Dear Editor,
Norrin, also known as the Norrie disease protein or X-linked exudative vitreoretinopathy 2 protein, is a secreted retinal growth factor with angiogenic and neuroprotective properties. Mutations in the NPD gene, which encodes the Norrin protein, are associated with Norrie disease, familial exudative vitreoretinopathy (FEVR), retinopathy of prematurity (ROP), and other retinal hypovascularization diseases1,2. Norrin has a weak homology with TGF-β, but no significant sequence homology with Wnt proteins. The crystal structure of Norrin revealed a dimeric complex of Norrin that folds more like TGF-β than Wnt3. However, Norrin does not activate the TGF-β pathway. Instead, despite its structure being distinct from that of Wnt proteins, Norrin functions predominantly through the activation of the canonical Wnt/β-catenin signaling pathway. Frizzled 4 (Fz4), but not other Frizzled family members, serves as the primary receptor for Norrin on the cell surface. The Norrin-Fz4 interaction is essential for Norrin signaling and function, as both NPD mutations and FZD4 mutations are associated with FEVR. How Norrin activates the Wnt/β-catenin signaling pathway through its interaction with Fz4 remains enigmatic.
The 133-residue Norrin forms a covalent homodimer, with three pairs of intermolecular disulfide bonds, in addition to four pairs of intramolecular disulfide bonds in each Norrin subunit3. This structure does not immediately explain that Norrin can specifically recognize Fz4, which is also a receptor for canonical Wnt proteins (e.g., Wnt8)4, and activate the canonical Wnt/β-catenin signaling pathway inside the cell. Frizzled proteins, along with smoothened, a hedgehog regulator, form the class F GPCR family. This family of receptors only weakly activates G-protein or arrestin pathways, the canonical signaling mechanisms downstream of the other GPCR classes. Like other Frizzled proteins, Fz4 contains a ∼120-residue N-terminal extracellular cysteine-rich domain (CRD) followed by the 7-helix transmembrane domain. Due to the technical difficulties generating soluble Norrin and Wnt proteins, the only complex structure available for the Frizzled family is that of the Wnt8/Fz8-CRD complex5. Structural information regarding Norrin-Fz4 interaction would provide key insights into many important aspects of cell signaling.
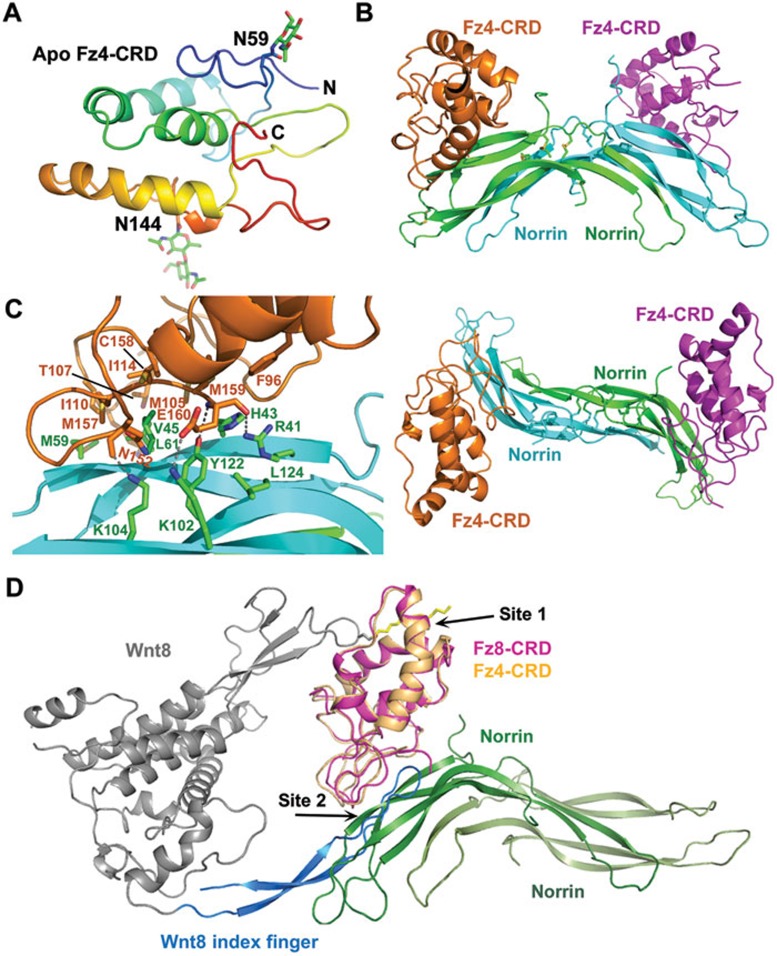
To understand how Fz4 recognizes Norrin and Wnt8, we first determined the crystal structure of the extracellular CRD of Frizzled 4 (Fz4-CRD) at 2.4 Å resolution (Figure 1A). Structural comparison of Fz4-CRD with previously solved Fz8-CRD and sFRP3-CRD structures indicates that all three CRDs share a very similar fold (Supplementary information, Figure S1). Therefore Fz4-CRD may interact with Wnt8 in a manner very similar to that of Fz8-CRD5. That is, Wnt8 may interact with Fz4-CRD at two distinct sites: the thumb with a covalently attached palmitoleic acid at Ser187 in the N-terminal saposin-like domain may contact the Fz4-CRD hydrophobic binding groove (site 1 interaction), and the tip of the index finger with an antiparallel β-hairpin structure in the C-terminal domain may contact the Fz4-CRD's shallow pocket on the opposite site (site 2 interaction). Notably, the site 1 interaction is mediated primarily through hydrophobic interactions with a lipid molecule whereas the site 2 interaction is mediated through specific protein-protein interactions.
Figure 1.
Crystal structures of apo Fz4-CRD and the Norrin/Fz4-CRD complex. (A) Structure overview of the apo Fz4-CRD in rainbow color scheme from N-terminus (blue) to C-terminus (red). The N-acetylglucosamine groups at the two glycosylation sites (N59 and N144) are shown in stick presentation. (B) Two orthogonal views of the overall structure of the Norrin/Fz4-CRD complex. Two separate Fz4-CRD molecules interact with one Norrin homodimer, which is stabilized by three pairs of intermolecular disulfide bonds. (C) Detailed contacts between Norrin and Fz4-CRD. The interface contains a hydrophobic core formed by Norrin residues V45, M59, L61 and L124, and Fz4-CRD residues F96, M105, I110, M157 and M159. In addition, R41 and K102 of Norrin interact with the main chain and side chain of Fz4 E160, respectively. K104 and Y122 of Norrin also form hydrogen bonds with Fz4-CRD main chain atoms. (D) Superposition of the Wnt8/Fz8-CRD structure (4F0A) with the Norrin/Fz4-CRD structure. Wnt8 is colored in blue for the index finger and gray for the rest of the molecule, and the lipid is depicted as a stick model in yellow. Norrin is colored in green and light green for the two monomers. Fz8-CRD is colored in pink whereas Fz4-CRD is colored in light orange. Note that Norrin and the index finger of Wnt8 bind to an overlapping interface on Fz-CRD (site 2).
We then determined the crystal structure of a MBP-Norrin/Fz4-CRD complex at 3.8 Å resolution, using the molecular replacement method with the Fz4-CRD, MBP, and Norrin structures as search models (Figure 1B and Supplementary information, Figure S2 and Table S1). The Fz4-CRD conformation was essentially identical in both structures (with Cα, root-mean-square deviation (rmsd) = 0.52 Å Supplementary information, Figure S3). MBP was used to maintain the solubility of Norrin and to promote crystallization, and MBP-Norrin is fully active in cell-based assays3. The relative orientation of MBP versus Norrin is not fixed as observed in our crystal structure. Previous studies3 and our size-exclusion chromatography (SEC) analysis results indicated that two Fz4-CRD molecules interact with one Norrin homodimer (2:2 complex). Consistent with this result, there are two Fz4-CRD molecules bound to two symmetric sites in the Norrin homodimer in each asymmetric unit (ASU). The structure of Norrin when it is bound to Fz4-CRD is largely identical to that of unbound Norrin, except for the loop regions connecting β-strands (with an rmsd of 1.27 Å Supplementary information, Figure S3).
The two Fz4-CRD molecules reside on two sides of the Norrin homodimer (Figure 1B and Supplementary information, Figure S4). The Norrin β-strands interact with the C-terminal tail of Fz4-CRD, as well as with the loops between Fz4-CRD helices (Figure 1C). The interface involves mostly hydrophobic residues, including residues V45, M59, L61, and L124 of Norrin, and residues F96, M105, I110, M157, and M159 of Fz4-CRD. Several polar interactions with a charge complementarity further enhance the Norrin/Fz4-CRD interaction affinity and specificity (Figure 1C and Supplementary information, Figures S4 and S5). The detailed interactions are summarized in Supplementary information, Table S2. The importance of this interface in the functions of Norrin and Fz4 has been validated by mutations in Norrin residues R41, H43, V45, L61, Y122, which affect the Norrin/Fz4 interaction and signaling3, as well as by a number of mutations in Norrin (R41K, R41T, H43R, H43Q, V45M, V45E, L61P, L61F, K104Q and L124F) and Fz4-CRD (M105V, M105T, I114T and M157V), all of which have been identified in Norrie disease and FEVR.
How does Fz4 interact with both Wnt and Norrin? Superposition of Fz4-CRD/Norrin and Fz8-CRD/Wnt8 structures demonstrates that both the Wnt8 C-terminal cysteine-rich cytokine-like domain and Norrin bind to the same C-terminal area of Fz4-CRD (Supplementary information, Figure S6). The palmitoleate moiety-binding site is located at an open surface opposite to the Fz4-CRD/Norrin interface. Norrin contacts Fz4-CRD in the shallow pocket opposite to the lipid-binding groove with a buried surface area of 1880 Å2, which is more extensive than the interaction between Wnt8 and Fz8-CRD at site 2, which has a buried surface area of only 397 Å2. Thus Fz4-CRD may recognize Wnt8 with a two-finger mechanism, and recognize Norrin with a single, larger interface. Detailed sequence analysis also indicates that Norrin-binding residues are conserved among Fz4 proteins from different species (Supplementary information, Figure S7). But several of the interface residues of Fz4 (e.g., K109, I110, and M157) are not conserved in other Frizzled proteins (Supplementary information, Figure S8), which explains the specificity of Norrin for the recognition of Fz4 at the cell surface.
How does the binding activate Wnt signaling? Wnt-mediated receptor oligomerization plays a critical role in Wnt/β-catenin signaling6,7,8,9. Previous work also indicated that Norrin induces clustering of Fz4 for downstream β-catenin signaling10,11. There is also evidence that Fz4 forms homodimers and the homodimerization involves the transmembrane domain of Fz49. In our crystal structure, the two Fz4-CRD molecules bound to the same Norrin homodimer are separated by a large distance (> 30 Å), and therefore more likely belong to two separate Fz4 dimers on the membrane. Our structure thus is consistent with a model in which Norrin promotes the clustering of Fz4.
An alternative, but not mutually exclusive, possibility is that Norrin induces a conformational change in the Fz4 transmembrane domain. In our crystal structures, Norrin forms a homodimer with a rigid scaffold that restrains the position of Fz4-CRD, which in turn may reposition the transmembrane domains in the Fz4 dimer. A potential Norrin-mediated conformational change in the Fz4 transmembrane domain may play a role in transducing the signal across the membrane. Future studies are needed to explore this possibility.
Understanding the structural basis of Norrin-induced Wnt pathway activation will not only reveal the Norrin signaling mechanism, but also help with the development of new approaches for prevention and/or treatment of retinal hypovascularization diseases. The poor solubility of Wnt proteins due to their essential lipid modifications has prevented their practical use in therapeutics. In contrast, recombinant Norrin protein lacks lipid modification and is fully functional in activating canonical Wnt signaling. Therefore, the Norrin-Fz4 complex structure may also provide a structural template for designing soluble Wnt mimetics useful for stem cell biology and regenerative medicine.
During the review of this paper, crystal structures of two Norrin/Fz4-CRD complexes as well as the apo Fz4-CRD were reported12. Despite the use of MBP-tagged Norrin in our structure, in contrast to untagged Norrin in the reported structure (PDB code: 5BQC)12, the two structures are exceedingly similar with the rmsd of 0.51 Å and 0.64 Å for the entire Cα atoms of Norrin (residues 34-133) and Fz4-CRD (residues 43-160), respectively (Supplementary information, Figure S9). Both complexes adopted a 2:2 configuration of Norrin and Fz4-CRD, suggesting that the MBP tag has little effect on Norrin's binding to Fz4. Together with previous functional assays3, the MBP-Norrin appears to be active in both the binding to Fz4 and the activation of downstream β-catenin signaling.
Accession numbers
The coordinates and structure factors for the apo Fz4-CRD and MBP-Norrin/Fz4-CRD structures have been deposited into the Protein Data Bank under the accession codes of 5CM4 and 5CL1, respectively.
Acknowledgments
We thank staff members of the Life Science Collaborative Access Team (ID-21) of the Advanced Photon Source (APS) for assistance in data collection at the beam lines of sector 21, which is in part funded by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). Use of APS was supported by the Office of Science of the US Department of Energy, under Contract No DE-AC02-06CH11357. We also thank staff members of the Berkeley Center for Structural Biology of the Advanced Light Source (ALS). Use of ALS was supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under Contract No DE-AC02-05CH11231. This work was supported by National Institute of Health (EY026044 to WX, DK071662 to HEX, and GM102545 and GM104212 to KM) and by the Van Andel Research Institute (HEX and KM).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Supplementary infomration, Figures, Tables and Data
References
- Ohlmann A, Tamm ER. Prog Retin Eye Res. 2012. pp. 243–257. [DOI] [PubMed]
- Ye X, Wang Y, Nathans J. Trends Mol Med. 2010. pp. 417–425. [DOI] [PMC free article] [PubMed]
- Ke J, Harikumar KG, Erice C, et al. Genes Dev. 2013. pp. 2305–2319. [DOI] [PMC free article] [PubMed]
- Smallwood PM, Williams J, Xu Q, et al. J Biol Chem. 2007. pp. 4057–4068. [DOI] [PubMed]
- Janda CY, Waghray D, Levin AM, et al. Science. 2012. pp. 59–64. [DOI] [PMC free article] [PubMed]
- Bilic J, Huang YL, Davidson G, et al. Science. 2007. pp. 1619–1622. [DOI] [PubMed]
- Carron C, Pascal A, Djiane A, et al. J Cell Sci. 2003. pp. 2541–2550. [DOI] [PubMed]
- Cong F, Schweizer L, Varmus H. Development. 2004. pp. 5103–5115. [DOI] [PubMed]
- Kaykas A, Yang-Snyder J, Heroux M, et al. Nat Cell Biol. 2004. pp. 52–58. [DOI] [PubMed]
- Junge HJ, Yang S, Burton JB, et al. Cell. 2009. pp. 299–311. [DOI] [PubMed]
- Xu Q, Wang Y, Dabdoub A, et al. Cell. 2004. pp. 883–895. [DOI] [PubMed]
- Chang TH, Hsieh FL, Zebisch M, et al. Elife 2015. Jul 9; doi: 10.7554/eLife.06554 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary infomration, Figures, Tables and Data