Abstract
Providencia alcalifaciens is an emerging bacterial pathogen known to cause acute gastroenteritis in children and travelers. In July 2013, P. alcalifaciens was isolated from four children appearing for diarrhea at Kiambu District Hospital (KDH) in Kenya. This study describes the outbreak investigation, which aimed to identify the source and mechanisms of infection. We identified seven primary and four secondary cases. Among primary cases were four mothers who had children and experienced mild diarrhea after eating mashed potatoes. The mothers reported feeding children after visiting the toilet and washing their hands without soap. P. alcalifaciens was detected from all secondary cases, and the isolates were found to be clonal by random amplified polymorphic DNA (RAPD) fingerprinting. Our study suggests that the outbreak was caused by P. alcalifaciens, although no fluid accumulation was observed in rabbit ileal loops. The vehicle of the outbreak was believed to be the mashed potato dish, but the source of P. alcalifaciens could not be confirmed. We found that lack of hygiene, inadequate food storage, and improper hand washing before food preparation was the likely cause of the current outbreak. This is the first report of a foodborne infection caused by P. alcalifaciens in Kenya.
Introduction
Providencia alcalifaciens is a gram-negative rod-shaped bacteria that belongs to the family Enterobacteriaceae and has been implicated as a causative agent of diarrhea.1 Several studies reported that P. alcalifaciens most frequently affects children, including travelers from developing countries.2–4 The etiological role of P. alcalifaciens has been established through case–control studies of diarrhea and pathogenicity studies of isolates.2,5 Both in vitro and in vivo studies demonstrated that P. alcalifaciens can invade cultured mammalian cells and intestinal tissues, cause diarrhea in rabbits, and induce fluid accumulation in rabbit ileal loop model.5–8 Furthermore, two large outbreaks of foodborne infection caused by P. alcalifaciens have been reported in Japan and the Czech Republic, thus providing the evidence of causing gastroenteritis.9,10
Providencia alcalifaciens is usually considered to be a commensal in the gastrointestinal tract, and most clinical laboratories do not recognize it as a potential cause of diarrhea. Therefore, it can be easily passed through the routine screening as pathogen, except for rare cases of pure culture isolation with no other recognized enteric pathogen. In July 2013, four cases of acute gastroenteritis were admitted to the Kiambu District Hospital (KDH) in Kenya. All cases were children less than 2 years of age. In our clinical studies, we have isolated P. alcalifaciens as the predominant organism from stool samples. The diagnosis of a foodborne disease should be considered when two or more persons experience a similar illness with gastrointestinal manifestation after ingestion of a common food.11 Therefore, an investigation was conducted to identify the vehicle and source of infection and other contributing factors that might help in the design of control measures. To our knowledge, this is the first report of a foodborne outbreak due to P. alcalifaciens in Kenya.
Materials and Methods
Study area and report of a case.
Kiambu District is a densely populated (638 people/km2) and rapidly urbanizing area north of Nairobi. In August 2008, we initiated hospital-based rotavirus and other enteric pathogen surveillance in KDH. On July 18, 2013, we isolated four P. alcalifaciens strains as the predominant organism from stool samples of children younger than 2 years of age with acute gastroenteritis from KDH, who live in suburban area with poor sanitation and water supply. All parents/guardians of the children were asked to fill up a questionnaire that covered personal information such as age, gender, place of residence, country of birth, contact with other infected persons, clinical symptoms, source of the meals consumed within the days before the onset of outbreak, living conditions, and sanitary behaviors. Following the collection of information that might suggest commonalities among the cases, we launched an epidemiological investigation of the outbreak. The study was approved by the Kenya Medical Research Institute Ethics Review Committee (KEMRI/SSC/1323).
Microbiological investigation.
Stool specimens were examined for bacterial and viral pathogens. Non–lactose-fermenting bacterial colonies on MacConkey agar (Becton, Dickinson and Company, Sparks, MD) were identified biochemically as outlined by Ewing,12 and confirmed by a VITEK2 automated analyzer (bioMérieux, Inc., Durham, NC). For genetic examination, bacterial genomic DNA was extracted by the phenol–chloroform extraction method. Polymerase chain reaction (PCR) was performed targeting dnaJ gene.13 The PCR mixture was prepared with the Takara Ex Taq Hot Start version kit (TAKARA BIO INC., Otsu, Shiga, Japan) according to the manufacturer's instructions. Enzyme-linked immunosorbent assay was performed for rotavirus identification.
Antimicrobial susceptibility tests.
Test organisms were included in plates of Mueller-Hinton agar (Becton, Dickinson and Company) as recommended by the Clinical and Laboratory Standards Institute.14 Minimal inhibitory concentrations (MICs) were determined by the Etest (BioMerieux) strips according to the manufacturer's instruction manual and all specimens were tested with the same lot number. Antibiotics tested included ampicillin, amoxicillin, amoxicillin/clavulanate, ceftriaxone, chloramphenicol, ciprofloxacin, doxycycline, erythromycin, kanamycin, nalidixic acid, ofloxacin, streptomycin, sulfamethoxazole–trimethoprim, and tetracycline.
Random amplified polymorphic DNA fingerprinting.
Random amplified polymorphic DNA (RAPD) was performed by the method described by Libisch and others15 using primer 5-(AACGCGCAAC) with some modification. Amplification products were separated by electrophoresis in 2% agarose and visualized by ethidium bromide staining.
Rabbit ileal loop test.
New Zealand white rabbit (2.5 kg) was used for the rabbit ileal loop test. Of an 18-hour tryptic soy broth (Becton, Dickinson and Company) culture medium containing 2 × 107 bacteria grown at 37°C, 1 mL was injected into an ∼10-cm-long small intestinal loop of an adult rabbit that had previously fasted for 24 hours. After 18 hours, the rabbit was killed, and the loop was examined for fluid accumulation and other gross pathological changes. A portion of the mucosa of the loop was fixed in buffered formal saline, and paraffin sections were stained with hematoxylin and eosin for light microscopy. Each bacterial isolate was tested in two rabbits, and a pathogenic Vibrio cholerae strain was included as a positive control while phosphate buffered saline (PBS) was used for negative.
Results
Descriptive findings.
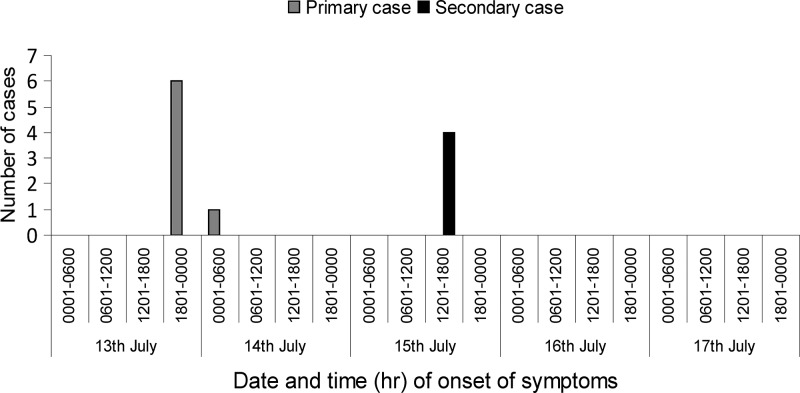
It was found that on July 13, 2013, a group of 12 females gathered in a church for a social event known locally as “chama” and ate mashed potatoes (mukimo), stew, fried bread (mandazi), and tea purchased from Kiambu municipal market. Among the 12 participants, three had children. We were able to interview seven adults including six chama participants for outbreak investigation and obtain stool samples from four of the children but not from the adults. The people we interviewed reported that on July 13 and 14 they experienced mild diarrhea, although none reported to the hospital. However, on July 15, three children (KDH/1666, KDH/1667, and KDH/1668) whose mothers had attended the event became sick with diarrhea. Initially, all the children experienced foul smelling, watery, and non-bloody diarrhea up to five times a day (Table 1). Later, on July 16, the children started vomiting and became severely dehydrated and were admitted to the KDH on July 17. The mother of the fourth child (KDH/1670) also purchased the mashed potatoes from the Kiambu market and reported experiencing similar symptoms. These findings support that the mashed potatoes could be the vehicle for the outbreak. This suggests that the primary cases were members of the social event who had eaten mashed potatoes for lunch on July 13, 2013 and experienced diarrhea after 10–12 hours (Figure 1 ). Subsequent contacts with the primary cases were defined as secondary cases. The period between the onset of diarrhea and admission for all the secondary cases, who reported to the hospital on July 17 was less than 48 hours. However, we could not sample any food items because of the long delay (5 days) between the gathering and the notification of the outbreak.
Table 1.
Age distribution and clinical characteristics of outbreak isolates
| Patient no. | Age (m) | Gender | Symptom | Isolated strain | Co-pathogen |
|---|---|---|---|---|---|
| KDH/1666 | 7 | F | Diarrhea (watery and non-bloody), vomiting, severe dehydration | Providencia alcalifaciens | Citrobacter freundii |
| KDH/1667 | 15 | M | Diarrhea (watery and non-bloody), vomiting, severe dehydration | P. alcalifaciens | Proteus mirabilis |
| KDH/1668 | 17 | F | Diarrhea (watery and non-bloody), vomiting, severe dehydration | P. alcalifaciens | C. freundii |
| KDH/1670 | 16 | M | Diarrhea (watery and non-bloody), vomiting, severe dehydration | P. alcalifaciens | C. freundii |
F = female; KDH = Kiambu District Hospital; m = month; M = male.
Figure 1.
Date of onset of diarrheal symptoms among mothers, including chama participants and children, defined as primary and secondary cases, respectively, July 2013.
Identification of etiological agent.
Providencia alcalifaciens were identified from all four stool samples as non-lactose fermenters and appeared as blue colonies on bromothymol blue medium (Eiken Chemical Co., Ltd., Shimotsuga, Tochigi, Japan) and pale colonies on Deoxycholate Hydrogen Sulfide Lactose agar (Eiken Chemical Co., Ltd., Shimotsuga, Tochigi, Japan). In addition, other enteropathogenic bacterial species were detected, including Proteus mirabilis from one and Citrobacter freundii from three other stool specimens. VITEK 2 automated analyzer and Providencia-specific partial dnaJ gene amplification confirmed the identity of the isolates as P. alcalifaciens (data not shown). We were unable to conclude that the isolated co-pathogens were the causative agents of the outbreak. Furthermore, we could not collect the blood samples to test antibody titer against outbreak-derived P. alcalifaciens since the outbreak investigation begun after patients discharged from the hospital. No rotavirus was detected from the stool specimens. The samples were kept at −80°C for further study.
Susceptibility to antibiotics, genotyping, and enteropathogenicity test.
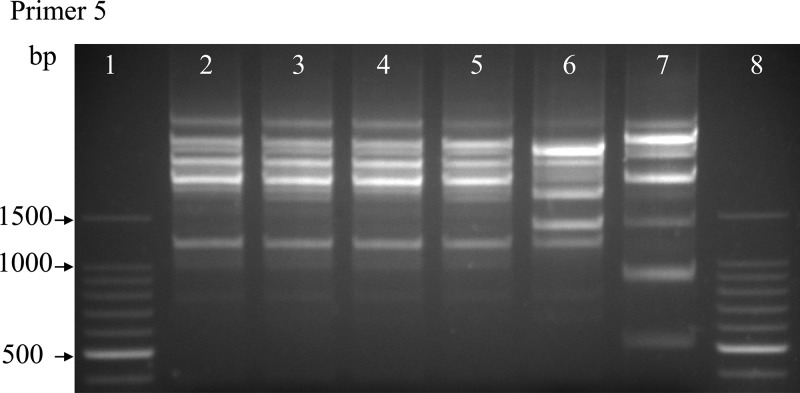
All Providencia isolates revealed similar antibiotic patterns showing susceptibility to ceftriaxone, chloramphenicol, ciprofloxacin, kanamycin, nalidixic acid, ofloxacin, and streptomycin and resistance to amoxicillin, ampicillin, doxycycline, erythromycin, sulfamethoxazole–trimethoprim, and tetracycline. Amoxicillin/clavulanate showed intermediate action (Table 2). In addition, the same genotyping was marked by the RAPD fingerprinting result (Figure 2 ). These findings suggest that the P. alcalifaciens from different children were clonal. None of the outbreak isolates included negative control-induced fluid accumulation in the loops of the two rabbits. In contrast, loops treated with V. cholerae strain (positive control) showed fluid accumulation (2.5 mL/cm) including mucosal necrosis (data not shown).
Table 2.
MIC (μg/mL) results of antimicrobial drugs of Providencia alcalifaciens strains
| Antimicrobial drugs | KDH/1666 | KDH/1667 | KDH/1668 | KDH/1670 |
|---|---|---|---|---|
| Ampicillin | 256 (R) | 256 (R) | 256 (R) | 256 (R) |
| Amoxicillin | 256 (R) | 256 (R) | 256 (R) | 256 (R) |
| Amoxicillin/clavulanate | 16 (I) | 16 (I) | 12 (I) | 12 (I) |
| Ceftriaxone | 0.002 (S) | 0.002 (S) | 0.002 (S) | 0.002 (S) |
| Chloramphenicol | 1.5 (S) | 2 (S) | 6 (S) | 2 (S) |
| Ciprofloxacin | 0.002 (S) | 0.002 (S) | 0.002 (S) | 0.002 (S) |
| Doxycycline | 256 (R) | 256 (R) | 256 (R) | 256 (R) |
| Erythromycin | 256 (R) | 256 (R) | 256 (R) | 256 (R) |
| Kanamycin | 0.125 (S) | 0.125 (S) | 0.125 (S) | 0.125 (S) |
| Nalidixic acid | 3 (S) | 4 (S) | 3 (S) | 3 (S) |
| Ofloxacin | 0.047 (S) | 0.047 (S) | 0.047 (S) | 0.047 (S) |
| Streptomycin | 3 (S) | 2 (S) | 3 (S) | 3 (S) |
| Sulfamethoxazole–trimethoprim | 32 (R) | 32 (R) | 32 (R) | 32 (R) |
| Tetracycline | 64 (R) | 48 (R) | 48 (R) | 48 (R) |
KDH = Kiambu District Hospital; MIC = minimum inhibitory concentration; I = intermediate; R = resistant; S = susceptible.
Figure 2.
Random amplified polymorphic DNA (RAPD) profiles obtained from genomic DNA of Providencia alcalifaciens and P. rettgeri strains. Lanes 1 and 8, 100 bp DNA marker (Takara); lanes 2–5, Kiambu outbreak isolates from different patients: KDH/1666–1668 and 1670; lane 6, P. alcalifaciens non-outbreak strain: KDH/1659; lane 7, P. rettgeri non-outbreak strain: KDH/1640.
Discussion
The data suggest that the mashed potato dish was the likely source of the outbreak at the social gathering of July 13 and that P. alcalifaciens was the pathogen most probably responsible. However, as no samples of food consumed at the social gathering were available for laboratory analysis, there was no direct evidence of P. alcalifaciens contamination of the mashed potatoes. We suspect that mashed potato dish was the vehicle of transmission because the mother of the fourth child was not a chama participant but became sick after consumption of mashed potatoes that was purchased on the same day from the same shop at the market.
Providencia alcalifaciens was identified as the etiological agent responsible for diarrhea among children and adults in other studies.2–4 The incubation period of the diarrhea was 1–4 days with the symptoms of non-bloody and mucus diarrhea that is partially in agreement with our study. Albert and others2 compared the isolation rate of P. alcalifaciens from stool samples of children and demonstrated a significant association between P. alcalifaciens and diarrhea. The epidemiology and clinical course of our study is similar to reported outbreaks in developed regions.9,10
Chlibek and others10 reported that gastroenteritis outbreaks among Czech army hospital personnel due to P. alcalifaciens have occurred as a result of food contamination originating from food handlers. The suspected food was potato salad, which was consumed 3–10 hours after preparation and indicated the possibility of multiplication of the infectious organisms at ambient temperature ≥ 25°C. Several studies also documented that consumption of long-stored mashed potato at room temperature has possible risk of bacterial multiplication.16–18 Unfortunately, we could not determine the time interval between mashed potato preparation and consumption because it was purchased as ready-made food.
We can speculate that all outbreak organisms that are biochemically and genetically identified as P. alcalifaciens are pathogenic in humans. We could not confirm the pathogenicity of P. alcalifaciens that showed no fluid accumulation. Albert and others (1992) also found the negative result in an ileal model even though P. alcalifaciens was the only recognized enteric pathogen causing diarrhea in three patients. This finding suggested that there could be differences in the disease-producing abilities of different strains of P. alcalifaciens.5 However, further analysis is needed to confirm the enteropathogenicity of P. alcalifaciens.
The specimens of outbreak patients from whom P. alcalifaciens were also confirmed to have co-pathogens. Previous studies showed that the isolation of multiple enteric pathogens from stool samples is common in many developing countries.19 This problem indicates the high degree of fecal contamination in the environments of developing countries. Antibiogram pattern and RAPD analysis showed that the isolated P. alcalifaciens from all patients are clonal, which indicates the common source of infection.
We concluded that a combination of lack of hygiene, improper hand washing before food preparation, inadequate food storage, and handling was the cause of the current outbreak. Therefore, promoting health education campaigns that emphasize basic household sanitary methods can improve the control of the foodborne outbreak. This report contributes to the epidemiologic knowledge concerning P. alcalifaciens, and there is much to learn about the local epidemiology, pathogenesis, and immunology of this emerging pathogen.
ACKNOWLEDGMENTS
We are grateful to the Director of Kenya Medical Research Institute for collaborative support and the staff members of the Kiambu District Hospital (KDH) and the NUITM-KEMRI project. We thank all the chama participants and their family members for their contribution to our study and Amina Galata, who assisted to fill up a questionnaire for outbreak investigation.
Footnotes
Financial support: This study was supported in part by Grant-in-Aid for Scientific Research, Japan (Research B: 15H05286) and the Program for Nurturing Global Leaders in TECD, Nagasaki University, Japan.
Authors' addresses: Mohammad Monir Shah, Peter S. Larson, and Yoshio Ichinose, Centre for Infectious Disease Research in Asia and Africa, Institute of Tropical Medicine Nagasaki University, Nagasaki, Japan, and Institute of Tropical Medicine-Kenya Medical Research Institute Project, Nairobi, Kenya, E-mails: shah@nagasaki-u.ac.jp, pslarson2@gmail.com, and ichinose@nagasaki-u.ac.jp. Erick Odoyo, Cyrus Kathiiko, and Gabriel Miringu, Nagasaki University Institute of Tropical Medicine-Kenya Medical Research Institute Project, Nairobi, Kenya, E-mails: e.odoyo@gmail.com, ckathiiko@gmail.com, and gbm900@yahoo.com. Ernest Apondi, Centre for Infectious Disease Research in Asia and Africa, Institute of Tropical Medicine Nagasaki University, Nagasaki, Japan, E-mail: wandesh2000@yahoo.com. Masahiro Nakashima, Department of Tumor and Diagnostic Pathology, Nagasaki University Graduate School of Biomedical Sciences, Nagasaki, Japan, E-mail: moemoe@nagasaki-u.ac.jp.
References
- 1.Guth BEC, Perrella E. Prevalence of invasive ability and other virulence-associated characteristics in Providencia alcalifaciens strains isolated in Sao Paulo, Brazil. J Med Microbiol. 1996;45:459–462. doi: 10.1099/00222615-45-6-459. [DOI] [PubMed] [Google Scholar]
- 2.Albert MJ, Faruque ASG, Mahalanabis D. Association of Providencia alcalifaciens with diarrhea in children. J Clin Microbiol. 1998;36:1433–1435. doi: 10.1128/jcm.36.5.1433-1435.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haynes J, Hawkey PM. Providencia alcalifaciens and travellers' diarrhoea. BMJ. 1989;299:94–95. doi: 10.1136/bmj.299.6691.94-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoh M, Matsuyama J, Ohnishi M, Takagi K, Miyagi H, Mori K, Park KS, Ono T, Honda T. Importance of Providencia species as a major cause of traveler's diarrhoea. J Med Microbiol. 2005;54:1077–1082. doi: 10.1099/jmm.0.45846-0. [DOI] [PubMed] [Google Scholar]
- 5.Albert MJ, Alam K, Ansaruzzaman M, Islam MM, Rahman ASMH, Haider K, Bhuiyan NA, Nahar S, Ryan N, Montanaro J, Mathan MM. Pathogenesis of Providencia alcalifaciens-induced diarrhea. Infect Immun. 1992;60:5017–5024. doi: 10.1128/iai.60.12.5017-5024.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathan M, Mathan VI, Albert MJ. Electron microscopic study of the attachment and penetration of rabbit intestinal epithelium by Providencia alcalifaciens. J Pathol. 1993;171:67–71. doi: 10.1002/path.1711710114. [DOI] [PubMed] [Google Scholar]
- 7.Albert MJ, Ansaruzzaman M, Bhuiyan NA, Neogi PKB, Faruque ASG. Characteristics of invasion of HEp-2 cells by Providencia alcalifaciens. J Med Microbiol. 1995;42:186–190. doi: 10.1099/00222615-42-3-186. [DOI] [PubMed] [Google Scholar]
- 8.Janda JM, Abbott SL, Woodward D, Khashe S. Invasion of HEp-2 and other eukaryotic cell lines by Providenciae: further evidence supporting the role of Providencia alcalifaciens in bacterial gastroenteritis. Curr Microbiol. 1998;37:159–165. doi: 10.1007/s002849900357. [DOI] [PubMed] [Google Scholar]
- 9.Murata T, Iida T, Shiomi Y, Tagomori K, Akeda Y, Yanagihara I, Mushiake S, Ishiguro F, Honda T. A large outbreak of food borne infection attributed to Providencia alcalifaciens. J Infect Dis. 2001;184:1050–1055. doi: 10.1086/323458. [DOI] [PubMed] [Google Scholar]
- 10.Chlibek R, Jirous J, Beran J. Diarrhea outbreak among Czech Army Field Hospital personnel caused by Providencia alcalifaciens. J Travel Med. 2002;9:151–152. doi: 10.2310/7060.2002.23190. [DOI] [PubMed] [Google Scholar]
- 11.Bean NH, Griffin PM, Goulding JS, Ivey CB. Foodborne disease outbreaks, 5-year summary, 1983–1987. MMWR CDC Surveill Summ. 1990;39:15–57. [PubMed] [Google Scholar]
- 12.Ewing WH. Edwards and Ewing's Identification of Enterobactericeae. 4th edition. New York, NY: Elsevier Science Publishing Co. Inc.; 1986. pp. 443–459. [Google Scholar]
- 13.Pham HN, Ohkusu K, Mishima N, Noda M, Shah MM, Sun X, Hayashi M, Ezaki T. Phylogeny and species identification of the family Enterobacteriaceae based on dnaJ sequences. Diagn Microbiol Infect Dis. 2007;58:153–161. doi: 10.1016/j.diagmicrobio.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standard Institute . Performance Standards for Antimicrobial Susceptibility Testing; Seventeenth Informational Supplement. Wayne, PA: Clinical and Laboratory Standard Institute; 2007. (Clinical and Laboratory Standard Institute (CLSI) document M100-S17). [Google Scholar]
- 15.Libisch B, Watine J, Balogh B, Gacs M, Muzslay M, Szabó G, Füzi M. Molecular typing indicates an important role for two international clonal complexes in dissemination of VIM-producing Pseudomonas aeruginosa clinical isolates in Hungary. Res Microbiol. 2008;159:162–168. doi: 10.1016/j.resmic.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Lee VJ, Ong AE, Auw M. An outbreak of Salmonella gastrointestinal illness in a military camp. Ann Acad Med Singapore. 2009;38:207–211. [PubMed] [Google Scholar]
- 17.Contzen M, Hailer M, Rau J. Isolation of Bacillus cytotoxicus from various commercial potato products. Int J Food Microbiol. 2014;174:19–22. doi: 10.1016/j.ijfoodmicro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 18.Doan CH, Davidson PM. Microbiology of potatoes and potato products: a review. J Food Prot. 2000;63:668–683. doi: 10.4315/0362-028x-63.5.668. [DOI] [PubMed] [Google Scholar]
- 19.Faruque ASG, Mahalanabis D, Islam A, Hoque SS. Severity of cholera during concurrent infections with other enteric pathogens. J Diarrhoeal Dis Res. 1994;12:214–218. [PubMed] [Google Scholar]