Abstract
The association between the G894T polymorphism (Glu298Asp) of nitric oxide synthase 3 (NOS3) and risk of Alzheimer’s disease (AD) was explored by performing a meta-analysis of case-control studies. Bibliographical searches were conducted in the MEDLINE, EMBASE, and China National Knowledge Infrastructure (CNKI) databases without any language limitations. Two investigators independently assessed abstracts for relevant studies, and reviewed all eligible studies. We adopted regrouping in accordance with the most probably appropriate genetic model. Odds ratios (ORs) with 95% confidence intervals (CIs) were used to assess the strength of this association. We performed a meta-analysis including 21 published articles with 23 case-control studies (5,670 cases and 5,046 controls). In the analyses, we found significant association between G894T polymorphism and AD risk under a complete overdominant model (GG + TT vs. GT) (OR = 1.18; 95%CI, 1.04–1.35; P = 0.010). When stratified by time of AD onset, we found the association between this polymorphism and AD susceptibility to be more substantial among late onset patients than among early onset patients (OR for late vs. early onset: 1.33 vs. 1.02, P interaction = 0.049). The meta-analysis showed that the polymorphism G894T of NOS3 was associated with risk of AD.
Alzheimer’s disease (AD), also called senile dementia of the Alzheimer type or primary degenerative dementia of the Alzheimer’s type, is a degenerative disease of the central nervous system characterized by a gradual decline in memory and cognition, which has been correlated with synaptic dysfunction and loss, and eventually to neuronal death1. According to data from the World Alzheimer Report, the number of people with AD is forecast to nearly double every 20 years from 36 million in 2010 to 115 million in 2050, and the costs associated with AD reached the total of US$604 billion, about 1% of global GDP2. Therefore, it is particularly urgent to gain an insight into the pathogenesis factors of the AD in order to discover different possibilities of preventive and effective treatment.
Although it was not until 1901 that the first case with AD was identified in a fifty-year-old woman by German psychiatrist Alois Alzheimer3, its pathological cause, like many other mental diseases, is unclear so far. Recently, it is reported that oxidative stress caused by nitric oxide (NO) may play a pathogenetic role in AD. The deposits of β-Amyloid (Aβ) in AD brains can lead to the production of superoxide radicals. These radicals combined with NO in endothelial cells to form peroxynitrite, which in turn induced cellular injury and caused lipid peroxidation to further accelerate neurodegenerative changes leading to AD incurrence4. NO in endothelial cells mainly was synthesized from L-arginine by NOS3. Therefore, NOS3 may be a pivotal protein involved in a complex cascade of events in the oxidative stress.
NOS3, gene encoding for nitric oxide synthase 3, is located on chromosome 7q355 and has been widely studied in several AD populations. Dahiyat et al.6 first found a significant effect of the G allele and the GG genotype of NOS3 G894T polymorphism on AD development. Subsequently, this association has been studied in many studies7,8, but with inconsistent findings. Therefore, we conducted a meta-analysis of the existing epidemiologic studies by using a comprehensive search strategy to determine whether there was an association between G894T polymorphism of NOS3 and risk of AD.
Material and Methods
Study selection
Methodology advocated by the MOOSE guideline was followed to perform this meta-analysis9. We conducted a systematically electronic search using the following terms “(Alzheimer* or AD) and (NOS* or nitric oxide synthase*) and (polymorphism* or genotype* or variant*)” in the PubMed database (from 1966 to May, 2015). We subsequently repeated this search in the EMBASE, CNKI (http://www.cnki.net) and GOOGLE Scholar (http://scholar.google.com/). Reference lists of relevant articles were reviewed manually to search for more studies.
For inclusion, studies included in the meta-analysis had to meet the following criteria: (1) studies were designed as the case–control type; (2) reported outcomes included AD; and (3) polymorphism included G894T (Glu298Asp) polymorphism. Studies were excluded if: (1) no detailed genotype frequency; and (2) insufficient information for data extraction. When multiple publications from the same patient population resource or overlapping data sets were available, only the most recent or largest sample size study was included in the meta-analysis.
Data extraction
The citations (titles and abstracts) search and data extraction were carried out independently by two reviewers, and disagreements were resolved by consensus. The following information was collected in a predefined data collection form: the first author’s name, year of publication, country of origin, ethnicity, AD diagnosis method, source of controls, proportion of men in cases and controls, total number of cases and controls, mean (range) age of cases and controls, time of AD onset, and number of cases and controls with different genotypes. The quality of each study selected for inclusion in the meta-analysis was assessed by quality assessment criteria, which was derived from previously published meta-analysis of molecular association studies10. Total scores range from 0 to 15, with 15 being the maximum. Quality was assigned as optimal with 10–15 points and suboptimal with 0–9 points11.
Quality scores of studies ranged from 0 (lowest) to 15 (highest). Studies with scores ≤9 were categorized into low quality, while those with scores >9 were considered as high quality11.
Genotype based mRNA expression analysis
We used the data of NOS3 G894T genotypes from the International HapMap Project (http://hapmap.ncbi.nlm.nih.gov/) for the 209 subjects, and their corresponding mRNA expression levels data were available from SNPexp (http://app3.titan.uio.no/biotools/help.php?app=snpexp) as described previously12,13.
Statistical analysis
We used crude Odds ratios (ORs) with their 95% confidence intervals (CIs) to assess the strength of association between G894T polymorphism and AD risks. OR1, OR2, and OR3 were calculated for the genotypes GG vs. TT, GT vs. TT, and GG vs. GT for G894T polymorphism, respectively. The most appropriate genetic model was determined by the above pairwise differences: If OR1 = OR3 ≠ 1 and OR2 = 1, then a recessive model (GG vs. GT + TT) is suggested; If OR1 = OR2 ≠ 1 and OR3 = 1, then a dominant model (GG + GT vs. TT) is suggested; If OR2 = 1/OR3 ≠ 1 and OR1 = 1, then a complete overdominant model (GG + TT vs. GT) is suggested; If OR1 > OR2 > 1 and OR1 > OR3 > 1 (or OR1 < OR2 < 1 and OR1 < OR3 < 1), then a codominant model (GG vs. GT vs. TT) is suggested14. We also performed subgroup analysis according to time of AD onset, APOE ε4 polymorphism, ethnicity, HWE in controls, control population source, year of publication, and quality score, respectively.
The departure of frequencies in controls from expectation under Hardy–Weinberg equilibrium (HWE) was assessed by chi-square test. We calculated the false-positive report probability (FPRP) to evaluate the significant findings as described previously15. 0.2 was set as an FPRP threshold and assigned a prior probability of 0.1 to detect an odds ratio (OR) of 0.67/1.50 (protective/risk effects) for an association with genotypes under investigation. An FPRP value <0.2 was considered as a noteworthy finding. Sensitivity analysis for the overall effect was performed by influence-analyses to evaluate whether a single or more studies might markedly affect the results16. Statistical heterogeneity between studies was determined by the Q-test and P < 0.05 was considered statistically significant. With lack of heterogeneity among studies, the pooled OR estimate was merged by the fixed effects model17. Otherwise, the random effects model was applied18. Publication bias was investigated with Egger’s and Begg’s regression asymmetry test and funnel plot19,20. We used independent t-test to test the differences of mRNA expression between genotypes. All statistical analyses were completed using Stata Version 11.0 (College Station, TX, USA).
Results
Studies characteristics
A total of 142 articles were achieved by literature search from the Pubmed, Embase, CNKI and other searching methods, using different combinations of key words (Fig. 1). 23 case-control studies from 21 publications2,6,7,8,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37 including 5,670 cases and 5,046 controls were used to evaluate the association of G894T polymorphism with AD risk (Table 1). Sample sizes ranged from 102 to 1,369 (median 365). 17 studies included populations of Caucasian, and 6 of Asian. The mean ages at study for cases and controls were 71.4 years and 73.7 years, and the mean ages at onset of AD cases were 75.4 years. Fewer men were observed for all individual studies (percentages of men range from 25.6% to 100%, median was 38.4%). The criteria for AD diagnosis for 17 studies was the National Institute of Neurological Disorders and Stoke–Alzheimer Diseases and Related Disorders Association criteria (NINCDS/ADRDA criteria), 1 was used the fourth Diagnostic and Statistical Manual of Mental Disorders criteria (DSM-III-R criteria), and 5 for both. 9 studies only included late-onset AD cases, 2 for early-onset, and 12 for both. For most studies (n = 19), controls came from population-based settings, and 4 studies from hospital-based. The control group in 5 studies was not in HWE (Table 2).
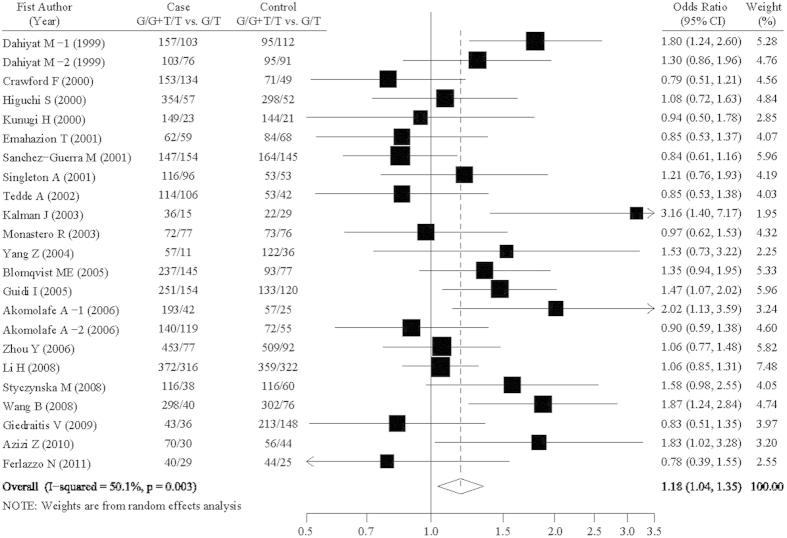
Figure 1. Flow chart of selection studies in our meta-analysis.
Table 1. Study characteristics from included studies in the meta-analysis.
| First author | Year | Country | Ethnicity | Cases |
Controls |
Criteria for AD diagnosis | Time of AD onset | Control type | Quality score | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Na | Ageb | Agec | Genderd | N | Ageb | Genderd | ||||||||
| Dahiyat M -1 | 1999 | UK | Caucasian | 260 | 57.0 | 79.8 | 35.6 | 207 | 71.0 | 47.1 | NINCDS/ADRDA | Mixed | PB | 10 |
| Dahiyat M -2 | 1999 | UK | Caucasian | 179 | 74.0 | 82.0 | 51.4 | 186 | 77.0 | 50.0 | NINCDS/ADRDA | Late | PB | 10 |
| Crawford F | 2000 | USA | Caucasian | 287 | 73.0 | — | 33.0 | 120 | 73.3 | 42.0 | NINCDS/ADRDA | Late | PB | 8 |
| Higuchi S | 2000 | Japan | Asian | 411 | — | 76.0 | 27.3 | 350 | 75.7 | 27.7 | NINCDS/ADRDA | Mixed | PB | 10 |
| Kunugi H | 2000 | Japan | Asian | 172 | — | 74.1 | 34.3 | 165 | 57.0 | 41.2 | NINCDS/ADRDA | Mixed | HB | 6 |
| Emahazion T | 2001 | Scotland | Caucasian | 121 | — | — | — | 152 | — | — | NINCDS/ADRDA | Early | PB | 11 |
| Sanchez-Guerra M | 2001 | Spain | Caucasian | 301 | 71.5 | 75.2 | 34.0 | 309 | 80.3 | 29.0 | NINCDS/ADRDA | Mixed | HB | 7 |
| Singleton A | 2001 | UK | Caucasian | 212 | 79.8 | — | 33.3 | 106 | 78.2 | 47.6 | NINCDS/ADRDA | Late | PB | 9 |
| Tedde A | 2002 | Italy | Caucasian | 220 | 65.0 | — | 36.4 | 95 | 80.8 | 42.1 | DSM-IV | Mixed | PB | 9 |
| Kalman J | 2003 | Hungary | Caucasian | 51 | — | 76.0 | 31.4 | 51 | 72.0 | 47.1 | NINCDS/ADRDA DSM-III | Late | PB | 7 |
| Monastero R | 2003 | Italy | Caucasian | 149 | 69.7 | 72.0 | 38.5 | 149 | 73.3 | 38.5 | NINCDS/ADRDA | Mixed | PB | 10 |
| Yang Z | 2004 | China | Asian | 68 | 72.8 | — | 38.2 | 158 | 71.2 | 50.0 | NINCDS/ADRDA DSM-III | Late | PB | 11 |
| Blomqvist ME | 2005 | Sweden | Caucasian | 382 | 76.2 | — | 37.1 | 170 | 73.2 | 46.7 | NINCDS/ADRDA | Mixed | HB | 7 |
| Guidi I | 2005 | Italy | Caucasian | 405 | 73.0 | — | 29.6 | 253 | 71.0 | 42.3 | NINCDS/ADRDA | Mixed | HB | 8 |
| Akomolafe A -1 | 2006 | USA | Caucasian | 235 | — | 71.4 | 24.0 | 82 | 72.3 | 30.0 | NINCDS/ADRDA | Mixed | PB | 13 |
| Akomolafe A -2 | 2006 | Mixede | Caucasian | 259 | — | 68.3 | 34.0 | 127 | 71.7 | 59.0 | NINCDS/ADRDA | Mixed | PB | 10 |
| Zhou Y | 2006 | China | Asian | 530 | — | 71.3 | 63.0 | 601 | 71.5 | 55.9 | NINCDS/ADRDA DSM-IV | Mixed | PB | 9 |
| Li H | 2008 | Canada | Caucasian | 688 | 77.8 | 71.9 | 42.4 | 681 | 73.4 | 35.6 | NINCDS/ADRDA | Mixed | PB | 15 |
| Styczynska M | 2008 | Poland | Caucasian | 154 | — | 71.5 | 33.8 | 176 | 72.7 | 31.8 | NINCDS/ADRDA | Late | PB | 11 |
| Wang B | 2008 | China | Asian | 338 | 77.6 | — | 51.0 | 378 | 72.9 | 58.0 | NINCDS/ADRDA DSM-III | Late | PB | 11 |
| Giedraitis V | 2009 | Sweden | Caucasian | 79 | — | 80.2 | 100 | 361 | 81.8 | 100 | NINCDS/ADRDA DSM- IV | Late | PB | 10 |
| Azizi Z | 2010 | Iran | Asian | 100 | 73.8 | — | 44.0 | 100 | 72.6 | 48.0 | NINCDS/ADRDA | Late | PB | 9 |
| Ferlazzo N | 2011 | Italy | Caucasian | 69 | 77.2 | 42.0 | 69 | 75.6 | 34.8 | NINCDS/ADRDA | Early | PB | 10 | |
DSM: the Diagnostic and Statistical Manual of Mental Disorders; NINCDS: the National Institute of Neurological Disorders and Stoke; ADRDA: Alzheimer Diseases and Related Disorders Association; MMSE: mini-mental state examination; NA: not applicable; SNP: Single Nucleotide Polymorphism; PB = population based control; HB = hospital based control.
aNumber.
bage at survey.
cage at onset of Alzheimer’s disease.
dpercentage of male.
eparticipants from four countries (Canada, Germany and Greece).
Table 2. NOS3 G894T genotype distribution among AD cases and controls in the included studies.
| First author | Cases |
Controls |
Pa (HWE) | ||||
|---|---|---|---|---|---|---|---|
| G/G | G/T | T/T | G/G | G/T | T/T | ||
| Dahiyat M -1 | 136 | 103 | 21 | 80 | 112 | 15 | 0.004 |
| Dahiyat M -2 | 95 | 76 | 8 | 74 | 91 | 21 | 0.376 |
| Crawford F | 129 | 134 | 24 | 61 | 49 | 10 | 0.971 |
| Higuchi S | 350 | 57 | 4 | 297 | 52 | 1 | 0.416 |
| Emahazion T | 48 | 59 | 14 | 70 | 68 | 14 | 0.664 |
| Kalman J | 30 | 15 | 6 | 22 | 29 | 0 | 0.005 |
| Kunugi H | 149 | 23 | 0 | 143 | 21 | 1 | 0.812 |
| Monastero R | 62 | 77 | 10 | 65 | 76 | 8 | 0.017 |
| Sanchez-Guerra M | 97 | 154 | 50 | 101 | 145 | 63 | 0.408 |
| Singleton A | 88 | 96 | 28 | 43 | 53 | 10 | 0.269 |
| Tedde A | 91 | 106 | 23 | 38 | 42 | 15 | 0.554 |
| Akomolafe A -1 | 193 | 42 | 0 | 56 | 25 | 1 | 0.326 |
| Akomolafe A -2 | 109 | 119 | 31 | 61 | 55 | 11 | 0.778 |
| Blomqvist ME | 199 | 145 | 38 | 81 | 77 | 12 | 0.270 |
| Guidi I | 210 | 154 | 41 | 110 | 120 | 23 | 0.228 |
| Yang Z | 56 | 11 | 1 | 121 | 36 | 1 | 0.334 |
| Zhou Y | 441 | 77 | 12 | 495 | 92 | 14 | <0.001 |
| Li H | 285 | 316 | 87 | 292 | 322 | 67 | 0.108 |
| Styczynska M | 106 | 38 | 10 | 100 | 60 | 16 | 0.120 |
| Wang B | 296 | 40 | 2 | 299 | 76 | 3 | 0.441 |
| Azizi Z | 67 | 30 | 3 | 54 | 44 | 2 | 0.039 |
| Ferlazzo N | 28 | 29 | 12 | 33 | 25 | 11 | 0.108 |
| Giedraitis V | 37 | 36 | 6 | 183 | 148 | 30 | 0.992 |
HWE: Hardy-Weinberg equilibrium.
aP value for HWE test in controls.
Quantitative synthesis
The OR1, OR2, and OR3 were 0.99 (95% CI = 0.83–1.19; P = 0.901), 0.91 (95% CI = 0.76–1.08; P = 0.289), and 1.19 (95% CI = 1.04–1.36; P = 0.011), respectively, suggesting a complete overdominant effect of the putative susceptibility allele T. Thus, in accordance with a complete overdominant model, the original grouping was collapsed, and GG and TT were merged in one group compared with the GT genotype group. The pooled odds ratios and 95% confidence intervals for the associations between G894T polymorphism and AD susceptibility was 1.18 (1.04–1.35; P = 0.010) under a complete overdominant model (GG + TT vs. GT) (Table 3, Fig. 2). For a prior probability of 0.25, the FPRP values was 0.046 with statistical power of 0.597.
Table 3. Total, stratified and sensitivity analysis of NOS3 G894T polymorphism and the AD susceptibility.
| Summary | Na | Cases/Controls | Q Test |
I2,% | Odds Ratio | 95% Confidence Interval | P for Z testc | P for interactiond | ||
|---|---|---|---|---|---|---|---|---|---|---|
| χ2 | df | P Valueb | ||||||||
| All studies | 23 | 5,670/5,046 | 44.07 | 22 | 0.003 | 50.1 | 1.18 | 1.04, 1.35 | 0.010 | - |
| Subgroup analysis | ||||||||||
| Time of AD onset | 0.049 | |||||||||
| Early | 8 | 706/1,525 | 4.17 | 7 | 0.760 | 0.0 | 1.02 | 0.83, 1.26 | 0.834 | |
| Late | 14 | 2,568/2,866 | 37.63 | 13 | <0.001 | 65.5 | 1.33 | 1.07, 1.66 | 0.010 | |
| Ethnic descent | 0.246 | |||||||||
| Caucasiane | 17 | 4,051/3,294 | 35.04 | 16 | 0.004 | 54.3 | 1.15 | 0.99, 1.34 | 0.075 | |
| Asian | 6 | 1,619/1,752 | 7.68 | 5 | 0.175 | 34.9 | 1.31 | 1.03, 1.67 | 0.030 | |
| APOE | 0.925 | |||||||||
| ε4+ | 5 | 347/161 | 5.74 | 4 | 0.220 | 30.3 | 1.25 | 0.82, 1.91 | 0.304 | |
| ε4- | 5 | 560/884 | 9.74 | 4 | 0.045 | 58.9 | 1.22 | 0.95, 1.56 | 0.116 | |
| HWE in controls | 0.065 | |||||||||
| Yes | 18 | 4,580/3,938 | 29.16 | 17 | 0.033 | 41.7 | 1.12 | 1.02, 1.23 | 0.021 | |
| No | 5 | 1,090/1,108 | 11.51 | 4 | 0.021 | 65.3 | 1.48 | 1.04, 2.12 | 0.030 | |
| Control type | 0.912 | |||||||||
| PB | 19 | 4,410/4,149 | 36.99 | 18 | 0.005 | 51.3 | 1.20 | 1.03, 1.39 | 0.018 | |
| HB | 4 | 1,260/897 | 7.07 | 3 | 0.070 | 57.5 | 1.15 | 0.85, 1.54 | 0.367 | |
| Date of publication | 0.139 | |||||||||
| ≤2003 | 11 | 2,363/1,890 | 21.51 | 10 | 0.018 | 53.5 | 1.09 | 0.90, 1.34 | 0.380 | |
| >2003 | 12 | 3,307/3,156 | 20.37 | 11 | 0.040 | 46.0 | 1.27 | 1.08, 1.50 | 0.005 | |
| Quality score | 0.599 | |||||||||
| Low | 10 | 2,660/1,970 | 20.04 | 9 | 0.018 | 55.1 | 1.16 | 0.94, 1.43 | 0.158 | |
| High | 13 | 3,010/3,076 | 23.75 | 12 | 0.022 | 49.5 | 1.21 | 1.02, 1.43 | 0.031 | |
| Sensitivity analyses | — | |||||||||
| Minimal | 22 | −/− | 38.46 | 21 | 0.011 | 45.4 | 1.16 | 1.02, 1.31 | 0.025 | |
| Maximal | 22 | −/− | 39.82 | 21 | 0.008 | 47.3 | 1.21 | 1.06, 1.38 | 0.004 | |
CI: Confidence interval; PB: population-based; HB: hospital-based.
aNumber of comparisons.
bP value of Q-test for between study heterogeneity test.
cP-value of Z-test for significant test.
dP value of Q-test for between sub-group heterogeneity test.
eAfrican descents were excluded from one study by Akomolafe A et al.27.
Figure 2. Forest plot of association between NOS3 G894T polymorphism (G/G+T/T vs. G/T) and AD risk.
When stratified by time of AD onset, we found the association between this polymorphism and AD susceptibility to be more substantial among participants with late onset of AD than among those with early onset (OR for late onset vs. early onset: 1.33 vs. 1.02, P interaction = 0.049) (Table 3). No significant interaction were observed for ethnic descent, APOE ε4 +/−, HWE in controls, control population source, time of publication, and quality score (all P interactions > 0.05).
Sensitivity analysis and bias diagnosis
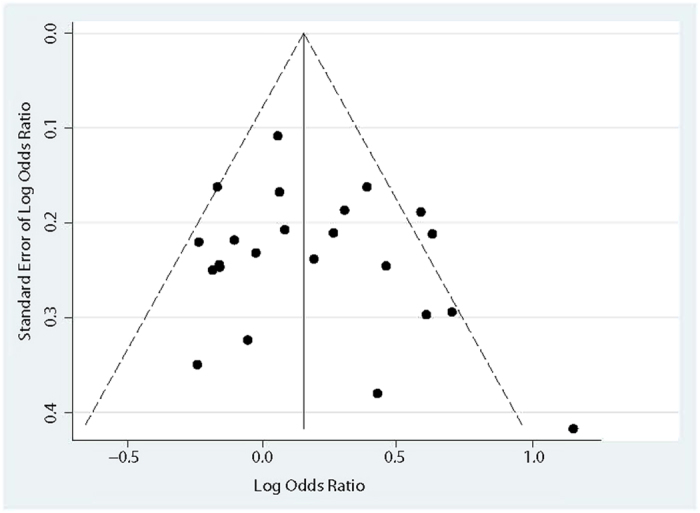
We did sensitivity analysis to explore whether modification of included studies in the meta-analysis could influence the overall effects, and these results were unchanged in terms of magnitude and significance. The OR (95% CI) ranged from 1.16 (1.02, 1.31) to 1.21 (1.06, 1.38), which implied no other single study affected the summary risks qualitatively. The Egger’s, Begg’s test, and funnel plot were performed to access the publication bias. Ultimately, Both Egger’s and Begg’s test revealed that no significant biases existed (P value: 0.335 and 0.597). And the funnel plot also indicated no evidence of publication bias (Fig. 3).
Figure 3. Funnel plot of association between NOS3 G894T polymorphism (G/G+T/T vs. G/T) and the risk of AD risk.
The mRNA expression by NOS3 G894T genotypes
The difference of mRNA expressions levels between genotypes were explored for three ethnic groups (i.e., CEU, YRI and Asian) and the total sample (Table 4). Significant higher mRNA expression was observed for individuals with GG genotype than those with GT genotype among YRIs (P = 0.007). No significant alteration in the mRNA expression levels was found for Caucasian, Asians, and total sample.
Table 4. mRNA expression by the NOS3 G894T genotypes, using data from the HapMapa.
| Population | G894T genotypesb | No. | Mean ± SD | Pc |
|---|---|---|---|---|
| CEU | GG+TT | 29 | 6.23±0.082 | 0.421 |
| GT | 31 | 6.24±0.075 | ||
| YRI | GG | 52 | 6.16±0.092 | 0.007 |
| GT | 8 | 6.20±0.031 | ||
| Asian | GG | 73 | 6.10±0.072 | 0.884 |
| GT | 16 | 6.07±0.070 | ||
| ALL | GG+TT | 154 | 6.15±0.094 | 0.603 |
| GT | 55 | 6.18±0.100 |
aGenotyping data and mRNA expression levels for MTHFR by genotypes were obtained from the HapMap phase II release 28 data from EBV-transformed lymphoblastoid cell lines from 209 individuals.
bOnly 5 cases among CEU were detected with TT genotype.
cP value of independent t-test for between sub-group heterogeneity.
Discussion
Our meta-analysis included a total of 5,670 AD case and 5,046 control subjects from 23 published case control studies for the G894T polymorphism. The results from the meta-analysis showed that there was a significant association between the polymorphism of NOS3 and risk of AD (overdominant model: P < 0.001, OR = 1.18, 95%CI = 1.08–1.29), which contrasts with the results from previous meta-analyses38,39 that observed a null association between the G894T polymorphism and AD risk. Several reasons may explain the different results: First, our meta-analysis included more case-control studies than the previous meta-analysis, thus our study was more powerful and the conclusion may be more reliable. Second, we selected an optimal genetic model to illustrate the inheritance in the complex disease gene by adopting an effective statistical method that did not a priori assume a genetic model and avoided the problem of multiple comparisons.
The production of basal vascular wall NO is mainly determined by the key NOS 3 enzyme40. NO may react with reactive oxygen species to produce peroxynitrate, which can cause oxidative stress associated with neurodegenerative diseases including Alzheimer’s disease41. Therefore, NOS 3 enzyme involved in oxidative stress plays an important role in the pathophysiology of AD development. Evidence has been obtained showing that the G894T polymorphism in the NOS3 influences NOS3 activity and basal NO production42,43. Increased expression of NOS3 results in altered mitochondrial function in neurons44. It may be explained that the polymorphism is involved in AD development through modifying the expression of NOS 3 that caused the excessive production of NO.
However, it might be argued that the identified association between NOS3 G894T polymorphism and AD risk is due to heterogeneity. Therefore, subgroup analyses were performed to explore the stability of the association.
It is well established that APOE є4 plays an important role in the pathogenic mechanism of AD by regulating the formation of Aβ and the APOE є4 is the only established genetic risk factor for AD45. Only 10 studies performed the stratification analysis to evaluate the association between NOS3 G894T polymorphism and AD risk by the APOE є4 status in the included studies. When stratified by APOE є4 status, the results from stratification analyses showed that the significant differences in genotype (overdominant model) of the polymorphism in the APOE є4(+) sample and the APOE є4(−) sample were disappeared between case group and the control group. This negative result may be because that few studies stratified by APOE є4 status were included in the meta-analysis or APOE є4 actually exerted an effect on association between NOS3 and AD. Subsequently, we performed a comparison on risk of NOS3 G894T polymorphism for AD development between APOE є4(+) group and APOE є4(−) group to explore the potential effect of APOE є4 status. However, the statistical difference for risk comparison between the two subgroup (P=0.925) was not found, which excluded the possibility that APOE є4 status exerted an effect on association between NOS3 and AD, and speculated that the negative result may be yielded by lowed power of few included studies.
Age is very important factors for AD development. Most often, AD is diagnosed in people over 65 years of age, and every five years after the age of 65, the risk of the disease incurrence approximately doubles46. Moreover, some authors also reported that AD mainly affected 10% of the population with a predilection for more than 65 years6. Results from the stratification analysis showed that the AD risk was associated with late onset with a significant overdominant model (OR = 1.33, 95%CI = 1.07, 1.66, P = 0.01). Further, the findings from heterogeneity analysis showed that there was a statistical difference among age groups (P = 0.049). Therefore, the NOS3 G894T polymorphism may be age-dependently associated with AD risk. In fact, a possible explanation for the age-dependent association could be the different plasma homocysteine level, an important factor in AD pathogenesis, as the onset time of AD was affected by the heterologous amino acid47. The findings from Italian study demonstrated that plasma homocysteine levels were significantly increased in late-onset AD compared with early-onset AD29. Moreover, plasma homocysteine levels were closely associated with NOS3 genotype29. Therefore, the mechanism by which the different genotypes may affect plasma homocysteine levels may explain the age-dependent association.
The different ethnic background is a well-known confounding factor in genetic studies. Variations in the frequency of NOS3 T allele among different ethnic groups have been reported. Evidence has been shown that NOS3 T allele was more common in Whites than in African Americans or Asians48. The results from our study showed that distribution frequency of the NOS3 T allele was 42.71–70.81% in Caucasian populations, which was lower than that in Asian populations (69.24–92.56%). However, the statistical difference was not found in Caucasian populations with an overdominant model (OR = 1.15, 95%CI = 0.99–1.34, P = 0.075) and in Asian populations with an overdominant model (OR = 1.31, 95% = 1.03–1.67, P = 0.03) for AD risk in the stratified analysis by ethnicity (P = 0.246).
There are some limitations: (i) we had no ability to ascertain whether studies included in our review had an adequate sample size. The Genetic Power Calculation often is a measure to evaluate the sample size in genetic analysis on association between polymorphisms and diseases while no study reported an a priori sample size calculation in the included studies. Inadequate choice of sample size may lead to chance and exaggerate (or dilute) the association between NOS3 and AD. (ii) It is a pity that the stratification by gender could not be performed in the meta-analysis because of unavailable information in the included case control studies. It is well known that the gender is as an important factor as the age, and gender-dependent genetic effects on risk of AD have been reported in many studies. Kim et al. reported that a significant increased risk of AD was found in female subjects (OR = 4.4, 95%CI = 2.4–8.0) with the APOE є4 variant genotypes while not in male subjects (OR = 0.6, 95%CI = 0.1–2.2)49. In addition, Zou et al. showed the rs688T/T genotype of LDLR gene was associated with increased AD odds in males (recessive model, OR = 1.49, 95%CI = 1.13–1.97, uncorrected p = 0.005), but not in females50. Therefore, it needs to explore the effect of gender on association between NOS3 and AD in the future studies.
There are some strengths: (i) the association on the NOS3 polymorphism and AD has been reported in many studies; however, because of limitation of sample size, the power may be lower to assess the NOS3 G894T polymorphism and AD risk. Therefore, the meta-analysis is a good measure to explicitly explore the effect of NOS3 G894T polymorphism on the AD incurrence. (ii) we assessed studies comparing the genotype and allele distribution of NOS3 G894T polymorphism in case group with these in various control types separately because individual control types may address different questions. Hospital-based population as a control is intended to be compared with the study group as a convenience sample. However, the limitation of the hospital-based population is inability to generalize the results to other populations and yield the selection bias. In our meta-analysis, population-based control type was used in 19 of 23 studies (83%) and a significantly statistical difference was found in the subgroup analysis (P = 0.018), which suggested the ability to generalize the results to other populations. Thus, the result has a robust stability to support the association. (iii) compared to other meta-analyses, more studies with larger sample size were included to make the result powerful and an effective method for meta-analysis of molecular association studies was adopted to pool data in a way that reflects the biology of gene effects and genetic models in our study.
In conclusion, result from our meta-analysis might provide evidence that the G894T polymorphism of NOS3 is associated with AD susceptibility, especially late onset AD susceptibility. The findings from the meta-analysis on the genetic polymorphism of NOS3 G894T may be used to potentially evaluate individual susceptibility and explore the effective measures of control and prevention for AD.
Additional Information
How to cite this article: Liu, S. et al. The nitric oxide synthase 3 G894T polymorphism associated with Alzheimer's disease risk: a meta-analysis. Sci. Rep. 5, 13598; doi: 10.1038/srep13598 (2015).
Acknowledgments
This work was supported by funding from the National Nature Science Foundation of China (grant number: 81302482), China Postdoctoral Science Foundation funded project (grant number 2013M531879), Medical Scientific Research Foundation of Guangdong Province (grant number: A2015387) and Nanshan Science and Technology Bureau Fund project (grant number: 2012007).
Footnotes
Author Contributions Dr. L.K.S. and Z.B. had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: L.S.Y., Z.F.F., L.K.S. and Z.B. Acquisition of data: L.S.Y., Z.F.F., W.C.Y. and C.Z.W. Analysis and interpretation of data: L.S.Y., Z.F.F., L.K.S. and Z.B. Drafting of the manuscript: L.S.Y., Z.F.F., L.K.S. and Z.B. Critical revision of the manuscript for important intellectual content: L.S.Y., Z.F.F., W.C.Y., C.Z.W., L.K.S. and Z.B. Statistical analysis: L.S.Y., Z.F.F. and W.C.Y. Administrative, technical, and material support: L.S.Y., Z.F.F., L.K.S. and Z.B.
References
- Querfurth H. W. & LaFerla F. M. Alzheimer’s disease. N Engl J Med 362, 329–344 (2010). [DOI] [PubMed] [Google Scholar]
- Wimo A. & Prince M. Alzheimer’s disease International. World Alzheimer Report 2009-2011. Available from: http://www.alz.co.uk/research/world-report, accessed on 11/02/2015 (2011).
- Berchtold N. C. & Cotman C. W. Evolution in the conceptualization of dementia and Alzheimer’s disease: Greco-Roman period to the 1960s. Neurobiol Aging 19, 173–189 (1998). [DOI] [PubMed] [Google Scholar]
- Galbusera C. et al. Increased susceptibility to plasma lipid peroxidation in Alzheimer disease patients. Curr Alzheimer Res 1, 103–109 (2004). [DOI] [PubMed] [Google Scholar]
- Marsden P. A. et al. Structure and chromosomal localization of the human constitutive endothelial nitric oxide synthase gene. J Biol Chem 268, 17478–17488 (1993). [PubMed] [Google Scholar]
- Dahiyat M. et al. Association between Alzheimer’s disease and the NOS3 gene. Ann Neurol 46, 664–667 (1999). [DOI] [PubMed] [Google Scholar]
- Crawford F. et al. No association between the NOS3 codon 298 polymorphism and Alzheimer’s disease in a sample from the United States. Ann Neurol 47, 687 (2000). [PubMed] [Google Scholar]
- Higuchi S. et al. NOS3 polymorphism not associated with Alzheimer’s disease in Japanese. Ann Neurol 48, 685 (2000). [PubMed] [Google Scholar]
- Stroup D. F. et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 283, 2008–2012 (2000). [DOI] [PubMed] [Google Scholar]
- Thakkinstian A. et al. Systematic review and meta-analysis of the association between complement component 3 and age-related macular degeneration: a HuGE review and meta-analysis. Am J Epidemiol 173, 1365–1379 (2011). [DOI] [PubMed] [Google Scholar]
- He J. et al. Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: evidence from a meta-analysis. Sci Rep 4, 6159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm K., Melum E., Franke A. & Karlsen T. H. SNPexp - A web tool for calculating and visualizing correlation between HapMap genotypes and gene expression levels. BMC Bioinformatics 11, 600 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J. et al. Associations of Lys939Gln and Ala499Val polymorphisms of the XPC gene with cancer susceptibility: a meta-analysis. Int J Cancer 133, 1765–1775 (2013). [DOI] [PubMed] [Google Scholar]
- Thakkinstian A., McElduff P., D’Este C., Duffy D. & Attia J. A method for meta-analysis of molecular association studies. Stat Med 24, 1291–1306 (2005). [DOI] [PubMed] [Google Scholar]
- He J. et al. Genetic variations of mTORC1 genes and risk of gastric cancer in an Eastern Chinese population. Mol Carcinog 52 Suppl 1, E70–79 (2013). [DOI] [PubMed] [Google Scholar]
- Tobias A. Assessing the influence of a single study in the meta-analysis estimate. Stata Tech Bull 8, 15–17 (1999). [Google Scholar]
- Mantel N. & Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22, 719–748 (1959). [PubMed] [Google Scholar]
- DerSimonian R. & Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials 28, 105–114 (2007). [DOI] [PubMed] [Google Scholar]
- Begg C. B. & Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101 (1994). [PubMed] [Google Scholar]
- Egger M., Davey Smith G., Schneider M. & Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emahazion T. et al. SNP association studies in Alzheimer’s disease highlight problems for complex disease analysis. Trends Genet 17, 407–413 (2001). [DOI] [PubMed] [Google Scholar]
- Kalman J. et al. The nitric oxide synthase-3 codon 298 polymorphism is not associated with late-onset sporadic Alzheimer’s dementia and Lewy body disease in a sample from Hungary. Psychiatr Genet 13, 201–204 (2003). [DOI] [PubMed] [Google Scholar]
- Kunugi H. et al. No evidence for an association between the Glu298Asp polymorphism of the NOS3 gene and Alzheimer’s disease. J Neural Transm 107, 1081–1084 (2000). [DOI] [PubMed] [Google Scholar]
- Monastero R. et al. No association between Glu298Asp endothelial nitric oxide synthase polymorphism and Italian sporadic Alzheimer’s disease. Neurosci Lett 341, 229–232 (2003). [DOI] [PubMed] [Google Scholar]
- Singleton A. B. et al. Nitric oxide synthase gene polymorphisms in Alzheimer’s disease and dementia with Lewy bodies. Neurosci Lett 303, 33–36 (2001). [DOI] [PubMed] [Google Scholar]
- Tedde A., Nacmias B., Cellini E., Bagnoli S. & Sorbi S. Lack of association between NOS3 poly morphism and Italian sporadic and familial Alzheimer’s disease. J Neurol 249, 110–111 (2002). [DOI] [PubMed] [Google Scholar]
- Akomolafe A. et al. Genetic association between endothelial nitric oxide synthase and Alzheimer disease. Clin Genet 70, 49–56 (2006). [DOI] [PubMed] [Google Scholar]
- Blomqvist M. E. et al. Towards compendia of negative genetic association studies: an example for Alzheimer disease. Hum Genet 119, 29–37 (2006). [DOI] [PubMed] [Google Scholar]
- Guidi I. et al. Influence of the Glu298Asp polymorphism of NOS3 on age at onset and homocysteine levels in AD patients. Neurobiol Aging 26, 789–794 (2005). [DOI] [PubMed] [Google Scholar]
- Yang Z. et al. Association of polymorphism of the endothelial nitric oxide synthase gene with Alzheimer disease. Chin I Geriatr 7, 468–471 (2004). [Google Scholar]
- Zhou Y., Zhang Z., Zhang J., He X. & Xu T. Association Between Nitric Oxide Synthase-III Polymorphism and Alzheimer’s Disease in Chinese Han Population. Chin J Clin Neurosci. 2006 14, 246–250 (2006). [Google Scholar]
- Li H. et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol 65, 45–53 (2008). [DOI] [PubMed] [Google Scholar]
- Styczynska M. et al. Simultaneous analysis of five genetic risk factors in Polish patients with Alzheimer’s disease. Neurosci Lett 344, 99–102 (2003). [DOI] [PubMed] [Google Scholar]
- Wang B. et al. Association between Alzheimer’s disease and the NOS3 gene Glu298Asp polymorphism in Chinese. J Mol Neurosci 34, 173–176 (2008). [DOI] [PubMed] [Google Scholar]
- Azizi Z., Noroozian M., Kaini-Moghaddam Z. & Majlessi N. Association between NOS3 gene G894T polymorphism and late-onset Alzheimer disease in a sample from Iran. Alzheimer Dis Assoc Disord 24, 204–208 (2010). [DOI] [PubMed] [Google Scholar]
- Ferlazzo N. et al. The 894G > T (Glu298Asp) variant in the endothelial NOS gene and MTHFR polymorphisms influence homocysteine levels in patients with cognitive decline. Neuromolecular Med 13, 167–174 (2011). [DOI] [PubMed] [Google Scholar]
- Giedraitis V. et al. Genetic analysis of Alzheimer’s disease in the Uppsala Longitudinal Study of Adult Men. Dement Geriatr Cogn Disord 27, 59–68 (2009). [DOI] [PubMed] [Google Scholar]
- Ntais C. & Polycarpou A. Association of the endothelial nitric oxide synthase (NOS3) Glu298Asp gene polymorphism with the risk of Alzheimer’s disease—a meta-analysis. J Neurol 252, 1276–1278 (2005). [DOI] [PubMed] [Google Scholar]
- Hua Y., Zhao H., Kong Y. & Lu X. Association between Alzheimer’s disease and the NOS3 gene Glu298Asp polymorphism. Int J Neurosci 124, 243–251 (2014). [DOI] [PubMed] [Google Scholar]
- Wang X. L. et al. Genetic contribution of the endothelial constitutive nitric oxide synthase gene to plasma nitric oxide levels. Arterioscler Thromb Vasc Biol 17, 3147–3153 (1997). [DOI] [PubMed] [Google Scholar]
- Patel V. P. & Chu C. T. Nuclear transport, oxidative stress, and neurodegeneration. Int J Clin Exp Pathol 4, 215–229 (2011). [PMC free article] [PubMed] [Google Scholar]
- Veldman B. A. et al. The Glu298Asp polymorphism of the NOS 3 gene as a determinant of the baseline production of nitric oxide. J Hypertens 20, 2023–2027 (2002). [DOI] [PubMed] [Google Scholar]
- Wang X. L. et al. Genotype dependent and cigarette specific effects on endothelial nitric oxide synthase gene expression and enzyme activity. FEBS Lett 471, 45–50 (2000). [DOI] [PubMed] [Google Scholar]
- Kapoor S. Close association between polymorphisms of the nitric oxide synthetase 3 gene and neurological disorders other than stroke. Int J Gen Med 5, 431–432 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni V. The effect of apolipoprotein E (ApoE) genotype on biomarkers of amyloidogenesis, tau pathology and neurodegeneration in Alzheimer’s disease. Clin Chem Lab Med 49, 375–383 (2011). [DOI] [PubMed] [Google Scholar]
- Brookmeyer R., Gray S. & Kawas C. Projections of Alzheimer’s disease in the United States and the public health impact of delaying disease onset. Am J Public Health 88, 1337–1342 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M. S. Homocysteine and Alzheimer’s disease. Lancet Neurol 2, 425–428 (2003). [DOI] [PubMed] [Google Scholar]
- Tanus-Santos J. E., Desai M. & Flockhart D. A. Effects of ethnicity on the distribution of clinically relevant endothelial nitric oxide variants. Pharmacogenetics 11, 719–725 (2001). [DOI] [PubMed] [Google Scholar]
- Kim K. W. et al. Association between apolipoprotein E polymorphism and Alzheimer’s disease in Koreans. Neurosci Lett 277, 145–148 (1999). [DOI] [PubMed] [Google Scholar]
- Zou F. et al. Sex-dependent association of a common low-density lipoprotein receptor polymorphism with RNA splicing efficiency in the brain and Alzheimer’s disease. Hum Mol Genet 17, 929–935 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]