Abstract
Owners of biobanks are in an unfortunate position: on the one hand, they need to protect the privacy of their participants, whereas on the other, their usefulness relies on the disclosure of the data they hold. Existing methods for Statistical Disclosure Control attempt to find a balance between utility and confidentiality, but come at a cost for the analysts of the data. We outline an alternative perspective to the balance between confidentiality and utility. By combining the generation of synthetic data with the automated execution of data analyses, biobank owners can guarantee the privacy of their participants, yet allow the analysts to work in an unrestricted manner.
Introduction
Biobanks are becoming more important in evidence-based medicine, and their potential applications in personalized healthcare are enormous. Unfortunately, their growing popularity confronts the institutes who own biobank data with a dilemma. On the one hand, they would like to share the data necessary to support further clinical and epidemiological research, but on the other hand, biobanks contain confidential information that cannot be made publicly available.
Various methods for Statistical Disclosure Control have been proposed to deal with this dilemma.1–5 These can be broadly categorized as: suppression, obfuscation, and the creation of synthetic data.
Suppression removes the obvious identifiers such as names and addresses from the dataset, but unfortunately this is often not enough to guarantee an acceptable level of protection against identification of participants. Often different fields (called quasi-identifiers) can be combined to uniquely identify an individual. For example, 87% of the American population can be identified based on a combination of their date of birth, gender, and 5-digit postal code.6 Similarly, it is estimated that only a few single nucleotide polymorphisms (SNPs) are needed to uniquely identify a person's DNA record.7
To reduce the amount of disclosure through quasi-identifiers, values for these can be generalized into discrete categories. For example, instead of reporting the exact birth date of the participant only his or her age range (e.g., 18–25 years) would be disclosed.
But even with suppression and generalization, the risk of disclosure through record linkage remains.8 To prevent the disclosure of confidential information, biobank owners will often use a different strategy for statistical disclosure control, called data enclaves. Data enclaves will only share data with authorized individuals in regulated settings, or will only reveal summary statistics to ensure no sensitive data is disclosed. This strategy is appealing to biobank owners: the privacy concerns are greatly reduced and it is easy to explain to all the parties involved.
However, data enclaves come at a cost for the analysts. Summary statistics involve a large amount of information loss, which makes it harder to detect outliers, and poses limits to exploratory data analysis. These points are famously illustrated by Anscombe's quartet,9 which shows that the same summary statistics can have dramatically different underlying datasets.
Furthermore, if the data cannot be accessed publicly the validity of claims, risks of bias, and data provenance in publications cannot be assessed. Restricting access limits the ways in which results can be reproduced, and could potentially invite bias. Indeed, the data in restricted platforms has very limited public scientific value.
The core issue is thus how to achieve wider and more usable dissemination of biobank data for the various beneficial forms of analytics while preserving the privacy of the participants. To address this we propose a hybrid system for Statistical Disclosure Control. This system consists of a platform that can guarantee data security and participant privacy, but gives unrestricted access by generating synthetic data.
Initially proposed by Rubin,10 the idea behind synthetic data is to build a model from the data, and then to draw samples from it. Instead of the original data, these draws will be released to the public as “synthetic data.” The synthetic data can attempt to preserve the relevant statistical properties, but will still protect participants' privacy.
Although synthetic data have been used by the U.S. Census Bureau (http://www.census.gov/ces/dataproducts/synlbd/), its potential application to biobanks is novel. Furthermore, the novel hybrid platform approach proposed here will allow for better sharing of methods and data produced during research using biobank data.
For the remainder of this article, we will focus on evaluating the potential and feasibility of this synthetic data, providing:
• A short review of existing methods for synthetic data generation.
• A proposal for a method that combines synthetic data generation with automated script execution to ensure participant privacy and ease-of-use.
• The outline of a possible implementation strategy for this proposal.
We will show that new methods for dealing with confidential information are needed, and that this proposal is a feasible alternative to current methods.
Hybrid System Perspective
Synthetic data consist of draws from models fitted to the original data, and those draws are released instead of the original microdata. If these models are a good representation of the data, the released synthetic data will preserve relevant statistical properties. The released synthetic data will then also protect participant privacy. This protection of privacy is twofold. First, one can safely say that no personal data are disclosed to the public. Second, the synthetic data are useless to somebody who attempts to identify individuals, because even if information is disclosed, there is no way of telling whether that information is truthful or synthetic (i.e., artificial).
Synthetic data techniques have been applied to census data, but are hard to apply to biobanks directly. The models are never a perfect description of reality. Only if the models could accurately represent the data-generation process itself, the draws would be able to fully mimic the real world. Unfortunately, models will always be an approximation. So, synthetic data methods might ascribe characteristics to individuals that are not possible in the real world. This lack of truthfulness diminishes the utility of the synthetic data.
To work around this issue of truthfulness, we propose a hybrid system. The steps involved in this system are:
1. The user requests the data needed for the desired analysis.
2. This request generates a synthetic data set that can be freely accessed.
3. The analysis is performed on the synthetic data set.
4. After completing the analysis on the synthetic data, the code necessary for doing the analysis can be submitted.
5. The system can then run exactly the same analysis on the original data and, optionally after review, send the results to the analyst.
In this system, the analyst can work in an unrestricted manner, but can also draw valid inferences. For the biobank owner, concerns about confidentiality are addressed, and they serve the public good by disseminating data. Furthermore, the remote execution can ensure the reproducibility of the methods used. Inferences on just the synthetic data could also be published, as these inferences would be philosophically similar to an aggregate data meta-analysis.
Methods for generation
We will briefly discuss the various methods for synthetic data generation, needed to realize this hybrid system.
Notationally we let 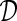 be the original biobank data on the individual level (microdata), with n records and m variables. Let
be the original biobank data on the individual level (microdata), with n records and m variables. Let 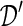 be the synthetic microdata set to be generated, with n′ records and m variables.
be the synthetic microdata set to be generated, with n′ records and m variables. 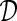 can be viewed as an n×m matrix and
can be viewed as an n×m matrix and 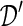 can be viewed as an n′×m matrix.
can be viewed as an n′×m matrix.
Each variable (column) can be categorical (e.g., the A,T,C,G denoting genotype data), ordinal (e.g., intensity questions), continuous (e.g., height and weight), or binary (e.g., yes/no questions). Each row in the matrix denotes an individual. Longitudinal data can be handled by converting the data from different measurement moments to a wide format, and, missing data could be modeled by extending 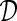 with a (binary) mask, so that reapplying this mask on the drawn samples will yield synthetic values for missing data.
with a (binary) mask, so that reapplying this mask on the drawn samples will yield synthetic values for missing data.
Multiple Imputation Using Conditional Parametric Distributions
By treating the sensitive values as missing data, we can use techniques for filling in the missing values, such as multiple imputation, to obtain synthetic data.10–12 Usually parametric (and conditional) regression models are used for multiple imputation. For example, Reiter12 uses a combination of logistic, multinominal logit, and linear regression, to account for the different types of variables. Each of these models is conditional on the other data (i.e., Y1 is an imputation from a regression on 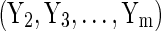 , Y2 a regression on
, Y2 a regression on 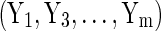 , and so on).13,14 This method is known as fully conditional specification (FCS) or the chained-equations approach. Synthetic data then constitute independent draws from the Bayesian posterior predictive distribution of these models.
, and so on).13,14 This method is known as fully conditional specification (FCS) or the chained-equations approach. Synthetic data then constitute independent draws from the Bayesian posterior predictive distribution of these models.
Multiple data sets can be constructed and a random sample from each of these sets could be released as a synthetic data set. This allows the multiple data sets to be pooled, allowing the user to draw frequency valid inferences using standard multiple imputation techniques. Multiple imputation models are attractive because they attempt to preserve conditional correlation structures, and give flexibility in the number of samples being drawn.
While multiple imputation has become more mature and there are now several software tools, for example MICE,15 it still requires modeling effort. For example, to account for different subpopulations, we might need different regression models, which might be hard to automate entirely. The modeling becomes more complex as the number of variables and their potential interactions increases.
Nonparametric Models
Because parametric models often require considerable calibration and manual supervision, some attention has been given to the use of nonparametric methods to increase the expressiveness of the models.
Drechsler and Reiter14 evaluate the use of Classification and Regression Trees (CART), bagging, random forests and Support Vector Machines (SVM) for generating synthetic data.
CART is the most promising in preserving the balance between disclosure and information loss with a minimum of supervision.16 CART is a nonparametric decision tree learning technique that produces either classification or regression trees, depending on whether the dependent variable is categorical or numeric. The trees essentially partition the predictor space into homogeneous clusters, with the most specific clusters at the leaf nodes. The trees can be pruned (e.g., by collapsing branches) to satisfy a balance between disclosure and information utility. Values from the fitted trees are obtained by Bayesian bootstrap from the leaf nodes, or draws from a (Gaussian) kernel density estimation over the leaves in case of numeric data.16
CART is also the method used in the recently released synthpop R package.17 This package can create synthetic data by conditionally fitting trees and sampling from the fitted models, thereby generating useful synthetic data. The synthpop package was developed for longitudinal census data by the SYLLS project (Synthetic Data Estimation for UK Longitudinal Studies), but can be applied to biobanks in a straight-forward manner.
Synthetic data generation with CART is the most mature and well understood method. But, there are some other promising methods. For example, Bayesian nonparametric methods such as infinite mixture models using Dirichlet processes.18 While these are computationally still challenging, they do offer flexible ways of capturing the nonlinearities in high-dimensional cases, and effective ways of synthesis using Markov chain Monte Carlo. In addition, neural network techniques such as stacked Restricted Boltzmann Machines (Deep Learning)19,20 could create a “memory” of the input. This memory can potentially also be used to create synthetic data. Neural networks are still an active, and promising, field of research that could also allow for fast generation of synthetic data.
Evaluation
We applied the synthpop package on data from the PREVEND (Prevention of Renal and Vascular End-stage Disease) study. PREVEND is a longitudinal cohort study based on the general population of the city of Groningen, the Netherlands, and included individuals between the ages of 28 and 75 years.21 Specifically we used the variables from the BioSHaRE Healthy Obese Project (HOP).22 Below we show plots comparing some variables in the real data set and in the synthetic data set. The full data set is available as a supplementary download. (Supplementary material is available in the online article at www.liebertpub.com/bio.)
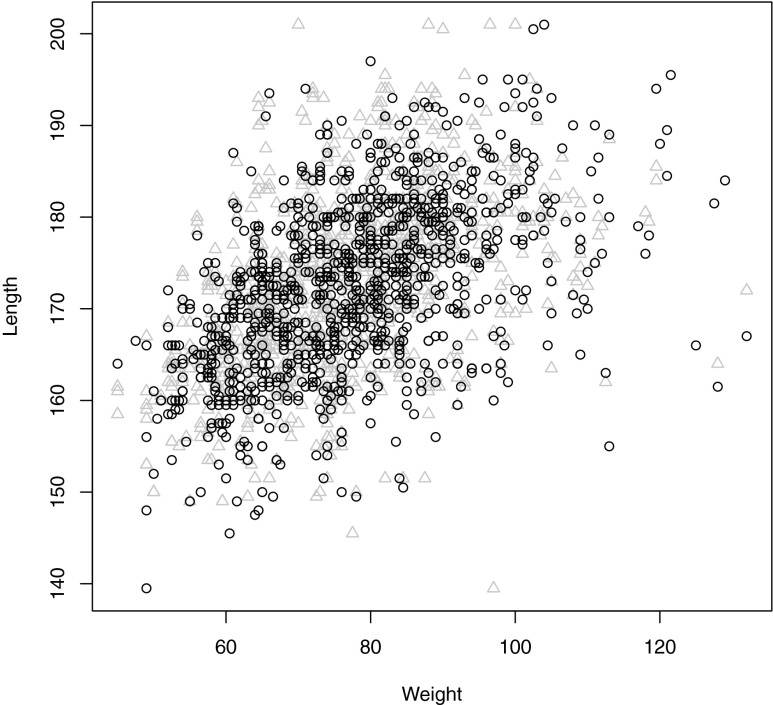
Figure 1 shows samples from the synthetic data and the original data, and shows that the synthetic data is useful for exploratory data analysis. Both the general correlation and its outliers can be synthesized.
FIG. 1.
Comparison of synthetic data (circles) and original data (triangles) across two correlated variables.
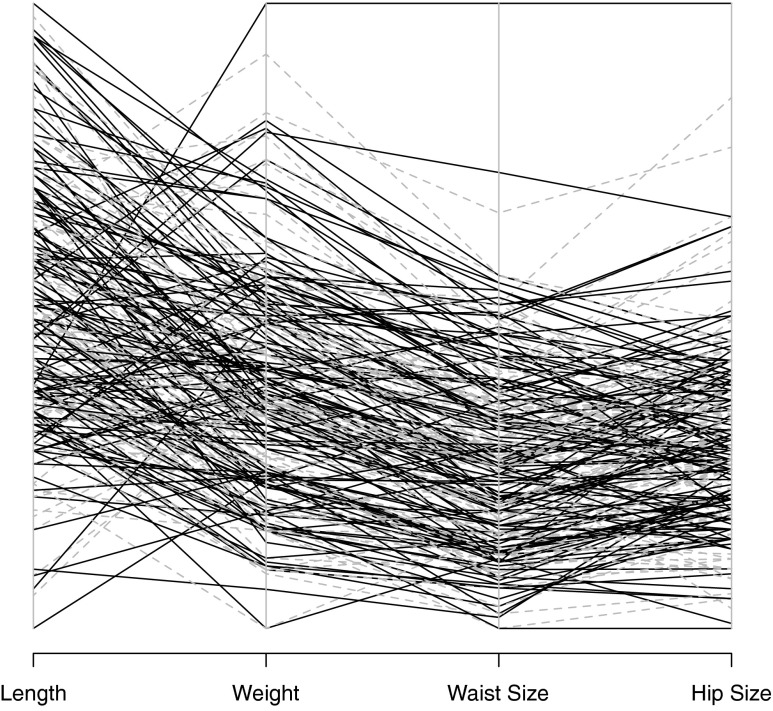
Figure 2 shows a parallel coordinates plot of 250 entries across several variables. This figure shows more clearly that correlations across variables are preserved, and that realistic values can be obtained.
FIG. 2.
Parallel coordinates plot of several variables. Synthetic data (solid) and original data (dashed).
The generated synthetic data is an accurate and useful representation of both the variables and their correlations. However, the evaluation of the synthetic data must be twofold as there is a trade-off between disclosure and utility. While the risk of re-identification (of a record or individual participant) might be virtually non-existent with synthetic data, one could predict unknown attributes of a known individual, given an ideal model of synthesis. In other words, an attacker could find unknown attributes of some individual with a certain probability by looking for the closest match in the synthetic data. This is known as attribute disclosure.
There are several methods for quantifying attribute disclosure, most notably t-closeness, which is defined as: An equivalence class is said to have t-closeness if the distance between the distribution of a sensitive attribute in this class and the distribution of the attribute in the whole table is no more than a thresholdt. A table is said to have t-closeness if all equivalence classes have t-closeness.
In short: the distribution of a particular sensitive value should not be further away than a distance t from the overall distribution.
Using the t-closeness metric circumvents issues associated with k-anonymity and ℓ-diversity. Briefly, k-anonymity states that a certain attribute class should be present in at least k records, which introduces ambiguity in the data set. However, if each of the k equivalence classes are the same, properties could still be resolved simply by elimination. The ℓ-diversity metric circumvents this problem by adding a further requirement: in addition to the class to being seen in k records, these records must have at least ℓ ‘well represented’ values. But if an attacker knows the real-world distribution of values, then attributes could still be disclosed with a certain probability, simply by combining different data sources.6,23,24
We did not attempt automated calculation of the t-closeness metric to quantify attribute disclosure. However, if the disclosure risk of a particular individual should exceed a predefined threshold, steps could be taken to mitigate that risk, for example, by smoothing the distribution of drawn values, or by regenerating the synthetic data while omitting the risk prone individuals.
Implementation Strategy
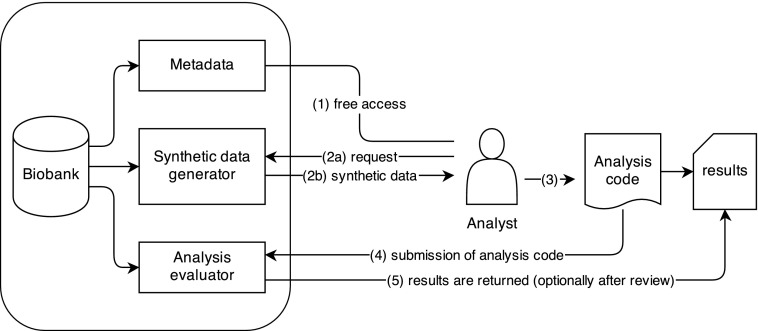
To realize a hybrid system, we propose the architecture outlined in Figure 3. The analyst has free access to the metadata (1) associated with a particular biobank. Through the metadata, the measured endpoints can be “ordered”, similar to the shopping cart metaphor in various web shops. This order might constitute various variables of interest or a specific subpopulation. The order triggers the generation of synthetic data (2a), which will be made available as a link specifically for the analysts' request (2b).
FIG. 3.
Implementation strategy for a hybrid synthetic data system.
From here on, the analysts can inspect the synthetic data and can build, or use, the tools necessary to answer their research questions (3). When these tools take the form of programming code, such as R or Python scripts, an analyst can then submit this code to the biobank owner (4), ideally through the same interface to which the initial request was made.
Without manual intervention the submitted code can then be evaluated against the original data. After evaluation, the results of the analysis can be put in a holding queue for further (manual) evaluation by the owners. This holding queue is especially useful when dealing with code and analysts who are not trusted (or intruders). After clearing the holding queue, the results can be accessed by the analyst, giving truthful results for the constructed analysis (5).
Concretely the synthetic data generation systems could be added to the existing data enclave systems such as DataSHIELD.25 However, unfortunately not all scripts can be fully automated. This is partly due to the increasing complexity of statistical analysis that requires custom software, and partly because some functionality might expose too much of the confidential data. Nonetheless, this does not diminish the utility of the vast majority of scripts that can be fully automated and which would benefit from the hybrid approach.
The synthetic data generation and code evaluation should be sand boxed using containers or virtual machines, thereby preventing unprivileged access to system resources. This point is crucial since it is trivial to engineer an “analysis” that would, when executed, immediately upload the raw data to a third party. Furthermore, all standard data security and guarantees for dealing with remote access and data “on the wire” should be firmly in place.
Discussion and Conclusion
We have outlined a hybrid system in which synthetic data are generated as a surrogate. This allows analysts to develop tools and methods without being restricted to a closed platform, while still ensuring that confidentiality concerns are addressed. Furthermore, by providing a remote execution platform, valid inferences can still be made, while the original data reside in a secure environment.
While merits of this system may be self-evident, it still remains a perspective. Implementation should determine the utility of the system as a whole. Important technical challenges remain, such as the automatic evaluation of disclosure risk and the fully automatic generation of high quality synthetic data. However, these technical challenges may well pale in comparison to legal, financial, and political hurdles.
Convincing biobank owners of the merits of implementing a synthetic data system, which always comes at a cost, may prove hard, especially considering that the system should be kept operational for a long period (preferably indefinitely) if it is to be useful in the permanent dissemination of scientific findings. Furthermore, synthetic data have met resistance from researchers because they need to be convinced of the merits of performing an analysis on “fake” data first.
It is also crucial to point out that while synthetic data protect the privacy of individual participants, it does not provide any guarantees about the confidentiality of information from whole or subpopulations. In fact, the whole point of the exercise is to mimic the population level correlations as closely as possible. But this also means that care must also be taken in releasing synthetic data: for example, disclosing air pollution data might not be detrimental to the privacy of individuals, but it could severely cut real estate value. Another example is how, disclosing correlations associated with certain professions, genetic mutations or lifestyles could unintentionally have severe consequences for certain individuals and groups, and these risks are not mitigated by the sue of synthetic data.
Despite this, we believe that a hybrid system is both useful and necessary for biobanks and other databases with privacy issues. Not only would a hybrid system ease the often cumbersome data acquisition and use of closed and proprietary platforms, it would also guarantee better accessibility to the data for verification and reproduction of results by other researchers.
This is a point that must be stressed even when dealing with confidential data. If there is no method for reproduction and verification by peers (or other third-parties) then publications that rely on that data have a severely diminished credibility.
As we come to rely more and more on large scale information retrieval in our day to day scientific and medical work, we have to realize that access to the data we have used must also be guaranteed. Methods such as the one outlined here could prove to be a valuable step in ensuring that research efforts remain credible, and thus relevant, in the future.
Supplementary Material
Acknowledgments
We thank Jackie Senior for her thorough copy-editing and proofreading.
Author Disclosure Statement
No conflicting financial interests exist. Funding: This research has received funding from the European Union Seventh Framework Programme [FP7/2007–2013] under grant agreement n° 261433 (Biobank Standardisation and Harmonisation for Research Excellence in the European Union - BioSHaRE-EU).
References
- 1.Chen B-C, Kifer D, LeFevre K, Machanavajjhala A. Privacy-preserving data publishing. Foundations Trends Databases 2009;2:1–167 [Google Scholar]
- 2.Matthews GJ, Harel O. Data confidentiality: A review of methods for statistical disclosure limitation and methods for assessing privacy. Statistics Surveys 2011;5:1–29 [Google Scholar]
- 3.Navarro-Arribas G, Torra V. Information fusion in data privacy: A survey. Inform Fusion 2012;13:235–244 [Google Scholar]
- 4.Malin B, Loukides G, Benitez K, Clayton EW. Identifiability in biobanks: Models, measures, and mitigation strategies. Human Genet 2011;130:383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prada SI, Gonzalez C, Borton J, et al. Avoiding Disclosure of Individually Identifiable Health Information: A Literature Review. University Library of Munich, Germany; 2011 [Google Scholar]
- 6.Sweeney L. K-anonymity: A model for protecting privacy. Intl J Uncertainty Fuzziness Knowledge-based System 2002;10:1–14 [Google Scholar]
- 7.Homer N, Szelinger S, Redman M, et al. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet 2008;4:e1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sweeney L. Weaving technology and policy together to maintain confidentiality. J Law Med Ethics 1997;25:98–110 [DOI] [PubMed] [Google Scholar]
- 9.Anscombe F. Graphs in statistical analysis. Am Statist 1973;27:17–21 [Google Scholar]
- 10.Rubin DB. Statistical disclosure limitation. J Official Statist 1993;9:461–468 [Google Scholar]
- 11.Raghunathan T. Multiple imputation for statistical disclosure limitation. J Official Statist 2003;19:1–16 [Google Scholar]
- 12.Reiter JP. Releasing multiply imputed, synthetic public use microdata: An illustration and empirical study. J Royal Statist Soc Series A (Statistics in Society) 2005;168:185–205 [Google Scholar]
- 13.Raghunathan T, Lepkowski J. A multivariate technique for multiply imputing missing values using a sequence of regression models. Survey Methodol 2001;27:85–95 [Google Scholar]
- 14.Drechsler J, Reiter JP. (2011) An empirical evaluation of easily implemented, nonparametric methods for generating synthetic datasets. Computat Statist Data Anal 2011;55:3232–3243 [Google Scholar]
- 15.Buuren S van, Groothuis-Oudshoorn K. MICE: Multivariate imputation by chained equations in R. J Statist Software VV: 2011;45:1–67 [Google Scholar]
- 16.Reiter JP. Using CART to generate partially synthetic, public use microdata. J Official Statist 2003;21:441–462 [Google Scholar]
- 17.Beata Nowok GMR, Dibben C. Synthpop: Generating synthetic versions of sensitive microdata for statistical disclosure control. 2014; http://cran.r-project.org/web/packages/synthpop/
- 18.Quintana Fa, Müller P. Nonparametric Bayesian data analysis. Statist Sci 2004;19:95–110 [Google Scholar]
- 19.Ackley DH, Hinton GE, Sejnowski TJ. A learning algorithm for Boltzmann machines. Cognit Sci 1985;9:147–169 [Google Scholar]
- 20.Hinton GE. Reducing the dimensionality of data with neural networks. Science 2006;313:504–507 [DOI] [PubMed] [Google Scholar]
- 21.Pinto-Sietsma S-J, Janssen WM, Hillege HL, Navis G, Zeeuw D de, Jong PE de. Urinary albumin excretion is associated with renal functional abnormalities in a nondiabetic population. J Am Soc Nephrol 2000;11:1882–1888 [DOI] [PubMed] [Google Scholar]
- 22.Vliet-Ostaptchouk J van, Nuotio M-L, Slagter S, et al. The prevalence of metabolic syndrome and metabolically healthy obesity in Europe: A collaborative analysis of ten large cohort studies. BMC Endoc Disorders 2014;14:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Emam K, Dankar FK. (2008) Protecting privacy using k-anonymity. J American Medical Informatics Association 2008;15:627–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machanavajjhala A, Kifer D, Gehrke J, Venkitasubramaniam M. L-diversity: Privacy beyond k-anonymity. ACM Transactions on Knowledge Discovery from Data (TKDD) 2007;1:24–36 [Google Scholar]
- 25.Wolfson M, Wallace SE, Masca N, et al. DataSHIELD: Resolving a conflict in contemporary bioscience–-Performing a pooled analysis of individual-level data without sharing the data. Intl J Epidemiol 2010;39:1372–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.