Abstract
Leishmania parasites isolated, between 1979 and 1988 by the late Bryce Walton, from Dominican Republic (DR) patients with diffuse cutaneous leishmaniasis, were characterized using a panel of 12 isoenzymes, 23 monoclonal antibodies, small subunit ribosomal DNA (SSu rDNA), and multilocus sequence analysis (MLSA). The isoenzyme and monoclonal antibody profiles and the MLSA results showed that the Dominican Republic parasites were distinct from other described Leishmania species. This new species belongs to the mexicana complex, which is distributed in central and parts of northern South America. It is suggested that the parasites uniqueness from other members of the mexicana complex is related to it being isolated on an island for millions of years. If Leishmania (Leishmania) waltoni fails to adapt to some imported mammal, such as the house rat, it will be the only Leishmania to be classified as an endangered species. The excessive destruction of habitats on Hispaniola threatens the survival of its vectors and presumed natural reservoirs, such as the rodent hutias and the small insectivorous mammal solenodon. The concept of Leishmania species is discussed in the light of recent evaluations on criteria for defining bacterial species.
Introduction
Diffuse cutaneous leishmaniasis in Latin America has been associated with three Leishmania species that all belong to the subgenus Leishmania (Leishmania)—L. (L.) amazonensis, L. (L.) mexicana, and L. (L.) pifanoi.1 In the Dominican Republic (DR), three cases of diffuse cutaneous leishmaniasis (DCL) were reported in 19752 and, during the following 10 years, 22 additional cases were diagnosed.3
An epidemiological investigation3 carried out between May 1981 and April 1984 lead to the incrimination of Lutzomyia christophei as the probable vector. This sandfly species was collected in the vicinity of several human cases and shown to be experimentally susceptible to infection with the parasite. The incrimination of an animal reservoir remained unsolved after this study, while four Rattus rattus out of 44 were found to be seropositive for antibodies against the DR parasite.
Five isolates were obtained from patients by the late Bryce Walton between 1979 and 1988, and given in parallel to both Jeffrey Shaw (Instituto Evandro Chagas, Belem, Brazil) and David Evans (London School of Tropical Medical and Hygiene, London, United Kingdom). Initial studies4,5 showed that the parasite belonged to the genus Leishmania. Profiles5 of excreted factor and the electrophoretic mobility of malate dehydrogenase distinguished them from L. (L.) amazonensis. However, nuclear and kinetoplastid DNA buoyant densities4 indicated that two DR isolates were closer to L. (L.) amazonensis but distinct from L. (L.) mexicana.
In this study, we report the characterization of five isolates of the DR Leishmania by isoenzymatic electrophoresis, multilocus sequence analysis (MLSA), and a panel of 23 monoclonal antibodies and examination of the small subunit ribosomal DNA (SSU rDNA) for two of them. Numerical taxonomic analysis, including cladistic study enabled us to determine the precise taxonomic position of this parasite, which we consider as a new taxon within the L. (L.) mexicana complex.
Materials and Methods
Studied strains.
Five strains isolated from DCL human cases from the DR were cryopreserved in both the Cryobank of the Department of Medical Protozoology, London School of Tropical Medicine and Hygiene (LSTMH), and the Instituto Evandro Chagas's cryobank, where monoclonal and rDNA examinations were performed.
Those obtained from the LSTMH collection are stored at the International Cryobank and Identification Center for Leishmania in Montpellier, under Biobank No. *BB-0033-00052 (Montpellier, France). These strains were studied using isoenzymatic electrophoresis, MLSA, and numerical taxonomic analysis.
The strain code numbers are as follows: MHOM/DO/79/CECILIO, MHOM/DO/79/CONSTANCIA, MHOM/DO/88/025, MHOM/DO/0000/452-A, and MHOM/DO/0000/450-B.
References strains for isoenzyme characterization and MLSA.
The above strains were compared with the following 18 MON zymodeme reference strains: MON-40 (MNYC/BZ/62/M379), MON-121(MHOM/MX/89/RIOS), MON-152(MHOM/MX/85/SOLIS), MON-153(MHOM/BZ/85/BEL65), MON-154(IYLE/GT/81/23L), MON-155(MHOM/PA/00/GML637), MON-156(MHOM/BZ/82/BEL21), MON-110(MHOM/EC/87/EC-103), MON-194 (MHOM/00/92/LPN88), and MON-195(MHOM/MX/93/CRE47) for L. (L.) mexicana; MON-41(IFLA/BR/67/PH8), MON-132(MHOM/BR/73/M2269), MON-157(IFLA/TT/71/71-110), MON-41(MHOM/PA/87/GML416), and MON-41(MHOM/CO/82/CELIS) for L. (L.) amazonensis; MON-133(MORY/PA/68/GML-3) for L. (L.) aristidesi; and MON-97(MCAV/BR/45/L88) and MON-227(MCAV/BR/95/CUR3) for L. enriettii. The enzymatic profiles of all these reference strains are given in Table 1.
Table 1.
Enzymatic profiles of the zymodeme reference strains used in the isoenzyme analysis
| Leishmania taxa | WHO code | NRCL*code | ME | PGD | G6PD | DIA | NP1 | NP2 | GOT1 | GOT2 | PGM | FH | MPI | GPI | ZYMODEMEMON |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L. amazonensis | IFLA/BR/67/PH8 | LEM 395 | 105 | 110 | 65 | 98 | 0 | 90 | 85 | 105 | 111 | 95 | 115 | 45 | 41 |
| MHOM/BR/73/M2269 | LEM 690 | 105 | 116-110 | 75-65 | 98 | 0 | 90 | 70 | 112 | 111 | 95 | 115 | 45 | 132 | |
| IFLA/TT/71/71-110 | LEM 2246 | 105 | 110 | 65 | 98 | 0 | 90 | 85 | 105 | 111 | 95 | 106 | 45 | 157 | |
| MHOM/PA/87/GML416 | LEM 2237 | 105 | 110 | 65 | 98 | 0 | 90 | 85 | 105 | 111 | 95 | 115 | 45 | 41 | |
| MHOM/CO/82/CELIS | LEM 2247 | 105 | 110 | 65 | 98 | 0 | 90 | 85 | 105 | 111 | 95 | 115 | 45 | 41 | |
| L. aristidesi | MORY/PA/68/GML-3 | LEM 693 | 95-85 | 110 | 65 | 98 | 0 | 87 | 85 | 100 | 111 | 95 | 80 | 45 | 133 |
| L. mexicana | MHOM/MX/89/RIOS | LEM 1871 | 100 | 110 | 65 | 75 | 295 | 90 | 80 | 105 | 133 | 95 | 135 | 35 | 121 |
| MHOM/MX/85/SOLIS | LEM 2223 | 106-100 | 110 | 70-65 | 75 | 295 | 90 | 80 | 105 | 133 | 95 | 135 | 35 | 152 | |
| MHOM/BZ/85/BEL65 | LEM 2224 | 100 | 110 | 65 | 75 | 295 | 90 | 80 | 105 | 133-123 | 95 | 135 | 35 | 153 | |
| IYLE/GT/81/23L | LEM 2225 | 100 | 110 | 65 | 75 | 295 | 90 | 80 | 105 | 133 | 95 | 145-120 | 35 | 154 | |
| MHOM/PA/00/GML637 | LEM 2263 | 100 | 115 | 70 | 75 | 295 | 87 | 80 | 105 | 109,4 | 95 | 105 | 35 | 155 | |
| MHOM/BZ/82/BEL21 | LEM 695 | 100 | 110 | 65 | 75 | 295 | 90 | 80 | 105 | 133 | 95 | 150 | 35 | 156 | |
| MHOM/EC/87/EC-103 | LEM 1221 | 109 | 115 | 70 | 75 | 290 | 87 | 75 | 114 | 109,4 | 95 | 115-103 | 38 | 110 | |
| MHOM/00/92/LPN88 | LEM 2573 | 100 | 110-95 | 65 | 75 | 295 | 90 | 80 | 105 | 133-117 | 95 | 135 | 35 | 194 | |
| MHOM/MX/93/CRE47 | LEM 2695 | 100 | 110-95 | 65 | 75 | 295 | 90 | 80 | 105 | 133 | 95 | 135 | 35 | 195 | |
| MNYC/BZ/62/M379 | LEM 280 | 100 | 110 | 65 | 75 | 295 | 90 | 80 | 105 | 133 | 95 | 118-110 | 35 | 40 | |
| L. enriettii | MCAV/BR/45/L88 | LEM 1120 | 65 | 110 | 83 | 20 | 0 | 135 | 120 | 120 | 111 | 75 | 137 | 79 | 97 |
| MCAV/BR/95/CUR3 | LEM 3045 | 65 | 110 | 92 | 20 | 0 | 135 | 120 | 120 | 111 | 75 | 137 | 79 | 227 |
Biochemical characterization.
Isoenzymatic characterization based on starch gel electrophoresis was used according to Rioux and others (1990), using the following 12 enzymes: malic enzyme (ME): EC1.1.1.40, phosphogluconate dehydrogenase (PGD): EC 1.1.1.44; glucose-6-phosphate dehydrogenase (G6PD): EC 1.1.1.49; diaphorase NADH, DIA: EC 1.6.2.2; nucleoside purine phosphorylase 1 (NP1) : EC 2.4.2.1 and 2 (NP2) EC 2.4.2.*; glutamate-oxaloacetate transaminase 1 and 2 (GOT1 and GOT2): EC 2.6.1.1; phosphoglucomutase (PGM): EC 5.4.2.2; fumarate hydratase (FH): EC 4.2.1.2; mannose-phosphate isomerase (MPI): EC 5.3.1.8; and glucose-phosphate isomerase (GPI): EC 5.31.9.
Cladistic analysis.
A cladistic analysis of the New World subgenus Leishmania was carried out to define the position of the DR isolates. The zymodemes were considered as operational taxonomic units and each enzymatic system as a multivalent character, each electromorph being a character state. The construction was based on Hennig's principles6 and parsimony using Felsenstein's MIX software (Difco B45 - Becton Dickinson, Franklin Lakes, NJ).
Reference strains for monoclonal and rDNA studies.
The DR strains were compared with the following species: L. (L.) mexicana (MNYC/BZ/62/M379 and MHOM/BZ/82/BEL21), L. (L.) pifanoi (MHOM/VE/76/ESTHER), L. (L.) garnhami (MHOM/VE/76/JAP78), L. (L.) amazonensis (IFLA/BR/67/PH8), L. forattinii (MDID/BR/82/RV288), L. (L.) aristidesi (MORY/PA/68/GML3), L. (L.) venezuelensis (MHOM/VE/81/PMH17), L. (L.) infantum chagasi (MHOM/BR/74/PP75), and Endotrypanum (MCHO/BR/79/M5725). These strains were chosen as they represent taxa that the DR strains need to be differentiated from.
Indirect antibody fluorescent protocol for leishmanial monoclonal antibodies.
Promastigotes of all strains were grown in blood agar base medium (Difco B45).7 Log phase parasite were washed in phosphate buffered saline (PBS) ph7.2 (2.5 mM NaH2PO4, 7.4 mM Na2HPO4, and 14 mM NaCl) three times by centrifugation at 5,000 G for 10 minutes at 4°C. The pellet was suspended in PBS (4°C) to give a final concentration of 104 parasites/mL. Ten microliter of this suspension was placed in each orifice of teflon-coated slides. They were air dried, fixed for 15 minutes in analytical grade acetone and stored at 20°C in plastic bags containing silica gel.
A total of 23 monoclonal antibodies were used (B2, B5, B12, B13, B18, B19, M2, M11, M12, CO1, CO2, CO3, L18,9; T3, D1310,11; WIC.79.312; N2, N3, LA2, WH1, WA2, V113,14). The B and N series react selectively with species of the subgenus L. (Viannia); M2, T3, D13, M11, M12, WIC.79.3, WA2, and V1 react selectively with parasites of the subgenus L. (Leishmania) and Endotrypanum; CO1, CO2, CO3, and L1 are group specific and react selectively with members of both Leishmania subgenera, Endotrypanum, and some species of Trypanosoma. The protocol followed a three-step biotin/avidin procedure15 that allows antibodies to be used at higher dilutions. After the application of the leishmanial monoclonal antibody biotinilated anti-mouse (H + L) is used and then fluorescence is obtained using fluorescent-labeled avidin.
Examination of SSU rDNA.
DNA was extracted from washed cultures and the SSU rDNA was amplified16 using the S1/S4 or S12/S4 primers and then with the S8, S9, and S10 rDNA probes.17 S8 reacts with L. (L.) amazonensis and L. (L.) major complex parasites, S9 with parasites of the L. (L.) mexicana and L. (L.) donovani complexes, and S10 with L. (Viannia) species and the Endotrypanum group that included L. colombiensis.
Multilocus sequence analysis.
In all, 15 Leishmania strains from South and central America (including the five L. waltoni strains) were analyzed using loci 03.0980, 12.0010, 14.0130, and 31.2610 of four housekeeping genes.18 Sequences were deposited into the GenBank database under the following accession numbers: KC158811, KC158589, KC159255, KC159699, KC849477-KC849479, KC849511-KC849513, KC849613-KC849615, KC849647-KC849649, KC960499, KC960504, KC960509, KC960513, and KM555296-KM555339. The four loci were concatenated and duplicated to avoid information loss due to ambiguous states. The Maximum likelihood tree was constructed using PhyML, version 3.019,20 with the generalized time reversible (GTR) model for nucleotide substitutions including a proportion of invariables sites and gamma-distributed rate variation across sites. One L. infantum strain (LEM 75) was used as an out-group and 1,000 bootstrap replicates were performed to estimate nodes support.
Results
Enzymatic characters.
The enzymatic profiles of the five DR Leishmania strains showed the existence of two different new zymodemes: MON-192 (four strains) and MON-259 (one strain) (Table 2).
Table 2.
Enzymatic profiles of the five Dominican Republic Leishmania strains
| WHO code | NRCL* code | ME | PGD | G6PD | DIA | NP1 | NP2 | GOT1 | GOT2 | PGM | FH | MPI | GPI | ZYMODEME MON- |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MHOM/DO/00/452A | LEM2579 | 109-87 | 110 | 70 | 260-70 | 180 | 90 | 80 | 0 | 118 | 84 | 135 | 32 | 192 |
| MHOM/DO/79/Cecilio | LEM2580 | 109-87 | 110 | 70 | 260-70 | 180 | 90 | 80 | 0 | 118 | 84 | 135 | 32 | 192 |
| MHOM/DO/79/Constancia | LEM2581 | 109-87 | 110 | 70 | 260-70 | 180 | 90 | 80 | 0 | 118 | 84 | 135 | 32 | 192 |
| MHOM/DO/00/450B | LEM2582 | 109-87 | 110 | 70 | 260-70 | 180 | 90 | 80 | 0 | 118 | 84 | 135 | 32 | 192 |
| MHOM/DO/88/025 | LEM3365 | 90 | 110 | 70 | 260-70 | 180 | 90 | 80 | 105 | 118 | 84 | 135 | 32 | 259 |
WHO = World Health Organization; ME = malic enzyme; PGD = phosphogluconate dehydrogenase; G6PD = glucose-6-phosphate dehydrogenase; DIA = diaphorase; NP1 = nucleoside purine phosphorylase 1; NP2 = nucleoside purine phosphorylase; GOT1 and GOT2 = glutamate-oxaloacetate transaminase 1 and 2; PGM = phosphoglucomutase; FH = fumarate hydratase; MPI = mannose-phosphate isomerase; GPI = glucose-phosphate isomerase.
National Reference center for Leishmania.
These strains were compared with the 10 zymodeme reference strains of L. (L.) mexicana complex: six L. (L.) mexicana electromorphs are found in the DR strains (PGD110, G6PD70, NP290, GOT180, GOT2105, and MPI135), three of them (PGD, NP2, and GOT1) regularly found in the majority of the zymodemes (Table 2).
The comparison with the three zymodemes of the L. (L.) amazonensis complex showed three common electromorphs: PGD110 and NP290, already found in L. (L.) mexicana, and GOT1105 found in two zymodemes of L. (L.) amazonensis. Concerning L. (L.) aristidesi and L. enriettii they both have a single electromorph in common with the DR strains, which is also common to L. (L.) mexicana and L. (L.) amazonensis (PGD110).
Three electromorphs are specific of the DR Leishmania zymodemes: DIA260-70, NP1180, and GPI32 in both zymodemes MON-192 and MON-259. Moreover, an autapomorph is present for the enzyme ME109-87 in MON-192.
Phylogenetic characters.
The cladistic analysis leads to the definition of a plesiomorphic state (PGD110) for the New World Leishmania subgenus: i.e., L. (L.) mexicana, L. (L.) amazonensis, L. (L.) aristidesi and L. enriettii complexes. Two states (NP290 and GOT2105) are common to the two complexes L. (L.) mexicana and L. (L.) amazonensis, and another state (GOT180) is found only in L. (L.) mexicana complex.21,22
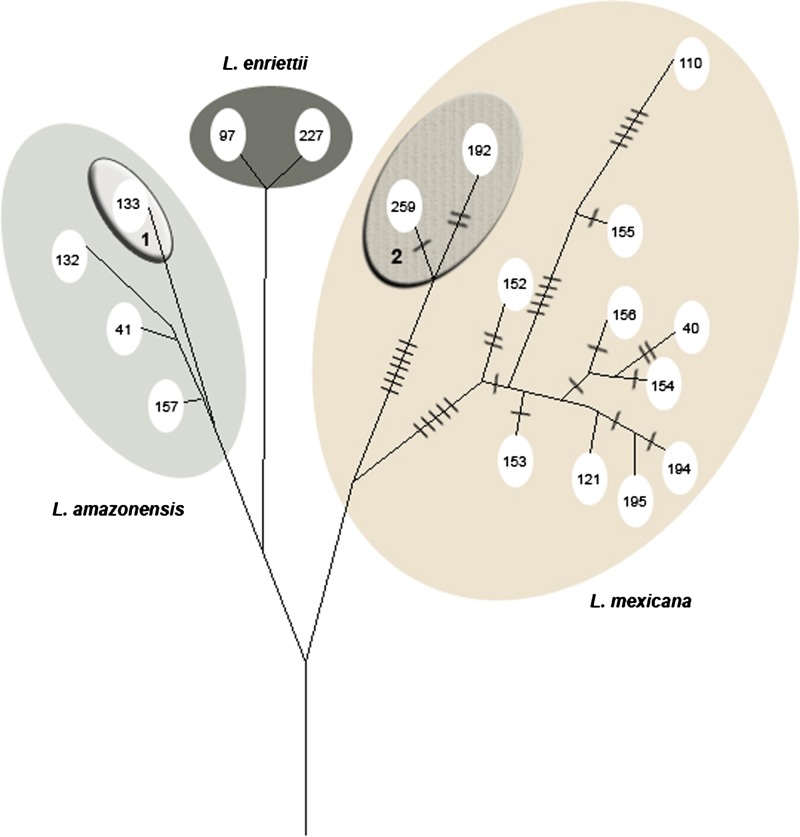
The phylogenetic tree shows the inclusion of the two DR zymodemes within the L. (L.) mexicana complex. However, they are located on a branch distinct from the L. (L.) mexicana sensu stricto species at 12 evolutive steps (Figure 1 ).
Figure 1.
Phylogenetic tree of the New World Leishmania sub-genus, with mention of the evolutive steps. (1: L. (L.) aristidesi, 2: L. (L.) waltoni).
Multilocus sequence analysis.
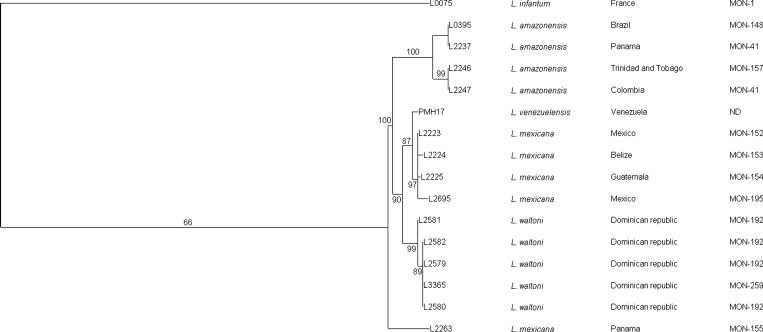
The four single copy-coding DNA sequences analyzed gave a 2,520 bp-long concatenated sequence. Excluding the L. infantum LEM75 reference strain used as an out-group, 36 polymorphic sites, including 23 informative parsimonious sites and 13 singletons, were identified among the 15 Leishmania (Viannia) strains analyzed. The five L. waltoni strains analyzed were almost genetically identical and only one polymorphism was detected in the LEM2581 strain among the 2,520 bp analyzed. The Maximum likelihood tree (Figure 2 ) has shown a well-supported L. waltoni strain's genetic cluster (BP = 99%).
Figure 2.
Maximum likelihood tree of the concatenated nucleotide sequences of 16 Leishmania strains. Significant bootstrap values are shown in percentage on each node. The species/taxonomic names, the geographical origins and the zymodemes are shown for each strain. The five L. (L.) waltoni strains fall into a highly supported cluster (BP 99%). The L numbers correspond to the National Reference Code Leishmania (NRCL) codes: for correspondence to the World Health Organization (WHO) codes, see Table 1. ND = not determined.
Monoclonal immunofluorescent antibody tests.
The results of the monoclonal immunofluorescent antibody (IFAT) tests are summarized in Table 3. The profile of the new parasite is distinct from those of the named South American species of the subgenus L. (L.) Leishmania. There were no reactions with the monoclonals M12 and WA2 that recognise L. (L.) mexicana or with M2 that has so far been shown to be specific for parasites of the L. (L.) amazonensis complex. This clearly differentiated the DR strains from parasites belonging to the L. (L.) amazonensis complex. The reaction with T3 puts it closer to L. (L.) mexicana and L. (L.) major. There are two distinct serodemes of the DR strains: one composing of strains 1137 and 1139 that do not react with the WIC-79.3 and CO2 and the other that includes the type strain that reacts with both these monoclonals.
Table 3.
Results of the indirect immunofluorescent test with a selected number of Leishmania-specific monoclonal antibodies that react with American parasites belonging to the subgenus L. (Leishmania) and the genus Endotrypanum
| Species | M2 | M11 | M12 | WA2 | T3 | WIC | CO2 | V1 | L1 | N2 |
|---|---|---|---|---|---|---|---|---|---|---|
| L. waltoni (1137) | − | − | − | − | + | − | − | − | + | − |
| L. waltoni (1139) | − | − | − | − | + | − | − | − | + | − |
| L. waltoni (452-A) | − | − | − | − | + | + | + | − | + | − |
| L. waltoni (450-B) | − | − | − | − | + | + | + | − | + | − |
| L. waltoni (Celio) | − | − | − | − | + | + | + | − | + | − |
| L. venezuelensis (PMH17) | − | − | − | − | − | − | − | − | + | − |
| L. mexicana (M379) | − | − | + | + | + | + | + | − | + | − |
| L. mexicana (BEL21) | − | + | + | + | + | + | + | − | + | − |
| L. pifanoi (ESTHER) | − | + | − | + | + | − | + | − | + | − |
| L. garnhami (JAP78) | + | + | − | - | + | − | + | − | + | − |
| L. amazonensis (PH8) | + | + | − | + | − | − | + | − | + | − |
| L. aristedesi (GML3) | + | + | − | + | − | − | − | − | + | − |
| L. forattinii (RV288) | + | + | − | − | − | − | − | − | + | − |
| L. major (P) | − | − | − | − | + | + | − | − | + | − |
| Endotrypanum (M5725) | − | − | − | − | + | + | − | − | + | + |
Examination of SSU rDNA.
The results of the SSU rDNA hybridizations are shown in Table 4. The L. (L.) amazonensis reference strain (M2269) hybridized only with S8, the L. (L.) mexicana reference strain (M7369) was positive only with S9. Two strains of L. (L.) major included in the study showed hybridization with S8, the same pattern observed for L. (L.) amazonensis. The L. (L.) infantum hybridization profile was the same as L. (L.) chagasi and L. (L.) donovani, being positive for S9 and S16. The reference strains not previously tested, L. (Viannia) lainsoni and L. (V.) shawi, of the L. (Viannia) subgenus, hybridized with S10. A strain of Endotrypanum schaudini hybridized with the S10 probe. The common reactivity between Viannia species and Endotrypanum means that it is impossible to use the S10 probe for identifying sylvatic reservoirs hosts and vectors. It is interesting that both have a peripylarian development yet antigenically Endotrypanum is closer to L. (L.) amazonensis.23 The two strains of the new parasite hybridized with the S9 probe but were negative with the S16 probe that is the same as parasites belonging to the mexicana complex.
Table 4.
Hybridisation of probes S8, S9, S10 and with DNA of strains considered as belonging the mexicana and amazonensis complexes
| Species (original code) | S8 | S9 | S10 | S16 |
|---|---|---|---|---|
| Leishmania (Leishmania) waltoni (1137) | − | + | − | − |
| L. (L.) waltoni (1139) | − | + | − | − |
| L. (L.) mexicana (M379) | − | + | − | − |
| L. (L.) mexicana (BEL21) | − | + | − | − |
| L. (L.) pifanoi (ESTHER) | − | + | − | − |
| L. (L.) garnhami (JAP78) | + | − | − | − |
| L. (L.) amazonensis (PH8) | + | − | − | − |
| L. (L.) major (P) | + | − | − | − |
| L. (L.) chagasi (PP75) | − | + | − | + |
| L. (V.) braziliensis (M2904) | − | − | + | − |
| Endotrypanum (M5725) | − | − | + | − |
Discussion
As a result of the phylogenetic analysis, the DR strains are characterized by three states (DIA260-70 NP1180 GPI32), which belong exclusively to all of them. These synapomorphic characters confirm the individualization of the group and its cohesion. Similarly, the monoclonal antibody profile clearly distinguishes the DR strains from other named species of the subgenus L. (Leishmania). These results lead us to consider the Dominican parasite as a new taxon, naming it Leishmania (Leishmania) waltoni, in honour to the late Bryce Walton, who isolated the DR strains that are the basis of this study. As a member of the American Armed Forces, he had unique access to leishmaniasis patients and his studies of these cases were important in developing clinical tests, such as the use of the immunofluorescent test for monitoring the treatment of cutaneous leishmaniasis.24 He was also one of the first researchers to draw attention to a link between the severity of leishmanial infections and genetic differences of human populations.25 Throughout his professional life he was interested in all aspects of leishmaniasis, especially in the New World, and was responsible for stimulating research on the disease within both military units and public health organizations, such as the World Health Organization and the Pan-American Health Organization, serving as a consultant to both.
Description of new taxon.
Leishmania (Leishmania) waltoni sp.n., Shaw JJ, Pratlong F and Dedet JP
Type host: man.
Locality in host: skin.
Type locality: Lagunita Cuchillo, El Seybo Province, Dominican Republic.
Strain designation: MHOM/DO/88/025
Promastigote measurements: body length: 7.8 ± 1.7 μ; body width: 1.9 ± 0.4 μ; flagellum length: 5.3 ± 1.6 μ
Suspected sandfly vector: Lutzomyia christophei
Reservoir(s): unknown
Enzyme profiles: ME90, PGD110, G6PD70, DIA260-70, NP1180, NP290, GOT180, GOT2105, PGM118, FH84, MPI135, GPI,32 corresponding to zymodeme MON-259.
Monoclonal antibodies: T3 + ve, WIC-79.3 + ve. CO1 + ve, L1 + ve, M2-ve, M11-,ve M12-ve, WA2-ve, CO2-vve, V1-ve, N2-ve.
rDNA Probes: Reactions with SSU rDNA probes: S9 + ve, S8-ve, S10-ve, S16-ve.
Multilocus Sequence Analysis (MLSA): GenBank Acc.no for loci 03.0980, 12.0010, 14.0130 and 31.2610 are KM555303, KM555314, KM555325, and KM555336 respectively.
Growth in vitro: easy to grow, doubling time 6 hours on SDM79 medium (PAA Laboratories GmbH, Austria) supplemented with 15% fetal calf serum and 7 μg /mL hemin.
Growth in vivo: produces a small lesion in the hamster (Mesocricetus auratus) similar in size to those of L. (L.) venezulensis and L. (Viannia) spp., unlike the large, parasite rich L. (L.) mexicana and L. (L.) amazonensis histiocytomata. There is no evidence of metastasis.
Pathology in man: producing multiple nodular skin lesions, with long-term evolution without spontaneous cure. Resistant to treatment by antimonials and amphotericin B.
Type material: cryopreserved promastigotes deposited in the International Cryobank and Identification Center for Leishmania, Laboratoire de Parasitologie, Montpellier (France).
The divergence between L. (L.) waltoni and L. (L.) mexicana is relatively recent, as the number of evolutive steps is reduced (12), but sufficient to individualize the taxon. It presumably results from island isolation, and poses several questions concerning the date of the appearance of leishmaniasis in Hispaniola Island. The most likely scenario would seem to be the introduction of a mexicana-like parasite from central America or the northern coastal region of Venezuela. This probably occurred well before man appeared in the region, which was only some 40,000 years ago.26 Sandflies have been recorded from Miocene (24Ma) in amber27 from the Dominican Republic and the island arch, of which present day Hispaniola is part, has been in place since the middle Eocene (49Ma).28 Fossil records indicate that the island had a rich mammalian fauna consisting of rodents, insectivores, monkeys, and sloths,29 suggesting periodic links with the mainland. Leishmania (L.) waltoni's original reservoir may have been native rodents such as hutias (Plagiodonta spp.) that were abundant in caves and forests. Four species are recorded from Hispaniola but only one or two have survived the post-Colombian colonization. Looking for evolutionary differences between L. (L.) waltoni and L. (L.) mexicana using molecular markers could serve as a basis for calibrating leishmanial evolutionary events. It is perhaps ironical that L. (L.) waltoni could become extinct if it remains limited to its endemic silvatic hosts and fails to adapt to non-endemic rodents such as domestic rats.
A 1380-bp fragment of the coding region of the heat shock protein 70 (Hsp70) gene that is commonly used in restriction fragment length polymorphism (RFLP) analysis was recently sequenced30 and its authors concluded “As such no subspecies can be defined based on Hsp70, and L. mexicana would be the single recognized species.” There are a number of valid reasons for not accepting this conclusion. The tree in this article showed good bootstrap values supporting the separation of the reference strain of L. (L.) mexicana from the L. (L.) amazonensis strains with the exception of one strain designated as L. (L.) mexicana from Peru. No other identification methods were used to validate its identity. In an article published in 199416 the authors could not distinguish between the standard strains of L. (L.) mexicana and L. (L.) amazonensis using probes based upon SSu rDNA sequences. However, in a later study,17 these same probes distinguished L. (L.) mexicana and L. (L.) amazonensis. In the second study, a new line of the same L. (L.) mexicana strain was obtained from a source where its identity had been validated. The problem was that somewhere the L. (L.) mexicana strain had become contaminated with a parasite that reacted with the S8 amazonensis probe. There is a constant danger of strains being contaminated by others. For instance, a L. (S.) adleri isolate turned out to be L. (L.) major31 and a strain of L. (L.) infantum was in fact Endotrypanum.32 Differences of this magnitude are easily detectable but may go unnoticed in dealing with closely related taxa such as ecologically isolated populations.
On the basis of SSu rDNA analysis, the mexicana clade as described in 198733 is clearly composed of two distinct genetic complexes.17 The first corresponds to L. (L.) mexicana and occurs throughout central America and the northern rim of South America and the second corresponds to L. (L.) amazonensis and is found throughout South America. Within these two groups some parasites have been given specific status. The validity of the mexicana and amazonensis complexes is supported by a number of characters such as functional differences in the domains of the rDNA promoter regions,34 isoenzyme analysis,22 and cytochrome b.35 A trend that should be avoided is embarking on taxonomic revisions using a single molecular character that may or may not reflect the true taxonomic status of the species.
The MLSA showed clearly that the L. (L.) waltoni forms a distinct group within the mexicana complex, which is supported by a high bootstrap value. Interestingly L. (L.) venezuelensis that shares biological similarities with L. (L.) waltoni is closer to L. (L.) mexicana. This suggests that this species may have diverged at a later date from the ancestral mexicana stock.
Statistically monoclonal antibodies have a greater discriminatory power than isoenzymes for distinguishing Leishmania species.36 This is confirmed in this study since with the exception of L. (L.) amazonensis and L. (L.) aristedesi all the named indigenous species of the subgenus L. (Leishmania) could be differentiated. They are perhaps some of the most useful tools for identifying Leishmania but since their epitopes are generally not completely known, they are of limited use in taxonomy. When WIC-79.3 was originally described, it was simply known as a promastigote surface antigen but it was later shown that it recognizes part of L. (L.) major lipophosphoglycan. There are monoclonals against other surface antigens such as GP63, and genetic studies have shown that polymorphism of its gene loci allow specific markers for subgenera, species, and geographical populations.37 There is therefore a solid link explaining the efficiency of monoclonals with genetic variation but it has yet to be exploited for identification.
It is questionable whether all the species described within the two clades merit specific status but there are notable biological differences between some of them and little is known of their individual zoonotic cycles. Before any decisions are reached both the biology and genetics of more strains from the different type areas from both animal and vectors need to be studied.
Recently it was suggested38 that going back to using subspecies could resolve present-day problems of leishmanial taxonomy. This is an interesting idea but one of the reasons that Lainson and Shaw33 abandoned the subspecies was that at that time it was becoming obvious that the genetic diversity was much greater than what had previously been expected. However, going back to using subspecies may be justified for parasites such as L. (V.) braziliensis that is composed of many distinct geographical populations. The basic building blocks of taxonomies are species but the definition of a Leishmania species causes great difficulties since the major form of their propagation is clonal.39 Hybrid strains have been found40–42 suggesting that occasionally genetic exchange does occur under certain ecological conditions. However, this does not negate the use of species names since they represent a stable method of communication and with modern computer technology they can be easily followed in name-based databases and infrastructures that collate biological information.43 Bacterial species are equally difficult to define44 since their reproduction, like the Leishmania, is clonal with occasional genetic exchange. In the present genomic era it has been suggested45 that a bacterial species should be a biologically meaningful cluster. For obligatory pathogens the ecological niches are very restricted and result in detectable clusters. This is perhaps a good starting point for us to begin examining the species concept for Leishmania and it is within this framework that we have given a specific name to the Dominican parasite.
The present observations confirm once again the importance that geographical isolation has on the evolution and diversity of living organisms. Its importance in the evolution of the genus Leishmania has received little attention due to the difficulty of knowing where such barriers begin and end. However, the importance of ecological niche barriers within an ecotope has been shown by the difference in sympatric arboreal and terrestrial cycles, such as those of L. (V.) guyanensis and L. (L.) amazonensis in Amazonia, that involve vectors and reservoirs whom have different ecological preferences.
Like many Leishmania, the reservoirs of L. (L.) waltoni are unknown, but it is amazing that this species has survived with the virtual elimination of the wild mammals of Hispaniola. Only two have survived, the insectivorous solenodon, that has venomous saliva and the rodent hutia that inhabit the forest of rough ravines.27 It is likely that one of them is the natural reservoir and if this is proven it is remarkable that a Leishmania has survived in such a reduced reservoir population. It also suggests that controlling human cutaneous leishmaniasis by reservoir management may be very difficult.
ACKNOWLEDGMENTS
We thank Geneviève Lanotte for the phylogenetic analysis, and Patrick Lami, Loic Talignani, Yves Balard and Ghyslène Serre for expert technical assistance.
Footnotes
Financial support: This study was financially supported by Wellcome Trust, The Brazilian Research Council (CNPq), São Paulo Research Foundation (FAPESP). The National Reference Centre for Leishmaniasis of Montpellier receives financial support from the Institut National de Veille sanitaire, French Ministry of Public Health.
Authors' addresses: Jeffrey Shaw, Parasitology Department, Biomedical Sciences Institute, São Paulo University, São Paulo, Brazil, E-mail: jayusp@hotmail.com. Francine Pratlong, Christophe Ravel and Jean-Pierre Dedet, French National Reference Centre on Leishmaniasis, CNRS 5290 - IRD 224, Montpellier University, 1&2 (UMR MiVEGEC), Montpellier, France, E-mails: f-pratlong@chu-montpellier.fr, christophe.ravel@univ-montp1.fr, and parasito@univ-montp1.fr. Lucile Floeter-Winter, Biology Department, BioSciences Institute, São Paulo University, São Paulo, Brazil, E-mail: lucile@ib.usp.br. Edna Ishikawa, Tropical Medicine Nucleus, Pará Federal University, Brazil, E-mail: ishikawa@ufpa.br. Fouad El Baidouri, School of Life Sciences, University of Lincoln, Lincoln, England, E-mail: elbaidourifouad@gmail.com.
References
- 1.Lainson R, Shaw JJ. 17 New World leishmaniasis. In: Cox FEG, Kreier JP, Wakelin D, editors. Topley and Wilson's Microbiology and Microbial Infections. London, Sydney, Auckland: Hodder Arnold; 2005. pp. 313–349. [Google Scholar]
- 2.Bogaert-Diaz H, Rojas RF, De Leon A, Martinez D, Quimones M. Leishmaniasis tegumentaria Americana. Reporte de los primeros tres casos. Forma anergica en tres hermanos. Revista Dominicana de Dermatologia. 1975;9:19–33. [Google Scholar]
- 3.Johnson RN, Young DG, Butler JF, Bogaert-Diaz H. Possible determination of the vector and reservoir of leishmaniasis in the Dominican Republic. Am J Trop Med Hyg. 1992;46:282–287. doi: 10.4269/ajtmh.1992.46.282. [DOI] [PubMed] [Google Scholar]
- 4.Barker DC, Arnot DE, Butcher J. Washington, DC: 1982. DNA characterization as a taxonomic tool for identification of kinetoplastic flagellate protozoans; pp. 139–180. Biochemical characterization of Leishmania: Proceedings of a Workshop held at the Pan American Health Organization, December 9–11, 1980. [Google Scholar]
- 5.Schnur LF, Walton BC, Bogaert-Diaz H. On the identity of the parasite causing diffuse cutaneous leishmaniasis in the Dominican Republic. Trans R Soc Trop Med Hyg. 1983;77:756–762. doi: 10.1016/0035-9203(83)90282-1. [DOI] [PubMed] [Google Scholar]
- 6.Hennig W. Phylogenetic systematics. Ann Rev Ent. 1965;10:97–116. [Google Scholar]
- 7.Walton BC, Shaw JJ, Lainson R. Observations on the in vitro cultivation of Leishmania braziliensis. J Parasitol. 1977;63:1118–1119. [PubMed] [Google Scholar]
- 8.McMahon Pratt D, Bennett E, Grimaldi G, Jaffe CL. Subspecies and species-specific antigens of Leishmania mexicana characterized by monoclonal antibodies. J Immunol. 1985;134:1935–1940. [PubMed] [Google Scholar]
- 9.McMahon-Pratt D, Bennett E, David JR. Monoclonal antibodies that distinguish subspecies of Leishmania braziliensis. J Immunol. 1982;129:926–927. [PubMed] [Google Scholar]
- 10.Jaffe CL, McMahon-Pratt D. Monoclonal antibodies specific for Leishmania tropica. I. Characterization of antigens associated with stage- and species-specific determinants. J Immunol. 1983;131:1987–1993. [PubMed] [Google Scholar]
- 11.Jaffe CL, Bennett E, Grimaldi G, Jr, McMahon-Pratt D. Production and characterization of species-specific monoclonal antibodies against Leishmania donovani for immunodiagnosis. J Immunol. 1984;133:440–447. [PubMed] [Google Scholar]
- 12.de Ibarra AA, Howard JG, Snary D. Monoclonal antibodies to Leishmania tropica major: specificities and antigen location. Parasitology. 1982;85:523–531. doi: 10.1017/s0031182000056304. [DOI] [PubMed] [Google Scholar]
- 13.Hanham CA, Shaw JJ, Lainson R. Monoclonal antibodies that react with Leishmania (Viannia) naiffi. J Parasitol. 1991;77:680–687. [PubMed] [Google Scholar]
- 14.Hanham CA, Shaw JJ, Lainson R, McMahon-Pratt D. Production of a specific monoclonal antibody for the identification of Leishmania (Leishmania) venezuelensis. Am J Trop Med Hyg. 1990;42:453–459. doi: 10.4269/ajtmh.1990.42.453. [DOI] [PubMed] [Google Scholar]
- 15.Shaw JJ, Ishikawa EA, Lainson R. A rapid and sensitive method for the identification of Leishmania with monoclonal antibodies using fluorescein-labelled avidin. Trans R Soc Trop Med Hyg. 1989;83:783–784. doi: 10.1016/0035-9203(89)90326-x. [DOI] [PubMed] [Google Scholar]
- 16.Uliana SR, Nelson K, Beverley SM, Camargo EP, Floeter-Winter LM. Discrimination amongst Leishmania by polymerase chain reaction and hybridization with small subunit ribosomal DNA derived oligonucleotides. J Eukaryot Microbiol. 1994;41:324–330. doi: 10.1111/j.1550-7408.1994.tb06085.x. [DOI] [PubMed] [Google Scholar]
- 17.Uliana SR, Ishikawa E, Stempliuk VA, de Souza A, Shaw JJ, Floeter-Winter LM. Geographical distribution of neotropical Leishmania of the subgenus Leishmania analysed by ribosomal oligonucleotide probes. Trans R Soc Trop Med Hyg. 2000;94:261–264. doi: 10.1016/s0035-9203(00)90314-6. [DOI] [PubMed] [Google Scholar]
- 18.El Baidouri F, Diancourt L, Berry V, Chevenet F, Pratlong F, Marty P, Ravel C. Genetic structure and evolution of the Leishmania genus in Africa and Eurasia: what does MLSA tell us. PLoS Negl Trop Dis. 2013;7:e2255. doi: 10.1371/journal.pntd.0002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 20.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 21.Armijos RX, Thomaz-Soccol V, Lanotte G, Racines J, Pratlong F, Rioux JA. Analyse chimiotaxonomique de vingt-deux souches de Leishmania isolées au Nord-ouest de l'Equateur. Parasite. 1995;2:301–305. [Google Scholar]
- 22.Thomaz-Soccol V, Lanotte G, Rioux JA, Pratlong F, Martini-Dumas A, Serres E. Phylogenetic taxonomy of New World Leishmania. Ann Parasitol Hum Comp. 1993;68:104–106. [PubMed] [Google Scholar]
- 23.Shaw JJ. Taxonomy of the genus Leishmania: present and future trends and their implications. Mem Inst Oswaldo Cruz. 1994;89:471–478. doi: 10.1590/s0074-02761994000300033. [DOI] [PubMed] [Google Scholar]
- 24.Walton BC, Brooks WH, Arjona I. Serodiagnosis of American leishmaniasis by indirect fluorescent antibody test. Am J Trop Med Hyg. 1972;21:296–299. doi: 10.4269/ajtmh.1972.21.296. [DOI] [PubMed] [Google Scholar]
- 25.Walton BC, Valverde L. Racial differences in espundia. Ann Trop Med Parasitol. 1979;73:23–29. doi: 10.1080/00034983.1979.11687222. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez S, Huddart D, Bennett MR, Gonzalez-Huesca A. Human footprints in Central Mexico older than 40,000 years ago. Quat Sci Rev. 2006;25:201–222. [Google Scholar]
- 27.Andrade Filho JD, Galati EA, Brazil RP. Review of American fossil Phlebotominae (Diptera: Psychodidae) with a description of two new species. J Med Entomol. 2009;46:969–979. doi: 10.1603/033.046.0501. [DOI] [PubMed] [Google Scholar]
- 28.Mann P, Draper G, Lewis JF. An overview of the geologic and tectonic development of Hispaniola. Spec Pap Geol Soc Am. 1991;262:1–28. [Google Scholar]
- 29.Woods CA. Last endemic mammals in Hispaniola. Oryx. 1981;16:146–152. [Google Scholar]
- 30.Fraga J, Montalvo AM, De Doncker S, Dujardin JC, Van der Auwera G. Phylogeny of Leishmania species based on the heat-shock protein 70 gene. Infect Genet Evol. 2010;10:238–245. doi: 10.1016/j.meegid.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Momen H, Cupolillo E. Speculations on the origin and evolution of the genus Leishmania. Mem Inst Oswaldo Cruz. 2000;95:583–588. doi: 10.1590/s0074-02762000000400023. [DOI] [PubMed] [Google Scholar]
- 32.Shaw JJ, Camargo EP. Are trees real? Parasitol Today. 1995;11:347. doi: 10.1016/0169-4758(95)80190-1. [DOI] [PubMed] [Google Scholar]
- 33.Lainson R, Shaw JJ. Evolution, classification and geographical distribution. In: Peters W, Killick-Kendrick R, editors. The Leishmaniases in Biology and Medicine, Volume I: Biology and Epidemiology. London, United Kingdom: Academic Press; 1987. pp. 1–120. [Google Scholar]
- 34.Stempliuk VA, Floeter-Winter LM. Functional domains of the rDNA promoter display a differential recognition in Leishmania. Mol Biochem Parasitol. 2002;32:437–447. doi: 10.1016/s0020-7519(01)00371-x. [DOI] [PubMed] [Google Scholar]
- 35.Asato Y, Oshiro M, Myint CK, Yamamoto Y, Kato H, Marco JD, Mimori T, Gomez EA, Hashiguchi Y, Uezato H. Phylogenic analysis of the genus Leishmania by cytochrome b gene sequencing. Exp Parasitol. 2009;121:352–361. doi: 10.1016/j.exppara.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 36.Cupolillo E, Grimaldi Junior EC, Momen H. Discriminatory ability of typing systems in Leishmania. Trans R Soc Trop Med Hyg. 1993;87:116–117. doi: 10.1016/0035-9203(93)90451-u. [DOI] [PubMed] [Google Scholar]
- 37.Victoir K, Banuls AL, Arevalo J, Llanos-Cuentas A, Hamers R, Noel S, De Doncker S, Le Ray D, Tibayrenc M, Dujardin JC. The gp63 gene locus, a target for genetic characterization of Leishmania belonging to subgenus Viannia. Parasitology. 1998;117:1–13. [PubMed] [Google Scholar]
- 38.Schonian G, Mauricio I, Cupolillo E. Is it time to revise the nomenclature of Leishmania? Trends Parasitol. 2010;26:466–469. doi: 10.1016/j.pt.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 39.Tibayrenc M, Ayala FJ. The clonal theory of parasitic protozoa: 12 years on. Trends Parasitol. 2002;18:405–410. doi: 10.1016/s1471-4922(02)02357-7. [DOI] [PubMed] [Google Scholar]
- 40.Evans DA, Kennedy WP, Elbihari S, Chapman CJ, Smith V, Peters W. Hybrid formation within the genus Leishmania? Parassitologia. 1987;29:165–173. [PubMed] [Google Scholar]
- 41.Belli AA, Miles MA, Kelly JM. A putative Leishmania panamensis/Leishmania braziliensis hybrid is a causative agent of human cutaneous leishmaniasis in Nicaragua. Parasitology. 1994;109:435–442. doi: 10.1017/s0031182000080689. [DOI] [PubMed] [Google Scholar]
- 42.Jennings YL, de Souza AAA, Ishikawa EA, Shaw J, Lainson R, Silveira F. Phenotypic characterization of Leishmania spp. causing region, western Pará state, Brazil, reveals a putative hybrid parasite, Leishmania (Viannia) guyanensis × Leishmania (Viannia) shawi shawi. Parasite. 2014;21:1–11. doi: 10.1051/parasite/2014039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patterson DJ, Cooper J, Kirk PM, Pyle RL, Remsen DP. Names are key to the big new biology. Trends Ecol Evol. 2010;25:686–691. doi: 10.1016/j.tree.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Cohan FM. What are bacterial species? Annu Rev Microbiol. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- 45.Konstantinidis KT, Ramette A, Tiedje JM. The bacterial species definition in the genomic era. Philos Trans R Soc Lond B Biol Sci. 2006;361:1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]