution via the petioles. Mean ± se of n = 3. ND, not detected.
(D) GC-MS chromatograms of unlabeled (m/z 266 and 324) and 13C9,15N-labeled (m/z 274 and 334) β-tyrosine fragments, derivatized with MSTFA.
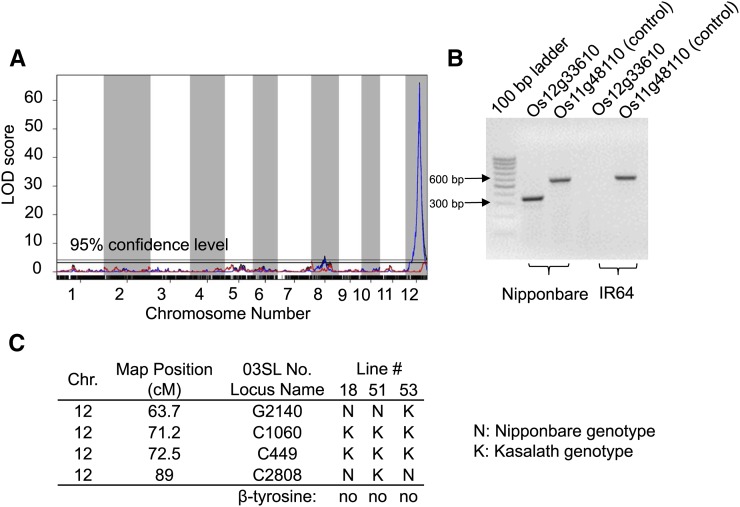
Recombinant inbred lines (RILs) derived from Nipponbare and IR64 were used to map β-tyrosine accumulation. Among 147 RILs derived from the Nipponbare x IR64 cross, 55 accumulated β-tyrosine, and among 147 RILs derived from the IR64 x Nipponbare cross, 47 accumulated β-tyrosine (Supplemental Figure 7). Genetic mapping using this population and scoring β-tyrosine as a binary trait (presence or absence) identified a highly significant QTL on chromosome 12 and a marginally significant QTL on chromosome 8 (Figure 3A). Crosses between indica and japonica rice almost always result in segregation distortion because of the numerous sterility genes that segregate when these two varietal groups are crossed (Spindel et al., 2013). Higher prevalence of the IR64 allele on chromosome 12 in the current mapping population (Supplemental Figure 8) can account for the fact that only about one-third of the RILs, rather than the expected 50%, contain β-tyrosine. Mapping β-tyrosine as a quantitative trait, using only the 102 RILs that contain β-tyrosine, did not identify any additional QTL (Figure 3A).
Figure 3.
Genetic Mapping of Rice β-Tyrosine QTL.
(A) β-Tyrosine QTL mapping with IR64 x Nipponbare recombinant inbred lines. Blue, binary model; red, quantitative model; black, combined.
(B) Detection of LOC_Os12g33610 by PCR of genomic DNA from Nipponbare and IR64.
(C) Three chromosome segment substitution lines have the Kasalath genotype at markers C1060 and C449 on the chromosome inserted in the Nipponbare genetic background. These three lines do not produce β-tyrosine.
The chromosome 12 QTL localized the presumed β-tyrosine biosynthetic gene to an interval containing 25 genes annotated by the Michigan State University Rice Genome Annotation Project (release 7; http://rice.plantbiology.msu.edu/; Kawahara et al., 2013; Supplemental Table 1). Among these genes, LOC_Os12g33610 (International Rice Genome Sequencing Project gene Os12g0520200), which was annotated as a PAL-like gene, stood out as a likely TAM candidate because of the well-established sequence similarity between MIO cofactor-containing aminomutases and aminolyases (Lohman and Shen, 2012). PCR amplification with primers designed to amplify LOC_Os12g33610 produced a product from Nipponbare genomic DNA, but not IR64 genomic DNA (Figure 3B).
Chromosome segment substation lines (CSSLs) that contain segments of Kasalath, which does not produce β-tyrosine (Figure 1A), integrated into the Nipponbare genome (Supplemental Figure 9; Sasaki et al., 2013) were used in a second genetic mapping approach. Among 53 tested CSSLs, 50 had a phenotype similar to that of Nipponbare, accumulating β-tyrosine and generally exhibiting higher accumulation in the leaves after jasmonic acid treatment (Supplemental Figure 9). Three CSSLs did not have any detectable foliar β-tyrosine (Figure 3C). Consistent with a role for LOC_Os12g33610 in β-tyrosine biosynthesis, all three of these CSSLs had an introgression of the Kasalath genome that covered the LOC_Os12g33610 region of chromosome 12.
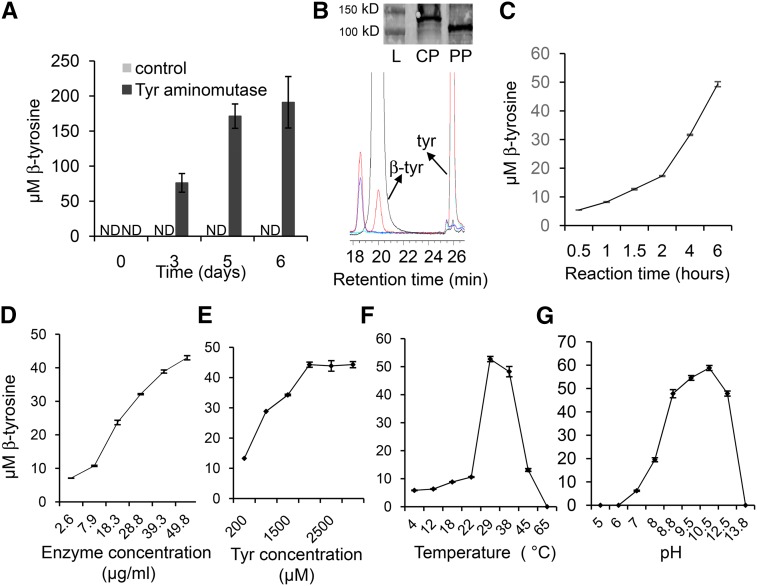
To confirm that LOC_Os12g33610 encodes TAM activity, the gene was transiently expressed in Nicotiana benthamiana, which normally does not contain β-tyrosine (Supplemental Figure 10). Transient expression over 6 d in N. benthamiana progressively increased the amount of β-tyrosine, whereas plants transformed with a control construct accumulated no detectable β-tyrosine in the same time period (Figure 4A). For in vitro enzyme assays, the LOC_Os12g33610 protein was purified from N. benthamiana and was shown to catalyze the conversion of tyrosine to β-tyrosine (Figure 4B). The amount of β-tyrosine that accumulated increased with the reaction time (Figure 4C), the enzyme concentration (Figure 4D), and the substrate concentration (Figure 4E). The temperature optimum of the in vitro reaction was 29°C (Figure 4F) and the pH optimum was 10.5 (Figure 4G). The Km for tyrosine was calculated to be 0.58 mM and the Vmax was 5.1 × 10−6 mM/s (Supplemental Figure 11). Several small deletions and point mutations that were found during the cloning and sequence confirmation of LOC_Os12g33610 eliminated TAM enzymatic activity in N. benthamiana (Table 1). Many of these point mutations are in regions of the protein that are highly conserved in other PAL-like enzymes (Lohman and Shen, 2012). No cinnamic acid or β-phenylalanine was detected using phenylalanine as the substrate, indicating that LOC_Os12g33610 does not encode a PAL or PAM enzyme (Supplemental Figure 12). Similarly, the lack of 4-hydroxycinnamic acid production from tyrosine showed that the enzyme is not a TAL. Therefore, we propose TAM1 as the name for the protein product of LOC_Os12g33610.
Figure 4.
The Rice Gene LOC_Os12g33610 Encodes a Functional Tyrosine Aminomutase That Produces β-Tyrosine.
(A) Accumulation of β-tyrosine in N. benthamiana with transient expression of LOC_Os12g33610. Control, empty vector; ND, not detected. Mean ± se of n = 3.
(B) HPLC chromatogram from in vitro tyrosine aminomutase activity assays. Black, β-tyrosine standard; red, Tris buffer + tyrosine + enzyme; blue, Tris buffer + tyrosine; green, Tris buffer+ enzyme. Inset picture shows an immunoblot. L, ladder; CP, crude protein; PP, His-tag purified protein.
(C) to (E) Accumulation β-tyrosine in vitro increases with the time of the reaction (C), the enzyme concentration (D), and the substrate concentration (E).
(F) and (G) The temperature optimum for the reaction is 29°C (F) and the pH optimum is 10.5 (G). Mean ± se of n = 3.
Table 1. Point Mutations in Rice TAM1 Eliminate β-Tyrosine Production in Vitro.
| Base Pair Change | Amino Acid Change | β-Tyrosine |
|---|---|---|
| Wild type | Wild type | Yes |
| G(371)A-A(400) | deletion | No |
| C-1246-T | Q-560-M | No |
| G(371)A-A(400); C-1246-T | deletion; Q-560-M | No |
| A-794-G | Glu-265-Gly | No |
| T-1409-C | Val-470-Ala | No |
| T-932-C | Met-311-Thr | No |
| A-464-G | Asn-155-Ser | No |
| T-249-C, T-353-C | Ser-83-Ser, Leu-118-Pro | No |
| G-640-A, C-724-T | Ala-214-Thr, Pro-242-Ser | No |
| C-2040-T | Ser-680-Ser | Yes |
| A-2068-G, T-2069-C | Asn-690-Ser | Yes |
To further confirm the in vivo function of TAM1, a TILLING (targeted induced localized lesions in genomes) approach (McCallum et al., 2000) was used to identify point mutations in the endogenous Nipponbare gene. TAM1 DNA segments from 1920 N-methyl-N-nitrosourea (MNU)-mutagenized plants were amplified by PCR and cleaved with CelI nuclease to identify mismatches. Three sites with the GC-to-AT transition that is typical of MNU mutagenesis were identified, causing the amino acid changes Thr-112-Ile, Gln-416-Stop, and Thr-560-Met (Supplemental Figure 13). Of the three point mutations, only Gln-416-Stop eliminated β-tyrosine accumulation in Nipponbare. Nevertheless, this targeted mutagenesis approach confirms that TAM1 is necessary for the production of β-tyrosine in rice.
The Nipponbare rice genome contains nine genes that are annotated as PALs or PAL-like genes (http://rice.plantbiology.msu.edu/; Supplemental Data Set 1; Figure 5). Protein sequence comparisons showed that LOC_Os11g48110 has 95% amino acid sequence identity to TAM1. However, in vitro assays with the LOC_Os11g48110 protein product did not show any TAM activity. Similarly, protein products of three other rice PAL genes (LOC_Os02g41680, LOC_Os04g43800, and LOC_Os02g41650) were tested in vitro and did not exhibit TAM activity.
Figure 5.
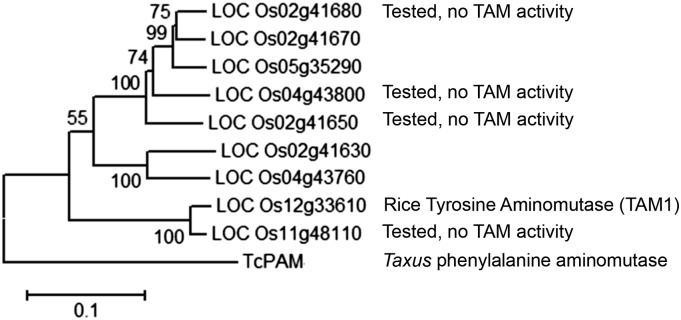
Cladogram of Nipponbare Proteins That Are Annotated as Phenylalanine Ammonia Lyases.
Gene names are from the Michigan State University rice annotation project (http://rice.plantbiology.msu.edu/). The phylogenetic tree was created using MEGA6.06. Numbers at branches indicate bootstrap values. For the amino acid sequence alignments, see Supplemental Data Set 1.
Alignment of available Kasalath genomic sequence data (Sakai et al., 2014) against the Nipponbare genome sequence (International Rice Genome Sequencing Project, 2005) showed that Kasalath has a 30-kb deletion that includes the TAM1 region on chromosome 12. For several of the tested cultivars from the Japanese and world rice cultivar collections, significant amounts of genomic sequence data are available in the NCBI short read archive (http://www.ncbi.nlm.nih.gov/sra; accession number numbers in Table 2). We analyzed these short-read sequencing data to determine the relative coverage of the TAM1 gene and the rest of the rice genome. Whereas rice cultivars that contain β-tyrosine have sequence coverage of TAM1 that is similar to the rest of the genome, those that do not contain β-tyrosine had few or no DNA sequencing reads that align with TAM1 (Table 2). As rice has nine PAL-like genes with similar DNA sequences (Figure 5), there is occasional spurious short-read alignment to TAM1, even in cultivars that do not have this gene. Similar to Kasalath, other rice cultivars that do not produce β-tyrosine have a deletion of the TAM1 gene. A similar analysis of 50 resequenced rice genomes (Xu et al., 2012b), which included all major groupings of cultivated rice and the wild ancestors Oryza rufipogon and Oryza nivara, showed that at least 11 are likely to contain a TAM1 gene (Supplemental Table 2). Seven of the cultivars with a predicted TAM1 are in the temperate japonica phylogenetic group. The others are two aus landraces, one tropical japonica landrace, and one isolate of O. rufipogon.
Table 2. TAM1 Gene Distribution and β-Tyrosine Production in Rice Cultivars.
| Cultivar | Variety Group | Origin | β-Tyrosine Present | TAM1 Present | TAM1 Coverage/Genome Coverage | NCBI/DDBJ Number |
|---|---|---|---|---|---|---|
| IR64 | indica | Philippines | No | No | 0.000 | SRR1328248 |
| Mansaku | japonica | Japan | Yes | Yes | 1.056 | SRR063630 |
| Kameji | japonica | Japan | Yes | Yes | 1.011 | DRR003658 |
| Omachi | japonica | Japan | Yes | Yes | 1.013 | DRR000720 |
| Nipponbare | japonica | Japan | Yes | Yes | 1.000 | SRR1043564 |
| Kasalath | indica | India | No | No | 0.134 | DRR008446 |
| Jena 035 | indica | Nepal | Yes | Yes | 1.117 | SRR1015932 |
| Keiboba | indica | China | No | No | 0.000 | SRR1015906 |
| Shoni | indica | Bangladesh | No | No | 0.131 | SRR1015933 |
| Tupa 121-3 | indica | Bangladesh | No | No | 0.037 | SRR1015927 |
| Surjamukhi | indica | India | No | No | 0.066 | SRR1015924 |
| Ratul | indica | India | No | No | 0.131 | SRR1015920 |
| Badari Dhan | indica | Nepal | No | No | 0.035 | SRR1015922 |
| Kluheenati | indica | Sri Lanka | No | No | 0.138 | SRR1015926 |
| Jaguary | japonica | Brazil | Yes | Yes | 1.045 | SRR1015930 |
| Rexmont | japonica | USA | No | No | 0.876 | SRR1015928 |
| Urasan 1 | japonica | Japan | No | No | 0.094 | SRR1015931 |
| Tupa 729 | japonica | Bangladesh | No | No | 0.064 | SRR1016472 |
| Milyang 23 | indica/japonica | Rep. Korea | Yes | Yes | 1.081 | SRR1239611 |
| Deejiaohualuo | indica | China | No | No | 0.000 | SRR1015904 |
| Hong Cheuh Zai | indica | China | No | No | 0.000 | SRR1015923 |
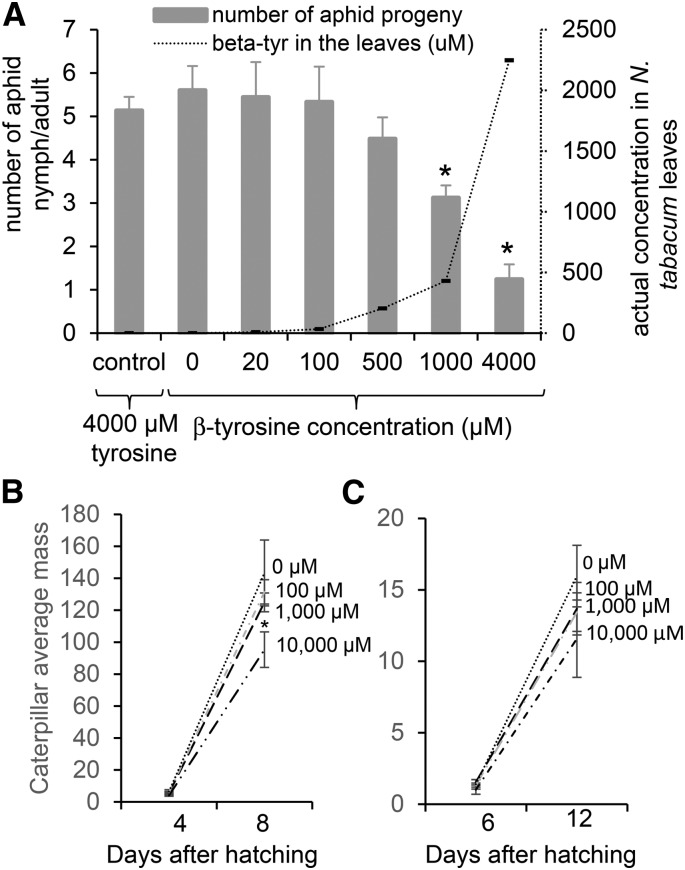
The induction of β-tyrosine by jasmonic acid in Nipponbare, as well as the sporadic distribution among the tested rice cultivars, suggested that this non-protein amino acid might have a defensive function. One hemipteran and two lepidopteran species were used to determine whether β-tyrosine has significant negative effects on insects. β-Tyrosine was taken up into tobacco (Nicotiana tabacum) leaves via the petioles, and aphid reproduction was measured on leaves containing varying amounts of β-tyrosine. Adult Myzus persicae (green peach aphid), a broad host range aphid species that feeds on many dicots and some monocots, were placed onto the tobacco leaves that had taken up β-tyrosine via the petioles. Aphid reproduction was inhibited, but only at 1 and 4 mM β-tyrosine concentrations (Figure 6A), which are significantly higher than that which was observed in rice foliar tissue (Figure 1C). When larvae of black cutworms (Agrotis ipsilon) and sugarcane borers (Diatraea saccharalis), two insects that are known to feed from rice, were placed on artificial diet containing β-tyrosine, no significant growth inhibition was observed (Figures 6B and 6C), even at concentrations that were much higher than those found in rice leaves. Thus, it is unlikely that β-tyrosine has an insect-deterrent function in rice.
Figure 6.
Effects of β-Tyrosine on Insect Growth.
(A) M. persicae reproduction on tobacco leaves taking up solutions containing 0, 20, 100, 500, 1000, or 4000 μM β-tyrosine or 4000 μM tyrosine (control) via the petioles. Aphid reproduction and actual concentration of β-tyrosine in the leaves on which aphids were feeding were measured. Mean ± se of n = 5, *P < 0.01, t test relative to 0 μM β-tyrosine.
(B) Black cutworm larval weight after 4 and 8 d on artificial diet containing 0, 100, 1000, or 10,000 μM β-tyrosine. Mean ± se of n = 6.
(C) Sugarcane borer larval weight after 6 and 12 d on artificial diet containing 0, 100, 1000, or 10,000 μM β-tyrosine. Mean ± se of n = 6, *P < 0.01, t test relative to 0 μM control.
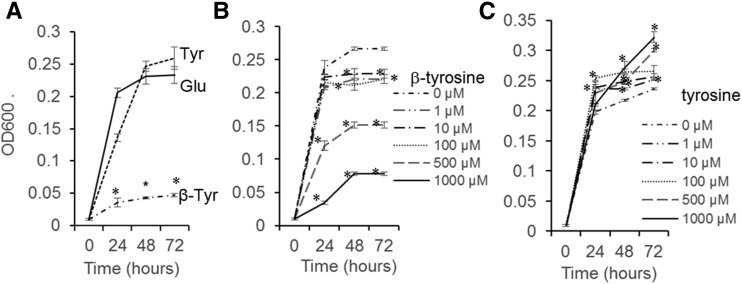
Pseudomonas syringae, a common pathogen of multiple plant species, was used as a representative bacterial species to study the effects of β-tyrosine. Whereas P. syringae was able to grow efficiently on minimal medium with tyrosine or glutamate as the only nitrogen source, the growth rate was significantly reduced when β-tyrosine was the only nitrogen source (Figure 7A). When β-tyrosine was added to minimal medium containing glutamate as the nitrogen source, P. syringae growth was significantly reduced at a concentration of 10 μM β-tyrosine (Figure 7B). As this concentration is lower than that of β-tyrosine in Nipponbare leaves (Figure 1C), it is possible that the in vivo rice β-tyrosine content would be inhibitory to bacterial growth. A similar concentration of tyrosine added to glutamate medium did not reduce P. syringae growth (Figure 7C).
Figure 7.
Effect of β-Tyrosine on P. syringae Strain DC3000 Growth.
(A) Growth of P. syringae in mannitol medium with 0.2 g L−1 of tyrosine, glutamate, or β-tyrosine as the only nitrogen source. Mean ± se of n = 3, *P < 0.001, t test relative glutamate at same day.
(B) Growth of P. syringae in mannitol medium with varying concentrations of β-tyrosine and 0.2 g L−1 glutamate as the nitrogen source. Mean ± se of n = 3, *P < 0.001, t test relative to 0 μm β-tyrosine at the same time point.
(C) Growth of P. syringae in mannitol medium with varying concentration of tyrosine and 0.2 g L−1 glutamate as the nitrogen source. Mean ± se of n = 3, *P < 0.001, t test relative to 0 μm tyrosine at the same time point.
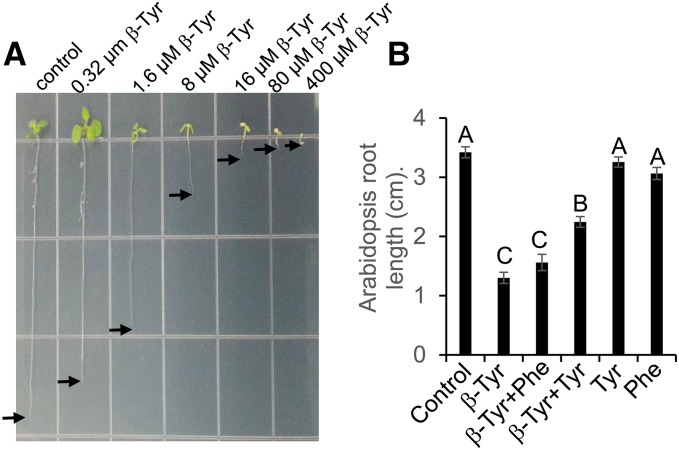
As allelopathic effects have been observed in rice, we conducted experiments to determine whether β-tyrosine could contribute to the growth inhibition of other plants. Arabidopsis thaliana root growth was significantly inhibited (Figure 8A), with an IC50 (50% inhibition of root growth) concentration of 4.4 μM β-tyrosine. Arabidopsis root growth inhibition by 5 μM β-tyrosine could be partially rescued by exogenous addition of 40 μM tyrosine (Figure 8B). β-Tyrosine root growth inhibition assays were conducted with several other plant species and the IC20 (20% inhibition of root growth) was calculated for each species by linear regression across a range of β-tyrosine concentrations (Table 3; Supplemental Figure 14). Among the tested species, dicots exhibited growth inhibition at an ∼100-fold lower β-tyrosine concentration than monocots. Rice itself, both Nipponbare and Kasalath, exhibited the highest β-tyrosine resistance, with no significant growth inhibition at any of the tested β-tyrosine concentrations. The β-tyrosine inhibitory concentrations for Arabidopsis and other dicots were similar to those that are found in the hydroponic medium surrounding Nipponbare roots. This suggests that the β-tyrosine that is found in Nipponbare and other rice cultivars could contribute to the allelopathic effects that have been observed in cultivated rice.
Figure 8.
Inhibition of Arabidopsis Root Growth by β-Tyrosine.
(A) Root elongation is inhibited by increasing β-tyrosine concentration.
(B) Root growth inhibition by 5 μM β-tyrosine is partially rescued by exogenous addition of tyrosine. Mean ± se of n = 11 to 15; different letters indicate significant differences, P < 0.05, Tukey’s HSD test.
Table 3. Root Growth Inhibition by β-Tyrosine.
| Species | IC20 (μM)a |
|---|---|
| Dicot species | |
| Arabidopsis | 0.6 |
| N. tabacum | 0.8 |
| Medicago truncatula | 2.7 |
| Brassica oleracea | 2.8 |
| N. benthamiana | 3.6 |
| Solanum lycopersicum | 4.8 |
| Monocot species | |
| Brachypodium distachyon | 105 |
| Hordeum vulgare | 154 |
| Setaria italica | 382 |
| O. sativa cv Nipponbare | >400 |
| O. sativa cv Kasalath | >400 |
Concentration at which root growth is inhibited by 20%.
DISCUSSION
Our results show that the non-protein amino acid β-tyrosine is an abundant secondary metabolite in some rice cultivars. The genome of Nipponbare, the first sequenced rice cultivar, contains the TAM1 gene, which is required for β-tyrosine production. Presence of the TAM1 gene and production of β-tyrosine occur together, but are sporadically distributed among rice cultivars (Supplemental Figure 4 and Supplemental Table 2). A recent resequencing of 50 rice genomes included five isolates each of the presumed wild ancestors of cultivated rice, O. rufipogon and O. nivara (Xu et al., 2012b). Whereas O. nivara clusters among the indica rice cultivars in a phylogeny of these sequenced genomes, O. rufipogon is more closely related to the japonica cultivars. Consistent with the observation that TAM1 and β-tyrosine are most prevalent in japonica cultivars, we found the TAM1 gene in one out of five sequenced O. rufipogon genomes, but not in any of the five sequenced O. nivara genomes (Supplemental Figure 2). Although the sample size is quite small and there are certainly other possible explanations, TAM1 may represent a presence/absence genomic polymorphism that predates indica and japonica rice domestication.
Rice TAM1 is a member of an MIO-containing class of aminolyases and aminomutases that has been investigated in Taxus and several bacterial species. By catalyzing the conversion of (S)-α-tyrosine to (R)-β-tyrosine, rice TAM1 stereospecificity is similar to the Taxus and C. crocatus aminomutases, which produce (R)-β-phenylalanine and (R)-β-tyrosine, respectively (Walker et al., 2004; Feng et al., 2011), and distinct from those of P. agglomerans and S. globisporus, which produce (S)-β-phenylalanine and (S)-β-tyrosine (Christenson et al., 2003; Ratnayake et al., 2011). MIO-containing aminomutase enzymes are of interest for synthetic biology studies because β-phenylalanine and β-tyrosine are required for the biosynthesis of several microbial antibiotics and anticancer drugs. Thus, our discovery of the rice TAM1 gene provides a new tool for the assembly of these biosynthetic pathways for the production of these medically relevant natural products.
Another rice gene, LOC_Os11g48110, encodes a predicted protein that is ∼95% identical to TAM1 at the amino acid sequence level (Figure 5). However, we were not able to detect any tyrosine aminomutase activity from the protein product, nor did any β-tyrosine QTL localize to LOC_Os11g48110 in the course of our genetic mapping. Given the sequence similarity of the two genes, it is possible that LOC_Os11g48110 also encodes an aminomutase, but perhaps with some other substrate. As MIO domain-containing aminomutases that produce β-phenylalanine have been identified in Taxus and several bacterial species (Feng et al., 2011), phenylalanine also is a possible substrate for this as yet uncharacterized rice gene. Future research will determine whether cultivated rice contains not only TAM but also PAM enzymatic activity.
In many plant species, the signaling molecule jasmonic acid serves as an inducer for the production of metabolites that provide protection against insect herbivores (Howe and Jander, 2008). Although we initially identified β-tyrosine as a jasmonate-induced amino acid in the Nipponbare cultivar, this inducible phenotype appears to be an exception. In almost all other tested cultivars, there was no significant increase in β-tyrosine abundance after jasmonic acid treatment (Supplemental Figure 4). Growth of three insect species, M. persicae, A. ipsilon, and D. saccharalis, was not inhibited by β-tyrosine at concentrations similar to those found in Nipponbare (Figure 6). Thus, β-tyrosine is not likely to have a significant protective effect against insect herbivory in rice.
P. syringae, a generalist bacterial pathogen of plants, was not able to use β-tyrosine as a nitrogen source (Figure 7A). Most likely, these bacteria do not contain the necessary enzymes to catabolize β-amino acids or perhaps amino acids with the (R) rather than the (S) configuration. Significant P. syringae growth reduction by 10 μM β-tyrosine in vitro (Figure 7B) suggests interference with endogenous bacterial metabolism. As the β-tyrosine concentration in rice leaves (Figure 1C) is considerably higher than the inhibitory concentration for P. syringae, there may be some protective effect for the rice plants. Pathogens such as Xanthomonas oryzae, which are more specialized for growing on rice, might be expected to exhibit a higher level of resistance to β-tyrosine.
Root exudates of some rice cultivars show significant allelopathic effects (Belz, 2007). Consistent with this observation, several classes of growth-inhibiting metabolites have been identified in rice exudates (Chung et al., 2001, 2002; Rimando et al., 2001; Kato-Noguchi and Ino, 2003; Kong et al., 2004; Kato-Noguchi et al., 2010), and it is likely that the effects of several of these compounds are combined in the most allelopathic cultivars. Based on the observation that β-tyrosine is found in Nipponbare hydroponic growth medium (Figure 1C) and can inhibit root growth of other plants, in particular dicots, at low concentrations (Table 3), there is some possibility that β-tyrosine contributes to the allelopathic potential of rice. Further research, for instance, field experiments with the TAM1 TILLING knockout mutant (Supplemental Figure 13), will be needed to determine whether β-tyrosine has an allelopathic function.
In a recent study, RNA interference targeting PAL (LOC_Os02g41630) transcription was shown to attenuate inhibition of barnyard grass root growth by a highly allelopathic rice cultivar (Fang et al., 2013). This effect was attributed to a general reduction in the abundance of rice phenolic compounds in root exudates. However, given the DNA sequence similarities of rice PAL and TAL genes, this expression silencing experiment also may have contributed to a reduction in β-tyrosine biosynthesis.
Further research is needed to determine how β-tyrosine inhibits plant growth. A different tyrosine isomer, meta-tyrosine, may provide allelopathic properties to Festuca rubra (Chewings fescue) (Bertin et al., 2007). Selection for Arabidopsis mutants that are resistant to meta-tyrosine identified a feedback-insensitive arogenate dehydratase and greatly increased phenylalanine accumulation (Huang et al., 2010), suggesting that meta-tyrosine interferes with aromatic amino acid metabolism. Although excess tyrosine can partially rescue the negative effects of β-tyrosine on Arabidopsis growth (Figure 8), it remains to be determined whether the improved growth is due to altered β-tyrosine uptake or direct competition between tyrosine and β-tyrosine in endogenous plant metabolism.
To date, β-tyrosine has been investigated most extensively as a component of bacterial polyketide antibiotics and anticancer drugs that have potential applications in human medicine (Lohman and Shen, 2012). Although we cannot rule out incorporation into other biologically active metabolites in rice, β-tyrosine itself certainly accumulates as a biosynthetic end product. With a concentration of ∼20 mg kg−1 (Figure 1B), the β-tyrosine abundance in rice seeds is comparable to that of other free amino acids (Saikusa et al., 1994). Thus, future research on rice β-tyrosine production should consider not only the likely antimicrobial and allelopathic functions in living plants, but perhaps also the as yet unknown metabolic role of this metabolite in human diets.
METHODS
Plants and Growth Conditions
Rice (Oryza sativa) seedlings were grown in Cornell Rice Mix (0.16 m3 peat, 0.34 m3 medium to coarse vermiculite, 2.3 kg lime, and 540 g Peters’ Unimix Plus III [Griffin Greenhouse Supply]). Seeds were soaked in 1.2% sodium hypochlorite solution with shaking for 15 to 20 min, rinsed with sterile deionized four times, and soaked in fresh water overnight. Seeds were planted ∼1 cm deep, and pots were placed in a greenhouse at 26°C with 60% humidity. For normal growth, plants were watered from below to maintain soil moisture. Unless otherwise noted, all plants were used for experiments when they were 2 weeks old, with three full leaves.
Nicotiana benthamiana and Nicotiana tabacum were grown in Cornell mix (by weight, 56% peat moss, 35% vermiculite, 4% lime, 4% Osmocoat slow-release fertilizer [Scotts], and 1% Unimix [Peters]) and were placed in Conviron growth chambers with a 16:8-h light:dark photoperiod, 180 μmol PAR m−2 s−1 light intensity, 23°C temperature, and 60% humidity.
Seeds from a world rice collection (Kojima et al., 2005) and Japanese rice collection (Ebana et al., 2008) were obtained from the National Institute of Agrobiological Sciences (NIAS) Genebank (https://www.gene.affrc.go.jp/databases-core_collections_en.php). Nipponbare/Kasalath CSSLs (http://www.rgrc.dna.affrc.go.jp/ineNKNCSSL48.html) were provided by the Rice Genome Research Center of the Japanese Ministry of Agriculture, Forestry, and Fisheries. Mutant seed stocks for TILLING were provided by the National Bio-Resource of the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan (Suzuki et al., 2008).
Chemicals and Solvents
[13C9 15N]Tyrosine was purchased from Cambridge Isotope Laboratories. Jasmonic acid, (R) and (S)-Boc-β-tyrosine, and O-phthaldehydialdehyde-N-acetyl-l-cysteine reagent were purchased from Cayman Chemical Company, Sigma-Aldrich, and Wako Pure Chemical Industries, respectively. Other chemicals and solvents were obtained from Sigma-Aldrich and Wako Pure Chemical Industries.
β-Tyrosine Structure Determination
Amino acid derivatives were analyzed using an HPLC-MS system. HPLC-MS was performed using an LCMS-2020 equipped with a Prominence HPLC system (Shimadzu) in electrospray ionization (ESI) positive-ion mode. A reversed-phase column (Mightysil RP-18 GP2.0 × 50 mm i.d.; Kanto Chemical Co.) was run at 0.2 mL/min with a gradient of acetonitrile containing 0.1% formic acid in water containing 0.1% formic acid: 1% (0 to 2 min), 1 to 15% (2 to 9 min), 15 to 30% (9 to 14 min), and 30 to 80% (14 to 20 min). The column temperature was maintained at 40°C. The MS was operated with nebulizer gas flow of 1.5 L/min, drying gas flow of 15 L/min, ESI voltage of 1.8 kV, and temperature of 250°C.
For GC-MS analysis, solutions were dried under a nitrogen stream and dissolved in 100 μL of water. Then, 80 μL of an ethanol and pyridine mixture (v/v, 4/1) was added. A 10-μL aliquot of ethyl chloroformate was added and the mixture was shaken for 30 s until the evolution of CO2 gas was complete. The derivatives were extracted with 200 μL of dichloromethane and subjected to GC-MS. GC-MS was performed with an Agilent 6890N GC linked to an Agilent 5975B (Agilent Technologies) operated at 69.9 eV with an HP-5ms capillary column (0.25 mm × 30 m, 0.25-μm film thickness) with helium carrier gas at 1.0 mL/min in the splitless mode. The oven temperature was programmed to increase from 178°C (10 min holding time) to 210°C at 10°C/min, from 210°C to 260°C at 30°C/min (10-min holding time), and then from 260°C to 290°C (5-min holding time) at 30°C/min. The injector temperature was maintained at 240°C, the ion source temperature at 300°C, and the quadrupole temperature at 150°C. ChemStation software (Agilent Technologies) was used for data acquisition. The amino acids were identified by comparing the retention times and fragmentation patterns with those of authentic samples.
The β-tyrosine stereoisomer structure was determined using O-phthaldialdehyde-N-acetyl-l-cysteine (OPA-NAC) reagent (Buck and Krummen, 1987). Thirty milligrams of OPA (Wako Pure Chemical Industries) was dissolved in 1 mL ethanol and diluted with 22 mL of sodium borate buffer (pH 10.0). Thirty milligrams of NAC was dissolved in this OPA solution. A 50-μL aliquot of the sample solution was mixed with 200 μL of OPA-NAC reagent for 10 min at room temperature and analyzed by LCMS-IT-TOF (Shimadzu) using ESI positive ion mode. The CDL temperature was 250°C, the block heater temperature was 200°C, voltage was 1.8 kV, nebulizer gas flow was 1.5 L/min, probe voltage was 4.50 kV, and ion accumulation time was 30 ms. HPLC separation of the reaction mixture was performed on a Mightysil RP-18GP column (50 × 2.0-mm inside diameter). The solvent system consisted of 20 mM ammonium formate (pH 6) (mobile phase A) and a 50% (v/v) mixture of acetonitrile and solution A (mobile phase B). The diastereomeric β-tyrosine derivatives were separated with a gradient of 10 to 30% B solution over 10 min and 99% B solution was maintained for 5 min. (R) and (S)-Boc-β-tyrosine was dissolved in dichloromethane:methanol (9:1) and cooled to 0°C. Trifluoroacetic acid (an amount equal amount to the solvent) was added and the solution was stirred at room temperature. After the starting material was consumed, the solution was concentrated in vacuo.
Stable Isotope Labeling and Detection
Rice plants were inserted into 15-mL tubes containing 1 mM [13C9 15N]tyrosine in water. Control plants were in tubes containing water only. The plants were allowed to stand for 2 h, after which they were sprayed with 1 mM jasmonic acid four times over 2 d. The plants were placed under dome covers and allowed to stand for 2 d. All leaves on each plant were combined as one replicate for derivatization and GC-MS analysis of amino acids.
Amino acid analysis by GC-MS was performed as described previously (Joshi and Jander, 2009), with minor modifications. Leaves were frozen in liquid nitrogen in 2-mL tubes and ground to fine powder with 3-mm steel balls using a Harbil model 5G-HD paint shaker. Ground tissue was extracted with methanol (500 μL per 100 mg fresh leaf tissue). The extracts were centrifuged at 18,000g for 10 min at room temperature. Supernatants (300 μL for each replicate) were dried to completion under nitrogen flow at 70°C. The residue was taken up in 25 μL N-methyl-N-(trimethylsilyl)trifluoroacetamide with 1% trimethylchlorosilane. The samples were heated for a further 50 min at 95°C, and GC-MS analysis was performed using a Varian 1200L GC-MS (Agilent Technologies) with a DB-17 capillary column. Spectra of known amino acids were assigned by reference to a spectral library of amino acid standards and the NIST (National Institute of Standards) mass library.
Amino Acid Assays of Plant Material
Rice seedlings were grown in growth chambers as described above. For elicitation, the leaves were sprayed with 1 mM jasmonic acid four times over 2 d. Control plants were treated with deionized water. About 100 mg of rice tissue was weighed, frozen in liquid nitrogen in 2-mL microcentrifuge tubes, and ground to fine powder with 3-mm steel balls using a Harbil model 5G-HD paint shaker (Fluid Management). Ground tissue was extracted with methanol (5 μL mg−1 of fresh tissue) containing 40 μM norleucine as an internal standard, the extracts were centrifuged at 18,000g for 10 min at 25°C, and the supernatants were saved for analysis.
For amino acid analysis, samples were derivatized using the AccQ-Fluor reagent kit (Waters) according to the manufacturer’s directions. Five-microliter plant extracts were mixed with 35 μL of borate buffer, and the reaction was initiated by the addition of 10 μL 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate reagent (AQC), followed by immediate mixing and incubation for 10 min at 55°C. Twenty microliters of each sample was injected into a Nova-Pak C18 column (4.6 × 150 mm; Waters) and separated using a Waters 2790 HPLC pump system. Solvent A (containing sodium acetate and triethanolamine at pH 5.05) was purchased from Waters; Solvent B was acetonitrile:water (60:40). The gradient used for rice leaf analysis was 0 to 4 min, 15% B; 4 to 14 min, linear gradient to 23.2% B; 14 to 15 min, linear gradient to 26% B; 15 to 22 min, linear gradient to 31.4% B; 22 to 22.5 min, linear gradient to 100% B; 22.5 to 26min, 100% B; 26 to 27 min, linear gradient to 15% B; 27 to 30 min, 15% B. The flow rate was 1.0 mL min−1, and the column temperature was 37°C. Eluted amino acid derivatives were detected using a Waters model 2475 fluorescence detector, with an excitation wavelength of 250 nm and an emission wavelength of 395 nm. Data were recorded and analyzed using Waters Empower software.
For analysis of β-tyrosine in different plant parts (Figure 1C), seedlings were grown hydroponically for 2 weeks and transferred to tubes filled with 4 mL of hydroponic medium. Induction was performed by spraying 1 mM jasmonic acid (Cayman Chemical) once every 12 h for 2 d. Before the amino acid extraction, seedlings were separated into leaves, seed, and roots and crushed with three stainless steel beads (3 min on a shaker, 3000 strokes/minute) in 300 μL of methanol. After centrifugation for 5 min at 3000 rpm, 5 μL of the supernatant was used in AQC derivatization as described above. The medium were dried up using a rotary evaporator and dissolved in a mixture of 5 μL of methanol and 70 μL of borate buffer. Dry seeds were extracted and the samples were derivatized in the same manner. The β-tyrosine AccQ derivative was analyzed with a Shimadzu 2020 HPLC-MS system, as described above.
Preparation of Genomic DNA, RNA, and cDNA
Rice leaf material was harvested from seedlings, flash-frozen in liquid nitrogen, and stored at −80°C until sample preparation. After grinding of the frozen leaf material to a fine powder in a mortar filled with liquid nitrogen, DNA and RNA were extracted using the TRIzol reagent (Life Technologies) and SV Total RNA Isolation System (Promega), respectively, according to the manufacturers’ instructions. Nucleic acid concentration, purity, and quality were assessed using a spectrophotometer (NanoDrop 2000c; Thermo Scientific). Single-stranded cDNA was prepared using SuperScript III reverse transcriptase and oligo(dT20) primers (Life Technologies).
Cloning and Expression of TAM Genes
The complete open reading frames of TAM1 and other rice PAL-like genes were amplified from cDNA with the primer pairs listed in Supplemental Table 3, with the exception of LOC_Os11g48110. The LOC_Os11g48110 could not be amplified from cDNA and instead was amplified from genomic DNA. Subsequently, the intron section was deleted using an overlap PCR method. PCR products were purified by Wizard SV Gel and PCR Clean-Up System (Promega), cloned as blunt fragments into the entry vector pDonr207, and then transferred to the destination vector pMDC32 using the Gateway recombination system (Life Technologies). Both strands were fully sequenced to confirm that there were no errors introduced during the PCR amplification. The constructs were introduced into the Escherichia coli strain DH5α. To overexpress rice TAM1 and PAL in N. benthamiana, the destination vectors with the transgenes were transfer into Agrobacterium tumefaciens strain GV3101. Liquid cultures of the bacteria harboring the expression constructs and a silencing suppressor (P38 carrying T-DNA constructs expressing the turnip crinkle virus capsid protein to reduce expression silencing; Thomas et al., 2003) were grown at 29°C to an OD600 of 0.6 to 0.7 with the antibiotics kanamycin (50 μg mL−1) and rifampicin (25 μg mL−1). Bacterial cultures were centrifuged at 3200g for 10 min, washed one time with infiltration buffer (10 mM MgCl2, 10 mM MES, and 200 μM acetosyringone), and resuspended in infiltration buffer to OD600 of 0.45 for expression constructs and OD600 of 0.9 for the silencing suppression constructs. Both cultures were incubated individually for 2 h. The construct-containing and P38-containing bacterial strains were mixed at a 1:1 ratio, and N. benthamiana leaves were infiltrated with the bacterial solution using a 1-mL syringe. Excess bacterial solution was wiped off with paper towel. Five days after infiltration, 8-mm-diameter leaf plugs were collected to confirm expression of measurement of amino acids by HPLC fluorescence detection using the same protocol as that used for rice tissue.
Protein Purification and in Vitro Assays
Three days after infiltration of destination vector pYL436 with the transgene into N. benthamiana, as described above, leaves were collected and ground in liquid nitrogen. About 1 cc 1-mm zirconia beads was added to each 15-mL tube, the 5 to 6 mL of ground tissue were extracted with 2 mL extraction buffer (100 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2 mM EDTA, 2 mM EGTA, 1% Triton-X, 0.1% BME, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 1 mM DTT, and 0.1% protease inhibitor cocktail [P8849; Sigma-Aldrich]) and shaken on a paint shaker 4 × 1 min, with 1 min incubation on ice between shaking periods. The extraction buffer was centrifuged at 21,000g, 1 mL of the supernatant was collected, and 40 μL IgG Sepharose 6 Fast Flow (GE Healthcare) were added. Samples were placed in 2-mL tubes on a roller drum for 2 h at 4°C. After 2 h, tubes were centrifuged at 400g for 2 min at 4°C, the supernatant was removed, and the beads were washed three times with 0.5 mL wash buffer (100 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2 mM EDTA, 2 mM EGTA, 0.5% Triton-X, 0.1% 2-mercaptoethanol, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 1 mM DTT, and 0.1% protease inhibitor). Beads were washed with 1 mL cleavage buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT), and 70 μL of cleavage buffer containing 1.4 μL Turbo3C cleavage protease was incubated for 10 h with rotation at 4°C. The supernatant was mixed with 5 μL washed GST beads (Clontech) and rotated at 4°C for 1 h. The supernatant was collected, mixed with 25% glycerol, and used immediate for enzyme activity assays or stored at −80°C. The protein concentration was determined using a Bradford protein assay kit (Bio-Rad). Proteins were transferred to a polyvinylidene fluoride membrane (Millipore) for protein gel blot analysis. Antibodies used were as follows: mouse anti-MYC and mouse anti-rat IgG peroxidase (Santa Cruz Biotechnology).
The in vitro enzymatic activity of rice TAM1 was measured in assays with purified recombinant protein and tyrosine or phenylalanine as the substrate. To measure time dependence, assays were performed with 0.1 mM Tris-HCl buffer, pH 8.8, 0.05 mM potassium chloride, 0.2 mM tyrosine, and 0.49 μg enzyme solution for 0.5, 1, 1.5, 2, 4, and 6 h. To measure temperature dependence, assays were performed with 0.1 mM Tris-HCl buffer, pH 8.8, 0.05 mM potassium chloride, 0.2 mM tyrosine, and 0.49 μg enzyme solution at 4, 12, 18, 22, 29, 38, 45, and 65°C. The pH dependence assays were performed with 0.05 mM potassium chloride, 0.2 mM tyrosine, and 0.49 μg enzyme solution at pH 5, 6, 7, 8, 8.8, 9.5, 10.5, 12.5, and 13.5 in 0.1 mM Tris-HCl buffer. To vary substrate concentration, 200, 500, 1500, 2000, 2500, and 3500 μM tyrosine were added to 0.1 mM Tris-HCl buffer, pH 8.8, 0.05 mM potassium chloride, and 0.49 μg enzyme, and 2.6, 7.9, 18.3, 28.3, 39.3, and 49.8 μg/mL enzyme was added to 0.1 mM Tris-HCl buffer, pH 8.8, 0.05 mM potassium chloride, and 0.2 mM tyrosine. All of the assays were performed in 0.2-mL tubes, with a 12.5-μL total reaction volume. All samples were incubated for 6 h at 29°C, with the exception of experiments in which the reaction times and temperatures were specifically varied. The reaction tubes were centrifuged at 2000g, and the supernatant was collected for amino acid analysis. Five microliters of each extract was derivatized for amino acid analysis using the AccQ-Fluor reagent kit (Waters). Reaction products were detected by LC using an approach similar to that used for measuring amino acids in rice tissue. The gradient conditions were 0 to 23 min, linear gradient to 14% B; 23.1 to 26 min, linear gradient to 25% B; 26.1 to 32 min, 100% B; 32.1 to 36 min, 14% B. The flow rate was 1.0 mL min−1, and the column temperature was 32°C.
Pseudomonas syringae Bioassays
For in vitro growth experiments, P. syringae strain DC3000 was cultured at 30°C in modified mannitol-glutamate medium (Kean et al., 1970; per liter: 10 g mannitol, 0.2 g glutamate, 0.5 g KH2PO4, 0.2 g NaCl, 0.2 g MgSO4-7H2O; pH adjusted to 7.0 with 1 n NaOH prior to autoclaving). To test growth with other amino acids as nitrogen sources, glutamate was replaced successively with 0.2 g L−1 β-tyrosine or tyrosine. Cultures were inoculated by diluting a fresh overnight culture of P. syringae 1:100 (50 μL into 5 mL). OD600 was measured at 0, 24, 48, and 72 h after treatment. For growth inhibition experiments, cultures were grown in mannitol-glutamate medium and 0, 1, 10, 100, 500, and 1000 μM β-tyrosine or tyrosine.
Insect Bioassays
Tobacco seedlings were used for Myzus persicae bioassays. The detached tobacco leaves were inserted into 2-mL microcentrifuge tubes containing 20, 100, 500, 1000, or 4000 μM β-tyrosine. Water and 4000 μM tyrosine were used as controls. After 3 h, five M. persicae adult aphids were put on the tobacco leaves. After 6 d, nymphs were counted and the tobacco leaves were harvested to determine the β-tyrosine and tyrosine content in the leaves on which the aphids had been feeding.
For black cutworm (Agrotis ipsilon) and sugarcane borer (Diatraea saccharalis) experiments, eggs were obtained from Benzon Research. Eggs were hatched at 29°C, and five neonate caterpillars were placed in each Petri dish (35 × 10 mm) with 0, 100, 1000, or 10,000 μM β-tyrosine added to artificial diet (Multiple Species Diet; Southland Products). After 4 d for black cutworms and 6 d for the sugarcane borer, caterpillars were weighed and moved to fresh diet with the same concentration of β-tyrosine. After an additional 4 or 6 d, respectively, the caterpillars were weighed again.
Agar Plate Bioassays with Arabidopsis and Other Seeds
To assess effects of β-tyrosine on the root growth of other plants, seeds were sterilized with 50% ethanol for 30 s, followed by 50% bleach for 2 min, and four rinses with sterile distilled water. Agar medium was prepared by mixing 250 mL water, 1.09 g Murashige and Skoog salts (Murashige and Skoog, 1962), and 2 g Phytagar, with pH adjusted to 5.7 with 1 M KOH. β-Tyrosine at concentrations ranging from 0 to 400 μM was added after autoclaving. Seeds were planted on the agar medium in square Petri dishes (100 × 100 × 15 mm), were placed at 4°C for 4 d, and then placed vertically in a growth chamber with a 16:8 h light:dark photoperiod, 180 μmol photons m−2 s−1 light intensity, 23°C temperature, and 60% humidity. Root lengths were measured after 5 d for Medicago truncatula, barley (Hordeum vulgare), and Brassica oleracea, 8 d for Arabidopsis thaliana, Brachypodium distachyon, Setaria italica, and Solanum lycopersicum, 11 d for N. tabacum and N. benthamiana, and 6 d for rice. In separate experiments, rescue of 5 μM β-tyrosine toxicity for Arabidopsis was assessed by adding 40 μM tyrosine or phenylalanine to the agar.
Quantitative Trait Mapping
A population of 322 F9 RILs was developed by single-seed descent from reciprocal crosses between Nipponbare and IR64. Total genomic DNA was isolated and purified using the 96-plex DNeasy kit (Qiagen). The RILs and the two parental lines were genotyped using 96-plex genotyping-by-sequencing (GBS) as described previously (Elshire et al., 2011; Spindel et al., 2013). The GBS data were analyzed using the TASSEL 3.0 GBS pipeline, and the results aligned to the MSU v.7.0 rice genome using Bowtie2, as described by Spindel et al. (2013). Data were imputed using Plaid impute (-m 15, -n 60, -w 5). PLUMAGE python scripts were used postimputation to perform a sequence error correction and remove singe-nucleotide polymorphisms (SNPs) with call rates ≤0.75, for a total SNP data set of 86,528 SNPs and a genetic map consisting of ∼1417 centimorgans. See Spindel et al. (2013) for details on the analysis pipeline and http://ricediversity.org/data for the molecular marker data set. A graphic representation of the genetic map, produced using r/qtl, confirmed that there were no distortions in the genetic map. β-Tyrosine content was measured by HPLC, as described above, in 147 RILs derived from Nipponbare x IR64 and 147 derived from IR64 x Nipponbare.
QTL mapping was performed using the above genotype and phenotype data sets in R version 3.0.1 using the r/qtl package (v. 1.27-10). Three QTL models were tested: a binary model, a parametric quantitative model, and a combined binary+quantitative model. For the binary model, phenotypes were coded as either 1 or 0, where 1 indicated presence of β-tyrosine and 0 indicated absence. The QTL mapping script published with Spindel et al. (2013) was used, except all references to the “normal” model were switched to “binary.” For the quantitative model, only individuals expressing β-tyrosine were included in the analysis, and the quantitative phenotypes were used. The same QTL mapping script was used to run the analysis, only with the model set to “normal.” The combined model (model = 2part) is recommended for use when there is a large spike at one phenotype, “0” in our case, and automates the process of testing both a quantitative and binary model (for details, see Broman and Sen, 2009). The QTL mapping script was modified to run the combined model in accordance with Broman and Sen (2009). Figure 3A shows the output of the combined model (rqtl plot () function). LOD significance thresholds were determined using 1000 permutations.
Chromosome segment substitution lines from an existing collection of lines with Kasalath genomic segments inserted in Nipponbare (Yano, 2001; Sasaki et al., 2013) were used for genetic mapping. Foliar β-tyrosine content was measured from seedlings with and without jasmonate treatment, as described above. Lines that did not produce β-tyrosine due to a Kasalath genomic insertion were identified based on the chromatograms.
Mutant Identification by TILLING
To identify point mutations in the LOC_Os12g33610 gene, DNA samples of 1920 M1 plants originating from the treatment of Nipponbare seeds with MNU was used for TILLING (Till et al., 2003). DNA pools and mutant seed stocks were provided by the National Bio-Resource of MEXT, Japan (Suzuki et al., 2008). PCR primer pairs that were used to amplify the LOC_Os12g33610 region are described in Supplemental Table 3. PCR was conducted using LA taq (with GC buffer) (Takara-Bio). Amplicons were digested with CelI nuclease and were electrophoresed to find single nucleotide polymorphisms. M1 plants that exhibited and extra band in the gels were sequenced to identify point mutations in LOC_Os12g33610.
Sequence Analysis and Phylogenetic Tree Reconstruction
Protein sequences for predicted Nipponbare rice PAL proteins were downloaded from http://rice.plantbiology.msu.edu/. The Tsuga canadensis PAM gene was downloaded from GenBank (GI:634380). A protein alignment was implemented using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Based on the ClustalW alignment, a phylogenetic tree was constructed with MEGA6 (http://www.megasoftware.net/) using neighbor joining. All positions with <90% site coverage were eliminated. Ambiguous bases were allowed at any position. A bootstrap resampling analysis with 1000 replicates was performed to evaluate the tree topology.
Bioinformatic Analyses of Rice Genomic Sequences
Illumina sequence reads were mapped to the Nipponbare genome with Bowtie 2 v 2.2.3 (Langmead and Salzberg, 2012). Coverage of the Nipponbare genome and the TAM gene were calculated using Bedtools (Quinlan and Hall, 2010).
Accession Numbers
DNA sequences used in this article can be found at http://rice.plantbiology.msu.edu/index.shtml and in the NCBI Short Read Archive (see Supplemental Table 2 for accession numbers).
Supplemental Data
Supplemental Figure 1. Detection of an unknown rice amino acid.
Supplemental Figure 2. Total ion chromatogram from GC-MS analysis of ethyl chloroformate derivatives.
Supplemental Figure 3. Identification of the β-tyrosine stereoconfiguration by LC-MS/MS.
Supplemental Figure 4. β-Tyrosine abundance in rice cultivars, with and without jasmonic acid treatment.
Supplemental Figure 5. GC-MS profile of tyrosine and β-tyrosine standards, derivatized with MSTFA.
Supplemental Figure 6. GC-MS profile confirms that tyrosine is converted to β-tyrosine in rice.
Supplemental Figure 7. Mapping β-tyrosine as a quantitative trait using recombinant inbred lines.
Supplemental Figure 8. Frequency of Nipponbare and IR64 molecular markers in the RIL population.
Supplemental Figure 9. Mapping β-tyrosine as a quantitative trait using chromosome segment substitution lines.
Supplemental Figure 10. β-Tyrosine production by transient expression in N. benthamiana.
Supplemental Figure 11. Enzymatic properties of rice TAM1.
Supplemental Figure 12. Rice TAM1 functions as a tyrosine aminomutase.
Supplemental Figure 13. Identification of TAM1 mutations from a rice TILLING population.
Supplemental Figure 14. β-Tyrosine inhibition of plant root growth.
Supplemental Table 1. Genes in the mapping region of the β-tyrosine QTL.
Supplemental Table 2. Presence of TAM1 in 50 resequenced rice genomes.
Supplemental Table 3. Primers used in this study.
Supplemental Data Set 1. Alignment of protein sequences that were used for the phylogenetic tree construction in Figure 5.
Supplementary Material
Acknowledgments
We thank Svetlana Temnykh, Sandra Harrington, and Fumio Onishi for assistance in generating recombinant inbred lines from crosses between Nipponbare and IR64. This research was funded by US National Science Foundation Awards IOS-1139329 to G.J. and PGRP-1026555 to S.R.M., by a grant from the Japan Science and Technology Agency to Y.O. and N.M., and by a fellowship from Science and Technology Star of Zhujiang, Guangzhou City (2013J2200082) to J.Y. S.R.S. was funded by the Boyce Thompson Institute.
AUTHOR CONTRIBUTIONS
J.Y. contributed to Figures 1A, 1B, 2, 3A, 3B, and 4 to 8, Tables 1 and 3, Supplemental Figures 1, 5, 6, 7, 10 to 12, and 14, Supplemental Tables 1 and 3, Supplemental Data Set 1, and article writing. T.A. contributed to Figures 1C, 1D, and 3C, Tables 2 and 3, and Supplemental Figures 2 to 4 and 9. M.T. contributed to Figure 3B and Supplemental Figures 9 and 13. S.R.S. contributed to Table 2 and Supplemental Table 2. J.E.S. and C.W.T. contributed to Figure 3A and Supplemental Figure 8. R.T. contributed to Figure 3B and Supplemental Figures 9 and 13. F.M. contributed to Figure 1C and Table 3. Y.M. contributed to Figure 1C and Supplemental Figure 3. S.R.M., Y.O., and N.M. contributed to experimental design and data analysis. G.J. contributed to experimental design, data analysis, and article writing.
Glossary
- PAL
phenylalanine aminolyase
- TAL
tyrosine aminolyase
- PAM
phenylalanine aminomutase
- TAM
tyrosine aminomutase
- MIO
3,5-dihydro-5-methylidene-4H-imidazol-4-one
- QTL
quantitative trait loci
- LC
liquid chromatography
- GC-MS
gas chromatography-mass spectrometry
- RIL
recombinant inbred line
- CSSL
chromosome segment substation line
- MNU
N-methyl-N-nitrosourea
- NIAS
National Institute of Agrobiological Sciences
- ESI
electrospray ionization
- OPA-NAC
O-phthaldialdehyde-N-acetyl-l-cysteine
- GBS
genotyping-by-sequencing
- SNP
singe-nucleotide polymorphism
References
- Bell E.A. (2003). Nonprotein amino acids of plants: significance in medicine, nutrition, and agriculture. J. Agric. Food Chem. 51: 2854–2865. [DOI] [PubMed] [Google Scholar]
- Belz R.G. (2007). Allelopathy in crop/weed interactions—an update. Pest Manag. Sci. 63: 308–326. [DOI] [PubMed] [Google Scholar]
- Bertin C., Weston L.A., Huang T., Jander G., Owens T., Meinwald J., Schroeder F.C. (2007). Grass roots chemistry: meta-tyrosine, an herbicidal nonprotein amino acid. Proc. Natl. Acad. Sci. USA 104: 16964–16969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K.W., Sen N. (2009). A Guide to QTL Mapping with R/qtl. (New York: Springer; ). [Google Scholar]
- Buck R.H., Krummen K. (1987). High-performance liquid chromatographic determination of enantiomeric amino acids and amino alcohols after derivatization with o-phthaldialdehyde and various chiral mercaptans. Application to peptide hydrolysates. J. Chromatogr. A 387: 255–265. [DOI] [PubMed] [Google Scholar]
- Chen W., et al. (2014). Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Genet. 46: 714–721. [DOI] [PubMed] [Google Scholar]
- Christenson S.D., Liu W., Toney M.D., Shen B. (2003). A novel 4-methylideneimidazole-5-one-containing tyrosine aminomutase in enediyne antitumor antibiotic C-1027 biosynthesis. J. Am. Chem. Soc. 125: 6062–6063. [DOI] [PubMed] [Google Scholar]
- Chung I.M., Ahn J.K., Yun S.J. (2001). Identification of allelopathic compounds from rice (Oryza sativa L.) straw and their biological activity. Can. J. Plant Sci. 81: 815–819. [Google Scholar]
- Chung I.M., Kim K.H., Ahn J.K., Chun S.C., Kim C.S., Kim J.T., Kim S.H. (2002). Screening of allelochemicals on barnyardgrass (Echinochloa crus-galli) and identification of potentially allelopathic compounds from rice (Oryza sativa) variety hull extracts. Crop Prot. 21: 913–920. [Google Scholar]
- Ebana K., Kojima Y., Fukuoka S., Nagamine T., Kawase M. (2008). Development of mini core collection of Japanese rice landrace. Breed. Sci. 58: 281–291. [Google Scholar]
- Elshire R.J., Glaubitz J.C., Sun Q., Poland J.A., Kawamoto K., Buckler E.S., Mitchell S.E. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6: e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C., Zhuang Y., Xu T., Li Y., Li Y., Lin W. (2013). Changes in rice allelopathy and rhizosphere microflora by inhibiting rice phenylalanine ammonia-lyase gene expression. J. Chem. Ecol. 39: 204–212. [DOI] [PubMed] [Google Scholar]
- Feng L., Wanninayake U., Strom S., Geiger J., Walker K.D. (2011). Mechanistic, mutational, and structural evaluation of a Taxus phenylalanine aminomutase. Biochemistry 50: 2919–2930. [DOI] [PubMed] [Google Scholar]
- Fowden L. (1981). Non-protein amino acids of plants. Food Chem. 6: 201–211. [Google Scholar]
- Garris A.J., Tai T.H., Coburn J., Kresovich S., McCouch S. (2005). Genetic structure and diversity in Oryza sativa L. Genetics 169: 1631–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G.A., Jander G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59: 41–66. [DOI] [PubMed] [Google Scholar]
- Huang T., Tohge T., Lytovchenko A., Fernie A.R., Jander G. (2010). Pleiotropic physiological consequences of feedback-insensitive phenylalanine biosynthesis in Arabidopsis thaliana. Plant J. 63: 823–835. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- Joshi V., Jander G. (2009). Arabidopsis methionine gamma-lyase is regulated according to isoleucine biosynthesis needs but plays a subordinate role to threonine deaminase. Plant Physiol. 151: 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato-Noguchi H., Ino T. (2003). Rice seedlings release momilactone B into the environment. Phytochemistry 63: 551–554. [DOI] [PubMed] [Google Scholar]
- Kato-Noguchi H., Hasegawa M., Ino T., Ota K., Kujime H. (2010). Contribution of momilactone A and B to rice allelopathy. J. Plant Physiol. 167: 787–791. [DOI] [PubMed] [Google Scholar]
- Kawahara Y., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean P.J., Kerr A., New P.B. (1970). Crown gall of stone fruit: II, Identification and nomenclature of Agrobacterium isolates. Aust. J. Biol. Sci. 23: 585–595. [Google Scholar]
- Kojima Y., Ebana K., Fukuoka S., Nagamine T., Kawase M. (2005). Development of an RFLP-based rice diversity research set of germplasm. Breed. Sci. 55: 431–440. [Google Scholar]
- Kong C., Xu X., Zhou B., Hu F., Zhang C., Zhang M. (2004). Two compounds from allelopathic rice accession and their inhibitory activity on weeds and fungal pathogens. Phytochemistry 65: 1123–1128. [DOI] [PubMed] [Google Scholar]
- Kong C.H., Chen X.H., Hu F., Zhang S.Z. (2011). Breeding of commercially acceptable allelopathic rice cultivars in China. Pest Manag. Sci. 67: 1100–1106. [DOI] [PubMed] [Google Scholar]
- Krug D., Müller R. (2009). Discovery of additional members of the tyrosine aminomutase enzyme family and the mutational analysis of CmdF. ChemBioChem 10: 741–750. [DOI] [PubMed] [Google Scholar]
- Kudo F., Miyanaga A., Eguchi T. (2014). Biosynthesis of natural products containing β-amino acids. Nat. Prod. Rep. 31: 1056–1073. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Christenson S.D., Standage S., Shen B. (2002). Biosynthesis of the enediyne antitumor antibiotic C-1027. Science 297: 1170–1173. [DOI] [PubMed] [Google Scholar]
- Lohman J.R., Shen B. (2012). 4-Methylideneimidazole-5-one-containing aminomutases in enediyne biosynthesis. In Methods in Enzymology, Hopwood D.A., ed (Amsterdam, The Netherlands: Elsevier; ), pp. 299–319. [DOI] [PubMed] [Google Scholar]
- Matsuda F., Nakabayashi R., Yang Z., Okazaki Y., Yonemaru J., Ebana K., Yano M., Saito K. (2015). Metabolome-genome-wide association study dissects genetic architecture for generating natural variation in rice secondary metabolism. Plant J. 81: 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum C.M., Comai L., Greene E.A., Henikoff S. (2000). Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 123: 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F.A. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Olofsdotter M. (2001a). Getting closer to breeding for competitive ability and the role of allelopathy - An example from rice (Oryza sativa). Weed Technol. 15: 798–806. [Google Scholar]
- Olofsdotter M. (2001b). Rice - A step toward use of allelopathy. Agron. J. 93: 3–8. [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachid S., Krug D., Weissman K.J., Müller R. (2007). Biosynthesis of (R)-beta-tyrosine and its incorporation into the highly cytotoxic chondramides produced by Chondromyces crocatus. J. Biol. Chem. 282: 21810–21817. [DOI] [PubMed] [Google Scholar]
- Ratnayake N.D., Wanninayake U., Geiger J.H., Walker K.D. (2011). Stereochemistry and mechanism of a microbial phenylalanine aminomutase. J. Am. Chem. Soc. 133: 8531–8533. [DOI] [PubMed] [Google Scholar]
- Rimando A.M., Olofsdotter M., Dayan F.E., Duke S.O. (2001). Searching for rice allelochemicals: An example of bioassay-guided isolation. Agron. J. 93: 16–20. [Google Scholar]
- Saikusa T., Horino T., Mori Y. (1994). Distribution of free amino acids in the rice kernel and kernel fractions and the effect of water soaking on the distribution. J. Agric. Food Chem. 42: 1122–1125. [Google Scholar]
- Sakai H., et al. (2014). Construction of pseudomolecule sequences of the aus rice cultivar Kasalath for comparative genomics of Asian cultivated rice. DNA Res. 21: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Kazama Y., Chae Y., Sato T. (2013). Confirmation of novel quantitative trait loci for seed dormancy at different ripening stages in rice. Rice Sci. 20: 207–212. [Google Scholar]
- Spindel J., Wright M., Chen C., Cobb J., Gage J., Harrington S., Lorieux M., Ahmadi N., McCouch S. (2013). Bridging the genotyping gap: using genotyping by sequencing (GBS) to add high-density SNP markers and new value to traditional bi-parental mapping and breeding populations. Theor. Appl. Genet. 126: 2699–2716. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Eiguchi M., Kumamaru T., Satoh H., Matsusaka H., Moriguchi K., Nagato Y., Kurata N. (2008). MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Mol. Genet. Genomics 279: 213–223. [DOI] [PubMed] [Google Scholar]
- Thomas C.L., Leh V., Lederer C., Maule A.J. (2003). Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology 306: 33–41. [DOI] [PubMed] [Google Scholar]
- Till B.J., et al. (2003). Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 13: 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lanen S.G., Oh T.J., Liu W., Wendt-Pienkowski E., Shen B. (2007). Characterization of the maduropeptin biosynthetic gene cluster from Actinomadura madurae ATCC 39144 supporting a unifying paradigm for enediyne biosynthesis. J. Am. Chem. Soc. 129: 13082–13094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lanen S.G., Dorrestein P.C., Christenson S.D., Liu W., Ju J., Kelleher N.L., Shen B. (2005). Biosynthesis of the beta-amino acid moiety of the enediyne antitumor antibiotic C-1027 featuring beta-amino acyl-S-carrier protein intermediates. J. Am. Chem. Soc. 127: 11594–11595. [DOI] [PubMed] [Google Scholar]
- Walker K.D., Klettke K., Akiyama T., Croteau R. (2004). Cloning, heterologous expression, and characterization of a phenylalanine aminomutase involved in taxol biosynthesis. J. Biol. Chem. 279: 53947–53954. [DOI] [PubMed] [Google Scholar]
- Wanninayake U., Walker K.D. (2013). A bacterial tyrosine aminomutase proceeds through retention or inversion of stereochemistry to catalyze its isomerization reaction. J. Am. Chem. Soc. 135: 11193–11204. [DOI] [PubMed] [Google Scholar]
- Xu M., Galhano R., Wiemann P., Bueno E., Tiernan M., Wu W., Chung I.M., Gershenzon J., Tudzynski B., Sesma A., Peters R.J. (2012a). Genetic evidence for natural product-mediated plant-plant allelopathy in rice (Oryza sativa). New Phytol. 193: 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., et al. (2012b). Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30: 105–111. [DOI] [PubMed] [Google Scholar]
- Yano M. (2001). Genetic and molecular dissection of naturally occurring variation. Curr. Opin. Plant Biol. 4: 130–135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.