Abstract
Coevolution between Drosophila and its endosymbiont Wolbachia pipientis has many intriguing aspects. For example, Drosophila ananassae hosts two forms of W. pipientis genomes: One being the infectious bacterial genome and the other integrated into the host nuclear genome. Here, we characterize the infectious and integrated genomes of W. pipientis infecting D. ananassae (wAna), by genome sequencing 15 strains of D. ananassae that have either the infectious or integrated wAna genomes. Results indicate evolutionarily stable maternal transmission for the infectious wAna genome suggesting a relatively long-term coevolution with its host. In contrast, the integrated wAna genome showed pseudogene-like characteristics accumulating many variants that are predicted to have deleterious effects if present in an infectious bacterial genome. Phylogenomic analysis of sequence variation together with genotyping by polymerase chain reaction of large structural variations indicated several wAna variants among the eight infectious wAna genomes. In contrast, only a single wAna variant was found among the seven integrated wAna genomes examined in lines from Africa, south Asia, and south Pacific islands suggesting that the integration occurred once from a single infectious wAna genome and then spread geographically. Further analysis revealed that for all D. ananassae we examined with the integrated wAna genomes, the majority of the integrated wAna genomic regions is represented in at least two copies suggesting a double integration or single integration followed by an integrated genome duplication. The possible evolutionary mechanism underlying the widespread geographical presence of the duplicate integration of the wAna genome is an intriguing question remaining to be answered.
Keywords: lateral gene transfer, horizontal gene transfer, whole-genome sequencing, endosymbiont
Introduction
The alpha-proteobacteria Wolbachia pipientis (Lo et al. 2007) is an intracellular bacterium that infects both arthropods and filarial nematodes (Werren et al. 2008). Wolbachia pipientis is an obligate symbiont in filarial nematodes; its elimination results in sterility or lethality of the nematodes (Taylor et al. 2005; Slatko et al. 2010). On the other hand, the symbiosis between W. pipientis and its arthropod hosts is mainly facultative (but see Hosokawa et al. 2010; Nikoh et al. 2014 for exceptions) and in most cases W. pipientis is parasitic toward its arthropod hosts. The parasitism causes harmful fitness consequences to its host mainly by controlling the host reproductive processes for its own benefit (Stouthamer et al. 1999).
Within a single species transmission of W. pipientis is usually maternal (mother to offspring) which requires the endosymbiont to localize within the host germline to be successfully transmitted to the next generation (Serbus et al. 2008). Due to the extensive association W. pipientis has with its host germline cells, opportunities of lateral gene transfer (LGT) are predicted to occur between the host eukaryotic genome and the endosymbiotic bacterial genome. LGT (sometimes referred as horizontal gene transfer) is the process of genetic exchange between two evolutionary divergent organisms. LGT between W. pipientis and its host has been identified both from the host to the endosymbiont (Woolfit et al. 2009) and from the endosymbiont to the host (Dunning Hotopp 2011).
After LGT, the recipient organism acquires novel genetic material and if the newly acquired genetic material improves the fitness of the recipient species, it will spread through the population ultimately fixing within the species (Long et al. 2013). For example, genes transferred from prokaryotes to eukaryotes have been postulated to be the source of novel genes that gave selective advantage to eukaryotes during adaptation to novel ecological niches (Keeling and Palmer 2008). However, the efficiency of the eukaryotic transcriptional machinery on a prokaryotic gene is questionable. Most LGT products from prokaryotes into eukaryotes are expected to have minimal functionality in the eukaryotic genome eventually accumulating mutations and becoming pseudogenes. Thus, the evolutionary consequence of these LGT elements is an intriguing question to examine.
Evidence of W. pipientis LGT (WLGT) into eukaryotic nuclear genomes, or sometimes referred as nuclear Wolbachia transfers, has been detected in various natural hosts of W. pipientis (Dunning Hotopp 2011). WLGT was discovered in the genome sequence of the filarial nematode Brugia malayi (Dunning Hotopp et al. 2007) where many of the WLGT genes from W. pipientis infecting B. malayi (wBm) were found to be degenerating suggesting their nonfunctionality. However, a recent study has found as many as 227 WLGT genes and genetic fragments with high sequence similarity to the wBm genome scattered across the host B. malayi genome (Ioannidis et al. 2013), and some of these WLGT were hypothesized to retain functionality in the new eukaryotic genome based on the detection of transcripts in different developmental stages of B. malayi. There are other examples of WLGT in nematodes, such as in the parasitic nematode Onchocerca volvulus, where the WLGT predates the speciation with its sister species Onchocerca ochengi (Fenn et al. 2006).
In arthropods, WLGT has been discovered in beetles (Kondo et al. 2002; Aikawa et al. 2009), fruit flies (Dunning Hotopp et al. 2007), mosquitoes (Klasson, Kambris, et al. 2009), parasitoid wasps (Werren et al. 2010), and the tsetse fly (Doudoumis et al. 2012; International Glossina Genome Initiative 2014). The WLGT in nematodes is characterized by small genetic fragments scattered across the host genome (Ioannidis et al. 2013). In contrast, arthropod WLGT is characterized by both large and small segments of genomic DNA integrated into the host genome. For arthropods, the functional consequences of these WLGT products are debatable. Transcription of some of the WLGT genes was detected by quantitative and reverse transcription polymerase chain reaction (PCR), albeit at very low expression levels compared with control genes (Dunning Hotopp et al. 2007; Nikoh et al. 2008). In addition, a recent RNA sequencing study detected only a few WLGT originating reads out of the total Drosophila ananassae RNA pool (Kumar et al. 2012), suggesting that the observed gene expression could be background transcriptional noise. Thus, with potential nonfunctionality, these WLGT elements were suggested as transient evolutionary phenomena that are ultimately decaying to become noncoding junk DNA in the host genome (Blaxter 2007). Indeed, in the case of the beetle Callosobruchus chinensis, many of the WLGT elements were found degenerated and pseudogenized (Nikoh et al. 2008). However, evidence of LGT from other bacterial endosymbionts has shown that the integrations of single genes or operons have become functional in host genomes, such as aphids (Nikoh et al. 2010), mealy bug (Husnik et al. 2013), and psyllids (Sloan et al. 2014).
The vinegar fly D. ananassae shows the most extreme case of WLGT; the whole genome of W. pipientis infecting D. ananassae (wAna) was estimated to have integrated into its host genome (Dunning Hotopp et al. 2007). Further, by sequencing several wAna loci, Choi and Aquadro (2014) have shown that the integrated wAna (wAnaITG) genome exists in D. ananassae lines sampled from much of the host’s geographic range. The study also noted that both infectious wAna (wAnaINF) and wAnaITG genes were highly similar to each other for the regions examined, suggesting that the wAna invasion and whole-genome integration might be relatively recent. However examination of the host D. ananassae mitochondrial DNA (mtDNA) variation, which is in complete linkage disequilibrium with W. pipientis due to their shared maternal inheritance (Hurst and Jiggins 2005), revealed the original wAna invasion more likely originated in the ancestral population of D. ananassae (Choi and Aquadro 2014). These results illustrated that sequencing of only a few loci does not reveal sufficient variation for resolving the joint evolution of wAnaINF and wAnaITG genomes within D. ananassae. However, studies utilizing whole-genome sequencing have been fruitful in elucidating the coevolutionary and population genomics of W. pipientis infecting Drosophila melanogaster (wMel) with its host (Richardson et al. 2012; Chrostek et al. 2013; Early and Clark 2013), suggesting that whole-genome sequencing is a better method for studying of W. pipientis genomics.
Here, we have described the evolution of the wAnaINF and wAnaITG genomes by whole-genome sequencing host D. ananassae flies that harbor either the wAnaINF or the wAnaITG genome. Using well-established computational pipelines, reads originating from the wAna genomes were separated out for further analysis. The wAnaINF genome comparisons, with additional data from mitochondrial genomic variation, reveal that wAnaINF is stably maternally transmitted by its D. ananassae host corroborating the findings of Choi and Aquadro (2014). In addition, we have discovered several nucleotide substitutions and large structural variations that distinguish several wAnaINF genome types. In contrast, the wAnaITG genome is pseudogene-like, accumulating large numbers of highly deleterious mutations. Interestingly, analysis of single nucleotide and copy number variation indicates that the majority of the wAnaITG regions has a minimum of two copies integrated into the D. ananassae genome consistent with independent results reported recently for two lines of D. ananassae by Klasson et al. (2014). The whole-genome integration and duplication of the wAna genome are surprising due to its wide geographic occurrence despite its nonfunctional pseudogenome like characteristics.
Materials and Methods
Genome-Sequenced D. ananassae Strains
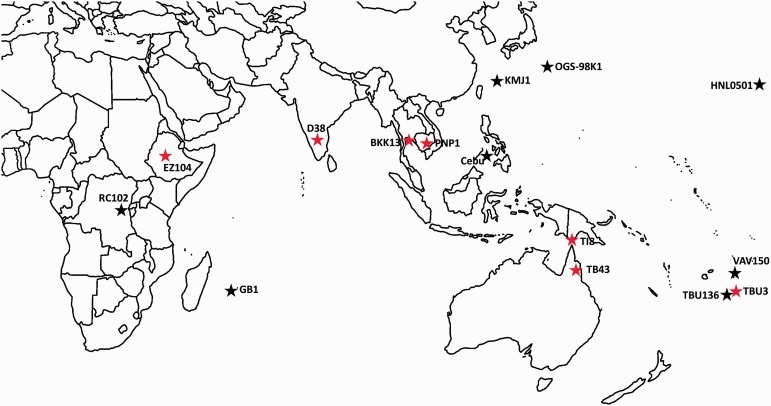
All D. ananassae strains examined in this study originated from the study of Choi and Aquadro (2014). Eight strains with evidence of the wAnaINF genome (Cebu, GB1, HNL0501, KMJ1, OGS-98K1, RC102, TBU136, and VAV150) and seven strains with evidence of only the wAnaITG genome (BKK13, D38, EZ104, PNP1, TB43, TBU3, and TI8) were examined (fig. 1). Each D. ananassae strain that was positive for wAna genes by PCR was treated with the antibiotic tetracycline with concentrations of 200 μl/ml to cure them of wAnaINF. Then, the cured D. ananassae strains were screened by PCR again for the wAna gene and from here we defined a D. ananassae strain to have the wAnaINF genome if no wAna genes were detected after curing, and as a strain having the wAnaITG genome if wAna genes were detected after curing. As it is possible for the D. ananassae strains with the wAnaITG genome to also have the wAnaINF genome, we avoided sequencing both genomes by only genome sequencing cured wAnaITG carrying D. ananassae strains.
Fig. 1.—
Geographic origin of samples analyzed in this study. Drosophila ananassae strains with wAnaINF genomes are indicated with black colored stars and the localities are Cebu (Cebu, Philippines), GB1 (Mauritius), HNL0501 (Oahu, USA), KMJ1 (Kumejima, Japan), OGS-98K1 (Ogasawara, Japan), RC102 (Rwanda), TBU136 (Tonga), and VAV150 (Vava’u, Tonga). Drosophila ananassae strains with wAnaITG genomes are indicated with red colored stars and the localities are BKK13 (Bangkok, Thailand), D38 (Coimbatore, India), EZ104 (Ethiopia), PNP1 (Phnom Pen, Cambodia), TB43 (Trinity Beach, Australia), TBU3 (Tonga), and TI8 (Thursday Island, Australia).
For each strain, DNA was extracted from whole bodies of 12–15 female D. ananassae flies using the Qiagen DNeasy blood and tissue kit. To prepare for genome sequencing, the purified DNA were processed by the Biotechnology Resource Center at Cornell University (http://www.biotech.cornell.edu/biotechnology-resource-center-brc, last accessed August 2015) using Illumina TruSeq DNA sample library prep kit as paired-end 2 × 150 bp samples. DNA libraries were barcoded and pooled for whole-genome sequencing using the Illumina HiSeq 2500 rapid run mode at the Biotechnology Resource Center Genomics Facility at Cornell University.
Reference Genome-Based Realignment and Alignment Statistics Calculation
The program Trimmomatic version 0.32 (Bolger et al. 2014) was used to preprocess the raw reads to trim any remaining adapter sequences and for quality control purposes (Parameters used: ILLUMINACLIP:2:30:10:5, LEADING:3, TRAILING:3, MINLEN:70, and SLIDINGWINDOW:4:15). The Program FastQC version 0.11.1 (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/, last accessed August 2015) was then used to heuristically check and visualize the quality of the raw reads.
The quality-controlled raw reads were then realigned to a reference genome using the program BWA-MEM version 0.7.10 (Li 2013) using default parameters. Currently, a high-quality wAna genome is unavailable, so we used the reference genome sequence of W. pipientis infecting Drosophila simulans strain Riverside (wRi) (Klasson, Westberg, et al. 2009). A previous study has shown high nucleotide identity between the genomes of wAna and wRi (Salzberg et al. 2005). The D. ananassae reference mitochondria genome (GenBank accession number BK006336.1; Montooth et al. 2009) was used to extract D. ananassae mitochondrial reads.
Genome-wide coverage was calculated by counting the number of reads aligned on a given site using the program genomeCoverageBed from the bedtools version 2.17.0 suite (Quinlan and Hall 2010). Total number of reads aligning to each reference genome was calculated using the program samtools version 1.0 (Li et al. 2009). For all wAna samples, read coverage per site was normalized against the mean read coverage of the longest scaffold assembled from the draft D. ananassae nuclear genome (scaffold 13340; GenBank accession number CH902617.1).
wAnaINF Copy Number Variation Analysis
The program CNVnator version 0.3 (Abyzov et al. 2011) was used to detect regions with significant changes in the copy number of the wAnaINF-aligned BAM files. CNVnator divides the whole genome into bins to calculate the mapping density and find regions with significantly different read depth. For the wAnaINF sample, genome-wide coverage was equal for most of the regions except for the few candidate regions with variation in the copy number (see Results section) so that CNVnator was appropriate to detect those copy number variations. In contrast, the wAnaITG samples had varying genome coverage throughout the genome making CNVnator an unsuitable method; thus, we used experimental methods to directly quantify the differences in the genome coverage (see below).
Quantitative PCR to Analyze wAnaITG Genome Copy Number
To verify the variable read coverage observed in the wAnaITG genomes (see Results section), quantitative PCR (qPCR) was conducted on three D. ananassae strains with the wAnaITG genome (BKK13, D38, and TI8). Based on our wAnaITG genome coverage results, a low coverage region (LOW) corresponding to gene YP_002726693.1 (coordinate: 40,653–41,915) and high coverage region (HIGH) corresponding to gene YP_002727436.1 (coordinate: 998,610–1,000,511) were selected as candidate regions and compared with the D. ananassae rp49 gene. qPCR primers were designed using the online version of the program primer3plus (Untergasser et al. 2012; http://primer3plus.com/cgi-bin/dev/primer3plus.cgi, last accessed August 2015), and primer sequences are provided in supplementary table S1, Supplementary Material online. qPCR reactions were conducted using iTaq Universal SYBR Green Supermix (Biorad), and fluorescence was detected using the ViiA7 Real-Time PCR system (Life Technology). PCR mixes were denatured at 95 °C for 10 min followed by a 40 cycle amplification stage, which consisted of 95 °C for 15 s followed by a data collection step of 53 °C for 1 min. A melt curve stage was conducted at the end with 95 °C for 15 s, then 60 °C for 1 min, followed by the data collection step where the temperature was raised 0.05 °C/s until 95 °C.
Single females from each of the three examined D. ananassae strains (BKK13, D38, and TI8) were combined and extracted for DNA using our previous methods of DNA extraction. This master mix was then serially diluted to fit a standard curve for the three genes (LOW, HIGH, and rp49) and estimate the efficiency of the primers. This standard curve was then used to convert the observed Ct values for each sample into its relative concentration. Afterwards, the HIGH and LOW relative concentrations were compared with the relative concentration of rp49. Three biological replicates and three technical replicates were conducted for each D. ananassae strains. All qPCR reactions were conducted in the same plate to keep the reaction condition and detection of fluorophores consistent.
Detection of Nucleotide and Structural Variations
BAM files generated from the previous reference genome-based raw read realignment step were then processed according to the guidelines of GATK’s BestPractice version 3.3 for downstream analysis (https://www.broadinstitute.org/gatk/guide/best-practices, last accessed August 2015). Briefly, this process involves removing duplicate reads using the program Picard version 1.115 (http://broadinstitute.github.io/picard/, last accessed August 2015), and realigning regions around insertion and deletions (INDELs) using the GATK suite version 3.2-2 (https://www.broadinstitute.org/gatk/, last accessed August 2015). The processed BAM file was then used to call INDEL and single nucleotide polymorphism (SNP) variants using the joint genotyping method of GATK. The program VariantFiltration from the GATK suite was then used to apply hard filters on the raw variant calls (parameters for INDEL filtering: QD < 2.0, FS > 200.0, ReadPosRankSum < −20.0; parameters for SNP filtering: QD < 2.0, FS > 60.0, MQ < 40.0, HaplotypeScore > 13.0, MappingQualityRankSum < −12.5, ReadPosRankSum < −8.0; please see https://www.broadinstitute.org/gatk/gatkdocs/index [last accessed August 2015] for details on parameters). Additionally for the SNP filtering stage, the previous INDEL calls were used to mask SNP calls that coincided with the INDEL position as these may cause false polymorphism calls because of alignment errors. SNP variants detected among our genomes and used for downstream analysis are reported in supplementary data, Supplementary Material online, in a tab-delimited format. Raw VCF files for the SNP and INDELs are available upon request.
To detect small- and large-sized structural variations, specifically INDELs and inversions, we used the program pindel version 0.2.5 (Ye et al. 2009), which uses a pattern growth algorithm to detect structural variations at base pair resolution. After the structural variations were predicted by pindel the pindel2vcf from the pindel suite was used to filter out variants that were predicted with less than four reads as well as any variants due to homopolymer and microsatellite repeats as these could represent sequencing errors (pindel2vcf parameters: -e 4 -ir 2 -il 10 -pr 1 -pl 10).
The functional effects of each SNPs and INDELs were predicted using the program snpEff version 4.0 e (Cingolani et al. 2012).
PCR Amplification of Pindel-Predicted Structural Variations
To verify the large deletions and inversions predicted by pindel, a subset of those predicted large structural variations were selected for PCR verification. We designed PCR primers that flank the predicted breakpoints of each of the nine large structural variant (primers are listed in supplementary table S2, Supplementary Material online). Each PCR reaction consisted of 5 μl of 5× GoTaq Reaction buffer, 0.5 μl of 10 mM dNTP, 2.5 μl of 10 mM upstream and downstream primer each, 0.25 μl of GoTaq DNA polymerase (Promega), and 13.75 μl of water. PCR for all primer pairs started with an initial denaturation step with 94 °C for 2 min for one cycle followed by 35 cycles of 94 °C for 30 s, then a primer annealing step for 45 s with the following annealing temperatures: 60 °C for SV4, SV7, SV8, SV9; 63 °C for SV1, SV2, SV3; and 65 °C for SV5, SV6. Annealing was followed by an extension step at 72 °C for 1 min for SV1, SV5, SV6; 1:30 min for SV9; 1:45 min for SV2, SV7, SV8; 2:30 min for SV4; and 3 min for SV3. PCR products were run on a 1% agarose and digital images processed using the program ImageJ (http://imagej.nih.gov/ij/, last accessed August 2015).
Phylogenetic Analysis
Nucleotide variants that were called using above methods were then used for phylogenetic analysis. Phylogenetic reconstruction was carried out using the maximum-likelihood method implemented in PhyML version 3.0 (Guindon et al. 2010), the HKY (Hasegawa–Kishino–Yano) model of DNA substitution (Hasegawa et al. 1985), and substitution rate variation across sites modeled with a gamma distribution with four rate categories (commonly known as the HKY + G model). Confidence of the inferred phylogenetic tree was estimated using 1,000 bootstrap replicates. Trees were plotted and manipulated using FigTree ver 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/, last accessed August 2015) and the R package phytools (Revell 2012).
Preparation of wAnaITG Genome for Phasing Heterozygous Genotypes
We initially assumed that the wAnaITG genome existed in a diploid state in D. ananassae strains with evidence of the integration, and thus attempted phasing sites called as heterozygotes by first making the cytotype of the wAnaITG genome haploid. Male D. ananassae strains with the wAnaITG genome were mated to virgin females of a D. ananassae strain that had neither the wAnaINF genome nor wAnaITG genome. Crossing males with the wAnaITG genome to females lacking both wAnaINF and wAnaITG genomes prevents any maternal factors (i.e., W. pipientis) from being inherited, and only the paternal nuclear DNA will be transmitted to the next generation. Both males and females of the cross were cured of W. pipientis with 200 μl/ml tetracycline for four generations to clear wAnaINF genomes. We collected only the F1 progeny females in case there were integrations in both the autosome and X chromosome, thus retaining equal coverage between the two different chromosomal backgrounds. Additionally to keep one copy of the haploid wAnaITG genome for downstream phasing analysis, we chose a single virgin female for DNA extraction and subsequent genome sequencing.
Single female flies were individually homogenized with a clean pestle in a lysis buffer (50 mM Tris–HCL ph8.2, 100 mM ethylenediaminetetraacetic acid, 100 mM NaCl, 1% SDS) then treated with Proteinase K and RNase I. DNA extraction was conducted using a standard phenol–chloroform–isoamyl protocol followed by an overnight ammonium acetate DNA precipitation. A detailed protocol is available at request. The purified DNA was then prepared for genome sequencing following the same protocol previously described.
In Silico Prediction of Breakpoints between wAnaITG and Host Nuclear Genome
In an effort to determine breakpoints between the wAna genomic fragments and the host genome, we treated those breakpoints as translocation events (i.e., exchange of chromosomal DNA between nonhomologous chromosomes) and used the program Lumpy ver 0.2.6 (Layer et al. 2014) to detect the breakpoints between the wAna genome and the host genome. Lumpy combines multiple signals from read-pair, split-reads, and read-depth of a sample to identify structural variations. We realigned our original raw reads to a reference genome that included a subset of the reference D. ananassae CAF1 assembly (Clark et al. 2007) and the wRi genome. Only a subset of scaffolds was selected because the D. ananassae reference genome is largely a draft genome that includes thousands of unassembled scaffolds. Further, it is possible that some scaffolds contain W. pipientis sequences that were accidently incorporated because the sequenced line of D. ananassae contained W. pipientis. In fact, Salzberg et al. (2005) were able to reassemble a draft wAna genome from the raw sequence repository of the D. ananassae reference genome. We selected large scaffolds that were identified as syntenic to the D. melanogaster Muller elements (Schaeffer et al. 2008) (see supplementary table S3, Supplementary Material online, for full list of scaffolds; GenBank assembly accession number: GCA_000005115.1). Using this custom reference genome, paired-end reads that contain both the bacterial and host sequences were aligned to their appropriate genome. Next, discordant paired-end and split-end reads were extracted from the BAM files using samtools. These reads were then processed using the pairend_distro.py script from the Lumpy suite to estimate the library size, mean, and standard deviation. The sample statistics of the library were then prepared as a configuration file with the following Lumpy parameters to detect translocation events using two different libraries: 1) Discordant paired-end library analysis: Read length = 150; min nonoverlap = 150; discordant z = 4; back distance = 20; weight = 1; id = 1; min mapping threshold = 40; and 2) split reads library analysis: Back distance = 20; weight = 1; id = 2; min mapping threshold = 40. Inferred breakpoints between D. ananassae and wAna were only used if they were supported by reads with a minimum mapping quality of 40, had evidence from both paired-end and split-end read libraries, and had a minimum of six supporting reads.
Our analysis revealed that Lumpy identified many breakpoints in which the host D. ananassae sequence corresponded to potential transposable elements (TEs) (described further in Results). To further characterize these breakpoints, we used the program RepeatMasker version open 4.0 (http://www.repeatmasker.org/, last accessed August 2015; developed by A.F.A. Smit, R. Hubley, and P. Green) to identify repetitive DNA and TEs across the reference D. ananassae genome. The Drosophila RepBase repeat library Update 20140131 (http://www.girinst.org/repbase/, last accessed August 2015) was used to identify specific TE families.
Results
Raw Read Alignment Statistics
Whole adult flies representing each of the 15 strains of D. ananassae that were previously screened for the presence of wAnaINF and wAnaITG genomes (Choi and Aquadro 2014) were genome sequenced, and all wAna originating reads were computationally extracted using the reference wRi genome (alignment statistics are shown in table 1). Raw reads from both wAnaINF and wAnaITG genomes comprised on average 2% of the total reads. However, the proportion of wAna reads was variable between the two genomes; the wAnaINF genomes had more variability among lines, ranging from 1.3% of the total reads in strain HNL0501 to 3.8% in strain TBU136. Mirroring this result, the average read depth (RDavg) of the wAnaINF genomes, normalized relative to the RDavg of the D. ananassae nuclear genome, also varied from 1.7-fold higher in HNL0501 wAnaINF to 5.2-fold higher in TBU136 wAnaINF. For wAnaITG, a single genome integration of wAna would be expected to have an RDavg equal to that of the D. ananassae nuclear genome. However, the RDavg for wAnaITG was roughly 2.6-fold higher.
Table 1.
Raw Read Realignment Statistics for mtDNA and wAna Genomes
| Strain | Total Reads | wAna Originating Reads | Proportion of wAna Reads | wAna RDavg | wAna Ratio | mtDNA Originating Reads | Proportion of mtDNA Reads | mtDNA RDavg | mtDNA Ratio |
|---|---|---|---|---|---|---|---|---|---|
| Raw reads from wAnaITG genome | |||||||||
| BKK13 | 35,914,931 | 721,253 | 0.020 | 74.4 | 2.5 | 238,401 | 0.003 | 2,376.8 | 78.5 |
| D38 | 40,631,930 | 987,102 | 0.024 | 101.8 | 3.1 | 299,310 | 0.007 | 2,977.6 | 89.6 |
| EZ104 | 29,830,655 | 613,044 | 0.021 | 63.2 | 2.6 | 216,173 | 0.007 | 2,150.7 | 86.7 |
| PNP1 | 90,416,482 | 1,766,984 | 0.020 | 180.3 | 2.4 | 291,938 | 0.003 | 2,681.3 | 35.1 |
| TB43 | 28,871,004 | 621,913 | 0.022 | 64.0 | 2.7 | 208,458 | 0.007 | 2,071.6 | 87.2 |
| TBU3 | 39,325,872 | 1,001,506 | 0.025 | 103.4 | 3.2 | 292,405 | 0.007 | 2,908.0 | 89.1 |
| TI8 | 37,948,414 | 853,466 | 0.022 | 88.0 | 2.8 | 265,647 | 0.007 | 2,645.9 | 83.2 |
| Median | 37,948,414 | 853,466 | 0.022 | 88.0 | 2.7 | 265,647 | 0.007 | 2,645.9 | 86.7 |
| Raw reads from wAnaINF genome | |||||||||
| Cebu | 141,458,302 | 2,971,843 | 0.021 | 304.7 | 2.6 | 528,382 | 0.004 | 5,262.5 | 44.6 |
| GB1 | 33,180,346 | 640,906 | 0.019 | 64.3 | 2.6 | 228,571 | 0.007 | 2,121.2 | 86.7 |
| HNL0501 | 19,121,957 | 243,774 | 0.013 | 24.9 | 1.7 | 101,483 | 0.005 | 1,008.5 | 68.3 |
| KMJ1 | 76,690,462 | 1,457,894 | 0.019 | 149.2 | 2.3 | 208,845 | 0.003 | 1,894.7 | 28.9 |
| OGS98-K1 | 30,352,531 | 709,801 | 0.023 | 72.1 | 3.3 | 78,842 | 0.003 | 784.6 | 35.6 |
| RC102 | 82,433,612 | 1,387,896 | 0.017 | 141.5 | 2.1 | 246,883 | 0.003 | 2,258.8 | 32.8 |
| TBU136 | 20,721,322 | 794,974 | 0.038 | 81.3 | 5.2 | 81,676 | 0.004 | 811.3 | 51.5 |
| VAV150 | 20,791,631 | 535,975 | 0.026 | 54.4 | 3.5 | 122,822 | 0.006 | 1,153.5 | 75.2 |
| Median | 31,766,439 | 752,388 | 0.020 | 76.7 | 2.6 | 165,834 | 0.004 | 1,524.1 | 48.1 |
Note.—RDavg, average read depth of a sample; Ratio, value obtained by dividing the mean depth of the wAna or mtDNA genome to the mean depth of D. ananassae nuclear genome (scaffold 13370).
Alignment statistics for the mtDNA genome indicated 0.3–0.7% of the total raw reads originated from the mtDNA for both D. ananassae strains with the wAnaINF and wAnaITG genomes. Deep genome coverage was observed for the mtDNA, and compared with the D. ananassae nuclear genome, the D. ananassae strains with the wAnaITG genomes had more mitochondrial genome copies than D. ananassae strains with the wAnaINF genomes (table 1). However, this difference is probably due to the tetracycline treatment of wAnaITG carrying D. ananassae to eliminate any residual wAnaINF genomes. A previous study has shown that tetracycline increases mtDNA copy in Drosophila that was not infected with W. pipientis (Ballard and Melvin 2007).
Analysis of wAnaINF and wAnaITG Genome Coverage
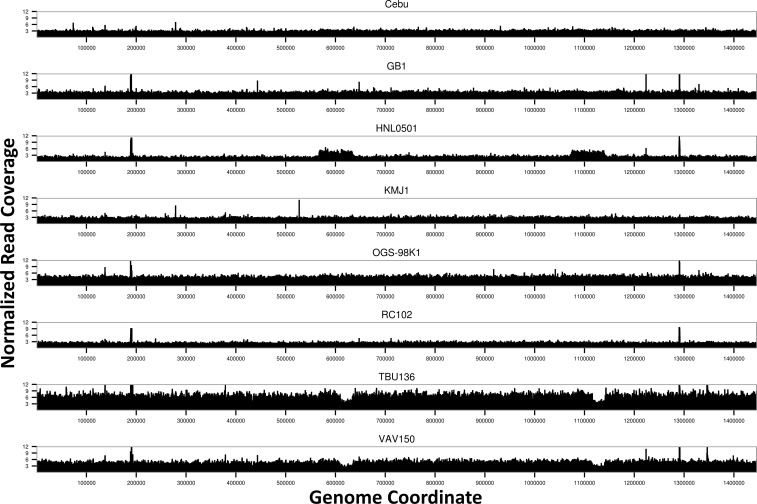
The read coverage for each wAna genome was visualized by plotting the per-site read depth normalized against the D. ananassae nuclear genome coverage. Most of the regions of the wAnaINF genomes had equal coverage across the genome (fig. 2). Spikes of increase in coverage across small regions were frequently observed (i.e., region around 0.18 and 1.28 Mb), and these regions mainly corresponded to the ribosomal RNA (rRNA) and transfer RNA (tRNA) genes of the wRi genome. As the extracted DNA were from well-fed adult flies, it is likely that these spikes in coverage were due to additional reads originating from the gut microbiota of the host. These regions were ignored for further downstream analysis.
Fig. 2.—
Genome-wide read coverage for the eight wAnaINF genomes. Per-site read depth normalized against the D. ananassae nuclear genome (scaffold 13340) coverage is shown against the reference wRi genome coordinate. Note that regions of tRNA and rRNA, comprising 0.48% of the genome, were expected to have spurious reads from other endosymbiotic bacteria, leading to spikes of read coverage and were ignored from subsequent analysis (see text).
The program CNVnator was used to detect regions with significant changes in the copy number among the wAnaINF samples. As expected from the wAnaINF genome coverage plots, no regions with significant changes in the copy number were detected for D. ananassae strains Cebu, GB1, KMJ1, OGS-98K1, and RC102. In the wAnaINF genomes of strains TBU136 and VAV150, regions between coordinates 0.61–0.63 and 1.12–1.14 Mb were estimated to have a 0.5-fold decrease (P < 0.0001) in copy number compared with their genome-wide background coverage. In the HNL0501 strain, the wAnaINF genome was predicted to have a significant 2-fold increase in copy number (P < 0.0001) between coordinates 0.57–0.63 and 1.07–1.14 Mb. The two regions between 0.61–0.63 and 1.12–1.14 Mb that showed copy number variation overlapped among wAnaINF genomes from strains HNL0501, TBU136, and VAV150.
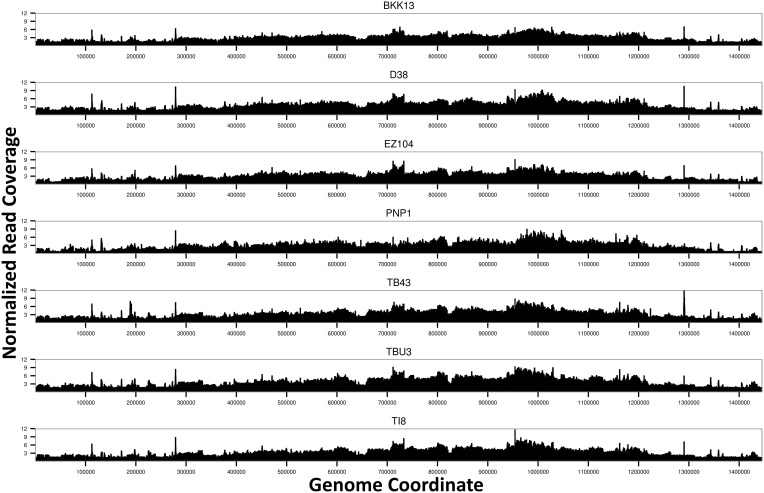
In contrast to the wAnaINF genomes, the wAnaITG genomes showed strikingly heterogeneous coverage across the genome (fig. 3), with low coverage in the beginning of the genome increasing to a maximum coverage around coordinates 1 Mb and subsequently decreasing. Regions with low and high coverage were also correlated across all seven wAnaITG genomes. The normalized read depth across the wAnaITG genomes indicated that the majority of the regions had at least double the read depth compared with the D. ananassae nuclear genome.
Fig. 3.—
Genome-wide read coverage for the seven wAnaITG genomes. Per-site read depth normalized against the D. ananassae nuclear genome (scaffold 13340) coverage is shown against the reference wRi genome coordinate. Note that regions of tRNA and rRNA, comprising 0.48% of the genome, were expected to have spurious reads from other endosymbiotic bacteria, leading to spikes of read coverage and were ignored from subsequent analysis (see text).
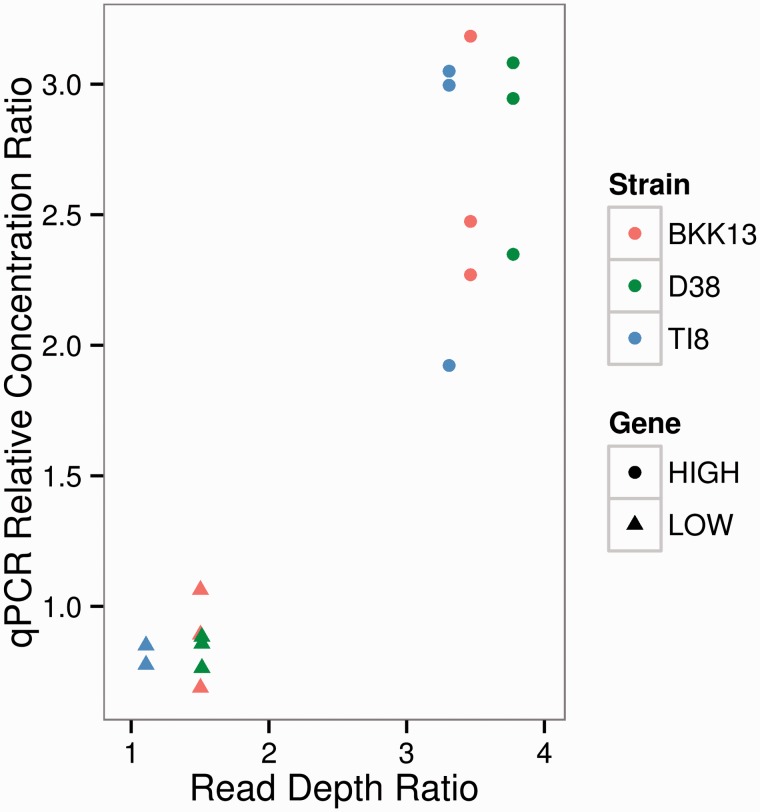
We verified that the high and low coverage regions of the wAnaITG segments represented actual differences in the genome sequence copy number using qPCR. A candidate high copy region (HIGH) at coordinate 998,610–1,000,511 and candidate low copy region (LOW) at coordinate 40,653–41,915 were each compared with the D. ananassae nuclear gene rp49 in three representative strains (BKK13, D38, and TI8). Based on genome coverage results compared with the D. ananassae rp49 gene, LOW was expected to have a 1.5-fold increase whereas HIGH was expected to have a 3.5-fold increase in copy number (fig. 4). Qualitatively, the qPCR results were concordant with the genome coverage results: For all three strains, copy number for LOW was 0.8-fold that of the rp49 gene, whereas HIGH was estimated to be 2.6-fold higher than rp49. Thus, the difference in read coverage across the wAnaITG genome was due to differences in the copy number of wAnaITG integrated into the D. ananassae nuclear genome.
Fig. 4.—
Computational and qPCR estimates of copy number for the LOW and HIGH regions of the wAnaITG genome. Relative copy number of LOW and HIGH region is compared with D. ananassae rp49. The x axis represents read depth of LOW and HIGH region divided by read depth of rp49. The y axis represents relative concentration of LOW and HIGH region divided by relative concentration of rp49.
Analysis of wAnaINF Genome Variants
Due to the relatively high read depth for both mtDNA and wAna genomes, variant detection programs were used to call SNPs, INDELs, and inversion variations for wAnaINF both within and among strains of D. ananassae. The program HaplotypeCaller from the GATK suite was used to call single nucleotide variants. This program assumes diploidy of the organism when calling each variants genotype. However, wAnaINF genomes are expected to be haploid, and most of the variants are predicted to be called as homozygous genotypes within a strain. Sites called as heterozygotes within a single strain could be due to infection by multiple wAna genomes, a duplicated region differing in one duplicate by a mutation, or a false variant call from sequencing error. Levels of variation among the eight wAnaINF genomes were assessed as follows: A site was considered monomorphic when all eight individuals have the same base call and polymorphic when the base calls for at least one line differ from the others. Polymorphic sites that had at least one line with a heterozygote call were also tabulated and were abbreviated as PSHET.
From the total 1,445,873-bp reference wRi genome, base calls were made for 1,194,063 bp (82.6%) for the sample of wAnaINF genomes (table 2). Initially base calls from the wAnaINF genomes were compared with the approximately 7,000-bp regions that Choi and Aquadro (2014) reported as monomorphic using Sanger sequencing, and we verified those regions to be monomorphic in our new data as well. Compared with the reference wRi genome from D. simulans, we detected ten fixed differences for the wAnaINF genomes suggesting that these are wAnaINF-specific mutations.
Table 2.
wAna and Mitochondrial Genome Variation
| wAna Status | N | Analyzed Sites | Fixed | STOTAL | SSINGL | SHET | π | INDELTotal | INDELHOM | INDELHET |
|---|---|---|---|---|---|---|---|---|---|---|
| wAna genomes | ||||||||||
| wAnaINF | 8 | 1,194,063 | 10 | 125 | 39 | 8 | 3.65E-05 | 10 | 6 | 4 |
| wAnaITG | 7 | 1,228,381 | 42 | 2,259 | 25 | 2,234 | NA | 122 | 8 | 114 |
| mtDNA genomes | ||||||||||
| Total | 15 | 14,905 | 5 | 165 | 59 | 11 | 2.84E-03 | 0 | — | — |
| wAnaINF | 8 | 14,905 | 5 | 127 | 34 | 11 | 3.02E-03 | 0 | — | — |
| wAnaITG | 7 | 14,905 | 25 | 109 | 67 | 0 | 2.45E-03 | 0 | — | — |
Note.—N, sample size; STOTAL and INDELTotal, total number of polymorphic sites with a single nucleotide and small sized INDEL (<100 bp) variants, respectively; SSINGL, total number of singletons; SHET and INDELHET, total number of polymorphic sites with at least one line with a heterozygote variant call (PSHET), respectively; π, number of pairwise differences per site; NA, not applicable as there were many heterozygous polymorphisms that appear due to the multicopy nature of the wAnaITG genome (see Results).
Among the eight wAnaINF genomes, PSHET were examined more closely, as other symbiotic bacterial reads could align to homologous regions of the reference genome and cause false heterozygote calls (as was observed in the rRNA and tRNA regions). We conservatively chose to discard PSHET sites that were clustered within 10-bp windows among the wAnaINF genomes. Among the eight wAnaINF genomes, there were 125 polymorphic sites where eight polymorphic sites had at least one line of wAnaINF called as a heterozygote. Among these eight PSHET, seven of the eight polymorphic sites had a single line called as a heterozygote whereas the remaining lines were called as the reference allele. This suggested that those lines with the heterozygote calls were likely to be false variant calls. A single PSHET had five of the eight lines called as a heterozygote but this was located within a pseudogenized gene (WRi_p10660). Thus, we infer that heteroplasmy was minimal and that only a single strain of wAna infected each D. ananassae strain. The average genome-wide number of pairwise nucleotide differences per site (π) was 3.65 × 10−5 among the eight wAnaINF genomes.
The program pindel was used to identify small-sized INDELs (<100 bp) across each wAnaINF genomes. Among the eight wAnaINF genomes, we detected a total of ten polymorphic sites with small-sized INDELs (table 2). Four of these polymorphic sites were a PSHET; however, for each PSHET the line with the heterozygote call had an INDEL with a single base pair deletion. This suggested that sequencing errors could have caused the false variant calls in those lines with a site called as a heterozygous INDEL.
We also used pindel to detect large structural variations, such as large deletions (>100 bp) and inversions, that were both polymorphic and fixed (compared with the reference wRi genome) among the wAnaINF genomes (supplementary tables S4 and S5, Supplementary Material online). The sizes of these predicted large deletions ranged from 619 to 2,544 bp. Examining the gene annotations of these large deletions, most involved the loss of a TE that existed in the reference wRi genome. Interestingly some of the inversions had predicted breakpoints that coincided with the same genome coordinates as for the large deletions. When the program lumpy was used to detect structural variations, no significant inversions or large deletions were found.
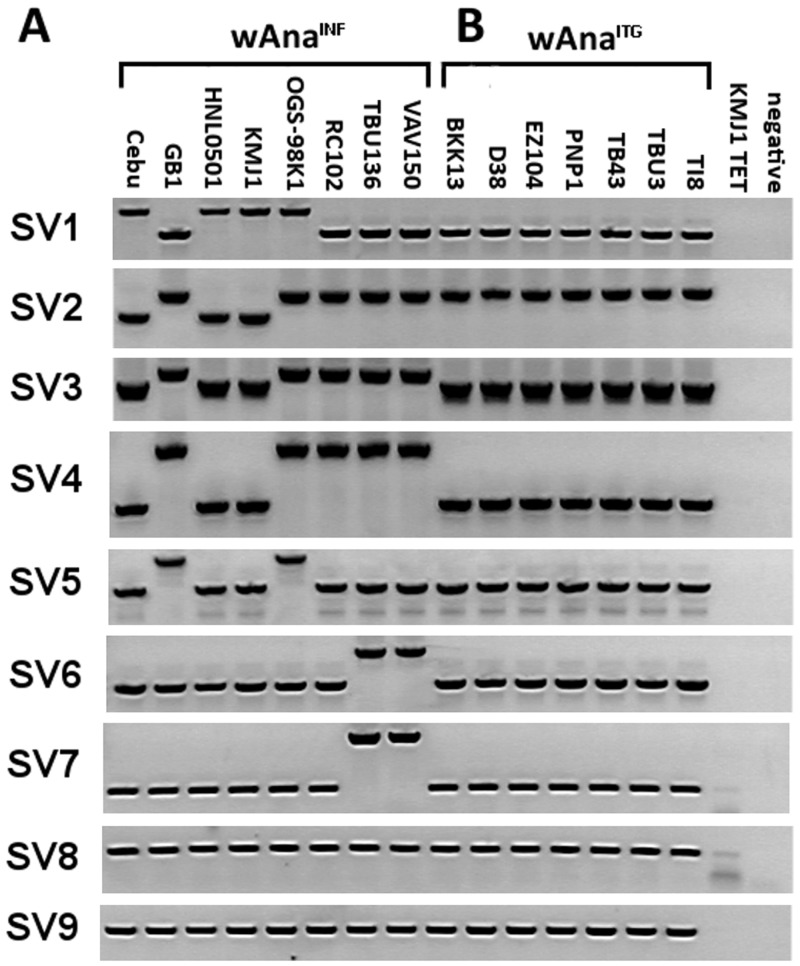
We designed PCR primers that flanked the breakpoints found by pindel and used PCR amplification to verify the computationally predicted structural variations (see supplementary table S2, Supplementary Material online, for genome coordinates of the PCR primers). Nine of the predicted large structural variations were PCR amplified and results are shown in figure 5A. There were seven large structural variations (SV1–SV7) that were polymorphic among the eight wAnaINF samples. The remaining two large structural variations (SV8 and SV9) were deletions of a TE in the reference wRi genome, and this deletion was observed in each of our wAnaINF samples as the PCR product was smaller than predicted from the wRi reference genome. When compared with the computational prediction, six of the large deletions detected by pindel (SV2, SV3, SV4, SV7, SV8, and SV9) were verified by PCR to be true large deletions. The three inversions computationally inferred by pindel (SV1, SV5, and SV6; supplementary table S2, Supplementary Material online) were examined by trying to PCR amplify one side of the predicted breakpoints. Results showed that these pindel-predicted inversions were not inversions but in fact were insertions of a large genetic sequence, likely from a TE.
Fig. 5.—
PCR amplifications of large structural variations found in (A) wAnaINF and (B) wAnaITG samples. PCR results for the nine large structural variations are shown according to the sample. KMJ1 TET, tetracycline-treated KMJ1 strain that is cured of W. pipientis infection and does not have evidence of the integration; Negative, negative control using water for the PCR reaction. Note SV8 and SV9 are large deletions that exist in all wAnaINF and wAnaITG samples.
Analysis of wAnaITG Genome Variants
In preparation of this manuscript, Klasson et al. (2014) reported evidence of extensive duplication of the wAnaITG genome from two samples of D. ananassae originating from Hawaii, the United States, and Mumbai, India. A third sample from Java, Indonesia, on the other hand, had no evidence of duplications for the wAnaITG genome. Further, they have conducted in situ hybridization experiments to determine the integration site of wAnaITG genome and suggested its localization to the fourth chromosome of D. ananassae (see “Investigating the integration site of the wAnaITG genome” for our computational analysis of verifying the fourth chromosome as the site of integration). Compared with Klasson et al. (2014) we have sampled from a wider geographic distribution and discovered that all of our samples had evidence of increased copy number throughout the wAnaITG genome (figs. 3 and 4). As Klasson et al. (2014) did not examine the nucleotide or structural variations of the wAnaITG genome, we next analyzed the genome variants existing in our sample of wAnaITG genomes.
Using HaplotypeCaller to call variant base calls for each wAnaITG genome of D. ananassae could result in a site being called as a heterozygote either because the two allelic copies of a specific wAnaITG sequence differ, or because the two duplicated regions on the same chromosome differ from each other (much like paralogs of a single gene might differ). Thus, among the wAnaITG genomes we also examined PSHET carefully.
Analysis of levels of nucleotide variation among the seven wAnaITG genomes compared with the eight wAnaINF genomes (table 2) revealed both greater differentiation from the wRi reference genome and more variation among wAnaITG genomes. Analyzing a total of 1,228,381 bp (85.0%), the wAnaITG genomes had 42 fixed differences from the reference wRi genome (compared with only 10 for wAnaINF).
Like the variant detection from the wAnaINF genomes, we took a conservative approach to filter out potentially false variants called as heterozygotes by examining each wAnaITG genome using a 10-bp sliding window, and masking windows with more than three variant base calls. These windows were considered as variant clusters likely due to misalignments and were thus ignored for downstream analysis. After our hard filtering we found 2,259 polymorphic sites among the 7 wAnaITG genomes, with 2,234 (98.9%) of these being PSHET (table 2). The remaining 25 polymorphic sites had a singleton variant. In contrast to results for 7 kb of the wAnaITG genome found to be invariant by Sanger sequencing (Choi and Aquadro 2014), our whole-genome analysis revealed a much higher number of polymorphic sites in other regions of the genome; this contrast will be addressed in the Discussion section.
The seven wAnaITG genomes had a higher number of polymorphic sites with small-sized INDELs (<100 bp) (table 2) compared with the eight wAnaINF genomes. Among the seven wAnaITG genomes the majority of the INDEL polymorphic sites was a PSHET (114 of 122 polymorphic sites). However, for each PSHET the line with the heterozygous INDEL call had a variant of size 13 bp on average. This suggested that in contrast to the heterozygous INDEL calls from wAnaINF genomes, those in wAnaITG were not false INDEL calls from sequencing errors.
Among the wAnaITG genomes, a total of ten large deletions (>100 bp) were detected using pindel and all were fixed between the wAnaITG samples and the wRi reference genome (supplementary table S4, Supplementary Material online). All of these large deletions were the same variants that were predicted in our wAnaINF genome analysis (fig. 5A). We verified these large deletions in the wAnaITG samples using the same primers from the PCR analysis of the wAnaINF large structural variation. Results showed that unlike the wAnaINF genomes, there were no variations in large deletions among the seven wAnaITG genomes (fig. 5B). Further, PCR results showed no large INDEL that was able to differentiate the wAnaINF or wAnaITG genomes and all large INDEL variants among the wAnaITG genomes existed within the wAnaINF genomes (fig. 5A and B).
Analysis of Mitochondrial Genome Variants
For the D. ananassae mitochondrial genomes, we analyzed 14,905 bp (99.9%) of the total 14,920 bp of the D. ananassae mtDNA genome (table 2). We discovered a total of 165 polymorphic sites, each with a single nucleotide variant among the mitochondrial genomes from both D. ananassae with the wAnaINF and wAnaITG genomes. We compared our computationally called polymorphic sites with the Sanger-sequenced mtDNA polymorphic sites of Choi and Aquadro (2014), and were able to verify 34 of the 35 segregating sites suggesting a high true positive rate of variant calling for our new results. Out of the total polymorphic sites, there were 11 PSHET where the lines called as a heterozygote were all from individuals with the wAnaINF genomes. However, all 11 PSHET were from only a single line called as a heterozygote suggesting that those sites were called as heterozygotes due to sequencing errors. Compared with the reference D. ananassae mitochondrial genome, individuals with the wAnaITG genomes had 25 fixed mtDNA differences whereas individuals with the wAnaINF genomes had 5 fixed differences; thus, the reference mitochondrial genome was more similar to the mtDNA haplotypes associated with the wAnaINF genomes. Drosophila ananassae strains with only the wAnaITG genomes had twice as many singletons as individuals with only the wAnaINF genomes, whereas the average mitochondrial genome-wide π across all D. ananassae strains was 2.84 × 10−3. No significant small- and large-sized INDELs or inversions were detected in the mtDNA.
Functional Classification of Polymorphic Site Variants among wAnaINF and wAnaITG Genomes
We used the program snpEff to infer functional consequences of the single nucleotide and small INDEL variants observed among the wAnaINF and wAnaITG genomes relative to the reference wRi genome (table 3). We found 52.8% of the polymorphic single nucleotide variant sites among the wAnaINF genomes segregated for nonsynonymous variants, all of which resulted in missense mutations. The remaining 47.2% of the single nucleotide polymorphic sites segregated variants of synonymous or intergenic sites. In contrast, among the wAnaITG genomes, 61.1% of polymorphic sites segregated for single nucleotide variants resulting in nonsynonymous changes, with 56.3% of those variants being missense mutations and 4.8% of those variants being nonsense mutations.
Table 3.
Functional Classification of Variants among the wAnaINF and wAnaITG Genomes
| Genome | Single Nucleotide Variant |
Small-Sized INDEL (<100 bp) |
|||||
|---|---|---|---|---|---|---|---|
| Coding Region |
Intergenic Region | Coding Region |
Intergenic Region | ||||
| NSynMis | NSynNon | Syn | Frameshift | Codon Δ | |||
| wAnaINF | 66 (52.8%) | 0 | 22 (17.6%) | 37 (29.6%) | 0 | 1 (10.0%) | 9 (90.0%) |
| wAnaITG | 1,271 (56.3%) | 108 (4.8%) | 408 (18.1%) | 472 (20.9%) | 67 (54.9%) | 19 (15.6%) | 36 (29.5%) |
Note.—NSynMis, number of polymorphic sites segregating on a nonsynonymous site with missense mutations; NSynNon, number of polymorphic sites segregating on a nonsynonymous site with nonsense mutations; Syn, number of polymorphic sites segregating on a synonymous site; Codon Δ, number of polymorphic sites with in-frame deletions. Proportions are indicated in parenthesis.
For small-sized INDELs, we observed ten polymorphic sites among the wAnaINF genomes (table 2) with 9 out of 10 of them occurring in intergenic regions. A single polymorphic site occurred in the coding region with a variant that caused an in-frame deletion (table 3). Among the wAnaITG genomes there were 122 polymorphic sites with small-sized INDELs (table 2), 60.5% of which occurred within a coding region. Of those, 54.9% had a variant resulting in a frameshift mutation and 15.6% had a variant causing an in-frame INDEL change (table 3). Thus, the wAnaITG genomes had an increased number of polymorphic sites with single nucleotide variants and small-sized INDELs that were predicted to cause a strongly deleterious effect (i.e., nonsense mutation and frameshift mutations) in a functioning wAna genome.
Phylogenomic Analysis of mtDNA and wAna Genomes
The single nucleotide variants observed for mtDNA and wAna were used to estimate the phylogeny of each genome we sampled, using a maximum-likelihood method from the program PhyML. The phylogenies of both the host D. ananassae mtDNA genome as well as wAna were examined because, like the W. pipientis genome, mtDNA is also maternally inherited yet has a higher mutation rate which provides more phylogenetically informative mutations for analysis (Richardson et al. 2012; Early and Clark 2013; this study). Many methods of phylogenetic reconstruction, however, are not suited to deal with heterozygous sites (Sota and Vogler 2003), which is problematic for the wAnaITG genomes where we found many sites to be called as heterozygotes within each line. Phasing the heterozygote calls using statistical methods was almost impossible as 98.9% of the total polymorphic sites among the wAnaITG genomes were a PSHET. Thus, only sites that were called as homozygous were examined from each wAnaITG genome, assuming that those were representative of the variants that existed in the wAna genome before integrating into the host genome as wAnaITG.
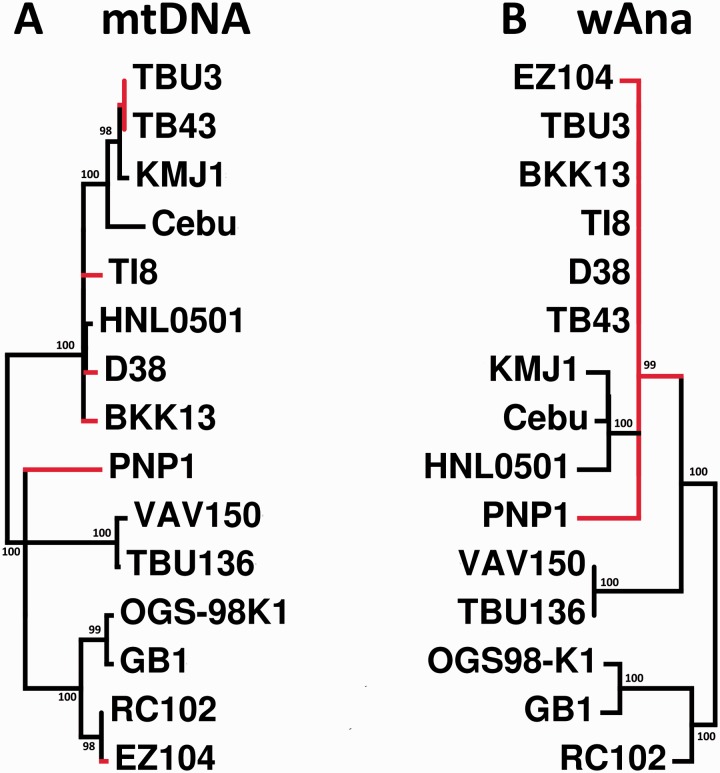
Qualitatively the mtDNA genome phylogeny (fig. 6A) was concordant with the mtDNA phylogeny from the study of Choi and Aquadro (2014), which was estimated using the same D. ananassae samples from this study but with Sanger sequences of only three mtDNA genes. No evidence of phylogenetic clustering was observed among the mtDNA haplotypes originating from D. ananassae strains with the wAnaINF or wAnaITG genomes.
Fig. 6.—
Maximum-likelihood phylogeny of the (A) host D. ananassae mtDNA and (B) wAna genomes. Bootstrap values of greater than 95% are shown on the nodes of the phylogeny. Drosophila ananassae strains with wAnaINF are indicated with black colored branches, whereas those with wAnaITG are indicated with red colored branches. Both trees are midpoint rooted.
As expected for maternally inherited organelles and endosymbionts, the branching patterns observed among the wAnaINF genomes were congruent with the phylogenetic relationship estimated for the mtDNA (fig. 6A and B). Interestingly, the copy number analysis of specific wAnaINF genomic regions (fig. 2), large structural variation analysis (fig. 5A), and the wAnaINF nucleotide phylogeny (fig. 6B), which were inferred independently from one another, gave congruent phylogenetic results. For example, wAnaINF genomes from strain TBU136 and VAV150 had a significant decrease in copy number in two regions of the genome (fig. 2), had shared large structural variations (SV6 and SV7; fig. 5A), and were phylogenetically the most closely related (fig. 6B). Additionally, the phylogenetic tree indicated wAnaINF genomes from strains Cebu, HNL0501, and KMJ1 as one group; and strains GB1 and OGS-98K1 as another group (fig. 6B), both of which were also consistent with the PCR-verified large structural variations (fig. 5A).
The wAna phylogenetic tree showed all of the wAnaITG genomes to be within a single monophyletic cluster consistent with a single original host nuclear genome-integration and subsequent inheritance as a biparentally transmitted nuclear gene. In contrast, the maternally inherited mtDNA for these wAnaITG strains of D. ananassae showed a much different pattern in that they were distributed across the full mtDNA phylogeny for all strains (fig. 6A and B). Interestingly the wAna phylogenetic tree indicated that the wAnaINF genomes from strains Cebu, HNL0501, and KMJ1 were phylogenetically most closely related to the wAnaITG genomes. This phylogenetic relationship showed both similarities and differences from those expected from the distribution of the PCR-verified large structural variations (fig. 5). SV1 and SV2 suggested that the wAnaITG genomes were different from wAnaINF of strains Cebu, HNL0501, and KMJ1; whereas all other large structural variants suggested that the wAnaITG genomes were most related to wAnaINF strains from Cebu, HNL0501, and KMJ1.
Analysis of Sites Called as Heterozygotes for Each wAnaITG Genome
Among the wAnaITG genomes the majority of the polymorphic sites with a single nucleotide variant or small-sized INDEL was PSHET (table 2) and filtered out for most of our downstream analysis. To analyze these heterozygous sites, for each wAnaITG genome we attempted to phase the variation by sequencing a single F1 offspring of each wAnaITG containing strain (while absent of wAnaINF) of D. ananassae that had been crossed to a strain lacking both integrated and infectious forms of wAna (see Materials and Methods for details). We expected these F1 progeny to have only a haploid complement of the wAnaITG genome that would have allowed phasing of sites with a heterozygote base call. Each site that was called as a heterozygote in the diploid wAnaITG genome was then examined in the haploid wAnaITG genome. Surprisingly, the majority of the sites initially called as heterozygotes in the diploid wAnaITG genome remained called as heterozygotes in the haploid wAnaITG genome F1 offspring (table 4).
Table 4.
Phasing Sites Called as Heterozygotes from the Diploid wAnaITG Genomes
| Sample | Diploid | Haploid |
|
|---|---|---|---|
| Het Variant | Unphased | Phased | |
| BKK13 | 511 | 480 (93.9%) | 31 (6.1%) |
| D38 | 466 | 435 (93.3%) | 31 (6.7%) |
| EZ104 | 493 | 441 (89.5%) | 52 (10.5%) |
| PNP1 | 374 | NA | |
| TB43 | 529 | 483 (91.3%) | 46 (8.7%) |
| TBU3 | 531 | 508 (95.7%) | 23 (4.3%) |
| TI8 | 525 | 501 (95.4%) | 24 (4.6%) |
Note.—Het Variant, total number of sites called as heterozygotes in the diploid wAnaITG genome; Unphased, total number of variants that were called heterozygotes in the diploid wAnaITG genome and were still called as heterozygotes in the haploid wAnaITG genome; Phased, total number of variants that were called heterozygotes in the diploid wAnaITG genome but homozygous as haploid wAnaITG genome; NA, not applicable as the average read coverage was too low to reliably call variants. Proportions are indicated in parenthesis.
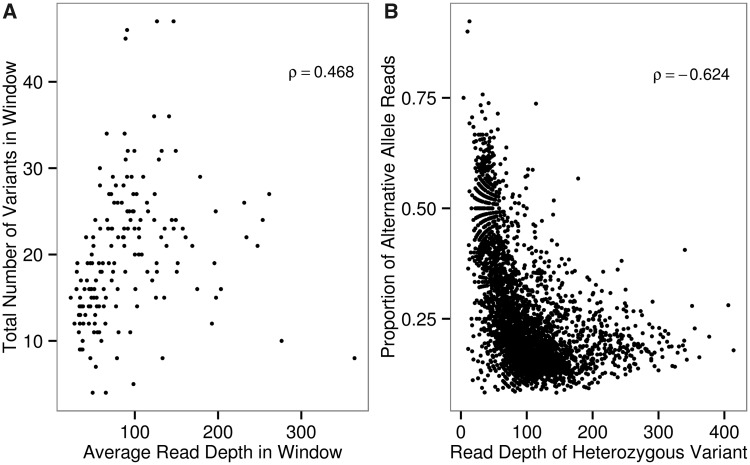
We next investigated whether for each wAnaITG genome the increased number of sites called as heterozygotes could be explained by an increase in copy number of specific regions of the wAnaITG genome (figs. 3 and 4). Duplication and triplication of regions would result in paralogs among which mutations could accumulate resulting in what would simply look like a heterozygote among Illumina reads. A sliding window analysis was conducted by dividing each wAnaITG genome into nonoverlapping 50,000 bp windows, and for each window the average genome coverage and total number of single nucleotide variants were counted. Across all seven wAnaITG genomes results showed a significant (P < 0.0001) positive correlation between the average genome coverage and the total number of single nucleotide variants in a given window (fig. 7A) as would be expected by varying numbers of paralogs of wAnaITG.
Fig. 7.—
Analysis of sites called as heterozygotes in the wAnaITG genomes. (A) For each wAnaITG genome, a nonoverlapping sliding window analysis of comparing average read depth and the total number of single nucleotide variants. (B) Each data point represents a site called as a heterozygote within a wAnaITG genome, with x axis representing the total read coverage and y axis representing the proportion of alternative alleles consisting the heterozygote variant. Spearman’s rho values are shown in top right corner of each figure and both correlations are highly significant (P < 0.0001).
Next, each site called as a heterozygote was individually examined by counting the total read coverage and the total number of reads with the alternative allele supporting that heterozygous site. For a diploid genomic region, a site called as a heterozygote from next-generation sequencing is expected to consist of reads where 50% are from the reference allele whereas the other 50% are from the alternative allele. These reads should remain 50:50 in proportion regardless of total read depth. In contrast to this expectation, analysis of our combined results from each of the seven wAnaITG genomes revealed that sites called as heterozygotes showed a significant (P < 0.0001) negative correlation between total read coverage and their proportion of reads originating from the alternative allele (fig. 7B). These results also support the hypothesis that variation in the number of paralogs of wAnaITG accounts for the large number of sites called as heterozygotes in strains of D. ananassae with the wAnaITG genome.
Investigating the Integration Site of the wAnaITG Genome
In an effort to localize the integration site of wAnaITG into the D. ananassae nuclear genome, we used the program lumpy to detect chimeric raw paired-end reads where one end mapped to the wAnaITG genome whereas the other end mapped to the D. ananassae nuclear genomes. Reads from wAnaINF were also examined as a potential negative control for our in silico experiment, expecting no evidence of integration in the D. ananassae nuclear genome, as previous PCR screens have not detected any evidence of wAnaITG genes for our wAnaINF samples (Choi and Aquadro 2014).
Four of the seven wAnaITG samples had significant evidence of a breakpoint existing between the wAnaITG and D. ananassae nuclear genome (supplementary table S6, Supplementary Material online). There were multiple wAna breakpoints with overlapping genome coordinates among the wAnaITG samples, whereas only a single breakpoint was detected at a single location around 23,595,400–23,595,600 bp at scaffold_13340 from chromosome 2 L (Muller element E) of the D. ananassae nuclear genome (supplementary table S6, Supplementary Material online). Due to its unique localization it was designated as the candidate site for the W. pipientis–D. ananassae integration (DanaITG). PCR primers were designed to flank the computationally predicted putative DanaITG region and amplified in all 15 D. ananassae samples examined in this study plus the reference D. ananassae strain. Surprisingly, PCR results showed the same size bands in the wAnaINF and wAnaITG samples; however, the reference strain did not show any PCR bands (supplementary fig. S1A, Supplementary Material online). Sanger sequencing the putative DanaITG region from strains TB43 (D. ananassae line with wAnaITG) and KMJ1 (D. ananassae line with wAnaINF) indicated that the reference genome contained a unique DNA sequence at this location (supplementary fig. S1B, Supplementary Material online). A BLAST (Basic Local Alignment Search Tool) search of this sequence matched the long terminal repeats (LTRs) of a retrotransposon (supplementary fig. S1C, Supplementary Material online) and the breakpoints predicted by lumpy spanned this LTR region (supplementary fig. S1B, Supplementary Material online). In fact, the program RepeatMasker indicated that this region between 23,595,477 and 23,595,672 at scaffold_13340 corresponds to the Pao family of LTR retrotransposons (Xiong et al. 1993). Thus, multiple regions of the wAnaITG genome had breakpoints matching with the LTR of the host D. ananassae retrotransposon.
The Cebu strain was the only wAnaINF genome-carrying D. ananassae line with a lumpy-predicted DanaITG region that was different from the wAnaITG genomes. However, PCR amplification using primers designed to flank the Cebu wAnaINF–DanaITG region could not verify the computational predictions (results not shown).
Klasson et al. (2014) have recently shown evidence that wAnaITG sequence could be detected on chromosome 4 (Muller element F) of D. ananassae by fluorescent in situ hybridization (FISH) to mitotic chromosomes. We thus reanalyzed our wAnaITG genome sequence data with lumpy to see whether we could identify potential breakpoints between the wAnaITG genome and D. ananassae scaffolds identified as Muller F from a previous study (Bhutkar et al. 2008; see supplementary table S7, Supplementary Material online, for names of scaffold). These Muller F scaffolds were not part of the reference D. ananassae genome sequence we initially used for our lumpy analysis. No significant breakpoints were detected in any of the eight wAnaINF genomes. However, all seven wAnaITG genomes had significant breakpoints (supplementary table S8, Supplementary Material online) with a total of 31 breakpoints found between the genomes of wAnaITG and D. ananassae Muller F scaffolds. Results from RepeatMasker indicated that for 28 of the 31 predicted breakpoints, the D. ananassae region of Muller F scaffolds corresponded to a TE or a retrotransposon (supplementary table S8, Supplementary Material online) and not a unique sequence.
Discussion
We have used next-generation sequencing to analyze the molecular evolution and phylogenomics of two different forms of W. pipientis associated with D. ananassae: 1) The bacterial originating and maternally transmitted wAnaINF genomes and 2) the host nuclear genome integrated wAnaITG genome. The wAnaINF and wAnaITG genomes showed distinct biological differences and characteristics.
Genomic Diversity within the wAnaINF Genomes
Our previous study of wAnaINF DNA sequence diversity based on Sanger sequencing failed to find any variability for 7 kb sampled from the wAnaINF genomes infecting eight geographically diverse strains of D. ananassae (Choi and Aquadro 2014). However, full genome sequencing reported here did find single nucleotide, INDEL, and copy number variation among these eight wAnaINF genomes, though for nucleotide variability at levels a 100-fold lower than those of the host mtDNA genomes. Congruent phylogenies for both the wAnaINF genomes and the maternally inherited host mtDNA suggest that the main mode of transmission for wAnaINF is through maternal transmission, an observation that has also been reported for W. pipientis in other species of Drosophila (e.g., Richardson et al. 2012; Early and Clark 2013).
Our analyses of copy number variation (fig. 2), large structural variation (fig. 5A), and nucleotide diversity (fig. 6) revealed several genomic strains of wAnaINF that could potentially differ functionally from each other. For example, wAnaINF from D. ananassae strain HNL0501 was the only strain to have a significant increase in copy number around a prophage region at genome coordinates 0.57–0.61 and 1.07–1.12 Mb (fig. 2). This prophage, named wRi-WOB (Klasson, Westberg, et al. 2009), exists as two identical duplicates in the wRi genome around genome coordinates 0.57–0.61 and 1.07–1.12 Mb, suggesting that the wAnaINF from D. ananassae strain HNL0501 has four copies of wRi-WOB. The functions of prophages in W. pipientis have not been fully described, but they can transpose and frequently horizontally transfer among divergent W. pipientis strains (Masui et al. 2000; Gavotte et al. 2007). Due to their lytic ability, the density of prophages has been suggested to control the strength of W. pipientis-mediated cytoplasmic incompatibility by decreasing the titer of W. pipientis infection (Bordenstein et al. 2006). Interestingly, wAnaINF from D. ananassae strain HNL0501 had the lowest wAna originating reads and normalized average read depth (table 1). Assuming this low read depth reflects a low wAnaINF titer in D. ananassae strain HNL0501, then the increased copy number of prophages may have caused the lowered bacterial load in this strain.
Another region with differences in copy number variation (0.61–0.63 and 1.12–1.14 Mb; fig. 2) had only half of the typical copy number in wAnaINF of D. ananassae strains TBU136 and VAV150 but double the typical copy number in D. ananassae strain HNL0501. The two regions are mostly identical in sequence and contain coding sequences that are repetitive (Klasson, Westberg, et al. 2009). The functional consequence of this variation in copy number is unknown; however, in wMel copy number variation of a 21-kb region has been suggested to cause pathogenicity in the virulent Popcorn strain wMelPop (Chrostek et al. 2013; Chrostek and Teixeira 2015). Because copy number variation in the W. pipientis coding region could lead to physiological responses in the host, future studies will be necessary to fully understand the potential host effects caused by the copy number variations observed across the wAnaINF genome.
As W. pipientis genomes have an unusually high abundance of mobile genetic elements compared with other endosymbionts (Moran and Plague 2004), it is not surprising that all of the large structural variations we detected in the wAnaINF genome involved genes with transposase ability (fig. 5; supplementary tables S2 and S3, Supplementary Material online). This variation can be used for genotyping variability within wAnaINF as has been reported in studies of wMel, where the absence or presence of these mobile elements has been used as genotyping tools to differentiate within wMel strains (Riegler et al. 2005; Woolfit et al. 2013). Here, we have identified several TE absences or presences that were both polymorphic and fixed among the wAnaINF genomes. The presence/absence of polymorphic TE insertions was generally consistent with the nucleotide substitution-inferred phylogenetic tree, suggesting that these TE polymorphisms also are phylogenetically informative and can be a useful fast way to genotype different wAnaINF strains.
Further, these variants can be used to describe W. pipientis strain variation in other hosts infected with strains highly similar to wAna. For example, wRi is not only very similar to wAna but also very similar to the W. pipientis infecting Drosophila suzukii (Siozios et al. 2013), suggesting a very recent horizontal transfer of W. pipientis among the three host species (D. ananassae, D. simulans, and D. suzukii). The phylogenetically informative variants found in our study could be used to characterize the W. pipientis diversity among these three host species, and potentially determine the coevolutionary history of the W. pipientis infection among those three species.
Finally, variation in the proportion of raw reads originating from the wAnaINF genomes and average read depth of wAnaINF genomes (table 1) suggest differences in bacterial titer among the D. ananassae hosts. The biological significance of this variation in wAnaINF load is unknown but previous studies have shown various effects that are W. pipientis density-dependent including level of cytoplasmic incompatibility (Clark and Karr 2002), male killing effect (Unckless et al. 2009), and differences in antiviral protection for its host (Osborne et al. 2012; Chrostek and Teixeira 2015). These phenotypes are likely to be under selection. Thus, the observed differences in bacterial titer among D. ananassae strains could have larger biological and evolutionary consequences. In addition, as the host genotype can also lead to differences in W. pipientis titer (Kondo et al. 2005; Serbus et al. 2011), it is important to understand the underlying coevolutionary history between W. pipientis and its host to understand the biology of W. pipientis (i.e., Chrostek et al. 2013). The genomic resources and results from this study can help advance future studies of the genetic interaction between wAnaINF and its host D. ananassae.
Evolution of the wAnaITG Genomes
In all seven of our D. ananassae samples with evidence of wAnaITG, we found not only the whole wAna genome integrated into the host genome but also that specific regions were present in as many as seven copies (fig. 3). During the preparation of this manuscript, Klasson et al. (2014) independently reported evidence of extensive duplication in the wAnaITG genome originating from their Hawaii and India strains of D. ananassae. In our study, however, we have genome sequenced D. ananassae samples from an even broader geographic range (Africa, southeast Asia, west Asia, and south Pacific islands) that encompasses both the ancestral and peripheral range of D. ananassae (Das et al. 2004; Schug et al. 2007). Additional phylogenetic analyses together with genotyping using large structural variation allowed us to demonstrate that all wAnaITG genomes were closely related to each other (figs. 5B and 6). These results suggest that a single wAna genotype initially integrated into the host genome and subsequently dispersed throughout much of the worldwide range of D. ananassae. Based on our phylogenetic and large structural variation results, the wAnaITG genomes are most closely related to the wAnaINF infecting D. ananassae strains Cebu, HNL0501, and KMJ1, suggesting that the common ancestor of the wAnaINF infecting these three strains was closely related to that which integrated into the host genome.
Interestingly the wAnaITG genomes examined in this study had on average twice the D. ananassae nuclear genome coverage (table 1), whereas Klasson et al. (2014) reported the wAnaITG samples from India and Indonesia to have far less coverage than the D. ananassae nuclear genome. In addition, the wAnaITG genome from Indonesia lacked the heterogeneous increased copy number, which was observed in all wAnaITG genomes in this study (fig. 3). This underrepresentation of wAnaITG DNA in the India and Indonesia samples (Klasson et al. 2014) could be due to temporal differences in the integration of the wAna genome. Through an unknown mechanism the initial wAnaITG genome could have accumulated increased copy number and overrepresentation of itself over time. Then, the wAnaITG genomes examined in this study and the Hawaii strain from Klasson et al. (2014) could represent an ancient integrated wAnaITG genome. The increased number of mutations observed in the wAnaITG genome compared with the wAnaINF genome further suggests it to be an ancient integration (table 2). On the other hand, the wAnaITG genomes from India and Indonesia (Klasson et al. 2014) could be from a more recent integration event. This further suggests that the original wAna genome type integrated into the host genome is polymorphic, and potentially is an ongoing process in D. ananassae occurring multiple times. Here, the structural variations identified from this study could test whether multiple wAna genome types have integrated into the host nuclear genome.
The wAnaITG genomes have many characteristics of a pseudogene (Li et al. 1981; Miyata and Hayashida 1981) with an elevated proportion of nonsynonymous relative to synonymous (and intergenic) variants, and many segregating variants were predicted to have strongly deleterious effects if present in a functional wAna genome (table 3). These observations lead us to conclude that the wAnaITG genome is becoming (if not already has become) a large pseudogenome after integrating into the host D. ananassae nuclear genome.
Regions with higher copy number in the wAnaITG genome also have more mutations (fig. 7A) due to the increased number of copies and sites accumulating mutations. In addition, our analysis of genome coverage (figs. 3 and 4) and unphasable sites computationally called as heterozygotes in the wAnaITG genome (table 4), further supports the interpretation that the majority of the wAnaITG genome exists in at least two copies in the host genome (Klasson et al 2014). Although copy number is variable across wAna genomic regions, the increased and decreased copy number regions were correlated across all seven wAnaITG genomes (fig. 3). This suggested that after the integration, specific regions of the wAnaITG first increased in copy number then subsequently dispersed worldwide. A recent study using FISH to mitotic chromosomes using the high copy region of the wAnaITG genome as a probe has found evidence of the integration only at a single location on the D. ananassae fourth chromosome (Klasson et al. 2014). The presence of only a single site of hybridization suggests that the increased copy number is likely to be due to increased tandem duplications of the wAnaITG regions.
wAnaITG regions with multiple copies could have a mutation occurring on one of the multiple paralogs, and this would be computationally called as a heterozygote as the multiple copies are collapsed into a single copy in the reference wRi genome. Compared with regions with low copy number, regions with higher copy number across the wAnaITG genome had more sites called as heterozygotes with lower proportions of reads from the alternative allele than from the reference allele (fig. 7B). This pattern would result if the rate of increase in copy number of specific wAnaITG regions is higher than the rate of nucleotide mutation.
The rapid increase in copy number also explains why our current full genome sequencing allowed us to detect numerous polymorphisms, whereas our previous analysis which had used Sanger sequencing of 7 kb from several wAnaITG gene regions found only three segregating sites (Choi and Aquadro 2014). The lower proportion of alternative to reference reads for many of the sites called as heterozygotes (fig. 7B) would be reflected in the Sanger sequencing chromatograms as different sized double peaks. We have reexamined the chromatograms of Choi and Aquadro (2014) and verified many PSHET identified in this study actually had double peaks in the chromatogram, albeit one peak was always much smaller than the other (results not shown), which normally would be discarded as noise. Thus, we believe that many of our computationally detected heterozygous base calls in the wAnaITG genome are not due to sequencing errors.
Using in silico analysis coupled with PCR verification, we found no convincing evidence of breakpoints between the wAnaITG and D. ananassae nuclear unique sequence from the euchromatic chromosomes X, 2, and 3. The only breakpoints discovered were between multiple wAnaITG regions and an LTR of a D. ananassae retrotransposon, suggesting that the host retrotransposons were actively integrating into various regions of the integrated wAnaITG genome. This has been noted previously (Dunning Hotopp et al. 2007) and our further analysis with the D. ananassae fourth chromosome scaffolds also corroborated this result. The fourth chromosomal scaffolds have a higher frequency of retrotransposons and TEs than D. ananassae scaffolds from chromosome X, 2, and 3 (Leung et al. 2015). Thus, many of the putative-predicted breakpoints between the D. ananassae fourth chromosome and wAnaITG genome (supplementary table S8, Supplementary Material online) are likely artifacts caused by D. ananassae TEs that have inserted into the wAnaITG genome. Consistent with the results previously reported by Dunning Hotopp et al. (2007), evidence of frequent integrations of D. ananassae TEs into the wAnaITG genome complicates the placement of wAnaITG genomic fragments in the D. ananassae genome. Long-read sequencing technology (e.g., Pacific Biosciences) and additional in situ hybridization experiments may in the future provide insight into the precise integration site for the wAnaITG genome.
Our analyses of the wAnaITG genomes thus lead to several intriguing conclusions: 1) Although pseudogenes are rare across Drosophila species (Clark et al. 2007), the wAnaITG of D. ananassae seems to have become a large pseudogenome after the integration; 2) Drosophila shows a strong bias toward deletion of nonfunctional genes (Petrov et al. 1996) but the wAnaITG genome shows evidence of extensive duplications despite its likely lack of functionality; and 3) more than 2% of the genomic reads from D. ananassae originated from the pseudogene-like wAnaITG genome (table 1) which we infer to have had minimal negative fitness consequences as evidenced by its wide geographic distribution (Choi and Aquadro 2014). Thus despite the apparent loss of functionality across the wAnaITG genome, some paralogs among the multiple wAnaITG copies could be under positive selection leading to the observed pattern of spreading across multiple D. ananassae populations.
Conclusion
The wAnaINF and wAnaITG genomes show drastically different evolutionary and population genomics. We discovered diverse nucleotide and structural variants among wAnaINF genomes that showed phylogenetic relationships consistent with a strict maternal inheritance after an initial single interspecific infection within D. ananassae. Similarly, the wAnaITG genome appears to have arisen from a single integration of one wAnaINF variant not long after the initial infection of the species. Subsequent to the initial integration, the majority of the wAnaITG genome had at least doubled its copy number within the D. ananassae host genome. Additionally after the integration of the wAnaITG genome, it appears to have become a large pseudogenome accumulating substantially more mutations than the cytoplasmic wAnaINF genome, including many of which would be predicted to be strongly deleterious in a functioning wAna genome. Although it is possible that the wAnaITG has spread across a wide geographic distribution through genetic drift and/or migration alone, further study of the wAnaITG genomes from additional geographically diverse strains of D. ananassae is needed to distinguish hypotheses as to what has apparently caused the geographically wide-spread distribution of wAnaITG despite its apparent loss of functionality.
Supplementary Material
Supplementary data, figures S1, and tables S1–S8 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the National Institute of Health grant number R01GM095793 to C.F.A. and Daniel A. Barbash, and by the Cornell Center for Comparative and Population Genomics (3CPG) through a Priming Grant to C.F.A. and J.Y.C. and a 3CPG Scholar Award to J.Y.C. The authors thank the Cornell Biotechnology Resource Center for helping with the preparation and genome sequencing our samples. They also thank Daniel Barbash, Vanessa Bauer DuMont, Angela M. Early, Sarah Elgin, Filip Husnik, Brian Lazzaro, Wilson Leung, John H. Werren, and the anonymous reviewer for their helpful discussions and comments.
Literature Cited
- Abyzov A, Urban AE, Snyder M, Gerstein M. 2011. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 21(6):974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aikawa T, et al. 2009. Longicorn beetle that vectors pinewood nematode carries many Wolbachia genes on an autosome. Proc Biol Sci. 276:3791–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard JW, Melvin RG. 2007. Tetracycline treatment influences mitochondrial metabolism and mtDNA density two generations after treatment in Drosophila. Insect Mol Biol. 16(6):799–802. [DOI] [PubMed] [Google Scholar]
- Bhutkar A, et al. 2008. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics 179(3):1657–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter M. 2007. Symbiont genes in host genomes: fragments with a future? Cell Host Microbe 2(4):211–213. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein S, Marshall M, Fry A, Kim U, Wernegreen J. 2006. The tripartite associations between bacteriophage, Wolbachia, and arthropods. PLoS Pathog. 2(5):e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Aquadro CF. 2014. The coevolutionary period of Wolbachia pipientis infecting Drosophila ananassae and its impact on the evolution of the host germline stem cell regulating genes. Mol Biol Evol. 31(9):2457–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrostek E, et al. 2013. Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: a phenotypic and phylogenomic analysis. PLoS Genet. 9(12):e1003896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrostek E, Teixeira L. 2015. Mutualism breakdown by amplification of Wolbachia genes. PLoS Biol. 13(2):e1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, et al. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450:203–218. [DOI] [PubMed] [Google Scholar]
- Clark ME, Karr TL. 2002. Distribution of Wolbachia within Drosophila reproductive tissue: implications for the expression of cytoplasmic incompatibility. Integr Comp Biol. 42(2):332–339. [DOI] [PubMed] [Google Scholar]
- Das A, Mohanty S, Stephan W. 2004. Inferring the population structure and demography of Drosophila ananassae from multilocus data. Genetics 168:1975–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudoumis V, et al. 2012. Detection and characterization of Wolbachia infections in laboratory and natural populations of different species of tsetse flies (genus Glossina). BMC Microbiol. 12(Suppl 1):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning Hotopp JC. 2011. Horizontal gene transfer between bacteria and animals. Trends Genet. 27(4):157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning Hotopp JC, et al. 2007. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317:1753–1756. [DOI] [PubMed] [Google Scholar]
- Early AM, Clark AG. 2013. Monophyly of Wolbachia pipientis genomes within Drosophila melanogaster: geographic structuring, titre variation and host effects across five populations. Mol Ecol. 22(23):5765–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenn K, et al. 2006. Phylogenetic relationships of the Wolbachia of nematodes and arthropods. PLoS Pathog. 2(10):e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavotte L, et al. 2007. A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol Biol Evol. 24(2):427–435. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 22(2):160–174. [DOI] [PubMed] [Google Scholar]
- Hosokawa T, Koga R, Kikuchi Y, Meng X, Fukatsu T. 2010. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc Natl Acad Sci U S A. 107(2):769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst GD, Jiggins FM. 2005. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc Biol Sci. 272:1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik F, et al. 2013. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153:1567–1578. [DOI] [PubMed] [Google Scholar]
- International Glossina Genome Initiative. 2014. Genome sequence of the tsetse fly (Glossina morsitans): vector of African trypanosomiasis. Science 344(6182):380–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis P, et al. 2013. Extensively duplicated and transcriptionally active recent lateral gene transfer from a bacterial Wolbachia endosymbiont to its host filarial nematode Brugia malayi. BMC Genomics 14(1):639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P, Palmer J. 2008. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 9(8):605–618. [DOI] [PubMed] [Google Scholar]
- Klasson L, et al. 2014. Extensive duplication of the Wolbachia DNA in chromosome four of Drosophila ananassae. BMC Genomics 15(1):1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L, Kambris Z, Cook P, Walker T, Sinkins S. 2009. Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. BMC Genomics 10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L, Westberg J, et al. 2009. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc Natl Acad Sci U S A. 106(14):5725–5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N, Nikoh N, Ijichi N, Shimada M, Fukatsu T. 2002. Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect. Proc Natl Acad Sci U S A. 99(22):14280–14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N, Shimada M, Fukatsu T. 2005. Infection density of Wolbachia endosymbiont affected by co-infection and host genotype. Biol Lett. 1(4):488–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, et al. 2012. Efficient subtraction of insect rRNA prior to transcriptome analysis of Wolbachia-Drosophila lateral gene transfer. BMC Res Notes. 5:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer RM, Chiang C, Quinlan AR, Hall IM. 2014. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 15(6):R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W, et al. 2015. Drosophila Muller F elements maintain a distinct set of genomic properties over 40 million years of evolution. G3 (Bethesda) 5:719–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Arxiv:1303 3997. [Google Scholar]
- Li H, et al. 2009. The sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH, Gojobori T, Nei M. 1981. Pseudogenes as a paradigm of neutral evolution. Nature 292(5820):237–239. [DOI] [PubMed] [Google Scholar]
- Lo N, et al. 2007. Taxonomic status of the intracellular bacterium Wolbachia pipientis. Int J Syst Evol Microbiol. 57:654–657. [DOI] [PubMed] [Google Scholar]
- Long M, VanKuren N, Chen S, Vibranovski M. 2013. New gene evolution: little did we know. Annu Rev Genet. 47(1):307–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S, Kamoda S, Sasaki T, Ishikawa H. 2000. Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J Mol Evol. 51(5):491–497. [DOI] [PubMed] [Google Scholar]
- Miyata T, Hayashida H. 1981. Extraordinarily high evolutionary rate of pseudogenes: evidence for the presence of selective pressure against changes between synonymous codons. Proc Natl Acad Sci U S A. 78(9):5739–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth K, Abt D, Hofmann J, Rand D. 2009. Comparative genomics of Drosophila mtDNA: novel features of conservation and change across functional domains and lineages. J Mol Evol. 69(1):94–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Plague GR. 2004. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 14(6):627–633. [DOI] [PubMed] [Google Scholar]
- Nikoh N, et al. 2008. Wolbachia genome integrated in an insect chromosome: evolution and fate of laterally transferred endosymbiont genes. Genome Res. 18(2):272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N, et al. 2010. Bacterial genes in the aphid genome: absence of functional gene transfer from Buchnera to its host. PLoS Genet. 6:e1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N, et al. 2014. Evolutionary origin of insect–Wolbachia nutritional mutualism. Proc Natl Acad Sci U S A. 111(28):10257–10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne S, Iturbe-Ormaetxe I, Brownlie J, O’Neill S, Johnson K. 2012. Antiviral protection and the importance of Wolbachia density and tissue tropism in Drosophila simulans. Appl Environ Microbiol. 78(19):6922–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA, Lozovskaya ER, Hartl DL. 1996. High intrinsic rate of DNA loss in Drosophila. Nature 384(6607):346–349. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ. 2012. Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3:217–223. [Google Scholar]
- Richardson MF, et al. 2012. Population genomics of the Wolbachia endosymbiont in Drosophila melanogaster. PLoS Genet. 8(12):e1003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegler M, Sidhu M, Miller WJ, O’Neill SL. 2005. Evidence for a global Wolbachia replacement in Drosophila melanogaster. Curr Biol. 15(15):1428–1433. [DOI] [PubMed] [Google Scholar]
- Salzberg SL, et al. 2005. Serendipitous discovery of Wolbachia genomes in multiple Drosophila species. Genome Biol. 6(3):R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer SW, et al. 2008. Polytene chromosomal maps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics 179(3):1601–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug M, Smith S, Tozier-Pearce A, McEvey S. 2007. The genetic structure of Drosophila ananassae populations from Asia, Australia and Samoa. Genetics 175:1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus L, Casper-Lindley C, Landmann F, Sullivan W. 2008. The genetics and cell biology of Wolbachia-host interactions. Annu Rev Genet. 42:683–707. [DOI] [PubMed] [Google Scholar]
- Serbus L, et al. 2011. A feedback loop between Wolbachia and the Drosophila gurken mRNP complex influences Wolbachia titer. J Cell Sci. 124:4299–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siozios S, et al. 2013. Draft genome sequence of the Wolbachia endosymbiont of Drosophila suzukii. Genome Announc. 1(1):e00032–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatko B, Taylor M, Foster J. 2010. The Wolbachia endosymbiont as an anti-filarial nematode target. Symbiosis 51(1):5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, et al. 2014. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol Biol Evol. 31:857–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sota T, Vogler AP. 2003. Reconstructing species phylogeny of the carabid beetles Ohomopterus using multiple nuclear DNA sequences: heterogeneous information content and the performance of simultaneous analyses. Mol Phylogenet Evol. 26(1):139–154. [DOI] [PubMed] [Google Scholar]
- Stouthamer R, Breeuwer J, Hurst G. 1999. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu Rev Microbiol. 54:71–102. [DOI] [PubMed] [Google Scholar]
- Taylor M, Bandi C, Hoerauf A. 2005. Wolbachia bacterial endosymbionts of filarial nematodes. Adv Parasitol. 60:245–284. [DOI] [PubMed] [Google Scholar]
- Unckless R, Boelio L, Herren J, Jaenike J. 2009. Wolbachia as populations within individual insects: causes and consequences of density variation in natural populations. Proc R Soc Lond B Biol Sci. 276(1668):2805–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, et al. 2012. Primer3—new capabilities and interfaces. Nucleic Acids Res. 40(15):e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J, Baldo L, Clark M. 2008. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 6:741–751. [DOI] [PubMed] [Google Scholar]
- Werren J, et al. 2010. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327:343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfit M, et al. 2013. Genomic evolution of the pathogenic Wolbachia strain, wMelPop. Genome Biol Evol. 5(11):2189–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfit M, Iturbe-Ormaetxe I, McGraw E, O’Neill S. 2009. An ancient horizontal gene transfer between mosquito and the endosymbiotic bacterium Wolbachia pipientis. Mol Biol Evol. 26(2):367–374. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Burke WD, Eickbush TH. 1993. Pao, a highly divergent retrotransposable element from Bombyx mori containing long terminal repeats with tandem copies of the putative R region. Nucleic Acids Res. 21(9):2117–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25(21):2865–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.