ASAT3 is a trichome-specific BAHD-type acyltransferase that adds aliphatic acyl chains to acylsucroses and contributes to phenotypic diversity of acylsugars among the Solanum tomato species.
Abstract
Glandular trichomes from tomato (Solanum lycopersicum) and other species in the Solanaceae produce and secrete a mixture of O-acylsugars (aliphatic esters of sucrose and glucose) that contribute to insect defense. Despite their phylogenetic distribution and diversity, relatively little is known about how these specialized metabolites are synthesized. Mass spectrometric profiling of acylsugars in the S. lycopersicum x Solanum pennellii introgression lines identified a chromosome 11 locus containing a cluster of BAHD acyltransferases with one gene (named Sl-ASAT3) expressed in tip cells of type I trichomes where acylsugars are made. Sl-ASAT3 was shown to encode an acyl-CoA-dependent acyltransferase that catalyzes the transfer of short (four to five carbons) branched acyl chains to the furanose ring of di-acylsucrose acceptors to produce tri-acylsucroses, which can be further acetylated by Sl-ASAT4 (previously Sl-AT2). Among the wild tomatoes, diversity in furanose ring acyl chains on acylsucroses was most striking in Solanum habrochaites. S. habrochaites accessions from Ecuador and northern Peru produced acylsucroses with short (≤C5) or no acyl chains on the furanose ring. Accessions from central and southern Peru had the ability to add short or long (up to C12) acyl chains to the furanose ring. Multiple ASAT3-like sequences were found in most accessions, and their in vitro activities correlated with observed geographical diversity in acylsugar profiles.
INTRODUCTION
Understanding the genetic basis of the evolution of form and function remains one of the grand challenges of biology. Plant metabolic networks create tremendous phenotypic diversity; in fact, it is estimated that the kingdom produces hundreds of thousands of structurally and functionally diverse compounds (Pichersky and Lewinsohn, 2011). The largest and most diverse general class of metabolites is the taxonomically restricted specialized (traditionally referred to as “secondary”) metabolites. While some of these pathways are well studied due to known defensive roles, such as glucosinolates in the Brassicaceae (Grubb and Abel, 2006), or pharmacological properties, such as alkaloids in Papaver (poppy) species (Winzer et al., 2012), the biosynthetic diversity of plants has barely been explored. Specialized metabolic networks that have been studied in sufficient detail often show interesting patterns of diversification (Pichersky and Lewinsohn, 2011), presumably due to reduced constraints compared with core metabolic processes such as carbon and nitrogen assimilation (Milo and Last, 2012), and their roles in mediating plant interactions with insects and microbes. This characteristic makes these pathways especially attractive for studies of evolution of genetic and phenotypic diversity.
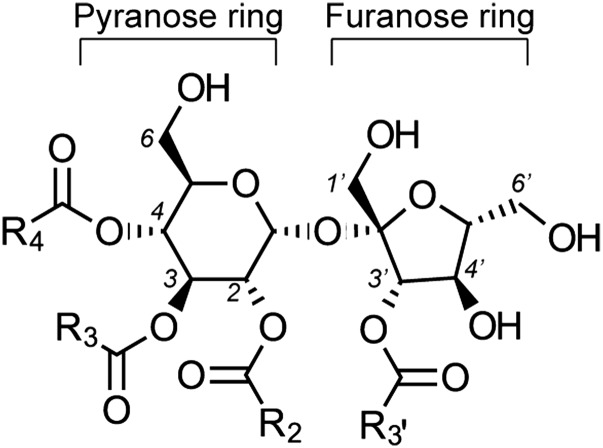
Glandular trichomes, present on the surface of many plants, serve as compartmentalized structures for high-level production of a variety of specialized metabolites (Werker, 2000; Schilmiller et al., 2008). The products can be stored, emitted as volatiles, or secreted to the plant surface where they participate in several aspects of plant physiology and ecology such as insect herbivory (Simmons and Gurr, 2005; Weinhold and Baldwin, 2011), pathogen attack (Shepherd et al., 2005), and attraction of pollinators (Werker, 2000). The glandular trichomes of many species in the nightshade family (Solanaceae), including Solanum pennellii, potato (Solanum berthaultii), tobacco (Nicotiana tabacum), petunia (Petunia hybrida), Physalis nicandroides, and Datura metel, produce and secrete sugar polyesters known as acylsugars (Fobes et al., 1985; Severson et al., 1985; King et al., 1986, 1988; King and Calhoun, 1988; Ohya et al., 1996; Maldonado et al., 2006). Characterized acylsugars are comprised of a sugar backbone, either glucose or sucrose, to which multiple short to medium chain length branched or straight aliphatic acids are esterified (Figure 1).
Figure 1.
S. lycopersicum Acylsucrose.
Acylsucroses from S. lycopersicum are typically either tri-acylated (R3, R4, and R3′) or tetra-acylated (R2, R3, R4, and R3′) with R2 being an acetyl group. Acyl chain lengths found on M82 acylsucroses include C2, C4, C5, C6, C10, C11, and C12.
Once secreted to the plant surface, the primary role of acylsugars is contributing to defense against a variety of insects including whiteflies, aphids, spider mites, and leaf miners (Hawthorne et al., 1992; Rodriguez et al., 1993; Liedl et al., 1995; Alba et al., 2009). Depending on the insect, the presence of acylsugars can be correlated with one or more modes of defense, including increased repellence, increased mortality, and reduced fecundity. Acylsucroses of Nicotiana attenuata are central to a tritrophic plant defense mechanism: following hydrolysis in the Manduca sexta caterpillar midgut, short-chain aliphatic acids recruit predators of herbivores (Weinhold and Baldwin, 2011). In addition to their role in plant defense, sucrose esters also have commercial value as emulsifiers for food and cosmetic products (Hill and Rhode, 1999).
Our understanding of the biosynthesis of acylsugars in the Solanaceae has largely come from studies using tobacco, tomato (Solanum lycopersicum), and related wild tomato species. The branched aliphatic acyl chains are derived from branched-chain amino acid metabolism (Kandra et al., 1990; Walters and Steffens, 1990). The action of branched-chain keto acid dehydrogenase on α-keto acids gives rise to CoA esters of 2-methyl-propanoic acid (iso-C4, from Val), 3-methyl-butanoic acid (iso-C5, from Leu), and 2-methyl-butanoic acid (anteiso-C5, from Ile) (Slocombe et al., 2008). Longer acyl chains (up to 12 carbons) that may be straight or branched chain are also present in acylsugars. Published evidence indicates that branched chains longer than C5 are produced through elongation via a fatty acid synthase mechanism in the case of S. pennellii LA0716 and D. metel (van der Hoeven and Steffens, 2000; Kroumova and Wagner, 2003) or by one carbon extension of α-keto acids through multiple cycles of isopropylmalate synthase, isopropylmalate dehydrogenase, and isopropylmalate dehydratase reactions in multiple Nicotiana species and petunia (Kroumova and Wagner, 2003). Collectively, the different acyl chain lengths (C2-C12) and the variety of iso-, anteiso-, and straight chains give rise to many different possible acylsugar structures. However, acylation of the sugar backbone is not random, as demonstrated by a series of NMR-resolved structures reported for acylsugars from S. lycopersicum cv M82 and Solanum habrochaites LA1392 and LA1777 (Ghosh et al., 2014). Therefore, the final profile of acylsugars for a given plant is likely controlled by the availability of acyl chains and activities of the enzymes that catalyze the transfer reactions for esterification to the sugar backbone.
Little is known about specific enzymes that control attachment of acyl chains to the sugar backbone. Earlier studies with S. pennellii LA0716 trichomes led to a proposal where acyl chains are first activated to 1-O-acylglucose via UDP-glucose and a glucosyltransferase (Ghangas and Steffens, 1993; Kuai et al., 1997). The 1-O-acylglucoses then act as donors in acyl transfer reactions catalyzed by a serine carboxypeptidase-like acyltransferase (Li and Steffens, 2000). However, no evidence was presented for acylation of sucrose by this mechanism. More recently, we characterized Sl-ASAT4 (formerly Sl-AT2, encoded by Solyc01g105580), a BAHD acyltransferase, named according to the first four characterized enzymes of the family: BEAT, AHCT, HCBT, DAT (D’Auria, 2006), which catalyzes the transfer of an acetyl group via acetyl-CoA to a tri-acylsucrose acceptor to form the acetylated tetra-acylsucroses produced by cultivated tomato trichomes (Schilmiller et al., 2012). A similar enzyme was also found in southern accessions of the wild tomato S. habrochaites, and its activity correlated with acetylation of structurally diverse acylsucroses in this species (Kim et al., 2012).
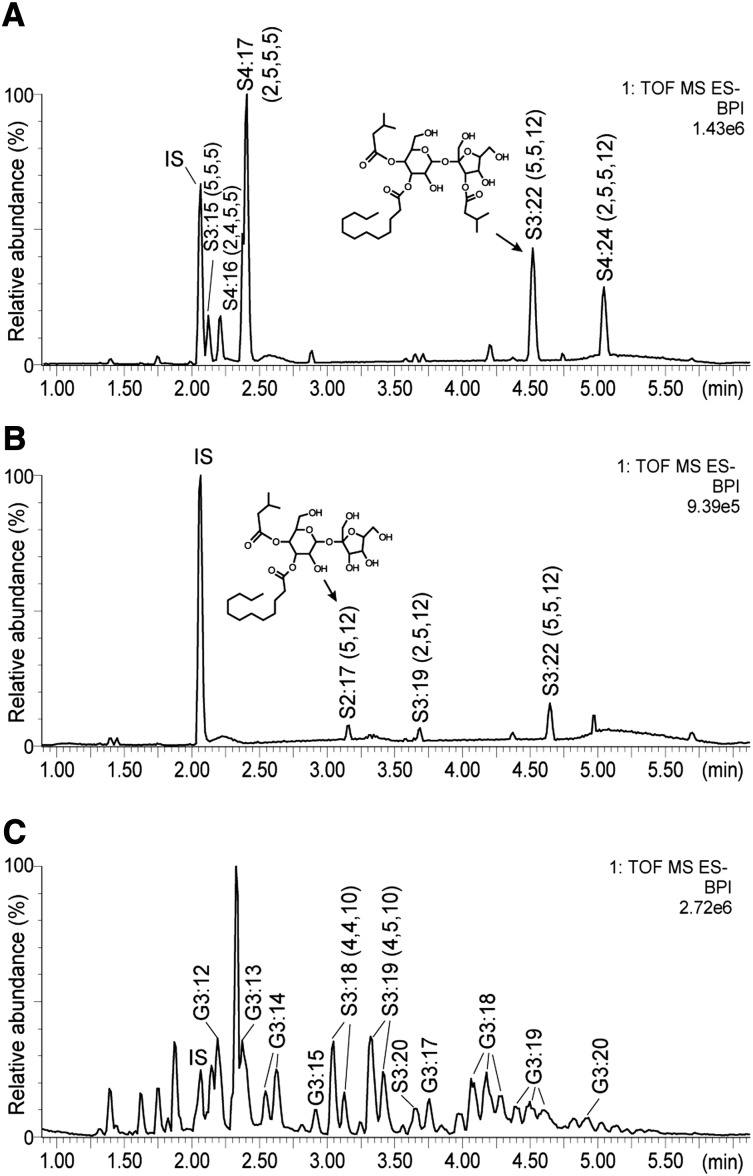
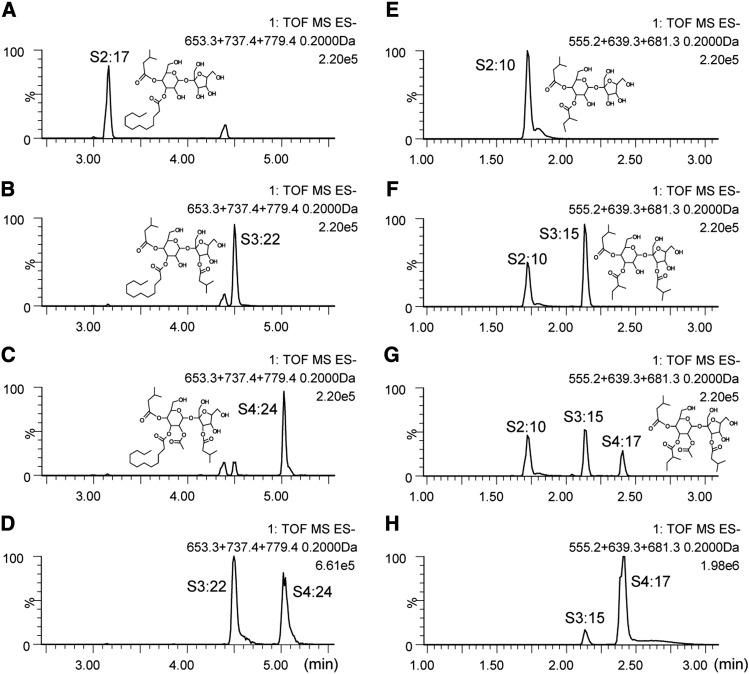
As previously reported (Schilmiller et al., 2010a; Ghosh et al., 2014), S. lycopersicum cultivar M82 plant extracts subjected to analysis using liquid chromatography coupled with time-of-flight mass spectrometry (LC-MS) show the presence of both tri- and tetra-acylsucroses with acyl chains of various lengths (Figure 2A). The nomenclature used here for acylsugars is as follows: “S” indicates an acylsucrose backbone, “3” corresponds to the number of acyl chains, “22” represents the total number of carbons in the acyl chains, and the length of each acyl chain is listed in parentheses. Additional information for each acyl chain, if known, is shown for branching (i, iso-branched; ai, anteiso; n, normal unbranched), and superscripts indicate position on the sucrose molecule. Thus, the structure shown in Figure 2A is named S3:22 (i5R4,n12R3,i5R3′). NMR spectra show that all of the major acylsucroses of M82 have a single acyl chain on the furanose ring of the sucrose backbone, in the R3′ position (Ghosh et al., 2014). The other acyl chains are located on the pyranose ring of the acylsucrose molecules.
Figure 2.
Base Peak Intensity LC-MS Chromatograms from Leaf-Dip Extractions Showing Major Acylsugars.
Chromatograms are shown for M82 (A), IL11-3 (B), and S. pennellii LA0716 (C) and are scaled to the abundance of the largest peak in each chromatogram. Acylsucroses S3:22 and S2:17 are known structures based on NMR. The S3:22 from IL11-3 has a different retention time compared with S3:22 from M82 because the IL11-3 isomer has all three acyl chains on the pyranose ring. Base peak intensity (BPI) chromatograms are a plot of the abundance of the most abundant ion in each scan. The numbers in the top right of each chromatogram (for instance 1.43e6 in [A]) indicate the ion counts represented by “100” relative abundance. IS, internal standard (propyl 4-hydroxybenzoate).
Here, we describe the identification of a BAHD-type acylsucrose acyltransferase in a region on chromosome 11 that was originally found to harbor a locus affecting total acylsugar levels (Schilmiller et al., 2010a). In the cultivated tomato, this acyltransferase, named ACYLSUGAR ACYLTRANSFERASE3 (ASAT3; Solyc11g067270), catalyzes the R3′ acylation on the five-member furanose ring of di-acylsucroses in the tip cells of acylsugar producing trichomes. The S. pennellii LA0716 allele of ASAT3 lacks this R3′ acylation activity, resulting in the accumulation of acylsucroses with no furanose ring acyl group in LA0716 and introgression line 11-3 (IL11-3) plants. A survey of acylsucrose furanose ring acylation in a selection of S. habrochaites accessions spanning the geographical range of occurrence in Ecuador and Peru demonstrates that northern accessions have only short chains (≤C5) or no acyl chain on the furanose ring, while southern accessions have the ability to add short or long acyl chains (up to C12). Sequencing of ASAT3 PCR products in S. habrochaites revealed that multiple ASAT3-like sequences are present in most accessions. The in vitro assay products of Sh-ASAT3 enzymes from northern and southern accessions correlates with the diversity of acylsucrose furanose ring acylation seen in plant extracts. This work extends our understanding of the acylsucrose biosynthetic network and illustrates the importance of gene duplication, neofunctionalization, and gene loss on generating specialized metabolic diversity in closely related populations and species.
RESULTS
IL11-3 Plants Accumulate Di- and Tri-Acylsucroses
A region on chromosome 11 regulating acylsugar levels was identified using a screen of chromosomal introgression lines derived by crossing S. lycopersicum cv M82 with S. pennellii LA0716 (Schilmiller et al., 2010a). Total acylsugar levels in the IL11-3 extracts from leaflets of 3-week-old plants were 17-fold lower than wild-type M82. The low abundance acylsugars produced by IL11-3 were quite different from the nearly isogenic M82, consisting of a di-acylsucrose, S2:17 (5,12), and two triacylsucroses, S3:19 (2,5,12) and S3:22 (5,5,12) (Figure 2B). Positive- and negative-ion mode mass spectrometry (MS) fragmentation patterns revealed that these acylsucroses are only acylated on the six-member pyranose ring (Supplemental Figures 1 and 2). Structural analysis by NMR confirmed that the furanose ring is not acylated and showed that the iso-C5 chain is in the R4 position and the normal (unbranched) C12 chain is in the R3 position for S2:17 (i5R4,n12R3) purified from IL11-3 plants (Supplemental Table 1).
The lack of furanose ring acylation of IL11-3 acylsucroses is similar to that found for S. pennellii LA0716, the parent from which the IL11-3 introgression region was derived. While LA0716 produces both acylglucoses and acylsucroses, none of the acylsucroses have an acyl group esterified to the furanose ring (Figure 2C; Supplemental Figure 1). This observation led to the hypothesis that the region of S. pennellii chromosome 11 introgressed in IL11-3 is missing an enzymatic activity responsible for R3′ acylation on the furanose ring of acylsucroses. This led us to search for acyltransferase genes in the IL11-3 region of S. lycopersicum.
Identification of Candidate Genes on IL11-3
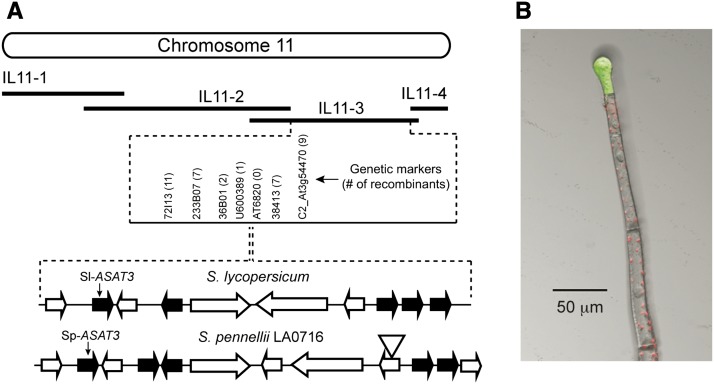
The IL11-3 introgression region is predicted to encode 1459 genes based upon recent high-resolution genotyping of the ILs (Chitwood et al., 2013), and the current S. lycopersicum Heinz 1706 genome assembly and annotation (Sol Genomics Network, http://solgenomics.net) (Tomato Genome Consortium, 2012). A map-based cloning approach was used to narrow down the region containing the locus responsible for the lack of furanose ring acylation in IL11-3. Recombinants from an F2 population derived by crossing M82 with IL11-3 positioned the locus to a region of ∼180 kb on chromosome 11 (Figure 3A). There are 20 predicted open reading frames in this region (Supplemental Table 2), including five candidate genes belonging to the BAHD family of acyltransferases. This is the same family of acyltransferases as the type I/IV gland cell expressed acylsugar acetyltransferase (Sl-ASAT4, formerly Sl-AT2) (Schilmiller et al., 2012).
Figure 3.
Sl-ASAT3 Is a Trichome-Expressed Gene Located in the Mapping Interval for the Locus Regulating the IL11-3 Acylsugar Phenotype.
(A) The map position of the locus regulating the reduced acylsugar levels of IL11-3 was narrowed to an ∼180-kb region. The predicted genes in a section of this region are shown for S. lycopersicum and S. pennellii LA0716 (the triangle indicates a transposon insertion in LA0716). Multiple BAHD acyltransferases genes were found (block arrows shaded black). Genetic marker information is in Supplemental Table 5.
(B) Fluorescence of GFP driven by the Sl-ASAT3 promoter is located in the tip cells of type I/IV trichomes where acylsugars are produced. Shown is an overlay of GFP fluorescence (green), chlorophyll autofluorescence (red), and a differential interference contrast image. Bar = 50 μm.
To narrow down this list of five candidates, we looked for evidence of trichome expression for each gene by searching the cultivated and wild tomato EST collections at Sol Genomics Network and our previously described M82 and LA0716 trichome RNA-seq data (Schilmiller et al., 2010b; McDowell et al., 2011). A gene controlling acylsucrose acylation is expected to be expressed in trichomes since acylsugars are synthesized in the tip cells of the type I/IV glands (Fobes et al., 1985; Schilmiller et al., 2012). Six of the 20 genes showed expression in trichomes of either M82 or LA0716 (Supplemental Table 2). Of those six, only Solyc11g067270 (Sl-ASAT3), which is one of the BAHD acyltransferases, was predicted to be trichome specific based on the lack of ESTs from any other tissue.
Trichome expression of Sl-ASAT3 was validated using two approaches. RT-PCR analysis was performed to test expression of this gene using RNA from intact stems and petioles, trichomes isolated from stems and petioles, and stems and petioles that had trichomes removed prior to RNA extraction. Transcript was detected in the intact stem and petiole tissues (“+”) and the isolated trichomes (“T”), but not in the stem and petioles after removal of the trichomes (“−”) (Supplemental Figure 3). To determine which trichome type and specific cells express Sl-ASAT3, a 1.6-kb fragment containing the promoter region was used to drive expression of a green fluorescent protein-β-glucuronidase (GFP-GUS) fusion protein in transgenic M82 plants. GFP signal was restricted to tip cells of the long slender (type I/IV) trichomes (Figure 3B). This cell-type-specific expression in the type I/IV trichomes is identical to that seen for the acylsucrose acetyltransferase Sl-ASAT4 (Schilmiller et al., 2012) and consistent with the expectation for an acylsugar metabolic enzyme (Fobes et al., 1985).
Sl-ASAT3 Expression Influences Acylation on the Furanose Ring of Acylsucroses
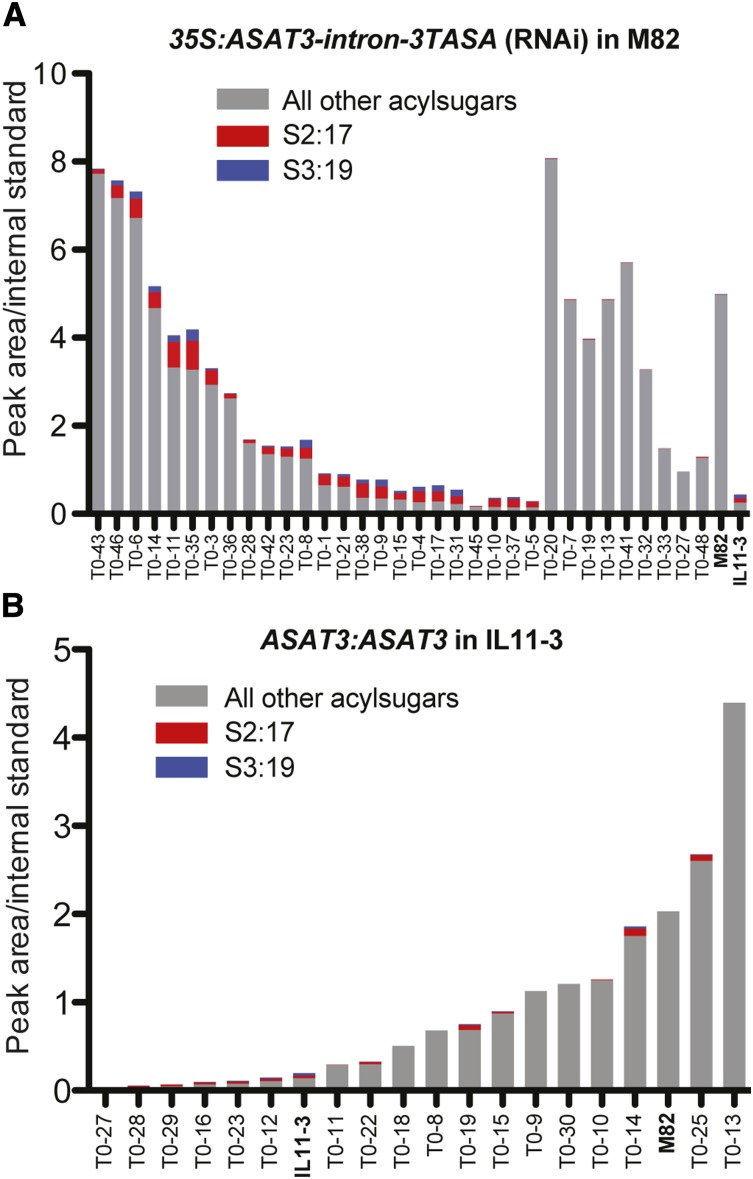
Two complementary transgenic plant approaches were taken to test the in vivo function of Sl-ASAT3. M82 plants with reduced gene expression were generated using RNA interference (RNAi). Acylsugar profiles were analyzed from 34 independent transgenic lines, and 27 showed an increase in the amount of S2:17 (5,12) and S3:19 (2,5,12) (Figure 4A; Supplemental Figure 4A). Among those 27 lines, we observed a range of total acylsugar levels with 10 lines having less than 10% of the M82 wild type. Because the S. pennellii LA0716 allele of ASAT3 is recessive to the S. lycopersicum allele, we predicted that transformation of Sl-ASAT3 into IL11-3 plants would restore acylsucrose R3′ acylation and result in an acylsugar profile similar to wild-type M82 plants. Out of 17 independent IL11-3 plants transformed with Sl-ASAT3 driven by its endogenous promoter, 12 showed various levels of complementation. Total acylsugar levels are increased due to accumulation of tri- and tetra-acylsucroses having a furanose ring acyl chain (Figure 4B; Supplemental Figure 4B). Interestingly, although IL11-3 plants do not accumulate S2:10 (5,5), the transgenic expression of Sl-ASAT3 in IL11-3 also restores some production of S3:15 (5,5,5) and S4:17 (2,5,5,5), consistent with the hypothesis that this enzyme also acylates the R3′ position of acylsucroses having only short acyl chains (<C10) in vivo.
Figure 4.
Transgenic Analysis of Sl-ASAT3.
RNAi silencing of Sl-ASAT3 in M82 plants (A) and expression of Sl-ASAT3 in IL11-3 plants (B). Total acylsugar levels are shown for each independent primary transgenic line and for M82 and IL11-3 controls. Amounts of S2:17 and S3:19, incomplete intermediate acylsugars lacking R3′ acylation, are shown in red and blue, respectively.
Sl-ASAT3 Acylates the Furanose Ring of Di-Acylsucroses in Vitro
Sl-ASAT3 enzymatic activity was tested using protein expressed in Escherichia coli. Accumulation of S2:17 (5,12) and S3:19 (2,5,12) in RNAi lines and IL11-3 plants led to the hypothesis that these are Sl-ASAT3 substrates. Enzyme assays were conducted using these di- and tri-acylsucroses collected from trichome extracts from IL11-3 and Sl-ASAT3 RNAi plants as acyl acceptors. The substrate iC5-CoA was tested first because S. lycopersicum acylsucroses typically have an iC5 acyl chain at the R3′ position (Ghosh et al., 2014). Consistent with expectation, recombinant Sl-ASAT3 enzyme acylated purified S2:17 (5,12) di-acylsucrose to produce S3:22 (5,5,12) (Figures 5A and 5B). Positive-ion mode MS fragment ions indicated that the newly added acyl chain was located on the furanose ring (Supplemental Figure 5). Activity could also be seen when using C2-CoA, iC4-CoA, and aiC5-CoA but not with the longer chain iC10-CoA and nC12-CoA (Supplemental Figure 6). Although the amount of product was too low for purification and NMR analysis, the S3:22 (5,5,12) in vitro assay product cochromatographed with the in vivo metabolite (Figure 5D); based on the known structures of S. lycopersicum acylsucroses, these results suggest that the enzyme acylated the di-acylsucrose at the R3′ position (Ghosh et al., 2014). In addition, the tomato Sl-ASAT4 tri-acylsucrose acetyltransferase (Schilmiller et al., 2012) acetylated the S3:22 (5,5,12) Sl-ASAT3 enzyme assay product to produce S4:24 (2,5,5,12) (Figure 5C; Supplemental Figure 5) with an identical reverse phase chromatography retention time as the M82 trichome metabolite (Figure 5D). Taken together, these results are consistent with the hypothesis that Sl-ASAT3 acylates di-acylsucroses at the R3′ position of the furanose ring.
Figure 5.
Results of Sl-ASAT3 Enzyme Activity Assays.
(A) to (D) Extracted ion chromatograms for m/z 653.3 (S2:17), 737.4 (S3:22), and 779.4 (S4:24).
(A) S2:17 substrate isolated from IL11-3.
(B) S2:17 + Sl-ASAT3 + iC5-CoA.
(C) S2:17 + Sl-ASAT3 + iC5-CoA followed by addition of Sl-ASAT4 + C2-CoA.
(D) S3:22 (5,5,12) and S4:24 (2,5,5,12) from M82 leaf.
(E) to (H) Extracted ion chromatograms for m/z 555.2 (S2:10), 639.3 (S3:15), and 681.3 (S4:17).
(E) S2:10 substrate.
(F) S2:10 + Sl-ASAT3 + iC5-CoA.
(G) S2:10 + Sl-ASAT3 + iC5-CoA followed by addition of Sl-ASAT4 + C2-CoA.
(H) S3:15 and S4:17 from M82 leaf.
The y axis indicates ion signal abundance relative to the highest signal in each chromatogram (%). The numbers in the top right of each chromatogram (for instance 2.2e5 in [A]) indicate the ion counts represented by “100” relative abundance. Structures of products are inferred based on mass spectra and cochromatography with acylsugars of known structure.
The failure of Sl-ASAT3 RNAi lines and IL11-3 plants to accumulate S3:15 (5,5,5) or S4:17 (2,5,5,5) acylsucroses having a C5 acyl chain on the furanose ring suggested that Sl-ASAT3 should also acylate the R3′ position of S2:10 (5,5). However, neither the RNAi lines nor the IL11-3 plants accumulated the di-acylsucrose precursor S2:10 (5,5). To test whether Sl-ASAT3 would also acylate S2:10 (5,5), we synthesized it in vitro and confirmed the structure by NMR to be S2:10 (i5R4,ai5R3) with iso-C5 at the R4 position and anteiso-C5 at the R3 position (Supplemental Table 3). This specific di-acylsucrose is predicted to be an in vivo substrate for R3′ acylation based on the known structure of S4:17 (2R2,i5R4,ai5R3,i5R3′) (Schilmiller et al., 2010a; Ghosh et al., 2014). Enzyme assays using Sl-ASAT3 together with S2:10 (5,5) and iC5-CoA showed that Sl-ASAT3 will add an iC5 chain to the furanose ring of S2:10 (5,5) to produce S3:15 (5,5,5) (Figures 5E and 5F). The S3:15 produced is also capable of being acetylated by Sl-ASAT4 to produce S4:17 (2,5,5,5) (Figure 5G). Both S3:15 and S4:17 produced in vitro had chromatographic retention times and mass spectra identical to the acylsucroses extracted from M82 trichomes (Figure 5H; Supplemental Figure 5).
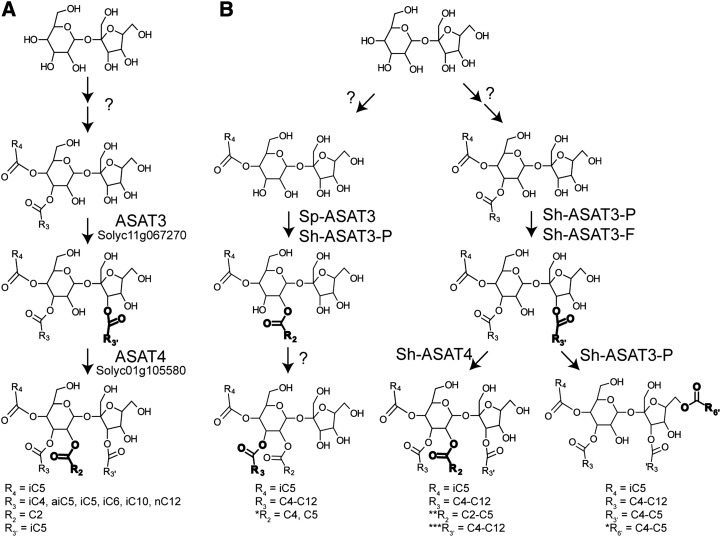
Acetylation of the acylsucrose R2 position prior to furanose ring acylation blocked Sl-ASAT3 enzyme activity. This was demonstrated using the acylsugars collected from an Sl-ASAT3-silenced S. lycopersicum RNAi line, which included both S2:17 (5,12) and S3:19 (2,5,12) as a mixed substrate. Either iC4 or iC5 acyl chains were added to the furanose ring of the di-acylsucrose substrate, but no acylation of the acetylated tri-acylsucrose S3:19 was observed with either acyl CoA donor (Supplemental Figure 7). We also tested whether Sl-ASAT3 would acylate unmodified sucrose or glucose directly; however, no activity was seen with either acceptor. These in vitro results, combined with accumulation of S2:17 (5,12) in IL11-3 and RNAi lines, are consistent with the hypothesis that Sl-ASAT3 normally uses a di-acylsucrose acceptor as substrate. Together, these results indicate that Sl-ASAT3 acylates di-acylsucroses at the R3′ position followed by acetylation by Sl-ASAT4 at the R2 position to produce the final acylsucrose products seen in vivo (Figure 6A).
Figure 6.
Models for Acylsucrose Biosynthesis.
(A) ASAT3 and ASAT4 acyltransferases catalyze the last two steps of S. lycopersicum acylsucrose biosynthesis. Sucrose is converted to di-acylsucrose with the R3 and R4 positions acylated by enzymes that are not yet described. Sl-ASAT3 (Solyc11g067270) adds short (≤C5) acyl chain to the R3′ position of these di-acylsucroses followed by acetylation at the R2 position by Sl-ASAT4 (Solyc01g105580, formerly AT2).
(B) S. pennellii LA0716 produces tri-acylsucroses through the action of Sp-ASAT3 on mono-acylsucrose followed by action of an unknown enzyme to add a third acyl chain to the pyranose ring. Acylsucrose biosynthesis in accessions of S. habrochaites results from either dual function Sh-ASAT3-P enzymes or the combined action of Sh-ASAT3-P and Sh-ASAT3-F enzymes. *The positions of the added acyl chains are inferred based on known NMR-resolved structures from Ghosh et al. (2014). **Activity of Sh-ASAT4 (Kim et al., 2012) has been shown for transfer of acetyl (C2) groups only. Transfer of acyl chains longer than C2 by Sh-ASAT4 is proposed here but not experimentally demonstrated. ***No acylsucroses having more than one long (≥C10) acyl chain have been detected in S. habrochaites plant extracts.
Diversity in Solanum Acylsucrose Furanose Ring Acylation
Acylsugar composition varies among different Solanum wild tomato species as well as within different accessions of a particular species (Shapiro et al., 1994; McDowell et al., 2011; Kim et al., 2012). To survey the diversity in furanose ring acylation, we chose seven different wild tomato species for which seed was available from the TGRC (Tomato Genetics Resource Center, UC Davis). For six of the species (Solanum neorickii, Solanum arcanum, Solanum huaylasense, Solanum peruvianum, Solanum corneliomulleri, and S. pennellii), five to seven accessions representing different locations in the geographical range of each species were grown. For S. habrochaites, 26 different accessions were grown, most of which were analyzed in a previous acylsugar screen (Kim et al., 2012). Acylsugars from leaves of 3-week-old plants were analyzed by LC-MS operating in positive-ion mode to identify acyl chains on the furanose ring (Supplemental Data Set 1).
Accessions from four of the tested species (S. neorickii, S. arcanum, S. corneliomulleri, and S. peruvianum) all accumulated acylsucroses with a single short acyl chain (C4 or C5) esterified to the furanose ring. This is the same phenotype observed for S. lycopersicum (Supplemental Figure 2). Of the five S. huaylasense accessions tested, four also produced acylsucroses with a single short acyl chain on the furanose ring and one accession (LA1982) accumulated acylsucroses with no furanose ring acyl chain. Accessions from the more distantly related species S. pennellii and S. habrochaites showed the most notable differences when compared with the other Solanum species. None of the S. pennellii accessions produced acylsucroses with a furanose ring acyl chain, and S. pennellii was the only species in this survey for which acylglucoses were found.
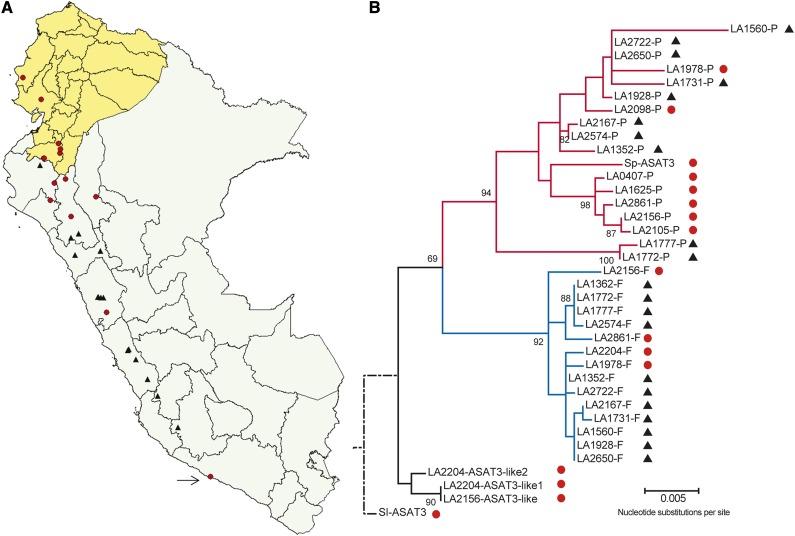
S. habrochaites accessions showed the most diversity in acylsucrose furanose ring acylation with molecules containing a single short or long (C10-C12) acyl chain, no furanose ring acyl chain, or two short acyl chains on the furanose ring (Supplemental Data Set 1). The furanose ring acylation variation in S. habrochaites accessions spanning the species geographical range could be separated into two groups. Fourteen accessions have at least one acylsugar with a long chain (≥C10) on the furanose ring and were collected at sites throughout Peru and up to the border with Ecuador (Figure 7A, black triangles). The other 12 accessions that have furanose ring acyl groups no longer than five carbons are predominately from Ecuador and northern Peru (Figure 7A, red circles) and showed little overlap with the other 14 accessions. This geographical separation of acylsugar phenotype overlaps to a great extent with the distribution of acylsugar acetylation in S. habrochaites. As previously shown, acetylation is absent in northern accessions and present in the southern accessions (Kim et al., 2012).
Figure 7.
Association between Geographical Distribution and Phylogenetic Relationships of S. habrochaites ASAT3 Loci.
(A) Geographical locations of the S. habrochaites accessions and S. pennellii LA0716 (arrow), shown on a map of Ecuador and Peru. Accessions producing at least one detectable acylsucrose with a long chain (≥C10) on the furanose ring are marked with a black triangle. Accessions that produce acylsucroses with only a short chain (≤C5) or no acyl chain on the furanose ring are marked with a red circle.
(B) ASAT3 phylogenetic tree obtained using 37 coding sequences from S. habrochaites (35), S. pennellii (1, Sp-ASAT3), and S. lycopersicum (1, Sl-ASAT3). The tree was inferred using maximum likelihood, based on the Tamura three-parameter model (T92+G) with shape parameter = 0.5, and was rooted using S. lycopersicum as the outgroup. The outgroup branch has been shortened for representational purposes. Values shown for the branches represent the bootstrap values obtained after 1000 replicates of bootstrapping. Except for the Sh-ASAT3-P/Sh-ASAT3-F ancestral branch with a bootstrap value of 69, only the bootstrap values >70 are shown. The bootstrap values for the -P and -F clades (94 and 92, respectively) provide strong support for each of the clades being monophyletic. The red and blue lines are colored to show the -P and -F clades, and the red circles and black triangles next to the locus names represent the furanose ring acylation phenotype of the accession as denoted in (A). The sequence alignment used to generate the tree is in Supplemental Data Set 2.
Most S. habrochaites Individuals Have Multiple ASAT3-Like Sequences
To test whether Sh-ASAT3 activity is responsible for the diversity in acyl chain length and number of acyl groups on the furanose ring, Sl-ASAT3-like sequences were isolated from selected accessions for expression in E. coli and activity testing. Using previously published S. habrochaites trichome RNA-seq data (Kim et al., 2012), PCR primers were designed to amplify the full-length Sh-ASAT3 open reading frame (ORF) from 18 of the 25 accessions. Sequence analysis of the Sh-ASAT3 PCR products revealed accessions with more than one Sh-ASAT3 sequence. Using separate forward PCR primers designed based upon two nucleotide differences in the first 18 bp of the two Sh-ASAT3 sequences, we could amplify two or more Sh-ASAT3-like sequences from the genomic DNA of all but four of the eighteen accessions tested (see Methods for details).
To assess the relationships between the Sh-ASAT3-like genes from different accessions, we constructed a phylogenetic tree using their nucleotide sequences (Figure 7B; Supplemental Data Set 2). The ASAT3-like sequences fell into three distinct groups. One set of sequences clustered with the S. pennellii LA0716 Sp-ASAT3 sequence, which we designated the “P” cluster (based on pyranose ring acylation activity; see below). Another set of ASAT3-like sequences formed a separate group designated as the “F” cluster (based on furanose ring acylation activity; see below). Both of these groups were monophyletic in a majority of the bootstrapped trees generated, consistent with the hypothesis that the ASAT3-P and ASAT3-F sequences were not a result of heterozygosity but represented two independent, ancestrally duplicated loci in the S. habrochaites genome. The four accessions for which only a P sequence was obtained (LA1625, LA0407, LA2098, and LA2105) are in the short chain furanose ring phenotypic group collected from the northern region. Two accessions (LA2204 and LA2156) had ASAT3-like sequences whose phylogenetic origins are uncertain given the low bootstrap values associated with these sequences across different evolutionary scenarios (Figure 7B; Supplemental Figure 8). The S. lycopersicum (both Heinz 1706 and M82) and the S. pennellii LA0716 genome assemblies contain a single ASAT3 sequence with other related BAHD genes present nearby on chromosome 11 (Figure 3A) (Tomato Genome Consortium, 2012; Bolger et al., 2014). When a phylogeny was constructed using the S. habrochaites ASAT3-like sequences along with the other chromosome 11 S. lycopersicum BAHD sequences, the Sh-ASAT3-like sequences clustered with Sl-ASAT3 and Sp-ASAT3 (Supplemental Figure 8G), suggesting that the Sh-ASAT3 sequences are true orthologs of Sl-ASAT3.
Diversity of ASAT3 Enzyme Activities Correlates with Trichome Acylsugars
The activities of seven Sh-ASAT3-P or -F enzymes from four different accessions (LA1777, LA1731, LA2861, and LA2098) were analyzed in vitro. These accessions were chosen because they represent the different furanose ring acyl group phenotypes observed: LA1777 has acylsugars with C4, C5, C10, C11, or a C12 on the furanose ring; LA1731 has C5, C10, C11, C12, two C5s, or no acyl chain; LA2861 has a C5 or no acyl chain; LA2098 has a C5, two C5s, or no acyl chain (Supplemental Data Set 1). Sh-ASAT3-F enzymes were given the “F” designation because they acylate the furanose ring of a di-acylsucrose. Both LA1777 and LA1731 Sh-ASAT3-F enzymes were able to add either a short (iC5) or a long chain (nC12) to the furanose ring of the S2:10 (5R3,5R4) substrate (Table 1; Supplemental Figures 9 and 10). This is consistent with the acylsucrose profile seen for these plants. The apparent Km values for LA1777 Sh-ASAT3-F were 6.8 ± 0.7 μM for iC5-CoA and 1.6 ± 1.1 μM for nC12-CoA, and for LA1731 Sh-ASAT3-F were 9.0 ± 2.7 μM for iC5-CoA and 5.2 ± 2.3 μM for nC12-CoA (Supplemental Figure 11). Substrate inhibition by the acyl-CoA substrates was observed for both enzymes with apparent Ki values of 5.5 ± 4.5 μM (nC12-CoA) for LA1777 (no inhibition observed for iC5-CoA), and 250 ± 160 μM (iC5-CoA) and 5.2 ± 2.6 μM (nC12-CoA) for LA1731 (Supplemental Figure 11). In contrast, LA2861 Sh-ASAT3-F adds iC5 to the furanose ring of S2:10 but did not use nC12-CoA as a substrate. This reflects the observation that the LA2861 accession produces acylsucroses with only a short chain on the furanose ring (Table 1; Supplemental Figures 9 and 10). No F sequence was amplified from LA2098.
Table 1. Summary of Enzyme Activities for S. pennellii LA0716 and S. habrochaites ASAT3 Sequences.
| Accession | Furanose Ring Phenotypes Seen in Plant Extracts | ASAT3 Variant | Acylsugar Products Detected Using the Indicated Substrates |
||
|---|---|---|---|---|---|
| S1:5 + iC5-CoA | S2:10 + iC5-CoA | S2:10 + nC12-CoA | |||
| LA0716 | No acyl chain | ShASAT3-P | S2:10 (i5R4,i5P) | no activity | No activity |
| LA2098 | C5, two C5′s, or no acyl chain | ShASAT3-P | S2:10 (i5R4,i5P) | S3:15 (i5R4, ai5R3,iC5F) & S4:20 (i5R4, ai5R3,iC5F,iC5F) | No activity |
| LA2861 | C5 or no acyl chain | ShASAT3-P | S2:10 (i5R4,i5P) | no activity | No activity |
| ShASAT3-F | no activity | S3:15 (i5R4, ai5R3,iC5F) | No activity | ||
| LA1731 | C5, C11, C12, two C5′s, or no acyl chain | ShASAT3-P | S2:10 (i5R4,i5P) | no activity | No activity |
| ShASAT3-F | no activity | S3:15 (i5R4, ai5R3,iC5F) | S3:15 (i5R4, ai5R3,nC12F) | ||
| LA1777 | C4, C5, C10, C11, or C12 | ShASAT3-P | S2:10 (i5R4,i5P) | S3:15 (i5R4, ai5R3,iC5F) and S4:20 (i5R4, ai5R3,iC5F,iC5F) | No activity |
| ShASAT3-F | No activity | S3:15 (i5R4, ai5R3,iC5F) | S3:15 (i5R4, ai5R3,nC12F) | ||
The activity of the Sh-ASAT3-P enzymes was also tested using S2:10 (5R3,5R4) as a substrate. The LA1777 and LA2098 Sh-ASAT3-P enzymes added iC5 to the furanose ring of S2:10 (5R3,5R4) to produce both S3:15 (5R3,5R4,5F) and S4:20 (5R3,5R4,5F,5F) (the “F” superscript here indicates the presence of an acyl chain somewhere on the furanose ring; Table 1; Supplemental Figures 9 and 10). S4:20, with two C5 acyl chains on the furanose ring, is one of the major acylsucroses that accumulated in LA2098 (Supplemental Data Set 1). In contrast, the LA1777 plant originally phenotyped did not accumulate detectable amounts of acylsucroses with two C5 acyl chains on the furanose ring even though the LA1777 Sh-ASAT3-P enzyme displays this activity in vitro. Quantitative RT-PCR analysis of Sh-ASAT3-P and -F gene expression in trichomes of this LA1777 individual revealed a 9-fold higher level of expression for Sh-ASAT3-F compared with Sh-ASAT3-P (Supplemental Figure 12). Two other LA1777 plants grown as biological replicates were analyzed for acylsugar phenotype and Sh-ASAT3 expression in trichomes. One individual had the same phenotype as the original LA1777 plant and had a 14-fold higher level of Sh-ASAT3-F expression compared with Sh-ASAT3-P. In contrast, the third LA1777 individual produced a small amount of detectable acylsucroses with two short acyl chains on the furanose ring (Supplemental Data Set 1) and the Sh-ASAT3-P and -F alleles were expressed at nearly equal levels (Supplemental Figure 12). Neither LA1731 nor LA2861 Sh-ASAT3-P enzymes had detectable activity with S2:10 and iC5-CoA or nC12-CoA, which is consistent with the lack of acylsucroses having two furanose ring acyl chains in these accessions. Taken together, the combination of in vitro enzyme activity and gene expression level correlates well with the observed acylsugar phenotypes.
Relationship between ASAT3 Enzyme Activities in S. habrochaites and S. pennellii LA0716
Despite the observation that IL11-3 has a recessive change in acylsucrose composition compared with M82, the Sp-ASAT3 open reading frame is predicted to encode a full-length enzyme, and expression of Sp-ASAT3 in LA0716 trichomes is high (Supplemental Table 2). The clustering of the Sp-ASAT3 (LA0716) sequence with S. habrochaites ASAT3-P (Figure 7B) and lack of furanose ring acylation in LA0716 and IL11-3 (Figure 2; Supplemental Figure 2) led us to hypothesize that the S. pennellii enzyme acylates the pyranose ring of an acylated sucrose substrate. No activity was detected with Sp-ASAT3 expressed in E. coli using either S2:10 (5,5) or S2:17 (5,12) as acceptor and acyl-CoA donors C2-, iC5-, aiC5-, iC10-, or nC12-CoA. We asked whether the enzyme could use the mono-acylated sucrose acceptor substrate S1:5 (5R4) (NMR data in Supplemental Table 4) because both M82 and LA1777 accumulate acylsucroses with the R4 position almost invariably substituted with an iso-C5 acyl chain (Ghosh et al., 2014). Indeed, LA0716 Sp-ASAT3 showed activity with S1:5 (5R4) and iC5-CoA, producing an S2:10 (5R4,5P) product that has a different retention time compared with the S2:10 (i5R4,ai5R3) used as a substrate for other ASAT3 reactions (Table 1; Supplemental Figure 9). Notably, the product was acylated on the pyranose ring, in contrast to transfer to the furanose ring catalyzed by Sl-ASAT3 and Sh-ASAT3-F enzymes (Supplemental Figure 10). Consistent with their close phylogenetic relationship to Sp-ASAT3 (Figure 7B), the LA1777, LA1731, LA2098, and LA2861 ShASAT3-P enzymes all added iC5 to the pyranose ring of S1:5 (5R4) (Table 1; Supplemental Figures 9 and 10). In contrast, none of the ASAT3-P enzymes showed in vitro activity with the longer chain donor nC12-CoA and S1:5 (5R4).
The in vitro activities of Sp-ASAT3 and the Sh-ASAT3-P and -F enzymes lead us to propose the model for the terminal steps of acylsucrose biosynthesis shown in Figure 6B. In this model, S. pennellii LA0716 produces relatively few different abundant tri-acylsucroses (Figure 2C), all lacking furanose ring acylation, due to the presence of a single Sp-ASAT3 gene (Figure 6B, left pathway). In the S. habrochaites accessions, the types of acylsucroses produced depends on the number of ASAT3 genes and activities of the enzymes that they encode (Figure 6B).
DISCUSSION
This work describes the identification and characterization of Sl-ASAT3, a type I/IV trichome apical cell-specific BAHD acyltransferase that acylates the furanose ring of acylsucroses in cultivated tomato. A survey of acylsucrose phenotypes in wild tomato species, which included analyses of 26 S. habrochaites accessions, provided several insights into acylsucrose diversity. First, S. habrochaites had the most furanose ring acyl chain diversity of the seven species studied. Second, multiple close homologs of Sl-ASAT3 are found in S. habrochaites individuals. Finally, the activities of Sh-ASAT3 enzymes correlate with the observed acylsugar phenotypes from which the gene sequences were isolated. Taken together, these results indicate that differences in ASAT3 enzyme activity contribute to the strong diversity in acylsugar structures seen in cultivated tomato and its wild relatives.
Enzymatic Activity Difference Enabled ASAT3 Identification
Sl-ASAT3 was identified using a forward genetic screen of S. lycopersicum M82 × S. pennellii LA0716 chromosome introgression lines, a strategy that previously led to the discovery of the ASAT4 sucrose acetyltransferase (originally named AT2 in Schilmiller et al., 2012). We demonstrated that the trichome tip cell-expressed Sl-ASAT3 enzyme, encoded by a gene on chromosome 11, is capable of acylating the furanose ring of di-acylsucroses S2:10 (5,5) and S2:17 (5,12) (Figure 5). Although both enzymes transfer short acyl chains from an acyl-CoA donor to an acylated sucrose acceptor, Sl-ASAT3 and Sl-ASAT4 share only 27% amino acid identity. This lack of sequence identity among BAHD acyltransferases that catalyze very similar reactions, outside of two short conserved domains, is common among BAHD enzymes (D’Auria, 2006).
In contrast to ASAT4, where LA0716 has a nonfunctional allele leading to acylsucroses without acetylation, the lack of R3′ acylation in LA0716 and IL11-3 results from an ASAT3 enzyme with altered activity. The genomic sequence of LA0716 Sp-ASAT3 has an open reading frame that is transcribed in trichomes and is predicted to encode a functional BAHD protein (Supplemental Table 2). In contrast to the Sl-ASAT3 enzyme, which uses di-acylsucrose substrates, activity was not detected with Sp-ASAT3 and di-acylsucrose acceptors. However, the recombinant enzyme added iC5 to the pyranose ring of mono-acylated S1:5 (i5R4) (Table 1; Supplemental Figures 9 and 10). The accumulation of S3:22 (5,5,12) in IL11-3 suggests that there is another uncharacterized ASAT in cultivated tomato type I/IV trichomes that can add a C12 acyl chain to the S2:10 (5,5) that is produced by Sp-ASAT3 in vivo to make S3:22 (5,5,12).
IL11-3 and Sl-ASAT3 RNAi lines accumulate the di-acylsucrose S2:17 (5R4,12R3) (Figure 2; Supplemental Figure 4), which was found to be a substrate of Sl-ASAT3. Despite the abundance of S3:15 (5,5,5) and S4:17 (2,5,5,5) in wild-type M82, no di-acylsucroses containing only short acyl chains (C5 or less) are detected in extracts of IL11-3 or the RNAi lines. Transformation of IL11-3 plants with Sl-ASAT3 under the control of its own promoter resulted in restoration of S3:15 (5,5,5) and S4:17 (2,5,5,5), indicating that Sl-ASAT3 adds the R3′ acyl group on the acylsucroses containing all short chains (Supplemental Figure 4). This activity of Sl-ASAT3 was confirmed in vitro with an S2:10 (5,5) substrate (Figure 5). The lack of accumulation of S2:10 (5,5) in IL11-3 and Sl-ASAT3 RNAi lines suggests that these intermediates are unstable and turned over or converted to one or more compounds that are not detected in our assays.
Diversity in Furanose Ring Acylation and ASAT3
S. lycopersicum M82 produces acylsucroses having a single C5 acyl chain on the furanose ring, while acylsucroses of S. pennellii LA0716 have no furanose ring acylation (Figure 2). In contrast, previous characterization of acylsucrose structures from two different S. habrochaites accessions showed that these plants had the ability to add a long acyl chain to the furanose ring (Ghosh et al., 2014). We screened multiple accessions spanning their geographical range from seven wild tomato species to examine the diversity in furanose ring acylation further and found that five of these species also primarily accumulate acylsucroses with a single short (C4 or C5) acyl chain on the furanose ring. Although we analyzed multiple accessions per species, we cannot rule out the possibility that other accessions might be able to add long chains or multiple acyl chains to the furanose ring.
Various acylation phenotypes were observed for the furanose ring within S. habrochaites, including no acyl chain present, a single acyl chain that could be C4, C5, C10, C11, or C12, and acylsucroses with two C5 acyl chains (Supplemental Data Set 1). Overlaying the phenotypic data on the geographical location of collection for each accession (Figure 7A) revealed a distribution where accessions from central and southern Peru produce acylsucroses with a long chain (C10-C12) on the furanose ring. In contrast, accessions from Ecuador and northern Peru accumulate acylsucroses having only short acyl chains, if any, on the furanose ring. The geographic separation of the phenotypes may be an indirect effect of population structure or the result of differential selective advantage (for example, in response to insect herbivory) conferred by a given phenotype at a particular location. This difference in phenotypes across the geographical distribution is reminiscent of acylsugar acetylation in the S. habrochaites accessions (Kim et al., 2012). With the exception of two accessions, those that lack acetylated acylsucroses were the same accessions that did not add a long acyl chain to the furanose ring. Further analysis of individual acylsugar structures is needed to determine if there is a mechanistic link between these two phenotypes.
In contrast to single-copy ASAT3 genes in the available full genome sequence assemblies of S. lycopersicum and S. pennellii LA0716 (Tomato Genome Consortium, 2012; Bolger et al., 2014), 14 of 18 accessions of S. habrochaites for which we analyzed ASAT3 sequences had more than one ASAT3-like sequence (Figure 7B), and two accessions had three ASAT3-like sequences. For these accessions, multiple genes likely exist since heterozygosity at a single gene cannot explain the two observed monophyletic clades of ShASAT3 sequences. The more parsimonious explanation for the observed pattern is that the gene duplication resulting in the P and F sequences occurred prior to the speciation of S. habrochaites and S. pennellii, followed by gene loss in the S. pennellii lineage. Currently there is genome sequence available for the S. habrochaites accession LYC4; however, the contig containing ASAT3 in the assembly appears to be a mixed assembly of the very similar ASAT3-P and -F sequences (The 100 Tomato Genome Sequencing Consortium, 2014). S. habrochaites genome assemblies that are able to deal with highly similar sequences for one or more of the accessions that we studied would be helpful for understanding the genetic basis for the sequence diversity of ASAT3.
The diversity of S. habrochaites ASAT3 activities is striking, and the sum of activities of the ASAT3 enzymes for each accession generally correlated well with the observed acylsugar phenotype. For example, LA2098 and LA2861 produce acylsucroses with short chains only on the furanose ring in vivo and their ASAT3 in vitro activities were limited to using short-chain acyl-CoAs (Table 1). Metabolic profiling of extracts of LA1777 and LA1731 showed that they accumulate acylsucroses having a long acyl chain on the furanose ring, and both have ASAT3-F enzymes with this activity (Table 1). In addition to having multiple ASAT3-like sequences, some S. habrochaites accessions have ASAT3 enzymes with dual functionality. Sh-ASAT3-P enzymes from LA1777 and LA2098 are notable because they can acylate S1:5 on the pyranose ring or add two short acyl chains to the furanose ring of S2:10 (ai5R3,i5R4) to make S4:20 (5,5,5,5) (Table 1). In contrast, we were unable to detect S2:10 (ai5R3,i5R4) acylation activity with LA1731 and LA2861 Sh-ASAT3-P enzymes. LA2861 extracts have low levels of only a single acylsucrose with two furanose ring acyl chains (Supplemental Data Set 1); therefore, Sh-ASAT3-P is not expected to have high activity with S2:10 to produce S4:20. Based on the presence of multiple acylsucroses with two furanose ring acyl chains in LA1731, we would expect LA1731 Sh-ASAT3-P to have this activity. It is possible that S2:10 (ai5R3,i5R4) is not the correct substrate for this enzyme. Although the LA1777 ASAT3-P enzyme had an enzymatic activity producing acylsucroses with two acyl chains on the furanose ring, this type of acylsucrose was not seen in two of the three LA1777 plants that were phenotyped. This lack of accumulation of ASAT3-P products was correlated with reduced expression of ASAT3-P compared with ASAT3-F in trichomes of those two LA1777 individuals (Supplemental Figure 12). The acylsugar phenotype for other S. habrochaites accessions presumably also could be a result of differential expression of ASAT3 sequences.
Acylsugar Biosynthesis in a Broader Context
Both Sl-ASAT3 and Sl-ASAT4 are BAHD-type acyltransferases that catalyze the transfer of acyl chains from acyl-CoAs to an acylsucrose acceptor. Our results are consistent with the hypothesis that all acylsucroses made in the cultivated tomato type I/IV trichomes are synthesized via BAHD acyltransferases starting from sucrose via sequential addition of each acyl group at specific positions (Figure 6A). The use of BAHD acyltransferases is consistent with the proposed role of the branched-chain keto-acid dehydrogenase complex, which generates acyl-CoAs, in acylsugar production (Slocombe et al., 2008). This type of pathway would not require the use of a glycosyltransferase or serine-carboxypeptidase-like acyltransferase, as was proposed for acylglucose production in S. pennellii LA0716 (Ghangas and Steffens, 1995; Li and Steffens, 2000).
Trichome-based biosynthesis of acylsugars in the Solanaceae is an attractive target for manipulation to improve natural pest resistance and reduce dependence on externally applied pesticides that have deleterious effects on the environment. To enable targeted breeding efforts, a better understanding of the genetic components necessary for acylsugar production and regulation is required. A significant effort has been made to identify quantitative trait loci (QTL) affecting acylsugar production in tomato using crosses with S. pennellii LA0716 (Mutschler et al., 1996; Lawson et al., 1997), a wild tomato species that produces high levels of acylsugars, including copious acylglucoses (Fobes et al., 1985). A QTL on chromosome 11 was identified that appears to colocalize with ASAT3 based on the markers used to define the QTL region (Leckie et al., 2012). Similar to our observation that total acylsugar levels decreased when plants harbored homozygous Sp-ASAT3 alleles, plants from multiple QTL mapping populations that were homozygous for this chromosome 11 region also showed low acylsucrose levels (Mutschler et al., 1996; Leckie et al., 2012). Interestingly, the QTL on chromosome 11 also had an effect on acylglucose production when combined with other introgressions (Leckie et al., 2013). It remains to be determined whether ASAT3 is the locus responsible for these changes in acylglucoses. Our results indicate that introduction of ASAT3 enzymes from S. habrochaites by transgenesis or interspecific crosses could be a productive strategy for generating diversity in S. lycopersicum trichome acylsugars.
What is the significance of adding a long acyl chain to the furanose ring of acylsucroses in accessions in the southern range of S. habrochaites in Peru? We currently do not know what kinds of acyl chains or what acylation patterns are important for the function of acylsugars, for example, in herbivore defense, except that volatile metabolic products of short acyl chain-containing acylsucroses can attract herbivore predators (Weinhold and Baldwin, 2011). Likewise, it is not known what biotic or abiotic factors influenced the evolution of acylsugar production in the Solanum. The data presented here on the biochemical and biosynthetic diversity of acylsugars across a range of accessions from the S. habrochaites species create opportunities to look more closely into ecological factors driving evolution of this specialized metabolic pathway. To date, production of acylsugars has been found only in species within the Solanaceae. However, there are now examples of plants from the Caryophyllaceae having glucose esters present in glandular trichome exudate that resemble Solanaceae acylsugars but with longer chain oxygenated fatty acids forming macrocyclic structures (Asai and Fujimoto, 2010; Asai et al., 2012). As more enzymes of the acylsugar biosynthetic pathway are discovered, the opportunities for acylsugar modification increases and allows for testing the effectiveness of specific acylsugar patterns. Similarly, discovery of novel alleles among the wild tomato species increases the potential for tomato breeders to effectively target acylsugar modification to produce a desired phenotype.
METHODS
Mapping of the IL11-3 Acylsugar Locus
An F2 mapping population was created by self-pollination of an M82 × IL11-3 F1 line provided by Dani Zamir (Hebrew University Faculty of Agriculture, Rehovot, Israel). Genotyping was performed using PCR-based cleaved-amplified polymorphic sequence markers (Supplemental Table 3) and phenotyping by LC-MS analysis of acylsugars is described below.
Acylsugar Mass Spectrometry
A combination of LC-MS approaches was taken to obtain structural information about acylsugars. Collision-induced dissociation MS in negative-ion mode provided information about the number and length of acyl chains present on each acylsugar based on masses of carboxylate fragment ions (Schilmiller et al., 2010a, 2012; Ghosh et al., 2014). Collision-induced dissociation MS in positive-ion mode, which results in cleavage of the glycosidic bond linking the six-membered pyranose and five-membered furanose rings, was used to determine the number and length of acyl chains present on each ring. Detailed LC-MS methods are available in the supplemental data.
Analysis of ASAT3 Gene Expression in Trichomes
Sl-ASAT3 expression was analyzed using total RNA isolated from stem and petiole tissue, trichomes collected from stem and petiole tissue, and stem and petioles after removal of trichomes using 3-week-old M82 plants. Trichomes were collected by gentle scraping from tissue frozen in liquid nitrogen. Reverse transcription was performed using 0.5 μg of total RNA followed by PCR (primers listed in Supplemental Table 5) with detection of products after various numbers of cycles. Sh-ASAT3-P and -F gene expression in LA1777 individuals was performed using isolated stem and petiole trichomes. Total RNA (0.25 μg) was used to synthesize first-strand cDNA using Superscript II (Invitrogen). The resulting cDNA was diluted 10-fold, and 1 μL was used as template for PCR amplification in 20-μL reactions using Power SYBR Green PCR Master Mix (Applied Biosystems) and gene-specific primers (Supplemental Table 5). Reactions were performed using an Applied Biosystems 7500 Fast Real Time PCR system and a final dissociation step was performed to assess the quality of amplification. Levels of Sh-ASAT3-P and -F expression were calculated using the standard curve method.
Tomato Transformation
Transformation of M82 and IL11-3 plants was performed using Agrobacterium tumefaciens strain AGL0 as previously described (McCormick, 1991). The construct for RNAi suppression of Sl-ASAT3 in M82 plants was created using the pHELLSGATE12 binary vector (Eamens and Waterhouse, 2011). A 701-bp sequence of Sl-ASAT3 was amplified from M82 genomic DNA using the primers 5′-CACCCATAAACGATTGGGCGTCTACAGC-3′ and 5′-CATTTAGCCCATCTCCACTTTGTTC-3′. The resulting product was cloned into pENTR/D-TOPO (Invitrogen) followed by recombination into pHELLSGATE12. To express Sl-ASAT3 under the control of its native promoter in IL11-3 plants, the full-length Sl-ASAT3 ORF with 1700 bp of sequence upstream of the translation start site was amplified from M82 genomic DNA using the primers 5′-CACCTATTACATCTTCTAGTGGCTGGTAT-3′ and 5′-CAGTTGGTTTATGTTGCTCTTTTATCTC-3′. The resulting 3111-bp product was subcloned into pENTR/D-TOPO followed by recombination into the binary vector pK7WG (Karimi et al., 2002). For in planta analysis of reporter gene expression driven by the Sl-ASAT3 promoter, a 1678-bp fragment was amplified using the primers 5′-CACCTATTACATCTTCTAGTGGCTGGTAT-3′ and 5′-ATAAATTAAGCTAAGTAATGTTCCA-3′ and cloned into pENTR-D/TOPO followed by recombination into pKGWFS7 (Karimi et al., 2002). The resulting construct was used to transform M82 plants and drive expression of a GFP-GUS fusion protein under the control of the Sl-ASAT3 promoter. GFP fluorescence was analyzed in multiple primary transformants as previously described (Schilmiller et al., 2012).
Wild Tomato Acylsugar Profiling and Sh-ASAT3 Sequencing
Accessions of Solanum neorickii, Solanum arcanum, Solanum huaylasense, Solanum peruvianum, Solanum corneliomulleri, Solanum pennellii, and Solanum habrochaites available at the C. M. Rick Tomato Genetic Resource Center (TGRC; tgrc.ucdavis.edu) were selected that covered the geographical range of the species. Plant growth conditions were as previously described (Schilmiller et al., 2010a). Leaf dip extracts were made from 3-week-old plants and acylsugars were analyzed as described below. Sh-ASAT3 sequences were amplified from genomic DNA of 18 S. habrochaites accessions using Sh-ASAT3-P and Sh-ASAT3-F primer pairs (Supplemental Table 5). The two-nucleotide difference in the P and F forward primers allowed for preferential amplification of one sequence or the other. In the accessions where only one ASAT3 (P or F) sequence was found, both primer sets amplified the same sequence. For most accessions, PCR products were sequenced directly and the consensus sequence from the Sanger sequencing reads were used for phylogenetic analysis (sequences in Supplemental Data Set 2). Eighteen sequences are from individual clones of PCR products and are deposited in the GenBank database (accession numbers listed below).
Phylogenetic Analyses
There are an average of 5, 15, and 29 pairwise nucleotide differences and 3, 9, and 22 pairwise amino acid differences between Sh-ASAT3-F/F, P/P, and F/P sequences, respectively. Although a rooted phylogenetic tree constructed using amino acid sequences produced a tree with similar topology (Supplemental Figure 8F), we present here the results of phylogenetic analyses performed using the ASAT3 coding sequences because more variation was modeled into this tree and the bootstrap values for older nodes in these trees were significantly better. The ASAT3 gene lacks introns; therefore, intronic sequences could not be used for phylogenetic reconstruction.
The ASAT3 coding sequences were aligned using the default MUSCLE algorithm with the MEGA6 software (Tamura et al., 2011). The best model for generating a phylogenetic tree (T92 three parameter model with gamma distributed rates) was chosen from among 24 competing models based on the lowest Bayesian Information Criterion calculated using maximum likelihood analysis of the aligned sequences. The maximum likelihood tree presented in Figure 7B represents the tree obtained using the above best model. We used the Sl-ASAT3 coding sequence as an outgroup since there are an average of 59 nucleotide differences between Sl-ASAT3 and Sh-ASAT3-P and -F pairs, compared with a maximum of 29 average pairwise differences within Sh-ASAT3 sequences. For determining whether the observed phylogenetic relationships are robust to the software, method, and model used, we performed additional validation using another software package (RaxML; Stamatakis, 2014), another method (neighbor joining), and other models (Tamura-Nei 93 with uniform rates, GTR with uniform rates, GTR+G and GTR+G+I). These results are presented in Supplemental Figure 8. The Sh-ASAT3-P and Sh-ASAT3-F sequences were independently found to be monophyletic in all instances. The clustering of ASAT3-like sequences had low bootstrap supports in all scenarios; hence, their evolutionary origins are not clear. Sequence alignments of nucleotide sequences are available in Supplemental Data Sets 2 to 4.
S2:17 (5,12) Purification for NMR and Enzyme Assays
Sixty IL11-3 plants were grown in a growth chamber using a cycle of 17 h light (200 to 220 μmol m−2 s−1 at 28°C) and 7 h dark (20°C) until 4 weeks postgermination. At that stage, plant leaf and petiole tissues were harvested and placed in a 5-liter beaker, to which 2000 mL of 100% methanol was added. The mixture was stirred for 2 min with a glass rod and then quickly transferred into two 1-liter glass bottles through a Buchner funnel fitted with filter paper. Solvent was evaporated to dryness under vacuum using a rotary evaporator, and the residue was redissolved in 3 mL of acetonitrile:water (4/1 [v/v]) with ultrasonication for 10 min followed by transfer to a polypropylene centrifuge tube and centrifugation at 2627g for 2 min at 25°C. Supernatants were collected in HPLC vials each with glass inserts (200 μL in each), and vials were sonicated for 10 min. Subsequent metabolite purification was performed using a Waters 2795 HPLC and a Thermo Scientific Acclaim 120 C18 semipreparative HPLC column (4.6 × 150 mm, 5 μm). The mobile phase consisted of 0.15% formic acid in water, pH 2.6 (Solvent A), and acetonitrile (Solvent B) using linear gradient elution of 1% B at 0 min and 48% B at 5 min and held at 48% B at 5 to 30 min, 62% B at 31 min, and 100% B at 55 min. Solvent composition was held at 100% B for 55 to 58 min and then brought back to 1% B at 59 min and held at 1% B for 59 to 60 min. The solvent flow rate was 1.5 mL/min, and the column temperature was 40°C. Eluted fractions were collected in a LKB fraction collector in 1-min fractions for 13 injections, using an injection volume of 200 μL for each injection. The desired compound was collected in 25 to 28 min.
NMR spectra were recorded of solutions in Shigemi solvent-matched tubes using a Bruker Avance 900 NMR spectrometer equipped with a TCI triple-resonance inverse detection cryoprobe at the Michigan State University Max T. Rogers NMR facility. The 1H spectra were recorded at 900 MHz, and 13C NMR spectra were recorded at 225 MHz.
Sl-ASAT3 RNAi Line Acylsucrose Collection for Enzyme Assays
Leaflets and petioles from five T1 plants from T0 line #10 were collected and dipped into 10 mL of extraction solvent (acetonitrile/isopropanol/water 3:3:2 [v/v/v] containing 0.1% [v/v] formic acid). To partially purify acylsugars, the extract was diluted 1:4 with water and then loaded onto a strong cation exchange solid phase extraction column (Supelco; 500 mg bed weight) preequilibrated with 10% (v/v) acetonitrile. Acylsugars were eluted with 50% acetonitrile, and the eluate was reduced to dryness under vacuum. The resulting residue was dissolved in 100 μL 50% ethanol (v/v) and was used directly in Sl-ASAT3 assays.
Production of aiC5-CoA and iC10-CoA
The 2-methylbutyric acid (aiC5) and 8-methylnonanoic acid (iC10) were purchased from Sigma-Aldrich and Cayman Chemical, respectively. CoA was from Adventbio. Acyl-CoAs were synthesized from the corresponding fatty acids using a carbonyldiimidazole method as previously described (Kawaguchi et al., 1981). The aiC5-CoA was purified using a Supelclean LC-18 SPE column (Sigma-Aldrich). Briefly, synthesized aiC5-CoA was loaded onto a column that was washed with 8 mL 100% methanol and preequilibrated with 8 mL distilled water. The column was washed with 8 mL water and then eluted with 4 mL 15% methanol in water. Fractions containing aiC5-CoA were combined and the methanol was evaporated under N2 gas. The pH was adjusted to 4.0 with formic acid and stored at −20°C before use. Purification of synthesized iC10-CoA was done by precipitation using 7% perchloric acid, and unreacted free fatty acid was removed by adding hexane. The iC10-CoA precipitate was washed with 1 mL 0.7% perchloric acid twice followed by solvent evaporation under N2 gas. The residue was dissolved in 0.01 M NaOAc/ethanol (1:1, pH 5.2). The concentration of synthesized acyl-CoAs was determined by UV spectroscopy using the CoA extinction coefficient of 16,400 at 260 nm.
ASAT3 Protein Expression and Enzyme Assay
Recombinant protein for enzyme assays was produced using Escherichia coli as the host. The full-length ORF for Sl-ASAT3, Sp-ASAT3, and various Sh-ASAT3 sequences were amplified using primers (Supplemental Table 5) that add NheI and XhoI restriction sites to the ends for subcloning into pET28b (EMD Millipore). The resulting construct, which adds the amino acid sequence “MGSSHHHHHHSSGLVPRGSH” to the N terminus of the protein, was transformed into BL21 Rosetta cells (EMD Millipore) for protein production. His-tagged protein was induced in cultures of E. coli using 0.05 mM isopropyl β-d-1-thiogalactopyranoside followed by growing overnight at 16°C and 120 rpm shaking. Soluble protein was partially purified by nickel-affinity chromatography as previously described (Schilmiller et al., 2012). ASAT3 enzyme assays were performed by incubating recombinant enzyme in 30 μL of 50 mM ammonium acetate (pH 6.0) buffer with 100 μM acyl-CoA and an acylsucrose acceptor. S2:17 (5,12) was purified from IL11-3 and the mixture of acylsucroses lacking a 3′ acyl group was from T1 plants from Sl-ASAT3 RNAi line #10. Assays were incubated at 30°C for 30 min followed by termination by addition of 60 μL acetonitrile/isopropanol/formic acid (1:1:0.001) containing 10 μM propyl-4-hydroxybenzoate internal standard and centrifugation at 13,000g for 10 min. Ten microliters of the reaction mixture was analyzed by LC-MS in both negative- and positive-ion mode as described below. Sl-ASAT4 reactions using Sl-ASAT3 products were performed by heating the Sl-ASAT3 reaction at 65°C for 5 min to inactivate Sl-ASAT3, cooling on ice, adding acetyl-CoA (100 μM) and Sl-ASAT4 (Schilmiller et al., 2012), and incubating at 30°C for 30 min before stopping the reaction as described above for Sl-ASAT3.
Enzyme reactions for determining apparent Km values of acyl-CoA substrates were performed using enzyme amount and incubation time where reaction velocities were in the linear range. The acylsucrose acceptor substrate S2:10 was held at saturating concentration while acyl-CoA concentration was varied. Assays were performed in triplicate and incubated at 30°C for 5 min in a 30-μL volume followed by termination as described above. Ten microliters of the reaction mixture was analyzed by LC-MS to determine velocities (peak area of the acylated product divided by peak area of the internal standard). Apparent Km and Ki values were determined by nonlinear regression using the standard Michaelis-Menten kinetics model or substrate inhibition model in GraphPad Prism5 (GraphPad Software).
Acylsugar LC-MS Analysis
Acylsugar extraction from plant tissues was done by incubating an intact leaflet from the youngest expanded leaf in 1 mL extraction solvent with gentle agitation for 2 min. The extraction solvent was acetonitrile/isopropanol/water (3:3:2) containing 0.1% formic acid and 10 μM propyl 4-hydroxybenzoate as internal standard.
Enzyme assay samples and samples extracted from plant tissues were analyzed using a Waters Xevo G2-S QToF LC-MS interfaced to a Waters Acquity UPLC system. Ten microliters of leaf dip extract was injected onto a fused core Ascentis Express C18 column (2.1 mm × 10 cm, 2.7-μm particle size; Sigma-Aldrich). Column temperature was maintained at 40°C. The flow rate was 0.3 mL/min with starting conditions at 95% solvent A (water + 0.15% formic acid) and 5% solvent B (acetonitrile). The 7-min gradient profile for elution was as follows: starting at 5% solvent B, then ramping to 40% B at 1 min, then to 100% B at 5 min, hold at 100% B to 6 min, at 6.01 min return to 95% A/5% B and maintain until 7 min. The MS settings were as follows: electrospray ionization in negative-ion mode, 2.14 kV capillary voltage, 90°C source temperature, 350°C desolvation temperature, 600 liters/h desolvation nitrogen gas flow rate, 10 V cone voltage, and mass range of m/z 50 to 1500 with spectra accumulated at 0.1 seconds/function. Three separate acquisition functions were set up to generate spectra at different collision energies (5, 25, and 60 eV) providing both nonfragmenting and fragmenting conditions.
Leaf dip samples from the accessions of various Solanum species were analyzed using a Shimadzu LC-20AD HPLC system connected to a Waters LCT Premier ToF-MS. Samples were injected onto a fused core Ascentis Express C18 column (2.1 mm × 10 cm, 2.7-μm particle size; Sigma-Aldrich) for reverse-phase separation. Column temperature was 40°C. The flow rate was set at 0.4 mL/min and starting conditions were 90% solvent A and 10% solvent B. The 22-min elution gradient was as follows: ramp to 40% B at 4 min, then to 100% B at 17 min, hold at 100% B to 20 min, at 20.01 min return to 90% A/10% B, and maintain until 22 min. The MS settings were as follows: electrospray ionization in negative mode, 2.5 kV capillary voltage, 100°C source temperature, 350°C desolvation temperature, 40 liters/h cone gas flow, 350 liters/h desolvation nitrogen gas flow, 10 V cone voltage, and mass range m/z 50 to 1500 with spectra accumulated at 0.1 seconds/function. Three separate acquisition functions were set up to generate spectra at different collision energies (10, 40, and 80 eV) providing both nonfragmenting and fragmenting conditions. Analyses of samples by electrospray ionization in positive-ion mode were performed using the Shimadzu/LCT system under the same conditions as negative-ion mode except the capillary voltage was set to 3.2 kV.
Acylsugar abundances were determined using the Waters Quanlynx analysis tool to integrate extracted ion peak areas relative to the peak area of the internal standard. Data collected using the lowest collision energy (5 eV) was used for analysis of intact acylsugars ionized using negative-ion mode. For analysis of acyl chains on the furanose ring of acylsucroses, positive-ion mode data collected with collision energy of 40 eV were used that resulted in fragmentation of the glycosidic bond allowing for determining the mass of acyl chains present on each hexose ring (Ghosh et al., 2014).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Sl-ASAT4 (JQ899258), Sl-ASAT3 (KM516150), Sp-ASAT3 (KM516151), LA1777-Sh-ASAT3-F (KM516152), LA1978-Sh-ASAT3-F (KM516153), LA2156-Sh-ASAT3-F (KM516154), LA2204-ASAT3-like1 (KM516155), LA2861-Sh-ASAT3-F (KM516156), LA2861-Sh-ASAT3-P (KM516157), LA2156-ASAT3-like (KM516158), LA1777-Sh-ASAT3-P (KM516159), LA1731-Sh-ASAT3-P (KM516160), LA1731-Sh-ASAT3-F (KM516161), LA2722-Sh-ASAT3-P (KM516162), LA2650-Sh-ASAT3-P (KM516163), LA2574-Sh-ASAT3-P (KM516164), LA2204-ASAT3-like2 (KM516165), LA1772-Sh-ASAT3-P (KM516166), and LA2098-Sh-ASAT3-P (KM524335).
Supplemental Data
Supplemental Figure 1. Mass spectra of abundant acylsucroses from M82, LA0716, and IL11-3.
Supplemental Figure 2. IL11-3 acylsugars lack the R3′ acyl group normally seen on M82 acylsucroses.
Supplemental Figure 3. RT-PCR analysis of Sl-ASAT3 expression in M82 plants.
Supplemental Figure 4. Base peak intensity LC-MS chromatograms for extracts of Sl-ASAT3 transgenic lines.
Supplemental Figure 5. Mass spectra of Sl-ASAT3 enzyme assay products when using S2:17 as the acceptor substrate.
Supplemental Figure 6. Sl-ASAT3 assays with S2:17 (5,12) and a variety of available acyl-CoA donors.
Supplemental Figure 7. Sl-ASAT3 activity using acylsugar acceptors collected from RNAi plants.
Supplemental Figure 8. Tests of phylogenetic relationships between different ASAT3 sequences.
Supplemental Figure 9. Extracted ion LC-MS chromatograms from analysis of Sh-ASAT3 enzyme assay products.
Supplemental Figure 10. Mass spectra of Sh-ASAT3 enzyme assay products.
Supplemental Figure 11. Kinetic analysis of LA777 and LA1731 Sh-ASAT3-F.
Supplemental Figure 12. qRT-PCR analysis of Sh-ASAT3 expression in trichomes of LA1777 individuals.
Supplemental Table 1. NMR chemical shifts of S2:17 (5,12) from IL11-3.
Supplemental Table 2. Genes in the IL11-3 mapping interval.
Supplemental Table 3. NMR chemical shifts of S2:10 (5,5) produced in vitro.
Supplemental Table 4. NMR chemical shifts of S1:5 (5) produced in vitro.
Supplemental Table 5. PCR primers used for mapping and cloning.
Supplemental Data Set 1. Furanose ring acyl chain phenotypes for various wild tomato species.
Supplemental Data Set 2. Clustal alignment of ASAT3 nucleotide sequences used for phylogenetic trees in Figure 7B and Supplemental Figures 8A to 8E.
Supplemental Data Set 3. Clustal alignment of ASAT3 protein sequences used for the phylogenetic tree in Supplemental Figure 8F.
Supplemental Data Set 4. Clustal alignment of selected ASAT3 nucleotide sequences and other chromosome 11 BAHD nucleotide sequences used for the phylogenetic tree in Supplemental Figure 8G.
Supplementary Material
Acknowledgments
We thank trichome project members for their contributions, especially Amanda Charbonneau, Karin Hanisch, Abigail Miller, Dan Lybrand, and Eran Pichersky for helpful comments on the article; the MSU Center for Advanced Microscopy and RTSF Mass Spectrometry and Metabolomics Core Facilities; Kermit Johnson and Dan Holmes at the MSU Max T. Rogers NMR facility; and Alisdair Fernie and Bjoern Usadel for early access to the LA0716 genomic sequence. Seed for M82, IL11-3, and accessions from other Solanum species was obtained from the C. M. Rick Tomato Genetics Resource Center (UC Davis, CA). The work was funded by National Science Foundation Grant IOS-1025636.
AUTHOR CONTRIBUTIONS
A.L.S. and R.L.L. designed the research. A.L.S., P.F., N.J., and B.G. performed the research. A.L.S., G.D.M., B.G., and A.D.J. analyzed data. A.L.S., G.D.M., and R.L.L. wrote the article.
Glossary
- LC-MS
liquid chromatography coupled with time-of-flight mass spectrometry
- MS
mass spectrometry
- RNAi
RNA interference
- QTL
quantitative trait loci
- ORF
open reading frame
Footnotes
Articles can be viewed online without a subscription.
References
- Alba J.M., Montserrat M., Fernández-Muñoz R. (2009). Resistance to the two-spotted spider mite (Tetranychus urticae) by acylsucroses of wild tomato (Solanum pimpinellifolium) trichomes studied in a recombinant inbred line population. Exp. Appl. Acarol. 47: 35–47. [DOI] [PubMed] [Google Scholar]
- Asai T., Fujimoto Y. (2010). Cyclic fatty acyl glycosides in the glandular trichome exudate of Silene gallica. Phytochemistry 71: 1410–1417. [DOI] [PubMed] [Google Scholar]
- Asai T., Nakamura Y., Hirayama Y., Ohyama K., Fujimoto Y. (2012). Cyclic glycolipids from glandular trichome exudates of Cerastium glomeratum. Phytochemistry 82: 149–157. [DOI] [PubMed] [Google Scholar]
- Bolger A., et al. (2014). The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 46: 1034–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Kumar R., Headland L.R., Ranjan A., Covington M.F., Ichihashi Y., Fulop D., Jiménez-Gómez J.M., Peng J., Maloof J.N., Sinha N.R. (2013). A quantitative genetic basis for leaf morphology in a set of precisely defined tomato introgression lines. Plant Cell 25: 2465–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Auria J.C. (2006). Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant Biol. 9: 331–340. [DOI] [PubMed] [Google Scholar]
- Eamens A.L., Waterhouse P.M. (2011). Vectors and methods for hairpin RNA and artificial microRNA-mediated gene silencing in plants. In Plant Chromosome Engineering, Vol. 701, J.A. Birchler, ed (New York: Humana Press), pp. 179–197. [DOI] [PubMed] [Google Scholar]
- Fobes J.F., Mudd J.B., Marsden M.P. (1985). Epicuticular lipid accumulation on the leaves of Lycopersicon pennellii (Corr.) D’Arcy and Lycopersicon esculentum Mill. Plant Physiol. 77: 567–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghangas G.S., Steffens J.C. (1993). UDPglucose: fatty acid transglucosylation and transacylation in triacylglucose biosynthesis. Proc. Natl. Acad. Sci. USA 90: 9911–9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghangas G.S., Steffens J.C. (1995). 1-O-acyl-β-D-glucoses as fatty acid donors in transacylation reactions. Arch. Biochem. Biophys. 316: 370–377. [DOI] [PubMed] [Google Scholar]
- Ghosh B., Westbrook T.C., Jones A.D. (2014). Comparative structural profiling of trichome specialized metabolites in tomato (Solanum lycopersicum) and S. habrochaites: acylsugar profiles revealed by UHPLC/MS and NMR. Metabolomics 10: 496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb C.D., Abel S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11: 89–100. [DOI] [PubMed] [Google Scholar]
- Hawthorne D.J., Shapiro J.A., Tingey W.M., Mutschler M.A. (1992). Trichome-borne and artificially applied acylsugars of wild tomato deter feeding and oviposition of the leafminer Liriomyza trifolii. Entomol. Exp. Appl. 65: 65–73. [Google Scholar]
- Hill, K., and Rhode, O. (1999). Sugar-based surfactants for consumer products and technical applications. Fett. Lipid 101: 25–33.
- Kandra G., Severson R., Wagner G.J. (1990). Modified branched-chain amino acid pathways give rise to acyl acids of sucrose esters exuded from tobacco leaf trichomes. Eur. J. Biochem. 188: 385–391. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kawaguchi A., Yoshimura T., Okuda S. (1981). A new method for the preparation of acyl-CoA thioesters. J. Biochem. 89: 337–339. [DOI] [PubMed] [Google Scholar]
- Kim J., Kang K., Gonzales-Vigil E., Shi F., Jones A.D., Barry C.S., Last R.L. (2012). Striking natural diversity in glandular trichome acylsugar composition is shaped by variation at the Acyltransferase2 locus in the wild tomato Solanum habrochaites. Plant Physiol. 160: 1854–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R.R., Calhoun L.A. (1988). 2,3-Di-O- and 1,2,3-tri-O-acylated glucose esters from the glandular trichomes of Datura metel. Phytochemistry 27: 3761–3763. [Google Scholar]
- King R.R., Calhoun L.A., Singh R.P. (1988). 3,4-Di-O- and 2,3,4-tri-O-acylated glucose esters from the glandular trichomes of nontuberous Solanum species. Phytochemistry 27: 3765–3768. [Google Scholar]
- King R.R., Pelletier Y., Singh R.P., Calhoun L.A. (1986). 3,4-Di-O-isobutyryl-6-O-caprylsucrose: the major component of a novel sucrose ester complex from the type B glandular trichomes of Solanum berthaultii hawkes (Pl 473340). J. Chem. Soc. Chem. Commun. 14: 1078–1079. [Google Scholar]
- Kroumova A.B., Wagner G.J. (2003). Different elongation pathways in the biosynthesis of acyl groups of trichome exudate sugar esters from various solanaceous plants. Planta 216: 1013–1021. [DOI] [PubMed] [Google Scholar]
- Kuai J.P., Ghangas G.S., Steffens J.C. (1997). Regulation of triacylglucose fatty acid composition - Uridine diphosphate glucose fatty acid glucosyltransferases with overlapping chain-length specificity. Plant Physiol. 115: 1581–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson D.M., Lunde C.F., Mutschler M.A. (1997). Marker-assisted transfer of acylsugar-mediated pest resistance from the wild tomato, Lycopersicon pennellii, to the cultivated tomato, Lycopersicon esculentum. Mol. Breed. 3: 307–317. [Google Scholar]
- Leckie B.M., De Jong D.M., Mutschler M.A. (2012). Quantitative trait loci increasing acylsugars in tomato breeding lines and their impacts on silverleaf whiteflies. Mol. Breed. 30: 1621–1634. [Google Scholar]
- Leckie B.M., De Jong D.M., Mutschler M.A. (2013). Quantitative trait loci regulating sugar moiety of acylsugars in tomato. Mol. Breed. 31: 957–970. [Google Scholar]
- Li A.X., Steffens J.C. (2000). An acyltransferase catalyzing the formation of diacylglucose is a serine carboxypeptidase-like protein. Proc. Natl. Acad. Sci. USA 97: 6902–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedl B.E., Lawson D.M., White K.K., Siiapiho J.A., Cohen D.E., Carson W.G., Trumble J.T., Mutschler M.A. (1995). Acylsugars of wild tomato Lycopersicon pennellii alters settling and reduces oviposition of Bemisia argentifolii (Homoptera: Aleyrodidae). J. Econ. Entomol. 88: 742–748. [Google Scholar]
- Maldonado E., Torres F.R., Martínez M., Pérez-Castorena A.L. (2006). Sucrose esters from the fruits of Physalis nicandroides var. attenuata. J. Nat. Prod. 69: 1511–1513. [DOI] [PubMed] [Google Scholar]
- McCormick S. (1991). Transformation of tomato with Agrobacterium tumefaciens. In Plant Tissue Culture Manual, Lindsey K., ed (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 1–9. [Google Scholar]
- McDowell E.T., et al. (2011). Comparative functional genomic analysis of Solanum glandular trichome types. Plant Physiol. 155: 524–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R., Last R.L. (2012). Achieving diversity in the face of constraints: lessons from metabolism. Science 336: 1663–1667. [DOI] [PubMed] [Google Scholar]
- Mutschler M.A., Doerge R.W., Liu S.C., Kuai J.P., Liedl B.E., Shapiro J.A. (1996). QTL analysis of pest resistance in the wild tomato Lycopersicon pennellii: QTLs controlling acylsugar level and composition. Theor. Appl. Genet. 92: 709–718. [DOI] [PubMed] [Google Scholar]
- Ohya I., Shinozaki Y., Tobita T., Takahashi H., Matsuzaki T. (1996). Sucrose esters from the surface lipids of Petunia hybrida. Phytochemistry 41: 787–789. [DOI] [PubMed] [Google Scholar]
- Pichersky E., Lewinsohn E. (2011). Convergent evolution in plant specialized metabolism. Annu. Rev. Plant Biol. 62: 549–566. [DOI] [PubMed] [Google Scholar]
- Rodriguez A.E., Tingey W.M., Mutschler M.A. (1993). Acylsugars of Lycopersicon pennellii deter settling and feeding of the green peach aphid. J. Econ. Entomol. 86: 34–39. [Google Scholar]
- Schilmiller A., Shi F., Kim J., Charbonneau A.L., Holmes D., Daniel Jones A., Last R.L. (2010a). Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J. 62: 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A.L., Last R.L., Pichersky E. (2008). Harnessing plant trichome biochemistry for the production of useful compounds. Plant J. 54: 702–711. [DOI] [PubMed] [Google Scholar]
- Schilmiller A.L., Charbonneau A.L., Last R.L. (2012). Identification of a BAHD acetyltransferase that produces protective acyl sugars in tomato trichomes. Proc. Natl. Acad. Sci. USA 109: 16377–16382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A.L., Miner D.P., Larson M., McDowell E., Gang D.R., Wilkerson C., Last R.L. (2010b). Studies of a biochemical factory: tomato trichome deep expressed sequence tag sequencing and proteomics. Plant Physiol. 153: 1212–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson R.F., Arrendale R.F., Chortyk O.T., Green C.R., Thome F.A., Stewart J.L., Johnson A.W. (1985). Isolation and characterization of the sucrose esters of the cuticular waxes of green tobacco leaf. J. Agric. Food Chem. 33: 870–875. [Google Scholar]
- Shapiro J.A., Steffens J.C., Mutschler M.A. (1994). Acylsugars of the wild tomato Lycopersicon pennellii in relation to geographic distribution of the species. Biochem. Syst. Ecol. 22: 545–561. [Google Scholar]
- Shepherd R.W., Bass W.T., Houtz R.L., Wagner G.J. (2005). Phylloplanins of tobacco are defensive proteins deployed on aerial surfaces by short glandular trichomes. Plant Cell 17: 1851–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons A.T., Gurr G.M. (2005). Trichomes of Lycopersicon species and their hybrids: effects on pests and natural enemies. Agric. For. Entomol. 7: 265–276. [Google Scholar]
- Slocombe S.P., Schauvinhold I., McQuinn R.P., Besser K., Welsby N.A., Harper A., Aziz N., Li Y., Larson T.R., Giovannoni J., Dixon R.A., Broun P. (2008). Transcriptomic and reverse genetic analyses of branched-chain fatty acid and acyl sugar production in Solanum pennellii and Nicotiana benthamiana. Plant Physiol. 148: 1830–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 100 Tomato Genome Sequencing Consortium; Aflitos, S., et al. (2014). Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. 80: 136–148. [DOI] [PubMed]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoeven R.S., Steffens J.C. (2000). Biosynthesis and elongation of short- and medium-chain-length fatty acids. Plant Physiol. 122: 275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters D.S., Steffens J.C. (1990). Branched-chain amino acid metabolism in the biosynthesis of Lycoperiscon pennellii glucose esters. Plant Physiol. 93: 1544–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold A., Baldwin I.T. (2011). Trichome-derived O-acyl sugars are a first meal for caterpillars that tags them for predation. Proc. Natl. Acad. Sci. USA 108: 7855–7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werker E. (2000). Trichome diversity and development. In Advances in Botanical Research, Vol. 31, D.L. Hallahan and J.C. Gray, eds (Waltham, MA: Academic Press), pp. 1–35. [Google Scholar]
- Winzer T., et al. (2012). A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336: 1704–1708. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.