Abstract
CYP2J2 epoxygenase is an extrahepatic, membrane bound cytochrome P450 (CYP) that is primarily found in the heart and mediates endogenous fatty acid metabolism. CYP2J2 interacts with membranes through an N-terminal anchor and various non-contiguous hydrophobic residues. The molecular details of the motifs that mediate membrane interactions are complex and not fully understood. To gain better insights of these complex protein-lipid interactions, we employed molecular dynamics (MD) simulations using a highly mobile membrane mimetic (HMMM) model that enabled multiple independent spontaneous membrane binding events to be captured. Simulations revealed that CYP2J2 engages with the membrane at the F-G loop through hydrophobic residues Trp-235, Ille-236, and Phe-239. To explore the role of these residues, three F-G loop mutants were modeled from the truncated CYP2J2 construct (Δ34) which included Δ34-I236D, Δ34-F239H and Δ34-I236D/F239H. Using the HMMM coordinates of CYP2J2 the simulations were extended to a full POPC membrane which showed a significant decrease in the depth of insertion for each of the F-G loop mutants. The CYP2J2 F-G loop mutants were expressed in E. coli and were shown to be localized to the cytosolic fraction at a greater percentage relative to construct Δ34. Notably, the functional data demonstrated that the double mutant, Δ34-I236D/F239H, maintained native-like enzymatic activity. The membrane insertion characteristics were examined by monitoring CYP2J2 Trp-quenching fluorescence spectroscopy upon binding pyrene phospholipids containing nanodiscs. Relative to the Δ34 construct, the F-G loop mutants exhibited lower Trp quenching and membrane insertion. Taken together, the results suggest that the mutants exhibit a different membrane topology in agreement with the MD simulations and provide important evidence towards the involvement of key residues in the F-G loop of CYP2J2.
Graphical Abstract
INTRODUCTION
The cytochrome P450 (CYP) superfamily is a class of heme-containing enzymes that are widespread throughout all kingdoms of life (1). In humans, CYPs mediate the biotransformation of both xenobiotics and endogenous substrates for homeostasis (2). Eukaryotic CYPs interact with membrane bilayers through an N-terminal anchor as well as various non-contiguous hydrophobic residues that constitute the monofacial membrane-binding domain (3, 4).
The CYP interaction with the membrane bilayer is a complex phenomenon that is an active area of both experimental and computational research. While seminal studies correctly identified the general nature of the protein-membrane binding and topology (5), more recent examples have elucidated the interactions at the molecular and atomistic levels (6-8). Notably, P450 interactions with membranes have been shown to regulate ligand binding mechanisms (9), membrane depth of insertion (10), redox potentials (11), enzyme stability (12), structure and orientation (6). Additionally, advanced computational approaches have revealed the dynamic nature of CYPs and how their hydrophobic motifs interact with the membrane bilayer (6, 8). Both experimental and computational approaches have revealed that CYPs are inserted within the membrane, with a partially imbedded active site and deeply immersed N-terminus and F-G loop (13, 14).
CYP2J2 is an extrahepatic human CYP that catalyzes the epoxidation of both omega-3 and omega-6 polyunsaturated fatty acids into a number of biologically active metabolites (15). Importantly, CYP2J2 is highly expressed in the myocardium and surrounding aortic epithelium where it has been shown to be a key regulator of cardiovascular homeostasis (16). The metabolism of arachidonic acid, an omega-6 polyunsaturated fatty acid, by CYP2J2 produces 8 unique regio- and stereoisomers known as epoxyeicostrienoic acids (EETs) that are collectively characterized as anti-inflammatory and vasodilatory lipid mediators (17).
CYP2J2 is anchored within the endoplasmic reticulum (2). The presence of the hydrophobic membrane binding domains of CYP2J2 complicates its use in many biochemical assays due to its inherent tendency to aggregate outside of the membrane environment. The disruption of the protein-membrane interactions affords solubility and enables structural elucidation through methods such as nuclear magnetic resonance (NMR) and high resolution x-ray crystallography. Currently, there are no reports of the successful solubilization and subsequent crystallization of any member of the CYP2J subfamily. However, this was achieved for other CYP2 family members including CYP2C5, CYP2A6, CYP2C8, CYP2C9 and CYP2D6 (4, 18-21). Specifically, soluble isozymes were engineered through a combination of deletions and hydrophilic substitutions of targeted membrane binding regions that resulted in increased expression yields and protein homogeneity. For example, the solubility of CYP2C5 was increased by substituting the N-terminus (Δ28) with MAKKTSSKG, adding a four-residue histidine tag at the C-terminus and incorporating five substitutions at the membrane binding interface with the mutations N202H, R206E, I207L, S209G, and S210T (4). Similarly, CYP2C9 N-terminal (Δ29) residues were replaced by MAKKTSSKGR, a four-histidine carboxy-tag and the seven amino acid mutations (K206E, I215V, C216Y, S220P, P221A, I222L and I223L) in the F-G loop (18). Soon after, CYP2D6 was successfully crystallized by truncating the N-terminus, adding a four-histidine tag at the C-terminus and mutating the F-G loop Leu-230 and Lue-231 with a combination of hydrophilic residues (Asn, Lys, His, Gln, Arg, and Ser) (21). Importantly, these changes afforded some of the first structural insights of mammalian P450s through high resolution x-ray crystallography. However, membrane dissociation while maintaining the native functional characteristics is an essential, yet difficult step, in the field of CYP protein engineering.
The truncation of the membrane-spanning CYP N-terminus is the most widely used strategy to disrupt protein-membrane interactions (19-23). Indeed, we previously demonstrated that the complete truncation of the CYP2J2 N-terminus results in a construct with significantly increased solubility. Importantly, this truncated construct (Δ34-CYP2J2) exhibited similar substrate turnover rates as the full length CYP2J2 construct in model membranes (24). Notably, Δ34 maintained its membrane insertion despite the N-terminal truncation (24). Accumulating evidence suggests that the F-G loop of CYPs (Figure 1A and 3A) is an essential mediator of membrane binding (25). Additionally, the F-G loop is positioned at the mouth of the substrate access tunnels and is thought to control the rate of substrate access to the buried active site (26). Unlike the N-terminal motif, the non-conservative mutations within the F-G loop region often results in significantly altered enzymatic activity (27). Thus far, strategies have typically included random mutagenesis and substitution of alternative CYP chimeras which are typically an exercise of trial-and-error (28).
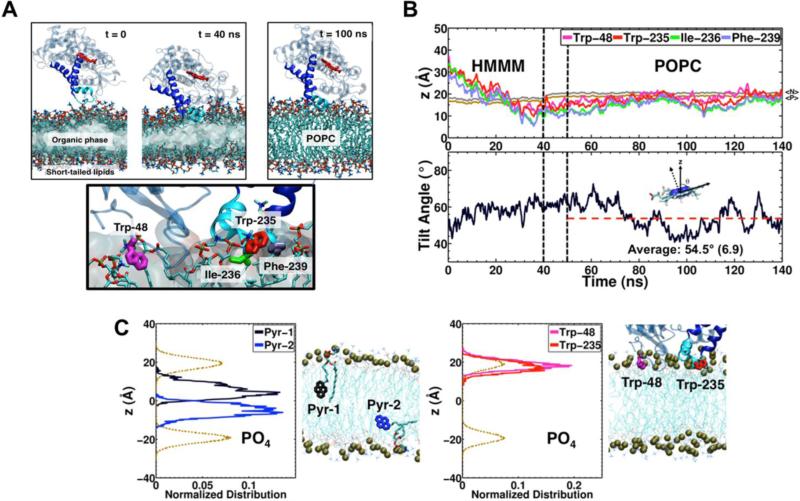
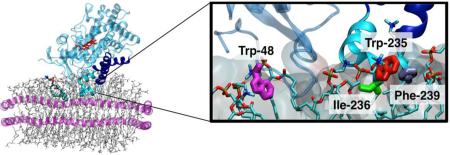
Figure 1. Insertion of CYP2J2 into Membrane.
(A) (Top) Membrane binding of CYP2J2 captured with the HMMM membrane in 40 ns. The membrane-bound model of CYP2J2 was extended to a POPC membrane and simulated for another 100 ns. The stick backbone of the heme is colored red, the F- and G- helices are blue with the remaining P450 colored light blue. Lipids are shown in stick representation. (Bottom) Residues Trp-48, Trp-235, Ile-236 and Phe-239, identified to interact with the membrane upon binding, are shown in stick representation. (B) (Top) Time series of the center of mass (COM) z-coordinate of the side chains Trp-48, Trp-235, Ile-236 and Phe-239 for the HMMM and POPC membranes. A side chain is assumed to insert into the membrane when its COM z-position is below the reference plane defined by the COM z-position of the PO4. The black dashed lines indicate the time allowed for the grown POPC membrane to relax. The positions of the phosphorous (PO4) and nitrogen (choline) atoms of the lipids are show in in brown and gray, respectively. (Bottom) Time series of the heme tilt angle of CYP2J2 in the HMMM and POPC membranes. The red dashed line indicates the average for the heme tilt angle in the POPC membrane. (C) Distribution of the z-coordinate of pyrene molecules attached to phospholipids and of the Trp-48 and Trp-235 side chains, obtained from separate simulations. The dashed brown lines indicate the distribution of the z-coordinate PO4 group of each leaflet. The location of pyrene and Trp side chains in the simulation is also shown.
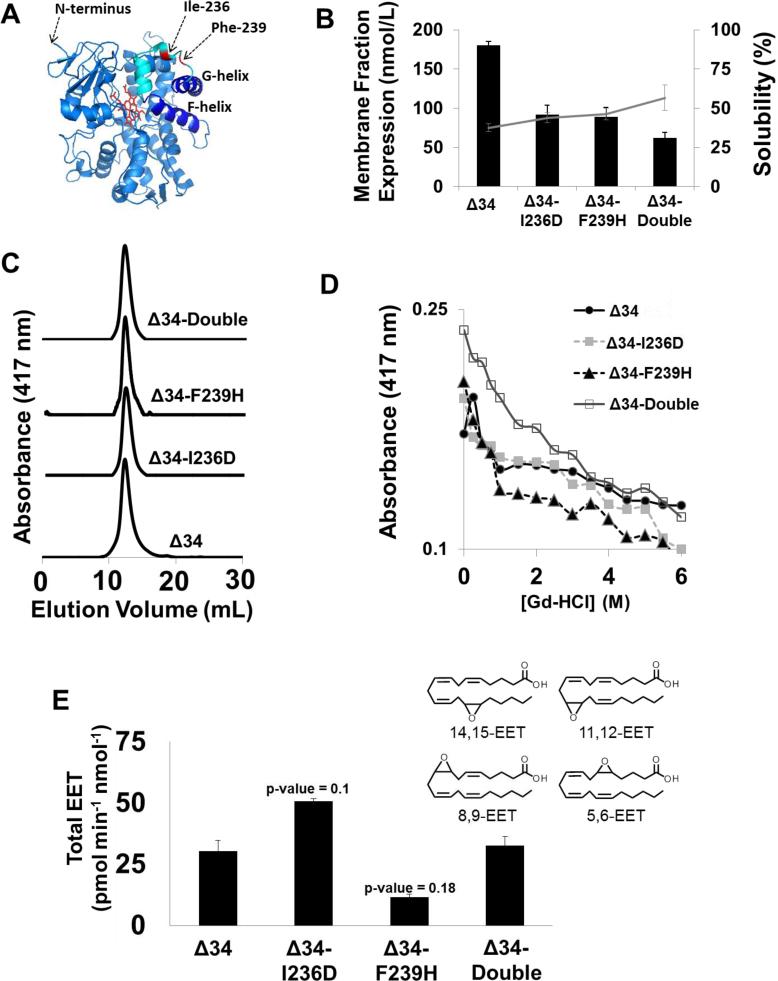
Figure 3. Biochemical characterization of the CYP2J2 F-G loop mutants.
(A) Representation of the CYP2J2 Δ34 homology model. The stick backbone of the heme is colored red, the F- and G- helices are dark blue with the remaining P450 protein fold and helices colored light blue. The two residues selected for mutation (I236 and F239) are colored red on the F-G loop represented by cyan. (B) The three CYP2J2 mutants Δ34-I236D, Δ34-F239H and Δ34-double were recombinantly expressed in E. coli. The bar graph denotes expression yields purified from the membrane fraction whereas the line plot signifies percentage of CYP found in the soluble fraction during purification. (C) The size exclusion chromatography purified CYP2J2 protein was examined for homogeneity and elution volume with a high resolution Superdex 75 10/30 column (D) Changes to the stability of each CYP2J2 construct was assessed by monitoring the loss of the prosthetic heme monitored at 417 nm using UV-Vis spectroscopy in the presence of increasing concentrations of guanidine hydrochloride. (E) F-G loop mutant functionality was gauged by monitoring the metabolism of arachidonic acid (40 μM) using LC/MS/MS in a phospholipid reconstituted system. The rate of the conversion of arachidonic acid to total EET consisting of 14,15-, 11,12-, 8,9- and 5,6-EET were determined for the wild-type construct and each of the three F-G loop mutants. In the arachidonic acid assay we did not observe statistical significance below p < 0.05 when using a paired, two-tailed student's t-test but did see a trend towards significance for Δ34-I236D and Δ34-F239H with p-values of 0.1 and 0.18, respectively.
In this study, we used molecular dynamics (MD) simulations in conjunction with experimental studies to evaluate the role of the F-G loop of CYP2J2 in membrane binding and insertion. Previously, MD simulations have been used to study protein-membrane interactions of membrane-bound CYPs (6-8, 13, 29, 30). Herein, we first employ a highly mobile membrane-mimetic (HMMM) model (31) to capture spontaneous binding of CYP2J2 to the membrane. The HMMM model employs short-tailed lipids while an organic solvent replaces the hydrophobic core of the membrane, resulting in increased lipid mobility. Due to the enhanced lipid mobility, the model accelerates the association of proteins with the lipid bilayer, and has been successfully employed to capture membrane binding of a variety of peripheral and membrane proteins including the GLA domain (31), cytochrome P450 3A4 (6), talin (32), α-synuclein (33), synaptotagmin (34), and synaptobrevin (35). This approach enabled us to capture atomic level interactions between CYP2J2 and the membrane. Importantly, these interactions were preserved when the short-tailed lipids were extended to a full POPC membrane. Secondly, after a membrane-bound model of CYP2J2 was obtained, we studied the effect of F-G loop mutations on membrane-binding and insertion depth, by taking advantage of the enhanced lipid dynamics of the HMMM model.
To experimentally support the MD findings, hydrophilic residues were engineered in the CYP2J2 F-G loop. Subsequent studies revealed the effects of the mutations on enzyme functionality, subcellular distribution and membrane binding characteristics. The details of the relative depth of insertion were explored by measuring the mutants’ intrinsic tryptophan fluorescence and energy transfer to pyrene phospholipids incorporated in lipid bilayers.
Determining the degree of the Trp-pyrene fluorescence resonance energy transfer (FRET) is a widely used method to probe the interaction of both peripheral and membrane proteins with membrane bilayers. Previously, liposomal systems were used to measure the Trp insertion of membrane proteins such as CYP3A4 (10) and CYP1A2 (36) as well as peripherally binding proteins such as cytochrome C (37) and 14-3-3γ (38). The use of nanodiscs in this application offers several advantages over traditional liposomal systems. Specifically, nanodiscs are nanoscale discoidal lipid bilayers that exhibit significantly enhanced stability and homogeneity relative to liposomal systems. Additionally, nanodiscs are more easily concentrated without aggregation and lack the typical scattering effects observed with liposomal systems (14, 24, 39).
Overall this work provides novel insights on the role of key residues in the CYP2J2 F-G loop and their effects on membrane binding and functional metabolism. The use of MD simulations provides high resolution interactions between the membrane and CYP2J2 F-G loop. Experimentally, the introduction of hydrophilic residues in the F-G loop provides essential functional data as well as an assay for membrane depth of insertion. Understanding the nuances of these interactions will enable new applications with other membrane bound CYPs.
METHODS
Materials
Ampicillin, arabinose, chloroamphenicol and IPTG were from Gold Biotechnology. NADPH and NADP were obtained from P212121.com. Protein standards—thyroglobulin, ferritin, bovine serum albumin, and cytochrome c, were purchased from Sigma. Phospholipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phospho-L serine (POPS) were purchased from Avanti Polar Lipids. The phospholipid 1-hexadecanoyl-2-(1-pyrenedecanoyl)-sn-glycero-3-phosphocholine (PYRPC) was obtained from Life Technologies. Amberlite XAD 2 was procured from Supelco. Nanosep MF (0.2 μM) and Amicon Ultra (10,000 MWCO) centrifugal filters were bought from Millipore. AA and ebastine were purchased from Cayman Chemical. All other materials and reagents used were purchased from Sigma-Aldrich and Fisher Scientific.
Molecular Dynamical Simulations: Preparation of HMMM membrane
The HMMM membrane was constructed starting from a solvated POPC membrane by shortening the lipid tails to only 5 carbons (31), while the remaining carbon atoms were converted to 1,1-dichloroethane (DCLE). This resulted in an HMMM membrane patch with two leaflets of short-tailed PC lipids at the interface of water and DCLE, as described in detail elsewhere (31). The solvated HMMM membrane contained 137 short-tailed PC lipids in each leaflet and 1,268 DCLE molecules, yielding a system with ~41,000 atoms. The solvated membrane mimetic system was energy minimized for 2,000 steps and simulated for 1 ns using an NPnAT ensemble with constant area, and with a target normal pressure and temperature of 1.0 atm and 310 K, respectively. A constant area of 10, 281 Å2 (101.4 × 101.4 Å2) was employed, yielding an area of 75 Å2 per lipid (AL), about 8% higher than the experimental AL for POPC membranes (40). Based on our experience with other peripheral proteins, a mild increase in AL (5-8%) can significantly accelerate membrane binding (31, 33). The resulting membrane was employed in all subsequent binding simulations of wild-type CYP2J2 and its mutants. To mimic the atomic distributions of a full lipid bilayer in the HMMM simulations more closely, a harmonic constraint along the z-axis (membrane normal) with a force constant k = 0.05 kcal·mol–1·Å–2 was applied to the carbonyl carbon atoms of the short-tailed PC lipids.
CYP2J2 model and initial configurations
A homology model of the catalytic domain of CYP2J2, generously provided by Dr. D. Mansuy (Université René Descartes, Paris France), was employed as the starting structure of CYP2J2 for this study. The homology model was derived using the available crystal structures of CYP2A6 (PDB: 1Z11), CYP2B4 (PDB: 2BDM), CYP2C5 (PDB: 1NR6), CYP2C8 (PDB: 1PQ2), and CYP2D6 (PDB: 2F9Q) as templates, as previously described (41). The sequence identity of the template structures with CYP2J2 ranged from 41% to 45%. The individual sequence identity of each template with CYP2J2 is as follows: CYP2A6: 42%; CYP2B4: 45%; CYP2C5: 41%; CYP2C8: 42%; CYP2D6: 42%. Initially, the CYP2J2 structure was relaxed by placing the enzyme in a water box with dimensions of 90 × 90 × 90 Å3, containing ~20,000 water molecules, generated with the SOLVATE plugin of VMD (42), and neutralized with 100 mM NaCl ions using the AUTOIONIZE plugin. The system was then energy minimized for 2,000 steps and simulated for 100 ps with the Cα atoms of the protein harmonically restrained (k = 1 kcal·mol−1·Å−2), and followed by 100 ns run with no constraints. The resulting structure of CYP2J2 was used as the initial structure for simulations.
For the membrane-binding simulations, wild-type CYP2J2 was placed ~10 Å above the surface of the HMMM membrane with different initial orientations (50° and 80° with respect to the z-axis). Three initial systems were prepared for simulation, two of which started from the same initial orientation (80°). The resulting structure was further solvated using the SOLVATE plugin, and neutralized with 100 mM NaCl using the AUTOIONIZE plugin of VMD (42). Each of the three resulting initial systems consisted of a box with dimensions of 101.4 × 101.4 × 122.0 Å3 and containing ~120,000 atoms. The systems were energy minimized for 2,000 steps and relaxed further for 100 ps with the Cα atoms of the protein harmonically restrained (k = 1 kcal·mol–1·Å–2), followed by 40 to 60 ns of production simulation.
To study the effect of F-G loop mutations on membrane binding of CYP2J2, the mutations I236D, F239H, and I236D/F239H were introduced in the last frame of each membrane-binding simulation of the wild-type CYP2J2. This approach allowed us to take advantage of the accelerated lipid dynamics to monitor changes in the membrane insertion of CYP2J2 induced by the mutations. The Asp residue in the mutants was modeled in its deprotonated (charged) form, while the His residue was modeled in its neutral form, to match the expected charge of these side chains at pH 7.4. The resulting systems were energy minimized for 1,000 steps and relaxed further for 100 ps with the Cα atoms of the protein, except for the mutated residues, harmonically restrained (k = 1 kcal·mol–1·Å–2), followed by 40 to 50 ns of production simulation.
The resulting models of membrane-bound CYP2J2 and its mutants in the HMMM membrane were extended to a full POPC membrane, to perform additional simulations. A membrane-bound model of the CYP2J2 for each case (wild-type and mutants) was adopted from the last frames of one of the HMMM membrane-binding simulations described above. The short-tailed lipid bilayer was transformed into a POPC bilayer by removing the DCLE molecules and adding the missing carbons of the lipid tails. The positions of the added lipid tail atoms were refined based on the coordinates of randomly selected lipid molecules obtained from a POPC bilayer equilibrated separately. During these steps, the positions of the lipid atoms already present in the HMMM membrane (headgroups and the initial five carbons of the lipid tails) were preserved, maintaining the original contacts established between the lipids and CYP2J2 during the HMMM simulations. The resulting POPC-bound CYP2J2 systems were then minimized for 2,000 steps and equilibrated for 100 ps while restraining all the protein and short-tailed lipids atoms with a force constant k = 1 kcal·mol–1·Å–2 to allow relaxation of the the newly added lipid tail atoms. Following this step, the resulting wild-type and mutants systems were simulated without restraints for 70 to 100 ns.
Pyrene phospholipid partitioning in the membrane
In order to gain a more detailed description of the partitioning and orientation of pyrene molecules in a lipid bilayer, we performed another set of MD simulations. A solvated POPC membrane containing one pyrene phospholipid in each leaflet was constructed to study the dynamics of pyrene in the membrane. A POPC molecule was employed as a template to generate the pyrene-attached phospholipid, by removing carbons from the acyl tail and replacing them by a pyrene moiety. The resulting system was then minimized for 1,000 step and simulated for 1 ns with the heavy atoms of the lipid head groups harmonically restrained (k = 1 kcal·mol–1·Å–2), except for the pyrene phospholipids. The step was then followed by a 50 ns production simulation.
Simulation conditions and protocols
All simulations were performed with NAMD2 (43) using the CHARMM27 (44) force field with cMAP corrections for the protein and CHARMM36 (45) for lipids. Parameters of the pyrene-attached phospholipid were derived by analogy from the CHARMM General Force Field (46, 47). The TIP3P model was used for water (48). The NPnAT ensemble at 1.0 atm and 310 K, with a constant area of 10,281 Å2 (101.4 × 101.4 Å2), was employed for all HMMM simulations. The POPC systems were simulated as an NPT ensemble at the same pressure and temperature. All simulations were performed with a time step of 2 fs. Constant pressure was maintained by the Nosé–Hoover Langevin piston method (49, 50) and constant temperature was maintained by Langevin dynamics with a damping coefficient γ of 0.5 ps–1 applied to all atoms. A cutoff of 12 Å was used for nonbonded interactions, with a smoothing function applied after 10 Å. The particle mesh Ewald (PME) method (51) was used for long-range electrostatic calculations with a grid density greater than 1 Å–3.
Heterologous expression and purification the CYP2J2 F-G loop constructs
The N-terminally truncated CYP2J2 construct (Δ34) was heterologously expressed and purified as described previously (24, 52). The mutations for the F-G loop plasmids were prepared using the reverse primers listed in Figure S3 and amplified by polymerase chain reaction (PCR) on the pCWori Δ34 plasmid in conjunction with the Δ34 forward primers. Each F-G loop mutant plasmid was co-transformed with the GroEL plasmid and grown in an overnight culture and used for inoculation of 1 L of terrific broth (TB) media supplemented with ampicillin (100 μg/mL) and chloramphenicol (20 μg/mL). The culture was grown at 37°C and 220 rpm for 2.5 hours and then supplemented with δ-aminolevulinic acid (0.5 mM) and maintained at 26°C and 160 rpm for 2 hours. Next the culture was induced with 1 mM of Isopropyl β-D-1-thiogalactopyranoside (IPTG) and 2g of arabinose and grown for an additional 44 hours. The cells were isolated via centrifugation for 10 min (2800 g) using a JA-10 rotor (Beckman Coulter, Brea CA) and sonicated in lysis buffer containing dithiothreitol (DTT), 0.2 mM phenylmethanesulfonylfluoride (PMSF), 5 mg DNase, and RNase. The membrane fraction was isolated via ultracentrifugation at 35,000 rpm from 30 min Ti-45 rotor (Beckman) and the soluble fraction was collected for analysis of the CYP content. Protein was extracted using 1% cholate (w/v) and the supernatant layer containing CYP2J2 was obtained by ultracentrifugation 35,000 rpm from 30 min Ti-45 rotor (Beckman). The histidine tagged protein was further purified by running the supernatant through a Ni-NTA column and eluted with 0.1mM DTT, 0.1% (w/v) cholate, 100 mM KPi, 200 mM imidazole and 20% glycerol. CYP2J2 concentration was measured by carbon monoxide difference spectra as described below.
Purification of oligomeric CYP2J2 by size-exclusion chromatography
For the membrane binding studies, the resulting Ni-NTA eluent was further purified using size exclusion - high performance liquid chromatography (SEC-HPLC) on a semi-preparative Superdex 200 10/300 column (GE Life Sciences, Piscataway, NJ) with a mobile phase consisting of 100 mM phosphate buffer (pH 7.4), 1 mM EDTA, 0.5 M NaCl, 2% glycerol and 0.1% cholate at a 0.5 ml/min flow rate. The predominant peak of each construct was collected and adjusted to contain 20% glycerol and concentrated using Amicon Ultra (10,000 MWCO) centrifugal filters (Millipore). The resulting protein concentration was quantified using a Micro BCA protein assay kit (Thermo Sci #23235). Next, the concentrated protein was examined for homogeneity (Figure 3C) by injecting 20 μL on a high resolution Superdex 75 10/30 SEC column (GE Life Sciences, Piscataway, NJ) with a mobile phase consisting of 100 mM phosphate buffer (pH 7.4), 1 mM EDTA, 0.5 M NaCl, 2% glycerol and 0.1% cholate at a 0.5 ml/min flow rate. The oligomeric state of the CYP2J2 constructs was estimated using the same mobile phase and Superdex 75 10/30 column and a standard curve plotted using thyroglobulin (669 kDa), ferritin (440 kDa), BSA (67 kDa), and cytochrome c (14 kDa) as protein standards (Figure S5).
Expression and purification of cytochrome P450 reductase
Cytochrome P450 reductase (CPR) from Ratticus norvegicus was expressed and purified as previously described (24).
Construction of ΔTrp-MSP1D1
In order to obtain ΔTrp-MSP1D1, the EcoR1-Sac1 gene fragment of MSP1D1 was replaced by the synthetic gene fragment in which two tryptophan codons (TGG) were substituted by phenylalanine codons (TTC). An additional silent mutation was added to create new restriction site (Xba1) in order to facilitate the screening. Briefly, the 300 bp G-block fragment (synthesized by IDT DNA, Inc.) and MSP1D1 plasmid were digested with Eco R1 and Sac1 restriction endonucleases. After separation on agarose gel, the 5.7 kBp fragment of MSP1D1 plasmid was ligated to synthetic insert using T4 ligase. After initial identification of the mutant clones by digestion mapping, the presence of the desired mutations and the absence of the unwanted ones were confirmed by DNA sequencing. MSP expression and purification was performed as described earlier (53).
Carbon monoxide binding assay
The carbon monoxide binding characteristics of the constructs were measured after Ni-NTA purification using a modified version of Omura and Sato's method (54) as described previously (24, 39, 55).
Guanidine hydrochloride induced denaturation of CYP2J2
The denaturation of CYP2J2 protein was studied by measuring the change in Soret at 417 nm as a function of increasing concentration of Gd-HCl (0 to 6 M) as previously described (56). Solutions containing varying concentrations of guanidinium hydrochloride were incubated with ~2 μM of CYP2J2 for 5 minutes before measuring their UV-vis spectra (800 – 300 nm). The resulting spectra were then processed using a MATLAB subroutine for determination of Soret absorbance changes.
Arachidonic acid metabolism assay for measurement of EETs
Incubations containing empty POPC-nanodiscs (10 μM), CYP2J2 (0.2 μM), CPR (0.6 μM) in 100 mM phosphate buffer (pH 7.4) were incubated with concentrations of arachidonic acid (40 μM) near the critical micelle concentration. The mixture was equilibrated at 37°C for 10 min before reaction initiation with 200 μM NADPH (total volume 500 μL) and was allowed to react for 30 min. CYP2J2-Δ34 AA metabolism linearity over a one hour time course was previously confirmed (39). Samples were quenched upon extraction with an equal volume of ethyl acetate (3 times), dried under a steady stream of nitrogen and reconstituted in ethanol. Samples were analyzed using a 1200 series HPLC (Agilent Technologies, Santa Clara, CA) coupled to a 5500 QTRAP LC-MS/MS system (AB Sciex, Foster City, CA) for EET quantification using LC-MS/MS as previously described (39).
Assembly of POPC nanodiscs
POPC was solubilized with cholate (50 mM) and mixed with the membrane scaffold protein ΔTrp-MSP1D1 (devoid of Trp ε280 = 10,040 mM−1 cm−1) in a 65:1 ratio before rocking the mixture at 4°C for one hour. Nanodisc assembly was initiated upon removal of detergents using of Amberlite Biobeads (Supelco). The homogenous nanodisc assembly was isolated using SEC-HPLC with a Superdex 200 10/300 column (GE Life Sciences, Piscataway, NJ) and a mobile phase consisting of 100 mM phosphate buffer (pH 7.4) and a 0.5 ml/min flow rate. The isolated nanodiscs were then concentrated with Amicon Ultra (10,000 MWCO) centrifugal filters (Millipore) to a final concentration of 500 μM.
Assembly of POPC-PYR nanodiscs
Nanodiscs containing PYR-PC were prepared with the same methodology as the POPC nanodiscs with a minor adaptation. Specifically, the PYR-PC was added to the POPC mixture in a 1.5:98.5 molar ratio. The phospholipids were then mixed with MSP1D1 in a 65:1 ratio and allowed to equilibrate for 1 hour at 4°C before detergent removal with Amberlite Biobeads. The assembled nanodiscs were isolated using SEC-HPLC on a Superdex 200 10/300 column (GE Life Sciences, Piscataway, NJ) with a mobile phase consisting of 100 mM phosphate buffer (pH 7.4) and a 0.5 ml/min flow rate (Figure 4B). Desired concentration of the incorporated PYR-PC was confirmed in solution spectrophotometrically at 342 nm using the molar extinction coefficient 42 mM−1 cm−1 (Figure 4B inset).
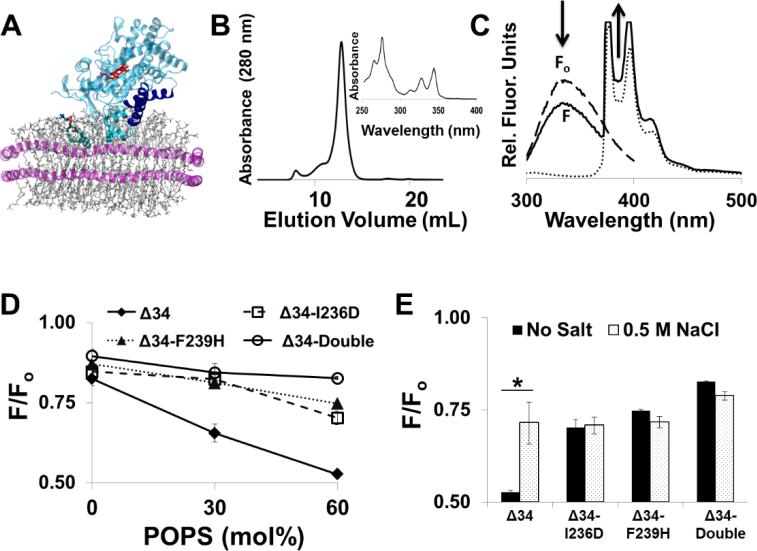
Figure 4. Fluorescence studies with CYP2J2 F-G loop constructs.
(A) Schematic of CYP2J2 insertion into pyrene nanodiscs (PYR-ND). The F-G loop is located between the CYP2J2 F- and G-helices (represented in blue) and contains the Trp 235 residue, which interacts with the pyrene phospholipids (shown in cyan stick model) incorporated in the membrane bilayer of nanodiscs (phospholipids are grey and membrane scaffold protein is purple). (B) Size-exclusion HPLC chromatography of the PYR-ND. (Inset) Spectra demonstrating successful PYR-PC phospholipids incorporation into nanodiscs. (C) The relative membrane insertion characteristics of each construct were determined by measuring CYP2J2 Trp quenching. Fo (dashed line) is the Trp-fluorescence maximum CYP2J2 in the presence of POPC nanodiscs (lacking pyrene). F (solid line) is the quenched CYP2J2 Trp fluorescence due to insertion in pyrene nanodiscs. The dotted line represents the pyrene nanodisc fluorescence spectra (excitation 295 nm; emission 300-600 nm) in the absence of protein. (D) Trp fluorescence signals of CYP2J2 constructs were monitored in the presence of empty nanodiscs (F0) and presence of PYR-ND (F) to calculate F/Fo. The F/Fo was measured using PYR-ND containing POPS/POPC molar ratios of 0:100, 30:70 and 60:40. (E) The effect of 0.5M NaCl on F/Fo ratio was measured for each CYP2J2 F-G loop mutant using 60 mol% POPS PYR-ND
Assembly of POPC-PYR-POPS nanodiscs
Nanodiscs containing PYR-PC were prepared as described for the POPC-PYR nanodiscs, albeit with the addition of differing ratios of anionic phospholipids (POPS). Nanodiscs containing 30% and 60% POPS were prepared by mixing POPC/PYR/POPS in a molar ratio of 68.5:1.5:30 and 38.5:1.5:60, respectively. Each POPC/PYR/POPS mixture was added to MSP1D1 in a 65:1 ratio and allowed to equilibrate for 1 hour at 4°C before detergent removal with Amberlite Biobeads. The assembled nanodiscs were isolated using SEC-HPLC on a Superdex 200 10/300 column (GE Life Sciences, Piscataway, NJ) with a mobile phase consisting of 100 mM phosphate buffer (pH 7.4) and a 0.5 ml/min flow rate
Steady state fluorescence titration experiments
Steady state fluorescence measurements were performed using a K2 multi-frequency phase and modulation fluorometer (ISS, Urbana, IL, USA). Fluorescence experiments utilized purified monomeric CYP (1 μM), nanodiscs (50 μM), 100 mM phosphate buffer (pH 7.4) and 1 mM Na-EDTA maintained at 25°C with a circulating water bath. Emission spectra of the intrinsic fluorescence of the CYP interacting with nanodiscs were recorded from 300-400 nm with an excitation of 295 nm and bandwidths of 8 nm for both excitation and emission measurements. The maximal initial fluorescence (Fo) was measured by preparing samples containing CYP and ΔTrp-MSP1D1 POPC nanodiscs (50 μM), incubating for 10 min and recording the emission spectra. The quenched fluorescence measurement (F) was recorded after an incubation (10 min) of individually preparing samples containing CYP with pyrene nanodiscs (50 μM) comprised of ΔTrp-MSP1D1 POPC, ΔTrp-MSP1D1 POPC/POPS (70:30), or ΔTrp-MSP1D1 POPC/POPS (40:60). A parallel sample devoid of CYP was recorded after each measurement and subsequently subtracted from the main spectra using Vinci 2 software (Urbana, IL, USA). Samples were analyzed for changes in the Trp fluorescent maxima (λmax) as well as for the ratio of changes in the initial fluorescent intensity (Fo) and final fluorescence (F).
Data analysis
Data were analyzed and presented as means ± standard error. Data were collected multiple times and were analyzed for statistical significance using a Student's paired t test where *p < 0.05, **p < 0.01, and ***p < 0.001.
RESULTS AND DISCUSSION
Molecular dynamics simulations - membrane-bound form of CYP2J2
In order to generate a membrane-bound model of CYP2J2 which would allow us to better understand the key lipid-protein interactions involved in membrane binding of the protein, we performed three independent MD simulations between 40 and 60 ns, starting with the Δ34-CYP2J2 placed above a phosphatidyl choline (PC) HMMM lipid bilayer. The aim was to capture the spontaneous membrane binding and insertion of the protein into lipid bilayer in unbiased simulations. Owing to the enhanced lipid dynamics of the membrane model, spontaneous membrane binding of the catalytic domain of CYP2J2 was consistently observed during these simulations (Figure 1A). The analysis of the three resulting simulations reveals a similar behavior in terms of depth of insertion and orientation in the membrane of CYP2J2 (Figure 1B and Figure S1). The resulting binding pose of CYP2J2 suggests that the enzyme can interact with the membrane through hydrophobic residues located in the F-G loop region, in particular residues Trp-235, Ille-236 and Phe-239. During the simulations, these side chains were observed to insert in the membrane (i.e., positioning below the reference plane defined by the PO4 level) (Figure 1B). The N-terminal loop formed containing residue Trp-48 is also observed to insert into the membrane after binding (Figure 1B).
Starting from the HMMM membrane-bound models of CYP2J2, the short-tailed lipids (with five carbons in each tail) were extended to full POPC lipids (see Methods), resulting in a model of CYP2J2 bound to a POPC membrane as shown in Figure 1A. The extended model simulation shows that the main interactions observed in the HMMM model are preserved, and Trp-48, as well as residues Trp-235, Ile-236 and Phe-239 located in the F-G loop remain inserted in the membrane (inserted below the PO4 level) (Figure 1B and Figure S1). In order to test the convergence of the membrane-bound models of CYP2J2, we characterized the orientation that the enzyme with respect to the lipid bilayer by calculating the heme tilt angle, defined as the angle between the heme plane (defined as the plane containing the four porphyrin nitrogen atoms) and membrane normal (z-axis). In the three extended POPC simulations, the heme tilt angle converges to values ~50°, suggesting a convergent membrane-bound pose for CYP2J2. The orientation that CYP2J2 adopts in the membrane is similar to that reported for other membrane-bound CYPs, measured both from experiments and from simulations, which are reported to be around 60 to 70° (6, 57).
Simulation of pyrene partitioning in the membrane
We performed experiments to evaluate the interaction of the mutated CYP2J2 proteins with membranes by measuring the Trp quenching by pyrene phospholipids inserted into the homogeneous lipid bilayer. Therefore, in order to identify potential interactions between a pyrene molecule attached to a phospholipid and the membrane-bound CYP2J2, we performed a separate simulation of a POPC membrane containing a single pyrene phospholipid in each leaflet for 50 ns in the absence of CYP2J2. The goal was to characterize the distribution of the pyrene molecules within the lipid bilayer and use it to infer information on the potential interaction of the side chains of CYP2J2 with pyrene upon membrane binding based on the location of this moiety in the membrane. The distribution of the z-position for each molecule revealed that the pyrene moiety is located mainly in the hydrophobic core of the membrane, below the phosphate groups (Figure 1C). Despite being attached to their respective phospholipid tail, pyrene molecules are able to sample a wide range of insertion depth within the hydrophobic core of the membrane, as indicated by their wide z-position distributions. Interestingly, the insertion depth of CYP2J2 in the POPC membrane would allow for the side chains of Trp-48 and Trp-235 to interact with the pyrene moiety located in the PYR-ND, as indicated by their z-position distributions (Figure 1C). The distribution of the z-position of each side chain shows that they both insert to a similar level around the PO4 level, with Trp-235 slightly more inserted than Trp-48. Therefore using PYR-ND one can delineate the membrane interactions of the CYP2J2 mutants with the lipid bilayer using experimental approaches as described later.
CYP2J2 construct design
The initial structural analysis and MD simulations suggested two hydrophobic residues in the F-G loop, Ile-236 and Phe-239, as putative membrane anchors (Figure 1A and 3A). These sites were mutated to hydrophilic residues with the aim of altering membrane-binding characteristics by decreasing the overall hydrophobicity of this region. To this end, two single mutations, I236D (Δ34-I236D) and F239H (Δ34-F239H), as well as a construct containing both I236D and F239H (Δ34-double) were designed. Apart from the N-terminal anchor, the CYP F-G loop has been hypothesized to be the primary membrane-binding motif. This assumption is further supported by the observation that the greatest structural differences between the soluble prokaryotic CYPs and the membrane bound mammalian CYPs are found in the F and G helices and the F-G loop (58). Notably, CYP2D6 was successfully solubilized and crystallized by truncating the N-terminus and mutating the F-G loop Leu-230 and Leu-231 with a combination of hydrophilic residues that included I230D and I231R (21). Following a similar design strategy we mutated Ile-236 and Phe-239 to hydrophilic residues to examine the effects on solubility and functionality. Additionally, the close proximity of the naturally occurring Trp-235 to the mutated sites enabled membrane-binding studies using fluorescence spectroscopy.
Molecular dynamics simulations of membrane binding modes of the F-G loop mutants
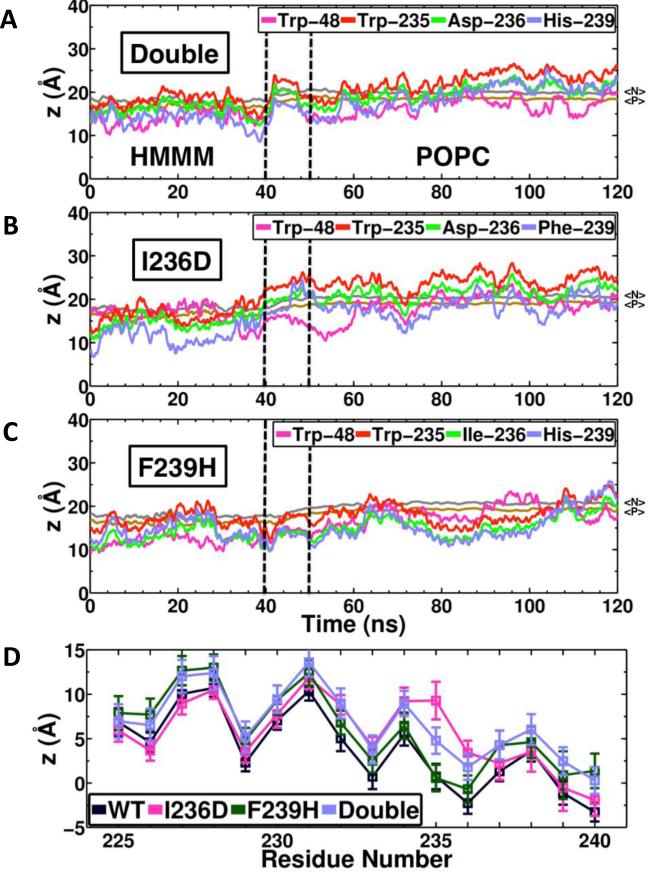
We studied the effect of these specific F-G loop mutations on membrane binding of CYP2J2 by introducing the mutations in the membrane-bound models obtained from MD simulations. To achieve this, we used the final snapshot of each of the three HMMM membrane-binding simulations, introduced the mutations and performed production simulations on the system. This approach allowed us to monitor changes in the membrane insertion depth of the protein due to mutations in the F-G loop. Single mutations Δ34-I236D and Δ34-F239H, as well as double mutation Δ34-double were introduced in the membrane-bound CYP2J2, resulting in 3 models for each mutant, which were simulated individually between 40 and 50 ns.
The results of these simulations show that the mutations affect the insertion of the F-G loop to various degrees (Figure 2). The I236D mutation promotes the detachment of Trp-235 from the membrane. In the cases were this mutation is introduced (I236D and 1236D/F239H), Trp-235 moves from a fully inserted position to an average position that is above the PO4 and choline levels, with occasional detachment of the residue from the membrane (Figure 2A and 2B). In both cases, the position of Trp-235 side chain (above the phosphate level) is maintained upon the conversion of the HMMM membrane with short-tail lipids to a full POPC membrane (Figure 2A and 2B). In the POPC membrane, the average position of Trp-235 resulting from the I236D mutation is between 5 to 10 Å above the reference PO4 plane, as shown in Figure 2D. The F239H mutation, on the other hand, seems to have a smaller effect on Trp-235 insertion, causing only transient detachment of Trp-235 both in the short-tail lipid membrane and in the POPC membrane (Figure 2C), resulting on an average relative position that is close to the Trp-235 insertion in the wild type case (Figure 2D). Taken together, the simulations indicate that introducing mutations in the F-G loop results in a shallower membrane insertion of Trp-235 in the F-G loop when compared to the wild-type simulations. As discussed later, this was corroborated by an increased percentage of the mutants in the cytosolic fraction and fluorescence experiments where we measured CYP2J2 Trp quenching by membranes containing pyrene lipids.
Figure 2. Effect of F-G loop mutations in membrane-binding of CYP2J2.
Time series of the COM z-position of side chains Trp-48, Trp-235, Ile-236 and Phe-239 for the HMMM and POPC membranes shown for (A) I236D/F239H mutant, (B) I236D mutant and (C) F239H mutant. For each case, the HMMM simulations were extended to a POPC membrane and simulated 80 ns. The black dashed lines indicate the time allowed for the extended POPC membrane to relax after conversion from HMMM. The positions of the phosphorous (PO4) and nitrogen (choline) atoms of the lipids are show in in brown and gray, respectively. (D) Average relative position of the COM z-coordinate of the side chains forming the F-G loop (residues 225 to 240) with respect to the reference plane defined by the COM z-position of the PO4. The average was taken from the last 20 ns of simulations for each case. The error bars represent one standard deviation.
Expression and purification of CYP2J2 F-G loop mutants
The growths of all constructs were performed in DH5alpha cells due their relatively slow growth and expression characteristics that improve the overall yields of functional P450 (52). We previously demonstrated that the N-terminus truncated CYP2J2 construct (Δ34) exhibited greater solubility, yet still maintained its ability to insert in the membrane bilayers of nanodiscs (24). Interestingly, the percentage of protein found in the soluble fraction during purification corresponded to the number of F-G loop mutations. Specifically, the percentage of protein in the soluble fraction of Δ34-double, Δ34-I236D, Δ34-F239H and Δ34 were 56.7%, 46.3%, 43.8% and 37.5%, respectively (Figure 3B). These results provide the expression and membrane binding characteristics of the CYP constructs in the E. coli membrane. The ratio of the membrane to soluble fraction of the protein only provides an indication of enzyme solubility and is not directly applicable to other systems such as model membranes and mammalian cells. Therefore we further explored the membrane insertion of these constructs using a Trp quenching assay in Nanodiscs as described later.
Spectral characterization, denaturation studies, and size-exclusion chromatography of the F-G loop mutants
The F-G loop mutants exhibited typical Soret and Q-bands similar to wild type CYP2J2 (data not shown). To investigate the potential heme-thiolate perturbations the mutants were reduced (FeII) and bound with carbon monoxide. All constructs exhibited a prominent 451 nm peak, albeit with some 420 nm peak, indicating that the charged mutations in the F-G loop did not largely perturb the heme-thiolate motif interactions and the common P450 fold (Figure S4). Furthermore, the stability of the mutants was assessed by performing guanidine hydrochloride denaturation studies using UV-Vis spectroscopy. The 417 nm Soret absorbance is attributed to the uniquely coordinated ferric (FeIII) heme. In the presence of increasing concentration of guanidine hydrochloride (Gd-HCl), the absorbance of the CYP at 417 nm decreased, indicating heme loss due to protein unfolding (56). The three constructs demonstrated comparable stability in Gd-HCl with a decrease in peak intensity of ~45%. The Δ34 construct exhibited less denaturation with an overall 30% decrease in the Soret peak intensity in the presence of 6M Gd-HCl (Figure 3D).
The range of oligomers formed from each mutant after Ni-NTA purification was examined using semi-preparative size exclusion chromatography All the constructs produced a predominant peak that eluted with a volume of ~15.5 ml on the Superdex 200 10/300 column (data not shown) using the conditions listed in the materials and methods. The primary oligomer was collected, concentrated and its homogeneity and molecular weight was assessed using a high resolution Superdex 75 10/30 column. As seen in Figure 3C, all the constructs show a single oligomer that corresponded to the monomeric form of the protein as measured using protein standards (Figure S5). Taken together, the results suggest that the non-conservative mutations did not change the protein in regards to P450 fold, oligomeric state and stability.
Arachidonic acid metabolism
Typically, random introduction of mutations in the F-G loop of CYPs alters their function as compared to the wild type construct (27). Therefore we examined the effect of the F-G loop mutation on the CYP2J2 mediated metabolism of arachidonic acid (AA) in the presence of a 50 μM concentration of empty POPC nanodiscs. As discussed in the steady state fluorescence section, a CYP to nanodisc stoichiometric ratio of 1:50 was chosen to ensure that the CYP construct was completely bound to nanodiscs. Under these conditions, the functional differences between CYP2J2 are results of differences in their ability to metabolize AA and not due to discrepancies between the membrane-bound and unbound states. AA is a slowly metabolized endogenous substrate that forms EET metabolites that are potent vasodilators and anti-inflammatory lipids (17, 59). The metabolism of AA is complex, as it produces four unique epoxide regio-isomers. Interestingly, the Δ34-I236D metabolism of AA was increased with a ~1.6 greater catalytic efficiency of all regio-isomers when compared to Δ34 (Figure 3E). Conversely, the function of Δ34-F239H was dramatically decreased relative to Δ34, indicating that the mutant's native function has been altered. Notably, the Δ34-double construct produced the EET metabolites with similar catalytic efficiency to Δ34 as shown in Figure 3E. Interestingly, none of the mutations altered the ratio of the EET products produced by CYP2J2 (data not shown), which suggests that the active site was not drastically altered.
In both mammalian and bacterial systems, the F-G loop is an important regulator of substrate access channels (60). Moreover, this region has been shown to be flexible with open/closed conformations and provides the roof of the internal substrate-binding pocket which mediates substrate recognition (61). Recent evidence suggests that the conformation of the F-G loop and ligand type may alter ligand egress routes. Specifically, MD simulations predicted that the non-helical CYP2C9 F-G loop would favor egress pathways through the membrane bilayer (62). In this work we demonstrate that single mutations in this region can alter the catalytic turnover characteristics of the epoxidation of arachidonic acid (Figure 3E). Interestingly, the enhanced catalytic functionality of Δ34-I236D or decreased activity of Δ34-F239H did not alter the the enzyme regioselectivity and therefore cannot simply be rationalized based on their enhanced solubility or active site alteration. Rather these changes suggest that the introduction of the acidic residue either alters the loop conformation dependent egress pathways and/or substrate recognition sites. Notably, these effects are abolished in the Δ34-double construct, which restores wild-type activity.
While only a small amount of histidine is charged at its physiological pH, this may be an important stabilizing residue. The combination of an anionic and cationic residue introduction was selected for the crystallization of CYP2D6 due to its restoration of wild-type like activity (21). Importantly, these mutations were chosen as a practice of trial-and-error. The rational introduction of the double mutation in CYP2J2 F-G loop corroborates this strategy. However, MD simulations demonstrated that although there is a close interaction of the side-chains we failed to observe a salt-bridge formation (data not shown).
CYP2J2 construct membrane insertion into pyrene nanodiscs
To investigate the CYP2J2 constructs’ insertion in the membrane bilayer, we measured the relative Trp quenching energy transfer between the protein monofacial Trp residues and pyrene phospholipids. To this end, we prepared nanodiscs with pyrene phospholipids (PYR-ND) using the ΔTrp-MSP1D1 construct in which Trp residues were mutated to Phe. As shown in Figure 4A, the Trp residues of the CYP2J2 membrane binding region are predicted to interact with pyrene phospholipids incorporated in the membrane bilayer. Using a POPC/PYR-PC molar ratio of 64:1 the expected PYR-PC was 1 pyrene molecule per nanodisc leaflet. Previously, pyrene-containing liposomes have been successfully employed to characterize membrane insertion properties of peripheral and integral CYP proteins (10, 63, 64). Here we demonstrate use of PYR-ND as a new iteration of this method with a few key advantages. Notably, the PYR-ND is a 10 nm discoidal lipid bilayer that is homogeneous (Figure 4B) and exhibits superior stability. Additionally, the nanodisc also enables stoichiometric control of the lipid compositions.
As previously discussed, MD simulations revealed that the pyrene moiety can sample a wide range of orientations just under the phospholipid heads in the membrane core. Upon insertion into the membrane bilayer the two residues, Trp48 and Trp235, were shown to interact with pyrene. Thus, in the presence of saturating concentrations of PYR-ND, the experiment is a measure of membrane depth of insertion of these fluorophores.
For the depth of insertion measurements, the CYP to PYR-ND (1:50) ratio was chosen based on fluorescence titrations that revealed that the overall Trp florescence was quenched with excess PYR-ND (data not shown). This has been previously detailed in other cases and is thought to reflect that there are two populations of Trp residues (65). Specifically, one population that interacts with the pyrene upon insertion and another population that is buried in the protein and is not substantially quenched (65). Therefore the relative extent of non-ideal Trp quenching is represented here by the quenched fluorescence (F) divided by the initial fluorescence (Fo).
The FRET transfer of energy is a dynamic quenching interaction. Evidence of a FRET between the Trp (donor) and pyrene (quencher) was observed upon titration of the CYP to a sample of PYR-ND. Specifically, a decrease in the Trp fluorescence of CYP is accompanied by an increase in the pyrene emission (Figure 4C). Using the PYR-ND system we examined the relative quenching of Trp fluorescence (F/Fo) as a function of both the mol% of anionic phospholipids (POPS) and the absence/presence of 0.5 M NaCl. The rationale of these two conditions used is explained below.
In the absence of anionic lipids, the differences among the CYP2J2 mutants F/Fo are subtle (Figure 4D). Specifically, Δ34, Δ34-I236D, Δ34-F239H and Δ34-double exhibited average F/Fo ratios of 0.82 ± 0.02, 0.848 ± 0.01, 0.845 ± 0.04 and 0.89 ± 0.01, respectively. These results suggest that the F-G loop mutants are oriented in a way that the Trp residues are not as deeply imbedded. Overall the quenching ratios reflect similar values observed with CYP3A4 binding to liposomes. Specifically, when the Trp quenching properties of CYP3A4 were examined with in phosphatidylcholine (PC) vesicles containing 1.5 mol% of PYR-PC the initial F/Fo values were ~0.85 (10). Conversely, in the case of CYP1A2, a negligible amount of quenching is observed with neutral phospholipid vesicles (36).
The presence of anionic lipids is known to induce both CYP conformational shifts as well as increased penetration into membrane bilayers (10, 36). Thus, we further examined the membrane insertion properties (relative F/Fo changes) of CYP2J2 when anionic lipids (POPS) were incorporated into the bilayers of nanodiscs (Figure 4D). When 30 mol% of POPS was substituted at the expense of POPC, we observed greatly enhanced differences among the F-G loop mutants and the Δ34 construct. Specifically, Δ34, Δ34-I236D, Δ34-F239H and Δ34-double exhibited average F/Fo ratios of 0.65 ± 0.03, 0.82 ± 0.002, 0.81 ± 0.01 and 0.84 ± 0.04, respectively. Interestingly, the differences among the F-G loop mutants and the Δ34 construct were the most pronounced when 60 mol% of POPS was incorporated into nanodiscs. For these experiments, Δ34, Δ34-I236D, Δ34-F239H and Δ34-double exhibited average F/Fo ratios of 0.52 ± 0.005, 0.74 ± 0.02, 0.7 ± 0.03 and 0.82 ± 0.002, respectively. Thus, using a high percentage of anionic lipids we were able to affect the greatest changes between the Δ34 construct and the F-G loop mutants. While these results are only qualitative, the different membrane binding interactions induced by the presence of anionic lipids suggest that the hydrophobicity and charge of the F-G loop is important for POPS mediated depth of insertion. Specifically, these differences support the assumption that the enzyme topology is altered when the F-G loop is mutated with these hydrophilic residues. This conclusion is supported by the MD data that demonstrate these residues are putative membrane anchors and hydrophilic mutations detach this region from the leaflet.
To further explore the nature of the membrane depth of insertion, we examined the effect of a high concentration of NaCl on the F/Fo ratio when the 60 mol% POPS PYR-ND was employed to achieve the maximum membrane interaction of CYP2J2 (Figure 4E). Importantly, high salt concentrations effectively solubilize membrane proteins, altering their nature from an irreversibly membrane bound protein to a state of protein that can bind and dissociate in a peripheral manner (4). This effect was observed when the Δ34G construct was examined, with a decrease in the Trp quenching (F/Fo) from 0.52 to 0.71, for the no salt buffer to 0.5M NaCl buffer, respectively. Surprisingly, the presence of a high concentration of NaCl had little effect on the extent of Trp quenching for the F-G loop mutants which demonstrates the membrane insertion depth is maintained despite the high salt concentration. Overall, the MD simulation results and sub-cellular localization in E. coli studies suggests that these proteins are peripherally binding in the no salt buffer.
Here we demonstrate that the subtle topology nuances between similar constructs and membrane depth of insertion can be measured using Trp quenching studies. While the fluorescence experiments only provide a relative measure of membrane insertion characteristics, this approach is an effective tool to understand how mutations, lipid composition and NaCl can alter the depth of insertion.
CONCLUSIONS
In this work, we demonstrate that the CYP2J2 F-G loop region is an important protein-membrane interface that controls membrane insertion and substrate metabolism characteristics. The HMMM model simulations revealed that CYP2J2 interacts at the F-G loop through the Trp-235, Ille-236 and Phe-239 residues. Importantly, these binding interactions and the protein orientation were preserved in the MD simulations when the short-tailed lipids were extended to POPC lipids. The effects of mutating the putative membrane anchors Ile-236 and Phe-239 with hydrophilic residues were also examined. These mutations significantly decrease the F-G loop depth of insertion when used in MD simulations. To experimentally corroborate these findings, four CYP2J2 constructs (Δ34, Δ34-I236D, Δ34-F239H and Δ34-double) were expressed in E. coli. Notably, the F-G loop mutant expression yields derived from the membrane fraction were substantially decreased and the protein was localized to the cytosolic fraction to a greater extent. While all constructs demonstrated sufficient P450 content and stability we observed significant differences in their functional metabolism. Wild-type activity was restored through the introduction of a double mutation F239H and I236D. Thus, we experimentally demonstrate that the insertion of CYP2J2 into the membrane bilayers of PYR-ND is influenced by the overall hydrophobicity of the F-G loop. The relative membrane binding characteristics were experimentally derived by monitoring CYP2J2 Trp quenching after insertion into the membrane bilayers of PYR-ND. The N-terminal truncated construct Δ34-CYP2J2 exhibited membrane binding to a greater extent than the F-G loop mutants (Δ34-I236D and Δ34-F239H) suggesting a different enzyme topology in agreement with the MD simulations.
Supplementary Material
Highlights.
Molecular dynamics simulations reveal atomistic details of CYP2J2 membrane binding and insertion.
CYP2J2-membrane interactions are mediated in part by residues of the F-G Loop.
F-G loop hydrophilic mutations of truncated CYP2J2 produces a soluble and functional mutant.
ACKNOWLEDGMENTS
We gratefully acknowledge the use of the Sligar lab fluorometer and helpful discussions on fluorescence measurements with Dr. Mark McLean. We want to thank Dr. Ko, Dr. Bagchi and Dr. Ferguson for allowing the use of their equipment. We appreciate the continued support of Dr. Kevin Tucker of the UIUC School of Chemical Sciences and Dr. Zhong Li at the Metabolomics lab of the UIUC Roy J. Carver Biotechnology Center.
Funding: This work was supported in part through the American Heart Predoctoral Fellowship [14PRE20130015] (DRM), American Heart Association Scientist Development Grant [15SDG25760064] (AD) and in part by the National Institutes of Health (R01-GM101048, U54-GM087519, and P41-GM104601 (ET), and in part by the National Institutes of Health (R01-GM101048, U54-GM087519, and P41-GM104601 to ET). All simulations have been performed using XSEDE resources (grant MCA06N060 to ET). The Roy J. Carver Biotechnology Center's 5500 QTrap MS was funded by the National Institutes of Health National Center for Research Resources [Grant S10-RR024516].
ABBREVIATIONS
- CYP
cytochrome P450
- ER
endoplasmic reticulum
- ND
nanodisc
- PYR
pyrene
- PYR-PC
pyrene phosphatidylcholine
- MD
molecular dynamics
- HMMM
highly mobile membrane mimetic
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPS
1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phospho-L-serine
- PYR-ND
pyrene nanodiscs
- Δ34-double
Δ34-I236D/F239H
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, Gunsalus IC, Nebert DW. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Anzenbacher P, Anzenbacherova E. Cytochromes P450 and metabolism of xenobiotics. Cell Mol Life Sci. 2001;58:737–747. doi: 10.1007/PL00000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pernecky SJ, Larson JR, Philpot RM, Coon MJ. Expression of truncated forms of liver microsomal P450 cytochromes 2B4 and 2E1 in Escherichia coli: influence of NH2-terminal region on localization in cytosol and membranes. Proc Natl Acad Sci U S A. 1993;90:2651–2655. doi: 10.1073/pnas.90.7.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol Cell. 2000;5:121–131. doi: 10.1016/s1097-2765(00)80408-6. [DOI] [PubMed] [Google Scholar]
- 5.Black SD. Membrane topology of the mammalian P450 cytochromes. FASEB J. 1992;6:680–685. doi: 10.1096/fasebj.6.2.1537456. [DOI] [PubMed] [Google Scholar]
- 6.Baylon JL, Lenov IL, Sligar SG, Tajkhorshid E. Characterizing the membrane-bound state of cytochrome P450 3A4: structure, depth of insertion, and orientation. J Am Chem Soc. 2013;135:8542–8551. doi: 10.1021/ja4003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denisov IG, Shih AY, Sligar SG. Structural differences between soluble and membrane bound cytochrome P450s. J Inorg Biochem. 2012;108:150–158. doi: 10.1016/j.jinorgbio.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berka K, Paloncyova M, Anzenbacher P, Otyepka M. Behavior of human cytochromes P450 on lipid membranes. Journal of Physical Chemistry B. 2013;117:11556–11564. doi: 10.1021/jp4059559. [DOI] [PubMed] [Google Scholar]
- 9.Nath A, Grinkova YV, Sligar SG, Atkins WM. Ligand binding to cytochrome P450 3A4 in phospholipid bilayer nanodiscs: the effect of model membranes. J Biol Chem. 2007;282:28309–28320. doi: 10.1074/jbc.M703568200. [DOI] [PubMed] [Google Scholar]
- 10.Kim KH, Ahn T, Yun CH. Membrane properties induced by anionic phospholipids and phosphatidylethanolamine are critical for the membrane binding and catalytic activity of human cytochrome P450 3A4. Biochemistry. 2003;42:15377–15387. doi: 10.1021/bi035280k. [DOI] [PubMed] [Google Scholar]
- 11.Das A, Varma SS, Mularczyk C, Meling DD. Functional investigations of thromboxane synthase (CYP5A1) in lipid bilayers of nanodiscs. Chembiochem. 2014;15:892–899. doi: 10.1002/cbic.201300646. [DOI] [PubMed] [Google Scholar]
- 12.Denisov IG, Grinkova YV, McLean MA, Sligar SG. The one-electron autoxidation of human cytochrome P450 3A4. The Journal of Biological Chemistry. 2007;282:26865–26873. doi: 10.1074/jbc.M704747200. [DOI] [PubMed] [Google Scholar]
- 13.Berka K, Hendrychova T, Anzenbacher P, Otyepka M. Membrane position of ibuprofen agrees with suggested access path entrance to cytochrome P450 2C9 active site. J Phys Chem A. 2011;115:11248–11255. doi: 10.1021/jp204488j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bayburt TH, Sligar SG. Single-molecule height measurements on microsomal cytochrome P450 in nanometer-scale phospholipid bilayer disks. Proc Natl Acad Sci U S A. 2002;99:6725–6730. doi: 10.1073/pnas.062565599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnold C, Markovic M, Blossey K, Wallukat G, Fischer R, Dechend R, Konkel A, von Schacky C, Luft FC, Muller DN, Rothe M, Schunck WH. Arachidonic acid-metabolizing cytochrome P450 enzymes are targets of {omega}-3 fatty acids. J Biol Chem. 2010;285:32720–32733. doi: 10.1074/jbc.M110.118406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delozier TC, Kissling GE, Coulter SJ, Dai D, Foley JF, Bradbury JA, Murphy E, Steenbergen C, Zeldin DC, Goldstein JA. Detection of human CYP2C8, CYP2C9, and CYP2J2 in cardiovascular tissues. Drug Metab Dispos. 2007;35:682–688. doi: 10.1124/dmd.106.012823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spector AA. Arachidonic acid cytochrome P450 epoxygenase pathway. J Lipid Res. 2009;50(Suppl):S52–56. doi: 10.1194/jlr.R800038-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams PA, Cosme J, Ward A, Angove HC, Matak Vinkovic D, Jhoti H. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424:464–468. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 19.Yano JK, Hsu MH, Griffin KJ, Stout CD, Johnson EF. Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat Struct Mol Biol. 2005;12:822–823. doi: 10.1038/nsmb971. [DOI] [PubMed] [Google Scholar]
- 20.Schoch GA, Yano JK, Wester MR, Griffin KJ, Stout CD, Johnson EF. Structure of human microsomal cytochrome P450 2C8. Evidence for a peripheral fatty acid binding site. J Biol Chem. 2004;279:9497–9503. doi: 10.1074/jbc.M312516200. [DOI] [PubMed] [Google Scholar]
- 21.Rowland P, Blaney FE, Smyth MG, Jones JJ, Leydon VR, Oxbrow AK, Lewis CJ, Tennant MG, Modi S, Eggleston DS, Chenery RJ, Bridges AM. Crystal structure of human cytochrome P450 2D6. J Biol Chem. 2006;281:7614–7622. doi: 10.1074/jbc.M511232200. [DOI] [PubMed] [Google Scholar]
- 22.Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 23.DeVore NM, Scott EE. Structures of cytochrome P450 17A1 with prostate cancer drugs abiraterone and TOK-001. Nature. 2012;482:116–119. doi: 10.1038/nature10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDougle DR, Palaria A, Magnetta E, Meling DD, Das A. Functional studies of N-terminally modified CYP2J2 epoxygenase in model lipid bilayers. Protein Science. 2013;22:964–979. doi: 10.1002/pro.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pikuleva IA. Putative F-G loop is involved in association with the membrane in P450scc (P450 11A1) Mol Cell Endocrinol. 2004;215:161–164. doi: 10.1016/j.mce.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Kingsley LJ, Lill MA. Ensemble generation and the influence of protein flexibility on geometric tunnel prediction in cytochrome P450 enzymes. PLoS One. 2014;9:e99408. doi: 10.1371/journal.pone.0099408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murtazina D, Puchkaev AV, Schein CH, Oezguen N, Braun W, Nanavati A, Pikuleva IA. Membrane-protein interactions contribute to efficient 27-hydroxylation of cholesterol by mitochondrial cytochrome P450 27A1. J Biol Chem. 2002;277:37582–37589. doi: 10.1074/jbc.M204909200. [DOI] [PubMed] [Google Scholar]
- 28.Kang JY, Ryu SH, Park SH, Cha GS, Kim DH, Kim KH, Hong AW, Ahn T, Pan JG, Joung YH, Kang HS, Yun CH. Chimeric cytochromes P450 engineered by domain swapping and random mutagenesis for producing human metabolites of drugs. Biotechnol Bioeng. 2014;111:1313–1322. doi: 10.1002/bit.25202. [DOI] [PubMed] [Google Scholar]
- 29.Cojocaru V, Balali-Mood K, Sansom MS, Wade RC. Structure and dynamics of the membrane-bound cytochrome P450 2C9. PLoS Comput Biol. 2011;7:e1002152. doi: 10.1371/journal.pcbi.1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lonsdale R, Rouse SL, Sansom MS, Mulholland AJ. A multiscale approach to modelling drug metabolism by membrane-bound cytochrome P450 enzymes. PLoS Comput Biol. 2014;10:e1003714. doi: 10.1371/journal.pcbi.1003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohkubo YZ, Pogorelov TV, Arcario MJ, Christensen GA, Tajkhorshid E. Accelerating membrane insertion of peripheral proteins with a novel membrane mimetic model. Biophys J. 2012;102:2130–2139. doi: 10.1016/j.bpj.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arcario MJ, Tajkhorshid E. Membrane-induced structural rearrangement and identification of a novel membrane anchor in talin F2F3. Biophys J. 2014;107:2059–2069. doi: 10.1016/j.bpj.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vermaas JV, Tajkhorshid E. Conformational heterogeneity of alpha-synuclein in membrane. Biochim Biophys Acta. 2014;1838:3107–3117. doi: 10.1016/j.bbamem.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Z, Schulten K. Synaptotagmin's role in neurotransmitter release likely involves Ca(2+)-induced conformational transition. Biophys J. 2014;107:1156–1166. doi: 10.1016/j.bpj.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blanchard AE, Arcario MJ, Schulten K, Tajkhorshid E. A highly tilted membrane configuration for the prefusion state of synaptobrevin. Biophys J. 2014;107:2112–2121. doi: 10.1016/j.bpj.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahn T, Guengerich FP, Yun CH. Membrane insertion of cytochrome P450 1A2 promoted by anionic phospholipids. Biochemistry. 1998;37:12860–12866. doi: 10.1021/bi980804f. [DOI] [PubMed] [Google Scholar]
- 37.Mustonen P, Virtanen JA, Somerharju PJ, Kinnunen PKJ. Binding of Cytochrome-C to Liposomes as Revealed by the Quenching of Fluorescence from Pyrene-Labeled Phospholipids. Biochemistry. 1987;26:2991–2997. doi: 10.1021/bi00385a006. [DOI] [PubMed] [Google Scholar]
- 38.Song Y, Yang Z, Ke Z, Yao Y, Hu X, Sun Y, Li H, Yin J, Zeng C. Expression of 14-3-3gamma in patients with breast cancer: correlation with clinicopathological features and prognosis. Cancer Epidemiol. 2012;36:533–536. doi: 10.1016/j.canep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 39.McDougle DR, Kambalyal A, Meling DD, Das A. Endocannabinoids Anandamide and 2-Arachidonoylglycerol Are Substrates for Human CYP2J2 Epoxygenase. J Pharmacol Exp Ther. 2014;351:616–627. doi: 10.1124/jpet.114.216598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kucerka N, Tristram-Nagle S, Nagle JF. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J Membr Biol. 2005;208:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 41.Lafite P, Andre F, Zeldin DC, Dansette PM, Mansuy D. Unusual regioselectivity and active site topology of human cytochrome P450 2J2. Biochemistry. 2007;46:10237–10247. doi: 10.1021/bi700876a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 43.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mackerell AD, Jr., Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 45.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD., Jr. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanommeslaeghe K, MacKerell AD., Jr. Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J Chem Inf Model. 2012;52:3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vanommeslaeghe K, Raman EP, MacKerell AD., Jr. Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J Chem Inf Model. 2012;52:3155–3168. doi: 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 49.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant-Pressure Molecular-Dynamics Simulation - the Langevin Piston Method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 50.Martyna GJ, Tobias DJ, Klein ML. Constant-Pressure Molecular-Dynamics Algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 51.Darden T, York D, Pedersen L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 52.Zelasko S, Palaria A, Das A. Optimizations to achieve high-level expression of cytochrome P450 proteins using Escherichia coli expression systems. Protein Expres Purif. 2013;92:77–87. doi: 10.1016/j.pep.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 53.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 54.Omura T, Sato R. The Carbon Monoxide-Binding Pigment of Liver Microsomes. Ii. Solubilization, Purification, and Properties. J Biol Chem. 1964;239:2379–2385. [PubMed] [Google Scholar]
- 55.Meling DD, McDougle DR, Das A. CYP2J2 epoxygenase membrane anchor plays an important role in facilitating electron transfer from CPR. J Inorg Biochem. 2014;142C:47–53. doi: 10.1016/j.jinorgbio.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 56.Yu XC, Shen S, Strobel HW. Denaturation of cytochrome P450 2B1 by guanidine hydrochloride and urea: evidence for a metastable intermediate state of the active site. Biochemistry. 1995;34:5511–5517. doi: 10.1021/bi00016a023. [DOI] [PubMed] [Google Scholar]
- 57.Bonthuis DJ, Netz RR. Beyond the Continuum: How Molecular Solvent Structure Affects Electrostatics and Hydrodynamics at Solid-Electrolyte Interfaces. Journal of Physical Chemistry B. 2013;117:11397–11413. doi: 10.1021/jp402482q. [DOI] [PubMed] [Google Scholar]
- 58.Otyepka M, Skopalik J, Anzenbacherova E, Anzenbacher P. What common structural features and variations of mammalian P450s are known to date? Bba-Gen Subjects. 2007;1770:376–389. doi: 10.1016/j.bbagen.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 59.Wu S, Moomaw CR, Tomer KB, Falck JR, Zeldin DC. Molecular cloning and expression of CYP2J2, a human cytochrome P450 arachidonic acid epoxygenase highly expressed in heart. J Biol Chem. 1996;271:3460–3468. doi: 10.1074/jbc.271.7.3460. [DOI] [PubMed] [Google Scholar]
- 60.Grahamlorence S, Amarneh B, White RE, Peterson JA, Simpson ER. A 3-Dimensional Model of Aromatase Cytochrome-P450. Protein Science. 1995;4:1065–1080. doi: 10.1002/pro.5560040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poulos TL. Cytochrome P450 flexibility. Proc Natl Acad Sci U S A. 2003;100:13121–13122. doi: 10.1073/pnas.2336095100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cojocaru V, Winn PJ, Wade RC. Multiple, ligand-dependent routes from the active site of cytochrome P450 2C9. Curr Drug Metab. 2012;13:143–154. doi: 10.2174/138920012798918462. [DOI] [PubMed] [Google Scholar]
- 63.Mustonen P, Virtanen JA, Somerharju PJ, Kinnunen PK. Binding of cytochrome c to liposomes as revealed by the quenching of fluorescence from pyrene-labeled phospholipids. Biochemistry. 1987;26:2991–2997. doi: 10.1021/bi00385a006. [DOI] [PubMed] [Google Scholar]
- 64.Somerharju P. Pyrene-labeled lipids as tools in membrane biophysics and cell biology. Chem Phys Lipids. 2002;116:57–74. doi: 10.1016/s0009-3084(02)00020-8. [DOI] [PubMed] [Google Scholar]
- 65.Eftink MR, Selvidge LA. Fluorescence quenching of liver alcohol dehydrogenase by acrylamide. Biochemistry. 1982;21:117–125. doi: 10.1021/bi00530a021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.