Abstract
Excessive exposure to estrogen is a well-established risk factor for endometrial cancer (EC), particularly for cancers of endometrioid histology. The physiological function of estrogen is primarily mediated by estrogen receptor alpha, encoded by ESR1. Consequently, several studies have investigated whether variation at the ESR1 locus is associated with risk of EC, with conflicting results. We performed comprehensive fine-mapping analyses of 3,633 genotyped and imputed single nucleotide polymorphisms (SNPs) in 6,607 EC cases and 37,925 controls. There was evidence of an EC risk signal located at a potential alternative promoter of the ESR1 gene (lead SNP rs79575945, P = 1.86 × 10−5), which was stronger for cancers of endometrioid subtype (P = 3.76 × 10−6). Bioinformatic analysis suggests that this risk signal is in a functionally important region targeting ESR1, and eQTL analysis found that rs79575945 was associated with expression of SYNE1, a neighbouring gene. In summary, we have identified a single EC risk signal located at ESR1, at study-wide significance. Given SNPs located at this locus have been associated with risk for breast cancer, also a hormonally driven cancer, this study adds weight to the rationale for performing informed candidate fine-scale genetic studies across cancer types.
Keywords: Endometrial cancer, ESR1, single-nucleotide polymorphisms, fine-mapping analysis
Introduction
Endometrial cancer is the most commonly diagnosed gynaecological malignancy in developed countries (http://globocan.iarc.fr/). Excessive endogenous and exogenous estrogen exposure or estrogen exposure unopposed by progesterone is a well-established risk factor for the development and progression of endometrial cancer (Kaaks, et al. 2002; Key and Pike 1988). Estrogen receptor alpha (encoded by ESR1) is the predominant receptor responsible for mediating the effects of estrogen in the endometrium.
A number of studies have previously been performed to investigate the hypothesis that variation at the ESR1 locus may be associated with predisposition to endometrial cancer (Ashton, et al. 2009, 2010; Einarsdottir, et al. 2009; Einarsdottir, et al. 2008; Iwamoto, et al. 2003; Li, et al. 2011; Sasaki, et al. 2002; Sliwinski, et al. 2010; Wedren, et al. 2008; Weiderpass, et al. 2000), but results from these relatively underpowered studies (maximum sample size 713 cases and 1567 controls) have been conflicting. However, comprehensive candidate gene and genome-wide association studies of breast cancer, which shares many risk factors with endometrial cancer, have identified cancer-associated risk variants at the ESR1 locus (Dunning, et al. 2009; Hein, et al. 2012; Turnbull, et al. 2010; Zheng, et al. 2009). These findings indicate a need for similar large-scale and comprehensive genetic analysis of endometrial cancer to elucidate the role of ESR1 variants in the risk of endometrial cancer. Here we present the results from fine-mapping of the ESR1 locus by dense SNP genotyping and imputation in 6,607 endometrial cancer cases and 37,925 controls of European descent within the Endometrial Cancer Association Consortium (ECAC).
Materials and Methods
Datasets
Genotyping of the fine-mapping dataset was performed on a custom Illumina Infinium iSelect array (“iCOGS”; designed by the Collaborative Oncological Gene-environment Study, details summarized in (Bahcall 2013)). All studies have the relevant IRB approval in each country in accordance with the principles embodied in the Declaration of Helsinki, and informed consent was obtained from all participants. Details of iCOGS genotyping of endometrial cancer cases and control samples can be found in Supplementary Table 1 and in Painter et al (Painter, et al. 2014). All cases and controls selected for analysis were of European ancestry, as defined by Identity-By-State (IBS) scores between study individuals and individuals in HapMap (http://hapmap.ncbi.nlm.nih.gov/). The final analysis of the iCOGS dataset included genotypes for 4,401 women with a confirmed diagnosis of endometrial cancer and 28,758 healthy female controls genotyped by the Breast Cancer Association Consortium (BCAC) or the Ovarian Cancer Association Consortium (OCAC). Additionally, three Caucasian GWAS datasets (ANECS, SEARCH and NSECG) were as previously described, totalling 2,206 cases and 9,167 controls after quality control.(Painter et al. 2014; Spurdle, et al. 2011). Overall, there were 6,607 endometrial cancer cases and 37,925 controls included in the meta-analysis of the four datasets (ANECS, SEARCH and NSECG GWAS datasets and the iCOGS dataset).
Fine-mapping
The study herein includes SNPs in a 1Mb region including ESR1 (chr6: 151,600,000–152,650,000; NCBI build 37 assembly). SNPs with a minor allele frequency > 2% using the 1000 Genomes Project (March 2010 Pilot version 60 CEU project data) were considered for inclusion for ESR1 fine-mapping on the iCOGS array by BCAC. In total 975 SNPs were selected, comprising 277 SNPs correlated (r2 > 0.1) with three previously reported breast cancer associated SNPs (rs2046210, rs3757318 and rs3020314), and a 698 SNP set tagging all remaining SNPs in the region with r2 > 0.9.
Regional Imputation
Genotypes for SNPs present in 1000 Genomes Phase 1 (April 2012 release) were imputed for the fine-mapping dataset and each GWAS dataset using IMPUTE V2.0 (Howie, et al. 2009). Imputation was performed separately for each dataset. SNPs with an imputation information score > 0.8 for all four datasets and minor allele frequency > 0.01 were included in analysis. Following quality control, a total of 3,633 genotyped and imputed SNPs were available across all four datasets (the three GWAS and iCOGS datasets).
Association Analysis
Odds ratios for each SNP were estimated for the four imputed datasets separately, using unconditional logistic regression with a per-allele (1 degree-of-freedom) model, based on the expected genotyped dosages for the imputed SNPs. The GWAS datasets were each analysed as a single stratum, with adjustment for the first two (ANECS and NSECG) and three (SEARCH) principal components. For the iCOGS dataset, analyses were performed adjusting for strata and for the first ten principal components, as previously described (Painter et al. 2014). The numbers of principal components included in the analyses were selected to adequately account for population stratification in each of the datasets. Results from the four studies were combined using standard fixed-effects meta-analysis, and between-study heterogeneity assessed by Q statistic (Higgins and Thompson 2002). Risk estimation was performed separately for each tested phenotype (endometrial cancer, endometrioid endometrial cancer, non-endometrioid endometrial cancer). To determine independently associated SNPs, we used forward stepwise logistic regression based on all SNPs with P < 0.05 in the single-SNP analysis; at each stage, SNPs were included in the model if they were significant at P < 0.05 after adjustment for other SNPs. To assess possible interaction with BMI group (≤30 kg/m2 or >30 kg/m2) for lead SNP rs79575945, the significance of multiplicative interaction was assessed by the change in the likelihood ratio estimate after inclusion of a BMI-by-genotype interaction term to a simpler model without this term. Analyses were conducted using R, including the GenABEL (Aulchenko, et al. 2007) and meta packages (Schwarzer 2010) and SNPTESTv2 (Ferreira and Marchini 2011). All statistical tests were 2-sided.
eQTL analysis
Data from endometrial tumours were accessed from The Cancer Genome Atlas (TCGA) (Cancer Genome Atlas Research, et al. 2013). Germline SNP genotypes (Affymetrix 6.0 arrays) were downloaded through the controlled access portal, while epidemiological data, normalized RNA-Seq data and copy-number information were downloaded through the public access TCGA portal. There were 290 TCGA patients (221 endometrioid histology) with complete genotype, RNA-Seq and copy-number data included in the analysis. Quality control was performed on the germline SNP genotypes as previously described (Carvajal-Carmona, et al. 2015). To increase the number of SNPs in the analysis, we imputed genotypes for SNPs present in the 1000 Genomes dataset v3 in the ESR1 region (chr6: 150,125,000–152,650,000, April 2012 release) which were not genotyped by the Affymetrix 6.0 platform using minimac (Fuchsberger, et al. 2014; Howie, et al. 2012) software. Haplotypes were phased using the MaCH program (Li, et al. 2009; Li, et al. 2010) before running minimac for genotype imputation, using the recommended parameters (20 iterations of the Markov sampler and 200 states). SNPs imputed with a RSQR (quality measure) > 0.8 and minor allele frequency > 0.01 were included in the eQTL analysis. RNA-Seq expression for genes 500kb upstream and downstream of ESR1 (SYNE1, ESR1, CCDC170, C6orf211, RMND1, ZBTB2, AKAP12, MYCT1) were adjusted for somatic copy number variation, as previously described by Li et al (Li, et al. 2013). The associations between genotype and adjusted expression for each gene were evaluated using linear regression models by the mach2qtl program (Li et al. 2009; Li et al. 2010). Associations were considered to be statistically significant after correction for the total number of genes analysed across the region (0.05/8 genes = 6.25 × 10−3).
Results
Meta-analysis performed on 3,633 SNPs that passed quality control criteria in the four studies (iCOGS, ANECS, SEARCH and NSECG) identified 401 SNPs associated with endometrial cancer risk with P < 0.05 (Supplementary Table 2), compared to 182 expected by chance. When analysis was restricted to endometrioid-only endometrial cancer, 411 mostly overlapping SNPs were identified to be associated with a P < 0.05 (Supplementary Table 2).
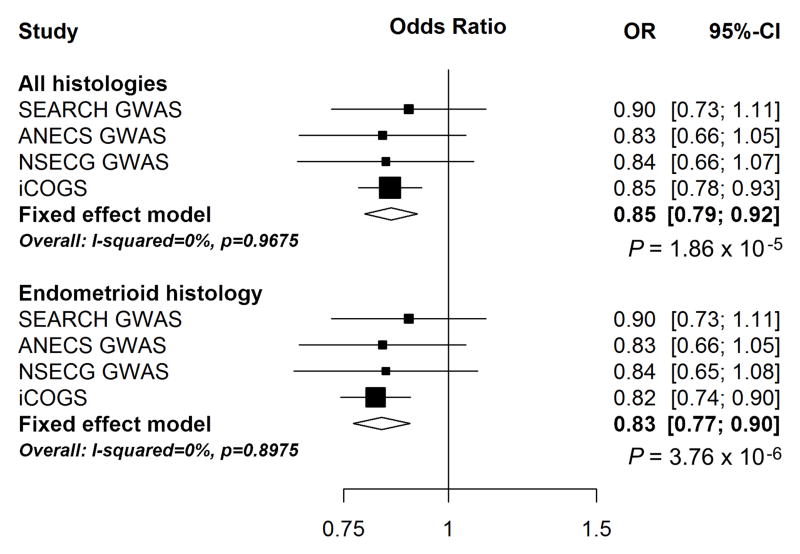
Imputed SNP rs79575945 displayed the strongest association for endometrial cancer risk (per A-allele OR 0.85, 95% CI 0.79–0.92, P = 1.85 × 10−5; Figure 1). The risk association was slightly stronger for endometrioid endometrial cancer (per A-allele OR 0.83, 95% CI 0.77–0.90, P = 3.76 × 10−6; 5,611 endometrioid cases and 37,926 controls). No other SNPs reached significance (P < 1.85 × 10−5) after conditioning on rs79575945, suggesting the presence of a single endometrial risk signal at this locus. Similar associations were observed for rs9341019 in the same linkage disequilibrium (LD) block as rs79575945, which was genotyped in all four datasets (rs9341019 OR 0.84, 95% CI 0.76–0.92, P = 2.2 × 10−4; r2 = 0.27 to rs79575945).
Fig. 1.
Forest plot of odds ratios for the GWAS and iCOGS fine-mapping datasets for SNP rs79575945 for all histologies and for endometrioid histology.
Supplementary Table 3 lists the 47 SNPs most likely to be the causal variant underlying the risk associations with most significant “lead” SNPs rs79575945. This SNP set was defined as the SNPs which were in LD (r2 > 0.2) and had a likelihood of association with endometrial cancer < 100:1 with the relevant lead SNP (Carvajal-Carmona et al. 2015; Glubb, et al. 2015).
Given BMI is a major epidemiological risk factor for endometrial cancer, analyses were repeated adjusting for BMI in the subset of cases (N = 4,088) and controls (N = 16,590) for whom BMI data were available, and also assessing the possible interaction of rs79575945 with BMI group (≤ 30 kg/m2 or > 30 kg/m2). There was no discernible difference in effect for rs79575945 (unadjusted OR = 0.86, P = 2.4 × 10−3; adjusted OR = 0.82, P = 3.7 × 10−4), and no significant evidence of interaction of rs79575945 with BMI (P-interaction=0.15).
SNP rs79575945 was not significantly associated with risk of non-endometrioid endometrial cancer (OR 0.94, 95% CI 0.80–1.13, P = 0.54), although there was reduced power to detect association due to the smaller case sample size (iCOGS fine-mapping and NSECG GWAS datasets only, case N = 887). No SNP reached study-wide significance for non-endometrioid endometrial cancer risk. Similarly, no significant associations were found in the case-only analysis, comparing endometrioid endometrial cancer patients to non-endometrioid patients (rs79575945 OR 1.08, 95% CI 0.89–1.30, P = 0.43).
None of the 47 potentially causal variants (Supplementary Table 3) showed evidence of an association with ESR1 expression, using genotype and RNA-Seq data from TCGA. The strongest association observed for any SNP in this region with ESR1 levels in endometrioid endometrial tumours was rs74575485 located upstream of the rs79575945 risk signal (r2 = 0.001), but this SNP was not associated with risk (eQTL P = 1.45 × 10−3, risk P = 0.77). We found evidence of an association between the top risk SNP rs79575945 and increased expression of SYNE1 in endometrioid endometrial tumour (eQTL P = 3.17 × 10−3). This association is considered to be statistically significant after correcting for the total number of genes analysed across the region (P for significance = 6.25 × 10−3).
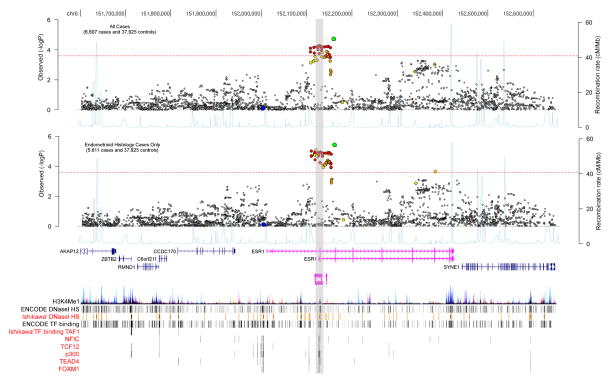
We integrated location of candidate causal SNPs with publicly available genomic data to assess likely functional relevance of SNPs. Candidate causal SNPs mapped to a potential regulatory element, which we defined by evidence of enhancer-specific histone modification (mono-methylation of H3 lysine 4 [H3K4Me1]), DNaseI hypersensitivity sites representative of open chromatin, and regions bound by transcription factors (Figure 2). Super-enhancers annotated in the study by Hnisz et al. 2013 were also found to overlap with candidate causal SNPs (Figure 2), indicating the functional importance of this region. Importantly, ENCODE data showed presence of DNaseI hypersensitivity sites and evidence for binding of transcription factors in Ishikawa endometrial cancer cells, indicating these regions may be active in endometrial tumours. The binding of these transcription factors were not found to be altered by the candidate causal SNPs, using two independent in silico prediction algorithms (Supplementary Table 4). Candidate causal SNP rs9340770 was predicted to alter binding of p300 by HaploReg, and ENCODE data have shown p300 binding to occur at this region in Ishikawa cells (Encode Project Consortium, et al. 2012).
Fig. 2.
Association results for all SNPs with endometrial cancer risk from the meta-analysis are shown in the first panel, and association with endometrioid histological subtype the second panel. There was the same number of genotyped or well-imputed samples available for the analysis of each SNP. Only SNPs passing quality control (information score > 0.8 and minor allele frequency > 0.01 across all datasets) are plotted as the negative log of the P value against relative position across the locus (base position [hg19] displayed across the top). SNPs genotyped in the iCOGS dataset are displayed as diamonds and SNPs imputed as circles. The lead SNP, rs79575945 is shown as a green filled circle and LD with surrounding SNPs indicated by colour (SNPs r2 ≥ 0.8 are red, r2 ≥ 0.5 and < 0.8 are orange, r2 ≥ 0.2 and < 0.5 are yellow and r2 < 0.2 are unfilled). The SNP most strongly associated with ESR1 expression in endometrial cancer tumours is shown as a filled blue circle. Red horizontal dashed lines denote study-wide significance thresholds (P = 2 × 10−4). The third panel shows a schematic of gene structures with exons (vertical boxes) joined by introns (lines). Enhancers predicted in Hnisz et al (Hnisz et al. 2013) which overlap SNPs associated with the three phenotypes are depicted as coloured bars, where the colour matches the schematic of its predicted target gene. Histone modification associated with promoters (H3K4Me1) from seven ENCODE Project cell types are indicated. DNaseI hypersensitivity sites (DHS) and transcription factor (TF) binding identified in 125 and 91 ENCODE Project cell types respectively, are displayed. DNaseI HS and transcription factor binding regions in Ishikawa endometrial cancer cells* are also shown. The grey vertical stripe indicates the putative promoter region overlapping the risk signal. *Note in 2015 ENCODE re-identified ECC-1 cells as Ishikawa (https://www.encodeproject.org/biosamples/ENCBS312UTV/)(Korch, et al. 2012).
Discussion
We have performed the largest and most comprehensive study assessing the association of SNPs across the ESR1 gene with endometrial cancer risk. We provide evidence of a study-wide significant association between endometrial cancer risk and imputed SNP rs79575945. Our study implemented parameters to reduce imputation errors and minimize false-positive associations, including rigorous pre-imputation quality control, excluding rare SNPs (minor allele frequency < 0.01) and using a high imputation quality score threshold (> 0.8) for analyses (Marchini and Howie 2010). These measures, and the similar association observed for the best genotyped SNP in the same LD block as imputed lead SNP rs79575945, increase our confidence for the observed association. Given the strong prior evidence for association of this region with a hormonal cancer, as well as with other hormone-related phenotypes (Estrada, et al. 2012; Perry, et al. 2014), we considered this a candidate-gene study. The consistency of SNP association with endometrial cancer risk between the four studies gives us confidence in this finding. Using tagger (de Bakker, et al. 2005), 246 SNPs were calculated to be required to tag our region of interest by pairwise-tagging (r2 ≥ 0.5). The most strongly associated SNP had a P-value an order of magnitude smaller than the Bonferroni-adjusted significance threshold based on the number of independent SNPs at the locus (P for significance = 0.05/246 = 2.0 × 10−4). Notably, there was a more significant association for the endometrioid histology subtype which is well-established to be estrogen driven (Kaaks et al. 2002).
Neither SNP rs79575945, nor any other in the risk-associated SNP set, has been previously reported to be associated with endometrial cancer risk. Reported associated SNPs from smaller candidate studies investigating the effect of genetic variation at the ESR1 locus on endometrial cancer risk are not in LD (r2 < 0.2) with rs79575945 and were not validated in our larger study (Table 1).
Table 1.
Associations of ESR1 SNPs previously reported to be associated with endometrial cancer.
| SNP Chr:Position (b37) | Effect/Reference Allele | Frequency of Effect Allele | Imputation information Score | Genotyped/Imputed | All Cases | Endometrioid Only | r2 with rs79575945 | Direction of effect relative to effect allele (Reference reporting association with endometrial cancer) a | ||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95 % CI) | P | OR (95 % CI) | P | |||||||
| rs2234693 6: 152163335 |
T/C | 0.46 | 1.00 | Genotyped | 0.98 (0.94–1.02) | 0.35 | 0.98 (0.94–1.02) | 0.32 | 0.12 | (↓) (Ashton et al. 2009) (↓) (Einarsdottir et al. 2009) (↑) (Iwamoto et al. 2003) (↑) (Wedren et al. 2008) |
| rs9340799 6: 152163381 |
A/G | 0.35 | 1.00 | Genotyped | 1.04 (1.0–1.08) | 0.08 | 1.04 (1.00–1.09) | 0.06 | 0.06 | (↓) (Ashton et al. 2009) (↓) (Einarsdottir et al. 2009) (↑) (Iwamoto et al. 2003) (↑) (Wedren et al. 2008) |
| rs3853250 6: 152159900 |
T/G | 0.46 | 0.99 | Imputed | 0.98 (0.94–1.02) | 0.35 | 0.98 (0.94–1.02) | 0.32 | 0.12 | (↓) (Einarsdottir et al. 2009) |
| rs1709181 6: 152175180 |
T/C | 0.26 | 0.98 | Imputed | 1.02 (0.98–1.07) | 0.40 | 1.03 (0.99–1.08) | 0.15 | 0.07 | (↓) (Einarsdottir et al. 2009) |
| rs4870057 6: 152171898 |
A/G | 0.33 | 0.91 | Imputed | 1.04 (0.99–1.08) | 0.11 | 1.04 (0.99–1.09) | 0.09 | 0.05 | (↓) (Einarsdottir et al. 2009) |
| rs2046210 6: 151948366 |
G/A | 0.34 | 1.00 | Genotyped | 1.03 (0.99–1.07) | 0.18 | 1.03 (0.99–1.08) | 0.17 | 0.0005 | (↓) (Li et al. 2011) |
| rs2077647 6: 152129077 |
T/C | 0.48 | 1.00 | Genotyped | 0.99 (0.96–1.04) | 0.78 | 1.00 (0.96–1.04) | 0.85 | 0.11 | (↑) (Sasaki et al. 2002) |
| rs1801132 6: 152265522 |
G/C | 0.22 | 0.98 | Imputed | 1.03 (0.98–1.08) | 0.27 | 1.01 (0.96–1.07) | 0.65 | 0.01 | (↑) (Sliwinski et al. 2010) |
SNPs previously reported as significantly associated with endometrial cancer risk were selected from a literature search. Significance thresholds were stated as P < 0.05 in all publications. Sample sizes for studies were as follows: Ashton - 191 cases and 291 controls, Einarsdottir - 713 cases and 1567 controls, Iwamoto - 92 cases and 65 controls, Li - 953 cases and 947 controls, Sasaki - 113 cases and 200 controls, Sliwinski - 100 cases and 100 controls, Wedren - 702 cases and 1563 controls.
SNPs associated with multiple phenotypes have been mapped to the ESR1 locus, notably breast cancer (Hein et al. 2012; Turnbull et al. 2010; Zheng et al. 2009), which shares many risk factors with endometrial cancer, and age-of-menarche (Perry et al. 2014) and bone mineral density (Estrada et al. 2012), which are both associated with estrogen exposure. However, none of the SNPs reported by these studies are correlated with any of the variants found to be associated with endometrial cancer risk (r2 < 0.2). The lack of overlap between risk variants for endometrial cancer, breast cancer and risk factors associated with estrogen exposure suggest that while these risks could be mediated through the same target gene, they are working via different regulatory mechanisms in different cell types.
Using log-likelihood ratios and LD, we have identified 47 candidate causal variants located at a potential alternative promoter of ESR1, represented by lead SNP rs79575945. Bioinformatics data provide evidence that these variants reside within a putative regulatory element for ESR1 and/or other genes in this region. By cross-reference to the catalogue created using 86 cell lines by Hnisz et al (Hnisz et al. 2013), we also provide evidence that candidate causal variants lie in a region encompassing super-enhancers that target ESR1. Super-enhancers consist of large clusters of transcriptional enhancers and are associated with genes that control and define cell identity (Loven, et al. 2013; Whyte, et al. 2013). The presence of super-enhancers overlapping the candidate causal variants indicates the functional importance of this region. Four candidate causal variants were predicted to alter transcription factor binding by two independent programs, is-rSNP (Macintyre, et al. 2010) and HaploReg (Ward and Kellis 2012); however none of these transcription factors identified have been examined by ENCODE. There was evidence of binding of transcription factors TAF1, NFIC, TCF12, p300, TEAD4 and FOXM1 overlapping candidate causal SNPs in Ishikawa cells by ENCODE. However, the binding of these transcription factors were not found to be altered by the candidate causal SNPs using is-rSNP and HaploReg. Given transcription factor binding frequently occurs in the absence of a known motif (Kheradpour and Kellis 2014), SNP effects may not have been correctly assessed in this analysis. Functional analysis would therefore be required to assess the impact of these SNPs on transcription factor binding. Using data from HaploReg alone, candidate causal SNP rs9340770 was predicted to alter binding of p300 and ENCODE data indicates that rs9340770 is in a region bound by p300 in Ishikawa cells. SNP rs9340770 is located upstream of an alternative transcript for ESR1, and the binding of p300 suggests this could be a putative promoter for these transcripts. Further functional work is required to uncover whether this SNP is affecting the expression of these alternative transcripts by disrupting p300 binding.
Although predicted to be the target gene bioinformatically, eQTL analysis using TCGA data did not find the candidate causal SNPs to be significantly associated with ESR1 expression. This is in line with previous fine-mapping studies performed for breast cancer, where candidate causal variants have not been found to act as eQTLs for predicted target genes in breast tissue samples (Ghoussaini, et al. 2014; Glubb et al. 2015). The reason for this is unclear; it is possible that the effect of candidate SNPs on expression levels cannot always be detected in tumour tissue due to tissue-heterogeneity. Furthermore, eQTLs are context-dependent and might only be expressed in certain stages of cancer development, or only when under particular stimuli.
We did find candidate causal SNPs to be significantly associated with SYNE1 (spectrin repeat containing, nuclear envelope 1) expression in endometrioid endometrial cancer tissue. SYNE1 encodes Nesprin-1 which is reported to be involved in a variety of cellular processes, including Golgi and nucleus organization and cytokinesis (Fan and Beck 2004; Gough, et al. 2003; Zhang, et al. 2001). Genetic variation in SYNE1 has been reported to be associated with increased risk of invasive ovarian cancer Doherty, et al. 2010. SYNE1 is frequently methylated in lung adenocarcinoma and colorectal cancer (Schuebel, et al. 2007; Tessema, et al. 2008) and mutations in SYNE1 have been reported in colorectal cancer (Sjoblom, et al. 2006). Downregulation of an N-terminal isoform of Nesprin-1, Drop1, has been observed in cancers of the uterus, cervix, kidney, thyroid, pancreas and lung (Marme, et al. 2008). Interestingly, a recent study has indicated a role for Nesprin-1 in the DNA damage response pathway, and identified Nesprin-1 as interacting with mismatch repair proteins MSH2 and MSH6 (Sur, et al. 2014). Given that mismatch repair deficiency is observed in up to 30% of endometrial tumours (Kanaya, et al. 2003), and the eQTL data from our study, the role of SYNE1 in endometrial cancer should be explored further.
In conclusion, we have identified a single endometrial cancer risk signal, at study-wide significance, located within a potential alternative promoter for ESR1. Lead SNP, rs79575945 is also reported to be associated with expression of SYNE1, adjacent to ESR1. Given SNPs at this locus have previously been identified as predisposing to breast cancer, also a hormonally driven cancer, this study adds weight to the rationale for performing informed candidate fine-scale genetic studies across cancer types (Carvajal-Carmona et al. 2015).
Supplementary Material
Acknowledgments
Funding
This work was supported by the National Health and Medical Research Council of Australia [ID#1031333 to ABS, DF, AMD, ID#39435 to ANECS, ID#552402, QIMR Controls]; National Health and Medical Research Council of Australia Fellowship Scheme [to ABS]; Principal Research Fellow of Cancer Research UK [to DFE]; Joseph Mitchell Trust [to AMD]; Oxford Comprehensive Biomedical Research Centre [to IT]; The European Community’s Seventh Framework Programme [grant agreement no 22175 [HEALTH-F2-2009-223175] [COGS]]; Cancer Research UK [C1287/A10118 to COGS and BCAC, C1287/A10710, C12292/A11174, C1281/A12014 to COGS and BCAC, C5047/A15007, C5047/A10692, C8197/A16565, C490/A10124 to SEARCH, CORGI - NSECG, to IT]; National Institutes of Health [CA128978, R01 CA122443 to MECS and MAY, P30 CA15083 to MECS, P50 CA136393 to MECS and MAY, CAHRES]; Post-Cancer GWAS Initiative [1U19 CA148537, 1U19 CA148065, 1U19 CA148112 - the GAME-ON initiative]; Department of Defence [W81XWH-10-1-0341]; Canadian Institutes of Health Research [CIHR] for the CIHR Team in Familial Risks of Breast Cancer; Komen Foundation for the Cure; The Breast Cancer Research Foundation; Ovarian Cancer Research Fund [to COGS]; Cancer Council Queensland [ID#4196615 to ANECS]; Council Cancer Tasmania [ID#403031, #ID457636 to ANECS]; Medical Research Council [G0000934 to the British 1958 Birth Cohort]; Wellcome Trust [068545/Z/02, 085475 to the British 1958 Birth Cohort]; Wellcome Trust Human Genetics Grant [090532/Z/09/Z to NSECG]; European Union [EU FP7 CHIBCHA to NSECG]; The University of Newcastle [to QIMR Controls, to NECS]; Gladys M Brawn Senior Research Fellowship [QIMR Controls]; The Vincent Fairfax Family Foundation [QIMR Controls]; Hunter Medical Research Institute [HCS, NECS]; Hunter Area Pathology Service [HCS]; ELAN fund of the University of Erlangen [BECS]; Verelst Foundation for endometrial cancer [LES]; Fred C and Katherine B Anderson Foundation [to MECS, to MAY]; Mayo Foundation [to MECS, to MAY]; Ovarian Cancer Research Fund with support of the Smith family, in memory of Kathryn Sladek Smith [MECS, PPD/RPCI.07 to OCAC]; Helse Vest Grant [MoMaTEC]; University of Bergen [MoMaTEC]; Melzer Foundation [MoMaTEC]; The Norwegian Cancer Society - Harald Andersens legat [MoMaTEC]; The Research Council of Norway [MoMaTEC]; Haukeland University of Hospital [MoMaTEC]; NBN Children’s Cancer Research Group [NECS]; Ms Jennie Thomas [NECS]; regional agreement on medical training and clinical research [ALF] between Stockholm County Council and Karolinska Institutet [20110222, 20110483, 20110141 and DF 07015 all to RENDOCAS, to KARBAC]; The Swedish Labor Market Insurance [100069 to RENDOCAS]; The Swedish Cancer Society [11 0439 to RENDOCAS]; Agency for Science, Technology and Research of Singapore [CAHRES]; Susan G Komen Breast Cancer Foundation [CAHRES]; UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge [OCAC]; Baden-Württemberg state Ministry of Science, Research and Arts [ESTHER]; Federal Ministry of Family Affairs, Senior Citizens, Women and Youth [ESTHER]; Federal Ministry of Education and Research [BMBF] Germany [01KW9975/5 to GENICA, 01KW9976/8 to GENICA, 01KW9977/0 to GENICA, 01KW0114 to GENICA, to ESTHER]; Robert Bosch Foundation [GENICA]; Deutsches Krebsforschungszentrum - DKFZ [GENICA]; Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum, IPA [GENICA]; Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus [GENICA]; Deutsche Krebshilfe e.V. [70-2892-BR I to MARIE]; Hamburg Cancer Society [MARIE]; German Cancer Research Center [MARIE]; Breast Cancer Research Foundation [MCBCS]; David F. and Margaret T. Grohne Family Foundation [MCBCS]; Ting Tsung and Wei Fong Chao Foundation [MCBCS]; VicHealth [MCCS]; Cancer Council Victoria [MCCS]; Breakthrough Breast Cancer [UKBGS]; Institute of Cancer Research [UKBGS]; and NHS funding to the NIHR Biomedical Research Centre [UKBGS/ICR].
The authors thank the many individuals who participated in this study and the numerous institutions and their staff that supported recruitment, detailed in full in the Supplementary Text. Control data was generated by the Wellcome Trust Case Control Consortium (WTCCC), and a full list of the investigators who contributed to the generation of the data is available from the WTCCC website. We acknowledge use of DNA from the British 1958 Birth Cohort collection. In addition, the results published here are based partly on data generated by TCGA, established by the NCI and the National Human Genome Research Institute. The authors also thank the specimen donors and relevant research groups associated with this project.
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Author contributions
AMD, DFE, PDPP, IT and ABS obtained funding for the study. DFE and ABS designed the study and ABS and TO’M drafted the manuscript. TO’M conducted all statistical analyses. DJT conducted genotype imputation. DMG and TO’M conducted bioinformatics analyses. TOM and JNP co-ordinated the endometrial cancer iCOGS genotyping, and associated data management. JD, KM and JPT co-ordinated quality control and data cleaning for the iCOGS datasets, and KM provided quality control for the SEARCH GWAS control set. MKB, QW, JPT and MS were responsible for data management. TO’M and ABS co-ordinated the ANECS GWAS genotyping; AMD co-ordinated the SEARCH GWAS genotyping; IT co-ordinated the NSECG GWAS genotyping. TC, JA, EGH, MMcE, RJS, KA, GO, TP, SA, CSH, MG, LM, SH, PAF, AH, MWB, ABE, PH, KC, HD, JL, JD, DA, FA, ELG, SCD, BLF, SJW, HBS, TSN, JT, HMFW, ET, TL, MM, JLH, JP, AJS, BB, HB, AM, HB, AL, JC-C, FJC, GGG, VNK, AC, ANECS, NSECG, RENDOCAS and the AOCS Group were involved in the co-ordination and/or extraction of phenotypic information for contributing studies. All authors provided critical review of the manuscript.
References
- Ashton KA, Proietto A, Otton G, Symonds I, McEvoy M, Attia J, Gilbert M, Hamann U, Scott RJ. Estrogen receptor polymorphisms and the risk of endometrial cancer. BJOG. 2009;116:1053–1061. doi: 10.1111/j.1471-0528.2009.02185.x. [DOI] [PubMed] [Google Scholar]
- Ashton KA, Proietto A, Otton G, Symonds I, McEvoy M, Attia J, Gilbert M, Hamann U, Scott RJ. Polymorphisms in genes of the steroid hormone biosynthesis and metabolism pathways and endometrial cancer risk. Cancer Epidemiol. 2010;34:328–337. doi: 10.1016/j.canep.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- Bahcall OG. iCOGS collection provides a collaborative model. Foreword. Nat Genet. 2013;45:343. doi: 10.1038/ng.2592. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Carmona LG, O’Mara TA, Painter JN, Lose FA, Dennis J, Michailidou K, Tyrer JP, Ahmed S, Ferguson K, Healey CS, et al. Candidate locus analysis of the TERT-CLPTM1L cancer risk region on chromosome 5p15 identifies multiple independent variants associated with endometrial cancer risk. Hum Genet. 2015;134:231–245. doi: 10.1007/s00439-014-1515-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Doherty JA, Rossing MA, Cushing-Haugen KL, Chen C, Van Den Berg DJ, Wu AH, Pike MC, Ness RB, Moysich K, Chenevix-Trench G, et al. ESR1/SYNE1 polymorphism and invasive epithelial ovarian cancer risk: an Ovarian Cancer Association Consortium study. Cancer Epidemiol Biomarkers Prev. 2010;19:245–250. doi: 10.1158/1055-9965.EPI-09-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning AM, Healey CS, Baynes C, Maia AT, Scollen S, Vega A, Rodriguez R, Barbosa-Morais NL, Ponder BA, et al. Search. Association of ESR1 gene tagging SNPs with breast cancer risk. Hum Mol Genet. 2009;18:1131–1139. doi: 10.1093/hmg/ddn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einarsdottir K, Darabi H, Czene K, Li Y, Low YL, Li YQ, Bonnard C, Wedren S, Liu ET, Hall P, et al. Common genetic variability in ESR1 and EGF in relation to endometrial cancer risk and survival. Br J Cancer. 2009;100:1358–1364. doi: 10.1038/sj.bjc.6604984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einarsdottir K, Darabi H, Li Y, Low YL, Li YQ, Bonnard C, Sjolander A, Czene K, Wedren S, Liu ET, et al. ESR1 and EGF genetic variation in relation to breast cancer risk and survival. Breast Cancer Res. 2008;10:R15. doi: 10.1186/bcr1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encode Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, Oei L, Albagha OM, Amin N, Kemp JP, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44:491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Beck KA. A role for the spectrin superfamily member Syne-1 and kinesin II in cytokinesis. J Cell Sci. 2004;117:619–629. doi: 10.1242/jcs.00892. [DOI] [PubMed] [Google Scholar]
- Ferreira T, Marchini J. Modeling interactions with known risk loci-a Bayesian model averaging approach. Ann Hum Genet. 2011;75:1–9. doi: 10.1111/j.1469-1809.2010.00618.x. [DOI] [PubMed] [Google Scholar]
- Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoussaini M, Edwards SL, Michailidou K, Nord S, Cowper-Sal Lari R, Desai K, Kar S, Hillman KM, Kaufmann S, Glubb DM, et al. Evidence that breast cancer risk at the 2q35 locus is mediated through IGFBP5 regulation. Nat Commun. 2014;4:4999. doi: 10.1038/ncomms5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glubb DM, Maranian MJ, Michailidou K, Pooley KA, Meyer KB, Kar S, Carlebur S, O’Reilly M, Betts JA, Hillman KM, et al. Fine-scale mapping of the 5q11.2 breast cancer locus reveals at least three independent risk variants regulating MAP3K1. Am J Hum Genet. 2015;96:5–20. doi: 10.1016/j.ajhg.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough LL, Fan J, Chu S, Winnick S, Beck KA. Golgi localization of Syne-1. Mol Biol Cell. 2003;14:2410–2424. doi: 10.1091/mbc.E02-07-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein R, Maranian M, Hopper JL, Kapuscinski MK, Southey MC, Park DJ, Schmidt MK, Broeks A, Hogervorst FB, Bueno-de-Mesquita HB, et al. Comparison of 6q25 breast cancer hits from Asian and European Genome Wide Association Studies in the Breast Cancer Association Consortium (BCAC) PLoS One. 2012;7:e42380. doi: 10.1371/journal.pone.0042380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto I, Fujino T, Douchi T, Nagata Y. Association of estrogen receptor alpha and beta3-adrenergic receptor polymorphisms with endometrial cancer. Obstet Gynecol. 2003;102:506–511. doi: 10.1016/s0029-7844(03)00578-7. [DOI] [PubMed] [Google Scholar]
- Kaaks R, Lukanova A, Kurzer MS. Obesity, endogenous hormones, and endometrial cancer risk: a synthetic review. Cancer Epidemiol Biomarkers Prev. 2002;11:1531–1543. [PubMed] [Google Scholar]
- Kanaya T, Kyo S, Maida Y, Yatabe N, Tanaka M, Nakamura M, Inoue M. Frequent hypermethylation of MLH1 promoter in normal endometrium of patients with endometrial cancers. Oncogene. 2003;22:2352–2360. doi: 10.1038/sj.onc.1206365. [DOI] [PubMed] [Google Scholar]
- Key TJ, Pike MC. The dose-effect relationship between ‘unopposed’ oestrogens and endometrial mitotic rate: its central role in explaining and predicting endometrial cancer risk. Br J Cancer. 1988;57:205–212. doi: 10.1038/bjc.1988.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradpour P, Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014;42:2976–2987. doi: 10.1093/nar/gkt1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch C, Spillman MA, Jackson TA, Jacobsen BM, Murphy SK, Lessey BA, Jordan VC, Bradford AP. DNA profiling analysis of endometrial and ovarian cell lines reveals misidentification, redundancy and contamination. Gynecol Oncol. 2012;127:241–248. doi: 10.1016/j.ygyno.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Xiang YB, Courtney R, Cheng JR, Huang B, Long JR, Cai H, Zheng W, Shu XO, Cai Q. Association of a single nucleotide polymorphism at 6q25.1,rs2046210, with endometrial cancer risk among Chinese women. Chin J Cancer. 2011;30:138–143. doi: 10.5732/cjc.010.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Seo JH, Stranger B, McKenna A, Pe’er I, Laframboise T, Brown M, Tyekucheva S, Freedman ML. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152:633–641. doi: 10.1016/j.cell.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macintyre G, Bailey J, Haviv I, Kowalczyk A. is-rSNP: a novel technique for in silico regulatory SNP detection. Bioinformatics. 2010;26:i524–530. doi: 10.1093/bioinformatics/btq378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- Marme A, Zimmermann HP, Moldenhauer G, Schorpp-Kistner M, Muller C, Keberlein O, Giersch A, Kretschmer J, Seib B, Spiess E, et al. Loss of Drop1 expression already at early tumor stages in a wide range of human carcinomas. Int J Cancer. 2008;123:2048–2056. doi: 10.1002/ijc.23763. [DOI] [PubMed] [Google Scholar]
- Painter JN, O’Mara TA, Batra J, Cheng T, Lose FA, Dennis J, Michailidou K, Tyrer JP, Ahmed S, Ferguson K, et al. Fine-mapping of the HNF1B multicancer locus identifies candidate variants that mediate endometrial cancer risk. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JR, Day F, Elks CE, Sulem P, Thompson DJ, Ferreira T, He C, Chasman DI, Esko T, Thorleifsson G, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Tanaka Y, Kaneuchi M, Sakuragi N, Dahiya R. Polymorphisms of estrogen receptor alpha gene in endometrial cancer. Biochem Biophys Res Commun. 2002;297:558–564. doi: 10.1016/s0006-291x(02)02248-9. [DOI] [PubMed] [Google Scholar]
- Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, Chan TA, Van Neste L, Van Criekinge W, van den Bosch S, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer G. meta: Meta-Analysis with R., edn R package 1.6–1 2010 [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Sliwinski T, Sitarek P, Stetkiewicz T, Sobczuk A, Blasiak J. Polymorphism of the ERalpha and CYP1B1 genes in endometrial cancer in a Polish subpopulation. J Obstet Gynaecol Res. 2010;36:311–317. doi: 10.1111/j.1447-0756.2009.01143.x. [DOI] [PubMed] [Google Scholar]
- Spurdle AB, Thompson DJ, Ahmed S, Ferguson K, Healey CS, O’Mara T, Walker LC, Montgomery SB, Dermitzakis ET, et al. Australian National Endometrial Cancer Study G. Genome-wide association study identifies a common variant associated with risk of endometrial cancer. Nat Genet. 2011;43:451–454. doi: 10.1038/ng.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur I, Neumann S, Noegel AA. Nesprin-1 role in DNA damage response. Nucleus. 2014;5:173–191. doi: 10.4161/nucl.29023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessema M, Willink R, Do K, Yu YY, Yu W, Machida EO, Brock M, Van Neste L, Stidley CA, Baylin SB, et al. Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23-25. Cancer Res. 2008;68:1707–1714. doi: 10.1158/0008-5472.CAN-07-6325. [DOI] [PubMed] [Google Scholar]
- Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, Seal S, Ghoussaini M, Hines S, Healey CS, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedren S, Lovmar L, Humphreys K, Magnusson C, Melhus H, Syvanen AC, Kindmark A, Landegren U, Fermer ML, Stiger F, et al. Estrogen receptor alpha gene polymorphism and endometrial cancer risk--a case-control study. BMC Cancer. 2008;8:322. doi: 10.1186/1471-2407-8-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiderpass E, Persson I, Melhus H, Wedren S, Kindmark A, Baron JA. Estrogen receptor alpha gene polymorphisms and endometrial cancer risk. Carcinogenesis. 2000;21:623–627. doi: 10.1093/carcin/21.4.623. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Skepper JN, Yang F, Davies JD, Hegyi L, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J Cell Sci. 2001;114:4485–4498. doi: 10.1242/jcs.114.24.4485. [DOI] [PubMed] [Google Scholar]
- Zheng W, Long J, Gao YT, Li C, Zheng Y, Xiang YB, Wen W, Levy S, Deming SL, Haines JL, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.