Abstract
Hydrozoans are known for their complex life cycles, which can alternate between an asexually reproducing polyp stage and a sexually reproducing medusa stage. Most hydrozoan species, however, lack a free-living medusa stage and instead display a developmentally truncated form, called a medusoid or sporosac, which generally remains attached to the polyp. Although evolutionary transitions in medusa truncation and loss have been investigated phylogenetically, little is known about the genes involved in the development and loss of this life cycle stage. Here, we present a new workflow for evaluating differential expression (DE) between two species using short read Illumina RNA-seq data. Through interspecific DE analyses between two hydractiniid hydrozoans, Hydractinia symbiolongicarpus and Podocoryna carnea, we identified genes potentially involved in the developmental, functional, and morphological differences between the fully developed medusa of P. carnea and reduced sporosac of H. symbiolongicarpus. A total of 10,909 putative orthologs of H. symbiolongicarpus and P. carnea were identified from de novo assemblies of short read Illumina data. DE analysis revealed 938 of these are differentially expressed between P. carnea developing and adult medusa, when compared with H. symbiolongicarpus sporosacs, the majority of which have not been previously characterized in cnidarians. In addition, several genes with no corresponding ortholog in H. symbiolongicarpus were expressed in developing medusa of P. carnea. Results presented here show interspecific DE analyses of RNA-seq data to be a sensitive and reliable method for identifying genes and gene pathways potentially involved in morphological and life cycle differences between species.
Keywords: RNA-Seq, transcriptomics, differential expression, comparative expression, Cnidaria, Hydrozoa
Introduction
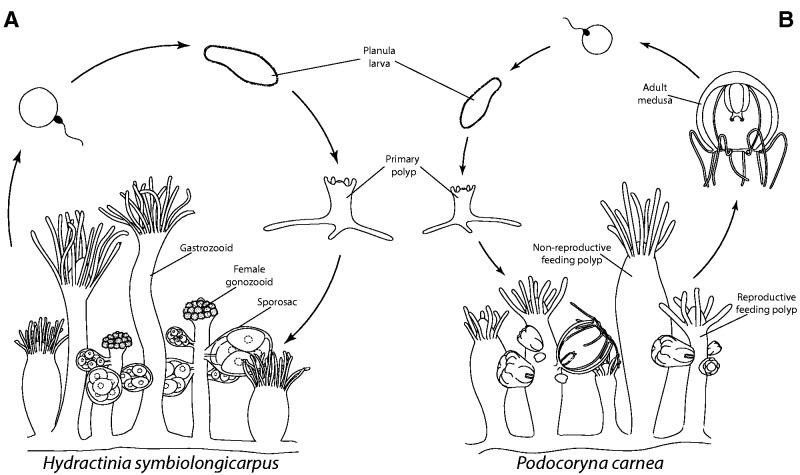
In Hydrozoa (phylum Cnidaria), many species exhibit an alternation of generations, where asexually reproducing polyps give rise to sexually reproducing jellyfish (medusae). However, across hydrozoans, there is much variation in this sexually reproducing life cycle stage. In most hydrozoan species (∼70%), development of the medusa bud (gonophore) is truncated to some degree or entirely absent (Leclère et al. 2009; Cartwright and Nawrocki 2010; Gibbons et al. 2010). In these taxa, sexual maturity is reached in a gonophore that resembles an early ontogenetic stage of medusae development. The degree of gonophore development ranges from completely reduced structures called sporosacs that lack any resemblance of the medusa (fig. 1A), to more developed forms called medusoids, that may or may not detach and swim, but lack the ability to feed (not shown), to the fully developed medusa stage that detaches from the hydroid polyp and can feed, swim, and sexually reproduce in the water column (fig. 1B).
Fig. 1.—
Illustration of hydrozoan life cycles. (A) In the life cycle of Hydractinia symbiolongicarpus, gonophores develop into sporosacs that lack all medusa features and remain attached to the colony on specialized reproductive polyps called gonozooids. Sexual reproduction occurs in the water column after the sporosacs release their gametes. Sexual reproduction results in a planula larva that eventually settles onto a suitable substrate and metamorphoses into a primary polyp. This polyp will asexually produce other polyps to form a colony and the cycle repeats. (B) Podocoryna carnea’s life cycle is similar to that of H. symbiolongicarpus except that medusae asexually bud from reproductive polyps and detach from the colony to sexually reproduce in the water column.
The evolution of this structure and its reduced forms has been a topic of investigation for the last 150 years (Allman 1864; Cornelius 1992; Cunningham and Buss 1993; Marques and Migotto 2001; Leclère et al. 2007, 2009; Miglietta et al. 2009, 2010; Cartwright and Nawrocki 2010; Miglietta and Cunningham 2012). Phylogenetic studies have revealed multiple independent losses of medusae (Cunningham and Buss 1993; Leclère et al. 2007, 2009; Cartwright and Nawrocki 2010; Miglietta and Cunningham 2012) and possibly even re-revolution (Leclère et al. 2009; Cartwright and Nawrocki 2010; Miglietta and Cunningham 2012). Although phylogenies are important for recognizing evolutionary patterns of character transitions, understanding complex patterns of character loss and possible re-gain will come from insight about their development. Specifically, maintenance of developmental regulatory pathways underlying medusae ontogeny in reduced forms could add support to arguments for medusae re-evolving in the Hydrozoa. The hydrozoan family Hydractiniidae provides an excellent system for identifying key components in medusa development and truncation, as the entire spectrum of gonophore development is exhibited within this group (Schuchert 2008). The hydractiniid species H. symbiolongicarpus and P. carnea exhibit either ends of this developmental spectrum, possessing a sporosac (fig. 1A) and medusa (fig. 1B), respectively.
Now that transcriptomes of nontraditional model systems can be readily obtained and characterized in different stages or parts of an organism (Hao et al. 2011; Siebert et al. 2011; Helm et al. 2013; Sanders et al. 2014; Schunter et al. 2014), comparing transcriptomes between species is the obvious next step. Dunn, Luo, and Wi (2013) extensively reviewed the utility of comparative expression across multiple species, as well as its challenges. Although not as abundant as intraspecific transcriptomic studies, interspecific analyses have proved illuminating on a diversity of topics (Yang and Wang 2013; Boyle et al. 2014; Pankey et al. 2014). These studies took a general approach to comparing whole transcriptomes but did not apply interspecific differential expression (DE) in an unbiased approach to identify genes potentially involved in differences between species.
Here, we present a workflow for performing DE analyses between two species from short read Illumina RNA-Seq data. Specifically, we use previously published RNA-Seq data from H. symbiolongicarpus (Sanders et al. 2014) and P. carnea (Sanders and Cartwright 2015), to identify genes and gene pathways that are potentially involved in the life cycle differences between truncated and fully developed medusae in the Hydractiniidae.
Materials and Methods
Animal Care
Transplanted colonies of P. carnea and H. symbiolongicarpus were kept on microscope slides, placed in slide racks, and kept in seawater (REEF CRYSTALS, Aquarium Systems) aquaria at room temperature (∼21 °C) with a salinity of 29 and 32 ppt, respectively. Colonies were fed 2–3-day-old nauplii of Artemia three times a week.
Transcriptome Assembly and Annotation
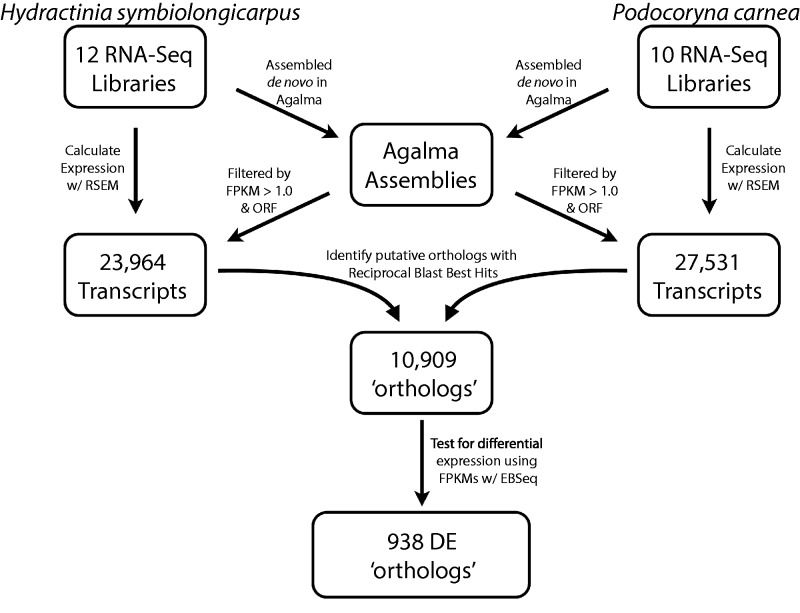
Figure 2 is a schematic of our bioinformatic pipeline for identifying differentially expressed orthologs. Raw Illumina RNAseq data for H. symbiolongicarpus and P. carnea were downloaded from the SRA archive (SRP038762 and SRP041583, respectively). Hydractinia symbiolongicarpus libraries included four replicated libraries of feeding (nonreproductive) polyps (gastrozooids), two replicated libraries of female reproductive polyps (gonozooids), two replicated libraries of male reproductive polyps (gonozooids), and four replicated libraries of defensive (nonreproductive) polyps (dactylozooids). Podocoryna carnea libraries included three replicated libraries of nonreproductive gastrozooids, four replicated libraries of female reproductive gastrozooids, and three replicated libraries of female adult medusa. These reads were pooled by species and then assembled with Trinity (Grabherr et al. 2011) through the automated bioinformatics pipeline, Agalma (Dunn, Howison, and Zapata 2013), under default settings.
Fig. 2.—
Schematic of bioinformatics workflow. Initial transcriptomes for Hydractinia symbiolongicarpus and Podocoryna carnea are assembled from 12 and 10,100 bp paired-end Illumina libraries, respectively, using the program Agalma. Each transcriptome is filtered for transcripts that meet the Transdecoder reading frame criteria (as implemented in Agalma) and have an FPKM ≥ 1.0. Expression values are estimated for these remaining transcripts from each library independently using RSEM. Orthologs are identified using one-to-one reciprocal BLAST best hits between the transdecoder protein translations of the subsetted transcriptome using BLASTp under default setting. DE analyses are performed with EBSeq using FPKM and TPM expression normalizations.
To perform gene ontology (GO) analyses, transcripts were blasted against the nr protein database using BLASTx with the “–outfmt 5” flag for xml formatted output. BLAST output was imported into BLAST2Go (Conesa et al. 2005; Götz et al. 2008) where GO mapping and annotations were performed. Conserved protein domains were also identified using with the PFAM (Punta et al. 2012) and TIGR (http://blast.jcvi.org/web-hmm/) databases using HMMER (http://hmmer.org/). The enriched GO terms and protein domains were assessed with the Fisher’s exact test and corrected for a false discovery rate (FDR) < 0.05 in R.
Ortholog Identification and DE Analysis
To identify putative orthologs, assemblies were filtered based on a minimum fragments per kilobase of transcript per million mapped reads (FPKM) value (≥1.0) and default Transdecoder (http://transdecoder.sourceforge.net/) reading frame criteria (fig. 2). An FPKM value was calculated for each transcript across all libraries used in the assemblies and were used as a means of assessing the relative coverage of each transcript. Transcripts that met both filtering criteria (filtered transcriptome) were then translated by their longest reading frame and blasted against the other filtered transcriptome using the BLASTp algorithm. One-to-one reciprocal best BLAST hits (RBBHs) with both e values > 1e-03 were treated as orthologous genes. As our study involved two closely related taxa, reciprocal BLAST best hits is an adequate means of establishing orthology and is a commonly used method (Yang and Wang 2013; Pankey et al. 2014). As the number of taxa considered increases, tree-based methods become necessary to identify orthologous genes.
Expression of orthologs was calculated with RSEM (Li and Dewey 2011) by remapping the raw reads from the individual libraries to the filtered transcriptome of the corresponding species, excluding only libraries specific to the H. symbiolongicarpus dactylozooids. RSEM calculates expression levels and computes three different expression values: expected counts, transcripts per million (TPM), and FPKM. Because fully annotated genomes were not available for both species, DE analyses were conducted between clusters of transcripts (as inferred by RSEM) as a proxy for a gene-level assessment. Separate DE analyses were performed with EBSeq (Leng et al. 2013) using the TPM and FPKM data sets. DE was not assessed using expected counts, as these do not include any normalization for library size. Results were filtered based on the inferred posterior probability that a gene was differentially expressed (PPDE; equal to one minus the FDR: 1 − FDR) for a particular expression pattern.
As the number of conditions increases, so do the number of possible expression patterns. With six conditions, there are 203 possible patterns (supplementary table S3, Supplementary Material online). To limit the results to those informative to our question, we identified 44 potentially informative expression patterns that are gonophore- and medusa specific. Transcripts marked by the remaining 159 expression patterns were ignored. Transcripts identified with a PPDE ≥ 0.95 along one of these 44 patterns (supplementary table S4, Supplementary Material online), with the highest expression observed in one of the gonophore/medusa containing conditions, were selected as candidates for further study in medusa evolution (supplementary table S5, Supplementary Material online).
Probe Synthesis and In Situ Hybridization
Sequences of transcripts listed in table 4 were identified from each assembly. The reading frames of each species copy were aligned, and primers (supplementary table S6, Supplementary Material online) were selected to encompass homologous regions of each transcript. These fragments were then amplified from cDNA, cloned using the Invitrogen TOPO-TA Cloning Kit, and DIG labeled riboprobes were synthesized from clones using the Invitrogen T7/T3 Megascript kit. In situ hybridization (ISH) protocol was adapted from Gajewsky et al. (1996). Hybridization was carried out at 50 °C for 16–18 h with a probe concentration of 0.1 ng/µl. Hybridization was detected by immunostaining with anti-DIG-Fab-AP (ROCHE) and NBT/BCIP.
Table 4.
Differentially Expressed Orthologs and Podocoryna carnea-Specific Genes Validated with ISH
| RBBH ID | Name | BLAST Hit | FPKM | TPM |
|---|---|---|---|---|
| RBBH_6358 | IF2B2 | Insulin-like growth factor 2 mRNA-binding protein 2 | ** | NS |
| RBBH_7273 | TF3C6 | General transcription factor 3C polypeptide 6 | ** | NS |
| RBBH_608 | Notch-like | Neurogenic locus notch homolog protein 1 | ** | ** |
| RBBH_8405 | KLF12 | Krueppel-like factor 12 | ** | ** |
| RBBH_2122 | PLST | Plastin-3 | ** | ** |
| RBBH_3474 | APLP | Apolipophorins | ** | ** |
| None | PDGF | Platelet-derived growth factor subunit A | NA | NA |
| None | Hox1 | Homeobox protein Hox-B1 | NA | NA |
Note.—NS, not significant; NA, not subject to DE analyses; none, ortholog not present in Hydractinia symbiolongicarpus.
**Orthologs that are significant in either data set at a PPDE = 1.00.
Molecular Phylogenetic Analyses
Cnidarian sequences belonging to homeobox, helix-loop-helix, and PDGF/VEGF gene families were mined from the nr NCBI database and subject to phylogenetic analysis as a means to quickly establish orthology with those genes in our data set. Podocoryna carnea and H. symbiolongicarpus amino acid sequences belonging to the gene families of interest were identified using the HMMER annotations, extracted from the assemblies, and subject to a clustering analysis using CD-HIT (Li and Godzik 2006; Fu et al. 2012) (under a 90% sequence similarity threshold) to remove redundant gene copies. Alignments were conducted with Mafft (Katoh et al. 2005) under the L-insi alignment algorithm. Maximum-likelihood estimates of the gene trees were then inferred using RAxML (Stamatakis et al. 2008) on the CIPRES portal (Miller et al. 2010) using the rapid bootstrapping (-f a) algorithm with 100 bootstrap replicates under the PROTGAMMA+WAG model (supplementary figs. S3 and S5, Supplementary Material online).
Results and Discussion
Transcriptome Assembly and Annotation, Enrichment Analyses, and Orthology Prediction
Raw Illumina RNA-Seq data for H. symbiolongicarpus and P. carnea were downloaded from NCBI (SRA archive no. SRP038762 and SRP041583, respectively). For H. symbiolongicarpus, these data were generated from four separate tissue sources: gastrozooids (feeding polyps), dactylozooids (defensive polyps), gonozooids (reproductive polyps) bearing male gonophores, and gonozooids bearing female gonophores (table 1). For P. carnea, data were generated from the following tissues: nonreproductive feeding polyps (gastrozooids), reproductive (medusa-budding) feeding polyps, and free-living medusae. All P. carnea data were generated from female tissues. Final assemblies, which combined data from all libraries of that species, consisted of 127,716 and 178,396 transcripts for H. symbiolongicarpus and P. carnea, respectively (table 2).
Table 1.
RNA-Seq Illumina Libraries
| Condition | No. Replicates |
|---|---|
| Hydractinia symbiolongicarpusa | |
| gastrozooid | 4 |
| Female gonozooidb | 2 |
| Male gonozooidb | 2 |
| Podocoryna carnea | |
| Nonreproductive polyp | 3 |
| Medusae-budding polypb | 4 |
| Adult medusaeb | 3 |
aThe other condition, dactylozooid, was included in the assembly but not in the DE analysis.
bConditions of interest.
Table 2.
Assembly Statistics Summary
|
Hydractinia symbiolongicarpus |
Podocoryna carnea |
|||||
|---|---|---|---|---|---|---|
| Initial | Filtered | RBBH | Initial | Filtered | RBBH | |
| No. transcripts | 127,716 | 23,964 | 10,909 | 178,396 | 27,531 | 10,909 |
| N25 (bp) | 3,960 | 4,389 | 4,538 | 3,342 | 3,906 | 4,341 |
| N50 (bp) | 2,459 | 2,895 | 2,991 | 1,977 | 2,626 | 2,882 |
| N75 (bp) | 1,290 | 1,929 | 2,016 | 945 | 1,796 | 1,959 |
| GC content | 35.40% | 36.49% | 36.71% | 38.60% | 38.12% | 36.56% |
Note.—Initial, transcriptomes assembled with Agalma; filtered, transcripts remaining after transcriptomes were filtered by FPKM and transdecoder reading frame criteria; RBBH, transcripts with a one-to-one RBBH match between the two filtered transcriptomes.
GO analyses identified 11,196 unique GO terms and 16,386 hidden Markov model (HMM) domains from at least one transcriptome. As a means of identifying candidate “medusa” genes, enrichment analyses (Fisher’s exact test) were performed on the abundance of each GO term and HMM domain in either assembly. GO enrichment analyses did not identify any over abundant GO terms in P. carnea’s transcriptome, when compared with the total number of GO terms for each species combined. Similarly, enrichment analyses of HMM domains did not identify any overrepresented domains in the P. carnea’s transcriptome. In contrast, 110 GO terms and 27 HMM domains were overrepresented in H. symbiolongicarpus’ transcriptome. This is most likely due to the inclusion of dactylozooids and both male and female gametic tissues in the assembly for H. symbiolongicarpus, whereas P. carnea does not have dactylozooids, and male gametic tissue were not sampled. Because neither sets of enrichment analyses yielded insight into gene and/or signaling pathways involved in life cycle differences between these two taxa, we performed interspecific DE analyses to detect quantitative differences in gene expression levels associated with the phenotypic differences between these species’ gonophores.
When comparing gene expression between species, the first critical step is to establish robust orthology assignments. To avoid artifactual differences due to different assembly methods, each transcriptome was assembled de novo by an automated bioinformatics pipeline, Agalma (Dunn, Howison, and Zapata 2013), under identical settings, as opposed to using a previously published genome-guided transcriptome for H. symbiolongicarpus (Sanders et al. 2014) and a de novo transcriptome for P. carnea (Sanders and Cartwright 2015). Of further concern is the effect of polymorphisms on transcript/gene redundancy in the assembly. Polymorphisms (common in data collected from noninbred lines) can lead to an increase in the number of paths to reconcile during the assembly process, thus increasing the number of fragmented and rare variants of a transcript/gene. To minimize the number of fragmented and redundant transcripts, each assembly was filtered for transcripts with a minimum FPKM expression value (≥1.0) and reading frame criteria prior to orthology prediction.
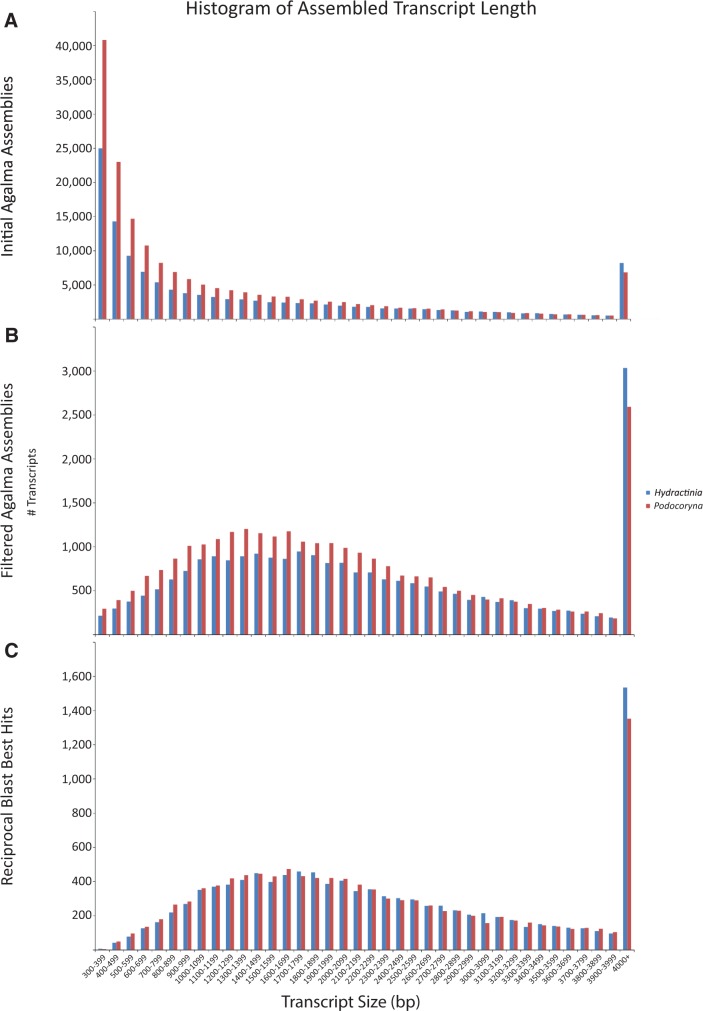
After initial filtering, approximately 24 K and 27.5 K transcripts remained (referred to from here on as the filtered transcriptomes) in the H. symbiolongicarpus (GCHW00000000) and P. carnea (GCHV00000000) assemblies, respectively (fig. 2, table 2). This reduction in transcript number greatly reduced the differences between each transcriptome assemblage characteristics, including the distribution of transcript size, N50, and GC content (table 2, fig. 3). Most importantly, removing incomplete transcripts and underrepresented variants increases our confidence in the transcripts remaining for orthology prediction and subsequently, the reliability of the inferred relative expression of each predicted ortholog. A total of 10,909 putative orthologs were identified (fig. 2, table 2; supplementary tables S1 and S2, Supplementary Material online) between our filtered assemblies. Not surprisingly, the resulting orthologous gene data set further decreased the disparity between the summary statistics for each species (table 2, fig. 3).
Fig. 3.—
Distributions of transcript size. Histograms of the (A) initial Agalma-assembled transcriptomes. (B) Assemblies filtered by FPKMs and reading frame criteria. (C) Transcripts with a one-to-one reciprocal BLAST best hit (orthologs). Red, Podocoryna carnea; blue, Hydractinia symbiolongicarpus. x axis is constant. y axis changes with each assembly.
DE Analyses
Two separate expression matrices, FPKM and TPM, were generated for the 10,909 orthologs using RSEM (Li and Dewey 2011) and analyzed with EBSeq (Leng et al. 2013). EBSeq is a well-suited software for assessing DE between species as EBSeq’s FDR and statistical power have been shown to be less sensitive to overdispersal of expression values between conditions, when compared with other DE software (Leng et al. 2013). Furthermore, EBSeq allows one to test for DE between multiple conditions (i.e., tissue types) simultaneously, whereas most other DE software only allow for individual pairwise comparisons between two conditions. EBSeq simultaneously assesses DE between multiple conditions by assigning a posterior probability to each possible expression pattern in an enumerated list of all possible expression patterns, given a set of conditions. These patterns are defined as the unique combination of significant differences in expression values between a given number of conditions. As more conditions are present, the number of possible patterns increases.
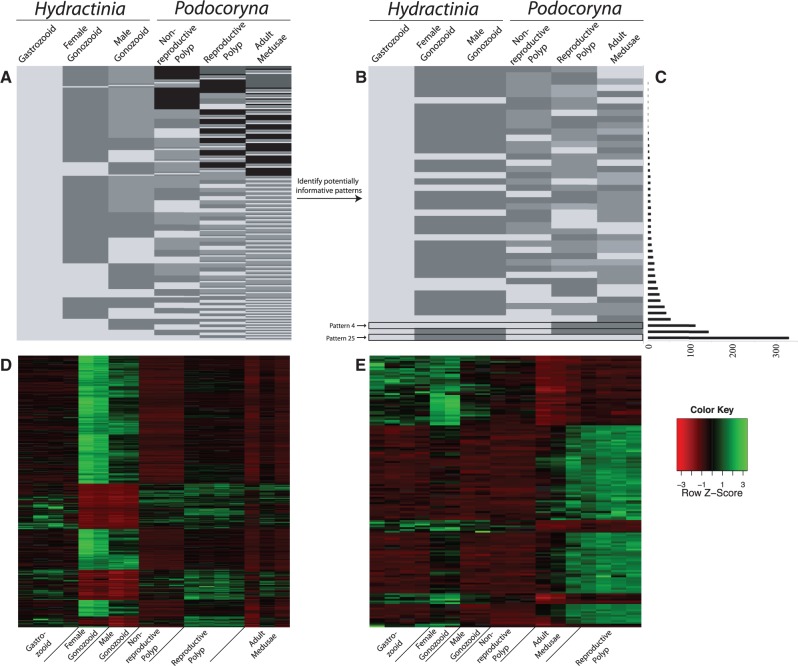
To take advantage of this feature, we included nonreproductive gastrozooids from each species (see description above) in addition to conditions that have a gonophore stage present (male and female gonozooids of H. symbiolongicarpus, medusa-budding polyps of P. carnea, and the fully developed medusae stage of P. carnea) (table 1). Inclusion of the nonreproductive tissue types increases the complexity of the expression landscape within and between each species (i.e., more patterns), effectively increasing the power of the DE analysis. With six conditions in the analyses (table 1), EBSeq identified a total of 203 possible expression patterns, although not all of which are informative to our question (fig. 4A; supplementary table S3, Supplementary Material online). This is another advantage of EBSeq, as the researcher can insert biologically relevant constraints on expression patterns a priori, retaining only those patterns that are specific to the conditions of interest for that particular study (fig. 4B).
Fig. 4.—
Pattern reduction to informative patterns. (A) With six conditions (columns) present in the DE analysis, EBSeq identifies 203 possible expression patterns (rows). (B) Using biologically relevant constraints on expression in an attempt to reduce the noise in the DE signal, the number of patterns is reduced to 44 potentially informative patterns. Colors in this schematic do not indicate magnitude of expression, just nondirectional levels of expression to show statistically equivalent, and nonequivalent levels of expression between conditions in the analysis. (C) Bar graph of the number of DE genes (FDR < 0.05) specific to each 44 of the potentially informative patterns. (D) Z-normalized heatmap of all orthologs whose expression is consistent with “Pattern 25” in the FPKM data set, an expression that should contain sporosac-specific orthologs. (E) Z-normalized heatmap of all orthologs whose expression is consistent with “Pattern 4” in the FPKM data set, an expression that should contain genes specific to Podocoryna carnea reproductive polyps and adult medusae.
Capitalizing on this aspect, we identified 44 expression patterns that were potentially informative, greatly reducing the number of potential results (fig. 4B; supplementary table S4, Supplementary Material online). These patterns were selected with an initial constraint that Hydractinia male and female gonozooid expressions are statistically equivalent. Following assumptions made by Sanders et al. (2014), transcripts differentially expressed between male and female gonozooids can be attributed to differences in gametogenesis (either spermatogenesis or oogenesis). Because only female gametic tissues were sampled in P. carnea, gene expression driven by maternal transcript generation during oogenesis will be highly expressed in the budding and adult medusae, potentially skewing the DE results. Assuming maternally loaded genes are conserved between closely related species, patterns where expression of H. symbiolongicarpus male and female sporosacs are not statistically equivalent were removed, thus reducing the number of patterns to 52. Further patterns that were not relevant to life cycle differences between species were also removed to increase the chance of finding differentially expressed genes in developing gonophore and/or adult medusae. For example, patterns where expression is statistically equivalent between nonhomologous, interspecific tissue samples (i.e., polyp tissue in Hydractinia and medusae tissue in Podocoryna) but are differentially expressed between intraspecific tissue samples (i.e., nonreproductive polyps and medusae tissue of Podocoryna) were removed from the analysis, resulting in a total of 44 potentially informative patterns (fig. 4B; supplementary table S4, Supplementary Material online).
Approximately 75% (8,210) of the putative orthologs are recovered as significantly differentially expressed in at least one of the data sets along one of the 203 expression patterns at a PPDE > 0.95 (table 3; supplementary table S2, Supplementary Material online). This high proportion of DE genes is not entirely surprising as only one condition needs to significantly vary from any of the others to be recovered as such (fig. 4). In either case, both data sets perform similarly, with only 686 and 858 of those DE transcripts unique to the FPKM and TPM data sets at this significance threshold, respectively. The percentage of transcripts identified as differentially expressed in both data sets remains roughly constant (between 79-81%), until the FDR decreases to 0.00 (PPDE = 1.00). At this threshold, FPKM performs more conservatively than TPM, with only 5.2% (266) of the total DE orthologs (5,120) specific to the FPKM data set, whereas 23.2% (1,187) are unique to the TPM data set (table 3; supplementary fig. S1, Supplementary Material online). This increased conservation can be attributed to the added scaling by transcript length for FPKM expression values.
Table 3.
Number of Differentially Expressed Transcripts
| FDR < 0.05 |
FDR = 0.00 |
|||||||
|---|---|---|---|---|---|---|---|---|
| FPKM | TPM | Shared | Total | FPKM | TPM | Shared | Total | |
| Tot. DE | 686 | 858 | 6,666 | 8,210 | 266 | 1,187 | 3,667 | 5,120 |
| Tot. PIT | 402 | 349 | 1,688 | 2,439 | – | – | – | – |
| Tot. DE PIT | 359 | 368 | 1,611 | 2,338 | 173 | 403 | 847 | 1,423 |
| Upreg. DE PIT | 181 | 152 | 605 | 938 | 80 | 168 | 244 | 492 |
Note.—FPKM and TPM columns correspond to the number of transcripts unique to that data set. Upreg. DE PIT, counts for transcripts specific to one of the conditions of interest shown in footnote b in table 1.
A total of 2,439 potentially informative transcripts (PIT; transcripts whose expression is consistent with one of the 44 potentially informative patterns) were identified in at least one of the two data sets (table 3). Of those 1,611 were found significantly differentially expressed in both data sets, whereas an additional 359 and 368 were specific to the FPKM and TPM sets, respectively (FDR < 0.05; table 3). Although 2,338 PIT are differentially expressed between the test conditions in at least one of the data sets, they are not necessarily expressed in a gonophore/medusa-specific manner since the predefined expression patterns do not contain information about the magnitude of expression for each condition (fig. 4D and E). Further examination of these DE PIT revealed that 938 are upregulated in one of the gonophore/medusa-containing conditions, of which 605 were significant in both (FDR < 0.05; table 3; fig. 4C; supplementary tables S2 and S5, Supplementary Material online).
Discrepancies between the different normalization methods on which putative orthologs are differentially expressed can be seen across all levels of the DE analysis and are explained by differences in the expression pattern assigned the highest posterior probability. This is largely due to disagreement on constraints imposed to identify potentially informative patterns such as scaling by transcript length in FPKM. Although Li and Dewey (2011) suggest that TPM is better expression measure for comparisons between species, there has been no comprehensive evaluation of normalization methods for RNA-Seq in an interspecific DE framework. Therefore, to minimize the effect that either normalization method has on the DE results, any putative ortholog identified in either data set are considered candidates for future study and ones shown to be significant in both data sets can be considered most reliable.
Spatial Expression of Differentially Expressed Orthologs
To further validate our unbiased interspecific DE analyses, several of the 492 transcripts identified as significantly upregulated in either Hydractinia sporosacs or Podocoryna developing and/or adult medusae in at least one data set (FDR = 0.00; table 3) were selected for spatial expression analysis by whole mount ISH (table 4; fig. 5; supplementary table S6, Supplementary Material online). Selection was based on relatively high levels of expression that could be detected with this method and because of potential biological interest. None of the candidates discussed below have been previously characterized in cnidarians. Spatial expression of each gene was examined with ISH in each of the tissues sampled for DE analyses. Medusa buds were examined across all 10 stages of medusa development as defined by Frey (1968) (fig. 5). Expression of the candidates discussed below was primarily restricted to tissues in the gonophores and adult medusae (fig. 5), except for APLP, which also exhibited expression in polyp tissues (supplementary fig. S2, Supplementary Material online).
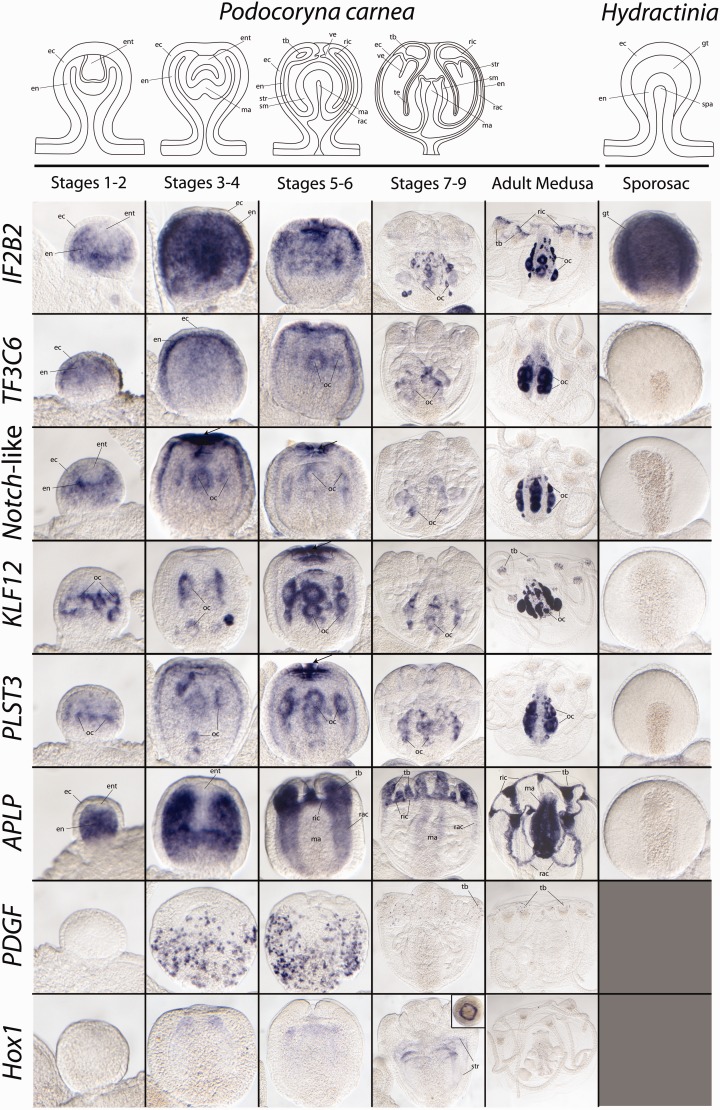
Fig. 5.—
Whole mount ISH results. Images position the oral end of the gonophores/medusa toward the top. Arrows mark regions of concentrated expression at the distal end of the bell axis or the oral end of the developing manubrium. Only male Hydractinia symbiolongicarpus sporosacs are shown as eggs in females block the view of the spadix (manubrium anlage). Inset in Hox1, stages 7–9 pane is a view from the oral end of the gonophore looking down. ec, ectoderm; en, endoderm; ent, entocodon; gt, gametic tissue; ma, manubrium, oc, oocytes; rac, radial canal; ric, ring canal; sm, smooth muscle; spa, spadix; str, striated muscle; tb, tentacle bulb; ve, velum.
Two of the candidates surveyed with ISH, IF2B2 and TF3C6 exhibited similar endodermal expression patterns in P. carnea gonophores. IF2B2 was recovered as significantly upregulated in all gonophore stages (including H. symbiolongicarpus sporosacs and P. carnea medusa buds and adult medusa), whereas TF3C6 was recovered as specific to P. carnea medusa tissues. ISH of both genes shows strong endodermal expression from stages 1 to 6 of medusa development in P. carnea, which then ceases by stage 7. After liberation, IF2B2 is also expressed the proximal portion of the tentacle bulbs of the adult medusae (fig. 5), a region corresponding to early stages of nematogenesis in hydrozoan medusae (Denker et al. 2008). Expression patterns here suggest that these genes might have a role inducing cell proliferation as they are expressed in highly proliferative regions of the developing and/or adult medusa (Spring et al. 2000; Seipel, Yanze, et al. 2004; Denker et al. 2008). These spatially restricted expression patterns were not observed in H. symbiolongicarpus sporosacs (fig. 5).
Furthermore, ISH of IF2B2 revealed expression consistent with a general role in gametogenesis in both H. symbiolongicarpus and P. carnea (fig. 5). IF2B2 is expressed around early- and late-stage oocytes in P. carnea. In H. symbiolongicarpus, IF2B2 is expressed around oocytes in the germinal zone (body column) of female gonozooids (not shown), where oogenesis begins (Berrill 1953; Müller 1964; Bunting 1894), and expression continues in the endoderm surrounding oocytes after it moves into the gonophores, where oocyte differentiation continues (Berrill 1953; Müller 1964; Bunting 1894). In males, expression is specific to the gametic tissues of mature sporosacs (fig. 5) (Bunting 1894; Berrill 1953). This expression pattern suggests that IF2B2 is only operating in gametogenesis and plays no role in patterning the sporosac. This is different from ISH results of TF3C6 where no expression of TF3C6 was seen in either male or female sporosacs, while expression was observed around early and late stage oocytes in P. carnea.
ISH of three other genes recovered as upregulated in P. carnea medusa libraries (Notch-like, KLF12, and PLST3) revealed similar spatially restricted expression patterns at the distal tip of the developing axes of medusa buds of P. carnea. For each of these genes, ISH shows minor endodermal expression at various stages of medusa development, but in each case, the prominent expression is seen at the distal end of the developing bell axis by stages 5 and 6. Notch-like expression precedes the expression of both KLF12 and PLST3 and is strongly expressed at the distal end of the gonophore prior to opening of the bell margin in stage four. Past stage 6, expression for all genes is specific to maturing oocytes, although after liberation, KLF12 is also expressed in the tentacle bulbs and along the manubrium (the structure bearing the gonads and mouth at its distal end). Similar to TF3C6 expression, ISH did not detect expression of these genes in the sporosacs of H. symbiolongicarpus (fig. 5), except for minor expression around early stage oocytes (not shown).
Although the top BLAST hit was a “neurogenic notch” gene, the ortholog, Notch-like, examined in this study are not orthologous to those notch genes examined in Hydra and Nematostella (Käsbauer et al. 2007; Marlow et al. 2012). Sequence analysis of Notch-like reveals that it lacks the LNR and NOD domains that characterizes “notch” genes but does possess the EGF domains which is present in, but not specific to notch genes (Käsbauer et al. 2007; Marlow et al. 2012). In each of the genes examined here, ISH reveals spatially restricted expression patterns in highly proliferative somatic regions during P. carnea development. These expression patterns, together with the lack of somatic expression of these genes in H. symbiolongicarpus sporosacs suggest a potential role or these genes in medusae morphogenesis and evolution. Future functional studies will need to be performed to confirm these results.
APLP is the only candidate selected for ISH that does not exhibit expression consistent with any role in oogenesis in P. carnea. Throughout gonophore development, ISH reveals APLP expression to be specific to the endodermal tissues that give rise to gastric structures in the adult medusa. Starting at stage one, strong APLP expression is detected in the endoderm of the newly formed gonophore. As development proceeds, APLP expression remains specific to the endodermal tissue beginning to form the radial canals in stages 3–6. APLP expression is excluded from and clearly outlines the entocodon, which is medusa-specific tissue layer formed through evagination of the distal ectoderm of the gonophore that gives rise to striated muscle (Avset 1961). By stages 5 and 6, strong expression is noted in the newly formed ring canal and tentacle bulbs but is excluded from the developing manubrium. This pattern continues through the later stage buds but seems to decrease in the strength of expression (especially in the radial canals) until the medusa is liberated from the colony, where expression strongly reappears in all digestive tissues; including the fully developed manubrium, radial and ring canals, and tentacle bulbs (fig. 5). APLP expression was not observed in the sporosacs of H. symbiolongicarpus.
Throughout medusa development, APLP appears to be expressed in a manner consistent with the patterning and development of the digestive tract of the medusa, whereas after liberation, it remains expressed in digestive tissues. This is consistent with APLP’s role in other animals, where it functions not only as a lipid trafficking molecule but also plays a critical role in patterning, through regulating hedgehog and Wnt signaling during wing development in Drosophila (Panáková et al 2005). Previous studies have implicated canonical Wnt signaling in medusae evolution (Duffy et al. 2010; Duffy 2011; Nawrocki and Cartwright 2013; Sanders and Cartwright 2015), yet, given the dual role of the Wnt pathway as both a maternal effect for larval development and in adult patterning (Plicket et al. 2006; Teo et al. 2006; Momose and Houliston 2007; Müller et al. 2007; Momose et al. 2008; Amiel and Houliston 2009; Duffy et al. 2010), DE expression of Wnt signaling genes in medusa would likely be obscured by high expression in female gametic tissue due to maternal loading. Yet even without many of the key Wnt signaling components recovered by our DE analyses, recovery of APLP further implicates the role of Wnt signaling in medusa development and evolution (Nawrocki and Cartwright 2013; Sanders and Cartwright 2015). Further ties between APLP and Wnt signaling come from the expression patterns of APLP in both P. carnea and H. symbiolongicarpus polyps (supplementary fig. S2, Supplementary Material online). In the feeding polyps of both species, APLP is expressed in a ring around the distal tip of the hypostome and in the endoderm at the tip of the tentacles (supplementary fig. S2, Supplementary Material online), consistent with observed Wnt3 expression in Hydra (Guder et al. 2005; Lengfeld et al. 2009; Gee et al. 2010), Hydractinia (Plickert et al. 2006; Müller et al. 2007; Duffy et al. 2010), Podocoryna (Sanders and Cartwright 2015), and Ectopleura (Nawrocki and Cartwright 2013). Similarly, ISH revealed APLP expression at the distal tip of H. symbiolongicarpus gonozooids (supplementary fig. S2, Supplementary Material online), consistent with Wnt3 expression in H. echinata (Müller et al. 2007; Duffy et al. 2010) and H. symbiolongicarpus (S. Sanders, unpublished data).
Previously Published Medusae-Specific Genes
Several previous studies, using a candidate gene approach, identified genes specific to medusae development in P. carnea (Schuchert et al. 1993; Aerne et al. 1995; Gröger et al. 1999; Masuda-Nakagawa et al. 2000; Müller et al. 1999; Yanze et al. 1999; Spring et al. 2000; Spring et al. 2002; Müller et al. 2003; Seipel, Yanze, et al. 2004; Seipel et al. 2004a–c; Stierwald et al. 2004; Torras et al. 2004; Galle et al. 2005; Reber-Müller et al. 2006). As additional validation of our DE results, we determined whether any of these genes were present in our pool of 938 candidates that were identified as significantly upregulated in developing gonophores and/or adult medusa (table 3). In our DE analysis, some, but not all, previously reported medusa-specific genes are differentially upregulated in developing and/or adult medusa stages of P. carnea relative to H. symbiolongicarpus sporosacs (table 5). These include orthologs of striated muscle-specific homeobox genes msx (Galle et al. 2005) and orthodenticle (otx) (Müller et al. 1999), a myosin heavy chain, myo1 (Schuchert et al. 1993), a tropomyosin, tpm2 (Gröger et al. 1999), and a zinc finger transcription factor snail (Spring et al. 2002).
Table 5.
Differentially Expressed Orthologs Consistent with Previously Published Studies in Podocoryna carnea
| RBBH ID | Name | Source | FPKM | TPM | Specificity |
|---|---|---|---|---|---|
| RBBH_4080 | Myo1 | Schuchert et al. (1993) | ** | ** | All medusa stages |
| RBBH_3250 | Tpm2 | Gröger et al. (1999) | ** | ** | All medusa stages |
| RBBH_5585 | Otx | Müller et al. (1999) | NS | * | Adult medusa |
| RBBH_6387 | Snail | Spring et al. (2002) | * | ** | All medusa stages |
| RBBH_540 | Msx | Galle et al. (2005) | * | * | Adult medusa |
Note.—NS, not significant.
*PPDE > 0.99.
**Orthologs significant in either data set at a PPDE = 1.00.
Moreover, several of the previously reported medusae-specific genes appear to be absent in the H. symbiolongicarpus transcriptome examined here. These include three homeobox genes, Hox1 (Aerne et al. 1995), six1/2 (Stierwald et al. 2004), and cnox2-Pc (Masuda-Nakagawa et al. 2000) (not orthologous to cnox-2 in H. symbiolongicarpus; Cartwright et al. 1999) (supplementary fig. S3, Supplementary Material online), as well as a helix-loop-helix transcription factor, jellyD1 (Müller et al. 2003). Interestingly, these genes have previously been shown to be expressed in medusa-specific structures, including striated muscle, the manubrium, the entocodon, and/or tentacle bulbs (Aerne et al. 1995; Masuda-Nakagawa et al. 2000; Stierwald et al. 2004). Gene tree analyses and searching unpublished genomic scaffolds of H. symbiolongicarpus reveal that Hox1 and jellyD1 lack an ortholog in H. symbiolongicarpus (supplementary figs. S3 and S4, Supplementary Material online), whereas the absence of six1/2 and cnox2-Pc in this H. symbiolongicarpus transcriptome appear to be instances of downregulated expression of these genes in H. symbiolongicarpus sporosacs.
Podocoryna carnea Genes that Lack a Corresponding Ortholog in H. symbiolongicarpus
Although differential regulation of orthologous genes does explain differences in homologous structures between species, evolutionary shifts between phenotypes are likely accompanied by gene gain or loss as well. To further explore the role of novel gene gain and loss in hydractiniid life cycle differences, we selected two P. carnea genes without a H. symbiolongicarpus ortholog (table 4; supplementary figs. S3 and S5, Supplementary Material online) that are also significantly upregulated in developing and adult medusae of P. carnea (Sanders and Cartwright 2015) for further study with ISH. These genes were, an unpublished growth factor, PDGF, and the previously published homeobox gene, Hox1 (Aerne et al. 1995) (table 4). Phylogenetic analysis of the platelet-derived/vascular endothelial growth factor family (PDGF/VEGF) (supplementary fig. S5, Supplementary Material online) further suggested the absence of a PDGF ortholog in H. symbiolongicarpus and was confirmed by its absence in the previously mentioned unpublished genomic scaffolds. Although a member of the same gene family, PDGF is not homologous to the VEGF gene previously characterized in P. carnea (Seipel et al. 2004c) (supplementary fig. S5, Supplementary Material online) whose expression is restricted to the endoderm of the medusae buds and its resulting tissues during medusae development. ISH of PDGF confirmed the specificity of this gene to developing and adult medusa stages with no expression detected in the polyp stage of P. carnea (not shown). PDGF expression begins at bud stages 3 and 4 of medusa development, revealing a speckled expression pattern (fig. 5). This pattern shifts as medusa development continues, and PDGF-positive cells appear to be evenly distributed except at the most distal tip of the gonophore by stages 5 and 6. By stages 7 and 8 of medusa development, expression is limited to just a few cells in the tentacle bulbs and this continues through the adult medusa. These cells most notably resemble differentiating stem cells called nematoblasts (Denker et al. 2008), most likely in some very early stage of nematogenesis as they appear to migrate toward the tentacle bulb, although further research is necessary to confirm this. Since this gene is not expressed in the known stem cell populations of hydrozoan polyps (Teo et al. 2006; Müller et al. 2007; Millane et al. 2011; Duffy et al. 2012; Hemmrich et al. 2012), it suggests a potential medusa-specific stem cell lineage.
Although members of the same gene family, PDGF and VEGF (Seipel et al. 2004c) display very different expression patterns throughout medusae development. Seipel et al. (2004c) showed VEGF expression to be consistent with morphogenesis in P. carnea, particularly during tube formation (both in the tentacles or the canal system of the medusae) and suggested that as the ancestral metazoan function of VEGF genes. This is consistent with the role of VEGF signaling in vasculogenesis and angiogenesis in vertebrates (Ferrara and Davis-Smyth 1997; Nasevicius et al. 2000). However, in Drosophila, VEGF signaling is involved in blood cell migration and differentiation (Duchek et al. 2001; Heino et al. 2001; Cho et al. 2002). Our expression patterns suggest that PDGF expression is more consistent with this role in cell migration and differentiation. These results would suggest that at least two different (and potentially conserved) PDGF/VEGF signaling pathways are operating during medusae development and that the alteration of these pathways has implications in medusae evolution.
ISH showed relatively little expression of Hox1 through medusa development with no expression detected in the polyp. Noticeable (although very faint) expression begins by stage 3 and continues through the later stages of gonophore development, until the medusa is fully developed, where expression ceases. At the earlier stages, Hox1 expression is seen as a ring-like pattern around the distal region of the differentiating entocodon (fig. 5). This pattern is maintained as gonophore development progresses, broadening the expression ring as the gonophore grows. By stages 7 and 8, the strongest expression is seen at the distal end of the expression domain yet with more minor expression dispersed more proximally along the striated muscle tissue of the developing medusa.
ISH of Hox1 is consistent with expression reported by Aerne et al. (1995), where expression was detected in bud stages with developing striated muscle. Later, Yanze et al. (2001) showed Hox1 expression throughout embryonic development and in the aboral end of the planula, consistent with expression of Hox1 orthologs (78% BS support; supplementary fig. S3, Supplementary Material online) reported in Clytia (Chiori et al. 2009) and Eleutheria (Jakob and Schierwater 2007), where it appears to play a role in the oral-aboral patterning. Two of three taxa (Clytia and Eleutheria) with a documented ortholog of Hox1 possess fully developed medusa and in each case Hox1 expression is not observed in the striated muscle. In Clytia hemisphaerica medusa, Hox1 expression is specific to the balancing organ (statocyst; not present in P. carnea medusa) (Chiori et al. 2009), whereas Eleutheria dichotoma benthic medusae exhibit no Hox1 expression (Jakob and Schierwater 2007).
Although results from the phylogenetic analyses suggest that Hox1 evolved earlier in Hydrozoa and was subsequently lost in H. symbiolongicarpus (supplementary fig. S3, Supplementary Material online), jellyD1 and PDGF appear to have be the result of duplication events in P. carnea (supplementary figs. S4 and S5, Supplementary Material online); however, this could be an artifact of limited sampling of these gene families in other cnidarians. The lack of an orthologous Hox1 gene in H. symbiolongicarpus is consistent with the loss of striated muscle during truncation of the medusa following a Hox1 deletion, this, however, does not explain the observed variability of Hox1 expression in the medusa of more distantly related hydrozoans. This variable expression of Hox1 across distantly related taxa suggests a potential evolutionary scenario where Hox1 was co-opted to be involved in striated muscle development during a transition toward fully developed medusa in P. carnea as no other hydrozoan medusa exhibits Hox1 expression in their striated muscle tissues (Jakob and Schierwater 2007; Chiori et al. 2009). Future areas of research should focus on sampling more intermediate levels of medusa truncation to determine whether changes in expression correlate with the development of striated muscle tissue.
Conclusion
These results show interspecific DE analyses to be a more sensitive method for identifying candidate genes and/or gene networks involved in the evolutionary transitions between different life history forms than more common comparative methods such as enrichment analyses. Our DE analysis revealed new candidate genes the may be involved in the evolutionary transition in medusae loss or re-evolution that have not been previously characterized in hydrozoa. Albeit more powerful, our method is reliant on identifiable orthologs. Further analyses of the genes PDGF and Hox1, which were absent in H. symbiolongicarpus, revealed expression consistent with an important role in medusa development in P. carnea. Thus, both up- and downregulation of orthologous genes and novel gene gain and loss appear important for life cycle differences between these two species and may play a role in reduction and possible re-evolution of the medusa life cycle stage in the Hydractiniidae. With nearly 100 million years of divergence between these two species (Miglietta and Cunningham 2012), which exhibit the “book-end” phenotypes of gonophore development, the differential regulation of orthologs, and gene duplication and loss, most likely accompanied the transitions between the fully developed and fully truncated medusa.
Addressing questions of parallel incidences of medusa loss, and even re-gain, requires increased taxonomic sampling. Increasing the number of taxa sampled adds a new layer of complexity to ensure the validity of the DE analysis. Dunn, Luo, and Wi (2013) extensively reviewed not only the utility of comparative expression across multiple species but also the numerous challenges it presents. As with any phylogenetic statistical analysis, the nonindependent nature of the data can have large effects on the results (Felsenstein 1985). Future studies sampling more than two species will need to expand current DE software to utilize independent contrasts.
Here, we have provided a new workflow with which one can effectively quantify cross-species differences in expression using short read Illumina data. DE results between these two hydractiniid species reveal 938 candidate orthologs correlated with hydractiniid life cycle variation. These can serve as a useful guide for future studies in spatial gene expression analyses and can potentially be combined with high-throughput functional assays (e.g., Varshney et al. 2015). Moreover, orthology assignments and phylogenetic analyses suggest multiple instances of novel gene loss and gain correlated with phenotypic differences of the gonophore in P. carnea and H. symbiolongicarpus. In addition, expanding this method to include more taxa and utilizing independent contrasts should provide significant insight into the role of these genes in medusa evolution in Hydrozoa.
Supplementary Material
Supplementary tables S1–S6 and figures S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Neil Blackstone for laboratory cultures of P. carnea, Clark Bloomer at KUMC-GSF for RNA isolation, library construction, and sequencing, Kirsten Jensen for the illustrations in figure 1, Susan Boyles for illustrations in figure 5, Casey Dunn and Felipe Zapata for help implementing the Agalma package, and Justin Blumenstiel and Uri Frank for discussions, and members of the Hileman Lab (University of Kansas) for comments on a previous version. This work was supported by NSF grant DEB-095357 to P.C.
Literature Cited
- Aerne BL, Baader CD, Schmid V. 1995. Life stage and tissue-specific expression of the homeobox gene cnox1-Pc of the hydrozoan Podocoryne carnea. Dev Biol. 169:547–556. [DOI] [PubMed] [Google Scholar]
- Allman GJ. 1864. On the construction and limitation of genera among the Hydroida. Ann Mag Nat Hist Ser. 13:345–380. [Google Scholar]
- Amiel A, Houliston E. 2009. Three distinct RNA localization mechanisms contribute to oocyte polarity establishment in the cnidarian Clytia hemisphaerica. Dev Biol. 327:191–203. [DOI] [PubMed] [Google Scholar]
- Avset K. 1961. The development of the medusa Podocoryne carnea. Nytt Mag Zool. 10:49–56. [Google Scholar]
- Berrill NJ. 1953. Growth and form in gymnoblastic hydroids; polymorphism within the Hydractiniidae. J Morphol. 92:241–272. [Google Scholar]
- Boyle AP, et al. 2014. Comparative analysis of regulatory information and circuits across distant species. Nature 512:453–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting M. 1894. The origin of the sex-cells in Hydractinia and Podocoryne; and the development of Hydractinia. J Morphol. 9:203–236. [Google Scholar]
- Cartwright P, Bowshier J, Buss LW. 1999. Expression of a Hox gene, Cnox-2, and the division of labor in a colonial hydroid. Proc Natl Acad Sci USA. 96:2183–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P, Nawrocki AM. 2010. Character evolution in Hydrozoa (phylum Cnidaria). Integr Comp Biol. 50:456–472. [DOI] [PubMed] [Google Scholar]
- Chiori R, et al. 2009. Are Hox genes ancestrally involved in axial patterning? Evidence from the hydrozoan Clytia hemisphaerica (Cnidaria). PLoS One 4:e4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho NK, et al. 2002. Developmental control of blood cell migration by the Drosophila VEGF pathway. Cell 108:865–876. [DOI] [PubMed] [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Cornelius PFS. 1992. Medusa loss in leptolid Hydrozoa (Cnidaria), hydroid rafting, and abbreviated life cycles among their remote island faunae: an interim review. Sci Mar. 56:245–261. [Google Scholar]
- Cunningham CW, Buss LW. 1993. Molecular evidence for multiple episodes of paedomorphosis in the family Hydractiniidae. Biochem Syst Ecol. 21:57–69. [Google Scholar]
- Denker E, Manuel M, Leclère L, Guyader H, Rabet N. 2008. Ordered progression of nematogenesis from stem cells through differentiation stages in the tentacle bulb of Clytia hemisphaerica (Hydrozoa, Cnidaria). Dev Biol. 315:99–113. [DOI] [PubMed] [Google Scholar]
- Duchek P, Somogyi K, Jekely G, Beccari S, Rorth P. 2001. Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell 107:17–26. [DOI] [PubMed] [Google Scholar]
- Duffy DJ. 2011. Modulation of Wnt signaling: a route to speciation? Commun Integr Biol. 4:59–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy DJ, Millane RC, Frank U. 2012. A heat shock protein and Wnt signaling crosstalk during axial patterning and stem cell proliferation. Dev Biol. 362:271–281. [DOI] [PubMed] [Google Scholar]
- Duffy DJ, Plickert G, Kuenzel T, Tilmann W, Frank U. 2010. Wnt signaling promotes oral but suppresses aboral structures in Hydractinia metamorphosis and regeneration. Development 137:3057–3066. [DOI] [PubMed] [Google Scholar]
- Dunn CW, Howison M, Zapata F. 2013. Agalma: an automated phylogenomics workflow. BMC Bioinformatics 14:330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, Luo X, Wi Z. 2013. Phylogenetic analysis of gene expression. Int Comp Biol. 53:847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Phylogenies and the comparative method. Am Nat. 125:1–15. [Google Scholar]
- Ferrara N, Davis-Smyth T. 1997. The biology of vascular endothelial growth factor. Endocr Rev. 18:4–12. [DOI] [PubMed] [Google Scholar]
- Frey J. 1968. Die Entwicklungsleistungen der Medusenknospen und Medusen von Podocoryne carnea M. Sars nach Isolation und Dissoziation. Wilhelm Roux’ Arch Entwicklungsmech Org. 168: 428–464. [DOI] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CH-HIT: accelerated for clustering the next generation sequencing data. Bioinformatics 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewsky M, Leitz T, Schlossherr J, Plickert G. 1996. LWamides from Cnidaria constitute a novel family of neuropeptide with morphogenetic activity. Roux’s Arch Dev Biol. 205:232–242. [DOI] [PubMed] [Google Scholar]
- Galle S, Yanze N, Seipel K. 2005. The homeobox gene Msx in development and transdifferentiation of jellyfish striated muscle. Int J Dev Biol. 49:961–967. [DOI] [PubMed] [Google Scholar]
- Gee L, et al. 2010. ß-catenin plays a central role in setting up the head organizer in hydra. Dev Biol. 340:116–124. [DOI] [PubMed] [Google Scholar]
- Gibbons MJ, Janson LA, Ismail A, Samaai T. 2010. Life cycle strategy, species richness and distribution in marine Hydrozoa (Cnidaria: Medusozoa). J Biogeogr. 37:441–448. [Google Scholar]
- Götz S, et al. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36:3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gröger H, Callearts P, Gehring WJ, Schmid V. 1999. Gene duplication and recruitment of a specific tropomyosin into striated muscle cells in the jellyfish Podocoryne carnea. J Exp Zool B Mol Dev Evol. 285:378–386. [PubMed] [Google Scholar]
- Guder C, et al. 2005. An ancient Wnt-Dickkopf anatagonism in Hydra. Development 133:901–911. [DOI] [PubMed] [Google Scholar]
- Hao DC, Ge G, Xiao P, Zhang Y, Yang L. 2011. The first insight into the tissue specific Taxus transcriptome via Illumina second generation sequencing. PLoS One 6:e21220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heino TI, et al. 2001. The Drosophila VEGF receptor homolog is expressed in hemocytes. Mech Dev. 109:69–77. [DOI] [PubMed] [Google Scholar]
- Helm RR, Siebert S, Tulin S, Smith J, Dunn CW. 2013. Characterization of differential transcript abundance through time during Nematostella vectensis development. BMC Genomics 14:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmrich G, et al. 2012. Molecular signatures of the three stem cell lineages in Hydra and the emergence of stem cell function at the base of multicellularity. Mol Biol Evol. 29:3267–3280. [DOI] [PubMed] [Google Scholar]
- Jakob W, Schierwater B. 2007. Changing hydrozoan bauplans by silencing Hox-like genes. PLoS One 8:e694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käsbauer T, et al. 2007. The Notch signaling pathway in the cnidarian Hydra. Dev Biol. 303:376–390. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in the accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclère L, Schuchert P, Cruaud C, Couloux A, Manuel M. 2009. Molecular phylogenetics of Thecata (Hydrozoa, Cnidaria) reveals long-term maintenance of life history traits despite high frequency of recent character changes. Syst Biol. 58:509–526. [DOI] [PubMed] [Google Scholar]
- Leclère L, Schuchert P, Manuel M. 2007. Phylogeny of the Plumularioidea (Hydrozoa, Leptothecata): evolution of colonial organisation and life cycle. Zool Scripta. 36:371–394. [Google Scholar]
- Leng N, et al. 2013. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29:1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengfeld T, et al. 2009. Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev Biol. 330:186–199. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. [DOI] [PubMed] [Google Scholar]
- Masuda-Nakagawa LM, Gröger H, Aerne BL, Schmid V. 2000. The Hox-like gene Cnox2-Pc is expressed in all life cycle stages of the jellyfish Podocoryne carnea. Dev Genes Evol. 210:151–156. [DOI] [PubMed] [Google Scholar]
- Marlow H, Roettinger R, Boekhout M, Martindale MQ. 2012. Functional roles of Notch signaling in the cnidarian Nematostella vectensis. Dev Biol. 362:295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Migotto AE. 2001. Cladistic analysis and new classification of the family Tubulariidae (Hydrozoa, Anthomedusae). Pap Avuls Zool. 41:465–88. [Google Scholar]
- Miglietta MP, Cunningham CW. 2012. Evolution of life cycle, colony morphology, and host-specificity in the family Hydractiniidae (Hydrozoa, Cnidaria). Evolution 66:3876–3901. [DOI] [PubMed] [Google Scholar]
- Miglietta MP, McNally L, Cunningham CW. 2010. Evolution of calcium-carbonate skeletons in the Hydractiniidae. Integr Comp Biol. 50:428–435. [DOI] [PubMed] [Google Scholar]
- Miglietta MP, Schuchert P, Cunningham CW. 2009. Reconciling genealogical and morphological species in a worldwide study of the Family Hydractiniidae (Cnidaria, Hydrozoa). Zool Scripta. 38:403–430. [Google Scholar]
- Millane RC, et al. 2011. Induced stem cell neoplasia in a cnidarian by ectopic expression of a POU domain transcription factor. Development 138:2429–2439. [DOI] [PubMed] [Google Scholar]
- Miller JR, Koren S, Sutton G. 2010. Assembly algorithms for next-generation sequencing data. Genomics 95:315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose T, Derelle R, Houliston E. 2008. A maternally localized Wnt ligand required for axial patterning in the cnidarian Clytia hemisphaerica. Development 135:2105–2113. [DOI] [PubMed] [Google Scholar]
- Momose T, Houliston E. 2007. Two oppositely localized frizzled RNAs as axis determinants in cnidarian embryo. PLoS Biol. 5:e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Yanze N, Schmid V, Spring J. 1999. The homeobox gene Otx of the jellyfish Podocoryne carnea: role of a head gene in striated muscle and evolution. Dev Biol. 216:582–594. [DOI] [PubMed] [Google Scholar]
- Müller P, et al. 2003. Evolutionary aspects of developmentally regulated helix-loop-helix transcription factors in striated muscle of jellyfish. Dev Biol. 255:216–229. [DOI] [PubMed] [Google Scholar]
- Müller W. 1964. Experimentelle Untersuchungen Über Stockentwicklung, Polypendifferenzierung, und sexualchimären bei Hydractinia echinata. Wilhelm Roux’ Arch Entwicklungsmech Org. 155:181–268. [DOI] [PubMed] [Google Scholar]
- Müller W, et al. 2007. Wnt signaling in hydroid development: ectopic heads and giant buds induced by GSK-3ß inhibitors. Int J Dev Biol. 51:211–220. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Larson J, Ekker SC. 2000. Distinct requirements for zebrafish angiogenesis revealed by a VEGF-A morphant. Yeast 17:294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki AM, Cartwright P. 2013. Expression of Wnt pathway genes in polyps and medusa-like structures of Ectopleura larynx (Cnidaria: Hydrozoa). Evol Dev. 15:373–384. [DOI] [PubMed] [Google Scholar]
- Panáková D, Sprong H, Marois E, Thiele C, Eaton S. 2005. Lipoprotein particles are required for Hedgehog and Wingless signaling. Nature 435:58–65. [DOI] [PubMed] [Google Scholar]
- Pankey MS, Mini VN, Imholte GC, Suchard MA, Oakley TH. 2014. Predictable transcriptome evolution in the convergent and complex bioluminescent organs of squid. Proc Natl Acad Sci USA. 111:E4736–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plickert G, Jacoby V, Frank U, Müller WA, Mokady O. 2006. Wnt signaling in hydroid development: formation of the primary body axis in embryogenesis and its subsequent patterning. Dev Biol. 298:368–378. [DOI] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. 2012. The Pfam protein families database. Nucleic Acid Res. 40:D290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber-Müller S, et al. 2006. BMP2/4 and BMP5-8 in jellyfish development and transdifferentiation. Int J Dev Biol. 50:377–384. [DOI] [PubMed] [Google Scholar]
- Sanders SM, Cartwright P. 2015. Patterns of Wnt signaling in the life cycle of Podocoryna carnea and its implications for medusae evolution in Hydrozoa (Cnidaria). Evol Dev. 17(5). [DOI] [PubMed] [Google Scholar]
- Sanders SM, Shcheglovitova M, Cartwright P. 2014. Differential gene expression between functionally specialized polyps of the colonial hydrozoan Hydractinia symbiolongicarpus (Phylum Cnidaria). BMC Genomics 15:406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchert P. 2008. The European athecate hydroids and their medusae (Hydrozoa, Cnidaria): Filifera part 3. Rev Suisse de Zool. 115:677–757. [Google Scholar]
- Schuchert P, Reber-Müller S, Schmid V. 1993. Life stage specific expression of a myosin heavy chain in the hydrozoan Podocoryne carnea. Differentiation 54:11–18. [DOI] [PubMed] [Google Scholar]
- Schunter C, Vollmer SV, Macpherson E, Pascual M. 2014. Transcriptome analyses and differential gene expression in a non-model fish species with alternative mating tactics. BMC Genomics 15:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipel K, Yanze N, Müller P, Streltwolf R, Schmid V. 2004. Basic leucine sipper transcripton factors C/EBP and Mafl in the hydrozoan jellyfish Podocoryne carnea. Dev Dynamics. 230:392–402. [DOI] [PubMed] [Google Scholar]
- Seipel K, Yanze N, Schmid V. 2004a. Developmental and evolutionary aspects of the basic helix-loop-helix transcription factors Atonal-like 1 and Achaete-scute homolog 2 in the jellyfish. Dev Biol. 269: 331–345. [DOI] [PubMed] [Google Scholar]
- Seipel K, Yanze N, Schmid V. 2004b. The germ line and somatic stem cell gene Cniwi in the jellyfish Podocoryne carnea. Int J Dev Biol. 48:1–7. [DOI] [PubMed] [Google Scholar]
- Seipel K, et al. 2004c. Homologs of vascular endothelial growth factor and receptor, VEGF and VEGFR, in the jellyfish Podocoryne carnea. Dev Dynamics. 231:303–3012. [DOI] [PubMed] [Google Scholar]
- Siebert S, et al. 2011. Differential gene expression in the siphonophore Nanomia bijuga (Cnidaria) assessed with multiple next-generation sequencing workflows. PLoS One 6:e22953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring J, et al. 2000. The mesoderm specification factor Twist in the life cycle of jellyfish. Dev Biol. 228:363–375. [DOI] [PubMed] [Google Scholar]
- Spring J, et al. 2002. Conservation of Brachyury, Mef2, and Snail in the myogenic lineage of jellyfish: a connection to the mesoderm of Bilateria. Dev Biol. 244:372–384. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A fast bootstrapping alogorithm for the RAxML web-servers. Syst Biol. 57:758–771. [DOI] [PubMed] [Google Scholar]
- Stierwald M, Yanze N, Bamert RP, Kammermeier L, Schmid V. 2004. The Sine oculis/Six class family of homeobox genes in jellyfish with and without eyes: development and eye regeneration. Dev Biol. 174: 70–81. [DOI] [PubMed] [Google Scholar]
- Teo R, Möhrlen F, Plickert G, Müller WA, Frank U. 2006. An evolutionary conserved role of Wnt-signaling in stem cell fate decision. Dev Biol. 289:91–99. [DOI] [PubMed] [Google Scholar]
- Torras R, Yanze N, Schmid V, Gonzalez-Crespo S. 2004. Nanos expression at the embryonic pole and the medusae phase in the hydrozoan Podocoryne carnea. Evol Dev. 6:362–371. [DOI] [PubMed] [Google Scholar]
- Varshney GK, et al. 2015. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 25:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Wang X. 2013. Organ evolution in angiosperms driven by correlated divergences of gene sequences and expressed patterns. Plant Cell 25:71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanze N, Gröger H, Müller P, Schmid V. 1999. Reversible inactivation of cell-type-specific regulatory and structural genes in migrating isolated striated muscle cells of jellyfish. Dev Biol. 213:194–201. [DOI] [PubMed] [Google Scholar]
- Yanze N, Spring J, Schmidli C, Schmid V. 2001. Conservation of Hox/ParaHox-related genes in the early development of a cnidarian. Dev Biol. 236:89–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.