Summary
Mechanosensitive channels protect bacteria against lysis caused by a sudden drop in osmolarity in their surroundings. Besides the channel of large conductance (MscL) and small conductance (MscS), Escherichia coli has five additional paralogs of MscS that are functional and widespread in the bacterial kingdom. Here, we present the structure of YnaI by cryo-electron microscopy to a resolution of 13 Å. While the cytosolic vestibule is structurally similar to that in MscS, additional density is seen in the transmembrane (TM) region consistent with the presence of two additional TM helices predicted for YnaI. The location of this density suggests that the extra TM helices are tilted, which could induce local membrane curvature extending the tension-sensing paddles seen in MscS. Off-center lipid-accessible cavities are seen that resemble gaps between the sensor paddles in MscS. The conservation of the tapered shape and the cavities in YnaI suggest a mechanism similar to that of MscS.
Keywords: bacteria, cryo-electron microscopy, fluorescence, osmotic stress, protein-lipid interaction
Graphical Abstract

Highlights
-
•
First structure of a large member of the MscS family of mechanosensitive channels
-
•
The density map indicates extended sensor paddles of tilted transmembrane helices
-
•
Lipids access the core of YnaI and fill cavities which are similar to gaps in MscS
-
•
These cavities and the tilted helices could lay the basis for tension sensing
Mechanosensitive channels of the MscS family are found in all kingdoms of life and protect bacteria against osmotic shock. Böttcher et al. provide the first structural insights into a larger family member, YnaI. Despite YnaI having additional transmembrane helices, important features are conserved, suggesting a common mechanism.
Introduction
Bacterial cells are directly exposed to the environment, and therefore must be able to adapt to sudden and drastic changes in external solute composition. Sudden decreases of osmolyte concentrations in the environment will cause a hypo-osmotic shock. Water is drawn into the cell with its higher solute concentrations, and consequently the internal pressure will increase to dangerous levels. In response, bacteria sense the increased turgor pressure as increased membrane tension and gate mechanosensitive (MS) channels release solutes. This relieves the pressure, preventing cell death.
Multiple MS channel genes are often found within a single species. Escherichia coli, for example, has seven MS channels: MscL, MscS, and five further homologs of MscS (Edwards et al., 2012; Li et al., 2002, 2007; Schumann et al., 2010). It is likely that this functional redundancy is required for a graded emergency response mechanism that minimizes the unspecific release of solutes and keeps the disturbance of homeostasis to a minimum. At tensions close to the rupture of the membrane, MscL is activated and forms the largest pore, whereas at lower tensions MscS is triggered, which has much smaller conductance.
Crystal structures of MscS (Bass et al., 2002; Lai et al., 2013; Pliotas et al., 2012; Wang et al., 2008; Zhang et al., 2012) show that the channel is formed by seven subunits. Each of the subunits contributes three transmembrane (TM) helices, TM1–3. TM1 and TM2 form a flexible paddle that is likely to be involved in tension sensing. TM3 is a long kinked helix, which consists of the TM pore-forming part, TM3a, followed by TM3b that links the membrane part with a large cytosolic domain. This domain forms an enclosed vestibule at the entrance of the pore.
Sequence alignments show that all homologs of MscS in E. coli have an N-terminal extension while the cytosolic domains have a similar size except for YbdG, which has a small insertion within the cytosolic domain (Schumann et al., 2010). Analysis of the sequences also indicates that these N-terminal extensions form TM helices in addition to the three found in MscS. YnaI and YbdG are predicted to contain two additional TM helices, whereas MscK, YjeP, and YbiO are predicted to have eight additional helices (Krogh et al., 2001). It is not fully understood whether these additional TM helices isolate the sensor paddle from direct contact to the lipids, provide access routes for lipids to the sensor paddle (Booth et al., 2011) or even extend the paddle.
Although it is generally agreed that the paddles must be important for tension sensing and rotationally rearrange giving rise to different conformations of MscS (Wang et al., 2008), it is not clear how they are actually sensing the tension in the membrane. Recently, we demonstrated that lipids can intercalate between the paddles in MscS, which may be important for the sensing mechanism (C. Pliotas et al., personal communication). In addition, it was proposed that gating could be driven by a change of local membrane curvature due to the tilted paddles (Phillips et al., 2009). However, it is unclear whether these mechanisms are transferable to the larger homologs.
Here, we present a first glimpse at the structure of one of the larger homologs, YnaI, which suggests that the additional N-terminal helices may induce membrane curvature and may move and act as an extended sensor paddle. Furthermore, we obtained evidence that the core of the membrane domain is fully accessible to lipids, and saw lipid-filled pockets comparable with the pockets seen between the sensor paddles in MscS. We found that the tryptophan W184 at the portals of the cytosolic domain is required for function.
Results
Cryo-Electron Microscopy of YnaI
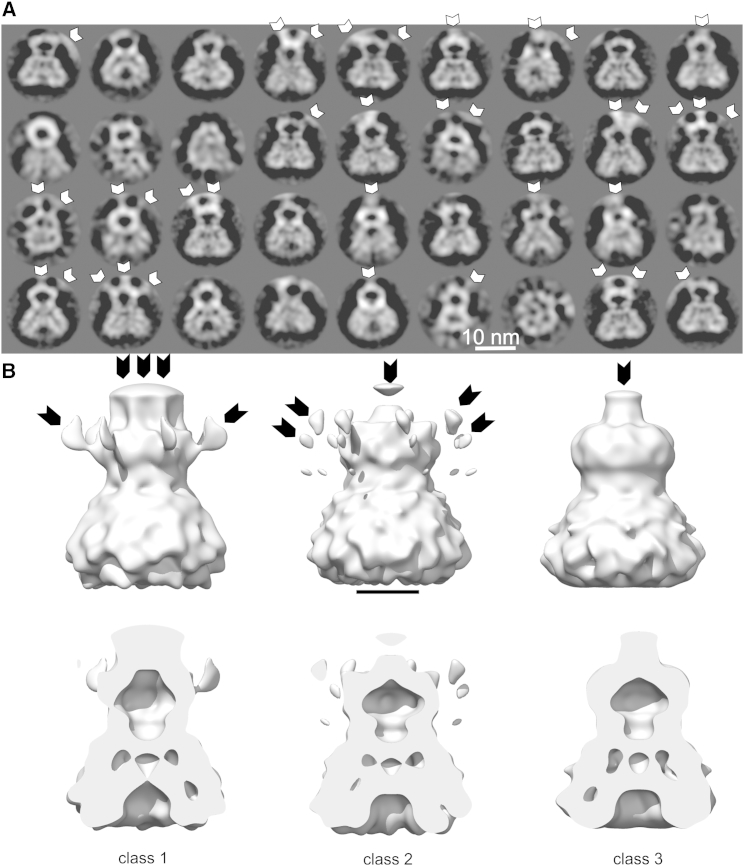
Electron micrographs showed that YnaI aggregated at one site and had very little overlap to neighboring particles in the smaller aggregates (Figure S1, arrows). We selected a total 34,700 of YnaI channels, of which 18,985 particle images grouped into well-defined classes in 2D classification. This subset was used in further analysis. The 2D class averages showed predominantly side views with two distinct moieties of different sizes (Figure 1). The smaller moiety had a distinct central cavity, which resembled the cytosolic domain of MscS and contained the major contact sites to other particles in the aggregates (Figure 1A, white arrowheads).
Figure 1.
Class Averages of Vitrified YnaI Samples
(A) 2D class averages of vitrified YnaI. The class averages were calculated with Relion. Only those class averages are shown in which the particle images could be aligned accurately. White arrowheads mark contact sites to neighboring particles in the aggregates.
(B) Three 3D class averages. The upper row shows a surface representation of the full class average; the lower row shows the same view, with the front half of the particle cut away. Arrows indicate extra density due to contacts with neighboring particles. The scale bar represents 5 nm.
See also Figure S1.
Next, we calculated a 3D map using a previously determined map of negatively stained particles as starting reference (see Supplemental Information; Figure S1). To identify those particles that were least disturbed by the aggregation, we performed a 3D classification of the aligned vitrified particles into three classes. All three 3D classes showed a similar shape of YnaI with extensions of different sizes at the top and at the side of the cytosolic domain (Figure 1B, arrows). These extensions were attributed to neighboring particles in the aggregates. On the inside (Figure 1B), all three classes had a similar distribution of cavities, with the cavity of the cytoplasmic vestibule clearly resolved and distinct cavities in the center and at the side of the membrane part. For further refinement, we used the particle images of class 3 (7,202 particles) that had the least additional density from aggregation (Figure 1B, black arrows). The refined map of class 3 (Figure 2; accession number EMDB: 3035) had a resolution of 12.6 Å (Figure S3).
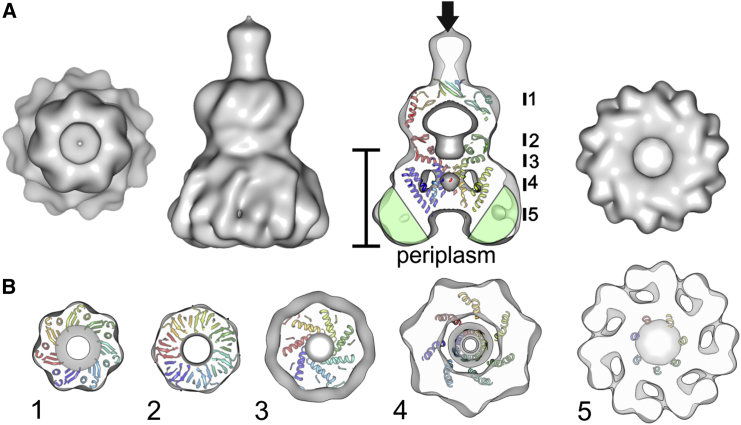
Figure 2.
Surface Representation of Vitrified YnaI with the Fitted Structure of MscS
(A) Different views of the surface representation of YnaI. From left to right: top view; side view; slice along the central, long axis of the side view; bottom view. The arrow indicates the major contact site in the aggregates. The ribbon presentation shows the fitted MscS (PDB: 2OAU; Bass et al., 2002). The green semi-circles with a diameter of 5 nm highlight the likely position of the detergent micelle. The upright scale bar represents 7.5 nm and highlights the length of the membrane part of YnaI.
(B) Several 1-nm thick slices of YnaI are shown parallel to the plane of membrane with the structure of MscS (PDB: 2OAU) fitted. The position of the slices is indicated with black bars in the central slice shown in (A).
See also Figure S4.
Structure of the Cytosolic Domain
The structure of YnaI shows on one side a large vestibule, which had an overall cross-correlation of 0.7 to the cytosolic domain of MscS (see Figure S4 for local cross-correlation). This domain provides in MscS stability (Rasmussen et al., 2007; Schumann et al., 2004) and selectivity (Maksaev and Haswell, 2012, 2013; Zhang et al., 2012), and is conserved between different channels of the MscS family. For structural comparison, we fitted the structure of MscS in the non-conducting state (PDB: 2OAU; Bass et al., 2002) into the electron microscopy (EM) map of YnaI (Figure 2). The fit showed excellent agreement of the cytosolic vestibule, but did not resolve the portals and the openings in the β-barrel structure of the vestibule. Furthermore, the EM map of YnaI had unaccounted density beyond the C-terminal β barrel of MscS (Figure 2A, arrow). This unaccounted density is most likely built by the seven C-terminal residues of MscS that are not resolved in the crystal structure plus the C-terminal purification tag of YnaI and density from the neighboring particles in the YnaI aggregates.
Structure of the Membrane Domain
The membrane domain in YnaI was funnel shaped. The cytosol-facing site was narrower and had the same diameter (8 nm) as that of MscS. The diameter at the periplasm was 12.5 nm, compared with 5.5 nm in MscS. The total length of the membrane domain perpendicular to the plane of membrane was 7.5 nm, which was much larger than the 4.5 nm in MscS (Figure 2A). This difference in size was probably due to the two additional TM helices of YnaI and the tightly bound detergent micelle (Figure 2A, green semi-circles of 5 nm diameter indicate the extent of a typical dodecyl maltoside micelle) that are absent in the crystal structure of MscS.
The EM map of YnaI revealed a hollow cylinder in the center of the membrane part, which formed a distinct pore at the inside that was obstructed at both the cytosolic (Figure 3, red arrow) and periplasmic side (Figure 3, blue arrow). In MscS the central pore is formed by the seven tightly packed TM3 helices, which fit well into the hollow cylinder in their closed conformation (PDB: 2OAU; Bass et al., 2002), but are not accommodated in their open conformation (PDB: 2VV5; Wang et al., 2008), suggesting that YnaI was in a closed conformation. In the closed state, the pore of MscS is sealed by the hydrophobic residues L105 and L109 (corresponding L154 and M158 in YnaI), which aligned well with the obstruction of the pore at the cytosolic site of YnaI.
Figure 3.
Slices of the EM map of YnaI in Comparison with Fitted Structures of Closed and Open MscS
The EM map of the fitted closed channel of MscS (PDB: 2OAU; Bass et al., 2002; magenta) and fitted open channel of MscS (PDB: 2VV5; Wang et al., 2008; blue) are shown. The left slice is parallel to the symmetry axis; the right slice is perpendicular to the symmetry axis at the position indicated on the right panel. The inner ring of helices, which forms the central pore of the open channel (blue) in MscS, is not accommodated by the EM density, whereas the inner ring of the closed channel (magenta) is accounted for by the EM density. The positions L105 and L109 of the hydrophobic seal of MscS are indicated by balls and sticks. The green arrow indicates the off-center membrane-embedded cavities that are also seen in the crystal structure of MscS. The red arrow highlights the cytosolic constriction, and the blue arrow the periplasmatic constriction of the pore.
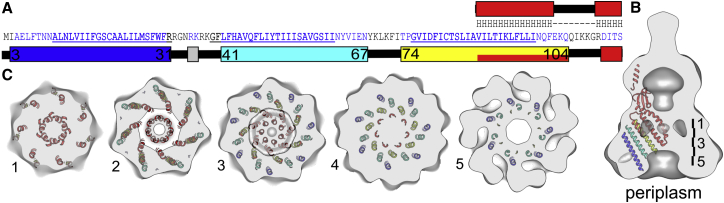
Sequence alignment of MscS and YnaI showed that YnaI has a 49-amino-acid long extension at the N terminus. Furthermore, structures of MscS lack the first 25 N-terminal amino acids. This N-terminal region of MscS does not align with the sequence of YnaI, and predictions of its structure have been made on the basis of spectroscopic data and simulations (Anishkin et al., 2008; Vásquez et al., 2008). Based on secondary structure prediction by JPRED, YnaI had two more N-terminal helices (residues 3–31 and 41–67; Figure 4A). These predicted helices largely overlapped with predictions of TM helices between 10–32 and 39–61 using TMHMM (Krogh et al., 2001).
Figure 4.
Structure Prediction for YnaI
(A) Secondary structure prediction of the N-terminal region (1–114) of YnaI. The positions of the predicted TM helices (TMHMM) are underlined. The prediction in the upper row is based on homology modeling with the Swiss-Model server. Prediction in the lower row is done with JPred. Longer helical regions are highlighted as squares.
(B) Homology model of YnaI (red) fitted into the EM map of YnaI. The predicted transmembrane helices in the N-terminal region are shown in blue (TM1), cyan (TM2), and yellow (TM3). The placement is arbitrary and aims at visualizing the possible extent of YnaI. The placement is based on assuming a tilt similar to the helices in the structure of MscS, keeping the helices within the density of the YnaI map and leaving continuous space at the likely position of the detergent micelle.
(C) 1-nm thick slices through the membrane part of YnaI with the symmetry-related copies. The approximate positions of the slices are indicated in (B).
To illustrate the possible contribution of the N-terminal helices to the structure of YnaI, we have placed a homology model of YnaI together with the predicted N-terminal helices in the EM map (Figures 4B and 4C). Since we cannot resolve the helices in our EM map, we have assumed that the additional helices were similarly staggered and tilted as the helices are in the structure of MscS, and that the ends of the N-terminal helices were close to the surface of the EM map of YnaI. Our illustration accounts for most of the observed EM density apart from a belt of fragmented density at the periplasmic side, which we think is accounted for by the detergent micelle.
The Core TM Helices Are Accessible to Lipids
The off-center cavities in the membrane domain of YnaI (Figure 3; green arrow) resembled crevices seen in the crystal structures of MscS between the sensor paddles and TM3. Recently, we proved lipid penetration into these crevices in MscS and hypothesized that this could also be important for the sensing of membrane tension (C. Pliotas et al., personal communication). In YnaI, these cavities are shielded by 14 more helices than in MscS. To test whether this had any impact on the accessibility to lipids, we used collisional quenching of tryptophan fluorescence by bromine atoms attached to fatty acid chains of the lipids (Kasha, 1952). Fluorescence quenching occurs if the tryptophan is in van der Waals contact with the bromine atoms in the lipids, and thus reports lipid contact for a particular location on a membrane protein (East and Lee, 1982). We constructed a mutant of YnaI where the five tryptophan residues were replaced by phenylalanine (Δ5W), allowing introduction of new tryptophan residues as probes. Based on sequence alignment of YnaI to MscS, we chose three residues that were likely to line the cavities: G152W, F168W, and the native tryptophan W201 (Δ4W). These positions were homologous to A103W (TM3a), A119W (TM3b), and I150W in E. coli MscS, respectively (Figures 5A and 5D). We showed that G152W and F168W were highly accessible to lipids (Figure 5C) as were the corresponding residues in MscS (C. Pliotas et al., personal communication). YnaI W201 had limited accessibility for lipids, similarly to the corresponding MscS I150W. Emission spectra of MscS and YnaI samples solubilized in n-dodecyl-β-D-maltopyranoside (DDM) showed peak positions that correlated well with the lipid accessibility (Figure 5B). The fatty acid tails of the phospholipids are predominantly located where the tryptophan probes indicate a low-polarity environment. There is good agreement of fluorescence properties between YnaI and MscS for these three residues, suggesting conservation of the lipid-filled cavities next to the pore-forming helices in these two proteins as indicated by the structural similarity of the EM map of YnaI. However, YnaI Δ5W L154W did not follow this trend. L154W was highly quenched, indicating greater lipid accessibility than its MscS counterpart, the pore-facing L105W (C. Pliotas et al., personal communication). Moving just two residues along the helix, however, restores the behavioral correlation, as G152W fluorescence data agree well with the MscS counterpart A103W. A possible explanation for the anomaly of the L154W data may be provided by the glycine-rich sequence GGIGG (Figure 5D) adjacent to L154W in YnaI. This sequence could introduce high flexibility and enable accessibility to lipids, e.g. by partial unwinding of the helix in YnaI.
Figure 5.
Lipid Accessibility of the TM Core Region of YnaI
(A) Crystal structure of MscS (PDB: 2OAU; Bass et al., 2002) showing the cavities between the “sensor paddles” and the pore-forming helices. For clarity only two subunits are shown (yellow and gray), but the pore-forming helices TM3a from all subunits to indicate the pore (light-blue; back), MscS A103W (red), and L105W (blue) on TM3a facing away or into the pore, respectively, are shown. A119W (green) on TM3b and I150W (magenta) protruding between TM3b helices into the cavities were used for fluorescence studies and homologs residues in YnaI identified by sequence alignment.
(B) Correlation of fluorescence properties between YnaI (red) and MscS (black). The peak position of emission spectra of detergent-solubilized samples (open circles) and the fractional quenching (F0 − F)/F0 of samples reconstituted into bilayers containing brominated or non-brominated lipids are shown (closed circles). Mean values with SD are indicated (n ≥ 3; see Table S1).
(C) Fluorescence emission spectra of YnaI mutants reconstituted into non-brominated lipids (black) or brominated lipids (colored according to homolog MscS residues in A).
(D) Sequence alignment between YnaI and MscS indicating the chosen residues for the fluorescence study. Conserved residues are shown in gray. An orange box indicates the position of tryptophan W184, which aligns with tyrosine Y135 of MscS.
Position of the First TM Relative to the Lipid Bilayer
Analysis of the sequence of YnaI (Krogh et al., 2001) and hydrophobicity plots suggested that YnaI had two additional TM helices N-terminal to the three conserved TM helices found in MscS. The fluorescence of one turn of the predicted first TM helix from V14W to F17W was investigated. All mutants showed high lipid accessibility, with the highest for V14W and the lowest for I16W (Figure 6A), confirming their location in the lipid bilayer. Emission peak positions for I15W and I16W were more red shifted than for V14W and F17W (Figure 6B), characteristic of a more hydrophilic environment. If a simple helical model is made (using PyMol v1.7.1), it can be seen that on both adjacent turns hydrophilic residues, N12 and S19, face in the same direction as I15W and I16W (Figure 6D). Therefore, it seemed likely that a helical structure was present and that this face of the helix was in contact with another TM helix, which is often promoted by formation of hydrogen bonds (Adamian and Liang, 2002). On this face of the helix, C20 and S27 may form additional hydrogen bonds with their side chains to neighboring helices. Lipids brominated at different positions along the fatty acid chain allow depth-dependent quenching experiments, which inform on the relative position of the tryptophan probe within the bilayer (Bolen and Holloway, 1990). Only F17W showed depth dependence in the quenching (Figure S5). The distribution analysis (Ladokhin, 1997, 2014) suggested that F17W was 7 ± 1 Å away from the membrane center (Figure 6C; dispersion σ = 3.4 ± 0.7 Ε; area parameter S = 12 ± 2), which agreed well with the predicted extend of the first TM helix from residues 10 to 32 (Figure 6D; Krogh et al., 2001).
Figure 6.
Fluorescence of YnaI V14W to F17W on the Proposed First TM Helix
(A) Lipid contacts were determined by reconstitution into brominated and non-brominated lipids. Fractional quenching (F0 − F)/F0 is shown as mean with SD.
(B) Emission peak positions of detergent-solubilized samples.
(C) Distribution analysis (blue line) of depth-dependent fluorescence quenching with three different brominated lipids for mutant F17W (black circles). Mean values with SD are shown (n = 5).
(D) Helical model (produced with PyMol) of the TM helix (residues 10–30) including the residues used for the fluorescence experiments. Hydrophilic residues are shown in orange. The prediction of this TM helix is in agreement with the results shown in (C) considering the positioning of F17W.
See also Figures S5, S6; Table S1.
Tryptophan W184 Is Required for Function
Wild-type (WT) YnaI provides less protection in osmotic downshock assays than MscS, requiring induction when expressed from the pTrc plasmid (Edwards et al., 2012). When assayed for function, the tryptophan mutants used for the fluorescence experiments in this study failed to protect cells against hypo-osmotic shock (Figure S6) while the corresponding MscS mutants retained functionality (C. Pliotas et al., personal communication). The similarity of spectroscopic properties between YnaI and MscS (Figure 5B) indicated that structural features are similarly independent of functionality. However, we sought to obtain evidence that structural conclusions drawn from our fluorescence experiments hold true for the active and native form of YnaI. Structures obtained from negatively stained particles of the mutants Δ5W G152W and F168W were similar to those of WT, indicating that these mutations do not cause a gross change in structure (Figure S7).
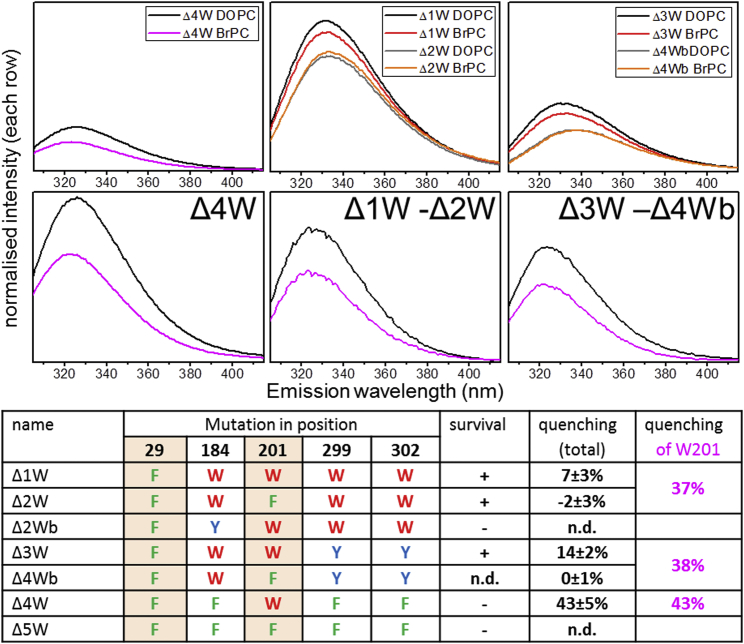
Several mutants of the native tryptophan residues were tested for functionality to better understand the cause of the loss of function. The double mutant W29F/W201F (Δ2W) is functional (Figure S6). These are the two tryptophan residues predicted to be located in the membrane domain. Δ2W shows no quenching by brominated lipids (Figure 7), consistent with the prediction that the three remaining tryptophan residues are located close together at the portals in the cytosolic domain away from the membrane. The related mutant W29F (Δ1W) is active and reports the quenching of tryptophan W201 by brominated lipids. The small quenching seen is consistent with that seen for W201 in the tryptophan-free background reported above (Δ4W) but with the constant, non-quenchable contributions from the three cytosolic tryptophan residues. Similarly, quenching of W201 is seen in the mutant Δ3W (Figure 7; see below).
Figure 7.
Quenching of W201 in Different Mutants and Properties of Tryptophan Mutants
Quenching of BrPC for the mutants Δ4W, Δ1W, Δ2W, Δ3W, and Δ4Wb are shown in the first row. For comparison of the quenching of W201 the constant, non-quenchable contributions of the cytosolic tryptophan residues were eliminated by subtracting spectra of Δ2W and Δ4Wb from Δ1W and Δ3W, respectively (second row). Fluorescence intensities in both rows are each normalized for protein concentration. The table on the bottom gives an overview of the used mutants, showing that W184 is required for function. The two tryptophan residues located in the membrane region have an orange background. The quenching by BrPC is given without and with correction for the fluorescence contributions of the cytosolic tryptophan residues.
See also Figures S6 and S7.
The mutant W29F/W184Y (Δ2Wb) is inactive while W29F/W299Y/W302Y (Δ3W) is active, which shows that W184 is required for function and cannot be substituted even by the conservative mutation to tyrosine (Figure S6). The role of tryptophan residues in YnaI and MscS are clearly different because the two residues of MscS in the cytosolic domain are located at the subunit interface and are important for complex stability (Rasmussen et al., 2007). Phenylalanine mutations can retain stability of the heptameric MscS complex, and function is not influenced (Rasmussen et al., 2007). On the other hand, the three tryptophan residues in the cytosolic domain of YnaI are not predicted to be at the subunit interface. All mutations we tested form stable heptameric complexes, but the function is lost by mutation of W184. The single tryptophan in MscS close to the membrane interface, W16, is functionally sensitive to mutation while this is not the case for the only tryptophan in YnaI on a TM helix, W29, as far as we can tell from our downshock assay. The lack of tryptophan residues at the membrane-water interfaces in MS channels is striking and has been proposed to be important for their function (Booth et al., 2007, 2011). Usually tryptophan residues serve as “anchors” to localize membrane proteins in the membrane (Killian and von Heijne, 2000).
In summary, we see similar quenching for W201 in two active mutants, although with constant fluorescence contributions from the cytosolic tryptophan residues overlaid, compared with W201 in the tryptophan-free background (Figure 7). Furthermore, the gross structure of two other mutants was similar to that of WT (Figure S7). Therefore, it seems reasonable to assume that the pockets seen in the membrane domain are also filled with lipids in the native YnaI. Interestingly, we saw a functional contribution of W184 at the portals of the cytosolic domain. MscS homologs often seem to have a tryptophan at this position while in the case of E. coli MscS itself, tyrosine Y135 is aligned (Figure 5D). W184 is far away from the membrane regions investigated by fluorescence in this study.
Discussion
We have reconstructed the 3D structure of YnaI at intermediate resolution. The map confirms that YnaI has a seven-fold symmetric organization similar to that of the related MscS, with an almost identical organization of the C-terminal cytosolic domain and a significantly larger membrane domain, which was consistent with the N-terminal extension of YnaI and the presence of a detergent micelle.
The size of the central pore in the EM map of YnaI agreed very well with the diameter of the pore formed by TM3a helices in closed structures of MscS. Furthermore, the seal region toward the cytosolic site of the pore has the highest sequence homology between YnaI and MscS, and the seal residues L154 and M158 are of similar size to L105 and L109 in MscS.
Electrophysiological characterization of YnaI showed very different properties of a conducting open state between YnaI and MscS (Edwards et al., 2012). YnaI opens less frequently and appears to require more pressure for activation, and the conductance is much smaller (Edwards et al., 2012). There may be several reasons for this. One could be the second constriction of the pore toward the periplasm in YnaI (Figure 3, blue arrow), which could stabilize the closed state and might restrict the pore size in the open state. For example, the large side chain of L142 could cause this second constriction. Another reason could be an extended sensor paddle (four TM instead of two in MscS), which forms a larger interaction interface with the lipids and so might require larger tension for changing the lipid content and, thus, for driving conformational switching. The off-center cavities in the EM map of YnaI agree in their position with cavities seen between TM3 and the sensor paddle in the crystal structures of MscS, suggesting that the inner core of the sensor paddle is structurally preserved despite the two additional TM helices in YnaI. Our fluorescence experiments on tryptophan mutants confirmed that three residues that are predicted to be exposed to the cavity in YnaI are similarly accessible to lipids as the corresponding residues in MscS.
Several MscS crystal structures from different organisms produced by different laboratories agree on structural features of the membrane domain (Bass et al., 2002; Lai et al., 2013; Wang et al., 2008; Zhang et al., 2012), which seem also be conserved in YnaI seen in our EM study. However, molecular dynamics (MD) simulations were performed by two independent groups who proposed that the gaps and tilted paddles are an artifact of crystallography and are not present in the active MscS in the membrane (Anishkin et al., 2008; Vásquez et al., 2008). One of these studies constrained its simulation on experimental continuous-wave electron spin resonance data, however, without distance measurements (Vásquez et al., 2008). Because of these contradicting views of the membrane domain structure, a follow-up study was performed in which distances of MscS reconstituted into membranes were measured by pulsed electron-electron double resonance (PELDOR) (Ward et al., 2014). These data agreed with the crystal structures and not with the MD simulations. Our recent fluorescence quenching data on MscS (C. Pliotas et al., personal communication) and YnaI in this study provide an independent experimental approach to the PELDOR experiments, showing that the gaps between the paddles are present and filled with lipids when these MS channels are located in the membrane. Thus, considering the currently available evidence, we believe that structural features suggested by the crystal structures are more likely to represent the active form of MscS and, indeed, homology of YnaI.
We noticed that the thickness of the membrane domain of YnaI perpendicular to the plane of the membrane was significantly larger than the typical width of a membrane and also as observed for MscS. We can exclude that this is caused by an extended N-terminal domain in the periplasm, as the predicted first TM helix showed strong lipid interaction and the depth-dependent fluorescence quenching agreed with the proposed location of this TM relative to the membrane. The highly charged loops between TM1 and TM2 and between TM3 and TM4 also give a good estimation for the length of the TM helices, which is similar to those in MscS. It therefore seemed unlikely that the membrane domain of the channel of YnaI had an extraordinarily large hydrophobic extent perpendicular to the plane of membrane that could lead to an atypical stretching of the membrane in the vicinity of the channels (Figure 8A). The likely position of the detergent micelle at the bottom of the channel suggested that the hydrophobic domain of the membrane-embedded part was further away from the cytosolic domain as in MscS (Figure 8B). This is probably caused by the outer ring of additional N-terminal helices in YnaI being vertically displaced toward the periplasm. A good indication for such a displacement is the fact that the diameter of the membrane part at the cytosol is the same in MscS and YnaI, which would be too small to accommodate the additional 14 N-terminal helices in YnaI at the same level close to the cytosol. However, toward the periplasm the diameter of YnaI increased, in contrast to MscS, providing plenty of space for an additional ring of helices, in agreement with a displacement toward the periplasm.
Figure 8.
Interaction of YnaI with the Membrane
Schematic drawing of how YnaI (gray) could shape the membrane. Size and shape of YnaI are based on the EM map. The hydrophobic core (orange) and headgroup regions (blue) of the membrane are indicated. The tensions in the membrane are likely to put forces onto the transmembrane part of YnaI. The direction of the forces is indicated by arrows.
(A) The large size of the membrane part could cause a local increase of the width of the membrane to bury the membrane part completely.
(B) However, the likely position of the detergent micelle suggests that the hydrophobic part is located further away from the cytosolic domain at the bottom of the channel.
(C) The tilt of the helices could induce a local curvature of the membrane as indicated.
The preserved off-center cavities of YnaI suggested that the TM helices were most likely tilted similarly to those in MscS. Such tilted helices would induce local tension in the membrane that could increase the local curvature of the membrane (Figure 8C), which is in agreement with simulations and models of MscS (C. Pliotas et al., personal communication) (Phillips et al., 2009; Sotomayor and Schulten, 2004).
In summary, we illustrate an unusual overall shape of the membrane domain of YnaI which is best explained with four similarly tilted TM helices that probably form an extended sensor paddle compared with that of MscS. The sensor paddle fully maintains lipid accessibility to the core of the channel. Similar to the proposal for MscS, changes in the curvature of the membrane could represent the tension-sensing mechanisms. The proposed extension of the sensor paddle would provide a larger lipid interaction interface, and thus would require larger tensions for changing the lipid content and thus for triggering the opening of the YnaI channel, as observed in patch-clamp experiments (Edwards et al., 2012). The presence of seven distinct “knobs” in this part of the EM map also supports the presence of seven defined sensor paddles, in contrast to an isolating ring of 14 evenly distributed TMs.
Experimental Procedures
Purification of YnaI
Purification of YnaI was performed as described in detail previously (Edwards et al., 2012). In brief, YnaI was expressed from a pTrc99A vector in E. coli strain MJF641 missing all seven genes of the known MS channels (Edwards et al., 2012). Expression was induced for 4 hr at 30°C with 0.8 mM isopropyl β-D-1-thiogalactopyranoside. Cell pellets were stored at −80°C until further use. Cells were lysed by a single passage through a French press, and membranes were collected by ultracentrifugation at 100,000 × g for 1 hr at 4°C. Membranes were then solubilized in 1% DDM (Glycon) for 1 hr at 4°C and were loaded onto a prepacked Ni-nitrilotriacetic agarose column (Sigma) using a C-terminal His6-tag on YnaI. After washing and elution, the peak fraction was further purified on a size-exclusion Superose 6 10/300 column (GE Healthcare). The MscS mutant YFF I150W was purified following a protocol very similar to that for YnaI and was described earlier (Rasmussen et al., 2010). A size-exclusion Superdex200 10/300 column (GE Healthcare) was used in this case.
Sample Preparation for EM
Quantifoil Grids (1.2/1.3) were glow discharged for 1 min in a Quorumtech mini sputter coater (Quorumtech SC7620) at 35 μA and used within 1 hr. Grids were mounted in a plunge freezer with an environmental chamber (Bellare et al., 1988). The chamber was humidified with two water-soaked sponges and kept at room temperature. 2 μl of concentrated YnaI solution (2.2 mg/ml) was applied to the grid and then blotted for 15 s with two layers of filter paper (Whatman No. 1). For consistent results the filter paper was kept in a humidified box for at least 1 hr before use. After blotting the grid was plunged into liquid ethane, which was cooled by a bath of liquid nitrogen. The ethane was prevented from freezing by a heating unit. Grids were transferred to grid boxes and stored in liquid nitrogen until use (within 2 months).
Electron Microscopy
EM was done with an FEI-F20 electron microscope with field emission gun and a TVIPS F416 4k × 4k CMOS camera. The microscope was operated at 200 kV, spot size 5, with a C2 condenser aperture and an objective aperture of 100 μm diameter. For data acquisition of negatively stained samples at room temperature, a standard room temperature holder was used. Vitrified samples were transferred with a Gatan 626 cryo-holder with a matching Gatan transfer station. Image acquisition was done semi-automatically with EM tools (TVIPS; see Supplemental Experimental Procedures for more details).
Image Processing
Particles were manually selected with e2boxer of the EMAN 2 package (Tang et al., 2007) using the auto-center option and windowed with a box size of 256 × 256 pixels. For each micrograph the defocus was determined with ctffind3 (Mindell and Grigorieff, 2003). Further image processing was done with Relion (Scheres, 2012) (for more details see Supplemental Information).
Fitting of Atomic Models to EM Maps
Fitting of the models of MscS (PDB: 2OAU [Bass et al., 2002] and 2VV5 [Wang et al., 2008]) was done with the “fit into map” option of Chimera (Pettersen et al., 2004). The homology model of YnaI was generated using the automated mode of the Swiss-Model server (Kiefer et al., 2009). The homology model of YnaI was placed into the EM map using the match maker option of Chimera and aligning the homology model to one of the chains of the fitted MscS cannel. The models of the individual helices (poly-Ala-chain of appropriate length) were generated with the “built” option of the structure-editing tool of Chimera. The helices were manually placed in the EM map.
Fluorescence Experiments
The five native tryptophan residues in YnaI were replaced by phenylalanine, and new tryptophan residues were introduced by site-directed mutagenesis (Stratagene). Furthermore, several intermediate mutants were obtained whereby some of the tryptophan residues were replaced by phenylalanine or tyrosine. A mutant with only one of the native tryptophan residues remaining, W201 (Δ4W), was also used. The mutant forms were purified as described for WT YnaI, and resulted in homogeneous heptameric YnaI complexes. Then YnaI was reconstituted into lipid bilayers in the same way as described earlier (Carney et al., 2006). In short, DDM-solubilized YnaI was mixed with cholate-solubilized lipids at a ratio of 1:100 (mol/mol) and incubated at room temperature for 15 min. Then YnaI was reconstituted into bilayers by diluting into a buffer containing 40 mM HEPES (pH 7.2), 100 mM KCl, and 1 mM EGTA. After 5 min of incubation at 20°C, fluorescence spectra were recorded using an FLS920 spectrometer (Edinburgh Instruments) with an excitation wavelength of 295 nm and emission from 300 to 420 nm. Spectra were compared for reconstitution into 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC; Avanti Polar Lipids) or 1,2-di-(9,10-dibromo)stearoyl-sn-glycero-3-phosphocholine (BrPC). The fractional quenching was calculated by (F0 − F)/F0, where F0 is the fluorescence intensity at 340 nm for samples reconstituted into DOPC and F for reconstitution into BrPC. Emission spectra were also recorded for DDM-solubilized YnaI samples. For depth-dependent quenching experiments, YnaI was reconstituted into 1-oleoyl-2-palmitoyl-sn-glycero-3-phosphocholine as non-brominated reference sample and into brominated lipids, 1-palmitoyl-2-(6,7-dibromo)stearoyl-sn-glycero-3-phosphocholine or similar lipids with bromination at 9,10- or the 11,12-positions (all obtained from Avanti Polar Lipids). Data were analyzed according to Ladokhin (1997) with distribution analysis using Gaussian functions and the program Origin 8.0 for fitting.
The quenching of the W201 fluorescence in the mutants W29F (Δ1W) and W29F/W299Y/W302Y (Δ3W) were determined by following procedure. The mutant forms were reconstituted in DOPC and BrPC as described above, and the fluorescence was measured. The intensity was normalized by the final protein concentration, ignoring the inner filter effect because of low absorption. The same was done for the corresponding control mutants that have the additional W210F mutation, resulting in W29F/W201F (Δ2W) and W29F/W201F/W299Y/W302Y (Δ4Wb), respectively. Emission spectra of the control mutants were subtracted from the former spectra and the fractional quenching was calculated as above. The concentration of the original samples before reconstitution were determined by UV-Vis spectroscopy using the extinction coefficients ε280nm = 38.29 mM−1 cm−1 (for Δ1W), ε280nm = 32.89 mM−1 cm−1 (for Δ2W), ε280nm = 27.39 mM−1 cm−1 (for Δ3W), and ε280nm = 21.89 mM−1 cm−1 (for Δ4W).
Author Contributions
B.B. and T.R. designed the study and analyzed data; all authors conducted experiments and prepared the manuscript.
Acknowledgments
We thank Chris Kennaway and Veselin Kukenski for their contributions in the early stages of the project, and Ian R. Booth and Samantha Miller for helpful discussions. Electron microscopy was done in the Cryo-EM facility of the University of Edinburgh, which was funded by contributions of the Wellcome Trust equipment grant WT087658 and the Scottish University Life Science Alliance (SULSA). The Aberdeen group was supported by the Wellcome Trust grant (WT092552/A/10/Z) awarded to I.R. Booth, S. Miller, T.R., J. Naismith (St. Andrews), and S. Conway (Oxford).
Published: August 6, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.str.2015.06.023.
Contributor Information
Bettina Böttcher, Email: bettina.boettcher@ed.ac.uk.
Tim Rasmussen, Email: t.rasmussen@abdn.ac.uk.
Accession Numbers
The accession number for the EM map reported in this paper is EMDB: 3035.
Supplemental Information
References
- Adamian L., Liang J. Interhelical hydrogen bonds and spatial motifs in membrane proteins: polar clamps and serine zippers. Proteins. 2002;47:209–218. doi: 10.1002/prot.10071. [DOI] [PubMed] [Google Scholar]
- Anishkin A., Akitake B., Sukharev S. Characterization of the resting MscS: modeling and analysis of the closed bacterial mechanosensitive channel of small conductance. Biophys. J. 2008;94:1252–1266. doi: 10.1529/biophysj.107.110171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass R.B., Strop P., Barclay M., Rees D.C. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science. 2002;298:1582–1587. doi: 10.1126/science.1077945. [DOI] [PubMed] [Google Scholar]
- Bellare J.R., Davis H.T., Scriven L.E., Talmon Y. Controlled environment vitrification system: an improved sample preparation technique. J. Electron. Microsc. Tech. 1988;10:87–111. doi: 10.1002/jemt.1060100111. [DOI] [PubMed] [Google Scholar]
- Bolen E.J., Holloway P.W. Quenching of tryptophan fluorescence by brominated phospholipid. Biochemistry. 1990;29:9638–9643. doi: 10.1021/bi00493a019. [DOI] [PubMed] [Google Scholar]
- Booth I.R., Edwards M.D., Black S., Schumann U., Miller S. Mechanosensitive channels in bacteria: signs of closure? Nat. Rev. Microbiol. 2007;5:431–440. doi: 10.1038/nrmicro1659. [DOI] [PubMed] [Google Scholar]
- Booth I.R., Rasmussen T., Edwards M.D., Black S., Rasmussen A., Bartlett W., Miller S. Sensing bilayer tension: bacterial mechanosensitive channels and their gating mechanisms. Biochem. Soc. Trans. 2011;39:733–740. doi: 10.1042/BST0390733. [DOI] [PubMed] [Google Scholar]
- Carney J., East J.M., Mall S., Marius P., Powl A.M., Wright J.N., Lee A.G. Fluorescence quenching methods to study lipid-protein interactions. Curr. Protoc. Protein Sci. 2006;Chapter 19 doi: 10.1002/0471142301.ps1912s45. Unit 19.12. [DOI] [PubMed] [Google Scholar]
- East J., Lee A. Lipid selectivity of the calcium and magnesium ion dependent adenosine triphosphatase, studied with fluorescence quenching by a brominated phospholipid. Biochemistry. 1982;21:4144–4151. doi: 10.1021/bi00260a035. [DOI] [PubMed] [Google Scholar]
- Edwards M.D., Black S., Rasmussen T., Rasmussen A., Stokes N.R., Stephen T.-L., Miller S., Booth I.R. Characterization of three novel mechanosensitive channel activities in Escherichia coli. Channels. 2012;6:272–281. doi: 10.4161/chan.20998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasha M. Collisional perturbation of spin-orbital coupling and the mechanism of fluorescence quenching. A visual demonstration of the perturbation. J. Chem. Phys. 1952;20:71. [Google Scholar]
- Kiefer F., Arnold K., Kunzli M., Bordoli L., Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian J.A., von Heijne G. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 2000;25:429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Ladokhin A.S. Distribution analysis of depth-dependent fluorescence quenching in membranes: a practical guide. Methods Enzymol. 1997;278:462–473. doi: 10.1016/s0076-6879(97)78024-8. [DOI] [PubMed] [Google Scholar]
- Ladokhin A.S. Measuring membrane penetration with depth-dependent fluorescence quenching: distribution analysis is coming of age. Biochim. Biophys. Acta. 2014;1838:2289–2295. doi: 10.1016/j.bbamem.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J., Poon Y., Kaiser J., Rees D. Open and shut: crystal structures of the dodecylmaltoside solubilized mechanosensitive channel of small conductance from Escherichia coli and Helicobacter at 4.4 Å and 4.1 Å resolutions. Protein Sci. 2013;22:502–509. doi: 10.1002/pro.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Moe P.C., Chandrasekaran S., Booth I.R., Blount P. Ionic regulation of MscK, a mechanosensitive channel from Escherichia coli. EMBO J. 2002;21:5323–5330. doi: 10.1093/emboj/cdf537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Edwards M.D., Jeong H., Jeong H., Roth J., Booth I.R. Identification of mutations that alter the gating of the Escherichia coli mechanosensitive channel protein, MscK. Mol. Microbiol. 2007;64:560–574. doi: 10.1111/j.1365-2958.2007.05672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksaev G., Haswell E.S. MscS-Like10 is a stretch-activated ion channel from Arabidopsis thaliana with a preference for anions. Proc. Natl. Acad. Sci. USA. 2012;109:19015–19020. doi: 10.1073/pnas.1213931109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksaev G., Haswell E. Recent characterizations of MscS and its homologs provide insight into the basis of ion selectivity in mechanosensitive channels. Channels. 2013;7:215–220. doi: 10.4161/chan.24505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell J.A., Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phillips R., Ursell T., Wiggins P., Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliotas C., Ward R., Branigan E., Rasmussen A., Hagelueken G., Huang H., Black S.S., Booth I.R., Schiemann O., Naismith J.H. Conformational state of the MscS mechanosensitive channel in solution revealed by pulsed electron-electron double resonance (PELDOR) spectroscopy. Proc. Natl. Acad. Sci. USA. 2012;109:E2675–E2682. doi: 10.1073/pnas.1202286109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A., Rasmussen T., Edwards M.D., Schauer D., Schumann U., Miller S., Booth I.R. The role of tryptophan residues in the function and stability of the mechanosensitive channel MscS from Escherichia coli. Biochemistry. 2007;46:10899–10908. doi: 10.1021/bi701056k. [DOI] [PubMed] [Google Scholar]
- Rasmussen T., Edwards M.D., Black S.S., Rasmussen A., Miller S., Booth I.R. Tryptophan in the pore of the mechanosensitive channel MscS: assessment of pore conformations by fluorescence spectroscopy. J. Biol. Chem. 2010;285:5377–5384. doi: 10.1074/jbc.M109.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann U., Edwards M.D., Li C., Booth I.R. The conserved carboxy-terminus of the MscS mechanosensitive channel is not essential but increases stability and activity. FEBS Lett. 2004;572:233–237. doi: 10.1016/j.febslet.2004.07.045. [DOI] [PubMed] [Google Scholar]
- Schumann U., Edwards M.D., Rasmussen T., Bartlett W., van West P., Booth I.R. YbdG in Escherichia coli is a threshold-setting mechanosensitive channel with MscM activity. Proc. Natl. Acad. Sci. USA. 2010;107:12664–12669. doi: 10.1073/pnas.1001405107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotomayor M., Schulten K. Molecular dynamics study of gating in the mechanosensitive channel of small conductance MscS. Biophys. J. 2004;87:3050–3065. doi: 10.1529/biophysj.104.046045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Vásquez V., Sotomayor M., Cortes D.M., Roux B., Schulten K., Perozo E. Three-dimensional architecture of membrane-embedded MscS in the closed conformation. J. Mol. Biol. 2008;378:55–70. doi: 10.1016/j.jmb.2007.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Black S.S., Edwards M.D., Miller S., Morrison E.L., Bartlett W., Dong C., Naismith J.H., Booth I.R. The structure of an open form of an E. coli mechanosensitive channel at 3.45 A resolution. Science. 2008;321:1179–1183. doi: 10.1126/science.1159262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward R., Pliotas C., Branigan E., Hacker C., Rasmussen A., Hagelueken G., Booth I.R., Miller S., Lucocq J., Naismith J.H. Probing the structure of the mechanosensitive channel of small conductance in lipid bilayers with pulsed electron-electron double resonance. Biophys. J. 2014;106:834–842. doi: 10.1016/j.bpj.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang J., Feng Y., Ge J., Li W., Sun W., Iscla I., Yu J., Blount P., Li Y. Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance. Proc. Natl. Acad. Sci. USA. 2012;109:18180–18185. doi: 10.1073/pnas.1207977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.