Analysis of the causal connections between environmental variation and the genotype-phenotype map revealed candidate genes for constitutive and plastic responses to soil moisture reduction in the context of QTL mapping.
Abstract
Soil water availability represents one of the most important selective agents for plants in nature and the single greatest abiotic determinant of agricultural productivity, yet the genetic bases of drought acclimation responses remain poorly understood. Here, we developed a systems-genetic approach to characterize quantitative trait loci (QTLs), physiological traits and genes that affect responses to soil moisture deficit in the TSUxKAS mapping population of Arabidopsis thaliana. To determine the effects of candidate genes underlying QTLs, we analyzed gene expression as a covariate within the QTL model in an effort to mechanistically link markers, RNA expression, and the phenotype. This strategy produced ranked lists of candidate genes for several drought-associated traits, including water use efficiency, growth, abscisic acid concentration (ABA), and proline concentration. As a proof of concept, we recovered known causal loci for several QTLs. For other traits, including ABA, we identified novel loci not previously associated with drought. Furthermore, we documented natural variation at two key steps in proline metabolism and demonstrated that the mitochondrial genome differentially affects genomic QTLs to influence proline accumulation. These findings demonstrate that linking genome, transcriptome, and phenotype data holds great promise to extend the utility of genetic mapping, even when QTL effects are modest or complex.
INTRODUCTION
Traits that drive adaptation in ecological and agricultural systems are typically affected by the allelic state at many loci, the environmental conditions, and the interaction of genes with the environment (Falconer and Mackay, 1996; Mackay, 2001). Elucidation of how genes and environments interact to produce complex phenotypes is a long-standing problem and “grand challenge” in modern biology as well as crop breeding (Araus et al., 2002, 2008; Rockman, 2012; Heslot et al., 2014). Among the most ecologically and agriculturally important environmental factors is variation in soil moisture availability, which has driven the evolution of morphological and physiological traits (Stebbins, 1952; Axelrod, 1972; Juenger, 2013) and directly affects agricultural productivity (Condon et al., 2004; Cattivelli et al., 2008; Richards et al., 2010).
Plants have evolved complex, diverse, and often highly inducible responses to soil moisture variation (Chaves et al., 2003; Chaves and Oliveira, 2004; Heschel and Riginos, 2005; Bogeat-Triboulot et al., 2007; Harb et al., 2010; Rosenthal et al., 2010; Pinheiro and Chaves, 2011; Mir et al., 2012). For example, drought-adapted genotypes may avoid cellular dehydration through plasticity of many traits, including stomatal conductance (Chater et al., 2011), root and shoot growth (MacMillan et al., 2006), leaf wilting or rolling (Kadioglu and Terzi, 2007), and phenology (Heschel and Riginos, 2005; Sherrard and Maherali, 2006). Upon exposure to drought, many plants accumulate high levels of the stress hormone abscisic acid (ABA). ABA-mediated signaling is important for the regulation of various drought-responsive traits, including stomatal conductance, gene expression (Cutler et al., 2010), and accumulation of the compatible solute proline.
Many plants accumulate high levels of proline upon exposure to drought. In Arabidopsis thaliana, expression of Δ1-PYRROLINE-5-CARBOXYLATE SYNTHETASE1 (P5CS1) is strongly induced by abiotic stress (Szabados and Savouré, 2010; Sharma et al., 2011). P5CS1 catalyzes the probable rate-limiting step in stress-induced proline biosynthesis. Conversely, expression of PROLINE DEHYDROGENASE1 (ProDH1), which encodes a mitochondrion-localized proline catabolism enzyme, is repressed by stress in many plant tissues. It is thought that induction of P5CS1 and repression of ProDH1 expression is important to suppress proline turnover and maximize the accumulation of proline for osmotic adjustment (Leprince et al., 2015). However, other studies have suggested that continued mitochondrial proline catabolism also contributes to drought resistance by balancing cellular redox status and maintaining a favorable ratio of oxidized versus reduced NADP (Sharma et al., 2011).
DNA sequence variation in both nuclear and cytoplasmic (mitochondrial and plastid) genomes may underlie variation in drought response, including proline and ABA accumulation. For example, biosynthesis of the carotenoid precursors of ABA as well as the rate-limiting carotenoid cleavage reaction catalyzed by the 9-cis-expoxycartenoid dioxygenase family of enzymes occur in the chloroplast. While nuclear genes encode the enzymes involved in ABA biosynthesis (Milborrow, 2001; Finkelstein, 2013), the later steps of ABA biosynthesis occur in the cytoplasm. Likewise, proline catabolism occurs in mitochondria but is catalyzed by nucleus-encoded enzymes. It is not established whether proline or ABA metabolism can be influenced by sequence variation of mitochondrion- or plastid-encoded genes.
Quantitative trait locus (QTL) mapping and global gene expression analyses are useful methods to assess the genetic basis of traits involved in drought adaptation (McKay et al., 2008; Hall et al., 2010; Juenger et al., 2010; Des Marais et al., 2012; Schmalenbach et al., 2014; El-Soda et al., 2015). In particular, the genomic perturbation of experimental crosses utilizes recombination to break up linkage disequilibrium and allows causal inference of how variation at a given locus leads to phenotypic variation. One of the goals of QTL mapping is the identification of the polymorphisms underlying heritable physiological variation. While broadly utilized for this goal, linkage-based QTLs do not provide a framework to distinguish among candidate genes without further fine-mapping and/or reverse genetics (Rockman, 2012). The most common method to define candidates underlying a QTL is to search for physically proximate genes with annotations or gene ontology reflecting the trait of interest (Al-Shahrour et al., 2005). While sometimes successful in model organisms, this approach may inhibit the discovery of new genes or candidates in species without annotated reference genomes.
Recently, several studies have combined gene expression and phenotypic trait QTL mapping in experimental populations (Chen et al. 2010; reviewed in Hansen et al., 2008; Cubillos et al., 2012). Through analysis of colocalization between differentially expressed genes and phenotypic trait QTLs, it is possible to produce lists of candidate genes (Swamy et al., 2013); however, the researcher is often left with long and unwieldy lists of candidates without direction regarding which genes to pursue further. Several approaches have been developed in mouse and human model systems to solve this problem by ranking candidate genes underlying QTL regions through the joint analysis of genome-wide transcript abundance data with trait and genotype data of QTL studies (Schadt et al., 2005; Drake et al., 2006; Farber et al., 2009).
Candidate gene effect analyses often seek to identify causation among predictor variables, such as genotype or gene expression polymorphism, and the phenotypic trait response variable (Al-Shahrour et al., 2005) and have been used as a post-hoc method to find candidate genes under QTL peaks. For example, by assessing the correlation structure of traits, transcripts, and alleles, causal inference testing (CIT) can be used to determine the statistical significance of potential candidate genes (Millstein et al., 2009). However, post-hoc tests of candidate genes like CIT take QTL candidate regions out of the multiple-QTL model in which they were generated and, in doing so, assume that additive and epistatic effects of other loci and covariates (e.g., sex, cytoplasm, or environment) do not affect the QTL peak. The power of multiple-QTL mapping is to incorporate all these variables simultaneously. As such, much of the power to determine the effects of genes underlying a QTL peak may be lost through current post-hoc approaches.
In QTL mapping, when a covariate explains residual variation, it can increase the researcher’s ability to define uncorrelated QTL peaks (Zeng, 1993; Broman and Sen, 2009). However, the opposite is also true. Correlated covariates can absorb QTL-specific variance, reducing the peak height and power to detect a local QTL (Supplemental Figure 1). Li et al. (2006) used this approach to infer causality among correlated phenotypic traits through structural equation modeling. In short, if using trait X as a covariate reduces the LOD score of a QTL for trait Y so that there is no longer statistical support for the QTL for trait Y, then X causes Y (Li et al., 2006). This methodology is commonly used to infer causality among phenotypic traits via partial regressions and directed networks (Broman and Sen, 2009; Neto et al., 2010). Here, we extended this approach to define the effect of candidate gene expression on the focal phenotypic trait. To do so, we iteratively refit multiple QTL models for physiological phenotypes (trait Y) with gene expression phenotypes (trait X) (of each candidate gene) as an additive covariate.
We used this approach in the context of a large-scale QTL analysis of drought physiology in Arabidopsis. The experimental population consisted of recombinant inbred lines (RILs) derived from a cross between the KAS-1 and TSU-1 ecotypes. These accessions originated from environments with very different water availability and differed in their water use efficiencies (McKay et al., 2003). Furthermore, strong ecological differentiation between TSU and KAS has been documented in life history (Lovell et al., 2013) and survival in drought conditions (McKay et al., 2008). To test which traits respond to drought and the genetic loci underlying this response, we conducted a quantitative genetic analysis of 39 total phenotypic traits related to drought adaptation, 18 of which had significant multiple-QTL models. Candidate genes were defined separately for each QTL-phenotypic trait combination as those genes within the QTL confidence interval with cis-acting transcript abundance QTLs (cis-eQTLs). We then applied our covariate method to assess the effects of each candidate gene on the local QTL peak morphology. We demonstrated the utility of this approach by recovering known causal genes underlying QTLs and developed ranked lists of candidate genes for each individual QTL. Finally, we combined the candidate gene search with observed epistatic and additive effects to document how known proline metabolism genes interact with newly discovered effects of mitochondrial natural variation to regulate proline accumulation in response to drought.
RESULTS
Quantitative Genetics of Drought Physiology
Utilizing a population of 341 KAS-1 × TSU-1 RILs, we examined the effects of a progressive drought treatment on a series of traits, including transcript abundance, metabolites, physiology, growth, and performance. Our experimental soil moisture treatment reduced soil water potential to −2 MPa (Figure 1). This low moisture level approximates water deficits often experienced by both wild and crop plant species.
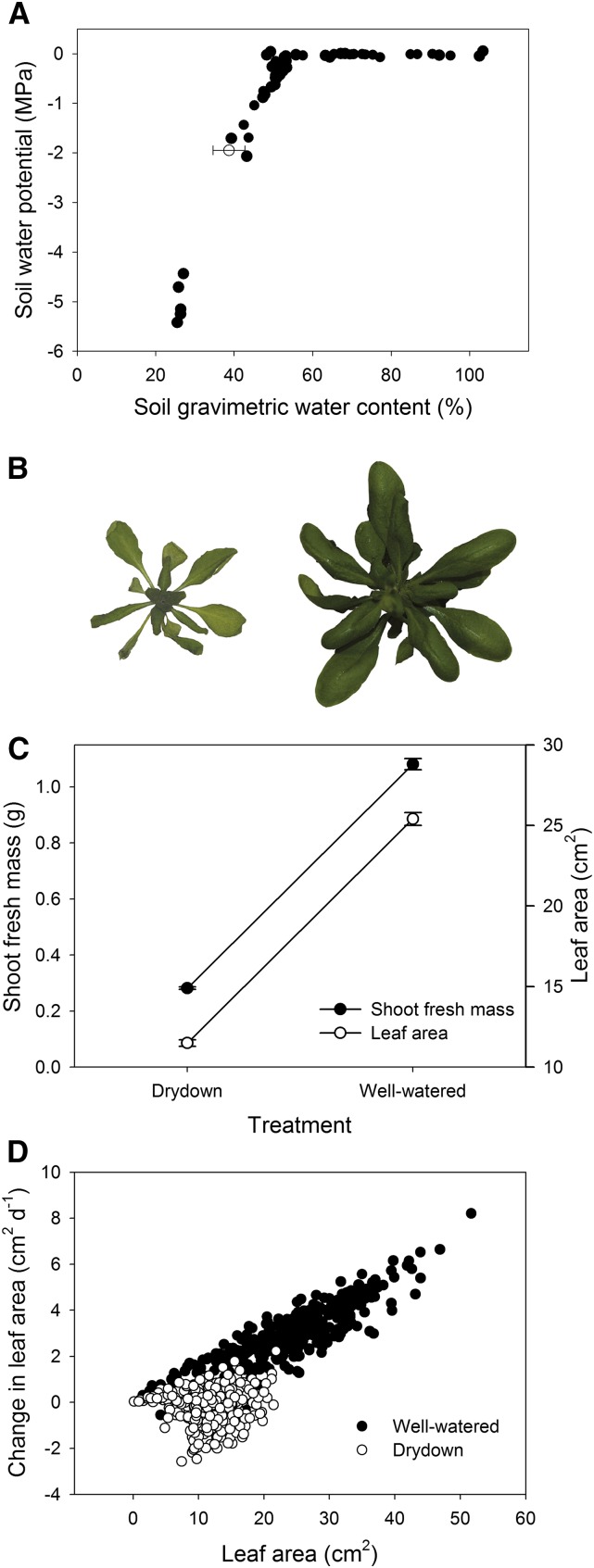
Figure 1.
Physiological Effects of the Experimental Drought on Leaf Growth.
(A) Soil moisture release curve. Closed circles represent measurements of soil water potential across a range of soil water content. The open circle shows mean soil moisture at the end of the dry-down treatment. These data were used to estimate water potential for the dry-down treatment.
(B) Isolated overhead image of a genotype (KT207) in drought (left) and well-watered (right) conditions. The images have the same scale and color adjustments.
(C) Reaction norms for shoot fresh mass under different water availability treatments.
(D) Dependence of the change in leaf area during the treatment on total leaf area. For the wet treatment, r = 0.87, and for the dry treatment, r = 0.06.
We measured 39 total phenotypic traits. Twelve phenotypic traits were measured in both well-watered and reduced water potential treatments (Table 1). These were related to growth, biomass partitioning, as well as key drought physiology traits. We also calculated and mapped QTLs for plasticity, which can also be considered a quantitative character (Falconer and Mackay, 1996). To assess plasticity for phenotypic traits measured in both environments, we calculated the RIL-specific difference between quantile normalized breeding values in wet and dry conditions. We quantified two leaf-level responses to drought stress only in the dry environment: change in leaf width (rolling) and leaf length (wilting). Finally, we examined days to flowering (FT) for each RIL, using previously reported data (Lovell et al., 2013).
Table 1. Variance Component Estimates and Summary Statistics for the Phenotypic Traits Measured.
| Phenotype (Units or Calculation) | Abbreviation | Environment | Mean | sd | VarEnv (%) | VarRILxEnv (%) | |
|---|---|---|---|---|---|---|---|
| Growth | Leaf area (cm2) | LA | Wet, dry, plast. | 18.40 | 9.160 | 66 | 1 |
| Growth rate (LAharvest/LApretreatment) | GR | Wet, dry, plast. | 5.44 | 7.660 | 72 | NA | |
| Relative GR [ln(LA)-ln(GR-LA)] | RGR | Wet, dry, plast. | 0.07 | 0.090 | 71 | NA | |
| Leaf wilting (%) | Wilt | Dry | 3.16 | 2.350 | NA | NA | |
| Leaf rolling (%) | Roll | Dry | 3.93 | 2.280 | NA | NA | |
| Biomass | Shoot fresh mass (g) | SFM | Wet, dry, plast. | 0.68 | 0.490 | 72 | 2 |
| Shoot dry mass (g) | SDM | Wet, dry, plast. | 0.06 | 0.026 | 36 | 0 | |
| Root dry mass (g) | RDM | Wet, dry, plast. | 0.01 | 0.004 | 5 | 0 | |
| Shoot:root ratio (SDM/RDM) | SR | Wet, dry, plast. | 6.02 | 1.860 | 42 | 0 | |
| Root mass ratio [RDM/(RDM+SDM)] | RMR | Wet, dry, plast. | 0.15 | 0.033 | 46 | 0 | |
| Grav. water content (SFM-SDM/SFM) | WC | Wet, dry, plast. | 87.80 | 5.540 | 83 | 2 | |
| Physiology | ABA conc. (μmol/g SDM) | ABA | Wet, dry, plast. | 5.92 | 5.960 | 1 | 0 |
| ABA (aqueous) (μmol/g SDM)* | Wet, dry, plast. | 0.72 | 1.100 | 35 | 0 | ||
| Water use efficiency (δ13C) | WUE | Wet, dry, plast. | −29.80 | 0.890 | 76 | 1 | |
| Proline conc. (μmol/g SFM) | Proline | Wet, dry, plast. | 93.60 | 102.500 | 85 | 4 | |
| Flowering time (days) | FT | Wet | 23.70 | 4.580 | NA | NA |
Genotype effects cannot be estimated for the growth rate traits because a single replicate was measured within each treatment. Environmental contributions cannot be estimated for the single environment phenotypes, wilting, rolling, and FT. Those traits that were not used for QTL mapping are indicated with an asterisk. SDM, shoot dry mass; SFM, shoot fresh mass.
As expected, drought stress had a substantial effect on most measured phenotypic traits (Table 1). Growth traits responded particularly strongly. Leaf area was reduced by ∼50% in the dry treatment, and shoot fresh mass in the dry treatment was only 25% of that in the wet treatment (Figures 1B and 1C). For several phenotypic traits, including growth rate, the degree of plasticity was strongly positively correlated with treatment-specific breeding values (Figure 1D). Physiological traits also exhibited strong stress responses. Leaf tissue concentrations of the compatible solute, proline, and carbon isotope ratio (δ13C), a measure of whole-plant, integrative water use efficiency (WUE), were 25.12-fold (t = −51.5, P < 0.001) and 1.24-fold (t = −36.4, P < 0.001) higher in dry conditions, respectively. The aqueous concentration of the drought-responsive hormone ABA was significantly higher in the dry treatment (t = 9.51, P < 0.001).
We observed significant negative genetic correlations between physiological and growth rate traits, especially in drought conditions (Table 2). The extent and direction of phenotypic plasticity to drought was also strongly correlated among growth traits (Supplemental Table 1; Figures 1C and 1D). Negative response correlations were observed between partitioning ratios (shoot:root ratio, root mass ratio), while root and shoot growth responses were positively correlated. Among physiological responses, only a strong negative correlation between WUE and proline response was significant (Supplemental Table 1), a pattern potentially driven by similar hormone signaling across these traits. Finally, the relatively weak signal of genotype*environment interactions (GxE; Table 1), except for proline concentration, indicated that physiological divergence between TSU and KAS was largely constitutive.
Table 2. Genetic Correlations among Traits.
| LA |
SFM |
SDM |
RDM |
RMR |
ABA |
WUE |
Proline |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dry | Wet | Dry | Wet | Dry | Wet | Dry | Wet | Dry | Wet | Dry | Wet | Dry | Wet | Dry | ||||
| LA | Wet | 0.25* | 0.91* | 0.19* | 0.85* | 0.18* | 0.74* | 0.10 | −0.07 | −0.02 | −0.33 | −0.07 | 0.02 | −0.05 | -0.12* | 0.11 | ||
| Dry | 0.22* | 0.89* | 0.28* | 0.59* | 0.20 | 0.55* | 0.04 | 0.11 | −0.04 | −0.04 | 0.03 | −0.24* | 0.04 | −0.04 | ||||
| SFM | Wet | 0.19* | 0.90* | 0.16* | 0.80* | 0.15 | −0.03 | 0.02 | −0.07 | −0.07 | 0.02 | −0.04 | -0.14* | 0.08 | ||||
| Dry | 0.23* | 0.69* | 0.25 | 0.68* | 0.16 | 0.20 | −0.03 | −0.05 | 0.01 | −0.29* | 0.02 | −0.11* | ||||||
| SDM | Wet | 0.23* | 0.71* | 0.08 | −0.19 | −0.15 | −0.05 | −0.07 | 0.12* | −0.04 | 0.00 | 0.14* | ||||||
| Dry | 0.38* | 0.61* | 0.18 | 0.07 | −0.06 | −0.11 | −0.02 | −0.33* | −0.02 | 0.13* | ||||||||
| RDM | Wet | 0.33* | 0.37* | 0.20 | −0.34* | 0.05 | 0.03 | −0.26* | −0.13 | 0.07 | ||||||||
| Dry | 0.30* | 0.59* | −0.16 | 0.29* | −0.05 | −0.48* | −0.03 | −0.20 | ||||||||||
| RMR | Wet | 0.37* | 0.05 | 0.13 | −0.07 | −0.31* | −0.02 | −0.17 | ||||||||||
| Dry | −0.26 | −0.15 | −0.21 | −0.35* | −0.10 | −0.24 | ||||||||||||
| ABA | Wet | 0.43* | −0.05 | 0.00 | −0.08 | 0.11 | ||||||||||||
| Dry | −0.04 | 0.02 | −0.05 | 0.08 | ||||||||||||||
| WUE | Wet | 0.21* | 0.17* | 0.04 | ||||||||||||||
| Dry | 0.02 | 0.29* | ||||||||||||||||
| Proline | Wet | −0.11 | ||||||||||||||||
Multiple-QTL Modeling
To define genomic regions associated with drought physiology, we implemented a stepwise model selection approach (Manichaikul et al., 2009) within the QTL mapping package R/qtl for all 39 traits (Broman et al., 2003). QTL mapping was conducted on the breeding values for each trait within each environment (nwet =13, ndry = 14) and the plasticity estimate for each of the 12 traits measured in both environments.
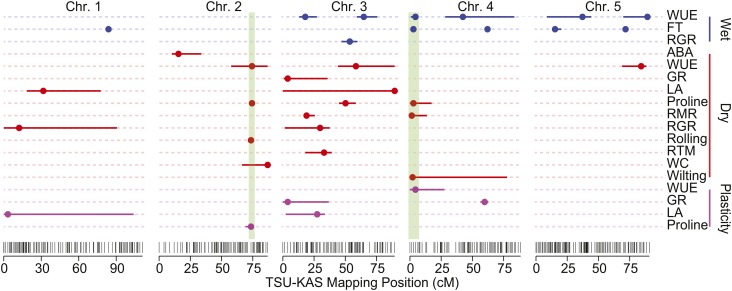
Single-trait multiple-QTL modeling revealed 36 significant QTLs across 18 of the 39 traits (Table 3, Figure 2; Supplemental Table 2). Strong QTL peaks were found for several traits on proximate chromosome 4 (Chr4) and distal Chr2. The Chr4 QTL was previously cloned as FRIGIDA for WUE (wet) and FT (Lovell et al. 2013); however, this marks the first documentation of colocalized QTLs for proline, root mass ratio (RMR), WUE plasticity, and wilting phenotypes at the same region. The QTL hot spot on Chr2 was centered on an extremely strong peak for proline (dry treatment, 24.9% variance explained) but also included single QTLs for WUE (dry), water content (dry), leaf rolling (dry), and proline plasticity. While correlations among traits with colocalized QTLs were generally weak, several pairs of traits were strongly correlated, including FT-WUE (wet) (r = 0.34, P < 0.0001; Supplemental Figure 2) and proline (dry)-WC (dry) (r = −0.35, P < 0.0001; Supplemental Figure 2B).
Table 3. Multiple-QTL Model Statistics.
| Trait | Treatment | Formula | %Var | P Value | pLOD |
|---|---|---|---|---|---|
| RGR | Wet | y ∼ cytoplasm + 3@54 | 7.754 | 0.016 | 2.418 |
| WUE | Wet | y ∼ cytoplasm + 3@18 + 3@65 + 4@4 + 4@42+ 5@37 + 5@89 | 34.301 | <0.001 | 8.621 |
| FT | Wet | y ∼ cytoplasm + 1@84 + 4@3 + 4@62+ 5@15+ 5@72 +4@3*4@62 | 70.998 | <0.001 | 63.283 |
| LA | Dry | y ∼1@32 + 3@90 | 3.974 | 0.121 | 0.493 |
| RDM | Dry | y ∼ cytoplasm + 3@33 | 14.674 | 0.281 | 0.438 |
| RMR | Dry | y ∼3@19 + 4@1 | 35.884 | 0.029 | 1.419 |
| GR | Dry | y ∼ cytoplasm + 3@4 | 1.965 | 0.360 | 0.818 |
| RGR | Dry | y ∼1@12 + 3@30 | 6.003 | 0.043 | 0.207 |
| Wilt | Dry | y ∼4@2 | 7.251 | 0.006 | 0.469 |
| Roll | Dry | y ∼ cytoplasm + 2@74 | 9.765 | 0.005 | 2.259 |
| WUE | Dry | y ∼2@74 + 3@58 + 5@84 | 6.222 | 0.088 | 1.118 |
| ABA | Dry | y ∼2@16 (+ 2@16*cytoplasm) | 4.349 | 0.034 | 1.324 |
| Proline | Dry | y ∼ cytoplasm + 2@74 + 3@50 + 4@3 + 2@74 *3@50 | 44.165 | <0.001 | 21.949 |
| WC | Dry | y ∼ cytoplasm + 2@87 | 7.731 | 0.017 | 0.130 |
| LA | Plasticity | y ∼ cytoplasm + 1@3 + 3@27 | 10.124 | 0.011 | 0.327 |
| GR | Plasticity | y ∼ cytoplasm + 3@4 + 4@60 (+ 4@60*cytoplasm) | 5.065 | 0.151 | 1.056 |
| WUE | Plasticity | y ∼ cytoplasm + 4@4 | 12.718 | 0.001 | 0.178 |
| Proline | Plasticity | y ∼ cytoplasm + 2@74 | 18.199 | <0.001 | 7.629 |
Degrees of freedom, percentage of variance explained by the model, and P values derived from χ2 tests were generated by fitting the QTL model with ANOVA. The penalized whole-model LOD score (pLOD) was derived from stepwise model selection, where models that increase pLOD are retained. Significant interactions between the cytoplasm and QTLs were indicated in the formula; statistics for the interactions are reported in Supplemental Table 4.
Figure 2.
Mapping Positions of Significant QTLs.
QTL point estimates (filled circles) and accompanying drop 1.5 (solid colored lines) confidence intervals for all phenotypes with significant multiple QTL models. Phenotypes in red were collected in the drought treatment. Blue-labeled phenotypes were from the “wet” treatment, and purple traits are plasticity estimates. The two focal regions for candidate gene method validation are highlighted in green.
Aside from the FRIGIDA region, WUE QTLs also colocalized with those for FT on distal Chr4, proximate Chr5, and distal Chr5 (Figure 2; Supplemental Figure 3). In general, allelic variation underlying each QTL peak produced a parallel response of WUE and FT, as indicated by similar QTL profiles across the genome for each trait (Supplemental Figure 3A). For three-quarters of the pleiotropic QTLs, the KAS allele conferred later FT and higher WUE (Supplemental Figure 3B). However, for the pleiotropic FT/WUE QTL on distal Chr5, the KAS allele was associated with earlier FT and lower WUE (Supplemental Figure 3B).
We found plasticity QTL for proline, WUE, leaf area, and shoot growth rate (Table 3). The QTL peaks for these traits largely aligned with the strongest QTL peaks for either the wet (WUE) or dry main-effect phenotype. In these cases, we observed “weak” interactions, where the magnitude but not the direction of the QTL effect changed with the environment.
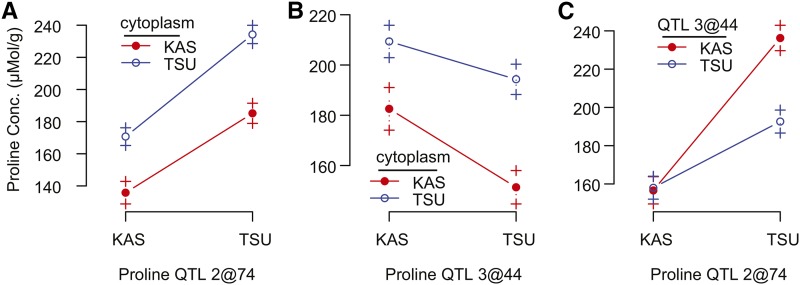
Since this population used reciprocal crosses, we were able to evaluate the effects of the cytoplasmic genomes. Adding cytoplasm as a factor improved model fit and was therefore retained in multiple QTL models for 12 of the phenotypic traits (Supplemental Table 2). In particular, there was a strong additive effect of cytoplasm on proline (10.6% of the total variation; Supplemental Tables 2 and 3; Figures 3A and 3B); however, for other phenotypic traits, cytoplasm generally explained little of the total variance (Supplemental Table 2). We tested the significance of QTL-cytoplasm epistasis post-hoc by iteratively fitting an interaction term between cytoplasm and each QTL in the final multiple-QTL model for each phenotypic trait where cytoplasm was retained in the model (Supplemental Table 4). This interaction was significant, improved model fit and was added to the model for two QTLs: plasticity of growth rate QTL 4@60 and WUE (dry) 3@58 (QTL identifier follows: phenotypic trait “QTL” Chr@cM position; Supplemental Figure 4). Interestingly, the WUE QTL 3@58 broadly colocalized with proline QTL 3@44, another QTL with a strong, albeit additive, effect of cytoplasm (Figure 3B).
Figure 3.
Interaction Plots for Proline QTLs 2@74 (Best Candidate P5CS1), 3@44 (ProDH), and the Effect of Cytoplasmic Variation.
Allelic means ± se are plotted.
QTL-QTL epistasis was found in only two phenotypic traits, proline and FT (Table 3; Supplemental Table 3). Proline was strongly increased in lines with KAS alleles at QTL 3@44, but TSU alleles at the main effect QTL 2@74 (Figure 3C). The latest FT phenotype was conferred by KAS alleles at both FT QTLs 4@3 and 4@62.
Candidate Gene Analysis
Our candidate gene discovery method used a three-step approach to determine the effect of gene expression on the peak QTL LOD score. First, all genes with significant gene expression polymorphism were extracted (for each QTL interval) by including only genes with significant cis-eQTLs (mapped in a recent study using the same gene expression data set; Lowry et al., 2013). Second, we ran QTL scans in which expression of each gene was iteratively added (and subsequently removed) as an additive covariate to the previously generated multiple-QTL model. This allowed us to determine the relative effect of transcript abundance of each gene on the focal QTL peak height. Finally, we ran a permutation test (10,000 permutations) for all genes in each QTL to determine significance of the effect (Figure 4).
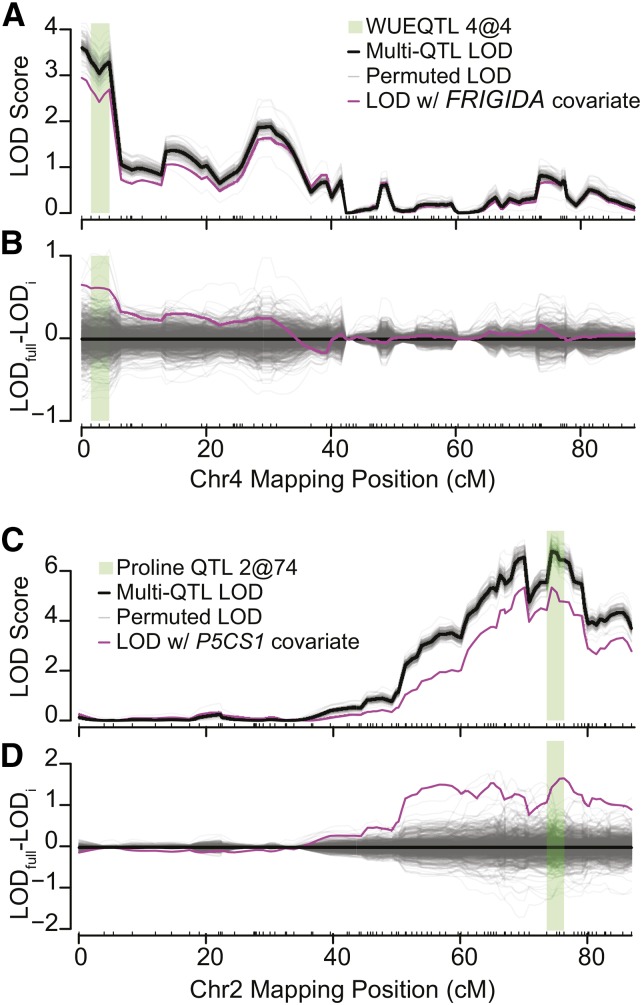
Figure 4.
Two Examples of the Candidate Gene Ranking Approach.
(A) The multiple-QTL model profile (solid black line) and the LOD profile with FRIGIDA as a covariate (solid magenta line) for WUE in the wet environment. The green highlighted region is the drop 1.5 LOD interval and matches that in Figure 2. Gray lines indicate LOD profiles for 100 permuted gene expression covariates.
(B) The difference between covariate scans (1000 permutations and FRIGIDA) and the full QTL model.
(C) and (D) The same data are displayed for the proline QTL 2@74 and P5CS1.
The WUE QTL 4@4 was previously cloned, and the phenotypic variation resulted from DNA sequence variation that caused an expression polymorphism at FRIGIDA. Within the WUE QTL 4@4 confidence interval, there were 92 genes. Our candidate selection approach returned FRIGIDA (AT4G00650) as the strongest candidate gene for WUE. We tested the significance of the estimates by permuting the gene expression covariate data and rerunning the QTL scans. This test resulted in an empirical P = 0.0057 for FRIGIDA (Figures 4A and 4B). For the pleiotropic FT QTL 4@3, FRIGIDA was the second strongest candidate. While not the strongest in the list, FRIGIDA had a significant effect on the FT QTL 4@3 LOD score (P = 0.01).
The proline QTL 2@74 contained 239 genes, many more than the WUE QTL 4@4. This QTL colocalized with a well-documented sequence polymorphism in the proline biosynthesis gene, P5CS1 (AT2G39800), described by Kesari et al. (2012). We ran the same covariate screening process with the proline QTL 2@74, with the additive effect of cytoplasm included in the model. Incorporation of cytoplasm effects into candidate selection resulted in three genes with significant gene expression covariate effects, including P5CS1 (Figures 4C and 4D, Table 4; Supplemental Table 5; P > 0.001). These results provided a proof of concept that our method could discover causal genes in moderately large QTL regions.
Table 4. List of the Top 10 Significant Candidate Genes for Each QTL Ranked by LOD Effect.
| Phenotype | QTL | %Var | Candidate Genes (Ordered by P Value) |
|---|---|---|---|
| LA.plast | 1@3 | (−) 3.13 | |
| RGR.dry | 1@12 | (−) 3.30 | |
| LA.dry | 1@32 | (−) 3.48 | |
| FT.wet | 1@84 | (+) 3.95 | AT1G65490, FAS1 |
| ABA.dry | 2@16 | (−) 5.16 | AT2G03140, AT2G04380**, ALDH6B2, AT2G04170*, UGT73B5, AT2G05915 |
| Roll.dry | 2@74 | (−) 6.23 | ACX5*, AT2G36460, CTF2A, GLTP1* |
| Proline.dry | 2@74 | (+) 24.8 | AT2G38800*, P5CS1, PROT1* |
| Proline_plast | 2@74 | (+) 13.6 | |
| WUE.dry | 2@74 | (+) 3.6 | |
| WC.dry | 2@87 | (−) 3.34 | MAC3B, ATTI3, AT2G43210, AT2G46220, IQD14, AT2G46100**, AT2G38800, AT2G32150, AT2G42490, AT2G32160 |
| GR.dry | 3@4 | (−) 4.56 | |
| GR_plast | 3@4 | (−) 4.38 | |
| WUE.wet | 3@18 | (+) 3.12 | CBSDUFCH1, RPL18AC*, AT3G18530, AT3G14595 |
| RMR.dry | 3@19 | (+) 19.7 | UGT88A1, AT3G14360**, AT3G18535, AT3G18530, CYP77A5P |
| LA.plast | 3@27 | (−) 4.18 | |
| RGR.dry | 3@30 | (−) 3.81 | |
| RDM.dry | 3@33 | (+) 19.2 | AT3G16750, AT3G25240, BRT1 (UGT84A2) |
| Proline.dry | 3@50 | (−) 4.91 | AT3G43230, AT3G28080, AT3G27250, ATCSLC04, AT3G30300*, AT3G26670*, PRODH, CAF1-9*, ATMYB30, |
| RGR.wet | 3@54 | (−) 6.69 | AT3G44430, emb2076, AT3G43430, AT3G43670, ATIVD (IVD), ATMLO3 (MLO3)*, AT3G45555 |
| WUE.dry | 3@58 | (−) 3.76 | |
| WUE.wet | 3@65 | (−) 5.14 | HR4, ABC2 Homolog 1 (ATATH1)*, scpl48, ALDH2B4**, AT3G47580, IVD, AT3G53730*, ATEXLA1, AT3G51470**, CSR1 |
| LA.dry | 3@90 | (+) 3.30 | |
| RMR.dry | 4@1 | (+) 15.9 | ATSTE24, SAM-2* |
| Wilt.dry | 4@2 | (−) 4.01 | |
| FT.wet | 4@3 | (−) 59.1 | AT4G00740, FRI, ECA2 |
| Proline.dry | 4@3 | (−) 3.46 | AT4G01130, SAM-2*, ATSTE24, AT4G00270, AT4G02540, MLO1 |
| WUE.wet | 4@4 | (−) 7.09 | FRI |
| WUE.plast | 4@4 | (+) 3.47 | |
| WUE.wet | 4@42 | (−) 2.59 | |
| GR.plast | 4@60 | (−) 4.82 | CRK22, CRK21, AT4G22990, AT4G24050, ISU1, AT4G21910, ATSBT3.12, TOM1* |
| FT.wet | 4@62 | (−) 5.19 | CRK22, CRK23, AT4G24340 |
| FT.wet | 5@15 | (−) 1.83 | CSA1, CHS3, AT5G18950, AT5G16890, AT5G17680, PAT1(TRP1), GDH1**, ATCBR* |
| WUE.wet | 5@37 | (−) 3.49 | |
| FT.wet | 5@72 | (+) 4.24 | ATATG18F, AT5G55180, AT5G54710, AT5G54720, PORA*, ATM2 |
| WUE.dry | 5@84 | (+) 3.53 | XYL4**, AGL62, AT5G61660, ABA1, LECRK110, AT5G62350, DAR5, AT5G55180, KCA2 |
| WUE.wet | 5@89 | (+) 6.33 | AT5G54710, AT5G55180, AT5G54720*, ARF2*, ATATG18F, AT5G63020, AT5G53700, SNRK2-3, AT5G60160**, AT5G61660 |
The QTL-specific percentage of variance explained is presented and is preceded by the direction of the QTL effect. Positive values indicate a higher mean of the TSU allele. Candidate genes for QTL with confidence intervals that spanned >25 cM are not listed. Bold font indicates those genes discussed in the text. Genes without DNA sequence or with neither DNA sequence nor protein divergence between TSU and KAS alleles are marked by one asterisk or two asterisks, respectively.
To explore the utility of this method, we conducted covariate candidate gene scans for all QTLs with intervals spanning <25 centimorgans (cM), including 20 main effect QTLs and two epistatic QTL regions. For the two epistatic QTLs, with significant interactive effects but small main effects (FT QTL 4@63 and proline QTL 3@50), gene expression polymorphism was defined by the interactive effect between gene expression at the local and interacting QTLs. We then ranked these lists of potential candidates by the relative proportion of the LOD score absorbed by the gene expression covariate (Table 4; Supplemental Table 5). Overall, we screened 652 genes, 169 of which had an empirical q-transformed P < 0.1. Mean gene expression was highly elevated across significant candidate genes relative to those without significant effects (t527.5 = 9.87, P < 0.0001).
Several of the candidate genes produced from our methods had particularly interesting gene annotations. The strongest candidate for the FT QTL5@15 was AT5G17880 (CSA1), which has been found to cause variation in stem elongation in response to a variety of environmental cues, including shading and red/far red ratio (Faigón-Soverna et al., 2006). Furthermore, transgenic lines of CSA1 have slight differences in FT. Indeed, we found that TSUxKAS NILs with introgressions in this region showed variation in shade avoidance traits (F3,25 = 6.031, P = 0.0031; Supplemental Figure 5). In addition, one of the significant candidates for proline QTL 3@50 was AT3G30775 (ProDH1). ProDH1 is a mitochondrion-localized proline oxidase that is an especially strong candidate because of the highly significant effect of both P5CS1 and the cytoplasm at proline QTL 3@50 (Figures 3B and 3C).
While all significant candidates warranted further consideration, we chose to examine two QTLs with candidates that were not annotated for the focal phenotype. The most significant gene for ABA QTL 2@16 was AT2G03140 (P < 0.0001; Table 4; Supplemental Table 5), a chloroplast-localized protease. Interestingly, the second strongest candidate gene AT2G04380 (Supplemental Table 5) had a gene expression phenotype that strongly covaried with AT2G03140 (Supplemental Figure 6), indicating the possibility of multiple strong candidates within this region. While there was substantial divergence between proteins encoded by the parental alleles of AT2G03140 (0.5%; Supplemental Table 5), both coding and untranslated region DNA sequences of AT2G04380 were monomorphic between TSU-1 and KAS-1. Combined, the stronger covariate effect and greater protein and sequence polymorphism made AT2G03140 a more likely candidate.
Distal Chr5 contained a pleiotropic QTL for FT, WUE (wet), and WUE (dry). The effects of this QTL on WUE in both conditions were parallel, indicating that a single constitutive locus may have caused the QTL. Of the 24 unique significant candidates among these three traits, one gene was found in all three: AT5G55180 encoding a plasma membrane-localized O-glycosyl hydrolase that had not been annotated for any physiological attributes. However, members of the glycosyl hydrolase family of proteins are involved in a diverse array of physiological functions in plants, including drought responses (Bray, 2004), signaling, and development (Minic, 2008), which presents the possibility of pleiotropic gene action across phenological and physiological traits.
Isolation of the Quantitative Trait Nucleotides in Cytoplasmic Genomes
Since the multiple-QTL model for proline (dry) was strongly influenced by an additive effect of cytoplasm (>10% variance explained), we explored DNA sequence variation in the cytoplasmic genomes. We downloaded high-depth resequencing of both parental lines (Lowry et al., 2013). Sequence comparisons revealed that the chloroplast genomes of TSU and KAS had identical DNA sequences. However, 16 single-nucleotide polymorphisms (SNPs) existed between the mitochondrial genomes. Of these SNPs, four were in gene-coding regions of ATMG00050 (unknown function), ATMG00070 (NADH DEHYDROGENASE SUBUNIT 9 [NAD-9]), ATMG00510 (NAD-7), and ATMG00710 (unknown function) (Supplemental Table 6). Interestingly, across a diverse panel of Arabidopsis accessions (218 genotypes from the Salk Institutes 1001 genome resequencing project, http://signal.salk.edu/atg1001/index.php), NAD-9 has two major haplotypes (I, 40%; II, 43%) that exist at relatively equal frequencies, a departure from neutral expectations that instead suggests balancing selection at the locus (Supplemental Figure 7). The other three candidate genes have single dominant haplotypes with a small proportion of independent, rare mutations (Supplemental Figure 7).
We verified the presence of the NAD-9 SNP through capillary sequencing. The TSU-1 (a high proline accumulator) NAD-9 allele differed from the allele common to KAS-1 (a low proline accumulator) and the Columbia-0 reference. This result was consistent with an effect of the KAS cytoplasm genotype in the RIL population (Figures 3A and 3B). While SNPs in introns and untranslated regions were found within several other mitochondrial genes, the NAD-9 SNP is a missense mutation that caused valine to be substituted for phenylalanine in the 19th codon.
DISCUSSION
Drought adaptation involves multivariate and often correlated evolution of physiological, developmental, and life history phenotypes. Underlying these physiological responses are diverse patterns of sequence and gene expression variation. Comparisons of gene expression and physiological traits have revealed a complex genetic basis of drought responses (Liu et al., 1998; Shinozaki et al., 2003; Des Marais et al., 2012). By exploiting the causal connections between environmental variation and the genotype-phenotype map, here, we present candidate genes for constitutive and plastic responses to soil moisture reduction in the context of QTL mapping.
Genetic Architecture of Drought-Responsive Traits
Our soil moisture reduction treatment imposed drought stress on the TSU-KAS RIL mapping population (Figures 1A and 1B) and induced physiological responses across many phenotypic traits (Table 1). For example, all lines increased proline concentration in drought conditions. Increased proline may contribute to osmotic adjustment and cellular redox balance (Szabados and Savouré, 2010; Verslues and Sharma, 2010), traits that may confer improved cellular dehydration tolerance.
Even though our experimental design was able to detect loci that explained as little as 1.8% of the total phenotypic variance (Table 3), for the majority of constitutive traits and plasticity estimates, we found no QTLs. The low number and small-effect sizes of QTLs determined in our analysis were indicative of a genomic architecture of drought-responsive traits that was decisively polygenic. While loci of small effect were the most common observation, there were several genomic regions that explained a very large proportion of phenotypic variance. In particular, the FT QTL 4@3 (FRIGIDA), RMR QTL 3@18, and proline QTL 2@74 (P5CS1) explained 59, 19.7, and 25% of the total variation, respectively (Table 3). In concert with observed (and unobserved) small-effect loci, the presence of these large-effect loci, and several moderate effect-size QTLs (e.g., RMR QTL4@1 and root mass QTL 3@33) provided evidence for an exponential distribution of allelic effects on potentially adaptive traits (Orr, 1998). This pattern has been observed in other recent physiological QTL mapping studies (Ågren et al., 2013; Joseph et al., 2013a).
Genetic Correlations Underlie Pleiotropic QTLs
Correlations among drought acclimation responses can directly affect the fitness (yield) of genotypes when challenged with low soil water potentials. For example, mild early season drought may simultaneously select for cellular dehydration avoidance through stomatal closure (Heschel et al., 2002) and reduced growth rates (Schmalenbach et al., 2014). Alternatively, strong late season drought may select for drought escape through early flowering and fast growth while soil water conditions are favorable (Meyre et al., 2001; Heschel and Riginos, 2005; Sherrard and Maherali, 2006). Genetic correlations among many of these traits have been observed both within (McKay et al., 2003; Lovell et al., 2013) and among (Angert et al., 2009) species, providing further evidence that selection acts on both the plasticity of and correlations among drought-associated phenotypes (Endler, 1986).
In our population, a “dehydration avoidance” drought adaptive strategy was conferred by increased water use efficiency, decreased growth rate, increased water foraging through root growth, and delayed flowering. Dehydration-avoidant phenotypic values and QTL effects were typical of KAS alleles, while TSU alleles conferred a drought escape strategy. Habitats with consistently limited precipitation throughout the growing season, such as Northwest India where the KAS genotype originated (McKay et al., 2008), may favor a dehydration-avoidant strategy (Heschel et al., 2002; Blum, 2005; Schmalenbach et al., 2014). By contrast, TSU originates from Southwestern Japan, where more mesic growing season conditions exist. In annual plants such as Arabidopsis, greater moisture availability usually favors rapid cycling and drought escape strategies (Sherrard and Maherali, 2006; Wilczek et al., 2009; Banta et al., 2012), an observation consistent with the effects of the TSU allele for many QTLs (Supplemental Table 2).
We found strong evidence of a genetic basis for correlations that conferred drought-adaptive syndromes, especially between WUE and phenology. QTLs for FT and WUE colocalized for 4/7 of the unique significant loci across the traits. Additionally at each locus, we found evidence for parallel responses where allelic effects for each trait produced phenotypic vectors of similar, positive orientation (Supplemental Figure 3). Even where the sign of the effects reversed (e.g., QTL 5@70), the correlation conferred by alleles at this QTL remained positive. This effect, which antagonized the degree of physiological differentiation between TSU and KAS, increased the strength of the drought adaptive trade-off between dehydration avoidance and drought escape in the RIL population. Finally, the genome-wide correlation between FT and WUE QTL locations suggests that pleiotropy (at the QTL level) between these traits is not unique to the FRIGIDA locus (Lovell et al., 2013).
Candidate Genes for Drought Adaptation
One of the goals of genetic mapping is to discover the regions, interactions, and ultimately the genes that underlie physiological variation. Many approaches permit inference of potential candidate genes underlying QTLs, including CIT (Schadt et al., 2005), differential expression analyses (Drake et al., 2006; Farber et al., 2009), and partial regressions (Bing and Hoeschele, 2005); however, it is difficult to rank or infer the effect of each without significant additional data, such as reverse genetics experiments. By combining gene expression data with genetic mapping approaches, we presented a method to define and rank sets of candidate genes for any QTL.
We tested the effects of 652 genes across all QTLs with intervals narrower than 25 cM. Transcript abundance of 169 of these genes significantly (Q-transformed P10000permutations < 0.1) affected the LOD score of the overlying QTL. Interestingly, these 169 represent a highly expressed subset of the total 25,662 genes with expression data.
We were able to recover the two a priori candidates for the main FT and proline QTLs, FRIGIDA and P5CS1, respectively. FRIGIDA is a vernalization-responsive transcription factor that affects flowering and pleiotropically drives variation in WUE (Lovell et al., 2013). In our RIL population, lines with the low-expression TSU alleles flowered earlier and had lower WUE, a drought escape life history strategy. Alternatively, KAS alleles were associated with dehydration avoidance physiology through increased WUE and FT. These effects were mediated by stomatal conductance and other upstream physiological traits (Lovell et al., 2013). P5CS1 catalyzes the rate-limiting step in proline biosynthesis. The induction of P5CS1 gene expression and subsequent increased P5CS1 protein abundance is required for high levels of proline accumulation (Kesari et al., 2012). Capillary sequencing revealed a functional polymorphism at P5CS1, where intronic sequence variation yielded a reduced function allele in KAS, which was nearly identical to the reduced function allele of P5CS1 previously described for the ecotype “Sha” (Kesari et al., 2012).
Aside from FRIGIDA and P5CS1, we were able to define candidate genes for all other narrow QTLs. Several of these were annotated to have similar effects as those shown in our physiological assays. For example, we found CSA1 (CONSTITUTIVE SHADE AVOIDANCE1) as a candidate for FT QTL 5@15. Furthermore, the much wider, but colocalized WUE QTL 5@37 also had CSA1 as a strong candidate (J.T. Lovell, unpublished data). CSA1 responds directly to shade and red/far red light ratios (Supplemental Figure 5), altering life history and vegetative growth structure (Faigón-Soverna et al., 2006). There is significant physiological crosstalk between shade avoidance, drought physiology, and phenology (Maliakal et al., 1999; Schmitt et al., 2003), raising the possibility that CSA1 pleiotropically affects phenological and physiological traits in the TSUxKAS population, a hypothesis that requires further testing.
Several studies have found that single QTL peaks can fractionate into multiple linked peaks, each caused by separate, but linked polymorphisms (Studer and Doebley, 2011; Johnson et al., 2012). To examine this possibility, we screened for QTLs with multiple strong candidate genes with correlated expression patterns. Several genes within the ABA 2@15 QTL returned strong covariate effects. Of these, two genes displayed gene expression phenotypes that were highly correlated, AT2G03140 and AT2G04380. To determine the relative strength of each candidate, we analyzed DNA sequence and protein polymorphism within the genes and compared TAIR-10 gene annotations. Only AT2G03140 had any sequence polymorphism between the TSU and KAS parental accessions (Supplemental Table 6). While gene function can easily be affected by polymorphism outside of the coding region, genes that are conserved between the widely divergent genomes of TSU and KAS may be less likely to contribute to quantitative genetic divergence than those with sequence, and especially protein, divergence (Lowry et al., 2013).
It is important to note that this approach does not definitively document functional effects of loci. Instead, genes such as AT2G03140, which lack functional annotation for drought response or ABA biosynthesis, necessitate further functional genetic analyses. With that said, AT2G03140 is a highly interesting candidate gene as it encodes a putative chloroplast-localized protein with similarity to CAAX amino terminal proteases involved in membrane anchoring of proteins (Choy et al., 1999). As the early steps of ABA biosynthesis occur within the chloroplast (Endo et al., 2008; Cutler et al., 2010; Lee et al., 2013) and involve lipid-soluble carotenoids and membrane-associated enzymes (Milborrow, 2001; Seo and Koshiba, 2002), the connection of AT2G03140 to ABA metabolism is plausible and highly promising for further analysis.
Documentation of Genetic Networks through QTL Mapping
Our candidate gene discovery method made use of the details of genetic architecture, through incorporation of epistasis, additive effects of cytoplasm (and other covariates of interest), and environmental interactions. For example, this approach validated the effects of P5CS1 and permitted inference of potential candidate quantitative trait nucleotides in the cytoplasmic genome. However, as with any other candidate gene selection approach, our method provides hypotheses and does not document functional variation of candidate genes.
Our analysis of the parental genomes (sequences were published in Lowry et al., 2013) revealed that the chloroplast genomes of TSU and KAS were identical and only 16 SNPs existed in the mitochondrial genome. These results differed from published TSU-1 (SRX246442) and KAS-1 (SRX246466) sequences (http://www.ncbi.nlm.nih.gov/sra/; Joseph et al., 2013b), where many more cytoplasm genomic SNPs were documented. However, despite having the same name, the KAS-1 and TSU-1 on the short read archive are not closely related to the KAS-1 and TSU-1 genomes that represent the parents of our mapping population. For example, over half of the published sequenome SNPs that are polymorphic in our mapping population are monomorphic between NCBI TSU-1 and KAS-1 (http://naturalvariation.org/hapmap). It is important to note that, while well suited for the characterization of SNPs, the short read sequencing and reference-based alignment used by Lowry et al. (2013) to sequence the mapping parents TSU and KAS may be unable to detect genomic rearrangements. As large-scale rearrangements are characteristic of the Arabidopsis mitochondrial genome (Davila et al., 2011), it is possible that these sequence variants are augmented by other undetected polymorphisms.
Proline concentration was strongly affected by cytoplasmic variation in our mapping population, indicating that sequence variation in the cytoplasmic genomes affected quantitative variation of adaptive traits. While the plastid genomes were monomorphic, there were 16 mitochondrial SNPs between the TSU and KAS parents of our mapping population, four of which were genic, including two in genes encoding NADH dehydrogenase subunits (NAD-7 and NAD-9). The observation that two of the mitochondrial polymorphisms were in genes for NADH dehydrogenase subunits is consistent with proposals that proline accumulation is tightly related to cellular redox status and that proline catabolism in the mitochondria is important in drought resistance (Sharma et al., 2011). For example, Szabados and Savouré (2010) and Verslues and Sharma (2010) found that proline metabolism is connected to oxidization/reduction status, and Sharma et al. (2011) showed that mitochondrial catabolism of proline is required to maintain growth under low water potential. We observed that p5cs1-4, which is blocked in stress-induced proline accumulation, was associated with strongly upregulated expression of a number of genes for NAD(P)H-dehydrogenases as well as additional genes related to mitochondrial respiration (P.E. Verslues, unpublished data).
To determine which of the NAD genes was the most likely candidate, we conducted capillary sequencing and downloaded sequence data from 218 natural accessions. Interestingly, while the SNP within NAD-7 was within an intron, the NAD-9 SNP was a missense mutation. Furthermore, there was evidence for historical balancing selection at NAD-9, but not NAD-7. While neutral evolution should yield many low-frequency haplotypes, and directional selection would reduce the number of variants, historical balancing selection should yield multiple haplotypes at elevated frequencies without a single dominant haplotype (reviewed in Nielsen, 2005). While three of the four candidate genes in the mitochondrial genome had a single dominant haplotype, the two main haplotypes of NAD-9 were maintained at >40% across a sample of 218 accessions. These results are consistent with the findings of Joseph et al. (2013b), who demonstrated that the mitochondrial genes for the NADH dehydrogenase complex harbor many more sequence polymorphisms than expected by neutral evolution. Given the nonsynonymous nature of the SNP, and evidence of historical balancing selection, it is possible that variation in NAD-9 affects proline catabolism in the mitochondria, a process that has consequences for redox balance and growth during drought.
To assay the effects of epistasis, we incorporated gene expression patterns of interacting loci (or covariates) into the candidate selection approach. Since many epistatic loci lacked strong additive effects, we fit a model where the expression of each gene underlying the epistatic QTL was a function of the local genotype and gene expression of the interacting QTL. Candidate determination of the proline epistatic locus QTL3@44 (with P5CS1) revealed a strong candidate: AT3G30775, ProDH1. These loci and the cytoplasm combined to affect proline levels both additively and interactively, where the TSU allele at P5CS1 and the KAS ProDH1 maximized the accumulation of proline in the TSU cytoplasmic background (Figure 3). This analysis provided evidence that proline metabolism is influenced by mitochondrial genes and that natural allelic variants in the mitochondria could have an evolutionarily significant effect on proline accumulation in terms of drought adaptation.
Finally, it is important to note that the genotype-phenotype cascade operates in many fashions. Our approach can discover only a distinct subset where transcript abundance is the causal phenotype underlying physiological traits. In particular, we expect our approach to be limited by the strength of correlations between physiological and gene expression phenotypes. For example, if genes are expressed at a similar rate, but alternative splicing or RNA sequence polymorphism causes protein and trait variation, we expect to have little power to detect signals of connections between candidate genes and QTLs. However, sequence-based gene expression quantification methods, such as RNA-seq, provide additional information that may improve the extensibility of our approach. For example, with information on alternative splicing, methylation patterns, and protein structure in hand, it would be possible to calculate a measure of gene functionality. As such, functionality, and not simply transcript abundance, could be used as a covariate in our method. While the data presented here come from microarray technology and do not permit such inference, we expect a combination of sequence and expression data to bolster our candidate gene approach in future analyses.
METHODS
Plant Materials and Growth Conditions
Seed of 341 RILs from reciprocal crosses between Arabidopsis thaliana accessions KAS (Kas-1; CS903) and TSU (Tsu-1; CS1640), along with the parents were sown on fritted clay (Profile Products) in 2.5-inch pots in duplicate in each of two blocks. The Tsu-1 x Kas-1 mapping population is publically available through the ABRC (ID: CS97026). Seeds were planted in a randomized complete block design, and then the pots were refrigerated at 4°C in darkness for 6 d to cold-stratify the seeds prior to commencement of a 12-h photoperiod in two Conviron ATC60 growth chambers (Controlled Environments), at 23°C and 40% humidity during the day and 18°C and 50% humidity during the dark period. Light intensity was ∼330 μmol m−2 s−1. After 4 weeks of growth, half of the plants were given a drought treatment, while the others remained fully watered. Two replicates of each RIL were randomly assigned to each treatment.
The drought treatment consisted of a slow decrease in soil moisture content over the course of 1 week. The treatment was imposed at the level of the flat (tray of 32 plants) and randomized within each chamber. Each day, all pots assigned to the drought treatment were weighed, and water was added to individual pots to bring them up to the target gravimetric water content. The target water content decreased each day, in the following series: 100, 90, 80, 70, 60, 45, and 40% of saturation. We had previously calculated the soil moisture release curve for fritted clay: 40% soil moisture content relates to approximately −2 MPa soil water potential (Figure 1A).
Phenotypic Analyses
At the end of the drought treatment, photographs were taken of each plant, and the shoots were excised at the hypocotyls and weighed to obtain shoot fresh mass. The shoots were then freeze-dried and their dry mass was measured. In a subset of 240 plants, root tissue was collected by rinsing away the fritted clay. Root tissue was then freeze-dried for dry mass determination.
Photographs of the plants were taken and used to calculate leaf area by summing pixels comprising the rosette image using the image processing software Scion Image (Scion). For half of the plants, a photograph was also taken prior to the onset of the dry-down treatment, so that we could calculate growth in leaf area during the treatment and relative growth rate.
ABA was assayed with a Phytodetek enzyme-linked immunosorbent assay kit from Agdia. Samples were prepared and measured according to the protocol from Agdia. Each sample and eight standards were run in duplicate on 32-well Phytodetek plates. A BioTek PowerWave HT spectrophotometer was used to quantify the absorbances (at 450 nm), which were fit to the standards of each plate using a logistic equation. To reduce residual variance caused by fresh mass variance among and within treatments, we performed all additional analyses on ABA concentrations standardized by the dry mass of the rosette.
Leaf tissue from each plant was crushed and lyophilized to quantify δ13C using a dual-inlet mass spectrometer at the Stable Isotope facility at University of California, Davis. Proline concentration was assayed by an acid ninhydrin assay adapted to 96-well plates (Bates et al., 1973; Verslues, 2010).
In addition to traits measured in both environments, we collected several environment-specific phenotypes. Flowering time for each line was measured in a separate experiment (Lovell et al., 2013) and reanalyzed here to make comparisons with all other phenotypes. Plants in the drought treatment were photographed both at the onset and the conclusion of the drought treatment. Fully expanded leaf characteristics were compared between the two time-points to determine the degree of rolling (Δwidth) and wilting (Δlength).
Quantitative Genetic Analyses
The phenotypic data set was analyzed with a linear mixed model, with RIL as a fixed effect and treatment, and the genotype-treatment interaction as random effects. These models and variance component estimates were calculated using PROC MIXED in the SAS software package (SAS Institute). Least square means of trait values were estimated for each RIL, and genetic correlations among traits were calculated as the standard Pearson pairwise correlations. Phenotypes were in general very normal; however, FT and ABA were both marginally skewed. Quantile normalizations of these traits did not strongly affect our QTL analysis (for detailed comparisons, see Supplemental Methods), so we opted to map the raw breeding values (Supplemental Data Set 1A).
A linkage map for this population was described previously (McKay et al., 2008). To this map we added eight additional simple sequence length polymorphism markers and an additional 276 single nucleotide polymorphism markers, based on Sequenom technologies (Supplemental Data Set 1B; Gabriel et al., 2009). The linkage map was reestimated using JoinMap4 (Van Ooijen, 2006) with the Kosambi mapping function, for a total of 450 markers.
Genotype probabilities were calculated for each locus and a set of pseudomarkers were placed in any region with a >1 cM gap in the map. The kosambi algorithm with an error probability of 0.01 was used to infer genotype probabilities. QTL mapping was performed using the Haley-Knott regression algorithm implemented in the R/qtl package within the R statistical computing environment (Broman et al., 2003; Broman and Sen, 2009). We developed multiple-QTL models via a penalized stepwise model selection approach (Manichaikul et al., 2009) where terms were included at α = 0.05. Significance was determined by 10,000 permutations. To test for QTL-by-environment interactions, we conducted stepwise model selection on plasticity of all traits that were measured in both wet and dry treatments. Plasticity breeding values were calculated as the difference between quantile-normalized wet and dry breeding values for each RIL (e.g., Figure 1C).
To achieve increased accuracy in our estimates of QTL peak means and breadth, we calculated confidence intervals (1.5 LOD drop) for each QTL point estimate separately by varying the position of the focal QTL while controlling for all other terms in the model. We also conducted multiple-QTL modeling using cytoplasm as an additive covariate and conducted post-hoc tests for cytoplasm-QTL interactions for all QTLs in those models with evidence for additive effects of cytoplasm.
It is important to note that we defined the significance of all covariate affects (e.g., gene expression and cytoplasm-QTL interactions) post-hoc. That is, the base multiple-QTL model was defined following Manichaikul et al. (2009). The QTL positions and interactions from this model were fixed for all other analyses. To determine significance of additional effects, we added and then removed a single term. Significance was determined by comparing the fit of the original and more complex QTL models.
All scripts and pipelines to conduct the QTL and covariate analyses have been posted on github: https://github.com/jtlovell/r-QTL_functions.
Candidate Gene Analysis
We downloaded gene expression and DNA sequence data from Lowry et al. (2013), who used Affymetrix atSNPTILE1.0 arrays (Zhang et al., 2007) to map eQTLs for the TSUxKAS population from RNA extracted from the experiment presented here. Gene expression data was available for a 104 RIL subset of our mapping population. To identify candidate genes, we conducted a three-step protocol that combined this gene expression data with QTL mapping results and covariates.
The first step of our candidate gene identification approach was to define a list of candidate genes that had significant gene expression polymorphism for each phenotypic trait QTL interval. For each QTL confidence interval for each trait, we extracted all genes that fell between the maximum and minimum physical positions of all markers within the interval. This approach was necessary because there are many small rearrangements throughout the population relative to the Columbia reference genome. Additionally, there is a single large inversion on Chr4 (Supplemental Figure 8). If the QTL interval was so narrow that it only included a single marker, the interval was expanded to the nearest bounding markers. For the majority of QTLs that had simple additive effects, we defined gene expression polymorphism as those genes with cis-eQTLs (Lowry et al., 2013). For other QTLs with strong additive effects of cytoplasm, differentially expressed genes were further culled to only those with significantly different expression between cytoplasmic backgrounds using fixed effects ANOVA. Finally, for QTLs with primarily epistatic effects, candidates were determined as those that have gene expression polymorphism that is significantly associated with gene expression of the primary candidate at the interacting QTL. In the latter two cases, significance was assessed with q-value estimation in the R package “q-value” (Dabney and Storey, 2014).
The second step of our analysis was to rank the candidate genes by their effect on the local phenotypic trait QTL. To accomplish this, we extracted RIL-specific gene expression values for all candidate genes. These expression values were iteratively added to the original QTL model (which may include several QTLs, cytoplasmic covariates, and epistasis) as a single additive term, or in the case of epistasis, as an interactive covariate with the epistatic QTL. To make comparisons with identical patterns of missing data, the entire genotype, gene expression, and phenotype matrices were culled so there was no missing data. This reduced the peak LOD scores for all QTLs, but made comparisons among models possible. Gene expression covariates that explained residual variance had higher LOD scores (negative difference) and those that were correlated with the phenotypic trait breeding values decreased the LOD score of the focal peak (Supplemental Figure 1). Therefore, we took the difference between LOD scores at the QTL point estimate in the original multiple-QTL model lacking the expression covariate and the new model with a gene-expression covariate as the estimated effect of that gene. We then obtained a ranked list for each QTL, where the strongest candidate gene had the most positive covariate effect.
The last step was to determine the significance of a subset of genes with the strongest covariate effect. To accomplish this, we permuted the gene expression data and reran the covariate scan 10,000 times and reported the LOD difference at the QTL point estimate. The number of permuted observations with a greater difference than the empirical data/nperm was used as our empirical P value.
Sequencing of NAD-9
PCR amplicons of ATMG00070 (NAD-9) from Col, KAS-1, and TSU-1 were sequenced using BigDye Terminator v3.1 sequencing chemistry at the Colorado State University Proteomics and Metabolomics Facility on an ABI 3130XL genetic analyzer, using the forward primer 5′-TCTGACAAGGCGGCTATCTT-3′ and the reverse primer 5′-CGAGTCGTCTAGGGCATCTC-3′.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Locus:2127013, AT4G00650, FRIGIDA; Locus:504954491, ATMG00070, NAD9; Locus:2063907, AT2G39800, P5CS1; Locus:2089706, AT3G30775, PRODH1; Locus:2161710, AT5G55180, O-GLYCOSYL HYDROLASE FAMILY 17 PROTEIN; Locus:2170333, AT5G17880, CSA1; and Locus:2056891, AT2G03140, α/β-HYDROLASE SUPERFAMILY PROTEIN.
Supplemental Data
Supplemental Figure 1. Visualization of the concept of the covariate scan approach.
Supplemental Figure 2. Correlation of phenotypes with colocalized QTL on proximate Chr4 and distal Chr2.
Supplemental Figure 3. Effect of allelic variation on the correlation between WUE and FT.
Supplemental Figure 4. Cytoplasmic interactions with genomic QTLs.
Supplemental Figure 5. Validation of the allelic effect of CSA1 using NILs.
Supplemental Figure 6. Hierarchical clustering of the covariance of all genes within each narrow QTL interval.
Supplemental Figure 7. Haplotype diversity of the four genes that contained SNPs in the mitochondrial genome.
Supplemental Figure 8. Comparison of the physical position (bp) for all TAIR10 gene models with the mapping position in cM.
Supplemental Table 1. Phenotypic correlations between plasticity and mean breeding values for all measured phenotypic traits.
Supplemental Table 2. Summary statistics for all terms in each QTL model.
Supplemental Table 3. T statistics for the additive effect of cytoplasm.
Supplemental Table 4. Significance of cytoplasm epistasis on each QTL.
Supplemental Table 5. Significance, effect, and divergence of each candidate gene in each narrow QTL.
Supplemental Table 6. List of cytoplasmic SNPs between TSU and KAS.
Supplemental Data Set 1A. Complete phenotypic trait data.
Supplemental Data Set 1B. Complete genotype matrix.
Supplemental Methods. Additional information pertaining to the analytical pipeline for candidate gene analyses, QTL methods, and materials.
Supplementary Material
Acknowledgments
Our work was supported by National Science Foundation Grants DEB 0420111 and DEB 0618347 to J.K.M., T.E.J., and J.H.R. and an Academia Sinica Career Development Award to P.E.V. J.T.L. was supported by NSF PRFB IOS-1402393. We thank A. Heiliger for assistance with phenotypic trait data collections and B. Gibson for laboratory assistance. K. Broman provided helpful feedback on the QTL analyses. Comments from D. Kliebenstein, J. Weber, D. Sloan, S. Schwartz, L. Milano, and two anonymous reviewers greatly improved earlier versions of the article.
AUTHOR CONTRIBUTIONS
All authors contributed extensively to this project. J.T.L., J.L.M., J.K.M., P.E.V., and T.E.J. wrote the article. J.T.L., D.B.L., J.L.M., and S.S. analyzed the data. J.L.M., K.A., J.H.R., and P.V. conducted physiological assays and provided reagents. J.L.M., K.A., T.E.J., J.H.R., and J.K.M. designed and executed the experiment.
Glossary
- ABA
abscisic acid
- QTL
quantitative trait locus
- cis-eQTL
cis-acting transcript abundance QTL
- RIL
recombinant inbred line
- WUE
water use efficiency
- FT
days to flowering
- RMR
root mass ratio
- cM
centimorgans
- SNP
single-nucleotide polymorphism
References
- Ågren J., Oakley C.G., McKay J.K., Lovell J.T., Schemske D.W. (2013). Genetic mapping of adaptation reveals fitness tradeoffs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 110: 21077–21082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F., Díaz-Uriarte R., Dopazo J. (2005). Discovering molecular functions significantly related to phenotypes by combining gene expression data and biological information. Bioinformatics 21: 2988–2993. [DOI] [PubMed] [Google Scholar]
- Angert A.L., Huxman T.E., Chesson P., Venable D.L. (2009). Functional tradeoffs determine species coexistence via the storage effect. Proc. Natl. Acad. Sci. USA 106: 11641–11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araus J.L., Slafer G.A., Reynolds M.P., Royo C. (2002). Plant breeding and drought in C3 cereals: what should we breed for? Ann. Bot. (Lond.) 89: 925–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araus J.L., Slafer G.A., Royo C., Serret M.D. (2008). Breeding for yield potential and stress adaptation in cereals. Crit. Rev. Plant Sci. 27: 377–412. [Google Scholar]
- Axelrod D.I. (1972). Edaphic aridity as a factor in angiosperm evolution. Am. Nat. 106: 311–320. [Google Scholar]
- Banta J.A., Ehrenreich I.M., Gerard S., Chou L., Wilczek A., Schmitt J., Kover P.X., Purugganan M.D. (2012). Climate envelope modelling reveals intraspecific relationships among flowering phenology, niche breadth and potential range size in Arabidopsis thaliana. Ecol. Lett. 15: 769–777. [DOI] [PubMed] [Google Scholar]
- Bates L.S., Waldren R.P., Teare I.D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39: 205–207. [Google Scholar]
- Bing N., Hoeschele I. (2005). Genetical genomics analysis of a yeast segregant population for transcription network inference. Genetics 170: 533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum A. (2005). Drought resistance, water-use efficiency, and yield potential—are they compatible, dissonant, or mutually exclusive? Crop Pasture Sci. 56: 1159–1168. [Google Scholar]
- Bogeat-Triboulot M.B., et al. (2007). Gradual soil water depletion results in reversible changes of gene expression, protein profiles, ecophysiology, and growth performance in Populus euphratica, a poplar growing in arid regions. Plant Physiol. 143: 876–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray E.A. (2004). Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. J. Exp. Bot. 55: 2331–2341. [DOI] [PubMed] [Google Scholar]
- Broman K.W., Wu H., Sen S., Churchill G.A. (2003). R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Broman K.W., Sen S. (2009). A Guide to QTL Mapping with R/qtl. (New York: Springer; ). [Google Scholar]
- Cattivelli L., Rizza F., Badeck F.W., Mazzucotelli E., Mastrangelo A.M., Francia E., Mare C., Tondelli A., Stanca A.M. (2008). Drought tolerance improvement in crop plants: An integrated view from breeding to genomics. Field Crops Res. 105: 1–14. [Google Scholar]
- Chater C., Kamisugi Y., Movahedi M., Fleming A., Cuming A.C., Gray J.E., Beerling D.J. (2011). Regulatory mechanism controlling stomatal behavior conserved across 400 million years of land plant evolution. Curr. Biol. 21: 1025–1029. [DOI] [PubMed] [Google Scholar]
- Chaves M.M., Oliveira M.M. (2004). Mechanisms underlying plant resilience to water deficits: prospects for water-saving agriculture. J. Exp. Bot. 55: 2365–2384. [DOI] [PubMed] [Google Scholar]
- Chaves M.M., Maroco J.P., Pereira J.S. (2003). Understanding plant responses to drought - from genes to the whole plant. Funct. Plant Biol. 30: 239–264. [DOI] [PubMed] [Google Scholar]
- Chen X., et al. (2010). An eQTL analysis of partial resistance to Puccinia hordei in barley. PLoS ONE 5: e8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy E., Chiu V.K., Silletti J., Feoktistov M., Morimoto T., Michaelson D., Ivanov I.E., Philips M.R. (1999). Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98: 69–80. [DOI] [PubMed] [Google Scholar]
- Condon A.G., Richards R.A., Rebetzke G.J., Farquhar G.D. (2004). Breeding for high water-use efficiency. J. Exp. Bot. 55: 2447–2460. [DOI] [PubMed] [Google Scholar]
- Cubillos F.A., Coustham V., Loudet O. (2012). Lessons from eQTL mapping studies: non-coding regions and their role behind natural phenotypic variation in plants. Curr. Opin. Plant Biol. 15: 192–198. [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679. [DOI] [PubMed] [Google Scholar]
- Dabney, A., and Storey, J. (2014). Q-value: Q-value estimation for false discovery rate control. R package version 1.38.0.
- Davila J.I., Arrieta-Montiel M.P., Wamboldt Y., Cao J., Hagmann J., Shedge V., Xu Y.Z., Weigel D., Mackenzie S.A. (2011). Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 9: 64–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais D.L., McKay J.K., Richards J.H., Sen S., Wayne T., Juenger T.E. (2012). Physiological genomics of response to soil drying in diverse Arabidopsis accessions. Plant Cell 24: 893–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake T.A., Schadt E.E., Lusis A.J. (2006). Integrating genetic and gene expression data: application to cardiovascular and metabolic traits in mice. Mamm. Genome 17: 466–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Soda M., Kruijer W., Malosetti M., Koornneef M., Aarts M.G. (2015). Quantitative trait loci and candidate genes underlying genotype by environment interaction in the response of Arabidopsis thaliana to drought. Plant Cell Environ. 38: 585–599. [DOI] [PubMed] [Google Scholar]
- Endo A., et al. (2008). Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol. 147: 1984–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler, J.A. (1986). Natural Selection in the Wild. (Princeton, NJ: Princeton University Press). [Google Scholar]
- Faigón-Soverna A., Harmon F.G., Storani L., Karayekov E., Staneloni R.J., Gassmann W., Más P., Casal J.J., Kay S.A., Yanovsky M.J. (2006). A constitutive shade-avoidance mutant implicates TIR-NBS-LRR proteins in Arabidopsis photomorphogenic development. Plant Cell 18: 2919–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D.S., Mackay T.F.C. (1996). Introduction to Quantitative Genetics, 4th ed. (Harlow, Essex, UK: Longmans Green; ). [Google Scholar]
- Farber C.R., et al. (2009). An integrative genetics approach to identify candidate genes regulating BMD: combining linkage, gene expression, and association. J. Bone Miner. Res. 24: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. (2013). Abscisic acid synthesis and response. The Arabidopsis Book 11: e0166, doi/10.1199/tab.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, S., Ziaugra, L., and Tabbaa, D. (2009). SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr. Protoc. Hum. Genet. 2: 2.12.1–2.12.18. [DOI] [PubMed]
- Hall M.C., Lowry D.B., Willis J.H. (2010). Is local adaptation in Mimulus guttatus caused by trade-offs at individual loci? Mol. Ecol. 19: 2739–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen B.G., Halkier B.A., Kliebenstein D.J. (2008). Identifying the molecular basis of QTLs: eQTLs add a new dimension. Trends Plant Sci. 13: 72–77. [DOI] [PubMed] [Google Scholar]
- Harb A., Krishnan A., Ambavaram M.M.R., Pereira A. (2010). Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 154: 1254–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heschel M.S., Riginos C. (2005). Mechanisms of selection for drought stress tolerance and avoidance in Impatiens capensis (Balsaminaceae). Am. J. Bot. 92: 37–44. [DOI] [PubMed] [Google Scholar]
- Heschel M.S., Donohue K., Hausmann N., Schmitt J. (2002). Population differentiation and natural selection for water‐use efficiency in Impatiens capensis (Balsaminaceae). Int. J. Plant Sci. 163: 907–912. [Google Scholar]
- Heslot N., Akdemir D., Sorrells M.E., Jannink J.L. (2014). Integrating environmental covariates and crop modeling into the genomic selection framework to predict genotype by environment interactions. Theor. Appl. Genet. 127: 463–480. [DOI] [PubMed] [Google Scholar]
- Johnson, E.B., Haggard, J.E., and Clair, D.A.S. (2012). Fractionation, stability, and isolate-specificity of QTL for resistance to Phytophthora infestans in cultivated tomato (Solanum lycopersicum). G3 (Bethesda) 2: 1145–1159. [DOI] [PMC free article] [PubMed]
- Joseph B., Corwin J.A., Züst T., Li B., Iravani M., Schaepman-Strub G., Turnbull L.A., Kliebenstein D.J. (2013a). Hierarchical nuclear and cytoplasmic genetic architectures for plant growth and defense within Arabidopsis. Plant Cell 25: 1929–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B., Corwin J.A., Li B., Atwell S., Kliebenstein D.J. (2013b). Cytoplasmic genetic variation and extensive cytonuclear interactions influence natural variation in the metabolome. eLife 2: e00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenger T.E. (2013). Natural variation and genetic constraints on drought tolerance. Curr. Opin. Plant Biol. 16: 274–281. [DOI] [PubMed] [Google Scholar]
- Juenger T.E., Sen S., Bray E., Stahl E., Wayne T., McKay J., Richards J.H. (2010). Exploring genetic and expression differences between physiologically extreme ecotypes: comparative genomic hybridization and gene expression studies of Kas-1 and Tsu-1 accessions of Arabidopsis thaliana. Plant Cell Environ. 33: 1268–1284. [DOI] [PubMed] [Google Scholar]
- Kadioglu A., Terzi R. (2007). A dehydration avoidance mechanism: leaf rolling. Bot. Rev. 73: 290–302. [Google Scholar]
- Kesari R., Lasky J.R., Villamor J.G., Des Marais D.L., Chen Y.-J.C., Liu T.-W., Lin W., Juenger T.E., Verslues P.E. (2012). Intron-mediated alternative splicing of Arabidopsis P5CS1 and its association with natural variation in proline and climate adaptation. Proc. Natl. Acad. Sci. USA 109: 9197–9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.H., Park J., Williams D.S., Xiong Y., Hwang I., Kang B.H. (2013). Defective chloroplast development inhibits maintenance of normal levels of abscisic acid in a mutant of the Arabidopsis RH3 DEAD-box protein during early post-germination growth. Plant J. 73: 720–732. [DOI] [PubMed] [Google Scholar]
- Leprince A.-S., Magalhaes N., De Vos D., Bordenave M., Crilat E., Clément G., Meyer C., Munnik T., Savouré A. (2015). Involvement of phosphatidylinositol 3-kinase in the regulation of proline catabolism in Arabidopsis thaliana. Front. Plant Sci. 5: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Tsaih S.-W., Shockley K., Stylianou I.M., Wergedal J., Paigen B., Churchill G.A. (2006). Structural model analysis of multiple quantitative traits. PLoS Genet. 2: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell J.T., Juenger T.E., Michaels S.D., Lasky J.R., Platt A., Richards J.H., Yu X., Easlon H.M., Sen S., McKay J.K. (2013). Pleiotropy of FRIGIDA enhances the potential for multivariate adaptation. Proc. Biol. Sci. 280: 20131043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D.B., Logan T.L., Santuari L., Hardtke C.S., Richards J.H., DeRose-Wilson L.J., McKay J.K., Sen S., Juenger T.E. (2013). Expression quantitative trait locus mapping across water availability environments reveals contrasting associations with genomic features in Arabidopsis. Plant Cell 25: 3266–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T.F.C. (2001). The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339. [DOI] [PubMed] [Google Scholar]
- MacMillan K., Emrich K., Piepho H.P., Mullins C.E., Price A.H. (2006). Assessing the importance of genotype x environment interaction for root traits in rice using a mapping population II: conventional QTL analysis. Theor. Appl. Genet. 113: 953–964. [DOI] [PubMed] [Google Scholar]
- Maliakal S.K., McDonnell K., Dudley S.A., Schmitt J. (1999). Effects of red to far‐red ratio and plant density on biomass allocation and gas exchange in Impatiens capensis. Int. J. Plant Sci. 160: 723–733. [Google Scholar]
- Manichaikul A., Moon J.Y., Sen S., Yandell B.S., Broman K.W. (2009). A model selection approach for the identification of quantitative trait loci in experimental crosses, allowing epistasis. Genetics 181: 1077–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay J.K., Richards J.H., Mitchell-Olds T. (2003). Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Mol. Ecol. 12: 1137–1151. [DOI] [PubMed] [Google Scholar]
- McKay J.K., Richards J.H., Nemali K.S., Sen S., Mitchell-Olds T., Boles S., Stahl E.A., Wayne T., Juenger T.E. (2008). Genetics of drought adaptation in Arabidopsis thaliana II. QTL analysis of a new mapping population, KAS-1 x TSU-1. Evolution 62: 3014–3026. [DOI] [PubMed] [Google Scholar]
- Meyre D., Leonardi A., Brisson G., Vartanian N. (2001). Drought-adaptive mechanisms involved in the escape/tolerance strategies of Arabidopsis Landsberg erecta and Columbia ecotypes and their F1 reciprocal progeny. J. Plant Physiol. 158: 1145–1152. [Google Scholar]
- Milborrow B.V. (2001). The pathway of biosynthesis of abscisic acid in vascular plants: a review of the present state of knowledge of ABA biosynthesis. J. Exp. Bot. 52: 1145–1164. [PubMed] [Google Scholar]
- Millstein J., Zhang B., Zhu J., Schadt E.E. (2009). Disentangling molecular relationships with a causal inference test. BMC Genet. 10: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minic Z. (2008). Physiological roles of plant glycoside hydrolases. Planta 227: 723–740. [DOI] [PubMed] [Google Scholar]
- Mir R.R., Zaman-Allah M., Sreenivasulu N., Trethowan R., Varshney R.K. (2012). Integrated genomics, physiology and breeding approaches for improving drought tolerance in crops. Theor. Appl. Genet. 125: 625–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto E.C., Keller M.P., Attie A.D., Yandell B.S. (2010). Causal graphical models in systems genetics: a unified framework for joint inference of causal network and genetic architecture for correlated phenotypes. Ann. Appl. Stat. 4: 320–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. (2005). Molecular signatures of natural selection. Annu. Rev. Genet. 39: 197–218. [DOI] [PubMed] [Google Scholar]
- Orr H.A. (1998). The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution 52: 935–949. [DOI] [PubMed] [Google Scholar]
- Pinheiro C., Chaves M.M. (2011). Photosynthesis and drought: can we make metabolic connections from available data? J. Exp. Bot. 62: 869–882. [DOI] [PubMed] [Google Scholar]
- Richards R.A., Rebetzke G.J., Watt M., Condon A.G., Spielmeyer W., Dolferus R. (2010). Breeding for improved water productivity in temperate cereals: phenotyping, quantitative trait loci, markers and the selection environment. Funct. Plant Biol. 37: 85–97. [Google Scholar]
- Rockman M.V. (2012). The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution 66: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal D.M., Stiller V., Sperry J.S., Donovan L.A. (2010). Contrasting drought tolerance strategies in two desert annuals of hybrid origin. J. Exp. Bot. 61: 2769–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E.E., et al. (2005). An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 37: 710–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalenbach I., Zhang L., Reymond M., Jiménez-Gómez J.M. (2014). The relationship between flowering time and growth responses to drought in the Arabidopsis Landsberg erecta x Antwerp-1 population. Front. Plant Sci. 5: 609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J., Stinchcombe J.R., Heschel M.S., Huber H. (2003). The adaptive evolution of plasticity: phytochrome-mediated shade avoidance responses. Integr. Comp. Biol. 43: 459–469. [DOI] [PubMed] [Google Scholar]
- Seo M., Koshiba T. (2002). Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7: 41–48. [DOI] [PubMed] [Google Scholar]
- Sharma S., Villamor J.G., Verslues P.E. (2011). Essential role of tissue-specific proline synthesis and catabolism in growth and redox balance at low water potential. Plant Physiol. 157: 292–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrard M.E., Maherali H. (2006). The adaptive significance of drought escape in Avena barbata, an annual grass. Evolution 60: 2478–2489. [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K., Seki M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6: 410–417. [DOI] [PubMed] [Google Scholar]
- Stebbins G.L. (1952). Aridity as a stimulus to plant evolution. Am. Nat. 86: 33–44. [Google Scholar]
- Studer A.J., Doebley J.F. (2011). Do large effect QTL fractionate? A case study at the maize domestication QTL teosinte branched1. Genetics 188: 673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamy B.P.M., et al. (2013). Genetic, physiological, and gene expression analyses reveal that multiple QTL enhance yield of rice mega-variety IR64 under drought. PLoS ONE 8: e62795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados L., Savouré A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15: 89–97. [DOI] [PubMed] [Google Scholar]
- Van Ooijen J.W. (2006). JoinMap 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. (Wageningen, The Netherlands: Kyazma; ). [Google Scholar]
- Verslues P.E. (2010). Quantification of water stress-induced osmotic adjustment and proline accumulation for Arabidopsis thaliana molecular genetic studies. In Plant Stress Tolerance: Methods in Molecular Biology 639, R. Sunkar, ed (New York: Springer Science+Business Media; ). [DOI] [PubMed] [Google Scholar]
- Verslues, P.E., and Sharma, S. (2010). Proline metabolism and its implications for plant-environment interaction. The Arabidopsis Book 8: e0140, doi/10.1199/tab.0140. [DOI] [PMC free article] [PubMed]
- Wilczek A.M., et al. (2009). Effects of genetic perturbation on seasonal life history plasticity. Science 323: 930–934. [DOI] [PubMed] [Google Scholar]
- Zeng Z.-B. (1993). Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc. Natl. Acad. Sci. USA 90: 10972–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Richards E.J., Borevitz J.O. (2007). Genetic and epigenetic dissection of cis regulatory variation. Curr. Opin. Plant Biol. 10: 142–148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.