Abstract
MicroRNAs (miRNAs) have emerged as promising diagnostic biomarkers. We introduce a kinetic fingerprinting approach called Single Molecule Recognition through Equilibrium Poisson Sampling (SiMREPS) for the amplification-free counting of single unlabeled miRNA molecules, which circumvents thermodynamic limits of specificity and virtually eliminates false positives. We demonstrate high-confidence single-molecule detection of synthetic and endogenous miRNAs in both buffer and minimally treated biological liquids, as well as >500-fold discrimination between single nucleotide polymorphisms.
Stable, diagnostically useful miRNAs have recently been detected in blood and other body fluids, but reproducible quantification of circulating miRNAs has proven challenging1. Standard assays based on amplification by polymerase chain reaction (PCR), although highly sensitive, require time-consuming extraction and amplification steps. Next-generation sequencing approaches enable high-throughput profiling of RNA transcripts, but cannot reliably quantify low-abundance analytes (see Supplementary Note 1). Although a number of sensitive, amplification-free nucleic acid assays have been reported2–5, these typically suffer from significant false positives and/or strict limits on target specificity imposed by the thermodynamics of hybridization6 (see Supplementary Note 1).
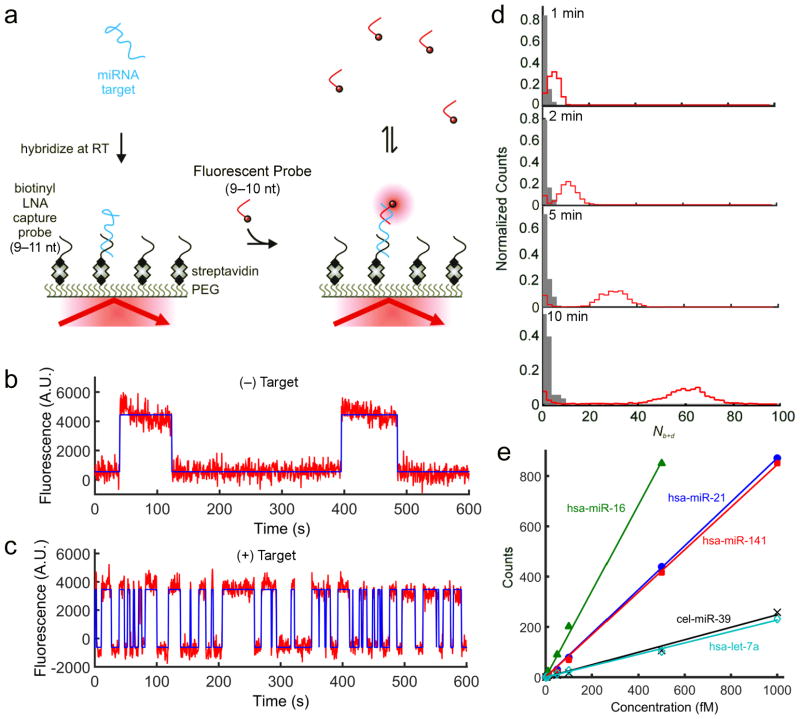
Here we present a technique for the amplification-free single-molecule detection of unlabeled RNA biomarkers that circumvents many of the above issues. The approach, which we call Single-Molecule Recognition through Equilibrium Poisson Sampling (SiMREPS), is inspired by the super-resolution imaging technique DNA-PAINT7 and exploits the direct binding of a short (9- to 10-nucleotide, nt) fluorescently labeled DNA probe to an unlabeled miRNA analyte immobilized on a glass surface (Fig. 1a). Using TIRF microscopy8,9, both specific binding to the immobilized target and non-specific surface binding are detected (Supplementary Fig. 1). However, the equilibrium binding of the probe to the target yields a distinctive kinetic signature, or fingerprint, that can be used to achieve ultra-high discrimination against background binding (Fig. 1b,c). Because the kinetics of exchange for probes of ~6–12 nt are highly sensitive to the number of complementary bases between the probe and target7,10,11, varying the length of the probe allows fine-tuning of the kinetic behavior to improve specificity of detection. For the probes used in this study, kinetics of binding and dissociation were found to be more closely correlated to probe length than to the melting temperature of the duplex (Supplementary Fig. 2).
Figure 1. High-confidence detection of miRNAs with SiMREPS.
a, Experimental approach of SiMREPS detection of miRNAs. An immobilized miRNA target is identified by fluorescence microscopy as the site of repeated transient binding by short fluorescent DNA probes. b,c, Binding of probes to the slide surface (b) exhibits kinetic behavior distinct from that of binding to a target molecule (c) (red curve = fluorescence intensity, blue curve = idealization from hidden Markov modeling). d, Histograms of the number of candidate molecules showing a given number of intensity transitions (Nb+d) in the absence (gray) or presence (red) of 1 pM miR-141 with varying acquisition time. e, Standard curves from SiMREPS assays of five miRNAs. Linear fits were constrained to a y-intercept of 0, yielding R2 values > 0.99.
As the transient binding of probes to an immobilized target can be idealized as a Poisson process, the standard deviation in the number of binding and dissociation events (Nb+d) is expected to increase only as , implying that the observation time can be lengthened to achieve arbitrarily high discrimination between target and off-target binding (see Supplementary Note 2). Consistent with this expectation, as the experimental acquisition time is increased, the signal and background peaks in histograms of Nb+d are progressively better resolved (Fig. 1d), and the width of the signal distribution increases only as (Supplementary Fig. 3). Note that the choice of probe length is critical to achieve this separation on convenient experimental time scales (Supplementary Fig. 4).
To test the generality of SiMREPS, we evaluated four human miRNAs that are dysregulated in cancer and other diseases12–14: hsa-let-7a, hsa-miR-21, hsa-miR-16, and hsa-miR-141; and one non-human miRNA from C. elegans: cel-miR-39 (Supplementary Fig. 5). Although the binding kinetics varied among the target-probe pairs, the signal and background peaks were well-separated for all targets (Supplementary Fig. 5b); by stipulating a threshold of Nb+d ≥ 15, empirically perfect discrimination (specificity = 1) was achieved (Supplementary Fig. 5e). Standard curves constructed using this threshold for the five miRNAs show a linear dependence on target concentration over 2–3 orders of magnitude (Fig. 1e).
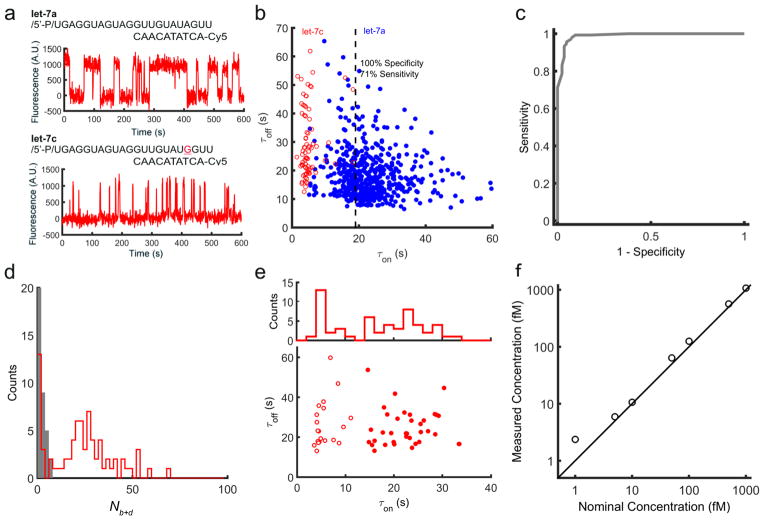
Because the lifetime of a short DNA duplex increases as an approximately exponential function of the number of base pairs7,10,11, we reasoned that SiMREPS might be used to achieve excellent single-base discrimination. To test this hypothesis, we used a single fluorescent probe to discriminate between two let-7 family members, hsa-let-7a and hsa-let-7c. We found that the lifetime of the probe-bound state τon differed by a factor of ~4.7 for the two targets, whereas the unbound-state lifetime τoff showed no target dependence (Fig. 2a,b). Photobleaching is much slower than probe dissociation under our illumination conditions (Supplementary Fig. 6). With the standard acquisition time of 10 min, let-7a and let-7c could be distinguished at the single-copy level with a discrimination factor > 100 at > 96% sensitivity, or with a discrimination factor > 570 (beyond the limit of quantification in this experiment) at ~70% sensitivity (Fig. 2b,c, Supplementary Fig. 7). Not only is this substantially larger than the typical discrimination factors of 2–100 reported for single mismatches using other hybridization-based probes6,2,15,4, but as SiMREPS achieves discrimination at the single-molecule level, it is possible to independently quantify a target and a point mutant with high confidence in a mixture containing both species.
Figure 2. Single-molecule mismatch discrimination and detection of RNAs in crude biological matrices.
a, The fluorescent probe for let-7a exhibits long lifetimes of binding to let-7a (τon = 23.3 ± 8.3 s) but much more transient binding to let-7c (τon = 4.7 ± 3.0 s) due to a single mismatch. b, Dwell time analysis enables high-confidence single-copy-level discrimination between let-7a and let-7c. c, Receiver operating characteristic (ROC) plot constructed by varying the τon threshold for discriminating between let-7a and let-7c. d, Nb+d histogram for the detection of let-7 in HeLa cell extract in the presence or absence of the miRCURY let-7 inhibitor. e, Dwell times for molecules detected in HeLa extract using the fluorescent and capture probes for let-7a. The filled and open circles represent two clusters of target molecules classified by k-means clustering of τon values. f, Quantification of synthetic miR-141 spiked into human serum together with proteinase K and SDS.
The high sensitivity and specificity of SiMREPS suggest that it may be capable of high-confidence RNA detection in complex biological matrices. To test this notion, we used SiMREPS to detect hsa-let-7a in HeLa whole-cell extract treated briefly with 1.67 % (w/v) SDS. The Nb+d histogram for endogenous hsa-let-7a showed a well-defined peak (Fig. 2d) similar to that observed for synthetic hsa-let-7a (Supplementary Fig. 5a) and hsa-let-7c (Supplementary Fig. 7) that vanished in the presence of an LNA let-7 inhibitor designed to bind and sequester let-7 family members. Notably, dwell time analysis of the fluorescent probe binding events yielded two populations of molecules that were readily resolved by k-means clustering of the τon values (Fig. 2e) and are consistent with the expected τon distributions for hsa-let-7a and hsa-let-7c.
To investigate whether SiMREPS can detect miRNAs of clinical interest in minimally treated biofluids, the assay for the prostate cancer biomarker hsa-miR-14116 was conducted in a serum sample from a healthy individual after spiking in varying concentrations of synthetic hsa-miR-141 together with 2% (w/v) SDS and 0.16 U/μL proteinase K to minimize degradation of the synthetic miRNA (Supplementary Fig. 8). The measured concentration (calibrated from the standard curve collected in buffer, Fig. 1e) was strongly correlated with the nominal spiked-in concentration (Fig. 2f, R > 0.999, slope = 1.07). Consistent with the expected low concentration (0.1–5 fM) of miR-141 in the serum of healthy individuals16, we measured a concentration of 0.4 ± 0.5 fM (s.e.m., n = 3) in this serum specimen in the absence of spiked-in synthetic miR-141.
We have presented a method for the rapid, high-confidence, direct detection and quantification of specific nucleic acid biomarkers with transiently binding probes. SiMREPS lends itself to miniaturization, multiplexing and extension to non-TIRF-based detection approaches17,18, so we expect it to find broad application in both clinical diagnostics and research.
Online Methods
Oligonucleotides
All miRNA samples were purchased from Integrated DNA Technologies with a 5′-phosphate modification and HPLC purification, except for let-7a and let-7c, which were purchased from Dharmacon with a 5′-phosphate modification, deprotected according to the supplier’s instructions, and purified by reverse-phase C18 HPLC (Varian ProStar 210, Waters SunFire C18). All DNA probes were purchased from IDT and HPLC-purified by the manufacturer. LNA capture probes were purchased from Exiqon with HPLC purification.
Single-molecule fluorescence microscopy
SiMREPS experiments were performed using either a previously described prism-type TIRF microscope19 (HeLa extract experiments) or a Olympus IX-81 objective-type TIRF microscope equipped with a 60× oil-immersion objective (APON 60XOTIRFM, 1.49NA) as well as Cell^TIRF and z-drift control modules (all other experiments). For prism-type TIRF experiments, fluidic sample cells were constructed using two pieces of double-sided tape sandwiched between a quartz slide and glass coverslip as previously described19 (Supplementary Fig. 10a). For objective-type TIRF measurements, sample cells were constructed by fixing a cut 1-cm length of a pipet tip (Eppendorf) to a coverslip using epoxy adhesive (Double Bubble, Hardman Adhesives; Supplementary Fig. 10b). In either case, imaging surface (quartz slide or coverslip) was coated with a 1:10 mixture of biotin-PEG-5000 and mPEG-5000 (Laysan Bio, Inc.) immediately prior to construction of the sample cell as previously described20. Prepared slides were stored in the dark for up to two weeks.
SiMREPS quantification of synthetic miRNAs
Quantification of synthetic miRNA targets by SiMREPS was performed as follows. All miRNA handling was performed in GeneMate low-adhesion 1.7-mL microcentrifuge tubes, and dilutions for standard curves were performed in the presence of 0.03 mg/mL oligo(dT10) (Integrated DNA Technologies) as a carrier. The slide surface was briefly incubated with T50 buffer (10 mM Tris-HCl, 1 mm EDTA, pH 8.0) followed by 1 mg/mL streptavidin. After 10 min, excess streptavidin was flushed out by 3 volumes of T50. The surface was then incubated with 20 nM of the appropriate biotinylated LNA capture probe (Exiqon, Inc.) in 1× PBS buffer for 10 min, and the excess flushed out by 3 volumes of 1× PBS. A 100-μL portion of target RNA (hsa-let-7a, hsa-miR-16, hsa-miR-21, cel-miR-39, or hsa-miR-141) was introduced into the sample chamber and incubated for 10 min (prism-type TIRF) or 60 min (objective-type TIRF). The longer incubation time for the objective-type TIRF measurements was necessary because of the tall (~1 cm) sample cell, which slowed the transport of analyte to the imaging surface. An imaging buffer containing 4× PBS, 2.5 mM 3,4-dihydroxybenzoate, 25 nM protocatechuate dioxygenase, 1 mM Trolox21, and 25 nM of the Cy3- or Cy5-labeled fluorescent probe was added to the sample chamber. The transient binding of probes to captured target molecules was monitored for 10 min under illumination by 532 nm and/or 640 nm laser light. Image acquisition was performed at a rate of 2 Hz using an iCCD (iPentamax:HQ Gen III, Roper Scientific, MCP gain 70) for measurements of let-7 in HeLa extract, and an EMCCD (IXon 897, Andor, EM gain 1000) for all other measurements.
Detection of endogenous let-7 in crude HeLa extract
A 5-μL aliquot of HeLa whole cell extract (from Thermo Scientific In-Vitro Protein Expression Kit, # 88881) was incubated for 5 minutes at room temperature in the presence of 0 or 1.7% (w/v) sodium dodecyl sulfate (SDS) and 0 or 140 nM miRCURY let-7 inhibitor (Exiqon). The lysate was vortexed, diluted 100-fold in 4× PBS imaging buffer containing 25 nM of the fluorescent probe for hsa-let-7a, and added to a microscope slide coated with an excess of the LNA capture probe. After a10-min incubation, the transient binding of the fluorescent probe was observed by prism-type TIRF microscopy for 10 min as described above.
Detection of synthetic miR-141 spiked into crude serum
50 uL of freshly thawed human serum (BioreclamationIVT, #BRH844152) was combined with SDS (final 2% w/v), proteinase K (New England BioLabs, Inc., P8107S; final concentration 0.16 units/μl), and synthetic hsa-miR-141, and incubated for 15 min at room temperature. Next, EDTA was added to a final concentration of 20 mM, and the sample heated to 90 °C in a copper bath for 2 min. After cooling to room temperature for 5 min, each sample was allowed to bind to the microscope coverslip surface for 1 h. Residual serum was removed, the surface washed with 1× PBS, and imaging carried out by objective-type TIRF microscopy as described above.
Analysis of SiMREPS data
Custom MATLAB code was used to identify sites of fluorescent probe binding and calculate intensity-versus-time trajectories from the CCD movie. Intensity trajectories were subjected to hidden Markov modeling (HMM) using QuB22 in order to identify the number of binding and/or dissociation events (Nb+d) and mean dwell times in the bound (τon) and unbound (τoff) states for each candidate molecule. Based on control measurements in absence of target, a threshold of Nb+d = 15 (6 standard deviations above background) was used to identify target molecules. Additional filtering criteria were used to reject spurious transitions detected by the HMM software: to be counted as a target molecule, a candidate must (1) have a mean bound-state intensity signal at least 2.5 standard deviations and 1000 counts above the mean background intensity, and (2) exhibit median values of τon and τoff of at least 4 s. In the case of let-7a and let-7c discrimination, an intensity threshold of 500 counts was used instead, and criterion (2) was not applied since it would have eliminated a large fraction of let-7c molecules from analysis.
Kinetic Monte Carlo simulations of probe binding
SiMREPS probing of an immobilized target was simulated in MATLAB with the assumption of (pseudo-)first-order kinetics for both binding and dissociation. A random-number generator was used to make stochastic decisions as to whether a molecule will undergo a transition within discrete time steps of 0.5 s. The matrix of per-frame transition probabilities was generated from specified first-order rate constants. The number of transitions was recorded for each molecule for subsequent analysis.
PAGE assay of miRNA degradation in human serum
Synthetic miR-16 (final concentration 1 μM) was combined with sodium dodecyl sulfate (SDS, final concentration 0 or 2% w/v), proteinase K (New England Biolabs, final concentration 0, 0.08, 0.16, or 0.32 units/μL), and deionized water, then mixed with either PBS, pH 7.4 (final concentration 1×, Gibco) or human serum (final 50% v/v BioreclamationIVT) to a total volume of 20 μL. After 15 min incubation, samples were either left at room temperature or spiked with 2 μL EDTA (final concentration 20 mM) and heated to 90 °C for 2 min in a copper bath, then brought back to room temperature. Samples were incubated for 1 h more at room temperature, then separated on a denaturing, 8 M urea, 20% (w/v) polyacrylamide gel. The gel was stained using SYBR gold (Life Technologies) and imaged on a Typhoon 9410 Variable Mode Imager (GE Healthcare Life Sciences).
Supplementary Material
Acknowledgments
This work was funded in part by the Department of Defense MURI Award W911NF-12-1-0420 (to N.G.W.) and US National Institutes of Health Transformative R01 grant R01DK085714 (to M.T.). X.S. acknowledges support from the China Scholarship Council. M.D.G. acknowledges initial support from a Rio Hortega Fellowship and later from a Martin Escudero Fellowship. The authors thank A. M. Chinnaiyan, M. Bitzer, A. Sahu, S. Pitchiaya, and L. A. Heinicke for helpful discussions.
Footnotes
Author contributions
N.G.W. and A.J.B. conceived the idea. A.J.B., N.G.W., and X.S. designed the experiments. X.S. and A.J.B. carried out experiments and analyzed the results. A.J.B., N.G.W., X.S., M.T., M.D.G., and M.Z. interpreted the results and wrote the paper.
Competing financial interests
The University of Michigan has filed a provisional patent (application serial number 14/589,467) on technologies described herein.
References
- 1.Schwarzenbach H, Hoon DSB, Pantel K. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 2.Gunnarsson A, Jonsson P, Marie R, Tegenfeldt JO, Hook F. Nano Lett. 2008;8:183–188. doi: 10.1021/nl072401j. [DOI] [PubMed] [Google Scholar]
- 3.Geiss GK, et al. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 4.Li L, Li X, Li L, Wang J, Jin W. Anal Chim Acta. 2011;685:52–57. doi: 10.1016/j.aca.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Ho SL, Chan HM, Ha AWY, Wong RNS, Li HW. Anal Chem. 2014;86:9880–9886. doi: 10.1021/ac5025182. [DOI] [PubMed] [Google Scholar]
- 6.Zhang DY, Chen SX, Yin P. Nat Chem. 2012;4:208–214. doi: 10.1038/nchem.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jungmann R, et al. Nano Lett. 2010;10:4756–4761. doi: 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- 8.Axelrod D, Burghardt TP, Thompson NL. Annu Rev Biophys Bioeng. 1984;13:247–268. doi: 10.1146/annurev.bb.13.060184.001335. [DOI] [PubMed] [Google Scholar]
- 9.Walter NG, Huang CY, Manzo AJ, Sobhy MA. Nat Methods. 2008;5:475–489. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cisse II, Kim H, Ha T. Nat Struct Mol Biol. 2012;19:623–627. doi: 10.1038/nsmb.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupuis NF, Holmstrom ED, Nesbitt DJ. Biophys J. 2013;105:756–766. doi: 10.1016/j.bpj.2013.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu M, et al. PLoS ONE. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruepp A, et al. Genome Biol. 2010;11:R6. doi: 10.1186/gb-2010-11-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russo F, et al. PLoS ONE. 2012;7:e47786. doi: 10.1371/journal.pone.0047786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Schwarz G, Santiago JG. Angew Chem Int Ed. 2013;52:11534–11537. doi: 10.1002/anie.201305875. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell PS, et al. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sorgenfrei S, et al. Nat Nanotechnol. 2011;6:126–132. doi: 10.1038/nnano.2010.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baaske MD, Foreman MR, Vollmer F. Nat Nanotechnol. 2014;9:933–939. doi: 10.1038/nnano.2014.180. [DOI] [PubMed] [Google Scholar]
- 19.Michelotti N, de Silva C, Johnson-Buck AE, Manzo AJ, Walter NG. Methods Enzymol. 2010;475:121–148. doi: 10.1016/S0076-6879(10)75006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abelson J, et al. Nat Struct Mol Biol. 2010;17:504–512. doi: 10.1038/nsmb.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aitken CE, Marshall RA, Puglisi JD. Biophys J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicolai C, Sachs F. Biophys Rev Lett. 2013;08:191–211. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.