Abstract
Amebiasis is an important cause of diarrheal disease worldwide and has been associated with childhood malnutrition. Traditional microscopy approaches are neither sensitive nor specific for Entamoeba histolytica. Antigen assays are more specific, but many cases are missed unless tested by molecular methods. Although polymerase chain reaction (PCR) is effective, the need for sophisticated, expensive equipment, infrastructure, and trained personnel limits its usefulness, especially in the resource-limited, endemic areas. Here, we report development of a recombinase polymerase amplification (RPA) method to detect E. histolytica specifically. Using visual detection by lateral flow (LF), the test was highly sensitive and specific and could be performed without additional equipment. The availability of this inexpensive, sensitive, and field-applicable diagnostic test could facilitate rapid diagnosis and treatment of amebiasis in endemic regions.
Introduction
Invasive amebiasis caused by Entamoeba histolytica is a major source of parasite infection-related mortality worldwide, accounting for 40,600–73,800 deaths annually.1 Diagnosis of invasive amebiasis remains challenging. Entamoeba histolytica is morphologically indistinguishable from other nonpathogenic Entamoeba species. Stool examination is also not sensitive enough for diagnosis.2 Detection of antigen by enzyme-linked immunosorbent assay (ELISA) and immunochromatic test3 has proven to be useful on fresh or frozen stool samples with good sensitivity and specificity for E. histolytica.4,5 However, fixation of the stool samples denatures the antigen, thus limiting testing to fresh or frozen samples.2 Some studies have identified problems with the specificity of antigen detection due to cross-reactions.2,3
Molecular methods, such as polymerase chain reaction (PCR) assays, have improved specificity and detect fewer organisms compared with antigen detection.6 Furthermore, real-time PCR has a quantitative advantage and improved sensitivity compared with standard PCR, with an ability to detect 0.1 cell per gram of feces.7,8 Despite the obvious advantage of molecular techniques, they are not widely used for several reasons. PCR detection requires the use of specialized equipment, trained technical staff, and thermostable reagents. Thus, costs are prohibitive for use in resource-poor areas. Therefore, PCR and real-time PCR have only been used in reference and research laboratories of wealthy countries and are not available to the population at highest risk. As a result, many practitioners resort to empiric therapy. The current practice of empiricism results in both overtreatment and undertreatment.9 Overtreatment with an antiprotozoal agent for all patients with cysts in feces is leading to increasing minimum inhibitory concentrations (MICs) against E. histolytica.10 It is clear that there is a critical need for a sensitive test for diagnosis of pathogenic E. histolytica that can be easily performed at the point-of-care (POC) in resource-limited settings.
Isothermal amplification methods provide robust signal amplification without requiring thermocyclers.11 Recombinase polymerase amplification (RPA) is a novel isothermal nucleic acid amplification method. In RPA, a recombinase and its cofactor form a nucleoprotein complex with oligonucleotide primers and scan for homologous sequences in a deoxyribonucleic acid (DNA) template. Recognition of a specific homologous sequence leads to the initiation of strand invasion and the opposing oligonucleotides are then extended by isothermal strand displacement amplification.12 RPA provides several advantages for POC of infectious diseases because it is fast, able to work at most ambient temperatures, and can be adapted for lateral flow (LF) detection. Recently, we have demonstrated the feasibility to detect protozoan parasites in stool samples.13,14 In this work, we have developed a specific and sensitive assay to detect E. histolytica that can be used at POC.
Methods
Parasites and DNA.
The parasites E. histolytica Schaudinn (ATCC® 50525™) and E. dispar Brumpt (ATCC PRA-353™) were purchased from ATCC (Manassas, VA). The genomic DNA of E. histolytica (ATCC 30459D), Cryptosporidium parvum (ATCC PRA-67D), and Giardia intestinalis (ATCC 50803D) was obtained from ATCC.
DNA extraction from stool samples.
Entamoeba parasites were quantified by microscopy using a Hemocytometer. After quantification, different amounts of parasites in the range of 4 up to 500,000 were spiked into 250 ng of human stool samples (from healthy donors in the United States) previously diluted in 1 mL of phosphate buffered saline (PBS). DNA was extracted using the mini DNA extractor QuickGene-Mini80 (Autogen, Holliston, MA) by using purification columns included in the Quick gene DNA tissue kit (Autogen). Briefly, 200 μL of stool samples was resuspended in 180 μL of tissue lysis buffer and 20 μL proteinase K buffer (included in the kit) and incubated at 55°C for 1 hour. Samples were centrifuged at 8,000 × g for 3 minutes at room temperature (RT) and the supernatant was transferred to a 1.5 mL tube, then 180 μL of lysis buffer was added and vortexed for 15 seconds. The sample was incubated at 70°C for 10 minutes, then was centrifuged at 8,000 × g for 1 minute. The supernatant was passed through DNA columns (contained in the kit) using the QuickGene-Mini extractor, the eluted were discarded and columns were washed two times with 350 μL of washing buffer included in the kit. The samples were then eluted with 200 μL of H2O and stored at −20°C until use. Stool samples from healthy donors were collected according to University of Texas Medical Branch Institutional Review Board (IRB) approved protocol 07-285.
Clinical samples.
DNA of clinical samples from patients infected with E. histolytica, E. dispar, and E. moshkovskii was obtained from the Universidad Nacional de Colombia as part of ongoing studies of amebiasis in Colombia, for these studies ethical standards for health research in force in Colombia were completed and approved by the Ethics Committee of the Faculty of Medicine of the National University of Colombia. Entamoeba histolytica-positive samples were identified by detecting the presence of Gal/GalNAc lectine in stools by ELISA using the commercial kit E. HISTOLYTICA II test™ (Techlab, Blacksburg, VA) and by PCR detecting 18s ribosomal ribonucleic acid (rRNA) using the primers and conditions described by Hamzah and others.15 DNA from clinical samples was extracted from 2 mL of stools fixed in ethanol 70% (1:4 w/v) following the instructions indicated in the DNA MP Isolation kit NORGEN (Thorold, Ontario, Canada).
Real-time PCR assays.
The samples were amplified using the SYBR Green PCR Kit (Life Technologies, Grand Island, NY). For each reaction, we used 2.5 μL of DNA as template obtained from 1:10 serial dilutions, concentrations were in the range of 250 ng to 0.025 fg. For DNA amplification, we targeted 18s rDNA using the primers described in Gonin and others. Reactions were performed with the ABI PRISM 7500 fast system (Life Technologies), and conditions were as follows: for cDNA synthesis, 50°C for 15 minutes and then 95°C for 5 minutes; for PCR amplification, 95°C for 30 seconds and then 60°C for 1 minute, for 40 cycles. Cycle threshold (Ct) values were determined using the 7500 fast system software. After real-time PCR, dissociation curves were analyzed to confirm the presence of specific amplicons.
RPA probe design.
For RPA assays, we amplified a 18s rDNA target (Figure 1 ) using primers designed to distinguish between E. histolityca and E. dispar that were previously described.16 Alignments conducted with Blastn software showed that these primers were able to detect all E. histolityca strains reported in the gene bank. For LF assays, we designed a 44-nt dual-labeled probe located between forward and reverse primers: 5′[FAM]AGTGAGTTAGGATGCCACGACAATTGTAGA[THF]CACACAGTGTTTAA[C3]3, the reverse primer used was modified as follows: 5′ [Biot] ACTACCAACTGATTGATAGATCAG.
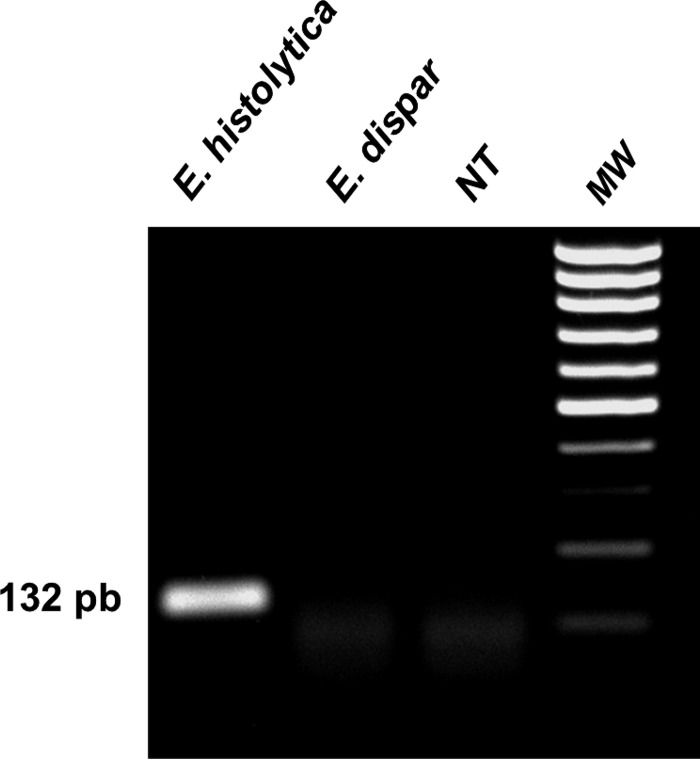
Figure 1.
Entamoeba histolytica recombinase polymerase amplification (RPA). Deoxyribonucleic acid (DNA) of E. histolytica and E. dispar and no template (NT) was used for RPA. After 40 minutes at 37°C, the reaction was analyzed by electrophoresis in agarose gels. Only RPA with E. histolytica DNA amplified the expected amplicon of 132 pb. Molecular weight markers are indicated as MW.
RPA assays.
The RPA assays were performed following the instructions indicated in the TWISTAMP® basic kit (TwistDx, Cambridge, United Kingdom). Briefly, the dried reagents contained in 0.6 μL-tubes were suspended in 29.5 μL of rehydration buffer and transferred to 1.5-μL tubes (reaction tube). Forward and reverse primers 2.4 μL (5 μM) and 0.6 μL of probe (5 μM) were mixed in a 1.5 mL tube and then incubated at 95°C for 1 minute. Then, 2.5 μL of DNA template, 5.4 μL of the primers-probe mixture, and water (up to 47.5 μL) were added to the reaction tube. The reaction was started adding 2.5 μL of MgCl2 (50 μM) and incubated for 40 minutes at 37°C in a dry bath. Five microliters of each RPA reaction was analyzed by electrophoresis in agarose gels (1%) and stained with ethidium bromide for visualization in an ultraviolet transilluminator.
Recombinase polymerase amplification-lateral flow.
RPA-LF assays were performed following the indications provided in the TWISTAMP NFO kit (TwistDx). Briefly, RPA reaction was assembled as above (basic kit RPA reaction) but 2.1 (5 μM) of each primer and 0.6 μL of the probe (5 μM) were added to the reaction tube. Amplified DNA was detected using LF strips (Milenia Hybridtech 1, Bad Nauheim, Germany) following the instructions indicated in the kit. Briefly, 5 μL of the RPA reaction was diluted with 95 mL of LF buffer in a 1.5-mL tube (LF reaction tube contained in the Milenia Kit). LF strips were introduced in the LF reaction tube, then a picture was taken 2 minutes after control bands (top in the strips) were observed and the result was interpreted by presence or absence of bands in the detection zone (low in the strips). For DNA amplification, we used 250 ng of DNA from E. histolytica (ATCC 30459D) diluted in water, for sensitivity assays we prepare 1:10 dilutions in the range of 250 ng to 0.025 fg. The limit of detection of RPA-LF was compared with SYBR Green real-time PCR conducted in our laboratory using the primers and conditions described by Gonin and others.16
Results
RPA and RPA-LF detection.
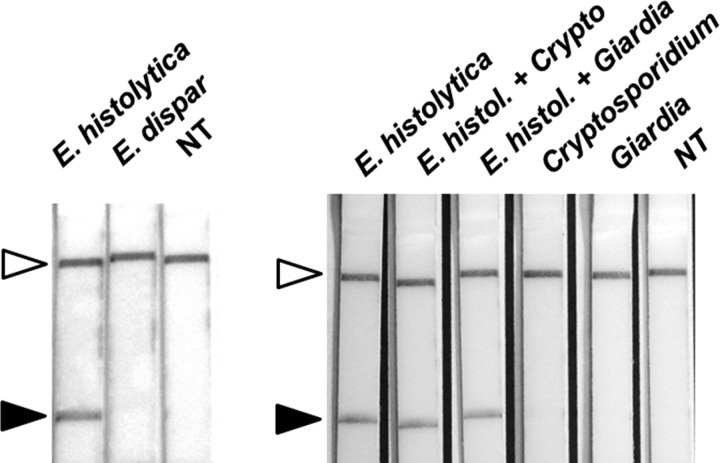
Our results showed that by targeting ribosomal 18s DNA from E. histolytica, RPA amplified a 132 pb amplicon after 40 minutes of incubation (Figure 1). No reaction was noted with E. dispar. By using a 44-nt dual-labeled probe, we were able to detect amplified DNA using paper strips. As observed in Figure 2 , DNA extracted from E. histolytica was detected by RPA-LF paper strips within 20 minutes and no cross-reaction was observed with E. dispar. Evidence of background or noise was not observed in samples with Cryptosporidium and Giardia (Figure 2), because these strips are not different than the negative control.
Figure 2.
Entamoeba histolytica recombinase polymerase amplification (RPA) in lateral flow (LF) strips. Entamoeba histolytica, E. dispar, Cryptosporidium, and Giardia deoxyribonucleic acid (DNA) and no template (NT) were used to evaluate the specificity of E. histolytica RPA in LF strips. RPA detects E. histolytica DNA but not E. dispar (left). RPA detects E. histolytica using DNA of E. histolytica spiked with DNA of Cryptosporidium and Giardia (Right). Cryptosporidium and Giardia DNA alone were negatives (Right). White arrows indicate control bands for the assay and black arrows show detection zone of RPA positives.
RPA-LF specificity.
RPA specificity was tested using nonpathogenic species as well as DNA from Cryptosporidium and Giardia (Figure 2). RPA-LF showed high specificity and no cross-reaction was observed against other parasites (Fasciola hepatica, Blastocistis hominis, Taenia solium, Toxoplasma gondii) neither with human nor with E. coli DNA (data no shown).
RPA-LF sensitivity.
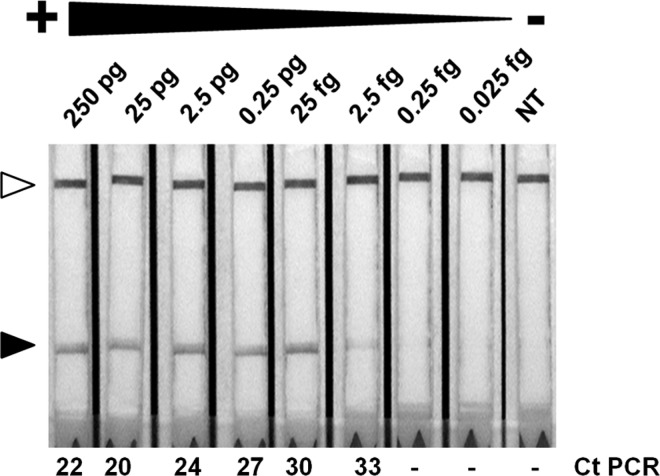
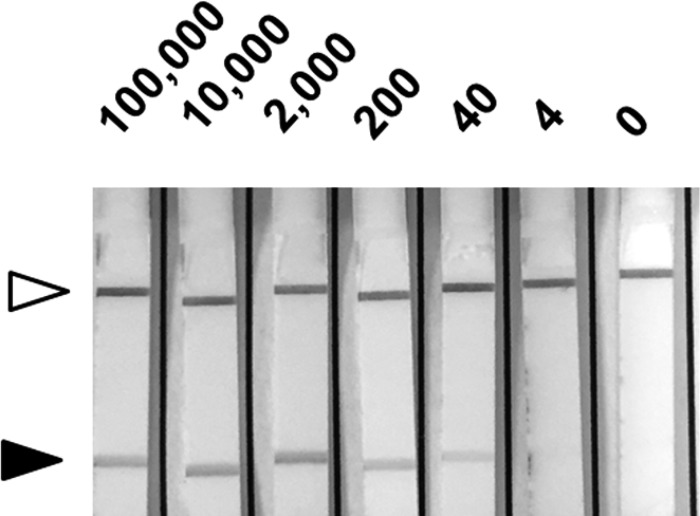
We tested the sensitivity of the assay by using serial dilutions of pure DNA extracted from parasites in a range of 250 pg to 0.025 fg. The results showed a limit of detection up to 2.5 fg (Figure 3 ). This limit of detection was the same as when we used SYBR Green real-time PCR with the same dilutions (Figure 3). We analyzed the sensitivity of RPA-LF with parasites spiked in stools, results shown a limit of detection of 40 parasites (Figure 4 ).
Figure 3.
Recombinase polymerase amplification (RPA) sensitivity. A total of 250 pg of Entamoeba histolytica deoxyribonucleic acid (DNA) was diluted (1:10) in phosphate buffered saline (PBS) and tested by RPA in lateral flow (LF) strips. Black arrows indicate detection zone and white arrows show control bands. Ct or negative values of PCR from dilutions are shown (bottom).
Figure 4.
Recombinase polymerase amplification (RPA) in stool samples. Different amounts of Entamoeba histolytica parasites (indicated in the top) were spiked with stool sample from a healthy donor diluted in phosphate buffered saline (PBS). After DNA extraction samples were analyzed by RPA in lateral flow (LF) strips. Black arrows indicate detection zone and white arrows show control bands.
RPA-LF in clinical samples.
To validate our method, we analyzed 32 samples of DNA extracted from clinical samples obtained from ongoing studies in Colombia. In the first experiment, using 2.5 μL of sample, we obtained an 86% of correlation with real-time PCR (data not shown). In a second experiment, using 5 μL of sample resulted in 100% correlation with PCR and ELISA results, as well as clinical presentation (Table 1).
Table 1.
RPA in DNA from clinical samples. DNA obtained from patients positives for Entamoeba histolytica, E. dispar, E moshkovskii, or negatives were analyzed by RPA in LF strips
| Sample | ELISA/PCR | RPA | Microscopy/symptoms |
|---|---|---|---|
| 1 | Entamoeba moshkovskii | Negative | Cysts in stools |
| 2 | Negative | Negative | Negative |
| 3 | Negative | Negative | Negative |
| 4 | Negative | Negative | Negative |
| 5 | E. histolytica | E. histolytica | Cysts in stools, tenesmus, stools with mucus and occasional blood, acute diarrhea, abdominal pain |
| 6 | E. moshkovskii | Negative | Cysts in stools |
| 7 | Negative | Negative | Negative |
| 8 | Negative | Negative | Negative |
| 9 | E. histolytica | E. histolytica | Cysts in stools, stools with mucus diarrhea, abdominal pain |
| 10 | Negative | Negative | Negative |
| 11 | E. moshkovskii | Negative | Cysts in stools |
| 12 | Negative | Negative | Negative |
| 13 | E. moshkovskii | Negative | Cysts in stools |
| 14 | Negative | Negative | Negative |
| 15 | E. moshkovskii/E.dispar | Negative | Cysts in stools |
| 16 | E. moshkovskii | Negative | Cyst in stools |
| 17 | E. dispar | Negative | Cyst in stools |
| 18 | E. histolytica | E. histolytica | Cysts in stools, soft stools with mucus and occasional blood, abdominal pain |
| 19 | E. dispar | Negative | Cysts in stools |
| 20 | E. histolytica | E. histolytica | Cysts in stools, tenesmus, abdominal pain |
| 21 | E. dispar | Negative | Cysts in stools |
| 22 | E. dispar | Negative | Cysts in stools |
| 23 | Negative | Negative | Negative, helminths infection |
| 24 | Negative | Negative | Negative |
| 25 | E. moshkovskii | Negative | Cyst in stools |
| 26 | E. dispar | Negative | Cyst in stools |
| 27 | E. moshkovskii | Negative | Cyst in stools |
| 28 | Negative | Negative | Negative |
| 29 | Negative | Negative | Negative |
| 30 | E. histolytica | E. histolytica | Cysts in stools, acute diarrhea, soft stools with mucus and occasional blood, tenesmus, abdominal pain |
| 31 | E. dispar | Negative | Cysts in stools |
| 32 | Negative | Negative | Negative |
| Total positives | 5 | 5 | |
| Total negatives | 27 | 27 |
The RPA results were obtained using 5 μL of template from each clinical sample. DNA = deoxyribonucleic acid; ELISA = enzyme-linked immunosorbent assay; LF = lateral flow; PCR = polymerase chain reaction; RPA = recombinase polymerase amplification.
Discussion
In this work, we have developed an RPA-LF test to detect E. histolytica in stool samples. Entamoeba species are morphologically identical and difficult to distinguish even at the molecular level. We selected the 18s ribosomal gene for RPA because of its high copy number and because PCR studies have demonstrated its usefulness to distinguish Entamoeba species.15–17 For the RPA reactions, we used a set of primers of 25 nt in size compatible with RPA assays that amplify specifically a 132 amplicon of E. histolytica but not E. dispar. We adapted the RPA for LF detection by using an internal dual-labeled probe that allowed the specific detection of the amplicon on LF paper strips. The sensitivity of the RPA-LF detection was similar to real-time PCR (Figure 3). The specificity of the assay was tested with the other E. dispar, as well as two other common protozoan causing diarrhea, and demonstrated 100% specificity. Because this technique does not require thermocyclers, refrigerators, or electrophoresis equipment, RPA represents a less expensive molecular diagnostic. The reduced cost, simplicity, and portability are valuable features that will allow this test to be more widely implemented than others.
Another important factor to consider in the developing of molecular diagnostic tests is the feasibility of its use at POC. We tested this by using a DNA mini-extractor to isolate DNA from parasites spiked in stool samples. This method did not require centrifugation and purified DNA from up to eight samples in 30 minutes. The mini-extractor is small and portable. We were even able to perform the assay with batteries, demonstrating that a local electric supply is not required. Therefore, the RPA method could be used to successfully extract DNA directly from patient stool at the POC, reducing the time and costs of the test.
To verify our ability to detect E. histolytica DNA in clinical samples, we evaluated the efficacy of the E. histolytica RPA with samples previously evaluated by ELISA and PCR. Using 2.5 μL of template, we noted an 86% and 98% of correlation for positive and negative cases, respectively. To improve the sensitivity of the assay, we evaluated the same samples using 5 μL of template and obtained 100% correlation with positive and negative cases (Table 1). The differences were mainly observed in samples with low parasite burden.
The usefulness of this assay at the POC would be improved by developing a multiplex assay designed to detect most of the common pathogenic intestinal protozoa. Studies are in progress by our group to combine this assay with the previously described RPA assays for Cryptosporidium and Giardia in a multiplex assay.13,14 In addition, simpler methods of DNA extraction will be optimal for widespread use. Future studies are needed to evaluate the feasibility of implementing RPA assays in endemic areas.
Footnotes
Financial support: This study was conducted with the support of the Institute for Translational Sciences at the University of Texas Medical Branch, supported in part by a Clinical and Translational Science Award (UL1TR000071) from the National Center for Advancing Translational Sciences, National Institutes of Health.
Authors' addresses: Gayatri Nair, Mauricio Rebolledo, A. Clinton White Jr., and Alejandro Castellanos-Gonzalez, Department of Internal Medicine, University of Texas Medical Branch, Glaveston, TX, E-mails: gvnair@utmb.edu, mereboll@utmb.edu, acwhite@utmb.edu, and alcastel@utmb.edu. Zachary Crannell and R. Rebecca Richards-Kortum, Department of Bioengineering, Rice University, Houston, TX, E-mails: zcrannell@gmail.com and rkortum@rice.edu. A. Elizabeth Pinilla, Departamento de Medicina Interna, and M. Consuelo López, Departamento de Salud Pública, Universidad Nacional de Colombia, Bogota, Colombia, E-mails: aepinillar@unal.edu.co and mclopezp@unal.edu.co. Juan David Ramírez, Facultad de Ciencias Naturales y Matemáticas, Universidad del Rosario, Grupo de Investigaciones Microbiológicas, Bogota, Colombia, E-mail: juand.ramirez@urosario.edu.co.
References
- 1.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais DC, Dharmaratne SD, Dorsey ER, Driscoll T, Duber H, Ebel B, Erwin PJ, Espindola P, Ezzati M, Feigin V, Flaxman AD, Forouzanfar MH, Fowkes FG, Franklin R, Fransen M, Freeman MK, Gabriel SE, Gakidou E, Gaspari F, Gillum RF, Gonzalez-Medina D, Halasa YA, Haring D, Harrison JE, Havmoeller R, Hay RJ, Hoen B, Hotez PJ, Hoy D, Jacobsen KH, James SL, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo JP, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz SE, Ohno SL, Mabweijano J, MacIntyre MF, Mallinger L, March L, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGrath J, Mensah GA, Merriman TR, Michaud C, Miller M, Miller TR, Mock C, Mocumbi AO, Mokdad AA, Moran A, Mulholland K, Nair MN, Naldi L, Narayan KM, Nasseri K, Norman P, O'Donnell M, Omer SB, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian JD, Rivero AP, Padilla RP, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope CA, 3rd, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm JT, Rein DB, Remuzzi G, Rivara FP, Roberts T, De León FR, Rosenfeld LC, Rushton L, Sacco RL, Salomon JA, Sampson U, Sanman E, Schwebel DC, Segui-Gomez M, Shepard DS, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor JA, Thomas B, Tleyjeh IM, Towbin JA, Truelsen T, Undurraga EA, Venketasubramanian N, Vijayakumar L, Vos T, Wagner GR, Wang M, Wang W, Watt K, Weinstock MA, Weintraub R, Wilkinson JD, Woolf AD, Wulf S, Yeh PH, Yip P, Zabetian A, Zheng ZJ, Lopez AD, Murray CJ, AlMazroa MA, Memish ZA. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fotedar R, Stark D, Beebe N, Marriott D, Ellis J, Harkness J. Laboratory diagnostic techniques for Entamoeba species. Clin Microbiol Rev. 2007;20:511–532. doi: 10.1128/CMR.00004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goni P, Martin B, Villacampa M, Garcia A, Seral C, Castillo FJ, Clavel A. Evaluation of an immunochromatographic dip strip test for simultaneous detection of Cryptosporidium spp., Giardia duodenalis, and Entamoeba histolytica antigens in human faecal samples. Eur J Clin Microbiol Infect Dis. 2012;31:2077–2082. doi: 10.1007/s10096-012-1544-7. [DOI] [PubMed] [Google Scholar]
- 4.Haque R, Ali IK, Akther S, Petri WA., Jr Comparison of PCR, isoenzyme analysis, and antigen detection for diagnosis of Entamoeba histolytica infection. J Clin Microbiol. 1998;36:449–452. doi: 10.1128/jcm.36.2.449-452.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pillai DR, Keystone JS, Sheppard DC, MacLean JD, MacPherson DW, Kain KC. Entamoeba histolytica and Entamoeba dispar: epidemiology and comparison of diagnostic methods in a setting of nonendemicity. Clin Infect Dis. 1999;29:1315–1318. doi: 10.1086/313433. [DOI] [PubMed] [Google Scholar]
- 6.Mirelman D, Nuchamowitz Y, Stolarsky T. Comparison of use of enzyme-linked immunosorbent assay-based kits and PCR amplification of rRNA genes for simultaneous detection of Entamoeba histolytica and E. dispar. J Clin Microbiol. 1997;35:2405–2407. doi: 10.1128/jcm.35.9.2405-2407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roy S, Kabir M, Mondal D, Ali IK, Petri WA, Jr, Haque R. Real-time-PCR assay for diagnosis of Entamoeba histolytica infection. J Clin Microbiol. 2005;43:2168–2172. doi: 10.1128/JCM.43.5.2168-2172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blessmann J, Buss H, Nu PA, Dinh BT, Ngo QT, Van AL, Alla MD, Jackson TF, Ravdin JI, Tannich E. Real-time PCR for detection and differentiation of Entamoeba histolytica and Entamoeba dispar in fecal samples. J Clin Microbiol. 2002;40:4413–4417. doi: 10.1128/JCM.40.12.4413-4417.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Townes JM, Quick R, Gonzales OY, Linares M, Damiani E, Bopp CA, Wahlquist SP, Hutwagner LC, Hanover E, Mintz ED, Tauxe RV. Etiology of bloody diarrhea in Bolivian children: implications for empiric therapy. Bolivian Dysentery Study Group. J Infect Dis. 1997;175:1527–1530. doi: 10.1086/516493. [DOI] [PubMed] [Google Scholar]
- 10.Bansal D, Sehgal R, Chawla Y, Mahajan RC, Malla N. In vitro activity of antiamoebic drugs against clinical isolates of Entamoeba histolytica and Entamoeba dispar. Ann Clin Microbiol Antimicrob. 2004;3:27. doi: 10.1186/1476-0711-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan L, Zhou J, Zheng Y, Gamson AS, Roembke BT, Nakayama S, Sintim HO. Isothermal amplified detection of DNA and RNA. Mol Biosyst. 2014;10:970–1003. doi: 10.1039/c3mb70304e. [DOI] [PubMed] [Google Scholar]
- 12.Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol. 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crannell ZA, Castellanos-Gonzalez A, Irani A, Rohrman B, White AC, Richards-Kortum R. Nucleic acid test to diagnose cryptosporidiosis: lab assessment in animal and patient specimens. Anal Chem. 2014;86:2565–2571. doi: 10.1021/ac403750z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crannell ZA, Cabada MM, Castellanos-Gonzalez A, Irani A, White AC, Richards-Kortum R. Recombinase polymerase amplification-based assay to diagnose Giardia in stool samples. Am J Trop Med Hyg. 2015;92:583–587. doi: 10.4269/ajtmh.14-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamzah Z, Petmitr S, Mungthin M, Leelayoova S, Chavalitshewinkoon-Petmitr P. Differential detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii by a single-round PCR assay. J Clin Microbiol. 2006;44:3196–3200. doi: 10.1128/JCM.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonin P, Trudel L. Detection and differentiation of Entamoeba histolytica and Entamoeba dispar isolates in clinical samples by PCR and enzyme-linked immunosorbent assay. J Clin Microbiol. 2003;41:237–241. doi: 10.1128/JCM.41.1.237-241.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamzah Z, Petmitr S, Mungthin M, Leelayoova S, Chavalitshewinkoon-Petmitr P. Development of multiplex real-time polymerase chain reaction for detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii in clinical specimens. Am J Trop Med Hyg. 2010;83:909–913. doi: 10.4269/ajtmh.2010.10-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]