Abstract
Hypoxia-inducible factors (HIFs) are master regulators of adaptive responses to low oxygen, and their α-subunits are rapidly degraded through the ubiquitination-dependent proteasomal pathway after hydroxylation. Aberrant accumulation or activation of HIFs is closely linked to many types of cancer. However, how hydroxylation of HIFα and its delivery to the ubiquitination machinery are regulated remains unclear. Here we show that Rho-related BTB domain-containing protein 3 (RHOBTB3) directly interacts with the hydroxylase PHD2 to promote HIFα hydroxylation. RHOBTB3 also directly interacts with the von Hippel-Lindau (VHL) protein, a component of the E3 ubiquitin ligase complex, facilitating ubiquitination of HIFα. Remarkably, RHOBTB3 dimerizes with LIMD1, and constructs a RHOBTB3/LIMD1-PHD2-VHL-HIFα complex to effect the maximal degradation of HIFα. Hypoxia reduces the RHOBTB3-centered complex formation, resulting in an accumulation of HIFα. Importantly, the expression level of RHOBTB3 is greatly reduced in human renal carcinomas, and RHOBTB3 deficiency significantly elevates the Warburg effect and accelerates xenograft growth. Our work thus reveals that RHOBTB3 serves as a scaffold to organize a multi-subunit complex that promotes the hydroxylation, ubiquitination and degradation of HIFα.
Keywords: RHOBTB3, HIF, PHD, VHL, hydroxylation, ubiquitination, the Warburg effect
Introduction
Oxygen is an essential element for the growth and survival of aerobic organisms. Metazoans have evolved sophisticated molecular mechanisms that sense insufficient supply of oxygen and induce adaptive physiological changes, including metabolic reprogramming, to maintain balance of bioenergetics and to eradicate excessive free radicals. Inadequate oxygen supply, or hypoxia, is a common feature of solid tumors. In order to sustain continuous growth and proliferation, tumor cells also modify their metabolism to adapt to challenging hypoxic environments1,2,3.
The hypoxia-inducible factors (HIFs) are master mediators of the cellular adaptive response to hypoxia1,2,4,5. HIFs are obligate heterodimers consisting of an oxygen-labile α-subunit and a stable β-subunit (ARNT)6,7. Mammals express three isoforms of HIFα, of which HIF1α and HIF2α (also known as EPAS1) are best characterized3,8,9. HIFs recognize and bind to hypoxia response elements (HREs) of target genes related to many aspects of cancer growth, including proliferation, angiogenesis and metastasis7,10,11,12,13,14,15,16,17. Notably, many of the HIF-upregulated genes also encode enzymes in metabolic pathways14,18,19. These include glucose transporters such as glucose transporter type 1 (GLUT1) and glycolytic enzymes such as hexokinase-2 (HK2) and lactate dehydrogenase a chain (LDHA), which promote glycolysis and lactate fermentation, respectively20,21,22,23. HIFs also upregulate the expression of pyruvate dehydrogenase kinase 1 (PDK1), which phosphorylates and inhibits the activity of pyruvate dehydrogenase, thus repressing the flux of pyruvate into the TCA cycle24,25,26. Exaggerated expression of HIFs therefore can result in a shift towards the Warburg effect characterized by a high rate of glycolysis27,28,29,30. In addition, abnormally stabilized HIF1α directs glucose-mediated ribose synthesis preferentially through the non-oxidative pentose phosphate pathway31. Not surprisingly, solid tumors often exhibit high levels of HIFα and increased HIFα expression correlates with poor clinical prognosis in many cancer types4,10,32,33. Overall, HIFs reprogram glucose metabolism from oxidative to anaerobic ATP production, lower the O2 consumption34, decrease the ROS level35,36 and suppress cancer cell death37,38.
The protein levels of HIFα isoforms are strictly regulated. Under normoxia, HIFα proteins are rapidly degraded7,39. The degradation of HIFα requires hydroxylation and subsequent ubiquitination of the proteins before they are targeted to the proteasomes. Hydroxylation of HIFα occurs on the conserved proline residues within the oxygen-dependent degradation (ODD) domain40,41, and is catalyzed by the dioxygenase prolyl hydroxylases (PHDs, also known as egg-laying-defective nine family members or EGLNs)42. Higher metazoans contain three PHD paralogs (PHD1-PHD3), among them PHD2 appears to be the primary HIFα hydroxylase43,44,45,46. Under normoxic condition, high intracellular oxygen tension directly stimulates PHD2-mediated hydroxylation1,45. Hydroxylation of HIFα can also be promoted or inhibited by various co-factors47,48,49; however, the regulatory mechanism is still poorly defined. The hydroxylation renders HIFα an increased affinity for the von Hippel-Lindau (VHL) protein, a tumor suppressor frequently mutated in a variety of cancers such as renal carcinoma50,51,52. As a component of ubiquitin E3 ligase complex, VHL mediates HIFα polyubiquitination for proteasomal degradation2,53,54. Chronic hypoxia or sustained activation of HIF signaling in turn promotes the expression of PHD2, which limits accumulation of HIF1α in the prolonged exposure to hypoxia and leads to accelerated degradation of HIF1α upon reoxygenation55,56,57. Importantly, it was previously shown that the LIM domain-containing protein 1 (LIMD1) reduces the stability of HIFα by promoting VHL-mediated ubiquitination of HIFα58. In addition, knockdown of LIMD1 promotes tumor growth, and it was thus proposed as a tumor suppressor59,60. However, how cells regulate HIFα signaling in normoxic and hypoxic conditions by coordinating the hydroxylation, ubiquitination and proteasomal degradation of HIFα remains unclear. Furthermore, recent reports have suggested a lysosome-dependent pathway as an alternative mechanism for HIFα degradation61,62,63.
Rho-related BTB domain-containing protein 3 (RHOBTB3), along with RHOBTB1 and RHOBTB2, is an atypical member of the RHO family. RHOBTB3 differs substantially from the other two members (∼48% identity)64 and was shown to function as a regulator controlling protein transport from endosome to the Golgi network. RHOBTB3 was also reported to be a component of CULLIN3 (CUL3)-dependent E3 ubiquitin ligase complex, which is responsible for the degradation of cyclin E and MUF-165,66,67. Unlike most of the other members of RHO family being small GTPases, RHOBTB3 is an ATPase and the ATPase activity is critical for its function68. Here we show that RHOBTB3 has an essential role in controlling the dynamic stability of HIFα. Mechanistically, RHOBTB3 can simultaneously interact with PHD2 and VHL, and these interactions stimulate PHD2's hydroxylase activity and facilitate the ubiquitination of HIFα. RHOBTB3 is able to form homodimers or interact with LIMD1 to form a heterodimer, with the latter being favored and more potent in interacting with PHD2 and VHL. Consistently, cells deficient in both RHOBTB3 and LIMD1 have higher levels of HIFα than cells lacking either protein alone. Intriguingly, the interaction between RHOBTB3 and HIFα-VHL-PHD2 is weakened in hypoxic condition, allowing for adaptive HIFα accumulation during hypoxia. In addition, we show that deficiency of RHOBTB3 promotes the Warburg effect. Furthermore, loss of RHOBTB3 significantly accelerates the growth of tumors in xenograft models. Collectively, our study identifies RHOBTB3 as a novel scaffolding protein for a multi-subunit complex that promotes HIFα degradation under both normoxic and hypoxic conditions, thereby suppressing the Warburg effect and preventing tumorigenesis.
Results
RHOBTB3 downregulates HIFα protein levels
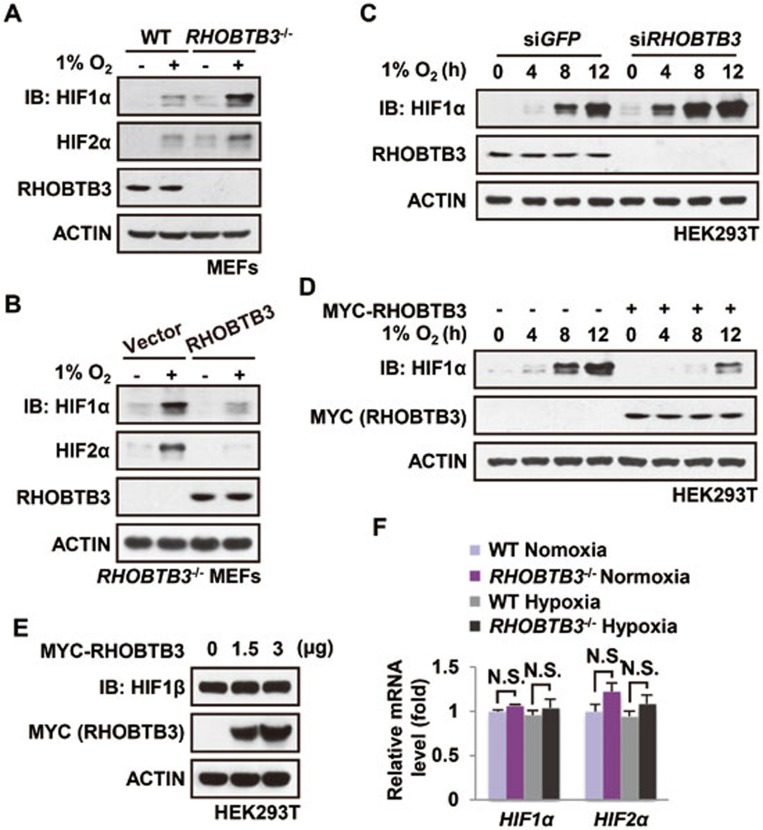
In an effort to study the regulation of HIFα, we identified RHOBTB3 as an interacting protein for VHL in a yeast two-hybrid screen (Supplementary information, Figure S1A). To explore the functional linkage, we first generated MEF cells from RHOBTB3−/− mice, and found that RHOBTB3 null cells had significantly elevated levels of all three HIFα isoforms (HIF1α-3α) under normoxic and hypoxic conditions and after cobalt chloride (CoCl2) treatment compared with WT MEFs (Figure 1A and Supplementary information, Figure S1B and S1C), suggesting that RHOBTB3 exerts a negative effect on HIFα expression. Transcriptional activities of HIF, measured in an HRE-luciferase reporter activity assay, were also significantly increased in the RHOBTB3-null MEFs (Supplementary information, Figure S1D). Consistently, reintroducing RHOBTB3 into RHOBTB3−/− MEFs restored the inhibition on both protein levels and transcriptional activities of HIFα (Figure 1B and Supplementary information, Figure S1D). We also knocked down RHOBTB3 in HEK293T cells, and detected an increase of HIF1α at the protein levels (Figure 1C). Conversely, ectopic expression of RHOBTB3 in HEK293T cells strongly reduced the protein levels and transcriptional activity of HIF1α (Figure 1D and Supplementary information, Figure S1E). Unlike HIF1α, the protein levels of HIF1β/ARNT were not changed by RHOBTB3 overexpression in HEK293T cells (Figure 1E). Compared with RHOBTB3, overexpression of RHOBTB1 or RHOBTB2 had no effect on the protein levels of HIF1α in HEK293T cells (Supplementary information, Figure S1F). Of note, there was no difference in the mRNA levels of HIF1α and HIF2α between WT and RHOBTB3−/− MEFs under both normoxic and hypoxic conditions (Figure 1F), indicating that RHOBTB3 selectively affects the protein levels of HIFα. Overall, these results suggest that RHOBTB3 strongly downregulates the basal protein levels of HIFα and its induction under hypoxia.
Figure 1.
RHOBTB3 downregulates HIF1α expression. (A) Protein levels of HIF1α and HIF2α are elevated in RHOBTB3−/− MEFs under both normoxic and hypoxic conditions. RHOBTB3−/− MEFs and control (WT) MEFs were maintained in normoxia or exposed to hypoxia (1% O2) for 8 h. Cells were then lysed and analyzed by immunoblotting with antibodies indicated. (B) Re-introduction of RHOBTB3 into RHOBTB3−/− MEFs reduces the HIFα levels. RHOBTB3−/− MEFs stably expressing GFP or RHOBTB3 were maintained in normoxia or exposed to hypoxia for 8 h, and were then lysed and analyzed as described in A. (C) Knockdown of RHOBTB3 increases the protein levels of HIF1α. HEK293T cells were infected with lentiviruses expressing siRNA targeting either GFP (control) or RHOBTB3. At 16 h post-infection, cells were exposed to hypoxia for different periods of time as indicated, and were then lysed and analyzed by immunoblotting with antibodies indicated. (D) Ectopic expression of RHOBTB3 in HEK293T cells downregulates HIF1α. HEK293T cells were transfected with pcDNA3.3-MYC-RHOBTB3 or pcDNA3.3-MYC vector as a control. At 16 h post-transfection, cells were exposed to hypoxia for the indicated periods of time and were then lysed, and the protein levels of HIF1α were analyzed. (E) RHOBTB3 does not affect the protein levels of HIF1β/ARNT. HEK293T cells were transfected with RHOBTB3. At 16 h post-transfection, cells were lysed and analyzed by immunoblotting with antibodies indicated. (F) RHOBTB3 has no effect on the mRNA levels of HIF1α or HIF2α under both normoxic and hypoxic conditions. RHOBTB3−/− MEFs and WT MEFs, maintained in normoxia or hypoxia, were homogenized in Trizol reagent, and total RNAs were purified, and were subjected to real-time PCR analysis for mRNA levels of HIF1α and HIF2α. Values are presented as mean ± SEM; n = 3 for each group; three replicate experiments. N.S., not significant. Statistical analysis was carried out by ANOVA followed by Tukey.
RHOBTB3 promotes HIFα hydroxylation and ubiquitination in a PHD2- and VHL-dependent manner
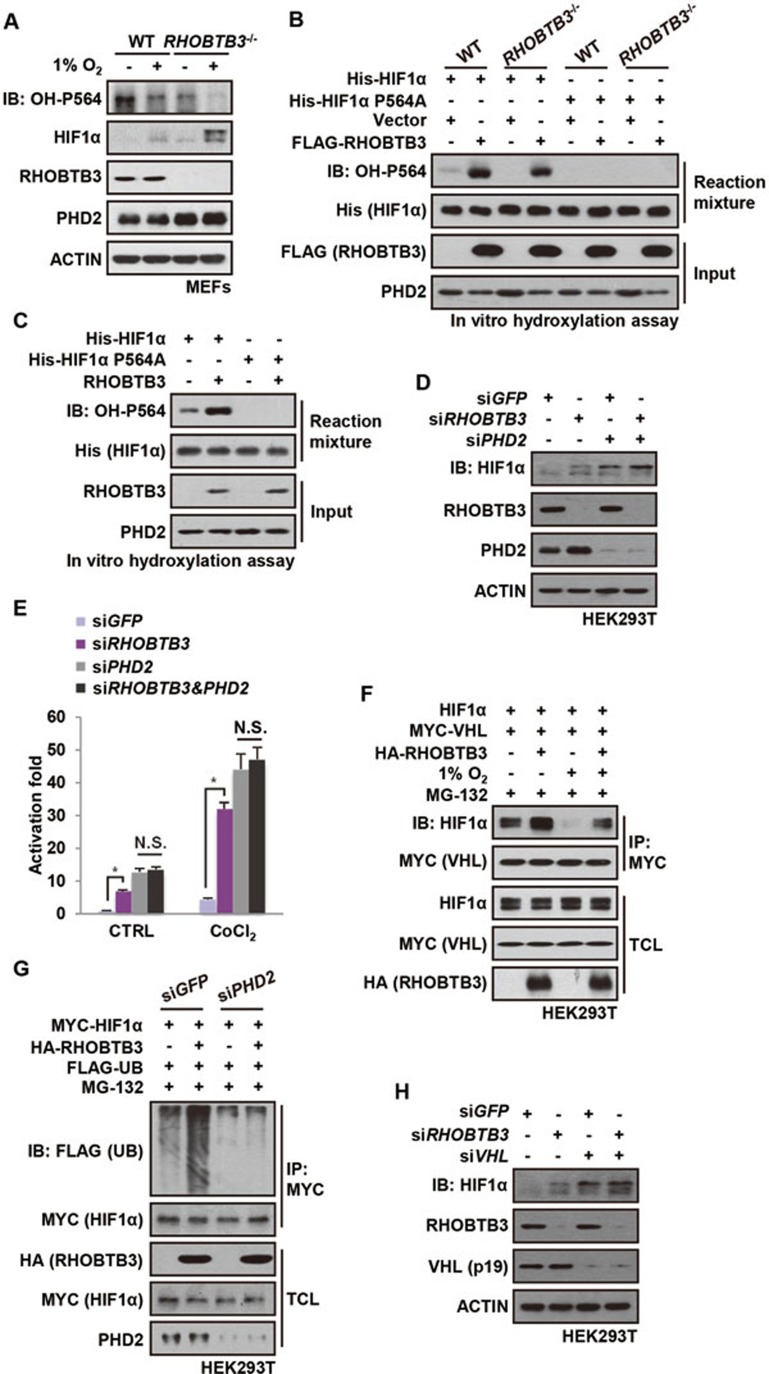
We then explored the mechanism by which RHOBTB3 downregulates the protein levels of HIFα. In the presence of lysosomal inhibitor chloroquine, RHOBTB3 could still suppress the protein levels of HIF1α, while addition of MG-132 strongly blocked RHOBTB3-mediated HIF1α degradation, suggesting that RHOBTB3 promotes HIF1α degradation in a proteasome-specific manner (Supplementary information, Figure S2A). We next explored the possibility that RHOBTB3 promotes HIFα hydroxylation and ubiquitination, two essential modifications prior to proteasomal degradation. While the total protein levels of HIF1α and its target gene, PHD1-3 was increased in RHOBTB3−/− MEFs, the level of its hydroxylation at proline-564 (OH-P564) was significantly reduced under normoxic or hypoxic conditions (Figure 2A and Supplementary information, Figure S2B), indicating that RHOBTB3 is important for the hydroxylation of HIF1α. Consistently, overexpression of RHOBTB3 effectively downregulated wild-type HIF1α, but had little effect on the hydroxylation-defective mutant P564A (Supplementary information, Figure S2C). To confirm the role of RHOBTB3 in HIF1α hydroxylation, we carried out in vitro hydroxylation assays. We mixed bacterially expressed ODD domain of HIF1α (aa 401-603 of human HIF1α) with different cell lysates, and found that lysate from RHOBTB3-overexpressing cells strongly stimulated the hydroxylation of HIF1α (OH-P564) (Figure 2B). Conversely, hydroxylation at P564 of HIF1α was significantly reduced when the cell lysate prepared from RHOBTB3−/− MEFs was added, even though higher protein levels of PHD2 hydroxylase were present in these cells, and the level of OH-P564 was increased by the lysate from RHOBTB3−/− MEFs supplemented with RHOBTB3 (Figure 2B). Moreover, adding in vitro translated RHOBTB3 protein into the lysates of RHOBTB3−/− MEFs markedly enhanced the OH-P564 levels of HIF1α (Figure 2C), excluding the possibility that RHOBTB3 regulates hydroxylation through affecting cellular concentrations of other factors such as co-factors for PHDs46,69. Depletion of PHD2 strongly impaired the RHOBTB3-induced hydroxylation and degradation of HIF1α in HEK293T cells (Supplementary information, Figure S2D and S2E). Moreover, double knockdown of RHOBTB3 and PHD2 did not significantly increase the protein levels or transcriptional activity of HIF1α compared with PHD2 single knockdown (Figure 2D and 2E), suggesting that RHOBTB3 and PHD2 function in the same pathway.
Figure 2.
RHOBTB3 promotes HIFα hydroxylation and ubiquitination in a PHD2- and VHL-dependent manner. (A) RHOBTB3 promotes hydroxylation of HIF1α in MEFs. RHOBTB3−/− MEFs and WT MEFs were maintained in normoxia or exposed to hypoxia for 8 h. Cells were then lysed and the hydroxylation on proline-564 of HIF1α (OH-P564) was analyzed by immunoblotting. As a consequence of sustained accumulation of HIF1α in RHOBTB3-null MEFs, the protein levels of PHD2 were increased. In contrast, the relatively short-term, 8-h hypoxic exposure did not change the protein levels of PHD2. (B) Ectopically expressed RHOBTB3 promotes hydroxylation of HIF1α in vitro. RHOBTB3−/− MEFs and WT MEFs were infected with blank lentiviruses or lentiviruses expressing FLAG-RHOBTB3. Following lysis, the cell lysates were incubated with nickel affinity resin-bound bacterially expressed His-HIF1α (aa 401-603) or the P564A mutant for 90 min at 30 °C. The mixtures were diluted twofold in a 2× SDS buffer, and analyzed by western blotting using antibodies indicated. (C) In vitro translated RHOBTB3 promotes hydroxylation of HIF1α. In vitro translated RHOBTB3 and His-HIF1α (aa 401-603) or the P564A mutant were separately added to cell lysates of RHOBTB3−/− MEFs, and the mixtures were incubated at 30 °C for 90 min. The mixtures were then analyzed for levels of HIF1α hydroxylation as in B. (D) Knockdown of PHD2 impairs RHOBTB3-induced degradation of HIF1α. HEK293T cells were infected by lentiviruses expressing control siRNA (GFP), or siRNA targeting RHOBTB3 or PHD2 or both. At 16 h post-infection, cells were exposed to hypoxia for 4 h, then lysed and analyzed by immunoblotting with antibodies indicated. (E) Double knockdown of RHOBTB3 and PHD2 does not significantly increase transcriptional activity of HIF1α in single knockdown of PHD2. HEK293T cells were infected with lentiviruses expressing different siRNAs as indicated. After 12 h, cells were treated with 200 μM CoCl2 for another 8 h and then lysed. The firefly luciferase reporter carrying HRE was measured and normalized against the Renilla luciferase activity in a dual luciferase assay system. Data are presented as mean ± SEM; n = 3 for each group; *P< 0.05 (ANOVA followed by Tukey); N.S., not significant. (F) RHOBTB3 promotes the interaction between HIF1α and VHL. HEK293T cells were transfected with different combinations of HIF1α, MYC-VHL and HA-RHOBTB3. At 16 h post-transfection, cells were treated with 10 μM MG-132 and maintained in normoxia or exposed to hypoxia for another 10 h, and were lysed. The protein extracts were immunoprecipitated with antibody against MYC for VHL, and were subjected to western blot analysis. TCL, total cell lysate. (G) Knockdown of PHD2 attenuates RHOBTB3-induced ubiquitination of HIF1α. HEK293T cells infected with lentiviruses expressing siRNA targeting GFP (control) or PHD2 were transfected with different combinations of MYC-HIF1α, HA-RHOBTB3 and FLAG-UB. At 16 h post-transfection, cells were treated with 10 μM MG-132 for another 10 h, and were then lysed with RIPA buffer containing 1% SDS and boiled. The protein extracts were diluted in RIPA buffer without SDS to a final concentration of 0.2% SDS, and were subjected to IP with antibody against MYC for HIF1α. The IP product was analyzed by immunoblotting. (H) Knockdown of VHL impairs RHOBTB3 deficiency-induced HIF1α accumulation. HEK293T cells were infected by lentiviruses expressing siRNA targeting GFP (control), RHOBTB3 and/or VHL. At 16 h post-infection, cells were exposed to hypoxia for 4 h, and analyzed by immunoblotting to determine HIF1α protein levels. Probably owing to the low expression levels of its 24 kDa isoform in HEK293T as described previously80 and the preferential affinity of antibody, only the 19 kDa isoform of VHL (VHL (p19)) could be detected and is shown here.
It is known that hydroxylated HIF1α exhibits increased affinity for VHL51,52,70. We thus carried out a co-immunoprecipitation experiment to determine whether the interaction between HIF1α and VHL could be affected by RHOBTB3. Indeed, the interaction was increased in HEK293T cells overexpressing RHOBTB3 under both normoxic and hypoxic conditions (Figure 2F), indicating that more HIFα is recruited to VHL in the presence of RHOBTB3. As a control, RHOBTB3 did not enhance the interaction between the mutant of HIF1α lacking hydroxylated proline residues (HIF1α P402AP564A) and VHL (Supplementary information, Figure S2F). Furthermore, in vitro translated RHOBTB3 did not enhance the interaction between bacterially expressed ODD domain (aa 401-603) of HIF1α (hydroxylation free) and VHL (Supplementary information, Figure S2G). These results strongly indicate that RHOBTB3 enhances HIF1α-VHL interaction by promoting hydroxylation. Elevated levels of ubiquitinated HIF1α were observed in RHOBTB3-overexpressing HEK293T cells (Supplementary information, Figure S2H). Furthermore, knockdown of PHD2 strongly dampened HIF1α ubiquitination enhanced by RHOBTB3 (Figure 2G). Moreover, overexpression or knockdown of RHOBTB3 had little effect on the protein levels of HIF1α in VHL knocked down HEK293T cells (Figure 2H and Supplementary information, Figure S2I). We also found that RHOBTB3 promoted the hydroxylation and ubiquitination of HIF2α (Supplementary information, Figure S2J). Together, these results suggest that RHOBTB3 downregulates the protein levels of HIFα through promoting hydroxylation and ubiquitination of HIFα, which depends on PHD2 and VHL, respectively.
RHOBTB3, PHD2 and VHL form a complex
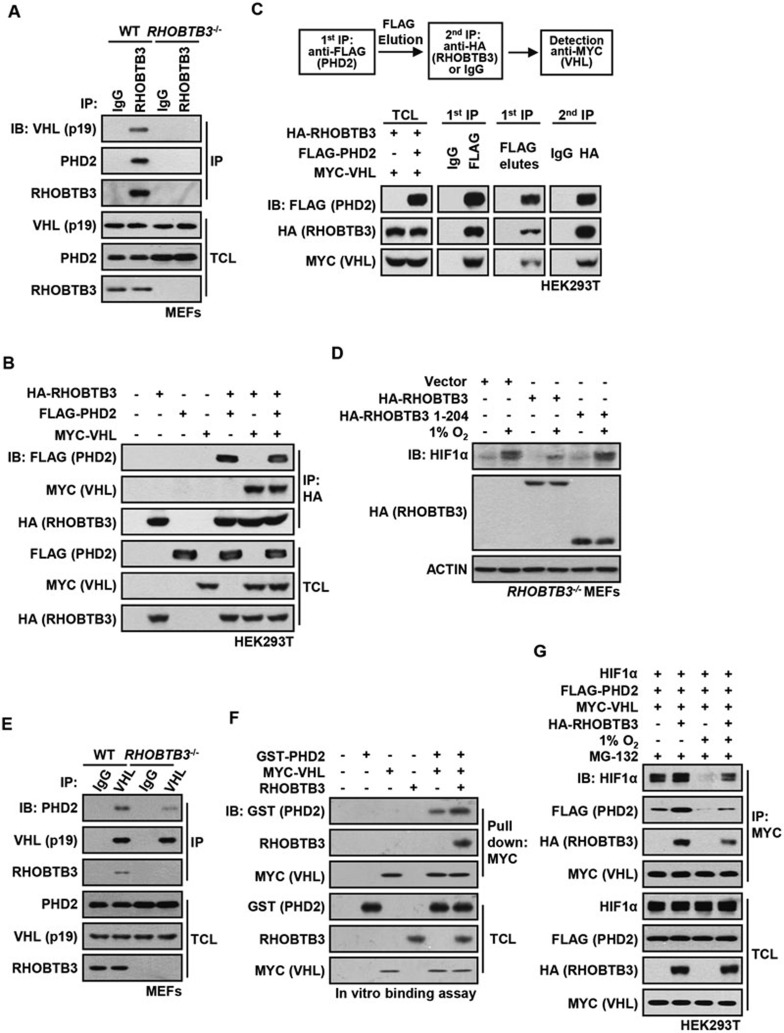
We next asked whether RHOBTB3 forms a complex with VHL and PHD2, particularly since RHOBTB3 and VHL showed interaction in our yeast two-hybrid screen (Supplementary information, Figure S1A) and that RHOBTB3 promoted PHD2-mediated HIFα hydroxylation. We found that endogenous or ectopically expressed RHOBTB3 was co-immunoprecipitated with VHL or vice versa, but not with the control IgG (Figure 3A and Supplementary information, Figure S3A). Consistently, immunofluorescent staining revealed the co-localization of RHOBTB3 with VHL (Supplementary information, Figure S3B). Similarly, PHD2 was readily co-precipitated with RHOBTB3 (Figure 3A and Supplementary information, Figure S3C). We next asked whether RHOBTB3, PHD2 and VHL co-exist in the same complex. It was observed that RHOBTB3 interacted simultaneously with PHD2 and VHL (Figure 3B). We further employed a two-step co-immunoprecipitation (IP) assay, and found that all three components were detected in the final immunoprecipitates (Figure 3C), indicating their co-existence in the same complex. Of note, HSP90, reported as a chaperone of HIFs71,72 and PHD273, was co-precipitated with RHOBTB3 only when HIF1α was present, suggesting that the HSP90 does not directly interact with RHOBTB3 (Supplementary information, Figure S3D). Although HSP90 has been shown to interact with PHD273 and RHOBTB3 interacts with PHD2 as shown in the present study, PHD2 could co-precipitate HSP90 and RHOBTB3 only when HIF1α was present, suggesting the PHD2-HSP90 complex depends on HIF1α to form a co-complex with RHOBTB3 (Supplementary information, Figure S3D). In the presence of ectopic HIF1α, more HSP90 was co-precipitated with RHOBTB3 under normoxia (Supplementary information, Figure S3E), consistent with a stronger interaction of HIF1α with RHOBTB3 under this condition (Figure 2F). These data indicate that HSP90 does not directly interact with RHOBTB3.
Figure 3.
RHOBTB3, PHD2 and VHL form a complex. (A) RHOBTB3 interacts with endogenous PHD2 and VHL. Protein extracts of WT MEFs and RHOBTB3−/− MEFs (control) were immunoprecipitated with antibody against RHOBTB3 or control IgG, and analyzed by immunoblotting with antibodies indicated. (B) Ectopically expressed RHOBTB3 interacts simultaneously with PHD2 and VHL. HEK293T cells were transfected with different combinations of HA-RHOBTB3, MYC-VHL and FLAG-PHD2. At 16 h post-infection, cells were lysed and the protein extracts were immunoprecipitated with antibody against HA, and the IP product was analyzed by western blotting. (C) RHOBTB3, VHL and PHD2 form a complex. HEK293T cells were transfected with HA-RHOBTB3, FLAG-PHD2 and MYC-VHL. After 16 h, cells were harvested. Two-step co-IP was performed by first using anti-FLAG antibody, followed by elution with the FLAG peptide. The eluates were subjected to a second round of IP with anti-HA or control IgG, and the final precipitated proteins were analyzed by immunoblotting. (D) The N-terminal region of RHOBTB3 (aa 1-204) does not have a role in the degradation of HIF1α. RHOBTB3−/− MEFs were infected with lentiviruses expressing HA-RHOBTB3 or HA-RHOBTB3 (aa 1-201). At 36 h post-infection, cells were maintained in normoxia or exposed to hypoxia for 8 h, and analyzed after lysis by immunoblotting. (E) RHOBTB3 strengthens the interaction between PHD2 and VHL. RHOBTB3−/− MEFs and WT MEFs were lysed and the endogenous VHL was immunoprecipitated. The IP product was analyzed by immunoblotting. (F) RHOBTB3 promotes the PHD2-VHL interaction in vitro. In vitro translated RHOBTB3 and bacterially expressed GST-PHD2 were incubated with anti-MYC-conjugated resin-bound MYC-tagged VHL. The mixtures were then pulled down by centrifugation, and analyzed by immunoblotting. (G) RHOBTB3 promotes the interaction between PHD2 and HIF1α. HEK293T cells were transfected with different combinations of HIF1α, FLAG-PHD2, MYC-VHL and HA-RHOBTB3. At 8 h post-transfection, cells were treated with 10 μM MG-132 and maintained in normoxia or exposed to hypoxia for 10 h. The protein extracts were immunoprecipitated with antibody against MYC (for VHL), and precipitated proteins were analyzed by immunoblotting.
Domain mapping using a series of deletion mutants revealed that the C-terminal region of RHOBTB3 (aa 205-611) is responsible for interacting with VHL and PHD2 (Supplementary information, Figure S4A-S4C). Consistently, expression of the N-terminal region of RHOBTB3 (aa 1-204) did not significantly affect the protein levels, transcriptional activities or levels of ubiquitination of HIF1α (Figure 3D and Supplementary information, Figure S4D and S4E). Previous studies reported that two residues of RHOBTB3, asparagine 138 (N138) and aspartic acid 532 (D532), are critical for its function in regulating protein transport to the Golgi network. The N138 residue is also required for the ATPase activity of RHOBTB368. We thus tested whether alteration of the two critical residues would affect RHOBTB3's ability to downregulate HIFα expression. We found that the D532E mutant, but not N138D mutant, was still able to promote HIF1α degradation (Supplementary information, Figure S5A and S5B), hydroxylation (Supplementary information, Figure S5C) and ubiquitination (Supplementary information, Figure S5D) and could suppress HIF1α's transcriptional activity (Supplementary information, Figure S5E), suggesting that the ATPase activity of RHOBTB3 is required. Consistently, N138D interacted with VHL and PHD2 with a much lower affinity compared with WT and D532E mutant (Supplementary information, Figure S5F and S5G).
We next asked whether RHOBTB3 could enhance the interaction between PHD2 and VHL by serving as a scaffold in the ternary complex. As shown in Supplementary information, Figure S6A, ectopically expressed PHD2 interacted weakly with VHL in the absence of RHOBTB3 in the HEK293T cells, and this interaction was enhanced upon ectopical expression of RHOBTB3. The interaction between PHD2 and VHL was significantly weaker in RHOBTB3−/− MEFs compared with WT MEFs (Figure 3E). We next performed an in vitro reconstitution experiment. In vitro translated RHOBTB3 was added to the mixture containing bacterially expressed GST-PHD2 and anti-MYC-conjugated resin-bound MYC-tagged VHL. The mixture was then centrifuged to pull-down resin-bound proteins. Addition of RHOBTB3 promoted the interaction between PHD2 and VHL (Figure 3F). These results together indicate that RHOBTB3 provides a scaffold for PHD2 and VHL interaction.
We last determined whether RHOBTB3 could serve as a scaffold for a larger complex that, in addition to PHD2 and VHL, also includes HIFα. Indeed, we found that HIF1α, VHL and PHD2 were co-precipitated with RHOBTB3 (Supplementary information, Figure S6B). Importantly, overexpression of RHOBTB3 could strengthen the interaction between VHL and PHD2/HIF1α (Figure 3G). These results support the idea that RHOBTB3 amalgamates the HIFα-VHL-RHOBTB3-PHD2 complex for HIFα degradation.
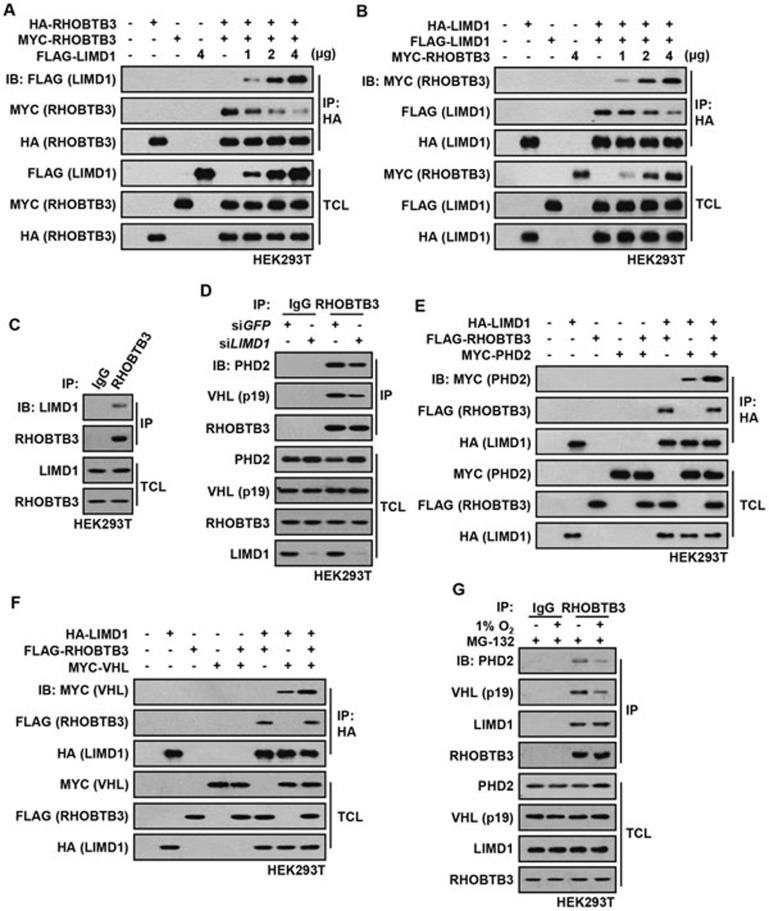
RHOBTB3 and LIMD1 form homodimers or heterodimers and cooperatively regulate HIF1α levels
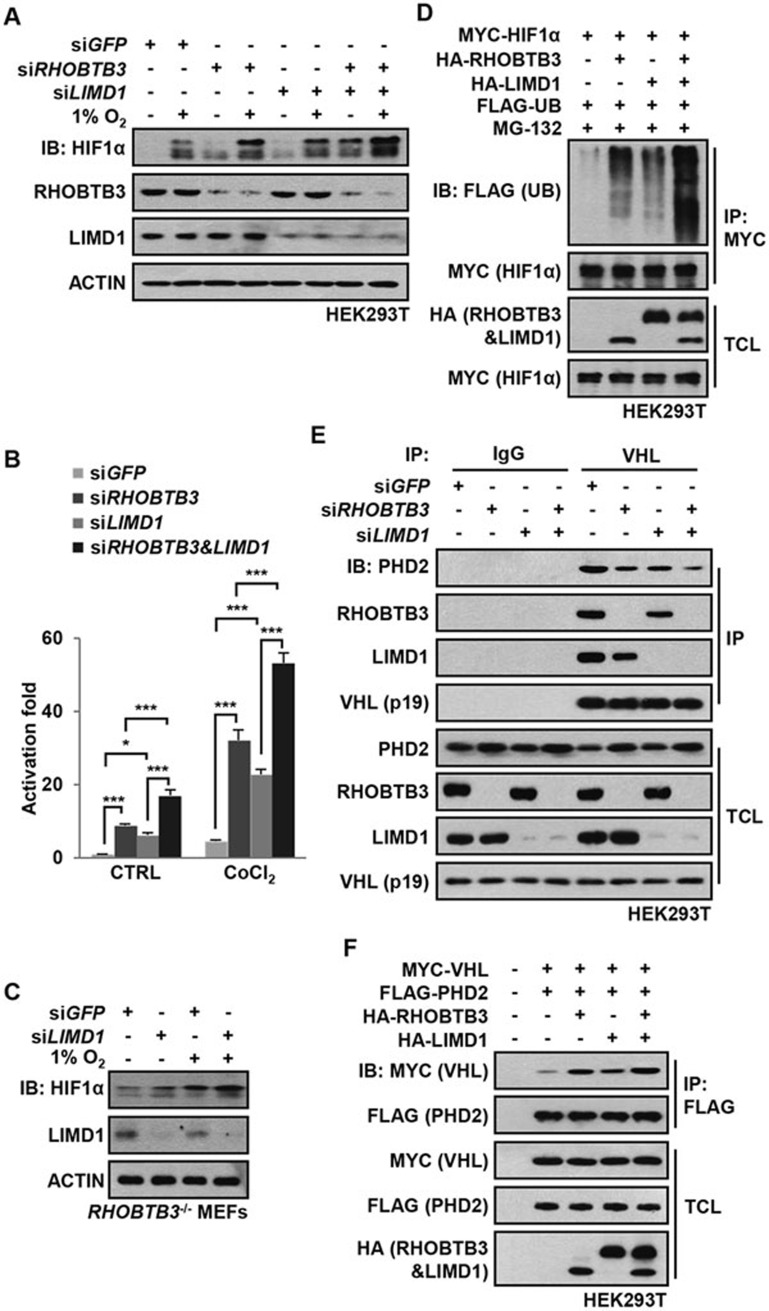
LIMD1 has been reported to serve as an adaptor protein for PHD2 and VHL in the degradation of HIF1α, although if it can enhance HIFα hydroxylation has not been demonstrated58. We thus explored whether there is a functional linkage between RHOBTB3 and LIMD1. Consistent with previous reports, we found that knockdown of LIMD1, such as RHOBTB3, elevated the protein levels and transcriptional activities of HIF1α, and simultaneous knockdown of RHOBTB3 and LIMD1 led to a further increase in protein levels and transcriptional activity of HIF1α (Figure 4A and 4B). Likewise, knockdown of LIMD1 in RHOBTB3−/− MEFs resulted in an increase of HIF1α above the level observed in control RHOBTB3−/− MEFs (Figure 4C). Consistently, co-expressing RHOBTB3 and LIMD1 produced an additive effect on HIF1α ubiquitination (Figure 4D). Moreover, knockdown of RHOBTB3 or LIMD1 decreased the interaction between endogenous PHD2 and VHL and knockdown of both RHOBTB3 and LIMD1 further dampened this interaction (Figure 4E). Conversely, overexpression experiment showed that RHOBTB3 and LIMD1 cooperatively strengthened this interaction (Figure 4F). These observations together suggest that both RHOBTB3 and LIMD1 are required for the suppression of HIF1α.
Figure 4.
RHOBTB3 and LIMD1 cooperatively regulate HIF1α. (A) RHOBTB3 and LIMD1 cooperatively suppress the protein level of HIF1α. HEK293T cells were infected with lentiviruses expressing siRNA targeting GFP, RHOBTB3 and/or LIMD1. At 16 h post-infection, cells were maintained in normoxia or exposed to hypoxia for 8 h before lysis and immunoblotting with antibodies indicated. (B) RHOBTB3 and LIMD1 cooperatively suppress the transcriptional activities of HIF1α. HEK293T cells were infected with different combinations of lentiviruses as indicated. Transcriptional activities of HIF1α were measured using a dual luciferase assay system as described in Figure 2E. Data are presented as mean ± SEM, n = 3 for each group, *P < 0.05, ***P < 0.001 (ANOVA followed by Tukey). (C) Knockdown of LIMD1 in RHOBTB3−/− MEFs further increases the protein levels of HIF1α. RHOBTB3−/− MEFs were infected with lentiviruses expressing siRNA targeting GFP or LIMD1. At 36 h post-infection, cells were maintained in normoxia or exposed to hypoxia for 8 h, before the western blot analysis. (D) RHOBTB3 and LIMD1 cooperatively promote the ubiquitination of HIF1α. HEK293T cells were transfected with different combinations of MYC-HIF1α, HA-RHOBTB3, HA-LIMD1 and FLAG-UB (ubiquitin). After treatment with 10 μM MG-132 for 10 h, the cells were lysed, and the lysates were subjected to IP with antibody against MYC (for HIF1α). The IP product was analyzed by western blotting to determine the ubiquitination levels of HIF1α. (E) Knockdown of RHOBTB3 and/or LIMD1 decreases PHD2-VHL interaction. HEK293T cells were infected with lentiviruses expressing siRNA targeting GFP, RHOBTB3 and/or LIMD1. At 16 h post-infection, cells were lysed and the endogenous VHL was immunoprecipitated, and the IP product was analyzed by immunoblotting. (F) Ectopically expressed RHOBTB3 and LIMD1 cooperatively promote PHD2-VHL interaction. HEK293T cells were transfected with different combinations of MYC-VHL, HA-RHOBTB3, HA-LIMD1 and FLAG-PHD2. Protein extracts from the transfected cells were subjected to IP with antibody against FLAG and analyzed by immunoblotting with antibodies indicated.
We were intrigued that RHOBTB3 and LIMD1, two proteins unrelated in sequence, could both enhance the interaction between PHD2 and VHL, and reduce HIFα in cells. We thus characterized the biochemistry of RHOBTB3 and LIMD1 in the context of HIF1α degradation. We found that ectopically expressed MYC-tagged RHOBTB3 interacted with HA-tagged RHOBTB3 (Figure 5A), suggesting that RHOBTB3 can form homodimers. Similarly, HA-tagged LIMD1 was co-precipitated with FLAG-tagged LIMD1 (Figure 5B), indicating that LIMD1 can homodimerize. More intriguingly, when co-expressed, RHOBTB3 could be co-precipitated with LIMD1 and vice versa, indicating that RHOBTB3 can form heterodimers with LIMD1 (Figure 5A, 5B and Supplementary information, Figure S6C). In addition, endogenous LIMD1 was readily co-immunoprecipitated with RHOBTB3 (Figure 5C). Moreover, the expression of MYC-RHOBTB3 disrupted the interaction between HA-LIMD1 and FLAG-LIMD1, leading to the formation of more LIMD1-RHOBTB3 heterodimers, and addition of FLAG-LIMD1 also reduced RHOBTB3 homodimerization (Figure 5A and 5B). These results suggest that RHOBTB3 and LIMD1 prefer to heterodimerize with each other, or form a higher order complex, than to homodimerize by themselves. Moreover, knockdown of RHOBTB3 or LIMD1 reduced the interaction between endogenous LIMD1/RHOBTB3 and PHD2/VHL (Figure 4E and 5D), whereas expression of RHOBTB3 strongly promoted the interaction between PHD2/VHL and LIMD1 (Figure 5E and 5F), suggesting that the RHOBTB3-LIMD1 heterodimers are more effective scaffolds for the formation of the HIFα degradation complexes that contain PHD2 and VHL. Furthermore, we observed that the interaction between PHD2 and VHL (Figure 3G), and the interaction between RHOBTB3-LIMD1 heterodimer and VHL/PHD2 were attenuated under hypoxic conditions (Figure 5G). The level of co-localization of RHOBTB3 and VHL was reduced under hypoxic condition (Supplementary information, Figure S3B). One obvious explanation is that RHOBTB3 and LIMD1 exert a tight control on the levels of HIF1α under normoxic condition, which is eased up in hypoxia to allow for appropriate HIFα accumulation. Notably, although LIMD1 and RHOBTB3 cooperatively regulate HIF1α levels, LIMD1, unlike RHOBTB3, does not promote the hydroxylation on the proline-564 residue of HIF1α as RHOBTB3 does (Supplementary information, Figure S6D and S6E).
Figure 5.
Dimerization of RHOBTB3 and LIMD1. (A) Ectopically expressed HA-tagged RHOBTB3 interacts with MYC-tagged RHOBTB3 or FLAG-tagged LIMD1. HEK293T cells were transfected with different combinations of HA-RHOBTB3, MYC-RHOBTB3 and FLAG-LIMD1. Cells were then lysed and the protein extracts were immunoprecipitated with antibody against HA. The IP product was analyzed by immunoblotting. (B) Ectopically expressed HA-tagged LIMD1 interacts with FLAG-tagged LIMD1 or MYC-tagged RHOBTB3. Lysates from transfected cells were subjected to IP with antibody against HA (for LIMD1), and analyzed by immunoblotting as in A. (C) RHOBTB3 interacts with endogenous LIMD1. Lysates of HEK293T cells were immunoprecipitated with antibody against RHOBTB3 or IgG (control), and analyzed by immunoblotting using antibodies indicated. (D) Knockdown of LIMD1 attenuates the interaction between RHOBTB3 and PHD2/VHL. HEK293T cells were infected with lentivirus expressing siRNA targeting GFP or LIMD1. At 16 h post-infection, cells were lysed and the endogenous RHOBTB3 was then immunoprecipitated, and analyzed by immunoblotting with antibodies indicated. (E, F) Ectopically expressed RHOBTB3 promotes the interaction between LIMD1 and PHD2 (E), and the interaction between LIMD1 and VHL (F). HEK293T cells were transfected with different combinations of HA-LIMD1, FLAG-RHOBTB3, MYC-PHD2 (E) and MYC-VHL (F). Protein extracts were immunoprecipitated and analyzed by immunoblotting. (G) Hypoxia attenuates the interaction between endogenous RHOBTB3/LIMD1 and PHD2/VHL. HEK293T cells were maintained in normoxia or exposed to hypoxia for 8 h in presence of 10 μM MG-132 to prevent the degradation of VHL under hypoxic condition as described previously80. Endogenous RHOBTB3 was then immunoprecipitated, and the IP product was analyzed by immunoblotting with antibodies indicated.
RHOBTB3 suppresses the Warburg effect and inhibits tumorigenesis
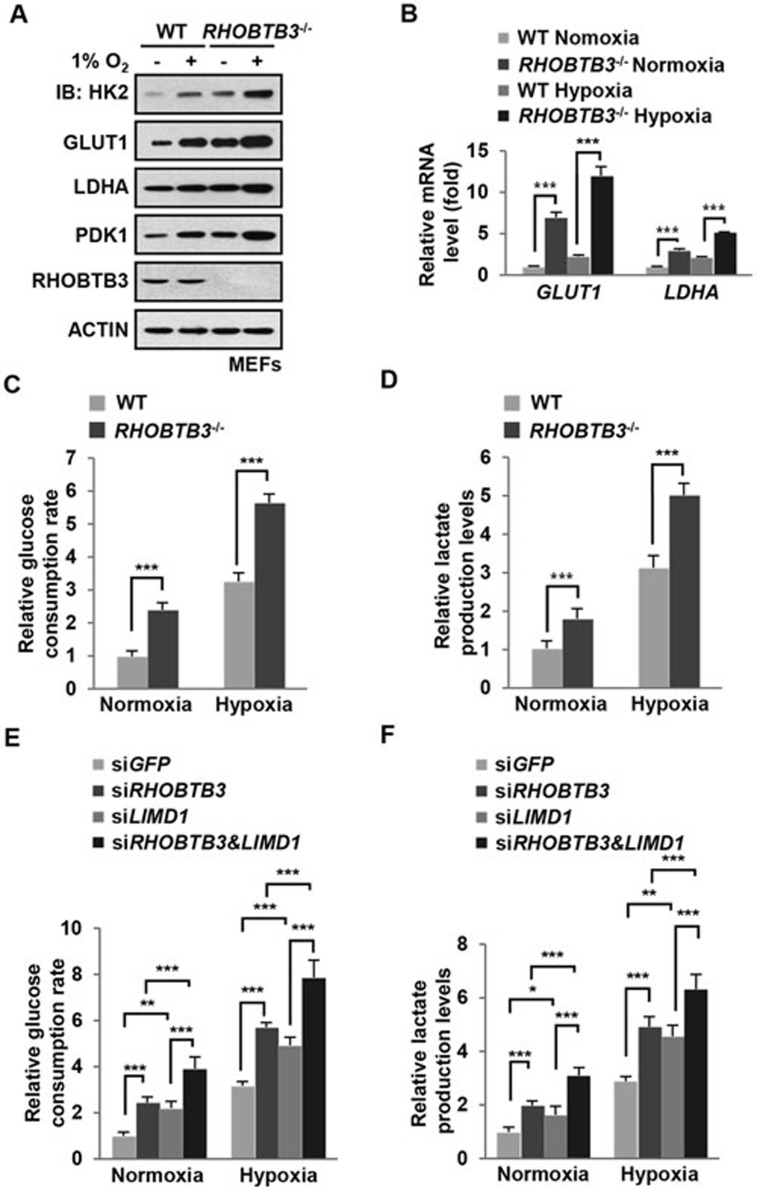
We next examined whether RHOBTB3 could suppress the Warburg effect since it could significantly reduce HIFα levels in cells, and levels of HK2, LDHA, GLUT1 and PDK1, known stream targets of HIFs, were elevated in RHOBTB3−/− MEFs under both normoxic and hypoxic conditions (Figure 6A and 6B). The Warburg effect is characterized by high rates of glucose uptake and lactate production regardless of oxygen concentration30. We thus measured the glucose consumption and lactate production rates, and found that they were increased in RHOBTB3−/− MEFs in both normoxia and hypoxia (Figure 6C and 6D). We also explored the possibility that RHOBTB3 and its partner LIMD1 could regulate the Warburg effect cooperatively, and found that knockdown of RHOBTB3 or LIMD1 promoted glucose consumption and lactate production under both normoxic and hypoxic conditions, and their double knockdown in HEK293T cells further enhanced these effects (Figure 6E and 6F). Taken together, our data suggest that RHOBTB3 inhibits the Warburg effect most likely through promoting HIF degradation.
Figure 6.
RHOBTB3 is a negative regulator of the Warburg effect. (A) Knockout of RHOBTB3 elevates the expression of HK2, LDHA GLUT1 and PDK1. RHOBTB3−/− MEFs and WT MEFs were maintained in normoxia or exposed to hypoxia for 16 h. Cells were then lysed and the protein extracts were analyzed by immunoblotting with antibodies indicated. (B) RHOBTB3 deficiency leads to increased mRNA levels of GLUT1 and LDHA. Total RNAs from RHOBTB3−/− MEFs and WT MEFs, maintained in normoxia or exposed to hypoxia for 16 h, were purified, and analyzed by real-time PCR analysis for the expression levels of GLUT1 and LDHA. Values are presented as mean ± SEM, n = 3 for each group, three replicate experiments. ***P< 0.001 (ANOVA followed by Tukey). (C, D) RHOBTB3 decreases rates of glucose consumption (C) and lactate production (D). RHOBTB3−/− and WT MEFs were maintained in normoxia or exposed to hypoxia for 8 h, and glucose consumption rates (C) and lactate production rates (D) were measured. Values are presented as mean ± SEM, n = 3 for each group, ***P< 0.001 (ANOVA followed by Tukey). (E, F) RHOBTB3 and LIMD1 cooperatively decrease glucose consumption (E) and lactate production (F). HEK293T cells were infected with lentiviruses expressing siRNAs targeting GFP, RHOBTB3 and/or LIMD1. At 16 h post-infection, cells were maintained in normoxia or exposed to hypoxia for 8 h and glucose consumption rates (E) and lactate production rates (F) were measured. Values are presented as mean ± SEM, n = 3 for each group, *P< 0.05, **P< 0.01, ***P< 0.001 (ANOVA followed by Tukey).
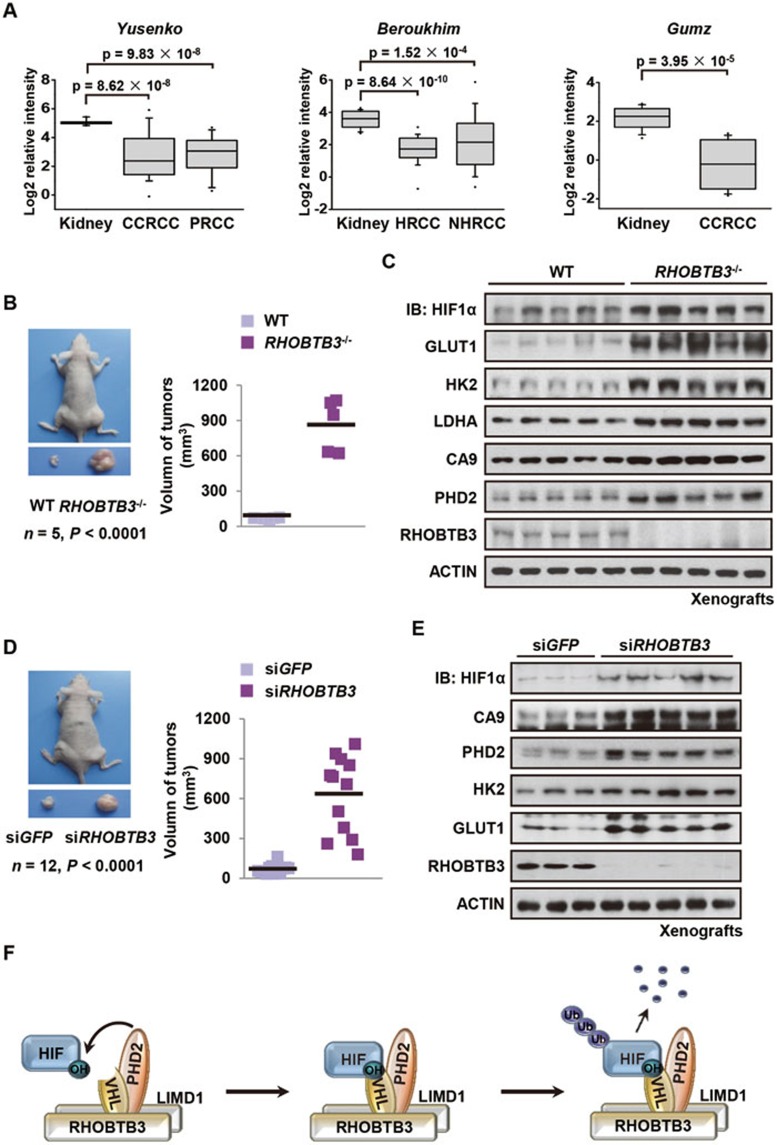
Finally, we explored whether regulation of HIFα by RHOBTB3 has any relevance to tumorigenesis. A search in three independent public data sets of kidney cancer: Yusenko Renal (GEO data set GSE11151)74, Beroukhim Renal (GEO data set GSE14994)75 and Gumz Renal (GEO data set GSE6344)76, summarized by The Oncomine Platform (Life Technologies, Ann Arbor, MI, USA) reveal that the mRNA levels of RHOBTB3 are significantly decreased in clear cell renal cell carcinoma, papillary renal cell carcinoma, hereditary clear cell renal cell carcinoma and non-hereditary clear cell renal cell carcinoma subtypes (Figure 7A). None of the data sets deposited in Oncomine show a significant upregulation of RHOBTB3 in kidney cancer (Figure 7A). These results suggest a potential role of RHOBTB3 in suppressing tumorigenesis. To test this hypothesis, we performed xenograft experiment implanting subcutaneously Ras V12/E1A H133-transformed RHOBTB3−/− MEFs and, as a control, WT MEFs, into nude mice. Xenografts derived from RHOBTB3−/− MEFs were significantly larger in both volume and weight when compared with those derived from control MEFs (Figure 7B and Supplementary information, Figure S7A). In addition, these xenografts had elevated levels of HIF1α and its targets including GLUT1, HK2, LDHA, PHD2 and carbonic anhydrase IX (CA9) (Figure 7C and Supplementary information, Figure S7B). Moreover, sections from RHOBTB3 null xenografts showed enhanced staining for downstream targets of HIF (Supplementary information, Figure S7C). Furthermore, the xenografts derived from RHOBTB3-deficient (siRHOBTB3) HeLa cells showed increased growth rates, tumor volumes, tumor weights and deregulated HIF signaling (Figure 7D, 7E and Supplementary information, Figure S7D-S7F), whereas HeLa cells overexpressing RHOBTB3 were unable to form xenograft tumors (Supplementary information, Figure S7G). We also detected higher proliferation rates in RHOBTB3−/− MEFs compared with WT MEFs, which could be reduced by the knockdown of HIF1α, suggesting that RHOBTB3 suppresses cell proliferation, at least in part, through HIFs (Supplementary information, Figure S7H). Thus, it is reasonable to conclude that RHOBTB3 suppresses tumorigenesis through downregulating HIFα levels.
Figure 7.
RHOBTB3 suppresses tumorigenesis. (A) Expression of RHOBTB3 in human renal cancer samples in public data sets summarized by the Oncomine Platform. (B) Xenografts derived from RHOBTB3−/− MEFs are significantly larger compared with those derived from control MEFs. Ras V12/E1A H133-transformed RHOBTB3−/− MEFs and control WT MEFs (1 × 106) were injected intradermally into each flank of nude mice. Tumors were allowed to develop for 35 days. Tumor volumes were then determined by direct measurement using a caliper and calculated by the formula: (widest diameter × smallest diameter2)/2. Data were presented as mean ± SEM, n = 5 for each group, P < 0.0001 (Student's t-test). (C) The protein levels of HIF1α and its targets are upregulated in xenografts derived from RHOBTB3−/− MEFs. Xenografts derived from RHOBTB3−/− and WT MEFs as described in B were homogenized and analyzed by immunoblotting with the indicated antibodies. (D) Xenografts derived from RHOBTB3 knocked down HeLa cells are significantly larger compared with those derived from control cells. Suspensions of HeLa cells expressing siGFP or siRHOBTB3 (1 × 106 each) were injected intradermally into each flank of nude mice. Tumors were allowed to develop for 37 days and their volumes were calculated as described in Figure 7B. The values of tumor volumes are presented as mean ± SEM, n = 12 for each group, P < 0.0001 (Student's t-test). (E) Protein levels of HIF1α and its target genes are upregulated in xenografts of RHOBTB3 knocked down HeLa cells. Xenografts derived from HeLa-siGFP or HeLa-siRHOBTB3 cells were homogenized and analyzed by immunoblotting with antibodies indicated. (F) Simplified model depicting that RHOBTB3 and LIMD1 promote the formation of the HIF1α degradation complex. In this scheme, RHOBTB3 and LIMD1 form a heterodimer, which interacts with PHD2 and VHL, and recruits HIFα, forming a RHOBTB3/LIMD1-PHD2-VHL-HIFα complex that promotes the hydroxylation, ubiquitination and degradation of HIFα. Hypoxia loosens the interaction between RHOBTB3-LIMD1 and HIFα-VHL-PHD2, allowing for the accumulation of HIFα in cells under hypoxia. Notably, RHOBTB3 can directly promote PHD2-mediated hydroxylation of HIFα, whereas LIMD1 cannot.
Discussion
In this study, we show that RHOBTB3 is essential for the downregulation of HIFα. RHOBTB3 promotes the hydroxylation, ubiquitination and degradation of HIFα. Mechanistically, RHOBTB3 simultaneously interacts with PHD and VHL, serving as a scaffold for the assembly of the RHOBTB3-PHD-VHL-HIFα degradation complex. Within the complex, PHD2 exhibits increased hydroxylase activity towards HIFα, resulting in enhanced interaction between HIFα and VHL, owing to the higher affinity of VHL for hydroxylated HIFα. This facilitates the channeling of hydroxylated HIFα to the VHL-E3 ligase complex for ubiquitination and subsequent degradation. Remarkably, RHOBTB3 is able to form heterodimer with LIMD1, resulting in maximal degradation of HIF (Figure 7F). Although PHD2 appears to have a major role in the RHOBTB3-enhanced hydroxylation of HIF1α, the other PHD isoforms may also participate in the process.
While RHOBTB3 is uniquely involved in facilitating hydroxylation of HIF, RHOBTB3 and LIMD1 are both critical for the degradation of HIFα by scaffolding PHD2 and VHL. Evidence for their mutual compensation is the observation that double knockdown of RHOBTB3 and LIMD1 increases the accumulation of HIF1α above the level in single knockdown under both normoxic and hypoxic conditions. In addition, knockdown of LIMD1 in RHOBTB3−/− MEFs further elevates the protein levels of HIF1α. Moreover, while overexpression of RHOBTB3 or LIMD1 alone can strengthen PHD2-VHL interaction and promote HIF1α degradation, overexpression of both proteins can further strengthen PHD2-VHL interaction and cause additional reduction in the level of HIFα. These observations also support the notion that RHOBTB3/LIMD1 heterodimers are more efficient than homodimers of each protein in strengthening PHD2-VHL interaction and degrading HIF1α. In physiological settings where both RHOBTB3 and LIMD1 are present, a RHOBTB3/LIMD1-PHD2-VHL-HIFα degradation complex is assembled to maximize the control of HIFα levels. It is also noteworthy that the interaction between RHOBTB3/LIMD1 and PHD2/VHL is loosened under hypoxic conditions, allowing the appropriate accumulation of HIFα that is required for the adaptive responses to low-oxygen stresses.
It has been established that RHOBTB3 is a component of a CUL3-dependent E3 ubiquitin ligase complex and required for degradation of cyclin E and MUF-165,66,67. The present study implicates RHOBTB3 in the degradation of HIFα, which is mediated by VHL, a member of CUL2-dependent E3 ubiquitin ligase complex53,54. It is interesting to note that RHOBTB3 can function in both CUL2- and CUL3-dependent ubiquitination pathways. These two pathways might be parallel, or intertwine; further investigation is needed to test whether CUL3 also cooperates with RHOBTB3/VHL in HIFα regulation. Furthermore, we show that HSP90, acting as a chaperon of HIF1α and PHD2 as reported previously71,72,73, can indirectly associate with the RHOBTB3-PHD2-VHL complex through HIF1α.
Also interesting is the reduction in the expression levels of RHOBTB3 in nearly all RCC data sets collected on Oncomine Platform. Our experimental data identify RHOBTB3 as a potent suppressor of the Warburg effect, repressing the production of lactic acid and glucose consumption rates. RHOBTB3 also markedly inhibits xenograft growth in nude mice. Taken together, our work demonstrates that RHOBTB3 assembles a degradation complex for HIFα, suppresses Warburg effect and functions as a critical tumor suppressor.
Materials and Methods
Generation of RHOBTB3−/− MEFs
RHOBTB3−/− mice were obtained from Knockout Mouse Project (KOMP) Repository (RHOBTB3tm1a(KOMP)Wtsi, project ID: CSD35571). RHOBTB3−/− MEFs and WT MEFs were immortalized by introducing SV40 T antigen (for cellular experiments) or Ras (V12) and E1A (H133) oncoproteins (for xenograft model) into primary MEFs from RHOBTB3−/− mice and their WT littermates.
Cell culture, transient transfection, and lentivirus packaging
HEK293T and MEFs were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM ℒ-glutamine, 100 IU penicillin, 100 mg/ml streptomycin at 37 °C in a humidified incubator containing 5% CO2. Polyethylenimine (Cat. #23966, Polysciences, Inc.) at a final concentration of 10 μM was used to transfect HEK293T cells. Total DNA for each plate was adjusted to the same amount by adding relevant empty vector. Lentiviruses for infection of the MEFs were packaged in HEK293T cells after transfection using Lipofectamine 2000 (Cat. 11668-027, Invitrogen). At 30 h post-transfection, medium was collected for further infection.
Immunoprecipitation and immunoblotting
Cell lysis and IP were carried out as previously described77. Briefly, cells were lysed in a lysis buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerolphosphate, 1 mM sodium orthovanadate and a protease inhibitor cocktail. Cell lysates were incubated with respective antibodies overnight. Protein aggregates resulting from the overnight incubation were removed by centrifugation, and protein A/G beads were then added into the lysates and incubated for another 3 h. Protein levels were analyzed on gels. For ubiquitination assays, cells were lysed in RIPA buffer containing 1% SDS and boiled. The protein extracts were diluted using RIPA buffer without SDS to a final concentration of 0.2% SDS, and were subjected to IP. The IP product was analyzed by immunoblotting (IB).
Luciferase reporter assays
Cells cultured in 12-well plates were transfected with the indicated plasmids, washed with phosphate-buffered saline, and lysed in Reporter Lysis Buffer (Invitrogen). Luciferase reporter activities were measured in triplicate using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's protocol and quantified using a GloMax 96-well plate luminometer (Promega). The firefly luciferase to Renilla luciferase ratios were determined and defined as the relative luciferase activity.
Xenograft model
Five-week-old male athymic BALB/c nude mice were housed under specific pathogen-free conditions in compliance with the Institutional Animal Care and Use Committee at Xiamen University. Single cell suspension of HeLa cells infected with lentivirus expressing siRNAs targeting RHOBTB3 or control (siGFP), HeLa cells stably expressing RHOBTB3, or Ras V12/E1A-transformed WT and RHOBTB3−/− MEFs in 0.1 ml DMEM without FBS were injected intradermally into each flank of nude mice. The xenografts were then measured as described in figure legends.
In vitro hydroxylation assay
In vitro hydroxylation assay was performed as described previously78,79. Briefly, cells were lysed in HEB buffer (20 mM Tris-HCl, pH 7.4, 5 mM KCl, 1.5 mM MgCl2) containing a protease inhibitor cocktail. Two-hundred microgram of total protein from each cell lysate was used for each reaction. The cell lysates were mixed with resin-bound HIF1α (aa 401-603) or its P564A mutant in NETN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40 and 1 mM phenylmethylsulfonyl fluoride) containing 2 mM ascorbic acid, 100 μM FeSO4 and 5 mM α-ketoglutarate in a final volume of 200 μl. After mild agitation (2 500× g) for 90 min at 30 °C, the reaction mixtures were centrifuged and washed three times in five volumes of NETN buffer. The final resin-bound proteins were dissolved in an equal volume of 2× SDS buffer, followed by IB with antibodies indicated.
Statistical analyses
ANOVA with Tukey's post-test was used to compare values among different experimental groups using the SPSS Statistics 17.0 program. For experiments with only two groups, Student's t-test was used as specified in the figure legends.
Acknowledgments
We thank colleagues and members of SCL laboratory for suggestions and technical assistance and Dr HR Wang for useful discussions. This work was supported by the National Natural Science Foundation of China (31130016, 31221065, 31370744 and 31300626), the National Basic Research Program of China (973 Program; 2014CB910602, 2011CB910800 and 2013CB530600) and the National Science Foundation for Fostering Talents in Basic Research of the National Natural Science Foundation of China (J1310027/J0106). S-CL is a Cheung Kong Scholar.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
(A) Verification of the interaction between VHL and RHOBTB3 by yeast two-hybrid screen as previously described[81]
(A) RHOBTB3 promotes HIF1α degradation in a proteasome-specific manner.
(A, C) Ectopically expressed RHOBTB3 interacts with VHL (A) and PHD2 (C).
(A) Schematic diagrams represent the full-length RHOBTB3 protein and its different deletion mutants used in the domain mapping experiments.
(A) The N138D mutant of RHOBTB3 fails to downregulate endogenous HIF1α.
(A) Ectopically expressed RHOBTB3 promotes the interaction between PHD2 and VHL.
(A) The sizes of xenografts derived from RHOBTB3−/− MEFs are overwhelmingly larger than those derived from control MEFs.
Full-length cDNAs encoding human RHOBTB3, LIMD1, PHD2 and VHL (VHL30, VHLp24) were obtained by PCR using cDNA purified from HEK293T cells.
References
- Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Oxygen sensing, homeostasis, and disease. N Engl J Med. 2011;365:537–547. doi: 10.1056/NEJMra1011165. [DOI] [PubMed] [Google Scholar]
- Semenza GL, Jiang BH, Leung SW, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Semenza GL. Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J Biol Chem. 1993;268:21513–21518. [PubMed] [Google Scholar]
- Jiang BH, Rue E, Wang GL, Roe R, Semenza GL. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J Biol Chem. 1996;271:17771–17778. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8:967–975. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erler JT, Bennewith KL, Nicolau M, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- Staller P, Sulitkova J, Lisztwan J, Moch H, Oakeley EJ, Krek W. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature. 2003;425:307–311. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- Shweiki D, Itin A, Soffer D, Keshet E. Vascular endothelial growth factor induced by hypoxia may mediate hypoxia-initiated angiogenesis. Nature. 1992;359:843–845. doi: 10.1038/359843a0. [DOI] [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer. 2008;8:851–864. doi: 10.1038/nrc2501. [DOI] [PubMed] [Google Scholar]
- Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe JA, Jiang BH, Iyer NV, et al. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin AT, Weidemann A, Fu Z, et al. Epidermal sensing of oxygen is essential for systemic hypoxic response. Cell. 2008;133:223–234. doi: 10.1016/j.cell.2008.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Semenza GL. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci USA. 1993;90:4304–4308. doi: 10.1073/pnas.90.9.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer NV, Kotch LE, Agani F, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert BL, Firth JD, Ratcliffe PJ. Hypoxia and mitochondrial inhibitors regulate expression of glucose transporter-1 via distinct Cis-acting sequences. J Biol Chem. 1995;270:29083–29089. doi: 10.1074/jbc.270.49.29083. [DOI] [PubMed] [Google Scholar]
- Firth JD, Ebert BL, Ratcliffe PJ. Hypoxic regulation of lactate dehydrogenase A. Interaction between hypoxia-inducible factor 1 and cAMP response elements. J Biol Chem. 1995;270:21021–21027. doi: 10.1074/jbc.270.36.21021. [DOI] [PubMed] [Google Scholar]
- Mathupala SP, Rempel A, Pedersen PL. Glucose catabolism in cancer cells: identification and characterization of a marked activation response of the type II hexokinase gene to hypoxic conditions. J Biol Chem. 2001;276:43407–43412. doi: 10.1074/jbc.M108181200. [DOI] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Sugden MC, Holness MJ. Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs. Am J Physiol Endocrinol Metab. 2003;284:E855–E862. doi: 10.1152/ajpendo.00526.2002. [DOI] [PubMed] [Google Scholar]
- Seagroves TN, Ryan HE, Lu H, et al. Transcription factor HIF-1 is a necessary mediator of the pasteur effect in mammalian cells. Mol Cell Biol. 2001;21:3436–3444. doi: 10.1128/MCB.21.10.3436-3444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest. 2013;123:3664–3671. doi: 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Mancuso A, Bui TV, et al. Imatinib resistance associated with BCR-ABL upregulation is dependent on HIF-1alpha-induced metabolic reprograming. Oncogene. 2010;29:2962–2972. doi: 10.1038/onc.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas J, Kaelin WG., Jr Dysregulation of HIF and VEGF is a unifying feature of the familial hamartoma syndromes. Cancer Cell. 2004;6:7–10. doi: 10.1016/j.ccr.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Rankin EB, Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008;15:678–685. doi: 10.1038/cdd.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimova T, Chandel NS. Mitochondrial complex III regulates hypoxic activation of HIF. Cell Death Differ. 2008;15:660–666. doi: 10.1038/sj.cdd.4402307. [DOI] [PubMed] [Google Scholar]
- Brunelle JK, Bell EL, Quesada NM, et al. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 2005;1:409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Guzy RD, Hoyos B, Robin E, et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature. 1998;392:405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- Graeber TG, Osmanian C, Jacks T, et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- Salceda S, Caro J. Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J Biol Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke JF, Tian YM, Ratcliffe PJ, Pugh CW. Oxygen-regulated and transactivating domains in endothelial PAS protein 1: comparison with hypoxia-inducible factor-1alpha. J Biol Chem. 1999;274:2060–2071. doi: 10.1074/jbc.274.4.2060. [DOI] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouyssegur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG. Proline hydroxylation and gene expression. Ann Rev Biochem. 2005;74:115–128. doi: 10.1146/annurev.biochem.74.082803.133142. [DOI] [PubMed] [Google Scholar]
- Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- Hagen T, Taylor CT, Lam F, Moncada S. Redistribution of intracellular oxygen in hypoxia by nitric oxide: effect on HIF1alpha. Science. 2003;302:1975–1978. doi: 10.1126/science.1088805. [DOI] [PubMed] [Google Scholar]
- Ivan M, Haberberger T, Gervasi DC, et al. Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc Natl Acad Sci USA. 2002;99:13459–13464. doi: 10.1073/pnas.192342099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarshan S, Sourbier C, Kong HS, et al. Fumarate hydratase deficiency in renal cancer induces glycolytic addiction and hypoxia-inducible transcription factor 1alpha stabilization by glucose-dependent generation of reactive oxygen species. Mol Cell Biol. 2009;29:4080–4090. doi: 10.1128/MCB.00483-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG., Jr The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8:865–873. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Tanimoto K, Makino Y, Pereira T, Poellinger L. Mechanism of regulation of the hypoxia-inducible factor-1 alpha by the von Hippel-Lindau tumor suppressor protein. EMBO J. 2000;19:4298–4309. doi: 10.1093/emboj/19.16.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo G, Duplan E, Boyer N, Vigne P, Frelin C. Hypoxia up-regulates prolyl hydroxylase activity: a feedback mechanism that limits HIF-1 responses during reoxygenation. J Biol Chem. 2003;278:38183–38187. doi: 10.1074/jbc.M302244200. [DOI] [PubMed] [Google Scholar]
- Marxsen JH, Stengel P, Doege K, et al. Hypoxia-inducible factor-1 (HIF-1) promotes its degradation by induction of HIF-alpha-prolyl-4-hydroxylases. Biochem J. 2004;381:761–767. doi: 10.1042/BJ20040620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Peso L, Castellanos MC, Temes E, et al. The von Hippel Lindau/hypoxia-inducible factor (HIF) pathway regulates the transcription of the HIF-proline hydroxylase genes in response to low oxygen. J Biol Chem. 2003;278:48690–48695. doi: 10.1074/jbc.M308862200. [DOI] [PubMed] [Google Scholar]
- Foxler DE, Bridge KS, James V, et al. The LIMD1 protein bridges an association between the prolyl hydroxylases and VHL to repress HIF-1 activity. Nat Cell Biol. 2012;14:201–208. doi: 10.1038/ncb2424. [DOI] [PubMed] [Google Scholar]
- Sharp TV, Al-Attar A, Foxler DE, et al. The chromosome 3p21.3-encoded gene, LIMD1, is a critical tumor suppressor involved in human lung cancer development. Proc Natl Acad Sci USA. 2008;105:19932–19937. doi: 10.1073/pnas.0805003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp TV, Munoz F, Bourboulia D, et al. LIM domains-containing protein 1 (LIMD1), a tumor suppressor encoded at chromosome 3p21.3, binds pRB and represses E2F-driven transcription. Proc Natl Acad Sci USA. 2004;101:16531–16536. doi: 10.1073/pnas.0407123101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira JV, Fofo H, Bejarano E, et al. STUB1/CHIP is required for HIF1A degradation by chaperone-mediated autophagy. Autophagy. 2013;9:1349–1366. doi: 10.4161/auto.25190. [DOI] [PubMed] [Google Scholar]
- Olmos G, Arenas MI, Bienes R, et al. 15-Deoxy-Delta(12,14)-prostaglandin-J(2) reveals a new pVHL-independent, lysosomal-dependent mechanism of HIF-1alpha degradation. Cell Mol Life Sci. 2009;66:2167–2180. doi: 10.1007/s00018-009-0039-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbi ME, Hu H, Kshitiz, Ahmed I, Levchenko A, Semenza GL. Chaperone-mediated autophagy targets hypoxia-inducible factor-1alpha (HIF-1alpha) for lysosomal degradation. J Biol Chem. 2013;288:10703–10714. doi: 10.1074/jbc.M112.414771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold J, Schenkova K, Rivero F. Rho GTPases of the RhoBTB subfamily and tumorigenesis. Acta Pharmacol Sin. 2008;29:285–295. doi: 10.1111/j.1745-7254.2008.00773.x. [DOI] [PubMed] [Google Scholar]
- Lu A, Pfeffer SR. Golgi-associated RhoBTB3 targets cyclin E for ubiquitylation and promotes cell cycle progression. J Cell Biol. 2013;203:233–250. doi: 10.1083/jcb.201305158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkova K, Lutz J, Kopp M, Ramos S, Rivero F. MUF1/leucine-rich repeat containing 41 (LRRC41), a substrate of RhoBTB-dependent cullin 3 ubiquitin ligase complexes, is a predominantly nuclear dimeric protein. J Mol Biol. 2012;422:659–673. doi: 10.1016/j.jmb.2012.06.016. [DOI] [PubMed] [Google Scholar]
- Berthold J, Schenkova K, Ramos S, et al. Characterization of RhoBTB-dependent Cul3 ubiquitin ligase complexes — evidence for an autoregulatory mechanism. Exp Cell Res. 2008;314:3453–3465. doi: 10.1016/j.yexcr.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa EJ, Calero M, Sridevi K, Pfeffer SR. RhoBTB3: a Rho GTPase-family ATPase required for endosome to Golgi transport. Cell. 2009;137:938–948. doi: 10.1016/j.cell.2009.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P, Lee S, Duncan CG, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, White SB, Zhao Q, Lee FS. HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc Natl Acad Sci USA. 2001;98:9630–9635. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs JS, Jung YJ, Mimnaugh EG, Martinez A, Cuttitta F, Neckers LM. Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1 alpha-degradative pathway. J Biol Chem. 2002;277:29936–29944. doi: 10.1074/jbc.M204733200. [DOI] [PubMed] [Google Scholar]
- Minet E, Mottet D, Michel G, et al. Hypoxia-induced activation of HIF-1: role of HIF-1alpha-Hsp90 interaction. FEBS Lett. 1999;460:251–256. doi: 10.1016/s0014-5793(99)01359-9. [DOI] [PubMed] [Google Scholar]
- Song D, Li LS, Heaton-Johnson KJ, Arsenault PR, Master SR, Lee FS. Prolyl hydroxylase domain protein 2 (PHD2) binds a Pro-Xaa-Leu-Glu motif, linking it to the heat shock protein 90 pathway. J Biol Chem. 2013;288:9662–9674. doi: 10.1074/jbc.M112.440552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusenko MV, Kuiper RP, Boethe T, Ljungberg B, van Kessel AG, Kovacs G. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer. 2009;9:152. doi: 10.1186/1471-2407-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Brunet JP, Di Napoli A, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumz ML, Zou H, Kreinest PA, et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:4740–4749. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- Rui Y, Xu Z, Lin S, et al. Axin stimulates p53 functions by activation of HIPK2 kinase through multimeric complex formation. EMBO J. 2004;23:4583–4594. doi: 10.1038/sj.emboj.7600475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callapina M, Zhou J, Schmid T, Kohl R, Brune B. NO restores HIF-1alpha hydroxylation during hypoxia: role of reactive oxygen species. Free Radic Biol Med. 2005;39:925–936. doi: 10.1016/j.freeradbiomed.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Patten DA, Lafleur VN, Robitaille GA, Chan DA, Giaccia AJ, Richard DE. Hypoxia-inducible factor-1 activation in nonhypoxic conditions: the essential role of mitochondrial-derived reactive oxygen species. Mol Biol Cell. 2010;21:3247–3257. doi: 10.1091/mbc.E10-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xin H, Eckert DT, Brown JA, Gnarra JR. Hypoxia and cell cycle regulation of the von Hippel-Lindau tumor suppressor. Oncogene. 2011;30:21–31. doi: 10.1038/onc.2010.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Verification of the interaction between VHL and RHOBTB3 by yeast two-hybrid screen as previously described[81]
(A) RHOBTB3 promotes HIF1α degradation in a proteasome-specific manner.
(A, C) Ectopically expressed RHOBTB3 interacts with VHL (A) and PHD2 (C).
(A) Schematic diagrams represent the full-length RHOBTB3 protein and its different deletion mutants used in the domain mapping experiments.
(A) The N138D mutant of RHOBTB3 fails to downregulate endogenous HIF1α.
(A) Ectopically expressed RHOBTB3 promotes the interaction between PHD2 and VHL.
(A) The sizes of xenografts derived from RHOBTB3−/− MEFs are overwhelmingly larger than those derived from control MEFs.
Full-length cDNAs encoding human RHOBTB3, LIMD1, PHD2 and VHL (VHL30, VHLp24) were obtained by PCR using cDNA purified from HEK293T cells.