Abstract
The winter moth (Operophtera brumata) belongs to one of the most species-rich families in Lepidoptera, the Geometridae (approximately 23,000 species). This family is of great economic importance as most species are herbivorous and capable of defoliating trees. Genome assembly of the winter moth allows the study of genes and gene families, such as the cytochrome P450 gene family, which is known to be vital in plant secondary metabolite detoxification and host-plant selection. It also enables exploration of the genomic basis for female brachyptery (wing reduction), a feature of sexual dimorphism in winter moth, and for seasonal timing, a trait extensively studied in this species. Here we present a reference genome for the winter moth, the first geometrid and largest sequenced Lepidopteran genome to date (638 Mb) including a set of 16,912 predicted protein-coding genes. This allowed us to assess the dynamics of evolution on a genome-wide scale using the P450 gene family. We also identified an expanded gene family potentially linked to female brachyptery, and annotated the genes involved in the circadian clock mechanism as main candidates for involvement in seasonal timing. The genome will contribute to Lepidopteran genomic resources and comparative genomics. In addition, the genome enhances our ability to understand the genetic and molecular basis of insect seasonal timing and thereby provides a reference for future evolutionary and population studies on the winter moth.
Keywords: winter moth, Lepidoptera, cytochrome P450, sexual dimorphism, phenology, circadian clock
Introduction
The winter moth (Operophtera brumata) is an insect species that belongs to the order of Lepidoptera (butterflies, moths, and skippers). It is a member of one of the largest families, the Geometridae, containing approximately 23,000 species (Scoble 2007). The vast majority of Lepidoptera are phytophagous and many geometrid moths are considered pests. They evolved in parallel to the evolution of the flowering plants (angiosperms) during and after the Cretaceous period (Wahlberg et al. 2013). This coevolutionary process, similar to other herbivorous insect groups, involved continuous adaptation to plant allelochemicals. A popular metaphor is that of an evolutionary arms race. Among the major groups of genes involved, both in plants and insects, are the ubiquitous P450 or CYP genes (Schuler 2011). Although evolution and ecology of host-plant choice and adaptation has been studied for over a century, the underlying genetic mechanisms are poorly understood.
The winter moth is widespread in Northern Europe and Asia and after 1930 it has become an invasive pest species in North America (Elkinton et al. 2010). Previous studies have described a shift in host-range in N. America compared with native Europe (Elkinton et al. 2010). When studying host selection, the P450 gene repertoire in winter moth is of special interest, as this gene family is involved in plant secondary metabolite detoxification. Another interesting adaptation displayed by the winter moth is female brachyptery, also known as wing reduction (fig. 1) (Meyer-Rochow and Lau 2008). This feature is not uncommon in moths (described in 26 of approximately 120 families; Viloria et al. 2003) and appears to be linked to adaptation to living in woods. The high degree of convergent evolution of brachyptery in moths due to adaptive forces suggests the existence of molecular pathways that may be altered in a convergent manner. Over the years, the winter moth phenology, that is, timing of biological events, has been studied extensively. Timing of egg hatching is known to have strong fitness consequences on winter moth survival and fecundity (van Asch et al. 2007). Synchronizing with the bud burst of its host plant, the Oak (Quercus robur) strongly affects the survival of newly hatched larvae and has reduces fecundity at the adult stage. An evolutionary response and a restoration of synchrony between the herbivore insect and its host plant have been identified (van Asch et al. 2013), but the molecular mechanism underlying the circannual clocks in both insect and plant is still unknown. A number of genes underlying circadian (daily) rhythms and their pathways have been identified (Bradshaw and Holzapfel 2007; Pegoraro and Tauber 2011). Although evidence is accumulating, the link between circadian rhythms and circannual rhythms is yet not well understood. In O. brumata the influence of photoperiod is still subject of investigation (van Asch et al. 2013), and the regulation of clock genes may also be moderated by ambient temperature (Chen et al. 2007).
Fig. 1.
—Male (left) and female (right) winter moth. The pictures show female brachyptery in winter moth.
Although the ecology of the winter moth is well studied, the current genomic knowledge on this species is limited, because genomic resources are scarce and little is known about the genome characteristics. The chromosome number in winter moth is unknown, as was, prior to this study, its genome size. Estimated genome sizes for geometrid species range from 400 to 500 Mb, but can reach up to 1.9 Gb, for example, Euchlaena irraria (Gregory and Hebert 2003). Related species have about 30 chromosome pairs (n = 28 in Bombyx mori; n = 31 in Biston betularia) (Van’t Hof et al. 2013). Females are the heterogametic sex in most Lepidoptera (WZ), whereas males are the homogametic sex (ZZ) (Sahara et al. 2012). In general, the Z chromosome is larger and contains more genes (Sahara et al. 2012). At the time of analysis, five other Lepidoptera genomes had been published: B. mori (Bombycidae) (Mita et al. 2004), Danaus plexippus (Nymphalidae) (Zhan et al. 2011), Heliconius melpomene (Nymphalidae) (Dasmahapatra et al. 2012), Plutella xylostella (Plutellidae) (You et al. 2013), and Melitaea cinxia (Nymphalidae) (Ahola et al. 2014). Genome properties such as genome size and repeat content from these species are rather diverse, although the number of protein-coding genes seems relatively conserved. Despite the presence of multiple Lepidopteran MT genomes, there is no MT genome available in the Larentiinae subfamily to which the winter moth belongs.
Here we present an annotated genome sequence for the winter moth, which fills a major taxonomic gap in Lepidopteran comparative genomics. The genome sequence provides valuable clues and testable hypotheses for understanding the winter moth’s morphology and phenology. Moreover, the genome may provide directions to pest control. Besides describing the genomic properties in general, we discuss in detail several aspects related to winter moth biology and its ecology. First, we describe the evolution of the cytochrome P450 gene family in relation to host-plant adaptation and coevolution. Then, we analyze the genomic elements that we hypothesize to be involved in the development of female brachyptery. Finally, we explore the circadian clock genes and discuss their implications in relation to seasonal timing. In this system, the combination of experimental approaches and genome sequencing offers novel and unexplored avenues into understanding the genetics of egg-hatching timing, and in the long term, the molecular mechanisms underlying seasonal timing.
Materials and Methods
Sequencing Strategy
A single adult winter moth female was used for the DNA extraction. The sample was collected on an oak tree in a forest in the Netherlands in December 2012. From the extracted DNA two paired-end libraries were constructed, and sequenced with 2*250 bp reads on an Illumina MiSeq instrument, resulting in overlapping forward and reverse reads. In addition three mate pair libraries were constructed with various insert sizes (1–2, 3, and 4–5 kb) and sequenced on an Illumina HiSeq2000 instrument using 101, 7, 101 flow cycles for forward, index and reverse reads, respectively. In total, we produced 27.14 Gb of raw genomic data (table 1). A more detailed description of the sequencing procedure can be found in supplementary section S1, Supplementary Material online.
Table 1.
Sequence Data: Raw and Preprocessed
| Library | Insert Size | Raw |
Preprocessed |
||||||
|---|---|---|---|---|---|---|---|---|---|
| # Reads (Million) | # Bases (Gb) | Read Length | Coverage | # Reads (Million) | # Bases(Gb) | Read Length | Coverage | ||
| PE400_1 | −145 bp | 58.8 | 14.5 | 246.5 | 22.7 | 56.1 | 13.8 | 246.4 | 21.6 |
| PE400_2 | −193 bp | 31.2 | 7.8 | 248.2 | 12.1 | 29.7 | 7.4 | 248.2 | 11.5 |
| MP1-2 kb | 1–2 kb | 22.9 | 2.3 | 101 | 3.6 | 14.7 | 1.3 | 86.4 | 2.0 |
| MP3kb | 3 kb | 15.1 | 1.5 | 101 | 2.4 | 4.0 | 0.3 | 85.9 | 0.53 |
| MP4-5 kb | 4–5 kb | 10.4 | 1.1 | 101 | 1.6 | 1.7 | 0.1 | 82.3 | 0.21 |
| Total | 138.5 | 27.1 | 42.4 | 106.1 | 22.9 | 35.8 | |||
Assembly Strategy
The winter moth reads were filtered for adapters, primers, quality (>10), and duplicates using Fastq-mcf software (v1.1.2) (Aronesty 2011). Read pairs shorter than 19 bp were discarded. From these reads, we first assembled the MT genome using MITObim (v1.6) (Hahn et al. 2013) with the MT genome of B. mori as reference. Manual curation was performed where alignment errors occurred. We merged gene annotations from DOGMA (Wyman et al. 2004) and MITOS (Bernt et al. 2013) to produce a consensus annotation in line with other geometrid species. To remove reads derived from the MT genome before the assembly of the nuclear genome, we aligned all reads to the O. brumata MT genome and set aside pairs of which one mate or both aligned to the MT sequence. Furthermore, the reads were filtered for Wolbachia contamination by mapping (Bowtie2 (v2.1.0); Langmead and Salzberg 2012) to all Wolbachia strains available in GenBank. The mapped reads were used to assemble the Wolbachia genome using the Celera assembler (v8.2beta) (Miller et al. 2008). This procedure was repeated iteratively by adding the assembly to the Wolbachia index. The preprocessing reduced the 27.1 Gb of raw data to 22.9 Gb of clean data (table 1). Finally, these clean reads were used to build a k-mer graph with Jellyfish (Marcais and Kingsford 2011) for k = 18. The size of the winter moth genome was estimated using a k-mer based method (Binghang et al. 2012).
We used the stand-alone error correction method from ALLPATHS-LG (release: 50721) to correct base-calling errors in the reads (Butler et al. 2008). SeqPrep was used to merge overlapping fragment data (John 2011). A de novo assembly was performed using the Celera assembler (v8.2beta) (Miller et al. 2008). We discarded all duplicate heterozygous contigs with Haplomerger (release: 20120810) (Huang et al. 2012), described in more detail in supplementary section S2, Supplementary Material online. We used SSPACE3 for additional scaffolding with Bowtie2 (v2.1.0) (Boetzer et al. 2011; Langmead and Salzberg 2012). Gapfiller (v1.10) was used to fill gaps (parameter -d 3500 to fill overestimated gap sizes due to improperly oriented paired ends) (Boetzer and Pirovano 2012). Potential contaminants were filtered using BLASTn (v2.2.8) (dc-megablast) on the nt database; sequences with exclusively prokaryotic alignments were discarded and not used in further downstream analysis. Potential nuclear MT DNA (numts) were detected using BLASTn (v2.2.8) with the MT DNA as subject. The completeness and structural consistency were assessed using the CEGMA (v2.4) pipeline and the alignment of 457 core Drosophila genes (Parra et al. 2007). In addition, the proteomes of two well-annotated insect species Drosophila melanogaster (Adams et al. 2000) and Tribolium castaneum (Richards et al. 2008) were aligned to the genome using genblastA (v1.0.4) (She et al. 2009). The coverage of the alignments on the scaffolds was calculated using a custom script.
Annotation Strategy
We estimated the repeat content of the winter moth genome by grouping the preprocessed PE400 genomic reads in repeat clusters using RepeatExplorer (Novak et al. 2013) and by estimating the single-copy region of the genome from the k-mer distribution (supplementary fig. S1, Supplementary Material online). A de novo repeat library was generated with RepeatModeler (v1.0.4) (Smit and Hubley 2014) using RECON (v1.0.7) (Bao and Eddy 2002), RepeatScout (v1.0.5) (Price et al. 2005), and TRF (v4.0.4) (Benson 1999). This library was used by RepeatMasker (v4.0.5) (Chen 2004) together with RepBase (Jurka 2000) for repeat annotation.
The structural annotation of protein-coding genes was done using the MAKER (v2.31.6) (Holt and Yandell 2011) pipeline with ab initio predictors SNAP (release: 2006-07-28) (Korf 2004) and AUGUSTUS (v3.0.2) (Stanke and Morgenstern 2005). We used H. melpomene (AUGUSTUS) and B. mori (SNAP) as model species in the ab initio predictors. We provided proteome data from H. melpomene, B. mori, D. plexippus, Plutella xylostella and Dr. melanogaster, and EST data from B. mori and D. plexippus as homology evidence in MAKER. We used tRNAscan-SE (v1.3.1) to identify tRNA-coding sequences (Lowe and Eddy 1997). Gene functions were assigned using BLASTp (E value = 0.1) (Altschul et al. 1990) on the SwissProt and TrEMBL databases (Apweiler et al. 2004). We assigned protein domains and gene-ontology terms using InterProScan (Quevillon et al. 2005) against the Superfamily (Wilson et al. 2009) and Pfam (Finn et al. 2010) protein databases. OrthoMCL (v2.0.9) was used to assign proteins to orthologous groups in the orthoMCL database (Li et al. 2003). We used SignalP (Petersen et al. 2011) to detect and WoLF PSORT (Horton et al. 2007) to localize signal peptides in the winter moth proteome. GOstat was used to find overrepresented GO terms in the secretome.
Genomic Properties
We used two methods to estimate genomic heterozygosity. First, we identified the volume of heterozygous k-mers from the k-mer distribution and divided those by the total volume of nonheterozygous k-mers (supplementary fig. S1, Supplementary Material online) (Binghang et al. 2012). Second, we aligned all reads to the genome using Bowtie2 (v2.1.0) (Langmead and Salzberg 2012), variants were called using freebayes (v0.9.18-25-g5781407) (Garrison and Marth 2012) and annotated with SnpEff (v4.0) (Cingolani et al. 2012). The same alignments were used to identify potential sex-chromosomal scaffolds (supplementary file S2, Supplementary Material online). A custom script was used to identify sex-scaffolds with a minimum length of 10 kb and a single nucleotide polymorphism (SNP) density less than 1 per kilobase, and genome coverage was estimated using bedtools genomecov (Quinlan and Hall 2010). We used tBLASTn to align the B. mori-annotated Z-chromosomal proteins to the genome.
Gene Family Analysis
To identify gene family contractions and expansions, we clustered the proteomes of 13 different species (12 insects, 1 mammal) into orthologous groups (supplementary table S13, Supplementary Material online) using orthoMCL. Single-copy orthologs and lineage-specific gene families were extracted using custom scripts. We identified specific insect order orthologs (supplementary fig. S4, Supplementary Material online). We built a general phylogeny from 526 single-copy orthologs (supplementary fig. S5, Supplementary Material online). Alignments were performed using ClustalW and concatenated using the Hal pipeline (Robbertse et al. 2011). A phylogenetic tree was constructed using PhyML with the LG substitution model (Guindon et al. 2009). Operophtera brumata-specific multicopy orthologous groups and O. brumata singletons were extracted using a custom script. We used GOstat (Beissbarth and Speed 2004) to identify overrepresented gene ontologies using a precomputed O. brumata gene ontology database. We identified gene family expansions and contractions using BadiRate (Librado et al. 2012) using the general species tree that was constructed, and with -anc, -start_value 1, and -outlier parameters. Gene family outliers were extracted using custom scripts.
To identify the P450 proteins in all species (IPR001128), we used InterProScan (Quevillon et al. 2005). Next, we used MAFFT (Katoh and Standley 2013) to align the Lepidoptera P450 proteins and FastTree2 (Price et al. 2010) to build the tree. We performed a phylogenetic analysis of the four orthologous groups containing genes homologous to the Drosophila rdx gene. We used MAFFT (Katoh and Standley 2013) to construct the alignment and Phyml (Guindon et al. 2010) to generate the phylogenetic tree using the LG substitution model. We used iTOL (Letunic and Bork 2007) for the visualization of the phylogenies. We used the orthoMCL groups to identify the winter moth clock genes. In addition, KEGG-KAAS (Moriya et al. 2007) was used to identify winter moth clock genes and assess the completeness of the pathway.
Results
The Winter Moth Genome
We have reconstructed the genome sequence of a female winter moth, yielding a total assembly size of 638 Mb, 98.9% of the estimated genome size of 645 Mb (supplementary table S1 and fig. S1, Supplementary Material online). The assembly comprises 25,801 scaffolds with an N50 scaffold length of 65.6 kb. The genome is predicted to encode 16,912 protein-coding genes. It has a GC content of 38.6% and an estimated repeat content of 53.5%. The heterozygosity rate (single individual) was estimated to be 0.72% based on the k-mer distribution (supplementary fig. S1, Supplementary Material online). In the assembly we find a slightly lower rate of 0.64%, corresponding to approximately 4.1 M heterozygous variants (SNP/indel) (supplementary tables S2–S4, Supplementary Material online). Based on read coverage and SNP density, we identified 875 potential sex-chromosomal scaffolds with a total length of 19.5 Mb, comparable to the B. mori Z chromosome (20.35 Mb) (Arunkumar et al. 2009) (supplementary file S2, Supplementary Material online). Scaffolds corresponding to the W chromosome are expected to be absent from this list, because of the high repeat content and low gene density, similar to the B. mori W chromosome (Fujii et al. 2010).
The genome assembly was built from 27.1 Gb of raw data (22.9 Gb after preprocessing), sequenced from five DNA libraries (table 1 and supplementary fig. S2, Supplementary Material online). Even though the fragmentation of the assembly is substantial, rigorous validation indicates that the gene space is largely covered, based on the CEGMA score and presence of related proteomes (table 2). In addition, the structural consistency with the genomic reads, that is, percentage mapped and properly paired, is high (supplementary table S5, Supplementary Material online). The high quality of the genome assembly allows for a comparison with published genomes of other Lepidoptera species.
Table 2.
Lepidopteran Genome Properties and Validation
| Species | Operophtera brumata (v1) | Bombyx mori (v2) (Xia et al. 2008) | Danaus plexippus (v3) (Zhan and Reppert 2013) | Heliconius melpomene (v1.1) (Dasmahapatra et al. 2012) | Plutella xylostella (v1) (You et al. 2013) | Melitaea cinxia (v1) (Ahola et al. 2014) |
|---|---|---|---|---|---|---|
| Genome assembly | ||||||
| Assembly size (Mb) | 638.2 | 431.7 | 248.6 | 269.7 | 394.0 | 393.3 |
| Number of scaffolds | 25,801 | 43,622 | 5,397 | 3,807 | 1,819 | 6,299 |
| N50 scaffold size (kb) | 65.6 | 3,717.0 | 715.6 | 277.0 | 737.2 | 258.3 |
| Min scaffold length (bp) | 502 | 11 | 300 | 124 | 1,842 | 1,512 |
| Max scaffold length (Mb) | 0.50 | 16.12 | 6.24 | 1.45 | 3.49 | 0.68 |
| Gene annotation | ||||||
| Protein-coding | 16,912 | 14,623 | 15,130 | 12,669 | 18,071 | 16,667 |
| InterPro domain | 11,793 | 9,892 | 10,034 | 9,803 | 12,877 | 8,529 |
| Genomic features | ||||||
| Repeat (%) | 53.5 | 43.6 | 10.2 | 24.9 | 34.0 | 27.9 |
| GC (%) | 38.6 | 38.8 | 31.6 | 32.8 | 38.0 | 32.6 |
| Coding (%) | 2.9 | 4.1 | 8.4 | 6.5 | 6.4 | 4.0 |
| Intron (%) | 17.7 | 16.3 | 28.1 | 25.8 | 30.8 | 30.5 |
| Gene length (bp) | 7,752 | 6,029 | 6,001 | 6,779 | 8,083 | 8,129 |
| tRNAs | 579 | 441 | 379 | 2,373 | 521 | 2,478 |
| Quality control | ||||||
| CEGMA partial (248) | 234 | 240 | 238 | 234 | 227 | 208 |
| Core Drosophilaa (%) | 46.6 | 47.7 | 54.0 | 51.2 | 48.8 | 41.6 |
| Drosophilaa (%) | 13.0 | 13.4 | 14.4 | 13.4 | 13.0 | 11.2 |
| Triboliuma (%) | 17.9 | 17.8 | 18.3 | 17.8 | 17.7 | 15.4 |
aPercentage of genes of which more than 90% of the sequence is found on a single scaffold.
The winter moth genome is the largest lepidopteran genome published, 48% larger than B. mori, its closest relative for which an assembly is available. The large genome size is not due to an increased number of protein-coding genes, which is comparable to that in other Lepidoptera, but is to a large extent explained by a higher repeat content (53.5% compared with 43.6% for B. mori). Long interspersed elements (mainly: CR1, RTE, L2, CRE elements) and Helitron transposons are more abundant in the O. brumata genome compared with B. mori (Osanai-Futahashi et al. 2008). Nevertheless, 48.6% of the annotated repeats remain unclassified in O. brumata (supplementary table S6, Supplementary Material online). For 60% of the repeat sequences in this unclassified category, we found homology in other Lepidoptera genomes.
In addition to the nuclear genome of winter moth, we have reconstructed and annotated the complete MT genome. It has a total length of 15,748 bp, and contains 13 protein-coding genes, 22 tRNA, and 2 ribosomal RNA genes (supplementary table S7 and fig. S3, Supplementary Material online). BLAST (Basic Local Alignment Search Tool) results show highest sequence identity (85%) and coverage (97%) with Apocheima cinerarium (Liu et al. 2014) and Phthonandria atrilineata (Yang et al. 2009), both geometrid moths (subfamily: Ennominae). The annotated genes are in the same order and orientation as in the mitogenomes of these geometrid moths. The A+T content is 79.97%, slightly lower than for A. cinerarium (80.83%) and P. atrilineata (81.02%). The control region (802 bp) is longer than in the other two geometrid species (A. cinerarium, 625 bp; P. atrilineata, 456 bp). In addition, we identified 185 scaffolds in the nuclear genome containing potential MT insertions (numts) (supplementary file S3, Supplementary Material online).
An interesting finding was the discovery of a Wolbachia infection in the sequenced individual. This bacterium commonly infects insects (Werren et al. 2008), but has not been described as an endosymbiont of winter moths. We found that 0.4% of the produced genomic reads were derived from Wolbachia. We assembled them into 120 scaffolds spanning 1.12 Mb, with an N50 of 15.6 kb (supplementary table S8, Supplementary Material online). The assembled sequence shows highest similarity with the wPip Wolbachia strain from Culex pipiens (Klasson et al. 2008), causing cytoplasmic incompatibility in this species. Even though further analysis was outside the scope of this study, this genome sequence could add to our understanding of the functioning and evolution of bacterial endosymbionts in insects.
Cytochrome P450
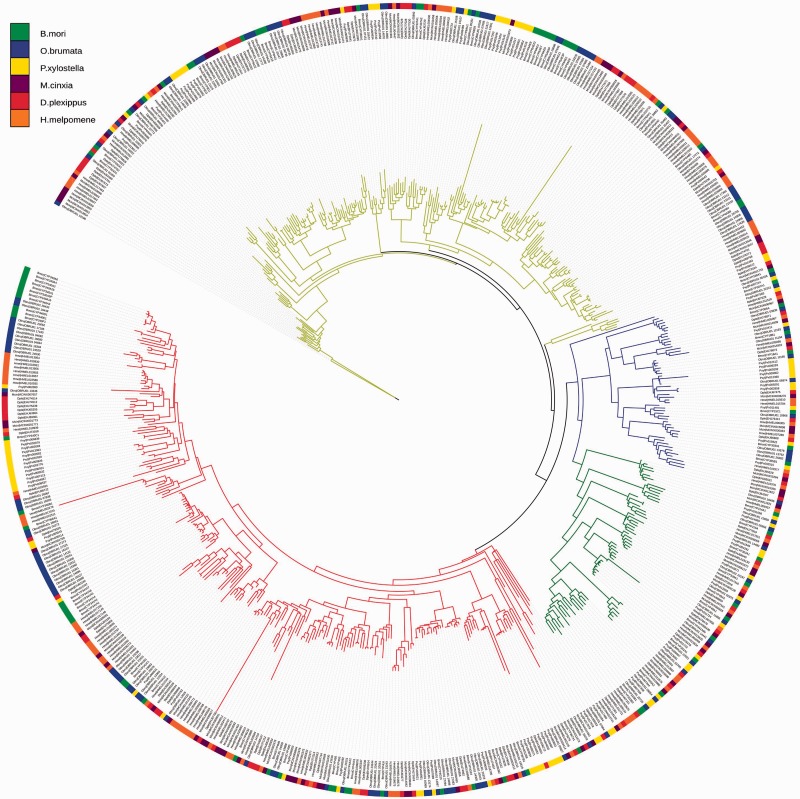
One of the main reasons to sequence the winter moth genome is to study the P450 gene family in relation to host-plant adaptation. P450 enzymes, of which large families are present in insect genomes, are involved in detoxification of plant toxins and play a central role in insecticide resistance (Feyereisen 1999). Insect P450s are subdivided into four clades, CYP2, CYP3, CYP4, and MT (Feyereisen 2006). Gene family analysis shows that winter moth contains 133 P450 genes (CYP2: 10, CYP3: 51, CYP4: 60, CYP-Mito: 12), and that members of the P450 protein family are overrepresented in winter moth-specific orthology groups. Specifically, we identified 52 O. brumata-specific cytochrome P450 proteins that were either unassigned or assigned to an O. brumata-specific group, meaning that we could not find an ortholog in any of the other five Lepidoptera (supplementary table S9, Supplementary Material online). These expansions mainly occurred in the larger CYP3 and CYP4 clades (fig. 2), with large expansions near the B. mori Cyp340A and Cyp341A genes (clade: CYP4). These expansions are likely representative for a specific detoxification gene repertoire in O. brumata. However, the fact that all species feed on different hosts is reflected by specific P450 gene family expansions in all species (fig. 2).
Fig. 2.
—Lepidoptera P450 gene family tree. The tree shows species-specific P450 gene family expansions in six Lepidoptera, with large winter moth expansions in CYP3 and CYP4. Branch colors represent the Cytochrome P450 clades: CYP2 (blue), CYP3 (yellow), CYP4 (red), CYP-Mito (green).
Female Brachyptery
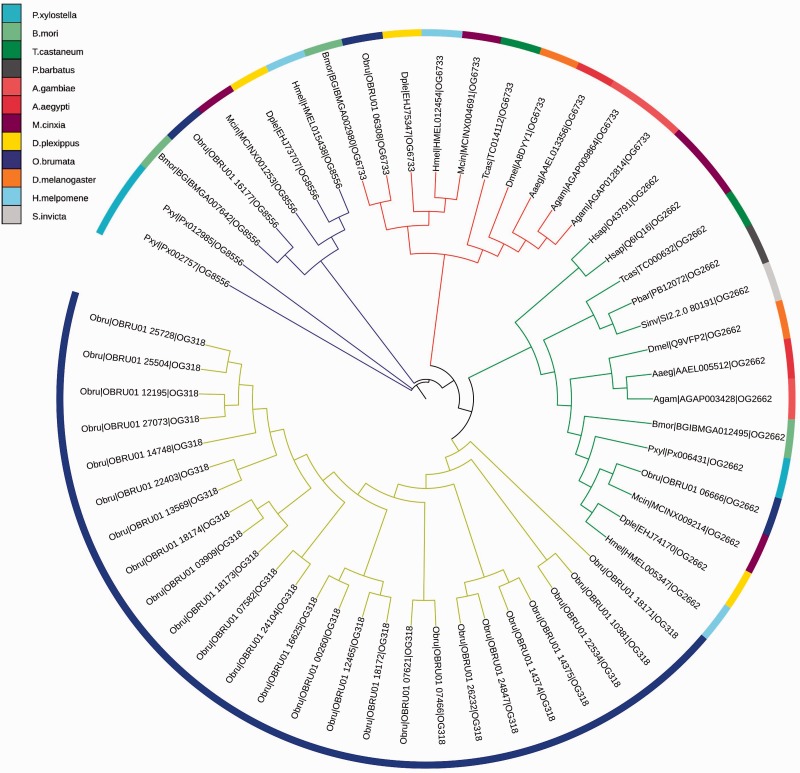
A second striking winter moth-specific gene-family expansion is in the rdx-like gene family. The members of this family are organized into four orthology groups, of which one (OG318) is specific to winter moth and contains 25 proteins (fig. 3). The proteins in this group show similarity to the Drosophila roadkill (rdx) gene (Flybase: FBgn0264493). Rdx forms a complex with Cullin 3 (through BTB domain) and attenuates Hedgehog responses through ubiquitination of cubitus interruptus (Ci) (Kent et al. 2006). Hedgehog (Hh) signaling regulates growth and patterning in many Drosophila organs assumed to be similar in Lepidoptera. In wing development, rdx and the CUL3-pathway modulate the amount of Ci (Kent et al. 2006). However, the Drosophila homolog contains both a BTB/POZ (interaction with Cul3) domain and MATH domain (OG2662) whereas the O. brumata proteins in this group lack the MATH domain (supplementary table S10, Supplementary Material online). The presence or absence of this MATH domain varies among different species in the gene family (TreeFam: TF313419, PhylomeDB: phy000Z4EB). In Drosophila, rdx mutants (overexpressed) led to smaller wing sizes and differential eye morphogenesis (Kent et al. 2006; Zhang et al. 2006). This suggests that this expanded gene family may play a role in sexual dimorphism in O. brumata, having very short winged females and a different eye form/shape compared with males (less facets, smaller diameter, and smaller clear zone) (Meyer-Rochow and Lau 2008).
Fig. 3.
—Phylogenetic tree of the four rdx(-like) orthology groups. Orthology group OG318 (yellow) contains 25 copies of an O. brumata specific rdx-like gene. The Drosophila ortholog is in group OG2662. All genes have a BZB-POZ domain to interact with CUL3 except for OG6733. Only proteins in group OG2662 also contain the MATH domain.
To further support this potential role in wing formation/reduction, we found four other insect species with a large rdx-like gene family: Nasonia vitripennis (Werren et al. 2010), Lygus hesperus (Hull et al. 2013), Microplitis demolitor (Burke and Strand 2012), Acyrthosiphon pisum (Richards et al. 2010), supported by BLAST results (supplementary table S11, Supplementary Material online). Interestingly, male brachyptery occurs in N. vitripennis, and both winged and wingless morphs occur in Ac. pisum, triggered by overcrowding or poor food quality.
Seasonal Timing
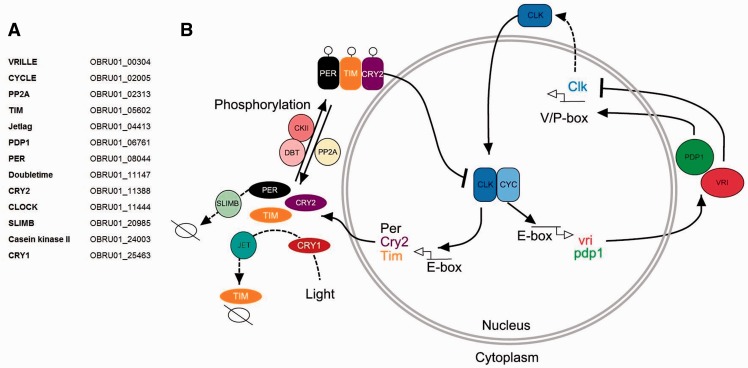
Because of its potential importance in seasonal timing, we studied the circadian clock mechanism in the winter moth genome. Our knowledge on this mechanism is based on Drosophila, where it comprises two transcriptional loops: A core negative transcriptional feedback loop, driving self-regulating daily rhythms and a second interlocking feedback loop (Allada and Chung 2010). Operophtera brumata, and all other sequenced Lepidoptera, contained all components from both loops (fig. 4 and supplementary table S12, Supplementary Material online). The critical clock genes from the core feedback loop are similar to those found in Drosophila; Cycle (Cyc), Clock (Clk), Period (Per), Timeless (Tim), and type-1 cryptochrome (Cry1). Interestingly, O. brumata also contains a cryptochrome 2 (Cry2) gene previously shown to be the main transcriptional repressor of Cyc and Clk in many other insects (Sandrelli et al. 2008), likely to be similar in winter moth. We found no evidence for clustering of this pathway in the genome. However, three of the core genes (Clk, Cyc, and Per) are located on the Z-chromosome in B. mori. Based on SNP density and coverage statistics we observe that Clk and Per are also located on the Z-chromosome in O. brumata, but Cycle is not. In addition, genes involved in posttranslational modifications of the Per and Tim gene are identified, as are the components from the secondary feedback loop that regulate the expression of Clk through Vrille (Vri) and Pdp1. These genes contain E-box elements, through which a Clk:Cyc complex could drive their transcription.
Fig. 4.
—Circadian clock mechanism in Lepidoptera. (A) List of clock genes, identified in the winter moth genome. (B) The mechanism comprises the core transcriptional/translational feedback loop (left) and the modulatory feedback loop (right). Clock (Clk) and Cycle (Cyc) form a heterodimer and drive the transcription of period (Per), timeless (Tim), and cryptrochrome 2 (Cry2) by binding their upstream E-box elements. Cry2 is responsible for the inhibition of Clk:Cyc transcription, that is, its own activators. Per and Tim bind through a PAS domain. Casein kinase II (CkII), discs overgrown (Dbt), and the protein phosphatase 2A (Pp2A) are involved in the posttranslational modifications and activation of Per and Tim. Jetleg (Jet) is responsible for Tim degradation, modulated by Cry1, activated by light. Slimb signals Per degradation. In insects, Per and Tim are transcribed mostly during the night. Vri (negative) and Pdp1 (positive) regulate the expression of Clk in the secondary feedback loop.
The Per and Tim genes are the main candidates for involvement in seasonal timing. The Per gene has already been found to have an effect on seasonal timing, egg-hatching, and egg-to-adult developmental time (Sauman et al. 1996; Itoh and Sumi 2000; Sandrelli et al. 2007). Also, two forms of the Tim gene have been found having different effects on (egg) diapause (Emerson et al. 2009). However, these effects seem to be driven by different photoperiods. In winter moth, egg-hatching is clearly influenced by temperatures, but there is no evidence of an effect of photoperiod on egg-hatching (van Asch et al. 2013). There is, however, evidence that increasing temperatures affect Tim expression or cause it to be alternatively spliced implicating the clock mechanism could be temperature driven (Chen et al. 2007).
Discussion
We have successfully sequenced and assembled the winter moth genome, including its MT genome. The genome contributes to Lepidopteran phylogenomics as this is the first geometrid genome to be characterized and the first Larentiinae (which may even be a distinct family; Ounap et al. 2008) MT genome. The larger genome size compared with other Lepidoptera is to a large extent explained by its higher repeat content. However, only part of the repeats can be classified and there is little overlap with B. mori repeats, suggesting minor commonality within the repetitive landscape of Lepidoptera.
An interesting finding during this sequencing effort was the discovery of a Wolbachia infection, of which we could reconstruct a partial genome sequence, in the sequenced individual. Wolbachia infections are known to play a large role in sexual differentiation of hosts through cytoplasmic incompatibility, feminization, and male killing (Werren et al. 2008). Cytoplasmic incompatibility is the most abundant phenotype among sequenced Wolbachia genomes (Werren et al. 2008). However, the phenotype of the strain in winter moth is difficult to determine because Wolbachia strains can switch phenotype depending on their host (Hornett et al. 2008).
We set out with a strong interest in the P450 gene family, which is known to play an important role in detoxification of plant allelochemicals and insecticides (Schuler 2011). Gene families that confer adaptations to fast-changing environmental circumstances are known to readily expand to generate diversity in the repertoire of that adaptation. In vertebrates, for instance, immune receptor and olfactory receptor genes are well-known examples. In insects, the P450s are among gene families that have the highest number of members. The importance of these genes relative to the total number of genes in genomes has led researchers to coin the term “CYPome” (Feyereisen 2011). The number of P450 genes identified in insect genomes ranges between 56 (honey bee; Weinstock et al. 2006) and 143 (red flour beetle; Richards et al. 2008). For the winter moth we identified 133 distinct genes, at the higher end of that range. Analysis of P450 evolution compared with other Lepidoptera reveals O. brumata specific, and perhaps geometrid specific, expansions of P450 groups. Interestingly, the comparative analysis shows specific expansions of certain P450 for all lepidopteran genomes sequenced to date, consistent with the “periodic blooming” model of P450 evolution (Feyereisen 2011). This model states that expansions of P450 genes will occur regularly, even though they may not constitute an immediate selective advantage. However, the expanded families may confer an adaptive advantage in changing environments, for example, in adaptation to changing allelochemical composition of host plants, in host-plant preference changes or in insecticide resistance. Expanding the P450 family, irrespective of the exact gene that is being duplicated, increases the range of substances which can be oxidatively altered. Our observation is highly consistent with that model. The exact function of the majority of P450 genes in the various insect species investigated to date is very poorly understood. The midgut-specific expression of the Cyp340 and Cyp341 families (Yu et al. 2015), largely expanded in winter moth, suggests a role in detoxification of plant allelochemicals and monooxygenase activities. Our comparative analysis highlights the importance of filling in the phylogenetic gaps in herbivorous insect taxa to provide a better understanding of the evolutionary dynamics and adaptive potential of insect CYPomes.
The gene family analysis revealed an unexpected expansion in the rdx-like gene family, which points to a potential negative feedback mechanism in insect brachyptery. Rdx forms a complex with CUL3, leading to ubiquitination of Ci. Ci is a transcription factor regulating hedgehog (hh) genes, involved in many key developmental processes including wing development. It is known to have an increased expression level during wing development in other insects and studies have shown that RNA interference of Ci resulted in wing-reduced phenotypes (Li et al. 2009). We suggest a role for this rdx-like gene family in sex-specific wing-size differentiation, although the mechanism is still unclear. There may be a common regulatory basis that interacts with genes that determine gender-specific traits. For example, the transcription factor doublesex is known to mediate sexual dimorphism in insects with a male and female splice variant (Kraaijeveld 2014), and could potentially target these rdx-like genes. Another explanation could be sex-specific methylation, a feature described earlier in insects, but not yet linked to sexual dimorphism (D’Avila et al. 2010). The genome sequence provides insights for further experimental validation, for example, measuring and comparing expression levels in pupa stages in males and females. More elaborate experiments might entail RNA silencing during wing developmental stages in female winter moths.
Finally, with the aim to study the genomic components involved in the winter moth’s phenology, we have described the clock mechanism in winter moth as the main target pathway underlying a genetic change in response to climate change. This pathway is well conserved within insects (Sandrelli et al. 2008) and we identified all components in O. brumata. There is no evidence for clustering of these genes in the genome. Of the four clock genes (Per, Clk, Cyc, and Pdp1) clustered on the Z-chromosome in B. mori, we only find two on a sex-chromosome-related scaffold in O. brumata (Per and Clk), suggesting the absence of coadaptation through colocalization of this pathway. The Period gene has been shown to affect egg-hatching time in other insects such as B. mori (Sandrelli et al. 2007; Allada and Chung 2010). The Per, Tim, and Cry2 genes are known to affect diapause in other insects (Xu et al. 2011; Yamada and Yamamoto 2011; Meuti et al. 2015). These genes will be the main candidates in population studies between early and late egg-hatchers to get more insight in the mechanisms behind this adaptation, or in expression studies over various temporal scales.
Conclusion
We present the first sequenced geometrid genome, which will contribute to comparative work within Lepidoptera and insects in general. The identified (species-specific) expansions in the P450 family in all sequenced Lepidoptera could provide leads to insect detoxification of, and adaptation to, host plants and should contribute to knowledge of insecticide resistance. In addition, we imply a novel mechanism for female brachyptery supported by large rdx-like gene families in winter moth and in other brachypterous insects. We studied and identified the circadian clock mechanism in winter moth and argue its link to seasonal timing, providing a reference for future population studies in winter moth to unravel its rapid genetic adaptation to climate change.
Supplementary Material
Supplementary files S1–S4, sections S1 and S2, references, figures S1–S7, and tables S1–S13 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgment
The authors thank Ken Kraaijeveld for discussions about the sexual dimorphism and Wolbachia infection in winter moth.
Literature Cited
- Adams MD, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185-2195. [DOI] [PubMed] [Google Scholar]
- Ahola V, et al. 2014. The Glanville fritillary genome retains an ancient karyotype and reveals selective chromosomal fusions in Lepidoptera. Nat Commun. 54737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Chung BY. 2010. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 72:605-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- Apweiler R, et al. 2004. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 32:D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronesty E. 2013. Comparison of sequencing utility programs. Open Bioinform J. 7:1-8. [Google Scholar]
- Arunkumar KP, Mita K, Nagaraju J. 2009. The silkworm Z chromosome is enriched in testis-specific genes. Genetics 182:493-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao ZR, Eddy SR. 2002. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 12:1269-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP. 2004. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 20:1464-1465. [DOI] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M, et al. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313-319. [DOI] [PubMed] [Google Scholar]
- Binghang L, et al. 2012. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. arXiv:1308.2012 [q-bio.GN]. [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27:578-579. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Pirovano W. 2012. Toward almost closed genomes with GapFiller. Genome Biol. 13(6)R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw WE, Holzapfel CM. 2007. Evolution of animal photoperiodism. Annu Rev Ecol Evol Syst. 38:1-25. [Google Scholar]
- Burke GR, Strand MR. 2012. Deep sequencing identifies viral and wasp genes with potential roles in replication of Microplitis demolitor Bracovirus. J Virol. 86:3293-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J, et al. 2008. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 18:810-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N. 2004. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. Chapter 4:Unit 4 10. [DOI] [PubMed] [Google Scholar]
- Chen WF, Low KH, Lim C, Edery I. 2007. Thermosensitive splicing of a clock gene and seasonal adaptation. Cold Spring Harb Symp Quant Biol. 72:599-606. [DOI] [PubMed] [Google Scholar]
- Cingolani P, et al. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w(1118); iso-2; iso-3. Fly 6:80-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avila MF, Garcia RN, Panzera Y, Valente VL. 2010. Sex-specific methylation in Drosophila: an investigation of the Sophophora subgenus. Genetica 138:907-913. [DOI] [PubMed] [Google Scholar]
- Dasmahapatra KK, et al. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkinton JS, et al. 2010. Survey for winter moth (Lepidoptera: Geometridae) in northeastern North America with pheromone-baited traps and hybridization with the native bruce spanworm (Lepidoptera: Geometridae) (vol 103, pg 135, 2010). Ann Entomol Soc Am. 103:Ii-Iii. [Google Scholar]
- Emerson KJ, Bradshaw WE, Holzapfel CM. 2009. Complications of complexity: integrating environmental, genetic and hormonal control of insect diapause. Trends Genet. 25:217-225. [DOI] [PubMed] [Google Scholar]
- Feyereisen R. 1999. Insect P450 enzymes. Annu Rev Entomol. 44:507-533. [DOI] [PubMed] [Google Scholar]
- Feyereisen R. 2006. Evolution of insect P450. Biochem Soc Trans. 34:1252-1255. [DOI] [PubMed] [Google Scholar]
- Feyereisen R. 2011. Arthropod CYPomes illustrate the tempo and mode in P450 evolution. Biochim Biophys Acta. 1814:19-28. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Abe H, Shimada T. 2010. Molecular analysis of sex chromosome-linked mutants in the silkworm Bombyx mori. J Genet. 89:365-374. [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 [q-bio.GN]. [Google Scholar]
- Gregory TR, Hebert PDN. 2003. Genome size variation in lepidopteran insects. Can J Zool. 81:1399-1405. [Google Scholar]
- Guindon S, Delsuc F, Dufayard JF, Gascuel O. 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 537:113-137. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307-321. [DOI] [PubMed] [Google Scholar]
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41(13)e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornett EA, et al. 2008. You can’t keep a good parasite down: evolution of a male-killer suppressor uncovers cytoplasmic incompatibility. Evolution 62:1258-1263. [DOI] [PubMed] [Google Scholar]
- Horton P, et al. 2007. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35:W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SF, et al. 2012. HaploMerger: reconstructing allelic relationships for polymorphic diploid genome assemblies. Genome Res. 22:1581-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull JJ, Geib SM, Fabrick JA, Brent CS. 2013. Sequencing and de novo assembly of the western tarnished plant bug (Lygus hesperus) transcriptome. PLoS One 8(1)e55105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh MT, Sumi Y. 2000. Circadian clock controlling egg hatching in the cricket (Gryllus bimaculatus). J Biol Rhythms. 15:241-245. [DOI] [PubMed] [Google Scholar]
- John J St. 2011. SeqPrep. Available from: https://github.com/jstjohn/SeqPrep. [Google Scholar]
- Jurka J. 2000. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 16:418-420. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent D, Bush EW, Hooper JE. 2006. Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development 133:2001-2010. [DOI] [PubMed] [Google Scholar]
- Klasson L, et al. 2008. Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol Biol Evol. 25:1877-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraaijeveld K. 2014. Reversible trait loss: the genetic architecture of female ornaments. Annu Rev Ecol Evol Syst. 45:159-177. [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127-128. [DOI] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Vieira FG, Rozas J. 2012. BadiRate: estimating family turnover rates by likelihood-based methods. Bioinformatics 28:279-281. [DOI] [PubMed] [Google Scholar]
- Liu SX, Xue DY, Cheng R, Han HX. 2014. The complete mitogenome of Apocheima cinerarius (Lepidoptera: Geometridae: Ennominae) and comparison with that of other lepidopteran insects. Gene 547:136-144. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcais G, Kingsford C. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27:764-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuti ME, Stone M, Ikeno T, Denlinger DL. 2015. Functional circadian clock genes are essential for the overwintering diapause of the Northern house mosquito, Culex pipiens. J Exp Biol. 218:412-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Rochow VB, Lau TF. 2008. Sexual dimorphism in the compound eye of the moth Operophtera brumata (Lepidoptera, Geometridae). Invertebr Biol. 127:201-216. [Google Scholar]
- Miller JR, et al. 2008. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics 24:2818-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita K, et al. 2004. The genome sequence of silkworm, Bombyx mori. DNA Res. 11:27-35. [DOI] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35:W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P, Neumann P, Pech J, Steinhaisl J, Macas J. 2013. RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29:792-793. [DOI] [PubMed] [Google Scholar]
- Osanai-Futahashi M, Suetsugu Y, Mita K, Fujiwara H. 2008. Genome-wide screening and characterization of transposable elements and their distribution analysis in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 38:1046-1057. [DOI] [PubMed] [Google Scholar]
- Ounap E, Vidalepp J, Saarma U. 2008. Systematic position of lythriini revised: transferred from Larentiinae to Sterrhinae (Lepidoptera, Geometridae). Zool Scr. 37:405-413. [Google Scholar]
- Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061-1067. [DOI] [PubMed] [Google Scholar]
- Pegoraro M, Tauber E. 2011. Animal clocks: a multitude of molecular mechanisms for circadian timekeeping. Wiley Interdiscip Rev RNA. 2:312-320. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 8:785-786. [DOI] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA. 2005. De novo identification of repeat families in large genomes. Bioinformatics 21:I351–I358. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, et al. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33:W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, et al. 2008. The genome of the model beetle and pest Tribolium castaneum. Nature 452:949-955. [DOI] [PubMed] [Google Scholar]
- Richards S, et al. 2010. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8(2)e1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbertse B, Yoder RJ, Boyd A, Reeves J, Spatafora JW. 2011. Hal: an automated pipeline for phylogenetic analyses of genomic data. PLoS Curr. 3:RRN1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara K, Yoshido A, Traut W. 2012. Sex chromosome evolution in moths and butterflies. Chromosome Res. 20:83-94. [DOI] [PubMed] [Google Scholar]
- Sandrelli F, Costa R, Kyriacou CP, Rosato E. 2008. Comparative analysis of circadian clock genes in insects. Insect Mol Biol. 17:447-463. [DOI] [PubMed] [Google Scholar]
- Sandrelli F, et al. 2007. Phenotypic effects induced by knock-down of the period clock gene in Bombyx mori. Genet Res. 89:73-84. [DOI] [PubMed] [Google Scholar]
- Sauman I, Tsai T, Roca AL, Reppert SM. 1996. Period protein is necessary for circadian control of egg hatching behavior in the silkmoth Antheraea pernyi. Neuron 17:901-909. [DOI] [PubMed] [Google Scholar]
- Schuler MA. 2011. P450s in plant-insect interactions. Biochim Biophys Acta. 1814:36-45. [DOI] [PubMed] [Google Scholar]
- Scoble MJ, Hausmann A. [updated 2007]: Online list of valid and available names of the Geometridae of the World. Available from: http://www.lepbarcoding.org/geometridae/species_checklists.php. [Google Scholar]
- She R, Chu JSC, Wang K, Pei J, Chen NS. 2009. genBlastA: enabling BLAST to identify homologous gene sequences. Genome Res. 19:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A, Hubley R. 2014. RepeatModeler. Available from: http://www.repeatmasker.org/RepeatModeler.html. [Google Scholar]
- Stanke M, Morgenstern B. 2005. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33:W465–W467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Asch M, Salis L, Holleman LJM, van Lith B, Visser ME. 2013. Evolutionary response of the egg hatching date of a herbivorous insect under climate change. Nat Clim Change. 3:244-248. [Google Scholar]
- van Asch M, Tienderen PH, Holleman LJM, Visser ME. 2007. Predicting adaptation of phenology in response to climate change, an insect herbivore example. Glob Chang Biol. 13:1596-1604. [Google Scholar]
- Van’t Hof AE, et al. 2013. Linkage map of the peppered moth, Biston betularia (Lepidoptera, Geometridae): a model of industrial melanism. Heredity 110:283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viloria AL, et al. 2003. A brachypterous butterfly? Proc Biol Sci. 270(Suppl. 1):S21–S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg N, Wheat CW, Pena C. 2013. Timing and patterns in the taxonomic diversification of Lepidoptera (butterflies and moths). PLoS One 8:e80875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock GM, et al. 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443:931-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 6:741-751. [DOI] [PubMed] [Google Scholar]
- Werren JH, et al. 2010. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327(5963):343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D, et al. 2009. SUPERFAMILY-sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 37:D380–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252-3255. [DOI] [PubMed] [Google Scholar]
- Xia QY, et al. 2008. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 38:1036-1045. [DOI] [PubMed] [Google Scholar]
- Xu L, et al. 2011. Timeless is a critical gene in the diapause of silkworm, Bombyx mori. Afr J Biotechnol. 10:16594-16601. [Google Scholar]
- Yamada H, Yamamoto MT. 2011. Association between circadian clock genes and diapause incidence in Drosophila triauraria. PLoS One 6(12)e27493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wei ZJ, Hong GY, Jiang ST, Wen LP. 2009. The complete nucleotide sequence of the mitochondrial genome of Phthonandria atrilineata (Lepidoptera: Geometridae). Mol Biol Rep. 36:1441-1449. [DOI] [PubMed] [Google Scholar]
- You MS, et al. 2013. A heterozygous moth genome provides insights into herbivory and detoxification. Nat Genet. 45:220-225. [DOI] [PubMed] [Google Scholar]
- Yu L, et al. 2015. Characterization and expression of the cytochrome P450 gene family in diamondback moth, Plutella xylostella (L.). Sci Rep. 5:8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S, Merlin C, Boore JL, Reppert SM. 2011. The monarch butterfly genome yields insights into long-distance migration. Cell 147:1171-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S, Reppert SM. 2013. MonarchBase: the monarch butterfly genome database. Nucleic Acids Res. 41:D758–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, et al. 2006. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 10:719-729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.