Abstract
Horizontal gene transfer (HGT) of operational genes has been widely reported in prokaryotic organisms. However, informational genes such as those involved in transcription and translation processes are very difficult to be horizontally transferred, as described by Woese’s complexity hypothesis. Here, we analyzed all of the completed prokaryotic genome sequences (2,143 genomes) in the NCBI (National Center for Biotechnology Information) database, scanned for genomes with high intragenomic heterogeneity of 16S rRNA gene copies, and explored potential HGT events of ribosomal RNA genes based on the phylogeny, genomic organization, and secondary structures of the ribosomal RNA genes. Our results revealed 28 genomes with relatively high intragenomic heterogeneity of multiple 16S rRNA gene copies (lowest pairwise identity <98.0%), and further analysis revealed HGT events and potential donors of the heterogeneous copies (such as HGT from Chlamydia suis to Chlamydia trachomatis) and mutation events of some heterogeneous copies (such as Streptococcus suis JS14). Interestingly, HGT of the 16S rRNA gene only occurred at intragenus or intraspecies levels, which is quite different from the HGT of operational genes. Our results improve our understanding regarding the exchange of informational genes.
Keywords: 16S rRNA genes, intragenomic heterogeneity, horizontal gene transfer, accumulation of substitutions
Introduction
Ribosomal RNA and protein constitute the ribosome, which is responsible for the translation of protein in a cell. Ribosomal RNA genes contain both conserved and variable regions, which make them a suitable biomarker for taxonomic identification. Thus, the 16S rRNA gene has been selected as a gold standard marker gene for prokaryotic classification (Woese 1987).
Horizontal gene transfer (HGT) was described in a virulent strain of Corynebacterium diphtheria in 1951 (Freeman 1951), and then HGT was demonstrated between bacteria and archaea (Nelson et al. 1999; Garcia-Vallve et al. 2000), prokaryotes and eukaryotes (Ros and Hurst 2009), and among eukaryotes (Andersson 2005). HGT has been considered to be an evolutionary mechanism for the development of adaptive features in the recipient of the transferred genes.
It is well known that informational genes are less likely to be horizontally transferred compared with operational genes (Woese et al. 1985; Rivera et al. 1998; Jain et al. 1999). The complexity hypothesis claimed that informational genes, such as those involved in transcription, translation and related processes, are members of large, complex systems, whereas operational genes (such as enzyme-encoding genes) are not as large and complex in most cases. Members of large complex systems are usually specific and conserved in terms of function, and they have a reduced potential to function normally in the systems of other species because they may not be compatible with other components of the system.
HGT and subsequent DNA recombination have been frequently detected in prokaryotes, including intracellular pathogens (such as Chlamydia [Gomes et al. 2004; DeMars et al. 2007; DeMars and Weinfurter 2008]), symbionts (such as symbiotic Synechocystis [Ponting et al. 1999]), and thermophilic bacteria and archaea (such as Aquifex aeolicus and Thermotoga maritima [Aravind et al. 1998, 1999]), among others. The transferred genes include genes that encode beneficial features, such as antibiotic resistance genes, toxins, cell surface components, and regulatory functions (Nakamura et al. 2004). However, HGT of ribosomal RNA genes has rarely been reported.
Recent advances have revealed the intragenomic heterogeneity of prokaryotic ribosomal RNA genes, mostly as a result of high-throughput genomic sequencing, which has supported the occurrence of HGT of ribosomal RNA genes. Two studies investigated intragenomic heterogeneity of multiple copies of 16S rRNA gene available in the NCBI (National Center for Biotechnology Information) database of complete prokaryotic genomes in 2004 (Acinas et al. 2004) and 2010 (Pei et al. 2010) using 81 and 883 finished genomes, respectively. The findings of these studies have generated great research interest. The intragenomic heterogeneity of the 16S rRNA gene could result in an overestimation of the community diversity (Sun et al. 2013). Studies of individual bacteria with heterogeneous copies of the rrn gene have also suggested that the ribosomal RNA genes were likely to have originated from closely related species (Mylvaganam and Dennis 1992; van Berkum et al. 2003).
There was also experimental evidence shedding light into the possibility of HGT of ribosomal RNA genes (Kitahara and Miyazaki 2013). Kitahara et al. (2012) cloned amplified 16S rRNA genes from soil metagenomic DNA into Escherichia coli and successfully acquired strains with foreign 16S rRNA gene horizontally transferred. Another case study showed that the 16S rRNA gene could be horizontally transferred by transformation in Helicobacter pylori (Trieber and Taylor 2002).
In this study, we scanned 2,143 complete prokaryotic genomes in the NCBI database in June 2014, searching for potential HGT of ribosomal RNA genes and potential sources of the transferred copies, to provide comprehensive evidence of HGT and, more importantly, the potential donor of the HGT of ribosomal RNA genes.
Materials and Methods
Retrieval of Genomic Data and 16S rRNA Gene Identification
The 2,143 finished prokaryotic genome sequences (including both chromosomes and plasmids) considered in this study were obtained from the NCBI database (ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria/, including both bacteria and archaea). The rRNA genes were identified by searching against a hidden Markov model of ribosomal RNA genes for bacteria and archaea using Meta_RNA (Huang et al. 2009).
Determination of the Intragenomic Heterogeneity of 16S rRNA Genes
The obtained 16S rRNA genes of each genome were submitted to reciprocal BLAST (Basic Local Alignment Search Tool), and the pairwise similarities were calculated using a custom Perl script. We then scanned the genomes with the lowest pairwise identity (alignment length > 1,000 bp, E value < 1e-20) of multiple copies of the 16S rRNA gene less than 98%, with the exclusion of one genome demonstrating ambiguous bases of greater than 2 Ns per 16S rRNA gene. The distances among the genes of each genome were displayed by multidimensional scaling (MDS), and the distance pattern of the multicopy 16S rRNA genes was categorized into four types based on the grouping patterns.
Construction of Phylogenetic Trees Containing Multiple Copies of 16S rRNA Genes and References
The multiple copies of the 16S rRNA gene of a single genome were submitted to a BLAST analysis against the NCBI nucleotide database (NT, version of December 2013), and 50 hits (>1,000 bp, E value < 1e-20) of each query with the highest identities (>95%) were extracted. The extracted sequences of each hit were merged, and repeated duplicates were removed using custom Perl scripts. The hits and queries were used to construct a maximum-likelihood (ML) phylogenetic tree using MEGA 6 (Tamura et al. 2013) using the general time-reversible model with correction for among site rate variation (Gamma distributed with Invariant sites, G+I). A bootstrap replication of 100 was used. For each ML tree, a neighbor-joining tree, using a maximum composition likelihood substitution model and a bootstrap replications of 100, was supplemented to confirm the phylogeny of the genes.
Genome Arrangement Comparison of 16S rRNA Genes and Adjacent Regions
Those genomes with intragenomic heterogeneous 16S rRNA genes were compared using a BLAST analysis (E value < e-20) with an output in table format, which was then imported into the Artemis comparison tool (Carver et al. 2005). The neighboring regions of the rRNA gene operons were also compared to provide evidence for HGT events of heterogeneous 16S rRNA genes.
Secondary Structure Comparison of 16S rRNA Genes
Pairwise 16S rRNA genes were imported into RNAalifold (RIBOSUM scoring) using the minimum free energy partition function (Bernhart et al. 2008). The mismatches that occurred in the stem (potentially influencing the secondary structure) and loop were then labeled (supplementary fig. S1, Supplementary Material online, shows a demonstration). When comparing two related secondary structures, a mismatch was defined as conserved if it occurred in a loop or a stem but caused GC:GU conversions or covariation that resulted in no changes in base pairing (Acinas et al. 2004; Pei et al. 2010). In contrast, a nonconserved mismatch that alters base pairing and structure was classified as a stem–loop transition (supplementary fig. S1, Supplementary Material online). The nonconserved substitution rate (NCSR) was calculated by counting the percentages of those mismatches in stems that caused stem–loop conversion among all of the mismatches. A low NCSR was a sufficient but not a necessary condition for the normal function of the 16S rRNA gene.
Results
Intragenomic Heterogeneity of Multicopy 16S rRNA Genes
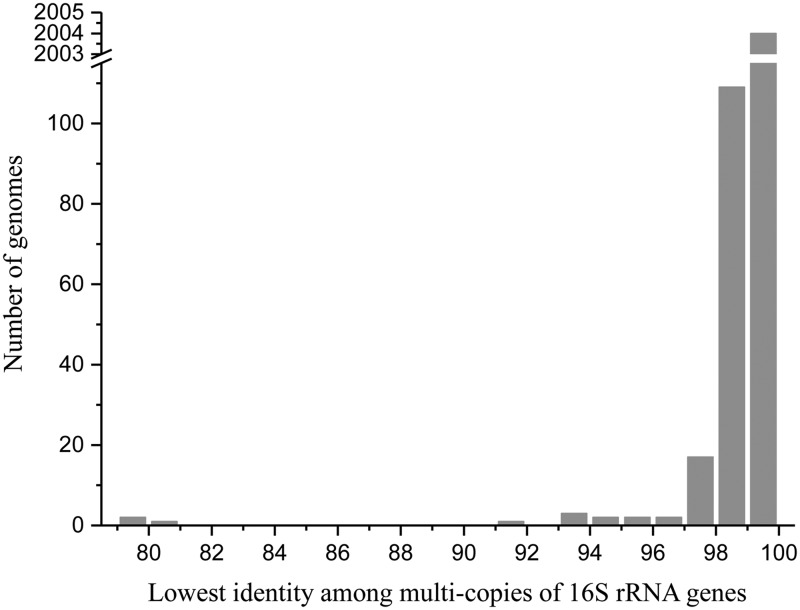
In total, 2,143 finished prokaryotic genomes (2,017 bacterial and 126 archaeal genomes) from the NCBI database (data collected up to June 22, 2014), encompassing 1,489 unique species, were assessed in this study. There were 446 genomes (20.1%) with single-copy 16S rRNA gene; the other genomes (1,697, 79.9%) contained 2–15 copies of the 16S rRNA gene. Among those containing multicopy 16S rRNA genes, 633 genomes (39.1%) had an identical 16S rRNA gene sequence, and 925 genomes (54.5%) had 16S rRNA gene sequences with the lowest intragenomic similarity between 99% and 100% (fig. 1). Using a threshold of 1–1.3% for the operational identification of species based on 16S rRNA genes (Thompson et al. 2005; Pei et al. 2010), 137 genomes demonstrated an intragenomic diversity equal to or higher than the operational threshold, revealing a borderline diversity (between 1% and 1.3%) in 57 genomes and ≥1.3% in 80 genomes. In this study, we focused on 28 genomes (table 1) that demonstrated the lowest identity among multicopy 16S rRNA genes less than 98%, which represented the genomes with the most diverse 16S rRNA genes.
Fig. 1.
—Summary of the lowest identity among the multicopy 16S rRNA genes of all of the analyzed prokaryotic genomes. This study focused on the genomes with the lowest identity among the multicopy 16S rRNA genes < 98%.
Table 1.
The Heterogeneity of Multiple 16S rRNA Gene Copies in 28 Genomes with an Intragenomic Dissimilarity of the 16S rRNA Gene >2% and Potential Gene Transfer Events
| Strain | Genome Accession Number | Number of rrn Gene Copy | Lowest Similarity | NCSR (%) | Putative Source of rrn HGT (Based on Similarity Pattern, Phylogeny, and Secondary Structure) |
|---|---|---|---|---|---|
| Chlamydia trachomatis J 6276tet1 | NC_021892 | 3 | 97.06 | 5.3 | Intraspecies HGT from host closely related to Chlamydia suis |
| Chlamydia trachomatis RC F s 342 | NC_021890 | 3 | 97.33 | 5.3 | Intraspecies HGT from host closely related to Chlamydia suis |
| Chlamydia trachomatis RC F s 852 | NC_021888 | 3 | 97.06 | 5.3 | Intraspecies HGT from host closely related to Chlamydia suis |
| Chlamydia trachomatis RC J s 122 | NC_021891 | 2 | 97.06 | 4.8 | Intraspecies HGT from host closely related to Chlamydia suis |
| Clostridium lentocellum DSM 5427 | NC_015275 | 11 | 96.88 | 2.4 | Intragenus HGT |
| Desulfotomaculum gibsoniae DSM 7213 | NC_021184 | 8 | 97.91 | 0.0 | Intragenus HGT |
| Escherichia coli APEC O1 | NC_008563 | 7 | 97.25 | 18.5a | Multiple intraspecies HGT |
| Escherichia coli CFT073 | NC_004431 | 7 | 97.57 | 3.6 | Multiple intraspecies HGT |
| Pectobacterium carotovorum PCC21 | NC_018525 | 7 | 97.59 | 8.1a | Intragenus HGT from host closely related to Erwinia aroideae |
| Photobacterium profundum SS9 | NC_006370 | 14 | 95.83 | Intragenus HGT | |
| Salmonella bongori NCTC 12419 | NC_015761 | 5 | 97.99 | 0.0 | Intragenus HGT |
| Thermoanaerobacter brockii finnii Ako 1 | NC_014964 | 4 | 93.68 | 0.0 | Intragenus HGT from host closely related to Thermoanaerobacter thermohydrosulfuricus |
| Thermoanaerobacter pseudethanolicus ATCC 33223 | NC_010321 | 4 | 93.77 | Intragenus HGT from host closely related to Thermoanaerobacter thermohydrosulfuricus | |
| Thermoanaerobacter tengcongensis MB4 | NC_003869 | 4 | 91.67 | 2.4 | Intragenus HGT |
| Shigella dysenteriae 1617 | NC_022912 | 7 | 97.64 | 0.0 | Insertion of 23-bp intervene sequence |
| Bacillus amyloliquefaciens plantarum AS43 3 | NC_019842 | 9 | 97.81 | 22.2 | Insertion of 17-bp intervene sequence |
| Bacteroides thetaiotaomicron VPI 5482 | NC_004663 | 5 | 97.91 | 15.2 | —b |
| Desulfotomaculum acetoxidans DSM 771 | NC_013216 | 10 | 97.33 | 2.9 | —b |
| Thermoanaerobacter mathranii A3 | NC_014209 | 4 | 96.81 | 38.9 | —b |
| Yersinia enterocolitica palearctica 105.5R r | NC_015224 | 7 | 97.09 | 7.0 | —b |
| Borrelia afzelii HLJ01 | NC_018887 | 2 | 80.03 | 12.3 | Accumulation of substitutions |
| Borrelia afzelii PKo | NC_017238 | 2 | 79.67 | 11.4 | Accumulation of substitutions |
| Borrelia afzelii PKo | NC_008277 | 2 | 79.67 | 12.5 | Accumulation of substitutions |
| Clostridium tetani 12124569 | NC_022777 | 4 | 93.41 | 10.5 | Accumulation of substitutions |
| Listonella anguillarum M3 | NC_022223 | 7 | 94.08 | 52.5 | Accumulation of substitutions |
| Streptococcus suis JS14 | NC_017618 | 4 | 95.66 | 16.0 | Accumulation of substitutions |
| Vibrio anguillarum 775 | NC_015633 | 7 | 97.88 | Accumulation of substitutions | |
| Exiguobacterium antarcticum B7 | NC_018665 | 9 | 94.32 | 0, 17.3 | One intragenus HGT and one accumulation of substitutions |
Note.—The corresponding phylogenetic analysis of each strain is shown in supplementary figure S6, Supplementary Material online.
aRelatively high NCSR of the normal 16S rRNA gene copy in comparison to the local copies, showing that the low NCSR is a sufficient but not a necessary condition for the normal function of the ribosomal RNA gene.
bEvidence for the phylogeny and secondary structure was not sufficient to determine the transfer source or the accumulation of substitutions.
Patterns of Intragenomic Heterogeneity of Multicopy 16S rRNA Genes
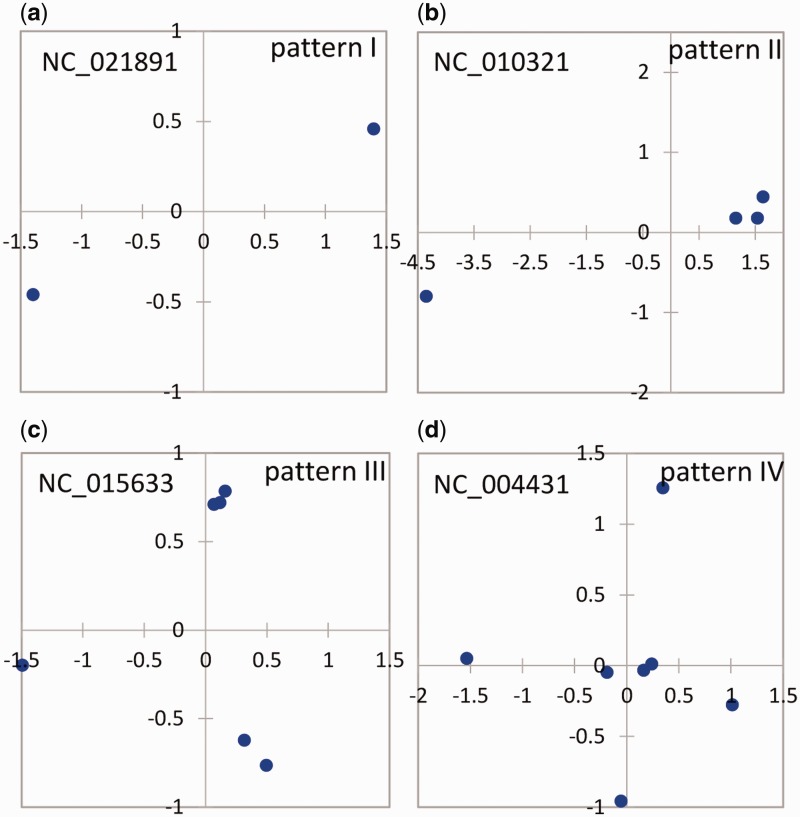
The pairwise similarity of multicopy 16S rRNA genes for each selected single genome was calculated and displayed in supplementary figure S2, Supplementary Material online. The pattern of distances among multicopy 16S rRNA genes within single genomes could be categorized into four patterns based on the grouping patterns of the multicopy genes (four typical patterns were selected and shown in fig. 2).
Fig. 2.
—MDS of four grouping patterns of the similarities of the multicopy 16S rRNA gene within single genomes with a high intragenomic heterogeneity of multiple 16S rRNA gene copies (lowest pairwise identity < 98.0%). Each dot indicates a copy of the 16S rRNA gene in the genome, and the distance between pairwise dots in the plot represents the dissimilarity between them.
The four patterns have different number of gene copies, and differed in terms of gene copy similarities. As shown in figure 2 and supplementary figure S2, Supplementary Material online, pattern I contained two copies of the 16S rRNA gene and demonstrated an identity less than 98% with the representative genome of Chlamydia trachomatis RC Js 122 (NC_021891, 2.94% dissimilarity between the two copies). Pattern II contained two groups of 16S rRNA genes, of which one might have been duplicated into several copies (identity > 99%); the identity of the two groups was less than 98%. The representative genome was obtained from Thermoanaerobacter pseudethanolicus ATCC33223 (NC_010321), and it contained three highly similar 16S rRNA gene copies with a lowest intragenomic identity of 99.6% and a heterogeneous copy with an identity less than 94.8% compared with the other three copies. Pattern III included three separate groups, among which one or two groups might have contained duplicated copies. The lowest identity between the pairwise groups was less than 98%. The representative genome of Vibrio anguillarum 775 (NC_015633) had a lowest pairwise identity of 2.1%. Pattern IV contained more than three separate groups, and some of the groups might have contained duplicated copies (identity > 99%). The representative genome E. coli CFT073 (NC_004431) contained four separated groups with a lowest pairwise identity of 97.6%.
Phylogenetic and Genomic Arrangement Analyses Reveal HGT of 16S rRNA Genes
Phylogenetic analysis of multiple copies of ribosomal RNA genes revealed the transfer of genes and, more importantly, the potential source of the transferred gene copies (the close relative of the real source). Phylogenetic tree of multicopy 16S rRNA genes from all of the selected genomes (supplementary fig. S6, Supplementary Material online) demonstrated the phylogenetic positions of native 16S rRNA gene copies that were in the lineage of the hosts, and foreign copies that were located on branches far from the host lineage. Some of those foreign 16S rRNA gene copies were closely related to the other lineages, whereas some of them were located on a long naked branch.
The Intragenus HGT of a 16S rRNA Gene Copy from Chlamydia suis to Chlamydia trachomatis
In this study, 94 genomes of Chlamydia were analyzed, including C. trachomatis (71 strains) (Harris et al. 2012; Jeffrey et al. 2013), C. muridarum (one strain), Chlamydia pecorum (four strains) and Chlamydia psittaci (18 strains), and only four strains of C. trachomatis contained intragenomic heterogenic 16S rRNA genes. In contrast, all of the other C. trachomatis strains contained one or two nearly identical (identity > 99.6%) 16S rRNA genes.
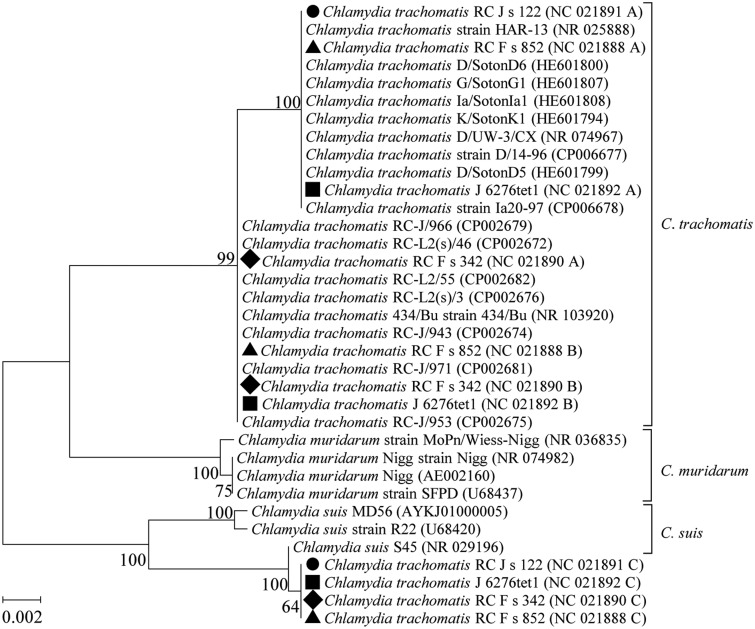
The phylogenetic tree of the four C. trachomatis strains (J 6276tet1, RC Fs 342, RC Fs 852, and RC Js 122) is shown in figure 3. The strain C. trachomatis RC Js 122 (NC_021891) possesses two copies of the 16S rRNA gene (copy A and copy B); copy A is located in the cluster of C. trachomatis, and copy B (97.1% identity to copy A) is closely related to Chlamydia suis S45 with only one mismatch over 1,520 bp (99.9% identity). These findings suggested that copy B was acquired from C. suis (an intragenus HGT event). The three other strains of C. trachomatis had very similar phylogenetic patterns and HGT events for multicopy 16S rRNA genes (fig. 3).
Fig. 3.
—Phylogenetic tree (ML, unrooted) of the 16S rRNA genes of four strains of Chlamydia trachomatis (J 6276tet1, three copies indicated by ▪; RC F s 342, three copies indicated by ♦; RC F s 852, three copies indicated by ▴; and RC J s 122, two copies indicated by •) and the closest relatives. The foreign 16S rRNA gene copies of the four strains are closely related to the C. suis. All of the 16S rRNA genes of the four strains are in operons.
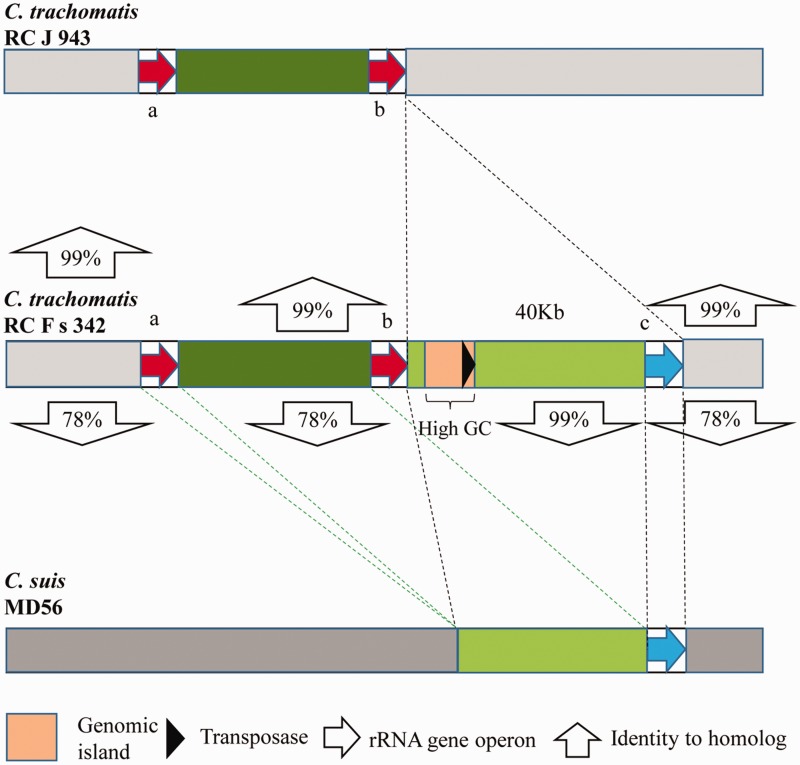
A comparison of the genomic structure further confirmed the HGT event of the ribosomal RNA gene operon with flanking sequences from C. suis to C. trachomatis (fig. 4 and supplementary fig. S2, Supplementary Material online). There was a 40-kb fragment present in the genome of C. trachomatis RC Fs 342 that was absent in the genome of C. trachomatis RC J 943 (close relative containing two identical ribosomal RNA gene operons). The inserted DNA fragment included an foreign rRNA gene operon (97.3% 16S rRNA gene identity to other copies) with a flanking fragment that was homologous to C. suis (in this comparison: Strain MD56, which is the only strain of C. suis with genome sequence available, 98.8% 16S rRNA gene identity to C. trachomatis RC Fs 342) and a genomic island with high GC content (56.1% in comparison to the average whole-genome GC content of 41.5%) from C. suis strain R19 (Donati et al. 2011) (AY428550, 99.9% identity over 13 kb). The genomic island contained a terminal transposase (the only transposase gene found throughout the genome), which may facilitate HGT of the genomic island containing tetracycline resistance genes enhancing the virulence of C. suis R19 (Donati et al. 2011). The four strains of C. trachomatis (J 6276tet1, RC Fs 342, RC Fs 852, and RC Js 122) were found to possess the same transferred rRNA operon and shared the same genomic organization in the region of the transferred rRNA operon (supplementary fig. S4, Supplementary Material online).
Fig. 4.
—Comparison of the genomic organization of C. trachomatis RC J 943 (with two copies of the ribosomal RNA gene operons), C. trachomatis RC F s 342 (with three copies of the ribosomal RNA gene operons), and C. suis MD56 (the potential source of the transferred ribosomal RNA gene operon in C. trachomatis RC F s 342). The original graph is shown in supplementary figure S3, Supplementary Material online. In comparison, the C. trachomatis RC F s 342 contains two native rRNA gene operons that are identical to the C. trachomatis RC J 943, and one more foreign rRNA gene operon in an inserted region (40 kb) that is closely related to that of C. suis MD56.
A comparison of the secondary structures of the transferred copy and the local copy of the 16S rRNA gene resulted in a relatively low NCSR of 2/45 (4.4%, supplementary fig. S5, Supplementary Material online, table 1 and supplementary table S1, Supplementary Material online), which indicated that most of the mismatches did not result in a loop–stem conversion. Also, the foreign 16S rRNA gene was transferred in the form of a complete operon, which was located in the 40-kb inserted region (fig. 4). These evidences suggested the normal functioning of the transferred copy in the recipient genomes.
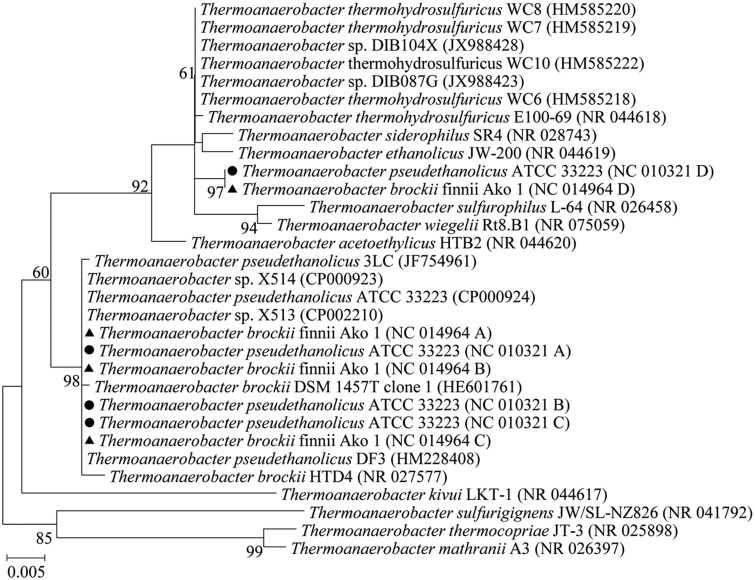
The Intragenus HGT of a Copy of the 16S rRNA Gene from Thermoanaerobacter thermohydrosulfuricus to T. pseudethanolicus and Thermoanaerobacter brockii
In correspondence to pattern II, the representative phylogenetic tree in figure 5 showed that the foreign copy of the 16S rRNA gene of T. pseudethanolicus ATCC 33223 was closely related to T. thermohydrosulfuricus (>99.3% similarity), whereas there were three identical copies (A, B, and C, 94.8% identity to the copy D) located in the lineage of the T. pseudethanolicus and the close relative species Thermoanaerobacter brockii. Closely related ortholog of the native copies was absent in T. thermohydrosulfuricus. A similar HGT also occurred in the strain T. brockii brockii finnii Ako 1, and the transferred copies (D) were identical whereas the native copies were 99.6% similar to those of T. pseudethanolicus ATCC 33223. The phylogenetic analysis indicated an intragenus HGT of the 16S rRNA gene in T. pseudethanolicus and T. brockii.
Fig. 5.
—Phylogenetic tree (ML, unrooted) of the 16S rRNA genes of T. pseudethanolicus ATCC 33223 and T. brockii finnii Ako 1 (with transferred rRNA gene copies from T. thermohydrosulfuricus) and the closest relatives. The copies A, B, and C are in the same cluster and are referred to as the native copy. The copy D of the two strains is closely related to the T. thermohydrosulfuricus (similarity > 99.3%). The 16S rRNA genes of T. pseudethanolicus ATCC 33223 are indicated by • and those of T. brockii finnii Ako 1 are indicated by ▴.
The Intraspecies HGT of Several Copies of the 16S rRNA Gene in E. coli Strains
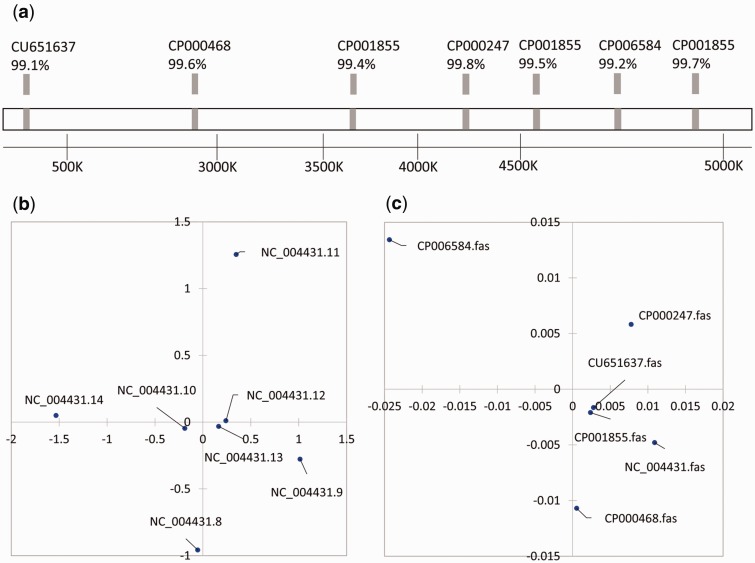
In pattern IV, there are more than three separate groups of 16S rRNA genes, and HGT of the gene from more than one donor was revealed by the phylogenetic analysis. In total, 62 genomes from different E. coli strains were analyzed, among which two strains (E. coli APEC O 1 NC_008563 and E. coli CFT073 NC_004431) have multiple copies of the 16S rRNA gene with the lowest identity less than 98%. The representative genome (E. coli CFT073) contains seven copies of the 16S rRNA gene dispersed throughout the chromosome (fig. 6) with a relatively low intragroup identity (lowest value of 97.6%, fig. 6b). The seven copies were closely related to the 16S rRNA genes of E. coli 536 (CP000247), E. coli NRG 857C (CP001855), E. coli APEC O1 (CP000468), E. coli LY180 (CP006584), and E. coli LF 82 (CU651637) (identity 99.1–99.8%, fig. 6a), indicating that they were potential HGT donors. It is difficult to determine the native 16S rRNA gene copy, because the putative HGTs are all at strain level, and there is no 16S rRNA gene copy closely related to other species. The potential HGT source strains were relatively distant strains with a whole-genome average nucleotide identity (ANI) of 96.4–98.8% to E. coli CFT073 (fig. 6c).
Fig. 6.
—Escherichia coli CFT073 with multicopy of the 16S rRNA gene dispersed in the genome and the pattern of dissimilarity of the 16S rRNA gene copies. (a) shows that these 16S rRNA gene copies were closely related to the 16S rRNA genes of E. coli 536 (CP000247), E. coli NRG 857C (CP001855), E. coli APEC O1 (CP000468), E. coli LY180 (CP006584), and E. coli LF 82 (CU651637) (identity 99.1–99.8%). (b) MDS result of the similarity of the multiple copies (with a relatively low intragroup similarity: Lowest value of 97.6%). (c) MDS result of similarity of the potential HGT donors based on the whole-genome ANI, which indicated that they were relatively distant strains with an ANI of 96.4–98.8% to E. coli CFT073.
HGT events of 16S RNA gene in other species and putative sources of the transfers based on a comprehensive consideration of similarity patterns, phylogeny, and secondary structure (based on NCSR) are listed in table 1 and supplementary table S1, Supplementary Material online.
Fixed Mutations in the 16S rRNA Gene Also Cause Intragenomic Heterogeneity
Phylogenetic analysis and genome arrangement comparison showed that the 16S rRNA gene intragenomic heterogeneity can be caused by HGT, but other events such as gene mutations may also result in such heterogeneity. The genome of Streptococcus suis JS14 (NC_017618) was found to have four 16S rRNA gene copies, one of which was highly dissimilar to the others with an identity less than 95.8% (supplementary fig. S2, Supplementary Material online). Phylogenetic analysis showed that the divergent copy was located in a long naked branch (supplementary fig. S6z, Supplementary Material online), indicating the lack of a close relative of the copy. The secondary structure analysis further indicated that the divergent 16S rRNA gene copy contained a high ratio of accumulation of substitutions in the stems that influenced the normal structures (supplementary fig. S7, Supplementary Material online), which would lead to malfunctions in the ribosome assemblage (with a relatively high NCSR of 16.0%). Putatively lethal mutations in other 16S rRNA genes have also been identified (shown in table 1 and supplementary table S1, Supplementary Material online) that appeared to have no phylogenetically close relatives. Most of them were located on a long naked branch in the phylogenetic tree (supplementary fig. S6, Supplementary Material online), forming a separate lineage from the closest relatives.
Discussion
In this study, we employed all of the currently available complete genomes to reveal the surprising HGT of heterogeneous 16S rRNA genes and their potential sources of transfer using methods of reciprocal BLAST of multicopy 16S rRNA genes, phylogenetic analysis and genome arrangement comparison. The pairwise similarities of multicopy 16S rRNA genes were visualized with MDS dot plots and the four similarity patterns could be clearly observed. Phylogenetic analysis showed the potential source of the foreign copies, and genome arrangement comparison confirmed the structures of the transferred regions.
It is well known that C. trachomatis is a pathogen that causes trachoma and sexual diseases, and C. suis causes conjunctivitis, enteritis, and pneumonia in swine. Chlamydia has recently attracted increasing research interest because of its harmful effects in clinical cases (Bachmann et al. 2014). In this study, more than 80 Chlamydia strains were analyzed, and four strains of C. trachomatis were found to have 16S rRNA genes that had been transferred from C. suis. HGT of functional genes such as antibiotic resistance genes between C. trachomatis and C. suis (Suchland et al. 2009) has been found, but never of ribosomal RNA genes. In this study, the four strains were generated by cross experiment using parental strains of C. trachomatis and C. suis (Jeffrey et al. 2013), and represented an experimental case among the other natural cases of HGT of 16S rRNA gene. The rRNA gene operon was transferred together with the genomic island containing the tetracycline resistance genes and a transposase coding gene. The insertion site was located immediately adjacent to one of the native rRNA operons (fig. 4), suggesting a recombination hot spot of the genome. The co-occurrence of the transfer of the rRNA gene operon and the tetracycline resistance genes suggested that the translation of the tetracycline resistance proteins might require a ribosome of the same host strain.
Thermoanaerobacter species are able to live in thermal environments such as hot springs. In this study, we analyzed T. brockii, Thermoanaerobacter italicus, Thermoanaerobacter mathranii, T. pseudethanolicus, Thermoanaerobacter wiegelii, T. X513 and X514, and the intragenus HGT of the 16S rRNA gene from T. thermohydrosulfuricus to both T. brockii and T. pseudethanolicus was detected, which also suggested that the HGT event occurred prior to the divergence of these two recipients.
Detection of current heterogeneity of 16S rRNA genes in a cell would neglect potential HGT of paralogs that have been homogenized through gene conversion, which results in concerted evolution of paralog genes. Concerted evolution of tandemly arrayed ribosomal genes in Xenopus has been described and subsequent studies have looked into the mechanisms for gene conversion (Arnheim et al. 1980). Gene conversion also drives concerted evolution of ribosomal RNA genes in prokaryotes (Liao 2000). In this study, we focused on those genomes with the most diverse copies of 16S rRNA genes, and would have underestimated potential HGT of those homogenized ribosomal RNA gene copies.
Our protocol might have underestimated the heterogeneity and potential HGT of 16S rRNA genes in some rare strains with multiple chromosomes, for example, the Haloarcula marismortui, by considering only the heterogeneity within replicons and some other minor technical issues. Also, it is important to know that current popular genome sequencing and assembling strategies may cause assemblage errors for those highly similar multicopy genes, especially for 16S rRNA gene. Without validation of Sanger resequencing, some genomes might have been finished by closing the gaps with an “average” 16S rRNA gene, which may also cause potential underestimation of the intragenomic heterogeneity.
In the study of Pei et al. (2010) on the 883 prokaryotic genomes, there were 24 genomes with the intragenomic heterogeneity of 16S rRNA genes greater than 1%, whereas our study scanned 2,143 genomes in the GenBank database of June 2014, and found more genomes with higher heterogeneity (28 genomes, >2%). Among them, 25 were newly sequenced genomes comparing to the study of Pei et al.. in 2010 (supplementary table S1, Supplementary Material online). The table 2 of the Pei et al. study showed the diversities of the 16S rRNA genes after masking (for the purpose of alignment) and the diversities of the masked hypervariable regions, whereas our study showed the diversities of full-length 16S rRNA genes for the purpose of HGT study.
Throughout all of the 28 genomes demonstrating an intragenomic heterogeneity of the 16S rRNA gene greater than 2%, HGT of the 16S rRNA gene only occurred at the intragenus or the intraspecies level (table 1), with a range of heterogeneity from 97.0% to 98.0% (fig. 1). This is very different from the HGT of functional genes, which can occur at all taxonomic levels, even between eukaryotes and prokaryotes. This difference can be explained by the complexity hypothesis that informational genes are complex and possess intrasystem compatibility. Informational genes that are transferred from highly distant organisms would not be compatible in a given system, and only the rare transfer of an informational gene from closely related organisms would function normally.
The HGT of the 16S rRNA gene would cause problems in the taxonomic classification of very few species. As shown in this study, 28 genomes demonstrated intragenomic heterogeneity (pairwise identity < 98%) of the 16S rRNA gene. Some of the heterogeneous copies were acquired from other organisms, and some of them might have been generated through accumulation of substitutions. The transferred copies could result in the misclassification of a species, and mutated copies could result in the misclassification of novel species. This problem could be solved by analyzing the phylogenetic locations of all of the 16S rRNA gene copies in a given genome. Based on the transfer level of 16S rRNA genes (intragenus and lower) found in this study, a classification at genus level or upper would be more reliable than at species level.
Previous studies have shed light on the intragenomic heterogeneity of 16S rRNA genes, revealing the divergence of ribosomal RNA genes (Acinas et al. 2004; Pei et al. 2010) and its impact on methods of classification based on the 16S rRNA gene: Overestimation of prokaryotic diversity (Sun et al. 2013). Our results demonstrated the occurrence of rare events of HGT of the 16S rRNA gene that only occurred at the intragenus and intraspecies levels. This study was unique because we considered a large amount of the available genome data and investigated the phylogeny of the multiple copies of the 16S rRNA genes and genomic structures to confirm HGT events, and more importantly, the potential source of the transfers.
In conclusion, we analyzed 2,143 prokaryotic genomes and investigated the HGT event of 16S rRNA genes and the potential sources of the transferred gene copies. Among them, 15 genomes were found to harbor 16S rRNA gene copies that were considered transferred at intraspecies and intragenus levels, based on analysis of gene similarity, phylogeny, secondary structure of rRNA genes, and genomic structure comparison. In contrast to HGT of functional genes, the HGT of 16S rRNA genes occurred at a low rate, and was only found occur between close taxa.
Supplementary Material
Supplementary figures S1–S7 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This study was supported by grants (SIDSSE-201206, XDB06010102, SIDSSE-201206) from the Sanya Institute of Deep Sea Science and Engineering, the Chinese Academy of Sciences (SIDSSE, CAS), a grant from China Ocean Mineral Resource R&D Association (COMRRDA12SC01), the National Basic Research Program of China (973 Program, No. 2012CB417304), and General Research Fund (661611) from HKSAR government to P.Y.Q.
Literature Cited
- Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol. 186:2629–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JO. 2005. Lateral gene transfer in eukaryotes. Cell Mol Life Sci. 62:1182–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Tatusov RL, Wolf YI, Walker DR, Koonin EV. 1998. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 14:442–444. [DOI] [PubMed] [Google Scholar]
- Aravind L, Tatusov RL, Wolf YI, Walker DR, Koonin E. 1999. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles (vol 14, pg 442, 1998). Trends Genet. 15:41. [DOI] [PubMed] [Google Scholar]
- Arnheim N, et al. 1980. Molecular evidence for genetic exchanges among ribosomal genes on non-homologous chromosomes in man and apes. Proc Natl Acad Sci U S A. 77:7323–7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann NL, Polkinghorne A, Timms P. 2014. Chlamydia genomics: providing novel insights into chlamydial biology. Trends Microbiol. 22:464–472. [DOI] [PubMed] [Google Scholar]
- Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. 2008. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics 9:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver TJ, et al. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423. [DOI] [PubMed] [Google Scholar]
- DeMars R, Weinfurter J. 2008. Interstrain gene transfer in Chlamydia trachomatis in vitro: mechanism and significance. J Bacteriol. 190:1605–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMars R, Weinfurter J, Guex E, Lin J, Potucek Y. 2007. Lateral gene transfer in vitro in the intracellular pathogen Chlamydia trachomatis. J Bacteriol. 189:991–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati M, et al. 2011. Antibody-neutralizing activity against all urogenital Chlamydia trachomatis serovars in Chlamydia suis-infected pigs. FEMS Immunol Med Microbiol. 61:125–128. [DOI] [PubMed] [Google Scholar]
- Freeman VJ. 1951. Studies on the virulence of bacteriophage-infected strains of Corynebacterium diphtheriae. J Bacteriol. 61:675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Vallve S, Romeu A, Palau J. 2000. Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res. 10:1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes JP, Bruno WJ, Borrego MJ, Dean D. 2004. Recombination in the genome of Chlamydia trachomatis involving the polymorphic membrane protein C gene relative to ompA and evidence for horizontal gene transfer. J Bacteriol. 186:4295–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SR, et al. 2012. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat Genet. 44:413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Gilna P, Li WZ. 2009. Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics 25:1338–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Rivera MC, Lake JA. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci U S A. 96:3801–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey BM, Suchland RJ, Eriksen SG, Sandoz KM, Rockey DD. 2013. Genomic and phenotypic characterization of in vitro-generated Chlamydia trachomatis recombinants. BMC Microbiol. 13: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahara K, Miyazaki K. 2013. Revisiting bacterial phylogeny: natural and experimental evidence for horizontal gene transfer of 16S rRNA. Mob Genet Elements. 3:e24210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahara K, Yasutake Y, Miyazaki K. 2012. Mutational robustness of 16S ribosomal RNA, shown by experimental horizontal gene transfer in Escherichia coli. Proc Natl Acad Sci U S A. 109:19220–19225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao DQ. 2000. Gene conversion drives within genic sequences: concerted evolution of ribosomal RNA genes in bacteria and archaea. J Mol Evol. 51:305–317. [DOI] [PubMed] [Google Scholar]
- Mylvaganam S, Dennis PP. 1992. Sequence heterogeneity between the two genes encoding 16S rRNA from the halophilic archaebacterium Haloarcula marismortui. Genetics 130:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Itoh T, Matsuda H, Gojobori T. 2004. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet. 36:760–766. [DOI] [PubMed] [Google Scholar]
- Nelson KE, et al. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399:323–329. [DOI] [PubMed] [Google Scholar]
- Pei AY, et al. 2010. Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl Environ Microbiol. 76:3886–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Aravind L, Schultz J, Bork P, Koonin EV. 1999. Eukaryotic signalling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J Mol Biol. 289:729–745. [DOI] [PubMed] [Google Scholar]
- Rivera MC, Jain R, Moore JE, Lake JA. 1998. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci U S A. 95:6239–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros VID, Hurst GDD. 2009. Lateral gene transfer between prokaryotes and multicellular eukaryotes: ongoing and significant? BMC Biol. 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchland RJ, Sandoz KM, Jeffrey BM, Stamm WE, Rockey DD. 2009. Horizontal transfer of tetracycline resistance among Chlamydia spp. In vitro. Antimicrob Agents Chemother. 53:4604–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun DL, Jiang X, Wu QL, Zhou NY. 2013. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl Environ Microbiol. 79:5962–5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, et al. 2005. Genotypic diversity within a natural coastal bacterioplankton population. Science 307:1311–1313. [DOI] [PubMed] [Google Scholar]
- Trieber CA, Taylor DE. 2002. Mutations in the 16S rRNA genes of Helicobacter pylori mediate resistance to tetracycline. J Bacteriol. 184:2131–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkum P, et al. 2003. Discordant phylogenies within the rrn loci of rhizobia. J Bacteriol. 185:2988–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR. 1987. Bacterial evolution. Microbiol Rev. 51:221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Stackebrandt E, Macke TJ, Fox GE. 1985. A phylogenetic definition of the major eubacterial taxa. Syst Appl Microbiol. 6:143–151 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.