Abstract
Sleep is an essential and conserved behavior, whose regulation at the molecular and anatomical level remains to be elucidated. Here we identify TARANIS (TARA), a Drosophila homolog of the Trip-Br (SERTAD) family of transcriptional coregulators, as a molecule that is required for normal sleep patterns. Through a forward-genetic screen, we isolated tara as a novel sleep gene associated with a marked reduction in sleep amount. Targeted knockdown of tara suggests that it functions in cholinergic neurons to promote sleep. tara encodes a conserved cell cycle protein that contains a Cyclin A (CycA)-binding homology domain. TARA regulates CycA protein levels and genetically and physically interacts with CycA to promote sleep. Further, decreased levels of Cyclin-dependent kinase 1 (Cdk1), a kinase partner of CycA, rescue the short-sleeping phenotype of tara and CycA mutants, while increased Cdk1 activity mimics the tara and CycA phenotypes, suggesting that Cdk1 mediates the role of TARA and CycA in sleep regulation. Finally, we describe a novel wake-promoting role for a cluster of ~14 CycA-expressing neurons in the pars lateralis (PL), previously proposed to be analogous to the mammalian hypothalamus. We propose that TARANIS controls sleep amount by regulating CycA protein levels and inhibiting Cdk1 activity in a novel arousal center.
Introduction
Most animals sleep, and evidence for the essential nature of this behavior is accumulating [1–3]. However, we are far from understanding how sleep is controlled at a molecular and neural level. The fruit fly, Drosophila melanogaster, has emerged as a powerful model system for understanding complex behaviors such as sleep [4, 5]. Mutations in several Drosophila genes have been identified that cause significant alterations in sleep [5–13]. Some of these genes were selected as candidates because they were implicated in mammalian sleep [10, 11]. However, others (such as Shaker and CREB) whose role in sleep was first discovered in Drosophila [6, 12] have later been shown to be involved in mammalian sleep [14, 15], validating the use of Drosophila as a model system for sleep research. Since the strength of the Drosophila model system is the relative efficiency of large-scale screens, we and other investigators have conducted unbiased forward-genetic screens to identify novel genes involved in sleep regulation [6–9, 16]. Previous genetic screens for short-sleeping fly mutants have identified genes that affect neuronal excitability [6, 7], protein degradation [9, 16], and cell cycle progression [8]. However, major gaps remain in our understanding of the molecular and anatomical basis of sleep regulation by these and other genes.
Identifying the underlying neural circuits would facilitate the investigation of sleep regulation. The relative simplicity of the Drosophila brain provides an opportunity to dissect these sleep circuits at a level of resolution that would be difficult to achieve in the more complex mammalian brain. Several brain regions, including the mushroom bodies, pars intercerebralis, dorsal fan-shaped body, clock neurons, and subsets of octopaminergic and dopaminergic neurons, have been shown to regulate sleep [17–23]. However, the recent discovery that Cyclin A (CycA) has a sleep-promoting role and is expressed in a small number of neurons distinct from brain regions detailed above [8] suggests the existence of additional neural clusters involved in sleep regulation.
From an unbiased forward-genetic screen, we discovered taranis (tara), a mutant that exhibits markedly reduced sleep amount. tara encodes a Drosophila homolog of the Trip-Br (SERTAD) family of mammalian transcriptional coregulators that are known primarily for their role in cell cycle progression [24–27]. TARA and Trip-Br proteins contain a conserved domain found in several CycA-binding proteins [26]. Our research shows that tara regulates CycA levels, and genetically interacts with CycA and its kinase partner Cyclin-dependent kinase 1 (Cdk1) [28] to regulate sleep. Furthermore, we show that a cluster of CycA-expressing neurons in the dorsal brain lies in the pars lateralis (PL), a neurosecretory cluster previously proposed to be analogous to the mammalian hypothalamus, a major sleep center [29, 30]. Knockdown of tara and increased Cdk1 activity in CycA-expressing PL neurons, as well as activation of these cells, reduces sleep. Collectively, our data suggest that TARA promotes sleep through its interaction with CycA and Cdk1 in a novel arousal center.
Results
Identification of tara as a Sleep Regulatory Gene in Drosophila
In an ongoing forward-genetic screen for sleep and circadian mutants in Drosophila [31], we identified a novel transposon insertion line (s132) that resulted in a substantial reduction in daily sleep (Figure 1A, B, Figure S1A, B). Sleep was reduced in both female and male mutants relative to background controls. Using inverse-PCR, we mapped the s132 P-element insertion to the tara locus (Figure 1C), which suggests that TARA has a previously unappreciated role in sleep regulation. The tara transcription unit generates two transcripts (A and B) with alternative transcriptional and translational start sites [26] (www.flybase.org; Figure 1C). The two protein isoforms are identical except for a small number of N-terminal amino acids, and appear to be functionally interchangeable [26].
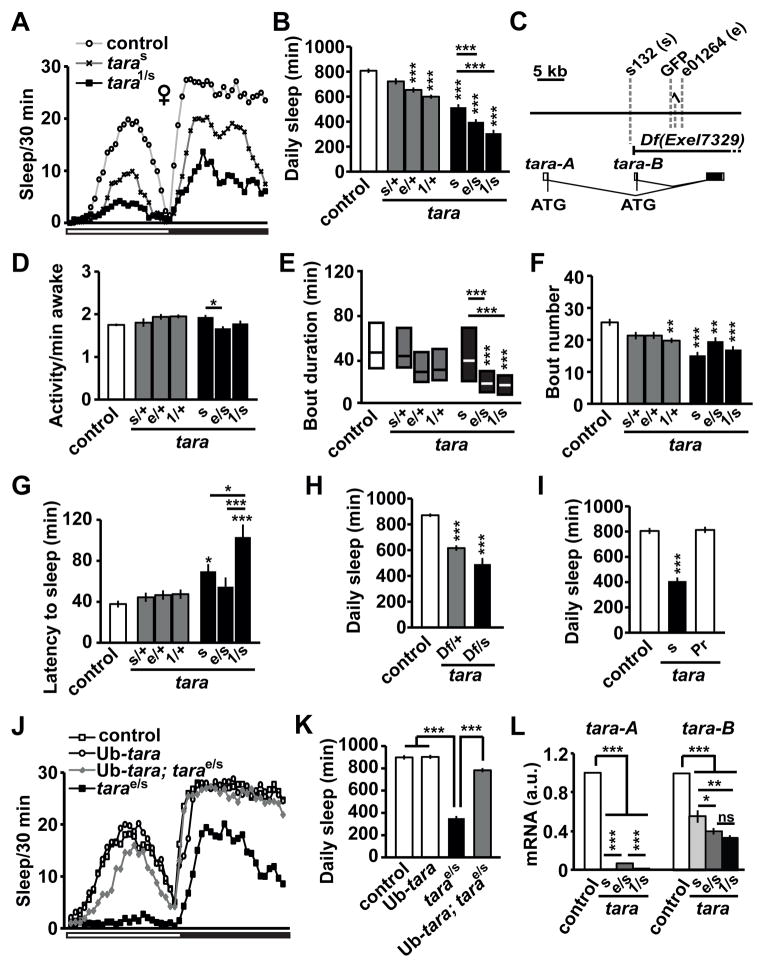
Figure 1. Sleep Phenotypes of tara Mutants.
(A) Sleep profile of background control (white circles), taras132 (taras, black X’s), and tara1/s132 (tara1/s, black squares) female flies (n=50–64) in 30 min bins. The white and black bars below the X-axis represent 12 h light and 12 h dark periods, respectively. (B) Total daily sleep amount for control and tara female flies of the indicated genotypes (n=44–72). In this and subsequent figures, s132 and e01264 alleles are referred to as s and e, respectively. (C) Schematic of the genomic region of the tara locus. Gray dashed lines indicate transposon insertion sites. The Exel7329 deficiency removes most of tara-A and all of tara-B coding regions as indicated. (D) Waking activity (activity counts per waking minute), (E) sleep bout duration, (F) sleep bout number, and (G) sleep latency (time from lights off to the first sleep bout) for the same female flies shown in (B). Sleep bout duration is not normally distributed, and is shown in simplified box plots, where the median and interquartile range are represented. (H) Total daily sleep amount of control and Df(3R)Exel7329 female heterozygotes in trans to either a wild type (Df/+) or taras132 (Df/s) allele (n=35–102). (I) Total daily sleep of control, taras132, and precise excision (taraPr) female flies (n=16–36). (J) Sleep profile of female flies of the indicated genotypes (n=53–58). The white and black bars below the X-axis represent 12 h light and 12 h periods, respectively. (K) Total daily sleep amount for the same flies showed in (J). (L) tara-A and tara-B mRNA levels relative to actin mRNA levels in head extracts of indicated genotypes (n=3–6). For each experiment, relative tara mRNA levels of the control flies were set to 1. Mean ± SEM is shown. *p < 0.05, **p < 0.01, ***p < 0.001, ns: not significant, one-way ANOVA followed by Tukey post hoc test (B, D, F, G, K, L) or Dunnett post hoc test relative to control flies (H, I); Kruskal-Wallis test (E). For simplicity, only significant differences between the control and each mutant genotype (above the bar for the mutant) and those between taras132, tarae01264/s132, and tara1/s132mutants (above the line for the mutant pair) are indicated. See also Figure S1.
For detailed characterization of the sleep phenotypes of tara mutants, we obtained two additional transposon insertions in the tara locus (tara1 and tarae01264) from Drosophila stock centers (Figure 1C). s132 homozygotes are viable, but tara1 and tarae01264 homozygotes are lethal, suggesting that 1 and e01264 are stronger alleles than s132. Consistent with this view, when combined in trans with s132, the lethal alleles exhibited a greater reduction in sleep than s132 (Figure 1A, B, Figure S1A, B). The strong tara alleles resulted in a significant reduction in sleep even as heterozygotes (Figure 1B, Figure S1B). Whereas waking activity (activity counts per minute awake) was slightly increased in some tara mutants, it was not increased in strong allelic combinations (Figure 1D, Figure S1C). Sleep bout duration in both females and males was reduced in strong allelic combinations (Figure 1E, Figure S1D), which suggests that TARA plays a role in sleep maintenance. The number of sleep bouts was markedly reduced in females with strong tara mutations (Figure 1F) and was unchanged in tara males (Figure S1E). In addition, sleep latency (time from lights off to the first sleep bout) was significantly increased in strong tara mutants (Figure 1G, Figure S1F), revealing a role for TARA in sleep initiation. Taken together, our data demonstrate that tara is a novel sleep gene essential for sleep initiation and maintenance.
We undertook additional experiments to rule out the possibility that secondary, background mutations are responsible for the sleep phenotype in tara mutants. First, a deficiency line deleting the tara locus did not complement the s132 allele (Figure 1H, Figure S1G). Second, precise excision of the s132 insertion by transposase-mediated mobilization restored normal sleep (Figure 1I, Figure S1H). Third, ubiquitous expression of tara-B [26] restored sleep to nearly normal levels (Figure 1J, K, Figure S1I). These data confirm that disruption of tara is indeed the underlying cause of the severe sleep reduction in tara mutants.
As shown in Figures 1B and S1B, three tara allelic combinations (s132, e01264/s132, and 1/s132) yielded varying degrees of sleep reduction, suggesting that tara1 is the strongest allele and taras132 is the weakest. To determine whether differences in tara mRNA levels mediate varying phenotypic strengths, we performed quantitative RT-PCR using primers designed to distinguish between the two tara isoforms. taras132 homozygous mutants had almost no detectable tara-A mRNA and a ~50% reduction in tara-B mRNA levels relative to control flies (Figure 1L). Like taras132 mutants, tara1/s132 flies had almost no detectable tara-A mRNA, but tara-B transcripts were further reduced, indicating that tara1 is a null or strongly hypomorphic allele. In tarae01264/s132 flies, tara-A mRNA levels were slightly higher than in tara1/s132 flies while tara-B mRNA levels were lower than in taras132 homozygous flies. These results demonstrate that the amount of daily sleep correlates with tara levels. Collectively, the above data establish tara as a novel sleep regulatory gene.
Sleep Loss in tara Mutants is Independent of the Circadian Clock and Light
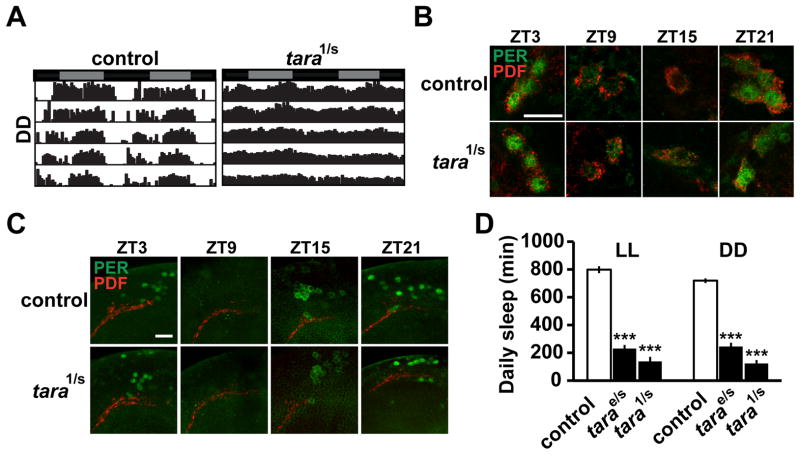
To examine whether tara mutants exhibit circadian phenotypes, we monitored their locomotor activity in constant darkness (DD). Most tara1/s132 mutants were arrhythmic or weakly rhythmic and the amplitude of their circadian rhythmicity was reduced, but the period length of all tara mutants was indistinguishable from that of control flies (Figure 2A, Figure S2A). Moreover, daily cycling of the core clock protein PERIOD (PER) in tara1/s132 mutants was similar to that in wild-type controls in two sets of clock neurons (Figure 2B, C), which suggests that dampened rhythmicity in these mutants is not due to a defect in the core molecular clock. Since arrhythmicity does not necessarily lead to short sleep (e.g., per and timeless mutants do not have reduced sleep [32]), the rhythm phenotype of tara mutants may not be the cause of the sleep phenotype. Our data showing that tarae01264/s132 mutants displayed almost as severe a sleep reduction as tara1/s132 but were largely rhythmic (Figure 1B, Figure S1B, Figure S2A) support the view that the sleep and circadian phenotypes in tara mutants may not be linked. To test whether the sleep phenotype in tara mutants was due to arrhythmicity, we assayed sleep in constant light (LL), in which both control and mutant flies are arrhythmic. Indeed, tara mutants had greatly reduced sleep compared with controls in LL, demonstrating that the short-sleeping phenotype is not caused by arrhythmicity (Figure 2D, Figure S2B). The short-sleeping phenotype was also observed in DD (Figure 2D, Figure S2B), suggesting that TARA’s role in sleep is independent of light. Of note, in both LL and DD, tara1/s132 mutants lost over 80% of sleep relative to control flies, which is one of the most severe phenotypes documented among sleep mutants. These data show that tara mutants exhibit a striking reduction in sleep amount, independent of the circadian clock and light conditions.
Figure 2. Circadian Phenotypes and Clock-Independent Sleep Loss of tara Mutants.
(A) Representative circadian actogram of individual control and tara1/s132 male flies in DD. Gray and black bars above the actogram indicate subjective day and night, respectively. (B) Cycling of PER protein (green) in small ventral lateral neurons (s-LNvs) is normal in tara1/s132 brains. Samples were dissected at indicated Zeitgeber Times (ZT) and stained for PER and PDF (red), which was used to identify s-LNvs. Scale bar: 10 μm. (C) Cycling of PER is also normal in a cluster of dorsal neurons (DN1s). Scale bar: 20 μm. (D) Total daily sleep amount in LL and DD for females of indicated genotypes (n=32–79 for LL; n=39–96 for DD). Sleep levels on the 3rd day in constant conditions are shown. Mean ± SEM is shown. ***p < 0.001, Dunnett post hoc tests relative to control flies (D). See also Figure S2.
TARA Is Required in Neurons to Control Sleep Levels
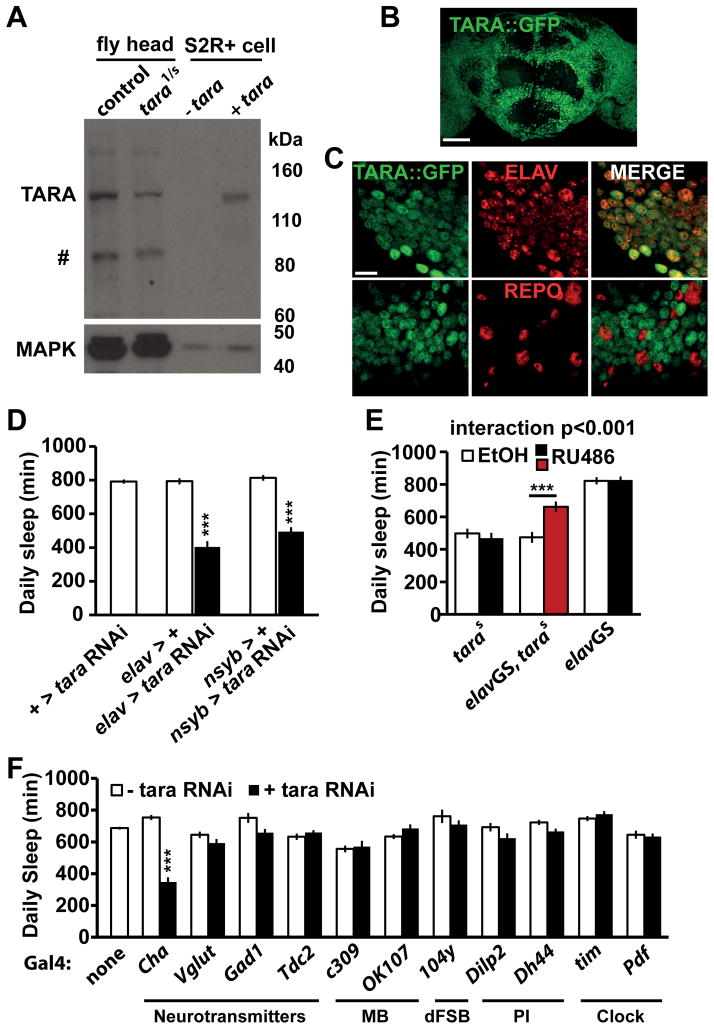
To examine the spatial requirements for TARA in regulating sleep, we generated a polyclonal antibody against the TARA protein (see Experimental Procedures). In Western blots, the antibody recognized a band that is upregulated when TARA is overexpressed in Drosophila S2 cells. As expected, this band was markedly downregulated in head extracts of tara1/s132 mutants compared with those of control flies (Figure 3A). The identity of the band was further examined by Western analysis of a previously generated GFP fusion trap in the tara locus (YB0035) [33], which we termed tara::GFP. The GFP exon is located upstream of the common second coding exon of both tara-A and tara-B isoforms (Figure 1C), and is expected to be incorporated into both isoforms close to the N terminus. The presumed TARA band in Western blots was shifted by the addition of GFP in head extracts of tara::GFP flies (Figure S3A), which confirms that the band indeed represents the TARA protein. Because the polyclonal antibody did not yield a specific signal when used for immunohistochemistry, we employed the TARA::GFP fusion protein to determine the expression pattern of TARA. Homozygotes for the tara::GFP allele did not exhibit altered sleep levels or circadian phenotypes (Figure S2A, Figure S3B, C), indicating that the TARA::GFP fusion protein is functional. Since the GFP coding region is inserted into the tara locus in the genome, the TARA::GFP expression pattern is likely to reflect endogenous TARA expression accurately. We thus examined the localization of TARA::GFP in the adult nervous system using an anti-GFP antibody. TARA::GFP was widely expressed throughout the adult brain (Figure 3B). Co-staining with neuronal and glial markers (ELAV and REPO, respectively) demonstrated that TARA is expressed in most, perhaps all, neurons but excluded from glial cells (Figure 3C).
Figure 3. TARA Regulates Sleep in Neurons.
(A) Western blot showing a marked reduction of TARA in tara1/s132 mutants compared with control flies. Head extracts of control flies and tara mutants (lanes 1 & 2) as well as S2 cell extracts transfected with an empty UAS vector or a UAS-tara construct under the control of actin-Gal4 (lanes 3 & 4) were probed with a polyclonal antibody to TARA. The band that corresponds to TARA can be readily recognized by the upregulation in S2 cells transfected with tara cDNA. # denotes non-specific labeling or a degradation product of TARA. MAPK was used to control for loading. (B) Maximal-intensity projection of confocal slices of the adult brain showing widespread expression of TARA::GFP. Scale bar: 50 μm. (C) Representative confocal sections of tara::GFP adult brains costained with antibodies to GFP and ELAV or REPO (neuronal or glial marker, respectively). Each panel shows a single confocal slice of a region ventral to the mushroom bodies. Scale bar: 10 μm. (D) Pan-neuronal knockdown of tara markedly reduces sleep. Pan-neuronal elav-Gal4 or nsyb-Gal4 was used to drive a combination of two UAS-tara RNAi constructs and UAS-dcr2 (elav > tara RNAi and nsyb > tara RNAi, respectively). Flies harboring the two UAS-tara RNAi constructs and UAS-dcr2 without a driver (+ > tara RNAi) and those harboring a driver and UAS-dcr2 (elav > + or nsyb > +) served as controls (n=31–58). (E) Adult-stage expression of tara partially rescues the tara short-sleeping phenotype. Daily sleep is presented for females of the indicated genotypes in the absence (white bar) or presence (black or red bar) of RU486 (n=24–32). Data from parental control flies show that RU486 by itself did not affect sleep. (F) Knockdown of tara in cholinergic neurons reduces sleep. For each Gal4, total daily sleep of females expressing tara RNAi under the control of the driver (black bar) was compared to parental controls (white bar) (n=30–173). The sleep phenotype of flies in which tara was knocked down in dopaminergic neurons was not determined due to lethality. Mean ± SEM is shown. ***p < 0.001, Dunnett post hoc test relative to both parental controls (D), t test with Bonferroni correction (E), Tukey post hoc test relative to both parental controls (F). See also Figure S3.
Given the expression pattern, we sought to demonstrate a role for neuronal TARA in regulating sleep. We used RNAi to reduce TARA expression specifically in neurons. As expected, driving tara RNAi with pan-neuronal drivers elav-Gal4 and nsyb-Gal4 resulted in a substantial reduction in daily sleep levels (Figure 3D, Figure S3D). Reduced TARA expression by RNAi-mediated knockdown was confirmed by Western analysis (Figure S3E). We next examined whether tara functions in the adult fly to regulate sleep by utilizing a UAS site in the s132 insertion to drive tara expression. We used GeneSwitch (GS), an RU486-dependent GAL4 protein that allows temporal control of transgenic expression [34]. Adult specific pan-neuronal expression of tara using the elav-GS driver partially rescued the short-sleeping phenotype of tara mutants (Fig. 3E), demonstrating an adult function of tara, although the incomplete adult-stage rescue suggests a potential developmental role as well.
To identify neuronal groups where TARA acts to control sleep, we utilized the Gal4/UAS system to target tara RNAi expression to subsets of neurons. We examined several neurotransmitter systems as well as brain regions involved in sleep regulation such as the mushroom bodies (MB), dorsal fan-shaped body (dFSB), pars intercerebralis (PI), and clock cells. Only tara knockdown by Cha-Gal4 produced a significant reduction in sleep (Figure 3F). These data suggest that cholinergic neurons likely mediate the effects of TARA on sleep.
tara Interacts with CycA to Control Sleep and Regulates CycA Levels
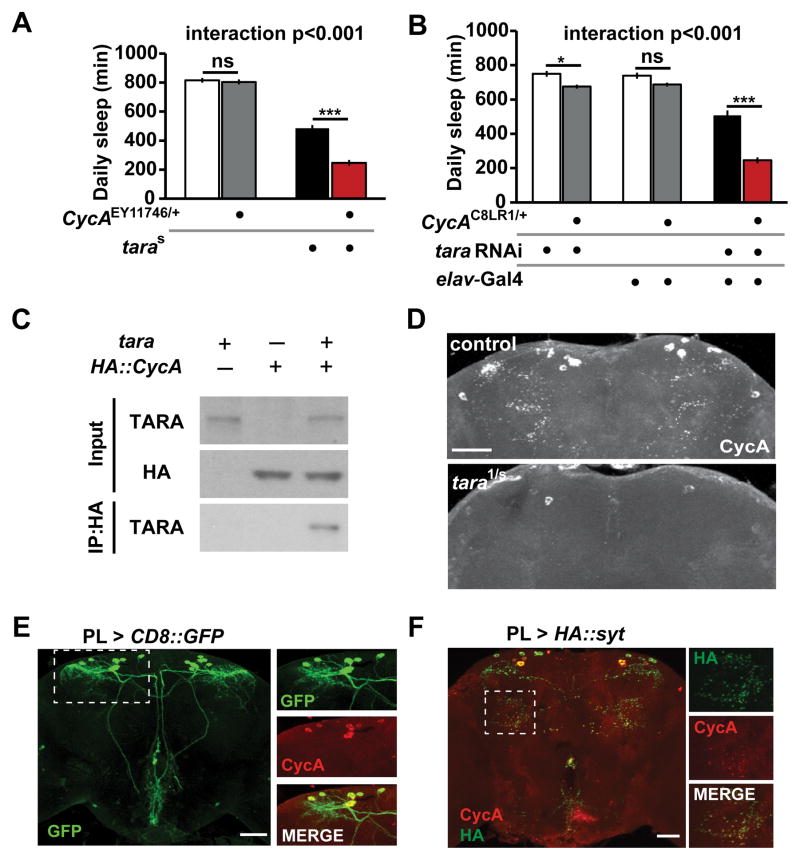
Since CycA has been shown to promote sleep in Drosophila [8], and since TARA contains a conserved CycA binding homology motif, we tested whether tara and CycA act in a common genetic pathway to regulate sleep. To do so, we generated double mutants and compared their sleep behavior with those of wild-type control and single mutant flies. The CycAEY11746/+ heterozygous mutation did not cause reduced sleep on its own, but it led to a significant reduction in sleep when combined with the taras132 hypomorphic mutation that has a moderate sleep phenotype (Figure 4A, Figure S4A). This interaction was confirmed using a second allele of CycA (CycAC8LR1/+) and tara RNAi (Figure 4B). Further, CycA did not exhibit a genetic interaction with the DATfmn short-sleeping mutant [13] (Figure S4B), demonstrating the specificity of the interaction between tara and CycA. These data reveal a synergistic interaction between tara and CycA and suggest they act in the same pathway to influence sleep.
Figure 4. tara Interacts with CycA and Regulates CycA Levels.
(A) Daily sleep for female flies of indicate genotypes demonstrating a synergistic interaction between cycAEY11746/+ and taras132 (n=43–60). (B) Daily sleep for female flies of the indicated genotypes. Pan-neuronal knockdown of tara by RNAi (elav > tara RNAi) was more effective at suppressing sleep in a CycAC8LR1/+ background than in a control background (n=46–52). (C) TARA and CycA form a complex in S2 cells. HA-tagged CycA was immunoprecipitated with an anti-HA antibody, and anti-TARA and anti-HA antibodies were used for Western blotting. The experiment was repeated 3 times with similar results. (D) Maximal-intensity confocal projections of the dorsal half of the central brain of representative control and tara1/s132 adult females immunostained with an antibody to CycA. Scale bar: 50 μm. (E) The central brain of an adult fly in which PL-Gal4 was used to express membrane-targeted CD8::GFP. Scale bar: 50 μm. Images on the right show colocalization of CycA and GFP driven by PL-Gal4 in the brain region indicated by the rectangle. (F) The central brain of a fly in which the synaptic marker HA::SYT was expressed using PL-Gal4. The brain was costained with anti-HA and anti-CycA. The rectangle indicates the region magnified in the images on the right. Scale bar: 50 μm. Mean ± SEM is shown. *p < 0.05; ***p < 0.001, ns: not significant, Tukey post hoc test. See also Figure S4.
Given the genetic interaction between tara and CycA, the presence of a CycA-binding homology domain in TARA, and the fact that Trip-Br1/2, mammalian homologs of TARA, have been shown to bind CycA [24], we tested whether TARA physically binds CycA in a co-immunoprecipitation assay. Indeed, we found that TARA co-immunoprecipitated with CycA in Drosophila S2 cells (Figure 4C), suggesting that they can form a complex.
We next asked whether CycA levels are altered in tara mutants. We performed whole-mount immunostaining of adult brains using a CycA antibody previously shown to detect a dorsal set of CycA-positive neurons [8] (another CycA antibody previously used to detect a few additional clusters of CycA-expressing neurons is no longer available). We found that CycA protein levels were greatly reduced in the adult brain of tara mutants (Figure 4D). In contrast, CycA protein levels were not reduced in DATfmn mutants (Figure S4C), which demonstrates the specificity of the regulation of CycA levels by TARA. CycA mRNA levels were not affected in tara mutants (Figure S4D), indicating that TARA regulates CycA levels post-transcriptionally. Our data suggest that TARA promotes sleep in part through regulation of CycA protein levels.
We noticed that the dorsal CycA cluster might correspond to the pars lateralis (PL) [35], so we drove expression of CD8::GFP using PL-Gal4, a driver expressed in the PL neurons [36], while simultaneously labeling brains for CycA. Both GFP and CycA were expressed in ~14 neurons with large cell bodies in the dorsal brain (Figure 4E and Figure S4E). The striking overlap seen between the GFP and CycA signals demonstrates that the dorsal CycA neurons indeed lie in the PL. This is significant because the PL, along with the pars intercerebralis, shares several features with the mammalian hypothalamus, a major sleep center [29, 30]. However, a possible contribution of the PL to sleep regulation has not been previously explored.
We employed the PL driver to determine whether the CycA-expressing cells were present in tara mutants. By examining flies expressing CD8::GFP under the control of PL-Gal4, we confirmed that the PL neurons were indeed present (Figure S4F). Interestingly, CycA protein was observed not only in cell bodies but also in discrete puncta that appeared to be synapses (Figure 4D). This is noteworthy because according to the synaptic homeostasis hypothesis, waking activity leads to a net increase in synaptic strength, whereas sleep leads to overall downscaling of synapses [37]. To determine whether these puncta represent synapses, we used PL-Gal4 to express a synaptic marker (HA::SYT) [38] and demonstrated that CycA indeed localized to synaptic regions (Figure 4F). We note that CycA protein levels were downregulated in both cell bodies and synaptic regions in tara mutants (Figure 4D). CycA levels and function at synapses, under the control of TARA, may be important for normal sleep.
TARA Regulates Sleep in CycA-Expressing PL Neurons, which Define a New Arousal Center
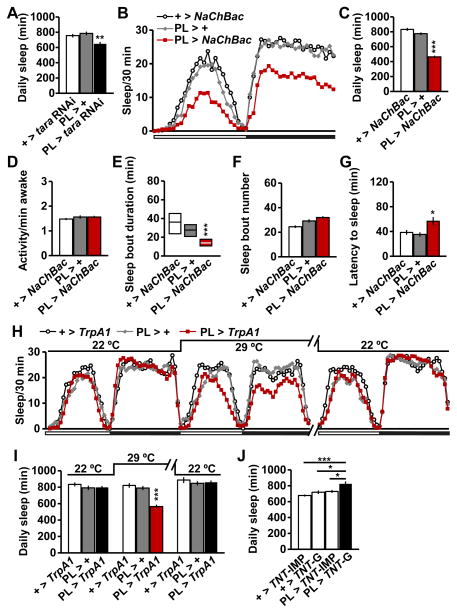
To address whether TARA is required in CycA-expressing cells for sleep regulation, we examined the sleep phenotype of flies in which tara was knocked down using the PL driver. We found that PL-specific tara knockdown significantly reduced sleep (Figure 5A). We note that this manipulation produced a weaker sleep reduction than pan-neuronal knockdown of tara (Figure 3D), which suggests that TARA likely functions in additional groups of neurons to regulate sleep.
Figure 5. TARA Regulates Sleep in CycA-Expressing PL Neurons that Define a New Arousal Center.
(A) Total daily sleep of female flies of the indicated genotypes (n=32). (B) Sleep profile for female flies expressing the NaChBac sodium channel under the control of PL-Gal4 (PL > NaChBac) and parental controls (n=48–63). (C–G) Total daily sleep (C), waking activity (D), sleep bout duration (E), sleep bout number (F), and latency to sleep after lights off (G) for the flies shown in (B). (H) Sleep profile for female flies expressing TrpA1 under the control of PL-Gal4 (PL > TrpA1) and parental controls (n=16–32). Flies were monitored at 29°C, which activates the TrpA1 channel, and at 22°C, which inactivates the TrpA1 channel. Sleep profile for the second day at 29C is omitted for simplicity. (I) Total daily sleep for flies shown in (H). (J) Female flies expressing functional tetanus toxin under the control of PL-Gal4 (PL > TNT-G) exhibited a significant increase in sleep relative to flies expressing inactive tetanus toxin (PL > TNT-IMP) or those carrying either form of tetanus toxin transgene without the PL driver (+ > TNT-G and + > TNT-IMP) (n = 30–32). Mean ± SEM is shown. * p < 0.05, **p < 0.01, ***p < 0.001, ns: not significant, Dunnett post hoc test relative to parental controls (A, C–D, F–G, I) or PL > TNT-G flies (J); Kruskal-Wallis test (E). See also Figure S5.
Our results pointed to a possible role of the PL neurons in sleep regulation. Indeed, we found that activation of the PL neurons via expression of the bacterial sodium channel NaChBac [39] led to decreased sleep (Figure 5B, C, Figure S5A, B). In contrast, activity levels during waking periods were not affected by PL activation (Figure 5D, Figure S5C). Sleep bout duration was markedly decreased while sleep bout number showed little change, and sleep latency was significantly increased in flies with activated PL neurons (Figure 5E–G, Figure S5D–F). These data suggest that activation of PL neurons promote wakefulness by delaying sleep onset and impairing sleep maintenance. Adult-stage specific activation of these neurons using the warmth-activated cation channel TrpA1 [40] also reduced sleep, demonstrating that this cell cluster functions in adult animals to promote wakefulness (Figure 5H, I, Figure S5G). Further, blocking the activity of PL neurons with tetanus toxin [41] significantly increased sleep (Figure 5J), which confirms the wake-promoting role of these neurons. The above data identify the PL neurons as a novel arousal center and demonstrate that TARA acts, at least in part, in CycA-positive PL neurons to promote sleep.
tara and Cdk1 Interact Antagonistically to Regulate Sleep
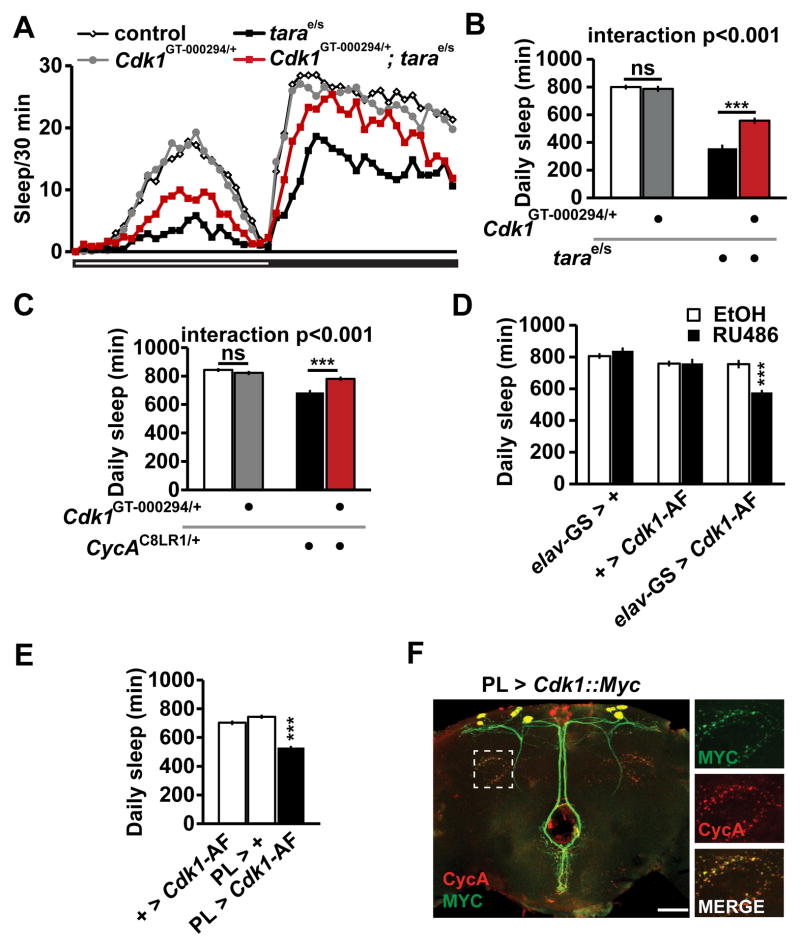
CycA has been shown to bind Cdk1, and can either increase or decrease Cdk1 activity depending on the cellular context [28, 42]. We therefore asked whether Cdk1 also interacts with tara for sleep regulation. We introduced a heterozygous Cdk1GT-000294/+ mutation (the GT-000294 insertion is in the coding region and is likely to be a null allele) into a tara mutant background, and compared their sleep with tara and Cdk1 single mutants as well as with wild-type control flies. We found that the Cdk1GT-000294/+ heterozygous mutation did not cause a sleep phenotype in a wild-type background, but it resulted in a substantial rescue of the tara sleep phenotype (Figure 6A, B). We confirmed the antagonistic interaction between tara and Cdk1 using a second allele of Cdk1 (Cdk1c03495/+) (Figure S6A). The Cdk1GT-000294/+ heterozygous mutation did not rescue the short-sleeping phenotype of insomniac (inc) mutants (Figure S6B) [9, 16], which demonstrates that the interaction between tara and Cdk1 is not due to additive effects. In contrast, the Cdk1 mutation did rescue the sleep phenotype of heterozygous CycA null mutants (Figure 6C), consistent with a model in which tara and CycA act together to antagonize Cdk1. Transcript levels of Cdk1 were not significantly affected in tara mutants (Figure S6C), suggesting that the interaction between tara and Cdk1 is not likely to be due to transcriptional regulation of Cdk1 by TARA. The antagonistic interaction between tara and Cdk1 suggests that Cdk1 has a previously unrecognized wake-promoting role.
Figure 6. Cdk1 Interacts Antagonistically with tara and CycA, and Increased Cdk1 Activity Suppresses Sleep.
(A) Sleep profile for control (white diamonds), Cdk1GT-000294/+ (gray circles), tarae01264/s132 (black squares), and Cdk1GT-000294/+; tarae01264/s132 (red squares) female flies (n=50–64). (B) Total daily sleep for the flies shown in (A). (C) Total daily sleep of female flies of the indicated genotypes (n = 81–88). (D) Adult-stage specific expression of Cdk1-AF induced by feeding flies food that contain RU486 diluted in ethanol (EtOH) reduced sleep (n=31–32). (E) Cdk1-AF expression specifically in CycA-expressing PL neurons resulted in reduced sleep (n=70–75). (F) The central brain of a fly in which UAS-Cdk1-myc was expressed using PL-Gal4. The brain was costained with anti-MYC and anti-CycA. The rectangle indicates the region magnified in the images on the right. Scale bar: 50 μm. Mean ± SEM is shown. ***p < 0.001, ns: not significant, Tukey post hoc test (B, C), Dunnett post hoc test relative to all other controls (D); relative to both parental controls (E). See also Figure S6.
To investigate the potential wake-promoting role of Cdk1, we assayed sleep in flies overexpressing wild-type Cdk1 (Cdk1-WT). Since activity of Cyclin-dependent kinases is tightly controlled by a number of regulatory molecules [42–44], we also examined flies overexpressing Cdk1-AF, a mutant Cdk1 protein that has elevated kinase activity due to mutations in inhibitory phosphorylation sites [42]. Because overexpression of Cdk1-AF under the control of elav-Gal4 resulted in lethality, we used the RU486 inducible elav-GS to express Cdk1 specifically in the adult stage. Whereas RU486 had little effect on control flies, flies in which cdk1-AF was expressed under the control of elav-GS exhibited significantly reduced sleep when fed RU486 (Figure 6D), which indicates that increased Cdk1 activity indeed promotes wakefulness. In contrast, overexpression of Cdk1-WT had little effect on sleep (Figure S6C), presumably because overexpression of wild-type Cdk1 alone was not sufficient to increase its kinase activity. To examine whether Cdk1 acts in CycA-expressing cells to regulate sleep, we assayed sleep in flies expressing the Cdk1-AF transgene under the control of PL-Gal4. These flies had significantly reduced sleep compared with parental control flies (Figure 6E). These data provide strong evidence for a novel role of Cdk1 in suppressing sleep.
Since CycA is expressed in synaptic regions (Figure 4F), we next asked whether Cdk1 colocalizes with CycA at synaptic regions. To address this question, we expressed MYC-tagged wild-type Cdk1 [45] in PL neurons, and found that Cdk1::MYC exhibited marked overlap with CycA puncta at synaptic regions (Figure 6F). Although the synaptic localization of Cdk1::MYC could be an artifact of overexpression, the potential colocalization of Cdk1 and CycA at synaptic regions raises the interesting possibility that synaptic Cdk1 activity may be important for maintaining normal sleep amount. Together, our data demonstrate that Cdk1 interacts antagonistically with TARA and CycA and acts in PL neurons to promote wakefulness.
Discussion
From an unbiased forward genetic screen, we have identified a novel sleep regulatory gene, tara. Our data demonstrate that TARA interacts with CycA to regulate its levels and promote sleep. We have also identified Cdk1 as a wake-promoting molecule that interacts antagonistically with TARA. Given the fact that TARA regulates CycA levels, the interaction between TARA and Cdk1 may be mediated by CycA. Our finding that Cdk1 and CycA also exhibit an antagonistic interaction supports this view. The previous discovery that CycE sequesters its binding partner Cdk5 to repress its kinase activity in the adult mouse brain [46] points to a potential mechanism; namely, that TARA regulates CycA levels, which in turn sequesters and inhibits Cdk1 activity. TARA and its mammalian homologs (the Trip-Br family of proteins) are known for their role in cell cycle progression [24–27]. However, recent data have shown that Trip-Br2 is involved in lipid and oxidative metabolism in adult mice [47], demonstrating a role beyond cell cycle control. Other cell cycle proteins have also been implicated in processes unrelated to the cell cycle. For example, CycE functions in the adult mouse brain to regulate learning and memory [46]. Based on the finding that CycA and its regulator Rca1 control sleep, it was hypothesized that a network of cell cycle genes has been appropriated for sleep regulation [8]. Our data showing that two additional cell cycle proteins, TARA and Cdk1, control sleep and wakefulness provide support for that hypothesis. Moreover, the fact that TARA and CycA, factors identified in two independent unbiased genetic screens, interact with each other highlights the importance of a network of cell cycle genes in sleep regulation.
There are two main regulatory mechanisms for sleep: the circadian mechanism that controls the timing of sleep and the homeostatic mechanism that controls the sleep amount [48]. We have shown that TARA has a profound effect on total sleep time. TARA also affects rhythmic locomotor behavior. Since TARA is expressed in clock cells (our unpublished data) whereas CycA is not [8], it is possible that TARA plays a non-CycA dependent role in clock cells to control rhythm strength. Our finding that tara mutants exhibit severely reduced sleep in constant light suggests that the effect of TARA on sleep amount is not linked to its effect on rhythmicity. Instead TARA may have a role in the sleep homeostatic machinery, which will be examined in our ongoing investigation.
To fully elucidate how sleep is regulated, it is important to identify the underlying neural circuits. Here we have shown that activation of the CycA-expressing neurons in the PL suppresses sleep while blocking their activity increases sleep, which establishes them as a novel wake-promoting center. Importantly, knockdown of tara and increased Cdk1 activity specifically in the PL neurons leads to decreased sleep. A simple hypothesis, consistent with our finding that both activation of PL neurons and increased Cdk1 activity in these neurons suppress sleep, is that Cdk1 affects neuronal excitability and synaptic transmission. Interestingly, large-scale screens for short-sleeping mutants in fruit flies and zebrafish have identified several channel proteins such as SHAKER, REDEYE, and ETHER-A-GO-GO [6, 49, 50], and channel modulators such as SLEEPLESS and WIDE AWAKE [51, 52]. Thus it is plausible that Cdk1 regulates sleep by phosphorylating substrates that modulate the function of synaptic ion channels or proteins involved in synaptic vesicle fusion, as has previously been demonstrated for Cdk5 at mammalian synapses [53].
Whereas our data mapped some of TARA’s role in sleep regulation to a small neuronal cluster, the fact that pan-neuronal tara knockdown results in a stronger effect on sleep than specific knockdown in PL neurons suggests that TARA may act in multiple neuronal clusters. PL-specific restoration of TARA expression did not rescue the tara sleep phenotype (data not shown), further implying that the PL cluster may not be the sole anatomical locus for TARA function. Given that CycA is expressed in a few additional clusters [8], TARA may act in all CycA-expressing neurons including those not covered by PL-Gal4. TARA may also act in non-CycA-expressing neurons. Our data demonstrate that tara knockdown using Cha-Gal4 produces as strong an effect on sleep as pan-neuronal knockdown (Figure 3D, F). This finding suggests that TARA acts in cholinergic neurons, although we cannot rule out the possibility that the Cha-Gal4 expression pattern includes some non-cholinergic cells. Taken together, our data suggest that TARA acts in PL neurons as well as unidentified clusters of cholinergic neurons to regulate sleep.
Based on genetic interaction studies, tara has been classified as a member of the trithorax group genes, which typically act as transcriptional coactivators [26, 54]. However, TARA and Trip-Br1 have been shown to up- or down-regulate the activity of E2F1 transcription factor depending on the cellular context, raising the possibility that they also function as transcriptional corepressors [24, 27]. Interestingly, TARA physically interacts with CycA and affects CycA protein levels but not its mRNA expression. These findings suggest a novel non-transcriptional role for TARA, although we cannot rule out an indirect transcriptional mechanism. The hypothesis that TARA plays a non-transcriptional role in regulating CycA levels and Cdk1 activity at the synapse may provide an exciting new avenue for future research.
Experimental Procedures
Details of experimental procedures are available in the online Supplemental Information.
Supplementary Material
Figure S1. Sleep Phenotypes and Genetic Analysis of tara: Male Data. (A) Sleep profile of homozygous taras132 (black X’s), transheterozygous tara1/s132 (black squares), and their background control males (white circles) (n=60–73). The white and black bars below the X-axis represent light and dark periods, respectively. (B) Control and tara mutant males of the indicated genotypes (n=41–73). (C) Waking activity, (D) sleep bout duration, (E) sleep bout number, and (F) sleep latency at lights off for the same male flies shown in (B). Sleep bout duration is shown in simplified box plots, where the median and interquartile range are represented. (G) Total daily sleep of control and Df(3R)Exel7329 male heterozygotes in trans to either a wild type (Df/+) or taras132 (Df/s) allele (n=35–70). (H) Total daily sleep of control, taras132, and precise excision male flies (n=15–34). (I) Total daily sleep of male flies of the indicated genotypes (n=35–42). Ubiquitous expression of tara-B (Ub-tara) rescued the tara sleep phenotypes. Mean ± SEM is shown. *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA followed by Dunnett post hoc test (G, H); Tukey post hoc test (B, C, E, F, I); Kruskal-Wallis test (D). For simplicity, only significant differences between each mutant and the control and those between taras132, tarae01264/s132, and tara1/s132 mutants are indicated. Related to Figure 1.
Figure S2. Circadian Phenotypes and Clock-Independent Sleep Loss of tara Mutant Males. (A) Free-running circadian phenotypes of male flies of the indicated genotypes in DD. χ2 periodogram analysis was performed for each fly using the FaasX software to determine the free-running period, tau. Power, a measure of rhythmicity, corresponds to peak – significance value at p = 0.05. (B) Total daily sleep amount in LL and DD for control, tarae01264/s132, and tara1/s132 males (n=29–77 for LL; n=55–94 for DD). Sleep levels on the 3rd day in constant conditions are shown. Mean ± SEM is shown. ***p < 0.001, one-way ANOVA followed by Dunnett post hoc test relative to control flies (B). Related to Figure 2.
Figure S3. Characterization of taraGFP Flies. (A) Western blot showing a band shift in homozygous taraGFP lysates. (B) Total daily sleep and (C) waking activity of taraGFP and control females (n=47–64). taraGFP flies did not exhibit sleep abnormalities, suggesting TARA::G-FP is functional. (D) Pan-neuronal tara knockdown using two independent Gal4 lines (elav and nsyb) resulted in a significant reduction of sleep in males (n=33–63 except for elav > tara RNAi, for which n=11). (E) Western blot shows a marked reduction of TARA levels in females in which tara was knocked down pan-neuronally (elav > tara RNAi). The experiment was performed three times with similar results. MAPK was used as loading control (A, E). Mean ± SEM is shown. ***p < 0.001, ns: not significant, Student’s t-test (B, C); one-way ANOVA followed by Dunnett post hoc test relative to controls (D). Related to Figure 3.
Figure S4. Genetic Interaction between tara and CycA. (A) Daily sleep for male flies of the indicated genotypes (n=54–64). (B) Daily sleep for female flies of the indicated genotypes (n=40–56). (C) Maximal-intensity confocal projections of the dorsal half of the central brain of representative control and DATfmn adult flies immunostained with an antibody to CycA. In contrast to tara mutants, CycA levels were not altered in DATfmn mutants. Scale bar: 100 μm. (D) CycA mRNA levels relative to actin mRNA levels were not significantly different between tara1/s132 and control flies (n=3). (E) Confocal projection of a female brain (top) and ventral nerve cord (bottom) expressing CD8::GFP under the control of PL-Gal4. GFP expression was observed in a small number of neurons in the dorsal brain. Scale bar: 100 μm. (F) Confocal projection of a representative control or tarad40/e01264 central brain expressing CD8::GFP under the control of PL-Gal4. The CycA-expressing PL neurons were present and grossly normal in morphology in tara mutants. d40 (d, for short) is an imprecise excision allele with a sleep phenotype similar to s132 (see Experimental Procedures). Scale bar: 100 μm. Mean ± SEM is shown. *p < 0.05, ***p < 0.001, ns: not significant, two-way ANOVA followed by Tukey post hoc test (A, B); Student’s t-test (D). Related to Figure 4.
Figure S5. Activation of CycA-Expressing PL Neurons Suppresses Sleep. (A) Sleep profile for male flies expressing the NaChBac sodium channel under the control of PL-Gal4 (PL > NaChBac) and parental controls (n=48–64). (B) Daily sleep, (C) waking activity, (D) sleep bout duration, (E) sleep bout number, and (F) sleep latency at ZT12 for the flies shown in (A). (G) Total daily sleep for male flies carrying both UAS-TrpA1 and PL-Gal4 (PL > TrpA1, n=32) relative to parental controls (n=16–32). Flies were monitored at 29°C, which activates the TrpA1 channel, and at 22°C, which inactivates the TrpA1 channel. Mean ± SEM is shown. *p < 0.05, **p < 0.01, and ***p < 0.001, one-way ANOVA followed by Dunnett post hoc test relative to controls (B–C, E–G); Kruskal-Wallis test (D). Related to Figure 5.
Figure S6. tara and Cdk1 interact antagonistically to control sleep. (A) Daily sleep for control, Cdk1c03495/+, tarae01264/s132, and Cdk1c03495/+; tarae02164/s132 females (n=23–32). (B) Daily sleep for male flies of the indicated genotypes (n=26–43). (C) Cdk1 mRNA levels of tara1/s132 mutants were comparable to those of control flies. (D) Adult-stage pan-neuronal overexpression of wild-type Cdk1 has little effect on sleep. Flies were fed either RU486 or vehicle (EtOH) (n=31–32). Mean ± SEM is shown. **p < 0.01, ns: not significant, one-way ANOVA followed by Tukey post hoc test (A, B, D); Student’s t-test (C). Related to Figure 6.
Acknowledgments
We thank Drs. Shelagh Campbell, Jae Park, Lynn Cooley, Henri-Marc Bourbon, Kazuhiko Kume, and Amita Seghal, the Bloomington Stock Center, National Institute of Genetics, and the Harvard (Exelixis) Stock Center for fly stocks; Dr. Ralf Stanewsky for the PER antibody; Drs. M. Boudinot and Francois Rouyer for the FaasX software; Dr. William Joiner for the Sleeplab software; Andrea Nam and Katelyn Kallas for technical assistance; and Jennifer Wilson, Drs. Amita Sehgal, Mi Shi, James Jaynes, and Angelique Lamaze for comments on the manuscript. This work was supported by a grant from the National Institutes of Health (R01GM088221 to K.K.) and predoctoral fellowships from the Portuguese Foundation for Science and Technology (SFRH/BD/51726/2011 to D.J.S.A and SFRH/BD/52321/2013 to D.R.M). Sequencing was performed at the Kimmel Cancer Center Nucleic Acid Facility, which is supported by a grant from the NIH (P30CA56036).
Footnotes
Author Contributions
K.K. conceived the study, and D.J.S.A. and K.K. designed the experiments and analyzed the data. D.J.S.A. performed the experiments with the help of D.L., D.R.M., J.E.C.J., and H.H.; and D.R. identified the dorsal CycA-expressing cells as the pars lateralis cluster. D.L. and D.R.M contributed equally to this work. The manuscript was written by K.K. and D.J.S.A. with editorial input from D.R. and J.E.C.J.
Inventory of Supplemental Information
Supplemental Information contains 6 Figures, Experimental Procedures, and References.
References
- 1.Cirelli C, Tononi G. Is sleep essential? PLoS Biol. 2008;6:216. doi: 10.1371/journal.pbio.0060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rechtschaffen A, Gilliland MA, Bergmann BM, Winter JB. Physiological correlates of prolonged sleep deprivation in rats. Science. 1983;221:182–184. doi: 10.1126/science.6857280. [DOI] [PubMed] [Google Scholar]
- 3.Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–291. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- 4.Cirelli C. The genetic and molecular regulation of sleep: from fruit flies to humans. Nat Rev Neurosci. 2009;10:549–560. doi: 10.1038/nrn2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crocker A, Sehgal A. Genetic analysis of sleep. Genes Dev. 2010;24:1220–1235. doi: 10.1101/gad.1913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 7.Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ, Sehgal A. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008;321:372–376. doi: 10.1126/science.1155942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogulja D, Young MW. Control of sleep by cyclin A and its regulator. Science. 2012;335:1617–1621. doi: 10.1126/science.1212476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stavropoulos N, Young MW. insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron. 2011;72:964–976. doi: 10.1016/j.neuron.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M, Griffith LC. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci. 2008;11:354–359. doi: 10.1038/nn2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan Q, Joiner WJ, Sehgal A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol. 2006;16:1051–1062. doi: 10.1016/j.cub.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 12.Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, Yin JC, Sehgal A. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–1115. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- 13.Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–7384. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douglas CL, Vyazovskiy V, Southard T, Chiu SY, Messing A, Tononi G, Cirelli C. Sleep in Kcna2 knockout mice. BMC Biol. 2007;5:42. doi: 10.1186/1741-7007-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graves LA, Hellman K, Veasey S, Blendy JA, Pack AI, Abel T. Genetic evidence for a role of CREB in sustained cortical arousal. J Neurophysiol. 2003;90:1152–1159. doi: 10.1152/jn.00882.2002. [DOI] [PubMed] [Google Scholar]
- 16.Pfeiffenberger C, Allada R. Cul3 and the BTB adaptor insomniac are key regulators of sleep homeostasis and a dopamine arousal pathway in Drosophila. PLoS Genet. 2012;8:e1003003. doi: 10.1371/journal.pgen.1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- 18.Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- 19.Crocker A, Shahidullah M, Levitan IB, Sehgal A. Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron. 2010;65:670–681. doi: 10.1016/j.neuron.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Q, Liu S, Kodama L, Driscoll MR, Wu MN. Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr Biol. 2012;22:2114–2123. doi: 10.1016/j.cub.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, Shaw PJ. Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science. 2011;332:1571–1576. doi: 10.1126/science.1202249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R. The GABA(A) Receptor RDL Acts in Peptidergic PDF Neurons to Promote Sleep in Drosophila. Curr Biol. 2009;19:386–390. doi: 10.1016/j.cub.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M, et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60:672–682. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu SI, Yang CM, Sim KG, Hentschel DM, O’Leary E, Bonventre JV. TRIP-Br: a novel family of PHD zinc finger- and bromodomain-interacting proteins that regulate the transcriptional activity of E2F-1/DP-1. The EMBO journal. 2001;20:2273–2285. doi: 10.1093/emboj/20.9.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sim KG, Zang Z, Yang CM, Bonventre JV, Hsu SI. TRIP-Br links E2F to novel functions in the regulation of cyclin E expression during cell cycle progression and in the maintenance of genomic stability. Cell Cycle. 2004;3:1296–1304. doi: 10.4161/cc.3.10.1157. [DOI] [PubMed] [Google Scholar]
- 26.Calgaro S, Boube M, Cribbs DL, Bourbon HM. The Drosophila gene taranis encodes a novel trithorax group member potentially linked to the cell cycle regulatory apparatus. Genetics. 2002;160:547–560. doi: 10.1093/genetics/160.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manansala MC, Min S, Cleary MD. The Drosophila SERTAD protein Taranis determines lineage-specific neural progenitor proliferation patterns. Dev Biol. 2013;376:150–162. doi: 10.1016/j.ydbio.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 28.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. The EMBO journal. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437:1257–1263. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- 30.de Velasco B, Erclik T, Shy D, Sclafani J, Lipshitz H, McInnes R, Hartenstein V. Specification and development of the pars intercerebralis and pars lateralis, neuroendocrine command centers in the Drosophila brain. Dev Biol. 2007;302:309–323. doi: 10.1016/j.ydbio.2006.09.035. [DOI] [PubMed] [Google Scholar]
- 31.Jepson JE, Shahidullah M, Lamaze A, Peterson D, Pan H, Koh K. dyschronic, a Drosophila homolog of a deaf-blindness gene, regulates circadian output and Slowpoke channels. PLoS Genet. 2012;8:e1002671. doi: 10.1371/journal.pgen.1002671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hendricks JC, Lu S, Kume K, Yin JC, Yang Z, Sehgal A. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms. 2003;18:12–25. doi: 10.1177/0748730402239673. [DOI] [PubMed] [Google Scholar]
- 33.Quinones-Coello AT, Petrella LN, Ayers K, Melillo A, Mazzalupo S, Hudson AM, Wang S, Castiblanco C, Buszczak M, Hoskins RA, et al. Exploring strategies for protein trapping in Drosophila. Genetics. 2007;175:1089–1104. doi: 10.1534/genetics.106.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao Z, Roman G, Zong L, Davis RL. Pharmacogenetic rescue in time and space of the rutabaga memory impairment by using Gene-Switch. Proc Natl Acad Sci USA. 2004;101:198–203. doi: 10.1073/pnas.0306128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiga S. Anatomy and functions of brain neurosecretory cells in Diptera. Microscopy Research and Technique. 2003;62:114–131. doi: 10.1002/jemt.10366. [DOI] [PubMed] [Google Scholar]
- 36.Choi SH, Lee G, Monahan P, Park JH. Spatial regulation of Corazonin neuropeptide expression requires multiple cis-acting elements in Drosophila melanogaster. J Comp Neurol. 2008;507:1184–1195. doi: 10.1002/cne.21594. [DOI] [PubMed] [Google Scholar]
- 37.Tononi G, Cirelli C. Sleep and the price of plasticity: from synaptic and cellular homeostasis to memory consolidation and integration. Neuron. 2014;81:12–34. doi: 10.1016/j.neuron.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson IM, Ranjan R, Schwarz TL. Synaptotagmins I and IV promote transmitter release independently of Ca(2+) binding in the C(2)A domain. Nature. 2002;418:336–340. doi: 10.1038/nature00915. [DOI] [PubMed] [Google Scholar]
- 39.Nitabach MN, Wu Y, Sheeba V, Lemon WC, Strumbos J, Zelensky PK, White BH, Holmes TC. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, Garrity PA. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454:217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sweeney ST, Broadie K, Keane J, Niemann H, O’Kane CJ. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14:341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 42.Ayeni JO, Varadarajan R, Mukherjee O, Stuart DT, Sprenger F, Srayko M, Campbell SD. Dual phosphorylation of cdk1 coordinates cell proliferation with key developmental processes in Drosophila. Genetics. 2014;196:197–210. doi: 10.1534/genetics.113.156281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su SC, Tsai LH. Cyclin-dependent kinases in brain development and disease. Annu Rev Cell Dev Biol. 2011;27:465–491. doi: 10.1146/annurev-cellbio-092910-154023. [DOI] [PubMed] [Google Scholar]
- 44.Suryadinata R, Sadowski M, Sarcevic B. Control of cell cycle progression by phosphorylation of cyclin-dependent kinase (CDK) substrates. Biosci Rep. 2010;30:243–255. doi: 10.1042/BSR20090171. [DOI] [PubMed] [Google Scholar]
- 45.Meyer CA, Jacobs HW, Datar SA, Du W, Edgar BA, Lehner CF. Drosophila Cdk4 is required for normal growth and is dispensable for cell cycle progression. The EMBO journal. 2000;19:4533–4542. doi: 10.1093/emboj/19.17.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Odajima J, Wills ZP, Ndassa YM, Terunuma M, Kretschmannova K, Deeb TZ, Geng Y, Gawrzak S, Quadros IM, Newman J, et al. Cyclin E constrains Cdk5 activity to regulate synaptic plasticity and memory formation. Dev Cell. 2011;21:655–668. doi: 10.1016/j.devcel.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liew CW, Boucher J, Cheong JK, Vernochet C, Koh HJ, Mallol C, Townsend K, Langin D, Kawamori D, Hu J, et al. Ablation of TRIP-Br2, a regulator of fat lipolysis, thermogenesis and oxidative metabolism, prevents diet-induced obesity and insulin resistance. Nat Med. 2013;19:217–226. doi: 10.1038/nm.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borbely AA, Achermann P. Sleep homeostasis and models of sleep regulation. J Biol Rhythms. 1999;14:557–568. doi: 10.1177/074873099129000894. [DOI] [PubMed] [Google Scholar]
- 49.Rihel J, Prober DA, Arvanites A, Lam K, Zimmerman S, Jang S, Haggarty SJ, Kokel D, Rubin LL, Peterson RT, et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science. 2010;327:348–351. doi: 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi M, Yue Z, Kuryatov A, Lindstrom JM, Sehgal A. Identification of Redeye, a new sleep-regulating protein whose expression is modulated by sleep amount. Elife. 2014;3:e01473. doi: 10.7554/eLife.01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu MN, Joiner WJ, Dean T, Yue Z, Smith CJ, Chen D, Hoshi T, Sehgal A, Koh K. SLEEPLESS, a Ly-6/neurotoxin family member, regulates the levels, localization and activity of Shaker. Nat Neurosci. 2010;13:69–75. doi: 10.1038/nn.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu S, Lamaze A, Liu Q, Tabuchi M, Yang Y, Fowler M, Bharadwaj R, Zhang J, Bedont J, Blackshaw S, et al. WIDE AWAKE mediates the circadian timing of sleep onset. Neuron. 2014;82:151–166. doi: 10.1016/j.neuron.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verstegen AM, Tagliatti E, Lignani G, Marte A, Stolero T, Atias M, Corradi A, Valtorta F, Gitler D, Onofri F, et al. Phosphorylation of synapsin I by cyclin-dependent kinase-5 sets the ratio between the resting and recycling pools of synaptic vesicles at hippocampal synapses. J Neurosci. 2014;34:7266–7280. doi: 10.1523/JNEUROSCI.3973-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gutierrez L, Zurita M, Kennison JA, Vazquez M. The Drosophila trithorax group gene tonalli (tna) interacts genetically with the Brahma remodeling complex and encodes an SP-RING finger protein. Development. 2003;130:343–354. doi: 10.1242/dev.00222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sleep Phenotypes and Genetic Analysis of tara: Male Data. (A) Sleep profile of homozygous taras132 (black X’s), transheterozygous tara1/s132 (black squares), and their background control males (white circles) (n=60–73). The white and black bars below the X-axis represent light and dark periods, respectively. (B) Control and tara mutant males of the indicated genotypes (n=41–73). (C) Waking activity, (D) sleep bout duration, (E) sleep bout number, and (F) sleep latency at lights off for the same male flies shown in (B). Sleep bout duration is shown in simplified box plots, where the median and interquartile range are represented. (G) Total daily sleep of control and Df(3R)Exel7329 male heterozygotes in trans to either a wild type (Df/+) or taras132 (Df/s) allele (n=35–70). (H) Total daily sleep of control, taras132, and precise excision male flies (n=15–34). (I) Total daily sleep of male flies of the indicated genotypes (n=35–42). Ubiquitous expression of tara-B (Ub-tara) rescued the tara sleep phenotypes. Mean ± SEM is shown. *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA followed by Dunnett post hoc test (G, H); Tukey post hoc test (B, C, E, F, I); Kruskal-Wallis test (D). For simplicity, only significant differences between each mutant and the control and those between taras132, tarae01264/s132, and tara1/s132 mutants are indicated. Related to Figure 1.
Figure S2. Circadian Phenotypes and Clock-Independent Sleep Loss of tara Mutant Males. (A) Free-running circadian phenotypes of male flies of the indicated genotypes in DD. χ2 periodogram analysis was performed for each fly using the FaasX software to determine the free-running period, tau. Power, a measure of rhythmicity, corresponds to peak – significance value at p = 0.05. (B) Total daily sleep amount in LL and DD for control, tarae01264/s132, and tara1/s132 males (n=29–77 for LL; n=55–94 for DD). Sleep levels on the 3rd day in constant conditions are shown. Mean ± SEM is shown. ***p < 0.001, one-way ANOVA followed by Dunnett post hoc test relative to control flies (B). Related to Figure 2.
Figure S3. Characterization of taraGFP Flies. (A) Western blot showing a band shift in homozygous taraGFP lysates. (B) Total daily sleep and (C) waking activity of taraGFP and control females (n=47–64). taraGFP flies did not exhibit sleep abnormalities, suggesting TARA::G-FP is functional. (D) Pan-neuronal tara knockdown using two independent Gal4 lines (elav and nsyb) resulted in a significant reduction of sleep in males (n=33–63 except for elav > tara RNAi, for which n=11). (E) Western blot shows a marked reduction of TARA levels in females in which tara was knocked down pan-neuronally (elav > tara RNAi). The experiment was performed three times with similar results. MAPK was used as loading control (A, E). Mean ± SEM is shown. ***p < 0.001, ns: not significant, Student’s t-test (B, C); one-way ANOVA followed by Dunnett post hoc test relative to controls (D). Related to Figure 3.
Figure S4. Genetic Interaction between tara and CycA. (A) Daily sleep for male flies of the indicated genotypes (n=54–64). (B) Daily sleep for female flies of the indicated genotypes (n=40–56). (C) Maximal-intensity confocal projections of the dorsal half of the central brain of representative control and DATfmn adult flies immunostained with an antibody to CycA. In contrast to tara mutants, CycA levels were not altered in DATfmn mutants. Scale bar: 100 μm. (D) CycA mRNA levels relative to actin mRNA levels were not significantly different between tara1/s132 and control flies (n=3). (E) Confocal projection of a female brain (top) and ventral nerve cord (bottom) expressing CD8::GFP under the control of PL-Gal4. GFP expression was observed in a small number of neurons in the dorsal brain. Scale bar: 100 μm. (F) Confocal projection of a representative control or tarad40/e01264 central brain expressing CD8::GFP under the control of PL-Gal4. The CycA-expressing PL neurons were present and grossly normal in morphology in tara mutants. d40 (d, for short) is an imprecise excision allele with a sleep phenotype similar to s132 (see Experimental Procedures). Scale bar: 100 μm. Mean ± SEM is shown. *p < 0.05, ***p < 0.001, ns: not significant, two-way ANOVA followed by Tukey post hoc test (A, B); Student’s t-test (D). Related to Figure 4.
Figure S5. Activation of CycA-Expressing PL Neurons Suppresses Sleep. (A) Sleep profile for male flies expressing the NaChBac sodium channel under the control of PL-Gal4 (PL > NaChBac) and parental controls (n=48–64). (B) Daily sleep, (C) waking activity, (D) sleep bout duration, (E) sleep bout number, and (F) sleep latency at ZT12 for the flies shown in (A). (G) Total daily sleep for male flies carrying both UAS-TrpA1 and PL-Gal4 (PL > TrpA1, n=32) relative to parental controls (n=16–32). Flies were monitored at 29°C, which activates the TrpA1 channel, and at 22°C, which inactivates the TrpA1 channel. Mean ± SEM is shown. *p < 0.05, **p < 0.01, and ***p < 0.001, one-way ANOVA followed by Dunnett post hoc test relative to controls (B–C, E–G); Kruskal-Wallis test (D). Related to Figure 5.
Figure S6. tara and Cdk1 interact antagonistically to control sleep. (A) Daily sleep for control, Cdk1c03495/+, tarae01264/s132, and Cdk1c03495/+; tarae02164/s132 females (n=23–32). (B) Daily sleep for male flies of the indicated genotypes (n=26–43). (C) Cdk1 mRNA levels of tara1/s132 mutants were comparable to those of control flies. (D) Adult-stage pan-neuronal overexpression of wild-type Cdk1 has little effect on sleep. Flies were fed either RU486 or vehicle (EtOH) (n=31–32). Mean ± SEM is shown. **p < 0.01, ns: not significant, one-way ANOVA followed by Tukey post hoc test (A, B, D); Student’s t-test (C). Related to Figure 6.