Abstract
Viruses rely completely on the hosts’ machinery for translation of viral transcripts. However, for most viruses infecting humans, codon usage preferences (CUPrefs) do not match those of the host. Human papillomaviruses (HPVs) are a showcase to tackle this paradox: they present a large genotypic diversity and a broad range of phenotypic presentations, from asymptomatic infections to productive lesions and cancer. By applying phylogenetic inference and dimensionality reduction methods, we demonstrate first that genes in HPVs are poorly adapted to the average human CUPrefs, the only exception being capsid genes in viruses causing productive lesions. Phylogenetic relationships between HPVs explained only a small proportion of CUPrefs variation. Instead, the most important explanatory factor for viral CUPrefs was infection phenotype, as orthologous genes in viruses with similar clinical presentation displayed similar CUPrefs. Moreover, viral genes with similar spatiotemporal expression patterns also showed similar CUPrefs. Our results suggest that CUPrefs in HPVs reflect either variations in the mutation bias or differential selection pressures depending on the clinical presentation and expression timing. We propose that poor viral CUPrefs may be central to a trade-off between strong viral gene expression and the potential for eliciting protective immune response.
Keywords: human viruses, codon usage preferences, mutation, translational selection, immune system, clinical presentation, genotype–phenotype, warts, cancer, chronic infection, acute infection
Introduction
Synonymous codons are not used at random (Aota and Ikemura 1986; Shields and Sharp 1987). Codon usage preferences (CUPrefs) vary between species, and between genes within the same genome (Marin et al. 1989). CUPrefs have arisen from a complex interplay between several evolutionary processes, essentially mutation and selection (Bulmer 1991). The mutational model postulates that the main factor influencing average codon usage is nucleotide composition (Guanine-Cytosine [GC] content) in the genome (Chen et al. 2004). This model considers changes in synonymous codon usage neutral: It assumes that no fitness effect is associated with the preferential use of a given synonymous codon (Plotkin and Kudla 2011). The selection-related model postulates coadaptation between synonymous codon usage and the translation machinery (e.g., differential transfer RNA [tRNA] abundance) to optimize translational speed and enhance translational accuracy (Sharp et al. 1995; Duret 2000; Rocha 2004). Hence, the selection model claims that synonymous mutations can indeed influence the fitness of an organism (Plotkin and Kudla 2011). The mutation model and the selection model are not mutually exclusive. In fast-growing organisms with large population sizes, such as Escherichia coli or Saccharomyces cerevisiae, experimental evidence supports the idea that translation selection is the main factor conditioning CUPrefs (Stenico et al. 1994; Moriyama and Powell 1997). In contrast, in slowly growing organisms with small population sizes, such as mammals, natural selection may be inefficient to strongly pattern CUPrefs, and its effect on codon usage remains controversial (Duret 2002). Besides selection for translational efficiency and accuracy, the choice of synonymous codons may also be under the selective pressure for optimal translation kinetics, to ensure correct messenger RNA (mRNA) structure and protein folding (Plotkin and Kudla 2011). Inappropriate CUPrefs may impair translation kinetics, thus leading to fitness costs associated to low quantity of functional protein, but also to waste of cellular resources incurred through accumulation of erroneous and misfolded protein, increased toxicity, and cleaning costs (Gingold and Pilpel 2011).
Codon usage in viruses seems to be shaped both by selection and mutation. On one side, all viruses depend on host translational machinery, in particular viruses that do not encode their own tRNAs (as it is the case for human viruses), and CUPrefs in viral genes tend to match protein-specific requirements (Akashi and Eyre-Walker 1998): proteins required in large amount are usually encoded by genes optimized to the host CUPrefs, while maladaptation of CUPrefs results in reduced protein production (Bahir et al. 2009). On the other side, genomic GC content is often a strong predictive variable for codon usage in viruses (Sharp and Li 1986; Karlin et al. 1990), revealing that genome-wide mutational pressures play an important role in patterning viral CUPrefs (Shackelton et al. 2006). Other studies suggest that additional selective factors such as fine-tuning selection on translation kinetics and escape from antiviral cellular responses may also underlie viral CUPrefs (Sugiyama et al. 2005; Aragones et al. 2008, 2010).
Papillomaviruses (PVs) are nonenveloped, double-stranded DNA viruses with a circular genome of approximately 8 kbp. PVs infect epithelia in a wide spectrum of vertebrates, at cutaneous and mucosal sites (Bravo and Felez-Sanchez 2015). The PV life cycle depends on keratinocyte differentiation (Bedell et al. 1991). Viral genomes are primarily present as nuclear episomes, which replicate in parallel to cell division. As the daughter cell migrates upwards and undergoes differentiation, the viral DNA is amplified and the viral expression pattern is modified, eventually leading to nonlytic virion release (Longworth and Laimins 2004). PV genomes typically contain eight well-defined open reading frames (ORFs) classified as early (E1, E2, E4, E5, E6, and E7) and late (L1 and L2) based on their temporal expression during viral life cycle. The early E5, E6, and E7 oncoproteins induce cell immortalization and transformation; E1 and E2 are associated with viral genome replication; and the E4 protein is associated with cytokeratin filament collapse. The late L1 and L2 genes encode for capsid proteins that are strongly immunogenic (Zheng and Baker 2006).
Human PVs belong to five monophyletic genera (supplementary fig. S1, Supplementary Material online): Alphapapillomaviruses (AlphaPVs), Betapapillomaviruses (BetaPVs), Gammapapillomaviruses (GammaPVs), Mupapillomaviruses (MuPVs), and Nupapillomaviruses (NuPVs) (Bernard et al. 2010). In PV taxonomy, two PV genomes sharing more than 60% nucleotide identity in the L1 gene belong to the same PV genera (de Villiers et al. 2004). Different human PV lineages have adapted to specific epithelial niches, with different types showing differences in cell tropism, natural history of the infection, prevalence, and association with disease (Bravo and Felez-Sanchez 2015). The large majority of the hitherto known human PVs, essentially Beta- and GammaPVs, cause asymptomatic infections and can be detected in healthy skin swabs or, for a reduced number of GammaPVs, also in mucosal rinses (Nindl et al. 2007). Mu- and NuPVs cause conspicuous, productive cutaneous lesions usually at palmar and plantar epithelial sites (Bernard et al. 2010). Finally, AlphaPVs are very diverse in terms of tropism and clinical manifestation of the disease. They include viruses with cutaneous tropism causing warts in the hands, lips, or eyelids; viruses with a very defined tropism and causing sexually transmitted warts and condylomas; and viruses causing less productive, long-lasting infections associated with certain human cancers, such as cervical cancer, other anogenital cancers, and a fraction of head and neck tumors (Bernard et al. 2010). Thus, there is no sharp correspondence between clinical manifestation and phylogenetic relationships for human PVs, as members from different genera could produce similar clinical presentations (e.g., Beta- and GammaPVs essentially causing asymptomatic mucocutaneous lesions [MucCutAsym]), but certain viruses belonging to the same genera could cause different clinical presentations (e.g., AlphaPVs can cause cutaneous warts [CutW], genital warts [GenW], or other mucosal lesions [MucL]).
PVs do not encode for any element of the transcription or the translation machinery. They rely on the host cellular apparatus for gene expression and it would be expected that PV CUPrefs match those of the host. It is thus puzzling, however, that CUPrefs in human PVs are different from CUPrefs in humans (Zhao et al. 2003; Bravo and Müller 2005): the preferred codons in human PV genes are the less-preferred codons in the average human genes (Bravo and Müller 2005) and show a strong bias toward codons ending with Adenine/Thymine [A/T] (Bravo and Müller 2005; Cladel et al. 2010). These compositional differences possibly reflect a bias in the mutation/selection evolutionary processes that still needs to be understood. Hitherto, two adaptive explanations for the biased CUPrefs in PV genes have been proposed. First, it has been suggested that PV CUPrefs have been selected for because they decrease viral protein synthesis, thereby lowering immune exposure (Tindle 2002; Cid-Arregui et al. 2003). Second, it has been postulated that PV CUPrefs may have evolved to differentially match the varying tRNA profile of the host cell in which viral protein actually occurs: the differentiating keratinocyte (Zhou et al. 1999; Gu et al. 2004; Aragones et al. 2010; Cladel et al. 2010).
We have analyzed here the CUPrefs for 156 human PVs from five distinct phylogenetic groups to determine whether variations in CUPrefs could be explained by differences in tissue tropism, association with disease, and/or timing of gene expression. Due to the high dimensionality of CUPrefs data, dimensionality reduction techniques were applied: Multidimensional scaling (MDS), correspondence analysis (CA), and cluster analyses.
Materials and Methods
Human PVs Gene Sequences
The ORFs of all human PVs available at the Papillomavirus Episteme Database (http://pave.niaid.nih.gov ) were collected between March and April 2013. Using an in-house PERL script, the ORFs were examined by checking the start codon, stop codon, and internal stop codons to guarantee that only true ORFs were used. The final data set included 156 HPV types: 63 AlphaPVs, 45 BetaPVs, 45 GammaPVs, 2 MuPVs, and 1 NuPV. Names, accession numbers, and other information are detailed in supplementary table S1, Supplementary Material online.
Clinical Manifestations of Human PV Infections
Human PVs were classified according to their phenotypic clinical presentation characteristics. This classification took into account both the nature of the infection and its tropism. As for the nature of the infection, most human PVs are recovered from healthy skin and healthy mucosa, and generate unapparent, nonproductive infections. Other PVs cause highly productive infections that cause self-limited benign proliferative lesions, chiefly warts. Finally, a few human PVs cause long-lasting, low productive infections that can lead to the development of malignant proliferative lesions, essentially anogenital cancers (Doorbar et al. 2012). As for tropism, human PV infections are either mucosal or cutaneous. The following four groups were defined (supplementary fig. S1, Supplementary Material online, and supplementary table S1, Supplementary Material online): Mucocutaneous asymptomatic (MucCutAsym), including Beta and GammaPVs that cause unapparent infections; GenW group, including AlphaPVs causing proliferative lesions at mucosal sites; MucL group contained AlphaPVs causing other lesions at mucosal sites and with potential for malignisation; finally, the CutW group included Alpha-, Mu-, and NuPVs causing proliferative cutaneous lesions.
Codon Usage Preferences Data
Detailed codon composition for each genus is provided in supplementary table S4, Supplementary Material online. Patterns of synonymous codon usage were analyzed in the E1, E2, E4, E6, E7, L1, and L2 genes. The E5 gene was excluded from the analysis because it is absent in most human PVs. The relative frequency (RF) distribution of 59 codons (excluding Met, Trp, and stop codons) was calculated using an in-house PERL script. The abundance of each codon in a gene was calculated and pondered by a factor corresponding to the sum of all synonymous codons:
where nac is the number of events in which the c-th codon for the a-th amino acid is used, and ta the total number of synonymous codons that encode the ath amino acid. The final representation of the codon usage data for each gene is thus a vector of 59 positions with values between 0 and 1.
We calculated the pairwise CUPrefs distances as the Euclidean distances between the RF vectors of the corresponding human PVs.
Codon Adaptation Index
To analyze the relationship of CUPrefs between human PVs and humans, we employed the codon adaptation index (CAI) (Sharp and Li 1987). This index evaluates the match between the CUPrefs of a particular gene and those in a reference set. In our case, the reference values were the average CUPrefs in the human genome, as retrieved from the Kazusa codon usage database, under http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=9606, last accessed April 2013) (Nakamura et al. 2000). CAI values were calculated for all human PV genes. CAI values were also calculated for the subset of human genes differentially expressed in the epithelium. Epithelial genes were retrieved from the UCSC browser in May 2015 (Kent et al. 2002) filtering skin genes by their expression (log2(tissue/reference) with a maximum value of 5 and minimum of −5). No differences in CUPrefs were identified in genes underexpressed or overexpressed in skin compared with the average CUPrefs. We therefore chose the CAI values for the subset of epithelial genes as a reference (supplementary fig. S2, Supplementary Material online). CAI was calculated using in-house PERL Scripts. For each gene, the output of the CAI calculation was a single value, between 0 and 1, with higher values reflecting a higher similarity in CUPrefs to the reference. The maximum value of 1 is only achieved if, for each synonymous codons set, all amino acids in the considered gene are encoded by the most used codon in the reference set.
Dimensionality Reduction Methods: Multidimensional Scaling and Correspondence Analysis
Our final data set for analysis was a matrix in which the rows correspond to the genes of one human PV genome and the columns to the 59 codons, such that each row has the codon usage information for a specific gene, in terms of relative frequencies. This data set was subjected to dimensionality reduction techniques to analyze similarities among codon usage of human PV genes, by applying MDS and CA.
MDS refers to a broad class of procedures that create low-dimensional representations of complex data with preservation of the similarities between data points (Cox TF and Cox MAA 1994). In an n-dimensional representation, samples with very similar codon usage profiles are displayed close together. We performed a nonmetric MDS with column wise Z-transformation of the variables using SPSS Statistics Version 17.0 (IBM, Chicago IL). For MDS analysis, the matrix based on codon RF values was used in order to avoid biases linked to amino acid composition. In order to determine the appropriate dimensionality in which data should be scaled, we used scree plots (data not shown), which display stress as a function of dimensionality. Based on the stress values and on interpretability, we chose two dimensions as the “best” solution to plot our data.
CA consists in a multivariate statistical method widely used to summarize the lack of independence between objects represented through rows and columns of a matrix (here genes and codons, respectively) as a small number of derived variables, called axes. By definition, the axes are ordered according to the amount of variance in the data explained by them. Data were plotted on the first two axes with the information on the amount of variance explained in these two-dimensional representations.
CA of codon usage data is a widely used method in sequence analysis, which can be refined through internal correspondence analysis (ICA) to account for the variability in amino acid composition between proteins as a confounding factor when one wants to analyze synonymous codon usage variability. ICA, which is basically a double within-between-CA, has been found to be the best method in generating axes that reflect variations in synonymous codon usage (Suzuki et al. 2008). This method further allows distinguishing within and between group variability with respect to genotype, gene, or clinical manifestations. ICA was run in R-3.0.1 with package ade4-1.5.2 and cross-checked with the implementations in vegan-2.0.9 and FactoMineR-1.25. We additionally used seqinr-3.0.7 for computing codon usage counts and boot1.3-11 for bootstrapping to assess the variance in each codon’s contribution to the first principal axis under sampling from a sequence population. Here, we sampled with replacement from the population of all PVs, performed the ICA, and obtained the projections of codons on the first axis of the ICA. Repeating this 1,000 times rendered a distribution of projections that informed us on how reliable the ICA analysis is with regards to sampling from a population.
Cluster Analysis
Statistical clustering was used to determine the optimal number of natural conglomerates within the data, and to classify each individual gene into one of the identified conglomerates (Kent and Kongsted 2012). We applied the statistical clustering technique implemented as two-step cluster analysis (SPSS Statistics; IBM). The first step groups the cases into many small subclusters. The second step groups the subclusters into the final, optimal number of clusters, estimated using the Bayesian information criterion (BIC).
Phylogenetic Analysis
Amino acid sequences were aligned with MUSCLE (Edgar et al. 2004) and back-translated into codon-aligned nucleotide sequences. Informative positions were filtered with GBLOCKS under nonstringent conditions (Castresana 2000). Phylogenetic relationships were inferred in a maximum likelihood framework using RAxML v.8 (http://www.exelixis-lab.org/) (Stamatakis 2014) at the nucleotide level. We used the GTR+ Γ4 model, considering three partitions (one per codon position). The number of required bootstrap cycles was determined with the –autoMRE command (Stamatakis 2014). Pairwise evolutionary distances between terminal taxa were estimated on the best-known maximum likelihood tree using RAxML.
Statistical Analysis
The Huber’s M-estimator calculated with R was used as robust central estimator for CAI values. The significance of the differences in CAI values between PV genes and human genes was tested with a Wilcoxon and Mann–Whitney (WMW) test, implemented in R package stats. The effect of clinical presentation and gene (and their interaction) on CAI values was tested through a two-way analysis of variance (ANOVA) followed by Tukey post hoc test (SPSS Statistics; IBM). Pearson’s correlations (implemented in R) were used to determine if there was a relationship between evolutionary and codon usage-based distances. Moreover, in order to identify major sources of variation among human PVs on the axes generated by ICA, we tested for the correlation between projections to the first principal axis with CAI and average GC content at the third position in 4-fold family codons and also for the 4-fold component in the 6-fold family codons, for each gene separately. Pearson’s correlation (r) was calculated. The square of r indicated the percentage of the variance in the first axis that is explained by the variance in the gene feature values.
Results
Genes in Human PVs Do Not Follow Human Codon Usage Preferences
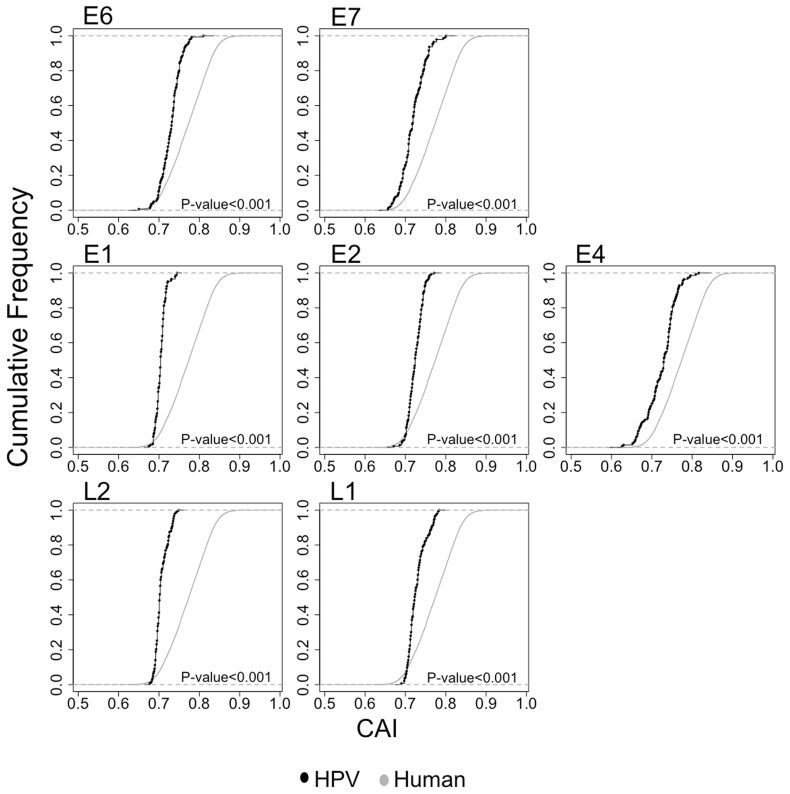
We first evaluated the global adaptation of CUPrefs in human PV genes to the human CUPrefs, by calculating the CAI (Sharp and Li 1987). This index evaluates the match between the CUPrefs of a particular gene and those in a reference set, in our case the human average CUPrefs. CAI values were calculated for each gene in the human PV data set. Because human PVs infect epithelial cells, CAI values were also calculated for the subset of human genes expressed in the epithelium. Figure 1 shows the cumulative frequency of CAI for human epithelial genes and for each gene in the human PVs. For all human PV genes, frequency distributions are shifted to lower CAI values compared with the reference human epithelial genes. WMW tests showed that for each of the seven human PV genes, the median CAI value is significantly lower than the median CAI value for the human epithelial genes (P < 0.001). A Kolmogorov–Smirnov (KS) test demonstrated further that for all human PV genes the distribution of the CAI values were significantly different from that of human genes (P < 0.001). All these results point toward a low level of adaptation of the human PV genes to the CUPrefs of their host.
Fig. 1.—
Cumulative frequency plot showing the distribution of CAI values of HPVs and human epithelial genes. CAI values for HPV genes were calculated for every gene separately, using the human mean codon usage as a reference set. CAI values of HPV genes are represented in black. CAI values for human genes expressed at epithelial cells are represented in grey. Statistical differences in CAI values between human and HPV genes were assessed by a WMW test.
Capsid Genes in Human PVs Causing Cutaneous Warts Present a Greater Adaptation to Human Codon Usage
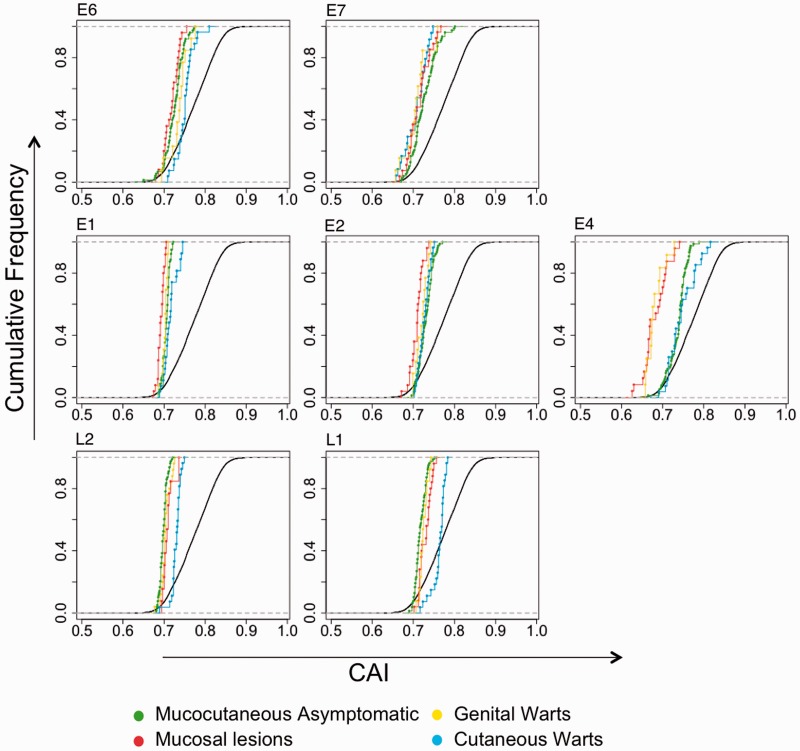
In order to understand the link between CUPrefs and virus clinical presentation, we analyzed CAI values stratifying human PVs by their clinical presentation, essentially in terms of productivity of the lesion and tropism. Four groups were defined (supplementary fig. S1, Supplementary Material online, and supplementary table S1, Supplementary Material online): MucCutAsym PVs including mucocutaneous human PVs typically associated with unapparent infections belonging to BetaPVs and GammaPVs; GenW group including AlphaPVs causing proliferative lesions at mucosal sites; MucL group including other AlphaPVs causing other lesions at mucosal sites and with potential for malignization; and finally, the cutaneous warts group (CutW) including Alpha-, Mu-, and NuPVs causing proliferative cutaneous lesions.
We observed that CAI values to the human average for all viral genes and for all viral clinical presentations were statistically lower than CAI values for human epithelial genes (WMW test, P < 0.001; KS test, P < 0.001). The only exception was the L1 capsid gene of human PVs causing cutaneous warts (WMW test, P > 0.5). CAI cumulative distributions for each gene and clinical presentation are represented in figure 2central values are given in supplementary table S2, Supplementary Material online. All genes in human PVs are thus poorly adapted to the average human CUPrefs, independently of their clinical presentation, to the exception of one of the capsid genes in the very productive group of viruses causing cutaneous warts.
Fig. 2.—
Cumulative frequency plot of CAI values for human and HPV genes stratified by clinical manifestation. CAI values of each HPV gene were computed on the basis of CUPrefs in human genes. In green are represented CAI values of mucocutaneous asymptomatic HPVs (both Beta- and GammaPVs). In red, HPVs that cause mucosal lesions. In yellow, HPVs that cause genital warts. In blue, HPVs causing cutaneous warts. In black, human epithelial genes.
A two-way ANOVA with clinical presentation, gene, and their interaction as factors was performed to analyze its effect on CAI values (table 1). This analysis revealed that all factors had a significant effect. The significant “clinical presentation × gene” interaction indicated that different genes from human PVs with different clinical presentation had different degrees of adaptation to the average human CUPrefs. Then a one-way ANOVA followed by Tukey test was performed for each gene with clinical presentation as factor (supplementary table S2, Supplementary Material online). For the E1, E6, L1, and L2 genes, human PVs causing cutaneous warts showed significantly greater adaptation to human CUPrefs (P < 0.005, Tukey post hoc test) than those with other clinical presentations. The lowest level of adaptation in L1 and L2 was found in the MucCutAsym group (P < 0.005, Tukey post hoc test). In contrast, for the E1 and E6 genes, the lowest level of adaptation was seen in mucosal PVs (P < 0.005, Tukey post hoc test). For E2, the highest level of adaptation was found in the MucCutAsym group, and the lowest in PVs causing MucLs (P < 0.005, Tukey post hoc test) (fig. 2supplementary table S2, Supplementary Material online). Finally, for E7 and E4, all cutaneous PVs—MucCutAsym and CutW—showed higher adaptation to the average human CUPrefs than mucosal PVs—GenW and MucL (fig. 2supplementary table S2, Supplementary Material online).
Table 1.
Effect of Clinical Manifestation and Gene on CAI
| Factor | df | F ratio | P value |
|---|---|---|---|
| Clinical manifestation | 3 | 74.52 | <0.001 |
| Gene | 6 | 33.16 | <0.001 |
| Clinical manifestation × gene | 18 | 16.69 | <0.001 |
Note.—A two-way ANOVA was performed to assess the influence of clinical manifestation (mucocutaneous asymptomatic, mucosal lesions, genital warts, and cutaneous warts) and gene (E6, E7, E1, E2, E4, L1, and L2) on CAI. Both factors and their interaction show a significant effect on CAI. df, degree of freedom.
We also performed a one-way ANOVA followed by Tukey test for each clinical manifestation with gene as a factor (fig. S2 and supplementary table S2, Supplementary Material online). For Human PVs causing CutW, GenW, and MucL, the L1 gene showed the highest level of adaptation to human CUPrefs (P < 0.005, Tukey post hoc test). In contrast, for MucCutAsym human PVs, late genes (L1 and L2) showed the lowest level of adaptation. The highest level of adaptation in MucCutAsym was found in E4, whereas for GenW and MucL, the E4 gene exhibited the lowest level of adaptation (P < 0.005, Tukey post hoc test) (supplementary fig. S3, Supplementary Material online, and supplementary table S2, Supplementary Material online).
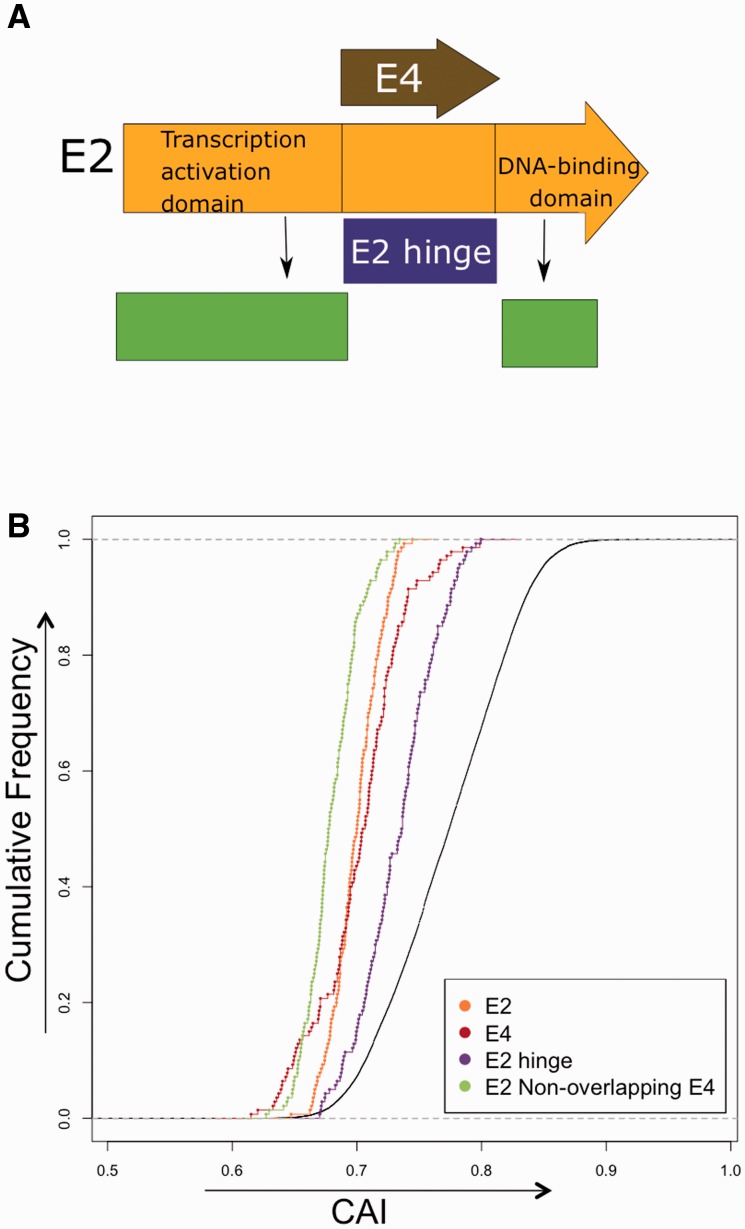
The E2 Hinge Region Shows Higher Adaptation to Human CUPrefs than the Overlapping E4 Gene
The E4 ORF is nested within the E2 sequence in a different reading frame, with the E4 coding sequence overlapping the so-called hinge region of the E2 gene (fig. 3A). To assess the influence of the presence of an overlapping region on CUPrefs, we calculated the CAI values for the E2 hinge region, containing E4, as well as for the nonoverlapping region (fig. 3B). We performed a WMW paired test in order to compare the adaptation of these two regions for each human PV. We found that the CAI values for the hinge region were significantly higher than those for the nonoverlapping region. Moreover, the E2 hinge region also showed significantly higher degree of adaptation compared with E4 (P < 0.005, WMW paired test). We performed the same analysis but stratifying each human PV by their clinical manifestation (supplementary fig. S4, Supplementary Material online). The results of the WMW paired test indicated again that for all clinical manifestations the CAI values for the E2 hinge region were significantly higher than those for the nonoverlapping region (supplementary fig. S4, Supplementary Material online). Also, for all clinical manifestations, the CAI values for E2 hinge region were significantly higher than for E4. Finally, we found that for MucL and GenW, E4 genes were significantly less adapted than E2 genes. On the contrary, for MucCutAsym and CutW, we found the opposite pattern, with E2 being significantly less adapted than E4 (supplementary fig. S4, Supplementary Material online).
Fig. 3.—
(A) Schematic map of the E2 and E4 genes. The E4 ORF overlaps the hinge region of the E2 gene. This region is a flexible, essentially disordered connector between the functionally conserved transcription activation domain and the DNA-binding domain. (B) Cumulative frequency plot of E2 and E4 genes, and the overlapping and nonoverlapping region of both genes. CAI values of each HPV gene were computed on the basis of CUPrefs in human genes. Code for genes: Orange, E2; brown, E4; violet, E2 hinge region; and green, E2 nonoverlapping region with E4.
Evolutionary Distances Explain only a Low Proportion of Codon Usage Differences
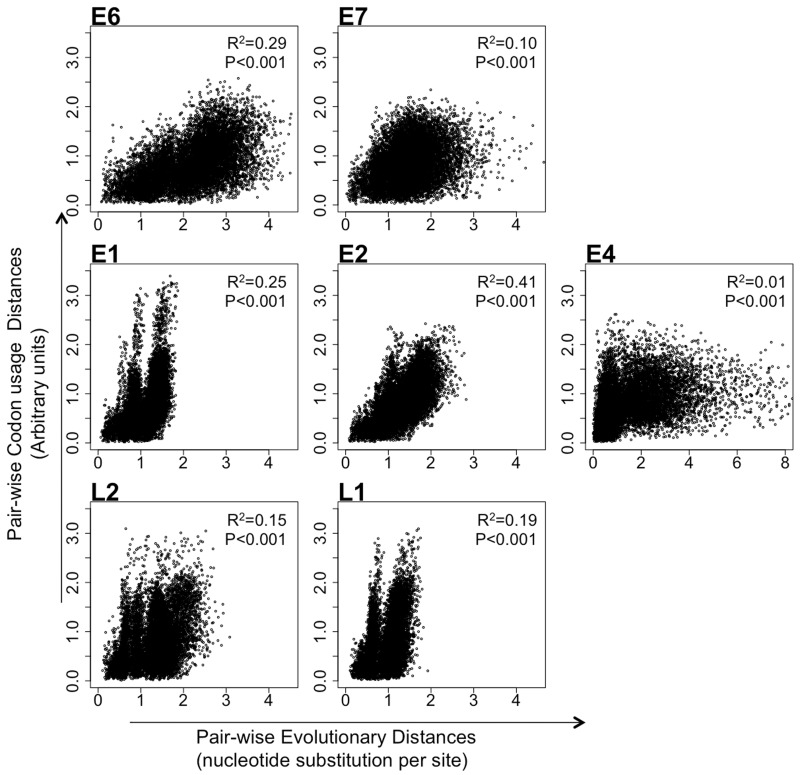
In order to address the relationship between CUPrefs in human PVs and their evolutionary history, we analyzed the correlation between pairwise evolutionary distances and pairwise CUPrefs distances for each of the seven genes studied (fig. 4). Pairwise evolutionary distances were calculated from the best-known tree reconstructed by maximum likelihood techniques (see Methods). Pairwise CUPrefs distances were calculated as the Euclidean distances between relative frequencies vectors of synonymous codon usage. They represent the similarities in terms of codon usage between pairs of PVs. The correlation between both variables was significant and positive for all genes, with r2 ranging from 0.01 (E4) to 0.41 (E2) with a median value of 0.19. This means that on average, evolutionary distances explained only a fifth of the variance in codon-based distance. No clear trend was obvious depending on gene expression timing, as early genes displayed both the highest and the lowest r2, for E2 and E4, respectively.
Fig. 4.—
Scatter plot of the evolutionary distances and the CUPref-based distances in the complete data set for each HPV gene. For all plots, x-axis represents evolutionary pairwise distances and y-axis represents CUPref-based pairwise distances. Phylogenetic distances were obtained with RAxML for every gene separately. The correlation coefficient and P value obtained after Pearson’s analysis are reported for each bivariate analysis.
Orthologous Genes in Viruses with Similar Clinical Presentation Display Similar Codon Usage Preferences
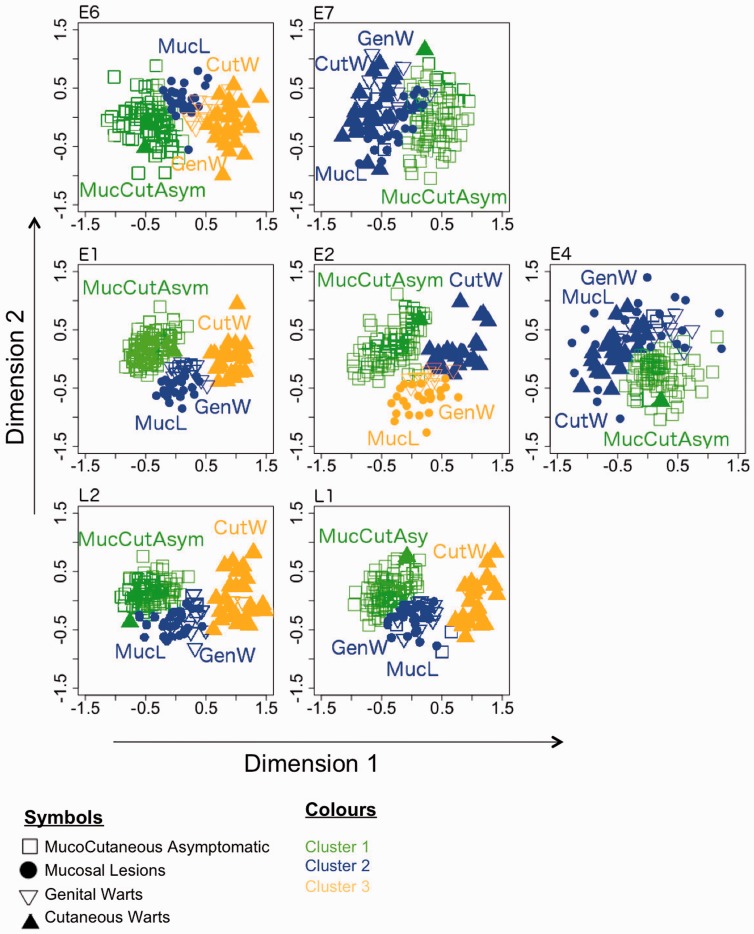
In order to elucidate common patterns on CUPrefs among human PVs, MDS in combination with cluster analysis was performed for each gene independently. The 59 RF variables in the CUPrefs matrix were reduced to two dimensions using MDS procedure (fig. 5). The closer to one another two points lie in this two-dimensional space, the more similar the two corresponding taxa are in terms of codon usage. However, MDS does not classify individuals into clusters, and it is impossible to capture all the variability of the multidimensional data in a lower dimensional display. Hence, in parallel and independently of MDS, we performed a tag-free two-step cluster analysis on the 59 relative frequencies variable matrix for each gene independently. The optimal number of clusters was also inferred blindly using the BIC. The results of the cluster analysis were incorporated into the MDS plot in figure 5 (see also clustering analysis results in table 2).
Fig. 5.—
MDS plot of codon usage differences among HPVs. The different HPVs are classified by an unsupervised two-step clustering algorithm, and visualized by MDS dimension reduction. The clusters are colored yellow (cluster1), blue (cluster 2), and green (cluster 3), respectively. Cluster analysis has been conducted for each gene separately; for this reason, “cluster 1” represents a totally independent set of PVs for each gene. Each symbol represents one individual HPV, and the distance between points is proportional to the overall dissimilarity of CUPrefs. Codes for phenotypic presentation of the infection: Squares, mucocutaneous asymptomatic; dots, mucosal lesions; inverse triangles, genital warts; triangles, cutaneous warts.
Table 2.
Distribution of HPVs by their Clinical Presentation in Each Conglomerate Obtained by Two-Step Cluster Analysis
| E6a, % |
E7, % |
||||||
|---|---|---|---|---|---|---|---|
| Cluster1 | Cluster2 | Cluster3 | Cluster1 | Cluster2 | |||
| MucCutAsym (n = 90) | 97.8 | — | 2.2 | 94.4 | 5.6 | ||
| MucL (n = 25) | — | 100 | — | — | 100 | ||
| GenW (n = 13) | — | — | 100 | — | 100 | ||
| CutW (n = 28) | 3.6 | — | 96.4 | 3.6 | 96.4 | ||
| E1, % |
E2, % |
E4, % |
||||||
|---|---|---|---|---|---|---|---|---|
| Cluster1 | Cluster2 | Cluster3 | Cluster1 | Cluster2 | Cluster3 | Cluster1 | Cluster2 | |
| MucCutAsym (n = 90) | 94.4 | 3.4 | 94.4 | 94.4 | 3.4 | 2.2 | 94.4 | 5.6 |
| MucL (n = 25) | — | 100 | — | — | 100 | — | — | 100 |
| GenW (n = 13) | — | 100 | — | — | 100 | — | — | 100 |
| CutW (n = 28) | 3.6 | — | 3.6 | 3.6 | — | 96.4 | 3.6 | 96.4 |
| L2, % |
L1, % |
|||||||
|---|---|---|---|---|---|---|---|---|
| Cluster1 | Cluster2 | Cluster3 | Cluster1 | Cluster2 | Cluster3 | |||
| MucCutAsym (n = 90) | 94.4 | 3.4 | 2.2 | 94.4 | 3.4 | 2.2 | ||
| MucL (n = 25) | — | 100 | — | — | 100 | — | ||
| GenW (n = 13) | — | 92.3 | 7.7 | — | 84.6 | 15.4 | ||
| CutW (n = 28) | 3.6 | — | 96.4 | 3.6 | — | 96.4 | ||
Table should be read as follows, taking E7 as an example: For E7, two-step cluster analysis identifies two different clusters: Cluster 1 spans 94.4% (n = 85) of MucCutAsym HPVs and 3.6% (n = 1) of HPVs causing CutW. Cluster 2 spans 100% (n = 25) of HPV causing mucosal lesions, 100% (n = 13) of HPVs causing GenW, 96.4% (n = 27) of HPVs causing CutW and 5.6% (n = 5) of HPVs causing MucCutAsym infections.
aCutA in E6, n = 87.
In line with our previous findings (fig. 4), common ancestry did not explain similarity in CUPrefs, as the cluster analysis did not identify the three main genera, Alpha-, Beta-, and GammaPVs (supplementary table S3, Supplementary Material online), for any gene. Instead, the main factor driving codon usage-based grouping was virus clinical presentation, as MucCutAsym including Beta- and GammaPVs appeared together for all genes, both for cluster and MDS analysis. Human PVs in the MuPV genus, which cause cutaneous warts, grouped together with very distant PVs in the AlphaPV genus and associated with similar clinical manifestations. Furthermore, HPV41, the only member in the NuPV genus, which has been associated with cutaneous warts, clustered together with the MucCutAsym group. HPV4 and HPV65, classified as GammaPVs but associated to cutaneous warts, also appeared together with CutW group. The analyses also revealed that for all genes (except for E6, due to the absence of this gene in certain GammaPVs) three GammaPVs (in species Gamma-6) grouped with the phylogenetically distant GenW group. Finally, we found that HPV32 and HPV54, belonging to the GenW group, appeared together with CutW group in L1 and L2 for HPV32 and L1 for HPV54.
Viruses in the AlphaPVs genus (encompassing viruses with different clinical presentations, namely GenW, CutW, and MucL) clustered together only for the E7 and E4 genes, but grouped separately for other genes, according to their clinical presentation (fig. 5). For E6, the main driving factor for CUPrefs was productivity of the infections, as CutW and GenW groups clustered together, separate from MucL. On the other hand, for E1, E2, L2, and L1, the main driving factor seemed to be tropism, as the GenW and MucL groups (both with mucosal tropism) clustered separately from viruses in the CutW group (cutaneous tropism).
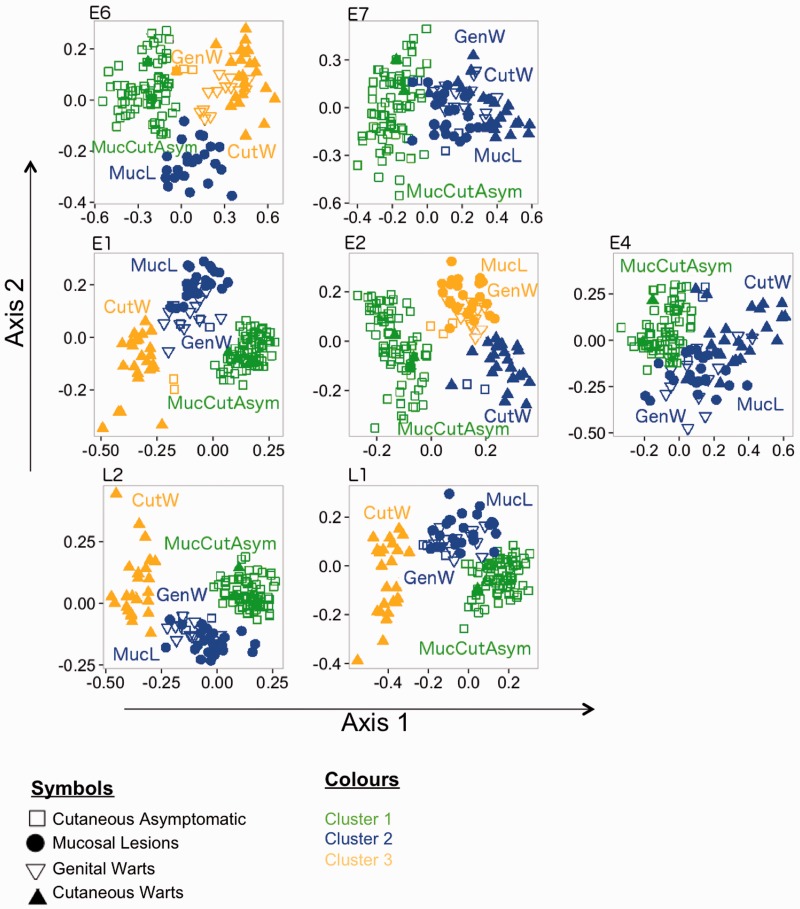
We also performed ICA, a different method of dimensionality reduction, in order to cross-validate the MDS results (fig. 6). ICA is a powerful CA method for the analysis of synonymous codon usage (Suzuki et al. 2008). It is able to dissociate the effects of amino acid composition from the effects directly related to synonymous codon usage. Both approaches, ICA and MDS, rendered globally similar results, further sustaining the claim that clinical presentation was the main driving factor of CUPrefs in human PVs.
Fig. 6.—
ICA plots of synonymous CUPrefs among HPVs. The different HPVs are classified by an unsupervised two-step clustering algorithm. Cluster analysis has been conducted for each gene separately; for this reason, “cluster 1” represents a totally independent set of PVs in for each gene. Each symbol represents an individual HPV, and the distance between points is proportional to the overall dissimilarity of CUPrefs. Codes for phenotypic presentation of the infection: Squares, mucocutaneous asymptomatic; dots, mucosal lesions; inverse triangles, genital warts; triangles, cutaneous warts.
Differences in GC Content in the Third Position and in Codon Adaptation Index Partly Account for Codon Usage Preferences
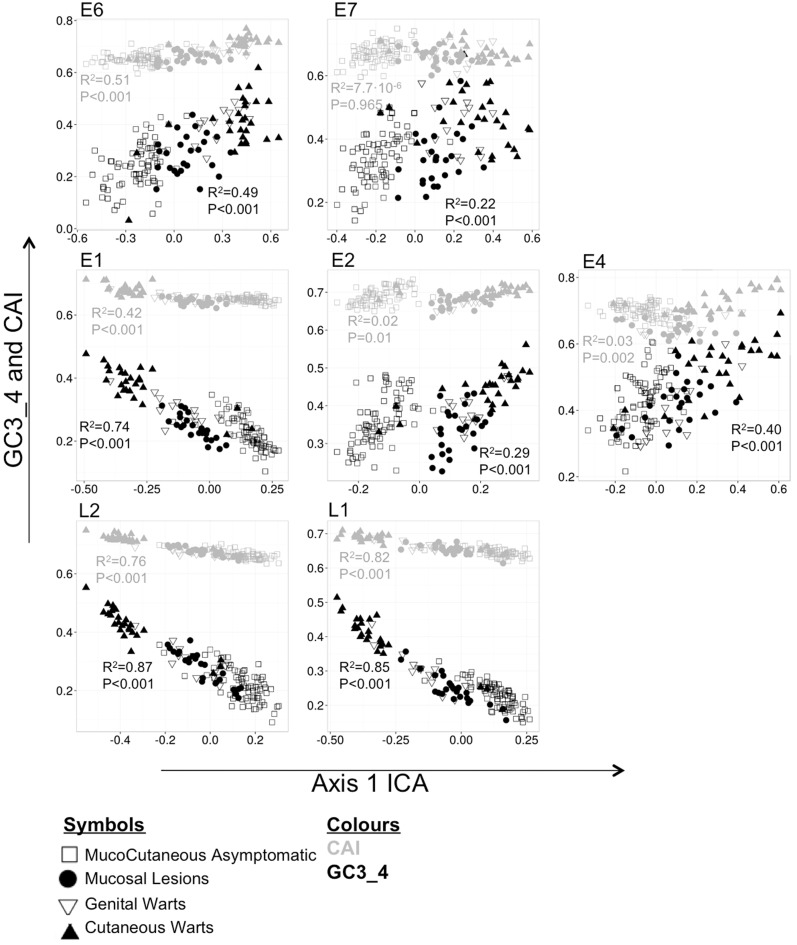
In order to identify major sources of variation among genes on the axes generated by ICA of codon usage data, we assessed separately for each gene the linear correlation between projections on the first principal axis and values of CAI and of average GC content in the third codon position of 4-fold degenerate codon families and in the 4-fold component of 6-fold degenerate codon families (GC3_4) (fig. 7supplementary figs. S5–S11, Supplementary Material online). Both CAI and average GC content appeared as important sources of variation in L1 and L2 genes. For E6 and E1 genes, only GC content correlated significantly with the first ICA axis. For E7, E2, and E4, none of these gene features correlated with the first axis of the ICA. These results demonstrate that both gene features contribute to variations in synonymous codon usage among genes, but their contributions vary among different genes as also may vary the correlations between GC3_4 content and CAI.
Fig. 7.—
Projections of the first principal axis in combination with either CAI or GC content at the third position of 4-fold degenerated codon families and at the 4-fold component of the 6-fold degenerated codon families (GC3_4). Codes for phenotypic presentation of the infection: Squares, mucocutaneous asymptomatic; dots, mucosal lesions; inverse triangles, genital warts; triangles, cutaneous warts.
Differences in Gene Temporal Expression Largely Determine Differential Codon Usage Preferences
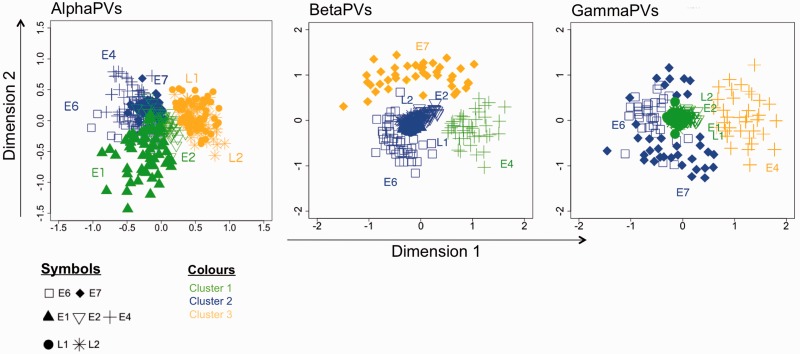
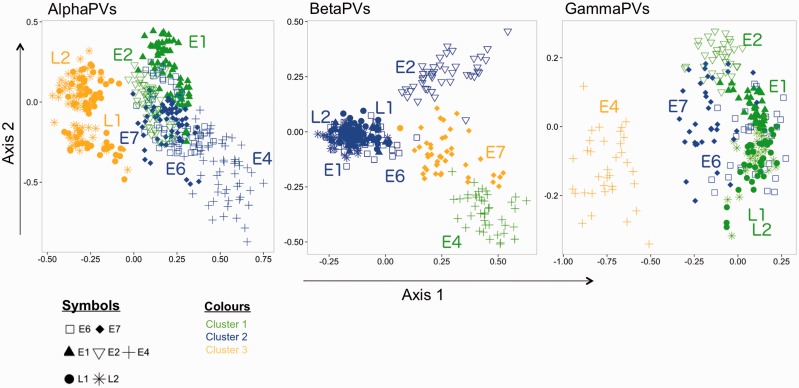
Finally, we approached the connection between CUPrefs of the different genes and the corresponding gene expression patterns, analyzing separately the three main genera, that is, Alpha-, Beta-, and GammaPVs, by means of unsupervised two-step clustering. For each genus, the optimal number of clusters was automatically determined using the BIC criterion. For every genus, gene temporal expression was the main factor driving data clustering (fig. 8table 3), and genes expressed simultaneously during virus life cycle showed similar CUPrefs and clustered together. For AlphaPVs, the late genes largely differed in CUPrefs from the early genes, which showed a larger variability. Two-step clustering also clustered late stage genes L1 and L2 together for BetaPVs and GammaPVs in combination with some early stage genes (fig. 8). We confirmed the results obtained by MDS using ICA, which however shows subtle differences in particular for the E2 gene (fig. 9).
Fig. 8.—
MDS plot of codon usage differences for HPV genes within each genus. HPVs are classified by an unsupervised two-step clustering algorithm and visualized by MDS dimension reduction. The clusters are colored yellow (cluster1), blue (cluster 2), and green (cluster 3), respectively. Codes for genes: Squares, E6; diamonds, E7; triangles, E1; inverse triangles, E2; crosses, E4; stars, L2; circles, L1.
Table 3.
Distribution of Genes Stratified by Genera in Each Conglomerate of the Two-Step Cluster Analysis
| AlphaPVs, % (n = 63) |
BetaPVs, % (n = 45) |
GammaPVsa, % (n = 45) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Cluster 1 | Cluster 2 | Cluster 3 | Cluster 1 | Cluster 2 | Cluster 3 | Cluster 1 | Cluster 2 | Cluster 3 | |
| E6 | 100 | — | — | — | 100 | — | 100 | — | — |
| E7 | 100 | — | — | 100 | — | — | 100 | — | — |
| E1 | — | 100 | — | — | 100 | — | — | 100 | — |
| E2 | — | 100 | — | — | 100 | — | — | 100 | — |
| E4 | 100 | — | — | — | — | 100 | — | — | 100 |
| L2 | — | — | 100 | — | 100 | — | — | 100 | — |
| L1 | — | — | 100 | — | 100 | — | — | 100 | — |
Note.—Table should be read as follows taking AlphaPVs as an example: For AlphaPVs, two-step cluster analysis distinguishes three different clusters. Cluster 1 groups E6, E7, and E4 genes. Cluster 2 groups E1 and E2 genes. Finally, cluster 3 groups L1 and L2 genes.
aFor E6, n = 42.
Fig. 9.—
ICA showing variance of synonymous CUPrefs between genes of HPV genera. The difference between genes in terms of synonymous codon usage is shown for Alpha-, Beta-, and GammaPVs (from left to right). Codes for genes: Squares, E6; diamonds, E7; triangles, E1; inverse triangles, E2; crosses, E4; stars, L2; circles, L1.
Discussion
We have sought to understand the differential contribution of the different evolutionary forces shaping human PV CUPrefs. We first assessed adaptation to the host’s CUPrefs, and showed that all genes in human PVs display poor adaptation to human CUPrefs. Our null, most parsimonious, hypothesis was that CUPrefs in human PVs should be close to CUPrefs in humans, because: 1) PVs do not encode for any element involved in translation and rely completely on their host’s cell machinery to translate their genes and 2) the relationship of these viruses with their amniote hosts lineage is ancient (Garcia-Vallve et al. 2005; Gottschling et al. 2011; Bravo and Felez-Sanchez 2015). Deviation from host’s CUPrefs may lead to inefficient translation, in terms of decreased amount of translated protein, decreased quality of the synthesized protein, and/or decreased amount of properly folded protein (Bravo and Müller 2005; Drummond and Wilke 2008).
We tried then to evaluate whether closely related PVs displayed closely related CUPrefs. Our analyses revealed however that evolutionary distances between PVs explain only a small fraction of CUPref variation among human PVs (fig. 4). Similar analyses in Herpesviruses had also shown that codon usage cannot be explicitly tied to species evolution (Roychoudhury and Mukherjee 2009). Furthermore, we found that Euclidean distances within AlphaPVs based on CUPrefs were larger than within BetaPVs or within GammaPVs distances (supplementary fig. S12, Supplementary Material online). We interpret that large Euclidean CUPrefs-based distances between AlphaPVs reflect the broad diversity in clinical presentations for these viruses, spanning viruses with cutaneous and mucosal tropisms. Our results suggest that clinical variables related to tropism, productivity, and immune exposure could be responsible for variation in CUPrefs in PVs. Indeed, detailed analyses by means of different methods of dimensionality reduction (MDS and ICA) showed that orthologous genes in viruses with similar clinical presentation display similar patterns of CUPrefs.
A major finding obtained through unsupervised cluster analysis of viral CUPrefs was an almost perfect match between groups of viruses showing similar CUPrefs and the clinical, phenotypic presentation of the infection in terms of tropism, productivity, and potential for malignization (figs. 56). A reduced number of human PVs were initially possible exceptions to this pattern, as they presented a tissue tropism different from the rest of their sister taxa. However, a closer analysis showed that indeed viruses with exceptional tropism also displayed exceptional CUPrefs compared with their genetically close counterparts: 1) Gamma-6 PVs phylogenetically group with asymptomatic cutaneous GammaPVs, but were isolated from cervical lesions (Chen et al. 2007; Nobre et al. 2008), and CUPrefs of Gamma-6 PVs are similar in all six genes to those of AlphaPVs causing GenW, and not to GammaPVs; 2) two members of Gamma-1 PVs (HPV2 and HPV65) exhibited similar CUPrefs to those of CutWarts, consistent with their association with skin warts (Chen et al. 1993; Iftner et al. 2003); and 3) MuPVs, causing cutaneous warts, shared common CUPrefs in all genes with AlphaPVs also causing cutaneous warts, although these viruses show large phylogenetic distances. Remarkably, HPV41, the only member of NuPV genus, showed similar CUPrefs to viruses causing MucCutAsym infections. Although this virus has been associated to cutaneous warts, it has also been associated with squamous cell carcinoma lesions as it has also been the case for some BetaPVs (Forslund et al. 2007).
We tried finally to evaluate whether gene expression pattern during the virus life cycle could also influence CUPrefs. We found a strikingly sharp pattern with genes expressed at similar stages of the viral infection cycle sharing similar CUPrefs (figs. 78). In AlphaPVs, genes clustered separately into three groups depending on their CUPrefs. Early genes involved in replication exhibited similar patterns of codon usage, which differ from codon usage patterns of oncogenes (E4, E6, and E7). Finally, structural genes (L1 and L2) expressed in differentiated keratinocytes also shared patterns of CUPrefs. It had been described that early genes (E1 and E2) in AlphaPVs differed in codon usage from late genes (L1 and L2) (Cladel et al. 2010). Our results thus suggest that in AlphaPVs, differential CUPrefs match differences in temporal expression. Such differences may reflect changes in tRNA availability, as it has been reported that keratinocytes express different tRNA profiles as they differentiate (Zhao et al. 2005). In Beta- and GammaPVs, the gene clustering based on CUPrefs was different from that found in AlphaPVs. Life cycle in Beta- and GammaPVs has not been studied in detail and little information is available about the temporal expression of the genes from these viruses (Doorbar et al. 2012). Their transcription patterns, life cycle, and other molecular characteristics are often inferred by homology with those of the best-described AlphaPVs. This might lead to an overgeneralization of the HPV biology, overlooking the fact that different PVs within a given genus are not genetically homogeneous and that different PVs infecting the same host (here human PVs within Alpha- and BetaPVs) might indeed present different biologies (Cladel et al. 2010; Bravo and Felez-Sanchez 2015). We propose that studying CUPrefs of PVs infecting the same host could allow inferring life-cycle characteristics that could then be experimentally explored.
The E4 gene is expressed following a splice event including a small number of amino acids from the E1 gene, while most of the coding sequence overlaps the hinge region of the E2 gene in a different reading frame. The E2 hinge region is a flexible connector between the functionally conserved transcription activation domain and the DNA-binding domain (fig. 3A) (Ham et al. 1991; Eriksson et al. 1999). We have evaluated the influence of the overlapping nature of the E2 and E4 reading frames on their CUPrefs (fig. 3). Our analyses revealed first that for all clinical manifestations the E4 gene was less adapted to human CUPrefs than the E2 hinge region; and second that the E2 nonoverlapping region was also significantly less adapted than the hinge region (fig. 3B). This is a striking result for two main reasons. On the one hand, because previous studies on the differential evolution of these overlapping ORFs had shown that the E4 region presents an excess of synonymous mutations, compared with and excess of nonsynonymous mutation in the E2 hinge region (Hughes AL and Hughes MA 2005; Narechania et al. 2005). Hence, diversifying selection in the hinge region of E2 coexists with purifying selection on the overlapping region of E4 (Narechania et al. 2005). In this context, our results suggest that purifying selection in E4 concurs with deadaptation to human CUPrefs, thus implying that lowering CAI values to the human average could indeed be adaptive. On the other hand because the E2 hinge region is essentially disordered but displays the highest CAI values to the human average (Rancurel et al. 2009)This is in sharp contrast with previous findings showing that disordered proteins tend to be encoded by genes with poor CUPrefs (Zhou et al. 2015). Overall our results for the E2/E4 gene pair suggest that we are still far from understanding the evolutionary interplay between overlapping genes, and that CUPrefs may play a major role that may not be captured by the standard dN/dS analyses.
The evolutionary forces shaping CUPrefs are basically mutation and/or selection, and they are not always easy to disentangle. Mutational pressures are produced by differential probability of different nucleotide exchange types, leading to nucleotide composition biases and eventually shaping CUPrefs (Shackelton et al. 2006; Belalov and Lukashev 2013). We have shown that nucleotide composition of human PV genes differs from that of human genes, in line with previous results (Bravo and Müller 2005; Cladel et al. 2010). Our analysis also revealed a correlation between GC composition and CUPrefs of different viral clinical presentations for the E1, E6, L1, and L2 genes (fig. 7). As some authors assume that GC3_4 composition reflects to some extent mutational pressure, this correlation would suggest that mutational pressures have a role patterning CUPrefs of human PVs genes, as it has been proposed for other DNA viruses (Shackelton et al. 2006). However, PVs do not encode for any element of the genome replication machinery and rely instead in cellular polymerases for replication (Park et al. 1994). Mutational biases associated to viral DNA replication should thus a priori be similar to those associated with cellular DNA replication, but it does not seem to be the case. Several mutation-based explanations between viral and cellular replication may account for the observed compositional differences. First, nucleotide composition biases in the human genome reflect mutational biases during replication in the germinal line, while nucleotide composition biases in PVs reflect mutational biases during replication in somatic cells (Martincorena et al. 2015). Analogously, the spectrum of mutations accumulated in human cancers is different from that arising through common ancestry (Temiz et al. 2015). Second, PV DNA is the target of the APOBEC3 internal mutators, a family of cellular cytidine deaminases that introduce directional C > T substitutions (Vartanian et al. 2008). The APOBEC3 locus has been under strong selection in the primate lineage, possibly reflecting an arms race between virus and host (Munk et al. 2012). In this sense, the decreased GC content in human PVs, but also in most human DNA viruses, could reflect the sustained edition pressure by the APOBEC3 proteins (Vartanian et al. 2010). Additionally, some experimental studies (Chen et al. 2010; Turabelidze et al. 2014) have shown differences in APOBEC3 expression patterns between oral mucosa and cutaneous skin, thus allowing for differential edition of PV genomes depending on their tropism. Third, viral genome replication in PVs infecting sun-exposed keratinocytes will be subject to a higher mutation rate via the error-prone mechanisms of resolution and repair of cyclobutane pyrimidine dimers (Protic-Sabljic et al. 1986). Thus, nucleotide bias in human PV genes with respect to human genes may be accounted for to some extent by differences in biochemical environment and in gene expression context in infected cells compared with the germinal line.
Regarding adaptation, CUPrefs can increase viral fitness by directly modulating protein production and by indirectly modulating immune exposure and expression timing. The counterintuitive result of selection on PV CUPrefs is that expression of PV genes in human cell culture from the wild-type gene sequence leads to very low protein amounts, independently of the promoter and the cell line used. Instead, high gene expression levels are only achieved when the gene sequence has been “humanized” by modification of CUPrefs (Liu et al. 2002; Cid-Arregui et al. 2003; Disbrow et al. 2003; Mossadegh et al. 2004; Samorski et al. 2006; Gruener et al. 2007; Cladel et al. 2008). We communicate here a correlation between the adaptation to human codon usage and CUPrefs of different viral clinical presentations in E6, L1, and L2 (fig. 7), suggesting an important role of adaptation in shaping CUPrefs of at least these genes in human PVs. Our analyses reveal that the only exceptions to the systematic CUPrefs maladaptation are the capsid genes (L1 and L2) of human PVs causing cutaneous warts, which show the highest similarity to human CUPrefs. We interpret that the observed variation in mismatch between human PV genes and human CUPrefs is related to differential virus clinical presentation. Cutaneous warts are very productive lesions, and the infected keratinocytes contain a large amount of virions that are released when dead cells shed off from skin surface (Doorbar et al. 2012). Productive lesions require large amounts of the capsid proteins and we propose that the increased similarity with human CUPrefs in late genes of PVs causing cutaneous warts enhances gene expression by facilitating capsid protein synthesis. For viruses in other viral families, the highest levels of adaptation of viral proteins to host’s CUPrefs are also observed for proteins appearing abundantly in the virion (Karlin et al. 1990; Bahir et al. 2009). Although GenW are also productive lesions, capsid genes in human PVs responsible for these infections do not show a higher adaptation to human CUPrefs. We suggest that differences on their human codon usage adaptation may arise from differences in productivity between the two wart types, as it has been demonstrated that human PVs causing GenW produce fewer virions compared with those producing cutaneous warts (Peh et al. 2002). In contrast, Beta- and GammaPVs also have a mucocutaneous tropism but different clinical presentation (asymptomatic infections) and showed lower adaptation to human CUPrefs in the capsid components of the virion (L1 and L2). CUPref maladaptation could be adaptive if decreased protein synthesis lead to a less intense host immune response (Tindle 2002). Because L1 and L2 are the most immunogenic proteins (Hibma 2012), we propose that limiting the expression of structural proteins by means of codon usage maladaptation allows these viruses to better escape immune surveillance for a prolonged period of time without compromising their life cycle. These findings are supported by previous reports showing that seroprevalence against Beta- and GammaPVs exhibits a delayed but long-lasting antibody response: the immune response was low in children and increased continuously with age (Iftner et al. 2010). In human PVs causing MucCutAsym infections, the highest level of adaptation to human CUPrefs was found in the E4 gene. In some cutaneous infections, E4 can be expressed at higher levels than the virion coat proteins, and can account for as much as 30% of the total protein content (Doorbar 2013).
We hypothesize that an evolutionary trade-off exists in virus clinical presentation between a potential for strong gene expression and a potential for eliciting strong immune responses. Modulation of viral CUPrefs with respect to the host’s CUPrefs may help push the equilibrium in one direction or another. For viruses associated to chronic infections, such as human PVs, the adaptive strategy could thus be to sacrifice virion productivity to avoid the generation of strong, protective immune responses, resulting in long-lasting infections and allowing for reinfection of a previously infected host. For viruses associated to acute infections, on the contrary, large virion production is accompanied by induction of a strong immune response that may eventually render the infected host nonsusceptible to subsequent reinfections by closely related viruses. Experimental evidence of innate immune activity of the schlafen 11 gene against viral infections further sustains our hypothesis. Expression of schlafen 11 is triggered by cellular exposure to interferon, as a response to viral infections (Sohn et al. 2007). The activity of schlafen 11 protein is to selectively inhibit translation from mRNAs enriched in A/T-ending codons (Li et al. 2012), and many viruses infecting mammals are enriched precisely in A/T-ending codons (Jenkins and Holmes 2003; Shackelton et al. 2006). The result is that, as a response against a viral infection, the cellular machinery shuts down specifically the translation of transcripts that are potentially of viral origin, using CUPrefs as a guide for pinpointing viral transcripts.
In summary, we have presented here a thorough analysis of CUPrefs in human PV genes, connecting codon preferences with virus infection clinical presentation and with gene expression patterns. We have shown that, for viruses with a well-characterized infection cycle, genes expressed simultaneously tend to show similar CUPrefs. Furthermore, closely related viruses did not necessarily display closely related CUPrefs, while orthologous genes in distantly related viruses but with similar tropism tend to show similar CUPrefs. Finally, we propose that modulation of viral CUPrefs, as a result of differential mutation and/or selection pressures, may have an adaptive value, as they may strongly condition expression efficiency, virion production, immune exposure, and propensity toward chronic/acute virus lifestyle. Comparative research, with insight into the different life-history traits of virus lifestyle and not remaining merely on descriptions of preferences, will be required to elucidate the role and the evolutionary forces fuelling the evolution of viral CUPrefs.
Supplementary Material
Supplementary tables S1–S4 and files S1–S12 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the disappeared Spanish Ministry for Science and Innovation (grant CGL2010-16713 to I.G.B.), and by grants by the IDIBELL (PhD fellowship to M.F.S.), by the Dexeus Foundation for Women's Health (to I.G.B.) and by the Adolf Messer Stiftung (personal grant to J.H.T.).
Literature Cited
- Akashi H, Eyre-Walker A. 1998. Translational selection and molecular evolution. Curr Opin Genet Dev. 8:688–693. [DOI] [PubMed] [Google Scholar]
- Aota S, Ikemura T. 1986. Diversity in G + C content at the third position of codons in vertebrate genes and its cause. Nucleic Acids Res. 14:6345–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragones L, Bosch A, Pinto RM. 2008. Hepatitis A virus mutant spectra under the selective pressure of monoclonal antibodies: codon usage constraints limit capsid variability. J Virol. 82:1688–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragones L, Guix S, Ribes E, Bosch A, Pinto RM. 2010. Fine-tuning translation kinetics selection as the driving force of codon usage bias in the hepatitis A virus capsid. PLoS Pathog. 6:e1000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahir I, Fromer M, Prat Y, Linial M. 2009. Viral adaptation to host: a proteome-based analysis of codon usage and amino acid preferences. Mol Syst Biol. 5:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell MA, et al. 1991. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J Virol. 65:2254–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belalov IS, Lukashev AN. 2013. Causes and implications of codon usage bias in RNA viruses. PLoS One 8:e56642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard HU, et al. 2010. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401:70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo IG, Felez-Sanchez M. 2015. Papillomaviruses: viral evolution, cancer and evolutionary medicine. Evol Med Public Health. 2015:32–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo IG, Müller M. 2005. Codon usage in papillomavirus genes: practical and functional aspects. Papillomavirus Rep. 16:63–72. [Google Scholar]
- Bulmer M. 1991. The selection-mutation-drift theory of synonymous codon usage. Genetics 129:897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Chen L, et al. 2010. Positional differences in the wound transcriptome of skin and oral mucosa. BMC Genomics 11:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Lee W, Hottes AK, Shapiro L, McAdams HH. 2004. Codon usage between genomes is constrained by genome-wide mutational processes. Proc Natl Acad Sci U S A. 101:3480–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Tsao YP, Lee JW, Sheu WC, Liu YT. 1993. Characterization and analysis of human papillomaviruses of skin warts. Arch Dermatol Res. 285:460–465. [DOI] [PubMed] [Google Scholar]
- Chen Z, Schiffman M, Herrero R, Desalle R, Burk RD. 2007. Human papillomavirus (HPV) types 101 and 103 isolated from cervicovaginal cells lack an E6 open reading frame (ORF) and are related to gamma-papillomaviruses. Virology 360:447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid-Arregui A, Juarez V, zur Hausen H. 2003. A synthetic E7 gene of human papillomavirus type 16 that yields enhanced expression of the protein in mammalian cells and is useful for DNA immunization studies. J Virol. 77:4928–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cladel NM, Bertotto A, Christensen ND. 2010. Human alpha and beta papillomaviruses use different synonymous codon profiles. Virus Genes 40:329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cladel NM, Hu J, Balogh KK, Christensen ND. 2008. CRPV genomes with synonymous codon optimizations in the CRPV E7 gene show phenotypic differences in growth and altered immunity upon E7 vaccination. PLoS One 3:e2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox TF, Cox MAA. 1994. Multidimensional scaling. London: Chapman & Hall. [Google Scholar]
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. 2004. Classification of papillomaviruses. Virology 324:17–27. [DOI] [PubMed] [Google Scholar]
- Disbrow GL, Sunitha I, Baker CC, Hanover J, Schlegel R. 2003. Codon optimization of the HPV-16 E5 gene enhances protein expression. Virology 311:105–114. [DOI] [PubMed] [Google Scholar]
- Doorbar J. 2013. The E4 protein; structure, function and patterns of expression. Virology 445:80–98. [DOI] [PubMed] [Google Scholar]
- Doorbar J, et al. 2012. The biology and life-cycle of human papillomaviruses. Vaccine 30(Suppl 5):F55–F70. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. 2008. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L. 2000. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 16:287–289. [DOI] [PubMed] [Google Scholar]
- Duret L. 2002. Detecting genomic features under weak selective pressure: the example of codon usage in animals and plants. Bioinformatics 18(Suppl 2):S91. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson A, Herron JR, Yamada T, Wheeler CM. 1999. Human papillomavirus type 16 variant lineages characterized by nucleotide sequence analysis of the E5 coding segment and the E2 hinge region. J Gen Virol. 80(Pt 3):595–600. [DOI] [PubMed] [Google Scholar]
- Forslund O, et al. 2007. Cutaneous human papillomaviruses found in sun-exposed skin: beta-papillomavirus species 2 predominates in squamous cell carcinoma. J Infect Dis. 196:876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Vallve S, Alonso A, Bravo IG. 2005. Papillomaviruses: different genes have different histories. Trends Microbiol. 13:514–521. [DOI] [PubMed] [Google Scholar]
- Gingold H, Pilpel Y. 2011. Determinants of translation efficiency and accuracy. Mol Syst Biol. 7:481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling M, et al. 2011. Quantifying the phylodynamic forces driving papillomavirus evolution. Mol Biol Evol. 28:2101–2113. [DOI] [PubMed] [Google Scholar]
- Gruener M, Bravo IG, Momburg F, Alonso A, Tomakidi P. 2007. The E5 protein of the human papillomavirus type 16 down-regulates HLA-I surface expression in calnexin-expressing but not in calnexin-deficient cells. Virol J. 4:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, et al. 2004. tRNASer(CGA) differentially regulates expression of wild-type and codon-modified papillomavirus L1 genes. Nucleic Acids Res. 32:4448–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham J, Dostatni N, Gauthier JM, Yaniv M. 1991. The papillomavirus E2 protein: a factor with many talents. Trends Biochem Sci. 16:440–444. [DOI] [PubMed] [Google Scholar]
- Hibma MH. 2012. The immune response to papillomavirus during infection persistence and regression. Open Virol J. 6:241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Hughes MA. 2005. Patterns of nucleotide difference in overlapping and non-overlapping reading frames of papillomavirus genomes. Virus Res. 113:81–88. [DOI] [PubMed] [Google Scholar]
- Iftner A, et al. 2003. The prevalence of human papillomavirus genotypes in nonmelanoma skin cancers of nonimmunosuppressed individuals identifies high-risk genital types as possible risk factors. Cancer Res. 63:7515–7519. [PubMed] [Google Scholar]
- Iftner T, et al. 2010. Prevalence of low-risk and high-risk types of human papillomavirus and other risk factors for HPV infection in Germany within different age groups in women up to 30 years of age: an epidemiological observational study. J Med Virol. 82:1928–1939. [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Holmes EC. 2003. The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Res. 92:1–7. [DOI] [PubMed] [Google Scholar]
- Karlin S, Blaisdell BE, Schachtel GA. 1990. Contrasts in codon usage of latent versus productive genes of Epstein-Barr virus: data and hypotheses. J Virol. 64:4264–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent P, Kongsted A. 2012. Identifying clinical course patterns in SMS data using cluster analysis. Chiropr Man Therap. 20:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. 2002. The human genome browser at UCSC. Genome Res. 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, et al. 2012. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 491:125–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, et al. 2002. Codon modified human papillomavirus type 16 E7 DNA vaccine enhances cytotoxic T-lymphocyte induction and anti-tumour activity. Virology 301:43–52. [DOI] [PubMed] [Google Scholar]
- Longworth MS, Laimins LA. 2004. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol Mol Biol Rev. 68:362–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin A, Bertranpetit J, Oliver JL, Medina JR. 1989. Variation in G + C-content and codon choice: differences among synonymous codon groups in vertebrate genes. Nucleic Acids Res. 17:6181–6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, et al. 2015. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348:880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama EN, Powell JR. 1997. Codon usage bias and tRNA abundance in Drosophila. J Mol Evol. 45:514–523. [DOI] [PubMed] [Google Scholar]
- Mossadegh N, et al. 2004. Codon optimization of the human papillomavirus 11 (HPV 11) L1 gene leads to increased gene expression and formation of virus-like particles in mammalian epithelial cells. Virology 326:57–66. [DOI] [PubMed] [Google Scholar]
- Munk C, Willemsen A, Bravo IG. 2012. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol Biol. 12:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Gojobori T, Ikemura T. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narechania A, Terai M, Burk RD. 2005. Overlapping reading frames in closely related human papillomaviruses result in modular rates of selection within E2. J Gen Virol. 86:1307–1313. [DOI] [PubMed] [Google Scholar]
- Nindl I, Gottschling M, Stockfleth E. 2007. Human papillomaviruses and non-melanoma skin cancer: basic virology and clinical manifestations. Dis Markers. 23:247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobre RJ, de Almeida LP, Martins TC. 2008. Complete genotyping of mucosal human papillomavirus using a restriction fragment length polymorphism analysis and an original typing algorithm. J Clin Virol. 42:13–21. [DOI] [PubMed] [Google Scholar]
- Park P, et al. 1994. The cellular DNA polymerase alpha-primase is required for papillomavirus DNA replication and associates with the viral E1 helicase. Proc Natl Acad Sci U S A. 91:8700–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peh WL, et al. 2002. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J Virol. 76:10401–10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G. 2011. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 12:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protic-Sabljic M, et al. 1986. UV light-induced cyclobutane pyrimidine dimers are mutagenic in mammalian cells. Mol Cell Biol. 6:3349–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancurel C, Khosravi M, Dunker AK, Romero PR, Karlin D. 2009. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J Virol. 83:10719–10736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP. 2004. Codon usage bias from tRNA’s point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 14:2279–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhury S, Mukherjee D. 2009. A detailed comparative analysis on the overall codon usage pattern in herpesviruses. Virus Res. 148:31–43. [DOI] [PubMed] [Google Scholar]
- Samorski R, Gissmann L, Osen W. 2006. Codon optimized expression of HPV 16 E6 renders target cells susceptible to E6-specific CTL recognition. Immunol Lett. 107:41–49. [DOI] [PubMed] [Google Scholar]
- Shackelton LA, Parrish CR, Holmes EC. 2006. Evolutionary basis of codon usage and nucleotide composition bias in vertebrate DNA viruses. J Mol Evol. 62:551–563. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Averof M, Lloyd AT, Matassi G, Peden JF. 1995. DNA sequence evolution: the sounds of silence. Philos Trans R Soc Lond B Biol Sci. 349:241–247. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH. 1986. Codon usage in regulatory genes in Escherichia coli does not reflect selection for ‘rare’ codons. Nucleic Acids Res. 14:7737–7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. 1987. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15:1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields DC, Sharp PM. 1987. Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases. Nucleic Acids Res. 15:8023–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn WJ, et al. 2007. Novel transcriptional regulation of the schlafen-2 gene in macrophages in response to TLR-triggered stimulation. Mol Immunol. 44:3273–3282. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenico M, Lloyd AT, Sharp PM. 1994. Codon usage in Caenorhabditis elegans: delineation of translational selection and mutational biases. Nucleic Acids Res. 22:2437–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, et al. 2005. CpG RNA: identification of novel single-stranded RNA that stimulates human CD14+CD11c+ monocytes. J Immunol. 174:2273–2279. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Brown CJ, Forney LJ, Top EM. 2008. Comparison of correspondence analysis methods for synonymous codon usage in bacteria. DNA Res. 15:357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temiz NA, et al. 2015. The somatic autosomal mutation matrix in cancer genomes. Hum Genet. 134(8):851–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindle RW. 2002. Immune evasion in human papillomavirus-associated cervical cancer. Nat Rev Cancer. 2:59–65. [DOI] [PubMed] [Google Scholar]
- Turabelidze A, et al. 2014. Intrinsic differences between oral and skin keratinocytes. PLoS One 9:e101480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian JP, Guetard D, Henry M, Wain-Hobson S. 2008. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science 320:230–233. [DOI] [PubMed] [Google Scholar]
- Vartanian JP, et al. 2010. Massive APOBEC3 editing of hepatitis B viral DNA in cirrhosis. PLoS Pathog. 6:e1000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao KN, Gu W, Fang NX, Saunders NA, Frazer IH. 2005. Gene codon composition determines differentiation-dependent expression of a viral capsid gene in keratinocytes in vitro and in vivo. Mol Cell Biol. 25:8643–8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao KN, Liu WJ, Frazer IH. 2003. Codon usage bias and A+T content variation in human papillomavirus genomes. Virus Res. 98:95–104. [DOI] [PubMed] [Google Scholar]
- Zheng ZM, Baker CC. 2006. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci. 11:2286–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu WJ, Peng SW, Sun XY, Frazer I. 1999. Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. J Virol. 73:4972–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Wang T, Fu J, Xiao G, Liu Y. 2015. Non-optimal codon usage influences protein structure in intrinsically disordered regions. Mol Microbiol. Advance Access published June 25, 2015; doi: 10.1111/mmi.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.