Abstract
Even though mitochondrial genomes, which characterize eukaryotic cells, were first discovered more than 50 years ago, mitochondrial genomics remains an important topic in molecular biology and genome sciences. The Phylum Alveolata comprises three major groups (ciliates, apicomplexans, and dinoflagellates), the mitochondrial genomes of which have diverged widely. Even though the gene content of dinoflagellate mitochondrial genomes is reportedly comparable to that of apicomplexans, the highly fragmented and rearranged genome structures of dinoflagellates have frustrated whole genomic analysis. Consequently, noncoding sequences and gene arrangements of dinoflagellate mitochondrial genomes have not been well characterized. Here we report that the continuous assembled genome (∼326 kb) of the dinoflagellate, Symbiodinium minutum, is AT-rich (∼64.3%) and that it contains three protein-coding genes. Based upon in silico analysis, the remaining 99% of the genome comprises transcriptomic noncoding sequences. RNA edited sites and unique, possible start and stop codons clarify conserved regions among dinoflagellates. Our massive transcriptome analysis shows that almost all regions of the genome are transcribed, including 27 possible fragmented ribosomal RNA genes and 12 uncharacterized small RNAs that are similar to mitochondrial RNA genes of the malarial parasite, Plasmodium falciparum. Gene map comparisons show that gene order is only slightly conserved between S. minutum and P. falciparum. However, small RNAs and intergenic sequences share sequence similarities with P. falciparum, suggesting that the function of noncoding sequences has been preserved despite development of very different genome structures.
Keywords: Symbiodinium, Plasmodium, mitochondrial genome expansion, RNA editing, gene map, noncoding
Introduction
Mitochondrial (mt) genomes are considered characteristic of eukaryotic cells (Lang et al. 1999; Gray et al. 2004). Although eukaryotic mt genomes are believed to have arisen from alpha-proteobacteria, extant eukaryotes possess either linear or circularized mtDNA with varied and reduced gene content (Lang et al. 1999). For example, most metazoan mt genomes are 13–20 kb, compact, circular molecules, encoding 12–13 proteins, 24–25 transfer RNAs (tRNAs), and 2 ribosomal RNAs (rRNAs). On the other hand, linear mtDNAs with terminal repeats (putative telomeres) have also been found in many species, such as the yeast, Candida, and the ciliate, Tetrahymena (Lang et al. 1999; Rycovska et al. 2004).
The ciliates, Paramecium aurelia, Tetrahymena pyriformis, and Tetrahymena thermophile, have linear mt genomes of 40–47 kb, which contain approximately 50 genes (Burger et al. 2000; Gray et al. 2004). In contrast, only three protein-coding genes (cox1 [cytochrome oxidase subunit I], cox3 [cytochrome oxidase subunit III], and cob [cytochrome b]) and fragmented rRNAs (LSU, large subunit; SSU, small subunit) have been identified in mt genomes of apicomplexans and dinoflagellates (Feagin 1992; Nash et al. 2008; Vaidya and Mather 2009). Recent work on the mt genome of the malaria parasite, Plasmodium falciparum, found additional fragmented rRNAs and uncharacterized small RNAs (Feagin et al. 2012). Diverse linear mt genomes have been reported in apicomplexans (Vaidya and Mather 2009). For example, Plasmodium has mt genomes in tandemly repeated arrays with a unit length of approximately 6 kb. On the other hand, Babesia and Theileria have monomeric mt genomes (Hikosaka et al. 2012). Previous work on dinoflagellate mt genomes has suggested complex organization, with extensive recombined and fragmented gene copies (Waller and Jackson 2009). Fragmented mt genomes and/or transcripts have been reported in at least 25 dinoflagellate taxa (table 1). The foregoing studies have confirmed that dinoflagellate mtDNA includes cox1, cox3, cob and fragmented rRNAs, and have detailed unusual mRNA characteristics (reviewed by Nash et al. 2008; Waller and Jackson 2009). Extensive RNA editing of the three protein-coding genes (Lin et al. 2002; reviewed in Lin et al. 2008) and trans-splicing of cox3 have been reported (Jackson et al. 2007; Imanian et al. 2012; Jackson and Waller 2013). However, transcripts from the basal dinoflagellates, Hematodinium sp. and Oxyyrhis marina, did not show RNA editing (Slamovits et al. 2007; Jackson et al. 2012). Trans-splicing of cox3 was not found in O. marina (Slamovits et al. 2007). Losses of canonical start and stop codons have also been suggested (Norman and Gray 1997; Jackson et al. 2012; reviewed in Nash et al. 2008). On the other hand, analyses of noncoding sequences have been frustrated by high recombination rates in these genomes (Patron et al. 2005; reviewed in Waller and Jackson 2009). In addition, some reports have suggested that the total dinoflagellate mt genome size is likely to be large (Waller and Jackson 2009; Shoguchi et al. 2013), and the dinoflagellate mt genome is thought to be one of the most complex (Nash et al. 2008). For example, it is estimated that 85% of the mt genome in Amphidinium carterae is noncoding (Nash et al. 2007). Although inverted repeat (IR) elements in intergenic regions have been reported, functions of these elements are unknown (Waller and Jackson 2009). Thus, it has been assumed that each alveolate lineage developed different mt genomic structure (Slamovits et al. 2007). Interestingly, recently reported mt genomes of colponemids, an early alveolate lineage, suggest that the ancestral alveolate genome encoded a typical mt gene set (Janouškovec et al. 2013).
Table 1.
Summary of the Papers Reporting mt Genomes and/or Transcriptomes in Dinoflagellates
amt genome sequences were reported.
Our previous work on the endosymbiotic dinoflagellate, Symbiodinium minutum, has confirmed the presence of unusual nuclear (Shoguchi et al. 2013) and plastid genomes (Mungpakdee et al. 2014). In addition, this species may possess high mt genome copy numbers (Shoguchi et al. 2013). In this study, by analyzing the wealth of sequence data, we characterized the Symbiodinium mt genome and transcriptomes, including many noncoding sequences, and we compared them with mt genomes of Plasmodium and dinoflagellates. Assembly of fragmented DNA in general is technically difficult, but physical link information from fosmid end sequencing greatly aided mt genome assembly. Our analysis reveals conserved, noncoding sequences during myzozoan (apicomplexans and dinoflagellates) mt genome evolution. In addition, Symbiodinium is a large genus, classified into nine major clades (Coffroth and Santos 2005; Pochon et al. 2014); therefore, the complete Symbiodinium mt genome will be an important resource to study populations and environmental adaptations using genomic approaches (Shinzato et al. 2014).
Results and Discussion
The De Novo Assembled mt Genome of S. minutum
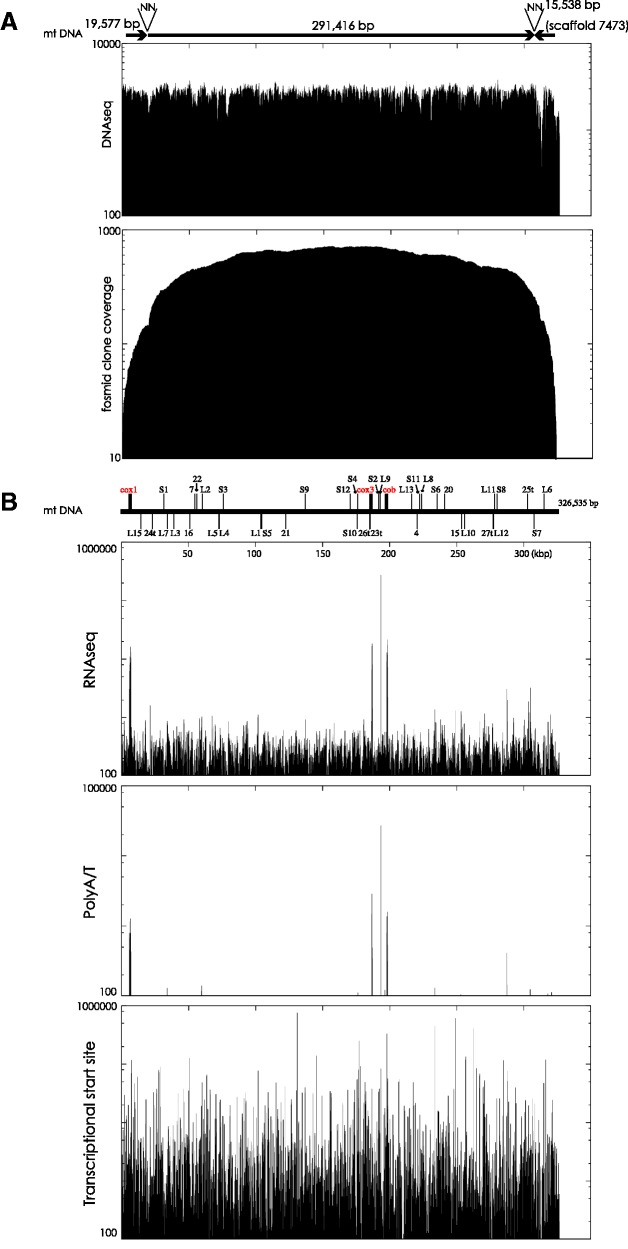
To reconstruct the mt genome of S. minutum, 20 analyses using only high coverage illumina paired-end reads (DNAseq) were performed (see also Materials and Methods). Two candidate mt contigs having more than 100× read coverage were obtained (19,577 and 291,416 bp) (accession numbers: LC002801 and LC002802) by 49-kmer assembly. Physical link information from fosmid paired-end sequences (FPESs) confirmed contig structures from computational sequence assembly (fig. 1A). In addition, joining of the 3′-end of the approximately 19-kb contig and the 5′-end of the approximately 291-kb contig was supported by FPES. BLAST (Basic Local Alignment Search Tool) searches showed that the approximately 19-kb contig contains the cox1 gene. The approximately 291-kb contig contains cob, cox3, and fragments of the LSU rRNA gene. Gene locations are explained in detail hereafter. Comparisons between the two contigs and the S. minutum genome assembly v1.0, using mapped FPES, showed that only scaffold 7473 (length: 15,538 bp) from genome assembly v1.0 (Shoguchi et al. 2013) was joined to the approximately 291-kb contig by more than 80 FPESs (fig. 1A). This suggested that nearly 40 kb of mtDNA had been identified. Estimation of the lengths of the two gaps was difficult. Accordingly, two bases (NN) were arbitrarily added between the two contigs and between the 291-kb contig and scaffold 7473 (see also fig. 1A). Comparison of the assembled mt genome with FPESs implies the presence of multiple recombinant mtDNA fragments, but our analysis suggests that S. minutum has a continuous mt genome of approximately 326 kb. Only simple repeats with fewer than 8 bp (∼1.49%) and low complexity (∼0.23%) were found in the mt genome assembly. The 49-bp repeats, which might be relevant to the assembly process, occurred fewer than four times in the approximately 326 kb.
Fig. 1.—
A mitochondrial genome and transcripts in S. minutum. (A) The assembled mt genome of S. minutum showing the high copy number. Arrows show two contigs and one scaffold (scaffold 7473), which are joined by paired-end sequences of fosmid clones and are labeled “NN” because of indeterminate distances. These constitute a scaffold of 326,535 kb. The upper graph indicates the high coverage of illumina reads that were mapped onto the scaffold. The lower plot shows clone coverage by fosmid paired-end mapping, partially supporting the accuracy of the assembly. (B) Transcriptomes from mtDNA and possible ends. The S. minutum mt genome with predicted genes is shown in upper region. Genes above or below the line indicate the transcription direction. Protein-coding genes are in red. Detailed gene map information is shown in supplementary figure S4, Supplementary Material online, and table 3. The upper graph shows coverage of RNAseq reads from illumina libraries that are enriched RNAs with polyA sequences. High coverage reads are found on cox1, cox3, fragment E of the ribosomal LSU, and cob. Only reads with poly A or T (more than four) are shown on middle graph, suggesting polyadenylated transcripts and potential 3′-ends. The lower graph displays reads from the TSS library, which is enriched RNA with 5′ cap structures, indicating the presence of multiple 5′-ends.
Transcriptomes of Symbiodinium mt Coding Genes
RNAseq reads were mapped onto the continuous genome (fig. 1B), revealing high coverage of cox1, cob, cox3, and the fragmented LSU gene (fig. 1B). Mapped data indicated the possibility of polycistronic expression. Mapping of reads with polyA or T in the 5′ sequence showed four major peaks for three protein-coding genes and the fragmented LSU. The highest peak is likely to be from the fragmented LSU gene, suggesting high expression and enhanced polyadenylation during RNA processing. Reads mapped from the transcription start site (TSS) library showed high coverage of multiple sites, suggesting multiple 5′ cleavage sites and transcripts with modified 5′-phosphate groups (fig. 1B). Symbiodinium minutum mt transcripts did not show evidence of RNA processing, such as 5′ oligo (U) caps of O. marina mt transcripts (Slamovits et al. 2007).
Edited RNA sites for transcripts of cox1, cob, and cox3 were investigated using comparisons between assembled genomes and transcripts. A to G editing was found in 61% of the 72 sites, showing conservation between dinoflagellates (table 2; Lin et al. 2008). In addition, patterns of RNA editing-mediated amino acid substitutions correspond to previous report about another species (supplementary fig. S1, Supplementary Material online; Lin et al. 2008).
Table 2.
RNA Editing Types in Three Mitochondrial Genes of Symbiodinium minutum
| Gene | Transcriptome ID | No. of Edits (%)a | Editing Type |
No. of Amino Acid Substitutions (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A/G | G/A | C/U | U/C | G/C | U/G | A/C | ||||
| cox1 | symbB1.comp234_c0_seq1 | 29/1,455 (2.0) | 18 | 0 | 3 | 4 | 2 | 1 | 1 | 24/485 (4.9) |
| cox3 | symbB1.comp4_c1_seq1, symbB1.EST_k37c20_2341 | 24/774 (3.1) | 18 | 1 | 1 | 4 | 0 | 0 | 0 | 23/258 (8.9)b |
| cob | symbB1.EST_k37c20_4808 | 19/1,062 (1.8) | 8 | 0 | 4 | 4 | 2 | 1 | 0 | 19/354 (5.4)b |
aEdits in predicted coding sequences were counted.
bIncluding a signal from stop codon. For cox3, adenylated sequences may be used as stop signals.
Another unusual feature of dinoflagellate mt genes is the lack of canonical start and stop codons to direct the initiation and termination of translation (Norman and Gray 2001; Jackson et al. 2012; reviewed in Nash et al. 2008). We have characterized start and stop codons of S. minutum mt genes using manual alignments between genomic and transcriptomic sequences (supplementary fig. S2, Supplementary Material online). We found AUA (Ile) and AUU (Ile) at the 5′-end of cox1 and AUU (Ile) in cox3. They are also candidates for start codons as mt genes in both ciliates and apicomplexans use AUA and AUU for this purpose (Feagin 1992; Edqvist et al. 2000). The cob gene in S. minutum contained both canonical start and stop codons; cox3 contained a canonical stop codon resulting from polyA addition (supplementary fig. S2, Supplementary Material online), as reported in other dinoflagellates (Waller and Jackson 2009). cox1 does not contain a stop codon (supplementary fig. S2, Supplementary Material online).
Noncoding RNA Genes and Gene Map
In the apicomplexan P. falciparum, 39 RNA genes, including fragmented rRNA LSUs (15), SSUs (12), and uncharacterized small RNAs (12), have been identified (Feagin et al. 2012). These rRNA fragments are not arranged linearly, but synteny was conserved in Plasmodium (Vaidya and Mather 2009). It is suggested that this fragmentation occurred in the common ancestor of apicomplexans and dinoflagellates (Slamovits et al. 2007; Jackson et al. 2012). To predict fragmented rRNAs in the S. minutum mt genome, the most similar regions from P. falciparum (Feagin et al. 2012) were surveyed and aligned (table 3 and supplementary fig. S3, Supplementary Material online). Their alignments with interspersed regions of the S. minutum mt genome showed more than 50% similarity, corresponding to RNAseq reads from TSS libraries, and indicating the presence of multiple RNAs (fig. 1B). Comparisons of secondary structures for aligned sequences of RNA genes showed that the majority of predicted genes have stem-loop structures (supplementary fig. S3B, Supplementary Material online). Thus, the assembled S. minutum genome contains orthologs to genes in the P. falciparum mt genome (table 3 and supplementary fig. S3, Supplementary Material online).
Table 3.
Predicted Genes in Mitochondrial Genome of Symbiodinium minutum
| Gene | Subunit Order | Predicted Location | Orientation to Scaffold | Similarity to Plasmodium falciparum Gene (%)a |
|---|---|---|---|---|
| cox1 | 5809–7248 | + | 916/1,441 (63) | |
| cox3 | 186587–187332 | + | 405/771 (52) | |
| cob | 197602–198718 | + | 688/1131 (60) | |
| SSUA | S4 | 177279–177354 | + | 57/80 (71) |
| SSUB | S6 | 236311–236394 | + | 48/86 (55) |
| SSUD | S10 | 176902–176959 | − | 37/63 (58) |
| SSUE | S11 | 221699–221724 | + | 19/26 (73) |
| SSUF | S12 | 170409–170456 | + | 31/48 (64) |
| LSUA | L1 | 105339–105493 | − | 89/158 (56) |
| LSUB | L3 | 38813–38831 | − | 17/19 (89) |
| LSUC | L4 | 73563–73580 | − | 17/18 (94) |
| LSUD | L8 | 222761–222836 | + | 55/76 (72) |
| LSUE | L9 | 193381–193573 | + | 149/195 (76) |
| LSUF | L11 | 279828–279907 | + | 55/80 (68) |
| LSUG | L12 | 278801–278900 | − | 74/100 (74) |
| RNA1 | L6 | 317063–317147 | + | 54/88 (61) |
| RNA2 | L2 | 60688–60729 | + | 26/42 (61) |
| RNA3 | L7 | 34514–34593 | − | 45/81 (55) |
| RNA4 | 220439–220506 | − | 39/68 (57) | |
| RNA5 | S9 | 138204–138280 | + | 48/80 (60) |
| RNA6 | L15 | 14596–14626 | − | 27/33 (81) |
| RNA7 | 56199–56266 | + | 53/69 (76) | |
| RNA8 | S5 | 106227–106279 | − | 30/53 (56) |
| RNA9 | S8 | 281819–281866 | + | 34/50 (68) |
| RNA10 | L13 | 217165–217255 | + | 59/92 (64) |
| RNA11 | L5 | 73434–73479 | − | 29/46 (63) |
| RNA12 | S2 | 191852–191892 | + | 30/41 (73) |
| RNA13 | L10 | 256591–256614 | − | 17/24 (70) |
| RNA14 | S1 | 31151–31177 | + | 21/27 (77) |
| RNA15 | 253421–253447 | − | 19/27 (70) | |
| RNA16 | 51961–51991 | − | 21/31 (67) | |
| RNA17 | S3 | 76949–76985 | + | 24/37 (64) |
| RNA18 | L14 | 300291–300312 | + | 17/22 (77) |
| RNA19 | S7 | 308063–308089 | − | 21/27 (77) |
| RNA20 | 242197–242225 | + | 20/29 (68) | |
| RNA21 | 122845–122864 | − | 16/20 (80) | |
| RNA22 | 57665–57699 | + | 24/35 (68) | |
| RNA23t | 186459–186487 | − | 20/29 (68) | |
| RNA24t | 23626–23665 | − | 27/40 (67) | |
| RNA25t | 302697–302717 | + | 16/21 (76) | |
| RNA26t | 186374–186415 | − | 29/43 (67) | |
| RNA27t | 278662–278712 | − | 36/52 (69) |
Note.—Gene names and subunit order refer to Feagin et al. (2012). L and S indicate LSU and SSU, respectively.
aGenes are from P. falciparum M76611 (Feagin et al. 2012). Alignments are shown in supplementary figure S3, Supplementary Material online.
tRNA genes were not found in the S. minutum mt genome using tRNA scan. So far no studies of dinoflagellate or apicomplexan mtDNAs have identified any tRNA genes, suggesting that tRNAs have been imported from the nuclear genome, as was reported for the apicomplexan, Toxoplasma gondii (Esseiva et al. 2004).
Interestingly, two LSU fragments, L4 and L5, map onto neighboring regions of the S. minutum mt genome with fewer than 100 bp between them (table 3 and supplementary fig. S4, Supplementary Material online). The L4–L5 arrangement in S. minutum, corresponding to the continuous large rRNA sequence order, appears to be evolutionarily conserved. Secondary structure prediction for L4 and L5 sequences yields a very stable, double-stranded form; however, the predicted structure varies depending on which secondary structure prediction program is used (supplementary fig. S3C and D, Supplementary Material online). Genes with unknown functions, RNA 23 t and RNA 26 t, are close, separated by only an approximately 40-bp intergenic sequence. Conserved, fragmented LSUs and SSUs may be cleaved accurately by small RNAs, such as RNA 23 t and RNA 26 t.
Comparisons of mt gene arrangements between Symbiodinium and Plasmodium showed only one microsyntenic region, which has the same gene arrangement on S12 and S10 (fig. 2 and supplementary fig. S4, Supplementary Material online), suggesting that the fragmentation occurred in the common ancestor of apicomplexans and dinoflagellates and that genome rearrangements in these lineages were very frequent. Thus, this basic information is valuable for possible functional analysis of dinoflagellate mt genomes.
Fig. 2.—
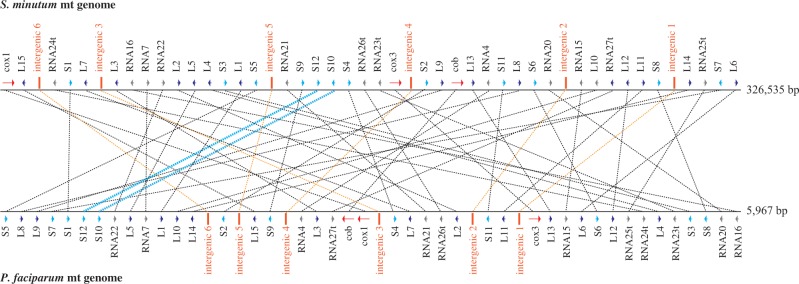
Mitochondrial gene order comparisons between S. minutum and P. falciparum. Genes from the S. minutum mt genome (∼326 kb) to the upper are joined to those of P. falciparum. The gene order of S10 and S12 was the same in mt genomes of both P. falciparum and S. minutum (aqua lines), showing minimal conservation of gene order. Sequence similarities from intergenic regions of the P. falciparum mt genome are indicated by orange lines. Details for the S. minutum mt genome map are shown in supplementary figure S4, Supplementary Material online.
Unknown Noncoding Regions and Possible Expansion in the Dinoflagellate Lineage
Our analysis confirms that noncoding sequences of the Symbiodinium mt genome have been expanded, raising the question as to where the expanded sequences originated. Enormous expansion of intergenic content in the mt genomes of seed plants (200–2,900 kb) has been reported (Mower et al. 2012), and repeated proliferation of “selfish” DNA has contributed overwhelmingly to these expansions (Chaw et al. 2008). Highly repetitive sequences were not found in the mt genome of S. minutum. In addition, when compared with other dinoflagellate mt sequences, the S. minutum mt sequences suggest additional RNA fragments or pseudogenes (supplementary fig. S5, Supplementary Material online), but these were not identified.
To find conserved secondary structures of potential RNA genes, each chopped 300-bp sequence from the mt genome was employed as a query sequence to perform RNA homology searches using Infernal (Nawrocki and Eddy 2013). Twenty-two sequences showed similarity to reported sequences in the Rfam database, including microRNAs and LSUs (Nawrocki et al. 2015) and suggest the presence of unknown RNA genes (supplementary table S1, Supplementary Material online). Unexpectedly, possible secondary structures for these genes included a stem of more than 20 bp (supplementary fig. S6, Supplementary Material online). Although we did not find sequence similarities to reported small, IRs of dinoflagellate mtDNA (reviewed in Waller and Jackson 2009) in the S. minutum mtDNA, the result suggests that structures comprising IRs are conserved characters among dinoflagellates mtDNAs.
Transcriptional control of the alveolate mt genome is not clear (Gray et al. 2004; Waller and Jackson 2009). Our RNAseq reads support the possibility of polycistronic expression. To detect conserved intergenic regions, similarities to the six intergenic sequences of P. falciparum and mtDNA sequences of dinoflagellates were surveyed. Interestingly, our analysis showed that intergenic sequences of P. falciparum have similarities to the mt genome of S. minutum at the same level as comparisons between rRNA sequences. (fig. 2 and supplementary fig. S7, Supplementary Material online).
It is very interesting to examine how organelle genomes of dinoflagellates evolved different structures, given the large variety of structures between plastid and mt genomes of Symbiodinium. Although the plastid genome has undergone reconfiguration to a compact DNA minicircle (∼1.8–3.0 kb) with its own regulatory regions (Mungpakdee et al. 2014), the mt genome appears greatly expanded and fragmented.
Materials and Methods
Genome Assembly
Genomic DNA sequences from cloned and cultured S. minutum Mf1.05 b.01 (clade type: B1) were obtained previously (Shoguchi et al. 2013). To obtain longer mt contigs, using Velvet (version 1.2.08) software (Zerbino and Birney 2008) for illumina paired-end reads (6.8 Gb from a polymerase chain reaction-free library, accession number: DRX003100), calculations of 20 patterns were performed on a combination of ten kmer parameters (kmer size: 27, 31, 35, 37, 41, 43, 45, 47, 49, and 51) using two coverage threshold values (>50 and >100). The 151,553 FPESs with approximately 2.1× coverage of the whole genome (Hattori et al. 2000; Shoguchi et al. 2013) (deposited in DNA Data Bank of Japan, accession numbers: GA453877–GA605429) were mapped onto mt contigs using BLASTN and relationships between contigs were examined.
The assembled mt genome was also compared with FPESs and assembly version 1 scaffolds from contigs based on Roche 454 reads (GenBank ID: BASF00000000.1; see also http://marinegenomics.oist.jp/genomes/gallery, last accessed September 1, 2014) (Koyanagi et al. 2013; Shoguchi et al. 2013).
Transcriptome Mapping
Transcriptome reads deposited at DRP000944 were mapped onto the assembled mt genome using Bowtie 2 (version 2.1.0) software (Langmead and Salzberg 2012). Detection of RNA editing for cob, cox1, and cox3 was basically performed in the same manner as for plastid transcripts (Mungpakdee et al. 2014). Differences between DNA and RNA were detected by aligning transcriptome contigs to a scaffold. RNAseq reads were mapped onto mt transcriptome contigs using TopHat (Trapnell et al. 2009) and accuracy was confirmed. Reads from a TSS library (Yamashita et al. 2011; Shoguchi et al. 2013) were mapped using Bowtie 2, as described in Mungpakdee et al. (2014).
Data Analysis Software and Sequences Used for Comparisons
Repeats in the mt genome assembly were detected using RepeatMasker in default mode (http://www.repeatmasker.org). tRNAscan-SE (with default parameters in organellar mode) (Schattner et al. 2005) was used to find tRNA genes in the mt genome and transcriptome contigs. Sequence alignments between Symbiodinium and Plasmodium were performed using GENETYX-MAC version 17 and BLASTN. RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi; Zuker and Stiegler 1981) and CentroidFold (http://www.ncrna.org/centroidfold/; Sato et al. 2009) with initial settings were used for prediction of RNA secondary structure. We prepared chopped 300-bp sequences from the mt genome with 100-bp overlap sequences. RNA homology searches for each of the 300-bp sequences were performed using infernal (INFERence of RNA ALignment) using default parameters (Nawrocki and Eddy 2013). The probability cutoff value (E value) was set at 0.001.
mt genome sequences used for comparisons have the following accession numbers: “HE610722–HE610773” for Hematodinium sp., “AB265207–AB265210 and AB374233–AB374251” for Alexandrium catenella, “JX001584–JX001600” for Durinskia baltica, “EF442995–EF443047 and AM773790–AM773803” for Karlodinium micrum, “JX001601–JX001608” for Kryptoperidinium foliaceum, “EF680822–EF680839” for O. marina, “M76611” for P. falciparum, “KF651061” for Alveolata sp. 1 JJ-2013 (colponemid-like Peru), which is a new species, Acavomonas peruviana (Tikhonenkov et al. 2014), and “AF396436” for T. thermophile.
Supplementary Material
Supplementary figures S1–S7 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank members of the OIST DNA sequencing section for whole performing genome sequencing. They gratefully acknowledge Dr Atsushi Toyoda, Dr Yoko Kuroki, and Dr Asao Fujiyama for construction of fosmid libraries and end sequencing. They are grateful to Dr Yutaka Suzuki and Dr Sumio Sugano, who performed TSS sequencing, and Dr Mary Alice Coffroth, who kindly provided the Symbiodinium minutum isolate. They also thank Dr Takeshi Kawashima for useful discussions of sequence analysis and Dr Steven D. Aird for editing the manuscript. This work was supported in part by Grants-in-Aids from MEXT (No. 25128712, No. 221S0002) and JSPS (No. 90378563 to E.S., No. 24241071 to N.S.), Japan. The authors also greatly appreciate support from OIST Graduate University to Nori Satoh.
Literature Cited
- Burger G, et al. 2000. Complete sequence of the mitochondrial genome of Tetrahymena pyriformis and comparison with Paramecium aurelia mitochondrial DNA. J Mol Biol. 297:365-380. [DOI] [PubMed] [Google Scholar]
- Chaput H, Wang Y, Morse D. 2002. Polyadenylated transcripts containing random gene fragments are expressed in dinoflagellate mitochondria. Protist 153:111-122. [DOI] [PubMed] [Google Scholar]
- Chaw SM, et al. 2008. The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Mol Biol Evol. 25:603-615. [DOI] [PubMed] [Google Scholar]
- Coffroth MA, Santos SR. 2005. Genetic diversity of symbiotic dinoflagellates in the genus Symbiodinium. Protist 156:19-34. [DOI] [PubMed] [Google Scholar]
- Edqvist J, Burger G, Gray MW. 2000. Expression of mitochondrial protein-coding genes in Tetrahymena pyriformis. J Mol Biol. 297:381-393. [DOI] [PubMed] [Google Scholar]
- Esseiva AC, Naguleswaran A, Hemphill A, Schneider A. 2004. Mitochondrial tRNA import in Toxoplasma gondii. J Biol Chem. 279:42363-42368. [DOI] [PubMed] [Google Scholar]
- Feagin JE. 1992. The 6-kb element of Plasmodium falciparum encodes mitochondrial cytochrome genes. Mol Biochem Parasitol. 52:145-148. [DOI] [PubMed] [Google Scholar]
- Feagin JE, et al. 2012. The fragmented mitochondrial ribosomal RNAs of Plasmodium falciparum. PLoS One 7:e38320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Burger G. 2004. Mitochondria of protists. Annu Rev Genet. 38:477-524. [DOI] [PubMed] [Google Scholar]
- Hattori M, et al. 2000. The DNA sequence of human chromosome 21. Nature 405:311-319. [DOI] [PubMed] [Google Scholar]
- Hikosaka K, et al. 2012. Novel type of linear mitochondrial genomes with dual flip-flop inversion system in apicomplexan parasites, Babesia microti and Babesia rodhaini. BMC Genomics 13:622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanian B, Keeling PJ. 2007. The dinoflagellates Durinskia baltica and Kryptoperidinium foliaceum retain functionally overlapping mitochondria from two evolutionarily distinct lineages. BMC Evol Biol. 7:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanian B, Pombert JF, Dorrell RG, Burki F, Keeling PJ. 2012. Tertiary endosymbiosis in two dinotoms has generated little change in the mitochondrial genomes of their dinoflagellate hosts and diatom endosymbionts. PLoS One 7:e43763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CJ, et al. 2007. Broad genomic and transcriptional analysis reveals a highly derived genome in dinoflagellate mitochondria. BMC Biol. 5:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CJ, Gornik SG, Waller RF. 2012. The mitochondrial genome and transcriptome of the basal dinoflagellate Hematodinium sp.: character evolution within the highly derived mitochondrial genomes of dinoflagellates. Genome Biol Evol. 4:59-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CJ, Waller RF. 2013. A widespread and unusual RNA trans-splicing type in dinoflagellate mitochondria. PLoS One 8:e56777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouškovec J, et al. 2013. Colponemids represent multiple ancient alveolate lineages. Curr Biol. 23:2546-2552. [DOI] [PubMed] [Google Scholar]
- Kamikawa R, Inagaki Y, Sako Y. 2007. Fragmentation of mitochondrial large subunit rRNA in the dinoflagellate Alexandrium catenella and the evolution of rRNA structure in alveolate mitochondria. Protist 158:239-245. [DOI] [PubMed] [Google Scholar]
- Kamikawa R, Nishimura H, Sako Y. 2009. Analysis of the mitochondrial genome, transcripts, and electron transport activity in the dinoflagellate Alexandrium catenella (Gonyaulacales, Dinophyceae). Phycol Res. 57:1-11. [Google Scholar]
- Koyanagi R, et al. 2013. MarinegenomicsDB: an integrated genome viewer for community-based annotation of genomes. Zool Sci. 30:797-800. [DOI] [PubMed] [Google Scholar]
- Lang BF, Gray MW, Burger G. 1999. Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet. 33:351-397. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Zhang H, Gray MW. 2008. RNA editing in dinoflagellates and its implications for the evolutionary history of the editing machinery. In: Smith HC, editor RNA and DNA editing: molecular mechanisms and their integration into biological systems. Hoboken (NJ): John Wiley & sons, Inc; p. 280–309. [Google Scholar]
- Lin S, Zhang H, Spencer DF, Norman JE, Gray MW. 2002. Widespread and extensive editing of mitochondrial mRNAS in dinoflagellates. J Mol Biol. 320:727-739. [DOI] [PubMed] [Google Scholar]
- Mower JP, Case AL, Floro ER, Willis JH. 2012. Evidence against equimolarity of large repeat arrangements and a predominant master circle structure of the mitochondrial genome from a monkeyflower (Mimulus guttatus) lineage with cryptic CMS. Genome Biol Evol. 4:670-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungpakdee S, et al. 2014. Massive gene transfer and extensive RNA editing of a symbiotic dinoflagellate plastid genome. Genome Biol Evol. 6:1408-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash EA, et al. 2007. Organization of the mitochondrial genome in the dinoflagellate Amphidinium carterae. Mol Biol Evol. 24:1528-1536. [DOI] [PubMed] [Google Scholar]
- Nash EA, Nisbet RE, Barbrook AC, Howe CJ. 2008. Dinoflagellates: a mitochondrial genome all at sea. Trends Genet. 24:328-335. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29:2933-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, et al. 2015. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43:D130-D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman JE, Gray MW. 1997. The cytochrome oxidase subunit 1 gene (cox1) from the dinoflagellate, Crypthecodinium cohnii. FEBS Lett. 413:333-338. [DOI] [PubMed] [Google Scholar]
- Norman JE, Gray MW. 2001. A complex organization of the gene encoding cytochrome oxidase subunit 1 in the mitochondrial genome of the dinoflagellate, Crypthecodinium cohnii: homologous recombination generates two different cox1 open reading frames. J Mol Evol. 53:351-363. [DOI] [PubMed] [Google Scholar]
- Patron NJ, Waller RF, Archibald JM, Keeling PJ. 2005. Complex protein targeting to dinoflagellate plastids. J Mol Biol. 348:1015-1024. [DOI] [PubMed] [Google Scholar]
- Pochon X, Putnam HM, Gates RD. 2014. Multi-gene analysis of Symbiodinium dinoflagellates: a perspective on rarity, symbiosis, and evolution. PeerJ. 2:e394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rycovska A, Valach M, Tomaska L, Bolotin-Fukuhara M, Nosek J. 2004. Linear versus circular mitochondrial genomes: intraspecies variability of mitochondrial genome architecture in Candida parapsilosis. Microbiology 150:1571-1580. [DOI] [PubMed] [Google Scholar]
- Sato K, Hamada M, Asai K, Mituyama T. 2009. CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res. 37:W277–W280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, Mungpakdee S, Satoh N, Shoguchi E. 2014. A genomic approach to coral-dinoflagellate symbiosis: studies of Acropora digitifera and Symbiodinium minutum. Front Microbiol. 5:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoguchi E, et al. 2013. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol. 23:1399-1408. [DOI] [PubMed] [Google Scholar]
- Slamovits CH, Saldarriaga JF, Larocque A, Keeling PJ. 2007. The highly reduced and fragmented mitochondrial genome of the early-branching dinoflagellate Oxyrrhis marina shares characteristics with both apicomplexan and dinoflagellate mitochondrial genomes. J Mol Biol. 372:356-368. [DOI] [PubMed] [Google Scholar]
- Sloan DB, et al. 2012. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 10:e1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 25:1105-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonenkov DV, et al. 2014. Description of Colponema vietnamica sp.n. and Acavomonas peruviana n. gen. n. sp., two new alveolate phyla (Colponemidia nom. nov. and Acavomonidia nom. nov.) and their contributions to reconstructing the ancestral state of alveolates and eukaryotes. PLoS One 9:e95467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya AB, Mather MW. 2009. Mitochondrial evolution and functions in malaria parasites. Annu Rev Microbiol. 63:249-267. [DOI] [PubMed] [Google Scholar]
- Waller RF, Jackson CJ. 2009. Dinoflagellate mitochondrial genomes: stretching the rules of molecular biology. Bioessays 31:237-245. [DOI] [PubMed] [Google Scholar]
- Yamashita R, et al. 2011. Genome-wide characterization of transcriptional start sites in humans by integrative transcriptome analysis. Genome Res. 21:775-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bhattacharya D, Maranda L, Lin S. 2008. Mitochondrial cob and cox1 genes and editing of the corresponding mRNAs in Dinophysis acuminata from Narragansett Bay, with special reference to the phylogenetic position of the genus Dinophysis. Appl Environ Microbiol. 74:1546-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lin S. 2002. Detection and quantification of Pfiesteria piscicida by using the mitochondrial cytochrome b gene. Appl Environ Microbiol. 68:989-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lin S. 2005. Mitochondrial cytochrome b mRNA editing in dinoflagellates: possible ecological and evolutionary associations? J Eukaryot Microbiol. 52:538-545. [DOI] [PubMed] [Google Scholar]
- Zuker M, Stiegler P. 1981. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 9:133-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.