Abstract
Background
African weakly-electric fishes of the family Mormyridae are able to produce and perceive weak electric signals (typically less than one volt in amplitude) owing to the presence of a specialized, muscle-derived electric organ (EO) in their tail region. Such electric signals, also known as Electric Organ Discharges (EODs), are used for objects/prey localization, for the identification of conspecifics, and in social and reproductive behaviour. This feature might have promoted the adaptive radiation of this family by acting as an effective pre-zygotic isolation mechanism. Despite the physiological and evolutionary importance of this trait, the investigation of the genetic basis of its function and modification has so far remained limited. In this study, we aim at: i) identifying constitutive differences in terms of gene expression between electric organ and skeletal muscle (SM) in two mormyrid species of the genus Campylomormyrus: C. compressirostris and C. tshokwe, and ii) exploring cross-specific patterns of gene expression within the two tissues among C. compressirostris, C. tshokwe, and the outgroup species Gnathonemus petersii.
Results
Twelve paired-end (100 bp) strand-specific RNA-seq Illumina libraries were sequenced, producing circa 330 M quality-filtered short read pairs. The obtained reads were assembled de novo into four reference transcriptomes. In silico cross-tissue DE-analysis allowed us to identify 271 shared differentially expressed genes between EO and SM in C. compressirostris and C.tshokwe. Many of these genes correspond to myogenic factors, ion channels and pumps, and genes involved in several metabolic pathways. Cross-species analysis has revealed that the electric organ transcriptome is more variable in terms of gene expression levels across species than the skeletal muscle transcriptome.
Conclusions
The data obtained indicate that: i) the loss of contractile activity and the decoupling of the excitation-contraction processes are reflected by the down-regulation of the corresponding genes in the electric organ’s transcriptome; ii) the metabolic activity of the EO might be specialized towards the production and turn-over of membrane structures; iii) several ion channels are highly expressed in the EO in order to increase excitability; iv) several myogenic factors might be down-regulated by transcription repressors in the EO.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-1858-9) contains supplementary material, which is available to authorized users.
Background
Bioelectrogenesis (i.e., the ability to produce strong or weak electric signals by specialized organs) has evolved several times independently in aquatic vertebrates [1]. In fact, it can be observed in the marine electric rays (Torpediniformes) and skates (Rajiformes), in the African freshwater Mormyridae and Gymnarchidae (Osteoglossiformes; Mormyroidea), in the South American knifefishes (Gymnotiformes), in several catfish species (Siluriformes), and in few marine stargazers (Perciformes; Uranoscopidae). In all the above-mentioned groups, electric organs originate from myogenic tissue; the only exception are members of the family Apteronotidae (Gymnotiformes), where the electric organs are formed by modified spinal motor neurons [2]. The amount of excitable cells within each electric organ determines the electric potential of an EOD, which can range from few millivolts to several hundreds of volts (e.g., in the electric eel Electrophorus electricus) [3].
African weakly-electric fishes of the family Mormyridae constitute a group of teleost fishes, formed by approximately 200 species [4], all endemic to African riverine and, partially, lacustrine systems. As their name suggests, they are able to produce only weak electric fields, in the order of millivolts to few volts, which are not used for predation or defence. The cells forming their electric organ are compressed disk-like cells commonly called electrocytes. In many species they are longitudinally stacked behind each other in order to form columns of cells embedded within tubes of isolating connective tissue. The synchronous activity of each electrocyte defines the output of the electric organ, known as Electric Organ Discharge (EOD) [5]. Such weak pulses are mainly employed for the localization and discrimination of objects in water (active electrolocation) [6], for the recognition of conspecific individuals [7, 8], and in social and reproductive behaviour [9, 10].
Besides these functional roles, the EODs of Mormyrids display remarkable levels of differentiation in terms of shape and length across different species [11]. Such differences, which can be dramatic even among closely related species, are considered to have acted as effective prezygotic isolation mechanisms, promoting thus the adaptive radiation observed in this family, facilitated by either ecological speciation [12–14] or speciation driven by sexual selection [15].
In all mormyrids, the adult electric organ is located in the caudal peduncle and is formed by four columns of electrocytes, two dorsal and two ventral ones. Each electrocyte is innervated by electromotoneurons originating in the spinal cord [3]. Electric organs arise in juvenile fishes from several myomeres of the deep lateral muscle; their myogenic origin is confirmed by the presence of disorganized myofibrils within the electrocytes [16, 17].
During the last decade, several studies have investigated the genetic basis of EOD function and evolution [18–22]. Some of them have stressed the importance of duplication in a class of sodium channel genes for the origin of EOD production and diversification in weakly-electric fishes, by revealing the presence of important functional substitutions across paralogs and by discovering their differential patterns of expression between electric organ (EO) and skeletal muscle (SM) [18, 19, 22]. More recent studies however [20, 21], based on high throughput genomic technologies (e.g., SSH; RNA-Seq) have identified many differentially expressed genes, belonging to multiple functional classes (e.g., transcription factors; ion channels; sarcomeric proteins), between skeletal muscle and electric organ in several weakly-electric species (including representatives of Mormyridae). All studies conducted so far have been focusing on gene expression differences between two tissues –i.e., skeletal muscle and electric organ- within a species (cross-tissue approach), overlooking possible differences in the same tissue across different species (cross-species approach). However, tissue-specific gene expression differences across several species might underlie important phenotypic differences [23] which, in the case of the electric organ of mormyrid fishes, could explain the species-specific variability of EODs.
The aim of the present work is twofold; first we aim at exploring the differential patterns of gene expression between skeletal muscle and electric organ (cross-tissue comparison) in adult specimens belonging to the mormyrid genus Campylomormyrus (C. compressirostris and C. tshokwe). We focus then, on the identification of the differentially expressed genes, that are in common between the two species, and that might be responsible for the functional differences between the two tissues, and compare them to the results obtained by previous studies. The second task is to find differences in gene expression among three mormyrid species (C.compressirostris, C.tshokwe, and the outgroup species Gnathonemus petersii; Fig. 1) for the skeletal muscle and electric organ separately (cross-species comparison), and identify genes potentially related to phenotypic differences in EOD shape and duration.
Fig. 1.
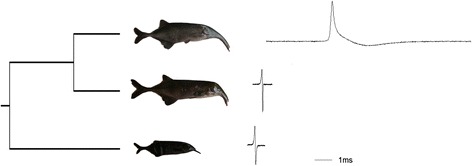
Analyzed species. The three species analysed in this study, with their relative EODs. From bottom to top: G. petersii, C. compressirostris, C. tshokwe
Results
Transcriptome sequencing and assembly
Sequencing of the twelve cDNA libraries produced a total amount of 371,043,357raw read pairs, resulting in 330,595,546 quality-filtered read pairs (89.1 %); see Additional file 1 for per library sequencing statistics. Trinity assembly resulted in 260,598 and 369,030 contigs for C. compressirostris and C.tshokwe cross-tissue transcriptomes respectively (Table 1); 357,832 and 399,878 contigs were obtained for the SM and EO cross-species assemblies respectively (Table 2). Contigs were then compared to the Danio rerio proteome, retrieving 18,458 and 19,363 unique proteins for C. compressirostris and C.tshokwe respectively; of these retrieved matches, 7971 (43.1 %) and 8993 (46.4 %) hits corresponded to full or nearly full-length coding sequences (Fig. 2a). For the cross-species assemblies, 20,023 and 20,352 contigs, for the SM and EO respectively, matched unique proteins in the D. rerio proteome, with 8662 (43.3 %) and 8768 (43.1 %) hits corresponding to full or nearly full-length coding sequences (Fig. 2b).
Table 1.
Assembly statistics for the cross-tissue comparison
| C. compressirostris | C. tshokwe | |
|---|---|---|
| Trinity contigs | 260,598 | 369,030 |
| # of retrieved ORFs | 139,963 | 228,306 |
| # of unique PFAM domains | 9,986 | 9,941 |
| # of contigs matching D. rerio proteome | 108,705 | 159,263 |
| # of unique hits to D. rerio proteome | 18,458 | 19,363 |
| N50 | 3,010 | 3,597 |
| Average contig length | 1,392.16 | 1,672.28 |
| Total assembled bases | 362,794,286 | 617,123,151 |
Table 2.
Assembly statistics for the cross-species comparison
| SM | EO | |
|---|---|---|
| Trinity contigs | 357,832 | 399,878 |
| # of retrieved ORFs | 150,068 | 182,682 |
| # of unique PFAM domains | 10,374 | 10,752 |
| # of contigs matching D. rerio proteome | 193,516 | 232,755 |
| # of unique hits to D. rerio proteome | 20,023 | 20,352 |
| N50 | 2,299 | 2,345 |
| Average contig length | 1,119.38 | 1,149.25 |
| Total assembled bases | 400,549,148 | 459,560,613 |
Fig. 2.
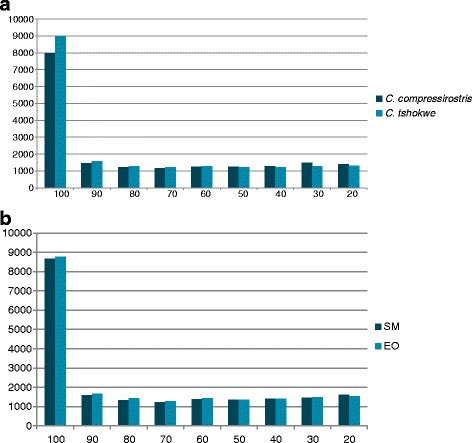
Distribution of length coverage between Campylomormyrus Trinity transcripts and corresponding top-blast hits (D. rerio proteome). Histogram showing the distribution of the percent in length of the sequences in the D. rerio proteome that aligns to the assembled Trinity contigs. Numbers on the x-axis indicate the upper limit of the binned interval (e.g., 100 is the upper value of the interval 100–91). a Cross-tissue comparison. b Cross-species comparison
Differential Expression (DE) analysis
Cross-tissue comparison
After transcript quantification with RSEM and DE-analysis with edgeR, 1313 transcripts resulted to be differentially expressed between EO and SM in C. compressirostris (356 up-regulated in EO and 957 down-regulated in EO) and 1002 in C. tshokwe (594 up-regulated in EO and 408 down-regulated in EO). Of all differentially expressed transcripts, 271 resulted to be shared between the two species (97 up-regulated in EO and 174 down-regulated in EO) (Fig. 3).
Fig. 3.
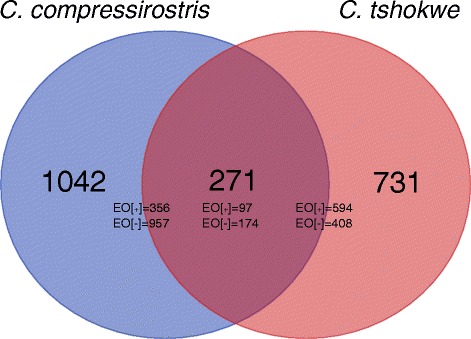
Number of differentially expressed genes (Cross-tissue). Venn diagram showing the amount of differentially expressed genes within each Campylomormyrus species’ transcriptome (full circles) and the amount of differentially expressed genes shared between the two Campylomormyrus species (overlapping area). The amount of genes that are up (EO[+])- or down (EO[−])-regulated in the electric organ are reported for each dataset
Cross-species comparison
In order to obtain an initial overview of transcriptome-wide gene expression patterns, we performed a principal component analysis on the expression levels of all assessed transcripts with non-zero levels in both assemblies (see methods for details). The results clearly separate the data according to tissue rather than species (Fig. 4). A distance matrix was then obtained from the same dataset and a neighbour-joining gene expression tree was built (see methods) in order to analyse global evolutionary trends in more detail. The obtained tree (Fig. 5) shows a clustering pattern for the EO data where species characterized by similar EODs (G. petersii, C. compressirostris) are grouped together, whereas the species with a rather different EOD (C. tshokwe) forms an isolated cluster. SM data, on the other hand, do not seem to form any particular clustering scheme.
Fig. 4.
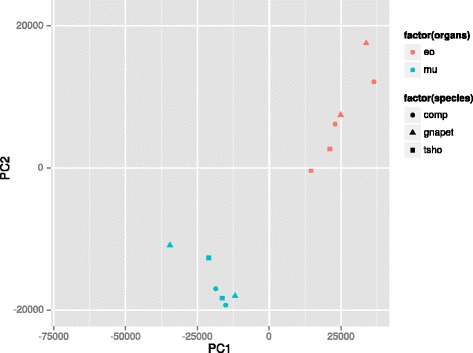
Principal component analysis of expression levels. Gnapet = G. petersii; comp = C. compressirostris; tsho = C. tshokwe; eo = electric organ; mu = skeletal muscle
Fig. 5.
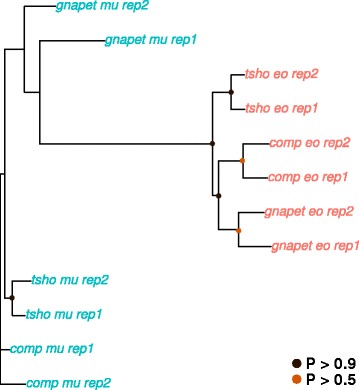
Neighbour-joining analysis of expression levels. Neighbour-joining tree based on pairwise distance matrix (1 – ρ, Spearman’s correlation coefficient) for EO and SM expression values. Bootstrap replicates = 10,000. Circles at nodes indicate bootstrap support. Gnapet = G. petersii; comp = C. compressirostris; tsho = C. tshokwe; eo = electric organ; mu = skeletal muscle; rep = replicate
After DE analysis, 166 and 950 genes resulted to be differentially expressed across the three analysed species for SM and EO, respectively. Within the skeletal muscle 78 genes are up-regulated in G. petersii, 16 in C. compressirostris, 17 in C. tshokwe, and 55 in C. compressirostris and C. tshokwe together (Fig. 6a). As far as the electric organ is concerned, 232 genes are up-regulated in G. petersii, 87 in C. compressirostris, and 631 in C. tshokwe (Fig. 7a).
Fig. 6.
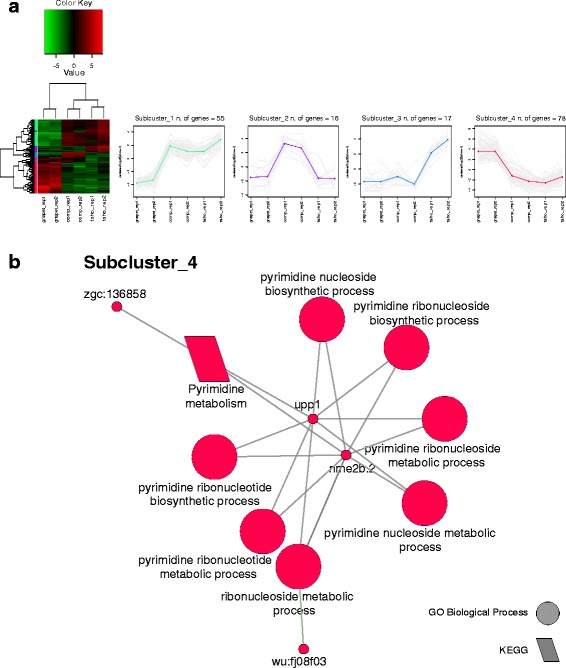
Cross-species analysis (SM). a Results of the DE analysis. Left: heatmap showing the differentially expressed genes clustered by expression levels. Expression sub-clusters obtained from k-mean clustering. Each cluster groups together genes characterized by similar expression levels. b Network showing significantly enriched terms and their relative genes for each sub-cluster
Fig. 7.
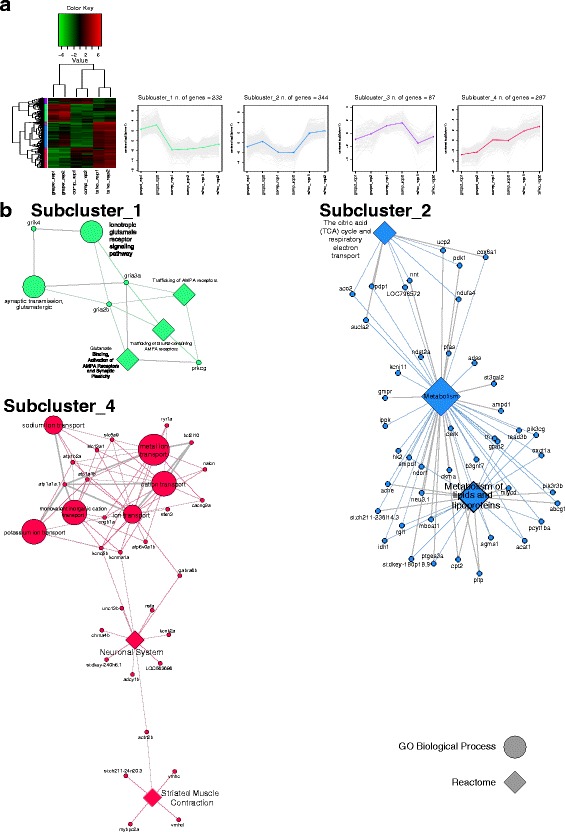
Cross-species analysis (EO). a Results of the DE analysis. Left: heatmap showing the differentially expressed genes clustered by expression levels. Expression sub-clusters obtained from k-mean clustering. Each cluster groups together genes characterized by similar expression levels. b Networks showing significantly enriched terms and their relative genes for each sub-cluster
Functional annotation
Cross-tissue comparison
In order to identify over-represented functional categories and pathways within the C. compressirostris and C. tshokwe transcriptomes, the sets of up- and down-regulated genes in the electric organ were subject to an enrichment analysis involving Gene Ontology categories, as well as Reactome and KEGG biological pathways (Fig. 8). The number of retrieved categories/pathways is proportional to the amount of differentially expressed genes present in each analysed set; with fewer categories for the EO up-regulated genes compared to the EO down-regulated genes in C. compressirostris, and a similar number of categories identified between the two sets in C. tshokwe. For both species the most represented categories within the EO up-regulated genes are related to ion channel transport (e.g., sodium ion transport), multicellular organismal development, development and patterning of nerves (branching morphogenesis of a nerve; semaphorin interactions; axon guidance). The genes found to be down-regulated in the EO regard mainly functional classes like: metabolic pathways specific to muscle tissue (oxidative phosphorylation, pyruvate metabolism, calcium signalling pathway), and muscle specific categories (muscle cell differentiation, striated muscle contraction, cardiac muscle contraction).
Fig. 8.
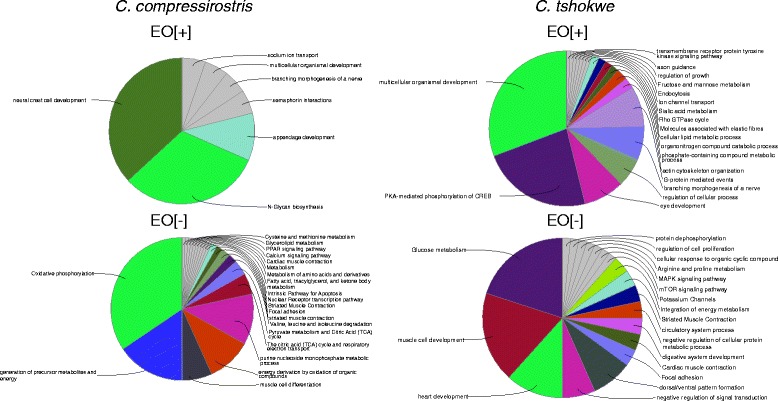
Functional annotation results (Cross-tissue). Pie charts showing the composition in terms of enriched functional categories (GO) and pathways (KEGG, Reactome) for each cluster of differentially expressed genes. EO[+] = up-regulated in the electric organ; EO [−] = down-regulated in the electric organ
Given the information provided by the category-based functional annotation, and to better understand the functional differences in terms of gene expression between EO and SM, independently from the species analysed, a literature search was performed on the shared set of differentially expressed genes between C. compressirostris and C. tshokwe. For each gene, phenotypic information consequent to its mis-expression (e.g., via knockdowns or non-sense mutations) in D. rerio was retrieved from the “Zebrafish Model Organism Database” (ZFIN; http://zfin.org/). All shared genes were divided into five “general” functional classes, which synthesize the categories reported in Fig. 8. The chosen categories are: “electrical activity” (genes responsible for the differential accumulation and transfer of ions across the plasma membrane), “muscular activity” (genes important for keeping a functional muscle phenotype), “metabolism” (genes involved in metabolic pathways), “transcription factors” (genes regulating gene expression) and “signal transduction” (molecules involved in signalling pathways) (Tables 2, 3, 4, 5 and 6). Many of the genes present in the category “electrical activity” are up-regulated in the EO (Table 2), they include genes coding for Na+/K+ pumps (atp1a2a), voltage-gated sodium (scn4aa) and potassium channels (kcnq5a) and cholinergic receptors (chrna7). However, other voltage-gated ion channels result to be down-regulated in the EO (kcna3, cacna2d2). There are then two members of the subfamily J of inwardly-rectifying potassium channels that show distinct patterns of expression, with one member (kcnj9) up-regulated in EO and the other (kcnj12) down-regulated. All the genes included in the class “muscular activity” are down-regulated in the EO (Table 3). As far as the “metabolism” genes are concerned (Table 4), most of the EO up-regulated transcripts are involved in the metabolism of fatty acids, glycerol, and phospholipids (e.g., acsl3b, gdpd4a, cds1), whereas the down-regulated transcripts are more involved in muscle-specific, energy production processes, like glycolysis (aldoab) and gluconeogenesis (gpib). Among transcription factors (Table 5), two of the four known myogenic factors (transcription factors that activate the expression of sarcomeric proteins), are down-regulated in the EO (myog, myf6), while the other two (myoD, myf5) do not show significant differences in expression between the two tissues. Two basic helix-loop-helix (bHLH) transcription factors (hey1, hes6) and one co-factor (her6) are up-regulated in the electric organ. Two myocyte enhancer factors (mef2aa, mef2b) show high levels of expression in the EO, whereas two regulators of SM cell proliferation (six1b, six4b) are lowly expressed in the EO. Most of the EO up-regulated genes involved in signal transduction (Additional file 9) belong to the G-protein coupled receptor (GPCR) signalling pathway (e.g., arhgef7a, arhgef7b, gpr22) and to the fibroblast growth-factor receptor (FGFR) signalling pathway (e.g., fgf8a, kal1b)
Table 3.
Electrical activity
| Gene | Protein name | Expression in EO | Pathway/Function | Disrupted phenotype | Reference |
|---|---|---|---|---|---|
| atp1a2a | ATPase, Na+/K+ transporting, alpha 2a polypeptide | + | Ion channel transport | Impaired depolarization of the resting membrane potential in slow-twitch fibers of skeletal muscles. | [50] |
| chrna7 | cholinergic receptor, nicotinic, alpha 7 (neuronal) | + | Activation of Nicotinic Acetylcholine Receptors | ||
| kcnj9 | potassium inwardly-rectifying channel, subfamily J, member 9 | + | Potassium Channels; GABA receptor activation | ||
| kcnq5a | potassium voltage-gated channel, KQT-like subfamily, member 5a | + | Potassium Channels; Synaptic transmission ion currents | ||
| grik3 | Glutamate Receptor, Ionotropic, Kainate 3 | + | Transmission across Chemical Synapses | ||
| scn4aa | sodium channel, voltage-gated, type IV, alpha, a | + | Ion channel transport; Axon guidance | ||
| kcna3 | potassium voltage-gated channel, shaker-related subfamily, member 3 | - | Potassium Channels; Transmission across Chemical Synapses | ||
| kcnj12 | potassium inwardly-rectifying channel, subfamily J, member 12 | - | Potassium Channels; GABA receptor activation | ||
| cacna2d2 | calcium channel, voltage-dependent, alpha 2/delta subunit 2 | - | Ion channel transport |
For each of the shared differentially expressed gene are reported: the gene and protein names obtained from the top hit blast results against the proteome of D. rerio; whether it is up(+)- or down(−)- regulated in the EO; its function or pathway (or both when available); the phenotypic effect on D. rerio of its mis-expression (when available)
Table 4.
Muscular activity
| Gene | Protein name | Expression in EO | Pathway/Function | Disrupted phenotype | Reference |
|---|---|---|---|---|---|
| atp2a1 | ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 | - | Muscle contraction | Abnormal locomotion | [51] |
| atp2a2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2a | - | regulation of heart contraction | Abnormal heart development | [31] |
| casq1a | calsequestrin 1a | - | Calcium homeostasis | ||
| jph1a | junctophilin 1a | - | structural constituent of muscle | ||
| jph1b | junctophilin 1b | - | structural constituent of muscle | ||
| myl2a | myosin, light chain 2a, regulatory, cardiac, slow | - | Striated Muscle Contraction | ||
| mybpc2a | myosin binding protein C, fast type a | - | Striated Muscle Contraction | ||
| mybpc3 | myosin binding protein C, cardiac | - | Cardiac muscle contraction | Abnormal heart development | [52] |
| myhb | myosin, heavy chain b | - | Striated Muscle Contraction | ||
| myl10 | myosin, light chain 10, regulatory | - | Regulation of actin cytoskeleton; Focal adhesion | ||
| myl12.2 | myosin, light chain 12, genome duplicate 2 | - | Striated Muscle Contraction | ||
| mylk2 | myosin light chain kinase 2 | - | Focal adhesion; Regulation of actin cytoskeleton | ||
| mylk3 | myosin light chain kinase 3 | - | Focal adhesion; Regulation of actin cytoskeleton | Cardiac sarcomere disruption | [53] |
| mylpfb | myosin light chain, phosphorylatable, fast skeletal muscle b | - | Focal adhesion; Regulation of actin cytoskeleton | ||
| myo18ab | myosin XVIIIAb | - | Signaling by FGFR | ||
| myoz3a | myozenin 3a | - | Calcineurin signaling | ||
| nexn | nexilin (F actin binding protein) | - | cardiac muscle fiber development | ||
| parvb | parvin, beta | - | Focal adhesion; Cell junction organization | Abnormal trunk musculature development | [54] |
| pdlim3b | PDZ and LIM domain 3b | - | |||
| pdlim5b | PDZ and LIM domain 5b | - | |||
| pvalb3 | parvalbumin 3 | - | calcium ion homeostasis | ||
| ryr1a | ryanodine receptor 1a (skeletal) | - | calcium ion channel transport | Abnormal trunk musculature development | [32] |
| ryr1b | ryanodine receptor 1b (skeletal) | - | calcium ion channel transport | Abnormal trunk musculature development | [33] |
| tnnc2 | troponin C type 2 (fast) | - | Striated Muscle Contraction | ||
| smpx | small muscle protein, X-linked | - | Striated Muscle Contraction | ||
| smyd1b | SET and MYND domain containing 1b | - | Muscle Development | Thick myosin filament disorganization | [55] |
| srl | sarcalumenin | - | calcium ion homeostasis | ||
| stac3 | SH3 and cysteine rich domain 3 | - | Striated Muscle Contraction | Excitation–contraction coupling disruption | [34] |
| tcap | titin-cap (telethonin) | - | Striated Muscle Contraction | Myofibril disorganization | [30] |
| tmod4 | tropomodulin 4 (muscle) | - | Muscle contraction | ||
| tnnc1b | troponin C type 1b (slow) | - | Muscle contraction | ||
| tnni2b.2 | troponin I type 2b (skeletal, fast), tandem duplicate 2 | - | Striated Muscle Contraction | ||
| tnnt1 | troponin T type 1 (skeletal, slow) | - | Muscle contraction | ||
| tnnt3b | troponin T type 3b (skeletal, fast) | - | Striated Muscle Contraction | ||
| tpm1 | tropomyosin 1 (alpha) | - | Striated Muscle Contraction | ||
| tpm2 | tropomyosin 2 (beta) | - | Striated Muscle Contraction | ||
| trdn | triadin | - | Muscle contraction | ||
| trim54 | tripartite motif containing 54 | - | Titin-kinase regulation | ||
| xirp1 | xin actin-binding repeat containing 1 | - | |||
| myl13 | myosin, light chain 13 | - |
For each of the shared differentially expressed gene are reported: the gene and protein names obtained from the top hit blast results against the proteome of D. rerio; whether it is up(+)- or down(−)- regulated in the EO; its function or pathway (or both when available); the phenotypic effect on D. rerio of its mis-expression (when available)
Table 5.
Metabolism
| Gene | Protein name | Expression in EO | Pathway/Function | Disrupted phenotype | Reference |
|---|---|---|---|---|---|
| acsbg2 | acyl-CoA synthetase bubblegum family member 2 | - | Fatty acid metabolism | ||
| acsl3b | acyl-CoA synthetase long-chain family member 3b | + | Fatty acid metabolism | ||
| acy1 | Aminoacylase-1 | - | Aminoacids metabolism | ||
| adssl1 | adenylosuccinate synthase like 1 | - | Purine metabolism | ||
| aldoab | Fructose-bisphosphate aldolase | - | Glycolysis | ||
| ampd1 | Adenosine monophosphate deaminase 1 (Isoform M) | - | Purine metabolism | ||
| aoc2 | amine oxidase, copper containing 2 | + | beta-Alanine metabolism | ||
| cds1 | CDP-Diacylglycerol Synthase 1 | + | Glycerophospholipid biosynthesis | Imperfect angiogenesis | [56] |
| ckma | creatine kinase, muscle a | - | Metabolism of amino acids and derivatives | ||
| ckmt2a | creatine kinase, mitochondrial 2a (sarcomeric) | - | Metabolism of amino acids and derivatives | ||
| cox4i2 | cytochrome c oxidase subunit IV isoform 2 | - | Oxidative phosphorylation | ||
| cpt2 | carnitine palmitoyltransferase 2 | + | Fatty acid beta-oxidation | ||
| cyp24a1 | cytochrome P450, family 24, subfamily A, polypeptide 1 | - | Steroid biosynthesis | ||
| dhrs9 | dehydrogenase/reductase (SDR family) member 9 | + | Retinol metabolism | ||
| gdpd4a | glycerophosphodiester phosphodiesterase domain containing 4a | + | Glycerol metabolism | ||
| gdpd5a | glycerophosphodiester phosphodiesterase domain containing 5a | + | Glycerol metabolism | ||
| gdpd5b | glycerophosphodiester phosphodiesterase domain containing 5b | + | Glycerol metabolism | ||
| glo1 | Glyoxalase 1 | - | Pyruvate metabolism | ||
| glud1b | glutamate dehydrogenase 1b | - | Nitrogen metabolism | ||
| got2a | glutamic-oxaloacetic transaminase 2a, mitochondrial | - | Glucose metabolism; aminoacids metabolism | ||
| gpib | glucose-6-phosphate isomerase b | - | Gluconeogenesis | ||
| idi1 | isopentenyl-diphosphate delta isomerase 1 | - | Cholesterol biosynthesis | ||
| man1a1 | mannosidase, alpha, class 1A, member 1 | + | N-Glycan biosynthesis | ||
| me3 | malic enzyme 1, NADP(+)-dependent, cytosolic | - | Pyruvate metabolism | ||
| pcyox1 | prenylcysteine oxidase 1 | - | Terpenoid backbone biosynthesis | ||
| pfkfb1 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | - | Glycolysis | ||
| pgam2 | phosphoglycerate mutase 2 (muscle) | - | Glycolysis and Gluconeogenesis | ||
| pgm5 | Phosphoglucomutase 5 | - | Glucuronidation | Failure in myofibril assembly | [57] |
| ucp3 | uncoupling protein 3 | - | Respiratory electron transport | ||
| ugp2b | UDP-glucose pyrophosphorylase 2b | - | Glucose metabolism | ||
| gyg1a | glycogenin 1a | - | Glycogen Metabolism | ||
| mid1ip1l | MID1 interacting protein 1, like | - | lipid metabolic process |
For each of the shared differentially expressed gene are reported: the gene and protein names obtained from the top hit blast results against the proteome of D. rerio; whether it is up(+)- or down(−)- regulated in the EO; its function or pathway (or both when available); the phenotypic effect on D. rerio of its mis-expression (when available)
Table 6.
Transcription factors
| Gene | Protein name | Expression in EO | Pathway/Function | Disrupted phenotype | Reference |
|---|---|---|---|---|---|
| eng1b | engrailed homeobox 1b | + | neuron fate commitment | ||
| her6 | hairy-related 6 | + | Notch signaling pathway | ||
| hes6 | hes family bHLH transcription factor 6 | + | Notch signaling pathway | ||
| hey1 | hes-related family bHLH transcription factor with YRPW motif 1 | + | Notch signaling pathway | ||
| hipk2 | homeodomain interacting protein kinase 2 | + | Wnt signaling pathway; p53 Signaling; ERK Signaling | Induced apoptosis | [58] |
| hoxc11a | homeobox C11a | + | |||
| hoxd11a | homeobox D11a | + | |||
| mef2aa | myocyte enhancer factor 2aa | + | Signaling by FGFR | Abnormal development of posterior somites | [29] |
| mef2b | myocyte enhancer factor 2b | + | miRs in Muscle Cell Differentiation | ||
| rb1 | retinoblastoma 1 | + | E2F mediated regulation of DNA replication; Cell cycle | Abnormal retina development | [59] |
| taf6 | TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor | + | GPCR Pathway | ||
| arxa | aristaless related homeobox a | - | Axon guidance | Abnormal dopaminergic neurons development | [60] |
| klf15 | Kruppel-like factor 15 | - | Adipogenesis | ||
| pbxip1 | pre-B-cell leukemia homeobox interacting protein 1 | - | |||
| myf6 | myogenic factor 6 | - | Myogenesis | Disrupted myogenesis | [28] |
| myog | myogenin | - | Myogenesis | Disrupted myogenesis | [27] |
| nfatc1 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 | - | Wnt signaling pathway | ||
| nr0b2a | nuclear receptor subfamily 0, group B, member 2a | - | Nuclear Receptor transcription pathway; NOD-like Receptor Signaling Pathways | ||
| pitx2 | paired-like homeodomain 2 | - | retinoic acid receptor signaling pathway | Abnormal eye and craniofacial development | [61] |
| rxrgb | retinoid X receptor, gamma b | - | steroid hormone receptor activity; retinoic acid receptor signaling pathway | ||
| six1b | SIX homeobox 1b | - | regulation of skeletal muscle cell proliferation | Abnormal trunk musculature development | [62] |
| six4b | SIX homeobox 4b | - | regulation of skeletal muscle cell proliferation | ||
| tbx15 | T-box 15 | - | regulation of transcription, DNA-templated |
For each of the shared differentially expressed gene are reported: the gene and protein names obtained from the top hit blast results against the proteome of D. rerio; whether it is up(+)- or down(−)- regulated in the EO; its function or pathway (or both when available); the phenotypic effect on D. rerio of its mis-expression (when available)
Cross-species comparison
The two sets of differentially expressed genes identified for SM and EO across the three analysed species were each partitioned into four sub-clusters with related expression patterns (see the methods section for details); each sub-cluster was then subjected to an enrichment analysis like the one described in the previous paragraph and in the methods section. Of the analysed sub-clusters for the SM dataset, one out of four showed significantly enriched terms, all related to nucleotides metabolic processes (Fig. 6b). Conversely, three out of four sub-clusters were significantly enriched in functional categories for the EO dataset (Fig. 7b). The most representative enriched functional categories are: glutamate receptor activity (sub-cluster 1); TCA cycle and fatty acid metabolism (sub-cluster 2); ion transport, neuronal system, and striated muscle contraction (sub-cluster 4).
For each of the analysed sub-clusters genes with known phenotypic effect in D. rerio or H. sapiens are reported in Table 7.
Table 7.
Cross-species differentially expressed genes with known phenotypic effect
| Gene | Protein | Tissue | Sub-cluster | Phenotype | Source |
|---|---|---|---|---|---|
| nme2b.2 | NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 2 | SM | 4 | GTP biosynthesis | ZFIN |
| prkcg | protein kinase C, gamma | EO | 1 | AMPA-R kinetics | ZFIN |
| aco2 | aconitase 2, mitochondrial | EO | 2 | Infantile cerebellar-retinal degeneration | OMIM |
| pdp1 | pyruvate dehyrogenase phosphatase catalytic subunit 1 | EO | 2 | Pyruvate dehydrogenase phosphatase deficiency | OMIM |
| nnt | nicotinamide nucleotide transhydrogenase | EO | 2 | Glucocorticoid deficiency 4 | OMIM |
| sucla2 | succinate-CoA ligase, ADP-forming, beta subunit | EO | 2 | Mitochondrial DNA depletion syndrome 5 | OMIM |
| kcnj11 | potassium inwardly-rectifying channel, subfamily J, member 11 | EO | 2 | Diabetes mellitus | OMIM |
| ippk | inositol 1,3,4,5,6-pentakisphosphate 2-kinase | EO | 2 | Craniofacial development | ZFIN |
| smpd1 | sphingomyelin phosphodiesterase 1, acid lysosomal | EO | 2 | Niemann-Pick disease | OMIM |
| th | tyrosine hydroxylase | EO | 2 | Adult brain function | ZFIN |
| ache | acetylcholinesterase | EO | 2 | Locomotion | ZFIN |
| oxct1a | 3-oxoacid CoA transferase 1a | EO | 2 | Succinyl CoA:3-oxoacid CoA transferase deficiency | OMIM |
| mlycd | malonyl-CoA decarboxylase | EO | 2 | Malonyl-CoA decarboxylase deficiency | OMIM |
| idh1 | isocitrate dehydrogenase 1 (NADP+), soluble | EO | 2 | Susceptibility to glioma | OMIM |
| cpt2 | carnitine palmitoyltransferase 2 | EO | 2 | Myopathy/Encephalopathy | OMIM |
| pltp | phospholipid transfer protein | EO | 2 | HDL cholesterol level | OMIM |
| acat1 | acetyl-CoA acetyltransferase 1 | EO | 2 | Alpha-methylacetoacetic aciduria | OMIM |
| atp1a1a.1 | ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 1 | EO | 3 | Brain development | ZFIN |
| ryr1a | ryanodine receptor 1a (skeletal) | EO | 3 | Myopathy | ZFIN |
| bcl2l10 | BCL2-like 10 (apoptosis facilitator) | EO | 3 | Cytoskeletal activity | ZFIN |
| nalcn | sodium leak channel, non-selective | EO | 3 | Hypotonia | OMIM |
| cngb1a | cyclic nucleotide gated channel beta 1a | EO | 3 | Retinitis pigmentosa 45 | OMIM |
| kcnma1a | potassium large conductance calcium-activated channel, subfamily M, alpha member 1a | EO | 3 | Hearing sensitivity | ZFIN |
| kcnq5b | potassium voltage-gated channel, KQT-like subfamily, member 5b | EO | 3 | Cell membrane excitability | ZFIN |
| nsfa | N-ethylmaleimide-sensitive factor a | EO | 3 | Axon development | ZFIN |
| chrna4b | cholinergic receptor, nicotinic, alpha 4b | EO | 3 | Epilepsy | OMIM |
| adcy1b | adenylate cyclase 1b | EO | 3 | Deafness | OMIM |
| actn2b | actinin, alpha 2b | EO | 3 | Cardiomyopathy | OMIM |
| vmhc | ventricular myosin heavy chain | EO | 3 | Cardiomyopathy | OMIM |
List of cross-species differentially expressed genes belonging to the terms obtained from the enrichment analysis. For each gene, we report: the analysed tissue; the relative sub-cluster as reported in Figs. 6 or 7; and the phenotypic effect of gene function disruption on D. rerio (ZFIN) or Homo sapiens (OMIM)
Discussion
Cross-tissue comparison
Functional annotation of the 271 differentially expressed genes that are shared between C. compressirostris and C. tshokwe has revealed marked differences within several functional categories, which are probably critical in determining the observed phenotypic differences between the electric organ and the skeletal muscle. Below, the functional implications of the differentially expressed genes, in the light of what is known from other fish models, are discussed.
Electrical activity
The up-regulation of the atp1a2a gene is explained by the fact that its product, the Na+/K+ ATP-ase, is fundamental for keeping the electrochemical gradient across the plasma membrane. Over-expression of this gene was already observed in the mormyrid fish Brienomyrus brachyistius [20], as well as in several species of south-American weakly-electric fishes (Gymnotiformes) [21]. Voltage-gated ion channels, on the other hand, are important for dissipating the electric potential generated by the ATP-ases and therefore for producing an EOD in response to an action potential. In the electric organ of the analyzed species, one gene coding for a voltage-gated sodium channel (scn4aa) is highly expressed in the electric organ; differential expression of this gene and of its paralog (scn4ab) between EO and SM was demonstrated by Zakon et al. [18] for mormyrid and gymnotiform fishes, and suggest the role of gene duplication followed by neo-functionalization as a main driver for the evolution of electric communication [19]. Other over-expressed genes that increase cell excitability are the potassium channels kcnq5a and kcnj9. The latter belongs to the family of inwardly rectifying potassium channels, a class of ion channels that favour the influx of K+ ions in the cell; up-regulation of members of this family was observed in the EO of the Electric eel (E. electricus) [24].
Repression of muscular phenotype in the EO
Many of the differentially expressed transcription factors retrieved in this study are fundamental for the regulation of myogenic development. In particular, we have found that two bHLH transcription factors: hey1 and hes6, in co-operation with her6, are up-regulated in the EO; these factors are known to negatively regulate the expression of myogenic factors in several model organisms [25, 26], including electric fish [21]. Two of the four known myogenic regulatory factors (MRFs: myog, myf6) are down-regulated in the EO, both genes are fundamental for muscle development and differentiation [27]; in particular, knock-down experiments on myf6 have shown the degradation of posterior somites in D. rerio [28], the region where the adult EO originates [17]. Another gene important for muscle development is the myocyte enhancer factor mef2aa. Unlike MRFs, this gene is up-regulated in the electric organ of the two species analysed here, as well as in other electric fish species [20, 21], and it is also important for the correct development of posterior somites in D. rerio [29]. The concerted activity of transcriptional repressors and co-repressors of the myogenic program results in the down-regulation of genes coding for muscle specific proteins (Table 4), which finally determine the non-muscle characteristics of the EO like: i) the presence of few, non-contractile, myofibrils [17] (e.g., tcap [30]); ii) loss of calcium compartmentalization activity (e.g., atp2a1, atp2a2, casq1a [31]); and iii) decoupling of the excitation-contraction process (e.g., ryr1, stac3, jph1 [32–34]).
Metabolic activity
The observed differences in terms of gene expression between EO and SM suggest that the metabolic machinery of the electric organ could be mainly devoted to the production and turn-over of membrane structures. Indeed, many of the metabolism-related genes up-regulated in the EO are involved in the metabolism of fatty acids (acsbg2, acsl3b, cpt2), glycerophospholipids (cds1, gdpd4a, gdpd5a, gdpd5b), and cholesterol (idi1). On the other hand, most of the SM up-regulated genes are involved in typical processes of muscle metabolism like: glycolysis (aldoab, glo1, me3); gluconeogenesis (gpib, pgam2); and aminoacids metabolism (acy1, ckma, ckmt2a).
Cross-species comparison
The grouping pattern emerging from the principal component analysis, where expression levels tend to group in an “organ-wise” rather than a “species-wise” fashion, is putatively due to the fact that expression levels are conserved for the same organ across different species for functional reasons. A similar pattern was already observed for more tissues across broader phylogenetic distances [23].
The clustering scheme obtained from the neighbour-joining analysis for the EO data might be indicative of the observed differences in terms of EOD among the three species, which may be reflected in the expression levels of a conspicuous part of the EO transcriptome. Previous studies [23, 35] have revealed that, for most tissues, gene expression levels tend to accumulate over evolutionary time, such that more closely related species have more similar expression levels. However, for tissues characterized by increased levels of adaptation (e.g., testis and liver in mammals), expression trees tend to group according to phenotypic similarity [23].
The results of the enrichment analysis conducted on the expression clusters for the EO have revealed interesting results. In particular, sub-cluster 2 and sub-cluster 4 (Fig. 7b) are enriched in terms which might underlie the observed EOD differences across the three species; both sub-clusters are characterized by genes which are mainly up-regulated in the EO of C. tshokwe. The terms relative to sub-cluster 2 are all related to metabolic pathways, the metabolism of fatty acids in particular. Many of the observed genes are involved in the production and turnover of cell membranes (e.g., smpd1, oxct1a, mlycd, cpt2). Sub-cluster 4 is mainly characterized by genes involved in ion transport and neuronal function; of particular importance here are sodium/potassium ATPases (atp1a1a, atp1a1b, atp1b2a), as their over-expression in the EO of C. tshokwe might explain the higher amplitude observed in its EOD [36]. Other genes which may potentially influence EOD features are potassium channels (kcnq5b, kcnma1a, kcnk2a, kcnj11).
Conclusions
The cross-tissue analysis of differentially expressed genes between skeletal muscle and electric organ in two species of African weakly-fishes suggests that: i) the loss of contractile activity and the decoupling of the excitation-contraction processes are reflected by the down-regulation of the corresponding genes in the electric organ; ii) the metabolic activity of the EO might be specialized towards the production and turn-over of membrane structures; iii) several ion channels are highly expressed in the EO in order to increase excitability; iv) several myogenic factors might be down-regulated by transcription repressors in the EO.
The cross-species analysis has revealed that the EO transcriptome is more variable in terms of gene expression levels across species than the SM transcriptome. The functional annotation indicates that the most diverging functional classes across species in the EO include “metabolism of fatty acids” and “ion transport”.
In order to better understand the role played by the differentially expressed gene identified in this study, the onset of transgenic experiments (e.g., knockdown) will be necessary either in fully established model organisms (D. rerio), or in emerging models for electric fish (E. electricus).
Methods
Specimen collection
Several specimens of C. compressirostris, C. tshokwe and G. petersii were collected in the wild during a sampling campaign conducted at the Congo River rapids south of Brazzaville (Republic of the Congo, August/September 2012). For the present study, adult female specimens for each species were selected, kept in captivity for a maximum period of 2 weeks and then euthanized for tissue sample collection. Gender and sexual maturity were assessed after dissection by checking for the presence of mature ovaries in the selected specimens. Electric organ and skeletal muscle tissue samples were dissected from the caudal peduncle and the posterior trunk musculature respectively and immediately transferred into RNAlater® (Life Technologies). The research followed internationally recognized guidelines and applicable national legislation. We received ethical approval from the deputy of animal welfare of the University of Potsdam.
RNA extraction and cDNA library preparation
The dissected tissues were processed at the University of Potsdam for RNA extraction: they were first removed from the RNAlater®-containing vials; shock frozen in liquid nitrogen and then homogenized into a buffer containing guanidine isothiocyanate and β-mercaptoethanol using a Mini-beadbeater-1 (Biospec). Total RNA was extracted using the RNeasy® Mini Kit (Qiagen), RNA quality and concentration was inspected using a Fragment Analyzer™ (Advanced Analytical Technologies, Inc.).
For the present study eight cDNA libraries were selected for sequencing (one library per species per tissue; two biological replicates), each library was obtained by pooling the total RNA from a minimum of two to a maximum of 4 different individuals (see Additional file 1). The paired-end (100 nt), strand-specific cDNA libraries were prepared using the NEXTflex™ Directional RNA-Seq Kit V2 (dUTP based) (Bioo Scientific); preparation was performed in six steps: i) mRNA enrichment from total RNA via polyA selection; ii) fragmentation; iii) first and second strand syntheses; iv) A-tailing; v) adapter and barcode ligation and vi) PCR amplification. Fragment size distribution and quality was estimated using an Agilent 2100 Bioanalyzer with the High Sensitivity DNA Chip.
Transcriptome sequencing, assembly and annotation
Transcriptome sequencing was performed at the Max Delbrück Center for Molecular Medicine; the multiplexed cDNA libraries were sequenced using one lane of an Illumina HiSeq2000 sequencing system. After sequencing, the resulting raw reads were subject to five processing steps using the program Flexbar v2.4 [37] : i) filtering reads with uncalled bases; ii) trimming of reads at 3′-end to get a minimum average Phred quality score of 20; iii) barcode detection, removal and reads separation; iv) adapter detection and removal, and v) filtering of reads shorter than 20 bp after trimming. Quality control of both raw and processed reads was performed with FastQC v0.10.1 (Babraham Bioinformatics).
The processed reads were assembled de novo (i.e., without using a reference genome) with Trinity r20131110 [38] (kmer length = 25). Two reference transcriptomes were produced from C. compressirostris and C. tshokwe respectively, by assembling together the reads obtained from the EO and SM libraries. Combining all reads across all tissues and all biological replicates for each species (cross-tissue assembly), or across all species and all replicates for each tissue (cross-species assembly) (Fig. 9) into a single RNA-seq dataset, allows to correctly compare transcript abundances from the analysed tissues or species by aligning the short reads from each library independently onto the same set of reference transcripts (see below and Additional files 2, 3, 4, 5, 6 and 7 for more details) [39].
Fig. 9.
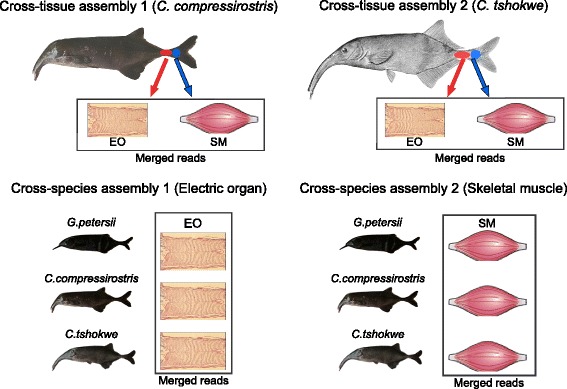
Assembly schemes for the cross-tissue and cross-species comparison
Transcriptome annotation was conducted using the stand-alone version of the blastx algorithm implemented in Blast + v2.2.29 [40] (E-value cutoff = 10−10) against the proteome of Danio rerio (Uniprot ID = UP000000437). Likely coding sequences were extracted from Trinity transcripts using TransDecoder (http://transdecoder.github.io/) and the longest translated Open Reading Frames (ORFs) were reported (Table 1). Protein domains were searched on the PFAM database (Pfam-A.hmm available at http://pfam.xfam.org/) using HMMER v3.1b1. The retrieved ORFs were later annotated by “blasting” them against the SwissProt (http://web.expasy.org/docs/swiss-prot_guideline.html) database using the blastp algorithm. Transcripts’ completeness was assessed by computing the proportion of transcripts and ORFs that matched to full-length top hits in their respective searches using the Perl script “analyze_blastPlus_topHit_coverage.pl” (provided with Trinity) (Fig. 2 and Additional file 8) [39].
The use of a single reference species for annotation is sub-optimal in terms of number of retrieved orthologs if compared to other methods like iterative BLAST searches to multiple species [41], however the use of a long-time established model organism (D. rerio) facilitates the enrichment analysis and the identification of experimental evidence for the functional role of a given gene.
Transcript abundance quantification and DE-analysis
Short reads were individually mapped to their respective transcriptome assemblies using Bowtie v1.0.0 [42] with default parameters. Gene expression levels were estimated using RSEM v1.2.12 [43]. Putative transcript artifacts and lowly expressed transcripts were filtered out using the Perl script “filter_fasta_by_rsem_values.pl” (provided with Trinity).
Principal component analyses of cross-species data was performed on a matrix of expression values of 14,436 genes with non-zero values in both assemblies, using the function “prcomp” in R. The same dataset was used for building a distance matrix of Spearman’s correlation coefficients (ρ) which was then subjected to neighbour-joining tree construction with the function “nj” in R (bootstrap = 10,000). Differential expression analysis was performed using the Bioconductor package edgeR [44] (minimum fold change = 4, p-value cutoff = 0.001 after FDR correction). The differentially expressed transcripts were then subject to an enrichment analysis using the Cytoscape plugin ClueGO v2.1.4 [45, 46], in order to identify over-represented functional categories from the Gene Ontology (GO) database [47] (http://geneontology.org), as well as over-represented biological pathways from KEGG (http://www.genome.jp/kegg/) and Reactome (http://www.reactome.org/) [48, 49]. Statistical significance was assessed using a Fisher’s exact test with FDR p-value correction (≤0.05). Cross-species data were partitioned into expression clusters using a k-means algorithm (k = 4), implemented in a perl script provided with Trinity.
Data availability
All the Illumina reads used for this study are available at the Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra), under the accession number SRP050174.
Acknowledgements
Funding was provided by the University of Potsdam and the Leibniz-SAW-project GENART. Computational resources were kindly provided by the Unit of Bioinformatics (J. Selbig, S. Hartmann) at the University of Potsdam.
The authors are immensely grateful to Dr. Victor Mamonekene (Université Marien Ngouabi, Brazzaville) for the logistic and scientific support provided during their stay in Congo.
Additional files
Sequencing statistics. For each of the eight produced libraries we report: the number of pooled individuals, the number of pre- and post- processing read pairs, and the percent of retained reads after quality filtering. (DOCX 17 kb)
Blast results of C. compressirostris transcriptome assembly. Tabular blast report showing the results of the blastx comparison between the transcriptome of C. compressirostris and the proteome of D. rerio. (TXT 14648 kb)
Blast results of C. tshokwe transcriptome assembly. Tabular blast report showing the results of the blastx comparison between the transcriptome of C. tshokwe and the proteome of D. rerio. (TXT 19887 kb)
Changes in gene expression between EO and SM for C. compressirostris transcriptome. Results of the edgeR DE-analysis. The table reports the fold-change in the expression levels and relative uncorrected and corrected p-values between EO and SM for all genes in the C. compressirostris transcriptome. (TXT 5878 kb)
Changes in gene expression between EO and SM for C. tshokwe transcriptome. Results of the edgeR DE-analysis. The table reports the fold-change in the expression levels and relative uncorrected and corrected p-values between EO and SM for all genes in the C. tshokwe transcriptome. (TXT 7336 kb)
Transcripts’ length distribution of the C. compressirostris Trinity assembly. (PDF 24 kb)
Transcripts’ length distribution of the C. tshokwe Trinity assembly. (PDF 5 kb)
Distribution of length coverage between retrieved ORFs and reference database (SwissProt). (PDF 44 kb)
List of differentially expressed genes belonging to the class “signal transduction”, obtained from the cross-tissue comparison. (DOCX 39 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
FL participated in specimen sampling, extracted the RNA, carried out the bioinformatic analyses and drafted the manuscript. FK participated in specimen sampling, determined sex and species for the collected samples and participated in manuscript drafting and supervision. IK produced the cDNA libraries and prepared them for sequencing. CD participated in experimental design and provided sequencing resources. RT participated in specimen sampling, conceived and supervised the study. All authors read and provided advice on the manuscript.
Contributor Information
Francesco Lamanna, Email: lamanna@uni-potsdam.de.
Frank Kirschbaum, Email: frank.kirschbaum@staff.hu-berlin.de.
Isabelle Waurick, Email: isabelle.waurick@mfn-berlin.de.
Christoph Dieterich, Email: christoph.dieterich@age.mpg.de.
Ralph Tiedemann, Email: tiedeman@uni-potsdam.de.
References
- 1.Bass A. Electric organs revisited: evolution of a vertebrate communication and orientation organ. In: Bullock TH, Heiligenberg W, editors. Electroreception. New York: Wiley; 1986. pp. 13–70. [Google Scholar]
- 2.Kirschbaum F. Myogenic electric organ precedes the neurogenic organ in apteronotid fish. Naturwissenschaften. 1983;70:205–7. doi: 10.1007/BF01047569. [DOI] [PubMed] [Google Scholar]
- 3.Moller P. Electric organs. In: Moller P, editor. Electr fishes hist behav. London: Chapman & Hall; 1995. pp. 385–402. [Google Scholar]
- 4.Daget J, Gosse JP. Thys van den audenaerde DFE: check list of the freshwater fishes of africa = catalogue Des poissons D’eau douce d’Afrique. Paris/Tervuren: ORSTOM/MRAC; 1984. [Google Scholar]
- 5.Lissmann HW. On the function and evolution of electric organs in fish. J Exp Biol. 1958;35:156–91. [Google Scholar]
- 6.Lissmann HW, Machin KE. The mechanism of object location in gymnarchus niloticus and similar fish. J Exp Biol. 1958;35:451–86. [Google Scholar]
- 7.Feulner PGD, Plath M, Engelmann J, Kirschbaum F, Tiedemann R. Electrifying love: electric fish use species-specific discharge for mate recognition. Biol Lett. 2009;5:225–8. doi: 10.1098/rsbl.2008.0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kramer B, Kuhn B. Species recognition by the sequence of discharge intervals in weakly electric fishes of the genus Campylomormyrus (Mormyridae, Teleostei) Anim Behav. 1994;48:435–45. doi: 10.1006/anbe.1994.1257. [DOI] [Google Scholar]
- 9.Bratton BO, Kramer B. Patterns of the electric organ discharge during courtship and spawning in the mormyrid fish, Pollimyrus isidori. Behav Ecol Sociobiol. 1989;24:349–68. doi: 10.1007/BF00293263. [DOI] [Google Scholar]
- 10.Crawford JD. Sex recognition by electric cues in a sound-producing mormyrid fish, pollimyrus isidori. Brain Behav Evol. 1991;38:20–38. doi: 10.1159/000114377. [DOI] [PubMed] [Google Scholar]
- 11.Alves-Gomes J, Hopkins CD. Molecular insights into the phylogeny of mormyriform fishes and the evolution of their electric organs. Brain Behav Evol. 1997;49:343–51. doi: 10.1159/000316291. [DOI] [PubMed] [Google Scholar]
- 12.Tiedemann R, Feulner P, Kirschbaum F. Evol action. Berlin Heidelberg: Springer; 2010. Electric organ discharge divergence promotes ecological speciation in sympatrically occurring African weakly electric fish (campylomormyrus) pp. 307–21. [Google Scholar]
- 13.Feulner PGD, Kirschbaum F, Mamonekene V, Ketmaier V, Tiedemann R. Adaptive radiation in African weakly electric fish (Teleostei: Mormyridae: Campylomormyrus): a combined molecular and morphological approach. J Evol Biol. 2007;20:403–14. doi: 10.1111/j.1420-9101.2006.01181.x. [DOI] [PubMed] [Google Scholar]
- 14.Feulner P, Kirschbaum F, Tiedemann R. Adaptive radiation in the Congo river: an ecological speciation scenario for African weakly electric fish (teleostei; mormyridae; campylomormyrus) J Physiol. 2008;102:340–6. doi: 10.1016/j.jphysparis.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Arnegard ME, McIntyre PB, Harmon LJ, Zelditch ML, Crampton WGR, Davis JK, Sullivan JP, Lavoué S, Hopkins CD. Sexual signal evolution outpaces ecological divergence during electric fish species radiation. Am Nat. 2010;176:335–56. doi: 10.1086/655221. [DOI] [PubMed] [Google Scholar]
- 16.Szabo T. Development of the electric organ of mormyridae. Nature. 1960;188:760–2. doi: 10.1038/188760b0. [DOI] [PubMed] [Google Scholar]
- 17.Denizot JP, Kirschbaum F, Westby GWM, Tsuji S. On the development of the adult electric organ in the mormyrid fish Pollimyrus isidori (with special focus on the innervation) J Neurocytol. 1982;11:913–34. doi: 10.1007/BF01148308. [DOI] [PubMed] [Google Scholar]
- 18.Zakon HH, Lu Y, Zwickl DJ, Hillis DM. Sodium channel genes and the evolution of diversity in communication signals of electric fishes: convergent molecular evolution. Proc Natl Acad Sci U S A. 2006;103:3675–80. doi: 10.1073/pnas.0600160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnegard ME, Zwickl DJ, Lu Y, Zakon HH. Old gene duplication facilitates origin and diversification of an innovative communication system--twice. Proc Natl Acad Sci U S A. 2010;107:22172–7. doi: 10.1073/pnas.1011803107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallant JR, Hopkins CD, Deitcher DL. Differential expression of genes and proteins between electric organ and skeletal muscle in the mormyrid electric fish Brienomyrus brachyistius. J Exp Biol. 2012;215:2479–94. doi: 10.1242/jeb.063222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallant JR, Traeger LL, Volkening JD, Moffett H, Chen PH, Novina CD, Phillips GN, Anand R, Wells GB, Pinch M, Guth R, Unguez GA, Albert JS, Zakon HH, Samanta MP, Sussman MR. Genomic basis for the convergent evolution of electric organs. Science (80-) 2014;344:1522–5. doi: 10.1126/science.1254432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novak AE, Jost MC, Lu Y, Taylor AD, Zakon HH, Ribera AB. Gene duplications and evolution of vertebrate voltage-gated sodium channels. J Mol Evol. 2006;63:208–21. doi: 10.1007/s00239-005-0287-9. [DOI] [PubMed] [Google Scholar]
- 23.Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, Albert FW, Zeller U, Khaitovich P, Grützner F, Bergmann S, Nielsen R, Pääbo S, Kaessmann H. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–8. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 24.Traeger LL, Volkening JD, Moffett H, Gallant JR, Chen P-H, Novina CD, Phillips GN, Anand R, Wells GB, Pinch M, Güth R, Unguez GA, Albert JS, Zakon H, Sussman MR, Samanta MP. Unique patterns of transcript and miRNA expression in the South American strong voltage electric eel (Electrophorus electricus) BMC Genomics. 2015;16:243. doi: 10.1186/s12864-015-1288-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer A, Gessler M. Delta-Notch--and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res. 2007;35:4583–96. doi: 10.1093/nar/gkm477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buas MF, Kabak S, Kadesch T. The Notch effector Hey1 associates with myogenic target genes to repress myogenesis. J Biol Chem. 2010;285:1249–58. doi: 10.1074/jbc.M109.046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinits Y, Osborn DPS, Hughes SM. Differential requirements for myogenic regulatory factors distinguish medial and lateral somitic, cranial and fin muscle fibre populations. Development. 2009;136:403–14. doi: 10.1242/dev.028019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y-H, Li C-K, Lee G-H, Tsay H-J, Tsai H-J, Chen Y-H. Inactivation of zebrafish mrf4 leads to myofibril misalignment and motor axon growth disorganization. Dev Dyn. 2008;237:1043–50. doi: 10.1002/dvdy.21478. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Qian L, Dong Y, Jiang Q, Gui Y, Zhong TP, Song H. Myocyte-specific enhancer factor 2A is essential for zebrafish posterior somite development. Mech Dev. 2006;123:783–91. doi: 10.1016/j.mod.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 30.Zhang R, Yang J, Zhu J, Xu X. Depletion of zebrafish Tcap leads to muscular dystrophy via disrupting sarcomere-membrane interaction, not sarcomere assembly. Hum Mol Genet. 2009;18:4130–40. doi: 10.1093/hmg/ddp362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ebert AM, Hume GL, Warren KS, Cook NP, Burns CG, Mohideen MA, Siegal G, Yelon D, Fishman MC, Garrity DM. Calcium extrusion is critical for cardiac morphogenesis and rhythm in embryonic zebrafish hearts. Proc Natl Acad Sci U S A. 2005;102:17705–10. doi: 10.1073/pnas.0502683102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jurynec MJ, Xia R, Mackrill JJ, Gunther D, Crawford T, Flanigan KM, Abramson JJ, Howard MT, Grunwald DJ. Selenoprotein N is required for ryanodine receptor calcium release channel activity in human and zebrafish muscle. Proc Natl Acad Sci U S A. 2008;105:12485–90. doi: 10.1073/pnas.0806015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dowling JJ, Arbogast S, Hur J, Nelson DD, McEvoy A, Waugh T, Marty I, Lunardi J, Brooks SV, Kuwada JY, Ferreiro A. Oxidative stress and successful antioxidant treatment in models of RYR1-related myopathy. Brain. 2012;135(Pt 4):1115–27. doi: 10.1093/brain/aws036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horstick EJ, Linsley JW, Dowling JJ, Hauser MA, McDonald KK, Ashley-Koch A, Saint-Amant L, Satish A, Cui WW, Zhou W, Sprague SM, Stamm DS, Powell CM, Speer MC, Franzini-Armstrong C, Hirata H, Kuwada JY. Stac3 is a component of the excitation-contraction coupling machinery and mutated in Native American myopathy. Nat Commun. 2013;4:1952. doi: 10.1038/ncomms2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perry GH, Melsted P, Marioni JC, Wang Y, Bainer R, Pickrell JK, Michelini K, Zehr S, Yoder AD, Stephens M, Pritchard JK, Gilad Y. Comparative RNA sequencing reveals substantial genetic variation in endangered primates Comparative RNA sequencing reveals substantial genetic variation in endangered primates. Genome Res. 2012;22:602–10. doi: 10.1101/gr.130468.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paul C, Mamonekene V, Vater M, Feulner PGD, Engelmann J, Tiedemann R, Kirschbaum F. Comparative histology of the adult electric organ among four species of the genus Campylomormyrus (Teleostei: Mormyridae) J Comp Physiol A. 2015;201(4):357–74. doi: 10.1007/s00359-015-0995-6. [DOI] [PubMed] [Google Scholar]
- 37.Dodt M, Roehr JT, Ahmed R, Dieterich C. FLEXBAR-flexible barcode and adapter processing for next-generation sequencing platforms. Biology (Basel) 2012;1:895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–52. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, Macmanes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, Leduc RD, Friedman N, Regev A. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tzika AC, Helaers R, Schramm G, Milinkovitch MC. Reptilian-transcriptome v1.0, a glimpse in the brain transcriptome of five divergent Sauropsida lineages and the phylogenetic position of turtles. Evodevo. 2011;2:19. doi: 10.1186/2041-9139-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman W-H, Pagès F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. Gene Ontology Consortium Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joshi-Tope G, Gillespie M, Vastrik I, D’Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L, Lewis S, Birney E, Stein L. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33(Database issue):D428–32. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doganli C, Kjaer-Sorensen K, Knoeckel C, Beck HC, Nyengaard JR, Honoré B, Nissen P, Ribera A, Oxvig C, Lykke-Hartmann K. The α2Na+/K + −ATPase is critical for skeletal and heart muscle function in zebrafish. J Cell Sci. 2012;125(Pt 24):6166–75. doi: 10.1242/jcs.115808. [DOI] [PubMed] [Google Scholar]
- 51.Friedrich T, Lambert AM, Masino MA, Downes GB. Mutation of zebrafish dihydrolipoamide branched-chain transacylase E2 results in motor dysfunction and models maple syrup urine disease. Dis Model Mech. 2012;5:248–58. doi: 10.1242/dmm.008383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y-H, Pai C-W, Huang S-W, Chang S-N, Lin L-Y, Chiang F-T, Lin J-L, Hwang J-J, Tsai C-T. Inactivation of Myosin binding protein C homolog in zebrafish as a model for human cardiac hypertrophy and diastolic dysfunction. J Am Heart Assoc. 2013;2:e000231. doi: 10.1161/JAHA.113.000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seguchi O, Takashima S, Yamazaki S, Asakura M, Asano Y, Shintani Y, Wakeno M, Minamino T, Kondo H, Furukawa H, Nakamaru K, Naito A, Takahashi T, Ohtsuka T, Kawakami K, Isomura T, Kitamura S, Tomoike H, Mochizuki N, Kitakaze M. A cardiac myosin light chain kinase regulates sarcomere assembly in the vertebrate heart. J Clin Invest. 2007;117:2812–24. doi: 10.1172/JCI30804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Postel R, Vakeel P, Topczewski J, Knöll R, Bakkers J. Zebrafish integrin-linked kinase is required in skeletal muscles for strengthening the integrin-ECM adhesion complex. Dev Biol. 2008;318:92–101. doi: 10.1016/j.ydbio.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 55.Li H, Zhong Y, Wang Z, Gao J, Xu J, Chu W, Zhang J, Fang S, Du SJ. Smyd1b is required for skeletal and cardiac muscle function in zebrafish. Mol Biol Cell. 2013;24:3511–21. doi: 10.1091/mbc.E13-06-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan W, Pham VN, Stratman AN, Castranova D, Kamei M, Kidd KR, Lo BD, Shaw KM, Torres-Vazquez J, Mikelis CM, Gutkind JS, Davis GE, Weinstein BM. CDP-diacylglycerol synthetase-controlled phosphoinositide availability limits VEGFA signaling and vascular morphogenesis. Blood. 2012;120:489–98. doi: 10.1182/blood-2012-02-408328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Molt S, Bührdel JB, Yakovlev S, Schein P, Orfanos Z, Kirfel G, Winter L, Wiche G, van der Ven PFM, Rottbauer W, Just S, Belkin AM, Fürst DO. Aciculin interacts with filamin C and Xin and is essential for myofibril assembly, remodeling and maintenance. J Cell Sci. 2014;127(Pt 16):3578–92. doi: 10.1242/jcs.152157. [DOI] [PubMed] [Google Scholar]
- 58.Bitomsky N, Conrad E, Moritz C, Polonio-Vallon T, Sombroek D, Schultheiss K, Glas C, Greiner V, Herbel C, Mantovani F, del Sal G, Peri F, Hofmann TG. Autophosphorylation and Pin1 binding coordinate DNA damage-induced HIPK2 activation and cell death. Proc Natl Acad Sci U S A. 2013;110:E4203–12. doi: 10.1073/pnas.1310001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gyda M, Wolman M, Lorent K, Granato M. The tumor suppressor gene retinoblastoma-1 is required for retinotectal development and visual function in zebrafish. PLoS Genet. 2012;8:e1003106. doi: 10.1371/journal.pgen.1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Filippi A, Jainok C, Driever W. Analysis of transcriptional codes for zebrafish dopaminergic neurons reveals essential functions of Arx and Isl1 in prethalamic dopaminergic neuron development. Dev Biol. 2012;369:133–49. doi: 10.1016/j.ydbio.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 61.Liu Y, Semina EV. pitx2 Deficiency results in abnormal ocular and craniofacial development in zebrafish. PLoS One. 2012;7:e30896. doi: 10.1371/journal.pone.0030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bessarab DA, Chong S-W, Srinivas BP, Korzh V. Six1a is required for the onset of fast muscle differentiation in zebrafish. Dev Biol. 2008;323:216–28. doi: 10.1016/j.ydbio.2008.08.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the Illumina reads used for this study are available at the Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra), under the accession number SRP050174.


