Hybridization of parasites is an emerging public health concern at the interface of infectious disease biology and evolution. Increasing economic development, human migration, global trade, and climate change are all shifting the geographic distribution of existing human, livestock, companion animal, and wildlife parasites [1–9]. As a result, human populations encounter new infections more frequently, and coinfection by multiple parasites from different lineages or species within individual hosts occurs. Coinfection may have a large impact on the hosts and parasites involved, often as a result of synergistic or antagonistic interactions between parasites [10]. Indeed, mixed-species coinfections have been found to influence parasite establishment, growth, maturation, reproductive success, and/or drug efficacy [11–13]. However, coinfections can allow for heterospecific (between-species or between-lineage) mate pairings, resulting in parthenogenesis (asexual reproduction in which eggs occur without fertilization), introgression (the introduction of single genes or chromosomal regions from one species into that of another through repeated backcrossing), and whole-genome admixture through hybridization [14].
Recent molecular developments have revealed instances of fertile hybridization and introgression in plants [15], animals [16,17], and humans [18]. However, modern genetics and genomics have also uncovered the first confirmed cases of introgression within eukaryotic parasites [14,19]. Examples for such successful reticulate evolution in parasites include, but are not exclusive to, causative agents of important diseases initiated by fungi (Cryptococcus [20]), helminthic worms (Schistosoma, Fasciola, Ascaris, and Trichinella [21,22–24]), and protozoa (Plasmodium, Leishmania, Toxoplasma, and Trypanosoma [25,26–31]), as well as their vectors. These cases involve introgression between members of the neglected tropical diseases (NTDs) and/or neglected zoonotic diseases (NZDs)—highly debilitating diseases infecting more than a sixth of the world’s human population (and their livestock), with devastating consequences for individuals and communities. Such diseases are predominantly diseases of the world’s poorest communities, afflicting those people who are most at risk of contracting mixed parasitic infections (and hence also at risk for potential novel introgressed parasite infections) and at the same time least likely to get proper medical care once infected [32].
Evidence gathered to date, mainly from studies on nonparasitic animals and plants, suggests that hybridization can have a major evolutionary impact [33]. While hybridization can result in lower average fitness due to F1 sterility and inviability, caused by negative epistatic interactions [34–36] and the disruption of beneficial gene complexes [37,38], hybridization is at the same time a rich source of new genetic variation [39]. This can provide the raw material for natural selection to shape the evolution of ecologically relevant traits [33,40,41]. Thus, while a hybrid offspring, under certain conditions, may be less fit than its purebred offspring counterparts from either parental line, hybridization broadens the “working surface” for selection by producing a whole range of potentially adaptive phenotypes, ranging from one parent to the other and beyond. In nonparasitic taxa, hybridization has already been shown to promote geographic range expansions of populations (for example, in spiders [42]) as well as adaptation to new environments (Darwin finches [43]). There is also increasing evidence that hybridization can lead to the functional diversification of a group, as shown in Helianthus sunflowers [44], and even to speciation (reviewed in [40,45,46]). One key example of this is the case of “transgressive hybrids,” i.e., hybrids expressing extreme phenotypes that do not reside in either of the parental populations. These are “hopeful monsters” with respect to their evolutionary potential because they can diverge from their parents ecologically and may quickly become reproductively isolated from them [47–49]. In addition to broadening the selection surface, hybridization can also speed up the process of adaptation. After hybridization events, new alleles or allelic combinations that happen to be beneficial in the new environment are available immediately. This is in contrast to nonhybrid populations in which adaptation relies on alleles brought in via immigration or de novo mutation, both processes that require relatively longer periods of time [50,51]. As a consequence, populations with a hybrid origin may survive rapid environmental change better than their nonhybrid parents, as recently shown in yeast [52].
We predict that parasites are one of the major groups of organisms in which hybridization can have major impacts on the evolution and diversification of a group [14,19] and can lead to speciation [53], affecting key pathogenic traits and transmission. Hybridization in parasites may thus pose a serious challenge for the prevention, effective control, and therapy of disease [19]. While it has been suggested that hybridization and introgression between parasites can potentially drive the emergence and rapid evolution of novel zoonotic diseases [14], most studies to date are descriptive and do not consider the evolutionary consequences. Here, we review some of the most recent advances in the detection of hybridization in eukaryotic parasites (and their vectors) and discuss the significance of parasite hybridization for adaptive evolution and public health supported by relevant case studies in Schistosoma spp. (Box 1), Leishmania and Trypanosoma (Box 2), and the malaria vector Anopheles (Box 3). Given the role of infectious agents in our changing world, particularly in terms of emerging parasitic disease in response to anthropogenic change [32], it is time for a new and integrative perspective. Here, we argue for the integration of parasitology, disease biology, and evolutionary biology to understand the consequences of parasite hybridization to aid in the management and prevention of disease.
Box 1. Case Example—Introgression in Helminths: Schistosomiasis
Schistosoma spp. are the causative agents of schistosomiasis, a prevalent, chronic, and debilitating helminthic disease of humans and animals that occurs across much of the developing world. As early as 1948, there have been phenotypic reports of eggs indicative of potential Schistosoma haematobium–S. mattheei hybrids in Rhodesia/Zimbabwe [75], S. bovis–S. curassoni hybrids in West African ruminants, S. haematobium–S. mattheei hybrids in Southern African ruminants, and S. haematobium–S. bovis hybrids in humans from West Africa [76–78]. More recently, ITS1+2 and cox1 barcoding studies of viable schistosome miracidial larvae hatched from the stool and urine of Senegalese school children confirmed bidirectional hybridization between human S. haematocium and livestock S. bovis [67], as well as for S. haematobium and S. curassoni [79]. Studies from infected snails in Kenya have observed hybrid cercariae between S. mansoni from humans and its sister species, S. rodhaini, from rodents [80]. These authors, using microsatellite markers (ribosomal DNA [rDNA] and mitochondrial DNA [mtDNA]), demonstrated that these hybrids produce viable offspring through first or successive generation backcrosses with S. mansoni. Unlike the S. haematobium and S. bovis or S. currassoni hybrids described above, the direction of introgression appeared highly asymmetric, causing unidirectional gene flow from the rodent S. rodhaini to the human S. mansoni [80]. Recent evidence from infected humans in Senegal has also revealed the potential for introgressions between the more phylogenetically distant pairings of the two major human schistosome species in Africa, S. haematobium and S. mansoni, a pairing previously thought to result in unviable eggs exclusively through parthogenesis [81]. These studies combined provide convincing evidence that schistosome species readily hybridize in nature, which may have major implications in light of the current global push for human disease control programmes to shift from controlling morbidity to halting transmission [32]. How such introgression may alter host range is perhaps the most pressing area for future research. Many schistosome species infecting livestock have a broader geographical range beyond Asia and Africa, with compatible snail intermediate hosts present. Novel zoonotic hybrids may therefore have the potential to be a global disease, particularly in our current climate of global warming and increased human and animal movement and transportation. This may be highlighted most clearly where novel introgressed hybrids between human S. haematobium and livestock S. bovis have recently been identified, with substantial ongoing transmission amongst both local residents and tourists, within Europe [82–84].
Box 2. Case Example—Introgression in Protozoans: Leishmaniosis and Trypanosomiasis
Introgressions can occur within the causative agents of protozoal diseases leishmaniasis and trypanosomiasis (Kinetoplastida: Trypanosomatidae) [85–88]. Approximately 30,000 people in 36 countries of sub-Saharan Africa suffer from human African trypanosomiasis (HAT), and Chagas disease has been classed as the most important vector-borne infection in Latin America, affecting an estimated 7–8 million humans, with around 21,000 deaths per year [89]. Leishmania parasites are another of the most important vector-borne pathogens in the developing world. Both Leishmania and Trypanosoma are also parasites with major zoonotic reservoirs. Two of the major lineages of Trypanosoma cruzi (discrete typing units [DTUs] III and IV) are now thought to have arisen by intraspecific hybridisation [87], despite their predominant mode of asexual reproduction, and introgression between subspecies has been associated with virulence [88]. Similarly, whilst asexual reproduction through clonal propagation has been proposed to be the major reproductive mechanism across the genus Leishmania, a hybridizing sexual cycle has been detected within its sand fly vector from across a range of geographical locations [90,91]. Whole-genome sequencing of Leishmania parasites isolated from sand flies from a Turkish endemic area indicated that variation in these parasites arose following a single cross between two phylogenetically distinct strains. Furthermore, whilst it appears that these populations do reproduce primarily clonally following this original hybridization event, subsequent recombination between the progeny does also occur [92]. The potentially large epidemiological consequences of such recombination events may be demonstrated by the observation that Leishmania infantum/L. major hybrids possess an enhanced host range, as hybrid offspring, and, unlike their parental single species, they are able to infect another vector, Phlebotomus papatasi [93].
Box 3. Case Example—Introgression in the Parasite Vector Anopheles: Malaria
Anopheles has become a model organism at the interface of speciation genomics and epidemiology [94], showcasing the potential perils of hybridization for public health but also the possible benefits of using controlled and induced hybridization as a means for disease control. Anopheles mosquitoes are vectors for malaria, which is caused by parasitic protozoans belonging to the genus Plasmodium. Malaria affects 200 million people a year worldwide, with an estimated 0.5–1 million deaths per year, mostly among young children in sub-Saharan Africa, where 90% of the world’s malaria deaths occur [95].
There are approximately 60 different species of Anopheles found worldwide [96], and genomic sequences of 16 Anopheles species (including vector and nonvector species) have recently become available [94,97]. These sequences have revealed fast and flexible evolutionary rates with respect to traits affecting their transmission potential, shown extensive introgression between Anopheles species, and suggested that enhanced vectorial capacity and adaptation to humans as primary hosts can result from interspecific genetic exchange. A multilocus single nucleotide polymorphism (SNP) genotyping panel is also at hand to detect F1 hybrids and backcrosses between the main vectors of malaria, Anopheles gambiae sensu stricto (S form) and A. coluzzii (M form) [98].
Control of Anopheles through insecticides, such as dichlorodiphenyltrichloroethane (DDT) and pyrethroids, has contributed to the prevention of malaria, but resistance to insecticides has recently emerged in Anopheles populations [99,100]. Recent studies have demonstrated rapid adaptive introgression of the insecticide resistance mutation Vgsc-L1014F from A. gambiae to A. coluzzii, in response to strong anthropogenic selection from increased insecticide use [101,102]. Others have found gene flow occurring at rates “far from inconsequential” between other species of Anopheles (e.g., between A. gambiae and A. arabiensis in Uganda [99,103] and A. sinensis and A. kleini in Korea [104]). Experimental interpopulation crosses of A. gambiae, monitored for traits determining their malaria transmission potential, scored higher for fecundity, body size, adult longevity, and average blood meal size, compared to both parental strains [105]. It is thus conceivable that the fitness-enhancing potential of hybridization may also apply to closely related interspecific hybrid crosses.
Hybridization between Anopheles species may have beneficial effects for disease control. Asymmetric introgression has recently been shown to transfer adaptive immunity and increased refractoriness to the Plasmodium pathogen from A. coluzzii to A. gambiae in Guinea [106]. Another opportunity for disease control is to modify the host preference of A. gambiae from humans to cattle by hybridizing it with A. quadriannulatus, a more zoophilic nonvector species, as a strategy to decrease its competence as malaria vector [107].
Consequences of Hybridization in Parasites
Since multidrug resistance became a worldwide problem in pathogenic bacteria in the 1950s [54], we know that the exchange of genetic material via horizontal gene transfer among bacterial taxa has contributed to their evolution and pathogenesis [55,56]. Horizontal gene transfer can be advantageous and confer higher fitness, for instance, through the acquisition of antibiotic drug resistance [57] or through the spread of virulence factors; a well-studied example is the Shiga toxin genes exchanged between Escherichia coli and Shigella bacterial pathogens [58]. There are also several examples of recombinant human viruses that have exchanged genes with other strains with detrimental effects (Spanish flu, human rotavirus, and dengue fever; [59,60,61]). Conversely, despite their equally negative impact on host populations, not much is known about the hybridization of eukaryotic parasites, their frequency in the wild, or how hybridization may affect their spread and pathogenicity. Modern molecular techniques, however, can expose the signature of hybridization in the genome more rapidly and accurately, thereby increasing the number of recent reports of parasite hybridization.
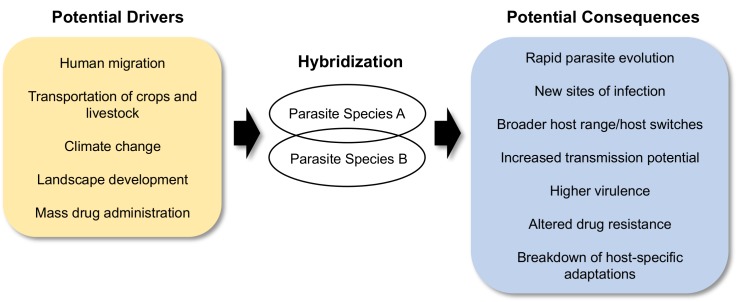
We outline what we believe will be the most important and/or potentially dangerous effects of hybridization in eukaryotic parasites: (1) the generation of novel and extreme infection phenotypes, (2) an increase in host range, being just one component of (3) an increase in transmission potential, (4) an increase in parasite evolutionary potential with consequences for host–parasite coevolution, (5) the breakdown of host-specific adaptations, and also (6) an altered response to drug therapy (Fig 1).
Fig 1. Schematic of the drivers and consequences of parasite hybridization.
As a result of new allelic combinations produced by hybridization, hybrid parasites may show enhanced phenotypic characteristics compared to the parents. Hybrids may be better at host exploitation, such that they may be more efficient at obtaining nutrition from the host, leading to higher fecundity or faster maturation time. Also, certain new hybrid parasite genotypes have been shown to be better at avoiding recognition and resistance from the host’s adaptive immune system, potentially leading to higher infectivity and unusual pathologies [29,62]. We thus predict that hybrid parasites are able to exploit novel resources and sites of infection within the host, which parental genotypes cannot utilize.
Hybrid parasites may be able to infect a greater range of host species [14,19,63]. While parental parasites are often restricted to one host species, hybrid parasites may be able to exploit both. Interspecific Schistosoma spp. hybrids, for instance, are able to infect both parental intermediate snail hosts (Box 1) [21]. In addition, just as some hybrids of nonparasitic taxa adapt to new ecological conditions and colonize novel environments that neither of the parents can survive [44,49], we would predict that hybrid parasites may be able to infect entirely new host species. As an indication that this indeed occurs in the wild, two plant pathogenic fungi (Phytophthora spp. infecting alder trees [64] and Zymoseptoria pseudotritici infecting a range of grasses [53]) and one amphibian fungal pathogen (a new lineage of Batrachochytrium dendrobatidis causing dramatic outbreaks of chytridiomycosis [65 ]), which all emerged via hybridization, have been found to live on host species neither of their parents are found on.
With increasing land use and current rates of anthropogenic change across both the environment and agricultural/livestock practices, we predict that recombinant parasite genotypes will be generated at an increasing rate. Encounters between previously isolated parasite species become more frequent when geographic and ecological barriers that previously existed between parasite species are lost. Taking together the potential impact of hybrid superiority and host range expansions (described in 2) and an increase in anthropogenically mediated parasite dispersal, the transmission of disease may become considerably facilitated [66,67]. This may lead to epidemics, threaten global food security, and endanger natural animal and plant populations. For instance, the increasing creation and alteration of water bodies for agriculture can lead to areas of overlap and co-occurrence of the different intermediate snail host species of Schistosoma parasites [66,68]. This generates ample opportunity for coinfections of the definitive host by different Schistosoma species, which may then hybridize. In addition, humans and livestock are drawn to the same water resources, where they can both release parasites and become infected, thus creating a “hotbed” for disease transmission between human and livestock.
Evolutionary theory predicts that elevated virulence and genetic diversity, both potential outcomes of parasite hybridization, can increase the evolutionary potential of parasites and alter the host–parasite coevolutionary process [69]. For instance, for Phytophthora, hybridization has led to a range of new species with particularly aggressive pathogenicity [64], and hybrid lineages of the amphibian zoosporic fungus Batrachochytrium, which cause dramatic outbreaks of chytridiomycosis, have been found to be hypervirulent to many hosts [65]. In the context of coevolutionary races, higher virulence of hybrid parasites may impose stronger selection for host resistance.
Hybridization in parasites could, however, also be maladaptive for the parasites because of fitness-reducing genetic incompatibilities (negative epistasis) and the breakdown of host-specific adaptations, potentially leading to reduced infectivity/virulence. While such outbreeding depression is well known from free-living organisms, studies on the fitness of hybrid parasites are scarce. Outbreeding depression has been reported in a snail–trematode host–parasite system (Potamopyrgus antipodarum–Microphallus sp.), in which hybrid parasites suffer from reduced infectivity in both parental host populations [70,71]. In such cases, we predict that hybrid parasites may be less likely to evade host immunity or they may fail to effectively colonize the correct organ or body site. Through recombination and segregation, hybrids may also lose beneficial alleles that the parents had acquired previously, such as those conferring drug resistance (see below). Hybrid parasite inferiority can also lead to unusual pathogensis, which may hamper diagnosis, particularly when two very different disease phenotype parasites are introgressed (Box 1). As such, hybridization between divergent parasite populations may both promote and alter pathogenesis, which has important implications on disease prevalence, pathology, and treatment.
Hybridization may have a wide range of effects on drug efficacy in parasites. On the one hand, zoonotic hybrids may exhibit enhanced susceptibility to drugs if resistance alleles circulating in the human host parasite population are swamped by introgression of drug-susceptible alleles from the animal host parasite population. Furthermore, if hybridization between human and animal parasites increases the host range from human host specificity to that of a large animal host range reservoir, this could act against the establishment and spread of drug resistance [32]. Alternatively, hybridization could potentiate adaptive evolution within certain parasites in response to drug treatment (Box 1). One example for this is Cryptococcus serotype hybrids that are resistant to an antifungal drug usually used to treat infections with the individual parental serotypes [20]. More evidence for the adaptive introgression of drug resistance genes from nonsusceptible strains or species comes from Haemonchus nematodes that gained resistance against the broad-spectrum drug ivermectin through hybridization [72,73] and the introgression of anticoagulant rodent poison resistance by hybridization between Old World mice [74].
Conclusions
Understanding and monitoring hybridization in animal and human parasites will be essential for optimizing and evaluating control strategies across potential hybrid zones. Relatively simple diagnostic procedures currently exist for detecting hybrids in some parasite groups (e.g., using a multilocus approach with mitochondrial cytochrome c oxidase subunit 1 (COX-1)/internal transcribed spacer (ITS) barcoding; [67]). However, the advancement of state-of-the-art genomic technologies will be helpful for more fine-scale detection of hybridization in parasites, to determine their evolutionary rates, and to identify patterns of cross transmission between host species (i.e., sources of transmission of zoonotic parasites to humans). Since hybrid parasites appear to be a particular problem in NTDs, there is a need to develop cheap, robust diagnostics appropriate to use in the field.
The circumstances producing increased opportunity for hybridization are the same ones that cause increased rates for disease distribution and transmission. Thus, the likelihood of parasite hybridization is increasing with the intensification of world trade of plants and animals, human migration, land use, and drug administration (Fig 1). Interspecific hybridization and introgression appear to be viable strategies for many parasites to maintain transmission, with potentially major impacts on the evolution of virulence, infection persistence, drug resistance, and host range, as well as on the clinical outcomes of disease. The combined effects of anthropogenic distribution and increased hybridization opportunities could have hazardous and catalytic effects on epidemiology, imposing further challenges and constraints for their effective control. More empirical work on the differential transmission, infectivity, drug efficacy, pathogenesis, and evolution of hybrid parasite populations is therefore urgently required to guide policies on their monitoring and management.
Funding Statement
This work was supported by a Royal Society Newton Fellowship (KCK), a Marie Curie Fellowship (RBS), and a European Research Council Starting Grant (MAB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Semenza JC, Menne B (2009) Climate change and infectious diseases in Europe. Lancet Infectious Diseases 9: 365–375. 10.1016/S1473-3099(09)70104-5 [DOI] [PubMed] [Google Scholar]
- 2. Nichols GL, Andersson Y, Lindgren E, Devaux I, Semenza JC (2014) European Monitoring Systems and Data for Assessing Environmental and Climate Impacts on Human Infectious Diseases. International Journal of Environmental Research and Public Health 11: 3894–3936. 10.3390/ijerph110403894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martens P, Kovats RS, Nijhof S, de Vries P, Livermore MTJ, et al. (1999) Climate change and future populations at risk of malaria. Global Environmental Change-Human and Policy Dimensions 9: S89–S107. [Google Scholar]
- 4. Lafferty KD (2009) The ecology of climate change and infectious diseases. Ecology 90: 888–900. [DOI] [PubMed] [Google Scholar]
- 5. Brooks DR, Hoberg EP (2007) How will global climate change affect parasite-host assemblages? Trends in Parasitology 23: 571–574. [DOI] [PubMed] [Google Scholar]
- 6. Patz JA, Graczyk TK, Geller N, Vittor AY (2000) Effects of environmental change on emerging parasitic diseases. International Journal for Parasitology 30: 1395–1405. [DOI] [PubMed] [Google Scholar]
- 7. Hales S, de Wet N, Maindonald J, Woodward A (2002) Potential effect of population and climate changes on global distribution of dengue fever: an empirical model. Lancet 360: 830–834. [DOI] [PubMed] [Google Scholar]
- 8. Astrom C, Rocklov J, Hales S, Beguin A, Louis V, et al. (2012) Potential Distribution of Dengue Fever Under Scenarios of Climate Change and Economic Development. Ecohealth 9: 448–454. 10.1007/s10393-012-0808-0 [DOI] [PubMed] [Google Scholar]
- 9. Moore S, Shrestha S, Tomlinson KW, Vuong H (2012) Predicting the effect of climate change on African trypanosomiasis: integrating epidemiology with parasite and vector biology. Journal of the Royal Society Interface 9: 817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nowak MA, May RM (1994) Superinfection and the evolution of parasite virulence. Proceedings of the Royal Society of London B: Biological Sciences 255: 81–89. [DOI] [PubMed] [Google Scholar]
- 11. Norton AJ, Webster JP, Kane R, Rollinson D (2008) Inter-specific parasite competition: mixed infections of Schistosoma mansoni and S. rodhaini in the definitive host. Parasitology 135: 1–12. [DOI] [PubMed] [Google Scholar]
- 12. Webster JP, Gower CM, Norton AJ (2008) Application of evolutionary concepts to predicting and evaluating the impact of mass-chemotherapy schistosomiasis control programmes. Evolutionary Applications 1: 66–83. 10.1111/j.1752-4571.2007.00012.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koukounari A, Donnelly CA, Sacko M, Keita A, Landoure A, et al. (2010) The impact of single versus mixed schistosome species infections on liver, spleen and bladder morbidity within Malian children pre- and post-praziquantel treatment. BMC Infectious Diseases 10: 227 10.1186/1471-2334-10-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Detwiler JT, Criscione CD (2010) An infectious topic in reticulate evolution: introgression and hybridization in animal parasites. Genes 210: 102–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baack EJ, Rieseberg LH (2007) A genomic view of introgression and hybrid speciation. Current Opinion in Genetics & Development 17: 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arnold M (2004) Natural hybridization and the evolution of domesticated, pest and disease organisms. Molecular Ecology 13: 997–1007. [DOI] [PubMed] [Google Scholar]
- 17. Mavarez J, Salazar CA, Bermingham E, Salcedo C, Jiggins CD, et al. (2006) Speciation by hybridization in Heliconius butterflies. Nature 441: 868–871. [DOI] [PubMed] [Google Scholar]
- 18. Hawks J, Cochran G (2006) Dynamics of adaptive introgression from archaic to modern humans. Paleoanthropology 2006: 101–115. [Google Scholar]
- 19. Arnold ML (2004) Natural hybridization and the evolution of domesticated, pest and disease organisms. Molecular Ecology 13: 997–1007. [DOI] [PubMed] [Google Scholar]
- 20. Li W, Averette AF, Desnos-Ollivier M, Ni M, Dromer F, et al. (2012) Genetic diversity and genomic plasticity of Cyptococcus neoformans AD hybrid strains. G3: Genes, Genomes, Genetics 2: 83–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Webster BL, Diaw OT, Seye MM, Webster JP, Rollinson D (2013) Introgressive hybridization of Schistosoma haematobium group species in Senegal: species barrier break down between ruminant and human schistosomes. PloS Neglected Tropical Diseases 7: e2110 10.1371/journal.pntd.0002110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Criscione CD, Anderson JD, Sudimack D, Peng W, Jha B, et al. (2007) Disentangling hybridization and host colonization in parasitic roundworms of humans and pigs. Proceedings of the Royal Society B-Biological Sciences 274: 2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Le TH, De NV, Agatsuma T, Thi Nguyen TG, Nguyen QD, et al. (2008) Human fascioliasis and the presence of hybrid/introgressed forms of Fasciola hepatica and Fasciola gigantica in Vietnam. International Journal for Parasitology 38: 725–730. [DOI] [PubMed] [Google Scholar]
- 24. Dunams-Morel DB, Reichard MV, Torretti L, Zarlenga DS, Rosenthal BM (2012) Discernible but limited introgression has occurred where Trichinella nativa and the T6 genotype occur in sympatry. Infection, Genetics and Evolution 12: 530–538. 10.1016/j.meegid.2012.01.004 [DOI] [PubMed] [Google Scholar]
- 25. Rogers MB, Downing T, Smith BA, Imamura H, Sanders M, et al. (2014) Genomic confirmation of hybridisation and recent inbreeding in a vector-isolated Leishmania population PLoS Genetics 10: e1004092 10.1371/journal.pgen.1004092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sturm NR, Vargas NS, Westenberger SJ, Zingales B, Campbell DA (2003) Evidence for multiple hybrid groups in Trypanosoma cruzi . International Journal for Parasitology 33: 269–279. [DOI] [PubMed] [Google Scholar]
- 27. Machado CA, Ayala FJ (2001) Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi . Proceedings of the National Academy of Sciences of the United States of America 98: 7396–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gaunt MW, Yeo M, Frame IA, Stothard JR, Carrasco HJ, et al. (2003) Mechanism of genetic exchange in American trypanosomes. Nature 421: 936–939. [DOI] [PubMed] [Google Scholar]
- 29. Grigg ME, Bonnefoy S, Hehl AB, Suzuki Y, Boothroyd JC (2001) Success and virulence in toxoplasma as the result of sexual recombination between two distinct ancestries. Science 294: 161–165. [DOI] [PubMed] [Google Scholar]
- 30. Akopyants NS, Kimblin N, Secundino N, Patrick R, Peters N, et al. (2009) Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science 324: 265–268. 10.1126/science.1169464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramiro RS, Khan SM, Franke-Faynard B, Janse CJ, Obbard DJ, et al. (2015) Hybridization and pre-zygotic reproductive barriers in Plasmodium. Proceedings of the Royal Society Biological Sciences Series B 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Webster JP, Molyneux D, Hotez PJ, Fenwick A (2014) The contribution of mass drug administration to global health—past, present and future. Philosophical Transactions of the Royal Society of London B Biological Sciences 369: 20130434 10.1098/rstb.2013.0434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arnold ML (2006) Evolution through genetic exchange; Press OU, editor. Oxford. [Google Scholar]
- 34. Dobzhansky T (1937) Genetic nature of species differences. American Naturalist 71: 404–420. [Google Scholar]
- 35. Müller HJ (1942) Isolating mechanisms, evolution and temperature. Biological Symposium 6: 71–125. [Google Scholar]
- 36. Coyne JA, Orr HA (2004) Speciation. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 37. Edmands S (2002) Does parental divergence predict reproductive compatibility? Trends in Ecology & Evolution 17: 520–527. [Google Scholar]
- 38. Lynch M (1991) The genetic interpretation of inbreeding depression and outbreeding depression. Evolution 45: 622–629. [DOI] [PubMed] [Google Scholar]
- 39. Barton NH (2001) The role of hybridization in evolution. Molecular Ecology 10: 551–568. [DOI] [PubMed] [Google Scholar]
- 40. Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, et al. (2013) Hybridization and speciation. Journal of Evolutionary Biology 26: 229–246. 10.1111/j.1420-9101.2012.02599.x [DOI] [PubMed] [Google Scholar]
- 41. Seehausen O (2004) Hybridization and adaptive radiation. Trends in Ecology & Evolution 19: 198–207. [DOI] [PubMed] [Google Scholar]
- 42. Krehenwinkel H, Tautz D (2013) Northern range expansion of European populations of the wasp spider Argiope bruennichi is associated with global warming correlated genetic admixture and population-specific temperature adaptations. Molecular Ecology 22: 2232–2248. 10.1111/mec.12223 [DOI] [PubMed] [Google Scholar]
- 43. Grant BR, Grant PR (2008) Fission and fusion of Darwin's finches populations. Philosophical Transactions of the Royal Society B-Biological Sciences 363: 2821–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, et al. (2003) Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301: 1211–1216. [DOI] [PubMed] [Google Scholar]
- 45. Mallet J (2007) Hybrid speciation. Nature 446: 279–283. [DOI] [PubMed] [Google Scholar]
- 46. Schumer M, Rosenthal GG, Andolfatto P (2014) How common is homoploid hybrid speciation? Evolution 68: 1553–1560. 10.1111/evo.12399 [DOI] [PubMed] [Google Scholar]
- 47. Dittrich-Reed DR, Fitzpatrick BM (2012) Transgressive hybrids as hopeful monsters. Evolutionary Biology 40: 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rieseberg LH, Archer MA, Wayne RK (1999) Transgressive segregation, adaptation and speciation. Heredity 83: 363–372. [DOI] [PubMed] [Google Scholar]
- 49. Stelkens RB, Brockhurst MA, Hurst GDD, Miller EL, Greig D (2014) The effect of hybrid transgression on environmental tolerance in experimental yeast crosses. Journal of Evolutionary Biology 27: 2507–2519. 10.1111/jeb.12494 [DOI] [PubMed] [Google Scholar]
- 50. Barrett RDH, Schluter D (2008) Adaptation from standing genetic variation. Trends in Ecology & Evolution 23: 38–44. [DOI] [PubMed] [Google Scholar]
- 51. Hedrick PW (2013) Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation. Molecular Ecology 22: 4606–4618. 10.1111/mec.12415 [DOI] [PubMed] [Google Scholar]
- 52. Stelkens RB, Brockhurst MA, Hurst GDD, Greig D (2014) Hybridization facilitates evolutionary rescue. Evolutionary Applications 7: 1209–1217. 10.1111/eva.12214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stukenbrock EH, Christiansen FB, Hansen TT, Dutheil JY, Schierup MH (2012) Fusion of two divergent fungal individuals led to the recent emergence of a unique widespread pathogen species. Proceedings of the National Academy of Sciences of the United States of America 109: 10954–10959. 10.1073/pnas.1201403109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Davies J (1996) Origins and evolution of antibiotic resistance. Microbiologia 12: 9–16. [PubMed] [Google Scholar]
- 55. Gyles C, Boerlin P (2014) Horizontally Transferred Genetic Elements and Their Role in Pathogenesis of Bacterial Disease. Veterinary Pathology Online 51: 328–340. [DOI] [PubMed] [Google Scholar]
- 56. Ochman H, Moran NA (2001) Genes lost and genes found: the molecular evolution of bacterial pathogenesis and symbiosis. Science 292: 1096–1098. [DOI] [PubMed] [Google Scholar]
- 57. Ochman H, Lawrence JG, Groisman EA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405: 299–304. [DOI] [PubMed] [Google Scholar]
- 58. Strauch E, Lurz R, Beutin L (2001) Characterization of a Shiga Toxin-Encoding Temperate Bacteriophage of Shigella sonnei . Infection and Immunity 69: 7588–7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gibbs MJ, Armstrong JS, Gibbs AJ (2001) Recombination in the hemagglutinin gene of the 1918 "Spanish flu". Science 293: 1842–1845. [DOI] [PubMed] [Google Scholar]
- 60. Laird AR, Ibarra V, Ruiz-Palacios G, Guerrero ML, Glass RI, et al. (2003) Unexpected detection of animal VP7 genes among common rotavirus strains isolated from children in Mexico. Journal of Clinical Microbiology 41: 4400–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Worobey M, Rambaut A, Holmes EC (1999) Widespread intra-serotype recombination in natural populations of dengue virus. Proceedings of the National Academy of Sciences of the United States of America 96: 7352–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schelkle B, Faria PJ, Johnson MB, van Oosterhout C, Cable J (2012) Mixed infections and hybridisation in monogenean parasites. Plos ONE 7: e39506 10.1371/journal.pone.0039506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Volf P, Benkova I, Myskova J, Sadlova J, Campino L, et al. (2007) Increased transmission potential of Leishmania major/Leishmania infantum hybrids. International Journal for Parasitology 37: 589–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brasier CM, Cooke DEL, Duncan JM (1999) Origin of a new Phytophthora pathogen through interspecific hybridization (vol 96, pg 5878, 1999). Proceedings of the National Academy of Sciences of the United States of America 96: 13589–13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Farrer RA, Weinert LA, Bielby J, Garner TWJ, Balloux F, et al. (2011) Multiple emergences of genetically diverse amphibian-infecting chytrids include a globalized hypervirulent recombinant lineage. Proceedings of the National Academy of Sciences 108: 18732–18736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tchuente LAT, Southgate VR, Njiokou F, Njine T, Kouemeni LE, et al. (1997) The evolution of schistosomiasis at Loum, Cameroon: replacement of Schistosoma intercalatum by S. haematobium through introgressive hybridization. Transactions of the Royal Society of Tropical Medicine and Hygiene 91: 664–665. [DOI] [PubMed] [Google Scholar]
- 67. Huyse T, Webster BL, Geldof S, Stothard JR, Diaw OT, et al. (2009) Birdirectional introgressive hybridization between a cattle and human schistosome species. PLoS Pathogens 5: e1000571 10.1371/journal.ppat.1000571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Webster BL, Tchuente LAT, Southgate VR (2007) A single-strand conformation polymorphism (SSCP) approach for investigating genetic interactions of Schistosoma haematobium and Schistosoma guineensis in Loum, Cameroon. Parasitology Research 100: 739–745. [DOI] [PubMed] [Google Scholar]
- 69. Lively CM (1999) Migration, virulence, and the geographic mosaic of adaptation by parasites. American Naturalist 153: S34–S47. [DOI] [PubMed] [Google Scholar]
- 70. Dybdahl MF, Jokela J, Delph LF, Koskella B, Lively CM (2008) Hybrid fitness in a locally adapted parasite. American Naturalist 172: 772–782. 10.1086/592866 [DOI] [PubMed] [Google Scholar]
- 71. Lively CM, Dybdahl MF (2000) Parasite adaptation to locally common host genotypes. Nature 405: 679–681. [DOI] [PubMed] [Google Scholar]
- 72. Redman E, Sargison N, Whitelaw F, Jackson F, Morrison A, et al. (2012) Introgression of Ivermectin resistance genes into a susceptible Haemonchus contortus strain by multiple backcrossing. PloS Pathogens 8: e1002534 10.1371/journal.ppat.1002534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chaudhry U, Redman E, Abbas M, Muthusamy R, Ashraf K, et al. (2015) Genetic evidence for hybridisation between Haemonchus contortus and Haemonchus placei in natural field populations and its implications for interspecies transmission of anthelmintic resistance. International Journal for Parasitology 45: 149–159. 10.1016/j.ijpara.2014.09.002 [DOI] [PubMed] [Google Scholar]
- 74. Song Y, Endepols S, Klemann N, Richter D, Matuschka F-R, et al. (2011) Introgression of anticoagulant rodent poison resistance by hybridization between old world mice. Current Biology 21: 1296–1301. 10.1016/j.cub.2011.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Alves W (1948) Transactions of the Royal Society of Tropical Medicine and Hygiene, 41: 430–439. [Google Scholar]
- 76.Bremond P, Campagne G, Sellin B, Labbo R, Garba A, et al. (1996) Les schistosomes anthropophiles et zoophiles au Niger et leur impact sur la santé publique: détermination du risque réel de contamination et de pathogénicité pour les populations humaines. Rapport CERMES 6/96 document: 22 pages.
- 77. Rollinson D, Southgate VR, Vercruysse J, Moore PJ (1990) Observations on natural and experimental interactions between Schistosoma bovis and Schistosoma curassoni from West-Africa. Acta Tropica 47: 101–114. [DOI] [PubMed] [Google Scholar]
- 78. Taylor MG (1970) Hybridisation experiments on five species of african schistosomes. Journal of Helminthology 17: 253–314. [DOI] [PubMed] [Google Scholar]
- 79. Webster BL, Diaw OT, Seye MM, Webster JP, Rollinson D (2013) Introgressive hybridization of Schistosoma haematobium group species in Senegal: species barrier break down between ruminant and human schistosomes. PLoS Neglected Tropical Diseases 7: e2110 10.1371/journal.pntd.0002110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Steinauer ML, Hanelt B, Mwangi IN, Maina GM, Agola LE, et al. (2008) Introgressive hybridization of human and rodent schistosome parasites in western Kenya. Molecular Ecology 17: 5062–5064. 10.1111/j.1365-294X.2008.03957.x [DOI] [PubMed] [Google Scholar]
- 81. Huyse T, Van den Broeck F, Hellemans B, Volckaert FAM, Polman K (2013) Hybridisation between the two major African schistosome species of humans. International Journal of Parasitology 43: 687–689. 10.1016/j.ijpara.2013.04.001 [DOI] [PubMed] [Google Scholar]
- 82. Berry A, Moné H, Iriart X, Mouahid G, Abbo O, et al. (2014) Schistosomiasis haematobium, Corsica, France [letter]. Emerging Infectious Diseases 20: 1595–1597. 10.3201/eid2009.140928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.ECDC (2014) Rapid risk assessment: Local transmission of Schistosoma haematobium in Corsica, France– 16 May 2014. Stockholm.
- 84. Boissier J, Moné H, Mitta G, Bargues MD, Molyneux DH, et al. (2015) Schistosomiasis reaches Europe. Lancet Infectious Diseases 15: 757–758. 10.1016/S1473-3099(15)00084-5 [DOI] [PubMed] [Google Scholar]
- 85. Messenger LA, Garcia L, Vanhove M, Huaranca C, Bustamante M, et al. (2015) Ecological host fitting of Trypanosoma cruzi TcI in Bolivia: mosaic population structure, hybridization and a role for humans in Andean parasite dispersal. Molecular Ecology 24: 2406–2422. 10.1111/mec.13186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Messenger LA, Llewellyn MS, Bhattacharyya T, Franzén O, Lewis MD, et al. (2012) Multiple mitochondrial introgression events and heteroplasmy in Trypanosoma cruzi revealed by Maxicircle MLST and Next Generation Sequencing. PLoS Neglected Tropical Diseases 6: e1584 10.1371/journal.pntd.0001584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Miles MA, Llewellyn MS, Lewis MD, al. e (2009) The molecular epidemiology and phylogeography of Trypanosoma cruzi and parallel research on Leishmania: looking back and to the future. Parasitology 136: 1509–1528. 10.1017/S0031182009990977 [DOI] [PubMed] [Google Scholar]
- 88. Goodhead I, Capewell P, Bailey JW, Beament T, Chance M, et al. (2013) Whole-genome sequencing of Trypanosoma brucei reveals introgression between subspecies that is associated with virulence. mBio 4: e00197–13. 10.1128/mBio.00197-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.WHO (2014) Global Burden of Disease Estimates for 2000–2012.
- 90. Kelly JM, Law JM, Chapman CJ, Van Eys GJ, Evans DA (1991) Evidence of genetic recombination in Leishmania. Molecular and Biochemical Parasitology 46: 253–263. [DOI] [PubMed] [Google Scholar]
- 91. Inbar E, Akopyants NS, Charmoy M, Romano A, Lawyer P, et al. (2013) The Mating Competence of Geographically Diverse Leishmania major Strains in Their Natural and Unnatural Sand Fly Vectors. PLoS Genetics 9: e1003672 10.1371/journal.pgen.1003672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Rogers MB, Downing T, Smith BA, Imamura H, Sanders M, et al. (2014) Genomic confirmation of hybridisation and recent Inbreeding in a vector-isolated Leishmania population. PLoS Genetics 10: e1004092 10.1371/journal.pgen.1004092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Volf P, Benkova I, Myskova J, Sadlova J, Campino L, et al. (2007) Increased transmission potential of Leishmania major/Leishmania infantum hybrids. Intenational Journal for Parasitology 37: 589–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, et al. (2015) Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347: 1258524 10.1126/science.1258524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.World Health Organization. Malaria 2015 [updated 2015; cited 2015 23 June]. http://www.who.int/malaria/en/.
- 96. Manguin S, Carnivale P, Mouchet J (2008) Biodiversity of Malaria in the World; John Libbey Eurotext, editor. Paris, France: [Google Scholar]
- 97. Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, et al. (2015) Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science 347: 1258522 10.1126/science.1258522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lee Y, Marsden CD, Nieman C, Lanzaro GC (2014) A new multiplex SNP genotyping assay for detecting hybridization and introgression between the M and S molecular forms of Anopheles gambiae . Molecular Ecology Resources 14: 297–305. 10.1111/1755-0998.12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mawejje HD, Wilding CS, Rippon EJ, Hughes A, Weetman D, et al. (2013) Insecticide resistance monitoring of field-collected Anopheles gambiae s.l. populations from Jinja, eastern Uganda, identifies high levels of pyrethroid resistance. Medical and Veterinary Entomology 27: 276–283. 10.1111/j.1365-2915.2012.01055.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wondji CS, Coleman M, Kleinschmidt I, Mzilahowa T, Irving H, et al. (2012) Impact of pyrethroid resistance on operational malaria control in Malawi. Proceedings of the National Academy of Sciences 109: 19063–19070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Clarkson CS, Weetman D, Essandoh J, Yawson AE, Maslen G, et al. (2014) Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nat Commun 5: 4248 10.1038/ncomms5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Norris LC, Main BJ, Lee Y, Collier TC, Fofana A, et al. (2015) Adaptive introgression in an African malaria mosquito coincident with the increased usage of insecticide-treated bed nets. Proceedings of the National Academy of Sciences 112: 815–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Weetman D, Steen K, Rippon E, Mawejje H, Donnelly M, et al. (2014) Contemporary gene flow between wild An. gambiae s.s. and An. arabiensis . Parasites & Vectors 7: 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Choochote W, Min G-S, Intapan P, Tantrawatpan C, Saeung A, et al. (2014) Evidence to support natural hybridization between Anopheles sinensis and Anopheles kleini (Diptera: Culicidae): possibly a significant mechanism for gene introgression in sympatric populations. Parasites & Vectors 7: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Menge D, Guda T, Zhong D, Pai A, Zhou G, et al. (2005) Fitness consequences of Anopheles gambiae population hybridization. Malaria Journal 4: 44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Mancini E, Spinaci MI, Gordicho V, Caputo B, Pombi M, et al. (2015) Adaptive Potential of Hybridization among Malaria Vectors: Introgression at the Immune Locus TEP1 between Anopheles coluzzii and A. gambiae in ‘Far-West’ Africa. PLoS ONE 10: e0127804 10.1371/journal.pone.0127804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Pates HV, Curtis CF, Takken W (2014) Hybridization studies to modify the host preference of Anopheles gambiae . Medical and Veterinary Entomology 28: 68–74. 10.1111/mve.12070 [DOI] [PubMed] [Google Scholar]