Abstract
Enhancers lie at the heart of transcriptional and developmental gene regulation. Therefore, changes in enhancer sequences usually disrupt the target gene expression and result in disease phenotypes. Despite the well-established role of enhancers in development and disease, evolutionary sequence studies are lacking. The current study attempts to unravel the puzzle of bony vertebrates’ conserved noncoding elements (CNE) enhancer evolution. Bayesian phylogenetics of enhancer sequences spotlights promising interordinal relationships among placental mammals, proposing a closer relationship between humans and laurasiatherians while placing rodents at the basal position. Clock-based estimates of enhancer evolution provided a dynamic picture of interspecific rate changes across the bony vertebrate lineage. Moreover, coelacanth in the study augmented our appreciation of the vertebrate cis-regulatory evolution during water–land transition. Intriguingly, we observed a pronounced upsurge in enhancer evolution in land-dwelling vertebrates. These novel findings triggered us to further investigate the evolutionary trend of coding as well as CNE nonenhancer repertoires, to highlight the relative evolutionary dynamics of diverse genomic landscapes. Surprisingly, the evolutionary rates of enhancer sequences were clearly at odds with those of the coding and the CNE nonenhancer sequences during vertebrate adaptation to land, with land vertebrates exhibiting significantly reduced rates of coding sequence evolution in comparison to their fast evolving regulatory landscape. The observed variation in tetrapod cis-regulatory elements caused the fine-tuning of associated gene regulatory networks. Therefore, the increased evolutionary rate of tetrapods’ enhancer sequences might be responsible for the variation in developmental regulatory circuits during the process of vertebrate adaptation to land.
Keywords: CNEs, enhancers, evolutionary rate, bony vertebrates, cis-regulatory evolution
Introduction
Vertebrate adaptation to land is a major breakthrough in metazoan evolutionary history, marked by extensive morphological diversification. Large-scale genome sequencing of metazoans greatly enhanced our understanding of their genome architecture, which in turn spearheaded efforts to unfold the potential mechanisms implicated in diversification of the animal form. Previously, gene duplication and protein evolution were envisaged as potential mechanisms underlying the morphological complexity (Ohno 1970). Earlier endeavors were largely focused on protein-coding genes, as they were easier to pinpoint, owing to their well-established genetic code and the properly annotated coding landscape. Later innovative findings by King and Wilson (1975) articulated the revolutionary theory of morphological evolution. According to this theory, changes in regulation of gene expression are central to phenotypic evolution between and within species.
Cis-regulatory elements, specifically enhancers, orchestrate gene expression in a spatiotemporal manner during early development (Maston et al. 2006). The precise control of gene expression is pivotal for key developmental processes during embryogenesis. Enhancers are central to transcriptional and developmental gene regulation, with transcriptional regulation accomplished through combinatorial interactions of cis-regulatory elements with specific transcription factors (Abbasi et al. 2007). Cis-regulatory elements lack a well-defined vocabulary and syntax, as opposed to the coding sequences, which restrains attempts to pinpoint enhancers based on their genomic sequence alone. However, several endeavors centered on the evolutionary conservation metric have proved their potential to discern putative cis-regulatory elements (Abbasi et al. 2010, 2013). For instance, pan-vertebrate genome comparisons have revealed some highly constrained genomic sequences across a wide phylogenetic span (Bejerano et al. 2004; Sandelin et al. 2004; Dermitzakis et al. 2005; Woolfe et al. 2005). Surprisingly, these elements are ubiquitous in the vertebrate noncoding genomic landscape, hereafter termed as the conserved noncoding elements (CNEs) (Dermitzakis et al. 2005). Their profound sequence conservation across divergent vertebrates implies functional implications and, in line with these speculations, in vivo testing of CNEs in transgenic mice assays has confirmed an enhancer potential for the majority of these elements (Pennacchio et al. 2006; Visel et al. 2007, 2009; Noonan and McCallion 2010). For the purpose of clarity, we termed them CNE enhancers. CNE enhancers are frequently found clustering around genes implicated in transcriptional regulation and development (Woolfe and Elgar 2008). The preponderance of enhancers around developmentally important genes, as well as their notable role in gene expression regulation, highlights them as a key component of vertebrate developmental regulatory networks.
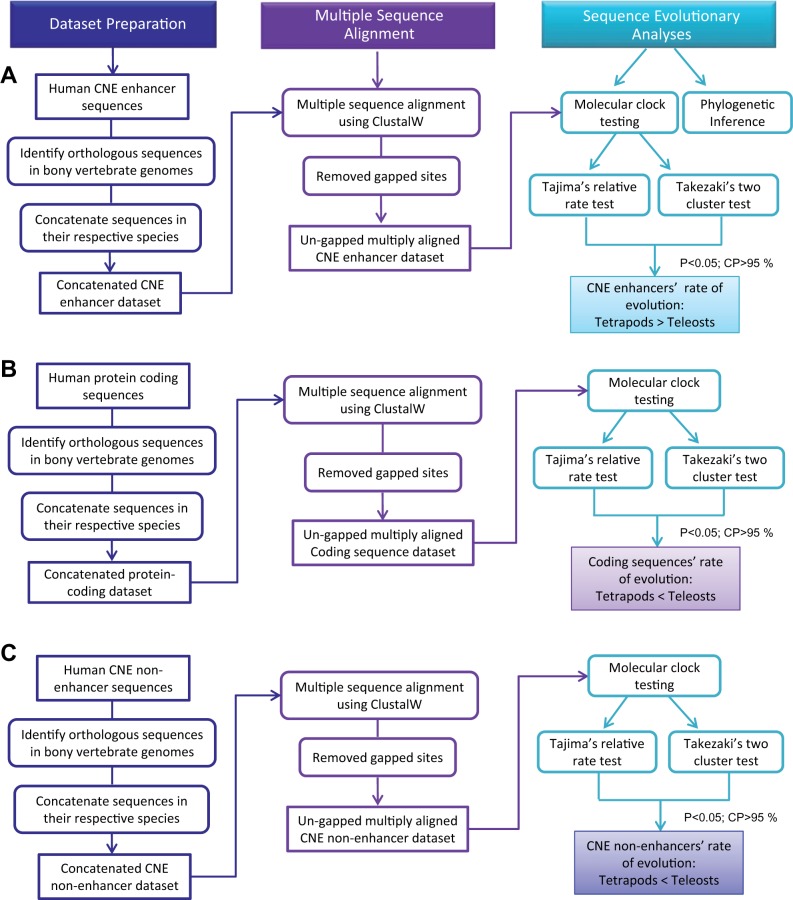
In light of evolutionary development claims, the evolution of gene regulatory elements, in addition to the gene itself, is seen as playing an important role in morphological evolution (Carroll 2005; Wray 2007). The main reason for this supposition is related to the modular scenario of enhancers, as any change in enhancer sequence is less pleiotropic, altering gene regulation in a particular tissue or developmental stage (Carroll 2008). In contrast, mutations in protein-coding sequences lead to deleterious pleiotropic effects. Therefore, this modularity of enhancers and their important role in developmental regulation underscore their candidacy for evolutionary consideration. Although significant progress has previously been made to understand enhancer evolution, this study aims to build on this by elucidating CNE enhancer evolution across a wide phylogenetic span. Furthermore, this study has also incorporated the protein-coding landscape to improve our understanding of relative evolutionary dynamics of the protein-coding and cis-regulatory landscapes (fig. 1).
Fig. 1.—
The flowchart depicts the complete hierarchy of analyses carried out in the current study. (A) Portrays the main steps to identify the bony vertebrate CNE enhancers and characterize the evolutionary trend of concatenated data set through phylogenetic analyses and molecular clock testing. (B) In the case of the protein-coding landscape, the bony vertebrates’ concatenated data set was subjected to tests of rate constancy. (C) Tajima’s relative rate and Takezaki’s two cluster tests were performed on a control data set (CNE-nonenhancer sequences) to provide unbiased estimates of the CNE enhancer evolution. The key steps involved to carry out the analyses are shown.
The current study provides an insight into the bony vertebrates’ cis-regulatory realm from an evolutionary perspective. A broad spectral exploration of a putative enhancer data set, based on experimentally verified human enhancers, provided an outlook on regulatory evolution, whereas Bayesian phylogenetics of enhancer sequences resolved the historical relationships between bony vertebrates. Furthermore, the present work serves to illustrate a paradigmatic shift in the evolutionary rates of protein-coding and cis-regulatory landscapes. Both the statistical and phylogenetic tests of the clock hypothesis unveiled a reduced rate of evolution for tetrapod coding sequences. These observations suggest a variety of evolutionary constraints underlying these diverse genomic landscapes (fig. 1). Specifically, rate changes in tetrapod enhancer sequences, as opposed to the coding sequences, have possible implications for the rewiring of regulatory circuits during the process of vertebrate adaptation to land.
Materials and Methods
Gene Ontology Analysis of the CNE Enhancer Target Genes
Highly constrained noncoding elements in the human genome, which are experimentally confirmed enhancers, were retrieved from Vista Enhancer Browser (Visel et al. 2007). For the purpose of our investigation, we focused on deeply conserved (>450 Myr) enhancer elements expressing in different anatomical regions of the brain (supplementary table S1, Supplementary Material online). These enhancers had previously been associated with their probable target genes by our group (Parveen et al. 2013). Here, we ascertained the biological themes of those enhancer target genes through the Gene Ontology (GO) analysis. To accomplish this task, CNE enhancer target genes were subjected to the PANTHER classification system’s binomial statistics tool (Cho and Campbell 2000). The program compared target genes with a reference set of all genes within the human genome and sought to probe their over- or underrepresentation for the PANTHER biological process and molecular function categories (Thomas et al. 2006).
Identifying the Bony Vertebrate CNE Enhancers and Their Phylogenetic Reconstruction
Genome-wide scanning of the bony vertebrates for putative orthologs of the human CNE enhancers was done by running BLAT (Kent 2002) against the tetrapod, lobe-finned fish and teleost genomes, available at the UCSC genome browser (Fujita et al. 2011). Orthologous sequences from the cartilaginous fish genome were obtained by comparing human CNE enhancers against the elephant shark genome, available at IMCB (http://esharkgenome.imcb.a-star.edu.sg/, last accessed 2014). Only the highest scoring BLAST hits with maximum percentage identity and longer query span were retained. The species that were incorporated in the current analysis comprised: Homo sapiens, Mus musculus, Rattus novergicus, Canis familiaris, Felis catus, Bos taurus, Equus caballus, Monodelphis domestica, Gallus gallus, Ornithorhynchus anatinus, Xenopus tropicalis, Anolis carolinensis, Latimeria chalumnae, Danio rerio, Gasterosteus aculeatus, Tetraodon nigroviridis, Oryzias latipes, and Callorhinchus milii.
Human, and their orthologous, bony vertebrate sequences were retrieved by similarity searches, and then concatenated using the head-to-tail approach in their respective species. The concatenated bony vertebrate CNE enhancer data set was subjected to multiple sequence alignment (MSA) using ClustalW (Thompson et al. 1994) under default parameters. The accuracy of this alignment was enhanced by deleting any gapped sites in the sequences. The resulting un-gapped alignment (available upon request) was operated upon in tree construction and forthcoming analyses. The phylogenetic inferences of the bony vertebrate CNE enhancer data set were accomplished to gain insights into the evolutionary account of the bony vertebrate regulatory realm. Evolutionary relationships were inferred, taking key tetrapod, lobe-finned fish and teleost lineages and by employing the neighbor-joining (NJ) (Saitou and Nei 1987) and the Bayesian tree inferential techniques. For the NJ phylogenetic inference, uncorrected p-distance was exploited as a nucleotide substitution model and tree construction was carried out in MEGA 5.0 (Tamura et al. 2011). The resulting tree topology was tested through bootstrapping, which provided a level of statistical confidence to the interior branches in a tree. A total of 1,000 bootstrap replicates were generated in the NJ tree.
Bayesian phylogenetic inference of the CNE enhancer data set was carried out through the BEAST software package (Drummond et al. 2012). BEAUti, a graphical user interface to BEAST, constructed the initial BEAST XML file of input data and enabled us to specify parameters for tree construction and for running the Markov Chain Monte Carlo algorithm. The best fitting nucleotide substitution model, GTR+G+I (General Time Reversible, with γ distributed rates and proportion of invariable sites), was established to construct the Bayesian phylogenetic tree. The forest of trees and log files, generated through independent runs of BEAST, were combined by LogCombiner. The resultant Bayesian tree along with its branch support values was visualized in FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Relative Rate Analyses of Diverse Genomic Landscapes
We carried out the rate analyses to detect significant rate changes across the bony vertebrate lineages by employing Tajima’s relative rate test (Tajima 1993) and Takezaki’s two-cluster test (Takezaki et al. 1995). These tests are fully equipped to decipher interspecific rate changes using the molecular clock as a null hypothesis. In both cases, the ungapped MSA of the CNE enhancer data set was utilized for this process, with C. milii being used as an outgroup. Tajima’s relative rate test compared molecular evolutionary rates among each pair of the bony vertebrate lineages. The resultant P values from rate analysis were corrected for multiple testing using the Benjamini and Yekutieli correction (Benjamini and Yekutieli 2001). Likewise, Takezaki’s two cluster test was employed by utilizing the LINTREE (Takezaki et al. 1995), which established a significant rate of heterogeneity between the two major clusters in our data set, that is, tetrapods and teleosts. The test results were presented in the tabular form (supplementary table S3, Supplementary Material online). To further understand the cis-regulatory evolution, we explored the sequence evolutionary trend of 10-mer DNA motifs, discovered in a subset of the CNE enhancer data set using the MEME motif discovery algorithm (Bailey and Elkan 1994). The resulting motif sequences were concatenated in their respective species and subjected to MSA using the ClustalW algorithm. The ungapped concatenated motif data set was analyzed by Tajima’s relative rate test to compute the interspecific rate differences among bony vertebrate lineages by utilizing the elephant shark as an outgroup.
Our aim to model unbiased enhancer evolution prompted us to investigate the evolutionary inclination of the coding landscape as well. Therefore, a data set of 50 human protein-coding genes was randomly selected (supplementary table S5, Supplementary Material online). The coding sequences belonging to these protein-coding genes were acquired from the Ensembl genome browser (Hubbard et al. 2009). These sequences were BLASTed against genomes of similar sets of species in the CNE enhancer data set. The resulting sequences were concatenated by species and subjected to MSA. The ungapped multiple alignment of the bony vertebrate coding sequence data set was further analyzed by Tajima’s relative rate test and Takezaki’s two cluster test. Both tests were employed to compare the evolutionary rates of each pair of the tetrapod and lobe-finned fish lineages as well as the tetrapod and teleost lineages, taking cartilaginous fish as an outgroup.
In addition, to uncover the dynamic evolution of diverse genomic landscapes, we performed the rate analysis on a control data set, comprising conserved noncoding nonenhancer sequences. To accomplish this task, we carefully selected a subset of UCEs and CNEs from the UCNEbase (Dimitrieva and Bucher 2013) and the CONDOR database (Woolfe et al. 2007), respectively. In addition, 22 CNEs (one element from each chromosome) were collected from previously published work (Lee et al. 2011; supplementary table S8, Supplementary Material online). None of the selected elements had shown any enhancer potential. Therefore, we have termed them as CNE nonenhancers. The selected CNE nonenhancers and their bony vertebrate orthologous sequences were concatenated by species and subjected to MSA. After this, Tajima’s relative rate and Takezaki’s two cluster tests were applied on the ungapped concatenated CNE nonenhancer data set. Evolutionary rate comparisons, by pair, of the tetrapod and teleost lineages were presented in supplementary tables S9 and S10, Supplementary Material online. The evolutionary rate inferences drawn from the coding sequence evolution as well as those from the CNE nonenhancer sequences were compared with those of the CNE enhancers to explore their relative evolutionary dynamics. The complete schematic result of analyses carried out on these diverse genomic landscapes is presented as a flowchart (fig. 1).
Results
CNE Enhancers Are Associated with Development-Related Genes
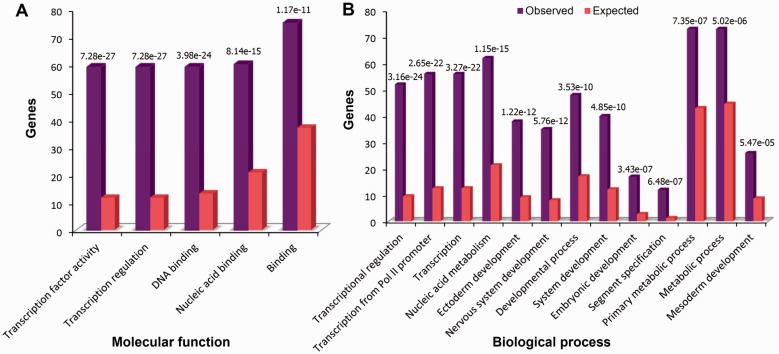
The primacy of enhancers in the human gene regulatory landscape makes them a promising target for evolutionary consideration. In this study we have presented a large-scale exploration of experimentally validated enhancers, approached from an evolutionary sequence perspective. Moreover, we have provided profound insights about biological themes that are most pertinent to CNE enhancer target genes. These target genes were previously pinpointed by our group based on their conserved syntenic association to enhancers over a longer evolutionary distance (>450 Myr) and their endogenous expression pattern in sync with the enhancer expression pattern (Parveen et al. 2013). Here, we conducted GO analyses on CNE enhancer target genes through the PANTHER classification system’s binomial statistics tool (Cho and Campbell 2000). Statistical analyses for PANTHER GO categories delineated significant (P < 0.01) functional enrichment of target gene bodies for transcriptional regulation, DNA binding, nucleic acid binding, and transcription factor activity (fig. 2A). Furthermore, in the biological process category, embryonic development, nervous system development, nucleic acid metabolism, and transcriptional processes were also significantly overrepresented (P < 0.01) along with other developmental processes (fig. 2B). Centered on these observations, we suggest that CNE enhancers are associated with an important group of genes which are involved in transcriptional regulation and are therefore master regulators of development, termed as the trans-dev genes (Woolfe et al. 2005).
Fig. 2.—
Statistical overrepresentation of the human CNE enhancer target genes for the molecular function and biological process categories. There were in total 109 target gene bodies for a data set of 100 human CNE enhancers analyzed in the present study. Statistical analysis of these gene bodies for particular GO categories, by using all genes in the human genome as a reference, was performed by using a binomial statistics tool (Thomas et al. 2006). (A) The test highlighted significant overrepresentation of enhancer target genes for transcriptional regulation, DNA binding, nucleic acid binding, and transcription factor activity. (B) In the biological process category, embryonic development, nervous system development, nucleic acid metabolism, and transcriptional processes were significantly overrepresented (P < 0.01) along with other developmental processes.
Bony Vertebrate CNE Enhancer Data Set
We used an experimentally verified catalog of human CNE enhancers (supplementary table S1, Supplementary Material online) as a baseline to detect evolutionarily conserved putative cis-regulatory elements in other bony vertebrate lineages (Parveen et al. 2013). The resulting human and orthologous sequences, obtained through BLAST (Basic Local Alignment Search Tool) searches, were concatenated in their respective lineages. The final data set consisted of human orthologous sequences in lobe-finned fishes (coelacanth plus tetrapods) and ray-finned fishes (teleosts), spanning 1,315,184 and 135,698 bp, respectively. The clade-wise length distribution of putative cis-regulatory sequences in tetrapods and teleosts highlighted overall that tetrapod CNE enhancers have a longer length distribution than those of teleosts. The average size of the CNE enhancers in tetrapods and teleosts is 1,042 (±270) and 339 (±28) bp, respectively (table 1). This marked difference in length distribution raises the question as to how the CNE enhancers might have evolved during the water to land transition of vertebrates. To provide a comprehensive answer to this question, we carried out an examination of the molecular phylogenetics of CNE enhancers along with the long-range evolutionary rate comparisons.
Table 1.
Species-Wise and Clade-Wise Length Distribution of Concatenated CNE Enhancers across the Bony Vertebrate Lineages
| Species/Clades | Total Length (bp)a | Average Length (bp)b |
|---|---|---|
| Homo sapiens | 162,080 | 1,620 |
| Mus musculus | 118,419 | 1,184 |
| Rattus novergicus | 112,459 | 1,124 |
| Canis familiaris | 129,521 | 1,295 |
| Felis catus | 95,000 | 950 |
| Bos taurus | 127,777 | 1,277 |
| Equus caballus | 137,213 | 1,372 |
| Monodelphis domestica | 91,926 | 919 |
| Gallus gallus | 79,006 | 790 |
| Ornithorhynchus anatinus | 74,744 | 747 |
| Xenopus tropicalis | 53,449 | 534 |
| Anolis carolinensis | 69,580 | 695 |
| Latimeria chalumnae | 64,010 | 640 |
| Danio rerio | 37,236 | 372 |
| Gasterosteus aculeatus | 36,278 | 362 |
| Tetraodon nigroviridis | 30,722 | 307 |
| Oryzias latipes | 31,462 | 314 |
| Tetrapods | 1,251,174 | 1,042 (±270) |
| Teleosts | 135,698 | 339 (±28) |
aTotal length of 100 concatenated CNE enhancer data set. These concatenated sequences were operated upon in phylogenetic inference.
bLength of a single CNE enhancer after averaging over the total number of enhancers involved.
The Evolution of Bony Vertebrate CNE Enhancers
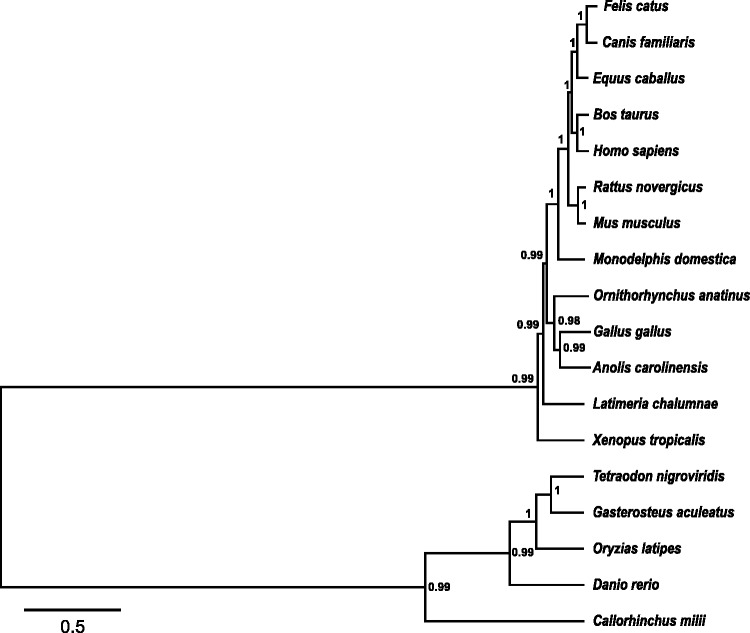
The evolution of the bony vertebrates’ CNE enhancer data set has never been modelled from a molecular phylogenetic standpoint. The current study addressed this by providing a thorough depiction of the CNE enhancer phylogeny across a wide phylogenetic span. A concatenated CNE enhancer data set from representative members of tetrapod and teleost lineages was subjected to NJ and Bayesian tree construction approaches by taking a model cartilaginous fish, the elephant shark, as an outgroup. Moreover, a recently sequenced lobe-finned fish, the African coelacanth, was also incorporated into the phylogenetic analysis, due to its genealogical placement between ray-finned fishes and tetrapods. The evolutionary branching which stemmed from NJ and Bayesian approaches for the CNE enhancer data set was highly congruent, so we chose the Bayesian tree for further examination (supplementary fig. S1, Supplementary Material online, and fig. 3).
Fig. 3.—
Bayesian phylogenetic inference of the bony vertebrates’ concatenated CNE enhancer data set. Tree construction was carried out by employing 100 CNE enhancers, concatenated in their respective lineages (supplementary table S1, Supplementary Material online). The Bayesian maximum clade credibility tree was inferred using the BEAST package (Drummond et al. 2012), under the “GTR+G+I” model of nucleotide substitution. The numbers on branches represent posterior values supporting that branch. The scale bar shows the number of nucleotide substitutions per site.
The CNE enhancer phylogeny delineated a placental mammal branching pattern (type: Rodents (Laurasiatherians, Primates)), which has placed rodents as basal to placental mammals. These findings suggest a resolution to the controversial trichotomy between placental mammals, by supporting Laurasiatherians and Primates as a monophyletic group and Rodents as the sister class to this group with significant posterior probability (100%). Moreover, Bayesian phylogenetics retrieved marsupials as the outgroup taxon to placental mammals with strong branch support value (100%). The platypus, which is a monotreme displaying a blend of mammalian, reptilian and avian features, clustered with birds and reptiles. Hence, monotremes, birds, and reptiles collectively made a perfect outgroup to therians. Furthermore, the coelacanth exhibited a basal relationship to amniotes and clustered more closely with tetrapods than its fish relatives (100% posterior probability). Based on these findings, it emerges that coelacanths are closer in an evolutionary sense to tetrapods than to ray-finned fishes. In the case of teleosts, the CNE enhancer phylogeny maintained a robust sister group relationship between G. aculeatus and T. nigroviridis with significant posterior probability support (99%). Danio rerio CNE enhancers appeared to be highly divergent and thus demonstrated a basal relationship to their teleost relatives. The tree branch which led to tetrapods was relatively long when compared with teleosts, highlighting a greater level of evolutionary changes in the tetrapod CNE enhancers during the process of bony vertebrate evolution.
Molecular Clock Testing and Long-Range Evolutionary Rate Comparisons
Since the inception of the molecular clock hypothesis, the rate of clock movement during the course of evolution has been discussed at length (Kumar and Hedges 1998; Peterson et al. 2004). Molecular clock hypothesis has played a pivotal role as a null model in the testing of rates of molecular evolution across different lineages. Here, we compared the molecular evolutionary rates of the concatenated CNE enhancer data set across the bony vertebrate lineages by clock testing. Variations in the rate of molecular evolution were modelled by employing a nonparametric relative rate approach, that is, Tajima’s relative rate test (Tajima 1993), and a phylogenetic test of the molecular clock, that is, two-cluster test (Takezaki et al. 1995), on a carefully curated CNE enhancers data set. Tajima’s relative rate test provided disparate substitution rates between ingroup species (osteichthyes) and reference outgroup species (chondrichthyes), thereby significantly (P < 0.01) rejecting the null hypothesis of the molecular clock. Evolutionary rate comparisons (supplementary table S2, Supplementary Material online) demonstrated a significantly reduced substitution rate for human lineage when compared with rodents, a process known as “hominid slowdown” or “fast rats,” respectively. Furthermore, we found that the CNE enhancers were accumulating substitutions at elevated levels in the tetrapod lineages. The coelacanth, proposed as a fish closer in relation to tetrapods than to ray-finned fishes, showed a substitution rate pattern which was parallel to its phylogenetic cousins. On the contrary, teleosts have been evolving at a relatively low pace, demonstrating significantly (P < 0.01) fewer substitutions per site than their land-dwelling relatives.
Takezaki’s two-cluster test (Takezaki et al. 1995) statistically rejected the clock hypothesis by demonstrating a significant rate of heterogeneity (CP > 95%) between two of the major clusters in our data set. Rate estimates drawn from the two-cluster test delineated that teleosts were experiencing markedly fewer substitutions (0.23 substitutions per site) than the lobe-finned fish plus tetrapods cluster (0.38 substitutions) when compared with the outgroup species, C. milii (supplementary table S3, Supplementary Material online). Furthermore, test results revealed a significantly faster evolution for zebrafish CNE enhancers than their teleost relatives. Likewise, stickleback CNE enhancers demonstrated a greater substitution rate than tetraodon and medaka. Therefore, both the statistical and phylogenetic measures of the evolutionary rate for bony vertebrate CNE enhancers were in corroboration.
To enrich our understanding of the cis-regulatory sequence evolution, we analyzed the evolutionary rate of concatenated DNA motifs that were specifically discovered in a subset of the CNE enhancer data set (supplementary table S4, Supplementary Material online). These results suggested a significantly elevated rate of substitution for tetrapods than for their teleost relatives (supplementary table S4, Supplementary Material online). Likewise, the coelacanth exhibited a significantly faster evolutionary trend than that of teleosts. Therefore, the evolutionary trends of concatenated DNA motifs confirm the “slowdown” of the molecular clock for the teleost cis-regulatory landscape.
Evolutionary Dynamics of Diverse Genomic Landscapes
Interspecific rate variation of the bony vertebrates’ CNE enhancer data set was found to be clearly in opposition to the previously obtained rate estimates for CNEs and ultraconserved noncoding elements (UCEs) (Stephen et al. 2008; Lee et al. 2011). These findings prompted us to further investigate the evolutionary rates of coding sequences, to unveil the evolutionary dynamics of coding versus regulatory landscapes, and to gauge their relative contributions in organismal evolution (supplementary table S5, Supplementary Material online). In addition, relative rate analysis was performed on a control data set (CNE nonenhancer elements) with the aim of providing unbiased estimates of the evolution of the CNE enhancer.
In the case of the multilocus coding sequence data set, evolutionary rate estimates drawn from Tajima’s relative rate test and Takezaki’s two-cluster test exhibited an entirely identical trend (supplementary tables S6 and S7, Supplementary Material online). The two-cluster test exploited Z-statistics to demonstrate a significant level of heterogeneity among both species pairs and major species clusters in our data set. However, Tajima’s relative rate test relied on chi-square statistics to determine interspecific rate changes in the coding sequence evolution. Both tests showed a dramatic decline in the evolutionary rates of the coding repertoire during the water to land transition of vertebrates. Teleosts exhibited an elevated level of coding sequence evolution (0.51 substitutions per site) to that of the lobe-finned fish plus tetrapods cluster (0.16 substitutions per site). Moreover, from the perspective of teleosts, the coelacanth, dubbed a “living fossil,” demonstrated a reduced rate of coding sequence evolution. This significantly faster coding sequence evolution in teleosts than in other vertebrate genomes (P < 0.01, CP > 95%) can be best explained by the documented “plasticity” of teleost genomes (Venkatesh 2003).
The relative rate analyses carried out on cis-regulatory as well as coding sequences, demonstrated strong disparities in their rates of molecular evolution during vertebrate adaptation to land. For instance, the land-dwelling vertebrates exhibited a significantly faster rate of regulatory sequence evolution compared with their protein-coding landscape. Furthermore, rate analyses on a control data set (nonenhancer-UCEs/CNEs) also revealed a significantly reduced evolutionary rate for land-dwelling vertebrates (supplementary tables S9 and S10, Supplementary Material online). Such discrepancies in the molecular evolutionary rate of enhancers and other genomic landscapes (protein-coding and noncoding nonenhancer landscapes) convincingly render fast evolution of tetrapods, a singular characteristic of the cis-regulatory landscape.
Discussion
CNEs are a substantial characteristic of vertebrate genomes. Their integral presence within divergent vertebrates suggests that they have made important functional contributions across the depth of evolutionary time. Accordingly, Pennacchio and colleagues experimentally validated the cis-regulatory potential for 1,154 CNEs (out of 2,192 tested elements) using an in vivo transgenic mice assay (Pennacchio et al. 2006; Visel et al. 2009). The availability of such a large-scale experimentally verified enhancer data set offers a platform to explore the significance of enhancers in disease and evolution. Enhancers are the key players in gene expression regulation; therefore, any change in enhancer sequences will usually result in target gene dysregulation and can underlie disease phenotypes, such as preaxial polydactyly, Hirschsprung’s disease, and various types of cancers (Lettice et al. 2003; Ahmadiyeh et al. 2010; Sribudiani et al. 2011). The pronounced role of enhancers in development and disease underscores their candidacy for evolutionary consideration. Previously, attempts have been made to characterize the evolution of CNEs and ultraconserved elements (Stephen et al. 2008; Wang et al. 2009; Lee et al. 2011). The major caveat to these studies was the dearth of experimental evidence for cis-regulatory potential of the elements under question. In this study, we have addressed this gap by employing the experimentally verified catalog of human enhancers (predominantly expressed in the brain) as a baseline to the current investigation. The study provides an unprecedented insight into the evolutionary trends of the bony vertebrates’ CNE enhancer data set in a phylogenetic context. Specifically, our data offer a valuable insight into the relative evolutionary dynamics of cis-regulatory and coding sequence landscapes during vertebrate adaptation to land.
Earlier studies have designated genes in the vicinity of enhancers or the genes harboring them as their probable targets (Pennacchio et al. 2006; Visel et al. 2008). Owing to the distal regulatory potential of enhancers, this association with nearby genes is impractical. Therefore, a previous report from our group pinpoints target gene bodies of the functionally characterized enhancer data set (Parveen et al. 2013). These enhancer–target gene associations were based on the syntenic conservation of the gene regulatory blocks over a longer evolutionary distance, coupled with endogenous expression analysis (Parveen et al. 2013). Here, GO analysis of enhancer target genes for the molecular function and biological process categories established an enhancer association to trans-dev genes, confirming earlier observations (Woolfe and Elgar 2008).
Molecular phylogenetics of the concatenated CNE enhancer data set successfully uncovered historical relationships among the bony vertebrate lineages, with strong branch support values (supplementary fig. S1, Supplementary Material online, and fig. 3). NJ and Bayesian tree topologies of the CNE enhancer data set corroboratively established the unique interordinal relationships for the contentious placental mammal clade. Our findings suggest a close evolutionary relationship among human and laurasiatherians while placing rodents at a basal position, and thereby provide a resolution to the much-hyped trichotomy between placental mammals (Murphy et al. 2001; Misawa and Nei 2003). Based on these findings, we can develop an assertion that the concatenation approach can yield a better phylogenetic resolution than single element phylogeny. Interestingly, the inferred placental mammal branching of enhancer sequences reconciles to the evolutionary pattern of microRNAs (Dolgin 2012). MicroRNAs, based on their presence in bilaterians and their ability to regulate development, are considered to be accountable for morphological complexity as well as being a source of morphological innovations (Peterson et al. 2009). Harmony among the microRNAs and the CNE enhancer evolutionary trajectories can be interpreted as a regulatory element exclusive evolutionary trend. The clustering of the platypus with reptiles and birds is in perfect harmony with earlier studies, which highlighted the fact that the platypus genome is a mixture of avian, reptilian, and mammalian characteristics (Warren et al. 2008). Furthermore, probing the coelacanth genome in the current study enriched our understanding of the vertical descent from ray-finned fish to tetrapods. The coelacanth maintained a closer evolutionary relationship to its land-living cousins than to its fish relatives, confirming earlier observations (Amemiya et al. 2013).
The clock-based evolutionary rate estimates of the concatenated CNE enhancer data set provide deep insights into the bony vertebrates’ cis-regulatory realm. Takezaki’s two cluster test and Tajima’s relative rate test demonstrated an analogous rate of enhancer evolution in the bony vertebrate lineages (supplementary tables S2 and S3, Supplementary Material online). Moreover, the concatenated 10-mer DNA motif data set delineated a parallel evolutionary trend as well (supplementary table S4, Supplementary Material online). Evolutionary rate comparisons demonstrated a significantly reduced rate of substitution for the human lineage when compared with rodents, thereby corroborating the phenomenon of “hominid slowdown” or “fast rats,” respectively (Bromham et al. 1996; Kumar 2005). Furthermore, rate analyses pinpointed a significantly faster rate of evolution for tetrapods than that of teleosts since their divergence from their last common ancestor (supplementary tables S2 and S3, Supplementary Material online). These clade-specific changes in tetrapods can be tracked down to their extensive morphological diversification in the course of vertebrate land adaptation. Therefore, the observed rate pattern of cis-regulatory evolution can be best explained by the “time scaling” phenomenon (Gingerich 2001; Rabosky and Adams 2012), which suggests slower morphological change in older lineages in relation to younger ones. Surprisingly, the coelacanth accumulated significantly (CP-value > 95%) more substitutions than its land-living relatives and ray-finned fishes. The evolutionary trend exhibited by the coelacanth cis-regulatory elements goes hand-in-hand with those of the transposable elements (Amemiya et al. 2013). This exceptional characteristic of the coelacanth putative cis-regulatory repertoire reinforces the theory that not all regions of the coelacanth genome are evolving slowly. Intriguingly, harmony among the evolutionary rates of the coelacanth transposable elements (thought to have an important role in gene regulation) and putative cis-regulatory elements establishes quicker evolution exclusive to the coelacanth regulatory landscape.
The modeling of enhancer evolution at the macroevolutionary scale and their rate analyses revealed an evolutionary inclination toward diverse genomic landscapes. Relative rate analyses of the multilocus coding sequence data set as well as the CNE nonenhancer data set (control data set) unveiled a dynamic picture of bony vertebrate evolution. Confirming previous observations, protein-coding sequences and the CNE non-enhancer sequences portrayed a significantly faster evolution among teleosts than for tetrapods (supplementary tables S6, S7, S9, and S10, Supplementary Material online) (Jaillon et al. 2004; Brunet et al. 2006; Steinke et al. 2006; Stephen et al. 2008; Lee et al. 2011). The pronounced upsurge of protein evolution shown in ray-finned fishes compared with the other vertebrate genomes reveals the diversity of fish genomes, previously proposed by Venkatesh (2003). The observed pattern of protein-coding and CNE nonenhancer sequence evolution was clearly at odds with the cis-regulatory landscape, specifically during vertebrate land adaptation. Accordingly, tetrapods and the coelacanth, which displayed a fast evolving regulatory landscape, depicted a slower rate for coding sequence evolution. These findings convincingly establish the quicker evolution of tetrapods and the coelacanth, a singular characteristic of the regulatory landscape. Quicker evolution of the regulatory elements in land-living vertebrates suggests that tetrapods have to endure many more changes in their cis-regulatory landscape to “brave land.” The variation in tetrapod cis-regulatory sequences might affect the expression of target genes and thereby alter the associated gene regulatory networks. Therefore, the elevated substitution rate in tetrapod cis-regulatory elements suggests the rewiring of regulatory circuits during the process of vertebrate adaptation to land (Abbasi 2011).
This observed variation in the relative evolutionary rates of coding and regulatory repertoires suggests that assorted evolutionary constraints determine the diverse genomic landscapes. The quicker evolution of tetrapod CNE enhancers is evidence of either adaptive evolution or a relaxation of constraints. Taking fast evolving regulatory repertoire of tetrapods as evidence of relaxation of selective constraints is not viable, as mutations in cis-regulatory elements have been associated with various genetic disorders (Sagai et al. 2005; Kleinjan and Coutinho 2009; VanderMeer and Ahituv 2011). This suggestion implicates enhancers as the prospective candidate for adaptive evolution. Therefore, cis-regulatory polymorphism studies probing archaic hominin genomes in concert with the contemporary human population will provide an enhanced dissection of the regulatory realm during the course of human evolution. Keeping in mind the well-established role of enhancer variation in eliciting disease phenotypes as well as in phenotypic variation, analyzing biologically eloquent cis-regulatory variants and pinpointing their disease association can be an important emphasis of future studies.
Conclusion
The inferred phylogeny of the CNE enhancers across teleostomi as well as the relative rate estimates of the bony vertebrates’ regulatory repertoire overwhelmingly enhances our understanding of vertebrate cis-regulatory evolution. The dramatic variation witnessed in the molecular evolutionary rates of regulatory, versus coding and noncoding, repertoires of the genome during vertebrate adaptation to land, suggests that a number of disparate selection constraints act on diverse genomic landscapes. The variations within tetrapod cis-regulatory sequences might affect the expression of target genes and thereby alter the associated gene regulatory networks. It is therefore speculated here that the evolutionary exclusiveness of cis-regulatory sequences underlies the rewiring of the regulatory circuit during vertebrate adaptation to land.
Supplementary Material
Supplementary figure S1 and tables S1–S10 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors are thankful to Muhammad Faisal for assisting with the motif analysis and to Nazia Parveen for her critical reading of the manuscript. This study was supported by Higher Education Commission (HEC) of Pakistan.
Footnotes
Associate editor: Greg Elgar
Literature Cited
- Abbasi AA. 2011. Evolution of vertebrate appendicular structures: insight from genetic and palaeontological data. Dev Dyn. 240:1005−1016. [DOI] [PubMed] [Google Scholar]
- Abbasi AA, et al. 2007. Human GLI3 intragenic conserved non-coding sequences are tissue-specific enhancers. PLoS One 2:e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbasi AA, et al. 2010. Human intronic enhancers control distinct sub-domains of Gli3 expression during mouse CNS and limb development. BMC Dev Biol. 10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbasi AA, Minhas R, Schmidt A, Koch S, Grzeschik KH. 2013. Cis-regulatory underpinnings of human GLI3 expression in embryonic craniofacial structures and internal organs. Dev Growth Differ. 55:699−709. [DOI] [PubMed] [Google Scholar]
- Ahmadiyeh N, et al. 2010. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci U S A. 107:9742−9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya CT, et al. 2013. The African coelacanth genome provides insights into tetrapod evolution. Nature 496:311−316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 2:28−36. [PubMed] [Google Scholar]
- Bejerano G, et al. 2004. Ultraconserved elements in the human genome. Science 304:1321−1325. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 29:1165−1188. [Google Scholar]
- Bromham L, Rambaut A, Harvey PH. 1996. Determinants of rate variation in mammalian DNA sequence evolution. J Mol Evol. 43:610−621. [DOI] [PubMed] [Google Scholar]
- Brunet FG, et al. 2006. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol Biol Evol. 23:1808−1816. [DOI] [PubMed] [Google Scholar]
- Carroll SB. 2005. Evolution at two levels: on genes and form. PLoS Biol. 3:e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134:25−36. [DOI] [PubMed] [Google Scholar]
- Cho RJ, Campbell MJ. 2000. Transcription, genomes, function. Trends Genet. 16:409−415. [DOI] [PubMed] [Google Scholar]
- Dermitzakis ET, Reymond A, Antonarakis SE. 2005. Conserved non-genic sequences—an unexpected feature of mammalian genomes. Nat Rev Genet. 6:151−157. [DOI] [PubMed] [Google Scholar]
- Dimitrieva S, Bucher P. 2013. UCNEbase—a database of ultraconserved non-coding elements and genomic regulatory blocks. Nucleic Acids Res. 41:D101–D109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin E. 2012. Phylogeny: rewriting evolution. Nature 486:460−462. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 29:1969−1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita PA, et al. 2011. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 39:D876–D882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingerich PD. 2001. Rates of evolution on the time scale of the evolutionary process. Genetica 112–113:127−144. [PubMed] [Google Scholar]
- Hubbard TJP, et al. 2009. Ensembl 2009. Nucleic Acids Res. 37:D690–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, et al. 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431:946−957. [DOI] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656−664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. 1975. Evolution at two levels in humans and chimpanzees. Science 188:107−116. [DOI] [PubMed] [Google Scholar]
- Kleinjan DJ, Coutinho P. 2009. Cis-ruption mechanisms: disruption of cis-regulatory control as a cause of human genetic disease. Brief Funct Genomic Proteomic. 8:317−332. [DOI] [PubMed] [Google Scholar]
- Kumar S. 2005. Molecular clocks: four decades of evolution. Nat Rev Genet. 6:654−662. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. 1998. A molecular timescale for vertebrate evolution. Nature 392:917−920. [DOI] [PubMed] [Google Scholar]
- Lee AP, Kerk SY, Tan YY, Brenner S, Venkatesh B. 2011. Ancient vertebrate conserved noncoding elements have been evolving rapidly in teleost fishes. Mol Biol Evol. 28:1205−1215. [DOI] [PubMed] [Google Scholar]
- Lettice LA, et al. 2003. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 12:1725−1735. [DOI] [PubMed] [Google Scholar]
- Maston GA, Evans SK, Green MR. 2006. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet. 7:29−59. [DOI] [PubMed] [Google Scholar]
- Misawa K, Nei M. 2003. Reanalysis of Murphy et al.’s data gives various mammalian phylogenies and suggests overcredibility of Bayesian trees. J Mol Evol. 57(Suppl. 1):S290–S296. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, et al. 2001. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294:2348−2351. [DOI] [PubMed] [Google Scholar]
- Noonan JP, McCallion AS. 2010. Genomics of long-range regulatory elements. Annu Rev Genomics Hum Genet. 11:1−23. [DOI] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Berlin/New York: Springer-Verlag. [Google Scholar]
- Parveen N, et al. 2013. Comparative genomics using teleost fish helps to systematically identify target gene bodies of functionally defined human enhancers. BMC Genomics 14:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, et al. 2006. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444:499−502. [DOI] [PubMed] [Google Scholar]
- Peterson KJ, Dietrich MR, McPeek MA. 2009. MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion. Bioessays 31:736−747. [DOI] [PubMed] [Google Scholar]
- Peterson KJ, et al. 2004. Estimating metazoan divergence times with a molecular clock. Proc Natl Acad Sci U S A. 101:6536−6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL, Adams DC. 2012. Rates of morphological evolution are correlated with species richness in salamanders. Evolution 66:1807−1818. [DOI] [PubMed] [Google Scholar]
- Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. 2005. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development 132:797−803. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406−425. [DOI] [PubMed] [Google Scholar]
- Sandelin A, et al. 2004. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics 5:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sribudiani Y, et al. 2011. Variants in RET associated with Hirschsprung’s disease affect binding of transcription factors and gene expression. Gastroenterology 140:572−582. [DOI] [PubMed] [Google Scholar]
- Steinke D, Salzburger W, Braasch I, Meyer A. 2006. Many genes in fish have species-specific asymmetric rates of molecular evolution. BMC Genomics 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen S, Pheasant M, Makunin IV, Mattick JS. 2008. Large-scale appearance of ultraconserved elements in tetrapod genomes and slowdown of the molecular clock. Mol Biol Evol. 25:402−408. [DOI] [PubMed] [Google Scholar]
- Tajima F. 1993. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 135:599−607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezaki N, Rzhetsky A, Nei M. 1995. Phylogenetic test of the molecular clock and linearized trees. Mol Biol Evol. 12:823−833. [DOI] [PubMed] [Google Scholar]
- Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731−2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, et al. 2006. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 34:W645–W650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673−4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderMeer JE, Ahituv N. 2011. cis-regulatory mutations are a genetic cause of human limb malformations. Dev Dyn. 240:920−930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B. 2003. Evolution and diversity of fish genomes. Curr Opin Genet Dev. 13:588−592. [DOI] [PubMed] [Google Scholar]
- Visel A, et al. 2008. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 40:158−160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457:854−858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. 2007. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 35:D88–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lee AP, Kodzius R, Brenner S, Venkatesh B. 2009. Large number of ultraconserved elements were already present in the jawed vertebrate ancestor. Mol Biol Evol. 26:487−490. [DOI] [PubMed] [Google Scholar]
- Warren WC, et al. 2008. Genome analysis of the platypus reveals unique signatures of evolution. Nature 453:175−183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, Elgar G. 2008. Organization of conserved elements near key developmental regulators in vertebrate genomes. Adv Genet. 61:307-338. [DOI] [PubMed] [Google Scholar]
- Woolfe A, et al. 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, et al. 2007. CONDOR: a database resource of developmentally associated conserved non-coding elements. BMC Dev Biol. 7:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 8:206−216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.