Abstract
Two red algal classes, the Florideophyceae (approximately 7,100 spp.) and Bangiophyceae (approximately 193 spp.), comprise 98% of red algal diversity in marine and freshwater habitats. These two classes form well-supported monophyletic groups in most phylogenetic analyses. Nonetheless, the interordinal relationships remain largely unresolved, in particular in the largest subclass Rhodymeniophycidae that includes 70% of all species. To elucidate red algal phylogenetic relationships and study organelle evolution, we determined the sequence of 11 mitochondrial genomes (mtDNA) from 5 florideophycean subclasses. These mtDNAs were combined with existing data, resulting in a database of 25 florideophytes and 12 bangiophytes (including cyanidiophycean species). A concatenated alignment of mt proteins was used to resolve ordinal relationships in the Rhodymeniophycidae. Red algal mtDNA genome comparisons showed 47 instances of gene rearrangement including 12 that distinguish Bangiophyceae from Hildenbrandiophycidae, and 5 that distinguish Hildenbrandiophycidae from Nemaliophycidae. These organelle data support a rapid radiation and surprisingly high conservation of mtDNA gene syntheny among the morphologically divergent multicellular lineages of Rhodymeniophycidae. In contrast, we find extensive mitochondrial gene rearrangements when comparing Bangiophyceae and Florideophyceae and multiple examples of gene loss among the different red algal lineages.
Keywords: Florideophyceae, mitochondrial genome, ordinal relationship, red algal evolution, rhodophytes, Rhodymeniophycidae
Introduction
Mitochondria (mt) supply chemical energy to cells and have a long evolutionary history that extends to the origin of eukaryotes (Gray et al. 1999; Burger et al. 2003; Koonin 2010). Most mt genomes are characterized by a compact structure (exceptions include plants that contain exceptionally large mt genomes) and maternal inheritance. Due to their ubiquitious distribution and conserved functions, complete mtDNA data have been used in diverse studies including population genetics, phylogenetics, and organellar genome evolution (Unseld et al. 1997; Turmel et al. 2002; Ogihara et al. 2005; Ratnasingham and Hebert 2007; Zink and Barrowclough 2008; Morin et al. 2010). For example, analysis of mtDNA data from land plants revealed gene replacements through horizontal gene transfer and endosymbiotic gene transfer to the nucleus (Adams and Palmer 2003; Bergthorsson et al. 2003, 2004). Similar to plants, Rhodophyta (red algal) mtDNA data have been used in systematic and population studies as molecular markers (Saunders 2005; Yang et al. 2008; Freshwater et al. 2010).
Red algae belong to an anciently diverged group (Supergroup Plantae or Archaeplastida) whose ancestor hosted the plastid primary endosymbiosis (Bhattacharya et al. 2004). This phylum is composed of seven classes (i.e., Bangiophyceae, Compsopogonophyceae, Cyanidiophyceae, Florideophyceae, Porphyridiophyceae, Rhodellophyceae, and Stylonematophyceae) with the most species-rich class Florideophyceae containing the subclasses Hildenbrandiophycidae, Nemaliophycidae, Corallinophycidae, Ahnfeltiophycidae, and Rhodymeniophycidae (Saunders and Hommersand 2004; Yoon et al. 2006; Le Gall and Saunders 2007; Le Gall et al. 2010). A putative member of the Bangiophyceae played a central role in the eukaryote tree of life as a donor through a single or more likely multiple secondary plastid endosymbioses that gave rise to chlorophyll-c containing groups, such as diatoms, dinoflagellates, haptophytes, and cryptophytes (Yoon et al. 2004; Chan et al. 2011; Bhattacharya et al. 2013). Despite the importance of red algae in eukaryote evolution, only a few representative genomes (nuclear and organellar) have been characterized thus far (Ohta et al. 2003; Hancock et al. 2010; Smith et al. 2012; Janouškovec et al. 2013). Although the complete mtDNA from 26 red algal species has been determined (two Cyanidiophyceae [Leblanc et al. 1995; Jain et al. 2015], ten Bangiophyceae [Burger et al. 1999; Mao et al. 2012; Smith et al. 2012; Hwang et al. 2013; Hughey et al. 2014; Kong et al. 2014; Silva and Hughey 2014; Bangia fuscopurpurea, Bi G, Cao M, Mao Y, unpublished data, NC_026905], and 14 Florideophyceae species [Leblanc et al. 1995; Hancock et al. 2010; Zhang et al. 2012; Campbell et al. 2014; DePriest et al. 2014; Kim, Yang, Yi, et al. 2014; Kim, Yang, Boo, et al. 2014; Tablizo and Lluisma 2014; Yang, Kim, Boo, et al. 2014; Yang, Kim KM, Kim SY, et al. 2014; Kim et al. 2015; Lee et al. 2015]), the broader evolutionary history of this genome has been challenging to reconstruct because of insufficient taxon sampling and the high level of structural variation seen in the available mt genomes. Apart from a recent report of the corallinophycidean (nongeniculate) species Sporolithon durum (Kim et al. 2015), the remaining mt genome data are primarily from the Rhodymeniophycidae.
Understanding mtDNA evolution in Rhodophyta, in particular as it relates to the sister taxa Bangiophyceae and Florideophyceae that comprise greater than 98% of red algal diversity (Guiry MD and Guiry GM 2015) is of great interest to understand mitochondrial evolution in general. The Bangiophyceae contains approximately 193 reported species, all with simple thalli, including the economically important Porphyra and Pyropia (source of sushi wrap, kim or nori) from which mtDNA from ten species has been determined (Mao et al. 2012; Smith et al. 2012; Hwang et al. 2013; Hughey et al. 2014). The Florideophyceae is the largest class of red algae containing greater than 95% (6,760 spp.) of reported species diversity in the phylum Rhodophyta [approximately 7,100 species (Guiry MD and Guiry GM 2015)]. Phylogenetic relationships of the five subclasses within the Florideophyceae are well-resolved based on molecular and ultrastructural data (e.g., Saunders and Hommersand 2004; Le Gall and Saunders 2007). The Hildenbrandiophycidae, the earliest diverging subclass (which likely lacks a gametophyte stage) has a sister relationship to the remaining subclasses (Pueschel and Cole 1982; Le Gall and Saunders 2007). Of the remaining well-supported subclasses, the Rhodymeniophycidae is the largest, containing ≥5,000 species (70% of red algae). In spite of the strongly supported monophyly of the subclass, relationships among the 12 rhodymeniophycidan orders remain unresolved (Saunders et al. 2004; Le Gall and Saunders 2007; Verbruggen et al. 2010).
Here, we report 11 mtDNA sequences, 3 from Ahnfeltia plicata (Ahnfeltiophycidae), Hildenbrandia rubra (Hildenbrandiophycidae), Palmaria palmata (Nemaliophycidae), and the remaining 8 from the following rhodymeniophycidan species: Asparagopsis taxiformis (Bonnemaisoniales), Ceramium japonicum (Ceramiales), Gelidium elegans (Gelidiales), Plocamium cartilagineum (Plocamiales), Riquetophycus sp. (Peyssonneliales), Schizymenia dubyi (Nemastomatales), Schimmelmannia schousboei (Acrosymphytales), and Sebdenia flabellata (Sebdeniales).
The aims of the present work were 1) to elucidate phylogenetic relationships at higher taxonomic levels and identify major evolutionary transitions in the Rhodophyta using a database of 37 mt genomes; 2) to compare the coding sequences (CDSs) content and synteny between the Bangiophyceae and Florideophyceae, and among the five florideophycean subclasses; and 3) to determine the extent of mtDNA gene synteny and loss within the Rhodymeniophycidae.
Materials and Methods
Taxon Sampling, DNA Isolation, Genome Sequencing, and Annotation
The red algal species used in this study were chosen to sample all 5 subclasses of the Florideophyceae, as well as 12 orders of the Rhodymeniophycidae (table 1). We collected nine red algal species from the field and two species from culture collections (UTEX; http://web.biosci.utexas.edu/utex/ and CCAP; http://www.ccap.ac.uk) (supplementary table S1, Supplementary Material online). Prior to processing, fresh samples or silica gel-preserved vouchers were transported to the lab and cleaned several times using sterilized seawater under the dissecting microscope.
Table 1.
Red Algal Taxonomy and General Features of the Mitochondrial Genome Used in the Present Study
| Taxa | Size (bp) | GC Content (%) | Number of Gene |
Accession | ||||
|---|---|---|---|---|---|---|---|---|
| CDSa | tRNA | rRNA | Intron | Intronic ORF | ||||
| Cyanidiophyceae | ||||||||
| Cyanidioschyzon merolae | 32,211 | 36.5 | 35 | 25 | 3 | — | — | NC_000887 |
| Galdieria sulphuraria | 21,428 | 44.0 | 19 | 7 | 2 | — | — | NC_024666 |
| Bangiophyceae | ||||||||
| Bangia fuscopurpurea | 43,517 | 33.0 | 31 | 23 | 2 | 5 | 6 | NC_026905 |
| Porphyra purpurea | 36,753 | 33.5 | 29 | 24 | 2 | 2 | 2 | NC_002007 |
| Porphyra umbilicalis | 29,123 | 31.9 | 25 | 24 | 2 | 1 | 1 | NC_018544 |
| Pyropia fucicola | 35,035 | 32.5 | 31 | 23 | 2 | 4 | 4 | NC_024288 |
| Pyropia haitanensis | 37,023 | 30.7 | 24 | 24 | 2 | 4 | 5 | NC_017751 |
| Pyropia kanakaensis | 39,300 | 30.0 | 30 | 25 | 2 | 5 | 3 | NC_024289 |
| Pyropia perforata | 40,042 | 31.8 | 24 | 23 | 2 | 5 | 5 | KF_515974 |
| Pyropia tenera | 42,269 | 32.8 | 25 | 23 | 2 | 5 | 6 | NC_021475 |
| Pyropia yezoensis | 41,688 | 32.7 | 27 | 27 | 2 | 5 | 5 | NC_017837 |
| Wildemania schizophylla | 29,156 | 33.2 | 26 | 23 | 2 | 2 | 2 | NC_024579 |
| Average: | 37,391 | 32.2 | 27.2 | 23.9 | 2 | 3.8 | 3.9 | |
| Florideophyceae | ||||||||
| Hildenbrandiophycidae | ||||||||
| Hildenbrandia rubrab | 33,066 | 36.4 | 25 | 18 | 2 | 2 | 2 | NC_026055 |
| Nemaliophycidae | ||||||||
| Palmaria palmatab | 29,735 | 32.2 | 26 | 22 | 2 | 2 | — | NC_026056 |
| Corallinophycidae | ||||||||
| Sporolithon durum | 26,202 | 28.4 | 23 | 20 | 2 | 1 | — | NC_023454 |
| Ahnfeltiophycidae | ||||||||
| Ahnfeltia plicatab | 32,878 | 33.4 | 24 | 24 | 2 | 3 | 2 | NC_026054 |
| Rhodymeniophycidae | ||||||||
| Asparagopsis taxiformisb | 26,097 | 26.7 | 25 | 24 | 2 | 1 | — | NC_024843 |
| Ceramium japonicumb | 26,110 | 28.5 | 22 | 24 | 2 | — | — | KJ398159 |
| Chondrus crispus | 25,836 | 27.9 | 29 | 24 | 4 | 1 | — | NC_001677 |
| Gelidium elegansb | 24,922 | 29.5 | 23 | 19 | 2 | 1 | — | NC_023053 |
| Gelidium vagum | 24,901 | 30.5 | 23 | 18 | 2 | 1 | — | NC_023077 |
| Gracilaria chilensis | 26,898 | 27.6 | 25 | 25 | 2 | 1 | — | NC_026831 |
| Gracilaria salicornia | 25,727 | 28.4 | 25 | 20 | 2 | 1 | — | NC_023784 |
| Gracilariophila oryzoides | 25,161 | 28.1 | 23 | 20 | 2 | 1 | — | NC_014771 |
| Gracilariopsis andersonii | 27,036 | 28.0 | 27 | 19 | 2 | 1 | — | NC_014772 |
| Gracilariopsis chorda | 26,543 | 27.6 | 26 | 24 | 2 | 1 | — | NC_023251 |
| Gracilariopsis lemaneiformis | 25,883 | 27.5 | 26 | 21 | 2 | 1 | — | JQ071938 |
| Grateloupia angusta | 27,943 | 30.2 | 25 | 19 | 2 | 2 | 1 | NC_023094 |
| Grateloupia taiwanensis | 28,906 | 31.4 | 25 | 24 | 2 | 2 | 1 | KM999231 |
| Kappaphycus striatus | 25,242 | 30.1 | 24 | 24 | 2 | 1 | — | NC_024265 |
| Plocamiocolax pulvinata | 25,894 | 26.0 | 25 | 20 | 2 | 2 | — | NC_014773 |
| Plocamium cartilagineumb | 26,187 | 23.6 | 24 | 25 | 2 | 2 | — | KJ398160 |
| Rhodymenia pseudopalmata | 26,166 | 29.5 | 24 | 21 | 2 | 1 | — | NC_023252 |
| Riquetophycus sp.b | 26,261 | 25.7 | 24 | 22 | 2 | 1 | — | KJ398161 |
| Schimmelmannia schousboeib | 25,816 | 26.7 | 23 | 21 | 2 | 1 | — | KJ398162 |
| Schizymenia dubyib | 26,347 | 25.8 | 25 | 24 | 2 | 1 | — | KJ398163 |
| Sebdenia flabellatab | 25,887 | 28.5 | 23 | 21 | 2 | 1 | — | KJ398164 |
| Average: | 26,866 | 28.7 | 24.6 | 21.7 | 2.1 | 1.3 | 0.08 | |
aNumber of protein-coding genes exclude intronic-orf.
bNewly determined mitochondrial genomes.
Total genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). We used the Ion Torrent Personal Genome Machine (PGM; Life Technologies, San Francisco, CA) to do shotgun DNA sequencing. About 100 ng of total DNA was used for library preparation (200 bp read) based on the manufacturer’s instructions. One or two independent libraries of each species were used for sequencing (supplementary table S2, Supplementary Material online). The high quality (Q20) data obtained from each species were assembled using MIRA (Chevreux et al. 1999) under default settings that were incorporated into the Ion Torrent PGM and the CLC de novo assembler under default settings in the CLC Genomics Workbench v.5.5.1 (CLC Bio, Aarhus, Denmark). The Riquetophycus sp. sample differed from the other samples in that the library was constructed and subsequently sequenced using the Illumina platform by Eureka Genomics (Hercules, CA). In this case, one paired-end, nonindexed library was constructed from the DNA of one vegetative individual (2 µg). Sequencing consisted of one lane, with a paired end run of 51 cycles. The resulting reads were assembled as above using the CLC de novo assembler. Contigs were scanned for organelle sequences using local Basic Local Alignment Search Tool (BLAST) (Altschul et al. 1997) with red algal mitochondrial genomes as search queries. The mitochondrial contigs were then used for scaffolding using Geneious Pro v.5.3.6 (http://www.geneious.com), followed by “read mapping to reference” implemented in the CLC Genomics Workbench. The average coverage of mitochondrial consensus sequences was higher than 74× for all samples (supplementary table S2, Supplementary Material online). Sanger sequencing was used for gap-filling with each mtDNA sample to produce complete genomes (supplementary fig. S1, Supplementary Material online).
Open-reading frame (ORF) finding and gene prediction were done using BLAST (BLASTn and BLASTx), GeneMarkS (Besemer et al. 2001), and Geneious Pro. Large and small subunits of ribosomal RNA (rRNA) were identified using BLASTn with published red algal rRNAs as queries, whereas transfer RNAs (tRNA) were identified using the tRNAscan-SE Search Server (http://lowelab.ucsc.edu/tRNAscan-SE/; Schattner et al. 2005). All protein-coding, rRNA, and tRNA genes, and intron boundaries within genes in the novel mitochondrial genomes were confirmed by multiple sequence alignment with published red algal data using the T-Coffee package (http://www.tcoffee.org; Di Tommaso et al. 2011). Novel mt genome data were deposited in GenBank (table 1).
Phylogenetic Analyses
To reduce tree artifacts due to high DNA divergence, amino acid sequences were used to reconstruct phylogenies. The inferred amino acid sequences of every protein-coding gene from 37 mtDNAs were aligned using the T-Coffee package (under the default setting of -mode fmcoffee) and then refined manually. This concatenated alignment comprised 6,436 amino acids (data set c24p, 24 CDS; gene no. 1–24 in supplementary table S3, Supplementary Material online). The alignment and tree files are available from TreeBase (http://treebase.org/treebase-web/search/study/summary.html?id=15150&x-access-code=7bdd870b7e5e820b3737098d36e3cec4&agreement=ok), the RedToL website (dbdata.rutgers.edu/redseq/), and from the corresponding author upon request.
The phylogeny and statistical support for monophyletic groups were inferred using the maximum-likelihood (ML and ML bootstrap analysis) method with RAxML v.8.1.17 (Stamatakis 2006) under the specified models. We used the best fit independent amino acid model (amino acid substitution matrix with empirical frequencies [F] + gamma distributed rate heterogeneity [G]) for each gene partition (24 genes) of the concatenated data set to reduce the heterogeneity effect among genes on the phylogeny. The specific amino acid substitution matrix for each partition was selected by using the protein model selection procedure provided in the RAxML program website (http://sco.h-its.org/exelixis/resource/download/software/ProteinModelSelection.pl). This script automatically determines the best-scoring model from all available (18 amino acid substitution matrices with/without amino acid frequencies estimation) to use with RAxML. The best-fit models for the genes were as follows: MTZOA for atp6 (ATP synthase F0 subunit 6), atp9, cob (apocytochrome b), cox2 (cytochrome c oxidase subunit 2), cox3, nad1 (NADH dehydrogenase subunit 1), nad2, nad3, nad4, nad5, JTTDCMUT for atp8, rpl16 (ribosomal protein subunit L16), rpl20, rps3 (ribosomal protein subunit S3), rps11, rps12, CPREV for cox1, nad4L, nad6, sdh3 (succinate:cytochrome c oxidoreductase subunit 3), FLU for sdh2, and JTT for sdh4, secY, ymf39. The independent evolutionary models for the partitions of each gene were unlinked by “-q” options. The “-f a” option was used in RAxML for the simultaneous search for the best likelihood tree and rapid bootstrap analysis with “-# 1,000” (1,000 bootstrap replications) with default options of “-i” (automatically optimized SPR branch rearrangement for heuristic search) and “-c” (25 distinct rate categories).
To test concordance of monophyletic groups between different data sets, we prepared 23 sub-data sets from the c24p (table S3) based on three criteria. First, based on the mean amino acid (AA) P-distance of each gene alignment that was calculated with PAUP* v4.0b10 (Swofford 2002), 24 CDSs were sorted by mean values (ranging from 0.046 [atp9] to 0.621 [rps3]), and then the genes with high mean P-distances were incrementally removed for the sub-data sets by 0.05 intervals (PV1–8). Second, 24 CDSs were ordered by AA length of gene (ranging from 156 [atp8] to 684 [nad5]), and the genes were incrementally removed for the sub-data sets by 50 AA length intervals (PL1-8). Third, 24 CDSs were sorted by translation products as listed in supplementary table S3, Supplementary Material online. The electron transport, oxidative phosphorylation protein (PE), and ribosomal protein (PR) genes were grouped as sub-data sets that include PE (atp-cob-cox-nad-sdh-secY-ymf39), PE1-5 (atp6-9, cob-cox1-3, nad1-6, sdh2-4, and secY-ymf39), and PR (rpl-rps). We compared the ML bootstrap values among the full c24p and 23 sub-data sets (100 resampling for bootstrap analysis; supplementary table S2 and fig. S2, Supplementary Material online).
Bayesian Inference (BI) was done using MrBayes v.3.2.4 (Ronquist and Huelsenbeck 2003) using the best-fit model for the data set of the c24p. Preliminary searches under the mixed aamodel (prset aamodel=mixed;) suggested the JONES amino acid substitution model as the best-fit for BI. The JONES + G model was used as a fixed model [prset aamodel=fixed(jones)] for all partitions. All model parameters were unlinked for each partition by using the program settings [prset applyto=(all) ratepr=variable; lset applyto=(all) rates=gamma ngammacat=4; unlink revmet=(all) statefreq=(all) shape=(all);]. Twenty million Metropolis-coupled Markov Chain Monte Carlo (MC3) simulations were completed with the following parameters: Two independent runs with different random start points, one cold chain and five heated chains for each run, and tree sampling at every 1,000th generation. The Bayesian Posterior Probabilities (BPP) of the monophyletic nodes were calculated using trees (total 2,002) post burn-in; that is, 10 million generations.
Alternative tree topologies were evaluated using the Approximately Unbiased (AU) and Shimodaira Hasegawa (SH) tests (Shimodaira 2002) implemented in the CONSEL v.1.20 package (Shimodaira and Hasegawa 2001). All 24-CDS combined data (c24p) were used for the paired-site test with the best phylogeny and alternative phylogenies. We selected five alternative relationships of rhodymeniophycidan orders based on sub-data set analyses that had more than 50% of nodes supported with greater than 80% maximum-likelihood bootstrap (MLB) (supplementary fig. S2, Supplementary Material online). The test was performed with 100,000 bootstrap replicates under the same amino acid model settings and partitions as used in the best ML search. We repeated the test three times to confirm the reproducibility of the analysis.
Results and Discussion
Characteristics of 11 Novel mt Genomes
We determined 11 mt genomes from representatives of the 4 florideophycean subclasses; that is, Ahnfeltiophycidae, Hildenbrandiophycidae, Nemaliophycidae, and Rhodymeniophycidae (table 1 and supplementary fig. S1, Supplementary Material online). These data were combined with existing mt genomes from 10 bangiophycean, 2 cyanidiophycean, and 14 florideophycean taxa, including 6 genomes recently determined by our group (Kim, Yang, Yi, et al. 2014; Kim, Yang, Boo, et al. 2014; Yang, Kim, Boo, et al. 2014; Yang, Kim KM, Kim SY, et al. 2014; Kim et al. 2015; Lee et al. 2015). This resulted in a database of 37 mt genomes from red algae for the phylogenetic and comparative analyses.
The newly determined mt genomes have a compact gene arrangement with 44–52 predicted genes (i.e., 22–26 protein-coding, 2 rRNAs, and 18–25 tRNAs; supplementary fig. S1, Supplementary Material online), which are shared among most red algae (Hancock et al. 2010; Zhang et al. 2012; Yang, Kim KM, Kim SY, et al. 2014). Differences were found in the absence of a few genes including atp8, nad4L, rpl20, sdh3, sdh4, secY, and ymf39, for example, the lack of atp8 in Ahnfeltia, Hildenbrandia, and Plocamium (see supplementary table S3, Supplementary Material online). The rrl (large subunit rRNA) of Ahnfeltia, Hildenbrandia, and Palmaria includes an intron (with/without an intronic ORF); the cox1 gene contains an intron with an ORF in Ahnfeltia and Hildenbrandia and without an ORF in Plocamium; and trnI in all species includes an intron except in Ceramium, Gelidium, and Hildenbrandia (supplementary fig. S1 and table S3, Supplementary Material online). The mt genomes of the Bangiophyceae were larger in size (i.e., min. = 29.1 kb, max. = 43.5 kb, average = 37.4 kb) and had a higher GC-content (min. = 30%, max. = 33.5%, average = 32.2%) than those of the Florideophyceae (size: min. = 24.9 kb, max. = 33.1 kb, average = 26.9 kb; GC-content: min. = 23.6%, max. = 36.4%, average = 28.7%). Within the Florideophyceae, H. rubra contained the largest mt genome and the highest GC content (33.1 kb, 36.4% GC; see table 1).
Mitochondrial Genome-Based Red Algal Phylogeny
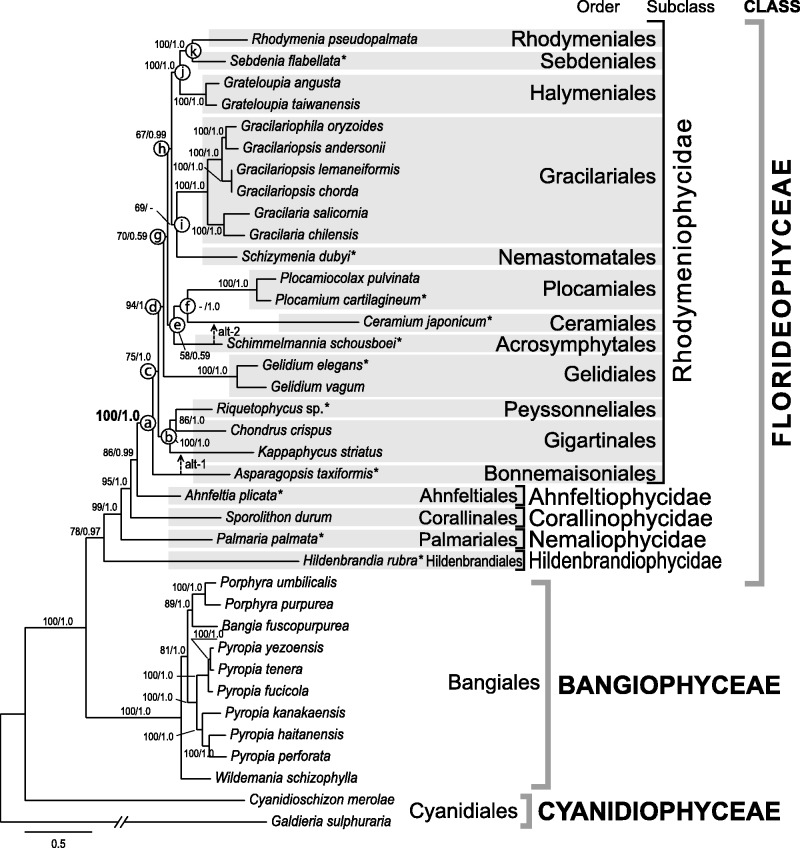
Phylogenetic analysis of concatenated mitochondrial data (c24p) from 37 red algal species strongly supports the currently accepted relationships of Rhodophyta (fig. 1). These include a close relationship of Bangiophyceae and Florideophyceae (Müller et al. 2001; Saunders and Hommersand 2004), the recognition of each of the five subclasses, and intersubclass relationships within the Florideophyceae (Le Gall and Saunders 2007; Verbruggen et al. 2010).
Fig. 1.—
Phylogeny of the red algae (Rhodophyta) based on mt genome data. Tree constructed using the ML method based on 24 concatenated genes (6,345 amino acids from 24 protein coding). The support values for each node are calculated from MLB and BPP. Asterisks after species names indicate newly determined mt genomes, followed by red algal order, subclass, and class system. Alternative tree branch position of species (alt) indicated by dot line with arrow, that is, alt-1 for Asparagopsis and alt-2 for Schimmelmannia. Alternative topologies are available in supplementary figure S3, Supplementary Material online.
The most notable finding of the phylogenetic analysis was the resolution of ordinal relationships among 21 species of Rhodymeniophycidae (fig. 1). The subclass Rhodymeniophycidae is the most morphologically diverse of the red algae and includes greater than 5,015 reported species under the current 12-order taxonomic system (Guiry MD and Guiry GM 2015). The lineages in this subclass likely underwent a rapid radiation as indicated by the mostly short and dense intermediate nodes among the orders, with only a few ordinal relationships resolved with weak to moderate statistical support (Le Gall and Saunders 2007; Verbruggen et al. 2010). Our mtDNA phylogeny nonetheless provides novel insights into the relationships of some of the rhodymeniophycidan groups. Within the subclass, the Bonnemaisoniales (A. taxiformis) diverged first and is sister to the remaining 11 orders with high support (i.e., 100% MLB and 1.0 BPP) in the c24p data set (node-a) and in the sub-data sets (15 sub-data sets support node-a with >91% MLB; supplementary fig. S2, Supplementary Material online). The Peyssonneliales (Riquetophycus sp.) and Gigartinales (Chondrus and Kappaphycus) grouped together with strong support (node-b in fig. 1, 100% MLB and 1.0 BPP) using c24p and 15 sub-data sets (>91% MLB in supplementary fig. S2, Supplementary Material online) in this restricted taxa data set of Gigartinales sensu lato. However, the monophyly of node-c (11 orders) was less well supported, that is, 75% MLB and 1.0 BPP in the c24p. The topology of sub-data sets (i.e., the sister relationship of Asparagopsis and Kappaphycus as shown in fig. 1 as alt-1) was not rejected in both AU and SH tests (supplementary fig. S3C, Supplementary Material online). A larger number of taxa are likely required to resolve the phylogenetic relationships of the Bonnemaisoniales, Gigartinales, and Peyssonneliales (Krayesky et al. 2009).
After the divergence of the Gelidiales (Gelidium spp.; 94% MLB and 1.0 BPP; node-d), two monophyletic clades (node-e and node-h) were found, that is, node-e: Plocamiales (Plocamiocolax pulvinata and P. cartilagineum) + Ceramiales (C. japonicum) (node-f) + Acrosymphytales (S. schousboei) clade (58% MLB and 0.59 BPP) and node-h: Rhodymeniales (Rhodymenia pseudopalmata) + Sebdeniales (Se. flabellata) (node-k) + Halymeniales (Grateloupia spp.) (node-j) + Gracilariales (Gracilaria spp., Gracilariopsis spp., and Gracilariophila oryzoides) + Nemastomatales (Schiz. dubyi) (node-i) clade (67% MLB and 0.99 BPP). Interordinal relationships within each clade are mostly congruent with results from prior studies (Le Gall and Saunders 2007; Le Gall et al. 2010; Verbruggen et al. 2010). However, interordinal relationships of the Acrosymphytales, Ceramiales, and Plocamiales were unresolved and an alternative topology (alt-2 in fig. 1) was not rejected by the tree topology tests (supplementary fig. S3B, Supplementary Material online). Because the Ceramiales is the largest group of red algae including greater than 2,655 species (Guiry MD and Guiry GM 2015), additional mt genome data from representative taxa of the order (such as an early branched Spyridia) may provide better resolution of this node.
Mt genome data included in this analysis resolved the relationship of 16 of the 25 florideophycean orders (Guiry MD and Guiry GM 2015). It has generally been difficult to resolve class-level relationships among red algae (i.e., Compsopogonophyceae, Porphyridiophyceae, and Stylonematophyceae) and intraclass relationships (i.e., Nemaliophycidae and Rhodymeniophycidae) using multigene data (Saunders and Hommersand 2004; Yoon et al. 2006; Le Gall and Saunders 2007; Verbruggen et al. 2010; Scott et al. 2011). However, our analysis using mtDNA data (data set c24p and 23 sub-data sets) provides moderate to strong support for intraclass relationships in the Rhodymeniophycidae. Based on these results, we expect that mt genome data from major representatives could be a promising tool to further resolve the relationships of the red algae.
Mitochondrial Genome Evolution in Red Algae
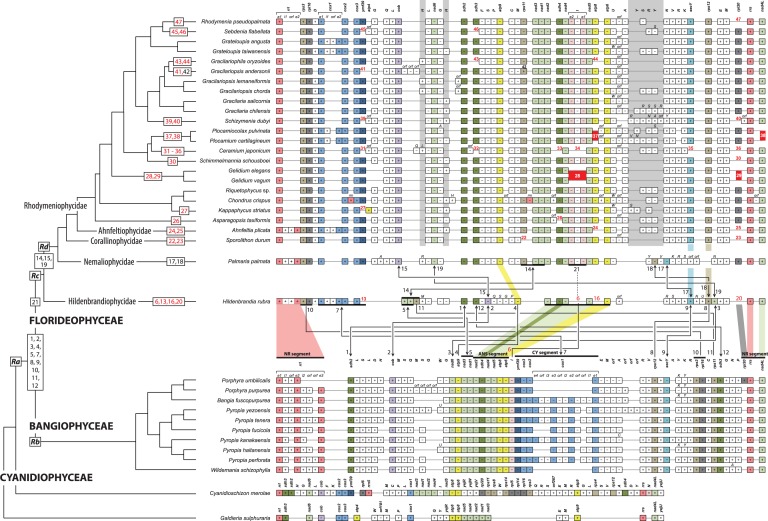
To infer mt genome evolution in the red algae, we compared gene synteny from 35 genomes (excluding the highly divergent extremophiles Cyanidioschyzon merolae and Galdieria sulphuraria) that represent two classes (Bangiophyceae and Florideophyceae), all 5 of the florideophycean subclasses, and all 12 rhodymeniophycidan orders (fig. 2). These data (supplementary fig. S1, Supplementary Material online) encompass the phylogenetic breadth of the Florideophyceae. The overall structure of mt genomes was mapped onto the best phylogeny (fig. 1) to infer the ancestral genome content, patterns of gene acquisition and loss, and types of rearrangements (i.e., numbers on fig. 2). Because of a conserved linear arrangement of three adjacent genes around mt rRNAs (rrs [small subunit rRNA]-nad4L-rrl, approximately 4,300 bp in length excluding intron regions), we used it as 5′- and 3′-anchor points of the mtDNA maps (NR segment in fig. 2). We infer that a total of 51 mitochondrial genes were present in the hypothetical last common ancestor of Bangiophyceae and Florideophyceae (Ra in fig. 2) including 24 protein-coding genes, 2 rRNAs, and 25 tRNAs (supplementary table S4, Supplementary Material online).
Fig. 2.—
Mitochondrial genome evolution in red algae. Mitochondrial gene order of 27 red algal species is illustrated along the best-supported phylogeny and associated taxonomy. Three classes (BANGIOPHYCEAE, CYANIDIOPHYCEAE, and FLORIDEOPHYCEAE) and five subclasses (Ahnfeltiophycidae, Corallinophycidae, Hildenbrandiophycidae, Nemaliophycidae, and Rhodymeniophycidae) of the red algal taxonomic system are indicated on the branches. Gene synteny is anchored by rrl (mt-rRNA large subunit) at the start and end rrs (mt-rRNA small subunit) at the end followed by nad4L, which shared an arrangement in all species (NR segment). Protein-coding and rRNA genes are abbreviated as shown in supplementary figure S1, Supplementary Material online. The tRNA genes are labeled using the one-letter code of their corresponding amino acid. The box indicates only gene existence, and box size does not correspond to gene size. The color code for genes corresponds to supplementary figure S1 and table S1, Supplementary Material online. Plus (+, forward) and minus (−, reverse) refer to the direction of gene transcription. The one letter code on the tRNA gene box indicates when alternative tRNA genes exist. Light-gray vertical boxes (three positions) point to variable tRNA gene positions in the Ahnfeltiophycidae, Corallinophycidae, and Rhodymeniophycidae. Evolutionary changes in the position of protein-coding genes are indicated with numbers (1–46) and mapped on the phylogeny; black numbers with blunt lines between genomes indicate gene rearrangement (e.g., no. 4); gene rearrangements with inversion are shown a line ended with arrowheads (e.g., no. 1); gene losses are designated with red numbers (e.g., no. 6); multigene rearrangements are marked with a thick horizontal line, that is, no. 5, 7, 10, and 14; and the trnI-intron region event (no. 21) is mapped on Palmaria and on the remaining phylogeny and corresponds to acquisition time. Strictly conserved regions are denoted with colored shadows between clades; that is, conserved ANS (nad2-sdh4-nad4-nad5-atp8-atp6), CY (ymf39-cox3-cox2-cox1), and NR (rpl20-rrs-nad4L-rrl) segments between Bangiophyceae and Florideophyceae, atp9 between Hildenbrandia and Palmaria, and rps12-secY between Hildenbrandia and Sporolithon.
The gene content of Bangiophyceae and Florideophyceae is nearly identical in terms of protein-coding genes (supplementary table S3, Supplementary Material online). However, two major syntenic differences involving inversion/translocation events were found between Bangiophyceae (e.g., Porphyra) and Hildenbrandiophycidae (Hildenbrandia) (see no. 1–12 in fig. 2) and between Hildenbrandiophycidae and Nemaliophycidae (Palmaria) (see no. 14, 15, 17–19). The majority of rearrangements occurred through gene inversions (12 inversions of 17 rearrangements, fig. 2) between Bangiophyceae and Hildenbrandiophycidae, and between Hildenbrandiophycidae and Nemaliophycidae. Rearrangements of a single gene were more frequent than simultaneous multigene rearrangements (i.e., more than two genes involved; i.e., genes no. 5, 7, 10, and 14).
The trnI gene (without intron; no. 6) exists between two large conserved regions, that is, nad2-sdh4-nad4-nad5-atp8-atp6 (ANS segment; approximately 6,500 bp) and ymf39-cox3-cox2-cox1 (no. 7 CY segment; approximately 4,000 bp in length when the intron and intronic ORF are excluded) in the bangiophycean lineage. In the Florideophyceae, the CY segment is inverted (5′-cox1-cox2-cox3-ymf39-3′), whereas the ANS segment is retained with the trnI-intron in all species (no. 21) but not in Hildenbrandia (no. 6; 5 bp intergenic region between nad4 and nad5), Gelidium (no. 28), and Ceramium (no. 34). This indicates that the trnI gene was present in the common ancestor of Bangiophyceae and Florideophyceae (Ra in fig. 2, supplementary table S4, Supplementary Material online). In the Bangiophyceae, the ANS and CY segments were present in all bangiophycean species with/without intron and intronic-ORF in cox1. However, in the Florideophyceae, the ANS segments acquired an intron in trnI and a reduced intron and intronic-ORF in cox1 of the CY segment. The trnI has gained an intron and it is inserted in the ANS segment (i.e., between nad4 and nad5) in the florideophycean ancestor (Rc). Thus, the trnI-intron was lost independently in Hildenbrandia, Gelidium, and Ceramium. This trnI-mediated intron acquisition is the first report of such an event in red algae; however, acquisition of heterogeneous DNA (protein-coding genes and introns) has been reported in the Viridiplantae, that is, in green algae and land plants (Palmer et al. 2000; Bergthorsson et al. 2004; Smith et al. 2013).
Since the split of the hildenbrandiophycidan ancestor (i.e., node Rd in fig. 2), gene synteny has been highly conserved, with changes only occurring in tRNAs and in intronic regions. Lineage-specific CDS gene losses were found in florideophycean species (red numbers in fig. 2). For example, there were losses of atp8 (no. 16), rpl20 (no. 20), trnI (no. 6), and ymf39 (no. 13) in Hildenbrandia, whereas losses of rps11 (no. 22) and rpl20 (no. 23) occurred in Sporolithon. In contrast to the conserved CDS gene content (except the rps11 inversion in Gracilariopsis andersonii; no. 42), there are several types of variation in tRNAs (i.e., gene duplication and insertion/deletion, see fig. 2 and supplementary table S3, Supplementary Material online) not only between the subclasses but also within the Rhodymeniophycidae in trnH (H—H gray regions around trnL-nad6-trnG) and/or trnY and one of each trnR and trnS (Y-S-R-Y gray region between trnA and trnN).
In this study, the new florideophycean mt genome data surprisingly show that overall gene synteny is conserved among subclasses, except for the Hildenbrandiophycidae. Based on the fossil record and molecular clock studies, the florideophycean ancestor (Rc) originated approximately 900 Ma and the nemaliophycidan ancestor (Rd) appeared at least 550 Ma (Corallinophycidae fossils) (Butterfield 2001; Xiao et al. 2004; Yoon et al. 2004). Despite the long evolutionary history of the florideophycean red algae, most are characterized by a highly conserved mtDNA synteny (fig. 2).
Reduction of Protein-Coding Genes
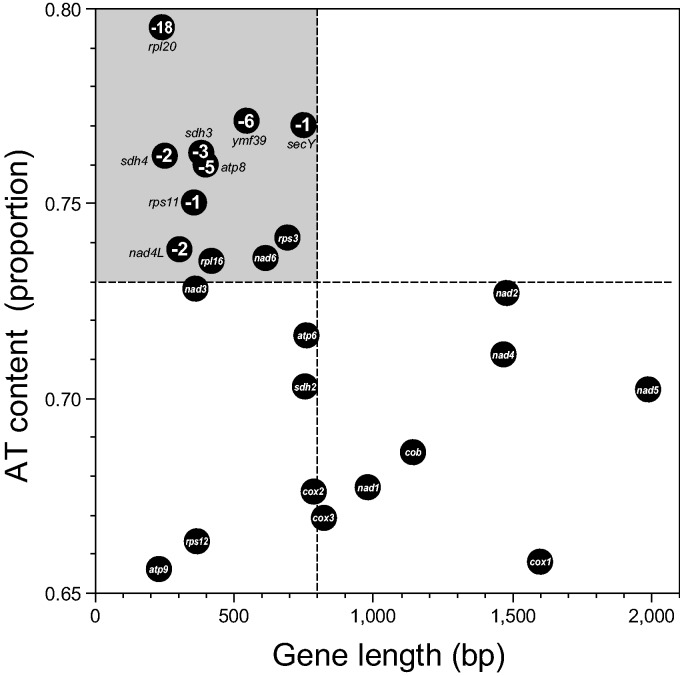
Based on our current collection of mt genomes, at least 51 genes (excluding hypothetical proteins) were present in the common ancestor (Ra) of Bangiophyceae and Florideophyceae (supplementary table S4, Supplementary Material online). We inferred a total of 29 lineage-specific gene losses during red algal evolution (no. 6, 13, 16, 20, 22–41, 43–47 in fig. 2). We marked these gene losses on the map of percent AT-content versus gene length, which shows a negative correlation between these values (fig. 3). Surprisingly, genes that were absent in some species were grouped in the top-left corner. These genes are small in size (<800 bp in mean length) and have a high AT content (>73% mean AT content). The rpl20 (relatively short with the highest AT content) was lost in 18 species among 35 investigated red algae (except Cyanidiophyceae), and this was followed by gene losses of ymf39 (6 species), atp8 (5 species), sdh3 (3 species), sdh4 and nad4L (2 species), and rps11 and secY (1 species). Gene loss appears however to be uncorrelated with phylogenetic relationships (see, red number on fig. 2). For example, within the strongly supported clade of the Halymeniales, Sebdeniales, and Rhodymeniales (100% MLB), rpl20 was lost only in R. pseudopalmata (no. 47), whereas it was retained in only three species. Interestingly, 9/10 bangiophycean species lost the rpl20 gene, except for Porphyra umbilicalis. It is reasonable to infer the rpl20 data as representing four independent losses in Porphyra purpurea, B. fuscopurpurea, the ancestor of six Pyropia species, and Wildemania schizophylla, respectively, as opposed to one loss in the ancestor of the Bangiophyceae (Rb) with a regain of rpl20 because it is extremely improbable to regain a gene in the correct reading frame after its loss.
Fig. 3.—
Protein-coding gene size reduction in red algal mitochondrial genomes. Correlation of AT contents (proportion) and sequence length (bp) of genes are illustrated with the minus number of species for each gene loss in bangiophycean and florideophycean red algae. Dark-gray circles with no number indicate that the gene is found in all taxa (35 species). The CDS of less than 800 bp in length and higher than 73% of AT content tend to be those lost.
There are three genes located near the boundary of the gene loss; that is, nad6, rps3 and rpl16, which are small in size (612, 691, and 419 bp mean length, respectively) and have a high AT-content (73.6%, 74.1%, and 73.5%, respectively; fig. 3) that are retained in all 35 bangio-florideophycean red algal mt genomes. Based on the absence of the ribosomal and succinate dehydrogenease genes (rpl, rps, and sdh) in land plants (Adams et al. 2002; Notsu et al. 2002; Adams and Palmer 2003), and the trend of gene loss in red algae, rpl16 and rps3 may be candidates for future gene loss (or perhaps already lost in some yet to be studied rhodophytes) than nad6. Meanwhile, many lost genes (rpl20, rps11, sdh4, secY, and ymf39) and candidates for loss (rpl16, rps2, rps3, and sdh2) were lost in G. sulphuraria but retained in Cy. merolae (Cyanidiophyceae; fig. 2, supplementary table S3, Supplementary Material online). The pattern that high AT-content genes are more frequently lost than low AT-content ones in red algae is an intriguing one, for which we currently have no explanation for the mechanisms. It may be a phenomenon restricted to this phylum, and further study is needed.
The lost genes have most likely been replaced by copies that were transferred to the nucleus (Lang et al. 1999). In red algae, G. sulphuraria is the only species with completed nuclear (Schönknecht et al. 2013) and mt genomes, and the mt genome has eight CDS losses (fig. 2, supplementary table S3, Supplementary Material online). We found one of the eight CDSs, mitochondrial-derived sdh2 gene, in the nuclear genome of G. sulphuraria (XM_005707240.1|G. sulphuraria succinate dehydrogenase [ubiquinone] iron-sulfur protein [Gasu_20110] mRNA, complete CDS, 275 AA; E value = 5e-105, 93% Query coverage, 59% identity) by using tBLASTn search with sdh2 of Cy. merolae (gi|8954361|ref|NP_059350.1; 265 AA) on the National Center for Biotechnology Information. However, other genes showed matches with corresponding plastid encoded genes (rpl16, rps3, rps11, rps12) or very short regions of similarity to nuclear genes (rpl20, shd4, ymf39). Additional genomes from Florideophyceae and mt-driven gene transfer to the nucleus are needed to test the genome reduction and gene transfer scenario presented here.
Among all reported mt genomes shown in figure 2, group II introns are present in all species except Ceramium, Cyanidioschyzon, Galdieria, and Gelidium (Dai et al. 2003; Mao et al. 2012; Smith et al. 2012; Yang, Kim, Boo, et al. 2014). It is interesting, however, that only three genes (rrl, cox1, and trnI) contain introns with or without intronic-ORFs (Leblanc et al. 1995; Hancock et al. 2010; Hwang et al. 2013; Kim, Yang, Boo, et al. 2014; Yang, Kim, Boo, et al. 2014). In the bangiophycean species (Bangia, Porphyra, Pyropia, and Wildemania), one or two copies of rrl-introns are present (Smith et al. 2012; Hwang et al. 2013), whereas these tend to be lost in most florideophycean taxa (i.e., only present in Hildenbrandia, Palmaria, and Ahnfeltia). A similar pattern is found for the cox1 intron, where 1–3 copies are present in bangiophycean taxa but sporadically present in the Florideophyceae (supplementary table S3, Supplementary Material online). In contrast, the florideophycean lineage acquired the trnI-intron without the intronic-ORF after divergence of the Hildenbrandiophycidae.
The distribution of introns in red algae together with the 11 novel mt genomes suggests that the majority of red algal introns (rrl and cox1) originated prior to the split of the two major classes (Ra in fig. 2; the last common ancestor of Bangiophyceae and Florideophyceae). Subsequent intronic-ORF loss resulted in pseudogenization of intronic-ORFs, followed by intron loss. Intron gain, however, may occur with an ORF-lacking intron, as seen for the trnI-intron, which is an apparent case of a recent intron gain.
Conclusions
Our study elucidates mt genome evolution in the Florideophyceae through analysis of this genome in Ahnfeltia, Hildenbrandia, Palmaria, and diverse rhodymeniophycidan species. The phylogeny based on concatenated amino acid mt genome data supports the current taxonomic system for Bangiophyceae and Florideophyceae. We demonstrate that all Florideophyceae mt genomes have compact and conserved architectures but they also contain several unique features, such as gene rearrangements that discriminate lineages and the gain of group II introns in trnI genes. Comparative analyses suggest that the mtDNA in Florideophyceae (except the Hildenbrandiophycidae), although representing a wide variety of morphologically divergent taxa, is surprisingly highly conserved with little variation in gene composition and with shared synteny across the class. This result is consistent with a rapid radiation of the class followed by strong constraints on mitochondrial gene content and genome evolution.
Supplementary Material
Supplementary figures S1–S3 and tables S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank to Ms Sun Jeong Lim for data analysis and Dr Ju Yun Bae and Dr Joon Park (Life Technologies) for technical support. This work was supported by grants from the Basic Science Research Program through the National Research Foundation of Korea (NRF) founded by the Ministry Science, ICT and Future Planning (NRF-2014R1A1A1006512) to E.C.Y., the Korean Rural Development Administration Next-generation BioGreen21 (PJ011121), the National Research Foundation of Korea (MEST: 2014R1A2A2A01003588), and Marine Biotechnology Program (PJT200620) funded by Ministry of Oceans and Fisheries, Korea to H.S.Y., National Science Foundation RedToL (DEB 0936884, 0936855, 1317114) to D.B., S.F., and H.S.Y., Korea Research Foundation (2012-0704) to S.M.B., a Marine and Extreme Genome Research Center Program grant of the Ministry of Oceans and Fisheries to J.H.L., a GoMRI-I (CWC) award to S.F., and NIWA Core funding to W.A.N. The authors declare that they have no competing interests.
Literature Cited
- Adams KL, Palmer JD. 2003. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol. 29:380–395. [DOI] [PubMed] [Google Scholar]
- Adams KL, Qiu Y-L, Stoutemyer M, Palmer JD. 2002. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci U S A. 99:9905–9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Adams KL, Thomason B, Palmer JD. 2003. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 424:197–212. [DOI] [PubMed] [Google Scholar]
- Bergthorsson U, Richardson AO, Young GJ, Goertzen LR, Palmer JD. 2004. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella . Proc Natl Acad Sci U S A. 101:17747–17752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29:2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Yoon HS, Hackett JD. 2004. Photosynthetic eukaryotes unite: endosymbiosis connects the dots. BioEssays 26:50–60. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, et al. 2013. Genome of the red alga Porphyridium purpureum. Nat Commun. 4:1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Gray MW, Lang BF. 2003. Mitochondrial genomes: anything goes. Trends Genet. 19:709–716. [DOI] [PubMed] [Google Scholar]
- Burger G, Saint-Louis D, Gary MW, Lang BF. 1999. Complete sequence of the mitochondrial DNA of the red alga Porphyra purpurea: cyanobacterial introns and shared ancestry of red and green algae. Plant Cell 11:1675–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield NJ. 2001. Paleobiology of the late Mesoproterozoic (ca 1200 Ma) hunting formation, Somerset Island, Arctic Canada. Precambrian Res. 111:235–256. [Google Scholar]
- Campbell MA, Presting G, Bennet MS, Sherwood AR. 2014. Highly conserved organellar genomes in the Gracilariales as inferred using new data from the Hawaiian invasive alga Gracilaria salicornia (Rhodophyta). Phycologia 53:109–116. [Google Scholar]
- Chan CX, et al. 2011. Red and green algal monophyly and extensive gene sharing found in a rich repertoire of red algal genes. Curr Biol. 21:328–333. [DOI] [PubMed] [Google Scholar]
- Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information. Vol. 99. Comput Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB). p. 45-56. [Google Scholar]
- Dai L, Toor N, Olson R, Keeping A, Zimmerly S. 2003. Database for mobile group II introns. Nucleic Acids Res. 31:424–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePriest M, Bhattacharya D, Lopez-Bautista J. 2014. The mitochondrial genome of Grateloupia taiwanensis (Halymeniaceae, Rhodophyta) and comparative mitochondrial genomics of red algae. Biol Bull. 227:191–200. [DOI] [PubMed] [Google Scholar]
- Di Tommaso P, et al. 2011. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39:W13–W17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshwater DW, Tudor K, O’Shaughnessy K, Wysor B. 2010. DNA barcoding in the red algal order Gelidiales: comparison of COI with rbcL and verification of the “barcoding gap.” Cryptogam Algol. 31:435–449. [Google Scholar]
- Gray MW, Burger G, Lang BF. 1999. Mitochondrial evolution. Science 283:1476–1481. [DOI] [PubMed] [Google Scholar]
- Guiry MD, Guiry GM. 2015. AlgaeBase world-wide electronic publication. Galway (Ireland): National University of Ireland. Available from: http://www.algaebase.org; searched on May 2015. [Google Scholar]
- Hancock L, Goff L, Lane C. 2010. Red algae lose key mitochondrial genes in response to becoming parasitic. Genome Biol Evol. 2:897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughey JR, et al. 2014. Minimally destructive sampling of type specimens of Pyropia (Bangiales, Rhodophyta) recovers complete plastid and mitochondrial genomes. Sci Rep. 4:5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang MS, Kim S-O, Ha D-S, Lee JE, Lee S-R. 2013. Complete sequence and genetic features of the mitochondrial genome of Pyropia tenera (Rhodophyta). Plant Biotechnol Rep. 7:435–443. [Google Scholar]
- Jain K, et al. 2015. Extreme features of the Galdieria sulphuraria organellar genomes: a consequence of polyextremophily? Genome Biol Evol. 7:367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouškovec J, et al. 2013. Evolution of red algal plastid genomes: ancient architectures, introns, horizontal gene transfer, and taxonomic utility of plastid markers. PLoS One 8:e59001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KM, Yang EC, Kim JH, Nelson WA, Yoon HS. 2015. Complete mitochondrial genome of rhodolith, Sporolithon durum (Sporolithales, Rhodophyta). Mitochondrial DNA 26:155–156. [DOI] [PubMed] [Google Scholar]
- Kim KM, Yang EC, Yi G, Yoon HS. 2014. Complete mitochondrial genome of sublittoral macroalga Rhodymenia pseudopalmata (Rhodymeniales, Rhodophyta). Mitochondrial DNA. 25:273–274. [DOI] [PubMed] [Google Scholar]
- Kim SY, Yang EC, Boo SM, Yoon HS. 2014. Complete mitochondrial genome of the marine red alga Grateloupia angusta (Halymeniales). Mitochondrial DNA 25:269–270. [DOI] [PubMed] [Google Scholar]
- Kong F, Sun P, Cao M, Wang L, Mao Y. 2014. Complete mitochondrial genome of Pyropia yezoensis: reasserting the revision of genus Porphyra. Mitochondrial DNA 25:335–336. [DOI] [PubMed] [Google Scholar]
- Koonin EV. 2010. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 11:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krayesky DM, Norris JN, Gabrielson PW, Gabriel D, Fredericq S. 2009. A new order of red algae based on the Peyssonneliaceae, with an evaluation of the ordinal classification of the Florideophyceae (Rhodophyta). Proc Biol Soc Wash. 122:364–391. [Google Scholar]
- Lang BF, Gray MW, Burger G. 1999. Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet. 33:351–397. [DOI] [PubMed] [Google Scholar]
- Le Gall L, Payri CE, Bittner L, Saunders GW. 2010. Multigene phylogenetic analyses support recognition of the Sporolithales ord . nov. Mol Phylogenet Evol. 54:302–305. [DOI] [PubMed] [Google Scholar]
- Le Gall L, Saunders GW. 2007. A nuclear phylogeny of the Florideophyceae (Rhodophyta) inferred from combined EF2, small subunit and large subunit ribosomal DNA: establishing the new red algal subclass Corallinophycidae. Mol Phylogenet Evol. 43:1118–1130. [DOI] [PubMed] [Google Scholar]
- Leblanc C, et al. 1995. Complete sequence of the mitochondrial DNA of the rhodophyte Chondrus crispus (Gigartinales): gene content and genome organization. J Mol Biol. 250:484–495. [DOI] [PubMed] [Google Scholar]
- Lee JM, Boo SM, Mansilla A, Yoon HS. 2015. Unique repeat and plasmid sequences in the mitochondrial genome of Gracilaria chilensis (Gracilariales, Rhodophyta). Phycologia 54:20–23. [Google Scholar]
- Mao Y, Zhang B, Kong F, Wang L. 2012. The complete mitochondrial genome of Pyropia haitanensis Chang et Zheng. Mitochondrial DNA 23:344–346. [DOI] [PubMed] [Google Scholar]
- Morin PA, et al. 2010. Complete mitochondrial genome phylogeographic analysis of killer whales (Orcinus orca) indicates multiple species. Genome Res. 20:908–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller KM, Oliveira MC, Sheath RG, Bhattacharya D. 2001. Ribosomal DNA phylogeny of the Bangiophycidae (Rhodophyta) and the origin of secondary plastids. Am J Bot. 88:1390–1400. [PubMed] [Google Scholar]
- Notsu Y, et al. 2002. The complete sequence of the rice (Oryza sativa L) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics. 268:434–445. [DOI] [PubMed] [Google Scholar]
- Ogihara Y, et al. 2005. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acid Res. 33:6235–6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta N, et al. 2003. Complete sequence and analysis of the plastid genome of the unicellular red alga Cyanidioschyzon merolae . DNA Res. 10:67–77. [DOI] [PubMed] [Google Scholar]
- Palmer JD, et al. 2000. Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates. Proc Natl Acad Sci U S A. 97:6960–6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pueschel CM, Cole KM. 1982. Rhodophycean pit plugs: an ultrastructural survey with taxonomic implications. Am J Bot. 69:703–720. [Google Scholar]
- Ratnasingham S, Hebert PD. 2007. BOLD: the barcode of life data systems ( http://wwwbarcodinglifeorg). Mol Ecol Notes. 7:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under the mixed models. Bioinformatics 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- Saunders GW. 2005. Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Philos Trans R Soc Lond B Biol Sci. 360:1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders GW, Chiovitti A, Kraft GT. 2004. Small-subunit rRNA gene sequences from representative of selected families of the Gigartinales and Rhodymeniales (Rhodophyta) 3. Recognizing the Gigartinales sensu stricto. Can J Bot. 82:43–74. [Google Scholar]
- Saunders GW, Hommersand MH. 2004. Assessing red algal supraordinal diversity and taxonomy in the contest of contemporary systematic data. Am J Bot. 91:1494–1507. [DOI] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönknecht G, et al. 2013. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339:1207–1210. [DOI] [PubMed] [Google Scholar]
- Scott J, et al. 2011. On the genus Rhodella, the emended orders Dixoniellales and Rhodellales with a new order Glaucosphaerales (Rhodellophyceae, Rhodophyta). Algae 26:277–288. [Google Scholar]
- Shimodaira H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 51:492–508. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246–1247. [DOI] [PubMed] [Google Scholar]
- Silva MY, Hughey JR. 2014. Complete mitochondrial genome of the holotype specimen of Wildemania schizophylla (Bangiales: Rhodophyta). Mitochondrial DNA. Advance Access published June 18, 2014, doi:10.3109/19401736.2014.926524 [DOI] [PubMed] [Google Scholar]
- Smith DR, et al. 2013. Organelle genome complexity scales positively with organism size in volvocine green algae. Mol Biol Evol. 30:793–797. [DOI] [PubMed] [Google Scholar]
- Smith DR, Hua J, Lee RW, Keeling PJ. 2012. Relative rates evolution among the three genetic compartment of the red alga Porphyra differ from those of green plants and do not correlate with genome architecture. Mol Phylogenet Evol. 65:339–344. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Swofford DL. 2002. PAUP*: Phylogenetic analysis using parsimony (*and other methods). Version 4.0b10 Sunderland (MA): Sinauer Associates. [Google Scholar]
- Tablizo FA, Lluisma AO. 2014. The mitochondrial genome of the red alga Kappaphycus striatus (“Green Sacol” variety): complete nucleotide sequence, genome structure and organization, and comparative analysis. Mar Genomics. 18:155–161. [DOI] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. 2002. The chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants. Proc Natl Acad Sci U S A. 99:11275–11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unseld M, Marienfeld JR, Brandt P, Brennicke A. 1997. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet. 15:57–62. [DOI] [PubMed] [Google Scholar]
- Verbruggen H, et al. 2010. Data mining approach identifies research priorities and data requirements for resolving the red algal tree of life. BMC Evol Biol. 10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Knoll AH, Yuan X, Pueschel CM. 2004. Phosphatized multicellular algae in the Neoproterozoic Doushantuo formation, China, and the early evolution of florideophyte red algae. Am J Bot. 91:214–227. [DOI] [PubMed] [Google Scholar]
- Yang EC, et al. 2008. Mitochondrial cox1 and plastid rbcL genes of Gracilaria vermiculophylla (Gracilariaceae, Rhodophyta). J Appl Phycol. 20:161–168. [Google Scholar]
- Yang EC, Kim KM, Boo GH, et al. 2014. Complete mitochondrial genome of the agarophyte red alga Gelidium vagum (Gelidiales). Mitochondrial DNA 25:267–268. [DOI] [PubMed] [Google Scholar]
- Yang EC, Kim KM, Kim SY, Yoon HS. 2014. Complete mitochondrial genome of agar-producing red alga Gracilariopsis chorda (Gracilariales). Mitochondrial DNA 25:339–341. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. 2004. A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol. 21:809–818. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Muller KM, Sheath RG, Ott FD, Bhattacharya D. 2006. Defining the major lineages of red algae (Rhodophyta). J Phycol. 42:482–492. [Google Scholar]
- Zhang L, et al. 2012. Complete sequence of the mitochondrial DNA of the wild Gracilariopsis lemaneiformis and two mutagenic cultivated breeds (Gracilariaceae, Rhodophyta). PLoS One 7:e40241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink RM, Barrowclough GF. 2008. Mitochondrial DNA under siege in avian phylogeography. Mol Ecol. 17:2107–2121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.