Abstract
Prostate Specific Antigen’s (PSA) role as a biomarker for prostate cancer is well established but the physiological role of its serine protease activity in the pathobiology of normal prostate and prostate carcinogenesis remains largely unknown. In light of recent studies that implicate PSA’s enzymatic activity in the initiation and/or progression of prostate cancer, we performed a molecular modeling study of substrate binding at the catalytic site of PSA wherein a PSA-selective substrate (HSSKLQ) was docked in an acyl-enzyme conformation to a 3-dimensional homology model of PSA. Additionally, virtual positional scanning studies were conducted in order to gain mechanistic insights into substrate recognition of PSA. Subsequently, 13 novel peptide substrates of 6-aa length and 4 peptide substrates with varying length were synthesized and assayed for PSA hydrolysis to evaluate the experimental validity of docking insights,. Additionally, six novel aldehyde-containing transition state analog inhibitors were synthesized and tested for their inhibitory potencies. The experimental data on the hydrolysis rates of the newly synthesized substrates and inhibitory potencies of the aldehyde peptides agreed with the docking predictions, providing validation of the docking methodology and demonstrating its utility towards the design of substrate-mimetic inhibitors that can be used to explore PSA’s role in the pathobiology of prostate cancer.
INTRODUCTION
PSA is a serine protease with chymotrypsin like specificity1 and a member of the human tissue kallikrein family comprising 12 tryspin-like and 3 chymotrypsin-like serine proteases2. Since the discovery of Prostate Specific Antigen (PSA) in seminal fluid in 19713, serological screening of PSA has become the most important tool to screen for prostate cancer, to detect recurrence and to follow response to local and systemic therapies. Consistent with its role as biomarker for prostate cancer, PSA levels are elevated in blood in both local adenocarcinoma of the prostate as well as metastatic disease, due to increased leakage of PSA into the circulation from the extracellular fluid surrounding invading prostate cancer cells.
While a significant amount of research has been conducted towards developing and validating PSA as a biomarker, the physiological role of the serine protease activity of PSA in normal prostate as well as in prostate carcinogenesis process remains ill defined4. One possible role of PSA’s enzymatic activity of PSA is in the process of liquefaction of human semen wherein PSA cleaves Semenogelin I and Semenogelin II, the major gel-forming proteins in human semen5,6. This hydrolysis of semen results in the release of trapped spermatozoa, thereby aiding in the fertilization process. Another normal physiological pathway in which the proteolytic activity of PSA might be involved is the activation of the latent form of transforming growth factor β (TGF-β) present abundantly in semen7. Recent studies also implicate PSA mediated activation of the TGF-β signaling cascade in the initiation and/or progression of prostate cancer.8. Prostate cancer cells express high levels of TGF-β which is shown to be critical for prostate cancer growth and metastasis. Therefore, through activation of TGF-β signaling in prostate cancer cells, PSAmight play an important role in stimulating angiogenesis and a variety of inflammatory processes. Similarly, PSA has been shown to directly cleave cytokine binding proteins (such as IGFBP) and release cytokines like IGF-1 that are involved in growth stimulation and inflammation9,10. Additional evidence suggests that PSA may play a significant role in osteoblastic bone metastasis11,12.
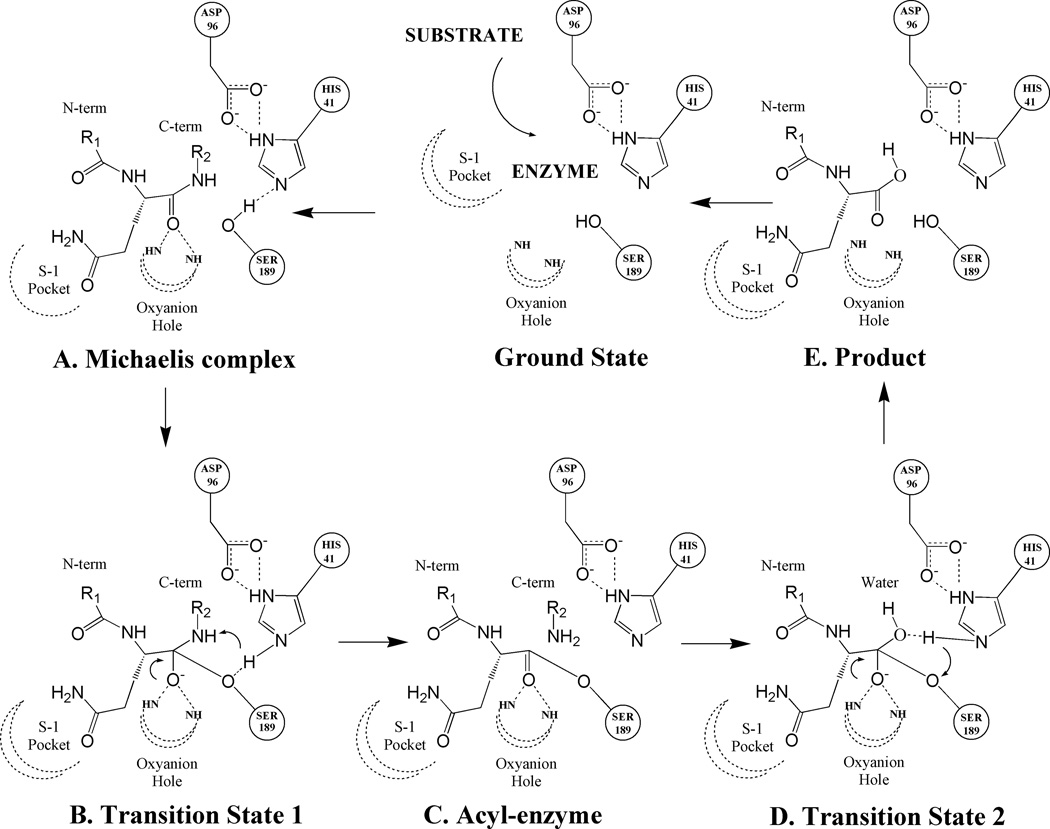
In spite of growing evidence, a direct role for the enzymatic activity of PSA in prostate pathobiology has not been established in the normal prostate or prostate cancer. PSA contains the classic serine protease triad of serine, histidine and asparatic acid residues in its catalytic pocket that are juxtaposed to each other in correct orientation to drive catalysis of substrate hydrolysis13–15 (Figure 1). The hydrolysis reaction is initiated by early non-covalent interactions between the substrate and the PSA catalytic pocket (Figure 1A) that leads to the formation of an intermediate acyl-enzyme complex between the peptide fragment on the N-terminal side of the peptide bond being cleaved (Figure 1C) via a tetrahedral transition state (Figure 1B). The acyl-enzyme complex is transient in nature and is quickly attacked by solvent water molecules resulting in the formation of a second tetrahedral transition state (Figure 1D) and subsequent release of the peptide product (Figure 1E) returning the enzyme to the ground state. The efficiency and specificity of the hydrolysis reaction is dictated by the nature of the energetic interactions between the peptide backbone/sidechains and the protease residues in the specific binding sites. PSA exhibits a low protease activity (four orders of magnitude less than chymotrypsin)16 but it is unique among serine proteases in that it can use glutamine as a P1 residue during substrate hydrolysis17. Thus by incorporating glutamine in the P1 position, PSA-specific substrate and inhibitors can be identified that lack activity against related homolog proteases such as chymotrypsin.
Figure 1.
Mechanistic scheme for the substrate hydrolysis reaction catalyzed by PSA. Illustrated in the scheme are catalytic triad residues (HIS41, SER189 and ASP96) and peptide substrate residue (with the P1 residue as glutamine) with its P1 residue docked at the catalytic site in the S-1 pocket. Dotted lines denote hydrogen bonds and curled arrows represent transfer of proton in a subsequent step. (A): Early non-covalent Michaelis complex (B): Initial tetrahedral transition state formed by the nucleophilic attack of catalytic serine residue (SER189) onto the carboxyl of the peptide bond being cleaved. (C): Acyl-enzyme complex formation between the substrate fragment on the N-terminal side of the cleaved peptide bond and the catalytic serine residue. (D): Second transition state formed upon the nucleophic attack of solvent water molecule onto the ester bond of acyl-enzyme complex. (E): Collapse of second transition state leads to the transfer of a water hydroxyl to the C-terminal carbon and a water proton to the catalytic serine residue. Ultimately, the product is released from the catalytic pocket and the enzyme returns to the ground state.
In this report we present a molecular docking study of substrate binding to the catalytic site of PSA in order to elucidate the molecular basis for the specificity of interactions involved in substrate recognition. The peptide substrate used in this study, HSSKLQ (residues numbered as P6P5P4P3P2P1) was discovered previously in our lab on the basis of a PSA cleavage map for Semenogelin I and Semenogelin II17. This peptide had a high degree of selectivity for PSA (i.e. no detectable hydrolysis by chymotrypsin) due to a glutamine residue in the P1 position and improved Km (470 µM) and Kcat (0.011s−1) parameters vs. chymotrypsin substrates (Km of 2–5 mM for PSA). This peptide was docked into a homology model of PSA utilized previously in gaining mechanistic insights into the inhibition of PSA by β-lactam class compounds18. The binding modes of the peptide substrate revealed through docking studies were analyzed to delineate the binding sites for the individual substrate residues and nature of key interactions taking place between substrate and protease residues. Subsequently, we performed a virtual positional scanning study in which P1, P2, P3 and P4 residues (Q, L, K, S respectively) were mutated to other 19 amino acids and re-docked in their respective binding pockets to assess the amino-acid preference of individual binding site. In the end, validity of the docking insights towards understanding PSA substrate specificity and inhibitor design was evaluated through experimental studies on newly synthesized peptide substrates and transition state analog inhibitors.
MATERIALS & METHODS
Homology model of PSA
In the absence of any crystal information on PSA till date, the 3-dimensional structural model of PSA was developed using the crystal structure of porcine kallikrein (PDB code: 2PKA) and human tissue kallikrein HK1 (PDB code: 1SPJ). The porcine kallikrein has 59% homology with the human PSA sequence and was used as the main template for building PSA’s structural model whereas the HK1 crystal structure was used as the secondary template to model the kallikrein loop which is missing in the main template. The technical details and the software packages used for model construction are described in the previous report from our group18. The homology model of PSA has its Kallikrein loop in a semi-open configuration which results in a catalytic site accessible to both inhibitors and substrates. This model is an improvement over previous models19,20 of PSA that were developed using a single template and de novo Kallikrein loop design which is less accurate than the loop design based on actual crystal structure templates such as the human tissue kallikrein structure (1SPJ) used in our study. This improved structural model of PSA was successfully utilized in our previous report18 on gaining mechanistic insights into the inhibition of Prostate specific antigen by the β-lactam class compounds.
Docking protocols
Ligand Preparation
Ligand preparation for the docking studies was performed within MOE software package (CCG, Montreal) as described previously15,21. All peptide substrates were built in an extended conformation using the Builder module within MOE software package and partial charges were assigned using CHARM22 force field. Also, the peptides were acetylated at the N-terminal to mimic the peptide backbone of a larger peptide and to remove the artificial effects of a charged amine at the N-terminal.
Docking Algorithm
The GOLD v3.0 program (CCDC, Cambridge, UK) was used to dock full substrate (HSSKLQ) and other peptides into the catalytic site of PSA in a covalent binding mode capturing the acyl-enzyme state formed during the substrate hydrolysis reaction (Figure 1C). GOLD docking program uses a genetic algorithm22,23 to explore the full range of ligand conformational flexibility while allowing partial flexibility for the protein. The catalytic site was defined by a 25Ǻ radius spherical space centered on the catalytic serine residue (Ser189). For each peptide substrate, 20 independent runs (GA=20) were performed resulting in 20 different docking poses ranked by their GOLD Score. For every independent run, the number of operations performed (Nop) was 100,000. For each peptide, the top scoring pose with the highest GOLD score was chosen as the most probable binding conformation.
Docking of full substrate
The covalent docking of the full substrate (HSSKLQ) was performed via a covalent bond constraint between the carboxyl carbon at the C-terminal of the P1 residue and the hydroxyl oxygen of the S189 residue. Past crystallographic studies on serine protease have revealed that approaching substrate has to form 3 crucial hydrogen bonds with 204–206 loop of protease14,24. These hydrogen bonds are critical to form the transition state and subsequent hydrolysis. To confirm with this experimental reality, the substrate (HSSKLQ) was docked in the catalytic site with their backbone constrained to form 3 hydrogen bonds with the 204–206 loop residues: the amide nitrogen of the P1 residue, the carbonyl group of the P3 residue and the amide nitrogen of the P3 residue were constrained to make hydrogen bonds with the carbonyl group of Ser204, the amide nitrogen of Trp205 and the carbonyl group of Gly206 respectively. The docking run with the full substrate resulted in 20 poses ranked based on their GOLD scores. These 20 poses were clustered into 7 sub-classes with unique conformations. The top pose with the highest was chosen as the most likely conformation.
Docking of Peptide Fragments
A novel virtual positional scanning approach was devised and utilized to explore P1, P2, P3 and P4 site specificity of PSA in a sequential manner using the GOLD docking protocols similar to the ones described in previous section. The scanning studies were performed in the covalent mode in which the C-terminal carboxyl of the peptide ligand was constrained to form an ester bond with the catalytic S189 residue. Additionally, based on the length of the peptide fragment, the amide nitrogen of the P1 residue, the carbonyl group of the P3 residue and the amide nitrogen of the P3 residue were constrained to make hydrogen bonds with the carbonyl group of Ser204, the amide nitrogen of Trp205 and the carbonyl group of Gly206 respectively. For every peptide fragment, the top pose with the highest was chosen as the most likely conformation.
For P1 scanning, single amino acids (all 20 amino acids) were docked in the catalytic pocket and their binding modes and GOLD scores were analyzed in order to evaluate the effect of each amino acid on the nature of the energetic interactions at the specificity pocket. For P2 scanning, a 2-mer virtual library of peptide substrates was constructed (denoted as XQ where X refers to all 20 amino acids) such that the P1 position was kept fixed as glutamine. For P3 scanning, a 3-mer virtual library was constructed (denoted as XLQ where X refers to all 20 amino acids) such that the P1 and P2 positions were kept fixed as glutamine and leucine respectively. For P4 scanning, a 4-mer virtual library was constructed (denoted as XKLQ where X refers to all 20 amino acids) such that the P1, P2 and P3 positions were kept fixed as Q, L and K respectively.
Substrate peptide synthesis
Peptides (“PepSpots”) were provided by Jerini Biotools (Berlin, Germany) and were synthesized on continuous cellulose assays using the SPOT-synthesis technique25. Each peptide contained an amino-benzoic acid (Abz) moiety at the N-terminus. Abz is a fluorescent molecule with optimal excitation at 325 nm and emission maxima at 420 nm. Peptides were synthesized on cellulose membranes then punched out as small disks into 96-well microtiter plates. Approximately 8 nmoles of peptide are synthesized per spot.
Aldehyde Inhibitor Synthesis
All peptide aldehydes inhibitors were synthesized by the method described by Barany and coworkers26 using the backbone amide linker (BAL) approach. Once protected, aldehydes were attached to the resin and standard Fmoc solid phase peptide synthesis conditions were used for peptide chain elongation on an APTECH Apex 396 40 well peptide synthesizer. The leucine peptide aldehydes were deprotected and cleaved from the resin support using a 95% triflouroactic acid / water mixture and purified by reverse phase-high pressure liquid chromatography (RP-HPLC). The purified fractions were lyophilized and stored at −20°C until further use. All peptide aldehydes were subjected to NMR and mass spectrometric analyses to confirm their compositions.
Substrate Hydrolysis Assays
To perform protease assays, peptide spots were rinsed first for 5 minutes with methanol in order to solubilize the peptides. The spots were then rinsed 4 times for 10 minutes under gentle agitation with a buffer consisting of 50 mM Tris and 0.1 M NaCl, pH 7.8 (Buffer A). Fresh buffer A was added to each well along with an aliquot of purified protease (i.e. PSA) or 50% human serum in buffer A. The plate was sealed with plastic and the reaction allowed to occur at room temperature without agitation. At described intervals (i.e. 1, 2, 4, 7 and 24 hrs) a 50 µl aliquot of the reaction mixture was transferred to a new 96-well microtiter plate. Fluorescence was then measured at room temperature using a 96-well fluorometric plate reader (Fluoroscan II) with excitation of 355 nm and emission 408 nm. Fluorescence at each point was plotted and reaction rates determined from the slope of the best-fit line. Rates are expressed in relative fluorescence units/hr/mg of protease.
Inhibitory Potency Assays
The protocol for evaluating the inhibitory activity of peptide inhibitors was based on similar protocols described previously18. Briefly, the assay for inhibition was performed in PSA activity buffer (50 mM Tris, 100 mM NaCl, pH 7.8, 10% DMSO) with an enzyme concentration of 2.5 µg/ml PSA (Calbiochem) and 300 µM of the PSA substrate Mu-SRKSQQY-AMC (California Peptide). All assays were performed at room temperature by the addition of inhibitor into the assay buffer containing enzyme and substrate with the final volume of 200 µl per assay. Subsequent dilutions were made from the highest concentration of inhibitor (1 mM) with each inhibitor tested at a minimum of ten different concentrations. Dilutions were plated in triplicate into a 96 well plate format with 50 µl per well. Substrate hydrolysis was monitored for 30 minutes with 3 minutes between each data point by measuring fluorescence change secondary to AMC release using a Fluoroskan 96-well fluorometric plate reader (excitation, 355 nm; emission, 460nm). Data were plotted as νo/νi -1 vs. [I], where νo is the rate of substrate hydrolysis in the absence of inhibitor, νi is the rate of substrate hydrolysis in the presence of inhibitor and [I] is the concentration of PSA inhibitor. The plots of νo/νi -1 vs. [I] were linear with the reciprocal of the slope of the line equal to the apparent inhibition constant, Ki(app). The inhibition constant, Ki, was then calculated using the equation Ki = Ki(app)/(1 + ([S]/Km)). The Km value of the PSA substrate Mu-SRKSQQY-AMC was 140 µM as determined by kinetic analysis using Sigma Plot software.
RESULTS & DISCUSSION
Overall binding mode of HSSKLQ
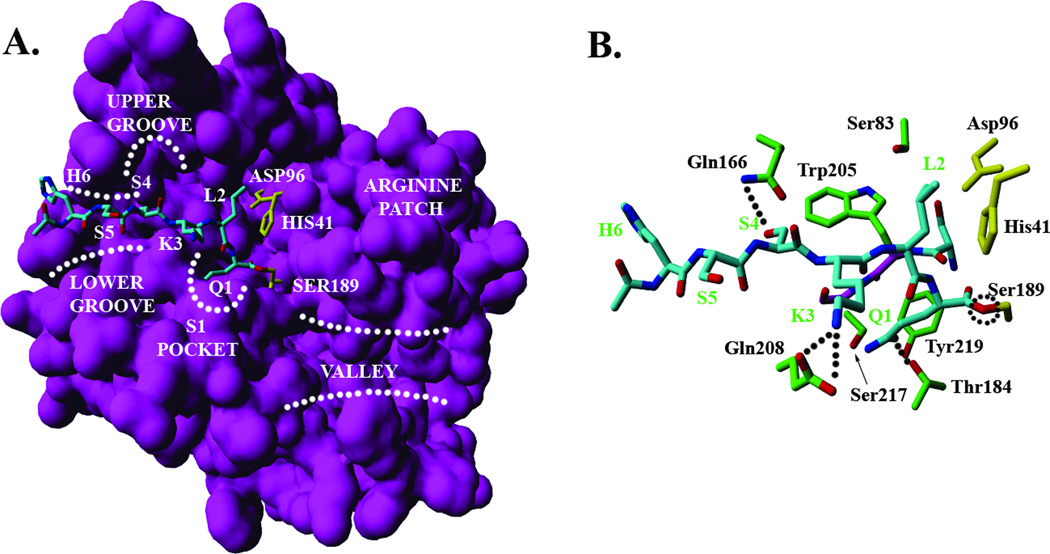
The overall binding orientation of the HSSKLQ peptide substrate (residues numbered as P6P5P4P3P2P1) in the acyl-enzyme state as illustrated by top docking pose is shown in Figure 2A. All residues except the P1 residue (Q1) are bound in an extended configuration which is consistent with the canonical view of how peptide substrates and inhibitors bind at the catalytic pocket of serine proteases27. The backbone atoms of P3-P1 residues are in the right orientation to form a short anti-parallel β sheet with the 204–206 residues of PSA (Figure 2B). Formation of this short β-sheet is required in all classes of serine proteases to correctly position the scissile peptide bond around the catalytic triad residues, i.e. His41, Ser189 and Asp96. In this orientation, the amide nitrogen of the P1 (Q1) residue, the carbonyl group of the P3 (K3) residue and the amide nitrogen of the P3 (K3) residue make hydrogen bonds with the carbonyl group of Ser204, the amide nitrogen of Trp205 and the carbonyl group of Gly206 respectively. The β-sheet conformation is interrupted at the P1 residue which adopts dihedral angles values in the right handed alpha helical (αR) region of the ramachandran plot.
Figure 2.
Binding mode of peptide substrate (H6S5S4K3L2Q1) making an acyl-enzyme complex with PSA. (A) Surface representation of the structural model of PSA and overall orientation of the substrate (shown in cyan) in the catalytic site. Catalytic triad residues (HIS41, ASP96 and SER189) are shown in yellow. Also illustrated are the major topological features of PSA structure, i.e. the S-1 pocket, upper groove, lower groove, arginine patch and valley region. (B) Interactions of substrate sidechains (shown in cyan) with the key residues in the catalytic pocket. Catalytic triad residues are shown in yellow, the protease residues in green and the backbone of 204–206 residues in navy blue. Dotted lines represent hydrogen bonds, and a dotted circle locates the ester bond between the C-terminal carbonyl of the hydrolyzed peptide bond of the substrate and the sidechain of catalytic SER189.
The importance of the conserved hydrogen bonds between the substrate backbone and 204–206 loop of the PSA was validated through the hydrolysis data on c8–12 peptides containing d-amino acid at the P1–5 positions respectively (Table I). A single d-amino acid in the peptide substrates is known to disrupt the formation of secondary structures such as β-sheets28 and its presence at the P1–3 positions is likely to have a drastic affect on the hydrolysis rates since the backbone of only these residues is involved in making crucial hydrogen bonds as part of the β-sheet formation. The d-amino stereochemistry at P4 and P5 is not likely to affect the backbone conformation of P1–3 residues, thus having little impact on overall hydrolysis rates. Almost complete loss of hydrolysis for c8–10 (single d-amino substitution at P1–3 respectively) with no change for c11–12 (single d-amino substitution at P4–5 respectively) is in direct support for this rationale.
Table I.
Amino acid sequence and relative hydrolysis rates of newly synthesized substrates.
| # | Peptide Substrate |
Relative Hydrolysis Rate |
|---|---|---|
| c0 | GSSKLQ | 100.0 |
| c1 | GSSKLH | 170.6 |
| c2 | GSSKLA | 31.5 |
| c3 | GSSKYQ | 256.9 |
| c4 | GSSKPQ | 7.2 |
| c5 | GSSSLQ | 52.6 |
| c6 | GSSALQ | 43.2 |
| c7 | GSAKLQ | 207.2 |
| c8 | GSSKLq | 14.2 |
| c9 | GSSKlQ | 10.7 |
| c10 | GSSkLQ | 5.0 |
| c11 | GSsKLQ | 103.8 |
| c12 | GsSKLQ | 104.1 |
The peptide GSSKLQ (c0) was used as a reference to calculate relative hydrolysis rates. The single amino acid difference between a peptide and the reference sequence is indicated by bold Italic font at the corresponding position. The small underlined letter in c8–12 peptides indicates d-amino acid at that position.
The application of covalent docking to model the acyl-enzyme conformation as presented above is a promising albeit a less studied technique. The covalent docking is less likely to suffer from limited sampling issues since conformation space of covalently-bound ligands is restricted and docking is more likely to produce correct binding orientations. Supporting this rationale, a recent report from our group presented the success of this methodology in explaining the stereochemistry dependent different in inhibitory potencies of β-lactams18.
Specificity of individual binding sites
To gain structural insights towards substrate recognition and the specificity of substrate-protease interactions, we analyzed the binding sites of individual substrate residues and the nature of their energetic interactions with the protease residues in the catalytic pocket. In the following sections, we present a detailed analysis of substrate binding sites in the catalytic pocket of PSA and the dominant hydrophobic and polar interactions taking place. We also present an analysis of results obtained from the virtual scanning studies that were performed in order to asses the amino acid preference of an individual binding site. In these scanning studies, the P1, P2, P3 and P4 positions of PSA substrate were sequentially mutated to all 20 amino acids and re-docked in their respective binding sites while the backbone heavy atoms of the substrate were constrained to the canonical conformation described in previous section. In this manner, we insured that observed differences in GOLD scores and sidechain orientations were not influenced by the unrealistic backbone movements and were solely due to the molecular interactions of sidechains within their binding pocket.
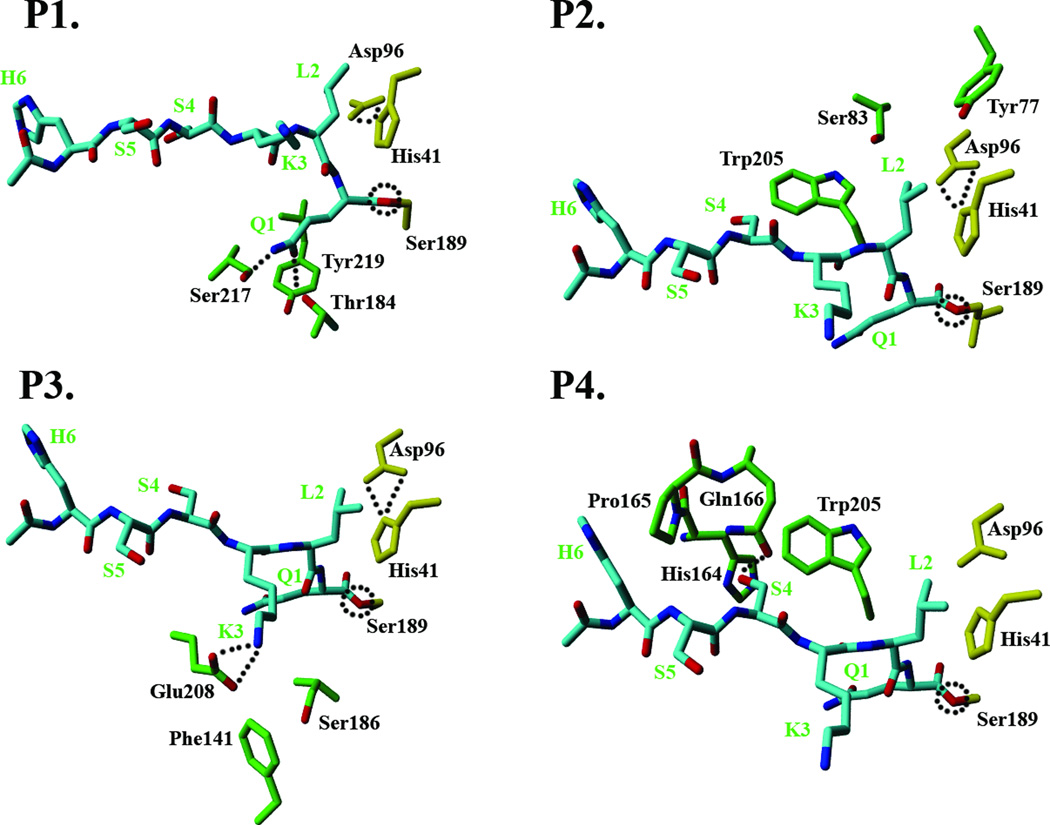
P1 binding site
The P1 residue (Q1) binds at the specificity pocket lined by polar residues with the partial hydrophobic sidechains such as Ser217, Thr184 and Tyr219 (Figure 3A). The presence of these residues imparts a hydrophobic character to the sides of the specificity pocket and at the same time, a polar character to the bottom of the pocket. This allows optimum binding of medium sized hydrophobic residues such as leucine but not large sized residues such as tryptophan. Additionally, residues such as glutamine and tyrosine can make hydrogen bonding interactions with the polar residues at the bottom of the pocket while preserving hydrophobic interactions through the hydrophobic part of the sidechains. These hydrogen bonds buried deep within the protease core are likely to be strong since the energetic penalty for an unsatisfied hydrogen bond buried in the protein core is very high. This molecular rationale was experimentally tested through hydrolysis data on the newly synthesized substrates c1 and c2 (Table I). The histidine residue at the P1 position in c1 has the ability to form hydrogen bonding interactions with polar residues at the bottom of specificity pocket while c2, with alanine at the P1 position, is unable to do so. Thus, c1, and not c2, is predicted to maintain good substrate activity for PSA. The hydrolysis data for c1 and c2 are in exact agreement with this assertion.
Figure 3.
The binding sites for individual residues of peptide substrate (HSSKLQ) and the key protease residues interacting with the sidechains of the substrate residue through hydrogen bonding or hydrophobic interactions. The covalently attached substrate is shown in cyan, catalytic triad residues in yellow, and the protease residues in green. (A): P1 residue binding site. (B): P2 residue binding site. (C): P3 residue binding site. (D). P4 residue binding site.
The virtual positional scanning at the P1 position revealed that while glutamine, leucine, phenylalanine, tyrosine and histidine are able to bind at the P1 pocket in a canonical conformation required for the inhibitory potency, residues such as isoleucine, valine and tryptophan bind less well. These docking results are in agreement with earlier studies which used experimental position scanning29 and phage display technique19 to explore the P1 specificity of PSA.
P2 binding Site
The P2 residue (L2) sidechain is docked against the face of the imidazole ring of the catalytic His41 residue (Figure 3B). On the opposite side, the γ-branched sidechain of leucine is in close contact with the hydrophobic Trp205 residue. Additionally, polar residues such as Ser93 and Tyr77 are also lining the farthest end of the P2 pocket, creating a predominantly negative electrostatic surface potential and providing a polar characteristic to the P2 pocket. While the nature of residues forming the P2 pocket is similar to the P1 pocket, an important distinction lies in the flexibility of the P2 pocket structure. P2 pocket residues are located at the edges of kallikrein loop and, as such, the shape of the pocket is strongly influenced by the mobility of the Kallikrein loop.
Consistent with the mixed polar-hydrophobic characteristics of the P2 pocket, virtual positional scanning revealed that the P2 pocket is permissive in allowing residues ranging from polar ones such as tyrosine and glutamine and hydrophobic residues such as leucine, norleucine, alanine and valine. Exceptions are isoleucine (negative GOLD score), proline and tryptophan that produced steric clashes with the P2 pocket residues. It was also noted that while valine binding to the P2 pocket was reasonably energetically favorable, its β-branched sidechains did not interact optimally in the positive electrostatic potential of His41. The validity of these structural insights was tested using hydrolysis data on substrates c3 (tyrosine at the P2 position) and c4 (proline at the P2 position) (Table I). The high substrate activity of c3 compared to the almost complete loss of hydrolysis activity for c4 is consistent with the structural rationale.
P3 binding site
The P3 residue (K3) acts as a lid to the specificity pocket and docks at a location just above the P1 binding site in the specificity pocket (Figure 3C). The aliphatic sidechain of K3 is positioned in the vicinity of the hydrophobic phenylalanine group of the Phe141 residue and also in close contact with the methylene group of the Ser186 sidechain. More importantly, the free amine group of K3 is within 3.5Ǻ distance of the carboxyl moiety of the Glu208 sidechain. While the orientation of the Glu208 sidechain is not ideal, there is a strong possibility for a salt bridge formation. It should be noted that Glu208 has been shown to be functionally significant20 for the binding of PSA with its physiological inhibitor α-antichymotrypsin (ACT). Also, the presence of an acidic residue at position 208 is not common in the chymotrypsin family of serine proteases. Taken together, these findings suggest an important role for Glu208 interactions in determining PSA specificity at the P3 residue.
While Glu208 salt bridge formation might be critical for providing the specificity of inhibition, virtual positional scanning of the P3 position suggests that this residue is not critical for the absolute inhibitory potency. Polar residues with a similar size as lysine (i.e. glutamine) that can make strong hydrogen bonds with Glu208 are also be permitted at the P3 position. Additionally, residues with relatively short sidechains such as serine, valine, and alanine are capable of initiating favorable interactions with Phe141 and Ser186 sidechains, though these interactions are not likely to be as strong as those for lysine and glutamine. Additionally, charged residues with short sidechains, such as asparatic acid and asparagine, are energetically unfavorable to dock in the area lined by Phe141 and Ser186 and are not predicted to be good candidates for P3 substitution. In order to test the structural rationale for P3 specificity, substrates c5 (serine at the P3 position) and c6 (alanine at the P3 position) were synthesized and tested for their hydrolysis efficiency (Table I). While serine and alanine are permitted at the P3 position, they do not have sidechains with adequate length to make polar interactions with Glu208. Thus c5 and c6 are predicted to be less efficient substrates compared to c0 with lysine at the P3 position. The hydrolysis rate data for these substrates concur with this structural prediction.
P4 binding site
The P4 pocket is located in the lower groove area, and mainly formed by His164, Pro165, Gln166 and Trp205 residues (Figure 3D). The P4 binding pocket is bounded by the Kallikrein loop from above but loop residues do not seem to be involved in any important energetic interactions. The Gln166 sidechain is ideally placed to form a hydrogen bond with the hydroxyl of the P4 serine. The presence of Gln166 at this position is unique for PSA and not common in other serine proteases from the kallikrein family. Previous studies20 have suggested that this residue interacts with the protease inhibitor ACT, suggesting its role in determining P4 specificity. The P4 pocket surface area is relatively small and the distance between the Gln166 and the backbone Cα atom of P4 residue is quite short, suggesting that only residues with small sidechains are likely to maintain substrate activity towards PSA.
To test these molecular insights on P4 amino acid preference, we synthesized and tested a substrate containing alanine at the P4 position (c7) for its hydrolysis activity. As predicted by docking studies, c7 possesses high activity for PSA hydrolysis. The 2-fold increase in the hydrolysis rate of c7 compared to c0 suggests that the alanine residue at the P4 position is more favorable compared to serine at the same position, possibly due to its smaller side-chain with high torsional degrees of freedom. The high substrate activity of this peptide also suggests that a hydrogen bonding interaction with Gln166 might not be critical for P4 site binding. Rather, the key determinant for P4 site specificity might be the impact of the P4 residue on the ability of P1–3 residues to make an anti-parallel β-sheet conformation with the backbone of the 204–206 protease residues. Smaller residues like serine and alanine allow formation of this conformation, which is crucial for hydrolysis, while residues with larger sidechains are likely to indirectly affect the β-sheet conformation through allosteric interactions.
P5 residue and beyond
The S5 and H6 sidechains are positioned in the lower groove area but they are exposed to the solvent and there are no specific interactions observed with protease residues. This seems to suggest that P5 and residues on its N-terminal do not provide a significant contribution towards the overall binding affinity of an inhibitor molecule. To test this assertion, we analyzed our previous studies on the hydrolysis rates for peptide substrates with varying length (Table II). As shown, shorter peptides (c13–15) containing three or less residues completely lacked the cleavage by PSA, and hydrolytic activity of PSA was seen only when serine at the P4 position was added (c16). While the addition of two more amino acids (c7 with 6 residues) produced slightly better hydrolysis, this experimental data agrees with the docking prediction in that the P1–4 residue are the most critical for the PSA’s substrate activity.
Table II.
Hydrolysis Rate of Substrates with Varying Sequence Length.
| # | Peptide substrate | Hydrolysis rate pmol/min per 100 pmol of PSA |
|---|---|---|
| c13 | NH2-Q-AMC | Undetectable |
| c14 | Mu-LQ-AMC | Undetectable |
| c15 | Mu-KLQ-AMC | Undetectable |
| c16 | Mu-SKLQ-AMC | 29.6 |
| c17 | Mu-HSSKLQ-AMC | 62.7 |
The data presented in this table has been taken from our previous study. All substrates had 7-amino-4-methyl coumarin (AMC) attached to their C-terminal, and 4-morpholinecarbonyl (Mu) capping group at the N-terminal (except c13). Each assay contained 200 pmol of PSA and 0.2 mM of substrate in assay buffer (50 mM Tris, 0.1 M NaCl).
Design of Transition State Analogs
To demonstrate the utility of molecular insights towards designing novel inhibitors of PSA, we synthesized six novel peptide sequences containing aldehyde functionality at the C-terminal (Table III). The aldehyde moiety at the C-terminal of peptide substrates is known to generate a transition state like conformation that is resistant to subsequent hydrolysis, thus resulting in the robust inhibition of protease activity30,31. The newly synthesized peptide differed only at the P2 position and rest of the peptide sequence was kept the same. The fluorescence-based inhibitory assays on these aldehyde peptides revealed that peptide c18 with leucine at the P2 position had the highest potency with a Ki value of 6.5 µM. When the P2 residue was replaced with tyrosine in c19, the potency was only slightly lower (13.09 µM). This is in agreement with the nature of the P2 binding site which suggests that tyrosine, with its polar hydroxyl at the end, can optimally interact with the polar area in the P2 binding pocket lined by Ser93 and Tyr77 (Figure 3B). Consistent with this observation, when the hydroxyl of the P2 tyrosine is replaced by bromine in the c20 peptide, there is a complete loss of inhibitory potency. The tryptophan at the P2 position as in peptide c21 does not act favorably with the electronegative potential in the P2 pocket, resulting in a Ki > 100 µM. Similarly, an asparatic residue at the P2 position is severely unfavorable due to electrostatic repulsion between the negatively charged carboxyl sidechain and the electronegative potential of the P2 binding pocket, explaining the complete loss of inhibitory potency for peptide c22. The proline residue at the P2 position in peptide c23 clashes with the catalytic His41 residue due to its irregular shape. This explains the poor inhibitory potency of c23 (> 500 µM).
Table III.
Amino Acid Sequence and Binding Potencies (Ki) of Newly Synthesized Transition State Analogs Containing Aldehyde (CHO) Functionality at the C-Terminal
| # | Transition state analog | Ki (µM) |
|---|---|---|
| c18 | Cbz-SSKLL-CHO | 6.51 |
| c19 | Cbz-SSKYL-CHO | 13.09 |
| c20 | Cbz-SSKF(Br)L-CHO | >500 |
| c21 | Cbz-SSKWL-CHO | 155.34 |
| c22 | Cbz-SSKDL-CHO | >1000 |
| c23 | Cbz-SSKPL-CHO | >500 |
The N-terminal of these compounds was capped with carboxybenzoyl (Cbz) group.
CONCLUSIONS
In this study, we have utilized a combination of substrate docking and a virtual positional scanning approach to elucidate the binding mode of peptide substrates within the catalytic pocket of PSA and determined amino acid preferences at the individual binding sites of peptide substrates. One of the conclusions drawn from these docking studies was that only P1–4 residue are major determinants of binding affinity and substrate specificity for PSA. The mechanistic insights revealed by the current modeling studies were experimentally validated through chemical synthesis and testing of new peptide substrates and aldehyde-based inhibitors. Currently, structural findings from this study are being utilized in ongoing work to design and develop potent substrate mimetic inhibitors of PSA through incorporation of aldehyde and boronic acid functionality at the C-terminus of substrate sequences. The small molecule inhibitors resulting from these efforts will serve as important chemical tools to explore PSA’s biological role and could potentially represent therapeutic agents given the growing evidence implicating a role for PSA activity in the pathogenesis of prostate cancer.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by NIH SPORE grant P50CA58236 and One-in-Six foundation.
REFERENCES
- 1.Watt KW, Lee PJ, M'Timkulu T, Chan WP, Loor R. Human prostate-specific antigen: structural and functional similarity with serine proteases. Proc Natl Acad Sci U S A. 1986;83(10):3166–3170. doi: 10.1073/pnas.83.10.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borgono CA, Diamandis EP. The emerging roles of human tissue kallikreins in cancer. Nat Rev Cancer. 2004;4(11):876–890. doi: 10.1038/nrc1474. [DOI] [PubMed] [Google Scholar]
- 3.Hara M, Koyanagi Y, Inoue T, Fukuyama T. [Some physico-chemical characteristics of " -seminoprotein", an antigenic component specific for human seminal plasma. Forensic immunological study of body fluids and secretion. VII] Nippon Hoigaku Zasshi. 1971;25(4):322–324. [PubMed] [Google Scholar]
- 4.Williams SA, Singh P, Isaacs JT, Denmeade SR. Does PSA play a role as a promoting agent during the initiation and/or progression of prostate cancer? Prostate. 2006;67(3):312–329. doi: 10.1002/pros.20531. [DOI] [PubMed] [Google Scholar]
- 5.Lilja H. A kallikrein-like serine protease in prostatic fluid cleaves the predominant seminal vesicle protein. J Clin Invest. 1985;76(5):1899–1903. doi: 10.1172/JCI112185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malm J, Hellman J, Hogg P, Lilja H. Enzymatic action of prostate-specific antigen (PSA or hK3): substrate specificity and regulation by Zn(2+), a tight-binding inhibitor. Prostate. 2000;45(2):132–139. doi: 10.1002/1097-0045(20001001)45:2<132::aid-pros7>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 7.Killian CS, Corral DA, Kawinski E, Constantine RI. Mitogenic response of osteoblast cells to prostate-specific antigen suggests an activation of latent TGF-beta and a proteolytic modulation of cell adhesion receptors. Biochem Biophys Res Commun. 1993;192(2):940–947. doi: 10.1006/bbrc.1993.1506. [DOI] [PubMed] [Google Scholar]
- 8.Robertson SA. Seminal plasma and male factor signalling in the female reproductive tract. Cell Tissue Res. 2005;322(1):43–52. doi: 10.1007/s00441-005-1127-3. [DOI] [PubMed] [Google Scholar]
- 9.Cohen P, Graves HC, Peehl DM, Kamarei M, Giudice LC, Rosenfeld RG. Prostate-specific antigen (PSA) is an insulin-like growth factor binding protein-3 protease found in seminal plasma. J Clin Endocrinol Metab. 1992;75(4):1046–1053. doi: 10.1210/jcem.75.4.1383255. [DOI] [PubMed] [Google Scholar]
- 10.Chan JM, Stampfer MJ, Giovannucci E, Gann PH, Ma J, Wilkinson P, Hennekens CH, Pollak M. Plasma insulin-like growth factor-I and prostate cancer risk: a prospective study. Science. 1998;279(5350):563–566. doi: 10.1126/science.279.5350.563. [DOI] [PubMed] [Google Scholar]
- 11.Nelson JB, Hedican SP, George DJ, Reddi AH, Piantadosi S, Eisenberger MA, Simons JW. Identification of endothelin-1 in the pathophysiology of metastatic adenocarcinoma of the prostate. Nat Med. 1995;1(9):944–949. doi: 10.1038/nm0995-944. [DOI] [PubMed] [Google Scholar]
- 12.Shah RB, Mehra R, Chinnaiyan AM, Shen R, Ghosh D, Zhou M, Macvicar GR, Varambally S, Harwood J, Bismar TA, Kim R, Rubin MA, Pienta KJ. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res. 2004;64(24):9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- 13.Otlewski J, Jaskolski M, Buczek O, Cierpicki T, Czapinska H, Krowarsch D, Smalas AO, Stachowiak D, Szpineta A, Dadlez M. Structure-function relationship of serine protease-protein inhibitor interaction. Acta Biochim Pol. 2001;48(2):419–428. [PubMed] [Google Scholar]
- 14.Perona JJ, Craik CS. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995;4(3):337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh P, Williams SA, Shah MH, Lectka T, Pritchard GJ, Isaacs JT, Denmeade SR. Mechanistic insights into the inhibition of prostate specific antigen by beta-lactam class compounds. Proteins. 2008;70(4):1416–1428. doi: 10.1002/prot.21676. [DOI] [PubMed] [Google Scholar]
- 16.Huang X, Knoell CT, Frey G, Hazegh-Azam M, Tashjian AH, Jr, Hedstrom L, Abeles RH, Timasheff SN. Modulation of recombinant human prostate-specific antigen: activation by Hofmeister salts and inhibition by azapeptides. Appendix: thermodynamic interpretation of the activation by concentrated salts. Biochemistry. 2001;40(39):11734–11741. doi: 10.1021/bi010364j. [DOI] [PubMed] [Google Scholar]
- 17.Denmeade SR, Lou W, Lovgren J, Malm J, Lilja H, Isaacs JT. Specific and efficient peptide substrates for assaying the proteolytic activity of prostate-specific antigen. Cancer Res. 1997;57(21):4924–4930. [PMC free article] [PubMed] [Google Scholar]
- 18.Singh P, Williams SA, Shah MH, Leckta T, Pritchard GJ, Isaacs JT, Denmeade SR. Mechanistic insights into the inhibition of prostate specific antigen by β-lactam class compounds. Proteins: Structure, Function, and Bioinformatics. 2007 doi: 10.1002/prot.21676. [DOI] [PubMed] [Google Scholar]
- 19.Coombs GS, Bergstrom RC, Pellequer JL, Baker SI, Navre M, Smith MM, Tainer JA, Madison EL, Corey DR. Substrate specificity of prostate-specific antigen (PSA) Chem Biol. 1998;5(9):475–488. doi: 10.1016/s1074-5521(98)90004-7. [DOI] [PubMed] [Google Scholar]
- 20.Villoutreix BO, Getzoff ED, Griffin JH. A structural model for the prostate disease marker, human prostate-specific antigen. Protein Sci. 1994;3(11):2033–2044. doi: 10.1002/pro.5560031116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeBeau AM, Singh P, Isaacs JT, Denmeade SR. Potent and selective peptidyl boronic acid inhibitors of the serine protease prostate-specific antigen. Chem Biol. 2008;15(7):665–674. doi: 10.1016/j.chembiol.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J Mol Biol. 1995;245(1):43–53. doi: 10.1016/s0022-2836(95)80037-9. [DOI] [PubMed] [Google Scholar]
- 23.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267(3):727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 24.Czapinska H, Helland R, Smalas AO, Otlewski J. Crystal structures of five bovine chymotrypsin complexes with P1 BPTI variants. J Mol Biol. 2004;344(4):1005–1020. doi: 10.1016/j.jmb.2004.09.088. [DOI] [PubMed] [Google Scholar]
- 25.Kramer A, Reineke U, Dong L, Hoffmann B, Hoffmuller U, Winkler D, Volkmer-Engert R, Schneider-Mergener J. Spot synthesis: observations and optimizations. J Pept Res. 1999;54(4):319–327. doi: 10.1034/j.1399-3011.1999.00108.x. [DOI] [PubMed] [Google Scholar]
- 26.Kappel JC, Barany G. Backbone amide linker (BAL) strategy for Nalpha-9-fluorenylmethoxycarbonyl (Fmoc) solid-phase synthesis of peptide aldehydes. J Pept Sci. 2005;11(9):525–535. doi: 10.1002/psc.614. [DOI] [PubMed] [Google Scholar]
- 27.Perona JJ, Craik CS. Evolutionary divergence of substrate specificity within the chymotrypsin-like serine protease fold. J Biol Chem. 1997;272(48):29987–29990. doi: 10.1074/jbc.272.48.29987. [DOI] [PubMed] [Google Scholar]
- 28.Lee DL, Powers JP, Pflegerl K, Vasil ML, Hancock RE, Hodges RS. Effects of single D-amino acid substitutions on disruption of beta-sheet structure and hydrophobicity in cyclic 14-residue antimicrobial peptide analogs related to gramicidin S. J Pept Res. 2004;63(2):69–84. doi: 10.1046/j.1399-3011.2003.00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Debela M, Magdolen V, Schechter N, Valachova M, Lottspeich F, Craik CS, Choe Y, Bode W, Goettig P. Specificity profiling of seven human tissue kallikreins reveals individual subsite preferences. J Biol Chem. 2006;281(35):25678–25688. doi: 10.1074/jbc.M602372200. [DOI] [PubMed] [Google Scholar]
- 30.Neidhart D, Wei Y, Cassidy C, Lin J, Cleland WW, Frey PA. Correlation of low-barrier hydrogen bonding and oxyanion binding in transition state analogue complexes of chymotrypsin. Biochemistry. 2001;40(8):2439–2447. doi: 10.1021/bi002535a. [DOI] [PubMed] [Google Scholar]
- 31.Krishnan R, Zhang E, Hakansson K, Arni RK, Tulinsky A, Lim-Wilby MS, Levy OE, Semple JE, Brunck TK. Highly selective mechanism-based thrombin inhibitors: structures of thrombin and trypsin inhibited with rigid peptidyl aldehydes. Biochemistry. 1998;37(35):12094–12103. doi: 10.1021/bi980840e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.