Abstract
Lice are among the oldest parasites of humans representing an excellent marker of the evolution and migration of our species over time. Here, we analyzed by real-time polymerase chain reaction (RT-PCR) developed in this study the mitochondrial DNA of seven ancient head louse eggs found on hair remains recovered from two sites in Israel: 1) five nits dating from Chalcolithic period (4,000 bc) were found in the Cave of the Treasure located at Nahal Mishmar, in the Judean Desert and 2) two nits dating from Early Islamic Period (ad 650–810) were found in Nahal Omer in the Arava Valley (between Dead Sea and Red Sea). Our results suggest that these eggs belonged to people originating from west Africa based on identification of the louse mitochondrial sub-clade specific to that region.
Head lice (Pediculus humanus capitis) and body lice (P. humanus humanus) are strict bloodsucking ectoparasites of humans, and each species lives in a specific ecological niche: hair for head lice, clothing for body lice.1 Hundreds millions of children worldwide are continually infested by head lice unrelated to hygienic conditions, resulting in insomnia and itching.2 Body lice exclusively infest populations exposed to stressful life conditions, such as the homeless, prisoners, and war refugees, and can serve as a vector for three serious humans diseases, namely, epidemic typhus, trench fever, and relapsing fever caused by Rickettsia prowazekii, Bartonella quintana, and Borrelia recurrentis, respectively.1 Body lice are also suspected to be able to host and transmit Yersinia pestis, the agent of plague.3 Genetically, human lice are distributed into three mitochondrial clades (A, B, and C), whereby only Clade A comprises head and body lice.4 Clade A is found on all continents, whereas Clade B, with an assignment of an American origin, is present in Europe and Australia and was recently characterized in north Africa. Finally, Clade C is to date limited to Africa and Asia.5
Lice are among the oldest parasites of humans, thus representing an excellent marker of the evolution and migration of the Homo species over time.4 For centuries, archaeologists excavating soil in different locations around the world have found nits, lice, and/or combs on mummies or human remains with ages varying between 300 and 10,000 years (see reference6 for a review). The most ancient specimens (nits from hair, 10,000 years old) were found in Brazil, South America.7 However, few molecular data on ancient lice are available. In 2008, Raoult and others8 showed that the most prevalent and well-distributed clade of lice (A) had a pre-Columbian presence on the American continent. In 2013, Boutellis and others9 confirmed this result and demonstrated that Clade B also had a presence of at least 4,000 years in America and that Clades A and B could live in sympatry.
In this work, we obtained and analyzed ancient head louse eggs recovered from Israel dating from two different periods, namely, the Chalcolithic (4,000 bc) and early Islamic periods (ad 650–810).
In 2014, two lots of ancient head louse nits recovered from human remains in Israel were sent to our laboratory for molecular analysis. A total of seven samples were examined. Five operculated nits were found on hair remains in the Cave of the Treasure located at Nahal Mishmar, in the Judean Desert, which date to the Chalcolithic period.10 The other two nit samples were found on human hair remains belonging to a population who lived in Nahal Omer in the Arava Valley (between Dead Sea and Red Sea) during the Early Islamic period (ad 650–810). The site appears to have been a way station on the north–south Arava route and on the Spice Route between Petra and Gaza.11,12 The operculum on the eggs was most likely detached during the centuries, but the embryos were still present inside the eggs.
The nits were photographed using an Axio Zoom V16 (Carl Zeiss AG, Oberkochen, Germany).
All precautions to avoid contamination by modern DNA templates were taken, and negative controls were used at each step of the study. Each experimental stage was performed in a clean room each of which was located in a different building free of P. humanus and its DNA. Each egg louse was rinsed twice in sterile water for 15 minutes, and then total genomic DNA was extracted from each egg louse by using a QIAamp Tissue Kit (Qiagen, Hilden, Germany) as described by the manufacturer.
To determine the clade of the specimens, an 89-bp fragment of the mitochondrial cytochrome b gene (cytb) was targeted. A sensitive tool based on the real-time polymerase chain reaction (PCR) with a hydrolysis probe that was useful to detect and amplify ancient DNA was developed. A pair of primers (the reverse primer was degenerated) that can amplify the different known clades of lice was used: cytbF (5′-AGTGTGAGGAGGGTTTTCAG-3′) and cytbR (5′-CAAACCCCAAYAAVAYAAACGG-3′). A TaqMan© FAM-labeled probe (Applied Biosystems, Courtaboeuf, France) that contained nonfluorescent quencher conjugated to a minor groove binder (MGB) at the 3′ was designed: FAM-CTACTTTAGAGCGGTTGTTTACTC-MGB.
Real-time PCR was performed using the CFX96 thermal cycler (Bio-Rad Laboratories, Foster City, CA). The final reaction volume of 20 μL contained 3 μL of the DNA template, 10 μL of 2× QuantiTect™ Probe PCR Master Mix (Qiagen), 0.5 μM of each primer and 0.2 μM of the FAM-labeled probe. The thermocycling parameters consisted of 95°C for 15 minutes and 40 cycles of 95°C for 10 seconds and 60°C for 30 seconds. No template controls (NTCs) were included in real time PCR (RT-PCR) assay.
The products of the real-time PCR amplifications were sequenced using the 3130XL genetic analyzer (Applied Biosystems) with the BigDye Terminator v1.1 cycle (Applied Biosystems, Foster City, CA). ChromasPro software (Technelysium Pty, Queensland, Australia) was used to analyze the obtained electropherograms for each sequence.
Nucleotide sequences were aligned using CLUSTAL X 2.0.11 (http://www.softpedia.com/get/Science-CAD/Clustal-X.shtml),13 and pairwise comparisons were conducted with MEGA6 (http://www.megasoftware.net/).14 Phylogenetic analyses were based on maximum-likelihood (ML) algorithms.
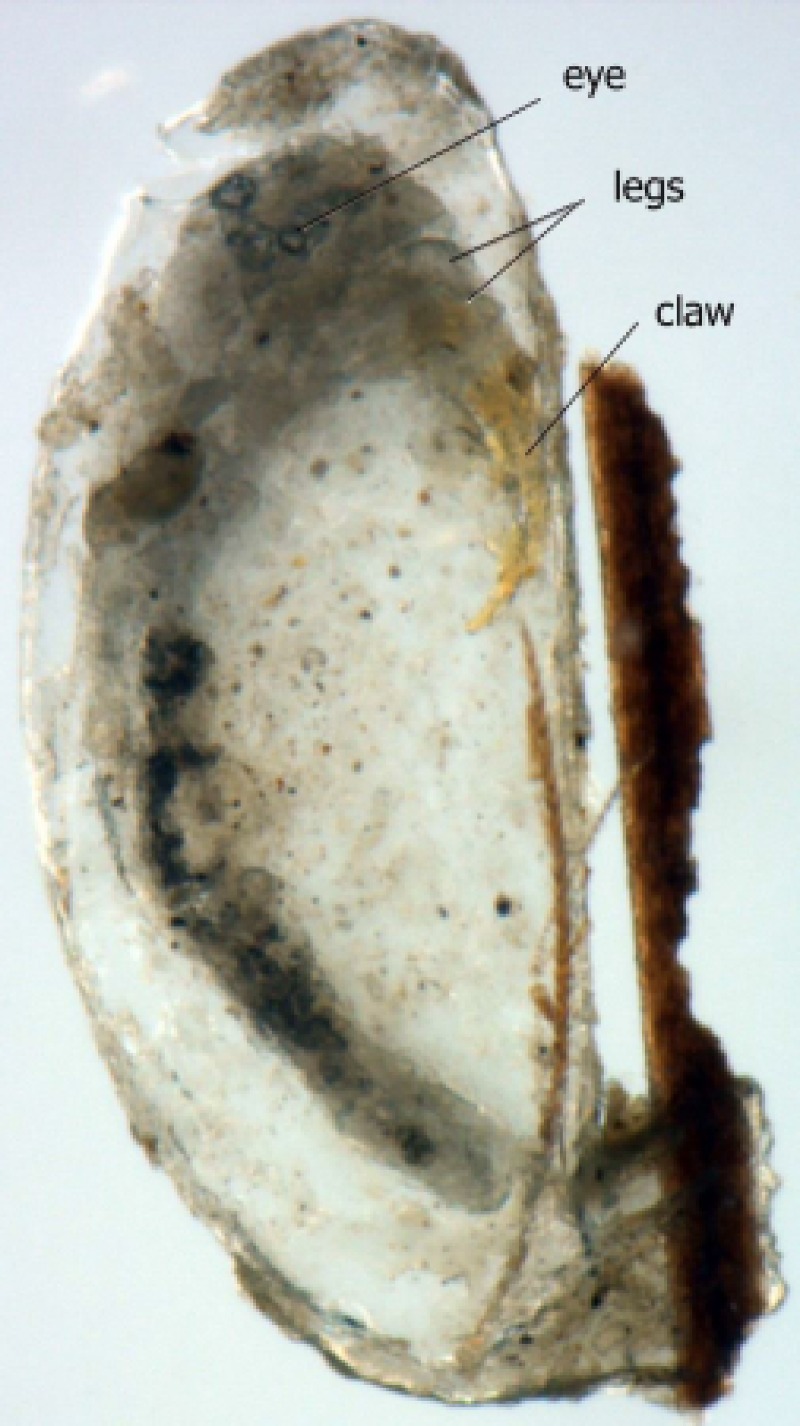
The appearance of nits under the microscope shows that they were well preserved despite the many centuries spent underground. As shown in Figure 1 , we can distinguish the eyes, legs, and claws of embryos still inside the operculated nits.
Figure 1.
Operculated head louse egg recovered from Israel dating from Chalcolithic period (4,000 bc).
RT-PCR was positive for the seven samples of ancient DNA tested (32 < Ct < 34). The Ct value was 20 in the positive control; no fluorescence was detected in the negative control.
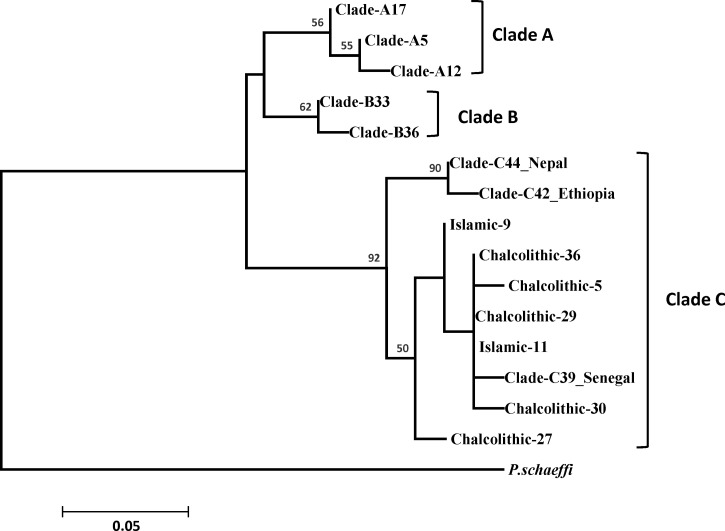
A ML phylogenetic analysis performed for the cytb gene showed that the ancient hair nits belonging to the Chalcolithic and early Islamic periods are part of Clade C. Within Clade C, these nits form a sub-clade with lice found in Senegal (Figure 2 ). Nonetheless, the ancient DNA sequences are quite unique and are different compared with known sequences of contemporary lice.
Figure 2.
Maximum-likelihood (ML) phylogram of cytochrome b gene. ML bootstrap that support values greater than 50 are located above the nodes. Mitochondrial clade memberships are indicated to the right of each tree.
In this work, the first molecular data on lice infesting the ancient inhabitants of the Near East were compiled. The tool implemented here based on the real-time PCR was effective in preventing the difficulties associated with the analysis of ancient DNA, which is often damaged or in a very low concentration.15
Interestingly, this study reveals the presence of a mitochondrial genotype, Clade C, in a Near East region that has thus far only been found in Africa (Ethiopia and Senegal) and Asia (Nepal).5 Specifically, these old nits are included in a subgroup in Clade C comprising contemporary lice that are characterized only in west Africa. The presence of this genotype in the Near East is necessarily linked to migration flows of humans through the ages. The slave trade practiced by Arab tribes to supply the Near and Middle East in manpower existed for a long time.16 The genetic information available today from humans suggests that significant gene flow most likely occurred within the past ∼2,500 years between sub-Saharan African and Near East populations.17
In conclusion, real-time PCR adopted in this study showed to be a fast, sensitive, and specific tool that is fully compatible with the analysis of ancient DNA. Through the results obtained in this work, we can affirm that the study of ancient lice is very useful to understand the migration patterns of humans, their lice, and therefore the flow of louse-borne pathogens.
ACKNOWLEDGMENTS
We thank Jean-Michel Berenger (Entomologist) and Abdul Karim Sangaré for their technical support.
Footnotes
Authors' addresses: Rezak Drali and Didier Raoult, Unité de Recherche sur les Maladies Infectieuses et Tropicales Emergentes (URMITE), Centre National de la Recherche Scientifique, Institut de Recherche pour le Développement, Institut National de la Santé et de la Recherche Médicale Unité, and Institut Hospitalo-Universitaire Méditerranée-Infection, Aix-Marseille Université, Marseille, France, E-mails: rezakdrali@hotmail.com and didier.raoult@gmail.com. Kosta Y. Mumcuoglu and Gonca Yesilyurt, Department of Microbiology and Molecular Genetics, The Kuvin Center for the Study of Infectious and Tropical Diseases, Hadassah Medical School, The Hebrew University, Jerusalem, Israel, E-mails: kostasm@ekmd.huji.ac.il and emine.gonca.yesilyurt@gmail.com.
References
- 1.Raoult D, Roux V. The body louse as a vector of reemerging human diseases. Clin Infect Dis. 1999;29:888–911. doi: 10.1086/520454. [DOI] [PubMed] [Google Scholar]
- 2.Chosidow O, Chastang C, Brue C, Bouvet E, Izri M, Monteny N, Bastuji-Garin S, Rousset JJ, Revuz J. Controlled study of malathion and d-phenothrin lotions for Pediculus humanus var capitis-infested schoolchildren. Lancet. 1994;344:1724–1727. doi: 10.1016/s0140-6736(94)92884-3. [DOI] [PubMed] [Google Scholar]
- 3.Houhamdi L, Lepidi H, Drancourt M, Raoult D. Experimental model to evaluate the human body louse as a vector of plague. J Infect Dis. 2006;194:1589–1596. doi: 10.1086/508995. [DOI] [PubMed] [Google Scholar]
- 4.Reed DL, Smith VS, Hammond SL, Rogers AR, Clayton DH. Genetic analysis of lice supports direct contact between modern and archaic humans. PLoS Biol. 2004;2:e340. doi: 10.1371/journal.pbio.0020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutellis A, Abi-Rached L, Raoult D. The origin and distribution of human lice in the world. Infect Genet Evol. 2014;23:209–217. doi: 10.1016/j.meegid.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 6.Mumcuoglu KY. Human lice: Pediculus and Pthirus. In: Raoult D, editor. Paleomicrobiology: Past Human Infections. Berlin, Germany: Springer-Verlag; 2008. pp. 215–222. [Google Scholar]
- 7.Araujo A, Ferreira LF, Guidon N, Maues Da Serra FN, Reinhard KJ, Dittmar K. Ten thousand years of head lice infection. Parasitol Today. 2000;16:269. doi: 10.1016/s0169-4758(00)01694-x. [DOI] [PubMed] [Google Scholar]
- 8.Raoult D, Reed DL, Dittmar K, Kirchman JJ, Rolain JM, Guillen S, Light JE. Molecular identification of lice from pre-Columbian mummies. J Infect Dis. 2008;197:535–543. doi: 10.1086/526520. [DOI] [PubMed] [Google Scholar]
- 9.Boutellis A, Drali R, Rivera MA, Mumcuoglu KY, Raoult D. Evidence of sympatry of clade A and clade B head lice in a pre-Columbian Chilean mummy from Camarones. PLoS One. 2013;8:e76818. doi: 10.1371/journal.pone.0076818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bar-Adon P. The Cave of the Treasure: The Finds from the Caves in Nahal Mishmar. Jerusalem, Israel: Israel Exploration Society; 1980. [Google Scholar]
- 11.Baginski A, Shamir O. Early Islamic textiles, basketry and cordage from Nahal Omer, Israel. Aticot. 1995;26:21–42. [Google Scholar]
- 12.Negev A. The date of the Petra–Gaza road. Palest Explor Q. 1966;98:89–98. [Google Scholar]
- 13.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 14.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raoult D, Dutour O, Houhamdi L, Jankauskas R, Fournier PE, Ardagna Y, Drancourt M, Signoli M, La VD, Macia Y, Aboudharam G. Evidence for louse-transmitted diseases in soldiers of Napoleon's Grand Army in Vilnius. J Infect Dis. 2006;193:112–120. doi: 10.1086/498534. [DOI] [PubMed] [Google Scholar]
- 16.Lewis B. Race and Slavery in the Middle East: An Historical Enquiry. New York, NY: Oxford University Press; 1992. [Google Scholar]
- 17.Richards M, Rengo C, Cruciani F, Gratrix F, Wilson JF, Scozzari R, Macaulay V, Torroni A. Extensive female-mediated gene flow from sub-Saharan Africa into near eastern Arab populations. Am J Hum Genet. 2003;72:1058–1064. doi: 10.1086/374384. [DOI] [PMC free article] [PubMed] [Google Scholar]