Abstract
Obligate bacterial symbionts are widespread in many invertebrates, where they are often confined to specialized host cells and are transmitted directly from mother to progeny. Increasing numbers of these bacteria are being characterized but questions remain about their population structure and evolution. Here we take a comparative genomics approach to investigate two prominent bacterial symbionts (BFo1 and BFo2) isolated from geographically separated populations of western flower thrips, Frankliniella occidentalis. Our multifaceted approach to classifying these symbionts includes concatenated multilocus sequence analysis (MLSA) phylogenies, ribosomal multilocus sequence typing (rMLST), construction of whole-genome phylogenies, and in-depth genomic comparisons. We showed that the BFo1 genome clusters more closely to species in the genus Erwinia, and is a putative close relative to Erwinia aphidicola. BFo1 is also likely to have shared a common ancestor with Erwinia pyrifoliae/Erwinia amylovora and the nonpathogenic Erwinia tasmaniensis and genetic traits similar to Erwinia billingiae. The BFo1 genome contained virulence factors found in the genus Erwinia but represented a divergent lineage. In contrast, we showed that BFo2 belongs within the Enterobacteriales but does not group closely with any currently known bacterial species. Concatenated MLSA phylogenies indicate that it may have shared a common ancestor to the Erwinia and Pantoea genera, and based on the clustering of rMLST genes, it was most closely related to Pantoea ananatis but represented a divergent lineage. We reconstructed a core genome of a putative common ancestor of Erwinia and Pantoea and compared this with the genomes of BFo bacteria. BFo2 possessed none of the virulence determinants that were omnipresent in the Erwinia and Pantoea genera. Taken together, these data are consistent with BFo2 representing a highly novel species that maybe related to known Pantoea.
Keywords: thrips, BFo, Frankliniella, symbiont, genome, evolution
Introduction
The Erwinia and Pantoea genera, within the Enterobacteriaceae, contain common human pathogens, insect symbionts, and phytopathogens (Baumler et al. 2013). There is increasing evidence that previously unknown bacteria isolated from a wide variety of insects belong to this family of bacteria (Husník et al. 2011). These include bacterial symbionts of insects found across a broad range of niches (Allen et al. 2007). However, due to the vastness of the many insect orders and the often highly rearranged genomes of even closely related bacterial species, a comprehensive understanding of these relationships remains to be defined.
Nevertheless, there are a limited number of studies reporting genetic information on the symbiotic relationships between bacteria and their arthropod hosts—helped by technical advances in whole-genome sequencing and analysis. Thus, researchers are now able to amass a significant volume of information on the evolution and relatedness of insect symbiotic lineages. For example, comparative genomics of heritable insect endosymbionts often reveals large scale reductive evolution and fast evolving genomes (Van Ham et al. 2003), often associated with endosymbioses. Despite the emerging picture that other symbiotic bacteria may also be undergoing similar evolutionary processes (Nikoh et al. 2011), many questions remain, particularly in reporting genomic features of gut residing bacteria (Kikuchi et al. 2009).
The western flower thrips (WFT), Frankliniella occidentalis (Pergande) (Thysanoptera: Thripidae), is a globally distributed insect pest causing significant damage to greenhouse-grown crops. Thrips infestations typically lead to reduced aesthetics and lowered yield (Kirk and Terry 2003), principally because their method of feeding causes damage to leaves and fruit, thus reducing the marketability of commercial crops. Moreover, WFT carry tospoviruses, including tomato spotted wilt virus (TSWV) (Jensen 2000a, 2000b; Pappu et al. 2009) and, although WFT are asymptomatic carriers of TSWV, it has been estimated that the annual loss to agriculture caused by TSWV alone amounts to $1 billion (Prins and Goldbach 1998). In addition, WFT also harbor two bacterial symbionts that have been shown to reside within the gut lumen. de Vries, Jacobs, et al. (2001) explored the hindgut of each life-stage of WFT and reported two predominant bacterial symbionts—designated BFo1 and BFo2 (Bacteria, F. occidentalis). Interestingly, transmission of these bacteria occurs during oviposition and probing during feeding; especially where the sites of feeding and egg laying are infected with BFo-contaminated thrips feces (de Vries, Jacobs, et al. 2001; de Vries 2010). However, for the latter, it is unclear how long, if at all, BFo bacteria are able to survive outside the host. Nevertheless, BFo bacteria appear to be important to F. occidentalis. Indeed, the fact that BFo bacteria have been isolated from geographically isolated, wild and greenhouse populations of F. occidentalis—including California, Germany, the Netherlands and the United Kingdom (Chanbusarakum and Ullman 2008, 2009; and this study)—provides strong evidence for this being a symbiotic relationship rather than simply a transient occurrence. However, the fact that these bacteria are culturable under laboratory conditions also suggests that they might not be entirely host dependent.
Molecular characterization of the BFo symbiotic bacteria using 16S rRNA and biochemical analysis using API 20E Biochemical tests (Chanbusarakum and Ullman 2008) showed that they had only 95% rRNA sequence identity and shared only 50% of the biochemical properties tested for including catalase, glucose, and mannitol fermentation (Chanbusarakum and Ullman 2008). This suggests divergent evolutionary histories and may indicate that they belong to separate genera. However, the exact classification of these two species has yet to be adequately addressed. For example, there is conflict as to their phylogeny, with de Vries, Breeuwer, et al. (2001) suggesting that BFo1 and BFo2 constitute a monophyletic group with Escherichia coli; yet Chanbusarakum and Ullman (2008) suggest that BFo1 groups within the genus Erwinia, whereas BFo2 lies separately. Thus, the exact classification of these species is a source of controversy. However, both previous studies on the classification of these symbionts have relied solely on single-gene, 16S rRNA phylogenies which often suffer from saturation in deeply diverged families.
We offer a phylogenomic analysis of the two prominent bacterial symbionts (BFo1 and BFo2) of F. occidentalis. Our study focuses first on the classification of these isolates using an in-depth phylogenetic approach and second on their genome evolution and relatedness to existing bacterial species. We reconstruct a core genome for the common ancestor to the Erwinia–Pantoea clade and compare this with both BFo genomes. Taken together, our analysis allows us to reconstruct a possible evolutionary history of the two prominent bacterial symbionts of F. occidentalis.
Materials and Methods
Isolation and Culturing of F. occidentalis Symbiotic Bacteria
Symbiotic bacteria, previously designated BFo1 and BFo2 (Chanbusarakum and Ullman 2008), were isolated from the following two populations of F. occidentalis: 1) A greenhouse population from the Netherlands and 2) a population that has been isolated and maintained at Keele University (Newcastle-under-Lyme, UK). This latter population was established in 1996 after collection of F. occidentalis from a UK commercial Chrysanthemum nursery in southern England (UK) and maintained since on flowering Chrysanthemum plants. Approximately 20 surface sterilized insects were homogenized in 1× TE using a micropestle. Sterilization was performed by the method outlined in de Vries, Breeuwer, et al. (2001). Serial dilutions of the homogenate were plated on LB agar and incubated at 30 °C overnight. Initial identification of isolated bacteria was performed using colony polymerase chain reaction (PCR) with the primers 27f and 1525r (Chanbusarakum and Ullman 2008). To identify BFo bacteria, sequenced amplicons were used, along with 16S sequences published previously for BFo bacteria (Chanbusarakum and Ullman 2008) to reconstruct a Neighbor-Joining (NJ) phylogenetic tree. Positive identification of BFo bacteria was assumed for those colonies that generated 16S amplicons that clustered together in the tree with previously published sequences from BFo bacteria. All strains isolated by ourselves and those used for genomic comparisons throughout this study are listed in table 1.
Table 1.
Genomic Features of BFo Bacteria and Closely Related Species
| Species | Estimated Genome Size (Mb) | Average Whole-Genome GC Content (% mol) | No. Genes | Predicted Pseudogenes | Total RNAs | Plasmids | Status | Source | GenBank Accession | Plasmid(s) Accession |
|---|---|---|---|---|---|---|---|---|---|---|
| BFo | ||||||||||
| BFo1 (Netherlands) | 5.13 | 51.82 | 4,829 | 98 | 76 | — | Draft | This study | JMS00000000 | — |
| BFo1 SwAb130 (Netherlands) | 5.17 | 54.59 | 4,914 | 264 | 75 | — | Draft | This study | LAGP00000000 | — |
| BFo1 Keele2 (UK) | 5.01 | 54.41 | 4,670 | 252 | 63 | — | Draft | This study | LAGQ00000000 | — |
| BFo2 Swan1 (Netherlands) | 3.10 | 45.72 | 3,068 | 101 | 53 | — | Draft | This study | JMSP00000000 | — |
| BFo2 Swan69 (Netherlands) | 3.24 | 46.34 | 3,136 | 185 | 61 | — | Draft | This study | LAGS00000000 | — |
| BFo2 Keele1 (UK) | 2.70 | 46.9 | 2,681 | 128 | 45 | — | Draft | This study | LAGR00000000 | — |
| Erwinia | ||||||||||
| E. amylovora ATCC49946 | 3.81 | 53.6 | 3,583 | 46 | 100 | 2 | Complete | Sebaihia et al. (2010) | NC_013971 | NC_013972.1; NC_13973.1 |
| E. amylovora CFBP1430 | 3.83 | 53.6 | 3,455 | 21 | 100 | 1 | Complete | Smits et al. (2010b) | NC_013961 | NC_013957.1 |
| E. amylovora LA637 | 3.87 | 53.7 | 3,565 | 2 | 58 | 2 | Draft | Unpublished | NZ_CBVU000000000 | NC_023056.1; NC_023072.1 |
| E. amylovora LA636 | 3.79 | 53.6 | 3,471 | 2 | 58 | — | Draft | Unpublished | NZ_CBVT000000000 | — |
| E. amylovora ACW56400 | 3.86 | 53.5 | 3,909 | 13 | 66 | 1 | Draft | Unpublished | NZ_AFHN00000000 | NC_018999.1 |
| E. pyrifoliae Ep1/96 | 4.07 | 53.4 | 3,728 | 50 | 97 | 4 | Complete | Kube et al. (2010) | NC_012214 | NC_013264.1/FP236827.1; NC_013265.1/FP236828.1; NC_013954.1/FP928999.1; NC_013263.1/FP236829.1 |
| E. pyrifoliae DSM 12163 | 4.07 | 53.4 | 3,725 | 75 | 97 | 4 | Complete | Smits et al. (2010a) | NC_017390 | NC_017391.1/FN392236.1; NC_017388.1/FN392237.2; NC_017392.1/FN392238.1; NC_017389.1/FN392239.1 |
| E. tasmaniensis Et1/99 | 4.07 | 53.4 | 3,721 | 57 | 103 | 5 | Complete | Kube et al. (2010) | NC_010694 | NC_010695.1; NC_010696.1; NC_10697.1; NC_010699.1; NC_010693.1 |
| E. toletana DAPP-PG 735 | 5.27 | 53.6 | 4,812 | 48 | 50 | — | Draft | Passos da Silva et al. (2013) | NZ_AOCZ00000000 | — |
| E. billingiae Eb661 | 5.37 | 55 | 4,928 | 39 | 98 | 2 | Complete | Kube et al. (2010) | NC_014306 | NC_014304.1; NC_014305.1 |
| E. billingiae Ep1/96 | 4.00 | 53.4 | 3,645 | 57 | 82 | 4 | Complete | Kube et al. (2010) | FP_236843 | FP_928999; FP236827; FP236828; FP236829 |
| E. tracheiphila PSU-1 | 4.72 | 50.1 | 4,314 | 487 | 54 | — | Draft | Unpublished | NZ_APJK00000000 | — |
| Pantoea | ||||||||||
| Pantoea dispersa EGD-AAK13 | 4.77 | 57.8 | 4,382 | 15 | 71 | — | Draft | Unpublished | NZ_AVSS00000000 | — |
| P. sp. At-9b | 6.31 | 54.3 | 6,008 | — | 107 | 5 | Complete | Unpublished | NC_014837 | NC_014838.1–NC_01482.1 |
| P. agglomerans IG1 | 4.83 | 55 | 4,443 | 32 | 65 | — | Draft | Matsuzawa et al. (2012) | NZ_BAEF00000000 | — |
| P. agglomerans 299R | 4.58 | 54.3 | 4,320 | 41 | 90 | — | Draft | Remus-Emsermann et al. (2013) | NZ_ANKX00000000 | — |
| P. agglomerans Tx10 | 4.86 | 55.1 | 4,482 | 17 | 95 | — | Draft | Smith et al. (2013a) | NZ_ASJI00000000 | — |
| P. agglomerans Eh318 | 5.04 | 54.8 | 4,791 | 55 | 101 | — | Draft | Unpublished | NZ_AXOF00000000 | — |
| P. agglomerans strain 190 | 5.00 | 55.1 | 4,621 | 58 | 101 | — | Draft | Lim et al. (2014) | JNGC00000000 | — |
| P. agglomerans RIT273 | 5.37 | 55.1 | 4,927 | 32 | 93 | — | Draft | Unpublished | NZ_JFOK00000000 | — |
| P. agglomerans DAPP-PG734 | 5.37 | 54.7 | 5,107 | 57 | 106 | — | Draft | Moretti et al. (2014) | NZ_JNVA00000000 | — |
| P. agglomerans strain 4 | 4.81 | 55.1 | 4,370 | 37 | 70 | — | Draft | Unpublished | JPOT00000000 | — |
| P. vagans C9-1 | 4.89 | 55.1 | 4,512 | 101 | 99 | 2 | Complete | Smits et al. (2010c) | NC_014562 | NC_014561.1/CP001893.1; NC_014563.1/CP001894.1 |
| P. vagans MP7 | 4.59 | 55.3 | 4,125 | — | — | — | Draft | Unpublished | N/A | — |
| P. ananatis LMG 5342 | 4.91 | 53.3 | 4,525 | 70 | 99 | 1 | Draft | De Maayer et al. (2012) | NC_016816 | NC_016817.1/HE617161.1 |
| P. ananatis LMG2665 | 4.98 | 53.4 | 4,624 | 47 | 88 | — | Draft | Adam et al. (2014) | NZ_JFZU00000000 | — |
| P. ananatis AJ13355 | 4.88 | 53.7 | 4,138 | 1 | Complete | Hara et al. (2012) | NC_017531.1 | NC_017533.1/AP012033.1 | ||
| P. ananatis BI-9 | 5.11 | 53.5 | 4,759 | 32 | 75 | — | Draft | Kim et al. (2012) | NZ_CAEI00000000 | — |
| P. ananatis DAR76143 | 5.25 | 53.4 | 4,914 | 792 | 64 | — | Draft | Unpublished | NZ_BATH00000000 | — |
| P. ananatis Sd-1 | 4.92 | 53.3 | 4,548 | 72 | 73 | — | Draft | Unpublished | GCF_000582575.1 | — |
| P. ananatis BRT175 | 4.85 | 53,.7 | 4,696 | 38 | 95 | — | Draft | Smith et al. (2013b) | NZ_ASJH00000000 | — |
| Tatumella | ||||||||||
| Tatumella ptyseos ATCC 33301 | 3.53 | 51.6 | 3,349 | — | — | — | Draft | Tracz et al. (2015) | ATMJ00000000.1 | — |
| Other | ||||||||||
| Plautia stali symbiont | 3.77 | 57 | 4,062 | 1,202 | 75 | 2 | Complete | Kobayashi et al. (2011) | NC_022546 | NC_022533.1; NC_022534.1 |
Note.—Genomic features of BFo bacteria and a comparison with representative individuals from closely related species. Table includes species from the genus Erwinia, Pantoea, and Tatumella and also includes the closely related symbiont from Plautia stali. Table shows genome size (Mb), number of plasmids (along with GenBank accession number), average whole-genome G+C content, number of genes, pseudogenes, and RNAs. Total RNAs do not include noncoding RNAs. An indication of the sequencing status of the genome is also given. For BFo bacteria, origin of F. occidentalis population, where isolated, is given in parenthesis along with GenBank accession numbers. “—” indicates value unknown.
De Novo Genome Sequencing
Bacteria isolated from F. occidentalis were grown in LB until mid-log phase at 30 °C with shaking (250 rpm). Genomic DNA was extracted from liquid cultures using the QIAmp mini kit (Qiagen). Genomic DNA libraries were prepared for sequencing using Illumina Nextera XT Sample Preparation technology. Libraries were sequenced using a combination of MiSeq runs utilizing V2 and V3 MiSeq reagent kits for 300, 500 (V2), and 600 (V3) cycle runs. The paired-end sequencing reads (approximately 1,650,000; BFo1 and 1,270,000; BFo2) were preprocessed using the FASTX-trimmer in the FASTX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/, last accessed July 8, 2015). Reads were trimmed such that 20 bases were removed from the 3′-ends. Low-quality base calls were removed prior to assembly of the reads into contigs using the Velvet genome assembler (Zerbino and Birney 2008). To check for sequencing errors and quality, isolates taken from the Netherlands population were resequenced independently at Aberystwyth University as follows: Nextera XT Libraries were prepared from 1 ng of genomic DNA. Libraries were sequenced on a MiSeq (Illumina) using 2 × 151 bp reads; generating 1,031,616 (BFo1) and 419,060 (BFo2) paired-end reads. De novo assembly of reads was performed in CLC Genomics Workbench version 6.5.1. Genome metrics for all isolates are shown in table 1. Genome sequencing was performed in duplicate on two separate isolates per symbiont, per population. Assembly and subsequent filtering for contamination with the PGAP pipeline resulted in the generation of the following contigs: BFo1 SwAb130 (130), Keele2 (191), Netherlands (528) and BFo2 Swan69 (68), Keele1 (201), Netherlands (Swan1) (757).
Contigs generated for each BFo isolate were ordered using Mauve Contig mover (Rissman et al. 2009). For BFo1, contig order was determined using E. pyrifoliae and E. tasmaniensis as reference genomes. Due to the fact that phylogenetic placement of BFo2 has been a source of conflict and our phylogeny was unable to identify a closely related species to use as a reference genome, contigs for this species were not reordered. Annotation of all draft genomes was undertaken using both RAST and the PGAP pipeline (Tatusova et al. 2013). Similarity within strains of BFo1 and BFo2 was calculated using in silico DNA–DNA hybridization (DDH) implemented with the Genome-to-Genome Distance Calculator (Auch et al. 2010).
Phylotyping of BFo Isolates
Closest relatives to BFo isolates were also identified with RAST (Aziz et al. 2008) using a comparison of 8–15 genes, by phylotyping using 31 protein-encoding genes in AMPHORANET (Wu and Eisen 2008) and by submitting the whole-genome sequences to the online ribosomal multilocus sequence typing (rMLST) database (Jolley et al. 2012). In the latter approach, 53 conserved genes encoding bacterial ribosome protein subunits (rps genes) are identified and compared with all bacterial sequences in the database. As of March 2015, the rMLST database contains approximately 113,000 whole bacterial genomes.
Phylogenetic Classification of BFo Bacteria
Previous studies that have relied solely on rRNA genes to classify species of Erwinia and Pantoea have had mixed success and it has been shown that 16S rRNA phylogenies of the genus Pantoea can be of poor resolution (Rezzonico et al. 2009). Thus, we took advantage of our whole-genome sequences of BFo1 and BFo2 to undertake a multifaceted classification. To improve on existing, single-gene classifications of BFo1 and BFo2 phylogenetic reconstructions were performed as follows. First, to confirm phylogenetic placement of both BFo isolates within the enterobacteriales, phylogenies were reconstructed using multilocus sequence analysis (MLSA). First, a phylogeny using a concatenation of five slowly evolving protein sequences (Frr, NusA, PgK, RpmA, and RpsS) was reconstructed. Concatenation was performed on BLASTP (Altschul et al. 1990) retrieved orthologs from 74 taxa within the enterobacteriales using protein sequences from BFo bacteria as queries. Individual protein sequences were aligned in MEGA (Molecular Evolutionary Genetic Analysis v.6; Tamura et al. 2013) and all alignments concatenated into a supermatrix using the Perl script FASCONCAT (Kuck and Meusemann 2010). The final datamatrix contained 74 taxa and an alignment of 1,379 amino acids. An amino acid phylogeny was chosen to circumvent the problems often associated with saturation of the third codon position in nucleotide phylogenies with deep branches. In addition, the five proteins were chosen because of their availability in many whole and partially sequenced enterobacterial genomes. To ensure gap-free alignments, species were removed if no orthologs for one of the proteins could be retrieved from its genome—however, when this occurred another representative from that genus was still present in the data set. Second, a four-gene MLSA using the housekeeping genes gyrB, rpoB, infB, and atpD was conducted from members of the Pantoea, Erwinia, and Tatumella genera (112 taxa; 2,620 nt). Gene sequences were chosen because of their previous use at elucidating relatedness among species in the Enterobacteriaceae (Brady et al. 2013). In both phylogenies, sequence alignments were performed using ClustalW, implemented in MEGA (Molecular Evolutionary Genetic Analysis v.6; Tamura et al. 2013) Concatenation of the individual alignments was performed using the Perl script FASCONCAT (Kuck and Meusemann 2010). Phylogenies were reconstructed using two algorithms—first a maximum-likelihood (ML) protein phylogeny was constructed in MEGA for the five-gene MLSA using the LG model (Le and Gascuel 2008) with a gamma distribution of five discrete gamma categories a +G parameter of 0.695 (LG + G). The branch leading to Dickeya was used to outgroup root the phylogeny. Reconstruction of the four-gene MLSA phylogeny was performed in MEGA using the general time reversible (GTR) model with a gamma distribution and proportion of sites invariable (G+I). Pectobacterium sp. (gamma-Proteobacteria) was used to outgroup root the phylogeny. In all cases, bootstrap analysis (1,000 pseudoreplicates) was used to infer nodal support. Second, a Bayesian Inference (BI) phylogeny was also reconstructed for each MLSA data set. Bayesian phylogenies were reconstructed using MrBayes (Ronquist et al. 2012) run on the Cipres Science Gateway Xsede server (Miller et al. 2015) using the following parameters: Protein model or nucmodel= 4by4, rates= equal, Nst=1, Nbetacat= 5, and default priors.
Finally, whole-genome-based phylogenies were constructed as a result of a reference pan-genome approach (Meric et al. 2014). Briefly, a preliminary BFo reference pan-genome was created containing “unique” loci from all strains of BFo1 and BFo2. To do so, automatic annotations of each individual BFo1 and BFo2 assembled genomes were obtained using RAST. To empirically determine the optimum percentage coding length to retrieve allelic variants without spurious hits, we used the following parameters: 70% nucleotide sequence identity over 1) 10%, 2) 50%, and 3) 70% of the coding sequence (CDS) length. Despite using three different parameters our core genomes differed by only nine genes. Thus, we opted for the more relaxed criteria with allelic variants being defined as genes with more than 70% nucleotide sequence identity on more than 10% of the coding length. Hits were subsequently filtered out using Basic Local Alignment Search Tool (BLAST) (Meric et al. 2014), thus creating a list of 7,090 genes. The prevalence of these genes was examined in the genomes of all BFo isolates, and to an initial list of 50 Erwinia, Dickeya, and Pantoea genomes using the tools implemented in BIGSdb (Jolley and Maiden 2010). Species of Dickeya were included in the analysis as these are reports of plant phytopathogens in this genus (Khayi et al. 2014) and an earlier reclassification of Erwinia chrysanthemi to Dickeya dadantii. A total of 1,040 homologous genes were identified between BFo1 and BFo2. Concatenated gene-by-gene alignments of these (Sheppard et al. 2012) were produced using MAFFT (Katoh et al. 2002) and used to create phylogenetic trees using the NJ algorithm implemented in FastTree (Price et al. 2010).
Construction of the Pantoea/Erwinia Last Common Ancestor Core Genome
To reconstruct the most probable evolutionary history of the BFo genomes after the split from Pantoea/Erwinia, core genomes for the Pantoea and Erwinia genera were reconstructed and compared with the genome of BFo2. Briefly, core genomes for five completely sequenced Erwinia genomes (E. amylovora, E. tasmaniensis, E. pyrifoliae, E. billingiae, and E. sp strain EJp617) and three completely sequenced Pantoea (P. ananatis, P. vagans, and P. sp. At-9b) were reconstructed independently using an all-against-all reciprocal BLAST approach. Briefly, each CDS was used as a BLAST query against a local database of CDS from all species outlined above. Presence of true orthologs of a particular gene was recorded if during pairwise reciprocal BLAST that gene returned as the best hit in both species. To avoid complications of gene duplication, we restricted the analysis to a presence-and-absence of orthologs approach—thus, in the case of gene duplication, the number of paralogous copies was not taken into account during the analysis. The putative ancestral core genome for the Last Common Ancestor (LCA) of Erwinia and Pantoea was reconstructed by retaining those genes that were conserved in these genus-specific core genomes. Comparisons were made between BFo genomes and the ancestral core genome.
Nucleotide Substitution Rates
Data sets of orthologous gene pairs between BFo strains and E. tasmaniensis and E. pyrifoliae were constructed using a reciprocal BLAST approach. Pairwise alignments were performed in MEGACC (Kumar et al. 2012) with low-quality, gap-rich alignments removed manually. Pairwise estimates of the synonymous (dS) and nonsynonymous (dN) substitution rates were obtained for all retained gene pairs using the program YN00 (PAML; Yang 1997). Plots of dN/dS were generated to compare intact orthologs among BFo and closely related species.
Results
Classification of BFo1 and BFo2 Using a Multifaceted Approach
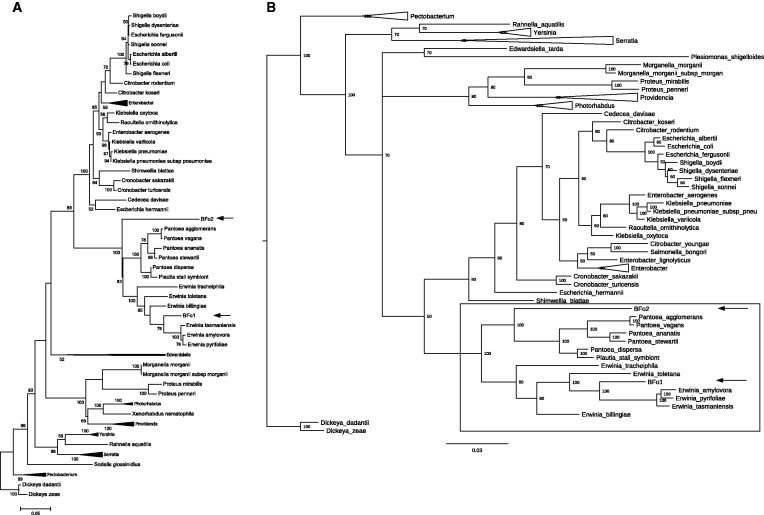
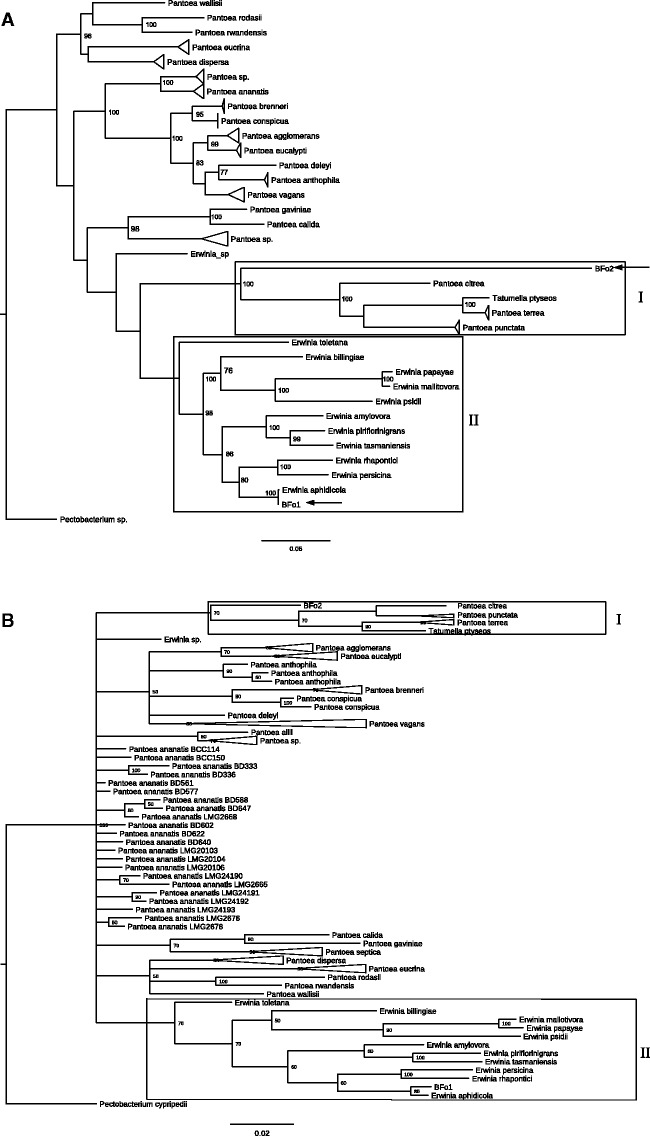
Consistent with previous studies, plating of the homogenate obtained from surface sterilized F. occidentalis in all populations revealed two predominant colony morphologies. A very small number of non-BFo colonies were also isolated, typing these by 16S PCR indicated that they were related to the endosymbiont of Nilaparvata lugens and bacteria isolated from the gut of Apis cerana japonica (GenBank accession numbers of 16S sequence: JQ975877 and AB668063, respectively). However, growth of these was not consistent between isolations and thus was not addressed in depth. Identification of BFo-like colonies was performed using a combinatorial approach of 16S colony PCR and reconstruction of an NJ phylogeny (supplementary fig. S1, Supplementary Material online). This showed that our isolates cluster together with previously identified BFo bacteria. All data analysis was performed on several strains of BFo1 and BFo2 (table 1); however, they all appeared to group together in our phylogenies, thus will be referred to collectively as BFo1 and BFo2 hereafter. Moreover, in silico DDH indicated that the probability that all BFo1 isolates being the same species and all BFo2 isolates being the same species (i.e., DDH > 70%) was always greater than 97%. Additional classification, using ML and BI phylogenies reconstructed from 833 aligned and concatenated amino acid positions from 83 species (fig. 1A and B, respectively) and ML and BI reconstructed phylogenies of four gene sequences (2,620 nt positions, 115 species; fig. 2A and B, respectively), shows that BFo1 clusters within the genus Erwinia. Evidence from both trees suggests a common ancestor with the phytopathogens E. amylovora, E. pyrilifoliae, and the nonpathogenic E. tasmaniensis. More specifically, the four-gene MLSA tree indicates that BFo1 may be closely related to E. aphidicola. In contrast, both data sets also show that BFo2 is a member of the Enterobacteriacae but both data sets also position BFo2 on a separate branch adjacent to the Pantoea clade. Interestingly, both data sets also position BFo2 as a sister branch to the “Japanese” Pantoea (rather than the “core” Pantoea). BFo species are also found in distinct sequence clusters from other phytopathogens and several other endosymbionts. The deep branching of BFo2 from Pantoea/Erwinia sensu stricto is consistent with a prolonged period of independent evolution.
Fig. 1.—
Phylogenetic reconstruction showing placement of BFo bacteria among closely related Enterobacteriales. Phylogenetic tree constructed using a 5-protein MLSA. Gap-free alignment of five protein sequences giving a total of 833 positions. (A) ML consensus tree was constructed using the LG+G model with five categories and a +G parameter of 0.695. The initial tree for heuristic searching was obtained using the NJ algorithm. Bootstrap analysis with 1,000 pseudoreplicates was performed to infer nodal support. Bootstrap values below 70% not shown. Homogenous clades have been collapsed. Arrows indicate BFo. (B) 50% majority rule consensus BI phylogeny using the model=protein, Nst=1, Nbetecat=5, and default priors. Numbers at nodes indicate percentage node probably. Probabilities <50% not shown. Shaded area indicates the positions of the Pantoea and Erwinia genera (+both BFo bacteria). Homogenous clades have been collapsed. Arrows indicate BFo. Both trees are outgroup rooted along the branch leading to Dickeya.
Fig. 2.—
Phylogenetic reconstruction using a four-gene MLSA of BFo bacteria and closely species of Pantoea and Erwinia. Phylogenetic reconstruction using a four-gene MLSA. Gap-free alignment of four housekeeping gene sequences giving a total of 2,620 nt positions. (A) ML tree was constructed using the GTR model with five categories and a +G parameter of 0.695. The initial tree for heuristic searching was obtained using the NJ algorithm. Bootstrap analysis with 1,000 pseudoreplicates was performed to infer nodal support. BFo2 species indicated by arrows. Japanese Pantoea (+Tatumella) indicated by box I. Erwinia indicated by box II. Branches with greater than two identical strains/species have been collapsed. Bootstrap values below 75 not shown. (B) 50% majority rule consensus BI phylogeny using the nucmodel 4by4, Nst=1, Nbetecat=5, and default priors. Numbers at nodes indicate percentage node probably (only probabilities >50% shown). Japanese Pantoea (+Tatumella) and Erwinia demarcated with boxes (I and II, respectively). Branches with greater than two identical strains/species are collapsed.
Despite being more polytomous, trees reconstructed using BI showed similar topologies to the ML reconstructions but with very minor rearrangements.
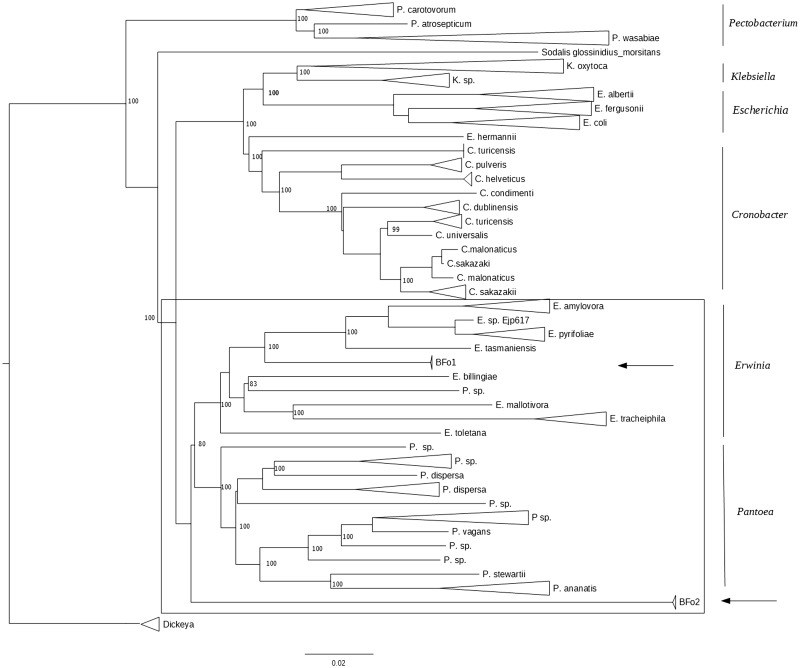
To support our initial findings, all BFo isolates were phylotyped to identify closely related species using online annotation and classification severs. Both AMPHORANET and RAST returned E. tasmaniensis as the most closely related species to BFo1 and Pantoea sp. At-9b as the most closely related species to BFo2. Additionally, a whole-genome NJ phylogeny of concatenated gene alignments from loci shared by 90% of 141 enterobacterial genomes, respectively, was also conducted (fig. 3). The topology of the resultant tree was consistent with that of both MLSA trees with BFo1 positioned within the Erwinia clade. Again, BFo2 was positioned at the end of a long branch outside of the Erwinia and Pantoea genera. Perhaps, the biggest difference was the positioning of BFo2 as an outgroup to the Erwinia/Pantoea genera in the five-gene MLSA and core genome phylogenies (figs. 1A, B, and 3) and the grouping of BFo2 more closely with species of Pantoea (four-gene MLSA; fig. 2A and B). Discrepancies between these data sets are likely the result of differing phylogenetic signal among the markers. rMLST analysis clustered BFo1 with Erwinia. In this case, all rps genes retrieved from the genomes of BFo1 returned best hit alignments to orthologous rps genes in E. pyrifoliae. In contrast, only a single rps gene produced any significant alignment (from P. ananatis; 100% query coverage; 90% identity) to orthologous rps genes in BFo2. None of the other rps genes in the BFo2 genome produced any significant hits, despite the database containing 105,000 whole bacterial genomes.
Fig. 3.—
Core genome phylogeny of BFo bacteria and closely related Enterobacteriales. NJ phylogenetic tree reconstructed using gene-by-gene alignments from genes shared by 90% of 141 whole bacterial genomes. Position of BFo1 and BFo2 strains are indicated with an arrow. Tree is rooted along the branch leading to Dickeya. Scale bar indicates genetic distance. Bootstrap values indicate nodal support from 1,000 pseudoreplicates. Boxed area shows the Erwinia/Pantoea sensu stricta.
Genome Comparisons of BFo Bacteria and Closely Related Species
The Whole-Genome Shotgun projects of BFo1-like and BFo2-like isolates described in this article have been deposited at DDBJ/EMBL/GenBank as individual Biosamples under the multispecies BioProject PRJNA23451 (table 1). The average genome size for BFo1 and BFo2 strains was 5.1 and 3.01 Mb, respectively. Within species genome sizes were consistent except for BFo2 isolated from the Keele (UK) population of F. occidentalis—which was slightly smaller. A comparison of genome sizes between BFo bacteria and closely related bacteria (table 1) shows that the genomes of the BFo1 strains in this study are consistent with those of many nonphytopathogenic Erwinia. More specifically, BFo1 shares similar genome sizes to E. amylovora and E. tasmaniensis. Conversely, average genome sizes for BFo2 strains are slightly smaller than those of the Erwinia and Pantoea genera.
Secretion Systems and Virulence Factors
Searches for virulence factors and pathogenicity determinants that are ubiquitous in Erwinia were performed. These included three secretion systems (T2SS, T3SS, and T6SS), the exopolysaccharide gene clusters of amylovoran (ams), stewartan (cps) and levansucrase (lscC), the sorbitol and sucrose operons, and two flagella biosynthesis loci. BFo1 isolate genomes in this study lacked both the ancestral Hrp-T3SS and Inv/Spa-type T3SS—evidenced by the absence of orthologs of hrpANW, dspE/A (Hrp) and the Inv–Spa type secretion systems found in PAI-2 and -3 of E. amylovora. However, the genomes of BFo1 are predicted to encode three previously described T6SS loci (T6SS-1, T6SS-2, and TSS-3). The three T6SS loci in BFo1 also show a high degree of gene order conservation when compared with other species of Erwinia. Additionally, the genome of BFo1 contains the exopolysaccharide gene cluster for amylovoran of which they share greatest amino acid similarity (>80%) to the glycoside transferases found in E. tasmaniensis. Gene clusters for levansucrase and stewartan were absent—despite their presence in E. amylovora and E. pyrifoliae. Moreover, the genome of BFo1 also encodes two flagella biosynthesis loci (flg-1 and flg-2). In comparison, BFo2 lacks the T6SS secretion systems, which is evidenced by the absence of orthologs for genes predicted to encode ImpDEFILM as well as the lipoprotein VasD. Moreover, the genome of BFo2 (unlike BFo1) lacks orthologs of impK (dotU) and vgrG. Similarly, BFo2 also lacks an ortholog of outO, which is a core component of the T2SS. The genomes of BFo2 strains also do not encode any of the exopolysaccharides commonly secreted by Erwinia and Pantoea species.
Comparison of the Putative LCA Core Genome and the BFo2 Genome
Our reconstructed core genomes highlighted 2,080 and 3,464 conserved genes in the Erwinia and Pantoea isolates in this study, respectively. Of these two core genomes, we uncovered 1,967 genes that were conserved among these two genera—thus representing a core putative genome of the Pantoea/Erwinia LCA (supplementary table S1, Supplementary Material online). A comparison of the conserved genes in the putative LCA and the BFo2 genome revealed that BFo2 retains 1,753 of these, indicating that since the split from the LCA, BFo2 has lost at least 214 genes from its core genome (supplementary table S2, Supplementary Material online). Of these, 75 were annotated as hypothetical and 37 were related to metabolism and transport of amino acids, carbohydrates, and coenzymes. In addition, losses of genes were noted that are associated with signal transduction (four genes), DNA replication, recombination and repair (four genes), virulence (ten genes), motility (seven genes), posttranslational modification (eight genes), and cell wall/membrane biogenesis and cell division (eight genes). This is in contrast to BFo1; comparisons of this genome with the core genome of the LCA show that BFo1 has a deficit of only 13 genes conserved in the putative LCA core genome (supplementary table S3, Supplementary Material online).
Molecular Evolution in BFo Bacteria
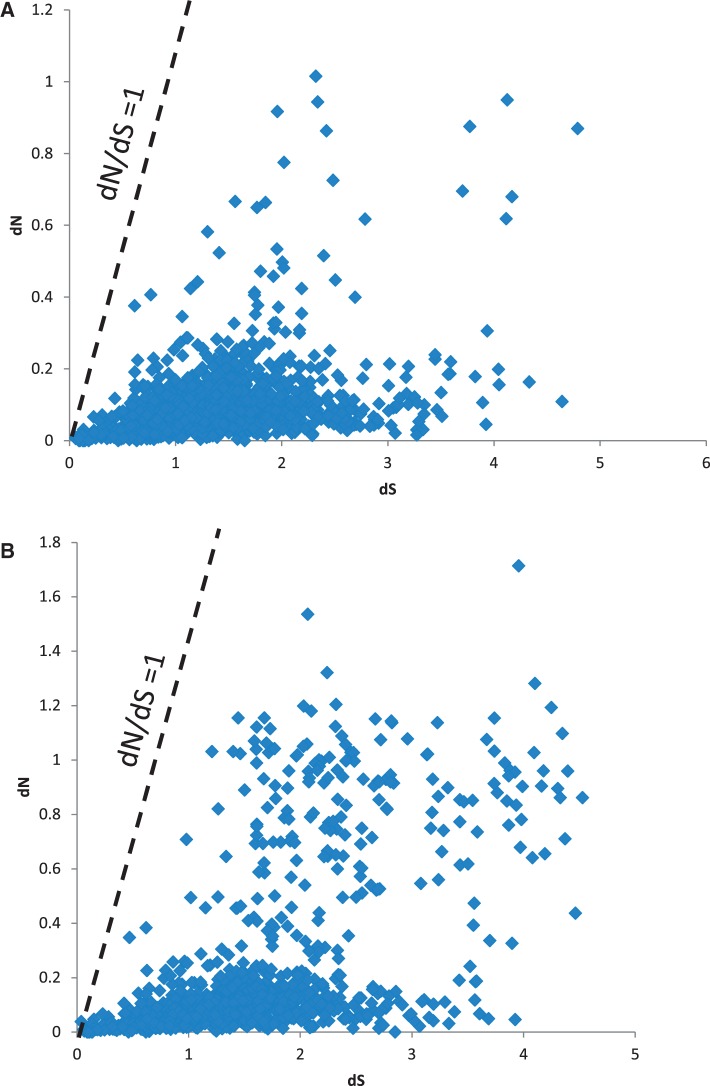
Genome-wide rates of synonymous (dS) and nonsynonymous (dN) substitutions for all BFo species and strains were calculated using pairwise alignments of orthologous genes from the closest nonpathogenic species (E. tasmaniensis and E. billingiae) and the closest plant pathogen (E. pyrifoliae) (fig. 4). Comparisons between BFo1 strains and E. tasmaniensis revealed an average dS of 1.5 (Netherlands BFo1) and 1.3 (UK BFo1) with similar numbers of genes in both BFo1 strains approaching saturation for dS (>3.0). Very few genes in BFo1 had dN/dS ratios greater than 1 indicating that the majority are under purifying selection. Comparisons of pairwise orthologs between BFo1 strains and E. pyrifoliae and E. billingiae revealed almost complete saturation of dS (>3.0). Similarly comparisons of the rate of nucleotide evolution between BFo2 and members of the Erwinia and Pantoea genera resulted in saturation of the synonymous nucleotide substitution rates (dS > 5).
Fig. 4.—
Plot of dN versus dS of orthologous genes between BFo1 and E. tasmaniensis. Plot of dN versus dS for all orthologous gene pairs between the nonpathogen, free living E. tasmiensis and BFo1. (A) BFo1 isolated from UK population of F. occidentalis. (B) BFo1 isolated from the Netherlands population of F. occidentalis. Each point represents an orthologous gene pair. Dotted line indicates neutrality (dN = dS).
Discussion
Phylogenetic Reconstruction Clusters BFo1 with Species of Erwinia
Previous studies have classified both F. occidentalis bacterial symbionts as members of the diverse bacterial family Enterobacteriaceae (Chanbusarakum and Ullman 2008). Their single-gene phylogeny, using 16S rRNA sequences, showed that BFo1 clustered with species of Erwinia, whereas BFo2 clustered more closely with E. coli. Here, based on multisequence concatenated phylogenies, whole-genome phylogeny and comparisons of ribosomal genes (rMLST), we have reanalyzed the phylogenetic placement of these symbionts. A five-gene MLSA using the amino acid sequences of slow evolving genes positions both symbionts within the Enterobacteriales. More, parochially, a four-gene MLSA using housekeeping genes indicated that BFo1 may be closely related to E. aphidicola—an insect symbiont previous isolated from the Pea Aphid (Acyrthosiphon pisum). This is interesting as E. aphidicola has been shown to infect and reduce yield of Phaseolus vulgaris (Harada et al. 1997; Marin et al. 2011). More specifically, E. aphidicola has been implicated in causing internervial chlorosis, necrotic pits, and rough roots (Marin et al. 2011). Such preliminary findings suggest that F. occidentalis might be carrying a symbiont related to a phytopathogen capable of reducing crop yield. Indeed such findings may have large agroeconomical implications considering the now pan-global biogeography of F. occidentalis. However, we advise caution when interpreting phylogenies of minimal gene sets—especially as their reliability has been questioned when comparing whole-genome equivalents (Stephan et al. 2014). Indeed, despite the damage caused by feeding and transmission of tospoviruses, F. occidentalis appears not to be implicated in the damage reported by Marin et al. (2011). Additionally, comparisons of biochemical properties between E. aphidicola and BFo1 conducted by Harada et al. (1997) and de Vries, Breeuwer, et al. (2001), respectively, indicate that E. aphidicola is able to utilize inositol, sorbitol and citrate, whereas BFo1 is not. Thus although BFo1 is perhaps closely related to E. aphidicola, inference of such a relationship based on a four-gene phylogeny coupled with discrepancies in biochemical data suggests that BFo1 is not E. aphidicola. However, a definitive answer lies in the sequencing the E. aphidicola genome.
The topology of our core genome phylogeny is consistent with previous work on F. occidentalis symbionts (de Vries, Breeuwer, et al. 2001). BFo1 clusters with plant phytopathogens in the genus Erwinia and is likely to have shared a common ancestor with E. pyrifoliae/E. amylovora and the nonpathogenic species E. tasmaniensis. Indeed, E. amylovora infects the leaves and flowers of plants in the Rosaceae (Vrancken et al. 2013) and these plants are common hosts of F. occidentalis (Rahman et al. 2010, 2011). The question of whether F. occidentalis continually acquires BFo1 from the environment is difficult to answer—however, the fact that both BFo bacteria have been isolated from populations of F. occidentalis across a wide geographical range and also from historical specimens (Chanbusarakum and Ullman 2008) suggests that they are both true symbionts and form a permanent association within the gut lumen of F. occidentalis.
BFo1 Virulence Determinants and Common Ancestry with Nonpathogenic E. billingiae and E. tasmaniensis
A comparison of the virulence and pathogenicity factors encoded by three BFo1 genomes and closely related species of Erwinia gives some insight to the phylogenetic placements of these strains. A predicted virulence genotype, for an erwinial ancestor, has been characterized and changes in the gene repertoire in different lineages in relation to Erwinia radiation (Smits et al. 2011). In this study, the plesiomorphic genotype for this genus was predicted to contain an Hrp-like T3SS, a single flagellal locus (Flg-1), two T6SS loci (T6SS-1 and T6SS-2), two expolysaccharides (lscC and stewartan), and the sorbitol metabolism (srl) operon. The genome of BFo1 does possess three T6SS loci, of which T6SS-1 and T6SS-2 but does not possess other elements of the secretion system; possibly because it is more associated with infection of plants and causation of disease (Zhao and Qi 2011; Mann et al. 2013). Of those three T6SS loci, the first two are common in Erwinia, whereas the third is likely resultant of Horizontal Gene Transfer (De Maayer et al. 2011). Furthermore, neither of the two Inv/Spa-type T3SS, thought to have been present early on in the radiation of the genus, is present in BFo1. This pattern of gene gain or loss is consistent with frequent horizontal gene transfer associated with the radiation of Erwinia and Pantoea (De Maayer et al. 2011). Species of Erwinia also possess several metabolic factors that are considered to be important in causing disease in plants (Kube et al. 2010). These include exopolysaccharides—such as amylovoran, levan production, and the ability to synthesize sorbitol and sucrose. The BFo1 genomes sequenced in this study possess the amylovoran biosynthesis cluster (ams) but lack the ability to produce levan and do not contain the sorbitol or sucrose metabolism operons. A comparison of the ams gene cluster in BFo1 shows that it is most similar to that found in E. tasmaniensis. Principally, the ams gene cluster has been shown to be distinguishable from the Stewartan biosynthesis gene cluster in other species of Erwinia and Pantoea principally by the exchange of two glycoside transferases (annotated as wbdN and cpsD) at the center of the cluster (Kamber et al. 2014). Taken together, details of the predicted BFo1 virulence genotype, and estimated genome size, closely mirror that reported for E. billingiae Eb661 (Kube et al. 2010) and support evidence from genomic analysis that BFo1 may share a common ancestor with this species. However, this is not consistent with our core genome phylogeny that separates BFo1 from E. billingiae. The lack of an Hrp type III secretion system and T2SS within the BFo1 genome is consistent with a nonpathogenic lifestyle—as both the T2SS and T3SS are important features acquired during pathoadaption in Erwinia (Smits et al. 2013). Further characterization of the population structure of multiple BFo1 isolates will be necessary to confirm if the absence of an Inv/Spa-type T3SS is an ancestral trait, as in E. pyrifoliae and E. tasmaniensis, where this secretion system has been lost (Smits et al. 2013), and if comparable genome metrics with E. toletana can be instructive in understanding the evolution of this lineage.
Phylogenetic and Genomic Analysis of BFo2
In contrast to the classification of BFo1, which clusters with the Erwinia genus, BFo2 isolated from the same host represents a relatively divergent lineage compared with the most closely related species based on protein and whole-genome phylogenies (figs. 1–3). Phylogenetic reconstruction with whole enterobacterial genomes indicates that it is somewhat divergent from known sequenced bacteria. Indeed, whole-genome alignments of BFo2 genomes and other enterobacteriales revealed very little genome conservation. Moreover, saturation of dS when comparing orthologous genes pairs in BFo2 and Pantoea and Erwinia provides some evidence of divergence of BFo2 from these two genera. The genome of BFo2 does not encode any of the commonly found secretion systems (T3SS and T6SS) in the Pantoea and Erwinia genera—the latter is evidenced by the lack of impK (dotU) and vgrG both of which are core components of the type VI secretion system (Broms et al. 2012). In addition BFo2 also appears also to lack genes of the T2SS—evidenced by the lack of an ortholog of outO which is a core component of the T2SS (Sandkvist 2001). The absence of these genes in BFo2 is perhaps consistent with gene loss and of ongoing evolution in an insect symbiont. The loss of secretion systems, including T2SS, T3SS and T6SS, can be beneficial to emerging symbionts—as their synthesis is energy demanding imposing a fitness cost and their absence could provide a selective advantage in environmental isolates (Gophna et al. 2003).
The BFo2 genomes also lacked 37 of the genes associated with transport and metabolism, 7 genes associated with motility, and 8 genes associated with cell wall/membrane biogenesis and cell division that we predicted in the Erwinia/Pantoea LCA core genome. The loss of so many genes involved in transport and metabolism of carbohydrates and amino acids in the BFo2 genome may indicate small-scale genome reduction as has been identified previously in a few insect symbionts such as Ishikawaella (Nikoh et al. (2011). Furthermore, it may suggest that BFo2 is reliant on BFo1 for full functionality. Consistent with this, BFo2 is rarely found in the gut of F. occidentalis without BFo1 (Chanbusarakum and Ullman 2009).
A comparative analysis of the genome metrics of BFo bacteria and members of the Erwinia and Pantoea genera reveal that although the genome size (in Mb) of BFo1 is consistent with closely related species, the genome of BFo2 is slightly smaller. A similar pattern is observed with %GC content, while BFo1 is consistent with the average Erwinia GC content, BFo2 is slightly lower. Such genome reduction and AT-bias in BFo2 may be indicative of an emerging facultative insect symbiont and is often associated with a long-term relationship with an eukaryotic host (Van Leuven and McCutcheon 2012). Indeed, although obligate insect symbionts, such as Buchnera aphidicola, with ancient relationships with their hosts often have reduced genomes, facultative symbionts, for example, Candidatus Regiella insecticola and Candidatus Hamiltonella defensa (Degnan et al. 2010), often have genomes that, while having undergone genome reduction, are much larger and have more dynamic genomes than endosymbiotic conspecifics (Engel and Moran 2013). Taken together, the reduced genome size, coding density, and reduced G+C content compared with closely related free-living species are consistent with BFo2 being a nascent symbiont, as observed in Serratia symbiotica (Burke and Moran 2011) and Candidatus Regiella insecticola in aphids (Degnan et al. 2010).
Conclusions
Our analysis suggests that two of the primary bacterial symbionts isolated from F. occidentalis may represent novel species. Based on phylogenetic analysis, BFo1 could be described as an Erwinia species sharing a common ancestor with the nonpathogenic E. tasmaniensis, the epiphytic E. billingiae, and the pathogenic E. pyrifoliae. The grouping of BFo1 with E. aphidicola using a four-gene MLSA could be significant from an agroeconomical standpoint. But disparity between biochemical data among these two species and the lack of a WGS for E. aphidicola means that this relationship cannot be accurately elucidated as yet. Genome analysis of BFo1 reveals a lack of key genomic signatures that are commensurate with pathogenicity within this genus, consistent with a commensal life-style for BFo1. Phylogenetic classification of BFo2 revealed that these bacteria do not cluster closely with any available bacterial species genomes.
Supplementary Material
Supplementary figure S1 and tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The work in this study was funded by the following grants: Bill and Melinda Gates Foundation Grand Challenges Explorations Grant Number OPP1068514; and ILS2020 Biomarkers and Clinical diagnostics, Academic Expertise for Business (A4B), part of a European regional Development Fund (ERDF) awarded though the Welsh Government; G.M. and S.K.S. are funded by the National Institute for Social Care and Health Research (NISCHR), the Biotechnology and Biological Sciences Research Council (BBSRC), the Medical Research Council (MRC), and the Wellcome Trust.
Literature Cited
- Allen JM, Reed DL, Perotti MA, Braig HR. 2007. Evolutionary relationships of “Candidatus Riesia spp.,” endosymbiotic enterobacteriaceae living within hematophagous primate lice. Appl Environ Microbiol. 3:1659–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Auch AF, von Jan M, Klenk HP, Goker M. 2010. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2:117–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, et al. 2008, The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumler DJ, Ma B, Reed JL, Perna NT. 2013. Inferring ancient metabolism using ancestral core metabolic models of enterobacteria. BMC Syst Biol. 7:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady C, Cleenwerck I, Venter S, Coutinho T, De Vos P. 2013. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst Appl Microbiol. 36:309–319. [DOI] [PubMed] [Google Scholar]
- Broms JE, Meyer L, Lavander M, Larsson P, Sjostedt A. 2012. DotU and VgrG, core components of type VI secretion systems, are essential for Francisella LVS pathogenicity. PLoS One 7:e34639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke GR, Moran NA. 2011, Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol Evol. 3:195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanbusarakum L, Ullman D. 2008, Characterization of bacterial symbionts in Frankliniella occidentalis (Pergande), Western flower thrips. J Invertebr Pathol. 99:318–325. [DOI] [PubMed] [Google Scholar]
- Chanbusarakum LJ, Ullman DE. 2009. Distribution and ecology of Frankliniella occidentalis (Thysanoptera: Thripidae) bacterial symbionts. Environ Entomol. 38:1069–1077. [DOI] [PubMed] [Google Scholar]
- De Maayer P, et al. 2011. Comparative genomics of the Type VI secretion systems of Pantoea and Erwinia species reveals the presence of putative effector islands that may be translocated by the VgrG and Hcp proteins. BMC Genomics 12:576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries EJ. 2010. Symbiosis of thrips and gut bacteria. [PhD Thesis] Institute for Biodiversity and Ecosystem Dynamics (IBED). [Google Scholar]
- de Vries EJ, Breeuwer JA, Jacobs G, Mollema C. 2001. The association of Western flower thrips, Frankliniella occidentalis, with a near Erwinia species gut bacteria: transient or permanent? J Invertebr Pathol. 77:120–128. [DOI] [PubMed] [Google Scholar]
- de Vries EJ, Jacobs G, Breeuwer JA. 2001. Growth and transmission of gut bacteria in the Western flower thrips, Frankliniella occidentalis. J Invertebr Pathol. 77:129–137. [DOI] [PubMed] [Google Scholar]
- Degnan PH, et al. 2010. Dynamics of genome evolution in facultative symbionts of aphids. Environ Microbiol. 12:2060–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel P, Moran NA. 2013. The gut microbiota of insects—diversity in structure and function. FEMS Microbiol Rev. 37:699–735. [DOI] [PubMed] [Google Scholar]
- Gophna U, Ron EZ, Graur D. 2003. Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events. Gene 312:151–163. [DOI] [PubMed] [Google Scholar]
- Harada H, Oyaizu H, Kosako Y, Ishikawa H. 1997. Erwinia aphidicola, a new species isolated from pea aphid, Acyrthosiphon pisum. J Gen Appl Microbiol. 43:349–354. [DOI] [PubMed] [Google Scholar]
- Husník F, Chrudimský T, Hypša V. 2011. Multiple origins of endosymbiosis within the Enterobacteriaceae (γ-Proteobacteria): convergence of complex phylogenetic approaches. BMC Biol. 9:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen SE. 2000a. Insecticide resistance in the Western flower thrips, Frankliniella occidentalis. Integr Pest Manag Rev. 5:131–146. [Google Scholar]
- Jensen SE. 2000b. Mechanisms associated with methiocarb resistance in Frankliniella occidentalis (Thysanoptera: Thripidae). J Econ Entomol. 93:464–471. [DOI] [PubMed] [Google Scholar]
- Jolley KA, et al. 2012. Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology 158:1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamber T, Smits THM, Rezzonico F, Duffy B. 2014. Genomics and current genetic understanding of Erwinia amylovora and the fire blight antagonist Pantoea vagans. Trees 26:227–238. [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayi S, et al. 2014. Genome sequence of the emerging plant pathogen Dickeya solani Strain RNS 08.23.3.1A. Genome Announc. 2(1):e01270–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, et al. 2009. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk DJW, Terry LI. 2003. The spread of the western flower thrips Frankliniella occidentalis (Pergande). Agric For Entomol. 5:301–310. [Google Scholar]
- Kube M, et al. 2010. Genome comparison of the epiphytic bacteria Erwinia billingiae and E. tasmaniensis with the pear pathogen E. pyrifoliae. BMC Genomics 11:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuck P, Meusemann K. 2010. FASconCAT: convenient handling of data matrices. Mol Phylogenet Evol. 56:1115–1118. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Peterson D, Tamura K. 2012. MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics 28:2685–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol. 25:1307–1320. [DOI] [PubMed] [Google Scholar]
- Mann RA, et al. 2013. Comparative genomics of 12 strains of Erwinia amylovora identifies a pan-genome with a large conserved core. PLoS One 8:e55644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin F, Santos M, Carretero F, Yau JA, Dianez F. 2011. Erwinia aphidicola isolated from commercial bean seeds (Phaseolus vulgaris). Phytoparasitica 39:483–489. [Google Scholar]
- Meric G, et al. 2014. A reference pan-genome approach to comparative bacterial genomics: identification of novel epidemiological markers in pathogenic Campylobacter. PLoS One 9:e92798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, et al. 2015. A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evol Bioinform Online. 11:43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N, Hosokawa T, Oshima K, Hattori M, Fukatsu T. 2011. Reductive evolution of bacterial genome in insect gut environment. Genome Biol Evol. 3:702–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu HR, Jones RA, Jain RK. 2009. Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res. 141:219–236. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins M, Goldbach R. 1998. The emerging problem of tospovirus infection and nonconventional methods of control. Trends Microbiol. 6:31–35. [DOI] [PubMed] [Google Scholar]
- Rahman T, Spafford H, Broughton S. 2010. Variation in preference and performance of Frankliniella occidentalis (Thysanoptera: Thripidae) on three strawberry cultivars. J Econ Entomol. 103:1744–1753. [DOI] [PubMed] [Google Scholar]
- Rahman T, Spafford H, Broughton S. 2011. Compatibility of spinosad with predaceous mites (Acari) used to control Frankliniella occidentalis (Pergande) (Thysanoptera: Thripidae). Pest Manag Sci. 67:993–1003. [DOI] [PubMed] [Google Scholar]
- Rezzonico F, Smits TH, Montesinos E, Frey JE, Duffy B. 2009. Genotypic comparison of Pantoea agglomerans plant and clinical strains. BMC Microbiol. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissman AI, et al. 2009. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics 25:2071–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandkvist M. 2001. Type II secretion and pathogenesis. Infect Immun. 69:3523–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, Jolley KA, Maiden MCJ. 2012. A gene-by-gene approach to bacterial population genomics: whole genome MLST of Campylobacter. Genes 3:261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits THM, et al. 2010a. Complete genome sequence of the fire blight pathogen Erwinia pyrifoliae DSM 12163T and comparative genomic insights into plant pathogenicity BMC Genomics 11:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits THM, et al. 2010b. Complete genome sequence of the fire blight pathogen Erwinia amylovora CFBP 1430 and comparison to other Erwinia spp. Mol Plant Microbe Interact. 23(4):384–393. [DOI] [PubMed] [Google Scholar]
- Smits THM, et al. 2010c. Genome Sequence of the Biocontrol Agent Pantoea vagans Strain C9-1 J Bacteriol. 192(24):6486–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits TH, Rezzonico F, Duffy B. 2011. Evolutionary insights from Erwinia amylovora genomics. J Biotechnol. 155:34–39. [DOI] [PubMed] [Google Scholar]
- Smits THM, et al. 2013. Phylogenetic position and virulence apparatus of the pear flower necrosis pathogen Erwinia piriflorinigrans CFBP 5888T as assessed by comparative genomics. Syst Appl Microbiol. 36:449–456. [DOI] [PubMed] [Google Scholar]
- Stephan R, et al. 2014. Re-examination of the taxonomic status of Enterobacter helveticus, Enterobacter pulveris and Enterobacter turicensis as members of the genus Cronobacter and their reclassification in the genera Franconibacter gen. nov. and Siccibacter gen. nov. as Franconibacter helveticus comb. nov., Franconibacter pulveris comb. nov. and Siccibacter turicensis comb. nov., respectively. Int J Syst Evol Microbiol. 64:3402–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T, et al. 2013. The NCBI Handbook [Internet]. Bethesda (MD): National Center for Biotechnology Information (US). [Google Scholar]
- Van Ham RC, et al. 2003. Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci U S A. 100:581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leuven JT, McCutcheon JP. 2012. An AT mutational bias in the tiny GC-rich endosymbiont genome of Hodgkinia. Genome Biol Evol. 4:24–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrancken K, Holtappels M, Schoofs H, Deckers T, Valcke R. 2013. Pathogenicity and infection strategies of the fire blight pathogen Erwinia amylovora in Rosaceae: state of the art. Microbiology 159:823–832. [DOI] [PubMed] [Google Scholar]
- Wu M, Eisen JA. 2008. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 9:R151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 13:555–556. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Qi M. 2011. Comparative genomics of Erwinia amylovora and related Erwinia species—what do we learn? Genes 2:627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.