Abstract
Streptococcus pneumoniae (Spneu) remains the most lethal bacterial pathogen and the dominant agent of community-acquired pneumonia. Treatment has perennially focused on the use of antibiotics, albeit scrutinized due to the occurrence of antibiotic-resistant Spneu strains. Immunomodulatory strategies have emerged as potential treatment options. Although promising, immunomodulation can lead to improper tissue functions either at steady state or upon infectious challenge. This argues for the availability of tools to enable a detailed assessment of whole pulmonary functions during the course of infection, not only those functions biased to the defense response. Thus, through the use of an unbiased tissue microarray and bioinformatics approach, we aimed to construct a comprehensive map of whole-lung transcriptional activity and cellular pathways during the course of pneumococcal pneumonia. We performed genome-wide transcriptional analysis of whole lungs before and 6 and 48 h after Spneu infection in mice. The 4,000 most variable transcripts across all samples were used to assemble a gene coexpression network comprising 13 intercorrelating modules (clusters of genes). Fifty-four percent of this whole-lung transcriptional network was altered 6 and 48 h after Spneu infection. Canonical signaling pathway analysis uncovered known pathways imparting protection, including IL17A/IL17F signaling and previously undetected mechanisms that included lipid metabolism. Through in silico prediction of cell types, pathways were observed to enrich for distinct cell types such as a novel stromal cell lipid metabolism pathway. These cellular mechanisms were furthermore anchored at functional hub genes of cellular fate, differentiation, growth and transcription. Collectively, we provide a benchmark unsupervised map of whole-lung transcriptional relationships and cellular activity during early and late pneumococcal pneumonia.
INTRODUCTION
Streptococcus pneumoniae (Spneu) remains the most lethal bacterial pathogen and the dominant agent of community-acquired pneumonia (1,2). Considerable research has been dedicated to defining risk factors and designing risk-scoring systems associated with mortality that could aid in important site-of-care decisions regarding the treatment patients with pneumonia (3). Treatment has relied heavily on the immediate initiation of empirical antibiotic therapy, which in recent years has received substantial scrutiny owing to the emergence of antibiotic-resistant Spneu strains (3). The development of new antibiotics has provided promising results (4); however, the ever-evolving nature of commensal bacteria coupled with their ability to suppress the normal host defense mechanisms by virtue of their virulome (5,6) dictate studies that provide more insight into the immune response to pneumococci that enter the airways. Modulating the immune responses to infection, either by suppression or activation, has emerged as a useful therapeutic approach in the prevention and treatment of bacterial infections (7). Despite such advantages, improper activation and/or suppression of innate immune reactions can result in detrimental inflammatory responses and tissue damage (8). Thus, it is important to assemble a detailed pattern of pangenomic responses and related cellular biological pathways representative of the whole lung and the host pulmonary immune response to pneumococcal infection.
Employing a mouse model of acute pneumococcal pneumonia, we analyzed the genome-wide transcriptional responses in the whole lung at the early (6 h) and late (48 h) stages of the host response. By combining linear modeling to assess differential gene expression and the concepts of scale-free network biology (9), we constructed a gene coexpression network based on the most variable genes across the early and late phases of the host pulmonary transcriptional response. This network was organized into transcriptional modules of highly significant cellular signaling pathways anchored at regulatory “driver” genes, which are understood to be fundamental components of biological response systems, oftentimes directly targeted by bacterial pathogens to favorably modulate host immunity (10). These findings represent a benchmark characterization of the modular properties underlying the whole lung and the host pulmonary response to early and late acute pneumococcal infection.
MATERIALS AND METHODS
Mice and Husbandry
C57BL/6 female mice (9–12 wks old) were purchased from Charles River (Maastricht, the Netherlands). All mice were maintained in pathogen-free conditions in the animal facility of the Academic Medical Center (Amsterdam, the Netherlands). Mice were supplied with the same rodent chow and water ad libitum and maintained on a 12-h light–dark cycle in a temperature- and humidity-controlled environment. The Animal Care and Use Committee of the University of Amsterdam approved all experiments.
Induction of Pneumonia and Bacterial Burden Assessment
The S. pneumoniae serotype 2 D39 strain was grown for 5 h to the midlogarithmic phase at 37°C using Todd–Hewitt broth (Difco, Detroit, MI) with yeast extract (0.5%), harvested by centrifugation at 2,683g for 15 min, and washed twice in isotonic saline. A suspension of 2 × 107 colony-forming units (CFUs) in 50 μL isotonic saline was used as the inoculum. Mice were lightly anaesthetized by inhalation of 2.0%–2.5% isoflurane (Upjohn, Ede, the Netherlands) mixed with O2 (1–2 L/min) and intranasally challenged as previously described (11,12). Mice were humanely euthanized at 6 or 48 h postinfection (n = 4 at each time point). Lung tissue was harvested and processed for the determination of bacterial outgrowth as described (11,12). CFUs in lung homogenates were determined from serial dilutions plated on blood agar plates incubated at 37°C for 16 h before colonies were counted. The lungs from 3 uninfected mice were used as controls.
Lung Histopathology
Lungs were fixed in formalin and embedded in paraffin. Sections (5 μm) were stained with hematoxylin and eosin (HE). Granulocyte staining was performed by anti-Ly6G hybdridization and quantified by means of ImageJ software (Bethesda, MD, USA; http://rsb.info.nih.gov/ij/) as previously described (12, 13).
RNA Preparation and Genome-Wide Transcriptional Profiling
Total RNA was isolated from lung homogenates using the Nucleospin RNA II kit (Machery-Nagel [BIOKÉ, Leiden, the Netherlands]). Yield and purity (260 nm:280 nm) were determined by use of the Nanodrop ND-1000 (Thermo Scientific [Thermo Fisher Scientific, Waltham, MA, USA]). The integrity (RNA integrity number [RIN] >7.0) of the resuspended total RNA was determined by using the RNA Nano Chip Kit on the Bioanalyzer 2100 and the 2100 Expert software (Agilent, Amstelveen, the Netherlands). The Illumina TotalPrep-96 RNA amplification kit (Illumina, Eindhoven, the Netherlands) was used to generate biotin-labeled (biotin-16-UTP) amplified cRNA starting from 200 ng total RNA. A total of 750 ng biotinylated cRNA was hybridized onto the Illumina MouseRef-8v2 Expression BeadChip. The samples were scanned using the Illumina iScan array scanner. Preparation of cRNA, chip hybridization, washing, staining and scanning were carried out at ServiceXS (Leiden, the Netherlands). The raw scan data were read using the BeadArray package (version 1.12.1) (14) available through Bioconductor (15) using the R statistical environment (version 2.13.2; R Foundation for Statistical Computing, Vienna, Austria). Estimated background was subtracted from the foreground for each bead. For replicate beads, outliers greater than 3 median absolute deviations from the median were removed and the average signal was calculated for the remaining intensities. For each probe a detection score was calculated by comparing its average signal with the summarized values for the negative control probes. Resulting data were neqc normalized (16). Quality control was performed both on the bead level and on bead summary data. The arrayQualityMetrics package v2.6.0 (17) was used for further quality assessment of the normalized bead summary data. Probes were reannotated using the package illuminaMousev2.db (18) from Bioconductor. All nonnormalized and normalized data are available at the gene expression omnibus of NCBI (GEO) with accession number GSE42464.
Differential Gene Expression Analysis and Modular Network Construction
Differential probe intensities on microarrays were identified using the R package limma (version 3.8.3) that implements linear models for microarray data (19). Empirical Bayesian methods are used to generate p values obtained from moderated t statistics or f statistics. These p values are then adjusted for multiple comparisons with Benjamini–Hochberg (BH) method to control the false discovery rate (20). The weighted gene coexpression network construction algorithm was used to build the lung gene coexpression network as described previously (21,22). Briefly, a pair-wise Spearman correlation matrix of the top 4,000 most variable unique genes (ranked by coefficient of variation) was transformed into an adjacency matrix by using a “soft” power function of 7 ensuring scale-free topology (21). The adjacency matrix was further transformed into a topological overlap matrix to enable the identification of modules (clusters) of highly correlating genes by implementing a dynamic tree cut algorithm (21). Each module represents a cluster of coregulated genes with a distinct expression pattern from other identified modules. To define module driver genes we made use of the module eigengene concept, defined as the first principal component of the module expression matrix and the module membership measure, k (21, 23). To provide robustness to our findings we performed weighted gene coexpression network analysis (WGCNA) on an independent publicly available lung transcriptome dataset generated from female mice (9–12 wks old) uninfected (n = 5) and 6 h after S. pneumoniae serotype 2 strain D39 intransal infection (n = 5) (GSE49533) (24).
Bioinformatics
Pathway analysis of the gene coexpression modules in connection with differential gene expression between experimental groups was performed using a comparison-based analysis in IPA (Ingenuity® Systems, Qiagen, Valencia, CA, USA; www.ingenuity.com). Specifically, biological function overrepresentation and canonical pathway enrichment were performed using default settings. Module genes presenting differential expression with multiple-comparison corrected p values <0.05 in the univariate analysis between uninfected controls and Spneu at 6 h and between uninfected controls and Spneu at 48 h were eligible. Coexpression network visualizations were mapped by using Cytoscape version 2.8.2 (25). In silico cell-specific coexpression predictions were performed by testing for overrepresentation in the coexpression atlas of the Immunological Genome Project (Immgen.org) (26) available in the ToppGene bioinformatics suite (27). This analysis was performed with default parameters. All probabilities were adjusted for multiple comparisons by the BH method. BioMart (Ensembl, Hinxton, UK) was used to identify network genes that coincided within the pneumonia susceptibility quantitative trait locus (QTL) support interval (28).
Statistics
Wilcoxon rank sum tests were performed to assess the HE pathology and anti-Ly6G+ histology scores. Logistic regression implemented in the globaltest R package (29) was used to assess the statistical significance of each module per infection time point. BH-adjusted p values <0.05 demarcated significant associations.
RESULTS
Differential Host Gene Expression during Pneumococcal Pneumonia
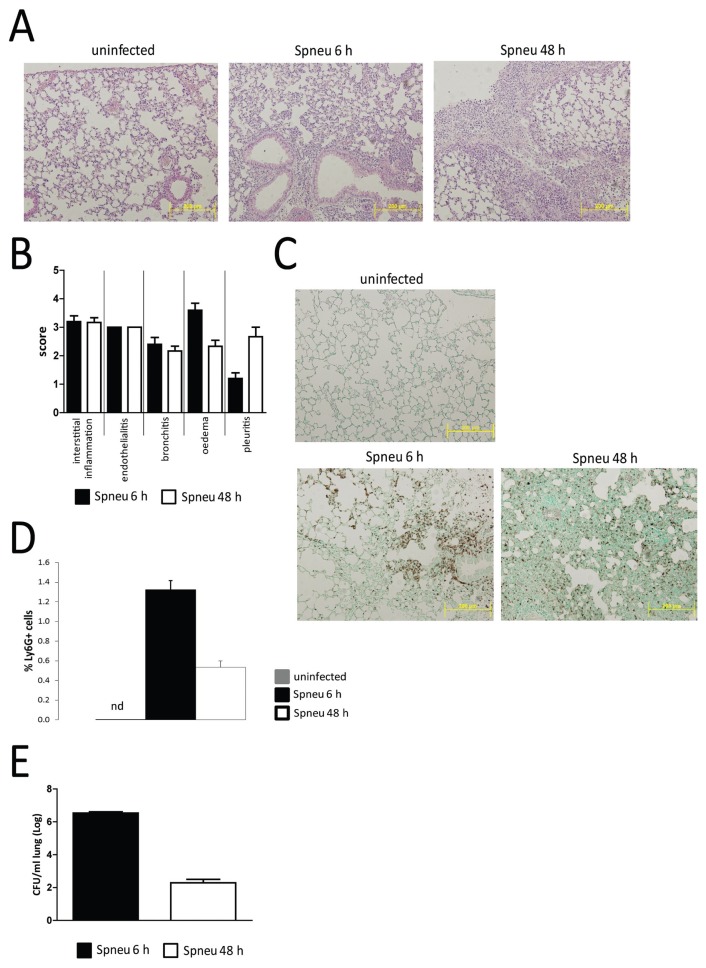
To confirm induction of pneumonia after infection with Spneu via the airways, we assessed histopathology of the lungs harvested 6 or 48 h postinoculation by HE stains and anti-Ly6G immunohistochemistry (Figure 1). As expected (11–13), Spneu induced microscopic features typical for pneumonia, characterized by neutrophil influx, interstitial damage, endothelialitis, bronchitis, edema and pleuritis. Consistent with previous reports (11–13), bacterial loads decreased between 6 and 48 h after infection.
Figure 1.
Immunohistochemical analysis of the early and late host pulmonary response to pneumococcal infection. C57BL/6 female mice were each intranasally inoculated with 2 × 107 CFUs S. pneumoniae D39. Mice were killed at 6 or 48 h (n = 4 each) and lungs were used for histological examination. (A) HE-stained representative lung sections of noninfected (C57BL/6; n = 3), 6-h postinfection (Spneu 6 h) and 48-h postinfection (Spneu 48 h) mice. (B) Histological scoring, determined as described in Materials and Methods, showed inflammatory pathology was largely similar between 6 h and 48 h. (C, D) Granulocyte abundance was higher at 6 h postinfection than at 48 h postinfection, evidenced by higher anti-Ly6G+ cell counts. (E) Bacterial burden measured as CFUs/mL was higher at 6 h postinfection than at 48 h postinfection. Data represent means ± SEM.
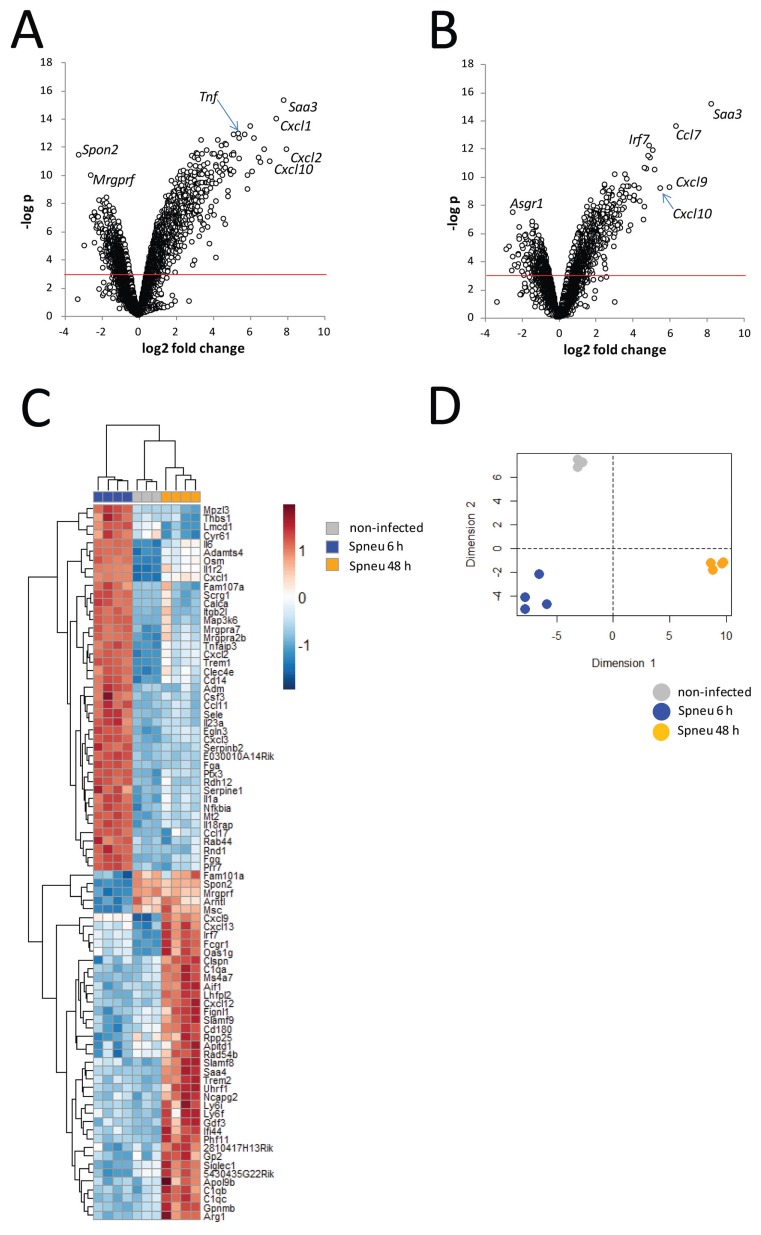
The temporal changes in host lung transcriptional responses to Spneu infection were assessed by pangenomic transcriptional arrays of whole lungs harvested 6 or 48 h after infection. After preprocessing and quality control, 23,742 probes were available for differential gene expression analysis. Compared with uninfected controls (moderated t test), the early (6-h) lung transcriptional response to Spneu infection revealed 2908 differentially abundant probes (BH p < 0.01). Considering a log2 fold change, ≥ 2,359 probes were detected (Figure 2A). Probes for Cxcl1, Cxcl2, Cxcl10, Saa3 and Tnf were prominent (Figure 2A). Considering a log2 fold change of ≤ −2, 19 probes were detected (Figure 2A). Spon2 and Mrgprfl were particularly decreased (Figure 2A). Analysis of the late (48-h) host lung transcriptional response to Spneu infection identified 1,431 differentially abundant probes (BH p < 0.01) compared with uninfected controls. Considering a log2 fold change, ≥ 2181 probes were detected, including Saa3, Ccl7, Cxcl9, Cxcl10 and Irf7 (Figure 2B). Considering a log2 fold change ≤−2, we uncovered 13 probes that included Asgr1 (Figure 2B). Direct comparison of the 6- and 48-h samples yielded 1,637 significant (BH p < 0.01) genes, including Irf7, Spon2, IL6, Ptx3, Osm, Arntl, C1qa, C1qb, C1qc (Figure 2C). Decomposing the 84 top differential genes (BH p < 0.01 and log2 fold change ≥2 or ≤−2) (Figure 2C) to 2 principal components resulted in a cumulative explainable variance of 95% (Dimension 1, 67%; Dimension 2, 28%) (Figure 2D). These results indicate that significant transcriptional alterations between our experimental groups capture a major part of the variance in this model.
Figure 2.
Genome-wide pulmonary gene expression analysis during early and late pneumococcal pneumonia. (A, B) Volcano plot representation (integrating probabilities and fold changes) of the transcriptional changes that occur at (A) 6 h postinfection (n = 4) and (B) at 48 h postinfection (n = 4), compared to noninfected controls (n = 3). Horizontal red line denotes BH-adjusted p < 0.05. (C) Heat-map representation of the 84 top differential genes (BH p < 0.01 and log2 fold change ≥2 or ≤−2) determined by direct comparison of the 6- and 48-h postinfection samples. (D) Principal component analysis by decomposing the 84 top differential genes to 2 principal components resulted in a cumulative explainable variance of 95% (Dimension 1, 67%; Dimension 2, 28%).
Modular Network Analysis Reveals Host Cellular Biological Pathways Shaping the Pulmonary Defense Response
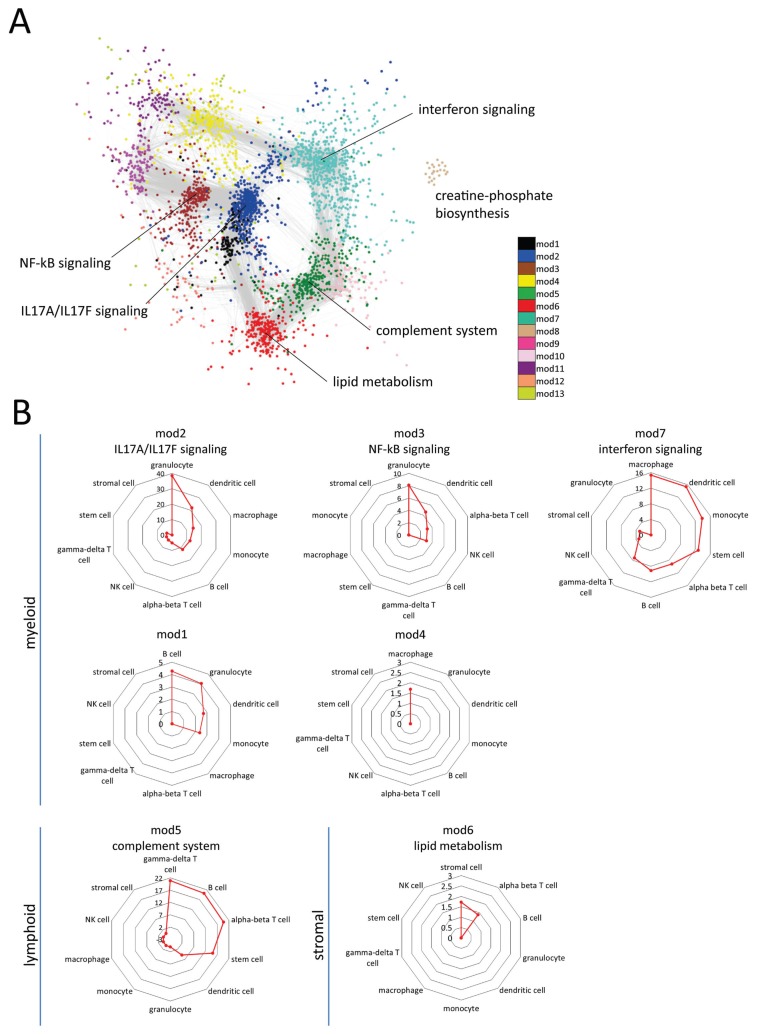
To better understand the organization of the host lung transcriptome during early (6-h) and late (48-h) pneumococcal pneumonia, we applied unsupervised WGCNA (21,22) to the top 4,000 most variable unique genes across all samples (ranked by coefficient of variation). In so doing, we sought to assemble a transcriptome representative of whole-lung tissue functions; thus, not only those functions biased toward the host defense response to pneumococcal pneumonia. On the basis of a Spearman correlation matrix a “weighted” network was constructed ensuring scale-free topology as described previously (see Methods and [22, 30]). Unsupervised hierarchical clustering applied to the resultant topological overlap matrix uncovered 13 modules (clusters), each harboring more than 100 genes. Modules were analyzed for association with biological signaling pathways and illustrated by an unsupervised Cytoscape yFiles organic layout of the weighted Spearman rho coefficients (Figure 3A). This analysis revealed that the lung coexpression modules at 6 h and 48 h postinfection were predominantly organized into functional biological units. The unsupervised network layout, depicted in Figure 3A, showed that IL17A/IL17F signaling was central to the host pulmonary response. Interferon signaling, NF-κB signaling, lipid metabolism, creatine-phosphate biosynthesis and complement system signaling modules were also identified (Figure 3A). Seven (out of 13) modules yielded no significant association to known biological pathways (IPA knowledgebase) (Figure 3A), which may constitute as yet unexplored biological components of the host pulmonary response. Throughout, we refer to these modules by their module number. Next, we performed cell type predictions of module genes by interrogating the sorted cell coexpression atlas of the Immunological Genome project (www.immgen.org) (26) available through the Toppgene bioinformatics suite (27). Considering BH-adjusted Fisher exact probabilities (BH p < 0.05), 7 transcriptional modules were significantly enriched for myeloid, lymphoid and stromal cell markers (Figure 3B). The most significant overrepresented cell types were granulocyte, macrophage, B, γ-delta T, α-β T and stromal cells (Figure 3B). IL17A/IL17F signaling, NF-κB signaling and interferon signaling modules were highly associated with myeloid cell types, namely granulocytes and macrophages, whereas complement system and lipid metabolism modules were associated with lymphoid (γ-delta T cells, B cells, α-β T cells) and stromal cell-types, respectively (Figure 3B).
Figure 3.
Modular organization of the pulmonary transcriptome during pneumococcal pneumonia. The top 4,000 most variable genes (coefficient of variation) across noninfected 6-h and 48-h postinfection samples were used for construction of a weighted pairwise Spearman correlation matrix and clustered into 13 transcriptional modules, each encompassing more than 100 genes. (A) Weighted correlation coefficients were imported into the Cytoscape software (www.cytoscape.org) for unsupervised visualization of the network geometry given module memberships (color-coded) by yFiles layout. Genes with high correlation coefficients (weight >0.1) were used. Each module was labeled by ingenuity pathway analysis (IPA)-derived biological pathways (BH-adjusted Fisher p value <0.05). Based on this layout IL17A/IL17F signaling module (mod2) was central to the host pulmonary response. (B) Modules were significantly associated (BH p < 0.05) to shared and distinct cell-types that allowed for their stratification into myeloid, lymphoid or stromal cell enriched modules. Axis in radar graphs denotes the negative log10 BH p-value. Note: 7 transcriptional modules yielded no significant association to known biological pathways, which may constitute as yet unexplored biological components of the host pulmonary response; these modules are indicated by their module number.
Temporal Patterns and Regulatory Drivers of the Host Modular Transcriptional Network during Pneumococcal Pneumonia
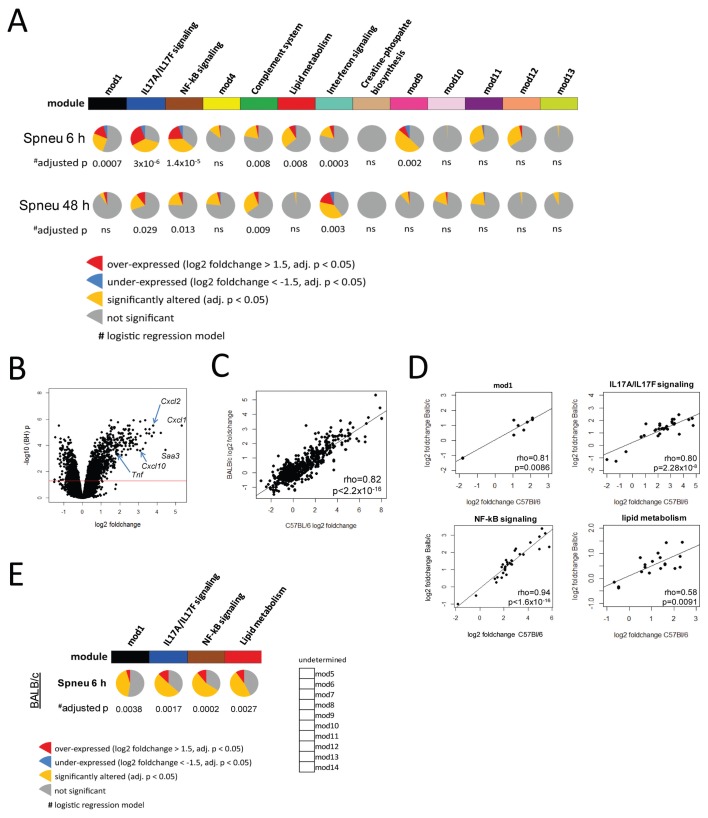
Thus far our scale-free network approach revealed a lung master network organized into 13 modules, wherein 6 modules were significantly associated with biological signaling pathways (Figure 3). Here, we sought to delineate modules by their significance to the host defense response during early and late pneumococcal pneumonia. We performed logistic regression analyses (29) of each module per time point (at 6 and 48 h postinfection) compared with uninfected mouse samples. Considering BH-adjusted p values (correcting for the 13 modules), 7 modules were significantly associated with the early (6-h) phase (Figure 4A), whereas 4 modules were significantly associated with the late (48-h) phase of the host lung response (Figure 4A). These results indicated 54% of the lung master network was significantly altered during pneumococcal pneumonia. By overlaying log2 fold changes and BH p values (pie charts in Figure 4A), we showed significantly altered genes per time point within most modules. For example, the granulocyte IL17A/IL17F signaling module (mod2), comprised of 621 genes, had 70% significantly different genes (BH p < 0.05) at 6 h postinfection compared with noninfected mice (yellow pie slice in Figure 4A). Twenty-eight percent of those significant genes were overexpressed by a more than 1.5 log2 fold change (red pie slice in Figure 4A), whereas 6% were underexpressed by less than a −1.5 log2 fold change (blue pie slice in Figure 4A). Thus, the early host response was characterized by heightened expression of myeloid cell (predominantly granulocyte) pathways, including IL17A/IL17F (mod2) and NF-κB signaling (mod3), as well as a stromal cell lipid metabolism module (mod6; Figure 4A). The latter stromal cell module (mod6) and a myeloid cell module of an undetermined biological signaling pathway (mod1) were identified as uniquely altered at 6 h postinfection (Figure 4A). At 48 h postinfection granulocyte-related functions (IL17A/IL17F and NF-κB signaling) were also significantly altered; however, they exhibited diminished activity compared with 6-h postinfection (Figure 4A). In contrast, higher expression of genes involved in the γ-delta T complement system and macrophage interferon signaling (mod9) modules were determined at 48 h postinfection (Figure 4A).
Figure 4.
Kinetic behavior of host pulmonary transcriptional modules during pneumococcal pneumonia. (A) Logistic regression was used to assess the significance of module activity given early (Spneu 6 h) and late (Spneu 48 h) pneumococcal pneumonia in C57BL/6 mice. Significance was demarcated by BH-adjusted p < 0.05. Seven modules were significantly associated with the early host pulmonary response phase, namely mod1, IL17a/IL17F signaling, NF-κB signaling, complement system, lipid metabolism, interferon signaling and mod9. Four modules were significantly associated with the late host pulmonary response phase, namely, IL17A/IL17F signaling, NF-κB signaling, complement system and interferon signaling. Pie charts depict the proportion of module genes with log2 fold changes <−1.5 or >1.5 that differed significantly (BH p < 0.05) at 6 h or 48 h postinfection, compared to noninfected mice. Note: 7 transcriptional modules yielded no significant association with known biological pathways; these modules are indicated by their module number. (B) Differential gene expression analysis was performed using publicly available data (GSE49533) of BALB/c mice, uninfected (n = 5) and 6 h after S. pneumoniae serotype 2 strain D39 intransal infection (n = 5). (C) Spearman correlation analysis of the BALB/c and C57BL/6 mouse pulmonary response (considering log2 fold changes) after 6 h pneumococcal infection showed a strong correlation. (D and E) Weighted gene coexpression and logistic regression analyses of the 6-h BALB/c response revealed 14 modules of closely correlating genes, of which four modules were significantly conserved between mouse strains. Pie charts depict the proportion of BALB/c module genes with log2 fold changes <−1.5 or >1.5 that differed significantly (BH p < 0.05) at 6 h postinfection, compared to noninfected mice.
To provide robustness to our findings we performed differential gene expression and unsupervised weighted gene coexpression network analysis using a publicly available gene expression dataset of pneumococcal lung infection in BALB/c mice (GSE49533) (24). Differential gene expression analysis between BALB/c mice discordant for pneumococcal pneumonia revealed a pronounced transcriptional response, with 2,141 significantly altered genes (adjusted p < 0.05) (Figure 4B). This BALB/c mouse lung response strongly correlated with our C57Bl/6 mouse lung transcriptional response after 6-h pneumococcal infection (Figure 4C). We applied the WGCNA method to the top 4,000 most variable transcripts and constructed a pair-wise Spearman correlation matrix, transformed into an adjacency matrix by using a soft power function of 9. In this way we identified 14 modules harboring >100 tightly correlating transcripts. Next, we sought to determine whether these BALB/c lung transcriptional modules and our previously delineated C57BL/6 modules were conserved. To this end we performed Spearman correlation tests using gene expression indices per module. Notwithstanding differences in normalization procedures and gene annotations, we found significant correlations in four modules. Of note, ingenuity pathway analysis of the BALB/c modules revealed that three of those four conserved modules were enriched for canonical signaling genes that matched the C57BL/6 module pathways, that is, IL17A/IL17F signaling, NF-κB signaling and lipid metabolism (Figure 4D). In line with our C57BL/6 data, logistic regression analysis showed that these four conserved modules were also significantly altered (adjusted p < 0.05) after 6 h of pneumococcal pneumonia in BALB/c mice (Figure 4E). Thus 57% of the significantly altered network modules in C57BL/6 mice (at 6 h postinfection) was conserved in the BALB/c pneumococcal pneumonia model. The lack of complement system and interferon signaling pathway (Figures 3 and 4) enrichment in the BALB/c model motivated us to evaluate network genes that coincide within a chromosome 7 quantitative trait locus that is understood to modulate the susceptibility to severe pneumococcal pneumonia, namely Spir1 (24). Using the Biomart application (28) we identified one IL17A/IL17F signaling module gene, two NF-κB signaling module genes, two complement system module genes and eight interferon signaling module genes that coincided within the Spir1 QTL support interval (Table 1). These findings provide robustness to the lung transcriptional network, at least in part, and represent a potentially important tool to prioritize candidate genes underlying pneumonia susceptibility QTLs.
Table 1.
Lung transcriptional network genes physically located within the chromosome 7 Spir1 quantitative trait locus support interval.
| Module | Gene symbol | Fold changea |
|---|---|---|
| IL17A/IL17F signaling | Zfp60 | −2.32 |
| NF-κB signaling | Zfp36 | 4.89 |
| NF-κB signaling | Nfkbib | 6.94 |
| Complement system | B3gnt8 | −2.04 |
| Complement system | Papln | 1.28 |
| Interferon signaling | Ltbp4 | −1.26 |
| Interferon signaling | Fbln1 | −1.95 |
| Interferon signaling | Zfp365 | −1.49 |
| Interferon signaling | Zfp607 | 1.13 |
| Interferon signaling | Mia3 | −1.63 |
| Interferon signaling | Fbxo17 | −1.01 |
| Interferon signaling | Sars2 | 0.87 |
| Interferon signaling | Spred3 | −1.09 |
Fold change determined by comparing lung transcriptional profiles of 6-h pneumococcal pneumonia to uninfected samples.
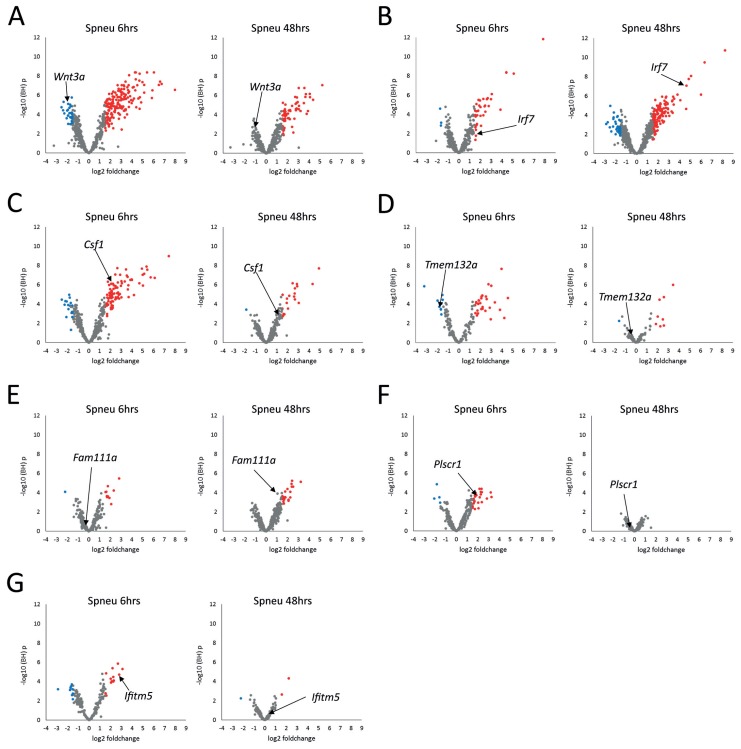
To gain unbiased insight into the underlying regulatory features of each module, we assessed the intramodular connectivity (number of pair-wise correlations) of each gene, as previously described (21,22,31). Genes presenting high intramodular connectivity are understood to represent focal points of module architecture and activity (32,33). The top intramodular driver genes coupled with panmodule expression kinetics at 6 h and 48 h postinfection are depicted in Figure 5. Myeloid cell IL17A/IL17F, interferon and NF-κB signaling modules were anchored at Wnt3a, encoding a cell fate and differentiation wingless-type MMTV integration site family member; Irf7, encoding an interferon response transcription factor; and Csf1, encoding a colony-stimulating factor that controls the production, differentiation and function of innate immune cells, respectively (Figures 5A–C). The lymphoid cell complement system module was anchored at Fam111a, encoding a family with sequence similarity 111 member and transcription regulator (Figure 5D). The stromal cell lipid metabolism module was anchored at the DNA-binding Plscr1, encoding phospholipid scramblase 1 (Figure 5F). Taken together these results suggest the early (6-h postinfection) host transcriptional response was substantially driven by granulocyte-associated modules directed by IL17A/IL17F and NF-κB signaling and regulated by cell fate, differentiation and proliferation driver genes (Wnt3a and Csf1, Figures 5A, C). The late (48-h postinfection) host transcriptional response was predominantly shaped by macrophage-associated interferon signaling and γ-delta T-cell–associated complement system activation, both putatively regulated at the pretransciptional level by transcription factors (Irf7 and Fam111a, Figure 5B, G).
Figure 5.
To gain insight into the regulatory features of the host pulmonary modular transcriptome, driver genes were identified on the basis of their intramodular connectivity (number of correlating genes within the same module). Driver genes were highlighted in their respective module-specific volcano plots (integrating BH p values and fold changes) at both 6 h (Spneu 6 h) and 48 h (Spneu 48 h) postinfection. (A) IL17A/IL17F signaling module (mod2); (B) interferon signaling module (mod7); (C) NF-κB signaling module (mod3); (D) undetermined pathway module (mod1); (E) complement system module (mod5); (F) lipid metabolism module (mod6); (G) undetermined pathway module (mod9). Red dots denote genes presenting BH p < 0.05 and log2 fold change >1.5; blue dots denote genes presenting BH p < 0.05 and log2 fold change <−1.5.
DISCUSSION
In this study a mouse model of early and late acute pneumococcal pneumonia was employed to construct a scale-free modular network of host pulmonary transcriptional responses to infection. This network encompassed modules of highly interconnected transcripts anchored at potentially crucial module driver genes. Modules were determined to be enriched for genes involved in distinct biological pathways, which included IL17A/IL17F signaling, NF-κB signaling, interferon signaling, lipid metabolism and the complement system pathway. These modules were predicted to constitute gene expression patterns attuned to shared and distinct cell-types including myeloid cells, such as granulocytes and macrophages, lymphoid cells, which included α-β and γ-delta T cells, as well as stromal cells. The early immune response phase was predominantly associated with an increase in granulocyte (IL17A/IL17F and NF-κB signaling) and stromal cell–associated (lipid metabolism) pathways, whereas the late phase of the host immune response was substantially impacted by macrophage (interferon signaling) and lymphoid cell–associated (complement system) pathways. Regulatory driver genes of the host pulmonary defense response included the cell fate, differentiation and proliferation genes, Wnt3a and Csf1, the transcriptional factors Irf7 and Fam111a, a glucose-regulated protein binding factor, Tmem132a, a phospholipid scramblase, Plscr1, and an interferon-induced transmembrane protein, Ifitm5. These findings suggest a previously undisclosed, yet potentially integral role of these network driver genes in the host lung response to Spneu infection.
Components of complex systems, such as the cellular and tissue responses to infectious pathogens, are not randomly connected. Signal transduction pathways that are activated upon pathogen recognition by the innate immune system follow an intricately coordinated path converging to specific nodes or hubs; therefore, they can be considered as scale free in character (9). This is exemplified by the toll-like receptor (TLR)–initiated signal transduction pathway that primarily culminates in the activation of the master transcription factor NF-κB. The biological importance of such networks has been highlighted by evidence of bacterial virulence factors directly targeting network hub components to suppress the hosts’ immune response (10). In the same fashion therapeutic agents, either in isolation or in combination with antibiotics as adjunctive therapies, have been proposed to treat bacterial infections; for example, the innate defense-regulator peptide-1 (IDR-1) for protection against gram-positive and gram-negative pathogens, including methicillin-resistant Staphylococcus aureus (MRSA) (34). The nonstochastic nature of the hosts’ defense mechanisms against bacterial infection (as well as other infectious pathogens) permits the emergence of immunomodulatory strategies. Although bacterial pathogens have evolved such immunomodulating mechanisms by selection pressures imposed by the hostile host environment, our knowledge of such host response topologies in the primary site of infection is lacking in comparison. To the aim of garnering a systems-based viewpoint of the host lung before and after 6- and 48-h pneumococcal infection, we utilized a weighted gene coexpression approach (21,22) to construct a scale-free network representative of the whole lung (Figure 3). This approach allowed us to define 13 transcriptional modules, of which 7 modules were significantly associated with distinct biological signaling pathways. Five of these functionally annotated modules, including the well-established protective host pathways IL17A/IL17F signaling (35), NF-κB signaling (36) and the complement system (37), were also, not surprisingly, significantly altered due to pneumococcal infection (Figure 4). Six network modules were not significantly associated with known biological pathways (Figure 3), which suggests biological interactions of these module genes have yet to be experimentally revealed. Moreover, two of these nonannotated modules (mod1 and mod9; Figures 5D, G) were significantly altered due to Spneu infection and predicted to be driven by Tmem132a (mod1) and Ifitm5 (mod9). These nonannotated modules (mod1 and mod9) and a stromal cell lipid metabolism module (Figure 5F) were uniquely significant to the early (6-h) host pulmonary response (Figure 4). Of note, the putative driver gene for the stromal cell lipid metabolism module, Plscr1, encoding phospholipid scramblase 1, was recently unmasked as a mediator of the type-I interferon–induced protection against staphylococcal α-toxin in the human epithelial A549 cell line (38).
Host responses to infectious or any other deleterious agent are characterized by an intricate pattern of cell mobilization, differentiation, sacrifice and, as important, cell-to-cell communication. Whole-tissue transcriptomics during infection, as employed in this study, certainly presents a challenge in interpretation because differential gene expression is often the result of cell influx into the tissue rather than a bona fide transcriptional activation or repression. However, tissue-wide transcriptional profiling coupled with current knowledge-based analytical tools can provide a unique opportunity to delineate biological functions and canonical signaling pathways pertaining to predicted cell-type–specific phenotypes in vivo. This approach, which circumvents artifacts that may occur as a consequence of cell sorting, has enabled us to map the transcriptional modules to various shared and distinct cell-types (Figure 3B). Although these results showed predominantly nonspecific mapping to cell-types, for example, the IL17A/IL17F signaling module was enriched for gene markers of granulocyte, dendritic cell, macrophage, monocyte and B-cell types (Figure 3B). Such non-specific mapping to cell subsets may be due to transcriptional network modules harboring submodules that might in turn map to specific cell-types, or certain biological pathways, for example NF-κB signaling (Figures 3A, B), are shared across cell subsets (39).
Host defense against pathogenic microbes (and other parasites) constitutes two primary components of conceptually different mechanisms (40). The first is defined as the ability of the host to limit microbial burden, termed resistance. The second is defined as the ability of the host to limit disease severity induced by a given microbial burden, which is termed tolerance (40). Notably, resistance has been defined as the inverse of pathogen burden (40,41). Through our systems-based approach we showed an inverse of lung bacterial burden (Figure 1E) and transcriptional modules enriched for interferon-signaling genes and, not surprisingly, complement system genes during the late phase of the host pulmonary response (Figure 4). Moreover, we predict macrophages (interferon signaling; Figures 3B and 5B) and γ-delta T cells (complement system pathway; Figures 3B and 5E) as key mediators. Monocytes, macrophages, dendritic cells and their precursors are understood to form networks of phagocytic cells, altogether called the mononuclear phagocyte system, which play important roles in development, inflammation and antipathogen defense (42, 43). γ-Delta T cells have been proposed as key mediators in the immune response by virtue of their interactions with antigen-presenting cells (44). Our results provide further indication for an intricate network of macrophage and γ-delta T-cell cross-talk, primarily via interferon signaling and complement system pathways, in the resistance phase of the host defense response, perhaps also driving the differentiation of progenitor/ precursor cells in the mononuclear phagocytic niche. Indeed, recent and past research highlighted the importance of complement factors in promoting the differentiation of myeloid-derived suppressor cells (45) and modulating T-cell responses (46, 47). Thus, our findings that show one module overrepresented for both the complement system and (γ-delta) T-cell markers (Figure 3) lends weight to the concept of nonconventional pleiotropic effects of complement system signaling.
Our study has limitations. The lung transcriptional network during early and late pneumococcal pneumonia was constructed using a single mouse strain (C57Bl/6). Although we provide initial validation of the transcriptional modules in a BALB/c mouse model, further exploration of the lung transcriptome during pneumococcal pneumonia in other mouse strains is needed. Moreover, our model relied on only one S. pneumoniae serotype strain. Evaluation of the lung transcriptional response to early and late pneumococcal pneumonia using other virulent S. pneumoniae strains is certainly warranted. Lastly, the mice used for this study were relatively young (9–12 wks old), considering that the majority of pneumococcal pneumonia cases are in elderly humans.
CONCLUSION
Here, we report an unbiased investigation of the host pulmonary transcriptional alterations that occur as a consequence of acute pneumococcal infection coupled with immunohistochemical analysis. By means of systems-based genomics and bioinformatics approaches we constructed a dynamic network of modular transcriptional programs that shape the early and late phases of the host defense response. Early host responses to pneumococcal pneumonia were characterized by heightened expression of granulocyte-associated IL17A/IL17F and NF-κB signaling, established as protective mechanisms during Spneu infection, as well as a novel stromal cell lipid metabolism pathway. The late host responses were characterized by a γ-delta T-cell complement system and macrophage interferon signaling.
ACKNOWLEDGMENTS
We thank Joost Daalhuisen and Regina de Beer for expert technical assistance and the animal caretakers of the AMC animal research core facility.
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors declare they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
Cite this article as: Scicluna BP, van Lieshout MH, Blok DC, Florquin S, van der Poll T. (2015) Modular transcriptional networks of the host pulmonary response during early and late pneumococcal pneumonia. Mol. Med. 21:430–41.
REFERENCES
- 1.O’Brien KL, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 2.Mandell LA, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44:S27–S72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramirez JA, Anzueto AR. Changing needs of community-acquired pneumonia. J Antimicrob Chemother. 2011;66(Suppl 3):iii3–iii9. doi: 10.1093/jac/dkr094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viasus D, Garcia-Vidal C, Carratalà J. Advances in antibiotic therapy for community-acquired pneumonia. Curr Opin Pulm Med. 2013;19:209–215. doi: 10.1097/MCP.0b013e32835f1c0b. [DOI] [PubMed] [Google Scholar]
- 5.van der Poll T, Opal SM. Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet. 2009;374:1543–1556. doi: 10.1016/S0140-6736(09)61114-4. [DOI] [PubMed] [Google Scholar]
- 6.Kadioglu A, Weiser JN, Paton JC, Andrew PW. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol. 2008;6:288–301. doi: 10.1038/nrmicro1871. [DOI] [PubMed] [Google Scholar]
- 7.Hancock RE, Nijnik A, Philpott DJ. Modulating immunity as a therapy for bacterial infections. Nat Rev Microbiol. 2012;10:243–254. doi: 10.1038/nrmicro2745. [DOI] [PubMed] [Google Scholar]
- 8.Opitz B, van Laak V, Eitel J, Suttorp N. Innate immune recognition in infectious and noninfectious diseases of the lung. Am J Respir Crit Care Med. 2010;181:1294–1309. doi: 10.1164/rccm.200909-1427SO. [DOI] [PubMed] [Google Scholar]
- 9.Barabási AL. Scale-free networks: a decade and beyond. Science. 2009;325:412–413. doi: 10.1126/science.1173299. [DOI] [PubMed] [Google Scholar]
- 10.Brodsky IE, Medzhitov R. Targeting of immune signalling networks by bacterial pathogens. Nat Cell Biol. 2009;11:521–526. doi: 10.1038/ncb0509-521. [DOI] [PubMed] [Google Scholar]
- 11.Dessing MC, Florquin S, Paton JC, van der Poll T. Toll-like receptor 2 contributes to antibacterial defence against pneumolysin-deficient pneumococci. Cell Microbiol. 2008;10:237–246. doi: 10.1111/j.1462-5822.2007.01035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lammers AJ, et al. Enhanced vulnerability for Streptococcus pneumoniae sepsis during asplenia is determined by the bacterial capsule. Immunobiology. 2011;216:863–870. doi: 10.1016/j.imbio.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 13.van Lieshout MH, Scicluna BP, Florquin S, van der Poll T. NLRP3 and ASC differentially affect the lung transcriptome during pneumococcal pneumonia. Am J Respir Cell Mol Biol. 2014;50:699–712. doi: 10.1165/rcmb.2013-0015OC. [DOI] [PubMed] [Google Scholar]
- 14.Dunning MJ, Smith ML, Ritchie ME, Tavare S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- 15.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi W, Oshlack A, Smyth GK. Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res. 2010;38:e204. doi: 10.1093/nar/gkq871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kauffmann A, et al. Importing ArrayExpress datasets into R/Bioconductor. Bioinformatics. 2009;25:2092–2094. doi: 10.1093/bioinformatics/btp354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbosa-Morais NL, et al. A re-annotation pipeline for Illumina BeadArrays: improving the interpretation of gene expression data. Nucleic Acids Res. 2010;38:e17. doi: 10.1093/nar/gkp942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 21.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 23.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 24.Jonczyk MS, et al. Genetic factors regulating lung vasculature and immune cell functions associate with resistance to pneumococcal infection. PLoS One. 2014;9:e89831. doi: 10.1371/journal.pone.0089831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heng TS, Painter MW Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasprzyk A. BioMart: driving a paradigm change in biological data management. Database. 2011;2011:bar049. doi: 10.1093/database/bar049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 30.Ravaz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 31.Scicluna BP, et al. Role of tumor necrosis factor-α in the human systemic endotoxin-induced transcriptome. PLoS One. 2013;8:e79051. doi: 10.1371/journal.pone.0079051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horvath S, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A. 2006;103:17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oldham MC, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott MG, et al. An anti-infective peptide that selectively modulates the innate immune response. Nat Biotechnol. 2007;25:465–472. doi: 10.1038/nbt1288. [DOI] [PubMed] [Google Scholar]
- 35.Tsai HC, Velichko S, Hung LY, Wu R. IL-17A and Th17 cells in lung inflammation: an update on the role of Th17 cell differentiation and IL-17R signaling in host defense against infection. Clin Dev Immunol. 2013;2013;267971 doi: 10.1155/2013/267971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang WY, Lim JH, Li JD. Synergistic and feedback signaling mechanisms in the regulation of inflammation in respiratory infections. Cell Mol Immunol. 2012;9:131–135. doi: 10.1038/cmi.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruyn GA, Zegers BJ, van Furth R. Mechanisms of host defense against infection with Streptococcus pneumoniae. Clin Infect Dis. 1992;14:251–262. doi: 10.1093/clinids/14.1.251. [DOI] [PubMed] [Google Scholar]
- 38.Lizak M, Yarovinsky TO. Phospholipid scramblase 1 mediates type i interferon-induced protection against staphylococcal α-toxin. Cell Host Microbe. 2012;11:70–80. doi: 10.1016/j.chom.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayden MS, Ghosh S. NF-κB in immunobiology. Cell Res. 2011;21:223–244. doi: 10.1038/cr.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider DS, Ayres JS. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol. 2008;8:889–895. doi: 10.1038/nri2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Råberg L, Sim D, Read AF. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science. 2007;318:812–814. doi: 10.1126/science.1148526. [DOI] [PubMed] [Google Scholar]
- 42.Gordon S. Pattern recognition receptors: doubling up for the innate immune response. Cell. 2002;111:927–930. doi: 10.1016/s0092-8674(02)01201-1. [DOI] [PubMed] [Google Scholar]
- 43.Jenkins SJ, Hume DA. Homeostasis in the mononuclear phagocyte system. Trends Immunol. 2014;35:358–367. doi: 10.1016/j.it.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Scotet E, Nedellec S, Devilder MC, Allain S, Bonneville M. Bridging innate and adaptive immunity through gammadelta T-dendritic cell crosstalk. Front Biosci. 2008;13:6872–6885. doi: 10.2741/3195. [DOI] [PubMed] [Google Scholar]
- 45.Hsieh CC, et al. The role of complement component 3 (C3) in differentiation of myeloid-derived suppressor cells. Blood. 2013;121:1760–1768. doi: 10.1182/blood-2012-06-440214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lalli PN, et al. Locally produced C5a binds to T cell-expressed C5aR to enhance effector T-cell expansion by limiting antigen-induced apoptosis. Blood. 2008;112:1759–1766. doi: 10.1182/blood-2008-04-151068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strainic MG, et al. Locally produced complement fragments C5a and C3a provide both costimulatory and survival signals to naive CD4+ T cells. Immunity. 2008;28:425–435. doi: 10.1016/j.immuni.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]