Abstract
The GATA family of transcription factors (GATA1–6) binds selected GATA sites in vertebrate genomes to regulate specific gene expression. Although vertebrate GATA factors have two highly conserved zinc finger motifs, how the two fingers act together to recognize functional DNA elements is not well understood. Here we determined the crystal structures of the C-terminal zinc finger (C-finger) of mouse GATA3 bound to DNA containing two variously arranged GATA-binding sites. Our structures and accompanying biochemical analyses reveal two distinct modes of DNA binding by GATA to closely arranged sites. One mode involves cooperative binding by two GATA factors that interact with each other through protein-protein interactions. The other involves simultaneous binding of the N-terminal zinc finger (N-finger) and C-finger of the same GATA factor. Our studies represent the first crystallographic analysis of GATA zinc fingers bound to DNA and provide new insights into the DNA recognition mechanism by the GATA zinc finger. Our crystal structure also reveals a dimerization interface in GATA that has previously been shown to be important for GATA self-association. These findings significantly advance our understanding of the structure and function of GATA and provide an important framework for further investigating the in vivo mechanisms GATA-dependent gene regulation.
Keywords: GATA, Zinc finger, DNA recognition, self-association, transcription
Introduction
Originally discovered as key regulators of erythroid-specific genes 1; 2; 3, the GATA family of proteins has now been established as an important class of eukaryotic transcription factors in a variety of cell types 4. Among the six vertebrate homologues, GATA1–3 play key roles in the development and maintenance of hematopoietic and immune cells 5, whereas GATA4–6 participate in the transcriptional regulation in tissues such as the heart, liver, and gonads 6. The diverse roles of GATA proteins in the development of vertebrates are particularly interesting, as uncovering the molecular basis of these roles may offer insights into the epigenetic mechanisms of cell differentiation and lineage specification 4; 7. GATA3, the subject of this study, is critical to the immune system and essential in the development of specific T help cell subset (Th2) from naïve T cells 8; 9; 10.
The most notable feature of GATA proteins at the sequence level is two highly conserved type IV zinc fingers 11, referred to as N-finger (N-terminal zinc finger) and C-finger (C-terminal zinc finger) hereafter. The sequences immediately following the two zinc fingers are also conserved whereas other sequence motifs, such as the putative transactivation domains, are less conserved in the GATA family. Overall, GATA proteins have a relatively compact and conserved domain structure and yet are able to achieve diverse functions in a variety of cellular processes.
The function of GATA depends critically on the two conserved zinc fingers and their flanking sequences, as suggested by mutagenesis in biochemical and transgenic animal studies, as well as disease-associated mutations found in human 12; 13; 14; 15; 16; 17. The C-finger and its adjacent C-terminal basic tail are necessary and sufficient for GATA to bind its cognate sequence (WGATAR, W=A/T; R=A/G) 1; 18; 19; 20; 21. The N-finger, especially that of GATA2 and GATA3, can also bind DNA independently, but with a slightly different sequence preference (GATC) 22; 23; 24. On certain sequences with two proximal GATA sites, including the palindromic GATA motif (ATC(A/T)GATAAG) found in the promoter of GATA1, the N-finger can also participate in DNA binding together with the C-finger, resulting in a GATA/DNA complex with markedly increased affinity 24; 25; 26; 27; 28; 29.
Both the N-finger and the C-finger can engage in protein-protein interactions leading to self-association and binding to other transcription factor. GATA factors have been shown to form homo- or hetero-oligomers in vivo and in vitro, and this self-association is thought to play important roles in the combinatorial and synergistic transcription regulation by GATA factors and in the assembly of high-order protein DNA complexes in locus control regions 30; 31. Recent studies with transgenic mice have indeed shown that self-association of GATA1 is important for proper mammalian erythroid development in vivo 32, but the structural basis of GATA self-association remains unclear. In addition to self-association, the interaction between the N-finger and Friend of GATA (FOG) has been shown to be essential in a broad range of GATA–dependent cellular functions 33; 34; 35; 36, whereas the C-finger of GATA interacts with NFAT to regulate transcription synergistically in T cells and muscle cells 37; 38; 39. Finally, GATA factors have been shown to bind and modulate chromatin structure, implicating a critical role of GATA in epigenetic control of chromosome structure during differentiation 4; 40; 41. The diverse cellular functions displayed by GATA may be attributed, at least in part, to its diverse biochemical modes of action in DNA binding, protein-protein interaction, and chromatin remodeling, but the molecular basis of these activities are not well understood.
One of the most intriguing questions regarding to the function of GATA factors is how they can locate and bind their functional sites in vivo given the high occurrence of the relatively small recognition motif (WGATAR) 4. Chromatin Immunoprecipitation (ChIP) analyses reveal that GATA binds only a small fraction of its cognate sites in the genome 42; 43; 44. Flanking DNA sequences and cellular context-specific factors, such as local chromatin structure and interacting proteins, may play important roles in binding site selection. The intrinsic DNA binding function of a given GATA member and its adaptability to environmental influences, however, also likely play key roles in this process. Despite the highly conserved nature of the zinc fingers, DNA binding by GATA proteins can be affected by subtle changes in amino acid sequences flanking the zinc core module, which may account for the different DNA binding properties of the N-finger and the C-finger, and between different GATA homologues 22; 29; 45; 46. Such adaptability in DNA binding may allow GATA proteins to achieve diverse functions in specific cellular contexts. Although the N-finger and the C-finger can bind certain sequences with two GATA sites (e.g. the palindromic GATA motif) simultaneously, there is no defined pattern of double GATA sites throughout vertebrate genomes. This raises another intriguing question as to why the two zinc fingers and their linker region are highly conserved in vertebrate GATA factors. It is possible that the N-finger and the C-finger interact with each other to determine specific DNA binding in a given genomic context 29, but exactly how the two zinc fingers of GATA bind DNA coordinately is not clear.
To better understand the diverse DNA recognition mechanisms by GATA proteins and how these recognition mechanisms may be affected by protein-protein interactions and chromatin environments, it is important to first characterize the detailed DNA binding interactions by GATA zinc fingers at high resolution. So far DNA binding by a single GATA zinc finger has been analyzed by NMR on the chicken GATA1 C-finger and the fungal homologue AREA 47; 48. Although these studies revealed the structure fold of GATA zinc finger and its general framework of DNA binding, many details of protein/DNA interactions are not fully defined 49. In the present study, we determined the crystal structure of mouse GATA3 C-finger bound to DNA containing two variously arranged GATA-binding sites. These are the first crystal structures of GATA zinc finger bound to DNA. As discussed below, our studies extend previous NMR analyses and provide high-resolution details to define the DNA recognition mechanism by the GATA zinc finger. Our crystal structures also reveal the atomic details of a dimerization interface in GATA3 C-finger that has previously been shown to be required for GATA self-association 30. Finally, our structure-guided biochemical analyses suggest that full-length GATA factors can bind closely arranged GATA sites in diverse modes depending on the arrangement of the binding sites, and more strikingly, the protein concentration of the GATA factors. These findings have important implications for understanding and further studying the in vivo functions GATA transcription factors.
Results
Crystallographic study of GATA3 C-finger bound to DNA
Our crystallographic analyses focused on the mouse GATA3 C-finger (amino acid residues 308–370) bound to DNA. Together with the previous NMR analyses of the chicken GATA1 C-finger bound to DNA and the fungal AREA zinc finger bound to DNA 47; 48, we hope to better understand the DNA binding mechanism of GATA zinc finger through comparison of structural data derived from different techniques and from different GATA homologues. Given the fact that vertebrate GATA factors contain two zinc fingers connected by a predicted loop and that GATA sites frequently occur as clusters in vertebrate genomes 29, another aim of our study is to explore how GATA binds closely arranged GATA sites, referred to as proximal GATA sites hereafter. We did so by using DNA sequences containing two GATA sites in our crystallization. Since GATA site clusters in genome have diverse orientation and spacing, there is no special design in the spacing and orientation of the two GATA sites on DNA. Instead, DNA fragments containing two GATA sites arranged differently were used for co-crystallization with GATA3 C-finger. Two DNA fragments, each having a distinct arrangement of two GATA sites, crystallized successfully with the GATA3 C-finger. On one DNA fragment, the two GATA sites are located on the opposite face of the DNA and the two bound zinc fingers make no direct contact to each other (Figure 1a). This complex is solved at 2.7Å resolution and referred to as OPP (opposite) hereafter. On the other DNA fragment, the two GATA sites are located on the same face of the DNA and the two adjacently bound zinc fingers interact with each other directly (Figure 1b). This structure is solved at 3.1Å resolution and referred to as ADJ (adjacent) hereafter. In total, we have observed four independent GATA3 C-finger/DNA complexes in two different crystal forms. Data statistics for both crystals are presented in Table 1.
Figure 1. Overall structure of GATA3 C-finger bound to DNA.
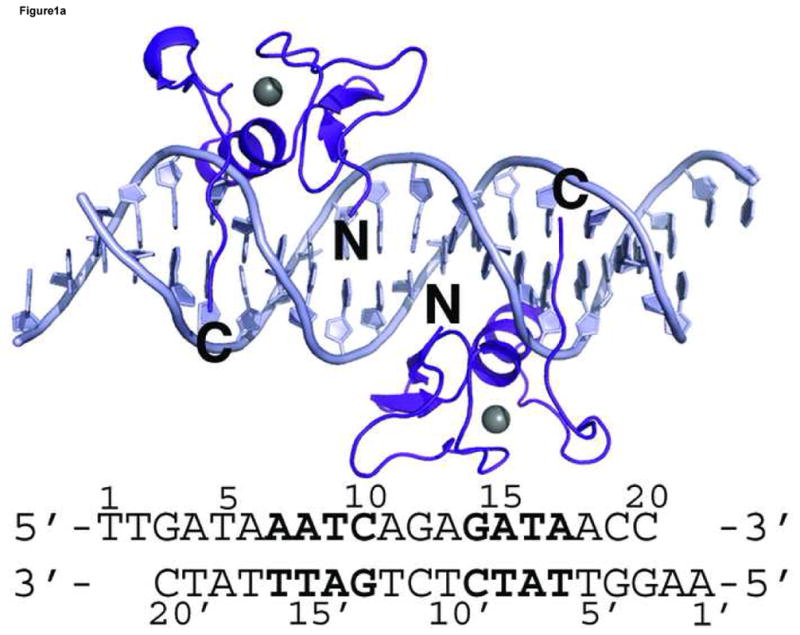
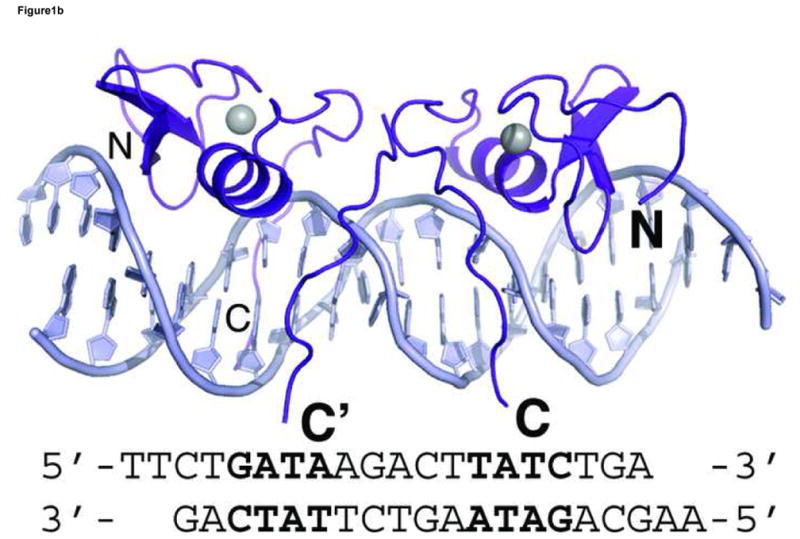
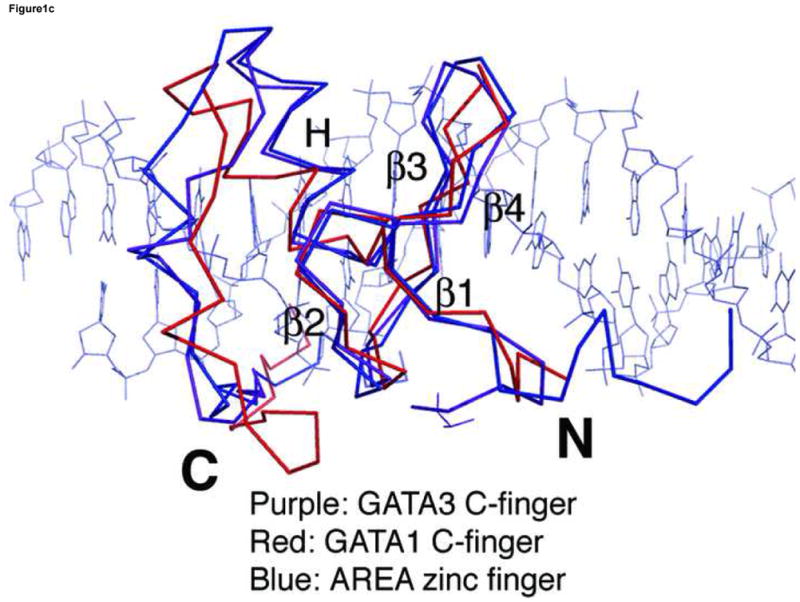
Crystal structures of GATA3 C-finger (purple, ribbon style) bound to the OPP (a) and ADJ (b) DNA (cyan, stick model); the zinc atom is shown in grey sphere. The same colour scheme is used throughout the illustration unless noted otherwise. The sequence of the DNA in each crystal structure is shown below the figure. The sequence of the OPP DNA in (a) is numbered for discussion in the text. The N- and C-terminus of the protein are labelled by bold letter N and C, respectively. The alternative C-terminal end of the right side GATA3 C-finger in (b) is indicated by C′. (c) C-alpha backbone superposition of the core zinc module between GATA3 C-finger (purple), chicken GATA1 C-finger (red), and the fungal AREA zinc finger (blue). The GATA3 C-finger bound to the consensus GATA site of the OPP DNA is used here. For clarity, only the DNA (stick model) from the GATA3 C-finger/DNA complex is shown.
Table 1.
Statistics of Crystallographic Analysis
| Data Collection | OPP SAD | OPP Nat | ADJ Nat |
|---|---|---|---|
| Resolution (Å) | 50-2.7 | 30-2.7 | 30-3.1 |
| Rsym (%)a | 0.178(0.472) | 0.179(0.475) | 0.078(0.164) |
| Completeness (%)b | 100 (100) | 100(100) | 99.9(100) |
| I/σb | 30.32 (6.22) | 30.49(6.15) | 23.18(7.75) |
| Refinement | |||
| Resolution (Å) | 30-2.7 | 30-3.1 | |
| R factor | 28 | 27.4 | |
| Rfree | 30 | 29.6 | |
| Rms deviations | |||
| Bond lengths (Å) | 0.01 | 0.009 | |
| Bond angles (Þ) | 1.7 | 1.3 | |
| Average B factor (Å2) | 62.2 | 50.2 | |
OPP SAD: single wavelength anomalous diffraction data collected on the OPP complex at the zinc edge;
OPP Nat: Native data set of the OPP complex;
ADJ Nat: Native data set of the ADJ complex;
Rsym = Σ|I−<I>|/ΣI, where I is the observed intensity, <I> is the statistically weighted average intensity of multiple observations of symmetry-related reflections;
Numbers in parentheses are for the outer shell (2.70–2.87Å for OPP SAD; 2.70–2.87Å for OPP Nat; 3.29–3.10Å for ADJ Nat)
R = Σ||Fo| − |Fc||/Σ|Fo|, where Fo and Fc are observed and calculated structure factor amplitudes, respectively. Rfree is calculated for a randomly chosen 10% of reflections.
Overall structure description
The structure of the GATA3 C-finger consists of a core zinc module (amino acid residues Ser308-Thr348) and a C-terminal basic tail (amino acid residues 349–365). The beginning of the C-terminal basic tail (residues 349–356) folds back onto the DNA-binding alpha helix through numerous hydrogen bonds and Van der Waals contacts (Supplemental Figure 1). The rest of the C-terminal basic tail inserts into the minor groove, where Arg364 makes base-specific hydrogen bonds to DNA (discussed further below). In the ADJ complex (Figure 1b), one copy of the GATA3 C-finger displays an alternative conformation wherein the second half of the C-terminal basic tail (amino acid residues 357–366) transverses the minor groove and reaches over to the major groove to interact with the C-terminal end of the alpha helix of the adjacently bound GATA3 C-finger (discussed further below). The overall structure of the GATA3 C-finger and its relative orientation to DNA are nearly identical in the four independent complexes observed in the crystal structures. The RMSD of Cα superposition is around 0.1 –0.6 Å among the four GATA3 C-fingers. The DNA fragments in both crystal forms are in approximately a straight B-form conformation.
Comparing the crystal structure of the mouse GATA3 C-finger bound to DNA with the corresponding NMR structure of chicken GATA1 (RMSD 1.1 Å for 25 Cα atoms) and the fungal AREA (RMSD 0.9Å for 25 Cα atoms) reveals many common structural features of DNA binding by the GATA zinc fingers. The fold of the core zinc module, the trajectory of the C-terminal basic tail, and the orientation of protein with respect to DNA, are very similar in the three superimposed structures (Figure 1c) 47; 48. The detailed interactions underlying the folding of the core zinc module, including the zinc coordination and the packing of numerous buried hydrophobic residues, are highly conserved (data not shown) 47. The backbone conformation and side chain orientation of the C-terminal basic tail, however, show marked differences in the three structures (Figure 1c). The differences between AREA zinc finger and the chicken GATA1 C-finger were thought to be due to different sequences in the C-terminal basic tail 48. However, our crystal structures showed that the conformation of the C-terminal basic tail is most likely determined by Tyr344, Tyr345, His348, Arg352, and Met356 that interact with each other and with the DNA backbone (Supplemental Figure 1). Most of these residues are conserved in AREA and the chicken GATA1 C-finger. Moreover, GATA1 and GATA3 have nearly identical sequence in the C-terminal basic tail. Thus, the apparent structural differences between GATA1 and GATA3 in the C-terminal basic tail are likely due to the inherent flexibility of this region that may affect structural analyses by NMR and X-ray crystallography differently (discussed further below).
A key limitation in structural analysis by NMR is the lack of restraints that can define long-range order. One way to overcome this problem is to use residual dipolar couplings. This was first demonstrated in the NMR analysis of the chicken GATA1 C-finger/DNA complex, wherein the orientation of the β3β4 loop and the alpha helix became much better defined when the residual dipolar couplings restraints were applied 49. Our crystal structures of the GATA3 C-finger/DNA complex superimpose much better with the chicken GATA1 C-finger/DNA complex refined with residual dipolar couplings (PDB code 2GAT.pdb, RMSD 0.8Å for 25 Cα atoms) than the one without (PDB code 1GAT.pdb, RMSD 1.1 Å for 25 Cα atoms) (Supplemental Figure 2), providing an independent confirmation for the utility of residual dipolar couplings in NMR structural analysis 49.
Protein DNA interactions
Of the four independent GATA3 C-finger/DNA complexes observed in the crystal structures (Figure 1a,b), three are bound to the consensus GATA site whereas one is bound to a fortuitous GATT site (see Materials and Methods for details) that is known to bind GATA zinc finger in site-selection studies and also found in natural promoters of GATA-regulated genes 18; 19; 50; 51; 52; 53. Since the three complexes bound to the consensus GATA site are nearly identical, we will focus our description on one of them (the one bound to the consensus site in the OPP complex) and compare it with the one on the GATT site and with the previously characterized chicken GATA1 C-finger/DNA complex and the fungal AREA/DNA complex 47; 48.
The GATA3 C-finger binds to DNA through the core zinc module in the major groove and the C-terminal basic tail in the minor groove. The N-terminal beta hairpin loop, the β3β4 anti-parallel beta sheet, and the alpha helix all contribute to the DNA binding surface in the major groove. Here a number of residues, including Thr326, Leu327, Arg329, Asn339, Leu343 and Leu347, make direct hydrogen bond and Van der Waals contacts to DNA bases (Figure 2a,b). On the consensus site of the OPP complex, Arg329 interacts with Gua14 through bidentate hydrogen bonds. In addition, Arg329 also forms a hydrogen bond to Thy8′. Asn339 forms a hydrogen bond with Arg339 as well as Ade7′ (Figure 2a), whereas Thr326 makes a hydrogen bond to Asn339 and Van der Waals contact to Thy8′. This network of interactions centers on the first three nucleotides (GAT) of the binding site (GATA) and plays a key role in sequence-specific recognition by GATA factors. The base-specific hydrogen bond interactions in the major groove are supplemented by numerous Van der Waals contacts. In addition to the Th326/Thy8′ interaction mentioned above, the C5 methyl group of Thy6′ is sandwiched by Leu343 and Leu347, which may explain the preference for Adenine at the fourth position of the GATA binding site (Figure 2b). Moreover, Leu327 makes Van der Waals contacts to Ade13 and Gua14 (not shown), suggesting that flanking sequences outside the core recognition site (GATA) may affect DNA binding by GATA. In addition to contacts to bases, numerous residues of the core zinc module, including Arg312, Thr326, Arg330, Asn339, Ala340, Tyr344, Lys346, and His348, interact with the sugar phosphate backbone of the DNA extensively (not shown). Residues from the first half of the C-terminal basic tail, including Arg352, Met356, Lys358 and Ile361 also contribute to the backbone binding (not shown), while Arg364 from the second half inserts deeply into the minor groove to form a hydrogen bond with the carbonyl of Thy6′. The aliphatic side chain of Arg364 makes extensive Van der Waals contacts to neighboring bases and sugar rings (Figure 2c). But it is the hydrogen bonding ability of the guanidino group of Arg364 that seems to enhance the sequence specificity at the forth position of the GATA site (see below).
Figure 2. DNA recognition by the GATA3 C-finger.
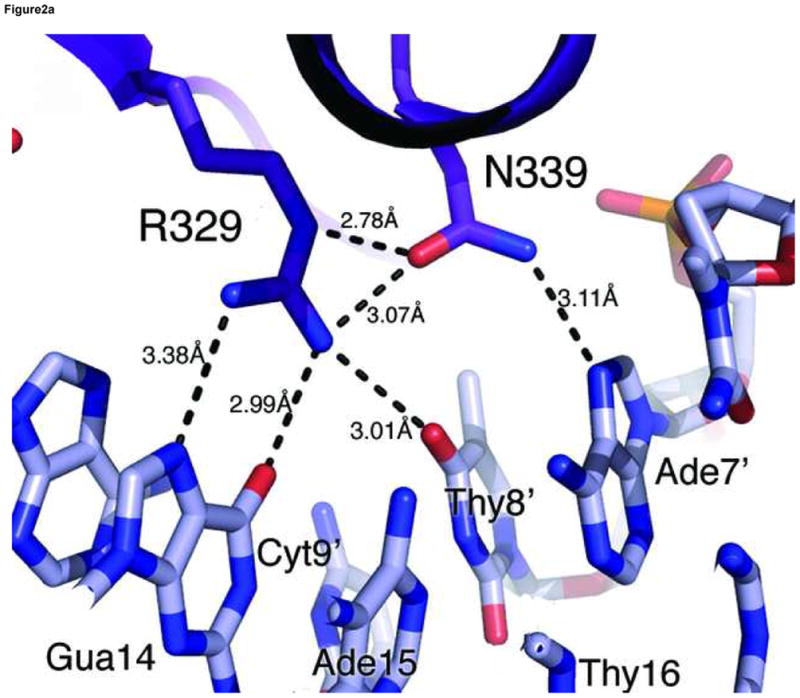
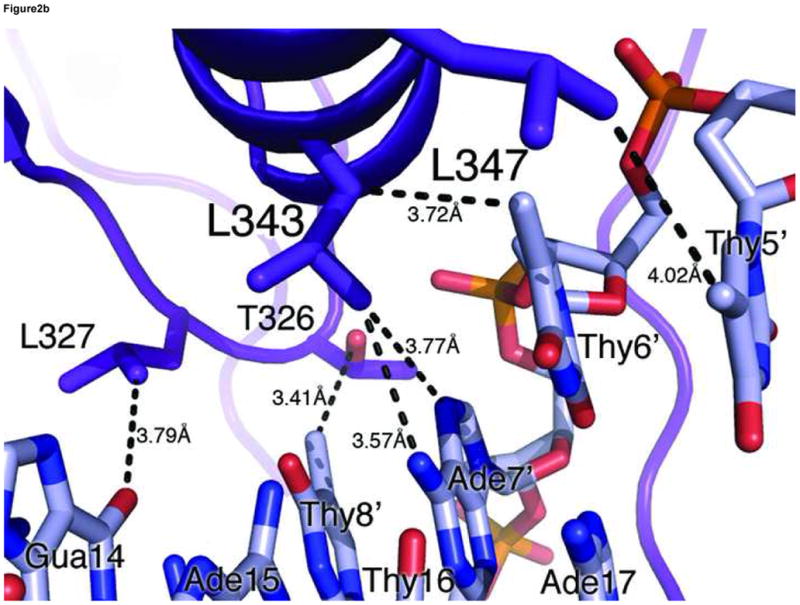
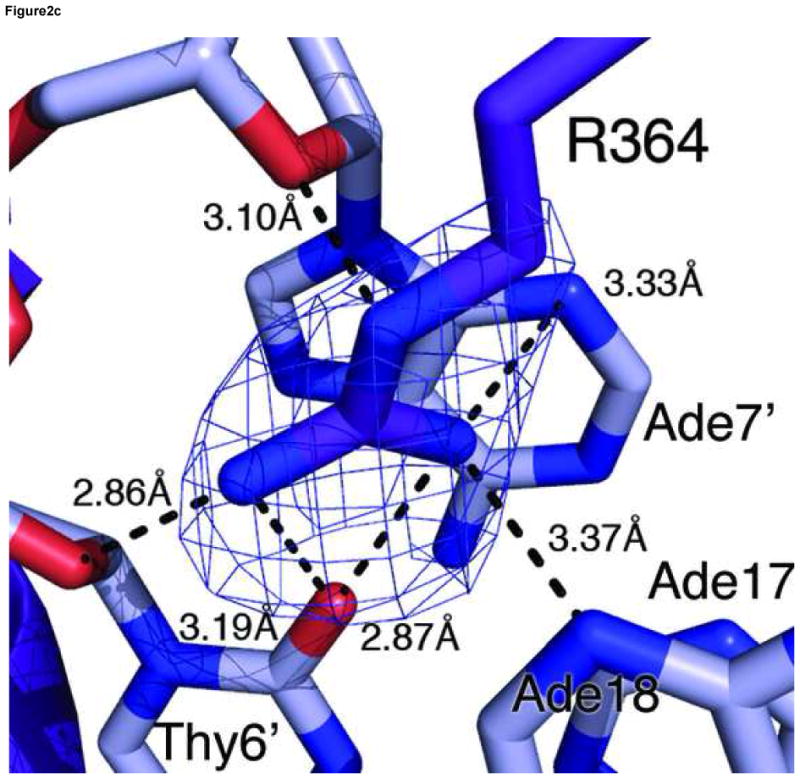
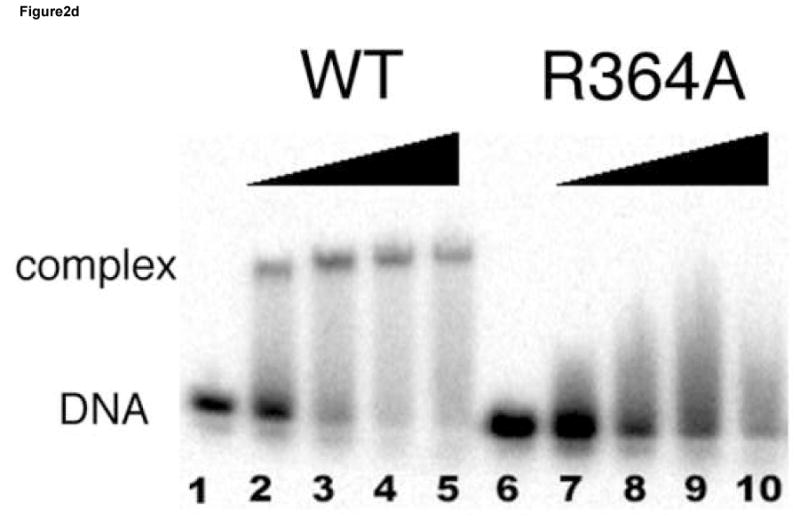
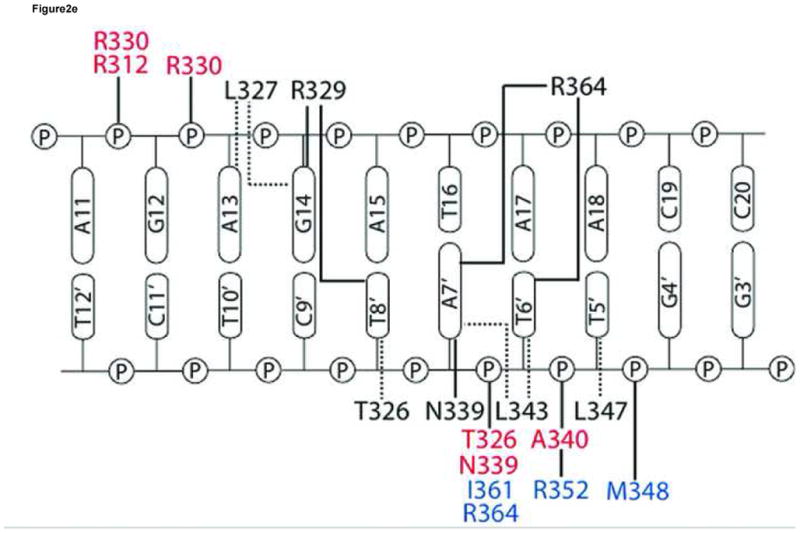
(a) Hydrogen bonding interactions between the conserved Arg329 and Asn339 and the first three nucleotides (GAT) in the binding site. Shown here is the consensus site in the OPP complex where the DNA base is numbered according to Figure 1a. Potential hydrogen bonds are indicated by dashed lines with corresponding distances. (b) Van der Waals contacts between conserved hydrophobic residues (Thr326, Leu327, Leu343, and Leu347) and DNA bases in the major groove. Several contacts are indicated by dashed lines to give a distance scale. Arg329 and Asn339 are omitted in this view for clarity. (c) DNA binding interactions by Arg364 in the minor groove. The electron density of Arg364 is calculated from simulated omit map and contoured at 2σ level. Several short distances are shown as potential hydrogen bonds and Van der Waals contacts. (d) Titration of the ADJ DNA with the wild type GATA3 DF (lanes 1–5) and the Arg364Ala (lanes 6–10) showing that Arg364 is important for DNA binding (see Materials and Methods for details). (e) Schematic summary of key DNA contacts by GATA3 C-finger. The region shown corresponds to the consensus site of the OPP complex. Solid lines to bases indicate hydrogen bond interactions; Dashed line to bases indicate Van der Waals contacts; Solid lines to DNA backbone denote contacts to the sugar phosphate backbone by residues from the core zinc module (blue font) and the C-terminal basic tails (red font), respectively.
On the GATT site in the OPP complex, the base change at the fourth position (GATA vs GATT) does not seem to affect the overall protein/DNA interaction. Binding in the major groove is conserved as Leu343 and Leu347 maintain Van der Waals contacts to the major groove though with different bases. In the minor groove, Arg364 donates a hydrogen bond to the N3 position of the Adenine paired with the fourth Thymine (GATT, underlined), which is similar to the hydrogen bond between Arg364 and the O2 position of Thymine on the GATA site (see above). Our structural analyses suggest that GATA factors should bind GATT similarly to GATA, which is consistent with previous biochemical and functional studies 18; 19; 50.
Our crystal structures suggest that Arg364 from the C-terminal basic tail of the GATA3 C-finger plays a key role in DNA binding. It inserts deeply into the minor groove and forms a hydrogen bond with an A/T or T/A base pair in the fourth position of the binding site and may thus contribute to the stability of DNA binding by GATA factors (Figure 2c). Arg364 apparently also plays a role to discriminate against Guanine or Cytosine at the fourth position in the GATA site since the guanidine group of Arg364 may clash with the exocyclic amino group (N2) of Guanine in the minor groove. Arg364 is highly conserved in the C-terminal basic tail of GATA factors. However, in the chicken GATA1 C-finger/DNA complex 47, the equivalent arginine (Arg54) does not insert into the minor groove but instead binds to the phosphate backbone. In the fungal AREA/DNA complex 48, the corresponding arginine residue (Arg59) projects toward the minor groove but not deep enough to make hydrogen bonds to bases. Whether the different structural roles of the conserved arginine in the three complexes reflect different DNA binding mechanisms by GATA3, GATA1 and AREA or uncertainty from different experimental studies is not clear. However, the same DNA binding interaction by Arg364 in the minor groove is observed four times in our crystallographic analyses here. To further examine the functional role of Arg364, we introduced a specific mutation (Arg364Ala) in the DNA binding domain of GATA3 and analyzed its effect on DNA binding (see Materials and Methods for details). The mutant behaved similarly to the wild type protein in expression and purification, but disrupted DNA binding to a consensus GATA site (Figure 2d). Although the C-terminal basic tail is known to be important for DNA binding by GATA factors 20; 46; 47; 48, our structural and biochemical analyses here reveal its detailed DNA binding interactions and identify Arg364 as one of the key DNA binding residues in the minor groove.
Overall, the four independent GATA3 C-finger/DNA complexes observed in the two crystal forms display nearly identical DNA binding interactions on three GATA sites and one GATT site, indicating that the observed protein-DNA interactions are maintained in different crystal packing environments (Figure 2e). Most of the DNA binding interactions observed here, especially those mediated by the core zinc module to the major groove and to the sugar phosphate, are shared by the chicken GATA1 C-finger/DNA complex and the fungal AREA/DNA complex 47; 48. But our structure and biochemical analyses also reveal new insights into the detailed DNA binding mechanism by the C-terminal basic tail, especially on the role of the conserved Arg364.
Binding of proximal GATA sites by the GATA3 C-finger
The vertebrate GATA factors (GATA1–6) contain two highly conserved zinc fingers connected by a linker region that is also conserved. The N-finger and C-finger share a homologous core zinc module and hence similar preference in the first three nucleotides in their binding sites (GATN, underlined) 19. The specificity for the fourth position and the overall affinity appear to be modulated by sequences flanking the core zinc module, especially the C-terminal basic tail following the zinc core 1; 18; 19; 20; 22; 23; 24; 46; 47; 48. While the C-finger of GATA factors binds the cognate site (GATA) with a high affinity (Kd~nM) throughout the family, the N-finger of different GATA members shows different DNA binding activity. The N-finger of chicken GATA1 binds DNA weakly (Kd~μM) but shows a preference for the GATC site, whereas the N-finger of GATA2 and GATA3 binds DNA with an affinity (Kd~nM) similar to that of the C-finger but prefers GATC, GATT, and GATG to GATA 22. However, the base identify at the fourth position (GATN, underlined) seems to have only limited discriminative power on the DNA binding specificity by the C-finger and N-finger. For example, the GATA1 C-finger binds a GATA site 3-fold better than a GATC site, whereas the N-finger of GATA2 binds a GATC site 5-fold better than a GATA site 22. These observations suggest that the N-finger and C-finger of a given GATA factor could simultaneously bind proximal consensus sites or sites containing the core GAT recognition motif. Although both zinc fingers are required to bind certain double GATA sites with high affinity 16; 25; 27; 28; 29, there is no defined pattern of double GATA sites found in vertebrate genomes that would suggest a singular mechanism of DNA binding by the two zinc fingers of GATA factors. Instead, paired GATA sites found in many GATA-regulated promoters occur with various spacing and orientation, suggesting that GATA factors may bind proximal sites in a variety of modes to regulate specific transcription in distinct promoter contexts.
Our two crystal structures represent two distinct arrangements of double GATA sites on DNA. In the OPP complex, the two sites are pointing away from each other and separated by 3 base pairs (AATCAGAGATA). This arrangement is reminiscent of the palindromic site (TATCAGATA) found in the promoter of GATA1 but with two additional bases in the spacer region 28. The two zinc fingers in the OPP structure are located on the opposite faces of the DNA and make no direct contact with each other. These structural features are consistent with the fact that GATA3 C-finger binds the two sites of the OPP DNA independently (Supplemental Figure 5). However in the OPP complex (Figure 1a), the C-terminus of one finger is only 19Å away from the N-terminus of the other and there is no structural hindrance in between. Given the length and apparent flexibility of the linker between the N-finger and C-finger, it is possible that double GATA sites resembling the OPP DNA may favor simultaneous DNA binding by the N-finger and C-finger of the same GATA factor (see below).
Dimerization of GATA3 C-finger
In the ADJ complex (Figure 1b), the two GATA sites point toward each other and are separated by 5 base pairs (TGATAAGACTTATCT). This arrangement of double GATA site is taken from the mouse GATA1 promoter and also found in other promoter contexts 28. In this configuration, the two zinc fingers bind adjacent major grooves on the same side of DNA, making direct protein-protein contacts to form an intimate dimer (Figure 3a). The protein-protein interaction is mediated mainly by a conserved motif consisting of Asn351, Arg352, Pro353, Leu354, and Thr355, also known as the NRPL motif 30(Figure 3b). Although the contacting surface (169Å2 buried surface area) is smaller than that seen in several higher-order transcription factor complexes 54; 55; 56; 57, this region of the C-terminal basic tail (amino acid residues His348-Lys358) of both fingers interact with the DNA backbone and minor groove extensively, thus forming an extended protein-DNA and protein-protein interaction interface (Figure 3a). The stabilization of the flexible C-terminal basic tail by DNA may contribute to the protein-protein interaction through reduced entropy cost of binding. In the ADJ complex, the C-terminal basic tail of one zinc finger shows an alternative (minor) conformation, wherein the second half of the C-terminal basic tail (amino acid residues Lys357-Arg366) crosses over the minor groove and interacts with the zinc finger bound to the adjacent major groove. Here residues from the C-terminal basic tail of one finger, including Lys357, Lys358, Glu359, and Gln362, interact with residues at the end of the recognition helix of the other finger, including Leu347, His348, Asn349 and Ile350 (Figure 3c). The GATA3 C-finger dimer interface observed in our crystal structure is in excellent agreement with biochemical data showing that residues in the C-terminal basic tail and near the end of the recognition helix are critical to GATA1 self-association 30. Most notably, mutations in the NRPL motif of GATA1, which corresponds to the major protein-protein interaction interface observed in the GATA3 C-finger dimer (Figure 3a), substantially reduced GATA1 self-association 30.
Figure 3. Structural basis of GATA3 C-finger dimerization.
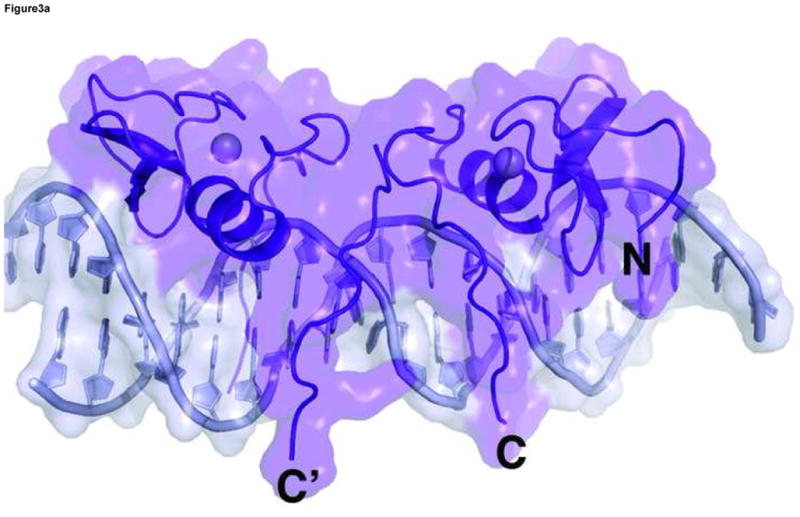
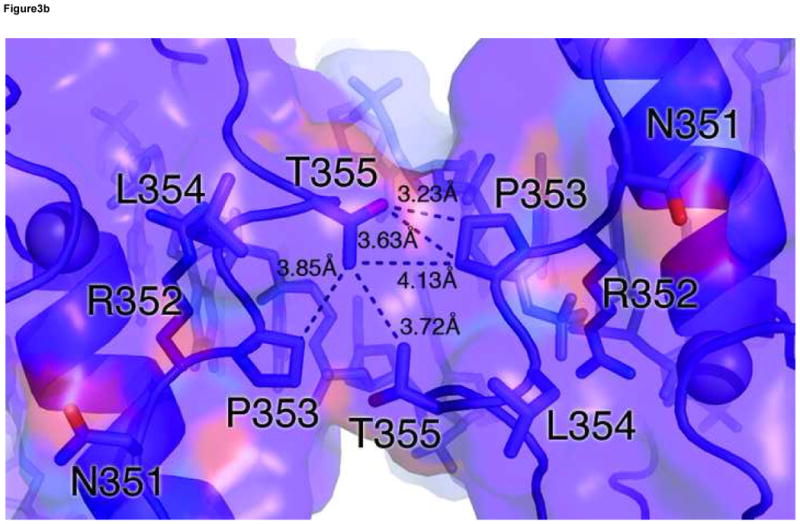
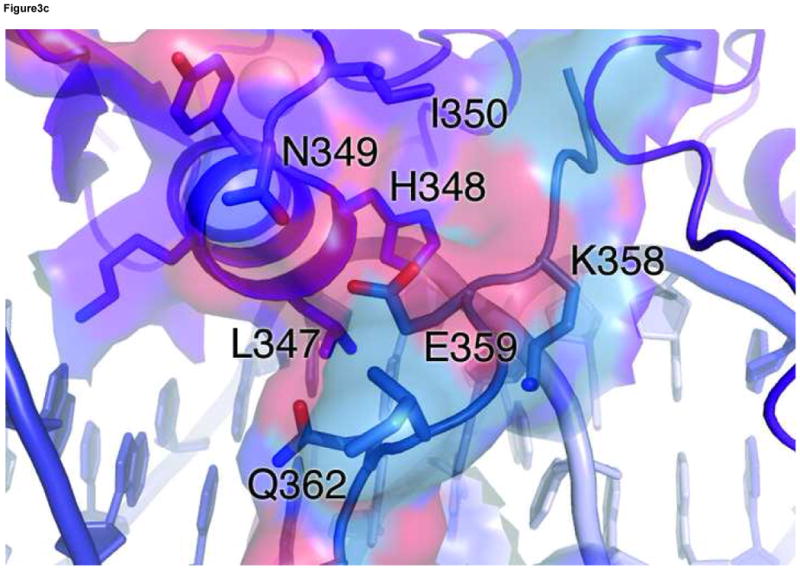
(a) Surface model of the GATA3 C-finger dimer bound to the ADJ DNA showing the extended protein-DNA and protein-protein interaction interfaces. The transparent surfaces are coloured according to the underling ribbon/stick/atom model. The orientation is similar to that of Figure 1b. (b) Detailed view of the main dimerization interface formed by the NRPL motif. Here Pro353 and Thr355 engage in extensive Van der Waals contacts, while Arg352 interacts with the DNA backbone to stabilize the conformation of the NRPL motif. Several contacts are indicated by dashed lines to give a distance scale. (c) Close contacts between the C-terminal basic tail of one zinc finger in alternative conformation (cyan) with the recognition helix of another zinc finger bound to the adjacent major groove.
Although we can not rule out crystal packing effects, the observation that the C-terminal basic tail can adopt different conformations (insert into the minor groove or cross over to the adjacent major groove) with distinct functional implications (DNA binding and protein-protein interaction) suggests that the C-terminal basic tail may play a key role in modulating the functions of GATA factors in different promoter contexts. Consistent with the structural features of the ADJ complex discussed above, GATA3 C-finger binds double GATA site with the ADJ configuration cooperatively (Supplemental Figure 5). In the ADJ complex, the C-terminus of one finger is far away from the N-terminus of the other (direct distance of 33Å and 42Å, respectively, for the minor and major conformation) and separated by the double stranded DNA, suggesting that this arrangement of double GATA sites may not allow simultaneous binding of the N-finger and C-finger from the same GATA factor, but rather favor dimerization of two GATA factors through protein-protein interactions between their C-fingers.
Binding of proximal GATA sites by full-length GATA DNA binding domain
Our structural analyses above suggest that the double GATA site in the OPP complex may favor simultaneous binding by the N-finger and C-finger of the same GATA factor whereas that in the ADJ complex may favor cooperative DNA binding by the C-finger of two GATA factors. To test this idea, we conducted a series of electrophoresis mobility shift assays (EMSA) using the full DNA binding domain of GATA3 that contains both the N-finger and C-finger (amino acid residues 260–370, referred to as DF hereafter). Titration of the ADJ DNA (DNA in the ADJ complex) with GATA3 DF yields only one complex throughout the entire concentration range (lanes 1–5, Figure 4a). The titration stoichiometry obtained at concentrations above the Kd (>10 nM) suggests that the complex corresponds to two GATA3 DFs bound to the ADJ DNA (Figure 5a). Similar to the binding of GATA3 C-finger to the ADJ DNA (Supplemental Figure 5), no monomer complex was observed under low protein/DNA ratio (molar ratio of protein:DNA <2), suggesting that the binding of the ADJ DNA by GATA3 C-finger and DF are highly cooperative. Titration of the OPP DNA (DNA from the OPP complex with the GATT site substituted by the GATA site) with GATA3 DF yields a fast mobility complex first (lanes 6–8, Figure 4a) and then a slow mobility complex when excess GATA3 DF is added (lanes 9–10, Figure 4a). This titration behavior seems to be similar to the independent binding of the GATA3 C-finger to the two GATA sites on the OPP DNA (Supplemental Figure 5). However, a close examination of the EMSA data reveals that GATA3 DF can shift all of the DNA at about 1:1 molar ratio (lane 8, Figure 4a) as if both the N-finger and the C-finger of GATA3 DF bind the two GATA sites in the OPP DNA (Figure 5b). This is likely the case since the GATA3 N-finger can bind the GATA site with a reasonable affinity (Kd ~ 28nM) though weaker than its C-finger (Kd ~ 5.2nM) 22. Similar to the binding of palindromic GATA sites by full length GATA factors observed before, the involvement of the N-finger in DNA binding here enhances the affinity of GATA3 DF for the OPP DNA and results in a fast mobility complex 16; 25; 28; 29. When excess GATA3 DF is added, the entropic advantage of intra-molecular DNA binding by the N-finger is balanced out by the high protein concentration. As a result, the C-finger of a second GATA3 DF, which binds the GATA site about five fold stronger than the N-finger, will compete off the N-finger of the first bound GATA3 DF (lanes 9–10, Figure 4a) 22. This will result in a slow mobility complex that contains two GATA3 DF bound the two GATA sites on the OPP DNA (Figure 5c). Consistent with this interpretation, when one of the GATA sites is substituted with the binding site preferred by the N-finger (GATC), the GATA3 DF not only binds the modified OPP DNA with increased affinity (compare lanes 12 and 7, Figure 4a), but also remained as the fast mobility complex even at higher protein concentrations (compare lanes 14–15 and lanes 9–10, Figure 4a). To further test our interpretation of the different DNA binding modes displayed by GATA3 DF, we made a point mutation (Arg275Glu) in GATA3 DF that is predicted to disrupt the DNA binding by the N-finger. As expected, this mutation had no apparent effect on the binding of GATA3 DF to the ADJ DNA (lanes 1–5, Figure 4b), consistent with our model that binding to the ADJ DNA only involves the C-finger of two neighboring DNA-bound GATA factors. However, this mutation significantly reduced the formation of the fast mobility complex on the OPP DNA (lanes 7–8, Figure 4b, compared with lanes 7–8, Figure 4a) and the modified OPP DNA (lanes 12–13, Figure 4b, compared with lanes 12–13, Figure 4a). The fact that we still observe some fast mobility complexes (lanes 7–8 and lanes 12–13, Figure 4b) could be simply attributed to the binding of the C-finger to one of the GATA sites. This mutation has also led to the formation of the slow mobility complex on the modified OPP DNA at high protein concentration (lanes 14–15, Figure 4b, compared with lanes 14–15, Figure 4a), presumably due to the binding of the C-finger to the GATC site at high protein concentrations. These EMSA studies support the two distinct modes of DNA binding to different double GATA sites by full-length GATA factors predicted by our structural analyses. Our structural and biochemical analyses suggest that the two zinc fingers of GATA can bind closely arrange GATA sites in distinct modes depending on the sequence and arrangement of the two sites and the protein concentration of the GATA factor. Such versatility of DNA binding by GATA may have important implications for its functional diversity in vivo.
Figure 4. Binding of proximal GATA sites by GATA factors.
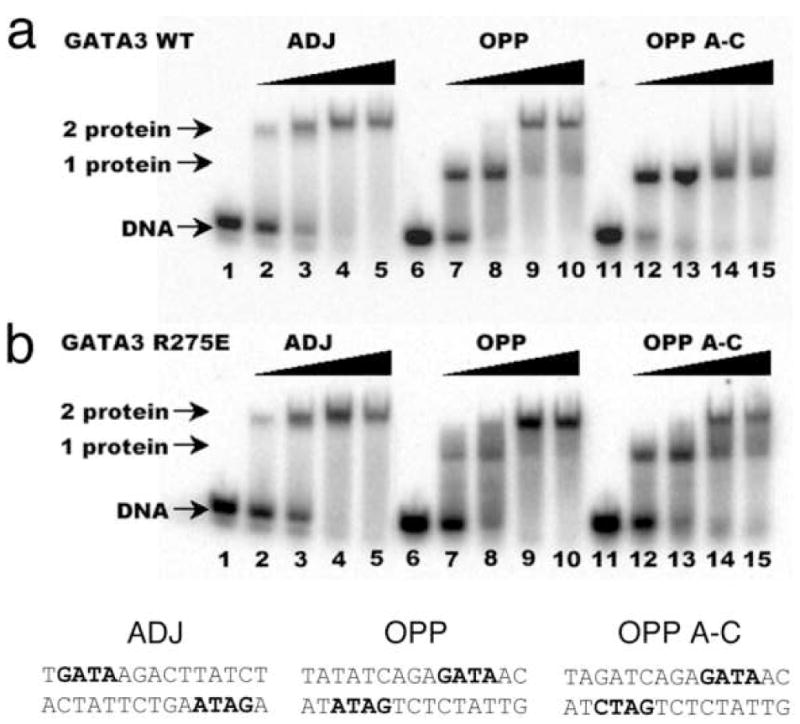
Electrophoresis mobility shift assay (EMSA) of wild type (a) and the Arg275Glu mutant (b) of GATA3 DF to three different DNA probes designed based on the crystal structures. The sequences of the three probes, ADJ, OPP and OPP A-C, are listed below the gel figure. The GATA site (preferred by the C-finger) and GATC site (preferred by the N-finger) in the sequences are highlighted by bold font. Only one strand is highlighted to indicate the orientation of the double site. For each set of five titrations, the DNA is held at 100 nM as increasing amount of protein is added (0, 50, 100, 200, 400 nM)(see Materials and Methods for details). DNA denotes free probe; “1 protein” denotes complexes of one GATA3 DF bound to DNA; “2 protein” denotes complexes of two GATA3 DF bound to DNA, either independently or cooperatively (see text for detailed discussions).
Figure 5. Diverse modes of DNA binding to different double GATA sites by GATA factors.
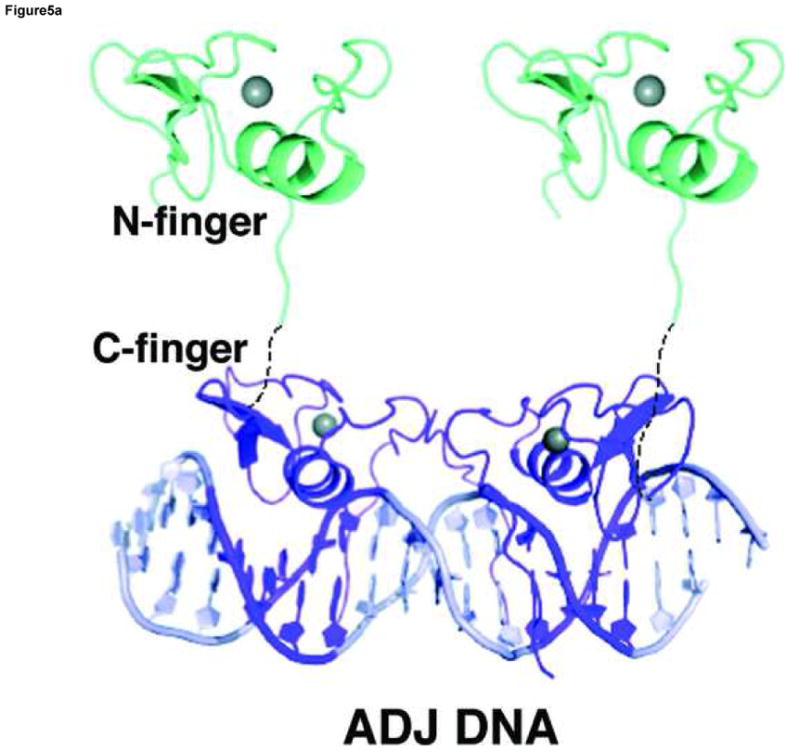
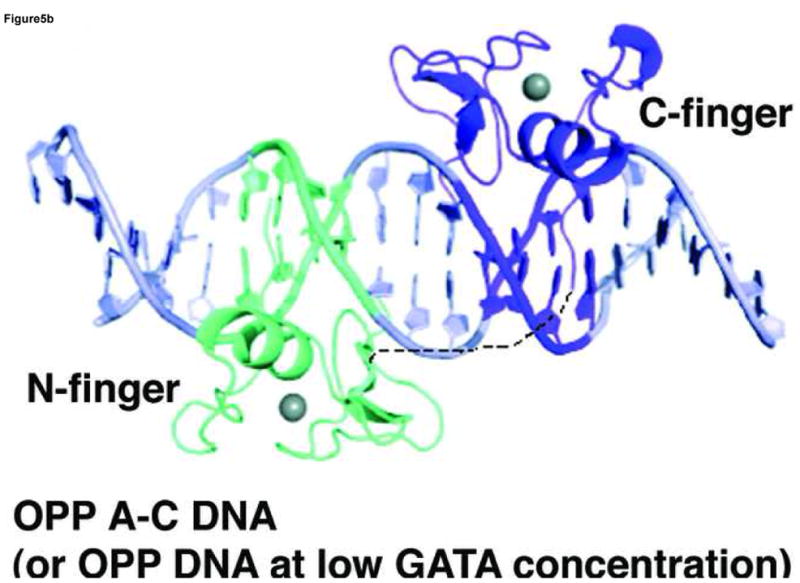
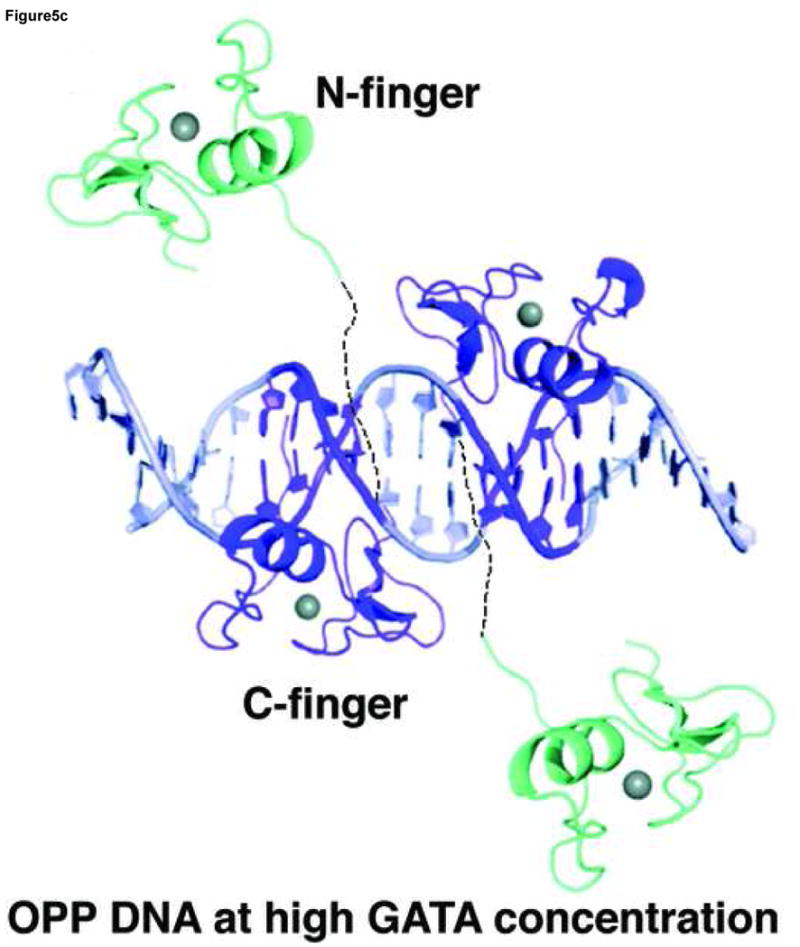
A model of GATA3 DF is built wherein the N-finger (cyan) is constructed by homology modelling based on the crystal structure of the C-finger (purple). The linker region is assumed to be flexible and may adopt different conformations depending on the arrangement of the double GATA sites. (a) Model of GATA3 DF bound to double GATA site resembling the ADJ DNA; (b) Model of GATA3 DF bound to the GATA/GATC composite site (OPP A–C) DNA) or palindromic double GATA site at low protein concentrations. (c) Model of GATA3 DF bound to palindromic double GATA site at high protein concentrations. The three models were constructed based on EMSA data of Figure 4 and were meant to interpret the specific DNA binding interactions by the N-finger and C-finger on different probes and under different conditions. The conformation of the linker region, and whether the N-finger interacts with the C-finger or DNA non specifically in (a) and (c), cannot be determined with current data and are therefore hypothetical in the figure.
Discussion
Our studies here represent one of the most comprehensive analyses of the DNA binding mechanisms by GATA zinc fingers at the structural level. DNA binding by the core zinc module in the major groove centers on a conserved arginine and asparagine (Arg329 and Asn339 in the GATA3 C-finger), which form base-specific hydrogen bonds with the first three nucleotides (GAT) in the GATA binding site. These hydrogen bonding residues are sandwiched by a number of conserved hydrophobic residues (Leu327, Leu343, and Leu347 in GATA3 C-finger) that make Van der Waals contacts to bases at the GAT tri-nucleotide motif and in the flanking region. These hydrogen bonding and Van der Waals interactions are highly conserved in the four independent GATA3 C-finger/DNA complexes and in the NMR structure of the chicken GATA1 C-finger/DNA complex and the fungal AREA/DNA complex, suggesting their critical roles in DNA binding by GATA proteins 47; 48. Consistent with the structural models presented here and published previously, a number of residues at the protein/DNA interface, such as Arg329, Leu343, and Leu347, have been shown to be functionally important in genetic and biochemical studies as well as analyses of disease-associated mutations 15; 48; 58; 59. It is noteworthy that the highly conserved Arg19 (corresponding to Arg329 in GATA3 C-finger) in chicken GATA1 was initially assigned to bind to DNA backbone but was later reassigned to bind the first Guanine in the GATA site together with the homologous Arg24 in AREA 47; 48. In our crystal structures, the corresponding arginine residue in the mouse GATA3 C-finger (Arg329) invariably make bidentate hydrogen bonds to the first Guanine (GATA, underlined) in all four independent complexes, establishing unambiguously the critical role of the conserved arginine in DNA binding by GATA factors. Our structural observations also suggest that DNA binding by GATA factors involves more hydrogen bonding interactions than initially realized, which is consistent with biochemical studies of DNA binding by GATA factors using base analogs 60.
In the minor groove, the C-terminal basic tail of GATA3 C-finger interacts with DNA bases and backbone extensively, where hydrogen bonding by Arg364 allows preferable binding to GATA/GATT sites over GATC/GATG sites. As discussed earlier, the specificity for the fourth position may be further enhanced by Leu343 and Leu347 in the major groove which make van der Waals contacts to the C5 methyl group of the fourth Thymine in the complementary strand, thus favoring a GATA site slightly over a GATT site. A sequence motif (QTRNRK) conserved in the C-finger but absent in the N-finger has been shown to be a critical DNA binding determinant 46. Replacing the corresponding motif (LVSKRA) in the N-finger of GATA1 with QTRNRK converts its DNA binding specificity to that of the C-finger. In our crystal structures, the QTRNRK motif is exactly where the C-terminal basic tail of the GATA3 C-finger binds DNA in the minor groove. Arg364, which contributes to sequence specificity to the fourth position of the binding site, is located right in the middle of this motif (QTRNRK, the underlined R corresponds to Arg364). Interestingly, replacing the QTRNRK motif in the C-finger of chicken GATA1 with LVSKRA did not convert its DNA binding specificity to that of the N-finger 46, suggesting that the DNA binding mechanism of the N-finger C-terminal basic tail may be different from that of the C-finger. The structural basis of the N-finger specificity (prefers GATC, GATG, GATT over GATA) is not clear without direct structural analysis of its complex to DNA.
It was thought that DNA recognition by GATA factors is dominated by hydrophobic residues (Leu327, Leu343, and Leu347 in the GATA3 C-finger), which make numerous Van der Waals contacts in the major groove 47; 48. However, these Van der Waals interactions are located at the periphery of the protein/DNA interface and do not seem to contribute directly to base-specific recognition of the GAT motif. It seems that the major role of the conserved hydrophobic residues is to enhance the stability of the protein/DNA complex 60. Nevertheless, these hydrophobic residues may modulate the DNA binding function of GATA factors through a number of mechanisms. First, they may mediate protein-protein interactions with a neighboring bound transcription factor partner. Second, they may impose sequence specificity in the flanking region where Van der Waals contacts by the conserved hydrophobic residues may favor some sequences. Finally, since Van der Waals interaction is sensitive to the shape complementarities of the binding interfaces, the conserved hydrophobic residues in the core zinc module may confer conformational specificity to DNA binding by GATA factors, i.e. favor binding to GATA sites embedded in DNA with certain conformations. Our crystal structures suggest that the Van der Waals interactions between Leu327, Leu343, and Leu347 of GATA3 C-finger and DNA could be enhanced if the major groove bends toward the protein surface. Although this favorable bend is not observed in our crystal structures, in vivo GATA sites may be bent in certain genomic contexts as suggested by biochemical analyses 61. This ability of shape recognition is reminiscent of that proposed for FOXP2 62 and may be particularly relevant when considering the mechanisms of selective binding of GATA sites in vivo. In this regard, it is interesting to note that GATA4 has been shown to bind in the linker region of a reconstituted nucleosomal array 41. Moreover, a mutation in GATA3 (Leu347Arg) linked to hypoparathyroidism-deafness-renal (HDR) dysplasia showed no apparent effect on the binding of GATA3 to an isolated consensus GATA site 58. According to our structural analyses above, this mutation could potentially alter the binding preference of GATA3 to sites in specific DNA conformations and hence the in vivo transcription targets, in a way similar to that proposed for the Leu22Val mutation found in AREA 63. Another GATA3 mutation involving the conserved hydrophobic residues, Leu343Phe, has been linked to human breast cancer and may affect the in vivo function of GATA3 by similar mechanisms 59. Although the hypothesis of shape recognition by hydrophobic residues in GATA factors is consistent with our structural analyses here, further studies will be needed to test it directly in vitro and in vivo.
Our structure-guided biochemical analyses reveal two distinct modes of DNA binding by GATA to proximal sites. These binding modes are likely to be used by the full-length protein since the rest of the GATA sequence seems to be unstructured and may not play a significant role in DNA binding. Previous studies have shown that the N-finger is required together with the C-finger to bind palindromic GATA sites (TATCAGATA) with high affinity and kinetic stability and that DNA binding by the N-finger is required for functions 12; 13; 16; 25; 27; 28; 29. The OPP complex presented here mimics the binding of the N-finger and C-finger of the same GATA factor to two proximal GATA sites because the C-terminus of one finger is near the N-terminus of the other. Our EMSA analyses of the binding of the wild type GATA3 DF and the Arg275Glu mutant to the OPP DNA suggest that the two fingers of the GATA3 DF indeed bind the two sites on the OPP DNA simultaneously at low protein concentration (Lanes 7–8, Figure 4a, Figure 5b). The arrangement of the two GATA sites in the OPP complex (TATCAGAGATA) resembles that of the palindromic GATA sites (TATCAGATA) but with two additional bases in the spacer region (bold). Given the long flexible linker between the N-finger and the C-finger, it is possible that the two zinc fingers of a given GATA factor may bind palindromic GATA sites with even a larger spacer. Indeed, recent studies have shown that GATA3 can bind a larger palindromic site (TATCTCATTGATA) on the FOXP3 promoter to inhibit the expression of FOXP3 and the formation of regulatory T Cells 64. Our preliminary EMSA studies indicate that GATA3 DF binds the FOXP3 palindromic site with high affinity and fast mobility, similar to its binding to the palindromic site from the GATA1 promoter (Supplemental Figure 3). With the current data, we propose that the palindromic GATA sites be at least expanded to ATC(N)1–5GATA (N refers to any nucleotide) which favor the simultaneous binding of the two zinc fingers of GATA factors 25; 27; 28; 29. The revised palindromic GATA motif defined here will guide future studies of such sites in genome and their functional relevance by combining ChIP-on-chip data and bioinformatics analysis. With the OPP configuration, it is interesting to note that the DNA binding mode of GATA3 DF changes at high protein concentration if the two sites are both GATA but remains the same if one of the sites is GATC (compare lanes 9–10 and lanes 14–15, Figure 4a) (Figure 5b,c). This observation further demonstrates the remarkable adaptability of the DNA binding mechanism of GATA factors to subtle sequence variations as well as change of protein concentration.
The ADJ complex mimics the cooperative binding of GATA factors to a different arrangement of double GATA site (GATAAGACTTATC). In this configuration, the N-terminus of one finger is far away from the C-terminus of the other, thus disfavoring the simultaneous binding of the N-finger and C-finger from the same GATA factor. Instead, this configuration of double GATA site supports direct protein-protein interaction between adjacently bound C-fingers of two GATA factors. GATA factors are known to self-associate and dimerize on DNA 30; 31. Recent studies also suggest that self-association of GATA factors may be functionally important 32. The protein elements of GATA self-association have previously been mapped to the C-terminal basic tail and the end of the recognition helix 30. In this study, our crystal structure of the GATA3 C-finger dimer/DNA complex not only confirms these biochemical data but also reveals the structural basis of GATA self-association at the atomic level. Our EMSA analyses show that GATA3 DF indeed binds the ADJ DNA as a cooperative dimer that depends on the C-finger but not the N-finger. In this DNA binding mode, the N-finger may bind DNA non-specifically or extend off DNA to interact with other proteins such as FOG (Figure 5a).
It is possible GATA factors may bind other GATA clusters in modes yet to be identified. However, our structural and biochemical studies here reveal two distinct DNA binding modes by GATA factors to different double GATA sites, demonstrating in principle the versatility of DNA binding by GATA factors. The different conformations of GATA complexes formed on different DNA elements may present distinct protein surfaces to interact with other factors in the assembly of the transcription complexes.
Materials and Methods
Protein expression, purification and mutagenesis
The C-terminal zinc finger of mouse GATA3 (amino acid residues 308–370) was subcloned into the expression vector pET28a (Novagen) as a histidine-tag fusion protein. A TEV protease cleavage site was introduced immediately before the N-terminus of the GATA3 C-finger. The expression construct was confirmed by sequencing and transformed into Rosetta (DE3) pLysS cells (Novagen, San Diego, CA) for protein expression. The expression of the GATA3 C-finger was induced by IPTG for 4 hours at 37 °C. The protein was first purified by Ni-NTA beads (Qiagen, Valencia, CA) and then digested by TEV protease for 12 hours at 4 °C to remove the histidine tag. The uncleaved protein was removed by incubating with Ni-NTA beads. The protein was further purified through a Mono S cation exchange column followed by a Superdex 75 size exclusion column (Amersham Biosciences, Piscataway, NJ). The protein sample was then concentrated to approximately 40 mg/ml in storage buffer (10 mM HEPES (pH 7.63), 5 mM β-mercaptoethanol, 0.5 μM Zinc Acetate, 100 mM NaCl, 200 mM NH4OAc, and 20% glycerol) and stored at −80°C. The mouse GATA3 double zinc finger fragment (amino acids 260–370, GATA3 DF) was expressed and purified similarly. All mutations (Arg364Ala and Arg275Glu) were made using the Quik-Change™ site-directed mutagenesis kit (Stratagene) and were confirmed by DNA sequencing. The mutants were also expressed and purified by the same protocol described above.
DNA preparation
DNA was purchased from Integrated DNA Technologies (Coralville, IA) at 1 μmole scale in the crude but desalted form. The crude DNA was dissolved in a buffer (100mM NaCl, 10mM NaOH, pH 12.0) and purified by a Mono Q cation exchange column on FPLC (Amersham biosciences, Piscataway, NJ). The peak fractions were pooled and neutralized to pH 7.0 by Hepes prior to over night dialysis against water. The desalted DNA sample was lyophilized to powder, resuspended in water, and quantified at 260 nm. Complementary DNA strands were annealed at 95°C in the annealing buffer (100mM NaCl, 5mM Hepes pH 7.6). The two double stranded DNA that crystallized successfully with GATA3 C-finger in our study are: 5′-TTCTGATAAGACTTATCTGC-3′ (Top strand of the ADJ DNA), 5′-AAGCAGATAAGTCTTATCAG-3′ (Bottom strand of the ADJ DNA), 5′-TTGATAAATCAGAGATAACC-3′, (Top Strand of the OPP DNA) and 5′-AAGGTTATCTCTGATTTATC-3′ (Bottom Strand of the OPP DNA). Note that in the OPP DNA, a GATA site (italicized) was originally introduced to create a tandem of two GATA sites. But in the crystal structure of the OPP complex, while one of the two GATA3 C-fingers binds the consensus GATA site at the 3′ half of the DNA, the other finger binds a fortuitous GATT site, resulting two GATA fingers binding to DNA in a palindromic orientation. The unoccupied GATA site was removed in EMSA studies (see below).
Crystallization, data collection, and structure determination
The GATA3 C-finger/DNA complexes were prepared by mixing protein and DNA at 2:1 molar ratio in storage buffer at a final concentration of 10 mg/ml. Crystals were grown by the hanging drop method at 18°C using a reservoir buffer of 20mM Mg(OAc)2, 20 mM Cacodylic acid pH 6.5, and 30% PEG 4K. Typically, crystals of the OPP and ADJ complexes grew to approximately 400 x 200 x 20 μm in 1–4 days. Crystals of the ADJ complex belong to the space group C2 with cell dimensions a = 137.977 Å, b = 35.756 Å, c = 54.487 Å, and β = 113.25°. Crystals of the OPP complex also belong to the space group C2 but with cell dimensions a = 128.882 Å, b = 30.370 Å, c = 75.648 Å, and β= 93.818°. Crystals were stabilized in the harvest/cryoprotectant buffer: 20 mM Mg(OAc)2, 20 mM Cacodylic acid pH 6.5, 30% PEG 4K, and 25% (w/v) glycerol and flash frozen with liquid nitrogen for cryo-crystallography. Data were collected at the ALS BL8.2.1 and BL8.2.2 beamlines at the Lawrence Berkeley National Laboratory. Data were reduced using DENZO and SCALEPACK 65. Initial phases for the OPP complex were determined by SAD phasing using Zinc anomalous signal. Phases for the ADJ complex were determined by molecular replacement using the GATA3 C-finger from the OPP structure as the search model. Molecular replacement, refinement, and final analysis were done with CNS 66. The structure determination of both GATA3 C-finger/DNA complexes is relatively straightforward. The refinement is carried out using standard strategies of energy minimization, grouped b-factor refinement, simulated annealing, and individual b-factor refinement. Temperature factors were first refined by groups (main and side chain for proteins; backbone and base for DNA), followed by restrained individual refinement at later and final stages. NCS restraints were applied to the entire protein at the initial simulated annealing but relaxed to only including the core zinc module in the final round of refinement. The test set for the molecular replacement of the ADJ complex was selected randomly by CNS.
Due to the limited resolution, we also applied B-DNA restraints throughout the refinement. The NCS and B-DNA restraints may account at least partly for the small separation of Rwork and Rfree. The diffraction spots of the OPP complex crystals show banana shape, which may lead to the relatively high Rsym in the reduced data. An unbiased electron density map for a portion of the DNA, calculated by simulated omitting map in CNS, is shown in Supplemental Figure 4. The map shows that the DNA has well defined electron density. The refinement is monitored by free R factor of 10% randomly selected test set of structural factors. Regions of interest have been checked by simulated omit map. The quality of the final model has also been analyzed by standard programs in CNS and CCP4. Specifically, the ADJ complex contains 57.1% of residues in the most favored region followed by 41.9% in the additional allowed and 1% in generously allowed. The OPP site complex separated into 80% in the most favored region and 20% in the additionally allowed region. Both structures satisfy the crystallographic standards for their resolutions. The statistics of crystallographic analysis are presented in Table 1. Figures of structure illustration were prepared using Pymol (DeLano Scientific, San Francisco, CA). Model building and structural comparisons were carried out in O 67.
Electrophoresis Mobility Shift Assay (EMSA)
DNA probes labeled with Cy3 were mixed with GATA3 double finger fragment (GATA3 DF) in a total of 20 μl binding buffer (5 mM Hepes, pH 7.63, 0.5 mM EDTA, 4 mM Mg(OAc)2, 50 mM KCl, 2mg/ml Bovine Calf thymus DNA, 10% glycerol, 1 mM DTT). The concentration of DNA probe was held at 100nM in each reaction, while the protein concentration was increased gradually in each set of titration (0, 50, 100, 200, 400 nM). The binding reactions were incubated at room temperature for 25 minutes. The samples were run on a native 6% (w/v) polyacrylamide gel in 0.5xTBE buffer for 3 hours at 4°C. The gel was transferred to blotting paper and dried for 1 hour in the gel drier. The gel was then exposed overnight onto a phosphoimage plate. The plate was scanned on a Typhoon Image Reader resulting in a digital format that was analyzed in Image Quant software. EMSA analyses of GATA3 mutants (Arg364Ala and Arg275Glu) were carried out in GATA3 DF. The Arg275Glu mutant was used to analyze the role of the N-finger in binding to various double GATA site, whereas the Arg364Ala mutant was created to test the role of Arg364 in binding to the cognate GATA site. In the latter case, we used the ADJ DNA as the probe since the binding of ADJ DNA by GATA3 DF only requires the C-finger.
Supplementary Material
Acknowledgments
The authors thank Robert Batey, James C. Stroud, Xiaojiang Chen, Reza Kalhor, and members of the Cech lab for helpful discussions. This research is supported by grants from NIH (L.C.). D.L.B and L.G are supported partly by NIH training grants
Footnotes
ACCESSION NUMBERS:
Coordinates and structure factors for the OPP complex and the ADJ complex have been deposited in the Protein Data Bank with accession code 3DFX and 3DFV, respectively.
Supplementary Information accompanies the paper
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Evans T, Reitman M, Felsenfeld G. An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc Natl Acad Sci U S A. 1988;85:5976–80. doi: 10.1073/pnas.85.16.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans T, Felsenfeld G. The erythroid-specific transcription factor Eryf1: a new finger protein. Cell. 1989;58:877–85. doi: 10.1016/0092-8674(89)90940-9. [DOI] [PubMed] [Google Scholar]
- 3.Tsai SF, Martin DI, Zon LI, D’Andrea AD, Wong GG, Orkin SH. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989;339:446–51. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- 4.Bresnick EH, Martowicz ML, Pal S, Johnson KD. Developmental control via GATA factor interplay at chromatin domains. J Cell Physiol. 2005;205:1–9. doi: 10.1002/jcp.20393. [DOI] [PubMed] [Google Scholar]
- 5.Weiss MJ, Orkin SH. GATA transcription factors: key regulators of hematopoiesis. Exp Hematol. 1995;23:99–107. [PubMed] [Google Scholar]
- 6.Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–52. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 7.Kim SI, Bresnick EH. Transcriptional control of erythropoiesis: emerging mechanisms and principles. Oncogene. 2007;26:6777–94. doi: 10.1038/sj.onc.1210761. [DOI] [PubMed] [Google Scholar]
- 8.Ho IC, Vorhees P, Marin N, Oakley BK, Tsai SF, Orkin SH, Leiden JM. Human GATA-3: a lineage-restricted transcription factor that regulates the expression of the T cell receptor alpha gene. Embo J. 1991;10:1187–92. doi: 10.1002/j.1460-2075.1991.tb08059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.George KM, Leonard MW, Roth ME, Lieuw KH, Kioussis D, Grosveld F, Engel JD. Embryonic expression and cloning of the murine GATA-3 gene. Development. 1994;120:2673–86. doi: 10.1242/dev.120.9.2673. [DOI] [PubMed] [Google Scholar]
- 10.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–96. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 11.Lowry JA, Atchley WR. Molecular evolution of the GATA family of transcription factors: conservation within the DNA-binding domain. J Mol Evol. 2000;50:103–15. doi: 10.1007/s002399910012. [DOI] [PubMed] [Google Scholar]
- 12.Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17:1642–51. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimizu R, Takahashi S, Ohneda K, Engel JD, Yamamoto M. In vivo requirements for GATA-1 functional domains during primitive and definitive erythropoiesis. Embo J. 2001;20:5250–60. doi: 10.1093/emboj/20.18.5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nichols KE, Crispino JD, Poncz M, White JG, Orkin SH, Maris JM, Weiss MJ. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet. 2000;24:266–70. doi: 10.1038/73480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nesbit MA, Bowl MR, Harding B, Ali A, Ayala A, Crowe C, Dobbie A, Hampson G, Holdaway I, Levine MA, McWilliams R, Rigden S, Sampson J, Williams AJ, Thakker RV. Characterization of GATA3 mutations in the hypoparathyroidism, deafness, and renal dysplasia (HDR) syndrome. J Biol Chem. 2004;279:22624–34. doi: 10.1074/jbc.M401797200. [DOI] [PubMed] [Google Scholar]
- 16.Yu C, Niakan KK, Matsushita M, Stamatoyannopoulos G, Orkin SH, Raskind WH. X-linked thrombocytopenia with thalassemia from a mutation in the amino finger of GATA-1 affecting DNA binding rather than FOG-1 interaction. Blood. 2002;100:2040–5. doi: 10.1182/blood-2002-02-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantor AB. GATA transcription factors in hematologic disease. Int J Hematol. 2005;81:378–84. doi: 10.1532/ijh97.04180. [DOI] [PubMed] [Google Scholar]
- 18.Ko LJ, Engel JD. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–22. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Omichinski JG, Trainor C, Evans T, Gronenborn AM, Clore GM, Felsenfeld G. A small single-“finger” peptide from the erythroid transcription factor GATA-1 binds specifically to DNA as a zinc or iron complex. Proc Natl Acad Sci U S A. 1993;90:1676–80. doi: 10.1073/pnas.90.5.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Visvader JE, Crossley M, Hill J, Orkin SH, Adams JM. The C-terminal zinc finger of GATA-1 or GATA-2 is sufficient to induce megakaryocytic differentiation of an early myeloid cell line. Mol Cell Biol. 1995;15:634–41. doi: 10.1128/mcb.15.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedone PV, Omichinski JG, Nony P, Trainor C, Gronenborn AM, Clore GM, Felsenfeld G. The N-terminal fingers of chicken GATA-2 and GATA-3 are independent sequence-specific DNA binding domains. Embo J. 1997;16:2874–82. doi: 10.1093/emboj/16.10.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newton A, Mackay J, Crossley M. The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J Biol Chem. 2001;276:35794–801. doi: 10.1074/jbc.M106256200. [DOI] [PubMed] [Google Scholar]
- 24.Martin DI, Orkin SH. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 1990;4:1886–98. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- 25.Schwartzbauer G, Schlesinger K, Evans T. Interaction of the erythroid transcription factor cGATA-1 with a critical auto-regulatory element. Nucleic Acids Res. 1992;20:4429–36. doi: 10.1093/nar/20.17.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans T, Felsenfeld G. trans-Activation of a globin promoter in nonerythroid cells. Mol Cell Biol. 1991;11:843–53. doi: 10.1128/mcb.11.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai SF, Strauss E, Orkin SH. Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes Dev. 1991;5:919–31. doi: 10.1101/gad.5.6.919. [DOI] [PubMed] [Google Scholar]
- 28.Trainor CD, Omichinski JG, Vandergon TL, Gronenborn AM, Clore GM, Felsenfeld G. A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high-affinity interaction. Mol Cell Biol. 1996;16:2238–47. doi: 10.1128/mcb.16.5.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trainor CD, Ghirlando R, Simpson MA. GATA zinc finger interactions modulate DNA binding and transactivation. J Biol Chem. 2000;275:28157–66. doi: 10.1074/jbc.M000020200. [DOI] [PubMed] [Google Scholar]
- 30.Crossley M, Merika M, Orkin SH. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol Cell Biol. 1995;15:2448–56. doi: 10.1128/mcb.15.5.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackay JP, Kowalski K, Fox AH, Czolij R, King GF, Crossley M. Involvement of the N-finger in the self-association of GATA-1. J Biol Chem. 1998;273:30560–7. doi: 10.1074/jbc.273.46.30560. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu R, Trainor CD, Nishikawa K, Kobayashi M, Ohneda K, Yamamoto M. GATA-1 self-association controls erythroid development in vivo. J Biol Chem. 2007;282:15862–71. doi: 10.1074/jbc.M701936200. [DOI] [PubMed] [Google Scholar]
- 33.Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–19. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 34.Svensson EC, Tufts RL, Polk CE, Leiden JM. Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. Proc Natl Acad Sci U S A. 1999;96:956–61. doi: 10.1073/pnas.96.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liew CK, Simpson RJ, Kwan AH, Crofts LA, Loughlin FE, Matthews JM, Crossley M, Mackay JP. Zinc fingers as protein recognition motifs: structural basis for the GATA-1/friend of GATA interaction. Proc Natl Acad Sci U S A. 2005;102:583–8. doi: 10.1073/pnas.0407511102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cantor AB, Orkin SH. Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins. Semin Cell Dev Biol. 2005;16:117–28. doi: 10.1016/j.semcdb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 37.Agarwal S, Avni O, Rao A. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity. 2000;12:643–52. doi: 10.1016/s1074-7613(00)80215-0. [DOI] [PubMed] [Google Scholar]
- 38.Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–51. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 39.Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–28. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyes J, Omichinski J, Clark D, Pikaart M, Felsenfeld G. Perturbation of nucleosome structure by the erythroid transcription factor GATA-1. J Mol Biol. 1998;279:529–44. doi: 10.1006/jmbi.1998.1783. [DOI] [PubMed] [Google Scholar]
- 41.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–89. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 42.Johnson KD, Grass JA, Boyer ME, Kiekhaefer CM, Blobel GA, Weiss MJ, Bresnick EH. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc Natl Acad Sci U S A. 2002;99:11760–5. doi: 10.1073/pnas.192285999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martowicz ML, Grass JA, Boyer ME, Guend H, Bresnick EH. Dynamic GATA factor interplay at a multicomponent regulatory region of the GATA-2 locus. J Biol Chem. 2005;280:1724–32. doi: 10.1074/jbc.M406038200. [DOI] [PubMed] [Google Scholar]
- 44.Im H, Grass JA, Johnson KD, Kim SI, Boyer ME, Imbalzano AN, Bieker JJ, Bresnick EH. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc Natl Acad Sci U S A. 2005;102:17065–70. doi: 10.1073/pnas.0506164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang HY, Evans T. Distinct roles for the two cGATA-1 finger domains. Mol Cell Biol. 1992;12:4562–70. doi: 10.1128/mcb.12.10.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghirlando R, Trainor CD. Determinants of GATA-1 binding to DNA: the role of non-finger residues. J Biol Chem. 2003;278:45620–8. doi: 10.1074/jbc.M306410200. [DOI] [PubMed] [Google Scholar]
- 47.Omichinski JG, Clore GM, Schaad O, Felsenfeld G, Trainor C, Appella E, Stahl SJ, Gronenborn AM. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 1993;261:438–46. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- 48.Starich MR, Wikstrom M, Arst HN, Jr, Clore GM, Gronenborn AM. The solution structure of a fungal AREA protein-DNA complex: an alternative binding mode for the basic carboxyl tail of GATA factors. J Mol Biol. 1998;277:605–20. doi: 10.1006/jmbi.1998.1625. [DOI] [PubMed] [Google Scholar]
- 49.Tjandra N, Omichinski JG, Gronenborn AM, Clore GM, Bax A. Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution. Nat Struct Biol. 1997;4:732–8. doi: 10.1038/nsb0997-732. [DOI] [PubMed] [Google Scholar]
- 50.Whyatt DJ, deBoer E, Grosveld F. The two zinc finger-like domains of GATA-1 have different DNA binding specificities. Embo J. 1993;12:4993–5005. doi: 10.1002/j.1460-2075.1993.tb06193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen ML, Kuo CL. A conserved sequence block in the murine and human T cell receptor Jalpha loci interacts with developmentally regulated nucleoprotein complexes in vitro and associates with GATA-3 and octamer-binding factors in vivo. Eur J Immunol. 2001;31:1696–705. doi: 10.1002/1521-4141(200106)31:6<1696::aid-immu1696>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 52.Zhang DH, Yang L, Ray A. Differential responsiveness of the IL-5 and IL-4 genes to transcription factor GATA-3. J Immunol. 1998;161:3817–21. [PubMed] [Google Scholar]
- 53.Takemoto N, Arai K, Miyatake S. Cutting edge: the differential involvement of the N-finger of GATA-3 in chromatin remodeling and transactivation during Th2 development. J Immunol. 2002;169:4103–7. doi: 10.4049/jimmunol.169.8.4103. [DOI] [PubMed] [Google Scholar]
- 54.Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–8. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 55.Stroud JC, Lopez-Rodriguez C, Rao A, Chen L. Structure of a TonEBP-DNA complex reveals DNA encircled by a transcription factor. Nat Struct Biol. 2002;9:90–4. doi: 10.1038/nsb749. [DOI] [PubMed] [Google Scholar]
- 56.Giffin MJ, Stroud JC, Bates DL, von Koenig KD, Hardin J, Chen L. Structure of NFAT1 bound as a dimer to the HIV-1 LTR kappa B element. Nat Struct Biol. 2003;10:800–6. doi: 10.1038/nsb981. [DOI] [PubMed] [Google Scholar]
- 57.Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF, Mathis D, Benoist C, Chen L, Rao A. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–87. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 58.Ali A, Christie PT, Grigorieva IV, Harding B, Van Esch H, Ahmed SF, Bitner-Glindzicz M, Blind E, Bloch C, Christin P, Clayton P, Gecz J, Gilbert-Dussardier B, Guillen-Navarro E, Hackett A, Halac I, Hendy GN, Lalloo F, Mache CJ, Mughal Z, Ong AC, Rinat C, Shaw N, Smithson SF, Tolmie J, Weill J, Nesbit MA, Thakker RV. Functional characterization of GATA3 mutations causing the hypoparathyroidism-deafness-renal (HDR) dysplasia syndrome: insight into mechanisms of DNA binding by the GATA3 transcription factor. Hum Mol Genet. 2007;16:265–75. doi: 10.1093/hmg/ddl454. [DOI] [PubMed] [Google Scholar]
- 59.Usary J, Llaca V, Karaca G, Presswala S, Karaca M, He X, Langerod A, Karesen R, Oh DS, Dressler LG, Lonning PE, Strausberg RL, Chanock S, Borresen-Dale AL, Perou CM. Mutation of GATA3 in human breast tumors. Oncogene. 2004;23:7669–78. doi: 10.1038/sj.onc.1207966. [DOI] [PubMed] [Google Scholar]
- 60.Mott BH, Bassman J, Pikaart MJ. A molecular dissection of the interaction between the transcription factor Gata-1 zinc finger and DNA. Biochem Biophys Res Commun. 2004;316:910–7. doi: 10.1016/j.bbrc.2004.02.142. [DOI] [PubMed] [Google Scholar]
- 61.Ghirlando R, Trainor CD. GATA-1 bends DNA in a site-independent fashion. J Biol Chem. 2000;275:28152–6. doi: 10.1074/jbc.M002053200. [DOI] [PubMed] [Google Scholar]
- 62.Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H, Chen L. Structure of the forkhead domain of FOXP2 bound to DNA. Structure. 2006;14:159–66. doi: 10.1016/j.str.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 63.Starich MR, Wikstrom M, Schumacher S, Arst HN, Jr, Gronenborn AM, Clore GM. The solution structure of the Leu22-->Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity. J Mol Biol. 1998;277:621–34. doi: 10.1006/jmbi.1997.1626. [DOI] [PubMed] [Google Scholar]
- 64.Mantel PY, Kuipers H, Boyman O, Rhyner C, Ouaked N, Ruckert B, Karagiannidis C, Lambrecht BN, Hendriks RW, Crameri R, Akdis CA, Blaser K, Schmidt-Weber CB. GATA3-driven Th2 responses inhibit TGF-beta1-induced FOXP3 expression and the formation of regulatory T cells. PLoS Biol. 2007;5:e329. doi: 10.1371/journal.pbio.0050329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 66.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 67.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47 (Pt 2):110–9. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


